Abstract
Transforming growth factor β (TGF-β) is a potent natural antiproliferative agent that plays an important role in suppressing tumorigenicity. In numerous tumors, loss of TGF-β responsiveness is associated with inactivating mutations that can occur in components of this signaling pathway, such as the tumor suppressor Smad2. Although a general framework for how Smads transduce TGF-β signals has been proposed, the physiological relevance of alterations of Smad2 functions in promoting tumorigenesis is still unknown. Here, we show that expression of Smad2.P445H, a tumor-derived mutation of Smad2 found in human cancer, suppresses the ability of the Smads to mediate TGF-β-induced growth arrest and transcriptional responses. Smad2.P445H is phosphorylated by the activated TGF-β receptor at the carboxy-terminal serine residues and associates with Smad3 and Smad4 but is unable to dissociate from the receptor. Upon ligand-induced phosphorylation, Smad2.P445H interacts stably with wild-type Smad2, thereby blocking TGF-β-induced nuclear accumulation of wild-type Smad2 and Smad2-dependent transcription. The ability of the Smad2.P445H to block the nuclear accumulation of wild-type Smad2 protein reveals a new mechanism for loss of sensitivity to the growth-inhibitory functions of TGF-β in tumor development.
The transformation or switch of normal cells into tumor cells that result in cancer can arise from a variety of alterations in normal cell function. In many cases, tumor cells develop when normal progenitor cells lose control of signaling pathways that regulate responses to negative growth-regulatory factors (17). The transforming growth factor β (TGF-β) is a potent antiproliferative factor that inhibits cell proliferation by arresting progression through the cell cycle or inducing programmed cell death (11, 26, 33). Because many cancers of epithelial and lymphoid origin develop resistance to the negative growth-regulatory effects of TGF-β, it has been postulated that one of the mechanisms whereby cells undergo neoplastic transformation and escape from normal growth control involves an altered response to TGF-β (11, 26).
TGF-β signaling is initiated when ligand induces formation of a heteromeric complex of two types of transmembrane serine/threonine kinases, which are known as type I and type II receptors (16, 26). The type II receptor (TβRII) can directly bind ligand but is incapable of mediating responses in the absence of a type I receptor (TβRI). Bound TGF-β is then recognized by TβRI, which is recruited into the complex and becomes phosphorylated by the receptor II kinase. Phosphorylation of the cytoplasmic domain of TβRI is believed to activate its kinase, thereby allowing propagation of the signal to downstream components (42). Genetic and biochemical studies have described a new family of intracellular effector molecules, known as the Smad proteins, acting downstream of TGF-β receptors (16, 26, 39). Upon ligand-induced activation of the TGF-β receptors, the receptor-regulated Smad2 and Smad3 proteins interact directly with TβRI and are phosphorylated within a conserved carboxy-terminal SSXS motif (6, 24, 28, 44). This phosphorylation event results in association of Smad2 and Smad3 with the shared partner Smad4 (21). The complexes then translocate to the nucleus, where they associate with DNA and other DNA-binding proteins, such as Fast-1, Fast-2, and c-Fos/c-Jun, and act as transcriptional activators for TGF-β-responsive genes (reviewed in references 12 and 39). In contrast to receptor-regulated Smads, the antagonistic Smads, which include Smad6 and Smad7, appear to block signal transduction by preventing access and phosphorylation of endogenous Smad2 or Smad3 to activated TβRI (15, 18, 29).
Cancer cells can acquire resistance to the antiproliferative effect of TGF-β by a number of different mechanisms, including defects in TGF-β cell surface receptors and mutational inactivation of downstream effector components of the TGF-β signaling pathways (reviewed in references 11 and 26). For example, TβRII mutations were found in sporadic cases of colon cancers with microsatellite instability and in colon and gastric cancer cells from individuals with hereditary nonpolyposis colon cancer (23, 25). Important support for a more general role of TGF-β signaling in tumor development came with the finding that human Smad2 and Smad4 are mutated in a variety of human tumors (11, 26). Smad4 (also called DPC4; deleted in pancreatic cancer) was originally identified as a candidate tumor suppressor gene in chromosome 18q21 that was somatically deleted or mutated in approximately half of human pancreatic carcinomas (14). In addition to pancreatic carcinomas, homozygous mutations of Smad4 were also found in up to 30% of colorectal cancers and in less than 10% of other human cancers (11, 26). In mouse constructs carrying a homozygous knockout mutation for the tumor suppressor gene APC, deficiency of Smad4 in intestinal adenomas causes an increase in the rate of tumor progression and invasion, supporting its role as a tumor suppressor (36). Genetic alterations and inactivating missense mutations have also been identified in Smad2 (MADR2 or JV-18) in some colorectal and lung cancers, although Smad2 is less frequently mutated than Smad4 (13, 32, 37). A homozygous targeted disruption of Smad2 is lethal to the mouse embryo, primarily due to severe developmental defects within the embryo (30, 38). The early embryonic lethality of these mice renders the functional analysis of this protein in the adult animals impossible, and therefore, these knockout mice did not shed light on the role of Smad2 in TGF-β signaling and carcinogenesis.
Two missense mutations of Smad2 (Smad2.P445H and Smad2.D450E) in human tumors were identified, suggesting that alteration of Smad2 may disrupt TGF-β signaling (13, 37). An indication that these mutations lead to inactivation of Smad2 protein functions came from overexpression studies of Xenopus embryos. Expression of wild-type Smad2 mimics the mesoderm-inducing effects of TGF-β on animal pole blastomeres, and these activities are abolished for mutant Smad2 proteins (13). Although these findings indicate that these tumor-derived mutations yield biologically inactive Smad2 protein during the development of Xenopus, the physiological functions of Smad2, particularly its potential involvement in tumor suppression, remain speculative.
In contrast to almost exclusive expression of mutant alleles in tumors carrying the missense mutation D450E (13, 37), comparison of mutant and wild-type alleles indicated that both P445H mutant and wild-type transcripts were expressed in the tumor sample (13). Interestingly, mice with inactivating mutations in the loci of Smad2 did not develop cancer when the mutation was present in the heterozygous state (38), thus ruling out any possibility of direct involvement of gene dosage of Smad2 in tumor development. In light of these observations, we investigated the possibility that the Smad2.P445H mutation can function as a dominant-negative mutation to block TGF-β signaling through endogenous Smad proteins. In this study, we demonstrate that expression of the tumor-derived mutation Smad2.P445H in TGF-β-responsive cells suppresses the ability of the Smads to mediate TGF-β-induced growth arrest and transcriptional responses. Smad2.P445H directly interferes with TGF-β-mediated activation of wild-type Smad2 by preventing its nuclear accumulation. These findings provide a description of a dominant inhibitory Smad protein isolated from a human tumor, suggesting a new paradigm for the inactivation of the Smad2 protein during the development of cancer.
MATERIALS AND METHODS
Cell lines and constructs.
MDCK, COS-7, and HepG2 cells were maintained in Dulbecco minimal essential medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS) and 5 mM glutamine. For stable transfectants, cells were transfected with expression vectors by Lipofectamine (GIBCO/BRL) and selected in G418- or hygromycin-containing growth medium and expanded as pools of stably transfected cells. Control cells were transfected with the appropriate vector alone and selected in parallel with the other cells. For the clonal growth assays, hygromycin-resistant colonies were stained with crystal violet 10 to 13 days after transfection.
Mammalian expression vectors for Smad2 mutants, pAR3-lux, and constitutively activated TβRI were a gift from J. Wrana. The p3TP-lux reporter construct was a gift from J. Massagué. Expression vector for Fast-1 was a gift from M. Whitman. Expression vectors for Smad4, Smad2, Smad3, GAL4-Smad2, and G5E1b-lux have been previously described (4, 31). The plasmid for GFP-Smad2.P445H was subcloned into pEGFP in frame with an amino-terminal green fluorescent protein (GFP) tag. The expression vector for GAL4-Smad3 was constructed using the PSG24 vector and Smad3 cDNA from pGEX-Smad3 (a gift from R. Derynck).
Gene expression analysis.
Luciferase assays were essentially carried out as previously described (5). Cells were plated to semiconfluency and 24 h later were transfected with expression vectors by Lipofectamine. Cells were subsequently treated with human TGF-β1 (Sigma) at the indicated concentrations for 16 h. Luciferase activity was measured using the luciferase assay system as described by the manufacturer (Promega) and was normalized for transfection efficiency by using a β-galactosidase-expressing vector (pCMV5.LacZ) and the Galacto-Star system (Perkin-Elmer).
Nuclear extracts, immunoprecipitation, and immunoblotting.
Nuclear extracts were prepared as described previously (2). For immunoprecipitation, cells were transfected by the Lipofectamine method and lysed at 4°C in lysis buffer (20 mM Tris-HCl [pH 8], 150 mM NaCl, 5 mM MgCl2, 10% glycerol, 0.5% NP-40, 1 mM sodium vanadate, 1 mM phenylmethylsulfonyl fluoride, 20 μg of aprotinin/ml, and 20 μg of leupeptin/ml). Lysates were subjected to immunoprecipitation with either monoclonal anti-Flag M2 antibody (Sigma) or monoclonal anti-c-Myc (9E10) antibody (Santa Cruz) for 2 h, followed by adsorption to Sepharose-protein G for 1 h. The beads were washed five times in lysis buffer, and samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting. For determination of total protein levels, aliquots of cell lysates were subjected to direct immunoblotting. Proteins were electrophoretically transferred to nitrocellulose membranes and probed with the indicated primary antibody. The bands were visualized by an enhanced chemiluminescence (ECL) detection system (Amersham) according to the manufacturer's instructions.
Apoptosis assays.
For flow cytometric determination of DNA degradation, adherent and floating cells were fixed in 70% ethanol and stained with propidium iodide. The stained cells were analyzed on a FACScan flow cytometer for relative DNA content.
[3H]thymidine incorporation.
[3H]thymidine incorporation was performed as previously described (3). Briefly, cells were seeded in six-well plates in DMEM supplemented with 10% FCS and then were incubated for 24 h with various concentrations of TGF-β1 in DMEM containing 0.2% FCS. During the last 4 h of incubation, the cells were labeled with 1 μCi of [3H]thymidine/ml. Cells were washed three times with cold phosphate-buffered saline and fixed with 5% trichloroacetic acid for 30 min at 4°C. The cells were then washed twice with 5% trichloroacetic acid and extracted with 0.5 M NaOH for 30 min. The extracts were collected and counted in a liquid scintillation spectrometer.
Immunofluorescence.
COS-7 cells were transfected with various combinations of 6xMyc-Smad2, GFP-Smad2.P445H, and wild-type or activated TβRI. After 48 h, the slides were washed twice in PBS, fixed in 4% paraformaldehyde for 30 min at room temperature and permeabilized in 0.1% Triton X-100. Cells were incubated overnight at 4°C with the primary monoclonal antibody anti-c-Myc. Cells were washed, incubated with Texas Red-conjugated goat anti-mouse immunoglobulin G, and examined on a Leica confocal microscope.
RESULTS
Expression of Smad2.P445H in MDCK cells results in loss of growth arrest by TGF-β.
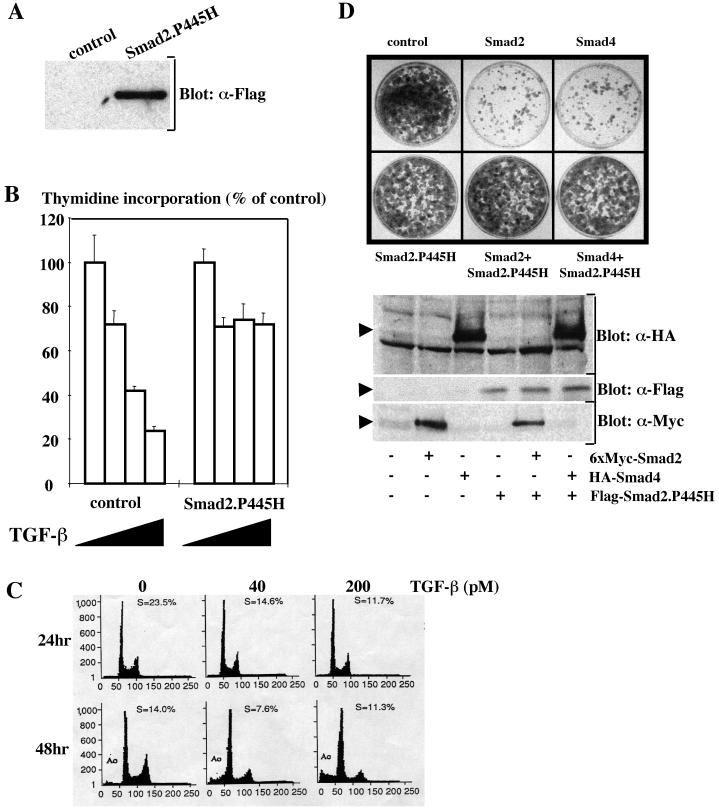
To investigate the potential effects of Smad2.P445H on TGF-β-mediated antiproliferative responses, we generated pools of epithelial Madin-Darby canine kidney (MDCK) cells stably expressing Flag-Smad2.P445H fusion protein (Fig. 1A). As shown in Fig. 1B, the proliferation of the control cell line was inhibited approximately 80% after 24 h of TGF-β treatment, as determined by [3H]thymidine incorporation into DNA. In contrast, this growth-inhibiting effect of TGF-β was largely lost (20%) in MDCK cells overexpressing Smad2.P445H. Under these experimental conditions, overexpression of the tumor-derived mutant Smad2.D450E had no effect on TGF-β-mediated growth arrest (31; data not shown), indicating that the inhibitory activity of Smad2.P445H is not a general phenomenon because of the accumulation of nonfunctional Smad2 proteins (13).
FIG. 1.
Overexpression of Smad2. P445H attenuates Smad–TGF-β-induced growth inhibition. (A) Characterization of MDCK cell transfectants. Cell lysates were prepared from control and MDCK cells stably expressing Flag-Smad2. P445H mutants were analyzed by immunoblotting using the monoclonal anti-Flag antibody M2. (B) Effects of TGF-β on the proliferation of MDCK cells stably transfected with empty vector (control) or Flag-Smad2.P445H (Smad2.P445H). (C) MDCK cells were transferred into medium containing 0.2% FCS and maintained in this medium for 24 or 48 h in the presence or absence of TGF-β. The cells were analyzed on a FACScan flow cytometer for relative DNA content. The percentage of cells in S phase of the cell cycle and the proportion of cells with subdiploid amounts of DNA (A0) indicative of apoptosis are indicated. (D) MDCK cells were transfected with the indicated vectors together with the pEMP4 vector containing a hygromycin resistance gene. Forty-eight hours after transfection, cells were selected in hygromycin-containing growth medium and hygromycin-resistant colonies were stained with crystal violet 10 to 13 days after transfection (top). Expression of transfected DNA was monitored by direct immunoblotting of total-cell lysates prepared 48 h after transfection (bottom).
To test directly whether the Smad2.P445H mutation might interfere with Smad-mediated growth inhibitory responses, we first investigated the possible contribution of Smad2 and Smad4 in specifying growth arrest and apoptotic responses by using the MDCK cell line. TGF-β addition to exponentially growing MDCK cells induced growth arrest, which preceded cell death (Fig. 1C). DNA fragmentation analysis confirmed that the death of MDCK cells induced by TGF-β occurred by an apoptotic mechanism (data not shown). Using a GFP cotransfection assay, we showed that MDCK cells overexpressing Smad4, but not Smad2, exhibited prominent apoptotic morphology, including cellular shrinkage, blebbing of plasma membranes, and generation of apoptotic bodies (4; data not shown). To quantitate these results, cells were scored in a blinded manner as healthy or apoptotic by cell morphology. Over 50% of green cells arising from cotransfection of Smad4 and GFP showed morphological changes consistent with apoptosis, whereas less than 5% of cells that had been transfected with the GFP plasmid or in combination with Smad2 exhibited such a phenotype. In a second experimental approach, cotransfections were performed with a vector encoding a hygromycin resistance gene instead of GFP and hygromycin-resistant colonies were visualized with crystal violet 10 to 13 days after transfection. As shown in Fig. 1D, expression of Smad4 significantly reduced the number of surviving colonies, reinforcing the role of Smad4 as a positive mediator of cell death (4, 9). Interestingly, a reduction in the number of colonies was also observed upon transfection with wild-type Smad2 (Fig. 1D), the expression of which is unable to induce apoptosis in the GFP assay (data not shown), indicating that Smad2 plays a critical role in mediating growth-inhibiting signals unrelated to cell death.
To determine whether the inhibitory activity of Smad2.P445H involved a direct effect on Smad-signaling pathways, we determined if it could block the ability of either wild-type Smad2 or Smad4 to reduce the number of surviving colonies in the clonal growth assay. As indicated previously, expression of wild-type Smad2 or Smad4 resulted in an almost complete arrest of cell proliferation in MDCK cells (Fig. 1D). This growth-inhibiting effect of wild-type Smad2 and Smad4 was lost in cells cotransfected with a similar amount of Smad2.P445H, indicating that expression of Smad2.P445H at a level similar to that of wild-type Smad2 or Smad4 can exert a dominant effect on Smad-induced growth inhibition. Expression of Smad2.P445H was unable to trigger growth arrest in MDCK cells (Fig. 1D). We note that expression of Smad2.P445H did not have any discernible effect on the amounts of cotransfected wild-type Smad2 and Smad4 (Fig. 1D), demonstrating the specificity of the inhibitory effect of Smad2.P445H. Taken together, these data suggest that Smad2.P445H may block TGF-β-mediated growth inhibition by interfering directly with the Smad signaling pathways.
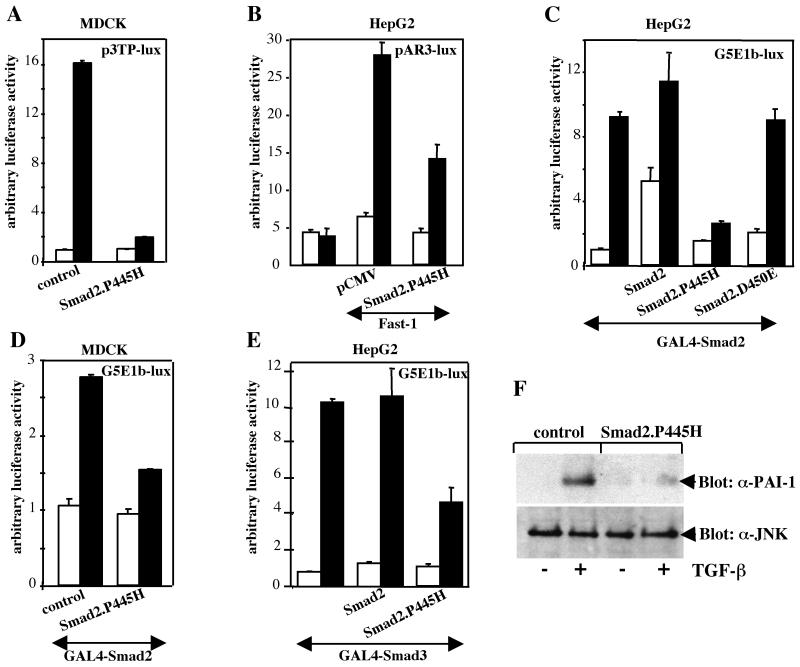
Smad2.P445H decreases TGF-β-dependent transcription.
To determine the basis for the inhibitory activity of Smad2.P445H, we investigated whether the loss of the growth-inhibiting effect of TGF-β might be related to Smad2.P445H inhibition of Smad transcriptional functions. Initially, we focused our analysis on the p3TP-lux reporter construct, which contains TGF-β elements from plasminogen activator inhibitor-1 (PAI-1) and collagenase promoters (41) and has been widely used to monitor TGF-β and Smad signaling. In wild-type MDCK cells, significantly increased (up to 16-fold) activation of this promoter was achieved upon TGF-β treatment (Fig. 2A). This activation was lost in MDCK cells stably expressing Smad2.P445H (Fig. 2A), suggesting that Smad signaling was impaired. A similar inhibition was also observed in cells cotransfected with Fast-1 and the pAR3-lux reporter construct (15), which contains three copies of the activin response element from the Xenopus Mix.2 promoter linked to a basic TATA box and a luciferase reporter gene (Fig. 2B). This activin response element is stimulated by TGF-β–activin signaling pathways, which induce assembly of a DNA-binding complex that is composed of the forkhead-containing DNA-binding protein Fast-1 and the complex Smad2-Smad4 (7, 8). Together, these data indicate that Smad2.P445H can function as a dominant-negative mutant to suppress TGF-β-dependent induction of transcription through endogenous Smad2 protein.
FIG. 2.
TGF-β-induced transcriptional activation is inhibited by Smad2.P445H. (A) Activation of the p3TP-lux reporter construct by TGF-β was analyzed in control MDCK cells (control) and MDCK cells stably expressing Flag-Smad2.P445H (Smad2.P445H). (B) HepG2 cells were transfected with pAR3-lux alone or with Fast-1 in the presence or absence of Smad2.P445H. (C) HepG2 cells were transfected with a luciferase reporter construct containing five upstream GAL4 binding sites (G5E1b-lux) and GAL4-Smad2 in either the absence or presence of wild-type Smad2, Smad2.P445H, or Smad2.D450E. (D) Activation of GAL4-Smad2 transcriptional activity by TGF-β was examined in MDCK cells stably transfected with empty vector (control) or Flag-Smad2.P445H (Smad2.P445H). (E) HepG2 cells were transfected with G5E1b-lux and GAL4-Smad3 in either the absence or presence of wild-type Smad2 or Smad2.P445H. In all cases, cells were treated with (black bars) or without (white bars) TGF-β for 16 h prior to lysis and then assayed for luciferase activity. Luciferase activity was normalized to β-galactosidase activity and was expressed as the mean ± standard deviation of triplicates from a representative experiment performed at least three times. (F) MDCK cells stably transfected with empty vector (control) or Flag-Smad2.P445H (Smad2.P445H) were treated with TGF-β for 1 h, and expression of PAI-1 was assessed by direct immunoblotting cell lysates with an anti-rabbit polyclonal antibody specific for PAI-1 (top). For comparison, the same membrane was reprobed with an anti-JNK1 antibody (bottom).
To directly test whether Smad2.P445H can interfere with TGF-β-induced Smad2-dependent transcription, we determined if Smad2.P445H could repress transcriptional activation by wild-type Smad2 when fused to the DNA-binding domain of the yeast transcription factor GAL4. In HepG2 cells, the transcriptional activity of GAL4-Smad2 from a heterologous GAL4 promoter (G5E1b-lux) was low but increased about 12-fold in response to TGF-β (Fig. 2C). Coexpression of wild-type Smad2 led to a marked increase in GAL4-Smad2 transcriptional activity in unstimulated cells, and addition of TGF-β resulted in a small but reproducible enhancement of this activity (Fig. 2C). In contrast, coexpression of Smad2.P445H resulted in an almost-complete block in TGF-β-dependent activation of the promoter G5E1b-lux by GAL4-Smad2 (Fig. 2C). This effect is specific to Smad2.P445H, since overexpression of the mutant Smad2.D450E had no effect in this assay (Fig. 2C). A similar effect on GAL4-Smad2 transcriptional activity was observed in MDCK cells stably expressing Smad2.P445H (Fig. 2D). We also tested for this inhibitory activity on GAL4-Smad3 and found a similar repression by Smad2.P445H (Fig. 2E). Under these conditions, expression of wild-type Smad2 had no appreciable effect on TGF-β-dependent activation of GAL4-Smad3 (Fig. 2E), indicating the specificity of the inhibition by Smad2.P445H.
To provide further evidence that Smad2.P445H suppressed TGF-β–Smad signaling, we tested the effect of TGF-β on endogenous PAI-1 expression in MDCK cells and MDCK cells stably expressing Smad2.P445H. We chose to focus our analysis on the PAI-1 gene as a target of the Smad signaling pathway because activation of the PAI-1 promoter by TGF-β requires the formation of a Smad3-Smad4 complex that binds to a sequence promoter known as the CAGA sequence (10). Western blotting analysis with a specific anti-PAI-1 antibody demonstrated that stable expression of Smad2.P445H blocked TGF-β-mediated expression of endogenous PAI-1 (Fig. 2F). Thus, in stably transfected cells and in transient-transfection assays, the presence of Smad2.P445H caused a loss not only of antiproliferative responses to TGF-β but also of Smad-dependent transcriptional responses.
Ligand-dependent association of Smad2.P445H with TβRI.
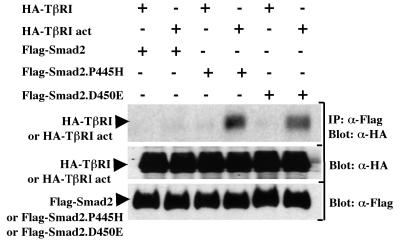
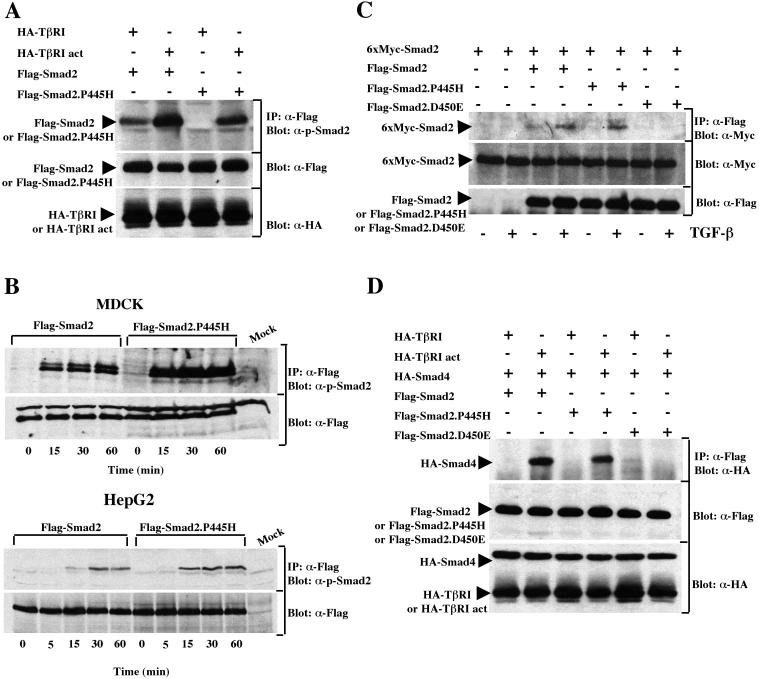
Activation of TGF-β signaling results in phosphorylation of Smad2 by TβRI on C-terminal serine residues (24). The phosphorylation of Smad2 is essential for the downstream signaling events that culminate in transcriptional activation of the target genes, as disruption of the phosphorylation sites abolished responses induced by TGF-β (1, 4, 35). Previous studies with the mutant Smad2.P445H showed that it is defective in its ability to be phosphorylated by activated TβRI (13). Since Smad2 interacts transiently with activated TβRI, we investigated whether Smad2.P445H might associate stably with the activated TβRI to block the interaction and phosphorylation of wild-type Smad2. COS-7 cells were transfected with Flag-tagged versions of wild-type Smad2 or Smad2.P445H together with either wild-type or constitutively activated TβRI (40). To detect the interaction, cell lysates were subjected to immunoprecipitation with anti-Flag antibody and the immunoprecipitates were analyzed by immunoblotting using a rabbit anti-hemagglutinin (HA) polyclonal antibody. As shown in Fig. 3, little or no interaction between the constitutively activated TβRI and wild-type Smad2 was detected, consistent with previous results (24). In contrast, we observed a strong interaction between constitutively activated TβRI and Smad2.P445H in cells expressing Smad2.P445H (Fig. 3). A similar association was also observed between the mutant Smad2.D450E and constitutively activated TβRI (Fig. 3), confirming earlier observations that the missense mutation Smad2.D450E enhances Smad2 binding to the receptor (22).
FIG. 3.
Smad2.P445H interacts stably with activated TβRI. COS-7 cells were transfected with wild-type Flag-Smad2, Flag-Smad2.P445H, or Flag-Smad2.D450E together with wild-type (HA-TβRI) or constitutively active (HA-TβRI act) TβRI. Cell lysates were subjected to anti-Flag immunoprecipitation (IP) and then were immunoblotted with anti-HA antibody (α-HA). The expression of transfected DNA was determined by immunoblotting total-cell lysates using anti-Flag (α-Flag) and anti-HA (α-HA) antibodies.
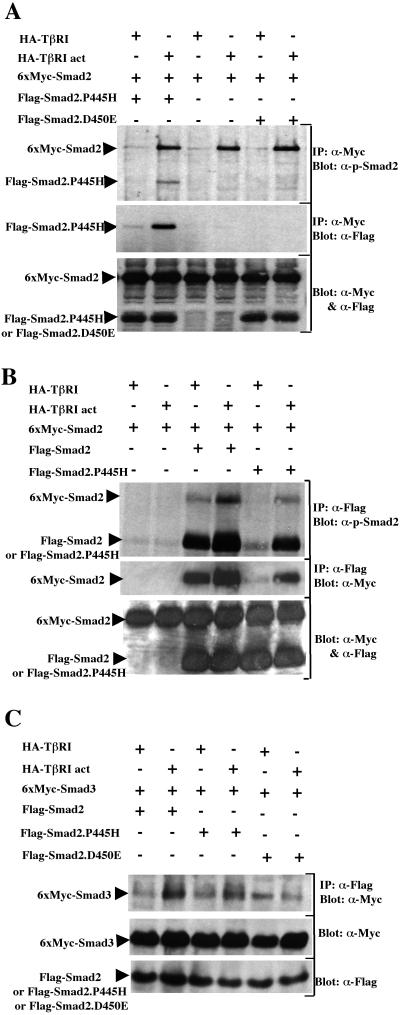
We next investigated if Smad2.P445H could block ligand-dependent phosphorylation of wild-type Smad2. For this, COS-7 cells were transfected with wild-type Myc-Smad2 in the presence or absence of Flag-Smad2.P445H and either wild-type or constitutively activated TβRI, and the extent of phosphorylation was assessed by anti-Myc immunoprecipitation followed by immunoblotting with anti-phospho-Smad2 antibody that specifically recognizes TGF-β receptor-phosphorylated Smad2 (20). In cells cotransfected with Flag-Smad2.P445H, wild-type Smad2 was phosphorylated by constitutively activated TβRI as strongly as it was in the absence of Smad2.P445H (Fig. 4A), indicating that expression of Smad2.P445H did not interfere with TGF-β receptor-mediated phosphorylation of wild-type Smad2 in COS-7 cells. A similar conclusion could be drawn when the tumor-derived mutation Smad2.D450E was cotransfected with wild-type Smad2 (Fig. 4A). Together, these results suggest that Smad2.P445H can exert an inhibitory effect without preventing access of wild-type Smad2 to TGF-β receptors.
FIG. 4.
Smad2.P445H interacts with wild-type Smad2 and Smad3. (A) COS-7 cells were transfected with the indicated combinations of wild-type 6xMyc-Smad2, HA-TβRI, and constitutively active TβRI (HA-TβRI act) with Flag-Smad2.P445H or Flag-Smad2.D450E. Cell lysates were subjected to anti-Myc immunoprecipitation (IP) and then were immunoblotted with anti-phospho-Smad2 antibody (top). Copre- cipitation of Flag-Smad2.P445H with wild-type 6xMyc-Smad2 was monitored by reprobing the same membrane with anti-Flag antibody (middle). Expression of transfected DNA was monitored by direct immunoblotting (bottom). (B) COS-7 cells were transfected with the indicated combinations of wild-type 6xMyc-Smad2, HA-TβRI, and HA-TβRI act with wild-type Flag-Smad2 or Flag-Smad2.P445H. Flag immunoprecipitates were subjected to immunoblotting with either antibodies against receptor-phosphorylated Smad2 (top) or anti-Myc antibodies (middle). Expression of transfected DNA was assessed by direct Western blotting (bottom). (C) Cell lysates from transiently transfected COS-7 cells were subjected to immunoprecipitation with anti-Flag antibody directed towards various Smad2 mutants and then were immunoblotted using anti-Myc (α-Myc) antibody that recognizes Smad3. The relative levels of transfected proteins were determined by direct Western immunoblotting of total-cell lysates.
In the course of these experiments, we found that when wild-type Myc-Smad2 was immunoprecipitated from cells coexpressing Flag-Smad2.P445H, a protein that was the size expected for Flag-Smad2.P445H specifically associated with wild-type Myc-Smad2 and became phosphorylated by activated TβRI (Fig. 4A). We hypothesized that this may reflect phosphorylation of Smad2.P445H and its association with wild-type Smad2 in response to TGF-β signaling. To examine this directly, cell lysates from COS-7 cells transfected as described above were subjected to anti-Flag immunoprecipitation followed by immunoblotting with anti-phospho-Smad2 antibody. Analysis of wild-type Smad2 revealed that coexpression of constitutively activated TβRI induced a significant increase in the phosphorylation of the protein (Fig. 4B) as described previously (13). Similarly, basal phosphorylation of Smad2.P445H was low in unstimulated cells and activation of TGF-β signaling resulted in a dramatic increase in Smad2.P445H phosphorylation, although Smad2.P445H was less phosphorylated than wild-type Smad2 (Fig. 4B). Consistent with our previous analysis, we also observed that coexpression of constitutively activated TβRI led to an increase in the association of Smad2.P445H with a phosphorylated protein that corresponds to wild-type Myc-Smad2 protein. Reprobing this membrane with anti-Myc antibody for the presence of wild-type Myc-Smad2 confirmed that Smad2.P445H associated with wild-type Smad2 protein in response to TGF-β signaling (Fig. 4B). In a reciprocal fashion, ligand-induced complex formation between wild-type Smad2 and Smad2.P445H could also be demonstrated when cell lysates were subjected to immunoprecipitation with the anti-Myc antibody directed against the tagged wild-type Smad2 protein followed by immunoblotting with anti-Flag antibody for the presence of Smad2.P445H (Fig. 4A). It should be noted that the ligand-dependent homodimerization of Smad2 was more pronounced when wild-type Flag-Smad2 was used instead of Flag-Smad2.P445H (Fig. 4B). However, this association was specific for Smad2.P445H, since we detected only weak interactions between Smad2.D450E and wild-type Smad2 that were unaffected by TGF-β signaling (Fig. 4A and 5C). Taken together, these data demonstrate that Smad2.P445H and wild-type Smad2 can form physical complexes, the levels of which can be enhanced by the activation of the TGF-β signal transduction pathway.
FIG. 5.
Phosphorylation of Smad2.P445H by the activated TβRI. (A) COS-7 cells were transfected with wild-type Flag-Smad2 or Flag-Smad2.P445H together with HA-TβRI or constitutively active TβRI (HA-TβRI act). Cell lysates were subjected to anti-Flag immunoprecipitation (IP) and then were immunoblotted with anti-phospho-Smad2 antibody (α-p-Smad2). The expression of transfected DNA was determined by direct Western immunoblotting of total-cell lysates. (B) MDCK (top) or HepG2 (bottom) cells were transfected with wild-type Flag-Smad2 or Flag-Smad2.P445H and treated with TGF-β for various time periods. Following immunoprecipitation (IP) with anti-Flag antibody, the phosphorylation of various Smad2 mutants was determined using the anti-phospho-Smad2 antibody (α-p-Smad2). Anti-Flag immunoblotting of whole extracts showed that similar amounts of wild-type Smad2 and Smad2.P445H were recovered in each sample (α-Flag). Mock cells were transfected with empty vector. (C) MDCK cells transfected with the indicated constructs were treated with or without TGF-β for 1 h. Cell lysates were subjected to anti-Flag immunoprecipitation (IP) and then were immunoblotted with anti-Myc antibody (α-Myc). The expression of transfected DNA was determined by immunoblotting total-cell lysates using anti-Flag (α-Flag) and anti-Myc (α-Myc) antibodies. (D) Flag-Smad2, Flag-Smad2.P445H, or Flag-Smad2.D450E were cotransfected in COS-7 cells with HA-Smad4 and with HA-TβRI or constitutively active TβRI (HA-TβRI act). Association of Smad4 with various Smad2 mutants was analyzed by blotting the Flag immunoprecipitates (IP) with an anti-HA antibody (α-HA). Expression of transfected proteins was monitored by direct Western blotting.
Smad2.P445H interacts with Smad3.
Smad2 and Smad3, which are structurally highly similar, can form heteromers with each other in response to TGF-β signaling (19, 28). Since Smad3 is centrally involved in mediating TGF-β responses, we also investigated whether Smad2.P445H might interact with Smad3 in response to TGF-β signaling. For this, we utilized the experimental approach developed for our studies on wild-type Smad2. Briefly, cell lysates from transiently transfected COS-7 cells were subjected to immunoprecipitation with anti-Flag antibody directed towards tagged Smad2 mutants, followed by immunoblotting with anti-Myc antibody for the presence of Smad3. Similar to wild-type Smad2, coexpression of activated TβRI with Flag-Smad2.P445H and Myc-Smad3 resulted in an increase in the amount of Smad3 present in Flag-Smad2.P445H immunoprecipitates, demonstrating that Smad2.P445H and Smad3 can form physical complexes upon activation of TGF-β signaling pathway (Fig. 4C).
Phosphorylation of Smad2.P445H by activated TβRI.
Because our biochemical analyses of Smad2.P445H phosphorylation and its interaction with wild-type Smad2 were performed by expressing both proteins in the cells, we sought to determine whether the phosphorylation of Smad2.P445H is direct or depends on its recruitment by wild-type Smad2 protein to activated TβRI. For this, COS-7 cells were transfected with Flag-tagged versions of wild-type Smad2 or Smad2.P445H together with either wild-type or constitutively activated TβRI. Relative phosphorylation levels were assessed by anti-Flag immunoprecipitation followed by immunoblotting with anti-phospho-Smad2 antibody. Analysis of wild-type Smad2 revealed that coexpression of constitutively activated TβRI induced a significant increase in the phosphorylation of the protein (Fig. 5A) as described previously (13). Interestingly, the P445H-substituted Smad2 retained its ability to be phosphorylated by activated TβRI, although the increase in phosphorylation in COS-7 cells was reproducibly 1.2- to 1.5-fold less than that of wild-type Smad2 (Fig. 5A). In contrast, Smad2.P445H was phosphorylated to a larger extent than wild-type Smad2 in transfected HepG2 and MDCK cells treated with TGF-β (Fig. 5B). To ascertain the relevance of this phosphorylation by the endogenous TGF-β receptor, we investigated whether complexes might be found between wild-type Myc-Smad2 and Flag-Smad2.P445H in MDCK cells treated with TGF-β. As shown in Fig. 5C, cotransfection of either wild-type Flag-Smad2 or Flag-Smad2.P445H with wild-type Myc-Smad2 and constitutively activated TβRI resulted in a similar increase in the amount of wild-type Myc-Smad2 present in Flag immunocomplexes. In contrast, no interaction between wild-type Myc-Smad2 and Flag-Smad2.D450E could be detected following exposure of cells to TGF-β (Fig. 5C), confirming that the missense mutation D450E disrupts regulation of Smad2 by TGF-β signaling (13, 22; data not shown). Therefore, we conclude that activated TβRI can effectively phosphorylate the Smad2.P445H mutant at C-terminal serine residues, allowing its association with wild-type Smad2.
Phosphorylation of Smad2 induces its association with Smad4 (21). To confirm the phosphorylation of Smad2.P445H by activated TβRI, we examined the formation of wild-type Smad2-Smad4 and Smad2.P445H-Smad4 complexes in response to TGF-β signaling. Cotransfection of either wild-type Flag-Smad2 or Flag-Smad2.P445H with HA-Smad4 and constitutively activated TβRI resulted in an increase in the amount of Smad4 present in Flag immunocomplexes (Fig. 5D). This association was specific to Smad2.P445H since overexpression of the mutant Smad2.D450E, which was not phosphorylated by activated TβRI, failed to interact with Smad4 in response to TGF-β signaling (Fig. 5D). Thus, it is likely that activation of TGF-β signaling induced phosphorylation of Smad2.P445H at the carboxy-terminal SSMS motif, allowing its interaction with Smad4.
Expression of Smad2.P445H does not affect the assembly of wild-type Smad2 and Smad4 in response to TGF-β signaling.
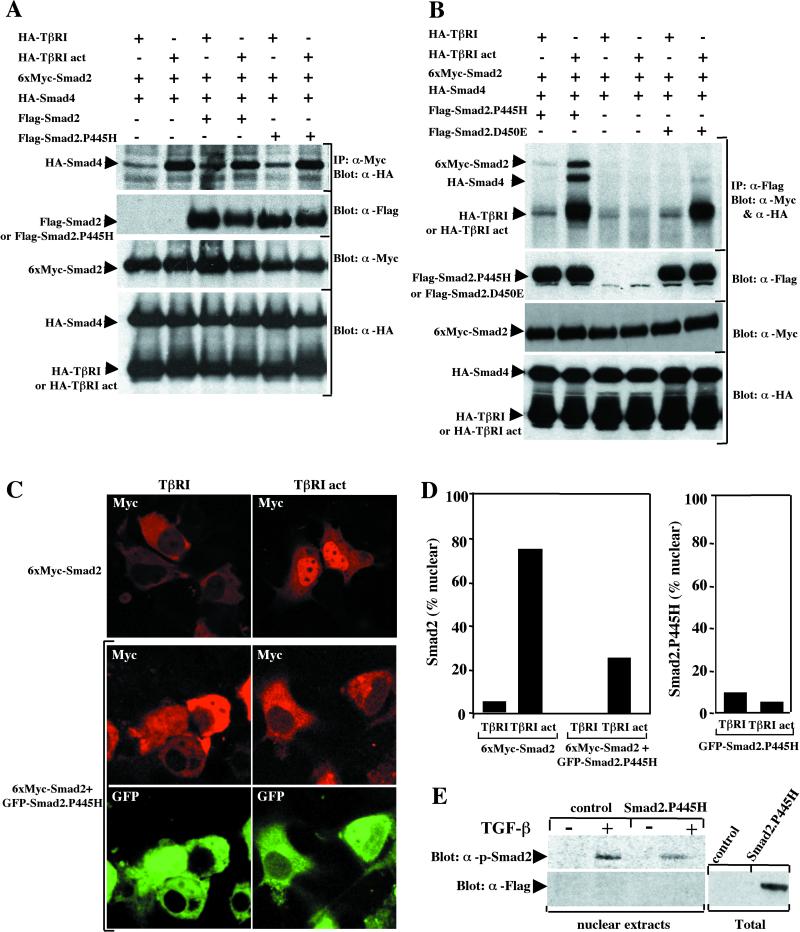
In light of our observations that Smad2.P445H did not interfere with TGF-β receptor-mediated phosphorylation of wild-type Smad2, we reasoned that the mechanism through which this mutant exerts its dominant-negative action must occur at downstream steps in the TGF-β-induced activation of Smad2. Key events in this process include the association of receptor-activated Smad2 with Smad4 and the translocation of this complex to the nucleus. To investigate heteromeric complex formation, COS-7 cells were transfected with wild-type Smad2 and Smad4 expression vectors in either the presence or absence of Flag-Smad2.P445H. Complexes were precipitated with anti-Myc antibody directed towards tagged wild-type Smad2 followed by immunoblotting with anti-HA antibodies for the presence of Smad4. As shown in Fig. 6A and in previous studies (21), the association of Smad2 with Smad4 was strongly increased by activated TβRI. This ligand-inducible interaction of wild-type Smad2 and Smad4 was not disrupted by the presence of Smad2.P445H (Fig. 6A). Taken together, these data indicate that the ligand-dependent complex formation between wild-type Smad2 and Smad2.P445H did not prevent the phosphorylation of wild-type Smad2 and its association with Smad4.
FIG. 6.
Smad2.P445H blocks TGF-β-induced nuclear accumulation of wild-type Smad2. (A) COS-7 cells were transfected with wild-type 6xMyc-Smad2 and HA-Smad4 together with HA-TβRI or constitutively active TβRI (HA-TβRI act) in either the absence or presence of wild-type Flag-Smad2 or Flag-Smad2.P445H. Cell lysates were subjected to anti-Myc immunoprecipitation (IP) and then were immunoblotted with anti-HA antibody (α-HA). The relative levels of transfected proteins were determined by direct Western immunoblotting of total-cell lysates. (B) COS-7 cells were transfected with wild-type 6xMyc-Smad2 and HA-Smad4 together with HA-TβRI or HA-TβRI act in either the absence or presence of Flag-Smad2.P445H or Flag-Smad2.D450E. Cell lysates were subjected to anti-Flag immunoprecipitation (IP) and then were immunoblotted with anti-HA (α-HA) and anti-Myc (α-Myc) antibodies. Expression of transfected proteins was monitored by direct Western blotting. (C) COS-7 cells were transfected with wild-type 6xMyc-Smad2 together with HA-TβRI or HA-TβRI act in either the presence or absence of GFP-Smad2.P445H. Forty-eight hours after transfection, cells were fixed and the localizations of wild-type 6xMyc-Smad2 (red) and GFP-Smad2.P445H (green) were visualized by a confocal microscope. (D) Determination of percentage of cells with wild-type 6xMyc-Smad2 or GFP-Smad2.P445H staining exclusively in the nucleus. (E) MDCK cells stably transfected with empty vector (control) or Flag-Smad2.P445H (Smad2.P445H) were treated with or without TGF-β for 1 h, and nuclear extracts were analyzed by immunoblotting with anti-phospho-Smad2 antibody (top) or anti-Flag antibody (bottom). Total, whole-cell lysates.
Inhibition of Smad2 nuclear accumulation by the Smad2.P445H mutant.
Since our previous results indicate that Smad2.P445H can interact with both the activated TGF-β receptor and wild-type Smad2, we wanted to know whether Smad2.P445H could coexist with wild-type Smad2 and activated TβRI or whether wild-type Smad2-Smad2.P445H and activated TβRI-Smad2.P445H complexes are mutually exclusive. Cotransfection of COS-7 cells with Flag-Smad2.P445H, wild-type Myc-Smad2, and constitutively activated TβRI revealed that wild-type Smad2 and activated TβRI were clearly detectable in complexes precipitated via the Flag epitope present on Smad2.P445H (Fig. 6B). As expected, wild-type Myc-Smad2, but not activated TβRI, was able to associate with wild-type Flag-Smad2 in response to TGF-β signaling (Fig. 3 and data not shown). This suggests that ligand-dependent association of Smad2.P445H and wild-type Smad2 might sequester wild-type Smad2 in the cytoplasm. To examine this directly, we tested for the association of wild-type Smad2 and activated TβRI with Flag-Smad2.D450E, which interacts stably with activated TβRI but is unable to associate with wild-type Smad2 in response to TGF-β signaling. In contrast to Smad2.P445H, we never detected wild-type Smad2 in Smad2.D450E immunocomplexes, despite the relatively high amount of activated TβRI that coprecipitated with Smad2.D450E (Fig. 6B).
The above observations raise the possibility that Smad2.P445H may function to sequester wild-type Smad2 in the cytoplasm. If this is so, then we should find that when wild-type Smad2 and Smad2.P445H are overexpressed, the nuclear accumulation of wild-type Smad2 by activated TβRI may be blocked. To examine this point, COS-7 cells were transfected with wild-type Myc-Smad2 in the presence or absence of GFP-Smad2.P445H and either wild-type or activated TβRI. As shown in Fig. 6C, in control cells, wild-type Myc-Smad2 was present mainly in the cytoplasm, whereas in cells expressing activated TβRI, predominantly nuclear staining was observed. In contrast, in cells coexpressing Smad2.P445H, the nuclear accumulation of wild-type Smad2 in response to TGF-β was inhibited (Fig. 6C). A quantitation of these results indicated that over 70% of COS-7 cells displayed prominent nuclear staining of wild-type Smad2 upon coexpression of activated TβRI, while less than 30% exhibited similar staining in the corresponding cells that have been cotransfected with Smad2.P445H (Fig. 6D). Visualization of GFP-Smad2.P445H revealed that Smad2.P445H colocalized with wild-type Smad2 in unstimulated cells and coexpression of activated TβRI did not appreciably affect the staining pattern. To ensure that GFP was not responsible for the localization of Smad2.P445H, we examined localization of Flag-Smad2.P445H. We found that Flag-Smad2.P445H showed the same subcellular localization as the GFP-Smad2.P445H construct (data not shown). Furthermore, the inhibition of ligand-dependent accumulation of wild-type Myc-Smad2 was not observed when wild-type GFP-Smad2 was used instead of GFP-Smad2.P445H (data not shown). Finally, we examined whether the stable expression of Smad2.P445H in MDCK cells could inhibit TGF-β-mediated nuclear accumulation of endogenous Smad2 protein. For this, MDCK and MDCK.Smad2.P445H cells were treated with TGF-β and nuclear extracts were immunoblotted with the anti-phospho-Smad2 antibody. As expected, only very little of the phospho-Smad2 was detected in the nucleus in the absence of a TGF-β signal, whereas exposure to TGF-β led to a large nuclear accumulation of phospho-Smad2 (Fig. 6E). Compared to MDCK cells, MDCK.Smad2.P445H cells respond to TGF-β with a limited accumulation of phosho-Smad2 in the nucleus. As expected, immunoblotting of the nuclear extracts with anti-Flag antibody showed no contamination with Flag-Smad2.P445H (Fig. 6E), confirming that Smad2.P445H is localized in the cytoplasm. Thus, the inhibitory activity of Smad2.P445H most likely takes place in the cytoplasm by a mechanism that might depend on its ability to engage wild-type Smad2 in a nonproductive complex.
DISCUSSION
In the present study, we have shown that expression of the tumor-derived mutation of Smad2.P445H in TGF-β-responsive cells suppresses the ability of the Smads to mediate TGF-β-induced growth arrest and transcriptional activation. Smad2.P445H directly interfered with TGF-β-mediated activation of wild-type Smad2 by preventing its nuclear accumulation and thus revealed a novel mechanism for the inactivation of Smad signaling during mammalian carcinogenesis. Together, the results outlined in the present study provide more evidence that Smad2 is a tumor suppressor gene, which, like other such genes, encodes a growth-inhibiting protein whose loss during tumorigenesis may lead to deregulated cell proliferation.
Mechanism of repression of TGF-β signaling pathway by the tumor-derived mutation Smad2.P445H.
Several lines of evidence support the conclusion that the tumor-derived mutation Smad2.P445H can suppress the TGF-β signaling pathway at the level of Smad. First, coexpression of Smad2.P445H repressed TGF-β-dependent induction of the pAR3-lux or p3TP-lux reporter construct in both HepG2 and MDCK cells. Second, similar to its effect on pAR3-lux and p3TP-lux reporters, expression of Smad2.P445H inhibited TGF-β-mediated activation of transcription by Smad2 or Smad3 when fused to the DNA binding domain GAL4. Third, this inhibition of Smad transcriptional activity was accompanied by a similar inhibition of the TGF-β antimitogenic responses, since stable expression of Smad2.P445H in the MDCK cell line rendered these cells resistant to TGF-β-induced growth arrest. Finally, in the clonal growth assay, Smad2.P445H was also able to restore the number of surviving colonies from MDCK cells expressing wild-type Smad2 or Smad4.
The phosphorylation of Smad2 by activated TβRI and its subsequent heterodimerization with Smad4 and translocation to the nucleus form the basis for a model of how Smad proteins work to transmit TGF-β signals from the plasma membrane to the nucleus (16, 26). We showed here that Smad2.P445H is phosphorylated by activated TβRI at the carboxy-terminal serines that serve as TGF-β receptor phosphorylation sites but fails to accumulate in the nucleus in response to TGF-β signaling. Because Smad2.P445H interacts with both wild-type Smad2 and Smad4 in a TGF-β-dependent manner, it may repress Smad-mediated transcriptional activation either by blocking the abilities of Smad2 and Smad4 to heterodimerize or by preventing the nuclear accumulation of wild-type Smad2. We found that the ability of wild-type Smad2 to heterodimerize with Smad4 was not affected by coexpression of Smad2.P445H. On the other hand, we showed that Smad2.P445H inhibited TGF-β-induced nuclear accumulation of wild-type Smad2, thereby resulting in repression of Smad-dependent transcription. We also tested for this inhibitory effect on nuclear translocation of Smad3 and found a similar suppression by Smad2.P445H (data not shown). Based on these observations and the finding that Smad2.P445H can associate stably with activated TβRI, we proposed a model in which Smad2.P445H engages wild-type Smad2 and Smad3 in nonproductive complexes in the cytoplasm. This situation for inactivating the TGF-β signaling pathway may not be restricted to the mutant P445H of Smad2 because another cancer-associated mutant of Smad4 has a similar effect on TGF-β-mediated nuclear translocation of Smad proteins (45).
Phosphorylation of Smad2.P445H by activated TGF-β receptor.
Our data differ significantly from data recently reported by Eppert et al. (13). Using 32P-labeled COS-1 cells, this group demonstrated that the mutant Smad2.P445H is defective in its ability to be phosphorylated by activated TβRI. What we have shown here—that Smad2 phosphorylation in response to TGF-β stimulation was not lost when the mutation P445H was introduced into wild-type protein—is intriguing. The apparent discrepancy between the previous findings and our present findings might be due to cell type differences or even to different experimental strategies. It should be noted that our analysis of Smad2 phosphorylation was performed using an anti-phospho-Smad2 antibody that specifically recognizes TGF-β receptor-phosphorylated carboxy-terminal serine residues, avoiding any interference in phosphorylation of Smad2 by other protein kinases, such as mitogen-activated protein kinase (20). Further evidence is provided by the ability of Smad2.P445H to form a heterocomplex with Smad3 or Smad4 in response to TGF-β signaling, processes that require phosphorylation of Smad2 protein by activated TβRI (19, 28). In addition, we found that Smad2.P445H was phosphorylated by endogenous TGF-β receptors to a larger extent than wild-type Smad2 in transfected HepG2 and MDCK cells treated with TGF-β1. At present, the basis for these different observations on the phosphorylation of Smad2.P445H in response to TGF-β signaling is not clear; clarification of the conditions under which Smad2.P445H may retain or lose its ability to be phosphorylated by activated TβRI will require further investigation. In any event, our data clearly indicate that the mutation P445H can disrupt the regulation of nuclear accumulation of Smad2 by TGF-β. By associating with Smads, Smad2.P445H inhibits the nuclear accumulation of the Smads and their ability to mediate TGF-β antiproliferative responses and other effects.
A structural basis for mutational inactivation of Smad2.
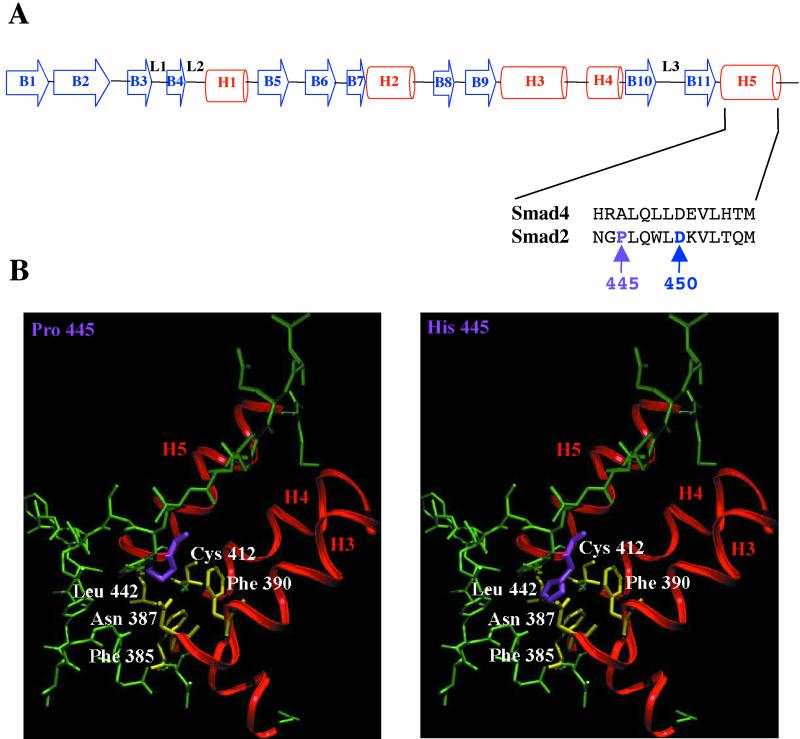
The tumor-derived mutations of Smad2.P445H and Smad2.D450E map to the MH2 domain that is involved in receptor recognition, homo- and hetero-oligomerization among Smads, and interaction with transcription factors (reviewed in reference 27). The structure of the MH2 domain consists of two β-sheets capped with a three-helix bundle (H3, H4, and H5) on one side and three large loops and an α-helix (L1, L2, and L3 loops and H1 helix) on the other side. The Smad4 MH2 domain assembles into a trimer, with the loop-helix region of one subunit packing with the three-helix bundle from the next subunit (34, 43). The similarity between Smad2 and Smad4 MH2 domains in terms of sequence and structural elements suggests that the Smad2 MH2 domain may form a homotrimer in solution (34, 43). The tumorigenic mutation P445H in Smad2 (corresponding to Ala of the Smad4 MH2 domain) maps to the helix H5 (Fig. 7A) of the three-helix bundle but does not have an apparent role in the trimer interface (34). However, Pro445 is in close proximity to several residues (Leu442, Cys412, Phe390, Asn387, and Phe385) in the crystal structure, and thus, these residues cannot accommodate the larger histidine side chain without disrupting the H5 helix and the packing between the three-helix bundle and the β-sandwich (Fig. 7B). Thus, the P445H mutation will likely perturb local structure but is unlikely to have a significant effect on the trimer interface. Although the exact nature of the structural defects is not clear, our biochemical studies suggest that the P445H mutant interacted stably with the TGF-β receptor, compared to wild-type Smad2. By sequestering wild-type Smad2 in the cytoplasm, Smad2.P445H acts as a dominant-negative mutant through endogenous Smad proteins in an important regulatory position in the pathway. This mechanism could potentially be involved in the loss of growth-inhibiting responses to TGF-β that are often observed during tumor progression.
FIG. 7.
Crystal structure of Smad2 MH2 domain. (A) Structure of Smad2 and Smad4 MH2 domains (adapted from references 34 and 43). In the MH2 domain structure, arrowheads B1 to B11 represent β-sheets, spaces L1 to L3 represent loops, and cylinders H1 to H5 represent α-helices. (B) Mapping of tumor-derived mutation of Smad2.P445H. Pro445 (purple) is in close proximity to residues Leu442, Cys412, Phe390, Asn387, and Phe385 (yellow) in the crystal structure; thus, introduction of His in position 445 (purple) could produce a steric clash with these residues.
ACKNOWLEDGMENTS
C. Prunier and N. Ferrand contributed equally to this work.
We thank G. Cherqui for helpful discussions and P. Fontange for assistance with immunofluorescence and confocal microscopy.
This work was supported by INSERM (Institut National de la Santé et de la Recherche Médicale), Centre National de la Recherche Scientifique (CNRS), la Ligue contre le Cancer Comité de Paris, and ARC (Association pour la Recherche sur le Cancer).
REFERENCES
- 1.Abdollah S, Macias-Silva M, Tsukazaki T, Hayashi H, Attisano L, Wrana J L. TβRI phosphorylation of Smad2 on Ser465 and Ser467 is required for Smad2-Smad4 complex formation and signaling. J Biol Chem. 1997;272:27678–27685. doi: 10.1074/jbc.272.44.27678. [DOI] [PubMed] [Google Scholar]
- 2.Andrews N C, Faller D V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atfi A, Lepage K, Allard P, Chapdelaine A, Chevalier S. Activation of a serine/threonine kinase signaling pathway by TGF-β. Proc Natl Acad Sci USA. 1995;92:12110–12114. doi: 10.1073/pnas.92.26.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atfi A, Buisine M, Mazars A, Gespach C. Induction of apoptosis by DPC4, a transcriptional factor regulated by TGF-β through SAPK/JNK signaling pathway. J Biol Chem. 1997;272:24731–24734. doi: 10.1074/jbc.272.40.24731. [DOI] [PubMed] [Google Scholar]
- 5.Atfi A, Djelloul S, Chastre E, Davis R, Gespach C. Evidence for a role of Rho-like GTPases SAPK/JNK in TGF-β-mediated signaling. J Biol Chem. 1997;272:1429–1432. doi: 10.1074/jbc.272.3.1429. [DOI] [PubMed] [Google Scholar]
- 6.Baker J C, Harland R M. A novel mesoderm inducer, Madr2, functions in the activin signal transduction pathway. Genes Dev. 1996;10:1880–1889. doi: 10.1101/gad.10.15.1880. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Rubock M J, Whitman M A. Transcriptional partner for MAD proteins in TGF-β signaling. Nature. 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- 8.Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- 9.Dai J L, Bansal R K, Kern S E. G1 cell cycle arrest and apoptosis induction by nuclear Smad4/Dpc4: phenotypes reversed by a tumorigenic mutation. Proc Natl Acad Sci USA. 1999;96:1427–1432. doi: 10.1073/pnas.96.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier J M. Direct binding of Smad3 and Smad4 to critical TGF-β-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derynck R, Feng X H. TGF-β receptor signaling. Biochim Biophys Acta. 1997;1333:105–150. doi: 10.1016/s0304-419x(97)00017-6. [DOI] [PubMed] [Google Scholar]
- 12.Derynck R, Zhang Y, Feng X H. Smads: transcriptional activators of TGF-β responses. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- 13.Eppert K, Scherer S W, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui L C, Bapat B, Gallinger S, Andrulis I L, Thomsen G H, Wrana J L, Attisano L. MADR2 maps to 18q21 and encodes a TGFβ-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell. 1996;86:543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 14.Hahn S A, Schutte M, Hoque A T, Moskaluk C A, daCosta L T, Rozenblum E, Weinstein C L, Fischer A, Yeo C J, Hruban R H, Kern S E. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350–353. doi: 10.1126/science.271.5247.350. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi H, Abdollah S, Qiu Y, Cai J, Xu Y Y, Grinnell B W, Richardson M A, Topper J N, Gimbrone M A, Jr, Wrana J L, Falb D. The MAD-related protein Smad7 associates with the TGFβ receptor and functions as an antagonist of TGFβ signaling. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 16.Heldin C H, Miyazono K, ten Dijke P. TGF-β signaling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 17.Hunter T. Oncoprotein networks. Cell. 1997;88:333–346. doi: 10.1016/s0092-8674(00)81872-3. [DOI] [PubMed] [Google Scholar]
- 18.Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. Smad6 inhibits signaling by the TGF-β superfamily. Nature. 1997;389:622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- 19.Kawabata M, Inoue H, Hanyu A, Imamura T, Miyazono K. Smad proteins exist as monomers in vivo and undergo homo- and hetero-oligomerization upon activation by serine/threonine kinase receptors. EMBO J. 1998;17:4056–4065. doi: 10.1093/emboj/17.14.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kretzschmar M, Doody J, Timokhina I, Massagué J. A mechanism of repression of TGFβ/Smad signaling by oncogenic Ras. Genes Dev. 1999;13:804–816. doi: 10.1101/gad.13.7.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagna G, Hata A, Hemmati-Brivanlou A, Massagué J. Partnership between DPC4 and SMAD proteins in TGF-β signaling pathways. Nature. 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 22.Lo R S, Chen U-G, Shi Y, Pavletich N P, Massagué J. The L3 loop: a structural motif determining specific interactions between Smad proteins and TGF-β receptors. EMBO J. 1998;17:996–1005. doi: 10.1093/emboj/17.4.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu S L, Zhang W C, Akiyama Y, Nomizu T, Yuasa Y. Genomic structure of the transforming growth factor β type II receptor gene and its mutations in hereditary nonpolyposis colorectal cancers. Cancer Res. 1996;56:4595–4598. [PubMed] [Google Scholar]
- 24.Macias-Silva M, Abdollah S, Hoodless P A, Pirone R, Attisano L, Wrana J L. MADR2 is a substrate of the TGFβ receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell. 1996;87:215–224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- 25.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B, Brattain M, Willson J K V. Inactivation of the type II TGF-β receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 26.Massagué J. TGF-β signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 27.Massagué J, Chen Y-G. Controlling TGF-β signaling. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- 28.Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J, Heldin C H, Miyazono K, ten Dijke P. TGF-β receptor-mediated signaling through Smad2, Smad3 and Smad4. EMBO J. 1997;16:5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakao A, Afrakhte M, Moren A, Nakayama T, Christian J L, Heuchel R, Itoh S, Kawabata M, Heldin N E, Heldin C H, ten Dijke P. Identification of Smad7, a TGF-β-inducible antagonist of TGF-β signaling. Nature. 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- 30.Nomura M, Li E. Smad2 role in mesoderm formation, left-right patterning and craniofacial development. Nature. 1998;393:786–790. doi: 10.1038/31693. [DOI] [PubMed] [Google Scholar]
- 31.Prunier C, Mazars A, Noe V, Bruyneel E, Mareel M, Gespach C, Atfi A. Evidence that Smad2 is a tumor suppressor implicated in the control of cellular invasion. J Biol Chem. 1999;274:22919–22922. doi: 10.1074/jbc.274.33.22919. [DOI] [PubMed] [Google Scholar]
- 32.Riggins G J, Kinzler K W, Vogelstein B, Thiagalingam S. Frequency of Smad gene mutations in human cancers. Cancer Res. 1997;57:2578–2580. [PubMed] [Google Scholar]
- 33.Roberts A B, Sporn M B. The transforming growth factors-βs. In: Roberts A B, Sporn M B, editors. Peptide growth factors and their receptors. Heidelberg, Germany: Springer-Verlag; 1990. pp. 421–472. [Google Scholar]
- 34.Shi Y, Hata A, Lo R S, Massagué J, Pavletich N P. A structural basis for mutational inactivation of the tumour suppressor Smad4. Nature. 1997;388:87–93. doi: 10.1038/40431. [DOI] [PubMed] [Google Scholar]
- 35.Souchelnytskyi S, Tamaki K, Engstrom U, Wernstedt C, ten Dijke P, Heldin C H. Phosphorylation of Ser465 and Ser467 in the C terminus of Smad2 mediates interaction with Smad4 and is required for transforming growth factor-β signaling. J Biol Chem. 1997;272:28107–28115. doi: 10.1074/jbc.272.44.28107. [DOI] [PubMed] [Google Scholar]
- 36.Takaku K, Oshima M, Miyoshi H, Matsui M, Seldin M F, Taketo M M. Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Apc genes. Cell. 1998;92:645–656. doi: 10.1016/s0092-8674(00)81132-0. [DOI] [PubMed] [Google Scholar]
- 37.Uchida K, Nagatake M, Osada H, Yatabe Y, Kondo M, Mitsudomi T, Masuda A, Takahashi T, Takahashi T. Somatic in vivo alterations of the JV18-1 gene at 18q21 in human lung cancers. Cancer Res. 1996;56:5583–5585. [PubMed] [Google Scholar]
- 38.Waldrip W R, Bikoff E K, Hoodless P A, Wrana J L, Robertson E J. Smad2 signaling in extraembryonic tissues determines anterior-posterior polarity of the early mouse embryo. Cell. 1998;92:797–808. doi: 10.1016/s0092-8674(00)81407-5. [DOI] [PubMed] [Google Scholar]
- 39.Whitman M. Smads and early developmental signaling by the TGF-β superfamily. Genes Dev. 1998;12:2445–2462. doi: 10.1101/gad.12.16.2445. [DOI] [PubMed] [Google Scholar]
- 40.Wieser R, Wrana J L, Massagué J. GS domain mutations that constitutively activate TβR-I, the downstream signaling component in the TGF-β receptor complex. EMBO J. 1995;14:2199–2208. doi: 10.1002/j.1460-2075.1995.tb07214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wrana J L, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang X F, Massagué J. TGF-β signals through a heteromeric protein kinase receptor complex. Cell. 1992;11:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- 42.Wrana J L, Attisano L, Wieser R, Ventura F, Massagué J. Mechanism of activation of the TGF-β receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 43.Wu G, Chen Y G, Ozdamar B, Gyuricza C A, Chong P A, Wrana J L, Massagué J, Shi Y. Structural basis of Smad2 recognition by the Smad anchor for receptor activation. Science. 2000;287:92–97. doi: 10.1126/science.287.5450.92. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Feng X, We R, Derynck R. Receptor-associated Mad homologues synergize as effectors of the TGF-β response. Nature. 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, Musci T, Derynck R. The tumor suppressor Smad4/DPC4 as a central mediator of Smad function. Curr Biol. 1997;7:270–276. doi: 10.1016/s0960-9822(06)00123-0. [DOI] [PubMed] [Google Scholar]