Abstract
Abiraterone acetate is a first-line therapy for castration-resistant prostate cancer. This prodrug is deacetylated in vivo to abiraterone, which is a potent and specific inhibitor of cytochrome P450 17A1 (CYP17A1). CYP17A1 performs two sequential steps that are required for the biosynthesis of androgens that drive prostate cancer proliferation, analogous to estrogens in breast cancer. Abiraterone can be further metabolized in vivo on the steroid A ring to multiple metabolites that also inhibit CYP17A1. Despite its design as an active-site–directed substrate analog, abiraterone and its metabolites demonstrate mixed competitive/noncompetitive inhibition. To understand their binding, we solved the X-ray structures of CYP17A1 with three primary abiraterone metabolites. Despite different conformations of the steroid A ring and substituents, all three bound in the CYP17A1 active site with the steroid core packed against the I helix and the A ring C3 keto or hydroxyl oxygen forming a hydrogen bond with N202 similar to abiraterone itself. The structure of CYP17A1 with 3-keto, 5α-abiraterone was solved to 2.0 Å, the highest resolution to date for a CYP17A1 complex. This structure had additional electron density near the F/G loop, which is likely a second molecule of the inhibitor and which may explain the noncompetitive inhibition. Mutation of the adjacent Asn52 to Tyr positions its side chain in this space, maintains enzyme activity, and prevents binding of the peripheral ligand. Collectively, our findings provide further insight into abiraterone metabolite binding and CYP17A1 function.
Keywords: cytochrome P450, CYP17A1, steroidogenesis, prostate cancer, abiraterone, abiraterone metabolism
Abbreviations: 3βHSD, 3β-hydroxysteroid dehydrogenase; PDB, Protein Data Bank
Even in the advanced, “castration-resistant” phase, prostate cancer typically remains fueled by androgen production and androgen signaling pathways (1). Regardless of the tissue or the steroidogenic route employed, synthesis of the initial androgenic sex steroid requires cytochrome P450 17A1 (CYP17A1) to perform a 17,20-lyase reaction on the steroid D ring, releasing acetate from the C17 position and forming a 17-keto androgen. This key role in human steroidogenesis has made CYP17A1 a valuable drug target for the treatment of prostate cancer. Abiraterone, the active form of the only Food and Drug Administration-approved selective CYP17A1 inhibitor, effectively blocks this 17,20-lyase reaction, resulting in a marked reduction in the levels of circulating androgens and improved overall survival in patients with castration-resistant prostate cancer, even those who have failed other therapies (2). Abiraterone was developed from the CYP17A1 substrate pregnenolone, primarily by adding a pyridine substituent at C17 (3). The nitrogen of this pyridine heterocycle coordinates directly to the CYP17A1 heme iron (4), preventing O2 binding to the iron as required for catalysis. A number of preclinical efforts and early-stage clinical trials explored various iron-coordinating heterocycles, but less is known of the structure-activity relationships for other parts of the steroidal scaffold.
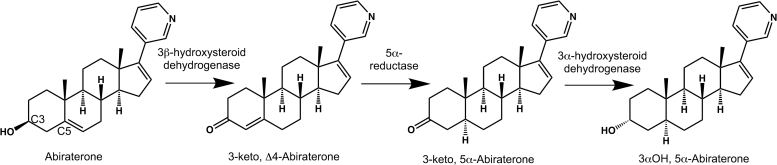
Such information has become increasingly important for several reasons. First, structures of CYP17A1 with abiraterone and other steroidal inhibitors (4) and steroidal substrates (5) demonstrated that 3β-hydroxy and 3-keto substituents found in the A ring of these steroids participate in hydrogen bonding networks in the “roof” of the active site. Second, in patients and in prostate cancer tumor cells, abiraterone is readily metabolized on the A ring to a variety of metabolites which can have differing effects on key enzymes in the androgen biosynthetic pathway (6, 7). Initially, 3β-hydroxysteroid dehydrogenase (3βHSD) converts the 3β−OH, Δ5 abiraterone to a 3-keto, Δ4 form also called Δ4−abiraterone (D4A, Fig. 1). D4A is then subject to the action of 5α- and 5β-reductases, which eliminate the A ring double bond, and further modifications by 3αHSD and 3βHSD (Fig. 1) to generate a total of six additional metabolites, varying in the composition and stereochemistry of the C3 substituent. Abiraterone, D4A, and these latter metabolites have varying effects on the inhibition of CYP17A1. Abiraterone metabolites show an unexplained mixed competitive-noncompetitive mode of inhibition which is not understood (8). Importantly, D4A was also shown to inhibit not only CYP17A1 but also two other enzymes required for androgen biosynthesis, 3β-hydroxysteroid dehydrogenase and 5α-reductase, as well as being an androgen receptor antagonist equally potent to the clinically employed enzalutamide (6). This four-way inhibition of androgen biosynthesis potentially represents a significantly improved multi-target approach for treatment of prostate cancer.
Figure 1.
Abiraterone metabolism pathway in vivo.
To discern how differences in the A ring of abiraterone might be accommodated in the CYP17A1 active site, human CYP17A1 was cocrystallized with three major abiraterone metabolites and structures determined by X-ray crystallography at resolutions of 2.0 to 2.7 Å. The structures demonstrate that such compounds bind CYP17A1 in the same general orientation as abiraterone, with alterations in the active site distal hydrogen bonding network to accommodate structural variation in the A ring. These findings support previous studies comparing the affinity and potency of CYP17A1 inhibition of these inhibitors compared to abiraterone. Furthermore, the 2.0 Å structure of CYP17A1 is the highest resolution structure of the enzyme to date and identifies a peripheral, second ligand binding site not clearly defined in previous CYP17A1 structures and which would be consistent with the mixed inhibition mode observed.
Results
Selection of compounds for analysis
Three abiraterone analogs (Table 1) were selected for structural studies based on their diversity of A-ring composition and stereochemistry and their potential in vivo relevance. The 3-keto, Δ4 (D4A), 3-keto, 5α, and 3α−OH and 5α versions of abiraterone are generated in tumor cells by the sequential action of 3βHSD, 5α-reductase, and 3αHSD (7). Prior spectral studies suggested that all three coordinate the CYP17A1 heme iron in a mode similar to abiraterone, while functional analysis revealed that all three inhibit CYP17A1 even better than abiraterone itself (8).
Table 1.
Data collection and refinement statistics for CYP17A1 X-ray structures
| Protein/Ligand complex | CYP17A1/3-keto, Δ4 (6WR0) | CYP17A1/3-keto, 5α (6WW0) | CYP17A1/3α−OH, 5α (5UYS) | CYP17A1 N52Y/Abiraterone (6WR1) |
|---|---|---|---|---|
| Data collection | ||||
| Beamline | SSRL BL 12-2 | SSRL BL 12-2 | SSRL BL 14-1 | SSRL BL 12-2 |
| Wavelength (Å) | 0.979 | 0.979 | 0.979 | 0.979 |
| Resolution (Å)a | 39.81–2.70 (2.85–2.70) | 38.68–2.00 (2.11–2.00) | 39.29–2.39 (2.52–2.39) | 39.23–1.85 (1.95–1.85) |
| Space group | P212121 | P212121 | P212121 | P1211 |
| Unit cell (Å) | 90.4, 153.0,168.5 | 85.7, 151.2, 169.9 | 89.8, 153.2, 168.9 | 48.1, 105.0, 108.6 |
| Completenessa | 99.7 (98.4) | 98.8 (93.5) | 99.8 (98.6) | 98.3 (97.4) |
| No. reflectionsa | 432,291 (58,652) | 1,503,586 (191,079) | 683,522 (97,537) | 567,619 (81,908) |
| Unique reflectionsa | 64,672 (9236) | 146,679 (20,040) | 92,392 (13,193) | 88,757 (12,773) |
| Multiplicitya | 6.7 (6.4) | 10.3 (9.5) | 7.4 (7.4) | 6.4 (6.4) |
| Mn (I/sd(I))a | 12.9 (1.7) | 8.5 (1.5) | 11.5 (1.5) | 14.1 (1.5) |
| Refinement | ||||
| Resolution (Å) | 39.81–2.70 | 38.68–2.01 | 39.29–2.40 | 39.23–1.85 |
| R/Rfree (%) | 20.1/24.7 | 18.5/21.8 | 19.0/24.5 | 19.0/21.8 |
| Ramachandran (%) favored/allowed/outliers | 97.0/3.0/0.0 | 98.1/1.9/0.0 | 96.3/3.6/0.1 | 97.6/2.4/0.0 |
| Rotamer outliers (%) | 1.9 | 0.6 | 2.7 | 0.36 |
| All-atom clash score | 5.0 | 3.1 | 1.6 | 2.2 |
| C-beta outliers | 0 | 0.06 | 0 | 0.11 |
| RMSD | ||||
| Bond lengths | 0.017 | 0.014 | 0.007 | 0.010 |
| Bond angles | 1.391 | 1.347 | 1.126 | 1.013 |
| # non-H Atoms/B-factors | ||||
| Protein | 14,905/57.1 | 14,879/34.6 | 15,015/43.0 | 7436/40.8 |
| Heme | 172/45.9 | 172/23.4 | 172/28.6 | 86/25.8 |
| Ligand | 104/47.1 | 104/24.8 | 104/30.2 | 52/29.4 |
| Waters | 118/47.7 | 967/37.1 | 339/36.0 | 494/40.2 |
| Ions | 3/55.8 | NA | NA | NA |
Abbreviation: NA, not applicable.
Statistics for the highest resolution shell.
Structural overview
CYP17A1 complexes with each of the three abiraterone analogs crystallized in the same space group (P212121) as previous structures of CYP17A1 (4, 5). In this space group, the four molecules (molecules A–D) composing the asymmetric unit adopt two slightly different protein conformations. Molecules A and B demonstrate high similarity to one another (RMSD over all Cα ranges from 0.16 to 0.27 Å), while molecules C and D are also similar to each other (RMSD over all Cα ranges from 0.26 to 0.41 Å), but there are greater disparities between the conformations of molecules A and B compared to molecules C and D (average RMSD over all Cα = 0.71 ± 0.04 Å), primarily in the N-terminus and the F/G region. CYP17A1 displays both conformations in all crystal structures reported to date regardless of the active-site substrate or inhibitor (4, 5).
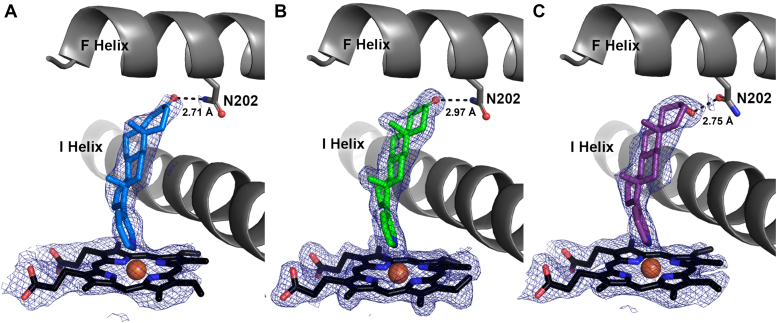
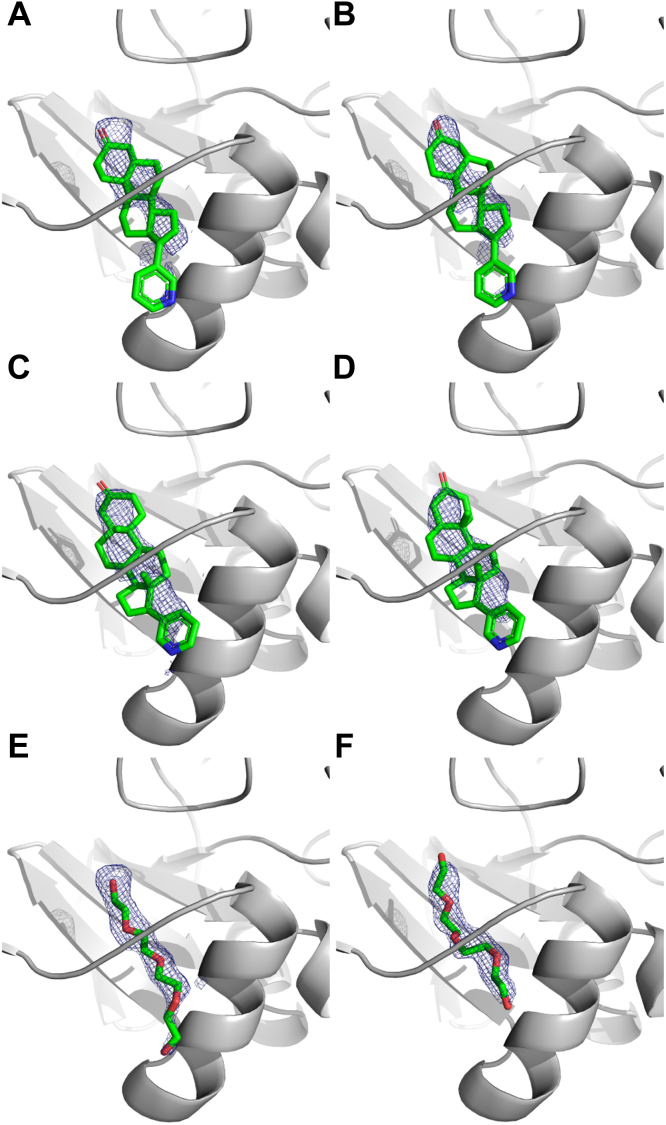
All three CYP17A1/abiraterone analog structures had clearly defined and consistent electron density in the active sites of all four copies of the protein (Fig. 2). Thus, further description of the active-site inhibitor binding modes will display only a representative copy of each CYP17A1/abiraterone analog complex. All three abiraterone analogs bind in the CYP17A1 active site with an overall orientation very similar to the parent inhibitor abiraterone (4). That is, the α-face of the steroid lies flat against the I helix at ∼60° angle from the plane of the heme. Consistent with type II spectral shifts observed during binding studies (8) and the structure of CYP17A1 with abiraterone (4), the pyridine nitrogen of each analog coordinates directly to the heme iron (Fig. 2). Despite variation in the structure of the A ring and C3 substituents of the different abiraterone analogs, all three also mimic abiraterone in forming some type of direct hydrogen bond between the C3 substituent and the F helix residue N202, although the nature of these interactions is variable.
Figure 2.
Abiraterone metabolites with variations in their A ring structures bind CYP17A1 in similar binding modes to each other and to abiraterone.A, 3-keto, Δ4 abiraterone (D4A, blue); B, 3-keto, 5α abiraterone (green); and C, 3α−OH, 5α abiraterone (purple) all coordinate to the heme iron via the conserved pyridine ring and have direct contacts between their respective C3 substituents and F helix residue N202. Distances shown to N202 are average distances among the four molecules of each complex but do not vary significantly. Average distances between the heme iron and ligand pyridine nitrogen are 2.46 Å, 2.38 Å, and 2.25 Å, espectively for the three structures. Simulated annealing composite omit maps (blue mesh) are contoured to 1.0 σ for the heme and ligand. Heme, black sticks with red sphere for iron.
Interactions between the A ring and the active-site roof
The 3-keto, Δ4 analog, and the 3-keto, 5α analogs both have a ketone as a C3 substituent, which can only serve as a hydrogen bond acceptor. Thus, the N202 side chain must be the hydrogen bond donor, adopting the rotamer with its amide nitrogen oriented toward the ligand (rather than with its side chain oxygen). In contrast, the C3 hydroxyl of the 3α−OH, 5α analog may serve as either a hydrogen bond donor or acceptor to interact with the N202 as either the rotamer presenting the amide oxygen or nitrogen to the ligand. It is modeled as the former in Figure 2C to illustrate this additional flexibility not seen in the 3-keto compounds. Hydrogen bond strength is related to both bond distances and angles. The average distance between the C3 substituent and N202 is relatively consistent, both among the molecules of the asymmetric unit and between structures (2.71 ± 0.24 Å for 3-keto-Δ4, 2.97 ± 0.05 Å for 3-keto-5α, and 2.75 ± 0.18 Å for 3αOH-5α), but the angles vary significantly for different ligands. Abiraterone and the 3α−OH, 5α analog have A rings with sp3 hybridization at C3. As a result, the A ring adopts a chair conformation, and both axial and equatorial C3 alcohols adopt a ∼95° bond angle from the heteroatom of N202. In contrast, the 3-keto analogs with sp2 hybridization at C3 adopt an A ring conformation closer to that of a half-chair. Consequently, the C3 keto substituent is projected more directly at N202 and forms an ∼130° degree angle with the nitrogen of that residue. This latter angle may be more favorable for CYP17A1 binding and inhibition.
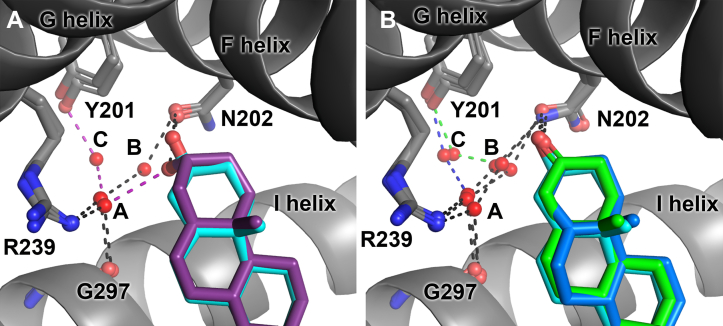
Differences in the A ring configurations also affect the indirect interactions of these inhibitors with active-site waters. The hydroxyl substituent of the 3α−OH, 5α abiraterone analog (Fig. 3A, purple) interacts with a water molecule (water A), which is in turn engaged in hydrogen bonds to both the side chain of arginine 239 on the G helix and the backbone carbonyl of glycine 297 in the I helix. This water also interacts with an additional water (water C) bridging to the side chain of tyrosine 201 on the F helix, forming a hydrogen bond network. In contrast, abiraterone (Fig. 3A, cyan), the 3-keto, Δ4 inhibitor (Fig. 3B, dark blue), and the 3-keto, 5α analog (Fig. 3B, green) do not directly hydrogen bond with water A. Instead, the C3 substituents of these compounds directly interact with N202, and N202 hydrogen bonds with water B, which interacts with water A to bridge to R239 and the G297 carbonyl. Both the 3-keto, Δ4 and 3-keto, 5α analogs also connect with the hydrogen bonding network involving water C and Y201, but water C was not apparent for the abiraterone complex.
Figure 3.
Abiraterone metabolites with modifications of the A ring participate in different hydrogen bond networks.A, the 3β-OH of abiraterone (cyan, from PDB 3RUK) hydrogen bonds directly to N202, while the abiraterone analog 3α-OH (purple) instead directly interacts with water A, which in turn is within a distance to interact with R239, G297, and water C. Water C bridges to Y201 for the 3α-OH complex but is not apparent in the abiraterone structure. B, like abiraterone (cyan, from PDB 3RUK), abiraterone 3-keto, Δ4 (blue), and 3-keto, 5α (green) analogs both form a hydrogen bond directly to N202, and this residue further engages in a hydrogen bonding network via two water molecules (A and B) bridging to R239 and the backbone carbonyl of G297. However, like the 3α-OH analog, a third water molecule (C) forms interactions with Y201 in the F helix. PDB, Protein Data Bank.
A second ligand binding site
Previous human CYP17A1 structures generated by cocrystallizing with various ligands include unmodeled electron density in the same two CYP17A1 molecules of the four in the asymmetric unit (molecules C and D). This continuous electron density occurs in a hydrophobic region between the N-terminal β strands and the F/G loop. The features of this electron density are not consistent with a chain of waters, and specific interactions with the polypeptide are not apparent. Of the known components of the crystallization solution, CYP17A1 ligands were always the best candidates, but at lower resolutions (2.4–3.0 Å) the electron density was previously not clear enough to unambiguously model.
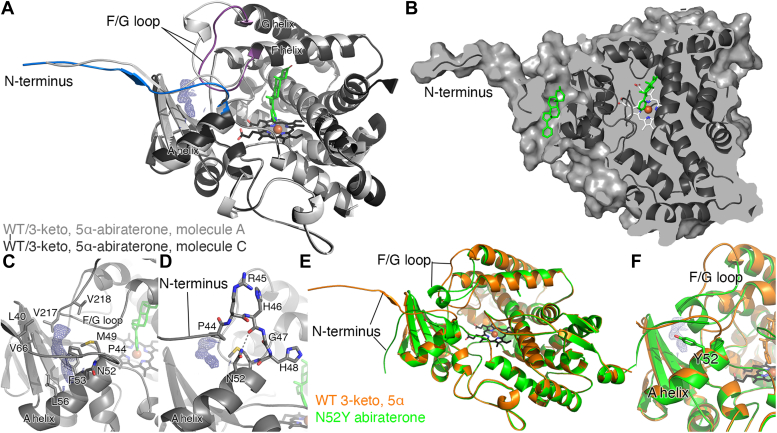
At 2.0 Å, CYP17A1 with the 3-keto, 5α abiraterone analog is the highest resolution structure to date, facilitating observation of elements that are less ordered and/or which have lower occupancy. Likely as a result, this previously unmodeled density is much better defined. This density is again present only in the conformation modeled for CYP17A1 molecules C and D and is not present in molecules A or B, which have a different conformation of the N-terminus and the F/G loop (Fig. 4A). The cavity for this ligand is separated from the active site (Fig. 4B). The peripheral ligand density is flanked by residues from the beta sheet system, the F/G loop, the N-terminus, and a portion of the A helix (Fig. 4C), but none of these seem to form specific interactions with the ligand.
Figure 4.
A peripheral ligand binding site in the CYP17A1 WT structure with the 3-keto, 5α abiraterone analog and comparison to the CYP17A1 N52Y mutant structure with abiraterone.A, overlay of the CYP17A1/3-keto, 5α-abiraterone conformations for molecule A (light grey) and molecule C (dark gray with blue N-terminus and purple F/G loop) highlights their conformational similarity except for these two regions. The peripheral ligand density (blue mesh) is only present in the conformation represented in molecules C and D and is flanked by the beta sheet system, the F/G loop, the N-terminus, and a portion of the A helix. B, surface view of the same CYP17A1 structure (dark gray, molecule C), using a slightly different rotation and cutaway to show the cavity for the peripheral ligand (green sticks) and its relationship to the copy in the active site (green sticks). C, detailed view of the peripheral binding site in the 3-keto, 5α abiraterone analog shows surrounding residues but does not indicate specific interactions with the ligand. D, in rationalizing a mutant to block occupation of the peripheral site, N52 appeared to be a good candidate since the favorable rotamer if mutated to a tyrosine would project into the peripheral binding site. E, comparison of the CYP17A1 WT cocrystallized with 3-keto, 5α abiraterone structure (chain C, orange) and the structure of the CYP17A1 N52Y mutant cocrystallized with abiraterone (chain B, green) reveals that the overall conformation is maintained, but differences in conformation are observed in the N-terminus and F/G loop. In all molecules, the N52Y mutant adopts a unique conformation for both of these elements, and electron density is not observed in the peripheral site. F, a detailed view of the peripheral binding site illustrating how the mutated tyrosine projects into the peripheral site as designed. All maps shown are 2Fo-Fc; maps generated without ligands and contoured at 1 σ (blue mesh).
In order to elucidate whether the occupation of the peripheral binding site affects CYP17A1 function or structure, a single mutation was designed with the intention of filling this site. Asparagine 52 lines the cavity of the secondary site, but the side chain is projected toward the peptide backbone of arginine 45 (Fig. 4D). Mutation of this residue to a bulkier tyrosine would provide a steric clash with the peptide backbone. Rotation of the tyrosine side chain to a common rotamer was predicted to project the hydrophobic side chain into the peripheral site.
The purified CYP17A1 N52Y protein was correctly folded and functional. It expressed a high yield and displayed typical absorption and reduced-CO difference spectra, indicating normal heme incorporation. Addition of either the hydroxylase substrate progesterone or the lyase substrate 17-hydroxypregnenolone produced the expected type I absorbance shift with high affinities similar to the WT enzyme. The N52Y mutant had a kcat for progesterone hydroxylation (0.94 ± 0.01 min−1) very similar to the WT enzyme (1.14 ± 0.03 min−1), while the Km was more than 3-fold lower (1.60 ± 0.09 μM) than WT (5.2 ± 0.5 μM). Thus the catalytic efficiency was actually ∼3-fold higher for the N52Y mutant (0.59 min−1 μM−1) than for the WT enzyme (0.22 min−1 μM−1). In contrast, the N52Y mutation had little effect on the lyase reaction converting 17-hydroxypregnenolone into dehydroepiandrosterone (kcat 0.35 ± 0.02 min−1; Km 0.6 ± 0.1 μM kcat/Km 0.58) compared to WT (kcat 0.41 ± 0.01 min−1; Km 0.8 ± 0.1 μM; kcat/Km 0.51 min−1 μM−1). Purified CYP17A1 and its N52Y mutant in the full-length forms were also active and had similar kinetic values to each other. Thus the N52Y enzyme was at least as active as the WT enzyme.
Cocrystallization of this N52Y mutation in the presence of abiraterone yielded a 1.85 Å structure (Table 1). Comparison with CYP17A1 WT cocrystallized with 3-keto-5α abiraterone structure reveals that the overall protein conformation is maintained, but differences in conformation are again observed in the N-terminus and F/G loop (Fig. 4E). The CYP17A1 N52Y crystals contained two molecules of CYP17A1 in the asymmetric unit, which are similar to each other but have a distinct conformation for the N-terminus and the F/G region compared to both CYP17A1 WT molecules A and B, in which no density is observed in the secondary site, and molecules C and D, in which ligand density is observed in the secondary site. Both regions of the N52Y mutant adopt a single unique conformation, and electron density is not observed in the peripheral site. Instead, the mutated tyrosine projects into the peripheral site as designed (Fig. 4F).
Peripheral ligand identification efforts
While the progress to date yielded versions of CYP17A1 that could bind a peripheral ligand (WT) and a version that did not (N52Y), the identity of the ligand is important. If the peripheral site represents an allosteric site for binding a second copy of steroidal inhibitor, this could explain the mixed inhibition observed functionally. Because the 3-keto, 5α-abiraterone inhibitor is present in relatively high concentrations during crystallization and generally compatible with the peripheral ligand density, a second copy of this molecule was modeled. The length and planarity to the steroidal scaffold is a good overall fit to the density, but likely because interactions between the inhibitor and the polypeptide are largely hydrophobic, it was not immediately apparent which end of the density was the best fit for the C3 end versus the pyridine ring. Thus, after the rest of the refinement was completed and had converged, the 3-keto, 5α abiraterone analog was modeled in two orientations—either with the pyridine ring directed either toward the interior of the protein or toward the solvent-exposed surface. Critical evaluation of fit to electron density maps and a 0.32% decrease in Rfree supported the orientation with the pyridine ring projected toward the interior of the protein. An alternate scenario in which both orientations were simultaneously present with partial occupancies was also explored but yielded no improvement in Rfree. Secondarily, the ligand was also modeled with the projecting C18 and C19 methyls directed either towards the N-terminus or away from the N-terminus (180° rotation), which fit similarly (Fig. 5, A and B versus Fig. 5, C and D). Examination of a Feature Enhanced Map (2Fo-Fc, (9)) did not provide additional details, but refinement of the peripheral ligand occupancies yielded values of 88% and 78% for molecules C and D, respectively.
Figure 5.
Modeling ligand into the peripheral binding site in the CYP17A1 3-keto, 5α abiraterone inhibitor structure.A and C (chain C) and B and D (chain D) show modeling of the inhibitor with methyls pointed away (A and B) or towards (C and D) the N-terminus. This density was also modeled as tetraethylene glycol (PDB ligand code PG4) in chains C (E) and D (F), because PEG3350 was present during crystallization, but the rest of the PEG molecule cannot be accommodated. 2Fo-Fc maps generated without the ligands are contoured to 1 σ and shown in blue mesh. PDB, Protein Data Bank.
The density in the peripheral binding site was also modeled as tetraethylene glycol (Protein Data Bank (PDB) ligand code PG4) in chains C and D (Fig. 5, E and F), because PEG3350 was present during crystallization, but there is no space for the rest of this molecule. Overall, the differences in Rfree were negligible for no peripheral ligand (22.05%), 3-keto,5α-abiraterone as the peripheral ligand (22.05%) and part of a PEG chain as the peripheral ligand (22.07%), but the density is convincing that a ligand is present.
Effects of filling the peripheral site on inhibition
For abiraterone inhibition of progesterone hydroxylation, there was very little difference in the IC50 value between WT enzyme (69 ± 5 nM) and the N52Y mutant (51 ± 2 nM). However, as both values are approximately half the enzyme concentration (100 nM), it is ultimately difficult to distinguish differences between the two enzymes. Regardless, these data suggest strong inhibition of both WT and mutant CYP17A1. As abiraterone is a similarly efficient inhibitor of the lyase reaction, subsequent studies were conducted with (R)-orteronel, which is a weaker inhibitor. When both WT and N52Y were evaluated for progesterone hydroxylation by (R)-orteronel, the IC50 value was ∼6-fold lower for the mutant (650 ± 30 nM) than for the WT enzyme (4000 ± 300 nM).
Discussion
Prostate cancer patients on abiraterone produce metabolites with modifications to the steroid nucleus in terms of the presence, absence, or placement of a double bond in the A ring and the type of C3 substituent (7). Several of these products demonstrate in vitro affinity for and inhibition of CYP17A1 similar to abiraterone itself (8), but no information has been available on how such compounds might interact structurally with the CYP17A1 active site. In the current work, the X-ray crystal structures of CYP17A1 with three such abiraterone analogs reveal their binding within the CYP17A1 active site with an overall orientation similar to abiraterone.
Despite inhibitor structural modifications, direct contacts with the enzyme via coordination of the pyridine nitrogen to the heme iron and hydrogen bonding between the C3 substituent and N202 were maintained. The coordinate–covalent interaction between the heme iron and the nitrogen lone pair of the pyridine heterocycle is very similar for each of the three compounds herein, as well as to abiraterone itself and to the interaction between the iron and the imidazole nitrogen of the steroidal galeterone (TOK-001, VN/124-1) CYP17A1 inhibitor (4).
Modifications to the A ring and its C3 substituent were accommodated by variable hydrogen bonding networks in this region. Like the 3β−OH, Δ5 of abiraterone itself (4), the C3 substituents of the 3-keto, 5α analog and the 3-keto, Δ4 analog participate in a hydrogen bonding network wherein the C3 substituent hydrogen bonds to N202, which in turn hydrogen bonds to two water molecules bridging to R239 and G297. In several cases, a third water molecule bridges from one or the other of these two water molecules to the side chain of Y201. This is the general configuration for C3 keto and C3-βOH substrates as well (5). Conversely, the 3α-OH, 5α abiraterone analog has its hydroxyl oriented such that simultaneous hydrogen bonds can occur with both N202 and directly with the water bridging to R239 and G297. In addition to the C3 substituent, the conformation of the A ring is also important. For example, a Δ4 double bond leads to a flattening of the A-ring, while the absence of a double bond confers a different ring conformation. Thus, different combinations of the ring conformation and the spatial orientation of the C3 substituent are accommodated by the flexible nature of the distal hydrogen bonding network.
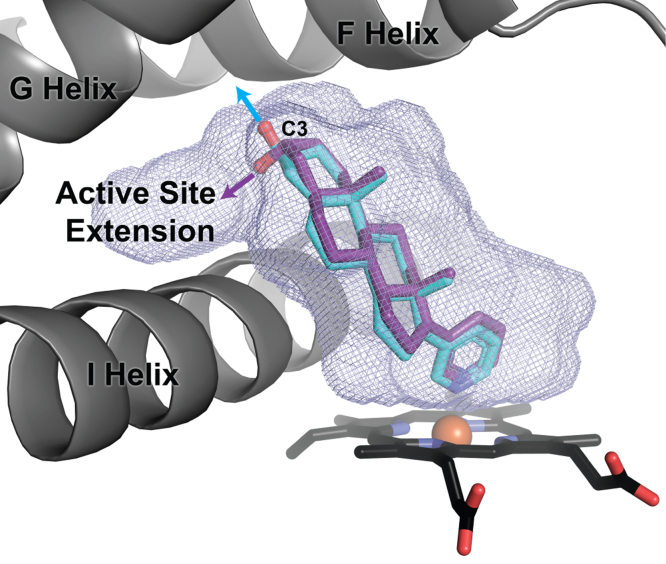
Since the C3 oxygen is one of the few heteroatoms available for modification on the steroidal scaffold, the direction in which it is projected could be useful in designing improved inhibitors accessing additional interactions with the CYP17A1 active site. For example, the CYP17A1 active-site cavity extends beyond the A ring and over the I helix (Fig. 6). This feature is not observed in other steroidogenic cytochrome P450s such as CYP21A2. CYP21A2 is also inhibited by abiraterone, and inhibitors that extend over the I helix may therefore demonstrate more selectivity for CYP17A1 versus CYP21A2. The 3α-hydroxyl is ideally projected over the I helix into this available space. Therefore, extending from this A ring configuration would likely be more effective than extending from the corresponding 3β-hydroxyl group in abiraterone, which is projected toward the F helix. The ability of CYP17A1 to accommodate 3α-OH compounds should not be surprising, given that the endogenous steroids allopregnanolone and 17α-hydroxyallopregnanolone have this structure and are excellent substrates for CYP17A1 in an alternate or “backdoor” steroidogenic pathway, leading to androsterone and dihydrotestosterone (10, 11). While structures are not available with these substrates, it would be reasonable to expect that their A ring would interact with N202 and water similarly to the 3α-OH, 5α abiraterone analog structure herein.
Figure 6.
The C3 hydroxyl of the 3α−OH, 5α abiraterone analog (purple) is directed into an extension of the CYP17A1 active-site void (blue mesh), whereas the C3 hydroxyl substituent of the 3β−OΗ, Δ5 abiraterone (cyan) is projected into the F helix.
Differences in A ring configuration and therefore hydrogen-bonding to the active-site water network do not appear to affect the potency of progesterone 17α-hydroxylase inhibition or 17,20-lyase inhibition (8). However, changes to the A ring configuration of abiraterone are reported to have significant effects on the inhibitory potency toward other prostate cancer targets. The 3-keto, Δ4 abiraterone analog (D4A) is a much more potent androgen receptor antagonist than abiraterone (6) and an effective inhibitor of 3β-hydroxysteroid dehydrogenase and 5α-reductase, both necessary enzymes in biosynthesis of the most potent androgen, 5α-dihydrotestosterone.
In addition to revealing the basis for CYP17A1 interactions with abiraterone with A ring modifications, the higher 2.0 Å resolution of the CYP17A1/3-keto, 5α abiraterone analog provides insight into a second ligand binding site some 20 Å distant from the inhibitor molecule in the active site. While electron density has been fairly consistently observed in the region between the N-terminal beta sheet region, A helix, and F/G loop in previous CYP17A1 crystal structures, it was too ambiguous to support modeling of an additional ligand. However, at 2.0 Å resolution, the map supports the idea that this density is due to binding of the steroidal ligand. The difference in density for the active-site copy and the peripheral copy suggest that this secondary site is probably a lower affinity site. Albeit more poorly defined, the fact that this density is also present in a variety of CYP17A1 complexes with steroidal substrates and inhibitors could suggest a broader functional role for this lower affinity, peripheral binding site. Crystal structures of other cytochrome P450 enzymes have revealed steroids interacting with similar regions of the enzyme.
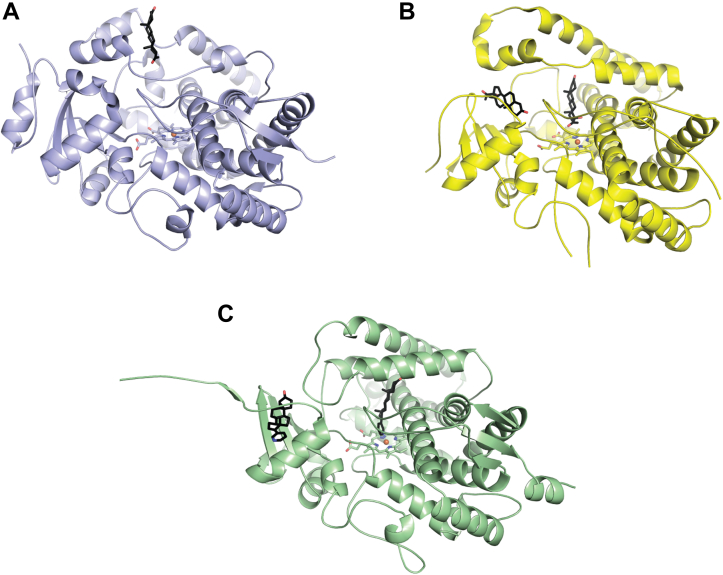
A structure of the steroidogenic bovine CYP21A2 revealed a second copy of the substrate 17α-hydroxyprogesterone at a site near the N-terminal β strands and the F′ helix (Fig. 7B) (12). This site lies between the active site of the enzyme and bulk solvent, suggesting that this channel may be a route for substrate access. The peripheral ligand in the structure of CYP17A1 with the 3-keto, 5α abiraterone analog interacts with very similar structural elements (Fig. 7C), but the conformation of the F/G region in bovine CYP21A2 is so dissimilar from that of human CYP17A1 that the second steroid is nearly 20 Å from the active-site steroid compared to ∼10 Å in the CYP21A2 structure. A more recent structure of human CYP21A2 with progesterone also identified a second binding site in the same region of the human form of the enzyme, albeit at lower occupancy (13).
Figure 7.
Cytochrome P450 enzymes with steroids (black sticks) binding in peripheral sites on the general membrane-binding surface.A, human CYP3A4 with a single copy of progesterone in the peripheral site. B, bovine CYP21A2 with two copies of 17α-hydroxyprogesterone, one in the active site and one in the peripheral site. C, human CYP17A1 with two copies of the 3-keto, 5α abiraterone inhibitor, one in the active site, and one in the peripheral site.
Additionally, one of the first structures of CYP3A4 revealed that progesterone, the intended active site ligand, was not located in the active site at all, but instead bound to a shallow pocket formed in the F/G loop (Fig. 7A) (14). While spectral binding studies primarily report on small molecule binding in the active site, progesterone binding to this secondary site is also supported by fluorescence resonance energy transfer studies. In CYP3A4, binding to this site is thought to participate in cooperative ligand binding by inducing a conformational change in CYP3A4 (15).
The functional relevance of this peripheral binding site in CYP17A1 is currently unknown, but there are several tantalizing pieces of information that suggest this influence would be worth exploring. First, this second binding site is associated with structural elements that are embedded in the membrane and whose conformational changes are thought to control substrate access from the membrane into the otherwise buried active site. As previously suggested for CYP21A2 (12), the peripheral site could simply be an initial loading site for substrates prior to movement into the active-site proper. Second, CYP17A1 performs two reactions, an initial 17α-hydroxylation followed by the 17,20-lyase reaction, necessary for androgen production. Facilitating release of the 17α-hydroxypregnenolone product of the first reaction, which is the substrate for the second reaction, could serve as a highly sought-after lyase-selective inhibitor strategy to reduce side effects associated with inhibition of the hydroxylase reaction. Current evidence suggests that the 17α-hydroxy intermediate is mostly released from the CYP17A1 active site and must rebind for the second reaction to occur (distributive mechanism), and only a relatively small amount of the 17α-hydroxy intermediate remains in the CYP17A1 active site for a second catalytic cycle (processive mechanism) (16). Thus, increasing release of the small proportion of 17α-hydroxy intermediate would be expected to have only a minor impact on lyase selective inhibition. However, a third possibility is that the peripheral site may allosterically affect inhibition. Filling the peripheral site by mutation reduced the IC50 for orteronel 6-fold. Allosteric mechanisms have already been implicated in a normal physiological control mechanism for CYP17A1. Cytochrome b5 binding to the proximal face of CYP17A1 allosterically and selectively augments the CYP17A1-mediated 17,20-lyase reaction but not 17α-hydroxylation (17). NMR studies have revealed that b5 binding on one face of CYP17A1 alters the backbone conformations of residues in the F and G helices on the opposite face of the enzyme (18). While it is possible that b5 promotes the lyase reaction by altering protein dynamics, coupling (19), or multiple steps in the catalytic cycle (16), overall, it appears there is intricate communication across the CYP17A1 structure, now including this peripheral binding site.
In conclusion, structures of CYP17A1 with abiraterone analogs modified in the A ring demonstrate these inhibitors make similar direct interactions with CYP17A1 and are accommodated by an adaptive network of water-mediated hydrogen bonding interactions. Additionally, the generation of the highest resolution CYP17A1 structure to date permitted definition of a second, peripheral ligand binding site. This latter binding site may be relevant to various aspects of CYP17A1 function and should be investigated further, potentially to develop more selective treatment of advanced prostate cancer.
Experimental procedures
Protein expression and purification
The human CYP17A1 pCWori+ construct employed herein bears a 19-residue truncation at the N-terminus, modification of the first five residues, and a 4x-histidine tag at the C-terminus (4). CYP17A1 was expressed and purified in the ligand-free form as described previously (5), with the following modifications. The antibiotic carbenicillin (100 μg/ml) was used to select for the pCWori+ plasmid as a more stable substitute for ampicillin (50 μg/ml). Overexpression cultures were initiated with 30 ml starter culture and subsequently grown for 48 to 72 h. During purification, membrane proteins were solubilized by 2% (v/v) Emulgen-913 for 60 min instead of 90 min.
Crystallization and structure determination
CYP17A1 was cocrystallized with inhibitors via hanging-drop vapor diffusion. CYP17A1 (30 mg/ml) in buffer (50 mM Tris–HCl, pH 7.4, 20% glycerol, 100 mM glycine, 500 mM NaCl, 0.5% (v/v) Emulgen-913) with 10 to 20 μM inhibitor was mixed in a 1:1 ratio with precipitant to form 2 μl drops. Drops were equilibrated against 750 μl precipitant containing 175 mM Tris–HCl, pH 8.5, 30% PEG-3350, 300 mM lithium sulfate, and 3% glycerol. Crystals were cryoprotected in a 7:3 mixture of mother liquor and 80% glycerol and flash cooled in liquid nitrogen. Data were collected on beamlines 14-1 and 12-2 at the Stanford Synchrotron Radiation Lightsource and processed using XDS (20). Statistics for data collection and refinement are shown in Table 1. Structures were solved by molecular replacement in Phaser (21) using a previous structure of CYP17A1 bound to an inhibitor (PDB 3SWZ) (4) as a search model. Model building and iterative refinement were performed using COOT (22) and PHENIX (23), respectively. Ligand coordinates were generated using eLBOW (24). Simulated-annealing composite omit maps were calculated in PHENIX (23). Probe-occupied active-site void volumes were calculated using VOIDOO (25) with a probe radius of 1.4 Å and grid spacing of 0.33. Structure alignments were generated in COOT (22) using the secondary structure matching algorithm. Figures were created using PyMOL (26).
Site-directed mutagenesis
The N52Y mutation was introduced to pCW17A1Δ19H vector using the QuikChange Lightning Site-Directed Mutagenesis kit (Agilent Technologies, Inc), according to the kit protocol with the forward primer 5′-CCT TTC TTA CCA CGA CAC GGT CAC ATG CAT AAT TAT TTC TTT AAA CTC-3′ and the complementary reverse primer. Bolded nucleotides were mutations, with the first mutation introducing a new Tth111I restriction site and the second introducing the desired N52Y mutation. An isolated plasmid with the correct Tth111I restriction pattern was sequenced to verify the intended mutations were present and there were no unintended mutations throughout the rest of the gene. Expression and purification proceeded as previously described for the WT construct. The N52Y purified protein displayed a typical absorbance spectrum and reduced CO difference spectrum, indicating normal heme incorporation.
Crystallization and structure determination of CYP17A1 N52Y
Purified CYP17A1 N52Y (230 μM in 50 mM Tris–HCl, pH 7.4, 20% glycerol, 100 mM glycine, and 300 mM NaCl) was diluted 8-fold in the same buffer supplemented with 25 μM abiraterone. The protein was concentrated 8-fold and was again diluted, abiraterone added, and concentrated as described two additional times before final concentration to 30 mg/ml. At this time, 0.5% (v/v) Emulgen-913 was added to the concentrated, ligand-bound protein.
All crystals were grown at 20 °C. CYP17A1 N52Y bound to abiraterone was first crystallized via sitting drop vapor diffusion in a 1.25 μl drop by mixing ligand-bound protein with well solution in a 1:1.5 mixture. The well solution contained 100 mM Tris–HCl, pH 8.5, 30% (v/v) PEG 4000, and 200 mM lithium sulfate. The resulting crystals were used to prepare a seed stock. Abiraterone-bound CYP17A1 N52Y was then crystallized in a 1 μl drop by hanging drop vapor diffusion by mixing protein (30 mg/ml in the aforementioned buffer) with well solution and seed stock in a 0.8:1:0.2 ratio. The well solution used for hanging drop vapor diffusion crystallization was 175 mM Tris–HCl, pH 8.5, 30% (v/v) PEG 3350, 250 mM lithium sulfate, 3% (v/v) glycerol with a layer of Al’s oil placed over the reservoir prior to incubation. Crystals were cryoprotected in silicone oil and flash-frozen in liquid nitrogen.
Diffraction data were collected on beamline 12-2 of the Stanford Synchrotron Radiation Lightsource, processed, and the structure as determined as indicated for the preceding WT structures.
Spectral ligand binding assay
The ligand binding to CYP17A1 N52Y was monitored via the change in UV-visible absorbance associated with the displacement of heme-coordinated water upon titration with progesterone or 17α-hydroxypregnenolone as described previously (5). Dissociation constants were determined by fitting data to the tight-binding equation in GraphPad Prism.
Catalysis of progesterone hydroxylation and 17α-Hydroxypregnenolone 17,20-lyase
Hydroxylation of progesterone (0.5–150 μM) by WT CYP17A1 and the N52Y mutant was conducted using a 1:4 ratio of CYP17A1 to human cytochrome P450 reductase bearing a 27-residue N-terminal truncation and a 3x glycine, 6x histidine tag at the C-terminus (27). All data points reflect at least two biological replicates comprised of 2-3 technical replicates. The product was separated from the remaining substrate by HPLC and quantified by UV-visible absorbance as described previously (5). Kinetic constants were extracted from fit to the Michaelis–Menten equation in GraphPad Prism.
Conversion of 17α-hydroxypregnenolone (0.1–15 μM) to the lyase product dehydroepiandrosterone was conducted using a 1:4:4 ratio of CYP17A1 to human cytochrome P450 reductase to rat cytochrome b5. All data points reflect at least four biological replicates comprised of two technical replicates. The product underwent solid-phase extraction and silylation as previously described in the presence of estriol (3.25 μg/ml) as an internal standard (5). Silylated product was normalized to silylated estriol and quantified via GC/MS using the specifications described previously. Data were fit to the Michaelis–Menten equation in GraphPad Prism.
Inhibition of progesterone hydroxylation and 17α-Hydroxypregnenolone 17,20-lyase activity
Abiraterone (10–500 nM) and R-orteronel (0.1–25 μM) were evaluated for inhibition of progesterone hydroxylation by CYP17A1 N52Y for comparison to WT. All data points reflect duplicate data from a single biological replicate. Inhibition was evaluated for WT and N52Y using a single concentration of progesterone equivalent to the Km value of each enzyme, 5 μM and 1.2 μM, respectively. The product was quantified by HPLC coupled with UV-vis absorbance as described above.
R-orteronel (50–5000 nM) was also evaluated for inhibition of 17α-hydroxypregnenolone 17,20-lyase activity by CYP17A1 N52Y compared to WT. Data points reflect two biological replicates with 2-3 technical replicates. Inhibition was evaluated using a 17α-hydroxypregnenolone concentration roughly equivalent to the Km (0.7 μM) for both N52Y and WT enzyme. Product was extracted, silylated, and quantified via GC/MS using estriol as an internal standard.
IC50 values, both progesterone hydroxylase and lyase, were determined using the equation for normalized activity versus log [inhibitor] (variable slope) in GraphPad Prism.
Expression, purification, and reconstituted assays with liposomal CYP17A1 N52Y
WT CYP17A1 and mutation N52Y (complementary DNA generated by site-directed mutagenesis as described with pCW17A1Δ19H) were expressed using pLW01 in C41(DE3) cells and purified as described previously (19). The purified P450 was reconstituted with human N-27 cytochrome P450 reductase (27) and cytochrome b5 (for 17,20-lyase reactions) with a 73:25:2 mixture of phosphatidyl choline, phosphatidyl ethanolamine, and phosphatidyl inositol. Assays and product analyses were performed as described previously (8, 19).
Data availability
All of the data is included in this article or the supplemental information statement, with the exception of the structure coordinates and structure factors. This latter information has been deposited with the PDB (http://www.rcsb.org/). The PDB codes are 6WR0, 6WW0, 5UYS, and 6WR1.
Conflict of interest
The authors declare that they have no conflicts of interest with the content of this article.
Acknowledgments
Crystallization employed instrumentation in the University of Kansas Protein Structure Lab, which was partially supported by NIH P20 RR17708. X-ray diffraction data were collected at the Stanford Synchrotron Radiation Lightsource. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health, National Institute of General Medical Sciences. We thank Dr Sunil Upadhyay for the synthesis of the abiraterone analogs as reported previously (8).
Author contributions
R. J. A. and E. E. S. conceptualization; R. J. A. resources; E. M. P., A. G. B., H.-M. P., and E. E. S. formal analysis; E. M. P. methodology; E. E. S. supervision; E. E. S. funding acquisition; E. M. P., A. G. B., and E. E. S. validation; E. M. P., H.-M. P., and E. E. S. investigation; E. M. P. and A. G. B. visualization; E. M. P., A. G. B., and E. E. S. writing – original draft; E. E. S. project administration; E. M. P., A. G. B., H.-M. P., R. J. A., and E. E. S. writing – review and editing.
Funding and additional information
E. M. P., A. G. B., and E. E. S. were supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM102505 (to E. E. S.). R. J. A. and H.-M. P. were supported by the Institutes of General Medical Sciences of the National institutes of Health under awards number R01GM086596 (to R. J. A.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Joseph Jez
References
- 1.Attard G., Reid A.H., Yap T.A., Raynaud F., Dowsett M., Settatree S., et al. Phase I clinical trial of a selective inhibitor of CYP17, abiraterone acetate, confirms that castration-resistant prostate cancer commonly remains hormone driven. J. Clin. Oncol. 2008;26:4563–4571. doi: 10.1200/JCO.2007.15.9749. [DOI] [PubMed] [Google Scholar]
- 2.de Bono J.S., Logothetis C.J., Molina A., Fizazi K., North S., Chu L., et al. Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Potter G.A., Barrie S.E., Jarman M., Rowlands M.G. Novel steroidal inhibitors of human cytochrome P45017α (17α-hydroxylase-C17,20-lyase): potential agents for the treatment of prostatic cancer. J. Med. Chem. 1995;38:2463–2471. doi: 10.1021/jm00013a022. [DOI] [PubMed] [Google Scholar]
- 4.DeVore N.M., Scott E.E. Structures of cytochrome P450 17A1 with prostate cancer drugs abiraterone and TOK-001. Nature. 2012;482:116–119. doi: 10.1038/nature10743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrunak E.M., DeVore N.M., Porubsky P.R., Scott E.E. Structures of human steroidogenic cytochrome P450 17A1 with substrates. J. Biol. Chem. 2014;289:32952–32964. doi: 10.1074/jbc.M114.610998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Z., Bishop A.C., Alyamani M., Garcia J.A., Dreicer R., Bunch D., et al. Conversion of abiraterone to D4A drives anti-tumour activity in prostate cancer. Nature. 2015;523:347–351. doi: 10.1038/nature14406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z., Alyamani M., Li J., Rogacki K., Abazeed M., Upadhyay S.K., et al. Redirecting abiraterone metabolism to fine-tune prostate cancer anti-androgen therapy. Nature. 2016;533:547–551. doi: 10.1038/nature17954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrido M., Peng H.M., Yoshimoto F.K., Upadhyay S.K., Bratoeff E., Auchus R.J. A-ring modified steroidal azoles retaining similar potent and slowly reversible CYP17A1 inhibition as abiraterone. J. Steroid Biochem. Mol. Biol. 2014;143:1–10. doi: 10.1016/j.jsbmb.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Afonine P.V., Moriarty N.W., Mustyakimov M., Sobolev O.V., Terwilliger T.C., Turk D., et al. FEM: feature-enhanced map. Acta Crystallogr. D Biol. Crystallogr. 2015;71:646–666. doi: 10.1107/S1399004714028132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta M.K., Guryev O.L., Auchus R.J. 5α-reduced C21 steroids are substrates for human cytochrome P450c17. Arch. Biochem. Biophys. 2003;418:151–160. doi: 10.1016/j.abb.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 11.Auchus R.J. The backdoor pathway to dihydrotestosterone. Trends Endocrinol. Metab. 2004;15:432–438. doi: 10.1016/j.tem.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 12.Zhao B., Lei L., Kagawa N., Sundaramoorthy M., Banerjee S., Nagy L.D., et al. Three-dimensional structure of steroid 21-hydroxylase (cytochrome P450 21A2) with two substrates reveals locations of disease-associated variants. J. Biol. Chem. 2012;287:10613–10622. doi: 10.1074/jbc.M111.323501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pallan P.S., Wang C., Lei L., Yoshimoto F.K., Auchus R.J., Waterman M.R., et al. Human cytochrome P450 21A2, the major steroid 21-hydroxylase: structure of the enzyme/progesterone substrate comples and rate-limiting C-H bond cleavage. J. Biol. Chem. 2015;290:13128–13143. doi: 10.1074/jbc.M115.646307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams P.A., Cosme J., Vinkovic D.M., Ward A., Angove H.C., Day P.J., et al. Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science. 2004;305:683–686. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- 15.Davydov D.R., Rumfeldt J.A., Sineva E.V., Fernando H., Davydova N.Y., Halpert J.R. Peripheral ligand-binding site in cytochrome P450 3A4 located with fluorescence resonance energy transfer (FRET) J. Biol. Chem. 2012;287:6797–6809. doi: 10.1074/jbc.M111.325654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez E., Guengerich F.P. Kinetic processivity of the two-step oxidations of progesterone and pregnenolone to androgens by human cytochrome P450 17A1. J. Biol. Chem. 2017;292:13168–13185. doi: 10.1074/jbc.M117.794917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Auchus R.J., Lee T.C., Miller W.L. Cytochrome b5 augments the 17,20-lyase activity of human P450c17 without direct electron transfer. J. Biol. Chem. 1998;273:3158–3165. doi: 10.1074/jbc.273.6.3158. [DOI] [PubMed] [Google Scholar]
- 18.Estrada D.F., Skinner A.L., Laurence J.S., Scott E.E. Human cytochrome P450 17A1 conformational selection: modulation by ligand and cytochrome b5. J. Biol. Chem. 2014;289:14310–14320. doi: 10.1074/jbc.M114.560144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng H.M., Im S.C., Pearl N.M., Turcu A.F., Rege J., Waskell L., et al. Cytochrome b5 activates the 17,20-lyase activity of human cytochrome P450 17A1 by increasing the coupling of NADPH consumption to androgen production. Biochemistry. 2016;55:4356–4365. doi: 10.1021/acs.biochem.6b00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moriarty N.W., Grosse-Kunstleve R.W., Adams P.D. Electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleywegt G.J., Jones T.A. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. D Biol. Crystallogr. 1994;50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- 26.Schrodinger L.L.C. 2010. The AxPyMOL Molecular Graphics Plugin for Microsoft PowerPoint, Version 1.0. [Google Scholar]
- 27.Sandee D., Miller W.L. High-yield expression of a catalytically active membrane-bound protein: human P450 oxidoreductase. Endocrinology. 2011;152:2904–2908. doi: 10.1210/en.2011-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All of the data is included in this article or the supplemental information statement, with the exception of the structure coordinates and structure factors. This latter information has been deposited with the PDB (http://www.rcsb.org/). The PDB codes are 6WR0, 6WW0, 5UYS, and 6WR1.