Abstract
Despite the development of cancer therapies, the success of most treatments has been impeded by drug resistance. The crucial role of tumor cell plasticity has emerged recently in cancer progression, cancer stemness and eventually drug resistance. Cell plasticity drives tumor cells to reversibly convert their cell identity, analogous to differentiation and dedifferentiation, to adapt to drug treatment. This phenotypical switch is driven by alteration of the transcriptome. Several pluripotent factors from the KLF and SOX families are closely associated with cancer pathogenesis and have been revealed to regulate tumor cell plasticity. In this review, we particularly summarize recent studies about KLF4, KLF5 and SOX factors in cancer development and evolution, focusing on their roles in cancer initiation, invasion, tumor hierarchy and heterogeneity, and lineage plasticity. In addition, we discuss the various regulation of these transcription factors and related cutting-edge drug development approaches that could be used to drug “undruggable” transcription factors, such as PROTAC and PPI targeting, for targeted cancer therapy. Advanced knowledge could pave the way for the development of novel drugs that target transcriptional regulation and could improve the outcome of cancer therapy.
Keywords: KLF4, KLF5, SOXs, pathogenesis, stemness, carcinogenesis and therapy
Introduction
Cancer cells almost invariably evolve resistance, leading to cancer recurrence, metastasis, and eventually patient mortality [1,2]. Historically, most studies focus on genetic drivers of drug resistance, that enable cancer cells to bypass the targeted pathway and then develop new therapies to overcome the resistance correspondingly [3]. But cancer cells can gain resistance to new treatments by similar mechanisms. More and more studies, especially data from single-cell profiling, show that non-mutational mechanisms, such as cell lineage plasticity, serve as key drivers for drug resistance in various cancers [1,2,4,5]. Cell plasticity enables cancer cells with a single genotype to switch between different phenotypic states (or cell identities) in response to external stimuli, including stresses in the adverse tumor microenvironment and anti-cancer drug treatments to survive and thrive (Fig. 1). Such a phenotypic switch can be reversible, as is demonstrated by the loss of resistance when cultured in the absence of the drug. Indeed, the subpopulation of cancer cells adapted to the treatment can serve as a reservoir to proliferate and gain stochastic mutations. Eventually, those cancer cells with genetic mutations that confer better resistance to the treatment will survive as the set of fittest clones under drug selection. Besides drug resistance, cancer cell plasticity also facilitates malignant progression, which requires phenotypic switches such as the epithelial-to-mesenchymal transition (EMT) for migration and mesenchymal-to-epithelial transition (MET) for colony formation [6]. In the context of cancer stem cell (CSC) theory, cancer cell plasticity can also be interpreted as the capacity of a differentiated cancer cell to revert to a stem cell-like state [2,7].
Figure 1. Transcription factors mediate cancer cell lineage plasticity, which enables the phenotypic switch of cancer cells and drives metastasis, stemness and drug resistance.
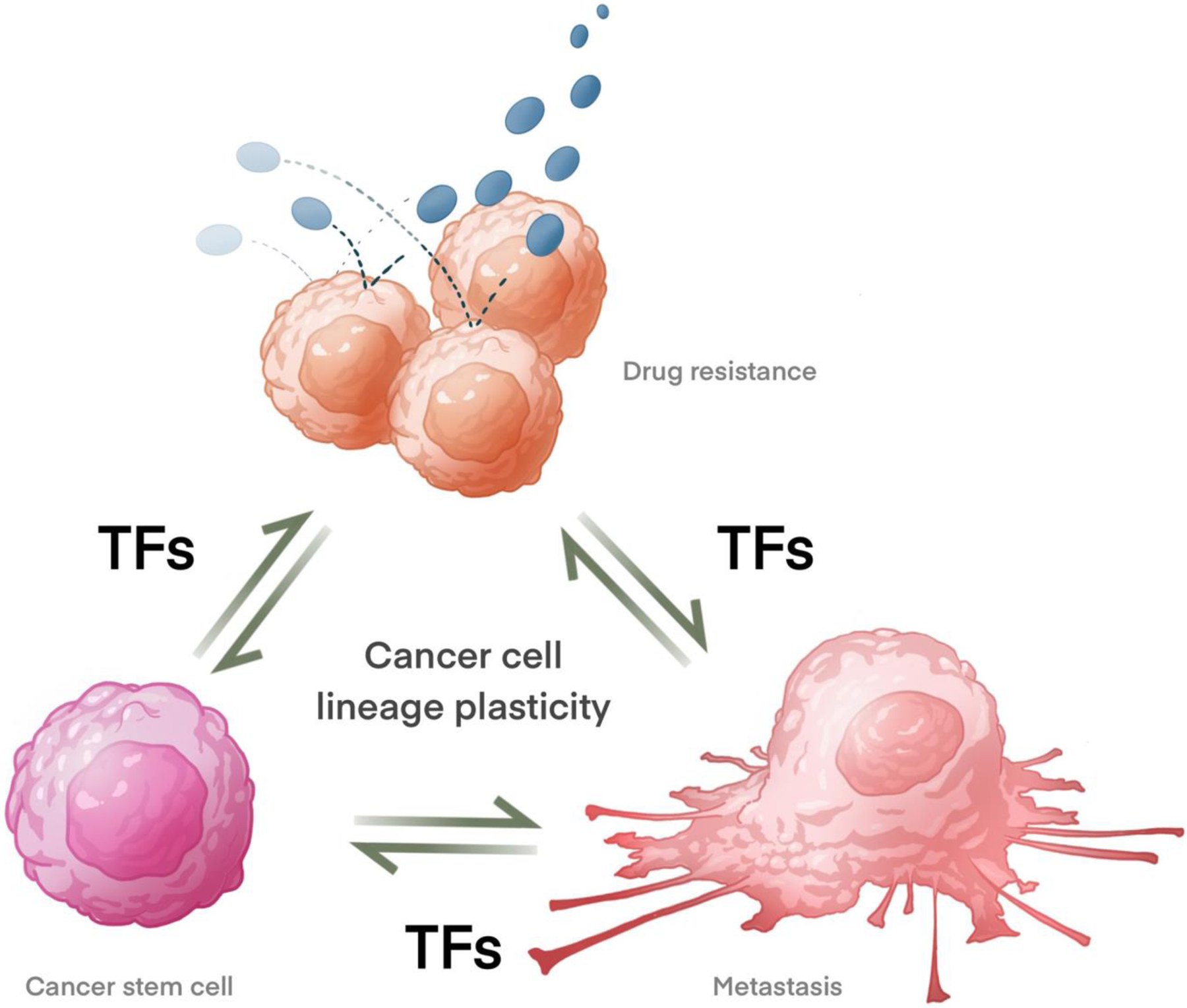
Resistant cancer cells surviving from anti-cancer treatment can undergo further transformation and leverage these malignant properties to promote cancer metastasis. TFs: transcription factors.
Transcription factors (TFs) are crucial in reprogramming cancer cells to control these phenotypic switches. Transcription factors bind to specific DNA sequences in the genome, and coordinate gene expression in response to signaling [8]. Transcription factors are the fundamental players in determining cell lineage during the development of multicellular organisms by decoding the same genetic code differentially. Furthermore, TFs are also essential for cell survival as they work as endpoints of cellular signaling pathways to coordinate the rapid transcriptional response to internal and external stimuli. Given their central regulatory role in biology, TFs are critical players in many diseases, including cancers [9–12]. It is hypothesized that dysregulated transcription leads to the fundamental features of cancer. Consistently, cancer cells have gene expression profiles distinct from their normal tissue of origin, and these gene expression profiles change during different stages of cancer development [13–18], highlighting the crucial roles of transcription factors. Furthermore, gene expression profiles can predict clinical outcomes of many cancers [19–22], indicating the critical roles of transcription factors in cancer prognosis.
Given their central regulatory roles in cancers, TFs represent potential therapeutic targets [9–12]. The therapeutical potential of modulating TF action has been well demonstrated by hormone therapies that target nuclear hormone receptors, a subtype of transcription factor, in cancer treatments. Anti-estrogen and anti-androgen compounds, such as tamoxifen, bicalutamide, and enzalutamide have been in clinical use to treat breast cancer and prostate cancer, respectively, for many years [23,24]. Despite the great potential of targeting transcription factors for cancer treatment, transcription factors (other than nuclear receptors with ligands) have been considered undruggable due to the lack of enzymatic sites or ligand binding pockets to develop small-molecule inhibitors or activators. However, new technologies, such as proteolysis-targeting chimeras (PROTACs) for targeted protein degradation and structure-based protein-protein interaction inhibitors, have made targeting transcription factors for cancer treatment into viable options [12,25]. In this review, we focus on the roles of several pluripotent transcription factors, KLF4, KLF5, SOX2, SOX4, and SOX9 in cancer progression, metastasis, stemness, stress response, and drug resistance, as well as their regulation to explore the potential and possible strategies of targeting these transcription factors in cancer treatment.
KLFs and SOXs in pluripotency
Human Krüppel-like factors (KLFs) are a family of DNA-binding transcription factors with 17 members, that is highly conserved among mammals from humans to mouse [26]. Structurally, all members of the KLF family have a highly conserved triple C2H2 zinc-finger DNA-binding domain at the carboxyl terminus regions [26]. Each zinc finger recognizes three base pairs in the DNA sequence, and thus KLFs can interact with nine base pairs in total. The N-terminus of KLFs, where an activation or repression domain is typically located, is highly divergent [26]. SRY-related high-mobility-group box (SOX) family is another group of transcriptional factors with over 20 members in vertebrates. SOX factors are defined by a highly conserved DNA-binding high-mobility group (HMG) domain homologous to the HMG domain of sex determining region Y protein (SRY) [27]. KLFs and SOXs play essential roles during development. They work as transcription factors to mediate responses to external and internal stimuli and regulate many cellular processes, including proliferation, apoptosis, differentiation, and migration [26,28,29]. Several members from the two families, especially KLF4, KLF5, SOX2, SOX4, and SOX9, are expressed in stem cells and function as pluripotent factors to regulate cell lineage and stemness (Fig. 2).
Figure 2.
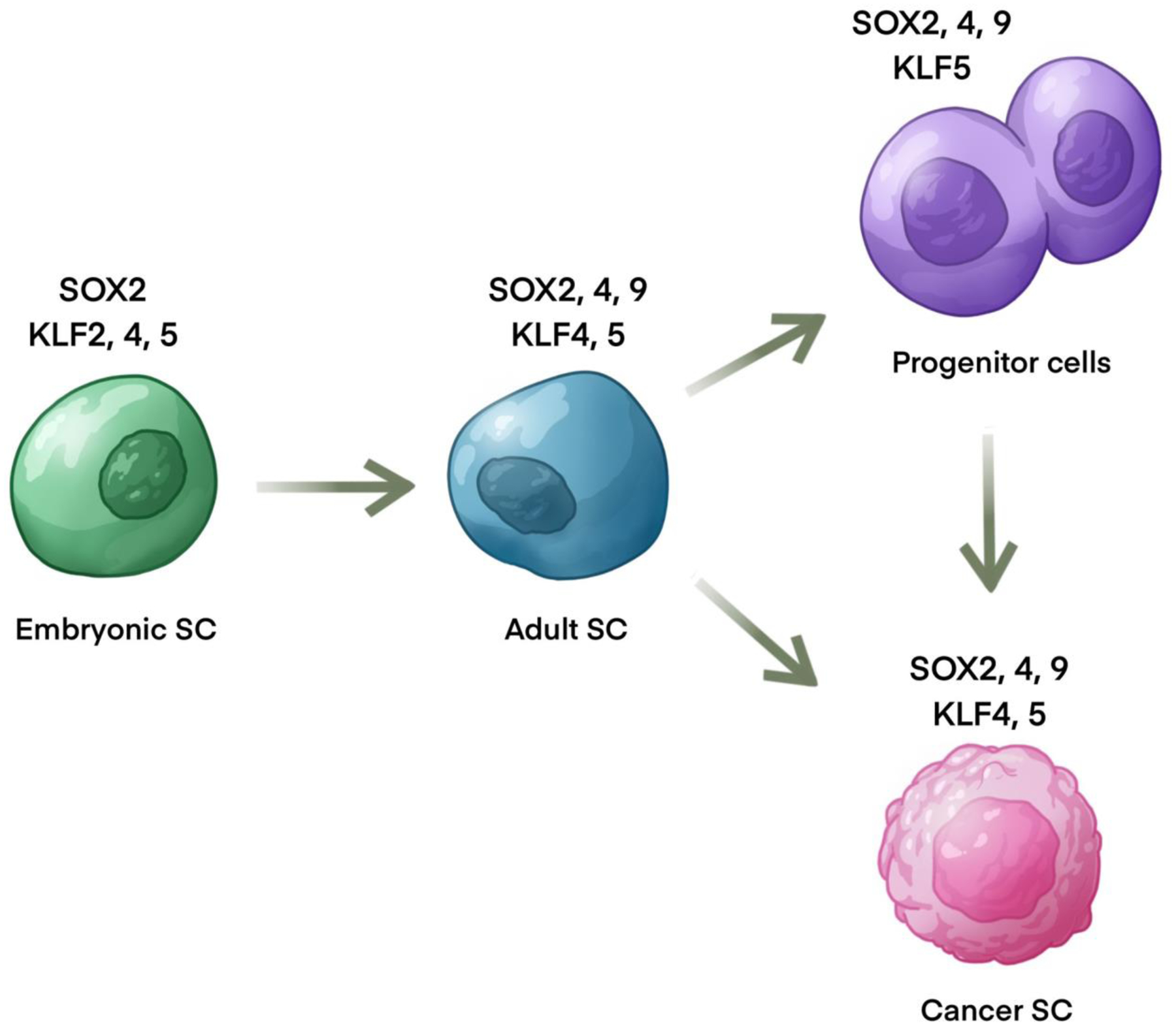
Expression of SOX and KLF factors in stem and progenitor cells.
KLF2, 4, and 5 are co-expressed in ESCs to maintain pluripotency and their combined knockdown leads to robust differentiation of ESCs [30,31]. The expression of KLF4 and KLF5 is induced through the LIF/STAT3 pathway, which is required to sustain pluripotency [32–34]. KLF4 and KLF5 activation leads to the expression of NANOG, SOX2, and OCT4 [35,36], while OCT4 further activates KLF2 [34] and reinforces SOX2 expression [37]. These downstream targets of KLF4 and KLF5: NANOG, SOX2, and OCT4, play a critical role in maintaining the pluripotency of embryonic stem cells [38]. SOX2, OCT4 and KLF4 are three of the four OSKM factors for induced pluripotent cells(iPSCs) [39,40]. The other OSKM factor is c-MYC, which promotes iPSCs but is strictly dispensable [41].
In addition, KLF4 and KLF5 also play a critical role in adult stem cells. Both KLF4 and KLF5 are highly expressed in the intestinal tract [42,43]. KLF4 modulates the intestinal stem cells marked by BMI1. In BMI1+ stem cells, KLF4 inhibits proliferation to maintain homeostasis under normal conditions while promoting the regenerative response upon ionizing radiation injury [44]. KLF5 is required to maintain intestinal stem cells. Conditional knockout of KLF5 suppresses the proliferation and survival of mouse intestinal stem cells and completely prevents the tumorigenesis induced by β-catenin [45]. KLF5 also regulates the maintenance and differentiation of basal progenitors in prostate development [46]. This differential regulation is mediated by the posttranslational regulation of KLF5: acetylated KLF5 is required to maintain basal progenitors while deacetylated KLF5 promotes basal-to-luminal differentiation partially through activating Notch signaling [46].
SOX factors also play critical roles in development and tissue homeostasis through maintenance of stem and progenitor cells. Besides the well documented role of SOX2 in induction of pluripotent stem cells [39], SOX2 and SOX9 play key roles in neuronal development and maintenance of neural progenitor cells [47]. SOX9 also is critical for maintenance of adult stem and progenitor cells in the intestine and hair follicles [48], and is essential for normal development of the pancreas [49] and mammary gland [50]. In addition to SOX9, SOX4 is also critical for development of the pancreas [51] and breast [52]. In fetal mammary stem cells, SOX4 inhibits differentiation by maintaining expression of a stem cell gene expression program [52]. SOX4 is essential to development of many tissues including neural crest [47], heart [53], B cells [54], and skeleton [55]. In adult stem cells, SOX4 is essential for stem cell reactivation in response to wounding of the skin [56].
KLFs and SOXs in cancer
Along with their roles in development, many studies have suggested that KLF4, KLF5, SOX2, SOX4 and SOX9 are associated with cancer stemness, progression, metastasis, drug resistance, and prognosis [28,57–65] (Fig. 4). Dysregulated KLF4 and KLF5 have been found in multiple cancer types including breast cancer, lung cancer, pancreatic cancer, prostate cancer, colon cancer, and stomach cancer [57]. SOX2, SOX4, and SOX9 also have increased expression in a wide variety of tumors [58–65].
Figure 4.
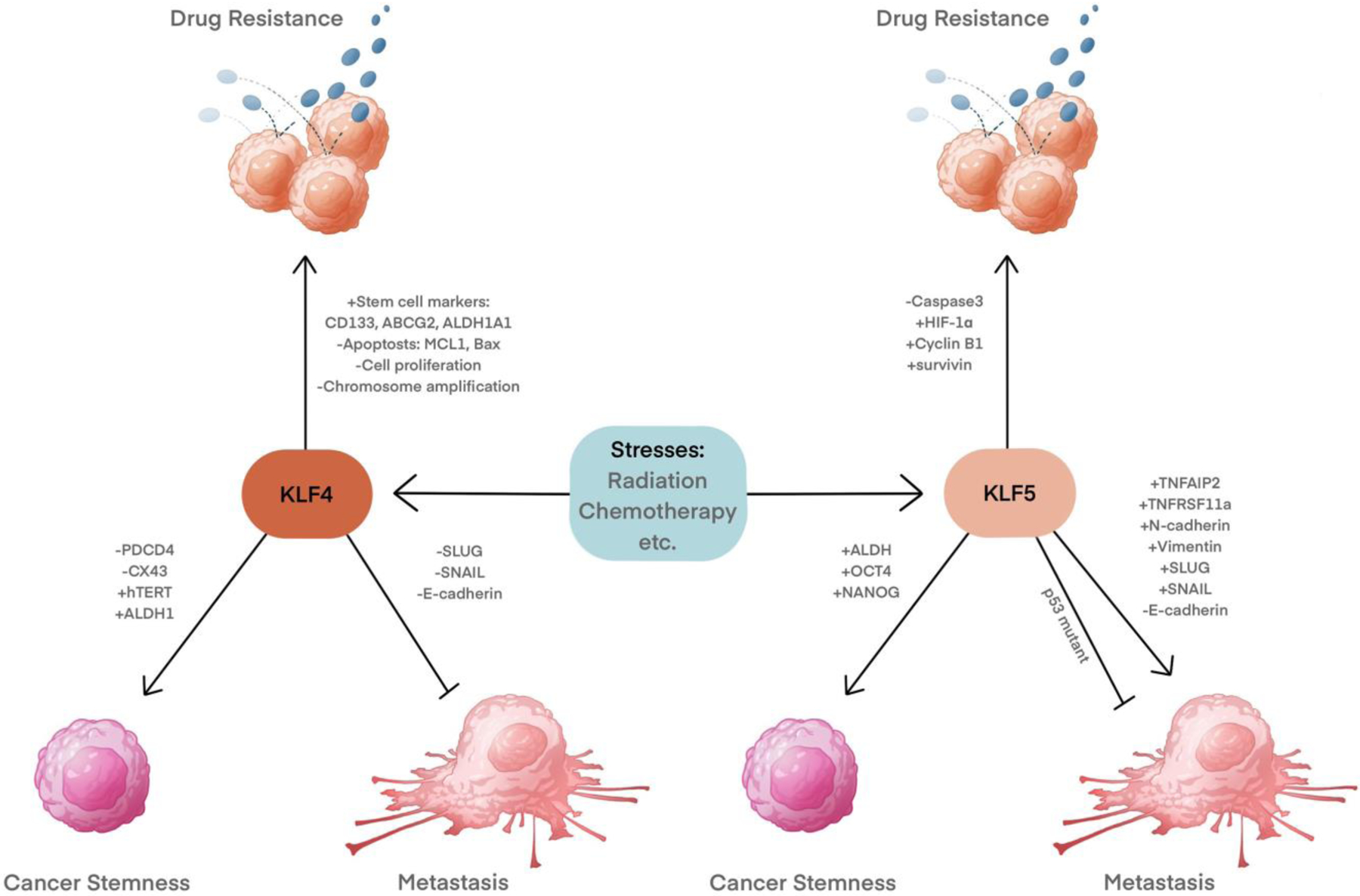
KLF4 and KLF5 regulate metastasis, cancer stemness and drug resistance.
Cancer pathogenesis
KLF4 and KLF5 exert contrasting effects on cell proliferation in many instances; while KLF4 is an inhibitor of cell growth, KLF5 stimulates proliferation [66] (Fig. 3). KLF4 inhibits cell proliferation by regulating the transcription of cell cycle genes. KLF4 transactivates the expression of cyclin-dependent kinase inhibitor 1B (CDKN1B, also known as p27 or Kip1) and cyclin-dependent kinase inhibitor 1A (CDKN1A, also known as p21 or Cip1) [67–70], and down-regulates cyclin D1 [68,69,71]. These actions of KLF4 lead to cell cycle arrest at the G1-S checkpoint. KLF4 generally plays a tumor-suppressive role in various cancers by inhibiting cell proliferation. In primary hepatocellular carcinoma, KLF4 has been demonstrated to suppress oncogenic TGF-β signaling by transactivating Smad7 and loss of KLF4 could lead to the activation of TGF-β signaling and tumor promotion [72]. In non-small cell lung cancer cells, KLF4 can also inhibit tumor cell proliferation by suppressing TGF-β1 mediated ERK/JNK/NF-κB signaling pathways [73].The absence or reduced expression of KLF4 has been found in a variety of cancers, including bladder cancer [74], pancreatic carcinoma [67,69], lung carcinoma [68], medulloblastomas [75], cervical carcinoma [70], colon cancer [76,77], hepatocellular carcinoma [78], and neuroblastoma [79]. However, KLF4 also plays an oncogenic role in breast cancer [80] and squamous cell carcinomas [81]. KLF4 overexpression was detected in about 70% of primary human breast cancer [80]. Ductal carcinoma in situ also exhibits similar KLF4 expression, suggesting that KLF4 overexpression may be an early event in breast carcinogenesis [80]. In addition, KLF4 shows stage-specific effects in some cancer types, such as esophageal squamous cell carcinogenesis (ESCC). The downregulation of KLF4 in early-stage ESCC, is required for esophageal tumorigenesis while in later stages, the expression of KLF4 is restored to promote tumor invasion and metastasis [82]. The contrasting roles of KLF4 in cancer cell proliferation can be partially explained through CDKN1A inactivation [81], the oncogenic RASV12 mutation, or cyclin-D1 overexpression (a common target of KLF4 and RAS) [83]. KLF5 can also partially contribute to this switch as the oncogenic RASV12 induces the expression of KLF5, which stimulates the proliferation of colorectal cancers [84]. In addition, KLF4α, an alternatively spliced isoform of KLF4 that is upregulated in pancreatic cancer, suppresses the expression of CDKN1A and CDKN1B to promote the cell cycle and enhance cancer progression [85].
Figure 3. KLF4 and KLF5 regulate cell proliferation in a context dependent manner.
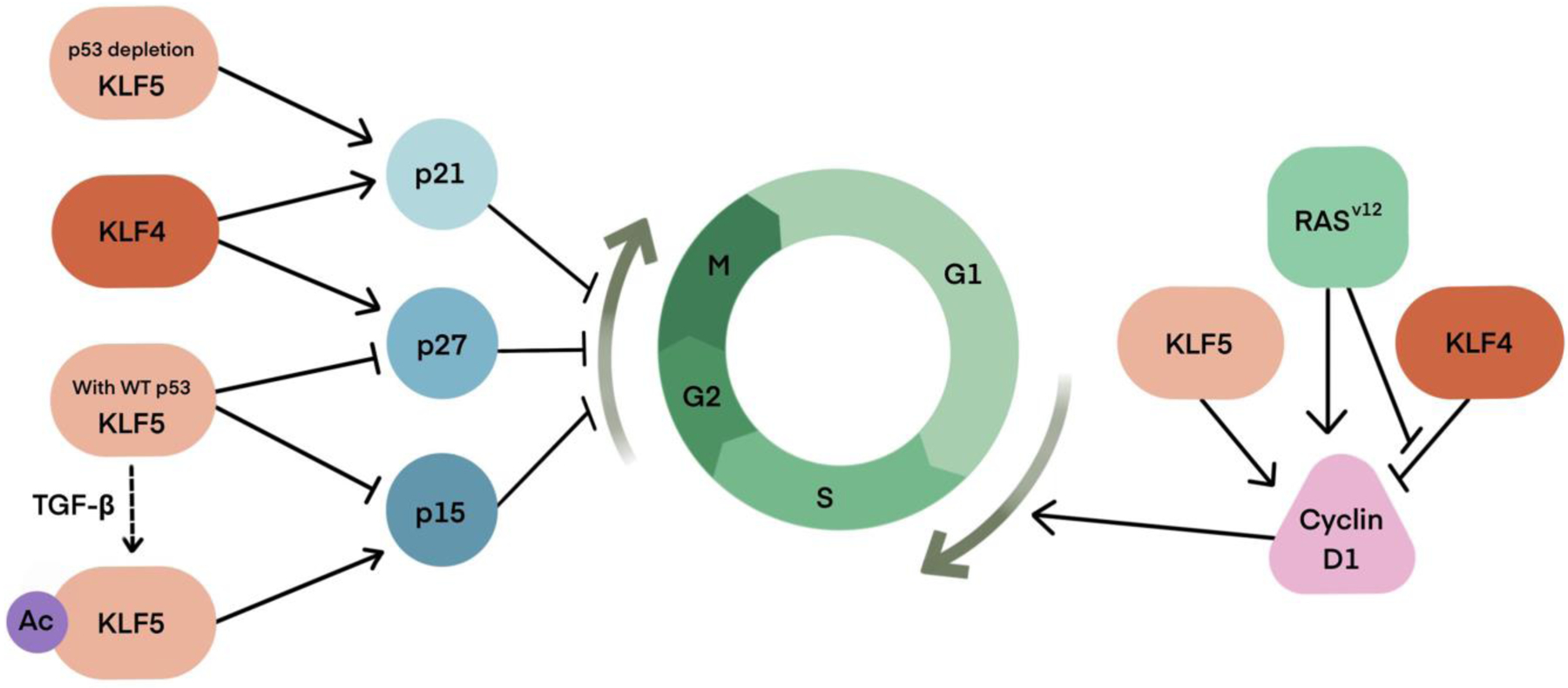
Generally, KLF4 inhibit cell cycle and KLF5 stimulates proliferation by regulating the transcription of cell cycle genes. But the cytostatic action of KLF4 can be neutralized by CDKN1A (p21) inactivation, the oncogenic RASV12 mutation, or cyclin-D1 overexpression (a common target of KLF4 and RAS). The cell cycle regulation function of KLF5 can be switched by the status of p53, which is a frequently inactivated tumor suppressor in cancers and posttranslational modification, especially acetylation. When p53 is mutated or KLF5 is acetylated at Lys369, which can be triggered by the TGF-β signaling, KLF5 becomes anti-proliferative through transactivating CDKN1A (p21) or CDKN2B (p15), respectively.
In contrast to KLF4, KLF5 is generally pro-proliferative in normal cells [26]. KLF5 promotes cell proliferation by accelerating the transition of G1/S and G2/M phases of the cell cycle. KLF5 transactivates the expression of cell cycle genes, such as cyclin D1 [86], cyclin B1, and Cdc2 [87], while repressing the expression of the cell cycle inhibitory proteins such as p27 and p15 [88]. KLF5 promotes the progression of various cancers, including pancreatic cancer, gastric cancer, and breast cancer by advancing the cell cycle through transcriptional regulation of cell cycle related genes [89–92].
However, KLF5 could be a potential tumor suppressor in other epithelial cancer types, such as prostate cancer and certain breast cancers. KLF5 deletion is frequently reported in breast [93] and prostate [94] cancers. KLF5 is frequently deleted or downregulated in prostate cancer and is associated with poor prognosis [89,95,96]. KLF5 knockdown increases prostate cancer cell proliferation in androgen receptor-positive cell models [97]. The context-dependent function of KLF5 in cancer progression could be partially determined by the status of p53, which is the most frequently inactivated tumor suppressor in cancers [98–100], and the switch of its transcriptional function by posttranslational modification [101–104]. In keratinocytes and ESCC cells, when p53 is wild-type, KLF5 is pro-proliferative. In addition, KLF5 and p53 work coordinately to induce the expression of BIRC5 (encoding survivin) and hypoxia-inducible factor (HIF1α), and therefore inhibit apoptosis [105]. However, when p53 is mutated, KLF5 becomes anti-proliferative through transactivating CDKN1A (p21) [98] and NOTCH1 [99], thus working as a potent tumor suppressor.
Recently, another KLF family member, KLF7, was found to play a oncogenic role in a variety of cancers, including pancreatic cancer [106], hepatocellular cancer [107], gastric cancer [108], endometrial cancer [109] and squamous carcinoma [110] and could be a potential therapeutic target. In high-grade serous ovarian cancer, KLF7 functions as an oncogene to promote tumor growth and serves as an unfavorable prognostic marker for overall survival in late-stage TCGA-OV and GSE26712 HGSOC cohorts. In addition, the downstream targets of KLF7 are involved in epithelial to mesenchymal transition, and the maintenance of pluripotency and self-renewal characteristics of cancer stem cells [111]. In pancreatic cancer, KLF7 promotes pancreatic cancer growth and metastasis by up-regulating ISG expression and maintaining Golgi complex integrity [106]. In hepatocellular cancer, the KLF7/VPS35 axis promoted HCC cell cycle progression by activating the Ccdc85c-medicated β-catenin pathway [107]. In gastric cancer, KLF7 promoted gastric carcinogenesis via upregulation of ANTXR cell adhesion molecule 1 (ANTXR1) [108].
The role of SOX factors in tumorigenesis and cancer progression has been the subject of several recent review articles [58–65]. SOX2, SOX4, and SOX9 are overexpressed in a wide variety of tumors, and SOX4 overexpression is associated with poor prognosis [112], metastasis [113], and EMT [114].
The roles of KLFs and SOXs in tumorigenesis have also been demonstrated in murine models. Generally, KLF4 deletion promotes tumorigenesis. Intestinal epithelium specific ablation in KLF4Loxp/Loxp/ AAV7-Cre-mCherry mice are more susceptible to DNA damage and increased genomic instability when challenged with genotoxic stress like radiation, suggesting KLF4 knockout may create a tumor permissive environment and facilitate gastric tumorigenesis [115]. Li et al also showed that conditional deletion of KLF4 in villin-positive antral mucosa cells (Villin-Cre(+);Klf4(fl/fl) mice) has greater chemical-induced gastric carcinogenesis and tumor promotion [116]. Similarly, Ghaleb et al. found that Klf4Δ/S/ApcMin/+ mice with in vivo KLF4 deletion leads to increased colonic adenomas as compared to the WT Klf4fl/fl/ApcMin/+ mouse [117]. It has also been shown by adoptive transfer studies that KLF4 deletion in CCR2+ myeloid-derived suppressor cells results in decreased lung metastases when mice are inoculated with melanoma or breast cancer cells [118].
In contrast to KLF4, KLF5 deletion tends to inhibit tumorigenesis in mouse model studies. Heterozygous deletion of KLF5 can inhibit intestinal tumor formation [45,119]. In one mouse model, tamoxifen-induced KLF5 deletion in Lgr5(+) stem cells suppressed cell proliferation and prevented oncogenic transformation [45]. In breast cancer, mammary gland-specific KLF5 conditional knockout mice showed decreased breast epithelial cell proliferation, reduced mammary cell survival and inhibited PyMT-induced tumorigenesis [120].
SOX2 was also found to regulate gastric stem cell self-renewal and epithelial homeostasis. Loss of SOX2 in gastric cells promotes Wnt-driven tumorigenesis. Sarkar et al generated APC knockout (APC KO) and APC/SOX2 double-knockout (DKO) mice and found significant increase in the number of adenomas in tamoxifen treated DKO mice compared to APC KO mice [121].
Cancer cell metastasis is a prime example of tumor cell plasticity, and most tumor cells have inherent plasticity [122]. Tumor cells often undergo the epithelial–mesenchymal transition (EMT) for migration for cancer metastasis. EMT is typically characterized by downregulation of the cell adhesion molecule E-cadherin. Transcriptional regulators, such as SNAIL and SLUG, drive EMT by suppressing the expression of E-cadherin [123]. KLF4 also plays a multi-faceted role in tumor metastasis. Some reports indicate that KLF4 works as a stem cell factor to maintain stemness required to promote EMT, as the expression of KLF4 along with other stem cell factors is altered by different factors regulating EMT, including ZEB1,p53, NF-kB and ALDH1 [124–127]. KLF4 is required to promote migration and invasion of the breast cancer cell lines, MCF-7 and MDA-MB-231, via the Notch pathway [128]. Consistently, KLF4 also promotes mesenchymal properties in colorectal cancer stem cells by upregulating EMT factors, such as SNAIL via the TGF-β1/SMAD pathway [129]. However, in mouse and human prostate cancer cells, KLF4 works with FOXA1, which directly inhibits the transcription of SLUG, the dominant regulator of TGF-β induced prostatic EMT [130]. In addition, KLF4 overexpression in 4T1 cells reduces the expression of SNAIL, a key mediator of EMT and metastasis in breast cancer [131].
Similar to KLF4, the role of KLF5 in metastasis also seems to be context-dependent. In some cases, KLF5 seems to promote metastasis. For example, KLF5 transactivates tumor necrosis factor-α (TNFα)-induced protein 2 (TNFAIP2), which in turn increases the activity of two GTPases: Rac1 and Cdc42, to change actin cytoskeleton and cell morphology, and thus promotes cell migration and invasion in breast cancer cells [132]. In agreement with this, KLF5 promotes cervical cancer proliferation, migration and invasion partly by transactivating tumor necrosis factor receptor superfamily member 11a (TNFRSF11a) [133]. KLF5 also promotes EMT in laryngeal carcinoma Hep-2 cells, where KLF5 knockdown increases E-cadherin, while decreasing N-cadherin, Vimentin, and the regulatory factors SNAIL and SLUG along with the attenuation of EMT [134]. In contrast, in the human non-small cell lung cancer (NSCLC) cell line, A549, KLF5 knock-down, decreases E-cadherin expression [135]. The function of KLF5 in EMT also seems to depend on the activity of p53, [98,99,105,136]. In the absence of p53, KLF5 inhibits EMT in liver cancer cells by suppressing ZEB2 protein expression via inducing miR-192 [136]. Reciprocally, in primary human keratinocytes harboring mutant p53, KLF5 depletion induces EMT as confirmed by the induction of SNAIL, TWIST, and the suppression of E-cadherin [99].
SOX4 is a master regulator of EMT [114] and regulates EMT in response to TGFβ signaling[137–140]. In gastric cancer cells, SOX4 promotes EMT by upregulating the expression of EMT transcription factors, including Twist1 and ZEB1 [138]. TGFβ signals can drive SOX4 to induce apoptosis, but KLF5 cooperates with SOX4 to prevent TGFβ-induced apoptosis [139] (Fig. 5). Both SOX4 and SOX9 have been shown to play critical roles in metastasis [113,137,141–144].
Figure 5.
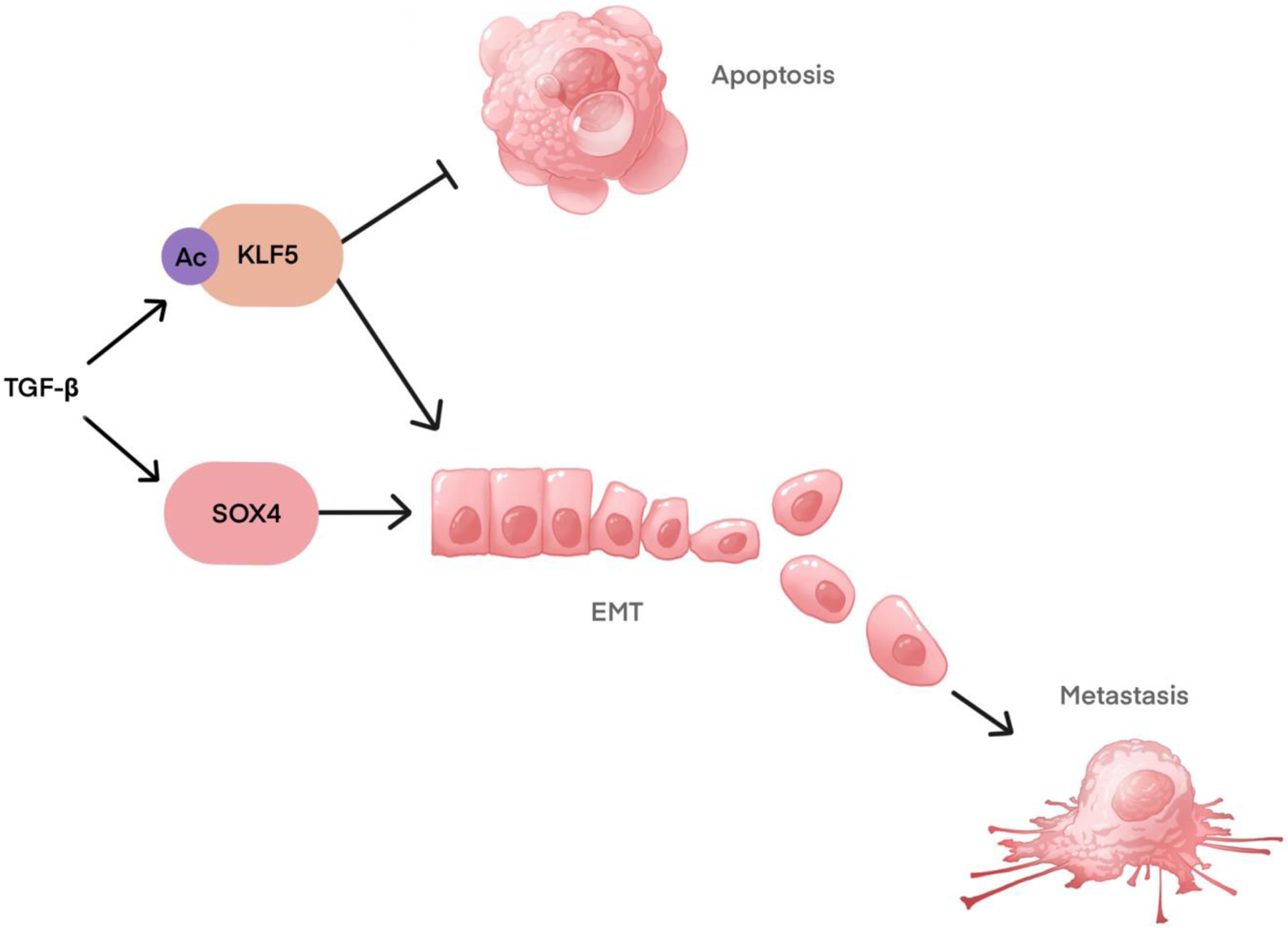
Interaction of SOX4 and KLF5 with TGFβ in apoptosis, EMT, and metastasis.
Cancer stem cells
In the cancer stem cell (CSC) theory, a subset of cells called CSCs are predisposed to drive self-renewal and differentiation to produce a diverse population of daughter cells [145]. Recently, pluripotency factors, including KLF4 and KLF5, have been rediscovered as the drivers of cancer stemness. KLF4 and KLF5 are preferentially expressed within cancer stem-like cells [146]. KLF4 is highly associated with the stemness of cancer cells. In both cancer cells and normal stem cells, KLF4 is required to maintain telomerase activity for the long-term proliferative potential of these cells through transactivating the expression of human telomerase reverse transcriptase (hTERT) [147]. In human osteosarcoma cells, KLF4 overexpression promotes tumorsphere formation, the transcription of stemness-associated genes, chemoresistance and distant metastasis [148]. KLF4 promotes stem cell-like characteristics partially by activating the p38 mitogen-activated protein kinase (MAPK) signaling pathway [148]. In breast cancer, KLF4 expression is increased selectively in the mammary cancer stem-like cells (MaCSCs) of cultured human triple-negative breast cancer (TNBC) cell lines, and the aldehyde dehydrogenase high MaCSCs derived from xenografted human mammary carcinomas [149]. In MaCSCs, KLF4 exerts pro-survival signaling by inhibiting pro-apoptotic signaling molecules like programmed cell death 4 (PDCD4) and connexin 43 (CX43) through its downstream effector, microRNA-206 [149]. In addition, KLF4 promotes the transcription of ALDH1, which induces and functionally marks CSCs [150]. In colorectal cancer, KLF4 is enriched in Lgr5+CD44+EpCAM+ cancer stem cells and required to maintain stemness via the TGF-β1 pathway [129]. In pancreatic acinar cells, KLF4 induces acinar-to-ductal cell reprogramming by inducing the ductal specific marker Krt19 expression and suppressing Amylase, Ptfa1, and CAII. In addition, KLF4 synergizes with K-Ras mutations to initiate the formation of premalignant pancreatic intraepithelial neoplasia (PanIN) [151]. Another study showed that KLF4 mediates pancreatic cancer cell stemness promoted by secreted mucin 5AC (MUC5AC) [152].
KLF5 has also been reported to promote stemness in various cancers, including lung cancer [153], hepatocellular carcinoma [154], and triple-negative breast cancer [155]. In lung cancer, KLF5 mediates cell stemness promoted by α-Catulin. Mechanistically, α-Catulin interacts with the C-terminal region of KLF5, and such an interaction inhibits WWP1-mediated KLF5 degradation and thus increases KLF5 protein levels [153]. KLF5 knockdown blocks α-Catulin-driven cancer stemness as measured by sphere formation, ALDH activity, and stemness factors OCT4 and NANOG [153]. In hepatocellular carcinoma, KLF5 overexpression enriches cancer stem-like cell populations, promotes colony-formation, and enhances drug resistance [154]. In triple-negative breast cancer (TNBC), KLF5 degradation that is triggered by metformin treatment reduces the percentage of stem cells marked with high ALDH activity [155]. Taken together, KLF4 and KLF5 could be potential therapeutic targets for cancer treatments to prevent cancer recurrence.
In breast cancer, a SOX2-SOX9 signaling axis is essential for maintenance of breast luminal progenitor cells, Wnt signaling, and expression of ALDH1A3 [156]. Consistent with these data, SOX2 and SOX9 were both top hits in a genome-wide CRISPR screen for genes essential for glioma stem cells [157]. Both SOX9 [158] and SOX4 [52] have been shown to be important in breast cancer stem cells [50]. In organoids derived from a PyMT genetic mouse model, SOX4 inhibits differentiation of breast cancer by reactivating a set of genes enriched in fetal mammary stem cells (fMaSC) [52]. In colorectal cancer, cells with high activity of the SOX2 promoter, indicated by Sox2 promoter dependent DsRed fluorescence, exhibit typical asymmetric cell division and high stem cell markers [159]. Consistently, SOX2 overexpression in colorectal cancer cells drives cancer stem characteristics [160]. In gastric cancer cells, SOX4 promotes cancer stemness through pluripotent factors SOX2 and OCT4 [138].
Drug resistance
Several studies have shown that conventional chemotherapy and ionizing radiation enhance the stemness of cancer cells and induce drug-resistant cancer stem cells (CSCs) [161–164]. This phenotypic switch is tightly interconnected with the concept of tumor cell plasticity and is associated with transcriptional reprogramming [2,7]. KLF4, KLF5, and SOX factors have been shown to modulate therapeutic responses in cancer.
In normal tissue, KLF4 and KLF5 function as stress response genes and are induced by various noxious stimuli to support cell survival. The expression of KLF4 is induced by shear stress [165] and pro-inflammatory stimuli [166] in endothelial cells. KLF4 can also be induced by injury in normal adult vascular smooth muscle cells [28]. Like KLF4, KLF5 also protects cells from various external cellular stresses. Part of this function of KLF5 is achieved by modulating apoptosis signaling. In pulmonary artery smooth muscle cells, KLF5 upregulates survivin, which hyperpolarizes mitochondria membrane potential and inhibits apoptosis [167,168]. In contrast, KLF5 knockdown increases apoptosis coupled with activation of caspase-3 and pro-apoptotic Bad in neurons [169].
Consistent with their pro-survival roles in normal tissues, KLF4 and KLF5 also protect cancer cells from a variety of harmful stimuli, including various drug treatments. KLF4 and KLF5 proteins are upregulated in human breast cancer cells after treatment with the HER2/EGFR inhibitor lapatinib and induce the anti-apoptotic factor, myeloid cell leukemia 1 (MCL1), to mediate resistance to lapatinib [170] (Fig. 4). Consistently, in human breast cancer, KLF4 deficient cancer cells show poorer survival under different stresses, even though they proliferate faster [149,170].
Recent studies have pinpointed the role of KLF4 in regulating DNA damage response and DNA repair, resulting in maintaining genome stability [171–175]. Upon DNA damage, KLF4 is arginine methylated by PRMT5, whose methylation reduces its basal ubiquitination, thus stabilizing KLF4 [174]. DNA damage-induced upregulation of KLF4 causes cell cycle arrest by transactivation of the cell cycle inhibitor p21Cip1/Waf1 that induces p53-dependent G1/S arrest [171–175], and suppressing the expression of cell cycle-promoting genes, including CCND1 (cyclin D1) [71] and CCNB1 (cyclin B1) [176]. In addition, KLF4 prevents chromosomal amplification following DNA damage by transcriptionally suppressing cyclin E expression [177]. In addition to regulating p21Cip1/Waf1, another critical function for KLF4 is modulating cellular apoptosis by inhibiting the expression of pro-apoptotic Bax [173–175]. In addition to ubiquitination in response to DNA damage, KLF4 is PARylated by PARP1, facilitating KLF4 recruitment from the soluble nucleus to the chromatin and its transcriptional activity [175]. Moreover, recent bioinformatics analyses have unraveled the previous undocumented role for KLF4 in transcriptionally regulating BRCA1 and further governing homologous recombination (HR) DNA repair [175]. Taken together, the DNA damage-induced ubiquitination and PARylation of KLF4 promote cellular survival under the circumstance of genotoxic stress through arresting cell cycle progression and enhancing HR DNA repair by regulating BRCA1, resulting in the maintenance of genome stability.
KLF4 also mediates doxorubicin-induced drug resistance in cancer cells. In osteosarcoma, doxorubicin treatment induces the upregulation of KLF4 along with a stem-like phenotype, shown as the increased transcriptional level of stem cell-related markers (CD133, ABCG2, and ALDH1A1) and high metastatic capacity (Fig. 4). KLF4 depletion with siRNA blocks these doxorubicin-induced phenotypes, showing that KLF4 is required to stimulate the formation of CSCs in response to doxorubicin treatment [178]. Consistently, KLF4 depletion sensitizes breast cancer cells to chemotherapy agents such as doxorubicin and cisplatin [175].
KLF5 is frequently overexpressed in non-small cell lung cancer (NSCLC), and increased KLF5 prevents cells from undergoing apoptosis induced by hypoxia and cisplatin treatment [179,180]. Consistently, high KLF5 expression is associated with a poor prognosis of NSCLC [179,180]. In NSCLC cells, KLF5 is upregulated by hypoxia. The increased KLF5 promotes the expression of hypoxia-inducible factor 1α (HIF-1α) and forms a complex with HIF-1α. The KLF5/HIF-1α complex upregulates cyclin B1 and survivin, while suppressing caspase-3 and therefore inhibiting apoptosis [179]. KLF5 can also protect cells from genotoxic stress by promoting DNA repair. In NSCLC, KLF5 affects the phosphorylation of H2AX at S139 to promote DNA repair after cisplatin treatment by regulating Chk1/Chk2 kinase levels [180]. In contrast, KLF5 knockdown attenuates the activation of checkpoint kinases Chk1 S345 and Chk2 T68, permitting cells with damaged DNA to enter mitosis and leading to mitotic catastrophe [180]. Thus, KLF5 promotes cell survival under stress by enhancing DNA repair and preventing cells with damaged DNA from entering mitosis.
Different treatments also modulate the expression of KLF5 to modulate therapeutic responses. KLF5 promotes resistance to endocrine therapy in advanced prostate cancer [181]. Endocrine therapy that inhibits the androgen receptor (AR) is frequently used in the treatment of recurrent prostate cancer but leads to the progression to castration-resistant prostate cancer (CRPC) in most cases. In these CRPC tumors, the expression of KLF5 is induced during endocrine therapy and works as an oncogene to promote cell migration and colony formation [181]. However, KLF5 is required to promote the sensitivity to docetaxel by inhibiting autophagy in prostate cancer cells and the expression of KLF5 is reduced by docetaxel treatment through the AMPK/mTOR/p70S6K signaling pathway [95]. Thus, the role of KLF5 in drug resistance could be treatment dependent. When targeting KLF5 for drug resistance the interaction of KLF5 with other therapies needs to be considered.
In nasopharyngeal carcinoma, loss of miR-34 can upregulate SOX4 and leads to cisplatin resistance [182]. SOX4 also mediates cisplatin resistance in non-small cell lung cancer cells [183,184], and 5-FU in colorectal cancer [185]. SOX9 also mediates cisplatin resistance in NSCLC via regulation of ALDH1A1 [186]. SOX9 can mediate resistance to tamoxifen in breast cancer [187], gefitinib in lung cancer [188], oxaliplatin in colon cancer 42[189], and temozolomide in glioblastoma [190]. SOX9 increases sorafenib resistance in liver cancer via upregulation of ABCG2 [191]. SOX2 mediates resistance to paclitaxel in melanoma via upregulation of ABCC1 [192]. Both SOX4 [193], SOX9 [194], and SOX2 [195] can promote resistance to T-cell mediated immune killing via multiple mechanisms.
The regulation of KLF4, KLF5 and SOXs
The expression of KLF4 can be regulated by epigenetic modifications and various signals. Reduced transcription of KLF4 due to hypermethylation of the KLF4 promoter region has been detected in primary lung carcinoma, cervical cancer, and medulloblastoma [68,75,196]. The expression of KLF4 is induced by the LIF/STAT3 pathway [32–34], RAS/RAF/MEK/ERK signaling [197], luteinizing hormone [198], shear stress [165] and proinflammatory stimuli [166]. Several microRNAs have been found to suppress the expression of KLF4 by directly targeting its 3’-untranslated region [199–203].
The protein level and transcription activity of KLF4 can be regulated by various post-translational modifications. Several E3 ligases, including VHL, the Cdh1 subunit of the anaphase promoting complex (APC), the F-box proteins βTrCP, and Mule can mediate the ubiquitination of KLF4 for proteolytic degradation [77,174,204–210]. ATXN3, a deubiquitinating cysteine enzyme, deubiquitinates KLF4 in breast cancer [208]. In addition, methylation by PRMT5 stabilizes KLF4 in triple negative breast cancer (TNBC) [211]. The transcriptional activity of KLF4 is regulated by several posttranslational modifications, including acetylation, SUMOylation, PARylation, and phosphorylation. The p300/CBP complex acetylates KLF4 to promote the transactivation function of KLF4 [81,212,213], while HDAC2 promotes KLF4 deacetylation [213]. SUMOylation is required for KLF4 to inhibit cell proliferation [214,215] and inhibit its transactivation of NANOG, which mediates the induction of iPSCs [216]. Upon DNA damage, KLF4 is PARylated, which enhances its binding to the CDKN1A and Bax promoters, resulting in cell cycle arrest [175]. TGF-β1 Induces KLF4 phosphorylation through Smad and p38 MAPK pathways in vascular smooth muscle cells [217].
Focal amplification of super enhancers near KLF5 leads to KLF5 expression in squamous cell carcinomas [218]. Various signaling pathways also regulate the transcription of KLF5. The expression of KLF5 is inhibited by retinoic acid receptor signaling [219], and upregulated by androgen receptor signaling [91,220], progesterone [92], tumor necrosis factor (TNF)-α [133], oncogenic KRAS mutation (KRASV12G) [84], and by lysophosphatidic acid (LPA) [221] in different cancers. In addition, the expression of KLF5 can also be inhibited by multiple microRNAs, including miR-153 [222], miR-217 [223], miR-5195–3p [224], miR-590–5p [225], miR-145–5p [226] and miR-320 [227] in different cancers.
KLF5 is also regulated by various posttranslational modifications, including ubiquitination, SUMOylation, phosphorylation and acetylation. KLF5 can be ubiquitinated by several E3 ubiquitin ligases for proteasomal degradation, including FBW7 [218,228,229], WWP1 [153,230], EFP [231], and SMURF2 [232]. In addition, three deubiquitinases (DUBs), ubiquitin-specific protease 3 (USP3), BRCA1-associated protein 1 (BAP1), and ataxin-3 like (ATXN3L), can stabilize KLF5 via deubiquitination [233–235]. SUMOylation is required for the nuclear accumulation of KLF5 [236]. Furthermore, SUMOylation also switches the transcriptional activity of KLF5 by regulating its interaction with transcription corepressors and coactivators [101]. Phosphorylation affects the binding of KLF5 to various effector proteins and therefore regulates its stability and transcriptional activity [229,237,238]. Acetylation of KLF5 can switch the transcriptional regulation effect of KLF5. For example, in epithelial cells, TGF-β recruits acetylase p300 to acetylate KLF5 at Lys369 [102,103]. Such acetylation in KLF5 reverses its transcriptional suppression of p15 (CDKN2B) to activation. In contrast, KLF5 transcriptional activation of MYC changes to repression, thus reversing the proliferation-promoting ability of KLF5 [102,103]. In addition, acetylation of KLF5 at Lys 369 can be deacetylated by histone deacetylase 1/2 (HDAC1/2), and the deacetylation promotes KLF5 degradation [239].
SOX9 has been shown to be directly transactivated by YAP, mediating stem-like properties in esophageal cancer cells [240]. SOX2 can also be transcriptional induced by YAP through its interaction with OCT4 in non-small cell lung cancer (NSCLC) cells [241]. SOX proteins are also regulated post-transcriptionally by many miRNAs [60,62] and post-translationally by multiple mechanisms [242]. Post-translational regulation of SOX proteins includes phosphorylation, acetylation, SUMOylation, methylation, and ubiquitylation and has been recently reviewed elsewhere [242]. SOX2 and SOX9 have been shown to undergo all of these modifications, whereas SOX4 PTMs are less well studied, and only acetylation of SOX4 has been demonstrated [243]. Acetylation of SOX factors promotes nuclear localization, methylation impacts pioneer factor activity, phosphorylation enables transcriptional activation, and SUMOylation reduces DNA binding activity [242].
In addition to the aforementioned regulations, the expression of KLF4, KLF5, SOX2, SOX4, SOX9 can be induced by hypoxia to promote cancer progression. Hypoxia is a common phenomenon in many solid tumors and strongly associated with metastasis, stemness, and drug resistance. The hypoxia-inducible factors (HIFs) are master regulators of hypoxia response through the transcriptional regulation of hypoxia associated genes [244]. Hypoxia promotes stemness of cancer cells through upregulating pluripotent factors, including KLF4 and SOX2 [245]. In glioblastoma, SOX2 and KLF4 are upregulated through HIF1α and HIF2α under hypoxia, contributing cancer stemness and chemotherapy resistance to temozolomide TMZ [246].
In human vascular smooth muscle cells, KLF4 is upregulated by HIF1α under hypoxia to mediate hypoxia-induced migration [247]. Interestingly, a KLF4 mutation, KLF4K409Q, found in meningioma patient samples, potentiates hypoxia signaling by inhibiting oxygen dependent degradation of HIF-1α [248].
Hypoxia induces the expression of KLF5 in various cancer cells to promote cancer survival [179,249,250]. In non-small cell lung cancer (NSCLC) cells, KLF5 promotes hypoxia-induced survival and inhibits apoptosis by interacting with HIF-1α and mediating its upregulation under hypoxia [179]. In addition, KLF5 also mediates hypoxia-induced cisplatin resistance by promoting the HIF-1α-dependent glycolysis via the PI3K/Akt/mTOR pathway in NSCLC cells [250]. Consistently, KLF5 also transactivates HIF-1 in colon cancer cells [105]. The expression of KLF5 can be regulated by HIF-1α reciprocally. In pancreatic cancer cells, KLF5 coimmunoprecipitated with HIF-1α and HIF-1α knock down reduces KLF5 [249]. Recent studies indicated that SOX2 could be an important mediator of hypoxia induced stemness and metastasis. SOX2 expression can be induced by hypoxia through HIF-1α and HIF-2α in prostate cancer cells [251], endometrial cancer stem cells [252], glioblastoma cells [246] and epithelial ovarian cancer cells [253] to promote malignant progression. In human embryonic stem cells (hESC), HIF-2α binds directly to predicted hypoxic response elements (HREs) in the proximal promoter of SOX2 under hypoxia [254]. Meanwhile, SOX2 has also been reported to mediate the upregulation of HIF-1α in various cancers including breast cancer [255], esophageal squamous cell carcinoma (ESCC) [256] and gastric cancer [257], promoting cancer metastasis. For example, in breast cancer cells, hypoxia induced SOX2 upregulates the expression of HIF-1α by transactivating NEDD9 and therefore promotes hypoxia-induced breast cancer cell migration [255].
The expression of SOX4 can be upregulated by HIF-1α via a lncRNA, CASC15. In NSCLC, HIF-1α transactivates CASC15, and CASC15 subsequently promotes the expression of its neighboring gene SOX4 [258]. The upregulation of SOX9 expression under hypoxia has been reported in various cancers, including ovarian cancer [259], colorectal cancer [260] and pancreatic cancer [261]. In ovarian cancer cell A2780, SOX9 promotes cell survival under hypoxia and depletion of HIF-2α abolishes the expression of SOX9 [259]. In colorectal cancer (CRC) cells, upregulated SOX9 under hypoxia promotes the EMT by stabilizing SNAIL via the transactivation of a deubiquitinating enzyme, ubiquitin-specific protease 47 (USP47) [260]. Interestingly, SOX9, induced by hypoxia in a nonmetastatic pancreatic cell line FG (LMET), becomes constitutively overexpressed in a corresponding metastatic derivate L3.6pl (HMET) cell line independently of hypoxia. In the metastatic cell line, SOX9 functions as an alternative transcription factor to induce the expression of hypoxia-associated genes independent of hypoxia to support growth, angiogenesis, and metastasis [261].
Targeting KLF4, KLF5, and SOXs in cancer treatment
Given their crucial roles in cancer pathogenesis, cancer cell stemness, and drug resistance, KLF4, KLF5 and SOXs are potential therapeutic targets. But for transcription factors without a ligand, direct targeting has been considered challenging. Thus, previous efforts have focused on targeting these transcription factors by modulating their upstream regulators, including transcription, mRNA stability, and protein degradation with small molecules.
In breast cancer cells, Kenpaullone inhibits the transcription of KLF4 [128]. Treatment with Kenpaullone suppresses cancer stem cell self-renewal and cell migration and sensitizes breast cancer cells and xenograft tumors to paclitaxel [150]. Application of PKA inhibitors, including H89 and 14–22 Amide (PKI), enhances the transcription of KLF4 expression [128,198]. Under these circumstances, when KLF4 works as a tumor suppressor, induction of KLF4 could have potential for cancer treatment. For example, APTO-253, initially discovered to be cytotoxic to human colon adenocarcinoma HT-29 cells, is a prominent inducer of KLF4 in various cancer cell lines. A Phase 1 study of APTO-253 in patients with advanced solid tumors showed stable disease for 3.6–8.4 months in 5 of 21 (24 %) patients, and APTO-253 toxicity was well tolerated in this clinical trial [262]. Three potential KLF5 inhibitors, wortmannin (a PI3K/AKT inhibitor), AG17, and AG879, were screened out from a library containing 1,280 biologically active compounds and found to be able to inhibit cell proliferation in vitro [263]. Mifepristone, a drug used for abortion, exerts its anti-tumor activity by inducing the expression miR-153 to down-regulate the expression of KLF5. Inhibition of KLF5 by mifepristone inhibits tumor growth of patient-derived xenografts, and reduces the population of CSCs [222]. Similarly, crocin also suppresses KLF5 expression via upregulation of a microRNA, miR-320, which mediates the inhibitory function of crocin on EMT, migration, and invasion of gastric cancer cells [227]. Short interfering RNA has also been applied to knock down KLF5. A “wrapsome” (WS) enveloped by a neutral lipid bilayer and hydrophilic polymers with siRNA targeting KLF5 and a cationic lipofection complex was used to transfect siRNA into tumors to knock down KLF5 and inhibit tumor angiogenesis in vivo [264].
Statins, the cholesterol-lowering agents, such as both simvastatin and atorvastatin, remarkably reduced the amount of KLF4 protein levels in osteosarcoma cells. The application of statins reduces CSC properties induced by doxorubicin treatment in osteosarcoma cells [178]. Similarly, curcumin, a natural compound with many anticancer properties, promotes the degradation of KLF5 by downregulating YAP/TAZ [265], which interacts with the PY motif of KLF5 and protects it from WWP1-mediated ubiquitination [266,267]. In addition, the degradation of KLF5 can be triggered by metformin, a first-line drug for type 2 diabetes mellitus. Metformin inhibits protein kinase A (PKA) activity, which in turn induces glycogen synthase kinase-3β (GSK3β)-mediated KLF5 phosphorylation and degradation [155]. Consistently, one of the direct target genes of KLF5, HNF4α, which is required for gastric cancer (GC) proliferation, is also downregulated by metformin treatment [268].
Targeting KLF4 could also modulate the efficacy of cancer immunotherapy as KLF4 is also implicated in the regulation of immunity. KLF4 was previously reported to be induced in response to proinflammatory signals, such as interferon-γ (IFN-γ), lipopolysaccharide (LPS) and tumor necrosis factor-α (TNF-α) to promote macrophage activation [269]. However, recent studies show that KLF4 plays an anti-inflammatory role through regulating macrophage polarization [270]. Macrophages are polarized into M1 or M2 states. M1 is considered pro-inflammatory and anti-tumorigenic and while M2 is anti-inflammatory and pro-tumorigenic. KLF4 suppresses inflammation by promoting M2 polarization macrophages while inhibiting M1 polarization [270]. Consistently, KLF4 deficient macrophages show increased expression of proinflammatory genes [270]. In cancer immunotherapy, a widely used immune checkpoint inhibitor, PD-L1 inhibitor, promotes polarization of macrophages to a proinflammation state partially by reducing the expression of KLF4 via upregulating the expression of KLF4-targeting miR-34a [271]. The high expression of KLF4 is also associated with an immunosuppressive tumor microenvironment in lung cancer [272]. In the Cancer Genome Atlas (TCGA)- lung adenocarcinoma (LUAD) cohort, KLF4 is significantly positively associated with the infiltration levels of immunosuppressive M2 macrophages [272]. In addition, a multikinase inhibitor, regorafenib, suppresses M2 polarization of macrophages and promotes CD8+ T cells partially by downregulating the transcription of KLF4, suggesting potential synergistic antitumor efficacy between KLF4 inactivation and cancer immunotherapy [273].
More recently, with the development of new technologies such as proteolysis-targeting chimeras (PROTACs) for degrading a protein of interest [274–276] and protein-protein interaction (PPI) inhibitors [277], targeting of TFs directly for cancer treatment may become a reality. Targeting TFs directly could provide a specific modulation of the transcriptional activity with fewer off-target effects and could reach the full therapeutic potential of TFs [10,12,23].
PROTACs degrade a protein of interest (POI) by hijacking the ubiquitin-proteasome system. A PROTAC is a bifunctional protein that contains the ligand of an E3 ligase and a molecule binding to POI. Therefore, it mediates the interaction of the POI with the E3 ligase, inducing its polyubiquitination, and finally promotes its degradation by the proteasomal machinery [278]. PROTAC-induced degradation offers a new strategy to target TFs with different kinds of baits, including analogs of known ligands for nuclear receptors, double-strand nucleotides containing the binding motif of TFs, and small molecules designed based on the structure of TFs [278]. Substrates of PROTACs were initially applied to target two nuclear receptors: estrogen receptor (ER) and androgen (AR) receptor, which have been implicated in the progression of breast and prostate cancer. The ability to degrade the POI has been demonstrated in vitro and ex vivo when injected into cells [279]. Taking advantage of the DNA binding ability of TFs, the challenge of lacking a defined ligand-binding pocket in most TFs can be bypassed by using double stranded oligonucleotides as a bait to recruit TFs for degradation. Nevertheless, the specificity of this strategy, known as TF-PROTAC [275], still needs to be demonstrated. In TF-PROTAC, the ligand of an E3 ligase is directly connected with a double strand oligonucleotide containing the binding motif of a TF. This method was successfully applied to induce the degradation of NF-kB and E2F, inhibiting proliferation of Hela cells [275]. Recently a highly potent PROTAC, SD-36, that targets STAT-3, has been developed and shows high selectivity for STAT3 over other STAT family members [280,281]. SD-36 is a cell-permeable small-molecule inhibitor based on the structure of SH2 domain of STAT3 that has the drug delivery advantage of a small molecule that can directly get into cells without additional assistance. In vivo studies with mouse xenograft models have shown that SD-36 is well tolerated and induces durable tumor regression [281]. Similar strategies can be expanded to design PROTAC for each TF, including KLF4, KLF5, and SOXs when suitable baits are created or screened out from chemical libraries. The structure of the DNA binding domain of KLF4 is now available and can be used to design baits binding to this domain [282].
To regulate transcription, TFs need to interact with co-activators or co-repressors to recruit or block the transcription machinery. In addition, the transcriptional activity and stability of TFs can be modulated by different posttranslational modifications, which is also mediated through PPIs with corresponding enzymes [8]. These PPIs are potential sites for drug targeting. PPI interfaces have been previously considered impossible to block with small molecules [283]. However, with the development of unbiased high throughput drug screening, huge chemical libraries, and broad cell-based reporter assays, more potential chemicals that modulate targeted PPI have been identified [277,284]. When KLF5 works as a tumor suppressor, a compound that disrupts the PPI between KLF5 and its ubiquitin ligases, such as FBW7 and WWP1 [218,230], could have the potential to suppress cancer progression. PPI inhibitors that block interactions of SOX factors with co-activator complexes could also have wide applicability for cancer therapies.
Conclusion
Considering the crucial roles of KLF4, KLF5 and SOXs in cancer progression, metastasis, cancer stemness, and drug resistance, targeting them could exert a synergic effect, provide longer-lasting clinical responses, and lead to survival benefits, which could surpass that of targeting downstream effectors of these TFs. Recently, the potential of modulating transcription factors to reprogram cancer cells to inhibit cancer metastasis has been demonstrated in preclinical models. EMT-derived breast cancer cells were differentiated into post-mitotic adipocytes with a combination of a potent agonist of peroxisome proliferator-activated receptor γ (PPARγ) and MEK inhibitors, resulting in loss their invasiveness [285]. In addition, significant progress in drug development has made targeting TFs from “undruggable” to reality [12]. In the future, new drugs targeting different TFs will fully exploit the power of transcription factors in reprogramming cancer cell plasticity. Reprogramming cancer cells by modulating sets of TFs with chemical cocktails that induce differentiation of cancer stem cells could hold significant promise for future cancer therapies.
Table 1.
The function of KLF4, KLF5, SOX2, SOX4 and SOX9 in cancers. “↑”: upregulation;” ↓” : down regulation.
| Function | Mechanism | |
|---|---|---|
| KLF4 | Inhibit cell proliferation (by inducing cell cycle arrest at the G1-S checkpoint) | ↑ CDKN1B (p27) [67–70] ↑ CDKN1A (p21) [67–70] ↓ cyclin D1 [68,69,71] |
| Promote cell proliferation (KLF4α) | ↓ CDKN1B (p27) ↓ CDKN1A (p21) |
|
| Promote tumor metastasis | ↑ Notch pathway [128] | |
| Inhibit tumor metastasis | ↓ SLUG [130] ↓ SNAIL [131] |
|
| Promote cancer stemness | ↑ hTERT [147] ↑ stemness-associated genes [148] ↑ MAPK signaling pathway [148] ↓ pro-apoptotic signaling molecules PDCD4 & CX43 through microRNA-206 [149] ↑ ALDH1 [150] ↑ MUC5AC [152] |
|
| Promote drug resistance | ↑ MCL1 (anti-apoptotic factor) [170] ↓ pro-apoptotic Bax [173–175] ↑ BRCA1 for HR DNA repair [175] Induce cell cycle arrest: ↑ p21 [171–175] ↓ CCND1 [71] ↓ CCNB1 [176] Prevent chromosomal amplification: ↓ cyclin E [177] |
|
| KLF5 | Promote cell proliferation (by accelerating the transition of G1/S and G2/M phases) | ↑ cyclin D1 [86] ↑ cyclin B1 [87] ↑ Cdc2 [87] ↓ CDKN1B (p27) [88] ↓ p15 [88] |
| Inhibit cell proliferation when p53 is mutated | ↑ CDKN1A (p21) [98] ↑ NOTCH1 [99] |
|
| Inhibit apoptosis | ↑ BIRC5 (survivin) [105] ↑ HIF1α [105,179] |
|
| Promote tumor metastasis | ↑ TNFAIP2 [132] ↑ TNFRSF11a [133] ↓ E-cadherin [134] ↑ N-cadherin [134] ↑ Vimentin [134] ↑ SNAIL [134] ↑ SLUG [134] |
|
| Inhibit tumor metastasis (in the absence of p53) | ↑ E-cadherin [99,135] ↓ ZEB2 [136] ↓ SNAIL [99] ↓ TWIST [99] |
|
| Promote cancer stemness | ↑ α-Catulin [153] | |
| Promote drug resistance | Inhibit apoptosis: ↑ MCL1 (anti-apoptotic factor) [170] ↑ cyclin B1 [179] ↑ survivin [179] ↓ caspase-3 [179] Promote DNA repair: ↑ Chk1/Chk2 kinase levels [180] |
|
| SOX2 | Paclitaxel resistance | ↑ ABCC1 [192] |
| Promote tumor metastasis | ↑ HIF1α [255–257] | |
| SOX4 | Promote tumor metastasis | ↑ Twist1 and ZEB1 [138] |
| Promote cancer stemness | ↑ SOX2 [138] ↑ OCT4 [138] |
|
| SOX9 | Cisplatin resistance | ↑ ALDH1A1 [186] |
| Sorafenib resistance | ↑ ABCG2 [191] |
Table 2.
Drugs targeting KLF4 and KLF5
| Mechanism | Compounds | |
|---|---|---|
| KLF4 | Inhibit KLF4 transcription | Kenpaullone [128] Regorafenib [273] |
| Enhance KLF4 transcription | PKA inhibitors: H89 and 14–22 Amide (PKI) [128,198] APTO-253 [262] |
|
| Reduce KLF4 protein | Simvastatin and atorvastatin [178] | |
| KLF5 | Inhibit KLF5 transcription | Wortmannin (a PI3K/AKT inhibitor), AG17, and AG879 [263] |
| Inhibit KLF5 expression via inducing miRNA targeting KLF5 | Mifepristone [222] Crocin [227] |
|
| Promotes KLF5 proteasome degradation | Curcumin [265] Metformin [155] |
Acknowledgements
We thank all members of Wan and Moreno laboratories for their helpful discussion. We thank Marco Moreno for figure artwork. This work was supported by NIH R21CA256375, NIH R01CA258857, NIH R01CA258765, NIH R01CA250110 and NIH R01CA202948.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest Statement
The authors declare that there are no conflicts of interest.
References:
- [1].Boumahdi S, de Sauvage FJ, The great escape: tumour cell plasticity in resistance to targeted therapy, Nat. Rev. Drug Discov 19 (2020) 39–56. 10.1038/s41573-019-0044-1. [DOI] [PubMed] [Google Scholar]
- [2].Quintanal-Villalonga Á, Chan JM, Yu HA, Pe’er D, Sawyers CL, Sen T, Rudin CM, Lineage plasticity in cancer: a shared pathway of therapeutic resistance, Nat. Rev. Clin. Oncol 17 (2020) 360–371. 10.1038/s41571-020-0340-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Redmond KM, Wilson TR, Johnston PG, Longley DB, Drug Resistance Laboratory, Centre for Cancer Research and Cell Biology, Queen’s University Belfast, Belfast, UK, (2008) 5138–5154. [Google Scholar]
- [4].Eyler CE, Matsunaga H, Hovestadt V, Vantine SJ, Van Galen P, Bernstein BE, Single-cell lineage analysis reveals genetic and epigenetic interplay in glioblastoma drug resistance, Genome Biol. 21 (2020) 1–21. 10.1186/s13059-020-02085-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Aissa AF, Islam ABMMK, Ariss MM, Go CC, Rader AE, Conrardy RD, Gajda AM, Rubio-Perez C, Valyi-Nagy K, Pasquinelli M, Feldman LE, Green SJ, Lopez-Bigas N, Frolov MV, Benevolenskaya EV, Single-cell transcriptional changes associated with drug tolerance and response to combination therapies in cancer, Nat. Commun 12 (2021) 1–25. 10.1038/s41467-021-21884-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Bakir B, Chiarella AM, Pitarresi JR, Rustgi AK, EMT, MET, Plasticity, and Tumor Metastasis, Trends Cell Biol. 30 (2020) 764–776. 10.1016/j.tcb.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Thankamony AP, Saxena K, Murali R, Jolly MK, Nair R, Cancer Stem Cell Plasticity – A Deadly Deal, Front. Mol. Biosci 7 (2020) 1–16. 10.3389/fmolb.2020.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, Weirauch MT, The Human Transcription Factors, Cell. 172 (2018) 650–665. 10.1016/j.cell.2018.01.029. [DOI] [PubMed] [Google Scholar]
- [9].Lee TI, Young RA, Transcriptional regulation and its misregulation in disease, Cell. 152 (2013) 1237–1251. 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Darnell JE, Transcription factors as targets for cancer therapy, Nat. Rev. Cancer 2 (2002) 740–749. 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- [11].Huilgol D, Venkataramani P, Nandi S, Bhattacharjee S, Transcription factors that govern development and disease: An achilles heel in cancer, Genes (Basel). 10 (2019) 1–35. 10.3390/genes10100794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bushweller JH, Targeting transcription factors in cancer — from undruggable to reality, Nat. Rev. Cancer 19 (2019) 611–624. 10.1038/s41568-019-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, Vogelstein B, Kinzler KW, Gene expression profiles in normal and cancer cells, Science (80-.) 276 (1997) 1268–1272. 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- [14].Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, Payette T, Pistone M, Stecker K, Zhang BM, Zhou YX, Varnholt H, Smith B, Gadd M, Chatfield E, Kessler J, Baer TM, Erlander MG, Sgroi DC, Gene expression profiles of human breast cancer progression, Proc. Natl. Acad. Sci. U. S. A 100 (2003) 5974–5979. 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cejalvo JM, De Dueñas EM, Galván P, García-Recio S, Gasión OB, Paré L, Antolín S, Martinello R, Blancas I, Adamo B, Guerrero-Zotano Á, Muñoz M, Nucíforo P, Vidal M, Pérez RM, López-Muniz JIC, Caballero R, Peg V, Carrasco E, Rojo F, Perou CM, Cortés J, Adamo V, Albanell J, Gomis RR, Lluch A, Prat A, Intrinsic subtypes and gene expression profiles in primary and metastatic breast cancer, Cancer Res. 77 (2017) 2213–2221. 10.1158/0008-5472.CAN-16-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Friederichs J, Rosenberg R, Mages J, Janssen KP, Maeckl C, Nekarda H, Holzmann B, Siewert JR, Gene expression profiles of different clinical stages of colorectal carcinoma: Toward a molecular genetic understanding of tumor progression, Int. J. Colorectal Dis 20 (2005) 391–402. 10.1007/s00384-004-0722-1. [DOI] [PubMed] [Google Scholar]
- [17].Zhou J, Zhao LQ, Xiong MM, Wang XQ, Yang GR, Qiu ZL, Wu M, Liu ZH, Gene expression profiles at different stages of human esophageal squamous cell carcinoma, World J. Gastroenterol 9 (2003) 9–15. 10.3748/wjg.v9.i1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kleivi K, Lind GE, Diep CB, Meling GI, Brandal LT, Nesland JM, Myklebost O, Rognum TO, Giercksky KE, Skotheim RI, Lothe RA, Gene expression profiles of primary colorectal carcinomas, liver metastases, and carcinomatoses, Mol. Cancer 6 (2007) 1–16. 10.1186/1476-4598-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hartmann LC, Lu KH, Linette GP, Cliby WA, Kalli KR, Gershenson D, Bast RC, Stec J, Iartcliouk N, Smith DI, Ross JS, Hoersch S, Shridhar V, Lillie J, Kaufmann SH, Clark EA, Damokosli AI, Gene expression profiles predict early relapse in ovarian cancer after platinum-paclitaxel chemotherapy, Clin. Cancer Res 11 (2005) 2149–2155. 10.1158/1078-0432.CCR-04-1673. [DOI] [PubMed] [Google Scholar]
- [20].Beer DG, Kardia SLR, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, Lizyness ML, Kuick R, Hayasaka S, Taylor JMG, Iannettoni MD, Orringer MB, Hanash S, Gene-expression profiles predict survival of patients with lung adenocarcinoma, Nat. Med 8 (2002) 816–824. 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- [21].Bertucci F, Nasser V, Granjeaud S, Eisinger F, Adelaïde J, Tagett R, Loriod B, Giaconia A, Benziane A, Devilard E, Jacquemier J, Viens P, Nguyen C, Birnbaum D, Houlgatte R, Gene expression profiles of poor-prognosis primary breast cancer correlate with survival, Hum. Mol. Genet 11 (2002) 863–872. 10.1093/hmg/11.8.863. [DOI] [PubMed] [Google Scholar]
- [22].Wang Y, Klijn JGM, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-Van Gelder ME, Yu J, Jatkoe T, Berns EMJJ, Atkins D, Foekens JA, Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer, Lancet. 365 (2005) 671–679. 10.1016/S0140-6736(05)70933-8. [DOI] [PubMed] [Google Scholar]
- [23].Gronemeyer H, åke Gustafsson J, Laudet V, PRINCIPLES FOR MODULATION OF THE NUCLEAR RECEPTOR SUPERFAMILY, 3 (2004) 950–964. 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- [24].Merseburger AS, Haas GP, Von Klot CA, An update on enzalutamide in the treatment of prostate cancer, Ther. Adv. Urol 7 (2015) 9–21. 10.1177/1756287214555336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yan C, Higgins PJ, Drugging the undruggable: Transcription therapy for cancer, Biochim. Biophys. Acta - Rev. Cancer 1835 (2013) 76–85. 10.1016/j.bbcan.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mcconnell BB, Yang VW, Mammalian Krüppel-Like Factors in Health and Diseases, (2022) 1337–1381. 10.1152/physrev.00058.2009.Mammalian. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bowles J, Schepers G, Koopman P, Phylogeny of the SOX Family of Developmental Transcription Factors Based on Sequence and Structural Indicators, 255 (2000) 239–255. 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- [28].Farrugia MK, Vanderbilt DB, Salkeni MA, Ruppert JM, Kruppel-like pluripotency factors as modulators of cancer cell therapeutic responses, Cancer Res. 76 (2016) 1677–1682. 10.1158/0008-5472.CAN-15-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].She Z-Y, Yang W-X, SOX family transcription factors involved in diverse cellular events during development, Eur. J. Cell Biol 94 (2015) 547–563. https://doi.org/ 10.1016/j.ejcb.2015.08.002. [DOI] [PubMed] [Google Scholar]
- [30].Bourillot PY, Savatier P, Krüppel-like transcription factors and control of pluripotency, BMC Biol. 8 (2010) 8–10. 10.1186/1741-7007-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Parisi S, Cozzuto L, Tarantino C, Passaro F, Ciriello S, Aloia L, Antonini D, De Simone V, Pastore L, Russo T, Direct targets of Klf5 transcription factor contribute to the maintenance of mouse embryonic stem cell undifferentiated state, BMC Biol. 8 (2010). 10.1186/1741-7007-8-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bourillot PY, Aksoy I, Schreiber V, Wianny F, Schulz H, Hummel O, Hubner N, Savatier P, Novel STAT3 target genes exert distinct roles in the inhibition of mesoderm and endoderm differentiation in cooperation with Nanog, Stem Cells. 27 (2009) 1760–1771. 10.1002/stem.110. [DOI] [PubMed] [Google Scholar]
- [33].Niwa H, Ogawa K, Shimosato D, Adachi K, A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells, Nature. 460 (2009) 118–122. 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- [34].Hall J, Guo G, Wray J, Eyres I, Nichols J, Grotewold L, Morfopoulou S, Humphreys P, Mansfield W, Walker R, Tomlinson S, Smith A, Oct4 and LIF/Stat3 Additively Induce Krüppel Factors to Sustain Embryonic Stem Cell Self-Renewal, Cell Stem Cell. 5 (2009) 597–609. 10.1016/j.stem.2009.11.003. [DOI] [PubMed] [Google Scholar]
- [35].Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, Robson P, Zhong S, Ng HH, A core Klf circuitry regulates self-renewal of embryonic stem cells, Nat. Cell Biol 10 (2008) 353–360. 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- [36].Parisi S, Passaro F, Aloia L, Manabe I, Nagai R, Pastore L, Russo T, Klf5 is involved in self-renewal of mouse embryonic stem cells, J. Cell Sci 121 (2008) 2629–2634. 10.1242/jcs.027599. [DOI] [PubMed] [Google Scholar]
- [37].Redmond LC, Dumur CI, Archer KJ, Grayson DR, Haar JL, Lloyd JA, Krüppel-like factor 2 regulated gene expression in mouse embryonic yolk sac erythroid cells, Blood Cells, Mol. Dis 47 (2011) 1–11. 10.1016/j.bcmd.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Pan G, Thomson JA, Nanog and transcriptional networks in embryonic stem cell pluripotency, Cell Res. 17 (2007) 42–49. 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- [39].Takahashi K, Yamanaka S, Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors, Cell. 126 (2006) 663–676. 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- [40].Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S, Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors, Cell. 131 (2007) 861–872. 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- [41].Apostolou E, Hochedlinger K, Chromatin dynamics during cellular reprogramming, (2013). 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Shields JM, Christy RJ, Yang VW, Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest, J. Biol. Chem 271 (1996) 20009–20017. 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Conkright MD, Wani MA, Anderson KP, Lingrel JB, A gene encoding an intestinal-enriched member of the Kruppel-like factor family expressed in intestinal epithelial cells, Nucleic Acids Res. 27 (1999) 1263–1270. 10.1093/nar/27.5.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kuruvilla JG, Kim C-K, Ghaleb AM, Bialkowska AB, Kuo CJ, Yang VW, Krüppel-like Factor 4 Modulates Development of BMI1(+) Intestinal Stem Cell-Derived Lineage Following γ-Radiation-Induced Gut Injury in Mice, Stem Cell Reports. 6 (2016) 815–824. 10.1016/j.stemcr.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Nakaya T, Ogawa S, Manabe I, Tanaka M, Sanada M, Sato T, Taketo MM, Nakao K, Clevers H, Fukayama M, Kuroda M, Nagai R, KLF5 regulates the integrity and oncogenicity of intestinal stem cells, Cancer Res. 74 (2014) 2882–2891. 10.1158/0008-5472.CAN-13-2574. [DOI] [PubMed] [Google Scholar]
- [46].Zhang B, Ci X, Tao R, Ni JJ, Xuan X, King JL, Xia S, Li Y, Frierson HF, Lee DK, Xu J, Osunkoya AO, Dong JT, Klf5 acetylation regulates luminal differentiation of basal progenitors in prostate development and regeneration, Nat. Commun 11 (2020). 10.1038/s41467-020-14737-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Stevanovic M, Drakulic D, Lazic A, Ninkovic DS, Schwirtlich M, Mojsin M, SOX Transcription Factors as Important Regulators of Neuronal and Glial Differentiation During Nervous System Development and Adult Neurogenesis, Front. Mol. Neurosci 14 (2021) 1–24. 10.3389/fnmol.2021.654031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jo A, Denduluri S, Zhang B, Wang Z, Yin L, Yan Z, Kang R, Shi LL, Mok J, Lee MJ, Haydon RC, The versatile functions of Sox9 in development, stem cells, and human diseases, Genes Dis. 1 (2014) 149–161. 10.1016/j.gendis.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Seymour PA, Freude KK, Tran MN, Mayes EE, Jensen J, Kist R, Scherer G, Sander M, SOX9 is required for maintenance of the pancreatic progenitor cell pool, Proc. Natl. Acad. Sci. U. S. A 104 (2007) 1865–1870. 10.1073/pnas.0609217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Liu Y, Guo W, SOX factors as cell-state regulators in the mammary gland and breast cancer, Semin. Cell Dev. Biol 114 (2021) 126–133. 10.1016/j.semcdb.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wilson ME, Yang KY, Kalousova A, Lau J, Kosaka Y, Lynn FC, Wang J, Mrejen C, Episkopou V, Clevers HC, German MS, The HMG Box Transcription Factor Sox4 Contributes to the Development of the Endocrine Pancreas, Diabetes. 54 (2005) 3402–3409. 10.2337/diabetes.54.12.3402. [DOI] [PubMed] [Google Scholar]
- [52].Roukens MG, Frederiks CL, Seinstra D, Braccioli L, Khalil AA, Pals C, De Neck S, Bornes L, Beerling E, Mokry M, de Bruin A, Westendorp B, van Rheenen J, Coffer PJ, Regulation of a progenitor gene program by SOX4 is essential for mammary tumor proliferation, Oncogene. 40 (2021) 6343–6353. 10.1038/s41388-021-02004-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ya J, Schilham MW, De Boer PAJ, Moorman AFM, Clevers H, Lamers WH, Sox4-deficiency syndrome in mice is an animal model for common trunk, Circ. Res 83 (1998) 986–994. 10.1161/01.RES.83.10.986. [DOI] [PubMed] [Google Scholar]
- [54].Sun B, Mallampati S, Gong Y, Wang D, Lefebvre V, Sun X, Sox4 Is Required for the Survival of Pro-B Cells, J. Immunol 190 (2013) 2080–2089. 10.4049/jimmunol.1202736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lefebvre V, Roles and regulation of SOX transcription factors in skeletogenesis, Curr. Top. Dev. Biol 133 (2019) 171–193. 10.1016/bs.ctdb.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Miao Q, Hill MC, Chen F, Mo Q, Ku AT, Ramos C, Sock E, Lefebvre V, Nguyen H, SOX11 and SOX4 drive the reactivation of an embryonic gene program during murine wound repair, Nat. Commun 10 (2019) 1–20. 10.1038/s41467-019-11880-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Tetreault MP, Yang Y, Katz JP, Krüppel-like factors in cancer, Nat. Rev. Cancer 13 (2013) 701–713. 10.1038/nrc3582. [DOI] [PubMed] [Google Scholar]
- [58].Grimm D, Bauer J, Wise P, Krüger M, Simonsen U, Wehland M, Infanger M, Corydon TJ, The role of SOX family members in solid tumours and metastasis, Semin. Cancer Biol 67 (2020) 122–153. 10.1016/j.semcancer.2019.03.004. [DOI] [PubMed] [Google Scholar]
- [59].Moreno CS, SOX4: The unappreciated oncogene, Semin. Cancer Biol 67 (2020) 57–64. 10.1016/j.semcancer.2019.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hanieh H, Ahmed EA, Vishnubalaji R, Alajez NM, SOX4: Epigenetic regulation and role in tumorigenesis, Semin. Cancer Biol 67 (2020) 91–104. 10.1016/j.semcancer.2019.06.022. [DOI] [PubMed] [Google Scholar]
- [61].Novak D, Hüser L, Elton JJ, Umansky V, Altevogt P, Utikal J, SOX2 in development and cancer biology, Semin. Cancer Biol 67 (2020) 74–82. 10.1016/j.semcancer.2019.08.007. [DOI] [PubMed] [Google Scholar]
- [62].Xu YR, Yang WX, SOX-mediated molecular crosstalk during the progression of tumorigenesis, Semin. Cell Dev. Biol 63 (2017) 23–34. 10.1016/j.semcdb.2016.07.028. [DOI] [PubMed] [Google Scholar]
- [63].Aguilar-Medina M, Avendaño-Félix M, Lizárraga-Verdugo E, Bermúdez M, Romero-Quintana JG, Ramos-Payan R, Ruíz-García E, López-Camarillo C, SOX9 Stem-Cell Factor: Clinical and Functional Relevance in Cancer, J. Oncol 2019 (2019). 10.1155/2019/6754040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Chen H, He Y, Wen X, Shao S, Liu Y, Wang J, SOX9: Advances in Gynecological Malignancies, Front. Oncol 11 (2021) 1–12. 10.3389/fonc.2021.768264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Mehta GA, Khanna P, Gatza ML, Emerging Role of SOX Proteins in Breast Cancer Development and Maintenance, J. Mammary Gland Biol. Neoplasia 24 (2019) 213–230. 10.1007/s10911-019-09430-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Yang VW, Krüppel-like factors 4 and 5 : the yin and yang regulators of cellular proliferation, 4 (2005) 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].V Lloyd R, Erickson LA, Jin L, Kulig E, Qian X, Cheville JC, Scheithauer BW, Review Human Cancers, Significance. 154 (1999) 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Hu W, Hofstetter WL, Li H, Zhou Y, He Y, Pataer A, Wang L, Xie K, Swisher SG, Fang B, Putative tumor-suppressive function of Krüppel-like factor 4 in primary lung carcinoma, Clin. Cancer Res 15 (2009) 5688–5695. 10.1158/1078-0432.CCR-09-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zammarchi F, Morelli M, Menicagli M, Di Cristofano C, Zavaglia K, Paolucci A, Campani D, Aretini P, Boggi U, Mosca F, Cavazzana A, Cartegni L, Bevilacqua G, Mazzanti CM, KLF4 is a novel candidate tumor suppressor gene in pancreatic ductal carcinoma, Am. J. Pathol 178 (2011) 361–372. 10.1016/j.ajpath.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Yang WT, Zheng PS, Krüppel-like factor 4 functions as a tumor suppressor in cervical carcinoma, Cancer. 118 (2012) 3691–3702. 10.1002/cncr.26698. [DOI] [PubMed] [Google Scholar]
- [71].Shie JL, Chen ZY, Fu M, Pestell RG, Tseng CC, Gut-enriched Kruppel-like factor represess cyclin D1 promoter activity through Sp1 motif, Nucleic Acids Res. 28 (2000) 2969–2976. 10.1093/nar/28.15.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Sun H, Peng Z, Tang H, Xie D, Jia Z, Zhong L, Zhao S, Ma Z, Gao Y, Zeng L, Luo R, Xie K, Loss of KLF4 and consequential downregulation of Smad7 exacerbate oncogenic TGF-β signaling in and promote progression of hepatocellular carcinoma., Oncogene. 36 (2017) 2957–2968. 10.1038/onc.2016.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Yu R, Han L, Ni X, Wang M, Xue P, Zhang L, Yuan M, Kruppel-like factor 4 inhibits non-small cell lung cancer cell growth and aggressiveness by stimulating transforming growth factor-β1-meidated ERK/JNK/NF-κB signaling pathways., Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med 39 (2017) 1010428317705574. 10.1177/1010428317705574. [DOI] [PubMed] [Google Scholar]
- [74].Ohnishi S, Ohnami S, Laub F, Aoki K, Suzuki K, Kanai Y, Haga K, Asaka M, Ramirez F, Yoshida T, Downregulation and growth inhibitory effect of epithelial-type Krüppel-like transcription factor KLF4, but not KLF5, in bladder cancer, Biochem. Biophys. Res. Commun 308 (2003) 251–256. 10.1016/S0006-291X(03)01356-1. [DOI] [PubMed] [Google Scholar]
- [75].Nakahara Y, Northcott PA, Li M, Kongkham PN, Smith C, Yan H, Croul S, Ra YS, Eberhart C, Huang A, Bigner D, Grajkowska W, Van Meter T, Rutka JT, Taylor MD, Genetic and epigenetic inactivation of Kruppel-like Factor 4 in medulloblastoma, Neoplasia. 12 (2010) 20–27. 10.1593/neo.91122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Yu T, Chen X, Zhang W, Colon D, Shi J, Napier D, Rychahou P, Lu W, Lee EY, Weiss HL, Evers BM, Liu C, Regulation of the potential marker for intestinal cells, Bmi1, by β-catenin and the zinc finger protein KLF4: Implications for colon cancer, J. Biol. Chem 287 (2012) 3760–3768. 10.1074/jbc.M111.316349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Gamper AM, Qiao X, Kim J, Zhang L, De Simone MC, Rathmell WK, Wan Y, Regulation of KLF4 Turnover Reveals an Unexpected Tissue-Specific Role of pVHL in Tumorigenesis, Mol. Cell 45 (2012) 233–243. 10.1016/j.molcel.2011.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Li Q, Gao Y, Jia Z, Mishra L, Guo K, Li Z, Le X, Wei D, Huang S, Xie K, Dysregulated Krppel-like factor 4 and vitamin D receptor signaling contribute to progression of hepatocellular carcinoma, Gastroenterology. 143 (2012) 799–810. 10.1053/j.gastro.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Shum CKY, Lau ST, Tsoi LLS, Chan LK, Yam JWP, Ohira M, Nakagawara A, Tam PKH, Ngan ESW, Krüppel-like factor 4 (KLF4) suppresses neuroblastoma cell growth and determines non-tumorigenic lineage differentiation, Oncogene. 32 (2013) 4086–4099. 10.1038/onc.2012.437. [DOI] [PubMed] [Google Scholar]
- [80].Foster KW, Frost AR, McKie-Bell P, Lin C-Y, Engler JA, Grizzle WE, Ruppert JM, Increase of GKLF Messenger RNA and Protein Expression during Progression of Breast Cancer1, Cancer Res. 60 (2000) 6488–6495. [PubMed] [Google Scholar]
- [81].Rowland BD, Peeper DS, KLF4, p21 and context-dependent opposing forces in cancer, Nat. Rev. Cancer 6 (2006) 11–23. 10.1038/nrc1780. [DOI] [PubMed] [Google Scholar]
- [82].Yang Y, Katz JP, KLF4 is downregulated but not mutated during human esophageal squamous cell carcinogenesis and has tumor stage-specific functions carcinogenesis and has tumor stage-specific functions, Cancer Biol. Ther 17 (2016) 422–429. 10.1080/15384047.2016.1156260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Rowland BD, Bernards R, Peeper DS, The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene, Nat. Cell Biol 7 (2005) 1074–1082. 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- [84].Nandan MO, McConnell BB, Ghaleb AM, Bialkowska AB, Sheng H, Shao J, Babbin BA, Robine S, Yang VW, Krüppel-Like Factor 5 Mediates Cellular Transformation During Oncogenic KRAS-Induced Intestinal Tumorigenesis, Gastroenterology. 134 (2008) 120–130. 10.1053/j.gastro.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Wei D, Wang L, Kanai M, Jia Z, Le X, Li Q, Wang H, Xie K, KLF4α up-regulation promotes cell cycle progression and reduces survival time of patients with pancreatic cancer, Gastroenterology. 139 (2010) 2135–2145. 10.1053/j.gastro.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Nandan MO, Yoon HS, Zhao W, Ouko LA, Yang VW, Krüppel-like factor 5 mediates the transforming activity of oncogenic, (2004) 3404–3413. 10.1038/sj.onc.1207397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Nandan MO, Chanchevalap S, Dalton WB, Yang VW, Krüppel-like factor 5 promotes mitosis by activating the cyclin B1 / Cdc2 complex during oncogenic Ras-mediated transformation, 579 (2005) 4757–4762. 10.1016/j.febslet.2005.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Chen C, Benjamin MS, Sun X, Otto KB, Guo P, Dong X, Bao Y, Zhou Z, Cheng X, Simons JW, Dong J, KLF5 promotes cell proliferation and tumorigenesis through gene regulation in the TSU-Pr1 human bladder cancer cell line, 1355 (2006) 1346–1355. 10.1002/ijc.21533. [DOI] [PubMed] [Google Scholar]
- [89].Li Y, Kong R, Chen H, Zhao Z, Li L, Li J, Hu J, Zhang G, Pan S, Wang Y, Wang G, Chen H, Sun B, Overexpression of KLF5 is associated with poor survival and G1/S progression in pancreatic cancer, Aging (Albany. NY) 11 (2019) 5035–5057. 10.18632/aging.102096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Chen W, Zhang J, Fu H, Hou X, Su Q, He Y, Yang D, Klf5 is activated by gene amplification in gastric cancer and is essential for gastric cell proliferation, Cells. 10 (2021). 10.3390/cells10051002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Takagi K, Miki Y, Onodera Y, Nakamura Y, Ishida T, Watanabe M, Inoue S, Sasano H, Suzuki T, Krüppel-like factor 5 in human breast carcinoma: A potent prognostic factor induced by androgens, Endocr. Relat. Cancer 19 (2012) 741–750. 10.1530/ERC-12-0017. [DOI] [PubMed] [Google Scholar]
- [92].Liu R, Zhou Z, Zhao D, Chen C, The induction of KLF5 transcription factor by progesterone contributes to progesterone-induced breast cancer cell proliferation and dedifferentiation, Mol. Endocrinol 25 (2011) 1137–1144. 10.1210/me.2010-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Chen C, Bhalala HV, Qiao H, Dong JT, A possible tumor suppressor role of the KLF5 transcription factor in human breast cancer, Oncogene. 21 (2002) 6567–6572. 10.1038/sj.onc.1205817. [DOI] [PubMed] [Google Scholar]
- [94].Chen C, Bhalala HV, Vessella RL, Dong JT, KLF5 is frequently deleted and downregulated but rarely mutated in prostate cancer, Prostate. 55 (2003) 81–88. 10.1002/pros.10205. [DOI] [PubMed] [Google Scholar]
- [95].Jia J, Zhang HB, Shi Q, Yang C, Bin Ma J, Jin B, Wang X, He D, Guo P, KLF5 downregulation desensitizes castration-resistant prostate cancer cells to docetaxel by increasing BECN1 expression and inducing cell autophagy, Theranostics. 9 (2019) 5464–5477. 10.7150/thno.33282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Bin Ma J, Bai JY, Zhang HB, Jia J, Shi Q, Yang C, Wang X, He D, Guo P, KLF5 inhibits STAT3 activity and tumor metastasis in prostate cancer by suppressing IGF1 transcription cooperatively with HDAC1, Cell Death Dis. 11 (2020) 1–14. 10.1038/s41419-020-2671-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Poluri RTK, Paquette V, Allain ÉP, Lafront C, Joly-Beauparlant C, Weidmann C, Droit A, Guillemette C, Pelletier M, Audet-Walsh É, KLF5 and NFYA factors as novel regulators of prostate cancer cell metabolism, Endocr. Relat. Cancer 28 (2021) 257–271. 10.1530/ERC-20-0504. [DOI] [PubMed] [Google Scholar]
- [98].Yang Y, Tarapore RS, Jarmel MH, Tetreault MP, Katz JP, P53 mutation alters the effect of the esophageal tumor suppressor KLF5 on keratinocyte proliferation, Cell Cycle. 11 (2012) 4033–4039. 10.4161/cc.22265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Yang Y, Nakagawa H, Tetreault MP, Billig J, Victor N, Goyal A, Sepulveda AR, Katz JP, Loss of transcription factor KLF5 in the context of p53 ablation drives invasive progression of human squamous cell cancer, Cancer Res. 71 (2011) 6475–6484. 10.1158/0008-5472.CAN-11-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Rivlin N, Brosh R, Oren M, Rotter V, Mutations in the p53 tumor suppressor gene: Important milestones at the various steps of tumorigenesis, Genes and Cancer. 2 (2011) 466–474. 10.1177/1947601911408889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Oishi Y, Manabe I, Tobe K, Ohsugi M, Kubota T, Fujiu K, Maemura K, Kubota N, Kadowaki T, Nagai R, SUMOylation of Krüppel-like transcription factor 5 acts as a molecular switch in transcriptional programs of lipid metabolism involving PPAR-δ, Nat. Med 14 (2008) 656–666. 10.1038/nm1756. [DOI] [PubMed] [Google Scholar]
- [102].Guo P, Dong XY, Zhao K, Sun X, Li Q, Dong JT, Opposing effects of KLF5 on the transcription of MYC in epithelial proliferation in the context of transforming growth factor β, J. Biol. Chem 284 (2009) 28243–28252. 10.1074/jbc.M109.036160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Guo P, Dong XY, Zhang X, Zhao KW, Sun X, Li Q, Dong JT, Pro-proliferative factor KLF5 becomes anti-proliferative in epithelial homeostasis upon signaling-mediated modification, J. Biol. Chem 284 (2009) 6071–6078. 10.1074/jbc.M806270200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Li X, Zhang B, Wu Q, Ci X, Zhao R, Zhang Z, Xia S, Su D, Chen J, Ma G, Fu L, Dong JT, Interruption of KLF5 acetylation converts its function from tumor suppressor to tumor promoter in prostate cancer cells, Int. J. Cancer 136 (2015) 536–546. 10.1002/ijc.29028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Lee SJ, No YR, Dang DT, Dang LH, Yang VW, Shim H, Yun CC, Regulation of hypoxia-inducible factor 1α (HIF-1α) by lysophosphatidic acid is dependent on interplay between p53 and Krüppel-like factor 5, J. Biol. Chem 288 (2013) 25244–25253. 10.1074/jbc.M113.489708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Gupta R, Malvi P, Parajuli KR, Janostiak R, Bugide S, Cai G, Zhu LJ, Green MR, Wajapeyee N, KLF7 promotes pancreatic cancer growth and metastasis by up-regulating ISG expression and maintaining Golgi complex integrity., Proc. Natl. Acad. Sci. U. S. A 117 (2020) 12341–12351. 10.1073/pnas.2005156117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Guo Y, Chai B, Jia J, Yang M, Li Y, Zhang R, Wang S, Xu J, KLF7/VPS35 axis contributes to hepatocellular carcinoma progression through CCDC85C-activated β-catenin pathway., Cell Biosci. 11 (2021) 73. 10.1186/s13578-021-00585-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Li Y, Wang Q, Wang D, Fu W, KLF7 Promotes Gastric Carcinogenesis Through Regulation of ANTXR1., Cancer Manag. Res 13 (2021) 5547–5557. 10.2147/CMAR.S308071. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [109].Wang J, Teng F, Chai H, Zhang C, Liang X, Yang Y, GNA14 stimulation of KLF7 promotes malignant growth of endometrial cancer through upregulation of HAS2., BMC Cancer. 21 (2021) 456. 10.1186/s12885-021-08202-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Yang J, Xie K, Wang Z, Li C, Elevated KLF7 levels may serve as a prognostic signature and might contribute to progression of squamous carcinoma., FEBS Open Bio. 10 (2020) 1577–1586. 10.1002/2211-5463.12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].De Donato M, Babini G, Mozzetti S, Buttarelli M, Ciucci A, Arduini G, De Rosa MC, Scambia G, Gallo D, KLF7: a new candidate biomarker and therapeutic target for high-grade serous ovarian cancer., J. Exp. Clin. Cancer Res 39 (2020) 265. 10.1186/s13046-020-01775-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Wang L, Zhang J, Yang X, Chang YWY, Qi M, Zhou Z, Zhang J, Han B, SOX4 is associated with poor prognosis in prostate cancer and promotes epithelial-mesenchymal transition in vitro, Prostate Cancer Prostatic Dis. 16 (2013) 301–307. 10.1038/pcan.2013.25. [DOI] [PubMed] [Google Scholar]
- [113].Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massagué J, Endogenous human microRNAs that suppress breast cancer metastasis, Nature. 451 (2008) 147–152. 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Tiwari N, Tiwari VK, Waldmeier L, Balwierz PJ, Arnold P, Pachkov M, Meyer-Schaller N, Schübeler D, vanNimwegen E, Christofori G, Sox4 Is a Master Regulator of Epithelial-Mesenchymal Transition by Controlling Ezh2 Expression and Epigenetic Reprogramming, Cancer Cell. 23 (2013) 768–783. 10.1016/j.ccr.2013.04.020. [DOI] [PubMed] [Google Scholar]
- [115].Zhou Z, Huang F, Shrivastava I, Zhu R, Luo A, Hottiger M, Bahar I, Liu Z, Cristofanilli M, Wan Y, New insight into the significance of KLF4 PARylation in genome stability, carcinogenesis, and therapy., EMBO Mol. Med 12 (2020) e12391. 10.15252/emmm.202012391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Li Q, Jia Z, Wang L, Kong X, Li Q, Guo K, Tan D, Le X, Wei D, Huang S, Mishra L, Xie K, Disruption of Klf4 in villin-positive gastric progenitor cells promotes formation and progression of tumors of the antrum in mice, Gastroenterology. 142 (2012) 531–542. 10.1053/j.gastro.2011.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Ghaleb AM, Elkarim EA, Bialkowska AB, Yang VW, KLF4 suppresses tumor formation in genetic and pharmacological mouse models of colonic tumorigenesis, Mol. Cancer Res 14 (2016) 385–396. 10.1158/1541-7786.MCR-15-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Shi Y, Ou L, Han S, Li M, Pena MMO, Pena EA, Liu C, Nagarkatti M, Fan D, Ai W, Deficiency of Kruppel-like factor KLF4 in myeloid-derived suppressor cells inhibits tumor pulmonary metastasis in mice accompanied by decreased fibrocytes., Oncogenesis. 3 (2014) e129. 10.1038/oncsis.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Nandan MO, Ghaleb AM, McConnell BB, Patel NV, Robine S, Yang VW, Krüppel-like factor 5 is a crucial mediator of intestinal tumorigenesis in mice harboring combined ApcMinand KRASV12mutations, Mol. Cancer 9 (2010) 1–14. 10.1186/1476-4598-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Liu R, Shi P, Zhou Z, Zhang H, Li W, Zhang H, Chen C, Krüpple-like factor 5 is essential for mammary gland development and tumorigenesis., J. Pathol 246 (2018) 497–507. 10.1002/path.5153. [DOI] [PubMed] [Google Scholar]
- [121].Sarkar A, Huebner AJ, Sulahian R, Anselmo A, Xu X, Flattery K, Desai N, Sebastian C, Yram MA, Arnold K, Rivera M, Mostoslavsky R, Bronson R, Bass AJ, Sadreyev R, Shivdasani RA, Hochedlinger K, Sox2 Suppresses Gastric Tumorigenesis in Mice, Cell Rep. 16 (2016) 1929–1941. 10.1016/j.celrep.2016.07.034. [DOI] [PubMed] [Google Scholar]
- [122].Rios AC, Capaldo BD, Vaillant F, Pal B, van Ineveld R, Dawson CA, Chen Y, Nolan E, Fu NY, Jackling FC, Devi S, Clouston D, Whitehead L, Smyth GK, Mueller SN, Lindeman GJ, Visvader JE, Intraclonal Plasticity in Mammary Tumors Revealed through Large-Scale Single-Cell Resolution 3D Imaging, Cancer Cell. 35 (2019) 618–632.e6. 10.1016/j.ccell.2019.02.010. [DOI] [PubMed] [Google Scholar]
- [123].Thiery JP, Acloque H, Huang RYJ, Nieto MA, Epithelial-Mesenchymal Transitions in Development and Disease, Cell. 139 (2009) 871–890. 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- [124].Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, Zur Hausen A, Brunton VG, Morton J, Sansom O, Schüler J, Stemmler MP, Herzberger C, Hopt U, Keck T, Brabletz S, Brabletz T, The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs, Nat. Cell Biol 11 (2009) 1487–1495. 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- [125].Pinho AV, Rooman I, Real FX, P53-Dependent Regulation of Growth, Epithelial-Mesenchymal Transition and Stemness in Normal Pancreatic Epithelial Cells, Cell Cycle. 10 (2011) 1312–1321. 10.4161/cc.10.8.15363. [DOI] [PubMed] [Google Scholar]
- [126].Wu A, Luo W, Zhang Q, Yang Z, Zhang G, Li S, Yao K, Aldehyde dehydrogenase 1, a functional marker for identifying cancer stem cells in human nasopharyngeal carcinoma, Cancer Lett. 330 (2013) 181–189. 10.1016/j.canlet.2012.11.046. [DOI] [PubMed] [Google Scholar]
- [127].Kumar M, Allison DF, Baranova NN, Wamsley JJ, Katz AJ, Bekiranov S, Jones DR, Mayo MW, NF-κB Regulates Mesenchymal Transition for the Induction of Non-Small Cell Lung Cancer Initiating Cells, PLoS One. 8 (2013) 1–10. 10.1371/journal.pone.0068597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Yu F, Li J, Chen H, Fu J, Ray S, Huang S, Zheng H, Ai W, Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion, Oncogene. 30 (2011) 2161–2172. 10.1038/onc.2010.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Leng Z, Li Y, Zhou G, Lv X, Ai W, Li J, Hou L, Krüppel-like factor 4 regulates stemness and mesenchymal properties of colorectal cancer stem cells through the TGF-β1/Smad/snail pathway, J. Cell. Mol. Med 24 (2020) 1866–1877. 10.1111/jcmm.14882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Liu Y-N, Abou-Kheir W, Yin JJ, Fang L, Hynes P, Casey O, Hu D, Wan Y, Seng V, Sheppard-Tillman H, Martin P, Kelly K, Critical and Reciprocal Regulation of KLF4 and SLUG in Transforming Growth Factor β-Initiated Prostate Cancer Epithelial-Mesenchymal Transition, Mol. Cell. Biol 32 (2012) 941–953. 10.1128/mcb.06306-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Yori JL, Seachrist DD, Johnson E, Lozada KL, Abdul-Karim FW, Chodosh LA, Schiemann WP, Keri RA, Krüppel-like factor 4 inhibits tumorigenic progression and metastasis in a mouse model of breast cancer, Neoplasia. 13 (2011) 601–610. 10.1593/neo.11260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Jia L, Zhou Z, Liang H, Wu J, Shi P, Li F, Wang Z, Wang C, Chen W, Zhang H, Wang Y, Liu R, Feng J, Chen C, KLF5 promotes breast cancer proliferation, migration and invasion in part by upregulating the transcription of TNFAIP2, Oncogene. 35 (2016) 2040–2051. 10.1038/onc.2015.263. [DOI] [PubMed] [Google Scholar]
- [133].Ma D, Chang LY, Zhao S, Zhao JJ, Xiong YJ, Cao FY, Yuan L, Zhang Q, Wang XY, Geng ML, Zheng HY, Li O, KLF5 promotes cervical cancer proliferation, migration and invasion in a manner partly dependent on TNFRSF11a expression, Sci. Rep 7 (2017) 1–13. 10.1038/s41598-017-15979-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Liu FF, Dong L, Yang X, Li DJ, Shen YY, Liu ZL, KLF5 silence attenuates proliferation and epithelial-mesenchymal transition induction in Hep-2 cells through NF-KB signaling pathway, Eur. Rev. Med. Pharmacol. Sci 23 (2019) 3867–3875. 10.26355/eurrev_201905_17814. [DOI] [PubMed] [Google Scholar]
- [135].Shimamura T, Imoto S, Shimada Y, Hosono Y, Niida A, Nagasaki M, Yamaguchi R, Takahashi T, Miyano S, A novel network profiling analysis reveals system changes in epithelial-mesenchymal transition, PLoS One. 6 (2011). 10.1371/journal.pone.0020804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Sun L, Zhou X, Li Y, Chen W, Wu S, Zhang B, Yao J, Xu A, KLF5 regulates epithelial-mesenchymal transition of liver cancer cells in the context of p53 loss through miR-192 targeting of ZEB2, Cell Adhes. Migr 14 (2020) 182–194. 10.1080/19336918.2020.1826216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Zhang J, Liang Q, Lei Y, Yao M, Li L, Gao X, Feng J, Zhang Y, Gao H, Liu DX, Lu J, Huang B, SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression, Cancer Res. 72 (2012) 4597–4608. 10.1158/0008-5472.CAN-12-1045. [DOI] [PubMed] [Google Scholar]
- [138].Peng X, Liu G, Peng H, Chen A, Zha L, Wang Z, SOX4 contributes to TGF-β-induced epithelial–mesenchymal transition and stem cell characteristics of gastric cancer cells, Genes Dis. 5 (2018) 49–61. 10.1016/j.gendis.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].David CJ, Huang YH, Chen M, Su J, Zou Y, Bardeesy N, Iacobuzio-Donahue CA, Massagué J, TGF-β Tumor Suppression through a Lethal EMT, Cell. 164 (2016) 1015–1030. 10.1016/j.cell.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Li L, Liu J, Xue H, Li C, Liu Q, Zhou Y, Wang T, Wang H, Qian H, Wen T, A TGF-β-MTA1-SOX4-EZH2 signaling axis drives epithelial–mesenchymal transition in tumor metastasis, Oncogene. 39 (2020) 2125–2139. 10.1038/s41388-019-1132-8. [DOI] [PubMed] [Google Scholar]
- [141].Shi S, Cao X, Gu M, You B, Shan Y, You Y, Upregulated Expression of SOX4 Is Associated with Tumor Growth and Metastasis in Nasopharyngeal Carcinoma, Dis. Markers 2015 (2015) 658141. 10.1155/2015/658141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Zhao Z, Liu W, Li J, MiR-140–5p inhibits cell proliferation and invasion in colorectal carcinoma by targeting SOX4, Oncol. Lett 17 (2019) 2215–2220. 10.3892/ol.2018.9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Ma Y, Shepherd J, Zhao D, Bollu LR, Tahaney WM, Hill J, Zhang Y, Mazumdar A, Brown PH, SOX9 is essential for triple-negative breast cancer cell survival and metastasis, Mol. Cancer Res 18 (2020) 1825–1838. 10.1158/1541-7786.MCR-19-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Yang LX, Guo HB, Liu SY, Feng HP, Shi J, ETS1 promoted cell growth, metastasis and epithelial-mesenchymal transition process in melanoma by regulating miR-16-mediated SOX4 expression, Melanoma Res. (2021) 298–308. 10.1097/CMR.0000000000000743. [DOI] [PubMed] [Google Scholar]
- [145].Prasetyanti PR, Medema JP, Intra-tumor heterogeneity from a cancer stem cell perspective, Mol. Cancer 16 (2017) 1–9. 10.1186/s12943-017-0600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Zhang Y, Hao J, Zheng Y, Jing D, Shen Y, Wang J, Zhao Z, Role of Krüppel-like factors in cancer stem cells, J. Physiol. Biochem 71 (2015) 155–164. 10.1007/s13105-015-0381-4. [DOI] [PubMed] [Google Scholar]
- [147].Wong CW, Hou PS, Tseng SF, Chien CL, Wu KJ, Chen HF, Ho HN, Kyo S, Teng SC, Krüppel-like transcription factor 4 contributes to maintenance of telomerase activity in stem cells, Stem Cells. 28 (2010) 1510–1517. 10.1002/stem.477. [DOI] [PubMed] [Google Scholar]
- [148].Qi X, Li Y, Zhang Y, Xu T, Lu B, Fang L, Gao J, Yu L, Zhu D, Yang B, KLF4 functions as an oncogene in promoting cancer stem cell- like characteristics in osteosarcoma cells, Acta Pharmacol. Sin (2019). 10.1038/s41401-018-0050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Lin CC, Sharma SB, Farrugia MK, McLaughlin SL, Ice RJ, Loskutov YV, Pugacheva EN, Brundage KM, Chen D, Ruppert JM, Kruppel-like factor 4 signals through microRNA-206 to promote tumor initiation and cell survival, Oncogenesis. 4 (2015) e155–13. 10.1038/oncsis.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Shi L, Tang X, Qian M, Liu Z, Meng F, Fu L, Wang Z, Zhu WG, Huang JD, Zhou Z, Liu B, A SIRT1-centered circuitry regulates breast cancer stemness and metastasis, Oncogene. 37 (2018) 6299–6315. 10.1038/s41388-018-0370-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Wei D, Wang L, Yan Y, Jia Z, Gagea M, Li Z, Zuo X, Kong X, Huang S, Xie K, KLF4 Is Essential for Induction of Cellular Identity Change and Acinar-to-Ductal Reprogramming during Early Pancreatic Carcinogenesis, Cancer Cell. 29 (2016) 324–338. 10.1016/j.ccell.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Ganguly K, Krishn SR, Rachagani S, Jahan R, Shah A, Nallasamy P, Rauth S, Atri P, Cox JL, Pothuraju R, Smith LM, Ayala S, Evans C, Ponnusamy MP, Kumar S, Kaur S, Batra SK, Secretory Mucin 5AC Promotes Neoplastic Progression by Augmenting KLF4-Mediated Pancreatic Cancer Cell Stemness, Cancer Res. 81 (2021) 91–102. 10.1158/0008-5472.CAN-20-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Tung CH, Huang MF, Liang CH, Wu YY, Wu JE, Hsu CL, Chen YL, Hong TM, α-Catulin promotes cancer stemness by antagonizing WWP1-mediated KLF5 degradation in lung cancer, Theranostics. 12 (2022) 1173–1186. 10.7150/thno.63627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Maehara O, Sato F, Natsuizaka M, Asano A, Kubota Y, Itoh J, Tsunematsu S, Terashita K, Tsukuda Y, Nakai M, Sho T, Suda G, Morikawa K, Ogawa K, Chuma M, Nakagawa K, Ohnishi S, Komatsu Y, Whelan KA, Nakagawa H, Takeda H, Sakamoto N, A pivotal role of Krüppel-like factor 5 in regulation of cancer stem-like cells in hepatocellular carcinoma, Cancer Biol. Ther 16 (2015) 1453–1461. 10.1080/15384047.2015.1070992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Shi P, Liu W, Tala T, Wang H, Li F, Zhang H, Wu Y, Kong Y, Zhou Z, Wang C, Chen W, Liu R, Chen C, Metformin suppresses triple-negative breast cancer stem cells by targeting KLF5 for degradation, Cell Discov. 3 (2017). 10.1038/celldisc.2017.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Domenici G, Aurrekoetxea-Rodríguez I, Simões BM, Rábano M, Lee SY, Millán JS, Comaills V, Oliemuller E, López-Ruiz JA, Zabalza I, Howard BA, Kypta RM, Vivanco M. d. M., A Sox2–Sox9 signalling axis maintains human breast luminal progenitor and breast cancer stem cells, Oncogene. 38 (2019) 3151–3169. 10.1038/s41388-018-0656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].MacLeod G, Bozek DA, Rajakulendran N, Monteiro V, Ahmadi M, Steinhart Z, Kushida MM, Yu H, Coutinho FJ, Cavalli FMG, Restall I, Hao X, Hart T, Luchman HA, Weiss S, Dirks PB, Angers S, Genome-Wide CRISPR-Cas9 Screens Expose Genetic Vulnerabilities and Mechanisms of Temozolomide Sensitivity in Glioblastoma Stem Cells, Cell Rep. 27 (2019) 971–986.e9. 10.1016/j.celrep.2019.03.047. [DOI] [PubMed] [Google Scholar]
- [158].Malhotra GK, Zhao X, Edwards E, Kopp JL, Naramura M, Sander M, Band H, Band V, The role of Sox9 in mouse mammary gland development and maintenance of mammary stem and luminal progenitor cells, BMC Dev. Biol 14 (2014) 1–11. 10.1186/s12861-014-0047-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Takeda K, Mizushima T, Yokoyama Y, Hirose H, Wu X, Qian Y, Ikehata K, Miyoshi N, Takahashi H, Haraguchi N, Hata T, Matsuda C, Doki Y, Mori M, Yamamoto H, Sox2 is associated with cancer stem-like properties in colorectal cancer, Sci. Rep 8 (2018) 1–9. 10.1038/s41598-018-36251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Lundberg IV, Edin S, Eklöf V, Öberg Å, Palmqvist R, Wikberg ML, SOX2 expression is associated with a cancer stem cell state and down-regulation of CDX2 in colorectal cancer, BMC Cancer. 16 (2016) 1–11. 10.1186/s12885-016-2509-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Abubaker K, Latifi A, Luwor R, Nazaretian S, Zhu H, Quinn MA, Thompson EW, Findlay JK, Ahmed N, Short-term single treatment of chemotherapy results in the enrichment of ovarian cancer stem cell-like cells leading to an increased tumor burden, (2013) 1–15. [DOI] [PMC free article] [PubMed]
- [162].Hu X, Ghisolfi L, Keates AC, Zhang J, Xiang S, Lee D, Li CJ, Hu X, Ghisolfi L, Keates AC, Zhang J, Xiang S, Lee D, Li CJ, chemotherapy Induction of cancer cell stemness by chemotherapy © 2012 Landes Bioscience. Do not distribute., 4101 (2012). 10.4161/cc.21021. [DOI] [PubMed] [Google Scholar]
- [163].Ghisolfi L, Keates AC, Hu X, Lee D, Li CJ, Ionizing Radiation Induces Stemness in Cancer Cells, 7 (2012) 1–11. 10.1371/journal.pone.0043628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Martins-Neves SR, Cleton-Jansen AM, Gomes CMF, Therapy-induced enrichment of cancer stem-like cells in solid human tumors: Where do we stand?, Pharmacol. Res 137 (2018) 193–204. 10.1016/j.phrs.2018.10.011. [DOI] [PubMed] [Google Scholar]
- [165].McCormick SM, Eskin SG, McIntire LV, Teng CL, Lu CM, Russell CG, Chittur KK, DNA microarray reveals changes in gene expression of shear stressed human umbilical vein endothelial cells, Proc. Natl. Acad. Sci. U. S. A 98 (2001) 8955–8960. 10.1073/pnas.171259298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Hamik A, Lin Z, Kumar A, Balcells M, Sinha S, Katz J, Feinberg MW, Gerszten RE, Edelman ER, Jain MK, Kruppel-like factor 4 regulates endothelial inflammation, J. Biol. Chem 282 (2007) 13769–13779. 10.1074/jbc.M700078200. [DOI] [PubMed] [Google Scholar]
- [167].Courboulin A, Tremblay VL, Barrier M, Meloche J, Jacob MH, Chapolard M, Bisserier M, Paulin R, Lambert C, Provencher S, Bonnet S, Krüppel-like Factor 5 contributes to pulmonary artery smooth muscle proliferation and resistance to apoptosis in human pulmonary arterial hypertension, Respir. Res 12 (2011) 1–10. 10.1186/1465-9921-12-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Hofmann AD, Takahashi T, Duess JW, Gosemann JH, Puri P, Increased pulmonary vascular expression of Krüppel-like factor 5 and activated survivin in experimental congenital diaphragmatic hernia, Pediatr. Surg. Int 30 (2014) 1191–1197. 10.1007/s00383-014-3606-7. [DOI] [PubMed] [Google Scholar]
- [169].Liu X, Yuan D, Nie X, Shen J, Yan Y, Zhang D, Gu J, BTEB2 Prevents Neuronal Apoptosis via Promoting Bad Phosphorylation in Rat Intracerebral Hemorrhage Model, J. Mol. Neurosci 55 (2015) 206–216. 10.1007/s12031-014-0305-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Farrugia MK, Sharma SB, Lin CC, McLaughlin SL, Vanderbilt DB, Ammer AG, Salkeni MA, Stoilov P, Agazie YM, Creighton CJ, Ruppert JM, Regulation of anti-apoptotic signaling by Kruppel-like factors 4 and 5 mediates lapatinib resistance in breast cancer, Cell Death Dis. 6 (2015) e1699. 10.1038/cddis.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Yoon HS, Chen X, Yang VW, Krüppel-like factor 4 mediates p53-dependent G1/S cell cycle arrest in response to DNA damage, J. Biol. Chem 278 (2003) 2101–2105. 10.1074/jbc.M211027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Zhang W, Geiman DE, Shields JM, Dang DT, Mahatan CS, Kaestner KH, Biggs JR, Kraft AS, Yang VW, The gut-enriched Kruppel-like factor (Kruppel-like factor 4) mediates the transactivating effect of p53 on the p21(WAF1)/(Cip)1 promoter, J. Biol. Chem 275 (2000) 18391–18398. 10.1074/jbc.C000062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Ghaleb AM, Katz JP, Kaestner KH, Du JX, Yang VW, Krüppel-like factor 4 exhibits antiapoptotic activity following γ-radiation-induced DNA damage, Oncogene. 26 (2007) 2365–2373. 10.1038/sj.onc.1210022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Hu D, Gur M, Zhou Z, Gamper A, Hung MC, Fujita N, Lan L, Bahar I, Wan Y, Interplay between arginine methylation and ubiquitylation regulates KLF4-mediated genome stability and carcinogenesis, Nat. Commun 6 (2015). 10.1038/ncomms9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Zhou Z, Huang F, Shrivastava I, Zhu R, Luo A, Hottiger M, Bahar I, Liu Z, Cristofanilli M, Wan Y, New insight into the significance of KLF4 PARylation in genome stability, carcinogenesis, and therapy, EMBO Mol. Med 12 (2020) 1–27. 10.15252/emmm.202012391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Yoon HS, Yang VW, Requirement of Krüppel-like Factor 4 in Preventing Entry into Mitosis following DNA Damage, J. Biol. Chem 279 (2004) 5035–5041. 10.1074/jbc.M307631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Yoon HS, Ghaleb AM, Nandan MO, Hisamuddin IM, Dalton WB, Yang VW, Krüppel-like factor 4 prevents centrosome amplification following γ-irradiation-induced DNA damage, Oncogene. 24 (2005) 4017–4025. 10.1038/sj.onc.1208576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Li Y, Xian M, Yang B, Ying M, He Q, Inhibition of KLF4 by Statins Reverses Adriamycin-Induced Metastasis and Cancer Stemness in Osteosarcoma Cells, Stem Cell Reports. 8 (2017) 1617–1629. 10.1016/j.stemcr.2017.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Li X, Liu X, Xu Y, Liu J, Xie M, Ni W, Chen S, KLF5 promotes hypoxia-induced survival and inhibits apoptosis in non-small cell lung cancer cells via HIF-1α, Int. J. Oncol 45 (2014) 1507–1514. 10.3892/ijo.2014.2544. [DOI] [PubMed] [Google Scholar]
- [180].Zhang H, Shao F, Guo W, Gao Y, He J, Knockdown of KLF5 promotes cisplatin-induced cell apoptosis via regulating DNA damage checkpoint proteins in non-small cell lung cancer, Thorac. Cancer 10 (2019) 1069–1077. 10.1111/1759-7714.13046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Che M, Chaturvedi A, Munro SA, Pitzen SP, Ling A, Zhang W, Mentzer J, Ku SY, Puca L, Zhu Y, Bergman AM, Severson TM, Forster C, Liu Y, Hildebrand J, Daniel M, Wang TY, Selth LA, Hickey T, Zoubeidi A, Gleave M, Bareja R, Sboner A, Tilley W, Carroll JS, Tan W, Kohli M, Yang R, Hsieh AC, Murugan P, Zwart W, Beltran H, Huang RS, Dehm SM, Opposing transcriptional programs of KLF5 and AR emerge during therapy for advanced prostate cancer, Nat. Commun 12 (2021). 10.1038/s41467-021-26612-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Bissey PA, Teng M, Law JH, Shi W, Bruce JP, Petit V, Tsao SW, Yip KW, Liu FF, MiR-34c downregulation leads to SOX4 overexpression and cisplatin resistance in nasopharyngeal carcinoma, BMC Cancer. 20 (2020) 597. 10.1186/s12885-020-07081-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Huang Q, Xing S, Peng A, Yu Z, NORAD accelerates chemo-resistance of non-small-cell lung cancer via targeting at miR-129-1-3p/SOX4 axis, Biosci. Rep 40 (2020) 1–12. 10.1042/BSR20193489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Hu B, Zhang H, Wang Z, Zhang F, Wei H, Li L, LncRNA CCAT1/miR-130a-3p axis increases cisplatin resistance in non-small-cell lung cancer cell line by targeting SOX4, Cancer Biol. Ther 18 (2017) 974–983. 10.1080/15384047.2017.1385679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Pan S, Bao D, Li Y, Liu D, Quan S, Wang R, SOX4 induces drug resistance of colorectal cancer cells by downregulating CYLD through transcriptional activation of microRNA-17, J. Biochem. Mol. Toxicol 36 (2022). 10.1002/jbt.22910. [DOI] [PubMed] [Google Scholar]
- [186].Voronkova MA, Rojanasakul LW, Kiratipaiboon C, Rojanasakul Y, The SOX9-Aldehyde Dehydrogenase Axis Determines Resistance to Chemotherapy in Non-Small-Cell Lung Cancer, Mol. Cell. Biol 40 (2020). 10.1128/mcb.00307-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Xue Y, Lian W, Zhi J, Yang W, Li Q, Guo X, Gao J, Qu H, Lin W, Li Z, Lai L, Wang Q, HDAC5-mediated deacetylation and nuclear localisation of SOX9 is critical for tamoxifen resistance in breast cancer, Br. J. Cancer 121 (2019) 1039–1049. 10.1038/s41416-019-0625-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Tripathi SK, Biswal BK, SOX9 promotes epidermal growth factor receptor-tyrosine kinase inhibitor resistance via targeting β-catenin and epithelial to mesenchymal transition in lung cancer, Life Sci. 277 (2021) 119608. 10.1016/j.lfs.2021.119608. [DOI] [PubMed] [Google Scholar]
- [189].Liu Y, Wu H, Luo T, Luo Q, Meng Z, Shi Y, Li F, Liu M, Peng X, Liu J, Xu C, Tang W, The SOX9-MMS22L Axis Promotes Oxaliplatin Resistance in Colorectal Cancer, Front. Mol. Biosci 8 (2021) 1–10. 10.3389/fmolb.2021.646542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Wang Z, Xu X, Nan-Liu Y. Cheng, Jin W, Zhang P, Wang X, Yang H, Liu H, Tu Y, SOX9-PDK1 axis is essential for glioma stem cell self-renewal and temozolomide resistance, Oncotarget. 9 (2018) 192–204. 10.18632/oncotarget.22773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Wang M, Wang Z, Zhi X, Ding W, Xiong J, Tao T, Yang Y, Zhang H, Zi X, Zhou W, Huang G, SOX9 enhances sorafenib resistance through upregulating ABCG2 expression in hepatocellular carcinoma, Biomed. Pharmacother 129 (2020) 110315. 10.1016/j.biopha.2020.110315. [DOI] [PubMed] [Google Scholar]
- [192].Si X, Gao Z, Xu F, Zheng Y, SOX2 upregulates side population cells and enhances their chemoresistant ability by transactivating ABCC1 expression contributing to intrinsic resistance to paclitaxel in melanoma, Mol. Carcinog 59 (2020) 257–264. 10.1002/mc.23148. [DOI] [PubMed] [Google Scholar]
- [193].Bagati A, Kumar S, Jiang P, Pyrdol J, Zou AE, Godicelj A, Mathewson ND, Cartwright ANR, Cejas P, Brown M, Giobbie-Hurder A, Dillon D, Agudo J, Mittendorf EA, Liu XS, Wucherpfennig KW, Integrin αvβ6–TGFβ–SOX4 Pathway Drives Immune Evasion in Triple-Negative Breast Cancer, Cancer Cell. 39 (2021) 54–67.e9. 10.1016/j.ccell.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Ashkenazi S, Ortenberg R, Besser M, Schachter J, Markel G, SOX9 indirectly regulates CEACAM1 expression and immune resistance in melanoma cells, Oncotarget. 7 (2016) 30166–30177. 10.18632/oncotarget.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Wu R, Wang C, Li Z, Xiao J, Li C, Wang X, Kong P, Cao J, Huang F, Li Z, Huang Y, Chen Y, Li X, Yang D, Zhang H, Mai J, Feng G, Deng R, Zhu X, SOX2 promotes resistance of melanoma with PD-L1 high expression to T-cell-mediated cytotoxicity that can be reversed by SAHA, J. Immunother. Cancer 8 (2020) 1–13. 10.1136/jitc-2020-001037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Yang WT, Zheng PS, Promoter hypermethylation of KLF4 inactivates its tumor suppressor function in cervical carcinogenesis, PLoS One. 9 (2014). 10.1371/journal.pone.0088827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Riverso M, Montagnani V, Stecca B, KLF4 is regulated by RAS/RAF/MEK/ERK signaling through E2F1 and promotes melanoma cell growth, Oncogene. 36 (2017) 3322–3333. 10.1038/onc.2016.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Choi H, Roh J, LH-induced transcriptional regulation of KLF4 expression in granulosa cells occurs via the camp/pka pathway and requires a putative sp1 binding site, Int. J. Mol. Sci 21 (2020) 1–14. 10.3390/ijms21197385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS, MicroRNA-145 Regulates OCT4, SOX2, and KLF4 and Represses Pluripotency in Human Embryonic Stem Cells, Cell. 137 (2009) 647–658. 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- [200].Tian Y, Luo A, Cai Y, Su Q, Ding F, Chen H, Liu Z, MicroRNA-10b promotes migration and invasion through KLF4 in human esophageal cancer cell lines, J. Biol. Chem 285 (2010) 7986–7994. 10.1074/jbc.M109.062877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Okuda H, Xing F, Pandey PR, Sharma S, Watabe M, Pai SK, miR-7 Suppresses Brain Metastasis of Breast Cancer Stem-Like Cells By Modulating KLF4, 73 (2013) 1434–1444. 10.1158/0008-5472.CAN-12-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Lv H, Zhang Z, Wang Y, Li C, Gong W, Wang X, MicroRNA-92a promotes colorectal cancer cell growth and migration by inhibiting KLF4, Oncol. Res 23 (2016) 283–290. 10.3727/096504016X14562725373833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Chen HY, Lin YM, Chung HC, Lang YD, Lin CJ, Huang J, Wang WC, Lin FM, Chen Z, Da Huang H, Shyy JYJ, Liang JT, Chen RH, MiR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4, Cancer Res. 72 (2012) 3631–3641. 10.1158/0008-5472.CAN-12-0667. [DOI] [PubMed] [Google Scholar]
- [204].Hu D, Zhou Z, Davidson NE, Huang Y, Wan Y, Novel insight into KLF4 proteolytic regulation in estrogen receptor signaling and breast carcinogenesis, J. Biol. Chem 287 (2012) 13584–13597. 10.1074/jbc.M112.343566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Hu D, Wan Y, Regulation of Krüppel-like factor 4 by the anaphase promoting complex pathway is involved in TGF-β signaling, J. Biol. Chem 286 (2011) 6890–6901. 10.1074/jbc.M110.179952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Kim MO, Kim SH, Cho YY, Nadas J, Jeong CH, Yao K, Kim DJ, Yu DH, Keum YS, Lee KY, Huang Z, Bode AM, Dong Z, ERK1 and ERK2 regulate embryonic stem cell self-renewal through phosphorylation of Klf4, Nat. Struct. Mol. Biol 19 (2012) 283–290. 10.1038/nsmb.2217. [DOI] [PubMed] [Google Scholar]
- [207].Hao Z, Sheng Y, Duncan GS, Li WY, Dominguez C, Sylvester J, Su YW, Lin GHY, Snow BE, Brenner D, You-Ten A, Haight J, Inoue S, Wakeham A, Elford A, Hamilton S, Liang Y, Zúñiga-Pflücker JC, He HH, Ohashi PS, Mak TW, K48-linked KLF4 ubiquitination by E3 ligase Mule controls T-cell proliferation and cell cycle progression, Nat. Commun 8 (2017). 10.1038/ncomms14003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Zou H, Chen H, Zhou Z, Wan Y, Liu Z, ATXN3 promotes breast cancer metastasis by deubiquitinating KLF4, Cancer Lett. 467 (2019) 19–28. 10.1016/j.canlet.2019.09.012. [DOI] [PubMed] [Google Scholar]
- [209].Ma B, Zhang L, Zou Y, He R, Wu Q, Han C, Zhang B, Reciprocal regulation of integrin β 4 and KLF4 promotes gliomagenesis through maintaining cancer stem cell traits, 1 (2019) 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Hu D, Wan Y, Regulation of Krüppel-like factor 4 by the anaphase promoting complex pathway is involved in TGF-beta signaling., J. Biol. Chem 286 (2011) 6890–6901. 10.1074/jbc.M110.179952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Zhou Z, Feng Z, Hu D, Yang P, Gur M, Bahar I, Cristofanilli M, Gradishar WJ, Xie X, Wan Y, EBioMedicine A novel small-molecule antagonizes PRMT5-mediated KLF4 methylation for targeted therapy, EBioMedicine. 44 (2019) 98–111. 10.1016/j.ebiom.2019.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Evans PM, Zhang W, Chen X, Yang J, Bhakat KK, Liu C, Krüppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation, J. Biol. Chem 282 (2007) 33994–34002. 10.1074/jbc.M701847200. [DOI] [PubMed] [Google Scholar]
- [213].min Jia Z, Ai X, fei Teng J, peng Wang Y, jun Wang B, Zhang X, p21 and CK2 interaction-mediated HDAC2 phosphorylation modulates KLF4 acetylation to regulate bladder cancer cell proliferation, Tumor Biol. 37 (2016) 8293–8304. 10.1007/s13277-015-4618-1. [DOI] [PubMed] [Google Scholar]
- [214].Du JX, McConnell BB, Yang VW, A small ubiquitin-related modifier-interacting motif functions as the transcriptional activation domain of Krüppel-like factor 4, J. Biol. Chem 285 (2010) 28298–28308. 10.1074/jbc.M110.101717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Kawai-Kowase K, Ohshima T, Matsui H, Tanaka T, Shimizu T, Iso T, Arai M, Owens GK, Kurabayashi M, PIAS1 mediates TGFβ-induced SM α-actin gene expression through inhibition of KLF4 function-expression by protein sumoylation, Arterioscler. Thromb. Vasc. Biol 29 (2009) 99–106. 10.1161/ATVBAHA.108.172700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Tahmasebi S, Ghorbani M, Savage P, Yan K, Gocevski G, Xiao L, You L, Yang XJ, Sumoylation of Krüppel-like factor 4 inhibits pluripotency induction but promotes adipocyte differentiation, J. Biol. Chem 288 (2013) 12791–12804. 10.1074/jbc.M113.465443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Li H, Han M, Bernier M, Zheng B, Sun S, Su M, Zhang R, Fu J, Wen J, Krüppel-like factor 4 promotes differentiation by transforming growth factor-beta receptor-mediated Smad and p38 MAPK signaling in vascular smooth muscle cells., J. Biol. Chem 285 (2010) 17846–17856. 10.1074/jbc.M109.076992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Zhang X, Choi PS, Francis JM, Gao GF, Campbell JD, Ramachandran A, Mitsuishi Y, Ha G, Shih J, Vazquez F, Tsherniak A, Taylor AM, Zhou J, Wu Z, Berger AC, Giannakis M, Hahn WC, Cherniack AD, Meyerson M, Somatic superenhancer duplications and hotspot mutations lead to oncogenic activation of the KLF5 transcription factor, Cancer Discov. 8 (2018) 108–125. 10.1158/2159-8290.CD-17-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Chanchevalap S, Nandan MO, Merlin D, Yang VW, All- trans retinoic acid inhibits proliferation of intestinal epithelial cells by inhibiting expression of the gene encoding Kru, 578 (2004) 99–105. 10.1016/j.febslet.2004.10.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Li J, Zhang B, Liu M, Fu X, Ci X, Jun A, Fu C, Dong G, Wu R, Zhang Z, Fu L, Dong JT, Klf5 is crucial for androgen-ar signaling to transactivate genes and promote cell proliferation in prostate cancer cells, Cancers (Basel). 12 (2020). 10.3390/cancers12030748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Zhang H, Bialkowska A, Rusovici R, Chanchevalap S, Shim H, Katz JP, Yang VW, Yun CC, Lysophosphatidic acid facilitates proliferation of colon cancer cells via induction of Krüppel-like factor 5, J. Biol. Chem 282 (2007) 15541–15549. 10.1074/jbc.M700702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Liu R, Shi P, Nie Z, Liang H, Zhou Z, Chen W, Chen H, Dong C, Yang R, Liu S, Chen C, Mifepristone suppresses basal triple-negative breast cancer stem cells by down-regulating KLF5 expression, Theranostics. 6 (2016) 533–544. 10.7150/thno.14315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Zhou W, Song F, Wu Q, Liu R, Wang L, Liu C, Peng Y, Mao S, Feng J, Chen C, MIR-217 inhibits triple-negative breast cancer cell growth, migration, and invasion through targeting KLF5, PLoS One. 12 (2017) 1–12. 10.1371/journal.pone.0176395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Jiang Z, Zhang Y, Cao R, Li L, Zhong K, Chen Q, Xiao J, MiR-5195–3p inhibits proliferation and invasion of human bladder cancer cells by directly targeting oncogene KLF5, Oncol. Res 25 (2017) 1081–1087. 10.3727/096504016X14831120463349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Cai W, Xu Y, Yin J, Zuo W, Su Z, miR-590–5p suppresses osteosarcoma cell proliferation and invasion via targeting KLF5, Mol. Med. Rep 18 (2018) 2328–2334. 10.3892/mmr.2018.9173. [DOI] [PubMed] [Google Scholar]
- [226].Zhou T, Chen S, Mao X, miR-145–5p affects the differentiation of gastric cancer by targeting KLF5 directly, J. Cell. Physiol 234 (2019) 7634–7644. 10.1002/jcp.27525. [DOI] [PubMed] [Google Scholar]
- [227].Zhou Y, Xu Q, Shang J, Lu L, Chen G, Crocin inhibits the migration, invasion, and epithelial-mesenchymal transition of gastric cancer cells via miR-320/KLF5/HIF-1α signaling, J. Cell. Physiol 234 (2019) 17876–17885. 10.1002/jcp.28418. [DOI] [PubMed] [Google Scholar]
- [228].Luan Y, Wang P, FBW7-mediated ubiquitination and degradation of KLF5., World J. Biol. Chem 5 (2014) 216–23. 10.4331/wjbc.v5.i2.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Zhao D, Zheng HQ, Zhou Z, Chen C, The Fbw7 tumor suppressor targets KLF5 for ubiquitin-mediated degradation and suppresses breast cell proliferation, Cancer Res. 70 (2010) 4728–4738. 10.1158/0008-5472.CAN-10-0040. [DOI] [PubMed] [Google Scholar]
- [230].Chen C, Sun X, Guo P, Dong XY, Sethi P, Cheng X, Zhou J, Ling J, Simons JW, Lingrel JB, Dong JT, Human Kruppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells, J. Biol. Chem 280 (2005) 41553–41561. 10.1074/jbc.M506183200. [DOI] [PubMed] [Google Scholar]
- [231].Zhao KW, Sikriwal D, Dong X, Guo P, Sun X, Dong JT, Oestrogen causes degradation of KLF5 by inducing the E3 ubiquitin ligase EFP in ER-positive breast cancer cells, Biochem. J 437 (2011) 323–333. 10.1042/BJ20101388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Du JX, Hagos EG, Nandan MO, Bialkowska AB, Yu B, Yang VW, The E3 ubiquitin ligase SMAD ubiquitination regulatory factor 2 negatively regulates Krüppel-like factor 5 protein, J. Biol. Chem 286 (2011) 40354–40364. 10.1074/jbc.M111.258707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Wu Y, Qin J, Li F, Yang C, Li Z, Zhou Z, Zhang H, Li Y, Wang X, Liu R, Tao Q, Chen W, Chen C, USP3 promotes breast cancer cell proliferation by deubiquitinating KLF5, J. Biol. Chem 294 (2019) 17837–17847. 10.1074/jbc.RA119.009102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Qin J, Zhou Z, Chen W, Wang C, Zhang H, Ge G, Shao M, You D, Fan Z, Xia H, Liu R, Chen C, BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5, Nat. Commun 6 (2015). 10.1038/ncomms9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Ge F, Chen W, Qin J, Zhou Z, Liu R, Liu L, Tan J, Zou T, Li H, Ren G, Chen C, Ataxin-3 like (ATXN3L), a member of the Josephin family of deubiquitinating enzymes, promotes breast cancer proliferation by deubiquitinating Krüppel-like factor 5 (KLF5), Oncotarget. 6 (2015) 21369–21378. 10.18632/oncotarget.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Du JX, Bialkowska AB, McConnell BB, Yang VW, SUMOylation regulates nuclear localization of Krüppel-like factor 5, J. Biol. Chem 283 (2008) 31991–32002. 10.1074/jbc.M803612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Zhang Z, Teng CT, Phosphorylation of Kruppel-like factor 5 (KLF5/IKLF) at the CBP interaction region enhances its transactivation function, Nucleic Acids Res. 31 (2003) 2196–2208. 10.1093/nar/gkg310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].He M, Han M, Zheng B, Shu YN, Wen JK, Angiotensin II stimulates KLF5 phosphorylation and its interaction with c-Jun leading to suppression of p21 expression in vascular smooth muscle cells, J. Biochem 146 (2009) 683–691. 10.1093/jb/mvp115. [DOI] [PubMed] [Google Scholar]
- [239].Tao R, Zhang B, Li Y, King JL, Tian R, Xia S, Schiavon CR, Dong JT, HDAC-mediated deacetylation of KLF5 associates with its proteasomal degradation, Biochem. Biophys. Res. Commun 500 (2018) 777–782. 10.1016/j.bbrc.2018.04.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Song S, Ajani JA, Honjo S, Maru DM, Chen Q, Scott AW, Heallen TR, Xiao L, Hofstetter WL, Weston B, Lee JH, Wadhwa R, Sudo K, Stroehlein JR, Martin JF, Hung MC, Johnson RL, Hippo coactivator YAP1 upregulates SOX9 and endows esophageal Cancer cells with stem-like properties, Cancer Res. 74 (2014) 4170–4182. 10.1158/0008-5472.CAN-13-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Bora-Singhal N, Nguyen J, Schaal C, Perumal D, Singh S, Coppola D, Chellappan S, YAP1 regulates OCT4 activity and SOX2 expression to facilitate self-renewal and vascular mimicry of stem-like cells, Stem Cells. 33 (2015) 1705–1718. 10.1002/stem.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Williams CAC, Soufi A, Pollard SM, Post-translational modification of SOX family proteins: Key biochemical targets in cancer?, Semin. Cancer Biol 67 (2020) 30–38. 10.1016/j.semcancer.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Jang SM, Kim JW, Kim CH, An JH, Johnson A, Song PI, Rhee S, Choi KH, KAT5-mediated SOX4 acetylation orchestrates chromatin remodeling during myoblast differentiation, Cell Death Dis. 6 (2015) 1–11. 10.1038/cddis.2015.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Muz B, de la Puente P, Azab F, Azab AK, The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy, Hypoxia. (2015) 83. 10.2147/hp.s93413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Emami Nejad A, Najafgholian S, Rostami A, Sistani A, Shojaeifar S, Esparvarinha M, Nedaeinia R, Haghjooy Javanmard S, Taherian M, Ahmadlou M, Salehi R, Sadeghi B, Manian M, The role of hypoxia in the tumor microenvironment and development of cancer stem cell: a novel approach to developing treatment, Cancer Cell Int. 21 (2021) 1–26. 10.1186/s12935-020-01719-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Wang P, Zhao L, Gong S, Xiong S, Wang J, Zou D, Pan J, Deng Y, Yan Q, Wu N, Liao B, HIF1α/HIF2α–Sox2/Klf4 promotes the malignant progression of glioblastoma via the EGFR–PI3K/AKT signalling pathway with positive feedback under hypoxia, Cell Death Dis. 12 (2021). 10.1038/s41419-021-03598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Shan F, Huang Z, Xiong R, Huang QY, Li J, HIF1α-induced upregulation of KLF4 promotes migration of human vascular smooth muscle cells under hypoxia, J. Cell. Physiol 235 (2020) 141–150. 10.1002/jcp.28953. [DOI] [PubMed] [Google Scholar]
- [248].Von Spreckelsen N, Waldt N, Poetschke R, Kesseler C, Dohmen H, Jiao HK, Nemeth A, Schob S, Scherlach C, Sandalcioglu IE, Deckert M, Angenstein F, Krischek B, Stavrinou P, Timmer M, Remke M, Kirches E, Goldbrunner R, Chiocca EA, Huettelmaier S, Acker T, Mawrin C, KLF4 K409Q-mutated meningiomas show enhanced hypoxia signaling and respond to mTORC1 inhibitor treatment, Acta Neuropathol. Commun 8 (2020) 1–11. 10.1186/s40478-020-00912-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Mori A, Moser C, Lang SA, Hackl C, Gottfried E, Kreutz M, Schlitt HJ, Geissler EK, Stoeltzing O, Up-regulation of Krüppel-like factor 5 in pancreatic cancer is promoted by interleukin-1β signaling and hypoxia-inducible factor-1α, Mol. Cancer Res 7 (2009) 1390–1398. 10.1158/1541-7786.MCR-08-0525. [DOI] [PubMed] [Google Scholar]
- [250].Gong T, Cui L, Wang H, Wang H, Han N, Knockdown of KLF5 suppresses hypoxia-induced resistance to cisplatin in NSCLC cells by regulating HIF-1α-dependent glycolysis through inactivation of the PI3K/Akt/mTOR pathway, J. Transl. Med 16 (2018) 1–10. 10.1186/s12967-018-1543-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Bae KM, Dai Y, Vieweg J, Siemann DW, Hypoxia regulates SOX2 expression to promote prostate cancer cell invasion and sphere formation, Am. J. Cancer Res 6 (2016) 1078–1088. [PMC free article] [PubMed] [Google Scholar]
- [252].Chen G, Liu B, Yin S, Li S, Guo Y, Wang M, Wang K, Wan X, Hypoxia induces an endometrial cancer stem-like cell phenotype via HIF-dependent demethylation of SOX2 mRNA, Oncogenesis. 9 (2020). 10.1038/s41389-020-00265-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Seo EJ, Kim DK, Jang IH, Choi EJ, Shin SH, Lee SI, Kwon SM, Kim KH, Suh DS, Kim JH, Hypoxia-NOTCH1-SOX2 signaling is important for maintaining cancer stem cells in ovarian cancer, Oncotarget. 7 (2016) 55624–55638. 10.18632/oncotarget.10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Petruzzelli R, Christensen DR, Parry KL, Sanchez-Elsner T, Houghton FD, HIF-2α regulates NANOG expression in human embryonic stem cells following hypoxia and reoxygenation through the interaction with an oct-sox cis regulatory element, PLoS One. 9 (2014) 1–11. 10.1371/journal.pone.0108309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Wang Y, Bibi M, Min P, Deng W, Zhang Y, Du J, SOX2 promotes hypoxia-induced breast cancer cell migration by inducing NEDD9 expression and subsequent activation of Rac1/HIF-1α signaling, Cell. Mol. Biol. Lett 24 (2019) 1–12. 10.1186/s11658-019-0180-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Gao H, Teng C, Huang W, Peng J, Wang C, SOX2 promotes the epithelial to mesenchymal transition of esophageal squamous cells by modulating slug expression through the activation of STAT3/HIF-α signaling, Int. J. Mol. Sci 16 (2015) 21643–21657. 10.3390/ijms160921643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Gan L, Meng J, Xu M, Liu M, Qi Y, Tan C, Wang Y, Zhang P, Weng W, Sheng W, Huang M, Wang Z, Extracellular matrix protein 1 promotes cell metastasis and glucose metabolism by inducing integrin β4/FAK/SOX2/HIF-1α signaling pathway in gastric cancer, Oncogene. 37 (2018) 744–755. 10.1038/onc.2017.363. [DOI] [PubMed] [Google Scholar]
- [258].Sun J, Xiong Y, Jiang K, Xin B, Jiang T, Wei R, Zou Y, Tan H, Jiang T, Yang A, Jia L, Wang L, Hypoxia-sensitive long noncoding RNA CASC15 promotes lung tumorigenesis by regulating the SOX4/β-catenin axis, J. Exp. Clin. Cancer Res 40 (2021) 1–14. 10.1186/s13046-020-01806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Raspaglio G, Petrillo M, Martinelli E, Li Puma DD, Mariani M, De Donato M, Filippetti F, Mozzetti S, Prislei S, Zannoni GF, Scambia G, Ferlini C, Sox9 and Hif-2α regulate TUBB3 gene expression and affect ovarian cancer aggressiveness, Gene. 542 (2014) 173–181. 10.1016/j.gene.2014.03.037. [DOI] [PubMed] [Google Scholar]
- [260].Choi BJ, Park SA, Lee SY, Cha YN, Surh YJ, Hypoxia induces epithelial-mesenchymal transition in colorectal cancer cells through ubiquitin-specific protease 47-mediated stabilization of Snail: A potential role of Sox9, Sci. Rep 7 (2017) 1–15. 10.1038/s41598-017-15139-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Camaj P, Jackel C, Krebs S, DeToni EN, Blum H, Jauch KW, Nelson PJ, Bruns CJ, Hypoxia-independent gene expression mediated by SOX9 promotes aggressive pancreatic tumor biology, Mol. Cancer Res 12 (2014) 421–432. 10.1158/1541-7786.MCR-13-0351. [DOI] [PubMed] [Google Scholar]
- [262].Cercek A, Wheler J, Murray PE, Zhou S, Saltz L, Phase 1 study of APTO-253 HCl, an inducer of KLF4, in patients with advanced or metastatic solid tumors, Invest. New Drugs 33 (2015) 1086–1092. 10.1007/s10637-015-0273-z. [DOI] [PubMed] [Google Scholar]
- [263].Bialkowska AB, Du Y, Fu H, Yang VW, Identification of novel small-molecule compounds that inhibit the proproliferative Krüppel-like factor 5 in colorectal cancer cells by high-throughput screening, Mol. Cancer Ther 8 (2009) 563–570. 10.1158/1535-7163.MCT-08-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Yagi N, Manabe I, Tottori T, Ishihara A, Ogata F, Jong HK, Nishimura S, Fujiu K, Oishi Y, Itaka K, Kato Y, Yamauchi M, Nagai R, A nanoparticle system specifically designed to deliver short interfering RNA inhibits tumor growth in vivo, Cancer Res. 69 (2009) 6531–6538. 10.1158/0008-5472.CAN-08-3945. [DOI] [PubMed] [Google Scholar]
- [265].Gao Y, Shi Q, Xu S, Du C, Liang L, Wu K, Wang K, Wang X, Chang LS, He D, Guo P, Curcumin Promotes KLF5 Proteasome Degradation through Downregulating YAP/TAZ in Bladder Cancer Cells, Int. J. Mol. Sci 15 (2014) 15173–15187. 10.3390/ijms150915173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Zhi X, Zhao D, Zhou Z, Liu R, Chen C, YAP promotes breast cell proliferation and survival partially through stabilizing the KLF5 transcription factor, Am. J. Pathol 180 (2012) 2452–2461. 10.1016/j.ajpath.2012.02.025. [DOI] [PubMed] [Google Scholar]
- [267].Zhao D, Zhi X, Zhou Z, Chen C, TAZ antagonizes the WWP1-mediated KLF5 degradation and promotes breast cell proliferation and tumorigenesis, Carcinogenesis. 33 (2012) 59–67. 10.1093/carcin/bgr242. [DOI] [PubMed] [Google Scholar]
- [268].Chia NY, Deng N, Das K, Huang D, Hu L, Zhu Y, Lim KH, Lee MH, Wu J, Sam XX, Tan GS, Wan WK, Yu W, Gan A, Tan ALK, Tay ST, Soo KC, Wong WK, Dominguez LTM, Ng HH, Rozen S, Goh LK, The BT, Tan P, Regulatory crosstalk between lineage-survival oncogenes KLF5, GATA4 and GATA6 cooperatively promotes gastric cancer development, Gut. 64 (2015) 707–719. 10.1136/gutjnl-2013-306596. [DOI] [PubMed] [Google Scholar]
- [269].Feinberg MW, Cao Z, Wara AK, Lebedeva MA, SenBanerjee S, Jain MK, Kruppel-like Factor 4 Is a Mediator of Proinflammatory Signaling in Macrophages*, J. Biol. Chem 280 (2005) 38247–38258. https://doi.org/ 10.1074/jbc.M509378200. [DOI] [PubMed] [Google Scholar]
- [270].Liao X, Sharma N, Kapadia F, Zhou G, Lu Y, Hong H, Paruchuri K, Mahabeleshwar GH, Dalmas E, Venteclef N, Flask CA, Kim J, Doreian BW, Lu KQ, Kaestner KH, Hamik A, Clément K, Jain MK, Krüppel-like factor 4 regulates macrophage polarization, 121 (2011) 2736–2749. 10.1172/JCI45444.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].Xia W, Zou C, Chen H, Xie C, Hou M, Immune checkpoint inhibitor induces cardiac injury through polarizing macrophages via modulating microRNA-34a / Kruppel-like factor 4 signaling, Cell Death Dis. (2020). 10.1038/s41419-020-02778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [272].Arora S, Singh P, Ahmad S, Ahmad T, Dohare R, Almatroodi SA, Alrumaihi F, Rahmani AH, Syed MA, Comprehensive Integrative Analysis Reveals the Association of KLF4 with Macrophage Infiltration and Polarization in Lung Cancer Microenvironment, Cells. 10 (2021). 10.3390/cells10082091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Ou D-L, Chen W, Lee S, Regorafenib enhances antitumor immunity via inhibition of p38 kinase / associated Creb1 / Klf4 axis in tumor- - macrophages, (2021) 1–14. 10.1136/jitc-2020-001657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Shao J, Yan Y, Ding D, Wang D, He Y, Pan Y, Yan W, Kharbanda A, yu Li H, Huang H, Destruction of DNA-Binding Proteins by Programmable Oligonucleotide PROTAC (O’PROTAC): Effective Targeting of LEF1 and ERG, Adv. Sci 8 (2021) 1–10. 10.1002/advs.202102555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Liu J, Chen H, Kaniskan HÜ, Xie L, Chen X, Jin J, Wei W, TF-PROTACs Enable Targeted Degradation of Transcription Factors, J. Am. Chem. Soc 143 (2021) 8902–8910. 10.1021/jacs.1c03852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Hu B, Zhou Y, Sun D, Yang Y, Liu Y, Li X, Li H, Chen L, PROTACs: New method to degrade transcription regulating proteins, Eur. J. Med. Chem 207 (2020) 112698. 10.1016/j.ejmech.2020.112698. [DOI] [PubMed] [Google Scholar]
- [277].Ivanov AA, Khuri FR, Fu H, Targeting protein-protein interactions as an anticancer strategy, Trends Pharmacol. Sci 34 (2013) 393–400. 10.1016/j.tips.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Hu Z, Crews CM, Recent Developments in PROTAC-Mediated Protein Degradation: From Bench to Clinic, ChemBioChem. 23 (2022). 10.1002/cbic.202100270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Sakamoto KM, Kim KB, Verma R, Ransick A, Stein B, Crews CM, Deshaies RJ, Development of Protacs to target cancer-promoting proteins for ubiquitination and degradation., Mol. Cell. Proteomics 2 (2003) 1350–1358. 10.1074/mcp.T300009-MCP200. [DOI] [PubMed] [Google Scholar]
- [280].Zhou H, Bai L, Xu R, Zhao Y, Chen J, McEachern D, Chinnaswamy K, Wen B, Dai L, Kumar P, Yang CY, Liu Z, Wang M, Liu L, Meagher JL, Yi H, Sun D, Stuckey JA, Wang S, Structure-Based Discovery of SD-36 as a Potent, Selective, and Efficacious PROTAC Degrader of STAT3 Protein, J. Med. Chem 62 (2019) 11280–11300. 10.1021/acs.jmedchem.9b01530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [281].Bai L, Zhou H, Xu R, Zhao Y, Chinnaswamy K, McEachern D, Chen J, Yang CY, Liu Z, Wang M, Liu L, Jiang H, Wen B, Kumar P, Meagher JL, Sun D, Stuckey JA, Wang S, A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo, Cancer Cell. 36 (2019) 498–511.e17. 10.1016/j.ccell.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [282].Schuetz A, Nana D, Rose C, Zocher G, Milanovic M, Koenigsmann J, Blasig R, Heinemann U, Carstanjen D, The structure of the Klf4 DNA-binding domain links to self-renewal and macrophage differentiation, Cell. Mol. Life Sci 68 (2011) 3121–3131. 10.1007/s00018-010-0618-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [283].Buchwald P, Small-molecule protein-protein interaction inhibitors: Therapeutic potential in light of molecular size, chemical space, and ligand binding efficiency considerations, IUBMB Life. 62 (2010) 724–731. 10.1002/iub.383. [DOI] [PubMed] [Google Scholar]
- [284].Berg T, Cohen SB, Desharnais J, Sonderegger C, Maslyar DJ, Goldberg J, Boger DL, Vogt PK, Small-molecule antagonists of Myc/Max dimerization inhibit Myc-induced transformation of chicken embryo fibroblasts, Proc. Natl. Acad. Sci. U. S. A 99 (2002) 3830–3835. 10.1073/pnas.062036999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [285].Ishay-Ronen D, Diepenbruck M, Kalathur RKR, Sugiyama N, Tiede S, Ivanek R, Bantug G, Morini MF, Wang J, Hess C, Christofori G, Gain Fat—Lose Metastasis: Converting Invasive Breast Cancer Cells into Adipocytes Inhibits Cancer Metastasis, Cancer Cell. 35 (2019) 17–32.e6. 10.1016/j.ccell.2018.12.002. [DOI] [PubMed] [Google Scholar]


