Abstract
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignant tumours worldwide. Clarification of the somatic mutational landscape of important genes could reveal new therapeutic targets and facilitate individualized therapeutic approaches for HCC patients. The mutation and expression changes in the ARID1A gene in HCC remain controversial.
Methods
First, cBioPortal was used to visualize genetic alterations and DNA copy number alterations (CNAs) in ARID1A. The relationships between ARID1A mutation status and HCC patient clinicopathological features and overall survival (OS) were also determined. Then, a meta-analysis was performed to evaluate the effect of ARID1A mutation or expression on the prognosis of HCC patients. Finally, the role of ARID1A in HCC progression was verified by in vitro experiments.
Results
ARID1A mutation was detected in 9.35% (33/353) of sequenced HCC cases, and ARID1A mutation decreased ARID1A mRNA expression. Patients with ARID1A alterations presented worse OS than those without ARID1A alterations. Meta-analysis and human HCC tissue microarray (TMA) analysis revealed that HCC patients with low ARID1A expression had worse OS and relapse-free survival (RFS), and low ARID1A expression was negatively correlated with tumour size. Then, ARID1A gain-of-function and loss-of-function experiments demonstrated the tumour suppressor role of ARID1A in HCC in vitro. In terms of the mechanism, we found that ARID1A could inhibit HCC progression by regulating retinoblastoma-like 1 (RBL1) expression via the JNK/FOXO3 pathway.
Conclusions
ARID1A can be considered a potential prognostic biomarker and candidate therapeutic target for HCC.
Keywords: ARID1A, Prognosis, Hepatocellular carcinoma, Prognostic biomarker
Abbreviations: HCC, Hepatocellular carcinoma; OS, overall survival; RFS, relapse-free survival; RBL1, regulating retinoblastoma-like 1; ARID1A, AT-rich interaction domain 1A; CCK-8, Cell Counting Kit-8; EdU, 5-Ethyl-2'-deoxyuridine
1. Introduction
Hepatocellular carcinoma (HCC) is one of the most prevalent and deadly cancers worldwide [1,2]. In recent decades, the treatment of HCC has been dramatically revolutionized by comprehensive treatments such as surgery, targeted therapies and immunotherapy [3]. Unfortunately, the overall prognosis of HCC patients is still unsatisfactory due to the highly invasive nature, strong metastatic ability and chemotherapy resistance of cancer cells [4]. There is an urgent need to identify the key molecular drivers that can regulate the comprehensive treatment effect to facilitate individualized treatment for HCC patients.
Genemutations play an important role in the development and progression of cancer [5,6]. A better understanding of the landscape of commonly mutated genes in tumour cells could facilitate drug development and personalized treatment of cancer patients. For example, as the most commonly mutated RAS isoform in human cancers, Kirsten rat sarcoma (KRAS) has been identified as a key oncogenic driver in tumorigenesis and cancer progression [7]. Recently, research on directly targeting KRAS by irreversible covalent binding has made exciting advances [8,9].
Genomic sequencing of cancer patient DNA has revealed the relative abundance of genetic mutations. Several commonly mutated genes, such as the p53 tumour suppressor gene (TP53) and CTNNB1, have been identified in HCC samples [[10], [11], [12]]. The high TP53 mutation rate in cancers makes this mutation a very attractive therapeutic target [13]. A description of the somatic mutational landscape of critical genes and clarification of its exact effects in HCC progression could contribute to the identification of new therapeutic targets and the application of individualized therapeutic approaches for HCC patients.
ARID1A (AT-rich interaction domain 1A) is a subunit of the chromatin remodelling complex SWI/SNF that is mutually exclusive with two homologous subunits, ARID1B and ARID2 [14]. Abnormal genetic and epigenetic ARID1A alterations have been identified in many types of cancers, such as bladder tumours [15], colorectal cancer [16], pancreatic neuroendocrine tumours [17], breast micropapillary carcinomas [18], and uterine endometrioid carcinoma [19,20]. Most ARID1A mutations are frameshift or inactivating mutations that lead to the loss of ARID1A expression [21]. ARID1A is considered to be a tumour suppressor [22]. Until now, the underlying mechanism of ARID1A mutation in HCC samples has not been reported, and the role of ARID1A expression in HCC patient prognosis assessment remains controversial [23].
As a member of the pocket protein family, retinoblastoma-like (RBL)1/p107 protein is a key factor that regulates the cell cycle [24]. Combined with its family members (retinoblastoma (RB)1/p105 proteins and RBL2/p130 proteins), RBL1 plays an important role in the regulation of cell proliferation, and its dysregulation is associated with cancer occurrence and development [25]. Multiple studies support the role of RBL1 as a tumour suppressor that can also regulate the cell cycle [[26], [27], [28]]. However, the mechanism of RBL1 regulation is still unclear.
Our study aimed to assess the somatic mutational landscape of the ARID1A gene in HCC samples and determine the relationship between ARID1A mutation status and its expression. Our data demonstrated that ARID1A mutations were frequent (present in 9.35% of HCC cases) and that ARID1A mutation resulted in decreased ARID1A mRNA expression. Patients with ARID1A mutations had worse OS than those without ARID1A mutations. Then, the impacts of ARID1A mutation on HCC patient prognosis and clinicopathological features were assessed. Furthermore, meta-analysis revealed that HCC patients with low ARID1A expression had worse OS and relapse-free survival (RFS). Gain-of-function and loss-of-function experiments were performed in vitro to verify the function of ARID1A in HCC progression. More importantly, we identified RBL1 as a crucial downstream molecule of ARID1A in regulating HCC progression.
2. Methods
2.1. Gene alterations of ARID1A in HCC samples from cBioPortal
ARID1A gene alterations were analysed using cBioPortal (http://www.cbioportal.org). The data used in cBioPortal are included in the liver hepatocellular carcinoma (TCGA, PanCancer Atlas) (353 samples with mutation or CAN data) dataset. The dataset is updated monthly based on the latest TCGA database update. Mutations, DNA copy-number alterations (CNAs), and the relationship between mRNA expression and mutations were assessed according to the online guide. The impacts of ARID1A changes on the OS of HCC patients were analysed using Kaplan‐Meier survival curves generated from TCGA data in cBioPortal.
2.2. Correlation analysis between clinical characteristics and ARID1A mutation status
We downloaded LIHC tumour mRNA-seq data from the TCGA database (https://portal.gdc.cancer.gov/). The correlations between ARID1A mutation status and clinical features were analysed using the R program v4.0.3 and visualized by the R software package. Statistical significance was determined by a P value less than 0.05.
2.3. Literature search and eligibility criteria
Our meta-analysis followed the preferred reporting item: the PRISMA statement [29]. We selected relevant studies from the Web of Science, Cochrane Library, Embase and PubMed from inception until July 15, 2022. We identified studies by searching (“ARID1A″) OR (“BAF250A″) AND (“hepatocellular carcinoma”) and their connected entry terms.
The selection criteria of our meta-analysis were as follows: (1) the included patients had pathologically confirmed HCC; (2) the article described the relationship between ARID1A and the survival outcome in HCC patients; and (3) the article included the hazard ratio (HR) and 95% confidence interval (95% CI) or survival curves could be used to calculate the combined HR. The following studies were excluded: (1) case reports, meeting abstracts, review articles, editorials, and letters; (2) non-English papers; (3) studies involving animal experiments; and (4) studies without Kaplan‐Meier curve data to calculate risk.
Two researchers (GX M and CC Y) screened the abstracts of all retrieved literature. The whole texts of the literature were read and screened, and the differences were resolved through in-depth discussion with another researcher (ZR D). We extracted the following items: author first name, year of publication, country, number of patients, ARID1A expression test methods, follow-up time, endpoints, and clinical characteristics, including sex, HBV infection, liver cirrhosis, TNM staging, recurrence, metastasis, histodifferentiation, and tumour size. If HRs were not directly provided in the study, we extracted the HRs from the provided Kaplan‐Meier curves with Get Data software.
Two investigators (GX M and CC Y) independently assessed the quality of each study according to the Newcastle‒Ottawa Scale (NOS). Each study was rated up to 9 points: the choice of the research groups (4 points), the comparison between the research groups (2 points), and the evaluation of results (3 points).
2.4. Quantitative reverse transcription polymerase chain reaction (qRT‒PCR)
RNA was extracted from the HCC cell lines using an RNA Fast 200 Kit (Fastagen, Shanghai). We used the Reverse Transcription Kit (Vazyme, China) to reverse transcribe RNA into cDNA. qRT‒PCR was performed using ChamQTM SYBR® qPCR Master Mix (Vazyme, China) on a Bio-Rad CFX Connect (Bio-Rad Laboratories, USA). The gene-specific primers were as follows: ARID1A-F, 5′-TTATCTCCGCGTCAGCCTTC-3′, ARID1A-R, 5′-ACTGGGGTAGTTGGCATTGG-3′, RBL1-F, 5′-ACTTGGCGAATCAGGACCAT-3′, RBL1-R, 5′-CCCATTCTTGTGCGGGGAAA-3′, β-actin-F, 5′-GAAGAGCTACGAGCTGCCTGA-3′, β-actin-R, 5′-CAGACAGCACTGTGTTGGCG-3′. qRT‒PCR analysis was performed according to previous studies [30].
2.5. Cell counting Kit-8 (CCK-8) and transwell assays
CCK‐8 and transwell assays were performed following a previous study [31].
2.6. 5-Ethyl-2′-deoxyuridine (EdU) assay
A total of 8000 transfected HCC cells were added to 96-well plates. The cells were incubated with EdU (Beyotime, China) for 1 h. After fixation with 3% paraformaldehyde (Beyotime, China) for 15 min, the cells were soaked with 0.3% Triton X-100 (Beyotime, China) for 15 min, and DAPI (1 μg/mL) was added for 15 min. The cells were observed and imaged under a fluorescence microscope.
2.7. Colony formation experiments
A total of 1500 transfected HCC cells were added evenly in 6-well plates. We used 4% formaldehyde to fix the cells and 0.1% crystal violet to stain the cells after 14 days.
2.8. Tissue microarray (TMA) and HCC patient follow-up
A TMA was constructed by Shanghai Outdo Biotech Company using archival specimens from 200 anonymous hepatocellular carcinoma patients. A follow-up process and clinicopathological information were performed following a previous study [30].
2.9. Western blotting (WB) and immunohistochemistry (IHC)
WB and IHC analyses were performed according to a previous study [31].
2.10. Gene correlation and functional enrichment analysis
We downloaded the LIHC tumour RNA-seq data from the TCGA database (https://portal.gdc.cancer.gov/). Spearman's correlation analysis was used to describe the correlation among quantitative variables without a normal distribution, and then the correlation genes with Spearman's correlation coefficient >0.5 were screened out. The ClusterProfiler package in R was used to analyse the Gene Ontology (GO) functions and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched in the related genes.
2.11. Statistical analysis
Statistical analyses were performed using GraphPad Prism 8.0 (GraphPad Software, USA) and IBM SPSS Statistics 20.0 (SPSS, Inc., Chicago, IL). Each experiment was independently performed in triplicate, and the data are presented as the mean ± standard deviation unless otherwise stated. T tests were used to analyse differences between the two groups, and the chi-square test was used for comparison of categorical variables. Kaplan–Meier curves and log-rank tests were used to achieve survival analysis. We extracted reported HRs with 95% CIs for OS and RFS from each reference. When results were present for both multivariate analysis and univariate analysis, we extracted the multivariate analysis results. We used the Q test to examine heterogeneity. When the P value < 0.1 and I2 > 50%, we considered that there was significant heterogeneity in the article. The pooled HR was then calculated using a random-effects model. Otherwise, we used the fixed-effects model to calculate the pooled HR. We performed Begg's and Egger's tests to evaluate publication bias. STATA 12.0 was used to complete our meta-analysis. Statistical significance was determined by a two-tailed P value < 0.05.
3. Results
3.1. The somatic mutational landscape of the ARID1A gene in HCC samples
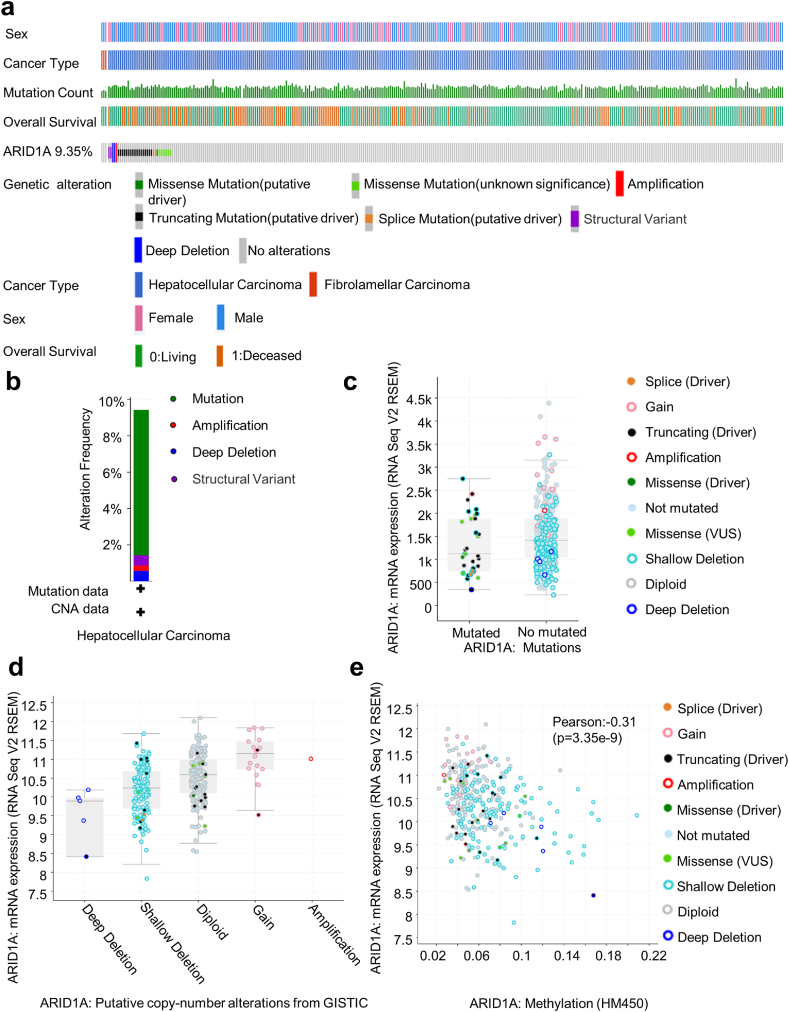
First, cBioPortal was used to analyse ARID1A genetic alterations in HCC samples. ARID1A mutation was found in 9.35% (33/353) of sequenced cases; the total case set included 350 cases of HCC and 3 cases of fibrolamellar carcinoma according to the OncoPrint schematic. All of the mutations occurred in HCC. Mutations occurred in 9 out of 114 cases in females and 24 out of 239 cases in males. Chi-square analysis showed that there was no statistically significant difference in ARID1A mutation between males and females (Fig. 1a). TCGA database analyses showed that missense mutation (8 cases, 2.27%), truncating mutation (18 cases, 5.10%), deep deletion (2 cases, 1.37%), amplification (1 case, 0.27%), and multiple alterations (2 cases, 0.55%) were the most frequent ARID1A mutation types in HCC samples (Fig. 1b).
Fig. 1.
Genetic alterations of ARID1A in HCC tissue (cBioPortal).
(a) OncoPrint of ARID1A alterations in HCC. OncoPrint provides an overview of genomic alterations (legend) in particular genes (rows) affecting particular individual samples (columns). (b) Summary of the frequency of ARID1A mutations, including mutation, amplification, deep deletion, and multiple alterations, in HCC. (c) The relationship between ARID1A mRNA expression and gene mutations. The mRNA expression of ARID1A in mutant cases was lower than that in nonmutant cases. (d) The relationship between ARID1A mRNA expression and different types of gene mutations. (e) The relationship between ARID1A mRNA expression and ARID1A methylation.
Then, we analysed the relationship between the ARID1A mRNA level and genetic mutations. We found that the ARID1A mRNA level in the mutant group was significantly lower than that in the nonmutant group (Fig. 1c). There were more ARID1A shallow deletions and diploids but fewer deep deletions, amplifications, and gains in HCC. Functional plotting of ARID1A indicated that the ARID1A mRNA level was associated with ARID1A genetic status (deletion or diploid) in HCC (Fig. 1d). In addition, ARID1A mRNA expression was negatively correlated with ARID1A methylation (Fig. 1e).
3.2. Correlation between ARID1A mutation status and clinical features
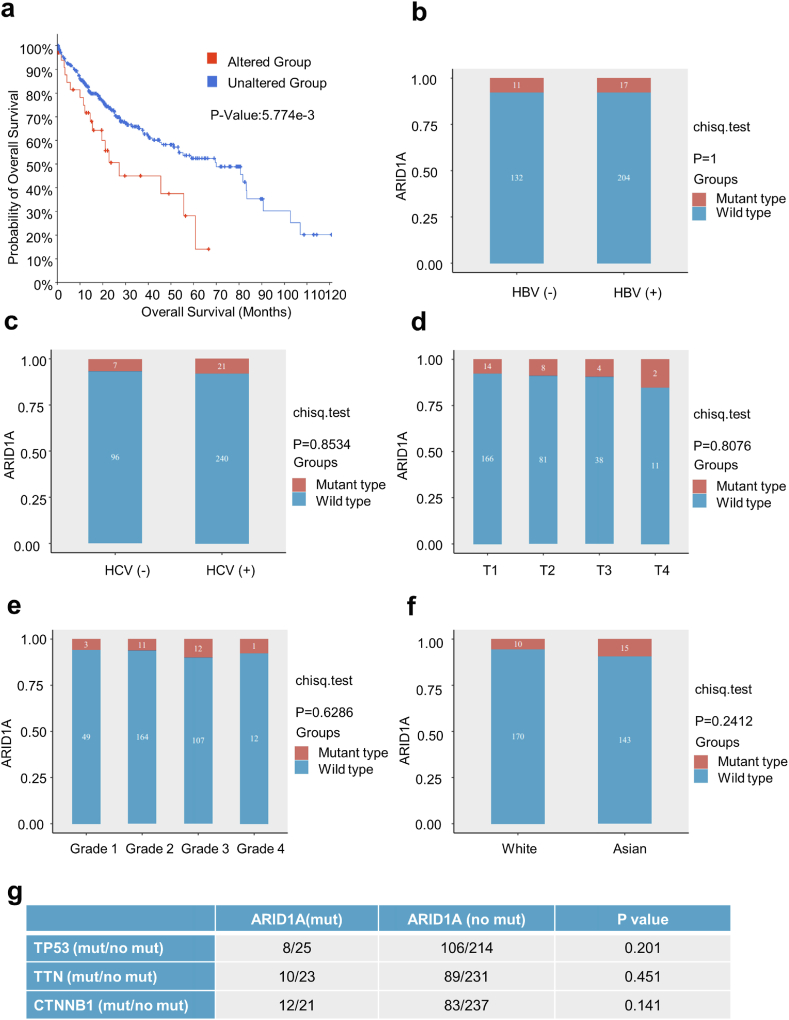
We examined the association between the gene alterations of ARID1A and HCC prognosis in the TCGA database. The results demonstrated that patients with ARID1A alterations had worse OS than those without ARID1A alterations (Fig. 2a).
Fig. 2.
Association between ARID1A gene mutation and the clinical features of HCC.
(a) Survival analysis based on the TCGA database. HCC patients with ARID1A alterations have worse OS than patients without ARID1A alterations. (b–f) The association between ARID1A gene mutation and HBV or HCV infection status, T stage, tumour grade, and race. Tumour grade represents the degree of differentiation in G1-G4, where G1 represents a high degree of differentiation and G4 represents a low degree of differentiation. (g) The relationship between ARID1A mutation status and mutations in other genes with higher HCC mutation rates.
Some important clinicopathological features have been shown to influence postoperative OS and RFS in HCC patients, and mutations of some commonly mutated genes, such as TP53, CTNNB1, and PIK3CA, in HCC samples are associated with HBV or HCV infection [32,33]. We further investigated the relationship between ARID1A gene mutations and important clinical features in HCC patients. We found that there was no significant relationship between ARID1A gene mutation and HBV or HCV infection status, tumour stage, tumour grade or race (Fig. 2b–f). However, the TCGA database showed that ARID1A mutation was not correlated with mutations in TP53, TTN, and CTNNB1 in HCC patients (Fig. 2g).
3.3. Correlation between ARID1A expression and HCC patient prognosis
A meta-analysis was conducted to investigate whether ARID1A expression was related to HCC patient prognosis. After the literature screening process (Supplementary Fig. 1a), we enrolled 1565 participants from 10 studies on ARID1A expression or mutation in HCC published between Mar. 2015 and Apr. 2022. There were 2 studies focusing on ARID1A gene mutations [11,34] and 7 studies concerning ARID1A expression [23,[35], [36], [37], [38], [39], [40], [41]]. The mean NOS score of the enrolled studies was 7.9 (Supplementary Table 1), which suggested that the quality of the enrolled studies was relatively high. Overall, the patients were distributed into a group with wild-type ARID1A or high expression and a group with ARID1A mutation or low expression. The features of the extracted studies are shown in Table 1. Supplementary Table 2 shows the data related to clinicopathological features and ARID1A expression in HCC patients.
Table 1.
Baseline characteristics of hepatocellular carcinoma and cholangiocarcinoma included in the studies.
|
Author(ref.) year |
Country | Methods | ARID1A(number) |
Median/range Of follow-up, m |
Survival analysis | Hazard ratio | 95% CI | p-value | Altered | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ALL | Low/Altered | High/Normal | |||||||||
| Abe [23]2015 | Japan | immunohistochemistry | 290 | 63 | 227 | NA | OS RFS |
1.41 1.21 |
0.72–2.48 0.93–1.56 |
0.99 0.78 |
expression |
| He [35]2015 | China | immunohistochemistry | 64 | 41 | 23 | NA | OS | 2.12 | 1.14–3.94 | 0.042 | expression |
| Zhao [36]2016 | China | immunohistochemistry | 115 | 14 | 101 | 38(3–72) | OS RFS |
1.19 1.26 |
0.58–2.43 0.57–2.79 |
0.612 0.57 |
expression |
| Zhou [37]2019 | China | immunohistochemistry | 60 | 41 | 19 | NA | OS | 2.31 | 1.36–3.92 | 0.021 | expression |
| Norifumi [38]2020 | Japan | immunohistochemistry | 255 | 174 | 81 | NA | OS RFS |
2.206 1.4979 |
1.438–3.385 1.053–2.13 |
0.0003 0.0245 |
expression |
| Yim [40] 2020 | Korea | immunohistochemistry | 242 | 121 | 121 | NA | OS | 2.46 | 1.90–3.17 | 0.001 | expression |
| Zhang [41] 2022 | China | immunohistochemistry | 263 | 160 | 83 | 70 | OS RFS |
1.69 1.51 |
1.32–2.16 1.22–1.89 |
0.001 0.0046 |
expression |
| Aly [39] 2022 | Egypt | immunohistochemistry | 50 | 22 | 26 | NA | RFS | 8.00 | 1.22–52.60 | 0.031 | expression |
| Kelly [34]2020 | US | institutional multigene assay | 64 | 11 | 53 | NA | OS | 2.12 | 1.01–4.43 | 0.046 | gene |
| Li [11]2020 | China | Next-generation sequencing | 362 | 25 | 337 | NA | OS | 2.577 | 1.42–4.675 | 0.037 | gene |
NA. Not available.
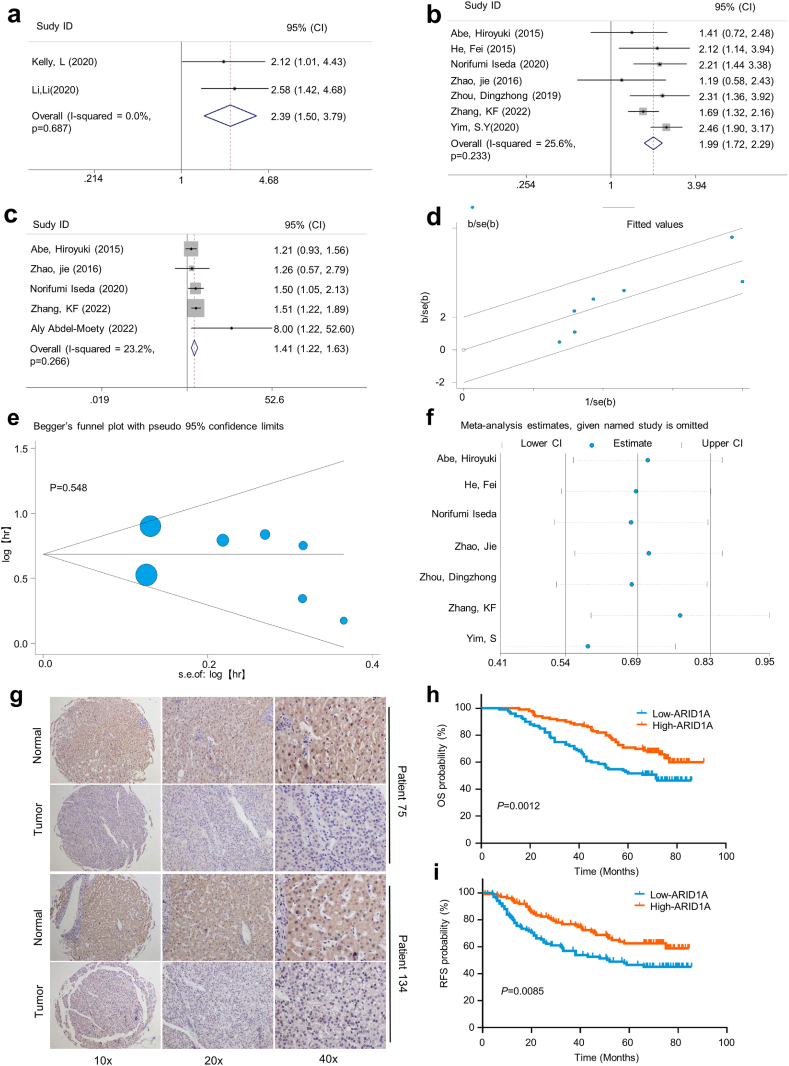
The meta-analysis results showed that patients with ARID1A genomic mutations or low ARID1A expression had worse OS and RFS (Fig. 3a–c). The fixed effect model was adopted due to substantial heterogeneity (I2 = 25.6%, P = 0.233). There was no substantial heterogeneity according to the Galbraith plot in this meta-analysis (Fig. 3d). We performed a subgroup analysis to clarify whether some potential confounding factors might influence the meta-analysis results (Table 2). We found that two studies centred on the impact of ARID1A genomic mutation on OS in HCC patients, and the pooled HR was 2.39 (95% CI, 1.50–3.79), while 7 studies focused on the impact of ARID1A expression on OS, and the pooled HR was 1.99 (95% CI, 1.72–2.29). After stratification according to sample size, the HR was 1.95 (95% CI = 1.67–2.28) in studies of >100 patients and 2.23 (95% CI = 1.49–3.33) in studies of <100 patients. After stratifying studies according to the HR estimation method, ARID1A mutation and low expression were found to be significantly related to unfavourable OS in the studies reporting HR (HR = 2.25, 95% CI = 1.61–3.13) and in the studies for which HR was estimated based on the Kaplan‒Meier curve (HR = 1.92, 95% CI = 1.93–2.27). Subgroup analysis showed no heterogeneity.
Fig. 3.
Prognostic value of ARID1A mutations and expression levels for patients with HCC.
(a) Forest plots of associations between ARID1A mutation status and OS in HCC. (b, c) Forest plots of the associations between low ARID1A expression and OS and RFS in HCC. (d) Galbraith plot for heterogeneity. There was no substantial heterogeneity among those studies. (e) Publication bias analysis Begg's test. There was no publication bias in this meta-analysis. (f) Sensitivity analysis of HCC studies. No studies had an impact on the pooled HR. (g) The expression of ARID1A in HCC tissues was assessed by IHC staining of a human HCC TMA. (h, i) Kaplan‒Meier analysis of TMA showed that low expression of ARID1A in HCC tissues was associated with worse OS and RFS.
Table 2.
Pooled HRs for OS and subgroup analysis of ARID1A mutation or low expression in HCC patients.
|
Categories |
Number of studies | Number of patients | Pooled HR | 95% CI | Heterogeneity | |
| I2 | PH value | |||||
| Altered Type | ||||||
| Gene mutation | 2 | 426 | 2.39 | 1.50–3.79 | 0.0% | 0.687 |
| expression | 7 | 1139 | 1.99 | 1.72–2.29 | 25.6% | 0.233 |
| Sample size | ||||||
| >100 | 5 | 1015 | 1.95 | 1.67–2.28 | 47.8% | 0.105 |
| <100 | 2 | 124 | 2.23 | 1.49–3.33 | 0.0% | 0.837 |
| Method to estimate HR | ||||||
| Reported in article | 2 | 315 | 2.25 | 1.61–3.13 | 0% | 0.894 |
| Estimate in Kaplan-Meier curve | 5 | 824 | 1.92 | 1.93–2.27 | 45.9% | 0.116 |
To assess publication bias in the meta-analysis, Egger's test and Begg's test were performed. Both Egger's test and Begg's test revealed that there was no publication bias (P = 0.519, P = 0.548, respectively) (Fig. 3e, Supplementary Fig. 1b). To determine how much each study influenced the meta-analysis results, we conducted a sensitivity analysis, which was performed by removing each included article sequentially to examine the impact of the removed article on the combined HR. Sensitivity analysis revealed that none of those studies had an influence on the combined HR (Fig. 3f). Furthermore, our results revealed that ARID1A expression was negatively correlated with HCC tumour size but was not correlated with HBV infection, cirrhosis, histological differentiation, TNM staging, recurrence, metastasis, or sex (Supplementary Fig. 2).
Next, we evaluated ARID1A expression in HCC tissues by IHC staining of a human HCC tissue microarray (TMA). Our results showed that ARID1A expression in HCC tissues was lower than that in adjacent liver tissues (Fig. 3g). Kaplan‒Meier survival analysis revealed that patients with low ARID1A expression had worse OS and RFS than those with high ARID1A expression (Fig. 3h and i). ARID1A expression and TMA clinical characteristics analysis showed that low ARID1A expression was significantly associated with younger age, larger tumour size, and lower tumour differentiation in HCC (Table 3). The Cox proportional hazard model confirmed that low ARID1A expression was an independent predictor of OS and RFS in HCC patients (Supplementary Tables 3 and 4).
Table 3.
Statistics for ARID1A and clinicopathologic features in HCC patients.
| Variables | ARID1A Expression |
χ2 | P-value | ||
|---|---|---|---|---|---|
| Total (n = 200) | Low(n = 100) | High(n = 100) | |||
| Age, years | 5.794 | 0.016 | |||
| ≤50 | 95 | 56 | 39 | ||
| >50 | 105 | 44 | 61 | ||
| Sex | 0.954 | 0.329 | |||
| Male | 169 | 82 | 87 | ||
| Female | 31 | 18 | 13 | ||
| Tumour number | 0.267 | 0.606 | |||
| 1 | 157 | 82 | 75 | ||
| ≥2 | 43 | 18 | 25 | ||
| Tumour size, cm | 5.050 | 0.025 | |||
| ≤5 | 133 | 59 | 74 | ||
| >5 | 67 | 41 | 26 | ||
| Tumour capsule | 1.070 | 0.301 | |||
| Yes | 71 | 39 | 32 | ||
| No | 129 | 61 | 68 | ||
| Tumour differentiation | 5.373 | 0.020 | |||
| I-II | 152 | 83 | 69 | ||
| III-IV | 48 | 17 | 31 | ||
| Vascular invasion | 1.503 | 0.220 | |||
| No | 159 | 76 | 83 | ||
| Yes | 41 | 24 | 17 | ||
| Liver cirrhosis | 0.000 | 1.000 | |||
| No | 34 | 17 | 17 | ||
| Yes | 166 | 83 | 83 | ||
| HBsAg | 0.767 | 0.381 | |||
| Negative | 41 | 23 | 18 | ||
| Positive | 159 | 77 | 82 | ||
| AFP, ng/ml | 3.089 | 0.079 | |||
| ≤400 | 74 | 31 | 43 | ||
| >400 | 126 | 69 | 57 | ||
| Recurrence | 1.628 | 0.202 | |||
| No | 93 | 42 | 51 | ||
| Yes | 107 | 58 | 49 | ||
3.4. ARID1A regulates HCC cell proliferation, colony formation ability, migration and invasion in vitro
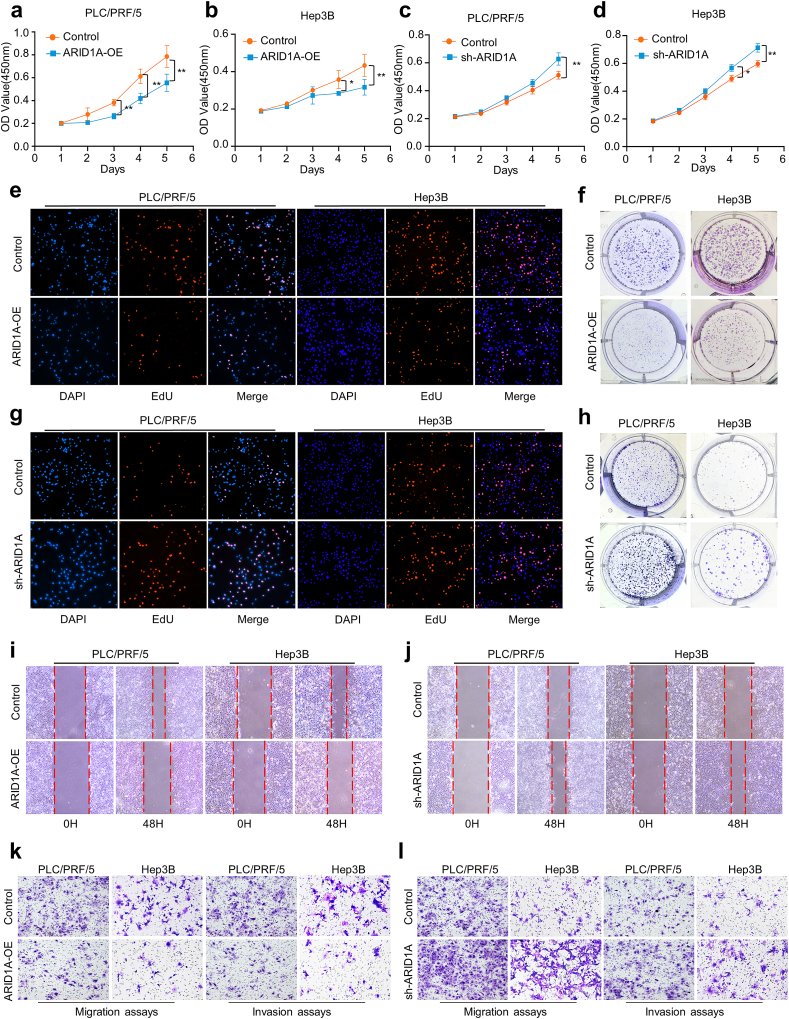
To determine the role of ARID1A expression in HCC progression, Sanger sequencing was used to detect whether ARID1A mutations existed in different HCC cell lines. Our results revealed that there was no ARID1A mutation in the Hep3B, Huh-7, PLC/PRF/5 and MHCC-97H genomes. Then, we performed transfection of the ARID1A overexpression plasmid and lentivirus-mediated transduction of ARID1A knockdown in PLC/PRF/5 cells and Hep3B cells to perform a series of functional experiments (Supplementary Figs. 3a–d). CCK-8, EdU and colony formation assays were used to assess the changes in HCC cell proliferation and clonogenic ability. We found that overexpression of ARID1A inhibited the proliferation and decreased the clonogenic ability of PLC/PRF/5 and Hep3B cells, and these abilities were enhanced when ARID1A was knocked down (Fig. 4a–h). Next, cell-based wound healing assays and transwell assays showed that overexpression of ARID1A significantly inhibited the migration and invasion of the PLC/PRF/5 and Hep3B cell lines, and knockdown induced the opposite effects (Fig. 4i-l). Quantitative analysis of all experiments is shown in Supplementary Fig. 3e-n.
Fig. 4.
ARID1A regulates HCC cell proliferation, clonogenic ability, migration and invasion in vitro.
(a–h) The viability of HCC cells was detected by CCK-8, EdU, and colony formation assays. (i–l) Cell wound scratch assays and transwell assays were used to detect the migration and invasion of HCC cells after ARID1A overexpression or knockdown. *P < 0.05, **P < 0.01, ***P < 0.001.
3.5. ARID1A regulates RBL1 expression in HCC via the JNK/FOXO3 pathway
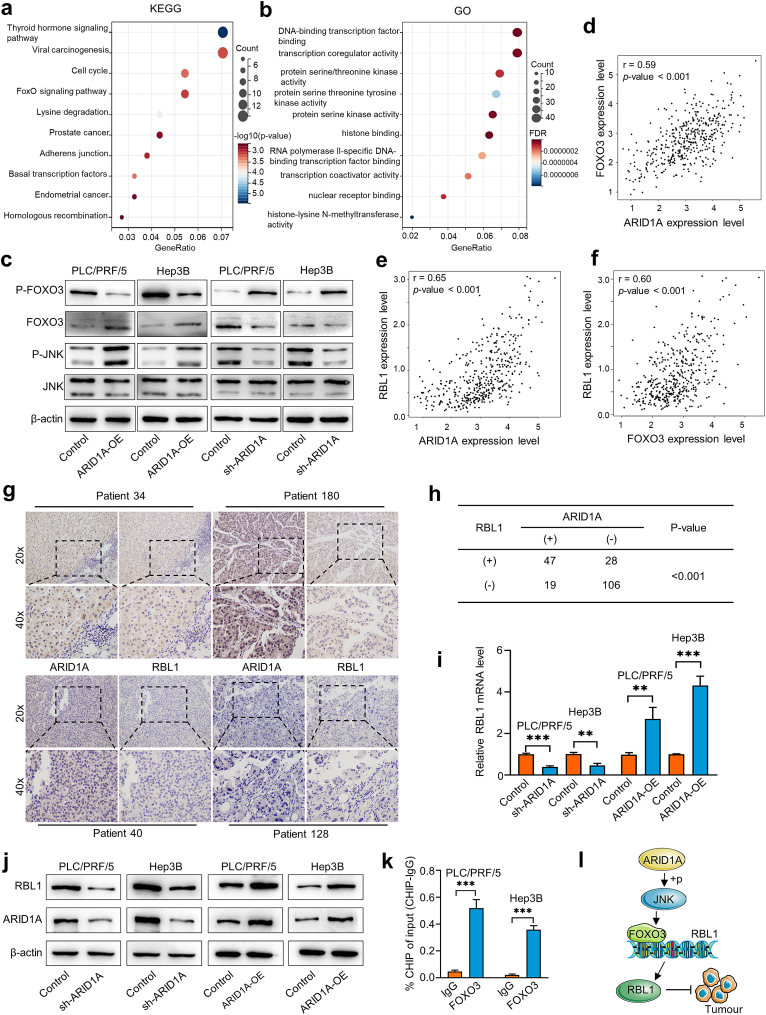
To elucidate the molecular mechanism by which ARID1A regulates HCC progression, we analysed the genes significantly associated with ARID1A expression in HCC samples in the TCGA database (Spearman's correlation >0.5, P < 0.05). A total of 536 positively and 199 negatively correlated genes were identified. KEGG analysis of positively correlated genes showed that the thyroid hormone signalling pathway, lysine degradation, the cell cycle, and the FoxO signalling pathway were the main enriched pathways (Fig. 5a). GO analyses of these genes revealed that the most enriched biological processes were DNA-binding transcription factor binding, transcription coregulator activity, protein serine/threonine kinase activity, and RNA polymerase II-specific DNA-binding transcription factor binding (Fig. 5b). The KEGG and GO enrichment analysis results of negatively correlated genes are shown in Supplementary Fig. 4.
Fig. 5.
ARID1A regulates HCC malignant behaviour by modulating RBL1 expression.
(a, b) GO function and KEGG pathway enrichment analysis was performed for genes significantly associated with ARID1A. (c) Western blot analysis confirmed the effects of ARID1A OE and KD on the JNK/FOXO3 signalling pathways in HCC. (d–f) The correlation between ARID1A, FOXO3 and RBL1 mRNA expression was analysed from the TCGA database. (g) The expression of ARID1A and RBL1 in HCC tissues was assessed by IHC staining of a human HCC TMA. (h) Statistical analysis showed that ARID1A expression was significantly correlated with RBL1 expression in HCC. (i, j) Changes in RBL1 mRNA levels and protein levels after knockdown or overexpression of ARID1A. (k) ChIP‒qPCR revealed that FOXO3 binds to the RBL1 promoter. (l) Schematic diagram of the mechanism by which ARID1A regulates HCC progression through the JNK/FOXO3/RBL1 axis. Unadjusted Western blot images are available in the Supplementary Material a, b.
The forkhead box O3 (FOXO3) transcription factors play a key role in regulating the cell cycle and cell death and mediating cytotoxic functions in cancer therapy. FOXO3 is stimulated by the upstream kinase c-Jun N-terminal kinase (JNK) to increase its nuclear localization and transcriptional activity, thereby preventing tumour cell proliferation [42]. Based on the results of the KEGG analysis, we hypothesized that ARID1A plays a tumour suppressor role in HCC by regulating the JNK/FOXO3 pathway. To verify our hypothesis, we detected the effect of ARID1A on the changes in JNK/FOXO3 pathway components. We found that ARID1A-OE induces the phosphorylation of JNK, which leads to the dephosphorylation of FOXO3 and the upregulation of FOXO3, while ARID1A-KD inhibits the phosphorylation of JNK and leads to the downregulation of FOXO3 (Fig. 5c). Moreover, TCGA data analysis revealed that the ARID1A mRNA level was positively associated with the forkhead box O3 (FoxO3) mRNA level in HCC samples (Fig. 5d), and the ARID1A and FOXO3 mRNA levels were positively associated with the retinoblastoma-like 1 (RBL1) mRNA level in HCC samples (Fig. 5e and f). As an important pocket protein family member, RBL1 has been demonstrated to have important tumour suppressor activity in some contexts. Therefore, we hypothesized that ARID1A inhibits HCC progression by regulating RBL1 expression.
To validate this hypothesis, we further analysed the relationship between ARID1A and RBL1 protein levels in HCC samples by IHC of a human HCC tissue microarray (Fig. 5g). A kappa consistency test was used to determine the correlation between ARID1A protein expression and RBL1 levels, and the results showed that ARID1A protein expression was consistent with RBL1 expression in HCC samples (Fig. 5h). More importantly, we found that ARID1A knockdown downregulated RBL1 at the mRNA and protein levels in HCC cells in vitro, while overexpression induced the opposite effects (Fig. 5i and j). In addition, ChIP‒qPCR revealed that FOXO3 binds to the RBL1 promoter in HCC cells (Fig. 5k). Taken together, our results show that ARID1A functions as a tumour suppressor gene that inhibits the malignant behaviour of HCC cells by regulating the expression of RBL1 via the JNK/FOXO3 pathway. Quantitative analysis of all western blots is shown in Supplementary Fig. 5a-l.
4. Discussion
As a frequently mutated SWI/SNF subunit, the ARID1A protein targets SWI/SNF complexes to tissue-specific enhancers and maintains the accessibility of chromatin [43]. Previous study showed that the loss of ARID1A may affect the function of the SWI/SNF complex and then impair cellular transcription by mechanisms such as disruption of nucleosome activity, abnormal assembly of the SWI/SNF complex, targeting of specific genomic sites, and recruitment of coactivators/corepressors [44]. Another study revealed that the formation of an abnormal SWI/SNF complex induced by the absence of ARID1A can result in a dysregulation of enhancer activity, impair differentiation processes and result in gene expression dysregulation that contributes to tumour formation [45]. Our findings suggest that the ARID1A gene has a relatively high mutation rate in patients with HCC and that most mutations are deletions that lead to the loss of ARID1A mRNA expression. As a tumour suppression, ARID1A is involved in the cell cycle/DNA damage checkpoint, regulation of P53 targets, and telomerase activation. Mutation of ARID1A in HCC could promote proliferation and migration [46], which may be the reason for the poor prognosis of HCC patients. Another study revealed that ARID1A deletions contribute to HCC cell proliferation, invasion, and migration by activating the PI3K/mTOR pathway and lncRNA MVIH [46,47]. In addition, Yao et al. reported that ARID1A deletion promotes hepatocellular carcinoma progression by promoting MYC transcription [48]. The ARID1A status can also be used to identify whether HCC is sensitive to PI3K/mTOR inhibitors [49,50]. Our results showed that mutation or low expression of ARID1A is correlated with worse OS in HCC patients. Therefore, detection of ARID1A mutation and levels in circulating tumour DNA samples or HCC samples can be regarded as a potential prognostic tool.
A number of factors have been demonstrated to influence HCC patient outcomes and prognosis [51]. Four of the most important factors determining HCC patient survival are the severity of the accompanying liver disease, tumour size/number, portal vein invasion, and presence of metastases [52,53]. Our study analysed the relationship between ARID1A status and HCC patient clinicopathologic features. The results of the meta-analysis and TMA analysis both showed that ARID1A status was negatively associated with tumour size. Since there are only six studies were considered for the analysis of clinicopathological features, it is necessary to conduct in-depth studies on ARID1A expression in cohorts with available data on clinicopathological features in the future. In addition, we confirmed that overexpression of ARID1A could inhibit the proliferation, migration and invasion of HCC cells in vitro, while ARID1A knockdown could promote HCC cell proliferation, migration and invasion, which was consistent with previous experimental results [35]. Therefore, our study demonstrated the role of ARID1A as a tumour suppressor in HCC.
Previous evidence suggested that the RBL1 protein is an important tumour suppressor [[26], [27], [28]]. Dysregulation of RBL1 is associated with the development and progression of tumours [25]. It is critical to identify the regulatory mechanism of RBL1 in HCC. FOXO has been demonstrated to be a key molecular trigger for Rb/E2F-induced apoptosis and expression of Bim and p73 proapoptotic genes in retinoblastoma [54]. The dephosphorylation and increased activity of FOXO3 can be regulated by the activation of JNK, mimicking paclitaxel activity [55]. Our study demonstrated that ARID1A could increase the expression of transcription factor FOXO3 by activating JNK, and then regulate the expression of RBL1. Besides, ARID1A expression was significantly correlated with FOXO3 and RBL1 expression in HCC samples. (Fig. 5l). These results indicate that RBL1 may be a potential downstream target of the ARID1A/JNK/FOXO3 axis in HCC.
There are some limitations to our study that cannot be ignored. First, owing to the limited number of enrolled studies, the relationship between ARID1A status and HCC patient prognosis needs to be further evaluated. Second, our study may have selection bias and inevitable recall bias. Third, ARID1A mutation is a loss-of-function mutation, and further experiments are needed to explore the roles of ARID1A and RBL1 to determine their exact mechanisms of action by using CRISPR/CRISPR-9 techniques. And the specific mechanism by which ARID1A regulates the FOXO3 signalling pathway and RBL1 expression should be further explored.
Declarations
Author contribution statement
Guang-Xiao Meng; Chun-Cheng Yang: Conceived and designed the experiments; Performed the experiments; Wrote the paper.Lun-Jie Yan; Ya-Fei Yang; Yu-Chuan Yan: Performed the experiments; Analysed and interpreted the data.Jian-Guo Hong: Analysed and interpreted the data; Contributed reagents, materials, analysis tools or data.Zhi-Qiang Chen: Performed the experiments.Zhaoru Dong; Tao Li: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
Tao Li was supported by Taishan Scholars Program of Shandong Province [tstp20221158], National Natural Science Foundation of China [82073200; 81874178], Major basic research of Shandong Provincial Natural Science Foundation [ZR2021ZD26], Funds for Independent Cultivation of Innovative Team from Universities in Jinan [2020GXRC023]. Zhaoru Dong was supported by National Natural Science Foundation of China [82172647], Postdoctoral Research Foundation of China [2020M682192 & 2022T150385], Natural Science Foundation of Shandong Province [ZR2021MH194].
Data availability statement
Data included in article/supp. material/referenced in article.
Declaration of interest's statement
The authors declare no competing interests.
Acknowledgements
We thank all of the original studies for sharing their data.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e14307.
Contributor Information
Zhao-Ru Dong, Email: dongzhaoru0911@163.com.
Tao Li, Email: litao7706@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Akinyemiju T., Abera S., Ahmed M., et al. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the global burden of disease study 2015. JAMA Oncol. 2017;3:1683–1691. doi: 10.1001/jamaoncol.2017.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2021;7:7. doi: 10.1038/s41572-021-00245-6. [DOI] [PubMed] [Google Scholar]
- 4.Chidambaranathan-Reghupaty S., Fisher P.B., Sarkar D. Hepatocellular carcinoma (HCC): epidemiology, etiology and molecular classification. Adv. Cancer Res. 2021;149:1–61. doi: 10.1016/bs.acr.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey M.H., Tokheim C., Porta-Pardo E., et al. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;173:371–385.e318. doi: 10.1016/j.cell.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park M.H., Yun H.M., Hwang C.J., et al. Presenilin mutation suppresses lung tumorigenesis via inhibition of peroxiredoxin 6 activity and expression. Theranostics. 2017;7:3624–3637. doi: 10.7150/thno.21408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malumbres M., Barbacid M. RAS oncogenes: the first 30 years. Nat. Rev. Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 8.Thein K.Z., Biter A.B., Hong D.S. Therapeutics targeting mutant KRAS. Annu. Rev. Med. 2021;72:349–364. doi: 10.1146/annurev-med-080819-033145. [DOI] [PubMed] [Google Scholar]
- 9.Uprety D., Adjei A.A. KRAS: from undruggable to a druggable Cancer Target. Cancer Treat Rev. 2020;89 doi: 10.1016/j.ctrv.2020.102070. [DOI] [PubMed] [Google Scholar]
- 10.Howell J., Atkinson S.R., Pinato D.J., et al. Identification of mutations in circulating cell-free tumour DNA as a biomarker in hepatocellular carcinoma. Eur. J. Cancer. 2019;116:56–66. doi: 10.1016/j.ejca.2019.04.014. [DOI] [PubMed] [Google Scholar]
- 11.Li L., Rao X., Wen Z., et al. Implications of driver genes associated with a high tumor mutation burden identified using next-generation sequencing on immunotherapy in hepatocellular carcinoma. Oncol. Lett. 2020;19:2739–2748. doi: 10.3892/ol.2020.11372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zucman-Rossi J., Villanueva A., Nault J.-C., Llovet J.M. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology. 2015;149:1226. doi: 10.1053/j.gastro.2015.05.061. [DOI] [PubMed] [Google Scholar]
- 13.Duffy M.J., Synnott N.C., Crown J. Mutant p53 as a target for cancer treatment. Eur. J. Cancer. 2017;83:258–265. doi: 10.1016/j.ejca.2017.06.023. [DOI] [PubMed] [Google Scholar]
- 14.Karachaliou N., Paulina Bracht J.W., Rosell R. ARID1A gene driver mutations in lung adenocarcinomas. J. Thorac. Oncol. 2018;13:e255–e257. doi: 10.1016/j.jtho.2018.07.099. [DOI] [PubMed] [Google Scholar]
- 15.Balbas-Martinez C., Rodriguez-Pinilla M., Casanova A., et al. ARID1A alterations are associated with FGFR3-wild type, poor-prognosis, urothelial bladder tumors. PLoS One. 2013;8 doi: 10.1371/journal.pone.0062483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cajuso T., Hanninen U.A., Kondelin J., et al. Exome sequencing reveals frequent inactivating mutations in ARID1A, ARID1B, ARID2 and ARID4A in microsatellite unstable colorectal cancer. Int. J. Cancer. 2014;135:611–623. doi: 10.1002/ijc.28705. [DOI] [PubMed] [Google Scholar]
- 17.Han X., Chen W., Chen P., et al. Aberration of ARID1A is associated with the tumorigenesis and prognosis of sporadic nonfunctional pancreatic neuroendocrine tumors. Pancreas. 2020;49:514–523. doi: 10.1097/MPA.0000000000001535. [DOI] [PubMed] [Google Scholar]
- 18.Onder S., Fayda M., Karanlık H., et al. Loss of ARID1A expression is associated with poor prognosis in invasive micropapillary carcinomas of the breast: a clinicopathologic and immunohistochemical study with long-term survival analysis. Breast J. 2017;23:638–646. doi: 10.1111/tbj.12823. [DOI] [PubMed] [Google Scholar]
- 19.Liu G., Xu P., Fu Z., et al. Prognostic and clinicopathological significance of ARID1A in endometrium-related gynecological cancers: a meta-analysis. J. Cell. Biochem. 2017;118:4517–4525. doi: 10.1002/jcb.26109. [DOI] [PubMed] [Google Scholar]
- 20.Xie H., Chen P., Huang H.W., Liu L.P., Zhao F. Reactive oxygen species downregulate ARID1A expression via its promoter methylation during the pathogenesis of endometriosis. Eur Rev Med Pharmaco. 2017;21:4509–4515. [PubMed] [Google Scholar]
- 21.Wu R.C., Wang T.L., Shih Ie M. The emerging roles of ARID1A in tumor suppression. Cancer Biol. Ther. 2014;15:655–664. doi: 10.4161/cbt.28411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J.N., Roberts C.W.M. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer Discov. 2013;3:35–43. doi: 10.1158/2159-8290.CD-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abe H., Hayashi A., Kunita A., et al. Altered expression of AT-rich interactive domain 1A in hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2015;8:2763–2770. [PMC free article] [PubMed] [Google Scholar]
- 24.Henley S.A., Dick F.A. The retinoblastoma family of proteins and their regulatory functions in the mammalian cell division cycle. Cell Div. 2012;7:10. doi: 10.1186/1747-1028-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schade A.E., Fischer M., DeCaprio J.A.R.B. p130 and p107 differentially repress G1/S and G2/M genes after p53 activation. Nucleic Acids Res. 2019;47:11197–11208. doi: 10.1093/nar/gkz961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lara M.F., Santos M., Ruiz S., et al. p107 acts as a tumor suppressor in pRb-deficient epidermis. Mol. Carcinog. 2008;47:105–113. doi: 10.1002/mc.20367. [DOI] [PubMed] [Google Scholar]
- 27.Naert T., Dimitrakopoulou D., Tulkens D., et al. RBL1 (p107) functions as tumor suppressor in glioblastoma and small-cell pancreatic neuroendocrine carcinoma in Xenopus tropicalis. Oncogene. 2020;39:2692–2706. doi: 10.1038/s41388-020-1173-z. [DOI] [PubMed] [Google Scholar]
- 28.Ventura E., Iannuzzi C.A., Pentimalli F., Giordano A., Morrione A. RBL1/p107 expression levels are modulated by multiple signaling pathways. Cancers. 2021;13 doi: 10.3390/cancers13195025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moher D., Liberati A., Tetzlaff J., Altman D.G. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6 doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong Z.R., Sun D., Yang Y.F., et al. TMPRSS4 drives angiogenesis in hepatocellular carcinoma by promoting HB-egf expression and proteolytic cleavage. Hepatology. 2020;72:923–939. doi: 10.1002/hep.31076. [DOI] [PubMed] [Google Scholar]
- 31.Wang C.H., Guo Z.Y., Chen Z.T., et al. TMPRSS4 facilitates epithelial-mesenchymal transition of hepatocellular carcinoma and is a predictive marker for poor prognosis of patients after curative resection. Sci. Rep. 2015;5 doi: 10.1038/srep12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong F., Kong D., Yang X., et al. Integrative analysis of highly mutated genes in hepatitis B virus-related hepatic carcinoma. Cancer Med. 2020;9:2462–2479. doi: 10.1002/cam4.2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tornesello M.L., Buonaguro L., Tatangelo F., et al. Mutations in TP53, CTNNB1 and PIK3CA genes in hepatocellular carcinoma associated with hepatitis B and hepatitis C virus infections. Genomics. 2013;102:74–83. doi: 10.1016/j.ygeno.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 34.Kelly L., Yarmohammadi H., Erinjeri J., et al. 4:03 PM Abstract No. 197 ARID1A mutations are associated with shorter time to local progression and worse overall survival after embolization of hepatocellular carcinomas. J. Vasc. Intervent. Radiol. 2020;31:S90. doi: 10.1016/j.jvir.2019.12.236. [DOI] [Google Scholar]
- 35.He F., Li J., Xu J., et al. Decreased expression of ARID1A associates with poor prognosis and promotes metastases of hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2015;34 doi: 10.1186/s13046-015-0164-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao J., Chen J., Lin H., et al. The clinicopathologic significance of BAF250a (ARID1A) expression in hepatocellular carcinoma. Pathol. Oncol. Res. 2016;22:453–459. doi: 10.1007/s12253-015-0022-9. [DOI] [PubMed] [Google Scholar]
- 37.Zhou D., Cao S., Long W., Luo L. Expression of ARID1A in hepatocellular carcinoma and its relation to the prognosis. Acta Med. Mediterr. 2019;35:2411–2415. doi: 10.19193/0393-6384_2019_5_376. [DOI] [Google Scholar]
- 38.Iseda N., Itoh S., Yoshizumi T., et al. ARID1A deficiency is associated with high programmed death ligand 1 expression in hepatocellular carcinoma. Hepatol Commun. 2021;5:675–688. doi: 10.1002/hep4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdel-Moety A., Baddour N., Salem P., Rady A., El-Shendidi A. ARID1A expression in hepatocellular carcinoma and relation to tumor recurrence after microwave ablation. Clin. Exp. Hepatol. 2022;8:49–59. doi: 10.5114/ceh.2022.114172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yim S.Y., Kang S.H., Shin J.H., et al. Low ARID1A expression is associated with poor prognosis in hepatocellular carcinoma. Cells. 2020;9 doi: 10.3390/cells9092002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang F.K., Ni Q.Z., Wang K., et al. Targeting USP9X-AMPK Axis in arid1a-deficient hepatocellular carcinoma. Cell Mol Gastroenterol Hepatol. 2022;14:101–127. doi: 10.1016/j.jcmgh.2022.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao X., Peng Y., Fang Z., et al. Inhibition of EZH2 ameliorates hyperoxaluria-induced kidney injury through the JNK/FoxO3a pathway. Life Sci. 2022;291 doi: 10.1016/j.lfs.2021.120258. [DOI] [PubMed] [Google Scholar]
- 43.Mullen J., Kato S., Sicklick J.K., Kurzrock R. Targeting ARID1A mutations in cancer. Cancer Treat Rev. 2021;100 doi: 10.1016/j.ctrv.2021.102287. [DOI] [PubMed] [Google Scholar]
- 44.Wu J.N., Roberts C.W. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer Discov. 2013;3:35–43. doi: 10.1158/2159-8290.Cd-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mathur R. ARID1A loss in cancer: towards a mechanistic understanding. Pharmacol. Ther. 2018;190:15–23. doi: 10.1016/j.pharmthera.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 46.Cheng S., Wang L., Deng C.H., Du S.C., Han Z.G. ARID1A represses hepatocellular carcinoma cell proliferation and migration through lncRNA MVIH. Biochem. Biophys. Res. Commun. 2017;491:178–182. doi: 10.1016/j.bbrc.2017.07.072. [DOI] [PubMed] [Google Scholar]
- 47.Sun X., Wang S.C., Wei Y., et al. Arid1a has context-dependent oncogenic and tumor suppressor functions in liver cancer. Cancer Cell. 2017;32:574. doi: 10.1016/j.ccell.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao Y., Liu G., Ouyang X., et al. Loss of ARID1A promotes hepatocellular carcinoma progression via up-regulation of MYC transcription. J Clin Transl Hepatol. 2021;9:528–536. doi: 10.14218/jcth.2021.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young Y.S., Shin J.H., Jeong Y.S., et al. Loss of arid1a serves as prognostic biomarker for hepatocellular carcinoma and has synthetic lethality with pi3k/mtor inhibition. Hepatology. 2018;68:553A. doi: 10.1002/hep.30257. [DOI] [Google Scholar]
- 50.Pópulo H., Lopes J.M., Soares P. The mTOR signalling pathway in human cancer. Int. J. Mol. Sci. 2012;13:1886–1918. doi: 10.3390/ijms13021886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marengo A., Rosso C., Bugianesi E. Liver cancer: connections with obesity, fatty liver, and cirrhosis. Annu. Rev. Med. 2016;67:103–117. doi: 10.1146/annurev-med-090514-013832. [DOI] [PubMed] [Google Scholar]
- 52.Tan D.J.H., Ng C.H., Lin S.Y., et al. Clinical characteristics, surveillance, treatment allocation, and outcomes of non-alcoholic fatty liver disease-related hepatocellular carcinoma: a systematic review and meta-analysis. Lancet Oncol. 2022;23:521–530. doi: 10.1016/s1470-2045(22)00078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giannini E.G., Bucci L., Garuti F., et al. Patients with advanced hepatocellular carcinoma need a personalized management: a lesson from clinical practice. Hepatology. 2018;67:1784–1796. doi: 10.1002/hep.29668. [DOI] [PubMed] [Google Scholar]
- 54.Xie C., Lu H., Nomura A., et al. Co-deleting Pten with Rb in retinal progenitor cells in mice results in fully penetrant bilateral retinoblastomas. Mol. Cancer. 2015;14:93. doi: 10.1186/s12943-015-0360-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sunters A., Madureira P.A., Pomeranz K.M., et al. Paclitaxel-induced nuclear translocation of FOXO3a in breast cancer cells is mediated by c-Jun NH2-terminal kinase and Akt. Cancer Res. 2006;66:212–220. doi: 10.1158/0008-5472.Can-05-1997. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supp. material/referenced in article.