Summary
Centrosomes are cellular structures that nucleate microtubules. At their core is a pair of centrioles that recruit pericentriolar material (PCM). Although centrosomes are considered membraneless organelles, in many cell types, including human cells, centrosomes are surrounded by endoplasmic reticulum-derived membranes of unknown structure and function. Using volume electron microscopy (vEM), we show that centrosomes in the C. elegans early embryo are surrounded by a three-dimensional (3D), ER-derived membrane reticulum that we call the centriculum, for centrosome-associated membrane reticulum. The centriculum is adjacent to the nuclear envelope in interphase and early mitosis, and fuses with the fenestrated nuclear membrane at metaphase. Centriculum formation is dependent on the presence of an underlying centrosome and on microtubules. Conversely, increasing centriculum size by genetic means led to expansion of the PCM, increased microtubule nucleation capacity and altered spindle width. The effect of the centriculum on centrosome function suggests that in the C. elegans early embryo, the centrosome is not membraneless. Rather, it is encased in a membrane meshwork that affects its properties. We provide evidence that the centriculum serves as a microtubule “filter”, preventing the elongation of a subset of microtubules past the centriculum. Finally, we propose that the fusion between the centriculum and the nuclear membrane contributes to nuclear envelope breakdown by coupling spindle elongation to nuclear membrane fenestration.
eTOC blurb
Centrosomes are considered membraneless organelles, but in some cell types they are surrounded by ER membrane. Maheshwari, Rahman et al. show that in C. elegans early embryos, the centrosome is surrounded by a membrane reticulum, the centriculum, that affects centrosome size and function, and limits microtubule extension and orientation.
Introduction
Centrosomes are considered membraneless organelles that serve as microtubule-organizing centers, such as at the poles of mitotic spindles. They are composed of a pair of centrioles surrounded by pericentriolar material (PCM) that promotes microtubule nucleation. In most organisms examined to date, centrioles originate from the sperm 1. Soon after fertilization, the genomes of the sperm and oocyte become encased in separate nuclei, called pronuclei. Each pronucleus is surrounded by a nuclear envelope that consists of two membranes, the outer and inner nuclear membranes, that are traversed by nuclear pore complexes (NPCs) that allow transport of material between the cytoplasm and nucleoplasm 2. After pronuclei form, the centrioles separate and recruit PCM proteins to form two centrosomes that are initially associated with the male pronucleus via the linker of nucleoskeleton and cytoskeleton (LINC) complex 3,4. The pronuclei then migrate toward each other, meet, and the embryo enters its first mitosis. Nuclear membrane-associated proteins dissociate from the NE and the pronuclear membranes themselves become fenestrated in a process known as NE breakdown 2,5. During this time, centrosomes continue to separate and start nucleating microtubules that extend towards the cell cortex or chromosomes, ultimately forming the mitotic spindle.
As the centrosomes separate, the PCM increases in size in a process known as centrosome maturation 6. During this process, recruitment of PCM proteins, such as the C. elegans SPD-5 or its functional homolog of CDK5RAP2/Cnn, depends on the centrosomal proteins SPD-2/CEP192, Aurora kinase and Polo-like kinase 7–9. The PCM, in turn, recruits proteins that promote microtubule nucleation, such as the γ-tubulin ring complex (γ -TuRC), and increases the local concentration of tubulin 10,11. Some C. elegans PCM components, as well as those of human cells, can form liquid-liquid phase-separated (LLPS) condensates in vitro 10,12–14, although it is not clear to what extent this happens in intact cells 15. If the PCM is phase separated in vivo, its size would be expected to be a function of the concentration of its components and the properties of the surrounding liquid environment 10,16.
Centrosomes in the C. elegans early embryo are surrounded by an ER-derived membrane (17 and Figure 1A). Centrosome-associated membranes were also observed in other systems, such as Drosophila, sea urchin, and the kidney epithelial cell lines PtK2 and LLC-PK2 18–23. The configuration of these membranes and their function are unknown. Using volume electron microscopy (vEM) analyses, we show here that the membrane around the C. elegans centrosomes forms a reticulum, leading us to name it the “centriculum”, for centrosome-associated endoplasmic reticulum. We found an interdependent relationship between the PCM and the centriculum. The centriculum requires the PCM for its formation. Conversely, PCM size and its microtubule-nucleating capacity are affected by the size of the centriculum, an unexpected finding given that the centrosome is considered a membraneless organelle. Our data also suggest that the centriculum acts as a “microtubule filter”, limiting the number of microtubules that can extend past the centriculum. Finally, during mitosis, the centriculum may play a role in promoting the fenestration of the nuclear membranes.
Figure 1: The centrosome is surrounded by ER membrane that contains some, but not all, NE proteins.
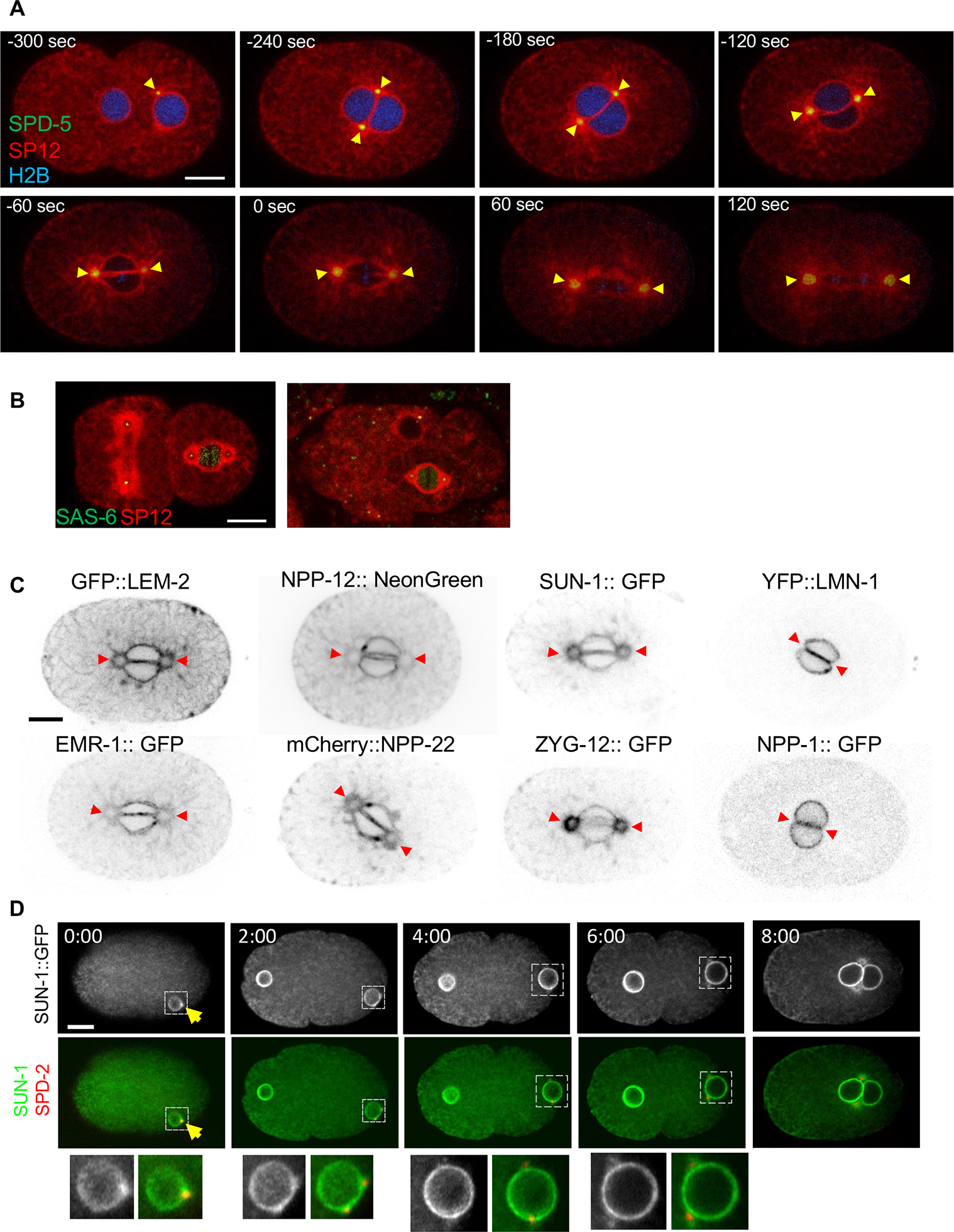
(A) Time lapse images of a C. elegans 1-cell embryo, expressing the resident ER signal peptidase, SP12, fused to mCherry (red), the PCM protein, SPD-5, fused to GFP (green), and histone H2B fused to mTurquoise2 (blue) (strain OCF164). Yellow arrowheads point to the membrane surrounding the centrosomes. Time points are relative to metaphase (t=0), as determined by the appearance of the membrane gap at the membrane interphase between the two pronuclei (note that the histone H2B signal is faint and not always visible in all embryos). In the first time point (−300 sec), the pronucleus containing the sperm DNA and associated with the centrosomes is on the right. (B) A 2-cell embryo (left) and a multi-cell stage embryo (right) expressing the centriolar protein SAS-6 fused to GFP (green) and SP12 fused to mCherry (red) (strain OCF124). The association of the centrosome with ER membrane in the vulval precursor cell from a L3 stage C. elegans larvae is shown in Figure S1A. (C) Localization pattern of the indicated NE proteins relative to the membrane around centrosomes (indicated by red arrowheads) in 1-cell embryos at prometaphase. The strains used were OCF46 (GFP::LEM-2), BN243 (EMR-1::GFP), JH3908 (NPP-12::mNeonGreen), WLP801 (mCherry::NPP-22), OCF145 (SUN-1::GFP) OCF85 (ZYG-12::GFP), OCF4 (YFP::LMN-1) and OCF3 (NPP-1::GFP). Scale bar in all panels=10 μm. For more examples see Figure S1B. (D) A time course of a representative embryo expressing SUN-1::GFP (grey scale images and green in the merged images) and SPD-2::mCherry (red in the merged images) (OCF178). Enlargements of the indicated nuclei (dashed lines) are in the bottom row. At time 0 only the male pronucleus is visible in the focal plane shown. The arrow points to the centrosome at time 0. Time points are in minutes. Time courses of LEM-2 and NPP-12 around centrosomes are shown in Figure S1C. Scale bar =10 μm. For related data see Figure S1.
Results
The centrosome is surrounded by an ER membrane that contains some, but not all, NE proteins.
After fertilization in C. elegans, the two pronuclei move towards each other and meet, forming two parallel pronuclear membrane surfaces at the interface between them. At the same time, the duplicated centrosomes increase in size and separate via microtubule-associated forces, until they reach opposite ends of the pronuclear membrane interface (Figure 1A, −240 sec, reviewed in 9). Throughout this process, centrosomes are surrounded by an ER-derived membrane system (17 and Figure 1A), the configuration and function of which were unknown. Centrosome-associated ER membranes are also present in multi-cellular embryos (Figure 1B). The presence of membranes around centrosomes in the adult was less obvious: Centrosomes in vulval precursor cells (VPC) were associated with membrane accumulation, but the presence of membrane around the entire centrosome was ambiguous (Figure S1A). Since the membranous structures surrounding the centrosome were the largest and most easily visualized in 1-cell embryos, we characterized them further at this developmental stage.
The proximity of the centrosome to the NE led us to examine which NE-associated proteins also localized around the centrosome. We found that membrane-embedded NE proteins, such as the inner nuclear membrane proteins LEM-2 and EMR-1 (Emerin), the LINC complex proteins SUN-1 and ZYG-12, and the transmembrane NPC subunits such NPP-12 (human homolog: gp210) and NPP-22 (NDC-1), were also present around the centrosome (Figures 1C and S1B). In contrast, the peripheral NE proteins LMN-1 and the NPC subunit NPP-1 (Nup54) did not localize around centrosomes (Figures 1C and S1B). Published data indicate that the ESCRT protein CHMP-7 also localizes around centrosomes 24, while other non-membrane embedded NPC subunits, such as NPP-3 (Nup205), NPP-8 (Nup155), NPP-19 (Nup35), and NPP-5 (Nup107), do not 25–28. Thus, membrane-embedded NE proteins localize to both the NE and around centrosomes, while peripheral NE proteins are confined to the NE. In the case of LEM-2, EMR-1, NPP-12 and NPP-22, the localization around the centrosome likely reflects a general ER localization at this embryonic stage. LEM-2 can be observed around centrosomes as early as prometaphase, while NPP-12 can be detected around centrosomes earlier, shortly after centrosome duplication (Figure S1C). In both cases, the protein fluorescence intensities around the centrosome were similar to those in the rest of the ER. In contrast, SUN-1 and ZYG-12 appear to be specifically enriched at centrosome-associated membranes during mitosis (Figures 1C and S1B). Interestingly, SUN-1::GFP accumulates at centrosomes even before centrosome separation (Figure 1D). The mechanism that enriches the LINC complex at this locus is not known.
The membrane around the centrosome forms a mesh-like, reticular structure, now named the centriculum
To analyze the ultrastructure of the membrane around centrosomes, we used a vEM technique called Focus Ion Beam - Scanning Electron Microscopy (FIB-SEM, 29–32). Briefly, individual 1- cell C. elegans embryos in mitosis were high-pressure frozen, followed by freeze substitution and embedding in resin. A trench was created near the area of interest, and an ion beam sequentially milled off 9 nm from the block, with a SEM image taken after each iteration (final voxel size= 9 nm3). In this study, we present 3D reconstruction of membranes around three prophase centrosomes and three metaphase centrosomes using previously published FIB-SEM data 31.
In a single SEM slice from the vEM image stack, centrosomes appeared as an electron-dense region at opposite poles of the interface between the two pronuclei (Figure 2A, B, C, F, G, K and Figures S2 and S3, panels i and ii). With our staining method, which was optimized for maximal membrane contrast, microtubules were observed only occasionally (see 31) and centrioles were never detected. The need for three-dimensional information becomes apparent when examining a single EM image, where membranes around the centrosome could have been interpreted as an aggregation of vesicles (Figure 2A). However, segmentation (i.e., 3D-reconstructions) of 400–500 nm above and below a center plane of centrosomes revealed that the membranes around centrosomes form a reticulum (Figure 2C–E, G–I and Figures S2 and S3; segmentations were done to the edge of the reticulum, beyond which only ER tubules were present). This configuration was also confirmed by electron tomography data (see below). Based on this, we named the spherical membrane structure that surrounds centrosomes the centriculum, for centrosome-associated membrane reticulum (plural= centricula). The membrane configuration of the centriculum, a 3D network of highly interconnected tubules, is distinct from previously reported membrane configurations of the ER 33.
Figure 2: The membrane around the centrosome forms a reticulum.
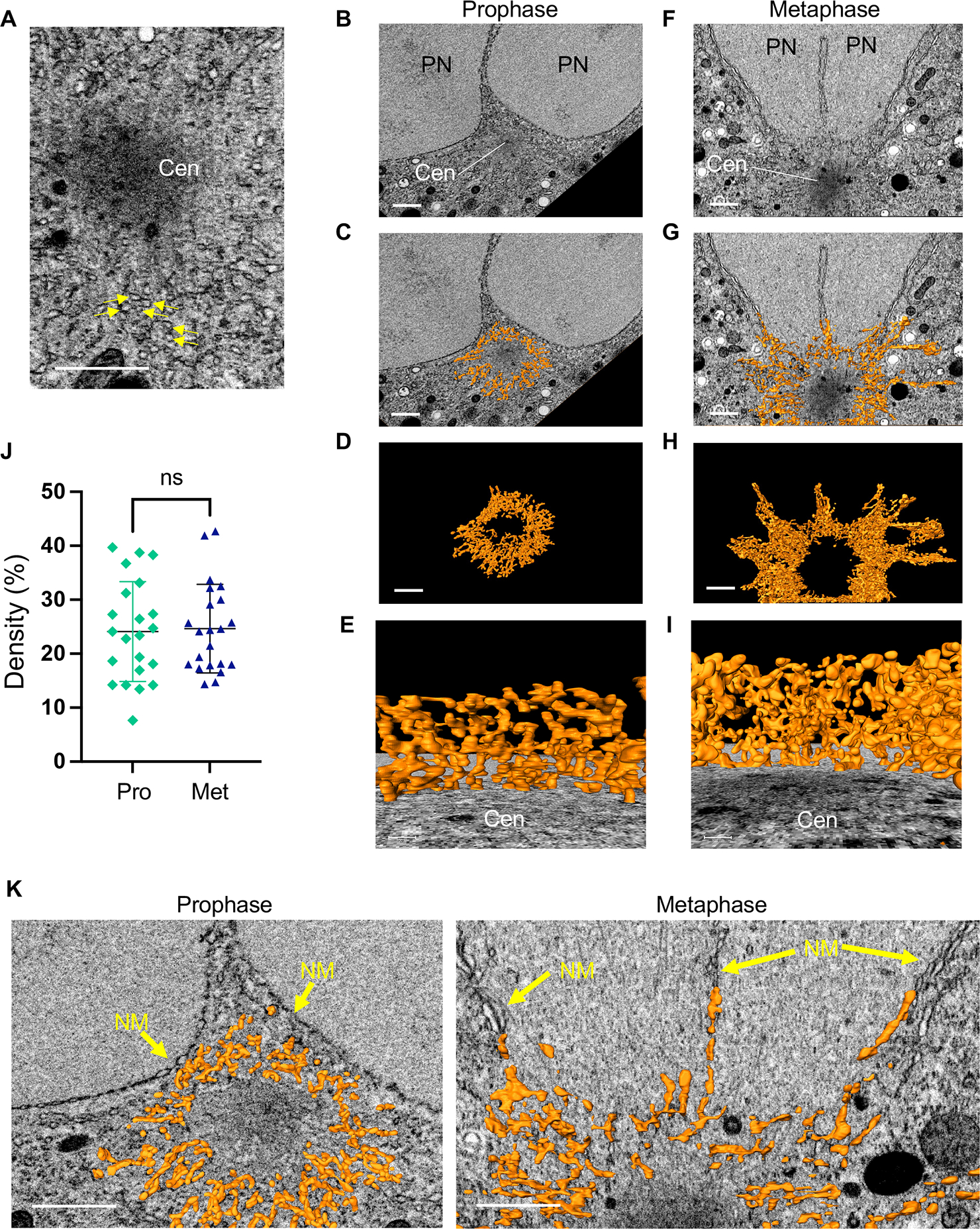
(A) A single SEM image from a region surrounding the centrosome in metaphase. Arrows point to membrane structures. The centrosome (Cen) appears as a dark zone that, when segmented using 3D FIB-SEM data, forms an irregular sphere (not shown). Scale bar= 1 μm. (B-I) 3D reconstructions using FIB-SEM data from 31 of the membranes around the centrosome in a 1-cell embryo in prophase (panels B-E) and metaphase (panels F-I). Additional examples of prophase and metaphase centrosomes and their associated membranes are shown in Figure S2 and S3, respectively. A portion of the two pronuclei (PN) is visible adjacent to the centrosome in panels B, C, F and G. (B, F) A SEM image from the centrosome’s mid-section. Scale bar= 1 μm. (C, G) The same SEM images as in panels B and F, superimposed with a 200 nm (in the z axis) of segmented membrane (orange) around the centrosome. Scale bar= 1 μm. (D, H) An 800 nm segment of the reconstructed membranes surrounding the centrosome from the same prophase (D) and metaphase (H) embryos as above. Scale bar= 1 μm. (E, I) A view from the centrosome towards the pronuclei/chromosomes (not shown) through the entire centriculum wall. Scale bar = 100 nm. (J) Centriculum density was determine by cropping at least 10 consecutive 250 nm3 cubes from the center portion of two centricula in 1-cell embryos in either prophase (Pro) or metaphase (Met), and determining the percent volume that is occupied by a membrane. Bars indicate means and standard deviations. Statistical analysis was done using Student t-test. (K) Enlarged images of the same centricula as in panels C and G, with a 100 nm slice of segmented membrane superimposed on the SEM image. Arrows point the nuclear membranes (prophase) or remnants thereof (metaphase). For additional images of the relationship between the centriculum and nuclear membrane see Figure S4. For related data see Figures S2, S3 and S4.
The density of the centriculum was the same in prophase and metaphase, as determined by the amount of membrane per unit volume (Figure 2J). However, the extent of centriculum association with the NE changed during the cell cycle. During prophase, the centriculum is distinct from the pronuclear membranes (Figure 2K, left panel, Figure S2, Ai and Bi, and Figure S4A). In contrast, during metaphase the centriculum is indistinguishable from the remnants of the pronuclear membranes (Figure 2K, right panel, Figure S3Ai and Bi and Figure S4B–D), which remain as perforated sheets around and between both pronuclei 31. Thus, the centriculum may play a role in associating the centrosome with mitotic remnants of the nuclear membrane.
The centrosome is encased by the centriculum
Having established that the membrane around the centrosome, the centriculum, has a reticular structure, we next investigated the spatial and functional relationship between centrosome components and the centriculum. We first examined the localization of centrosomal proteins with respect to the centriculum at metaphase, starting with an inner most centrosomal protein, the centriolar protein SAS-6, and working our way outwards 34: the inner PCM protein, SPD-2, the outer PCM protein, SPD-5, the Aurora A kinase, AIR-1, and finally the α-tubulin subunit, TBA-2. Based on images and traces of fluorescence intensities (Figure S5A), SAS-6 and SPD-2 were clearly within the confines of the centriculum (Figure 3A and B), while SPD-5 abuts the centriculum (Figure 3C). AIR-1 and TBA-2 partially overlap with the centriculum (Figure 3D and E). The peak intensities of the centriculum were outside the peak intensities of both AIR-1 and TBA-2, suggesting that the centriculum surrounds the outermost part of the centrosome.
Figure 3: The centrosome is encased by the centriculum.
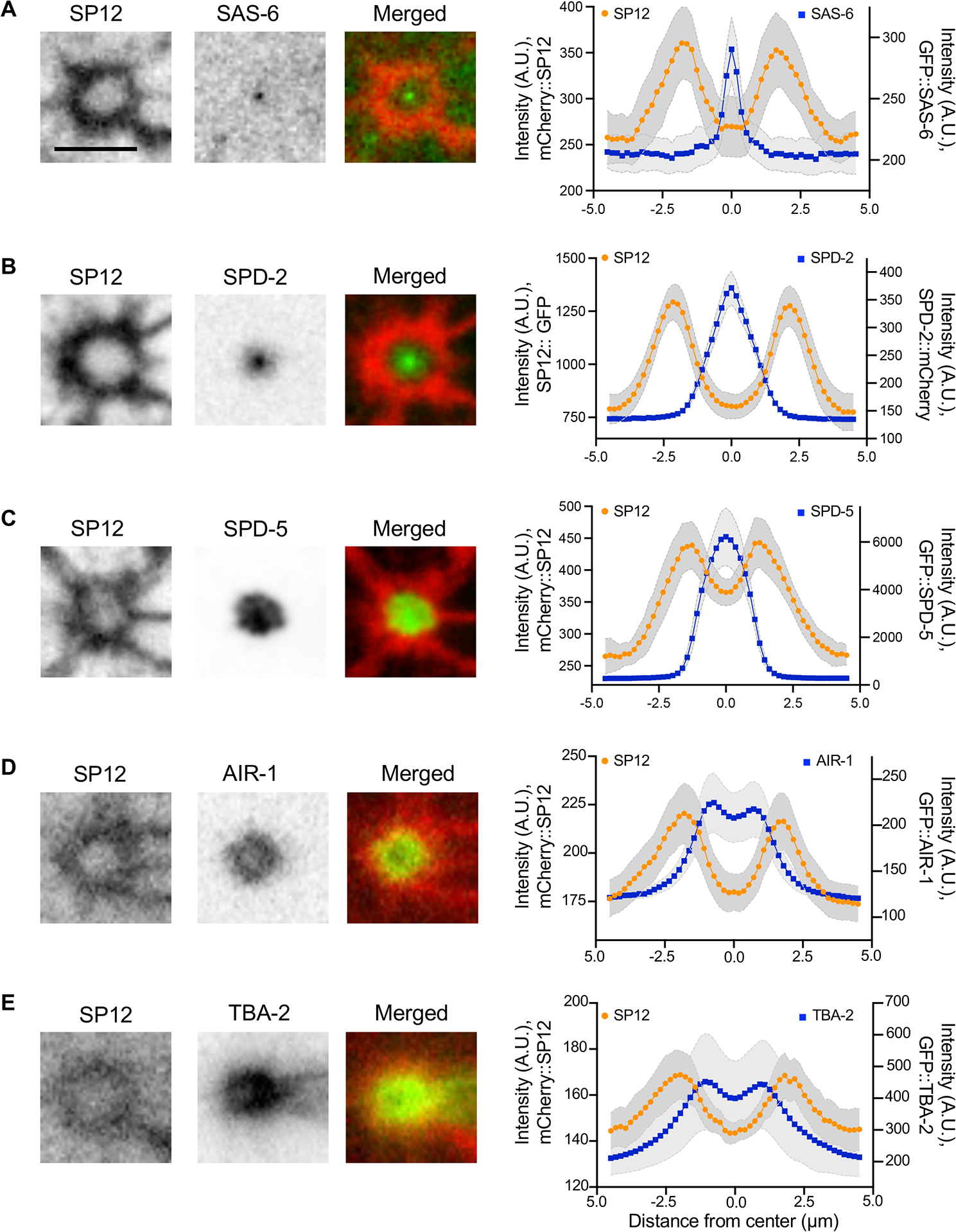
(A-E) The spatial relationship between the centriculum (detected using mCherry::SP12 or SP12::GFP) and the following components of the centrosome in a 1-cell stage embryo at metaphase: GFP::SAS-6 (strain OCF124, n=6 centrosomes), SPD-2::mCherry (OCF127, n=10 centrosomes), GFP::SPD-5 (OCF176, n=6 centrosomes), GFP::AIR-1 (OCF158, n=16 centrosomes) and GFP::TBA-2 (MSN146, n=8 centrosome). Representative images are shown. Scale bar = 5 μm. Graphs show the fluorescence intensities along lines (Figure S5A) that traverse the centrosomal protein (blue) or the centriculum (orange). Error (in grey) around the mean represents 95% confidence interval. A.U. = arbitrary units. For related data see Figure S5.
Centriculum size is dependent on the integrity of the centrosome and on microtubules
Given that SUN-1 accumulates at centrosomes immediately after fertilization (Figure 1D), and because the LINC complex is required for tethering the centrosome to the NE 4, we tested whether the LINC complex is required for the formation of the centriculum, by down-regulating ZYG-12 via RNAi. Despite their detachment, centricula were still observed in embryos from zyg-12 RNAi treated worms, and their size at metaphase (as determined by NE breakdown) was equivalent to centricula in control metaphase embryos (Figure 4A). We cannot exclude the possibility that low levels of ZYG-12, below the threshold needed for centriculum tethering to the NE, are still present. Nonetheless, our data are consistent with the possibility that the LINC complex is not essential for centriculum formation.
Figure 4: Centriculum size depends on the integrity of the centrosome.
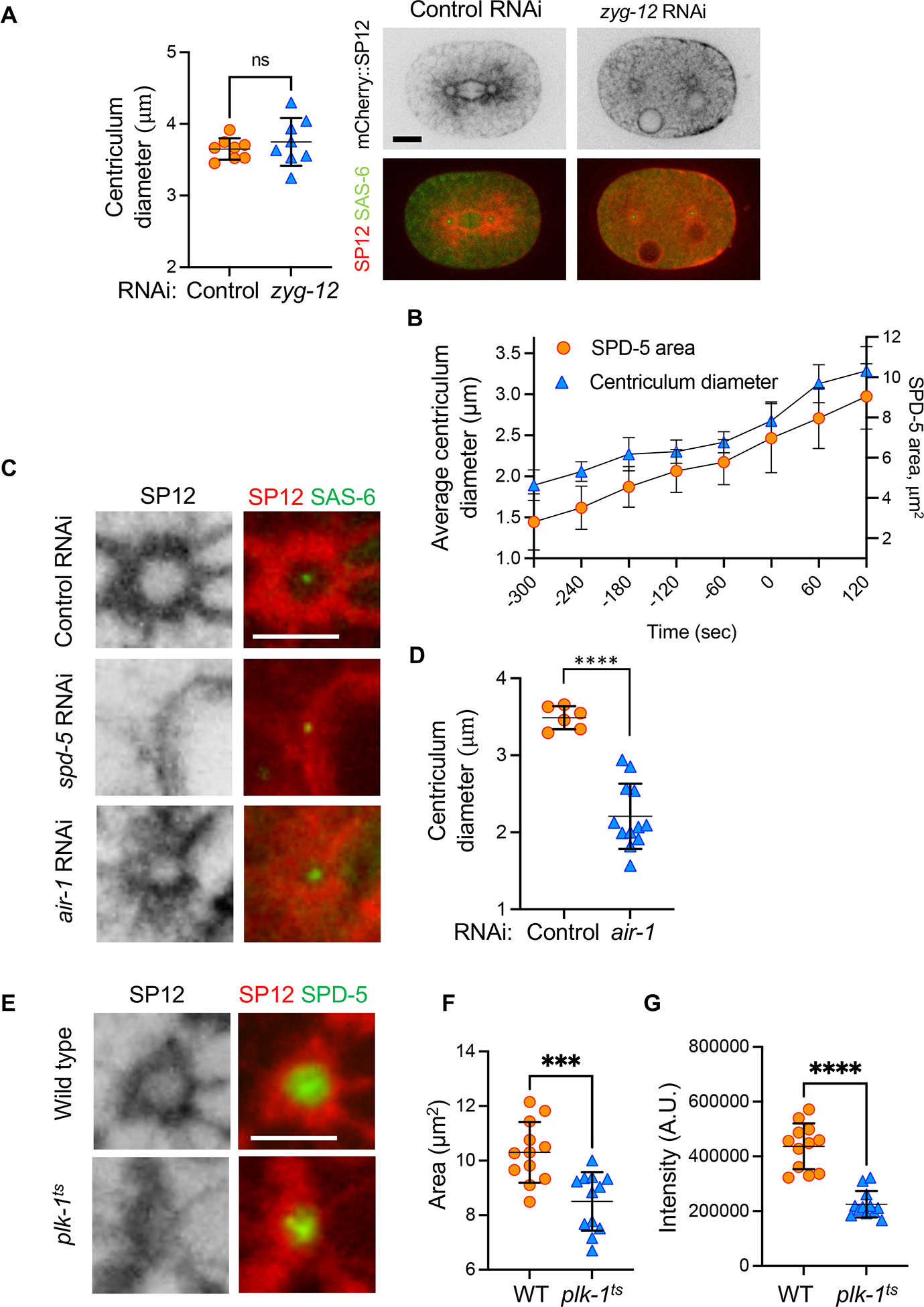
(A) Worms expressing mCherry::SP12 and GFP::SAS-6 (to identify centrosome location; strain OCF124) were treated with either control or zyg-12 RNAi. Shown are representative 1-cell embryos at metaphase from either condition. In the case of RNAi against zyg-12, metaphase was determined as the last time point before NE breakdown, when nuclei become deformed. Centriculum diameter was determined as described under STAR methods. Statistical analysis was done using two-tailed Mann Whitney test. Scale bar= 10 μm. (B) Centriculum diameter and SPD-5 area were determine during the development of 1-cell embryos (n=8 centricula/centrosomes) expressing mCherry::SP12 and GFP::SPD-5 (OCF164) at the indicated time points (t=0 is at metaphase). Error bars indicated standard deviation. (C) Representative images of centricula, as detected by mCherry::SP12, in 1-cell embryos at metaphase in a strain also expressing GFP::SAS-6 (OCF124) that was treated with the following RNAi conditions: Control RNAi (against smd-1 61, top row) by feeding, injection of double stranded RNA (dsRNA) against spd-5 (middle row), and feeding RNAi against air-1 (bottom row). Control injection of dsRNA against smd-1 was indistinguishable from feeding control RNAi (not shown). Scale bar = 5 μm. (D) Quantification of centriculum diameters of control and air-1 RNAi as shown in panel (C). Bars represent means and standard deviation. Statistical analysis was done using two-tailed Mann Whitney test, P=0.0001. Note that because there is ambiguity in determining the cell cycle stage of embryos from air-1 RNAi treated worms due to lack of chromosome alignment, the size of the centriculum is only an estimate. (E) Centricula in 1-cell embryos at metaphase from wild type (WT, OCF176) and plk1-(or638ts) (OCF170) worms expressing mCherry::SP12 and GFP::SPD-5 and grown at the semi-permissive temperature for the plk-1 mutant. Representative images are shown. Scale bar = 5 μm. (F, G) Quantification of GFP::SPD-5 area (panel F) and total SPD-5 fluorescence intensity (panel G) at a central plane of 1-cell metaphase embryos from wild type and plk-1(or638ts) worms as shown in panel E. Statistical analyses were done using unpaired t-test. p= 0.0006 (panel F) and <0.0001 (panel G).
Previous studies showed that the PCM increases in size as cells progress through the cell cycle 35,36. The same is true for the centriculum: the increase in centriculum diameter was proportional to the increase in SPD-5 area at the centrosome central plane (Figure 4B). We thus hypothesized that centriculum size could be dependent on the size of the underlying PCM. To test this, we depleted the PCM component, SPD-5, by dsRNA injection into worms expressing the SP12 ER marker fused to mCherry (mCherry::SP12) and the SAS-6 centriole marker fused to GFP (GFP::SAS-6). This allowed us to determine the location of the centrosome even in the absence of the PCM. Consistent with our hypothesis, SPD-5 depletion led to the disappearance of the centriculum (Figure 4C, n=14), suggesting that the centriculum requires the PCM for its formation. A similar phenomenon was reported by Audhya et al 37. To further address this possibility, we depleted another PCM component, AIR-1, by feeding RNAi. AIR-1 depletion led to a significant decrease in centriculum size (Figure 4C and D), although not as dramatically as SPD-5 depletion. This is likely because depletion of AIR-1 does not have as dramatic an effect as SPD-5 depletion on centrosome components 35,37. In addition, AIR-1 depletion does not affect an already assembled PCM 38, and AIR-1 depletion by RNAi feeding may not have been complete as injection of dsRNA, further dampening the observed effect. Nevertheless, these results support the idea that there is a spatial relationship between the PCM and the centriculum. Consistent with this, downregulation of the Polo-like kinase PLK-1 using the conditional plk-1(or683ts) mutant was previously shown to severely affect the ER-derived membrane in the vicinity of the centrosomes 39. When examining the PCM using GFP::SPD-5 in plk-1(or683ts) mutants at the semi-permissive temperature, we found that the centriculum was disorganized but the PCM was still there (Figure 4E). However, the amount of SPD-5 and the area that it occupied were smaller in plk-1(or683ts) mutants grown at semi-permissive temperatures (Figure 4F and G), consistent with the role of PLK-1 in C. elegans centrosome maturation 36. Taken together, these data show that the presence and morphology of the centriculum are dependent on the underlying centrosome.
Finally, we examined whether the establishment and maintenance of the centriculum are dependent on microtubules. To do so, we treated permeabilized embryos with the microtubule depolymerizing drug nocodazole (or DMSO alone, as a control) and examined the effect on the centriculum shortly thereafter. For centriculum establishment, 1-cell embryos expressing GFP::TBA-2 and mCherry::SP12 were followed until they completed cytokinesis and were then exposed to either DMSO or nocodazole (Figure 5A). The embryos were imaged 6-minute later, when they were in the 2-cell stage (Figure 5B). In 2-cell embryos, the AB cell (on the left) enters mitosis before the P1 cell. In control treated embryos, centricula were clearly visible, as expected. In nocodazole-treated embryos, however, no centricula were detected (n=6 embryos) and none formed even at later time points. Despite the absence of centricula, the centrosomes in nocodazole treated embryos were still associated with an accumulation of ER membrane near the NE. Because microtubules are not needed for PCM formation 35, this suggests that microtubules play a direct role in establishing the centriculum (see also below).
Figure 5: The centriculum depends on microtubules and affects centrosome function and microtubule organization.
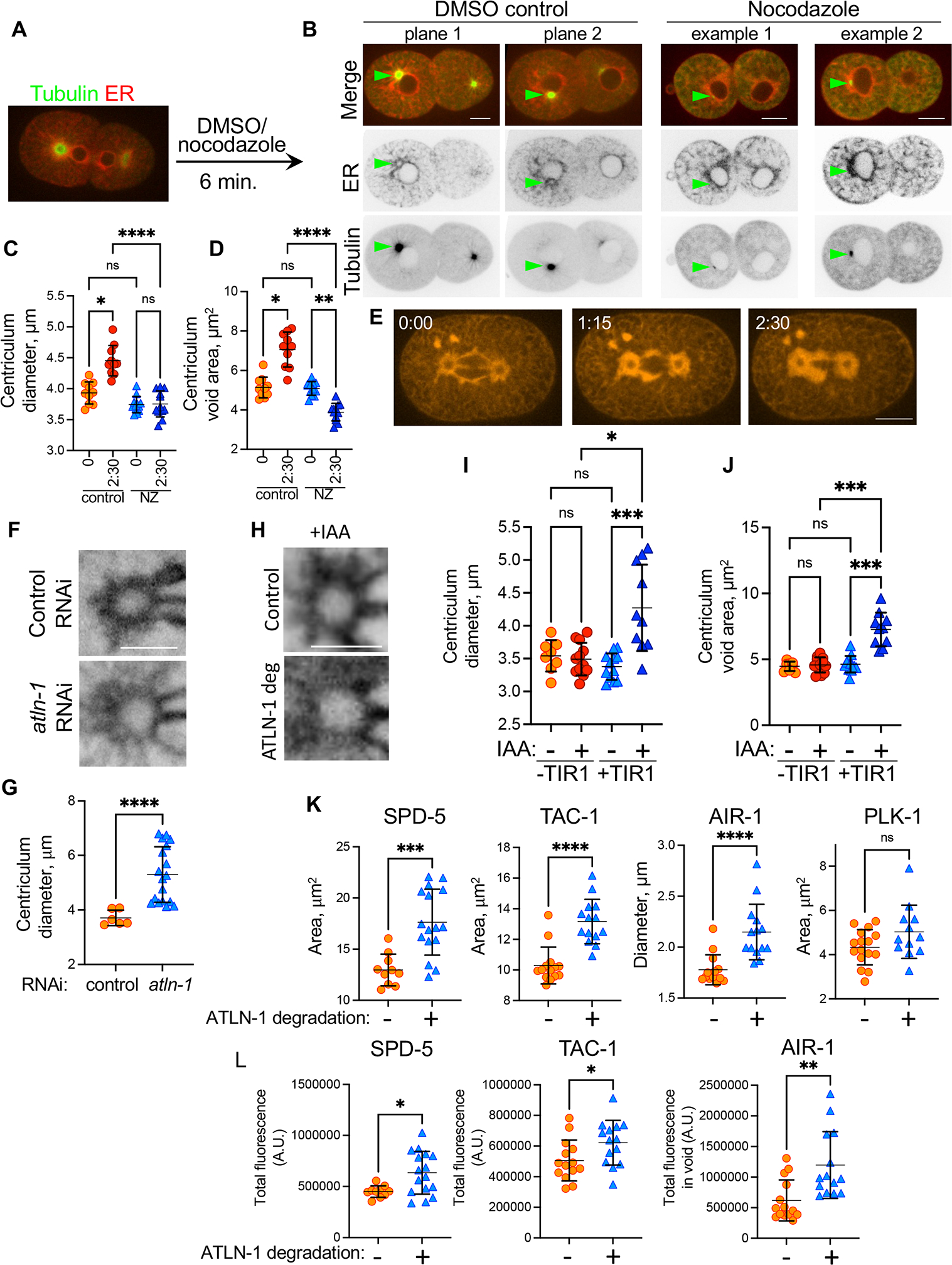
(A) Design of centriculum establishment experiments: embryos expressing GFP::TBA-2 and mCherry::SP12 (strain MSN146) from worm that were treatment with perm-1 RNAi to permeabilize the eggshell were imaged as they completed cytokinesis and then exposed to either DMSO (control) or nocodazole, as detailed under STAR methods. Embryos were imaged again after 6 minutes, as shown in (B). (B) Two focal planes from a control treated embryo and two examples of nocodazole treated embryos as described in panel (A). In these 2-cell embryos, the AB cell is on the left. Green arrows point to centrosome location as determined by the focal point of the tubulin signal. Scale bars = 10 μm. (C, D) Diameter (C) and void area (D) of centricula from metaphase embryos that were treated with DMSO (control) or nocodazole (NZ) for 0 and 2:30 minutes. Centriculum void area was measured as described in Figure S6A. Measurements were based on images of embryos expressing mCherry::SP12 (OCF124), shown in panel (E). n=10 and 12 for control and nocodazole treatment, respectively. Error bars indicate mean and standard deviation. Statistical analyses were done using Kruskal Wallis test with multiple comparisons (panel C: *, p=0.035; ****, p<0.0001. Panel D: *, p=0.0301, **, p=0.0058, ****, p<0.0001). (E) A representative example of a metaphase 1-cell embryo expressing mCherry::SP12 that was treated at time 0 with nocodazole. Indicated times are in minutes. Scale bar= 10 μm. (F) Representative examples of centricula from 1-cell embryos at metaphase expressing SP12::GFP (OCF5), following control (top panel) or partial atln-1 RNAi treatment (bottom panel). The effect of several RNAi treatment is shown in Figure S5C. Scale bar= 5 μm. (G) Quantification of average centriculum diameter from the experiment shown in panel F (n=6 and 18 for control (orange) and atln-1 partial RNAi (blue), respectively). p<0.0001 using the Mann-Whitney test. Error bars indicate mean and standard deviation. (H-J) The effect of ATLN-1 down-regulation using the auxin degradation system: ATLN-1 was tagged with an auxin inducible degron (AID; atln-1::degron) tag and introduced into worm strains without (OCF116) or with (OCF118) the TIR1 ubiquitin ligase. These strains also expressed the ER marker SP12::GFP. Centriculum diameter (panel I) and void area (panel J) were measured in the presence or absence of the auxin analog IAA. Panel H shows representative examples of centricula from control (OCF116) or atln-1::degron strain (OCF118) in the presence of IAA. Scale bar= 5 μm. Error bars indicate mean and standard deviation. Statistical analyses were done using Kruskal Wallis test with multiple comparisons (panel I: *, p=0.0131; ***, p=0.0004. Panel J: ***, p=0.0004 and 0.0005 for +TIR1 − vs. + IAA and − vs + TIR1 both with IAA, respectively). (K) Areas occupied by the indicated fluorescently-tagged centrosomal proteins, as determined at the centrosome mid-plane, in 1-cell embryos at metaphase expressing atln-1::degron, without (orange symbols) or with (blue symbols) IAA treatment. The effect of the treatment on centriculum diameter is shown in Figure S5E. For AIR-1, because it forms a ring, the diameter of the ring, rather than the area occupied by AIR-1, was determined. The number of centricula analyzed (−/+ IAA) and the p values are as follows: SPD-5 (OCF164): 14/18, p=0.0003; TAC-1 (OCF167): 14/14, p<0.0001; AIR-1 (OCF172): 14/14, p<0.0001; PLK-1 (OCF166): 18/12, p=0.29. Statistical analyses were done using unpaired t-test (SPD-5) or non-parametric Mann-Whitney test (TAC-1, AIR-1 and PLK-1). Representative images for all conditions are shown in Figure S5F and quantification of PLK-1 distribution is shown in Figure S5G. (L) Intensity of the indicated GFP-tagged centrosomal proteins in the same centricula as in panel K. For AIR-1, which resides both in and out of the centrosome, total fluorescence intensity was determined in the area encompassed by the centriculum, referred to as the centriculum “void” (Figure S6A). p values, as determined by unpaired t-test (SPD-5, TAC-1) or non-parametric Mann-Whitney test (AIR-1), were as follows: SPD-5: p=0.0125; TAC-1: p=0.0372; AIR-1: p=0.0021. For related data see Figures S5 and S6.
For centriculum maintenance, 1-cell metaphase embryos expressing mCherry::SP12 and GFP::SAS-6 were imaged (t=0:00), treated with either DMSO or nocodazole and imaged again 1:15 and 2:30 minutes later (Figure 5C–E). In control embryos, centriculum diameter increased (Figure 5C), as expected (Figure 4B). In contrast, microtubule depolymerization abolished centriculum expansion (Figure 5C). In addition, the space that is surrounded by the centriculum, which we define as the centriculum “void” area (Figure S6A), increased in the control embryos but shrunk significantly following nocodazole treatment (Figure 5D and E). Our data suggest that microtubules push the centriculum membrane away from the centrosome.
Increased centriculum size leads to increased PCM size and microtubule nucleating capacity
We next examined whether the centriculum affects centrosome size. To date, the only conditions that completely abolish the centriculum is by eliminating either the SPD-5 PCM component (Figure 4C) or microtubules (Figure 5A–E), and neither condition can be used to study the effect of the centriculum on centrosome size or function. However, by mild manipulation of ER structure we were able to alter centriculum size, which then allowed us to examine the consequences to the underlying centrosome. Specifically, the reticular nature of the centriculum suggested that the dynamin-related GTPase, ATLN-1, the C. elegans homolog of atlastin, could play a role in centriculum structure. Atlastin is known for its role in ER-ER fusion 40, and in particular for creating junctions between ER tubules. Tagging endogenously expressed C. elegans ATLN-1 with GFP at its C-terminus revealed that it localizes to the endoplasmic reticulum (Figure S5B), similar to the localization of atlastin in other organisms 41–43. Importantly, ATLN-1::GFP also localizes to the centriculum (Figure S5B). RNAi against atln-1 led to complete disruption of the ER and embryonic lethality before centricula could be formed (Figure S5C). A milder RNAi treatment (using a mix of E. coli expressing double stranded RNA against altn-1 (20%) and the control smd-1 (80%)) allowed most embryos to progress through the first mitosis. Under these conditions, centricula in atln-1 downregulated embryos at metaphase were significantly larger than the control treatment (Figure 5F and G). However, because the partial RNAi treatment still resulted in embryos that failed to reach mitosis, we turned to reducing the levels of ATLN-1 using the auxin-induced degron system 44, which allowed for a more precise temporal down-regulation of the protein. To this end, we created an auxin-inducible atln-1::degron allele and introduced it into worm strains that either had or did not have the TIR1 ubiquitin ligase that is necessary for auxin-mediated degradation. The requirement of ATLN-1 for cell viability necessitated the use of a short exposure (20–25 minute) to the auxin analog indole-3 acetic acid (IAA), meaning that under these conditions, ATLN-1 was present in lower amounts but not eliminated. Treatment of worms with IAA in the absence of TIR1 had no effects on centriculum size (Figure 5I and J). However, the addition of IAA in the presence of TIR1 led to a significant increase in centriculum diameter and void area (Figure 5H–J). It was previously shown that centrosome size is proportional to embryo size 36, raising the possibility that ATLN-1 downregulation affected centriculum size indirectly by affecting cell size. However, the average size of control and ATLN-1 downregulated embryos was the same (Figure S5D). Thus, downregulation of ATLN-1 leads to an increase in centriculum size.
The ability to increase the size of the centriculum allowed us to explore the effect of the centriculum on centrosome size and function. To do so, the localization of GFP-tagged SPD-5, TAC-1, AIR-1, and PLK-1 was analyzed in 1-cell embryos at metaphase from IAA-treated and untreated worms expressing ATLN-1::degron and the TIR1 ubiquitin ligase. ATLN-1 downregulation consistently led to larger centricula for each of these strains (Figure S5E). Unexpectedly, the increase in centriculum size was accompanied by an increase in the area occupied by SPD-5, TAC-1 and AIR-1 (Figures 5K and S5F and G; because AIR-1 appears at as a ring in the central focal plane of the centriculum, we measured its diameter rather than area). Moreover, when ATLN-1 was downregulated, the total amount of SPD-5, TAC-1 and AIR-1 at the centrosome was increased (Figure 5L). This increase in PCM components may be due to recruitment from cytoplasmic pools as more space becomes available following the increase to centriculum size. The localization and intensity of PLK-1::GFP, which resides in the inner sphere of the PCM 34, remained unchanged when ATLN-1 was downregulated (Figure S5G). Taken together, these results show that increased centriculum size leads to a larger PCM, suggesting that the size of the centrosome in C. elegans is affected by the membrane that surrounds it.
We next tested whether the change in PCM size due to centriculum enlargement affected the centrosome’s capacity to nucleate microtubules. To do so, we examined the abundance of TBA-2, an α-tubulin subunit, and EBP-2, a plus-end microtubule-binding protein, under conditions of centriculum expansion (i.e. ATLN-1 downregulation) in 1-cell metaphase embryos. As before, downregulation of ATLN-1 resulted in centricula that had larger diameters and void areas (Figure S6B and C). In 2-dimentional images, both TBA-2 and EBP-2 formed ring like structures within the centriculum (Figure 6A and B and Figure S6D and E). Down-regulation of ATLN-1 led to an increase in TBA-2 and EBP-2 “ring” diameter and to a greater abundance of both proteins in the centriculum void area (Figure 6Ci, ii and 6Di, ii). Since EBP-2 binds microtubule + ends, these results suggest that when the centriculum is expanded, the centrosome is capable of nucleating more microtubules, consistent with the increase in the amount of PCM proteins under these conditions (Figure 5L). We next determined whether enlarged centricula affect the spindle in 1-cell metaphase embryos by examining the abundance of TBA-2 and EBP-2 at the spindle midzone (Figure S6A). We also determined spindle width at the spindle midzone, as well as spindle length (Figure S6A). When ATLN-1 was down regulated the amount of TBA-2 and EBP-2 at the spindle mid-zone was greater (Figure 6Ciii), and the spindle was wider (Figure 6Diii). Spindle length, however, was unchanged (Figure 6Cv and Dv). The wider spindle when centricula are larger is consistent with microtubules radiating from a larger volume. This also raised the possibility that the centriculum affects the orientation of microtubules emanating from the centrosome.
Figure 6: Centriculum expansion increases the microtubule nucleating capacity of the centrosome.
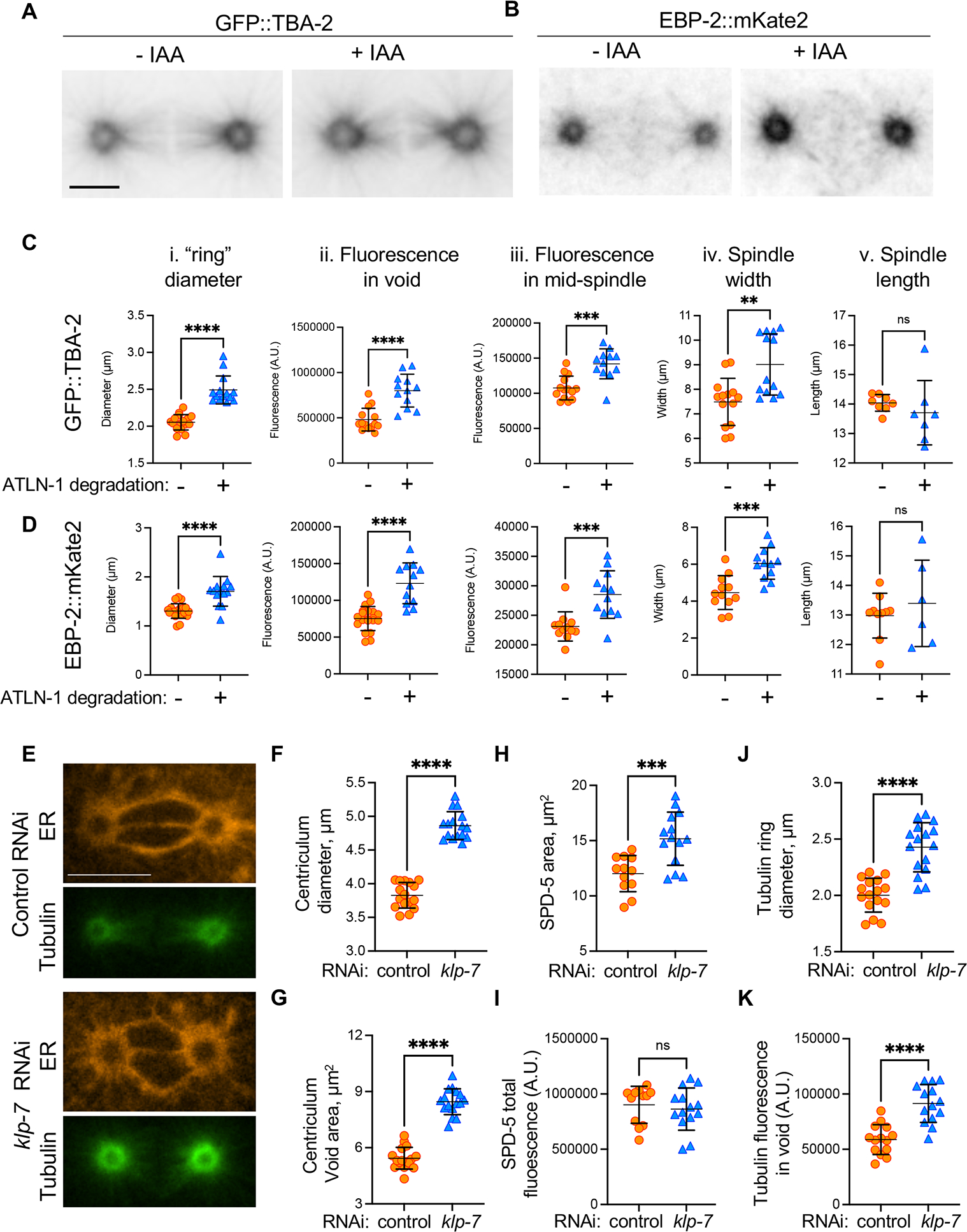
(A) A typical example of tubulin distribution in metaphase 1-cell embryo expressing GFP::TBA-2, mCherry::SP12, ATLN-1::degron and TIR1 (strain OCF183), in the absence or presence of IAA. Scale bar= 10 μm. (B) The same as panel A except cells were expressing EBP-2::mKate2 and SP12::GFP (strain OCF162). (C and D) quantification of TBA-2 or EBP-2 “ring” diameter (panel i), TBA-2 or EBP-2 fluorescence intensities in the centriculum void area (ii) and at the mid-spindle (iii), spindle width (iv), and spindle length (v) for the same strains as in panels (A) and (B), in the absence (orange symbols) or presence (blue symbols) of IAA. The methodology to quantify these parameters is shown in Figure S6A and centriculum diameters and void areas are shown in Figure S6B and C. Examples of TBA-2 and EBP-2 localization relative to the centriculum are shown in Figure S6D and E. Error bars represent mean and standard deviation. Statistical analyses were done using unpaired t test. The number of centrosomes analyzed (−/+ IAA) and p values for each panel were as follows: Ci: n=16/14, p<0.0001; Cii: n=14/12, p<0.0001; Ciii: n=14/12, p=0.0001; Civ: n=14/13, p= 0.0014; Cv: n=8/7, p=0.4205; Di: n=10/14, p<0.0001; Dii:20/12, p<0.0001; Diii: n=12/12, p=0.0007; Div: n=12/12, p=0.0002; Dv: n=10/6, p=0.4615. (E) Representative images of 1-cell metaphase embryos expressing mCherry::SP12 (orange) and GFP::TBA-2 (green) (strain MSN146) treated with control RNAi (top two panels) or RNAi against klp-7 (bottom two panels). Scale bar= 10 μm. (F-K) Measurements of centriculum diameter (F), void area (G), SPD-5 area (H), SPD-5 fluorescence in the centriculum void (I), tubulin ring diameter (J), and tubulin intensity in the centriculum void (K), in a strain expressing mCherry::SP12 and GFP::TBA-2 (MSN146, panels F, G, J and K) or mCherry::SP12 and GFP::SPD-5 (OCF176, panels H and I) that were treated with either control RNAi (orange symbols) or RNAi against klp-7 (blue symbols). Error bars represent mean and standard deviation. Statistical analyses were done using unpaired t test. The number of centrosomes analyzed (control/klp-7 RNAi) and p values for each panel were as follows: panel F: n=16/16, p<0.0001; G: n=16/16, p<0.0001; H: n=12/12, p= 0.0008; I: n=12/12, p=0.5604; J: n=16/16, p<0.0001; K: n=12/14, p<0.0001. For related data see Figure S6.
To determine whether the effect of the enlarged centriculum on the centrosome is a general phenomenon and not specific to ATLN-1 downregulation, we sought an alternative method for centriculum enlargement. Based on the effect of microtubule on the centriculum, we hypothesized that it should be possible to expand the centriculum by either elongating or increasing the number of microtubules at the centrosome. To test this possibility, we treated worms with RNAi against klp-7. KLP-7 is a kinesin, and its down-regulation results in an increase in microtubule outgrowth from centrosomes 45. If microtubules “push” the centriculum membrane away from the centrosome, then the increase in microtubule outgrowth following downregulation of KLP-7 should lead to a larger centrosome. Indeed, 1-cell metaphase embryos from worms treated with RNAi against klp-7 exhibited larger centricula than control embryos (Figure 6E and F). Moreover, the area occupied by both SPD-5 and TBA-2 was significantly higher (Figure 6H and J). The expansion of the TBA-2 area could be attributed to KLP-7’s role in microtubule dynamics. However, since SPD-5 recruitment to the PCM is independent of microtubules 35, the expansion of the PCM when KLP-7 is down-regulated is likely due to the enlargement of the centriculum caused by an increase in amount of microtubules nucleated by the centrosome (Figure 6K). While SPD-5 occupied a larger area when KLP-7 was downregulated, its amount in the centrosome did not increase (Figure 6I). We speculate that this strain may not have SPD-5 reserves in the cytoplasm. Overall, our results suggest that under normal conditions, the centriculum restricts the size of the centrosome, and consequently its microtubule nucleating capacity.
The centriculum may serve as a microtubule “filter”
During mitosis in C. elegans embryos, microtubules originating from centrosomes must somehow traverse the centriculum to reach the chromosomes. To visualize both microtubules and membranes, we analyzed published electron tomography data of 1-cell C. elegans embryos at metaphase 46. Microtubules in this data set had been traced previously 46, and we segmented the centriculum between the centrosome and nucleoplasm in selected regions of the serial sections (Figure 7A). Within the centrosome, microtubules were oriented randomly (Figure 7A, right panel and enlargement i). However, after passing through the centriculum, on the nucleoplasm side, the microtubules radiated away from the centrosome, roughly in the same direction (Figure 7A, right panel and enlargement ii). Moreover, our data suggest that the centriculum allowed only a fraction of the microtubules to pass, blocking microtubules that “hit” the membrane (Figure 7B and C, Figure S7 and Video S1). In the example shown in Figure 7B, C and Video S1, the microtubules labeled in yellow and blue terminate inside the centriculum, while the microtubule labeled in green passes through the centriculum. In fact, the existence of the centriculum could explain the ring-like structures of tubulin and EBP-2 (Figure 6A and B, and Figure S6D and E) around the centrosome that have also been seen by others (for example, 46–50). While the length distribution of microtubules is determined by their inherent dynamic instability, microtubules associated proteins and the availability of tubulin heterodimers 45,51–53, the presence of the centriculum may contribute to the abundance of short microtubules in the vicinity of the centrosome 11,46. Thus, the centriculum may serve as a filter by limiting the number of microtubules that are allowed to extend out of the centrosome.
Figure 7: The centriculum may serve as a microtubule filter and assist in NE breakdown.
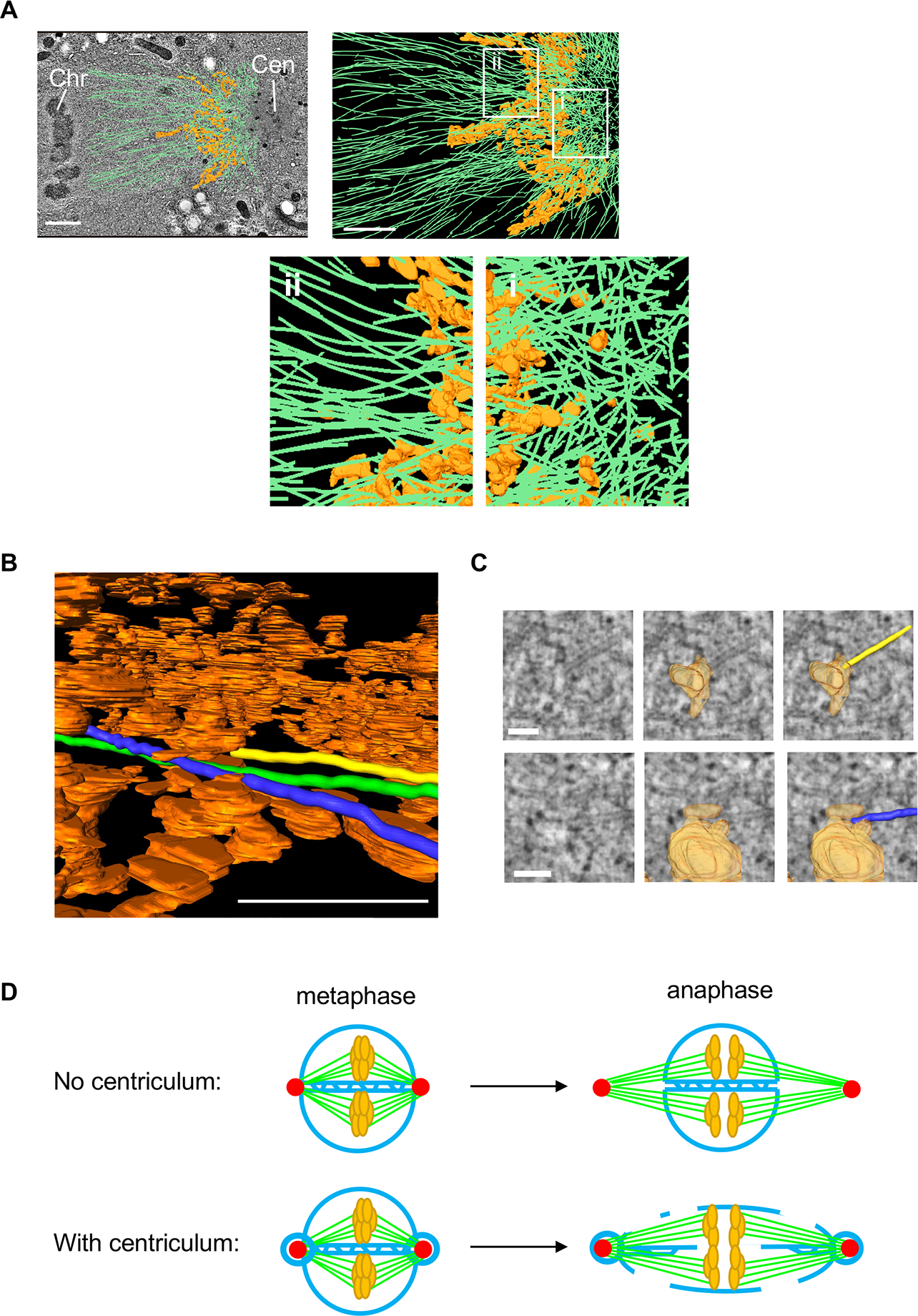
(A) Top left panel: microtubule vectors (green), and a segment of the centriculum (orange) between the centrosome (Cen) and nucleoplasm in a 1-cell embryo in metaphase, superimposed on a single TEM image. Tomography datasets, including microtubule assignments, are from 46. Metaphase chromosomes (Chr) can be seen on the far left of the image. Scale bar= 5 μm. Top right: the same microtubule and centriculum reconstructions as in the left panel, but without the TEM image, which obscures structures below the plane of the image. The thickness of this segment, along the z axis, is ~ 60 nm. Regions i and ii, on the centrosome and chromosome sides of the centriculum, respectively, are enlarged below. Scale bar= 5 μm. (B) Segmentation of three microtubules (marked in yellow, blue and green) shown as they traverse the centriculum (orange). Note that the yellow microtubule terminates at a membrane. See Video S1 for the entire route of these three microtubules through the centriculum. Scale bar = 0.5 μm. (C) TEM images (left column) overlayed with membrane reconstruction (middle column) and membrane + microtubules (right column) of the yellow and blue microtubules from panel B and Video S1 at the sites where they hit the membrane. Scale bar= 100 nm. See additional examples in Figure S7. (D) A diagram explaining the possible role of the centriculum in nuclear envelope fenestration during mitosis. Centrosomes are indicated in red, membrane in blue, microtubules in green and chromosomes in orange. See text for more detail. For related data see Figure S7.
Discussion
The centrosome is considered a membraneless organelle. The existence of ER-derived membranes around centrosomes has been known for decades through studies using light microscopy 17–23, but their configuration and functional importance were unknown. Using two vEM approaches, we show that centrosome-associated membranes in C. elegans form a reticulum, leading us to name it the centriculum. The identification of the centriculum underscores the importance of vEM analysis; while the centriculum can be easily detected by conventional EM, in a single imaging plane it appears as an accumulation of vesicles. The reticular nature of the centriculum, on the other hand, provided insight into its possible functions. At present, we are unaware of other cellular organelles that are surrounded by a membrane reticulum. Our data show that the centriculum in the early C. elegans embryo affects centrosome function, suggesting that the centrosome is not as membraneless as previously assumed, although the membrane around them is not contiuous as in other organelles (e.g. mitochondria, the nucleus).
Our study sheds new light on the association of the centrosome with the NE, which was shown to be mediated by the LINC complex. The two components of the LINC complex, the SUN domain protein SUN-1 and the KASH domain protein ZYG-12, traverse the inner and outer nuclear membranes, respectively. ZYG-12 was previously shown to localize around the centrosome 4, but it was not clear how it could do so, given that ZYG-12 is an integral membrane protein. The presence of the centriculum provides an explanation; indeed, both components of the LINC complex, SUN-1 and ZYG-12, localize to the centriculum (Figures 1C and S1B). It should be noted, however, that unlike the NE, the centriculum does not have obvious membrane structures that are equivalent to an inner or outer nuclear membrane. Thus, the configuration of the LINC complex in the centriculum remains to be determined. In C. elegans, centrosomes play an important role in NE breakdown and the preferential loss of the NPC subunit NPP-3 from the NE adjacent to centrosomes in prometaphase 26,54. We observed that the centriculum is adjacent to the NE in prophase and fused to it in metaphase (Figures 2, S2 and S3). We speculate that the ability of microtubules to enter the nucleoplasm, and loss of NPP-3 from the NE near centrosomes, coincide with centriculum-NE fusion. This step is also reminiscent of the insertion of the fission yeast Schizosaccharomyces pombe microtubule organizing center, the spindle pole body, into the NE, which is dependent on the LINC complex 55. Whether the LINC complex is required not only for tethering the centrosome to the NE, as noted above, but also for its insertion once centrosome migration is completed, is currently under investigation.
To fully understand the physiological role(s) of the centriculum we will need to identify conditions that abolish its formation without affecting other pertinent structures and processes, such as centrosome and spindle assembly. Thus far, the only conditions that abolished centriculum formation also disrupted the centrosome (Figure 4C–F) and/or microtubule assembly (Figure 5A–E); the latter was also seen in Drosophila 23. Nonetheless, we were able to identify a condition that altered centriculum size, namely the downregulation of atlastin (Figure 5F–J). How atlastin downregulation affects the structure of the centriculum is currently not known. Since atlastin is required for homotypic ER-ER fusion, we imagine that when it is down regulated, the centriculum contains fewer junctions and is thus less rigid. Given that centriculum formation depends on the PCM and microtubules, and that PCM size is affected by the centriculum, we speculate that as the centrosome matures there is a balance between outward forces exerted by the growing PCM and its associated microtubules, and resistance applied by the centriculum. When atlastin is downregulated, the centriculum may be less able to resist centrosome-associated forces, leading to an increase in centrosome size.
The relationship between the PMC and the centriculum introduces a new element when considering centrosome size control. Whether and to what extent the PCM is a phase-separated condensate is still up for debate (reviewed in 15). The C. elegans SPD-5 can form a liquid-liquid phase separated condensate in vitro 10. However, in vivo, SPD-5 and its analogous protein in Drosophila, centrosomin (Cnn), do not spread throughout the entire PCM once incorporated, suggesting that they are in a gel or solid-like state 56,57. An emerging model is that SPD-5 starts out as a condensate and then matures to a more solid state 10. We observed that increasing centriculum size results in a larger PCM that nucleates more microtubules, leading to a wider spindle (Figure 6C, D). At face value, our observations argue against a PCM that is purely liquid-liquid phase-separated, as the size of these condensates, and the fraction of condensate material that phase separates, is a function of the properties and concentration of condensate components and the liquid environment in which they reside 16, independent of a juxtaposed membrane. Interactions of a condensate with a membranes have been observed previously (reviewed in 58), and in these cases the membrane often serves as a platform for phase separated condensate nucleation. It is therefore possible that an early SPD-5 condensate forms on a “young” centriculum.
What might be the function(s) of the centriculum? It was previously suggested that the accumulation of ER around centrosomes in Drosophila serves to ensure that daughter cells receive adequate amounts of ER 22. We think that this is unlikely to be the case in C. elegans given that during mitosis, the ER is distributed throughout the cell and the fraction of the ER around centrosomes appears relatively small (Figure 1A, B). More recently, Araujo et al suggested that in Drosophila embryos, the ER affects spindle size and forces, possibly through membranes at the spindle poles 18. Our study suggests that the centriculum contributes to centrosome assembly and function. Moreover, the centriculum may limit the number of fully extended microtubules in two ways: by restricting the size of the PCM (Figures 5 and 6), thus limiting its microtubule nucleation capacity, and by blocking a fraction of microtubules from extending past the centriculum (Figures 7A–C and Figure S7). These microtubules, presumably, are not in the correct orientation to make it through the centriculum. We imagine that in the absence of a centriculum, a larger fraction of microtubules would be able to extend. Taken together, the centriculum may serve as a microtubule filter that affects microtubule orientation, length and number.
The centriculum may also be important for tethering the centrosome to the NE during mitosis. As discussed above, the fusion of the centriculum with the NE may facilitate early stages in nuclear envelope breakdown in the vicinity of the centrosomes. Subsequently, NE proteins dissociate from the rest of the NE, but the nuclear membranes remain and become highly fenestrated 31. We proposed that the fusion between the centriculum and the nuclear membrane contributes to this subsequent nuclear membrane fenestration as the spindle begins to elongate at metaphase, before sister chromatid separation at anaphase 48. Had the centriculum not existed (Figure 7D), the force generated by the elongating microtubules would not have been transmitted to the nuclear membrane, except by individual microtubules directly attaching to the nuclear membrane, as has been proposed previously 59,60. The centriculum, on the other hand, provides a mechanism for transmitting the force generated by spindle elongation to the nuclear membrane: as the centrosomes move apart, they pull the centricula with them, and the centricula, in turn, pull on the rest of the nuclear membrane. Assuming that there is no net increase in membrane at this stage, this pulling action will cause the membrane to fenestrate, as is observed during NE breakdown.
In summary, in this study we characterized a centrosome-associated membrane reticulum, the centriculum, and showed that there is a mutual dependency between the centriculum and the centrosome: The centriculum depends on the presence of the centrosome, and the size of the PCM is affected by the size of the centriculum. Our data suggested that the centriculum also affects centrosome function by orienting microtubules and limiting the fraction of microtubules that can fully elongate, and perhaps contributes to nuclear membrane fenestration. Given the conservation in centrosome components and ER organization, centricula are likely to exist in other organisms.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Orna Cohen-Fix (ornac@niddk.nih.gov).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact without restriction.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All experiments described in this study were done with the nematode Caenorhabditis elegans. C. elegans strains expressing SPD-2::mCherry 62, GFP::SAS-6 63, PLK-1::sGFP 25, GFP::AIR-1 37, GFP::TAC-164, EBP-2::mKate2 65, GFP::SPD-5 38, mTurquoise2::H2B 66, SP12::GFP and mCherry::SP12 67, NPP-1::GFP 61, YFP::LMN-1 68, EMR-1::GFP, LEM-2::GFP and LEM-2::mCherry 39,69, ZYG-12::GFP 4, SUN-1::mRuby 31, NPP-12::mNeonGreen 70, and the temperature sensitive mutant plk-1 (or683ts) 71 have been described previously and were used in creating strains for this study, listed in the Key Resources Table. Strains expressing endogenously tagged ATLN-1::GFP, 3XFLAG::degron tagged ATLN-1 and SUN-1::GFP were generated by CRISPR/Cas9-induced homologous recombination 72. FIB-SEM experiments were done with strain N2. All strains were maintained at 20°C (except strain OCF170, containing the plk-1 temperature sensitive allele or683ts, which was maintained at 16°C), on MYOB plates (in 1 liter: 2.0 g NaCl, 0.55 g TrisHCl, 0.24 g TrisOH, 4.6 g Bactotryptone, 8 mg Cholesterol, 20 g Agar) seeded with OP50 bacteria.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| None | ||
| Bacterial and virus strains | ||
| Escherichia coli OP50 | CGC | OP50 |
| Escherichia coli clone for C. elegans AIR-1 K07C11.2 | RNAi feeding library (Open Biosystems, Huntsville, AL) | N/A |
| Escherichia coli clone for C. elegans ZYG-12 ZK546.1 | RNAi feeding library (Open Biosystems, Huntsville, AL) | N/A |
| Escherichia coli clone for C. elegans SMD-1 F47G4.7 | RNAi feeding library (Open Biosystems, Huntsville, AL) | N/A |
| Escherichia coli clone for C. elegans PERM-1 T01H3.4 | RNAi feeding library (Open Biosystems, Huntsville, AL) | N/A |
| Escherichia coli clone for C. elegans KLP-7 K11D9.1 | RNAi feeding library (Open Biosystems, Huntsville, AL) | N/A |
| Biological samples | ||
| None | ||
| Chemicals, peptides, and recombinant proteins | ||
| Indole-3-acetic acid (auxin) | Alfa Aesar | Cat# A10556 |
| Nocodazole | Sigma Aldrich | Cat# 1404 |
| DMSO | Sigma Aldrich | Cat# 41648 |
| IPTG | Sigma Aldrich | Cat# I6758 |
| L15 buffer | Thermo Fisher Scientific | Cat# 21083027 |
| Qiagen MinElute Reaction Cleanup Kit | Qiagen | Cat# 28206 |
| MEGAscript™ T7 Transcription Kit | Invitrogen | Cat# AM1333 |
| Phenol:CHCl3:Isoamyl Alcohol | Invitrogen | Cat# 15593031 |
| Ethanol 200 proof | The Warner-Graham company | Cat# 64-17-5 |
| Levamisole | Sigma Aldrich | Cat# L9756 |
| TE buffer (10 mM, pH 8) | Quality biological | Cat# 351-011-131 |
| Noble agar | Sigma-Aldrich | Cat# A5431 |
| Critical commercial assays | ||
| None | ||
| Deposited data | ||
| None | ||
| Experimental models: Cell lines | ||
| None | ||
| Experimental models: Organisms/strains | ||
| C. elegans N2: wild isolate | CGC | N2 |
| C. elegans OCF3: unc-119(ed3) III; jjIs1092 [pNUT1::npp-1::GFP + unc-119(+)]; ltIs37 [pAA64: pie-1p::mCherry::his-58 + unc-119(+)] | Golden et al, 2009 61 | OCF3 |
| C. elegans OCF4: unc-119(ed3) III; qaIs3502 [pie-1p::YFP::lmn-1 + pie-1p::CFP:H2B + unc-119(+)]; ltIs37 [pAA64: pie-1p::mCherry::his-58 + unc-119(+)] | Golden et al, 2009 61 | OCF4 |
| C. elegans OCF5: unc-119(ed3) III; ojIs23 [pie-1p::SP12::GFP + unc-119(+)]; ltIs37 [pAA64: pie-1p::mCherry::his-58 + unc-119(+)] | Golden et al, 2009 61 | OCF5 |
| C. elegans OCF46: unc-119(ed3) III; qaIs3507 [pie-1p::GFP::lem-2 + unc-119(+)] III; ltIs37 [pAA64: pie-1p::mCherry::his-58 + unc-119 (+)] | Rahman et al 2015 39 | OCF46 |
| C. elegans OCF85: unc-119(ed3) III; ojIs9 [pie-1p::zyg-12(all)::GFP + unc-119(+)]; ieSi21 [sun-1p::sun-1::mRuby::sun-1 3' UTR + Cbr-unc-119(+)] IV | Rahman et al 2020 31 | OCF85 |
| C. elegans OCF108: ocf102[atln-1::GFP::3xFLAG] IV (CRISPR); unc-119(ed3) III; ocfIs2 [pie-1 p::mCherry::SP12::pie-1 3' UTR + unc119 (+)] | This paper | OCF108 |
| C. elegans OCF116: ocf101[atln-1::3xFLAG::degron] IV (CRISPR); unc-119(ed3) III; ojIs23 [pie-1p::SP12::GFP + unc-119(+)]; mTurquoise2::H2B (his-72::linker::mTurquoise2) III (CRISPR) | This paper | OCF116 |
| C. elegans OCF118: ocf101[atln-1::3xFLAG::degron] IV (CRISPR); TIR1::mRuby IV; unc-119(ed3) III; ojIs23 [pie-1p::SP12::GFP + unc-119(+)]; mTurquoise2::H2B (his-72::linker::mTurquoise2) III (CRISPR) | This paper | OCF118 |
| C. elegans OCF124: unc119(ed3) III; ItIs33 [pOD224; pie-1/GFPTEV-stag::sas-6 genomic; unc119(+)genomic]; unc-119(ed3) III; ocfIs2 [pie-1p::mCherry::SP12::pie-1 3' UTR + unc119 (+)] | This paper | OCF124 |
| C. elegans OCF127: unc-119(ed3) III; bsSi15 [pKO109: spd-2p-spd-2::mCherry::spd-2 3' UTR, unc-119(+)], ojIs23 [pie-1p::SP12::GFP + unc-119(+)] | This paper | OCF127 |
| C. elegans OCF145: ocf103[sun-1::GFP::3xFLAG] V (CRISPR) | This paper | OCF145 |
| C. elegans OCF158: unc-119(ed3) III; ItIs78 [pKO5:pie-1p::GFP::air-1; unc-119(+)]; ocfIs2 [pie-1p::mCherry::SP12::pie-1 3' UTR + unc119 (+)] | This paper | OCF158 |
| C. elegans OCF162: ebp-2(or1954[ebp-2::mKate2]) II (CRISPR); unc-119(ed3) III; ojIs23 [pie-1p::SP12::GFP + unc-119(+)]; ocf101[atln-1::3xFLAG::degron] IV (CRISPR); TIR1::mRuby IV; mTurquoise2::H2B (his-72::linker::mTurquoise2) III (CRISPR) | This paper | OCF162 |
| C. elegans OCF164: spd-5(vie26[GFP::spd-5 +loxP]) I (CRISPR); unc-119(ed3) III; ocfIs2 [pie-1 p::mCherry::SP12::pie-1 3' UTR + unc119 (+)]; ocf101[atln-1::3xFLAG::degron] IV (CRISPR); TIR1::mRuby IV; mTurquoise2::H2B (his-72::linker::mTurquoise2) III (CRISPR) | This paper | OCF164 |
| C. elegans OCF165: spd-5(vie26[GFP::spd-5 +loxP]) I (CRISPR); unc-119(ed3) III; ocfIs2 [pie-1 p::mCherry::SP12::pie-1 3' UTR + unc119 (+)]; ocf101[atln-1::3xFLAG::degron] IV (CRISPR); mTurquoise2::H2B (his-72::linker::mTurquoise2) III (CRISPR) | This paper | OCF165 |
| C. elegans OCF166: plk-1(lt18[plk-1::sGFP::loxp] III); ocf101[atln-1::3xFLAG::degron] IV (CRISPR); TIR1::mRuby IV; unc-119(ed3) III; ocfIs2 [pie-1 p::mCherry::SP12::pie-1 3’ UTR + unc119 (+)] | This paper | OCF166 |
| C. elegans OCF167: tac-1(or1955[GFP::tac-1]) II (CRISPR); unc-119(ed3) III; ocfIs2 [pie-1 p::mCherry::SP12::pie-1 3’ UTR + unc119 (+)]; ocf101[atln-1::3xFLAG::degron] IV (CRISPR); TIR1::mRuby IV; mTurquoise2::H2B (his-72::linker::mTurquoise2) III (CRISPR) | This paper | OCF167 |
| C. elegans OCF170: plk-1(or683ts) III, unc-119(ed3) III; ocfIs2 [pie-1p::mCherry::SP12::pie-1 3’ UTR + unc119 (+)]; spd-5(vie26[GFP::spd-5 +loxP]) I (CRISPR) | This paper | OCF170 |
| C. elegans OCF172: unc-119(ed3) III; ItIs78 [pKO5:pie-1p::GFP::air-1; unc-119(+)]; ocfIs2 [pie- 1p::mCherry::SP12::pie-1 3’ UTR + unc119 (+)]; ocf101[atln-1::3xFLAG::degron] IV (CRISPR); TIR1::mRuby IV; mTurquoise2::H2B (his- 72::linker::mTurquoise2) III (CRISPR) | This paper | OCF172 |
| C. elegans OCF173: spd-5(vie26[GFP::spd-5 +loxP]) I (CRISPR); unc-119(ed3) III; bqSi226 [lem-2p::lem-2::mCherry + unc-119(+)] IV | This paper | OCF173 |
| C. elegans OCF174: unc-119(ed3) III; bsSi15 [pKO109: spd-2p-spd-2::mCherry::spd-2 3’ UTR + unc-119(+)] I; bqSi210 [lem-2p::lem-2::GFP + unc-119(+)] II | This paper | OCF174 |
| C. elegans OCF176: unc-119(ed3) III; ocfIs2 [pie-1p::mCherry::SP12::pie-1 3’ UTR + unc119 (+)]; spd-5(vie26[GFP::spd-5 +loxP]) I (CRISPR); mTurquoise2::H2B (his-72::linker::mTurquoise2) III (CRISPR) | This paper | OCF176 |
| C. elegans OCF178: ocf103[sun-1::GFP::3xFLAG] V (CRISPR) ); unc-119(ed3) III; bsSi15 [pKO109: spd-2p-spd-2::mCherry::spd-2 3’ UTR + unc-119(+)] I | This paper | OCF178 |
| C. elegans OCF179: ax4539[npp-12::mNeonGreen] I (CRISPR); unc-119(ed3) III; bsSi15 [pKO109: spd-2p-spd-2::mCherry::spd-2 3’ UTR + unc-119(+)] I | This paper | OCF179 |
| C. elegans OCF183: ocf101[atln-1::3xFLAG::degron] IV (CRISPR); TIR1::mRuby IV; mTurquoise2::H2B (his-72::linker::mTurquoise2) III (CRISPR); unc-119(ed3) III; ocfIs2 [pie-1p::mCherry::SP12::pie-1 3’ UTR + unc119 (+)]; ltIs25 [pAZ132; pie-1p::GFP::tba-2 + unc-119 (+)] | This paper | OCF183 |
| C. elegans WLP718: GFP-LMN-1 WT (CRISPR) | Pintard lab | WLP718 |
| C. elegans WLP801: mCherry::NPP-22 (CRISPR) | Pintard lab | WLP801 |
| C. elegans JH3908: ax4539[npp-12::mNeonGreen] I (CRISPR); bqSi189[lmn-1p::mCherry::his-58::pie-1 3’ UTR] II (MosSCI)) | Thomas et al 2022 70 | JH3908 |
| C. elegans MSN146: unc-119(ed3) III; ltIs76 [pAA178: pie-1p::mCherry::SP12 + unc-119(+)]; ltIs25 [pAZ132; pie-1p::GFP::tba-2 + unc-119(+)] | Audhya lab | MSN146 |
| C. elegans BN243: unc-119(ed3) III; bqSi235 [emr-1p::emr-1::GFP + unc-119(+)] II; bqSi226 [lem-2p::lem-2::mCherry + unc-119(+)] IV | CGC | BN243 |
| Oligonucleotides | ||
| See Table S1 | ||
| Recombinant DNA | ||
| Plasmid for synthesis of dsRNA of atln-1: pRM01 | This paper | N/A |
| Plasmid for synthesis of dsRNA of spd-5: pRM02 | This paper | N/A |
| Plasmid for atln-1::GFP::3xFLAG CRISPR tagging: pAP973 | Seydoux lab | N/A |
| Plasmid for atln-1::3xFLAG::degron CRISPR tagging: pKO132 | O’Connell lab | N/A |
| Vector for expressing dsRNA: pL4440 | Addgene | #1654 |
| Software and algorithms | ||
| FIJI (ImageJ release 2.1.0) | Schindelin et al., 2012 74 | https://imagej.nih.gov/ij/ |
| GraphPad Prism [version 9.1.2 (255)] | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Nikon Elements Software | Nikon USA | N/A |
| IMOD or 3dmod [version 4.11.1] | https://bio3d.colorado.edu | N/A |
| AMIRA 6.5.0 (release 2018-03-07) | Thermo Fisher Scientific | N/A |
| Amira ZIB edition Version 2020.1 | Zuse Institute Berlin Dep. Visual Data Analysis, Takustr.7, Berlin, Germany. | N/A |
| Adobe Photoshop CC (release 23.1) | Adobe | N/A |
| Other | ||
| none | ||
METHOD DETAILS
Experimental growth conditions
Temperature Shift Experiments
For temperature shift experiments, plk-1ts animals expressing GFP::SPD-5; mCherry::SP12 (OCF170) were maintained at 16°C. plk-1ts adult hermaphrodites were shifted to 23°C and early embryos were imaged 30–60 min later.
RNA-Mediated Interference: Feeding RNAi
For atln-1, air-1, zyg-12, klp-7, perm-1 or smd-1 (control) feeding RNAi, a 5 ml Luria Broth (LB) with 50 mg/ml ampicillin were inoculated using 1:100 inoculum from a 2 ml overnight saturated culture (at 37 °C) of E. coli carrying the gene of interest expressed from both ends. RNAi clones are from the RNAi feeding library (Open Biosystems, Huntsville, AL) except atln-1 for which approximately 0.6 kb of the coding region was cloned in plasmid pL4440. Once the culture grew to OD600 of around 0.5 (~ 4 h at 37 °C), 0.5 M IPTG (1 mM final concentration) was used to induce the bidirectional transcription of the relevant gene for another 4 h. The culture was centrifuged at 5000 g for 5 min at room temperature, and the pellet was resuspended in 1 ml of fresh LB + ampicillin (50 mg/ml) media. 200 μl of this culture was spread on each RNAi plate (MYOB with 4 mM IPTG and ampicillin 50 mg/ml). For feeding RNAi treatment, 15–20 L4-stage larvae were transferred to RNAi plates, and after 48 h (for atln-1, air-1 and klp-7), 24 h (for zyg-12), 16–24 h (for partial atln-1 RNAi), or 16 h (for perm-1), the RNAi treated worms were dissected on a glass slide (Cat #EF15978A, Daigger Scientific), mounted on a 2% agar pad and imaged as described below. Control RNAi treatments (smd-1) were done for the same amount of time as the experimental ones. For embryonic viability scoring, a few worms were allowed to lay embryos for 3–4 h on a fresh RNAi plate, and embryonic viability was scored after 24 h of RNAi treatment.
RNA-Mediated Interference: Injection of double stranded RNA
For double stranded RNA (dsRNA) against spd-5, approximately 1 kb of the coding region was cloned in plasmid pL4440 (Addgene). The plasmid containing the spd-5 coding DNA was linearized at one end of the coding region using KpnI restriction enzyme and subsequently cleaved by XbaI. For dsRNA against smd-1, around 1 kb was PCR amplified from pL4440+smd-1 plasmid from the RNAi feeding library (Open Biosystems, Huntsville, AL). For the forward strand, a forward primer upstream of the first T7 promoter site and a reverse primer at the end specific to the smd-1 were used. Similarly, for the reverse strand, a reverse primer downstream of the T7 promoter site at the other end and a forward primer at the beginning specific to the smd-1 were used. Digested or PCR amplified fragments were purified using the Qiagen MinElute Reaction Cleanup Kit (Cat# 28206, Qiagen). To prepare dsRNA for injection, in vitro RNA synthesis was carried out using “MEGAscript™ T7 Transcription Kit (Cat# AM1333, Invitrogen) followed by purification using Phenol:CHCl3:Isoamyl Alcohol (25:24:1, v/v) (Cat #15593031, Invitrogen) and precipitated using 100% ethanol (Cat #64-17-5, The Warner-Graham company). The RNA pellet was dissolved in ~100 to 150 μl of TE buffer (10 mM, pH 8) (Cat #351-011-131, Quality biological). To prepare dsRNA, about 100 μl (1 μg/μl) of the ssRNA was mixed and kept at 85 °C for 3 min in an aluminum heat block incubator followed by slow cooling to RT for annealing. Injection of dsRNA was done according to Ohta laboratory protocol 73. L4s (15–20 worms) were injected with ~1 μg/ μl dsRNA. These worms were maintained at 16 °C for 48 h prior to live imaging of early embryos by confocal microscopy.
Auxin-mediated degradation
For auxin mediated degradation, the gene coding for the protein of interest, atln-1, was tagged with an auxin-inducible degron tag (atln-1::degron), and cells either expressed TIR1, an exogenous F-box protein 44. One of two types of controls was used: either worms with or without TIR1, both treated with the auxin analog indole-3 acetic acid (IAA), or atln-1::degron TIR1 worms with and without IAA treatment. Worms were transferred to bacteria seeded IAA plates (MYOB plates with 4mM IAA (Alfa Aesar, #A10556)) for 20–25 minutes, and embryos were imaged immediately thereafter. IAA plates (4 mM) were prepared from a 400 mM IAA stock (in 100% ethanol and stored at 4°C for up to a month). IAA was added to the MYOB media at ~50°C before pouring plates. The following day, IAA plates were seeded with a thick layer of OP50 cells and left to dry in a dark place. Based on 44.
Nocodazole treatment
15–20 L4-stage larvae were transferred to perm-1 RNAi plates to permeabilize embryos to nocodazole. After 16 h, the RNAi-treated worms were dissected in 20 μl of L15 buffer (Cat# 21083027, Thermo Fisher Scientific) in a 0.17 mm glass-bottom, open-top Delta T® dish (Cat#10199-956, Avantor VWR), and 1-cell embryos were identified. For centriculum establishments, once the embryo completed cytokinesis, an image was taken, and then 20 μl of 4% DMSO (control) or 20 μl of 40 μg/ml nocodazole (Cat# 1404, Sigma Aldrich) were added. After 6 minutes of drug treatment the embryo was imaged again. For centriculum maintenance, 10 μl of 4% DMSO (control) or 10 μl of 40 μg/ml nocodazole were added and embryos were imaged after 1:15 and 2:30 minutes.
Confocal microscopy
Imaging
Unless indicated otherwise, images were taken using a Nikon confocal Ti2 with Yokagawa CSU-X1 spinning disk and a photometrix Prime 95B camera using a Nikon water/oil 60X 1.2-NA Apo Plan objective. Images were captured using Nikon Elements software version 5.20.00. Images in Figure S1A were taken using a custom-assembled spinning disk confocal microscope consisting of a Zeiss Axio Imager A2 frame, a Borealis modified Yokogawa CSU10 spinning disk, an ASI 150-micron piezo stage controlled by a MS2000, and an ASI filter wheel and a Hamamatsu ImagEM X2 EM-CCD camera using a Plan Apochromat 100x/1.4 NA DIC objective (Carl Zeiss). Images in Figure 5F were taken using a Nikon confocal Ti2 with Yokagawa CSU-X1 spinning disk with a Prime BSI camera, using a 60X Plan Apo VC objective, NA 1.2. Images in Figure S5C were taken using Nikon Eclipse TE2000U spinning-disk confocal microscope with Metamorph software version 7.8.6.0 (Molecular Devices). The microscope was equipped with a 60X 1.4-NA Apo objective, an LMM5 laser merge module with four diode lasers (excitation at 405, 491, 561, and 655 nm) from Applied Research, a Yokogawa CSU10 spinning disk, and a Hamamatsu C9100-13 EM-CCD camera. For imaging, embryos were mounted on 2% agarose (Cat #214010, Invitrogen) pads prepared in standard M9 buffer (Cat# 11006-517, IPM scientific). Images were taken at z = 1 μm intervals unless otherwise mentioned. Images were processed with Fiji (74 ImageJ release 2.1.0; http://imagej.nih.gov/ij) and Adobe Photoshop CC (release 23.1).
For imaging adult somatic cells, we used a custom-built spinning disk confocal microscope with a 100x oil objective 75. First, worms at the L3 larval stage were immobilized using 5 mM levamisole (Cat# L9756, Sigma-Aldrich) in M9 buffer. Subsequently, the immobilized worms were mounted on a 5% Noble agar (Cat #A5431, Sigma-Aldrich) pad split into two asymmetric halves with worms placed on the larger half. A coverslip (Cat #12-541B, ThermoFisher Scientific) was gently placed on the worms. We used VALAP (http://cshprotocols.cshlp.org/content/2015/2/pdb.rec082917) to secure the coverslip onto the microscope slide, leaving two diagonal openings. We flooded the chamber with M9 buffer to prevent desiccation. Finally, we set the step size at 1 μm and imaged every 2.5 minutes for 3 hours using 488 and 561 nm lasers.
Image analysis
All images were analyzed using Fiji (74, http://imagej.nih.gov/ij). For diameter of structures that appeared as rings in the central plane of focus (e.g. the centriculum, TBA-2, AIR-1 and EBP-2), two perpendicular lines (line # 7 in Fiji, width = 1 μm) were drawn across the central plane of focus and through the estimated center of the centrosome, and intensity profiles of the plotted lines were obtained (see Figure S5A for an example). A rectangle tool was used to calculate the distance between the two peaks of fluorescence intensity, resulting in two diameters (d1 and d2). The diameters were then used to determine the average diameter (d1+d2)/2 of the protein structures in question.
For localization of centrosomal proteins relative to the centriculum, two lines, 9 μm in length and 1 μm in width, were drawn through the centrosome as described above. Intensity profiles were plotted for the centriulum channel (typically SP12 fused to GFP or mCherry) and the centrosomal protein channel. The average values from multiple centrosomes across the line for both channels were superimposed.
For embryo size, a freehand selection tool in Fiji was used to trace the edges of the central plane of the 1-cell embryos at metaphase stage. The software provided the area of the traced region.
To determine the area and total fluorescence intensity of PCM proteins (e.g. SDP-5 and TAC-1), the mean background fluorescence was determined using Fiji between the two centrosomes at the central focal plane of the centrosome being measured. The lowest pixel value of the entire image was added to the mean fluorescence, and the resulting value was used as the lower threshold limit to create an area of the fluorescent protein examined. The areas and total fluorescence (Raw Intensity or RawInt) were measured in the defined area using Fiji.
For proteins that were present both at the centrosome and outside of it (e.g. TBA-2, AIR-1, EBP-2), their total intensity inside the centriculum was determined by determining the centriculum void area, defined as the area inside the perimeter of the centriculum at its mid-section. To mark the void area, a circle (area constant throughout each analysis) was drawn in the center of the centriculum void. The integrated fluorescence intensity (IntDen value) in Fiji of this circle was used as the upper threshold limit, thus excluding pixels with centriculum fluorescence. The void area was traced, the area was measured, and the total fluorescence intensity (RawInt) of the aforementioned proteins in this area was determined.
For EBP-2::mKate2 and GFP::TBA-2 fluorescence intensity in the spindle 0–0.5 μm from the chromosomes, a sum projection was generated. Then, 0.5 μm wide boxes were drawn to the left and right of the chromosomes, visualized using mTurquoise2::H2B. The boxes spanned from the top to the bottom of the spindle, as measured by the SP12 signal of the nuclear envelope, and the total fluorescence of the microtubule marker was measured. See also Figure S6A.
For spindle length, a line, 0.5 μm in width, was drawn through both centrosomes/centricula, and a fluorescence intensity profile was generated. The distance between the minima of the two centriculum in the trace was used to calculate spindle length (center of one centrosome to center of the other centrosome). For spindle width 0–0.5 μm from the chromosomes, a 0.5 μm line was draw through the pronuclei. A fluorescence intensity profile based on SP12 fluorescence was generated, and the distance between peaks was measured. See also Figure S6A.
Focused Ion Beam-Scanning Electron Microscopy (FIB-SEM)
Embryo collection and sample processing
We dissected wild-type N2 hermaphrodites (maintained at 20°C) in 20% Bovine Serum Albumin (BSA) and collected single 1-cell embryos in a cellulose capillary tube 76 (Cat# 16706869, Leica Microsystems, Vienna, Austria) under a stereomicroscope (SMZ645, 50X total magnification, Nikon). We followed the zygotes under the same stereoscope until they reached the desired stage in mitosis, i.e., prophase, prometaphase, metaphase. Then we froze the embryo immediately using gold-coated planchettes (Cat #16770152, Cu-Au 3.0 × 0.5 mm, Leica Microsystems, Vienna, Austria) using a high-pressure freezer (model EM ICE; Leica Microsystems, Vienna, Austria). We then applied quick-freeze substitution (QFS) 77 with a cocktail of 0.2 g Osmium tetroxide (OsO4) powder (Cat# 19100, Electron Microscopy Sciences in 9 ml of acetone (Cat #10000, Electron Microscopy Sciences), 0.5 ml of double deionized water, and 0.5 ml of 2% uranyl acetate (UA) solution (Cat #22400, Electron Microscopy Sciences). Then we washed the samples for 10 minutes (x3) with 100 % acetone at room temperature and infiltration in Polybed 812 resin (Cat #08791; Polysciences: 14.6 g Polybed, 8.4 g DDSA, 7.0 g NMA, and 0.42 ml DMP30) followed by resin: acetone ratios of 1:2, 1:1, and 2:1 each for an hour. Finally, we transferred the samples into 100% resin for ~16 hours prior to curing at 55°C oven for 40 to 60 hrs. The cured resin was trimmed using a jeweler’s saw and razor blades and sectioned in an ultramicrotome until the embryos were tangentially exposed for FIB-SEM imaging. For a step-wise protocol, please refer to 78.
Data acquisition and postprocessing
Prior to imaging, the embryos were sputter coated with a 10 nm carbon coat to increase overall conductivity, and a thicker ~300 nm carbon pad was deposited with the FIB to cover the buried embryo. FIB-SEM acquisition was done using a Crossbeam 540 (Carl Zeiss). The front edge of a sample was milled with FIB beam (at 65 or 30 nA) followed by a reduced FIB current (3 nA). Samples were milled until the embryos were detected at the cliff face. At this point, a 1.5-μm-thick patterned platinum and carbon pad was laid over the carbon pad 79. Milling with a FIB current of 1.5 nA was resumed across the entire embryo, with intermittent SEM imaging, until a recognizable biological feature such as the hazy pericentriolar material was revealed. Next, imaging was initiated using ATLAS 5 (Fibics) at high-resolution for a region of interest in the embryo. The SEM was operated at 1.5 kV accelerating voltage and 1.0 nA current. FIB parameters were 30 kV with a 1.5 nA or 700 pA current. High-resolution, high-m/z contrast signal was recorded using an in-column energy selective back-scatter detector with a grid voltage of 1000 V. Images at 3 nm pixel size, dwell time of 3 μs, and 9 nm FIB step size or “z thickness” were obtained. Several thousand high-resolution images were captured in 40–60 h. After dataset acquisition, the image stack was cropped and processed using IMOD-based scripts 80 to produce aligned, inverted, and binned mrc-image volumes with 9 nm isotropic voxels.
Segmentation of FIB-SEM datasets
We used Amira 6.5.0 (release 2018-03-07; Thermo Fisher Scientific) software with XIMagePAQ, XMesh, and XSkeleton extension packages for a threshold-based semi-automated segmentation. Segmentation using Amira was done following a general scheme unless specified elsewhere. We made a threshold-based selection of membranes first, followed by three rounds of slice-by-slice visual inspection (through XY, YZ, and XZ planes separately). Upon careful visual inspection, we manually added all undetected areas (about 1–2% of the total area) to the segmented volume. We removed any unrelated structures that were automatically selected due to a similar threshold level, (e.g., ER membranes beyond centriculum; nuclear envelope, lipid droplets, mitochondria, and other organelles in the proximity of centriculum). In our current study, we included FIB-SEM data from four wild-type embryos: two prophase (P1 and P2), and two metaphase (M1 and M2) embryos.
Electron tomography of serial sections
Embryo collection and sample processing
We used samples that were previously obtained for a 3D reconstruction of the first mitotic spindle in C. elegans 46. Briefly, C. elegans N2 (wild type) gravid adults were dissected in M9 buffer, and zygotes in early mitosis were collected in cellulose capillary tubes (Cat# 16706869, Leica Microsystems) (Pelletier et al., 2006). The embryos were observed under a stereoscope until metaphase was reached and then immediately frozen using an EMPACT2 high-pressure freezer equipped with a rapid transfer system (RTS, Leica Microsystems). Freeze substitution of the cryo-immobilized embryos was done over three days in anhydrous acetone containing 1% OsO4 and 0.1% UA using freeze-substitution equipment (EM AFS, Leica Microsystems, (Pelletier et al., 2006)). Epon/Araldite infiltration was followed by thin-layer embedding and polymerization for three days at 60°C. After remounting the specimens on dummy blocks, serial semi-thick sections (300 nm) were cut using an ultramicrotome (Ultracut UCT, Leica Microsystems). Sections were collected on a Formvar-coated copper slot grids (EMS) and post-stained with 2% UA (in 70% methanol) followed by brief exposure to Reynold’s lead citrate Error! Hyperlink reference not valid..
Data acquisition and postprocessing
Prior to imaging, 15 nm colloidal gold particles (Cat# 777137, Sigma-Aldrich) were added to both sides of the 300 nm semi-thick sections to serve as fiducial markers for the calculation of electron tomograms. Next, a series of tilted images was taken by using a TECNAI F30 transmission electron microscope (Thermo Fisher Scientific) operated at 300 kV. TEM images were taken every 1° over a ±60° range using a Gatan US1000 CCD camera (2k × 2k). The pixel size of the images was 2.3 nm. Tilted views/images were aligned using the fiducial gold markers and later back-projected and combined to a super montage using IMOD software package or 3dmod software package 80. To cover the pole-to-pole region of each mitotic spindle, on average, 24 consecutive serial sections per spindle were imaged and processed accordingly 46.
Segmentation of tomogram datasets
The software Amira ZIB (Zuse Institute Berlin) version 2020.1 was used for manual segmentation of membranes around centrosome. Segmented membranes were visualized following the Amira default scheme unless mentioned otherwise. Segmented membrane sections were inspected twice slice-by-slice through visual inspection of the XY (imaging plane). Any undetected area (about 1–2% of the total segmented volume) left during segmentation in the XY plane was manually added to the final volume when inspecting the XZ and YZ planes. Any unrelated area, for example, round and isolated vesicles less than 300 nm in diameter 81, were removed manually.
The Amira 6.5.0 (release 2018-03-07; Thermo Fisher Scientific) software with XIMagePAQ, XMesh, and XSkeleton extension packages was used for microtubule segmentation. Microtubules were segmented in individual sections prior to serial stitching based on a previously published template matching and stitching algorithms 82,83. Here we show previously segmented microtubules data from sections #6 and #7 of the tomogram dataset described in 46.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
All analyses were done using GraphPad Prism. Prior to analysis, the distribution of the datapoint was examined for normality. For comparison between two normally distributed datasets, Student’s t-test was used; otherwise two-tailed Mann Whitney test was used. For comparison between multiple datasets, one-way ANOVA non-parametric Kruskal-Wallis test with correction for multiple comparisons was used. In all cases, the statistical tests used, the number of samples analyzed, and the p-values are indicated in the figure legends. The criterion for statistical significance was set at P < 0.05. Results are represented as mean ± SEM unless indicated otherwise.
Supplementary Material
Video S1. Not all centrosome-nucleated microtubules pass through the centriculum. Related to Figure 7.
3D reconstruction of the microtubules and membrane shown in Figure 7B. The movie, done using Amira software, tracks the microtubules as the pass through the centriculum. The yellow and blue microtubules end where they encounter a membrane, while the green microtubule goes all the way through the centriculum.
Highlights.
C. elegans embryo centrosomes are surrounded by a membrane reticulum, the centriculum
Centriculum formation depends on microtubules and the pericentriolar material (PCM)
Increasing centriculum size leads to a larger PCM and more spindle microtubules
The centriculum may couple spindle elongation to nuclear membrane fenestration
Acknowledgements
We thank Alexander Dammermann (University of Vienna, Austria), Lionel Pintard (Institut Jacques Monod, France), Jessica Feldman (Stanford University, USA), Jon Audyha (University of Wisconsin-Madison), Geraldine Seydoux and Laura Thomas (Johns Hopkins University School of Medicine) and the Caenorhabditis Genetics Center (CGC, University of Minnesota, USA) for strains and reagents, and Tetsunari Fukushige for help with generating dsRNA. We also thank Will Prinz and Kevin O’Connell for valuable discussion and comments on the manuscript. R.M., M.M.R., S.D., M.O. and O.C.-F. were funded by an Intramural NIDDK grant number DK069012. G.F. was funded by the German Research Foundation (DFG grant MU 1423/8-1 and 8-2 to T. M.R.). M.A.Q.M. and D.Q.M. were funded by the NCI (F30CA257383) and NIGMS (R01GM121597).
Inclusion and Diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQIA+ community. We support inclusive, diverse, and equitable conduct of research.
Footnotes
Declaration of Interests
D.Q.M. is a paid employee of Arcadia Science.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Palazzo RE, Vogel JM, Schnackenberg BJ, Hull DR, and Wu X (2000). Centrosome maturation. Curr. Top. Dev. Biol. 49, 449–470. [DOI] [PubMed] [Google Scholar]
- 2.Cohen-Fix O, and Askjaer P (2017). Cell biology of the Caenorhabditis elegans nucleus. Genetics 205, 25–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyerzon M, Gao Z, Liu J, Wu J-C, Malone CJ, and Starr DA (2009). Centrosome attachment to the C. elegans male pronucleus is dependent on the surface area of the nuclear envelope. Dev. Biol. 327, 433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malone CJ, Misner L, Le Bot N, Tsai M-C, Campbell JM, Ahringer J, and White JG (2003). The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell 115, 825–836. [DOI] [PubMed] [Google Scholar]
- 5.Guttinger S, Laurell E, and Kutay U (2009). Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol. 10, 178–191. [DOI] [PubMed] [Google Scholar]
- 6.Gomes Pereira S, Dias Louro MA, and Bettencourt-Dias M (2021). Biophysical and quantitative principles of centrosome biogenesis and structure. Annu. Rev. Cell Dev. Biol. 37, 43–63. [DOI] [PubMed] [Google Scholar]
- 7.Kemp CA, Kopish KR, Zipperlen P, Ahringer J, and O’Connell KF (2004). Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev. Cell 6, 511–523. [DOI] [PubMed] [Google Scholar]
- 8.Pelletier L, Özlü N, Hannak E, Cowan C, Habermann B, Ruer M, Müller-Reichert T, and Hyman AA (2004). The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr. Biol. 14, 863–873. [DOI] [PubMed] [Google Scholar]
- 9.Pintard L, and Bowerman B (2019). Mitotic cell division in Caenorhabditis elegans. Genetics 211, 35–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodruff JB, Gomes BF, Widlund PO, Mahamid J, Honigmann A, and Hyman AA (2017). The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell 169, 1066–1077. [DOI] [PubMed] [Google Scholar]
- 11.Baumgart J, Kirchner M, Redemann S, Bond A, Woodruff J, Verbavatz J-M, Jülicher F, Müller-Reichert T, Hyman AA, and Brugués J (2019). Soluble tubulin is significantly enriched at mitotic centrosomes. J. Cell Biol. 218, 3977–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zwicker D, Decker M, Jaensch S, Hyman AA, and Jülicher F (2014). Centrosomes are autocatalytic droplets of pericentriolar material organized by centrioles. Proc. Natl. Acad. Sci. U.S.A. 111, E2636–E2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahn JI, Park J-E, Meng L, Zhang L, Kim T-S, Kruhlak MJ, Kim BY, and Lee KS (2020). Phase separation of the Cep63• Cep152 complex underlies the formation of dynamic supramolecular self-assemblies at human centrosomes. Cell Cycle 19, 3437–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang X, Ho DBT, Mahe K, Mia J, Sepulveda G, Antkowiak M, Jiang L, Yamada S, and Jao L-E (2021). Condensation of pericentrin proteins in human cells illuminates phase separation in centrosome assembly. J. Cell Sci. 134, jcs258897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raff JW (2019). Phase Separation and the Centrosome: A Fait Accompli? Trends Cell Biol. 29, 612–622. [DOI] [PubMed] [Google Scholar]
- 16.Shin Y, and Brangwynne CP (2017). Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382. [DOI] [PubMed] [Google Scholar]
- 17.Poteryaev D, Squirrell JM, Campbell JM, White JG, and Spang A (2005). Involvement of the actin cytoskeleton and homotypic membrane fusion in ER dynamics in Caenorhabditis elegans. Mol. Biol. Cell 16, 2139–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Araujo M, Tavares A, Vieira DV, Telley IA, and Oliveira RA (2023). Endoplasmic reticulum membranes are continuously required to maintain mitotic spindle size and forces. Life Sci Alliance 6. 10.26508/lsa.202201540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris P (1975). The role of membranes in the organization of the mitotic apparatus. Exp. Cell Res. 94, 409–425. [DOI] [PubMed] [Google Scholar]
- 20.Waterman-Storer CM, Sanger JW, and Sanger JM (1993). Dynamics of organelles in the mitotic spindles of living cells: membrane and microtubule interactions. Cell motil. cytoskelet. 26, 19–39. [DOI] [PubMed] [Google Scholar]
- 21.Bergman ZJ, Mclaurin JD, Eritano AS, Johnson BM, Sims AQ, and Riggs B (2015). Spatial reorganization of the endoplasmic reticulum during mitosis relies on mitotic kinase cyclin A in the early Drosophila embryo. PLoS ONE 10, e0117859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karabasheva D, and Smyth JT (2019). A novel, dynein-independent mechanism focuses the endoplasmic reticulum around spindle poles in dividing Drosophila spermatocytes. Sci. Rep. 9, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diaz U, Bergman ZJ, Johnson BM, Edington AR, de Cruz MA, Marshall WF, and Riggs B (2019). Microtubules are necessary for proper Reticulon localization during mitosis. PLoS ONE 14, e0226327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shankar R, Lettman MM, Whisler W, Frankel EB, and Audhya A (2022). The ESCRT machinery directs quality control over inner nuclear membrane architecture. Cell Rep 38, 110263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martino L, Morchoisne-Bolhy S, Cheerambathur DK, Van Hove L, Dumont J, Joly N, Desai A, Doye V, and Pintard L (2017). Channel nucleoporins recruit PLK-1 to nuclear pore complexes to direct nuclear envelope breakdown in C. elegans. Dev. Cell 43, 157–171. e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hachet V, Busso C, Toya M, Sugimoto A, Askjaer P, and Gönczy P (2012). The nucleoporin Nup205/NPP-3 is lost near centrosomes at mitotic onset and can modulate the timing of this process in Caenorhabditis elegans embryos. Mol. Biol. Cell 23, 3111–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ródenas E, Klerkx EP, Ayuso C, Audhya A, and Askjaer P (2009). Early embryonic requirement for nucleoporin Nup35/NPP-19 in nuclear assembly. Dev. Biol. 327, 399–409. [DOI] [PubMed] [Google Scholar]
- 28.Franz C, Askjaer P, Antonin W, Iglesias CL, Haselmann U, Schelder M, de Marco A, Wilm M, Antony C, and Mattaj IW (2005). Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates. EMBO J 24, 3519–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narayan K, and Subramaniam S (2015). Focused ion beams in biology. Nat. Methods 12, 1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kizilyaprak C, Stierhof Y-D, and Humbel BM (2019). Volume microscopy in biology: FIB-SEM tomography. Tissue Cell 57, 123–128. [DOI] [PubMed] [Google Scholar]
- 31.Rahman M, Chang IY, Harned A, Maheshwari R, Amoateng K, Narayan K, and Cohen-Fix O (2020). C. elegans pronuclei fuse after fertilization through a novel membrane structure. J. Cell Biol. 219, e201909137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu CS, Hayworth KJ, Lu Z, Grob P, Hassan AM, García-Cerdán JG, Niyogi KK, Nogales E, Weinberg RJ, and Hess HF (2017). Enhanced FIB-SEM systems for large-volume 3D imaging. eLife 6, e25916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H, and Hu J (2016). Shaping the endoplasmic reticulum into a social network. Trends Cell Biol. 26, 934–943. [DOI] [PubMed] [Google Scholar]
- 34.Magescas J, Zonka JC, and Feldman JL (2019). A two-step mechanism for the inactivation of microtubule organizing center function at the centrosome. eLife 8. 10.7554/eLife.47867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamill DR, Severson AF, Carter JC, and Bowerman B (2002). Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev. Cell 3, 673–684. [DOI] [PubMed] [Google Scholar]
- 36.Decker M, Jaensch S, Pozniakovsky A, Zinke A, O’Connell KF, Zachariae W, Myers E, and Hyman AA (2011). Limiting amounts of centrosome material set centrosome size in C. elegans embryos. Curr. Biol. 21, 1259–1267. [DOI] [PubMed] [Google Scholar]
- 37.Portier N, Audhya A, Maddox PS, Green RA, Dammermann A, Desai A, and Oegema K (2007). A microtubule-independent role for centrosomes and aurora a in nuclear envelope breakdown. Dev. Cell 12, 515–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cabral G, Laos T, Dumont J, and Dammermann A (2019). Differential requirements for centrioles in mitotic centrosome growth and maintenance. Dev. Cell 50, 355–366. e356. [DOI] [PubMed] [Google Scholar]
- 39.Rahman MM, Munzig M, Kaneshiro K, Lee B, Strome S, Muller-Reichert T, and Cohen-Fix O (2015). Caenorhabditis elegans polo-like kinase PLK-1 is required for merging parental genomes into a single nucleus. Mol. Biol. Cell 26, 4718–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu J, and Rapoport TA (2016). Fusion of the endoplasmic reticulum by membrane-bound GTPases. (Elsevier; ), pp. 105–111. [DOI] [PubMed] [Google Scholar]
- 41.Orso G, Pendin D, Liu S, Tosetto J, Moss TJ, Faust JE, Micaroni M, Egorova A, Martinuzzi A, and McNew JA (2009). Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature 460, 978–983. [DOI] [PubMed] [Google Scholar]
- 42.Hu J, Shibata Y, Zhu P-P, Voss C, Rismanchi N, Prinz WA, Rapoport TA, and Blackstone C (2009). A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell 138, 549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen S, Novick P, and Ferro-Novick S (2012). ER network formation requires a balance of the dynamin-like GTPase Sey1p and the Lunapark family member Lnp1p. Nat. Cell Biol. 14, 707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L, Ward JD, Cheng Z, and Dernburg AF (2015). The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 142, 4374–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srayko M, Kaya A, Stamford J, and Hyman AA (2005). Identification and characterization of factors required for microtubule growth and nucleation in the early C. elegans embryo. Dev. Cell 9, 223–236. [DOI] [PubMed] [Google Scholar]
- 46.Redemann S, Baumgart J, Lindow N, Shelley M, Nazockdast E, Kratz A, Prohaska S, Brugués J, Fürthauer S, and Müller-Reichert T (2017). C. elegans chromosomes connect to centrosomes by anchoring into the spindle network. Nat. Commun. 8, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O’Toole E, Greenan G, Lange KI, Srayko M, and Müller-Reichert T (2012). The role of γ-tubulin in centrosomal microtubule organization. PLoS ONE 7, e29795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Velez-Aguilera G, Ossareh-Nazari B, Van Hove L, Joly N, and Pintard L (2022). Cortical microtubule pulling forces contribute to the union of the parental genomes in the Caenorhabditis elegans zygote. eLife 11, e75382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Delattre M, Canard C, and Gonczy P (2006). Sequential protein recruitment in C. elegans centriole formation. Curr. Biol. 16, 1844–1849. [DOI] [PubMed] [Google Scholar]
- 50.Le Bot N, Tsai MC, Andrews RK, and Ahringer J (2003). TAC-1, a regulator of microtubule length in the C. elegans embryo. Curr. Biol. 13, 1499–1505. [DOI] [PubMed] [Google Scholar]
- 51.Mitchison T, and Kirschner M (1984). Dynamic instability of microtubule growth. Nature 312, 237–242. [DOI] [PubMed] [Google Scholar]
- 52.Marshall WF (2015). Subcellular size. Cold Spring Harb. Perspect. Biol. 7, a019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reber S, and Hyman AA (2015). Emergent Properties of the Metaphase Spindle. Cold Spring Harb. Perspect. Biol. 7, a015784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hachet V, Canard C, and Gönczy P (2007). Centrosomes promote timely mitotic entry in C. elegans embryos. Dev. Cell 12, 531–541. [DOI] [PubMed] [Google Scholar]
- 55.Fernández-Álvarez A, Bez C, O’Toole ET, Morphew M, and Cooper JP (2016). Mitotic nuclear envelope breakdown and spindle nucleation are controlled by interphase contacts between centromeres and the nuclear envelope. Dev. Cell 39, 544–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Conduit PT, and Raff JW (2010). Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in Drosophila neuroblasts. Curr. Biol. 20, 2187–2192. [DOI] [PubMed] [Google Scholar]
- 57.Laos T, Cabral G, and Dammermann A (2015). Isotropic incorporation of SPD-5 underlies centrosome assembly in C. elegans. Curr. Biol. 25, R648–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao YG, and Zhang H (2020). Phase separation in membrane biology: the interplay between membrane-bound organelles and membraneless condensates. Dev. Cell 55, 30–44. [DOI] [PubMed] [Google Scholar]
- 59.Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner J, and Burke B (2002). Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell 108, 97–107. [DOI] [PubMed] [Google Scholar]
- 60.Beaudouin J, Gerlich D, Daigle N, Eils R, and Ellenberg J (2002). Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell 108, 83–96. [DOI] [PubMed] [Google Scholar]
- 61.Golden A, Liu J, and Cohen-Fix O (2009). Inactivation of the C. elegans lipin homolog leads to ER disorganization and to defects in the breakdown and reassembly of the nuclear envelope. J. Cell Sci. 122, 1970–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peel N, Iyer J, Naik A, Dougherty MP, Decker M, and O’Connell KF (2017). Protein phosphatase 1 down regulates ZYG-1 levels to limit centriole duplication. PLoS Genet. 13, e1006543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dammermann A, Maddox PS, Desai A, and Oegema K (2008). SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the γ-tubulin–mediated addition of centriolar microtubules. J. Cell Biol. 180, 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chuang C-H, Schlientz AJ, Yang J, and Bowerman B (2020). Microtubule assembly and pole coalescence: early steps in Caenorhabditis elegans oocyte meiosis I spindle assembly. Biol. Open 9, bio052308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sugioka K, Fielmich L-E, Mizumoto K, Bowerman B, van den Heuvel S, Kimura A, and Sawa H (2018). Tumor suppressor APC is an attenuator of spindle-pulling forces during C. elegans asymmetric cell division. Proc. Natl. Acad. Sci. U.S.A.115, E954–E963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sloan DE, and Bembenek JN (2021). Endogenous expression and localization of HIS-72:: mTurquoise2 in C. elegans. microPubl., biol. 10.17912/micropub.biology.000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Joseph-Strauss D, Gorjánácz M, Santarella-Mellwig R, Voronina E, Audhya A, and Cohen-Fix O (2012). Sm protein down-regulation leads to defects in nuclear pore complex disassembly and distribution in C. elegans embryos. Dev. Biol. 365, 445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Galy V, Antonin W, Jaedicke A, Sachse M, Santarella R, Haselmann U, and Mattaj I (2008). A role for gp210 in mitotic nuclear-envelope breakdown. J. Cell Sci. 121, 317–328. [DOI] [PubMed] [Google Scholar]
- 69.Morales-Martínez A, Dobrzynska A, and Askjaer P (2015). Inner nuclear membrane protein LEM-2 is required for correct nuclear separation and morphology in C. elegans. J. Cell Sci. 128, 1090–1096. [DOI] [PubMed] [Google Scholar]
- 70.Thomas L, Askjaer P, and Seydoux G (2022). Multiple mechanisms prevent ectopic condensation of FG nucleoporins in the cytoplasm bioRxiv 2022.08.15.504042. [Google Scholar]
- 71.O’Rourke SM, Carter C, Carter L, Christensen SN, Jones MP, Nash B, Price MH, Turnbull DW, Garner AR, and Hamill DR (2011). A survey of new temperature-sensitive, embryonic-lethal mutations in C. elegans: 24 alleles of thirteen genes. PLoS ONE 6, e16644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dickinson DJ, Ward JD, Reiner DJ, and Goldstein B (2013). Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10, 1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ohta M, Zhao Z, Wu D, Wang S, Harrison JL, Gómez-Cavazos JS, Desai A, and Oegema KF (2021). Polo-like kinase 1 independently controls microtubule-nucleating capacity and size of the centrosome. J. Cell Biol. 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adikes RC, Kohrman AQ, Martinez MA, Palmisano NJ, Smith JJ, Medwig-Kinney TN, Min M, Sallee MD, Ahmed OB, and Kim N (2020). Visualizing the metazoan proliferation-quiescence decision in vivo. eLife 9, e63265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pelletier L, O’Toole E, Schwager A, Hyman AA, and Müller-Reichert T (2006). Centriole assembly in Caenorhabditis elegans. Nature 444, 619–623. [DOI] [PubMed] [Google Scholar]
- 77.McDonald KL, and Webb RI (2011). Freeze substitution in 3 hours or less. J. Microsc. 243, 227–233. [DOI] [PubMed] [Google Scholar]
- 78.Rahman MM, Chang IY, Cohen-Fix O, and Narayan K (2021). A workflow for high-pressure freezing and freeze substitution of the Caenorhabditis elegans embryo for ultrastructural analysis by conventional and volume electron microscopy. Bio-protoc. 11, e3981–e3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Narayan K, Danielson CM, Lagarec K, Lowekamp BC, Coffman P, Laquerre A, Phaneuf MW, Hope TJ, and Subramaniam S (2014). Multi-resolution correlative focused ion beam scanning electron microscopy: applications to cell biology. J. Struct. Biol. 185, 278–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kremer JR, Mastronarde DN, and McIntosh JR (1996). Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76. [DOI] [PubMed] [Google Scholar]
- 81.Hammerling BC, Shires SE, Leon LJ, Cortez MQ, and Gustafsson ÅB (2020). Isolation of Rab5-positive endosomes reveals a new mitochondrial degradation pathway utilized by BNIP3 and Parkin. Small GTPases 11, 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weber B, Möller M, Verbavatz J-M, Baum D, Hege H-C, and Prohaska S (2011). Fast Tracing of Microtubule Centerlines in Electron Tomograms. BioVis 2011 Abstracts, 1st IEEE Symposium on Biological Data Visualization. [Google Scholar]
- 83.Lindow N, Brünig FN, Dercksen VJ, Fabig G, Kiewisz R, Redemann S, Müller-Reichert T, Prohaska S, and Baum D (2021). Semi-automatic stitching of filamentous structures in image stacks from serial-section electron tomography. J. Microsc. 284, 25–44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Not all centrosome-nucleated microtubules pass through the centriculum. Related to Figure 7.
3D reconstruction of the microtubules and membrane shown in Figure 7B. The movie, done using Amira software, tracks the microtubules as the pass through the centriculum. The yellow and blue microtubules end where they encounter a membrane, while the green microtubule goes all the way through the centriculum.
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


