Abstract
The ADP-ribosylation factor (Arf) GTPases and their regulatory proteins are implicated in cancer progression. NAV-2729 was previously identified as a specific inhibitor of Arf6 that reduced progression of uveal melanoma in an orthotopic xenograft. Here, our goal was to assess the inhibitory effects of NAV-2729 on the proliferation of additional cell types. We found NAV-2729 inhibited proliferation of multiple cell lines, but Arf6 expression did not correlate with NAV-2729 sensitivity, and knockdown of Arf6 affected neither cell viability nor sensitivity to NAV-2729. Furthermore, binding to native Arf6 was not detected; however, we determined that NAV-2729 inhibited both Arf exchange factors and Arf GTPase-activating proteins. ASAP1, a GTPase-activating protein linked to cancer progression, was further investigated. We demonstrated that NAV-2729 bound to the PH domain of ASAP1 and changed ASAP1 cellular distribution. However, ASAP1 knockdown did not fully recapitulate the cytoskeletal effects of NAV-2729 nor affect cell proliferation. Finally, our screens identified 48 other possible targets of NAV-2729. These results illustrate the complexities of defining targets of small molecules and identify NAV-2729 as a model PH domain–binding inhibitor.
Keywords: ASAP1, ADP-ribosylation factor, GTPase, GTPase-activating protein, guanine nucleotide exchange factor, enzyme inhibitor
Abbreviations: Arf, ADP-ribosylation factor; GAP, GTPase-activating protein; GEF, guanine nucleotide exchange factor; LUV, large unilamellar vesicle; NAV-2729, 3-(4-chlorophenyl)-5-(4-nitrophenyl)-2-(phenylmethyl)-pyrazolo[1,5-a]pyrimidin-7)4H)-one; PH, pleckstrin homology; PIP2, phosphatidylinositol 4,5-bisphosphate; PIP3, phosphatidylinositol 3,4,5-trisphosphate; TMT, tandem mass tag
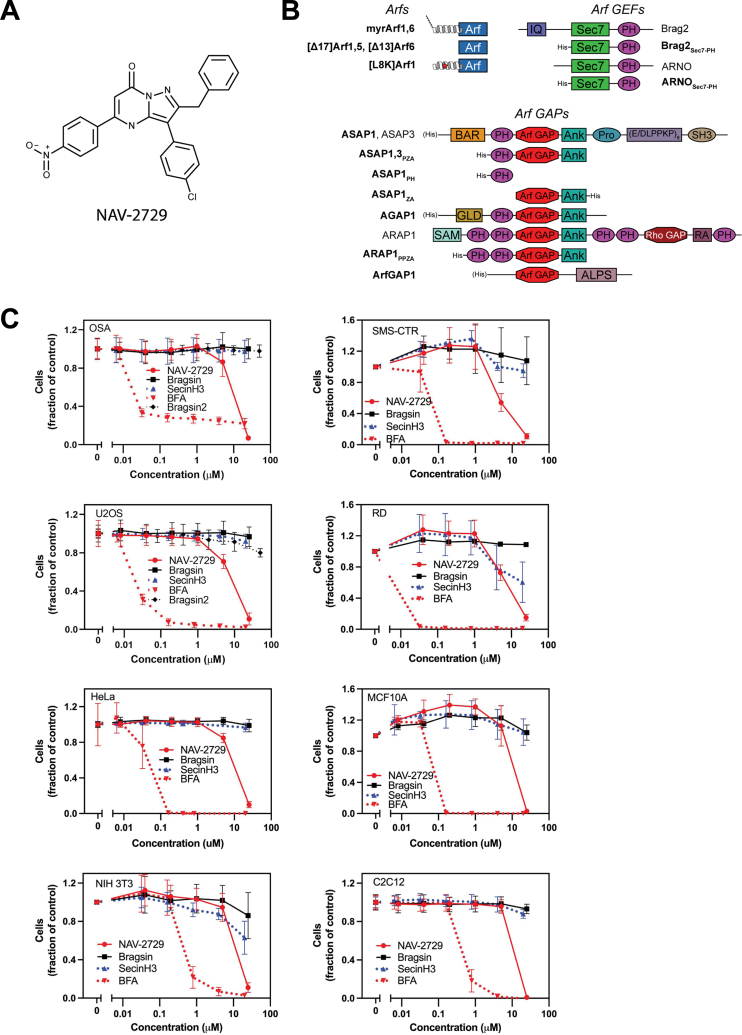
Arfs are part of the Arf family of GTPases. There are five Arfs in humans (1, 2). With slow spontaneous exchange and no detectable intrinsic GTPase activity (3), Arf function depends on guanine nucleotide exchange factors (GEFs), encoded by 15 genes in human, and GTPase-activating proteins (GAPs), encoded by 28 genes (2). Arf•GTP binds an eclectic array of effectors to regulate membrane traffic and actin cytoskeleton remodeling. Effects on proliferation, differentiation, and cell survival signaling have also been identified. Dysregulation of the pathways affected by Arfs are being investigated in cancer progression (4, 5, 6).
One of the first reports suggesting the Arf pathway contributes to cancer progression was the finding that amplification of the gene ASAP1, which encodes an Arf GAP, correlated with metastasis in uveal melanoma (7). Subsequently, ASAP1 amplification and/or expression was found to correlate with poor prognosis and metastasis in several carcinomas (8, 9, 10). Other elements of the Arf pathway have also been implicated in cancer. For instance, Arf exchange factor Brag2/GEP100/IqSec1 expression correlated with invasiveness and metastasis in breast and prostate cancer (11, 12). High levels of Arf6 expression are associated with poor prognosis in uveal melanoma; the finding motivated the screen for small molecules that bind directly to Arf6 and prevent its activation, leading to the identification of NAV-2729 (3-(4-chlorophenyl)-5-(4-nitrophenyl)-2-(phenylmethyl)-pyrazolo[1,5-a]pyrimidin-7)4H)-one, Figure 1A) (13).
Figure 1.
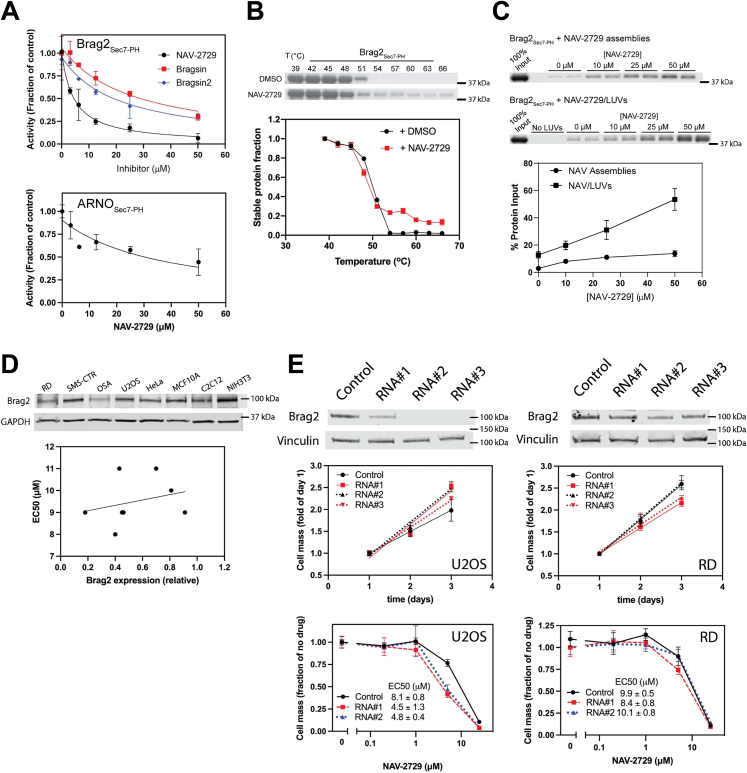
NAV-2729 reduces proliferation of sarcomas, carcinomas, and nontransformed cells with similar potency and efficiency.A, the structure of the small molecule NAV-2729. B, proteins and constructs used in this paper. Domains of each protein are shown (schematics are not to scale). Bolded text indicates proteins or constructs that were recombinantly purified. N-terminal helices are shown for myrArf1, myrArf6, and [L8K]Arf1; the former are myristoylated (shown as lines), whereas [L8K]Arf1 (mutation point shown as red star) is not. When present, His tags used for purification are shown on the proteins’ N- or C-termini; His tags on full-length proteins are shown in parentheses to indicate that they are not normally present in the natural protein sequence. C, effect of Arf GEF inhibitors on proliferation. The effect of NAV-2729, Brefeldin A, Bragsin, SecinH3, or Bragsin2 on relative cell mass was determined using Cell Titer Glo reagent (Promega). Summaries of three (2 for Bragsin2) experiments are shown. Arf, ADP-ribosylation factor; GEF, guanine nucleotide exchange factor.
NAV-2729 was reported to reduce proliferation of uveal melanoma cells in vitro and reduced progression of tumors in an orthotopic xenograft model of uveal melanoma in mice (13). The precise mechanism of action is still being discovered. Initial work indicated that NAV-2729 bound specifically to Arf6, blocking spontaneous exchange and exchange catalyzed by Brag2 and ARNO (13). Subsequent studies indicated that it inhibited Brag2 exchange on Arf1 (14). These latter results raise the possibility that NAV-2729 might bind to multiple Arf isoforms, affecting their interaction with GEFs. Furthermore, if NAV-2729 functions by binding to Arfs, a plausible hypothesis is that it affects Arf interaction with other Arf targets.
Here, we set out to determine whether NAV-2729 might affect proliferation of pediatric sarcomas, some of which have amplifications of Arf pathway genes. After finding that NAV-2729 affected proliferation of a variety of cell types with similar potency that did not correlate with levels of Arf6, nor did Arf6 knockdown affect cell proliferation or NAV-2729 sensitivity, we examined other potential mechanisms of action. We found that NAV-2729 directly bound to and inhibited both Arf GEFs and Arf GAPs, including the Arf GAP ASAP1 (Fig. 1B shows proteins studied here). Although some of the effects of NAV-2729 on cell morphology could be attributed to the inhibition of ASAP1, additional targets of NAV-2729 likely account for effects on other morphological changes and cell viability. Our preliminary screens also identified potential targets outside of the Arf pathway.
Results
NAV-2729 decreases proliferation of unrelated cell lines with similar potency
We determined the effect of Arf exchange factor inhibitors, including NAV-2729, on proliferation of two pediatric osteosarcomas cell lines (U2OS and OSA), two fusion negative rhabdomyosarcoma cell lines (RD and SMS-CTR), HeLa cervical carcinoma cells, a breast epithelium cell line (MCF 10A), a mouse myoblast cell line (C2C12), and a mouse fibroblast cell line (NIH 3T3) (Fig. 1C). NAV-2729 was toxic with similar potency for all cells examined with EC50s in the range of 8 to 11 μM (Table 1). In contrast, Bragsin, an inhibitor of Brag2 (15), had no effect on the sarcomas but a small effect on NIH 3T3 fibroblasts and C2C12 myoblasts. We tested the effect of a more stable Bragsin analog, Bragsin2 (15), on the osteosarcoma cell lines and similarly observed little or no toxicity. SecinH3, an inhibitor of Cytohesins (16), had no effect on OSA, U2OS, or SMS-CTR cells but had a small effect at the highest concentration tested on NIH 3T3 fibroblasts, C2C12 myoblasts, and RD cells. As previously described, Brefeldin A, which inhibits the Arf1 exchange factors GBF1 and BIG1/2 (17, 18), was toxic to all cells examined with 10-fold greater potency for the human than mouse cells examined (Fig. 1C and Table 1). Thus, the effect of NAV-2729 was distinct from other Arf exchange factor inhibitors (Fig. 1C and Table 1).
Table 1.
Relative protein expression and effect of inhibitors on cell lines and recombinant Arf GEFs and Arf GAPs used in this study
| Cell line or recombinant protein | Arf6 expression (relative to U2OS) | Brag2 expression (relative to NIH3T3) | ASAP1 expression (relative to NIH3T3) | EC50 for cell killing (μM) |
||||
|---|---|---|---|---|---|---|---|---|
| NAV-2729 | Brefeldin A | Bragsin | Bragsin2 | SecinH3 | ||||
| OSA | 0.17 | 0.20 | 0.03 | 9 ± 2 (n = 3) | 0.026 ± 0.004 | >50 | >50 | >50 |
| U2OS | 1.00 | 0.51 | 0.37 | 9 ± 1 (n = 3) | 0.024 ± 0.002 | >50 | >50 | >50 |
| RD | 0.11 | 0.44 | 0.20 | 8 ± 4 (n = 3) | 0.006 ± 0.002 | >50 | >50 | |
| SMS-CTR | 0.71 | 0.77 | 0.14 | 11 ± 5 (n = 3) | 0.016 ± 0.002 | >50 | >50 | |
| HeLa | 0.30 | 0.49 | 0.02 | 9 ± 2 (n = 3) | 0.04 ± 0.01 | >50 | >50 | |
| MCF 10A | 0.58 | 0.89 | 0.05 | 10 ± 4 (n = 3) | 0.11 ± 0.04 | >50 | >50 | |
| NIH3T3 | 0.71 | 1.00 | 1.00 | 11 ± 3 (n = 3) | 0.44 ± 0.14$$ | >50 | >50 | |
| C2C12 | 0.43 | 0.43 | 0.95 | 11 ± 2 (n = 3) | 0.46 ± 0.07$$ | >50 | >50 | |
| IC50 for protein inhibition (μM) | ||||||||
| Brag2Sec7-PH with LUV | 7.1 ± 2.5 (n = 3) | 31 ± 12 (n = 3)∗ | 25 ± 10 (n = 4) | |||||
| ARNOSec7-PH with LUV | 37 ± 11 (n = 4)∗∗ | |||||||
| AGAP1 with LUV | 2.7 ± 1.4 (n = 5) | |||||||
| ARAP1PPZA with LUV | >50 (n = 3) | |||||||
| ArfGAP1 with LUV | >50 (n = 3) | |||||||
| ASAP1PZA with LUV | 4.6 ± 0.9 (n = 4) | |||||||
| ASAP3PZA with LUV | 9.1 ± 3.2 (n = 4)#,&& | |||||||
| ASAP1PZA with diC8-PIP2, L8K | 29 ± 6.7 (n = 6)#### | |||||||
| ASAP1 with LUV | 12.2 ± 1.4 (n = 4) | |||||||
| ASAP1PZA with LUV (2.5% PIP2), L8K | 13.4 ± 2.3 (n = 4) | |||||||
Top. $$, different than human cells, p < 0.01. ANOVA with Tukey post hoc multiple comparison test. Proteins were detected by immunoblotting. Signal was normalized to GAPDH or vinculin and is expressed as a fraction of the maximum signal observed. Twenty micrograms protein of the total cell lysate were loaded on the gels. Bottom. IC50s for recombinant protein are means ± SDs. ∗, different than NAV-2729 for inhibition of Brag2, p < 0.05; ∗∗, different than inhibition of Brag2, p < 0.01 by ANOVA analysis of Brag2 and ARNO data with Tukey post hoc multiple comparison test. #, greater than ASAP1 with LUV, p < 0.05, ####, greater than ASAP1 with LUV, p < 0.0001, &&, greater than AGAP1 with LUV, p < 0.01, determined by one way ANOVA followed by a Tukey multi comparison test for AGAP1 with LUV, ASAP1 with LUV, ASAP1 with diC8-PIP2, and ASAP3 with LUV.
NAV-2729 affects the actin cytoskeleton and cell morphology
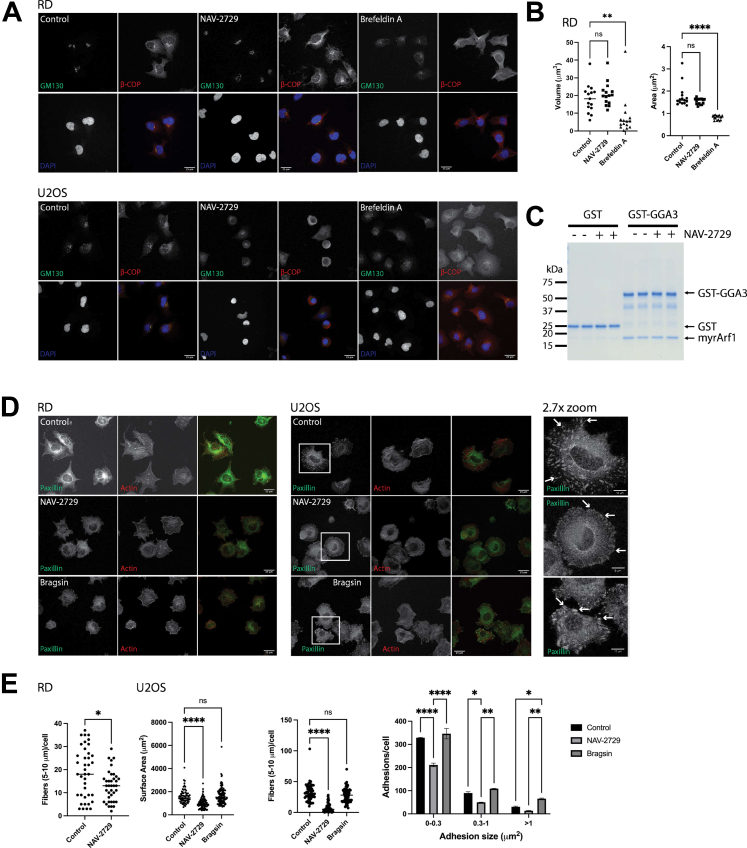
NAV-2729, by binding to Arfs, might affect Arf-dependent structures in cells, including the Golgi apparatus and the actin cytoskeleton. We tested the predictions in two cell lines, U2OS and RD cells (Fig. 2). The Golgi apparatus was visualized by immunofluorescence of GM130 and β-COP (19, 20). Treatment of RD cells with Brefeldin A caused the compact perinuclear GM130 signal to disperse in numerous puncta through the cell and the β-COP signal to be cytoplasmic (Fig. 2A), with a decrease in Golgi surface area and volume (Fig. 2B). In contrast, GM130 and β-COP were unaffected by NAV-2729 (Fig. 2, A and B). NAV-2729 caused a reduction of cell surface area of U2OS cells—see below—limiting quantification of effects on the Golgi apparatus. To further examine the possibility that NAV-2729 interfered with Arf function, we determined whether NAV-2729 affected Arf binding to the coat protein GGA3, an Arf effector (21). Arf1•GTP binding to GGA3 can be measured using a pull-down assay (21). Fifty micromolar NAV-2729 had no effect, with the signal indistinguishable from the control (Fig. 2C). At least in this case, NAV-2729 does not block Arf interaction with an effector, further supporting the idea that NAV-2729 does not directly interfere with Arf regulation of membrane trafficking proteins (Fig. 2C).
Figure 2.
NAV-2729 does not affect the Golgi apparatus but does affect the actin cytoskeleton.A, effect of NAV-2729 and Brefeldin A on RD and U2OS cells. Cells were treated with DMSO, NAV-2729, or Brefeldin A, then stained using DAPI and immunostained for GM130 and β-COP. Scale bars are 25 μm. B, NAV-2729 does not affect the Golgi in RD cells. Surface area and volume of the Golgi in the cells treated as described in (A) were determined using GM130 as a marker. C, NAV-2729 does not affect Arf1•GTP binding to GGA3. myrArf1•GTP binding to the VHS-GAT tandem from GGA3 fused to GST in the presence or absence of 50 μM NAV-2729 was determined. GST was the negative control. Representative experiment of 3 is shown. D, NAV-2729 and Bragsin effects on the actin cytoskeleton. RD and U2OS cells were plated on fibronectin-coated coverslips and treated with either DMSO, NAV-2729, or Bragsin. Cells were immunostained for paxillin and for filamentous actin. Scale bars are 25 μm for all images except the far right images, which are an approximately 2.7-factor zoom of the images for paxillin within the white boxes. The scale bars are 10 μm in length for the zoomed images. Arrows point to example focal adhesions. E, quantification of actin stress fibers (RD and U2OS), focal adhesions (U2OS), and surface area (U2OS) of the cells shown in (D). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p <0.0001. Arf, ADP-ribosylation factor.
Arf pathways also regulate actin stress fibers and integrin adhesions, including focal adhesions. Knockdown of Brag2, a putative target of NAV-2729 (13, 14), increases the number of large focal adhesions in HeLa cells (22). To determine whether NAV-2729 affected actin cytoskeleton structures, RD and U2OS cells were plated on fibronectin-coated coverslips in either complete (RD) or serum-free (U2OS) medium in the presence or the absence of 25 μM NAV-2729 or Bragsin. After 2 h, the cells were fixed and stained for paxillin and actin filaments. Untreated RD cells did not form robust focal adhesions, so measuring an effect of NAV-2729 was inconclusive. However, NAV-2729 reduced actin stress fibers (Fig. 2, D and E). U2OS cells formed robust focal adhesions and actin stress fibers; we found that NAV-2729 reduced focal adhesions and actin stress fibers (Fig. 2, D and E). In contrast, Bragsin, an inhibitor of Brag2, increased the number of large focal adhesions and did not affect actin stress fibers (Fig. 2, D and E). NAV-2729 and Bragsin also had different effects on the surface area of U2OS cells: NAV-2729 decreased the surface area while Bragsin had no effect (Fig. 2E). We conclude that although NAV-2729 might bind to Arfs, it disrupts only a subset of Arf-dependent cellular structures.
Arf6 is not the primary NAV-2729 target in the cell lines examined
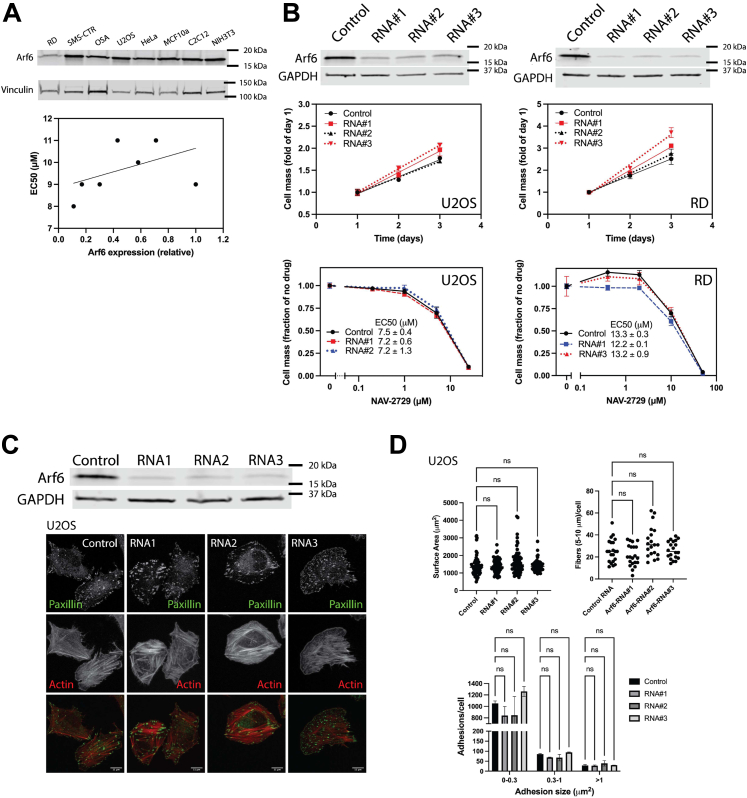
With similar sensitivity to NAV-2729, the cell lines we examined might be expected to have similar levels of the putative target of NAV-2729, Arf6. Arf6 expression was quantified by immunoblotting and levels plotted against EC50s for inhibition of cell proliferation (Figs. 1C and 3A and Table 1). Arf6 expression varied among the cell lines with no correlation with EC50s (r2 = 0.21, p = 0.24; Fig. 3A and Table 1). We determined the effect of reduced expression of Arf6 on cell proliferation in RD and U2OS. Arf6 knockdown, accomplished by transfection with three dicer substrate RNA (diRNA), was efficient for both cell lines but had no detectable effect on proliferation (Fig. 3B). If Arf6 was a target of NAV-2729, then knockdown of Arf6 might affect NAV-2729 sensitivity. Contrary to the prediction, there was no change in sensitivity in either cell line (Fig. 3B). Furthermore, in contrast to NAV-2729 treatment, which reduced actin stress fibers and focal adhesions (Fig. 2, D and E), reduced Arf6 expression affected neither (Fig. 3, C and D). Similarly, cell surface area was reduced by NAV-2729 (Fig. 2E) but was not affected by decreased Arf6 expression (Fig. 3D). We conclude that although Arf6 might be a target, it is not the primary target responsible for the effect of NAV-2729 on proliferation of these cells.
Figure 3.
Arf6 might not be the primary target of NAV-2729.A, expression of Arf6 and sensitivity to NAV-2729 do not correlate. Relative expression levels were plotted against EC50 values for inhibition of cell proliferation (Fig. 1C and Table 1). B, knockdown of Arf6 affected neither proliferation nor NAV-2729 sensitivity of RD and U2OS cells. Three different diRNA targeting Arf6 were used to knockdown Arf6 for proliferation assays and two to test NAV-2729 sensitivity. Relative cells mass was determined as in Figure 1C. Figures show summarized results from two experiments. C, knockdown of Arf6 does not affect actin stress fibers or focal adhesions. U2OS cells treated with three different diRNA targeting Arf6 were plated and immunostained as described in Figure 2D. Scale bars are 11 μms in length. D, quantification of the surface area, actin stress fibers, and focal adhesions in U2OS cells shown in (C). Arf, ADP-ribosylation factor.
Full-length Arf1 and Arf6 do not bind to NAV-2729
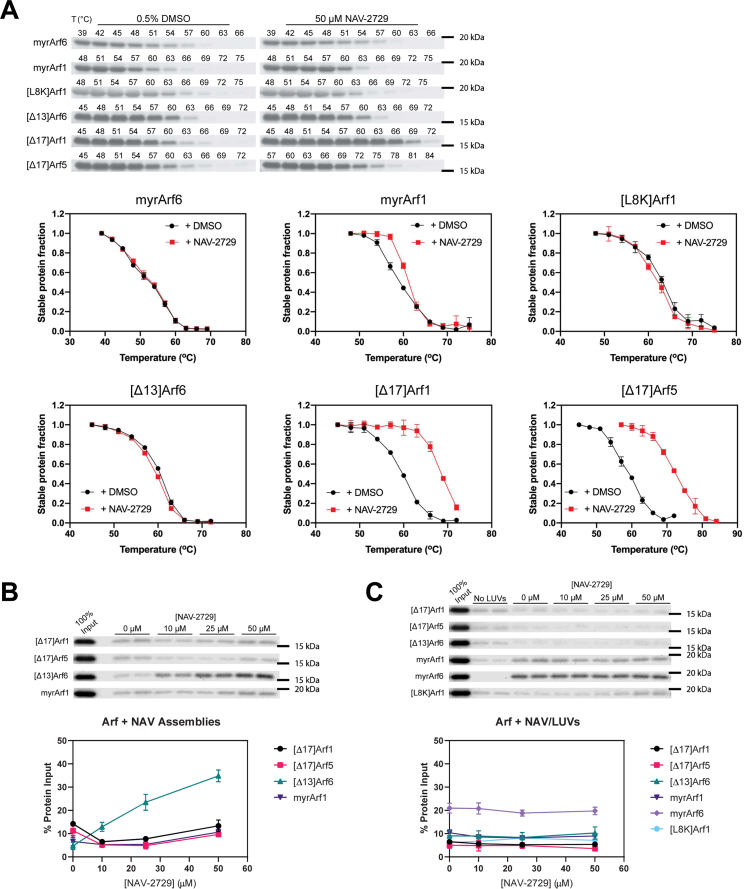
As described in Supplemental Materials under “Determination of optical and physical properties of NAV-2729” and in Figs. S1–S3, NAV-2729 forms supramolecular assemblies in aqueous solution and partitions into phospholipid membranes. While previous biochemical analysis has been performed under conditions in which NAV-2729 forms supramolecular assemblies (13, 14), NAV-2729 likely partitions efficiently into the cellular membrane (Fig. S3). We performed binding studies using both supramolecular assemblies and NAV-2729 integrated into large unilamellar vesicles (LUVs). We first examined the effect of NAV-2729 on thermal stability of the Arfs. Ligand binding can increase the thermal stability of proteins and, therefore, might be detected as an increase in the temperature required to denature a protein (23). We determined the extent of denaturation by measuring protein remaining in solution after heating.§ Fifty micromolar NAV-2729 had no effect on either myrArf6, the native form of the protein (myristoylated, full-length protein), or [Δ13]Arf6 (Fig. 4A). By contrast, NAV-2729 caused a shift of approximately 3 degrees for myrArf1, greater than 9 degrees for [Δ17]Arf1, and 12 to 15 degrees for [Δ17]Arf5 (Fig. 4A). We also tested an Arf mutant, [L8K]Arf1, which retains N-terminal residues but is not myristoylated (25, 26). NAV-2729 did not affect the thermal stability of [L8K]Arf1 (Fig. 4A). Binding to [L8K]Arf1, myrArf6, and [Δ13]Arf6 could not be excluded, as a thermal shift requires a change in heat capacity upon ligand binding, which does not necessarily occur (27). Nevertheless, the data support the conclusion that NAV-2729 might bind to at least some Arf proteins.
Figure 4.
NAV-2729 binds to Arf1, Arf5, and Arf6 in different contexts.A, thermal shift assays. The effect of NAV-2729 on thermal stability of the indicated proteins was determined and described in Experimental procedures. Representative gels and summaries of three experiments are shown. B, cosedimentation of Arfs with NAV-2729 supramolecular assemblies. NAV-2729 supramolecular assemblies were incubated with protein, then pelleted in an ultracentrifuge. Protein in the pellet was analyzed by SDS-PAGE. Representative gels and summary of two experiments, three experiments for [Δ13]Arf6, are shown. C, cosedimentation of Arfs with LUVs containing NAV-2729. Proteins associating with NAV-2729 containing LUVs were analyzed as described for supramolecular assemblies in (B). Representative gels and summary of two experiments with myrArf1, myrArf6, and [L8K]Arf1 and three with truncated Arfs are shown. Arf, ADP-ribosylation factor; LUV, large unilamellar vesicle.
Arf binding to NAV-2729 supramolecular assemblies was also determined by isolating supramolecular assemblies by centrifugation (Fig. S2C). Associated protein was fractionated by SDS-PAGE, visualized with Coomassie blue stain, and quantified (Fig. 4B). The truncated mutants of Arfs which bound NAV-2729 as assessed by thermal shift did not associate with NAV-2729 supramolecular assemblies (Fig. 4B). MyrArf1•GDP, which bound to NAV-2729 in thermal shift assays, did not cosediment with supramolecular assemblies (Fig. 4B). Although binding to NAV-2729 was not detected in thermal shift analysis, [Δ13]Arf6 cosedimented with NAV-2729 supramolecular assemblies (Fig. 4B).
We measured the association of Arf with LUVs into which NAV-2729 was partitioned (Fig. 4C). Arf proteins were incubated with LUVs containing up to 50 μM NAV-2729. The vesicles were separated from bulk solution by centrifugation, after which associated proteins were fractionated by SDS-PAGE, visualized with Coomassie blue stain, and quantified. None of the Arfs and Arf mutants bound to NAV-2729 in LUVs (Fig. 4C). Based on these results, we conclude Arfs have Kds >50 μM for NAV-2729 in a lipid bilayer.
NAV-2729 inhibits the Arf GEFs Brag2 and ARNO
We reexamined the effect of NAV-2729 on the Arf exchange factors Brag2 and ARNO. NAV-2729 has been reported to inhibit Brag2 and ARNO (13, 14), in assays with truncated Arfs and no phospholipids. We incubated myrArf1 with Brag2Sec7-PH or ARNOSec7-PH, [35S]GTPγS, and the indicated concentration of NAV-2729 in a reaction mixture containing LUVs. Arf1•[35S]GTPγS complex was trapped on nitrocellulose and quantified by scintillation counting. Brag2Sec7-PH-catalyzed exchange was inhibited with greater potency than ARNOSec7-PH-catalyzed exchange (Fig. 5A and Table 1). Bragsin and Bragsin2, included as positive controls, also inhibited Brag2Sec7-PH-induced nucleotide exchange on Arf1 (Fig. 5A and Table 1). We did not test SecinH3 because of its poor solubility (14).
Figure 5.
NAV-2729 binds to and inhibits Brag2.A, inhibition of Brag2 and ARNO. GEF activity was measured as described in Experimental procedures. Bragsin and Bragsin2 were used as positive controls. IC50 values are reported in Table 1. A representative experiment of three is shown for Brag2 and four for ARNO. B, thermal shift to determine binding to Brag2. The steps for the reaction are the same as described in Figure 4A. Representative gels and summary of two experiments are shown. C, cosedimentation with NAV-2729 supramolecular assemblies and with LUVs containing NAV-2729. The steps for the reaction are the same as described in Figure 4, B and C. Representative gels and summary of three experiments for each condition are shown. D, expression of Brag2 does not correlate with sensitivity to NAV-2729. The indicated cell lines were probed for their expression of Brag2 using immunoblotting, and the relative expression levels were plotted against EC50 values for cell killing (Fig. 1C and Table 1). E, effect of reduced expression of Brag2 on proliferation and NAV-2729 sensitivity in RD and U2OS cells. Three different diRNA targeting Brag2 were used to knockdown Brag2 for proliferation assays and two to test NAV-2729 sensitivity. Relative cells mass was determined as in Figure 1C. Results shown are the summary of two experiments. GEF, guanine nucleotide exchange factor; LUV, large unilamellar vesicle.
NAV-2729 binds to Brag2
Given that Arf proteins did not bind to LUVs containing NAV-2729 (Fig. 4C), we considered that the inhibitory effect of NAV-2729 could be due to binding to Brag2Sec7-PH. We measured the association of Brag2Sec7-PH to both NAV-2729 supramolecular assemblies and LUVs containing NAV-2729 (Fig. 5, B and C). In thermal shift assays, NAV-2729 increased the fraction of soluble protein at higher temperatures (Fig. 5B). Brag2 cosedimented with NAV-2729 supramolecular assemblies and with LUVs dependent on NAV-2729 concentration (Fig. 5C).
Brag2 is not the primary target of NAV-2729
Several lines of evidence indicate that although Brag2 is inhibited by NAV-2729, it might not be the primary target mediating the cellular effects. First, cells treated with Bragsin, a bona fide inhibitor of Brag2, had more large focal adhesions than untreated cells, while NAV-2729-treated had fewer focal adhesions (Fig. 2, D and E). Second, Bragsin and Bragsin2 were not toxic to cells that were sensitive to NAV-2729 (Fig. 1C and Table 1). Third, Brag2 expression in cells did not correlate with NAV-2729 sensitivity (r2 = 0.076, p = 0.51) (Figs. 1C and 5D and Table 1). Fourth, Brag2 knockdown did not affect cell proliferation (Fig. 5E). Fifth, a reduction in Brag2 expression only had a small effect on NAV-2729 sensitivity in U2OS cells. The RD cells were not affected, but the knockdown of Brag2 was not efficient in these cells (Fig. 5E). We conclude that although Brag2 is not likely to be the only target of NAV-2729, it might be one target necessary for effects on cell proliferation.
NAV-2729 binds to and inhibits Arf GAPs including ASAP1
Several Arf GAPs also affect focal adhesions and actin stress fibers (28, 29, 30, 31, 32, 33, 34). We examined five Arf GAPs representing four subtypes: ArfGAP1, AGAP1, ARAP1, ASAP1, and ASAP3 (Fig. 1B). Full-length ArfGAP1 and AGAP1 and recombinant proteins composed of the active fragment of ARAP1 (ARAP1PPZA), ASAP1 (ASAP1PZA), and ASAP3 (ASAP3PZA) (Fig. 1B) were used. GAP activity was determined using myrArf1 and LUVs, the latter necessary to stabilize myrArf1•GTP and to present phosphatidylinositol 4,5-bisphosphate (PIP2) and phosphatidylinositol 3,4,5-trisphosphate (PIP3), activating ligands for some of the Arf GAPs we examined (26, 35, 36, 37, 38, 39). NAV-2729 inhibited ASAP1PZA, ASAP3PZA, and AGAP1 (Fig. 6A and Table 1). ArfGAP1 and ARAP1PPZA were not inhibited (Fig. 6A and Table 1). NAV-2729 also inhibited full-length ASAP1 (Fig. S4A and Table 1).
Figure 6.
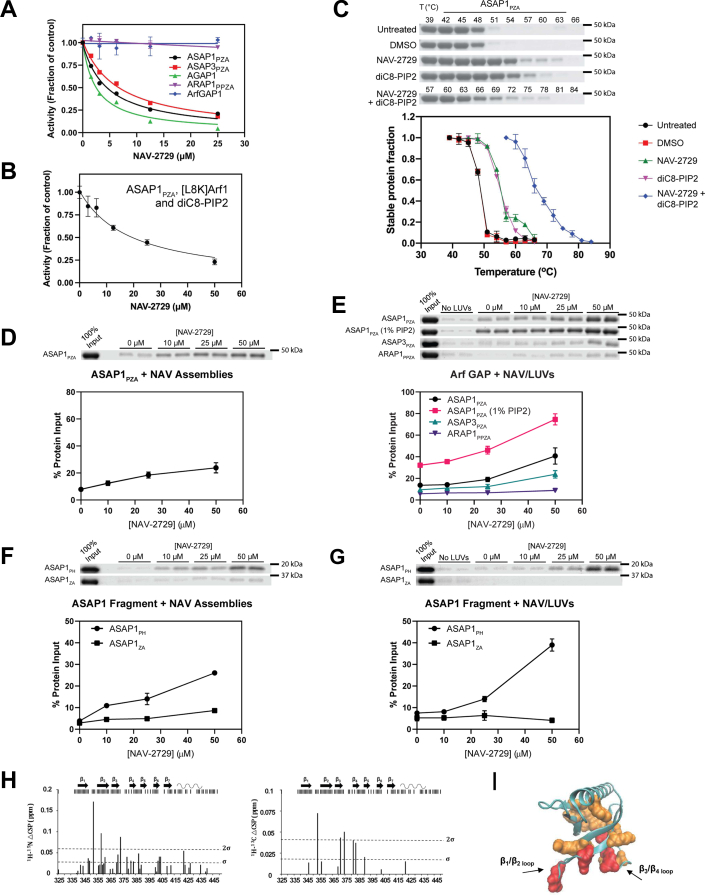
NAV-2729 binds to and inhibits specific Arf GAPs.A, effect on GAP activity. NAV-2729 was titrated into reactions containing the indicated Arf GAPs, myrArf1•GTP, and LUVs as described in Experimental procedures. A representative experiment of 4 with ASAP1 and ASAP3, and 5 with AGAP1, are shown. B, NAV-2729 inhibits ASAP1 in the absence of a membrane. GAP activity was determined using ASAP1 as the GAP, [L8K]Arf1•GTP as the substrate and diC8-PIP2 as the activating ligand. Results are representative of six experiments. C, ASAP1 binds to NAV-2729 and diC8-PIP2. Thermal shift assays were conducted as described in Figure 4A. Representative gels and the summary of six experiments are shown. D, ASAP1 binds NAV-2729 supramolecular assemblies. ASAP1 was tested for binding as described in Figure 4B. Representative gel and summary of four experiments are shown. E, ASAP1 and ASAP3, but not ARAP1, bind to LUVs containing NAV-2729. These assays were the same as described in Figure 4C. In addition to the 0% PIP2 condition, ASAP1 was tested with LUVs containing 1% PIP2. Summaries of ≥3 experiments are shown. F, the PH domain, but not Arf GAP or ankyrin repeats (ZA construct) of ASAP1, binds to NAV-2729 supramolecular assemblies. Binding was determined as described in Figure 4B. Summary of three experiments is shown. G, the PH domain, but not Arf GAP or ankyrin repeats (ZA construct) of ASAP1, binds to NAV-2729 in LUVs. These assays were the same as described in Figure 4C. Three experiments are summarized. H, interaction between NAV-2729 and ASAP1PH at the membrane interface. The top of each chart shows a summary of data collected. Black bars correspond to residues where data could be collected. Most of the missing residues come from exposed HN exchanging with the solvent at pH 7.4 (unstructured N-terminal stretch between residues 325 and 339 and loops). Charts show chemical shift perturbation differences (△CSP) between 1H-15N CS (Left) or 1H-13C CS (Right) for ASAP1 PH bound to nanodiscs with NAV-2729 and nanodiscs without NAV-2729. Nanodiscs with NAV-2729 contained four NAV-2729 molecules per nanodisc. I, △CSPs are mapped on the crystal structure of the ASAP1 PH domain (PDB 5C79) (43). Residues colored in red have △CSP larger than 2σ and residues colored in orange have △CSP larger than σ, where σ is the SD calculated over all △CSPs. Arf, ADP-ribosylation factor; CSP, chemical shift perturbation; GAP, GTPase-activating protein; LUV, large unilamellar vesicle; PH, pleckstrin homology; PIP2, phosphatidylinositol 4,5-bisphosphate.
We considered that NAV-2729 might disrupt the structure of LUVs such that the GAP could not bind; three results excluded this mechanism. First, 50 μM NAV-2729 did not prevent ASAP1PZA binding to sucrose-loaded PIP2-containing LUVs (Fig. S4B). Second, NAV-2729 inhibited ASAP1PZA when assayed in the absence of LUVs (Fig. 6B). Using [L8K]Arf1, which is stable in the GTP bound form while in solution (25, 26), and a soluble PIP2 analog, dioctanoyl PIP2 (diC8-PIP2) for measuring GAP activity (26), NAV-2729 inhibited ASAP1PZA activity, although less potently than with LUVs and myrArf1. (Fig. 6B and Table 1). The difference in potency could be due to differences in availability and orientation of NAV-2729 in supramolecular assemblies and in LUVs. ASAP1PZA activity using [L8K]Arf1 as a substrate and LUVs-containing PIP2 was inhibited with potency closer to that using myrArf1 on LUVs (Fig. S4C and Table 1). Third, NAV-2729 did not have a detectable effect on LUVs observed by EM (Fig. S3, D and E).
NAV-2729 binds specific Arf GAPs
NAV-2729 binding to ASAP1PZA was detected in the three assays we used for Arfs and BragSec7-PH. First, 50 μM NAV-2729 caused a >6 degree shift in thermal stability (Fig. 6C). NAV-2729 also stabilized ASAP1PZA at a fixed temperature (51 °C) with a half-maximum effect at ∼25 μM (Fig. S4D). We also included diC8-PIP2, which binds to ASAP1 and, therefore, would be predicted to increase thermal stability. At a concentration of 200 μM, diC8-PIP2 alone caused a ∼6 degree shift in the thermal stability of ASAP1PZA (Fig. 6C). When mixed together, diC8-PIP2 and NAV-2729 induced a shift of greater than 15 degrees, indicating that ASAP1PZA might bind PIP2 and NAV-2729 simultaneously (Fig. 6C). Second, ASAP1PZA cosedimented with NAV-2729 supramolecular assemblies (Fig. 6D). Third, ASAP1PZA bound to LUVs containing NAV-2729 (Fig. 6E). The effect was additive with PIP2 in the LUVs (Fig. 6E). At saturating concentrations of PIP2 with maximum binding of ASAP1PZA to the LUVs observed, NAV-2729 had no further effect (Fig. S4B). ASAP3PZA, which is also inhibited by NAV-2729, bound to LUVs containing NAV-2729 while ARAP1PPZA, which is not inhibited by NAV-2729, did not (Fig. 6, A and E). Thus, NAV-2729 binding to GAPs correlated with NAV-2729–dependent inhibition of GAP activity.
NAV-2729 binds to the ASAP1 PH domain
The GEFs and GAPs that depend on a PIP2-binding PH domain for activity (namely Brag2, ARNO, ASAP1, ASAP3, and AGAP1) (26, 35, 36, 38, 40, 41) were inhibited by NAV-2729, while the Arf GAP without a PH domain (ArfGAP1) and one where the critical PH domain is atypical (ARAP1) (37) were not inhibited (Figs. 1B, 5A and 6A). We considered the possibility that NAV-2729 binds to PH domains. The isolated PH domain of ASAP1 (ASAP1PH) bound to NAV-2729 supramolecular assemblies and to LUVs containing NAV-2729 while the ASAP1 Arf GAP and ankyrin repeat tandem (ASAP1ZA) did not (Fig. 6, F and G). NAV-2729 also affected the PIP2 and diC8-PIP2 dependence of ASAP1PZA activity (Fig. S4, E and F).
NMR was used to further define the binding site of NAV-2729 on ASAP1. Since NAV-2729 partitions into lipid bilayers, we used nanoscale lipid domains (nanodiscs) as a well-established membrane mimetic to facilitate solution-state NMR spectroscopy of membrane surface complexes (39, 42). Interaction of NAV-2729 with membrane bound U2H-15N, ILV 13C methyl labeled-ASAP1PZA, and -myrArf1•GTP was probed by following changes in chemical shifts when proteins were bound to NAV-2729-free or NAV-2729-containing nanodiscs. Note that all nanodiscs, with or without NAV-2729, contained PIP2. While chemical shift perturbations for myrArf1•GTP were minimal, binding of ASAP1PZA to nanodiscs containing NAV-2729 induced statistically significant 1H-13C chemical shift perturbations. Those perturbations could be assigned to resonances belonging to residues of the PH domain of ASAP1PZA and not the ZA portion. However, superimposition of resonances from the PH domain and ZA portion of ASAP1PZA prevented us from delineating a precise location for the interaction (Data not shown). To go further, we therefore repeated the experiment using the PH domain of ASAP1 only (ASAP1PH) and monitored 1H-13C and 1H-15N chemical shift perturbations upon addition of NAV-2729 to the lipid bilayer. Binding of ASAP1PH to NAV-2729–containing nanodiscs induced large perturbations for residues in the β1/β2 loop (residues G352, I353, Q358) and β3/β4 loop (residues I371, H373, A374), which are involved in the binding of PI(4,5)P2 headgroups at the membrane (Fig. 6H). Mapping the changes onto the PH domain structure suggested that NAV-2729 and the phosphoinositide bind to a similar location (Fig. 6I) (43, 44).
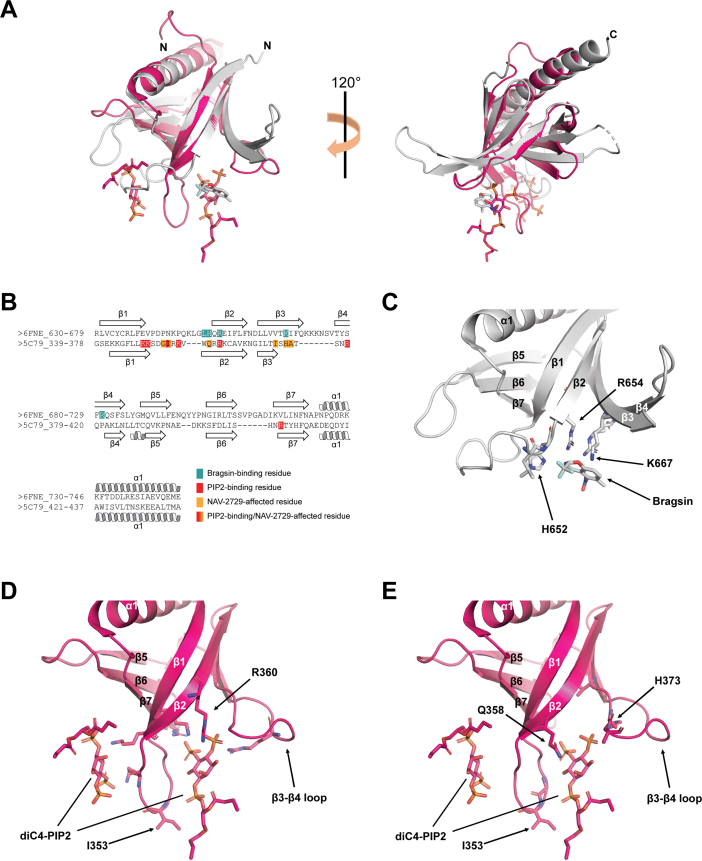
The Brag2 inhibitor Bragsin requires a membrane to inhibit Arf GEF activity and the structure was previously solved in complex with a Brag2Sec7-PH. Bragsin bound between the PH domain of Brag2 and the lipid bilayer (15). Because NAV-2729 bound the PH domain of ASAP1, we compared the crystal structure of Brag2Sec7-PH:Bragsin (15) with that of ASAP1PH:diC4-PIP2 (43) (Fig. 7). We chose to use the structure of ASAP1PH in complex with diC4-PIP2 for our analyses, as our data indicated that ASAP1PH might bind to both PIP2 and NAV-2729 simultaneously (Fig. 6, C, E and H). In superimposed structures of Brag2PH:Bragsin and ASAP1PH:diC4-PIP2, Bragsin and one of the diC4-PIP2 monomers bind to Brag2PH and ASAP1PH in the same region of each PH domain and overlap between the β2 and β3 strands (Fig. 7A). In primary sequence alignment, R654 of Brag2 and R360 of ASAP1 overlapped when comparing Brag2 residues that bind to Bragsin or ASAP1 residues that bind to PIP2 (Fig. 7B); indeed, examination of the crystal structures suggests that these residues bind to Bragsin/diC4-PIP2 in similar poses (Fig. 7, C and D). When comparing Brag2 residues that bind to Bragsin to ASAP1 residues that exhibit chemical shift perturbations upon binding to nanodiscs containing NAV-2729, we observed that Brag2 residues H652 and K667 overlapped with ASAP1 residues Q358 and H373 (Fig. 7, B, C and E). ASAP1 I353 was the sole residue involved in binding to PIP2 that is also affected by NAV-2729 (Figs. 6H and 7, B, D and E).
Figure 7.
Superposition and primary sequence alignment of Brag2PH:Bragsin and ASAP1PH:diC4-PIP2 structures.A, overall superposition of Brag2PH:Bragsin (gray) and ASAPPH:diC4-PIP2 (pink). Bragsin is shown as gray sticks and the two diC4-PIP2 molecules are shown as pink sticks. N- and C-termini are labeled. B, alignment of Brag2PH (6FNE_630-746) and ASAP1PH (5C79_339-437) residues based on structural superposition. Brag2PH residues that bind to Bragsin are colored teal. ASAP1PH residues that bind to PIP2 are shown in red, while those that exhibit chemical shift perturbations upon binding to nanodiscs containing NAV-2729 are shown in orange. The single ASAP1PH residue that binds to both PIP2 and exhibits chemical shift perturbations upon binding NAV-2729 nanodiscs (I353) is shown in a red/orange gradient. Secondary structure elements are labeled. C, Brag2PH residues that bind to Bragsin. R654 is labeled to indicate its positioning relative to R360 in (D). H652 and K667 are labeled to indicate their positioning relative to Q358 and H373 in (E). Secondary structure elements are labeled. D, ASAP1PH residues that bind to diC4-PIP2. R360 is labeled to indicate its positioning relative to R654 in (C). I353 is labeled as it is the sole residue that binds diC4-PIP2 and exhibited chemical shift perturbations upon binding nanodiscs containing NAV-2729. Secondary structure elements are labeled. E, ASAP1PH residues that exhibit chemical shift perturbations upon binding to nanodiscs containing NAV-2729. Q358 and H373 are labeled to indicate their positioning relative to H652 and K667 in (C). All other features are labeled as described in (D). PH, pleckstrin homology; PIP2, phosphatidylinositol 4,5-bisphosphate.
ASAP1 is not the primary target of NAV-2729
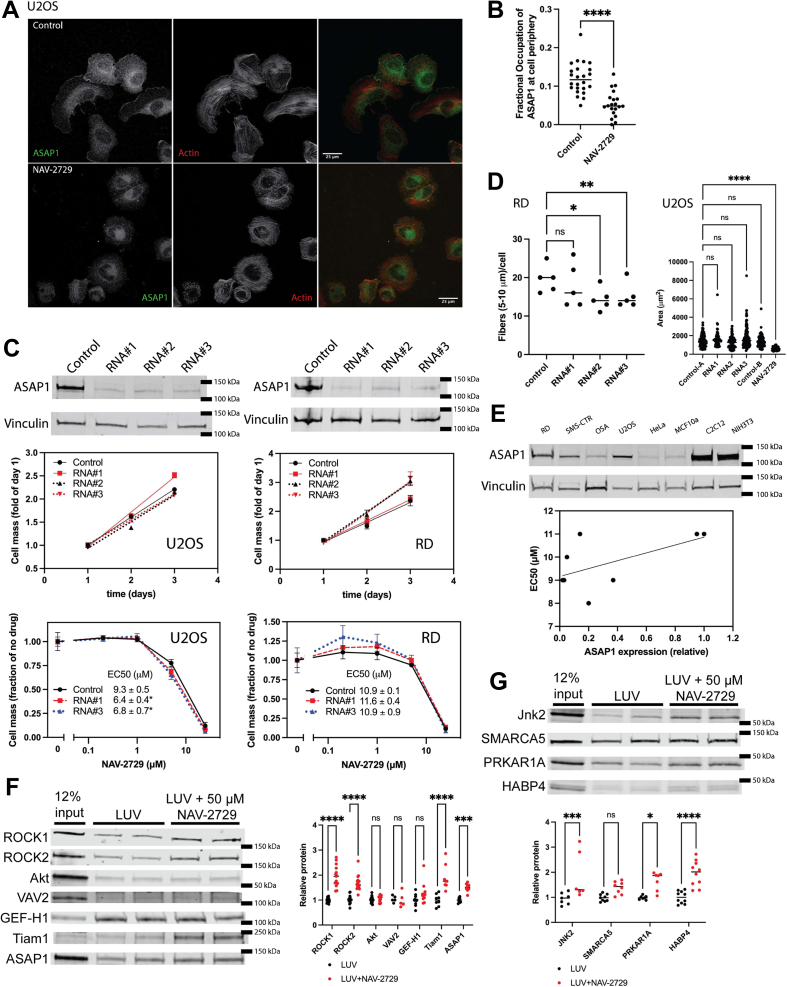
Three lines of evidence indicated that ASAP1 might be one target of NAV-2729. First, NAV-2729 affected the cellular distribution of ASAP1 in U2OS cells. In untreated U2OS cells, some ASAP1 signal localized to the cell edge in membrane ruffles; NAV-2729 treatment reduced the ASAP1 signal at the edge of the cell (Fig. 8, A and B). Second, reducing ASAP1 expression increased NAV-2729 sensitivity of U2OS cells (Fig. 8C). Third, the effect of NAV-2729 on the actin cytoskeleton partially overlapped that of reduced ASAP1 expression with a reduction in actin stress fibers in RD cells (Figs. 2, D and E and 8D) and in U2OS cells (45). However, there are also several lines of evidence that ASAP1 cannot be the sole or primary target. ASAP1 expression in the cell lines tested did not correlate with NAV-2729 EC50 for inhibition of proliferation (r2 = 0.35, p = 0.12) (Figs. 1C and 8 E and Table 1). ASAP1 knockdown had no effect on cell proliferation in either cell line tested nor on NAV-2729 sensitivity in RD cells, and the effect on U2OS was minimal (Fig. 8C). Additionally, different than NAV-2729, ASAP1 knockdown did not cause a reduction in U2OS cell surface area (Fig. 8D).
Figure 8.
ASAP1 is a target of NAV-2729 but NAV-2729 may bind to additional proteins.A, NAV-2729 reduces ASAP1 in ruffles. U2OS cells were stained with Alexa568-phalloidin, as described in Figure 2D, and immunostained for ASAP1. Scale bars are 25 μms in length. B, fractional occupancy of ASAP1 in the cells shown in (A). The intensity of the ASAP1 signal at the cell edge was compared to total ASAP1. C, reduced expression of ASAP1 did not affect proliferation in RD or U2OS cells. Three diRNA targeting ASAP1 were used to knockdown ASAP1 for proliferation assays, and two were used to test NAV-2729 sensitivity. Relative cells mass was determined as in Figure 1C. Figures summarize two experiments. D, quantification of the number of actin fibers in RD cells and the cell surface area in U2OS cells plated and immunostained as described in Figure 2D. E, ASAP1 expression and NAV-2729 sensitivity are not correlated. Relative expression of ASAP1 was plotted against the EC50 for inhibition of cell proliferation. (Fig, 1C and Table 1). F, candidate approach identifies ROCK1, ROCK2, and Tiam1 as possible binding partners. Binding of proteins from lysates of RD cells to LUVs with and without NAV-2729 was determined as described in Experimental procedures. Each protein was measured in ≥3 experiments performed in duplicate. All data points were plotted. The data were analyzed by two-way ANOVA followed by Sidak’s multiple comparison test implemented in GraphPad Prism. G, validation of proteins identified in cellular thermal shift assay screen. Experiment performed as described in (F). Each protein was measured in ≥3 experiments performed in duplicate. A similar analysis was performed as described in (F). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. LUV, large unilamellar vesicle.
NAV-2729 binds to proteins outside of the Arf pathway
Given the promiscuity of NAV-2729 targeting PH domain–containing proteins within the Arf pathway, we considered that NAV-2729 might bind to proteins with PH domains outside of the Arf pathway. Because NAV-2729 disrupts the actin cytoskeleton, we examined Rho pathway proteins that have PH domains. Rho GTPases are important regulators of the actin cytoskeleton (46, 47, 48, 49, 50, 51, 52, 53). Lysates of RD cells were mixed with LUVs with or without NAV-2729. Proteins that bound to the LUVs were analyzed by immunoblotting for ROCK1, ROCK2, Akt1/2, VAV2, GEF-H1, Tiam1, and ASAP1. Of the proteins examined, ROCK1, ROCK2, Tiam1, and ASAP1 were enriched in the NAV-2729–containing LUVs (Fig. 8F).
We also used a cellular thermal shift assay followed by mass spectrometry proteome analysis (54), to identify additional targets. In these experiments, lysates from RD cells were mixed with NAV-2729 (in the absence of LUVs) and subjected to a temperature ramp as described for the purified proteins. The treated lysates were cleared by centrifugation. The proteins remaining in solution were analyzed by mass spectrometry. Forty-five proteins were identified as having significant shifts in the temperature of denaturation (Table S1). For 20 proteins, the temperature was increased, consistent with NAV-2729 binding the proteins. For twenty-five, the temperature of apparent denaturation was decreased. The decrease might indicate the proteins were binding to the supramolecular assemblies of NAV-2729 and being removed by centrifugation meant to remove denatured protein. The 45 proteins considered to have a significant shift included Jnk2 and multiple transcription factors and other nucleic acid–binding proteins (see Table S1). Of the proteins with a negative shift, 19 bound either nucleotide or nucleic acid. We determined if a subset of the identified proteins from RD cell lysates cosedimented with LUVs containing NAV-2729. Jnk2, PRKAR1A, and HABP4 were enriched in the pellets when NAV-2729 was present (Fig. 8G).
In summary, NAV-2729 was found to inhibit the Arf GEFs Brag2 and ARNO and the Arf GAPs ASAP1, ASAP3, and AGAP1, which are active against Arf1 and Arf5, but not Arf6. Although Arfs interacted with NAV-2729 in aqueous solution where NAV-2729 formed supramolecular assemblies, binding was not detected to NAV-2729 in lipid bilayers, where NAV-2729 efficiently partitions. The proteins in the Arf pathway that were inhibited contain PH domains, and candidate proteins from the Rho pathway with PH domains bound NAV-2729 in lipid bilayers. Proteins without PH domains were identified in a cellular thermal shift assay screen. We conclude that NAV-2729 targets the Arf pathway by inhibiting PH domain–dependent Arf GEFs and GAPs, but cannot exclude additional cellular targets that mediate effects on cell viability and proliferation.
Discussion
With accumulating evidence for a role of the Arf pathway in cancer progression, a number of inhibitors have been developed (5). Recently, one promising small molecule was discovered, NAV-2729 (13). Initial studies indicated NAV-2729 functioned by specifically binding to Arf6 to prevent spontaneous and GEF-catalyzed nucleotide exchange, resulting in selective toxicity for cancers that are dependent on Arf6 (13). We tested NAV-2729 for cytotoxic activity against a number of nontransformed and transformed cells, including pediatric sarcomas that have amplification of genes in the Arf pathway. After finding that NAV-2729 blocked proliferation of all cell lines examined with similar potency, but that other Arf GEF inhibitors had little or no effect, we reexamined the mechanism of action of NAV-2729. We discovered that NAV-2729 (monodisperse in a membrane) did not bind full-length Arfs in our hands but targeted multiple regulators of Arf proteins, likely binding through their PH domains, and that the Arf pathway proteins are not the sole or primary targets of NAV-2729. Screens indicate that NAV-2729 might bind to numerous other proteins, including proteins in the Rho GTPase pathway. Goals of ongoing studies are the identification of other targets and determination of the mechanism of inhibition of Arf regulators.
Although we did not detect binding to full-length Arfs, Arf GEFs and GAPs were inhibited by NAV-2729. We considered that inhibition of the GEF Brag2 might mediate the cytotoxic effects as Bragsin inhibits Brag2 and has been reported to be active against cancer cell lines (15); however, several lines of evidence indicated Brag2 was not the target in the cells we examined. Because of the low potency of NAV-2729 against ARNO, ARNO was not investigated. We also considered that the Arf GAP ASAP1 might be the target of NAV-2729 mediating cytotoxic effects given it has been implicated in cancer progression. Several lines of evidence supported ASAP1 being one target of NAV-2729. NAV-2729 inhibited ASAP1 GAP activity, directly bound to ASAP1, and altered the cellular localization of ASAP1. In addition, NAV-2729 reduced the size and number of focal adhesions and the associated actin stress fibers similar to that observed with reduced ASAP1 expression (30, 45). However, reduction of ASAP1 expression did not affect proliferation (see next paragraph). ASAP1 might be just one of multiple targets. Also interesting is that NAV-2729 was found active against uveal melanoma, the first cancer in which amplification of the ASAP1 gene was found to correlate with metastasis (7). Plausibly, part of the cytotoxic effect in uveal melanoma cell lines was due to inhibition of ASAP1. Regardless, in the cell lines we examined, ASAP1 was not the sole or primary target of NAV-2729.
A cumulative effect due to inhibition of multiple targets, as suggested by Benabdi et al. (14) for the small molecules M-COPA and NAV-2729, might be relevant for the cells we examined. NAV-2729 inhibited three of the five Arf GAPs examined and the Arf GEFs Brag2 and ARNO. We only examined five of the 28 Arf GAPs and two of the 15 Arf GEFs. We consider it likely that NAV-2729 inhibits ASAP2 in addition to other AGAP family members. ACAPs, which have PH domains like those in the ASAPs, might also be targets. Forty-eight potential targets, any of which could either contribute to a cumulative effect or might be the primary target explaining the cytotoxic effects, were found by candidate approaches and cellular thermal shift assay screens. Identifying which are relevant is the subject of ongoing studies.
NAV-2729 is an example of a small molecule that may inhibit an enzyme by binding to PH domain. The mechanistic basis for NAV-2729 inhibition through binding PH domains remains to be defined. One mechanism for a PH-binding inhibitor has been described for Akt. The PH domain of Akt binds PIP3 to recruit the protein to plasma membranes where it is activated. Triciribine, identified in screens for inhibitors of proliferation of Akt2-transformed NIH 3T3 fibroblasts (55), is phosphorylated to form triciribine phosphate, which binds to the PIP3-binding site in the PH domain of Akt, displacing Akt from membranes and preventing activation (56). P-Rex, a Rho family exchange factor, is also inhibited by small molecules that bind to its PH domain. The mechanism of inhibition for P-Rex has not yet been examined, however it seems unlikely to be displacement from the membrane as the PH domain of P-Rex does not drive membrane recruitment. Similarly, displacement of ASAP1 from a membrane unlikely explains inhibition by NAV-2729. NAV-2729 partitioned into lipid bilayers and bound the PH domain while in the bilayers, and did not displace ASAP1PZA from LUVs containing PIP2. In addition, NMR revealed that NAV-2729 and PIP2 bound simultaneously. NAV-2729 may function similarly to the small molecule Bragsin, an interfacial inhibitor, that binds to the Brag2 PH domain while associated with a lipid bilayer (15). There were differences between Bragsin and NAV-2729. First, whereas Bragsin does not disrupt Brag2 interactions with liposomes containing PIP2 (15), NAV-2729 promoted ASAP1PZA binding to LUVs with or without PIP2. Second, Bragsin did not function as an inhibitor in the absence of a membrane (15), whereas NAV-2729 did. Third, Bragsin was a specific inhibitor of Brag2 when compared to a subset of other Arf GEFs (15), whereas NAV-2729 lacks specificity and inhibits multiple Arf GEFs and GAPs. In ongoing studies, we are further defining the structural determinants of NAV-2729 for binding to PH domains as well as the molecular mechanism for NAV-2729 inhibition of ASAP1. These studies might provide general insights into mechanisms by which PH domains regulate adjacent catalytic domains, the structural features required of PH domain inhibitors, and consequently, the possibility of predicting which PH domains might be NAV-2729 targets and identifying small molecules that might more specifically bind to PH domain–containing proteins.
Our results highlight challenges in identifying cellular targets for small molecules: e.g., the physical chemistry of the small molecule of interest might be complex. NAV-2729 formed supramolecular assemblies in solution unless a membrane was present, in which case it partitioned into the lipid bilayer. Proteins might interact differently with the supramolecular assemblies than with drug monodisperse in a lipid bilayer. Furthermore, integration into membranes might bias binding to transmembrane and peripheral membrane proteins. Consequently, assays of proteins in aqueous solution might not be suitable for identifying and studying targets. Thermal shift assays, used to screen for binding partners, might be confounded by the chemistry of the small molecule in a couple ways. First, when using intact cells, if the drug partitions into membranes, proteins that bind might be removed in the first step of preparing samples. Second, in the case of drugs like NAV-2729, when adding the drug to cell lysates, proteins that bind to supramolecular assemblies might precipitate with the denatured proteins, which would be analyzed as a reduction in thermal stability (i.e., less soluble protein present at a given temperature). Of the 45 proteins we detected as having a significant shift in thermal stability, 25 had a reduction. Other assays could similarly be affected by compound chemistry. NAV-2729 supramolecular assemblies introduce light scatter, which could confound assays based on optical methods, e.g., fluorescence. Light absorption in the UV and VIS regions might also confound some optical-based assays. Finally, in aqueous solution, the concentration of the active form of NAV-2729 would be difficult to determine: one would expect an equilibrium of monodisperse NAV-2729 and supramolecular assemblies. These complexities are likely to be relevant to other small molecules.
Our work may have led to a different conclusion from that of previous studies for a number of reasons. First, we have used native proteins whereas previous work was restricted to using truncated forms of the GTPases. Although it is perfectly reasonable to use the truncated proteins and indeed many important advances have been made using them as a model, in this particular instance there appears to be some differences from native proteins that could have led to different conclusions. Second, membrane mimetics could be utilized with the assays we used. Previous work did not use membrane mimetics, although Benabdi et al. (14) did attempt to use them for some assays and concluded that the kinetics were too complex to warrant further investigation. Again, it is perfectly reasonable to avoid membrane mimetics, as they can confound assays. For example, they are not compatible with the nucleotide analog used for exchange assays by Yoo et al. (13), as they would increase background to mask signal. However, in this instance, the membrane mimetics were important for the solubility of NAV-2729. Third, some of the properties of NAV-2729, such as light scatter and absorbance, may have confounded the fluorescence-based assays used by Yoo et al. (13). Finally, we considered the possibility that NAV-2729 might interact with proteins other than GTPases. Previous work restricted tests of specificity to GTPases or only a subset of Arf GEFs (13, 14). Given that knocking down Arf6 reduced proliferation of the cell lines being examined (13), there was no compelling reason to look beyond Arf6. Our findings with multiple cell lines motivated the reexamination of specificity. Although we cannot exclude that some effects of NAV-2729 are mediated by inhibiting Arf6 as described in the first paper about NAV-2729 (13), the possibility that other Arf pathways are affected was raised by Benabdi et al. (14), in which NAV-2729 was found to inhibit Arf1 exchange catalyzed by Brag2 and not to affect spontaneous exchange on either Arf1 or Arf6. Our expanded analysis indicates that the Arfs themselves might not be direct targets, but rather that their regulators are and, further, there are likely other targets of NAV-2729.
Identification of NAV-2729 is a step towards establishing whether Arf pathways are therapeutic targets in cancer. To extend the work reported here, (i) we will determine the molecular basis for inhibition of ASAP1 by NAV-2729 and (ii) identify other binding proteins that might be more important for the cellular effects of NAV-2729, which might provide insights into therapeutic targeting of cancer.
Experimental procedures
Chemicals
NAV-2729 was purchased from Tocris Bioscience and Sigma-Aldrich. SecinH3 and Brefeldin A were from Sigma-Aldrich. Bragsin2 was from MedChem Express. Bragsin was a kind gift from Mahel Zeghouf and Jacqueline Cherfils. Alexa568-phalloidin was from Thermo Fisher. MemGlow560 was from Cytoskeleton. Cell Titer Glo was from Promega. TMT10plex Isobaric Label Reagent Set was from Thermo Fisher. [α32P]GTP and [35S]GTPγS were from PerkinElmer.
Antibodies
A monoclonal mouse anti-ASAP1 antibody was purchased from Abnova. A polyclonal rabbit anti-ASAP1 serum was raised in our lab as previously described (31). Anti-paxillin and anti-GM130 antibodies were purchased from BD Biosciences, anti-ROCK2 antibody from BD Transduction Laboratories, anti-HABP4 antibody from Bioss USA, anti-Akt, anti-Arf6, anti-GEF-H1, anti-JNK2, anti-PRKAR1A (PKA RI-α/β), anti-ROCK1, anti-SMARCA5 (SNF2H), anti-TIAM1, and anti-VAV2 from Cell Signaling Technology, anti-β-COP antibody from Invitrogen, IRDye680 anti-mouse IgG and IRDye800 anti-rabbit IgG were from LI-COR, and anti-Brag2 (IqSec1) antibody was from Sigma Precision Antibodies.
Cell culture
RD, SMS-CTR, C2C12, HeLa, and NIH 3T3 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum and penicillin and streptomycin. U2OS cells were cultured in McCoy’s 5A media with 10% fetal calf serum and penicillin and streptomycin. OSA cells were cultured in RPMI with 10% fetal calf serum and penicillin and streptomycin. MC10a cells were cultured in DMEM/F12 media with 10% fetal calf serum.
Knockdown with dicer substrate RNA duplex
Dicer substrate RNA (diRNA) duplexes were obtained from IDT (sold as Trifecta Kits). 300,000 cells/well in 6-well plates (35 mm diameter wells) were transfected with 10 nM nontargeting (control) diRNA or diRNA targeting Arf6, Brag2, or ASAP1 using DharmaFECT 1 transfection reagent (Dharmacon). The cells were harvested 3 days later for use in proliferation, drug sensitivity, or immunofluorescence experiments.
Immunofluorescence
Twenty thousand cells were incubated on fibronectin-coated 12 mm glass coverslips in McCoys 5A media without serum (U2OS) or DMEM with 10% fetal calf serum (RD cells) for 2 to 3 h prior to fixing with 4% paraformaldehyde and either immunostaining for paxillin and incubating with Alexa568-phalloidin to visualize filamentous actin or immunostaining for GM130 and β-COP. Confocal microscopy was performed on a Leica TCS SP8 confocal laser scanning microscope using system-optimized z-stack parameters with pinhole of 1.00 Airy Unit, an HC PL APO CS 63×/1.4 oil objective at resolution 512 × 512 pixels. Adhesions were analyzed as previously described (32, 57). Actin fibers were analyzed using the Ridge detector plug in ImageJ (https://imagej.nih.gov/ij/download.html) as described (45), counting fibers between 5 and 10 μm in length. Golgi quantification from z-stack images were done using Fiji software (https://imagej.nih.gov/ij/download.html) (58). Images of individual cells were cropped out from the original z-stacks, subjected to intensity thresholding using IsoData algorithm, converted to a mask, and applied back to the images to determine the area and volume (taking in step-size of z-stack) of GM130 signal patches.
Proliferation assays
2500 to 5000 cells per well were seeded in white bottom 96-well cell culture plates. Relative cell mass 3 days after plating was determined using the CellTiter-Glo assay (Promega).
Immunoblotting
Proteins were fractionated by SDS-PAGE and transferred onto nitrocellulose membranes. The nitrocellulose membranes were incubated in LI-COR proprietary TBS blocking buffer for 1 h and subsequently incubated overnight at 4 °C with primary antibodies diluted in the LI-COR proprietary antibody diluent buffer. After three washes in TBS with 0.2% Tween-20, the membranes were incubated for 1 h with IRDye-labeled (LI-COR) secondary antibodies at room temperature, washed 3 times in TBS with 0.2% Tween-20, and visualized using a LI-COR Odyssey scanner.
Preparation of proteins
Myristoylated Arf (59), truncated Arf proteins, [L8K]Arf1 (60), the Sec7 and PH domains (Sec7-PH construct) of Brag2 and ARNO (40), the PH, Arf GAP, and ankyrin repeats (PZA construct) of ASAP1 and ASAP3 (35, 61), full-length ASAP1 (26), full-length ArfGAP1 and AGAP1 (62), the tandem PH domains, Arf GAP, and ankyrin repeats of ARAP1 (63), the PH domain of ASAP1 (36), and the Arf GAP and ankyrin repeats (ZA construct) of ASAP1 (36) were expressed and purified as described. MSP△H5 was expressed and purified as previously described (64).
Preparation of labeled myrArf1, ASAP1 PZA, and ASAP1 PH domain
Labeling was performed as previously described (65). Briefly, for the production of [U-2H], [U-15N]-methyl specifically labeled protein, NH4Cl is substituted by ammonium chloride (15N ≥99%), D-glucose is replaced by D-Glucose-1,2,3,4,5,6,6-d7 (2H ≥97%), and 13CH3-methyl specifically labeled precursors are added as described below. For a cell culture of 500 ml, a few freshly transformed colonies of BL-21(DE3) cells were picked to inoculate 5 ml of M9/H2O minimal media for overnight growth at 37 °C in a shaking incubator (250 rpm). One milliliter of the overnight culture (typical optical density at 600 nm [A600] 1–1.2) was then used to inoculate 4 ml of fresh M9/H2O medium to achieve a starting A600 of 0.25. At A600 ∼0.5, 5 ml of M9/D2O minimal media was added and cell growth continued until an A600 of ∼0.5 is reached. Cells were diluted again by a factor of 2 and growth followed to A600 ∼0.5. This cycle was repeated until a D2O/H2O ratio of 3:1 (20 ml total) is reached. Cells were then harvested by centrifugation (3000g for 30 min) and resuspended in 25 ml of M9/D2O, and growth was continued in a 100 ml baffled flask until an A600 of 0.5 is reached, before an additional 25 ml of M9/D2O was added for overnight growth at 37 °C. When the overnight A600 was between 1.3 and 1.5, the overnight cell suspension (50 ml) was added to 500 ml of M9/D2O and growth followed at 37 °C, up to A600 ∼0.6. For selective I-[13CH3]δ1, L-[13CH3]proS, V-[13CH3]proS methyl labeling of ASAP1 PZA and ASAP PH, the TLAM-Iδ1LVproS kit was used (NMR-Bio). For labeling of ASAP1 PH, it was supplemented by 3-[13C]-2-[2H]-L-Alanine for selective methyl labeling of A-[13CH3]β. After the addition of the precursor according to the manufacturer’s protocol, cell growth continued until an A600 of approximately 0.7 at 25 °C is reached, at which time protein expression was induced with the addition of 1 mM IPTG. Note that for Arf1, instead of kits, two 13C-labeled keto acid precursors for Ile, Leu/Val (Sigma-Aldrich) were added to the cell culture 30 min before induction. Cultures were then grown overnight at 25 °C after induction.
Preparation of nanodiscs
All lipids in chloroform solutions were air-dried with nitrogen flow and resolubilized with cholate in aqueous buffer (20 mM Tris–HCl pH 7.4, 150 mM NaCl, and 75 mM sodium cholate). Nanodiscs were assembled by mixing MSP△H5 with solubilized lipids at a ratio of 1:50, followed by the removal of sodium cholate from the mixture with Bio-Beads SM2 resin (Bio-Rad), under overnight rocking at 22 °C. Assembled nanodiscs were then purified via a Superdex-200 size-exclusion column (GE Healthcare) and concentrated on a centrifugal concentrator (10 kDa molecular weight cutoff, Thermo Fisher Scientific). The concentration of nanodiscs was determined by UV spectroscopy (ε280 = 18,450 M−1 cm−1).
Preparation of LUVs
LUVs were prepared by extrusion. Briefly, 1 μmol lipids (molar ratio, 40% PC, 25% PE, 15% PS, 10% cholesterol, and 10% total phosphoinositide) dissolved in chloroform, in a siliconized glass tube were dried under a nitrogen stream for 30 min to 1 h, followed by lyophilization for at least 1 h. The dried lipids were resuspended in 200 μl of either PBS (for GEF and GAP assays) or in a sucrose solution (20 mM Hepes pH 7.4, 20 mM KCl, 0.2 M sucrose, 0.01% sodium azide) for cosedimentation assays, for a final concentration of 5 mM. The solution was vortexed, subjected to five rounds of freeze/thaw, and extruded using a lipid extruder (Avanti Polar Lipids) through a Whatman Nuclepore Track-Etched membrane with 1 μM pores. The LUVs were stored at 4 °C and were used within a week for activity assays and within 3 days for LUV cosedimentation assays.
GGA pull-down assay
The binding of myrArf1•GTP with GGA3 was performed as previously described (21). MyrArf1•GDP was incubated in 20 mM Hepes, pH 7.4, 100 mM NaCl, 1 mM DTT, 0.5 mM MgCl2, 1 mM EDTA, and 100 μM GTP. Purified GST or GST-VHSGAT-GGA3 at 1 μM was mixed with 0.5 μM of myrArf1•GTP, 1 mM DTT, 5% DMSO, or 50 μM NAV-2729 in 50 μl and incubated at room temperature for 5 min. Fifteen microliters of 50% glutathione sepharose 4B slurry (Cytiva) in PBS were added. The tubes were incubated with gentle agitation at room temperature for 30 min, chilled on ice, and spun at 2300 rpm for 1 min. The supernatants were removed, and the glutathione beads were washed two times with 100 μl of ice-cold PBS. The proteins bound to the glutathione beads were eluted with Laemmli SDS-PAGE sample buffer. The samples were heated to 95 °C for 5 min and separated by SDS-PAGE.
Thermal shift assays
The thermal stability assay was performed as described (45) with minor modifications. Purified proteins (10 μM in 25 μl volume) with 0.5% DMSO or 50 μM NAV-2729/0.5% DMSO were distributed into thin-walled PCR tubes in duplicates. In the case of ASAP1 PZA, the conditions also included with or without 200 μM diC8-PIP2. The tubes were heated in a thermocycler at a temperature gradient with 3 °C increments, with a 3 min hold at each temperature. Samples were cooled to room temperature and centrifuged to sediment precipitated protein. Fifteen microliters of supernatant were transferred into microcentrifuge tubes, mixed with 5 μl of 4× sample buffer, and separated on 4 to 20% SDS-PAGE gel. Densitometric quantification was performed using ImageJ (NIH).
GEF activity assays
GEF activity assays were performed as described previously (40). Reaction mixtures contained 25 mM Hepes, pH 7.4, 100 mM NaCl, 1 mM DTT, 2 mM MgCl2, 1 mM EDTA, 5 μM GTPγS, and [35S]GTPγS (specific activity of 10,000 cpm/pmol), 0.5 mM LUVs, 0.5 μM Arf•GDP ,and either 1 nM of Brag2 or 30 nM of ARNO. The reactions were incubated at 30 °C for 3 min and terminated with 2 ml of ice-cold 20 mM Tris, pH 8.0, 100 mM NaCl, 10 mM MgCl2, and 1 mM DTT. Protein-bound nucleotide was trapped on nitrocellulose, and the bound radioactivity was quantified by liquid scintillation counting.
GAP activity assays
GAP-induced conversion of myrArf1•GTP to myrArf1•GDP (or [L8K]Arf1•GTP to [L8K]Arf1•GDP) was determined as described previously (26, 43). The concentration of GAPs required to hydrolyze 50 to 70% of the Arf-bound GTP in 3 min was included in the reaction. The LUVs were included in the myrArf1 GTP-loading reaction.
NAV-2729 supramolecular assembly binding assay
5, 12.5, and 25 mM NAV-2729 in DMSO were prepared as 500× stocks. The stocks (or pure DMSO) were diluted 1:450 by addition of 1× PBS supplemented with 0.334% DMSO. Forty five microliters were added to polypropylene ultracentrifuge tubes (8 × 34 mm, Beckman Coulter). Five microliters of 10 μM proteins of interest in PBS were added in duplicate. The mixture was incubated at 10 min at RT and then centrifuged in a pre-chilled S100-AT3 rotor (Thermo Fisher Scientific) for 15 min at 75,000 rpm. The supernatant was removed, and the pellet was resuspended in 20 μl of 2× sample buffer supplemented with β-mercaptoethanol. A standard curve was included on each gel. Densitometry analysis was performed using ImageJ (NIH).
LUV cosedimentation-binding assay
5, 12.5, and 25 mM NAV-2729 in DMSO or pure DMSO was diluted 1:400 by addition of 1× PBS supplemented with 0.376% DMSO. Forty microliters of the stocks were mixed with 5 μl LUV (5 mM stock in buffer with sucrose) in polypropylene ultracentrifuge tubes (8 × 34 mm, Beckman Coulter) and incubated at room temperature for 5 min. Five microliters of 10 μM protein of interest in PBS were added and the mixture incubated for 10 min at RT. Control reactions lacking LUVs were run to ensure that the proteins were not aggregating. The mixtures were centrifuged in a prechilled S100-AT3 rotor (Thermo Fisher Scientific) for 15 min at 75,000 rpm, the supernatant was removed, and the pellet was resuspended in 20 μl of 2× sample buffer. Analysis was performed as described for the supramolecular assembly binding.
A similar protocol was used for identifying binding proteins in cell lysates. RD cells cultured in 15 cm plates were harvested with trypsin/EDTA. Cells were collected by centrifugation, washed with PBS, and resuspended at 1 to 2 × 107 cells/ml in PBS with a protease inhibitor cocktail (Cytoskeleton). The cells were rapidly frozen and thawed 5 times, then passed through a 27 gauge needle three times. The lysate was clarified by centrifugation at 14,000g for 20 min at 4 °C followed by a centrifugation at 250,000g for 15 min at 4 °C. Either DMSO or NAV-2729 in DMSO was added to LUVs for a final DMSO concentration of 0.2% and a final NAV-2729 concentration of 250 μM. Hundred microliters of LUVs with or without NAV-2729 were added to 400 μl of cell lysate. The mixture was incubated for 30 min at room temperature, chilled to 4 °C, and centrifuged at 250,000g for 15 min at 4 °C. Supernatants were aspirated. The pellet was resuspended in 140 μl of Laemmli sample buffer. Proteins were fractionated on a 4 to 20% gradient polyacrylamide gel and transferred to nitrocellulose for blotting with the indicated antibodies.
NMR measurements and analysis
NMR experiments were collected at 25 °C or 40 °C on Bruker Avance III 800 or 850 MHz spectrometers that were equipped with TCI triple resonance cryoprobes. All spectra were processed and analyzed using TopSpin 3.6 and NMRpipe (66). 1H -15N TROSY HSQC (67) and 1H-13C HMQC (68) experiments were carried out with selective 1H excitation pulses to improve water suppression (69). The experiments were typically carried out with 2-s recycle delay, 128 scans, 256 complex points in the 15N (or 13C) dimension (36 [20] parts per million [ppm]), and 2048 complex points in the 1H dimension (14 ppm).
Chemical shift perturbations were calculated using:
where are changes in the observed 1H or heteroatom chemical shift, respectively. A is a scale factor equal to 0.17 (0.185) when X is 15N (13C).
Nanodiscs were composed of 14:0-14:0-PC and 18:1-18:1-PI(4,5)P2 lipids (95:5 mol: mol). DMSO fraction in buffer was 0.4 % v:v. 1H-13C and 1H-15N chemical shift perturbations were measured at 25 °C and 40 °C. ASAP1 PH and nanodisc concentrations were 10 and 50 μM to ensure full binding. The higher temperature used to record amide resonances was necessary to reduce the rotational correlation time of the ∼120 kDa complex (39, 44), while concentration of proteins and nanodiscs were chosen to minimize DMSO concentration while maximizing NAV-2729 to protein ratio.
Mass spectrometry
Thermal stability assay was performed with 80 μg protein of RD cell lysate with either 0.5% DMSO or 50 μM NAV-2729/0.5% DMSO in 90 μl. The tubes were heated to 45, 50, 55, 60, and 65 °C, with a 3 min hold. The samples were centrifuged, and the supernatants were snap frozen and later analyzed by mass spectrometry. Before sample processing, protein concentrations in thermally treated cell lysates were measured. Proteins were then captured in S-Trap micro spin columns (PROTIFI) and digested with trypsin following the manufacturer’s protocol. Briefly, 50 ml of each cell lysate containing ∼100 mg (37 °C) or lower amounts (higher temperatures) was denatured with 5% SDS, reduced with 20 mM tris(2-carboxylethyl)phosphine, and alkylated with 20 mM iodoacetamide. The proteins were then loaded on the S-trap and digested overnight with trypsin. Peptides were then extracted by separate washes with 50 mM TEAB, 0.2% formic acid, and 50% acetonitrile and lyophilized. Dried peptides were labeled with one each of the 10-plex tandem mass tag (TMT) reagents (Thermo Fisher Scientific) for quantitation; reactions were performed using the manufacturer’s protocol. TMT-labeled lyophilized pooled peptides were dissolved in 0.1% TFA and fractionated using high pH reversed-phase peptide fractionation columns (Thermo Fisher Scientific) by batch elution method using the manufacturer’s protocol. The eluted fractions containing the less-complex peptide mix were quickly frozen and lyophilized before identification by LC-MS.
Fractionated peptides were separated on an UltiMate 3000 RSLCnano HPLC coupled to an Exploris 480 Orbitrap mass spectrometer (both Thermo Fisher Scientific). Samples were loaded onto a PepMap 100, 2 cm × 75 μm trap column (Thermo Fisher Scientific) at a flow rate of 3 μl/min using 0.1% formic acid as mobile phase, which was switched in-line with the PepMap 100250 mm × 75 μm analytical column (Thermo Fisher Scientific) after 10 min. Peptides were eluted at a flow rate of 300 nl/min across a 120 min gradient from 4% B to 35% B (A: 0.1% formic acid in water, B: 0.1% formic acid in acetonitrile). The mass spectrometer was operated in a data-dependent mode, with parent full-scan m/z range 350-1800 and nominal resolution of 120,000. MS/MS spectra were acquired following HCD fragmentation using 30% normalized collision energy and resolution of 45,000. Proteome Discoverer 2.4 (Thermo Fisher Scientific) was used to search the data against human proteins from the UniProt database using SequestHT. The search was limited to tryptic peptides, with maximally two missed cleavages allowed. Cysteine carbamidomethylation and TMT modification were set as fixed modifications; methionine oxidation was set as a variable modification. The precursor mass tolerance was 10 ppm; the fragment mass tolerance was 0.02 Da. The Percolator node was used to score and rank peptide matches using a 1% false discovery rate. Quantitation of TMT reporter ions was performed using the Minora node in Proteome Discoverer and corrected for lot-specific isotope impurities. Data normalization, calculation of melting curves, and significance testing was performed using the TPP package (version 3.24.0) (70) in R, as described previously (71).
Comparison of Brag2PH:Bragsin and ASAP1PH:diC4-PIP2 structures
The crystal structures of human Brag2Sec7-PH in complex with Bragsin (PDB: 6FNE) (15) as well as murine ASAP1PH in complex with two diC4-PIP2 molecules (PDB: 5C79) (43) were visualized in PyMOL (72). Specifically, residues 628–757 of chain A and Bragsin in 6FNE were aligned with residues 334–436 of chain B and the two diC4-PIP2 monomers in 5C79 using the “super” command. Note that amino acid positions are from UniProt (73) entries Q6DN90-2 and Q9QWY8 for Brag2 and ASAP1. Due to low sequence similarity between Brag2 and ASAP1, the primary sequences of each protein were aligned based on structural superposition using the Pairwise Structure Alignment analysis tool hosted by the PDB (https://www.rcsb.org/alignment) (74). Chain A of each structure (6FNE and 5C79) were selected for the pairwise structure alignment using the jFATCAT method (75, 76). The resulting alignment of residues 630–746 of Brag2 and residues 339–437 of ASAP1 was manually examined, and residues P710 and G711 (which are missing in 6FNE chain A) were reintroduced. Residues of Brag2 that bind to Bragsin and residues of ASAP1 that bind to PIP2 are based on previous studies (15, 43). Residues of ASAP1 that are affected by binding to nanodiscs containing NAV-2729 are from this work (see Fig. 6H).
Statistics
Most results were confirmed in n ≥2 independent experiments with replicates as indicated in the figure legends. The EM and cellular thermal shift assay experiments were performed once. Details of statistical analyses are indicated in the figure legends and relevant Experimental procedures sections.
Data availability
All data are available in the main text or the supporting information.
Supporting information
This article contains supporting information (77, 78).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Erin Gladu for analyzing EM images. We thank Mahel Zeghouf and Jacqueline Cherfils for the generous contribution of Bragsin. We thank Kayla Rosenberg for wisdom in graphical presentation.
Author contributions
E. M. R., X. J., O. S., R. A. B., and P. A. R. conceptualization; E. M. R., X. J., O. S., P.-W. C., L. M. J., and P. A. R. methodology; E. M. R., X. J., O. S., H.-Y. Y., M. P. Y., I. A., L. M. J., M. E. Y., and P. A. R. investigation; E. M. R., X. J., O. S., S. H., T. K. M., and P. A. R. visualization; R. A. B. and P. A. R. funding acquisition; P. A. R. project administration; R. A. B. and P. A. R. supervision; E. M. R., O. S., and P. A. R. writing–original draft; E. M. R., X. J., O. S., H.-Y. Y., S. P., I. A., L. M. J., M. E. Y., R. A. B., and P. A. R. writing–review and editing.
Funding and additional information
This work was supported by National Institutes of Health intramural program: National Institutes of Health project # ZIA BC007365 (P. A. R.), National Institutes of Health project # ZIA BC 011419 (R. A. B.), and National Institutes of Health project # ZIA BC 011132 (R. A. B.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Donita Brady
Footnotes
Present address for Mukesh P. Yadav: University of Maryland School of Medicine, Department of Diagnostic Radiology; Baltimore, MD, USA.
We were not able to use the thermal shift assay that detects protein unfolding as an increase in fluorescence from SYPRO Orange (24) because NAV-2729 interfered with the signal (Data not shown).
Supporting information
References
- 1.Kahn R.A., Cherfils J., Elias M., Lovering R.C., Munro S., Schurmann A. Nomenclature for the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. J. Cell Biol. 2006;172:645–650. doi: 10.1083/jcb.200512057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sztul E., Chen P.W., Casanova J.E., Cherfils J., Dacks J.B., Lambright D.G., et al. ARF GTPases and their GEFs and GAPs: concepts and challenges. Mol. Biol. Cell. 2019;30:1249–1271. doi: 10.1091/mbc.E18-12-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.East M.P., Kahn R.A. Models for the functions of arf GAPs. Semin. Cell Dev. Biol. 2011;22:3–9. doi: 10.1016/j.semcdb.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'Souza-Schorey C., Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 5.Casalou C., Ferreira A., Barral D.C. The role of ARF family proteins and their regulators and effectors in cancer progression: a therapeutic perspective. Front. Cell Dev. Biol. 2020;8:217. doi: 10.3389/fcell.2020.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen P.W., Gasilina A., Yadav M.P., Randazzo P.A. Control of cell signaling by Arf GTPases and their regulators: focus on links to cancer and other GTPase families. Biochim. Biophys. Acta Mol. Cell Res. 2022;1869:119171. doi: 10.1016/j.bbamcr.2021.119171. [DOI] [PubMed] [Google Scholar]
- 7.Ehlers J.P., Worley L., Onken M.D., Harbour J.W. DDEF1 is located in an amplified region of chromosome 8q and is overexpressed in uveal melanoma. Clin. Cancer Res. 2005;11:3609–3613. doi: 10.1158/1078-0432.CCR-04-1941. [DOI] [PubMed] [Google Scholar]
- 8.Lin D., Watahiki A., Bayani J., Zhang F., Liu L., Ling V., et al. ASAP1, a gene at 8q24, is associated with prostate cancer metastasis. Cancer Res. 2008;68:4352–4359. doi: 10.1158/0008-5472.CAN-07-5237. [DOI] [PubMed] [Google Scholar]
- 9.Müller T., Stein U., Poletti A., Garzia L., Rothley M., Plaumann D., et al. ASAP1 promotes tumor cell motility and invasiveness, stimulates metastasis formation in vivo, and correlates with poor survival in colorectal cancer patients. Oncogene. 2010;29:2393–2403. doi: 10.1038/onc.2010.6. [DOI] [PubMed] [Google Scholar]
- 10.Li M., Tian L., Yao H., Lu J., Ge J., Guo Y., et al. ASAP1 mediates the invasive phenotype of human laryngeal squamous cell carcinoma to affect survival prognosis. Oncol. Rep. 2014;31:2676–2682. doi: 10.3892/or.2014.3150. [DOI] [PubMed] [Google Scholar]
- 11.Morishige M., Hashimoto S., Ogawa E., Toda Y., Kotani H., Hirose M., et al. GEP100 links epidermal growth factor receptor signalling to Arf6 activation to induce breast cancer invasion. Nat. Cell Biol. 2008;10:85–92. doi: 10.1038/ncb1672. [DOI] [PubMed] [Google Scholar]
- 12.Nacke M., Sandilands E., Nikolatou K., Román-Fernández Á., Mason S., Patel R., et al. An ARF GTPase module promoting invasion and metastasis through regulating phosphoinositide metabolism. Nat. Commun. 2021;12:1623. doi: 10.1038/s41467-021-21847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoo J.H., Shi D.S., Grossmann A.H., Sorensen L.K., Tong Z., Mleynek T.M., et al. ARF6 is an actionable node that orchestrates oncogenic GNAQ signaling in uveal melanoma. Cancer Cell. 2016;29:889–904. doi: 10.1016/j.ccell.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benabdi S., Peurois F., Nawrotek A., Chikireddy J., Cañeque T., Yamori T., et al. Family-wide analysis of the inhibition of arf guanine nucleotide exchange factors with small molecules: evidence of unique inhibitory profiles. Biochemistry. 2017;56:5125–5133. doi: 10.1021/acs.biochem.7b00706. [DOI] [PubMed] [Google Scholar]
- 15.Nawrotek A., Benabdi S., Niyomchon S., Kryszke M.H., Ginestier C., Cañeque T., et al. PH-domain-binding inhibitors of nucleotide exchange factor BRAG2 disrupt Arf GTPase signaling. Nat. Chem. Biol. 2019;15:358–366. doi: 10.1038/s41589-019-0228-3. [DOI] [PubMed] [Google Scholar]
- 16.Hafner M., Schmitz A., Grüne I., Srivatsan S.G., Paul B., Kolanus W., et al. Inhibition of cytohesins by SecinH3 leads to hepatic insulin resistance. Nature. 2006;444:941–944. doi: 10.1038/nature05415. [DOI] [PubMed] [Google Scholar]
- 17.Niu T.K., Pfeifer A.C., Lippincott-Schwartz J., Jackson C.L. Dynamics of GBF1, a brefeldin A-sensitive Arf1 exchange factor at the Golgi. Mol. Biol. Cell. 2005;16:1213–1222. doi: 10.1091/mbc.E04-07-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pacheco-Rodriguez G., Moss J., Vaughan M. BIG1 and BIG2: Brefeldin A-inhibited guanine nucleotide-exchange proteins for ADP-ribosylation factors. Methods Enzymol. 2002;345:397–404. doi: 10.1016/s0076-6879(02)45032-x. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura N., Rabouille C., Watson R., Nilsson T., Hui N., Slusarewicz P., et al. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffiths G., Pepperkok R., Locker J.K., Kreis T.E. Immunocytochemical localization of beta-COP to the ER-Golgi boundary and the TGN. J. Cell Sci. 1995;108:2839–2856. doi: 10.1242/jcs.108.8.2839. [DOI] [PubMed] [Google Scholar]
- 21.Yoon H.Y., Bonifacino J.S., Randazzo P.A. In vitro assays of Arf1 interaction with GGA proteins. Methods Enzymol. 2005;404:316–332. doi: 10.1016/S0076-6879(05)04028-0. [DOI] [PubMed] [Google Scholar]
- 22.Dunphy J.L., Moravec R., Ly K., Lasell T.K., Melancon P., Casanova J.E. The Arf6 GEF GEP100/BRAG2 regulates cell adhesion by controlling endocytosis of beta1 integrins. Curr. Biol. 2006;16:315–320. doi: 10.1016/j.cub.2005.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cimmperman P., Baranauskiene L., Jachimoviciūte S., Jachno J., Torresan J., Michailoviene V., et al. A quantitative model of thermal stabilization and destabilization of proteins by ligands. Biophys. J. 2008;95:3222–3231. doi: 10.1529/biophysj.108.134973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huynh K., Partch C.L. Analysis of protein stability and ligand interactions by thermal shift assay. Curr. Protoc. Protein Sci. 2015;79:28.9.1–28.29.14. doi: 10.1002/0471140864.ps2809s79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoon H.Y., Jacques K., Nealon B., Stauffer S., Premont R.T., Randazzo P.A. Differences between AGAP1, ASAP1 and arf GAP1 in substrate recognition: Interaction with the N-terminus of Arf1. Cell Signal. 2004;16:1033–1044. doi: 10.1016/j.cellsig.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 26.Roy N.S., Jian X., Soubias O., Zhai P., Hall J.R., Dagher J.N., et al. Interaction of the N terminus of ADP-ribosylation factor with the PH domain of the GTPase-activating protein ASAP1 requires phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 2019;294:17354–17370. doi: 10.1074/jbc.RA119.009269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matulis D., Kranz J.K., Salemme F.R., Todd M.J. Thermodynamic stability of carbonic anhydrase: measurements of binding affinity and stoichiometry using ThermoFluor. Biochemistry. 2005;44:5258–5266. doi: 10.1021/bi048135v. [DOI] [PubMed] [Google Scholar]
- 28.Turner C.E., West K.A., Brown M.C. Paxillin-ARF GAP signaling and the cytoskeleton. Curr. Opin. Cell Biol. 2001;13:593–599. doi: 10.1016/s0955-0674(00)00256-8. [DOI] [PubMed] [Google Scholar]
- 29.Randazzo P.A., Inoue H., Bharti S. Arf GAPs as regulators of the actin cytoskeleton. Biol. Cell. 2007;99:583–600. doi: 10.1042/bc20070034. [DOI] [PubMed] [Google Scholar]
- 30.Vitali T., Girald-Berlingeri S., Randazzo P.A., Chen P.W. Arf GAPs: a family of proteins with disparate functions that converge on a common structure, the integrin adhesion complex. Small GTPases. 2019;10:280–288. doi: 10.1080/21541248.2017.1299271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Randazzo P.A., Andrade J., Miura K., Brown M.T., Long Y.Q., Stauffer S., et al. The Arf GTPase-activating protein ASAP1 regulates the actin cytoskeleton. Proc. Natl. Acad. Sci. U. S. A. 2000;97:4011–4016. doi: 10.1073/pnas.070552297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen P.W., Jian X., Heissler S.M., Le K., Luo R., Jenkins L.M., et al. The arf GTPase-activating protein, ASAP1, binds nonmuscle Myosin 2A to control remodeling of the actomyosin network. J. Biol. Chem. 2016;291:7517–7526. doi: 10.1074/jbc.M115.701292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gasilina A., Vitali T., Luo R., Jian X., Randazzo P.A. The ArfGAP ASAP1 controls actin stress fiber organization via its N-BAR domain. iScience. 2019;22:166–180. doi: 10.1016/j.isci.2019.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoon H.Y., Miura K., Cuthbert E.J., Davis K.K., Ahvazi B., Casanova J.E., et al. ARAP2 effects on the actin cytoskeleton are dependent on Arf6-specific GTPase-activating-protein activity and binding to RhoA-GTP. J. Cell Sci. 2006;119:4650–4666. doi: 10.1242/jcs.03237. [DOI] [PubMed] [Google Scholar]
- 35.Ha V.L., Bharti S., Inoue H., Vass W.C., Campa F., Nie Z., et al. ASAP3 is a focal adhesion-associated Arf GAP that functions in cell migration and invasion. J. Biol. Chem. 2008;283:14915–14926. doi: 10.1074/jbc.M709717200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kam J.L., Miura K., Jackson T.R., Gruschus J., Roller P., Stauffer S., et al. Phosphoinositide-dependent activation of the ADP-ribosylation factor GTPase-activating protein ASAP1 - evidence for the pleckstrin homology domain functioning as an allosteric site. J. Biol. Chem. 2000;275:9653–9663. doi: 10.1074/jbc.275.13.9653. [DOI] [PubMed] [Google Scholar]
- 37.Miura K., Jacques K.M., Stauffer S., Kubosaki A., Zhu K., Hirsch D.S., et al. ARAP1: a point of convergence for Arf and rho signaling. Mol. Cell. 2002;9:109–119. doi: 10.1016/s1097-2765(02)00428-8. [DOI] [PubMed] [Google Scholar]
- 38.Nie Z., Stanley K.T., Stauffer S., Jacques K.M., Hirsch D.S., Takei J., et al. AGAP1, an endosome-associated, phosphoinositide-dependent ADP-ribosylation factor GTPase-activating protein that affects actin cytoskeleton. J. Biol. Chem. 2002;277:48965–48975. doi: 10.1074/jbc.M202969200. [DOI] [PubMed] [Google Scholar]
- 39.Li Y., Soubias O., Li J., Sun S., Randazzo P.A., Byrd R.A. Functional expression and characterization of human myristoylated-Arf1 in nanodisc membrane mimetics. Biochemistry. 2019;58:1423–1431. doi: 10.1021/acs.biochem.8b01323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jian X., Gruschus J.M., Sztul E., Randazzo P.A. The pleckstrin homology (PH) domain of the Arf exchange factor Brag2 is an allosteric binding site. J. Biol. Chem. 2012;287:24273–24283. doi: 10.1074/jbc.M112.368084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macia E., Paris S., Chabre M. Binding of the PH and polybasic C-terminal domains of ARNO to phosphoinositides and to acidic lipids. Biochemistry. 2000;39:5893–5901. doi: 10.1021/bi992795w. [DOI] [PubMed] [Google Scholar]
- 42.McLean M.A., Gregory M.C., Sligar S.G. Nanodiscs: a controlled bilayer surface for the study of membrane proteins. Annu. Rev. Biophys. 2018;47:107–124. doi: 10.1146/annurev-biophys-070816-033620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jian X., Tang W.K., Zhai P., Roy N.S., Luo R., Gruschus J.M., et al. Molecular basis for cooperative binding of Anionic phospholipids to the PH domain of the arf GAP ASAP1. Structure. 2015;23:1977–1988. doi: 10.1016/j.str.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soubias O., Pant S., Heinrich F., Zhang Y., Roy N.S., Li J., et al. Membrane surface recognition by the ASAP1 PH domain and consequences for interactions with the small GTPase Arf1. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abd1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gasilina A., Yoon H.Y., Jian X., Luo R., Randazzo P.A. A lysine-rich cluster in the N-BAR domain of ARF GTPase-activating protein ASAP1 is necessary for binding and bundling actin filaments. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2022.101700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Etienne-Manneville S., Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 47.Nobes C.D., Hall A. RHO, RAC, and CDC42 GTPASES regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 48.Ridley A.J. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006;16:522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 49.Spiering D., Hodgson L. Dynamics of the Rho-family small GTPases in actin regulation and motility. Cell Adh. Migr. 2011;5:170–180. doi: 10.4161/cam.5.2.14403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall A. The cytoskeleton and cancer. Cancer Metastasis Rev. 2009;28:5–14. doi: 10.1007/s10555-008-9166-3. [DOI] [PubMed] [Google Scholar]
- 51.Jaffe A.B., Hall A. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 52.Raftopoulou M., Hall A. Cell migration: rho GTPases lead the way. Dev. Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- 53.Ridley A.J., Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 54.Jafari R., Almqvist H., Axelsson H., Ignatushchenko M., Lundbäck T., Nordlund P., et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc. 2014;9:2100–2122. doi: 10.1038/nprot.2014.138. [DOI] [PubMed] [Google Scholar]
- 55.Yang L., Dan H.C., Sun M., Liu Q., Sun X.M., Feldman R.I., et al. Akt/protein kinase B signaling inhibitor-2, a selective small molecule inhibitor of Akt signaling with antitumor activity in cancer cells overexpressing Akt. Cancer Res. 2004;64:4394–4399. doi: 10.1158/0008-5472.CAN-04-0343. [DOI] [PubMed] [Google Scholar]
- 56.Berndt N., Yang H., Trinczek B., Betzi S., Zhang Z., Wu B., et al. The Akt activation inhibitor TCN-P inhibits Akt phosphorylation by binding to the PH domain of Akt and blocking its recruitment to the plasma membrane. Cell Death Differ. 2010;17:1795–1804. doi: 10.1038/cdd.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen P.W., Jian X., Luo R., Randazzo P.A. Approaches to studying arf GAPs in cells: in vitro assay with isolated focal adhesions. Curr. Protoc. Cel. Biol. 2012 doi: 10.1002/0471143030.cb1713s55. Chapter 17:17.13.1-17.13.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ha V.L., Thomas G.M., Stauffer S., Randazzo P.A. Preparation of myristoylated Arf1 and Arf6. Methods Enzymol. 2005;404:164–174. doi: 10.1016/S0076-6879(05)04016-4. [DOI] [PubMed] [Google Scholar]
- 60.Kahn R.A., Randazzo P., Serafini T., Weiss O., Rulka C., Clark J., et al. The amino terminus of ADP-ribosylation factor (ARF) is a critical determinant of ARF activities and is a potent and specific inhibitor of protein transport. J. Biol. Chem. 1992;267:13039–13046. [PubMed] [Google Scholar]
- 61.Brown M.T., Andrade J., Radhakrishna H., Donaldson J.G., Cooper J.A., Randazzo P.A. ASAP1, a phospholipid-dependent arf GTPase-activating protein that associates with and is phosphorylated by Src. Mol. Cell Biol. 1998;18:7038–7051. doi: 10.1128/mcb.18.12.7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Che M.M., Nie Z., Randazzo P.A. Assays and properties of the arf GAPs AGAP1, ASAP1, and Arf GAP1. Methods Enzymol. 2005;404:147–163. doi: 10.1016/S0076-6879(05)04015-2. [DOI] [PubMed] [Google Scholar]
- 63.Campa F., Yoon H.Y., Ha V.L., Szentpetery Z., Balla T., Randazzo P.A. A PH domain in the Arf GTPase-activating protein (GAP) ARAP1 binds phosphatidylinositol 3,4,5-trisphosphate and regulates Arf GAP activity independently of recruitment to the plasma membranes. J. Biol. Chem. 2009;284:28069–28083. doi: 10.1074/jbc.M109.028266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hagn F., Etzkorn M., Raschle T., Wagner G. Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J. Am. Chem. Soc. 2013;135:1919–1925. doi: 10.1021/ja310901f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li J., Byrd R.A. A simple protocol for the production of highly deuterated proteins for biophysical studies. J. Biol. Chem. 2022;298:102253. doi: 10.1016/j.jbc.2022.102253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 67.Pervushin K., Riek R., Wider G., Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc. Natl. Acad. Sci. U. S. A. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tugarinov V., Kay L.E. Quantitative NMR studies of high molecular weight proteins: Application to domain orientation and ligand binding in the 723 residue enzyme malate synthase G. J. Mol. Biol. 2003;327:1121–1133. doi: 10.1016/s0022-2836(03)00238-9. [DOI] [PubMed] [Google Scholar]
- 69.Schanda P., Brutscher B. Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of seconds. J. Am. Chem. Soc. 2005;127:8014–8015. doi: 10.1021/ja051306e. [DOI] [PubMed] [Google Scholar]
- 70.Childs D., Kurzawa N., Franken H., Doce C., Savitski M., Huber W. 2022. TPP: Analyze Thermal Proteome Profiling (TPP) Experiments. R Package Version 3.24.0. [Google Scholar]
- 71.Franken H., Mathieson T., Childs D., Sweetman G.M., Werner T., Tögel I., et al. Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat. Protoc. 2015;10:1567–1593. doi: 10.1038/nprot.2015.101. [DOI] [PubMed] [Google Scholar]
- 72.Schrodinger, LLC . 2015. The PyMOL Molecular Graphics System, Version 1.8. [Google Scholar]
- 73.Consortium T.U. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2020;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., et al. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ye Y., Godzik A. Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics. 2003;19:ii246–ii255. doi: 10.1093/bioinformatics/btg1086. [DOI] [PubMed] [Google Scholar]
- 76.Li Z., Jaroszewski L., Iyer M., Sedova M., Godzik A. Fatcat 2.0: towards a better understanding of the structural diversity of proteins. Nucleic Acids Res. 2020;48:W60–W64. doi: 10.1093/nar/gkaa443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lakowicz J.R. Principles of Fluorescence Spectroscopy. Springer; Berlin: 2006. [Google Scholar]
- 78.Senning E.N., Collins M.D., Stratiievska A., Ufret-Vincenty C.A., Gordon S.E. Regulation of TRPV1 ion channel by phosphoinositide (4,5)-bisphosphate: the role of membrane asymmetry. J. Biol. Chem. 2014;289:10999–11006. doi: 10.1074/jbc.M114.553180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supporting information.