Abstract
Iron is required by most living systems. A great variety of means of acquisition, avenues of uptake, and methods of storage are used by pathogenic fungi to ensure a supply of the essential metal. Solubilization of insoluble iron polymers is the first step in iron assimilation. The two methods most commonly used by microorganisms for solubilization of iron are reduction and chelation. Reduction of ferric iron to ferrous iron by enzymatic or nonenzymatic means is a common mechanism among pathogenic yeasts. Under conditions of iron starvation, many fungi synthesize iron chelators known as siderophores. Two classes of compounds that function in iron gathering are commonly observed: hydroxamates and polycarboxylates. Two major responses to iron stress in fungi are a high-affinity ferric iron reductase and siderophore synthesis. Regulation of these two mechanisms at the molecular level has received attention. Uptake of siderophores is a diverse process, which varies among the different classes of compounds. Since free iron is toxic, it must be stored for further metabolic use. Polyphosphates, ferritins, and siderophores themselves have been described as storage molecules. The iron-gathering mechanisms used by a pathogen in an infected host are largely unknown and can only be posited on the basis of in vitro studies at present.
Iron is required by most living systems (14, 37, 100). The metal has two readily available ionization states, Fe(II) and Fe(III), and is thus often used as a cofactor for oxidation-reduction enzymes (14, 94). While iron is the second most abundant metal on Earth (after aluminum), it is present in very insoluble compounds (oxides-hydroxides) in aerobic environments (37, 87a, 94). Fungi overcome this problem of unavailability in a variety of ways, and that variety is the major theme of this review.
Iron is toxic in uncontained situations because it catalyzes the production of free radicals (14, 32). Therefore, after uptake, storage of the accessed iron becomes essential in fungal metabolism of the metal to prevent repolymerization (87a) and toxicity (14). A number of different storage mechanisms are known. Polyphosphates may serve as vacuolar storage compounds in Saccharomyces cerevisiae (54). Among the zygomycetes, ferritin-like proteins function as iron storage compounds (61, 63). Such proteins have not been observed in ascomycetes or basidiomycetes; hydroxamate siderophores (iron chelators) serve instead as storage molecules in these phyla (61, 63).
This review is designed to cover the acquisition, transport, and storage of iron by pathogenic fungi. The major emphasis is on zoopathogens, but especially interesting or instructive examples among phytopathogens and nonpathogens are included.
ACQUISITION OF IRON
The various means by which fungi acquire iron are listed in Table 1. Included are methods of acquisition of iron from a variety of ferric chelates in iron-replete media [e.g., low-affinity Fe(III) reduction] and those regulated by iron concentration (e.g., methods involving the siderophores). The different means are not exclusive. For example, S. cerevisiae expresses a high-affinity Fe(III) reductase under conditions of low iron availability and may also utilize siderophores produced by other microorganisms in its environment. The use of siderophores which the yeast cannot itself synthesize may occur by uptake of the entire iron-ligand complex with intracellular release of iron by reduction or by extracellular reduction of iron and transport of the Fe(II) ion. Thus, iron mobilization is potentially a multifaceted process whose details vary in accordance with iron availability. It should be noted that some reports (Table 1) on siderophore formation by fungi are based solely on color reactions and that such reactions are not necessarily specific for iron-regulated siderophores (29, 75, 80).
TABLE 1.
Mechanisms of iron acquisition by pathogenic fungia
| Mechanism | Examples | Reference(s) |
|---|---|---|
| Reduction of ferric to ferrous iron | Candida albicans | 72 |
| Cryptococcus neoformans | 44, 78, 95 | |
| Geotrichum candidum | 71 | |
| Saccharomyces cerevisiaeb | 28, 49, 54, 88 | |
| Siderophore acquisition of ferric iron | ||
| Hydroxamates (families) | ||
| Rhodotorulic acid | Blastomyces dermatitidis | 13 |
| Epicoccum purpurescens | 33 | |
| Histoplasma capsulatum | 12 | |
| Stemphilium botryosum | 57 | |
| Coprogens | Blastomyces dermatitidis | 13 |
| Curvularia lunata | 94 | |
| Epicoccum purpurescens | 33 | |
| Fusarium dimerum | 94 | |
| Histoplasma capsulatum | 12 | |
| Neurospora crassa | 94 | |
| Stemphilium botryosum | 57 | |
| Ferrichromes | Aspergillus spp. | 20, 94 |
| Epicoccum purpurescens | 33 | |
| Microsporum spp. | 8, 70 | |
| Neurospora crassa | 94 | |
| Trichophyton spp. | 70 | |
| Ustilago maydis | 4, 5 | |
| Fusarinines | Aspergillus spp. | 94 |
| Epicoccum purpurescens | 33 | |
| Fusarium spp. | 94 | |
| Histoplasma capsulatum | 12 | |
| Paecilomyces spp. | 94 | |
| Unidentified in report referenced | Absidia corymbiferac | 40 |
| Candida albicansc | 40, 42, 90 | |
| Madurella mycetomatisc | 66 | |
| Pseudallescheria boydii | 26 | |
| Rhizopus arrhizusc | 40 | |
| Rhizopus oryzaec | 40 | |
| Scedosporium prolificansd | 26 | |
| Sporothrix schenckiic | 40 | |
| Polycarboxylates (rhizoferrin) | Zygomycetes | 94 |
| Phenolates-catecholates (chemical structures not identified) | Candida albicanse | 42 |
| Wood-rotting fungif | ||
| Miscellaneous iron resources | ||
| Hemin | Candida albicans | 69 |
| Histoplasma capsulatum | 102 | |
| β-Keto aldehydes (phytotoxins) | Stemphylium botryosum | 6, 58 |
| Acidification and mobilization | Neurospora crassa | 98 |
| Saccharomyces cerevisiae | 54 |
Examples given are those discussed in the review; both zoopathogens and phytopathogens are included. In some examples, general references are given in which additional reports about the listed fungus are cited.
Some strains are reported to be human pathogens (51).
Exact chemical structures not provided. Identified as hydroxamates by the Neilands method (40).
Assay based on color formation on CAS medium; no further work reported (26).
Exact chemical structures not provided; phenolates identified by the Arnow assay (42).
Reduction of Ferric Iron
S. cerevisiae.
Although S. cerevisiae is only rarely involved in human disease (51), it is chosen as the leading example of this type of iron acquisition because of the extent of the work done with it over the last 12 years (54). There are six ways in which S. cerevisiae can acquire iron: (i) iron reduction by a low-affinity (Km, 40 μM) Fe(III) reductase with transport of the Fe(II) ion; (ii) iron reduction by a high-affinity (Km, 0.15 μM) Fe(III) reductase followed by oxidation of the Fe(II) back to the Fe(III) form by a copper oxidase and transport of the Fe(III) ion; (iii) utilization of an iron-bearing siderophore (produced by some other microorganism) by Fe(III) reductase-mediated removal and transport of Fe(II); (iv) uptake of an iron-bearing siderophore with intracellular release of iron by a Fe(III) reductase; (v) nonenzymatic reduction of Fe(III) and transport of Fe(II); and (vi) acidification of a medium and deposition of iron onto cell walls of the fungus with subsequent mobilization by hydroxy acids.
The low-affinity system (Km, 40 μM) is used under the conditions of an iron-replete culture medium and also transports magnesium and calcium (54). The high-affinity transport system (Km, 0.15 μM) is regulated by iron; i.e., it occurs at very low iron concentrations (54). The high-affinity system is accompanied by a member of the copper oxidase family, which serves to oxidize the iron to the ferric [Fe(III)] form (28, 49, 88). The question naturally arises why S. cerevisiae should use opposing enzyme functions, reductase and oxidase, to transport iron under iron-limiting conditions. One suggested answer is that the low-affinity reductase system transports other ions [for example, Mg(II) and Ca(II)] and that oxidation would lend greater substrate specificity to the uptake process (28). But why, then, does the yeast not simply transport Fe(III) without bothering with the preliminary reduction step? The probable answer is that Fe(III) is bound to chelators in most environments and is relatively unavailable (28). However, some of these iron chelates can be recruited by S. cerevisiae, as discussed in the next paragraph.
The uptake of iron by S. cerevisiae in natural niches may be accomplished by other mechanisms. The yeast has been shown to utilize siderophores (iron chelators) synthesized by other fungi (e.g., rhodotorulic acid) or bacteria (e.g., ferrioxamine B) (54). Such utilization may be either by direct uptake of the iron-bearing ligand or by extracellular reductive release of iron from the ligand. In addition, S. cerevisiae is apparently able to mobilize iron by acidification of the environment with recruitment of iron, deposited onto its cell walls by citric or other hydroxy acids (37, 54). These alternate opportunities for iron acquisition are discussed in subsequent sections of this review.
Microorganisms and plants are known to excrete low-molecular-weight small phenolic compounds under conditions of low iron availability (54). These compounds could reduce ferric molecules to release Fe(II) for uptake, they could help to maintain a reduced environment and prolong the survival of Fe(II) at the fungal membrane, or they could solubilize Fe(III) (54). In S. cerevisiae the main compounds of this sort are anthranilate and 3-hydroxyanthranilate (54). However, these compounds do not appear in increased amounts in an Fe-depleted medium, and their role in uptake by the yeast is considered to be limited (54). The main component of yeast cell reducing activity is the cell membrane-bound reductase activity. The results so far achieved indicate that the cell surface has several reductase systems or that a multicomponent reductase system exists (54).
C. neoformans.
Although Cryptococcus neoformans is a basidiomycetous yeast and S. cerevisiae is an ascomycetous one, there is great similarity in their methods of acquiring iron. Neither of the yeasts synthesizes siderophores, and both utilize Fe(III) reductases as major means of iron acquisition. Moreover, both kinds of yeasts can mobilize iron from siderophores formed by other microorganisms and both can use nonenzymatic means of reducing iron (43, 44, 78, 95).
Fe(III) reduction is used by C. neoformans for the uptake of iron. Both high-affinity (Km, 0.16 μM) and low-affinity systems have been described (44). The Km value for the low-affinity system was not reported (44). Copper starvation drastically decreased the high-affinity system suggesting that, as in S. cerevisiae, copper metabolism may be involved (44). Nonenzymatic reduction of Fe(III) with transport of Fe(II) ions was observed with secreted 3-hydroxyanthranilic acid and melanin (78). The latter compound is produced by C. neoformans by the oxidation of catecholamines in vitro and thus represents a natural means of iron recruitment that could be visualized as occurring in vivo (22, 78; however, see reference 55a). In contrast to the situation with S. cerevisiae, 3-hydroxyanthranilic acid secretion by C. neoformans was regulated by the iron concentration in the medium (78).
C. neoformans can also utilize the siderophore ferrioxamine B, but there is no evidence that it can recruit iron from host proteins such as transferrin (43). Although the yeast can utilize ferric citrate as an iron source, there is no evidence that citrate alone can act as an iron chelator useful to C. neoformans (43). Likewise, while the capsule of the fungus is polyanionic and could serve as a nucleus for ferric hydroxide polymers, there is no evidence that it does so (43). Moreover, the acidic pH required for this acquisition mechanism by hydroxy acids (see above) would be unlikely in cultures of the nonfermentative C. neoformans or in vivo.
C. albicans.
The commensal Candida albicans is phylogenetically closely related to S. cerevisiae, and genes with homologous functions are highly conserved (89). Therefore, one would anticipate a similarity in iron uptake between the two yeasts, and, indeed, recent studies establish those similarities. A cell-associated Fe(III) reductase which is regulated by iron concentration has been revealed in C. albicans (72). A copper reductase has also been found that is both iron and copper regulated (72). Thus, copper metabolism appears to be involved in the ferric reductase activity (72).
C. albicans can mobilize Fe(III) from siderophores (ferrochrome and ferrioxamine B), but the mechanisms involved have not been studied (67). The somewhat controversial topic of whether C. albicans synthesizes siderophores and, if so, of what chemical sort will be discussed in a subsequent section, as will the utilization of other iron sources, e.g., hemin.
G. candidum.
Geotrichum candidum, like S. cerevisiae, does not form siderophores under conditions of iron starvation (71). Both Fe(II) and Fe(III) uptake systems are displayed by the fungus under conditions of iron starvation (71). The two systems were independent, and there was no evidence that the Fe(III) was reduced before uptake (71). In addition, an uptake system for ferrioxamine B was detected (71).
H. capsulatum.
The dimorphic zoopathogen Histoplasma capsulatum exists in the tissue of a host or in appropriate culture media at 37°C as a unicellular yeast (51). Moreover, the fungus is a facultative intracellular parasite of mononuclear phagocytes, a location that presents special problems of iron acquisition (34, 79). Uptake systems for both ferrous and ferric iron are suggested by the inhibitory effects of chelators, specific for each form of iron, on the intracellular growth of H. capsulatum (76, 77).
Production of Siderophores
Under conditions of extreme iron stress, fungi produce low-molecular-weight (Mr <1,500) ferric iron chelators known collectively as siderophores (37, 39, 86, 91, 94, 100). Most of the fungal siderophores are hydroxamates. However, the zygomycetes form iron-regulated polycarboxylates and there are well-documented reports of phenolate-catecholates in species of the wood-rotting fungi. The compounds were identified by comparison to known phenolic siderophores separated by paper chromatography (30, 31). There is also an unconfirmed report of phenolates being formed by C. albicans, but this report was based solely on color reactions (42). Siderophores are named on the basis of their iron-charged forms, while the deferrated form (the one that gathers iron) is called deferri-siderophore or desferri-siderophore (30, 31, 40, 42, 90).
Detection of fungal siderophores.
Detection methods have been covered in detail in several monographs (29, 75, 80), but since some of the ensuing discussion involves methods employed by investigators, a brief summary is given here.
As one investigator has commented (84), those of us working on fungal siderophores are singularly blessed: the compounds are produced in large excess by fungi in iron-starved cultures and turn red upon capture of Fe(III). Thus, we need only design a limited-iron medium for the fungus of our choice (23), grow it in the limited-iron medium, periodically add iron salts to a centrifuged sample, and, when a sample turns red, harvest the culture medium for further processing.
The iron salts procedure commonly used to detect siderophores is the method of Arkin et al. (75, 80). The culture supernatants are mixed with a solution containing 5 mM Fe(ClO4)3 in 0.1 M HClO4. The method is semiquantitative, and the amount of siderophore can be estimated by measurement of the optical density at 510 nm. The test may be used only as a rapid screening method because it lacks both specificity and sensitivity (75). Another colorimetric assay involves the use of the dye Chrome Asurol S (CAS) complexed with hexadecyltrimethylammonium bromide (HDTMA) (29, 75, 80). The removal of iron from CAS-HDTMA complex by a siderophore turns the dye yellow. The dye may also be included in a nutrient medium to measure the ability of an organism to remove iron from the dye complex (i.e., to make siderophores) by direct cultivation, a use that can be adapted to screening for non-siderophore-producing mutants (74, 82). A caveat is, of course, that the medium must support the growth of the organism—sometimes a bit tricky because of the potential toxicity of the HDTMA used to form the dye complex (29, 74, 75, 80). Unfortunately, the CAS assay does have drawbacks. For example, phosphate strips iron off the dye, as does cysteine (see reference 75 for a more complete discussion). Since there are several iron-binding substances that are not functional siderophores but nevertheless give positive color reactions with iron salts or CAS (75), the presence of hydroxamates or phenolates must be tested for (carboxylates are a different matter [94]). Hydroxamates may be detected by the Csáky assay (75, 80), in which the end product detected is nitrite. The test is highly specific for hydroxamates (75). The Neilands assay (75) for hydroxamates identifies a cis-nitroso alkali dimer formed by periodic acid oxidation that is detected at 264 nm. The Neilands assay has a drawback of questionable specificity: the originator has said, “By working so deep in the ultraviolet it is difficult to have confidence that the material being measured is actually the desired dimer” (75). Phenolates-catecholates are identified by the Arnow assay (75, 80), in which the centrifugate is treated with nitrous acid, molybdate, and alkali, which yields a pink compound that is detected at 515 nm. All of the color assays are subject to some uncertainties (75, 94), and chemical characterization is required to identify the assay reactivity as that of a siderophore (see the next section, on chemical structure).
The biological activity of a hydroxamate may be measured in a bioassay that employs a bacterium, Aureobacterium (Arthrobacter) flavescens JG.9 (ATCC 29091), which requires hydroxamate siderophores for mobilization of iron, as reflected by its growth (11, 29, 75). The bacterium uses a wide range of hydroxamates for its growth needs (75). Of course, A. flavescens may not have an appropriate receptor for a given compound. Thus, an assay procedure should eventually be developed that uses the siderophore-generating fungus to assess the biologic reactivity of the putative siderophore for the fungus from which it is isolated (80).
There are bioassays for phenolates. These involve strains of Salmonella typhimurium or Escherichia coli defective in the biosynthesis of enterochelin (for details, see reference 75).
Once it has been determined that a presumed siderophore is a hydroxamate (or phenolate), an excess of iron salts (often FeCl3) is added to the harvested culture medium and the presence of red color is monitored through various procedures designed to isolate, purify, and characterize the compound(s) giving reactivity. This last step, chemical characterization, is essential. There are reports of siderophores for a given fungus that are based on the formation of a red color in the presence of iron salts or a yellow color in the presence of CAS. These initial color reactions for iron chelation may be followed by an assay for hydroxamates or phenolates that are again based on color formation, but no chemical structure studies have been presented. It has been pointed out that “a variety of cellular materials from lysed cells may react positively in tests based on color formation” (94). Thus, precise chemical characterization is required for further work with the putative siderophore. Such chemical characterization may be performed by thin-layer chromatography, high-performance liquid chromatography, nuclear magnetic resonance, and mass spectroscopy (94).
Hydroxamates.
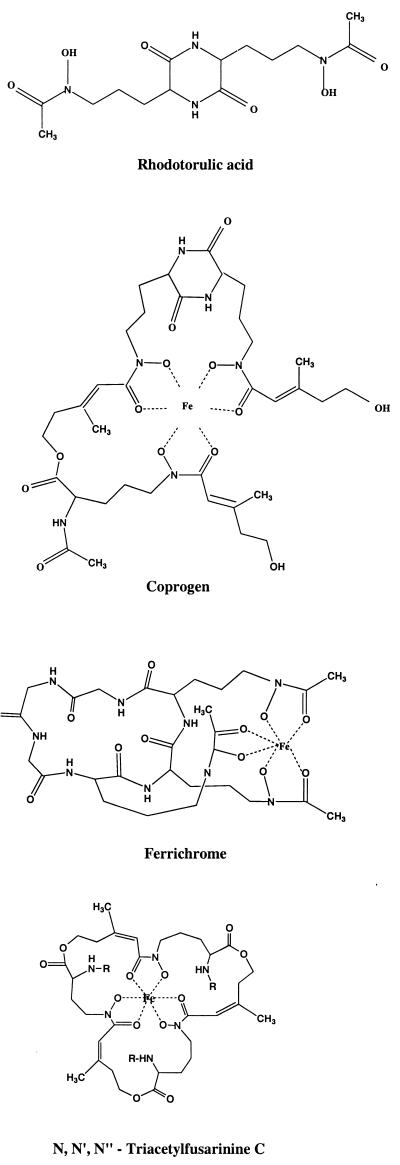
The hydroxamates all contain an N-δ-hydroxyornithine moiety. My descriptions of chemical structures are those given by Höfte (39). More complete consideration of the chemistry of the hydroxamates has been presented on a number of occasions (45, 60, 91, 94, 100).
There are four recognized families of hydroxamates, as well as other hydroxamates reported to be produced by certain fungi.
(i) Rhodotorulic acid.
Rhodotorulic acid is the diketopiperazine of N-δ-acetyl-l-N δ-hydroxyornithine (Fig. 1). It has been found mainly in basidiomycetous yeasts (94). A derivative of rhodotorulic acid is a dihydroxamate named dimerum acid; 3 mol of dimerum acid (DA) is combined with 2 mol of iron (Fe2DA3) to form the iron-bearing ligand. The binding of iron is weaker than that with the other three hydroxamate families. Dimerum acid is produced by some phytopathogens (e.g., Stemphylium botryosum and Epicoccum purpurescens [33, 57]) and by H. capsulatum (12) and Blastomyces dermatitidis (13) among the medically important fungi.
FIG. 1.
Representative hydroxamate siderophores. Adapted from reference 100 with permission of the publisher.
(ii) Coprogens.
Coprogens are composed of 3 mol of N-δ-acyl-N-δ-hydroxy-l-ornithine, 3 mol of anhydromevalonic acid, and 1 mol of acetic acid (Fig. 1). Unlike the situation with the rhodotorulic acid family, 1 mol of iron combines with one ligand in the coprogen, ferrichrome, and fusarinine families (94). The coprogens are produced by a number of plant pathogens (39, 57), by H. capsulatum (12), by B. dermatitidis (13), and by the occasional human pathogens Fusarium dimerum and Curvularia lunata (94).
The coprogens may be considered trihydroxamate derivatives of rhodotorulic acid with a linear structure, and by such reckoning the number of hydroxamate families would be reduced to three (the view of the separateness of the rhodotorulic acid family has been adopted to keep the presentation [Table 1] less cluttered). It is known that the hydrolysis of the ester group of coprogen B results in 1 mol of dimerum acid and 1 mol of trans-fusarinine (100). In the original report on DA (referred to as “dimerumic acid” in that report) from H. capsulatum (12), the title implied that it was a degradation product of coprogen B. In fact, hydrolysis of the ester bond of coprogen gives rise to DA and trans-fusarinine (100), but DA is the predominant hydroxamate in liquid shake cultures of H. capsulatum (12).
(iii) Ferrichromes.
Ferrichromes are cyclic peptides containing a tripeptide of N-δ-acyl-N-δ-hydroxyornithine and combinations of glycine, serine, or alanine (Fig. 1). Among the pathogenic fungi, ferrichromes are produced by some phytopathogenic fungi (39) and by Microsporum spp. (8, 70), Trichophyton spp. (70), and Aspergillus spp. including the important pathogen A. fumigatus (53, 94). Another function of ferrichromes is the intracellular storage of iron (see below).
(iv) Fusarinines.
Fusarinines, also called fusigens, may be either linear or cyclic hydroxamates. Fusarinine is a compound in which N-hydroxyornithine is N acylated by anhydromevalonic acid (Fig. 1). Among zoopathogens, various fusarinines are found in Fusarium spp., Paecilomyces spp., and Aspergillus spp. (94). Compounds identified as trans-fusarinine and an unidentified monohydroxamate were observed in culture filtrates of H. capsulatum, but they did not have biological activity (i.e., they did not stimulate growth of the fungus) (12). The work on siderophores of H. capsulatum was initiated because of the well-known fact that the fungus does not clone in its yeast cell phase of growth on most culture media in vitro (12, 102). Burt used hydroxamates isolated from culture filtrates to relieve the growth restriction (12). The same strategy, i.e., growth stimulation, was used by Castaneda et al. (19) in a study of Paracoccidioides brasiliensis. The siderophores coprogen B and DA isolated from B. dermatitidis by Burt (13) and sent to Castaneda et al. were used to improve the plating efficiency of P. brasiliensis (19).
(v) Unidentified hydroxamates.
There are unconfirmed reports of formation of hydroxamates by C. albicans (40, 42, 90). No exact chemical structures were presented, and it has been suggested by others (94) that long-term cultivation (20 days) of a fungus will result in materials that react positively in tests based on color formation. Cutler and Han (25) have reported their failure to display siderophores in C. albicans. The synthesis of hydroxamate siderophores by C. albicans has not been proven by chemical characterization.
Hydroxamates have been reported in other zoopathogens: Absidia corymbifera, Madurella mycetomatis, Pseudallescheria boydii, Rhizopus arrhizus, R. oryzae, Scedosporium prolificans, and Sporothrix schenckii. The report of hydroxamates from the zygomycetes A. cormybifera, R. arrhizus, and R. oryzae is unconfirmed (94). It appears that an altogether different class of siderophores, the polycarboxylates, are formed by zygomycetes (94). The report on P. boydii and S. prolificans was based solely on a positive CAS reaction. No studies of siderophore function or chemical characterization were performed (26). The report on M. mycetomatis was based on the CAS reaction (66). The report on S. schenckii was based on a reaction with iron salts and the use of the Neilands method for hydroxamates (40).
Diversity in siderophore synthesis is observed. Sometimes this diversity is with regard to a number of representatives in a single family, and at other times it is reflected in the synthesis of a number of representatives in several families. Frederick et al. provided evidence for synthesis of siderophores belonging to all four families of hydroxamates by Epicoccum purpurescens (33), and Höfte reported that Aspergillus ochraceus “…can produce up to 10 or more different siderophores…” (39). The reason for diversity is thought to be the ability of organisms to adapt to a wide variety of environmental situations (39, 100). This idea is especially interesting for pathogens for which a host immune response may be involved (see “Siderophores as pathogenic factors,” below). Among bacteria there are notable examples of diversity of molecular detail in siderophores. For example, Mycobacterium tuberculosis has been shown to synthesize 15 molecular varieties of exochelin (35).
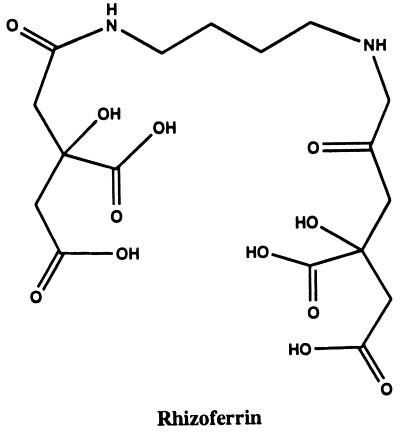
Polycarboxylates.
Although there is one incomplete report of hydroxamates among the zygomycetes (40), these fungi have not been observed by others to synthesize siderophores of the hydroxamate sort. Instead, a citric acid-containing polycarboxylate called rhizoferrin has been isolated from Rhizopus microsporus var. rhizopodiformis (94). The molecule contains two citric acid units linked to diaminobutane (Fig. 2). Rhizoferrin is widely distributed among the members of the phylum Zygomycota, having been observed in the order Mucorales (families Mucoraceae, Thamndidiaceae, Choanephoraceae, and Mortierellaceae) and in the order Entomophthorales (94).
FIG. 2.
The polycarboxylate rhizoferrin. Adapted from reference 100 with permission of the publisher.
Phenolates-catecholates.
Most investigators consider that the phenolate-catecholate class of siderophores is not produced by fungi (see e.g., references 39 and 94). Only two exceptions have been reported. The first exception is C. albicans (42), and this study was flawed by a lack of chemical structure studies. Phenolate siderophores were not found by two other groups of investigators (26, 40) who did, however, report hydroxamates in C. albicans. The other exception involves certain wood-rotting fungal species (30, 31). The siderophores were detected by the CAS dye and identified as phenolates by comparison to standard phenolates (e.g., enterochelin), and both the siderophores and the standard phenolates were separated by paper chromatography and visualized by UV fluorescence (31). Further chemical structure studies were not reported, and the authors report that the compounds “…appear to be phenolate in character” (48).
Miscellaneous Iron Resources
Hemin.
The use of hemin as a source of iron by bacteria is well known (see, e.g., reference 83). Iron-containing products of hemoglobin degradation are known to stimulate two pathogenic fungi, C. albicans and H. capsulatum.
Normal human serum inhibits the growth of C. albicans, and this fungistasis is thought to be largely due to the presence of transferrin, which binds the iron and reduces its concentration to levels that will not support growth of the fungus (41). This is a general phenomenon observed in vitro with many fungi (15, 41). Moors et al. (69) have shown that hemoglobin or hemin restores the growth of C. albicans in a serum-containing medium to which they are added. Moreover, these workers have detected Candida surface proteins that are homologues of mammalian complement receptors and can rosette complement-coated erythrocytes, thereby obtaining erythrocyte-derived iron for growth. These studies indicate an iron-gathering ability by C. albicans that may obviously have in vivo significance (18, 69). It should be pointed out that while an antifungal effect is attributable to transferrin, its primary function in the animal is to solubilize iron and effect its transport into the cell, where it is metabolized (92, 96).
The growth of H. capsulatum is also stimulated by addition of hemin to culture media (102). However, the fungus does not produce a hemolysin for human erythrocytes (87), and the in vivo significance would be limited to situations where a supply of heme, heme proteins, or hemin was available.
β-Ketoaldehyde phytotoxin.
Barash et al. (6) have isolated and chemically characterized two phytotoxins from Stemphylium botryosum f. sp. lycopersici. The synthesis of these toxins is iron regulated, and the ligands chelate Fe(III). The stemphylotoxins share some of the properties with the hydroxamate siderophores also synthesized by S. botryosum (Table 1), namely, regulation by iron and preferred binding of Fe(III). However, the toxins differ from the hydroxamates (dimerum acid and coprogen B) in their dependence on low but not extremely low iron concentrations for biosynthesis and a much lower affinity for Fe(III). Hence, the stemphylotoxins may act as chelators at low iron concentrations whereas DA and coprogen B come into play only at extreme levels of iron deficiency (6).
Bacterial chelators not known to be used by zoopathogenic fungi.
(i) Cysteine.
The amino acid cysteine is well known to bind iron (1 mol of iron to four cysteine residues). Legionella pneumophila does not produce hydroxamate siderophores, but culture filtrates were observed to decolorize CAS plates (55). The activity was related to the cysteine content of the medium.
(ii) α-Keto acids.
The α-keto acids are excreted by bacteria and may serve a siderophore function (27). This topic has not been extensively explored with pathogenic fungi. Although one example has been reported (i.e., α-keto glutarate was excreted by C. neoformans), no consistent relationship to iron limitation was observed (78).
(iii) Hydroxy acids.
Many fungi excrete Krebs cycle intermediates such as citric, malic, and succinic acids. The concentration of these hydroxy acids in a growth medium can be quite high (>1 μM) (54). These compounds, especially citric acid, are potential ligands of iron. One way in which they might do so is explained in “Acidification and mobilization,” below.
Utilization of Siderophores by Nonproducers
In a number of instances, pathogenic fungi have been shown to use siderophores even though they cannot synthesize them. Some examples are given below.
S. cerevisiae.
Ferrioxamine B, ferricrocin, and rhodotorulic acid are iron sources for S. cerevisiae. Two mechanisms for utilization of ferrioxamine B have been described: (i) at relatively high concentrations (360 μM), the iron is made available by reductive dissociation, while (ii) at low iron concentrations (7 μM), yeasts transport the entire iron-bearing ligand with iron into the cells and reductively remove the iron internally (54).
C. neoformans.
Growth of C. neoformans was stimulated by ferrioxamine B (160 μM) added to an iron-depleted medium (43). Although diffusion of the siderophore would dilute the concentration from that applied to the paper disc used, the level of siderophores close to the disc would probably be high enough to indicate a reductive mobilization of Fe2+ from the ligand (by analogy to S. cerevisiae). Studies on very low concentrations of the siderophore were not conducted, but ligand uptake is also possible.
C. albicans.
Ferrichrome and several of its constitutive peptides stimulated the growth of C. albicans in an iron-depleted medium (67). There is no information on whether utilization involves reductive mobilization or ligand uptake or concentration-dependent utilization of one or the other mechanisms for iron recruitment.
U. maydis.
Studies of the phytopathogen Ustilago maydis have provided fascinating evidence for utilization of both synthesized and nonsynthesized siderophores. Under iron-limiting conditions, U. maydis produces ferrichrome and ferrichrome B. However, under laboratory conditions, it also uses the bacterial siderophore ferrioxamine B (4, 5). Ferrichrome was taken up by entire-ligand transport, while the ferrioxamine B was utilized by reductive removal of the Fe(III) and transport of the Fe(II) (4). Nonenzymatic NADH- and flavin mononucleotide-dependent reduction of ferric siderophores has also been recorded (1). Although utilization of an extraneous bacterial siderophore would not affect in vivo recruitment of iron by the phytopathogenic fungus, its saprophytic existence in soil could be influenced. The interest in such an occurrence is heightened by the fact that the concentration of ferrioxamine B in the soil can be as high as 0.1 μM (54). The same arguments can be raised with regard to zoopathogenic fungi with a saprophytic form of growth that constitutes the infectious phase of the pathogen.
Rhizopus spp.
The occurrence of mucormycosis in patients being treated with Desferal (a methanesulfonate salt of desferrioxamine) clearly indicates the utilization of this bacterial siderophore by Rhizopus spp. (9).
P. brasiliensis.
The growth of Paracoccidioides brasiliensis is stimulated by coprogen B and DA synthesized by B. dermatitidis (19). P. brasiliensis may synthesize its own siderophores under conditions of iron stress, but appropriate studies have not been conducted.
Host iron proteins.
In a mammalian host, iron is bound to transferrin and lactoferrin or stored in ferritin. No fungus has been reported to be able to remove iron from transferrin or lactoferrin, although several bacteria and some animal parasites can do so (79, 97).
Acidification and Mobilization
Many fungi can grow anaerobically and will acidify their growth medium. It has been suggested for Neurospora crassa that under acidic conditions iron could accumulate at the cell surface and be mobilized by excreted hydroxy acids to supply iron to the cell (98). This means of iron acquisition has been mentioned for S. cerevisiae in its ecological locations in nature (54). However, no evidence was found for participation of citric acid or other polycarboxylic acids in iron uptake by C. neoformans (43).
TRANSPORT OF SIDEROPHORES
Transport Proteins
In bacterial systems, iron-regulated proteins occur and are expressed under conditions of iron depletion, often in concert with siderophore synthesis (24, 81). Proteins from the plasma membrane of N. crassa that had been grown in iron-replete and iron-depleted media were compared (94). There were no significant differences in the sodium dodecyl sulfate SDS-gel electrophoretic profiles of the two sources of proteins. There was no observed overproduction of membrane proteins like that seen in bacterial systems (81). Because at least fivefold differences in transport rates were observed, the authors suggested that the membrane siderophore transport system is constitutively expressed (94). In contrast to the work with N. crassa, iron-regulated outer membrane proteins have been found in the mycetoma-causing fungus, Madurella mycetomatis (66). One unique protein and three amplified ones were observed under iron-limiting conditions (66). The situation with other zoopathogens has not been explored.
Mechanisms of Hydroxamate-Iron Transport
There are four described mechanisms of siderophore iron uptake across the cytoplasmic membrane of fungi. The following summary is modified from descriptions given by others (16, 21, 59, 62, 64, 73, 94, 99, 101).
Shuttle mechanism.
The intact siderophore-iron complex is taken into the cell. The iron is released by a reductase or by direct ligand exchange in which the recipient siderophore becomes the storage molecule. The gathering ligand is released to capture another iron molecule. This mechanism is the one used for uptake by siderophores of the coprogen and ferrichrome families (61, 101).
Direct-transfer mechanism.
Iron is taken up without entrance of the ligand into the cell. The iron transfer is not a membrane-reductive event (73) but is a membrane-mediated exchange between the gathering siderophore and an internal chelating agent (16). The transfer mechanism may be by ligand exchange (nonenzymatic) to an internal pool of the chelating agent, which then serves as the storage compound (61). This type of transfer has been reported with the rhodotorulic acid family of siderophores.
Esterase-reductase mechanism.
The esterase-reductase mechanism was shown to operate with the ferric triacetylfusarinine C (99, 100). The ester bonds of the iron ligand are split after uptake of the ligand, the fusarinine moieties are excreted, and the ferric iron is reduced and stored (by an unknown mechanism).
Reductive mechanism.
Another reductive mechanism appears to be involved in the transport of some ferrichromes, which were shown not to enter cells but, rather, to give up ferric iron by reduction with transport of the ferrous iron (99, 100). The storage mechanisms are not known. This sort of transport is commonly expressed by fungi utilizing siderophores that they, themselves, do not synthesize (see “Miscellaneous iron resources,” above).
Mechanisms of Carboxylate Transport
The uptake of rhizoferrin has been studied. Transport of the entire ligand is observed, and a Km value of 8 mM has been determined (94). It is interesting that both ferrioxamines B and C are taken up at similar rates by Rhizopus microsporus var. rhizopodiformis, the species used to study rhizoferrin. This observation probably accounts for the cases of mucormycosis seen in patients treated with Desferal (desferrioxamine mesylate) (9, 93).
Phenolate transport has not been studied in either of the instances in which it was reported to occur in fungi (30, 42).
STORAGE OF IRON
Importance
Free iron is a devastating metal. In the Fenton reaction, Fe(II) reacts with H2O2 (a normal metabolite in aerobic organisms) to form a hydroxy radical, which binds to critical molecules found in living cells: sugars, amino acids, phospholipids, DNA bases, and organic acids (14, 32, 96). Should the supply of Fe(II) dwindle, more is generated by the reduction of Fe(III) by superoxide anions (14, 32). In S. cerevisiae, the action of an Fe(III) reductase is followed by that of an Fe(II) oxidase, which generates Fe(III) for uptake. The Fe(III) would tend to polymerize to an insoluble form were it not prevented from so doing (14). Therefore, some method of iron storage must be used to escape the toxic carnage that would result from the occurrence of free iron and to prevent repolymerization of the iron transported into the fungal cell.
Storage Molecules
The need for iron storage is universal. Soon after uptake, iron is found in the vacuoles of S. cerevisiae, where it is perhaps bound to polyphosphates (54). The vacuoles serve as “major” storage compartments for iron in the yeast (54). The reduced iron within the compartment is kept in the ferrous form and serves as the substrate for ferrochelatase, which is the enzyme involved in the insertion of iron into heme (52). It is believed that intracellular movement of iron could be effected by intracellular citric and malic acids (54). Polyphosphate ferrous iron storage has also been revealed in the low-molecular-mass iron pool of Escherichia coli (10).
Iron-rich proteins have been discovered in animals (ferritin), plants (phytoferritins), and bacteria (bacterioferritins) (92). However, surprisingly few such molecules have been described in fungi. Those that have been are found in members of the Zygomycota (61). Three types of ferritins have been described: (i) mycoferritin, which resembles mammalian ferritins; (ii) zygoferritin, a unique form of ferritin found only in the zygomycetes; and (iii) a bacterioferritin found in Absidia spinosa (17).
Ferritin-like molecules have not been discovered among members of the phyla Ascomycota and Basidiomycota. Work on the ascomycetes has focused on N. crassa and Aspergillus ochraceus. N. crassa forms the two predominant hydroxamate siderophores coprogen and ferrichrocin. The major extracellular siderophore formed under conditions of iron depletion is desferricoprogen. Desferriferricrocin is found mostly intracellularly. After uptake of coprogen (the iron-charged desferricoprogen), iron is released and transferred to desferricrocin by ligand exchange (which is not necessarily enzymatically mediated [64]), and the desferricrocin thereby becomes the ferricrocin and serves as the main intracellular iron storage compound (65). Ferrocrocin also serves as a long-term iron storage siderophore in A. ochraceus (94).
The basidiomycetes that have been studied are Rhodotorula minuta and Ustilago sphaerogena. Biosynthesis of rhodotorulic acid is characteristic of the heterobasidiomycetous yeasts. Other members of the Ustilagnaceae produce ferrichrome-type siderophores. Rhodotorulic acid (RA) forms a complex with iron, Fe2RA3, that is not transported across the membrane; rather, the iron is transferred by ligand exchange to an internal pool of rhodotorulic acid that then functions as the iron storage molecule (61). It should be noted that energy-dependent ligand exchange occurs at the membrane. It is not a simple exchange between Fe2(RA)3 and internal desferrirhodotorulic acid; an additional mediator is required (61).
In U. sphaerogena, iron is transported by ferrichrome A, which does not accumulate. The iron storage compound is ferrichrome (61). In the ascomycetes and basidiomycetes that have been studied, hydroxamate-type siderophores are the iron storage molecules.
In summary, iron storage ferritin-like compounds have been found only in the zygomycetes. The iron storage function in ascomycetes and basidiomycetes is performed by polyphosphates and hydroxamates.
SIDEROPHORES AS PATHOGENETIC FACTORS
Activities
Phytopathogens.
Many siderophores are produced by both bacterial and fungal plant pathogens, but their role in pathogenesis is largely unknown (56). The siderophore of the bacterial phytopathogen Erwinia chrysanthemi, chrysobactin, has been identified as a virulence factor (86). However, the fungal phytopathogen Verticillium dahlia produces the hydroxamate siderophores coprogen B and DA, but efforts to detect these compounds in plants grown under iron-limited conditions were unsuccessful (7).
Phytotoxins.
Only a single example of phytotoxins is discussed. Stemphylotoxin I and II are produced by Stemphilium botryosum f. sp. lycopersici (6, 58) and are ferric iron chelators. Their formation depends on iron concentration but is not as stringently iron regulated as are the hydroxamates, and their siderophore activity is thus secondary to their primary phytotoxicity (58).
Wood-rotting fungi.
The wood-rotting fungus Gloeophyllum trabeum produces iron-binding compounds “that appear to be phenolate in character” (46). Partially purified iron-binding compounds were conjugated with bovine serum albumin and injected into rabbits (29). The antisera were then used to immunolocalize siderophore molecules in slices of wood infected with G. trabeum (29, 46–48). Siderophores are indeed antigenic, and the approach by these workers is appealing in its potential application to mammalian systems.
Zoopathogenic fungi.
Studies to reveal the in vivo elaboration of siderophores by zoopathogens have not been performed, but the methods for revealing them directly (see the preceding section) or indirectly (see “Host response,” below) are at hand.
Bacterial siderophores.
There are a few reports of finding in vivo expression of siderophores by bacterial zoopathogens. For example, siderophores have been detected in sputum samples from the lungs of cystic fibrosis patients with infections due to Pseudomonas aeruginosa (38) and enterochelin has been found in peritoneal washings of guinea pigs infected with Escherichia coli (36). Immunoglobulins to siderophores have been detected in some instances (see “Host response,” below). Such a host response is indicative of in vivo synthesis of iron chelators by some pathogens (85).
Host Response
The siderophores are known to be antigenic (e.g., see “wood-rotting fungi,” above). In fact it has been suggested that siderophores may not be effective iron scavengers in vivo because they bind to serum proteins and elicit an immune response (85). Such immunogenicity might compromise the efficiency of siderophores in iron gathering. The immune response is, however, also evidence of in vivo synthesis. For example, enterochelin-specific immunoglobulins are found in normal human serum (68). This siderophore is produced by a number of enteric bacteria. Therefore, it is not certain whether the occurrence of anti-enterochelin activity relates to a previous infection (e.g., with Salmonella) or is a response to E. coli resident in the bowel.
CONCLUSIONS
More is known about the microbial metabolism of iron and copper than that of any of the other biologically important transition metals (50). In this review, I have focused on the acquisition, uptake, and storage of iron by pathogenic fungi. Some of the conclusions that emerge are as follows.
A great variety of means of acquisition, avenues of uptake, and methods of storage are used by pathogenic fungi to ensure a ready supply of the essential element iron. This diversity is expressed both by groups of different fungi and by individual species. The common explanation for such diversity is that it provides the organism with the ability to deal with a variety of environmental challenges, which in the case for pathogens includes an infected host.
Reduction of ferric iron to ferrous iron by enzymatic or nonenzymatic means is a common theme among pathogenic yeasts and includes the extracellular and/or intracellular acquisition of iron from chelates such as the siderophores which the utilizing species cannot itself synthesize. The ability of fungi to utilize chelated iron has not been observed to extend to host transfer proteins such as transferrin and lactoferrin or the storage protein ferritin.
Under conditions of iron starvation, many fungi synthesize low-molecular-weight iron chelators known as siderophores. Three common classes, phenolates, hydroxamates, and polycarboxylates, are observed. Some phytopathogenic fungi produce unique compounds that function as phytotoxins but also chelate iron. Methods for detection of siderophore activity that rely on color formation are subject to error because compounds other than siderophores may give positive reactions. The simple color reactions may be used to detect potentially interesting activities, but full chemical characterization is required to confirm siderophore detection.
The two major responses to iron stress in fungi are (i) a high-affinity ferric reductase and (ii) siderophore synthesis. These two responses are manifested by different species. The regulation of the two responses at a molecular level has been studied in S. cerevisiae (reductase) (53, 54) and U. maydis (siderophore) (2, 3, 53).
Uptake of siderophores is a diverse process which varies among the different classes of compounds. Moreover, utilization of distinct siderophores is dictated by the class of molecule involved even when several classes are excreted by the same species. Hence, the iron ligated by DA is transported by direct membrane transfer to another carrier (a second intracellular molecule of DA), while the entire iron-ligand of coprogen B is transported across the membrane and the iron is released internally by reduction or by ligand exchange (which need not be enzymatic) with an internal storage molecule.
Since iron is a toxic ion in the free state, it must be stored (e.g., by heme synthesis) for further metabolic use. Several molecules with this function have been described, including polyphosphates, ferritins, and the siderophores themselves.
The cardinal question involves the iron-gathering mechanisms of pathogenic fungi in a host. At present, suggestions can be posited only on the basis of in vitro studies. For hemolytic pathogens such as C. albicans, hemin might be a likely option. However, for a nonhemolytic fungus such as H. capsulatum, such an option is available only in certain in vivo locations. Since siderophores are antigenic, studies of host response-specific immunoglobulins might provide a ready source of information on in vivo synthesis of the compounds.
ACKNOWLEDGMENTS
During the time of the literature review conducted for the preparation of this article, I was supported in part by funds from USPHS grant AI 40011-2 from NIAID, National Institutes of Health.
I gratefully acknowledge the cooperation of many investigators who provided reprints and preprints of their work. I apologize to anyone whose work I may have overlooked. I thank my wife for the careful typing and retyping a work of this sort requires. I thank Anupama Tiwari and Uyen Truong for their assistance in the library searches for “iron” articles. The formulae in Fig. 1 and 2 were drawn on a computer with CSC ChemDraw Plus 3.0.1 (Cambridge Scientific Computing, Inc.) by Houman Langroodi. I am indebted to J. Domer, who invited me to prepare the review and thus gave me an opportunity to air views on an aspect of fungal physiology that has occupied my attention for the last several years.
REFERENCES
- 1.Adjimani J P, Owusu E. Nonenzymatic NADH/FMN-dependent reduction of ferric siderophores. J Inorg Biochem. 1997;66:247–252. [Google Scholar]
- 2.An Z, Mei B, Yuan W M, Leong S A. The distal GATA sequences of the sid1 promoter of Ustilago maydis mediate iron repression of siderophore production and interact directly with Urbs1, a GATA family transcription factor. EMBO J. 1997;16:1742–1750. doi: 10.1093/emboj/16.7.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An Z, Zhao Q, McEvoy J, Yuan W M, Markley J L, Leong S A. The second finger of Urbs1 is required for iron-mediated repression of sid1 in Ustilago maydis. Proc Natl Acad Sci USA. 1997;94:5882–5887. doi: 10.1073/pnas.94.11.5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ardon O, Nudelman R, Caris C, Libman J, Shanzer A, Chen Y, Hadar Y. Iron uptake in Ustilago maydis: tracking the iron path. J Bacteriol. 1998;180:2021–2026. doi: 10.1128/jb.180.8.2021-2026.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ardon O, Weizman H, Libman J, Shanzer A, Chen Y, Hadar Y. Iron uptake in Ustilago maydis: studies with fluorescent ferrichrome analogues. Microbiology. 1997;143:3625–3631. doi: 10.1099/00221287-143-11-3625. [DOI] [PubMed] [Google Scholar]
- 6.Barash I, Pupkin G, Netzer D, Kashman Y. A novel enolic β-ketoaldehyde phytotoxin produced by Stemphylium botryosum f. sp. lycopersici. Plant Physiol. 1982;69:23–27. doi: 10.1104/pp.69.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barash I, Dori S, Mor H, Manulis S. Iron chelation in plants and soil microorganisms. New York, N.Y: Academic Press, Inc.; 1993. Role of iron in fungal phytopathologies; pp. 251–267. [Google Scholar]
- 8.Bentley M D, Anderegg R J, Szaniszlo P J, Davenport R F. Isolation and identification of the principal siderophore of the dermatophyte Microsporum gypseum. Biochemistry. 1986;25:1455–1457. doi: 10.1021/bi00354a040. [DOI] [PubMed] [Google Scholar]
- 9.Boelaert J R, de Locht M, Van Cutsem J, Kerrels V, Cantinieaux B, Verdonck A, Van Landuty H W, Schneider Y-J. Mucormycosis during deferoxamine therapy is a siderophore-mediated infection. J Clin Investig. 1993;91:1979–1986. doi: 10.1172/JCI116419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Böhnke R, Matzanke B F. The mobile ferrous iron pool in Escherichia coli is bound to a phosphorylated sugar derivative. BioMetals. 1995;8:223–230. doi: 10.1007/BF00143380. [DOI] [PubMed] [Google Scholar]
- 11.Burnham B F, Neilands J B. Studies on the metabolic function of the ferrichrome compounds. J Biol Chem. 1961;236:554–559. [PubMed] [Google Scholar]
- 12.Burt W R. Identification of coprogen B and its breakdown products from Histoplasma capsulatum. Infect Immun. 1982;35:990–996. doi: 10.1128/iai.35.3.990-996.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burt W R. Abstracts of the Annual Meeting of the American Society for Microbiology 1983. Washington, D.C: American Society for Microbiology; 1983. Isolation of hydroxamic acids from Blastomyces dermatitidis, abstr. F17; p. 385. [Google Scholar]
- 14.Byers B R, Arceneaux J E L. Microbial iron transport: iron acquisition by pathogenic microorganisms. Metal Ions Biol Syst. 1998;35:37–66. [PubMed] [Google Scholar]
- 15.Caldwell C W, Sprouse R F. Iron and host resistance in histoplasmosis. Am Rev Respir Dis. 1982;125:674–677. doi: 10.1164/arrd.1982.125.6.674. [DOI] [PubMed] [Google Scholar]
- 16.Carrano C J, Raymond K N. Coordination chemistry of microbial iron transport compounds: rhodotorulic acid and iron uptake in Rhodotorula pilimanae. J Bacteriol. 1978;136:69–74. doi: 10.1128/jb.136.1.69-74.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrano C J, Böhnke R, Matzanke B F. Fungal ferritins: the ferritin from mycelia of Absidia spinosa is a bacterioferritin. FEBS Lett. 1996;390:261–264. doi: 10.1016/0014-5793(96)00667-9. [DOI] [PubMed] [Google Scholar]
- 18.Casanova M, Cervera A M, Gozalbo D, Martinez P. Hemin induces germ tube formation in Candida albicans. Infect Immun. 1997;65:4360–4364. doi: 10.1128/iai.65.10.4360-4364.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castaneda E, Brummer E, Perlman A M, McEwen J G, Stevens D A. A culture medium for Paracoccidioides brasiliensis with high plating efficiency, and the effect of siderophores. J Med Vet Mycol. 1988;26:351–358. [PubMed] [Google Scholar]
- 20.Charlang G, Ng B, Horowitz N H, Horowitz R M. Cellular and extracellular siderophores of Aspergillus nidulans and Penicillium chrysogenum. Mol Cell Biol. 1981;1:94–100. doi: 10.1128/mcb.1.2.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung T D Y, Matzanke B F, Winkelmann G, Raymond K N. Inhibitory effect of the partially resolved coordination isomers of chromic desferricoprogen on coprogen uptake in Neurospora crassa. J Bacteriol. 1986;165:283–287. doi: 10.1128/jb.165.1.283-287.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coulanges V, Andere P, Ziegler O, Buchheit L, Vidon D J-M. Utilization of iron-catecholamine complexes involving ferric reductase activity in Listeria monocytogenes. Infect Immun. 1997;65:2778–2785. doi: 10.1128/iai.65.7.2778-2785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox C D. Deferration of laboratory media and assays for ferric and ferrous iron. Methods Enzymol. 1994;235:315–329. doi: 10.1016/0076-6879(94)35150-3. [DOI] [PubMed] [Google Scholar]
- 24.Crosa J H. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cutler J E, Han Y. Fungal factors implicated in pathogenesis. In: Esser K, Lemke P A, editors. The Mycota. VI. Human and animal relationships. Berlin, Germany: Springer-Verlag KG; 1996. pp. 3–29. [Google Scholar]
- 26.de Hoog G S, Marvin-Sikkema F D, Lahpoor G A, Gottschall J G, Prins R A, Guého E. Ecology and physiology of the emerging opportunistic fungi Pseudallescheria boydii and Scedosporium prolificans. Mycoses. 1994;37:71–78. doi: 10.1111/j.1439-0507.1994.tb00780.x. [DOI] [PubMed] [Google Scholar]
- 27.Drechsel H, Thieken A, Reissbrodt R, Jung G, Winkelmann G. α ketoacids are novel siderophores in the genera Proteus, Providencia, and Morganella and are produced by amino acid diaminases. J Bacteriol. 1993;175:2727–2733. doi: 10.1128/jb.175.9.2727-2733.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eide D, Guerinot M L. Metal ion uptake in eukaryotes. ASM News. 1997;63:199–205. [Google Scholar]
- 29.Fekete F A. Iron chelation in plants and soil microorganisms. New York, N.Y: Academic Press, Inc.; 1993. Assays for microbial siderophores; pp. 399–417. [Google Scholar]
- 30.Fekete F A, Chandhoke V, Jellison J. Iron-binding compounds produced by wood-decaying basidiomycetes. Appl Environ Microbiol. 1989;55:2720–2722. doi: 10.1128/aem.55.10.2720-2722.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fekete F A, Spence J T, Emery T. A rapid and sensitive paper electrophoresis assay for detection of microbial siderophores elicited in solid-plating culture. Anal Biochem. 1983;131:516–519. doi: 10.1016/0003-2697(83)90207-5. [DOI] [PubMed] [Google Scholar]
- 32.Fleischmann J, Lehrer R I. Phagocytic mechanisms in host response. In: Howard D H, editor. Fungi pathogenic for humans and animals. B. Pathogenicity and detection: II. New York, N.Y: Marcel Dekker, Inc.; 1985. pp. 123–149. [Google Scholar]
- 33.Frederick C B, Szaniszlo P J, Vickrey P E, Bentley M D, Shive W. Production and isolation of siderophores from the soil fungus Epicoccum purpurescens. Biochemistry. 1981;20:2432–2436. doi: 10.1021/bi00512a010. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-del Portillo F, Foster J W, Maguire M E, Finlay B B. Characterization of the micro-environment of Salmonella typhimurium-containing vacuoles within MDCK epithelial cells. Mol Microbiol. 1992;6:3289–3297. doi: 10.1111/j.1365-2958.1992.tb02197.x. [DOI] [PubMed] [Google Scholar]
- 35.Gobin J, Moore C H, Reeve J R, Jr, Wong D K, Gibson B W, Horwitz M A. Iron acquisition by Mycobacterium tuberculosis: isolation and characterization of a family of iron-binding exochelins. Proc Natl Acad Sci USA. 1995;92:5189–5193. doi: 10.1073/pnas.92.11.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griffiths E, Humphreys J. Isolation of enterochelin from the peritoneal washings of guinea pigs lethally infected with Escherichia coli. Infect Immun. 1980;28:286–289. doi: 10.1128/iai.28.1.286-289.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guerinot M L. Microbial iron transport. Annu Rev Microbiol. 1994;48:743–772. doi: 10.1146/annurev.mi.48.100194.003523. [DOI] [PubMed] [Google Scholar]
- 38.Haas B, Kraut J, Marks J, Zanker S C, Castignetti D. Siderophore presence in sputa of cystic fibrosis patients. Infect Immun. 1991;59:3997–4000. doi: 10.1128/iai.59.11.3997-4000.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Höfte M. Classes of microbial siderophores. In: Barton L L, editor. Iron chelation in plants and soil microorganisms. New York, N.Y: Academic Press, Inc.; 1993. pp. 3–26. [Google Scholar]
- 40.Holzberg M, Artis W M. Hydroxamate siderophore production by opportunistic and systemic fungal pathogens. Infect Immun. 1983;40:1134–1139. doi: 10.1128/iai.40.3.1134-1139.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howard D H. Mechanisms of resistance in the systemic mycoses. In: Nahmias A J, O’Reilly R J, editors. Immunology of human infection. New York, N.Y: Plenum Press; 1981. pp. 475–494. [Google Scholar]
- 42.Ismail A, Bedell G W, Lupan D M. Siderophore production by the pathogenic yeast, Candida albicans. Biochem Biophys Res Commun. 1985;130:885–891. doi: 10.1016/0006-291x(85)90499-1. [DOI] [PubMed] [Google Scholar]
- 43.Jacobson E S, Petro M J. Extracellular iron chelation in Cryptococcus neoformans. J Med Vet Mycol. 1987;25:415–418. [PubMed] [Google Scholar]
- 44.Jacobson E S, Goodner A P, Nyhus K J. Ferrous iron uptake in Cryptococcus neoformans. Infect Immun. 1998;66:4169–4175. doi: 10.1128/iai.66.9.4169-4175.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jalal M A F, van der Helm D. Isolation and spectroscopic identification of fungal siderophores. In: Winkelmann G, editor. Handbook of microbial iron chelates. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 235–269. [Google Scholar]
- 46.Jellison J, Goodell B. Immunological detection of decay in wood. Wood Sci Technol. 1988;22:293–297. [Google Scholar]
- 47.Jellison J, Goodell B. Inhibitory effects of undecayed wood and the detection of Postia placenta using the enzyme-linked immunosorbent assay. Wood Sci Technol. 1989;23:13–20. [Google Scholar]
- 48.Jellison J, Chandhoke V, Goodell B, Fekete F A. The isolation and immunolocalization of iron-binding compounds produced by Gloeophyllum trabeum. Appl Biotechnol Microbiol. 1991;35:805–809. [Google Scholar]
- 49.Kaplan J, O’Halloran T V. Iron metabolism in eukaryotes: Mars and Venus at it again. Science. 1996;271:1510–1512. doi: 10.1126/science.271.5255.1510. [DOI] [PubMed] [Google Scholar]
- 50.Kosman D J. Transition metal ion uptake in yeasts and filamentous fungi. In: Winkelmann G, Winge D R, editors. Metal ions in fungi. Vol. 11. New York, N.Y: Marcel Dekker, Inc.; 1994. pp. 1–38. [Google Scholar]
- 51.Kwon-Chung K J, Bennett J E. Medical mycology. Philadelphia, Pa: Lea & Febiger; 1992. [Google Scholar]
- 52.Labbe-Bois R, Camadro J-M. Ferrochelatase in Saccharomyces cerevisiae. In: Winkelmann G, Winge D R, editors. Metal ions in fungi. Vol. 11. New York, N.Y: Marcel Dekker, Inc.; 1994. pp. 413–453. [Google Scholar]
- 53.Leong S A, Winkelmann G. Molecular biology of iron transport in fungi. Metal Ions Biol Syst. 1998;35:147–186. [PubMed] [Google Scholar]
- 54.Lesuisse E, Labbe P. Reductive iron assimilation in Saccharomyces cerevisiae. In: Winkelmann G, Winge D R, editors. Metal ions in fungi. Vol. 11. New York, N.Y: Marcel Dekker, Inc.; 1994. pp. 149–178. [Google Scholar]
- 55.Liles M R, Cianciotto N P. Absence of siderophore-like activity in Legionella pneumophila supernatants. Infect Immun. 1996;64:1873–1876. doi: 10.1128/iai.64.5.1873-1875.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55a.Liu L, Wakamatsu K, Iton S, Williamson P. Catecholamine oxidative products, but not melanin, are produced by Cryptococcus neoformans during neuropathogenesis in mice. Infect Immun. 1999;67:108–112. doi: 10.1128/iai.67.1.108-112.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loper J E, Buyer J S. Siderophores in microbial interactions on plant surfaces. Mol Plant-Microbe Interact. 1991;4:5–13. [Google Scholar]
- 57.Manulis S, Kashman Y, Barash I. Identification of siderophores and siderophore-mediated uptake of iron in Stemphylium botryosum. Phytochemistry. 1987;26:1317–1320. [Google Scholar]
- 58.Manulis S, Kashman Y, Netzer D, Barash I. Phytotoxins from Stemphylium botryosum: structural determination of stemphyloxin II, production in culture and interaction with iron. Phytochemistry. 1984;23:2193–2198. [Google Scholar]
- 59.Matzanke B F. Mossbauer spectroscopy of microbial iron uptake and metabolism. In: Winkelmann G, van der Helm D, Neilands J B, editors. Iron transport in microbes, plants and animals. Weinheim, Germany: VCH Publishers; 1987. pp. 252–284. [Google Scholar]
- 60.Matzanke B F. Structures, coordination chemistry and functions of microbial iron chelates. In: Winkelmann G, editor. Handbook of microbial iron chelates. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 15–64. [Google Scholar]
- 61.Matzanke B F. Iron storage in fungi. In: Winkelmann G, Winge D R, editors. Metal ions in fungi. Vol. 11. New York, N.Y: Marcel Dekker, Inc.; 1994. pp. 179–214. [Google Scholar]
- 62.Matzanke B F. Iron transport: siderophores. In: King R B, editor. Encyclopedia of inorganic chemistry. 4, Iro–Met. New York, N.Y: John Wiley & Sons, Inc.; 1994. pp. 1915–1933. [Google Scholar]
- 63.Matzanke B F. Iron storage in Microorganisms. In: Winkelmann G, Carrano C J, editors. Transition metals in microbial metabolism. Australia: Harwood Academic Publishers; 1997. pp. 117–157. [Google Scholar]
- 64.Matzanke B F, Bill E, Muller G I, Trautwein A X, Winkelmann G. Metabolic utilization of 57Fe-labeled coprogen in Neurospora crassa. An in vivo Mossbauer study. Eur J Biochem. 1987;162:643–650. doi: 10.1111/j.1432-1033.1987.tb10686.x. [DOI] [PubMed] [Google Scholar]
- 65.Matzanke B F, Bill E, Trautwein A X, Winkelmann G. Ferricrocin functions as the main intracellular iron-storage compound in mycelia of Neurospora crassa. Biol Metals. 1988;1:18–25. doi: 10.1007/BF01128013. [DOI] [PubMed] [Google Scholar]
- 66.Mezence M I B, Boiron P. Studies on siderophore production and effect of iron deprivation on the outer membrane proteins of Madurella mycetomatis. Curr Microbiol. 1995;31:220–223. doi: 10.1007/BF00298377. [DOI] [PubMed] [Google Scholar]
- 67.Minnick A A, Eizember L E, McKee J A, Dolence E K, Miller M J. Bioassay for siderophore utilization by Candida albicans. Anal Biochem. 1991;194:223–229. doi: 10.1016/0003-2697(91)90171-o. [DOI] [PubMed] [Google Scholar]
- 68.Moore D G, Yancey R J, Lankford C E, Earhart C F. Bacteriostatic enterochelin-specific immunoglobulin from normal human serum. Infect Immun. 1980;27:418–423. doi: 10.1128/iai.27.2.418-423.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moors M A, Stull T L, Blank K J, Buckley H R, Mosser D M. A role for complement receptor-like molecules in iron acquisition by Candida albicans. J Exp Med. 1992;175:1643–1651. doi: 10.1084/jem.175.6.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mor H, Kashman Y, Winkelmann G, Barash I. Characterization of siderophores produced by different species of dermatophytic fungi Microsporum and Trichophyton. BioMetals. 1992;5:213–216. [Google Scholar]
- 71.Mor H, Pasternak M, Barash I. Uptake of iron by Geotrichum candidum, a non-siderophore producer. BioMetals. 1988;1:99–105. [Google Scholar]
- 72.Morrissey J A, Williams P H, Cashmore A M. Candida albicans has a cell-associated ferric reductase activity which is regulated in response to levels of iron and copper. Microbiology. 1996;142:485–492. doi: 10.1099/13500872-142-3-485. [DOI] [PubMed] [Google Scholar]
- 73.Müller G, Barclay S J, Raymond K N. The mechanism and specificity of iron transport in Rhodotorula pilimanae probed by synthetic analogs of rhodotorulic acid. J Biol Chem. 1985;260:13916–13920. [PubMed] [Google Scholar]
- 74.Neilands J B. Identification and isolation of mutants defective in iron acquisition. Methods Enzymol. 1994;235:352–357. doi: 10.1016/0076-6879(94)35153-8. [DOI] [PubMed] [Google Scholar]
- 75.Neilands J B, Nakamura K. Detection, determination, isolation, characterization and regulation of microbial iron chelates. In: Winkelmann G, editor. Handbook of microbial iron chelates. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 1–14. [Google Scholar]
- 76.Newman S L, Gootee L, Brunner G, Deepe G S., Jr Chloroquine induces human macrophage killing of Histoplasma capsulatum by limiting the availability of intracellular iron and is therapeutic in a murine model of histoplasmosis. J Clin Investig. 1994;93:1422–1429. doi: 10.1172/JCI117119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Newman S L, Gootee L, Stroobant V, van der Goot H, Boelaert J R. Inhibition of growth of Histoplasma capsulatum yeast cells in human macrophages by the iron chelator VUF 8514 and comparison of VUF 8514 with deteroxamine. Antimicrob Agents Chemother. 1995;39:1824–1829. doi: 10.1128/aac.39.8.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nyhus K J, Wilborn A T, Jacobson E S. Ferric iron reduction by Cryptococcus neoformans. Infect Immun. 1997;65:434–438. doi: 10.1128/iai.65.2.434-438.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Payne S M. Iron acquisition in microbial pathogenesis. Trends Microbiol. 1993;1:66–68. doi: 10.1016/0966-842x(93)90036-q. [DOI] [PubMed] [Google Scholar]
- 80.Payne S M. Detection, isolation, and characterization of siderophores. Methods Enzymol. 1994;235:329–344. doi: 10.1016/0076-6879(94)35151-1. [DOI] [PubMed] [Google Scholar]
- 81.Payne S M. Effects of iron deprivation in outer membrane protein expression. Methods Enzymol. 1994;235:344–352. doi: 10.1016/0076-6879(94)35152-x. [DOI] [PubMed] [Google Scholar]
- 82.Payne S M. Identification and isolation of mutants defective in iron acquisition. Methods Enzymol. 1994;235:352–356. doi: 10.1016/0076-6879(94)35153-8. [DOI] [PubMed] [Google Scholar]
- 83.Perry R D. Acquisition and storage of inorganic iron and hemin by the yersiniae. Trends Microbiol. 1993;1:142–147. doi: 10.1016/0966-842x(93)90129-f. [DOI] [PubMed] [Google Scholar]
- 84.Plattner H J, Diekmann H. Enzymology of siderophore biosynthesis in fungi. In: Winkelmann G, Winge D R, editors. Metal ions in fungi. Vol. 11. New York, N.Y: Marcel Dekker, Inc.; 1994. pp. 99–116. [Google Scholar]
- 85.Reissbrodt R, Kingsley R, Rabsch W, Beer W, Roberts M, Williams P H. Iron-regulated excretion of α-keto acids by Salmonella typhimurium. J Bacteriol. 1997;179:4538–4544. doi: 10.1128/jb.179.14.4538-4544.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Riquelme M. Fungal siderophores in plant-microbe interactions. Microbiologia. 1996;12:537–546. [PubMed] [Google Scholar]
- 87.Salvin S B. Hemolysin from the yeastlike phases of some pathogenic fungi. Proc Soc Exp Biol Med. 1951;76:852–854. doi: 10.3181/00379727-76-18653. [DOI] [PubMed] [Google Scholar]
- 87a.Spiro T G, Allerton S F, Renner J, Terzis A, Bils R, Saltman P. The hydrolytic polymerization of iron (III) J Am Chem Soc. 1966;88:2721–2725. [Google Scholar]
- 88.Stearman R, Yuan D S, Yamaguchi-Iwai Y, Klausner R D, Dancis A. A permease-oxidase complex involved in high affinity iron uptake in yeast. Science. 1996;271:1552–1557. doi: 10.1126/science.271.5255.1552. [DOI] [PubMed] [Google Scholar]
- 89.Sudbery P. The non-Saccharomyces yeasts. Yeast. 1994;10:1707–1726. doi: 10.1002/yea.320101305. [DOI] [PubMed] [Google Scholar]
- 90.Sweet S P, Douglas L J. Effect of iron concentration on siderophore synthesis and pigment production by Candida albicans. FEMS Microbiol Lett. 1991;80:87–92. doi: 10.1016/0378-1097(91)90214-u. [DOI] [PubMed] [Google Scholar]
- 91.Telford J R, Raymond K N. Siderophores. In: Latwood J, Davis J E D, MacNicol D D, Vogtle F, editors. Comprehensive supramolecular chemistry. Vol. 1. Oxford, United Kingdom: Elsevier Science Ltd.; 1996. pp. 245–266. [Google Scholar]
- 92.Theil E C. Ferritin: structure, gene regulation, and cellular function in animals, plants, and microorganisms. Annu Rev Biochem. 1987;56:289–315. doi: 10.1146/annurev.bi.56.070187.001445. [DOI] [PubMed] [Google Scholar]
- 93.Tilbrook G S, Hider R C. Iron chelators for clinical use. Metal Ions Biol Syst. 1998;35:691–730. [PubMed] [Google Scholar]
- 94.van der Helm D, Winkelmann G. Hydroxamates and polycarboxylates as iron transport agents (siderophores) in fungi. In: Winkelmann G, Winge D R, editors. Metal ions in fungi. Vol. 11. New York, N.Y: Marcel Dekker, Inc.; 1994. pp. 39–98. [Google Scholar]
- 95.Vartivarian S E, Cowart R E, Anaisse E J, Tashiro T, Sprigg H A. Iron acquisition by Cryptococcus neoformans. J Med Vet Mycol. 1995;33:151–156. [PubMed] [Google Scholar]
- 96.Weinberg E D. The iron-withholding defense system. ASM News. 1993;59:559–562. [Google Scholar]
- 97.Wilson M E, Vorhies R W, Anderson K A, Britigen B E. Acquisition of iron from transferrin and lactoferrin by the protozoan Leishmania chagasi. Infect Immun. 1994;62:3262–3269. doi: 10.1128/iai.62.8.3262-3269.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Winkelmann G. Surface iron polymers and hydroxy acids. A model of iron supply in sideramine-free fungi. Arch Microbiol. 1979;121:43–51. [Google Scholar]
- 99.Winkelmann G. Specificity of iron transport in bacteria and fungi. In: Winkelmann G, editor. Handbook of microbial iron chelates. Boca Raton, Fla: CRC Press, Inc.; 1991. pp. 65–105. [Google Scholar]
- 100.Winkelmann G. Kinetics, energetics, and mechanisms of siderophore iron transport in fungi. In: Barton L L, editor. Iron chelation in plants and soil microorganisms. New York, N.Y: Academic Press, Inc.; 1993. pp. 219–239. [Google Scholar]
- 101.Winkelmann G, Zahner H. Stoffwechselprodukte von Mikroorganismen. 115, Mitteilung. Eisenaufnahme bei Neurospora crassa. I. Zur Spezifitat des Eisentransportes. Arch Mikrobiol. 1973;88:49–60. [PubMed] [Google Scholar]
- 102.Worsham P L, Goldman W E. Quantitative plating of Histoplasma capsulatum without addition of conditioned medium or siderophores. J Med Vet Mycol. 1988;6:137–143. [PubMed] [Google Scholar]