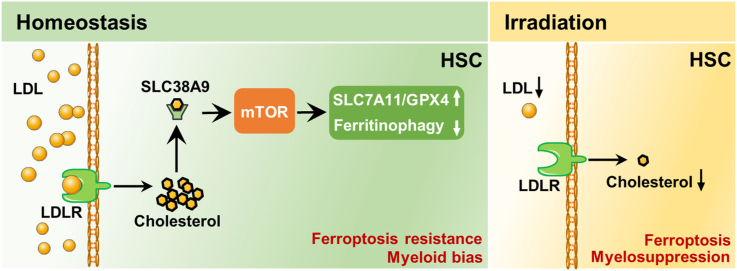
Abstract
There is growing appreciation that hematopoietic alterations underpin the ubiquitous detrimental effects of metabolic disorders. The susceptibility of bone marrow (BM) hematopoiesis to perturbations of cholesterol metabolism is well documented, while the underlying cellular and molecular mechanisms remain poorly understood. Here we reveal a distinct and heterogeneous cholesterol metabolic signature within BM hematopoietic stem cells (HSCs). We further show that cholesterol directly regulates maintenance and lineage differentiation of long-term HSCs (LT-HSCs), with high levels of intracellular cholesterol favoring maintenance and myeloid bias of LT-HSCs. During irradiation-induced myelosuppression, cholesterol also safeguards LT-HSC maintenance and myeloid regeneration. Mechanistically, we unravel that cholesterol directly and distinctively enhances ferroptosis resistance and boosts myeloid but dampens lymphoid lineage differentiation of LT-HSCs. Molecularly, we identify that SLC38A9–mTOR axis mediates cholesterol sensing and signal transduction to instruct lineage differentiation of LT-HSCs as well as to dictate ferroptosis sensitivity of LT-HSCs through orchestrating SLC7A11/GPX4 expression and ferritinophagy. Consequently, myeloid-biased HSCs are endowed with a survival advantage under both hypercholesterolemia and irradiation conditions. Importantly, a mTOR inhibitor rapamycin and a ferroptosis inducer imidazole ketone erastin prevent excess cholesterol-induced HSC expansion and myeloid bias. These findings unveil an unrecognized fundamental role of cholesterol metabolism in HSC survival and fate decisions with valuable clinical implications.
Keywords: Hematopoietic stem cell, Cholesterol, Ferroptosis, Myeloid bias, Ionizing radiation, Myelosuppression
Graphical abstract
Highlights
-
•
HSCs exhibit a distinct and heterogeneous cholesterol metabolic signature.
-
•
Cholesterol directly enhances ferroptosis resistance and myeloid bias of HSCs.
-
•
SLC38A9–mTOR axis mediates cholesterol sensing and signal transduction in HSCs.
-
•
mTOR upregulates SLC7A11/GPX4 and inhibits ferritinophagy to regulate ferroptosis sensitivity of HSCs.
Abbreviations
- BM
bone marrow
- FACS
fluorescence-activated cell sorting
- GPX4
glutathione peroxidase-4
- GSH
reduced glutathione
- GSSG
oxidized glutathione
- HCD
high-cholesterol diet;
- HSC
hematopoietic stem cell
- HSPC
hematopoietic stem and progenitor cell
- IKE
imidazole ketone erastin
- IR
ionizing radiation
- LDL
low-density lipoprotein
- LDL-C
LDL-cholesterol
- LDLR
LDL receptor
- LT-HSC
long-term
- HSC
MCD, methyl-beta-cyclodextrin
- mTOR
mechanistic target of rapamycin
- PB
peripheral blood
- SLC38A9
solute carrier 38, family A member 9
- SLC7A11
solute carrier family 7, family A member 11
1. Introduction
Cholesterol is a unique lipid that plays an essential role in membrane biogenesis. In mammals, cholesterol is principally obtained from diet and de novo biosynthesis by the liver and is distributed throughout the body via low-density lipoprotein (LDL) and high-density lipoprotein [1]. Nowadays, due to overnutrition and sedentary lifestyle of modern humans, serum cholesterol levels trend to rise at a young age. Unfortunately, high levels of cholesterol are associated with increased risks of multiple diseases such as cardiovascular disease, immune disorder, and cancer [2]. Besides, there is a substantial unmet clinical need for prophylactic and therapeutic strategies for these diseases.
Most recently, increasing studies have spotlighted on hematopoietic alterations as an initial event of deregulation of systemic organs especially during aging [[3], [4], [5]]. Interestingly, bone marrow (BM) hematopoiesis is particularly vulnerable to perturbations of cholesterol metabolism. Cholesterol metabolism is tightly regulated by the concerted regulation of cholesterol biosynthesis, uptake, export, and esterification [6]. However, excess cholesterol such as that results from hypercholesterolemia or defective cholesterol export always accompanies hematopoietic stem and progenitor cell (HSPC) expansion and myeloid-biased hematopoiesis that is manifested by overrepresentation of myeloid cells (neutrophils/monocytes) in the peripheral blood (PB) and BM [[7], [8], [9]]. Moreover, myeloid-biased hematopoiesis always underlies the pathogenesis of innate immune inflammation and adaptive immunodeficiency, which render people susceptible to cardiovascular disease, immune disorder, and cancer [10,11]. These lines of evidence infer that targeting BM hematopoiesis holds promise in preventing the ubiquitous detrimental effects of excess cholesterol on the body. Nevertheless, there remains gap of knowledge regarding the impact and mechanism of excess cholesterol on BM hematopoiesis.
Life-long hematopoiesis is maintained by a rare group of hematopoietic stem cells (HSCs) residing within the BM. At steady state, the pool size of HSCs is stably maintained and the lineage differentiation of HSCs is balanced [10]. Recently, accumulating studies including our own have revealed the distinct regulatory roles of nutrient metabolism, such as glucose [12], amino acid [[12], [13], [14], [15]], calcium [16], and phosphate metabolism [17,18], in HSC pool size and fate decisions. However, the intrinsic properties of cholesterol metabolism in HSCs are nearly undefined. Meanwhile, although it is well documented that excess cholesterol influences HSC maintenance [[7], [8], [9]], the cellular and molecular mechanisms by which HSCs adapt to perturbations of cholesterol metabolism remain poorly understood.
Ferroptosis is a unique modality of cell death driven by iron-dependent lipid peroxidation that is provoked by redox imbalance during diverse pathophysiological stresses such as ionizing radiation (IR), inflammation, and aging [19,20]. Ferroptosis is regulated by various signaling pathways and by multiple cellular metabolic pathways, including iron metabolism, energy metabolism and metabolism of amino acids, lipids and sugars [21,22]. Recent studies have suggested a close link between cholesterol metabolism and ferroptosis sensitivity [[23], [24], [25], [26]], while the direct regulatory role of cholesterol in ferroptosis remain elusive. In addition, due to the metabolic specificality, HSCs are particularly vulnerable to redox imbalance [27]. However, the pathophysiological significance of ferroptosis in HSC biology and its interplay with cholesterol metabolism remain largely unknown.
In this study, we discover that long-term HSCs (LT-HSCs) are maintained at a state of cholesterol deficiency among HSPCs at homeostasis, whereas myeloid-biased HSCs have relatively higher levels of intracellular cholesterol. Using mouse models of hypercholesteremia and irradiation-induced myelosuppression, we show that cholesterol favors maintenance and myeloid bias of LT-HSCs. Mechanistically, cholesterol signal is sensed and transduced by the solute carrier 38, family A member 9 (SLC38A9)– mechanistic target of rapamycin (mTOR) axis to instruct lineage differentiation and ferroptosis sensitivity of HSCs. Overall, these findings unmask a distinct and heterogeneous cholesterol metabolic signature of HSCs as well as a pivotal regulatory role of cholesterol metabolism in HSC survival and fate decisions.
2. Materials and methods
2.1. Animals
Normal C57BL/6 mice were purchased from the Institute of Zoology (Chinese Academy of Sciences, Beijing, China). B6.129P2-apoEtm1Unc/J (ApoE−/−; C57BL/6 background) and B6; 129S7-Ldlrtm1Her/J (Ldlr−/−; C57BL/6 background) mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). GFP-LC3 mice were kindly gifted by Dengqun Liu (University of Electronic Science and Technology of China). To induce hypercholesterolemia, normal C57BL/6 mice were fed a high-cholesterol diet (HCD) (D12108C, Research Diets, New Brunswick, NJ, USA) for 3 months. For rapamycin and imidazole ketone erastin (IKE) administration, mice were treated with a dose of 4 mg/kg rapamycin (MedChem Express, Monmouth Junction, NJ, USA) or 40 mg/kg imidazole ketone erastin (IKE, MedChem Express) body weight by intraperitoneal injection every other day for 1 month from the third month of HCD treatment. Mice were fed with autoclaved food and housed in specific pathogen-free conditions. All mice used were male, background-matched and age-matched (8–10 weeks of age). All animal procedures were performed in accordance with National Institutes of Health guidelines [28] and were approved by the Animal Care Committee of the Army Medical University (AMUWE2019092).
2.2. Irradiation
To induce sublethal or lethal myelosuppression, mice were subjected to a total body irradiation of 5 Gy or 7.5 Gy respectively by using a60Co γ-ray source (Irradiation Center, Army Medical University, Chongqing, China). The dose rate was 92.8–95.5 cGy/min. For HCD treatment, mice were adapted to HCD for one week before irradiation and then subjected to total body irradiation, followed by one week of HCD post irradiation.
2.3. Flow cytometry and cell sorting
Mice were anesthetized by intraperitoneal injection with sodium pentobarbital (50 mg/kg) and sacrificed by cervical dislocation, and femur and tibia were isolated. Bone marrow cells (BMCs) were flushed from the femur and tibia, followed by RBC lysis using a red cell lysis buffer (StemCell Technologies, Vancouver, BC, Canada). For mouse hematopoietic cell phenotypic analysis, a lineage cocktail was used, including CD3, Mac-1, Gr-1, B220 and Ter-119 (all eBioscience, San Diego, CA, USA). Mouse long-term HSCs (LT-HSCs; Lineage−c-Kit+Sca1+CD34−CD135−), short-term HSCs (ST-HSCs; Lineage−c-Kit+Sca1+CD34+CD135−), multipotent progenitors (MPPs; Lineage−c-Kit+Sca1+CD34+CD135+), LT-HSC subsets (Fraction 2/Fr2; CD41+CD150+ LT-HSC), Fraction1 (Fr1; CD41−CD150+ LT-HSC), and Fraction3 (Fr3; CD41−CD150− LT-HSC)], common myeloid progenitors (CMPs; Lineage−c-Kit+Sca1−CD127−CD16/32−CD34+), common lymphoid progenitors (CLPs; Lineage−CD127+Sca1medc-Kit+), granulocyte-monocyte progenitors (GMPs; Lineage−CD127−Sca1−c-Kit+CD16/32hiCD34+), megakaryocyte-erythroid progenitors (MEPs; Lineage−CD127−Sca1−c-Kit+CD16/32−CD34−), myeloid cell (CD45+Mac-1+Gr-1+), B cell (CD45+B220+), and T cell (CD45+CD3+) were analyzed using monoclonal antibodies as indicated. For surface LDL receptor (LDLR) expression analysis, BMCs were stained with HSC markers and anti-LDLR (eBioscience) for 30 min at 4 °C. For protein expression detection, BMCs were stained with HSC markers and carefully washed. Cells were firstly fixed with IC Fixation buffer (eBioscience) at room temperature for 20 min and then permeabilized with Permeabilization buffer (eBioscience) in the presence of anti-Transferrin Receptor (TfR1), anti-p-AKTSer473, anti-p-mTORSer2448 (all eBioscience), anti-ATF4, anti-SLC7A11, anti-ACSL4, anti-ALOX15, anti-SREBP1 (all Thermo Fisher Scientific, Carlsbad, CA, USA), anti-GPX4, anti-SCD1, anti-LPCAT3 (all Abcam, Cambridge, UK) at room temperature for another 30 min. If necessary, cells were stained with fluorescent dye conjugated secondary antibodies (Thermo Fisher Scientific) and finally analyzed by flow cytometry. LT-HSCs were sorted using a FACSAriaⅡ (BD Biosciences, San Jose, CA, USA) and analyzed using a FACSVerse (BD Biosciences) or a Sony ID7000™ Spectral Cell Analyzer (Sony Corporation, Tokyo, Japan). Data analysis was conducted using FlowJo software (Treestar Inc, San Carlos, CA, USA). The detailed information of used antibodies was listed in Table S1.
2.4. HSC culture
Mouse HSCs were cultured as reported [29]. For analysis LT-HSC differentiation and cellular expansion, cell cultures were initiated with 50 fluorescence-activated cell sorting (FACS)-sorted LT-HSCs in 96-well plates with Ham’s F-12 Nutrient Mix liquid medium (Gibco, Grand Island, NY, USA) as the basal media, supplemented with 10 ng/mL recombinant mouse SCF (PeproTech, Rocky Hill, NJ, USA), 100 ng/mL recombinant mouse TPO (PeproTech), penicillin-streptomycin-glutamine (PSG; 100 × ; Gibco), insulin-transferrin-selenium-ethanolamine (ITSX; 100 × ; Gibco), 1 mg/mL polyvinyl alcohol (PVA; 87–90%, hydrolyzed; Sigma-Aldrich, St. Louis, MO, USA), 10 mM HEPES (Gibco). Cell numbers per well were counted every other day using a hematocytometer (Countess™ II FL; Thermo Fisher Scientific) and dead cells were excluded by trypan blue. For ferroptosis resistance analysis, 5000 FACS-sorted LT-HSCs were cultured in 24-well plates for 48 h in the presence of 5 μM erastin (MedChem Express), 250 nM RSL3 (MedChem Express) or 20 μM dihydroartemisinin (DHA). For LDL and/or methyl-beta-cyclodextrin (MCD) treatment, LT-HSC cultures were respectively supplemented with 50 μg/mL LDL (Yiyuan biotechnology, Guangzhou, China) and/or 0.5% MCD (Sigma-Aldrich). The LDL (>98% purity, as determined by agarose gel electrophoresis) used in our study was dissolved in PBS and PBS was used as vehicle control for LDL. For free cholesterol treatment, precomplexed MCD and cholesterol at a 1:1 M ratio (50 μM) was used. For oxidized LDL (oxLDL) treatment, LT-HSC cultures were supplemented with 20 μg/mL oxLDL (Solarbio, Beijing, China).
2.5. Serum LDL-cholesterol (LDL-C) measurement
Blood samples from mice were obtained from cardiac puncture of mice with an EDTA-coated syringe and centrifuged at 3000 rpm (4 °C) for 10 min. Following centrifugation, serum LDL-C levels were assessed using an LDL Cholesterol Mouse Assay Kit (Crystal Chem, Elk Grove Village, IL, USA) according to the manufacturer’s instructions.
2.6. Intracellular GSH and GSSG measurement
Freshly FACS-sorted LT-HSCs were rinsed with PBS and collected with centrifugation. Intracellular glutathione (GSH) and oxidized glutathione (GSSG) contents in LT-HSCs were measured by a GSH/GSSG Ratio Detection Assay Kit (Abcam) according to the manufacturer’s instructions. The GSH and GSSG concentrations were evaluated by a standard curve and normalized to the total protein level in each sample.
2.7. Intracellular total iron, ATP, NADP+ and NADPH measurement
Freshly FACS-sorted LT-HSCs were rinsed with PBS and collected with centrifugation. Intracellular total iron, ATP, NADP+ and NADPH contents in LT-HSCs were respectively measured by an Iron Assay Kit (Abcam), a Luminescent ATP Detection Assay Kit (Abcam) or a NADP+/NADPH Assay Kit (Abnova, Taiwan) according to the manufacturer’s instructions. Data were normalized to the total protein level in each sample.
2.8. Hematological parameter test
Hematological parameter test of mice was conducted as we previously reported [30]. In brief, 30 μL PB were collected from the tail veins of mice and diluted in 1% EDTA solution, finally counted automatically by a Sysmex XT-2000i hematology analyzer (Sysmex Corporation, Kobe, Japan).
2.9. Colony-formation assay
FACS-sorted LT-HSCs (50 cells/1 mL in a 3 cm dish) were plated and grown in methylcellulose media (M3434; StemCell Technologies) containing 50 μg/mL LDL or vehicle and cultured at 37 °C. Colony numbers were determined at day 14.
2.10. Intracellular cholesterol measurement
To measure intracellular cholesterol contents in LT-HSCs by flow cytometry, BMCs were isolated, stained with LT-HSC markers, and fixed with IC Fixation buffer (eBioscience) at room temperature for 30 min. Then, cells were permeabilized with Permeabilization buffer (eBioscience) in the presence of 0.05 mg/mL Filipin III (Sigma-Aldrich) at room temperature for another 120 min and finally analyzed by flow cytometry. Intracellular cholesterol contents were also verified using an Amplex™ Red Cholesterol Assay Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Data were normalized to the total DNA level in each sample.
2.11. Ferroptosis assay
For ferroptosis analysis, BMCs were firstly stained with HSC markers and carefully washed. Cell death analysis was performed by suspending cells in a 7-amino-actinomycin D (7-AAD) staining solution (eBioscience) and incubating for 15 min at room temperature. Lipid peroxidation was measured by suspending cells in prewarmed (37 °C) PBS with 10 μM Liperfluo (Dojindo Molecular Technologies, Kumamoto, Japan) and incubating for 30 min at 37 °C. Ferrous ion deposition was measured by suspending cells in prewarmed (37 °C) PBS with 2 μM FerroOrange (Dojindo Molecular Technologies) and incubating for 30 min at 37 °C. For detection of 4-hydroxynonenal (4-HNE) and malondialdehyde (MDA), cells were fixed with IC Fixation buffer (eBioscience) at room temperature for 30 min. Then, cells were permeabilized with Permeabilization buffer (eBioscience) in the presence of anti-4-HNE or anti-MDA antibodies (all Abcam) at room temperature for 45 min and stained with fluorescent dye conjugated secondary antibodies (Thermo Fisher Scientific) at room temperature for another 30 min and finally analyzed by flow cytometry. 4-HNE and MDA levels LT-HSCs were also respectively determined using 4-HNE and MDA ELISA Kits (all Elabscience, Texas, USA) according to the manufacturers’ protocols.
2.12. Ferritinophagy assay
For ferritinophagy analysis, BMCs of GFP-LC3 mice were stained with HSC markers and followed by flow cytometric detection of GFP-LC3. For ferritin detection, cells were permeabilized with Permeabilization buffer (eBioscience) in the presence of anti-Ferritin (Abcam) antibody at room temperature for 45 min and stained with fluorescent dye conjugated secondary antibodies (Thermo Fisher Scientific) at room temperature for another 30 min and finally analyzed by flow cytometry.
2.13. Transplantation studies
For competitive BM transplantation assay, CD45.1+CD45.2+ recipient mice were lethally irradiated with a split dose totaling 10.0 Gy by using a60Co γ-ray source (92.8–95.5 cGy/min). Then, 5 × 105 BM cells from CD and HCD mice (CD45.2+), together with 5 × 105 competitor BM cells (CD45.1+) were transplanted into lethally irradiated recipient mice. PB was collected from the tail veins of mice and multi-lineage reconstitution for myeloid cells, B cells, T cells were analyzed 16 weeks after transplantation using flow cytometry.
2.14. Lentiviral transduction
Lentiviral transduction of HSCs was performed as we previously reported [18]. Briefly, the shRNA against SLC38A9 was cloned into pLKO.1 vector. Then the recombinant plasmids were transfected into HEK-293T cells together with psPAX2 and pMD2.G plasmids to generate lentiviral particles. For lentivirus infections, FACS-sorted LT-HSCs (2 × 104) were seeded in 24 well culture plates to be infected with recombinant lentivirus-transducing units in the presence of 8 μg/mL polybrene (Sigma-Aldrich). Infected LT-HSCs were purified with GFP expression through FACS sorting.
2.15. Immunofluorescence
Freshly FACS-sorted LT-HSCs were stained with 5 μM BODIPY 581/591C11 (37 °C, 30 min) or 2 μM FerroOrange (37 °C, 30 min) and placed onto poly-l-lysine coating slides with 10 μL HBSS (Gibco). Then, cells were photographed under a Zeiss LSM800 NLO confocal microscope (Carl Zeiss, Jena, Germany) as soon as possible.
2.16. Quantitative RT-PCR (qRT-PCR)
RNA was extracted from FASC-sorted HSPCs using a RNeasy® Micro Kit (QIAGEN, Hilden, Germany) and was reverse-transcribed into cDNA using a PrimeScript™ RT reagent Kit (TaKaRa, Shiga, Japan) according to the manufacturer’s instructions. The mRNA expression levels of indicated genes were measured by a CFX96™ Real-Time system (BioRad, Hercules, CA, USA) with a GoTaq® qPCR Master Mix (Promega, Madison, WI, USA). The primers were listed in Table S2. Data was normalized relative to HPRT.
2.17. RNA-seq
The concentration and integrity of extracted RNA were evaluated using an Agilent 2100 Bioanalyzer using an Agilent RNA 6000 Nano Kit. Library construction and RNA sequencing were performed at the Beijing Genomics Institute (BGI, Shenzhen, China). An Illumina HiSeq 2500 was used to sequence the produced library. SOAPnuke (version: v1.5.2, https://github.com/BGI-flexlab/SOAPnuke) was used to filter all RNA-seq raw readings to get clean reads. Following sequence alignment, gene expression was assessed by RSEM (version: v1.2.12, http://deweylab.biostat.wisc.edu/RSEM) and differential gene expression was evaluated using DEseq2. Gene set enrichment analysis (GSEA, Broad Institute) was performed using GSEA version 4.0.3 (http://www.broadinstitute.org/gsea). Ingenuity Pathway Analysis (IPA, Ingenuity Systems; QIAGEN; https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/) was used to analyze alterations of molecular and cellular functions.
2.18. Data and code availability
All data are available in the main text or the supplementary materials. Transcriptome datasets generated or analyzed in this study are available at Gene Expression Omnibus (GSE214035; RNA-seq) and ArrayExpress (E-MTAB-3079; RNA-seq).
2.19. Statistical analysis
For experimental data, we used Prism 9.0 (GraphPad Software, La Jolla, CA, USA) software to analyze data from biological experiments. All results are expressed as mean ± standard deviation (SD). n represents the number of independent experiments, as described in figure legends. Normal distribution was determined with Shapiro-Wilk test according to the sample size. Comparisons between two groups were determined by two-tailed paired or unpaired Student’s t-test. Three or more groups were compared by one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. For survival analysis, Kaplan-Meier curves and Log-rank test were used. p < 0.05 was considered statistically significant.
3. Results
3.1. HSC displays a distinct cholesterol metabolic signature
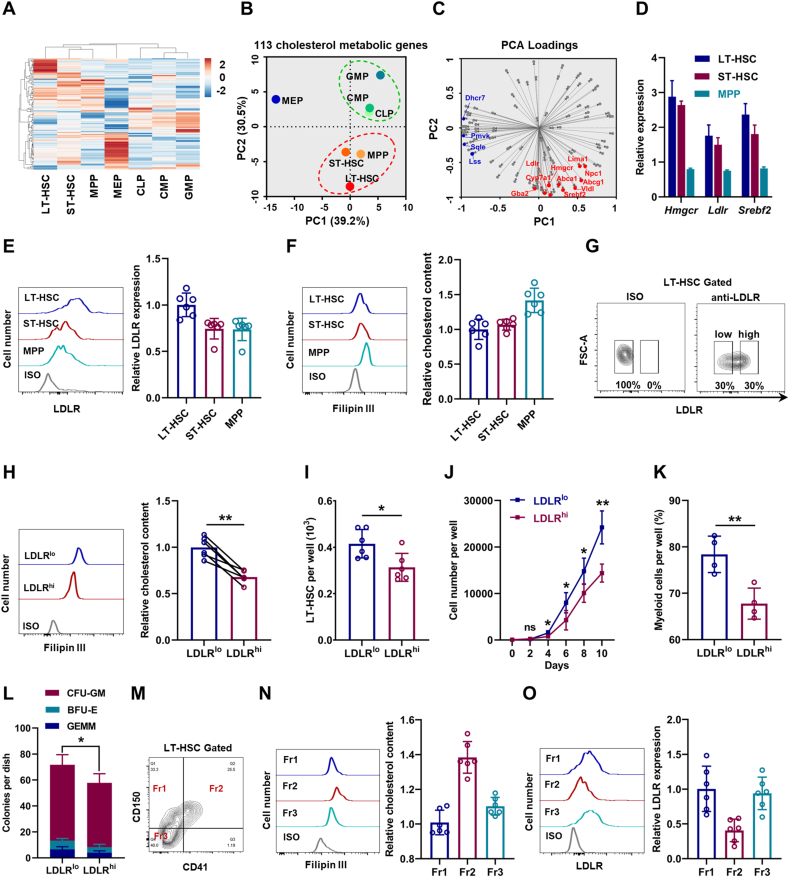
Initially, we determined the cholesterol metabolic landscape based on a published transcriptome database of murine HSPCs [31]. Interestingly, the transcriptome data revealed a great heterogeneity of cholesterol metabolism among HSPCs (Fig. 1A and Table S3). Principal component analysis (PCA) confirmed the heterogeneity and that HSPC population displayed three discrete clusters based on cholesterol metabolic genes (Fig. 1B). Especially, LT-HSCs, which have the strongest self-renewal capacity, were mainly distinguished by PC2, in which genes related to cholesterol biosynthesis [such as 3-hydroxy-3-methylglutaryl-CoA reductase (Hmgcr)] and uptake [such as LDL receptor (Ldlr)], as well as their key regulators [such as sterol regulatory element binding transcription factor 2 (Srebf2)] were enriched (Fig. 1C). Especially, we noticed that Ldlr was among the genes contributing largest loading index to PC2 (Fig. 1C). Besides, the high expression of Hmgcr, Ldlr, and Srebf2 in LT-HSCs was validated by qPCR (Fig. 1D). Using flow cytometry, we confirmed relatively high surface expression of LDLR protein on LT-HSCs (Fig. 1E). Unexpectedly, using Filipin III staining and a fluorometric method, we detected relatively low intracellular free cholesterol contents in LT-HSCs (Fig. 1F and Fig. S1A). This might reflect the tight and negative regulation of cholesterol biosynthesis and uptake by intracellular cholesterol contents [6]. Overall, these results highlight a distinct cholesterol metabolic signature of HSCs.
Fig. 1.
High levels of cholesterol favor maintenance and myeloid bias of HSCs. (A) Heat map of cholesterol metabolic genes based on a published transcriptome database of HSPCs (ArrayExpress: E-MTAB-3079). (B) PCA of cholesterol metabolic genes shown in two principal components (PCs 1–2). (C) Loading matrix of the two principal components (PCs 1–2). (D) Relative mRNA expression of Hmgcr, Ldlr, and Srebf2 in LT-HSCs, S T-HSCs, and MPPs (n = 3). (E) Flow cytometric analysis of surface expression of LDLR on LT-HSCs, ST-HSCs, and MPPs (n = 6). (F) Flow cytometric analysis of intracellular cholesterol contents in LT-HSCs, ST-HSCs, and MPPs (n = 6). (G) Flow cytometric gating strategy for LDLRlo and LDLRhi LT-HSCs. (H) Flow cytometric analysis of intracellular cholesterol contents in LDLRlo and LDLRhi LT-HSCs (n = 6). (I) LT-HSC numbers per well in LDLRlo and LDLRhi LT-HSC cultures (n = 6). (J) Total cell numbers per well in LDLRlo and LDLRhi LT-HSC cultures (n = 4). (K) Frequencies of myeloid cell output in LDLRlo and LDLRhi LT-HSC cultures (n = 4). (L) Colony numbers per well in LDLRlo and LDLRhi LT-HSC cultures (n = 4). (M) Flow cytometric gating strategy for LT-HSC subpopulations. (N and O) Flow cytometric analysis of intracellular cholesterol contents (N) and LDLR expression (O) in LT-HSC subpopulations (n = 6). Data are mean ± SD. *p < 0.05, **p < 0.01. Two-tailed unpaired Student’s t-test unless stated otherwise. Two-tailed paired Student’s t-test (H). GEMM, granulocyte/erythroid/megakaryocyte/macrophage; BFU-E, burst-forming units-erythroid.
3.2. High levels of cholesterol favor maintenance and myeloid bias of HSCs
Notably, we observed continuous surface LDLR expression on LT-HSCs (Fig. 1E) and intracellular cholesterol contents in LT-HSCs (Fig. 1F), further reflecting a heterogeneity of cholesterol metabolism within LT-HSCs. Then, we partitioned LT-HSCs into LDLRlow (LDLRlo) and LDLRhigh (LDLRhi) subpopulations based on the top 30% and bottom 30% of LDLR expression (Fig. 1G). Correspondingly, intracellular free cholesterol contents in LDLRlo LT-HSCs were much higher than those in their LDLRhi counterparts (Fig. 1H and Fig. S1B). To determine the functional significance of the heterogeneous cholesterol metabolism, LDLRlo and LDLRhi LT-HSCs were isolated and cultured in a serum- and cholesterol-free liquid medium. Comparing to LDLRhi LT-HSCs, LDLRlo LT-HSCs exhibited enhanced LT-HSC maintenance (Fig. 1I), more cellular expansion (Fig. 1J), and a higher fraction of myeloid output (Fig. 1K). In methylcellulose medium, LDLRlo LT-HSCs also exhibited stronger colony-forming ability with significantly increased fraction of granulocyte-monocyte colony forming unit (CFU-GM) (Fig. 1L). As reported, HSCs with distinct lineage potencies can be partially distinguished by CD150 and CD41 expression (Fig. 1M), with CD41+CD150+ (Fr2) subpopulation being most myeloid biased [32]. Intriguingly, intracellular free cholesterol contents were much higher (Fig. 1N) and LDLR expression was much lower (Fig. 1O) in CD41+CD150+ LT-HSCs than those in other LT-HSC subpopulations. Collectively, these data inform that high levels of cholesterol favor maintenance and myeloid bias of HSCs, and LDLR expression could mark cholesterol homeostasis and functionality of HSCs. Notably, due to the redundancy of LDL uptake and competent cholesterol export [33], almost no alteration in intracellular cholesterol contents in LT-HSCs and BM hematopoiesis was observed in Ldlr−/− mice (Figs. S1C–F).
3.3. Cholesterol directly regulates maintenance and lineage differentiation of HSCs
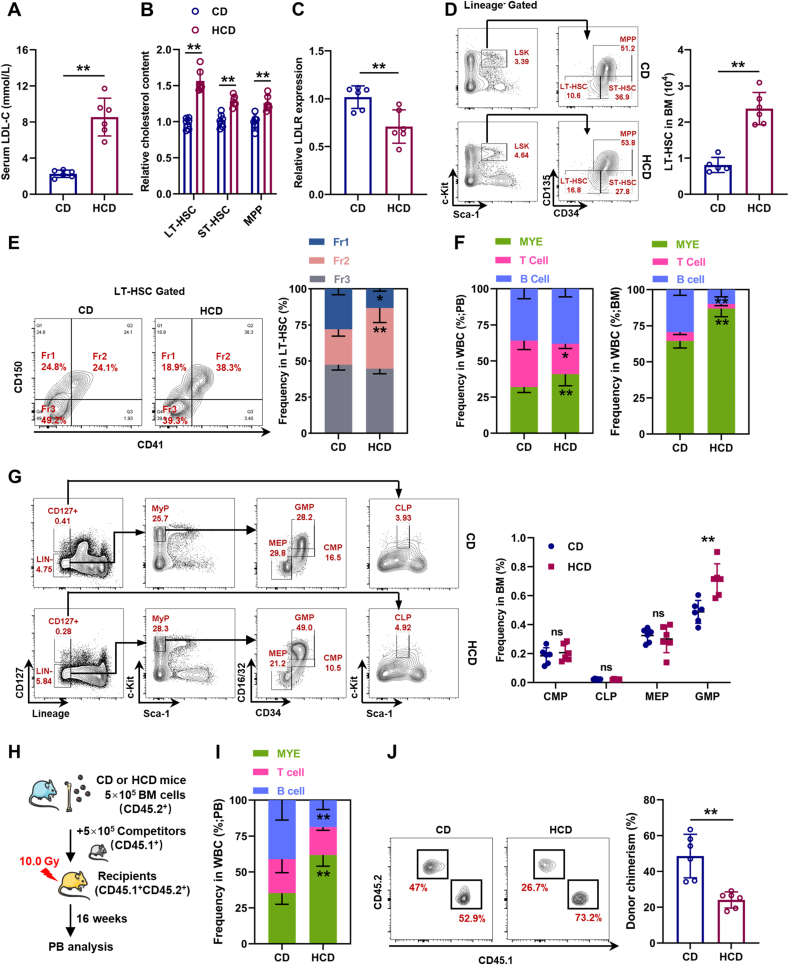
To further verify this, mice were fed HCD for 3 months to induce hypercholesterolemia, which was manifested by strikingly increased serum LDL-C levels (Fig. 2A). Parallelly, we observed that cholesterol was particularly accumulated in LT-HSCs (Fig. 2B) and surface LDLR expression was significantly downregulated on LT-HSC (Fig. 2C). Consistent with previous studies [[7], [8], [9]], LT-HSC numbers were remarkably increased in the BM of HCD mice (Fig. 2D). Meanwhile, we also observed a considerable increase in the fraction of CD41+CD150+ LT-HSCs in HCD mice (Fig. 2E), along with significantly increased frequency of myeloid cells in both PB and BM (Fig. 2F). At the hematopoietic progenitor cell level, significantly increased frequency of GMPs was detected in the BM of HCD mice (Fig. 2G). Competitive transplantation also confirmed that LT-HSCs from HCD mice were more myeloid-biased than those from CD mice (Fig. 2H, I), albeit their reconstitution ability was declined (Fig. 2J). Analogously, mice with ApoE knockout (ApoE−/−), which resulted in defective efflux of intracellular cholesterol [8], exhibited cholesterol accumulation (Fig. S2A), as well as expansion (Fig. S2B) and myeloid bias (Figs. S2C–E) of LT-HSCs.
Fig. 2.
Cholesterol directly regulates maintenance and lineage differentiation of HSCs in vivo. (A) Serum LDL-C levels in mice with control diet (CD) or HCD (n = 6). (B) Flow cytometric analysis of intracellular cholesterol contents in BM HSPCs of CD and HCD mice (n = 6). (C) Relative LDLR expression on BM LT-HSCs of CD and HCD mice (n = 6). (D) Flow cytometric analysis of BM LT-HSC pool size in CD and HCD mice (n = 6). (E) Flow cytometric analysis of frequency of LT-HSC subpopulations in CD and HCD mice (n = 6). (F) Frequencies of T cells, B cells, and myeloid cells in PB and BM of CD and HCD mice (n = 6). (G) Flow cytometric analysis of frequencies of CMPs, CLPs, GMPs, and MEPs in BM of CD and HCD mice (n = 6). (H) Schematic of competitive BM transplantation. (I) Fraction of donor-derived T cells, B cells, and myeloid cells in PB of recipient mice (n = 6). (J) Frequency of donor-derived cells in PB of recipient mice (n = 6). Data are mean ± SD. n.s., not significant. *p < 0.05, **p < 0.01. Two-tailed unpaired Student’s t-test.
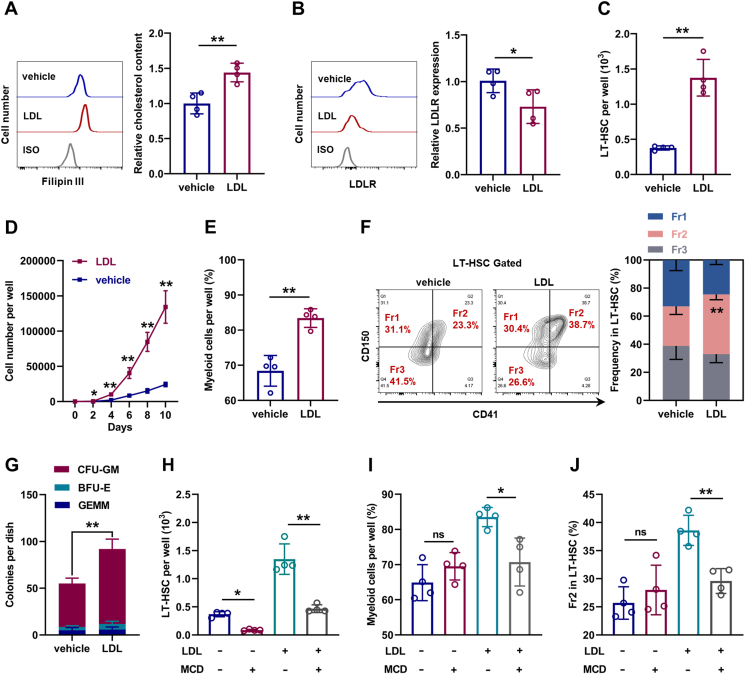
Subsequently, LT-HSC culture was supplemented with LDL to mimic hypercholesterolemia in vitro. Compared with LT-HSCs cultured in control medium, LT-HSCs cultured with LDL exhibited strikingly increased intracellular cholesterol contents (Fig. 3A) and downregulated LDLR expression (Fig. 3B). Meanwhile, the maintenance of LT-HSCs was dramatically enhanced in the presence of LDL (Fig. 3C), accompanied by more cellular expansion (Fig. 3D) and myeloid bias (Fig. 3E, F). In methylcellulose medium, LDL treatment remarkably increased the colony-forming ability and CFU-GM explosion of LT-HSCs (Fig. 3G). However, depletion of intracellular cholesterol with MCD nearly restored the maintenance and lineage differentiation of LDL-treated (Fig. 3H–J) and ApoE−/− LT-HSCs (Figs. S3A–C), indicating that the effects of LDL and ApoE deficiency on LT-HSCs depended on cholesterol. Analogously, LT-HSCs cultured with free cholesterol also exhibited cholesterol accumulation, LDLR downregulation, enhanced maintenance, and myeloid bias (Figs. S3D–H). However, LDL only moderately affected the intracellular cholesterol contents, maintenance, and lineage differentiation of Ldlr−/− LT-HSCs (Fig. S3I–L), indicating that LDL directly affected LT-HSCs at least in part via LDLR-mediated cholesterol uptake. Notably, MCD treatment alone nearly depleted LT-HSCs in the culture (Fig. 3H), indicating that LT-HSC maintenance was dampened upon cholesterol depletion. Taken together, these data indicates that HSC is sensitive to perturbations of cholesterol metabolism and cholesterol directly regulates maintenance and lineage differentiation of HSCs.
Fig. 3.
Cholesterol directly regulates maintenance and lineage differentiation of HSCs in vitro. (A and B) Flow cytometric analysis of relative intracellular cholesterol contents (A) and surface expression of LDLR (B) in LT-HSCs with or without LDL treatment (n = 4). (C) LT-HSC numbers per well in LT-HSC cultures with or without LDL treatment (n = 4). (D) Total cell numbers per well in LDLRlo and LDLRhi LT-HSC cultures with or without LDL treatment (n = 4). (E) Frequencies of myeloid cell output in LT-HSC cultures with or without LDL treatment (n = 4). (F) Frequency of LT-HSC subpopulations in LT-HSC cultures with or without LDL treatment (n = 4). (G) Colony numbers per well in LT-HSC cultures with or without LDL treatment (n = 4). (H) LT-HSC numbers per well in LT-HSC cultures with indicated treatment (n = 4). (I) Frequencies of myeloid cell output in LT-HSC cultures with indicated treatment (n = 4). (J) Frequency of Fr2 LT-HSCs in LT-HSC cultures with indicated treatment (n = 4). Data are mean ± SD. n.s., not significant. *p < 0.05, **p < 0.01. Two-tailed unpaired Student’s t-test unless stated otherwise. One-way ANOVA (H–J).
3.4. Cholesterol safeguards HSC maintenance and myeloid regeneration during irradiation-induced myelosuppression
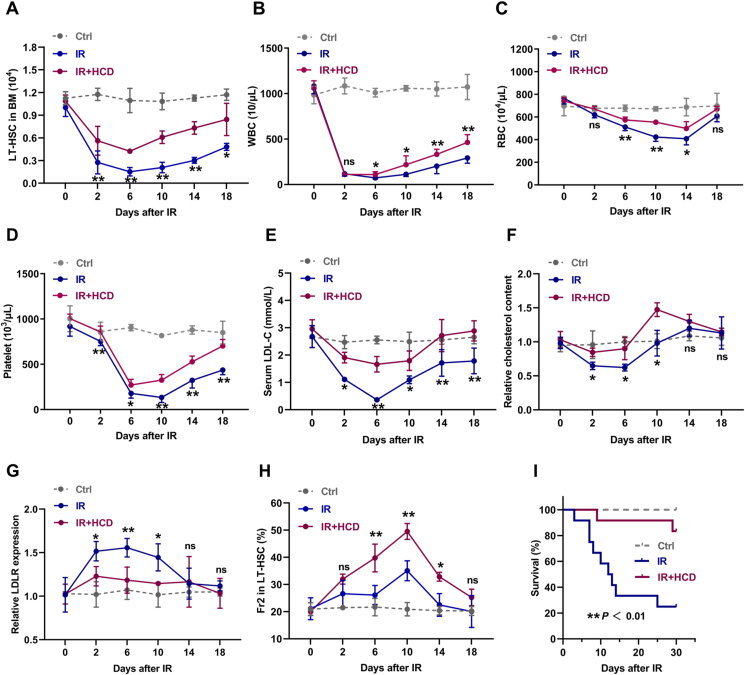
Next, we explored the role of cholesterol metabolism in BM hematopoiesis under stress condition. Mice were exposed to sublethal IR at a dose of 5 Gy to induce myelosuppression, which was manifested by rapid and dramatic shrink of LT-HSC pool (Fig. 4A) and pancytopenia in PB (Fig. 4B–D) and BM (Fig. S4A). Concurrently, profound hypocholesterolemia (Fig. 4E) and cholesterol deficiency in LT-HSCs (Fig. 4F) were rapidly occurred in mice post IR, accompanied by dramatic LDLR upregulation on LT-HSCs (Fig. 4G). Especially, we observed that cholesterol deficiency was exclusively presented in LT-HSCs among the HSPC compartment (Fig. S4B). Meanwhile, the change dynamics of serum cholesterol (Fig. 4E), intracellular cholesterol of LT-HSCs (Fig. 4F), and LT-HSC pool (Fig. 4A) were similar post IR, further indicating a close link between cholesterol metabolism and LT-HSC maintenance. Then, mice were adapted to HCD for one week before IR, followed by one week of HCD post IR, to preserve cholesterol homeostasis in LT-HSCs post IR (Fig. 4E–G). It was observed that LT-HSC pool was dramatically preserved in HCD mice post IR (Fig. 4A). Of note, CD41+CD150+ LT-HSCs seemed to be more resistant to IR and HCD further promoted the preponderance of CD41+CD150+ LT-HSCs post IR (Fig. 4H). Consequently, HCD significantly accelerated the overall hematopoietic regeneration particularly myeloid regeneration (Fig. 4B–D and Figs. S4A and C) and increased the survival of mice with lethal myelosuppression (Fig. 4I). These results further emphasize the strong sensitivity of LT-HSCs to perturbations of cholesterol metabolism and that cholesterol safeguards HSC maintenance and myeloid regeneration post IR.
Fig. 4.
Cholesterol safeguards HSC maintenance and myeloid regeneration during irradiation-induced myelosuppression. (A) LT-HSC numbers in BM of CD and HCD mice post IR (n = 4). (B–D) White blood cell (WBC), red blood cell (RBC), and platelet counts in PB of CD and HCD mice post IR (n = 6). (E) Serum LDL-C levels in CD and HCD mice post IR (n = 3). (F and G) Flow cytometric analysis of intracellular cholesterol contents and LDLR surface expression in BM LT-HSCs of CD and HCD mice post IR (n = 3). (H) Frequency of Fr2 LT-HSCs in BM of CD and HCD mice post IR (n = 4). (I) Survival analysis of CD and HCD mice after lethal IR (7.5 Gy) exposure (n = 10). Data are mean ± SD. n.s., not significant. *p < 0.05, **p < 0.01, IR + HCD group compared with IR group. Two-tailed unpaired Student’s t-test unless stated otherwise. Kaplan–Meier survival curves and log-rank test (I). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.5. Cholesterol enhances ferroptosis resistance and unbalances myeloid/lymphoid lineage differentiation of HSCs
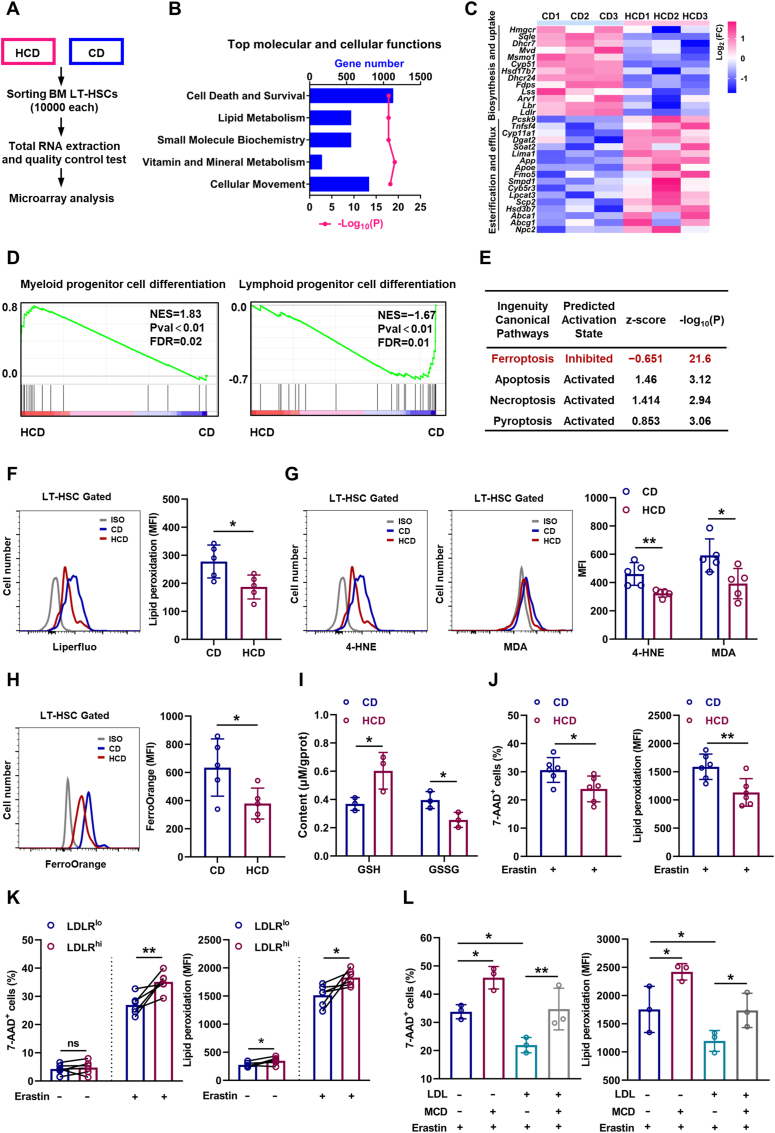
To understand the underlying mechanisms, we performed RNA-seq analysis of LT-HSCs from HCD mice (Fig. 5A). Consistently, based on Ingenuity Pathway Analysis (IPA) of molecular and cellular functions, we observed robust alteration of lipid metabolism in LT-HSCs (Fig. 5B), involving suppressed cholesterol uptake/biosynthesis and augmented cholesterol esterification/efflux (Fig. 5C), further verifying the feedback regulation of cholesterol metabolism by intracellular cholesterol contents. Simultaneously, LT-HSCs of HCD mice exhibited a transcriptional signature of enhanced myeloid lineage differentiation but dampened lymphoid lineage differentiation (Fig. 5D), which molecularly explains the myeloid bias of LT-HSCs induced by excess cholesterol. Interestingly, we noticed that the cell death and survival signaling pathway was predominantly altered in LT-HSCs of HCD mice (Fig. 5B). Among the various death signaling pathways, we noticed that ferroptosis signaling pathway was prominently inhibited in LT-HSCs of HCD mice, while other death signaling pathways were only mildly affected (Fig. 5E). In vivo, although LT-HSC death as detected by 7-AAD staining in HCD mice was negligible and comparable to that in CD mice (Fig. S5A), membrane lipid peroxidation (Fig. 5F), toxic lipid peroxidation end products including 4-HNE and MDA (Fig. 5G and Fig. S5B), and Fe2+ pool (Fig. 5H) was significantly reduced in LT-HSCs of HCD mice, paralleled by increased GSH and decreased GSSG contents (Fig. 5I). Moreover, the freshly isolated LT-HSCs from HCD mice were more resistant to ferroptosis induced by erastin ex vivo (Fig. 5J), informing that the ferroptosis resistance of LT-HSCs was enhanced in HCD mice. Furthermore, the ferroptosis resistance of LDLRlo LT-HSCs (Fig. 5K) and CD41+CD150+ LT-HSCs (Fig. S5C) was significantly enhanced comparing to their respective counterparts. In vitro, although LDL and free cholesterol treatment had no noticeable impact on LT-HSC death and lipid peroxidation (Figs. S5D and E), they remarkably counteracted erastin-induced LT-HSC ferroptosis (Fig. 5L and Fig. S5E). Conversely, cholesterol depletion by MCD not only increased the ferroptosis sensitivity of LT-HSCs but also offset the ferroptosis resistance of LT-HSCs conferred by LDL treatment (Fig. 5L). These data demonstrate a direct link of cholesterol to ferroptosis sensitivity of HSCs. Therefore, excess cholesterol enhances ferroptosis resistance and unbalances myeloid/lymphoid lineage differentiation of LT-HSCs, thereby contributing to expansion and myeloid bias of LT-HSCs.
Fig. 5.
Cholesterol enhances ferroptosis resistance and unbalances myeloid/lymphoid lineage differentiation of HSCs. (A) Workflow of the RNA-seq analysis. (B) Ingenuity Pathway Analysis (IPA) showing the top changed molecular and cellular functions in LT-HSCs of HCD mice relative to CD mice. (C) Heat map showing fold change of cholesterol uptake, biosynthesis, esterification, and efflux genes in BM LT-HSCs of HCD mice. (D) GSEA of indicated gene sets in BM LT-HSCs of HCD mice. (E) IPA of cell death signaling pathways in BM LT-HSCs of HCD mice. (F–H) Flow cytometric analysis of membrane lipid peroxidation (F), 4-HNE and MDA contents (G), and ferrous ion deposition (H) in BM LT-HSCs of CD and HCD mice (n = 5). (I) Intracellular glutathione (GSH) and oxidized glutathione (GSSG) contents in BM LT-HSCs of CD and HCD mice (n = 3). (J) Ferroptosis resistance analysis of BM LT-HSCs from CD and HCD mice (n = 6). (K) Ferroptosis resistance analysis of LDLRlo and LDLRhi LT-HSCs (n = 6). (L) Ferroptosis resistance analysis of LT-HSCs with indicated treatment (n = 3). Data are mean ± SD. n.s., not significant. *p < 0.05, **p < 0.01. Two-tailed unpaired Student’s t-test unless stated otherwise. Two-tailed paired Student’s t-test (K). One-way ANOVA (L).
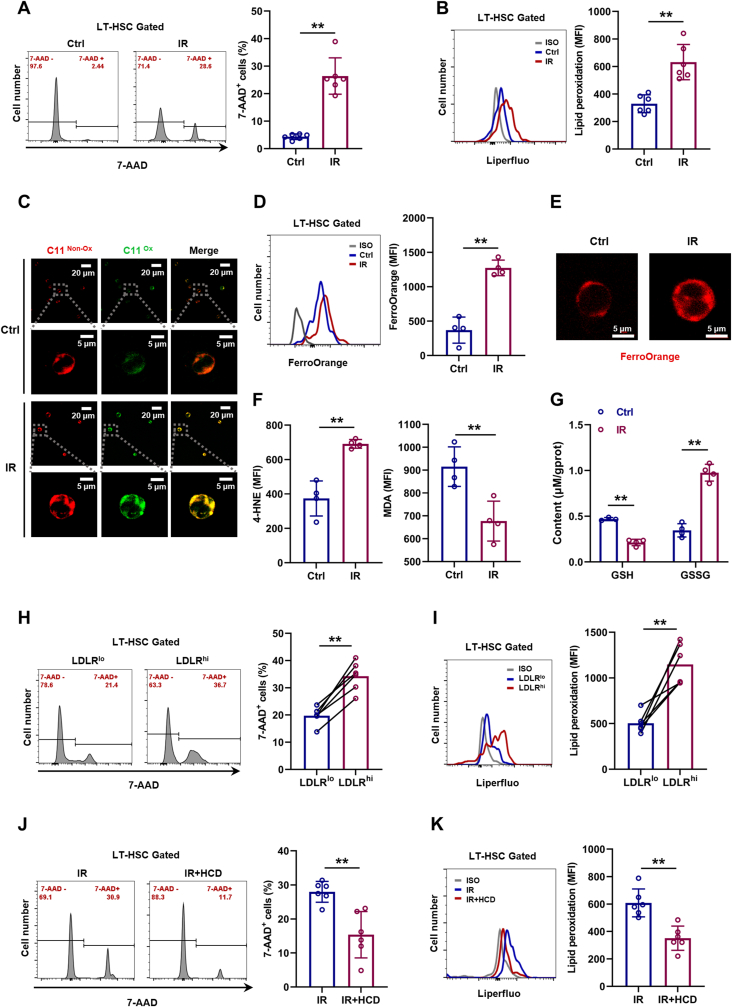
IR is well demonstrated as an inducer of ferroptosis [20]. In consistent with this, dramatic cell death (Fig. 6A) and lipid peroxidation (Fig. 6B, C) were incurred in LT-HSCs especially by 24 h post IR (Fig. S6A), along with increased Fe2+ pool (Fig. 6D, E), 4-HNE and MDA levels (Fig. 6F and Fig. S6B), as well as increased GSSG and decreased GSH (Fig. 6G). Meanwhile, both LDLRlo (Fig. 6H, I) and CD41+CD150+ LT-HSCs (Figs. S6C and D) were more resistant to IR-induced ferroptosis than their respective counterparts. However, excess cholesterol strikingly mitigated IR-induced LT-HSC ferroptosis (Fig. 6J, K). These data could explain the enhanced HSC maintenance and accelerated myeloid regeneration in HCD mice post IR.
Fig. 6.
Cholesterol enhances ferroptosis resistance of HSCs during irradiation-induced myelosuppression. (A and B) Flow cytometric analysis of cell death (A) and membrane lipid peroxidation (B) in BM LT-HSCs at 24 h post IR (n = 6). (C) Immunofluorescence analysis of membrane lipid peroxidation by BODIPY 581/591C11 staining in LT-HSCs at 24 h post IR. (D) Flow cytometric analysis of ferrous ion deposition in BM LT-HSCs at 24 h post IR (n = 4). (E) Immunofluorescence analysis of ferrous ion deposition by FerroOrange staining in LT-HSCs at 24 h post IR. (F) Flow cytometric analysis of 4-HNE and MDA contents in BM LT-HSCs at 24 h post IR (n = 4). (G) GSH and GSSG contents in BM LT-HSCs at 24 h post IR (n = 4). (H and I) Flow cytometric analysis of cell death (H) and membrane lipid peroxidation (I) in LDLRlo and LDLRhi LT-HSCs at 24 h post IR (n = 6). (J and K) Flow cytometric analysis of cell death (J) and membrane lipid peroxidation (K) in BM LT-HSCs of IR mice with CD or HCD at 24 h post IR (n = 6). Data are mean ± SD. *p < 0.05, **p < 0.01. Two-tailed unpaired Student’s t-test unless stated otherwise. Two-tailed paired Student’s t-test (H, I).
3.6. SLC38A9–mTOR axis mediates cholesterol sensing and signal transduction in HSCs
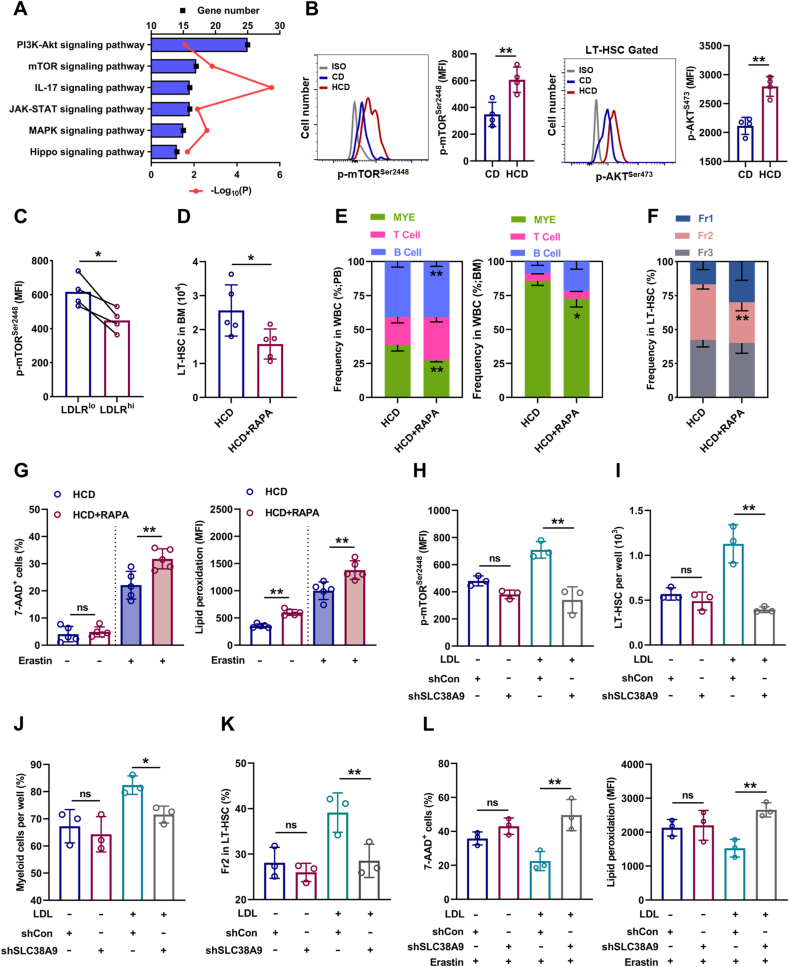
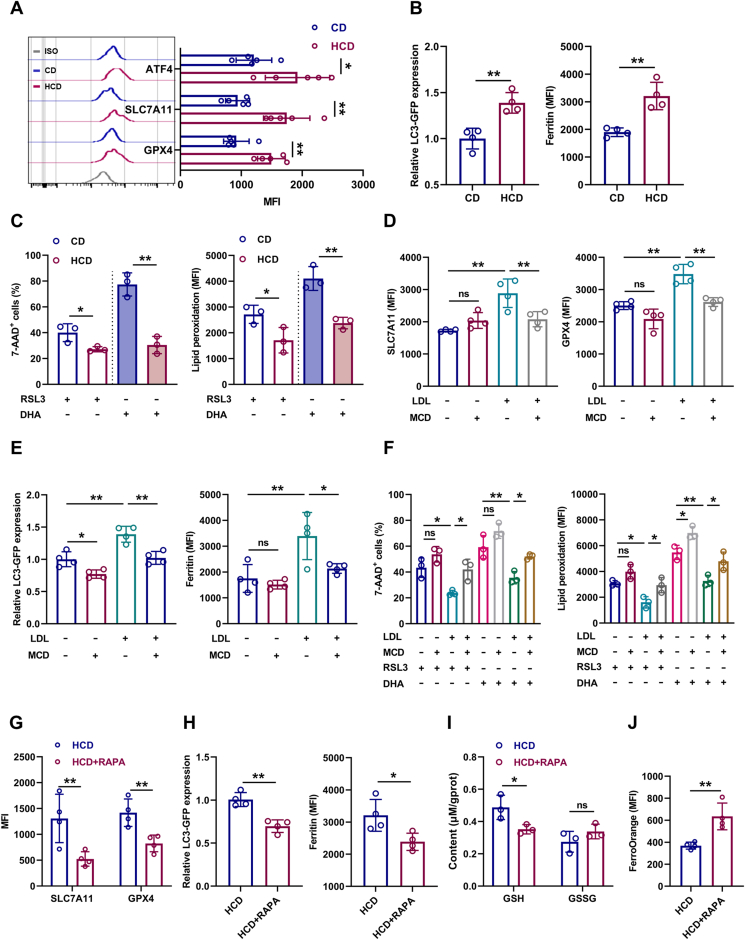
We further sought to determine the molecular basis underlying the adaptive responses of HSCs to excess cholesterol. Enrichment analysis of signaling pathways revealed that mTOR signaling, which is well recognized to unbalance myeloid/lymphoid lineage differentiation of HSCs [34], is predominantly activated in LT-HSCs of HCD mice (Fig. 7A). Meanwhile, the activation of mTOR signaling in LT-HSCs of HCD mice was further validated by flow cytometry (Fig. 7B). Furthermore, LDLRlo LT-HSCs were characterized by augmented mTOR activity comparing to their LDLRhi counterparts (Fig. 7C). In vitro, LDL treatment significantly enhanced mTOR activation in LT-HSCs, while MCD treatment significantly suppressed mTOR activity in LT-HSCs cultured with or without LDL (Fig. S7A). Conversely, mTOR inhibition by rapamycin remarkably reversed HCD-induced expansion (Fig. 7D), myeloid bias (Fig. 7E, F), and ferroptosis resistance (Fig. 7G) of LT-HSCs. These data demonstrate that mTOR plays a central and integrate role in HSC adaption to cholesterol accumulation. Besides, it is reported that SLC38A9 senses intracellular cholesterol to stimulate mTOR activation in mammalian cells [35]. Interestingly, we observed higher expression of SLC38A9 in LT-HSCs (Fig. S7B). Moreover, SLC38A9 knockdown by lentivirus-mediated shRNA transfection (Figs. S7C and D) reversed LDL-induced mTOR activation (Fig. 7H), expansion (Fig. 7I), myeloid bias (Fig. 7J, K), and ferroptosis resistance (Fig. 7L) of LT-HSCs. Overall, these results demonstrate that SLC38A9–mTOR axis mediates cholesterol sensing and signal transduction to orchestrate ferroptosis resistance and myeloid bias of HSCs.
Fig. 7.
SLC38A9–mTOR axis mediates cholesterol sensing and signal transduction in HSCs. (A) KEGG pathway enrichment analysis showing the top changed signaling pathways in BM LT-HSCs of HCD mice relative to CD mice. (B) Flow cytometric analysis of p-mTORSer2448 and p-AKTSer473 protein levels in BM LT-HSCs of CD and HCD mice (n = 4). (C) Flow cytometric analysis of p-mTORSer2448 protein levels in LDLRlo and LDLRhi LT-HSCs (n = 4). (D) Flow cytometric analysis of LT-HSC pool size in HCD mice with or without rapamycin treatment (n = 5). (E) Frequencies of T cells, B cells, and myeloid cells in PB and BM of HCD mice with or without rapamycin treatment (n = 5). (F) Frequency of LT-HSC subpopulations in BM of HCD mice with or without rapamycin treatment (n = 5). (G) Ferroptosis resistance analysis of LT-HSCs from HCD mice with or without rapamycin treatment (n = 5). (H) Flow cytometric analysis of p-mTORSer2448 protein levels in LDL-treated LT-HSCs after shSLC38A9 lentiviral transduction (n = 3). (I) LT-HSC numbers per well in LT-HSC cultures with or without LDL treatment after shSLC38A9 lentiviral transduction (n = 3). (J) Frequencies of myeloid cell output in LT-HSC cultures with or without LDL treatment after shSLC38A9 lentiviral transduction (n = 3). (K) Frequency of Fr2 LT-HSCs in LT-HSC cultures with or without LDL treatment after shSLC38A9 lentiviral transduction (n = 3). (L) Ferroptosis resistance analysis of LT-HSCs with or without LDL treatment after shSLC38A9 lentiviral transduction (n = 3). Data are mean ± SD. n.s., not significant. *p < 0.05, **p < 0.01. Two-tailed unpaired Student’s t-test unless stated otherwise. Two-tailed paired Student’s t-test (C). One-way ANOVA (H–L).
3.7. mTOR distinctively upregulates SLC7A11/GPX4 and inhibits ferritinophagy in LT-HSCs upon cholesterol accumulation
The ferroptosis resistance of LT-HSCs upon cholesterol accumulation informed that cyst(e)ine metabolism, iron metabolism, fatty acid metabolism and generation of reducing equivalent NADPH, which are closely regulated by mTOR, might be affected.
mTOR may regulate ferroptosis sensitivity from several aspects: 1) upregulation of GPX4 expression [36]; 2) upregulation of ATF4-mediated SLC7A11 expression [37]; 3) downregulation of TfR1-mediated iron import [38] or inhibition of ferritinophagy-mediated Fe2+ release [39]; 4) upregulation of SREBP1/SCD1-mediated monounsaturated fatty acid (MUFA) generation and pentose phosphate pathway which respectively displace PUFA from plasma membrane and generate NADPH [40], as well as downregulation of ACSL4/LPCAT3/ALOX15-mediated lipid peroxide generation [41]; 5) remodeling of energy metabolism that culminates in increased NADPH generation [42]. Using flow cytometry, we detected that the protein expression of TfR1, SREBP1/SCD1 and ACSL4/LPCAT3/ALOX15 was nearly unperturbed in LT-HSCs of HCD mice (Figs. S8A–C). Based on our RNA-seq data, GSEA revealed no noticeable influence of HCD on glucose, fatty acid, and energy metabolism in LT-HSCs (Fig. S8D). Correspondingly, the total iron pool as well as intracellular ATP, NADP+ and NADPH contents were comparable in LT-HSCs of HCD mice to those of CD mice (Fig. S8E). Excitingly, we found that protein expression of ATF4/SLC7A11 and GPX4 was remarkably upregulated in LT-HSCs of HCD mice (Fig. 8A). Using GFP-LC3 reporter mice, we observed that autophagy was significantly inhibited in LT-HSCs of HCD mice, along with increased ferritin protein (Fig. 8B), indicating a state of ferritinophagy inhibition. Accordingly, besides erastin, which is a SLC7A11 blocker, LT-HSCs from HCD mice were also more resistant to ferroptosis induced by a GPX4 inhibitor RSL3 and a ferritinophagy inducer DHA ex vivo (Fig. 8C). Similar results were verified in vitro with LDL treatment (Fig. 8D–F). Inversely, MCD treatment significantly downregulated GPX4/SLC7A11 expression (Fig. 8D) and boosted autophagy (Fig. 8E) in LT-HSCs cultured with or without LDL, and sensitized them to RSL3-and DHA-induced ferroptosis (Fig. 8F). Furthermore, rapamycin treatment remarkably reversed HCD-induced GPX4/SLC7A11 upregulation (Fig. 8G) and ferritinophagy inhibition (Fig. 8H), finally restoring the redox homeostasis of LT-HSCs in HCD mice (Fig. 8I, J). Thus, mTOR distinctively induces SLC7A11/GPX4 upregulation and ferritinophagy inhibition in HSCs upon cholesterol accumulation, which synergistically confer ferroptosis resistance.
Fig. 8.
mTOR distinctively upregulates SLC7A11/GPX4 and inhibits ferritinophagy in LT-HSCs upon cholesterol accumulation. (A) Flow cytometric analysis of ATF4, SLC7A11 and GPX4 protein levels in BM LT-HSCs of CD and HCD mice (n = 5). (B) Relative LC3-GFP expression and ferritin protein levels in BM LT-HSCs of CD mice and HCD mice (n = 4). (C) Ferroptosis analysis of LT-HSCs from CD and HCD mice with indicated treatment (n = 3). (D) Flow cytometric analysis of SLC7A11/GPX4 protein levels in LT-HSCs with indicated treatment (n = 4). (E) Relative LC3-GFP expression and ferritin protein levels in LT-HSCs with indicated treatment (n = 4). (F) Ferroptosis resistance analysis of LT-HSCs with indicated treatment (n = 3). (G) Flow cytometric analysis of SLC7A11/GPX4 protein levels in BM LT-HSCs of HCD mice with or without rapamycin treatment (n = 4). (H) Relative LC3-GFP expression and ferritin protein levels in BM LT-HSCs of HCD mice with or without rapamycin treatment (n = 4). (I) GSH and GSSG contents in BM LT-HSCs of HCD mice with or without rapamycin treatment (n = 3). (J) Flow cytometric analysis of ferrous ion deposition in BM LT-HSCs of HCD mice with or without rapamycin treatment (n = 4). Data are mean ± SD. n.s., not significant. *p < 0.05, **p < 0.01. Two-tailed unpaired Student’s t-test unless stated otherwise. One-way ANOVA (D–F).
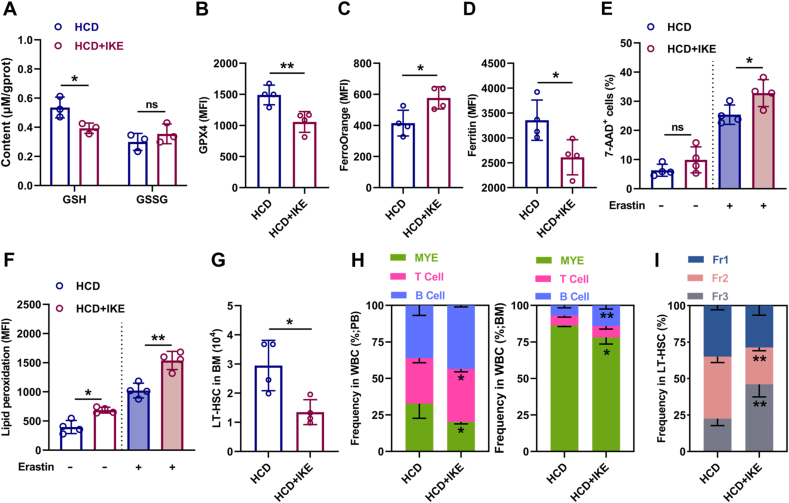
3.8. Ferroptosis inducer administration restores maintenance and differentiation balance of HSCs in HCD mice
Finally, we interrogated that whether pharmacological intervention of redox homeostasis could restore ferroptosis sensitivity of LT-HSCs. To achieve this, we treated HCD mice with IKE, which has been proven in vivo as a suitable ferroptosis inducer that can potently block SLC7A11 [43], downregulate GPX4 expression [36], and trigger autophagy [44]. As shown, IKE treatment significantly restored the redox homeostasis of LT-HSCs in HCD mice, as manifested by reduced GSH contents (Fig. 9A) and GPX4 expression (Fig. 9B), as well as increased Fe2+ pool (Fig. 9C) and decreased ferritin protein (Fig. 9D). Consequently, the ferroptosis sensitivity of LT-HSCs was significantly enhanced in HCD mice (Fig. 9E, F), resulting in apparent recovery of maintenance and differentiation balance of LT-HSCs (Fig. 9G–I).
Fig. 9.
Ferroptosis inducer administration restores maintenance and differentiation balance of HSCs in HCD mice. (A) GSH and GSSG contents in BM LT-HSCs of HCD mice with or without IKE treatment (n = 3). (B–D) Flow cytometric analysis of GPX4 expression (B), ferrous ion deposition (C) and ferritin expression (D) in BM LT-HSCs of HCD mice with or without IKE treatment (n = 4). (E–F) Ferroptosis resistance analysis of LT-HSCs from HCD mice with or without IKE treatment (n = 4). (G) Flow cytometric analysis of LT-HSC pool size in HCD mice with or without IKE treatment (n = 4). (H) Frequencies of T cells, B cells, and myeloid cells in PB and BM of HCD mice with or without IKE treatment (n = 4). (I) Frequency of LT-HSC subpopulations in BM of HCD mice with or without IKE treatment (n = 4). Data are mean ± SD. n.s., not significant. *p < 0.05, **p < 0.01. Two-tailed unpaired Student’s t-test.
4. Discussion
Perturbations of cholesterol metabolism always accompany hematopoietic alterations, while the underlying mechanisms remain largely unknown. Here, we show that cholesterol metabolism is heterogenous among HSPCs and LT-HSCs are particularly sensitive to perturbations of cholesterol metabolism. Excess cholesterol directly enhances ferroptosis resistance and unbalances myeloid/lymphoid lineage differentiation of HSCs, resulting in HSC expansion and myeloid-biased hematopoiesis. Molecularly, we identify that SLC38A9–mTOR axis mediates cholesterol sensing and signal transduction to instruct lineage differentiation of HSCs as well as to orchestrate ferroptosis sensitivity of HSCs through regulating SLC7A11/GPX4 expression and ferritinophagy. Moreover, a mTOR inhibitor rapamycin and a ferroptosis inducer IKE prevent HSC alterations induced by excess cholesterol. Above all, this is the first study as far as we know to illustrate the direct and distinctive link among cholesterol, ferroptosis and HSC homeostasis as well as the underlying molecular mechanism. Given the close link between myeloid-biased hematopoiesis and the deregulation of systemic organs especially atherosclerosis [45], our findings highlight the great clinical significance of early monitoring and intervention of hematopoietic alterations during hypercholesterolemia.
Myeloid-biased hematopoiesis such as monocytosis and neutrophilia is frequently observed in mammals with atherosclerosis [7]. However, the interplay between hypercholesterolemia, hematopoietic alterations, and atherosclerosis remains obscure and is the subject of ongoing research [46]. Meanwhile, although cholesterol metabolic signaling pathways have been well delineated, the intrinsic properties of cholesterol metabolism in BM hematopoietic system are almost undefined. In this study, we discover that cholesterol metabolism in LT-HSCs is quite distinct from their progenies, characterized by a cholesterol-deficient state with high expression of cholesterol sensing and metabolic machineries. This trait may render LT-HSCs particularly susceptible to the mild elevation of cholesterol levels before atherosclerosis develops. Besides, cholesterol metabolism is also heterogenous among LT-HSCs, with those having high levels of cholesterol tending to be endowed with survival advantage and myeloid bias. These findings argue that hypercholesterolemia provokes hematopoietic alterations through directly acting at the level of LT-HSCs. Nevertheless, further studies are warranted to decipher the intrinsic and/or extrinsic molecular programs that coordinately determine the heterogeneities of cholesterol metabolism and functionality among HSCs.
LDLR is well known as a main player responsible for LDL uptake from the circulation. In this study, we further identify that LDLR can mark cholesterol metabolism and functionality of HSCs. Notably, unlike ApoE−/− mice, the intracellular cholesterol contents in LT-HSCs and the BM hematopoiesis of Ldlr−/− mice were nearly unperturbed, which could be explained by the competency of cholesterol efflux and/or uptake via other LDL receptors in Ldlr−/− mice [33]. This discrepancy reflects the rigorous control of cholesterol metabolism in HSCs through concerted regulation of cholesterol biosynthesis, uptake, and export. However, relative to ApoE−/− mice, BM hematopoiesis of Ldlr−/− mice respond bluntly to HCD-induced hypercholesterolemia, albeit blood LDL levels were even higher in Ldlr−/− mice than ApoE−/− mice [8]. These data further verify our findings that LDL directly trigger HSC alterations via LDLR-mediated cholesterol uptake. Besides, in contrast to LDL, oxLDL, which is also accumulated in hypercholesterolemic human and animals [47], promoted ferroptosis and impaired maintenance of LT-HSCs in vitro (Fig. S9), highlighting a dominant role of LDL in driving HSC alterations upon hypercholesterolemia. Therefore, LDLR could be considered as a candidate target for monitoring cholesterol metabolism and functionality of HSCs as well as for treatment of hypercholesterolemia-associated hematopoietic disorders.
The direct regulatory role of cholesterol in HSC proliferation has been well documented to promote HSC expansion in the face of excess cholesterol [[7], [8], [9]]. However, proliferative stress always exhausts rather than expands HSC pool due to oxidative stress-induced HSC differentiation and/or cell death [[48], [49], [50]]. In this study, we uncover that excess cholesterol distinctively enhances the resistance of HSCs to ferroptosis, a well-known cell death modality triggered by oxidative stress [21]. Therefore, enhanced proliferation and ferroptosis resistance synergistically contribute to HSC expansion in response to excess cholesterol. Meanwhile, excess cholesterol also enhances myeloid and dampens lymphoid lineage differentiation of HSCs. Consequently, myeloid-biased HSCs expand and dominate BM hematopoiesis, resulting in myeloid-biased hematopoiesis during hypercholesterolemia. Notably, HSCs also act as a primary responder to diverse physiopathological stresses such as genotoxicity, infection, and inflammation, which frequently trigger ferroptosis and myeloid-biased hematopoiesis [11,19,20]. Based on our findings, the survival advantage of myeloid-biased HSCs due to high levels of intracellular cholesterol may be implicated in these scenarios.
Ferroptosis is featured by iron-dependent phospholipid peroxidation and is regulated by multiple cellular metabolic pathways [22]. Cholesterol as a fundamental membrane lipid has been reported to protect phospholipid bilayer from oxidative damage. Currently, this protective effect is largely attributed to the unique physicochemical properties of cholesterol [51,52]. In this study, we further observe that excess cholesterol dampens ferroptotic signaling pathway, informing an intersection between cholesterol metabolic and ferroptotic signaling pathways. Intracellular cholesterol homeostasis is tightly regulated by cholesterol sensing and metabolic signaling pathways [6]. We here identify that SLC38A9 mediates the sensing of intracellular cholesterol by HSCs, whereas the activation of downstream mTOR enhances ferroptosis resistance of HSCs through upregulating SLC7A11/GPX4 and inhibiting ferritinophagy. In addition, excess cholesterol can also enhance IL-3/GM-CSF signaling in HSCs [9], which also signals to mTOR [53]. Since that mTOR activation also promotes proliferation and myeloid lineage differentiation of HSCs [34], these lines of evidence argue a central role of mTOR in orchestrating HSC alterations induced by excess cholesterol. Overall, these findings substantially advance our understanding of the regulatory actions of cholesterol metabolism in HSC survival and fate decisions. It is worth noting that excess cholesterol as an inducer of oxidative stress also sensitizes cells such as endothelial cells and smooth muscle cells to ferroptosis [23]. This may reflect the diverse activities of cholesterol sensing and metabolic pathways in different kinds of cells.
In addition, HSC aging is also featured by survival advantage, autophagy inhibition, and myeloid bias of HSCs [54,55]. Since that cholesterol usually accumulates in cells with age [56] and that mTOR is activated in aged HSCs [57], it is reasonable to expect that cholesterol accumulation may also contribute to HSC aging. Of note, mTOR also serves as a central signaling hub of various nutrients such as glucose, amino acid, and phosphate [18,58]. As reported, overnutrition, which is prevalent in our current lifestyle, always accompany alterations of HSC survival and fate decisions [[12], [13], [14], [15],18], raising concerns regarding our dietary habit and its influence on HSC fitness. Meanwhile, our findings could also serve as a common mechanism to interpret the similar hematopoietic alterations observed in obesity and metabolic syndrome [8]. Moreover, dysregulated cholesterol homeostasis is closely associated with enhanced tumorigenicity, metastasis, and therapy resistance of cancers [26,59]. Cancer stem cells (CSCs), which refer to tumor-initiating cell populations, now serve as a pivotal framework to understand aggressive and therapeutically resistant cancers [60]. Given the many similarities shared by CSCs and HSCs, it is plausible that ferroptosis resistance conferred by high levels of cholesterol may also be implicated. On the other hand, cancer therapies such as radiotherapy and chemotherapy are extremely toxic to BM hematopoiesis, and the resultant myelosuppression is always life-threatening and is a primary cause for the discontinuation of cancer therapies [17]. Based on our findings, fine-tuning cholesterol metabolism represents a promising avenue for treating cancers and/or preventing cancer therapy-associated hematopoietic toxicity.
In conclusion, this study for the first time unveils the distinct cholesterol metabolic signature of HSCs as well as its intersection with HSC survival and fate decisions. The findings of our study not only provide novel insights into the regulatory role of niche nutrients in stem cell survival and fate decisions, but also substantially extend our understanding of the effects and mechanisms of dyslipidemia on human health. Meanwhile, our study also provides a clinically feasible strategy for therapeutic intervention of dyslipidemia-related hematopoietic disorders by mTOR inhibitors and ferroptosis inducers as well as genotoxicity-associated hematopoietic toxicity by fine-tuning cholesterol metabolism.
Author contributions
C.L. and W.L. designed and performed experiments, analyzed data, and wrote the manuscript, J.C., K.Y., and Y.W. contributed to flow cytometric analysis and animal experiments. S.Z. and M.C. contributed to mouse model creation and animal experiments. F.C. contributed to the ex vivo experiment and data analysis. S.W. contributed to bioinformatics analysis and assisted with writing the manuscript. T.C., J.W., and C.D. conceived and supervised the study, interpreted data, revised the manuscript.
Declarations of competing interest
None.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 81874256, 82273571), the Key Program of the Natural Science Foundation of China (No. 81930090), the National Science Fund for Distinguished Young Scholars (No. 81725019), Chongqing Natural Science Foundation (cstc2015jcyjys10001). All authors would like to thank the professor Dengqun Liu for the gift of GFP-LC3 mice.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102661.
Contributor Information
Junping Wang, Email: wangjunping@tmmu.edu.cn.
Changhong Du, Email: changhongdu@tmmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Silvente-Poirot S., Poirot M. Cholesterol and cancer, in the balance. Science. 2014;343:1445–1446. doi: 10.1126/science.1252787. [DOI] [PubMed] [Google Scholar]
- 2.Song Y., Liu J., Zhao K., Gao L., Zhao J. Cholesterol-induced toxicity: an integrated view of the role of cholesterol in multiple diseases. Cell Metabol. 2021;33:1911–1925. doi: 10.1016/j.cmet.2021.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Yousefzadeh M.J., Flores R.R., Zhu Y., Schmiechen Z.C., Brooks R.W., Trussoni C.E., Cui Y., Angelini L., Lee K.-A., McGowan S.J., Burrack A.L., Wang D., Dong Q., Lu A., Sano T., O’Kelly R.D., McGuckian C.A., Kato J.I., Bank M.P., Wade E.A., Pillai S.P.S., Klug J., Ladiges W.C., Burd C.E., Lewis S.E., LaRusso N.F., Vo N.V., Wang Y., Kelley E.E., Huard J., Stromnes I.M., Robbins P.D., Niedernhofer L.J. An aged immune system drives senescence and ageing of solid organs. Nature. 2021;594:100–105. doi: 10.1038/s41586-021-03547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogeska R., Mikecin A.-M., Kaschutnig P., Fawaz M., Büchler-Schäff M., Le D., Ganuza M., Vollmer A., Paffenholz S.V., Asada N., Rodriguez-Correa E., Frauhammer F., Buettner F., Ball M., Knoch J., Stäble S., Walter D., Petri A., Carreño-Gonzalez M.J., Wagner V., Brors B., Haas S., Lipka D.B., Essers M.A.G., Weru V., Holland-Letz T., Mallm J.-P., Rippe K., Krämer S., Schlesner M., McKinney Freeman S., Florian M.C., King K.Y., Frenette P.S., Rieger M.A., Milsom M.D. Inflammatory exposure drives long-lived impairment of hematopoietic stem cell self-renewal activity and accelerated aging. Cell Stem Cell. 2022;29:1273–1284.e1278. doi: 10.1016/j.stem.2022.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeon O.H., Mehdipour M., Gil T.-H., Kang M., Aguirre N.W., Robinson Z.R., Kato C., Etienne J., Lee H.G., Alimirah F., Walavalkar V., Desprez P.-Y., Conboy M.J., Campisi J., Conboy I.M. Systemic induction of senescence in young mice after single heterochronic blood exchange. Nature Metabolism. 2022;4:995–1006. doi: 10.1038/s42255-022-00609-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo J., Yang H., Song B.-L. Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell Biol. 2020;21:225–245. doi: 10.1038/s41580-019-0190-7. [DOI] [PubMed] [Google Scholar]
- 7.Heyde A., Rohde D., McAlpine C.S., Zhang S., Hoyer F.F., Gerold J.M., Cheek D., Iwamoto Y., Schloss M.J., Vandoorne K., Iborra-Egea O., Muñoz-Guijosa C., Bayes-Genis A., Reiter J.G., Craig M., Swirski F.K., Nahrendorf M., Nowak M.A., Naxerova K. Increased stem cell proliferation in atherosclerosis accelerates clonal hematopoiesis. Cell. 2021;184:1348–1361.e1322. doi: 10.1016/j.cell.2021.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy A.J., Akhtari M., Tolani S., Pagler T., Bijl N., Kuo C.-L., Wang M., Sanson M., Abramowicz S., Welch C., Bochem A.E., Kuivenhoven J.A., Yvan-Charvet L., Tall A.R. ApoE regulates hematopoietic stem cell proliferation, monocytosis, and monocyte accumulation in atherosclerotic lesions in mice. J. Clin. Investig. 2011;121:4138–4149. doi: 10.1172/JCI57559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yvan-Charvet L., Pagler T., Gautier E.L., Avagyan S., Siry R.L., Han S., Welch C.L., Wang N., Randolph G.J., Snoeck H.W., Tall A.R. ATP-binding cassette transporters and HDL suppress hematopoietic stem cell proliferation. Science. 2010;328:1689–1693. doi: 10.1126/science.1189731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinho S., Frenette P.S. Haematopoietic stem cell activity and interactions with the niche. Nat. Rev. Mol. Cell Biol. 2019;20:303–320. doi: 10.1038/s41580-019-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao W., Du C., Wang J. The cGAS-STING pathway in hematopoiesis and its physiopathological significance. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.573915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oburoglu L., Tardito S., Fritz V., de Barros Stéphanie C., Merida P., Craveiro M., Mamede J., Cretenet G., Mongellaz C., An X., Klysz D., Touhami J., Boyer-Clavel M., Battini J.-L., Dardalhon V., Zimmermann Valérie S., Mohandas N., Gottlieb E., Sitbon M., Kinet S., Taylor N. Glucose and glutamine metabolism regulate human hematopoietic stem cell lineage specification. Cell Stem Cell. 2014;15:169–184. doi: 10.1016/j.stem.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Taya Y., Ota Y., Wilkinson A.C., Kanazawa A., Watarai H., Kasai M., Nakauchi H., Yamazaki S. Depleting dietary valine permits nonmyeloablative mouse hematopoietic stem cell transplantation. Science. 2016;354:1152–1155. doi: 10.1126/science.aag3145. [DOI] [PubMed] [Google Scholar]
- 14.Li C., Wu B., Li Y., Chen J., Ye Z., Tian X., Wang J., Xu X., Pan S., Zheng Y., Cai X., Jiang L., Zhao M. Amino acid catabolism regulates hematopoietic stem cell proteostasis via a GCN2-eIF2α axis. Cell Stem Cell. 2022;29:1119–1134.e1117. doi: 10.1016/j.stem.2022.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Kalaitzidis D., Lee D., Efeyan A., Kfoury Y., Nayyar N., Sykes D.B., Mercier F.E., Papazian A., Baryawno N., Victora G.D., Neuberg D., Sabatini D.M., Scadden D.T. Amino acid–insensitive mTORC1 regulation enables nutritional stress resilience in hematopoietic stem cells. J. Clin. Investig. 2017;127:1405–1413. doi: 10.1172/JCI89452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luchsinger L.L., Strikoudis A., Danzl N.M., Bush E.C., Finlayson M.O., Satwani P., Sykes M., Yazawa M., Snoeck H.-W. Harnessing hematopoietic stem cell low intracellular calcium improves their maintenance in vitro. Cell Stem Cell. 2019;25:225–240.e227. doi: 10.1016/j.stem.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y., Liao W., Chen J., Liu C., Zhang S., Yu K., Wang X., Chen M., Wang S., Ran X., Su Y., Cheng T., Wang J., Du C. Phosphate metabolic inhibition contributes to irradiation-induced myelosuppression through dampening hematopoietic stem cell survival. Nutrients. 2022;14:3395. doi: 10.3390/nu14163395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du C., Wang X., Wu Y., Liao W., Xiong J., Zhu Y., Liu C., Han W., Wang Y., Han S., Chen S., Xu Y., Wang S., Wang F., Yang K., Zhao J., Wang J. Renal Klotho and inorganic phosphate are extrinsic factors that antagonistically regulate hematopoietic stem cell maintenance. Cell Rep. 2022;38 doi: 10.1016/j.celrep.2022.110392. [DOI] [PubMed] [Google Scholar]
- 19.Chen X., Kang R., Kroemer G., Tang D. Ferroptosis in infection, inflammation, and immunity. J. Exp. Med. 2021;218 doi: 10.1084/jem.20210518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lei G., Zhang Y., Koppula P., Liu X., Zhang J., Lin S.H., Ajani J.A., Xiao Q., Liao Z., Wang H., Gan B. The role of ferroptosis in ionizing radiation-induced cell death and tumor suppression. Cell Res. 2020;30:146–162. doi: 10.1038/s41422-019-0263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang X., Stockwell B.R., Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 2021;22:266–282. doi: 10.1038/s41580-020-00324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng J., Conrad M. The metabolic underpinnings of ferroptosis. Cell Metabol. 2020;32:920–937. doi: 10.1016/j.cmet.2020.10.011. [DOI] [PubMed] [Google Scholar]
- 23.Ouyang S., You J., Zhi C., Li P., Lin X., Tan X., Ma W., Li L., Xie W. Ferroptosis: the potential value target in atherosclerosis. Cell Death Dis. 2021;12:782. doi: 10.1038/s41419-021-04054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ouled-Haddou H., Messaoudi K., Demont Y., Lopes dos Santos R., Carola C., Caulier A., Vong P., Jankovsky N., Lebon D., Willaume A., Demagny J., Boyer T., Marolleau J.-P., Rochette J., Garçon L. A new role of glutathione peroxidase 4 during human erythroblast enucleation. Blood Adv. 2020;4:5666–5680. doi: 10.1182/bloodadvances.2020003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma J., Ma H.-M., Shen M.-Q., Wang Y.Y., Bao Y.-X., Liu Y., Ke Y., Qian Z.-M. The role of iron in atherosclerosis in apolipoprotein E deficient mice. Front. Cardiovascular Med. 2022;9 doi: 10.3389/fcvm.2022.857933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu W., Chakraborty B., Safi R., Kazmin D., Chang C.-y., McDonnell D.P. Dysregulated cholesterol homeostasis results in resistance to ferroptosis increasing tumorigenicity and metastasis in cancer. Nat. Commun. 2021;12:5103. doi: 10.1038/s41467-021-25354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura-Ishizu A., Ito K., Suda T. Hematopoietic stem cell metabolism during development and aging. Dev. Cell. 2020;54:239–255. doi: 10.1016/j.devcel.2020.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Council N.R. 2010. Guide for the Care and Use of Laboratory Animals. [Google Scholar]
- 29.Wilkinson A.C., Ishida R., Nakauchi H., Yamazaki S. Long-term ex vivo expansion of mouse hematopoietic stem cells. Nat. Protoc. 2020;15:628–648. doi: 10.1038/s41596-019-0263-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du C., Xu Y., Yang K., Chen S., Wang X., Wang S., Wang C., Shen M., Chen F., Chen M., Zeng D., Li F., Wang T., Wang F., Zhao J., Ai G., Cheng T., Su Y., Wang J. Estrogen promotes megakaryocyte polyploidization via estrogen receptor beta-mediated transcription of GATA1. Leukemia. 2017;31:945–956. doi: 10.1038/leu.2016.285. [DOI] [PubMed] [Google Scholar]
- 31.Qian P., He X.C., Paulson A., Li Z., Tao F., Perry J.M., Guo F., Zhao M., Zhi L., Venkatraman A., Haug J.S., Parmely T., Li H., Dobrowsky R.T., Ding W.X., Kono T., Ferguson-Smith A.C., Li L. The dlk1-gtl2 locus preserves LT-HSC function by inhibiting the PI3K-mTOR pathway to restrict mitochondrial metabolism. Cell Stem Cell. 2016;18:214–228. doi: 10.1016/j.stem.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamoto R., Wilkinson A.C., Ooehara J., Lan X., Lai C.Y., Nakauchi Y., Pritchard J.K., Nakauchi H. Large-scale clonal analysis resolves aging of the mouse hematopoietic stem cell compartment. Cell Stem Cell. 2018;22:600–607.e604. doi: 10.1016/j.stem.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tolani S., Pagler T.A., Murphy A.J., Bochem A.E., Abramowicz S., Welch C., Nagareddy P.R., Holleran S., Hovingh G.K., Kuivenhoven J.A., Tall A.R. Hypercholesterolemia and reduced HDL-C promote hematopoietic stem cell proliferation and monocytosis: studies in mice and FH children. Atherosclerosis. 2013;229:79–85. doi: 10.1016/j.atherosclerosis.2013.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandes H., Moura J., Carvalho E. mTOR signaling as a regulator of hematopoietic stem cell fate. Stem Cell Rev. Rep. 2021;17:1312–1322. doi: 10.1007/s12015-021-10131-z. [DOI] [PubMed] [Google Scholar]
- 35.Castellano B.M., Thelen A.M., Moldavski O., Feltes M., van der Welle R.E., Mydock-McGrane L., Jiang X., van Eijkeren R.J., Davis O.B., Louie S.M., Perera R.M., Covey D.F., Nomura D.K., Ory D.S., Zoncu R. Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science. 2017;355:1306–1311. doi: 10.1126/science.aag1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y., Swanda R.V., Nie L., Liu X., Wang C., Lee H., Lei G., Mao C., Koppula P., Cheng W., Zhang J., Xiao Z., Zhuang L., Fang B., Chen J., Qian S.-B., Gan B. mTORC1 couples cyst(e)ine availability with GPX4 protein synthesis and ferroptosis regulation. Nat. Commun. 2021;12:1589. doi: 10.1038/s41467-021-21841-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park Y., Reyna-Neyra A., Philippe L., Thoreen C.C. mTORC1 balances cellular amino acid supply with demand for protein synthesis through post-transcriptional control of ATF4. Cell Rep. 2017;19:1083–1090. doi: 10.1016/j.celrep.2017.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bayeva M., Khechaduri A., Puig S., Chang H.-C., Patial S., Blackshear Perry J., Ardehali H. mTOR regulates cellular iron homeostasis through tristetraprolin. Cell Metabol. 2012;16:645–657. doi: 10.1016/j.cmet.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo H., Ouyang Y., Yin H., Cui H., Deng H., Liu H., Jian Z., Fang J., Zuo Z., Wang X., Zhao L., Zhu Y., Geng Y., Ouyang P. Induction of autophagy via the ROS-dependent AMPK-mTOR pathway protects copper-induced spermatogenesis disorder. Redox Biol. 2022;49 doi: 10.1016/j.redox.2021.102227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yi J., Zhu J., Wu J., Thompson C.B., Jiang X. Oncogenic activation of PI3K-AKT-mTOR signaling suppresses ferroptosis via SREBP-mediated lipogenesis. Proc. Natl. Acad. Sci. USA. 2020;117:31189–31197. doi: 10.1073/pnas.2017152117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu L., Li Y., Cao D., Qiu S., Li Y., Jiang C., Bian R., Yang Y., Li L., Li X., Wang Z., Ju Z., Zhang Y., Liu Y. SIRT3 inhibits gallbladder cancer by induction of AKT-dependent ferroptosis and blockade of epithelial-mesenchymal transition. Cancer Lett. 2021;510:93–104. doi: 10.1016/j.canlet.2021.04.007. [DOI] [PubMed] [Google Scholar]
- 42.Hoxhaj G., Manning B.D. The PI3K–AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer. 2020;20:74–88. doi: 10.1038/s41568-019-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y., Tan H., Daniels J.D., Zandkarimi F., Liu H., Brown L.M., Uchida K., O'Connor O.A., Stockwell B.R. Imidazole ketone erastin induces ferroptosis and slows tumor growth in a mouse lymphoma model. Cell Chem.l. Biol. 2019;26:623–633.e629. doi: 10.1016/j.chembiol.2019.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park E., Chung S.W. ROS-mediated autophagy increases intracellular iron levels and ferroptosis by ferritin and transferrin receptor regulation. Cell Death Dis. 2019;10:822. doi: 10.1038/s41419-019-2064-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Winther M.P.J., Lutgens E. The link between hematopoiesis and atherosclerosis. N. Engl. J. Med. 2019;380:1869–1871. doi: 10.1056/NEJMcibr1901397. [DOI] [PubMed] [Google Scholar]
- 46.Lusis A.J. A vicious cycle in atherosclerosis. Cell. 2021;184:1139–1141. doi: 10.1016/j.cell.2021.02.005. [DOI] [PubMed] [Google Scholar]
- 47.Owens A.P., III, Passam F.H., Antoniak S., Marshall S.M., McDaniel A.L., Rudel L., Williams J.C., Hubbard B.K., Dutton J.-A., Wang J., Tobias P.S., Curtiss L.K., Daugherty A., Kirchhofer D., Luyendyk J.P., Moriarty P.M., Nagarajan S., Furie B.C., Furie B., Johns D.G., Temel R.E., Mackman N. Monocyte tissue factor–dependent activation of coagulation in hypercholesterolemic mice and monkeys is inhibited by simvastatin. J. Clin. Investig. 2012;122:558–568. doi: 10.1172/JCI58969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dobbelstein M., Sørensen C.S. Exploiting replicative stress to treat cancer. Nat. Rev. Drug Discov. 2015;14:405–423. doi: 10.1038/nrd4553. [DOI] [PubMed] [Google Scholar]
- 49.Flach J., Bakker S.T., Mohrin M., Conroy P.C., Pietras E.M., Reynaud D., Alvarez S., Diolaiti M.E., Ugarte F., Forsberg E.C., Le Beau M.M., Stohr B.A., Méndez J., Morrison C.G., Passegué E. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 2014;512:198–202. doi: 10.1038/nature13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh S.K., Singh S., Gadomski S., Sun L., Pfannenstein A., Magidson V., Chen X., Kozlov S., Tessarollo L., Klarmann K.D., Keller J.R. Id1 ablation protects hematopoietic stem cells from stress-induced exhaustion and aging. Cell Stem Cell. 2018;23:252–265.e258. doi: 10.1016/j.stem.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parasassi T., Giusti A.M., Raimondi M., Ravagnan G., Sapora O., Gratton E. Cholesterol protects the phospholipid bilayer from oxidative damage. Free Radic. Biol. Med. 1995;19:511–516. doi: 10.1016/0891-5849(95)00038-y. [DOI] [PubMed] [Google Scholar]
- 52.Zhang X., Barraza K.M., Beauchamp J.L. Cholesterol provides nonsacrificial protection of membrane lipids from chemical damage at air–water interface. Proc. Natl. Acad. Sci. USA. 2018;115:3255–3260. doi: 10.1073/pnas.1722323115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guglielmelli P., Barosi G., Rambaldi A., Marchioli R., Masciulli A., Tozzi L., Biamonte F., Bartalucci N., Gattoni E., Lupo M.L., Finazzi G., Pancrazzi A., Antonioli E., Susini M.C., Pieri L., Malevolti E., Usala E., Occhini U., Grossi A., Caglio S., Paratore S., Bosi A., Barbui T., Vannucchi A.M., i A.-G.I.M.M. On behalf of the, Safety and efficacy of everolimus, a mTOR inhibitor, as single agent in a phase 1/2 study in patients with myelofibrosis. Blood. 2011;118:2069–2076. doi: 10.1182/blood-2011-01-330563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gutierrez-Martinez P., Hogdal L., Nagai M., Kruta M., Singh R., Sarosiek K., Nussenzweig A., Beerman I., Letai A., Rossi D.J. Diminished apoptotic priming and ATM signalling confer a survival advantage onto aged haematopoietic stem cells in response to DNA damage. Nat. Cell Biol. 2018;20:413–421. doi: 10.1038/s41556-018-0054-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dong S., Wang Q., Kao Y.-R., Diaz A., Tasset I., Kaushik S., Thiruthuvanathan V., Zintiridou A., Nieves E., Dzieciatkowska M., Reisz J.A., Gavathiotis E., D’Alessandro A., Will B., Cuervo A.M. Chaperone-mediated autophagy sustains haematopoietic stem-cell function. Nature. 2021;591:117–123. doi: 10.1038/s41586-020-03129-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rivnay B., Bergman S., Shinitzky M., Globerson A. Correlations between membrane viscosity, serum cholesterol, lymphocyte activation and aging in man. Mech. Ageing Dev. 1980;12:119–126. doi: 10.1016/0047-6374(80)90088-3. [DOI] [PubMed] [Google Scholar]
- 57.Chen C., Liu Y., Liu Y., Zheng P. mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci. Signal. 2009;2 doi: 10.1126/scisignal.2000559. ra75-ra75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim J., Guan K.-L. mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Biol. 2019;21:63–71. doi: 10.1038/s41556-018-0205-1. [DOI] [PubMed] [Google Scholar]
- 59.Martínez-Reyes I., Chandel N.S. Cancer metabolism: looking forward. Nat. Rev. Cancer. 2021;21:669–680. doi: 10.1038/s41568-021-00378-6. [DOI] [PubMed] [Google Scholar]
- 60.Bayik D., Lathia J.D. Cancer stem cell–immune cell crosstalk in tumour progression. Nat. Rev. Cancer. 2021;21:526–536. doi: 10.1038/s41568-021-00366-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary materials. Transcriptome datasets generated or analyzed in this study are available at Gene Expression Omnibus (GSE214035; RNA-seq) and ArrayExpress (E-MTAB-3079; RNA-seq).