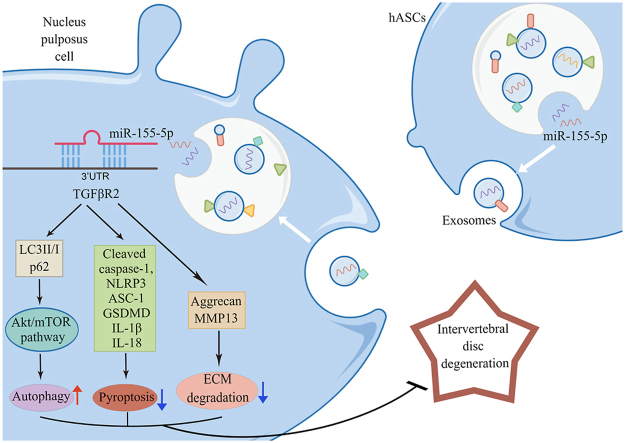
Abstract
Background
Intervertebral disc degeneration (IDD) is a complex chronic disease involving nucleus pulposus cells (NPCs) senescence, apoptosis, autophagy and extracellular matrix (ECM) degradation. In this study, we aimed to investigate the role of human adipose tissue stem cells (hASCs)-derived exosomal miR-155-5p targeting TGFβR2 in IDD and the mechanisms involved. Then miRNA sequencing was performed, and hASCs-derived Exo (hASCs-Exo) was extracted and characterized.
Methods
First, NPCs were treated with different concentrations of LPS. Then miRNA sequencing was performed, and hASCs-Exo was extracted and characterized. NPCs were treated with PBS or autophagy inhibitor 3-MA. NPCs were transfected with miR-155-5p mimic, si-TGFβR2 and negative control. Cell viability, apoptosis, ROS, caspase-1+PI, pyroptosis markers, inflammatory cytokines, autophagy markers, Aggrecan, MMP13, and Akt/mTOR pathway-related factors were measured. Bioinformatics prediction and dual-luciferase were performed to verify the binding sites of miR-155-5p to TGFβR2. Finally, we validated the role of hASCs-derived exosomal miR-155-5p on IDD in vivo.
Results
LPS promoted pyroptosis of NPCs, and inhibited autophagy and ECM synthesis. MiR-155-5p was characterized as an inflammation-related miRNA in NPCs. HASCs-derived exosomal miR-155-5p inhibited pyroptosis of NPCs and promoted autophagy and ECM synthesis. After bioinformatics prediction and verification, it was found that miR-155-5p targeted TGFβR2. Moreover, miR-155-5p targeted TGFβR2 to promote autophagy and inhibit pyroptosis in NPCs. In vivo experiments revealed that hASCs-derived exosomal miR-155-5p alleviated IDD in rats.
Conclusions
HASCs-derived exosomal miR-155-5p alleviated IDD by targeting TGFβR2 to promote autophagy and reduce pyroptosis. Our study may provide a new therapeutic target for IDD.
Translational Potential of this Article
HASCs-derived exosomal miR-155-5p is expected to be a biomarker for clinical treatment of IDD. Our study may provide a new therapeutic target for IDD.
Keywords: hASCs-exo, miR-155-5p, TGFβR2, Autophagy, Pyroptosis, IDD
Graphical abstract
1. Introduction
Intervertebral disc degeneration (IDD) is an usual degenerative disease that can cause the nucleus pulposus (NP) to collapse or protrude, leading to radiculopathy [1], which is the main cause of low back pain [2]. IDD severely reduces patients' quality of life and brings massive economic pressure on their families [3]. Currently, the main way of curing IDD is surgery, such as discectomy and vertebral body fusion [4]. Molecular biological therapy is the basic strategy for IDD treatment, but there is still needing to find novel markers for the treatment of IDD.
More and more evidence shows that the dysregulation of the autophagy pathway highlights many skeletal pathologies that affect the spine, joints, and growth plate cartilage [[5], [6], [7]]. Autophagy limited the activation of inflammasomes and alleviated the secretion of inflammatory cytokines [8]. Ge Y et al. assumed that autophagy regulated inflammasome activation and affected the outcome of cell pyroptosis [9]. Pyroptosis is a newly discovered form of programmed cell death, which is also involved in NP cells (NPCs) during IDD progression [10]. Zhang J et al. reported that mesenchymal stem cells (MSCs)-derived exosomes (Exos) inhibited pyroptosis of NPCs [11]. Zhou Y et al. revealed that morin attenuated pyroptosis of NPCs to ameliorate IDD [12]. However, the comprehensive roles of pyroptosis in IDD have not been fully established.
Stem cell-based regeneration strategies are very promising in IDD [13]. It was reported that SOD2 and catalase could improve the pathological condition of IDD by modifying human adipose tissue stem cells (hASCs) [14]. Exo is a tiny membrane vesicle with a 40–100 nm diameter containing protein and nucleic acid, which can be released from many cell types into the extracellular space and can be non-immunogenic and capable of crossing the blood–brain barrier [15]. The multiple applications of hASCs-derived Exo (hASCs-Exo) have been considered as a new cell-free treatment strategy in the field of regenerative and aesthetic medicine [16]. hASCs-Exo could increase the migration and proliferation of NPCs and inhibit inflammatory activity, suggesting that it could be used to treat IDD [17]. The paracrine function of MSCs works by secreting soluble factors and releasing extracellular vesicles such as Exo and microvesicles. They contain many microRNAs (miRNAs) transferred from their original cells to target cells. In many animal disease models, single extracellular vesicles are responsible for the therapeutic effect of MSCs [18]. For example, MSCs-derived exosomal miR-21 activated the PI3K/Akt pathway by suppressing PTEN, inhibiting NPCs apoptosis and alleviating IDD [19]. In addition, it has been reported that serum miR-155-5p was notably reduced in degenerative disc disease patients, which may be a diagnostic marker for degenerative spinal diseases [20]. However, there is no research report on hASCs-derived exosomal miR-155-5p in IDD.
According to reports, activating the transforming growth factor beta (TGFβ) pathway has a good therapeutic prospect for IDD, and over-activation of the TGFβ signaling pathway may lead to the progression of IDD [21]. TGFβ receptor 2 (TGFβR2) is a cell membrane protein involved in TGFβ receptor signaling and an antigen associated with vascular endothelial cell proliferation. Previous studies have shown that TGFβR2 has been identified as one of the miR-155-5p targets [22,23]. We found through bioinformatics prediction that miR-155-5p has a binding site with TGFβR2. Therefore, we want to explore the study of miR-155-5p and TGFβR2 in IDD. In this study, we tried to explore the role of hASCs-derived exosomal miR-155-5p targeting TGFβR2 in IDD through in vivo and in vitro experiments and bioinformatics predictions and whether the mechanisms involved are related to autophagy and pyroptosis. Our research may provide new treatment strategies and targets for IDD.
2. Materials and methods
2.1. Cell culture and treatment
Human intervertebral disc NPCs were provided by Procell (CP-H097, Wuhan, China). To find the appropriate concentration of lipopolysaccharide (LPS) for NPCs, we set up different concentration gradients of LPS (0, 0.1, 1, 10, 100, 200, 500, and 1000 μg/mL) to treat NPCs. The experiment was divided into the Control group (NPCs) and the LPS group.
HASCs were purchased from Procell (CP-H202, Wuhan, China), and hASCs-Exo were extracted and characterized. NPCs were treated with LPS (500 μg/mL) for 24 h. NPCs were added with an equal volume of PBS. HASCs-Exo (Exo secreted by hASCs without any treatment), NC-Exo (Exo secreted by hASCs transfected with miR-155-5p NC plasmids), and miR-155-5p-Exo (Exo secreted by hASCs transfected with miR-155-5p mimic plasmids) were incubated with LPS-induced NPCs for 24 h, and the concentration of Exo in each treatment was 100 μg/mL [24]. The experimental groups were the PBS group, hASCs-Exo group, NC-Exo group, and miR-155-5p-Exo group. Cells were then collected for further experiments.
NPCs were treated with LPS (500 μg/mL) for 24 h. Before adding hASCs-Exo, NPCs were pretreated with PBS or 10 mM 3-MA (autophagy inhibitor) for 2 h [25], corresponding to the LPS group and the 3-MA group, respectively. Cells in the 3-MA group were added with equal volumes of PBS. HASCs-Exo (Exo secreted by hASCs without any treatment), NC-Exo (Exo secreted by hASCs transfected with miR-155-5p NC plasmids), and miR-155-5p-Exo (Exo secreted by hASCs transfected with miR-155-5p mimic plasmids) were incubated with LPS-induced NPCs for 24 h, and the concentration of Exo in each treatment was 100 μg/mL [24], corresponding to the 3-MA + hASCs-Exo group, 3-MA + NC-Exo group and 3-MA + miR-155-5p-Exo group. The cells were then collected for further experiments.
NPCs were treated with LPS (500 μg/mL) for 24 h, and then NPCs were pretreated with 10 mM 3-MA for 2 h [25]. Then NPCs were added NC-Exo or miR-155-5p-Exo and incubated with NPCs for 24 h. On this basis, the cells were seeded in a 6-well plate for 24 h, and the cells were transfected with recombinant lentivirus si-TGFβR2 or negative control (si-NC) [26]. The experimental groups were the 3-MA + NC-Exo + si-NC group, 3-MA + NC-Exo + si-TGFβR2 group, 3-MA + miR-155-5p-Exo + si-NC group, and 3-MA + miR-155-5p-Exo + si-TGFβR2 group.
2.2. Cell counting Kit-8 (CCK-8) assay
Cells were digested and seeded in a 96-well plate (5 × 103), and corresponding intervention treatments were grouped according to the experiment, and cultured for 24 h. After incubating for a while, 10 μL of CCK8 working solution was added. Cells were incubated for 4 h in a 37 °C, 5% CO2 incubator. Elx800 (BioTek, USA) was applied to measure the absorbance at 450 nm. Cell proliferation ability was detected at 24, 48, and 72 h.
2.3. TUNEL
TUNEL Apoptosis Detection Kit (#40306ES50, YEASEN, Shanghai) was utilized to detect NPCs apoptosis. The slices were fixed with 4% paraformaldehyde. 100 μL Proteinase K working solution and 100 μL 1 × Equilibration Buffer were added. TdT incubation Buffer was prepared. Most of the 100 μL 1 × Equilibration Buffer was washed with absorbent paper around the Equilibration area, and then 50 μL TdT incubation Buffer was added. DAPI working solution was stained. The tablets were sealed with buffered glycerin and observed under the fluorescence microscope.
2.4. Reactive oxygen species (ROS)
ROS detection kit (S0033S, Beyotime) was performed to measure ROS levels. 1 × 106 cells were taken and centrifuged at 400 g for 5 min. DCFH-DA was diluted with the serum-free medium at 1:1000. The diluted DCFH-DA was added to suspend the cells. The cell precipitate was collected by centrifugation at 400g and washed with serum-free medium twice for flow cytometry (A00-1-1102, Beckman) detection.
2.5. Cell pyroptosis
The caspase-1+PI expression was detected using the caspase-1 pyroptosis assay kit (AB219935, Abcam). First, the dye solution was prepared, and 300 μL Washing buffer was added with 2 μL FAM-YVAD-FMK stock solution. 300 μL dye solution was added. They were centrifuged at 1000 rpm for 5 min, and washed twice with 200 μL Washing buffer. The PI working solution was dyed. Flow cytometry was used to observe and detect cell pyroptosis.
2.6. Flow cytometry
Cells were re-suspended with 200 μL PBS, and primary antibodies CD73 (12-0739-42, eBioscience), CD44 (12-0441-82, eBioscience), CD45 (12-0459-42, eBioscience), and CD34 (12-0349-42, eBioscience) were added. 200 μL PBS was added to suspend cells. Cells were filtered by nylon mesh, and CD73, CD44, CD45, and CD34 expressions were measured by flow cytometry.
2.7. Immunofluorescence (IF)
Pyroptosis markers (Cleaved caspase-1, NLRP3, ASC-1, and GSDMD) expressions were tested by IF. The slices were fixed with 4% paraformaldehyde and permeabilized with 0.5% TritonX-100. Cells were incubated with primary antibodies Cleaved caspase-1 (PA5-38099, 1:50, ThermoFisher), NLRP3 (19771-1-AP, 1:50, Proteintech), ASC-1 (ab283684, 1:50, Abcam), and GSDMD (20770-1-AP, 1:50, Proteintech) at 4 °C overnight. Then secondary antibody was incubated, and the DAPI working solution (Wellbio, China) was stained. Tablets were sealed with buffered glycerol and observed under the fluorescence microscope.
2.8. Enzyme linked immunosorbent assay (ELISA)
IL-1β and IL-18 levels were detected using IL-1β (CSB-E08053h or CSB-E08055r, CUSABIO) and IL-18 (CSB-E07450h or CSB-E04610r, CUSABIO) quantitative ELISA kits according to the instructions. Bio-Tek microplate reader (MB-530, Heales, China) was applied to test the concentrations of IL-1β and IL-18 by forming a standard curve with the provided values.
2.9. Western blot
Total protein was extracted from cells and tissues using RIPA lysis buffer (#P0013B, Beyotime). SDS-PAGE loading buffer (#MB2479, Meilunbio) was mixed with adsorbing protein onto the PVDF membrane by gel electrophoresis. Aggrecan (13880-1-AP, 1:1000, Proteintech), MMP13 (18165-1-AP, 1:800, Proteintech), TGFβR2 (ab186838, 1:1000, Abcam), Akt/mTOR pathway-related factors Akt (10176-1-AP, 1:1000, Proteintech), p-Akt (ab38449, 1:1000, Abcam), mTOR (66888-1-Ig, 1:10,000, Proteintech) and p-mTOR (67778-1-Ig, 1:10,000, Proteintech), and pyroptosis markers Cleaved caspase-1 (4199, 1:1000, CST), NLRP3 (19771-1-AP, 1:800, Proteintech), ASC-1 (PA5-90403, 1:1000, Invitrogen), and GSDMD (PA5-30823, 1:5000, Invitrogen) and autophagy markers LC3 (14600-1-AP, 1:2500, Proteintech), p62 (66184-1-Ig, 1:4000, Proteintech) primary antibodies, and β-actin (66009-1-IG, 1:1000, Proteintech) were incubated overnight at 4 °C. Then secondary antibodies were incubated. ECL chromogenic exposure was performed, protein bands were detected, and β-actin acted as the internal reference.
2.10. miRNA sequencing
Cells in the Control group and LPS group were selected for miRNA sequencing. First, UMI tools were used to extract and label the sequencing data in the UMI sequence, and then the sequencing data was removed by cutadapt to remove the adapter sequence, and the length was filtered. Double-end merging of the sequences was performed by bbmerge to verify, avoid sequencing errors, obtain clean data, and generate quality control reports for raw and clean data by fastqc. Then comparative analysis was conducted, and a differentially expressed miRNAs heatmap was drawn. Volcano plots showed changes in miRNA expression between the two groups.
2.11. Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted by the Trizol method and reverse transcribed into cDNAs by cDNA reverse transcription kit (#CW2569, Beijing ComWin Biotech, China). Ultra SYBR Mixture (#CW2601, Beijing ComWin Biotech, China) was utilized to detect gene expression on quantitative PCR (QuantStudio1, Thermo, USA). Using U6 or GAPDH as an internal reference gene, gene expression was calculated by the 2-ΔΔCt method. Table 1 shows the primer sequences.
Table 1.
The sequence used in this study.
| Name | Sequences |
|---|---|
| hsa-miR-92b-5p-F | AGGGACGGGACGCGGTGCAGTG |
| hsa-miR-92b-5p-R | GCTGTCAACGATACGCTACGTA |
| hsa-miR-376a-3p-F | ATCATAGAGGAAAATCCACGT |
| hsa-miR-376a-3p-R | GCTGTCAACGATACGCTACGTA |
| hsa-miR-154-3p-F | AATCATACACGGTTGACCTATT |
| hsa-miR-154-3p-R | GCTGTCAACGATACGCTACGTA |
| hsa-miR-155-3p-F | TTAATGCTAATCGTGATAGGGGTT |
| hsa-miR-155-3p-R | GCTGTCAACGATACGCTACGTA |
| hsa-miR-7-5p-F | TGGAAGACTAGTGATTTTGTTGTT |
| hsa-miR-7-5p-R | GCTGTCAACGATACGCTACGTA |
| hsa-miR-3614-5p-F | CCACTTGGATCTGAAGGCTGCCC |
| hsa-miR-3614-5p-R | GCTGTCAACGATACGCTACGTA |
| hsa-miR-4773-F | CAGAACAGGAGCATAGAAAGGC |
| hsa-miR-4773-R | GCTGTCAACGATACGCTACGTA |
| hsa-miR-155-5p-F | TTAATGCTAATCGTGATAGGGGTT |
| hsa-miR-155-5p-R | GCTGTCAACGATACGCTACGTA |
| hsa-miR-12136-F | GAAAAAGTCATGGAGGCC |
| hsa-miR-12136-R | GCTGTCAACGATACGCTACGTA |
| hsa-miR-132-5p-F | ACCGTGGCTTTCGATTGTTACT |
| hsa-miR-132-5p | GCTGTCAACGATACGCTACGTA |
| U6-F | CTCGCTTCGGCAGCACA |
| U6-R | AACGCTTCACGAATTTGCGT |
| TGFβR2-F | CGTGAAGAACGACCTAACC |
| TGFβR2-R | CCACCTGCCCACTGTTAG |
| GAPDH-F | ACAGCCTCAAGATCATCAGC |
| GAPDH-R | GGTCATGAGTCCTTCCACGAT |
2.12. Oil-red O staining
The slices were washed with PBS 2–3 times. The slices were fixed with 4% paraformaldehyde. 100 μL Oil-red O staining solution was added. 70% alcohol was added. Hematoxylin was stained for 1–3 min. The slices were rinsed with distilled water and returned to blue with PBS. Tablets were sealed with buffered glycerin and observed under the microscope.
2.13. Alizarin red staining
2 mL 4% neutral formaldehyde solution was added and fixed for 30 min. A neutral formaldehyde solution was sucked up. 1 mL alizarin red dye was added for 3–5 min. Alizarin red dye was sucked up and observed by microscope.
2.14. Alcian blue staining
As mentioned above, 2 mL 4% neutral formaldehyde solution was added and fixed for 30 min. A neutral formaldehyde solution was sucked up. 1 mL alcian blue dye was added for 30 min. Alcian blue dye was sucked up and observed by the microscope.
2.15. Extraction and identification of hASCs-Exo
Exos were extracted from cell culture supernatant using an exo extraction kit (EXOQ5A-1, SBI) according to instructions. HASCs were cultured in DMEM, and the supernatant was collected at 48–72 h to extract exos for subsequent detection. The extracted exos were re-suspended in PBS in an appropriate volume. Particle size distribution of hASCs-Exos was measured by nanoparticle tracking analysis (NTA). The morphology and size of hASCs-Exos were displayed by transmission electron microscopy (TEM). Western blot analysis was performed using CD63 (25682-1-AP, 1:500, Proteintech), CD81(66866-1-Ig, 1:3000, Proteintech), TSG101 (14497-1-AP, 1:1000, Proteintech), GM130 (11308-1-AP, 1:5000, Proteintech) and Calnexin (10427-1-AP, 1:8000, Proteintech) and exosome uptake experiment was performed according to the previous study [27].
2.16. Bioinformatics prediction
MiR-155-5p and TGFβR2 binding sites were predicted by Starbase. Three IDD-related datasets GSE15227, GSE23130 and GSE67567 were selected. GSE15227 and GSE23130 were used for differences in TGFβR2 between high and low grades in IDD. GSE67567 was used for differences in TGFβR2 between IDD and Normal patients. All data were normalized. Difference analysis R language limma package was applied. The up-regulated TGFBR2 was screened in all three datasets. TGFβR2_PPI was plotted, and GO and KEGG enrichment analyses were performed. Venn diagram and TGFβR2_miRNA_network diagram were drawn.
2.17. Dual-luciferase reporter gene assay
To verify the binding of TGFβR2 to miR-155-5p, a wild-type (WT) or mutant (MUT) TGFβR2 fragment was constructed and inserted into the pmirGLO vector (Promega). Lipofectamine 3000 reagents (Invitrogen) were performed to transfect the recombinant vector, and mimics NC and miR-155-5p mimics were transferred into cells. Luciferase activity was measured by Nano-Glo dual luciferase reporter method.
2.18. Animals model
This study was approved by the animal experiment ethics committee of China-Japan Friendship Hospital and conducted in strict accordance with the national institutes of health guidelines for the care and use of experimental animals (zryhyy61-21-05-01). The treatment of animals during the experiment conforms to the standards of ‘‘Guiding Opinions on Being kind to Experimental Animals’’ by the Ministry of Science and Technology in 2006.
First, 30 male Sprague–Dawley rats (8 weeks old) were randomly divided into the Control group (n = 6) and the Model group (n = 24). The rats in the Model group were anesthetized with isoflurane and a needle (27G) was applied to completely pierce entire annulus fibrosus (Co7/8) via tail skin, as confirmed by experimental X-ray photographs. The needle was left in the intervertebral disc for about 1 min, and the length of the needle was about 5 mm [11,23,25]. In the Control group, needles (27G) were not used to pierce the entire annulus fibrosus (Co7/8), and other operations were the same as in the Model group. After seven days of modeling, the Model group was randomly divided into the IDD, hASCs-Exo, NC-Exo, and miR-155-5p-Exo groups. IDD group no longer does processing. The hASCs-Exo, NC-Exo, and miR-155-5p-Exo groups were treated as follows: the rats were anesthetized and made a small incision on the left side to expose the previously punctured intervertebral disc [28]. About 1.5 × 106 purified hASCs-Exo (Exo secreted by hASCs without any treatment), NC-Exo (Exo secreted by hASCs transfected with miR-155-5p NC plasmids), and miR-155-5p-Exo (Exo secreted by hASCs transfected with miR-155-5p mimic plasmids) were slowly injected into the punctured intervertebral disc using no. 33 needle (Hamilton, Bonaduz, Switzerland) and connected with a microliter syringe (Hamilton) [19,22,24]. Repeated injections were given after 4 weeks (the intervertebral disc was collected 4 weeks later after the second injection) or once every two weeks for 2 months or once a week until 9 weeks. After the intervertebral disc tissues were collected for fixation and decalcification. NP tissues were collected and stored at −80 °C for qRT-PCR and Western blot detection.
2.19. HE staining
The sections were baked at 60 °C for 12 h, dewaxed to water, and stained with hematoxylin for 1 min. The eosin was stained for 0.5 min. Gradient alcohol (95–100%) was used for dehydration. Tablets were sealed with neutral gum and observed under the microscope.
2.20. SafraninO-Fast Green Staining
The intervertebral disc tissue sections were dewaxed to water. After dyeing with fast green dye, they were dyed with SafraninO. The sections were blow-dried with a hair dryer, placed in xylene, and sealed with neutral gum. Then intervertebral disc tissue was observed and photographed under the microscope to evaluate the disc injury.
2.21. Statistical analysis
GraphPad Prism 8.0 software was used for statistical analysis. The measurement data were expressed by Mean ± Standard deviation (SD). Student's t-test was performed between two groups. One-way analysis of variance (ANOVA) was performed for multiple groups. P < 0.05 indicated statistically significant.
3. Results
3.1. LPS promoted pyroptosis of NPCs, inhibited autophagy and ECM synthesis
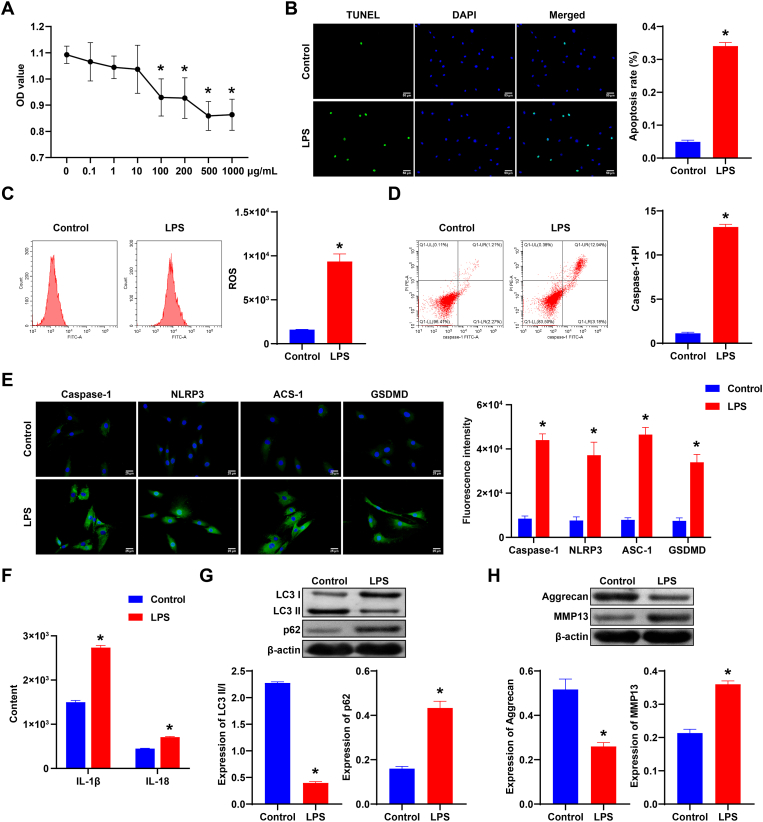
First, to find the appropriate concentration of LPS, we set different concentration gradients of LPS (0, 0.1, 1, 10, 100, 200, 500, and 1000 μg/mL) to treat NPCs. Cell viability decreased notably with increased LPS concentration (Fig. 1A). LPS treated with 500 μg/mL or 1000 μg/mL showed higher inhibition of NPCs activity than other concentrations, suggesting that 500 μg/mL LPS may be the maximum effective dose to inhibit NPCs activity. Therefore, 500 μg/mL LPS was selected for subsequent experiments. Next, we detected the apoptosis of NPCs. NPCs apoptosis of the LPS group was notably increased compared to the Control group (Fig. 1B). ROS and caspase-1+PI levels were increased after LPS treatment (Fig. 1C and D). In addition, Caspase-1, NLRP3, ASC-1, and GSDMD expressions were increased after LPS treatment (Fig. 1E). ELISA results showed that IL-1β and IL-18 levels were increased after LPS treatment (Fig. 1F). After LPS treatment, p62 expression was elevated, while LC3II/I expression was reduced (Fig. 1G). MMP13 expression was promoted, while Aggrecan expression was inhibited in the LPS group (Fig. 1H). Our results indicated LPS promoted pyroptosis of NPCs, and inhibited autophagy and ECM synthesis.
Fig. 1.
LPS promoted pyroptosis of NPCs, and inhibited autophagy and ECM synthesis. A. CCK-8 was applied to measure NPCs viability. ∗, P < 0.05. B. The apoptosis of NPCs. C. ROS expression in each group. D. caspase-1+PI expression. E. Caspase-1, NLRP3, ASC-1, and GSDMD expressions were detected by IF in each group. F. The expression of IL-1β and IL-18 in NPCs was measured by ELISA. G. Western blot was applied to measure autophagy markers (LC3, p62) expressions in each group. H. Western blot detection of Aggrecan and MMP13 expressions in NPCs of each group. ∗, P < 0.05 compared with the Control group.
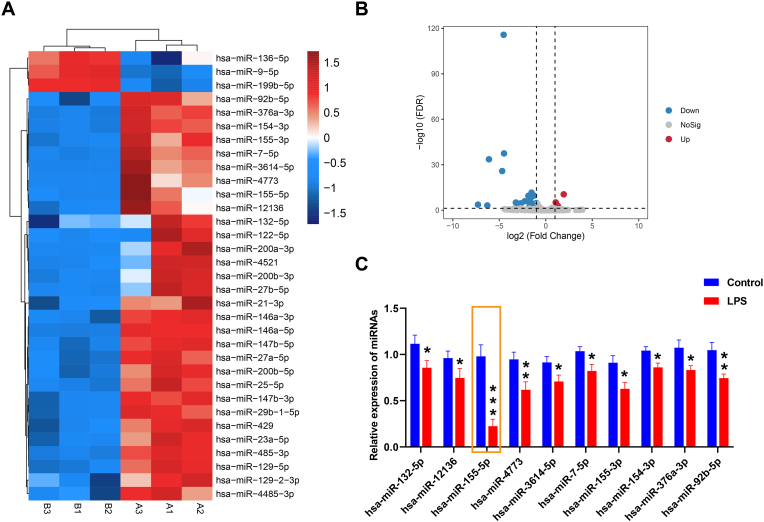
3.2. Differentially expressed miRNAs were identified in LPS-induced NPCs
Next, miRNA sequencing analysis was performed on NPCs. Fig. 2A showed the heatmap of differentially expressed miRNAs in the Control group and the LPS group. The volcanic map showed changes in miRNA expression between the two groups (Fig. 2B). Finally, qRT-PCR was performed to detect the top ten differentially expressed miRNA expressions in two groups. Among them, the expressions of miR-155-5p, miR-4773, and miR-92b-5p were significantly decreased in the LPS group compared with the Control group (P < 0.01, P < 0.001, Fig. 2C), so miR-155-5p, miR-4773, and miR-92b-5p were selected for subsequent validation.
Fig. 2.
Differentially expressed miRNAs were identified in LPS-induced NPCs. A. Heat map of differentially expressed miRNAs. B. Volcanic map of changes in miRNA expression between the two groups. C. qRT-PCR detection of top ten differentially expressed miRNAs expressions in the Control group and LPS group. ∗, P < 0.05 compared with the Control group.
3.3. Extraction and identification of hASCs-Exos
Next, we purchased hASCs and identified them. As shown in Fig. S1A, hASCs were shuttle type. After induction and differentiation, Oil Red, Alizarin Red and Alcian blue staining were all positive. Flow cytometry revealed that CD73 and CD44 were positive, and CD45 and CD34 were negative (Fig. S1B). This suggested hASCs identification was successful. Next, we extracted and characterized hASCs-Exos. As shown in Fig. S2A, the particle size of exos was about 100–200 nm. TEM results showed that exos were oval vesicles (Fig. S2B). CD63, CD81 and TSG101 were expressed in hASCs and hASCs-Exos, and the expression was more obvious in hASCs-Exos. GM130 and Calnexin were not obvious in hASCs-Exos (Fig. S2C). These results indicated that hASCs-Exos had been successfully extracted and identified.
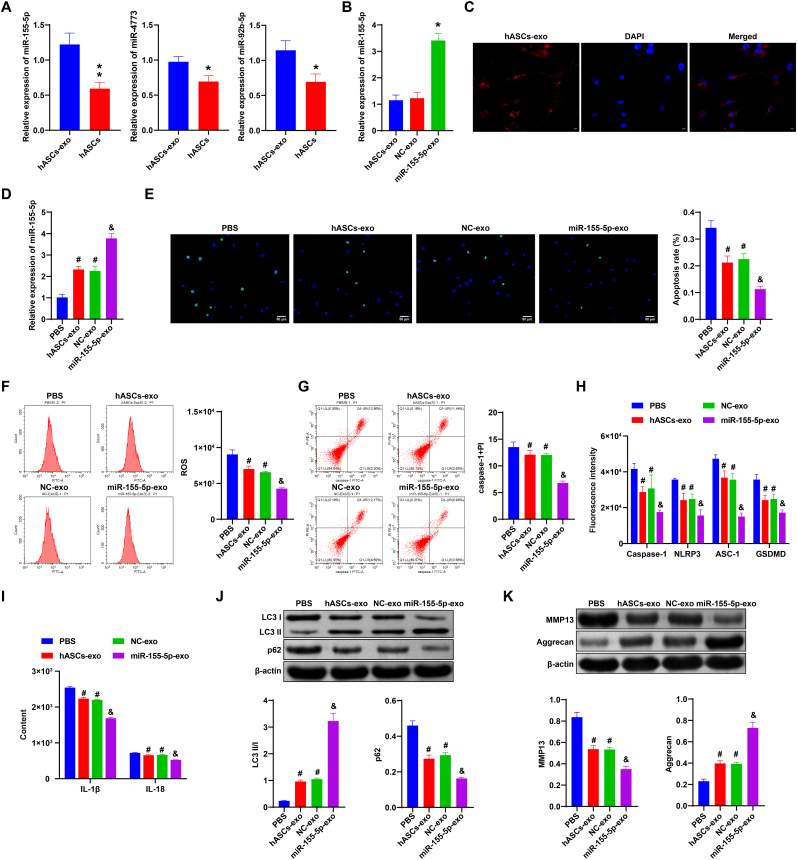
3.4. hASCs-derived exosomal miR-155-5p inhibited pyroptosis of NPCs and promoted autophagy and ECM synthesis
Then we verified the expression of miR-155-5p, miR-4773 and miR-92b-5p in hASCS-Exos and hASCs. As shown in Fig. 3A, the expression of miR-155-5p, miR-4773, and miR-92b-5p in the hASCs group was significantly decreased compared with the hASCs-exo group (P < 0.05, P < 0.01). Among them, the decrease of miR-155-5p was more obvious. Therefore, miR-155-5p was selected for subsequent study. In addition, the transfection efficiency of the miR-155-5p mimic was determined by qRT-PCR. Fig. 3B showed that hASCs plasmids were successfully transfected. The results of the exosome uptake experiment showed that the presence of exos in NPCs was observed (Fig. 3C). In addition, qRT-PCR was performed to verify whether NPCs affected miR-155-5p expression through uptaking exos. Fig. 3D showed that miR-155-5p expression was elevated in hASCs-Exo and miR-155-5p-Exo groups compared to PBS and NC-Exo groups, respectively. Compared with the PBS group and NC-Exo group, NPCs apoptosis, ROS and caspase-1+PI levels were repressed in the hASCs-Exo group and miR-155-5p-Exo group, respectively (Fig. 3E–G).
Fig. 3.
hASCs-derived exosomal miR-155-5p inhibited pyroptosis of NPCs and promoted autophagy and ECM synthesis. A. qRT-PCR detection of miR-155-5p level in hASCs and hASCs-Exos. B. Transfection efficiency of miR-155-5p mimic. C. Exosome uptake experiment. D. miR-155-5p expression was detected by qRT-PCR. E. Apoptosis of NPCs in each group was detected by TUNEL. F and G. Flow cytometry detection of ROS and caspase-1+PI expressions. H. The expression of Caspase-1, NLRP3, ASC-1, and GSDMD was detected by IF. I. The expressions of IL-1β and IL-18 in NPCs were tested by ELISA. J and K. Western blot was applied to measure the expression of autophagy markers (LC3, p62), Aggrecan and MMP13. ∗, P < 0.05 compared with the PBS group. #, P < 0.05 compared with the NC-Exo group.
Next, IF was performed to determine the expression of pyroptosis markers Caspase-1, NLRP3, ASC-1, and GSDMD in each group. Pyroptosis markers expressions were decreased in the hASCs-Exo group and miR-155-5p-Exo group compared to the PBS group and NC-Exo group, respectively (Fig. 3H, Fig. S3). ELISA results showed that IL-1β and IL-18 levels were decreased in the hASCs-Exo group and miR-155-5p-Exo group compared with the PBS group and NC-Exo group, respectively (Fig. 3I). In addition, we detected the expression of autophagy markers (LC3, p62), Aggrecan, and MMP13 in each group. p62 expression was reduced, and LC3II/I expression was elevated in the hASCs-Exo group and miR-155-5p-Exo group in contrast with the PBS group and NC-Exo group, respectively. Compared with the PBS group and NC-Exo group, MMP13 expression in the hASCs-Exo group and miR-155-5p-Exo group decreased, while Aggrecan expression increased (Fig. 3J–K). These results revealed hASCs-derived exosomal miR-155-5p inhibited pyroptosis of NPCs and promoted autophagy and ECM synthesis.
3.5. hASCs-derived exosomal miR-155-5p promoted autophagy and inhibited pyroptosis in NPCs
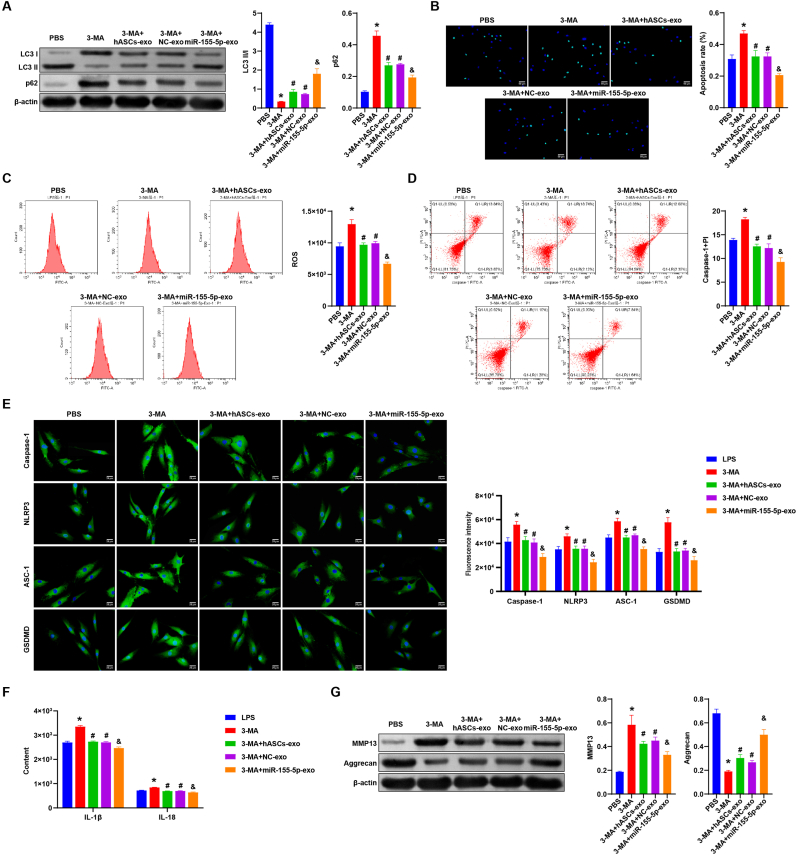
Then we added the autophagy inhibitor 3-MA to further study the effect of hASCs-derived exosomal miR-155-5p on NPCs pyroptosis and ECM degeneration. Firstly, Western blot was performed to measure the expression of autophagy markers (LC3, p62) in each group. Compared with the LPS group, p62 expression was elevated, and LC3II/I expression was repressed in the 3-MA group. However, p62 expression was inhibited, and LC3II/I expression was facilitated in the 3-MA + hASCs-Exo group and 3-MA + NC-Exo group. Similarly, p62 expression was suppressed, and LC3II/I expression was promoted in the 3-MA + miR-155-5p-Exo group compared to the 3-MA + NC-Exo group (Fig. 4A). NPCs apoptosis, ROS and caspase-1+PI levels were increased in the 3-MA group and decreased in the 3-MA + hASCs-Exo group and 3-MA + NC-Exo group than the LPS group. Compared with the 3-MA + NC-Exo group, NPCs apoptosis, ROS and caspase-1+PI levels in the 3-MA + miR-155-5p-Exo group were also reduced (Fig. 4B–D).
Fig. 4.
hASCs-derived exosomal miR-155-5p promoted autophagy and inhibited pyroptosis in NPCs. A. Western blot detection of the protein expression of autophagy markers (LC3, p62) in each group. B. Apoptosis of NPCs in each group was detected by TUNEL. C and D. Flow cytometry detection of ROS and caspase-1+PI levels in each group. E. The expression of Caspase-1, NLRP3, ASC-1, and GSDMD was detected by IF in all groups. F. The level of IL-1β and IL-18 in NPCs was detected by ELISA. G. Western blot detection of Aggrecan and MMP13 expressions in NPCs of each group. ∗, P < 0.05 compared with the LPS group. #, P < 0.05 compared with the 3-MA group. @ P < 0.05 compared with the 3-MA + NC-Exo group.
Next, IF was utilized to test the expression of pyroptosis markers Caspase-1, NLRP3, ASC-1, and GSDMD in each group. The results showed that pyroptosis markers levels were increased in the 3-MA group and decreased in the 3-MA + hASCs-Exo and 3-MA + NC-Exo group compared to the LPS group. Pyroptosis markers levels were also decreased in the 3-MA + miR-155-5p-Exo group than the 3-MA + NC-Exo group (Fig. 4E). ELISA results revealed that IL-1β and IL-18 levels were promoted in the 3-MA group and decreased in the 3-MA + hASCs-Exo and the 3-MA + NC-Exo group compared to the LPS group. IL-1β and IL-18 levels were also decreased in the 3-MA + miR-155-5p-Exo group in contrast with the 3-MA + NC-Exo group (Fig. 4F). Finally, the expression of Aggrecan and MMP13 was detected by Western blot. Compared with the LPS group, MMP13 expression was promoted, and Aggrecan expression was repressed in the 3-MA group. However, MMP13 expression was reduced, and Aggrecan expression was elevated in the 3-MA + hASCs-Exo group and 3-MA + NC-Exo group. MMP13 expression in the 3-MA + miR-155-5p-Exo group was also decreased, and Aggrecan expression was also increased than the 3-MA + NC-Exo group (Fig. 4G). Our results further illustrated that hASCs-derived exosomal miR-155-5p promoted autophagy and inhibited pyroptosis in NPCs.
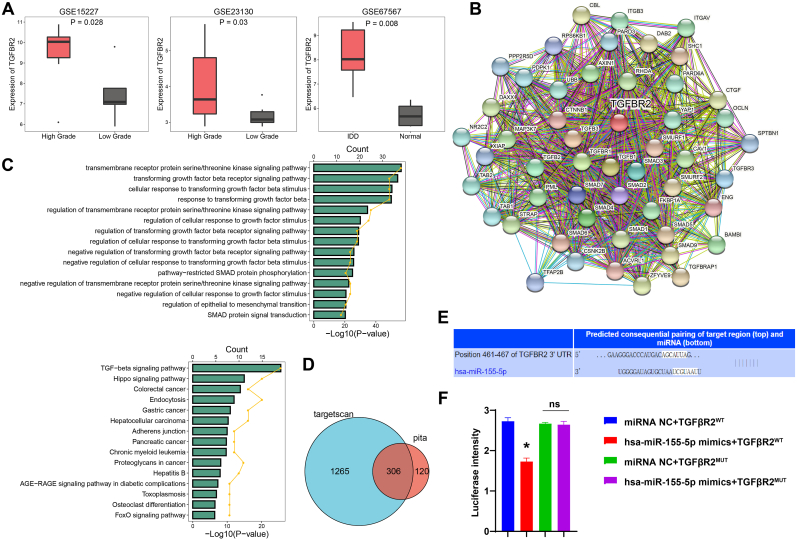
3.6. miR-155-5p targeted TGFβR2
Studies have shown that TGFβR2 was identified as one of the miR-155-5p targets [22,23]. Boxplots showed differences in TGFβR2 between high and low grades in IDD and between IDD and Normal patients (Fig. 5A). Fig. 5B showed the TGFβR2_PPI map. In addition, GO showed that it was mainly enriched in the transmembrane receptor protein serine/threonine kinase signaling pathway and transforming growth factor beta (TGFβ) receptor signaling pathway. KEGG showed that it was mainly enriched in the TGF-beta signaling pathway and hippo signaling pathway (Fig. 5C). Venn diagram showed that TGFβR2 targeted different miRNAs (Fig. 5D). Next, through bioinformatics prediction, we found miR-155-5p had binding sites with TGFβR2 (Fig. 5E). In addition, dual-luciferase assay verified the targeting relationship of has-miR-155-5p and TGFβR2 (Fig. 5F).
Fig. 5.
miR-155-5p targeted TGFβR2. A. Boxplots showed differences in TGFβR2 between high and low grades in IDD and between IDD and Normal patients. B. TGFβR2_PPI figure. C. GO and KEGG enrichment analysis. D. Venn diagram showed TGFβR2 targeting miRNAs. E. Bioinformatics prediction of the binding sites of miR-155-5p and TGFβR2. F. Dual-luciferase detection of the targeting relationship between miR-155-5p and TGFβR2. ∗, P < 0.05.
3.7. miR-155-5p targeted TGFβR2 to promote autophagy and inhibit pyroptosis in NPCs
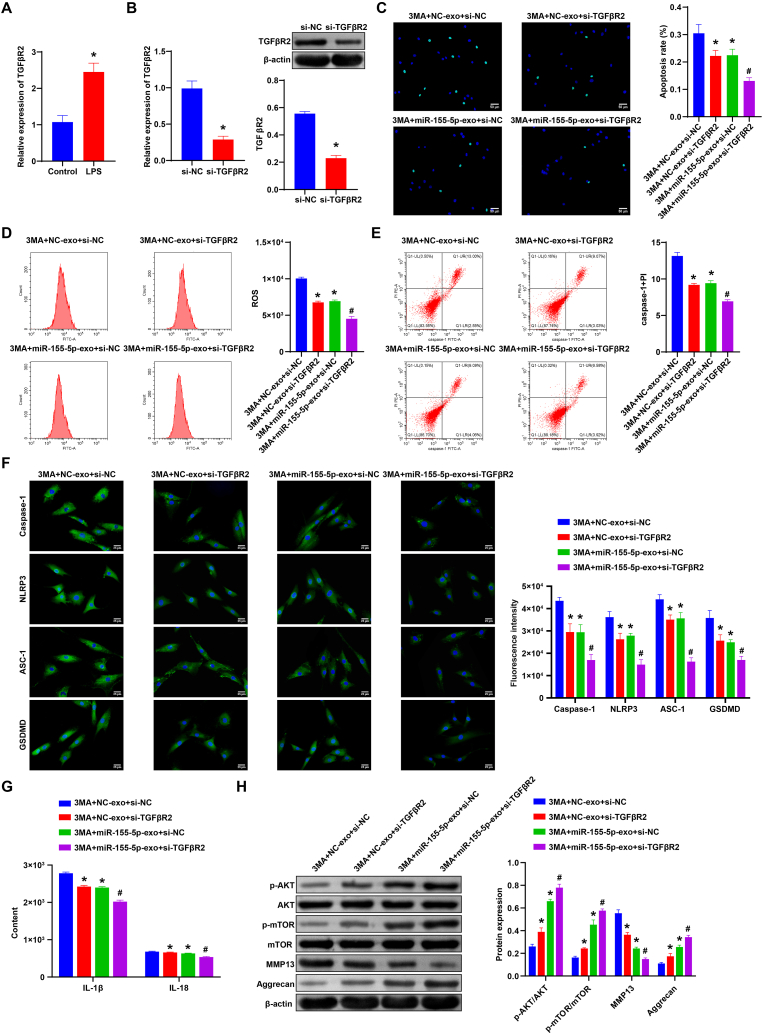
To study the role of miR-155-5p and TGFβR2 in IDD, miR-155-5p-Exo was added, and TGFβR2 was knocked down. First, TGFβR2 expression was detected in the Control group and LPS group. After LPS treatment, TGFβR2 expression was significantly increased (Fig. 6A). qRT-PCR and western blot verification revealed that si-TGFβR2 was successfully transfected (Fig. 6B). Compared with the NC-Exo + si-NC group and miR-155-5p-Exo + si-NC group, NPCs apoptosis was decreased, and ROS and caspase-1+PI levels were decreased in the NC-Exo + si-TGFβR2 group and miR-155-5p-Exo + si-TGFβR2 group, respectively (Fig. 6C–E). Caspase-1, NLRP3, ASC-1, and GSDMD expression were decreased in the NC-Exo + si-TGFβR2 group and miR-155-5p-Exo + si-TGFβR2 group than the NC-Exo + si-NC group and miR-155-5p-Exo + si-NC group, respectively (Fig. 6F). ELISA results showed that IL-1β and IL-18 levels were decreased in the NC-Exo + si-TGFβR2 group and miR-155-5p-Exo + si-TGFβR2 group compared to the NC-Exo + si-NC group and miR-155-5p-Exo + si-NC group, respectively (Fig. 6G).
Fig. 6.
miR-155-5p targeted TGFβR2 to promote autophagy and inhibit pyroptosis in NPCs. A. qRT-PCR was operated to detect TGFβR2 expression in the Control group and LPS group. B. qRT-PCR and western blot were used to verify the transfection efficiency of si TGFβR2. C. Apoptosis of NPCs in each group was detected by TUNEL. D and E. Flow cytometry was applied to detect the ROS and caspase-1+PI levels in each group. F. The expression of Caspase-1, NLRP3, ASC-1, and GSDMD was detected by IF. G. The expression of IL-1β and IL-18 in NPCs was detected by ELISA. H. Western blot was used to measure the expression of Akt/mTOR pathway-related factors (Akt, p-Akt, mTOR, and p-mTOR), Aggrecan and MMP13 in NPCs of each group. ∗, P < 0.05 compared with the NC-Exo + si-NC group. #, P < 0.05 compared with the miR-155-5p-Exo + si-NC group.
In addition, we detected the expression of Aggrecan, MMP13, and Akt/mTOR pathway-related factors (Akt, p-Akt, mTOR, and p-mTOR). Compared with the NC-Exo + si-NC group and miR-155-5p-Exo + si-NC group, the expression of MMP13 in the NC-Exo + si-TGFβR2 group and miR-155-5p-Exo + si-TGFβR2 group decreased, while the expression of Aggrecan increased. There was no significant difference in the expression of Akt and mTOR. The expressions of p-Akt and p-mTOR were increased in the NC-Exo + si-TGFβR2 group and miR-155-5p-Exo + si-TGFβR2 group compared to the NC-Exo + si-NC group and miR-155-5p-Exo + si-NC group, respectively (Fig. 6H). Our results suggested miR-155-5p targeted TGFβR2 to promote autophagy and inhibit pyroptosis in NPCs.
3.8. hASCs-derived exosomal miR-155-5p alleviated IDD in rats
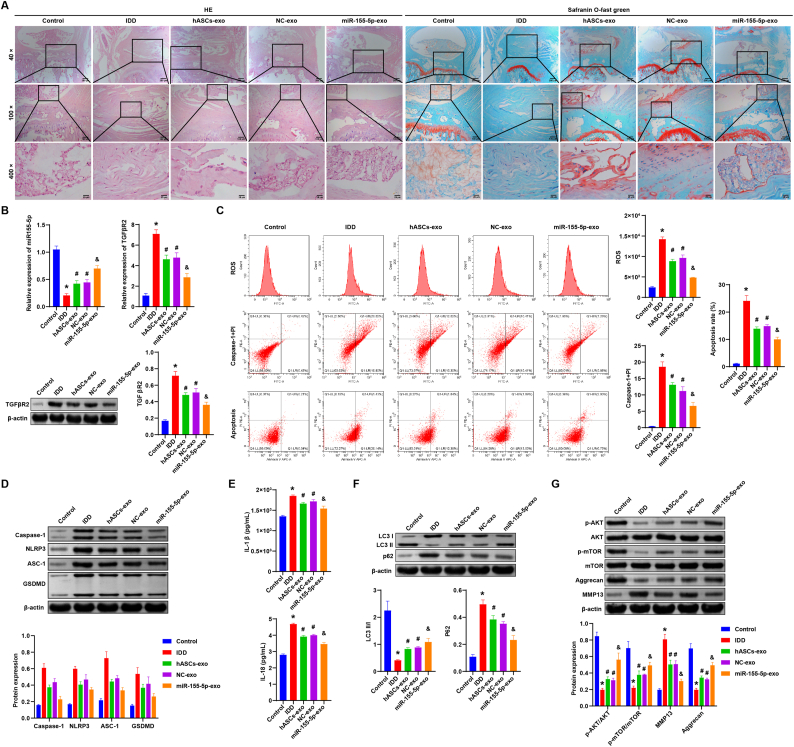
Finally, we verified the role of hASCs-derived exosomal miR-155-5p on IDD in vivo. First, HE staining showed that the morphology and structure of the intervertebral disc in the Control group were normal without lesions. The structure of the intervertebral disc in the IDD group showed degeneration. NP disappeared and was replaced by disordered fibrous tissues, while the orderly arrangement of annulus fibrosus was disrupted, and part of the endplate disappeared. Compared with the NC-Exo group, the miR-155-5p-Exo group had a more significant effect on alleviating the symptoms of intervertebral disc tissue degeneration (Fig. 7A). SafraninO-Fast Green Staining showed that there was no damage in the Control group, and the SafraninO coloration was uniform. The cartilage damage in the IDD group was severe, and the coloration of SafraninO was uneven. Compared with the NC-Exo group, the miR-155-5p-Exo group was more effective in relieving the symptoms of intervertebral disc injury (Fig. 7A). In addition, miR-155-5p expression in the IDD group was decreased, and TGFβR2 expression was increased than the Control group. Compared with the NC-Exo group, the miR-155-5p-Exo group had elevated miR-155-5p expression and decreased TGFβR2 expression (Fig. 7B). Flow cytometry results showed IDD group had higher caspase-1+PI and ROS levels and increased apoptosis. Compared with the NC-Exo group, caspase-1+PI and ROS levels decreased by 60% and 50% in the miR-155-5p-Exo group, and apoptosis decreased by 67% (Fig. 7C).
Fig. 7.
hASCs-derived exosomal miR-155-5p alleviated IDD in rats. A. HE staining was applied to detect the degeneration of intervertebral disc tissue. Safranin O-Fast Green Staining assessment of intervertebral disc injury. B. qRT-PCR detection of miR-155-5p and TGFβR2 expression in NP tissue of each group. Western blot detection of the expression of TGFβR2 in NP tissue of each group. C. Flow cytometry detection of ROS and caspase-1 levels and the apoptosis of NPCs in each group. D. Western blot detection of the protein expressions of pyroptosis markers (Caspase-1, NLRP3, ASC-1, and GSDMD) in the NP tissue of each group. E. The level of IL-1β and IL-18 in NP tissue was tested by ELISA. F. The expression of autophagy markers (LC3, p62) in the NP tissue was detected by Western blot. G. Western blot detection of the expression of Akt/mTOR pathway-related factors (Akt, p-Akt, mTOR, p-mTOR), Aggrecan and MMP13 in the NP tissue of each group. ∗, P < 0.05 compared with the Control group. #, P < 0.05 compared with the IDD group. @ P < 0.05 compared with the NC-Exo group. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Next, Western blot was applied to measure the expression of pyroptosis markers (Caspase-1, NLRP3, ASC-1, and GSDMD) in each group. The levels of pyroptosis markers increased in the IDD group in contrast with the Control group. Compared with the NC-Exo group, pyroptosis markers Caspase-1, NLRP3, ASC-1, and GSDMD levels were decreased by 52%, 74%, 69%, and 63% in the miR-155-5p-Exo group (Fig. 7D). ELISA results showed the expressions of IL-1β and IL-18 in the IDD group were increased than Control group. Compared with the NC-Exo group, the expression of IL-1β and IL-18 was reduced by 90% and 86% in the miR-155-5p-Exo group (Fig. 7E).
Additionally, we examined the expression of autophagy markers (LC3, p62). As shown in Fig. 7F, compared with the Control group, the LC3II/I expression was inhibited and the p62 expression was promoted in the IDD group. LC3II/I expression was increased by 123% and p62 expression was repressed by 66% in the miR-155-5p-Exo group than the NC-Exo group. There was no significant difference in the expression of Akt and mTOR among the groups. The expressions of p-Akt and p-mTOR were repressed in the IDD group differently from the Control group. Compared with the NC-Exo group, the expression of p-Akt and p-mTOR was increased by 180% and 129% in the miR-155-5p-Exo group. Aggrecan expression was repressed, and MMP13 expression was more elevated in the IDD group than in the control group. Aggrecan expression was promoted by 152%, and MMP13 expression was suppressed by 59% in the miR-155-5p-Exo group in contrast with the NC-Exo group (Fig. 7G). Our study demonstrated that hASCs-derived exosomal miR-155-5p alleviated IDD in rats.
4. Discussion
IDD is a complex chronic disease involving NPCs senescence, apoptosis, autophagy and ECM degradation [29]. In this study, we found through in vivo and in vitro verification that hASCs-derived exosomal miR-155-5p alleviated IDD by targeting TGFβR2 to promote autophagy and reduce pyroptosis. This is the first time we have reported the study of hASCs-derived exosomal miR-155-5p in IDD.
Autophagy is the process of cells self-digesting and recovering damaged components under stress conditions, especially under nutrient deprivation conditions, and is an important mechanism for cell survival [30]. There are autophagy markers in the intervertebral disc tissue, and intervertebral disc cells can respond to cellular stressors in vitro to regulate autophagy [31]. Most of the results of previous studies have shown that by promoting autophagy of NPCs inhibited apoptosis and thus improved IDD [23,[32], [33], [34]]. There are also a small number of previous studies indicating that inhibiting autophagy improved IDD [6,29]. Moreover, studies have reported that ROS relied on NLRP3 and PYCARD to induce pyroptosis of NPCs, and increased NFE2L2, Nrf2 and autophagy of NPCs, and these two factors could alleviate pyroptosis [35]. This indicates that in the IDD model, there is an interaction between NPCs autophagy and pyroptosis. During IDD process, multiple inflammatory factors and catabolic factors accumulate in NP tissue, subsequently affects the viability and function of NPCs [36]. Currently, many studies have reported that NPCs death was a crucial event during IDD formation. Various types of programmed cell death contributed to the pathogenesis of IDD, including cell apoptosis, pyroptosis, and necroptosis [37]. Studies have shown that LPS could induce inflammatory factor production and ECM degeneration in NPCs [38]. Zhang J et al. developed an IDD model using LPS-treated NPCs and evaluated the therapeutic ability of MSCs and related exos on NPCs pyroptosis [11]. Their results suggested that MSCs treatment inhibited LPS-induced pyroptosis in NPCs. Liao Z et al. also treated NPCs with LPS to establish the cell pyroptosis model and investigated the effects of autophagy intervention on NPCs in vitro [10]. Their results suggested that LPS induced the pyroptotic phenotype in NPCs in a dose-dependent manner and impaired autophagy accelerated the pyroptotic NPCs death induced by LPS. Therefore, we also performed LPS as a stimulating factor in our in vitro experiments of IDD. In the present study, we found that LPS induced pyroptosis of NPCs and inhibited autophagy and ECM synthesis. Our data showed that NPCs pyroptosis played a vital role in the IDD formation. In addition, hASCs-based treatment is considered to be a promising way for treating NP degeneration [39]. Exos is reported to inhibit NPCs apoptosis and attenuate IDD by activating the AKT and autophagy pathways [40]. After we successfully extracted and identified hASCs-Exo, we found that hASCs-derived exosomal miR-155-5p inhibited NPCs pyroptosis and promoted autophagy and ECM synthesis.
Previous research has shown that overexpression of miR-155-5p alleviated lung injury in ventilator-induced lung injury mice following reduced apoptosis and increased autophagy. Bone-marrow-derived MSCs might protect lung tissues from mechanical ventilation-induced injury, inhibit lung inflammation, and promote autophagy by up-regulating miR-155-5p [41]. Chen LJ et al. reported that miR-155-5p was downregulated in BEAS-2B cells induced by LPS. Overexpression of miR-155-5p increased cell proliferation and suppressed inflammatory factors levels and cell apoptosis [42]. Furthermore, lower expression of miR-155-5p was observed in exosomes from LPS-stimulated periodontal ligament stem cells compared with exosomes from normal periodontal ligament stem cells. Exos from periodontal ligament stem cells alleviated the inflammatory microenvironment through Th17/Treg/miR-155-5p/SIRT1 regulatory network [43]. Xu W et al. revealed that downregulation of miR-155-5p enhanced the anti-tumor effect of cetuximab on triple-negative breast cancer cells via inducing cell apoptosis and pyroptosis [44]. These studies suggested that miR-155-5p played an active role in diseases and reduced pyroptosis. However, miR-155 was also reported to induce pyroptosis. Li C et al. reported that miR-155 promoted macrophage pyroptosis induced by Porphyromonas gingivalis via regulating the NLRP3 inflammasome [45]. In a similar study, Wang B et al. reported that macrophage-derived miR-155-containing exosomes promoted cardiomyocyte pyroptosis [46]. These studies suggested that the pyroptosis involved in miR-155 had two sides in the disease. After adding the autophagy inhibitor 3-MA, we further illustrated that hASCs-derived exosomal miR-155-5p promoted autophagy and inhibited pyroptosis in NPCs. This indicates that hASCs-derived exosomal miR-155-5p also played an active role in IDD.
There is increasing evidence that non-coding RNA is involved in IDD. It was reported that miR-29a was demonstrated to effectively silence MMP-2 expression, repress the fibrosis process, and reverse IDD via blocking the β-catenin translocation pathway from cytoplasm to nucleus [47]. Zhu J et al. reported the lncRNA/circRNA-miRNA-mRNA ceRNA network in IDD. These determined ceRNA interaction axes may be the key targets for treating IDD, including miR-155-5p and TGFβ1 [48]. We have predicted and verified through bioinformatics that miR-155-5p targeted and regulated TGFβR2. Moreover, previous studies have shown that abnormal TGFβ activation damaged NPCs and cartilage endplate cells, which could lead to IDD [49]. Tian Y et al. reported that TGFβ controlled Galectin-3 expression in NPCs through Smad3, which may be related to IDD [50]. We found miR-155-5p targeted TGFβR2 to promote autophagy and inhibit pyroptosis in NPCs. This is the first time we reported miR-155-5p targets TGFβR2 in IDD.
In addition, there are few studies on miR-155-5p involved in AKT/mTOR pathway in diseases. Ghafouri-Fard S et al. reported that miR-155-5p had been recognized as a target of non-coding RNA activated by DNA damage in different cell lines. Furthermore, non-coding RNA activated by DNA damage also interacted with the Akt/mTOR pathway [51]. In nephroblastoma, miR-155-5p targeted IGF2 directly to inactivate the PI3K/AKT/mTOR pathway and exert tumor suppressor effects [52]. However, research on the miR-155-5p and AKT/mTOR pathway in IDD have not been published yet. We found through in vivo experiments that the expression of p-Akt and p-mTOR in the IDD group decreased. P-Akt and p-mTOR expressions increased after adding miR-155-5p-Exo. Our research showed that hASCs-derived exosomal miR-155-5p alleviated rat IDD, which involved AKT/mTOR pathway.
However, although our study suggests that miR-155-5p may target TGFβR2, how TGFβR2 affects autophagy remains unclear. In addition, how autophagy may relate to pyroptosis is unknown. Due to the limitation of time and funds, we cannot solve this problem well at present. With sufficient time and funds in the future, we will further study how TGFβR2 affects autophagy and how autophagy may relate to pyroptosis.
In conclusion, our research revealed that hASCs-derived exosomal miR-155-5p alleviated IDD by targeting TGFβR2 to promote autophagy and reduce pyroptosis. This is our first report on the study of hASCs-derived exosomal miR-155-5p in IDD. This is the first time we report that miR-155-5p targets TGFβR2 in IDD. Our research may provide new treatment strategies and targets for IDD.
Ethical statement
This study was approved by the animal experiment ethics committee of China-Japan Friendship Hospital and conducted in strict accordance with the national institutes of health guidelines for the care and use of experimental animals (zryhyy61-21-05-01). The treatment of animals during the experiment conforms to the standards of “Guiding Opinions on Being kind to Experimental Animals” issued by the Ministry of Science and Technology in 2006.
Funding
This Work is Supported by Beijing Natural Science Foundation (L212047). Research on the Mechanism of 3D Printing Degradable Spinal Interbody Fusion Cage's Osteogenic Activity.
Author contributions
Dong Chen: conceptualization, methodology, investigation, validation, project administration, writing – original draft. Xin Jiang: supervision, funding acquisition, resources, writing – review and editing. Haibo Zou: data aggregation, validation, analysis of experimental results, resources, writing – review and editing. All authors contributed to the article and approved the submitted version.
Data availability statement
The data used to support the findings of this study are available from the corresponding author upon request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank the Laboratory Animal of the China-Japan Friendship Hospital for their technical assistance technical support. The authors would like to acknowledge the FiGDRAW software. The Graphical abstract is drawn using FiGDRAW software.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jot.2023.02.004.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhao Y., Qiu C., Wang W., Peng J., Cheng X., Shangguan Y., et al. Cortistatin protects against intervertebral disc degeneration through targeting mitochondrial ROS-dependent NLRP3 inflammasome activation. Theranostics. 2020;10(15):7015–7033. doi: 10.7150/thno.45359. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shao Z., Wang B., Shi Y., Xie C., Huang C., Chen B., et al. Senolytic agent Quercetin ameliorates intervertebral disc degeneration via the Nrf2/NF-κB axis. Osteoarthritis Cartilage. 2021;29(3):413–422. doi: 10.1016/j.joca.2020.11.006. [eng] [DOI] [PubMed] [Google Scholar]
- 3.Zhang G.Z., Deng Y.J., Xie Q.Q., Ren E.H., Ma Z.J., He X.G., et al. Sirtuins and intervertebral disc degeneration: roles in inflammation, oxidative stress, and mitochondrial function. Clin Chim Acta. 2020;508:33–42. doi: 10.1016/j.cca.2020.04.016. [eng] [DOI] [PubMed] [Google Scholar]
- 4.Chang Y., Yang M., Ke S., Zhang Y., Xu G., Li Z. Effect of platelet-rich plasma on intervertebral disc degeneration in vivo and in vitro: a critical review. Oxid Med Cell Longev. 2020;2020 doi: 10.1155/2020/8893819. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madhu V., Guntur A.R., Risbud M.V. Role of autophagy in intervertebral disc and cartilage function: implications in health and disease. Matrix Biol. 2021;100-101:207–220. doi: 10.1016/j.matbio.2020.12.002. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin L.Y., Lv Z.D., Wang K., Qian L., Song X.X., Li X.F., et al. Estradiol alleviates intervertebral disc degeneration through modulating the antioxidant enzymes and inhibiting autophagy in the model of menopause rats. Oxid Med Cell Longev. 2018;2018 doi: 10.1155/2018/7890291. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y., Lin J., Chen J., Huang C., Zhang Z., Wang J., et al. Mfn2 is involved in intervertebral disc degeneration through autophagy modulation. Osteoarth Cartil. 2020;28(3):363–374. doi: 10.1016/j.joca.2019.12.009. [eng] [DOI] [PubMed] [Google Scholar]
- 8.Claude-Taupin A., Bissa B., Jia J., Gu Y., Deretic V. Role of autophagy in IL-1β export and release from cells. Semin Cell Dev Biol. 2018;83:36–41. doi: 10.1016/j.semcdb.2018.03.012. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ge Y., Chen Y., Guo C., Luo H., Fu F., Ji W., et al. Pyroptosis and intervertebral disc degeneration: mechanistic insights and therapeutic implications. J Inflamm Res. 2022;15:5857–5871. doi: 10.2147/JIR.S382069. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao Z., Li S., Liu R., Feng X., Shi Y., Wang K., et al. Autophagic degradation of gasdermin D protects against nucleus pulposus cell pyroptosis and retards intervertebral disc degeneration in vivo. Oxid Med Cell Longev. 2021;2021 doi: 10.1155/2021/5584447. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J., Zhang J., Zhang Y., Liu W., Ni W., Huang X., et al. Mesenchymal stem cells-derived exosomes ameliorate intervertebral disc degeneration through inhibiting pyroptosis. J Cell Mol Med. 2020;24(20):11742–11754. doi: 10.1111/jcmm.15784. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y., Chen Z., Yang X., Cao X., Liang Z., Ma H., et al. Morin attenuates pyroptosis of nucleus pulposus cells and ameliorates intervertebral disc degeneration via inhibition of the TXNIP/NLRP3/Caspase-1/IL-1β signaling pathway. Biochem Biophys Res Commun. 2021;559:106–112. doi: 10.1016/j.bbrc.2021.04.090. [eng] [DOI] [PubMed] [Google Scholar]
- 13.Hodgkinson T., Wignall F., Hoyland J.A., Richardson S.M. High BMPR2 expression leads to enhanced SMAD1/5/8 signalling and GDF6 responsiveness in human adipose-derived stem cells: implications for stem cell therapies for intervertebral disc degeneration. J Tissue Eng. 2020;11 doi: 10.1177/2041731420919334. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao L., Xu S.J., Liu C., Wang J., Hu B., Xu H.G. Sod2 and catalase improve pathological conditions of intervertebral disc degeneration by modifying human adipose-derived mesenchymal stem cells. Life Sci. 2021;267 doi: 10.1016/j.lfs.2020.118929. [eng] [DOI] [PubMed] [Google Scholar]
- 15.Zhang J., Li S., Li L., Li M., Guo C., Yao J., et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Dev Reprod Biol. 2015;13(1):17–24. doi: 10.1016/j.gpb.2015.02.001. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon H.H., Yang S.H., Lee J., Park B.C., Park K.Y., Jung J.Y., et al. Combination treatment with human adipose tissue stem cell-derived exosomes and fractional CO2 laser for acne scars: a 12-week prospective, double-blind, randomized, split-face study. Acta Derm Venereol. 2020;100(18) doi: 10.2340/00015555-3666. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z., Zhang L., Yang J., Huang J., Cai J., Zhang S., et al. Influence of extracellular nanovesicles derived from adipose-derived stem cells on nucleus pulposus cell from patients with intervertebral disc degeneration. Exp Ther Med. 2021;22(6):1431. doi: 10.3892/etm.2021.10866. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keshtkar S., Azarpira N., Ghahremani M.H. Mesenchymal stem cell-derived extracellular vesicles: novel frontiers in regenerative medicine. Stem Cell Res Ther. 2018;9(1):63. doi: 10.1186/s13287-018-0791-7. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng X., Zhang G., Zhang L., Hu Y., Zhang K., Sun X., et al. Mesenchymal stem cells deliver exogenous miR-21 via exosomes to inhibit nucleus pulposus cell apoptosis and reduce intervertebral disc degeneration. J Cell Mol Med. 2018;22(1):261–276. doi: 10.1111/jcmm.13316. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Divi S.N., Markova D.Z., Fang T., Guzek R., Kurd M.F., Rihn J.A., et al. Circulating miR-155-5p as a novel biomarker of lumbar degenerative disc disease. Spine (Phila Pa 1976. 2020;45(9):E499–e507. doi: 10.1097/BRS.0000000000003322. [eng] [DOI] [PubMed] [Google Scholar]
- 21.Chen S., Liu S., Ma K., Zhao L., Lin H., Shao Z. TGF-β signaling in intervertebral disc health and disease. Osteoarthritis Cartilage. 2019;27(8):1109–1117. doi: 10.1016/j.joca.2019.05.005. [eng] [DOI] [PubMed] [Google Scholar]
- 22.Xie L., Chen Z., Liu M., Huang W., Zou F., Ma X., et al. MSC-derived exosomes protect vertebral endplate chondrocytes against apoptosis and calcification via the miR-31-5p/ATF6 Axis. Mol Ther Nucleic Acids. 2020;22:601–614. doi: 10.1016/j.omtn.2020.09.026. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie L., Huang W., Fang Z., Ding F., Zou F., Ma X., et al. CircERCC2 ameliorated intervertebral disc degeneration by regulating mitophagy and apoptosis through miR-182-5p/SIRT1 axis. Cell Death Dis. 2019;10(10):751. doi: 10.1038/s41419-019-1978-2. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao Z., Luo R., Li G., Song Y., Zhan S., Zhao K., et al. Exosomes from mesenchymal stem cells modulate endoplasmic reticulum stress to protect against nucleus pulposus cell death and ameliorate intervertebral disc degeneration in vivo. Theranostics. 2019;9(14):4084–4100. doi: 10.7150/thno.33638. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang D., He X., Wang D., Peng P., Xu X., Gao B., et al. Quercetin suppresses apoptosis and attenuates intervertebral disc degeneration via the SIRT1-autophagy pathway. Front Cell Dev Biol. 2020;8 doi: 10.3389/fcell.2020.613006. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qu Y., Zhang H., Sun W., Han Y., Li S., Qu Y., et al. MicroRNA-155 promotes gastric cancer growth and invasion by negatively regulating transforming growth factor-β receptor 2. Cancer Sci. 2018;109(3):618–628. doi: 10.1111/cas.13472. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He L., Zhu W., Chen Q., Yuan Y., Wang Y., Wang J., et al. Ovarian cancer cell-secreted exosomal miR-205 promotes metastasis by inducing angiogenesis. Theranostics. 2019;9(26):8206–8220. doi: 10.7150/thno.37455. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y., Lin J., Wu X., Guo X., Sun H., Yu B., et al. Aspirin-mediated attenuation of intervertebral disc degeneration by ameliorating reactive oxygen species in vivo and in vitro. Oxid Med Cell Longev. 2019;2019 doi: 10.1155/2019/7189854. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhan S., Wang K., Xiang Q., Song Y., Li S., Liang H., et al. lncRNA HOTAIR upregulates autophagy to promote apoptosis and senescence of nucleus pulposus cells. J Cell Physiol. 2020;235(3):2195–2208. doi: 10.1002/jcp.29129. [eng] [DOI] [PubMed] [Google Scholar]
- 30.Yurube T., Ito M., Kakiuchi Y., Kuroda R., Kakutani K. Autophagy and mTOR signaling during intervertebral disc aging and degeneration. JOR Spine. 2020;3(1) doi: 10.1002/jsp2.1082. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kritschil R., Scott M., Sowa G., Vo N. Role of autophagy in intervertebral disc degeneration. J Cell Physiol 2022; 237(2): 1266-1284. [eng]. [DOI] [PMC free article] [PubMed]
- 32.Chen D., Xia D., Pan Z., Xu D., Zhou Y., Wu Y., et al. Metformin protects against apoptosis and senescence in nucleus pulposus cells and ameliorates disc degeneration in vivo. Cell Death Dis. 2016;7(10) doi: 10.1038/cddis.2016.334. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z., Shao Z., Chen S., Huang D., Peng Y., Chen S., et al. TIGAR impedes compression-induced intervertebral disc degeneration by suppressing nucleus pulposus cell apoptosis and autophagy. J Cell Physiol. 2020;235(2):1780–1794. doi: 10.1002/jcp.29097. [eng] [DOI] [PubMed] [Google Scholar]
- 34.Tang Z., Hu B., Zang F., Wang J., Zhang X., Chen H. Nrf2 drives oxidative stress-induced autophagy in nucleus pulposus cells via a Keap1/Nrf2/p62 feedback loop to protect intervertebral disc from degeneration. Cell Death Dis. 2019;10(7):510. doi: 10.1038/s41419-019-1701-3. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai Z., Liu W., He D., Wang Y., Yi W., Luo C., et al. Protective effects of autophagy and NFE2L2 on reactive oxygen species-induced pyroptosis of human nucleus pulposus cells. Aging (Albany NY) 2020;12(8):7534–7548. doi: 10.18632/aging.103109. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shang P., Tang Q., Hu Z., Huang S., Hu Y., Zhu J., et al. Procyanidin B3 alleviates intervertebral disc degeneration via interaction with the TLR4/MD-2 complex. J Cell Mol Med. 2020;24(6):3701–3711. doi: 10.1111/jcmm.15074. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao C.Q., Jiang L.S., Dai L.Y. Programmed cell death in intervertebral disc degeneration. Apoptosis. 2006;11(12):2079–2088. doi: 10.1007/s10495-006-0290-7. [eng] [DOI] [PubMed] [Google Scholar]
- 38.Guo Y., Tian L., Liu X., He Y., Chang S., Shen Y. ERRFI1 inhibits proliferation and inflammation of nucleus pulposus and is negatively regulated by miR-2355-5p in intervertebral disc degeneration. Spine (Phila Pa 1976. 2019;44(15):E873. doi: 10.1097/BRS.0000000000003011. e81. [eng] [DOI] [PubMed] [Google Scholar]
- 39.Zhou X., Wang J., Fang W., Tao Y., Zhao T., Xia K., et al. Genipin cross-linked type II collagen/chondroitin sulfate composite hydrogel-like cell delivery system induces differentiation of adipose-derived stem cells and regenerates degenerated nucleus pulposus. Acta Biomater. 2018;71:496–509. doi: 10.1016/j.actbio.2018.03.019. [eng] [DOI] [PubMed] [Google Scholar]
- 40.Luo L., Jian X., Sun H., Qin J., Wang Y., Zhang J., et al. Cartilage endplate stem cells inhibit intervertebral disc degeneration by releasing exosomes to nucleus pulposus cells to activate Akt/autophagy. Stem Cell. 2021; 39(4): 467-481 doi: 10.1002/stem.3322. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin X., Yu T., Luo J., Chen L., Liu Y., Xu J., et al. BMSCs mediates endothelial cell autophagy by upregulating miR-155-5p to alleviate ventilator-induced lung injury. J Biochem Mol Toxicol. 2022;36(7) doi: 10.1002/jbt.23060. [eng] [DOI] [PubMed] [Google Scholar]
- 42.Chen L.J., Li J.M., Zhang W.D., Liu W., Li X.Y., Ouyang B., et al. LncRNA NEAT1 activates MyD88/NF-κB pathway in bronchopneumonia through targeting miR-155-5p. Autoimmunity. 2021;54(2):104–113. doi: 10.1080/08916934.2021.1891534. [eng] [DOI] [PubMed] [Google Scholar]
- 43.Zheng Y., Dong C., Yang J., Jin Y., Zheng W., Zhou Q., et al. Exosomal microRNA-155-5p from PDLSCs regulated Th17/Treg balance by targeting sirtuin-1 in chronic periodontitis. J Cell Physiol. 2019;234(11):20662–20674. doi: 10.1002/jcp.28671. [eng] [DOI] [PubMed] [Google Scholar]
- 44.Xu W., Song C., Wang X., Li Y., Bai X., Liang X., et al. Downregulation of miR-155-5p enhances the anti-tumor effect of cetuximab on triple-negative breast cancer cells via inducing cell apoptosis and pyroptosis. Aging (Albany NY) 2021;13(1):228–240. doi: 10.18632/aging.103669. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li C., Yin W., Yu N., Zhang D., Zhao H., Liu J., et al. miR-155 promotes macrophage pyroptosis induced by Porphyromonas gingivalis through regulating the NLRP3 inflammasome. Oral Dis. 2019;25(8):2030–2039. doi: 10.1111/odi.13198. [eng] [DOI] [PubMed] [Google Scholar]
- 46.Wang B., Wang Z.M., Ji J.L., Gan W., Zhang A., Shi H.J., et al. Macrophage-derived exosomal mir-155 regulating cardiomyocyte pyroptosis and hypertrophy in uremic cardiomyopathy. JACC Basic Transl Sci. 2020;5(2):148–166. doi: 10.1016/j.jacbts.2019.10.011. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng G., Zha Z., Huang Y., Li J., Wang Y., Ke W., et al. Sustained and bioresponsive two-stage delivery of therapeutic miRNA via polyplex micelle-loaded injectable hydrogels for inhibition of intervertebral disc fibrosis. Adv Healthc Mater. 2018;7(21) doi: 10.1002/adhm.201800623. [eng] [DOI] [PubMed] [Google Scholar]
- 48.Zhu J., Zhang X., Gao W., Hu H., Wang X., Hao D. lncRNA/circRNA-miRNA-mRNA ceRNA network in lumbar intervertebral disc degeneration. Mol Med Rep. 2019;20(4):3160–3174. doi: 10.3892/mmr.2019.10569. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu S., Cheng Y., Tan Y., Dong J., Bian Q. Ligustrazine prevents intervertebral disc degeneration via suppression of aberrant TGFβ activation in nucleus pulposus cells. BioMed Res Int. 2019;2019 doi: 10.1155/2019/5601734. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tian Y., Yuan W., Li J., Wang H., Hunt M.G., Liu C., et al. TGFβ regulates Galectin-3 expression through canonical Smad3 signaling pathway in nucleus pulposus cells: implications in intervertebral disc degeneration. Matrix Biol. 2016;50:39–52. doi: 10.1016/j.matbio.2015.11.008. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghafouri-Fard S., Azimi T., Hussen B.M., Abak A., Taheri M., Dilmaghani N.A. Non-coding RNA activated by DNA damage: review of its roles in the carcinogenesis. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.714787. [eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo X., Dong J., He X., Shen L., Long C., Liu F., et al. MiR-155-5p exerts tumor-suppressing functions in Wilms tumor by targeting IGF2 via the PI3K signaling pathway. Biomed Pharmacother. 2020;125 doi: 10.1016/j.biopha.2020.109880. [eng] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.