Abstract
Understanding the detailed mechanism behind every human disease, disorder, defect, and deficiency is a daunting task concerning the clinical diagnostic tools for patients. Hence, a closely resembling living or simulated model is of paramount interest for the development and testing of a probable novel drug for rectifying the conditions pertaining to the various ailments. The animal model that can be easily genetically manipulated to suit the study of the therapeutic motive is an indispensable asset and within the last few decades, the zebrafish models have proven their effectiveness by becoming such potent human disease models with their use being extended to various avenues of research to understand the underlying mechanisms of the diseases. As zebrafish are explored as model animals in understanding the molecular basis and genetics of many diseases owing to the 70% genetic homology between the human and zebrafish genes; new and fascinating facts about the diseases are being surfaced, establishing it as a very powerful tool for upcoming research. These prospective research areas can be explored in the near future using zebrafish as a model. In this review, the effectiveness of the zebrafish as an animal model against several human diseases such as osteoporosis, atrial fibrillation, Noonan syndrome, leukemia, autism spectrum disorders, etc. has been discussed.
Keywords: Human disease models, Zebrafish model, Teleost disease model, Zebrafish translational research, Experimental disease models
Graphical abstract
1. Introduction
With the increasing complexities arising from the disease being caused by rapidly mutating pathogens or new infectious strains in modern times, the importance of understanding their pathogenicity is becoming the need of the hour. These studies leading to the understanding of the cellular and molecular mechanisms of such infections can only be undertaken in a suitable animal model. These animal models not only help researchers understand the progression of such diseases and their mechanisms, but also allow them to design various targeted therapies and drugs to help cure the disease [1]. One such popular animal model is that of the Zebrafish (Danio rerio), a teleost found in freshwater systems, which has been at the forefront of several research fields since the 1980s [2]. When zebrafish models started to be used in research, the primary focus was to contribute to the field of developmental biology, with thousands of zebrafish mutant models being identified with highly conserved disease genes among different species [3]. This finding made researchers aware of the importance of the zebrafish model in other research fields and subsequently led to gene cloning through rigorous expansion of gene mapping resources.
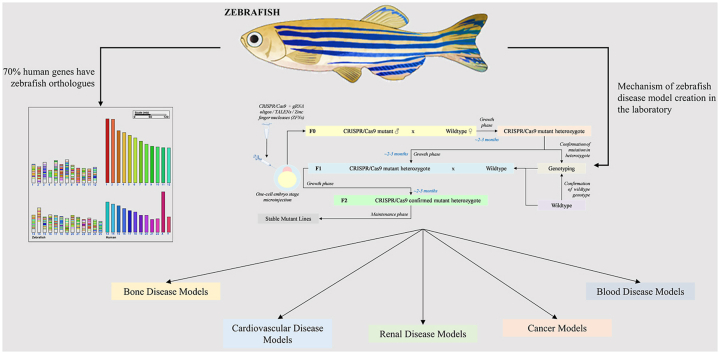
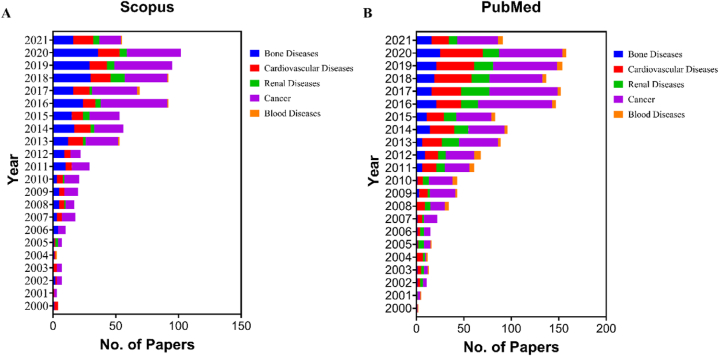
Today, zebrafish are being seen as a powerful model of acquired and genetic human diseases with increasing utility in translational research. The freshwater teleost is a popular animal model choice because of its high embryo yield, rapid reproduction rate, transparent embryos, economical maintenance, and fewer space requirements for setting up these zebrafish husbandries. Additionally, zebrafish embryos develop outside of the uterus, so they can be easily accessed post-fertilization for various studies. After the zebrafish genome sequencing project provided a comprehensive gene set in 2013, it was found that the genome possessed considerable homology with that of the human genome and that 70% of human genes had zebrafish orthologues [4]. The growth of its use in translational research can be extrapolated from Fig. 1A-B, which shows the increase of the use in zebrafish disease models over the years with the data being collected over the available literature from PubMed and Scopus databases (2000–2021).
Fig. 1.
The figure represents the progressive use of zebrafish disease models over the years from data that has been curated from [A] Scopus and [B] PubMed databases showing few of the broadly categorized fields where the zebrafish models are of great value.
In this review, we highlight the effectiveness of zebrafish as an animal model in understanding human diseases, genetic or otherwise, compiled around the various studies undertaken over the years bringing new knowledge of disease mechanisms previously unknown to the man with the help of different zebrafish models – mutant, knockdown, transgenic or otherwise. We are aware that some detailed articles specific to certain disease and disorder areas in relation to the modeling of zebrafish as a disease model already exist, but our goal here is to present a broad overview of the pathogenies associated to the modeling of the diseases in zebrafish models over the past two decades. Instead of providing a general overview of the entire field related to disease modeling in zebrafish, we have chosen to focus on a subsection of diseases in the fields of bone health, cardiovascular research, renal diseases, cancer, and blood ailments aimed at providing a glance understanding of the molecular mechanisms that lead to the diseases and specific zebrafish models that can be modeled around these molecular mechanisms to explore the disease condition. This article also discusses future areas of study that can be undertaken by using zebrafish as a model organism while also stating a number of exhaustive uses of the model organism in other fields of study not discussed at lengths in the paper.
2. Zebrafish–the making of an interesting translational model
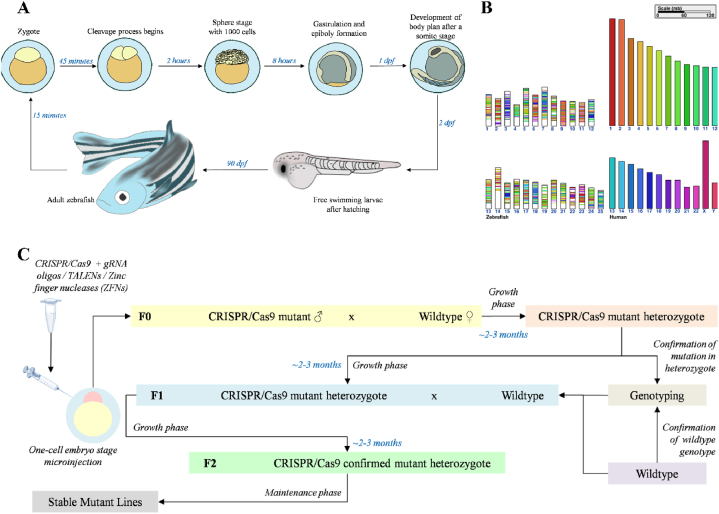
One of the most interesting aspects of a zebrafish model is the rapid breeding amongst the models to generate stable disease model lines within 3–4 months. It is highly fertile, allowing it to produce at least 200 eggs per clutch every 2–3 days (Fig. 2A), which can be used for experiments as soon as 3 days post fertilization (dpf) [5,6]. The lucrative part about working with zebrafish larvae is the fact that the eggs are optically transparent in addition to having a diameter of about 0.7 mm making genetic manipulations through all the developmental stages feasible for the researchers [7]. This optical clarity observed during the initial stages of embryogenesis permits the researchers to image the detailed developmental processes concerning drug discovery, tissue regeneration, or pathogenesis of an infection, notably due to the homology of the zebrafish genome with that of humans (shown in Fig. 2B). The project undertaken to sequence zebrafish genome further calls to our attention that about 82% of the known human disease genes have zebrafish orthologs making it a highly interesting prospect for designing disease models within the last decade [4] driven by the modern technological advances (Fig. 2). This also allows us to presume that a considerably similar portion of the loci associated with genome-wide association studies (GWAS) having certain disease-causing genes can be tested using mutant disease models in zebrafish, suggesting a comparability of 368 BMD loci in mutant zebrafish to 393 BMD loci testable in a bigger model organism such as mutant mice as an example [8]. Few studies have been conducted in the past that have shown the potential of zebrafish to easily verify GWAS data [9,10]. Precision genome editing with the help of clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 and transcription activator-like effector nucleases (TALENS) are a few of these recent technological marvels that grant for the swift and streamlined production of site-specific genetic mutants in zebrafish of the tissue-specific study being performed [11] during a research (process shown in Fig. 2C). In one of the more recent studies performed for the efficient modeling of human diseases in zebrafish, it was shown that very precise point mutations can be introduced with high efficiency over multiple genes simultaneously using a variant of CBE4max-SpRY, which recognizes a variety of different PAMs, thus expanding on the base editor techniques for human disease model creation [12]. The reverse genetics techniques such as ‘Targeting Induced Local Lesions In Genomes’ (TILLING) might be used in zebrafish with ease to identify the loss-of-function or gain-of-function of the alleles in any gene in theory, thereby making it a well-rounded model for studying human diseases [13].
Fig. 2.
This figure shows the schematic representations of [A] The life cycle of zebrafish which make it one of the most lucrative propositions as a model organism for diseases, [B] the comparison of the human and zebrafish genome with the help of Cinteny web server (http://cinteny.cchmc.org/) where the chromosomes are segregated and arranged with a particular color code which encodes for various syntenic regions between the two and [C] the mechanisms through which stable zebrafish mutant lines are created in the laboratory.
Moreover, zebrafish further give rise to two variants of models, namely, the larval and the adult models. The maintenance of these models is both space and cost-effective and in addition to this, their extended life spans in comparison to mice make it further interesting to scientists for developing new and effective disease models. The extent of its utility over rodent models is listed in Table 1.
Table 1.
| Basis | Zebrafish Models | Rodent Models |
|---|---|---|
| Size | Very small | Relatively bigger |
| Animal Care Costs | Quiet low | Quite high |
| Housing Requirements | Less space needed | Takes up a lot of space |
| Development | External | Internal |
| Number of offspring | ∼100–300 per week | ∼5–12 per month |
| Life span | ∼3–5 years | ∼2–3 years |
| F0 reverse genetic screens | Easily available and highly affordable | Not readily available and difficult to afford |
| F3 reverse genetic screens | Moderately available | Rarely available |
| External visibility of skeletal development | Easily viewable due to transparency at developmental stages | Not possible due to development in the uterus |
| Possibility of in-vivo fluorescence imaging | Highly possible | Low possibility |
| Orthologues count per human gene | ∼1 or >1 | 1 |
| Genome-wide screening | Possible | Not possible |
| Genetic manipulation | Can be done easily | Harder process |
| Phenotyping | Rapid | Cumbersome |
| Neuroregeneration | Possible | Not possible |
| Telomere size | Short like humans with the age-related progression of decline | Long |
| Drug administration | Easier | Relatively difficult |
| 3R strategy | Developed | Yet to be developed |
| Xenograft engraftment | Rapid | Slow |
| Experimental cycle | Short | Long |
| Ethical constraints | Very low | Quite a lot |
Zebrafish have another very important tool in their immensely diverse but medically relevant toolkit; their potential in personalized regenerative medicine. Regenerative medicine fuelled by stem cell therapies has stepped into a dimension of personalized medicine that caters to any individual's need for targeted therapies based on the available medical data on them, such as pharmacogenomics and other genetic parameters. Due to the high regenerative potential of the zebrafish and its accessibility for genetic manipulations, zebrafish are being considered the future of personalized regenerative treatments, especially because the drug discoveries and their associated regenerative values found using zebrafish take lesser time compared to other models and can be made available to a greater number of patients at a lower cost owning to the homology between zebrafish and humans [17]. All these reasons and the fact that zebrafish make for a potent small system for screening multiple drugs during drug discovery make it one of the most powerful proponents of the 'tank-to-bedside' line of thought for in vivo models, which is at the core of any translational research [18]. The drug delivery is comparatively easier with zebrafish as water-soluble drugs can be directly administered by dissolving them in the water to be absorbed by the fish through their gills and skin [19] making for a non-invasive technique which reduces the stress associated with injections often seen in the murine systems. However, this does not limit researchers from using drug administration techniques such as intraperitoneal injections [20] or oral drug delivery [21], which allows the understanding of the bioavailability of the drug and other parameters critical to drug discovery and translational research. The extrapolation of the drug doses established using zebrafish models into higher mammals such as mice and subsequently humans would lead to further validation of the translational models of zebrafish and provide us with an affirmative nod towards the translational prowess of the models themselves, etching a new revolution in disease modeling and treatment discoveries in the near future. In such instances to develop precision medicine and the need to replace long and cumbersome complex trials to pace the proper conduction of drug trials, zebrafish models have stood out as a viable disease modeling system [22].
3. Effective zebrafish disease models
3.1. Bone diseases
One of the most important and unique features of the zebrafish model is that the models allow real-time live imaging of various biological processes like skeletal development and repair. This allows researchers to follow the skeletal development processes in zebrafish models for specifically designed bone diseases with ease. The embryonal stages of the zebrafish are considered to be one of the most powerful models for bone research with researchers reportedly using them for studying the process of osteogenesis since the first mineralized vertebrae can already be visualized after five days of the fertilization of the egg [23]. Table 2 lists the different zebrafish models developed for various bone-related diseases along with their effectiveness.
Table 2.
Zebrafish models and their effectiveness in the study of bone diseases.
| Disease Studied | Zebrafish Models | Effectiveness of the Study | Reference |
|---|---|---|---|
| Osteogenesis imperfecta (OI) | Chihuahua (chi) | Understanding the role of collagen in bone development through stress study of the endoplasmic reticulum in fibroblasts and osteoblasts | [25,26] |
| frilly fins (frf) | Studying the less common form of OI caused by bmp1 mutation affecting the formation of mature collagen I | [28,29] | |
| Osteoporosis | atp6v1h mutant | Finding a new bone formation pathway as well as identifying a possible therapy | [30] |
| Osteopetrosis | panther (fms) | Provides an osteopetrosis disease study model | [31] |
| Osteoarthritis | prg4a−/−; prg4b−/−double mutant | Studying wear and tear of cartilages present in joints due to depletion of lubricating proteins of synovial joints | [32] |
| Fibrodysplasia Ossificans Progressiva (FOP) | acvr1-alk2 mutant | Studying the role of acvr in dorsoventral patterning and understanding its role in the disease | [33] |
| Scoliosis | fused somites (fss) | Understanding the role of the Delta-Notch pathway in the progression of the disease | [34] |
| leviathan | Study of the possibility of genetic factors playing a role in the disease | [35] | |
| zmynd10, ift88, kif3b, uts2ra mutant | Establishing a correlation between scoliosis and Parkinson's disease | [36] | |
| Ankylosing spondylitis (AS) Klippel-Feil syndrome (KFS) | dolphin, stocksteif | Understanding the possible role of cyp26b1 in AS and KFS | [37] |
| Craniosynostosis | stocksteif | Understanding that osteoblast differentiation defects can induce the disease | [44] |
| tcf12/twist1b double mutant | Using CRISPR technology for understanding the disease progression | [38] |
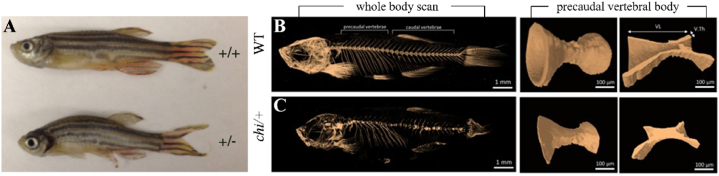
One such bone disease of great relevance is Osteogenesis imperfecta (OI) which is also called the brittle bone disease caused by mutations in the collagen genes col1a1 or col1a2 [24]. The first zebrafish mutant model characterized in the year 2003 for the bone disease was known as chihuahua (chi) [25], which mapped for the mutation to collagen1a1 (col1a1) and presented similar symptoms of OI (as depicted in Fig. 3B–C). It also led to a better understanding of the role of collagen in bone development through a stress study of the endoplasmic reticulum in fibroblasts and osteoblasts [26]. The study with the chi model of the zebrafish designed to mimic the classical dominant OI symptoms found that the deformities and fractures observed in the zebrafish model were a result of altered bone quality brought about by a heterozygous glycine substitution in α1 chain of the type I collagen. Zebrafish mutants for col1a2 and col1a1b were identified as presenting the disease phenotype during recent studies [27]. Zebrafish models with mutant bmp1 are known as frilly fins (frf) [28] which facilitates the study of a less common form of Osteogenesis imperfecta which results due to mutation of the bone morphogenetic protein (BMP1), which is known to generate mature collagen I [29] causing alteration in the osteoblast morphology which appears cuboidal in shape. The assumptions that it was a result of reduced adhesion and not merely a defect in the osteoblast was verified later through their in-vitro studies where the osteoblast cultured on collagen-deficient bone matrix showcased somewhat rounded morphology instead of its flattened morphology when viewed on a purified mineralized collagen matrix. Through this study, it was understood that autosomal recessive OI with an increased bone matrix is generally the result of insufficient removal of the C-propeptide from collagen-I.
Fig. 3.
The figure shows two models of zebrafish, used effectively to treat bone diseases. [A] The zebrafish having the atp6V1h mutation (±) shows a curved phenotype when compared to its wild-type counterpart (+/+) due to the curvature of the spine. [B] Shows the scan of wild-type zebrafish alongside the longitudinal cut of the vertebral body while [C] represents the scan of the chi/+ mutant showing deformities of the fins and ribs along with heterogeneous skull mineralization while also demonstrating an odd geometry and mineralization of hard tissues. Adapted with permission from references [26,30].
Osteoporosis is a bone disease characterized by porous bones due to the increased osteoclastic action or the inadequacy of osteoblasts in replacing resorbed bone. To study this disease, a mutant zebrafish atp6v1h model (represented in Fig. 3A) was created that showcased a new bone formation pathway along with identifying a possible therapy for such patients [30]. This study allowed for the understanding of the crucial role of activation of mmp9/mmp13 with respect to osteoporosis and led to the proposal of a novel model of action related to therapeutic entry points for osteoporosis and related diseases. Although no orthologues are available in zebrafish for the known human osteopetrosis genes that cause dense but brittle bones, a zebrafish mutant model for the c-fms oncogene, panther has been identified to provide an osteopetrosis model [31]. The study outcome also suggested that the dysfunction of osteoclasts can lead to an abnormal vein formation through the gathering of mononuclear precursor cells in congested spaces of haemal arches and blood vessels suggesting a role of certain factors triggering osteoclastogenesis at these sites.
To study osteoarthritis which is caused by the wear and tear of cartilage present in the joints, a genetic zebrafish double mutant model for prg4a and prg4b was developed in which both knockout genes are known to encode essential lubricating proteins of synovial joints [32] and the severity of the disease aggravates with aging, just like in humans. The study found that the jaw and fin joints of the model require lubricin for their maintenance which is a molecular lubricant similar to synovial joints making it a prospective model for the study of synovial specializations including the process of cavitation and in-depth understanding of the development and progression of synovial joint diseases.
Models such as acvr1 zebrafish mutants presented phenotypes associated with Fibrodysplasia Ossificans Progressiva (FOP) which is a rare autosomal disorder where connective tissues, as well as muscles, transform into bone [33], allowing researchers to use the model in better understanding the disease and mechanisms related to the pathophysiology of the disease. The development of this model allows for future investigation of injury models, building a better understanding of the progression of the disease. There are models designed to study spine curvature diseases such as scoliosis which are presented primarily due to gene mutations in the Delta-Notch pathway. The zebrafish mutant model fused somites (fss), which contains a mutant of the tbx6 gene of the Delta-Notch pathway [34], and the zebrafish model called leviathan (shown in Fig. 3) has a mutation of the col8a1a gene but completely unrelated to the Delta-Notch pathway [35] are used to study scoliosis as of late. Leviathan was used to probe into the possibility of genetic factors playing a role in scoliosis while at the same time proposing a novel mechanism for the formation of vertebral columns arising from the aberrant deposition of bone in misshapen notochord tissue regions independent of somitogenesis defects. The former study with the model helped identify the relationship of certain gene mutations involved in the patterning of somites through signaling pathways arising from the notochord. Quite recently, using zebrafish mutant models for zmynd10, ift88, kif3b, and uts2ra, a correlation was established between Parkinson's and scoliosis [36]. The research showed that the severity of the spine curvature has a correlation with the severity of the zebrafish phenotype and that urotensin provides adrenergic signals through the cerebrospinal fluid (CSF) activating the slow-twitch muscles portraying a role in the straightening of the axis, allowing us to speculate on the aforementioned correlation.
A cyp26b1 mutant zebrafish model called dolphin and stocksteif has been successfully shown to be useful for studies of bone diseases such as Ankylosing spondylitis (AS) and Klippel-Feil syndrome (KFS) [37] with stocksteif being used as a model of craniosynostosis [38,39] as well. The studies helped to understand that the skeletogenesis of zebrafish is closely regulated by adequate levels of retinoic acid (RA) and critically controlled cyp26b1 activity and that cyp26b1-dependent RA restriction is required to properly control osteoblast biology and bone formation. A tcf12/twist1b double mutant model has also been designed to study craniosynostosis disease using CRISPR technology [38]. The study helped to easily detect cells expressing tcf12 in cranial sutures, skull bone, and neural tissues in the zebrafish model, signifying that these heterozygous mutations can affect maintaining the patency of sutures. Zebrafish can also be used to provide insight into transgenesis. This is because transgenic reporter models have found great use as of late to visualize the skeletal system growth and regeneration as well as the cellular behaviors based on genetic context to gain an understanding of the molecular mechanisms related to the disease etiology [40].
In addition to these, there have been several transgenic models of zebrafish which have been used to study the various genetic pathways in association with the cell types involved in the disease pathogenesis of their targets. Osteoblasts have been studied using Tg(Ola.sp7:mCherry)zf131 and Tg (Ola.sp7:NLS-GFP)zf132 with osterix (osx) as a genetic marker [39] while Tg(-2.2col10a1a:GFP)ck3 that has been known to express GFP in the presence of col10a1 promoter which unlike mammals is not limited to chondrocytes in zebrafish and thereby makes for an excellent model for studying molecular mechanisms indicative of both chondrogenesis as well as osteogenesis [41]. The osteoclast studies in relation to multiple diseases and skeletal regeneration have been investigated using transgenic zebrafish models such as Tg(ctsk:DsRed) [42] and Tg(trap:GFP-CAAX)◦u2031 [43] that use ctsk and trap as markers for osteoclastogenesis.
3.2. Cardiovascular diseases
The embryonic zebrafish heart can be easily inspected in live zebrafish due to its position on the ventral side, thus allowing researchers to understand the mechanisms involved in the development and functioning of the heart. This is not the only attractive feature of the zebrafish model, as its effectiveness as a disease model is increased by the fact that they are not dependent on circulation initially during its early developmental phase, but rather remains oxygenated by passive diffusion. This provides the researchers an opportunity to probe into the disease progressions of the various cardiac phenotypes. Table 3 describes the various zebrafish models that have been used effectively to study many of the different types of cardiovascular disease.
Table 3.
Zebrafish models and their effectiveness in the study of cardiovascular diseases and renal diseases.
| Renal Diseases | |||
|---|---|---|---|
| Glomerular diseases | Tg(podocin:GFP) | Studies related to podocytes and their role in the progression of glomerular diseases | [64] |
| Noonan syndrome | sos2-acp1mutant | Studying the role of sos2 and acp1 in renal tubule alterations and insufficient glomerular functions | [65] |
| Nephrotic syndrome disease | plce1 knockdown | Studying the role of plce1 in causing oedemas and effacement of podocytes along with the progression of the disease conditions | [66] |
| Steroid-resistant nephrotic syndrome | Tg(wt1b:EGFP) | Understanding the role of FAT1 in the onset of the disease conditions | [67] |
| Diabetic neuropathy | elmo1 mutant | Establishing the role of the elmo1 gene in the diabetic conditions | [68] |
| Fanconi syndrome | ctns mutant | Undertaking various other nephropathic cystinosis studies | [69] |
| Autosomal dominant polycystic kidney disorder (ADPKD) | double bubble, fleer | Studying the disease pathophysiology and understanding the role of Na+/K+ ATPase mislocation in the disease progression | [70] |
Out of the many effective phenotypes of heart-specific mutant lines obtained for studying various cardiovascular diseases, one of the most useful lines is the silent heart (sih) model [45] which is perfect for the study of various molecular pathways involved in cardiac disorders as well as the structural changes occurring due to these disease conditions. The study with the sih model provided the first animal model for tnnt2 deficiency that is responsible for the proper assembly of the cardiac sarcomere making the heart muscle non-functional. Identification of the molecule SB216763 allowed researchers to understand the role of the Wnt signaling pathway in the pathophysiology of arrhythmogenic cardiomyopathy studied in the zebrafish model obtained with a mutated plakoglobin [46]. The study with the plakoglobin mutated model (as shown in Fig. 4A) helped to elucidate that aberrant trafficking of proteins related to the functioning of intercalated discs is the main cause of myocyte injury in arrhythmogenic cardiomyopathy (ACM) along with various electrical abnormalities.
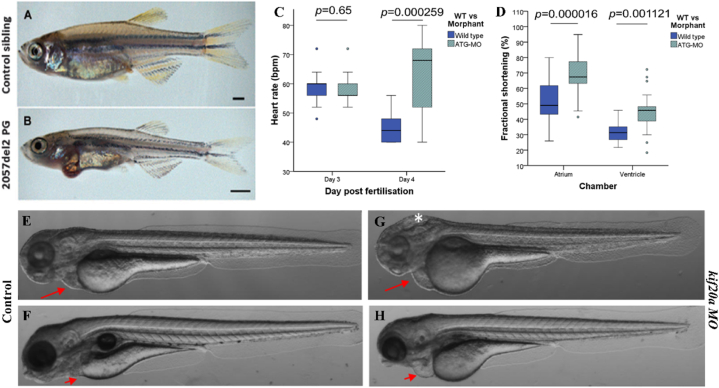
Fig. 4.
The figure shows zebrafish models used in treating cardiac-related issues. [A-B] Shows the morphology of the [A] 5-week-old control zebrafish and the [B] zebrafish with a mutated plakoglobin demonstrating arrhythmogenic cardiomyopathy (scaled to 1 mm); [C-D] Represents the cardiac function analysis performed in the control and kif20a morphants where [C] the heart rate is compared between the control and the kif20a morphants at 3 days post fertilization (dpf) and 4 dpf resulting in an appreciable rise in the heart rate at 4 dpf associated with progressive cardiac failure [D] A significant fractional shortening can be observed at 4dpf in both atrium and ventricle in morphants of the morphants when compared to the control but based on more outliers and systolic failure could be predicted; [E-H] Represents the control and kif20a mutant models where it can be observed that [G-H] pronounced cardiac edema (denoted by the red arrows) can be observed in morphants at both 3 and 4 dpf while cerebral edema in morphants (denoted by *) could only be seen at 3 dpf which was not present in the [E-F] controls at 3 and 4 dpf. Adapted with permission from references [46,52]
The potassium interacting channel 1 gene (kcip1) which is associated with atrial fibrillation (AF) was shown to play a role in the modulation of atrial rate in the heart of zebrafish, indicating a possible target for AF treatment [10]. The results obtained using zebrafish mutated kcip1 showed that the mRNA level of the kcip1 gene and its protein determined by the intronic CNV present in the kcip1 gene can be linked to high atrial rates and could be a possible future target for the disease treatment. Long QT syndrome type 2 (LQT2) zebrafish models with a mutated potassium voltage-gated channel subfamily H member 2 (kcnh2) have been used to understand severe cardiac repolarization and long ventricular depolarization associated [47] with the loss of kcnh2 activity [48]. The results obtained using Long QT syndrome type 2 zebrafish models demonstrated the functional abnormalities of E637K mutations in terms of cardiac polarizations. Mutations in prrx1a and prrx1b in the zebrafish model for human prrx1, a transcription factor associated with AF, showed that the duration of the atrial potential is modulated by the slightest of changes in prrx1 expression [49]. The study helped identify a prrx1 upstream enhancer for its promoter and was found to be modulated by single-nucleotide polymorphism (SNP) that has an inverse action on prrx1 expression. The conclusions drawn from the use of the zebrafish model implicated prrx1 as the causative gene of AF.
Dilated cardiomyopathies can be studied with the help of the zebrafish heat shock protein, beta 7 (hspb7) mutant model [50] and integrin-linked kinase (ilk) mutant model, main squeeze (msq) [51]. The study undertaken with the hspb7 zebrafish model provided the basic understanding of how the hspb7 gene regulated the proteostatic mechanisms functioning in the cardiomyocytes giving rise to a new stress-dependent model for cardiomyopathy in humans while the zebrafish model msq contributed to the scientific knowledge about cardiomyopathy by finding a relationship between the laminin-integrin-ilk mutations with the disease condition and heart failure. The models of mutant kif20a of zebrafish, as demonstrated in Fig. 4C–H, are associated with studies of congenital cardiomyopathy and the studies suggest that kif20a is conserved in developing the heart through evolution [52]. Through the study with the zebrafish model, it was shown that translational blocking of the kif20a gene promoted a cardiomyopathy-like phenotype, which showed the functionality of the gene for proper heart function. Zebrafish models obtained by genome editing using the CRISPR-Cas9 technique of the gene pbx4 are associated with congenital heart defect studies [53] where it acts as a 'genetic modifier' that promotes the disease condition caused by loss of function of the gene hand2. Valvulopathies are a form of cardiovascular disease that can cause fatal heart failure in patients. Studies in zebrafish lmcd1 and tensin1 mutant models have strengthened our understanding of mitral valve prolapse (MVP), which is one of the most common valvulopathy observed [9]. The study with these two zebrafish models provided proof that both the genes which have been previously implicated in the proliferation and migration of cells, contribute to the degeneration of the mitral valve during valve development. The genes lcmd1 and tensin1 are now considered important genes in the pathogenesis of MVP.
The zebrafish gridlock (grl) mutant models are very effective in studying aortic coarctations [54,55] and their causative mechanisms [56]. The studies with the grl model of zebrafish showed that the gene hlh is necessary for the proper functioning of the aorta which otherwise would form a phenotype resembling coarctation of the aorta. The zebrafish slow mo mutant is a model for bradycardia [57] whereas models such as tremblor, santa, and reggae are used to study diseases related to the rhythmicity of the heart [58]. The slow mo (smo) model helped to understand the role of a form of current associated with the nervous system, Ih, and its contribution towards the pacemaking ability of the heart. The potential of cardiovascular disease modeling in zebrafish also makes it a lucrative model to lead into studies dealing with precision medicine for diseases, thus letting us study the pharmacological responses associated with novel drugs awaiting translational mobility [59].
3.3. Renal diseases
The zebrafish has pronephros which comprises two nephrons characterized by fused glomerulus along with paired bilateral ducts [60] which have established zebrafish models for studying renal diseases that affects the development of nephrons and their physiology [61]. The podocyte epithelial cells of the glomerulus are a potent target of many causative agents of renal diseases and hence a lot of the more recent research employing renal disease zebrafish models include podocytopathies [62]. The various models of zebrafish developed to investigate renal diseases can be found in Table 3.
The transgenic models of zebrafish give an outstanding prospect for the development and regeneration of podocytes in modeled zebrafish [63]. The zebrafish transgenic line model Tg(podocin:GFP) has shown that the morphology between mammalian and zebrafish podocytes is conserved [64] which has led to the use of zebrafish models to understand the progression of various glomerular diseases. According to the study, the model is potentially equipped to facilitate real-time imaging, sorting through FACS and molecular profiling of zebrafish podocytes, as well as HTS of functional genes by injecting morpholino. Noonan syndrome observed in humans and kidney abnormalities can be studied in the zebrafish mutant models for SOS Ras/Rho guanine nucleotide exchange factor 2 (sos2) and acid phosphatase 1 (acp1), which results in morphological alterations in the renal tubule and low dextran clearance suggesting insufficient glomerular function and damaged tubular flow [65]. The study led to the discovery of eight loci associated with kidney functionality, as well as to establish the roles of sos2 and acp1 in relation to kidney development in embryos. Furthermore, the zebrafish model gave rise to functional systemic models for studying gene perturbations with respect to the embryonic renal system. The knockdown of the phospholipase C epsilon (plce1) gene in zebrafish embryos has been implicated in causing edema in phenotypes along with podocyte effacement observed in the disease condition of nephrotic syndrome [66]. Through the study, a conclusion was drawn stating that the plce1 based arrest of the development of glomerulus was reversible through glucocorticoid or cyclosporine A treatment although the mechanism of its action couldn't be figured out. An interaction was also established between IQ motif–containing GTPase-activating protein 1 (IQGAP1) and plce1 through this multifaceted study, which further indicated that plce1 could serve the purpose of an ‘assembly scaffold’ necessary for the development of the glomerulus through its role in organizing a complex of molecules necessary for the proper glomerular morphogenetic processes.
Tg(wt1b:EGFP) zebrafish model with a knocked down FAT atypical cadherin 1 (fat1) has been developed for studies of steroid-resistant nephrotic syndrome [67]. Researchers through the study model of the transgenic zebrafish line demonstrated that in case of a low expression of fat1, there is impairment in the activity of rac1 and cdc42 suggesting that the activation of the two GTPases would partially restore functionality to the defective cell migration caused by fat1 knockdown. This mechanistic theory tested out to be true when rac1/cdc42 activators were introduced in the study mitigating the disease phenotype.
The zebrafish elmo1 mutant model (as can be observed in Fig. 5E and F) is a model of diabetic neuropathy that has established the role of the elmo1 gene under diabetic conditions [68]. The study findings clearly define the protective role of elmo1 through the anti-apoptotic route in hyperglycaemic conditions, thus predicting that favorable up-regulation of the elmo1 gene would be beneficial in the study of diabetic nephropathy. Nephropathic cystinosis zebrafish models (Fig. 5C and D) with a mutant cystinonin (ctns) gene have been used to study renal Fanconi syndrome [69]. The model was validated through their in vivo studies recording the various phenotypic similarities of the model to the human disease condition, providing the basis for future therapeutic screening for drugs. The disease condition autosomal dominant polycystic kidney disorder (ADPKD) is often associated with the Na+/K+ ATPase mislocation, which was studied using the double bubble [60] model of zebrafish. This study also shows the role of the model in assessing cell polarity. This study allowed the zebrafish model for various other studies related to the disease. Elipsa and fleer can be used as zebrafish disease mutant models in the study of another disease condition called Senior-Loken syndrome [60].
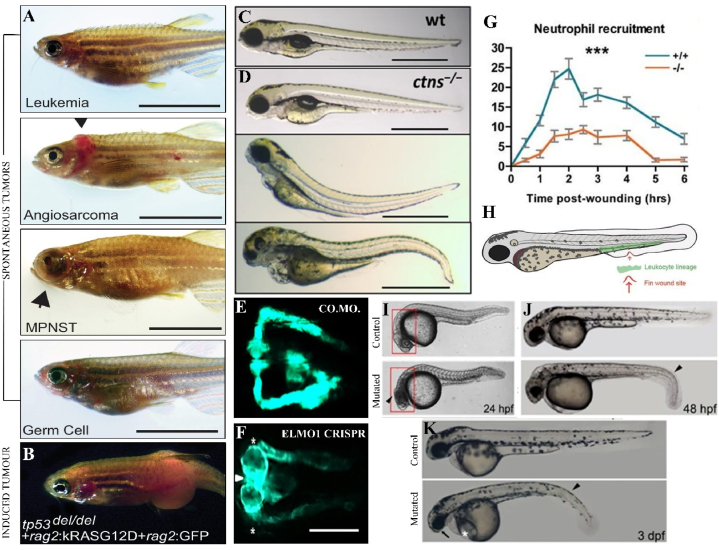
Fig. 5.
This figure demonstrates the zebrafish models utilized in understanding human diseases. [A-B] Shows the zebrafish model, CG1 tp53del/del used for studying spontaneous and induced tumors; [C-D] Even though the wild-type zebrafish larva shows a normal morphology, the ctns−/− mutant shows deformities such as growth retardation [D, upper] denoted by the bigger yolk, [D, middle and lower] bent head, and bulging eyes with mild and severe deformities; [E-F] In comparison to the [E] control morphant (Co.Mo.), [F]ELMO1 crispant (ELMO1 CRISPR) showed an enlarged glomerulus (depicted by the white arrowhead) and shortened pronephric neck (denoted by white asterisk) suggesting an adverse effect of hyperglycemia upon the pronephric structure of zebrafishes; [G] The effectiveness of the WASp1−/− model in the timed recruitment of neutrophils when compared to wild-type zebrafish larvae and [H] the schematic diagram of the WASp1−/− model; [I–K] Shows the various models and experimental results obtained for studies related to blood disease models. The different phenotypes of rps19 homozygous mutants at different timed stages – [I] 24hpf, [J] 48hpf, and [K] 3dpf are shown. Adapted with permission from references [68,69,77,89,95].
3.4. Cancer
Over the years, cancer has been a looming topic in the minds of researchers across the globe, with one of the biggest challenges associated with cancer studies being intratumor heterogeneity. Cancer is considered to be a collection of genetically variable diseases with certain commonalities at the molecular level, and hence it is important to understand the molecular mechanisms at play to address any progressing malignancy. Such studies require models, and few of the most important models for cancer studies are created in zebrafish due to the relative ease in gene manipulations that can be done in it for the studying of a variety of cancers as well as the initial transparency exhibited by the zebrafish embryos and larvae. This transparency quotient led to the development of a zebrafish model casper, that remains transparent throughout adulthood allowing the visualization of cancer formation and its gradual progression [71].
The zebrafish-mutated model tp53M214K is one of the first mutation models found by genetic screening techniques and it helped to study multiple cancer types such as peripheral nerve sheath tumor (PNST), breast cancer, brain tumor and leukemia [72]. A similar PNST study can also be carried out in the zebrafish model brca2Q658X-tp53M214K [73] while a cancer model was created using the zebrafish-mutated model BRAFV600E-tp53−/− [74] that lost its malignancy in the absence of tp53−/− mutations [75]. The findings from zebrafish model brca2Q658X-tp53M214K studies showed that brca2 genotype and sex of the model act as independent variables affecting the outcome of survival in fishes bearing the form of cancer while the studies with BRAFV600E-tp53−/− models showed for the first time a ‘cooperative genetic interaction’ which resulted in the cancer of zebrafish posing the models potential for understanding the progression of the development of cancer genetically adding to the pathological relevance of the BRAF mutation.
Zebrafish mutated models rag2:KRASG12D and CG1-tp53−/− (Fig. 5A-B) are implicated in the study of rhabdomyosarcoma (RMS) [76] and a combination of sarcoma and leukemia studies [77] respectively. The study undertaken with the rag2:KRASG12D mutated model of zebrafish suggested that the molecular pathways involved in embryonal rhabdomyosarcoma (ERMS) are conserved in both zebrafish and humans in addition to being potential isolation models for cancer stem cells for the disease thus allowing for a better understanding of the molecular and regulatory pathways associated in the onset of the disease. On the other hand, the work done with the zebrafish mutated model CG1-tp53−/− showed that ERMS metastasis is greatly affected by the loss of tp53 that accounts for the aggressive invasiveness of RMS in the disruption of the tp53 pathway. A model designed to study Costello syndrome called HRASG12V (represented in Fig. 5) can also be used to study a variety of lymphomas, melanomas, and sarcomas [78]. The results of the study show that HRAS activation induced by the absence of tp53 causes overgrowths in the CNS that are often associated with simultaneous inactivation of p53 leading to age-related aggravation of the disease phenotype. A zebrafish model ptfa1:KRASG12V has been developed to study pancreatic adenocarcinoma [79] showed upregulation of hedgehog pathway components, indicating that the disease phenotype is affected by the hedgehog pathway. Another model tg:BRAFV600E has been implicated in the study of thyroid carcinomas [80] whose study results showed for the first time the role of TWIST gene expression downstream of BRAFV600E gene mutations.
Models like rag2:mMyc are used for the study of T-cell acute lymphoid leukemia (T-ALL) [81], spi1:tel-jak2a for studies of T-ALL and chronic myeloid leukemia (CML) [82,83] and spi1:MYST3/NOCA2 for studies of acute lymphoid myeloma (AML) [84]. The use of the spi1:tel-jak2a model for T-ALL and CML provided insight into the etiology of the disease condition arising from alternative fusions of TEL-JAK2 exerting its oncogenic effect in a cell lineage-specific manner caused by probable downstream signaling differences. The outcome of the spi1:MYST3/NOCA2 zebrafish model study confirmed the transforming properties of MYST3/NOCA2 previously observed in murine species and hematopoietic stem cells. A list of the effective zebrafish models implicated in the translation of cancer research can be found in Table 4.
Table 4.
Zebrafish models and their effectiveness in the study of cancer and blood diseases.
| Disease Studied |
Zebrafish Models |
Effectiveness of the Study |
Reference |
|---|---|---|---|
| Cancer | |||
| Peripheral nerve sheath tumor (PNST) | tp53M214Kmutant | Studying the molecular mechanisms leading to cancer formation and its progression | [72] |
| brca2Q658X-tp53M214Kmutant | Undertaking the studies related to PNST | [73] | |
| Melanoma | BRAFV600E-tp53−/−mutant | Malignancy studies in the absence or presence of tp53 | [74,75] |
| Rhabdomyosarcoma (RMS) | rag2:KRASG12Dmutant | Undertaking tumorogenesis studies to understand the progression of the disease better | [76] |
| Pancreatic adenocarcinoma | ptfa1:KRASG12Vmutant | Undertaking studies related to the human pancreatic adenocarcinoma | [79] |
| Thyroid carcinoma | tg:BRAFV600Emutant | Studying a novel potential target responsible for BRAFV600E-driven thyroid follicle transformation | [80] |
| Leukemia | rag2:mMyc mutant | Undertaking studies related to T-ALL | [81] |
| spi1:tel-jak2a mutant | Undertaking studies related to T-ALL and CML | [82,83] | |
| spi1:MYST3/NOCA2 mutant | Undertaking studies related to AML | [84] | |
| Blood Diseases | |||
| Sideroblastic anemia | sauternes | Studying the role of alas2 in the progression of sideroblastic anemia | [86] |
| Porphyrias | yquem | Understanding the role of the urod gene in the heme biosynthesis pathway and the progression of the disease | [87] |
| dracula | Studying the effect of ferrochelatase gene mutations in the heme biosynthesis pathway and the progression of the disease | [88] | |
| Diamond-Blackfan anemia (DBA) | rps19-rpl11 mutant | Understanding the correlation between protein reduction and defects observed in the erythrocytes in the disease condition | [89] |
| Hypochromic anemia | weiβherbst | Understanding the role of ferropontin1gene in the disease | [90] |
| Thalassemia | zinfandel | Providing a model for the studies related to thalassemia | [91,92] |
| Congenital dyserythropoietic anemia type II (HEMPAS) | retsina | Undertaking studies related to HEMPAS and understanding the role of certain genes related to it | [93] |
| Systemic mastocytosis (SM) | c-KIT-D816V mutant | Studying mast cell accumulation and its effect on multiple organs | [94] |
3.5. Blood diseases
Zebrafish form great animal models for studying diseases related to blood and blood cells since mutants obtained through various screening and transgenic processes present an almost accurate model of human hematopoietic disease. The blood system and the pathways related to the synthesis of blood are very similar and more or less conserved in both mammals and zebrafish which also boosts the translational potential of these models greatly [85]. During the forward genetic screening, the first animal model for congenital sideroblastic anemia disease was discovered and the zebrafish mutant model for δ-aminolevulinate synthase (ALAS-2) was called sauternes [86]. The study showed that the sauternes model of zebrafish is heme deficient probably caused by a defect in the heme biosynthesis pathway and suggested a role of the alas2 gene in the differentiation of adult blood cells. The severity of the heme-deficient phenotype of the zebrafish model was shown to depend on the expression of the globin gene. The zebrafish yquem model with mutations in the uroporphyrinogen decarboxylase (UROD) gene [87] and the dracula model with mutations in the ferrochelatase gene [88] are considered good models to study human porphyrias, both of which relate to faulty heme biosynthesis pathways. The study with the yquem model of zebrafish provides insight into the first attempt at in vivo gene therapy for the disease condition of porphyria. The study also suggested there was a need for a urod promoter or enhancer in the study to completely alleviate the disease phenotype. The research involving the dracula model of zebrafish proposed the presence of maternal deposition of heme, ferrochelatase activity, or fch gene in the yolk of the embryo accounting for the survival of the fch gene mutants studied with the help of the zebrafish model of porphyria.
For studies related to Diamond-Blackfan anemia (DBA), zebrafish genetic models were developed for deficiencies of the ribosomal protein S19 (rps19) (as shown in Fig. 5I–K) and the ribosomal protein L11 (rpl11), leading to a significant contribution to the study undertaken [89]. The outcome of the study interestingly showed that translational defects of the globin gene were partially rescued by treatment with l-leucine which is known to work in correlation with the mTOR pathway in the rps19 and rpl11 mutant models, suggesting a role for l-leucine in activating the translational processes of erythroid cells allowing for a better understanding of the clinical manifestations of DBA. Hypochromic anemia studies have been conducted in zebrafish models weiβherbst with a mutated ferropontin1 gene [90] and zinfandel with mutant globin gene, which can also be useful for thalassemia studies [91,92]. The studies with ferropontin1 in the weiβherbst model suggested that ferropontin1 is required for intestinal iron transportation in zebrafish and that it could play a role in the export of iron from macrophages that is necessary for the recycling of iron from senescent erythrocytes, while also suggesting based on the results of the globin gene studies that teleosts have very well defined erythrocyte lineages that express various other globins as well, which can be used for generating more intriguing models of hemoglobin switching during the evolution of vertebras.
The study with Zebrafish mutant model retsina with a defect in the erythroid anion exchanger 1/band3 gene has been used to study a rare blood disease called congenital dyserythropoietic anemia type II (HEMPAS) [93]. The results of the study showed that the retsina mutation is caused by defects in the zebrafish band 3 gene based on certain criteria derived from gene expression studies. A zebrafish transgenic model with the c-KIT-D816V mutation was recently developed to study systemic mastocytosis (SM) whose severity is characterized by mast cell accumulation in multiple organs [94]. It was understood from the study that the transgenic zebrafish model was capable of providing ‘embryonic surrogate markers’ having the potential of being used for future drug testing. It is also the first model for human mast cell disease. Studies quite recently led to the development and characterization of a zebrafish null mutant allele model for the wasp gene, represented in Fig. 5G and H, implicated in the study of Wiskott-Aldrich syndrome (WAS) and also X-linked severe congenital neutropenia (XNS) [95]. The study showed the importance of the wasp function and the potential of the model in testing with live imaging techniques the various effects of wasp mutations on the model system. It also linked the susceptibility of the model to bacterial infection as similar to those seen in patients with WAS and XNS. To facilitate studies of leukocyte adhesion deficiency (LAD), Rac2D57N zebrafish mutation models were developed [96] that suggested the role of rac2 signaling in neutrophil retention by hematopoietic tissues. The results from this study helped provide a deeper understanding of rac2 signaling which might be associated with its modulation leading to an increase in the mobilization of neutrophils since the role of rac2 signaling is predominantly seen in neutrophils where they function by active retention of neutrophils thus limiting its mobility. Table 4 lists the various zebrafish models developed to investigate blood diseases.
3.6. Other diseases
Apart from all the human disease studies previously discussed that are effective in zebrafish models, there are many other diseases for which models have been developed or are being developed to gain insight into the disease conditions. Some of these are diseases such as Duchene muscular dystrophy which is a fatal muscle-wasting disease that is being studied using the zebrafish model dmd-MO developed by knocking down Dystrophin using anti-sense morpholino oligomers (MO) [97], and retinal diseases in humans using the zebrafish model no optokinetic response c [98]. There have been studies that use zebrafish as model animals for understanding schizophrenia [99] and in various other addiction studies [100]. Recently, zebrafish models have been used to study autism spectrum disorders (ASD) [101] since zebrafish are considered highly social and for depression studies [102]. Zebrafish have also been used as models for many microgravity-related studies [[103], [104], [105]] in order to understand the effects of microgravity on humans as we take bold strides in space exploration over the next few decades.
4. Limitations associated with zebrafish disease modeling
Even though zebrafish disease models are very powerful models in terms of their potential towards building scientific knowledge, it doesn't come without a few of their inherent flaws. To fully comprehend the advantages of using zebrafish as a model organism in the future we must study and try to work around the limitations associated with zebrafish in general and while modeling. These hindrances have been highlighted in Table 5.
Table 5.
Limitations associated with disease modeling zebrafish.
| Type of Limitations | Description | Reference |
|---|---|---|
| General Limitations | Presence of duplicate copies of mammalian genes despite the high similarity in the homology of zebrafish to rodents and human beings with the genomic duplication being events that are independent and continuous. | [108] |
| Availability of genetically characterized strains of zebrafish is constrained due to its loss of fertility upon inbreeding and those that have been developed aren't easy to maintain. | [109] | |
| Divergence of a few functional genes due to genomic duplication limit the accuracy of disease modeling in zebrafish | [110] | |
| Zebrafish don't have sex chromosomes unlike humans and hence there is a difference in the sex ratio of the teleosts in the majority of research designs leading to varying levels of sexual dimorphism in comparison to mammals. | [6] | |
| Water-insoluble drug administration is a problem and requires the administration of the drug through various other mechanisms including the use of vehicles or techniques such as developing oral gavage or microinjections through the oesophagus. | [18] | |
| Despite having a good amount of physiological homology, the environmental conditioning between zebrafish and humans remains with the teleosts being maintained at about a temperature of 28 °C. | [5] | |
| Even though zebrafish have a blood-brain barrier (BBB) that is substantially alike to its human counterpart, due to the effect of differences between species, the action of drugs keeps changing due to the change of the BBB permeability causing a hindrance to drug testing. Similar effects due to species differences are also observed in cases of metabolism and thermoregulation. This accounts for one of the more serious limitations associated with the teleost models. | [18] | |
| The dosage cannot be easily translated directly from zebrafish into rodent or human doses and vice versa due to differences in the physiology between the species. | ||
| Limitations in Bone Disease Modelling | Mineral ossification pathway is widely different in zebrafish in comparison to mammals especially terrestrial mammals due to the variance in the uptake and excretion of calcium. | [111] |
| The development of the teleost skeletal system is prone to environmental influences which is not the case for most placental organisms. | [112] | |
| Limitations in Cardiovascular Disease Modelling | Studying septal development including blood pressure and metabolism is a hindrance. | [113] |
| There are differences in cardiac electrophysiology due to the variation in the cycling of calcium and ionic currents. | [114] | |
| There is a possibility of ventricular amputation due to the smaller size of zebrafish during ECG studies affecting the heart rate and the heart rate variability. | [115] | |
| Limitations in Renal Disease Modelling | Disruption of the functionality of the glomerulus in the kidneys of zebrafish embryos causes the death of the embryos within a few days. | [116] |
| Limitations in Cancer Modelling | Most of the present zebrafish disease models for cancer are made by knocking out certain genes deviating from their corresponding syndromes which usually show missense mutations failing to truncate proteins. This doesn't allow for a proper investigation into the protein interactions or the genetic basis of the disease. | [117] |
| Limitations in Blood Disease Modelling | Studying lymphocyte subtypes and the biology of leukocytes is difficult due to the scarcity of well-characterized monoclonal antibodies acting on the leukocyte surface molecule. | [118] |
| The difference in the morphology of some blood cells like erythrocytes and thrombocytes which are nucleated in zebrafish models might cause a hindrance in drawing parallels with humans and translating research. |
One of the other research areas which have gone back and forth over the years in agreeing to the utility of zebrafish in the field of research is neurosciences. This stems from the fact that, although there is a fair amount of conservation to the planning of the body and brain of vertebrates between zebrafish and humans, there is a lack of conservation in terms of brain anatomy that manifests itself in the form of an expanded telencephalon and a lack of prefrontal cortex [6]. In addition to that, the behavioral characteristics and the modeling of several developmental disorders are hindered as there is a lack of information about parental care among zebrafish. The difficulty in mapping CNS structures in zebrafish to draw parallels with mammals has also contributed to the modeling of complex brain diseases in the comparatively simple zebrafish neural system. These drawbacks however have not overshadowed the utility of the animal model in deriving the disease models as zebrafish have most of the major neurotransmitters, transporters, receptors, and a well-developed subcortical neural circuitry – allowing for the complex cognition functionality to be conserved with its subsequent human diseases of the CNS [106]. Thus, even though problems with the models remain in the field, it isn't ruled out as a future alternative for complex brain and neuron disease modeling with many zebrafish disease models for the brain and CNS being designed and available at present already for a wide range of diseases and disorders such as – anxiety, depression, pain, ADHD, epilepsy, neurodegeneration and many more [107].
However, all these limitations should not deter the field from gaining scientific traction, as although many of the aforementioned concerns regarding the zebrafish modeling are rational, most of them are relative and can be worked around with comparatively easier solutions and a probable scientific advancement in the future on the animal disease modeling front. Finding comparatively workable solutions for these limitations would aid the translational value gained from the studies conducted in zebrafish disease models.
5. Conclusion and future prospects
In this review, we have discussed the ever-expanding role of zebrafish models for human diseases in the avenue of research especially in the fields discussed earlier; helping us in understanding the cause, pathogenicity, mechanisms, and modes of infection, and figuring out the various molecular pathways associated with the disease condition allowing an effective research development into finding a cure for the disease by aiming for non-toxic but high efficacy drug designing. It also discusses the various zebrafish models that led to a successful as well as an effective study into several of the human diseases along with the general and disease modeling-specific limitations associated with zebrafish.
Although many of the diseases remain elusive, scientific pursuits toward understanding such diseases are gradually catching up with the creation of effective models that will revolutionize the medical field soon. Scientific research would always require animal models as they are integral to the understanding, development, characterization, reliability, and legitimacy of the hypothesized concepts and drug designs related to the disease study. The standout amongst these models will most likely be zebrafish owing to its genetic tractability and many more advantages related to it already discussed before in the introductory section of the review. With the advent of CRISPR technology, genetic manipulations have become easier and since such manipulations can easily be done in the zebrafish models to mimic human disease conditions, these teleost fish are becoming a major player in the myriad research fields.
One such prospective future initiation is studies related to microphthalmia, andophthalmia, and coloboma (MAC). Although zebrafish models for studying conditions have been around for more than a decade now [119], new zebrafish mutants discovered through various targeted mutagenesis and synthetic enhancer screens would guarantee the identification of new candidate genes for the condition, thereby improving our knowledge about eye developmental pathways while also shedding light on observed variabilities related to ocular malformation phenotypes [120,121]. Another fast-growing prospect already steadily compiling knowledge in its frontier is that of the effective use of zebrafish models to study neurodevelopmental diseases through the recognition of pathophysiologically relevant risk genes associated with them imparting translational relevancy [122]. Research has also begun dedicated to inflammasomes with the help of zebrafish models associated with the increasing number of autoinflammatory diseases which have thus far yielded a lot of knowledge related to disease conditions, but more perspective about them will be achieved in the future when cellular aspects will be recorded and therapeutic initiations are undertaken [123]. More prospects of the effectiveness of zebrafish models in understanding human diseases will be seen in the fields of oncology, Alzheimer's disease, immunology, diabetic studies, regenerative medicine studies, aging-related studies, food safety, nutrigenomics, and its related disease studies.
These translational researches will pave the way hopefully for a paradigm shift in medical research and add therapeutic values to the product of those researches. The teleost animal model continues showing vast potential in terms of its translational potential which can probably be capitalized on in the near future to develop probable novel therapeutic interventions.
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Data availability statement
Copyright permission obtained for used figures and images accordingly.
Data search strategy
The articles presented and cited in the review have been searched in PubMed, Google Scholar, ZFIN, Scopus engines. The articles were screened using the keywords “zebrafish” in addition to the name of the broad category of diseases. Both review and research papers were considered for studying the work associated with zebrafish over the span of the last two decades. The references in previously published reviews were also screened, searched and read before revising the articles in this review.
Funding source
This work hasn't been funded.
Declaration of competing interest
The authors declare that there is no conflict of interest.
References
- 1.Elson C.O., Balfour Sartor R., Tennyson G.S., Riddell R.H., Ii SPECIAL reports and reviews experimental models of inflammatory bowel disease. Gastroenterology. 1995;109:1344–1367. doi: 10.1016/0016-5085(95)90599-5. [DOI] [PubMed] [Google Scholar]
- 2.Zizioli D., Mione M., Varinelli M., Malagola M., Bernardi S., Alghisi E., Borsani G., Finazzi D., Monti E., Presta M., Russo D. Zebrafish disease models in hematology: highlights on biological and translational impact. Biochim. Biophys. Acta, Mol. Basis Dis. 2019;1865:620–633. doi: 10.1016/j.bbadis.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 3.Driever W., Solnica-Krezel L., Schier A.F., Neuhauss S.C.F., Malicki J., Stemple D.L., Stainier D.Y.R., Zwartkruis F., Abdelilah S., Rangini Z., Belak J., Boggs C. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.5167/uzh-215. [DOI] [PubMed] [Google Scholar]
- 4.Howe K., Clark M.D., Torroja C.F., Torrance J., Berthelot C., Muffato M., Collins J.E., Humphray S., McLaren K., Matthews L., McLaren S., Sealy I., Caccamo M., Churcher C., Scott C., Barrett J.C., Koch R., Rauch G.J., White S., Chow W., Kilian B., Quintais L.T., Guerra-Assunção J.A., Zhou Y., Gu Y., Yen J., Vogel J.H., Eyre T., Redmond S., Banerjee R., Chi J., Fu B., Langley E., Maguire S.F., Laird G.K., Lloyd D., Kenyon E., Donaldson S., Sehra H., Almeida-King J., Loveland J., Trevanion S., Jones M., Quail M., Willey D., Hunt A., Burton J., Sims S., McLay K., Plumb B., Davis J., Clee C., Oliver K., Clark R., Riddle C., Eliott D., Threadgold G., Harden G., Ware D., Mortimer B., Kerry G., Heath P., Phillimore B., Tracey A., Corby N., Dunn M., Johnson C., Wood J., Clark S., Pelan S., Griffiths G., Smith M., Glithero R., Howden P., Barker N., Stevens C., Harley J., Holt K., Panagiotidis G., Lovell J., Beasley H., Henderson C., Gordon D., Auger K., Wright D., Collins J., Raisen C., Dyer L., Leung K., Robertson L., Ambridge K., Leongamornlert D., McGuire S., Gilderthorp R., Griffiths C., Manthravadi D., Nichol S., Barker G., Whitehead S., Kay M., Brown J., Murnane C., Gray E., Humphries M., Sycamore N., Barker D., Saunders D., Wallis J., Babbage A., Hammond S., Mashreghi-Mohammadi M., Barr L., Martin S., Wray P., Ellington A., Matthews N., Ellwood M., Woodmansey R., Clark G., Cooper J., Tromans A., Grafham D., Skuce C., Pandian R., Andrews R., Harrison E., Kimberley A., Garnett J., Fosker N., Hall R., Garner P., Kelly D., Bird C., Palmer S., Gehring I., Berger A., Dooley C.M., Ersan-Ürün Z., Eser C., Geiger H., Geisler M., Karotki L., Kirn A., Konantz J., Konantz M., Oberländer M., Rudolph-Geiger S., Teucke M., Osoegawa K., Zhu B., Rapp A., Widaa S., Langford C., Yang F., Carter N.P., Harrow J., Ning Z., Herrero J., Searle S.M.J., Enright A., Geisler R., Plasterk R.H.A., Lee C., Westerfield M., De Jong P.J., Zon L.I., Postlethwait J.H., Nüsslein-Volhard C., Hubbard T.J.P., Crollius H.R., Rogers J., Stemple D.L. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldsmith J.R., Jobin C. Think small: zebrafish as a model system of human pathology. J. Biomed. Biotechnol. 2012:1–12. doi: 10.1155/2012/817341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meshalkina D.A., Kysil E.V., Warnick J.E., Demin K.A., Kalueff A.V. Adult zebrafish in CNS disease modeling: a tank that's half-full, not half-empty, and still filling. Lab Anim. (NY) 2017;46:378–387. doi: 10.1038/laban.1345. [DOI] [PubMed] [Google Scholar]
- 7.Spence R., Gerlach G., Lawrence C., Smith C. The behaviour and ecology of the zebrafish, Danio rerio. Biol. Rev. 2008;83:13–34. doi: 10.1111/j.1469-185X.2007.00030.x. [DOI] [PubMed] [Google Scholar]
- 8.Kwon R.Y., Watson C.J., Karasik D. Using zebrafish to study skeletal genomics. Bone. 2019;126:37. doi: 10.1016/J.BONE.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dina C., Bouatia-Naji N., Tucker N., Delling F.N., Toomer K., Durst R., Perrocheau M., Fernandez-Friera L., Solis J., Le Tourneau T., Chen M.H., Probst V., Bosse Y., Pibarot P., Zelenika D., Lathrop M., Hercberg S., Roussel R., Benjamin E.J., Bonnet F., Lo S.H., Dolmatova E., Simonet F., Lecointe S., Kyndt F., Redon R., Le Marec H., Froguel P., Ellinor P.T., Vasan R.S., Bruneval P., Markwald R.R., Norris R.A., Milan D.J., Slaugenhaupt S.A., Levine R.A., Schott J.J., Hagege A.A., Jeunemaitre X. Genetic association analyses highlight biological pathways underlying mitral valve prolapsed. Nat. Genet. 2015;47:1206–1211. doi: 10.1038/ng.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsai C.T., Hsieh C.S., Chang S.N., Chuang E.Y., Ueng K.C., Tsai C.F., Lin T.H., Wu C.K., Lee J.K., Lin L.Y., Wang Y.C., Yu C.C., Lai L.P., Den Tseng C., Hwang J.J., Chiang F.T., Lin J.L. Genome-wide screening identifies a KCNIP1 copy number variant as a genetic predictor for atrial fibrillation. Nat. Commun. 2016;7 doi: 10.1038/ncomms10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patton E.E., Tobin D.M. Spotlight on zebrafish: the next wave of translational research. DMM Dis. Model. Mech. 2019;12 doi: 10.1242/dmm.039370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosello M., Serafini M., Mignani L., Finazzi D., Giovannangeli C., Mione M.C., Concordet J.P., Del Bene F. Disease modeling by efficient genome editing using a near PAM-less base editor in vivo. Nat. Commun. 2022;13 doi: 10.1038/s41467-022-31172-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raby L., Völkel P., Le Bourhis X., Angrand P.O. Genetic engineering of zebrafish in cancer research. Cancers. 2020;12:1–36. doi: 10.3390/CANCERS12082168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veinotte C.J., Dellaire G., Berman J.N. Hooking the big one: the potential of zebrafish xenotransplantation to reform cancer drug screening in the genomic era. DMM Dis. Model. Mech. 2014;7:745–754. doi: 10.1242/dmm.015784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ali S., Champagne D.L., Spaink H.P., Richardson M.K. Zebrafish embryos and larvae: a new generation of disease models and drug screens. Birth Defects Res. Part C Embryo Today - Rev. 2011;93:115–133. doi: 10.1002/bdrc.20206. [DOI] [PubMed] [Google Scholar]
- 16.Kwon R.Y., Watson C.J., Karasik D. Using zebrafish to study skeletal genomics. Bone. 2019;126:37–50. doi: 10.1016/j.bone.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arjmand B., Tayanloo-Beik A., Foroughi Heravani N., Alaei S., Payab M., Alavi-Moghadam S., Goodarzi P., Gholami M., Larijani B. Zebrafish for personalized regenerative medicine; A more predictive humanized model of endocrine disease. Front. Endocrinol. 2020;11:396. doi: 10.3389/fendo.2020.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalueff A.V., Echevarria D.J., Stewart A.M. Gaining translational momentum: more zebrafish models for neuroscience research. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2014;55:1–6. doi: 10.1016/j.pnpbp.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 19.Goldsmith P. Zebrafish as a pharmacological tool: the how, why and when. Curr. Opin. Pharmacol. 2004;4:504–512. doi: 10.1016/j.coph.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 20.Parker M.O., Gaviria J., Haigh A., Millington M.E., Brown V.J., Combe F.J., Brennan C.H. Discrimination reversal and attentional sets in zebrafish (Danio rerio) Behav. Brain Res. 2012;232:264–268. doi: 10.1016/j.bbr.2012.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulkarni P., Chaudhari G.H., Sripuram V., Banote R.K., Kirla K.T., Sultana R., Rao P., Oruganti S., Chatti K. Oral dosing in adult zebrafish: proof-of-concept using pharmacokinetics and pharmacological evaluation of carbamazepine. Pharmacol. Rep. 2014;66:179–183. doi: 10.1016/j.pharep.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 22.Fazio M., Ablain J., Chuan Y., Langenau D.M., Zon L.I. Zebrafish patient avatars in cancer biology and precision cancer therapy Maurizio. Nat. Rev. Cancer. 2020;20:263–273. doi: 10.1038/s41568-020-0252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carnovali M., Banfi G., Mariotti M. Zebrafish models of human skeletal disorders: embryo and adult swimming together. BioMed Res. Int. 2019:1–13. doi: 10.1155/2019/1253710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Körkkö J., Ala-Kokko L., De Paepe A., Nuytinck L., Earley J., Prockop D.J. Analysis of the COL1A1 and COL1A2 genes by PCR amplification and scanning by conformation-sensitive gel electrophoresis identifies only COL1A1 mutations in 15 patients with osteogenesis imperfecta type I: identification of common sequences of null-allele. Am. J. Hum. Genet. 1998;62:98–110. doi: 10.1086/301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher S., Jagadeeswaran P., Halpern M.E. Radiographic analysis of zebrafish skeletal defects. Dev. Biol. 2003;264:64–76. doi: 10.1016/S0012-1606(03)00399-3. [DOI] [PubMed] [Google Scholar]
- 26.Fiedler I.A.K., Schmidt F.N., Wölfel E.M., Plumeyer C., Milovanovic P., Gioia R., Tonelli F., Bale H.A., Jähn K., Besio R., Forlino A., Busse B. Severely impaired bone material quality in chihuahua zebrafish resembles classical dominant human osteogenesis imperfecta. J. Bone Miner. Res. 2018;33:1489–1499. doi: 10.1002/jbmr.3445. [DOI] [PubMed] [Google Scholar]
- 27.Henke K., Daane J.M., Brent Hawkins M., Dooley C.M., Busch-Nentwich E.M., Stemple D.L., Harris M.P. Genetic screen for postembryonic development in the zebrafish (Danio rerio): dominant mutations affecting adult form. Genetics. 2017;207:609–623. doi: 10.1534/genetics.117.300187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Eeden F.J.M., Granato M., Schach U., Brand M., Furutani-Seiki M., Haffter P., Hammerschmidt M., Heisenberg C.P., Jiang Y.J., Kane D.A., Kelsh R.N., Mullins M.C., Odenthal J., Warga R.M., Nüsslein-Volhard C. Genetic analysis of fin formation in the zebrafish, Danio rerio. Development. 1996;123:255–262. doi: 10.1242/dev.123.1.255. [DOI] [PubMed] [Google Scholar]
- 29.Asharani P.V., Keupp K., Semler O., Wang W., Li Y., Thiele H., Yigit G., Pohl E., Becker J., Frommolt P., Sonntag C., Altmüller J., Zimmermann K., Greenspan D.S., Akarsu N.A., Netzer C., Schönau E., Wirth R., Hammerschmidt M., Nürnberg P., Wollnik B., Carney T.J. Attenuated BMP1 function compromises osteogenesis, leading to bone fragility in humans and Zebrafish. Am. J. Hum. Genet. 2012;90:661–674. doi: 10.1016/j.ajhg.2012.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., Huang H., Zhao G., Yokoyama T., Vega H., Huang Y., Sood R., Bishop K., Maduro V., Accardi J., Toro C., Boerkoel C.F., Lyons K., Gahl W.A., Duan X., Malicdan M.C.V., Lin S. ATP6V1H deficiency impairs bone development through activation of MMP9 and MMP13. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatani M., Takano Y., Kudo A. Osteoclasts in bone modeling, as revealed by in vivo imaging, are essential for organogenesis in fish. Dev. Biol. 2011;360:96–109. doi: 10.1016/j.ydbio.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 32.Askary A., Smeeton J., Paul S., Schindler S., Braasch I., Ellis N.A., Postlethwait J., Miller C.T., Gage Crump J. Ancient origin of lubricated joints in bony vertebrates. Elife. 2016;5 doi: 10.7554/eLife.16415.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Labonty M., Pray N., Yelick P.C. A zebrafish model of human Fibrodysplasia Ossificans Progressiva. Zebrafish. 2017;14:293–304. doi: 10.1089/zeb.2016.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Eeden F.J.M., Granato M., Schach U., Brand M., Furutani-Seiki M., Haffter P., Hammerschmidt M., Heisenberg C.P., Jiang Y.J., Kane D.A., Kelsh R.N., Mullins M.C., Odenthal J., Warga R.M., Allende M.L., Weinberg E.S., Nüsslein-Volhard C. Mutations affecting somite formation and patterning in the zebrafish, Danio rerio. Development. 1996;123:153–164. doi: 10.1242/dev.123.1.153. [DOI] [PubMed] [Google Scholar]
- 35.Gray R.S., Wilm T.P., Smith J., Bagnat M., Dale R.M., Topczewski J., Johnson S.L., Solnica-Krezel L. Loss of col8a1a function during zebrafish embryogenesis results in congenital vertebral malformations. Dev. Biol. 2014;386:72–85. doi: 10.1016/j.ydbio.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X., Jia S., Chen Z., Chong Y.L., Xie H., Feng D., Wu X., Song Z., Roy S., Zhao C. Cilia-driven cerebrospinal fluid flow directs expression of urotensin neuropeptides to straighten the vertebrate body axis. Nat. Genet. 2018;50:1666–1673. doi: 10.1038/s41588-018-0260-3. https://www.nature.com/articles/s41588-018-0260-3 [DOI] [PubMed] [Google Scholar]
- 37.Laue K., Jänicke M., Plaster N., Sonntag C., Hammerschmidt M. Restriction of retinoic acid activity by Cyp26b1 is required for proper timing and patterning of osteogenesis during zebrafish development. Development. 2008;135:3775–3787. doi: 10.1242/dev.021238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bluemel R., Klopocki E., Liedtke D. Zebrafish as model organism for craniosynostosis. Bone Abstr. 2017;6:P019. doi: 10.1530/boneabs.6.p019. [DOI] [Google Scholar]
- 39.Spoorendonk K.M., Peterson-Maduro J., Renn J., Trowe T., Kranenbarg S., Winkler C., Schulte-Merker S. Retinoic acid and Cyp26b1 are critical regulators of osteogenesis in the axial skeleton. Development. 2008;135:3765–3774. doi: 10.1242/dev.024034. [DOI] [PubMed] [Google Scholar]
- 40.Busse B., Galloway J.L., Gray R.S., Harris M.P., Kwon R.Y. Zebrafish: an emerging model for orthopedic research. J. Orthop. Res. 2020;38:925–936. doi: 10.1002/JOR.24539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Il Kim Y., Lee S., Jung S.H., Kim H.T., Choi J.H., Lee M.S., You K.H., Yeo S.Y., Yoo K.W., Kwak S., Lee J.N., Park R., Choe S.K., Kim C.H. Establishment of a bone-specific col10a1:GFP transgenic zebrafish. Mol. Cells. 2013;36:145–150. doi: 10.1007/s10059-013-0117-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caetano-Lopes J., Henke K., Urso K., Duryea J., Charles J.F., Warman M.L., Harris M.P. Correction: unique and non-redundant function of csf1r paralogues in regulation and evolution of post-embryonic development of the zebrafish. Dev. 2020;147 doi: 10.1242/dev.192211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kobayashi-Sun J., Yamamori S., Kondo M., Kuroda J., Ikegame M., Suzuki N., ichiro Kitamura K., Hattori A., Yamaguchi M., Kobayashi I. Uptake of osteoblast-derived extracellular vesicles promotes the differentiation of osteoclasts in the zebrafish scale. Commun. Biol. 2020;3 doi: 10.1038/s42003-020-0925-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laue K., Pogoda H.M., Daniel P.B., Van Haeringen A., Alanay Y., Von Ameln S., Rachwalski M., Morgan T., Gray M.J., Breuning M.H., Sawyer G.M., Sutherland-Smith A.J., Nikkels P.G., Kubisch C., Bloch W., Wollnik B., Hammerschmidt M., Robertson S.P. Craniosynostosis and multiple skeletal anomalies in humans and zebrafish result from a defect in the localized degradation of retinoic acid. Am. J. Hum. Genet. 2011;89:595–606. doi: 10.1016/j.ajhg.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sehnert A.J., Huq A., Weinstein B.M., Walker C., Fishman M., Stainier D.Y.R. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat. Genet. 2002;31:106–110. doi: 10.1038/ng875. [DOI] [PubMed] [Google Scholar]
- 46.Asimaki A., Kapoor S., Plovie E., Arndt A.K., Adams E., Liu Z., James C.A., Judge D.P., Calkins H., Churko J., Wu J.C., Macrae C.A., Kléber A.G., Saffitz J.E. Identification of a new modulator of the intercalated disc in a zebrafish model of arrhythmogenic cardiomyopathy. Sci. Transl. Med. 2014;6:240ra74. doi: 10.1126/scitranslmed.3008008. www.ScienceTranslationalMedicine.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanaka Y., Hayashi K., Fujino N., Konno T., Tada H., Nakanishi C., Hodatsu A., Tsuda T., Nagata Y., Teramoto R., Yoshida S., Nomura A., aki Kawashiri M., Yamagishi M. Functional analysis of KCNH2 gene mutations of type 2 long QT syndrome in larval zebrafish using microscopy and electrocardiography. Heart Ves. 2019;34:159–166. doi: 10.1007/s00380-018-1231-4. [DOI] [PubMed] [Google Scholar]
- 48.Splawski I., Shen J., Timothy K.W., Lehmann M.H., Priori S., Robinson J.L., Moss A.J., Schwartz P.J., Towbin J.A., Vincent G Michael, Keating M.T. Spectrum of mutations in long-QT syndrome genes KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation. 2000;102:1178–1185. doi: 10.1161/01.cir.102.10.1178. www.circulationaha.org [DOI] [PubMed] [Google Scholar]
- 49.Tucker N.R., Dolmatova E.V., Lin H., Cooper R.R., Ye J., Hucker W.J., Jameson H.S., Parsons V.A., Weng L.C., Mills R.W., Sinner M.F., Imakaev M., Leyton-Mange J., Vlahakes G., Benjamin E.J., Lunetta K.L., Lubitz S.A., Mirny L., Milan D.J., Ellinor P.T. Diminished PRRX1 expression is associated with increased risk of atrial fibrillation and shortening of the cardiac action potential. Circ. Cardiovasc. Genet. 2017;10 doi: 10.1161/CIRCGENETICS.117.001902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mercer E.J., Lin Y.F., Cohen-Gould L., Evans T. Hspb7 is a cardioprotective chaperone facilitating sarcomeric proteostasis. Dev. Biol. 2018;435:41–55. doi: 10.1016/j.ydbio.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knöll R., Postel R., Wang J., Krätzner R., Hennecke G., Vacaru A.M., Vakeel P., Schubert C., Murthy K., Rana B.K., Kube D., Knöll G., Schäfer K., Hayashi T., Holm T., Kimura A., Schork N., Toliat M.R., Nürnberg P., Schultheiss H.P., Schaper W., Schaper J., Bos E., Den Hertog J., Van Eeden F.J.M., Peters P.J., Hasenfuss G., Chien K.R., Bakkers J. Laminin-α4 and integrin-linked kinase mutations cause human cardiomyopathy via simultaneous defects in cardiomyocytes and endothelial cells. Circulation. 2007;116:515–525. doi: 10.1161/CIRCULATIONAHA.107.689984. [DOI] [PubMed] [Google Scholar]
- 52.Louw J.J., Nunes Bastos R., Chen X., Verdood C., Corveleyn A., Jia Y., Breckpot J., Gewillig M., Peeters H., Santoro M.M., Barr F., Devriendt K. Compound heterozygous loss-of-function mutations in KIF20A are associated with a novel lethal congenital cardiomyopathy in two siblings. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Farr G.H., Imani K., Pouv D., Maves L. Functional testing of a human PBX3 variant in zebrafish reveals a potential modifier role in congenital heart defects. DMM Dis. Model. Mech. 2018;11:dmm035972. doi: 10.1242/dmm.035972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weinstein B.M., Stemple D.L., Driever W., Fishman M.C. Gridlock, a localized heritable vascular patterning defect in zebrafish. Nat. Med. 1995;1:1143–1147. doi: 10.1038/nm1195-1143. https://www.nature.com/articles/nm1195-1143 [DOI] [PubMed] [Google Scholar]
- 55.Towbin J.A., Mcquinn T.C., gridlock A model for coarctation of the aorta? Nat. Med. 1995;1:1141–1142. doi: 10.1038/nm1195-1141. https://www.nature.com/articles/nm1195-1141#article-info [DOI] [PubMed] [Google Scholar]
- 56.Fishman M.C. gridlock, an HLH gene required for assembly of the aorta in zebrafish. Science. 2000;287:1820–1824. doi: 10.1126/science.287.5459.1820. [DOI] [PubMed] [Google Scholar]
- 57.Baker K., Warren K.S., Yellen G., Fishman M.C. Defective “‘pacemaker’” current (I h) in a zebrafish mutant with a slow heart rate. Genetics. 1997;94:4554–4559. doi: 10.1073/pnas.94.9.4554. www.pnas.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nguyen C.T., Lu Q., Wang Y., Chen J.N. Zebrafish as a model for cardiovascular development and disease. Drug Discov. Today Dis. Model. 2008;5:135–140. doi: 10.1016/j.ddmod.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bowley G., Kugler E., Wilkinson R., Lawrie A., van Eeden F., Chico T.J.A., Evans P.C., Noël E.S., Serbanovic-Canic J. Zebrafish as a tractable model of human cardiovascular disease. Br. J. Pharmacol. 2022;179:900–917. doi: 10.1111/BPH.15473. [DOI] [PubMed] [Google Scholar]
- 60.Drummond Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development. 1998;125:4655–4667. doi: 10.1242/dev.125.23.4655. [DOI] [PubMed] [Google Scholar]
- 61.Ebarasi L., Oddsson A., Hultenby K., Betsholtz C., Tryggvason K. Zebrafish: a model system for the study of vertebrate renal development, function, and pathophysiology. Curr. Opin. Nephrol. Hypertens. 2011;20:416–424. doi: 10.1097/MNH.0b013e3283477797. [DOI] [PubMed] [Google Scholar]
- 62.Gehrig J., Pandey G., Westhoff J.H. Zebrafish as a model for drug screening in genetic kidney diseases. Front. Pediatr. 2018;6:183. doi: 10.3389/FPED.2018.00183/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poureetezadi S.J., Wingert R.A. Little fish, big catch: zebrafish as a model for kidney disease. Kidney Int. 2016;89:1204–1210. doi: 10.1016/j.kint.2016.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He B., Ebarasi L., Hultenby K., Tryggvason K., Betsholtz C. Podocin-green fluorescence protein allows visualization and functional analysis of podocytes. J. Am. Soc. Nephrol. 2011;22:1019–1023. doi: 10.1681/ASN.2010121291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li M., Li Y., Weeks O., Mijatovic V., Teumer A., Huffman J.E., Tromp G., Fuchsberger C., Gorski M., Lyytikäinen L.P., Nutile T., Sedaghat S., Sorice R., Tin A., Yang Q., Ahluwalia T.S., Arking D.E., Bihlmeyer N.A., Böger C.A., Carroll R.J., Chasman D.I., Cornelis M.C., Dehghan A., Faul J.D., Feitosa M.F., Gambaro G., Gasparini P., Giulianini F., Heid I., Huang J., Imboden M., Jackson A.U., Jeff J., Jhun M.A., Katz R., Kifley A., Kilpeläinen T.O., Kumar A., Laakso M., Li-Gao R., Lohman K., Lu Y., Mägi R., Malerba G., Mihailov E., Mohlke K.L., Mook-Kanamori D.O., Robino A., Ruderfer D., Salvi E., Schick U.M., Schulz C.A., Smith A.V., Smith J.A., Traglia M., Yerges-Armstrong L.M., Zhao W., Goodarzi M.O., Kraja A.T., Liu C., Wessel J., Boerwinkle E., Borecki I.B., Bork-Jensen J., Bottinger E.P., Braga D., Brandslund I., Brody J.A., Campbell A., Carey D.J., Christensen C., Coresh J., Crook E., Curhan G.C., Cusi D., De Boer I.H., De Vries A.P.J., Denny J.C., Devuyst O., Dreisbach A.W., Endlich K., Esko T., Franco O.H., Fulop T., Gerhard G.S., Glümer C., Gottesman O., Grarup N., Gudnason V., Hansen T., Harris T.B., Hayward C., Hocking L., Hofman A., Hu F.B., Husemoen L.L.N., Jackson R.D., Jørgensen T., Jørgensen M.E., Kähönen M., Kardia S.L.R., König W., Kooperberg C., Kriebel J., Launer L.J., Lauritzen T., Lehtimäki T., Levy D., Linksted P., Linneberg A., Liu Y., Loos R.J.F., Lupo A., Meisinger C., Melander O., Metspalu A., Mitchell P., Nauck M., Nürnberg P., Orho-Melander M., Parsa A., Pedersen O., Peters A., Peters U., Polasek O., Porteous D., Probst-Hensch N.M., Psaty B.M., Qi L., Raitakari O.T., Reiner A.P., Rettig R., Ridker P.M., Rivadeneira F., Rossouw J.E., Schmidt F., Siscovick D., Soranzo N., Strauch K., Toniolo D., Turner S.T., Uitterlinden A.G., Ulivi S., Velayutham D., Völker U., Völzke H., Waldenberger M., Wang J.J., Weir D.R., Witte D., Kuivaniemi H., Fox C.S., Franceschini N., Goessling W., Köttgen A., Chu A.Y. SOS2 and ACP1 loci identified through large-scale exome chip analysis regulate kidney development and function. J. Am. Soc. Nephrol. 2017;28:981–994. doi: 10.1681/ASN.2016020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hinkes B., Wiggins R.C., Gbadegesin R., Vlangos C.N., Seelow D., Nürnberg G., Garg P., Verma R., Chaib H., Hoskins B.E., Ashraf S., Becker C., Hennies H.C., Goyal M., Wharram B.L., Schachter A.D., Mudumana S., Drummond I., Kerjaschki D., Waldherr R., Dietrich A., Ozaltin F., Bakkaloglu A., Cleper R., Basel-Vanagaite L., Pohl M., Griebel M., Tsygin A.N., Soylu A., Müller D., Sorli C.S., Bunney T.D., Katan M., Liu J., Attanasio M., O'Toole J.F., Hasselbacher K., Mucha B., Otto E.A., Airik R., Kispert A., Kelley G.G., Smrcka A.V., Gudermann T., Holzman L.B., Nürnberg P., Hildebrandt F. Positional cloning uncovers mutations in PLCE1 responsible for a nephrotic syndrome variant that may be reversible. Nat. Genet. 2006;38:1397–1405. doi: 10.1038/ng1918. [DOI] [PubMed] [Google Scholar]
- 67.Gee H.Y., Sadowski C.E., Aggarwal P.K., Porath J.D., Yakulov T.A., Schueler M., Lovric S., Ashraf S., Braun D.A., Halbritter J., Fang H., Airik R., Vega-Warner V., Jee Cho K., Chan T.A., Morris L.G.T., Ffrench-Constant C., Allen N., McNeill H., Büscher R., Kyrieleis H., Wallot M., Gaspert A., Kistler T., Milford D.V., Saleem M.A., Keng W.T., Alexander S.I., Valentini R.P., Licht C., Teh J.C., Bogdanovic R., Koziell A., Bierzynska A., Soliman N.A., Otto E.A., Lifton R.P., Holzman L.B., Sibinga N.E.S., Walz G., Tufro A., Hildebrandt F. FAT1 mutations cause a glomerulotubular nephropathy. Nat. Commun. 2016;7 doi: 10.1038/ncomms10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharma K.R., Heckler K., Stoll S.J., Hillebrands J.L., Kynast K., Herpel E., Porubsky S., Elger M., Hadaschik B., Bieback K., Hammes H.P., Nawroth P.P., Kroll J. ELMO1 protects renal structure and ultrafiltration in kidney development and under diabetic conditions. Sci. Rep. 2016;6 doi: 10.1038/srep37172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Elmonem M.A., Khalil R., Khodaparast L., Khodaparast L., Arcolino F.O., Morgan J., Pastore A., Tylzanowski P., Ny A., Lowe M., De Witte P.A., Baelde H.J., Van Den Heuvel L.P., Levtchenko E. Cystinosis (ctns) zebrafish mutant shows pronephric glomerular and tubular dysfunction. Sci. Rep. 2017;7 doi: 10.1038/srep42583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilson P.D. Epithelial cell polarity and disease. Am. J. Physiol. Ren. Physiol. 1997;272:434–442. doi: 10.1152/ajprenal.1997.272.4.f434. [DOI] [PubMed] [Google Scholar]
- 71.White R.M., Sessa A., Burke C., Bowman T., LeBlanc J., Ceol C., Bourque C., Dovey M., Goessling W., Burns C.E., Zon L.I. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2:183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Phane Berghmans S., Murphey R.D., Wienholds E., Neuberg D., Kutok J.L., Fletcher C.D.M., Morris J.P., Liu T.X., Schulte-Merker S., Kanki J.P., Plasterk R., Zon L.I., Look A.T. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc. Natl. Acad. Sci. USA. 2005;102:407–412. doi: 10.1073/pnas.0406252102. www.pnas.orgcgidoi10.1073pnas.0406252102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mensah L., Ferguson J.L., Shive H.R. Genotypic and phenotypic variables affect meiotic cell cycle progression, tumor ploidy, and cancer-associated mortality in a brca2-mutant zebrafish model. JAMA Oncol. 2019;2019:1–15. doi: 10.1155/2019/9218251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Patton E.E., Zon L.I. Taking human cancer genes to the fish: a transgenic model of melanoma in zebrafish. Zebrafish. 2005;1:363–368. doi: 10.1089/zeb.2005.1.363. [DOI] [PubMed] [Google Scholar]
- 75.Patton E.E., Widlund H.R., Kutok J.L., Kopani K.R., Amatruda James F., Murphey R.D., Berghmans S., Mayhall E.A., Traver D., Fletcher C.D.M., Aster J.C., Granter S.R., Look A.T., Lee C., Fisher D.E., Zon L.I. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr. Biol. 2005;15:249–254. doi: 10.1016/j.cub.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 76.Langenau D.M., Keefe M.D., Storer N.Y., Guyon J.R., Kutok J.L., Le X., Goessling W., Neuberg D.S., Kunkel L.M., Zon L.I. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 2007;21:1382–1395. doi: 10.1101/gad.1545007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ignatius M.S., Hayes M.N., Moore F.E., Tang Q., Garcia S.P., Blackburn P.R., Baxi K., Wang L., Jin A., Ramakrishnan A., Reeder S., Chen Y., Nielsen G.P., Chen E.Y., Hasserjian R.P., Tirode F., Ekker S.C., Langenau D.M. tp53 deficiency causes a wide tumor spectrum and increases embryonal rhabdomyosarcoma metastasis in zebrafish. Elife. 2018;7 doi: 10.7554/eLife.37202.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Santoriello C., Deflorian G., Pezzimenti F., Kawakami K., Lanfrancone L., Di Fagagna F.D.A., Mione M. Expression of H-RASV12 in a zebrafish model of Costello syndrome causes cellular senescence in adult proliferating cells. DMM Dis. Model. Mech. 2009;2:56–67. doi: 10.1242/dmm.001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Park S.W., Davison J.M., Rhee J., Hruban R.H., Maitra A., Leach S.D. Oncogenic KRAS induces progenitor cell expansion and malignant transformation in zebrafish exocrine pancreas. Gastroenterology. 2008;134:2080–2090. doi: 10.1053/j.gastro.2008.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anelli V., Villefranc J.A., Chhangawala S., Martinez-Mcfaline R., Riva E., Nguyen A., Verma A., Bareja R., Chen Z., Scognamiglio T., Elemento O., Houvras Y. Oncogenic BRAF disrupts thyroid morphogenesis and function via twist expression. Elife. 2017;6 doi: 10.7554/eLife.20728.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Langenau D.M., Traver D., Ferrando A.A., Kutok J.L., Aster J.C., Kanki J.P., Lin S., Prochownik E., Trede N.S., Zon L.I., Look A.T. Myc-induced T cell leukemia in transgenic zebrafish. Science. 2003;299:887–890. doi: 10.1126/science.1080280. [DOI] [PubMed] [Google Scholar]
- 82.Onnebo S.M., Condron M.M., McPhee D.O., Lieschke G.J., Ward A.C. Hematopoietic perturbation in zebrafish expressing a tel-jak2a fusion. Exp. Hematol. 2005;33:182–188. doi: 10.1016/j.exphem.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 83.Onnebo S.M.N., Rasighaemi P., Kumar J., Liongue C., Ward A.C. Alternative TEL-JAK2 fusions associated with T-cell acute lymphoblastic leukemia and atypical chronic myelogenous leukemia dissected in zebrafish. Haematologica. 2012;97:1895–1903. doi: 10.3324/haematol.2012.064659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhuravleva J., Paggetti J., Martin L., Hammann A., Solary E., Bastie J.N., Delva L. MOZ/TIF2-induced acute myeloid leukaemia in transgenic fish. Br. J. Haematol. 2008;143:378–382. doi: 10.1111/j.1365-2141.2008.07362.x. [DOI] [PubMed] [Google Scholar]
- 85.Konantz M., Schürch C., Hanns P., Müller J.S., Sauteur L., Lengerke C. Modeling hematopoietic disorders in zebrafish. DMM Dis. Model. Mech. 2019;12 doi: 10.1242/dmm.040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brownlie A., Donovan A., Pratt S.J., Paw B.H., Oates A.C., Brugnara C., Witkowska H.E., Sassa S., Zon L.I. Positional cloning of the zebrafish sauternes gene: a model for congenital sideroblastic anaemia. Nat. Genet. 1998;20:244–250. doi: 10.1038/3049. http://genetics.nature.com [DOI] [PubMed] [Google Scholar]
- 87.Wang H., Long Q., Marty S.D., Sassa S., Lin S. A zebrafish model for hepatoerythropoietic porphyria. Nat. Genet. 1998;20:239–243. doi: 10.1038/3041. https://www.nature.com/articles/ng1198_239 [DOI] [PubMed] [Google Scholar]
- 88.Childs S., Weinstein B.M., Mohideen M.-A.P., Donohue S., Bonkovsky H., Fishman M.C. Zebrafish dracula encodes ferrochelatase and its mutation provides a model for erythropoietic protoporphyria. Curr. Biol. 2000;10:1001–1004. doi: 10.1016/s0960-9822(00)00653-9. [DOI] [PubMed] [Google Scholar]
- 89.Zhang Y., Ear J., Yang Z., Morimoto K., Zhang B., Lin S. Defects of protein production in erythroid cells revealed in a zebrafish diamond-blackfan anemia model for mutation in RPS19. Cell Death Dis. 2014;5:e1352. doi: 10.1038/cddis.2014.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Donovan Adriana, Brownlie Alison, Zhou Yi, Shepard Jennifer, Pratt Stephen J., Moynihan John, Paw Barry H., Anna Drejer, Barut Bruce, Zapata Agustin, Law Terence C., Brugnara Carlo, Lux Samuel E., Pinkus Geraldine S., Pinkus Jack L., Kingsley Paul D., James Palis, Fleming Mark D., Andrews Nancy C., Zon Leonard I. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature. 2000;403:776–781. doi: 10.1038/35001596. www.nature.com [DOI] [PubMed] [Google Scholar]
- 91.Dooley K., Zon L.I. Zebrafish: a model system for the study of human disease. Curr. Opin. Genet. Dev. 2000;10:252–256. doi: 10.1016/S0959-437X(00)00074-5. [DOI] [PubMed] [Google Scholar]
- 92.Chan F.-Y., Robinson J., Brownlie A., Shivdasani R.A., Donovan A., Brugnara C., Kim J., Lau B.-C., Witkowska H.E., Zon L.I. Characterization of adult a-and b-globin genes in the zebrafish. Blood. 1997;89:688–700. http://ashpublications.org/blood/article-pdf/89/2/688/1642219/688.pdf [PubMed] [Google Scholar]
- 93.Paw B.H. Cloning of the zebrafish retsina blood mutation: a genetic model for dyserythropoiesis and erythroid cytokinesis, Blood Cells. Mol. Dis. 2001;27:62–64. doi: 10.1006/bcmd.2000.0354. [DOI] [PubMed] [Google Scholar]
- 94.Balci T.B., Prykhozhij S.V., Teh E.M., Da’as S.I., Mcbride E., Liwski R., Chute I.C., Leger D., Lewis S.M., Berman J.N. A transgenic zebrafish model expressing KIT-D816V recapitulates features of aggressive systemic mastocytosis. Br. J. Haematol. 2014;167:48–61. doi: 10.1111/bjh.12999. [DOI] [PubMed] [Google Scholar]
- 95.Jones R.A., Feng Y., Worth A.J., Thrasher A.J., Burns S.O., Martin P. Modelling of human Wiskott-Aldrich syndrome protein mutants in zebrafish larvae using in vivo live imaging. J. Cell Sci. 2013;126:4077–4084. doi: 10.1242/jcs.128728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Deng Q., Yoo S.K., Cavnar P.J., Green J.M., Huttenlocher A. Dual roles for Rac2 in neutrophil motility and active retention in zebrafish hematopoietic tissue. Dev. Cell. 2011;21:735–745. doi: 10.1016/j.devcel.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Johnson N.M., Farr G.H., Maves L. The HDAC inhibitor TSA ameliorates a zebrafish model of duchenne muscular dystrophy. PLoS Curr. 2013;5 doi: 10.1371/currents.md.8273cf41db10e2d15dd3ab827cb4b027. ecurrents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Van Epps H.A., Yim C.M., Hurley J.B., Brockerhoff S.E. Investigations of photoreceptor synaptic transmission and light adaptation in the zebrafish visual mutant nrc. Invest. Ophthalmol. Vis. Sci. 2001;42:868–874. [PubMed] [Google Scholar]
- 99.Langova V., Vales K., Horka P., Horacek J. The role of zebrafish and laboratory rodents in schizophrenia research. Front. Psychiatr. 2020;11:703. doi: 10.3389/fpsyt.2020.00703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gerlai R., Lahav M., Guo S., Rosenthal A. Drinks like a fish: zebra fish (Danio rerio) as a behavior genetic model to study alcohol effects. Pharmacol. Biochem. Behav. 2000;67:773–782. doi: 10.1016/S0091-3057(00)00422-6. [DOI] [PubMed] [Google Scholar]
- 101.Meshalkina D.A., Kizlyk M.N., Kysil E.V., Collier A.D., Echevarria D.J., Abreu M.S., Barcellos L.J.G., Song C., Warnick J.E., Kyzar E.J., Kalueff A.V. Zebrafish models of autism spectrum disorder. Exp. Neurol. 2018;299:207–216. doi: 10.1016/j.expneurol.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 102.Fonseka T.M., Wen X.Y., Foster J.A., Kennedy S.H. Zebrafish models of major depressive disorders. J. Neurosci. Res. 2016;94:3–14. doi: 10.1002/jnr.23639. [DOI] [PubMed] [Google Scholar]
- 103.Aceto J., Nourizadeh-Lillabadi R., Bradamante S., Maier J.A., Alestrom P., van Loon J.J.W.A., Muller M. Effects of microgravity simulation on zebrafish transcriptomes and bone physiology—exposure starting at 5 days post fertilization. Npj Microgravity. 2016;2 doi: 10.1038/npjmgrav.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aceto J., Nourizadeh-Lillabadi R., Marée R., Dardenne N., Jeanray N., Wehenkel L., Aleström P., Van Loon J.J.W.A., Muller M. Zebrafish bone and general physiology are differently affected by hormones or changes in gravity. PLoS One. 2015;10 doi: 10.1371/journal.pone.0126928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Edsall S.C., Franz-Odendaal T.A. An assessment of the long-term effects of simulated microgravity on cranial neural crest cells in zebrafish embryos with a focus on the adult skeleton. PLoS One. 2014;9 doi: 10.1371/journal.pone.0089296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stewart A.M., Braubach O., Spitsbergen J., Gerlai R., Kalueff A.V. Zebrafish models for translational neuroscience research: from tank to bedside. Trends Neurosci. 2014;37:264–278. doi: 10.1016/j.tins.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kalueff A.V., Stewart A.M., Gerlai R. Zebrafish as an emerging model for studying complex brain disorders. Trends Pharmacol. Sci. 2014;35:63–75. doi: 10.1016/j.tips.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lu J., Peatman E., Tang H., Lewis J., Liu Z. Profiling of gene duplication patterns of sequenced teleost genomes: evidence for rapid lineage-specific genome expansion mediated by recent tandem duplications. BMC Genom. 2012;13:246–256. doi: 10.1186/1471-2164-13-246. http://www.biomedcentral.com/1471-2164/13/246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nasiadka Andrzej, Clark Matthew D. Zebrafish breeding in the laboratory environment. ILAR J. 2012;53:161–168. doi: 10.1093/ilar.53.2.161. [DOI] [PubMed] [Google Scholar]
- 110.Tonelli F., Bek J.W., Besio R., De Clercq A., Leoni L., Salmon P., Coucke P.J., Willaert A., Forlino A. Zebrafish: a resourceful vertebrate model to investigate skeletal disorders. Front. Endocrinol. 2020;11:489. doi: 10.3389/fendo.2020.00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lleras-Forero L., Winkler C., Schulte-Merker S. Zebrafish and medaka as models for biomedical research of bone diseases. Dev. Biol. 2020;457:191–205. doi: 10.1016/j.ydbio.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 112.Witten P.E., Hall B.K. Teleost skeletal plasticity: modulation, adaptation, and remodelling. Copeia. 2015;103:727–739. doi: 10.1643/CG-14-140. [DOI] [Google Scholar]
- 113.Giardoglou P., Beis D. On zebrafish disease models and matters of the heart. Biomedicines. 2019;7:15. doi: 10.3390/biomedicines7010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gut P., Reischauer S., Stainier Y.R., Arnaout R. Little fish, big data: zebrafish as a model for cardiovascular and metabolic disease. Physiol. Rev. 2017;97:889–938. doi: 10.1152/physrev.00038.2016.-The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ling D., Chen H., Chan G., Lee S.M.Y. Quantitative measurements of zebrafish heartrate and heart rate variability: a survey between 1990–2020, Comput. Biol. Med. 2022;142 doi: 10.1016/j.compbiomed.2021.105045. [DOI] [PubMed] [Google Scholar]
- 116.Cirio M.C., de Caestecker M.P., Hukriede N.A. Zebrafish models of kidney damage and repair. Curr. Pathobiol. Rep. 2015;3:163–170. doi: 10.1007/s40139-015-0080-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kobar K., Collett K., Prykhozhij S.V., Berman J.N. Zebrafish cancer predisposition models. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.660069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Carradice D., Lieschke G.J., Hall E. Zebrafish in hematology: sushi or science? Blood. 2008;111:3331–3342. doi: 10.1182/blood-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Richardson R., Tracey-White D., Webster A., Moosajee M. The zebrafish eye—a paradigm for investigating human ocular genetics. Eye. 2016;31:68–86. doi: 10.1038/eye.2016.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cavodeassi F., Wilson S.W. Looking to the future of zebrafish as a model to understand the genetic basis of eye disease. Hum. Genet. 2019;138:993–1000. doi: 10.1007/s00439-019-02055-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Young R.M., Hawkins T.A., Cavodeassi F., Stickney H.L., Schwarz Q., Lawrence L.M., Wierzbicki C., Cheng B.Y.L., Luo J., Ambrosio E.M., Klosner A., Sealy I.M., Rowell J., Trivedi C.A., Bianco I.H., Allende M.L., Busch-Nentwich E.M., Gestri G., Wilson S.W. Compensatory growth renders Tcf7l1a dispensable for eye formation despite its requirement in eye field specification. Elife. 2019;8 doi: 10.7554/ELIFE.40093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sakai C., Ijaz S., Hoffman E.J. Zebrafish models of neurodevelopmental disorders: past, present, and future. Front. Mol. Neurosci. 2018;11:294. doi: 10.3389/fnmol.2018.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Forn-Cuní G., Meijer A.H., Varela M. Zebrafish in inflammasome research. Cells. 2019;8:901. doi: 10.3390/cells8080901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Copyright permission obtained for used figures and images accordingly.