Abstract
Downstream target genes of p53 are thought to mediate its tumor-suppressive activity, but it is unknown whether differential transactivation of these genes is regulated at the level of p53 binding to their promoters. To address this issue, p53 binding in vivo to consensus sites in the p21Waf1, MDM2, and PIG3 promoters was investigated in cells exposed to adriamycin (ADR) or ionizing radiation as well as in an inducible p53 cell line. p53-DNA complexes were cross-linked in vivo by treating the cells with formaldehyde and processed by chromatin immunoprecipitation-PCR. This methodology allowed for the analysis of relevant p53-DNA complexes by preventing redistribution of cellular components upon collection of cell extracts. Increased p53 binding to the p21Waf1, MDM2, and PIG3 promoters occurred within 2 h after p53 activation; however, significant increases in PIG3 transcription did not occur until 15 h after p53 binding. Gel shift analyses indicated that p53 had lower affinity for the consensus binding site in the PIG3 promoters compared to its consensus sites in the p21 and MDM2 genes, which suggests that additional factors may be required to stabilize the interaction of p53 with the PIG3 promoter. Further, acetylated p53 (Lys382) was found in chemically cross-linked complexes at all promoter sites examined after treatment of cells with ADR. In summary, the kinetics of p53 binding in vivo to target gene regulatory regions does not uniformly correlate with target gene mRNA expression for the p53 target genes examined. Our results suggest that target genes with low-affinity p53 binding sites may require additional events and will have delayed kinetics of induction compared to those with high-affinity binding sites.
Cells are capable of altering their physiology in response to environmental signals. One mechanism by which this is accomplished is through modulation of cellular gene expression. Changes in gene expression are achieved by the convergence of biochemical signaling pathways on transcription factors, presumably governing their transactivation potential at specific target genes. p53 is one such transcription factor that is downstream of stress-activated biochemical pathways and plays a critical role in coordinating the response of cells to a diverse range of environmental stresses.
The ability of p53 to bind DNA in a sequence-specific manner and activate transcription is integral for its tumor-suppressive properties. In normal cells, the p53-induced G1/S cell cycle arrest is mediated by p21Waf1, a downstream target gene product (9, 10). p21Waf1 mediates cell cycle arrest by binding to and inhibiting cyclin–cyclin-dependent kinase complexes; the kinase activity of these complexes is essential for the coordinated transitions between cell cycle phases (8, 14, 15, 41). In addition to cell cycle arrest, p53 can also initiate apoptosis by transactivation of certain genes. These genes include those encoding Bax, a proapoptotic Bcl-2 family member (24), Killer/DR5 (death receptor), which is upstream in the activation of the apoptotic caspase cascade (29), and p53AIP1, a mitochondrial protein that when overexpressed induces apoptosis (26). Other genes transactivated in a p53-dependent manner in cells undergoing apoptosis are the PIGs (p53-inducible genes) (27).
In addition to stimulating growth arrest and apoptosis through its transcriptional properties, p53 can also regulate its own activity and turnover through transactivation of MDM2 (4). The MDM2 protein can bind to the amino-terminal transactivation domain of p53 and block the association of p53 with the basal transcription machinery (16, 40). Additionally, MDM2 can target p53 for ubiquitin-mediated proteolysis (17, 19).
Even with the ever-increasing number of p53 downstream target genes identified, it remains unclear what mechanism(s) dictates the in vivo selectivity of p53 for a given target gene under a specific physiological condition. It is possible that distinct posttranslational modifications of p53, such as acetylation or phosphorylation, are required for the differential binding to, and activation of, specific promoters (13, 20, 28, 37, 38). After cells are treated with ionizing or UV radiation, p53 is phosphorylated at several serine and threonine residues (23, 28, 30, 31) and acetylated at several lysine residues (20, 28). Phosphorylation of p53 at amino-terminal residues may block MDM2 binding (31) and may promote protein stabilization and stimulate the acetylation of p53 at its carboxyl terminus (3, 7, 28, 31, 34). Acetylation increases p53 sequence-specific DNA binding in vitro (13), suggesting that this modification may be required for p53-mediated transactivation.
As the number of posttranslational modifications described for p53 increases, the task of defining the role that these modifications play in the in vivo selectivity or kinetics of p53 binding to promoter regions will become more complex. Technologies will be required that enable us to stably trap p53 in vivo at a specific time under a given condition, isolate the protein, and study the biochemical properties as well as the protein and DNA interactions of p53. Toward this end, in the present study we treated cultured cells with formaldehyde and used chromatin immunoprecipitation PCR (CHIP-PCR) to analyze p53 binding to consensus binding sites in select target gene promoters including p21Waf1, MDM2, and PIG3. Furthermore, we explored whether acetylated p53 (Lys382) was found in covalently cross-linked complexes at these promoters in cultured cells. We discovered that acetylated p53 was found in cross-linked complexes after adriamycin (ADR) treatment at all the promoter sites analyzed. Further, the kinetics of p53 binding to these target gene promoters were differential and in the case of p21Waf1 and MDM2 correlated with observed increases in expression of target RNA and protein. However, significant increases in PIG3 mRNA were delayed compared to p53 binding to the PIG3 promoter. The differential kinetics observed were in accord with gel shift analyses showing higher affinity of p53 for the p21Waf1 and MDM2 response elements compared to the element present in the PIG3 promoter, and we hypothesize that additional factors are required for p53-mediated PIG3 transactivation.
MATERIALS AND METHODS
Cell culture and treatment.
The human colorectal carcinoma cell line RKO was grown in McCoy's 5A medium (Gibco BRL, Gaithersburg, Md.) supplemented with 10% fetal calf serum (Gemini Bio-Products, Inc., Calabasas, Calif.) and 1% penicillin-streptomycin (Sigma, St. Louis, Mo.). The generation of the HIp53 ponasterone A (PonA)-inducible p53 cell line is described in reference 11. All cells were grown at 37°C with 5% CO2 in a humidified incubator.
Cells were treated with 0.4 μM ADR or 10 Gy of ionizing radiation (IR). IR was delivered at room temperature (RT) with a 137Cs irradiator (J. L. Shepherd and Associates).
Transfections were performed by mixing 5 μg of the indicated plasmids and 24 μl of Lipofectamine (Gibco BRL) in a final volume of 1 ml of Optimem (Gibco BRL) and allowing the DNA-lipid complexes to form for 20 min at RT. H1299 cells were rinsed twice with unsupplemented F-12 medium prior to addition of the DNA-lipid complexes in 4 ml of unsupplemented F-12 medium. The DNA-lipid mixture was removed from the cells after 4 h, and the cells were allowed to recover overnight prior to treatment as indicated. The p53 (S15A) phosphorylation site mutant (1) was a generous gift from Karen Vousden (National Cancer Institute, Frederick, Md.).
Western and immunoprecipitation analyses.
For Western analysis, 40 μg of protein was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 10 and 12% polyacrylamide gels, transferred onto Immobilon-P membrane (Millipore, Bedford, Mass.), and blocked with 5% (wt/vol) nonfat dry milk in TTBS (100 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% [vol/vol] Tween 20). Membranes were incubated with the following primary antibodies: anti-p53 PAb1801 and PAb421 (Oncogene Research Products, Cambridge, Mass.), anti-p21Waf1/Cip1 EA10 (Oncogene Research Products), anti-MDM2 SMP14 (Santa Cruz Biotechnology, Santa Cruz, Calif.), anti-PIG3 7F8 (Oncogene Research Products), anti-phospho-p53 α-Ser15-p53 (Ser15) (New England Biolabs, Beverly, Mass.), and anti-acetylated p53 (Lys382) α-AC p53 (Oncogene Research Products). Subsequently, membranes were incubated with goat anti-mouse- or goat anti-rabbit-horseradish peroxidase (Pierce, Rockford, Ill.) and analyzed by enhanced chemiluminescence. For immunoprecipitation of acetylated p53, α-Ac p53 (Oncogene Research Products) was chemically cross-linked to protein A-Sepharose (PAS) with 52 mM dimethyl pimelimidate (Pierce). Immunoprecipitations were performed with 2 mg of cellular protein as previously described (11, 32, 33).
Formaldehyde cross-linking.
Growth medium was aspirated from 107 cells and replaced with a 1% formaldehyde (EM Science, Gibbstown, N.J.) solution in phosphate-buffered saline. Cells were incubated in formaldehyde for 10 min at room temperature, after which the cross-linking was stopped by the addition of glycine to a final concentration of 0.125 M. Glycine remained on the cells for 5 min. Monolayers were washed twice with phosphate-buffered saline. Extracts were prepared by scraping cells in 1 ml of radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% [vol/vol] Nonidet P-40, 0.5% [wt/vol] deoxycholate, 0.1% [wt/vol] SDS, 50 mM Tris [pH 8], 5 mM EDTA) containing the protease inhibitors antipain (10 μg/ml), leupeptin (10 μg/ml), pepstatin A (10 μg/ml), chymostatin (10 μg/ml) (Sigma), and 4-(2-aminoethyl)benzenesulfonylfluoride (200 μg/ml; Calbiochem-Novabiochem Corp., La Jolla, Calif.). Phosphatase inhibitors (50 mM NaF and 0.2 mM sodium orthovanadate) and the deacetylase inhibitor trichostatin A (5 μM; Calbiochem) were also added to the RIPA buffer. Cell lysates were sonicated to yield chromatin fragments of approximately 600 bp as assessed by agarose gel electrophoresis. Debris was pelleted by centrifugation for 10 min at 13,000 × g, and 2 mg of protein extract was precleared with 50 μg of rabbit immunoglobulin G or 10 μg of mouse immunoglobulin G bound to PAS (Pharmacia Biotech, Piscataway, N.J.) for 1 h with rocking at 4°C. After centrifugation for 2 min at 13,000 × g, supernatants were transferred to a new tube. Then a 15-μl bed volume of PAS and 2 μg of appropriate antibody were added to precleared extract. Immunoprecipitation was performed by rocking overnight at 4°C. Of note, similar results were obtained using either PAb1801 or PAb421 for the p53 immunoprecipitations.
Immunocomplexes were washed twice with RIPA buffer, four times with IP wash buffer (100 mM Tris [pH 8.5], 500 mM LiCl, 1% [vol/vol] Nonidet P-40, 1% [wt/vol] deoxycholic acid), and twice more with RIPA buffer. Between washes, samples were rocked end-over-end for 5 min; 300 μl of cross-linking reversal buffer (125 mM Tris [pH 6.8], 10% [vol/vol] β-mercaptoethanol 4% [wt/vol] SDS) was added to the washed PAS pellet. Samples were boiled for 30 min to reverse the formaldehyde cross-links. DNA was phenol-chloroform extracted, ethanol precipitated, allowed to air dry, and dissolved in sterile H2O.
PCR amplification.
MDM2 PCRs were performed using Ready-To-Go PCR beads (Amersham Pharmacia, Uppsala, Sweden) according to the manufacturer's directions with a 68°C annealing temperature and 30 cycles. p21Waf1 and PIG3 PCRs were performed in 16.6 mM (NH4)2SO4–67 mM Tris (pH 8.8)–6.7 mM MgCl2–10 mM β-mercaptoethanol, 10% (vol/vol) dimethyl sulfoxide–1.5 mM nucleotides. Each primer was used at 350 ng per 50-μl reaction. For p21Waf1, 30 PCR cycles were performed, each cycle consisting of a 1-min 95°C denaturation and 1-min annealing at the indicated temperature, followed by a 2-min extension at 72°C. For PIG3, 40 cycles were performed, each cycle consisting of denaturation at 20 s for 94°C and 45-s annealing at 63°C, followed by a 25-s extension at 72°C. The primers used are as follows. for PIG3 (63°C anneal), 5′-CAGGACTGTCAGGAGGAGGCGAGTGATAAGG-3′ (forward) and 5′-GTGCGATTCTAGCTCTCACTTCAAGGAGAGG-3′ (reverse); for p21Waf1 (61°C anneal), 5′-CCGCTCGAGCCCTGTCGCAAGGATCC-3′ (forward) and 5′-GGGAGGAAGGGGATGGTAG-3′ (reverse); and for MDM2 (62°C anneal), 5′-GGATTGGGCCGGTTCAGTGG-3′ (forward) and 5′-GGTCTACCCTCCAATCGCCAC-3′ (reverse).
PCRs were resolved using 8% polyacrylamide gels (acrylamide:bis acrylamide, 19:1) in 1× Tris acetate-EDTA buffer. Gels were stained with ethidium bromide. Relative levels of DNA were determined using QuantifyOne software (Bio-Rad Laboratories, Inc., Hercules, Calif.).
mRNA preparation and Northern analysis.
Cells were harvested in RNA lysis buffer (10 mM Tris [pH 7.5], 100 mM NaCl, 2 mM EDTA, 1% [wt/vol] SDS) and lysed by eight passages through an 18-gauge needle. Proteinase K was added to a final concentration of 100 μg/ml, and the lysate was incubated at 37°C for 1 h. Following proteinase K digestion, the NaCl concentration was adjusted to 400 mM. The samples were heated at 65°C for 5 min with constant agitation followed by immediate cooling in an ice bath for 30 s. mRNA was isolated using oligo(dT)-cellulose (Ambion, Inc., Austin, Tex.) with rocking at RT for 2 h. The mRNA–oligo(dT)-cellulose mixture was washed twice with high-salt buffer (10 mM Tris) [pH 7.5], 400 mM NaCl, 1 mM EDTA, 0.2% [wt/vol] SDS) and packed with high-salt buffer on a Poly-Prep chromatography column (Bio-Rad). The column was washed once with high-salt buffer and once with low-salt buffer (10 mM Tris [pH 7.5], 100 mM NaCl, 1 mM EDTA, [wt/vol] 0.2% SDS). The mRNA was eluted from the column in no-salt buffer (5 mM Tris [pH 7.5], 1 mM EDTA, 0.2% [wt/vol] SDS) that was heated to 55°C prior to elution. mRNA was precipitated at −20°C overnight with the addition of sodium acetate (pH 5.2) to a final concentration of 220 mM and 2 volumes of 95% ethanol. mRNA was recovered by centrifugation at 12,000 × g for 30 min, and the pellet was rinsed once with 70% ethanol. The pellet was air dried and dissolved in sterile H2O.
mRNA (5 μg for PIG3 and 4 μg for MDM2 and p21Waf1) was dried under vacuum, resuspended in sample buffer (1× morpholinepropanesulfonic acid [MOPS] [0.1 M MOPS {pH 7.0}, 40 mM sodium acetate, 5 mM EDTA {pH 8.0}], 50% [vol/vol] formamide, 6.5% [vol/vol] formaldehyde), and heated at 55°C for 15 min. Then 1× loading buffer (10× loading buffer is 50% [vol/vol] glycerol, 1 mM EDTA, 0.25% [wt/vol] bromophenol blue, 0.25% [wt/vol] xylene cyanol, and 0.3 mg of ethidium bromide/ml) was added, and mRNA was resolved by electrophoresis on a 1% agarose gel containing 2% (vol/vol) formaldehyde and 1× MOPS. The gel was washed twice in 10× SSC (20× SSC contains 3 M NaCl, 0.12 M sodium citrate, and 0.02 M Tris) buffer, and mRNA was transferred to supported nitrocellulose membrane (Gibco BRL). p21Waf1, MDM2, PIG3, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNAs were labeled with [α-32P]dCTP using Rediprime II (Amersham). After a 2-h prehybridization in Express Hybe (Clontech Laboratories, Inc., Palo Alto, Calif.), membranes were probed with the indicated [32P]cDNA (2 × 106 cpm/ml) in Express Hybe at 42°C overnight. Membranes were washed twice at RT in 2× SSC–0.1% (wt/vol) SDS and then twice at 42°C in 0.2× SSC–0.1% (wt/vol) SDS. Levels of mRNA were quantified using an Instant Imager (Packard Instruments, Meriden, Conn.).
Purification of p53 protein.
Infected Sf9 cells were harvested in lysis buffer (50 mM Tris-HCl [pH 8], 150 mM NaCl, 0.5% [vol/vol] Nonidet P-40, 1 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride, 1 μM E-64, protease inhibitors [listed above in the description of formaldehyde cross-linking]), sonicated, and incubated on ice for 30 min. The lysate was centrifuged at 40,000 × g for 20 min at 4°C. The supernatant was precipitated by the addition of ammonium sulfate to 50% (g/ml) saturation, followed by centrifugation at 40,000 × g for 20 min at 4°C. The pellet was resuspended in lysis buffer and passed over a PAb1801 immunoaffinity column, and the column was washed extensively with buffer containing 20 mM Tris-HCl (pH 8), 1 mM EDTA, 100 mM NaCl, 1% (vol/vol) Nonidet P-40, 10% (vol/vol) glycerol, and 1 mM DTT followed by a buffer containing 0.5 M NaCl in buffer B (5× buffer B contains 100 mM Tris-HCl [pH 8], 5 mM EDTA, and 50% [vol/vol] glycerol). The p53 was eluted from the column using 55% (vol/vol) ethylene glycol–0.5 M NaCl in buffer B. Fractions were collected and separated by SDS-PAGE on 10% gels, and protein was visualized by silver stain. Fractions with relatively high concentrations of p53 purified to 95 to 98% homogeneity were dialyzed in 10 mM HEPES (pH 7.5)–5 mM NaCl–0.1 mM EDTA–10% (vol/vol) glycerol–1 mM DTT at 4°C for 8 h with three buffer changes. Following dialysis, p53 protein was concentrated by submerging the dialysis bag in polyethylene glycol (molecular weight, 15,000 to 20,000) until the volume decreased by 50%.
Acetylation of recombinant p53.
Protein acetyltransferase assays were performed as described by Gu and Roeder (13) with recombinant glutathione S-transferase (GST)–p53 produced in DH10B cells and Flag-p300 (amino acids 1195 to 1810) (13) produced in Escherichia coli BL21(Lys) cells. The p53 was purified on glutathione-Sepharose, and p300 (amino acids 1195 to 1810) was purified on M2 agarose (Kodak).
EMSAs.
Oligonucleotide duplexes representing the following p53 response elements were used (p53 consensus sequences are underlined): p21Waf1, 5′TGGCCATCAGGAACATGTCCCAACATGTTGAGCTCTGGCA; PIG3, 5′TAGCAGCACCCAGCTTGCCCACCCATGCTCAAGATGGGCG; and MDM2, 5′GAGCTGGTCAAGTTCAGACACGTTCCGAAACTGCAGTAAAAGGAGTTAAGTCCTGACTTGTCTCCAGC. Oligonucleotides were end labeled using T4 polynucleotide kinase (New England BioLabs). Electromobility shift assays (EMSAs) were performed in 30 μl containing 20 mM Tris-HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.5 mM EDTA, 0.5 mg of bovine serum albumin/ml, 1 ng of 32P-labeled DNA, and 50 ng of pure p53 protein. After a 20-min incubation at RT, the indicated amounts of several different competitor DNAs were added, and reactions were incubated for another 20 min. Reactions were loaded on a 4% polyacrylamide (acrylamide: bisacrylamide, 30:1) gel containing 0.5× Tris-borate-EDTA buffer, prerun at 150 V for 40 min at 4°C. Samples were electrophoresed in 0.5× Tris-borate-EDTA at room temperature at 150 V for 2.5 h. Gels were dried and exposed for autoradiography. Quantification of protein-DNA complexes was performed using a Molecular Dynamics (Sunnyvale, Calif.) PhosphoRimager and ImageQuant software.
RESULTS
Formaldehyde treatment of cells generates higher-molecular-weight complexes containing p53.
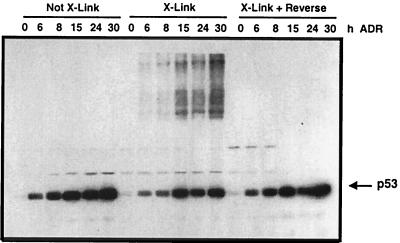
To determine if p53 could be covalently cross-linked into higher-molecular-weight complexes within the cell, monolayers of RKO human colon carcinoma cells (wild-type [wt] p53) were first treated with ADR for 0 to 30 h to elevate endogenous p53 levels and then exposed to formaldehyde as described in Materials and Methods. Cell extracts were analyzed for p53 protein migration and levels by immunoblotting as shown in Fig. 1. The hallmark increase in p53 protein levels after ADR treatment was observed in cells that were not exposed to formaldehyde. In ADR-treated cells exposed to formaldehyde, the level of monomeric p53 was lower than in cells not treated with formaldehyde; however, higher-molecular-weight complexes containing p53 became apparent and increased through the time course of ADR treatment. The p53 that remained monomeric after formaldehyde treatment may represent p53 protein that was not bound to other cellular material. Alternatively, 100% of the p53 may not have been cross-linked with this treatment; the time of exposure and concentration of formaldehyde used in the experiments described herein were optimized to yield maximum protein-DNA cross-linking without the formation of large aggregates of higher-molecular-weight complexes that were neither soluble nor reversible. The higher-molecular-weight covalent complexes that formed after formaldehyde treatment were reversible after heat treatment, as evidenced by the loss of slower-migrating p53-containing complexes and the presence of monomeric p53 protein at levels comparable to that observed in the cells that were not treated with formaldehyde (Fig. 1).
FIG. 1.
Formaldehyde treatment of cells generates high-molecular-weight complexes of p53. RKO cells were treated with ADR (0.4 μM) for the indicated times. Before collection of the cell extracts, cells were treated with 1% formaldehyde (X-Link) or processed without formaldehyde treatment as described in Materials and Methods. Cell extracts were analyzed by Western blotting with a p53-specific antibody. Formaldehyde treatment causes cross-links, as evidenced by the generation of p53-containing complexes with retarded migration in SDS-PAGE. The cell extracts collected from formaldehyde-treated cells were heat treated to reverse the formaldehyde cross-links (X-Link + Reverse).
Isolation of p53-DNA complexes from formaldehyde-treated cells.
Using the in vivo cross-linking procedure described above, we investigated p53 binding to known DNA response elements in the genome. Endogenous p53 was activated by treatment of cells with genotoxic agents. The cells were exposed to 1% formaldehyde prior to harvest, p53 was immunoprecipitated, and the DNA to which it was bound was purified. The DNA was PCR amplified using primers specific for sequences that flank the p53 response elements in each promoter studied.
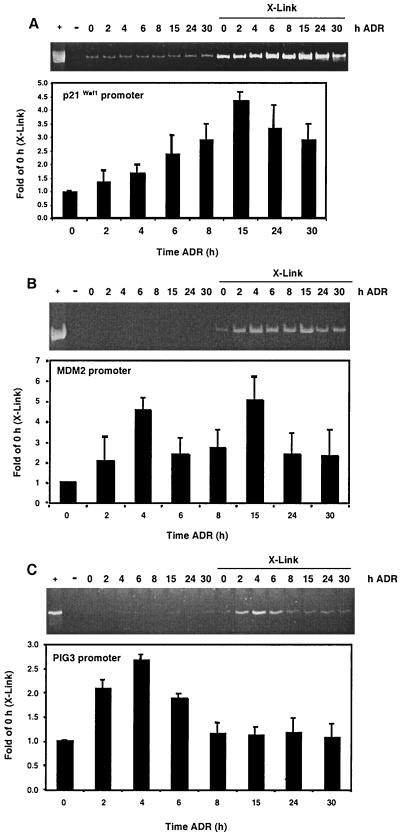
The kinetics of p53 occupancy at the p21Waf1, MDM2, and PIG3 promoters were initially investigated in cells exposed to ADR. In parallel, two sets of RKO cells were treated with ADR for 0 to 30 h. After ADR treatment, one set was cross-linked with formaldehyde, while the other was processed similarly but not exposed to formaldehyde. p53 immunocomplexes were isolated, the cross-links were reversed, and equivalent aliquots of the p53-immunoprecipitated DNA were PCR amplified using primers that flank response elements as described in Materials and Methods. The PCR products were resolved by PAGE, stained with ethidium bromide, and quantified. The primers specific for the p21Waf1 promoter generated a PCR product of 230 bp representing nucleotides −2280 to −2050 (with respect to the TATA box). A low, background level of PCR products was generated from DNA coimmunoprecipitated with p53 in cells not exposed to formaldehyde, presumably due to low levels of genomic DNA binding nonspecifically to the immunocomplexes as previously reported (5, 39). By 4 to 6 h after ADR treatment, the p21Waf1 PCR product amplified from DNA template derived from cross-linked p53 immunocomplexes increased twofold over control levels (Fig. 2A). The level of product continued to increase up to 15 h of ADR treatment. After this time, however, the PCR signal decreased to threefold of the control level and remained at this level for the remainder of the time course. To assess the relative amount of cellular p21Waf1 promoter immunoprecipitated with p53, total genomic DNA from ADR-treated cells was isolated from the same number of cells that were processed at each experimental time point. The PCR product amplified from an equal aliquot of this genomic DNA template represented approximately 32 times the PCR product amplified from the cross-linked DNA template immunoprecipitated with p53 from untreated cells (Fig. 2A, compare + lane to 0-h, ADR X-link lane).
FIG. 2.
Kinetics of p53 binding promoters in RKO cells exposed to ADR. RKO cells were incubated in ADR (0.4 μM) and collected at the indicated times. At the time of harvest, a set of RKO cells was treated with formaldehyde (X-Link) and processed as described in Materials and Methods. A duplicate set was not treated with formaldehyde and processed identically. The DNA derived from p53-specific immunocomplexes was PCR amplified using primers for the p21Waf1 (A), MDM2 (B), or PIG3 (C) promoter. PCRs were resolved with PAGE, the gels were stained with ethidium bromide, and the PCR products were quantified by densitometry. Data are expressed as fold of the 0-h (X-Link) PCR signal. For p21Waf1, MDM2, and PIG3, + indicates PCR products that were generated using DNA template derived from total genomic DNA harvested from ADR-treated cells. Each ethidium bromide-stained gel shows one representative result of at least nine independent experiments that are quantified in the graphs below.
The kinetics of p53 binding to the MDM2 promoter after ADR treatment were investigated next. The MDM2-specific primers generated a 172-bp PCR product spanning nucleotides −119 to +53 with respect to the TATA box in the first intron of the gene. The PCR signal appeared biphasic, with a fivefold elevation over control apparent at 4 h after ADR treatment followed by a decrease in signal at 6 and 8 h and a second fivefold elevation at 15 h (Fig. 2B). Again, to assess the relative amount of cellular MDM2 promoter immunoprecipitated with p53, total genomic DNA was isolated from the same number of cells that were processed at each time point. The PCR product amplified from an equal aliquot of this genomic DNA template represented approximately 28 times the PCR product amplified from the cross-linked DNA template immunoprecipitated with p53 from untreated cells (Fig. 2B, compare + lane to 0-h, ADR X-link lane).
Finally, the DNA in p53 immunocomplexes was analyzed for the PIG3 promoter. The primers specific for the PIG3 promoter generated a 275-bp PCR product representing nucleotide positions −441 to −166 relative to the transcriptional start site. Levels of DNA amplified from the immunocomplexes with the PIG3-specific primers were elevated by 2 h, peaked at 2.5-fold over control by 4 h, and declined throughout the remainder of the time course (Fig. 2C). To assess the relative amount of cellular PIG3 promoter immunoprecipitated with p53, total genomic DNA was isolated from the same number of cells that were processed at each time point. The PCR product amplified from a 1:200 dilution of this genomic DNA template represented approximately seven times the PCR product amplified from the cross-linked DNA template immunoprecipitated with p53 from untreated cells (Fig. 2B, compare + lane to 0-h, ADR X-link lane). Also, note that more cycles of amplification were used to generate the PIG3 PCR signal than the other genes analyzed (see Materials and Methods). Thus, significantly greater amounts of cross-linked, immunoprecipitated DNA template were required to generate similar levels of the PIG3 PCR product compared to the p21Waf1 and MDM2 analyses.
Linearity of the PCRs was verified by analyzing serial dilutions of the 24-h DNA sample from cross-linked cells used in Fig. 2A as well as two and four times the amount. The different concentrations of template were PCR amplified and resolved with PAGE, and the PCR products were quantified. The results of this experiment verified that the PCRs were performed in a linear range; the reactions exhibited a linear increase in PCR signal with up to a twofold increase of input template (data not shown).
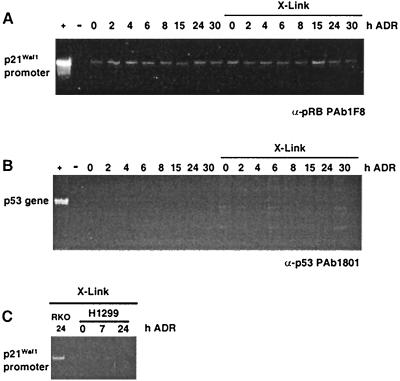
The specificity of the assay was verified by several approaches. In the first, we determined if an antibody that was not p53 specific, such as a pRb- or cyclin B-specific antibody, could immunoprecipitate p21Waf1 promoter-containing DNA fragments. Figure 3A shows that only background levels of PCR products were generated when the indicated p21Waf1 promoter primers were used with the DNA isolated from pRb immunoprecipitates. The same result was observed using a cyclin B1-specific antibody (data not shown). In the second approach, we investigated whether PCR primers that would amplify a region of the genome that does not contain a p53 consensus binding site (nucleotides 1082 to 1227 of the p53 gene, encoding amino acids 279 to 326) could be immunoprecipitated with p53-specific antibodies. While a significant level of PCR product was generated from total genomic DNA (Fig. 3B, + lane), the indicated region of the p53 gene was not amplified from DNA present in the complex immunoprecipitated with p53-specific antibodies. In a third approach, we determined whether p21-specific primers would amplify a product from DNA template generated from p53 immunoprecipitations performed on cross-linked protein harvested from the H1299 cell line (p53 null). As shown in Fig. 3C, compared to the PCR signal generated with RKO cells, we were unable to generate a significant signal above background from the H1299 cells. Finally, for each PCR product studied, we transferred the amplified DNA to nitrocellulose and performed Southern analysis to verify the identity of PCR products (data not shown).
FIG. 3.
Verifying the formaldehyde cross-linking assay. (A and B) Verification of the specificity of the assay using two sets of RKO cells treated with ADR for the times indicated. At the time of harvest, one set was treated with formaldehyde (X-Link) and processed as described in Materials and Methods. The other set was not treated with formaldehyde and processed identically. Immunocomplexes from the extracts were generated using an antibody specific for the pRb protein (A) or the p53 protein (B). PCRs were performed with primers specific for the p21Waf1 promoter (A) or the p53 gene (B). PCR-amplified DNA was resolved by PAGE, and the gels were stained with ethidium bromide. + indicates an amplification performed using genomic DNA as a template; -indicates an amplification performed without genomic DNA. (C) Further analysis to verify specificity. RKO (wt p53-containing) and H1299 (p53-null) cells were treated with ADR for the indicated times. At the time of harvest, the cells were treated with formaldehyde (X-Link) and processed as described in Materials and Methods using PAb1801. The PCRs were performed with primers specific for the p21Waf1 promoter.
Kinetics of p53 binding endogenous promoters after IR.
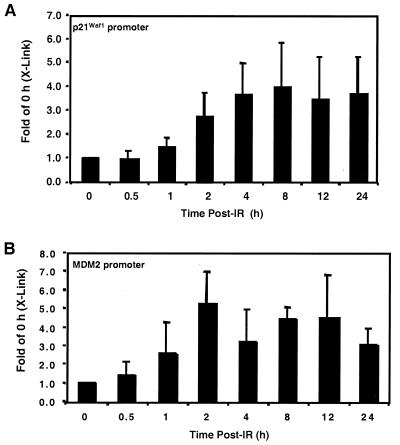
The association of p53 with the p21Waf1 and MDM2 promoters in cells exposed to IR (10 Gy) was examined next. At 2 h after IR, the p21Waf1 promoter PCR product generated from DNA purified from a p53 immunocomplex was threefold greater than that from untreated cells. The PCR product increased fourfold over control by 4 h and remained relatively constant for all subsequent time points examined (Fig. 4A).
FIG. 4.
Kinetics of p53 binding downstream promoters in RKO cells exposed to IR. RKO cells were exposed to IR (10 Gy) and collected at the indicated times after IR. The DNA derived from p53-specific immunocomplexes was PCR amplified using primers for the p21Waf1 (A) or MDM2 (B) promoter. PCRs were resolved by PAGE, the gels were stained with ethidium bromide, and the PCR products were quantified by densitometry. Data are expressed as fold of the 0-h (X-Link) PCR signal. The data are representative of at least three independent experiments.
The same DNA templates were used for PCR amplification of the MDM2 promoter. Two hours after IR, the amount of PCR product generated from the samples was approximately five times the control level. At subsequent times, the DNA amplified from the p53 immunocomplex with the MDM2-specific primers exhibited an approximate fourfold increase over the control level (Fig. 4B).
Finally, the p53-immunoprecipitated DNA from cells exposed to IR was analyzed by PCR for the PIG3 promoter. It was difficult to detect PCR products that exceeded the background signal, precluding analysis of p53 binding to this promoter after IR. This result was consistent with low levels of PIG3 mRNA detected after IR treatment as presented below.
p53 downstream target gene mRNA levels increase after genotoxic stress.
To determine whether the kinetics of p53 binding to promoter sites correlated with the elevation of target gene mRNA, we exposed RKO cells to ADR (0.4 μM) or IR (10 Gy) for the time course evaluated in the previous experiments and isolated mRNA for Northern analyses of p21Waf1, MDM2, and PIG3.
p21Waf1 mRNA was detectable at a low basal level in RKO cells and increased by 4 h after ADR and 2 h after IR treatment and remained elevated throughout the time courses (Fig. 5). Like p21Waf1, MDM2 mRNA was elevated by 4 h after ADR and 2 h after IR treatment (Fig. 5). After both ADR and IR treatment, the MDM2 mRNA expression appeared biphasic, with an elevation of expression apparent at 4 to 6 h and a second elevation observable at 24 to 30 h (Fig. 5); this biphasic trend was also evident in the PCR analyses of the MDM2 promoter in p53 immunocomplexes harvested from ADR-treated cells (Fig. 2B).
FIG. 5.
Increased p21Waf1, MDM2, and PIG3 mRNAs after ADR and IR treatment of RKO cells. RKO cells were treated with ADR (0.4 μM) or IR (10 Gy) for the times indicated. (A and C) Cells were harvested, mRNA was isolated, and Northern analyses were performed for p21Waf1, MDM2, PIG3, and GAPDH mRNAs. (B and D) Graphic representation of the levels of mRNA in the Northern blots in panels A and C, normalized to GAPDH levels. The data are representative of two independent experiments.
Compared to p21Waf1 and MDM2, the kinetics of PIG3 mRNA expression did not correlate with that of p53 binding to the response element in the PIG3 promoter. A 2.5-fold increase in p53 binding to the PIG3 promoter was observed by 4 h after ADR treatment (Fig. 2C); and although this increase in binding was accompanied by an increase in PIG3 mRNA, it was modest at 1.5-fold of the control level (Fig. 5A and B). Later in the time course between 24 to 30 h after ADR treatment, a four- to sixfold increase in PIG3 mRNA levels was observed (Fig. 5A and B), a time period during which we were unable to detect appreciable p53 binding to the PIG3 promoter by CHIP-PCR (Fig. 2C). Of note, after ADR treatment, a slower-migrating band hybridizes in the PIG3 Northern blot and may represent either an alternatively spliced form of PIG3 or a transcript that shares homology with PIG3. After IR treatment, only low levels of PIG3 mRNA were detectable and only one species of mRNA was readily apparent between 8 to 12 h after treatment (Fig. 5C and D).
p53 has differential affinity for response elements in downstream promoters.
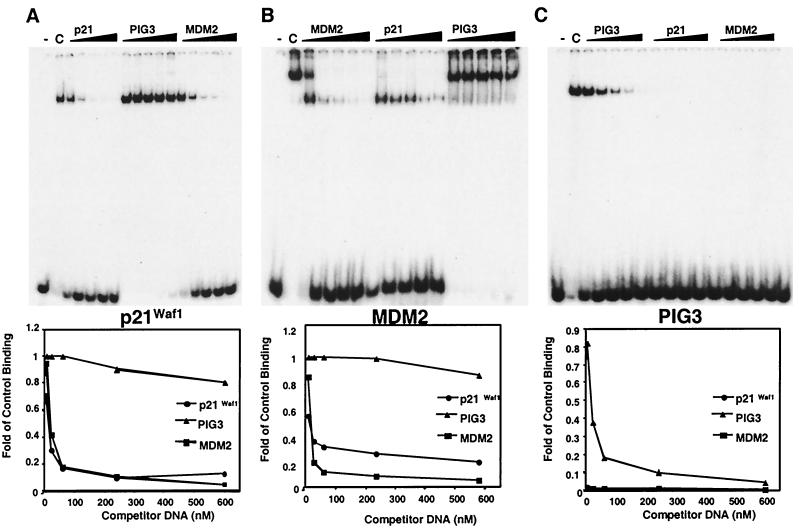
We hypothesized that the observed difference in kinetics of p53 binding to the p21Waf1 and MDM2 promoters compared to the PIG3 promoter was due to differential p53 binding affinity for the response elements found in these promoters. To test this hypothesis, we performed EMSAs using p53 protein purified from baculovirus-infected insect cells and oligonucleotide duplexes containing the p53 response elements found within the promoter regions of these genes. Purified p53 bound to all the response elements, as shown in the control lanes in Fig. 6. The observation of two shifted bands with the MDM2 consensus site is consistent with previous studies showing that p53 bound to either one or both of the response elements found in the MDM2 intron (42).
FIG. 6.
Differential affinity of p53 for response elements. Gel shifts were performed to analyze the relative affinity of p53 binding the consensus sites in the p21Waf1 (A), MDM2 (B), and PIG3 (C) promoters. For competition assays, a 5-, 20-, 50-, 200-, or 500-fold excess of the indicated unlabeled oligonucleotide was added. “C” in each panel represents the binding of p53 to the indicated response element in the absence of competitor DNA. Bands were quantified using a PhosphorImager, and the graphed results are shown below each gel. Results are representative of three independent experiments.
Each p53-DNA complex was competed with a range of 5 to 500 nM excess unlabeled competitor DNA representing either the same response element or the consensus sites found in the other two promoters. When p53 was bound to either the p21Waf1 or the MDM2 response element, the complexes were competed with similar efficiencies with the addition of either p21Waf1 or MDM2 binding site unlabeled competitor DNA, and the 50% effective concentration for each of these competitions was approximately 30 nM (Fig. 6A and B).
The affinity of p53 for the response element in the PIG3 promoter appeared to be much lower than that of the p21Waf1 and MDM2 consensus sites. The complexes formed between p53 and the p21Waf1 or MDM2 response elements were not efficiently competed with a 500-fold excess of unlabeled PIG3 consensus site competitor DNA (Fig. 6A and B). However, unlabeled p21Waf1 or MDM2 consensus site competitor DNA could completely inhibit p53 binding to radiolabeled PIG3 consensus sites at the lowest concentration of competitor DNA examined (Fig. 6C). Thus, p53 has low affinity for the binding site present in the PIG3 promoter and likely requires other factors and/or posttranslational modifications to increase its on rate or decrease its off rate. These factors may be recruited to the PIG3 promoter at later times after ADR treatment and stabilize p53 binding at this promoter site. Based on our CHIP-PCR results, we hypothesize that the binding of other factors or later posttranslation modifications block antibody epitopes on p53. Thus, at times of greatest p53-mediated PIG3 transactivation, p53 protein is bound by other factors or differentially modified and after chemical cross-linking, not accessible for immunoprecipitation. Of note, we obtained identical results immunoprecipitating protein-DNA complexes with PAb1801 (epitope is amino acids 46 to 55; this region includes and is near sites of phosphorylation) and PAb421 (epitope is amino acids 371 to 380; this region includes and is near sites of phosphorylation and acetylation).
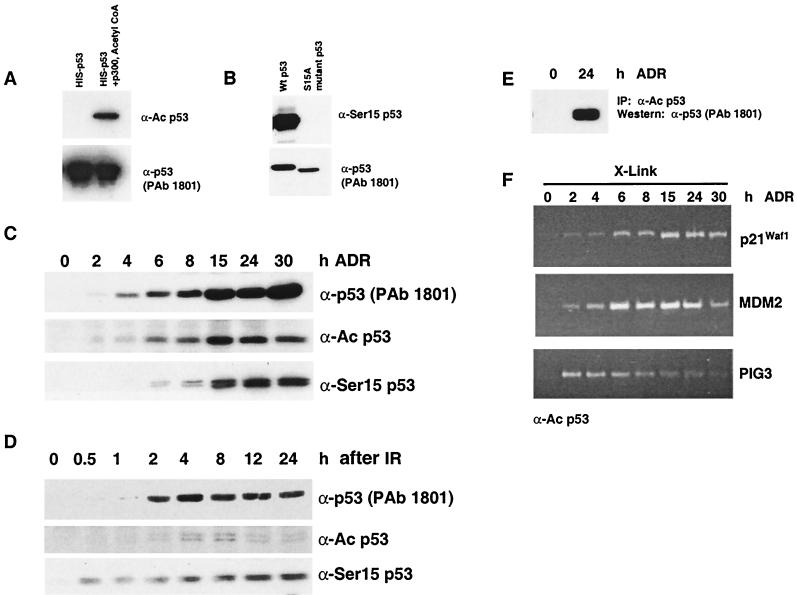
Acetylated p53 is detectable and bound at target promoters after genotoxic stress.
Previously it has been shown that p53 is acetylated and phosphorylated after cells are exposed to ionizing or UV radiation (20, 28, 30, 31); however, no study has addressed whether acetylated or phosphorylated protein occupies promoter sites in vivo. Thus, the next aim was to use the formaldehyde cross-linking protocol described above to investigate whether acetylated p53 (Lys382) or phosphorylated p53 (Ser15) is bound to promoter sites in cultured cells. Initially, we verified that p53 proteins with these posttranslational modifications were detectable after treatment of RKO cells with the doses of IR or ADR used in the previous experiments and that the antibodies were specific for the select modifications under examination. First, we demonstrated that the acetylation-specific antibody (α-Ac p53) recognizes only acetylated p53. Recombinant p53 was purified from E. coli and either untreated or incubated with p300 and acetyl coenzyme A (acetyl-CoA). The control and treated proteins were analyzed by Western with α-Ac p53 and PAb1801. Whereas both proteins were recognized by PAb1801, only p53 incubated with p300 and acetyl-CoA was recognized by α-Ac p53 (Fig. 7A). To test the specificity of the Ser15 phosphospecific antibody (α-Ser15 p53), we transfected H1299 cells with expression vectors containing either wt p53 or p53 with a Ser-to-Ala mutation at residue 15 (1). One day after transfection, protein was harvested and Western analysis with α-Ser15 p53 and PAb1801 was performed. Both proteins were recognized by PAb1801; however, only wt p53 was recognized by α-Ser15 p53 (Fig. 7B). As shown in Fig. 7C and D, acetylated and phosphorylated (Ser 15) p53 proteins were detectable in RKO cells after ADR and IR treatment.
FIG. 7.
Analysis of p53 posttranslational modifications after treatment of RKO cells with ADR or IR. (A and B) Verification of α-Ac p53 and α-Ser15 p53 antibody specificity. (A) Purified recombinant p53 produced in E. coli was either untreated or incubated with p300 and acetyl-CoA. The proteins were analyzed by Western blotting with the indicated antibodies. (B) H1299 cells were transfected with expression vectors containing wt p53 or p53 with a Ser-to-Ala mutation at residue 15. One day after transfection, protein was prepared and analyzed by Western blotting with the indicated antibodies. RKO cells were exposed to ADR (0.4 μM) (C) or IR (10 Gy) (D), and protein extracts were collected at the time points indicated. Posttranslational modifications of p53 were analyzed by Western blotting using antibodies specific for acetylated p53 (Lys382), phosphorylated p53 (Ser15), or all forms of p53 (PAb1801). (E) To verify the immunoprecipitation capability of the α-Ac p53 antibody, protein extracts from control and ADR-treated RKO cells were immunoprecipitated with α-Ac p53 and analyzed by Western blotting with PAb1801. (F) RKO cells were treated with ADR for the times indicated and treated with formaldehyde (X-Link) at the time of harvest. Cell extracts were processed as outlined in Materials and Methods. Immunocomplexes from the extracts were generated using the antibody specific for acetylated p53 at Lys382. The DNA isolated from each immunocomplex was PCR amplified using primers for the p21Waf1, MDM2, and PIG3 promoters. All results are representative of two independent experiments.
To investigate whether acetylated p53 (Lys 382) was bound to downstream promoters, ADR-treated RKO cells were incubated with formaldehyde and processed as outlined in Materials and Methods with the acetylation-specific antibody. The ability of this antibody to immunoprecipitate p53 was confirmed by an immunoprecipitation-Western blotting experiment (Fig. 7E). p53 protein from control and ADR-treated cells was immunoprecipitated with the acetylation-specific antibody, immunoprecipitates were analyzed by Western blotting with an antibody that recognizes all forms of p53 (PAb1801), and p53 was immunoprecipitated successfully (Fig. 7E). Cross-linked immunocomplexes were analyzed for the presence of p21Waf1, MDM2, and PIG3 promoter DNAs. Similar to the results observed for total p53 immunoprecipitates, the p21Waf1 PCR signal increased at 4 h after ADR treatment and remained elevated through 30 h (Fig. 7F). MDM2 promoter DNA was not detectable in immunocomplexes derived from control cell extracts, but PCR products were evident throughout the time course, with a decreased signal apparent at the 30 h time point (Fig. 7F). An increase in amplified PIG3 promoter DNA was detected early in the time course, with a decline in the signal at the later time points. The results indicate that acetylated p53 is found at all of the promoters examined (Fig. 7F). However, we could not determine whether acetylated p53 was directly bound to the consensus binding site or if it was heterodimerized with unacetylated p53 monomers. It was not possible to efficiently immunoprecipitate acetylated p53 from cells exposed to IR due to the low levels of modified p53 protein (Fig. 7D), nor was it possible to efficiently immunoprecipitate phosphorylated p53 (Ser15) from cells after either genotoxic treatment, and thus sufficient amounts of DNA template were not obtained for PCR analyses (data not shown).
Gu and Roeder have shown that the acetylated p53 (Lys382) has an increased affinity for its DNA consensus site (13), suggesting that acetylation plays an important role in p53 promoter recognition or binding after genotoxic stress. However, based on the results of numerous p53 overexpression experiments (12, 22, 35), we hypothesized that in the absence of genotoxic stress, ectopically expressed p53 must still undergo modifications that allow the protein to bind to target gene promoters with similar kinetics if a level of ecoptic p53 protein comparable to that of the endogenous protein after genotoxic stress could be achieved. To test this hypothesis, the levels of acetylated and phosphorylated p53 and p53 promoter occupancy were examined in a cell system in which the levels of p53 could be conditionally regulated in the absence of genotoxic stress.
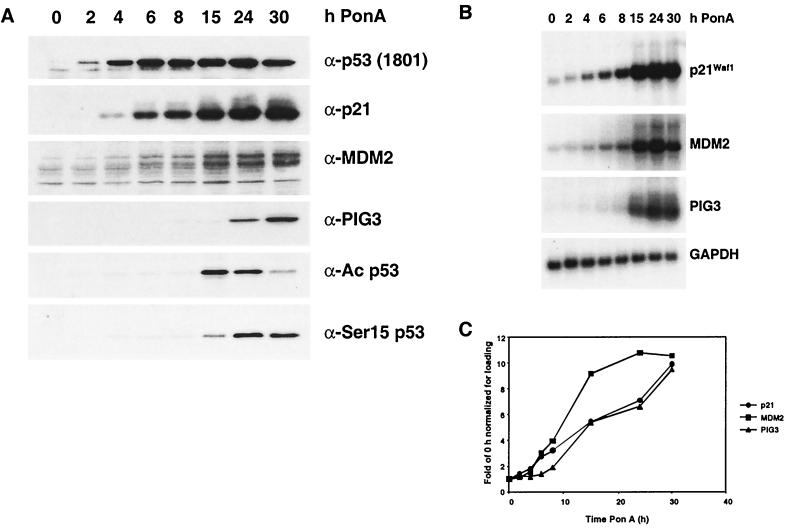
The HIp53 cell line was used for this line of experimentation. HIp53 cells were derived from the human lung carcinoma cell line H1299, which is null for endogenous p53. The cells were stably transfected with an ecdysone-inducible p53 expression vector, and treatment of HIp53 cells with the ecdysone analog PonA induces p53 expression (previously described [11]). As shown in Fig. 8A, p53 was elevated in HIp53 cells by 2 h after PonA incubation. p21Waf1 and MDM2 protein and mRNA were readily detectable at 4 and 6 h, respectively, and the levels continue to increase through the time course (Fig. 8). Similar to the results obtained with the ADR- and IR-treated RKO cells, detectable increases in PIG3 mRNA and protein were delayed, occurring between 15 and 30 h. Of note, acetylated (Lys382) and phosphorylated p53 (Ser15) were detected under these conditions (Fig. 8A).
FIG. 8.
Analysis of p53 target gene protein and mRNA levels as well as p53 posttranslational modifications after induction of ectopic p53. HIp53 cells were treated with PonA for the indicated times to induce the expression of p53. (A) Protein extracts were resolved by SDS-PAGE, and the proteins were analyzed by immunoblotting with antibodies shown. The 1801 antibody recognizes all forms of p53, and the other p53-specific antibodies recognize an acetylated form of p53 (Lys382) or a phosphorylated form of p53 (Ser15). (B) Cells were harvested, mRNA was isolated, and Northern analyses were performed for p21Waf1, MDM2, PIG3, and GAPDH mRNAs. (C) Graphic representation of the levels of mRNA in the Northern blots in panel B, normalized to GAPDH levels. The data are representative of two independent experiments.
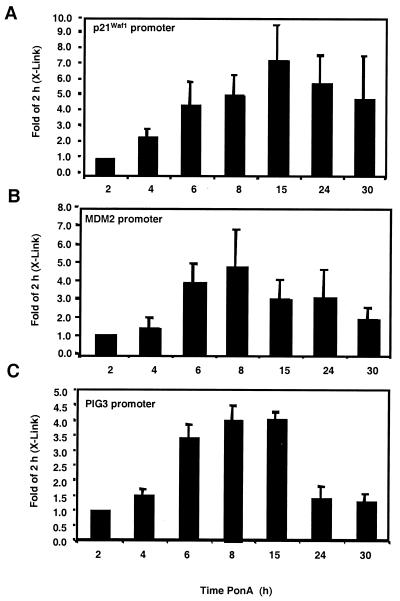
Using the HIp53 cells, we next examined the in vivo binding of p53 to the p21Waf1, MDM2, and PIG3 promoters in the absence of apparent genotoxic stress. The DNA templates obtained from p53 immunocomplexes were analyzed by PCR using primers specific for the p21Waf1, MDM2, and PIG3 promoters. Results across time courses were compared to those for the 2-h time point since PCR products were not detectable in control samples. The DNA from the p21Waf1 PCR amplification exhibited a steady increase above the 2-h signal, peaking at 15 h with a sevenfold increase over the 2-h signal (Fig. 9A). The PCR signal for the MDM2 promoter peaked at 8 h after p53 induction with a PCR signal fivefold higher than that observed at the 2-h time point (Fig. 9B).
FIG. 9.
Kinetics of p53 binding to downstream promoters in the HIp53 cell line. Exogenous p53 was induced with the addition of PonA for the indicated times. The DNA derived from p53-specific immunocomplexes was PCR amplified using primers for the p21Waf1 (A), MDM2 (B), or PIG3 (C) promoter. PCRs were resolved by PAGE, the gels were stained with ethidium bromide, and the PCR products were quantified by densitometry. Data are expressed as fold of the 2-h (X-Link) PCR signal. The level of PCR products derived from control cells was negligible. The results are representative of at least three independent experiments.
Similar to the results with the RKO cells, PIG3 promoter-specific PCR products were elevated at earlier time points than the significant increases in PIG3 mRNA were observed. After 6 to 8 h of PonA treatment, a fourfold increase in the PCR signal over the 2-h time point was apparent (Fig. 9C). This level remained relatively constant through 15 h and decreased by 24 h. Thus, p53 binding to the PIG3 promoter precedes PIG3 transactivation, and we predict that in the HIp53 model system, additional transcription factors are required for PIG3 transactivation.
DISCUSSION
In this study, the in vivo binding of p53 to its consensus site(s) in the p21Waf1, MDM2, and PIG3 promoters was investigated. p53-DNA complexes were trapped by treating the cells with formaldehyde. This approach allowed analysis of relevant complexes by preventing redistribution of cellular components upon collection of cell extracts. The level of PCR product generated from p21Waf1, MDM2, and PIG3 promoter DNA isolated from p53 immunocomplexes began to increase by 2 to 4 h after both genotoxic stress and ectopic p53 expression. p53 binding to the p21Waf1 and MDM2 regulatory regions correlated with increases in corresponding mRNA levels. Of note, for the time courses investigated, p53 binding at the MDM2 promoter appeared to be biphasic after genotoxic stress. This oscillation of promoter occupancy may be due to the displacement of p53 by RNA polymerase, as it generates transcripts initiated at the upstream transcription initiation site in the MDM2 promoter. In cells treated with ADR, an increase in p53 binding to the PIG3 promoter was detectable at early time points; however, detectable binding occurred prior to the significant increase in PIG3 mRNA production. Similarly, in HIp53 cells, the PCR products generated from PIG3 promoter DNA in p53 immunocomplexes were evident as early as 4 h after p53 induction; however, a marked increase in PIG3 mRNA was not apparent until 15 h.
The observed difference in kinetics of detectable p53 binding to the various promoters and production of target gene mRNA suggests that p53-dependent transactivation may be dictated by differential p53 affinity for the consensus sites in promoters and the contribution of other transcription factors working in concert with p53. The gel shift analyses show that p53 has a low affinity for the PIG3 response element compared to the p21Waf1 and MDM2 promoter binding sites. This low affinity may be due to a slow on rate or fast off rate or combination of both. Consistent with these observations, we hypothesize that p53 may require other factors or posttranslational modification to stabilize binding at the PIG3 promoter and allow transactivation. These factors may be recruited to the PIG3 promoter at later times after a stress treatment, stabilize p53 binding at the promoter site, and facilitate acquisition of the basal transcriptional machinery. However, we predict that the binding of these other factors or differential posttranslational modifications mask antibody epitopes on p53. Thus, at times of highest p53-mediated transactivation, p53 protein is bound by other factors or differentially modified, causing it to be inaccessible for immunoprecipitation after chemical cross-linking. Consistent with our hypothesis are the findings of Venot et al., who demonstrated that the proline-rich domain in the amino terminus of p53 is required for transactivation of PIG3 but not p21Waf1 and MDM2 (36). It is possible that this domain is involved in recruiting a transcriptional coactivator that is specifically required for PIG3 transactivation.
Despite compelling published data, the in vivo physiological relevance of p53 phosphorylation and acetylation remains unclear. The transcriptional coactivator p300 binds p53, and this interaction is proposed to be necessary for p53-mediated G1/S arrest and apoptosis (2). p300 has been shown to acetylate p53 in vitro at Lys382, increasing the sequence-specific binding activity of p53 (13). Therefore, acetylation of p53 may be required for transcriptional activation by p53. The posttranslational modification status of p53 may also account for discrimination by p53 for different promoters. For example, Wang and Prives reported that phosphorylation of p53 by cyclin B1-Cdc2 and cyclin A-Cdk2 complexes enhanced p53 binding to the p21Waf1 and GADD45 promoter sites in in vitro assays but had virtually no effect on its binding to another consensus site, the ribosomal gene cluster (37). Using an antibody specific for p53 acetylated at Lys382, the acetylation status of p53 at the p21Waf1, MDM2, and PIG3 promoters was examined. Acetylated p53 was found in cross-linked complexes at all these promoter sites after treatment of cells with ADR. Further, inducible expression of p53 in the HIp53 cells resulted in p53 protein that was still acetylated and phosphorylated (Ser15) in the absence of apparent genotoxic stress, suggesting that these modifications are constitutive.
Some studies have shown that DNA binding activity of p53 is not linked to its transactivation ability. Although, a correlation between p53 occupancy at p21Waf1 and MDM2 promoters and the mRNA levels of these gene products was observed, this was not the case for PIG3. It is possible that with certain posttranslational modifications, p53 may retain DNA binding activity but be incompetent for transactivation. For example, point mutations in the amino-terminal serines of murine p53 cause decreased transactivation, while stabilization, localization, and DNA binding activity of the protein are unaffected (22). Also, okadaic acid treatment of cells results in hyperphosphorylated, stabilized p53; this p53 is competent for DNA binding as shown by gel shift assays but exhibits decreased transcriptional activation in reporter assays (43). Under such conditions, identification of the phosphorylation sites which allow p53 DNA binding but interfere with its transactivation would contribute to our understanding of the transcriptional regulation of p53 target genes. Finally, select posttranslational modification may be required for p53 transactivation at a promoter region. Recently, Oda et al. have shown that p53-mediated transactivation of p53AIP1, requires serine 46 phosphorylation (26). Similar to PIG3, p53AIP1 is another potential mediator of p53-dependent apoptosis and has delayed kinetics of induction relative to p21Waf1 (26).
The use of reversible cross-linking agents as described in this study can be a valuable tool in linking p53 biochemistry to biology. It is conceivable that after cross-linking, complexes containing p53 can be dissected to reveal the components of the transcriptional activation complex at different promoters. Such an approach was used for defining the role of the corepressor mSin3a in p53-mediated transcriptional repression and probing the chromatin structure at endogenous, repressed promoter sites (25). In some cases, repression may be difficult to evaluate using exogenous reporter genes which may not be decorated with chromatin-associated proteins to the extent of endogenous promoters (6, 21). Furthermore, use of a cross-linking approach may lend insight to the regulation and promoter selectivity of the p53 homologs, p73 and p63 (18).
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
We are grateful to the P. J. Farnham laboratory for helpful suggestions regarding the formaldehyde cross-linking procedure. We thank D. Hill for provision of phosphorylation- and acetylation-specific antibodies, L. J. Tang for expert technical assistance, and members of the Pietenpol laboratory and Vanderbilt-Ingram Cancer Center for critical reading of the manuscript.
This work was supported by NIH grants CA70856 (to J.A.P.) and CA68485 (core services), a Burroughs Wellcome New Investigator in Toxicology Award (to J.A.P.), NIEHS grant ES00267 (core services), and Department of the Army grant DAMD 17-97-1-7267 (to S.T.S.).
REFERENCES
- 1.Ashcroft M, Kubbutat M H G, Vousden K H. Regulation of p53 function and stability by phosphorylation. Mol Cell Biol. 1999;19:1751–1758. doi: 10.1128/mcb.19.3.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 3.Banin S, Moyal L, Shieh S Y, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 4.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd K E, Wells J, Gutman J, Bartley S M, Farnham P J. c-Myc target gene specificity is determined by a post-DNA-binding mechanism. Proc Natl Acad Sci USA. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 7.Canman C E, Lim D S, Cimprich K A, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan M B, Siliciano J D. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 8.Deng C X, Zhang P M, Harper J W, Elledge S J, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 9.El-Deiry W S, Harper J W, O'Connor P M, Velculescu V E, Canman C E, Jackman J, Pietenpol J A, Burrell M, Hill D E, Wang Y, Wiman K G, Mercer W E, Kastan M B, Kohn K W, Elledge S J, Kinzler K W, Vogelstein B. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 10.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 11.Flatt P M, Tang L J, Scatena C D, Szak S T, Pietenpol J A. p53 regulation of G2 checkpoint is retinoblastoma protein dependent. Mol Cell Biol. 2000;20:4210–4223. doi: 10.1128/mcb.20.12.4210-4223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs B, O'Connor D, Fallis L, Scheidtmann K H, Lu X. p53 phosphorylation mutants retain transcription activity. Oncogene. 1995;10:789–793. [PubMed] [Google Scholar]
- 13.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 14.Gu Y, Turck C W, Morgan D O. Inhibition of CDK2 activity in vivo by an associated 20K regulatory subunit. Nature. 1993;366:707–710. doi: 10.1038/366707a0. [DOI] [PubMed] [Google Scholar]
- 15.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 16.Haupt Y, Barak Y, Oren M. Cell type-specific inhibition of p53-mediated apoptosis by mdm2. EMBO J. 1996;15:1596–1606. [PMC free article] [PubMed] [Google Scholar]
- 17.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 18.Kaelin W G., Jr The emerging p53 gene family. J Natl Cancer Inst. 1999;91:594–598. doi: 10.1093/jnci/91.7.594. [DOI] [PubMed] [Google Scholar]
- 19.Kubbutat M H G, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 20.Liu L, Scolnick D M, Trievel R C, Zhang H B, Marmorstein R, Halazonetis T D, Berger S L. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 22.Mayr G A, Reed M, Wang P, Wang Y, Schwedes J F, Tegtmeyer P. Serine phosphorylation in the NH2 terminus of p53 facilitates transactivation. Cancer Res. 1995;55:2410–2417. [PubMed] [Google Scholar]
- 23.Meek D W. Mechanisms of switching on p53: a role for covalent modification? Oncogene. 1999;18:7666–7675. doi: 10.1038/sj.onc.1202951. [DOI] [PubMed] [Google Scholar]
- 24.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 25.Murphy M, Ahn J, Walker K K, Hoffman W H, Evans R M, Levine A J, George D L. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 1999;13:2490–2501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, Taya Y. p53AlP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 2000;102:849–862. doi: 10.1016/s0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- 27.Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 28.Sakaguchi K, Herrera J E, Saito S, Miki T, Bustin M, Vassilev A, Anderson C W, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheikh M S, Burns T F, Huang Y, Wu G S, Amundson S, Brooks K S, Fornace A J, Jr, El-Deiry W S. p53-dependent and -independent regulation of the death receptor KILLER/DR5 gene expression in response to genotoxic stress and tumor necrosis factor α. Cancer Res. 1998;58:1593–1598. [PubMed] [Google Scholar]
- 30.Shieh S-Y, Taya Y, Prives C. DNA damage-inducible phosphorylation of p53 at N-terminal sites including a novel site, Ser20, requires tetramerization. EMBO J. 1999;18:1815–1823. doi: 10.1093/emboj/18.7.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shieh S Y, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 32.Stewart Z A, Leach S D, Pietenpol J A. p21Waf1/Cip1 inhibition of cyclin E/Cdk2 activity prevents endoreduplication after mitotic spindle disruption. Mol Cell Biol. 1999;19:205–215. doi: 10.1128/mcb.19.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szak S T, Pietenpol J A. High affinity insertion/deletion lesion binding by p53—evidence for a role of the p53 central domain. J Biol Chem. 1999;274:3904–3909. doi: 10.1074/jbc.274.6.3904. [DOI] [PubMed] [Google Scholar]
- 34.Unger T, Juven-Gershon T, Moallem E, Berger M, Sionov R V, Lozano G, Oren M, Haupt Y. Critical role for Ser20 of human p53 in the negative regulation of p53 by Mdm2. EMBO J. 1999;18:1805–1814. doi: 10.1093/emboj/18.7.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Unger T, Sionov R V, Moallem E, Yee C L, Howley P M, Oren M, Haupt Y. Mutations in serines 15 and 20 of human p53 impair its apoptotic activity. Oncogene. 1999;18:3205–3212. doi: 10.1038/sj.onc.1202656. [DOI] [PubMed] [Google Scholar]
- 36.Venot C, Maratrat M, Dureuil C, Conseiller E, Bracco L, Debussche L. The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. EMBO J. 1998;17:4668–4679. doi: 10.1093/emboj/17.16.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Prives C. Increased and altered DNA binding of human p53 by S and G2/M but not G1 cyclin-dependent kinases. Nature. 1995;376:88–91. doi: 10.1038/376088a0. [DOI] [PubMed] [Google Scholar]
- 38.Waterman M J F, Stavridi E S, Waterman J L F, Halazonetis T D. ATM-dependent activation of p53 involves dephosphorylation and association with 14-3-3 proteins. Nat Genet. 1998;19:175–178. doi: 10.1038/542. [DOI] [PubMed] [Google Scholar]
- 39.Wells J, Boyd K E, Fry C J, Bartley S M, Farnham P J. Target gene specificity of E2F and pocket protein family members in living cells. Mol Cell Biol. 2000;20:5797–5807. doi: 10.1128/mcb.20.16.5797-5807.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu X, Bayle J H, Olson D, Levine A J. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 41.Xiong Y, Hannon G J, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 42.Zauberman A, Flusberg D, Haupt Y, Barak Y, Oren M. A functional p53-responsive intronic promoter is contained within the human mdm2 gene. Nucleic Acids Res. 1995;23:2584–2592. doi: 10.1093/nar/23.14.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang W, McClain C, Gau J, Guo X-Y D, Deisseroth A B. Hyperphosphorylation of p53 induced by okadaic acid attenuates its transcriptional activation function. Cancer Res. 1994;54:4448–4453. [PubMed] [Google Scholar]