Abstract
It has been reported previously that the 5′ untranslated region of the mRNA encoding Apaf-1 (apoptotic protease-activating factor 1) has an internal ribosome entry site (IRES), whose activity varies widely among different cell types. Here it is shown that the Apaf-1 IRES is active in rabbit reticulocyte lysates, provided that the system is supplemented with polypyrimidine tract binding protein (PTB) and upstream of N-ras (unr), two cellular RNA binding proteins previously identified to be required for rhinovirus IRES activity. In UV cross-linking assays and electrophoretic mobility shift assays with individual recombinant proteins, the Apaf-1 IRES binds unr but not PTB; however, PTB binding occurs if unr is present. Over a range of different cell types there is a broad correlation between the activity of the Apaf-1 IRES and their content of PTB and unr. In cell lines deficient in these proteins, overexpression of PTB and unr stimulated Apaf-1 IRES function. This is the first example where an IRES in a cellular mRNA has been shown to be functionally dependent, both in vitro and in vivo, on specific cellular RNA binding proteins. Given the critical role of Apaf-1 in apoptosis, these results have important implications for the control of the apoptotic cascade.
The balance between cell proliferation and cell death is essential for the development and maintenance of multicellular organisms. The mechanisms for controlling the expression of proteins required for cell death to proceed are as complex as those required for cell proliferation, and in addition to control of transcription, it has been shown that regulation of translation is important (4).
In eukaryotic cells there are two major mechanisms by which protein synthesis can be initiated, cap-dependent scanning and internal ribosome entry. The former mechanism is the more commonly used and requires the binding of the eukaryotic initiation factor complex 4F (eIF4F) (which is composed of the cap binding protein eIF4E, the RNA helicase eIF4A, and the scaffold protein eIF4G to which they both bind) to the 7-methyl G at the 5′ end of the mRNA. This is followed by recruitment of the 40S ribosomal subunit and scanning to the first AUG codon in good context (for reviews, see references 8 and 26). For internal ribosome entry to occur, a complex structural element is formed in the 5′ untranslated region of an mRNA, and this allows the recruitment of the ribosome (8). This system has been extensively studied for the picornavirus family, and most of these viruses render cellular translation cap independent by the production of a protease that cleaves eIF4G, separating the eIF4E and eIF3 binding sites (for reviews, see references 17 to 19). Many picornavirus internal ribosome entry sites (IRESs) function efficiently in rabbit reticulocyte lysates, but others show high activity only in translation-competent extracts of nucleated cells (e.g., HeLa cells) or reticulocyte lysates supplemented with HeLa cell cytoplasmic extract. This difference is believed to be due to different protein factor requirements, some of which have been identified. Some viral IRESs, e.g., the encephalomyocarditis virus (EMCV) IRES, do not appear to require proteins other than canonical translation initiation factors for function (29), while others require an additional complex set of factors for activity. Such factors include polypyrimidine tract binding protein (PTB), which binds specifically to several viral IRESs, although the absolute requirement of viral IRESs for this factor differs. For example, PTB stimulates the initiation of translation by internal ribosome entry from hepatitis C and A virus RNA in vivo (7) and from the human rhinovirus (HRV) and poliovirus IRESs in vitro (15) but is not necessary for the activity of wild-type EMCV (21). The autoantigen La appears to be necessary for hepatitis C virus and poliovirus (16, 36), and poly(rC) binding protein 2 (PCBP-2) is required for poliovirus and rhinovirus IRESs (38) and is associated with the hepatitis C virus IRES (31). upstream of N-ras (unr, an RNA binding protein that contains five cold shock domains) is required for HRV IRES function, and a novel protein unrip (unr-interacting protein [14]) may also be important in this regard. More recently a 45-kDa protein termed ITAF45 has been cloned and found to be necessary for initiation to occur on the foot-and-mouth disease virus IRES but not on the Theiler's murine encephalomyelitis virus IRES (30).
Many examples now exist of eukaryotic cellular mRNAs that contain IRESs (8), and these are similarly used under conditions where cap-dependent translation is inhibited. One area that has been of interest is the control of protein synthesis during apoptosis, since when an apoptotic trigger is applied to cells there is a large reduction in global protein synthesis rates. This mimics viral infection of cells since the inhibition is also due to the cleavage of eukaryotic initiation factors (eIFs) (including eIF4G, eIF2α, and eIF3) and the eIF4E binding partners 4E-BP1 and 4E-BP2 by proteases, but in apoptosis it is members of the caspase family that cause the cleavage (3, 5, 23, 25, 33). Several of the genes whose protein products are associated with apoptosis contain IRESs, including XIAP (12), DAP5 (10), c-myc (33, 34), and Apaf-1 (6), and can therefore be translated in a cap-independent manner. Of particular interest is the translational regulation of Apaf-1, since this protein (the mammalian homologue of CED4) is pivotal to the caspase cascade. Thus, in the presence of cytochrome c (released from the mitochondria), dATP, and caspase 9 (27, 41, 42), Apaf-1 is able to promote the activation of caspase 9, which in turn cleaves and activates caspase 3, thus triggering apoptosis via the caspase cascade (24). Initiation of protein synthesis via the Apaf-1 IRES is not increased during the late stages of apoptosis (our unpublished data), and this probably reflects the fact that Apaf-1 is required for one of the first steps of this process. It has been proposed that Apaf-1 is involved in mammalian cell death pathways which are initiated by Bax (22). However, it is also required for apoptosis that is initiated by the tumor necrosis factor-related apoptosis-inducing ligand (27) and UV light (32).
Despite the large number of cellular IRESs that have now been identified, neither the mechanism(s) that they use to initiate translation nor the protein requirements for eukaryotic IRESs have been discovered. The data on the use of cellular IRESs during apoptosis would suggest that these IRESs do not require intact eIF4G or fully formed eIF4F complex (6, 10, 12, 33). In addition to canonical initiation factors, two proteins have been identified which bind to cellular IRESs; thus, the autoantigen La is an essential component of the XIAP IRES ribonuclear protein complex (11), and PTB interacts with the vascular endothelial growth factor IRES element (13). Studies on the protein requirements for eukaryotic IRESs have proved to be difficult since very few have been shown to function efficiently (if at all) in any in vitro translation system. This is in part because many of these IRES-containing mRNAs appear to need to be transcribed in the nuclei of eukaryotic cells before they function in the cytoplasm (35, 40). Whether the inactivity of these IRESs when solely transfected into the cytoplasm of cells is due to the requirement for an RNA modification or the recruitment of nuclear specific protein factors to the RNA is unknown.
We have investigated the noncanonical trans-acting protein factors that are required for the function of the Apaf-1 IRES. We show that both unr and PTB are required for internal ribosome entry on the Apaf-1 IRES and that they stimulate its internal ribosome entry in vitro. Moreover, in cell lines that lack or have reduced levels of these proteins, internal ribosome entry mediated via the Apaf-1 IRES can be stimulated by cotransfection of plasmids encoding these factors.
MATERIALS AND METHODS
Materials.
Media and serum were purchased from GIBCO BRL; luciferase assay kits “Stop & Glo” and rabbit reticulocyte lysates were purchased from Promega. The Galactolight Plus assay system was purchased from Tropix. The cell lines used, HeLa, SY5Y, HepG2, MCF7, MRC5, COS7, and BALB/c, were all obtained originally from the American Tissue Type Culture Collection. FuGene 6 was purchased from Roche Molecular Biochemicals. All other chemical were purchased from Sigma (Poole, United Kingdom).
Plasmid constructs.
Plasmid pSKAL is a Bluescript-based vector which contains the Apaf-1 IRES (see reference 6 for details) fused in frame with the firefly luciferase gene [Fig. 1A (i)]; pRAF, pRHRVF and pRF are as described [Fig. 1B (i)] (6). The cDNAs encoding unr and PTB were either present in PET28a vectors, which enabled protein to be expressed in Escherichia coli and purified, subcloned into pCDNA3.1 for expression in tissue culture cells, or subcloned into the vector pBlueBac4 (Invitrogen) for expression in insect cells.
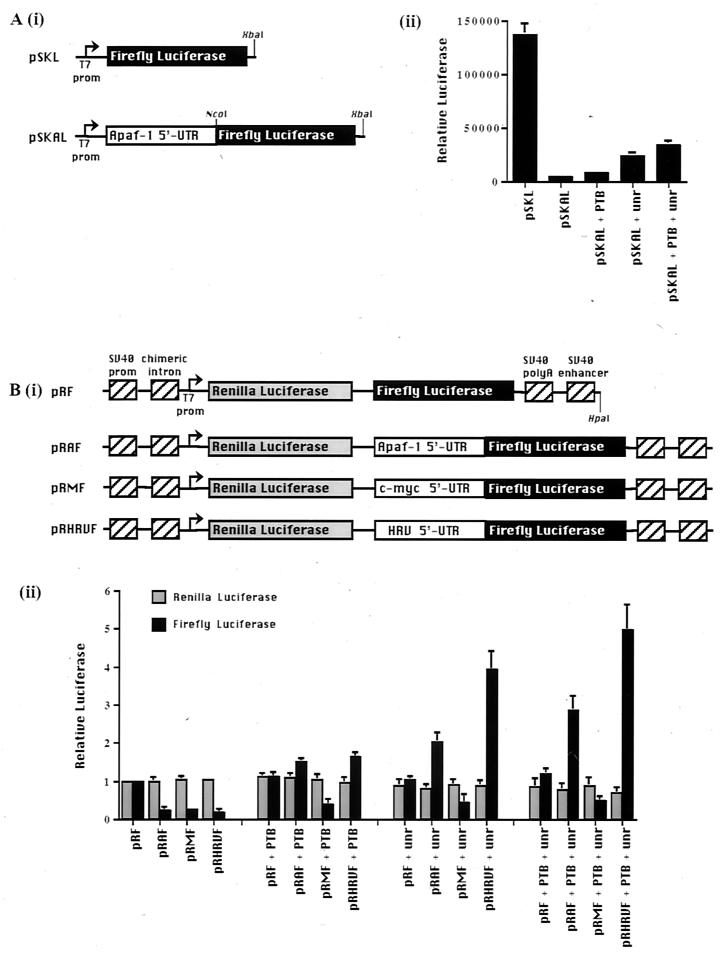
FIG. 1.
Effect of addition of PTB and unr proteins to in vitro assays for IRES activity. (A) (i) Schematic diagram of the monocistronic plasmid showing pSKL and pSKAL, which contains the Apaf-1 IRES fused in frame with the luciferase gene. (ii) Luciferase reporter levels can be increased up to sixfold on the addition of 200 ng of unr and 100 ng of PTB, individually and together, in rabbit reticulocyte lysates primed with capped monocistronic pSKAL. (B) (i) Schematic diagram of dicistronic reporter constructs, pRF, pRAF, pRMF, and pRHRVF. (ii) Addition of unr (200 ng) and PTB (100 ng) to RRL increases the levels of firefly luciferase produced from the capped dicistronic reporter vectors pRAF and pRHRVF but has no effect on the c-myc IRES.
Protein expression.
PTB and unr were overexpressed in E. coli from the PET28a vector by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to the growth medium. The proteins that contained a His tag were purified using a nickel affinity column. unr, which also had a C-terminal His tag, was purified from cultures of Sf9 cells that had been infected with a recombinant baculovirus expressing unr-His, as specified by the supplier (Invitrogen). Cells were harvested and lysed in phosphate-buffered saline containing 0.1% Triton X-100, and the tagged protein was again purified on a nickel affinity column.
Cell culture and transient transfections.
All cells with the exception of SY5Y were grown in Dulbecco's modified Eagle's medium (GIBCO-BRL) containing 10% fetal calf serum, under humidified atmosphere containing 5% CO2. SY5Y cells required a mixture of 50% Dulbecco's modified Eagle's medium and 50% Ham's F12 medium containing 10% fetal calf serum. SY5Y cells were differentiated by the addition of retinoic acid for 24 h. Calcium phosphate-mediated DNA transfection of mammalian cells was performed essentially as described by Jordan et al. (20), with minor modifications as described by Stoneley et al. (34). Alternatively, cells were transfected using FuGene 6 (Roche) as specified by the manufacturer. All transfections were performed in triplicate on at least three independent occasions.
The activities of firefly and Renilla luciferases in lysates prepared from transfected cells were measured using a Dual-Luciferase reporter assay system (Promega), and light emission was measured over 10 s using an OPTOCOMP I luminometer. The activity of β-galactosidase (which was used as a transfection control) in lysates prepared from cells transfected with pcDNA3.1/HISB/LacZ (Invitrogen) was measured using a Galactolight Plus assay system (Tropix).
In vitro runoff transcription and in vitro translation.
Vector DNA (pSKAL or pRAF) was linearized by restriction digestion using a site downstream of the sequence of interest (XbaI or HpaI, respectively). Transcripts were synthesized in a reaction mixture containing 1 × transcription buffer (40 mM HEPES-KOH [pH 7.9], 6 mM MgCl2, 2 mM spermidine, 10 mM dithiothreitol [DTT], 10 mM NaCl), 40 U of RNasin, 1 mM ATP, 1 mM UTP, 1 mM CTP, 1 mM GTP, 1 mM 7methyl-GTP, 1 μg of DNA template, and 20 U of T7 or T3 RNA polymerase in a final volume of 50 μl. For radiolabeled RNAs, 50 μCi of [α-32P]CTP was included in the reaction mixtures. After incubation of the reaction mixture for 1 h at 37°C, the RNA was isolated. RNA (5 ng/μl) was used to prime the Promega rabbit reticulocyte flexi-lysate in vitro translation system as specified by the manufacturer. The final volume of the reaction was 12.5 μl. Either 0.2 μg of unr or 0.1 μg of PTB was added to the translation reaction mixtures where indicated. Luciferase activities were determined (as described above), and the firefly and Renilla values are expressed relative to that of the control plasmid pRF, which was assigned a value of 1. All experiments were performed in triplicate on at least three independent occasions.
Immunoblotting.
For analysis of unr and PTB expression, cell pellets were solubilized by sonication in electrophoresis buffer (50 mM Tris-HCl [pH 6.8], 4% sodium dodecyl sulfate [SDS], 10% 2-mercaptoethanol, 1 mM EDTA, 10% glycerol, 0.01% bromophenol blue), supplemented with 1% aprotinin, 1 μg of leupeptin per ml, and 1 μg of N-α-p-tosyl-l-lysine chloromethylketone (TLCK) per ml immediately before use. Cell extracts (equal cell numbers per lane) were then analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and electroblotted as described previously (39). The blots were probed as described (39). Anti-unr and anti-PTB polyclonal antibodies were generated in the Jackson laboratory and were used at dilutions of 1:2,000 and 1:5,000, respectively. Anti-La monoclonal antibody (a gift from Mike Clemens, St George's Hospital Medical School, London, United Kingdom) was used at a 1:60 dilution. Anti-actin monoclonal antibody was purchased from Sigma and used at 1:2,000. The blots were then incubated with peroxidase-conjugated secondary antibodies raised against mouse or rabbit immunoglobulin and developed using the chemiluminescent reagent Illumin 8 (generated by M. Murray, Department of Genetics, Leicester University).
UV cross-linking assay.
Approximately 2.5 pmol (5 × 105 cpm) of radiolabeled RNA transcript made from pSKAL linearized with NcoI was incubated with 0.5 μg of unr and 0.25 μg of PTB in 30 μl of buffer mix (containing 10 mM HEPES [pH 7.4], 3 mM MgCl2, 100 mM KCI, 5 mM creatine phosphate, 1 mM DTT, 1 mM ATP, 6% glycerol, 0.1 μg of tRNA per μl), in the presence or absence of unlabeled competitor transcripts, for 10 min at room temperature in a 96-well microtiter plate (Falcon). The samples were then incubated for a further 10 min with 0.2 mg of heparin per ml. Samples were UV irradiated on ice for 15 min using a 305-nm UV light source. Then 0.2 mg of RNase A per ml was added to each of the samples, which were incubated at 37°C for 30 min to allow the degradation of any unprotected RNA species. An equal volume of 2× SDS sample buffer was added to the samples prior to separation by SDS-PAGE (10% polyacrylamide gels). The gels were then dried, and the results were visualized on a Molecular Dynamics PhosphorImager.
Electrophoretic mobility shift assays (EMSAs).
Approximately 23,000 cpm of labeled transcript was added to 10 μl of buffer mix containing 2 μl of 5× transcription buffer (200 mM Tris-HCl [pH 8.0], 40 mM MgCl2, 10 mM spermidine, 250 mM NaCl), 0.75 μl of 1 M DTT, 1.5 μl of tRNA (10 mg/ml), 1 μl of 10 mM rATP, and 40 U of RNAsin. unr (0.2 μg) and/or PTB (0.1 μg) was then incubated with the mixture at room temperature for 10 min. Loading buffer was added and samples were loaded directly onto 5 or 10% acrylamide gels made using 1× TBE (Tris-borate-EDTA) filter-sterilized buffer. Samples were then electrophoresed at 150 V for 1 h in 1× TBE filter-sterilized buffer. The gels were dried under vacuum at 60°C for 2 h and exposed on a phosphorimager.
RESULTS
unr and PTB stimulate the activity of Apaf-1 IRES in vitro.
We have recently shown that the 5′ UTR of Apaf-1 mRNA has an IRES which is active in transient transfection assays of a wide variety of cell types. Although the majority of cellular IRESs are inactive in cell-free translation systems, we decided to test whether the Apaf-1 IRES was functional in the rabbit reticulocyte lysate system and, if not, whether it could be activated by any of the cellular RNA binding proteins which potentiate the activity of viral IRESs in this system.
Accordingly, rabbit reticulocyte lysates were primed with either monocistronic RNAs containing the Apaf-1 IRES fused to firefly luciferase RNA [pSKAL, Fig. 1A (i)] or dicistronic RNAs containing the Apaf-1 IRES between RNAs encoding Renilla and firefly luciferases [pRAF, Fig. 1B (i)] (for further details, see reference 6). In the monocistronic RNAs the presence of the Apaf-1 IRES caused a large inhibition of translation in vitro, presumably because scanning ribosomes are unable to read through this highly structured region [Fig. 1A (ii)]. Addition of unr alone and to a lesser extent PTB alone activates the translation of the Apaf-1 5′ untranslated region, while in the presence of both PTB and unr there is at least an additive effect. Two sources of recombinant unr were used in these experiments (purified from Sf9 cells or E. coli); however, no difference in activity of these proteins was detected. The effect of these proteins on IRES-dependent translation was confirmed by assays performed using the RNA derived from the dicistronic constructs. Again, both PTB alone and unr alone caused an increase in translation from the downstream cistron [Fig. 1B (ii)]. The smaller activation observed with PTB alone is likely to reflect the fact that reticulocyte lysates contain no detectable unr but do contain a small amount of PTB (14). Again, an additive effect was observed when both PTB and unr were included in the assays. Stimulation of the Apaf-1 IRES was not as great as that of the HRV IRES, although their activities in vivo are similar (6), suggesting that other, unidentified factors are required for full activity of the Apaf-1 IRES. In control experiments performed with dicistronic c-myc IRES RNA, there was no stimulation of IRES function with unr and/or PTB [Fig. 1B (ii)], suggesting that either these proteins are not required for c-myc IRES function or additional, unidentified proteins are also necessary. In addition, our data suggest that PTB and unr stimulate translation rather than increase the fidelity of translation initiation since we observed only one band that corresponded to luciferase on SDS-PAGE (data not shown).
In view of these in vitro translation results, we proceeded to investigate the interaction of PTB and unr with the Apaf-1 5′ UTR by using UV cross-linking assays and EMSAs.
unr but not PTB is able to bind directly to the Apaf-1 IRES.
It has been shown that PTB binds to and can be cross-linked to the poliovirus IRES and to the EMCV IRES (9, 21) and that unr can be cross-linked to the HRV IRES (14). To determine whether unr and PTB interact directly with the Apaf-1 IRES, radiolabeled Apaf-1 RNA was incubated with unr or PTB in the presence of excess unlabeled competitor Apaf-1 IRES, HRV IRES, or glyceraldehyde-3-phosphate dehydrogenase (G3PDH) mRNA. Samples were exposed to UV light, treated with RNases, and then separated by PAGE (Fig. 2). It can be seen that the Apaf-1 IRES binds tightly to unr (Fig. 2A) although not as strongly as the HRV-IRES does, since a lower molar excess of this unlabeled RNA was required to compete for binding (Fig. 2B).
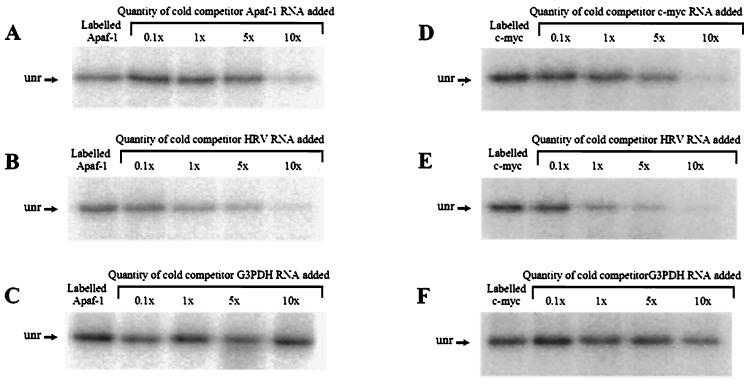
FIG. 2.
Cross-linking of unr protein to the Apaf-1 IRES. (A) unr is able to bind to radiolabeled Apaf-1 IRES RNA. This binding is effectively competed by unlabeled Apaf-1 IRES RNA when added at a 10-fold excess. (B) HRV IRES RNA has a higher affinity for unr and is able to compete in the binding reaction at equal molarity. (C) G3PDH RNA used as a control does not compete in this reaction at any concentration used. (D) unr protein is able to bind to radiolabeled c-myc IRES RNA. This binding is effectively competed by unlabeled c-myc IRES RNA when added at a fivefold excess. (E) HRV IRES RNA has a higher affinity for unr and is able to compete in the binding reaction at equal molarity. (F) G3PDH RNA used as a control does not compete in this reaction at any concentration used.
To test whether unr was able to interact with other cellular IRESs, these experiments were repeated using the c-myc IRES as a control. It can be seen that unr is able to interact with c-myc IRES RNA with an affinity for binding similar to that for the Apaf-1 IRES (Fig. 2D), even though this protein does not stimulate the activity of internal ribosome entry via the c-myc IRES in vitro. The unlabelled HRV IRES RNA was again more efficient in competing for binding to the unr than was the unlabeled c-myc IRES RNA (Fig. 2E); the unlabelled control G3PDH mRNA did not dissociate the unr from the radiolabeled c-myc IRES or Apaf-1 IRES RNA (Fig. 2C and F). No PTB was found to bind directly to either IRES RNA using this technique (Fig. 3A, lane 2). This would suggest either that PTB is not able to interact directly with these IRES RNAs or that these RNAs are not in the correct conformation for this protein to bind.
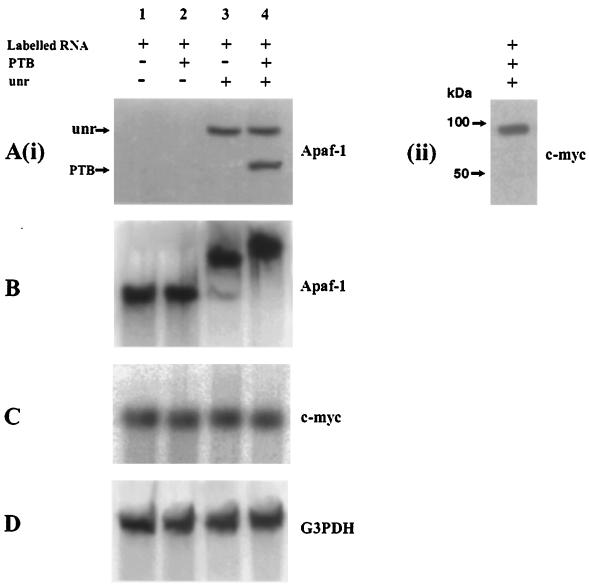
FIG. 3.
PTB is able to bind to the Apaf-1 IRES only in the presence of unr. (A) (i) Cross-linking assay. Lanes 1, no protein addition to the radiolabeled Apaf-1 RNA; 2, addition of 0.25 μg of PTB; 3, addition of 0.5 μg of unr; 4, addition of both unr and PTB. (ii) Cross-linking assay with c-myc RNA on the addition of unr and PTB. (B) EMSA with radiolabeled Apaf-1 RNA. Lanes 1, no protein addition; 2, addition of 0.1 μg of PTB; 3, addition of 0.2 μg of unr; 4, addition of both unr and PTB. (C) EMSA of c-myc IRES RNA does not show any binding to unr or PTB. (D) EMSA of G3PDH mRNA used a control does not show any binding to unr or PTB.
PTB binds to the Apaf-1 IRES in the presence of unr.
It has been shown that unr and PTB were able to act in synergy to stimulate HRV IRES function on the addition of both proteins to rabbit reticulocyte lysates, leading to a greater increase in translation than when they were added individually (14). Therefore, to test whether PTB and unr could act in concert on Apaf-1 IRES RNA, we carried out UV cross-linking and EMSA using radiolabeled Apaf-1 RNA and purified unr and PTB (Fig. 3A). It can clearly be seen from the UV cross-linking results that PTB alone is not cross-linked to the Apaf-1 IRES [Fig. 3A (i), lane 2]. However, in the presence of unr, this protein now interacts with the Apaf-1 RNA so that both proteins are labeled [Fig. 3A (i), lane 4]. A different result was obtained with the c-myc IRES RNA, since this case no PTB was found to bind in the presence of unr [Fig. 3A (ii)].
These data obtained with the Apaf-1 IRES RNA were confirmed by the EMSAs. Thus, there was no alteration in the migration of the radiolabeled Apaf-1 IRES RNA in the presence of PTB alone, although there was a distinct shifted band is the presence of unr alone (Fig. 3B, lanes 2 and 3). However when both proteins were added to the reaction mixture the radiolabeled Apaf-1 RNA migrated even more slowly on the gel, showing that the PTB was now binding in addition to unr (lane 4). In this case it also appears that the presence of PTB stimulates the binding of unr, since more of the radiolabeled Apaf-1 IRES RNA was shifted in the presence of PTB. This is a very important result because it demonstrates for the first time that PTB and unr interact with a cellular IRES. These data suggest that when unr binds to the IRES, the RNA then attains the correct conformation for PTB to interact.
These EMSAs were repeated using the c-myc IRES to determine whether unr and PTB could similarly interact with this RNA (Fig. 3C). It can be seen that there was not a shift in the position of the radiolabeled c-myc IRES RNA on the gel in the presence of unr or PTB, showing that by this technique the c-myc IRES does not bind to these proteins. The difference between the results obtained using these two techniques to analyze the c-myc IRES RNA probably reflects the fact that UV cross-linking is able to detect more transient interactions while tighter protein binding is required to cause a mobility shift on a gel. However, the data would suggest that these two cellular IRESs examined have different protein factor requirements.
Radiolabeled G3PDH mRNA was used as a control, and this also does not bind either unr or PTB (Fig. 3D).
Location of the unr and PTB binding sites in the Apaf-1 IRES.
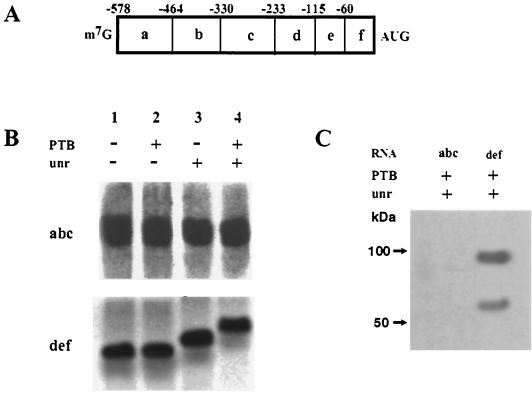
To address whether unr and PTB were binding to a similar region of RNA or whether PTB was recognizing a longer-range interaction, UV cross-linking and EMSAs were performed using deletion constructs of the Apaf-1 IRES (as described in reference 6). These deletions divided the Apaf-1 IRES into two segments termed abc and def (Fig. 4A). Radiolabeled RNAs were generated from these plasmids and used in EMSAs. The data clearly show that the unr binds only to fragment def that contains the 3′-terminal region of the IRES RNA from -233 to 1 (Fig. 4B). These results are in agreement with our data published previously which show that when this region of RNA is used in a dicistronic assay in vivo, 75% of the IRES function is maintained (6). This would imply that the minimum region for Apaf-1 IRES function resides in this fragment, which, although rather small compared to IRESs of viral origin, is consistent with the sizes of IRESs observed in other eukaryotic genes, e.g., the Bip and FGF-2 genes (37, 40). The data also show that in the presence of unr, PTB also binds to this region (Fig. 4B), although further experimentation would be necessary to more precisely define their binding sites on the RNA. In agreement with these data, UV cross-linking analysis also demonstrated that unr and PTB interacted solely with this 233-nucleotide fragment of the Apaf-1 IRES (Fig. 4C).
FIG. 4.
Identification of the unr binding region within the Apaf-1 IRES. (A) Schematic diagram of Apaf-1 IRES deletions. (B) EMSAs with radiolabeled Apaf-1 IRES RNA fragments in the presence of 0.2 μg of unr and 0.1 μg of PTB. Only fragment def shows a retardation of RNA mobility in the presence of the proteins. (C) UV cross-linking analysis of fragments of the Apaf-1 IRES with 0.5 μg of unr and 0.25 μg of PTB. Again, these data show that unr and PTB bind to fragment def only.
The activity of the Apaf-1 IRES in vivo correlates with the cellular expression of PTB and unr.
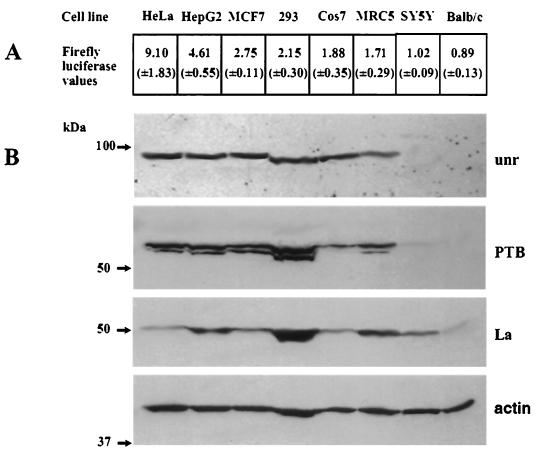
Since we have shown previously that the ability of the Apaf-1 IRES to mediate internal ribosome entry in transient-transfection assays varies considerably between cell lines of different origin (6), we next examined the correlation between IRES activity and the PTB and unr content of these different cell lines. To do this, cells were transfected with the dicistronic plasmids pRF and pRAF and subsequently harvested and assayed for luciferase activity (Fig. 5A). In parallel experiments, cell samples were separated by SDS-PAGE and immunoblotted. The blots were then probed for PTB, stripped, and reprobed for unr and additionally La, since this protein is necessary for XIAP IRES function (11); actin was used as a loading control (Fig. 5B). There was no correlation between the expression of La and Apaf-1 IRES activity, and, indeed, in HeLa cells where the Apaf-1 IRES has the greatest activity, this protein was expressed at very low levels. However, the function of the Apaf-1 IRES activity in vivo correlated with the protein expression of both unr and PTB. For example, there was a ninefold difference between HeLa and BALB/c cell lines in the ability of the Apaf-1 IRES to initiate synthesis of firefly luciferase, and it can be seen that there was no detectable unr and PTB in BALB/c cells. Indeed, in the cell lines where the relative expression of firefly luciferase was less than 2, there was a marked reduction in the PTB and/or unr levels (Fig. 5B). The reduced level of unr and PTB in the murine cell lines is not likely to be due to a reduced cross-reactivity of the antibodies since it has been found that the anti-PTB antibody reacts equally well with mouse, rat, and sheep PTB (C. Gooding, M. Wollerton, and C. Smith, personal communication). Moreover, the rat and human unr proteins are very highly conserved, so that good cross-reactivity of the antibody used with the murine form of the protein would seem very probable (2).
FIG. 5.
Apaf-1 IRES activity in a range of cell lines can be correlated with the presence of endogenous PTB and unr. (A) The human Apaf-1 IRES in plasmid pRAF was transfected into the eight cell lines shown. Firefly luciferase activities from the dicistronic vector are presented normalized against “readthrough,” i.e., the level of firefly luciferase produced from the control plasmid pRF. Transfections were performed in triplicate on at least three separate occasions; standard errors are shown in parentheses. (B) Western blots of cell lysates probed for the presence of endogenous unr, PTB, La, and β-actin as a loading control.
unr and PTB increase the activity of the Apaf-1 IRES in vivo.
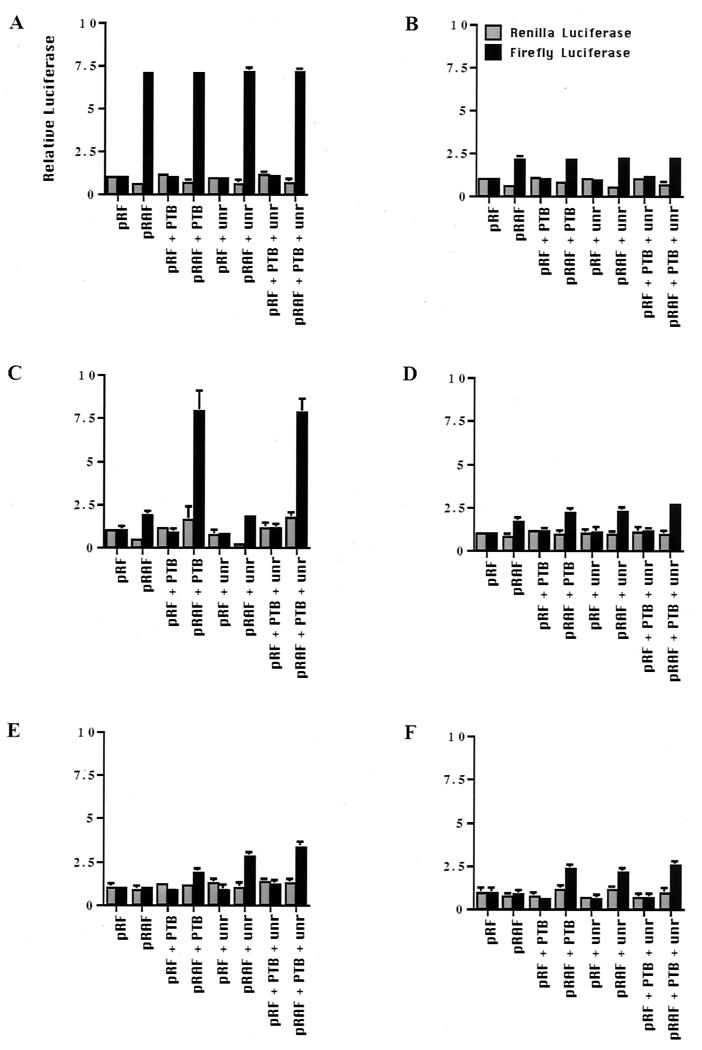
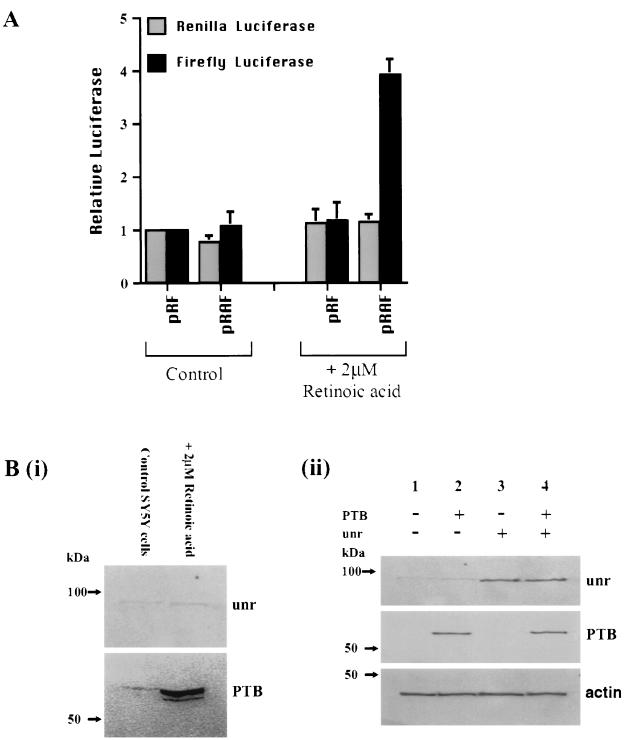
To determine whether the inactivity of the Apaf-1 IRES in the cell lines shown was due to their low expression of PTB and unr, cotransfections were performed with the dicistronic plasmids pRAF and plasmids harboring PTB and/or unr (Fig. 6). In each case the firefly and Renilla activities shown are expressed relative to the activity in the cells without the addition of PTB or unr. It appears that it is possible to increase the activity of the Apaf-1 IRES by cotransfections, suggesting that in some cases the cellular levels of PTB and unr are limiting. Thus, in HeLa cells, where the IRES is maximally active, no additional stimulation of cap-independent translation was observed (Fig. 6A). With MRC5 and BALB/c cells, PTB and unr independently stimulated Apaf-1 IRES-mediated translation (Fig. 6D and F). However, in COS7 cells, which have very low levels of endogenous PTB, cotransfection with PTB (but not unr) caused an increase in the amount of firefly luciferase produced so that it was equivalent to that observed in HeLa cells (Fig. 6C). Similarly, in the neuronal SY5Y cells, which lack both PTB and unr, cotransfection with plasmids harboring DNA encoding these proteins caused an additive increase in the level of firefly luciferase produced (Fig. 6E). The expression of the proteins that result from the transfected PTB and unr cDNAs was detected by Western blotting; it is clear that the levels of these two proteins were greatly increased in SY5Y cells [Fig. 7B (ii)]. However, it is clear that other protein factors must be required or that an IRES-specific inhibitor is active in some cell types, since in most cases (except in COS7 cells) cotransfection of the unr- and PTB-containing plasmids did not restore IRES function to that observed in HeLa cells in vivo. To test this hypothesis, we have additionally performed the transfection experiments with La and ITAF45; however, in these cases we observed no stimulation in IRES-mediated translation (data not shown). Moreover, in this regard, differentiation of SY5Y cells down the neuronal pathway with retinoic acid resulted in a large increase in PTB expression, which was greater than that observed with the transfected PTB DNA (Fig. 7B), and this correlated with a very large increase in Apaf-1 IRES activity (Fig. 7A). There was only a very slight increase in the cellular levels of unr (Fig. 7B), so that clearly in this case there is also increased expression of an as yet undetermined IRES trans-acting factor.
FIG. 6.
Cotransfection of unr and PTB in cell lines increases Apaf-1 IRES activity. The cell lines HeLa (A), HEK293 (B), COS7 (C), MRC5 (D), SY5Y (E), and BALB/c (F) were cotransfected with the dicistronic plasmid harboring the Apaf-1 IRES and those containing PTB and/or unr. There is no increase in IRES function in cell lines which already contain high levels of unr and/or PTB (A and B), while in those that lack one of these proteins there is a significant increase in luciferase produced by internal ribosome entry (C to F).
FIG. 7.
Differentiation of SY5Y down the neuronal pathway increases PTB expression and Apaf-1 IRES activity. (A) SY5Y cells (with or without retinoic acid) were transfected with pRAF, and luciferase activities were determined. (B) (i) Western blot of lysates from control and differentiated SY5Y cells probed with anti-PTB and anti-unr antibodies. Differentiated SY5Y cells show a large increase in the levels of endogenous PTB but little change in the levels of unr. (ii) Western blot of lysate from transfected SY5Y cells probed with anti-PTB and anti-unr antibodies. Actin is shown as a loading control. Lanes: 1, cells transfected with pCDNA3.1 as a control, showing very low levels of endogenous unr and almost no detectable PTB; 2, cells transfected with pCDNA-PTB; a clear band that corresponds to PTB is observed, but the level is lower than that produced by differentiation [see panel (i)]; 3, cells transfected with pCDNA-unr; a clear band that corresponds to unr is observed; 4, cells transfected with both pCDNA-PTB and pCDNA-unr; both proteins can be observed.
DISCUSSION
Although considerable progress has been made in recent years toward the discovery of the proteins that are necessary for internal ribosome entry on viral IRESs, little is known about the trans-acting factors used by cellular eukaryotic IRESs. It is clear that different classes of viral IRESs have distinct sets of proteins that they use to stimulate internal ribosome entry, although in each case these are produced by the host cell. The viral IRESs range from those like the EMCV IRES which appears to need only cleaved eIF4G and eIF4A of the eIF4F complex for function (28, 29), to the hepatitis A virus IRES, where the proteins needed for optimal activity are substantially different from other picornavirus IRESs, especially since it requires intact eIF4G (1). The wide spectrum of protein usage by viral IRESs to initiate internal ribosome entry probably in part reflects the fact that they have evolved to infect specific cell types.
An important question relating to cellular IRESs is whether each IRES has the requirement for a specific yet different group of proteins or whether there are some proteins that they have in common. To address this, we have investigated the protein factor requirements for the Apaf-1 IRES. In vitro translation assays demonstrated that unr and PTB alone each activated the Apaf-1 IRES and that when they were used together there was at least an additive stimulation of internal ribosome entry. By UV cross-linking analysis and EMSAs, we show that both unr and PTB interact with the Apaf-1 IRES. Interestingly, PTB was able to bind to Apaf-1 only in the presence of unr, suggesting that unr facilitates the folding of the Apaf-1 IRES RNA into the correct tertiary structure for PTB to interact (Fig. 3). The region to which these proteins bind has been further defined by deletion analysis and correlates with a 233-nucleotide fragment that still has 75% activity in vivo (Fig. 4) (6). We show that there is a direct correlation between IRES activity and the cellular expression of unr and PTB (Fig. 5), and by cotransfection of unr and PTB into cell lines which expressed low levels of these proteins, we were able to stimulate internal ribosome entry in vivo (Fig. 6). However, our data also suggest that factors other than unr and PTB are required for full activity of the Apaf-1 IRES since (i) levels of firefly luciferase in vitro were not as high as those observed in vivo in HeLa cells (Fig. 1 and 6), (6), (ii) cotransfection of unr and PTB into neuronal SY5Y cells or BALB/c cells does not increase the level of Apaf-1 IRES activity to that observed in HeLa cells (Fig. 6E and F), and (iii) in MCF7 and HEK293 cells which express similarly high levels of PTB and unr to HeLa cells, the Apaf-1 IRES does not have an equivalently high activity (Fig. 5).
It has been shown recently that the XIAP IRES requires the autoantigen La for activity (11). However, no correlation was observed between the cellular expression of the autoantigen La and Apaf-1 IRES activity (Fig. 5).
Our data would also suggest that, as with viral IRESs, cellular IRESs are cell type specific. The lack of activity of Apaf-1 IRES in certain cell types would suggest that cellular IRESs function in a cell-specific fashion (Fig. 5 and 6). Moreover, there is a wide variation in c-myc IRES activity between cell types, although it functions in all cell types examined (35). In addition, we observe a variation in the cell types in which the cellular IRESs c-myc and Apaf-1 show greatest activity (Fig. 5), (6, 35). This is probably due to differential expression of the trans-acting factors that are required for each IRES to function. In agreement with this, we have shown that there is a surprising variation in the cellular expression of unr and PTB required for activity of the Apaf-1 IRES (Fig. 5). Moreover, differentiation of SY5Y cells caused a large increase in PTB expression, which corresponds to a fourfold increase in Apaf-1 IRES activity (Fig. 7).
In conclusion, we have identified two proteins that interact with the Apaf-1 IRES, namely, PTB and unr. These are the first proteins shown to directly stimulate the activity of a cellular IRES in vitro and in vivo, and this will enable us to search for the other factors required for the Apaf-1 IRES to function. Our data would suggest that cellular IRESes have different trans-acting protein factor requirements, some of which are cell type specific.
ACKNOWLEDGMENTS
This work was funded by project grants from the Wellcome Trust (S.A.M. and R.J.J.) and the BBSRC (advanced fellowship held by A.E.W.). M.J.C. and E.C.B. were supported by MRC studentships.
REFERENCES
- 1.Borman A, LeMercier P, Girard M, Kean K M. Comparison of picornaviral IRES-drived internal initiation of translation in cultured cells of different origins. Nucleic Acids Res. 1997;25:925–932. doi: 10.1093/nar/25.5.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boussadia O, Jacquemin-Sablon H, Dautry F. Exon skipping in the expression of the gene immediately upstream of N-ras. Biochim Biophys Acta. 1993;1172:64–72. doi: 10.1016/0167-4781(93)90270-n. [DOI] [PubMed] [Google Scholar]
- 3.Bushell M, Poncet D, Marissen W E, Flotow H, Lloyd R E, Clemens M J, Morley S J. Cleavage of polypeptide chain initiation factor eIF4GI during apoptosis in lymphoma cells: characterisation of an internal fragment generated by caspase-3-mediated cleavage. Cell Death Differ. 2000;7:628–636. doi: 10.1038/sj.cdd.4400699. [DOI] [PubMed] [Google Scholar]
- 4.Clemens M J, Bushell M, Jeffrey I W, Pain V M, Morley S J. Translation initiation factor modifications and the regulation of protein synthesis in apoptotic cells. Cell Death Differ. 2000;7:603–615. doi: 10.1038/sj.cdd.4400695. [DOI] [PubMed] [Google Scholar]
- 5.Clemens M J, Bushell M, Morley S J. Degradation of eIF4G in response to induction of apoptosis in human lymphoma cell lines. Oncogene. 1998;17:2921–2931. doi: 10.1038/sj.onc.1202227. [DOI] [PubMed] [Google Scholar]
- 6.Coldwell M J, Mitchell S A, Stoneley M, MacFarlane M, Willis A E. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene. 2000;19:899–905. doi: 10.1038/sj.onc.1203407. [DOI] [PubMed] [Google Scholar]
- 7.Gosert R, Chang R, Rijnbrand R, Yi M, Sangar D, Lemon S. Transient expression of cellular PTB binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites in vivo. Mol Cell Biol. 2000;20:1583–1585. doi: 10.1128/mcb.20.5.1583-1595.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray N, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 9.Hellen C U T, Pestova T V, Wimmer E. The cellular polypeptide p57 (polypyrimdine tract binding protein) binds to multiple sites in the poliovirus 5′ non-translated region. J Virol. 1994;68:941–950. doi: 10.1128/jvi.68.2.941-950.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henis-Korenblit S, Levy Strumpf N, Goldstaub D, Kimchi A. A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol Cell Biol. 2000;20:496–506. doi: 10.1128/mcb.20.2.496-506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holcik M, Korneluk R G. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol Cell Biol. 2000;20:4648–4657. doi: 10.1128/mcb.20.13.4648-4657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holcik M, Lefebvre C, Yeh C, Chow T, Korneluk R G. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat Cell Biol. 1999;1:190–192. doi: 10.1038/11109. [DOI] [PubMed] [Google Scholar]
- 13.Huez I, Creancier L, Audiger S, Gensac M-C, Prats A-C, Prats H. Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol Cell Biol. 1998;18:6178–6190. doi: 10.1128/mcb.18.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunt S L, Hsuan J J, Totty N, Jackson R J. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of human rhinovirus RNA. Genes Dev. 1999;13:437–448. doi: 10.1101/gad.13.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunt S L, Jackson R J. Polypyrimidine tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA. 1999;5:344–359. doi: 10.1017/s1355838299981414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isoyama T, Kamoshita N, Yasui K, Iwai A, Shiroki K, Toyoda H, Yamada A, Takasaki Y, Nomoto A. Lower concentration of La protein is required for internal ribosome entry on hepatitus C virus RNA than on poliovirus RNA. J Gen Virol. 1999;80:2319–2327. doi: 10.1099/0022-1317-80-9-2319. [DOI] [PubMed] [Google Scholar]
- 17.Jackson R J, Hunt S L, Gibbs C L, Kaminski A. Internal initiation of translation of picornavirus RNAs. Mol Biol Rep. 1994;19:147–159. doi: 10.1007/BF00986957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson R J, Hunt S L, Reynolds J E, Kaminski A. Cap-dependent and cap-independent translation: operational distinctions and mechanistic interpretations. Curr Top Microbiol Immunol. 1995;203:1–29. doi: 10.1007/978-3-642-79663-0_1. [DOI] [PubMed] [Google Scholar]
- 19.Jackson R J, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- 20.Jordan M, Schallhorn A, Wurm F. Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 1996;24:596–601. doi: 10.1093/nar/24.4.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaminski A, Jackson R. The polypyrimidine tract binding protein requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA. 1998;4:626–638. doi: 10.1017/s1355838298971898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 23.Marissen W, Lloyd R E. Eukaryotic translation initiation factor 4G is targetted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol Cell Biol. 1998;18:7565–7574. doi: 10.1128/mcb.18.12.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mignotte B, Vayssiere J-L. Mitochondria and apoptosis. Eur J Biochem. 1998;252:1–15. doi: 10.1046/j.1432-1327.1998.2520001.x. [DOI] [PubMed] [Google Scholar]
- 25.Morley S J, McKendrick L, Bushell M. Cleavage of translation initiation factor 4G during anti-fas IgM induced apoptosis does not require signalling through the p38 mitogen-activated protein kinase. FEBS Lett. 1998;438:41–48. doi: 10.1016/s0014-5793(98)01269-1. [DOI] [PubMed] [Google Scholar]
- 26.Pain V M. Initiation of protein synthesis in eukaryotic cells. Eur J Biochem. 1996;236:747–771. doi: 10.1111/j.1432-1033.1996.00747.x. [DOI] [PubMed] [Google Scholar]
- 27.Pan G, O'Rourke K, Dixit V M. Caspase-9, Bcl-XL and Apaf-1 form a ternary complex. J Biol Chem. 1998;273:5841–5845. doi: 10.1074/jbc.273.10.5841. [DOI] [PubMed] [Google Scholar]
- 28.Pestova T V, Hellen C U T, Shatsky I N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pestova T V, Shatsky I N, Hellen C U T. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pilipenko E V, Pestova T V, Kolupaeva V G, Khitrina E V, Poperechnaya A N, Agol V I, Hellen C U T. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 31.Spangberg K, Schwartz S. Poly(C) binding protein interacts with the hepatitis C virus 5′ UTR J. Gen Virol. 1999;80:1371–1376. doi: 10.1099/0022-1317-80-6-1371. [DOI] [PubMed] [Google Scholar]
- 32.Srinivasula S M, Ahmad M, Fernandes-Alnemri T, Alnemri E S. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- 33.Stoneley M, Chappell S A, Jopling C L, Dickens M, MacFarlane M, Willis A E. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol Cell Biol. 2000;20:1162–1169. doi: 10.1128/mcb.20.4.1162-1169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stoneley M, Paulin F E M, Le Quesne J P C, Chappell S A, Willis A E. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 35.Stoneley M, Subkhankulova T, Le Quesne J P C, Coldwell M J, Jopling C L, Belsham G J, Willis A E. Analysis of the c-myc IRES: a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 2000;28:687–694. doi: 10.1093/nar/28.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svitkin Y V, Meerovitch K, Lee H S, Dholakia J N, Kenan D J, Agol V I, Sonenberg N. Internal translation initiation on poliovirus RNA; Further characterization of La function in poliovirus translation in vitro. J Virol. 1994;68:1544–1550. doi: 10.1128/jvi.68.3.1544-1550.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vagner S, Gensac M-C, Maret A, Bayard F, Amalric F, Prats H, Prats A-C. Alternative translation of human fibroblast growth factor 2 mRNA occurs by internal entry of ribosomes. Mol Cell Biol. 1995;15:35–44. doi: 10.1128/mcb.15.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walter B, Nguyen J H C, Ehrenfeld E, Semler B L. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA. 1999;5:1570–1587. doi: 10.1017/s1355838299991483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.West M J, Stoneley M, Willis A E. Translational induction of the c-myc oncogene via activation of the FRAP/TOR signalling pathway. Oncogene. 1998;17:769–780. doi: 10.1038/sj.onc.1201990. [DOI] [PubMed] [Google Scholar]
- 40.Yang G, Sarnow P. Location of the internal ribosome entry site in the 5′ non-coding region of the immunoglobulin heavy-chain binding protein (BiP) mRNA: evidence for specific RNA-protein interactions. Nucleic Acids Res. 1997;25:2800–2807. doi: 10.1093/nar/25.14.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zou H, Henzel W J, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 42.Zou H, Li Y, Liu X, Wang X. An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J Biol Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]