Abstract
Fission yeast Cds1 is phosphorylated and activated when DNA replication is interrupted by nucleotide starvation or DNA damage. Cds1 enforces the S-M checkpoint that couples mitosis (M) to the completion of DNA synthesis (S). Cds1 also controls replicational stress tolerance mechanisms. Cds1 is regulated by a group of proteins that includes Rad3, a kinase related to human checkpoint kinase ATM (ataxia telangiectasia mutated). ATM phosphorylates serine or threonine followed by glutamine (SQ or TQ). Here we show that in vitro, Rad3 and ATM phosphorylate the N-terminal domain of Cds1 at the motif T11Q12. Substitution of threonine-11 with alanine (T11A) abolished Cds1 activation that occurs when DNA replication is inhibited by hydroxyurea (HU) treatment. The cds1-T11A mutant was profoundly sensitive to HU, although not quite as sensitive as a cds1− null mutant. Cds1T11A was unable to enforce the S-M checkpoint. These results strongly suggest that Rad3-dependent phosphorylation of Cds1 at threonine-11 is required for Cds1 activation and function.
Inheritance of complete and accurate copies of the genome is the singular goal of cell division. Precise genome duplication is an intrinsically difficult process that can be strained further by external agents that interfere with DNA replication or damage DNA. Genome surveillance mechanisms exist to cope with these problems (16, 21). These systems serve two primary purposes. One purpose is to prevent mitosis when DNA replication is interrupted or DNA is damaged. These cell cycle checkpoints actively couple the onset mitosis to the completion of DNA replication and repair. The other purpose of genome surveillance mechanisms is to regulate various repair and replication systems that help cells survive replicational stress and DNA damage.
The fission yeast Schizosaccharomyces pombe has served as a valuable model system for the discovery and investigation of genome integrity checkpoint mechanisms (35). Genetic studies of fission yeast have uncovered a group of genes that are required for arresting cell division when DNA is damaged or when replication is inhibited with the drug hydroxyurea (HU). The products of these checkpoint RAD genes include Rad1, Rad3, Rad9, Rad17, Rad26, and Hus1 (13). Other proteins such as Cut5 (also known as Rad4) and Rfc3, which are necessary for DNA replication, are also important for both the DNA replication (S-M) and G2-M DNA damage checkpoints (38, 40). The most intriguing checkpoint protein may be Rad3, a very large protein (2,386 amino acids) that is related to phosphatidylinositol kinases (PIKs) (6). Other PIK-like proteins include human ATM (ataxia telangiectasia mutated), ATR (ATM and Rad3 related) and DNA-dependent protein kinase (20, 39, 43). Although related to phosphatidylinositol 3-kinases, these enzymes function as protein kinases in vivo. In common with fission yeast Rad3, ATM is required for arrest in G2 phase of the cell cycle in response to DNA damage caused by ionizing radiation and for slowed replication of damaged DNA (16, 24, 37). ATM is thought to control G2 arrest in part by activating Cds1 (also known as Chk2) (7, 8, 26), the mammalian homolog of the budding yeast Rad53 and fission yeast Cds1 checkpoint kinases. ATR has been implicated in the checkpoint response to UV damage and the inhibition of DNA replication (11, 43). Recently, Chk1 phosphorylation was found to be ATR dependent, suggesting that Chk1 is regulated by ATR (19, 25).
Rad3 and the other checkpoint Rad proteins control two downstream protein kinases in fission yeast. When DNA is damaged during G2 phase, Chk1 becomes phosphorylated in a Rad3-dependent manner (42). Chk1 prevents the onset of mitosis by regulation of Cdc25 and Mik1, two proteins that control the inhibitory phosphorylation of the cyclin-dependent kinase Cdc2 (2, 17, 18, 34, 36). The significance of Chk1 phosphorylation is uncertain, but it correlates with the requirement for Chk1 to arrest cell division in response to DNA damage. It is unknown if Chk1 is a direct physiological substrate of Rad3.
Fission yeast Cds1 becomes phosphorylated and activated by a Rad3-dependent mechanism when DNA is damaged during S phase or when DNA replication is interrupted with HU or mutations of several essential genes (9, 24). Cds1 enforces the S-M checkpoint by regulating Cdc25 and Mik1 (9, 17). In cds1 mutants treated with HU, the onset of mitosis is prevented by Chk1, but these cells are inviable (9, 10, 24). This fact demonstrates that Cds1 has replicational stress recovery functions that are distinct from its cell cycle checkpoint activity. How Cds1 is regulated is unknown.
Fission yeast Cds1 and its homologs are recognizable by similar kinase domains, an N-terminal Ser-Gln/Thr-Gln (SQ/TQ) cluster domain, and a forkhead-associated (FHA) domain (8, 26). SQ and TQ sequences are the preferred sites of phosphorylation by ATM in p53, c-Abl, Brca1, and Nbs1 (3, 4, 12, 14, 23). FHA domains are believed to act as protein-protein interaction domains and in some instances can bind to phosphorylated partners (22, 41). Budding yeast Rad53 is unique in possessing a second C-terminal FHA domain (1).
Several findings link Rad3 to Cds1. As mentioned above, Rad3 is necessary for phosphorylation and activation of Cds1 in vivo (9, 24). Active Cds1 associates with overproduced Rad3 in vivo (30). Furthermore, Cds1 associates with Rad26 when both proteins are overproduced, and Rad26 forms a protein complex with Rad3 (15, 24). These findings suggested that Rad3 might directly activate Cds1 in vivo. Experiments designed to test this hypothesis are described in this report. We show that Rad3 and human ATM phosphorylate fission yeast Cds1 at threonine-11. Threonine-11 forms part of a conserved TQ motif. We report that threonine-11 is crucially important for Cds1 activation and function in vivo. These studies provide strong support for the model that Rad3 directly controls Cds1 activity by phosphorylating Cds1 at threonine-11.
MATERIALS AND METHODS
Fission yeast strains, media, and general techniques.
Yeast strains used in this study are listed in Table 1. Fission yeast strains were grown and used as described by Moreno et al. (29). Growth media, general biochemical and genetic methods for fission yeast, and procedures for staining with 4′,6-diamidino-2-phenylindole (DAPI) have been described elsewhere (29). Yeast cultures were grown at 32°C in YES medium (0.5% yeast extract, 3% glucose, supplements). HU (Sigma) was used at the concentrations described.
TABLE 1.
S. pombe strains described in this report
| Straina | Genotype | Origin or reference |
|---|---|---|
| PR109 | h− | Our lab stock |
| NR1604 | h− chk1::ura4+ ade6-M704 | Our lab stock |
| KT2751 | h− cds1::ura4+ | 31 |
| KT2752 | h− cds1-T8A | This work |
| KT2753 | h− cds1-T11A | This work |
| KT2754 | h− cds1-T8AT11A | This work |
| KT2755 | h+ cds1-T8AT11A | This work |
| KT2777 | h+ cds1-T8A chk1::ura4+ | This work |
| KT2778 | h+ cds1-T11A chk1::ura4+ | This work |
| KT2756 | h+ cds1-T8AT11A chk1::ura4+ | This work |
| KT2779 | h− cds1::ura4+ pREP1-GST-Rad3 | This work |
| BF1916 | h+ nmt1:GST:cds1:leu1+ | Our lab stock |
| BF2504 | h+ nmt1:GST:cds1-D312E:leu1+ | Our lab stock |
| KT2780 | h− nmt1:GST:cds1-T8AT11A:leu1+ | This work |
All are leu1-32 ura4-D18.
Site-directed mutagenesis of Cds1.
The 3.3-kb PstI-SpeI genomic fragment containing the cds1 gene was cloned from pAL-cds1 (31) into pBluescript II KS(+) to give plasmid pBS-cds1. The cds1 genomic fragment with an NdeI site at its first ATG was constructed in pBluescript II KS(+) by PCR to give plasmid pBS-cds1N. Site-directed mutagenesis by PCR changed threonine to alanine at codons 8 and 11. All mutations were confirmed by DNA sequencing. The NdeI-SphI fragment containing the N terminus of Cds1 was replaced with the same fragment derived from site-directed mutagenesis. The resulting plasmids were pBS-cds1-T8A, pBS-cds1-T11A, and pBS-cds1-T8AT11A.
Expression and purification of glutathione S-transferase (GST) fusion proteins in bacteria.
Wild-type and mutant cds1 genes were amplified by PCR to remove the first ATG and create a BamHI site. BamHI-SphI fragments containing the N-terminal domain (amino acids 2 to 41) of Cds1 were cloned into pUC28 (5) and sequenced. Plasmids pGST-Cds1ND, pGST-Cds1T8AND, pGST-Cds1T11AND, and pGST-Cds1T8AT11AND were created by ligation of BamHI-EcoRI fragments from pUC28-Cds1ND, pUC28-Cds1T8AND, pUC28-Cds1T11AND, and pUC28-Cds1T8AT11AND, respectively, into pGEX-2T. Each plasmid was transformed into Escherichia coli DH5α.
GST-Cds1ND protein expression was induced by the addition of isopropyl-β-d-thiogalactopyranoside (0.2 mM) to exponentially growing cells at an absorbance at 600 nm of 0.6 in Luria-Bertani medium plus ampicillin (100 μg/ml) at 25°C. After 16 h at 25°C, cells were collected by centrifugation. Cells were lysed in Y-PER yeast protein extraction reagent (Pierce) by rotating at room temperature for 1 h and centrifuged at 10,000 rpm for 20 min at 4°C. Glutathione (GSH)-Sepharose (Pharmacia) was added to the supernatants and incubated at 4°C for 1 h. GSH-Sepharose was washed three times with ice-cold phosphate-buffered saline.
Integration of site-directed mutagenized cds1 genes into the cds1 locus.
Site-directed mutagenized cds1 genes flanked by endogenous chromosomal promoter and terminator sequences were isolated by digestion of plasmids pBS-cds1-T8A, pBS-cds1-T11A, and pBS-cds1-T8AT11A with PstI and SpeI. The PstI-SpeI fragments containing mutated cds1 genes were transformed into the cds1::ura4+ strain (31). After 5-fluoro-orotic acid selection, stable integrants of mutagenized cds1 were identified and further confirmed by colony PCR analysis followed by the direct sequencing of PCR products.
Preparation of S. pombe cell extracts and immunoblotting.
Logarithmically growing cells (about 5 × 106/ml) were harvested. Pellets were washed with water and then with STOP buffer (29). Cells were disrupted with glass beads in lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM EDTA, 10% glycerol, 0.2% NP-40, leupeptin-aprotinin-pepstatin A [5 μg/ml], 1 mM phenylmethylsulfonyl fluoride [PMSF]). Protein extract was cleared at 13,000 rpm in a microcentrifuge at 4°C for 10 min and either used fresh or frozen. Cell extracts were boiled in Sodium dodecyl sulfate (SDS) sample buffer and subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to Immobilon transfer membranes (Millipore). The membranes were blocked with TBSM (Tris-buffered saline plus 5% milk) and incubated in Cds1 antiserum (1:3,000 dilution in TBSM), washed in TBST (Tris-buffered saline plus 0.2% Tween 20), and incubated with horseradish peroxidase-conjugated rabbit immunoglobulin G secondary antibody (1:10,000 dilution; Promega). For the detection of immunoprecipitated Cds1, horseradish peroxidase-conjugated protein A secondary antibody (1:1,000 dilution in TBST–0.3% fetal bovine saline; Amersham) was used. Enhanced chemiluminescence detection (Pierce) was used to visualize proteins.
Cds1 kinase assay.
Cds1 kinase assay with immunoprecipitated Cds1 was carried out as described elsewhere (9), with the following modification. Cds1 antibody was incubated with protein A-Sepharose (Pharmacia) in lysis buffer at 4°C for 1 h; 2 mg of total extract in 0.5 ml of lysis buffer was incubated with protein A-Sepharose-bound Cds1 antibody at 4°C for 2 h. Protein A-Sepharose was washed three times with lysis buffer, collected, and processed for Cds1 kinase activity. Protein A-Sepharose was mixed with GSH-Sepharose-bound GST-Wee1152 substrates as described elsewhere (9) and washed two times in kinase buffer (50 mM Tris-HCl [pH 7.5], 10 mM MgCl2). Mixtures of beads were incubated with 50 μl of kinase buffer containing 0.25 μl of [γ-32P]ATP and 100 μM ATP at 30°C for 30 min. The reaction was stopped by the addition of 20 μl of 4 × SDS sample buffer. Samples were boiled and subjected to SDS-PAGE (10% gel).
In vitro Rad3 and ATM kinase assays.
ATM immunoprecipitates were produced essentially as described elsewhere (7, 28). Briefly, 1 mg of HeLa cell lysate was incubated with 1 μg of anti-ATM antibody (H-248; Santa Cruz) and with protein A-Sepharose (Pharmacia). The precipitated beads were washed three times with lysis buffer and three times with kinase buffer (50 mM imidazole [pH 7.4], 50 mM NaCl, 10 mM MnCl2, 10 mM MgCl2, 1 mM dithiothreitol [DTT]). Kinase reactions were initiated by addition of [γ-32P]ATP. After 15 min at 30°C, reactions were stopped by addition of SDS sample buffer and analyzed by autoradiography following SDS-PAGE. GST-Cds1ND derivatives purified from E. coli were used as substrates. ATM and GST-Rad3 activities were sensitive to the PIK inhibitor wortmannin.
GST-Rad3 was expressed from the full-strength nmt1 promoter. Yeast cells were disrupted with glass beads in lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 10% glycerol, 1% NP-40, 50 mM NaF, 1 mM Na3VO4, 0.2 mM 4-amidino-PMSF, 1 mM PMSF, 1 mM DTT, protease inhibitor cocktail complete [Roche]. Cell extracts were incubated with GSH-Sepharose at 4°C for 2 h. GSH-Sepharose was washed three times with lysis buffer and twice with kinase buffer (25 mM HEPES-KOH [pH 7.5], 50 mM KCl, 10 mM MgCl2, 10 mM MnCl2, 2% glycerol, 0.1% NP-40, 1 mM Na3VO4, 1 mM DTT); 50 μl of kinase buffer containing 10 μM ATP, [γ-32P]ATP, 10 mM GSH, 1 mM PMSF, leupeptin-aprotinin-pepstatin A (5 μg/ml), and substrate was added to GSH-Sepharose-bound GST-Rad3. After 25 min at 30°C, reactions were stopped by the addition of 20 μl of 4× SDS sample buffer. Samples were boiled and subjected to SDS-PAGE (15% gel).
RESULTS
ATM phosphorylates threonine-11 in the N-terminal domain of Cds1.
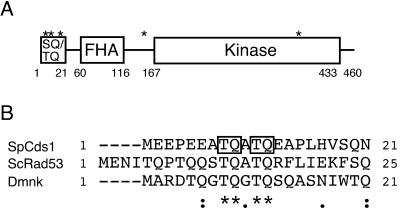
PIK-like kinases such as ATM prefer to phosphorylate serine or threonine residues followed by glutamine (SQ or TQ). Cds1 has five SQ/TQ motifs: three in the first 20 amino acids at the N terminus of the protein, one located between the FHA and kinase domains, and one in the kinase domain (Fig. 1A). Alignment to Cds1 homologs from budding yeast (Rad53) (1) and Drosophila melanogaster (Dmnk) (32) indicated weak homology in the N-terminal SQ/TQ cluster domains (Fig. 1A). A GST fusion protein that contained the Cds1 N-terminal SQ/TQ cluster domain (GST-Cds1ND; amino acids 2 to 41) was produced in E. coli. GST-Cds1ND was tested as an in vitro substrate of ATM. In our initial studies, ATM was used instead of Rad3 because immunoprecipitated ATM has a much more robust in vitro kinase activity.
FIG. 1.
(A) Domain structure of S. pombe Cds1. ∗, SQ and TQ motifs. (B) Alignment of the N-terminal SQ/TQ cluster domains of the budding yeast Rad53, fission yeast Cds1, and D. melanogaster Dmnk. Alignment was performed by the CLUSTAL W program. Asterisks and dots show identical and similar amino acids, respectively. T8Q9 and T11Q12 of Cds1 are boxed.
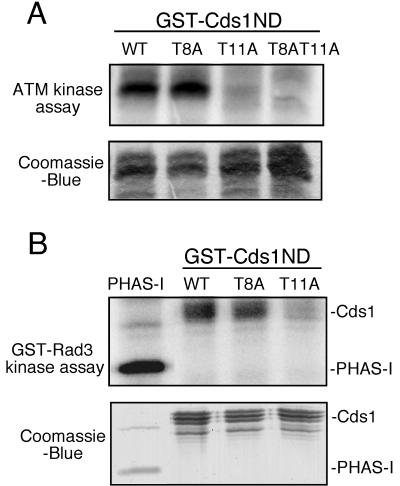
ATM immunoprecipitated from HeLa cells phosphorylated GST-Cds1ND (Fig. 2A). This result prompted us to determine the phosphorylation site(s). T8Q9 and T11Q12 appeared to be conserved among the Cds1 homologs aligned in Fig. 1. Therefore, the relevant threonine codons were mutated to alanine to produce T8A, T11A, and T8AT11A constructs. These mutated proteins were expressed as GST-Cds1ND fusion proteins and tested in the ATM kinase assays. The T8A mutation did not reduce 32P incorporation into GST-Cds1ND (Fig. 2A). In contrast, the T11A and T8AT11A substitutions appeared to eliminate 32P incorporation into GST-Cds1ND (Fig. 2A). These findings indicated that threonine-11 of Cds1 was phosphorylated by ATM in vitro.
FIG. 2.
Phosphorylation of the N-terminal SQ/TQ cluster domain of Cds1 by ATM and Rad3. (A) Immunoprecipitate with anti-ATM antibodies from HeLa cells was incubated with [γ-32P]ATP and the N-terminal SQ/TQ cluster domain of Cds1 fused to GST (GST-Cds1ND, GST-Cds1T8A, GST-Cds1T11A, and GST-Cds1T8AT11A). Proteins were resolved by SDS-PAGE and visualized with a Phosphorlmager (Molecular Dynamics). GST-Cds1ND substrates were stained with Coomassie blue. (B) GST-Rad3 was purified from fission yeast and incubated with PHAS-I and the GST-Cds1ND, GST-Cds1T8A, and GST-Cds1T11A substrates as described above.
Rad3 phosphorylates threonine-11 of Cds1.
Having obtained evidence that ATM phosphorylated Cds1 at threonine-11, we investigated whether Rad3 possessed the same activity. GST-Rad3 was expressed from the strong nmt1 promoter and then purified with GSH-Sepharose. GST-Rad3 was expressed in a cds1− strain to eliminate the possibility of Cds1 copurifying with GST-Rad3 (30). Consistent with our previous studies of hemagglutininin epitope-tagged Rad3 expressed and purified from fission yeast (30), we found that the GST-Rad3 precipitate phosphorylated the model substrate PHAS-I (Fig. 2B). GST-Rad3 phosphorylated the wild-type form GST-Cds1ND (Fig. 2B). The T8A mutation did not significantly diminish phosphorylation of GST-Cds1ND by GST-Rad3 (Fig. 2B). In contrast, the T11A mutation appeared to eliminate phosphorylation of GST-Cds1ND by GST-Rad3. Thus, in these assays, Rad3 and ATM possessed the same substrate specificity for phosphorylation of threonine-11 in the N-terminal domain of Cds1.
Threonine-11 is required for Cds1 phosphorylation in vivo.
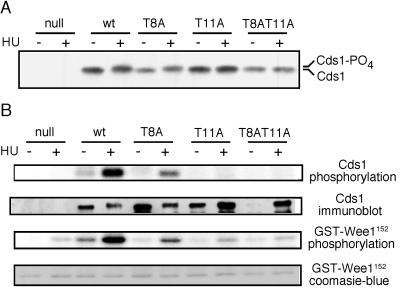
To evaluate if threonine-11 is important for Cds1 phosphorylation in vivo, the T8A, T11A, and T8AT11A mutations were cloned into full-length cds1. These constructs were used to replace the genomic copy of cds1. HU-induced phosphorylation of Cds1 is observed as a mobility shift in SDS-PAGE (24). The significance of this phosphorylation is unknown; it correlates with Cds1 activation and may result from autophosphorylation. An HU-induced mobility shift was readily detected with wild-type Cds1 and mutant Cds1T8A protein (Fig. 3A). In contrast, the mobility of Cds1T11A and Cds1T8AT11A mutant proteins was unaltered by HU treatment (Fig. 3A). These findings demonstrated that threonine-11 is required for the phosphorylation-induced mobility shift of Cds1 that occurs in HU-treated cells.
FIG. 3.
Phosphorylation and activation of Cds1 kinase of cds1 mutant cells. Logarithmically growing cds1+, cds1−, cds1-T8A, cds1-T11A, and cds1-T8AT11A cells were mock treated or treated with 20 mM HU for 4 h. Cells were collected, and extract was prepared. (A) Cds1 mobility was analyzed by immunoblotting. (B) Cds1 immunoprecipitates were analyzed for Cds1 kinase activity in vitro. The autophosphorylation of Cds1 and phosphorylation of GST-Wee1152 were subjected to Phosphorlmager analysis. Cds1 immunoprecipitates were also analyzed by Cds1 immunoblotting. Cds1 was strongly activated by HU, whereas Cds1T8A activation was intermediate. The activity of Cds1T11A and Cds1T8AT11A from HU-treated cells was negligible. Recovery of Cds1 by immunoprecipitation was variable and appeared to fail in the −HU sample from cds1-T8AT11A cells. The very small increase in activity in the HU-treated samples of Cds1T11A and Cds1T8AT11A might be attributed to better recovery of Cds1 in these samples or contamination with an HU-stimulated kinase that is not Cds1 (note the small increase of GST-Wee1152 phosphorylation in the samples from cds1− cells).
Threonine-11 is required for Cds1 activation in vivo.
To understand the significance of threonine-11 for Cds1 activity, kinase assays were performed with wild-type and mutant Cds1 proteins. These proteins were immunoprecipitated from HU-treated and mock-treated cells. GST-Wee1152, a fusion of GST to amino acids 11 to 152 of Wee1 protein, was used as the Cds1 substrate (9). Autophosphorylation of Cds1 was also monitored. As noted previously, wild-type Cds1 was highly activated when immunoprecipitated from HU-treated cells (Fig. 3B). In contrast, Cds1T11A and Cds1T8AT11A mutant proteins exhibited little or no activation in response to HU (Fig. 3B). These findings established that threonine-11 is required for Cds1 activation in vivo. Curiously, an intermediate level of activity was seen in the Cds1T8A sample (Fig. 3B), although an HU-induced mobility shift of Cds1T8A was readily detected (Fig. 3A).
HU sensitivity of the cds1-T11A mutant.
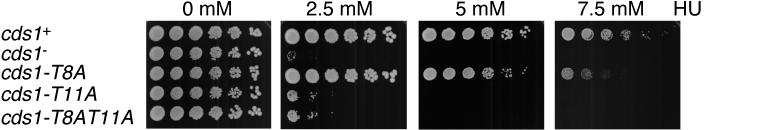
Cds1 is required for proper recovery from a DNA replication arrest (31). If threonine-11 phosphorylation is important for Cds1 activation in vivo, then replacement of threonine-11 with alanine should render cells sensitive to HU. Serial dilutions of wild-type and mutant cds1 strains were spotted on media containing different concentrations of HU (Fig. 4). Relative to wild-type cells, the cds1-T11A and cds1-T8AT11A cells showed severe sensitivity to HU. The mutant strains were unable to form colonies on media containing 5 mM HU, whereas wild-type cells readily formed colonies on this medium (Fig. 4). However, on 2.5 mM HU medium, the cds1-T11A and cds1-T8AT11A cells grew slightly better than cds1− cells. These findings suggested that the cds1-T11A and cds1-T8AT11A strains retained very weak Cds1 activity. The cds1-T8A cells were not abnormally sensitive to 2.5 mM HU (Fig. 4), indicating that Cds1 function was substantially retained in these cells. However, in 7.5 mM HU medium, the cds1-T8A cells grew poorly compared to wild-type cds1+ cells. This observation correlated with the reduced kinase activity of Cds1T8A protein (Fig. 3B). Threonine-8 might be a secondary site of phosphorylation, or mutation of threonine-8 might impair threonine-11 phosphorylation or its regulatory effect.
FIG. 4.
HU sensitivity of cds1 mutant cells. Fivefold serial dilutions of cds1+, cds1−, cds1-T8A, cds1-T11A, and cds1-T8AT11A cells were plated in YES medium supplemented with 0, 2.5, 5, or 7.5 mM HU and incubated for 5 days at 32°C.
S-M checkpoint defect of the cds1-T8AT11A mutant.
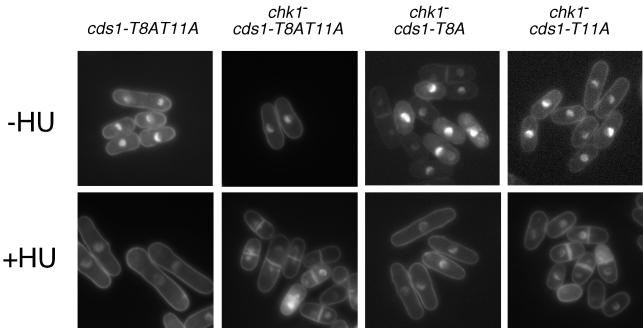
Genetic and biochemical experiments have indicated that Cds1 has primary responsibility for enforcing the S-M checkpoint in cells treated with HU. However, cds1 mutants arrest division in response to HU due to the function of Chk1 (9, 24, 44). A chk1 mutant arrests division in response to HU in a Cds1-dependent manner. We therefore examined the replication checkpoint in cds1-T8AT11A and cds1-T8AT11A chk1 cells (Fig. 5). HU arrested division in cds1-T8AT11A cells. The arrest was evident from the appearance of elongated cells and the absence of septated cells. In contrast, cds1-T8AT11A chk1− double-mutant cells underwent division after addition of HU (Fig. 5). The checkpoint defect was apparent from the appearance of cut cells, in which the DNA visualized with DAPI appeared to be bisected by the division septum (Fig. 5). Thus, the S-M checkpoint appeared to be abolished in cds1-T8AT11A chk1− double-mutant cells. The relative importance threonine-8 and threonine-11 was evaluated by examining the response of cds1-T8A chk1− and cds1-T11A chk1− cells to HU. We found that cds1-T8A chk1− cells underwent checkpoint arrest in HU, whereas cds1-T11A chk1− cells displayed a profound checkpoint defect (Fig. 5). These results demonstrated that Cds1T11A is unable to enforce the S-M checkpoint.
FIG. 5.
Mutant cds1-T11A and cds1-T8AT11A cells undergo a Chk1-dependent cell cycle arrest when exposed to HU. Cells of the indicated genotypes were stained with DAPI after being grown in the absence or presence of 12 mM HU for 4 h at 32°C.
GST-Cds1T8AT11A arrests division when overexpressed.
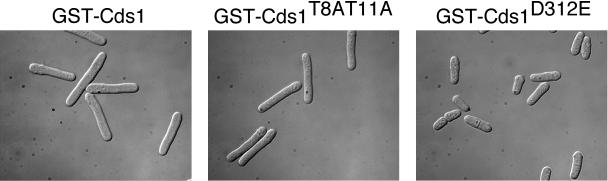
High expression of GST-Cds1 arrests division in fission yeast (9). This arrest is suppressed in a strain that expresses Cdc2Y15F, a mutant form of Cdc2 that is insensitive to checkpoint regulation. GST-Cds1 expression arrests division in a rad3− strain, a result that implies that GST-Cds1 has a basal amount of activity in the absence of regulation by Rad3 (9). If Cds1T11A were defective solely because it cannot be phosphorylated by Rad3, we would expect GST-Cds1T11A and GST-Cds1T8AT11A to arrest division when overexpressed. Accordingly, strains that expressed wild-type and mutant forms of Cds1 fused to GST and regulated by the nmt1 promoter were constructed. As observed previously (9), expression of GST-Cds1 caused a cell cycle arrest, as demonstrated by the appearance of elongated cells that were not septated (Fig. 6). Expression of GST-Cds1T8AT11A caused an identical cell cycle arrest phenotype (Fig. 6). In contrast, expression of GST-Cds1D312E, which encodes a kinase-inactive form of Cds1 (24), had no ability to arrest division (Fig. 6). These data support the conclusion that Cds1T8AT11A retains intrinsic kinase activity but cannot be activated by Rad3.
FIG. 6.
Overexpression of GST-Cds1T8AT11A causes cell cycle arrest. Strains that expressed GST fused to Cds1, Cds1T8AT11A, or Cds1D312E (kinase inactive) from the thiamine-repressible nmt1 promoter were grown in medium lacking thiamine for 18 h. Cells that expressed Cds1 and Cds1T8AT11A constructs elongated and arrested division as unseptated cells, whereas cells that expressed GST-Cds1D312E continued division.
DISCUSSION
Genetic and biochemical studies in fission yeast have mapped a signal transduction pathway in which replication interruption caused by HU results in Cds1 activation. Rad3 and at least five other proteins are required for Cds1 activation. In this report we have explored exactly how Cds1 is activated, starting with the hypothesis that activation requires direct phosphorylation by Rad3. The findings in this paper strongly support this hypothesis. Rad3 phosphorylates threonine-11 of Cds1 in vitro. Cds1T11A mutant protein cannot be activated by HU, nor can it enforce the S-M checkpoint. Moreover, HU survival of cds1-T11A cells is highly deficient. We have not yet formally demonstrated that threonine-11 is phosphorylated in vivo, but threonine-11 is required the phosphorylation-induced electrophoretic mobility shift of Cds1. Importantly, mutant Cds1T8AT11A retains the ability to cause a cell cycle arrest when overexpressed as a GST fusion protein. This result demonstrates that mutation of the Rad3-directed phosphorylation site does not ablate a basal activity of Cds1 but does prevent the activation of Cds1. Taken together, these findings strongly suggest that S-M checkpoint and replicational stress signal transduction pathways require direct phosphorylation of Cds1 by Rad3.
In our initial studies we used ATM as a Rad3 surrogate in the in vitro kinase assays because it is much easier to purify active ATM from HeLa cells than to purify active Rad3 from fission yeast. The fact that ATM phosphorylates a site that is required for Cds1 activation and function in vivo, and which is also phosphorylated by Rad3 in vitro, reinforces the notion that ATM and Rad3 are to a significant extent structural and functional homologs. This idea is further supported by two recent studies showing that human Cds1 is phosphorylated and activated by ATM (27, 28). Threonine-68, which is located in the N-terminal SQ/TQ cluster domain of human Cds1, was identified as the important phosphorylation site. Activation of mutant Cds1T68A by ionizing radiation was substantially diminished in transfected HeLa cells, consistent with the behavior of fission yeast Cds1T11A mutant protein. Interestingly, threonine-11 in fission yeast Cds1 and threonine-68 in human Cds1 are found in the motif TQE, which bears some similarity to the LSQE motif that was identified as the optimum ATM substrate motif by a peptide library approach (33). It is probably significant that these sites in fission yeast and human Cds1 are almost exactly the same distance from the FHA domain in each protein. These findings suggest that threonine-11 in fission yeast Cds1 and threonine-68 in human Cds1 are functionally and structurally conserved sites of regulatory phosphorylation by Rad3 and ATM, respectively.
The T11A mutation causes an HU-sensitive phenotype that is not as severe as that caused by a complete deletion of the cds1+ open reading frame. These data suggest that Cds1 that is not phosphorylated at threonine-11 has a weak basal activity. It is possible that Rad3 performs some phosphorylation at threonine-8. This possibility is consistent with the very mild HU-sensitive phenotype of a cds1-T8A mutation and the apparent partial decrease in Cds1T8A kinase activity relative to wild-type Cds1. However, the T8A mutation does not appear to significantly decrease phosphorylation of GST-Cds1ND by Rad3 and ATM in vitro, and the cds1-T8AT11A and cds1-T11A mutants appear to be equally HU sensitive. Thus, the weak phenotypes of cds1-T8A cells might be attributed to a small effect on the amount of phosphorylation at threonine-11, or the T8A mutation might weakly impair the activity of Cds1 that has been phosphorylated at threonine-11.
Roles for human Cds1 in enforcing cell cycle checkpoints or promoting survival of DNA damage or replication inhibition have not been clearly established. In comparison, the biological consequences of loss of Cds1 activity in fission yeast are better understood. Hence, it was possible to more directly assess the biological consequences of mutation of threonine-11 in fission yeast Cds1. Our studies demonstrated that Cds1T11A was ineffective as an enforcer of the S-M checkpoint and was highly defective at promoting cell viability in medium that contains HU. Thus, our findings not only confirm that a site that can be phosphorylated by Rad3 is important for Cds1 activation but also extend this analysis to show that this site is essential for Cds1 to carry out its biological functions. These findings, and the recent studies linking ATM to phosphorylation of Cds1 in human cells, provide a more precise understanding of checkpoint signaling.
Many questions that concern the interaction between Rad3 and Cds1 remain to be answered. Activation of Cds1 appears to require phosphorylation catalyzed by Rad3, but is this phosphorylation sufficient for Cds1 activation? Thus far, there have been no reports of in vitro activation of Cds1 by immunoprecipitated Rad3 or ATM. A second question concerns the cell cycle specificity of Cds1 activation. In fission yeast, Cds1 is activated by DNA damage only during S phase. It seems likely that Cds1 activation requires a protein, a protein complex, or a protein activity that exists only during S phase. This entity must somehow link Rad3 to Cds1. Our future experiments will be aimed at answering these questions.
ACKNOWLEDGMENTS
We are grateful to Teresa Wang for the gift of anti-Cds1 antibody, H. Murakami and H. Okayama for the gift of plasmid pAL-cds1 and the cds1::ura4 strain, and Beth Baber-Furnari for plasmid pREP1-GST-Rad3. Antonia Lopez-Girona made helpful comments and suggestions. Members of the Scripps Cell Cycle Groups provided support and encouragement.
K.T. was supported by The Naito Foundation. This work was funded by NIH grants awarded to C.H.G. and P.R.
REFERENCES
- 1.Allen J B, Zhou Z, Siede W, Friedberg E C, Elledge S J. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 2.Baber-Furnari B A, Rhind N, Boddy M N, Shanahan P, Lopez-Girona A, Russell P. Regulation of mitotic inhibitor Mik1 helps to enforce the DNA damage checkpoint. Mol Biol Cell. 2000;11:1–11. doi: 10.1091/mbc.11.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banin S, Moyal L, Shieh S, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 4.Baskaran R, Wood L D, Whitaker L L, Canman C E, Morgan S E, Xu Y, Barlow C, Baltimore D, Wynshaw-Boris A, Kastan M B, Wang J Y. Ataxia telangiectasia mutant protein activates c-Abl tyrosine kinase in response to ionizing radiation. Nature. 1997;387:516–519. doi: 10.1038/387516a0. [DOI] [PubMed] [Google Scholar]
- 5.Benes V, Hostomsky Z, Arnold L, Paces V. M13 and pUC vectors with new unique restriction sites for cloning. Gene. 1993;130:151–152. doi: 10.1016/0378-1119(93)90360-f. [DOI] [PubMed] [Google Scholar]
- 6.Bentley N J, Holtzman D A, Flaggs G, Keegan K S, DeMaggio A, Ford J C, Hoekstra M, Carr A M. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- 7.Blasina A, Price B D, Turenne G A, McGowan C H. Caffeine inhibits the checkpoint kinase ATM. Curr Biol. 1999;9:1135–1138. doi: 10.1016/s0960-9822(99)80486-2. [DOI] [PubMed] [Google Scholar]
- 8.Blasina A, Van de Weyer I, Laus M C, Luyten W H M L, Parker A E, McGowan C H. A human homolog of the checkpoint kinase Cds1 directly inhibits Cdc25. Curr Biol. 1999;9:1–10. doi: 10.1016/s0960-9822(99)80041-4. [DOI] [PubMed] [Google Scholar]
- 9.Boddy M N, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinases Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- 10.Brondello J M, Boddy M N, Furnari B, Russell P. Basis for the checkpoint signal specificity that regulates Chk1 and Cds1 protein kinases. Mol Cell Biol. 1999;19:4262–4269. doi: 10.1128/mcb.19.6.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown E J, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 12.Canman C E, Lim D S, Cimprich K A, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan M B, Siliciano J D. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 13.Carr A M. Analysis of fission yeast DNA structure checkpoints. Microbiology. 1998;144:5–11. doi: 10.1099/00221287-144-1-5. [DOI] [PubMed] [Google Scholar]
- 14.Cortez D, Wang Y, Qin J, Elledge S J. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- 15.Edwards R J, Bentley N J, Carr A M. A Rad3-Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat Cell Biol. 1999;1:393–398. doi: 10.1038/15623. [DOI] [PubMed] [Google Scholar]
- 16.Elledge S J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 17.Furnari B, Blasina A, Boddy M N, McGowan C H, Russell P. Cdc25 inhibited in vivo and in vitro by checkpoint kinases Cds1 and Chk1. Mol Biol Cell. 1999;10:833–845. doi: 10.1091/mbc.10.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furnari B, Rhind N, Russell P. Cdc25 mitotic inducer targeted by chk1 DNA damage checkpoint kinase. Science. 1997;277:1495–1497. doi: 10.1126/science.277.5331.1495. [DOI] [PubMed] [Google Scholar]
- 19.Guo Z, Kumagai A, Wang S X, Dunphy W G. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartley K O, Gell D, Smith G C, Zhang H, Divecha N, Connelly M A, Admon A, Lees-Miller S P, Anderson C W, Jackson S P. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 21.Hartwell L H, Weinert T A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 22.Hofmann K, Bucher P. The FHA domain: a putative nuclear signalling domain found in protein kinases and transcription factors. Trends Biochem Sci. 1995;20:347–349. doi: 10.1016/s0968-0004(00)89072-6. [DOI] [PubMed] [Google Scholar]
- 23.Lim D S, Kim S T, Xu B, Maser R S, Lin J, Petrini J H, Kastan M B. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- 24.Lindsay H D, Griffiths D J, Edwards R J, Christensen P U, Murray J M, Osman F, Walworth N, Carr A M. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Q, Guntuku S, Cui X S, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, Donehower L A, Elledge S J. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuoka S, Huang M, Elledge S J. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 27.Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge S J. Ataxia telangiectasia-mutated phosphorylates chk2 in vivo and in vitro. Proc Natl Acad Sci USA. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melchionna R, Chen X-B, Blasina A, McGowan C H. Threonine 68 is required for radiation-induced phosphorylation and activation of Cds1. Nat Cell Biol. 2000;10:762–765. doi: 10.1038/35036406. [DOI] [PubMed] [Google Scholar]
- 29.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 30.Moser B A, Brondello J-M, Baber-Furnari B, Russell P. Mechanism of caffeine-induced checkpoint override in fission yeast. Mol Cell Biol. 2000;20:4288–4294. doi: 10.1128/mcb.20.12.4288-4294.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murakami H, Okayama H. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature. 1995;374:817–819. doi: 10.1038/374817a0. [DOI] [PubMed] [Google Scholar]
- 32.Oishi I, Sugiyama S, Otani H, Yamamura H, Nishida Y, Minami Y. A novel Drosophila nuclear protein serine/threonine kinase expressed in the germline during its establishment. Mech Dev. 1998;71:49–63. doi: 10.1016/s0925-4773(97)00200-1. [DOI] [PubMed] [Google Scholar]
- 33.O'Neill T, Dwyer A J, Ziv Y, Chan D W, Lees-Miller S P, Abraham R H, Lai J H, Hill D, Shiloh Y, Cantley L C, Rathbun G A. Utilization of oriented peptide libraries to identify substrate motifs selected by ATM. J Biol Chem. 2000;275:22719–22727. doi: 10.1074/jbc.M001002200. [DOI] [PubMed] [Google Scholar]
- 34.Rhind N, Furnari B, Russell P. Cdc2 tyrosine phosphorylation is required for the DNA damage checkpoint in fission yeast. Genes Dev. 1997;11:504–511. doi: 10.1101/gad.11.4.504. [DOI] [PubMed] [Google Scholar]
- 35.Rhind N, Russell P. Mitotic DNA damage and replication checkpoints in yeast. Curr Opin Cell Biol. 1998;10:749–758. doi: 10.1016/s0955-0674(98)80118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhind N, Russell P. Roles of the mitotic inhibitors Wee1 and Mik1 in the G2 DNA damage and replication checkpoints. Mol Cell Biol. 2001;21:1499–1508. doi: 10.1128/MCB.21.5.1499-1508.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rhind N, Russell P. The Schizosaccharomyces pombe S-phase checkpoint differentiates between different types of DNA damage. Genetics. 1998;149:1729–1737. doi: 10.1093/genetics/149.4.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saka Y, Yanagida M. Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+ Cell. 1993;74:383–393. doi: 10.1016/0092-8674(93)90428-s. [DOI] [PubMed] [Google Scholar]
- 39.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle D A, Smith S, Uziel T, Sfez S, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 40.Shimada M, Okuzaki D, Tanaka S, Tougan T, Tamai K K, Shimoda C, Nojima H. Replication factor C3 of Schizosaccharomyces pombe, a small subunit of replication factor C complex, plays a role in both replication and damage checkpoints. Mol Biol Cell. 1999;10:3991–4003. doi: 10.1091/mbc.10.12.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun Z, Hsiao J, Fay D S, Stern D F. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- 42.Walworth N C, Bernards R. rad-dependent response of the chk1-encoded protein kinase at the DNA damage checkpoint. Science. 1996;271:353–356. doi: 10.1126/science.271.5247.353. [DOI] [PubMed] [Google Scholar]
- 43.Wright J A, Keegan K S, Herendeen D R, Bentley N J, Carr A M, Hoekstra M F, Concannon P. Protein kinase mutants of human ATR increase sensitivity to UV and ionizing radiation and abrogate cell cycle checkpoint control. Proc Natl Acad Sci USA. 1998;95:7445–7450. doi: 10.1073/pnas.95.13.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng Y, Forbes K C, Wu Z, Moreno S, Piwnica-Worms H, Enoch T. Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature. 1998;395:507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]