Abstract
Many flaviviruses are well-known pathogens, such as dengue, Zika, Japanese encephalitis, and yellow fever viruses. Among them, dengue viruses cause global epidemics and threaten billions of people. Effective vaccines and antivirals are in desperate need. In this review, we focus on the recent advances in understanding viral non-structural (NS) proteins as antiviral drug targets. We briefly summarize the experimental structures and predicated models of flaviviral NS proteins and their functions. We highlight a few well-characterized inhibitors targeting these NS proteins and provide an update about the latest development. NS4B emerges as one of the most promising drug targets as novel inhibitors targeting NS4B and its interaction network are entering clinical studies. Studies aiming to elucidate the architecture and molecular basis of viral replication will offer new opportunities for novel antiviral discovery. Direct-acting agents against dengue and other pathogenic flaviviruses may be available very soon.
Flaviviruses and Dengue virus
The Flavivirus genus, of the virus family Flaviviridae, is a group of positive-sense single-stranded RNA viruses (+ssRNA). Most flaviviruses are arboviruses - transmitted through ticks and mosquitoes. Notably, the genus includes the dengue virus (DENV), Zika virus (ZIKV), Japanese encephalitis virus (JEV), yellow fever virus (YFV), West Nile virus (WNV), and tick-borne encephalitis virus (TBEV). The combined burden of these viruses is immense and continues to escalate, with millions of people suffering from the diseases they cause yearly.
More than a quarter of the human population lives within Dengue-endemic areas today, with nearly half a billion people infected with DENV every year, of which hundreds of thousands develop severe disease with a fatality rate of ~5% in treated patients [1, 2]. JEV is the most significant cause of encephalitis globally, impacting 30–50 thousand cases a year [3], with most clinical cases occurring in children and infants. Recently, the re-emergence of other flaviviruses such as ZIKV and WNV has also caused significant transmission globally and presents a global health threat [4].
DENV causes an acute febrile disease named dengue fever, however, many infected people, including a large proportion of children, develop life-threatening forms of the disease known as dengue haemorrhagic fever and dengue shock syndrome [5]. Severe dengue is characterized by excessive inflammation, with studies showing that viral antigens induce the pro-inflammatory responses that underpin pathogenesis.
There is a lack of approved treatments that effectively combat most flaviviral diseases, with no drugs currently approved for use [6, 7]. A major impediment in developing drugs is that both the morphology and composition of the virus replication complex (RC), the interplay between its molecular constituents as well as the precise molecular mechanisms remain elusive [8]. Several non-structural proteins of the RC constitute validated drug targets because of their crucial functions during viral replication [6, 7].
Flavivirus Replication and Replicase Proteins
The +ssRNA flaviviral genome consists of a single open reading frame as well as 5’ and 3’ end untranslated regions (UTR). Due to the positive-sense nature of the flaviviral genome, it can hijack host cell translation machinery to produce a single polyprotein. This polyprotein, which is anchored to the ER membrane [9], is co and post-translationally cleaved by both viral and host proteases into the structural proteins - capsid (C), precursor membrane (prM), and envelope (E) proteins, as well as non-structural (NS) proteins NS1, NS2A, NS2B, NS3, NS4A, 2k peptide, NS4B and NS5 (Figure 1).
Figure 1. Molecular and structural flavivirology.
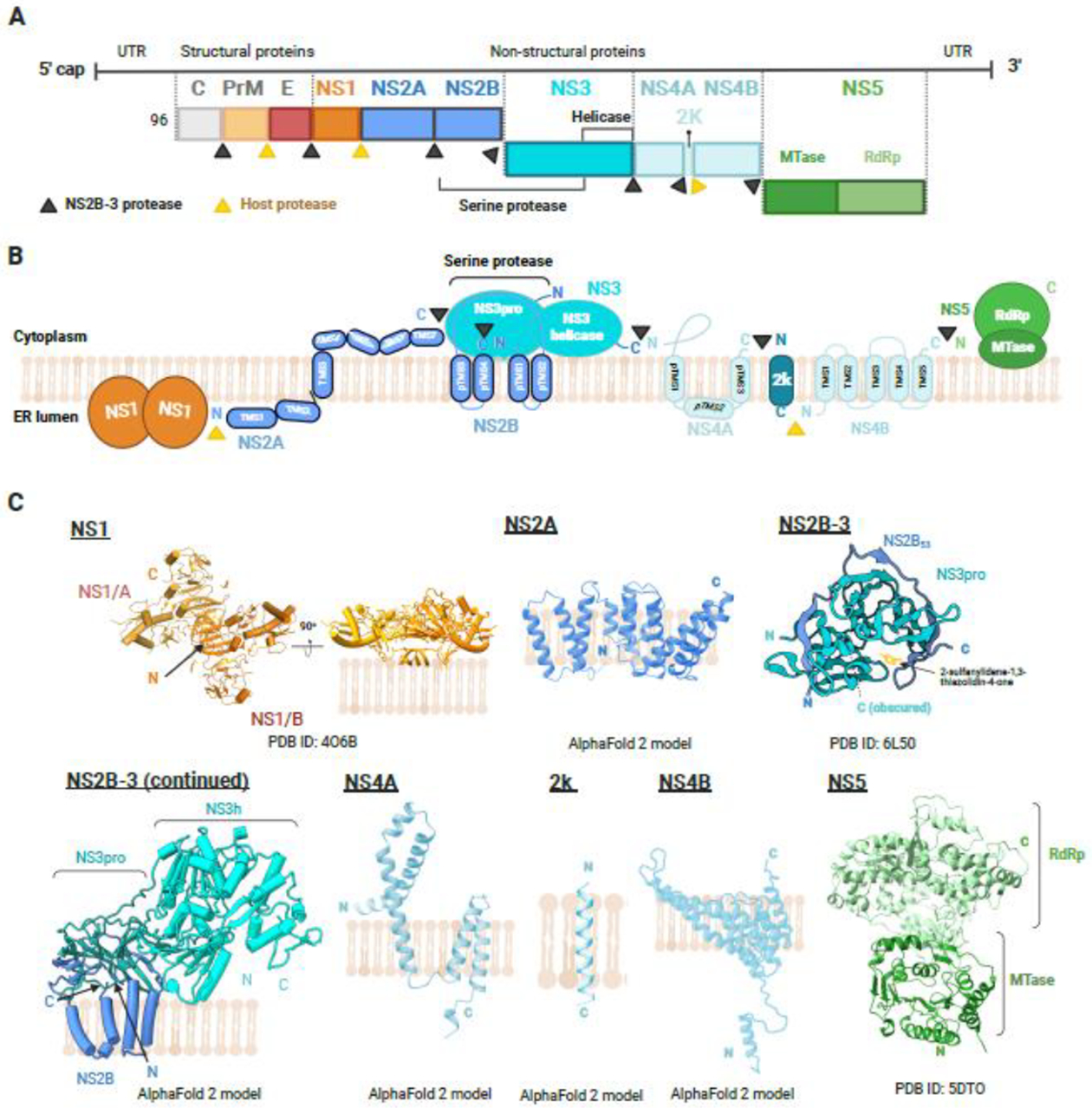
(A) Genome organization of the flavivirus genome. Flaviviral RNA is composed of a 5′UTR, one single open reading frame, and a 3′UTR. Translation of the flavivirus genomes produces a single polyprotein, later cleaved to structural and non-structural proteins by both host (in yellow) and viral NS2B-3 (in black) proteases. Capsid protein (C protein), pre-membrane protein (prM protein), and envelope protein (E protein) are structural proteins, whereas the remaining (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) are non-structural proteins (B) Diagrammatic representation of the membrane topology of the flaviviral polyprotein, as well as the polyprotein cleavage sites. NS2A, NS2B, NS4A, and NS4B contain transmembrane domains, while NS1 contains conserved residues that interact with the ER membrane [19]. NS5 is indirectly associated with the ER membrane through binding to NS3, which is in turn bound to transmembrane proteins NS2A, NS4A, and NS4B [124]. (C) Respective solved protein structures predicted models, and their structural organization in the ER membrane are illustrated. Each structure is annotated with a representative PDB ID, where appropriate, and visualized in cartoon. AlphaFold 2 (AF2) was used to predict the structures for NS2A, NS4A, and NS4B transmembrane proteins as they remain unresolved. NS1 (PDB: 4O6B) consists of a homodimer, annotated as NS1/A and NS1/B. The diagram is rotated 90 degrees to depict NS1 interaction with the membrane. NS2B-NS3 protease (NS3pro) interacts with E60 (2-sulfanylidene-1,3-thiazolidin-4-one) (PDB: 6L50). NS2B-3, NS4A, 2k peptide and NS4B were predicted with high confidence by AF2 (pLDDT score > 70). NS2B anchors the NS2B-3 complex to the ER membrane. While NS4A has been predicted with high confidence, the structural organization within the membrane appears to differ from the literature, with the C-terminus of AF2-predicted NS4A residing in the ER lumen. NS5 (PDB: 5DTO) consists of an N-terminal MTase and a C-terminal RNA-dependent RNA polymerase (RdRp). Figures were created with Biorender.
The NS proteins then assemble to form the viral replication complex or replicase, resulting in the formation of replication organelles (RO) through ER membrane invagination. These non-structural proteins perform crucial functions in viral replication. Elements of their structures are highly conserved among different flaviviruses or serotypes of flaviviruses. Targeting these elements, rather than the less conserved envelope protein, may provide the key to finding treatments for flaviviral infections. With known structures and predicted models of these NS proteins and protein complexes, we provide a structural proteomic view of the non-structural proteins and introduce a few highlights of the recent progress in anti-flaviviral drug development. We utilised AlphaFold 2 (AF2) [10] to predict the structures of unknown flaviviral transmembrane protein structures, NS2A, NS4A and NS4B, and discussed the prediction accuracy and differences to biochemical characterization studies that were reported.
NS1 structures and antibody-based protection.
NS1 is a ~48-kDa multi-functional glycoprotein that is found circulating in high levels during acute infection, is highly immunogenic, and exerts tissue-specific pathogenic effects of vascular permeability and pro-inflammatory responses [11] consistent with the flaviviral disease tropism [12]. Hence, secreted NS1 (sNS1) as a prognostic and diagnostic marker of different flavivirus infections using monoclonal antibodies or detection of anti-NS1 antibodies is well established. More recently, a new generation of NS1-based diagnostics based on DNA aptamers that can detect sub-serotype DENV variants was reported [13]. However, it does not serve well as a marker of disease severity due to confounding reports such as sNS1 levels being serotype-dependent in a mouse model [14], and no significant difference being observed between sNS1 levels in DENV2-infected patients with disease state in a study in Mexico [15] and Vietnam [16]. In the case of sNS1 biotherapeutic potential using small molecules, anti-NS1 antibodies, and in NS1-based vaccines, they remain vigorously pursued with a rapidly changing landscape of NS1 structures and the mechanistic basis for antibody-mediated protection.
The crystal structure of NS1 revealed a three-domain architecture, a hydrophobic β-roll (residues 1–29), an a/b wing (38–151), and a β-ladder (181–352). The connector segments between the wing and β-ladder domains, residues 30–37 and 152–180, form a 3-stranded β-sheet [17–19]. The dimer has a distinct crossed shape with the wings extending from the central β-ladder which has an extended β-sheet that faces the β-roll as the membrane-associated surface and a “spaghetti loop” on the opposite hydrophilic outer face that lacks structured elements (Figure 2A). Upon synthesis, It is translocated into the ER lumen where it is glycosylated and dimerizes [20, 21] and drives the biogenesis of the viral replication complex with known interactions to the NS4A-2K-4B precursor for viral RNA replication [22–24]. Extracellularly, the dimers are found associated with lipid rafts on the plasma membrane [25] and secreted in a presumed hexameric form (Figure 2C) that is barrel-shaped with a lipid-cargo held together by hydrophobic interactions based on results from native PAGE gel, analytical size-exclusion chromatography, electron microscopy models at low-resolution, and crystallographic contact analysis [26–28]. Recent virological studies show that secreted NS1 (sNS1) is associated with HDL which can trigger pro-inflammatory responses [29, 30] and uses scavenger receptor B1 (SRB1) as a cell receptor in cultured cells [31]. There is now definitive and direct evidence that sNS1 are found predominantly as dimers in complex with HDL (Figure 2D) and unlikely exists as hexamers in infected supernatants or patients’ blood as reported in our preprint [32]. This view is supported by the first direct evidence of hexameric sNS1 but they were only found in low numbers (~3.1%, 37,422 out of 1,219,277 particles) and the major form is tetrameric (Figure 2B) in their sample, his-tagged NS1 expressed in Expi293 HEK cells, which is reflective of the recombinant form of NS1 [33]. The direct interaction of sNS1 with SRB1 or indirectly as a complex with HDL for its internalisation to support viral replication and disruption of endothelial glycocalyx could help explain altered lipoprotein metabolism observed in Dengue patients [34] and hence offer new therapeutic routes. While it is unclear how the sNS1-SRB1 interaction may influence flaviviral disease tropism, a recent study identified the NS1 wing domain (residues 91 to 93) as the primary determinant for cell binding specificity while the β-ladder is involved in inducing endothelial hyperpermeability [35]. Correspondingly, the use of monoclonal antibodies against NS1 were able to counter NS1 pathogenesis and their binding to the β-ladder domain sterically hinders NS1 from interacting with the lipid membrane [36, 37] (Figure 2E). The development of anti-NS1 monoclonal antibodies for treatment is promising.
Figure 2. The landscape of NS1 structures.
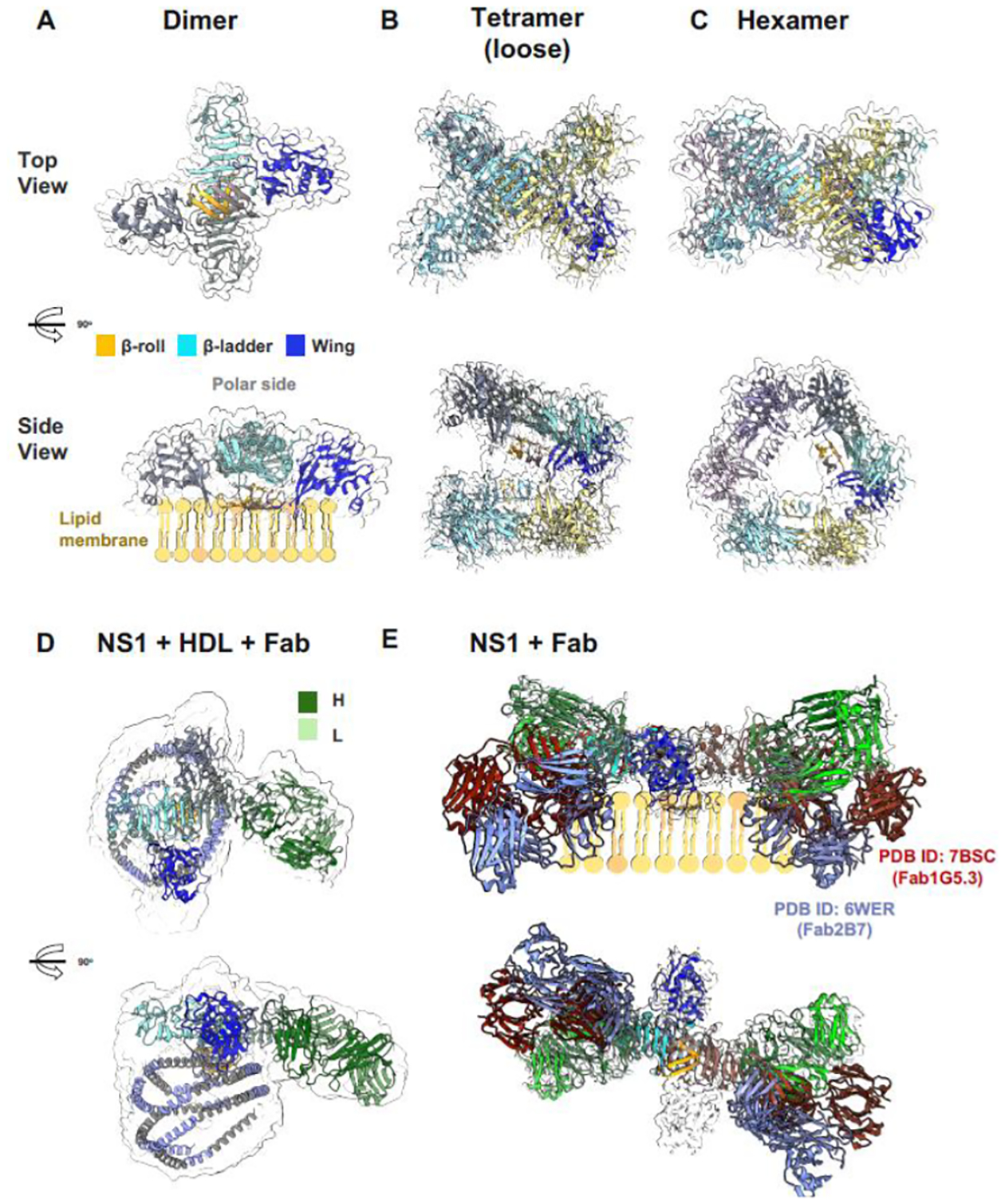
(A) Dimeric NS1 cartoon model in a simulated transparent map at 2.5 Å at a low contour of 0.2. One of the monomers is colored by its three-domain architecture, a hydrophobic β-roll (residues 1–29, orange), an a/b wing (residues 38–151, blue), and a rigid β-ladder (residues 181–352, cyan). The dimer has a distinct crossed shape with the wings extending from the central β-ladder which has an extended β-sheet that faces the β-roll as the membrane-associated surface (side view) and a “spaghetti loop” on the opposite polar outer face that lacks structured elements. (B) Tetrameric and (C) hexameric recombinant secreted NS1 (sNS1) forms as confirmed by Bo et al. (2021) cryoEM structures [1], modelled using the dimeric NS1 structure coupled with a simulated transparent map at 2.5 Å at the high contour of 1.0 and at 6.0 Å at the low contour of 0.3 respectively. The second NS1 dimer is coloured in yellow and blue for each monomer chain. The third NS1 dimer for the hexamer model is coloured in light and dark purple. (D) Immunoaffinity-purified sNS1 form from infected cell cultures as reported by Chew et al (2022) [2]. Model of sNS1wt: Fab56.2 predicted structures rigid body fitted in the CryoEM map (transparent grey, contoured at 0.14) with a correlation value of 0.75 to the fitted regions (map simulated from atoms at 5 Å). The antibody Fab56.2 domain is coloured in dark and light green for the Heavy (H) and Light (L) chains respectively. The predicted apoA-I, major protein component of HDL, model is coloured in intervals of grey and light purple representing its 11 and 22 residue alpha helical repeats. ApoA-I model is fitted in the spherical map region with its orientation informed by cross-linking mass spectrometry. (E) Comparison of the NS1 binding by anti-Denv Fab56.2 to earlier published NS1:Fab structures namely 2B7 (PDB ID: 6WER) [3] and 1G5.3 (PDB ID: 7BSC) overlayed on the lipid membrane cartoon for the side view and the top view shown at the bottom depicting the differing angles that they bind to the β-ladder. Figure prepared using ChimeraX.
Small molecules.
Celgosivir, an iminosugar, that causes NS1 to misfold and accumulate in the ER [38] did not reduce the disease burden in DENV patients in a Phase I proof-of-concept trial [39]. A Phase II clinical trial (NCT02569827) was subsequently approved when further dose regimen optimization of Celgosivir from 2 to 4 times daily was shown to reduce viremia significantly even on day 2 or 3 post-infection [40]. Unfortunately, the clinical trial has been withdrawn due to the lack of funding.
NS1-based vaccines.
There is evidence of autoimmunity from cross-reactive anti-NS1 antibodies especially in the case of Zika virus infection [41], and inflammatory activation and apoptosis induction via a proposed molecular mimicry mechanism [42], recently reviewed by Carpio and Barrett (2021) [43]. Overall, the use of NS1 for vaccine development remains doubtful.
NS2A
NS2A is the first NS protein that has multiple transmembrane helixes (Figure 1C). Its N-terminus is liberated from NS1 by a host protease and is therefore localized on the endoplasmic reticulum (ER) lumen. Its C-terminus is cytoplasmic as it is cleaved by viral NS2B-3 protease. A detailed model of the NS2A topology that was proposed [44] differed from the AF2-predicted model which was also of low confidence scores, with predicted Local Distance Difference Test (plDDT) scores ranging from 50–90. Existing literature has placed the third transmembrane segment (TMS) of NS2A within the membrane, with the other 6 TMS associated peripherally [45], with the N-terminus region including the first 2 TMS residing in the ER lumen, and TMS4-TMS7 at the C-terminus residing in the cytoplasm. This is in contrast to the AF2-predicted structures, which proposed multiple membrane-spanning segments (Figure 1C).
It is well understood that NS2A functions in both flaviviral RNA synthesis and virion assembly [46–49]. NS2A may act as a membrane-embedded platform to bring viral RNA and viral structural/nonstructural proteins to the virion assembly site. NS2A is not only an essential component of the viral replication complex but also a viral antagonist of the host immune response [50–52]. Given its critical role in the virus life cycle, it could be a good drug target. However, there has not been a promising compound targeting NS2A reported so far.
Inhibitors targeting NS2B-3 protease-helicase
NS3 is an essential protein in both polyprotein processing and viral replication. It has an MW of ~69kDa and its structure and function have been extensively studied and are well characterized [53]. It is multifunctional and contains 2 key domains: protease and helicase. Additionally, NS3 plays a role in viral assembly and the production of infectious particles [54–56]. The protease domain, of which the NS2B is a crucial cofactor, is instrumental in structural polyprotein processing and cleaves junctions between many of the viral proteins [57]. The helicase unwinds the RNA [58] secondary structures of the duplex formed during replication to preface the synthesis of new strands of viral RNA [59]. Finally, the 5’terminal RNA triphosphatase subdomain is presumably required for the synthesis of the 5’ cap structure which is a structure on the terminus of the RNA that mimics that of the host. This prevents degradation and allows for efficient translation by cellular machinery [60]. These two proteins and the interaction between them perform pivotal roles in the replication of flaviviruses and are therefore potential targets for therapeutics [61, 62]. High-resolution structures of the NS2B-NS3 protease structures from DENV, WNV, and ZIKV have been determined with various ligands and inhibitory compounds [63–65]. Many protease inhibitors have been reported, yet none has advanced into clinical stages [7, 8, 62, 66–68]. Additionally, several promising compounds have been identified by the Christian Klein group, but haven’t yet been tested in vivo [69–72]. The NS3 helicase domain, given its dynamic nature upon RNA binding, translocation, and unwinding activities coupling to ATP binding and hydrolysis, is considered a less attractive target to develop specific inhibitors [61, 73, 74].
A drug screening and chemical synthesis produced a range of compounds with antiviral properties targeting the flaviviral protease. One of the most potent of these compounds, which also had minimal activities against human proteases, was designated compound 9 (Figure 3A) [75]. This compound showed activity against the proteases of Zika virus, Dengue virus 2 and 3, and West Nile virus. The authors determined a crystal structure of the molecule bound to DV2pro, revealing that it binds to an allosteric pocket (Figure 3A). This acts to ‘lock’ NS3 in an open conformation, blocking it from binding NS2B or the substrate to perform its function. However, the crystal structure suffers from poor refinement, probably due to largely disordered regions in the structure. Compound 9 was shown to have strong antiviral activity against the aforementioned viruses in cell culture experiments, inhibiting viral replication at a high level with no significant cytotoxicity. The Compound had an IC50 as low as 120nM and an EC68 of 300–600nM in a mouse model of ZIKV infected cells, with no significant toxicity. In-vivo experiments were also performed in mice, showing good tolerance and a significant survival benefit [75]. Similar ideas of targeting the open conformation using allosteric inhibitors have also yield some new compounds of different chemical classes [76, 77]. Further analysis to confirm the binding model of these compounds is required.
Figure 3. Selected flavivirus Protease Inhibitors.
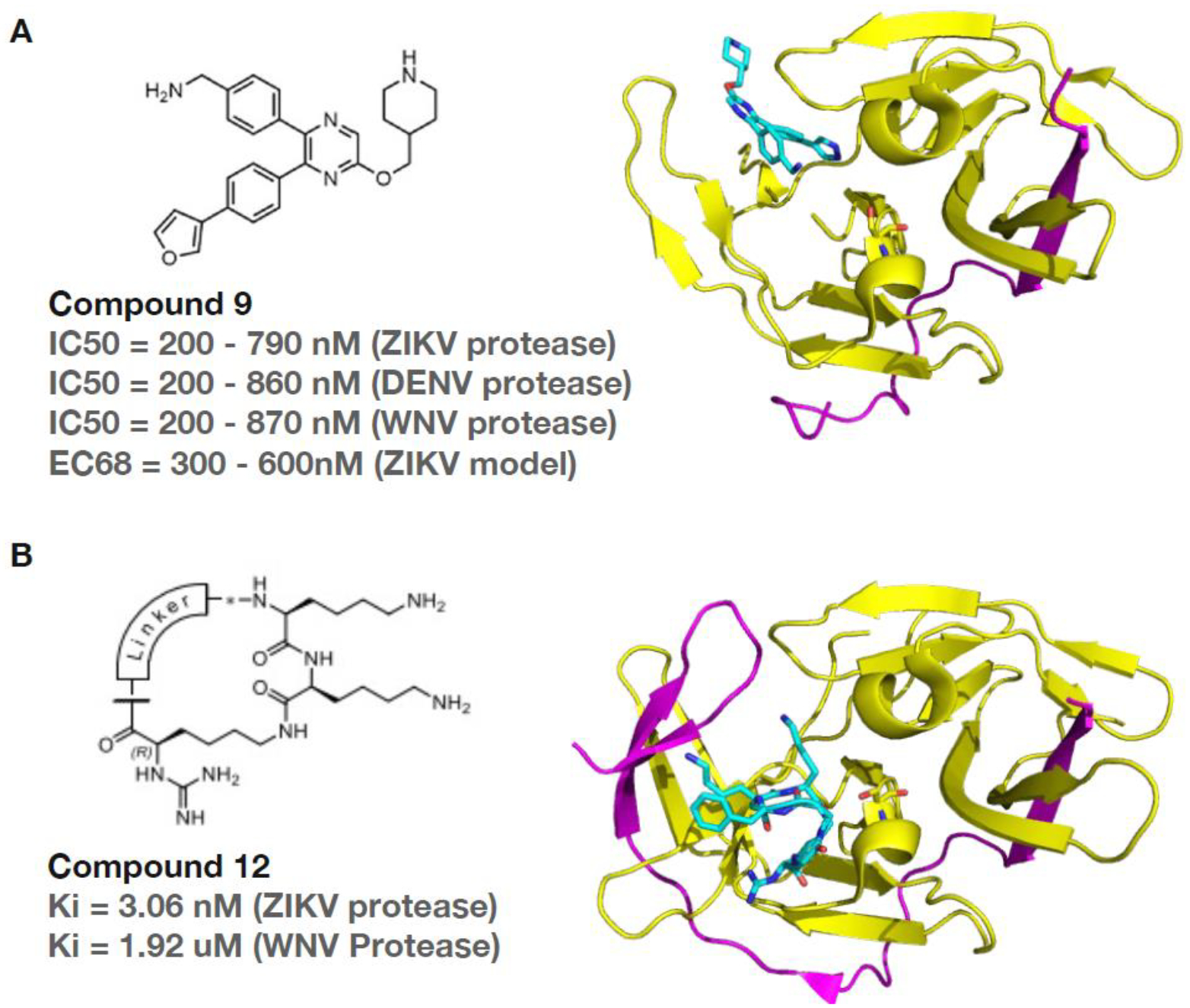
(A) Structure of a small molecule compound 9, formula 4-(NH2CH2)-Ph, in complex with the NS2B-NS3 protease from Dengue 2 virus. (B) Structure of a cyclic peptide compound 12 in complex with the NS2B-NS3 protease from Zika virus. Colour code: compounds light blue, partially unfolded NS2B cofactor magenta, and NS3 protease domain yellow.
Flaviviral proteases share a similar dibasic peptide substrate recognition pocket next to the active site. Substrate peptide-based protease inhibitors reported so far suffer from weak cellular activities due to the positive charges. Nonetheless, some most potent cyclic peptide inhibitors have been reported recently following a series of structure-based peptide chemistry studies [78–81]. Compound 12 shown in Figure 3B is one of the best characterised cyclic peptide inhibitors. These highly charged inhibitors suffer from poor membrane permeability and weak antiviral efficacy in cell-based Zika virus infection assays. Future efforts should focus on the chemical modification of substrate-like peptide scaffolds to produce potent, specific, cellularlyactive non-peptidic inhibitors [71, 82].
NS4A
NS4A is a small membrane protein (~16kDa) that has crucial roles in RNA replication and the formation of the RO [24, 83–87]. It acts as a scaffold for the replication complex, and is sufficient to induce membrane curvature alone, likely through a wedge mechanism. AlphaFold2 model shows this wedge-shaped structure. However, this prediction also contains inconsistencies when compared to the biochemical data. Notably, the protein’s C-terminus is located on the luminal side of the ER membrane in the AF2 prediction (Figure 1C), which is incompatible with the fact that NS3 cleavage is known to occur on the cytoplasmic side.
Several NS4A inhibitors have been identified and present a promising avenue for further development, considering the importance of NS4A in the viral life cycle. In particular, the drug SBI-0090799 is a potent and selective inhibitor of ZIKV and has been shown to block the formation of ROs de novo [88]. Currently, no flavivirus NS4A inhibitors has progressed to clinical trials.
NS4B inhibitors/ polyprotein processing and RC
NS4B is the largest transmembrane protein (~27kDa). The expected topology of NS4b from a range of biochemical studies and bioinformatic predictions disagree on the number of transmembrane helices and their arrangement [86]. The AF2 models also differ significantly from the biochemical data, predicting 9 helices rather than around 5 helices (Figure 4A). These discrepancies may be due to the intrinsically disordered nature of NS4b. The extra helices may unfold when the protein interacts with lipids or other proteins and exhibit a dynamic structure. This is consistent with molecular modelling, which predicts that the N-terminus has a disordered structure, the C-terminus order is dependent on the presence of lipids and the cytosolic loop may adopt a partially ordered state [89]. In situ structural studies are needed to clarify the structures of these proteins in conjunction with lipids, RNA and binding partners to be more certain of their topologies. NS4b is known to dimerise in vivo and when purified [90]. It has been demonstrated that the cytoplasmic loop is critical for dimerisation. The C-terminus (TM4–5) alone can dimerise, which may also mediate dimerisation alongside the cytoplasmic loop, although deletion of this region does not prevent dimerisation. It too colocalizes with other proteins of the RC, specifically NS3 and NS4A, as well as the viral RNA. It is involved in dissociating the RNA from the NS3 helicase domain, working cooperatively with NS4A [91–93]. Additionally, NS4B putatively inhibits interferon signalling, seriously impeding the host’s immune response to the virus. Inducing alterations in NS4B hampers replication of viral RNA significantly, indicating that its function is much more than just structural [94].
Figure 4. Predicted protein structure of NS4B and its inhibitors.
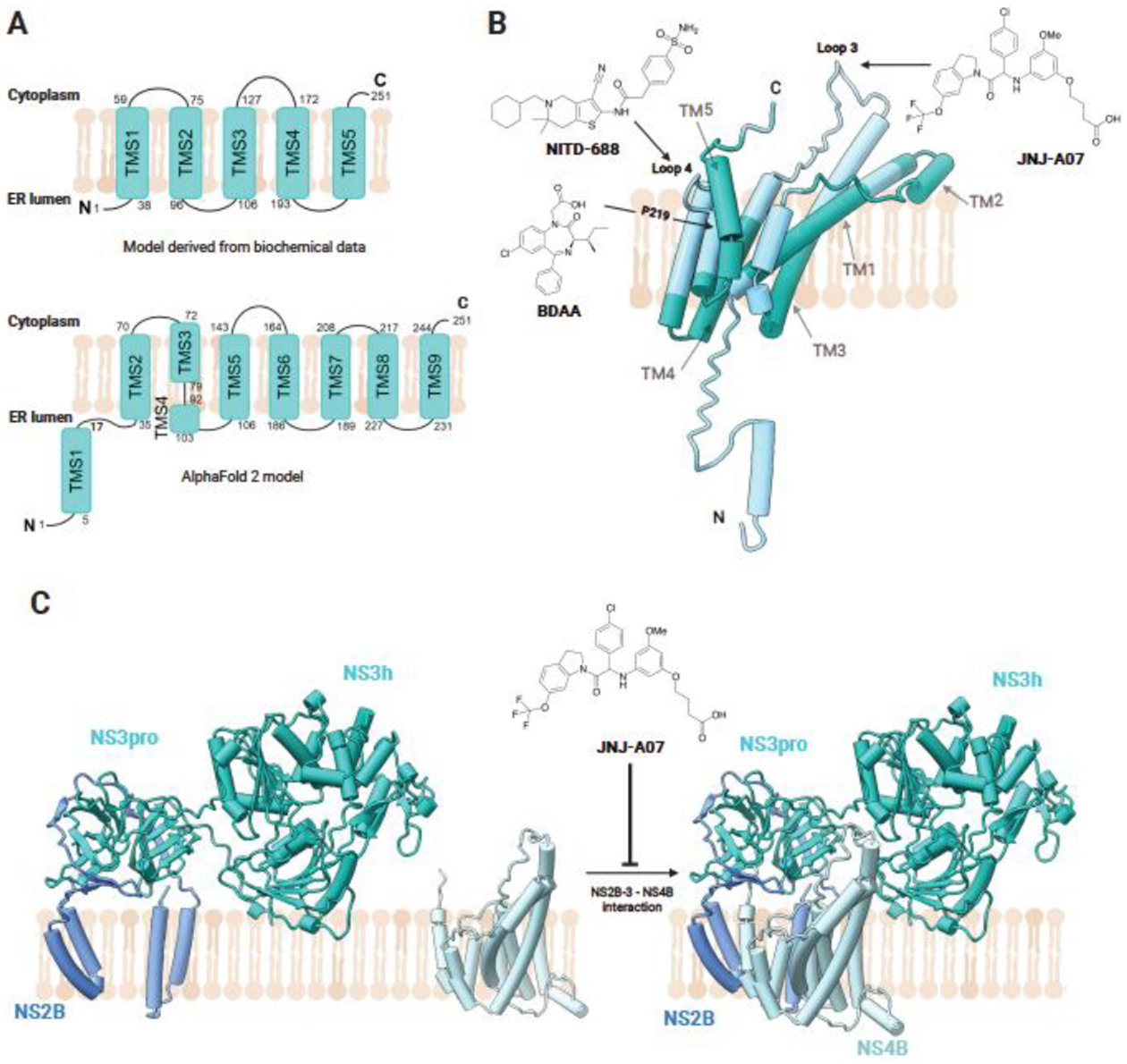
(A) Comparison between NS4B topology derived from biochemical data, adapted from Bhardwaj, et al., and topology of the AF2 model. (B) Schematic of AF2-predicted structural organisation of NS4B in the ER membrane, with proposed transmembrane domains based on biochemical studies labelled and colored in dark green and cytoplasmic facing regions coloured in cyan. Sites of interaction between NS4B and the compounds JNJ-A07, NITD-688, and BDAA are annotated. JNJ-A07 alters the conformation of loop 3 of NS4B, occurring between TM3 and TM4. BDAA, a small molecule inhibitor of YFV, binds P219 in TM5 of NS4B. NITD-688 has also been identified to bind to NS4B. (C) Effect of JNJ-A07 binding on the interaction of NS2B3-NS4B. JNJ-A07 blocks NS3-NS4B interaction and complex formation but does not disrupt existing NS3-NS4B complexes. Created with Biorender.
Previous studies have identified several compounds which showed activity against NS4b and some showing effectiveness in vivo. These inhibitors generally had EC50s in the micromolar range. The more recently identified compounds discussed below have EC50s in the nanomolar range, all show strong potency in vivo and pan-flaviviral or pan-serotype activity [95–97]. Understanding NS4B and its crucial functions in viral replication better could shed light on several viral processes that are currently beyond our knowledge. It is also an attractive target for antiviral development as it is well-conserved among flaviviruses and takes a central role in the replication complex.
JNJ-A07.
A recent study identified a new compound in an anti-DENV screen, JNJ-A07, which is shown to be a potent inhibitor of DENV replication in all serotypes, with nanomolar to picomolar activity [98]. The study demonstrated that the compound blocks the interaction between NS4B and NS3, preventing the formation of NS3-NS4B complexes but does not disrupt after its formation, therefore attenuating the cleavage of the polyprotein (Figure 4B). JNJ-A07 was found to have a high barrier to resistance, with three mutations in conjunction being necessary to attain a prominent resistance. Additionally, these drug-resistant mutants are no longer able to replicate in mosquito cells, suggesting that transmission would be abolished. NS4B is known to dissociate NS3 from the viral RNA and increase the NS3 helicase activity [92]. The findings of the study suggest that JNJ-A07 allosterically alters the conformation of the cytosol facing loop 3 of NS4B, blocking the formation of the complex. In-vivo experiments showed that the drug is well tolerated and was efficacious in lowering viral load and disease[98].
NITD-688.
There is a strong necessity for antivirals that can target all four dengue serotypes, and currently, no approved drugs meet this need. In 2021, Moquin et al [99] reported the discovery of NITD-688, a potent inhibitor of pan-serotype dengue replication introduced by NOVARTIS. Using NMR, the authors demonstrated that the molecule binds to NS4b, further reinforced by resistance mutational studies which found that resistance-conferring mutations take place in NS4b (Figure 4A). These mutations are contained in TM helices 4 and 5, a possible indication that the drug affects dimerisation, which would need to be demonstrated by further study. These residues are all conserved across dengue serotypes, accounting for the compound’s effectiveness in all four. The drug has a good pharmacokinetic profile, is well tolerated and reduced viremia in in vivo studies.
BDAA.
A small molecule inhibitor of the yellow fever virus, BDAA, has also been found to also target NS4B [100]. This is a benzodiazepine compound that was discovered to present significant antiviral activity against the yellow fever virus. The mutational resistance profile showed that the compound targets P219 located in TM5 of NS4B (Figure 4A). This may indicate that NITD-688 shares a similar mechanism of action. The compound was found to inhibit viral replication in infected cells with high potency with corresponding significant increase in the innate immune system response via cytokine release. The use of electron microscopy images of the infected and treated cells reveal a significantly altered morphology of the ROs when compared to the infected-untreated cells. NS4b is thought to influence the morphology of the RO and may induce curvature in the ER membrane to produce these, indicating this action may be compromised. BDAA was also reported to have potent antiviral activity in vivo and was well tolerated. Subsequent reported mechanistic results led to a dual antiviral model whereby BDAA inhibits viral RNA replication by disrupting the NS4B-driven integrity of YFV ROs which also causes the release of viral RNA from ROs resulting in RIG-I and MDA5 activation [101].
NS5 MTase-RdRp and the RdRp inhibitors
NS5 is the largest protein (~104kDa) and is highly conserved across flaviviruses. The N-terminal methyltransferase (MTase) domain is globular with 3 sub domains (Figures 1 and 5). These include a GTP-binding pocket, SAM binding pocket, and MTase activity [102]. This domain is involved in the capping of the viral RNA alongside NS3 [103]. The C terminal RNA dependant RNA polymerase (RdRp) domain is the final portion of the viral polyprotein and has a critical function [104]. It catalyses the synthesis of the viral RNA strand from the duplex RNA templates. It has a structure that is common among many RdRp proteins with a cupped right-hand architecture with subdomains termed: the fingers, palm, and thumb [104]. It also contains a GDD motif for incorporating nucleotides. The structure of NS5 as well as its functioning has been well characterised [105]. Among several known full-length NS5 structures, there are different relative conformations between MTase and RdRp [104, 106–111]. The MTase and RdRp are connected via a flexible and less conserved linker, which seems to help regulate the virus replication process [112] There is no structure of NS5 RdRp bound with RNA and the molecular understanding of the RNA polymerization process is based on the homology of flaviviral RdRp to the RdRp of the related positive sense RNA viruses.
Figure 5. Structure of NS5 MTase-RdRp and its inhibitors.
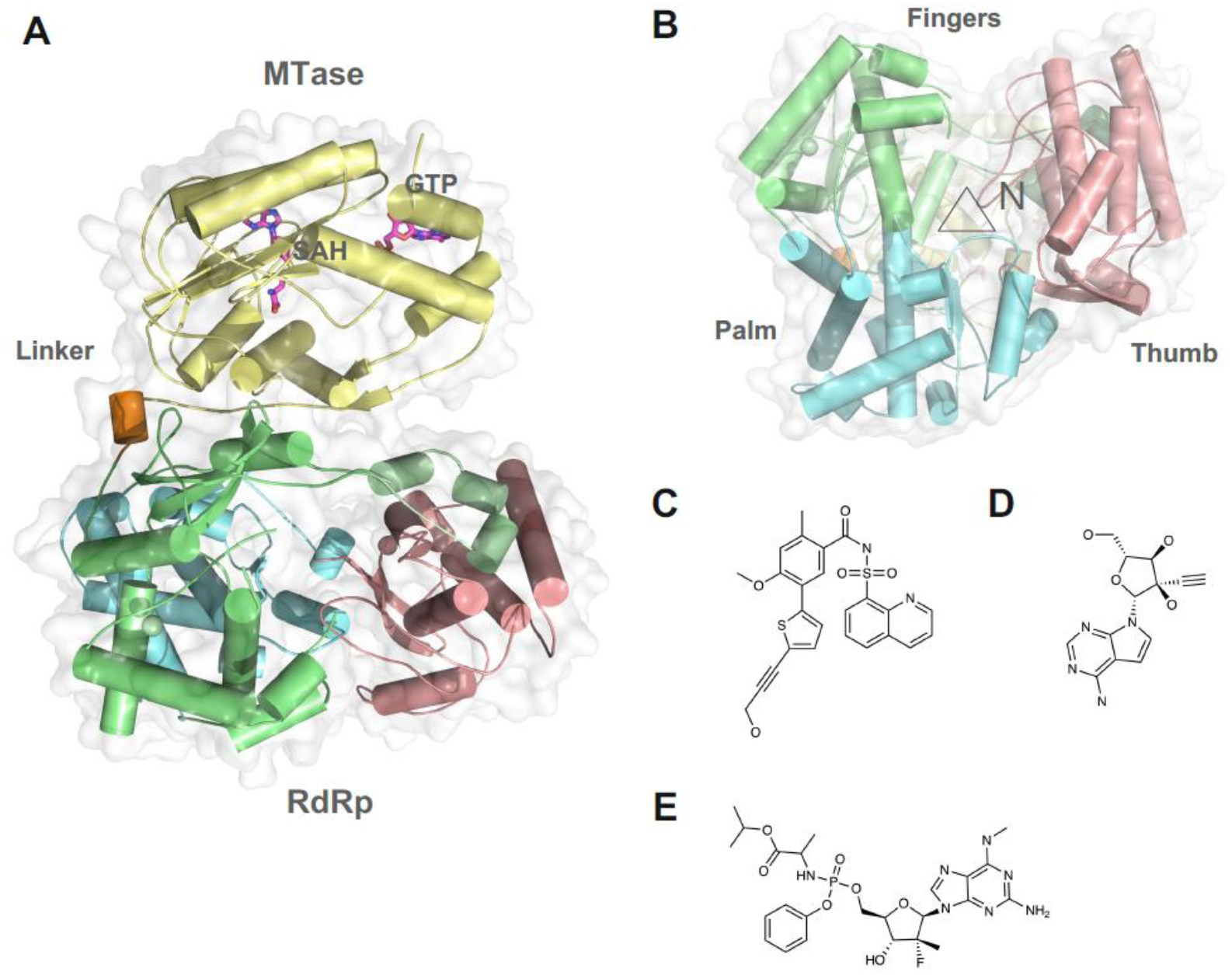
(A) Overall structure of DENV3 full length NS5. MTase domain is colored in yellow. Fingers Palm and Thumb domains of the RdRp are colored in green, cyan, and pink. Linker region is colored in orange. (B) View of the RdRp active site, marked with a triangle. The allosteric inhibitors binds a novel pocket on the Thumb domain next to the active site. Chemical structure of the (C) Compound 29 [114], (D) NITD008. (E) AT-752.
Using a structure-guided fragment-based screening method, allosteric inhibitors targeting the N pocket of DENV/ZIKV NS5 RdRp have been developed [113–116] (Figures 5B–C). These compounds represent a novel class of pan-serotype and cell-active Non-nucleoside Inhibitors against NS5 RdRp (Figures 5C). Enzyme kinetics study suggests these compounds may be uncompetitive inhibitors and primarily inhibit the de novo initiation step of RNA synthesis. Interestingly, resistance mutations have been identified using a replicon cellline, which suggests that the N pocket may also affect the replication complex formation. Given the subtle difference of the N pocket among flaviviral NS5, the authors suggest that it is possible to develop selective N pocket inhibitors for difference subgroups of flaviviruses [117].
Nucleotide analogue inhibitors remain the most attractive candidate for an RdRp, given the successful track record in inhibiting RNA polymerases of several other viruses such as HIV, HCV, and SARS-COV-2.
NITD008
NITD008 is an adenosine analog inhibitor against pan-flaviviruses that is developed by Novartis (Figures 5D) [118, 119] It functions as a chain terminator during viral RNA synthesis. However, NITD008 is tested to be too toxic in pre-clinical animal testing. As a proof-of-concept inhibitor to treat flavivirus infections, it is also commonly used as a research tool for antiviral research for emerging viral diseases.
AT-752.
In 2021, Good et al reported the discovery of a guanosine nucleotide analogue with potent anti-dengue activity, AT-752 9 (Figures 5E). In vitro, the compound was potent in its activity against both DENV 2 and 3, as well as HCV. It inhibited viral replication with an EC50 of 0.48 and 0.77μM in DENV2 and 3 respectively and was able to inhibit all other flaviviruses potently. The drug is orally available and was found to significantly increase the longevity of infected mice in-vivo and lower viremia. As a nucleotide analogue, the drug is presumed to target the viral RdRp. To confirm this and establish the mechanism of action, the authors performed an incorporation and elongation assay. They found that, during RNA synthesis, AT-752 is incorporated into the viral RNA by the RdRp, consequently terminating synthesis [120]. Atea Pharmaceuticals, the company which developed AT-752, is conducting clinical trials to access the safety, antiviral activity, and pharmacokinetics of the drug in dengue-endemic regions.
Conclusion and Future Perspectives
This review summarises recent advances in flavivirus antivirals, with a focus on those that target non-structural proteins. These proteins have great potential as drug targets, as their functions are crucial to viral replication and their structures are often conserved. The above studies highlight the possibility of a pan-dengue and even a pan-flavivirus antiviral due to the structural similarity between key non-structural proteins. In particular, NS4B seems to be a lucrative target, with high similarity among flaviviruses and its importance in viral replication. Additionally, pharmacological targeting of the NS3 protease and the RdRp NS5 have shown to be promising in arresting viral replication. Targeting the non-structural proteins gives a key advantage; the NS proteins are well conserved, and therefore, the activity of drugs that bind to them is generally less affected by resistance mutations. Structural studies provide crucial information that can guide the development of drug candidates and further structural studies are needed to produce safe and potent flavivirus antivirals. As outlined above, the structures and interactions of the non-structural proteins provide key information to producing new drugs and understanding how they work. The structural details of the full replication complex would be instrumental in achieving a complete understanding of the arranging of the viral RNA, viral and host proteins, and their interactions, which would provide the key to producing molecules that block these interactions and effectively halt replication across flaviviruses [24, 88, 121–123].
Highlights.
Molecular structures and predicated models of flaviviral NS proteins.
Recent advance in antiviral development targeting NS proteins.
Inhibitors targeting NS4B entering clinical studies.
Acknowledgments
We apologize to all colleagues whose contributions could not be highlighted or discussed within this manuscript. This research is supported by 1) Singapore Ministry of Education under its Singapore Ministry of Education Academic Research Fund Tier 2 (T2EP30220-0020) to DL; 2) Singapore Ministry of Education under its Singapore Ministry of Education Academic Research Fund Tier 1 Award 2021-T1-002-021 to DL; 3) National Institute of Allergy and Infectious Disease grant U19-AI171954 to DL. 4) LKCMedicine Dean’s Postdoctoral Fellowship to C.B.L.A.; 5) A*STAR Singa Scholarship and the Lee Kong Chian School of Medicine to K.v.d.E. We wish to acknowledge the funding support to H.J.S. for this project from Nanyang Technological University under the URECA Undergraduate Research Programme.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
References
- 1.Bhatt S, et al. , The global distribution and burden of dengue. Nature, 2013. 496(7446): p. 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gyawali N, Bradbury RS, and Taylor-Robinson AW, The epidemiology of dengue infection: Harnessing past experience and current knowledge to support implementation of future control strategies. J Vector Borne Dis, 2016. 53(4): p. 293–304. [PubMed] [Google Scholar]
- 3.Solomon T and Vaughn DW, Pathogenesis and clinical features of Japanese encephalitis and West Nile virus infections. Curr Top Microbiol Immunol, 2002. 267: p. 171–94. [DOI] [PubMed] [Google Scholar]
- 4.Pierson TC and Diamond MS, The emergence of Zika virus and its new clinical syndromes. Nature, 2018. 560(7720): p. 573–581. [DOI] [PubMed] [Google Scholar]
- 5.WHO, Dengue guidelines, for diagnosis, treatment, prevention and control. 2009, World Health Organization: Geneva. p. 157. [PubMed] [Google Scholar]
- 6.Lim SP, Dengue drug discovery: Progress, challenges and outlook. Antiviral Res, 2019. 163: p. 156–178. [DOI] [PubMed] [Google Scholar]
- 7.Lim SP, et al. , Ten years of dengue drug discovery: progress and prospects. Antiviral Res, 2013. 100(2): p. 500–19. [DOI] [PubMed] [Google Scholar]
- 8.Lescar J, et al. , The Dengue Virus Replication Complex: From RNA Replication to Protein-Protein Interactions to Evasion of Innate Immunity. Adv Exp Med Biol, 2018. 1062: p. 115–129. [DOI] [PubMed] [Google Scholar]
- 9.Westaway EG, et al. , Flaviviridae. Intervirology, 1985. 24(4): p. 183–92. [DOI] [PubMed] [Google Scholar]
- 10.Mirdita M, et al. , ColabFold: making protein folding accessible to all. Nature Methods, 2022. 19(6): p. 679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glasner DR, et al. , The Good, the Bad, and the Shocking: The Multiple Roles of Dengue Virus Nonstructural Protein 1 in Protection and Pathogenesis. Annu Rev Virol, 2018. 5(1): p. 227–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puerta-Guardo H, et al. , Flavivirus NS1 Triggers Tissue-Specific Vascular Endothelial Dysfunction Reflecting Disease Tropism. Cell Rep, 2019. 26(6): p. 1598–1613 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsunaga KI, et al. , High-affinity five/six-letter DNA aptamers with superior specificity enabling the detection of dengue NS1 protein variants beyond the serotype identification. Nucleic Acids Res, 2021. 49(20): p. 11407–11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe S, et al. , The magnitude of dengue virus NS1 protein secretion is strain dependent and does not correlate with severe pathologies in the mouse infection model. J Virol, 2012. 86(10): p. 5508–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de la Cruz-Hernandez SI, et al. , Determination of viremia and concentration of circulating nonstructural protein 1 in patients infected with dengue virus in Mexico. Am J Trop Med Hyg, 2013. 88(3): p. 446–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chau TN, et al. , Clinical and virological features of Dengue in Vietnamese infants. PLoS Negl Trop Dis, 2010. 4(4): p. e657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akey DL, et al. , Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system. Science, 2014. 343(6173): p. 881–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X, et al. , Contribution of intertwined loop to membrane association revealed by Zika virus full-length NS1 structure. EMBO J, 2016. 35(20): p. 2170–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown WC, et al. , Extended surface for membrane association in Zika virus NS1 structure. Nat Struct Mol Biol, 2016. 23(9): p. 865–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winkler G, et al. , Newly synthesized dengue-2 virus nonstructural protein NS1 is a soluble protein but becomes partially hydrophobic and membrane-associated after dimerization. Virology, 1989. 171(1): p. 302–5. [DOI] [PubMed] [Google Scholar]
- 21.Winkler G, et al. , Evidence that the mature form of the flavivirus nonstructural protein NS1 is a dimer. Virology, 1988. 162(1): p. 187–96. [DOI] [PubMed] [Google Scholar]
- 22.Scaturro P, et al. , Dengue Virus Non-structural Protein 1 Modulates Infectious Particle Production via Interaction with the Structural Proteins. PLoS Pathog, 2015. 11(11): p. e1005277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindenbach BD and Rice CM, Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J Virol, 1999. 73(6): p. 4611–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plaszczyca A, et al. , A novel interaction between dengue virus nonstructural protein 1 and the NS4A-2K-4B precursor is required for viral RNA replication but not for formation of the membranous replication organelle. PLoS Pathog, 2019. 15(5): p. e1007736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noisakran S, et al. , Association of dengue virus NS1 protein with lipid rafts. J Gen Virol, 2008. 89(Pt 10): p. 2492–2500. [DOI] [PubMed] [Google Scholar]
- 26.Flamand M, et al. , Dengue virus type 1 nonstructural glycoprotein NS1 is secreted from mammalian cells as a soluble hexamer in a glycosylation-dependent fashion. J Virol, 1999. 73(7): p. 6104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gutsche I, et al. , Secreted dengue virus nonstructural protein NS1 is an atypical barrel-shaped high-density lipoprotein. Proc Natl Acad Sci U S A, 2011. 108(19): p. 8003–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller DA, et al. , Structure of the dengue virus glycoprotein non-structural protein 1 by electron microscopy and single-particle analysis. J Gen Virol, 2012. 93(Pt 4): p. 771–779. [DOI] [PubMed] [Google Scholar]
- 29.Coelho DR, et al. , ApoA1 Neutralizes Proinflammatory Effects of Dengue Virus NS1 Protein and Modulates Viral Immune Evasion. J Virol, 2021. 95(13): p. e0197420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benfrid S, et al. , Dengue virus NS1 protein conveys pro-inflammatory signals by docking onto high-density lipoproteins. EMBO Rep, 2022. 23(7): p. e53600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alcala AC, et al. , Dengue Virus NS1 Uses Scavenger Receptor B1 as a Cell Receptor in Cultured Cells. J Virol, 2022. 96(5): p. e0166421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chew BLA, et al. , Secreted dengue virus NS1 is predominantly dimeric and in complex with high-density lipoprotein. 2022, Cold Spring Harbor Laboratory. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shu B, et al. , CryoEM structures of the multimeric secreted NS1, a major factor for dengue hemorrhagic fever. Nature Communications, 2022. 13(1): p. 6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farfan-Morales CN, et al. , Anti-flavivirus Properties of Lipid-Lowering Drugs. Front Physiol, 2021. 12: p. 749770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lo NTN, et al. , Molecular Determinants of Tissue Specificity of Flavivirus Nonstructural Protein 1 Interaction with Endothelial Cells. J Virol, 2022. 96(19): p. e0066122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Modhiran N, et al. , A broadly protective antibody that targets the flavivirus NS1 protein. Science, 2021. 371(6525): p. 190–194. [DOI] [PubMed] [Google Scholar]
- 37.Biering SB, et al. , Structural basis for antibody inhibition of flavivirus NS1-triggered endothelial dysfunction. Science, 2021. 371(6525): p. 194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rathore AP, et al. , Celgosivir treatment misfolds dengue virus NS1 protein, induces cellular pro-survival genes and protects against lethal challenge mouse model. Antiviral Res, 2011. 92(3): p. 453–60. [DOI] [PubMed] [Google Scholar]
- 39.Low JG, et al. , Efficacy and safety of celgosivir in patients with dengue fever (CELADEN): a phase 1b, randomised, double-blind, placebo-controlled, proof-of-concept trial. Lancet Infect Dis, 2014. 14(8): p. 706–715. [DOI] [PubMed] [Google Scholar]
- 40.Watanabe S, et al. , Optimizing celgosivir therapy in mouse models of dengue virus infection of serotypes 1 and 2: The search for a window for potential therapeutic efficacy. Antiviral Res, 2016. 127: p. 10–9. [DOI] [PubMed] [Google Scholar]
- 41.Cavazzoni CB, et al. , The immunodominant antibody response to Zika virus NS1 protein is characterized by cross-reactivity to self. J Exp Med, 2021. 218(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen CL, et al. , Anti-dengue virus nonstructural protein 1 antibodies cause NO-mediated endothelial cell apoptosis via ceramide-regulated glycogen synthase kinase-3beta and NF-kappaB activation. J Immunol, 2013. 191(4): p. 1744–52. [DOI] [PubMed] [Google Scholar]
- 43.Carpio KL and Barrett ADT, Flavivirus NS1 and Its Potential in Vaccine Development. Vaccines (Basel), 2021. 9(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xie X, et al. , Membrane topology and function of dengue virus NS2A protein. J Virol, 2013. 87(8): p. 4609–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, et al. , Genetic and biochemical characterizations of Zika virus NS2A protein. Emerg Microbes Infect, 2019. 8(1): p. 585–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leung JY, et al. , Role of nonstructural protein NS2A in flavivirus assembly. J Virol, 2008. 82(10): p. 4731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kummerer BM and Rice CM, Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J Virol, 2002. 76(10): p. 4773–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie X, et al. , Dengue NS2A Protein Orchestrates Virus Assembly. Cell Host Microbe, 2019. 26(5): p. 606–622 e8. [DOI] [PubMed] [Google Scholar]
- 49.Bateman A, et al. , UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Research, 2021. 49(D1): p. D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu RH, et al. , Mutagenesis of Dengue Virus Protein NS2A Revealed a Novel Domain Responsible for Virus-Induced Cytopathic Effect and Interactions between NS2A and NS2B Transmembrane Segments. J Virol, 2017. 91(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Melian EB, et al. , West Nile virus NS2A protein facilitates virus-induced apoptosis independently of interferon response. J Gen Virol, 2013. 94(Pt 2): p. 308–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Munoz-Jordan JL, et al. , Inhibition of interferon signaling by dengue virus. Proc Natl Acad Sci U S A, 2003. 100(24): p. 14333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luo D, et al. , Crystal structure of the NS3 protease-helicase from dengue virus. J Virol, 2008. 82(1): p. 173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patkar CG and Kuhn RJ, Yellow Fever virus NS3 plays an essential role in virus assembly independent of its known enzymatic functions. J Virol, 2008. 82(7): p. 3342–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu WJ, et al. , Complementation analysis of the flavivirus Kunjin NS3 and NS5 proteins defines the minimal regions essential for formation of a replication complex and shows a requirement of NS3 in cis for virus assembly. J Virol, 2002. 76(21): p. 10766–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gebhard LG, et al. , A Proline-Rich N-Terminal Region of the Dengue Virus NS3 Is Crucial for Infectious Particle Production. J Virol, 2016. 90(11): p. 5451–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeidler JD, et al. , Non-Canonical Roles of Dengue Virus Non-Structural Proteins. Viruses, 2017. 9(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang X, et al. , Zika Virus NS2A-Mediated Virion Assembly. mBio, 2019. 10(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swarbrick CMD, et al. , NS3 helicase from dengue virus specifically recognizes viral RNA sequence to ensure optimal replication. Nucleic Acids Res, 2017. 45(22): p. 12904–12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu L, et al. , Flavivirus RNA cap methyltransferase: structure, function, and inhibition. Front Biol (Beijing), 2010. 5(4): p. 286–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luo D, Vasudevan SG, and Lescar J, The flavivirus NS2B-NS3 protease-helicase as a target for antiviral drug development. Antiviral Res, 2015. 118: p. 148–58. [DOI] [PubMed] [Google Scholar]
- 62.Kang C, Keller TH, and Luo D, Zika Virus Protease: An Antiviral Drug Target. Trends Microbiol, 2017. 25(10): p. 797–808. [DOI] [PubMed] [Google Scholar]
- 63.Phoo WW, et al. , Structure of the NS2B-NS3 protease from Zika virus after self-cleavage. Nat Commun, 2016. 7: p. 13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Z, et al. , Crystal structure of unlinked NS2B-NS3 protease from Zika virus. Science, 2016. 354(6319): p. 1597–1600. [DOI] [PubMed] [Google Scholar]
- 65.Lei J, et al. , Crystal structure of Zika virus NS2B-NS3 protease in complex with a boronate inhibitor. Science, 2016. 353(6298): p. 503–5. [DOI] [PubMed] [Google Scholar]
- 66.Rassias G, et al. , Cell-active carbazole derivatives as inhibitors of the zika virus protease. Eur J Med Chem, 2019. 180: p. 536–545. [DOI] [PubMed] [Google Scholar]
- 67.Nitsche C, Proteases from dengue, West Nile and Zika viruses as drug targets. Biophys Rev, 2019. 11(2): p. 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boldescu V, et al. , Broad-spectrum agents for flaviviral infections: dengue, Zika and beyond. Nat Rev Drug Discov, 2017. 16(8): p. 565–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kühl N, et al. , Beyond Basicity: Discovery of Nonbasic DENV-2 Protease Inhibitors with Potent Activity in Cell Culture. Journal of Medicinal Chemistry, 2021. 64(8): p. 4567–4587. [DOI] [PubMed] [Google Scholar]
- 70.Nitsche C, et al. , De Novo Discovery of Nonstandard Macrocyclic Peptides as Noncompetitive Inhibitors of the Zika Virus NS2B-NS3 Protease. ACS Medicinal Chemistry Letters, 2019. 10(2): p. 168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kühl N, et al. , A New Class of Dengue and West Nile Virus Protease Inhibitors with Submicromolar Activity in Reporter Gene DENV-2 Protease and Viral Replication Assays. J Med Chem, 2020. 63(15): p. 8179–8197. [DOI] [PubMed] [Google Scholar]
- 72.Drazic T, et al. , Peptide-beta-lactam Inhibitors of Dengue and West Nile Virus NS2B-NS3 Protease Display Two Distinct Binding Modes. J Med Chem, 2020. 63(1): p. 140–156. [DOI] [PubMed] [Google Scholar]
- 73.Luo D, et al. , Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J, 2008. 27(23): p. 3209–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tian H, et al. , Structural basis of Zika virus helicase in recognizing its substrates. Protein Cell, 2016. 7(8): p. 562–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yao Y, et al. , Discovery, X-ray Crystallography and Antiviral Activity of Allosteric Inhibitors of Flavivirus NS2B-NS3 Protease. J Am Chem Soc, 2019. 141(17): p. 6832–6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brecher M, et al. , A conformational switch high-throughput screening assay and allosteric inhibition of the flavivirus NS2B-NS3 protease. PLoS Pathog, 2017. 13(5): p. e1006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Millies B, et al. , Proline-Based Allosteric Inhibitors of Zika and Dengue Virus NS2B/NS3 Proteases. J Med Chem, 2019. 62(24): p. 11359–11382. [DOI] [PubMed] [Google Scholar]
- 78.Li Y, et al. , Structural Dynamics of Zika Virus NS2B-NS3 Protease Binding to Dipeptide Inhibitors. Structure, 2017. 25(8): p. 1242–1250 e3. [DOI] [PubMed] [Google Scholar]
- 79.Phoo WW, et al. , Structures of Zika virus NS2B-NS3 protease in complex with peptidomimetic inhibitors. Antiviral Res, 2018. 160: p. 17–24. [DOI] [PubMed] [Google Scholar]
- 80.Braun NJ, et al. , Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses. ChemMedChem, 2020. 15(15): p. 1439–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huber S, et al. , Structure-Based Optimization and Characterization of Macrocyclic Zika Virus NS2B-NS3 Protease Inhibitors. J Med Chem, 2022. 65(9): p. 6555–6572. [DOI] [PubMed] [Google Scholar]
- 82.Kuhl N, et al. , Beyond Basicity: Discovery of Nonbasic DENV-2 Protease Inhibitors with Potent Activity in Cell Culture. J Med Chem, 2021. 64(8): p. 4567–4587. [DOI] [PubMed] [Google Scholar]
- 83.Stern O, et al. , An N-terminal amphipathic helix in dengue virus nonstructural protein 4A mediates oligomerization and is essential for replication. J Virol, 2013. 87(7): p. 4080–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee CM, et al. , Determinants of Dengue Virus NS4A Protein Oligomerization. J Virol, 2015. 89(12): p. 6171–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roosendaal J, et al. , Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J Virol, 2006. 80(9): p. 4623–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miller S, et al. , The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J Biol Chem, 2007. 282(12): p. 8873–82. [DOI] [PubMed] [Google Scholar]
- 87.Cortese M, et al. , Determinants in Nonstructural Protein 4A of Dengue Virus Required for RNA Replication and Replication Organelle Biogenesis. J Virol, 2021. 95(21): p. e0131021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Riva L, et al. , The Compound SBI-0090799 Inhibits Zika Virus Infection by Blocking De Novo Formation of the Membranous Replication Compartment. J Virol, 2021. 95(22): p. e0099621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bhardwaj T, Kumar P, and Giri R, Investigating the conformational dynamics of Zika virus NS4B protein. Virology, 2022. 575: p. 20–35. [DOI] [PubMed] [Google Scholar]
- 90.Zou J, et al. , Dimerization of flavivirus NS4B protein. J Virol, 2014. 88(6): p. 3379–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zou J, et al. , Mapping the Interactions between the NS4B and NS3 proteins of dengue virus. J Virol, 2015. 89(7): p. 3471–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Umareddy I, et al. , Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. J Gen Virol, 2006. 87(Pt 9): p. 2605–2614. [DOI] [PubMed] [Google Scholar]
- 93.Zmurko J, Neyts J, and Dallmeier K, Flaviviral NS4b, chameleon and jack-in-the-box roles in viral replication and pathogenesis, and a molecular target for antiviral intervention. Rev Med Virol, 2015. 25(4): p. 205–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Naik NG and Wu HN, Mutation of Putative N-Glycosylation Sites on Dengue Virus NS4B Decreases RNA Replication. J Virol, 2015. 89(13): p. 6746–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van Cleef KW, et al. , Identification of a new dengue virus inhibitor that targets the viral NS4B protein and restricts genomic RNA replication. Antiviral Res, 2013. 99(2): p. 165–71. [DOI] [PubMed] [Google Scholar]
- 96.Xie X, et al. , Inhibition of dengue virus by targeting viral NS4B protein. J Virol, 2011. 85(21): p. 11183–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang QY, et al. , Discovery of Dengue Virus NS4B Inhibitors. J Virol, 2015. 89(16): p. 8233–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kaptein SJF, et al. , A pan-serotype dengue virus inhibitor targeting the NS3-NS4B interaction. Nature, 2021. 598(7881): p. 504–509. [DOI] [PubMed] [Google Scholar]
- 99.Moquin SA, et al. , NITD-688, a pan-serotype inhibitor of the dengue virus NS4B protein, shows favorable pharmacokinetics and efficacy in preclinical animal models. Sci Transl Med, 2021. 13(579). [DOI] [PubMed] [Google Scholar]
- 100.Guo F, et al. , A Novel Benzodiazepine Compound Inhibits Yellow Fever Virus Infection by Specifically Targeting NS4B Protein. J Virol, 2016. 90(23): p. 10774–10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gao Z, et al. , A yellow fever virus NS4B inhibitor not only suppresses viral replication, but also enhances the virus activation of RIG-I-like receptor-mediated innate immune response. PLoS Pathog, 2022. 18(1): p. e1010271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Potisopon S, et al. , The methyltransferase domain of dengue virus protein NS5 ensures efficient RNA synthesis initiation and elongation by the polymerase domain. Nucleic Acids Res, 2014. 42(18): p. 11642–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhao Y, et al. , Molecular basis for specific viral RNA recognition and 2′-O-ribose methylation by the dengue virus nonstructural protein 5 (NS5). Proceedings of the National Academy of Sciences, 2015. 112(48): p. 14834–14839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhao Y, et al. , A crystal structure of the Dengue virus NS5 protein reveals a novel inter-domain interface essential for protein flexibility and virus replication. PLoS Pathog, 2015. 11(3): p. e1004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Klema VJ, et al. , Dengue Virus Nonstructural Protein 5 (NS5) Assembles into a Dimer with a Unique Methyltransferase and Polymerase Interface. PLoS Pathog, 2016. 12(2): p. e1005451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.El Sahili A, et al. , NS5 from Dengue Virus Serotype 2 Can Adopt a Conformation Analogous to That of Its Zika Virus and Japanese Encephalitis Virus Homologues. J Virol, 2019. 94(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dubankova A and Boura E, Structure of the yellow fever NS5 protein reveals conserved drug targets shared among flaviviruses. Antiviral Res, 2019. 169: p. 104536. [DOI] [PubMed] [Google Scholar]
- 108.Lu G and Gong P, Crystal Structure of the full-length Japanese encephalitis virus NS5 reveals a conserved methyltransferase-polymerase interface. PLoS Pathog, 2013. 9(8): p. e1003549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhao B, et al. , Structure and function of the Zika virus full-length NS5 protein. Nat Commun, 2017. 8: p. 14762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Duan W, et al. , The crystal structure of Zika virus NS5 reveals conserved drug targets. EMBO J, 2017. 36(7): p. 919–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Upadhyay AK, et al. , Crystal structure of full-length Zika virus NS5 protein reveals a conformation similar to Japanese encephalitis virus NS5. Acta Crystallogr F Struct Biol Commun, 2017. 73(Pt 3): p. 116–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Flory C, et al. , Optimal flexibility of the linker region of Zika virus NS5 methyltransferase-polymerase is critical for virus replication. Antiviral Research, 2021. 195: p. 105194. [DOI] [PubMed] [Google Scholar]
- 113.Noble CG, et al. , A Conserved Pocket in the Dengue Virus Polymerase Identified through Fragment-based Screening. J Biol Chem, 2016. 291(16): p. 8541–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yokokawa F, et al. , Discovery of Potent Non-Nucleoside Inhibitors of Dengue Viral RNA-Dependent RNA Polymerase from a Fragment Hit Using Structure-Based Drug Design. J Med Chem, 2016. 59(8): p. 3935–52. [DOI] [PubMed] [Google Scholar]
- 115.Lim SP, et al. , Potent Allosteric Dengue Virus NS5 Polymerase Inhibitors: Mechanism of Action and Resistance Profiling. PLoS Pathog, 2016. 12(8): p. e1005737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gharbi-Ayachi A, et al. , Non-nucleoside Inhibitors of Zika Virus RNA-Dependent RNA Polymerase. J Virol, 2020. 94(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lim SP, et al. , Discovery of Potent Non-nucleoside Inhibitors of Dengue Viral RNA-Dependent RNA Polymerase from Fragment Screening and Structure-Guided Design. Adv Exp Med Biol, 2018. 1062: p. 187–198. [DOI] [PubMed] [Google Scholar]
- 118.Deng Y-Q, et al. , Adenosine Analog NITD008 Is a Potent Inhibitor of Zika Virus. Open Forum Infectious Diseases, 2016. 3(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yin Z, et al. , An adenosine nucleoside inhibitor of dengue virus. Proceedings of the National Academy of Sciences, 2009. 106(48): p. 20435–20439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Good SS, et al. , Evaluation of AT-752, a Double Prodrug of a Guanosine Nucleotide Analog with In Vitro and In Vivo Activity against Dengue and Other Flaviviruses. Antimicrob Agents Chemother, 2021. 65(11): p. e0098821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.van den Elsen K, Quek JP, and Luo D, Molecular Insights into the Flavivirus Replication Complex. Viruses, 2021. 13(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cerikan B, et al. , A Non-Replicative Role of the 3’ Terminal Sequence of the Dengue Virus Genome in Membranous Replication Organelle Formation. Cell Rep, 2020. 32(1): p. 107859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Paul D and Bartenschlager R, Flaviviridae Replication Organelles: Oh, What a Tangled Web We Weave. Annu Rev Virol, 2015. 2(1): p. 289–310. [DOI] [PubMed] [Google Scholar]
- 124.Yu L, Takeda K, and Markoff L, Protein-protein interactions among West Nile non-structural proteins and transmembrane complex formation in mammalian cells. Virology, 2013. 446(1–2): p. 365–77. [DOI] [PubMed] [Google Scholar]


