We thank Dr. Sorscher for bringing attention to the lead-time aspects, as a commentary to our manuscript on circulating tumor (ctDNA) in patients undergoing loco-regional treatment of colorectal cancer (CRC) metastases.1 In this, we presented results of a systematic review and meta-analysis addressing the prognostic value of ctDNA detection prior to or after local treatment for CRC metastases.
In the presented dataset and thus the resulting discussion, we used aggregated data provided by the respective authors of the individual analyses, and since the majority do not draw attention to the aspects on any lead-time data, this aspect has consequently also not been brought to discussion in the review.
We do however, agree that lead-time data, as they are currently presented in the literature, can in fact be somehow misleading, and that this is another important subject for discussion.
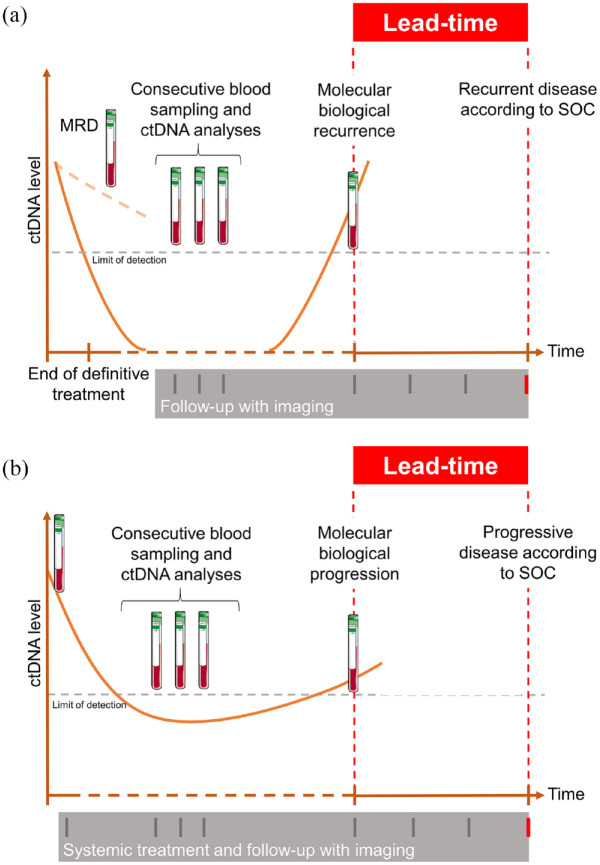
ctDNA presence after a curatively intended treatment for solid tumors is a strong prognostic marker, indicating a markedly high (if not 100%) risk of relapse of the disease.2 This is commonly referred to as ctDNA minimal residual disease (MRD). In studies where ctDNA analysis is done later, during the follow-up period, the same prognostic value is reported3,4 – but in addition, data can be used to describe a potential lead-time between the ctDNA-based definition of relapse and the standard assessment of recurrence, by clinical and/or imaging-based examinations. Figure 1 shows our suggestions for definitions of lead-time in curative and palliative situations. The magnitude of the lead-time reported in the literature can indeed be misinterpreted if the study is not prospectively designed to address the topic. The true sensitivity of ctDNA for detection of early recurrence can only be assessed from studies with direct comparisons with current standards (radiological and clinical definition of recurrence) at similar timepoints. It is natural to assume that imaging procedures in most retrospective observational studies are performed according to clinical practice, but there is still value in analyzing the intermittent ctDNA testing to describe the potential of such new observations.
Figure 1.
Schematic overview of definition of lead-time in the curative (a) and palliative (b) setting.
Clinical lead-time can be defined as the time between blood-based detected and clinical-detected recurrence/progression. The biological lead-time can only be defined if scans and blood samples are done at the same time.
MRD, minimal residual disease; SOC, standard of care.
An overview of the included studies in a previous review and meta-analysis in advanced disease reveals that in a significant number of the studies blood samples were drawn more frequently than the clinical/radiological evaluations, whereas other studies failed to report on the frequency of scans.5 Review of our present overview regarding loco-regional treatment of metastases from CRC revealed that only few of the studies included presentation of a lead-time aspect, and this primarily as case reports on a limited number of patients.6–13 The frequency of scans was presented in only three studies.9,12,13 In two of these studies, scans were less frequent than ctDNA analysis.9,12 The studies mentioned by the author show similar tendency. Tarazona et al. reported a median lead-time of 11.5 months based on 14 patients who underwent 6-monthly scans and blood sampling every 4 months.14 Henriksen et al. reported a median lead-time of 9.8 months in 21 patients, and adequately addressed the issue of less frequently performed scans (i.e. 6-monthly or at 12- and 36-month compared to 3-monthly blood samples).15 A shorter lead-time of 5.5 months was reported by Tie et al. but in this study both scans and sampling were performed with 3-monthly intervals.16 To compare the results between studies, standardization in definitions and reporting of results is mandatory.
In summary, the true clinical value must be investigated in prospective carefully designed studies. Prospective observational studies should first be designed to compare the ctDNA analysis and imaging results at the same timepoints, blinded to the results from each modality. This will allow for a reliable quantification of the lead-time and results can thus be used to calculate the most relevant sample size for prospective randomized trials. A few randomized studies have already been designed to investigating ctDNA as adjunct to standard follow-up programs, for example, in the Nordic trial of ctDNA guided follow-up in anal cancer (NOAC9 clinical trial.gov NCT05572801) or direct comparison of ctDNA based follow-up with standard of care in the IMPROVE-IT 2 study.17 The results of these trials will provide valuable information to the field.
A natural consequence of the lead-time observations is the need for relevant intervention in ctDNA-positive patients. The first step should be additional and advanced clinical and radiological investigations, to confirm recurrence and, if possible, to identify the site of the recurrence. More advanced imaging, such as PET-based scans and/or artificial intelligence supported scan algorithms could add value in this situation. This may also prompt into a decision model, whether a local treatment strategy and/or systemic treatment should be the therapeutic consequence. However, in the case of no radiographic findings, the use of systemic treatment to eliminate ctDNA is also yet to be established.
In case of MRD, several studies have been designed to analyze the value of postoperative adjuvant chemotherapy in ctDNA-positive patients,18 and data have shown successful ctDNA clearance in a fraction of patients during treatment.16,19 Whether a ctDNA clearance is a true elimination or merely results in postponed tumor activity is also yet to be elucidated, but data are encouraging.
Finally, it is of high importance to liaise with patients’ representatives in the design of ctDNA-based studies and to analyze the consequences in quality of life and fear of cancer aspects in such complex new clinical approaches. In addition, cost–benefit analysis should be performed when possible.
In conclusion, ctDNA analysis has shown immense potential in this disease and observational studies on lead-time aspects all point in the same direction toward a high sensitivity for early detection of recurrences. The use of consecutive blood sampling is attractive from both a patient and physicians’ perspective and could contribute to better and more individualized follow-up strategies. However, the true clinical utility will only be established from carefully designed prospective observational studies and subsequent randomized trials.
Acknowledgments
None.
Footnotes
ORCID iD: Louise Bach Callesen  https://orcid.org/0000-0003-2799-1492
https://orcid.org/0000-0003-2799-1492
Contributor Information
Karen-Lise Garm Spindler, Department of Oncology, Research 1, Aarhus University Hospital, Palle Juhl Jensens Boulevard 99, Århus N, 8200, Denmark.
Louise Bach Callesen, Department of Oncology, Aarhus University Hospital, Denmark and Department of Clinical Medicine, Aarhus University, Denmark.
Dirk Arnold, Department of Oncology and Hematology, Asklepios Tumourzentrum Hamburg, AK Altona, Hamburg, Germany.
Declarations
Ethics approval and consent to participate: Not applicable.
Consent for publication: Not applicable.
Author contribution(s): Karen-Lise Garm Spindler: Conceptualization; Investigation; Methodology; Supervision; Writing – original draft; Writing – review & editing.
Louise Bach Callesen: Conceptualization; Investigation; Methodology; Writing – review & editing.
Dirk Arnold: Conceptualization; Methodology; Supervision; Writing – review & editing.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
The authors declare that there is no conflict of interest.
Availability of data and material: All data/statements in this perspective are either referenced in the text or the opinion of the authors. The data that support the findings/statements in this perspective are openly available in the references provided.
References
- 1. Callesen LB, Takacova T, Hamfjord J, et al. Circulating DNA in patients undergoing loco-regional treatment of colorectal cancer metastases: a systematic review and meta-analysis. Ther Adv Med Oncol 2022; 14: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Faulkner LG, Howells LM, Pepper C, et al. The utility of ctDNA in detecting minimal residual disease following curative surgery in colorectal cancer: a systematic review and meta-analysis. Br J Cancer Epub ahead of print 2022. DOI: 10.1038/s41416-022-02017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Garcia-Murillas I, Schiavon G, Weigelt B, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015; 7: 302ra133. [DOI] [PubMed] [Google Scholar]
- 4. Reinert T, Schøler L V., Thomsen R, et al. . Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016; 65: 625–634. [DOI] [PubMed] [Google Scholar]
- 5. Callesen LB, Hamfjord J, Boysen AK, et al. Circulating tumour DNA and its clinical utility in predicting treatment response or survival in patients with metastatic colorectal cancer: a systematic review and meta-analysis. Br J Cancer 2022; 127: 500–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yamada T, Iwai T, Takahashi G, et al. Utility of KRAS mutation detection using circulating cell-free DNA from patients with colorectal cancer. Cancer Sci 2016; 107: 936–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Scholer L V, Reinert T, Orntoft MW, et al. Clinical implications of monitoring circulating tumor DNA in patients with colorectal cancer. Clin Cancer Res 2017; 23: 5437–5445. [DOI] [PubMed] [Google Scholar]
- 8. Beagan JJ, Sluiter NR, Bach S, et al. Circulating tumor DNA as a preoperative marker of recurrence in patients with peritoneal metastases of colorectal cancer: a clinical feasibility study. J Clin Med 2020; 9: 1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tie J, Wang Y, Cohen J, et al. Circulating tumor DNA dynamics and recurrence risk in patients undergoing curative intent resection of colorectal cancer liver metastases: a prospective cohort study. PLoS Med 2021; 18: e1003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bolhuis K, van ’t Erve I, Mijnals C, et al. Postoperative circulating tumour DNA is associated with pathologic response and recurrence-free survival after resection of colorectal cancer liver metastases. EBioMedicine 2021; 70: 103498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parikh AR, Van Seventer EE, Siravegna G, et al. Minimal residual disease detection using a plasma-only circulating tumor DNA assay in patients with colorectal cancer. Clin Cancer Res 2021; 27: 5586–5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reinert T, Petersen LMS, Henriksen TV, et al. Circulating tumor DNA for prognosis assessment and postoperative management after curative-intent resection of colorectal liver metastases. Int J Cancer 2022; 150: 1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Øgaard N, Reinert T, Henriksen T V, et al. Tumour-agnostic circulating tumour DNA analysis for improved recurrence surveillance after resection of colorectal liver metastases: a prospective cohort study. Eur J Cancer 2022; 163: 163–176. [DOI] [PubMed] [Google Scholar]
- 14. Tarazona N, Gimeno-Valiente F, Gambardella V, et al. Targeted next-generation sequencing of circulating-tumor DNA for tracking minimal residual disease in localized colon cancer. Ann Oncol 2019; 30: 1804–1812. [DOI] [PubMed] [Google Scholar]
- 15. Henriksen T V, Tarazona N, Frydendahl A, et al. Circulating tumor DNA in stage III colorectal cancer, beyond minimal residual disease detection, toward assessment of adjuvant therapy efficacy and clinical behavior of recurrences. Clin Cancer Res 2022; 28: 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 2016; 8: 346ra92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nors J, Henriksen TV, Gotschalck KA, et al. IMPROVE-IT2: implementing noninvasive circulating tumor DNA analysis to optimize the operative and postoperative treatment for patients with colorectal cancer - intervention trial 2. Study protocol. Acta Oncologica 2020; 59: 336–341. [DOI] [PubMed] [Google Scholar]
- 18. Masfarré L, Vidal J, Fernández-Rodríguez C, et al. CtDNA to guide adjuvant therapy in localized colorectal cancer (CRC). Cancers (Basel) 2021; 13: 2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tie J, Cohen JD, Wang Y, et al. Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol 2019; 5: 1710–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]