Abstract
Six4 is a member of the Six family genes, homologues of Drosophila melanogaster sine oculis. The gene is thought to be involved in neurogenesis, myogenesis, and development of other organs, based on its specific expression in certain neuronal cells of the developing embryo and in adult skeletal muscles. To elucidate the biological roles of Six4, we generated Six4-deficient mice by replacing the Six homologous region and homeobox by the β-galactosidase gene. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside staining of the heterozygous mutant embryos revealed expression of Six4 in cranial and dorsal root ganglia, somites, otic and nasal placodes, branchial arches, Rathke's pouch, apical ectodermal ridges of limb buds, and mesonephros. The expression pattern was similar to that of Six1 except at the early stage of embryonic day 8.5. Six4-deficient mice were born according to the Mendelian rule with normal gross appearance and were fertile. No hearing defects were detected. Six4-deficient embryos showed no morphological abnormalities, and the expression patterns of several molecular markers, e.g., myogenin and NeuroD3 (neurogenin1), were normal. Our results indicate that Six4 is not essential for mouse embryogenesis and suggest that other members of the Six family seem to compensate for the loss of Six4.
Six family genes are homologues of Drosophila melanogaster sine oculis, one of the homeobox genes essential for compound eye formation (5). They are characterized by the presence of the Six domain and Six-type homeodomain in the encoded proteins, which confer specific DNA binding activity and function as transcription factors (12, 28). In mammals, six members of the family have so far been identified (2–4, 7, 8, 10, 12, 13, 31, 32, 34, 41; for a review, see reference 14). Each member of the family is expressed in a spatiotemporally regulated manner during embryogenesis. In mice, Six3 and Six6 are exclusively expressed in the developing forebrain and eyes (10, 22, 31, 40). In contrast, Six1, Six2, and Six5 show relatively broader expression patterns (15, 32). Six1 is expressed in the cranial and dorsal root ganglia, somites, otic and nasal placodes, branchial arches, limbs, Rathke's pouch, and nephrogenic cords (32). Six2 is expressed in head mesenchyme, branchial arches, limbs, and some mesenchymal regions surrounding the gastrointestinal tract (32). Six5 shows a broad expression in branchial arches, limb buds, telencephalon, eye, sclerotomes, and cartilages (15). Such distribution suggests that these genes play specific roles in embryogenesis.
Overexpression and misexpression experiments showed that Six3 and Six6 genes are involved in forebrain and eye organogenesis (18, 21, 30, 48). Consistently, SIX3 mutations cause holoprosencephaly, a severe malformation of the brain in humans (42). SIX5 is located immediately downstream of the CTG trinucleotide repeats whose expansion causes myotonic dystrophy (3). Downregulation of the gene was observed in myotonic dystrophy patients and is thought to be responsible for some of the symptoms of the disease such as cataracts (16, 44). In agreement with this, Six5-heterozygous and -homozygous mutant mice develop cataracts (15, 35).
Six4 was originally identified as a binding factor to the positive regulatory element of the Na+,K+-ATPase α1 subunit gene (Atp1a1) (12, 38). Immunostaining showed the presence of Six4 protein in some populations of neuronal cells in developing mouse embryos and developing retina (27, 29). In adult mice, Six4 protein is localized in skeletal muscles, as demonstrated by gel retardation assay (37). These observations suggest that this gene is involved in neurogenesis, myogenesis, and probably the development of other organs. In myogenesis, myogenin plays an important role along with other myogenic genes such as MyoD, Myf5, and MRF4 (33). Promoter analysis with transgenic mice demonstrated that the MEF3 site in the promoter region is essential for myogenin expression (37). Mouse Six1 and Six4 proteins are present in the developing somites in which myogenin expression is activated and bind to the MEF3 site in the myogenin promoter, as shown by gel retardation assay (37). Furthermore, Six4 can activate the myogenin gene promoter alone or in synergy with the specific cofactor, Eya, through direct binding to the MEF3 site in cultured cells (28). These results support the notion that Six4 is one of the genes that control myogenesis through activation of myogenin. In addition, Eya1, one of the specific cofactors of Six4, is implicated in branchio-oto-renal syndrome, a dominantly inherited disorder characterized by hearing loss and branchial arch and renal anomalies in humans (1). Eya1-deficient mice lack ears and kidneys, and heterozygous mutant mice show hearing loss and renal anomalies, as seen in human branchio-oto-renal syndrome (46). Because immunostaining showed the presence of Six4 protein in acoustic ganglia and otic vesicles (29), it is possible that Six4 is involved in the development of the ear in association with Eya1. Nevertheless, the biological function of Six4 in development is not clear, due to the lack of natural mutants, knockout models, and overexpression experiments. To access the biological function of Six4, we generated Six4-deficient mice and analyzed phenotypic changes both in adults and in embryos. We found no apparent changes in morphology and expression patterns of some marker genes in Six4-deficient mice. Functionally, hearing ability was normal. The reason for the lack of phenotype in Six4-deficient mice and the possible compensation among Six family genes is discussed.
MATERIALS AND METHODS
Construction of Six4 gene targeting vector.
The complete murine Six4 locus was cloned from a 129/SvJ genomic library (Stratagene, La Jolla, Calif.) and partially sequenced, and the exon-intron organization was determined (34). PCR mutagenesis using AmpliTaq Gold DNA polymerase (Perkin Elmer-Cetus, Foster City, Calif.) was performed to introduce a KpnI site immediately downstream of the initiation codon to allow the insertion of an in-frame lacZ gene, with the forward primer (9705; 5′-CAA AAG GAG GAG TCA CGT T-3′) and reverse primer (9706; 5′-CGG GGT ACC CTT TCC ATC CCA TTC TC-3′). The PCR products were sequenced with Sequencing Pro kits (Toyobo, Osaka, Japan). The targeting vector was constructed as follows. The lacZ fragment (KpnI-BamHI) from pCH110 (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) was ligated to the 3′ end of a 5′ homology region (XbaI-KpnI; 5.1 kb), the PGKneobpA cassette (XhoI-PvuII) from pPGKneobpA (36) was ligated to the 5′ end of a 3′ homology region (SmaI-SalI; 2.5 kb), and the resulting two inserts were ligated together. Finally, the diphtheria toxin A cassette (XhoI-NotI) from pMC1DTpA (47) was ligated to the 3′ end of the 3′ homology region. The plasmid was linearized with SalI at the 5′ end. In this construct, the homeobox and the Six homologous region, which together encode a specific DNA binding domain, were completely removed.
ES cell screening and chimeric mouse production.
The linearized targeting vector (60 μg) was electroporated (250 V; 500 μF) into 107 E14-1 embryonic stem (ES) cells (19), and transformants were selected with 250 μg (active form) G418 (Gibco/BRL) per ml for 7 to 10 days. Homologous recombinants were screened by PCR as follows. The forward primer in the PGKneobpA cassette was 5′-CTC TAT GGC TTC TGA GGC GGA AAG-3′, and the reverse primer was 5′-GGC AAG GTC TGC TAG AAA CGG TAC-3′. PCR was carried out with LA Taq DNA polymerase (TaKaRa, Kyoto, Japan) for 35 cycles at 94°C for 1 min, 60°C for 2 min, and 72°C for 3 min in a volume of 36 μl. Homologous recombination was further confirmed by Southern blot hybridization as follows: 15 μg of DNA from PCR-positive clones was digested with BamHI (to confirm 5′ homologous recombination) or SacI (to confirm 3′ homologous recombination), electrophoresed through a 0.7% agarose gel, and transferred to Hybond-N+ membranes (Amersham Pharmacia Biotech). Hybridization was carried out with a buffer containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 2.5× Denhardt's solution, 0.5% sodium dodecyl sulfate, 0.1 mg of heat-denatured herring testis DNA per ml, and radiolabeled probes specific to either the 5′ or 3′ restriction fragment (see below). Two ES clones that yielded hybridization bands of the correct size gave germ line chimeras by the aggregation method (26). Once homologous recombination and germ line transmission were confirmed, mouse genotyping was carried out by PCR as follows. The forward primer in exon 1 was 5′-ACA TCA AGC AGG AGA ATG GGA TGG-3′. The reverse primer specific to lacZ in the mutant allele was 5′-CCG TAA TGG GAT AGG TTA CGT TGG-3′. The reverse primer for the wild-type allele was 5′-AGA AGT TCC GAG TGG AGT TGT ACC-3′. PCR was carried out with AmpliTaq Gold DNA polymerase for 35 cycles at 95°C for 58 s, 63°C for 28 s, and 72°C for 55 s in a volume of 9 μl. The germ line chimeras were backcrossed to C57BL/6 mice. F2 or F3 mice were backcrossed to C57BL/6 mice or intercrossed, and the resulting founder mice were used in the following experiments. Mice were maintained under specific-pathogen-free conditions in environmentally controlled clean rooms at the Laboratory Animal Research Center, Institute of Medical Science, University of Tokyo, and the Center for Experimental Medicine, Jichi Medical School. The experiments were conducted according to the institutional ethical guidelines for animal experiments and safety guidelines for gene manipulation experiments.
Northern blot analysis.
Total RNA was extracted with Isogen (Nippon Gene, Tokyo, Japan) from adult tissues or embryos, electrophoresed through a 1.2% denatured agarose gel containing 2.2 M formaldehyde, and transferred to Hybond-N+ membranes (Amersham Pharmacia Biotech). Hybridization was carried out under the same conditions used for Southern blot analysis described above.
Preparation of probes.
For Southern and Northern blot analyses, 25 ng of the following DNA fragments was used to synthesize 32P-labeled probes with a Megaprime DNA labeling kit (Amersham Pharmacia Biotech): 0.6-kb BamHI-HindIII fragment 1.8 kb upstream of the 5′ end of the 5′ homology region (for the 5′ probe in Southern analysis), a 0.8-kb KpnI-XbaI fragment immediately downstream of the 3′ end of the 3′ homology region (for the 3′ probe in Southern analysis), a 2.3-kb XhoI-XbaI fragment of mouse Six4 cDNA (SM type, for Six4) (12), a 0.7-kb PstI-PstI fragment of Six1-LZ8 (for Six1) (32), a 1.1-kb EcoRI-Sau3AI fragment of pfSix2 (for Six2) (28), a 2.4-kb NotI-BglII fragment of pfSix5 (for Six5) (28), a 2.2-kb NcoI-NcoI fragment of rat Atp1a1 cDNA (for Atp1a1) (9), a 1.5-kb EcoRI-EcoRI fragment of pEMSV2α-MGN (for myogenin) (45), and a 0.8-kb fragment amplified from mRNAs extracted from HeLa cells by reverse transcription-PCR using primers 5′-TGGTGGGAATGGGTCAGA-3′ and 5′-AGGGAGGAAGAGGATGCG-3′ (for β-actin).
X-Gal staining of mouse embryo.
Embryos were removed from the uterus in ice-cold phosphate-buffered saline (PBS). Genotyping was carried out by PCR using DNA extracted from yolk sac. For whole-mount staining, embryos were fixed in the fixing solution (1% formaldehyde, 0.2% glutaraldehyde, and 0.02% Nonidet P-40 in PBS) on ice for 30 min, washed twice in PBS at room temperature for 30 min, and then stained in the 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining solution [1 mg/ml X-Gal, 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, and 2 mM MgCl2 in PBS] at 30°C overnight. After being stained, embryos were washed and stored in PBS at 4°C. For sections, embryos were embedded in freezing medium and frozen on dry ice. The embedded embryos were sectioned at a 30-μm thickness at −15°C. Each section was transferred onto a silanized slide, allowed to dry, and fixed in a fixing solution (0.2% glutaraldehyde, 2 mM MgCl2, and 5 mM EGTA in PBS). After being washed three times in a washing solution (2 mM MgCl2, 0.01% sodium deoxycholate, and 0.02% Nonidet P-40 in PBS), each section was stained in the X-Gal staining solution as above.
Hematoxylin-eosin staining.
Skeletal muscles of the hindlimb of wild-type and Six4-deficient mice were fixed in 10% formalin. After being washed with water, fixed samples were dehydrated by sequentially increasing concentrations of ethanol, cleared in xylene, and then embedded in paraffin. The embedded samples were sectioned at a 5-μm thickness, and each section was transferred onto a slide, dewaxed in xylene, rehydrated by sequentially decreasing concentrations of ethanol, stained in hematoxylin solution [0.1% hematoxylin, 5% K2Al2(SO4)4, 0.02% NaIO3, 5% chloral hydrate, 0.1% citric acid, and 20% glycerol] for 15 min, and differentiated in water. Then, the sections were counterstained in eosin solution (0.25% eosin Y, 0.55% acetic acid, and 60% ethanol) for 30 min; differentiated sequentially in 70, 80, and 90% ethanol solutions; dehydrated in absolute ethanol; cleared in xylene; and coverslipped.
In situ hybridization.
Embryos were removed from the uterus in ice-cold PBS. Genotyping was carried out by PCR using DNA extracted from yolk sac. For whole-amount in situ hybridization, embryos were fixed in a fixing solution (4% paraformaldehyde in PBS) at 4°C overnight. After being washed twice in PBT (0.1% Tween 20 in PBS), embryos were dehydrated by being washed sequentially in 25, 50, and 75% methanol solutions in PBT and 100% methanol and then rehydrated by being washed sequentially in 75, 50, and 25% methanol solutions in PBT and then in PBT alone. After being bleached in 6% H2O2 in PBT for 1 h, embryos were treated in 10 μg of proteinase K per ml in PBT for 15 min, washed with 2 mg of glycine per ml in PBT and in PBT alone, and refixed in 0.2% glutaraldehyde and 4% formaldehyde in PBT for 20 min. After being washed in PBT, embryos were prehybridized in a prehybridization buffer (50% formamide, 5× SSC [pH 5.0], 50 μg of yeast tRNA per ml, 1% sodium dodecyl sulfate, and 50 μg of heparin per ml) at 70°C for 1 h and then hybridized in a hybridization buffer containing 1 μg of digoxigenin (DIG)-labeled antisense RNA probe (see below) at 70°C overnight. After being washed, embryos were treated twice in 100 μg of RNaseA per ml in 0.5 M Tris-HCl (pH 7.5) and 0.1% Tween 20 at 37°C for 30 min, followed by blocking in 10% fetal calf serum in TBST (0.14 M NaCl, 2.7 mM KCl, 25 mM Tris-HCl [pH 7.5], and 0.1% Tween 20) for 90 min. Then, embryos were treated in TBST containing 1% fetal calf serum and alkaline phosphatase-labeled anti-DIG antibody (Boehringer GmbH, Mannheim, Germany) at 4°C overnight. After being washed in TBST and NTMT (100 mM NaCl, 100 mM Tris-HCl [pH 9.5], 50 mM MgCl2, and 0.1% Tween 20), embryos were stained in NTMT containing nitroblue tetrazolium and X-phosphate (Boehringer). Frozen sections (10-μm thick) were cut on a cryostat and attached to slides coated with Vectabond reagent (Vector Laboratories, Burlingame, Calif.). Samples were treated with proteinase K (1 mg/ml) at 37°C for 10 min, refixed in 4% paraformaldehyde, and hybridized overnight with the DIG-labeled RNA probe. The hybridized mRNA was detected by alkaline phosphatase-conjugated anti-DIG Fab fragments (Boehringer) according to the procedure described by Wilkinson (43).
DIG (Boehringer)-labeled antisense RNA probes were prepared from the following linearized plasmids with DIG RNA Labeling mixture (Boehringer) and T3 or T7 RNA polymerase according to the instructions provided by the manufacturer: pKSMGN1, which contains a 1.5-kb EcoRI-EcoRI fragment of pEMSV2α-MGN in pBulescript KS(+) (for myogenin) (45); pKS-ngn1/E2 (for NeuroD3) (23); mouse NeuroD1 pSK P/P350#5 (for NeuroD1) (25); and pSK, which contains a 630-bp PstI(1545)-PstI(2175) fragment from Six4 cDNA SM type (for Six4) (12).
ABR.
Hearing was assessed by recording auditory brainstem response (ABR) as described previously (39). Acoustic stimuli, consisting of tone bursts at frequencies of 10, 20, 30, and 40 kHz, with a rise-and-fall time of 1 ms, a 5-ms duration, and repetition every 70 ms, were delivered to each mouse with a sound stimulator (DPS-725; Diamedical System) and a speaker (PT-RIII; Pioneer) in an open field. A microcomputer (Synapac 1100; NEC Sanei) was used to analyze the response. For each time point, 500 responses for each mouse were recorded and filtered for bandwidths of 100 to 3,000 Hz.
RESULTS AND DISCUSSION
Generation of Six4-deficient mice.
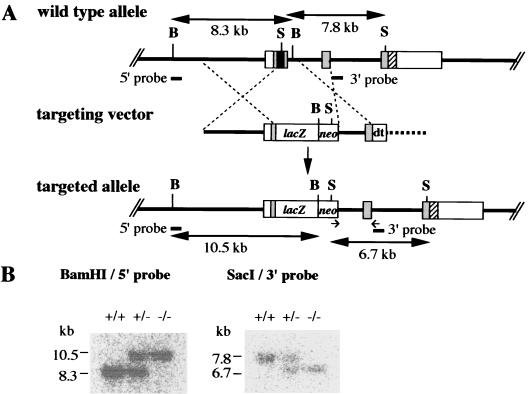
To inactivate Six4, an in-frame β-galactosidase (lacZ) reporter and a neomycin-resistant cassette (neo) were introduced for monitoring the expression of endogenous Six4 and for positive selection, respectively, which replaced the Six homologous region and the homeobox in exon 1 (Fig. 1).
FIG. 1.
Targeted disruption of mouse Six4. (A) Structures of the wild-type allele, targeting vector, and targeted allele. Boxes represent exons. Gray shading indicates coding regions, and black shading indicates the Six homologous region and homeobox. The hatched region marks the region encoding the transactivation domain. The targeting vector consisted of the 5′ homology region, lacZ, neo, 3′ homology region, and at the 3′ end the diphtheria toxin A gene (dt) for negative selection. Arrows beneath the target allele represent PCR primers (9705 and 9706) for screening. Restriction fragments detected by Southern blot analysis are shown by horizontal arrows with their sizes in kilobases. B, BamHI; S, SacI. (B) Southern blot analysis of mouse tail DNA isolated from the founder mice from a mating of heterozygous parents. DNAs were digested with BamHI or SacI and hybridized with the probes indicated in panel A. +/+, wild-type mouse; +/−, heterozygous mutant mouse; −/−, homozygous mutant mouse.
No obvious phenotype was apparent in heterozygous mutants. When heterozygous mutants were intercrossed, wild-type offspring, heterozygotes, and homozygotes were born according to the Mendelian rule (Table 1). The heterozygotes and homozygotes had a normal appearance, and both male and female homozygotes were fertile.
TABLE 1.
Genotypic analysis of founder mice at 3 or 4 weeks of age from heterozygous × heterozygous intercross
| Sex | No. (%) of genotype
|
||
|---|---|---|---|
| Wild type | Heterozygous | Homozygous | |
| Male | 33 (31) | 56 (53) | 17 (16) |
| Female | 31 (26) | 59 (49) | 31 (26) |
| Combined | 64 (28) | 115 (51) | 48 (21) |
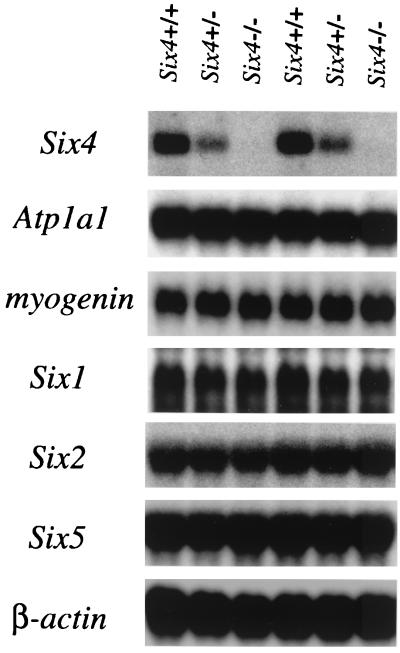
In our targeting strategy, exon 3, which encodes a transcriptional activation domain (12), was left intact. To confirm that exon 3 was not transcribed irregularly to produce aberrant Six4 molecules with some activity, Northern blot analysis of the total RNA from embryonic day 11.5 (E11.5) whole embryos (Fig. 2) and from adult skeletal muscle (data not shown) was performed, using a probe that covered the 3′ part of exon 1, exon 2, and the coding region of exon 3. The amount of Six4 transcripts was proportional to the gene dosage, and in homozygous mutants, no Six4 transcripts of correct size or irregular transcripts were detected. Thus, we concluded that the Six4 gene was functionally inactivated in the targeted allele.
FIG. 2.
Analysis of gene expression in wild-type and mutant mouse embryos. Ten micrograms of total RNA from E11.5 embryos of the indicated genotypes was analyzed by Northern hybridization with the indicated probes. Three independent experiments yielded essentially the same results, and two representative hybridization patterns are shown here. In spite of complete lack of Six4 mRNA in Six4−/− embryos, the expression levels of the genes analyzed except Six4 were not altered.
Expression pattern of Six4lacZ in heterozygous mutant embryos.
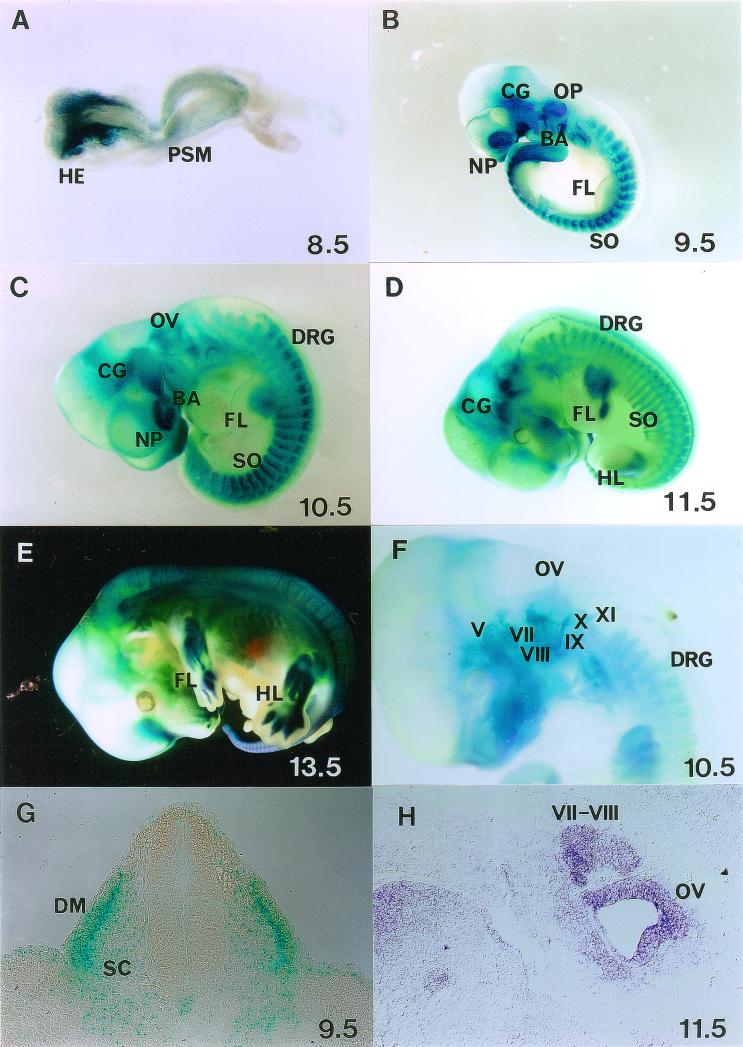
So far, Six4 expression has been analyzed by immunostaining, gel retardation assay, and Northern analysis in restricted areas of embryonic tissue and at restricted stages of development (12, 27, 29, 37). To analyze the expression pattern of Six4 during the entire developmental process, X-Gal staining was performed on heterozygous embryos (Fig. 3). Six4lacZ expression was detected in various ganglia, somites, nasal and otic placodes, branchial arches, and several other tissues, as summarized in Table 2. To our knowledge, this is the first comprehensive analysis of the Six4 expression pattern in mice. In a previous report, Six4 protein was detected mainly in the cranial and dorsal root ganglia by immunostaining (29). This is probably because of the higher level of expression of Six4 and/or a higher number of Six4-expressing cells in these ganglia than in other sites in which X-Gal staining was confirmed in our heterozygous mutant embryos.
FIG. 3.
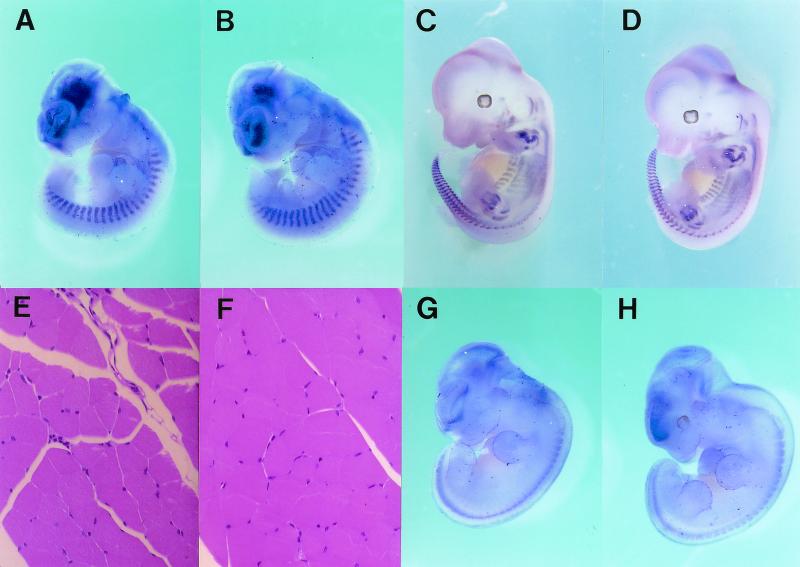
X-Gal staining of Six4 heterozygous mutant embryos showing spatiotemporally regulated expression of Six4 in somites and myotomes (SO), cranial and dorsal root ganglia (V to XI) (DRG), sensory placodes (otic [OP] and nasal [NP]), and some other restricted areas. (A) At E8.5, Six4 expression commences in the surface ectoderm of the head region (HE) and presomitic mesoderm (PSM). (B) At E9.5, note the expression of Six4 in OP, NP, SO, branchial arches (BA), and cranial ganglia (CG). (C) At E10.5, Six4 expression is evident also in DRG. (D) At E11.5, Six4 is expressed also in mesenchymal tissues of fore- (FL) and hindlimb (HL) buds at the posterior margin. (E) At E13.5, Six4 expression in digits becomes evident. (F) The embryo at E10.5 was cleared with benzylbenzoate-benzylalcohol after staining. Note the staining of cranial ganglia V and VII-XI, DRG, and otic vesicles (OV). (G) X-Gal staining of a transverse section of an embryo at E9.5 at the hindlimb level shows strong staining in dermamyotomes (DM) and weak staining at sclerotomes (SC) of somites. (H) In situ hybridization of a sagittal section of an embryo (Jcl:ICR strain) at E11.5, showing Six4 expression at cranial ganglia (VII and VIII) and OV. As analyzed, in situ hybridization to Six4 transcripts and X-Gal staining of heterozygotes showed essentially the same pattern. One of the typical hybridization results are shown.
TABLE 2.
Expression pattern of Six4 determined by X-Gal staining of heterozygotes
| Stage | Tissuea |
|---|---|
| Embryonic | |
| E8.5 | Surface ectoderm outside the neural folds, somites, presomitic mesoderm |
| E9.5 | Nasal and otic placodes, cranial ganglia, branchial arches, somites (dermamyotomes and sclerotomes), AER |
| E10.5–11.5 | Nasal pits, otic vesicles, cranial ganglia (V, VII–XI), dorsal root ganglia, branchial arches, somites, myotomes, limb mesenchyme, AER, notochord, mesonephros |
| E12.5–E13.5 | Skeletal muscles, mesenchyme in limbs and digits, nasal epithelium, inner ear |
| Adult | Skeletal muscle, nasal epithelium, cochlea, parathyroid, salivary gland |
AER, apical ectodermal ridges.
In chickens, Six4 is expressed in a pattern similar to that of mouse Six4, although chick Six4 is expressed in additional tissues such as the optic placodes and motoneurons in the spinal cord (6). Moreover, in zebra fish two orthologues of mammalian Six4, Six4.1 and Six4.2, exhibit essentially the same expression pattern with mouse Six4 in combination (17). Thus, the Six4 expression pattern is essentially conserved through vertebrate evolution. In addition, the expression pattern of Six4 in the mouse was strikingly similar to that of Six1, except in the head region at E8.5, immediately after the onset of their expression (Six4 in surface ectoderm outside the neural folds; Six1 in head mesenchyme) (Fig. 3A) (32). Because Six1 and Six4 are located in tandem on mouse chromosome 12 (H. Ozaki, unpublished data), these two genes might share common cis-regulatory elements. Alternatively, cis-regulatory elements that control the expression of each gene might be well conserved between these two genes, although their protein structure itself was different in that Six4 protein, but not Six1 protein, has a large C-terminus region containing a transactivation domain (12, 32).
Eya1 and Eya2, the putative coactivators of Six4 and other Six family genes, are expressed in an extensively overlapping pattern with Six4, for example, in cranial ganglia, cranial placodes, and somites, indicating the possible interaction of Six4 with Eya1 and/or Eya2 in these organs, as shown by transient transfection assays (28).
Hearing ability in Six4-deficient mice.
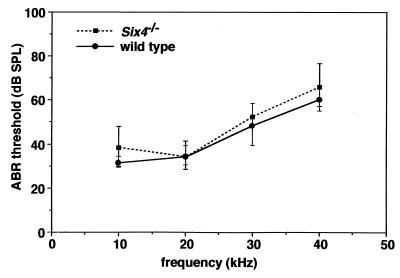
Six4 showed overlapping expression with Eya1 in the otic vesicle and in the acoustic ganglion (11). Considering the functional cooperativity between Six4 and Eya1 in target gene activation (28), we suspected that Six4-deficient mice might have hearing defects. However, hearing ability was normal in Six4-deficient mice as tested by ABR (Fig. 4).
FIG. 4.
ABR thresholds in wild-type and Six4-deficient mouse ears. Data are mean threshold ± standard deviation of five wild-type and five Six4-deficient mice at 7 to 9 weeks of age. The result shows that hearing function determined by ABR is normal in Six4-deficient mice (repeated-measure analysis of variance test; P > 0.05).
Morphological analysis and X-Gal staining of Six4 homozygous mutant embryos.
Six4-deficient mice seemed normal in appearance and in anatomical aspects after birth. We then assessed the morphological abnormalities in Six4 homozygous mutant embryos, focusing on the sites of Six4lacZ expression. We compared the expression pattern of Six4 in heterozygous and homozygous mutant embryos by X-Gal staining. The overall expression pattern was the same except that staining was stronger in homozygotes than in heterozygotes, probably reflecting the gene dosage (data not shown).
Expression of putative targets of Six4 and markers for somites, muscle, and cranial and dorsal root ganglia.
Because Six4 was previously reported to activate the myogenin promoter through the MEF3 site (28, 37), we analyzed the expression of myogenin by in situ hybridization. The staining pattern was exactly the same in wild-type and Six4-deficient embryos (Fig. 5A to D). Furthermore, Northern analysis revealed that the myogenin expression level was similar in wild-type mice, heterozygotes, and homozygotes (Fig. 2). In accordance with these findings, skeletal muscles of adult mice (Fig. 5E and F) and of embryos at E16.5 (data not shown) of each genotype were normal as tested by hematoxylin-eosin staining.
FIG. 5.
Expression of marker genes for somite-myotome (myogenin) and neuronal cells (neuroD3) in wild-type (A, C, and G) and Six4-deficient (B, D, and H) embryos. E11.5 (A and B) and E12.5 (C and D) embryos were analyzed for myogenin expression by in situ hybridization. Adult skeletal muscles of wild-type (E) and Six4-deficient (F) mice were also analyzed by hematoxylin-eosin staining. The expression of neuroD3, one of the neural marker genes, in E11.5 embryos was also analyzed by in situ hybridization (G and H). For both probes, the pattern and intensity of the staining were not different between wild-type and Six4-deficient embryos.
The cranial and dorsal root ganglia are also major sites of Six4 expression, and therefore we analyzed the expression of specific molecular markers in these ganglia. In situ hybridization for NeuroD3 (neurogenin1) (24) (Fig. 5G and H) and NeuroD1 (neuroD) (data not shown) (20) revealed no difference in the expression pattern between wild-type and homozygous mutant embryos (strong staining in the telencephalon of the homozygous mutant embryo was not reproducible).
We also assessed the effect of loss of Six4 on Atp1a1 expression. As shown in Fig. 2, the expression level of Atp1a1 was not altered. It has been shown that the regulatory region of Atp1a1 is composed of multiple elements, but no single element mutation reduced the expression of the gene, at least in several cultured cells (38). As such, although compensation by other Six proteins may exist, the presence of several other binding factors may be sufficient to activate the promoter up to the normal level.
Compensation among Six genes.
Compensation among Six genes is the most likely explanation for the lack of morphological and functional abnormalities and changes in marker gene expression in Six4-deficient mice. Of the six Six genes identified, Six3 and Six6 are expressed in restricted areas of the forebrain, in which Six4 is not expressed (10, 22, 31, 40). Six3 has a different DNA binding specificity and is unable to cooperate with Eya (13, 28). Thus, it is not plausible that Six3 and Six6 compensate for Six4 function. On the other hand, Six1, Six2, and Six5 proteins share a DNA binding specificity with Six4 protein (13, 28, 37). Six1 shows mostly the same expression pattern with Six4 (32), and Six5 mostly resembles Six4 with respect to the molecular architecture with its large C-terminal portion and overall amino acid sequence similarity (13, 14). Thus, these two members are the probable candidates to compensate for the loss of Six4. To gain insight into the compensation mechanism, the expression of Six1, Six2, and Six5 in Six4-deficient mice was analyzed by Northern hybridization. The expression levels of these genes were not altered among different genotypes (Fig. 2). This finding suggests that the normal expression levels of Six1, Six2, and Six5 are sufficient to compensate for the loss of Six4 and to activate common target genes.
Similarly, the Six5-deficient mouse manifests limited phenotypes such as a higher rate of cataract formation (15, 35), compared to the relatively broad expression of Six5 (15), suggesting compensation for the loss of Six5 by Six1, Six2, and/or Six4 in these tissues. Because of such compensatory mechanisms among Six1, Six2, Six4, and Six5, knockout mouse models deficient in a single Six gene are unlikely to contribute to our understanding of their biological functions. Generation of Six1-Six4 and Six4-Six5 double knockout mice should allow us to understand the compensation between them and the biological function of Six4.
ACKNOWLEDGMENTS
We thank F. Relaix for discussion and critical reading of the manuscript, S. J. Tapscott for providing mouse NeuroD cDNA, Q. Ma for providing neurogenin1 cDNA, and T. Yagi for providing pMC1DTpA. We also thank M. Yamakado, K. Ikeda, and S. Sato for discussion and H. Ohto and M. Kikuchi for technical assistance.
This work was supported by grants from the Ministry of Education, Science, Sports, and Culture of Japan and from the Ministry of Health and Welfare of Japan and by the Jichi Medical School Young Investigator Award.
REFERENCES
- 1.Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Weil D, Cruaud C, Sahly I, Leibovici M, Bitner-Glindzicz M, Francis M, Lacombe D, Vigneron J, Charachon R, Boven K, Bedbeder P, Van Regemorter N, Weissenbach J, Petit C. A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat Genet. 1997;15:157–164. doi: 10.1038/ng0297-157. [DOI] [PubMed] [Google Scholar]
- 2.Boucher C A, Carey N, Edwards Y H, Siciliano M J, Johnson K J. Cloning of the human SIX1 gene and its assignment to chromosome 14. Genomics. 1996;33:140–142. doi: 10.1006/geno.1996.0172. [DOI] [PubMed] [Google Scholar]
- 3.Boucher C A, King S K, Carey N, Krahe R, Winchester C L, Rahman S, Creavin T, Meghji P, Bailey M E, Chartier F L, Brown S D, Siciliano M J, Johnson K J. A novel homeodomain-encoding gene is associated with a large CpG island interrupted by the myotonic dystrophy unstable (CTG)n repeat. Hum Mol Genet. 1995;4:1919–1925. doi: 10.1093/hmg/4.10.1919. [DOI] [PubMed] [Google Scholar]
- 4.Boucher C A, Winchester C L, Hamilton G M, Winter A D, Johnson K J, Bailey M E. Structure, mapping and expression of the human gene encoding the homeodomain protein, SIX2. Gene. 2000;247:145–151. doi: 10.1016/s0378-1119(00)00105-0. [DOI] [PubMed] [Google Scholar]
- 5.Cheyette B N, Green P J, Martin K, Garren H, Hartenstein V, Zipursky S L. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 6.Esteve P, Bovolenta P. cSix4, a member of the six gene family of transcription factors, is expressed during placode and somite development. Mech Dev. 1999;85:161–165. doi: 10.1016/s0925-4773(99)00079-9. [DOI] [PubMed] [Google Scholar]
- 7.Gallardo M E, Lopez-Rios J, Fernaud-Espinosa I, Granadino B, Sanz R, Ramos C, Ayuso C, Seller M J, Brunner H G, Bovolenta P, Rodriguez de Cordoba S. Genomic cloning and characterization of the human homeobox gene SIX6 reveals a cluster of SIX genes in chromosome 14 and associates SIX6 hemizygosity with bilateral anophthalmia and pituitary anomalies. Genomics. 1999;61:82–91. doi: 10.1006/geno.1999.5916. [DOI] [PubMed] [Google Scholar]
- 8.Granadino B, Gallardo M E, Lopez-Rios J, Sanz R, Ramos C, Ayuso C, Bovolenta P, Rodriguez de Cordoba S. Genomic cloning, structure, expression pattern, and chromosomal location of the human SIX3 gene. Genomics. 1999;55:100–105. doi: 10.1006/geno.1998.5611. [DOI] [PubMed] [Google Scholar]
- 9.Hara Y, Urayama O, Kawakami K, Nojima H, Nagamune H, Kojima T, Ohta T, Nagano K, Nakao M. Primary structures of two types of alpha-subunit of rat brain Na+,K+-ATPase deduced from cDNA sequences. J Biochem. 1987;102:43–58. doi: 10.1093/oxfordjournals.jbchem.a122039. [DOI] [PubMed] [Google Scholar]
- 10.Jean D, Bernier G, Gruss P. Six6 (Optx2) is a novel murine Six3-related homeobox gene that demarcates the presumptive pituitary/hypothalamic axis and the ventral optic stalk. Mech Dev. 1999;84:31–40. doi: 10.1016/s0925-4773(99)00068-4. [DOI] [PubMed] [Google Scholar]
- 11.Kalatzis V, Sahly I, El-Amraoui A, Petit C. Eya1 expression in the developing ear and kidney: towards the understanding of the pathogenesis of branchio-oto-renal (BOR) syndrome. Dev Dyn. 1998;213:486–499. doi: 10.1002/(SICI)1097-0177(199812)213:4<486::AID-AJA13>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 12.Kawakami K, Ohto H, Ikeda K, Roeder R G. Structure, function and expression of a murine homeobox protein AREC3, a homologue of Drosophila sine oculis gene product, and implication in development. Nucleic Acids Res. 1996;24:303–310. doi: 10.1093/nar/24.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawakami K, Ohto H, Takizawa T, Saito T. Identification and expression of six family genes in mouse retina. FEBS Lett. 1996;393:259–263. doi: 10.1016/0014-5793(96)00899-x. [DOI] [PubMed] [Google Scholar]
- 14.Kawakami K, Sato S, Ozaki H, Ikeda K. Six family genes—structure and function as transcription factors and their roles in development. Bioessays. 2000;22:616–626. doi: 10.1002/1521-1878(200007)22:7<616::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 15.Klesert T R, Cho D H, Clark J I, Maylie J, Adelman J, Snider L, Yuen E C, Soriano P, Tapscott S J. Mice deficient in Six5 develop cataracts: implications for myotonic dystrophy. Nat Genet. 2000;25:105–109. doi: 10.1038/75490. [DOI] [PubMed] [Google Scholar]
- 16.Klesert T R, Otten A D, Bird T D, Tapscott S J. Trinucleotide repeat expansion at the myotonic dystrophy locus reduces expression of DMAHP. Nat Genet. 1997;16:402–406. doi: 10.1038/ng0897-402. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi M, Osanai H, Kawakami K, Yamamoto M. Expression of three zebrafish Six4 genes in the cranial sensory placodes and the developing somites. Mech Dev. 2000;98:151–155. doi: 10.1016/s0925-4773(00)00451-2. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi M, Toyama R, Takeda H, Dawid I B, Kawakami K. Overexpression of the forebrain-specific homeobox gene six3 induces rostral forebrain enlargement in zebrafish. Development. 1998;125:2973–2982. doi: 10.1242/dev.125.15.2973. [DOI] [PubMed] [Google Scholar]
- 19.Kuhn R, Rajewsky K, Muller W. Generation and analysis of interleukin-4 deficient mice. Science. 1991;254:707–710. doi: 10.1126/science.1948049. [DOI] [PubMed] [Google Scholar]
- 20.Lee J K, Cho J H, Hwang W S, Lee Y D, Reu D S, Suh-Kim H. Expression of neuroD/BETA2 in mitotic and postmitotic neuronal cells during the development of nervous system. Dev Dyn. 2000;217:361–367. doi: 10.1002/(SICI)1097-0177(200004)217:4<361::AID-DVDY3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 21.Loosli F, Winkler S, Wittbrodt J. Six3 overexpression initiates the formation of ectopic retina. Genes Dev. 1999;13:649–654. doi: 10.1101/gad.13.6.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez-Rios J, Gallardo M E, Rodriguez de Cordoba S, Bovolenta P. Six9 (Optx2), a new member of the six gene family of transcription factors, is expressed at early stages of vertebrate ocular and pituitary development. Mech Dev. 1999;83:155–159. doi: 10.1016/s0925-4773(99)00017-9. [DOI] [PubMed] [Google Scholar]
- 23.Ma Q, Chen Z, del Barco Barrantes I, de la Pompa J L, Anderson D J. neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;20:469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- 24.Ma Q, Sommer L, Cserjesi P, Anderson D J. Mash1 and neurogenin1 expression patterns define complementary domains of neuroepithelium in the developing CNS and are correlated with regions expressing notch ligands. J Neurosci. 1997;17:3644–3652. doi: 10.1523/JNEUROSCI.17-10-03644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCormick M B, Tamimi R M, Snider L, Asakura A, Bergstrom D, Tapscott S J. neuroD2 and neuroD3: distinct expression patterns and transcriptional activation potentials within the neuroD gene family. Mol Cell Biol. 1996;16:5792–5800. doi: 10.1128/mcb.16.10.5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder J C. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niiya A, Ohto H, Kawakami K, Araki M. Localization of Six4/AREC3 in the developing mouse retina; implications in mammalian retinal development. Exp Eye Res. 1998;67:699–707. doi: 10.1006/exer.1998.0562. [DOI] [PubMed] [Google Scholar]
- 28.Ohto H, Kamada S, Tago K, Tominaga S, Ozaki H, Sato S, Kawakami K. Cooperation of Six and Eya in activation of their target genes through nuclear translocation of Eya. Mol Cell Biol. 1999;19:6815–6824. doi: 10.1128/mcb.19.10.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohto H, Takizawa T, Saito T, Kobayashi M, Ikeda K, Kawakami K. Tissue and developmental distribution of Six family gene products. Int J Dev Biol. 1998;42:141–148. [PubMed] [Google Scholar]
- 30.Oliver G, Loosli F, Koster R, Wittbrodt J, Gruss P. Ectopic lens induction in fish in response to the murine homeobox gene Six3. Mech Dev. 1996;60:233–239. doi: 10.1016/s0925-4773(96)00632-6. [DOI] [PubMed] [Google Scholar]
- 31.Oliver G, Mailhos A, Wehr R, Copeland N G, Jenkins N A, Gruss P. Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development. 1995;121:4045–4055. doi: 10.1242/dev.121.12.4045. [DOI] [PubMed] [Google Scholar]
- 32.Oliver G, Wehr R, Jenkins N A, Copeland N G, Cheyette B N, Hartenstein V, Zipursky S L, Gruss P. Homeobox genes and connective tissue patterning. Development. 1995;121:693–705. doi: 10.1242/dev.121.3.693. [DOI] [PubMed] [Google Scholar]
- 33.Olson E N, Klein W H. bHLH factors in muscle development: dead lines and commitments, what to leave in and what to leave out. Genes Dev. 1994;8:1–8. doi: 10.1101/gad.8.1.1. [DOI] [PubMed] [Google Scholar]
- 34.Ozaki H, Yamada K, Kobayashi M, Asakawa S, Minoshima S, Shimizu N, Kajitani M, Kawakami K. Structure and chromosomal mapping of human SIX4 and mouse Six4 genes. Cytogenet Cell Genet. 1999;87:108–112. doi: 10.1159/000015407. [DOI] [PubMed] [Google Scholar]
- 35.Sarkar P S, Appukuttan B, Han J, Ito Y, Ai C, Tsai W, Chai Y, Stout J T, Reddy S. Heterozygous loss of Six5 in mice is sufficient to cause ocular cataracts. Nat Genet. 2000;25:110–114. doi: 10.1038/75500. [DOI] [PubMed] [Google Scholar]
- 36.Soriano P, Montgomery C, Geske R, Bradley A. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- 37.Spitz F, Demignon J, Porteu A, Kahn A, Concordet J P, Daegelen D, Maire P. Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proc Natl Acad Sci USA. 1998;95:14220–14225. doi: 10.1073/pnas.95.24.14220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki-Yagawa Y, Kawakami K, Nagano K. Housekeeping Na,K-ATPase α1 subunit gene promoter is composed of multiple cis elements to which common and cell type-specific factors bind. Mol Cell Biol. 1992;12:4046–4055. doi: 10.1128/mcb.12.9.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahashi K, Osawa N, Ohmura M, Kitamura K. Evaluation of inner ear histology and auditory brainstem response in Wriggle Mouse Sagami. Acta Otolaryngol. 1999;119:767–772. doi: 10.1080/00016489950180405. [DOI] [PubMed] [Google Scholar]
- 40.Toy J, Sundin O H. Expression of the optx2 homeobox gene during mouse development. Mech Dev. 1999;83:183–186. doi: 10.1016/s0925-4773(99)00049-0. [DOI] [PubMed] [Google Scholar]
- 41.Toy J, Yang J M, Leppert G S, Sundin O H. The optx2 homeobox gene is expressed in early precursors of the eye and activates retina-specific genes. Proc Natl Acad Sci USA. 1998;95:10643–10648. doi: 10.1073/pnas.95.18.10643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wallis D E, Roessler E, Hehr U, Nanni L, Wiltshire T, Richieri-Costa A, Gillessen-Kaesbach G, Zackai E H, Rommens J, Muenke M. Mutations in the homeodomain of the human SIX3 gene cause holoprosencephaly. Nat Genet. 1999;22:196–198. doi: 10.1038/9718. [DOI] [PubMed] [Google Scholar]
- 43.Wilkinson D G. Whole mount in situ hybridization of vertebrate embryos. In: Wilkinson D G, editor. In situ hybridization: a practical approach. New York, N.Y: Oxford IRL Press; 1992. pp. 75–83. [Google Scholar]
- 44.Winchester C L, Ferrier R K, Sermoni A, Clark B J, Johnson K J. Characterization of the expression of DMPK and SIX5 in the human eye and implications for pathogenesis in myotonic dystrophy. Hum Mol Genet. 1999;8:481–492. doi: 10.1093/hmg/8.3.481. [DOI] [PubMed] [Google Scholar]
- 45.Wright W E, Sassoon D A, Lin V K. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell. 1989;56:607–617. doi: 10.1016/0092-8674(89)90583-7. [DOI] [PubMed] [Google Scholar]
- 46.Xu P X, Adams J, Peters H, Brown M C, Heaney S, Maas R. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999;23:113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- 47.Yagi T, Nada S, Watanabe N, Tamemoto H, Kohmura N, Ikawa Y, Aizawa S. A novel negative selection for homologous recombinants using diphtheria toxin A fragment gene. Anal Biochem. 1993;214:77–86. doi: 10.1006/abio.1993.1459. [DOI] [PubMed] [Google Scholar]
- 48.Zuber M E, Perron M, Philpott A, Bang A, Harris W A. Giant eyes in Xenopus laevis by overexpression of XOptx2. Cell. 1999;98:341–352. doi: 10.1016/s0092-8674(00)81963-7. [DOI] [PubMed] [Google Scholar]