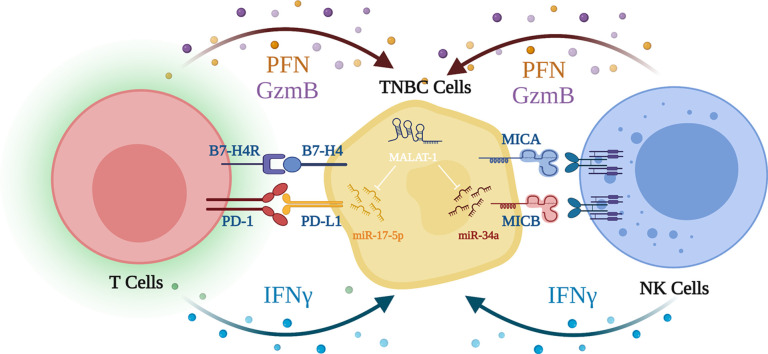
Abstract
Background
Triple negative breast cancer (TNBC) is known as hot immunogenic tumor. Yet, it is one of the most aggressive BC subtypes. TNBC evolve several tactics to evade the immune surveillance phenomena, one of which is shedding of natural killer (NK) cells activating immune ligands such as MICA/B and/or by inducing the expression of the immune checkpoints such as PD-L1 and B7-H4. MALAT-1 is an oncogenic lncRNA. MALAT-1 immunogenic profile is not well investigated.
Aim
The study aims at exploring the immunogenic role of MALAT-1 in TNBC patients and cell lines and to identify its molecular mechanism in altering both innate and adaptive immune cells present at the tumor microenvironment of TNBC
Methods
BC patients (n = 35) were recruited. Primary NK cells and cytotoxic T lymphocytes were isolated from normal individuals using the negative selection method. MDA-MB-231 cells were cultured and transfected by several oligonucleotides by lipofection technique. Screening of ncRNAs was performed using q-RT-PCR. Immunological functional analysis experiments were performed upon co-culturing primary natural killer cells and cytotoxic T lymphocytes using LDH assay. Bioinformatics analysis was performed to identify potential microRNAs targeted by MALAT-1.
Results
MALAT-1 expression was significantly upregulated in BC patinets with a profound expression in TNBC patients compared to their normal counterparts. Correlation analysis revealed a positive correlation between MALAT-1, tumor size and lymph node metastasis. Knocking down of MALAT-1 in MDA-MB-231 cells resulted in a significant induction of MICA/B, repression of PD-L1 and B7-H4 expression levels. Enhancement of cytotoxic activity of co-cultured NK and CD8+ cells with MALAT-1 siRNAs transfected MDA-MB-231 cells. In silico analysis revealed that miR-34a and miR-17–5p are potential targets to MALAT-1; accordingly, they were found to be downregulated in BC patients. Forcing the expression of miR-34a in MDA-MB-231 cells resulted in a significant induction in MICA/B levels. Ectopic expression of miR-17–5p in MDA-MB-231 cells significantly repressed the expression of PD-L1 and B7-H4 checkpoints. Validations of MALAT-1/miR-34a" and "MALAT-1/miR-17–5p axes were performed by a series of co-transfections and functional assessment of cytotoxic profile of primary immune cells.
Conclusion
This study proposes a novel epigenetic alteration exerted by TNBC cells mainly by inducing the expression of MALAT-1 lncRNA. MALAT-1 mediates innate and adaptive immune suppression events partially via targeting miR-34a/MICA/B and miR-175p/PD-L1/B7-H4 axes in TNBC patients and cell lines.
Keywords: MALAT-1, Natural killer cells, Cytotoxic t lymphocytes, Breast cancer, PD-L1, B7-H4, MICA/B, miR-34a, miR-17–5p
Graphical abstract
Introduction
Triple-negative breast cancer (TNBC) is a vastly heterogeneous malignancy that is highly associated with chemo-resistance, high frequencies of relapse, and metastasis [1,2]. TNBC patients do not benefit from endocrine or targeted therapy due to lack in the expression of human epidermal growth factor receptor 2 (HER2), estrogen and progesterone receptors [2], leaving chemotherapy as the mainstay of treatment for TNBC patients [3]. Consequently, immunotherapy represents a clinical promise for TNBC patients, notably immune checkpoint inhibitors that showed promising results in decreasing the episodes of relapse among TNBC patients [4]. The IMpassion130 trial, which combined atezolizumab (anti-PD-L1) with standard-of-care nab-paclitaxel to treat TNBC patients, was the first to show that immune checkpoint inhibitors benefit might be achieved in treating BC patients [5]. Followed by, the KEYNOTE-355 trial, which used pembrolizumab (anti-PD-1) in conjunction with various neoadjuvant chemotherapies (paclitaxel, nab-paclitaxel, or gemcitabine and carboplatin), improved progression-free survival of metastatic TNBC patients [6]. However, it has been recently reported that some of the TNBC patients started to develop either innate or acquired resistance to immune checkpoint inhibitors [7,8]. Resistance to immunotherapy might be explained by the numerous defense mechanisms that malignancies use to either thwart immunotherapy sensitivity (primary resistance) or evade an initial positive response (acquired resistance). Among these immunotherapy resistance mechanisms, is the ability of the tumor cells to reduce immune recognition via modifying their capability for proliferation and decreasing the production of neoantigens. On the other hand, tumor cells have the ability to increase the tumorigenic and immunosuppressive functions via changing the stromal and immune cells in the TME. Nevertheless, immune checkpoint inhibitors have been reported to cause a condition aptly known as “hyper-progressive disease” (HPD) which results in unanticipated and opposing consequences in specific patient subgroups of various tumor types, whose tumor burden rapidly advances [9]. The exact mechanism underlying HPD is still not comprehensively elucidated but some research linked it to the mutagenic overexpression of EGFR, MDM2 and FGF [10]. The multiple resistance mechanism by tumor cells to immunotherapy rendered it inevitable to elucidate the molecular mechanisms that underlie immunologic resistance in an attempt to improve the efficacy of immunotherapy and overcome resistance.
Recently, our research group has focused on a hot-topic in the immunoncology field where we found that vital non-cellular components of the tumor microenvironment (TME) of TNBC patients such as IL-10 and TNF-α might pose as a barrier to immune checkpoint inhibitors efficiency in some patients [11]. However, in order to have the full picture we thought of a study to decipher the cellular components of the TME of TNBC patients as well.
The immunosuppressive nature of the TME highlights the diminished role of the immune system where it lacks the tumor recognition and elimination processes yet participates in tumor progression through several processes of immune editing [12,13]. The immune editing processes occurs through a diverse array of mechanisms that involves evasion of cytotoxic response of both the innate cytotoxic arm; natural killer (NK) cells and the adaptive cytotoxic arm; CD8+ T lymphocytes. Among the tumor evasion tactics, is down-regulation of NK cells activation ligand MHC class I-related chain A and B (MICA, MICB) [1] and upregulation of immune checkpoint proteins programmed death ligand-1 (PD-L1) and B7-H4 on TNBC cells [14]. Such strategies represent a vicious evasion mechanism that obverts immune responses against tumors [15]. Another tactic that has been extensively studied by our research group is hijacking the immune-surveillance by TNBC tumors via genetic and epigenetic alterations which modulate several components of TME, consequently suppressing the infiltrating immune cellular activity [16], [17], [18], [19], [20], [21], [22]. Accordingly, re-sensitizing the cytotoxic immune cells at the TME through reversing these immunoediting processes is considered a milestone towards the development of more effective immunotherapeutic therapeutic modality.
Non-coding RNAs (ncRNAs) including long noncoding RNAs (lncRNAs) and short microRNAs (miRNAs), although do not encode for proteins but evidence showed that they act as key regulators in many biological processes [23,24]. Deregulation of lncRNAs in several types of cancers emphasizes its association with tumor progression and carcinogenesis [25]. Accordingly, this rendered lncRNAs prognostic biomarkers in many cancers including BC [26], [27], [28], [29]. Yet, the immunoncological side of several lncRNAs and their specific role at the tumor cell-immune cell synapse at the TME has been rarely investigated.
One of the most conserved and extremely abundant lncRNAs is Metastasis-associated lung adenocarcinoma transcript 1 (MALAT-1) which is approximately 8000 nucleotides in length [30]. MALAT-1 dysregulation has been associated with tumor development, cellular migration and invasion in many cancers [31]. MALAT-1 has been characterized as oncogenic lncRNA in BC and a vital promoter of epithelial-mesenchymal transition in BC cell lines [29,32]. High expression of MALAT-1 was highly correlated with poor prognosis, and metastasis in TNBC patients [33]. The interplay between lncRNAs and microRNAs (miRNAs) in modulating tumorigenesis has recently gained scientists attention; several studies concluded that MALAT-1 might exert its function via targeting/sponging miRNAs [34].
Recently, our research group showed that knockdown of MALAT-1, through its regulatory role on miRNAs, alleviated the immune suppressive TME of TNBC by inhibiting immune suppressor cytokines IL-10 and TNF-α produced by TNBC cells. miR-34a has been demonstrated to exhibit a tumor suppressor function; where its down-regulation has been associated with high proliferation and migration in TNBC [28,35]. Moreover, miR-17 5p was proven to possess an anti-tumor activity in TNBC through suppressing tumor invasion, migration and proliferation in-vitro and in-vivo [36]. Nonetheless, a positive immunomodulatory role of miR-34a and miR-17–5p was reported in several solid malignancies [37,38]. Yet, the exact immunomodulatory role of miR-34a and miR-17–5p in TNBC is still to be investigated. Thereby, in this study we attempt to unravel the impact of MALAT-1 on primary NK cells and CD8+ T cells cytotoxicity through investigating its regulatory role on an array of immune ligands and immune checkpoints expressed by TNBC cells.
Materials and procedures
Collection of human tissue samples
Thirty five (n=35) female BC patients participated in this study. The average age of the female participants was 48.8 years (range: 28–71). Samples were collected from Kasr Al-Aini hospitals. The samples were collected from March 2019 till September 2021. Female BC patients who underwent conservative mastectomy or lumpectomy had their tumors and the surrounding non-cancerous tissues resected. Following resection, tissues were immediately frozen in liquid nitrogen and preserved at -80 °C. The pathological examination of all samples was confirmed by pathology specialists. All patients included in this study gave their approval and signed an informed consent form. Ethical committee of Kasr Al-Aini hospital ethically approved the current study, which was conducted in compliance with the Declaration of Helsinki's ethical criteria. The clinicopathological characteristics of the patients included in this study is summarized in Table 1.
Table 1.
Clinico-pathological features of the recruited cohort of BC patients.
| Category | Frequency | Percent (%) |
|---|---|---|
| Age | ||
| ≤ 40 | 20 | 57.1% |
| > 40 | 15 | 42.8% |
| Histological Type | ||
| Invasive ductal carcinoma | 29 | 82.85% |
| Invasive lobular carcinoma | 7 | 20% |
| Both | 1 | 2.85% |
| Histologic grade | ||
| G1 | 1 | 2.85% |
| G2 | 27 | 77.14% |
| G3 | 7 | 20% |
| ER status | ||
| Negative | 17 | 48.5% |
| Positive | 18 | 51.4% |
| PR status | ||
| Negative | 17 | 48.5% |
| Positive | 18 | 51.4% |
| HER2 status | ||
| Negative | 28 | 80% |
| Positive | 7 | 20% |
| Ki-67 | ||
| Low (<14) | 9 | 25.7% |
| High (≥14) | 26 | 74.2% |
| Molecular subtype | ||
| Luminal A | 9 | 25.7% |
| Luminal B | 9 | 25.7% |
| HER2-enriched | 0 | 0% |
| Triple-negative | 17 | 48.5% |
| Tumor stage | ||
| Stage 1 | 7 | 20% |
| Stage 2 | 24 | 68.5% |
| Stage 3 | 3 | 8.57% |
Cell culture
For cell culture experiments, MDA-MB-231 TNBC cell line was purchased from VACSERA, Cairo, Egypt. Cells were cultured in DMEM (Dulbecco's Modified Eagle Medium) (Serox GmbH, Germany) containing 10% Fetal bovine serum (FBS) and 1% penicillin/streptomycin at 37 °C and 5% CO2 [39].
Oligonucleotides transfection in MDA-MB-231 cells
MDA-MB-231 cells were transfected with MALAT-1 siRNAs, miR-17–5p, and miR-34a mimics and antagomirs (Qiagen, Germany) using HiPerfect® Transfection Reagent (Qiagen, Germany) according to the manufacturer's protocol. When 80–90% cellular confluence was achieved, MDA-MB-231 cells were trypsinized, counted, and seeded into 24-well plates the day before transfection as previously described [15]. Transfection was done at least three times and repeated in quadruplicates for each experiment.
Total RNA extraction from MDA-MB-231 cells and BC tissues
To extract the total RNA from BC patient tissues and TNBC cell lines, Biazol reagent was purchased from Hangzhou Bioer Technology Co., China. Total RNA was extracted according to manufacturer protocol. The yield of the extracted DNA was quantified using spectrophotometer.
Quantitative real-time polymerase chain reaction analysis (qRT-PCR)
The following messenger RNA (mRNA): MALAT-1, MICA, MICB, PD-L1 and B7-H4, β-actin, and 18 s rRNA was reversely transcribed into complementary DNA (cDNA) using a high-capacity cDNA Reverse-Transcription Kit (ABI, California, and USA). Specific primers were utilized to reversibly transcribe the isolated miRNAs; hsa-miR 34a, hsa-miR-17–5p, and RNU6B into single stranded cDNA. The relative expressions of MALAT-1, MICA, MICB, PD-L1 and B7-H4 were determined by normalization to β-actin and 18s rRNA as reference genes. The relative expressions of miR-34a, miR-17–5p were determined by normalization to the RNU6 as reference gene. StepOneTM Systems from ABI in California was used to determine the relative expression [40]. Relative expression was calculated using the 2–ΔΔCT method. All PCR reactions were run in triplicates and repeated at least three times.
NK cell isolation
Peripheral blood mononuclear cells (PBMCs) were extracted from healthy donors' peripheral blood using Ficoll-Hypaque centrifugation (Axis-Shield PoC AS, Norway) within 4 h of blood collection. NK cells were isolated from PBMCs by using MACS NK cell isolation kit through the negative selection method (Miltenyi Biotec, Cologne, Germany). To ensure proper NK isolation, flow cytometric analysis was performed.
CD8+ T cell isolation
For CB8+ T cell isolation, PBMCs from healthy donors were used to isolate CB8+ T cells using CD8+ T Cell Isolation Kit, an LS Column, and a MidiMACS™ Separator (Miltenyi Biotec, Cologne, Germany). To ensure the purity of the primary CD8+ T Cell isolated from PBMCs, flow cytometric analysis was performed.
Lactate dehydrogenase (LDH) activity assay
The in-vitro cytotoxicity was assessed using the lactate dehydrogenase (LDH) activity assay kit (Lactate dehydrogenase (LDH) –Liquizyme, Spectrum, Egypt) in accordance with the manufacturer's instructions. Briefly, MDA-MB-231 cells, at a cell density of 20,000 cells/well, were transfected with different oligonucleotides and then seeded into a U-shaped 96-well plate. After 4 h, either NK cells or CD8+ T cells were introduced to MDA-MB-231 cells with a 5:1 or 3:1 effector to target (E:T) ratio, respectively. Effector and target cells were kept co-cultured for 10 h. This was followed by assessment of the in-vitro cytotoxicity. The following equation:% cytotoxicity = (target maximum release – experimental release)/ (target maximum release) × 100 was used to assess the percentage of lysis. The experiment was done in triplicate and repeated at least 3 times [41].
Statistical analysis
The mean and standard error of the mean (SEM) of at least three independent experiments are used to express data. The Student's t-test was used for the statistical analysis when comparison between 2 groups was performed. One-way ANOVA was carried out to compare two or more groups. Correlation analyses were evaluated using Spearman rank correlation analysis. P < 0.05 was considered statistically significant. Analysis was performed using the GraphPad Prism 5.0 software. All experiments were conducted at least 3 times and in triplicates or quadruplicates.
Results
MALAT-1 lncRNA is over-expressed in BC patients
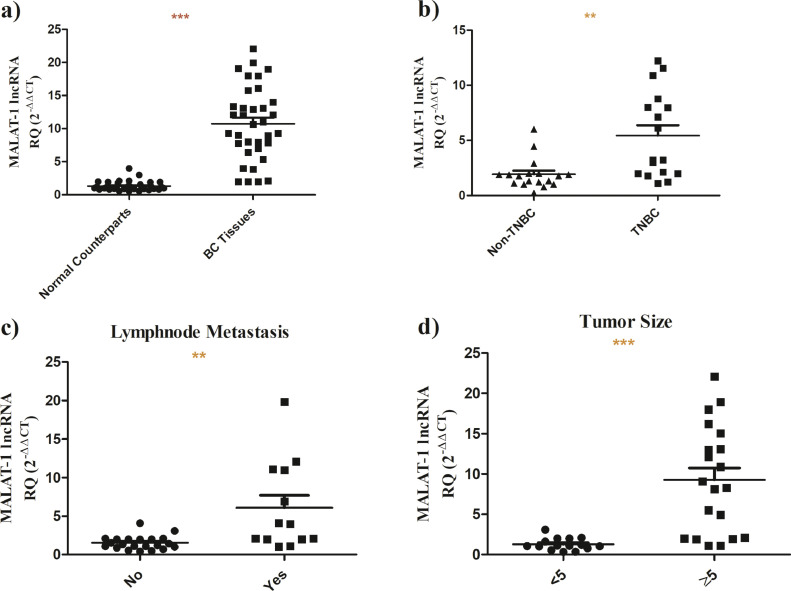
Screening of MALAT-1 revealed that it exhibits a higher expression in BC tissues compared to normal counterparts (n = 35, p<0.0001) (Fig. 1a). Upon stratification of BC patients, it was shown that MALAT-1 is strikingly over-expressed in TNBC tissues (n = 17) compared to its other BC subtypes (n = 18, P = 0.001) (Fig. 1b). Nonetheless, it was found that MALAT-1 is highly upregulated in BC patients with lymhnode metstasis (P = 0.0011) (Fig. 1c) and with bigger tumor size (P<0.0001) (Fig. 1d).
Fig. 1.
Screening of MALAT-1 lncRNA in BC patients.
MALAT-1 expression profile was analyzed in BC patients using 18srRNA as a housekeeping gene using q-RT-PCR. a) MALAT-1 expression is highly expressed in BC tissues compared to normal counterparts b) MALAT-1 is remarkably overexpressed in TNBC tissues compared to its other BC subtypes. c) MALAT-1 is significantly overexpressed in patients BC with lymph node metastasis. d) MALAT-1 is substantially elevated in BC patients with larger tumor sizes.
MALAT-1 lncRNA boosts NK cells and CD8+ T cells cytotoxic activities
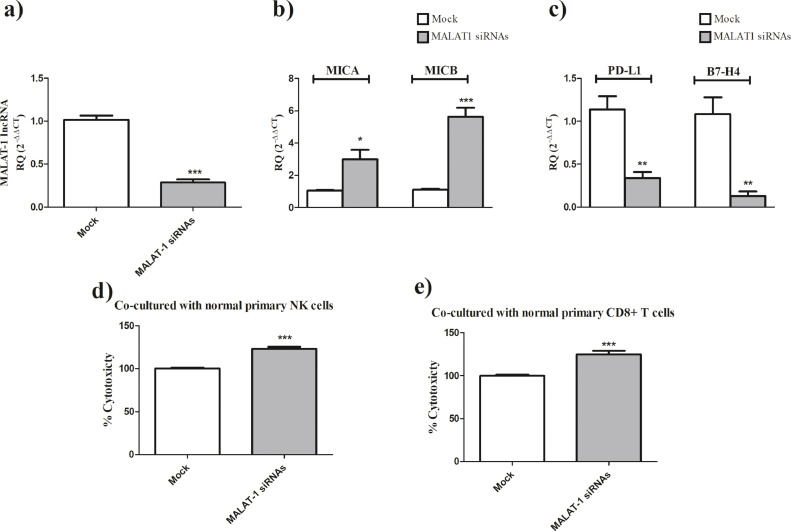
Upon knocking down of MALAT-1 in MDA-MB-231 cells using MALAT-1 siRNAs (Fig. 2a, P<0.0001), a marked increase in the expression of the activating immune ligands; MICA (P = 0.0114) and MICB (P<0.0001) (Fig. 2b) and repression of immune checkpoints PD-L1 (P = 0.0016) and B7-H4 (P = 0.0019) (Fig. 2c). On the functional level, MALAT-1 knockdown in MDA-MB-231 cells resulted in an augmented increase in NK cells-mediated killing (P<0.0001) (Fig. 2d) and CD8+ T cells-mediated cytotoxic activity (P = 0.0003) (Fig. 2e).
Fig. 2.
MALAT-1 siRNAs augments NK cells and CD8+ T cells cytotoxic activities.
(a) Transfection efficiency of MALAT-1 siRNAs in MDA-MB-231 cells was confirmed. MALAT-1 siRNAs significantly decreased the expression of MALAT-1 compared to mock cells. (b) Efficient knocking down of MALAT-1 in MDA-MB-231 cells resulted in a significant induction of the NK cell activating immune ligands MICA and MICB. (c) knock down of MALAT-1 in MDA-MB-231 cells, resulted in the repression of immunological checkpoints PD-L1 and B7-H4. d) Primary NK cells co-cultured with MDA-MB-231 cells transfected MALAT-1 siRNAs showed an enhanced cytotoxicity compared to mock cells. (e) Similarly, primary CD8+ cells co-cultured with MDA-MB-231 cells transfected MALAT-1 siRNAs showed an enhanced cytotoxicity compared to mock cells.
miR-34a and miR-17–5p are down-regulated in bc patients and inversely correlated with MALAT-1 expression
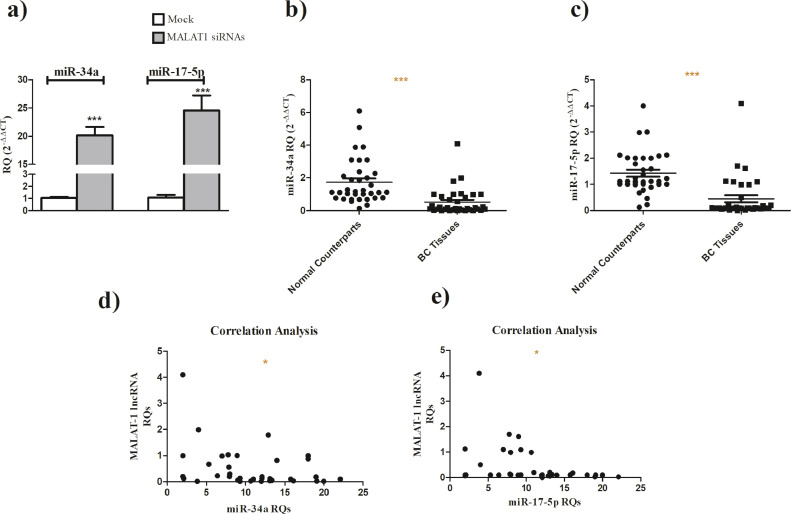
Knockind down of MALAT-1 in MDA-MB-231 cells resulted in a significant increase in miR-34a and miR-17-5p expression levels (Fig. 3a). Screening of miR-34a (Fig. 3b, P<0.0001) and miR-17–5p (Fig. 3c, P<0.0001) in BC patients revealed that both miRNAs are down-regulated in BC tissues when compared to their normal counterparts. Moreover, results showed that miR-34a (P = 0.0238, Spearman r =−0.3813) (Fig. 3d) and miR-17–5p (P = 0.0303, Spearman r =−0.3666) (Fig. 3e) expression levels are inversely correlated with MALAT-1 expression level in the same patients.
Fig. 3.
miR-34a and miR-17–5p in BC patients and their correlation with MALAT-1 levels.
(a) miR-34a and miR-17–5p expression is upregulated following knockind down of MALAT-1 using MALAT-1 siRNAs when compared to mock cells. (b) miR-34a expression in BC patients tissues is downregulated compared to normal counterparts. (c) miR-17–5p expression in BC patients tissues is downregulated compared to normal counterparts. (d) Expression level of miR-34a was inversely correlated with MALAT-1 expression levels in the same patients. (e) Expression of miR-17-5p is inversely correlated with MALAT-1 expression levels in the same patients.
miR-34a is an activator of NK cells mediated cytotoxicity
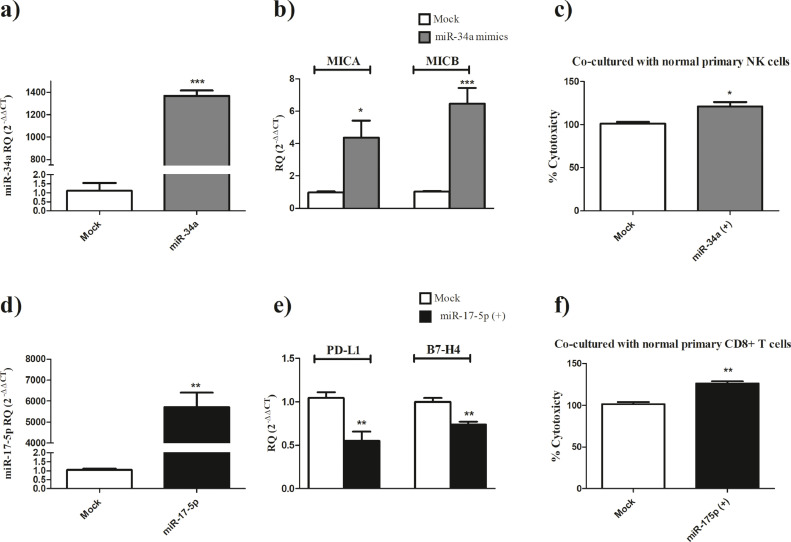
Ectopic expression of miR-34a in MDA-MB-231 cells (Fig. 4a, P<0.0001) resulted in a significant induction of MICA (P = 0.0124) and MICB (P = 0.0005) expression levels (Fig. 4b) and consequently an increase in NK cells mediated cytotoxicity against MDA-MB-231 cells transfected with miR-34a mimics compared to mock cells (P = 0.0129) (Fig. 4c).
Fig. 4.
Immunomodulatory role of miR-34a and miR-17–5p in TNBC
a) Ectopic expression of miR-34a using miR-34a mimics in MDA-MB-231 showed more than 1300 fold increase in mimicked cells compared to mock cells. b) MICA and MICB expression levels are induced following ectopic expression of miR-34a in MDA-MB-231 cells compared to mock cells c) Cellular-mediated-cytotoxicity of primary NK cells is enhanced when co-cultured with MDA-MB-231 cells that were transfected with miR-34a mimics compared to mock cells. d) Transfection efficiency of miR-17–5p mimics was confirmed in MDA-MB-231 cells transfected with miR-17–5p mimics showing more than 5000 fold increase in mimicked cells compared to mock cells. e) PD-L1 and B7-H4 expression levels are repressed following ectopic expression of miR-17–5p in MDA-MB-231 cells compared to mock cells consequently d) CD8+ T cell cytotoxicity is enhanced when co-cultured with MDA-MB-231 cells that were transfected with miR-17–5p mimics compared to mock cells.
miR-17–5p is an activator of CD8+ T cells mediated cytotoxicity
Ectopic expression of miR-17–5p in MDA-MB-231 cells (Fig. 4c, P = 0.0012) resulted in a significant repression of PD-L1 (P = 0.0075) and B7-H4 (P = 0.0041) expression levels (Fig. 4d) and consequently a marked increase in CD8+ T cells mediated cytotoxicity against MDA-MB-231 cells transfected with miR-17–5p mimics compared to mock cells (P = 0.0024 Fig. 4e).
Involvement of miR-34a and miR-17–5p in mediating MALAT-1 induced effects of innate and adaptive immune arms
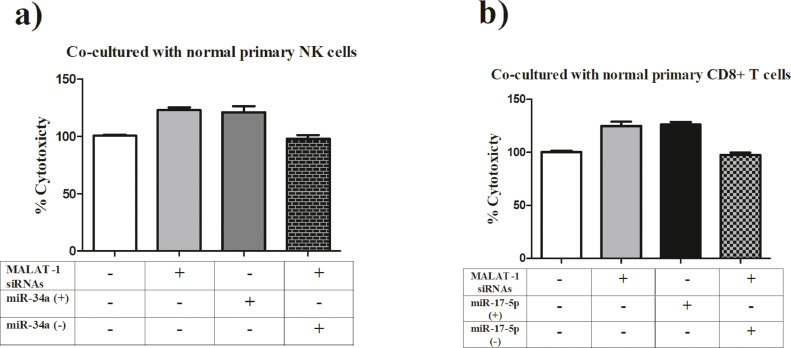
To validate that MALAT-1 mediates its effect on NK cells-mediated cytotoxic activity via miR-34a, MDA-MB-231 cells were co-transfected with MALAT-1 siRNAs and miR-34a antagomirs, consequently this abrogated the cytotoxic activity of NK cells compared to MDA-MB-231 cells treated with either MALAT-1 siRNAs or miR-34a mimics (Fig. 5a). On the adaptive immune arm, to validate involvement of miR-17–5p in mediating MALAT-1 effects on CD8+ T cells, MDA-MB-231 cells were co-transfected with MALAT-1 siRNAs and miR-17–5p antagomirs which resulted in a total abrogation of CD8+ T cells cytotoxic effects compared to MDA-MB-231 cells treated with either MALAT-1 siRNAs or miR-17–5p mimics (Fig. 5b).
Fig. 5.
Involvement of miR-34a and miR-17–5p in mediating MALAT-1 induced effects of innate and adaptive immune arms.
a) To verify that MALAT-1 mediates its effects on NK cell-mediated cytotoxic activity through miR-34a, MDA-MB-231 cells were co-transfected with both MALAT-1 siRNAs and miR-34a antagomirs was prepared. This abolished NK cytotoxic activity in comparison to MDA-MB-231 cells treated with MALAT-1 siRNAs or miR-34a mimics. b) To verify the involvement of miR-17–5p in mediating MALAT-1 effects on CD8+ T cells, MDA-MB-231 cells were co-transfected with MALAT-1 siRNAs and miR-17–5p antagomirs, this resulted in a significant reduction of the cytotoxic effects of CD8+ T cells compared with MDA-MB-231 cells treated with MALAT-1 siRNAs or miR-17–5p mimics.
Discussion
The majority of TNBC patients suffer from poor treatment outcomes and chemo-resistance as a result [3]. Although it has long been believed that immunotherapeutic approaches, particularly immune checkpoint inhibitors, offer TNBC patients a clinical advantage by reducing relapse occurrences, many patients have begun to show signs of innate and adaptive immunological resistance to immune checkpoint inhibitors [7,8]. Patients' resistance to immune checkpoint inhibitors appears to be caused by an unknown mechanism. Our research team recently identified a novel mechanism by which IL-10 and TNF-α reduce immune checkpoint inhibitors effectiveness in some individuals in an effort to unravel this mystery [11]. In the present study, we unlocked another piece in the puzzle by probing the cytotoxic potential of other cellular components at the TME of TNBC patients.
Scarce data exists regarding the role of ncRNAs in shaping the TME behavior. Therefore, in this study the authors focused on investigating the immunoregulatory role of MALAT-1 in modulating the expression of immune ligands, in particular MICA, MICB, PD-L1 and B7-H4 in TNBC cells. Moreover, we attempted in this study to unravel the mechanism by which MALAT-1 mediates its immunomodulatory effect. Screening of MALAT-1 revealed that it exhibits a higher expression in BC tissues compared to its normal counterparts. We also found that MALAT-1 is significantly upregulated in TNBC patients in particular compared to non-TNBC patients. Such elevated expression of MALAT-1 in an aggressive subtype such as TNBC confirms the role of MALAT-1 as a poor prognostic biomarker in BC [42,43].
It is also worth mentioning that in our pool of patients MALAT-1 is significantly elevated in BC patients with lymph node metastasis and larger tumor size. This goes in line with some recent studies highlighting the direct correlation of MALAT-1 expression and distal metastasis as well as reduced overall survival in HR+ and HR−BC patients [44,45]. However, as far as our knowledge goes, this study is the first to show the association of MALAT-1 expression with BC tumor size.
Tumor immunoediting continues to be an inevitable barrier to immune-surveillance rendering TNBC patients to become resistant to novel immunotherapeutic therapies. Noteworthy, MALAT-1 has been recently casted as a multi-tasker lncRNA that could modulate the sensitivity of cancer cells to several anti-cancer agents such as Herceptin [46] and Metformin [47]. However, its immunomodulatory role and involvement at the tumor-immune cell synapse has been rarely investigated. Among the tumor evasion tactics is evading NK cells through shedding of NK-activating ligands MICA and MICB [1,40]. Nonetheless, tumor cells over-express immune checkpoints such as B7-H4 and PD-L1 to provide inhibitory signals to T cells and thus evading both arms of immune system; innate and adaptive arms. Yet, the exact mechanisms by which tumor cells can escape immune-surveillance through modulating these immune ligands are still not fully understood. Several reports have recently shed the light onto the central role of ncRNAs in modulating immune ligands [48], [49], [50]. However, MALAT-1 lncRNA has been rarely investigated in such context.
This study showed that upon knocking down of MALAT-1 in MDA-MB-231 cells, a marked increase in the expression of the activating immune ligands; MICA and MICB and dual-repression of immune checkpoints PD-L1 and B7-H4 have been observed. On the immunological level, MALAT-1 knockdown in MDA-MB-231 cells resulted in an augmented increase in NK cells-mediated killing and CD8+ T cells-mediated cytotoxic activity against transfected TNBC cells. This goes in line with a recent study showing that knocking down of MALAT-1 in hepatocellular carcinoma cell lines resulted in a repression of PD-L1 expression [51]. However, our study was the first to show the modulatory role of MALAT-1 on NK cells immune ligands and B7-H4 in TNBC cells.
It was essential to unravel the mechanism by which MALAT-1 could multi-strike the respective immune ligands reported in this study. It is noteworthy that MALAT-1 has been highly involved in several ceRNAs circuits in several oncological contexts such as MALAT-1/miR-1914 regulating YAP oncogenic pathway [52], and MALAT-1/miR-124 modulating STAT3 transcription factor [53] in non-small cell lung cancer and MALAT-1/miR-155/miR-146a regulating nitric oxide production in BC [7]. Accordingly, we have hypothesized a possible ceRNA circuit underneath the immunomodulatory role of MALAT-1 reported in this study. Based on previous literature and in-silco analysis, miR-34a and miR-17–5p were selected as possible candidates for proposed ceRNA circuit. This was supported by evidence that was reported by Li et al. that MALAT-1 could directly sponge miR-34a in melanoma cells [54] and directly target miR-17–5p in macrophages [55]. Nonetheless, our group and others have recently proven the tumor suppressor effects of both miRNAs in TNBC cells [28,56] and their possible immunomodulatory role in several malignancies [57,58]. To confirm our hypothesis, expression profiling of the selected miRNAs was performed in the same cohort of BC patients recruited in the current study. The results showed that both miR-34a and miR-17–5p are downregulated in BC tumors compared to its normal counterparts. Moreover, the expression of both miR-34a and miR-17–5p were inversely correlated with MALAT-1 expression level in the same BC patients. Furthermore, we confirmed that miR-34a is a potent immune activator of NK cells cytotoxicity mainly by inducing the expression of the NKG2D activating immune ligands MICA/B expression on TNBC cells. This goes in line with recent study reporting the inducing effects of miR-34a mimics on the expression of MICB in hepatocellular carcinoma cells as well as precancerous hepatocytes [59]. miR-34a was also found to induce the expression of another NKG2D ligand known as ULBP2 in melanoma cells [60]; thus supporting the innate immune activator role of miR-34a in several oncological and non-oncological contexts. On the other hand, our research group has recently highlighted the immunomodulatory role of miR-17–5p in TNBC mainly through alleviating the immune suppressive TME upon dampening the release of IL-10 and TNF-α from TNBC cells at the TME [11]. Ohno et.al. suggested that transfection of miR-17–5p could enhance the efficacy of immunotherapy through preventing antigen escape [61]. In the present study, the authors showed that ectopic expression of miR-17–5p in MDA-MB-231 cells resulted in a significant repression of PD-L1 and B7-H4 expression levels and consequently a marked increase in CD8+ T cell mediated cytotoxicity against MDA-MB-231 cells transfected with miR-17–5p mimics. These findings support the potential utilization of miR-17–5p mimics to enhance the efficacy of immune therapeutic modalities and revert the tumor-immune escape.
Finally, the involvement of miR-34a and miR-17–5p in MALAT-1 regulatory effect on immune ligands was validated when co-transfection of MDA-MB-231 with MALAT-1 siRNAs and miR-34a and/or miR-17–5p mimics and antagomirs. Our data suggests a novel mechanism by which MALAT-1 paralyzes the immune system and aids in tumor escape. In the current study, the authors explained how MALAT-1 manipulates the expression of MICA/MICB and PD-L1/B7-H4 by highlighting a novel role of miR-34a and miR-17–5-p downstream the immunomodulatory MALAT-1. This study gives a robust evidence that combination of efficient knocking down of MALAT-1 together with miR-34a or miR-17–5p might have a beneficial effect if combined with immune checkpoint inhibitors as they might decrease the chance of resistance in TNBC patients.
In conclusion, our study helped in a better understanding of the process of immune escape through shedding the light on a novel mechanism by which MALAT-1 diminishes immune responses in TNBC patients via manipulating the expression of miR-34a and miR-17–5p and consequently modulating MICA//MICB, PDL1 and B7-H4 expression on TNBC cells. This study paves the road for the future implications of RNA-based therapeutics in future clinical implication as it highlights a potential re-sensitizing potential of several ncRNAs for the innate and adaptive immunity against TNBC.
CRediT authorship contribution statement
Radwa Y. Mekky: Investigation, Formal analysis, Methodology, Writing – original draft. Mai F. Ragab: Data curation, Formal analysis, Validation, Writing – review & editing. Tamer Manie: Supervision, Data curation, Formal analysis, Writing – review & editing. Abdelrahman A. Attia: Investigation, Formal analysis, Methodology, Validation, Writing – review & editing. Rana A. Youness: Investigation, Data curation, Formal analysis, Methodology, Supervision, Validation, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
All authors declare no conflict of interest.
References
- 1.Abdel-Latif M., Youness R.A. Why natural killer cells in triple negative breast Cancer? World J. Clin. Oncol. 2020;11:464–476. doi: 10.5306/wjco.v11.i7.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fahad Ullah M. Breast Cancer: current perspectives on the disease status. Adv. Exp. Med. Biol. 2019;1152:51–64. doi: 10.1007/978-3-030-20301-6_4. [DOI] [PubMed] [Google Scholar]
- 3.Won K.A., Spruck C. Triple‑negative breast cancer therapy: current and future perspectives (Review) Int. J. Oncol. 2020;57:1245–1261. doi: 10.3892/ijo.2020.5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keenan T.E., Tolaney S.M. Role of immunotherapy in triple-negative breast Cancer. J. Natl. Compr. Canc. Netw. 2020;18:479–489. doi: 10.6004/jnccn.2020.7554. [DOI] [PubMed] [Google Scholar]
- 5.Schmid P., Adams S., Rugo H.S., Schneeweiss A., Barrios C.H., Iwata H., Diéras V., Hegg R., Im S.A., Shaw Wright G., Henschel V., Molinero L., Chui S.Y., Funke R., Husain A., Winer E.P., Loi S., Emens L.A. Atezolizumab and nab-paclitaxel in advanced triple-negative breast Cancer. N. Engl. J. Med. 2018;379:2108–2121. doi: 10.1056/NEJMoa1809615. [DOI] [PubMed] [Google Scholar]
- 6.Cortes J., Cescon D.W., Rugo H.S., Nowecki Z., Im S.A., Yusof M.M., Gallardo C., Lipatov O., Barrios C.H., Holgado E., Iwata H., Masuda N., Otero M.T., Gokmen E., Loi S., Guo Z., Zhao J., Aktan G., Karantza V., Schmid P. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): a randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet. 2020;396:1817–1828. doi: 10.1016/s0140-6736(20)32531-9. [DOI] [PubMed] [Google Scholar]
- 7.Abdel-Latif M., Riad A., Soliman R.A., Elkhouly A.M., Nafae H., Gad M.Z., Motaal A.A., Youness R.A. MALAT-1/p53/miR-155/miR-146a ceRNA circuit tuned by methoxylated quercitin glycoside alters immunogenic and oncogenic profiles of breast cancer. Mol. Cell. Biochem. 2022;477:1281–1293. doi: 10.1007/s11010-022-04378-4. [DOI] [PubMed] [Google Scholar]
- 8.Kim H., Choi J.M., Lee K.M. Immune checkpoint blockades in triple-negative breast Cancer: current state and molecular mechanisms of resistance. Biomedicines. 2022;10 doi: 10.3390/biomedicines10051130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adashek J.J., Subbiah I.M., Matos I., Garralda E., Menta A.K., Ganeshan D.M., Subbiah V. Hyperprogression and immunotherapy: fact, fiction, or alternative fact? Trends Cancer. 2020;6:181–191. doi: 10.1016/j.trecan.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato S., Goodman A., Walavalkar V., Barkauskas D.A., Sharabi A., Kurzrock R. Hyperprogressors after immunotherapy: analysis of genomic alterations associated with accelerated growth rate. Clin. Cancer Res. 2017;23:4242–4250. doi: 10.1158/1078-0432.Ccr-16-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soliman R.A., Youness R.A., Manie T.M., KHALLAF E., El-Shazly M., Abdelmohsen M., Handoussa H., Gad M.Z. Uncoupling tumor necrosis factor-α and interleukin-10 at tumor immune microenvironment of breast Cancer through miR-17-5p/MALAT-1/H19 circuit. Biocell. 2022;46:769–783. [Google Scholar]
- 12.Deepak K.G.K., Vempati R., Nagaraju G.P., Dasari V.R., Nagini S., Rao D.N., Malla R.R. Tumor microenvironment: challenges and opportunities in targeting metastasis of triple negative breast cancer. Pharmacol. Res. 2020;153 doi: 10.1016/j.phrs.2020.104683. [DOI] [PubMed] [Google Scholar]
- 13.O'Sullivan T., Saddawi-Konefka R., Vermi W., Koebel C.M., Arthur C., White J.M., Uppaluri R., Andrews D.M., Ngiow S.F., Teng M.W., Smyth M.J., Schreiber R.D., Bui J.D. Cancer immunoediting by the innate immune system in the absence of adaptive immunity. J. Exp. Med. 2012;209:1869–1882. doi: 10.1084/jem.20112738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Din G.S., Youness R.A., Assal R.A., Gad M.Z. miRNA-506-3p directly regulates rs10754339 (A/G) in the immune checkpoint protein B7-H4 in breast cancer. Microrna. 2020;9:346–353. doi: 10.2174/2211536609666201209152949. [DOI] [PubMed] [Google Scholar]
- 15.Awad A.R., Youness R.A., Ibrahim M., Motaal A.A., El-Askary H.I., Assal R.A., Gad M.Z. An acetylated derivative of vitexin halts MDA-MB-231 cellular progression and improves its immunogenic profile through tuning miR- 20a-MICA/B axis. Nat. Prod. Res. 2021;35:3126–3130. doi: 10.1080/14786419.2019.1686372. [DOI] [PubMed] [Google Scholar]
- 16.Selem N., Nafae H., Youness R.A., Gad M.Z. 32P Immunoregulatory loop between let-7a and CCAT1 lncRNA coordinated by c-Myc underlies the PD-1/PD-L1 immunoresistance in triple negative breast cancer patients. Ann. Oncol. 2021;32:S1355. doi: 10.1016/j.annonc.2021.08.2028. [DOI] [Google Scholar]
- 17.Elkhouly A., Youness R.A., Gad M. miR-486-5p counteracts the shedding of MICA/B and CD155 immune-ligands in TNBC Patients. Ann. Oncol. 2019;30:xi60–xi61. doi: 10.1093/annonc/mdz450.009. [DOI] [Google Scholar]
- 18.Soliman R.A., Youness R.A., Handoussa H., El-Shazly M., Gad M.Z. Interplay between miR-17-5p and MALAT-1 shapes the cytokine storm in triple negative breast cancer (TNBC) tumor microenvironment. Ann. Oncol. 2019;30:v769. doi: 10.1093/annonc/mdz268.024. [DOI] [Google Scholar]
- 19.Soliman R., Youness R.A., El-Shazly M., Handoussa H., Gad M. Regulatory interacting network between the immunomodulatory non-coding RNAs: miR-17-5p, MALAT1 and H19 lncRNAs in modulating the tumour microenvironment in TNBC. Ann. Oncol. 2019;30:xi57. doi: 10.1093/annonc/mdz452.032. [DOI] [Google Scholar]
- 20.Abdel-Latif M., Afifi A., Soliman R., Elkhouly A., Abdelmotaal A., Youness R.A. A new quercetin glycoside enhances TNBC immunological profile through TP53/miR-155/MICA/ULBP2. Ann. Oncol. 2019;30:vii7–vii8. doi: 10.1093/annonc/mdz413.028. [DOI] [Google Scholar]
- 21.Youness R.A., Abdelmotaal A. 13P A mitigation of breast cancer-induced immune-suppressive tumor microenvironment through curbing miR-155/IL-10/TNF-α loop using a novel quercetin derivative. Ann. Oncol. 2020;31:S4. doi: 10.1016/j.annonc.2020.01.061. [DOI] [Google Scholar]
- 22.Selem N., Nafae H., Youness R.A., Gad M.Z. 28P Hijacking CCAT1/miR-17-5p axis alleviates immune checkpoint blockers resistance in PDL1+ TNBC patients. Ann. Oncol. 2021;32:S12. doi: 10.1016/j.annonc.2021.01.042. [DOI] [Google Scholar]
- 23.Youness R.A., Gad M.Z. Long non-coding RNAs: functional regulatory players in breast cancer. Noncoding RNA Res. 2019;4:36–44. doi: 10.1016/j.ncrna.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.ElKhouly A.M., Youness R.A., Gad M.Z. MicroRNA-486-5p and microRNA-486-3p: multifaceted pleiotropic mediators in oncological and non-oncological conditions. Noncoding RNA Res. 2020;5:11–21. doi: 10.1016/j.ncrna.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 26.Selem N.A., Youness R.A., Gad M.Z. What is beyond LncRNAs in breast cancer: a special focus on colon cancer-associated transcript-1 (CCAT-1) Noncoding RNA Res. 2021;6:174–186. doi: 10.1016/j.ncrna.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abdallah R.M., Elkhouly A.M., Soliman R.A., El Mechawy N., El Sebaei A., Motaal A.A., El-Askary H., Youness R.A., Assal R.A. Hindering the synchronization between miR-486-5p and H19 lncRNA by hesperetin halts breast cancer aggressiveness through tuning ICAM-1. Anticancer Agents Med. Chem. 2022;22:586–595. doi: 10.2174/1871520621666210419093652. [DOI] [PubMed] [Google Scholar]
- 28.Youness R.A., Hafez H.M., Khallaf E., Assal R.A., Abdel Motaal A., Gad M.Z. The long noncoding RNA sONE represses triple-negative breast cancer aggressiveness through inducing the expression of miR-34a, miR-15a, miR-16, and let-7a. J. Cell. Physiol. 2019;234:20286–20297. doi: 10.1002/jcp.28629. [DOI] [PubMed] [Google Scholar]
- 29.Nafea H., Youness R.A., Abou-Aisha K., Gad M.Z. LncRNA HEIH/miR-939-5p interplay modulates triple-negative breast cancer progression through NOS2-induced nitric oxide production. J. Cell. Physiol. 2021;236:5362–5372. doi: 10.1002/jcp.30234. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X., Hamblin M.H., Yin K.J. The long noncoding RNA malat1: its physiological and pathophysiological functions. RNA Biol. 2017;14:1705–1714. doi: 10.1080/15476286.2017.1358347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhuo M., Yuan C., Han T., Cui J., Jiao F., Wang L. A novel feedback loop between high MALAT-1 and low miR-200c-3p promotes cell migration and invasion in pancreatic ductal adenocarcinoma and is predictive of poor prognosis. BMC Cancer. 2018;18:1032. doi: 10.1186/s12885-018-4954-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y., Zhou Y., Yang Z., Chen B., Huang W., Liu Y., Zhang Y. MiR-204/ZEB2 axis functions as key mediator for MALAT1-induced epithelial-mesenchymal transition in breast cancer. Tumour Biol. 2017;39 doi: 10.1177/1010428317690998. [DOI] [PubMed] [Google Scholar]
- 33.Ou X., Gao G., Bazhabayi M., Zhang K., Liu F., Xiao X. MALAT1 and BACH1 are prognostic biomarkers for triple-negative breast cancer. J. Cancer Res. Ther. 2019;15:1597–1602. doi: 10.4103/jcrt.JCRT_282_19. [DOI] [PubMed] [Google Scholar]
- 34.Zuo Y., Li Y., Zhou Z., Ma M., Fu K. Long non-coding RNA MALAT1 promotes proliferation and invasion via targeting miR-129-5p in triple-negative breast cancer. Biomed. Pharmacother. 2017;95:922–928. doi: 10.1016/j.biopha.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Li L., Yuan L., Luo J., Gao J., Guo J., Xie X. MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin. Exp. Med. 2013;13:109–117. doi: 10.1007/s10238-012-0186-5. [DOI] [PubMed] [Google Scholar]
- 36.Li J., Lai Y., Ma J., Liu Y., Bi J., Zhang L., Chen L., Yao C., Lv W., Chang G., Wang S., Ouyang M., Wang W. miR-17-5p suppresses cell proliferation and invasion by targeting ETV1 in triple-negative breast cancer. BMC Cancer. 2017;17:745. doi: 10.1186/s12885-017-3674-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yong H., Fu J., Gao G., Shi H., Zheng D., Zhou X. MiR-34a suppresses the proliferation and invasion of gastric cancer by modulating PDL1 in the immune microenvironment. Mol. Cell. Probes. 2020;53 doi: 10.1016/j.mcp.2020.101601. [DOI] [PubMed] [Google Scholar]
- 38.Cui Z.J., Xie X.L., Qi W., Yang Y.C., Bai Y., Han J., Ding Q., Jiang H.Q. Cell-free miR-17-5p as a diagnostic biomarker for gastric cancer inhibits dendritic cell maturation. Onco Targets Ther. 2019;12:2661–2675. doi: 10.2147/ott.S197682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Youness R.A., Assal R.A., Abdel Motaal A., Gad M.Z. A novel role of sONE/NOS3/NO signaling cascade in mediating hydrogen sulphide bilateral effects on triple negative breast cancer progression. Nitric Oxide. 2018;80:12–23. doi: 10.1016/j.niox.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Youness R.A., Rahmoon M.A., Assal R.A., Gomaa A.I., Hamza M.T., Waked I., El Tayebi H.M., Abdelaziz A.I. Contradicting interplay between insulin-like growth factor-1 and miR-486-5p in primary NK cells and hepatoma cell lines with a contemporary inhibitory impact on HCC tumor progression. Growth Factors. 2016;34:128–140. doi: 10.1080/08977194.2016.1200571. [DOI] [PubMed] [Google Scholar]
- 41.Youness R.A., Gad A.Z., Sanber K., Ahn Y.J., Lee G.J., Khallaf E., Hafez H.M., Motaal A.A., Ahmed N., Gad M.Z. Targeting hydrogen sulphide signaling in breast cancer. J. Adv. Res. 2021;27:177–190. doi: 10.1016/j.jare.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y., Zhang Y., Hu K., Qiu J., Hu Y., Zhou M., Zhang S. Elevated long noncoding RNA MALAT-1 expression is predictive of poor prognosis in patients with breast cancer: a meta-analysis. Biosci. Rep. 2020;40 doi: 10.1042/bsr20200215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jadaliha M., Zong X., Malakar P., Ray T., Singh D.K., Freier S.M., Jensen T., Prasanth S.G., Karni R., Ray P.S., Prasanth K.V. Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget. 2016;7:40418–40436. doi: 10.18632/oncotarget.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang N.S., Chi Y.Y., Xue J.Y., Liu M.Y., Huang S., Mo M., Zhou S.L., Wu J. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) interacts with estrogen receptor and predicted poor survival in breast cancer. Oncotarget. 2016;7:37957–37965. doi: 10.18632/oncotarget.9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shao J., Zhang Q., Wang P., Wang Z. LncRNA MALAT1 promotes breast Cancer progression by sponging miR101-3p to mediate mTOR/PKM2 signal transmission. Am. J. Transl. Res. 2021;13:10262–10275. [PMC free article] [PubMed] [Google Scholar]
- 46.Yang C., Zhu H., Tan Y., Zhu R., Wu X., Li Y., Wang C. MALAT1 promotes tumorigenesis and increases cellular sensitivity to herceptin in HER2-positive breast Cancer. Curr. Cancer Drug Targets. 2021;21:860–869. doi: 10.2174/1568009621666210618164300. [DOI] [PubMed] [Google Scholar]
- 47.Huang Y., Zhou Z., Zhang J., Hao Z., He Y., Wu Z., Song Y., Yuan K., Zheng S., Zhao Q., Li T., Wang B. lncRNA MALAT1 participates in metformin inhibiting the proliferation of breast Cancer cell. J. Cell. Mol. Med. 2021;25:7135–7145. doi: 10.1111/jcmm.16742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kataoka K., Shiraishi Y., Takeda Y., Sakata S., Matsumoto M., Nagano S., Maeda T., Nagata Y., Kitanaka A., Mizuno S., Tanaka H., Chiba K., Ito S., Watatani Y., Kakiuchi N., Suzuki H., Yoshizato T., Yoshida K., Sanada M., Itonaga H., Imaizumi Y., Totoki Y., Munakata W., Nakamura H., Hama N., Shide K., Kubuki Y., Hidaka T., Kameda T., Masuda K., Minato N., Kashiwase K., Izutsu K., Takaori-Kondo A., Miyazaki Y., Takahashi S., Shibata T., Kawamoto H., Akatsuka Y., Shimoda K., Takeuchi K., Seya T., Miyano S., Ogawa S. Aberrant PD-L1 expression through 3′-UTR disruption in multiple Cancers. Nature. 2016;534:402–406. doi: 10.1038/nature18294. [DOI] [PubMed] [Google Scholar]
- 49.Xia R., Geng G., Yu X., Xu Z., Guo J., Liu H., Li N., Li Z., Li Y., Dai X., Luo Q., Jiang J., Mi Y. LINC01140 promotes the progression and tumor immune escape in lung cancer by sponging multiple microRNAs. J. Immunother. Cancer. 2021;9 doi: 10.1136/jitc-2021-002746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Omar H.A., El-Serafi A.T., Hersi F., Arafa E.A., Zaher D.M., Madkour M., Arab H.H., Tolba M.F. Immunomodulatory MicroRNAs in Cancer: targeting immune checkpoints and the tumor microenvironment. FEBS J. 2019;286:3540–3557. doi: 10.1111/febs.15000. [DOI] [PubMed] [Google Scholar]
- 51.Atwa S.M., Handoussa H., Hosny K.M., Odenthal M., El Tayebi H.M. Pivotal role of long non-coding ribonucleic acid-X-inactive specific transcript in regulating immune checkpoint programmed death ligand 1 through a shared pathway between miR-194-5p and miR-155-5p in hepatocellular carcinoma. World J. Hepatol. 2020;12:1211–1227. doi: 10.4254/wjh.v12.i12.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jin D., Guo J., Wu Y., Du J., Yang L., Wang X., Di W., Hu B., An J., Kong L., Pan L., Su G. m(6)A mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J. Hematol. Oncol. 2021;14:32. doi: 10.1186/s13045-021-01048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li S., Mei Z., Hu H.B., Zhang X. The lncRNA MALAT1 contributes to non-small cell lung cancer development via modulating miR-124/STAT3 axis. J. Cell. Physiol. 2018;233:6679–6688. doi: 10.1002/jcp.26325. [DOI] [PubMed] [Google Scholar]
- 54.Li F., Li X., Qiao L., Liu W., Xu C., Wang X. MALAT1 regulates miR-34a expression in melanoma cells. Cell Death. Dis. 2019;10:389. doi: 10.1038/s41419-019-1620-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu L., Tan L., Yao J., Yang L. Long non‑coding RNA MALAT1 regulates cholesterol accumulation in ox‑LDL‑induced macrophages via the microRNA‑17‑5p/ABCA1 axis. Mol. Med. Rep. 2020;21:1761–1770. doi: 10.3892/mmr.2020.10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soliman R., Youness R.A., El-Shazly M., Handoussa H., Gad M. 161P - Regulatory interacting network between the immunomodulatory non-coding RNAs: miR-17-5p, MALAT1 and H19 lncRNAs in modulating the tumour microenvironment in TNBC. Ann. Oncol. 2019;30:xi57. doi: 10.1093/annonc/mdz452.032. [DOI] [Google Scholar]
- 57.Taheri F., Ebrahimi S.O., Shareef S., Reiisi S. Regulatory and immunomodulatory role of miR-34a in T cell immunity. Life Sci. 2020;262 doi: 10.1016/j.lfs.2020.118209. [DOI] [PubMed] [Google Scholar]
- 58.Soliman R.A., Youness R.A., Handoussa H., Gad M.Z. 25P Promising immuno-oncological role of rosemary against breast Cancer through altering miR-17-5p, MALAT-1, H19 and tumour microenvironment. Ann. Oncol. 2021;32:S11. doi: 10.1016/j.annonc.2021.01.039. [DOI] [Google Scholar]
- 59.Zhou M.T., Zhao C., Chen X., Zhang H.C., Li G., Lou H., Huang W.J., Wei L.J., Li D.W., Wu X., Zhang Z.C., Liu H., Ou R., Yang W.J., Hu S., Xu Y., Tang K.F. MicroRNA-34a promotes MICB expression in hepatocytes. Carcinogenesis. 2018;39:1477–1487. doi: 10.1093/carcin/bgy128. [DOI] [PubMed] [Google Scholar]
- 60.Heinemann A., Zhao F., Pechlivanis S., Eberle J., Steinle A., Diederichs S., Schadendorf D., Paschen A. Tumor Suppressive microRNAs miR-34a/c control cancer cell expression of ULBP2, a stress-induced ligand of the natural killer cell receptor NKG2D. Cancer Res. 2012;72:460–471. doi: 10.1158/0008-5472.Can-11-1977. [DOI] [PubMed] [Google Scholar]
- 61.Ohno M., Ohkuri T., Kosaka A., Tanahashi K., June C.H., Natsume A., Okada H. Expression of miR-17-92 enhances anti-tumor activity of T-cells transduced with the anti-EGFRvIII chimeric antigen receptor in mice bearing human GBM xenografts. J. Immunother. Cancer. 2013;1:21. doi: 10.1186/2051-1426-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]