Sir,
Acinetobacter baumannii is an emerging nosocomial pathogen of concern because it persists in hospital environments and can acquire exogenous DNA via horizontal genetic transfer (HGT) mechanisms [1]. The World Health Organization recently classified A. baumannii as a priority one pathogen, indicating there is a great need for research to combat this species [2]. Although it is well established that A. baumannii is naturally competent and that natural transformation, one of the HGT mechanisms, plays an important role in the extensive and rapid development of multidrug- and extensively drug-resistant strains of A. baumannii, research focusing on this phenomenon is scarce [1]. Our recent publications show that human serum albumin significantly increases transformation frequencies in two strains of A. baumannii, while other host human products show no such effects [1,3]. We aimed to identify additional inducers of competence in A. baumannii by testing the effect of sub-inhibitory concentrations of three antibiotics on transformation frequencies in this species using previously described methods [1,3,4].
Briefly, four kanamycin-susceptible A. baumannii strains (A118, ATCC 17978, ATCC 19606, and A42) were grown in the presence of sub-inhibitory concentrations of antibiotics (Mitomycin C [MMC] or Nalidixic Acid [NAL], both of which are DNA-damaging antibiotics used in previous studies [5,6], or Meropenem [MEM]) and then transformed with plasmid DNA (pDSredAK, conferring kanamycin resistance) or genomic DNA (from strain A. baumannii 144, known to carry a kanamycin resistance gene) and plated on LB agar supplemented with 10 μg/mL kanamycin. Colony-forming units (CFUs) were quantified in parallel by plating serial dilutions on LB agar plates and transformation events were determined by calculating the number of kanamycin-resistant colonies per CFU. Experiments were performed in at least triplicate, statistical analysis (Mann-Whitney Test) was performed using PrismPad (GraphPad software, San Diego, CA, USA) and a P-value < 0.05 was considered significant.
MMC functions by cross-linking complimentary DNA strands and has been shown to induce the DNA repair (SOS) response pathway in several bacteria, to increase transformation frequencies in Legionella pneumophila and to increase genes associated with stress response in A. baumannii and Acinetobacter baylyi [7]. The minimum inhibitory concentration (MIC) of MMC on A. baumannii A118 cells was determined to be 3.125 μg/mL and a concentration of 0.2 μg/mL was used during transformation assays. While transformation frequencies increased with all strains and DNA types under MMC induction, strain ATCC 17978 and ATCC 19606 showed significant increases, with ATCC 17978 showing the most dramatic increases of 19.83-fold with plasmid DNA and 13.04-fold with genomic DNA (Fig. 1A). NAL is a synthetic quinolone antibiotic that functions by inhibiting DNA gyrase and/or topoisomerase. The MIC of NAL was determined as 5 μg/mL and it was tested at a concentration of 1.25 μg/mL. As with MMC, NAL increased transformation frequencies in all strains with both DNA types but only significantly increased transformation frequencies in strain A42, transformed with plasmid DNA (Fig. 1B). MEM is a carbapenem that functions by inhibiting the formation of peptidoglycan in the cell wall and is considered the last line of defense in treating infections caused by Gram-negative multidrug-resistant bacteria. The MIC of MEM was determined to be 0.25 μg/mL and a concentration of 0.063 μg/mL was used for transformation. As with the previously described antibiotics, MEM increased transformation frequencies in all strains tested with both DNA types. However, it significantly increased transformation frequencies by 12.12-fold in ATCC 17978, when transformed with plasmid DNA, and 3.14-fold in A42, when transformed with genomic DNA (Fig. 1C).
Fig. 1.
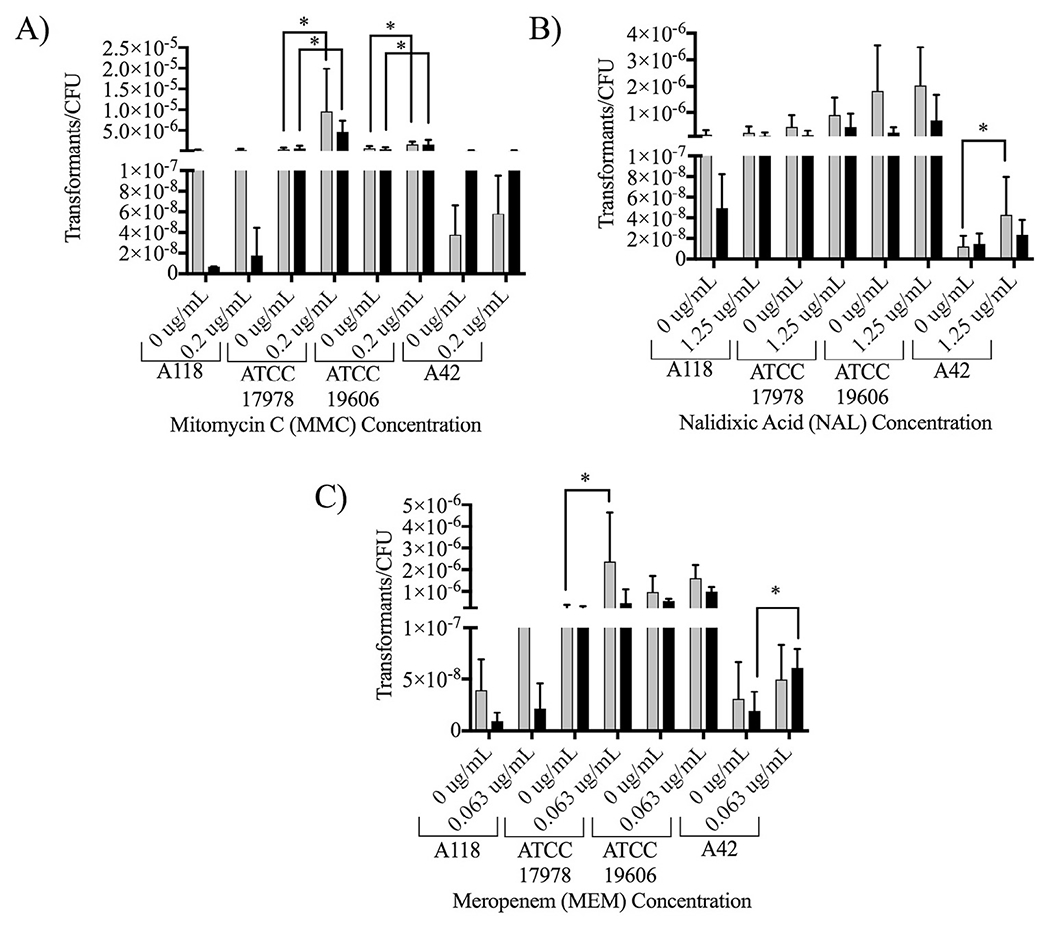
Natural transformation frequencies with sub-inhibitory antibiotics. Transformation assays were performed in LB broth with A) 0.2 μg/mL MMC, B) 1.25 μg/mL NAL, or C) 0.063 μg/mL MEM. Cultures were transformed with plasmid DNA (grey) or genomic DNA (black) and plated on LB agar supplemented with 10 μg/mL kanamycin and CFUs were plated on LB agar. Data are presented as the mean and error bars represent the standard deviation. At least three independent replicates were performed and P < 0.05 was considered significant (Mann Whitney t test, n = 3 to 6).
As MMC and NAL target the bacterial genome, it is likely that they induce competence via the unique DNA damage stress response pathway of A. baumannii. In many bacterial species, DNA damage triggers RecA-dependent autoproteolysis of the LexA repressor, but this does not occur in A. baumannii because of the absence of several crucial genes, including lexA [8]. Instead, we hypothesize that A. baumannii, like A. baylyi, responds to DNA damage in a RecA-independent manner, in which RecA is involved in DNA repair as a member of the recombination repair pathway, and that competence developed as a DNA damage response mechanism in this species. The role of MEM is not as clear. Although carbapenems like MEM have been shown to increase plasmid stability in Klebsiella pneumoniae by inducing expression of the CRISPR-cas activity regulator, H-NS, the CRISPR-cas system was not identified in any of the tested A. baumannii strains [9]. As such, carbapenems may interact with the H-NS regulator to induce competence via a novel pathway. Together, these results indicate there are multiple pathways to competence induction in A. baumannii and that further analysis of these pathways is crucial to understanding how competence functions in this species.
Funding:
BQ was supported by grant MHIRT 2T37MD001368 from the National Institute on Minority Health and Health Disparities, National Institute of Health.
Footnotes
Competing interests: No.
Ethical approval: Not required.
References
- [1].Traglia GM, Quinn B, Schramm ST, Soler-Bistue A, Ramirez MS. Serum albumin and Ca2+ are natural competence inducers in the human pathogen Acinetobacter baumannii. Antimicrob Agents Chemother 2016;60:4920–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Tacconelli E, Magrini N. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organization; 2017. [Google Scholar]
- [3].Quinn B, Traglia GM, Nguyen M, Martinez J, Liu C, Fernandez JS, et al. Effect of host human products on natural transformation in Acinetobacter baumannii. Curr Microbiol 2018;doi: 10.1007/s00284-017-1417-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ramirez MS, Don M, Merkier AK, Bistue AJS, Zorreguieta A, Centron D, et al. Naturally competent Acinetobacter baumannii clinical isolate as a convenient model for genetic studies. J Clin Microbiol 2010;48:1488–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Charpentier X, Kay E, Schneider D, Shuman HA. Antibiotics and UV radiation induce competence for natural transformation in Legionella pneumophila. J Bacteriol 2011;193:1114–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 2006;313:89–92. [DOI] [PubMed] [Google Scholar]
- [7].Charpentier X, Polard P, Claverys JP. Induction of competence for genetic transformation by antibiotics: convergent evolution of stress responses in distant bacterial species lacking SOS? Curr Opin Microbiol 2012;15:570–6. [DOI] [PubMed] [Google Scholar]
- [8].Robinson A, Brzoska AJ, Turner KM, Withers R, Harry EJ, Lewis PJ, et al. Essential biological processes of an emerging pathogen: DNA replication, transcription, and cell division in Acinetobacter spp. Microbiol Mol Biol Rev 2010;74:273–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lin TL, Pan YJ, Hsieh PF, Hsu CR, Wu MC, Wang JT. Imipenem represses CRISPR-Cas interference of DNA acquisition through H-NS stimulation in Klebsiella pneumoniae. Sci Rep 2016;6:31644. [DOI] [PMC free article] [PubMed] [Google Scholar]


