Highlights
-
•
Seven ferroptosis-related genes that might play a role in periodontitis were identified.
-
•
The ferroptosis-related ceRNA network was established.
-
•
A new role of interleukin 1 beta in pathogenesis was found.
Key words: Ferroptosis, Periodontitis, ceRNA network, Noncoding RNA
Abstract
Objectives
Periodontitis is a chronic inflammatory illness that may lead to tooth loosening and even loss, and its pathogenesis is not fully understood. Ferroptosis is an iron-dependent, regulated cell death. The present study aims to find the key ferroptosis-related genes (FRGs) in periodontitis and develop an mRNA-miRNA-lncRNA network to deeply explore the pathogenesis of periodontitis.
Methods
Data from the Gene Expression Omnibus (GEO) database and FerrDb database were downloaded to discover the differentially expressed mRNA, miRNA, and FRGs. Functional enrichment analysis was conducted for the differentially expressed FRGs (DE-FRGs), including gene ontology, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, and protein–protein interaction (PPI) network analysis. Targetscan and miRtarbase were used to estimate the miRNAs that DE-FRGs may interact with, whilst StarBase v3.0 was used for lncRNA-miRNA interaction.
Results
Seven DE-FRGs were identified through differential expression analysis. Interleukin 1 beta (IL1B) interacted with XBP1 and MMP13 in the PPI network. After taking the intersection between DE-miRNAs and predicted miRNAs, a ceRNA network containing IL1B, has-miR-185, has-miR-204, has-miR-211, has-miR-4306, and 28 lncRNAs was established.
Conclusions
Seven FRGs in periodontitis were identified, which might promote deeper understanding of ferroptosis in periodontitis.
Introduction
Periodontitis is a chronic inflammatory illness that causes the deterioration of periodontal soft and hard tissues all over the world.1 It is one of the most common oral disorders and is triggered by a myriad of factors.2 It causes gradual loss of attachment and alveolar bone degradation, which can lead to tooth loosening, displacement, and even loss.3 Periodontitis is thought to have a bidirectional link with a number of systemic disorders, including heart disease and diabetes, and causes substantial harm to overall health and well-being, including edentulousness, masticatory dysfunction, low patient self-esteem, increased risk of poor glycemic control, and unfavourable pregnancy outcomes.4 Microbial infection and the host immune inflammatory response are important components in periodontitis pathogenesis, and genetic and epigenetic factors can influence periodontitis progression by modifying the host immunologic response evoked by periodontal pathogens.5 However, there are still numerous issues in the molecular mechanism of periodontitis pathogenesis that need to be investigated further.
Ferroptosis is an iron-dependent, regulated cell death characterised by the accumulation of iron-dependent reactive oxygen species (ROS), leading to excessive lipid peroxidation and cell death.6 Intracellular glutaminolysis and disruption of glutathione peroxidase 4 (GPX4) result in diminished cellular antioxidant capacity and inadequate degradation of ROS and peroxidised lipids during ferroptosis.7 Generally, intracellular iron is mainly stored in ferritin, and the cargo receptor nuclear receptor coactivator 4 can transport ferritin-bound iron to autophagosomes, a process known as ferritinophagy.8 Irons released in free form by ferritin enters the transiently labile iron pool to further join the Fenton reaction, leading to increased levels of ROS.6 Ferroptosis seems to be an ancient frailty engendered by polyunsaturated fatty acid incorporation into cell membranes, and cells have evolved dynamic networks to manipulate and fortify against this vulnerability in a variety of environments.9 Many biological processes, including the metabolism of amino acids, iron, and polyunsaturated fatty acids, are linked to ferroptosis sensitivity. Ferroptosis has been reported to be linked to pathologic cell death in Alzheimer's disease, cancer, traumatic brain damage, ischemia-reperfusion damage, and renal degeneration.10
Sequencing technologies and bioinformatics have evolved significantly in recent decades and are now frequently employed to detect illness biomarkers.11 Sequencing or microarray technologies can be used to obtain differently expressed mRNA, miRNA, and lncRNA in diverse disease samples, and bioinformatics can be used to further investigate the role of the differentially expressed molecules in disease development.12 RNA is the most adaptable biomolecule, and it can be protein-coding (coding) or nonprotein-coding (noncoding). Noncoding RNAs are not translated into proteins and instead impacts gene expression, which are further classified into 2 types based on their length: micro noncoding RNAs (miRNAs) and long noncoding RNAs (lncRNAs).13 MicroRNAs, which range in length from 20 to 23 nucleotides, are short noncoding RNAs that act as posttranscriptional blockers by adhering to mRNAs and silencing certain target genes.14 Yet according to previous studies, miRNAs are differentially expressed between healthy and periodontitis gingiva and can influence proliferation and osteogenic differentiation of periodontal stem cells, implying that miRNAs might play an important role in periodontal homeostasis and gingival histopathology.15 With lengths surpassing 200 nucleotides, lncRNAs are a huge and varied family of transcriptional RNA molecules that have garnered considerable attention in recent years. They have indeed been linked to the pathophysiology of a variety of disorders and have the capacity to influence gene expression at both the transcriptional and posttranscriptional stages. Studies have highlighted the critical role of lncRNAs in pathologic states associated with periodontitis, such as mediating the activation of periodontal cell autophagy and regulating the osteogenic differentiation of periodontal stem cells.15,16 miRNAs and lncRNAs are involved in various processes of the immune response by regulating the expression of immune-related genes.17
The competing endogenous RNA (ceRNA) hypothesis was proposed in 2011, revealing a new mechanism of RNA interactions, whereby lncRNAs could compete for miRNA-mRNA binding and thus affect the negative regulation of gene expression by miRNAs.18 Several periodontitis-associated ceRNA networks have been established.11,19,20 Notably, further mechanistic studies have shown that the ceRNA network partially participates in the regulation of inflammatory cytokine expression in gingival fibroblasts.21 To a certain extent, the periodontitis-related ceRNA network had some impact on the development of periodontitis via influencing inflammation response.22 Nevertheless, few studies have explored the ferroptosis-related ceRNA networks and periodontitis.
In the current study, we analysed the Gene Expression Omnibus (GEO) data set GSE10334 and GSE54710 to identify differences in mRNA expression profiles between periodontitis and healthy gingival tissues and took intersections with ferroptosis-related genes (FRGs) to obtain ferroptosis-related differentially expressed genes (DE-FRGs) in periodontitis. Then we used multiple databases to identify the molecular interaction network of lncRNA, miRNA, and mRNA to construct the ferroptosis-related ceRNA network implicated in the pathogenesis of periodontitis. This discovery might lead to a new understanding of the role of ferroptosis in periodontitis and create the theoretical groundwork for further research into the disease's molecular process.
Materials and methods
Source of data
The data set of transcriptomes in healthy and diseased gingival tissues microarray (GSE10334, GSE54710) was downloaded from the GEO database in the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/geo/). GEO is a public genomics data repository that contains microarray and sequencing data for a wide range of diseases and provides online tools to help users perform analysis. GSE10334 represented mRNA expression profiles in GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array, including 90 nonsmokers, with periodontitis and 247 gingival samples (183 from diseased and 64 from healthy sites). GSE54710 represented miRNA expression profiles in GPL15159 Agilent-031181 Unrestricted_Human_miRNA_V16.0_Microarray, including 68 individuals with periodontitis, contributing 198 gingival samples (158 from diseased and 40 from healthy sites). The diseased gingival papilla was defined as papillae with bleeding on probing (BOP), probing pocket depth (PPD) ≥4 mm, and clinical attachment loss (CAL) ≥3 mm. The healthy papillae were defined as papillae with no BOP, PPD ≤4 mm, and CAL ≤2 mm. All donors had signed informed consent in the original research.23,24 The FerrDb database (datjar.com) was screened to find 388 FRGs. The FerrDb database was the first manually collated database of ferroptosis for the management and identification of ferroptosis-related markers and regulators and ferroptosis-associated diseases.
Differentially expressed genes analysis
The GEO Online Analysis Tool, GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/), was applied to identify differentially expressed genes (DEGs) in GSE10334 and differentially expressed miRNAs (DE-miRNAs) in GSE54710. DEGs were defined by the evaluation criteria of adjusted P value <.01 and |log2(FC)| >1.0. The threshold P value <.05 and |log2(FC)| >.5 was for DE-miRNAs. Then Venn diagram was constructed to find the overlapping parts of the DEGs and FRGs, which were the critical ferroptosis-related DEGs (DE-FRGs). Spearman correlation coefficient between the DE-FRGs was calculated. The correlation coefficient with P < .05 was considered statistically significant. The expression value of the 7 DE-FRGs were extracted, and then the Shapiro-Wilk test and homogeneity of variance test were applied. If the data were normally distributed and the variance was homogeneous, a t test was used. If not, the Mann–Whitney U test was applied. The receiver operating characteristic curve of each DE-FRGs was drawn.
Functional enrichment analysis of DE-FRGs
To identify the biological function of DE-FRGs, the Metascape database (https://metascape.org/) was used to perform gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment, in which ferroptosis-related DEGs were involved. The Metascape database integrates more than 40 bioinformatics databases, providing biologists with easy access to comprehensive data analysis through a simple interface for quick one-click analysis.
Protein–protein interaction (PPI) network analysis
STRING (https://string-db.org/) was used to construct of protein interactions. Nodes in the protein network that did not interact with other proteins were removed. The STRING database was a searchable database of known protein–protein and predicted protein–protein interactions that could be applied to 2031 species and contained 9.6 million proteins and 13.8 million protein interactions.
Establishment of the ceRNA network
Targetscan (https://www.targetscan.org/)25 and miRtarbase (https://mirtarbase.cuhk.edu.cn/)26 were used to estimate the miRNAs with which DE-FRGs may interact. With DE-miRNAs intersection set, the results predicted by both databases were taken. Targetscan and miRtarbase were databases for predicting miRNA binding sites and were very good at predicting miRNA binding sites in mammals. The StarBase v3.0 database could identify more than 1.1 million miRNA-ncRNAs, 2.5 million miRNA-mRNAs, 2.1 million RBP-RNAs, and 1.5 million RNA–RNA interactions from multidimensional sequencing data. The lncATLAS database is based on high-throughput sequencing data from 15 cell lines and contains 6768 lncRNAs from GENCODE annotations. The database was evaluated by relative concentration index for specific localisation. The lncRNAs that interact with the aforementioned DE-miRNAs were then predicted in the StarBase v3.0 database, with CLIP data set to high stringency (≥3). Because lncRNAs can only function as nodes in the ceRNA network in the cytoplasm, we utilised the lncATLAS database (https://lncatlas.crg.eu/)27 to look into the intracellular localisation of lncRNAs, selecting those detected in the cytoplasm. The lncRNA-miRNA-mRNA ceRNA network was built using the Cytoscape programme (v3.8.2; www.cytoscape.org).
Results
The DE-FRGs of periodontitis
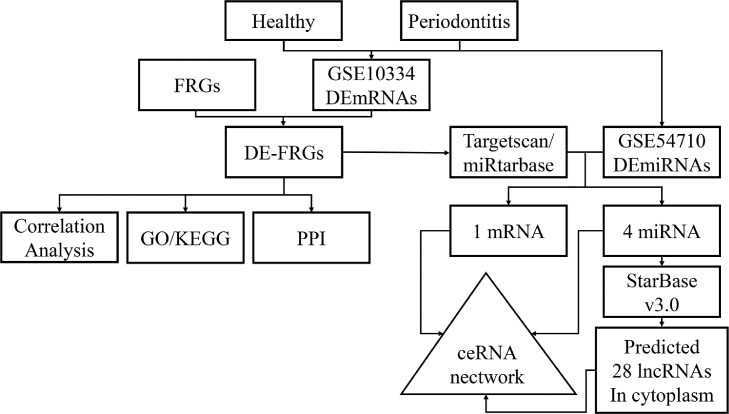
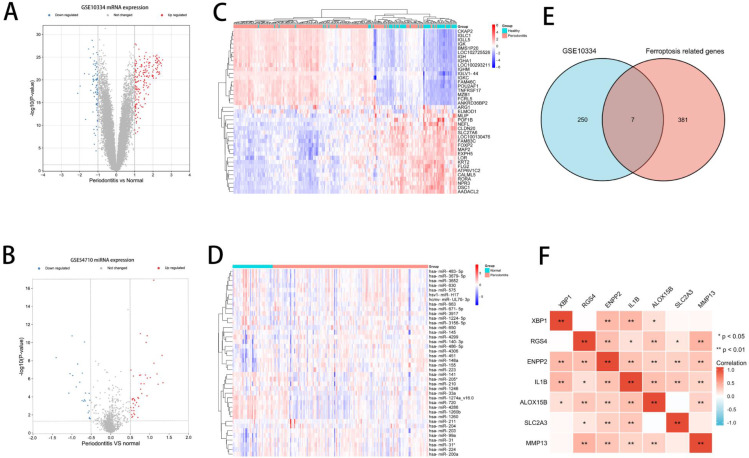
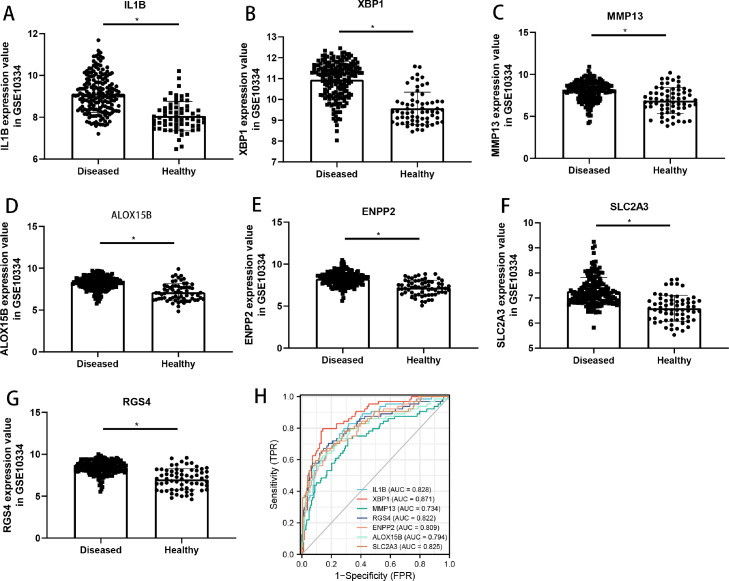
Figure 1 depicts a diagram of the flowchart for this investigation. Based on the criterion of DEGs and DE-miRNAs, 182 up-regulated genes and 75 down-regulated genes were identified in GSE10334, and 42 up-regulated miRNAs and 23 down-regulated miRNAs were identified in GSE54710. The volcano plot showed the DEGs of GSE10334 and DE-miRNAs of GSE54710. (Figure 2A and 2B) The heatmaps showed the expression and cluster analysis of the top 20 up-regulated and down-regulated mRNAs and miRNAs (Figure 2C and 2D). The FerrDb database was screened to find 388 FRGs. Then a Venn diagram was constructed to find the overlapping parts of the DEGs and FRGs, and it identified 7 DE-FRGs (Figure 2E). The correlation between the DE-FRGs is shown in Figure 2F. The up-regulated expression level of the 7 DE-FRGs in periodontitis are shown in Figure 3. Moreover, XBP1 had the highest accuracy in distinguishing periodontitis from health (area under curve = 0.871). The details of the DE-FRGs are shown in Table 1. Heatmap and volcano plot were plotted by http://www.bioinformatics.com.cn, a free online platform for data analysis and visualisation.
Fig. 1.
Experimental flowchart.
Fig. 2.
Differential expression analysis of GSE10334 and GSE54710. A, Volcano plot of mRNA expression level in GSE10334, mRNAs with adjusted P value <.01 and log2(FC) >1.0 are shown by the red dot, whereas mRNAs with adjusted P value <.01 and log2(FC) <−1 are shown by the blue dot. B, Volcano plot of miRNAs expression level in GSE54710, miRNAs, with P value <.05 and log2(FC) >0.5 are shown by the red dot, whereas miRNAs with P value <.05 and log2(FC) <−0.5, are shown by the blue dot. C, The heatmap shows the expression of the top 20 up-regulated mRNAs and down-regulated mRNAs. D, The heatmap shows the expression of the top 20 up-regulated miRNAs and down-regulated miRNAs. E, The Venn diagram shows the overlapping parts of the differentially expressed genes (DEGs) and ferroptosis-related genes (FRGs), identifying 7 differentially expressed ferroptosis-related genes (DE-FRGs). F, Spearman correlation analysis of the 7 DE-FRGs.
Fig. 3.
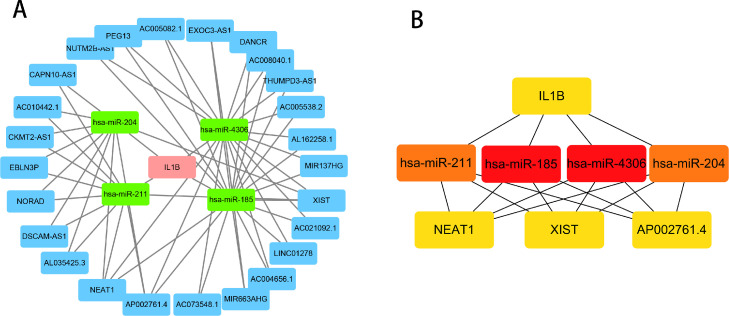
ceRNA network of IL1B, has-miR-185, has-miR-204, has-miR-211, has-miR-4306, and 28 lncRNAs. A, The pink square represents mRNA, the green square represents miRNA, and the blue square represents lncRNA. B, The top 8 hub nodes were identified through the CytoHubba.
Table 1.
Differentially expressed ferroptosis-related genes in GSE10334.
| Gene symbol | Gene title | Adjusted P value | logFC | Expression |
|---|---|---|---|---|
| XBP1 | X-box binding protein 1 | 2.13E-21 | 1.374 | UP |
| RGS4 | Regulator of G-protein signaling 4 | 2.51E-19 | 1.424 | UP |
| ENPP2 | Ectonucleotide pyrophosphatase/phosphodiesterase 2 | 5.04E-18 | 1.072 | UP |
| IL1B | Interleukin 1 beta | 1.49E-14 | 1.037 | UP |
| ALOX15B | Arachidonate 15-lipoxygenase, type B | 3.08E-14 | 1.072 | UP |
| SLC2A3 | Solute carrier family 2 member 3 | 1.35E-13 | 1.057 | UP |
| MMP13 | Matrix metallopeptidase 13 | 4.42E-09 | 1.190 | UP |
Functional enrichment analysis and PPI network construction of DE-FRGs
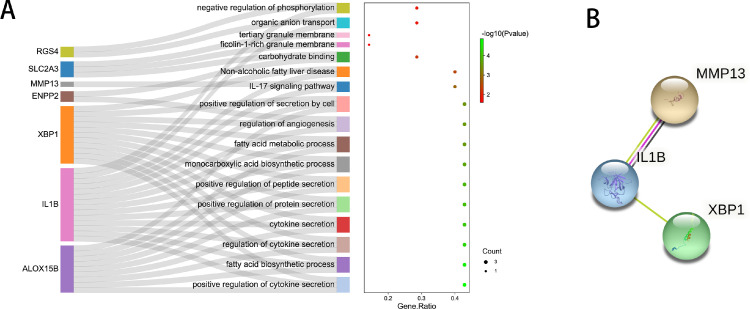
The GO/KEGG pathway analysis of the 7 DE-FRGs was counducted. GO analysis found that the DE-FRGs were enriched in positive regulation of cytokine secretion, positive regulation of protein secretion, negative regulation of phosphorylation, regulation of angiogenesis, and so on. The interleukin (IL)-17 signaling pathway and non-alcoholic fatty liver disease were identified in the KEGG pathway analysis (Figure 4A). PPI analysis was used to determine the interaction of the DE-FRGs. It was hypothesised by the STRING database that IL1B would interact with XBP1 and MMP13 (Figure 4B). DE-FRGs that did not interact with other proteins were not displayed.
Fig. 4.
Expression levels and receiver operating characteristic (ROC) analysis of the 7 differentially expressed ferroptosis-related genes (DE-FRGs) in GSE10334. A–G, Expression levels of IL1B, XBP1, MMP13, ALOX15B, ENPP2, SLC2A3, and RGS4. H, ROC results of IL1B, XBP1, MMP13, ALOX15B, ENPP2, SLC2A3, and RGS4.
Establishment of ceRNA network
There were no interacting miRNAs with XBP1 and MMP13 in miRtarbase. The DE-FRG IL1B was regarded as the core to establish the ferroptosis-related ceRNA network. We retrieved 231 related miRNAs by searching the miRtarbase and Targetscan databases for IL1B-interacting miRNAs, which we intersected with the DE-miRNAs acquired from GSE54710 to obtain 4 miRNAs (Table 2). lncRNAs interacting with 4 miRNAs were downloaded from the Starbase database, and Cytoscape was used to visualise the ceRNA network (Figure 5A). In addition, the top 8 hub nodes were identified through the CytoHubba, comprising IL1B, has-miR-185, has-miR-204, has-miR-211, has-miR-4306, NEAT1, XIST, and AP002761.4 (Figure 5B).
Table 2.
Interleukin 1 beta (IL1B)-interacting differentially expressed miRNAs, the overlap with miRtarbase, Targetscan. lncRNAs interacting with 4 miRNAs were downloaded from Starbase database. The subcellular localisation of the lncRNAs was obtained from lncATLAS database.
| miRNA_ID | P value | logFC | lncRNAs |
|||
|---|---|---|---|---|---|---|
| In both nuclear and cytoplasm | Only in nuclear | Only in cytoplasm | No data | |||
| hsa-miR-211 | 4.43E-05 | −0.5528 | CAPN10-AS1, AC010442.1, CKMT2-AS1, EBLN3P, NEAT1, AP002761.4, NORAD, DSCAM-AS1, XIST, AL035425.3 |
MIR29B2CHG, AC073335.2, KCNQ1OT1, MALAT1, AC073896.4, AC010542.4, AC009022.1, AC007952.4, ASB16-AS1, AC018628.1, INE1 |
None | AP000553.1 |
| hsa-miR-204 | 4.02E-05 | −0.6448 | CAPN10-AS1, AC010442.1, CKMT2-AS1, EBLN3P, NEAT1, AP002761.4, NORAD, DSCAM-AS1, XIST, AL035425.3 |
MIR29B2CHG, AC073335.2, KCNQ1OT1, MALAT1, AC073896.4, AC010542.4, AC009022.1, AC007952.4, ASB16-AS1, AC018628.1, INE1 |
None | AP000553.1 |
| hsa-miR-4306 | 3.97E-06 | 0.7837 | MIR137HG, AL162258.1, AC005538.2 THUMPD3-AS1, AC008040.1, DANCR, EXOC3-AS1, AC005082.1, PEG13, NUTM2B-AS1, NEAT1, AP002761.4, AC073548.1, MIR663AHG, AC004656.1, LINC01278, XIST |
KCNQ1OT1, SNHG1,MALAT1, AC020658.3, DNAAF4-CCPG1, MCM3AP-AS1 |
AC021092.1 |
AC118758.3, GABPB1-IT1, AC130462.1, AC020978.7, |
| hsa-miR-185 | 1.73E-03 | 0.5881 | MIR137HG, AL162258.1, AC005538.2 THUMPD3-AS1, AC008040.1, DANCR, EXOC3-AS1, AC005082.1, PEG13, NUTM2B-AS1, NEAT1, AP002761.4, AC073548.1, MIR663AHG, AC004656.1, LINC01278, XIST |
KCNQ1OT1, SNHG1,MALAT1, AC020658.3, DNAAF4-CCPG1, MCM3AP-AS1 |
AC021092.1 |
AC118758.3, GABPB1-IT1, AC130462.1, AC020978.7, |
Fig. 5.
Functional enrichment analysis of differentially expressed ferroptosis-related genes (DE-FRGs). A, The gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the 7 DE-FRGs. B, The protein–protein interaction (PPI) network of the DE-FRGs. DE-FRGs that did not interact with other proteins are not displayed.
Discussion
Periodontitis is a common noncommunicable oral illness that impairs periodontal health and occlusal function by damaging periodontal supporting tissues and triggering systemic inflammation via periodontal bacteria and inflammatory mediators. Genetic factors have been linked to periodontitis, and epigenetic mechanisms impact periodontitis progression by regulating pathogen-induced host immune responses and altering host susceptibility to periodontitis. Noncoding RNAs, such as lncRNAs and miRNAs, have exciting implications by forming ceRNA crossover networks with mRNAs through miRNA response elements, a gene regulatory mechanism that affects posttranscriptional levels in a variety of physiologic and pathophysiologic processes. As a result, it is worthwhile to investigate the etiology of periodontitis from a genetic standpoint, utilizing ceRNA network assembly as a starting point.
A number of studies have been carried out to build ceRNA networks linked to periodontitis. Lin et al16 constructed an interaction network comprising 10 lncRNAs, 11 miRNAs, and 83 mRNAs by using periodontitis-related public data sets GSE80715 and GSE54710 and discovered that the elevated mRNAs were substantially correlated with inflammation through GO analysis. The RGS4 was also identified in our network, indicating that it played a role both in the inflammatory response and ferroptosis. Jin et al11 utilised lncRNA as an entry point to identify 3 important lncRNAs (LINC00687, LBX2-AS1, and LINC01566) and then created ceRNA networks featuring LBX2-AS1 as the centre, predicting 573 differentially expressed genes. Respectively, Lai et al4 and Gu et al28 constructed ceRNA networks for their regulatory functions in periodontal stem cell osteogenic differentiation, and both studies discovered that the differential mRNAs were functionally enriched in osteogenic differentiation and MAPK signaling pathways. Bian et al29 explored the relationship between autophagy and periodontitis and identified 4 autophagy-related genes (LAMP2, NFE2L2, NCKAP1, and EGFR), 3 miRNAs (hsa-miR-140-3p, hsa-miR-142-5p, and hsa-miR-671-5p), and 30 lncRNAs that were significantly expressed in periodontitis, contributing to further understanding of the mechanism of autophagy in periodontitis. Tang et al30 created a network of immune-related ceRNAs in periodontitis, including lncRNA MIAT, has-miR-3652, has-miR-4286, has-miR-1246, has-miR-1260b, and 27 mRNAs. Their functions were mainly enriched in the proliferation, differentiation, and activation of B cells. Amongst the 27 identified mRNAs, XBP1 was also found in our present study, indicating the both roles of XBP1 in immune and ferroptosis. However, bioinformatic analyses on the role of ferroptosis in the development of periodontitis were scarce. Our present study established a ferroptosis-related ceRNA network, including IL1B, hsa-miR-185, hsa-miR-204, hsa-miR211, hsa-miR4306, and 28 lncRNAs, amongst which lncRNA NEAT1, XIST, and AP002761.4 were identified as the hub lncRNAs.
Periodontal homeostasis was characterised by a unique harmony between cell death and cytokinesis, cell differentiation in the periodontal biological system. Studies have revealed various cell demise modalities, like apoptosis, autophagy, and pyroptosis, which were related to periodontitis development.1 Ferroptosis is an oxidative cell demise actuated by small molecules with iron particle reliance, portrayed by an overabundance of intracellular ROS and lipid peroxidation on the cell film. As a particular inducer of iron demise, erastin might repress the cystine-glutamate adversarial framework Xc and block the vehicle of cystine into the cytoplasm. Inability to lessen extreme intracellular ROS because of consumption of the cancer-prevention agent glutathione may prompt deadly lipid peroxidation in the cell film.7 Ferroptosis was involved in the immune response. Lipopolysaccharide (LPS) from Porphyromonas gingivalis, one of the main pathogens of periodontitis, might cause mitochondrial dysfunction, high ROS production, and oxidative stress.31 In addition, one study found that periodontitis-grade sodium butyrate treatment was able to induce ferroptosis in periodontal ligament cells.32 In addition, the occurrence of sodium butyrate–induced ferroptosis was closely related to the activation of NCOA4-mediated ferritin phagocytosis. Phospholipids on cell membranes served as the first line of defense against microbial stressors and helped to keep host cells in a state of equilibrium. Lipoxygenase-producing microorganisms could target the lipid content of cellular membrane. Pseudomonas aeruginosa was a gram-negative, rod-shaped bacteria that causes cell death through ferritin.33
Our study indicated that IL1B might influence the progression of periodontitis partially by regulating ferroptosis. IL1B was a member of the IL1 cytokine family. IL1B could be produced by activated macrophages, gingival fibroblasts, and so on.34 It was an important mediator of the inflammatory response and participated in a dynamic biological processes, such as cell proliferation, differentiation, and apoptosis.35 IL1B was reported to induce ferroptosis by promoting the production of ROS, reducing the GXP4 level, and improving the MMP13 level in chondrocytes.36 The role of IL1B in the development of periodontitis had been studied extensively. Periodontal pathogens and their metabolites stimulated gingival epithelial cells, causing them to release additional pro-inflammatory cytokines such as IL1B, resulting in more periodontal tissue deterioration.37 Salivary IL1B levels could help distinguish gingivitis from periodontitis.38 The level of IL1B in saliva was much higher than that in patients with periodontitis and had the potential to be used as a biomarker for periodontitis.39 X-box binding protein (XBP) 1 functioned as a transcription factor during endoplasmic reticulum stress by regulating the unfolded protein response. XBP1 was reported to be highly expressed in periodontitis, and it might play an important role in the interaction between periodontitis and diabetes mellitus.40 Matrix metalloproteinase 13 (MMP13) played a role in the degradation of extracellular matrix proteins, one of the main features of periodontitis.41 Arachidonate 15-lipoxygenase type B (ALOX15B) might regulate cytokine secretion by macrophages and function in the immune response, which was also highly expressed in periodontitis.42 Regulator of G protein signaling 4 (RGS4) prevented signal transmission through increasing the GTPase activity, and RGS4 was the marker of ferroptosis.43 Ectonucleotide pyrophosphatase/phosphodiesterase 2 (ENPP2) could act on sphingosylphosphorylcholine producing sphingosine-1-phosphate.44 Solute carrier family 2 member 3 (SLC2A3) participated in glucose transmembrane transporter activity. There were few studies about the correlation amongst RGS4, ENPP2, SLC2A3, and periodontitis. Our study provided new research perspectives for future mechanism research of periodontitis. has-miR-185-5p exhibited anti-inflammatory function through the cell division cycle 42/JNK pathway in LPS-stimulated macrophages.45 has-miR‑4306 was found to be decreased in colorectal cancer, and it could inhibit the malignant behaviours of colorectal cancer by regulating FoxD2‑adjacent opposite strand RNA 1.46 has-miR-204 and has-miR-211 were shown to participate in the chondrogenesis of periodontal ligament stem cells.47 Overexpression of has-miR-211 could significantly decrease the production of ILB by the inhibition of the nuclear factor kappa B signaling pathway in LPS-induced endometritis.48 lncRNA NEAT1 was proven to interact with has-miR-211 by luciferase reporter and further activate MAPK1 to promote the expression of the inflammasomes.49 Our present study predicted the interaction between lncRNA NEAT1 and has-miR-211-IL1B, of which the function in periodontitis needed to be studied.
There are still some limitations in this study. First, the data set on the expression of lncRNA in periodontitis was not included, because the mRNA data set GSE10334 and GSE54710 both have more than 100 samples, and the inflamed gums and healthy gums were collected at the same time. The existing periodontitis-related lncRNA data set GSE80715. The sample size of GSE80715 is only 20, and healthy gums and inflamed gums are seen in different patients; the expression of lncRNA predicted in this study in periodontitis is not clear, so the established ceRNA network needs further verification. Also, due to the lack of clinical and prognostic data, clinical correlation studies and prognostic analyses were not possible in this study. Although there are limitations, this study provides a different perspective for understanding the role and mechanism of ferroptosis in periodontitis.
Conclusions
A ferroptosis-related ceRNA network was established in periodontitis, including IL1B, has-miR-185, has-miR-204, has-miR-211, has-miR-4306, and 28 lncRNAs, and the ferroptosis-related function of IL1B in periodontitis might play an important role in periodontitis progression. However, the role of ferroptosis in periodontitis remains to be elucidated.
Author contributions
Churen Zhang and Penxin Xue: conceptualisation, methodology, data curation, software, and writing–original draft preparation. Qiaoling Cai: visualisation, investigation, supervision, validation, and article revision. Jianguo Ke: conceptualisation, writing–reviewing and editing.
Conflict of interest
None disclosed.
Acknowledgments
Funding
The study is funded by the Start-up fund for introducing talents to the hospital of The First Affiliated Hospital of Xiamen University (XYJ2021006).
Acknowledgements
The authors thank TCGA and GEO for providing the public data available for us. The authors thank a free online platform (http://www.bioinformatics.com.cn) for data analysis and visualisation.
Contributor Information
Churen Zhang, Email: 18311321821@163.com.
Pengxin Xue, Email: 2020027648@qq.com.
Jianguo Ke, Email: 13666009910@163.com.
Qiaoling Cai, Email: caiqiaoling12003@163.com.
REFERENCES
- 1.Slots J. Periodontitis: facts, fallacies and the future. Periodontol 2000. 2017;75:7–23. doi: 10.1111/prd.12221. [DOI] [PubMed] [Google Scholar]
- 2.Jepsen S, Suvan J, Deschner J. The association of periodontal diseases with metabolic syndrome and obesity. Periodontol 2000. 2020;83:125–153. doi: 10.1111/prd.12326. [DOI] [PubMed] [Google Scholar]
- 3.Papapanou PN, Sanz M, Buduneli N, et al. Periodontitis: consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J Clin Periodontol. 2018;45(Suppl 20):S162–S170. doi: 10.1111/jcpe.12946. [DOI] [PubMed] [Google Scholar]
- 4.Lai L, Wang Z, Ge Y, et al. Comprehensive analysis of the long noncoding RNA-associated competitive endogenous RNA network in the osteogenic differentiation of periodontal ligament stem cells. BMC Genomics. 2022;23:1. doi: 10.1186/s12864-021-08243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartold PM, Van Dyke TE. Periodontitis: a host-mediated disruption of microbial homeostasis. Unlearning learned concepts. Periodontol 2000. 2013;62:203–217. doi: 10.1111/j.1600-0757.2012.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang WS, SriRamaratnam R, Welsch ME, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imai H, Matsuoka M, Kumagai T, Sakamoto T, Koumura T. Lipid peroxidation-dependent cell death regulated by GPx4 and ferroptosis. Curr Top Microbiol Immunol. 2017;403:143–170. doi: 10.1007/82_2016_508. [DOI] [PubMed] [Google Scholar]
- 9.Stockwell BR, Friedmann Angeli JP, Bayir H, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu Y, Cao Y, Cao W, Jia Y, Lu N. The application of ferroptosis in diseases. Pharmacol Res. 2020;159 doi: 10.1016/j.phrs.2020.104919. [DOI] [PubMed] [Google Scholar]
- 11.Jin SH, Zhou RH, Guan XY, Zhou JG, Liu JG. Identification of novel key lncRNAs involved in periodontitis by weighted gene co-expression network analysis. J Periodontal Res. 2020;55:96–106. doi: 10.1111/jre.12693. [DOI] [PubMed] [Google Scholar]
- 12.Inamoto T, Uehara H, Akao Y, et al. A panel of microRNA signature as a tool for predicting survival of patients with urothelial carcinoma of the bladder. Dis Markers. 2018;2018 doi: 10.1155/2018/5468672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atianand MK, Caffrey DR, Fitzgerald KA. Immunobiology of long noncoding RNAs. Annu Rev Immunol. 2017;35:177–198. doi: 10.1146/annurev-immunol-041015-055459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S, Fan M, Zheng Q, et al. MicroRNAs in Alzheimer's disease: potential diagnostic markers and therapeutic targets. Biomed Pharmacother. 2022;148 doi: 10.1016/j.biopha.2022.112681. [DOI] [PubMed] [Google Scholar]
- 15.Sanchez-Munoz F, Martinez-Coronilla G, Leija-Montoya AG, et al. Periodontitis may modulate long-non coding RNA expression. Arch Oral Biol. 2018;95:95–99. doi: 10.1016/j.archoralbio.2018.07.023. [DOI] [PubMed] [Google Scholar]
- 16.Lin Y, Jin L, Tong WM, Leung YY, Gu M, Yang Y. Identification and integrated analysis of differentially expressed long non-coding RNAs associated with periodontitis in humans. J Periodontal Res. 2021;56:679–689. doi: 10.1111/jre.12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manco G, Lacerra G, Porzio E, Catara G. ADP-ribosylation post-translational modification: an overview with a focus on RNA biology and new pharmacological perspectives. Biomolecules. 2022;12 doi: 10.3390/biom12030443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li S, Liu X, Li H, et al. Integrated analysis of long noncoding RNA-associated competing endogenous RNA network in periodontitis. J Periodontal Res. 2018;53:495–505. doi: 10.1111/jre.12539. [DOI] [PubMed] [Google Scholar]
- 20.Zhou H, Chen D, Xie G, Li J, Tang J, Tang L. LncRNA-mediated ceRNA network was identified as a crucial determinant of differential effects in periodontitis and periimplantitis by high-throughput sequencing. Clin Implant Dent Relat Res. 2020;22:424–450. doi: 10.1111/cid.12911. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Wang M, Song L, Wang X, Lai W, Jiang S. LncRNA MALAT1 regulates inflammatory cytokine production in lipopolysaccharide-stimulated human gingival fibroblasts through sponging miR-20a and activating TLR4 pathway. J Periodontal Res. 2020;55:182–190. doi: 10.1111/jre.12700. [DOI] [PubMed] [Google Scholar]
- 22.Huang N, Li C, Sun W, Wu J, Xiao F. Long non-coding RNA TUG1 participates in LPS-induced periodontitis by regulating miR-498/RORA pathway. Oral Dis. 2021;27:600–610. doi: 10.1111/odi.13590. [DOI] [PubMed] [Google Scholar]
- 23.Demmer RT, Behle JH, Wolf DL, et al. Transcriptomes in healthy and diseased gingival tissues. J Periodontol. 2008;79:2112–2124. doi: 10.1902/jop.2008.080139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stoecklin-Wasmer C, Guarnieri P, Celenti R, Demmer RT, Kebschull M, Papapanou PN. MicroRNAs and their target genes in gingival tissues. J Dent Res. 2012;91:934–940. doi: 10.1177/0022034512456551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pottoo FH, Barkat MA, Harshita, et al. Nanotechnological based miRNA intervention in the therapeutic management of neuroblastoma. Semin Cancer Biol. 2021;69:100–108. doi: 10.1016/j.semcancer.2019.09.017. [DOI] [PubMed] [Google Scholar]
- 26.Huang HY, Lin YC, Li J, et al. miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020;48:D148–D154. doi: 10.1093/nar/gkz896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mas-Ponte D, Carlevaro-Fita J, Palumbo E, Hermoso Pulido T, Guigo R, Johnson R. LncATLAS database for subcellular localization of long noncoding RNAs. RNA. 2017;23:1080–1087. doi: 10.1261/rna.060814.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu X, Li M, Jin Y, Liu D, Wei F. Identification and integrated analysis of differentially expressed lncRNAs and circRNAs reveal the potential ceRNA networks during PDLSC osteogenic differentiation. BMC Genet. 2017;18:100. doi: 10.1186/s12863-017-0569-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bian M, Wang W, Song C, Pan L, Wu Y, Chen L. Autophagy-related genes predict the progression of periodontitis through the ceRNA network. J Inflamm Res. 2022;15:1811–1824. doi: 10.2147/JIR.S353092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang H, Yuan S, Chen T, Ji P. Development of an immune-related lncRNA-miRNA-mRNA network based on competing endogenous RNA in periodontitis. J Clin Periodontol. 2021;48:1470–1479. doi: 10.1111/jcpe.13537. [DOI] [PubMed] [Google Scholar]
- 31.Bullon P, Cordero MD, Quiles JL, Morillo JM, del Carmen Ramirez-Tortosa M, Battino M. Mitochondrial dysfunction promoted by Porphyromonas gingivalis lipopolysaccharide as a possible link between cardiovascular disease and periodontitis. Free Radic Biol Med. 2011;50:1336–1343. doi: 10.1016/j.freeradbiomed.2011.02.018. [DOI] [PubMed] [Google Scholar]
- 32.Zhao Y, Li J, Guo W, Li H, Lei L. Periodontitis-level butyrate-induced ferroptosis in periodontal ligament fibroblasts by activation of ferritinophagy. Cell Death Discov. 2020;6:119. doi: 10.1038/s41420-020-00356-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dar HH, Tyurina YY, Mikulska-Ruminska K, et al. Pseudomonas aeruginosa utilizes host polyunsaturated phosphatidylethanolamines to trigger theft-ferroptosis in bronchial epithelium. J Clin Invest. 2018;128:4639–4653. doi: 10.1172/JCI99490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohammadian Haftcheshmeh S, Momtazi-Borojeni AA. Berberine as a promising natural compound for the treatment of periodontal disease: a focus on anti-inflammatory properties. J Cell Mol Med. 2021;25:11333–11337. doi: 10.1111/jcmm.17019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Papathanasiou E, Conti P, Carinci F, Lauritano D, Theoharides TC. IL-1 superfamily members and periodontal diseases. J Dent Res. 2020;99:1425–1434. doi: 10.1177/0022034520945209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao X, Sun K, Yu S, et al. Chondrocyte ferroptosis contribute to the progression of osteoarthritis. J Orthop Translat. 2021;27:33–43. doi: 10.1016/j.jot.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chompunud Na Ayudhya C, Roy S, Thapaliya M, Ali H. Roles of a mast cell-specific receptor MRGPRX2 in host defense and inflammation. J Dent Res. 2020;99:882–890. doi: 10.1177/0022034520919107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kc S, Wang XZ, Gallagher JE. Diagnostic sensitivity and specificity of host-derived salivary biomarkers in periodontal disease amongst adults: systematic review. J Clin Periodontol. 2020;47:289–308. doi: 10.1111/jcpe.13218. [DOI] [PubMed] [Google Scholar]
- 39.Arias-Bujanda N, Regueira-Iglesias A, Blanco-Pintos T, et al. Diagnostic accuracy of IL1beta in saliva: the development of predictive models for estimating the probability of the occurrence of periodontitis in non-smokers and smokers. J Clin Periodontol. 2020;47:702–714. doi: 10.1111/jcpe.13285. [DOI] [PubMed] [Google Scholar]
- 40.Li M, Huang S, Zhang Y, et al. Regulation of the unfolded protein response transducer IRE1alpha by SERPINH1 aggravates periodontitis with diabetes mellitus via prolonged ER stress. Cell Signal. 2022;91 doi: 10.1016/j.cellsig.2022.110241. [DOI] [PubMed] [Google Scholar]
- 41.Luchian I, Goriuc A, Sandu D, Covasa M. The role of matrix metalloproteinases (MMP-8, MMP-9, MMP-13) in periodontal and peri-implant pathological processes. Int J Mol Sci. 2022;23 doi: 10.3390/ijms23031806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin L, Yu W, Zhang W, et al. Expression profile of lipoxygenases in gingival tissues of human periodontitis. Oral Dis. 2021;27:567–576. doi: 10.1111/odi.13558. [DOI] [PubMed] [Google Scholar]
- 43.Dixon SJ, Patel DN, Welsch M, et al. Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. Elife. 2014;3:e02523. doi: 10.7554/eLife.02523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clair T, Aoki J, Koh E, et al. Autotaxin hydrolyzes sphingosylphosphorylcholine to produce the regulator of migration, sphingosine-1-phosphate. Cancer Res. 2003;63:5446–5453. [PubMed] [Google Scholar]
- 45.Ma X, Liu H, Zhu J, et al. miR-185-5p regulates inflammation and phagocytosis through CDC42/JNK pathway in macrophages. Genes (Basel) 2022;13 doi: 10.3390/genes13030468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye J, Liu J, Tang T, Xin L, Bao X, Yan Y. miR4306 inhibits the malignant behaviors of colorectal cancer by regulating lncRNA FoxD2AS1. Mol Med Rep. 2021:24. doi: 10.3892/mmr.2021.12362. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Wang P, Li Y, Meng T, et al. KDM6A promotes chondrogenic differentiation of periodontal ligament stem cells by demethylation of SOX9. Cell Prolif. 2018;51:e12413. doi: 10.1111/cpr.12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang C, Yang C, Zhang J, et al. MicroRNA-211 regulates the expression of TAB1 and inhibits the NF-kappaB signaling pathway in lipopolysaccharide-induced endometritis. Int Immunopharmacol. 2021;96 doi: 10.1016/j.intimp.2021.107668. [DOI] [PubMed] [Google Scholar]
- 49.An Q, Zhou Z, Xie Y, Sun Y, Zhang H, Cao Y. Knockdown of long non-coding RNA NEAT1 relieves the inflammatory response of spinal cord injury through targeting miR-211-5p/MAPK1 axis. Bioengineered. 2021;12:2702–2712. doi: 10.1080/21655979.2021.1930925. [DOI] [PMC free article] [PubMed] [Google Scholar]