Abstract
Cancer/testis antigens (CTAs) are reproductive tissue-restricted genes, frequently ectopic expressed in tumors. CTA genes associate with a poor prognosis in some solid tumors, due to their potential roles in the tumorigenesis and progression. However, whether CTAs relate with hepatocellular carcinoma (HCC) remains unclear. In this study, the prognostic signatures based on CTA genes were investigated and validated in three cohorts including Chinese HCC patients with hepatitis B virus infection (CHCC-HBV), International Cancer Genome Consortium (ICGC) and The Cancer Genome Atlas (TCGA) cohorts. Univariate, LASSO, and multivariate Cox regression analyses were used to screen prognostic genes and develop the prognostic gene signature. A prognosis model was established with six CTA genes (SSX1, CTCFL, OIP5, CEP55, NOL4, and TPPP2) in CHCC-HBV cohort, and further validated in the ICGC and TCGA cohorts. The CTA signature was an essential prognostic predictor independent of other clinical pathological factors. High-risk group exhibited up-regulated cell cycle-related and tumor-related pathways and more M0 macrophage, activated mast cell, activated memory CD4+ T cell, and memory B cell infiltration. Furthermore, CTA signature correlated with the sensitivity to multiple chemotherapy drugs. Our results highlighted that the CTA gene profiling was a prognostic assessment tool for HCC patients.
Keywords: Hepatocellular carcinoma, Cancer-testis antigens, Bioinformatics, Risk score, Prognosis
Highlights
-
•
We identified a list of cancer-testis antigens and developed a signature for hepatocellular carcinoma prognosis prediction.
-
•
The signature based on cancer-testis antigens may serve as an independent prognostic factor.
-
•
A predictive nomogram was constructed.
-
•
Our study regarding cancer-testis antigens genes provides new insight for the immunotherapy of hepatocellular carcinoma.
1. Introduction
Hepatocellular carcinoma (HCC) is the most common primary liver cancer, ranking the sixth major contributor to cancer-related mortality on a global scale [1]. Since most patients with HCC are asymptomatic in the early stage, they are usually diagnosed at the advanced stage [2]. Despite the rapid advancement in the target therapy and immunotherapy of HCC, the prognosis of HCC patients remains grim, with the average five-year survival rate of 19.6% [3]. The Barcelona Clinic Liver Cancer (BCLC) classification for HCC provides an efficient decision-making guide for clinicians. However, patients with the same BCLC stage undergoing the same therapy frequently have different prognoses, highlighting that HCC is a highly heterogeneous malignancy [4]. Consequently, it is extremely important to discover novel prognostic biomarkers which could improve the risk stratification among HCC patients. Cancer-testis antigens (CTAs) belong to the tumor-associated antigens [5]. Due to their potential roles in tumor progression, CTAs could be considered as prognosis biomarkers of some tumors [6]. Abnormal CTA reactivation may facilitate tumor progression through epigenetic regulation, angiogenesis promotion, and immune escape [7]. Recent studies revealed that synovial sarcoma X-2 (SSX2) and synaptonemal complex protein 1 (SCP-1) expressions led to cellular genomic instability in multiple cancers [7,8]. Melanoma-associated antigen A1(MAGEA1) overexpression correlated with the poor overall survival in esophageal squamous cell carcinoma and lactate dehydrogenase C4 (LDH-C4) expression positively correlated with the tumor size, the epidermal growth factor receptor (EGFR) and Ki-67 expressions in breast cancer [9,10]. In this study, we investigated whether CTA signature was a prognostic biomarker for survival among HCC patients. Prognostic signature based on six CTA genes was established in CHCC-HBV cohort [11] and then validated in the The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC) datasets. To further elucidate the possible biological mechanisms underlying the signature, we examined the association between the signature and signaling pathways, immune heterogeneity, and drug sensitivity in HCC. The prognostic significance of CTAs was highlighted in HCC and this CTA panel may promote patient stratification in clinical practice.
2. Materials and methods
2.1. Cancer-testis antigens
276 CTA genes were extracted in the CTA database (http://www.cta.lncc.br/).
2.2. Patient cohorts
Our group previously performed the multi-omics analysis of hepatitis B virus (HBV)-related HCC using paired tumor and adjacent liver tissues from 159 patients [https://www.biosino.org/node,OEP000321] [11]. The RNA-seq data and clinical information of 365 patients with HCC with were collected from TCGA (https://portal.gdc.cancer.gov/) and another 231 HCC patients were collected from ICGC (https://dcc.icgc.org/).
2.3. Screening of cancer-testis antigens
To acquire the differentially expressed CTA genes, the univariate Cox proportional hazard regression model was constructed using the R package ‘coxph’ (http://rstudio.com). In this study, log rank P < 0.05 was selected as the threshold. The least absolute shrinkage and selection operator (LASSO) was performed using the R package ‘glmet’ to identify the CTA signature gene.
2.4. Establishment and validation of the cancer-testis antigen signature
Multivariate Cox survival analysis was performed to construct the prognostic risk model. According to the median of risk score, all patients from the validation datasets were divided into two groups: the low-risk and high-risk groups. The R package ‘survival’ was performed on the two groups. The R package ‘timeROC’ was used to perform a receiver operating characteristic (ROC) analysis of the classification of this risk score. The univariate and the multivariate Cox proportional hazards regression were used to decipher the relevant clinic pathological variables. The model was represented in the form of a nomogram and the plotting of calibration curves by R language (version: 4.1.0).
2.5. Analysis of gene set enrichment
To identify the differences of the biological functions between the two groups, GSEA (version: 4.1.0, http://www.gsea-msigdb.org/gsea/index.jsp) and the package ‘GSVA Bioconductor’ by the enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway were performed.
2.6. Immune heterogeneity underlying the CTA signature
CIBERSORT analysis of our cohorts identifies relative scores of immune cell subpopulations. An analysis was performed on the two groups differences by performing the Wilcoxon matched-pairs test. P < 0.05 was selected as the threshold. Heatmap and violin plots were plotted with R packages ‘pheatmap’ and ‘vioplot,’ respectively. Calculation and visualization on the infiltrating immune cells and risk score were performed with the R package ‘corrplot’.
2.7. Analysis of protein-protein interaction
Construction and analysis of protein-protein interaction networks of the signature gene was carried out through the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (http://string-db.org).
2.8. Analysis of immunohistochemical
Protein expressions of 6 CTA signature genes by immunohistochemical staining were acquired from Human Protein Atlas (https://www.proteinatlas.org/).
2.9. Analysis of the correlation between drug susceptibility and the CTA signature
The correlation between the CTA signature and antitumor drug sensitivity/resistance was assessed with gene expression data and chemical compound activity (DTP NCI-60) data obtained from the CellMiner database. R packages including ‘impute’, ‘limma’, ‘ggplot2’, and ‘ggpubr’ were utilized to assess the association between gene expression levels and drug susceptibility. Notably, all the drugs used in the present research were subjected to approval by the US Food and Drug Administration.
3. Results
3.1. Establishment of a cancer-testis antigen risk model
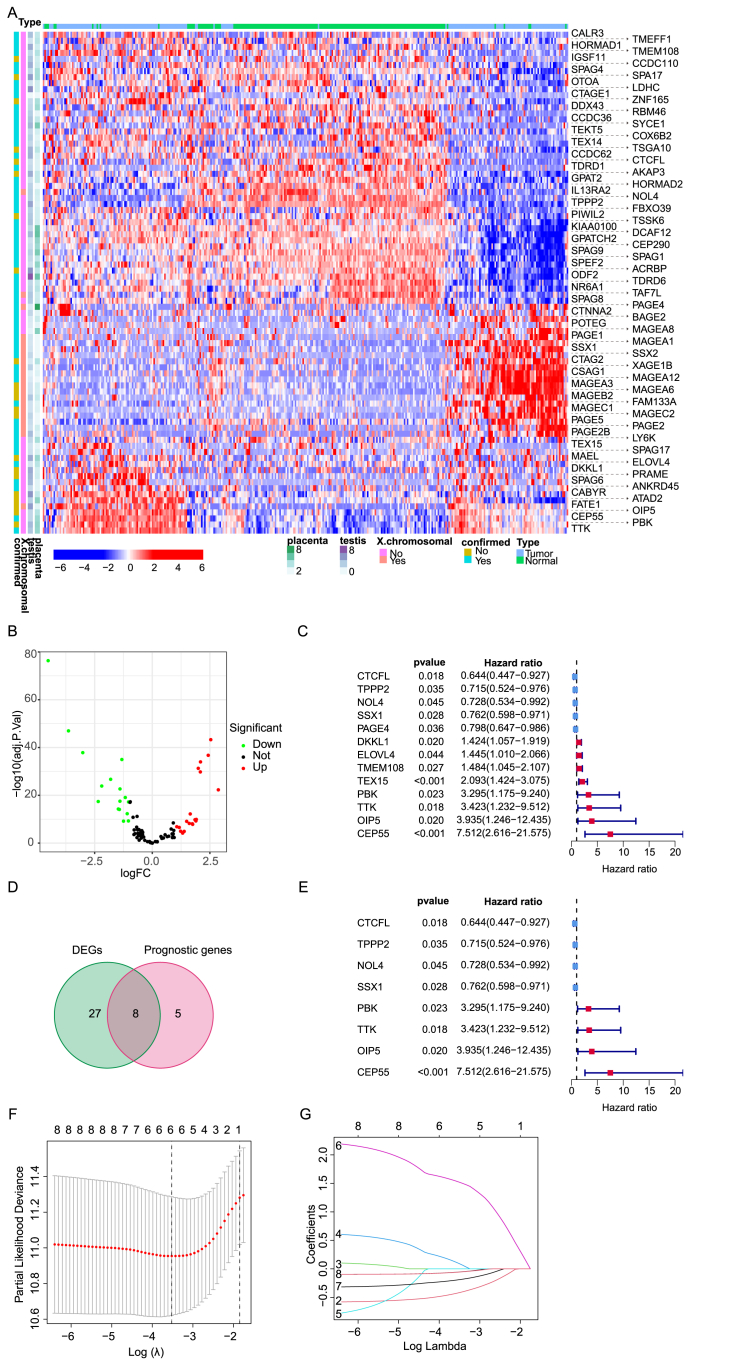
A total of 755 HCC patients (Table 1) were collected from three cohorts (159 patients in CHCC-HBV cohort as the training set, 365 patients in TCGA cohort and 231 patients in ICGC cohort as the validation data sets). We obtained 276 CTAs from CT database (http://www.cta.lncc.br), and 95 genes were excluded based on these criteria below: (1) not matched to the Ensembl database; (2) lack protein-coding transcripts; (3) not expressed in any normal or HCC samples; (4) were double annotated in CT database; (5) Considering expression in at least 60% of HCC patients in the CHCC-HBV cohort. Totally, 148 genes were filtered out from the database. Hierarchical cluster analysis demonstrated the pair-wise correlation between all 159 HCC cases based on mRNA expression of 57,820 putative protein-coding genes (Fig. 1A). A total of 16 and 19 CTAs were downregulated and upregulated in HCC, respectively (Fig. 1B). Then, Cox regression analysis revealed that 13 genes were associated with overall survival (OS) among the list of above 84 CTAs (P < 0.05). Eight genes had a high hazard ratio (HR > 1) and were defined as high risk factors, while five genes had a low hazard ratio (HR < 1) and were defined as protective factors (Fig. 1C). Following stepwise Cox proportional hazards regression analysis, eight CTA genes were chosen to construct the risk model (Fig. 1D).Ultimately, following LASSO logistic regression analysis six prognostic genes were selected including TPPP2, NOL4, OIP5, CEP55, CTCFL, and SSX1 (Fig. 1E–G; risk score = (−0.057 × expression level of TPPP2) + (−0.191 × expression level of NOL4) + (0.097 × expression level of OIP5) + (1.525 × expression level of CEP55) + (−0.419 × expression level of CTCFL) + (−0.055 × expression level of SSX1)).
Table 1.
Clinical characteristics of HCC patients by three cohorts
| CHCC-HBV cohort | TCGA cohort | ICGC cohort | |
|---|---|---|---|
| No. of patients | 159 | 365 | 231 |
| Age (median, range) | 54(20-81) | 61(16-90) | 69(31-89) |
| Gender (%) | |||
| Female | 31(19.5%) | 119(32.6%) | 61(26.4%) |
| Male | 128(80.5%) | 246(67.4%) | 170(72.6%) |
| AFP (ng/ml) | |||
| <200 | 89(56.0%) | NA | NA |
| >200 | 70(44.0%) | NA | NA |
| unknown | 0(0.0%) | NA | NA |
| Grade (%) | |||
| Grade 1 | NA | 55(15.1%) | NA |
| Grade 2 | NA | 175(47.9%) | NA |
| Grade 3 | NA | 118(32.3%) | NA |
| Grade 4 | NA | 12(3.3%) | NA |
| unknown | NA | 5(1.4%) | NA |
| Vascular Invasion | |||
| Yes | 122(76.7%) | NA | NA |
| No | 37(23.3%) | NA | NA |
| unknown | 0(0.0%) | NA | NA |
| TNM Stage (%) | |||
| I | 91(57.2%) | 170(46.6%) | 36(15.6%) |
| II | 14(8.8%) | 84(23.0%) | 105(45.5%) |
| III | 52(32.7%) | 83(22.7%) | 71(30.7%) |
| IV | 2(1.3%) | 4(1.1%) | 19(8.2%) |
| unknown | 0(0.0%) | 24(6.6%) | 0(0.0%) |
| Survival status | |||
| OS days (median) | 1020.9 | 556 | 780 |
| Censored (%) | 0(0.0%) | 126(34.5) | 42(18.2) |
TCGA, The Cancer Genome Atlas; ICGC, International Cancer Genome Consortium; AFP, Alpha Fetoprotein; TNM, Tumor Node Metastasis; NA, Not Applicable.
Figure.1.
The transcriptomic landscape of 83 CTA and the identification of CTA genes in prognostic model construction in the training cohort. (A) The expression of 83 CTAs after screening and filtering in each patient excluding 95 genes which were not matched to the Ensembl database, or lack protein-coding transcripts, or was not expressed in any normal or HCC samples, and the certain genes were double annotated in the CT database. (B) Volcano plot of differential expression of CTA between tumor and adjacent normal tissues of 159 HCC patients. (C) Forest plot of 13 CTAs related to prognosis selected by single factor Cox regression analysis. (D, E) VENN map and forest map of 8 prognostic-related differentially expressed CTAs. (F) The CvFit Line chart shows that the model with the smallest cross-validation error is composed of 6 CTAs. (G) Lambda chart of CTAs that might not be an independent indicator of prognostic monitoring.
The expression status of signature proteins in human HCC tumors and normal liver tissues by IHC staining were acquired from Human Protein Atlas (https://www.proteinatlas.org/). The proteins encoded by risk-related genes CEP55 and OIP5 were evidently highly expressed in HCC tissues (Fig. S1A). According to PPI network analysis, the signature genes were centered around CEP55 and OIP5 (Fig. S1B-C)..
3.2. Evaluation of six CTA genes-panel for predicting prognosis in HCC
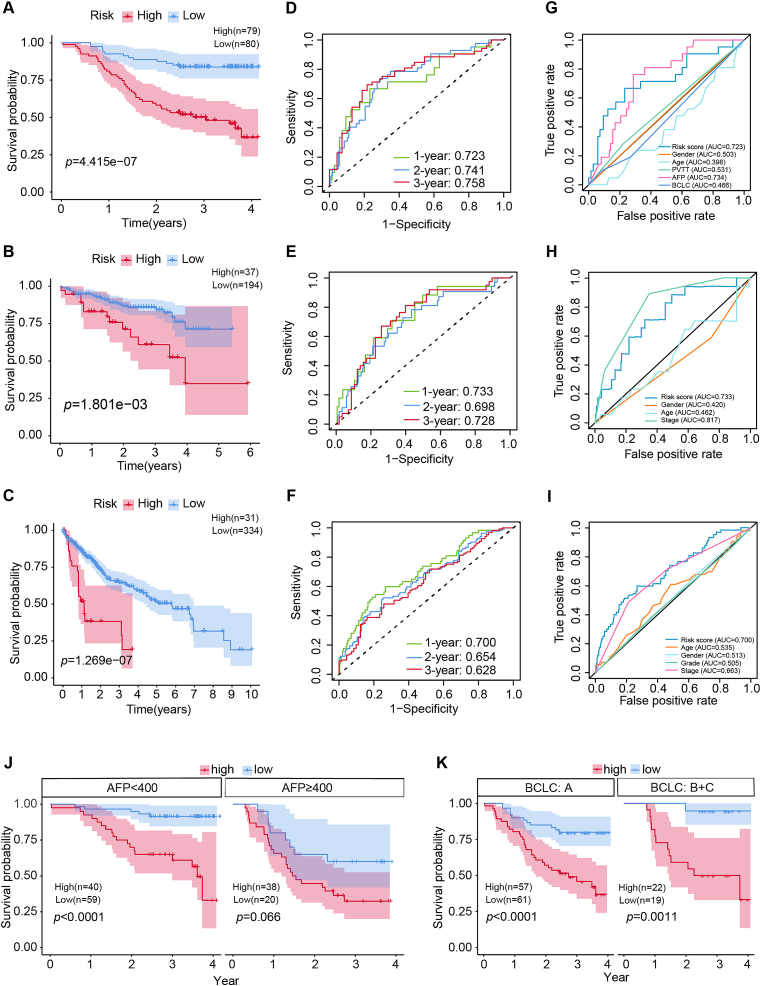
Patients were assigned to either high- or low-risk group using the median risk score as the cutoff value. The distribution of risk scores was plotted and higher risk scores were corresponded to greater mortality (Fig. S2A-F). PCA and t-SNE analysis were used to evaluate the efficiency of discrimination between the high-risk and low-risk groups, and the distribution of patients in the different risk groups extended in two directions, suggesting that the model could classify patients effectively (Fig. S2G-L). Survival analysis indicated that patients in the high-risk group exhibited worse OS in training and validating datasets (P < 0.05) (Fig. 2A–C). The ROC areas under the curve (AUC) were all greater than 0.6 in both the training data set as well as validation data sets for OS at one, two, and three years (Fig. 2D–F). The CTA gene signature was validated in the internal validation series in subgroups with AFP < 400 ng/mL, early-stage (BCLC stage A), and advanced stage (BCLC stages B and C) (P < 0.05). Moreover, the signature was substantially better than majority of clinicopathological parameters, with the AUC being 0.723, 0.733, and 0.700 for one-year OS in the CHCC-HBV, TCGA, and ICGC cohorts, respectively (Fig. 2G–I). Nevertheless, AUC was slightly lower than AFP (AUC = 0.734) and TNM stage (AUC = 0.817) in CHCC-HBV and TCGA cohorts, respectively. Moreover, the signature was of great importance to AFP false-negative (AFP < 20 ng/mL) HCC patients.
Figure.2.
The clinical significance and accuracy verification of CTAs in different cohorts and subgroups. Survival analysis of CTAs in HCC patients in training cohort (A), ICGC validation cohort (B), TCGA validation cohort (C), different AFP groups (J), and different BCLC stages (K). (D, E, F) Time-dependent ROC analysis of CTAs signature used to predict 1-, 2- and 3-year survival rates in the training, ICGC and TCGA cohort. (G, H, I) ROC curves of CTA signatures and other clinicopathological indicators in training, ICGC and TCGA cohorts.
3.3. Clinical value of six CTA genes-panel
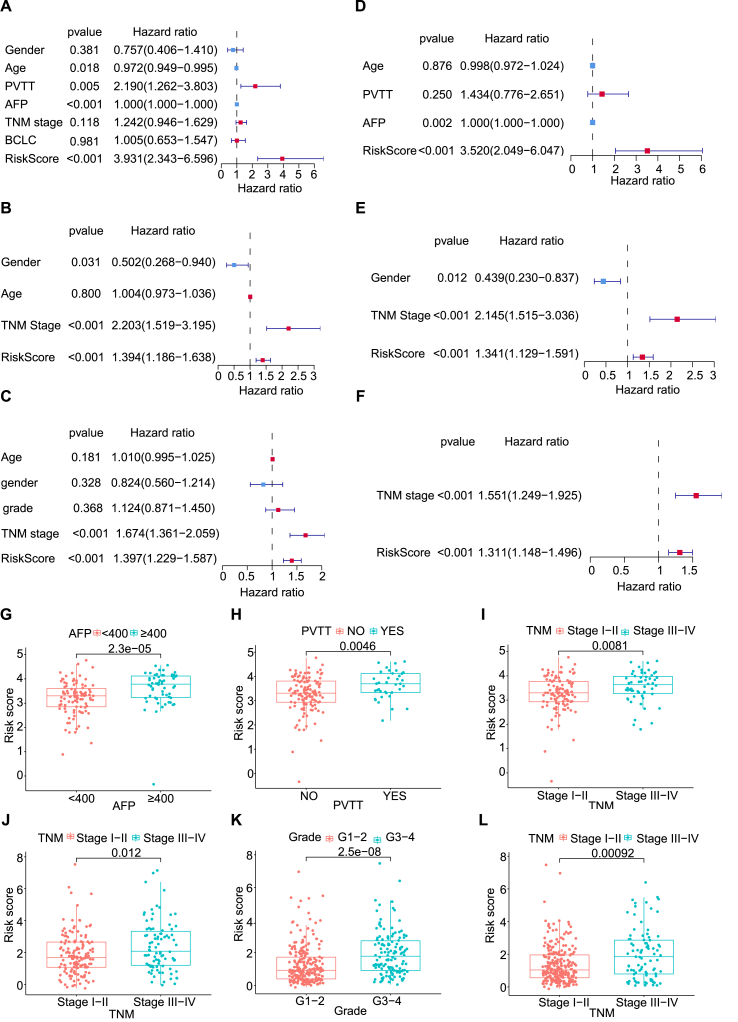
To assess the independence of the CTA signature in the three cohorts, univariate and multivariate Cox regression analyses were performed on the clinicopathological characteristics and the risk score. Potential confounders, such as TNM stage, Edmondson-Steiner grade, portal vein tumor thrombus (PVTT), AFP, BCLC stage, age, and sex, which were most likely to be related to clinical outcomes, were subjected to univariate and multivariate analyses. Age, PVTT, AFP, and risk score were statistically significant risk factors in the training set (P < 0.05) through the univariate analysis (Fig. 3A). Likewise, Sex, TNM stage, and risk score were risk factors in the ICGC validation data set (P < 0.05) (Fig. 3B), and TNM stage and risk score were risk factors in the TCGA validation data set (P < 0.05) (Fig. 3C). Following the multivariate Cox regression analysis, the risk score was an independent prognostic factor in three data sets (P < 0.05) (Fig. 3D–F). Our results showed that this CTA panel had superior predictive capacity over the conventional clinicopathological variables. In addition, an exploratory data analysis on the association of risk score and the clinicopathological features was performed to further evaluate the clinical significance of the six CTA panel. Different AFP levels, TNM stages, and PVTT were correlated with different stratification of risk scores in the training data set (Fig. 3G–I), so were TNM stages in the ICGC validation data set (Fig. 3J), and different grades and TNM stages in the TCGA validation data set (Fig. 3K-L). The above mentioned analysis indicated that the higher risk scores might be associated with the severe TNM stage and Edmondson-Steiner grade.
Figure.3.
Univariate and multivariable cox analysis of CTAs and the relationship between risk scores and clinical factors. (A, B, C) Forest plot of univariate cox analysis of CHCC-HBV, ICGC and TCGA cohort. (D, E, F) Forest plot of multivariate cox analysis of CHCC-HBV, ICGC and TCGA cohort. (G, H, I) Scatter plot between AFP, PVTT, TNM stage and risk score in the CHCC-HBV cohort. (J) Scatter plot between TNM stage and risk score of the ICGC cohort. (K, L) Scatter plot between grade, TNM stage and risk score of the TCGA cohort.
3.4. Construction a nomogram for predicting the prognosis of HCC
A nomogram was developed based on the training cohort which integrated the CTA gene signature, AFP to predict the OS of HCC patients (Fig. 4A). The calibration curves for three-year OS closely aligned with the diagonal, indicating good alignment between the observed and predicted possibility for the three-year OS (Fig. 4B and C). However, the accuracy of the model for predicting the OS at one year and two years was not same as at three years.
Figure.4.
The establishment of a noval nomogram based on CTAs. (A) Nomogram used to predict the 1-year, 2-year, and 3-year OS. (B, C, D) A nomogram calibration curve for the consistency between the predicted and observed 1-year, 2-year, and 3-year survival rates.
3.5. CTA related biological processes and pathway analysis
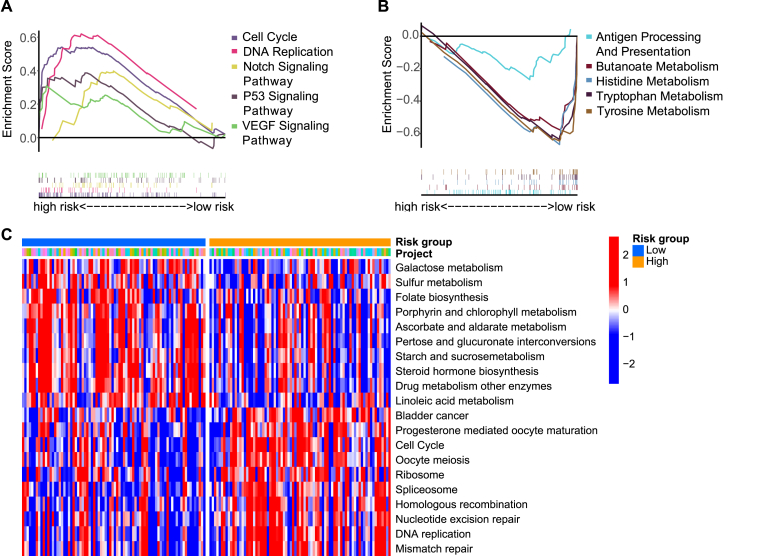
GSEA and GSVA were performed to explore the mechanism of potential molecular mechanisms in high- and low-risk groups in the training cohort. The KEGG pathways were enriched in Fig. 5A–C. As expected, GSEA indicated that cancer-related pathways including Notch pathway, P53 pathway, and VEGF pathway were subjected to an enrichment in the high-risk group. Both GSVA and GSEA pathway analyses indicated that in the high-risk group, cell cycle-related pathways were activated, whereas pathways that were metabolism-related were upregulated in low-risk group. An interesting finding was that antigen processing as well as the presentation pathway were stimulated in low-risk patients.
Figure.5.
Enrichment analysis of KEGG pathway based on CTAs signature. (A) Five representative upregulated KEGG pathways in the low-risk group by GSEA analysis. (B) Five representative down-regulated KEGG pathways in the low-risk group by GSEA analysis. (C) Ten representative upregulated and down-regulated KEGG pathways in two risk group by GSVA analysis
3.6. CTAs predicted immune infiltration and responses of immunotherapy
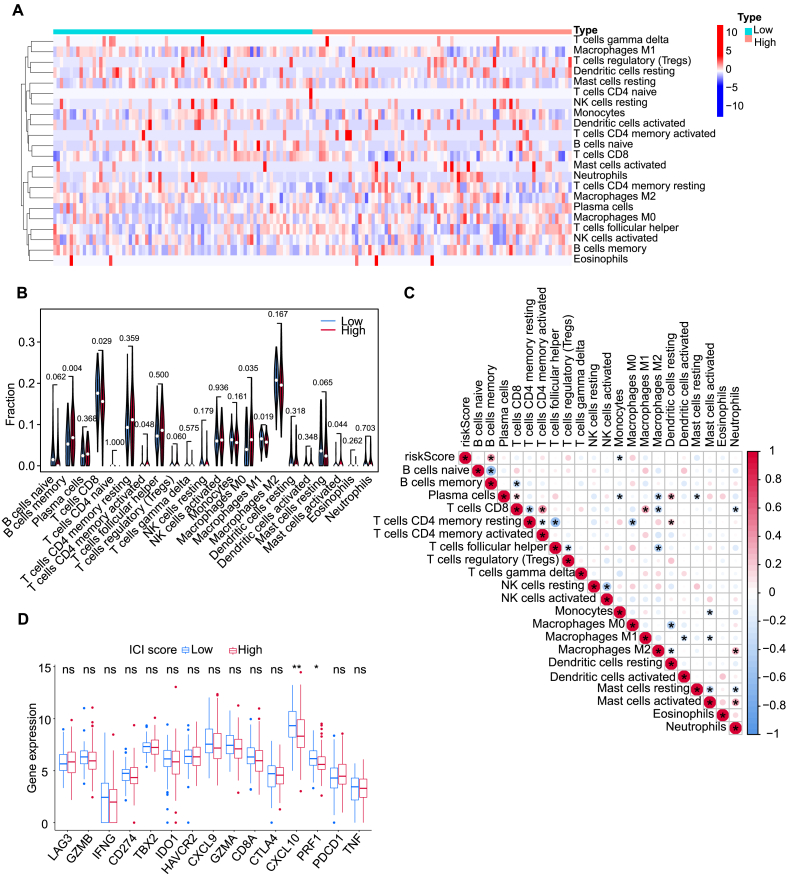
Despite a plethora of advances in treatment, most patients with HCC respond poorly to immunotherapy [12].Whether CTA expression in HCC is associated with immune infiltration is substantive but still ill-defined. Using CIBERSORT, a deconvolution method which deduces the immune cell subpopulation abundance from the data of bulk-tissue transcriptome, the immune cell subpopulations were identified. There were more activated memory CD4+ T cells, memory B cells, activated mast cells, and M0 macrophages infiltrated in the high-risk group, whereas more M1 macrophages and CD8+ T cells were infiltrated in the low-risk group (Fig. 6A-B). Increased expression of CXCL10 and PRF1 were detected in the low-risk group compared to those in the high-risk group (Fig. 6D). Accordingly, the immune response of the low-risk group might be significantly activated. The correlation between the infiltration degrees of 22 immune cells and the risk score was obtained (Fig. 6C). A positive correlation was found between memory B cells and risk score, while a negative correlation was found between macrophages and the risk score, indicating that the low-risk group had a stronger innate immune response. Taken together, these analyses underscore a potentially important role of the CTA signature in predicting immune response.
Figure.6.
Analysis of immune heterogeneity. (A) Distribution of immune cells in high-risk and low-risk groups. (B) Differences in tumor-infiltrating immune cells in different risk groups. (C) The Correlations between immune cells and risk score. (D) Differences in immune-related gene expression between low- and high-risk groups.
3.7. Drug sensitivity features underlying the CTA signature
Using the CellMiner dataset, we interpreted the correlation between cell sensitivity to anticancer drugs with the CTA signature genes in the model. Different expression of the signature genes had different sensitivity to antitumor drugs, with the top 16 drugs with the smallest p-values demonstrated (Fig. S3). The sensitivity of most antitumor drugs was negatively correlated with the expression of CTCFL and CEP55. Conversely, the sensitivity of most antitumor drugs was positively correlated with NOL4 and OIP5 expressions. It was worth noting that the expression of CTCFL had a positive correlation with resistance to adriamycin and epirubicin, frequently used in transcatheter arterial chemoembolization (TACE). It was illustrated that the expression of CTCFL might be helpful for choosing appropriate chemotherapy drugs for TACE in HCC patients.
4. Discussion
Tumor antigens are the optimal targets for cancer immunotherapy, especially CTAs encoded by genes expressed in tumors but not in most normal tissues [13]. Melanoma antigen 1 was the antigen firstly discovered by van der Bruggen through cytolytic T lymphocytes and DNA-cloning [14]. Subsequently, several tumor antigens, such as SSX, SCP1, and NY-ESO-1, was identified by means of screening the cDNA expression library. Due to the growing list of testis-related genes expressed in cancer, the term ‘CTA’ was named to cover all of these genes [15]. CTAs are classified into two types: CT-X antigens which are encoded in the X chromosomes and those not encoded in the X chromosomes (non-X CT antigens). The high expression of multiple CTAs, such as PRAME, MAGE family, NY-ESO-1, and CT45 family, accelerates tumor growth, epithelial-mesenchymal transition, drug resistance, metastasis, and recurrence [7,16]. The expression of the most CTA genes increases in cancers, and are positively correlated with the clinical stage and predicated poor prognosis of patients with tumors, indicating their potential role in tumor development, progression, and prognosis [17,18].
Our work reveals a transcriptome landscape of CTA genes expression in HCC and provides potential biomarkers for cancer prognostic prediction. We also developed a composite prognostic nomogram based on the six identified CTA genes signature and the clinical characteristics and could be considered as a potential prognostic prediction model. The six CTA genes panel comprising SSX1, CTCFL, OIP5, CEP55, NOL4, and TPPP2 might play an important role in HCC development, progression and metastasis. OIP5 promotes the metastasis and progression of HCC via the signaling pathways of mTORC1 and β-catenin [19]. CEP55, functioning as an oncogene, enhanced cell migration and invasion of HCC via upregulating JAK2-STAT3-MMPs signal pathway [20]. Identification of mRNA microarray expression profiling detect the high expression of CEP55 and CEP55 is a candidate prognostic indicator of tumor, which is consistent with our CTA gene signature [21,22]. The protein expression of OIP5 and CEP55 are more abundant in human HCC tissues than the normal liver tissues through the immunohistochemistry from public database. OIP5 and CEP55 are the critical nodes of signature interconnection.
Tumor cells commonly underwent the atavisms, such as increased expression of embryonic and CTA genes [23]. Based on the published researches, the overwhelming majority CTAs acting as the oncogenic genes facilitated the tumorigenicity and metastasis of tumors. However, some CTAs were regarded as the tumor suppressor genes, such as SSX1, CTCFL, NOL4, and TPPP2, the deeper molecular mechanism of regulating HCC remains elusive and need further exploration. This study showed the CTA signature was associated with the pathway activation, immune heterogeneity, protein expression, and drug response in HCC.
The antigen processing and presentation pathways are highly enriched in the low-risk group through functional enrichment analysis. Tumor cells could reduce immune recognition by targeted antigen processing and presentation to achieve immune escape. The defects within the antigen processing and presentation pathways could drive tumorigenesis and malignant transformation [24]. In tumor microenvironment, the activity of indoleamine 2, 3-dioxygenase (IDO) suppresses the immune response by decreasing the levels of tryptophan which is necessary for T cell proliferation, and meanwhile increasing the levels of kynurenine which promotes CD8+T-cell exhaustion [25]. A preclinical study illuminated that targeting IDO in rodent models inhibits tumor growth and elicits an antitumor immune response [26]. Histidine degradation affects the sensitivity of cancer cells to methotrexate, which may be exploited by histidine supplementation to increase methotrexate efficacy [27]. Concurrently, GSEA analysis reveals that the metabolism-related pathway is enriched in the low-risk group, suggesting intervention that exploit metabolism to improve tumor immunotherapy.
The CTA genes exert a crucial function in modulating tumor immunity. According to the published studies, the number of MAGE-A1- and MAGE-A3-specific T cells boosted following vaccination in melanoma patients [28,29]. In our study, more infiltration of memory B cells, M0 macrophages, activated memory CD4+ T cells, and activated mast cells were found in the high-risk group. In addition, CXCL10 and PRF1, the two immune activity-related genes, were higher in the low-risk group. We inferred that the immune response was activated to enhance the antitumor response in the low-risk group, thereby improving the clinical outcome of HCC patients. The innate immune cells also modulate the tumor progression [30]. Interestingly, we observed that the innate immune response negatively correlated with the risk score. We have performed analysis between CTA signature and immune cell infiltration, while the precise mechanism through which CTA genes regulates tumor microenvironment still remained poorly understood. Lastly, we observed that CTAs were correlated to doxorubicin and adriamycin resistance, indicating that our CTA signature might serve as a guide for choosing chemotherapy drugs.
In conclusion, this work delineates the landscape of CTA gene expression in HCC, several key predictions require further functional validation. Larger clinical samples are needed to verify the accuracy of the model. The six CTA genes-panel for HCC patients may be a reliable prognostic evaluation tool for clinical practice.
Declarations
Author contribution statement
Yingyu Xu: Performed the experiments; Analyzed and interpreted the data; Wrote the paper. Xu Xin, Xiaojian Ni: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data. Youpei Lin: Analyzed and interpreted the data. Jiaomeng Pan, MaoPei Chen: Contributed reagents, materials, analysis tools or data. Zhiying Zhao, Lan Zhang, Ningling Ge: Conceived and designed the experiments. Guohe Song, Juan Zhang: Conceived and designed the experiments; Analyzed and interpreted the data.
Funding statement
Juan Zhang and MaoPei Chen were supported by National Natural Science Foundation of China [81400768 & 81802360].Guohe Song was supported by Science and Technology Innovation Plan Of Shanghai Science and Technology Commission [22YF1407200].
Data availability statement
Data will be made available on request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e13269.
Contributor Information
Guohe Song, Email: docsong2013@163.com.
Juan Zhang, Email: zhang.juan@zs-hospital.sh.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. CA A Cancer J. Clin. 2022;72(1):7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Kulik L., El-Serag H.B. Epidemiology and management of hepatocellular carcinoma. Gastroenterology. 2019;156(2):477–491.e1. doi: 10.1053/j.gastro.2018.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chidambaranathan-Reghupaty S., Fisher P.B., Sarkar D. Hepatocellular carcinoma (HCC): epidemiology, etiology and molecular classification. Adv. Cancer Res. 2021;149:1–61. doi: 10.1016/bs.acr.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llovet J.M., Zucman-Rossi J., Pikarsky E., Sangro B., Schwartz M., Sherman M., Gores G. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2016;2 doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 5.Meng X., Sun X., Liu Z., He Y. A novel era of cancer/testis antigen in cancer immunotherapy. Int. Immunopharm. 2021;98 doi: 10.1016/j.intimp.2021.107889. [DOI] [PubMed] [Google Scholar]
- 6.Salmaninejad A., Zamani M.R., Pourvahedi M., Golchehre Z., Hosseini Bereshneh A., Rezaei N. Cancer/testis antigens: expression, regulation, tumor invasion, and use in immunotherapy of cancers. Immunol. Invest. 2016;45(7):619–640. doi: 10.1080/08820139.2016.1197241. [DOI] [PubMed] [Google Scholar]
- 7.Whitehurst A.W. Cause and consequence of cancer/testis antigen activation in cancer. Annu. Rev. Pharmacol. Toxicol. 2014;54:251–272. doi: 10.1146/annurev-pharmtox-011112-140326. [DOI] [PubMed] [Google Scholar]
- 8.Türeci O., Sahin U., Zwick C., Koslowski M., Seitz G., Pfreundschuh M. Identification of a meiosis-specific protein as a member of the class of cancer/testis antigens. Proc. Natl. Acad. Sci. U.S.A. 1998;95(9):5211–5216. doi: 10.1073/pnas.95.9.5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu Y., Huang C., Li Z., Zhao F., Zhou Y., Li J., Zhu C., Li Q., Zhuang Y., Xu J., Luo J., Chen L., Wang W. Expressions of melanoma-associated antigen A1 as a prognostic factor in Chinese patients with resectable oesophageal squamous cell carcinoma. Interact. Cardiovasc. Thorac. Surg. 2019;29(4):510–516. doi: 10.1093/icvts/ivz141. [DOI] [PubMed] [Google Scholar]
- 10.Cui Z., Chen Y., Hu M., Lin Y., Zhang S., Kong L., Chen Y. Diagnostic and prognostic value of the cancer-testis antigen lactate dehydrogenase C4 in breast cancer, Clinica chimica acta. international journal of clinical chemistry. 2020;503:203–209. doi: 10.1016/j.cca.2019.11.032. [DOI] [PubMed] [Google Scholar]
- 11.Gao Q., Zhu H., Dong L., Shi W., Chen R., Song Z., Huang C., Li J., Dong X., Zhou Y., Liu Q., Ma L., Wang X., Zhou J., Liu Y., Boja E., Robles A.I., Ma W., Wang P., Li Y., Ding L., Wen B., Zhang B., Rodriguez H., Gao D., Zhou H., Fan J. Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma. Cell. 2019;179(2):561–577.e22. doi: 10.1016/j.cell.2019.08.052. [DOI] [PubMed] [Google Scholar]
- 12.Sangro B., Sarobe P., Hervás-Stubbs S., Melero I. Advances in immunotherapy for hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2021;18(8):525–543. doi: 10.1038/s41575-021-00438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krishnadas D.K., Bai F., Lucas K.G. Cancer testis antigen and immunotherapy. ImmunoTargets Ther. 2013;2:11–19. doi: 10.2147/ITT.S35570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Bruggen P., Traversari C., Chomez P., Lurquin C., De Plaen E., Van den Eynde B., Knuth A., Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254(5038):1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y.T., Scanlan M.J., Sahin U., Türeci O., Gure A.O., Tsang S., Williamson B., Stockert E., Pfreundschuh M., Old L.J. A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc. Natl. Acad. Sci. U.S.A. 1997;94(5):1914–1918. doi: 10.1073/pnas.94.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang P., Meng M., Zhou Q. Oncogenic cancer/testis antigens are a hallmarker of cancer and a sensible target for cancer immunotherapy, Biochimica et biophysica acta. Reviews on cancer. 2021;1876(1) doi: 10.1016/j.bbcan.2021.188558. [DOI] [PubMed] [Google Scholar]
- 17.Sani S.A., Forghanifard M.M., Sharifi N., Bidokhti M.H., Bagherpoor A.J., Abbaszadegan M.R. Investigation of melanoma-associated antigen A4 cancer/testis antigen clinical relevance in esophageal squamous cell carcinoma. J. Cancer Res. Therapeut. 2018;14(5):1059–1064. doi: 10.4103/0973-1482.183180. [DOI] [PubMed] [Google Scholar]
- 18.Karia B.T.R., Zamuner F.T., Carlin V., de Oliveira C.Z., Carvalho A.L., Vettore A.L. Expression and prognostic relevance of GAGE1 and XAGE1 cancer/testis antigens in head and neck squamous cell carcinoma. Curr. Mol. Med. 2017;17(10):707–717. doi: 10.2174/1566524018666180322162145. [DOI] [PubMed] [Google Scholar]
- 19.Li H., Zhang J., Lee M.J., Yu G.R., Han X., Kim D.G. OIP5, a target of miR-15b-5p, regulates hepatocellular carcinoma growth and metastasis through the AKT/mTORC1 and β-catenin signaling pathways. Oncotarget. 2017;8(11):18129–18144. doi: 10.18632/oncotarget.15185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M., Gao J., Li D., Yin Y. CEP55 promotes cell motility via JAK2⁻STAT3⁻MMPs cascade in hepatocellular carcinoma. Cells. 2018;7(8) doi: 10.3390/cells7080099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones J., Otu H., Spentzos D., Kolia S., Inan M., Beecken W.D., Fellbaum C., Gu X., Joseph M., Pantuck A.J., Jonas D., Libermann T.A. Gene signatures of progression and metastasis in renal cell cancer, Clinical cancer research : an. official journal of the American Association for Cancer Research. 2005;11(16):5730–5739. doi: 10.1158/1078-0432.CCR-04-2225. [DOI] [PubMed] [Google Scholar]
- 22.Xiang X.H., Yang L., Zhang X., Ma X.H., Miao R.C., Gu J.X., Fu Y.N., Yao Q., Zhang J.Y., Liu C., Lin T., Qu K. Seven-senescence-associated gene signature predicts overall survival for Asian patients with hepatocellular carcinoma. World J. Gastroenterol. 2019;25(14):1715–1728. doi: 10.3748/wjg.v25.i14.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas F., Ujvari B., Renaud F., Vincent M. Cancer adaptations: atavism, de novo selection, or something in between? Bioessays. 2017;39(8) doi: 10.1002/bies.201700039. [DOI] [PubMed] [Google Scholar]
- 24.Reeves E., James E. Antigen processing and immune regulation in the response to tumours. Immunology. 2017;150(1):16–24. doi: 10.1111/imm.12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takenaka M.C., Gabriely G., Rothhammer V., Mascanfroni I.D., Wheeler M.A., Chao C.C., Gutiérrez-Vázquez C., Kenison J., Tjon E.C., Barroso A., Vandeventer T., de Lima K.A., Rothweiler S., Mayo L., Ghannam S., Zandee S., Healy L., Sherr D., Farez M.F., Prat A., Antel J., Reardon D.A., Zhang H., Robson S.C., Getz G., Weiner H.L., Quintana F.J. Control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39. Nat. Neurosci. 2019;22(5):729–740. doi: 10.1038/s41593-019-0370-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Naour J., Galluzzi L., Zitvogel L., Kroemer G., Vacchelli E. Trial watch: Ido inhibitors in cancer therapy. OncoImmunology. 2020;9(1) doi: 10.1080/2162402X.2020.1777625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanarek N., Keys H.R., Cantor J.R., Lewis C.A., Chan S.H., Kunchok T., Abu-Remaileh M., Freinkman E., Schweitzer L.D., Sabatini D.M. Histidine catabolism is a major determinant of methotrexate sensitivity. Nature. 2018;559(7715):632–636. doi: 10.1038/s41586-018-0316-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carrasco J., Van Pel A., Neyns B., Lethé B., Brasseur F., Renkvist N., van der Bruggen P., van Baren N., Paulus R., Thielemans K., Boon T., Godelaine D. Vaccination of a melanoma patient with mature dendritic cells pulsed with MAGE-3 peptides triggers the activity of nonvaccine anti-tumor cells. J. Immunol. 2008;180(5):3585–3593. doi: 10.4049/jimmunol.180.5.3585. [DOI] [PubMed] [Google Scholar]
- 29.Chianese-Bullock K.A., Pressley J., Garbee C., Hibbitts S., Murphy C., Yamshchikov G., Petroni G.R., Bissonette E.A., Neese P.Y., Grosh W.W., Merrill P., Fink R., Woodson E.M., Wiernasz C.J., Patterson J.W., Slingluff C.L., Jr. MAGE-A1-, MAGE-A10-, and gp100-derived peptides are immunogenic when combined with granulocyte-macrophage colony-stimulating factor and montanide ISA-51 adjuvant and administered as part of a multipeptide vaccine for melanoma. J. Immunol. 2005;174(5):3080–3086. doi: 10.4049/jimmunol.174.5.3080. [DOI] [PubMed] [Google Scholar]
- 30.Vesely M.D., Kershaw M.H., Schreiber R.D., Smyth M.J. Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 2011;29:235–271. doi: 10.1146/annurev-immunol-031210-101324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.