Abstract
The evolution of advanced cognition in vertebrates is associated with two independent innovations in the forebrain: the six-layered neocortex in mammals and the dorsal ventricular ridge (DVR) in sauropsids (reptiles and birds). How these innovations arose in vertebrate ancestors remains unclear. To reconstruct forebrain evolution in tetrapods, we built a cell type atlas of the telencephalon of the salamander Pleurodeles waltl. Our molecular, developmental, and connectivity data indicate that parts of the sauropsid DVR trace back to tetrapod ancestors. In contrast, the salamander dorsal pallium is devoid of cellular and molecular characteristics of the mammalian neocortex, yet shares similarities with entorhinal cortex and subiculum. Our findings chart the series of innovations that resulted in the emergence of the sauropsid DVR, and the mammalian six-layered neocortex.
One-Sentence Summary:
Evolutionary innovations in the vertebrate forebrain are revealed by multimodal analyses of salamander neurons.
The transition from water to land was a pivotal moment in vertebrate history that exposed the first tetrapods to new environmental and cognitive challenges, which may have accelerated adaptive innovations in the nervous system (1). After the divergence of mammals and sauropsids (reptiles and birds) about 320 million years ago, innovations in the pallium (i.e. dorsal telencephalon) paved the way for advanced cognition. In mammals, the neocortex, with its characteristic six-layers, evolved from a simpler ancestral cortex located in the dorsal pallium (2). In sauropsids, an expansion of the ventral pallium produced a large set of nuclei called the dorsal ventricular ridge (DVR) (Fig. 1A). Although neocortex and DVR develop from different parts of the pallium, they bear extensive similarities in terms of gene expression, connectivity, and function (3–5). A model centered on brain connectivity proposes the homology of neocortex and DVR, implying that differences in neocortex and DVR development and topological positions arose secondarily (6). Developmental studies (2) and adult transcriptomics data (7, 8) challenge this view, suggesting that DVR and neocortex have separate evolutionary origins in amniote ancestors, and therefore similar functions were acquired independently. However, the origin of innovations that led to the DVR and neocortex remain poorly understood at the molecular and cellular levels.
Fig. 1. Neuronal diversity in the Pleurodeles telencephalon.
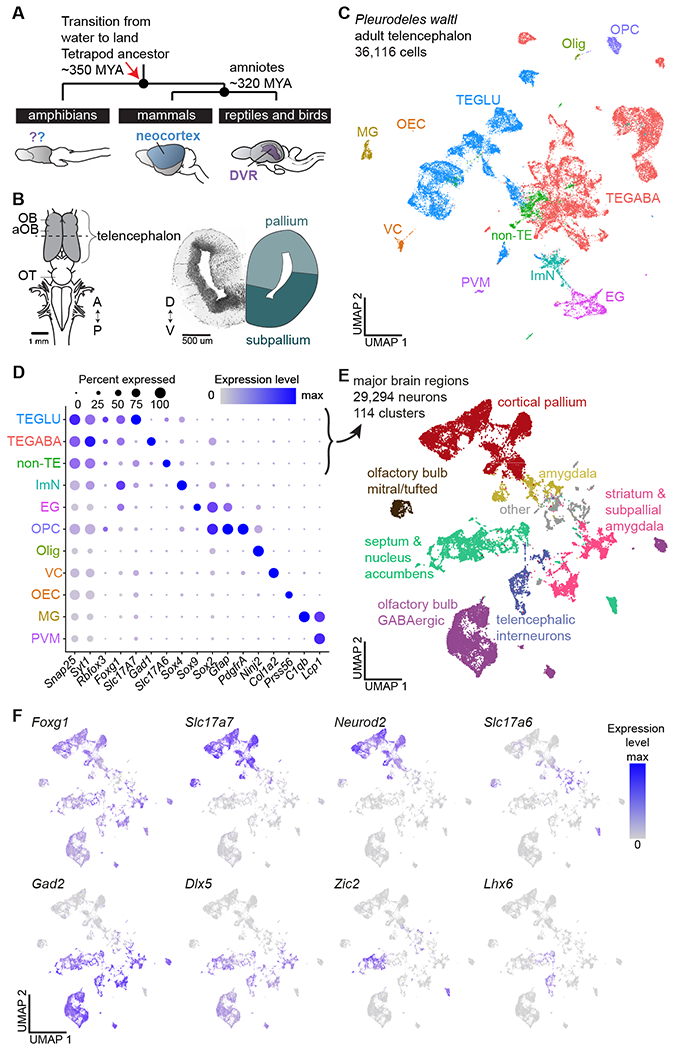
(A) Schematic highlighting the phylogenetic position of amphibians, the mammalian neocortex, and the reptilian DVR. (B) Left: schematic of the Pleurodeles waltl brain (dorsal view). Dotted line indicates section plane for coronal slice on the right. (C) UMAP (Uniform Manifold Approximation and Projection) plot of 36,116 salamander single-cell transcriptomes, colors indicate cell classes. (D) DotPlot showing the expression of marker genes used to annotate the telencephalic dataset in (C). (E) UMAP plot of 29,294 single-cell transcriptomes of salamander neurons, colors indicate major brain regions. (F) UMAP plots showing expression of key markers of glutamatergic and GABAergic neurons in the neuronal dataset.
Abbreviations: A, anterior; aOB, accessory olfactory bulb; D, dorsal; DVR, dorsal ventricular ridge; EG, ependymoglia; GLU, glutamatergic; ImN, immature neurons; MG, microglia; MYA, million years ago; OB, olfactory bulb; OEC, olfactory ensheathing cells; Olig, oligodendrocytes; OPC, oligodendrocyte precursor cells; OT, optic tectum; P, posterior; PVM, perivascular macrophages; TE, telencephalic; V, ventral; VC, vascular cells
We reasoned that if neocortex and DVR have separate origins, they may trace back to pallial regions that existed in a pre-amniote ancestor. Amphibians, which diverged from other tetrapods ~350 million years ago, have a seemingly simple telencephalic architecture, devoid of obvious layering or large brain nuclei (Fig. 1B). Both a dorsal and a ventral pallium exist in amphibians (9), but it is unclear whether they are related in any way to neocortex and DVR. Here, we analyzed the telencephalon of Pleurodeles waltl, a salamander species with a true adult (post-metamorphic) stage, to ask the following: (i) Are there neuron types in the amphibian pallium with transcriptomic similarity to neocortical or DVR neurons? (ii) If yes, how do these neurons develop? (iii) Do these neurons display patterns of connectivity similar to neocortex or DVR?
Results
A cell-type atlas of the salamander telencephalon
To build a cell type atlas of the salamander telencephalon, we profiled entire brains and microdissected telencephali of adult Pleurodeles (brain atlas in Fig. S1, Movie S1). After single-cell RNA sequencing (scRNAseq, 10x Genomics), reads were mapped on a new long-read de novo reference transcriptome (see Methods). Following quality filtering, we obtained 36,116 single-cell transcriptomes, performed Louvain clustering, and identified 11 major populations of neuronal and non-neuronal cells (Fig. 1C; Fig. S2).
We annotated clusters of differentiated neurons, immature neurons, ependymoglial cells, microglia, oligodendrocytes, oligodendrocyte precursors, and other non-neuronal cells based on well-established marker genes (Fig. 1D, Fig. S2C). Differentiated neurons (29,294 cells), identified by the expression of pan-neuronal markers such as Snap25, Syt1, and Rbfox3 (i.e., NeuN), were subclustered to classify neuron types. This revealed 47 clusters of glutamatergic neurons and 67 clusters of GABAergic neurons, which we annotated on the basis of marker genes with conserved expression across species, in situ hybridization for cluster-specific markers, and existing amphibian literature (reviewed in (10, 11)) (Fig. 1E,F; Figs. S3–8).
In the telencephalon, hierarchical clustering revealed four distinct groups of glutamatergic clusters (Fig. S4A). One expressed Neurod2 and Slc17a7 (Vglut1) at high levels; we named this group cortical pallium for its molecular similarity to the cerebral cortex of mammals and reptiles (12, 13). The remaining groups included olfactory bulb mitral and tufted cells, expressing the transcription factor Tbx21 (14); amygdala (Amy) neurons, expressing lower levels of Slc17a7 and Neurod2 and high levels of Slc17a6 (Vglut2); and glutamatergic neurons in the septum, expressing Slc17a6, Zic2, and Isl1 (Fig. 1E,F; Fig. S4A).
Telencephalic GABAergic neurons express markers of the subpallium such as Dlx5, Gad1 and Gad2. We found that the amphibian subpallium includes not only neurons from lateral and medial ganglionic eminences (LGE and MGE) as previously shown (15, 16), but also from the caudal ganglionic eminence (CGE). We identified several types of striatal and septal neurons, nucleus accumbens, bed nucleus of the stria terminalis, and diagonal band neurons, as well as olfactory bulb LGE-derived GABAergic interneurons (Fig. S4A; Fig. S5A–E). Telencephalic GABAergic interneurons, scattered throughout the pallium, included MGE- and CGE-derived cells (Fig. S5F). These data indicate that, despite its anatomical simplicity, the amphibian telencephalon harbors a greater degree of cell type complexity than anticipated.
Spatial distribution of pallial glutamatergic neurons
Literature indicates that the amphibian pallium is organized along the medio-lateral axis in four regions, called medial, dorsal, lateral, and ventral pallium (MP, DP, LP, and VP) (9, 17), but precise boundaries and further subdivisions are a matter of dispute (18, 19) (see Supplementary text about nomenclature). To clarify the organization of the amphibian telencephalon including the pallium, we associated clusters from scRNAseq to their spatial origin, and built a transcriptomics-based map of the telencephalon in Pleurodeles.
Hierarchical clustering of average cluster expression profiles indicates a clear distinction between cortical pallium and amygdala, and the existence of four groups of cortical pallium clusters (Fig. 2A,B, Fig. S4A,B). As shown by in situ hybridization for specific marker genes (Fig. 2C), the four groups largely correspond to MP, DP, LP, and VP. In mid-telencephalic sections, the MP, which is comparable to the hippocampus for its position and connectivity, expressed hippocampal transcription factors such as Fezf2, Lhx9, Zbtb20, and Etv1 (7, 20). The DP, anatomically distinct from the MP, expressed low levels of MP markers but high levels of Etv1. The LP, a narrow band of densely-packed neurons, expressed Lhx2, Satb1, Rorb, and Reln, markers of olfactory-recipient cells in the mammalian piriform cortex (semilunar cells) (21). Most of the ventral pallium expresses the transcription factor Sox6 and is molecularly diverse, in line with its anatomical heterogeneity (9, 22). Subdivisions of VP include a Nos1-negative anterior VP (VPa) and a Nos1+ posterior VP (VPp), see also (16). Along the anterior-posterior axis, Slc17a6 is expressed only in the most ventral portion of the VP, suggesting the existence of further diversity along the medio-lateral axis; Slc17a6 is expressed at higher levels in the adjacent amygdala, as described in (23) (Fig. S6–8; Movies S2,3). Besides these subdivisions of VP, we found expression of Sox6, Znf536 and Grm3 in an anterior pallial region, which is not continuous with VP, but instead is nested between the olfactory bulb and the septum, and corresponds to the amphibian post-olfactory eminence (POE) (24, 25).
Fig. 2. Spatial mapping of pallial neurons in Pleurodeles.
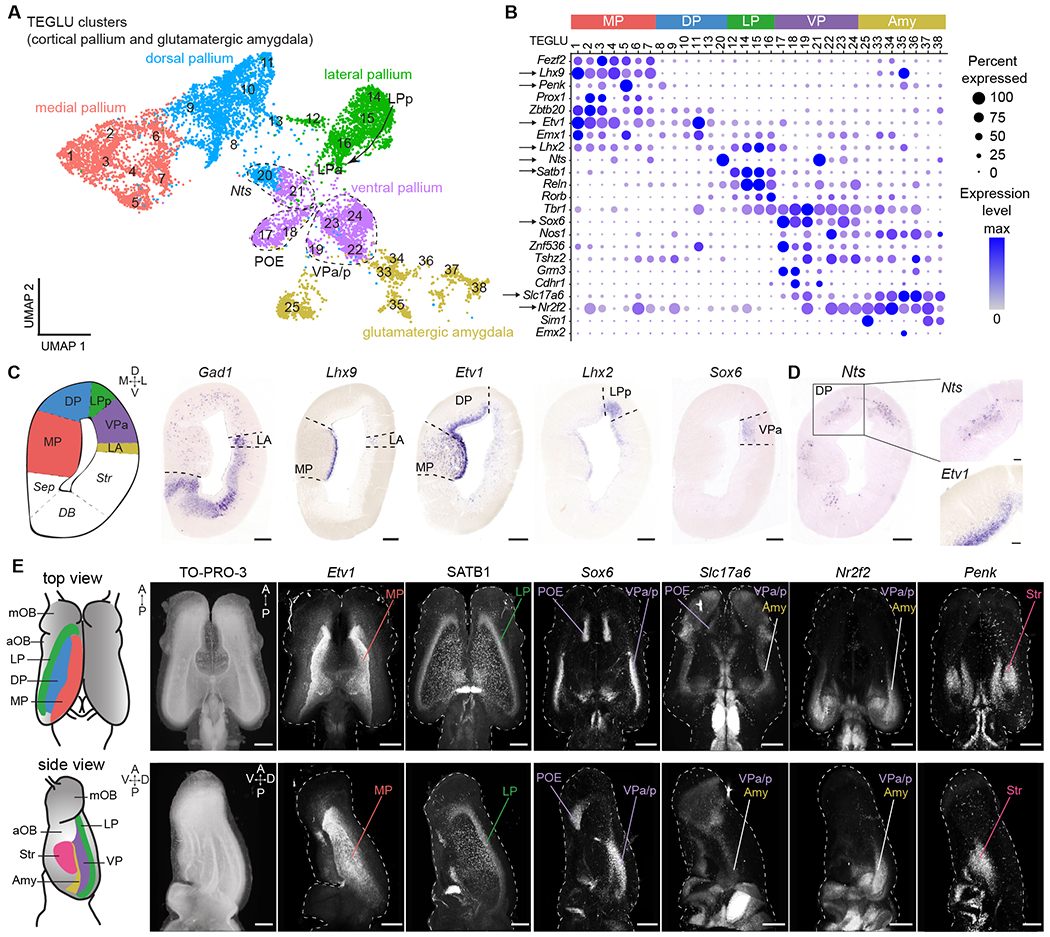
(A) UMAP plot of clusters from cortical pallium and amygdala, annotated by the inferred pallial region. (B) DotPlot showing the expression of key marker genes defining distinct pallial regions. Arrows: genes shown in C-E. (C) Left to right: schematic of a coronal section at mid-telencephalic level, expression of Gad1, marker of the subpallium, and of transcription factors labeling distinct pallial regions along the mediolateral axis. Scale bars: 200 um. (D) Expression of Nts and Etv1 in layers, boxed areas indicate magnifications on the right. Scale bars in right panels: 50 um. (E) Left: schematics of dorsal and lateral surfaces of the salamander telencephalon. Right: dorsal and lateral views of whole-mount immunohistochemistry or HCR stainings for telencephalic markers. Panels show maximum intensity projections of brains after clearing and volumetric light-sheet imaging. Scale bars: 500 um. See methods for specifics on SATB1 antibody. For full list of abbreviations, see Fig. S1; Amy, amygdala; TEGLU, telencephalic glutamatergic.
A closer look at Nts, a marker expressed at high levels in two clusters (Fig. 2B), revealed differential expression along the radial axis. Visualization of Nts expression in tandem with genes expressed in cells closer to the ventricle (e.g. Etv1 in MP/DP) demonstrates that Nts demarcates a discrete, superficial layer of the pallium (Fig. 2D; Fig. S6H). These results suggest that the amphibian pallium contains at least two separate layers of distinct neuron types.
To resolve the 3D organization of the pallium, we exploited the relatively small size of the salamander brain to combine whole-mount hybridization chain reaction (HCR) in situ hybridization, brain clearing (iDISCO), and light-sheet imaging, creating a 3D molecular map (Fig. 2E; Fig. S7; Movies S2,3). This revealed that the cortical pallium is organized in adjacent longitudinal stripes, running the length of the telencephalic vesicle (Fig. 2E). For example, the LP extends from the most rostral tip of the pallium, where it contacts the main olfactory bulb (mOB) and the POE, to the caudal tip of the telencephalon. Instead, the amygdala is localized caudally and is demarcated by Slc17a6 and Nr2f2 expression, and absence of Sox6 (Fig. 2E; Figs. S7–8; Movies S2,3). Together, these data represent a transcriptomics-based map of the amphibian pallium, and support the existence of distinct regions along the mediolateral axis and distinct layers along the radial axis.
Developmental trajectories of dorsal and ventral pallium
How regions of the amphibian pallium compare to distinct regions of the mammalian and sauropsid pallium, including hippocampus, olfactory cortex, and amygdala, remains debated (10, 26, 27). Current models postulate that pallial regions are homologous when they develop from homologous progenitor domains (17, 28, 29). To trace the developmental history of Pleurodeles pallial neuron types, we collected scRNAseq data from stage 36, 41, 46, and 50 larvae (Fig. 3A,B; Fig. S9A) (see also (30)). After unsupervised clustering, we identified radial glia and telencephalic glutamatergic and GABAergic developing neurons (Fig. 3C,D; Fig. S9B–C). To assign developing neurons to their terminal fate in the adult telencephalon, we mapped adult scRNAseq data on developmental data using the Seurat label transfer algorithm (see Methods) (31). This showed that our larval dataset included differentiating neurons from all major pallial and subpallial subdivisions (Fig. 3C Fig. S9C). We then inferred developmental trajectories for differentiation into olfactory bulb mitral and tufted cells, amygdala, VP, LP, DP and MP with Slingshot (32) (Fig. 3E). After reordering cells according to their pseudotime score, we compared gene expression along the VP and DP trajectories (Fig. S9D). Transcription factors upregulated along the dorsal trajectory included Lhx2, Sox8, and Nfix. Transcription factors upregulated along the ventral trajectory included Pbx3 and Sox6, which are also expressed in the developing mouse ventral pallium (33) (Fig. 3F,G). This indicates that neurons in the dorsal and ventral pallium are specified by distinct gene regulatory cascades, possibly controlled by medial wnt signaling and ventrolateral wnt antagonists (29). This analysis thus highlights clear differences in the specification of dorsal and ventral pallium neurons, demonstrating that the distinct VP and DP clusters identified in the adult data have their own developmental programs which rely on evolutionarily conserved transcription factor programs.
Fig. 3. Developmental trajectories in the Pleurodeles telencephalon.
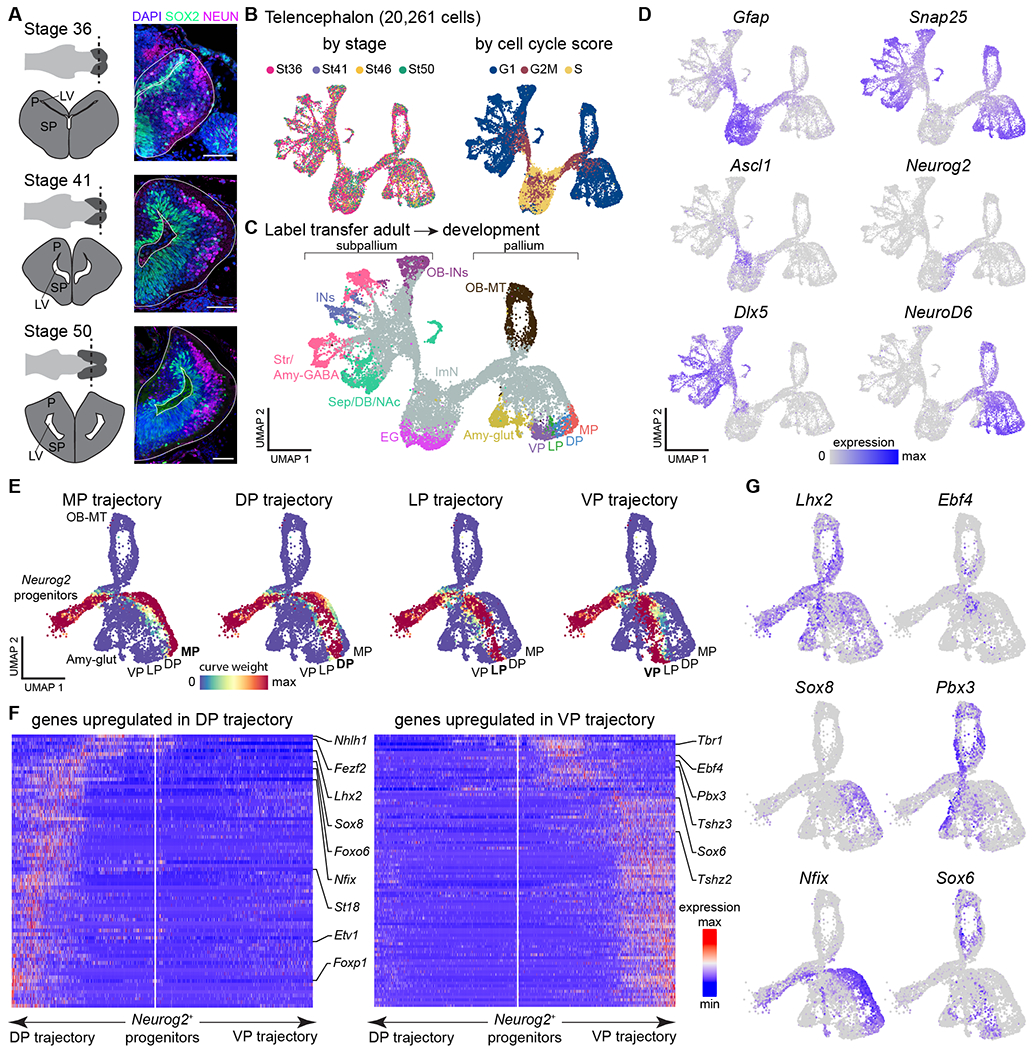
(A) Overview of telencephalic development in Pleurodeles. Right: coronal sections through the telencephalon showing SOX2+ radial glia and interneurons, and NEUN+ differentiated neurons. Scale bars: 100 um. (B) UMAP plots of 20,261 telencephalic cells colored by developmental stage (left) and cell cycle score (right). (C) UMAP plot of the developing telencephalon, colored according to cell classes after label transfer from the adult dataset. (D) UMAP plots colored by the expression of Gfap (radial glia), Snap25 (differentiated neurons), Ascl1 and Neurog2 (committed subpallial and pallial progenitors), and Dlx5 and NeuroD6 (postmitotic subpallial and pallial neurons). (E) UMAP plots showing the assignment of cells to each of the trajectories based on curve weights, which represent the likelihood that a cell belongs to a given principle curve calculated by Slingshot. (F) Heatmaps of genes differentially expressed along the trajectories of the dorsal and ventral pallium, with transcription factors highlighted on the side. White line in the middle of each panel indicates the position of the Neurog2+ progenitors and arrows to the left and right represent the two trajectories. Gene expression levels for each gene are scaled by root mean square ranging from −2 to 6. (G) UMAP plots showing pallial single cells color-coded by expression of transcription factors upregulated in the dorsal (left) or ventral (right) trajectory.
Abbreviations: see Figs. S1, S3; EG, ependymoglia; ImN, immature neurons; LV, lateral ventricle; OB-MT, olfactory bulb mitral and tufted cells; P, pallium; SP, subpallium.
Comparison of salamander VP and reptilian aDVR
To identify neuron types with similar gene expression profiles in salamanders, reptiles, and mammals, we compared scRNAseq datasets using manifold integration algorithms. We compared several data integration algorithms (Fig. S11, see Supplementary text) and present here the results obtained using Seurat, an algorithm based on the identification of mutual nearest-neighbors across single-cell datasets (see Methods) (31). For consistency and to facilitate data analysis, we limited data integration to single cells sampled from the same brain regions. We integrated our salamander dataset with data from the telencephalon of the agamid lizard Pogona vitticeps (34, 35) and from the pallium of the red-eared slider turtle Trachemys scripta (which also includes cells from the neighboring subpallium (7)). Clustering of the Seurat integrated data yielded 65 clusters, which we refer to as integrated clusters. Results from the Seurat integration were largely recapitulated by using alternative parameters in the integration pipeline, as well as alternative integration algorithms (Harmony, SAMap, and scVI; Supplementary text and Figs. S11, S12). Hierarchical clustering of average gene expression in integrated clusters produced a cross-species taxonomy of telencephalic neuron types (Fig. 4A–C; Fig. S13).
Fig. 4. Salamander and reptile telencephalon cross-species comparison.
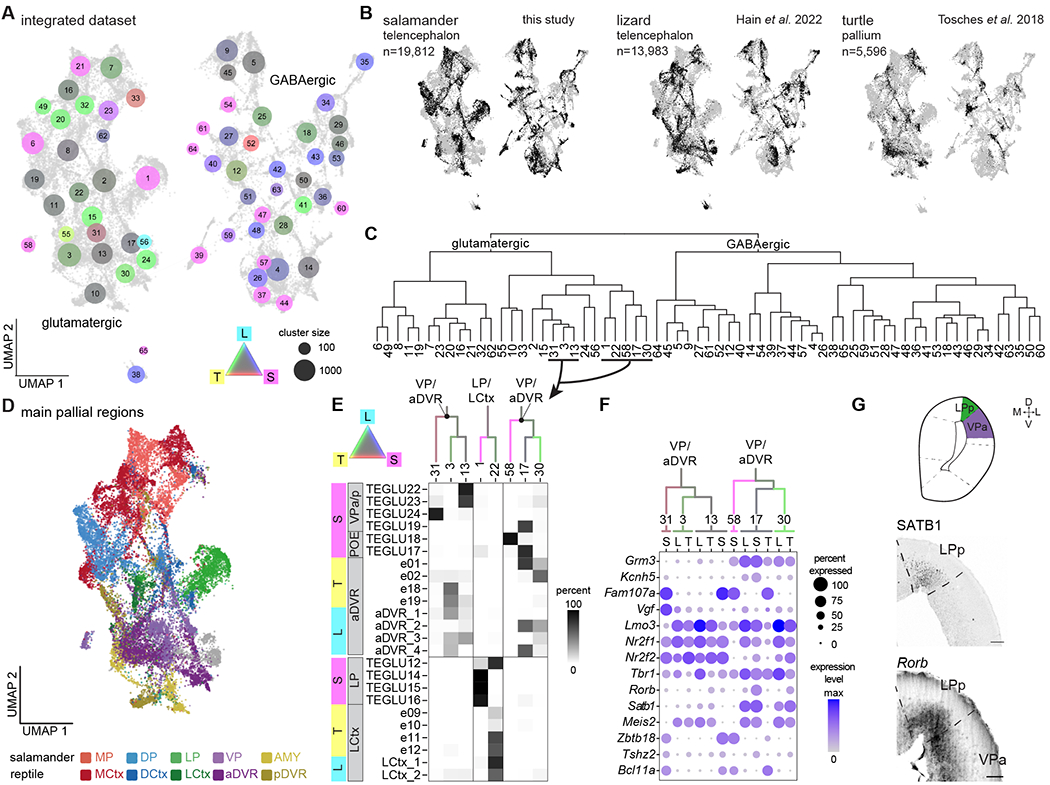
(A) UMAP plot after integration of scRNAseq data from the salamander and lizard telencephalon, and from the turtle pallium. Dot colors indicate species mixture in each integrated cluster (gray represents equal proportion of cells from each species). Dot size indicates the number of cells in each cluster. (B) UMAP plots of the integrated dataset showing cells from each species highlighted in black. (C) Hierarchical clustering of average expression profiles of the integrated clusters shown in (A). (D) UMAP plot of the glutamatergic clusters from the integrated dataset, colored by pallial region. (E) Top: ventrolateral portion of the dendrogram in C, with branches colored by species mixture. Bottom: percentage of cells from the original species-specific clusters (rows) in the integrated clusters (columns). (F) DotPlot showing the expression of molecular markers in aDVR or VP in the integrated clusters, with cells from each integrated cluster split by species (L, lizard; S, salamander; T, turtle). (G) Top: schematic of a coronal section at mid-telencephalic level in the Pleurodeles brain. Bottom: presence of SATB1 and expression of Rorb in the salamander lateral and ventral pallium. Scale bars: 100um.
This kind of analysis is built on molecular similarities of cells, that result from either homology or the convergent use of the same effector genes. Here, we observed co-clustering of salamander and reptilian cells from pallial regions that are considered homologous, on the basis of independent criteria such as their relative position in the pallium (2, 17). For example, we find co-clustering of salamander MP and the reptilian medial cortex, and of salamander LP and the reptilian lateral cortex. The salamander pallial amygdala (23) and reptilian posterior DVR (pDVR), putative homologs of the mammalian pallial amygdala (36), also co-clustered (Fig. 4D, Fig. S13). For its position in the pallium and its connectivity, the amphibian VP is a putative homolog of the reptilian ventrolateral pallium, including the anterior DVR (aDVR) (37). We found that reptilian aDVR and salamander VP neurons co-clustered in two distinct neighborhoods, segregating into a total of six clusters (3, 13, 31, 58, 17 and 30). Integrated cluster 13 included salamander cells from the VPa/p, and turtle and lizard cells from the centromedial aDVR, an area heavily connected with the hypothalamus (Fig. 4E) (7, 37) Integrated cluster 3, in the same neighborhood, included more cells from the lizard and turtle centromedial aDVR. Turtle, lizard, and salamander cells in clusters 13 and 3 shared expression of several transcription factors, including Tbr1, Nr2f2, Nr2f1, and Lmo3 (Fig. 4F), supporting the hypothesis that centromedial aDVR and VP have a shared evolutionary history.
In the second neighborhood, we found cells from the rostral part of the turtle and lizard aDVR (integrated clusters 17 and 30) and from the salamander post-olfactory eminence (POE, integrated clusters 58 and 17). The rostral aDVR is an area receiving sensory inputs (visual, somatosensory, auditory) relayed by the thalamus (7, 37, 38), and expresses the transcription factors Rorb and Satb1 at high levels (7), as well as specific effector genes such as the glutamate receptor Grm3 and the potassium channel Kcnh5 (Fig. 4E,F). The salamander POE is a pallial region primarily involved in olfaction, as suggested by its proximity to and inputs from the olfactory bulb (24, 25). In Pleurodeles, we found that Rorb and Satb1 are co-expressed in POE (weakly) and LP, both pallial regions with prominent olfactory inputs, but not in the VPa/p (Fig. 4F,G). Furthermore, other transcription factors with specific expression in the salamander POE are not expressed in the lizard or in the turtle rostral aDVR (Fig. S14), indicating that the co-clustering of neurons from these regions is driven by effector genes. The lack of transcription factor conservation between these cell types may indicate convergent use of effector genes, as only transcription factors are believed to form core regulatory complexes which track cell types as evolutionary units (39). Taken together, these results indicate that the salamander ventrolateral pallium comprises neuron types with molecular similarity to reptilian lateral cortex and centromedial aDVR. Furthermore, they suggest that the sensory-recipient neurons in the rostral aDVR may have evolved by recruiting effector genes involved in sensory processing in other pallial areas.
Molecular innovations in dorsal cortex and neocortex
To extend our molecular comparisons to mammals, we computed gene expression correlations between each salamander telencephalic cluster with digitized in situ hybridization data from the Allen Brain Atlas (40) (Fig. 5A). This confirmed the molecular similarity of salamander subpallial regions with their mouse counterparts. Results for the pallium were more ambiguous. For example, salamander LP clusters correlated with hippocampus, neocortex, piriform cortex, and lateral amygdala; pallial amygdala clusters with the entire mouse pallial amygdala and piriform cortex; and VP clusters with mouse piriform cortex and lateral amygdala (Fig. 5A). This is consistent with the observation that VP expresses transcription factors with specific or enriched expression in the mouse piriform cortex, such as Znf536 and Tshz2 (Fig. 2B, Fig. S8) (40, 41). Using an alternative approach where we mapped scRNAseq data from the mouse telencephalon (42, 43) on our salamander single-cell dataset (see Methods), we also found correspondence between subpallial and hippocampal regions. However, using this method, mouse cortical pyramidal types could not be mapped to single salamander clusters, suggesting a high degree of transcriptional divergence (Fig. S15).
Fig. 5. Salamander, reptile and mouse cross-species comparison.

(A) Correlations of the transcriptome of selected Pleurodeles clusters with in situ hybridization data from the Allen Adult Mouse Brain Atlas. (B) Integration of scRNAseq data from the salamander medial and dorsal pallium, the turtle and lizard (“reptile”) medial and dorsal cortex, and the mouse hippocampus and cortex. Left: UMAP of the integrated data with dots colored by species mixture, dot size indicates cluster size. Right: UMAP plots of the integrated dataset showing cells from each species highlighted in black. (C) Top: Hierarchical clustering of average expression profiles of integrated GABAergic clusters, branches colored by species mixture (gray represents equal proportion of cells from each species). Bottom: percentage of cells from the original species-specific clusters (rows) in the integrated clusters (columns). (D) DotPlot showing expression of differentiation markers and of transcription factors (TFs) in Lamp5 interneurons (integrated clusters 34, 17, and 49). Cells from each integrated cluster split by species (L, lizard; M, mouse; S, salamander; T, turtle). (E) Top: Hierarchical clustering of average expression profiles of integrated glutamatergic clusters, branches colored by species mixture. Bottom: percentage of mouse cells in each integrated cluster (columns); mouse cells grouped by projection identity (rows). Integrated clusters including selected mouse neuron types are highlighted (ENT, entorhinal cortex; L4 IT CTX, thalamorecipient L4 neurons; SUB, subiculum). (F) Top: percentage of mouse cells in each integrated cluster (columns); mouse cells grouped by cortical area (rows), columns reordered by cortical area. Bottom: percentage of salamander DP cells and turtle dorsal cortex cells (rows) in each integrated cluster (columns). (G-H) Close-up on part of the UMAP in (B), showing cells in cluster 10 (G) or cluster 12 (H) colored by species. (I) DotPlot showing expression of differentiation markers and transcription factors (TFs) in integrated clusters 12, 38, 30, and 32, split by species.
We reasoned that the integration of scRNAseq data was better suited than mapping approaches for the identification of high-level similarities between salamander, reptilian, and mouse neuron types. However, complete scRNAseq data from the entire telencephalon of a mammal are not available yet. In light of our results on development, we decided to focus on the derivatives of the dorsomedial pallium, which in mammals ranges from the hippocampus medially to the insular and entorhinal cortices laterally; complete mouse data are available for all these cortical areas (43) (Fig. 5B). Telencephalic interneurons (44) were also included in this analysis. Salamander GABAergic interneurons co-clustered with amniote MGE-derived (Pvalb and Sst) and CGE-derived (Lamp5, Sncg, and Vip) interneuron classes (Fig. 5C; Fig. S15B), indicating that these interneuron classes trace back to tetrapod ancestors. At deeper levels of interneuron classification, we found interneuron types conserved in tetrapods, such as long-range projecting Sst Chodl neurons (cluster 13), and mammalian-specific types, such as Pvalb Vipr2 Chandelier cells (cluster 50) (Fig. 5C). Lamp5 interneurons included a non-mammalian subclass (cluster 34), a conserved subclass (cluster 17, Lamp5 Ndnf neurogliaform cells in mouse) and a mouse-specific subclass (cluster 49, Lamp5 Lhx6 cells) (45). The transcription factors that differentiate between mammalian Lamp5 Ndnf and Lamp5 Lhx6 interneurons are coexpressed in non-mammalian Lamp5 cells (cluster 34), suggesting that amniote or mammalian-specific Lamp5 types evolved by diversification of ancestral Lamp5 interneurons (Fig. 5D).
In the cross-species taxonomy of glutamatergic neurons, major splits corresponded to neurons with distinct projection identities in mouse (IT: intratelencephalic; ET: extratelencephalic, including L5 pyramidal tract neurons; CT: corticothalamic neurons) (Fig. 5E; Fig. S16). Several integrated clusters included neurons from mouse only, indicating a greater diversity of mouse pyramidal types. These results were largely recapitulated by integration analyses with different algorithms (Fig. S17). Among the mouse-specific clusters, we identified neocortical CT and L5 pyramidal tract (PT) neurons, indicating that these neuron types have unique gene expression profiles and suggesting that they are mammalian innovations (46). In line with this, the transcription factor combinatorial codes that specify these types are found neither in the turtle dorsal cortex (7) nor in the Pleurodeles dorsal pallium (Fig. S16B). In Figure 5F, we plot the same integrated clusters reordered by mouse cortical region, and the proportions of salamander DP and turtle dorsal cortex neurons within these clusters. The large majority of salamander MP and DP neurons co-clustered with mammalian neurons from the hippocampus, entorhinal cortex, and subiculum (Fig. 5F–H, Fig. S16A). The same pattern was observed for neuron types from the reptilian cortex, but with one exception: reptiles also have neurons that co-cluster with the neocortical thalamorecipient L4 IT neurons (Fig. 5F). The analysis of transcription factor expression points to key differences that may underlie the diversity of pyramidal neurons in tetrapods. Some of the transcription factors instructing neuronal identity in the reptilian dorsal cortex and mammalian neocortex, such as Satb2 and Rorb (47, 48), are not expressed at all in the salamander DP (Fig. 2B, Fig. S16B). In other cases, there are differences in transcription factor combinatorial codes. For example, we find that mouse L5 PT neurons (cluster 30) are grouped together with L5 neurons from the pre-, para- and postsubiculum (L5 PPP, cluster 38) and with other neurons from the prosubiculum and subiculum (clusters 32 and 12); cluster 12 also includes neuron types from the salamander DP and the reptilian dorsal cortex. All these neurons share the expression of the transcription factors Bcl11b and Sox5, but differ for the expression of others (Fig. 5I), supporting the concept that neuronal diversity evolves through changes of transcription factor regulatory programs. Taken together, this analysis indicates that the salamander DP lacks cellular and molecular characteristics of the mammalian neocortex, and that DP neuron types are molecularly more similar to pyramidal neurons in the mammalian cortical areas intercalated between neocortex and hippocampus (13).
Similarities and innovations in vertebrate telencephalic connectivity
The results of our comparative analysis prompted us to ask whether neuron types with similar transcriptomes have similar connectivity across species. Expanding on previous findings in salamanders (reviewed in (26)), we conducted retrograde tracing experiments in adult Pleurodeles. We confirmed that both anterior and posterior VP regions project to the putative ventromedial hypothalamus (VMH) homolog (49) (Fig. S18A). The anterior VP receives afferents from mOB, LP and DP (Fig. 6A, Fig. S19), suggesting a function in olfactory processing. Interestingly, and in line with previous findings (22), VPa also sparsely receives projections from the central thalamus (Fig. 6A). The central thalamus expresses Slc17a6 and Calb2, and relays multimodal inputs to the telencephalon. Therefore, it is considered the amphibian homolog of amniote first-order sensory nuclei (50). In reptiles, aDVR receives inputs from the dorsal cortex, but not from olfactory areas, and is organized in subregions innervated by thalamic visual, auditory, and somatosensory nuclei (4, 37, 51, 52). While sensory inputs are processed separately by modality in aDVR, there is no indication that this is the case in the salamander VP. Projections to aDVR also include the striatum, the pDVR, and the VMH (4, 37, 53). Taken together, the molecular and connectivity data suggest that reptilian aDVR neurons might have evolved from olfactory-recipient ventral pallium neurons that lost their connections to the olfactory system and became specialized in the processing of sensory inputs relayed by the thalamus (Fig 6D, Fig. S19B).
Fig. 6. Connectivity of the Pleurodeles pallium.
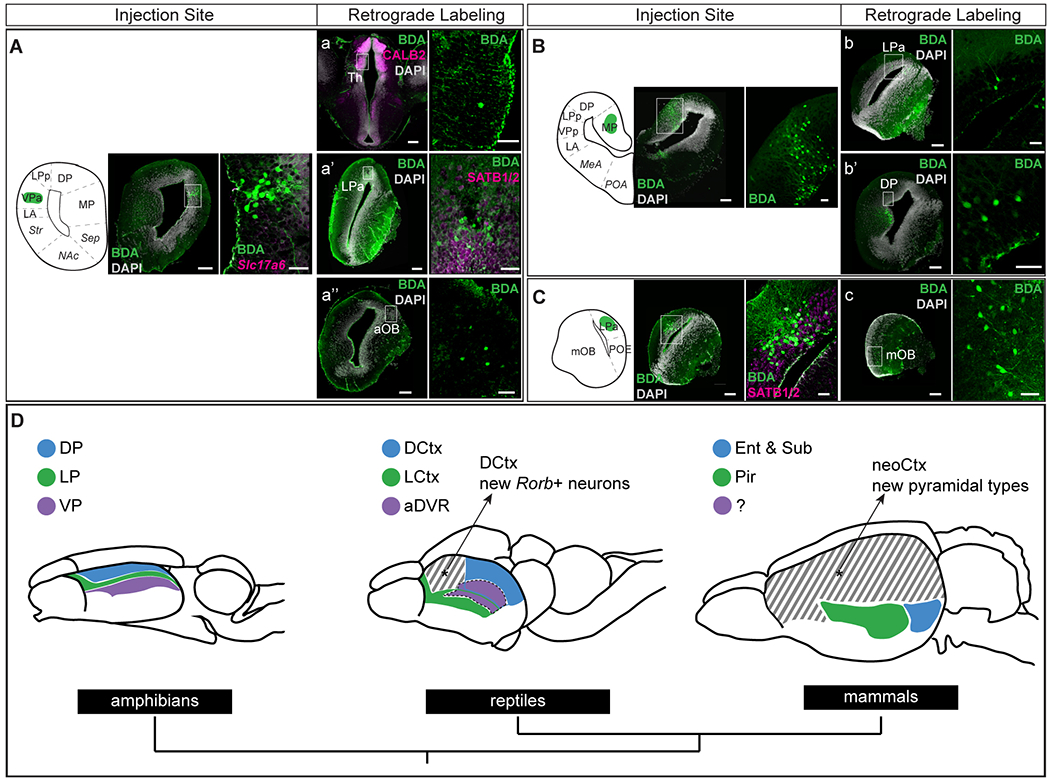
(A-C) Left: injection sites of the retrograde tracer BDA (biotinylated dextran amine). Scale bars: 200 um. Injection of 3 kD BDA into (A) VPa (n= 4), (B) MP (n=2), (C) LPa (n=2). Right: magnification of injection site with immunostaining or HCR in situ hybridization of relevant molecular markers. Scale bars: 50 um. (a-c) Left: representative coronal sections where retrogradely labeled cells were identified, with immunostaining or HCR in situ hybridization of relevant molecular markers when applicable. Right: magnification of retrogradely labeled cells, with immunostaining of relevant markers when applicable. (D) Top: schematic representation of amphibian, reptile, and mammalian brains. Colors indicate molecular and connectivity similarities of neuron types across species. Cross hatching denotes areas with cell type innovations. Medial pallium/medial cortex and hippocampus not shown; mammalian subiculum not shown in the drawing. Bottom: phylogenetic tree.
For full list of abbreviations, see Fig. S1; aDVR, anterior dorsal ventricular ridge; BDA, biotinylated dextran amine; DCtx, dorsal cortex; Ent, entorhinal cortex; LCtx, lateral cortex; Pir, piriform cortex; Sub, subiculum.
Informed by our molecular data, we also compared patterns of pallial connectivity in Pleurodeles to mammalian olfactory-entorhinal-hippocampal circuits. In mammals, the primary input to the hippocampal formation is the entorhinal cortex (EC), which includes a lateral EC strongly connected to olfactory areas, and a medial EC, which processes spatial and contextual information (54). The subiculum is the primary output region of the hippocampus. Motivated by our molecular data (Fig. 5), we asked whether the connections between salamander MP and DP are broadly analogous to the connections of mammalian hippocampus, entorhinal cortex, and subiculum, as suggested by previous literature (9, 22, 26). Retrograde tracer injections confirmed that DP and MP are reciprocally connected (Fig. 6B, Fig. S19A, E). MP and DP also receive direct projections from the Satb1+/Reln+ LP region, an area that receives strong mOB inputs –the lateral olfactory tract runs along this region (24) (Fig. 6B, Fig. S18C, Fig. S19E). Thus, LP neurons are similar to mammalian semilunar cells in the piriform cortex and fan cells in the entorhinal cortex (layer 2), both for their molecular profile (Satb1, Reln, Lhx2, Tbr1; Fig. 2 and (21)) and their connectivity (inputs from the mOB, projections to entorhinal deeper layers). Together, these findings suggest that components of mammalian olfactory-entorhinal-hippocampal circuits may trace back to tetrapod ancestors (Fig. 6D, Fig. S19C).
Discussion
Our molecular data show that, despite its anatomical simplicity, the salamander telencephalon harbors a complex repertoire of neuron types. The combined analysis of their molecular identity, development, and connectivity clarifies the evolution of two innovations in amniotes: the sauropsid aDVR and the mammalian neocortex.
The comparison of the reptilian aDVR and the salamander VP reveals similar and species-specific neuron types. The molecular similarities of salamander VPa/p neurons and neurons in the centromedial aDVR, together with the origin of these cells from a ventral pallium progenitor domain (distinct from the dorsal pallium) and the partial similarities of their connectivity (e.g. connections with the hypothalamus), suggest that the amphibian VPa/p and parts of the reptilian aDVR descend from a common set of neurons in tetrapod ancestors. We also identified reptilian-specific aDVR neurons that do not co-cluster with salamander VP neurons. These rostral aDVR neurons express a unique set of transcription factors (Rorb, Satb1) and receive thalamic inputs segregated by sensory modality (visual, somatosensory, auditory, but not olfactory) (38). In light of this, we propose that neuron types in the aDVR specialized in processing sensory inputs relayed by the thalamus are an evolutionary innovation in the sauropsid lineage.
The homologs of these ventral pallium neurons in mammals remain ambiguous. Connectivity data point to similarities of the salamander VPa, aDVR and the mammalian lateral amygdala, a region that receives sensory inputs relayed by the thalamus (37), and expresses some marker genes found in VPa and aDVR, such as Rorb (7, 26, 55). However, molecular data indicate similarities of the salamander VPa, bird HVC (part of aDVR (8)), and mammalian piriform cortex (21) (Fig. 5A). A kinship of VPa and aDVR with parts of the mammalian piriform cortex is not surprising, given that the aDVR and the sauropsid olfactory cortex develop sequentially from the same embryonic progenitors (56). Further molecular and developmental studies on the mammalian piriform cortex and pallial amygdala, whose cellular diversity remains poorly explored, are needed to clarify their evolutionary relationships with aDVR and VPa.
Our data shed light on the nature of the amphibian dorsal pallium. This region is molecularly distinct from the medial pallium, but does not express many of the markers that define the reptilian dorsal cortex, the area typically compared to the mammalian neocortex for its position, molecular makeup, and connectivity. Our cross-species analysis shows that salamander dorsal pallium and several reptilian dorsal cortex neurons co-cluster with neurons of the mammalian subiculum and entorhinal cortex. The input-output connectivity of the salamander dorsal pallium suggests that these molecular similarities may correspond in part to conserved circuit motifs. The Reln-expressing neurons in the lateral pallium occupy a peculiar position in this circuit, analogous to Reln neurons in the reptilian olfactory cortex and mammalian piriform (semilunar cells) and entorhinal cortex (fan cells) (21, 57). We suggest that mammalian piriform and entorhinal Reln-expressing cells are serial homologs (as sister cell types (58)), with the implication that neuron types in layer 2 and in deeper layers of entorhinal cortex may have two distinct evolutionary origins (from the lateral and the dorsal pallium of a tetrapod ancestor, respectively) (20, 59). This scenario can be tested with molecular data from the mammalian piriform cortex.
In sum, our findings chart the series of innovations that resulted in the emergence of a six-layered neocortex in mammals (Fig. 5). We propose that neocortical L4 Rorb-expressing neurons receiving sensory inputs from the thalamus evolved first, either in amniote ancestors (if salamanders retained the tetrapod ancestral state) (27, 60) or in earlier vertebrate ancestors (with secondary loss in salamanders or amphibians) (61). Neocortical corticothalamic (L6) and pyramidal tract neurons (L5B) emerged later, in mammalian ancestors (46). Expansion of the neocortex led to functional innovations, such as the transition from distributed to columnar information processing (62) and the direct top-down control of locomotion (46). How these molecular and cellular novelties supported such functional innovations within sensory-associative pallial regions remains to be explored.
Materials and methods summary
Animals
Adult Pleurodeles waltl were obtained from breeding colonies established at Columbia University and Karolinska Institute. All experiments were conducted in accordance with the NIH guidelines and Columbia University IACUC policies governing animal use and welfare.
Single-cell RNA sequencing library preparation
Telencephali were dissociated from either adult or larval salamanders and prepared for single-cell RNA sequencing using 10X Genomics Chromium Next GEM Single Cell 3’ Kits v3.1. Sequenced libraries were aligned to a Pleurodeles waltl reference transcriptome (see Supplementary methods for details).
Analysis of single-cell RNA sequencing data
After quality filtering, scRNAseq datasets were clustered and analyzed using the R package Seurat. For the adult data, high-level neuronal and non-neuronal clusters were first identified. Afterwards, the neuronal dataset was subsetted and re-clustered to identify subclusters. Final clusters were annotated, based on expression of established marker genes.
The quality-filtered developmental data were merged into a single object and non-neuronal cells were filtered out. These data were regressed for cell cycle score in addition to RNA count, stage and percent mitochondrial genes, before cluster annotation.
Cross-species comparisons of transcriptomics data
Using Seurat’s integration pipeline, we generated two integrated datasets using single-cell RNAseq data from the turtle pallium (7), the lizard telencephalon (35) and mouse cortex and hippocampal formation (43). Average cluster expression profiles were computed, distances were computed as 1-cor(x) (Spearman correlation), and this distance matrix was used for hierarchical clustering with the Ward.D2 method to generate dendrograms in Figures 4 and 5. Dendrograms were color-coded according to the proportion of each species’ cells in the integrated cluster. Additional annotation of the integrated clusters was carried out by analyzing the identities of each species’ cells that were contained in each integrated cluster (see Supplementary methods for details).
Trajectory Inference
Trajectories were calculated using Slingshot, to endpoints defined by label transfer against adult neurons. Genes that define the dorsal and ventral trajectories were calculated using Seurat’s FindMarkers function. Pseudotime values provided by Slingshot were used to generate heatmaps of differentially expressed genes along each trajectory.
Immunohistochemistry, colorimetric and fluorescent in situ hybridization
Immunohistochemistry and colorimetric in situ hybridization were performed on floating sections of the adult brain, or frozen sections of larval brains following standard protocols. The hybridization chain reaction protocol from Molecular Instruments was implemented and combined with iDISCO tissue clearing and light sheet imaging to generate 2D and 3D representations of gene expression (see Supplementary methods for details).
Axonal tracing
Dextran amine tracer injections were performed ex vivo and incubated for 24–48 hrs. The tracer injection site, and retrogradely or anterogradely labeled cells were visualized on floating sections and annotated using co-staining for known molecular markers and anatomical landmarks (see supplementary methods for details).
Supplementary Material
Acknowledgments:
The authors are grateful to J. Barber, S. Cook, and the Columbia University Institute of Comparative Medicine for animal care; B. Jekely for help with cloning; E. Subramanian for contributing to the IsoSeq reference; A. Matheson for the diagram in Fig. S19 B–C, and A. Matheson, L. Xu, G. Laurent, O. Hobert, and R. Satija for critical feedback on the manuscript. Light-sheet imaging was performed with support from L. Hammond and the Zuckerman Institute’s Cellular Imaging platform (NIH 1S10OD023587-01); computing resources were provided by Columbia University’s Shared Research Computing Facility (NIH 1G20RR030893-01 and NYSTAR Contract C090171). The authors acknowledge support of the National Genomics Infrastructure (NGI)/Uppsala Genome Center and UPPMAX for providing assistance in massive parallel sequencing and computational infrastructure. Work performed at NGI / Uppsala Genome Center has been funded by RFI/VR and Science for Life Laboratory, Sweden. Storage and handling of IsoSeq data were enabled by resources provided by the Swedish National Infrastructure for Computing (SNIC) at UPPMAX partially funded by the Swedish Research Council through grant agreement no. 2018-05973.
Funding:
McKnight Foundation (MAT)
National Institutes of Health grant RM1HG011014 (MAT)
National Science Foundation, Graduate Research Fellowship (DGE 2036197) (ECJ)
Knut and Alice Wallenberg Foundation (AS)
European Research Council (AS)
Footnotes
Competing interests: Authors declare that they have no competing interests.
Supplementary Materials
Data and materials availability:
The scRNA sequencing data used in this study have been deposited in the Gene Expression Omnibus (GEO), with accession numbers GSE197701, GSE197722, GSE197796, GSE197807, GSE198363, GSE198364, GSE198366, GSE198367, GSE198365, GSE206163. All other data are included in the main paper or supplement. Code used for the analysis of scRNAseq data is available at https://doi.org/10.5281/zenodo.6780577. A website to access the data and search genes of interest is available at https://toscheslab.shinyapps.io/salamander_telencephalon/.
References and Notes
- 1.MacIver MA, Finlay BL, The neuroecology of the water-to-land transition and the evolution of the vertebrate brain. Philos. Trans. R. Soc. Lond. B Biol. Sci 377, 20200523 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Striedter GF, The telencephalon of tetrapods in evolution. Brain Behav. Evol 49, 179–213 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Butler AB, Molnár Z, Development and evolution of the collopallium in amniotes: a new hypothesis of field homology. Brain Res. Bull 57, 475–479 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Butler AB, Reiner A, Karten HJ, Evolution of the amniote pallium and the origins of mammalian neocortex. Ann. N. Y. Acad. Sci 1225, 14–27 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jarvis ED, in Evolution of nervous systems (Elsevier, 2007), pp. 213–227. [Google Scholar]
- 6.Karten HJ, Neocortical evolution: neuronal circuits arise independently of lamination. Curr. Biol 23, R12–5 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Tosches MA, Yamawaki TM, Naumann RK, Jacobi AA, Tushev G, Laurent G, Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science. 360, 881–888 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Colquitt BM, Merullo DP, Konopka G, Roberts TF, Brainard MS, Cellular transcriptomics reveals evolutionary identities of songbird vocal circuits. Science. 371 (2021), doi: 10.1126/science.abd9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Northcutt RG, Kicliter E, in Comparative neurology of the telencephalon, Ebbesson SOE, Ed. (Springer US, Boston, MA, 1980), pp. 203–255. [Google Scholar]
- 10.González A, López JM, Morona R, Moreno N, in Evolutionary Neuroscience (Elsevier, 2020), pp. 125–157. [Google Scholar]
- 11.Smeets WJ, Marín O, González A, Evolution of the basal ganglia: new perspectives through a comparative approach. J. Anat 196 ( Pt 4), 501–517 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bormuth I, Yan K, Yonemasu T, Gummert M, Zhang M, Wichert S, Grishina O, Pieper A, Zhang W, Goebbels S, Tarabykin V, Nave K-A, Schwab MH, Neuronal basic helix-loop-helix proteins Neurod2/6 regulate cortical commissure formation before midline interactions. J. Neurosci 33, 641–651 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Puelles L, Alonso A, García-Calero E, Martínez-de-la-Torre M, Concentric ring topology of mammalian cortical sectors and relevance for patterning studies. J. Comp. Neurol 527, 1731–1752 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Faedo A, Ficara F, Ghiani M, Aiuti A, Rubenstein JLR, Bulfone A, Developmental expression of the T-box transcription factor T-bet/Tbx21 during mouse embryogenesis. Mech. Dev 116, 157–160 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Moreno N, González A, Rétaux S, Evidences for tangential migrations in Xenopus telencephalon: developmental patterns and cell tracking experiments. Dev. Neurobiol 68, 504–520 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Moreno N, González A, Regionalization of the telencephalon in urodele amphibians and its bearing on the identification of the amygdaloid complex. Front. Neuroanat 1, 1 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brox A, Puelles L, Ferreiro B, Medina L, Expression of the genes Emx1, Tbr1, and Eomes (Tbr2) in the telencephalon of Xenopus laevis confirms the existence of a ventral pallial division in all tetrapods. J. Comp. Neurol 474, 562–577 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Laberge F, Mühlenbrock-Lenter S, Grunwald W, Roth G, Evolution of the amygdala: new insights from studies in amphibians. Brain Behav. Evol 67, 177–187 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Moreno N, González A, The common organization of the amygdaloid complex in tetrapods: new concepts based on developmental, hodological and neurochemical data in anuran amphibians. Prog. Neurobiol 78, 61–90 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Abellán A, Desfilis E, Medina L, Combinatorial expression of Lef1, Lhx2, Lhx5, Lhx9, Lmo3, Lmo4, and Prox1 helps to identify comparable subdivisions in the developing hippocampal formation of mouse and chicken. Front. Neuroanat 8, 59 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diodato A, Ruinart de Brimont M, Yim YS, Derian N, Perrin S, Pouch J, Klatzmann D, Garel S, Choi GB, Fleischmann A, Molecular signatures of neural connectivity in the olfactory cortex. Nat. Commun 7, 12238 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.<b>Neary TJ, in Comparative structure and evolution of cerebral cortex, part I, Jones EG, Peters A, Eds. (Springer US, Boston, MA, 1990), vol. 8A of Cerebral Cortex, pp. 107–138. [Google Scholar]
- 23.Deryckere A, Woych J, Jaeger ECB, Tosches MA, Glutamatergic neuron types in the amygdala of the urodele amphibian Pleurodeles waltl. BioRxiv (2022), doi: 10.1101/2022.06.15.496313. [DOI] [Google Scholar]
- 24.Laberge F, Roth G, Connectivity and cytoarchitecture of the ventral telencephalon in the salamander Plethodon shermani. J. Comp. Neurol 482, 176–200 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Endepols H, Roden K, Walkowiak W, Hodological characterization of the septum in anuran amphibians: II. Efferent connections. J. Comp. Neurol 483, 437–457 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Joven A, Simon A, Homeostatic and regenerative neurogenesis in salamanders. Prog. Neurobiol 170, 81–98 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Striedter GF, Northcutt RG, The independent evolution of dorsal pallia in multiple vertebrate lineages. Brain Behav. Evol, 1–12 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Medina L, Abellán A, Desfilis E, Evolving views on the pallium. Brain Behav. Evol, 1–19 (2021). [DOI] [PubMed] [Google Scholar]
- 29.García-Moreno F, Molnár Z, Variations of telencephalic development that paved the way for neocortical evolution. Prog. Neurobiol 194, 101865 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joven A, Wang H, Pinheiro T, Hameed LS, Belnoue L, Simon A, Cellular basis of brain maturation and acquisition of complex behaviors in salamanders. Development. 145 (2018), doi: 10.1242/dev.160051. [DOI] [PubMed] [Google Scholar]
- 31.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R, Comprehensive Integration of Single-Cell Data. Cell. 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, Dudoit S, Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 19, 477 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreau MX, Saillour Y, Cwetsch AW, Pierani A, Causeret F, Single-cell transcriptomics of the early developing mouse cerebral cortex disentangle the spatial and temporal components of neuronal fate acquisition. Development. 148 (2021), doi: 10.1242/dev.197962. [DOI] [PubMed] [Google Scholar]
- 34.Norimoto H, Fenk LA, Li H-H, Tosches MA, Gallego-Flores T, Hain D, Reiter S, Kobayashi R, Macias A, Arends A, Klinkmann M, Laurent G, A claustrum in reptiles and its role in slow-wave sleep. Nature. 578, 413–418 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Hain D, Gallego-Flores T, Klinkmann M, Macias A, Ciirdaeva E, Arends A, Thum C, Tushev G, Kretschmer F, Tosches MA, Laurent G, Molecular diversity and evolution of neuron types in the amniote brain. Science. [DOI] [PubMed] [Google Scholar]
- 36.Moreno N, González A, Evolution of the amygdaloid complex in vertebrates, with special reference to the anamnio-amniotic transition. J. Anat 211, 151–163 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruce LL, Neary TJ, The limbic system of tetrapods: a comparative analysis of cortical and amygdalar populations. Brain Behav. Evol 46, 224–234 (1995). [DOI] [PubMed] [Google Scholar]
- 38.Manger PR, Slutsky DA, Molnár Z, Visual subdivisions of the dorsal ventricular ridge of the iguana (Iguana iguana) as determined by electrophysiologic mapping. J. Comp. Neurol 453, 226–246 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Arendt D, Musser JM, Baker CVH, Bergman A, Cepko C, Erwin DH, Pavlicev M, Schlosser G, Widder S, Laubichler MD, Wagner GP, The origin and evolution of cell types. Nat. Rev. Genet 17, 744–757 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen T-M, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Jones AR, Genome-wide atlas of gene expression in the adult mouse brain. Nature. 445, 168–176 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Caubit X, Tiveron M-C, Cremer H, Fasano L, Expression patterns of the three Teashirt-related genes define specific boundaries in the developing and postnatal mouse forebrain. J. Comp. Neurol 486, 76–88 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, Marques S, Munguba H, He L, Betsholtz C, Rolny C, Castelo-Branco G, Hjerling-Leffler J, Linnarsson S, Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 347, 1138–1142 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Yao Z, van Velthoven CTJ, Nguyen TN, Goldy J, Sedeno-Cortes AE, Baftizadeh F, Bertagnolli D, Casper T, Chiang M, Crichton K, Ding S-L, Fong O, Garren E, Glandon A, Gouwens NW, Gray J, Graybuck LT, Hawrylycz MJ, Hirschstein D, Kroll M, Zeng H, A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell. 184, 3222–3241.e26 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fishell G, Kepecs A, Interneuron types as attractors and controllers. Annu. Rev. Neurosci 43, 1–30 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gouwens NW, Sorensen SA, Baftizadeh F, Budzillo A, Lee BR, Jarsky T, Alfiler L, Baker K, Barkan E, Berry K, Bertagnolli D, Bickley K, Bomben J, Braun T, Brouner K, Casper T, Crichton K, Daigle TL, Dalley R, de Frates RA, Zeng H, Integrated morphoelectric and transcriptomic classification of cortical gabaergic cells. Cell. 183, 935–953.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shim S, Kwan KY, Li M, Lefebvre V, Sestan N, Cis-regulatory control of corticospinal system development and evolution. Nature. 486, 74–79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jabaudon D, Fate and freedom in developing neocortical circuits. Nat. Commun 8, 16042 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nomura T, Yamashita W, Gotoh H, Ono K, Species-Specific Mechanisms of Neuron Subtype Specification Reveal Evolutionary Plasticity of Amniote Brain Development. Cell Rep. 22, 3142–3151 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Moreno N, Morona R, López JM, González A, in Evolution of nervous systems (Elsevier, 2017), pp. 409–426. [Google Scholar]
- 50.Joven A, Morona R, Moreno N, González A, Regional distribution of calretinin and calbindin-D28k expression in the brain of the urodele amphibian Pleurodeles waltl during embryonic and larval development. Brain Struct. Funct 218, 969–1003 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Lanuza E, Belekhova M, Martínez-Marcos A, Font C, Martínez-García F, Identification of the reptilian basolateral amygdala: an anatomical investigation of the afferents to the posterior dorsal ventricular ridge of the lizard Podarcis hispanica. Eur. J. Neurosci 10, 3517–3534 (1998). [DOI] [PubMed] [Google Scholar]
- 52.Cordery P, Molnár Z, Embryonic development of connections in Turtle Pallium. Journal of Comparative Neurology (1999). [PubMed] [Google Scholar]
- 53.Voneida TJ, Sligar CM, Efferent projections of the dorsal ventricular ridge and the striatum in the Tegu lizard. Tupinambis nigropunctatus. J. Comp. Neurol 186, 43–64 (1979). [DOI] [PubMed] [Google Scholar]
- 54.Witter MP, Doan TP, Jacobsen B, Nilssen ES, Ohara S, Architecture of the Entorhinal Cortex A Review of Entorhinal Anatomy in Rodents with Some Comparative Notes. Front. Syst. Neurosci 11, 46 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Leary TP, Sullivan KE, Wang L, Clements J, Lemire AL, Cembrowski MS, Extensive and spatially variable within-cell-type heterogeneity across the basolateral amygdala. eLife. 9 (2020), doi: 10.7554/eLife.59003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnston JB, The development of the dorsal ventricular ridge in turtles. J. Comp. Neurol 26, 481–505 (1916). [Google Scholar]
- 57.Leitner FC, Melzer S, Lütcke H, Pinna R, Seeburg PH, Helmchen F, Monyer H, Spatially segregated feedforward and feedback neurons support differential odor processing in the lateral entorhinal cortex. Nat. Neurosci 19, 935–944 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Tosches MA, From cell types to an integrated understanding of brain evolution: the case of the cerebral cortex. Annu. Rev. Cell Dev. Biol 37, 495–517 (2021). [DOI] [PubMed] [Google Scholar]
- 59.Luzzati F, A hypothesis for the evolution of the upper layers of the neocortex through co-option of the olfactory cortex developmental program. Front. Neurosci 9, 162 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bloch S, Hagio H, Thomas M, Heuzé A, Hermel J-M, Lasserre E, Colin I, Saka K, Affaticati P, Jenett A, Kawakami K, Yamamoto N, Yamamoto K, Non-thalamic origin of zebrafish sensory nuclei implies convergent evolution of visual pathways in amniotes and teleosts. eLife. 9 (2020), doi: 10.7554/eLife.54945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suryanarayana SM, Robertson B, Wallén P, Grillner S, The lamprey pallium provides a blueprint of the mammalian layered cortex. Curr. Biol 27, 3264–3277.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Fournier J, Müller CM, Schneider I, Laurent G, Spatial Information in a Non-retinotopic Visual Cortex. Neuron. 97, 164–180.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Joven A, Kirkham M, Simon A, Husbandry of Spanish ribbed newts (Pleurodeles waltl). Methods Mol. Biol 1290, 47–70 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Gallien L, Durocher M, Table chronologique du développement chez Pleurodeles waltlII Michah. Bulletin biologique. 2, 1–19 (1957). [Google Scholar]
- 65.Matsunami M, Suzuki M, Haramoto Y, Fukui A, Inoue T, Yamaguchi K, Uchiyama I, Mori K, Tashiro K, Ito Y, Takeuchi T, Suzuki K-IT, Agata K, Shigenobu S, Hayashi T, A comprehensive reference transcriptome resource for the Iberian ribbed newt Pleurodeles waltl, an emerging model for developmental and regeneration biology. DNA Res. 26, 217–229 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, Regev A, De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J, eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol 38, 5825–5829 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C, Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, Satija R, Integrated analysis of multimodal single-cell data. Cell. 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martynoga B, Morrison H, Price DJ, Mason JO, Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev. Biol 283, 113–127 (2005). [DOI] [PubMed] [Google Scholar]
- 71.Osório J, Mueller T, Rétaux S, Vernier P, Wullimann MF, Phylotypic expression of the bHLH genes Neurogenin2, Neurod, and Mash1 in the mouse embryonic forebrain. J. Comp. Neurol 518, 851–871 (2010). [DOI] [PubMed] [Google Scholar]
- 72.Ma Q, Kintner C, Anderson DJ, Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 87, 43–52 (1996). [DOI] [PubMed] [Google Scholar]
- 73.Gu Z, ComplexHeatmap. Bioconductor (2017), doi: 10.18129/b9.bioc.complexheatmap. [DOI] [Google Scholar]
- 74.Choudhary S, Satija R, Comparison and evaluation of statistical error models for scRNA-seq. Genome Biol. 23, 27 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh P-R, Raychaudhuri S, Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lopez R, Regier J, Cole MB, Jordan MI, Yosef N, Deep generative modeling for single-cell transcriptomics. Nat. Methods 15, 1053–1058 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tarashansky AJ, Musser JM, Khariton M, Li P, Arendt D, Quake SR, Wang B, Mapping single-cell atlases throughout Metazoa unravels cell type evolution. eLife. 10 (2021), doi: 10.7554/eLife.66747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, Codeluppi S, Furlan A, Lee K, Skene N, Harris KD, Hjerling-Leffler J, Arenas E, Ernfors P, Marklund U, Linnarsson S, Molecular architecture of the mouse nervous system. Cell. 174, 999–1014.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kuehn E, Clausen DS, Null RW, Metzger BM, Willis AD, Özpolat BD, Segment number threshold determines juvenile onset of germline cluster expansion in Platynereis dumerilii. J. Exp. Zool. B Mol. Dev. Evol (2021), doi: 10.1002/jez.b.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, Cunha A, Pierce NA, Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development. 145 (2018), doi: 10.1242/dev.165753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M, iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 159, 896–910 (2014). [DOI] [PubMed] [Google Scholar]
- 82.Tanaka N, Kanatani S, Kaczynska D, Fukumoto K, Louhivuori L, Mizutani T, Kopper O, Kronqvist P, Robertson S, Lindh C, Kis L, Pronk R, Niwa N, Matsumoto K, Oya M, Miyakawa A, Falk A, Hartman J, Sahlgren C, Clevers H, Uhlén P, Three-dimensional single-cell imaging for the analysis of RNA and protein expression in intact tumour biopsies. Nat. Biomed. Eng 4, 875–888 (2020). [DOI] [PubMed] [Google Scholar]
- 83.Reiner A, Honig MG, in Neuroanatomical Tract-Tracing 3, Zaborszky L, Wouterlood FG, Lanciego JL, Eds. (2006), pp. 304–335. [Google Scholar]
- 84.Reiner A, Veenman CL, Medina L, Jiao Y, Del Mar N, Honig MG, Pathway tracing using biotinylated dextran amines. J. Neurosci. Methods 103, 23–37 (2000). [DOI] [PubMed] [Google Scholar]
- 85.Puelles L, Ayad A, Alonso A, Sandoval JE, MartÍnez-de-la-Torre M, Medina L, Ferran JL, Selective early expression of the orphan nuclear receptor Nr4a2 identifies the claustrum homolog in the avian mesopallium: Impact on sauropsidian/mammalian pallium comparisons. J. Comp. Neurol 524, 665–703 (2016). [DOI] [PubMed] [Google Scholar]
- 86.Musser JM, Wagner GP, Character trees from transcriptome data: Origin and individuation of morphological characters and the so-called “species signal”. J. Exp. Zool. B Mol. Dev. Evol 324, 588–604 (2015). [DOI] [PubMed] [Google Scholar]
- 87.Luecken MD, Büttner M, Chaichoompu K, Danese A, Interlandi M, Mueller MF, Strobl DC, Zappia L, Dugas M, Colomé-Tatché M, Theis FJ, Benchmarking atlas-level data integration in single-cell genomics. Nat. Methods 19, 41–50 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bakken TE, Jorstad NL, Hu Q, Lake BB, Tian W, Kalmbach BE, Crow M, Hodge RD, Krienen FM, Sorensen SA, Eggermont J, Yao Z, Aevermann BD, Aldridge AI, Bartlett A, Bertagnolli D, Casper T, Castanon RG, Crichton K, Daigle TL, Lein ES, Comparative cellular analysis of motor cortex in human, marmoset and mouse. Nature. 598, 111–119 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Striedter GF, Northcutt RG, Brains through time: A natural history of vertebrates (Oxford University Press, 2019). [Google Scholar]
- 90.Moreno N, López JM, Morona R, Lozano D, Jiménez S, González A, Comparative Analysis of Nkx2.1 and Islet-1 Expression in Urodele Amphibians and Lungfishes Highlights the Pattern of Forebrain Organization in Early Tetrapods. Front. Neuroanat 12, 42 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Joven A, Morona R, González A, Moreno N, Expression patterns of Pax6 and Pax7 in the adult brain of a urodele amphibian, Pleurodeles waltl. J. Comp. Neurol 521, 2088–2124 (2013). [DOI] [PubMed] [Google Scholar]
- 92.Joven A, Morona R, González A, Moreno N, Spatiotemporal patterns of Pax3, Pax6, and Pax7 expression in the developing brain of a urodele amphibian, Pleurodeles waltl. J. Comp. Neurol 521, 3913–3953 (2013). [DOI] [PubMed] [Google Scholar]
- 93.Chen Y, Chen X, Baserdem B, Zhan H, Li Y, Davis MB, Kebschull JM, Zador AM, Koulakov AA, Albeanu DF, Wiring logic of the early rodent olfactory system revealed by high-throughput sequencing of single neuron projections. BioRxiv (2021), doi: 10.1101/2021.05.12.443929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


