Summary
Upon infection, HIV disseminates throughout the human body within 1-2 weeks. However, its early cellular targets remain poorly characterized. We used a single-cell approach to retrieve the phenotype and TCR sequence of infected cells in blood and lymphoid tissue from individuals at the earliest stages of HIV infection. HIV initially targeted a few proliferating memory CD4+ T cells displaying high surface expression of CCR5. The phenotype of productively infected cells differed by Fiebig stage and between blood and lymph nodes. The TCR repertoire of productively infected cells was heavily biased, with preferential infection of previously expanded and disseminated clones, but composed almost exclusively of unique clonotypes, indicating that they were the product of independent infection events. Latent genetically intact proviruses were already archived early in infection. Hence, productive infection is initially established in a pool of phenotypically and clonotypically distinct T cells and latently infected cells are generated simultaneously.
Keywords: HIV, acute infection, lymph nodes, productive infection, memory CD4+ T cells, proliferative cells, HIV reservoir, clonal expansion, latency, inducibility
Graphical Abstract
eTOC blurb
The early cellular targets of HIV and the mechanisms of viral dissemination in humans remain elusive. Gantner et al. demonstrate that the early targets of HIV rapidly change during acute infection and differ between blood and lymph node. They also observe that latent and genetically intact proviruses persisting during therapy are archived from the earliest stages of acute infection.
Main Text:
Studying acute HIV is key to understand initial infection events and to identify the mechanisms leading to the establishment of the persistent viral reservoir. More than 40 years after its discovery 1, the early cellular targets of HIV within the first days/weeks of infection remain uncharacterized. Since accessing lymphoid tissues in people who recently acquired HIV is difficult, viral dissemination is hard to study in humans and most observations have been made in SIV-infected non-human primate models: Upon mucosal challenge, SIV rapidly establishes a small pool of productively infected cells that disseminate within days to lymphoid tissues and peripheral blood 2, following local replication of transmitted founders at mucosal sites 3,4. This early phase of SIV infection is characterized by a profound depletion of CD4+ T cells in blood and tissues 5, particularly in the intestinal tract 6. During chronic infection, the bulk of SIV replication is thought to occur in gut and lymph nodes 7. However, several parameters differ between non-human primate models and HIV-infected individuals (e.g. viruses, routes of infection, inoculum), hence the mechanisms of HIV dissemination in humans remain elusive.
In humans, we previously reported that maximal copy numbers of HIV DNA are reached in lymph nodes as early as Fiebig stages II/III 8. Although it is well established that T follicular helper (Tfh) cells are preferential targets for HIV in chronic infection 9, their contribution to the early dissemination of HIV is currently unknown. While productively infected cells usually do not survive for more than 2 days 10,11, they are rapidly replaced and maintain the rate of new infections which causes a rapid rise in plasma viremia during the first few weeks of infection 12. Little is known about the nature and the location of the cells in which HIV establishes productive infection in lymphoid tissues and blood during the initial phase of infection (i.e. before seroconversion, Fiebig stages I and II), and how these cells contribute to viral dissemination. Although duplications of HIV integration sites within the pool of HIV DNA-bearing cells are rarely detected during late stages of acute infection 13,14, it remains unclear whether antigen-driven expansions (particularly those resulting from HIV antigens) contribute to the initial dissemination of productively infected cells.
HIV reservoirs are established in the form of latently infected CD4+ T cells during acute infection 2,15 and persist for decades on ART through clonal expansion 16-19. The cellular and molecular mechanisms contributing to the establishment of HIV latency in vivo are still poorly elucidated. In vitro, activated CD4+ T cells transitioning to resting state in a relatively narrow time window are highly permissive for latent infection of R5-tropic HIV 20. An alternative and non-exclusive mechanism for the establishment of HIV latency is provided by the ability of specific chemokines to increase the permissiveness of resting CD4+ T cells to HIV infection up to the proviral integration step 21,22. The concept that latency is established during acute infection essentially stems from observational studies showing that viral rebound is observed upon treatment interruption even when ART is initiated during acute HIV infection 2,15,23 However, whether genetically intact genomes are already archived in the latent reservoir at this early stage has not yet been reported.
We used a combination of single-cell approaches in blood and lymph nodes from individuals at the earliest stages of HIV infection to investigate the location and mechanisms by which productive infection is established in acute infection. As we previously reported that most HIV genomes are not integrated during acute infection and cannot contibute to viral dissemination 8, we focused our analysis on cells producing the HIV capsid protein p24. In addition, we obtained near full-length HIV genomes from longitudinal samples of early treated individuals to characterize the fate, inducibility, and genetic intactness of proviruses archived early in infection.
Results
Productively infected cells are detected in blood and lymph nodes from the earliest stages of acute HIV infection
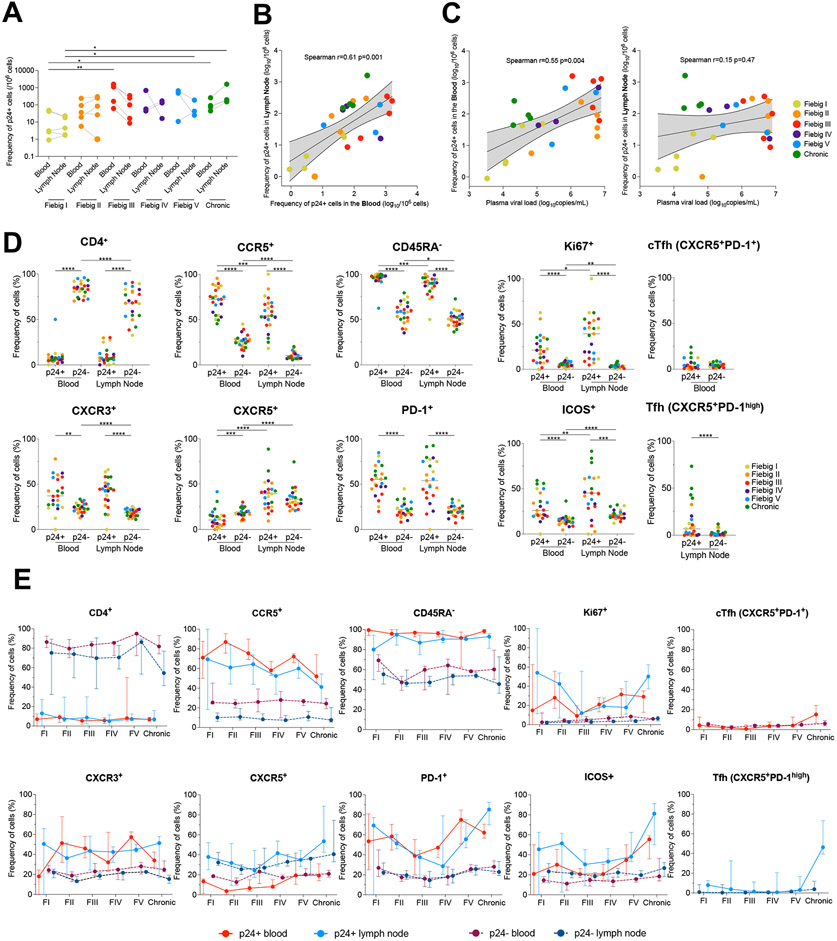
The RV254/SEARCH 010 cohort enrolls acutely infected individuals in Fiebig stages I-V who initiate ART within a median time of 2 days after diagnosis in Bangkok, Thailand 8,24. To quantify and characterize the initial pool of productively infected cells in blood and tissues, leukapheresis and inguinal lymph nodes biopsies were obtained at the time of diagnosis in acute infection prior to ART initiation in a subset of consenting participants. Paired PBMCs (peripheral blood mononuclear cells) and LNMCs (lymph node mononuclear cells) from 21 acutely infected participants of the RV254/SEARCH 010 study were obtained (Table S1). PBMCs and LNMCs from 4 chronically infected and ART-naïve Thai individuals enrolled in the RV304/SEARCH 013 study were also obtained. To measure the frequency of productively infected cells and assess their phenotype, we isolated CD4+ T cells by negative magnetic selection and used HIV-Flow 25, which captures cells expressing the p24 capsid protein (Fig. S1a). Productively infected cells were readily detected in blood and lymph node samples from all participants (Fig. 1a), including in all 5 individuals at Fiebig stage I (11 to 16 days post infection 26, median frequencies = 3.1 and 4.5 p24+ cells / million cells in blood and lymph nodes, respectively). Median frequencies of productively infected cells were higher during Fiebig stage II, maximal at Fiebig stage III (medians, 1,100 and 100 p24+ cells / million cells in blood and lymph nodes, respectively) and were similar afterwards, including during chronic infection. Frequencies of p24+ cells strongly correlated between blood and lymph nodes (Fig. 1b) and the frequency of p24+ measured in the blood, but not in the lymph nodes, correlated with plasma viremia (Fig. 1c). These results indicate that the frequency of productively infected cells rapidly increases during acute infection and that up to 1/1,000 circulating CD4+ T cells is productively infected when plasma viremia peaks (Fiebig stage III).
Fig. 1. The phenotype of productively infected cells differs between blood and lymph nodes and changes over time.
A. Frequencies of p24+ cells in CD4+ T cells measured by HIV-Flow in paired blood and lymph node samples from participants at different Fiebig stages of acute infection and in chronically infected controls. B. Correlation between p24+ cells frequencies in blood and lymph nodes. C. Correlations between p24+ cells frequencies in blood or lymph nodes with plasma viremia. D. Phenotype of productively infected cells. Frequencies of p24+ and p24− cells from blood and lymph node expressing each marker or combination of markers (CD4−, CCR5+, CD45RA−, Ki67+, CXCR3+, CXCR5+, PD-1+, ICOS+, circulating T follicular helpers (cTfh) cells and Tfh cells) are depicted for each participant (n=23). E. The frequency of p24+ (solid lines) and p24− (dotted lines) cells from blood (red) and lymph node (blue) expressing each marker or combination of markers (CD4+, CCR5+, CD45RA−, Ki67+, CXCR3+, CXCR5+, PD-1+, ICOS+, circulating T follicular helpers (cTfh) cells and Tfh cells) is depicted for each Fiebig stage (I to V and Chronic infection). Median values are plotted with 95% CI.
Several cellular markers are preferentially expressed by productively infected cells from the earliest stages of acute infection
To identify markers specifically expressed by productively infected cells, we analyzed the phenotypes of p24+ cells in blood and lymph nodes from all participants (Fig. S1b). As expected, most p24+ cells expressed low cell surface expression of the CD4 receptor when compared to their uninfected or latently infected counterparts (Fig. 1d), which likely resulted from its downregulation by Nef, Vpu and Env 27 Productively infected cells from both blood and lymph nodes frequently expressed CCR5 (the major HIV-co-receptor), displayed a memory phenotype (CD45RA−), and were enriched in subsets of activated (Ki67+, ICOS+, PD-1+) and Th1-like (CXCR3+) cells (Fig. 1d). Preferential expression of these markers by HIV-infected cells was maintained when memory CD4+ T cells rather than total CD4 T+ cells were used as comparators (Fig. S2a). The phenotypic signature of productively infected cells changed over time (Fig. 1e): whereas the contribution of lymph node T follicular helper cells (Tfh, CXCR5+PD-1high) to the initial pool of infected cells was modest at all stages of acute infection (<15%), Tfh cells were major HIV producers in lymph nodes from chronically infected participants 9 and encompassed 45% of all productively infected cells (Fig. 1d-e and Fig. S2b). The frequencies of p24+ cells expressing CCR5 tended to decrease in the blood over time, whereas proportions of productively infected cells expressing CXCR5, PD-1 and ICOS increased in both blood and lymph nodes, which resulted in increases in the fraction of p24+ cells displaying cTfh and Tfh cell phenotypes (Fig. 1e and Fig. S2b). We also measured the fold enrichment of specific markers at the surface of productively infected cells (Fig. S3a): When compared to their uninfected counterparts, the initial pool of infected cells in the lymph nodes from acutely infected participants (Fiebig I/II) were 27 times more likely to express Ki67 and 6 times more likely to express CCR5, whereas productively infected cells from participants at a more advanced stage of infection (Fiebig V/Chronic) were 11 times more likely to display a Tfh cell phenotype (Fig. S3b). Altogether, these observations indicate that during acute infection, the majority of productively infected cells are memory Th1-like T cells that frequently express activation markers and the HIV co-receptor CCR5 and do not display a cTfh or Tfh cell phenotype. Of note, most productively infected cells do not express CD4, which likely results from its downregulation from the cell surface by viral accessory proteins.
The phenotype of infected cells changes over time and varies between blood and lymph nodes
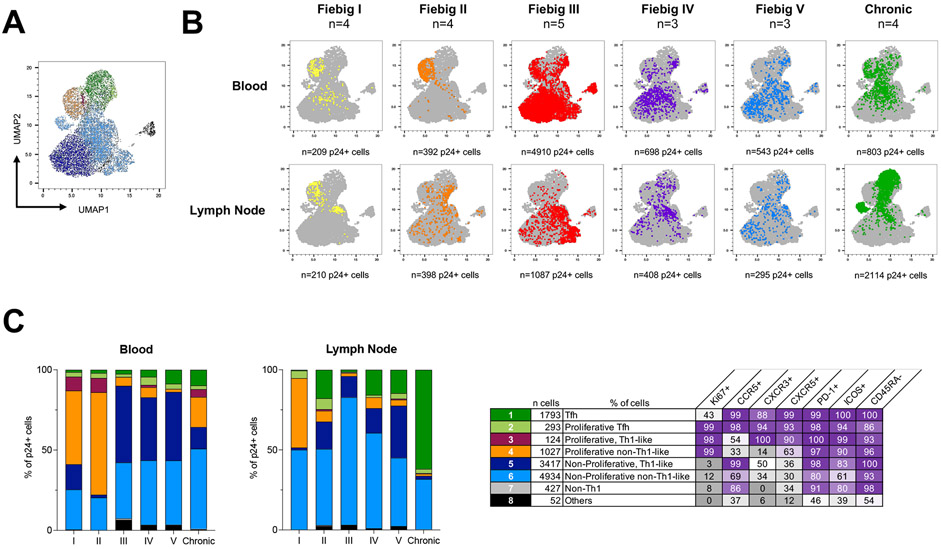
We next sought to compare the phenotype of p24+ cells between blood and lymph nodes at all stages of HIV infection. A UMAP analysis revealed that infected cells were distributed in 8 cell clusters (Fig. 2a and Fig. S4a-b). The clustering of p24+ cells was consistent across different participants from a defined Fiebig stage, although some interindividual variations at Fiebig stages IV and V were noted in both blood and lymph nodes (Fig. S5a-b, respectively). The relative contribution of each cluster to the overall pool of infected cells changed rapidly during acute infection and differed between blood and lymph nodes (Fig. 2b). Proliferating T-cells (Ki67+, clusters 2, 3 and 4) were the major contributors to the pool of productively infected cells in both blood and lymph node during early Fiebig stages (I and II). There was a rapid shift to non-proliferating cells (cluster 5 and 6) in both blood and lymph node, some of which expressed high frequencies of CCR5 (cluster 5), especially in blood (Fiebig III, IV, V; Fig. 2c). During chronic infection, Tfh (cluster 1) emerged as the major contributor to the pool of infected cells in the lymph node, representing an average of 62% of all productively infected cells, while the phenotypes of p24+ cells in the blood were more diverse. Altogether, these results indicate that the cell subsets that support productive HIV infection rapidly change throughout acute HIV infection, differ between blood and lymph nodes, and that in contrast to chronic infection, Tfh cells marginally contribute to the dissemination of HIV during acute infection.
Fig. 2. Temporal changes in the cell subsets contributing to viral production during HIV infection.
A. p24+ cells phenotypic data were integrated in a UMAP analysis and generated 8 cell clusters. B. Dot plots of the UMAP analysis of all p24+ cells (in grey) as shown in panel A, on which the p24+ cells by stage of infection and compartment are overlaid in colors. Numbers of samples analyzed are indicated at the top of the dot plots. C. The frequency of each of these clusters among p24+ cells is depicted in bar graphs according to the stage of infection for both blood and lymph node. Numbers of samples analyzed are as in B. The table shows the phenotype of each cell cluster. Statistically significant differences are highlighted (Wilcoxon; p<0.05, *; p<0.01**; p<0.001, ***; p<0.0001 ****).
Proviral diversity is limited in productively infected cells during acute infection
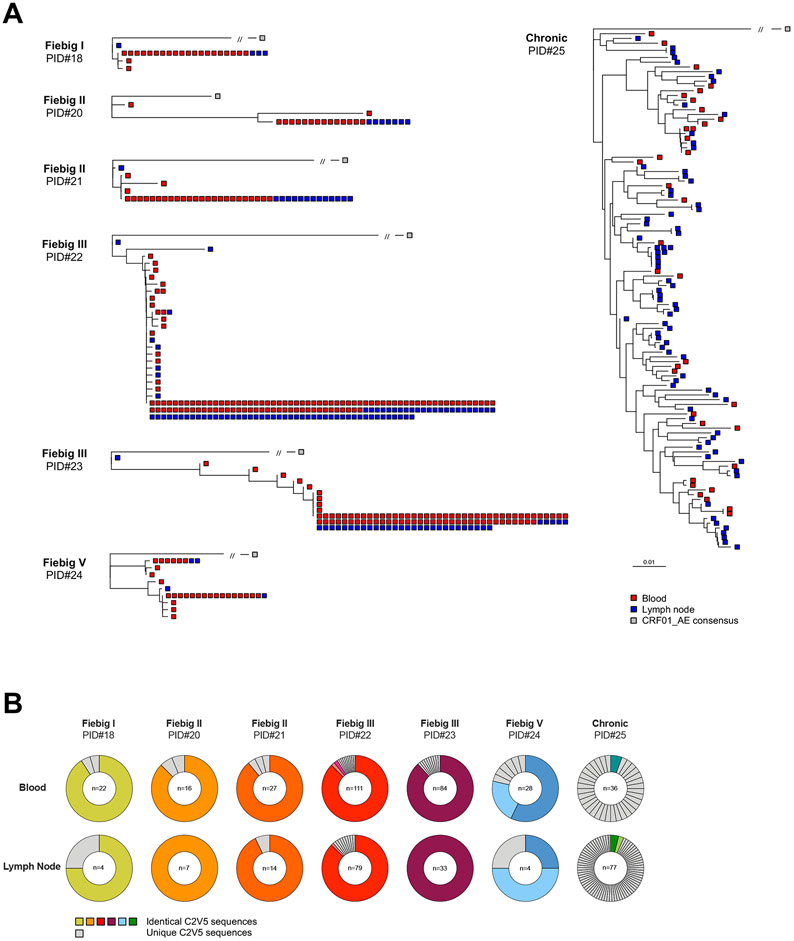
We then investigated if the diversity observed in the phenotype of productively infected cells was accompanied by genetic diversity in the proviral populations. We obtained env (C2-V5) sequences from 542 single HIV-infected cells from n=7 participants representing all stages of infection (Fig. 3a). As expected in recently infected participants, the majority of p24+ cells harbored a single C2-V5 env sequence (all CCR5-tropic) that was shared between blood and lymph nodes (mean, 87.4%, Fig. 3b). In sharp contrast, only a small proportion of p24+ cells isolated during chronic infection harbored identical proviral sequences (6.2%), reflecting the diversification of viral populations during untreated progressive HIV infection. Altogether, these results suggest that a small number of viral quasispecies initially infects a small pool of phenotypically diverse cells during acute HIV infection.
Fig. 3. Proviral diversity is low in productively infected cells during acute HIV infection.
A. Phylogenetic trees representing HIV C2-V5 env proviral sequences obtained from single-sorted p24+ cells from n=7 participants (PID#18 to #25). Trees were rooted on a CRF01_AE consensus (grey square) and env sequences from each single p24+ cell are depicted in red or blue according to their compartments (blood or lymph node, respectively). Identical sequences are shown as aggregated on the same branch of the tree. B. Pie charts representing the relative proportion of each C2V5 env sequence for each participant in both blood and lymph node. The number of p24+ cells analyzed is indicated in the center of the pie. Identical sequences are depicted in colors, whereas unique sequences are depicted in grey.
Productive HIV infection is established in clonotypically distinct T cells from the earliest stages of HIV infection
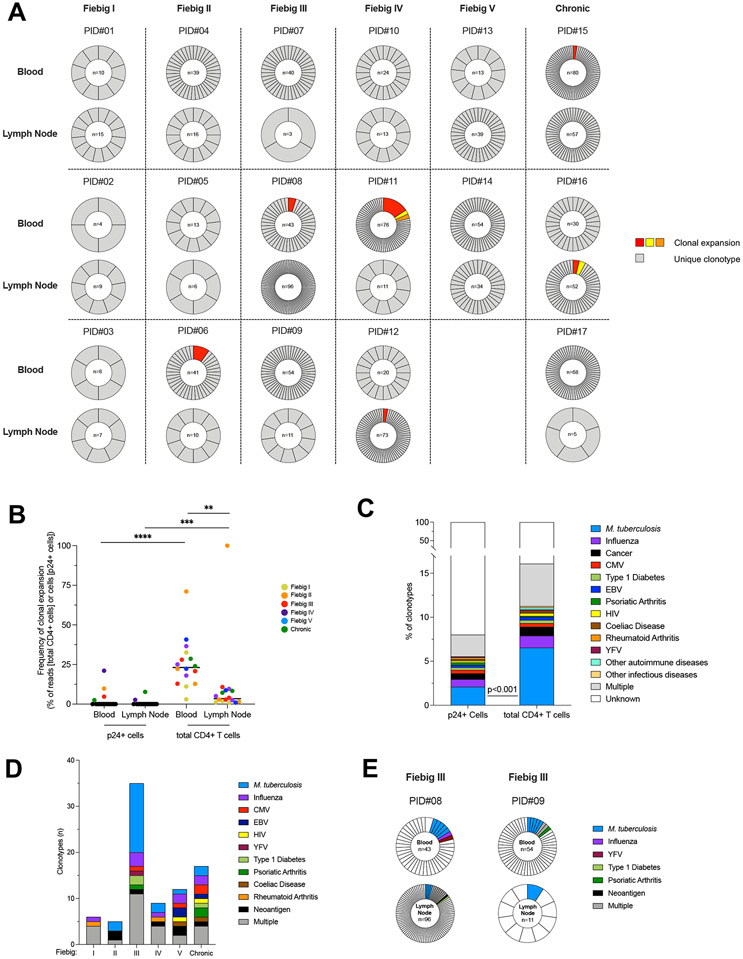
To determine if the limited proviral diversity observed during acute infection was attributed to clonal expansions of “founder” infected T cells or to independent infection events with identical HIV variants, we combined single-cell sorting of HIV-infected cells with sequencing of the V-J junction of the TCRβ chain, which can be used as a measure of T cell clonality 16. Clonotypes obtained from 1,048 single HIV-infected cells isolated from 17 participants were defined either as expanded (i.e. detected in at least two cells) or unique (i.e. detected in no more than one cell). In sharp contrast to the restricted viral diversity described above, unique TCRβ clonotypes were retrieved in the overwhelming majority of productively infected cells (median, >99%), at all stages of infection and both in blood and lymph nodes (Fig. 4a). Small clonal expansions of productively infected cells (encompassing 2 to 12 p24+ cells) were detected in 6/17 participants (PID# 6, 8, 11, 12, 15 and 16), with an overall mean proportion of clonally expanded p24+ cells of 2.2% in the blood (95% CI, 0-5.1) and 0.6% in the lymph node (95% CI, 0-1.6, Fig. 4b). Of note, shared p24+ clonotype between blood and lymph nodes were not observed in any participant. Altogether, these data indicate that multiple and independent infection events by homogeneous viral quasispecies, rather than clonal expansions of infected cells, are driving the early dissemination of HIV. In an attempt to predict if these highly diverse clonotypes could be specific to common antigens, we compared our sequences to a CDR3 database of TCR sequences with known specificities 16,28. Various antigen specificities were inferred for a small fraction (8%) of p24+ clonotypes (Fig. 4c), with no obvious temporal association (Fig. 4d). Interestingly, two participants at Fiebig stage III displayed p24+ clonotypes with different CDR3 sequences sharing a same M. tuberculosis antigenic specificity in both blood and lymph node (Fig. 4e), suggesting that M. tuberculosis-specific cells may contribute to HIV dissemination in these participants. However, M. tuberculosis-specific cells were underrepresented in the pool of p24+ cells when compared to total CD4+ T cells (Fig. 4c). Altogether, these data indicate that HIV-infection is established by a limited number of HIV variants infecting a large pool of clonotypically distinct T cells in both blood and lymph nodes and suggest that CD4+ T cells specific to common antigens or vaccines may constitute early targets for HIV during acute infection.
Fig. 4. Productive HIV infection is established in clonotypically distinct T cells since the earliest stages of HIV infection.
A. Frequencies of TCRβ clonotypes in p24+ cells are represented for n=17 participants (PID#01 to #17) and ordered according to the stage of acute infection (Fiebig I to V) followed by chronic controls (columns) and according to the compartment (i.e. blood and lymph node, rows). For each sample, the proportion of each clonotype in the pool of p24+ cells is represented in a pie chart. The number of p24+ cells analyzed is indicated in the center of the pie. Expanded clonotypes are depicted in colors; Unique clonotypes are depicted in grey. B. The proportion of clonal expansions in the pool of p24+ or total CD4+ T cells is depicted for each participant. Frequencies were calculated as follows: (1) for p24+ cells, the proportion of expanded cells within the pool of p24+ cells; and (2) for total CD4+ T cells, the number of reads of expanded cells within the total number of reads in CD4+ T cells. C. The frequency of predicted antigen specificities is represented in the pool of p24+ or total CD4+ T clonotypes recovered from 17 participants. The frequency of M. tuberculosis specific clonotypes was lower in p24+ cells than in total CD4+ T cells (Fisher’s exact Test; p<0.001). D. The number of predicted antigen specificities for p24+ clonotypes is represented according to the stage of infection (n=34 samples). E. Example of two participants who harbored distinct p24+ clonotypes with common antigenicity (M. tuberculosis). Significant differences are highlighted (Wilcoxon; p<0.05, *; p<0.01**; p<0.001, ***; p<0.0001 ****).
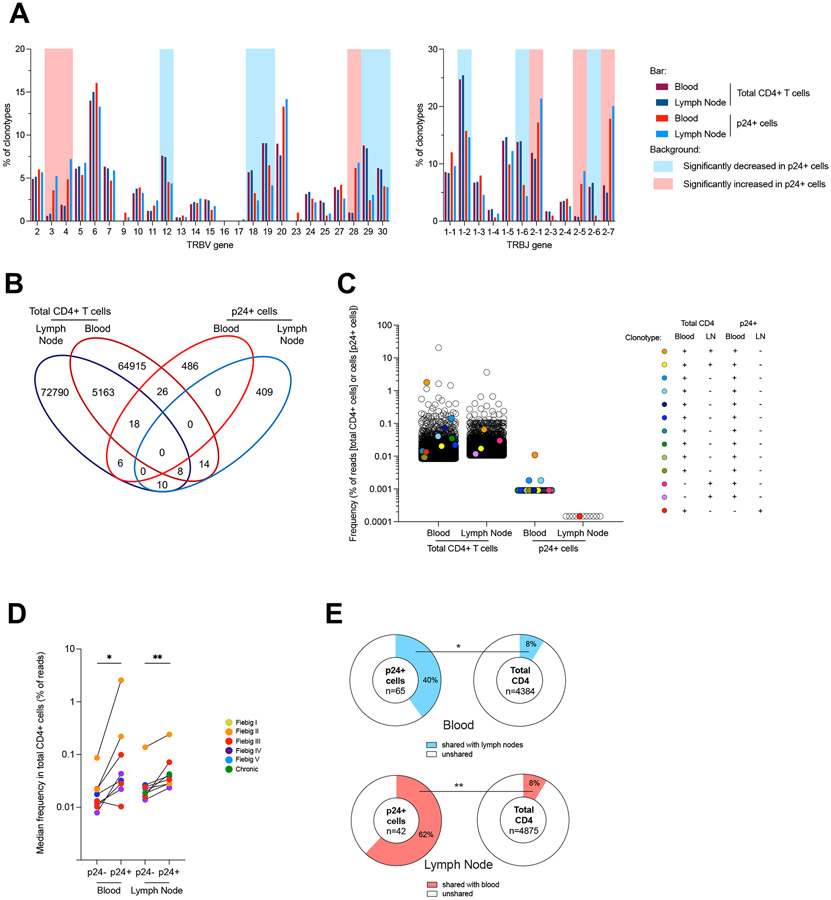
HIV preferentially infects cells expressing specific Vβ and Jβ motifs
To identify potential biases in the TCR repertoire of productively infected cells, we compared the TRBV and TRBJ usage in p24+ cells with the global CD4+ T cell repertoire from the same participants. To achieve this, we applied the same TCRβ chain PCR amplification approach to 100,000 blood and lymph node CD4+ T cells from 16 participants, followed by MiSeq sequencing and analyzed by MiXCR 29. There was no systematic bias in the total numbers of reads and clonotypes between blood and lymph node, allowing us to compare the repertoires between these two compartments (Fig. S6a). TRBV and TRBJ usage in total CD4+ T cells were highly conserved between participants and across compartments, whereas they were more variable in p24+ cells, probably due to lower number of cells analyzed (Fig. S6b). TRBV3, 4 and 28 families and TRBJ2-1, 2-5 and 2-7 were more frequently used by p24+ cells compared to total CD4+ T cells (Fig. 5a and Fig. S6c). Conversely, TRBV12, 18, 19, 29 and 30 families and TRBJ1-2, 1-6 and 2-6 were underrepresented in p24+ cells. Collectively, these results indicate that the TCR repertoire of productively infected cells is biased when compared to the repertoire of all CD4+ T cells (Fig. S6d).
Fig. 5. Productive HIV infection is preferentially established in previously expanded and disseminated clonotypes.
A. Frequency of TRBV and TRBJ segment usage for the clonotypes identified by TCRβ sequencing in p24+ cells and in total CD4+ cells in both blood and lymph nodes from 17 participants. Significant differences between p24+ and total CD4+ T cells are highlighted with a red or blue background depending on the trend (see Fig. S6c). B. Venn diagrams showing the number of unique and shared clonotypes in the four subsets: blood and lymph node total CD4+ T cells and p24+ cells for all participants C. Example (Participant PID#11, data from other participants are shown in Fig. S7d) of frequency distribution (based on bulk deep sequencing data and single-cell sorting/Sanger sequencing) of the clonotypes corresponding to the clones that were found in a single subset (empty circles) and in multiple subsets including p24+ cells (colored circles). Frequencies are shown as percentage of total reads for total CD4+ T cells and as the frequency of cells determined by HIV-Flow for p24+ cells. D. Median frequency of reads of CD4+ T cells clonotypes from blood and lymph node in which p24+ cells were identified (p24+) or not (p24−) (data from n=9 participants with at least n=3 p24+ clonotypes shared with blood or lymph nodes are shown). E. Pie charts representing the median frequency of p24+ cells and total CD4+ T cells clonotypes from a given compartment (blood or lymph node) that were shared with the other compartment (lymph node or blood), respectively (data from n=9 participants). The total number of cells (p24+ cells) and median number of reads (total CD4+ T cells) per condition is indicated at the center of the pie. Significant differences are highlighted (Wilcoxon or Fisher’s exact test; p<0.05, *; p<0.01**; p<0.001, ***; p<0.0001 ****).
Productive HIV infection is preferentially established in previously expanded and disseminated clonotypes
We then examined the distribution of the TCR repertoires and searched for shared clonotypes between p24+ and total CD4+ T cells. The distribution of the CD4+ T cell repertoire revealed that expanded clones were usually larger in the blood than in lymph nodes (Fig. S7a-b). Shared clonotypes were observed between compartments, accounting for approximately 8% of clonotypes in both compartments, and were also more expanded in the blood (median, 23% of reads) than in the lymph node (median, 11% of reads, p=0.0017, Fig. S7c). Among those expanded T cell clones shared between blood and lymph node, 82 clonotypes contained p24+ cells and were identified in 14/16 participants (Fig. 5b). Fig. 5c shows data from a representative example (participant #11): 13 clonotypes retrieved from p24+ cells were also detected in total CD4+ T cells (data from the other participants are presented in Fig. S7d). Interestingly, the T cell clones in which p24+ cells were detected were significantly larger than those in which p24+ were not retrieved (Fig. 5d), indicating that productive infection is preferentially established in unique or few CD4+ T cells belonging to relatively large T cell clones. In addition, the CD4+ T cells clonotypes in which p24+ cells were detected were more likely to be shared between blood and lymph nodes (Fig. 5e). Collectively, these observations indicate that previously expanded and disseminated CD4+ T cell clones are preferential targets for productive HIV infection.
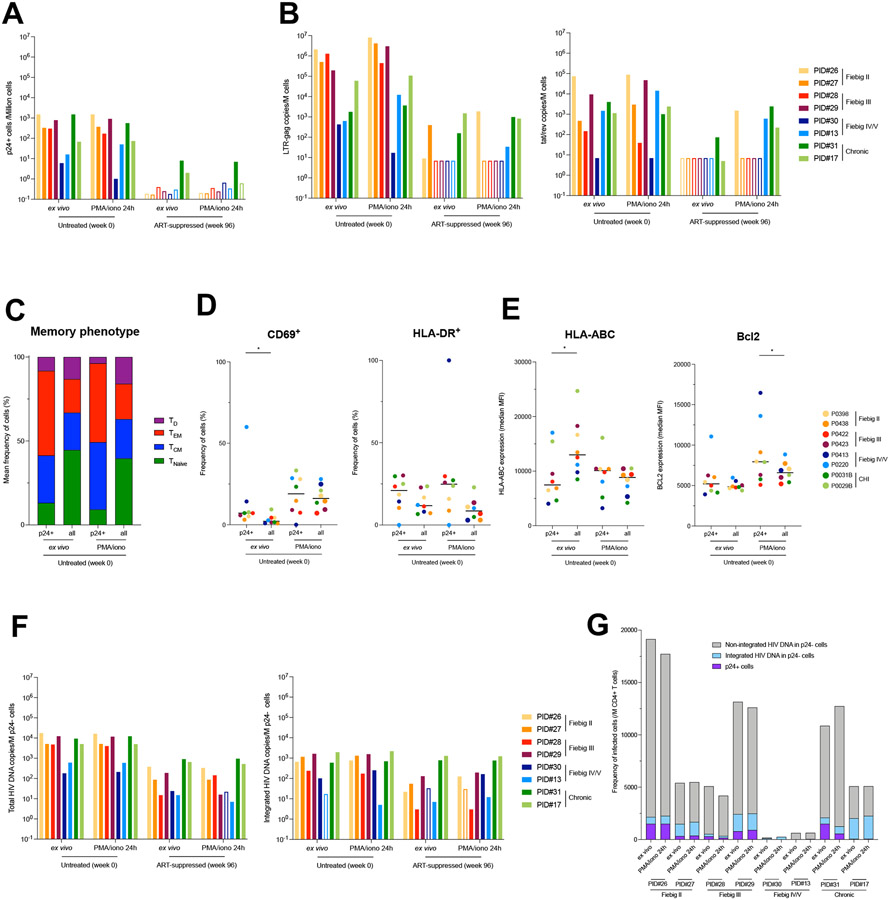
Intact and non-inducible proviruses are established during acute infection
Having demonstrated that productive HIV infection is rapidly established in phenotypically and clonotypically diverse CD4+ T cells, we sought to determine if latently infected cells could also be generated during the acute phase of infection. We used longitudinal blood samples from individuals initiating ART during acute (n=6) or chronic (n=2) infection collected before ART initiation and after 96 weeks of suppressive ART. CD4+ T cells isolated by negative magnetic selection were rested or stimulated with PMA/ionomycin to induce viral gene and protein expressions. In samples collected before ART, stimulation did not significantly increase the frequency of cells producing the viral protein p24 (Fig. 6a), nor the amount of cell-associated LTR-gag or tat/rev transcripts (Fig. 6b), indicating that latent and inducible proviruses were rare during untreated infection. To determine if productively infected cells display cellular features that may favor their long-term persistence during ART, we analyzed their phenotype (Fig. S1c). Most p24+ cells displayed a central memory (TCM, mean, 28.3%) or an effector memory (TEM, mean, 50.4%) phenotype (Fig. 6c) and were in a resting state as shown by low to moderate frequencies of expression of the activation markers CD69 and HLA-DR (median, 7.1% and 21.0%, respectively; Fig. 6d). In addition, productively infected cells displayed lower mean fluorescence intensity (MFI) of HLA class I and similar MFI of the pro-survival molecule Bcl-2 than their uninfected counterparts, suggesting potential escape/survival mechanisms (Fig. 6e). These differences were maintained when p24+ cells were compared to uninfected TCM and TEM cells (Fig. S7e). Upon stimulation, the memory phenotype of p24+ and p24− cells was maintained and both subsets upregulated CD69 to the same extent (Fig. 6d). HLA-DR expression frequencies were not significantly modified by the stimulation (Fig. 6d), whereas the lower MFI of HLA class I expression in p24+ cells compared to p24− cells observed ex vivo were not maintained upon stimulation (Fig. 6e). Interestingly, Bcl-2 MFI increased in both p24− and p24+ cells upon stimulation, but to a higher extent in p24+ cells (Fig. 6e), suggesting that infected cells are endowed with a pro-survival program. However, after 96 weeks of ART, none of the participants who initiated ART during acute infection displayed detectable translation-competent reservoir cells, even after stimulation (Fig. 6a), which was consistent with low to undetectable copy numbers of cell-associated viral transcripts in these samples (Fig. 6b).
Fig. 6. Limited inducibility of HIV genomes archived during acute infection and persisting on ART.
A. Frequency of p24+ cells measured by HIV-Flow in longitudinal blood samples of participants enrolled at different Fiebig stages (week 0) and after 96 weeks of ART. Frequencies were measured ex vivo to detect productively infected cells and after stimulation with PMA/ionomycin for 24h to detect the inducible reservoir. B. Total RNA from unstimulated or stimulated CD4+ T cells was used to quantify unspliced (LTR-gag) and multiply spliced RNA (tat/rev). HIV transcripts were normalized to the number of input cells. C. Proportion of each memory subset in p24+ cells and total CD4+ T cells at the viremic and on ART time points, both ex vivo or after stimulation with PMA/ionomycin (n = 16 samples). Infected cells were overrepresented in the TEM subset. D. Activation status of infected cells. The frequency of each subset (CD69+, HLA-DR+) is depicted ex vivo or after stimulation with PMA/ionomycin for each participant according to the cell population (p24+ and total CD4+ T cells). E. Phenotype of infected cells. The median fluorescence intensity (MFI) for each marker (HLA-ABC, Bcl2) is depicted ex vivo or after stimulation with PMA/ionomycin for each participant according to the cell population (p24+ and total CD4+ T cells). F. p24− cells were bulk sorted for total (LTR-gag) and integrated (alu/LTR-gag) HIV DNA quantification. Frequency of HIV DNA+ cells are presented in bar charts. Empty bars represent undetectable measures, and the limit of detection is plotted. G. Frequencies of CD4+ T cells harboring unintegrated HIV DNA, integrated HIV DNA and producing p24 proteins during untreated HIV infection. Integrated HIV DNA was measured by alu/LTR-gag PCR. Unintegrated HIV DNA was evaluated by subtracting the measure of integrated HIV DNA from the measure of total HIV DNA. Frequencies of p24+ cells were measured by HIV-Flow. All samples from participants from whom NFL HIV genome sequences were obtained are shown. For each sample, measures performed ex vivo and after 24h stimulation with PMA/ionomycin are represented.
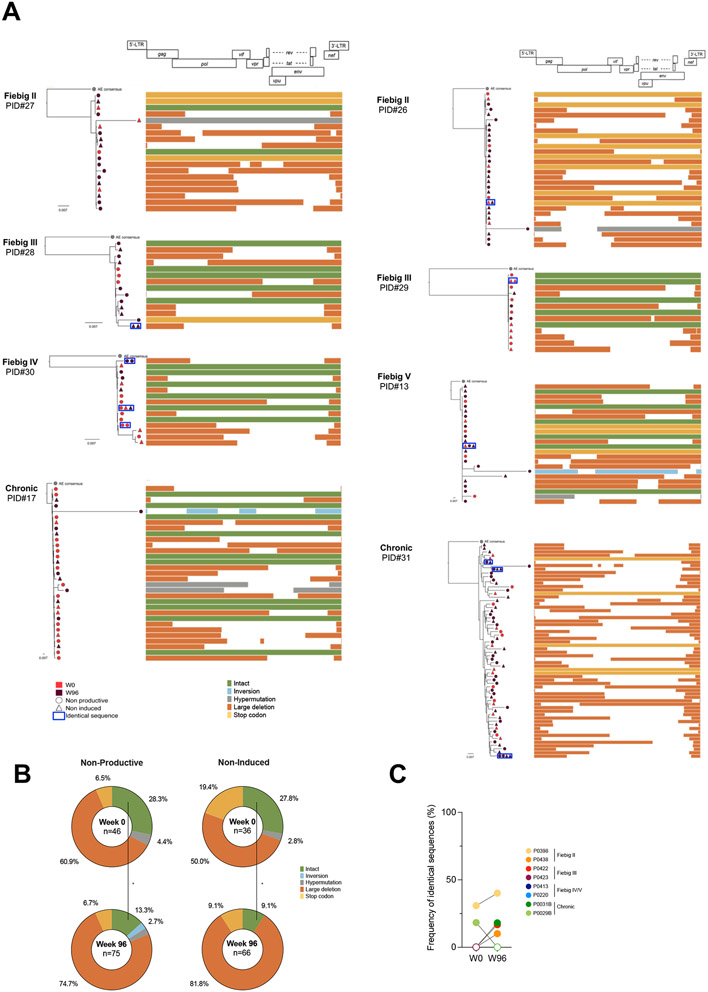
To determine if non-inducible proviruses could have been archived during acute infection, we bulk-sorted p24− cells at both time points and analyzed their proviral contents. Total and integrated HIV DNA were readily detected in almost all samples both during viremia and after 96 weeks of ART (Fig. 6f). Collectively, these data indicate that despite the fact that most cells infected during the first weeks of infection harbor unintegrated genomes that are rapidly cleared (Fig. 6g and 8), rare non-inducible proviruses are archived since the earliest stages of acute infection and can persist on ART. To assess the genetic integrity and persistence of these archived and non-induced proviruses, we performed near full-length genome amplification in limiting dilutions of p24− cells lysates followed by PacBio sequencing. A total of 223 proviral sequences were obtained from the ex vivo and stimulated conditions, representing the non-productive (no spontaneous production of p24) and non-induced (no induction of p24 production upon stimulation) HIV-reservoirs, respectively. Intact non-induced proviruses were observed in 6/8 participants (PID #13, 17, 27-30; Fig. 7a) whereas only defective proviruses were detected in the last 2/8 (PID #26 and 31; Fig. 7a). During untreated infection, a third of the viral genomes retrieved from both non-induced and non-productively infected cells were genetically intact (Fig. 7b), indicating that deeply latent intact proviruses are archived since the earliest stage of infection. Although the proportion of non-induced intact genomes diminished after 2 years of ART (from 27.8% to 9.1%), they were still detected in most participants, demonstrating that early ART does not prevent the establishment of latent and genetically intact HIV genomes. Notwithstanding the limitation associated with the relatively small number of near full-length genomes analyzed, identical sequences were rare both before ART initiation and after 2 years of suppressive therapy (median, 0 and 5%, respectively; Fig. 7c), suggesting that clonal expansions minimally contribute to the early dissemination and persistence of latent HIV genomes (p24− cells), similar to what we observed using TCR sequencing (Fig. 4a). Nonetheless, identical and genetically intact non-induced proviruses were found both in the pre- and post-ART samples from two participants (Participants #30 and #13, who initiated ART during Fiebig stages IV and V, respectively; Fig. 7a), indicating that genetically intact genomes that are refractory to viral reactivation are established early in infection and can persist during ART.
Fig. 7. Intact and non-inducible proviruses are established during acute infection and persist on ART.
A. Near-full length genome amplification was performed in p24− cells to assess the intactness of latent genomes. To assess the inducibility of these proviruses, p24− cells from both the ex vivo and stimulation conditions were included (n=8 participants). Phylogenetic trees representing the proviral landscape of five participants are depicted. Sequences obtained before (week 0, red) and during suppressive ART time point (week 96, burgundy) are included; Sequences obtained from p24− cells sorted directly ex vivo (circle) or after stimulation (triangle), representing the non-productive and the non-induced latent HIV reservoirs, respectively, are also included. Identical sequences are framed and represented on the same tree branch. On the right of the graph, proviral sequences are mapped to the HIV genome, and color-coded as follows: intact sequences in green, inversions in blue, hypermutations in grey, large deletions in orange and stop codons in yellow. B. Pie charts summarizing the frequency of defects and intact proviruses obtained from p24− cells sorted ex vivo (non-productive) and after stimulation (non-induced) before (week 0, n=8) and after ART initiation (week 96, n =8). The total number of sequences per condition is indicated at the center of the pie (sequence color categories are as in A.). C. The frequency of identical proviral sequences before and after ART in p24- cells is represented for each participant. Significant differences are highlighted (Wilcoxon or Fisher’s exact test; p<0.05, *; p<0.01**; p<0.001, ***; p<0.0001 ****).
Discussion
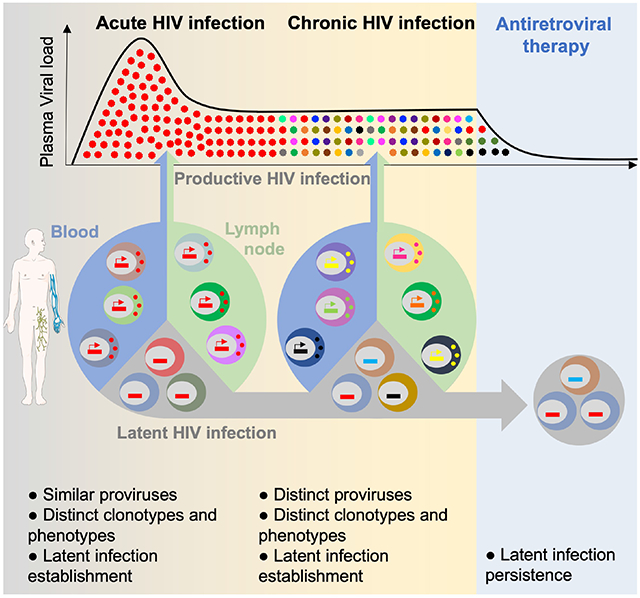
Using a combination of single-cell approaches, we extensively characterized the landscape of productively infected cells at the earliest stages of HIV infection. We observed that the frequency of these cells rapidly rises concomitant with plasma viremia and that their phenotype changes throughout the different stages of acute infection. Using TCR and near-full length HIV genome sequencing, we demonstrate that multiple independent infection events both in blood and lymph nodes drive the production of a relatively homogeneous viral population during acute infection and that a latent pool of cells harboring intact HIV genomes is established early in infection and can persist during ART.
Using HIV-Flow, we measured the frequencies of productively infected cells in blood and lymph node during acute infection and observed that these frequencies rise from less than 10 to up to 1,000 per million CD4+ T cells (i.e. 100-fold) in a short interval (less than a week during the transition from Fiebig stage I to III26,30), demonstrating the extremely fast dissemination of HIV and reflecting the kinetics of plasma viral load 26. This is in line with the results from a previous study conducted in individuals infected for a median time of 4 months in whom around 2,000 RNA+ cells/per million CD4+ T cells were quantified in lymph node 31. In our study, frequencies of p24+ cells in blood and lymph node were relatively similar, suggesting a possible recirculation of productively infected cells between these compartments 32, which is supported by the shared env sequences between the two sites.
When we analyzed the phenotype of productively infected cells, we observed that the vast majority of p24+ cells displayed a memory phenotype. Low cell surface expression frequencies of CD4 and MHC-I molecules suggest that nef was functional in most productively infected cells. The observation that not all p24+ cells expressed CCR5 suggests that this chemokine receptor may also be downregulated from the cell surface as previously proposed 33 or that HIV may use alternate coreceptors during acute infection 34. Unexpectedly, we observed that only 20-40% of p24+ cells expressed the proliferation marker Ki67, suggesting that HIV can productively infect cells that have not entered the cell cycle, as reported previously in several studies performed in vitro 35,36. The contribution of Tfh cells to the pool of productively infected cells was minimal throughout all stages of acute infection. While we cannot exclude that these cells were actually infected and immediately depleted, our observations argue against this model: We observed extremely low frequencies of Tfh in lymph nodes in acute infection, and their infection was concomitant with their development during chronic infection. Hence, our results are consistent with a model in which Tfh cells are scarce during acute infection, expand during early chronic infection when they become major targets for HIV replication. Therefore, our results suggest that Tfh cells have a limited role when plasma viremia peaks but are major contributors to the set point viremia. This is also supported by the delayed formation of germinal centers in SIV-infected macaques 37 and in HIV-infected individuals 38.
While we cannot exclude that changes in the phenotype of p24+ cells over time could be attributed to the modulation of the expression of cellular markers by HIV proteins, our data are consistent with a model in which HIV initially infects a small pool of proliferating cells expressing high surface expression of CCR5 in lymph nodes and then disseminates to other, possibly less permissive cell subsets. This is unlikely to be attributed to changes in the tropism of the viral populations since envelope sequences remain largely conserved throughout all stages of acute infection.
An unexpected finding was the fact that the overwhelming majority of productively infected cells expressed distinct TCRs during acute infection. This observation excludes two models: One in which an infected cell proliferates to disseminate HIV and one in which a clone proliferating actively at the time of infection supports viral dissemination. Instead, HIV targets individual cells belonging to distinct clonotypes that can be specific for several pathogens including HIV 39, M. tuberculosis 40 or influenzae 41. Since the repertoire of p24+ cells was heavily biased, it is likely that cells reactive to specific antigens and expressing specific TRBV and/or TRBJ regions were more susceptible or, on the contrary, more resistant to infection. For instance, in individuals from our cohort who had either been vaccinated or who have latent tuberculosis, we found that M. tuberculosis-specific cells were less frequently infected than cells from other specificities. This relative resistance to HIV infection may be attributed to high amounts of MIP1-β production by M. tuberculosis-specific cells in this particular context 39,40,42, as previously reported for CMV specific cells 43.
The fact that HIV preferentially infects cells belonging to large clones 44 that recirculate between blood and tissues is consistent with the preferential infection of effector memory CD4+ T cells during acute infection, as we previously reported 8,45. Indeed, when compared to less differentiated central memory cells, effector memory cells are known to display restricted TCR repertoire as a result of their expansions 46,47, to contain clones shared between anatomical sites 48, and to express higher surface expression of CCR5.
An outstanding question in the field of HIV reservoirs is how viral latency is established. While some studies have suggested that HIV latency can be directly established in resting CD4+ T cells (pre-activation latency) 49,50, others have proposed that latency is primarily established when HIV infects cells that transition from an activated phenotype to a resting memory state 20. Although it is likely that both models coexist, our observations are consistent with a model in which latency can be directly established in a minority of the CD4+ T cells initially targeted by the virus: we found translationally inactive and genetically intact proviruses even at the earliest stage of infection (Fiebig II; before peak viremia). The vast majority of these latently infected cells resisted reactivation in vitro, suggesting that “deep latency” was already established at this early stage. Within the pool of cells harboring non-inducible proviruses, those with intact proviruses decreased after 2 years of ART, indicating that they were preferentially cleared 51. This could suggest that a fraction of these intact proviruses underwent reactivation and were eliminated. Alternatively, intact non-induced genomes may have integrated into the chromatin of cells that are relatively short-lived compared to those in which defective proviruses were archived 52. As we observed in p24+ cells, duplication of HIV genomes was also rare in latently infected cells. We conclude that clonal expansion has a minimal contribution to the establishment of both the latently and productively infected pools of cells.
This study provides insight into the nature of the cells and the initial events that contribute to the dissemination of HIV in humans. We also report on the simultaneous establishment of two pools of productively and hard-to-reactivate latently infected cells. Therefore, our findings suggest that unlike pre- or post-exposure strategies which block early replication events, early ART will not prevent the establishment of CD4+ T cells harboring genetically intact and deeply latent proviruses.
Limitations of the study
Our study has several limitations. First we did not analyze the early dissemination of HIV in the gut, which is thought to be very important for the establishment of productive infection and persistence on ART 7,53. In addition, even after stimulation in vitro, we did not detect p24+ cells in samples from these early treated individuals after 2 years of ART: This precluded the study of the reactivatable reservoirs on ART. Although our data suggest that most of the latently infected cells persisting in these early treated individuals carry proviruses that are difficult to reactivate, we previously observed that 8/8 Fiebig I participants experienced viral rebound upon treatment cessation 15. Our in vitro stimulation assay may not be sensitive enough to recapitulate this rebound. Larger amounts of blood cells or access to tissue cells may be required to mimic viral reactivation. Alternatively, our in vitro stimulation may not have recapitulated the complex mechanisms that are causing viral resurgence in vivo and it is possible that proviral genomes of the CRF01_AE clade may require different stimuli to be optimally reactivated.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr Nicolas Chomont (nicolas.chomont@umontreal.ca).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files). Source data are provided with this paper within Supplementary Files. TCR sequences from CD4+ T cells and p24+ cells are provided as an Auxiliary Supplementary file. HIV env and HIV near-full length sequences have been deposited in GenBank and are publicly available from the date of publication. Accession numbers are listed in the key resources table. External databases used in this study are available online: IMGT® database (IMGT®, the international ImMunoGeneTics Information system®) [http://www.imgt.org]); McPAS-TCR database ([http://friedmanlab.weizmann.ac.il/McPAS-TCR/]).
All codes used were downloaded from Github and are publicly available.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| p24 KC57-PE | Beckman Coulter | Cat#6604667 |
| p24 28B7-APC | MediMabs | Cat#MM-0289-APC |
| CD4-APC-H7 (Clone: RPA-T4) | BD Bioscience | Cat#560168 |
| CD45RA-A700 (Clone: HI100) | BD Bioscience | Cat#560673 |
| CXCR5-BB515 (Clone: RF8B2) | BD Bioscience | Cat#564624 |
| CXCR3-PerCP-Cy5.5 (Clone: 1C6/CXCR3) | BD Bioscience | Cat#560832 |
| Ki67-PE-Cy7 (Clone: B56) | BD Bioscience | Cat#561283 |
| CCR5-BV421 (Clone: 2D7/CCR5) | BD Bioscience | Cat#562576 |
| CD45RA-BV786 (Clone: HI100) | BD Bioscience | Cat#563870 |
| HLA-DR-BV605 (Clone: G46-6) | BD Bioscience | Cat#562845 |
| CCR7-BB515 (Clone: 3D12) | BD Bioscience | Cat#565870 |
| PD-1-BV605 (Clone: EH12.2H7) | BioLegend | Cat#329923 |
| ICOS-BV785 (Clone: C398.4a) | BioLegend | Cat#313534 |
| HLA-ABC-A700 (Clone: W6/32) | BioLegend | Cat#311437 |
| CD69-PerCP-Cy5.5 (Clone: FN50) | BioLegend | Cat#310926 |
| Bcl-2-BV421 (Clone: 100) | BioLegend | Cat#658709 |
| Live/Dead Aqua Cell Stain (405nm) | ThermoFisher Scientific | Cat#L34957 |
| Biological samples | ||
| Human serum | Atlanta Biologicals | Cat#540110 |
| Demographics of study participants, see Table S1 | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Lamivudine (3TC) | NIH HIV Reagent Program | ARP-8146 |
| Raltegravir (RAL) | NIH HIV Reagent Program | HRP-11680 |
| Brefeldin A (BFA) | Sigma | Cat#B2651 |
| ionomycin | Sigma | Cat#I9657 |
| PMA | Sigma | Cat#P8139 |
| DirectPCR Lysis Reagent | Viagen Biotech | Cat#301-C |
| Proteinase K | Wisent | Cat#25530–015 |
| Proteinase K | Invitrogen | Cat#25530-015, |
| Critical commercial assays | ||
| EasySep Human CD4+ T Cell Enrichment Kit | StemCell Technology | Cat#19052 |
| FoxP3 Transcription Factor Staining Buffer Set | eBioscience | Cat#00-5523-00 |
| Phusion Master Mix | ThermoFisher Scientific | Cat#F531S |
| Taq DNA Polymerase kit | Invitrogen | Cat#18038-042, |
| Qiagen Multiplex PCR kit | Qiagen | Cat#206143 |
| QIAquick 96 PCR Purification kit | Qiagen | Cat#28181 |
| Platinum SuperFi II Master Mix | Life Tech | Cat#12368010 |
| AMPure XP | Beckman Coulter | Cat#A63881 |
| Deposited data | ||
| HIV env | GenBank | Genbank accession numbers ON500682 to ON500846 |
| HIV near full length sequences | GenBank | Genbank accession numbers ON500847 to ON501069 |
| TCRβ sequences | This paper | N/A |
| Oligonucleotides | ||
| TCRβ HIV env and HIV near full length amplification and sequencing primers, see Table S2 | This paper | N/A |
| Software and algorithms | ||
| MiXCR version 3.0.13 | Bolotin et al.29 | https://github.com/milaboratory/mixcr |
| IMGT® database | IMGT®, the international ImMunoGeneTics information system® | http://www.imgt.org |
| McPAS-TCR database | Friedman lab | http://friedmanlab.weizmann.ac.il/McPAS-TCR/ |
| Prism version 9.1.0 | Graphpad | https://www.graphpad.com |
| FlowJo version 10.7.1 | FlowJo | https://www.flowjo.com/solutions/flowjo |
| iqtree2, version 2.1.2 | IQTREE | http://www.iqtree.org |
| FigTree version 1.4.4 | Figtree | http://tree.bio.ed.ac.uk/software/figtree/ |
| HIVDatabase QCTool | HIV Los Alamos | https://www.hiv.lanl.gov/content/sequence/QC/index.html |
| ProseqIT | NIH | https://psd.cancer.gov/tools/nvs_annot.php |
| GeneCutter tool | HIV Los Alamos | https://www.hiv.lanl.gov/content/sequence/GENE_CUTTER/cutter.html |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Participants and sample collection
The RV254/SEARCH 010 cohort study (clinicaltrials.gov NCT00796146) enrolls participants at the earliest stages of acute HIV infection at the Thai Red Cross AIDS Research Centre in Bangkok. High-risk volunteers are screened for acute HIV infection (AHI) in real time with pooled nucleic acid testing and sequential immunoassay (IA). Individuals with AHI were enrolled if they had a positive HIV RNA with or without a reactive IA. Participants were then categorized as Fiebig stages I to V as follows: Fiebig I - positive HIV RNA, negative p24 antigen, non-reactive 3rd generation IA; Fiebig II – positive HIV RNA, positive p24 antigen, non-reactive 3rd generation IA; Fiebig III - positive HIV RNA, positive p24 antigen, reactive 3rd generation IA, negative western blot; Fiebig IV - positive HIV RNA, positive or negative p24 antigen, reactive 3rd generation IA, indeterminate western blot; Fiebig V - positive HIV RNA, positive or negative p24 antigen, reactive 3rd generation IA, positive western blot except p3126. ART was voluntary and offered to all participants and was initiated at a median (IQR) of 2 (0-5) days after enrollment 8 under a separate protocol (NCT00796263). Samples from chronically infected naïve individuals enrolled in the RV304/SEARCH 013 8 (NCT01397669) studies were also obtained.
For the present study, paired PBMCs and LNMCs samples from 25 untreated individuals were analyzed. Eight participants also provided before and during suppressive ART (96 weeks) PBMCs samples. Both PBMCs and LNMCs were isolated by Ficoll density gradient centrifugation and were cryopreserved in liquid nitrogen. Participants’ characteristics are presented in Table S1.
Ethics statement
All clinical studies were approved by the Institutional Review Boards (IRBs) of Chulalongkorn University in Thailand, Walter Reed Army Institute of Research (WRAIR) and the Centre Hospitalier de l’Université de Montréal in Canada. All participants gave written informed consent.
METHOD DETAILS
Cell culture
The HIV-Flow assay was used to quantify and analyze the phenotype of cells expressing p24 protein 25. Briefly, CD4+ T cells were isolated by negative magnetic selection using the EasySep Human CD4+ T Cell Enrichment Kit (StemCell Technology, Cat#19052). Purity was typically >98%. 5-15x106 CD4+ T cells were resuspended at 2x106 cells/mL in RPMI + 10% Fetal Bovine Serum and antiretroviral drugs were added to the culture medium (200nM raltegravir, 200nM lamivudine). Cells were then rested (18h) or stimulated, depending on the experiment. Stimulation included a pre-incubation of 1h with 5μg/mL Brefeldin A (BFA, Sigma, Cat#B2651) before stimulation in order to prevent the upregulation of cell surface markers, and BFA was maintained in the culture until the end of the stimulation. Cells were then stimulated with 1μg/mL ionomycin (Sigma, Cat#I9657) and 162nM PMA (24h) (Sigma, Cat#P8139).
Flow cytomety
After resting or stimulation, cells were collected, resuspended in PBS and stained with the Aqua Live/Dead staining kit for 20min at 4°C. Cells were then stained with antibodies against extracellular molecules in PBS + 4% human serum (Atlanta Biologicals, Cat#540110) for 20min at 4°C. After a 45min fixation/permeabilization step was performed with the FoxP3 Transcription Factor Staining Buffer Set (eBioscience, Cat#00-5523-00) following the manufacturer’s instructions, cells were then stained with anti-p24 KC57 and anti-p24 28B7 antibodies for an additional 45min at room temperature in the FoxP3 Buffer. Cells were then washed and resuspended in PBS for subsequent cell sorting.
p24 KC57-PE was purchased from Beckman Coulter (Cat#6604667, Dilution 1/1000) and p24 28B7-APC was purchased from MediMabs (Cat#MM-0289-APC, Dilution 1/1000). CD4-APC-H7 (Clone: RPA-T4, Cat#560168, Dilution 1/100), CD45RA-A700 (Clone: HI100, Cat#560673, Dilution 1/50), CXCR5-BB515 (Clone: RF8B2, Cat#564624, Dilution 1/50), CXCR3-PerCP-Cy5.5 (Clone: 1C6/CXCR3, Cat#560832, Dilution 1/25), Ki67-PE-Cy7 (Clone: B56, Cat#561283, Dilution 1/50), CCR5-BV421 (Clone: 2D7/CCR5, Cat#562576, Dilution 1/25), CD45RA-BV786 (Clone: HI100, Cat#563870, Dilution 1/50), HLA-DR-BV605 (Clone: G46-6, Cat#562845, Dilution 1/50), CCR7-BB515 (Clone: 3D12, Cat#565870, Dilution 1/25), were purchased from BD Bioscience. PD-1-BV605 (Clone: EH12.2H7, Cat#329923, Dilution 1/25), ICOS-BV785 (Clone: C398.4A, Cat#313534, Dilution 1/25), HLA-ABC-A700 (Clone: W6/32, Cat#311437, Dilution 1/100), CD69-PerCP-Cy5.5 (Clone: FN50, Cat#310926, Dilution 1/50), Bcl-2-BV421 (Clone: 100, Cat#658709, Dilution 1/50), were purchased from BioLegend. Live/Dead Aqua Cell Stain (405nm) was purchased from ThermoFisher Scientific (Cat#L34957). Flow cytometry data of p24+ cells were analyzed using FlowJo version 10.7.1. To gather an overview of the immune populations present in the pool of p24+ cells from our samples, flow cytometry data were visualized using uniform manifold approximation and projection (UMAP)-dimensionality reduction on the total concatenated pool of p24+ cells from each Fiebig stage and chronically-infected participants 54. The total number of cells depicted per UMAP visualization could not be normalized per Fiebig group. p24+ cells were then further grouped using FlowSOM, a self-organizing map that clusters cells depending on their overall similarities in marker expression 55.
Flow cytometry cell sorting
The frequency of p24 double positive cells (KC57+, 28B7+) was determined by flow cytometry in gated viable T cells. Examples of gating strategies are represented in Fig. S1 and S9. In all experiments, CD4+ T cells from an HIV-uninfected control were included to set the threshold of positivity. Single p24 double positive (p24+ cells) were indexed-sorted and p24 double negative (p24− cells) were bulk sorted on a BD FACS ARIA III. Single-cells were sorted in 96-wells PCR plates containing 7.6μL of DirectPCR Lysis Reagent (Viagen Biotech, Cat#301-C) and 0.4 μL of 10mg/mL proteinase K (Wisent, Cat#25530–015). The PCR plates were subsequently incubated at 55°C for 1 hour for cell lysis followed by 10 min at 95°C to inactivate proteinase K. Bulk-sorted and pelleted cells were digested in 30μL of Proteinase K (Cat#25530-015, Invitrogen; final concentration 400 μg/ml, TriS HCl (10mM) and KCl (50nM) overnight at 55°C followed by 10 min at 95°C to inactivate proteinase K. Cell lysates were then used for TCR sequencing or near-full length HIV genome sequencing.
Env C2-V5 amplification on proviral DNA
To amplify the Env C2-V5 region, we developed a semi-nested PCR method that amplifies a 600bp fragment of the env gene. Sequences of primers are listed in Table S2. The first PCR reaction was performed using the Phusion Master Mix (Cat#F531S, Thermo Fisher Scientific), in a total volume of 50 μL: 25 μL of Phusion master mix, 2.5 μL of Env7 primer and 2.5 of μL OutV5R_AE primer (each at a concentration of 10 μM, providing a final concentration of 500 nM per primer), 1.5 μL of DMSO, and 10 μL of the single-cell lysate. First PCR conditions were as follows: 2 min at 98°C followed by 40 cycles of 30 s at 98°C, 30 s at 60°C and 30 s at 72°C with a final elongation for 5 min at 72°C. A semi-nested PCR reaction was performed using the Taq DNA Polymerase kit (Cat#18038-042, Invitrogen), in a total volume of 50 μL: 5 μL of 10X PCR buffer, 3 μL of MgCl2 (50 mM), 1.5 μL of dNTPs (10 mM), 2 μL of Env7 primer and 2 μL of InV5R_AE primer (each at a concentration of 10 μM, providing a final concentration of 400 nM per primer), 0.5 μl Taq DNA Polymerase (5 U/ μL), 26 μL H2O and 10 μL of the first PCR products diluted 1:10. The amplification conditions for the second PCR reaction were as follows: 3 min at 94°C followed by 40 cycles of 45 s at 94°C, 30 s at 55°C and 45 s at 72°C with a final elongation of 10 min at 72°C. Successful amplification of the env C2V5 region was verified by electrophoresis on a 2% agarose gel and purification and Sanger sequencing were performed by Eurofins Genomics, with Env7 and InV5R_AE as sequencing primers.
TCR amplification on genomic DNA
We developed a two-step PCR method to amplify a portion of approximately 260bp of the TCRβ encompassing: (1) the end of the V segment, (2) the CDR3, and (3) the J segment, on genomic DNA from lysed single-cells or 100,000 bulk CD4+ T cells. We used a set of 22 forward primers complementary to the 23 functional V segments families, and 13 reverse primers complementary to the 13 functional J segments, to amplify the target portion of the TCRβ in a first multiplex PCR reaction, as previously described 16. M13 forward and reverse tags were added to the 5’ end of these primers, to allow a second PCR amplification, which was followed by Sanger sequencing. Sequences of all primers are listed in Table S2. The first PCR reaction was performed using the Qiagen Multiplex PCR kit (Cat#206143, Qiagen), in a total volume of 50 μL: 25 μL of Qiagen Multiplex PCR master mix, 10 μL of a mix of all primers (each primer at a concentration of 1.25 μM in the mix, providing a final concentration of 250 nM per primer), 5 μL of Q-Solution, and 10 μL of the single-cell lysate. First PCR conditions were as follows: 15 min at 95°C followed by 40 cycles of; 30 s at 95°C, 90 s at 68°C and 20 s at 72°C; with a final elongation for 5 min at 72°C. A second round of PCR reaction was performed using the M13F and M13R primers (see Table S2) and the Taq DNA Polymerase kit (Cat# 18038-042, Invitrogen), in a total volume of 50 μL: 5 μL of 10X PCR buffer, 3 μL of MgCl2 (50 mM), 1.5 μL of dNTPs (10 mM), 2 μL of M13F primer and 2 μL of M13R primer (each at 10 μM, providing a final concentration of 400 nM per primer), 0.5 μl Taq DNA Polymerase (5 U/ μL), 26 μL H2O and 10 μL of the first PCR products. The amplification conditions for the second PCR reaction were as follows: 3 min at 94°C followed by 40 cycles of; 45 s at 94°C, 60 s at 55°C and 30 s at 72°C; with a final elongation of 10 min at 72°C. A third round PCR adding next-generation sequencing adaptors to the TCR amplicons was performed for bulk cells to allow for further MiSeq sequencing. The third PCRs were performed using the same amplification conditions as for the second one (see MiSeq adaptors primers in Table S2).
TCR sequencing and analysis
Successful amplification of the TCRβ region was verified by electrophoresis on a 2% agarose gel and followed by gel purification of the TCRβ bands using the Buffer QG and the QIAquick 96 PCR Purification kit (Cat#28181, Qiagen), according to the manufacturer’s instructions. Sanger sequencing was performed by Eurofins Genomics, with M13F and M13R as sequencing primers. TCRβ sequences were re-constructed using both forward and reverse sequences, and were analyzed using the V-QUEST tool of the IMGT® database (IMGT®, the international ImMunoGeneTics information system® [http://www.imgt.org]56) to retrieve TCRβ information, including V and J segments usage and junction/CDR3 analysis. Next-generation sequencing (MiSeq, Illumina) was performed by Genome Quebec. TCR reads were aligned, identified and grouped using the regular pipeline of MiXCR version 3.0.13 29. Clonotypes with less than 10 reads were removed, and clonotypes representing at least 0.1% of the total read count for each sample were considered as expanded. TCR sequences were analyzed using an algorithm to predict antigen specificity: CDR3 sequences were compared to the McPAS-TCR database of TCRs of known antigenic specificity ([http://friedmanlab.weizmann.ac.il/McPAS-TCR/]57) and sequence similarities were identified. We predicted TCR specificity using the three criteria described by Meysman et al. 28: 1) CDR3 sequences should have identical length, 2) CDR3 sequences should be long enough and 3) CDR3 sequences should not differ by more than one amino acid. Among all CDR3 sequences, those fulfilling these three criteria with matched CDR3 sequences from the database were considered at high probability of sharing the same specificity.
Near-full length HIV genome amplification on genomic DNA
We developed a two-step PCR method to amplify a portion of approximately 9,000bp of the HIV genome on lysed bulk cells. Sequences of primers are listed in Table S2. Both PCR reactions were performed using the Platinum SuperFi II Master Mix (Cat#12368010, Life Tech). The first PCR was prepared in a total volume of 40 μL: 20 μL of Platinum SuperFi II master mix, 0.8 μL of each primer (each primer at a concentration of 10 μM in the mix, providing a final concentration of 200 nM per primer), 8.4 μL H2O. and 10 μL of the cell lysate. First PCR conditions were as follows: 30 s at 98°C followed by 25 cycles of; 30 s at 98°C, 10 s at 60°C and 5 min at 72°C; with a final elongation for 5 min at 72°C. A nested PCR, with PacBio barcoded primers was then performed prior to PacBio sequencing. The nested PCR was prepared in a total volume of 30 μL: 15 μL of Platinum SuperFi II master mix, 0.6 μL of each primer (each primer at a concentration of 10 μM in the mix, providing a final concentration of 200 nM per primer), 8.8 μL H2O. and 5 μL of the first PCR products diluted 1/3. Nested PCR conditions were as follows: 30 s at 98°C followed by 30 cycles of; 30 s at 98°C, 10 s at 60°C and 5 min at 72°C; with a final elongation for 5 min at 72°C.
Near-full length HIV genome sequencing and analysis
Successful amplification of the HIV genomes was verified by electrophoresis on a 0.5% agarose gel followed by AMPure XP (Cat#A63881, Beckman Coulter) purification with a beads/PCR products ratio of 1.6. Purified amplicons were then quantified using Nanodrop (Thermo Fisher Scientific) and pooled. PacBio sequencing was performed by Genome Quebec on a PacBio Sequel II instrument. Obtained sequences were then demultiplexed and those blasting the HIV genome, with at least 30 reads, containing both primers sequences on their ends and clustering together within the same individual were then aligned. Alignments were then used for phylogeny and clonality analysis using Maximum-Likelihood tree GTR+I+G model, with 1000 bootstraps in iqtree2, version 2.1.2. Trees were annotated with FigTree version 1.4.4. Sequences were also analyzed for integrity using HIVDatabase QCTool (https://www.hiv.lanl.gov/content/sequence/QC/index.html) and the ProseqIT (https://psd.cancer.gov/tools/pvs_annot.php). Start and stop codons were confirmed using GeneCutter tool (https://www.hiv.lanl.gov/content/sequence/GENE_CUTTER/cutter.html), and the integrity of the packaging signal was manually determined. Defects were evaluated in the following order: inversion, hypermutation, length (large deletion, i.e. <8800 pb), stop codon, frameshift, packaging signal (ψ), as previously described 58. If no defects were found (excluding defects in nef and in tat2), the sequence was considered genetically intact.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data were analyzed and represented using Graphpad Prism version 9.1.0. Results were represented as median or mean values, with interquartile range or minimum and maximum values, as indicated in the figure legends. Correlations were determined using nonparametric Spearman’s test. For group comparisons, non-parametric Wilcoxon matched-pairs signed rank or Mann-Whitney or Fisher’s exact tests were used. P values of less or equal to 0.05 were considered statistically significant.
Supplementary Material
Highlights.
Analyzed phenotype and TCR of single HIV-infected cells at earliest stages of infection.
The phenotype of productively HIV-infected cells rapidly evolves in acute infection.
The initial pool of HIV-infected cells is the product of independent infection events.
Latent and genetically intact proviruses are archived early and persist on therapy.
Acknowledgements
The study team is grateful to the individuals who volunteered to participate in this study and the staff at SEARCH/IHRI, the Thai Red Cross AIDS Research Centre and the Department of Retrovirology, U.S. Army Medical Component, Armed Forces Research Institute of Medical Sciences (AFRIMS). We thank the RV254/SEARCH 010 and RV304/SEARCH Protocols team members. We thank the flow cytometry core at the CRCHUM, managed by Dominique Gauchat, Philippe St-Onge and Gael Dulude for cell sorting, Corentin Richard for bioinformatics analysis as well as the NC3 core (Olfa Debbeche). We are grateful to the Thai Government Pharmaceutical Organization (GPO), ViiV Healthcare, Gilead Sciences and Merck for providing the antiretroviral medications for the RV254/SEARCH 010 study.
Funding
The empirical component of this study was funded by the US Military HIV Research Program, Walter Reed Army Institute of Research, Rockville, Maryland, USA, under a cooperative agreement (WW81XWH-18-2-0040) between the Henry M. Jackson Foundation for the Advancement of Military Medicine Inc., and the US Department of Defense (DOD) and by an intramural grant from the Thai Red Cross AIDS Research Centre and, in part, by the Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institute of Health (DAIDS, NIAID, NIH) (grant AAI20052001). The scientific component of this study was supported by the Foundation for AIDS Research (amfAR Research Consortium on HIV Eradication 108687-54-RGRL and 108928-56-RGRL) and by the Canadian Institutes for Health Research (CIHR; operating grant #364408 and the Canadian HIV Cure Enterprise (CanCURE) Team Grant HB2 - 164064), and the Réseau SIDA et maladies infectieuses du Fonds de Recherche du Québec - Santé (FRQ-S). P.G. is supported by a postdoctoral fellowship from CIHR (#415209). N.C. is supported by Research Scholar Career Awards of the FRQ-S (#253292). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. One or more of the authors of this paper self-identifies as a member of the LGBTQIA+ community. We avoided “helicopter science” practices by including the participating local contributors from the region where we conducted the research as authors on the paper.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare no competing interests. The views expressed are those of the authors. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, U.S. Army or Department of Defense, nor the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government including the U.S. National Institutes of Health. The investigators have adhered to the policies for protection of human subjects as prescribed in AR-70-25.
Data and Materials Availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files). Source data are provided with this paper within Supplementary Files. TCR sequences from CD4+ T cells and p24+ cells are provided as an Auxiliary Supplementary file. HIV env and HIV near full length sequences are available on GenBank under the accession numbers ON500682 to ON500846 and ON500847 to ON501069, respectively. External databases used in this study are available online: IMGT® database (IMGT®, the international ImMunoGeneTics information system® [http://www.imgt.org]); McPAS-TCR database ([http://friedmanlab.weizmann.ac.il/McPAS-TCR/]).
References
- 1.Barre-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, et al. (1983). Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220, 868–871. 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 2.Whitney JB, Hill AL, Sanisetty S, Penaloza-MacMaster P, Liu J, Shetty M, Parenteau L, Cabral C, Shields J, Blackmore S, et al. (2014). Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature 512, 74–77. 10.1038/nature13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller CJ, Li Q, Abel K, Kim EY, Ma ZM, Wietgrefe S, La Franco-Scheuch L, Compton L, Duan L, Shore MD, et al. (2005). Propagation and dissemination of infection after vaginal transmission of simian immunodeficiency virus. J Virol 79, 9217–9227. 10.1128/JVI.79.14.9217-9227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang ZQ, Wietgrefe SW, Li Q, Shore MD, Duan L, Reilly C, Lifson JD, and Haase AT (2004). Roles of substrate availability and infection of resting and activated CD4+ T cells in transmission and acute simian immunodeficiency virus infection. Proc Natl Acad Sci U S A 101, 5640–5645. 10.1073/pnas.0308425101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gordon SN, Klatt NR, Bosinger SE, Brenchley JM, Milush JM, Engram JC, Dunham RM, Paiardini M, Klucking S, Danesh A, et al. (2007). Severe depletion of mucosal CD4+ T cells in AIDS-free simian immunodeficiency virus-infected sooty mangabeys. J Immunol 179, 3026–3034. 10.4049/jimmunol.179.5.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Rasmussen T, Pahar B, Poonia B, Alvarez X, Lackner AA, and Veazey RS (2007). Massive infection and loss of CD4+ T cells occurs in the intestinal tract of neonatal rhesus macaques in acute SIV infection. Blood 109, 1174–1181. 10.1182/blood-2006-04-015172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estes JD, Kityo C, Ssali F, Swainson L, Makamdop KN, Del Prete GQ, Deeks SG, Luciw PA, Chipman JG, Beilman GJ, et al. (2017). Defining total-body AIDS-virus burden with implications for curative strategies. Nat Med 23, 1271–1276. 10.1038/nm.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leyre L, Kroon E, Vandergeeten C, Sacdalan C, Colby DJ, Buranapraditkun S, Schuetz A, Chomchey N, de Souza M, Bakeman W, et al. (2020). Abundant HIV-infected cells in blood and tissues are rapidly cleared upon ART initiation during acute HIV infection. Sci Transl Med 12. 10.1126/scitranslmed.aav3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, and Pantaleo G (2013). Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med 210, 143–156. 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perelson AS, Neumann AU, Markowitz M, Leonard JM, and Ho DD (1996). HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271, 1582–1586. 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 11.Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, Deutsch P, Lifson JD, Bonhoeffer S, Nowak MA, Hahn BH, and et al. (1995). Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373, 117–122. 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 12.Robb ML, Eller LA, Kibuuka H, Rono K, Maganga L, Nitayaphan S, Kroon E, Sawe FK, Sinei S, Sriplienchan S, et al. (2016). Prospective Study of Acute HIV-1 Infection in Adults in East Africa and Thailand. N Engl J Med 374, 2120–2130. 10.1056/NEJMoa1508952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu VH, Nobles CL, Kuri-Cervantes L, McCormick K, Everett JK, Nguyen S, Del Rio Estrada PM, Gonzalez-Navarro M, Torres-Ruiz F, Avila-Rios S, et al. (2020). Assessment of HIV-1 integration in tissues and subsets across infection stages. JCI Insight 5. 10.1172/jci.insight.139783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffin JM, Wells DW, Zerbato JM, Kuruc JD, Guo S, Luke BT, Eron JJ, Bale M, Spindler J, Simonetti FR, et al. (2019). Clones of infected cells arise early in HIV-infected individuals. JCI Insight 4. 10.1172/jci.insight.128432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colby DJ, Trautmann L, Pinyakorn S, Leyre L, Pagliuzza A, Kroon E, Rolland M, Takata H, Buranapraditkun S, Intasan J, et al. (2018). Rapid HIV RNA rebound after antiretroviral treatment interruption in persons durably suppressed in Fiebig I acute HIV infection. Nat Med 24, 923–926. 10.1038/s41591-018-0026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gantner P, Pagliuzza A, Pardons M, Ramgopal M, Routy JP, Fromentin R, and Chomont N (2020). Single-cell TCR sequencing reveals phenotypically diverse clonally expanded cells harboring inducible HIV proviruses during ART. Nat Commun 11, 4089. 10.1038/s41467-020-17898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Gurule EE, Brennan TP, Gerold JM, Kwon KJ, Hosmane NN, Kumar MR, Beg SA, Capoferri AA, Ray SC, et al. (2018). Expanded cellular clones carrying replication-competent HIV-1 persist, wax, and wane. Proc Natl Acad Sci U S A 115, E2575–E2584. 10.1073/pnas.1720665115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mendoza P, Jackson JR, Oliveira TY, Gaebler C, Ramos V, Caskey M, Jankovic M, Nussenzweig MC, and Cohn LB (2020). Antigen-responsive CD4+ T cell clones contribute to the HIV-1 latent reservoir. J Exp Med 217. 10.1084/jem.20200051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simonetti FR, Zhang H, Soroosh GP, Duan J, Rhodehouse K, Hill AL, Beg SA, McCormick K, Raymond HE, Nobles CL, et al. (2021). Antigen-driven clonal selection shapes the persistence of HIV-1-infected CD4+ T cells in vivo. J Clin Invest 131. 10.1172/JCI145254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shan L, Deng K, Gao H, Xing S, Capoferri AA, Durand CM, Rabi SA, Laird GM, Kim M, Hosmane NN, et al. (2017). Transcriptional Reprogramming during Effector-to-Memory Transition Renders CD4(+) T Cells Permissive for Latent HIV-1 Infection. Immunity 47, 766–775 e763. 10.1016/j.immuni.2017.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saleh S, Solomon A, Wightman F, Xhilaga M, Cameron PU, and Lewin SR (2007). CCR7 ligands CCL19 and CCL21 increase permissiveness of resting memory CD4+ T cells to HIV-1 infection: a novel model of HIV-1 latency. Blood 110, 4161–4164. 10.1182/blood-2007-06-097907. [DOI] [PubMed] [Google Scholar]
- 22.Cameron PU, Saleh S, Sallmann G, Solomon A, Wightman F, Evans VA, Boucher G, Haddad EK, Sekaly RP, Harman AN, et al. (2010). Establishment of HIV-1 latency in resting CD4+ T cells depends on chemokine-induced changes in the actin cytoskeleton. Proc Natl Acad Sci U S A 107, 16934–16939. 10.1073/pnas.1002894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okoye AA, Hansen SG, Vaidya M, Fukazawa Y, Park H, Duell DM, Lum R, Hughes CM, Ventura AB, Ainslie E, et al. (2018). Early antiretroviral therapy limits SIV reservoir establishment to delay or prevent post-treatment viral rebound. Nat Med 24, 1430–1440. 10.1038/s41591-018-0130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Souza MS, Phanuphak N, Pinyakorn S, Trichavaroj R, Pattanachaiwit S, Chomchey N, Fletcher JL, Kroon ED, Michael NL, Phanuphak P, et al. (2015). Impact of nucleic acid testing relative to antigen/antibody combination immunoassay on the detection of acute HIV infection. AIDS 29, 793–800. 10.1097/QAD.0000000000000616. [DOI] [PubMed] [Google Scholar]
- 25.Pardons M, Baxter AE, Massanella M, Pagliuzza A, Fromentin R, Dufour C, Leyre L, Routy JP, Kaufmann DE, and Chomont N (2019). Single-cell characterization and quantification of translation-competent viral reservoirs in treated and untreated HIV infection. PLoS Pathog 15, e1007619. 10.1371/journal.ppat.1007619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiebig EW, Wright DJ, Rawal BD, Garrett PE, Schumacher RT, Peddada L, Heldebrant C, Smith R, Conrad A, Kleinman SH, and Busch MP (2003). Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 17, 1871–1879. 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 27.Lama J, Mangasarian A, and Trono D (1999). Cell-surface expression of CD4 reduces HIV-1 infectivity by blocking Env incorporation in a Nef- and Vpu-inhibitable manner. Curr Biol 9, 622–631. 10.1016/s0960-9822(99)80284-x. [DOI] [PubMed] [Google Scholar]
- 28.Meysman P, De Neuter N, Gielis S, Bui Thi D, Ogunjimi B, and Laukens K (2019). On the viability of unsupervised T-cell receptor sequence clustering for epitope preference. Bioinformatics 35, 1461–1468. 10.1093/bioinformatics/bty821. [DOI] [PubMed] [Google Scholar]
- 29.Bolotin DA, Poslavsky S, Mitrophanov I, Shugay M, Mamedov IZ, Putintseva EV, and Chudakov DM (2015). MiXCR: software for comprehensive adaptive immunity profiling. Nat Methods 12, 380–381. 10.1038/nmeth.3364. [DOI] [PubMed] [Google Scholar]
- 30.Ananworanich J, Fletcher JL, Pinyakorn S, van Griensven F, Vandergeeten C, Schuetz A, Pankam T, Trichavaroj R, Akapirat S, Chomchey N, et al. (2013). A novel acute HIV infection staging system based on 4th generation immunoassay. Retrovirology 10, 56. 10.1186/1742-4690-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schacker T, Little S, Connick E, Gebhard K, Zhang ZQ, Krieger J, Pryor J, Havlir D, Wong JK, Schooley RT, et al. (2001). Productive infection of T cells in lymphoid tissues during primary and early human immunodeficiency virus infection. J Infect Dis 183, 555–562. 10.1086/318524. [DOI] [PubMed] [Google Scholar]
- 32.Murooka TT, Deruaz M, Marangoni F, Vrbanac VD, Seung E, von Andrian UH, Tager AM, Luster AD, and Mempel TR (2012). HIV-infected T cells are migratory vehicles for viral dissemination. Nature 490, 283–287. 10.1038/nature11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toyoda M, Ogata Y, Mahiti M, Maeda Y, Kuang XT, Miura T, Jessen H, Walker BD, Brockman MA, Brumme ZL, and Ueno T (2015). Differential Ability of Primary HIV-1 Nef Isolates To Downregulate HIV-1 Entry Receptors. J Virol 89, 9639–9652. 10.1128/JVI.01548-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Isaacman-Beck J, Hermann EA, Yi Y, Ratcliffe SJ, Mulenga J, Allen S, Hunter E, Derdeyn CA, and Collman RG (2009). Heterosexual transmission of human immunodeficiency virus type 1 subtype C: Macrophage tropism, alternative coreceptor use, and the molecular anatomy of CCR5 utilization. J Virol 83, 8208–8220. 10.1128/JVI.00296-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen A, Baker JJ, Scott GL, Davis YP, Ho YY, and Siliciano RF (2013). Endothelial cell stimulation overcomes restriction and promotes productive and latent HIV-1 infection of resting CD4+ T cells. J Virol 87, 9768–9779. 10.1128/JVI.01478-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agosto LM, Herring MB, Mothes W, and Henderson AJ (2018). HIV-1-Infected CD4+ T Cells Facilitate Latent Infection of Resting CD4+ T Cells through Cell-Cell Contact. Cell Rep 24, 2088–2100. 10.1016/j.celrep.2018.07.079. [DOI] [PubMed] [Google Scholar]
- 37.Hong JJ, Amancha PK, Rogers K, Ansari AA, and Villinger F (2012). Spatial alterations between CD4(+) T follicular helper, B, and CD8(+) T cells during simian immunodeficiency virus infection: T/B cell homeostasis, activation, and potential mechanism for viral escape. J Immunol 188, 3247–3256. 10.4049/jimmunol.1103138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell JL, Pollara J, Dietze K, Edwards RW, Nohara J, N'Guessan K F, Zemil M, Buranapraditkun S, Takata H, Li Y, et al. (2022). Anti-HIV antibody development up to 1 year after antiretroviral therapy initiation in acute HIV infection. J Clin Invest 132. 10.1172/JCI150937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, et al. (2002). HIV preferentially infects HIV-specific CD4+ T cells. Nature 417, 95–98. 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 40.Geldmacher C, Ngwenyama N, Schuetz A, Petrovas C, Reither K, Heeregrave EJ, Casazza JP, Ambrozak DR, Louder M, Ampofo W, et al. (2010). Preferential infection and depletion of Mycobacterium tuberculosis-specific CD4 T cells after HIV-1 infection. J Exp Med 207, 2869–2881. 10.1084/jem.20100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones RB, Kovacs C, Chun TW, and Ostrowski MA (2012). Short communication: HIV type 1 accumulates in influenza-specific T cells in subjects receiving seasonal vaccination in the context of effective antiretroviral therapy. AIDS Res Hum Retroviruses 28, 1687–1692. 10.1089/AID.2012.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, and Paxton WA (1996). HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381, 667–673. 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 43.Casazza JP, Brenchley JM, Hill BJ, Ayana R, Ambrozak D, Roederer M, Douek DC, Betts MR, and Koup RA (2009). Autocrine production of beta-chemokines protects CMV-Specific CD4 T cells from HIV infection. PLoS Pathog 5, e1000646. 10.1371/journal.ppat.1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collora JA, Liu R, Pinto-Santini D, Ravindra N, Ganoza C, Lama JR, Alfaro R, Chiarella J, Spudich S, Mounzer K, et al. (2022). Single-cell multiomics reveals persistence of HIV-1 in expanded cytotoxic T cell clones. Immunity. 10.1016/j.immuni.2022.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grau-Exposito J, Serra-Peinado C, Miguel L, Navarro J, Curran A, Burgos J, Ocana I, Ribera E, Torrella A, Planas B, et al. (2017). A Novel Single-Cell FISH-Flow Assay Identifies Effector Memory CD4(+) T cells as a Major Niche for HIV-1 Transcription in HIV-Infected Patients. mBio 8. 10.1128/mBio.00876-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bretschneider I, Clemente MJ, Meisel C, Guerreiro M, Streitz M, Hopfenmuller W, Maciejewski JP, Wlodarski MW, and Volk HD (2014). Discrimination of T-cell subsets and T-cell receptor repertoire distribution. Immunol Res 58, 20–27. 10.1007/s12026-013-8473-0. [DOI] [PubMed] [Google Scholar]
- 47.Boritz EA, Darko S, Swaszek L, Wolf G, Wells D, Wu X, Henry AR, Laboune F, Hu J, Ambrozak D, et al. (2016). Multiple Origins of Virus Persistence during Natural Control of HIV Infection. Cell 166, 1004–1015. 10.1016/j.cell.2016.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miron M, Meng W, Rosenfeld AM, Dvorkin S, Poon MML, Lam N, Kumar BV, Louzoun Y, Luning Prak ET, and Farber DL (2021). Maintenance of the human memory T cell repertoire by subset and tissue site. Genome Med 13, 100. 10.1186/s13073-021-00918-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evans VA, Kumar N, Filali A, Procopio FA, Yegorov O, Goulet JP, Saleh S, Haddad EK, da Fonseca Pereira C, Ellenberg PC, et al. (2013). Myeloid dendritic cells induce HIV-1 latency in non-proliferating CD4+ T cells. PLoS Pathog 9, e1003799. 10.1371/journal.ppat.1003799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chavez L, Calvanese V, and Verdin E (2015). HIV Latency Is Established Directly and Early in Both Resting and Activated Primary CD4 T Cells. PLoS Pathog 11, e1004955. 10.1371/journal.ppat.1004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinzone MR, VanBelzen DJ, Weissman S, Bertuccio MP, Cannon L, Venanzi-Rullo E, Migueles S, Jones RB, Mota T, Joseph SB, et al. (2019). Longitudinal HIV sequencing reveals reservoir expression leading to decay which is obscured by clonal expansion. Nat Commun 10, 728. 10.1038/s41467-019-08431-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neidleman J, Luo X, Frouard J, Xie G, Hsiao F, Ma T, Morcilla V, Lee A, Telwatte S, Thomas R, et al. (2020). Phenotypic analysis of the unstimulated in vivo HIV CD4 T cell reservoir. Elife 9. 10.7554/eLife.60933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Q, Duan L, Estes JD, Ma ZM, Rourke T, Wang Y, Reilly C, Carlis J, Miller CJ, and Haase AT (2005). Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature 434, 1148–1152. 10.1038/nature03513. [DOI] [PubMed] [Google Scholar]
- 54.Becht E, McInnes L, Healy J, Dutertre CA, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 55.Van Gassen S, Callebaut B, Van Helden MJ, Lambrecht BN, Demeester P, Dhaene T, and Saeys Y (2015). FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645. 10.1002/cyto.a.22625. [DOI] [PubMed] [Google Scholar]
- 56.Lefranc MP, Giudicelli V, Ginestoux C, Bodmer J, Muller W, Bontrop R, Lemaitre M, Malik A, Barbie V, and Chaume D (1999). IMGT, the international ImMunoGeneTics database. Nucleic Acids Res 27, 209–212. 10.1093/nar/27.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tickotsky N, Sagiv T, Prilusky J, Shifrut E, and Friedman N (2017). McPAS-TCR: a manually curated catalogue of pathology-associated T cell receptor sequences. Bioinformatics 33, 2924–2929. 10.1093/bioinformatics/btx286. [DOI] [PubMed] [Google Scholar]
- 58.Hiener B, Horsburgh BA, Eden JS, Barton K, Schlub TE, Lee E, von Stockenstrom S, Odevall L, Milush JM, Liegler T, et al. (2017). Identification of Genetically Intact HIV-1 Proviruses in Specific CD4(+) T Cells from Effectively Treated Participants. Cell Rep 21, 813–822. 10.1016/j.celrep.2017.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its supplementary information files). Source data are provided with this paper within Supplementary Files. TCR sequences from CD4+ T cells and p24+ cells are provided as an Auxiliary Supplementary file. HIV env and HIV near-full length sequences have been deposited in GenBank and are publicly available from the date of publication. Accession numbers are listed in the key resources table. External databases used in this study are available online: IMGT® database (IMGT®, the international ImMunoGeneTics Information system®) [http://www.imgt.org]); McPAS-TCR database ([http://friedmanlab.weizmann.ac.il/McPAS-TCR/]).
All codes used were downloaded from Github and are publicly available.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| p24 KC57-PE | Beckman Coulter | Cat#6604667 |
| p24 28B7-APC | MediMabs | Cat#MM-0289-APC |
| CD4-APC-H7 (Clone: RPA-T4) | BD Bioscience | Cat#560168 |
| CD45RA-A700 (Clone: HI100) | BD Bioscience | Cat#560673 |
| CXCR5-BB515 (Clone: RF8B2) | BD Bioscience | Cat#564624 |
| CXCR3-PerCP-Cy5.5 (Clone: 1C6/CXCR3) | BD Bioscience | Cat#560832 |
| Ki67-PE-Cy7 (Clone: B56) | BD Bioscience | Cat#561283 |
| CCR5-BV421 (Clone: 2D7/CCR5) | BD Bioscience | Cat#562576 |
| CD45RA-BV786 (Clone: HI100) | BD Bioscience | Cat#563870 |
| HLA-DR-BV605 (Clone: G46-6) | BD Bioscience | Cat#562845 |
| CCR7-BB515 (Clone: 3D12) | BD Bioscience | Cat#565870 |
| PD-1-BV605 (Clone: EH12.2H7) | BioLegend | Cat#329923 |
| ICOS-BV785 (Clone: C398.4a) | BioLegend | Cat#313534 |
| HLA-ABC-A700 (Clone: W6/32) | BioLegend | Cat#311437 |
| CD69-PerCP-Cy5.5 (Clone: FN50) | BioLegend | Cat#310926 |
| Bcl-2-BV421 (Clone: 100) | BioLegend | Cat#658709 |
| Live/Dead Aqua Cell Stain (405nm) | ThermoFisher Scientific | Cat#L34957 |
| Biological samples | ||
| Human serum | Atlanta Biologicals | Cat#540110 |
| Demographics of study participants, see Table S1 | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Lamivudine (3TC) | NIH HIV Reagent Program | ARP-8146 |
| Raltegravir (RAL) | NIH HIV Reagent Program | HRP-11680 |
| Brefeldin A (BFA) | Sigma | Cat#B2651 |
| ionomycin | Sigma | Cat#I9657 |
| PMA | Sigma | Cat#P8139 |
| DirectPCR Lysis Reagent | Viagen Biotech | Cat#301-C |
| Proteinase K | Wisent | Cat#25530–015 |
| Proteinase K | Invitrogen | Cat#25530-015, |
| Critical commercial assays | ||
| EasySep Human CD4+ T Cell Enrichment Kit | StemCell Technology | Cat#19052 |
| FoxP3 Transcription Factor Staining Buffer Set | eBioscience | Cat#00-5523-00 |
| Phusion Master Mix | ThermoFisher Scientific | Cat#F531S |
| Taq DNA Polymerase kit | Invitrogen | Cat#18038-042, |
| Qiagen Multiplex PCR kit | Qiagen | Cat#206143 |
| QIAquick 96 PCR Purification kit | Qiagen | Cat#28181 |
| Platinum SuperFi II Master Mix | Life Tech | Cat#12368010 |
| AMPure XP | Beckman Coulter | Cat#A63881 |
| Deposited data | ||
| HIV env | GenBank | Genbank accession numbers ON500682 to ON500846 |
| HIV near full length sequences | GenBank | Genbank accession numbers ON500847 to ON501069 |
| TCRβ sequences | This paper | N/A |
| Oligonucleotides | ||
| TCRβ HIV env and HIV near full length amplification and sequencing primers, see Table S2 | This paper | N/A |
| Software and algorithms | ||
| MiXCR version 3.0.13 | Bolotin et al.29 | https://github.com/milaboratory/mixcr |
| IMGT® database | IMGT®, the international ImMunoGeneTics information system® | http://www.imgt.org |
| McPAS-TCR database | Friedman lab | http://friedmanlab.weizmann.ac.il/McPAS-TCR/ |
| Prism version 9.1.0 | Graphpad | https://www.graphpad.com |
| FlowJo version 10.7.1 | FlowJo | https://www.flowjo.com/solutions/flowjo |
| iqtree2, version 2.1.2 | IQTREE | http://www.iqtree.org |
| FigTree version 1.4.4 | Figtree | http://tree.bio.ed.ac.uk/software/figtree/ |
| HIVDatabase QCTool | HIV Los Alamos | https://www.hiv.lanl.gov/content/sequence/QC/index.html |
| ProseqIT | NIH | https://psd.cancer.gov/tools/nvs_annot.php |
| GeneCutter tool | HIV Los Alamos | https://www.hiv.lanl.gov/content/sequence/GENE_CUTTER/cutter.html |
All data generated or analyzed during this study are included in this published article (and its supplementary information files). Source data are provided with this paper within Supplementary Files. TCR sequences from CD4+ T cells and p24+ cells are provided as an Auxiliary Supplementary file. HIV env and HIV near full length sequences are available on GenBank under the accession numbers ON500682 to ON500846 and ON500847 to ON501069, respectively. External databases used in this study are available online: IMGT® database (IMGT®, the international ImMunoGeneTics information system® [http://www.imgt.org]); McPAS-TCR database ([http://friedmanlab.weizmann.ac.il/McPAS-TCR/]).