Abstract
This review is an update on the transforming genes of human cytomegalovirus (HCMV) and human herpesvirus 6 (HHV-6). Both viruses have been implicated in the etiology of several human cancers. In particular, HCMV has been associated with cervical carcinoma and adenocarcinomas of the prostate and colon. In vitro transformation studies have established three HCMV morphologic transforming regions (mtr), i.e., mtrI, mtrII, and mtrIII. Of these, only mtrII (UL111A) is retained and expressed in both transformed and tumor-derived cells. The transforming and tumorigenic activities of the mtrII oncogene were localized to an open reading frame (ORF) encoding a 79-amino-acid (aa) protein. Furthermore, mtrII protein bound to the tumor suppressor protein p53 and inhibited its ability to transactivate a p53-responsive promoter. In additional studies, the HCMV immediate-early protein IE86 (IE2; UL122) was found to interact with cell cycle-regulatory proteins such as p53 and Rb. However, IE86 exhibited transforming activity in vitro only in cooperation with adenovirus E1A. HHV-6 is a T-cell-tropic virus associated with AIDS-related and other lymphoid malignancies. In vitro studies identified three transforming fragments, i.e., SalI-L, ZVB70, and ZVH14. Of these, only SalI-L (DR7) was retained in transformed and tumor-derived cells. The transforming and tumorigenic activities of SalI-L have been localized to a 357-aa ORF-1 protein. The ORF-1 protein was expressed in transformed cells and, like HCMV mtrII, bound to p53 and inhibited its ability to transactivate a p53-responsive promoter. HHV-6 has also been proposed to be a cofactor in AIDS because both HHV-6 and human immunodeficiency virus type 1 (HIV-1) have been demonstrated to coinfect human CD4+ T cells, causing accelerated cytopathic effects. Interestingly, like the transforming proteins of DNA tumor viruses such as simian virus 40 and adenovirus, ORF-1 was also a transactivator and specifically up-regulated the HIV-1 long terminal repeat when cotransfected into CD4+ T cells. Finally, based on the interactions of HCMV and HHV-6 transforming proteins with tumor suppressor proteins, a scheme is proposed for their role in oncogenesis.
Members of the herpesvirus family have been implicated in the etiology of several human cancers. These include the Epstein-Barr virus (EBV), a candidate etiological agent in nasopharyngeal carcinoma and African Burkitt’s lymphoma (198); herpes simplex virus type 2 (HSV-2), linked by serologic and molecular studies to cervical carcinoma (119, 120, 139); human cytomegalovirus (HCMV), associated with cervical carcinoma (71, 133, 172), adenocarcinomas of the prostate (15, 150) and colon (63, 73), and Kaposi’s sarcoma (KS) (16, 59, 60); human herpesvirus 6 (HHV-6), associated with lymphoproliferative disorders (1, 148); and Kaposi’s sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8 (HHV-8), associated with KS (54, 123, 125). The postulated involvement of these herpesviruses in the etiology of human cancer has led to the development of experimental systems in which transforming DNA fragments and genes of these viruses have been identified and characterized.
This review is an update of our current knowledge about the transforming genes of HCMV and HHV-6. The section on HCMV transformation contains an overview of HCMV (including the association of HCMV with human malignancies and transformation of mammalian cells in vitro) and descriptions of the morphological transforming regions (mtr) of HCMV, the mtrII oncogene (ORF 79; UL111A) of HCMV (including localization of mtrII activity to an open reading frame [ORF] encoding a 79-amino-acid [aa] protein and interaction of mtrII with the tumor suppressor protein, p53, i.e., binding and inhibition of p53-activated transcription), and the HCMV immediate-early (IE) genes (including interactions between IE proteins and p53 and interactions between IE proteins and retinoblastoma [Rb] protein).
The section on HHV-6 transformation contains an overview of HHV-6 and describes the identification of HHV-6 transforming fragments, the identification and characterization of the ORF-1 transforming gene (DR7) (including localization of ORF-1 within SalI-L, characteristics of the ORF-1 gene, association of ORF-1 with human malignancies, and the interaction between ORF-1 and p53 proteins), and the transactivation of the human immunodeficiency type 1 (HIV-1) long terminal repeat (LTR) promoter by ORF-1.
HUMAN CYTOMEGALOVIRUS
Overview
The human herpesviruses are grouped into three subfamilies: the Alphaherpesvirinae, which includes HSV-1, HSV-2, and varicella-zoster virus; the Betaherpesvirinae which includes HCMV, HHV-6, and HHV-7; and the Gammaherpesvirinae, which includes Epstein-Barr virus (EBV) and KSHV, also known as HHV-8 (145). Although the human herpesviruses generally cause asymptomatic infections in normal individuals, they can cause severe, life-threatening infections in immunocompromised individuals. Moreover, these virus infections can be reactivated following primary exposure.
Most primary HCMV infections are inapparent. When symptomatic, primary HCMV infections in children and adults can cause polyneuritis, myelitis, and heterophile-negative mononucleosis syndrome, as well as carditis and hepatitis (90). About 0.5 to 2.5% of all newborns are infected at birth (39, 163), with a majority of these infections being asymptomatic. As a result, HCMV is also the most common cause of viral birth defects in congenitally infected babies. Both deafness and mental retardation can result from congenital HCMV infections.
In immunocompromised individuals, HCMV can cause severe disseminated disease characterized by chorioretinitis, pneumonia, esophagitis, colitis, myelitis, meningitis, encephalitis, leukopenia, lymphocytosis, and hepatitis (145). In addition, HCMV can cause severe retinitis in AIDS patients, which can lead to blindness if not treated (78).
Reactivated HCMV infections can result following blood transfusions, pregnancy, solid-organ or bone marrow transplantation, immunosuppressive therapy, or other viral infections (137, 164). The virus infects leukocytes, endothelial cells, connective tissue cells, and epithelial cells (67, 128, 137) and is transmitted through milk, semen, urine, saliva, and cervical secretions (33, 41, 42, 97, 98, 136). HCMV infection is acquired via the transplacental, perinatal, and sexual routes and through blood transfusion and organ or bone marrow transplantation.
Association with human malignancies.
Because of the ubiquitous distribution of HCMV and the high seroconversion rates, an etiological association between HCMV infection and human cancer has been difficult to establish. However, evidence based on virologic, epidemiologic, and molecular studies which have demonstrated the presence of viral DNA or antigens in tumor tissues suggests its involvement in specific cancers.
While HCMV has been isolated from cervical cancer biopsy specimens and their derived cell cultures (72, 122), seroepidemiologic studies linking HCMV infection to cervical cancer have yielded conflicting results. Some investigators have found significantly higher levels of antibodies to HCMV in patients with cervical carcinoma than in controls (133, 172, 184), while other groups have found no correlation (53, 62, 94, 162). Huang et al. (71) and Fletcher et al. (51) have detected HCMV DNA in cervical cancer specimens. However, DNAs of several other viruses, including HSV-2 and human papillomavirus (HPV), have also been detected in these tumors (120). It is possible that synergistic interactions among these viruses in the infected cell leads to the development of cervical cancer.
In other studies, a large percentage of prostatic cancer patients exhibited high antibody titers against HCMV (150). Lymphocytes from these patients were cytotoxic to both HCMV-infected and -transformed cells, indicating the presence of HCMV-specific membrane antigen (149). Furthermore, HCMV nuclear antigens and DNA have been detected in prostatic carcinoma cells (25), and one HCMV strain, Mj, has been isolated from a primary culture of human prostatic tissue (138).
HCMV persistently infects (15) and has been isolated from the gastrointestinal tracts of patients with regional enteritis and ulcerative colitis. It has also been isolated from cell cultures derived from adenocarcinomas of the colon (63). While Huang and Roche (73) have detected HCMV DNA in adenocarcinomas of the colon, no HCMV DNA sequences were found in tumor biopsy specimens from adenocarcinomas of the colon and rectum in other studies (22, 61).
HCMV infection has also been linked to KS by (i) high levels of HCMV antibodies in patients with KS (59), (ii) detection of HCMV-related nuclear antigens in KS biopsy specimens and KS-derived lines (16, 60), and (iii) demonstration of HCMV-specific DNA and RNA (16, 50, 60, 80) in some KS biopsy specimens. KSHV (HHV-8) has recently been identified (27, 125, 147) and has been detected in over 85% of the KS lesions studied (34, 40, 166). This virus is now considered by most investigators to be the etiologic agent of KS (54, 123, 125). Whether HCMV also has some involvement in the development of KS has yet to be established.
Transformation of mammalian cells in vitro.
While the association of HCMV with specific cancers cited above is suggestive of a possible etiologic role, the data are far from conclusive. The ubiquitous nature of HCMV makes serological studies hard to interpret. Furthermore, the large size of HCMV (230 kbp) makes the detection of small viral DNA sequences (<1,000 bp) associated with the initiation or progression of cancer very difficult when a whole genomic probe is used. To overcome this difficulty, researchers have worked to identify viral transforming genes that could be used to screen human cancer tissues.
The oncogenic potential of HCMV was originally demonstrated by the ability of both infectious and UV-inactivated virus to transform a variety of rodent and human cells in vitro. Albrecht and Rapp (3) first observed that UV-inactivated HCMV transformed hamster embryo fibroblasts. The transformed cells induced poorly differentiated malignant fibrosarcomas after subcutaneous injection into newborn and weanling golden Syrian hamsters. Although HCMV-specific antigens were demonstrated in the transformed and tumor-derived lines, HCMV DNA sequences were not detected. Similarly, Boldogh et al. (17) transformed hamster embryo fibroblasts with UV-irradiated HCMV and found that both the transformed and tumor-derived lines retained HCMV specific antigens.
The transformation of human embryo lung cells was observed after infection with HCMV strain Mj (55), BT1757 (72), or Towne (72). Although the expression of HCMV-specific antigens in the transformed and tumor-derived lines decreased with increasing passage, the cell lines exhibited enhanced tumorigenicity in nude mice (56). Human endothelial cells were transformed to anchorage-independent growth by infection with strains Towne and K9V (156). UV-inactivated HCMV strain Towne also transformed human endothelial cells but at a reduced frequency compared to infectious virus.
HCMV infection has also been reported to modulate a number of cellular properties often associated with the malignant phenotype (for a review, see reference 35). Transcriptional activation of the proto-oncogenes fos, jun, and myc has been observed after HCMV infection with both laboratory strains and clinical isolates (11–14). Because this activation occurred in the absence of viral protein synthesis, including expression of the IE viral regulatory proteins, it is likely that up-regulation of these genes was triggered by the binding of HCMV to its host cell receptor with subsequent activation of a signal transduction pathway by a biologically active virion protein. Jault et al. (83) reported that HCMV infection increased the levels of several cell cycle-regulatory proteins such as cyclins, p53, and phosphorylated Rb and caused cell cycle arrest at the G2/M boundary. Bresnahan et al. (20, 21) have reported that HCMV infection induced cyclin E expression and altered the subcellular localization of a cyclin E-associated kinase, Cdk2, in G0 cells, resulting in cell cycle progression into the G1 and S phases. Moreover, Zhu et al. (197) showed that HCMV infection of human cells blocked the induction of apoptosis (programmed cell death) and that this block was mediated by the viral IE regulatory genes.
In summary, both infectious and UV-inactivated HCMV were shown to transform a variety of mammalian cells in vitro, and these transformed cells were tumorigenic in nude mice. In addition, HCMV infection was shown to modulate the expression of various proteins involved in cell cycle regulation and apoptosis, providing a rationale for studying specific viral genes and their role in cellular transformation.
Morphological Transforming Regions
The transformation of rodent cells with intact or UV-irradiated HCMV DNA suggested the presence of one or more possible transforming genes. To identify these, researchers have tested HCMV restriction fragments for their ability to convert normal cells to a transformed phenotype to permit growth in soft agar and/or to induce tumors when inoculated into appropriate rodent model systems. Initially, Nelson et al. (129) identified the mtrI sequence within the approximately 5.0-kbp Xba-N fragment in strain AD169 (Fig. 1A). However, mtrI was not retained in the transformed cells, suggesting that its retention was not essential for the transformed phenotype. This led to the proposition of a “hit-and-run” mechanism for transformation. Subsequently, Clanton et al. (36) identified a 20-kbp Xba-E fragment in strain Towne (Fig. 1A) based on its hybridization to the BglII-C and BglII-N transforming regions of HSV-2 under nonstringent conditions (36) and observed that it immortalized primary diploid Syrian hamster embryo (SHE) cells and transformed established NIH 3T3 cells. Because BamHI digestion of Towne XbaI-E did not affect the transforming activity of the mixture of resulting fragments, El-Beik et al. (46) studied the BamHI and XbaI-BamHI subclones of Towne XbaI-E to localize the transforming region. Subclones containing either the 3.0-kbp XbaI-BamHI EM fragment (mtrII) or the 7.6-kbp EJ fragment (mtrIII) were capable of transforming NIH 3T3 and Rat-2 cells (Fig. 1A). Interestingly, upon further subcloning of the EJ fragment, Thompson et al. (180) showed that the 2.1-kbp SalI-XbaI subfragment containing the mtrIII region retained transforming activity whereas the 5.5-kbp BamHI-SalI fragment containing the IE72 (UL123) gene did not, thereby confirming the transforming potential of mtrIII and excluding any role that IE72 may play in HCMV transformation. Furthermore, all Rat-2 lines transfected with either mtrII or mtrIII were tumorigenic in immunocompetent 5-week-old Fisher rats (46). Cooperation between mtrII and mtrIII was demonstrated by Jariwalla et al. (81). Rat-2 cells transfected with mtrII plus mtrIII exhibited a sevenfold-greater transformation frequency than did cells transfected with either mtrII or mtrIII alone. The doubly transfected cell lines produced tumors in syngeneic rats at a much higher rate (5 to 7 days) than did those transformed by either mtr alone (25 to 35 days). Southern blot hybridization showed multiple and amplified mtrII sequences in mtrII-plus-mtrIII-induced lines, as was observed for mtrII-induced lines. Consistent with previous results (46), mtrIII sequences were not detected in either the single or double transformants. Retention of mtrII suggested that it was required for the maintenance of the transformed phenotype.
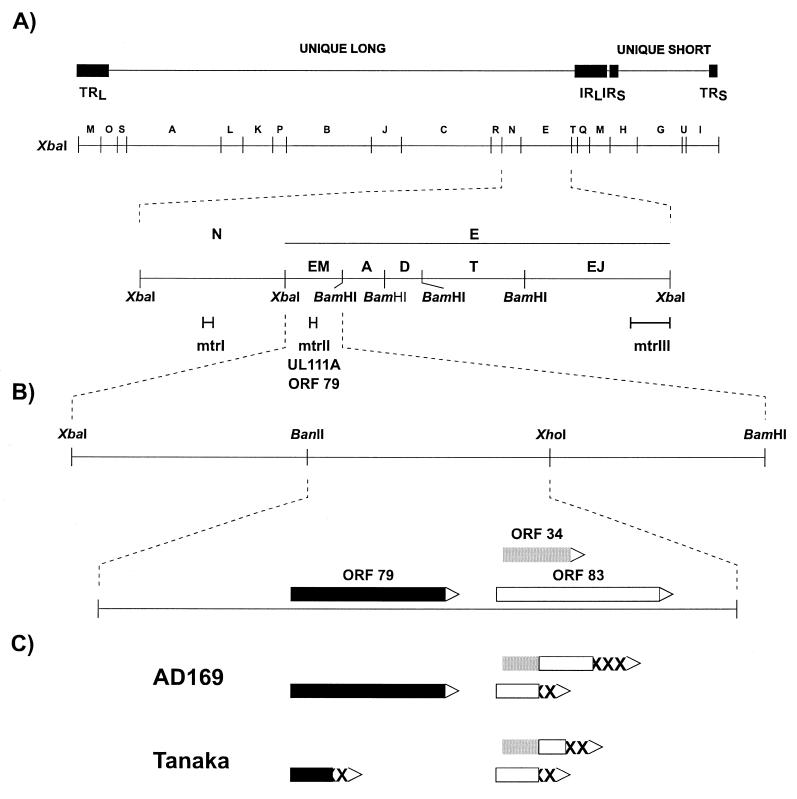
FIG. 1.
(A) Restriction map of HCMV strain Towne showing the location of the morphological transforming regions, mtrI, mtrII, and mtrIII. mtrI has been identified in strain AD169. mtrII, also designated UL111A, contains an ORF encoding the 79-aa protein. Adapted from reference 127 with permission of the publisher. (B) The XbaI-BamHI EM fragment of strain Towne showing the BanII-XhoI (mtrII 980) fragment with its encoded proteins. The solid open, and shaded rectangles represent the 79-, 83-, and 34-aa proteins, respectively. (C) ORFs of the mtrII 980 colinear regions of strains AD169 and Tanaka. The analogous amino acid sequences of strains AD169 and Tanaka are shown (the same rectangle symbols are used as those shown for the proteins of strain Towne). XXX, sequences not found in the proteins of strain Towne.
mtrII Oncogene
Localization of mtrII activity to ORF 79 (mtrII oncogene; UL111A).
Razzaque et al. (141) further localized mtrII to a 980-bp BanII-XhoI subfragment of the EM fragment which induced transformation in both NIH 3T3 and Rat-2 cells. The nucleotide sequence of this subfragment (mtrII 980) revealed three ORFs encoding proteins of 79, 83, and 34 aa (Fig. 1B). The sequences at the 5′ terminus of the ORFs contained regulatory elements that included CAAT boxes, Sp1 binding sites, and TACAAA and ATA transcriptional initiation signals, suggesting a promoter region capable of transcribing the above ORFs. In addition, six copies of the heptanucleotide sequence GGTG(A/G)TC which has similarity to the simian virus (SV40) enhancer core consensus sequence were observed. All of these were found in the first 300 nucleotides of mtrII upstream of the ORFs. The ORFs encoding the 79- and 34-aa products also contained motifs which have 55 to 78% homology to the Kozak consensus translational initiation sequence.
To determine if the mtrII region is transcriptionally active in HCMV-infected cells, expression of mtrII in HCMV infected human fibroblasts was studied by S1 nuclease analysis (141). A major 410-base transcript was expressed from the 3′ end of mtrII 980 and coded for the 34- and 83-aa proteins, and a minor 720-base mRNA also encoded the 79-aa protein. Both mRNAs were observed at 24 h postinfection but not at 14 h postinfection, suggesting they code for early HCMV gene products.
To analyze the transforming activity of the mtrII region, colinear regions in HCMV strains AD169 and Tanaka were examined. Jahan et al. (79) observed that the colinear mtrII region of HCMV strain AD169 exhibited similar transforming activity to mtrII from strain Towne. In contrast, mtrII from strain Tanaka showed a 75% reduction in transforming activity. Sequence analysis revealed frameshift differences in both Tanaka and AD169, resulting in the fusion of the N-terminal end of the 34-aa protein to the middle of the 83-aa protein and C-terminal amino acids not encoded by the ORFs of strain Towne (Fig. 1B). On the other hand, the 79-aa protein was intact in strain AD169 but truncated in strain Tanaka, suggesting that the reduced transforming activity of Tanaka mtrII resulted from the interruption of this 79-aa protein.
To ascertain whether transformation by mtrII resulted from promoter insertion or from expression of the 79-aa protein, Inamdar et al. (76) investigated the presence of functional promoters within mtrII 980. The identification of two early RNA transcripts expressed from Towne mtrII 980 suggested the presence of functional promoters which were identified by linking mtrII 980 subfragments to the chloramphenicol acetyltransferase (CAT) reporter gene. CAT activity was observed only in cells transfected with mtrII fragments cloned in front of the CAT gene in the sense orientation with reference to the ORFs. The existence of a functional promoter for the mtrII (the ORF encoding the 79-aa protein) was confirmed by the report of Razzaque et al. (142). Inamdar et al. (76) also observed CAT activity in experiments with similar constructs cloned with Tanaka mtrII subfragments in the sense orientation, showing that while the mtrII promoter was functional in this nontransforming colinear region, it was not sufficient to cause transformation. Moreover, the Towne promoter subfragments, in which the mtrII ORF was interrupted, exhibited no transforming activity. These data eliminated promoter insertion as the mechanism of mtrII transformation.
To determine if there was any homology between mtrII and other known proteins, a BLAST search (4) was performed with the mtrII amino acid sequence. No significant similarity to other known proteins in this database was revealed. However, a Kyte-Doolittle analysis (95) of mtrII (Fig. 2) predicted that the N-terminal region is highly hydrophobic, suggesting the presence of a membrane-anchoring domain.
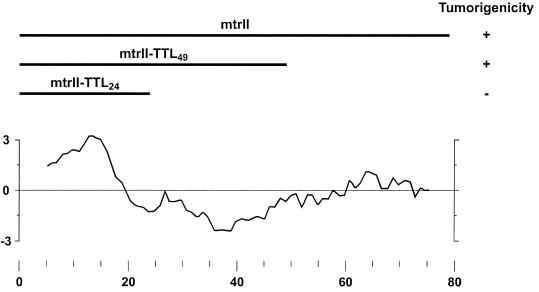
FIG. 2.
Hydropathy profile of mtrII. The hydropathy value was determined by the method of Kyte and Doolittle (95) with a window of 9 aa. Hydrophobic regions are indicated by positive values. Depicted above the hydropathy profile are wild-type and mutant peptides of mtrII and their tumorigenicity. Adapted from reference 179 with permission of the publisher.
Direct evidence for the role of ORF 79 in transformation was reported by Thompson et al. (179). They demonstrated the ability of a plasmid, pCHCmtrII, containing this ORF expressed from the HCMV IE promoter to transform NIH 3T3 cells. Permanent cell lines established after transfection with pCHCmtrII expressed mtrII mRNA and were tumorigenic after inoculation into immunodeficient athymic mice. Similar stable cell lines were developed after transfection with pCHCmtrII-TTL24 or pCHCmtrII-TTL49, which expressed mutant mtrII that terminated translation after aa 24 or 49, respectively. MtrII-TTL24 cells failed to produce tumors, suggesting that the hydrophilic N terminus alone was not sufficient for tumorigenesis. However, the mtrII-TTL49 cell line produced tumors, but with less efficiency than the wild-type mtrII cell line did (Fig. 2). The results demonstrated the transforming ability of ORF 79 (hereafter in this work referred to as mtrII) in the absence of the ORFs encoding the 34- and 83-aa proteins.
Binding of mtrII protein to p53 protein and inhibition of p53-activated transcription.
Understanding the mechanisms of mtrII transformation will help elucidate the role of HCMV in human malignancy. Studies of other DNA tumor virus oncogenes, like those encoding SV40 T-antigen, adenovirus E1A and E1B, and HPV-16 and -18 E6 and E7, have shown that their viral oncoproteins bind to the cellular tumor suppressor proteins, p53 and/or Rb (44, 151, 152, 191, 193). These interactions resulted in the inactivation of required checkpoints in the cell cycle, giving rise to uncontrolled cellular DNA replication and transformation.
The p53 protein is functionally pleiotropic and plays a role in G1 arrest, apoptosis, and DNA repair. In normal proliferating cells, p53 is expressed at low levels and has a short half-life of 6 to 20 min (132, 143, 144). In response to stress signals (such as hypoxia and DNA damage), p53 levels become elevated, predominantly due to its phosphorylation, which prevents its rapid degradation. Phosphorylation also activates p53, which then transcriptionally up-regulates a number of genes, such as mdm2, bax, GADD45, and p21(CIP1/WAF1), which contain p53 responsive elements (9, 29, 47, 124). The p21 protein (CIP1/WAF1), is an inhibitor of cyclin D- and E-dependent kinases. Inhibition of these kinases results in accumulation of hypophosphorylated Rb protein, which binds and hence inactivates the transcriptional activator E2F (130). Because E2F is crucial for the expression of genes involved in the G1- to S-phase transition, inactivation of E2F results in a G1 block. Hyperphosphorylation of Rb by cyclin D- and E-dependent kinases releases E2F, allowing the S phase to proceed (190). Thus, activation of p53 ultimately results in a G1 block (38). Viral regulatory proteins, such as SV40 T-antigen, that stabilize and inactivate p53 can relieve the G1 block by causing the release of E2F, which in turn cycles the cells into the S phase (151, 152).
To ascertain if expression of mtrII protein was essential for the transformed phenotype and to study the mechanisms of transformation, Muralidhar et al. (127) investigated the expression of mtrII protein in mtrII-transformed cells and its interaction with p53. To accomplish this, an anti-mtrII polyclonal antibody was produced by immunizing rabbits with the C-terminal 20-mer peptide of mtrII. The efficacy of anti-mtrII antibody was demonstrated by its ability to detect mtrII protein expressed in either bacteria or transiently transfected human T cells by Western blot analysis. Next, mtrII protein was detected in lysates of stably transformed NIH 3T3 cells established after transfection with mtrII cloned in the sense orientation in a selectable mammalian expression vector. mtrII protein was not detected in cells transfected with vector sequences or mtrII cloned in the antisense orientation. Expression of mtrII in transformed cells and HCMV-infected human embryonic lung fibroblasts was demonstrated by immunostaining, and the mtrII was localized predominantly to the perinuclear region in the transformed cells.
The interaction between mtrII and p53 was investigated by using in vitro-transcribed and -translated 35S-labeled mtrII and p53 proteins. mtrII was found to coimmunoprecipitate in the presence of p53 but not in its absence by using two monoclonal anti-p53 antibodies. By using p53 deletion mutant constructs in this assay, the mtrII binding domain was mapped to the N-terminal region of p53, residues 1 to 106, with a critical region from residues 1 to 44 (Fig. 3). This domain of p53 is required for (i) binding to mdm2, TATA-binding protein, and the adenovirus E1B 55-kDa protein; (ii) degradation of the HPV E6 protein; and (iii) transcriptional activation. Conversely, testing C-terminally truncated mtrII peptides and wild-type p53 protein showed that the p53-binding domain of mtrII protein was located in the first 49 aa. The interaction between mtrII and p53 was also confirmed in vivo by coimmunoprecipitation analysis of cell extracts.
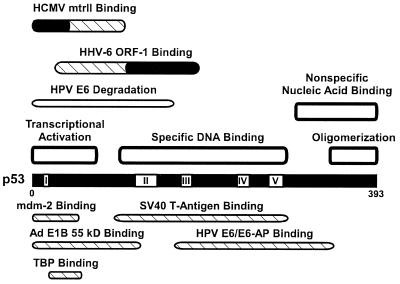
FIG. 3.
Map of the human wild-type p53 protein. Shown are the locations of the mtrII and ORF-1-binding domains, with the critical regions for binding indicated in black (86, 127). Also depicted are the DNA-binding domains (189), transcriptional activation and TATA-binding protein (TBP) binding domains (106), oligomerization domain (188), conserved regions (160), E6/E6-AP-binding and degradation domains (116), mdm-2- and adenovirus E1B (55-kDa)-binding domains (103), and SV40 T-antigen-binding domain (146). Adapted from reference 127 with permission of the publisher.
To study the functional consequences of the mtrII-p53 interaction, the effects of mtrII expression on p53-activated transcription were analyzed. mtrII inhibited p53-activated transcription both in transient-transfection assays and in stably transformed cells. When a CAT reporter construct driven by a promoter containing two p53-responsive elements (p53G5BCAT) was cotransfected with pCMV/p53, which expressed p53 protein, CAT activity increased over 70-fold. The addition of increasing amounts of the mtrII construct pCHCmtrII resulted in a dose-dependent decrease in CAT activity. Cotransfection with equal amounts of pCMV/p53 and pCHCmtrII decreased transactivation by 40%. The inhibition of p53-activated transcription reached approximately 85% with a 25-fold excess of pCHC/mtrII. No inhibition of basal p53G5BCAT transcription (transfection without pCMV/p53) by mtrII was found. Moreover, mtrII did not inhibit tax-activated transcription of the human T-cell virus promoter-directed CAT expression or tat-activated transcription of the HIV-1 promoter-directed CAT expression. This demonstrated the specificity of inhibition of p53-activated transcription by mtrII.
When similar experiments were performed with mtrII TTL mutants, only high levels of pCHC mtrII-TTL49 (20-fold in excess of pCMV/p53) inhibited p53-activated CAT activity whereas pCHC mtrII-TTL24 caused only a small reduction in activity. Moreover, full-length and TTL49-truncated mtrII proteins bound to p53 whereas TTL24 did not. This correlated with p53-activated transcription and transformation studies. Importantly, both wild-type and mtrII-TTL49 transformed rodent cells were tumorigenic in nude mice whereas mtrII-TTL24 cells were not (179). The concordance of these results suggests a causal relationship between mtrII binding to p53, mtrII inhibition of p53 function, and mtrII tumorigenic potential.
Viral oncoproteins such as SV40 T antigen not only bind to p53 and functionally inactivate it but also stabilize the level of p53 in transformed cells by making it inaccessible to the ubiquitin degradation pathway (151, 152). Interestingly, the level of p53 protein in stably transformed NIH 3T3 cells expressing mtrII was 10- to 20-fold greater than in parental cells or cells transfected with vector plasmid or a construct with mtrII cloned in the antisense orientation (127). This was the result of a 15-fold increase in the half-life of p53 in mtrII-transformed cells. In spite of the high steady-state levels of p53, the level of CAT activity in transformed cells was only 25 to 30% of that observed in control cells after transfection with p53G5BCAT. Therefore, despite the high steady-state levels, p53 was functionally inactive in mtrII-transformed cells. Elevated p53 levels were also observed after HCMV infection of human embryonic lung cells (126) and in HCMV-infected human smooth muscle cells (161). Considering the interaction between mtrII and p53, mtrII could contribute to the effects of HCMV infection on p53.
In summary, the mtrII gene has been identified as the HCMV oncogene. Not only are its sequences retained in transformed cells, but also the mtrII protein is expressed in transformed cells. Similar to the oncoproteins of the small DNA tumor viruses, mtrII oncoprotein binds to p53 in vitro and in vivo and inhibits p53-activated transcription in both transiently transfected and stably transformed cells. These observations characterize mtrII as a human herpesvirus oncogene. Further studies are needed to elucidate the complete mechanisms by which mtrII transforms cells. The binding of p53 by the mtrII oncoprotein may alter one or more of the functions of p53 in the cell. The wild-type p53 gene can exhibit both growth and transformation suppressor activities, which result in a G1 block in the cell cycle (93). The transcriptional transactivator function of p53 is required for growth arrest (38). While p53 transactivated WAF1/Cip1 (47), GADD45 (88), mdm2 (9, 29), and the human bax gene (124), it also repressed the basal transcriptional machinery and certain viral promoters (58, 154, 174). p53 is also involved in DNA replication and repair processes (43, 157) and mediates DNA damage-induced apoptosis (107). Thus, disruption of p53 function either by mutation or by binding to other proteins has been implicated in several human cancers (68, 114). The effects of mtrII expression and binding to p53 on all of the above pathways remain to be analyzed. In vitro transcription studies are needed to determine if mtrII inhibition of p53-activated transcription is direct or indirect. The effect of p53 inactivation on other cellular changes occurring during transformation, such as loss of contact inhibition of cell growth, remains to be determined. Finally, the presence and expression of the mtrII transforming gene in human tumors remain to be investigated. The identification of mtrII as a transforming gene and an understanding of its interaction with p53 will make this search possible.
Immediate-Early Genes
The HCMV IE gene locus encodes several proteins because of differential splicing of the primary transcript (Fig. 4) (168, 170, 171). Of these, the two major proteins, IE72 (IE1; UL123) and IE86 (IE2; UL122), are the best characterized. Although these IE proteins have not been demonstrated to be transforming in vitro, they have been shown to interact with the important cell cycle-regulatory proteins p53 and Rb. Shen et al. (155) have shown that IE72 and IE86 cooperated with the adenovirus E1A oncoprotein to transform baby rat kidney cells. In addition, the transformed cells exhibited mutations in cellular genes such as the p53 gene, suggesting the mutagenic potential of the IE proteins. However, expression of the IE proteins was transient, and neither IE DNA nor IE proteins were detected in the transformed cells. Based on these observations, the investigators proposed that the IE genes mediate a “hit-and-run” mechanism of transformation by inducing mutations in cellular genes. Zhu et al. (197) have shown that the IE proteins can inhibit the induction of apoptosis by either tumor necrosis factor alpha or the adenovirus E1A protein and could therefore presumably promote the replication and persistence of the virus.
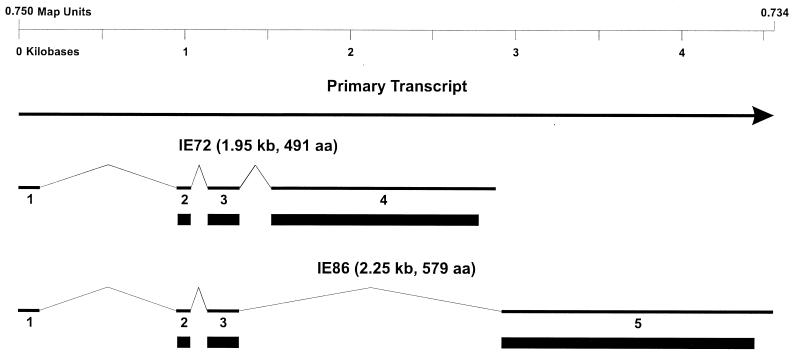
FIG. 4.
HCMV IE locus showing alternate splicing patterns used for the synthesis of the two major IE proteins, IE72 and IE86. The primary transcript and exons 1 to 5 are shown by the thick lines, and the introns are shown by thin lines. The solid rectangles indicate the protein-coding sequences.
Several studies have clearly demonstrated that the IE genes can function as viral transactivators. IE72 and IE86 proteins have the identical N-terminal 85 aa encoded by exons 2 and 3. The C-terminal 406 aa of IE72 is encoded by exon 4, while the C-terminal 494 aa of IE86 are encoded by exon 5. The IE proteins are the major regulators of viral early- and late-gene transcription. Moreover, IE86 is a strong transactivator of both viral and host genes (135, 195) while IE72 up-regulates the activity of its own promoter and cooperates synergistically with IE86 in its transactivation function (31, 115, 169). High concentrations of IE86 inhibit IE mRNA synthesis (66, 135), and this inhibition is mediated by a specific DNA sequence overlapping the RNA start site (30, 32, 96, 105, 113, 134). The transactivation of other promoters by IE86 has not been linked to the presence of specific enhancer DNA-binding sequences (158). Thus, IE proteins may mediate their effects through association with host transcription-regulatory proteins, as is the case for other viral transactivating genes such as HSV-1 and HSV-2 VP16 (104, 173) and adenovirus E1A (69, 100).
Interactions between IE proteins and p53.
Several studies have investigated the effect of HCMV infection and IE protein expression on the levels and functional status of p53. Muganda et al. (126) demonstrated that HCMV infection of human embryonic lung fibroblasts led to elevated levels of p53. In other studies, Speir et al. (161) proposed that the induction of p53 following HCMV infection played a role in coronary artery restenosis, characterized by hyperproliferation of smooth muscle cells. Latent HCMV has been detected in arterial walls of patients suffering from atherosclerosis (65, 121, 159). The authors found that a majority of the restenosis lesions that exhibited immunodetectable p53 were also positive for HCMV DNA (as determined by PCR). Furthermore, when smooth muscle cells from restenosis lesions were cultured, four of the seven cultures were immunopositive for both p53 and IE86. As a result, Speir et al. have proposed that activation of latent HCMV infection by physical injury during angioplasty may lead to elevated levels of functionally inactivated p53.
To study the effects of an interaction between the IE86 protein and p53, Speir et al. (161) infected normal human coronary smooth muscle cell cultures with HCMV. The kinetics of expression of p53 and IE86 proteins in these cells were strikingly similar, and double immunostaining indicated that both p53 and IE86 were expressed in the same cells. The physical interaction between IE86 and p53 was demonstrated by the specific coimmunoprecipitation of IE86 by anti-p53 in extracts of insect cells infected with baculovirus vectors expressing the two proteins. Finally, when human smooth muscle cells were transfected with a CAT reporter plasmid containing p53-responsive elements, CAT activity was greatly enhanced when the cells were cotransfected with a plasmid expressing p53. The observed enhancement of CAT activity was abrogated when p53 was coexpressed with IE86. These data suggested that HCMV infection of smooth muscle cells could lead to enhanced cell proliferation through the inactivation of p53 function by IE86.
To determine whether the interaction of IE proteins with p53 abrogated G1 checkpoint function, Bonin and McDougall (19) established stable clonal human foreskin fibroblast cell lines expressing IE72 or IE86. Expression of IE proteins was confirmed by Western blot analysis. IE86 protein, but not IE72, was coimmunoprecipitated with p53 from cell lysates demonstrating the binding of IE86 to p53 in these cells. However, upon treatment with a DNA-damaging agent such as actinomycin D, no difference was observed in the percentage of cells in G1/S phase in the parental and IE72- and IE86-expressing cells as determined by fluorescence flow cytometry. Furthermore, no differences were found in levels of p53, p21 (CIP1/WAF1), and mdm2 proteins and the phosphorylation status of Rb after actinomycin D treatment of IE protein-expressing cells. These data demonstrated that neither IE72 or IE86 abrogated the p53-mediated G1 block induced by DNA damage.
In conclusion, although a physical interaction between the HCMV IE86 and p53 has been demonstrated, there is no direct evidence for inhibition of p53-mediated cell cycle growth arrest by IE86. Further studies along these lines are necessary to elucidate the role of the interactions of IE proteins with p53 in transformation.
Interactions between IE proteins and Rb.
Like p53, Rb also interacts with a number of viral oncoproteins such as SV40 T-antigen and adenovirus E1A (8, 70, 74, 193). Sommer et al. (158) demonstrated the interactions of IE86 with Rb by using wild-type and deletion mutants of IE86 expressed in bacteria as glutathione S-transferase (GST)-IE86 fusion proteins. Binding of the GST-IE86 fusion protein to in vitro transcribed or translated 35S-labeled Rb was assessed by immobilization onto glutathione-coated beads. The authors showed that IE86 bound to wild-type Rb and mapped three internal domains of IE86, i.e., residues 85 to 135, 136 to 290, and 291 to 364, that could independently bind to Rb.
A complementary study by Fortunato et al. (52) determined the domains of Rb required for binding to IE86. More than one IE86-binding domain was found within Rb, i.e., the C-terminal domain from aa 768 to 926 and the Rb A/B pocket from aa 379 to 776. Most other proteins such as SV40 T-antigen and adenovirus E1A that bind Rb require the A/B pocket region or the C-terminal end or both (177, 186, 187). The authors also investigated the functional interaction between IE86 with Rb in vivo by employing a human osteosarcoma cell line, Saos-2, that expressed a nonfunctional form of Rb that localized to the cytoplasm rather than the nucleus. These cells were arrested in G1 when wild-type Rb was restored. Coexpression of IE86 in these cells did not reverse the Rb-induced G1 arrest, suggesting that interaction between IE86 and Rb did not affect the ability of Rb to cause cell cycle arrest at G1. In a second assay, Saos-2 cells were cotransfected with plasmids expressing wild-type Rb and puromycin resistance. Puromycin-resistant cells failed to divide (they were presumably arrested at G1) but continued to carry out protein synthesis. Such cells appeared as large flat cells. When IE86 was coexpressed in these cells, a 90% reduction in the frequency of large flat cells was observed, suggesting that IE86 had the ability to interfere with Rb function. Since conflicting results were obtained from two different assays, no conclusions could be drawn about the functional interactions between IE86 and Rb.
In summary, IE86 binds to both Rb and p53. However, there is no evidence that IE86 interferes with Rb or p53 cell cycle regulation, thus leading to an extended life span or transformation (as in the case of SV40 T-antigen). Further studies are necessary to determine if the IE genes play a role in HCMV transformation.
HUMAN HERPESVIRUS 6
Overview
Similarities between HCMV and HHV-6 in genetic sequence (45, 99), restricted host range, and other biological properties including association with human malignancies led to studies to determine the oncogenes of HHV-6. HHV-6 is a T-cell-tropic virus that was originally isolated from AIDS patients with lymphoproliferative disorders (85, 148) and later detected in various AIDS-related and other human lymphoid malignancies (2, 111, 112, 175). HHV-6 isolates are grouped into variants A and B and have been detected in the saliva and peripheral blood leukocytes of healthy adults (37) and in peripheral blood leukocytes of children with exanthem subitum (176). HHV-6 infection causes exanthem subitum (194) and has been linked to meningoencephalitis (77), infectious mononucleosis (10, 167), persistent lymphadenopathy (131), fulminant hepatitis with atypical lymphocytosis and lymphadenopathy (5), autoimmune disorders (92), chronic fatigue syndrome (23), Kikuchi syndrome (a form of necrotizing lymphadenopathy) (89), pneumonitis (24), and multiple sclerosis (26). HHV-6 was also detected by in situ hybridization in Rosai-Dorfman disease, a form of massive, generally benign lymphadenopathy affecting children and young adults (102).
HHV-6 has been proposed to be a cofactor in AIDS progression because both HHV-6 and HIV-1 coinfect CD4+ human T cells, resulting in accelerated cytopathic effects (110). HHV-6 may play a role in the reactivation of latent HIV-1 as well as in the up-regulation of HIV-1 expression, as has been observed by Ensoli et al. (49). Additionally, Lusso et al. (109) have demonstrated that HHV-6 infection of CD8+ human T cells induced CD4 expression, rendering these cells susceptible to HIV-1 infection. Therefore, HHV-6 may augment AIDS progression either by increasing HIV-1 production or by increasing the population of HIV-1-susceptible cells.
HHV-6 DNA sequences have been identified in various human cancers including African Burkitt’s lymphoma, Hodgkin’s lymphoma, and EBV-negative B-cell lymphoma (48, 82, 84, 183). In fact, integration of the HHV-6 genome into the 17p13 region of chromosome 17 has been demonstrated in peripheral blood mononuclear cells isolated from three individuals, one with Hodgkin’s disease, one with non-Hodgkin’s lymphoma, and one with multiple sclerosis (108, 182). Whether the integration of HHV-6 contributed to the etiology of the above lymphomas remains to be determined. These observations have encouraged researchers to identify HHV-6 transforming gene(s) and evaluate the potential of HHV-6 to be an oncogenic virus.
Identification of Transforming Fragments
Razzaque (140) detected focal transformation of NIH 3T3 cells with either total genomic DNA or HHV-6A strain GS or genomic DNA containing each of two nonoverlapping subfragments, ZVH14 (8.7 kbp) or ZVB70 (21 kbp) (Fig. 5A). The frequency of transformed foci was similar for genomic DNA and ZVH14 but was about fourfold lower for ZVB70. Independent cell lines, established from isolated foci of cells transfected with either genomic DNA or ZVH14, exhibited both anchorage-independent growth in agarose and tumorigenicity. Additionally, NIH 3T3 cell lines containing either genomic HHV-6 DNA or ZVH14 and a neomycin (G418) resistance gene were established. These cell lines also exhibited anchorage independence and tumorigenicity. Interestingly, no ZVH14 DNA was detected by Southern blot analysis in either genomic DNA or ZVH14 focally transformed cells or their tumor-derived lines. In contrast, the G418-selected lines contained intact or rearranged ZVH14 sequences. All but one of the selected lines developed after transfection with genomic HHV-6 DNA also retained ZVB70 sequences. The data suggested the presence of one or more transforming regions within the HHV-6 genome, specifically within ZVH14. Since the ZVH14 transforming region was not retained except under selective pressure, it may not be required for maintenance of the transformed phenotype. No further report has localized the ZVH14 transformation to a specific ORF.
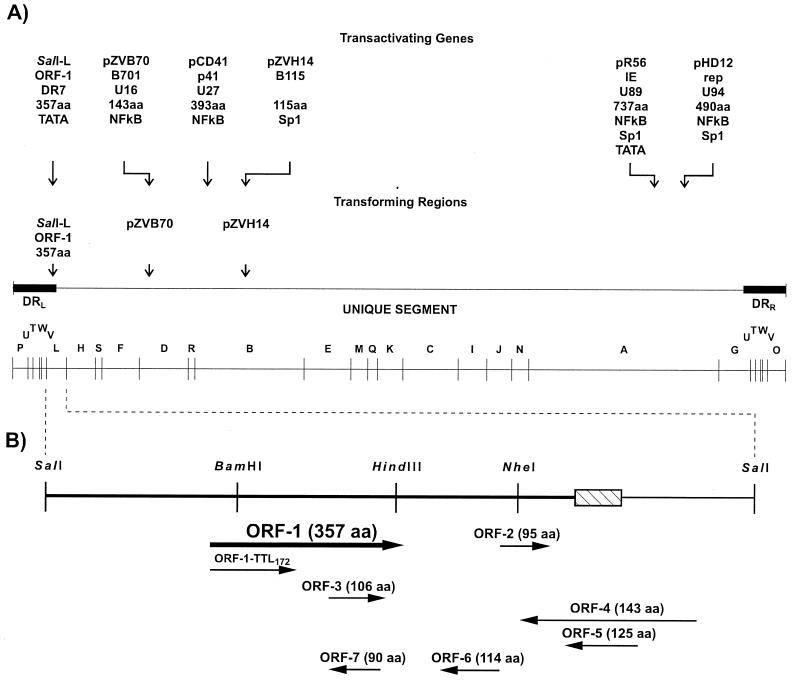
FIG. 5.
(A) SalI restriction map of the HHV-6 strain U1102 genome showing the locations of transforming regions (86, 140, 178) and genes that transactivate the HIV-1 LTR promoter (57, 87, 117, 181, 185, 196). Below each transactivating gene are indicated (i) the fragment in which the gene was originally identified, (ii) the common gene name, (iii) the HHV-6 systematic gene name (where known), (iv) the protein size, and (v) the HIV-1 promoter elements required for transactivation. Also shown are the unique segment and the left and right direct repeat regions (DRL and DRR, respectively). Adapted from reference 87 with permission of the publisher. (B) Restriction map of HHV-6 strain U1102 SalI-L showing the locations and sizes of the ORFs (178). The hatched rectangle represents the junctional telomeric repeats ([TAACCC]43) which divide the left direct repeat (DRL) (heavy line) from the unique segment (light line). Depicted below the ORF-1 is the truncated peptide translated from the ORF-1–TTL172 mutant construct. Adapted from reference 86 with permission of the publisher.
In another study, Thompson et al. (178) tested five fragments of HHV-6 strain U1102 for focal transformation of NIH 3T3 cells. Of these, only the 3.9-kbp SalI-L fragment caused transformation (Fig. 5A). The number of transformed foci observed for SalI-L was similar to that obtained with the HCMV mtrII oncogene. SalI-L-derived focal cell lines exhibited anchorage-independent growth in agarose and produced tumors with a latency period of under 2 weeks in athymic mice. Southern blot analysis revealed rearranged SalI-L DNA sequences in the tumor-derived cell lines. The data establish the malignant transforming activity of HHV-6 SalI-L and suggest that it is required for the maintenance of the transformed phenotype.
To determine the putative ORFs responsible for transformation, SalI-L was sequenced. SalI-L contains seven ORFs encoding proteins of 75 aa or more (Fig. 5B). A search of the DNA and protein sequence data bases revealed no similarity of these proteins to other known proteins. In addition, 43 tandem copies of the repeat motif TAACCC, which demarcates the junction between the direct repeat and unique sequences of HHV-6, were identified.
Identification and Characterization of the ORF-1 Transforming Gene
Localization of SalI-L transforming activity to ORF-1 (DR7).
To determine the transforming regions within the SalI-L fragment, Kashanchi et al. (86) subcloned six subfragments and tested them for focal transformation of NIH 3T3 cells. The results showed that only the SalI-HindIII subfragment containing ORF-1, ORF-3, and ORF-7 (Fig. 5B) induced morphological transformation. The individual ORF-1, ORF-3, and ORF-7 were then subcloned in a mammalian expression vector and tested for their ability to induce focal transformation. Only ORF-1 was found to induce foci above background levels, and the ORF-1 focal lines were tumorigenic in athymic nude mice. To demonstrate that ORF-1 translation was required for transformation, ORF-1 with a translation termination linker (TTL) inserted after codon 172 (TTL172) was constructed. The location of the TTL was upstream of ORF-3, leaving the ORF-3 protein intact (Fig. 5B). Cell lines transfected with wild-type ORF-1 exhibited morphological transformation, while those transfected with the TTL172 mutant did not. Expression of wild-type and mutant ORF-1 proteins in these cell lines was confirmed by Western blot analysis with rabbit polyclonal antibody raised against purified bacterially expressed ORF-1 protein. When wild-type ORF-1 and TTL172 cell lines were tested for tumorigenicity, wild-type ORF-1 cells produced fibrosarcomas in nude mice whereas TTL172 mutant cells did not. Furthermore, ORF-1 protein was detected in tumor tissue by Western blot analysis, suggesting that the expression of ORF-1 protein was required for tumorigenesis.
Characteristics of the ORF-1 gene.
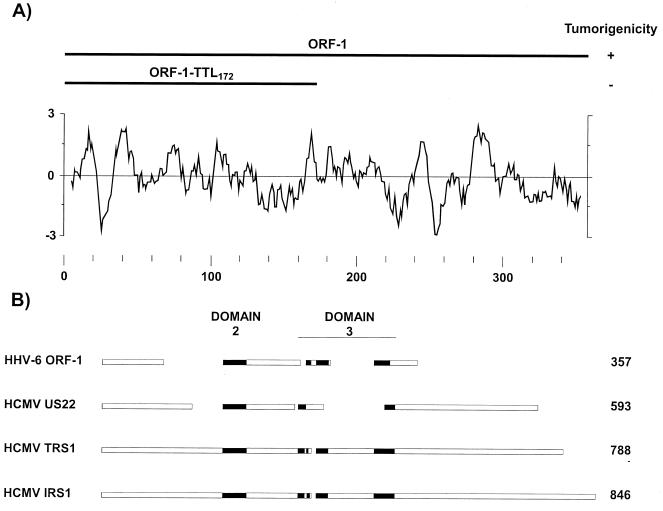
A Kyte-Doolittle analysis (95) of ORF-1 protein (Fig. 6A) revealed several hydrophobic and hydrophilic domains, indicating that it could be membrane associated. Interestingly, a search with the PROSITE protein motif library (6, 7) elucidated sequences (aa 31 to 41 and 183 to 193) that matched the consensus sequence for procaryotic membrane lipoprotein lipid attachment sites (64). Whether these sites are functional in ORF-1 remains to be determined. However, their presence suggested that ORF-1 protein could be a membrane-associated protein.
FIG. 6.
Features of the ORF-1 protein. (A) Hydropathy profile of ORF-1. The hydropathy value was determined by the method of Kyte and Doolittle (95) with a window of 9 aa. Hydrophobic regions are indicated by positive values. Depicted above the hydrophobicity profile are the wild-type ORF-1 and ORF-1-TTL172 mutant peptides and their tumorigenicity. Adapted from reference 87 with permission of the publisher. (B) Alignment of ORF-1 protein amino acid sequences with domains 2 and 3 of members of the HCMV US22 family of proteins. The schematic shows regions of alignment (solid rectangles) that were detected as statistically significant (P ≤ 0.003) by the MACAW software (153).
As previously described, a BLAST search (4) revealed no similarity in amino acid sequence between ORF-1 protein and other known proteins. However, the MACAW software (153) revealed sequences in ORF-1 protein that aligned with the HCMV US22 family of proteins as well as two HCMV transactivators, IRS1 and TRS1 (Fig. 6B) (28, 91, 165, 192), suggesting functionally analogous regions.
To determine if ORF-1 was expressed during HHV-6 infection, ORF-1 mRNA and protein expression in HHV-6-infected human T cells were examined by reverse transcription-PCR and Western blotting with anti-ORF-1 antibody, respectively (86). Both RNA and protein were observed from 18 to 48 h, suggesting that ORF-1 is an HHV-6 early gene.
Association of ORF-1 with human malignancies.
With the in vitro identification of the ORF-1 oncogene, studies were performed to determine if there was any association of ORF-1 with human malignancies (86). Glioblastomas and pathologic lymph nodes from patients with angioimmunoblastic lymphadenopathy, angioimmunoblastic lymphadenopathy-like lymphoma, Hodgkin’s disease, and both B- and T-cell-lineage non-Hodgkin’s lymphoma were examined by PCR. ORF-1 sequences were rarely detected in any of the malignancies. In contrast, ORF-1 sequences were found in 5 of 12 lymph nodes from patients with angioimmunoblastic lymphadenopathy, while nonmalignant lymph nodes and normal brain tissue specimens were negative. Thus, ORF-1 sequences, which exhibited oncogenic properties in vitro, were retained at variable frequency in various human tumor tissues. These observations, while not proof per se, are a necessary prerequisite for establishing the role of ORF-1 in human malignancies.
Interaction between the ORF-1 and p53 proteins.
The expression of ORF-1 protein in transformed cells and tumor tissues may suggest a maintenance function for ORF-1. Because binding to p53 is characteristic of several viral oncoproteins, including HCMV mtrII (see above) (75, 101), ORF-1 protein was also tested for binding to p53. In GST pulldown assays, ORF-1 was observed to bind specifically to GST-p53. Furthermore, anti-ORF-1 serum coimmunoprecipitated the p53–ORF-1 complex. Coimmunoprecipitation experiments performed with truncated p53 proteins demonstrated that the ORF-1-binding domain of p53 was between residues 28 and 187, with a critical region between 107 and 187 (Fig. 3). This overlapped the specific DNA and SV40 T-antigen-binding sites of p53.
Because the ORF-1 and specific DNA-binding domains of p53 overlapped, the ability of ORF-1 to affect p53-activated transcription was examined by using a p53-responsive reporter construct (p53G5BCAT). Wild-type ORF-1 cells transfected with p53G5BCAT exhibited a seven- to eightfold reduction in CAT activity compared to parental NIH 3T3 cells expressing the ORF-1-TTL172 mutant or cells containing a construct with no p53-binding sites (pSV2CAT). Thus, ORF-1 specifically suppressed p53-activated transcription. The binding of ORF-1 protein to p53 may alter the ability of p53 to regulate cellular genes important for growth control.
Transactivation of the HIV-1 Promoter by ORF-1
Proteins expressed from the transforming genes of many DNA tumor viruses are often transactivators. Therefore, Thompson et al. (178) tested the ability of the SalI-L fragment to transactivate the HIV-1 LTR promoter. It has been previously reported that coinfection of human T-cell lymphocytes with HHV-6 and HIV-1 increased HIV-1 replication (110). Moreover, HHV-6 infection was further shown to up-regulate the HIV-1 LTR promoter. When an HIV-1 LTR CAT reporter plasmid was cotransfected with SalI-L into either monkey fibroblasts or human lymphocytes, CAT activity increased by up to 15-fold in a dose-dependent manner as compared to the CAT activity observed after transfection with HIV-1 LTR CAT alone. Thus, SalI-L contained a transactivator gene that up-regulated the HIV-1 promoter. Because the transactivation occurred in human T lymphocytes, which are permissive for both HIV-1 and HHV-6 replication, a clinically relevant association may exist in coinfected cells.
Kashanchi et al. (87) identified the ORF within SalI-L responsible for HIV-1 LTR transactivation. Specifically, subfragments of SalI-L containing different ORFs were each tested for transactivation activity in both T-cell lymphocytes and monocytoid cells. These studies identified ORF-1, ORF-3, ORF-6, and ORF-7 as candidates (Fig. 5B). Of the individual ORFs that were cloned into a mammalian expression vector, only the ORF-1 construct exhibited transactivation of HIV-1 LTR that was comparable to that of SalI-L.
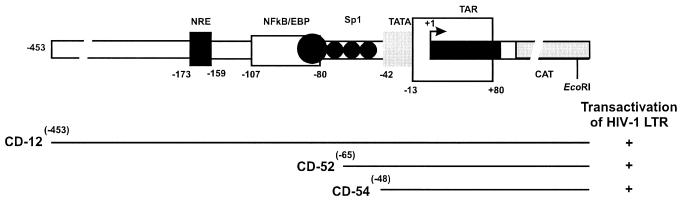
To determine which elements of the HIV-1 promoter were required for ORF-1 transactivation, the HIV-1 promoter deletion mutant CAT reporter constructs were tested for transactivation (Fig. 7) (87). ORF-1 was capable of transactivating both deletion mutant constructs (16- to 24-fold), including the one containing the minimal HIV-1 promoter with only the TATA box element but no other upstream enhancer elements. On the other hand, HPV-16 and HPV-18 promoter CAT reporter constructs, which each contain a TATA box initiation element, were not transactivated by the SalI-L fragment. These data demonstrated the specificity of ORF-1 transactivation and suggested that although HIV-1 upstream enhancer elements were not required, elements other than the TATA box were involved in transactivation.
FIG. 7.
Schematic diagram of the complete HIV-1 LTR promoter, indicating its regulatory sequences and showing deletion mutant CAT reporter constructs (CD52 and CD54) used to study sequences required for transactivation by ORF-1. The ability of SalI-L to transactivate each HIV-1 LTR construct is shown on the right. Adapted from reference 87 with permission of the publisher.
To study the role of ORF-1 protein in the enhancement of HIV-1 expression, purified ORF-1 protein was produced in bacteria. The ORF-1 gene was cloned in the bacterial expression vector, pET17b, which resulted in expression of ORF-1 protein fused to the pET17b 11-aa Tag sequence containing an epitope for the anti-T7.Tag monoclonal antibody (87). This fusion allowed affinity column purification of ORF-1 protein. When purified ORF-1 was cotransfected into human T cells with the HIV-1 LTR CAT construct, CAT activity increased up to fivefold compared to that obtained by transfection of the reporter gene alone. This transactivation was inhibited by anti-T7.Tag antibody. Thus, ORF-1 protein was responsible for the observed transactivation of the HIV-1 promoter by the ORF-1 gene.
Purified ORF-1 protein also up-regulated the HIV-1 promoter in an in vitro transcription assay. All necessary transcription factors were provided by a HeLa cell extract. Basal levels of the HIV-1 LTR CAT mRNA were increased fourfold upon addition of ORF-1 fusion protein to the reaction. This increase in transcription was specifically inhibited by anti-T7.Tag antibody. These in vitro studies demonstrated that ORF-1 protein acted directly on the HIV-1 LTR.
Because ORF-1 was shown to up-regulate the HIV-1 promoter, its ability to activate HIV-1 replication was investigated by using a human cervical carcinoma cell line which expresses the CD4 cell surface marker and contains integrated tat-defective HIV-1 provirus. Transfection of these cells with either SalI-L or ORF-1 constructs resulted in the release of significant levels of HIV-1 into the medium as measured by the HIV-1 p24 Gag antigen capture assay. Although the level of viral release was 50- to 100-fold lower than that achieved by transfection with tat, the level released by ORF-1 was sufficient to initiate a productive HIV-1 infection from latently infected cells. Thus, ORF-1 may be one of the specific transactivators of HHV-6 responsible for the up-regulation of HIV-1 expression (49).
In addition to ORF-1 (87), five other HHV-6 genes that transactivate the HIV-1 promoter have been identified (Fig. 5) (57, 117, 181, 185, 196). Each of these HHV-6 transactivators requires Sp1 and/or NF-κB enhancer sites, whereas ORF-1 requires only the TATA box element. This suggests that ORF-1 may function by recruiting basal transcription factors to the promoter. Initial studies have demonstrated binding of ORF-1 to the basal transcription factor TATA-binding protein (unpublished data). Further studies are essential to elucidate the mechanism of transactivation by ORF-1. Furthermore, the ability of ORF-1 to transactivate other HHV-6 genes or cellular genes remains to be determined. Further studies along these lines are necessary to define the role of ORF-1 in HHV-6 infection as well as to assess the role of HHV-6 as a cofactor in AIDS progression.
CONCLUSION
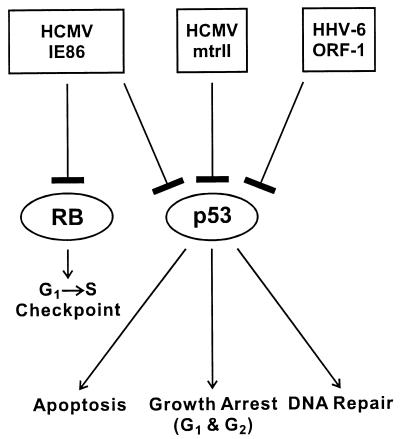
Human herpesviruses have been investigated for the past several decades as possible oncogenic agents. Studies cited in this review demonstrate that HCMV and HHV-6 contain in vitro transforming genes which are retained in both transformed and tumor-derived cell lines. These genes may be in part responsible for the tumorigenic phenotype observed in some human cancers. In the case of HCMV, the mtrII oncoprotein was expressed in transformed and tumor derived cell lines. Furthermore, mtrII protein bound to p53 and inhibited p53-activated transcription. In the case of HHV-6, both transactivating and transforming activities were localized to HHV-6 ORF-1. Like mtrII, ORF-1 also bound to p53 and inhibited p53-activated transcription. These activities, coupled with the detection of ORF-1 in malignant tissues, may indicate a role of ORF-1 in human cancer. Both the HCMV and HHV-6 oncoproteins exhibit the same characteristics as the oncoproteins of several DNA tumor viruses such as SV40 and polyomavirus T-antigens (191), adenovirus E1B (193), and HPV-16 E6 (44) because of their ability to bind to and inactivate host tumor suppressor proteins such as p53. This binding may uncouple normal growth control processes and lead to cellular transformation. A schematic summarizing the interactions of HCMV and HHV-6 proteins with host tumor suppressor proteins and the potential pathways that may be disrupted is shown in Fig. 8. Although the HCMV mtrII and IE86, as well as the HHV-6 ORF-1 proteins, bind to and inhibit p53-activated transcription, the effects on specific p53-mediated pathways such as apoptosis, stress-induced G1 and G2 growth arrest, and DNA repair have not yet been characterized. The IE86 protein also interacts with Rb. However, data showing the effect of this interaction on G1 growth arrest is inconclusive.
FIG. 8.
Schematic showing the interactions of HCMV and HHV-6 proteins with the host tumor suppressors p53 and Rb. The HCMV mtrII and IE86 proteins and the HHV-6 ORF-1 protein bind to p53 and inhibit p53-activated transcription. HCMV IE86 also binds to Rb. Shown below p53 and Rb are the potential cellular pathways that may be deregulated by the above interactions, such as apoptosis, G1 and G2 growth arrest, and DNA repair for p53 and control of the G1-to-S-phase transition for Rb.
The acquisition of a fully malignant phenotype by normal cells is thought to require several mutations or dysfunctions in a number of cellular genes and proteins. Inhibition of antioncogene functions, as discussed above, could be one mechanism by which herpesviruses modulate the malignant potential of cells. In the case of HCMV infection, a number of other mechanisms such as stimulation of growth factors or proto-oncogenes and inhibition of cellular apoptotic pathways may play a role in altering cell growth. Boldogh et al. (11–14, 18) have demonstrated that HCMV infection activated a number of proto-oncogenes, such as c-myc, c-fos, and c-jun, and signal-transducing proteins such as H-ras. Jault et al. (83) have shown that HCMV infection also resulted in the activation of a number of cell cycle-regulatory proteins such as p53, phosphorylated Rb, and cyclins. In this regard, Bresnahan et al. (20, 21) have reported that HCMV infection induced cyclin E expression and altered the subcellular localization of a cyclin E-associated kinase, Cdk2, in G0 cells, resulting in cell cycle progression into the G1 and S phases. Furthermore, Zhu et al. (197) showed that HCMV infection blocked the induction of apoptosis and that this block was mediated by the IE proteins. Thus, the ability of HCMV infection to alter several important cellular regulatory pathways may clearly play a role in determining its oncogenic potential.
In recent years, it has become increasingly clear that viral evasion of host immune responses plays an important role not only in disease but also in transformation and tumor development. In this regard, a number of viruses and viral oncogenes have been shown to modulate the expression of major histocompatibility complex genes (118). Loss of major histocompatibility complex antigen expression in infected or transformed cells may serve as a mechanism of survival and escape from the host immune system. Further analysis of the above mechanism(s) of transformation by herpesvirus oncogenes, together with their detection in human cancers, will provide insights into the multistep process of malignant transformation.
ACKNOWLEDGMENTS
The work on HHV-6 ORF-1 and HCMV mtrII was supported by Public Health Service grant CA 60577 from the National Institutes of Health and by a contract from the National Foundation for Cancer Research (Bethesda, Md.). Assistance with the tumorigenicity assays was provided by the Lombardi Cancer Research Center Animal Care Facility supported by Public Health Service grant P30 CA51008-09.
We thank Chemicon International Inc. for generating antibodies against HCMV mtrII and HHV-6 ORF-1 proteins. We thank John N. Brady, Laboratory of Receptor Biology and Gene Expression, National Cancer Institute, for his support and collaboration. L.J.R. thanks all the students and postdoctoral fellows who contributed to these studies over the past 15 years.
REFERENCES
- 1.Ablashi D V, Josephs S F, Buchbinder A, Hellman K, Nakamura S, Llana T, Lusso P, Kaplan M, Dahlberg J, Memon S, Imam F, Ablashi K L, Markham P D, Kramarsky B, Krueger G R F, Biberfeld P, Wong-Staal F, Salahuddin S Z, Gallo R C. Human B-lymphotropic virus (human herpesvirus-6) J Virol Methods. 1988;21:29–48. doi: 10.1016/0166-0934(88)90050-x. [DOI] [PubMed] [Google Scholar]
- 2.Ablashi D V, Salahuddin S Z, Josephs S F, Imam F, Lusso P, Gallo R C, Hung C, Lemp J, Markham P D. HBLV (or HHV-6) in human cell lines. Nature. 1987;329:207. doi: 10.1038/329207a0. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 3.Albrecht T, Rapp F. Malignant transformation of hamster embryo fibroblasts exposure to ultraviolet-irradiated human cytomegalovirus. Virology. 1973;55:53–61. doi: 10.1016/s0042-6822(73)81007-4. [DOI] [PubMed] [Google Scholar]
- 4.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Asano Y, Yoshikawa T, Suga S, Yazaki T, Kondo K, Yamanishi K. Fatal fulminant hepatitis in an infant with human herpesvirus-6 infection. Lancet. 1990;335:862–863. doi: 10.1016/0140-6736(90)90983-c. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 6.Bairoch A. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1992;20:2013–2018. doi: 10.1093/nar/20.suppl.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bairoch A. The PROSITE dictionary of sites and patterns in proteins, its current status. Nucleic Acids Res. 1993;21:3097–3103. doi: 10.1093/nar/21.13.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bandara L R, La Thangue N B. Adenovirus E1A prevents the retinoblastoma gene product from complexing with a cellular transcription factor. Nature. 1991;351:494–497. doi: 10.1038/351494a0. [DOI] [PubMed] [Google Scholar]
- 9.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild-type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertram G, Dreiner N, Krueger G R, Ramon A, Ablashi D V, Salahuddin S Z, Balachandran N. Frequent double infection with Epstein-Barr virus and human herpesvirus-6 in patients with acute infectious mononucleosis. In Vivo. 1991;5:271–279. [PubMed] [Google Scholar]
- 11.Boldogh I, Abu Bakar S, Fons M P, Deng C Z, Albrecht T. Activation of cellular oncogenes by clinical isolates and laboratory strains of human cytomegalovirus. J Med Virol. 1991;34:241–247. doi: 10.1002/jmv.1890340409. [DOI] [PubMed] [Google Scholar]
- 12.Boldogh I, Abu Bakar S, Millinoff D, Deng C Z, Albrecht T. Cellular oncogene activation by human cytomegalovirus. Lack of correlation with virus infectivity and immediate early gene expression. Arch Virol. 1991;118:163–177. doi: 10.1007/BF01314027. [DOI] [PubMed] [Google Scholar]
- 13.Boldogh I, AbuBakar S, Albrecht T. Activation of proto-oncogenes: an immediate early event in human cytomegalovirus infection. Science. 1990;247:561–564. doi: 10.1126/science.1689075. [DOI] [PubMed] [Google Scholar]
- 14.Boldogh I, AbuBakar S, Deng C Z, Albrecht T. Transcriptional activation of cellular oncogenes fos, jun, and myc by human cytomegalovirus. J Virol. 1991;65:1568–1571. doi: 10.1128/jvi.65.3.1568-1571.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boldogh I, Baskar J F, Mar E, Huang E. Human cytomegalovirus and herpes simplex type 2 virus in normal and adenocarcinomatous prostate glands. JNCI. 1983;70:819–826. [PubMed] [Google Scholar]
- 16.Boldogh I, Beth E, Huang E, Kyalwazi S K, Giraldo G. Kaposi’s sarcoma. IV. Detection of CMV DNA, CMV RNA and CMNA tumor biopsies. Int J Cancer. 1981;28:469–474. doi: 10.1002/ijc.2910280412. [DOI] [PubMed] [Google Scholar]
- 17.Boldogh I, Gonczol E, Vaczi L. Transformation of hamster embryonic fibroblast cells by UV-irradiated human cytomegalovirus. Acta Microbiol Acad Sci Hung. 1978;25:269–275. [PubMed] [Google Scholar]
- 18.Boldogh I, Huang E S, Rady P, Arany I, Tyring S, Albrecht T. Alteration in the coding potential and expression of h-ras in human cytomegalovirus-transformed cells. Intervirology. 1994;37:321–329. doi: 10.1159/000150396. [DOI] [PubMed] [Google Scholar]
- 19.Bonin L R, McDougall J K. Human cytomegalovirus IE2 86-kilodalton protein binds p53 but does not abrogate G1 checkpoint function. J Virol. 1997;71:5861–5870. doi: 10.1128/jvi.71.8.5861-5870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bresnahan W A, Albrecht T, Thompson E A. The cyclin E promoter is activated by human cytomegalovirus 86-kDa immediate early protein. J Biol Chem. 1998;273:22075–22082. doi: 10.1074/jbc.273.34.22075. [DOI] [PubMed] [Google Scholar]
- 21.Bresnahan W A, Thompson E A, Albrecht T. Human cytomegalovirus infection results in altered Cdk2 subcellular localization. J Gen Virol. 1997;78:1993–1997. doi: 10.1099/0022-1317-78-8-1993. [DOI] [PubMed] [Google Scholar]
- 22.Brichacek B, Hirsch I, Zavadova H, Prochazka M, Faltyn J, Vonka V. Absence of cytomegalovirus DNA from adenocarcinoma of the colon. Intervirology. 1980;14:223–227. doi: 10.1159/000149187. [DOI] [PubMed] [Google Scholar]
- 23.Buchwald D, Cheney P R, Peterson D L, Henry B, Wormsley S B, Geiger A, Ablashi D V, Salahuddin S Z, Saxinger C, Biddle R, Kikinis R, Jolesz F A, Folks T, Balachandran N, Peter J B, Gallo R C, Komaroff A L. A chronic illness characterized by fatigue, neurologic and immunologic disorders, and active human herpesvirus type 6 infection. Ann Intern Med. 1992;116:103–113. doi: 10.7326/0003-4819-116-2-103. [DOI] [PubMed] [Google Scholar]
- 24.Carrigan D R, Drobyski W R, Russler S K, Tapper M A, Knox K K, Ash R C. Interstitial pneumonitis associated with human herpesvirus-6 infection after marrow transplantation. Lancet. 1991;338:147–149. doi: 10.1016/0140-6736(91)90137-e. [DOI] [PubMed] [Google Scholar]
- 25.Centifanto Y M, Kaufman H E, Zam Z S, Drylie D M, Deardourff S L. Herpesvirus particles in prostatic carcinoma cells. J Virol. 1973;12:1608–1611. doi: 10.1128/jvi.12.6.1608-1611.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Challoner P B, Smith K T, Parker J D, MacLeod D L, Coulter S N, Rose T M, Schultz E R, Bennett J L, Garber R L, Chang M, Schad P A, Stewart P M, Nowinski R C, Brown J P, Burmer G C. Plaque-associated expression of human herpesvirus 6 in multiple sclerosis. Proc Natl Acad Sci USA. 1995;92:7440–7444. doi: 10.1073/pnas.92.16.7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 28.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison III C A, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 29.Chen C Y, Oliner J D, Zhan Q, Fornace A J, Jr, Vogelstein B, Kastan M B. Interactions between p53 and MDM2 in a mammalian cell cycle checkpoint pathway. Proc Natl Acad Sci USA. 1994;91:2684–2688. doi: 10.1073/pnas.91.7.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cherrington J M, Khoury E L, Mocarski E S. Human cytomegalovirus ie2 negatively regulates alpha gene expression via a short target sequence near the transcription start site. J Virol. 1991;65:887–896. doi: 10.1128/jvi.65.2.887-896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cherrington J M, Mocarski E S. Human cytomegalovirus IE1 transactivates the alpha promoter-enhancer via an 18-base-pair repeat element. J Virol. 1989;63:1435–1440. doi: 10.1128/jvi.63.3.1435-1440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiou C J, Zong J, Waheed I, Hayward G S. Identification and mapping of dimerization and DNA-binding domains in the C terminus of the IE2 regulatory protein of human cytomegalovirus. J Virol. 1993;67:6201–6214. doi: 10.1128/jvi.67.10.6201-6214.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chretien J H, McGinniss C G, Muller A. Venereal causes of cytomegalovirus mononucleosis. JAMA. 1977;238:1644–1645. [PubMed] [Google Scholar]
- 34.Chuck S, Grant R M, Katongole-Mbidde E, Conant M, Ganem D. Frequent presence of a novel herpesvirus genome in lesions of human immunodeficiency virus-negative Kaposi’s sarcoma. J Infect Dis. 1996;173:248–251. doi: 10.1093/infdis/173.1.248. [DOI] [PubMed] [Google Scholar]
- 35.Cinatl J, Jr, Cinatl J, Vogel J U, Rabenau H, Kornhuber B, Doerr H W. Modulatory effects of human cytomegalovirus infection on malignant properties of cancer cells. Intervirology. 1996;39:259–269. doi: 10.1159/000150527. [DOI] [PubMed] [Google Scholar]
- 36.Clanton D J, Jariwalla R J, Kress C, Rosenthal L J. Neoplastic transformation by a cloned human cytomegalovirus DNA fragment uniquely homologous to one of the transforming regions of herpes simplex virus type 2. Proc Natl Acad Sci USA. 1983;80:3826–3830. doi: 10.1073/pnas.80.12.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cone R W, Huang M L, Ashley R, Corey L. Human herpesvirus 6 DNA in peripheral blood cells and saliva from immunocompetent individuals. J Clin Microbiol. 1993;31:1262–1267. doi: 10.1128/jcm.31.5.1262-1267.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crook T, Marston N J, Sara E A, Vousden K H. Transcriptional activation by p53 correlates with suppression of growth but not with transformation. Cell. 1994;79:817–827. doi: 10.1016/0092-8674(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 39.Demmler G J. Infectious Diseases Society of America and Centers for Disease Control. Summary of a workshop on surveillance for congenital cytomegalovirus disease. Rev Infect Dis. 1991;13:315–329. doi: 10.1093/clinids/13.2.315. [DOI] [PubMed] [Google Scholar]
- 40.Dictor M, Rambech E, Way D, Witte M, Bendsoe N. Human herpesvirus 8 (Kaposi’s sarcoma-associated herpesvirus) DNA in Kaposi’s sarcoma lesions, AIDS Kaposi’s sarcoma cell lines, endothelial Kaposi’s sarcoma simulators, and the skin of immunosuppressed patients. Am J Pathol. 1996;148:2009–2016. [PMC free article] [PubMed] [Google Scholar]
- 41.Drew W L, Conant M A, Miner R C, Huang E, Ziegler J L, Groundwater J R, Gullett J H, Volberding P, Abrams D I, Mintz L. Cytomegalovirus and Kaposi’s sarcoma in young homosexual men. Lancet. 1982;ii:125–127. doi: 10.1016/s0140-6736(82)91092-3. [DOI] [PubMed] [Google Scholar]
- 42.Drew W L, Mintz L, Miner R C, Sands M, Ketterer B. Prevalence of cytomegalovirus infection in homosexual men. J Infect Dis. 1981;143:188–192. doi: 10.1093/infdis/143.2.188. [DOI] [PubMed] [Google Scholar]
- 43.Dutta A, Ruppert J M, Aster J C, Winchester E. Inhibition of DNA replication factor RPA by p53. Nature. 1993;365:79–82. doi: 10.1038/365079a0. [DOI] [PubMed] [Google Scholar]
- 44.Dyson N, Howley P M, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 45.Efstathiou S, Lawrence G L, Brown C M, Barrell B G. Identification of homologues to the human cytomegalovirus US22 gene family in human herpesvirus 6. J Gen Virol. 1992;73:1661–1671. doi: 10.1099/0022-1317-73-7-1661. [DOI] [PubMed] [Google Scholar]
- 46.El-Beik T, Razzaque A, Jariwalla R, Cihlar R L, Rosenthal L J. Multiple transforming regions of human cytomegalovirus DNA. J Virol. 1986;60:645–652. doi: 10.1128/jvi.60.2.645-652.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 48.Ellinger K, Neipel F, Seidl S, Borisch-Chappius B, Muller-Hermelink K, Fleckenstein B. Detection of HHV-6 genomes in lymphoproliferative diseases. In: Ablashi D V, Krueger G R F, Salahuddin S Z, editors. Human herpesvirus-6: epidemiology, molecular biology, and clinical pathology. New York, N.Y: Elsevier Science Publishing; 1992. pp. 209–226. [Google Scholar]
- 49.Ensoli B, Lusso P, Schachter F, Josephs S F, Rappaport J, Negro F, Gallo R C, Wong-Staal F. Human herpes virus-6 increases HIV-1 expression in co-infected T cells via nuclear factors binding to the HIV-1 enhancer. EMBO J. 1989;8:3019–3027. doi: 10.1002/j.1460-2075.1989.tb08452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fenoglio C M, Oster M W, Lo Gerfo P, Reynolds T, Edelson R, Patterson J A, Madeiros E, McDougall J K. Kaposi’s sarcoma following chemotherapy for testicular cancer in a homosexual man: demonstration of cytomegalovirus RNA in sarcoma cells. Hum Pathol. 1982;13:955–959. doi: 10.1016/s0046-8177(82)80063-4. [DOI] [PubMed] [Google Scholar]
- 51.Fletcher K, Cordiner J W, Macnab J C. Detection of sequences that hybridize to human DNA in cervical neoplastic tissue. Dis Markers. 1986;4:219–229. [PubMed] [Google Scholar]
- 52.Fortunato E A, Sommer M H, Yoder K, Spector D H. Identification of domains within the human cytomegalovirus major immediate-early 86-kilodalton protein and the retinoblastoma protein required for physical and functional interaction with each other. J Virol. 1997;71:8176–8185. doi: 10.1128/jvi.71.11.8176-8185.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fuccillo D A, Sever J L, Moder F L, Chen T C, Catalano L W, Johnson L D. Cytomegalovirus antibody in patients with carcinoma of the uterine cervix. Obstet Gynecol. 1971;38:599–601. doi: 10.1097/00006250-197110000-00016. [DOI] [PubMed] [Google Scholar]
- 54.Gao S J, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo C R, Saah A, Phair J, Detels R, Chang Y, Moore P S. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi’s sarcoma. Nat Med. 1996;2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 55.Geder L, Lausch R, O’Neill F, Rapp F. Oncogenic transformation of human embryo lung cells by human cytomegalovirus. Science. 1976;192:1134–1137. doi: 10.1126/science.179143. [DOI] [PubMed] [Google Scholar]
- 56.Geder L, Laychock A M, Gorodecki J, Rapp F. Alterations in biological properties of different lines of cytomegalovirus-transformed human embryo lung cells following in vitro cultivation. IARC Sci Publ. 1978;24:591–601. [PubMed] [Google Scholar]
- 57.Geng Y Q, Chandran B, Josephs S F, Wood C. Identification and characterization of a human herpesvirus 6 gene segment that trans activates the human immunodeficiency virus type 1 promoter. J Virol. 1992;66:1564–1570. doi: 10.1128/jvi.66.3.1564-1570.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ginsberg D, Mechta F, Yaniv M, Oren M. Wild-type p53 can down-modulate the activity of various promoters. Proc Natl Acad Sci USA. 1991;88:9979–9983. doi: 10.1073/pnas.88.22.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giraldo G, Beth E, Henle W, Henle G, Mike V, Safai B, Huraux J M, McHardy J, De-The G. Antibody patterns to herpesviruses in Kaposi’s sarcoma. II Serological association of American Kaposi’s sarcoma with cytomegalovirus. Int J Cancer. 1978;22:126–131. doi: 10.1002/ijc.2910220204. [DOI] [PubMed] [Google Scholar]
- 60.Giraldo G, Beth E, Huang E. Kaposi’s sarcoma and its relationship to cytomegalovirus (CMV). III. CMV DNA and CMV early antigens in Kaposi’s sarcoma. Int J Cancer. 1980;26:23–29. doi: 10.1002/ijc.2910260105. [DOI] [PubMed] [Google Scholar]
- 61.Hart H, Neill W A, Noval M. Lack of association of cytomegalovirus with adenocarcinoma of the colon. Gut. 1982;23:21–30. doi: 10.1136/gut.23.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hart H, Springbett A, Norval M. Lack of association of cytomegalovirus antibody level with carcinoma of the uterine cervix. Gynecol Obstet Investig. 1982;14:300–308. doi: 10.1159/000299470. [DOI] [PubMed] [Google Scholar]
- 63.Hashiro G M, Horikami S, Loh P C. Cytomegalovirus isolations from cell cultures of human adenocarcinomas of the colon. Intervirology. 1979;12:84–88. doi: 10.1159/000149072. [DOI] [PubMed] [Google Scholar]
- 64.Hayashi S, Wu H C. Lipoproteins in bacteria. J Bioenerg Biomembr. 1990;22:451–471. doi: 10.1007/BF00763177. [DOI] [PubMed] [Google Scholar]
- 65.Hendrix M G R, Salimans M M, van Boven C P, Bruggeman C A. High prevalence of latently present cytomegalovirus in arterial walls of patients suffering from grade III atherosclerosis. Am J Pathol. 1990;136:23–28. [PMC free article] [PubMed] [Google Scholar]
- 66.Hermiston T W, Malone C L, Stinski M F. Human cytomegalovirus immediate-early two protein region involved in negative regulation of the major immediate-early promoter. J Virol. 1990;64:3532–3536. doi: 10.1128/jvi.64.7.3532-3536.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hirsch M S. Cytomegalovirus-leukocyte interactions. Birth Defects Orig Artic Ser. 1984;20:161–173. [PubMed] [Google Scholar]
- 68.Hollstein M, Sidransky D, Vogelstein B, Harris C C. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 69.Horikoshi N, Maguire K, Kralli A, Maldonado E, Reinberg D, Weinmann R. Direct interaction between adenovirus E1A protein and the TATA box binding transcription factor IID. Proc Natl Acad Sci USA. 1991;88:5124–5128. doi: 10.1073/pnas.88.12.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu Q, Dyson N, Harlow E. The regions of the retinoblastoma protein needed for binding to adenovirus E1A or SV40 large T antigen are common sites for mutation. EMBO J. 1990;9:1147–1155. doi: 10.1002/j.1460-2075.1990.tb08221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang E, Boldogh I, Mar E. Human cytomegaloviruses: evidence for possible association with human cancer. In: Philips L A, editor. Viruses associated with human cancer. New York, N.Y: Marcel Dekker, Inc.; 1983. pp. 161–193. [Google Scholar]
- 72.Huang E, Davis M G, Baskar J F, Huong S. Molecular epidemiology and oncogenicity of human cytomegalovirus. In: Harris C C, editor. Biochemical and molecular epidemiology of cancer. New York, N.Y: Alan R. Liss, Inc.; 1986. pp. 323–344. [Google Scholar]
- 73.Huang E, Roche J K. Cytomegalovirus DNA and adenocarcinoma of the colon: evidence for latent viral infection. Lancet. 1978;i:957–960. doi: 10.1016/s0140-6736(78)90248-9. [DOI] [PubMed] [Google Scholar]
- 74.Huang S, Wang N P, Tseng B Y, Lee W-H, Lee E Y-H P. Two distinct and frequently mutated regions of retinoblastoma protein are required for binding to SV40 T antigen. EMBO J. 1990;9:1815–1822. doi: 10.1002/j.1460-2075.1990.tb08306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huibregtse J M, Scheffner M, Howley P M. E6-AP directs the HPV E6-dependent inactivation of p53 and is representative of a family of structurally and functionally related proteins. Cold Spring Harbor Symp Quant Biol. 1994;59:237–245. doi: 10.1101/sqb.1994.059.01.028. [DOI] [PubMed] [Google Scholar]
- 76.Inamdar A, Thompson J, Kashanchi F, Doniger J, Brady J N, Rosenthal L J. Identification of two promoters within human cytomegalovirus morphologic transforming region II. Intervirology. 1992;34:146–153. doi: 10.1159/000150275. [DOI] [PubMed] [Google Scholar]
- 77.Ishiguro N, Yamada S, Takahashi T, Takahashi Y, Togashi T, Okuno T, Yamanishi K. Meningo-encephalitis associated with HHV-6 related exanthem subitum. Acta Paediatr Scand. 1990;79:987–989. doi: 10.1111/j.1651-2227.1990.tb11369.x. [DOI] [PubMed] [Google Scholar]
- 78.Jabs D A, Enger C, Bartlett J G. Cytomegalovirus retinitis and acquired immunodeficiency syndrome. Arch Ophthalmol. 1989;107:75–80. doi: 10.1001/archopht.1989.01070010077031. [DOI] [PubMed] [Google Scholar]
- 79.Jahan N, Razzaque A, Brady J, Rosenthal L J. The human cytomegalovirus mtrII colinear region in strain Tanaka is transformation defective. J Virol. 1989;63:2866–2869. doi: 10.1128/jvi.63.6.2866-2869.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jahan N, Razzaque A, Greenspan J, Conant M A, Josephs S F, Nakamura S, Rosenthal L J. Analysis of human KS biopsies and cloned cell lines for cytomegalovirus, HIV-1, and other selected DNA virus sequences. AIDS Res Hum Retroviruses. 1989;5:225–231. doi: 10.1089/aid.1989.5.225. [DOI] [PubMed] [Google Scholar]
- 81.Jariwalla R J, Razzaque A, Lawson S, Rosenthal L J. Tumor progression mediated by two cooperating DNA segments of human cytomegalovirus. J Virol. 1989;63:425–428. doi: 10.1128/jvi.63.1.425-428.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jarrett R F, Gledhill S, Qureshi F, Crae S H, Madhok R, Brown I, Evans I, Krajewski A, O’Brien C J, Cartwright R A, Venables P, Onions D E. Identification of human herpesvirus 6-specific DNA sequences in two patients with non-Hodgkin’s lymphoma. Leukemia. 1988;2:496–502. [PubMed] [Google Scholar]
- 83.Jault F M, Jault J M, Ruchti F, Fortunato E A, Clark C, Corbeil J, Richman D D, Spector D H. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J Virol. 1995;69:6697–6704. doi: 10.1128/jvi.69.11.6697-6704.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Josephs S F, Buchbinder A, Streicher H Z, Ablashi D V, Salahuddin S Z, Guo H G, Wong-Staal F, Cossman J, Raffeld M, Sundeen J, Levine P, Biggar R, Krueger G R F, Fox R I, Gallo R C. Detection of human B-lymphotropic virus (human herpesvirus 6) sequences in B cell lymphoma tissues of three patients. Leukemia. 1988;2:132–135. [PubMed] [Google Scholar]
- 85.Josephs S F, Salahuddin S Z, Ablashi D V, Schachter F, Wong-Staal F, Gallo R C. Genomic analysis of the human B-lymphotropic virus (HBLV) Science. 1986;234:601–603. doi: 10.1126/science.3020691. [DOI] [PubMed] [Google Scholar]
- 86.Kashanchi F, Araujo J C, Doniger J, Muralidhar S, Khleif S, Mendleson E, Thompson J, Azumi N, Brady J N, Luppi M, Torelli G, Rosenthal L J. Human herpesvirus type 6 (HHV-6) ORF-1 transactivating gene exhibits malignant transforming activity and its protein binds to p53. Oncogene. 1997;14:359–367. doi: 10.1038/sj.onc.1200840. [DOI] [PubMed] [Google Scholar]
- 87.Kashanchi F, Thompson J, Sadaie M R, Doniger J, Duvall J, Brady J N, Rosenthal L J. Transcriptional activation of minimal HIV-1 promoter by ORF-1 protein expressed from the SalI-L fragment of human herpesvirus 6. Virology. 1994;201:95–106. doi: 10.1006/viro.1994.1269. [DOI] [PubMed] [Google Scholar]
- 88.Kastan M B, Zhan Q, el-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 89.Kikuchi M, Sumiyoshi Y, Minamishima Y. Kikuchi’s disease (histiocytic necrotizing lymphadenitis) In: Ablashi D V, Krueger G R F, Salahuddin S Z, editors. Human herpesvirus-6: epidemiology, molecular biology, and clinical pathology. New York, N.Y: Elsevier Science Publishing; 1992. pp. 175–184. [Google Scholar]
- 90.Klemola E. Cytomegalovirus infection in previously healthy adults. Ann Intern Med. 1973;79:267–268. doi: 10.7326/0003-4819-79-2-267. [DOI] [PubMed] [Google Scholar]
- 91.Kouzarides T, Bankier A T, Satchwell S C, Preddy E, Barrell B G. An immediate early gene of human cytomegalovirus encodes a potential membrane glycoprotein. Virology. 1988;165:151–164. doi: 10.1016/0042-6822(88)90668-x. [DOI] [PubMed] [Google Scholar]
- 92.Krueger G R, Sander C, Hoffmann A, Barth A, Koch B, Braun M. Isolation of human herpesvirus-6 (HHV-6) from patients with collagen vascular diseases. In Vivo. 1991;5:217–225. [PubMed] [Google Scholar]
- 93.Kuerbitz S J, Plunkett B S, Walsh W V, Kastan M B. Wild-type p53 is a cell cycle checkpoint determinant following irradiation. Proc Natl Acad Sci USA. 1992;89:7491–7495. doi: 10.1073/pnas.89.16.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kumar A, Selim M S, Madden D L, Wallen W C, Vasquez H H, Nankervis G A. Humoral and cell-mediated immune responses to herpesvirus antigens in patients with cervical carcinoma. Gynecol Oncol. 1980;10:18–25. doi: 10.1016/0090-8258(80)90057-8. [DOI] [PubMed] [Google Scholar]
- 95.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 96.Lang D, Stamminger T. The 86-kilodalton IE-2 protein of human cytomegalovirus is a sequence-specific DNA-binding protein that interacts directly with the negative autoregulatory response element located near the cap site of the IE-1/2 enhancer-promoter. J Virol. 1993;67:323–331. doi: 10.1128/jvi.67.1.323-331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lang D J, Kummer J F. Demonstration of cytomegalovirus in semen. N Engl J Med. 1972;287:756–758. doi: 10.1056/NEJM197210122871508. [DOI] [PubMed] [Google Scholar]
- 98.Lang D J, Kummer J F. Cytomegalovirus in semen: observations in selected populations. J Infect Dis. 1975;132:472–473. doi: 10.1093/infdis/132.4.472. [DOI] [PubMed] [Google Scholar]
- 99.Lawrence G L, Chee M, Craxton M A, Gompels U A, Honess R W, Barrell B G. Human herpesvirus 6 is closely related to human cytomegalovirus. J Virol. 1990;64:287–299. doi: 10.1128/jvi.64.1.287-299.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lee W S, Kao C C, Bryant G O, Liu X, Berk A J. Adenovirus E1A activation domain binds the basic repeat in the TATA box transcription factor. Cell. 1991;67:365–376. doi: 10.1016/0092-8674(91)90188-5. [DOI] [PubMed] [Google Scholar]
- 101.Levine A J. The tumor suppressor genes. Annu Rev Biochem. 1993;62:623–651. doi: 10.1146/annurev.bi.62.070193.003203. [DOI] [PubMed] [Google Scholar]
- 102.Levine P H, Jahan N, Murari P, Manak M, Jaffe E S. Detection of human herpesvirus 6 in tissues involved by sinus histiocytosis with massive lymphadenopathy (Rosai-Dorfman disease) J Infect Dis. 1992;166:291–295. doi: 10.1093/infdis/166.2.291. [DOI] [PubMed] [Google Scholar]
- 103.Lin J, Chen J, Elenbaas B, Levine A J. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kDa protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 104.Lin Y S, Ha I, Maldonado E, Reinberg D, Green M R. Binding of general transcription factor TFIIB to an acidic activating region. Nature. 1991;353:569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- 105.Liu B, Hermiston T W, Stinski M F. A cis-acting element in the major immediate-early (IE) promoter of human cytomegalovirus is required for negative regulation by IE2. J Virol. 1991;65:897–903. doi: 10.1128/jvi.65.2.897-903.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu X, Miller C W, Koeffler P H, Berk A J. The p53 activation domain binds the TATA box-binding polypeptide in Holo-TFIID, and a neighboring p53 domain inhibits transcription. Mol Cell Biol. 1993;13:3291–3300. doi: 10.1128/mcb.13.6.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lowe S W, Schmitt E M, Smith S W, Osborne B A, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 108.Luppi M, Marasca R, Barozzi P, Ferrari S, Ceccherini-Nelli L, Batoni G, Merelli E, Torelli G. Three cases of human herpesvirus-6 latent infection: integration of viral genome in peripheral blood mononuclear cell DNA. J Med Virol. 1993;40:44–52. doi: 10.1002/jmv.1890400110. [DOI] [PubMed] [Google Scholar]
- 109.Lusso P, De Maria A, Malnati M, Lori F, DeRocco S E, Baseler M, Gallo R C. Induction of CD4 and susceptibility to HIV-1 infection in human CD8+ T lymphocytes by human herpesvirus 6. Nature. 1991;349:533–535. doi: 10.1038/349533a0. [DOI] [PubMed] [Google Scholar]
- 110.Lusso P, Ensoli B, Markham P D, Ablashi D V, Salahuddin S Z, Tschachler E, Wong-Staal F, Gallo R C. Productive dual infection of human CD4+ T lymphocytes by HIV-1 and HHV-6. Nature. 1989;337:370–373. doi: 10.1038/337370a0. [DOI] [PubMed] [Google Scholar]
- 111.Lusso P, Garzino-Demo A, Crowley R W, Malnati M S. Infection of gamma/delta T lymphocytes by human herpesvirus 6: transcriptional induction of CD4 and susceptibility to HIV infection. J Exp Med. 1995;181:1303–1310. doi: 10.1084/jem.181.4.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lusso P, Markham P D, Tschachler E, DiMarzo Veronese F, Salahuddin S Z, Ablashi D V, Pahwa S, Krohn K, Gallo R C. In vitro cellular tropism of human B-lymphotropic virus (human herpesvirus-6) J Exp Med. 1988;167:1659–1670. doi: 10.1084/jem.167.5.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Macias M P, Stinski M F. An in vitro system for human cytomegalovirus immediate early 2 protein (IE2)-mediated site-dependent repression of transcription and direct binding of IE2 to the major immediate early promoter. Proc Natl Acad Sci USA. 1993;90:707–711. doi: 10.1073/pnas.90.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Malkin D, Li F P, Strong L C, Fraumeni J F, Jr, Nelson C M, Kim D H, Kassel J, Gryka M A, Bischoff F Z, Tainsky M A, Friend S H. Germline mutations in familial syndrome of breast cancer, sarcomas and other neoplasias. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 115.Malone C L, Vesole D H, Stinski M F. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J Virol. 1990;64:1498–1506. doi: 10.1128/jvi.64.4.1498-1506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mansur C P, Marcus B, Dalal S, Androphy E J. The domain of p53 required for binding HPV 16 E6 is separable from the degradation domain. Oncogene. 1995;10:457–465. [PubMed] [Google Scholar]
- 117.Martin M E, Nicholas J, Thomson B J, Newman C, Honess R W. Identification of a transactivating function mapping to the putative immediate-early locus of human herpesvirus 6. J Virol. 1991;65:5381–5390. doi: 10.1128/jvi.65.10.5381-5390.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Maudsley D J, Pound J D. Modulation of MHC antigen expression by viruses and oncogenes. Immunol Today. 1991;12:429–431. doi: 10.1016/0167-5699(91)90013-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McDougall J K, Galloway D A, Fenoglio C M. Cervical carcinoma: detection of herpes simplex virus RNA in cells undergoing neoplastic change. Int J Cancer. 1980;25:1–8. doi: 10.1002/ijc.2910250102. [DOI] [PubMed] [Google Scholar]
- 120.McDougall J K, Nelson J A, Myerson D, Beckmann A M, Galloway D A. HSV, CMV, and HPV in human neoplasia. J Investig Dermatol. 1984;83:72s–76s. doi: 10.1111/1523-1747.ep12281204. [DOI] [PubMed] [Google Scholar]
- 121.Melnick J L, Hu C, Burek J, Adam E, DeBakey M E. Cytomegalovirus DNA in arterial walls of patients with atherosclerosis. J Med Virol. 1994;42:170–174. doi: 10.1002/jmv.1890420213. [DOI] [PubMed] [Google Scholar]
- 122.Melnick J L, Lewis R, Wimberly I, Kaufman R H, Adam E. Association of cytomegalovirus (CMV) infection with cervical cancer: isolation of CMV from cell cultures derived from cervical biopsy. Intervirology. 1978;10:115–119. doi: 10.1159/000148975. [DOI] [PubMed] [Google Scholar]
- 123.Miller G, Rigsby M O, Heston L, Grogan E, Sun R, Metroka C, Levy J A, Gao S J, Chang Y, Moore P S. Antibodies to butyrate-inducible antigens of Kaposi’s sarcoma-associated herpesvirus in patients with HIV-1 infection. N Engl J Med. 1996;334:1292–1297. doi: 10.1056/NEJM199605163342003. [DOI] [PubMed] [Google Scholar]
- 124.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 125.Moore P S, Gao S J, Dominguez G, Cesarman E, Lungu O, Knowles D M, Garber R, Pellett P E, McGeoch D J, Chang Y. Primary characterization of a herpesvirus agent associated with Kaposi’s sarcoma. J Virol. 1996;70:549–558. doi: 10.1128/jvi.70.1.549-558.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Muganda P, Mendoza O, Hernandez J, Qian Q. Human cytomegalovirus elevates levels of the cellular protein p53 in infected fibroblasts. J Virol. 1994;68:8028–8034. doi: 10.1128/jvi.68.12.8028-8034.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Muralidhar S, Doniger J, Mendelson E, Araujo J C, Kashanchi F, Azumi N, Brady J N, Rosenthal L J. Human cytomegalovirus mtrII oncoprotein binds to p53 and down-regulates p53 activated transcription. J Virol. 1996;70:8691–8700. doi: 10.1128/jvi.70.12.8691-8700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Myerson D, Hackman R C, Nelson J A, Ward D C, McDougall J K. Widespread presence of histologically occult cytomegalovirus. Hum Pathol. 1984;15:430–439. doi: 10.1016/s0046-8177(84)80076-3. [DOI] [PubMed] [Google Scholar]
- 129.Nelson J A, Fleckenstein B, Jahn G, Galloway D A, McDougall J K. Structure of the transforming region of human cytomegalovirus AD169. J Virol. 1984;49:109–115. doi: 10.1128/jvi.49.1.109-115.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nevins J R. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 131.Niederman J C, Liu C R, Kaplan M H, Brown N A. Clinical and serological features of human herpesvirus-6 infection in three adults. Lancet. 1988;ii:817–819. doi: 10.1016/s0140-6736(88)92783-3. [DOI] [PubMed] [Google Scholar]
- 132.Oren M, Maltzman W, Levine A J. Posttranslational regulation of the 54K cellular tumor antigen in normal and transformed cells. Mol Cell Biol. 1981;1:101–110. doi: 10.1128/mcb.1.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pacsa A S, Kummerländer L, Pejtsik B, Pali K. Herpesvirus antibodies and antigens in patients with cervical anaplasia and in controls. J Natl Cancer Inst. 1975;55:775–781. doi: 10.1093/jnci/55.4.775. [DOI] [PubMed] [Google Scholar]
- 134.Pizzorno M C, Hayward G S. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target sequence located near the cap site. J Virol. 1990;64:6154–6165. doi: 10.1128/jvi.64.12.6154-6165.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Pizzorno M C, O’Hare P, Sha L, La Femina R L, Hayward G S. trans-activation and autoregulation of gene expression by the immediate-early region 2 gene products of human cytomegalovirus. J Virol. 1988;62:1167–1179. doi: 10.1128/jvi.62.4.1167-1179.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Quinnan G V, Jr, Masur H, Rook A H, Armstrong G, Frederick W R, Epstein J, Manischewitz J F, Macher A M, Jackson L, Ames J, Smith H A, Parker M, Pearson G R, Parrillo J, Mitchell C, Strauss S E. Herpesvirus infections in the acquired immune deficiency syndrome. JAMA. 1984;252:72–77. [PubMed] [Google Scholar]
- 137.Rapp F. Cytomegalovirus and carcinogenesis. JNCI. 1984;72:783–787. [PubMed] [Google Scholar]
- 138.Rapp F, Geder L, Murasko D, Lausch R, Ladda R, Huang E, Webber M M. Long-term persistence of cytomegalovirus genome in cultured cells of prostatic origin. J Virol. 1975;16:982–990. doi: 10.1128/jvi.16.4.982-990.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rawls W E, Bacchetti S, Graham F L. Relation of herpes simplex viruses to human malignancies. Curr Top Microbiol Immunol. 1977;77:71–95. doi: 10.1007/978-3-642-66740-4_3. [DOI] [PubMed] [Google Scholar]
- 140.Razzaque A. Oncogenic potential of human herpesvirus-6 DNA. Oncogene. 1990;5:1365–1370. [PubMed] [Google Scholar]
- 141.Razzaque A, Jahan N, McWeeney D, Jariwalla R J, Jones C, Brady J, Rosenthal L J. Localization and DNA sequence analysis of the transforming domain (mtrII) of human cytomegalovirus. Proc Natl Acad Sci USA. 1988;85:5709–5713. doi: 10.1073/pnas.85.15.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Razzaque A, Zhu F, Jones C. Functional analysis of human cytomegalovirus morphological transforming region II (mtrII) Virology. 1991;181:399–402. doi: 10.1016/0042-6822(91)90513-b. [DOI] [PubMed] [Google Scholar]
- 143.Reich N C, Levine A J. Growth regulation of a cellular tumor antigen, p53, in nontransformed cells. Nature. 1984;308:199–201. doi: 10.1038/308199a0. [DOI] [PubMed] [Google Scholar]
- 144.Rogel A, Popliker M, Webb C G, Oren M. p53 cellular tumor antigen: analysis of mRNA levels in normal adult tissues, embryos, and tumors. Mol Cell Biol. 1985;5:2851–2855. doi: 10.1128/mcb.5.10.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Roizman B. Herpesviridae. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2221–2230. [Google Scholar]
- 146.Ruppert J M, Stillman B. Analysis of a protein-binding domain of p53. Mol Cell Biol. 1993;13:3811–3820. doi: 10.1128/mcb.13.6.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Salahuddin S Z, Ablashi D V, Markham P D, Josephs S F, Sturzenegger S, Kaplan M, Halligan G, Biberfeld P, Wong-Staal F, Kramarsky B, Gallo R C. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science. 1986;234:596–601. doi: 10.1126/science.2876520. [DOI] [PubMed] [Google Scholar]
- 149.Sanford E J, Dagen J E, Geder L, Rohner T J, Jr, Rapp F. Lymphocyte reactivity against virally transformed cells in patients with urologic cancer. J Urol. 1977;118:809–810. doi: 10.1016/s0022-5347(17)58203-8. [DOI] [PubMed] [Google Scholar]
- 150.Sanford E J, Geder L, Laychock A, Rohner T J, Jr, Rapp F. Evidence for the association of cytomegalovirus with carcinoma of the prostate. J Urol. 1977;118:789–792. doi: 10.1016/s0022-5347(17)58194-x. [DOI] [PubMed] [Google Scholar]
- 151.Sarnow P, Ho Y S, Williams J, Levine A J. Adenovirus E1B-58kd tumor antigen and SV40 large T antigen are associated with the same 54kd cellular protein in transformed cells. Cell. 1982;28:387–394. doi: 10.1016/0092-8674(82)90356-7. [DOI] [PubMed] [Google Scholar]
- 152.Schreier A A, Gruber J. Viral T antigen interactions with cellular proto-oncogene and anti-oncogene products. J Natl Cancer Inst. 1990;582:354–360. doi: 10.1093/jnci/82.5.354. [DOI] [PubMed] [Google Scholar]
- 153.Schuler G D, Altschul S F, Lipman D J. A workbench for multiple alignment construction and analysis. Proteins Struct Funct Genet. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 154.Seto E, Usheva A, Zambetti G P, Momand J, Horikoshi N, Weinmann R, Levine A J, Shenk T. Wild-type p53 binds to the TATA-binding protein and represses transcription. Proc Natl Acad Sci USA. 1992;89:12028–12032. doi: 10.1073/pnas.89.24.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Shen Y, Zhu H, Shenk T. Human cytomegalovirus IE1 and IE2 proteins are mutagenic and mediate “hit-and-run” oncogenic transformation in cooperation with the adenovirus E1A proteins. Proc Natl Acad Sci USA. 1997;94:3341–3345. doi: 10.1073/pnas.94.7.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Smiley M L, Mar E, Huang E. Cytomegalovirus infection and viral-induced transformation of human endothelial cells. J Med Virol. 1988;25:213–226. doi: 10.1002/jmv.1890250212. [DOI] [PubMed] [Google Scholar]
- 157.Smith M L, Chen I T, Zhan Q, Bae I, Chen C Y, Gilmer T M, Kastan M B, O’Conner P M, Fornace A J., Jr Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science. 1994;266:1376–1380. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- 158.Sommer M H, Scully A L, Spector D H. Transactivation by the human cytomegalovirus IE2 86-kilodalton protein requires a domain that binds to both the TATA box-binding protein and the retinoblastoma protein. J Virol. 1994;68:6223–6231. doi: 10.1128/jvi.68.10.6223-6231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sorlie P D, Adam E, Melnick S L, Folsom A, Skelton T, Chambless L E, Barnes R, Melnick J L. Cytomegalovirus/herpesvirus and carotid atherosclerosis: the ARIC study. J Med Virol. 1994;42:33–37. doi: 10.1002/jmv.1890420107. [DOI] [PubMed] [Google Scholar]
- 160.Soussi T, Caron de Fromentel C, May P. Structural aspects of the p53 protein in relation to gene evolution. Oncogene. 1990;5:945–952. [PubMed] [Google Scholar]
- 161.Speir E, Modali R, Huang E, Leon M B, Shawl F, Finkel T, Epstein S E. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science. 1994;265:391–394. doi: 10.1126/science.8023160. [DOI] [PubMed] [Google Scholar]
- 162.Sprecher-Goldberger S, Thiry L, Lefebvere N, Dekegel D, DeHalleux F. Complement fixation antibodies to adeno-associated viruses, cytomegaloviruses, and herpes simplex viruses in patients with tumors and in control individual. Am J Epidemiol. 1971;94:351–358. doi: 10.1093/oxfordjournals.aje.a121330. [DOI] [PubMed] [Google Scholar]
- 163.Stagno S, Pass R F, Cloud G, Britt W J, Henderson R E, Walton P D, Veren D A, Page F, Alford C A. Primary cytomegalovirus infection in pregnancy. Incidence, transmission to fetus, and clinical outcome. JAMA. 1986;256:1904–1908. [PubMed] [Google Scholar]
- 164.Stagno S, Reynolds D W, Pass R F, Alford C A. Breast milk and the risk of cytomegalovirus infection. N Engl J Med. 1980;302:1073–1076. doi: 10.1056/NEJM198005083021908. [DOI] [PubMed] [Google Scholar]
- 165.Stasiak P C, Mocarski E S. Transactivation of the cytomegalovirus ICP36 gene promoter requires the alpha gene product TRS1 in addition to IE1 and IE2. J Virol. 1992;66:1050–1058. doi: 10.1128/jvi.66.2.1050-1058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Staskus K A, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson D J, Ganem D, Haase A T. Kaposi’s sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Steeper T A, Horwitz C A, Ablashi D V, Salahuddin S Z, Saxinger C, Saltzman R, Schwartz B. The spectrum of clinical and laboratory findings resulting from human herpesvirus-6 (HHV-6) in patients with mononucleosis-like illnesses not resulting from Epstein-Barr virus or cytomegalovirus. Am J Clin Pathol. 1990;93:776–783. doi: 10.1093/ajcp/93.6.776. [DOI] [PubMed] [Google Scholar]
- 168.Stenberg R M, Depto A S, Fortney J, Nelson J A. Regulated expression of early and late RNAs and proteins from the human cytomegalovirus immediate-early gene region. J Virol. 1989;63:2699–2708. doi: 10.1128/jvi.63.6.2699-2708.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Stenberg R M, Fortney J, Barlow S W, Magrane B P, Nelson J A, Ghazal P. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J Virol. 1990;64:1556–1565. doi: 10.1128/jvi.64.4.1556-1565.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Stenberg R M, Thomsen D R, Stinski M F. Structural analysis of the major immediate early gene of human cytomegalovirus. J Virol. 1984;49:190–199. doi: 10.1128/jvi.49.1.190-199.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Stenberg R M, Witte P R, Stinski M F. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J Virol. 1985;56:665–675. doi: 10.1128/jvi.56.3.665-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Stoian M, Hozoc M, Iosipenco M, Bolocan J, Nastac E. Presence of antibodies to human cytomegalovirus in patients with different forms of cancer and in other categories of subjects. Virologie. 1982;33:153–161. [PubMed] [Google Scholar]
- 173.Stringer K F, Ingles C J, Greenblatt J. Direct and selective binding of an acidic transcriptional activation domain to the TATA-box factor TFIID. Nature. 1990;345:783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- 174.Subler M A, Martin D W, Deb S. Inhibition of viral and cellular promoters by human wild-type p53. J Virol. 1992;66:4757–4762. doi: 10.1128/jvi.66.8.4757-4762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Takahashi K, Sonoda S, Higashi K, Kondo T, Takahashi H, Takahashi M, Yamanishi K. Predominant CD4 T-lymphocyte tropism of human herpesvirus 6-related virus. J Virol. 1989;63:3161–3163. doi: 10.1128/jvi.63.7.3161-3163.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Takahashi K, Sonoda S, Kawakami K, Miyata K, Oki T, Nagata T, Okuno T, Kamanishi K. Human herpesvirus 6 and exanthem subitum. Lancet. 1988;i:1463. doi: 10.1016/s0140-6736(88)92275-1. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 177.Taya Y. RB kinases and RB-binding proteins: new points of view. Trends Biochem Sci. 1997;22:14–17. doi: 10.1016/s0968-0004(96)10070-0. [DOI] [PubMed] [Google Scholar]
- 178.Thompson J, Choudhury S, Kashanchi F, Doniger J, Berneman Z, Frenkel N, Rosenthal L J. A transforming fragment within the direct repeat region of human herpesvirus type 6 that transactivates HIV-1. Oncogene. 1994;9:1167–1175. [PubMed] [Google Scholar]
- 179.Thompson J, Doniger J, Rosenthal L J. A 79 amino acid oncogene is responsible for human cytomegalovirus mtrII induced malignant transformation. Arch Virol. 1994;136:161–172. doi: 10.1007/BF01538825. [DOI] [PubMed] [Google Scholar]
- 180.Thompson J, Inamdar A, Jahan N, Doniger J, Rosenthal L J. Localization and sequence analysis of morphological transforming region III within human cytomegalovirus strain Towne. Intervirology. 1993;36:121–127. doi: 10.1159/000150330. [DOI] [PubMed] [Google Scholar]
- 181.Thomson B J, Weindler F W, Gray D, Schwaab V, Heilbronn R. Human herpesvirus 6 (HHV-6) is a helper virus for adeno-associated virus type 2 (AAV-2) and the AAV-2 rep gene homologue in HHV-6 can mediate AAV-2 DNA replication and regulate gene expression. Virology. 1994;204:304–311. doi: 10.1006/viro.1994.1535. [DOI] [PubMed] [Google Scholar]
- 182.Torelli G, Barozzi P, Marasca R, Cocconcelli P, Merelli E, Ceccherini-Nelli L, Ferrari S, Luppi M. Targeted integration of human herpesvirus 6 in the p arm of chromosome 17 of human peripheral blood mononuclear cells in vivo. J Med Virol. 1995;46:178–188. doi: 10.1002/jmv.1890460303. [DOI] [PubMed] [Google Scholar]
- 183.Torelli G, Marasca R, Luppi M, Selleri L, Ferrari S, Narni F, Mariano M T, Federico M, Ceccherini-Nelli L, Bendinelli M, Montagnani G, Montorsi M, Artusi T. Human herpesvirus-6 in human lymphomas: identification of specific sequences in Hodgkin’s lymphomas by polymerase chain reaction. Blood. 1991;77:2251–2258. [PubMed] [Google Scholar]
- 184.Vestergaard B F, Hornsleth A, Pedersen S N. Occurrence of herpes- and adenovirus antibodies in patients with carcinoma of the cervix uteri. Measurement of antibodies to herpesvirus hominis (types 1 and 2), cytomegalovirus, EB-virus, and adenovirus. Cancer. 1972;30:68–74. doi: 10.1002/1097-0142(197207)30:1<68::aid-cncr2820300111>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 185.Wang J, Jones C, Norcross M, Bohnlein E, Razzaque A. Identification and characterization of a human herpesvirus 6 gene segment capable of transactivating the human immunodeficiency virus type 1 long terminal repeat in an Sp1 binding site-dependent manner. J Virol. 1994;68:1706–1713. doi: 10.1128/jvi.68.3.1706-1713.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wang J Y. Retinoblastoma protein in growth suppression and death protection. Curr Opin Genet Dev. 1997;7:39–45. doi: 10.1016/s0959-437x(97)80107-4. [DOI] [PubMed] [Google Scholar]
- 187.Wang J Y, Knudsen E S, Welch P J. The retinoblastoma tumor suppressor protein. Adv Cancer Res. 1994;64:25–85. doi: 10.1016/s0065-230x(08)60834-9. [DOI] [PubMed] [Google Scholar]
- 188.Wang P, Reed M, Wang Y, Mayr G, Stenger J E, Anderson M E, Schwedes J F, Tegtmeyer P. p53 domains: structure, oligomerization, and transformation. Mol Cell Biol. 1994;14:5182–5191. doi: 10.1128/mcb.14.8.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Wang Y, Reed M, Wang P, Stenger J E, Mayr G, Anderson M E, Schwedes J F, Tegtmeyer P. p53 domains: identification and characterization of two autonomous DNA-binding regions. Genes Dev. 1993;7:2575–2586. doi: 10.1101/gad.7.12b.2575. [DOI] [PubMed] [Google Scholar]
- 190.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 191.Werness B A, Levine A J, Howley P M. Association of human papillomavirus type 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 192.Weston K, Barrell B G. Sequence of the short unique region, short repeats, and part of the long repeats of human cytomegalovirus. J Mol Biol. 1986;192:177–208. doi: 10.1016/0022-2836(86)90359-1. [DOI] [PubMed] [Google Scholar]
- 193.Whyte P, Buchkovich K J, Horowitz J M, Friend S H, Raybuck M, Weinberg R A, Harlow E. Association between an oncogene and an antioncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature. 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- 194.Yamanishi K, Okuno T, Shiraki K, Takahashi M, Kondo T, Asano Y, Kurata T. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet. 1988;i:1065–1067. doi: 10.1016/s0140-6736(88)91893-4. [DOI] [PubMed] [Google Scholar]
- 195.Yeung K C, Stoltzfus C M, Stinski M F. Mutations of the human cytomegalovirus immediate-early 2 protein defines regions and amino acid motifs important in transactivation of transcription from the HIV-1 LTR promoter. Virology. 1993;195:786–792. doi: 10.1006/viro.1993.1431. [DOI] [PubMed] [Google Scholar]
- 196.Zhou Y, Chang C K, Qian G, Chandran B, Wood C. Trans-activation of the HIV promoter by a cDNA and its genomic clones of human herpesvirus 6. Virology. 1994;199:311–322. doi: 10.1006/viro.1994.1129. [DOI] [PubMed] [Google Scholar]
- 197.Zhu H, Shen Y, Shenk T. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.zur Hausen H, Schulte-Holthausen H, Klein G, Henle W, Henle G, Clifford P, Santesson L. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature. 1970;228:1056–1058. doi: 10.1038/2281056a0. [DOI] [PubMed] [Google Scholar]