Abstract
Three-dimensional (3D) bioprinting demonstrates technology that is capable of producing structures comparable to native tissues in the human body. The freeform reversible embedding of suspended hydrogels (FRESH) technique involves hydrogel-based bio-inks printed within a thermo-reversible support bath to provide mechanical strength to the printed construct. Smaller and more uniform microsphere sizes of FRESH were reported to aid in enhancing printing resolution and construct accuracy. Therefore, we sought to optimize the FRESH generation protocol, particularly by varying stir speed and stir duration, in hopes to further improve microsphere size and uniformity. We observed optimal conditions at a stir speed of 600 rpm and stir duration for 20 h that generated the smallest microspheres with the best uniformity. Comparison of using the optimized FRESH to the commercial FRESH LifeSupport to bioprint single filament and geometrical constructs revealed reduced single filament diameters and higher angular precision in the optimized FRESH bio-printed constructs compared with those printed in the commercial FRESH. Overall, our refinement of the FRESH manufacturing protocol represents an important step toward enhancing 3D bioprinting resolution and construct fidelity. Improving such technologies allows for the fabrication of highly accurate constructs with anatomical properties similar to native counterparts. Such work has significant implications in the field of tissue engineering for producing accurate human organ model systems.
Impact statement
Freeform reversible embedding of suspended hydrogels (FRESH) is a method of sacrificial three-dimensional (3D) bioprinting that offers support to reinforce bio-ink extrusion during printing. During FRESH generation, the stir speed and stir duration of the mixture can significantly impact FRESH microsphere characteristics. In this study, we optimized FRESH microspheres to significantly improve resolution and accuracy in bioprinting. This advancement in FRESH-based 3D bioprinting technologies allows for the fabrication of highly accurate constructs with anatomical properties similar to native counterparts and has significant implications in the field of tissue engineering and translational medicine.
Keywords: FRESH, freeform reversible embedding of suspended hydrogel, microsphere, optimization
Introduction
Previous work in three-dimensional (3D) bioprinting technologies has proven to be promising for the fabrication of structurally accurate organ models.1 Current methods of bioprinting include vat polymerization using photo-initiators,2 material jetting technology,3,4 and material extrusion to print constructs comparable to native materials' biological properties,5 such as with hydrogel bio-ink.6 The mechanism behind extrusion-based printing involves a network between individual polymers to support the construct.7
With potential implications in regenerative medicine and tissue engineering, extrusion-based bioprinting allows for depositing bio-inks composed of hydrogel-based materials with or without living cells into 3D constructs.8–10 Following post-printing processing, such as chemical cross-linking of the bio-ink polymers, these printed constructs are comparable to their natural counterparts.11 However, challenges within this field persist, including the construct's fidelity1 and the choice of materials as bio-ink.12 Specifically, deposited biomaterials often deform when printed in air due to lack of physical support.13
One method that addresses this issue involves sacrificial bioprinting using freeform reversible embedding of suspended hydrogels (FRESH) composed of gelatin microspheres from the complex coacervation formation of negatively-charged gelatin and positively-charged gum arabic.14 During bioprinting, FRESH acts as a thermo-reversible gelatin microsphere support bath that offers mechanical strength for bio-inks, preventing constructs from collapsing during printing. With this method, highly complex constructs ranging from vessels to full-organ scale have been fabricated with high fidelity.15
The novel FRESH bioprinting protocol proposed by Hinton et al and Lee et al demonstrated significantly improved FRESH microspheres. The first version of FRESH consisted of irregularly shaped, large microspheres created by mechanical blending of a large gelatin block14; in the second version of FRESH, a coacervation approach was developed to generate gelatin microspheres with uniform spherical morphology and decreased particle diameter.15
However, several details of the FRESH manufacture protocol remain unclear.14,15 Factors such as specific pH, mixture temperature, stirring speed, and stirring duration may play a role in the size of gelatin microspheres. Reducing microsphere size can enhance printing resolution of smaller-scale hydrogel constructs with increased cell permeability.16
In this study, we aimed at further optimizing the FRESH manufacturing protocol, specifically examining the effect of stir speed and duration on FRESH microspheres. We hypothesized that there was an optimal stir speed and stir duration that produced the smallest-sized and most uniform microspheres.
Materials and Methods
FRESH manufacturing
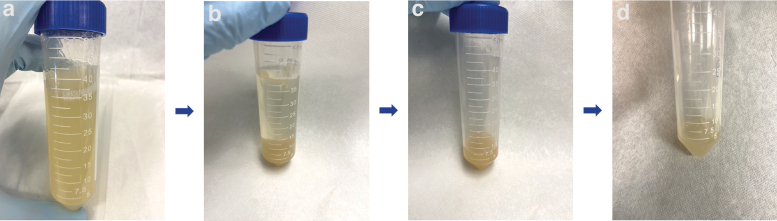
To manufacture FRESH using our protocol (Fig. 1), ethanol solution at 50% (w/v) was first heated to 40°C in a 1-L cylinder glass beaker. Next, 2.0% (w/v) gelatin type B (catalog no. 9000-70-8; Fisher Chemical, Hampton, NH, USA), 0.5% (w/v) Pluronic (catalog no. 9003-11-6; Sigma-Aldrich, St. Louis, MO, USA), and 3.0% (w/v) gum arabic (catalog no. 9000-01-5; Sigma-Aldrich) were dissolved in the ethanol solution, using a magnetic stirrer with an 80 mm magnetic cylindrical stir bar (catalog no. 16-800-513; Fisher Chemical) at 260 rpm, to 55°C. After removing the magnetic stir bar (catalog no. 16-800-513; Fisher Chemical), the beaker was sealed with parafilm to reduce evaporation and placed under an IKA overhead stirrer RW20 (catalog no. 3593000; IKA, Wilmington, NC, USA) overnight at 22°C to form the gelatin microsphere slurry.
FIG. 1.
FRESH manufacturing procedure. (a) Gelatin slurry portioned into a 50-mL conical tube before centrifugation. (b) Post-centrifuge showing water and ethanol supernatant on top and deep-yellow colored compacted gelatin gel at the bottom of the conical tube. (c) Compacted gelatin gel in tube after decanting supernatant. (d) FRESH microsphere bath used for bioprinting after three rounds of centrifugation, washing, and decantation. FRESH, freeform reversible embedding of suspended hydrogel.
Our experiment repeated the protocol with the slurry stirred at speeds ranging from 400 to 800 rpm in 100 rpm increments. For each stir speed, we allocated a 40 mL aliquot in 50-mL conical tubes after 14, 16, 18, 20, and 22 h of stirring. The mixtures were adjusted to a pH of 5.0 ± 0.1 with the addition of ∼225 μL hydrochloric acid. The conical tubes were centrifuged at 300 g for 5 min to compact the gelatin. After centrifugation, the supernatant was completely decanted.
Next, 0.1% (w/v) calcium chloride (CaCl2) solution (catalog no. 10043-52-4; Sigma-Aldrich) dissolved in deionized water at 300 rpm with a magnetic stirrer and stir bar (catalog no. 16-800-513; Fisher Chemical) for 2 min was used for washing. All washing steps involved the CaCl2 solution kept under magnetic stirring at 300 rpm for at least 1 min before use. The compacted gelatin mixture was washed with CaCl2 by adding washing solution until the total volume reached 40 mL.
The resulting slurry was centrifuged again at 1000 g for 2 min before its supernatant was decanted; then, the compacted gelatin was washed with 0.1% (w/v) CaCl2 to 40 mL again. After a third centrifugation at 1000 g for 2 min and supernatant decantation, the compacted gelatin, otherwise known as FRESH microspheres, was stored at 4°C for no more than 24 h until further use.
The recommended protocol from Lee et al's FRESH v2.0 was followed for producing the commercial FRESH as a comparison control.14 A 0.1% (w/v) CaCl2 solution was formed through 20 mg of sterile CaCl2 (catalog no. 10043-52-4; Sigma-Aldrich) dissolved in 20 mL of 50 mM HEPES (catalog no. 15630080; Gibco, Waltham, MA, USA) and added into a 50-mL microcentrifuge conical tube. This tube contained 1 g of sterile LifeSupport (catalog no. LIFES; Allevi, Inc., Philadelphia, PA, USA) powder composed of dehydrated gelatin microparticles of defined size and shape.
The LifeSupport powder was resuspended and dissolved in the CaCl2 solution through 1 min of vigorous mixing with a metal spatula. For full rehydration, the LifeSupport solution was stored at 4°C. Then, the LifeSupport solution underwent centrifugation for 7 min at 2000 g until the LifeSupport was compacted in the conical tube. All resulting supernatants were discarded. This FRESH support bath was then transported to well plates for 3D bioprinting. All support baths were used within 12 h of generation.
FRESH microsphere imaging and analysis
The compacted gelatin was washed and resuspended with 0.1% (w/v) CaCl2 to 50 mL, and it was then centrifuged at 1000 g for 2 min. After supernatant decantation, the compacted gelatin was washed and diluted with 0.1% (w/v) CaCl2 with a 1:5 gel-to-washing solution ratio. Next, 50–60 μL of the slurry was placed on 25 × 75 mm glass slides.
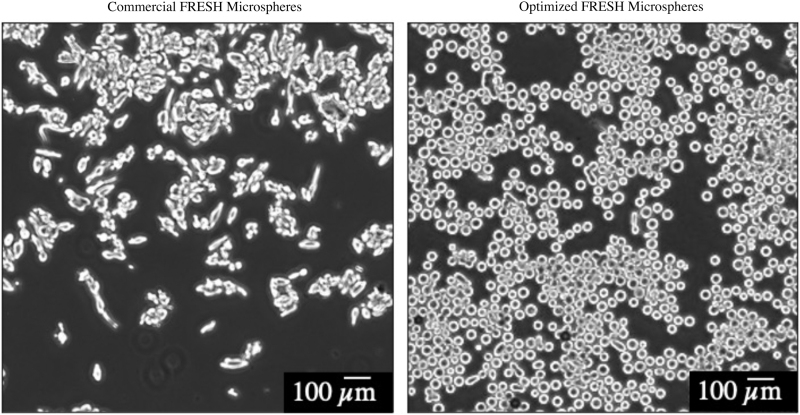
Within 5 min of depositing onto the slides, live images from each of three slides were taken using the Keyence microscopy phase contrast at 4 × magnification, with frame dimensions of 640 × 480 (Fig. 2). Microsphere analysis was completed using ImageJ Fiji (NIH, Bethesda, MD, USA) by measuring Ferret diameter, with a size threshold of 200 to 8000 μm and a circularity threshold of 0.8 to 1.0 μm.
FIG. 2.
Comparison of microsphere shape and size in the commercial FRESH LifeSupport and the optimized FRESH manufactured using our protocol. Images were taken under microscopy and analyzed with ImageJ's Analyze Particles function. Note the large-sized, non-uniform, rhomboid-shaped microspheres of the commercial FRESH microspheres and the significantly smaller-sized, uniform morphology, circular-shaped microspheres of the optimized FRESH microspheres. Scale bar = 100 μm.
Bio-ink preparation
Prior to bioprinting, native sodium alginate (catalog no. ALG; Allevi, Inc.) was sterilized using ethylene oxide. After sterilization, the alginate (catalog no. ALG; Allevi, Inc.) was dissolved in 50 mM HEPES (catalog no. 15630080; Gibco) while stirring at 60°C to create 5% (w/v) native sodium alginate. After dissolving, the alginate solution was cooled to 37°C under continued stirring. If not used immediately, alginate solutions were stored at 4°C for up to 48 h.
Before printing, 2 mL of the alginate solution were loaded into a 5-mL syringe (catalog no. PSYR5; Allevi, Inc.) under sterile conditions. Using a second 5-mL sterile syringe (catalog no. PSYR5; Allevi, Inc.) with a syringe coupler (catalog no. SYRCOUP; Allevi, Inc.), 1% (w/v) Alcian Blue 8GX (catalog no. J60122.14; Thermo Scientific, Waltham, MA, USA) and 1% (w/v) Alizarin Red (catalog no. A5533; Sigma-Aldrich) were mixed into the alginate solutions. Alcian Blue (catalog no. J60122.14; Thermo Scientific) provides visualization of the constructs before cross-linking, preventing any loss throughout post-print processing.
Alizarin Red (catalog no. A5533; Sigma-Aldrich) stains the calcium cross-linked alginates and illustrates final confirmation of the completed construct. After thorough mixing, the alginate solution was kept in one 5-mL sterile syringe (catalog no. PSYR5; Allevi, Inc.), the printing syringe (catalog no. PSYR5; Allevi, Inc.), with the other syringe (catalog no. PSYR5; Allevi, Inc.) discarded.
After de-airing the printing syringe by covering it with a syringe cap (catalog no. SYRCAP; Allevi, Inc.), centrifuging at 300 g for 1 min, and careful manual air extrusion using the plunger, the syringe (catalog no. PSYR5; Allevi, Inc.) was then stored at 4°C until printing if not used immediately. In addition, 100 mM of sterile CaCl2 solution, used for alginate cross-linking, was produced by dissolving CaCl2 (catalog no. 10043-52-4; Sigma-Aldrich) in 50 mM HEPES (catalog no. 15630080; Gibco). If not immediately used, the cross-linking CaCl2 solution was also stored in 4°C for up to 48 h.
3D bioprinting
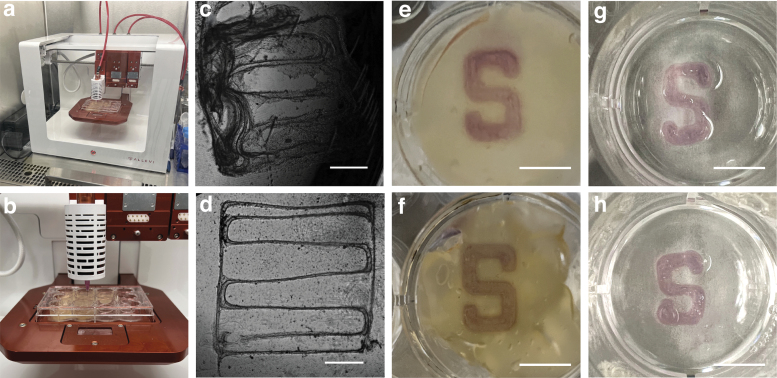
The Allevi 3 Bioprinter (Allevi, Inc.) was used for alginate bioprinting in FRESH. The pre-loaded alginate syringe prepared, described earlier, was attached to an extruder with a 30 G, 0.25-inch plastic tipped needle attached (Fig. 3a, b). Automatic calibration was performed following the standard protocol. Extrusion pressure was tested, and any remaining air was further removed by applying compressed air pressure to the syringe until the alginate bio-ink was dispensed.
FIG. 3.
3D bioprinting in FRESH for single filament printing resolution assessment and construct fidelity testing. (a, b) Allevi 3 Bioprinter used for construct printing. (c) Single filament construct printed using the commercial FRESH and (d) optimized FRESH taken under microscopy using phase contrast. Scale bar (c, d) = 1 mm. (e) Printed “S” construct using the commercial FRESH and (f) optimized FRESH before cross-linking. Photo taken from bottom of well for better visualization of construct. (g) Printed construct using the commercial FRESH and (h) optimized FRESH 24 h after cross-linking. Photo taken from top of well. Scale bar (e–h) = 5 mm.
The optimized FRESH based on our analysis described earlier was used for bioprinting. A metal spatula was used to transfer and compact the FRESH onto 12-well plates (catalog no. 351143; Corning, Inc., Corning, NY, USA) at 22°C.
For single filament bioprinting to assess printing resolution, both the optimized and commercial FRESH were used for one well each. To achieve a smaller single filament diameter, a higher print speed of 10 mm/s was used with 15 psi. In each well, two 5 × 5 mm boxes with zigzag single filament infill were printed with a 1 mm offset in the z-axis (Fig. 3c, d).
To allow for construct fidelity evaluation, the Stanford “S” was printed (Fig. 3e–h). Specifically, the “S” dimensions were 10 mm in length and 6.2 mm in width with a 1.6 mm middle thickness. Both the optimized and commercial FRESH were loaded in four wells each. The construct was printed with an extrusion pressure of 10 psi at a print speed of 6 mm/s with a 1 mm offset in the z-axis as our standard printing protocol.17
To bioprint complex hollow 3D constructs, hollow boxes of 12 × 12 × 12 mm were printed with triangle infills of 3 mm in distance into optimized FRESH. The same printing parameters described earlier were used.
Post-bioprinting, 1 mL cross-linking CaCl2 solution was used to cross-link structures for 30 min at 37°C. Next, the cross-linking solution was carefully removed using a pipette, and the constructs were washed twice using 1 mL of 50 mM HEPES (catalog no. 15630080; Gibco) at 22°C. The constructs were cross-linked once more using 1 mL CaCl2 cross-linking solution for 30 min at 37°C, and they were then washed twice with 50 mM HEPES (catalog no. 15630080; Gibco) at 22°C. A final cross-linking was performed using 1 mL of cross-linking solution for 7 min at 37°C. The constructs were washed a last time, then stored at 37°C overnight before imaging.
Printing resolution and construct fidelity analysis
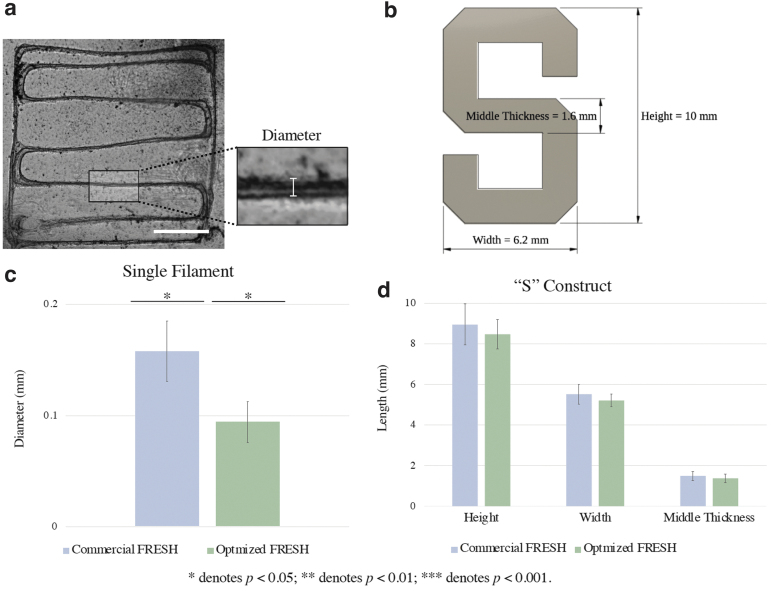
To assess printing resolution and construct fidelity, the single filament and Stanford “S” constructs were imaged using the Keyence microscope at 2 × magnification 24 h after bioprinting. All images were analyzed using ImageJ. Specifically, for the single filament constructs, line widths of the zigzag infill were measured at five separate points in each box (Fig. 4a). For the “S” construct, construct length, width, and middle section thickness (Fig. 4b) were measured for each construct.
FIG. 4.
Printing resolution and construct fidelity evaluation using the commercial FRESH LifeSupport and the optimized FRESH. (a) Illustration of measurement locations on the single filament construct for printing resolution analysis. Scale bar = 1 mm. (b) The Stanford “S” construct designed for fidelity testing. The preset dimensions were 10 mm in height, 6.2 mm in width, and 1.6 mm in middle thickness for bioprinting. (c) Bar graph comparison of single filament bioprinting results using the commercial FRESH and optimized FRESH. Optimized FRESH single filament has significantly reduced diameter compared with the commercial FRESH single filament. (d) Bar graph comparison of “S” construct bioprinting results using the commercial FRESH and optimized FRESH. “S” constructs printed using the commercial FRESH and optimized FRESH have similar dimensions with no significant difference. Error bars represent standard deviation. *p < 0.05.
Statistical analysis
Statistical analyses were performed using one-way analysis of variance with post-hoc Tukey honestly significant difference test to compare among different stir speeds and stir durations. To evaluate printing resolution and construct fidelity using the commercial versus optimized FRESH, the two-tailed Student's t-test was used. Sample variance was assessed using the F-test. A p-value of <0.05 was considered statistically significant. Continuous variables were reported as mean ± standard deviation.
Results
FRESH microspheres at different stir conditions
Effect of stir condition on FRESH microsphere shape
Shown in Supplementary Figure S1, an increase in stir speed to the optimum condition of 600 rpm corresponds to visually smaller-shaped and further uniform microspheres across all stir durations. Further increasing stir speeds above the optimum condition results in larger and less uniform microspheres. Under all stir durations, microsphere appearances revealed increased circularity with increased stir speeds to 18 h. Compared with the commercial FRESH (Fig. 2), the optimized FRESH yielded microspheres of smaller sizes, uniform morphology, and increased circularity.
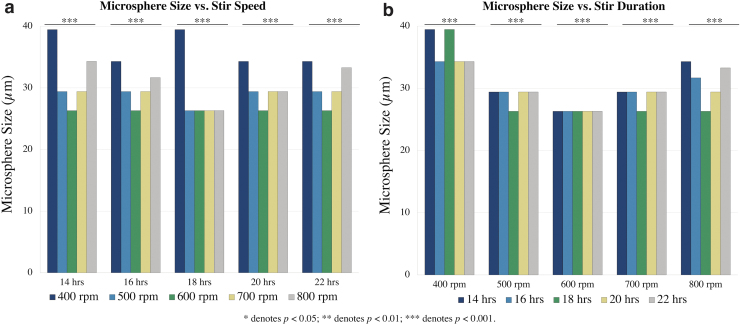
Effect of stir speed on FRESH microsphere size
A U-shaped distribution was observed between microsphere size as a function of stir speed for all stir durations except for 18 h (Fig. 5a; Tables 1 and 2). Six hundred rpm was shown to yield the smallest microsphere sizes, with a median diameter of 26.3 μm for all stir durations. The microsphere sizes at each stir duration under different stir speeds were significantly different from each other, with post-hoc adjustment showing that 600 rpm was associated with the smallest microsphere size (p < 0.001).
FIG. 5.
FRESH microsphere sizes under varied stir conditions. (a) Median microsphere size as a function of stir speed. U-shaped distribution across stir speeds with nadir occurring at 600 rpm. (b) Median microsphere size as a function of stir duration. U-shaped distribution across stir speeds with nadir occurring at 18 h. ***p < 0.001.
Table 1.
Median Microsphere Sizes with Interquartile Range (in μm) of the Optimized FRESH Support Bath Under Different Stir Speeds and Stir Durations
| 400 rpm | 500 rpm | 600 rpm | 700 rpm | 800 rpm | p-Value | |
|---|---|---|---|---|---|---|
| 14 h | 39.5 [33.3–45.9] | 29.4 [26.3–34.3] | 26.3 [25.0–29.4] | 29.4 [26.3–33.3] | 34.3 [29.4–39.5] | <0.0001 |
| 16 h | 34.3 [29.4–42.4] | 29.4 [26.3–31.7] | 26.3 [24.3–29.4] | 29.4 [26.3–33.3] | 31.7 [26.3–34.3] | <0.0001 |
| 18 h | 39.5 [33.3–45.9] | 26.3 [25.0–31.7] | 26.3 [24.3–29.4] | 26.3 [25.0–29.4] | 26.3 [25.0–29.4] | <0.0001 |
| 20 h | 34.3 [29.4–42.4] | 29.4 [26.3–31.7] | 26.3 [21.2–29.4] | 29.4 [25.0–31.7] | 29.4 [25.0–34.3] | <0.0001 |
| 22 h | 34.3 [29.4–42.4] | 29.4 [26.3–34.3] | 26.3 [24.3–29.4] | 29.4 [26.3–33.3] | 33.3 [29.4–37.7] | <0.0001 |
| p-Value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
p-Values in this table were all obtained via the one-way analysis-of-variance test. p < 0.05 denotes statistical significance.
FRESH, freeform reversible embedding of suspended hydrogels.
Table 2.
Median Variances with Interquartile Range (in μm) of the Optimized FRESH Support Bath Under Different Stir Speeds and Stir Durations
| 400 rpm | 500 rpm | 600 rpm | 700 rpm | 800 rpm | p-Value | |
|---|---|---|---|---|---|---|
| 14 h | 18.1 [4.9–48.8] | 12.6 [1.9–20.3] | 2.9 [0.3–16.9] | 6.2 [1.7–11.8] | 18.0 [2.1–43.1] | <0.0001 |
| 16 h | 19.3 [4.3–48.9] | 4.6 [2.7–12.3] | 4.3 [0.01–13.9] | 6.0 [1.8–11.6] | 14.1 [2.4–22.2] | <0.0001 |
| 18 h | 16.9 [5.6–62.5] | 5.4 [2.2–17.4] | 4.9 [0.0007–12.8] | 2.8 [0.5–17.7] | 10.5 [2.0–19.3] | <0.0001 |
| 20 h | 22.1 [5.8–48.9] | 5.6 [0.6–12.3] | 5.7 [0.03–11.7] | 4.6 [2.7–16.0] | 8.6 [4.9–40.0] | <0.0001 |
| 22 h | 16.1 [3.0–54.4] | 7.6 [0.8–14.8] | 5.2 [0.006–12.4] | 6.4 [1.9–14.8] | 9.9 [1.2–34.1] | <0.0001 |
| p-Value | 0.0006 | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
p-Values in this table were all obtained via the one-way analysis-of-variance test. p < 0.05 denotes statistical significance.
Comparison of microsphere size average variances revealed significant differences across all stir speeds in microsphere size uniformity (p < 0.001). Stir speeds of 500, 600, and 700 rpm are associated with the smallest variances compared with 400 and 800 rpm for all stir durations (p < 0.001).
Effect of stir duration on FRESH microsphere size
As for stir duration, seen in Figure 5b (Tables 1 and 2), a U-shaped distribution was observed for the median microsphere sizes stirred for 14, 16, 18, 20, and 22 h across all stir speeds, except for 600 rpm, which had consistent microsphere sizes for all stir durations. Under all stir durations, the median variances of microsphere sizes at all stir speeds were significantly different (p < 0.001).
Effect of stir condition on FRESH microsphere yield
Visual assessment of the solution post-stir included gelatin adhering to the stir blade and sides of the beaker as stir speed increased, most notably in mixtures stirred at 700 and 800 rpm. In addition, with increased stir duration, a decrease in yield of compacted gelatin post-centrifugation from the 40 mL aliquot was observed.
Overall, the aggregated trends of stir speed and stir duration revealed the smallest microsphere sizes with the narrowest interquartile range of variance (Tables 1 and 2; Supplementary Fig. S2) in the FRESH generated after stirring at 600 rpm for 20 h. This was used as the optimized FRESH protocol for the remainder of the study.
Single filament print resolution
Figures 3c and 3d shows the single filament construct printed using the commercial FRESH and optimized FRESH imaged under the Keyence microscopy, respectively. Qualitative observations revealed that constructs printed in the commercial FRESH tend to disassemble into separate fragments post cross-linking (Fig. 3c). This was not observed in constructs printed in the optimized FRESH, which held structural integrity throughout the printing and cross-linking processes (Fig. 3d).
In addition, constructs printed using the optimized FRESH compared with the commercial FRESH had higher uniformity in single filament diameter as reflected by Figure 4c and by the standard deviations of the single filament diameter measurements. Measurements of constructs printed in these two support baths revealed the average single filament diameter of 0.2 ± 0.03 mm in the commercial FRESH and 0.09 ± 0.02 mm in the optimized FRESH (p = 0.002; Fig. 4c).
“S” construct print resolution
The height, width, and middle thickness of the Stanford “S” shown in Figure 4b, was compared between constructs printed using the commercial and optimized FRESH. Figure 3e and 3f show the constructs printed in the commercial FRESH and optimized FRESH respectively, before alginate cross-linking. Figure 3g and 3h show the constructs respectively printed in the commercial and optimized FRESH after cross-linking.
Qualitatively, the “S” constructs printed using the optimized FRESH yielded higher angular precision in comparison to constructs printed using the commercial FRESH. Quantitatively (Fig. 4d), the average height of the “S” constructs printed was 9.0 ± 1.0 mm using the commercial FRESH and 8.5 ± 0.7 mm using the optimized FRESH (p = 0.3). The constructs' average width was 5.5 ± 0.5 mm using the commercial FRESH and 5.2 ± 0.3 mm using the optimized FRESH (p = 0.2). The average middle thickness of the “S” construct was 1.5 ± 0.2 mm using the commercial FRESH and 1.4 ± 0.2 mm using the optimized FRESH (p = 0.7).
Complex hollow 3D construct fidelity
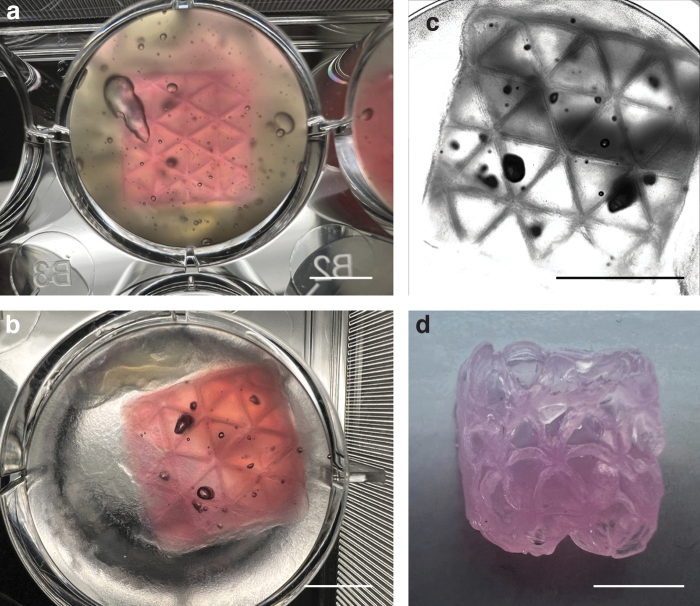
Optimized FRESH adequately supported complex hollow 3D constructs for bioprinting (Fig. 6). The printed constructs maintained extremely high geometric fidelity immediately after bioprinting and after cross-linking. The phase-contrast image also demonstrated clear triangular infills as well as a squared overall box geometry while the construct was submersed in aqueous solution. After the construct was removed from the liquid, it still maintained relatively high geometric fidelity, even though the material is soft and can be easily collapsed in air.
FIG. 6.
Complex hollow 3D constructs printing using the optimized FRESH. (a) A 12 × 12 × 12 mm box with 3 mm triangular infill immediately after printing before cross-linking. (b) The same box 24 h after cross-linking. (c) Phase contrast microscopy image of the same box 24 h after cross-linking. (d) The same box 24 h after cross-linking in air. Scale bar (a–d) = 5 mm.
Discussion
Within tissue engineering, 3D bioprinting involves the fabrication of constructs with anatomical geometry and native properties using hydrogel bio-inks.18 Extrusion-based bioprinting works through a computer-controlled syringe pump, where the extruder can deposit hydrogel, operate at high printing speeds, and create structurally complex constructs.19 Using the FRESH technique, constructs are physically reinforced with biocompatible support during the printing process to improve construct fidelity.14
Various geometries and organs have been successfully bioprinted, including capillaries, cardiac ventricles, and other components of the human heart.15 An aim to further improve FRESH-based bioprinting involves increasing the printing resolution of hydrogel constructs. Though previous studies have shown techniques for post-print shrinking technologies,20 in this study, we proposed a substantially improved protocol for FRESH generation pre-print in an effort to improve printing resolution.
FRESH is generated through complex coacervate formation.15,21 This process involves a solution's spontaneous immiscible separation driven by the electrostatic attraction of oppositely charged macroions or polymers.22–24 In this method, we utilize the negatively-charged gelatin protein biopolymer and positively-charged gum arabic polysaccharide for FRESH generation.25 The resulting support bath is composed of densely packed individual microspheres.
Previously, characteristics such as nanoparticle size have been accurately quantified.26 Smaller and more uniform microsphere sizes of FRESH were reported to aid in enhancing printing resolution and construct accuracy.15 Thus, we aimed at finding optimum conditions to further reduce microsphere size and improve uniform morphology. We specifically examined the microsphere size as a function of the stirring speed and stirring duration during initial mixture generation.
In a study on chitosan-tripolyphosphate nanoparticles, Hussain and Sahudin examined the impact that formulation conditions, including concentration, mass ratios, solution pH, ultra-sonication, and ultra-centrifugation, as well as stirring speed and duration, have on nanoparticle sizes.27 Hussain and Sahudin noted optimum conditions for a minimal particle size at a stirring speed of 700 rpm for 10 min. In Marinho et al's study on the polymerization of vinyl chloride monomers, a stir speed of 900 rpm was observed to result in the most uniform distribution of particle size.
Marinho et al also noted increasing particle size uniformity with increasing time.28 In terms of FRESH generation, the protocol proposed by Lee et al noted an overnight stir duration with no specifications on stir speed.15
As such, we proposed stir speeds between 400 and 800 rpm in 100 rpm increments and stir durations between 14 and 22 h in 2-h increments in our work. Following repeated generations and analyses of FRESH under different conditions, we found optimum conditions of FRESH microsphere size at 600 rpm for 20 h, which yielded the smallest microspheres with the narrowest interquartile range of variance. This optimal stir speed is comparable with previous studies; the slight variation in our microsphere sizes can be accounted for by the inherent biological difference of the FRESH gelatin particles in comparison to other studies' nanoparticles.27,28
Our optimal stir duration of 20 h for minimal size and most uniformity is similar to Lee et al's FRESH v2.0 protocol's overnight stirring, while also adhering to Marinho et al's findings regarding increased stir duration. The optimal conditions we observed in this study to improve FRESH microsphere size and uniformity represent an advancement toward increasing construct print resolution.
Qualitative observations during FRESH generation produced under different stir speeds revealed an effect on the gelatin. Specifically, gelatin increasingly adhered to the stirrer's stir blade and sides of the beaker at higher stir speeds of 700 and 800 rpm. This supports previous studies' observations of an increased stir speed associated with increased solution viscosity,29,30 as high turbulence results in particles' solidification. A decreased volume of solid gelatin in the 40 mL solution aliquot after centrifugation was similarly observed with increasing stir speed. This is due to initially decreased solid gelatin, associated with the aforementioned gelatin's adherence to the stir blade and beaker, which manifests post-centrifugation.
Comparison of the commercial FRESH and optimized FRESH revealed significant differences in microsphere characteristics. We found the optimized FRESH microspheres to have an increased uniformity compared to reported Lee v2.0 FRESH microspheres. In addition, the commercial FRESH showed larger and rhomboid-shaped microspheres whereas the optimized FRESH revealed smaller and circular-shaped microspheres. We identified a few likely key variables in the FRESH manufacturing protocol that could account for such disparities between the Lee v2.0 and the commercial FRESH microspheres.
We further hypothesized that the lyophilization process that is required to generate the commercial LifeSupport powder likely has an impact on microsphere shape, size, and uniformity, as previous studies revealed a large particle size and increased leakage,31,32 as well as a larger poly-dispersity index,33 after the freeze-drying process. Thus, lyophilizing the commercial LifeSupport powder pre-print may contribute to the differences observed between the Lee v2.0 and the commercial FRESH microspheres.
Under the same printing parameters, the single filament constructs printed using the optimized FRESH held high fidelity in comparison to disassembling observed on filaments printed using the commercial FRESH. In their study, Lee et al noted an improved printing resolution of FRESH v2.0 compared with the larger-sized, irregularly shaped, and polydisperse FRESH v1.0 microspheres.15 Similarly, our differences in construct fidelity and print resolution may also be attributed to the commercial FRESH microspheres' larger size and non-uniformity.
For future studies, single filament diameters may be further reduced by adjusting bio-ink concentration, operating temperature, extrusion pressure, and print speed.34 Following this work, the smallest single filament diameter using the optimized FRESH will be examined by further adjusting print parameters. Our findings in printing resolution enhancement present implications of bioprinting micron-scale constructs with high precision and accuracy. Such techniques can be utilized in tissue engineering for printing microvascular networks composed of thin micro-vessels to enable and promote nutrient diffusion and waste removal.35,36
The bioprinted Stanford “S” constructs using the optimized and the commercial FRESH achieved high construct fidelity. No significant difference in dimensions was found between the two constructs, likely due to their large sizes amid the smaller-scale LifeSupport microspheres. Despite no significant quantitative differences, we observed qualitatively higher angular fidelity and sharper angles in the optimized FRESH printed constructs. In the complex hollow 3D construct bioprinting experiment, we again confirmed high fidelity in bioprinting using the optimized FRESH.
Also note that a lower print speed was used for these experiments compared with the single filament experiment. As a future experiment, we will quantify angular precision using geometric methodology such as print circularity to determine constructs' printability values.37 We will continue examining construct fidelity by varying printing parameters.
As all construct images were taken 24 h post-printing, our measured results account for the alginates' swelling property,17 where preset construct dimensions are smaller than constructs' measurements. Yet, “S” constructs printed using the optimized FRESH still achieved smaller dimensions than those of the commercial FRESH, confirming enhanced printing resolution of our improved FRESH.
Overall, future work to further minimize microsphere size or increase uniform morphology in FRESH generation may involve the comparison of gelatin type A versus gelatin type B during complex coacervation formation, decreasing the mixture's initial pH with hydrochloric acid or increasing with sodium hydroxide, or adjusting the dilution of the optimized FRESH with CaCl2 used for bioprinting to aid needle extrusion in the support bath.
During bioprinting, parameters including bio-ink concentration, print temperature, extrusion pressure, and extrusion speed may be modified for further improved print resolution. Construct fidelity should be assessed over time in physiologic conditions. Assessing angular precision of the “S” constructs may also improve quantifications. Despite these areas for future refinement, our work presented an optimum condition for minimal microsphere size and highest uniformity through the examination of the FRESH manufacturing parameters such as stir speed and stir duration.
Conclusion
In summary, this study comprehensively demonstrated the effect of stir speed and stir duration on microsphere characteristics in FRESH. Specifically, we observed a U-shaped distribution between stir speed and resulting microsphere size, as well as between stir duration and microsphere size. Overall, our methodology to generate the optimized FRESH has significant implications in the field of 3D bioprinting given its capabilities in supporting bioprinting at significantly higher resolution and excellent construct fidelity compared with the commercial FRESH. Future evaluation of the optimized FRESH should be explored to assess its application in the field of tissue engineering and regenerative medicine.
Supplementary Material
Acknowledgment
The authors would like to thank Kevin Taweel for the generous donation for this research.
Authors' Contributions
C.A.W.: Conceptualization, methodology, validation, investigation, analysis, writing-original draft, and visualization. Y.Z.: Conceptualization, methodology, validation, investigation, and writing-review and editing. A.V.: Conceptualization, methodology, investigation, and writing-review and editing. C.J.S.: Methodology, investigation, and writing-review and editing. S.H.L.: Resources, investigation, and writing-review and editing. Y.J.W.: Conceptualization, supervision, funding acquisition, and writing-review and editing.
Disclaimer
The manuscript contents are solely the responsibility of the authors and do not necessarily represent the official views of the funders.
Disclosure Statement
The authors declare that they have no competing interests.
Funding Information
This work was supported by the National Institutes of Health (NIH R01 HL089315, Y.J.W.; NIH F32 HL158151, Y.Z.) and the Thoracic Surgery Foundation Resident Research Fellowship (Y.Z.).
Supplementary Material
References
- 1. Mirdamadi E, Tashman JW, Shiwarski DJ, et al. . FRESH 3D bioprinting a full-size model of the human heart. ACS Biomater Sci Eng 2020;6(11):6453–6459; doi: 10.1021/acsbiomaterials.0c01133 [DOI] [PubMed] [Google Scholar]
- 2. Ng WL, Lee JM, Zhou M, et al. . Vat polymerization-based bioprinting-process, materials, applications and regulatory challenges. Biofabrication 2020;12(2):022001; doi: 10.1088/1758-5090/ab6034 [DOI] [PubMed] [Google Scholar]
- 3. Pardo L, Wilson WC, Boland T. Characterization of patterned self-assembled monolayers and protein arrays generated by the ink-jet method. Langmuir 2003;19(5):1462–1466; doi: 10.1021/la026171u [DOI] [Google Scholar]
- 4. Li X, Liu B, Pei B, et al. . Inkjet bioprinting of biomaterials. Chem Rev 2020;120(19):10793–10833; doi: 10.1021/acs.chemrev.0c00008 [DOI] [PubMed] [Google Scholar]
- 5. Jiang T, Munguia-Lopez JG, Flores-Torres S, et al. . Extrusion bioprinting of soft materials: An emerging technique for biological model fabrication. Appl Phys Rev 2019;6(1):011310; doi: 10.1063/1.5059393 [DOI] [Google Scholar]
- 6. You F, Eames BF, Chen X. Application of extrusion-based hydrogel bioprinting for cartilage tissue engineering. Int J Mol Sci 2017;18(7):1597; doi: 10.3390/ijms18071597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xin S, Deo KA, Dai J, et al. . Generalizing hydrogel microparticles into a new class of bio-inks for extrusion bioprinting. Sci Adv 2021;7(42):eabk3087; doi: 10.1126/sciadv.abk3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Behre A, Tashman JW, Dikyol C, et al. . 3D bioprinted patient-specific extracellular matrix scaffolds for soft tissue defects. Adv Healthc Mater 2022;11(24):e2200866; doi: 10.1002/adhm.202200866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jia W, Gungor-Ozkerim PS, Zhang YS, et al. . Direct 3D bioprinting of perfusable vascular constructs using a blend bio-ink. Biomaterials 2016;106:58–68; doi: 10.1016/j.biomaterials.2016.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pi Q, Maharjan S, Yan X, et al. . Digitally tunable microfluidic bioprinting of multilayered cannular tissues. Adv Mater 2018;30(43):e1706913; doi: 10.1002/adma.201706913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cui X, Li J, Hartanto Y, et al. Advances in extrusion 3D bioprinting: A focus on multicomponent hydrogel-based bio-inks. Adv Healthc Mater 202;9(15):e1901648; doi: 10.1002/adhm.201901648 [DOI] [PubMed] [Google Scholar]
- 12. Matai I, Kaur G, Seyedsalehi A, et al. . Progress in 3D bioprinting technology for tissue/organ regenerative engineering. Biomaterials 2020;226:119536; doi: 10.1016/j.biomaterials.2019.119536 [DOI] [PubMed] [Google Scholar]
- 13. Ribeiro A, Blokzijl MM, Levato R, et al. . Assessing bio-ink shape fidelity to aid material development in 3D bioprinting. Biofabrication 2017;10(1):014102; doi: 10.1088/1758-5090/aa90e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hinton TJ, Jallerat Q, Palchesko RN, et al. . Three-dimensional printing of complex biological structures by freeform reversible embedding of suspended hydrogels. Sci Adv 2015;1(9):e1500758; doi: 10.1126/sciadv.1500758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee A, Hudson AR, Shiwarski DJ, et al. . 3D bioprinting of collagen to rebuild components of the human heart. Science 2019;365(6452):482–487; doi: 10.1126/science.aav9051 [DOI] [PubMed] [Google Scholar]
- 16. Fan C, Wang DA. Macroporous hydrogel scaffolds for three-dimensional cell culture and tissue engineering. Tissue Eng Part B Rev 2017;23(5):451–461; doi: 10.1089/ten.teb.2016.0465 [DOI] [PubMed] [Google Scholar]
- 17. Zhu Y, Stark CJ, Madira S, et al. . Three-dimensional bioprinting with alginate by freeform reversible embedding of suspended hydrogels with tunable physical properties and cell proliferation. Bioengineering (Basel) 2022;9(12):807; doi: 10.3390/bioengineering9120807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Duan B, Kapetanovic E, Hockaday LA, et al. . Three-dimensional printed trileaflet valve conduits using biological hydrogels and human valve interstitial cells. Acta Biomater 2014;10(5):1836–1846; doi: 10.1016/j.actbio.2013.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pusch K, Hinton TJ, Feinberg AW. Large volume syringe pump extruder for desktop 3D printers. HardwareX 2018;3:49–61; doi: 10.1016/j.ohx.2018.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gong J, Schuurmans CCL, Genderen AMV, et al. . Complexation-induced resolution enhancement of 3D-printed hydrogel constructs. Nat Commun 2020;11(1):1267; doi: 10.1038/s41467-020-14997-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shiwarski DJ, Hudson AR, Tashman JW, et al. . Emergence of FRESH 3D printing as a platform for advanced tissue biofabrication. APL Bioeng 2021;5(1):010904; doi: 10.1063/5.0032777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kizilay E, Kayitmazer AB, Dubin PL. Complexation and coacervation of polyelectrolytes with oppositely charged colloids. Adv Colloid Interface Sci 2011;167(1–2):24–37; doi: 10.1016/j.cis.2011.06.006 [DOI] [PubMed] [Google Scholar]
- 23. Blocher WC, Perry SL. Complex coacervate-based materials for biomedicine. Wiley Interdiscip Rev Nanomed Nanobiotechnol 2017;9(4):e1442; doi: 10.1002/wnan.1442 [DOI] [PubMed] [Google Scholar]
- 24. Rocha-Selmi GA, Theodoro AC, Thomazini M, et al. . Double emulsion stage prior to complex coacervation process for microencapsulation of sweetener sucralose. J Food Eng 2013;119(1):28–32; doi: 10.1016/j.jfoodeng.2013.05.002 [DOI] [Google Scholar]
- 25. Rousi Z, Malhiac C, Fatouros DG, et al. . Complex coacervates formation between gelatin and gum arabic with different arabinogalactan protein fraction content and their characterization. Food Hydrocoll 2019;96:577–588; doi: 10.1016/j.foodhyd.2019.06.009 [DOI] [Google Scholar]
- 26. Rasmussen MK, Pedersen JN, Marie R. Size and surface charge characterization of nanoparticles with a salt gradient. Nat Commun 2020;11(1):1–8; doi: 10.1038/s41467-020-15889-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hussain ZA, Sahudin SH. Preparation, characterisation and colloidal stability of chitosan-tripolyphosphate nanoparticles: Optimisation of formulation and process parameters. Int J Pharm Pharm Sci 2016;8(3):297–308. [Google Scholar]
- 28. Marinho R, Horiuchi L, Pires CA. Effect of stirring speed on conversion and time to particle stabilization of poly (vinyl chloride) produced by suspension polymerization process at the beginning of reaction. Braz J Chem Eng 2018;35:631–640; doi: 10.1590/0104-6632.20180352s20160453 [DOI] [Google Scholar]
- 29. Ng SK, Nyam KL, Nehdi IA, et al. . Impact of stirring speed on β-lactoglobulin fibril formation. Food Sci Biotechnol 2016;25(Suppl 1):15–21; doi: 10.1007/s10068-016-0093-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Adigun OD, Oni A, Obadele BA. Effect of stirring on solid solution hardening. Mater Today: Proc 2021;46:7740–7744; doi: 10.1016/j.matpr.2021.02.247 [DOI] [Google Scholar]
- 31. Glavas-Dodov M, Fredro-Kumbaradzi E, Goracinova K. The effects of lyophilization on the stability of liposomes containing 5-FU. Int J Pharm 2005;291(1–2):79–86; doi: 10.1016/j.ijpharm.2004.07.045 [DOI] [PubMed] [Google Scholar]
- 32. Maa YF, Nguyen PA, Sweeney T. Protein inhalation powders: Spray drying vs spray freeze drying. Pharm Res 1999;16(2):249–254; doi: 10.1023/a:1018828425184 [DOI] [PubMed] [Google Scholar]
- 33. Shokri M, Tavallaie M, Hosseini SM. Effect of lyophilization on the size and polydispersity of unilamellar and multilamellar liposomes. JMSN 2016;3:37–40; doi: 10.15436/2377-1372.16.1139 [DOI] [Google Scholar]
- 34. Yao Y, Molotnikov A, Parkington HC, et al. . Extrusion 3D bioprinting of functional self-supporting neural constructs using a photoclickable gelatin bio-ink. Biofabrication 2022;14(3):035014; doi: 10.1088/1758-5090/ac6e87 [DOI] [PubMed] [Google Scholar]
- 35. Liu X, Yue T, Kojima M, et al. . Bio-assembling and bioprinting for engineering microvessels from the bottom up. Int J Bioprint 2021;7(3):366; doi: 10.18063/ijb.v7i3.366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yue T, Zhao D, Phan DTT, et al. . A modular microfluidic system based on a multilayered configuration to generate large-scale perfusable microvascular networks. Microsyst Nanoeng 2021;7:4; doi: 10.1038/s41378-020-00229-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Deo KA, Singh KA, Peak CW, et al. . Bioprinting 101: Design, fabrication, and evaluation of cell-laden 3D bioprinted scaffolds. Tissue Eng Part A 2020;26(5–6):318–338; doi: 10.1089/ten.TEA.2019.0298 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.