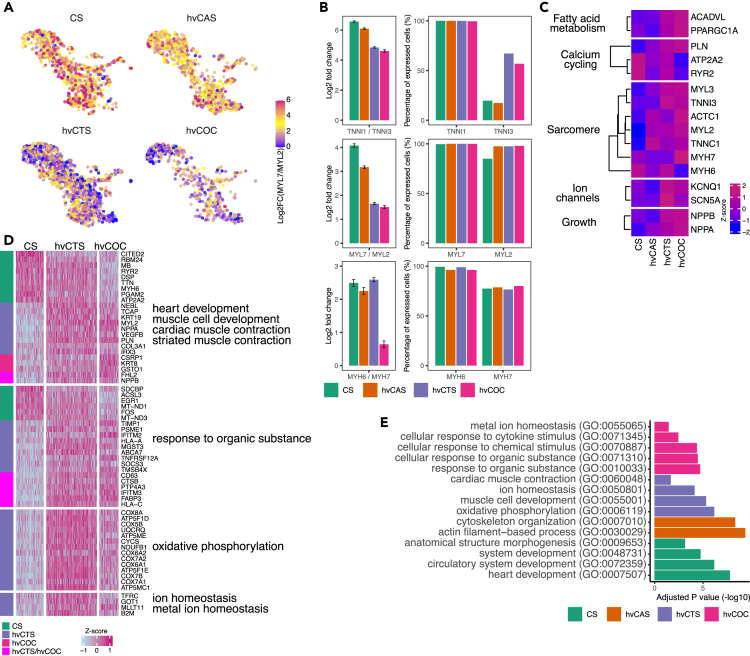
Figure 3.
Comparison of CM expression between CS and other tissue configurations hvCAS, hvCTS, and hvCOC
(A) UMAP of each tissue configuration with relative expression level of myosin light chain 7 to 2 (MYL7/MYL2) represented as log2 scale.
(B) The left panel shows Log2 ratio (mean+/−SE) of expression of troponin I isoform 1 to 3 (TNNI1/TNNI3), MYL7/MLY2, and myosin heavy chain 6 to 7 (MYH6/MYH7) for each tissue type. The right panel shows the percentage of cells expressing the myofibril isoforms for each tissue types.
(C) Heatmap showing mean scaled expression (mean Z score) of the selected of CM-associated genes in the key categories for the four tissue types. All genes in the table are DE between at least two tissue configurations.
(D) Heatmap showing the scaled expression values of the DEGs between CS and hvCTS/hvCOC belonging to enriched GO categories. Vertical bar indicates the higher expressed tissue construct.
(E) Top enriched GO terms for the DEGs between CS and other tissue configurations hvCAS, hvCTS, and hvCOC.