Abstract
Single-pass transmembrane receptors (SPTMRs) represent a diverse group of integral membrane proteins that are involved in many essential cellular processes, including signal transduction, cell adhesion and transmembrane transport of materials. Dysregulation of the SPTMRs is linked with many human diseases. Despite extensive efforts in the past decades, the mechanisms of action of the SPTMRs remain incompletely understood. One major hurdle is the lack of structures of the full-length SPTMRs in different functional states. Such structural information is difficult to obtain by traditional structural biology methods such as X-ray crystallography and nuclear magnetic resonance (NMR). The recent rapid development of single-particle cryo-electron microscopy (cryo-EM) has led to exponential surge in the number of high-resolution structures of integral membrane proteins, including SPTMRs. Cryo-EM structures of SPTMRs solved in the past few years have tremendously improved our understanding of how SPTMRs function. In this review, we will highlight these progresses in the structural studies of SPTMRs by single-particle cryo-EM, analyze important structural details of each protein involved, and discuss their implications on the underlying mechanisms. Finally, we also briefly discuss remaining challenges and exciting opportunities in the field.
1. Introduction
Membrane proteins play essential roles in a wide variety of cellular functions in all kingdoms of life1-3. They account for approximately one-third of all proteins identified in eukaryotes and prokaryotes proteomes and more than half of available drug targets4,5. One important type of membrane proteins is single-pass transmembrane (TM) receptors (abbreviated as SPTMRs hereafter)5, which typically contain an extracellular region that responds to external stimuli, one transmembrane helix and a cytosolic region that relays the external signal to intracellular downstream pathways. SPTMRs represent the most abundant and diverse group of integral membrane proteins6, with about 1300 such proteins in the human proteome (encoded by ~6% of genes in the human genome)7,8. These proteins exhibit remarkable structural and functional diversity and contribute to many cellular processes. Dysregulation of SPTMRs is associated with numerous human diseases, such as arteriosclerosis, diabetes, and cancer7,9.
SPTMRs typically comprise three domains: the extracellular domain (ECD), the single-pass transmembrane domain (TMD), and the intracellular domain (ICD). For SPTMRs involved in cell signaling, the external stimuli are first sensed by the ECD, and the signal is then transmitted to the ICD through the membrane embedded TMD. In general, SPTMRs respond to extracellular stimuli through oligomerization and/or conformational changes10-12. The most well-known mechanism for SPTMR activation is ligand-induced dimerization, where unliganded receptors are in monomeric apo-state, and ligand binding induces receptor dimerization and activation13-15. The activated receptor is able to function as a signaling hub that recruits cytosolic effector proteins, and thereby triggering the downstream signaling cascade, such as PI3K/AKT and MAPK/ERK pathways16-18. Unliganded SPTMRs may exist as autoinhibited dimers on the cell membrane, as exemplified by the two members of insulin receptor family (insulin receptor (IR) and insulin-like growth factor 1 receptor (IGF1R))19-23. In these cases, ligand binding induces dramatic conformational changes to ECD, which triggers rearrangement and activation of ICDs24. Many receptors form higher-order oligomers upon ligand binding, such as B-cell receptor, Eph receptors, EGFR and integrins25-29. The clustering plays a critical role in regulating the activities of the receptors, by driving the fully active state and/or increasing the local concentrations of the receptors and downstream signaling proteins at the membrane30. These mechanistic insights were mostly derived from the “divide and conquer” approach in the past, in which the structures of ECDs, TMDs and ICDs solved separately by X-ray crystallography or NMR were combined to infer the structure and mechanisms of intact receptors31. This approach, however, cannot fully address exactly how signal across the cell membrane is controlled by the structural coupling among the ECD, TMD and ICD. Answering this key question requires structural analyses of larger pieces of SPTMRs, and ultimately full-length SPTMRs as well as the complexes with their binding partners in the context of lipid membrane environment. These analyses could provide complete views of the proteins in question and directly elucidate the mechanisms. Even when complete views of SPTMRs cannot be obtained as a result of their structural flexibility, partial structures from intact protein samples are expected to be more informative because the resolved parts are not taken out of the context of the entire proteins or protein complexes. Large multi-domain SPTMRs and their complexes are however notoriously difficult to study with traditional structural approaches, due to technical difficulties such as low expression yield, large size, high conformational and compositional heterogeneity.
Recent hardware and software advances have ushered in the “resolution revolution” in single-particle cryo-electron microscopy (cryo-EM)32-40. As a result, there has been an exponential growth in the number of high-resolution membrane protein structures in the past few years, leading to tremendous expansion of our understandings of the functions and underlying mechanisms of these membrane proteins41-51. Single-particle cryo-EM also opens up new opportunities for the structural investigation of SPTMRs, due to the following reasons52,53: (1) as compared to X-ray crystallography, single-particle cryo-EM requires much less sample, can tolerate certain degrees of impurity, and more importantly does not need protein crystallization, which together greatly reduce the difficulty in sample preparation; (2) single-particle cryo-EM has no upper mass limit of target proteins, enabling structural investigation of large oligomers of SPTMRs; (3) the 3D classification tools in single-particle cryo-EM are capable of separating different conformational states in the sample, which allows dealing with structural heterogeneity. Cryo-EM structural analyses of SPTMRs have also benefited from many new methods that enhance sample preparation and image processing, including various lipid membrane mimetics for stabilizing the TMD54,55, antibodies and Fab fragments for reducing conformational heterogeneity and facilitating image alignment56, and various coated cryo-EM grids for enriching protein particles and alleviating exposure to air/water interface which could cause damage to proteins57. Table 1 summarizes the remarkable progress in structural studies of SPTMRs driven by the “resolution revolution” in cryo-EM in the past few years. These high-resolution structures provide unprecedented details into the molecular interactions that control the activity of SPTMRs.
Table 1.
| Receptor Family |
SPTMR | Protein Complex | Organism | Domainc | PDB ID | Year | Resolution (Å) |
|---|---|---|---|---|---|---|---|
| Receptor Tyrosine Kinase | IR | IR/insulin (WT) | Homo sapiens | FL | 6PXV | 2019 | 3.2 |
| IR | IR/insulin (WT) | Homo sapiens | ECD | 6SOF | 2020 | 4.3 | |
| IR | IR/insulin (WT) | Homo sapiens | ECD | 6HN5 | 2018 | 3.2 | |
| IR | IR/insulin (A-V3E) | Mus musculus | FL | 7SL1 | 2022 | 3.4 | |
| IR | IR/insulin (A-L13R) symmetric | Mus musculus | FL | 7SL3 | 2022 | 3.4 | |
| IR | IR/insulin (A-L13R) asymmetric | Mus musculus | FL | 7SL2 | 2022 | 3.6 | |
| IR | IR/insulin (B-L17R) symmetric | Mus musculus | FL | 7SL6 | 2022 | 3.7 | |
| IR | IR/insulin (B-L17R) asymmetric | Mus musculus | FL | 7SL4 | 2022 | 5.0 | |
| IR | IR/insulin (A-V3E/A-L13R) | Mus musculus | FL | 7SL7 | 2022 | 3.1 | |
| IR | IR/insulin (WT) unsaturated symmetric | Mus musculus | FL | 7STH | 2022 | 3.5 | |
| IR | IR/insulin (WT) unsaturated asymmetric | Mus musculus | FL | 7STI | 2022 | 4.9 | |
| IR | IR/insulin (WT) unsaturated asymmetric | Mus musculus | FL | 7STJ | 2022 | 4.4 | |
| IGF1R | IGF1R/IGF1 | Homo sapiens | FL | 6PYH | 2019 | 4.3 | |
| IGF1R | IGF1R IGF1 | Homo sapiens | FL | 6JK8 | 2019 | 5.0 | |
| IGF1R | IGF1R IGF2 | Homo sapiens | ECD | 6VWG | 2020 | 3.2 | |
| c-Met | c-Met/NK1 | Homo sapiens | ECD | 7MOB | 2021 | 5.0 | |
| c-Met | c-Met/HGF | Homo sapiens | ECD | 7MO9 | 2021 | 4.0 | |
| RET | RET/GDF15/GFRAL | Homo sapiens | ECD | 6Q2J | 2019 | 4.1 | |
| RET | RET/GDNF/GFRα1 | Homo sapiens | ECD | 6Q2N | 2019 | 4.4 | |
| RET | RET/NRTN/GFRα2 | Homo sapiens | ECD | 6Q2R | 2019 | 4.3 | |
| RET | RET/ARTN/GFRα3 | Homo sapiens | ECD | 6Q2S | 2019 | 3.8 | |
| RET | RET/NRTN/GFRα2 | Homo sapiens | ECD | 6GL7 | 2019 | 6.3 | |
| RET | RET/GDNF/GFRα1 | Danio rerio | ECD | 7AML | 2021 | 3.5 | |
| HER2 | HER2/Trastuzumab/ Pertuzumab | Homo sapiens | ECD | 6OGE | 2019 | 4.4 | |
| HER2 HER3 | HER2 (WT)/HER3 /NRG1β | Homo sapiens | FL | 7MN5 | 2021 | 2.9 | |
| HER2 HER3 | HER2 (S310F)/HER3 /NRG1β | Homo sapiens | FL | 7MN6 | 2021 | 3.1 | |
| HER2 HER3 | HER2/HER3/NRG1β/ Trastuzumab | Homo sapiens | FL | 7MN8 | 2021 | 3.5 | |
| EGFR | EGFR (WT)/EGF juxtaposed | Homo sapiens | FL | 7SYD | 2021 | 3.1 | |
| EGFR | EGFR (WT)/EGF separated | Homo sapiens | FL | 7SYE | 2021 | 3.3 | |
| EGFR | EGFR (WT)/TGF-α juxtaposed | Homo sapiens | FL | 7SZ7 | 2021 | 3.4 | |
| EGFR | EGFR (WT)/TGF-α separated | Homo sapiens | FL | 7SZ5 | 2021 | 3.6 | |
| EGFR | EGFR(L834R)/EGF juxtaposed | Homo sapiens | FL | 7SZ0 | 2021 | 3.3 | |
| EGFR | EGFR(L834R)/EGF separated | Homo sapiens | FL | 7SZ1 | 2021 | 3.3 | |
| Plexin | PlexinA4 | Sema3A/PlexinA4/Nrp1 | Mus musculus | ECD | 7M0R | 2021 | 3.7 |
| PlexinC1 | PlexinC1/A39R | Homo sapiens | ECD | 6VXK | 2020 | 3.1 | |
| Integrin | Integrin αvβ8 | αvβ8 | Homo sapiens | ECD | 6DJP | 2018 | 4.8 |
| Integrin αvβ8 | αvβ8/TGF-β1 | Homo sapiens | ECD | 6UJA | 2020 | 3.3 | |
| Integrin α5β1 | α5β1/fibronectin | Homo sapiens | FL | 7NWL | 2021 | 3.1 | |
| Integrin α5β1 | α5β1 | Homo sapiens | FL | 7NXD | 2021 | 4.6 | |
| Toll-like receptor | TLR3 | TLR3/UNC93B1 | Homo sapiens | FLd | 7C76 | 2021 | 3.4 |
| TLR3 | TLR3/UNC93B1 | Mus musculus | FLd | 7C77 | 2021 | 3.3 | |
| TLR7 | TLR7/UNC93B1 | Homo sapiens | FLd | 7CYN | 2021 | 4.2 | |
| TLR7 | TLR7/Cpd-7 | Macaca mulatta | ECD | 6LW1 | 2020 | 2.8 | |
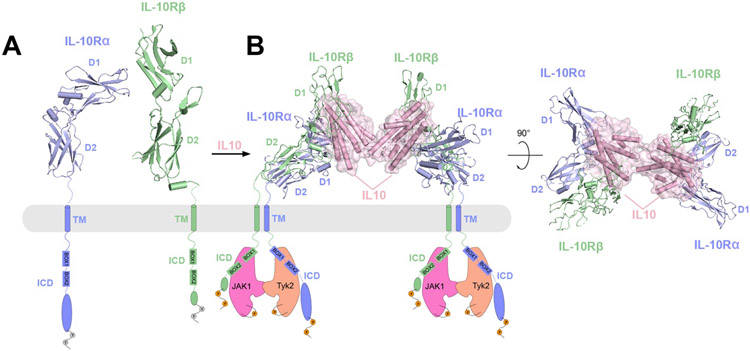
| Cytokine receptor | IL-10Rα IL-10Rβ |
IL-10Rα/IL-10Rβ/IL-10 | Homo sapiens | ECD | 6X93 | 2021 | 3.5 |
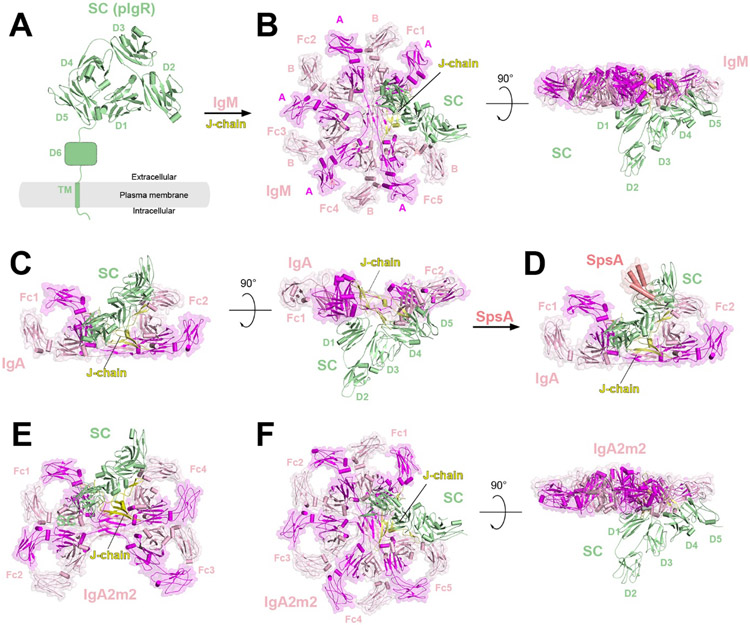
| Fc Receptor | PIgR | IgM/J-chain/SC | Homo sapiens | SC | 6KXS | 2020 | 3.4 |
| PIgR | IgM/J-chain/SC | Homo sapiens | SC | 7K0C | 2021 | 3.3 | |
| PIgR | IgA/J-chain/SC | Homo sapiens | SC | 6LX3 | 2020 | 3.2 | |
| PIgR | IgA/J-chain/SC/SpsA | Homo sapiens | SC | 6LXW | 2020 | 3.3 | |
| PIgR | IgA2M2/J-chain/SC | Homo sapiens | SC | 6UE7 | 2020 | 2.9 | |
| PIgR | IgA2M2/J-chain/SC | Homo sapiens | SC | 6UE8 | 2020 | 3.0 | |
| PIgR | IgA2M2/J-chain/SC | Homo sapiens | SC | 6UEA | 2020 | 3.0 | |
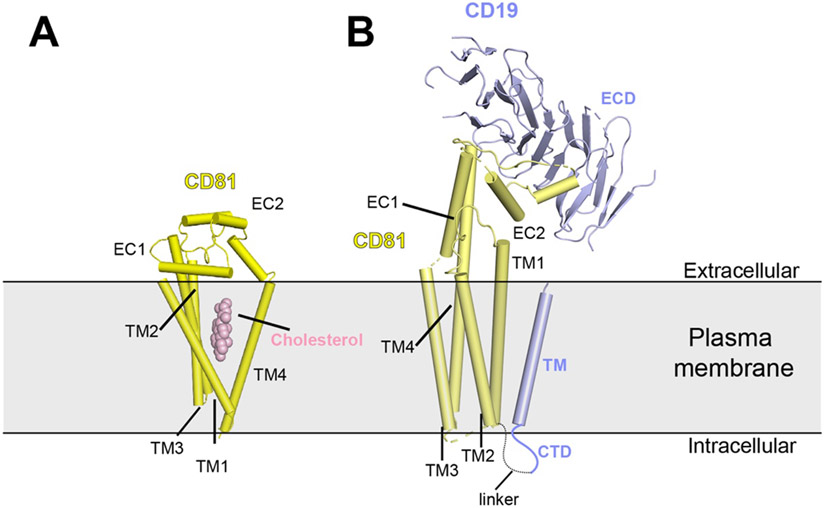
| B cell Coreceptor | CD19 | CD19/CD81 | Homo sapiens | ECD+ TMDd |
7JIC | 2021 | 3.8 |
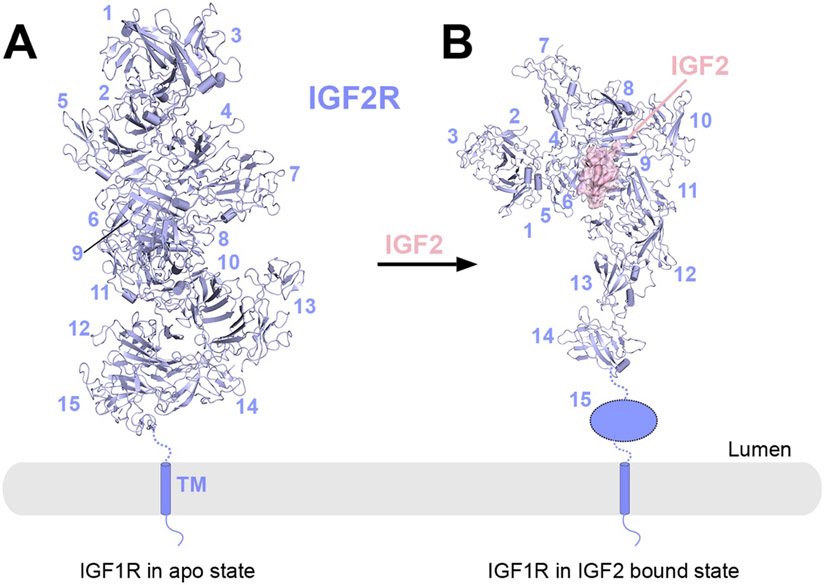
| IGF2 Receptor | IGF2R | IGF2R | Bos taurus | FL | 6UM1 | 2020 | 3.5 |
| IGF2R | IGF2R/IGF2 | Bos taurus | FL | 6UM2 | 2020 | 4.3 | |
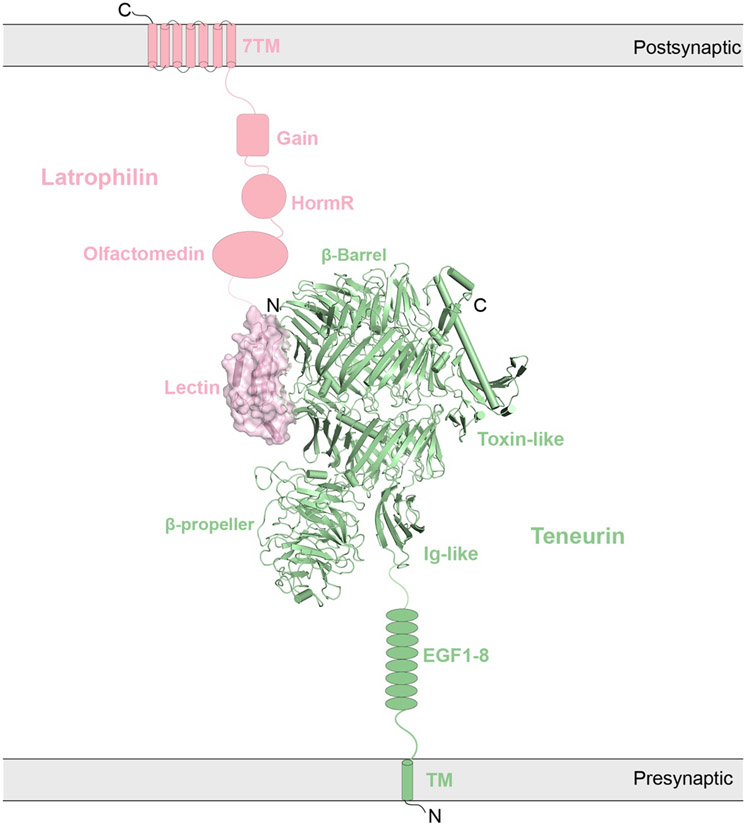
| Cell Adhesion receptor | Teneurin | Teneurin/Latrophilin | Homo sapiens | ECD | 6VHH | 2020 | 3.0 |
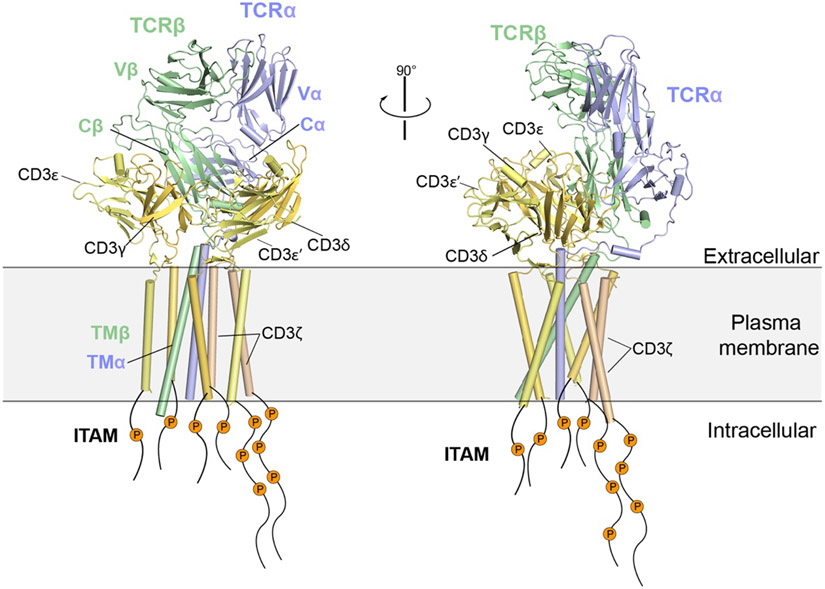
| T-cell receptor | TCR | TCR/CD3 | Homo sapiens | FLd | 6JXR | 2019 | 3.7 |
| TCR | TCR/CD3/cholesterol | Homo sapiens | FLd | 7FJF | 2022 | 3.2 |
This table was last updated in April, 2022.
Abbreviations: SPTMR, single pass transmembrane receptor; FL, full-length; ECD, extracellular domain; SC, secretory component; TMD, transmembrane; WT, wild-type.
The domain of the constructs of receptors used in the studies. e.g. FL indicates full-length constructs of receptors were used for structure determination.
Indicates the structures in which the TMDs of the receptors are resolved. In other structures where FL constructs were used for structure determination, TMDs were not resolved due to missing densities.
The literature of SPTMRs is vast and virtually impossible to be comprehensively reviewed in one article. This review is focused on the recent high-resolution cryo-EM structures of SPTMRs, especially those involved in transmembrane signal transduction, including receptor tyrosine kinases (RTKs), plexins, integrins, Toll-like receptors, Fc receptors, cytokine receptors, IGF2R receptor, cell adhesion receptors and T cell receptor. The purpose of this review is to provide a summary of the most recent progress in the field made possible by cryo-EM. Crystal structures, NMR structures and low-resolution cryo-EM structures that are of insufficient quality for model building are therefore not discussed. We will briefly describe the cryo-EM structures, but emphasize the mechanistic insights emerged from the structures. In addition, we propose the future research directions by putting forward the challenges and perspectives of this field.
2. Receptor tyrosine kinases
2.1. Introduction
Receptor tyrosine kinases (RTKs) represent a large family of SPTMRs for which more than 58 different members and 20 sub-families have been identified in humans17. RTKs play key roles in regulating normal cellular processes, such as cell proliferation, differentiation, and metabolism. Dysregulation of RTKs is linked to many diseases, including cancer and diabetes58. Therefore, RTKs are important therapeutic targets for these diseases58-60. RTKs share a common overall domain structure that contains an extracellular region (ECD) that binds ligands, a single-pass transmembrane helix (TM), and an intracellular domain (ICD) that includes the tyrosine kinase domain necessary for intracellular signaling61. Distinct to GPCR whose ligands are normally small molecules or peptides62, the ligands for RTK are folded proteins. In general, the ligand binding to the ECD of RTKs induces receptor dimerization, which subsequently leads to lateral TM-TM interactions and trans-autophosphorylation between two intracellular kinase domains in the receptor dimer. Following receptor activation, the phosphorylated C-terminal tail of the kinase domains recruits cytoplasmic effector proteins and thereby initiates the downstream signaling. For many RTKs, such as PDGFR, VEGFR, TrkA and Tie, their ligands form stable homodimers63-66. The dimeric ligand brings two receptor molecules to close proximity in a symmetric manner to promote receptor activation. The basis for such activation mechanism has been elucidated by crystal structures of many 2:2 symmetric receptor/ligand complexes. This ligand-induced dimerization model is a common mechanism for RTK activation17,67,68.
Although the common mechanism of RTK activation that involves ligand-induced dimerization has been revealed in detail, the working mechanisms for several RTKs remain poorly understood due to the lack of structural information. Solving the cryo-EM structures of different members of RTK family in the ligand-bound functional conformation would provide fundamental insights into the general mechanisms of RTK activation, and more importantly, reveal the diversification in the activation mechanisms of RTKs. Given that virtually all RTK family members have been linked to development and progression of cancer, these understandings could lead to new strategies to selectively inhibit/modulate individual pathways for cancer therapy or concurrently target multiple pathways which have been suggested to improve therapeutic effectiveness58.
Structural information on full-length RTKs, either in their apo- or ligand-bound multimeric states, has been elusive because these elongated and conformationally dynamic single-pass TM proteins are difficult to crystalize. Thus, the central question of how information is relayed between the extracellular and intracellular regions remains incompletely understood. Cryo-EM structures of full-length RTKs/ligands complex could reveal how ligand-induced conformational changes in the extracellular region leads to TM dimerization as well as conformational changes in the intracellular kinase domain that promote auto-phosphorylation and downstream signaling. The following sections summarize the recent progress in cryo-EM studies of several RTKs in the ligand bound, active states.
2.2. Full activation of insulin receptor requires the binding of multiple insulins to 2 distinct sites
Insulin receptor (IR), one of the most well-studied RTKs, plays essential roles in glucose metabolism and cell growth22,69. Dysregulation of IR signaling is linked to many human diseases, such as diabetes and cancers70,71. Structurally, each IR protomer consists of L1, Cysteine rich (CR), L2, FnIII-1, FnIII-2 and FnIII-3 domains in the ECD, a single-pass TM and the kinase domain in the intracellular region. Different from all other families of RTKs, IR forms a stable disulfide-linked dimer independent of ligand binding19,20,22,72. Previous studies have proposed that the insulin binding triggers large structural rearrangements of the IR dimer, resulting in optimal juxtaposition of the two kinase domains for efficient trans-autophosphorylation23,24,73. In addition, IR binds its ligand insulin with complex kinetics characterized by a curvilinear Scatchard plot, indicating the presence of two classes of binding sites with different affinities and/or negative cooperativity74-76. Such insulin binding result indicates that the IR activation may require the binding of multiple insulins to two distinct types of sites (sites 1 and 2)77. Notably, the residues in insulin involved in the binding to sites 1 and 2 have been extensively mapped by alanine scanning mutagenesis many years ago77,78.
In the past decades, X-ray crystallography has played a major role in revealing the molecular mechanism underlying the IR activation in response to insulin. The structure of the ECD of IR in the absence of insulin was solved first by X-ray crystallography in 2006, revealing an ‘Λ’-shaped architecture (Figure 1A, left panel)72. The crystal structure of the truncated version of the IR ECD (containing only L1 and CR domains of IR) bound to insulin was subsequently determined in 2013, showing how insulin engages site 1 of IR that consists of the L1 domain and the C-terminal helix of the α chain (α-CT)79. These structures represent major breakthroughs in the structure biology of IR. The next major advance came from cryo-EM. In 2018, the cryo-EM structures of the intact IR ECD in complex with insulin were determined by two different groups, at overall ~4 Å resolution80,81. These structures show that, upon the binding of two insulin molecules to the two copies of site 1 of the IR dimer, the overall architecture of the IR dimer is converted from the autoinhibited ‘Λ’-shape to the active ‘T’-shape. It has been further proposed that this large structural conversion brings the two kinase domains of the IR dimer into close proximity to enable efficient trans-autophosphorylation and activation. Nevertheless, none of these studies were able to identify site 2, leaving the open question why the activation of IR requires the binding of insulin to two types of sites. As insulin binding to full-length IR exhibits much stronger affinity relative to the isolated extracellular domain23,75, structural studies of the full-length IR in complex with insulin would be critical for capturing the fully liganded state of IR and identifying insulin binding site 2.
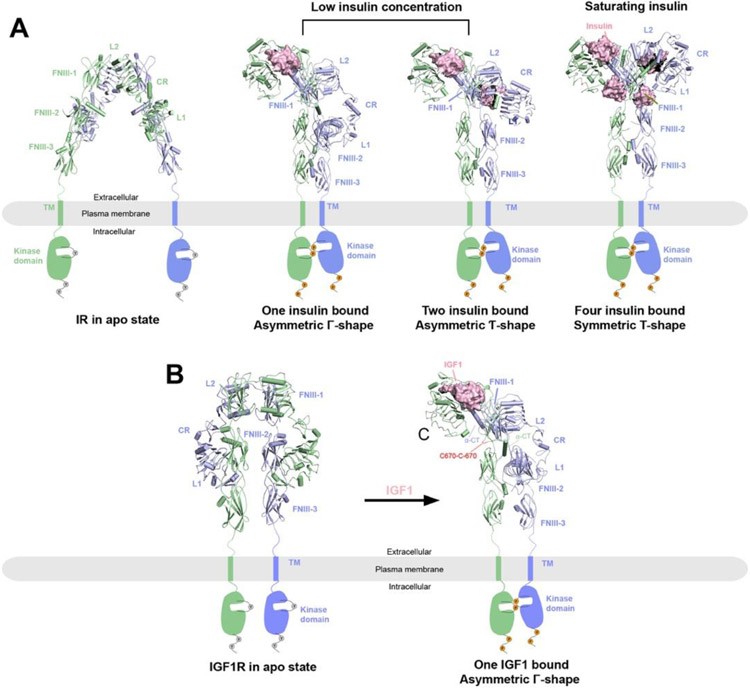
Figure 1.
Overall structures of IR and IGF1R in apo and ligand bounds states reveal activation mechanisms. The two protomers in the receptor dimers are shown in cartoon (blue and green) representations, whereas the ligands are shown in surface (pink) representations. The missing TM and kinase domains in the structures are indicated by schematic drawings. Binding of ligands (insulin or IGF1) leads to conformational changes to the ECD of the receptors, with subsequent dimerization of the TM and kinase domains and activation of the receptors as indicated by the phosphorylation of the kinase domains and C-terminal tails.
A) Structures of IR at different states. Left, IR in apo state as an auto-inhibited, Λ-shaped homodimer with lower domains separated (PDB: 4ZXB); middle, full-length 2:1 or 2:2 IR/insulin complexes as Γ- or Ƭ-shaped asymmetric dimers respectively at low insulin concentrations (PDBs: 7STI and 7STJ); right, full-length 2:2 IR/insulin complex as a T-shaped symmetric dimer at saturating insulin concentrations (PDB: 6PXV).
B) Left, crystal structure of apo-IGF1R in apo state as an auto-inhibited, Λ-shaped dimer (PDB: 5U8R). Right, cryo-EM structure of full-length 2:1 IGF1R/IGF1 complex (PDB: 6PYH) showing a Γ-shaped asymmetric dimer with two α-CT motifs covalently linked by disulfide bonds.
The cryo-EM structures of full-length IR/insulin complex at 3.2 Å resolution (PDB: 6PXV) as well as IR ECD bound with saturated insulins at 4.3 Å resolution (PDB: 6SOF) were reported in 2019 and 2020, respectively82,83 (Figure 1A, right panel). The dimerized TM domain of IR was partially resolved in the full-length IR/insulin complex, showing a crossover conformation82. This structural observation suggests the TM-TM interaction plays a role in stabilizing the active conformation of IR, which in part explains why the cryo-EM structure of full-length IR/insulin complex was resolved at higher resolution than that of the IR-ECD/insulin complex. However, the intracellular kinase was completely unresolved due to large structural dynamics. Intriguingly, these structures reveal that each ‘T’-shaped IR active dimer binds 4 insulins at four sites (site 1, 1’, 2 and 2’) that are related by two-fold symmetry (Figure 1A, right panel). Sites 1 is similar to that observed in the previous crystal structure of the truncated IR-L1-α-CT/insulin complex. However, in the ‘T’-shaped active conformation of the IR/insulin complex, insulin bound at L1/α-CT of one IR protomer (namely site 1a) also makes simultaneous contact with the top surface of the FnIII-1 domain of the second IR protomer (namely site 1b)82. With this binding mode, the site 1 insulins crosslink two IR protomers, thus stabilizing the active conformation of IR. Moreover, insulin binding to site 1 of IR can release the autoinhibited conformation by disrupting the inter-promoter L1/FnIII-2 interaction82,84. A comparison of the apo and active structures of IR further shows that, once the auto-inhibited IR dimer is disrupted by insulin site 1 binding, the two IR protomers are free to undergo both the intra-protomer hinge motion and inter-protomer rotation required for converting to the active ‘T’-shaped conformation. These structural rearrangements reduce the distance between the two intracellular kinase domains and thereby promote their autophosphorylation24,82.
Importantly, the cryo-EM structure of full-length IR/insulin complex for the first time revealed the binding mode between IR and the site 2 insulin82. The binding site for the site 2 insulin is located at the side surface of the FnIII-1 domain in IR. Insulin uses a total of 14 residues from both A and B chains to engage this binding site. Structure-based mutagenesis, binding and cell-based assays confirmed the functional importance of insulin binding at site 282. Nevertheless, this cryo-EM structure of IR at saturated insulin concentration cannot explain how insulin binding to sites 1 and 2 cooperatively promotes full activation of IR. Therefore, the activation mechanisms of IR are still incompletely understood.
2.3. Cryo-EM structures of IR bound with insulin site selective mutants or at unsaturated insulin concentrations
The cryo-EM structures of a series of full-length IR/insulin site selective mutants complexes were determined recently85. Intriguingly, these new structural data show that IR with insulin variants only bound at site 1 predominantly adopts an ‘Ƭ’-shaped asymmetric conformation (Figure 1A, middle panel). In this asymmetric IR dimer, one insulin is bound in the top part of the ‘Ƭ’, while another insulin is bound in the middle region of the ‘Ƭ’, simultaneously engaging the sites 1 and 2 from two neighboring protomers (namely hybrid site)85. In addition, the cryo-EM analyses of the full-length IR bound with unsaturated wild-type insulin also show that IR with only two insulins bound predominantly adopts an asymmetric conformation, similar to that of IR with insulin variants only bound at site 185,86.
These new cryo-EM results provide a structural explanation for the functional role of insulin site 2 binding. As described above, IR is trapped as ‘Ƭ’-shaped asymmetric conformation when only site 1 is bound with insulin (Figure 1A, middle panel). In the middle part of such asymmetric IR dimer, the site 1 bound insulin in one protomer also weakly contacts the site 2 of another protomer. At high insulin concentrations, the binding of another insulin to the site 2 in the middle part of asymmetric dimer requires the delocalization of the lower L1/α-CT domain to prevent the steric clashes between the two insulin molecules bound to sites 1 and 2, respectively. The bumping of two insulins in this hybrid site therefore promotes the lower L1/α-CT domains together with bound insulin to move upward to reach the top surface of the FnIII-1 domain. This structural rearrangement converts IR from asymmetric to symmetric, leading to the stable, ‘T’-shaped conformation. This explains why the binding of 4 insulins to both sites 1 and 2 is critical for the structural transition of IR from the ‘Ƭ’- to ‘T’-shape85.
2.4. Binding of one IGF1 molecule is sufficient to fully activate IGF1R.
Type 1 insulin-like growth factor receptor (IGF1R) plays an essential role in controlling cellular growth, proliferation, differentiation and migration, and also regulates the ageing process87-90. Aberrant IGF1R signaling causes a number of human diseases, including Laron's syndrome, acromegaly, and cancers91,92. Although IGF1R and IR are close relatives sharing high sequence and structural similarity, the two receptors have different ligand binding characteristics, indicating different regulatory mechanisms93.
The first IGF1-bound structure of IGF1R, determined by soaking crystals of IGF1R-ECD with IGF1, revealed that IGF1 binds site 1 of IGF1R in a manner similar to the binding of insulin to the site 1 of IR21. The crystal lattice, however, prevented IGF1 from inducing large conformational changes of IGF1R-ECD. Therefore, this crystal structure of the IGF1R-ECD/IGF1 complex may represent an intermediate state of IGF1R, rather than the active state.
In 2019, the first cryo-EM structure of the full-length IGF1R/IGF1 complex in the active state was solved at 4.3 Å resolution (PDB: 6PYH)94 (Figure 1B). Similar to the IR/insulin structure, the TM domains of IGF1R dimer were only resolved at ~8 Å resolution in the cryo-EM map, while the ICD was largely unresolved. Different from the 2:4 stoichiometry of the symmetric IR/insulin complex, only one IGF1 is bound at the site 1 of the IGF1R dimer, resulting an asymmetric active conformation of IGF1R. This 2:1 stoichiometry of the IGF1R/IGF1 structure is consistent with previous biochemical results showing that binding of one IGF1 molecule to site 1 of the IGF1R dimer hampers the binding of a second IGF1 to site 1’ (i.e., negative cooperativity)93,95. This structural model provides important insights into the activation mechanism of IGF1R: the binding of one IGF1 to IGF1R partially releases the structural constraints that stabilize the apo form of IGF1R84,94, and thereby triggers the conformational rearrangement in one half of the IGF1R dimeric complex. As a result, the overall conformation of IGF1R dimer is converted from a symmetric, auto-inhibited ‘Λ’-shape to an asymmetric, active ‘Γ’-shape (Figure 1B). During this structural conversion, the distance between two membrane proximal stalk regions of the IGF1R dimer becomes much shorter, which would facilitate intra-dimer autophosphorylation of the intracellular kinase domains and promote receptor activation.
Moreover, this cryo-EM work provides structural information regarding the source of negative cooperativity in the binding of IGF1 to IGF1R94. It has been well established that structural flexibility of α-CT in IGF1R is required for ligand binding21. In the apo structure of IGF1R, only the N-terminal part of α-CT is folded as a short α-helix21,96, whereas the rest of this motif is disordered. This partially folded α-CT does not have any structural constraints in either N- and C-termini, thus allowing for the conformational change of α-CT that is required for IGF1 binding. In contrast, two α-CT motifs in the active IGF1R dimer bound to one IGF1 are physically coupled because of the lengthening of the two α-CT helices and the tethering of their N-termini by multiple disulfide bonds. This specific structural configuration restricts the conformational change of the unliganded α-CT, preventing the binding of a second IGF1 molecule.
In 2020, the cryo-EM structure of a C-terminal leucine-zippered IGF1R-ECD in complex with IGF2 was determined at overall 4 Å resolution (PDB: 6VWG), showing a similar asymmetric conformation as observed in the structure of full-length IGF1R/IGF1 complex97. The major difference between these two structures is the conformation and role of the C-domain loop of the ligand in receptor binding. In the active state of the IGF1R/IGF1 complex, the C-domain loop of IGF1 adopts an extended conformation that contacts L1, CR and L2 domains of IGF1R. By contrast, the C-domain loop in IGF2 is shorter and thus cannot reach the CRD. Therefore, the C-domain loop of IGF2 only makes contacts with L1 and L2 domains of IGF1R, providing a structural explanation for the weaker affinity between IGF1R and IGF2 than that between IGF1R and IGF1. In addition, the conformation of C-domain loop of IGF2 is stabilized by the intra-molecular interaction between its C-terminus and the N-terminal region of its A domain. The C-terminus of the C-domain loop of IGF1, however, does not contact its A domain, and thus is flexible.
Notably, the activation mechanism of IGF1R is significantly different from the structurally related RTK – IR (Figure 1). Firstly, the binding of multiple ligands to two distinct sites is required for the full activation of IR82,85. In sharp contrast, one IGF1 binding is sufficient to break the autoinhibited apo-IGF1R dimer and promote its conformational change for activation. This 2:1 stoichiometry is ensured by the negative cooperativity in the binding of IGF1 to IGF1R. Secondly, IR and IGF1R utilize different strategies to position the membrane proximal domains for efficient kinase auto-phosphorylation. In the active state of IR, the proximity of the stalk regions of IR dimer that comprise FnIII-2 and -3 domains is achieved by the weak homotypic interactions between two long loops in FnIII-2 domains82. In the active state of IGF1R, however, the unliganded L1 domain bridges the two stalks of IGF1R dimer into close proximity and further stabilizes the active conformation94. It is possible that the differences in the arrangements of membrane proximal stalk regions between IR and IGF1R may provide a mechanism for defining their signaling strength and specificity, allowing these two closely related receptors to generate distinct signaling outcomes.
2.5. Activation of RET requires both ligand and co-receptor
RET (rearranged during transfection) is a RTK that plays essential roles in controlling the development of the kidney and nervous system98,99. RET with gain-of-function mutations is associated with multiple endocrine neoplasia type 2 (MEN2)100,101, while RET with loss-of-functions mutation is linked to Hirschsprung’s disease102-104. RET is among the few RTKs that require not only their cognate ligands but also co-receptors for activation105 (another example is MuSK106,107). Glia cell-derived growth factor (GDNF), Neurturin (NRTN), Artemin (ARTN) and Persephin are four ligands that induce dimerization of RET through assembling 2:2:2 ternary complexes with RET and the co-receptors GDNF receptor-α family proteins (GFRα1–4)102,108-111. In 2017, four pharmaceutical companies independently identified GDF15 and GFRAL as a novel ligand/co-receptor pair that activates RET in the central nervous system, leading to loss of appetite and other aversive responses112-115. These discoveries explain the role of GDF15 in body-weight regulation, hyperemesis gravidarum (server nausea and vomiting caused by pregnancy) and cancer-induced cachexia, suggesting new potential therapeutic targets for treating these conditions116,117.
Both RET and the co-receptors are expressed on the cell surface and use their ECDs to interact with the ligands. The extracellular region of RET contains four atypical cadherin-like domains (CLD1-4) followed by a cysteine-rich domain (CRD), which have all been implicated in interacting with ligands or co-receptors118,119. The CLD1 and CLD2 domains of RET can form a stable dimer on its own, as suggested by X-ray crystallographic studies120. However, this dimeric conformation of RET may represent its auto-inhibited state, as the two kinases of RET are separated by long distance in such dimer (Figure 2A). In 2014, the overall batwing-like shape of the intact RET extracellular region in complex with GDNF and GFRα1 was revealed by negative-stain EM to 24 Å resolution118. This structure, however, does not clearly resolve the individual domains in RET and their interactions with GDNF and GFRα1.
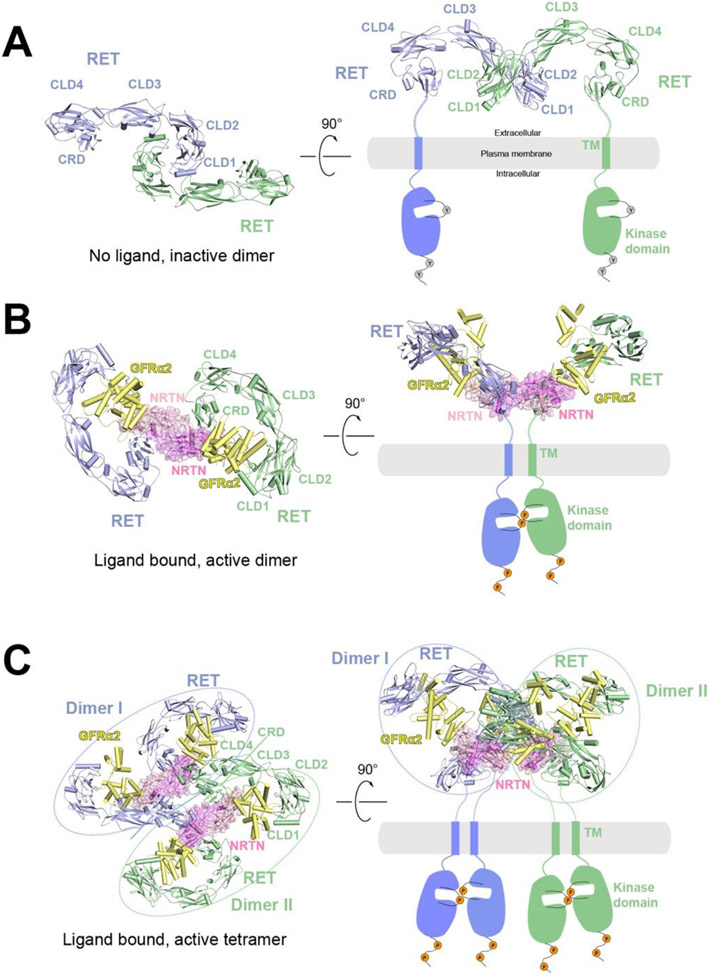
Figure 2.
Overall structures of RET at different oligomerization states and proposed activation mechanisms. The protomers in the receptor dimer or tetramer (blue and green) as well as the co-receptor (yellow) are shown in cartoon representations, whereas the ligands are shown in cartoon and surface (pink) representations with semi-transparency. The TM and kinase domains, not present in the structure, are indicated by schematic drawings.
A) Structural model of RET in apo state as an inactive dimer in two different views. The structural model is generated based on the crystal structure of CLD1/CLD2 domains (PDB: 2X2U). In this structural model, the two CRD domains are far apart, suggesting an autoinhibited state.
B) Cryo-EM structure of the 2:2:2 NRTN/GFRα2/RET complex in two different views (PDB: 6Q2O). In this structure, the two CRD domains are close to each other, consistent with being an active dimer.
C) Cryo-EM structure of the 4:4:4 NRTN/GFRα2/RET complex in two different views (PDB: 6Q2R). Dimer I (blue) and Dimer II (green) are structurally identical to the 2:2:2 NRTN/GFRα2/RET complex.
In 2019 and 2021, multiple cryo-EM structures of human and zebrafish RET ternary extracellular complexes—GDF15/GFRAL/RET, GDNF/GFRα1/RET, NRTN/GFRα2/RET and ARTN/GFRα3/RET in their active states were determined at 3 - 5 Å resolution using single particle cryo-EM (PDB: 6Q2J, 6Q2N, 6Q2R, 6Q2S, 6GL7, 7AMK, 7AB8 and 7AML)121-123 (Figure 2B). The symmetry expansion method124,125 together with focused refinement were utilized to further improve the resolution of each half of the 2:2:2 complex, facilitating accurate model building. In symmetry expansion, a duplicate set of the aligned particles were rotated along the 2-fold symmetry axis and combined with the original set. The following 3D refinement was focused on one half of the complex, eliminating the detrimental effect on resolution caused by relative motions between the two halves of the complex. These structures reveal the interfaces formed between RET and the different ligands/co-receptors in detail. In particular, in each half of the 2:2:2 RET signaling complex, the ligand is sandwiched between the co-receptor and RET, forming a stable 1:1:1 oval-shaped complex. This structural feature explains why the engagement of co-receptor is critical for the high affinity binding between RET and its ligands. In all the ternary RET complexes, the batwing shape is arranged to have the two CRDs from the two RET molecules placed into close proximity, facilitating the dimerization and activation of the intracellular kinase domain. However, the angle between the two wings shows substantial differences among these ternary complexes121. Therefore, different ligands and co-receptors may lead to different juxtaposition of the TM and intracellular kinase domain in the active dimer of RET, which may underlie different signaling patterns of RET induced by different ligands/co-receptors in vivo.
In addition, cryo-EM analyses from Li et. al’s work revealed that the 2:2:2 NRTN/GFRα2/RET complex can further pack into a 4:4:4 tetrameric assembly121 (Figure 2C). The cell-based assays suggest that the formation of the 4:4:4 NRTN/GFRα2/RET complex delays RET endocytosis, which may lead to more intense and sustained signaling. Previous studies have shown that RET endocytosis regulates both the duration and choice of pathway of signaling126. Therefore, the formation of high-order RET complexes provides an additional mechanism for regulating both the strength and pathway of RET signaling. Notably, this is the first example showing the structure of a high-order RTK complex with near-atomic details (overall resolution 4.3 Å). Further evidence of high-order complex formation of RET is provided in the work by Adams et. al, which shows that GDNF/GFRα1/RET can form even larger one-dimension array on the cryo-EM grid through side-by-side packing123. Together, these results reveal the common activation mechanism of RET in detail as well as the unique features in the ternary complexes formed by the different ligand/co-receptor pairs, paving the way for designing therapeutics targeting RET to treat diseases such as obesity and cancer.
2.6. One HGF, using two distinct interfaces, recruits two c-MET receptors for activation
The RTK c-MET (mesenchymal epithelial transition factor) plays essential role in controlling numerous cellular processes such as cell proliferation, survival, motility, which are critical for morphogenesis in development127,128. In adults, c-MET signaling is critical for the regeneration of liver, skin and heart, as well as wound repair127,129. Aberrant c-MET signaling causes several human diseases, including a variety of cancers, making it an important target for the development of inhibitors/antibodies for cancer therapy130-134. c-MET can be activated by its cognate ligand HGF (hepatocyte growth factor) as well as NK1, a naturally occurring splicing variant of HGF that only contains the N-terminal and the first kringle domain (K1 domain)135,136. Structurally, c-MET receptor is composed of a Semaphorin (SEMA) domain, a plexin-semaphorin-integrin (PSI) domain and four consecutive immunoglobulin-plexin-transcription (IPT1-4) domains in the ECD, a single TM, and an intracellular kinase domain137 (Figure 3A). HGF is a multi-domain protein, remarkedly larger than most other ligands of RTKs138. It is secreted as a single chain 83 kDa precursor protein containing a N-terminal domain, four consecutive kringle (K1–K4) domains, and a serine protease homology (SPH) domain. Proteolytic cleavage between Arg494 and Val495 of HGF generates the α and β subunits, which are linked by a disulfide bond138. The interaction between the SPH domain of HGF and c-MET has been well characterized by X-ray crystallography, showing HGF-SPH makes close contacts with the bottom surface of c-MET-SEMA139. Furthermore, it has been shown that glycosaminoglycan, such as heparin, can promote HGF induced c-MET activation through enhancing the binding affinity between HGF and c-MET140-142.
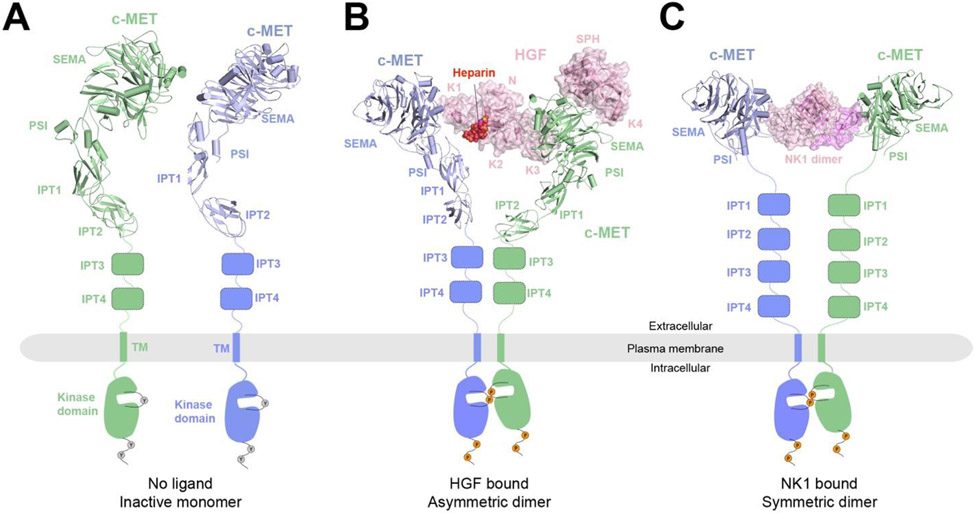
Figure 3.
Overall structures of c-MET in the apo and different ligand-bounds states and the proposed activation mechanisms. The protomers in the receptor dimer (blue and green) are shown in cartoon representations, whereas the ligands are shown in cartoon and surface (pink) representations with semi-transparency. The domains not present in the structures are represented by schematic drawings. Binding of either HGF or the NK1 dimer leads to dimerization and activation of c-MET receptor.
A) Structural model of c-MET in the apo state as inactive monomers. The structural model is taken from the dimer structure of the c-MET/HGF complex (PDB: 7MO7).
B) Cryo-EM structure of the c-MET/HGF complex. c-MET forms an asymmetric dimer when bound to full-length HGF and heparin (PDB: 7MO7). Heparin is shown in sphere representation.
C) Cryo-EM structure of 2:2 c-MET/NK1 complex showing a symmetric c-MET dimer bridged by the NK1 dimer (PDB: 7MOB).
In 2021, two structures of the c-MET extracellular complexes in the active state — c-MET/HGF and c-MET/NK1 (PDB: 7MO7 and 7MOB), were determined by using single particle cryo-EM, at an overall 4 - 5 Å resolution143 (Figure 3B, C). Notably, a GCN4 leucine zipper was introduced to the C- terminus of c-MET to stabilize the c-MET/HGF complex that enables the structural determination. Similar strategy has been applied in the cryo-EM studies of other RTK/ligand complexes80. Although the cryo-EM structures were resolved at modest resolution, the model building could be achieved by rigid-body fitting of the crystal structures of each domain of c-MET and HGF144,145. All the interfaces formed between c-MET and HGF, which are essential for HGF induced c-MET activation, are revealed in detail. Strikingly, the cryo-EM structure of the c-MET/HGF complex shows that, by utilizing two completely distinct interfaces, a single HGF molecule is able to bridge two c-MET molecules together for receptor activation (Figure 3B). In one half of the complex, 4 domains of HGF, including N, K2, K3 and SPH, simultaneously contact the SEMA domain of one c-MET; while, in the other half of the complex, the K1 domain of the same HGF exclusively recruits the second c-MET molecule through interacting with its SEMA domain. This minimal 2:1 c-MET:HGF active complex is further stabilized by a second HGF molecule, leading to a more stable 2:2 complex. A heparin molecule is sandwiched between N domain of HGF and IPT1 of c-MET, contributing to the high affinity binding between c-MET and HGF, which provides the structural basis for the critical functional role of heparin in c-MET activation143. This “single ligand induced dimerization model” represents a distinct mechanism underlying the activation of RTKs.
NK1 binds c-MET in a different mode. In contrast to the asymmetric conformation observed in the c-MET/HGF complex, the NK1 dimer recruits two c-MET in a symmetric manner for activation (Figure 3C). The formation of 2:2 c-MET/NK1 is mostly driven by the interaction between the K1 domain of NK1 and SEMA domain of c-MET143. The different dimeric configurations of c-MET triggered by HGF and NK1 may allow c-MET to preferentially activate different downstream signaling pathways through engaging distinct sets of downstream effectors135.
2.7. Cryo-EM structures of full-length EGFR bound with different ligands
Epidermal growth factor receptor (EGFR), a member of ErbB family receptors, plays critical roles in cell growth, division, and survival146. EGFR was among the first identified RTKs and has been extensively studied over several decades. The paradigm of ligand-induced dimerization in RTK activation was established initially in the studies of the activation of EGFR by its EGF ligand. Aberrant activation of EGFR is associated with many different cancers, making it an important drug target for cancer therapy147-150. At least seven ligands of EGFR have been identified, which all bind EGFR in similar modes151. However, different ligands show different binding affinities and kinetics to EGFR and in some cases trigger functionally different signal outcomes, the mechanisms of which are incompletely understood67,152.
Compared with other RTKs, the activation mechanisms of EGFR and other ErbB family members have several unique features. First, the ligands are monomeric and do not directly mediate the dimerization of EGFR. Instead, the ligand binds domains I and III in the ECD of EGFR, leading to a conformational change that exposes the dimerization arm in domain II of the EGFR-ECD, which mediates the dimerization of EGFR153,154. In the absence of the ligand, the ECD of EGFR adopts a closed autoinhibited conformation, in which the dimerization arm is concealed by domain IV155 (Figure 4A). The second unique feature is that the activation of the kinase activity is driven by the formation of an asymmetric dimer of the intracellular kinase domain, where one kinase domain acts as a cyclin-like allosteric activator for the second kinase domain156. The TM helix plays an active role in regulating the activity of EGFR by switching between two dimeric configurations. The active conformation of the TM dimer involves interactions at the N-terminal region of the TM helix, leading to a larger distance between the two C-termini of the TM helices that promotes the formation of the asymmetric kinase domain dimer157,158. In contrast, the inactive dimer of the TM has the C-terminal ends juxtaposed to each other, while the N-termini are further apart157,158. A critical remaining question is how the ligand-induced dimer of the ECD of EGFR is able to precisely control the switch of the TM from the inactive to the active dimer, given that the membrane proximal domain IV appears to be rather flexible relative to the other domains.
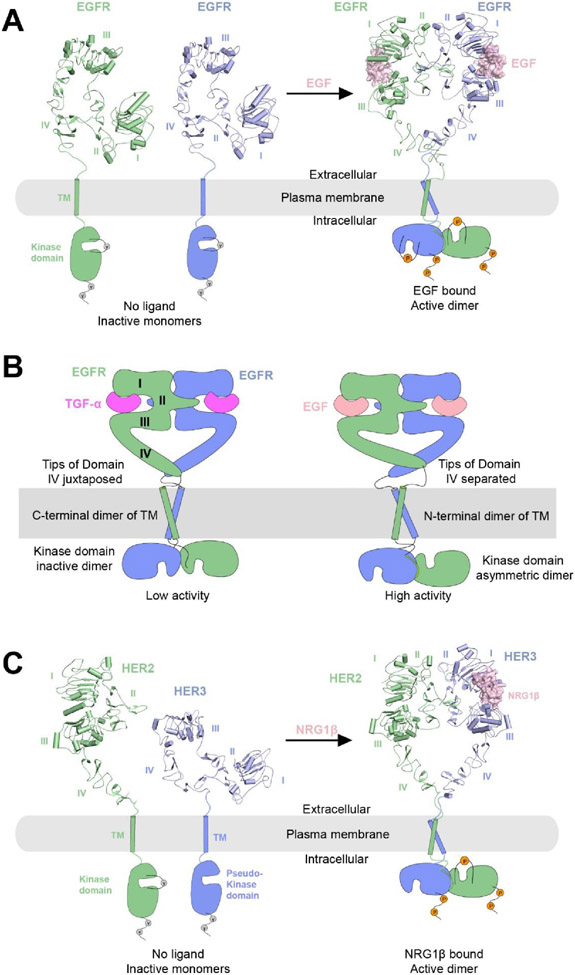
Figure 4.
Overall structures of EGFR and HER2-HER3 at their respective apo and ligand-bound states and the proposed activation mechanisms. The protomers in the receptor dimers (blue and green) are shown in cartoon representations, whereas the ligands are shown in cartoon and surface (pink) representations with semi-transparency. The TM and kinase domains are not resolved in the structures, and therefore indicated by schematic representations. Binding of ligands (EGF or NRG1β) leads to conformational changes to EGFR or HER3 that exposes the dimerization interface, enabling the dimerization and activation of receptors.
A) Left, crystal structure of apo-EGFR in the inactive monomeric state, in which EGFR shows an autoinhibited configuration with the dimerization interface buried (PDB: 1NQL). Right, cryo-EM structure of the 2:2 EGFR/EGF complex (PDB: 7SYD).
B) Schematic representations of the two dimeric conformations of EGFR induced by different type of ligands. Left, the low activity state induced by TGF-α with juxtaposed domain IV, C-terminal TM dimerization and kinase domains forming an inactive dimer. Right, the high activity state induced by EGF, characterized by separated domain IV tips, N-terminal TM dimerization and kinase domains forming an asymmetric dimer.
C) Left, crystal structures of HER2 (PDB: 1N8Z) and Her3 (PDB: 1M6B) in their inactive monomeric forms. HER2 lacks the ligand-binding site, and HER3 has a catalytically impaired pseudo-kinase domain. Right, cryo-EM structure of 1:1:1 HER2:HER3:NRG1β complex (PDB: 7MN5).
In 2021, Huang et. al reported a series of cryo-EM structures of full-length EGFR bound to either EGF or TGF-α in detergent or several membrane mimetic environments (lipid nanodiscs, amphipols and peptidiscs)159 (Figure 4A, B). The membrane mimetics have been used widely in cryo-EM structural analyses of membrane proteins, often better at stabilizing the TMD of the proteins because a belt of lipids remains to surround the TMD and mimics the native environment of the proteins160. In all the structures, the ligand-bound ECD of the EGFR dimer was resolved to high resolution (resolution ranges from 3.1 to 3.6 Å), but the TMD and ICD of EGFR were largely disordered, suggesting lack of rigid coupling between the ECD and the TMD. Interestingly, 3D classification of the cryo-EM data revealed multiple conformations of the EGFR-ECD dimer bound to either EGF or TGF-α. A comparison with previous crystal structures suggests that this structural ensemble from cryo-EM analyses represents an intrinsic conformational variability of the ligand-bound EGFR-ECD, characterized by a scissor-like rotation between the two EGFR subunits. Different rotation angles lead to different distances between the membrane proximal tips of the two protomers of domain IV in EGFR (Figure 4B). Close juxtaposition of the tips favors the inactive, C-terminal dimer of the TM helix, corresponding to the low activity state. On the other hand, larger distance between the domain IV tips supports the formation of the active, N-terminal dimer of the TM helix, promoting the formation of the asymmetric dimer and activation of the kinase domain. This mechanism is dependent on the conformational coupling of the TM helix with the short linker between domain IV and the TM (Figure 4B). Further analyses of the 3D class distributions suggest that EGF more strongly induces the tip-separated conformation of EGFR than TGF-α, providing a plausible explanation for the higher activating effect of EGF on EGFR, despite that the two ligands bind EGFR in a similar model with similar affinity. A cryo-EM structure of EGF in complex with EGFR with the oncogenic L834R mutation in the kinase domain suggests that the stabilization of the asymmetric kinase domain dimer promotes the tip separated conformation of the EGFR-ECD. These results further support the model that the ECD, TMD and ICD in EGFR are coupled through changes in distance in various parts of the EGFR dimer, even though the junctions among the different domains appear flexible. This study highlights the power of cryo-EM in revealing structural ensembles that may play essential roles in the regulatory mechanisms of proteins, especially SPTMRs, in which flexibility at two ends of the TM helix seems a common feature.
It is worth noting that, since the TM domains were unsolved in the cryo-EM maps of both the EGFR/EGF and EGFR/TGF-α complexes, the proposed model that the conformation of TM domain is coupled to that of ECD remains speculative. Resolving the full-length structure of an EGFR/ligand complex in its entirety with cryo-EM would provide more direct experimental evidence to support this model.
2.8. The cryo-EM structures of full-length HER2/HER3 complex bound with NRG1β
HER2 and HER3 (also known as ErbB2 and ErbB3), are two peculiar members in the ErbB family that cannot be activated by homodimerization161. HER2 is considered an orphan receptor as no ligand has been identified for it. HER3 has a cognate ligand neuregulin, but its kinase domain is catalytically impaired and incapable of autophosphorylation. However, HER2 and HER3 bound to neuregulin can form an active heterodimeric complex, which is potently pro-oncogenic162-165. In the HER2/HER3 complex, the pseudo-kinase domain of HER3 serves as the cyclin-like allosteric activator for the kinase domain of HER2, similar to that of the asymmetric kinase dimer of EGFR166,167. The crystal structure of HER3 in its unliganded apo form revealed an inactive closed conformation that is similar to that of EGFR at apo state168. Intriguingly, the ECD of HER2 adopts the open conformation in all previous crystal structures, despite the absence of any ligand (Figure 4C)169. This conformation has also been observed in a cryo-EM structure of HER2 ECD bound to two therapeutic antibodies trastuzumab and pertuzumab170. With its dimerization arm in domain II constitutively exposed, it was not clear why HER2 did not homodimerize in those structures. In 2021, the first structure of near full-length HER2/HER3 heterodimer bound to neuregulin-1β (NRG1β) was solved by cryo-EM to 2.9 Å resolution171 (Figure 4C). Notably, due to the low expression yield of the proteins, graphene oxide coated grids were used for cryo-EM sample preparation to increase the particles number in each image172. The overall structure of the HER2/HER3/NRG1β complex is similar to the EGFR/ligand complexes, with the TMD and ICD unresolved presumably due to flexibility. A key insight emerged from the HER2/HER3/NRG1β complex structure is that the dimerization arm in domain II of HER3 is not present in the cryo-EM map. The heterodimerization between HER2 and HER3 is mediated by the dimerization arm of HER2, which engages the binding pocket on HER3 in the same manner as in the structures of the homodimeric ErbB family members. Further analyses suggest that the dimerization arm in HER3 is disordered because its binding pocket on HER2 between domains I and III is not fully formed. The partially open conformation between domains I and III in HER2 is likely due to lack of ligand binding. These analyses therefore point to an unanticipated allosteric connection between the ligand-binding site and the binding pocket for the dimerization arm, which may explain the lack of homodimerization of HER2. Interestingly, the cryo-EM structure of heterodimeric complex with the S310F mutant of HER2 showed that the dimerization arm in HER3 was well resolved. S310F is one of the most frequent oncogenic missense mutations found in HER2, although the mechanism of its oncogenic effect was not clear. The cryo-EM structures show that F310 in the HER2 mutant forms a stable π-π stacking interaction with the Y265 in the dimerization arm of HER3. Therefore, the S310F mutation promotes the HER2/HER3 heterodimerization by forming a compensatory binding site on HER2 for the HER3 dimerization arm. Further analyses of the interactions between the HER2(S310F)/HER3 heterodimeric complex with two therapeutic antibodies suggest that these antibodies also exploit the conformational dynamics in HER2 and HER3 for their binding.
3. Plexin, Neuropilin and Semaphorin
3.1. Introduction
Plexin is the major receptor for semaphorin, the largest family of axon guidance molecules173,174. The interactions between some plexin family members and some secreted semaphorins are weak, requiring the assistance of neuropilin as the co-receptor175-177. Signaling through semaphorin-activated plexin is best known for its repulsive guidance function for developing neurons. Additional functions of plexin signaling include regulation of cardiovascular development, immunity regulation and wound healing173,174,178.
The semaphorin family members are multi-domain proteins either secreted to the extracellular space or attached to the cell surface. The N-terminal region of all semaphorins contains the conserved Sema domain, which is followed by a Plexin-Semaphorin-Integrin (PSI) domain and an Ig-like domain. Both the Sema and Ig-like domains mediate interactions for semaphorin dimerization, while the Sema also mediates the binding to plexin and neuropilin179-181. The C-terminal region following the PSI domain is relatively long and largely unstructured. In some semaphorin family members, the C-terminal region contains a cysteine residue that forms an inter-chain disulfide to further stabilize the dimer182,183. The C-terminal region of class 3 semaphorins contains multiple R/K-X-X-R (single-letter amino acid code; X, any residue) motifs, which upon cleavage by furin proteases expose a C-terminal arginine residue that binds tightly to a pocket in neuropilin (See below)184,185.
Plexins are all type I transmembrane proteins with an N-terminal ligand-binding ECD, a single TM helix and a C-terminal ICD. The ECDs of class A, B and D plexins all contain ten domains, starting with the N-terminal Sema domain that is responsible for interacting with the semaphorin ligand186-190. The Sema domain is followed by three PSI domains and six Ig-like Integrin-plexin-transcription factor (IPT) domains, arranged as PSI1-IPT1-PSI2-IPT2-PSI3-IPT4-IPT5-IPT6 (Figure 5A). The only class C plexin, PlexinC1, is unique in that it only contains seven domains in the extracellular region (Sema-PSI1-IPT1-PSI2-IPT2-IPT3-IPT4) (Figure 5B)191. In contrast, the ICD of all the plexin family members share a conserved domain structure, containing a juxtamembrane helix, a split GTPase Activating Protein (GAP) domain and a RhoGTPase binding domain (Figure 5A)192-198.
Figure 5.
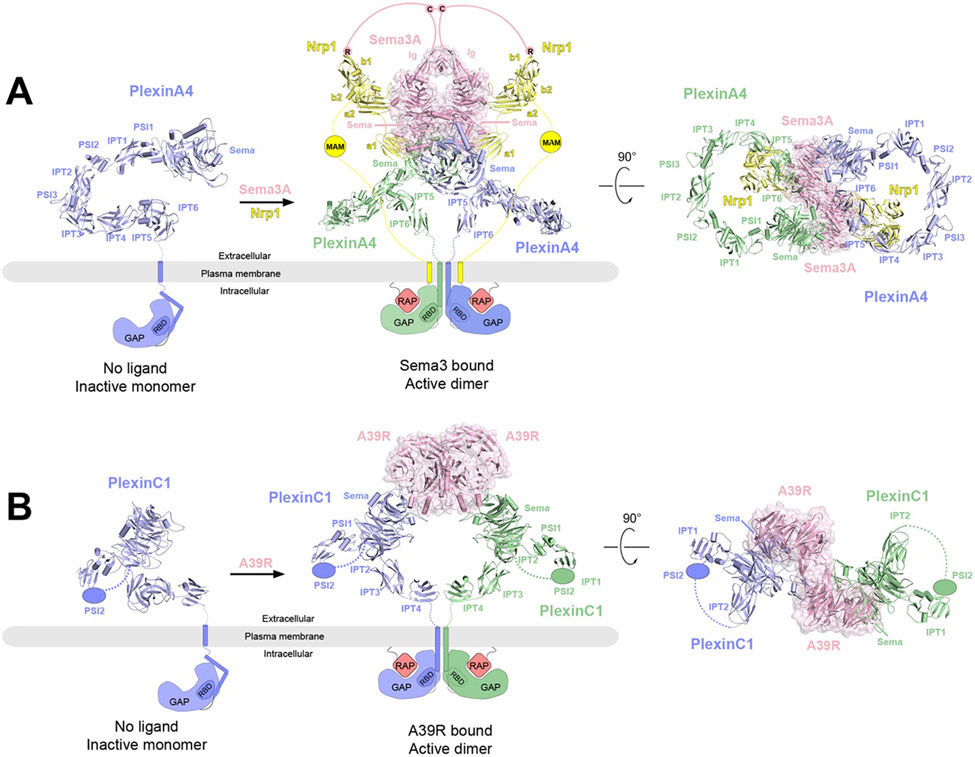
Overall structures of PlexinA4 and PlexinC1 in their respective apo and ligand-bound forms and the proposed activation mechanisms. The protomers in the receptor dimers (blue and green) as well as the coreceptor Nrp1 (yellow) are shown in cartoon representations, whereas the ligands are shown in cartoon and surface (pink) representations with semi-transparency. The missing domains and linkers in each structure are shown as schematic representations. In the apo state, the GAP domains of PlexinA4 or PlexinC1 are in the autoinhibited state with the Rap-binding site inaccessible. Binding of the ligands (Sema3A or A39R) induces plexin dimerization, opening up the GAP active site to allow Rap binding and GTP hydrolysis.
A) Left, crystal structure of ECD of PlexinA4 in its apo state (PDB: 5L5K). Middle and right, cryo-EM structure of 2:2:2 PlexinA4/Nrp1/Sema3A complex in two different views (PDB: 7M0R). The structure of the ECD of PlexinA4 in both structures are essentially the same. The lines represent the various linker domains between different protein components that are important to the formation of the complex. The pink lines represent the linker between C723 and the Ig-like domains of Sema3A as well as that between C723 and the C-terminal R770, which engages the binding pocket in the b1 domain of Nrp1. The yellow lines represent the b2-MAM and MAM-TM linkers in Nrp1.
B) Left, structural model of monomeric PlexinC1 in its apo state taken from the structure of the PlexinC1/A39R complex (PDB: 6VXK). Middle and right, the cryo-EM structure of the 2:2 PlexinC1/A39R complex (PDB: 6VXK) in two different views.
Neuropilins (Nrp1 and Nrp2) are also type I transmembrane proteins whose ECDs are composed of five domains (a1, a2, b1, b2 and c)199-202. The a1 and a2 domains both belong to the Ca2+-binding CUB domain family with a β-sandwich fold. The c domain also binds Ca2+, but adopts a jellyroll fold and belong to the MAM domain family (Meprin, A5 and Mu-phosphatase)203. The b1 domain adopts a coagulation factor V/VIII fold, containing a pocket at one end that binds the C-terminal arginine motif of class 3 semaphorins with high affinity185,202,204. The short cytoplasmic tail of neuropilin possesses a C-terminal SEA motif, which interacts with the GIPC adaptor proteins and thereby mediates the endocytosis 205-208.
Structures of various parts of semaphorin, neuropilin and plexin and partial complexes of these proteins have been solved mostly by X-ray crystallography and some by NMR181,188-190. These structures have provided many important insights, including the conformations of the individual proteins and how they interact with one another. These studies together with functional studies led to the general model of plexin activation in which semaphorin-induced dimerization of the plexin ECD promotes the formation of the active dimer of the ICD, which then relays the signal to downstream pathways (Reviewed in 209,210). There are some interesting exceptions to this general model of activation, such as monomeric semaphorin and semaphorin-plexin cis interactions211,212. However, lack of a whole picture of the proteins and their complexes left many questions unanswered. For example, what is the overall arrangement of semaphorin, plexin and neuropilin in the tripartite complex that underlies plexin activation? What roles does each of the multiple domains in the proteins play in the regulation of plexin signaling? How do the three proteins form multiple interfaces to ensure specificity and high affinity?
3.2. Cryo-EM structure of Sema3A/PlexinA4/Nrp1 complex
A cryo-EM structure of the near intact extracellular complex of Sema3A, PlexinA4 and Nrp1 published in 2021 reveals the overall architecture of this large assembly of ~600 kDa187 (Figure 5A). The complex has an expected 2:2:2 stoichiometry, with the overall shape resembling a frog when viewed from the side of the plasma membrane. The two copies of the a1 domain of Nrp1 and the Sema domains of both PlexinA4 and Sem3A in the dimeric complex constitute the body of the frog, while the two Ig-like domains of Sema3A converge at the top to form the head. The a2-b1-b2 domains of Nrp1 and the domains following the Sema domain of PlexinA4 are the front and rear limbs, respectively. The overall highly curved ring shape of the PlexinA4 extracellular region is similar to the crystal structures of PlexinA4 and PlexinA1 in the apo form reported previously186. Based on this ring shape and the known binding mode between Semaphorin and plexin, a model of the semaphorin/plexin complex has been constructed previously to explain how binding of semaphorin to the Sema domain of plexin induce the formation of the intracellular active dimer. The cryo-EM structure confirms that in the 2:2:2 complex, the membrane proximal domains of the two plexin molecules are placed into close proximity, poised to induce the dimerization of the transmembrane and intracellular regions 192,213.
More importantly, the cryo-EM structure of the Sema3A/PlexinA4/Nrp1 complex reveals several mechanistically critical structural features that were previously unknown (Figure 5A). Firstly, the cryo-EM structure reached adequate resolution (3.7 Å overall, with substantially higher local resolution at the center) for resolving the specific interactions in several binding interfaces among the three proteins. The interface between PlexinA4 and Nrp1 is dominated by the interaction between a lysine residue from the PlexinA4-Sema domain and the negatively charged surface patch near the Ca2+-binding site in the Nrp1-a1 domain. A similar interface is formed between Sema3A-Sema and Nrp-a2, mediated by a lysine residue from the Sema domain and the negatively charged surface near the Ca2+-binding site in the Nrp1-a2 domain. In both cases, the lysine residues are present in the plexin and semaphorin family members that interact with neuropilin, but absent in those that do not bind neuropilin. The lysine/Ca2+-site interface therefore provides a unified mechanism for the specificity of neuropilin towards both plexin and semaphorin. The placement of the triangle-shaped a2-b1-b2 module of neuropilin at the front limb position is dictated by the Nrp1-a2/Sema3A-Sema interaction. This arrangement leaves a large distance between the Nrp-b1 domain and the C-terminal region following the Ig-like domain of Sema3A. A long linker is therefore required for the C-terminal arginine residue of Semaphorin, exposed by furin protease cleavage, to engage the binding pocket in neuropilin. The structure therefore provides a basis for the previous observation that semaphorin cleaved at distal cleavage sites are more potent activators of plexin signaling184. Likewise, a linker following the b2 domain of neuropilin is required to span the large distance to the plasma membrane where the transmembrane region is anchored. Such a linker role is fulfilled by the MAM domain as well as the two long flexible regions flanking the MAM domain. The structure explains the fact that the total length of two flanking flexible regions is similar in neuropilin from different species, but the sequences of these two regions are not conserved.
3.3. Cryo-EM structure of full-length PlexinC1 bound with A39R dimer
The unique domain structure of the PlexinC1-ECD suggested that its conformation must differ from the ring shape of class A plexins. This difference has raised the question how its ligand Sema7A, which forms a similar dimer as other semaphorins, induces the formation of the active dimer of PlexinC1. This question has been addressed by the cryo-EM structure of the complex between full-length PlexinC1 and the A39R dimer, a viral mimic of Sema7A, reconstituted in the peptidisc, which organized into multiple helical elements that wrap around the PlexinC1 TM domain191 (Figure 5B). Distinct from the ring shape of class A plexins, the 7-domain ECD of PlexinC1 is composed of two curved rods arranged in an orthogonal fashion. The first rod contains the Sema, PSI1, IPT1 and PSI2 domains, while the second is formed by the membrane proximal IPT2-4 domains. There is a gap between the first and second rod, which is in fact connected by the flexible linker between the PSI2 and IPT2 domains. Despite the flexible linkage, the orientation of the second rod relative to the first is fixed by the extensive interactions between the two rods. Notably, an inter-domain disulfide bond is formed between a cysteine residue from the Sema domain and a cysteine residue from the IPT2 domain, effectively gluing the two rods together. This particular conformation of the PlexinC1-ECD ensures that the two membrane-proximal IPT4 domains in the ligand-induced dimeric complex are close to each other for inducing the dimerization of the TM and ICD of PlexinC1. Therefore, PlexinC1 and other plexin family members use different structural features to achieve the same goal of activating the intracellular signaling domains.
Similar to the structures described above, the TM and ICD of PlexinC1 are not resolved in the cryo-EM structure of the PlexinC1/A39R complex (Figure 5B). As a result, it is not known whether and how the two TM helices in the PlexinC1 dimer interact with each other and contribute to the induction of the ICD active dimer. It is also not clear how the ICD dimer is arranged to the lipid membrane, which may be important for regulating the GAP activity and binding to other signal proteins inside the cell. Similar questions remain open for the semaphorin/plexin/neuropilin tripartite complex. In this case, another interesting question is whether and how the TM and ICD of plexin and neuropilin make functionally important interactions.
4. Integrin
4.1. Introduction
Integrins are a large family of cell surface proteins that mediate adhesion by binding to their ligands expressed on other cells or extracellular matrix (ECM), such as fibronectin, laminin and collagen214. Ligand-activated integrins trigger intracellular signaling pathways and serve as hubs for the formation of the focal adhesion215. Conversely, binding of intracellular signaling proteins to the ICD of integrins can induce inside-out signal to enhance the binding affinity of the ECDs of integrins for their ligands216. Integrin-mediated adhesion and signaling play critical roles in the development and organization of various tissues217. Particularly, integrins are essential for the functions of immune cells, as reaching sites of antigen accumulation and establishing proper contacts with infected cells are prerequisites for launching robust immune responses218-223.
Integrins are heterodimeric proteins formed by two different subunits (α and β), both of which are single-pass TM proteins. In total, 18 α and 8 β subunits have been identified to date, forming at least 24 distinct heterodimeric receptors, which display distinct characteristics in ligand binding and cell signaling224. The ECDs of both α and β subunits contain multiple domains with flexible linkers between them218,219,225. The ECD of α subunit consists of four or five domains, including the α-head domain formed by a seven-bladed β-propeller, a thigh domain, and two calf domains with similar immunoglobulin (Ig)-like fold; while the ECD of β subunit consists of β-I, Hybrid, PSI, four consecutive I-EGF and β-tail domains. The α-head domain and β-I domain interact with each other, forming the head piece that serves as the ligand-binding site223,225-228. The integrin head piece also contains three conserved cation binding sites (namely SyMBS, MIDAS, and ADMIDAS), which are localized at the ligand binding pocket of β-I domain. MIDAS is a Mg2+ binding site while SyMBS and ADMIDAS are capable of Ca2+ binding228-230. The binding of cations plays key role in the ligand binding and conformational dynamics of integrins, as it is shown to increase the ligand binding affinity and prime the integrin for ligand binding228.
In the canonical ‘open-close’ model for integrin activation229-234, the activation of integrins requires large ligand-induced conformational rearrangement. According to this model, integrins adopt the ‘bent’ conformation in the apo-state 230,235-237, in which the head pieces point towards the plasma membrane, leading to occluded ligand binding site and low ligand-binding affinity. Moreover, the legs in this “bent” conformation are near to each other. Upon cations binding (such as Mn2+), integrins become ‘primed’ and adopt ‘extended-closed’ conformation, in which the head pieces swing upward, pointing away from the plasma membrane. Such conformational change exposes the ligand-binding groove and thereby significantly increases ligand-binding affinity, while the legs remain in close contact. Upon ligand binding, the β hybrid domain undergoes a ‘swing-out’ conformational change, causing the separation of the membrane proximal domains, hence the ‘extended-open’ conformation. Although all three states have been observed in negative staining EM238, until recently, no high-resolution structures were available for the extended conformations229-234. The lack of high-resolution structures of the extended conformation limited the understanding of the molecular details of the integrin activation. Several recent high-resolution cryo-EM structures of integrins αvβ8 and α5β1 determined in either the apo or ligand-bound state provide important structural details of the extended conformations and offer insights into alternative activation mechanisms239-241.
4.2. The cryo-EM structure of the ectodomain of αvβ8 integrin
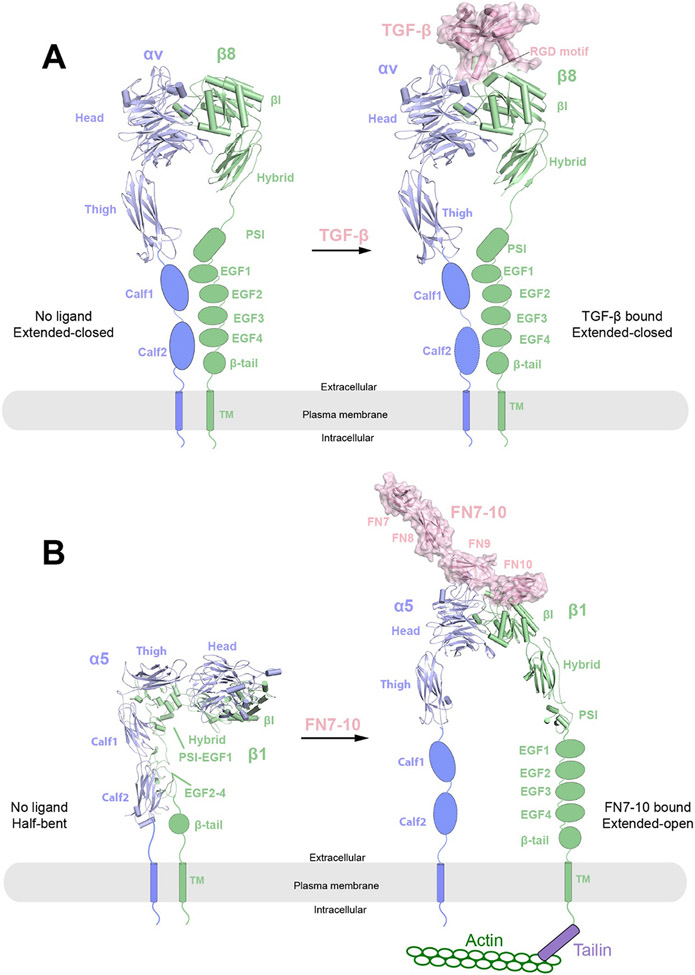
In 2018, the cryo-EM structure of the ECD of αvβ8 integrin was determined by using two different monoclonal Fabs – 8B8 and 68, directed at the α head and β-I domains of the αvβ8 integrin, respectively241. The binding of Fabs to integrin increases its molecular mass and thereby facilitate image alignment during the processing of cryo-EM data. The overall resolution for the αvβ8 integrin was 6.4 Å, while the headpiece (α head and β-I domains) was better resolved than the leg domains due to variations in local resolution. The resolution was largely limited by the inherent conformational flexibility between the headpiece and leg domains of integrin. Focused refinement on the headpiece improved the local resolution to 4.8 Å by masking out the flexible leg domains and the constant domains of the Fabs. All four domains of the αv subunit (α head, Thigh, Calf-1 and Calf-2) are visible in the map, whereas only the β-I and Hybrid domains are clearly visible in the β8 subunit. The structure shows that the integrin adopts the ‘extended-closed’ conformation, in which the head piece points away from the plasma membrane and therefore can bind the ligand, albeit with lower affinity than the extended-open conformation. Moreover, 3D classification analysis focused on the head piece revealed six classes. Structural comparison among the 6 classes suggest that the headpiece may undergo rotation of 30° relative to the leg domains. This structural variability revealed by 3D classification led to the “sunflower” model for the αvβ8 integrin where the head piece can sample an elliptical conic space to facilitate surveillance of the extracellular space for ligand detection. Further analyses of the different 3D classes show that weaker density of β8 leg is correlated with larger extension of the head piece, suggesting that loss of β8 leg contacts with the αv leg favors the extended conformation. Nevertheless, truncation of the β-leg domains (EGF2-EGF4 and β tail) did not lead to substantial increase in the extended conformation as shown by negative staining EM, suggesting the ‘extended-closed’ conformation is primarily maintained by the αv leg domains. Similar to the case of the EGFR/EGF complex structure, this study showcases the ability of cryo-EM in revealing the full dynamic conformational range of the protein of interest.
4.3. Cryo-EM structure of ECD of αvβ8 integrin in complex with L-TGF-β
Unlike other integrins, which are promiscuous with the ligands, integrin αvβ8 binds exclusively to latent-TGF-β (L-TGF-β)222,242. During the maturation of TGF-β, the pro-domain (also called latency-associated protein (LAP)) is cleaved by furin-like proteases, but it would remain bound with TGF-β that forms L-TGF-β243. Integrin αvβ8 binding is essential for activating L-TGF-β, a process that allows mature TGF-β to bind and activate its receptor by releasing the inhibition imposed by the pro-domain244,245. A previous study has shown that integrin αvβ6 in the extended-open conformation binds L-TGF-β in a specific orientation, which allows tensile force generated by actin cytoskeleton to transmit through integrin β6 to the pro-domain of TGF-β, leading to physical release of mature TGF-β246. However, this mechanism does not apply to integrin αvβ8, because it lacks the ability of directly interacting with actin cytoskeleton and appears to exist exclusively in the extended-closed conformation241,247. A cryo-EM structure of the ECD of αvβ8 integrin in complex with L-TGF-β reported in 2020 has led to a distinct model of TGF-β activation240 (Figure 6A). The structure reveals that αvβ8 integrin adopts an extended-closed conformation, similar to that of its unliganded state241. The binding interface between αvβ8 integrin and L-TGF-β is well resolved, showing that the ligand-binding cleft of αvβ8 integrin engaging the RGDLXXI/L integrin-binding motif of L-TGF-β in a manner similar to that in the complex between αvβ6 integrin and L-TGF-β. However, comprehensive classification of the cryo-EM dataset showed that the orientation between L-TGF-β and the headpiece of αvβ8 integrin displays a wide range of variation, which is likely a result of the flexible linkages between the integrin-binding motif and the rest of L-TGF-β. This flexible nature of L-TGF-β in complex with αvβ8 integrin led the authors to propose that αvβ8 integrin-mediated TGF-β activation does not require actin-cytoskeleton force or release/diffusion of mature TGF-β from the pro-domain, because it can directly engage its receptor in the confinement of the pro-domain.
Figure 6.
Overall structures of integrins αvβ8 and α5β1 in their respective apo and ligand-bound states. The protomers in the receptor dimers (blue and green) are shown in cartoon representations, whereas the ligands (TGF-β or FN7-10) are shown in cartoon and surface (pink) representations with semi-transparency. The missing domains in each structure are shown as schematic representations.
A) Cryo-EM structures of apo state (left) and TGF-β bound state (right) of integrin αvβ8. In both structures, integrin αvβ8 adopts an extended-closed conformation with the leg domains close to each other.
B) Cryo-EM structures of apo state (left) and FN7-10 bound state (right) of integrin α5β1. In the apo state, integrin α5β1 adopts a half-bent conformation with the α5 and β1 subunits parallel and leg domains close to each other. In the FN7-10 bound state, integrin α5β1 adopts an extended-open conformation where the leg domains become wide open. The conformational changes allow for the activation of integrin and binding of downstream signaling components such as Talin and Actin.
4.4. Cryo-EM structures of α5β1 integrin in resting state and in complex with FN7-10
Fibronectin (FN), a large protein containing repetitive modules of type I, II, and III FN domains, is a major component of ECM248,249. Integrin α5β1 is the primary FN receptor that is critical for cell migration and adhesion, by both serving as the hub of the intracellular focal adhesion and modulating the ECM organization239,250. FN interacts with α5β1 via its RGD motif located on the FN10 domain. It has been suggested that the activation of α5β1 integrin by FN may not utilize the canonical close-open mechanism as in β3 integrins, because the α5β1 ectodomain does not adopt the fully bent conformation in the resting state and does not undergo large conformational changes upon the binding to the isolated RGD motif251,252.
A cryo-EM structure of α5β1/FN7-10 complex reported in 2021 presents the first high-resolution view of integrin at the extended-open state and provides molecular details for the binding and activation of α5β1 integrin in response to FN239 (Figure 6B). In this study, full-length native integrin α5β1/FN was obtained from a natural source (human placenta), assembled into nanodiscs, and purified in the presence of FN7-10 and a stabilizing antibody fragment253. Single particle cryo-EM analysis of the complex yielded a final map at an overall nominal resolution of 3.5 Å, with the head piece better resolved at 3.1 Å. The flexible lower leg domains including the two Calf domains in the α subunit, as well as the four EGF-like domains and the β-tail domain in the β subunit were resolved at lower resolution. The structure shows that integrin adopts the ‘extended-open’ conformation, with the head pieces of α and β subunits in close contact with each other and the legs wide open. FN makes dual contacts with integrin: (1) FN9 interact with the α5 head; (2) FN10 contacts both the α5 and the β1 head domains, with the RGD loop in FN10 inserted into the ligand binding groove formed by α5 and the β1 heads. Furthermore, as demonstrated by binding assays, the interaction between α5β1 and FN is also enhanced by the presence of Mn2+ and glycosylation (glycanN275) of the α5 head. The simultaneous engagement of the multiple binding interfaces seen here is required for inducing the open conformation of α5β1 integrin, explaining why the RGD motif alone fails to do so.
In 2021, the cryo-EM structure of α5β1 at its unliganded state was determined in the presence of both Ca2+ and Mg2+ ions at overall 4.6 Å resolution (Figure 6B)239. Unlike other known resting structures of integrins such as αvβ3 or αIIbβ3, which show an acute bent conformation230,234, the structure of α5β1 in its resting state adopts a half-bent conformation. In this state, the α5 and β1 subunits, each of which adopts a ‘L’-shaped configuration251, are parallel to each other. The lower leg domains, including the two Calf domains of α5 and PSI-EGF domains of β1, were all resolved in the cryo-EM structure. The stabilization of the membrane-proximal domains is likely a result of the parallel interactions between the α and β legs. Notably, although Mn2+ binding to ADMIDAS significantly increases the binding affinity between FN and α5β1, it does not obviously affect the conformation of the resting state α5β1. This is distinct from αvβ3, for which Mn2+ is sufficient to induce the transition from the bent to the extended conformation, suggesting different activation mechanism among these closely related receptors251.
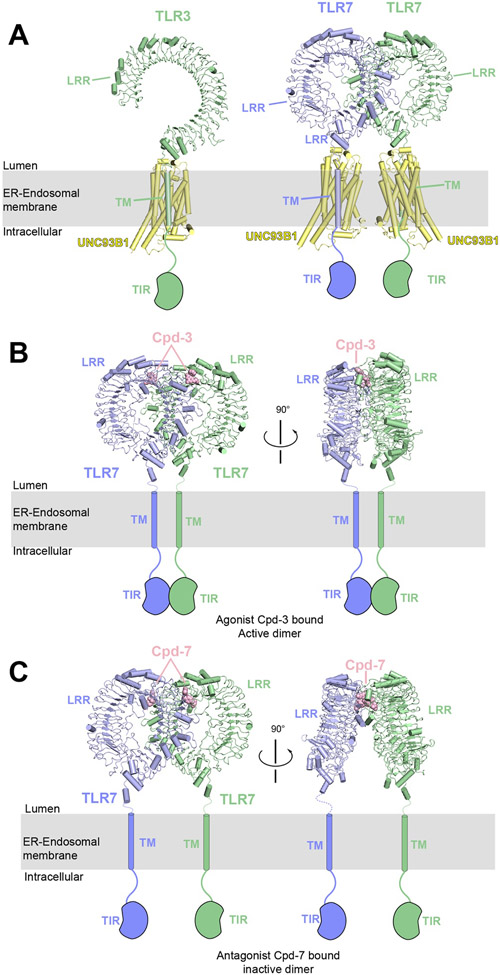
5. Toll-like receptors
5.1. Introduction
Toll-like receptors (TLRs) are a class of pattern recognition receptors (PRRs) that trigger innate immunity upon recognizing distinct pathogen-associated molecule patterns (PAMPs) or cellular damage-associated molecule patterns (DAMPs)254,255. TLRs signal through the recruitment of specific adaptor molecules, leading to activation of the transcription factors such as NF-κB and IRFs, which eventually promote the expression of cytokines, chemokines and type I interferons to protect the host from the invading pathogen254,256,257. TLRs are SPTMRs comprised of an ECD, a single TM and a cytoplasmic Toll/interleukin-1 receptor (TIR) domain adopting a conserved fold with a β-sheet surrounded by α helices258,259. The ECD contains a horseshoe-shaped leucine-rich repeat (LRR) domain (containing 19-25 leucine-rich repeats) that is responsible for ligand binding258,260-262. Ligand recognition by the LRR domain leads to TLR dimerization, which then induces the recruitment of downstream adaptor proteins through the TIR domain263-265. The TLR family consists of 10 members (TLR1-10) in human and 12 in mouse (TLR1-TLR9, TLR11-TLR13), which can be divided into two groups: the cell surface TLRs and endosomal TLRs. Cell surface TLRs are located on the cell membrane and recognize bacterial cell-surface components, whereas the endosomal TLRs (TLR3, TLR7, TLR8 and TLR9) are compartmentalized in endosomes and responsible for sensing microbial nucleic acids266. Each TLR recognizes its cognate ligand in a different manner: (1) hydrophobic ligands that can activate TLR1, TLR2, and TLR4, such as bacterial lipoproteins and lipopolysaccharides, bind to internal protein pockets; (2) hydrophilic ligands that activate TLR3, TLR7 and TLR8, such as double-stranded RNAs (dsRNAs) and single-stranded RNAs (ssRNAs), engage solvent-exposed surfaces of the LRR domains263,267.
TLR3 and TLR7 are two endosomal nucleic acids-sensing TLRs that recognize viral dsRNAs and ssRNAs, respectively268-271. TLR7 can also be activated by small chemical ligands, such as imidazoquinoline compounds and nucleotide analogues, suggesting its role as a multifunction receptor269,272,273. The crystal structure of TLR3/dsRNA complex reveal that the dsRNA binds both N- and C-terminal sites on the lateral side of the convex surface of TLR3 homodimer274. Crystal structures of TLR7 in complex with uridine-containing ssRNA or guanosine agonists unveil that TLR7 possesses two distinct ligand-binding sites: one for small chemical ligands such as guanosine located at the TLR7 homodimer interface, and the other for ssRNA located at the concave surface of the N-terminal part of the LRR horseshoe-like structure (LRR1–5)275.
Given that pathogen-derived and endogenous nucleic acids share common chemical and structural features, the activation of nucleic acid-sensing TLRs must be strictly regulated to prevent autoimmune diseases. Indeed, excessive activation of nucleic acid-sensing TLRs is involved in a variety of autoimmune or inflammatory diseases, including systemic lupus erythematosus (SLE), heart failure, arthritis and non-alcoholic steatohepatitis276-278. Endosome localized nucleic acid-sensing TLRs is believed to act as a safeguard to prevent undesirable immune responses caused by endogenous RNA-mediated TLR activation279,280. UNC93B1, a member of the atypical solute carriers of major facilitator superfamily (MFS)281, is an important regulatory factor that controls the trafficking and compartmentalization of TLRs280,282-284. UNC93B1 is also involved in other TLR functions including balancing TLR7 and TLR9 trafficking and activation285, recruiting syntenin-1 to dampen TLR7 signaling and prevent autoimmunity286, and regulating TLR7 maturation276.
5.2. Cryo-EM structures of TLR3 and TLR7 in complex with UNC93B1
In 2021, cryo-EM structures of the TLR3/UNC93B1 and TLR7/UNC93B1 complexes were determined, providing structural details for the direct interaction between UNC93B1 and TLRs287 (Figure 7A). The protein complexes were obtained by co-expression of full-length TLRs with UNC93B1 in mammalian cells, which were subsequently solubilized and purified in the detergent digitonin. Single-particle cryo-EM analyses yielded maps at overall resolutions of 3.4 Å, 3.3 Å and 4.2 Å for human TLR3/UNC93B1 (PDB: 7C76), mouse TLR3/UNC93B1 (PDB: 7C77), and human TLR7/UNC93B1 (PDB: 7CYN), respectively. The structures of the human and mouse TLR3/UNC93B1 complexes are essentially the same, with TLR3 and UNC93B1 being assembled in a stoichiometry of 1:1. The ECD and TM of TLR3 are well resolved, but the TIR domain is not resolved, possibility caused by the flexible linker between the TM and TIR domain. The horseshoe-like structure of TLR3 ECD is almost identical to previously determined crystal structures274. The 12 TMs of UNC93B1 can be divided into two pseudo-symmetrical six-helical bundles (TM1-TM6 and TM7-TM12). The binding between TLR3 and UNC93B1 is mostly maintained by the antiparallel packing between the single TM of TLR3 and TM3 of UNC93B1. TM6 of UNC93B1 also contributes to the complex formation. In addition, the C-terminal part of the LRR and the juxtamembrane region of TLR3 form a small interface with the luminal loops of UNC93B1. Importantly, structural comparison between the TLR3/dsRNA and TLR3/UNC93B1 complexes shows that the binding with UNC93B1 is incompatible with the formation of the ligand-bound active TLR3, which provides a structural explanation for the previous results that TLR3 must be released from UNC93B1 prior to ligand binding and activation288.
Figure 7.
Overall structures of Toll-like receptors TLR3 and TLR7 in complex with UNC93B1 or small molecule agonist/antagonist. The protomers in the TLR receptor dimers (blue and green) as well as UNC93B1 (yellow) are shown in cartoon representations, whereas the ligands (Cpd3 or Cpd7) are shown as spheres. The missing domains (TM and TIR) in each structure are shown as schematic representations.
A) Cryo-EM structure of 1:1 TLR3/UNC93B1 (left, PDB: 7C76) and 2:2 TLR7/UNC93B1 (right, PDB: 7CYN) complexes.
B-C) Cryo-EM structure of 2:2 TLR7/Cpd3 (PDB: 6LVZ) and 2:2 TLR7/Cpd7 (PDB: 6LW1). Cpd3 is an agonist for TLR7, which induces the formation of closed dimer, promoting TIR dimerization and TLR7 activation. In contrast, Cpd7 is an antagonist for TLR7, which induces the formation of open dimer, consequently inhibiting TIR dimerization and TLR7 activation.
The binding interface between TLR7 and UNC93B1 is similar to that in the TLR3/UNC93B1 complex. However, the TLR7 and UNC93B1 assembles into a 2:2 dimeric complex. The dimerization is driven by both TLR7-TLR7 and UNC93B1-UNC93B1 interactions. LRR11 from the middle region of the TLR7 ECD is responsible for the dimerization of TLR7-ECD. The short cytosolic helix connecting TM6 and TM7 in UNC93B1 mediate the UNC93B1-UNC93B1 interaction through antiparallel packing. The dimerization mode of TLR7 in the TLR7/UNC93B1 complex is completely different from that in the ligand-induced active dimer of TLR7, suggesting that the TLR7 dimer with UNC93B1 bound is not in the active state. The function of the dimerization of the TLR7/UNC93B1 complex remains unclear.
Collectively, this study elucidates the structural basis for the interaction of TLRs with UNC93B1, explaining the specificity of UNC93B1 towards endosomal TLRs. In addition, it presents some of the rare cases where the single-pass TM of SPTMRs is well resolved, owing to the stabilization effect from UNC93B1.
5.3. Cryo-EM studies of TLR7 in complex with designed agonists and antagonists
Aberrant activation of TLR7 has been implicated in several autoimmune diseases such as systemic lupus erythematosus (SLE), making TLR7 inhibitors promising therapeutic agents against SLE278,289,290. Tojo et al developed a series of TLR7-specific antagonists by modifying existing TLR7 agonistic 8-oxoadenine derivatives291,292. Surprisingly, the crystal structure of TLR7 bound to one of these antagonists Cpd-6 showed an active dimer, contradicting its inhibitory effect on TLR7 signaling. This discrepancy is reconciled by cryo-EM structures of TLR7 in complexes with Cpd-6 and related compounds. As expected, the cryo-EM structure of the TLR7 ECD in complex with the agonist Cpd-3 adopted the active dimer conformation291 (Figure 7B). In this active dimer, the C-terminal tips of the two ECDs are placed close to each other and therefore capable of inducing the dimerization of the TMD and the TIR domains. In contrast, the cryo-EM data of the TLR7/Cpd-6 complex contained two major types of TLR7 dimers. One of the forms is the active dimer, whereas the other called the “open” dimer represents the inactive dimeric state because the C-terminal tips of the two ECD protomers are far apart and likely incompatible with the dimerization of the TMD and TIR domain. Therefore, Cpd-6 appears to act as a weak antagonist by partially inducing the open dimer conformation of TLR7. Consistent with this idea, Cpd-7, a stronger antagonist, exclusively induces the formation of the open dimer of TLR7.
Notably, several techniques were used in this study to overcome technical difficulties in cryo-EM analyses of TLR7 bound to the antagonists. To stabilize the dimeric complexes, the samples were subjected to chemical crosslinking with glutaraldehyde. The samples showed severe preferred orientation on cryo-EM grids, which limited the resolution of the 3D reconstructions. This problem was alleviated by using TLR7 protein with partially uncleaved purification tags. These approaches helped achieve 2.8 Å resolution of the TLR7/Cpd-7 complex (Figure 7C). The structure shows that TLR7/Cpd-7 complex adopts an open dimer conformation and provides atomic details of the Cpd-7 binding site, which will be of great value for further modification of the compound for stronger binding and antagonism. With the achievable resolution of cryo-EM continuously improving, it will soon rival X-ray crystallography in aiding rational drug design.
6. IGF2R
6.1. Introduction
Insulin-like growth factor 2 receptor (IGF2R), also known as cation-independent mannose-6-phosphate receptor (CI-MPR), is a large SPTMR with a molecular mass of ~300 kDa. IGF2R comprises a large ECD containing 15 homologous β strand-rich repeat domains, a single-pass TM helix and a small ICD293. Intriguingly, IGF2R is a multifunctional receptor that can recognize both IGF2 and mannose-6-phosphate (M6P)294. IGF2 is a protein hormone that shares similar structural features with insulin and IGF1. Through the binding to IGF1R or IR isoform A (IR-A), IGF2 plays critical roles in regulating various cellular processes, such as cell proliferation, skeletal muscle development and regeneration295. Multiple lines of evidence have shown that excessive IGF2 signaling is linked to tumor growth296,297, therefore its signaling is strictly regulated through its interaction with IGF2R. At cell surface, IGF2R serves as an IGF2 “sweeper” that removes IGF2 via the receptor-mediated endocytosis to prevent excessive IGF2 signaling298.
M6P is a key signal molecule that could be recognized by IGF2R and is critical for the IGF2R mediated translocation of acid hydrolase precursor proteins to lysosomes. Particularly, M6P is added exclusively to the N-linked oligosaccharides of lysosomal acid hydrolases to enable the recognition by IGF2R over many other proteins in the trans-Golgi network299. Totally ~50 different M6P-tagged cargo proteins can be recognized by IGF2R to facilitate their transportation from the trans-Golgi to prelysosomes via the early endosome. The acidic environment of the endosome leads to release of cargoes from IGF2R. Subsequently, IGF2R is recycled to the Golgi apparatus or cell surface for another cycle of transportation293,300.
6.2. Cryo-EM structures of the full-length IGF2R in apo- and IGF2-bound form
Lack of structural information on full-length IGF2R limited the understanding of how the 15 β strand-rich repeat domains interact with one another and regulate ligand recognition. This knowledge gap was filled by a study published in 2020 reporting cryo-EM structures of the full-length IGF2R in both apo- and IGF2-bound states301 (Figure 8). Full-length IGF2R was purified directly from bovine liver at acidic pH to ensure release of endogenous cargoes. The IGF2R/IGF2 complex was then prepared by incubating IGF2 with IGF2R sample in a buffer at neutral pH. In the apo state, the β strand-rich repeat domains of IGF2R pack together into a compact structure (Figure 8A). Interestingly, fourteen out of the fifteen domains form seven pairs, which pack into structurally similar two-domain subgroups. These subgroups stack one after another with a similar rotational angle along the long axis of the protein, resulting in an overall helical shape.
Figure 8.
Overall structure of full-length IGF2R in the apo and IGF2-bound states.
A) Cryo-EM structure of apo-IGF2R (PDB: 6UM1). The 15 domains in ECD are organized into an elongated helix-like assembly.
B) Cryo-EM structure of the 1:1 IGF2R/IGF2 complex (PDB: 6UM2). Domains 1-14 rearrange into a pistol-like conformation. Domain 15 becomes disordered and is missing in the EM density. IGF2R (blue) is shown in cartoon representation, whereas the ligand IGF2 is shown in surface representation with semi-transparency. The missing domains in each structure are shown as schematic representations.
IGF2R undergoes a large conformational change and domain rearrangement upon binding the ligand IGF2, which transforms IGF2R from the helical apo-form to the pistol-like IGF2-bound form (Figure 8B). In the IGF2R/IGF2 complex, IGF2 binds IGF2R at the intersection of thes ‘grip’ and the ‘barrel’ of the pistol, making simultaneous interactions with domains 6, 8 and 11 of IGF2R. The binding interfaces bury a total solvent-accessible surface area of 4300 Å2. Some key structural differences between the apo- and IGF2-bound states are important for the IGF2 binding. For instance, in the apo-structure, domains 6 and 9 form one subgroup via their β7 to β11 sheets. In the IGF2-bound state, however, domain 9 is replaced by IGF2. Domain 13, which is on the edge of ECD in the apo-IGFR2R structure, is rearranged to interact with domain 11 in the structure of the IGF2R/IGF2 complex, potentially stabilizing the interaction between IGF2 and domain 11 of IGF2R.
7. Polymeric immunoglobulin receptor
7.1. Introduction
Polymeric immunoglobulin receptor (pIgR) is a glycosylated SPTMR and belongs to the Fc receptor family302. The ECD of pIgR comprises five immunoglobulin (Ig)-like domains (D1-D5), which are responsible for binding polymeric immunoglobins M and A (IgM and IgA), and a non-homologous domain (D6) that contains the site for proteolytic cleavage302-304 (Figure 9A). pIgR is expressed on the basolateral surface of ciliated epithelial cells in the mucosal epithelium and mediates the transcytosis of IgA and IgM from the basolateral to the apical mucosal epithelial cell surface, where they act as the first line of immune defense against mucosal pathogens. The transcytosis can be divided into several major steps303,305-307. It is initiated by the engagement of pIgR with the joining chain (JC or J chain), a small glycoprotein that mediates the polymerization of IgA and IgM on the basolateral surface of the epithelial layer. The pIgR/JC interaction triggers the internalization of pIgR/Ig complex and their subsequent transport from the epithelial cell to the mucosa via the clathrin-mediated endocytosis. Upon arriving at the apical surface, the ECD of pIgR is cleaved off by a host serine proteinase at D6 domain in ECD, releasing the secretory component (SC) that consists of the D1-D5 domains of the pIgR ECD or Ig-bound SC (SIg). The crystal structure of SC in its apo form show that SC adopts a triangular closed conformation with D1 and D5 close to each other (Figure 9A)308. Secretory polymeric IgA and IgM (SIgA and SIgM) are dimeric or higher-order oligomers, which form complexes with JC and SC of pIgR through both non-covalent and covalent interactions. The structures of these highly complicated molecular assembles remained mysterious until the reports of several high-resolution cryo-EM structures recently.
Figure 9.
Overall structures of pIgR (SC) in apo and ligand bound states. pIgR receptor (SC, green) and J-chain (yellow) are shown in cartoon representations, whereas the ligands (IgM, IgA and IgA2m2) are shown in surface representation with semi-transparency.
A) Crystal structure of SC in its apo state (PDB: 5D4K), in which SC adopts a closed triangle-shaped structure with D1 and D5 close to each other. The missing D6 and TM domains of pIgR are shown as schematic representations.
B) Cryo-EM structure of the pentameric IgM in complex with J-chain and SC (SIgM), in which the D2-D5 domains of SC rearrange into a liner conformation (PDB:7K0C). The subunits A and B for each homodimer are defined based on the previous literature.
C) Cryo-EM structure of dimeric SIgA complex (PDB: 6LX3) in two different views, which shows a boomerang-like structure with D2-D5 domains of SC adopting a liner conformation, similar to that of SIgM.
D) Cryo-EM structure of dimeric SIgA in complex with SpsA. The structure of SIgA remains unchanged while SpsA binds to the D3-D4 junction of SC.
E-F) Cryo-EM structures of tetrameric (E) and pentameric (F) SIgA2m2 complexes.
7.2. Cryo-EM structures of IgM and IgA in complex with SC and JC
In 2020 and 2021, two cryo-EM structures of the Fc domains of IgM (IgM-Fc) in complex with SC and JC, were determined by two groups at overall resolutions of 3.58 Å (PDB: 6KXS) and 3.25 Å (PDB: 7K0C) (Figure 9B), respectively309,310. Notably, in the case of 7K0C, additional data at a tilt angle of 40° was collected to overcome particle preferential orientation, a relatively common issue in cryo-EM that causes missing data and limits achievable resolution311. The structures show that IgM forms a pentameric (Fc1-5) complex with one JC and one SC molecule bound. Each Fc domain is a homodimer consisting of three subdomains (Cμ2, Cμ3 and Cμ4). The pentamerization interface of IgM is mediated by the 10 β-strands on the tailpieces, which are arranged into two five-stranded β-sheets that pack with each other in an antiparallel fashion. JC, clamped between Fc1 and Fc5, also contributes to the formation of the pentamer by stacking onto the tailpieces of Fc5B and Fc1A. In addition, two disulfide bonds formed by JC with Fc5B and Fc1A further stabilize their interaction. D1-D5 domains of SC (pIgR), which are well resolved in both cryo-EM structures, form a plane that is nearly perpendicular to the IgM pentamer plane. In the absence of ligands, the SC adopts a triangular closed conformation, similar to a previous crystal structure308. When bound to pentameric IgM, SC D2-D5 are rearranged from the triangular shape into a head-to-tail linear mode, and the linker between D1 and D2 is turned almost 180° to position D1 to interact with IgM. The D1 domain of SC is responsible for the binding to IgM-Fc1 and JC through its three CDR-like loops.
In 2020, the structure of the Fc domains of IgA1 (IgA1-Fc) in complex with SC and JC was determined at 3.2 Å resolution (PDB: 6LX3) 312 (Figure 9C), showing that two IgA1-Fcs assemble into a boomerang-like dimer in the presence of SC and JC, which bears high resemblance to a portion of the IgM-Fc/SC/JC structures. The IgA1-Fc dimeric interface is mediated by the bundling of the four β-strands of IgA1-Fc tailpiece. Similar to that in IgM-Fc/SC/JC structures, JC stabilizes the dimer by stacking onto the IgA1 tailpiece β-strands as well as by contributing to the formation of two disulfide bonds between JC and IgA-Fc. However, compared to that of IgM-Fc/SC/JC, a more complete structure of the JC is resolved in IgA1-Fc/SC/JC, due to the more extensive interactions between the IgA1-Fc dimer and the JC in the IgA1-Fc/SC/JC complex. Two IgA isotypes — IgA1 and IgA2, exist in humans, with the former favoring the dimeric state and the latter having a higher propensity to form higher-order oligomers. Indeed, cryo-EM analyses of IgA2m2-Fc in complex with SC and JC have revealed the tetrameric and pentameric structures 313 (Figure 9E, F). The IgA2m2 pentamer structure resembles that of the IgM pentamer and the tetrametric IgA2m2 structure resembles part of the pentamer. These structural comparisons suggest the JC-stabilized dimer core serves as a building block for high-order oligomerization of IgA2m2-Fc.
7.3. Cryo-EM structure of IgA in complex with the SpsANTD
Various pathogens have evolved strategies to disrupt the function of IgA as a mean of immune evasion306. For example, Streptococcus pneumoniae, an opportunistic pathogen that causes severe diseases, possesses an adhesin IgA-binding protein named SpsA314,315. The N-terminal domain (NTD) of SpsA contains two leucine zipper motifs termed R1 and R2, with R1 containing a highly conserved YRNYPT motif that is critical for its binding to IgA316. The details of the interaction have been revealed by a cryo-EM structure of the complex between the NTD of SpsA and IgA312 (Figure 9D). In this structure, the IgA-Fc/SC/JC core remains the same as seen in the previous structures. The YRNYPT hexapeptide-containing loop between the α1 and α2 helices in the R1 motif of SpsANTD docks into a pocket at the D3–D4 junction of SC, forming the major binding interface between SpsANTD and SC. Interestingly, the D3–D4 pocket is not present in the SC in the apo state, suggesting that SpsA specifically targets SC in the context of the IgA complex. Many residues in human SC involved in the interaction with the YRNYPT hexapeptide are not conserved in SC from other species, consistent with the fact that SpsA only binds to human IgA and S. pneumoniae is a human-specific pathogen.
8. IL-10 receptor
8.1. Introduction
Cytokines represent a diverse group of small soluble proteins that are secreted by a wide range of cells including immune cells, endothelial cells, fibroblasts, and various stromal cells317,318. Cytokines play important roles in many aspects of cell signaling including induction of immune responses, hematopoiesis, cell proliferation, differentiation, and apoptosis319-322. Interleukin-10 (IL-10) is best known as an anti-inflammatory cytokine that plays a central role in terminating inflammatory responses323-325. IL-10 is a member of the Type II cytokines, which possesses a characteristic α-helical fold consisting of six helices, with a four α-helical bundle core326. Functional IL-10 receptors are composed of two subunits – α (IL-10Rα) and β (IL-10Rβ)327. IL-10Rα is the high-affinity, IL-10-specific subunit, whereas IL-10β is the low-affinity, shared receptor subunit that is also used by other cytokines. IL-10Rα and IL-10Rβ are both SPTMRs that consist of an ECD that binds IL-10, a single-pass TM, and a cytoplasmic domain. Despite the low sequence similarity between the ECDs of IL-10Rα and IL-10Rβ, their structures are strikingly similar, i.e., both consist of two fibronectin type III domains (FN domains D1 and D2), forming an L-shaped structure (Figure 10A)328,329. The IL-10 signaling cascade is initiated by secreted IL-10 homodimer binding to both IL-10Rα and IL-10Rβ, inducing the heterodimerization of IL-10Rα and IL-10Rβ and bringing two copies of the heterodimeric receptor together. JAK1 and Tyk2 simultaneously binds to the ICDs of IL-10Rα and IL-10Rβ, respectively, and phosphorylate each other upon receptor heterodimerization. This leads to activation of the JAK/STAT3 signaling pathway, which triggers a specific transcriptional program that largely defines the IL-10–mediated anti-inflammatory response330. In addition to JAK/STAT3, activated IL-10 receptors could also trigger other signaling cascades, such as PI3K/Akt/GSK3β, in macrophages or primary monocytes331,332.
Figure 10.
Overall structure of IL-10Rα and IL-10Rβ complex in apo or IL-10 bound states and the proposed activation mechanism. Receptors IL-10Rα (blue) and IL-10Rβ (green) are shown in cartoon representations, whereas the ligand IL-10 is shown as surface representations with semi-transparency. The missing domains (TM and TIR) in each structure are shown as schematic representations.
A) Crystal structures of IL-10Rα and IL-10Rβ in their respective apo states as inactive monomers (PDBs: 1Y6K and 3LQM). The ECDs of both proteins consist of D1 and D2 domains. IL-10Rα has a longer ICD than IL-10Rβ, both contains two regions (named Box1 and Box2) that interact with the JAK1 or Tyk2 kinases.
B) Cryo-EM structure of 2:2:2 IL-10/IL-10Rα/IL-10Rβ in two different views (PDB: 6X93). Dimerization of IL-10Rα/IL-10Rβ induced by IL-10 leads to engagement of JAK1 and Tyk2 kinases, and phosphorylation of JAK1 and Tyk2 as well as the ICDs of IL-10Rα and IL-10Rβ.
Defects of IL-10 signaling has been implicated in a variety of diseases, including inflammatory bowel disease (IBD) and cancer333-335. Therefore, there is substantial interest in targeting IL-10 and IL-10R as therapeutics for these diseases336,337. However, the therapeutic utility of IL-10 is limited by its pleotropic and sometimes paradoxical functions. For example, its potential as an anti-inflammatory drug has not been fully realized, since it often elicits proinflammatory effects when administered systematically. To this end, a cryo-EM structure of IL-10/IL-10R complex has guided design of IL-10 variants with decreased binding affinity to IL-10Rβ, which appear to maintain the anti-inflammatory function of IL-10 but greatly reduce its proinflammatory activity 338,339.
8.2. Cryo-EM structure of IL-10/IL-10Rα/IL-10Rβ hexameric complex
Structural study of the complete IL-10/IL-10R complex is challenging partly due to the low binding affinity between IL-10 and IL-10Rβ323. Saxton et al overcame this obstacle by selecting IL-10 variants with stronger affinity to IL-10Rβ using yeast surface display-based directed evolution338. The best variant, dubbed ‘super 10’, contains four point-mutations (N18Y, N92Q, T100D, and R104W) and shows 10,000-fold increased affinity to IL-10Rβ compared to wild type IL-10. Super 10 forms a stable 2:2:2 complex with IL-10Rα and IL-10Rβ, allowing high-resolution cryo-EM structural determination. The structure shows that the IL-10/IL-10Rα/IL-10Rβ complex forms an elongated star-shape with the domain-swapped IL-10 homodimer localized at the junction between the two IL-10Rα/IL-10Rβ units (Figure 10B). Three distinct contact sites among the IL-10, IL-10Rα and IL-10Rβ proteins are essential for the formation of the hexametric complex. IL-10 interacts with D1 domain of IL-10Rα through α1 and α5 at site one, similar to that in the IL-10/IR-10Rα crystal structure329. The α1-α3 helices from IL-10 interact with the D1 domain of IL-10Rβ at site two. The D2 domains of IL-10Rα and IL-10Rβ interact with each other, forming a relatively small interface at site three. Notably, the IL-10/IL-10Rβ interaction at site two is primarily mediated by the hydrophobic contact between Met22 in α1 of IL-10 and Tyr82 in L3 of IL-10Rβ. Such interaction pattern suggests that the increased affinity of ‘super 10’ IL-10 variant for IL-10Rβ binding likely results from the N18Y and R104W mutations, which are placed close to Met22 and potentially enhance the interaction between of IL-10 and IL-10Rβ by increasing the hydrophobicity of site 2.
Based on the structure, the authors generated IL-10 variants with mutations at the IL-10/IL-10Rβ interface in order to modulate the pleotropic functions of IL-10. Remarkably, some of these variants showed reduced affinity for IL-10Rβ and behaved as partial agonists as they could selectively suppress monocyte and macrophage activity without activating inflammatory CD8+ T cells. These partial agonists have exciting potential as therapeutic agents for autoimmune diseases.
9. Single-pass transmembrane receptors in complex with tetraspanins
9.1. Introduction
Some SPTMRs form co-receptor complexes with tetraspanins, a family of integral membrane proteins characterized by the presence of four transmembrane helices340-342. Tetraspanins have been implicated in several biological processes, including development, reproduction, immunity341,343. However, the molecular mechanisms of these functions of tetraspanins and their roles in the regulation of SPTMRs remain largely unclear. CD81 represents one of the better understood tetraspanins in terms of the functional role (Figure 11A). Particularly, CD81 interacts with CD19, a SPTMR specifically expressed on the surface of B cells344,345. CD19, CD81 and the complement receptor CD21 together form the co-receptor complex for the B cell receptor, by which they play a critical role in regulating B cell maturation and activation346. CD19 acts as the signaling subunit of the co-receptor complex by using its ICD to recruit intracellular signaling components, while CD81 functions as a chaperone to facilitate the maturation through the secretory pathway and the cell surface presentation of CD19347. CD81 may also regulate the signaling of CD19 on the cell surface by controlling its localization in the plasma membrane348. The interaction between CD19 and CD81 has been elucidated by a cryo-EM structure in 2021349. In addition, two other structures of SPTRMs in complex with tetraspanins, EWI-2/CD9 and EWI-F/CD9, have also been solved by cryo-EM in 2020350,351. A general mode of interaction between SPTRMs and tetraspanins, along with some unique features in each case, has emerged from these studies349-351.
Figure 11.
Cartoon representations of the overall structures of tetraspanin CD81 alone and in complex with the B-cell coreceptor CD19.
A) Crystal structure of CD81 (PDB: 5TCX) reveals a cholesterol binding pocket.
B) Cryo-EM structure of the CD81/CD19 complex (PDB: 7JIC). The CTD of CD19 and the flexible linker between CTD of CD19 and TM1 of CD81 are shown as schematic representations.
9.2. Cryo-EM studies of EWI-2/CD9 and EWI-F/CD9
EWI-2 (also known as Immunoglobulin superfamily member 8 or IGSF8) and EWI-F (also known as Prostaglandin F2 receptor negative regulator or PTGFRN) are SPTRMs that comprise an ECD, a single-pass TM, and a short C-terminal cytoplasmic domain. As two members of immunoglobulin superfamily (IgSF), the ECDs of EWI-2 and EWI-F comprise four and six highly glycosylated Ig-like C2 domains, respectively. Both EWI-2 and EWI-F proteins have been shown to be major partners for tetraspanins CD9 and CD81352. They connect these two tetraspanins to the cytoskeleton through their direct association with ezrin-radixin-moesin proteins and play key roles in diverse functions of CD81 and CD9, such as oocyte fertilization353-356. In addition, EWI-2 has been shown to moderate TGF-β and integrin signaling via its interaction with CD9 and CD81357,358. High EWI-F expression has been shown to be associated with glioblastoma multiforme359.
Two independent cryo-EM studies on the EWI-2/CD9 and EWI-F/CD9 complexes were published in 2020 350,351. Although neither of the studies achieved high resolution, the EM maps provide valuable information on the interactions between EWI-2 or EWI-F and the tetraspanin. In the cryo-EM study of EWI-2/CD9351, the full-length EWI-2/CD9 complex was co-expressed and purified in the detergent digitonin. The cryo-EM map was resolved at an overall resolution of 8.2 Å and slightly improved to 7.2 Å when an anti-CD9 Fab was used to improve the particle alignment. The complex displays a 2:2 hetero-tetrameric architecture, with two protomers of EWI-2 forming a tight dimer at the center, accompanied by two CD9 protomers sandwiching the EWI-2 dimer. The four Ig-like C2 domains and the membrane-spanning region of EWI-2 could be roughly fit into the density map, showing a ‘L’ shaped conformation. The two TMs of the EWI-2 dimer are far apart, only making minimal contacts at the intracellular end, while the dimerization of EWI-2 is mediated primarily by the Ig-like domains D3 and D4. The arrangement of the four TM helices of CD9 is in good agreement with that in the crystal structure of CD9 alone.
It has been suggested that tetraspanins are able to self-assemble into high-order complexes, called ‘tetraspanin-enriched microdomains’352. In the same paper reporting the structure of EWI-2/CD9, the authors proposed that, due to the particular cone-like shape, tetraspanins in the higher-order oligomeric state could induce membrane curvature, and thereby facilitate the formation of exosomes vesicles. The tetraspanin cluster on the membrane is capable of recruiting different types of receptors and other critical functional proteins into proximity, which might represent an important initial step of cargo sorting in exosomes.
In the cryo-EM study of the EWI-F/CD9 complex350, a truncated EWI-F (EWI-FΔIg1-5) was co-expressed with full-length CD9 and purified in digitonin in the presence of a CD19-specific nanobody 4C8. Single-particle cryo-EM analysis of the EWI-F/CD9/4C8 complex yielded an EM map with an overall resolution of 8.6 Å. The reconstruction of EWI-F/CD9/4C8 revealed a hetero-tetrameric complex with strikingly similarity to that of EWI-2/CD9, i.e., a central EWI-FΔIg1-5 dimer flanked by 4C8-bound CD9 on each side. CD9 adopts a semi-open conformation, and TM3-4 helices of CD9 are in direct contact with the TM of EWI-2, similar to the binding model between EWI-2 and CD9. Notably, the TM helices of EWI-F are even further apart in EWI-F/CD9/4C8 than that in the EWI-2/CD9 complex, which might be caused by the presence of two palmitoylation sites in EWI-F360.
9.3. Cryo-EM structure of the B cell co-receptor CD19 bound to the tetraspanin CD81
CD19 is a SPTMR that is consisted of two Ig-like C2 domains in the extracellular region. Susa et al used several strategies to stabilize the CD19/CD81 complex for cryo-EM analyses349. Firstly, the C-terminal end of CD19 was fused to the N-terminal end of CD81 with a flexible linker. In addition, the Fab fragment of the therapeutic anti-CD19 antibody was bound to the purified CD19/CD81 fusion protein in order to increase the size of the complex for better image alignment. These strategies allowed the authors to solve the cryo-EM structure of the CD19/CD81 complex to an overall resolution of 3.8 Å 349 (Figure 11B). The structure shows that the interaction between CD19 and CD81 involves both the TM and ECD regions of both proteins. The interface at the TM is mediated by the single TM helix of CD19 with both TM1 and TM2 of CD81. The ECD interface is formed between a large hydrophobic surface of the ECD of CD19 and one face of the EC2 domain of CD81. Interestingly, a comparison with the crystal structure of apo-CD81340 shows that the binding of CD19 induces several concerted conformational changes to CD81. The most notable one is an upward swing of the EC2 domain of CD81 away from the TMD, which is accompanied by the disorder-to-order transition of the EC1 domain. This conformational change is necessary to expose the hydrophobic residues in EC2 for interacting with the ECD of CD19. CD19 also induces a dramatic conformational change to the TMD of CD81. In the apo-state, TM1/TM2 and TM3/TM4 form two pairs of helices that interact with each other extensively at the intracellular side of the membrane but diverge at the extracellular side, resulting in a central cavity that has been shown to bind cholesterol340. CD19 binding induces an inward movement of the TM1-TM2 and TM3-TM4 pairs relative to each other, eliminating the central cavity and precluding cholesterol binding. These observations suggest that cholesterol binding may play a regulatory role in the interaction between CD19 and CD81. Specifically, in ER membrane which has low level of cholesterol, CD81 is able to interact with CD19, and the binding of CD81 would facilitate the translocation of CD19 from ER to plasma membrane. However, at the plasma membrane where the concentration of cholesterol is much higher, the binding of cholesterol to CD81 would promote the dissociation of CD81 from CD19, allowing the CD19 to interact with the B cell receptor, a step that is essential for the functionality of the B cell receptor. Related to this point, antibodies that specifically recognize the CD19/CD81 complex may be able to trap CD19 in the complex form and suppress the signaling, which may be exploited for treating diseases caused by hyper activation of B cells.
10. Teneurins
10.1. Introduction
Teneurins are type-II single-pass transmembrane proteins, with their N-terminal and C-terminal parts constituting the intracellular and extracellular regions, respectively. They function as cell adhesion receptors that are involved in the development of the nervous system by regulating processes such as neuronal axon guidance and synapse formation361,362. The C-terminal extracellular region binds latrophilin, an adhesion G-protein-coupled receptor (GPCR), to form a complex that regulates synaptogenesis and axon guidance363-366. The functions of teneurins are not limited to adhesion, as their intracellular domain can be proteolytically cleaved and transduce signal through acting as a transcriptional repressor367. Conversely, it has been suggested that teneurins can trigger latrophilin intracellular signal and alter intracellular cAMP levels368. Teneurins are very large SPTMRs (~2800 residues), containing small N-terminal intracellular region, a single-pass TM and a multi-domain C-terminal extracellular region. Teneurins form constitutive dimers by inter-chain disulfide bonds formed in the membrane proximal epidermal growth factor-like (EGF) domains369,370. The extracellular region of teneurins does not contain common adhesion domains such as Ig-like or cadherin-like domains. Sequence analyses have shown that the C-terminal portion of this region is homologous to the tyrosine-aspartate (YD) repeat-containing toxins widespread in bacteria, suggesting that teneurins are originated from a horizontal gene transfer from bacteria371.
10.2. Cryo-EM structures of Teneurins
Several structures of the teneurins extracellular region either in the apo state or with latrophilin bound have been solved by X-ray crystallography and cryo-EM368,372-374 (Figure 12). The structures confirm that teneurins do show strong resemblance to bacterial Tc-toxins, although they have lost the actual nuclease activity for toxicity374. Therefore, it appears that this structural fold of the bacterial Tc-toxins has been repurposed for protein-protein interactions in higher organisms during evolution. The most characteristic feature of the teneurin structures is the 78-stranded β-barrel shell formed by the YD-repeats. One end of the YD-shell is sealed by a fibronectin-plug domain and downstream 6-bladed β-propeller domain. Strikingly, the linker following the YD shell mostly resides in the interior of the shell itself, while its C-terminal part threads out of the shell through a gap on the shell wall. The region downstream of the linker contains the domain that shows homology to bacterial toxins with nuclease activities, however, this domain in teneurins has lost the catalytic residues and unlikely to possess nuclease activities. The toxin-like domain sits on the side of the YD shell and appears loosely attached. In the cryo-EM structure of mouse Teneurin-3, the C-terminal region including the nuclease-like domain is not resolved, suggesting that it is flexible in relation to the YD shell. This flexibility has been suggested to facilitate the proteolytic cleavage event in this region, which releases a neuropeptide that has its own signaling function in the nervous system375-377. In this regard, structural flexibilities, which could be critical for functions for many proteins, could be more easily observed in cryo-EM structures as the proteins are not confined in crystal lattices. Moreover, multiple conformations could be resolved through 3D classification from one cryo-EM dataset, potentially allowing the conformational transition pathway to be inferred.
Figure 12.
Cryo-EM structure of 1:1 Teneurin (TEN2)/Latrophilin (LPHN3) complex (PDB: 6VHH). Four domains of TEN2 (Toxin-like, β-propeller, β-Barrel, and Ig-like) and one domain of LPHN3 (Lectin) are shown in the structure. The domains that are missing in the structure are shown as schematic representations. TEN2 (green, shown in cartoon representation) is localized to the presynaptic membrane, whereas LPHN3 (pink, shown in surface representation with semi-transparency) is localized to the postsynaptic membrane.
Latrophilin uses its N-terminal lectin domain to bind one side of the YD shell of teneurin, opposite to the side where the C-terminal toxin-like domain resides372,373. The binding mode is consistent with the trans-cellular interaction between teneurin and latrophilin. It has been shown that latrophilin can also use its olfactomedin domain, which follows immediately the lectin domain, to interact with the leucin-rich repeat domain of another cell surface receptor FLRT378. Modelling based on the two binary complex structures has suggested that the three proteins can form a ternary complex372,373. This observation is consistent with the idea that both teneurin and FLRT expressed on one cell surface serve as the ligands simultaneously for latrophilin expressed on an opposed cell surface, allowing latrophilin to act as a coincidence detector and transduce signals required for excitatory synapse formation366.
The N-terminal intracellular region, TM region and the EGF-like domains in teneurin are not present in any of the solved structures. It is therefore not clear how binding of latrophilin may lead to signaling of the teneurin intracellular region. Likewise, the full-length structures of latrophilin and FLRT are not available, and therefore it is not known how complex formation induces intracellular signaling of latrophilin and FLRT. Cryo-EM structures of the complex of full-length teneurin, latrophilin and FLRT will help address these questions.
11. T-cell receptor
11.1. Introduction
T cell-mediated immunity is a critical arm of the adaptive immune system379. Activation of T cells is governed by the αβ heterodimeric T cell receptor (TCR), which contains the extracellular variable domains that recognize antigenic peptides presented by the major histocompatibility complex (MHC) on the antigen presenting cells (APCs)380,381. The activation of the TCR-CD3 complex leads to the phosphorylation of intracellular immunoreceptor-tyrosine-based activation motifs (ITAMs) localized in the cytoplasmic region of the CD3 subunits, and thereby triggering the downstream signaling cascade382. The αβ TCR forms a holo-complex with the CD3 signaling hexamer composed of CD3γε, CD3δε and CD3ζζ dimeric units383. All the subunits of the TCR-CD3 complex are SPTMRs, which together organize into a complicated and precisely controlled signaling machinery that is highly sensitive to foreign antigenic peptides but tolerant to self-peptides. Due to its central role in T cell immunity, many structural studies have been devoted to understanding the mechanisms of assembly and signaling of the TCR-CD3 complex, but the whole picture of the holo-complex remained unknown until the application of cryo-EM to the field.
11.2. Cryo-EM structure of full-length TCR-CD3 complex
The cryo-EM structure of a complete version of full-length TCR-CD3 complex in detergent digitonin was determined in 2019384, representing a major milestone in the structural biology of TCR. Consistent with previous biochemical data379, the cryo-EM structure shows that the TCR-CD3 complex is assembled among dimeric TCRαβ, CD3γε, CD3δε and CD3ζζ in a stoichiometry of 1:1:1:1 (Figure 13). The assembly of TCR-CD3 is mainly driven by the tight packing of the 8 single-pass TMs, which can be considered the bottom layer of the receptor complex. The structure clearly resolves both the hydrophobic and more notably charge-complementary interactions among the TM helices that underlie the specific barrel-like structure of TMγε, TMδε and TMζζ that embraces TMαβ at the center. The ECDs of CD3γε and CD3δε and the membrane proximal constant domains of TCRαβ (Cα and Cβ) also contribute to the stability of the holo-complex by forming an approximately three-fold symmetric assembly at the middle layer of the receptor. The heterodimer of the variable domains Vα and Vβ of TCRαβ constitutes the top layer that protrudes away from the T cell surface, poised to engage the peptide/MHC complex on the opposed APC. Overall, this cryo-EM structure provides valuable information for defining the precise assembly mechanism of the TCR-CD3 complex. In 2022, the same group reported several cryo-EM structures of the same TCR-CD3 complex bound to cholesterol or cholesterol sulfate385. Based on these structures, the authors proposed that cholesterol serves as a negative regulator of TCR by locking the TM helix of CDζ in an inactive conformation in the plasma membrane.
Figure 13.
Cryo-EM structure of the TCR-CD3 holo-complex. CD3 contains 4 chains- δ, γ, ε and ζ that forms three dimers. Each of δ, γ and ε chains contains an ECD, a TM and an ICD contains an immunoreceptor-tyrosine-based activation motif (ITAM), whereas CD3ζ contains a short extracellular loop, a TM and an ICD with three ITAMs.
The TCRα/TCRβ/CD3 complex is assembled by the TCRαβ, CD3γε, CD3δε, and CD3ζζ dimers in a 1:1:1:1 stoichiometry. Activation of the complex by the antigenic peptide/MHC complex leads to phosphorylation of ITAMs of CD3.
However, a comparison of the cryo-EM structures of the holo-TCR complex in the apo state with the previous ECD structures of the TCRαβ either in the apo state or bound to a peptide/MHC complex shows no substantial conformational changes in the ECD of TCR that may trigger the activation of the receptor386,387. In addition, the intracellular ITAMs domains of CD3 are not resolved in the cryo-EM map, indicating their structural flexibility and lack of rigid coupling with the ECD and TM of the receptor. Therefore, the structures do not provide a clear answer to the question how exactly the receptor is activated by the antigenic peptide/MHC complex. In the absence of substantial ligand-induced conformational changes, several models have been proposed to explain TCR activation, including receptor clustering, orientational change relative to the membrane and mechanical force imposed on the TCR by the peptide/MHC complex. These mechanisms may only be fully operational when the TCR and peptide/MHC complex are located in the T cell membrane and APC membrane, respectively, which are arranged to form the immunological synapse with the facilitation of adhesion molecules and other auxiliary proteins. Therefore, solving the structure of the peptide/MHC/TCR-CD3 holo-complex in the membranes may provide insights into the fine structural changes in the ECDs and TMDs of the TCR-CD3 complex that are required for its activation. Nevertheless, mechanical force cannot be applied during cryo-EM sample preparation. Employing integrative approaches that combine cryo-EM/ET, optical tweezers, NMR and atomic force microscopy might be needed to fully reveal the activation mechanism of TCR. Finding mutations in TCR-CD3 that could stabilize its active conformation could become an alternative approach to obtain the structure of the TCR-CD3 complex in the activated functional state.
12. Future Perspectives
The structures discussed in this review showcase the power of cryo-EM in advancing our understanding of the structures and mechanisms of SPTMRs. Recent development in both software and hardware has allowed cryo-EM to reach true atomic resolution (around 1.2 Å resolution) for highly stable proteins, especially those with high symmetry39,40. However, it remains challenging to achieve atomic resolution with SPTMRs with cryo-EM for more detailed mechanistic analyses and drug design, largely due to the inherent conformational flexibility of these proteins. It is also difficult to capture weakly associated complexes with cryo-EM as it typically requires low sample concentrations, under which weakly associated proteins tend to dissociate, as apposed to in crystallography where proteins reach high concentrations in crystals to promote complex formation. In the foreseeable future, mechanistic studies of SPTMRs will continue to benefit from combining insights from cryo-EM structures of large complexes, high-resolution X-ray structures of smaller complexes or individual proteins as well as computational models. Another challenge in cryo-EM studies of SPTRMs is that a direct view of the coupling between the ECD and ICD by the TM helix remains elusive. To date, none of the cryo-EM structures was able to resolve a complete structure of any SPTMR, due to the flexible linkages of the TM helix with both the ECD and ICD. Going forward, stabilization of the junction at the two ends of the TM by means of accessory proteins such as tetraspanins388 and TM-associating peptides10,389 might be needed to allow for the visualization of the SPTMRs in their intact forms by cryo-EM. It is possible that reconstituting the receptor/ligand complexes into native-like lipid membrane may stabilize the conformation of the receptors55,160,390,391, allowing cryo-EM structural analyses to reveal the entire full-length structure of receptor/ligand complexes. In addition, many SPTMRs such as integrins and Eph receptors form large clusters upon activation392, of which the underlying mechanisms are poorly understood. Cryo-EM structural analyses of SPTMRs in clusters will help elucidate the protein-protein and protein-lipid interactions underlying their formation, paving the way for in depth analyses of their roles in regulating the function of these proteins. Another exciting frontier is to study SPTMRs from native source by cryo-EM, which is made possible by the fact that cryo-EM requires very small amounts of sample and can tolerate certain levels of impurity. Most SPTMRs have multiple protein partners, some of which may be at low abundance and/or only transiently interact with SPTMRs. Analyzing SPTMRs from native sources with a combination of biochemical (e.g., mass spectrometry) and structural techniques (e.g., cryo-EM with 3D classification) will make it possible to identify these obscure effectors and capture their interactions with SPTMRs, resulting in a more complete picture of the working mechanisms. This approach has been used successfully for several large protein complexes in the past few years393,394.
Recent development of another cryo-EM technique – cryo-electron tomography (cryo-ET) has opened up another exciting frontier that is to investigate SPTMRs structures in their native environment within the cellular context395,396. The structures and functions of SPTMRs on the cell surface are regulated by many molecules in the environment, including other proteins, lipids, and carbohydrates. Traditional approaches to retrieve the structural information of SPTMRs by using purified proteins inevitability lack this important cellular context. Cryo-ET, in contrast, does not require the isolation of proteins and therefore makes it possible to visualize proteins in their native conditions. The combination of cryo-correlative light and electron microscopy (cryo-CLEM)397 and cryo-focus ion-beam (cryo-FIB) milling398 would allow for precise localization of SPTMRs on the cell membranes and subsequent sample preparation for cryo-ET imaging. Subtomogram averaging, an averaging technique analogous to single particle analysis, can be used to improve the signal-to-noise ratio and achieve near-atomic resolution399. Cryo-ET can also be used to investigate the structures and mechanisms of receptor clusters on native cell membranes or reconstituted in liposomes, as exemplified by the in situ structural study of the IRE1α kinase400. Some SPTMRs interact with their transmembrane ligands at specific cell-cell contact sites (e.g., immunological synapse401), which play critical roles in regulating signaling. The powerful combination of cryo-CLEM, cryo-FIB and cryo-ET opens up exciting possibilities to study SPTMRs at the cell-cell contact sites and uncover their structures in the physiological context. High-resolution cryo-EM structures of SPTMRs in native conditions will greatly advance our understanding of their functions and the underlying mechanisms, which will assist development of novel therapeutics for diseases related to dysregulation of SPTMRs.
Funding sources:
This work is supported in part by grants from the National Institutes of Health (R35GM130289 to X.Z., R01GM136976 and R01GM143158 to X.-c.B.) and the Welch foundation (I-1702 to X.Z. and I-1944 to X.-c.B.). X.-c.B. and X.Z. are Virginia Murchison Linthicum Scholars in Medical Research at UTSW.
Biographies
Kai Cai Ph.D. obtained his B.S. in chemistry in Nanjing University, China. He obtained his Ph.D.in Biochemistry at University of Wisconsin-Madison under the supervision of Prof. John L. Markley, where he used nuclear magnetic resonance (NMR) to study proteins involved in human mitochondrial Iron-sulfur cluster biogenesis. He is currently a postdoctoral researcher at UT Southwestern Medical Center, in the research group Prof. Xiaochen Bai's lab, where his research interest is focused on investigating the structures of full-length receptor tyrosine kinases using cryo-electron microscopy.
Xuewu Zhang, Ph.D., obtained his bachelor degree in biology from Wuhan University in China in 1995. He got his Ph.D. degree at Albert Einstein College of Medicine in 2003, majored in structural biology. He then did his postdoctoral training with John Kuriyan at University of California, Berkeley, from 2003 to 2007. He joined the faculty of University of Texas Southwestern Medical Center in 2007 and currently a tenure professor in the Pharmacology department.
Xiao-chen Bai Ph.D. has been working on cryo-EM method development and structural determination for more than a decade. When he was a Ph.D. student in Prof. Sen-fang Sui lab at Tsinghua University, he was among the first to use single-particle cryo-EM for structural studies in China. While he was a postdoc in Sjors Scheres lab at MRC-LMB in UK, he was involved in the development of entirely new techniques that dramatically improved the capabilities of cryo-EM technology. After starting his independent lab at UTSW in 2017, he has established a highly productive research program on the structural and functional studies of receptor tyrosine kinases.
References:
- (1).Almen MS; Nordstrom KJ; Fredriksson R; Schioth HB Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol. 2009, 7, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Reis R; Moraes I Structural biology and structure-function relationships of membrane proteins. Biochem. Soc. Trans 2018, DOI: 10.1042/BST20180269 10.1042/BST20180269. [DOI] [PubMed] [Google Scholar]
- (3).Engel A; Gaub HE Structure and mechanics of membrane proteins. Annu. Rev. Biochem 2008, 77, 127–148. [DOI] [PubMed] [Google Scholar]
- (4).Overington JP; Al-Lazikani B; Hopkins AL How many drug targets are there? Nat. Rev. Drug Discov 2006, 5, 993–996. [DOI] [PubMed] [Google Scholar]
- (5).Bugge K; Lindorff-Larsen K; Kragelund BB Understanding single-pass transmembrane receptor signaling from a structural viewpoint-what are we missing? FEBS J. 2016, 283, 4424–4451. [DOI] [PubMed] [Google Scholar]
- (6).Hubert P; Sawma P; Duneau JP; Khao J; Henin J; Bagnard D; Sturgis J Single-spanning transmembrane domains in cell growth and cell-cell interactions: More than meets the eye? Cell Adh. Migr 2010, 4, 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Pahl MC; Askinazi OL; Hamilton C; Cheng I; Cichewicz K; Kuhn J; Manohar S; Deppmann CD Eng. Life Sci, DOI: 10.1002/9780470015902.a0025160 10.1002/9780470015902.a0025160. [DOI] [Google Scholar]
- (8).Fagerberg L; Jonasson K; von Heijne G; Uhlen M; Berglund L Prediction of the human membrane proteome. Proteomics 2010, 10, 1141–1149. [DOI] [PubMed] [Google Scholar]
- (9).Kirkbride KC; Ray BN; Blobe GC Cell-surface co-receptors: emerging roles in signaling and human disease. Trends Biochem. Sci 2005, 30, 611–621. [DOI] [PubMed] [Google Scholar]
- (10).Westerfield JM; Barrera FN Membrane receptor activation mechanisms and transmembrane peptide tools to elucidate them. J. Biol. Chem 2020, 295, 1792–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Maruyama IN Activation of transmembrane cell-surface receptors via a common mechanism? The "rotation model". Bioessays 2015, 37, 959–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Heldin CH; Lu B; Evans R; Gutkind JS Signals and receptors. Cold Spring Harb. Perspect. Biol 2016, 8, a005900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Schlessinger J Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell 2002, 110, 669–672. [DOI] [PubMed] [Google Scholar]
- (14).Heldin CH Dimerization of cell surface receptors in signal transduction. Cell 1995, 80, 213–223. [DOI] [PubMed] [Google Scholar]
- (15).Yarden Y; Schlessinger J Epidermal growth factor induces rapid, reversible aggregation of the purified epidermal growth factor receptor. Biochemistry 1987, 26, 1443–1451. [DOI] [PubMed] [Google Scholar]
- (16).Pawson T Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell 2004, 116, 191–203. [DOI] [PubMed] [Google Scholar]
- (17).Lemmon MA; Schlessinger J Cell signaling by receptor tyrosine kinases. Cell 2010, 141, 1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Citri A; Yarden Y EGF-ERBB signalling: towards the systems level. Nat. Rev. Mol. Cell Biol 2006, 7, 505–516. [DOI] [PubMed] [Google Scholar]
- (19).Ebina Y; Ellis L; Jarnagin K; Edery M; Graf L; Clauser E; Ou JH; Masiarz F; Kan YW; Goldfine ID et al. The human insulin receptor cDNA: the structural basis for hormone-activated transmembrane signalling. Cell 1985, 40, 747–758. [DOI] [PubMed] [Google Scholar]
- (20).Croll TI; Smith BJ; Margetts MB; Whittaker J; Weiss MA; Ward CW; Lawrence MC Higher-resolution structure of the human insulin receptor ectodomain: multi-modal inclusion of the insert domain. Structure 2016, 24, 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Xu Y; Kong GK; Menting JG; Margetts MB; Delaine CA; Jenkin LM; Kiselyov VV; De Meyts P; Forbes BE; Lawrence MC How ligand binds to the type 1 insulin-like growth factor receptor. Nat. Commun 2018, 9, 821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Ullrich A; Bell JR; Chen EY; Herrera R; Petruzzelli LM; Dull TJ; Gray A; Coussens L; Liao YC; Tsubokawa M et al. Human insulin receptor and its relationship to the tyrosine kinase family of oncogenes. Nature 1985, 313, 756–761. [DOI] [PubMed] [Google Scholar]
- (23).De Meyts P; Whittaker J Structural biology of insulin and IGF1 receptors: implications for drug design. Nat. Rev. Drug Discov 2002, 1, 769–783. [DOI] [PubMed] [Google Scholar]
- (24).Lawrence MC Understanding insulin and its receptor from their three-dimensional structures. Mol. Metab 2021, 52, 101255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Maity PC; Blount A; Jumaa H; Ronneberger O; Lillemeier BF; Reth M B cell antigen receptors of the IgM and IgD classes are clustered in different protein islands that are altered during B cell activation. Sci. Signal 2015, 8, ra93. [DOI] [PubMed] [Google Scholar]
- (26).Klasener K; Maity PC; Hobeika E; Yang J; Reth M B cell activation involves nanoscale receptor reorganizations and inside-out signaling by Syk. Elife 2014, 3, e02069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Miyamoto S; Akiyama SK; Yamada KM Synergistic roles for receptor occupancy and aggregation in integrin transmembrane function. Science 1995, 267, 883–885. [DOI] [PubMed] [Google Scholar]
- (28).Wimmer-Kleikamp SH; Janes PW; Squire A; Bastiaens PI; Lackmann M Recruitment of Eph receptors into signaling clusters does not require ephrin contact. J. Cell Biol 2004, 164, 661–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Huang Y; Bharill S; Karandur D; Peterson SM; Marita M; Shi X; Kaliszewski MJ; Smith AW; Isacoff EY; Kuriyan J Molecular basis for multimerization in the activation of the epidermal growth factor receptor. Elife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Bray D; Levin MD; Morton-Firth CJ Receptor clustering as a cellular mechanism to control sensitivity. Nature 1998, 393, 85–88. [DOI] [PubMed] [Google Scholar]
- (31).Endres NF; Das R; Smith AW; Arkhipov A; Kovacs E; Huang Y; Pelton JG; Shan Y; Shaw DE; Wemmer DE et al. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell 2013, 152, 543–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Nygaard R; Kim J; Mancia F Cryo-electron microscopy analysis of small membrane proteins. Curr. Opin. Struct. Biol 2020, 64, 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Cheng Y Membrane protein structural biology in the era of single particle cryo-EM. Curr. Opin. Struct. Biol 2018, 52, 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Vinothkumar KR Membrane protein structures without crystals, by single particle electron cryomicroscopy. Curr. Opin. Struct. Biol 2015, 33, 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Bai XC; McMullan G; Scheres SH How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci 2015, 40, 49–57. [DOI] [PubMed] [Google Scholar]
- (36).Bai XC Seeing atoms by single-particle cryo-EM. Trends Biochem. Sci 2021, 46, 253–254. [DOI] [PubMed] [Google Scholar]
- (37).Bai XC; Fernandez IS; McMullan G; Scheres SH Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. Elife 2013, 2, e00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Li X; Mooney P; Zheng S; Booth CR; Braunfeld MB; Gubbens S; Agard DA; Cheng Y Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 2013, 10, 584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Yip KM; Fischer N; Paknia E; Chari A; Stark H Atomic-resolution protein structure determination by cryo-EM. Nature 2020, 587, 157–161. [DOI] [PubMed] [Google Scholar]
- (40).Nakane T; Kotecha A; Sente A; McMullan G; Masiulis S; Brown P; Grigoras IT; Malinauskaite L; Malinauskas T; Miehling J et al. Single-particle cryo-EM at atomic resolution. Nature 2020, 587, 152–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Bai XC; Yan C; Yang G; Lu P; Ma D; Sun L; Zhou R; Scheres SHW; Shi Y An atomic structure of human gamma-secretase. Nature 2015, 525, 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Chen Q; She J; Zeng W; Guo J; Xu H; Bai XC; Jiang Y Structure of mammalian endolysosomal TRPML1 channel in nanodiscs. Nature 2017, 550, 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Guo J; She J; Zeng W; Chen Q; Bai XC; Jiang Y Structures of the calcium-activated, non-selective cation channel TRPM4. Nature 2017, 552, 205–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).She J; Guo J; Chen Q; Zeng W; Jiang Y; Bai XC Structural insights into the voltage and phospholipid activation of the mammalian TPC1 channel. Nature 2018, 556, 130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Shang G; Zhang C; Chen ZJ; Bai XC; Zhang X Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP. Nature 2019, 567, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Zhang C; Shang G; Gui X; Zhang X; Bai XC; Chen ZJ Structural basis of STING binding with and phosphorylation by TBK1. Nature 2019, 567, 394–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Yin J; Chen KM; Clark MJ; Hijazi M; Kumari P; Bai XC; Sunahara RK; Barth P; Rosenbaum DM Structure of a D2 dopamine receptor-G-protein complex in a lipid membrane. Nature 2020, 584, 125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Yu H; Bai XC; Wang W Characterization of the subunit composition and structure of adult human glycine receptors. Neuron 2021, 109, 2707–2716 e2706. [DOI] [PubMed] [Google Scholar]
- (49).Liao M; Cao E; Julius D; Cheng Y Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 2013, 504, 107–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Vinothkumar KR; Zhu J; Hirst J Architecture of mammalian respiratory complex I. Nature 2014, 515, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Zhu S; Noviello CM; Teng J; Walsh RM Jr.; Kim JJ; Hibbs RE Structure of a human synaptic GABAA receptor. Nature 2018, 559, 67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Cheng Y; Grigorieff N; Penczek PA; Walz T A primer to single-particle cryo-electron microscopy. Cell 2015, 161, 438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Cheng Y Single-particle cryo-EM at crystallographic resolution. Cell 2015, 161, 450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Efremov RG; Gatsogiannis C; Raunser S Lipid Nanodiscs as a Tool for High-Resolution Structure Determination of Membrane Proteins by Single-Particle Cryo-EM. Methods Enzymol. 2017, 594, 1–30. [DOI] [PubMed] [Google Scholar]
- (55).Carlson ML; Young JW; Zhao Z; Fabre L; Jun D; Li J; Li J; Dhupar HS; Wason I; Mills AT et al. The Peptidisc, a simple method for stabilizing membrane proteins in detergent-free solution. Elife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Wu S; Avila-Sakar A; Kim J; Booth DS; Greenberg CH; Rossi A; Liao M; Li X; Alian A; Griner SL et al. Fabs enable single particle cryoEM studies of small proteins. Structure 2012, 20, 582–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).D'Imprima E; Floris D; Joppe M; Sanchez R; Grininger M; Kuhlbrandt W Protein denaturation at the air-water interface and how to prevent it. Elife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Du Z; Lovly CM Mechanisms of receptor tyrosine kinase activation in cancer. Mol. Cancer 2018, 17, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Bennasroune A; Gardin A; Aunis D; Cremel G; Hubert P Tyrosine kinase receptors as attractive targets of cancer therapy. Crit Rev Oncol Hematol 2004, 50, 23–38. [DOI] [PubMed] [Google Scholar]
- (60).Regad T Targeting RTK Signaling pathways in cancer. Cancers (Basel) 2015, 7, 1758–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Ullrich A; Schlessinger J Signal transduction by receptors with tyrosine kinase activity. Cell 1990, 61, 203–212. [DOI] [PubMed] [Google Scholar]
- (62).Wacker D; Stevens RC; Roth BL How ligands illuminate GPCR molecular pharmacology. Cell 2017, 170, 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Wehrman T; He X; Raab B; Dukipatti A; Blau H; Garcia KC Structural and mechanistic insights into nerve growth factor interactions with the TrkA and p75 receptors. Neuron 2007, 53, 25–38. [DOI] [PubMed] [Google Scholar]
- (64).Shim AH; Liu H; Focia PJ; Chen X; Lin PC; He X Structures of a platelet-derived growth factor/propeptide complex and a platelet-derived growth factor/receptor complex. Proc. Natl. Acad. Sci. U S A 2010, 107, 11307–11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Leppanen VM; Saharinen P; Alitalo K Structural basis of Tie2 activation and Tie2/Tie1 heterodimerization. Proc. Natl. Acad. Sci. U S A 2017, 114, 4376–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Markovic-Mueller S; Stuttfeld E; Asthana M; Weinert T; Bliven S; Goldie KN; Kisko K; Capitani G; Ballmer-Hofer K Structure of the full-length VEGFR-1 extracellular domain in complex with VEGF-A. Structure 2017, 25, 341–352. [DOI] [PubMed] [Google Scholar]
- (67).Trenker R; Jura N Receptor tyrosine kinase activation: From the ligand perspective. Curr. Opin. Cell Biol 2020, 63, 174–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Hubbard SR; Till JH Protein tyrosine kinase structure and function. Annu. Rev. Biochem 2000, 69, 373–398. [DOI] [PubMed] [Google Scholar]
- (69).Haeusler RA; McGraw TE; Accili D Biochemical and cellular properties of insulin receptor signalling. Nat. Rev. Mol. Cell Biol 2018, 19, 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Malaguarnera R; Belfiore A The insulin receptor: a new target for cancer therapy. Front Endocrinol. (Lausanne) 2011, 2, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Olefsky JM The insulin receptor: its role in insulin resistance of obesity and diabetes. Diabetes 1976, 25, 1154–1162. [DOI] [PubMed] [Google Scholar]
- (72).McKern NM; Lawrence MC; Streltsov VA; Lou MZ; Adams TE; Lovrecz GO; Elleman TC; Richards KM; Bentley JD; Pilling PA et al. Structure of the insulin receptor ectodomain reveals a folded-over conformation. Nature 2006, 443, 218–221. [DOI] [PubMed] [Google Scholar]
- (73).Lee J; Pilch PF The insulin receptor: structure, function, and signaling. Am. J. Physiol 1994, 266, C319–334. [DOI] [PubMed] [Google Scholar]
- (74).de Meyts P; Roth J; Neville DM Jr.; Gavin JR 3rd; Lesniak MA Insulin interactions with its receptors: experimental evidence for negative cooperativity. Biochem. Biophys. Res. Commun 1973, 55, 154–161. [DOI] [PubMed] [Google Scholar]
- (75).Schaffer L A model for insulin binding to the insulin receptor. Eur. J. Biochem 1994, 221, 1127–1132. [DOI] [PubMed] [Google Scholar]
- (76).Gavin JR 3rd; Gorden P; Roth J; Archer JA; Buell DN Characteristics of the human lymphocyte insulin receptor. J. Biol. Chem 1973, 248, 2202–2207. [PubMed] [Google Scholar]
- (77).De Meyts P Insulin/receptor binding: the last piece of the puzzle? What recent progress on the structure of the insulin/receptor complex tells us (or not) about negative cooperativity and activation. Bioessays 2015, 37, 389–397. [DOI] [PubMed] [Google Scholar]
- (78).Kristensen C; Kjeldsen T; Wiberg FC; Schaffer L; Hach M; Havelund S; Bass J; Steiner DF; Andersen AS Alanine scanning mutagenesis of insulin. J. Biol. Chem 1997, 272, 12978–12983. [DOI] [PubMed] [Google Scholar]
- (79).Menting JG; Whittaker J; Margetts MB; Whittaker LJ; Kong GK; Smith BJ; Watson CJ; Zakova L; Kletvikova E; Jiracek J et al. How insulin engages its primary binding site on the insulin receptor. Nature 2013, 493, 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Weis F; Menting JG; Margetts MB; Chan SJ; Xu Y; Tennagels N; Wohlfart P; Langer T; Muller CW; Dreyer MK et al. The signalling conformation of the insulin receptor ectodomain. Nat. Commun 2018, 9, 4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Scapin G; Dandey VP; Zhang Z; Prosise W; Hruza A; Kelly T; Mayhood T; Strickland C; Potter CS; Carragher B Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis. Nature 2018, 556, 122–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Uchikawa E; Choi E; Shang G; Yu H; Bai XC Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex. Elife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Gutmann T; Schafer IB; Poojari C; Brankatschk B; Vattulainen I; Strauss M; Coskun U Cryo-EM structure of the complete and ligand-saturated insulin receptor ectodomain. J. Cell Biol 2020, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Kavran JM; McCabe JM; Byrne PO; Connacher MK; Wang Z; Ramek A; Sarabipour S; Shan Y; Shaw DE; Hristova K et al. How IGF-1 activates its receptor. Elife 2014, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Li J; Park J; Mayer JP; Webb KJ; Uchikawa E; Wu J; Liu S; Zhang X; Stowell MHB; Choi E et al. Synergistic activation of the insulin receptor via two distinct sites. Nat. Struct. Mol. Biol 2022, 29, 357–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Nielsen J; Brandt J; Boesen T; Hummelshoj T; Slaaby R; Schluckebier G; Nissen P Structural investigations of full-Length Insulin Receptor dynamics and signalling. J. Mol. Biol 2022, 434, 167458. [DOI] [PubMed] [Google Scholar]
- (87).Chitnis MM; Yuen JS; Protheroe AS; Pollak M; Macaulay VM The type 1 insulin-like growth factor receptor pathway. Clin. Cancer. Res 2008, 14, 6364–6370. [DOI] [PubMed] [Google Scholar]
- (88).Adams TE; Epa VC; Garrett TP; Ward CW Structure and function of the type 1 insulin-like growth factor receptor. Cell. Mol. Life Sci 2000, 57, 1050–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Kenyon C The first long-lived mutants: discovery of the insulin/IGF-1 pathway for ageing. Philos. Trans. R. Soc. Lond. B Biol. Sci 2011, 366, 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Ullrich A; Gray A; Tam AW; Yang-Feng T; Tsubokawa M; Collins C; Henzel W; Le Bon T; Kathuria S; Chen E et al. Insulin-like growth factor I receptor primary structure: comparison with insulin receptor suggests structural determinants that define functional specificity. EMBO J. 1986, 5, 2503–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Arcaro A Targeting the insulin-like growth factor-1 receptor in human cancer. Front. Pharmacol 2013, 4, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Riedemann J; Macaulay VM IGF1R signalling and its inhibition. Endocr. Relat. Cancer 2006, 13 Suppl 1, S33–43. [DOI] [PubMed] [Google Scholar]
- (93).De Meyts P The structural basis of insulin and insulin-like growth factor-I receptor binding and negative co-operativity, and its relevance to mitogenic versus metabolic signalling. Diabetologia 1994, 37 Suppl 2, S135–148. [DOI] [PubMed] [Google Scholar]
- (94).Li J; Choi E; Yu H; Bai XC Structural basis of the activation of type 1 insulin-like growth factor receptor. Nat. Commun 2019, 10, 4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Christoffersen CT; Bornfeldt KE; Rotella CM; Gonzales N; Vissing H; Shymko RM; ten Hoeve J; Groffen J; Heisterkamp N; De Meyts P Negative cooperativity in the insulin-like growth factor-I receptor and a chimeric IGF-I/insulin receptor. Endocrinology 1994, 135, 472–475. [DOI] [PubMed] [Google Scholar]
- (96).Garrett TP; McKern NM; Lou M; Frenkel MJ; Bentley JD; Lovrecz GO; Elleman TC; Cosgrove LJ; Ward CW Crystal structure of the first three domains of the type-1 insulin-like growth factor receptor. Nature 1998, 394, 395–399. [DOI] [PubMed] [Google Scholar]
- (97).Xu Y; Kirk NS; Venugopal H; Margetts MB; Croll TI; Sandow JJ; Webb AI; Delaine CA; Forbes BE; Lawrence MC How IGF-II binds to the human type 1 Insulin-like growth factor receptor. Structure 2020, 28, 786–798 e786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Takahashi M; Buma Y; Iwamoto T; Inaguma Y; Ikeda H; Hiai H Cloning and expression of the ret proto-oncogene encoding a tyrosine kinase with two potential transmembrane domains. Oncogene 1988, 3, 571–578. [PubMed] [Google Scholar]
- (99).Manie S; Santoro M; Fusco A; Billaud M The RET receptor: function in development and dysfunction in congenital malformation. Trends Genet. 2001, 17, 580–589. [DOI] [PubMed] [Google Scholar]
- (100).Lodish MB; Stratakis CA RET oncogene in MEN2, MEN2B, MTC and other forms of thyroid cancer. Expert Rev. Anticancer Ther 2008, 8, 625–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Mulligan LM RET revisited: expanding the oncogenic portfolio. Nat. Rev. Cancer 2014, 14, 173–186. [DOI] [PubMed] [Google Scholar]
- (102).Ibanez CF Structure and physiology of the RET receptor tyrosine kinase. Cold Spring Harb. Perspect. Biol 2013, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Amiel J; Sproat-Emison E; Garcia-Barcelo M; Lantieri F; Burzynski G; Borrego S; Pelet A; Arnold S; Miao X; Griseri P et al. Hirschsprung disease, associated syndromes and genetics: a review. J. Med. Genet 2008, 45, 1–14. [DOI] [PubMed] [Google Scholar]
- (104).Emison ES; McCallion AS; Kashuk CS; Bush RT; Grice E; Lin S; Portnoy ME; Cutler DJ; Green ED; Chakravarti A A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature 2005, 434, 857–863. [DOI] [PubMed] [Google Scholar]
- (105).Airaksinen MS; Titievsky A; Saarma M GDNF family neurotrophic factor signaling: four masters, one servant? Mol. Cell Neurosci 1999, 13, 313–325. [DOI] [PubMed] [Google Scholar]
- (106).Kim N; Stiegler AL; Cameron TO; Hallock PT; Gomez AM; Huang JH; Hubbard SR; Dustin ML; Burden SJ Lrp4 is a receptor for Agrin and forms a complex with MuSK. Cell 2008, 135, 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Zhang B; Luo S; Wang Q; Suzuki T; Xiong WC; Mei L LRP4 serves as a coreceptor of agrin. Neuron 2008, 60, 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Milbrandt J; de Sauvage FJ; Fahrner TJ; Baloh RH; Leitner ML; Tansey MG; Lampe PA; Heuckeroth RO; Kotzbauer PT; Simburger KS et al. Persephin, a novel neurotrophic factor related to GDNF and neurturin. Neuron 1998, 20, 245–253. [DOI] [PubMed] [Google Scholar]
- (109).Baloh RH; Tansey MG; Lampe PA; Fahrner TJ; Enomoto H; Simburger KS; Leitner ML; Araki T; Johnson EM Jr.; Milbrandt J Artemin, a novel member of the GDNF ligand family, supports peripheral and central neurons and signals through the GFRalpha3-RET receptor complex. Neuron 1998, 21, 1291–1302. [DOI] [PubMed] [Google Scholar]
- (110).Trupp M; Arenas E; Fainzilber M; Nilsson AS; Sieber BA; Grigoriou M; Kilkenny C; Salazar-Grueso E; Pachnis V; Arumae U Functional receptor for GDNF encoded by the c-ret proto-oncogene. Nature 1996, 381, 785–789. [DOI] [PubMed] [Google Scholar]
- (111).Kotzbauer PT; Lampe PA; Heuckeroth RO; Golden JP; Creedon DJ; Johnson EM Jr.; Milbrandt J Neurturin, a relative of glial-cell-line-derived neurotrophic factor. Nature 1996, 384, 467–470. [DOI] [PubMed] [Google Scholar]
- (112).Mullican SE; Lin-Schmidt X; Chin C-N; Chavez JA; Furman JL; Armstrong AA; Beck SC; South VJ; Dinh TQ; Cash-Mason TD et al. GFRAL is the receptor for GDF15 and the ligand promotes weight loss in mice and nonhuman primates. Nature Medicine 2017, 23, 1150–1157. [DOI] [PubMed] [Google Scholar]
- (113).Hsu J-Y; Crawley S; Chen M; Ayupova DA; Lindhout DA; Higbee J; Kutach A; Joo W; Gao Z; Fu D et al. Non-homeostatic body weight regulation through a brainstem-restricted receptor for GDF15. Nature 2017, 550, 255–259. [DOI] [PubMed] [Google Scholar]
- (114).Emmerson PJ; Wang F; Du Y; Liu Q; Pickard RT; Gonciarz MD; Coskun T; Hamang MJ; Sindelar DK; Ballman KK et al. The metabolic effects of GDF15 are mediated by the orphan receptor GFRAL. Nat. Med 2017, 23, 1215–1219. [DOI] [PubMed] [Google Scholar]
- (115).Yang L; Chang C-C; Sun Z; Madsen D; Zhu H; Padkjær SB; Wu X; Huang T; Hultman K; Paulsen SJ et al. GFRAL is the receptor for GDF15 and is required for the anti-obesity effects of the ligand. Nature Medicine 2017, 23, 1158–1166. [DOI] [PubMed] [Google Scholar]
- (116).Emmerson PJ; Duffin KL; Chintharlapalli S; Wu X GDF15 and growth control. Front. Physiol 2018, 9, 1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).Wang D; Day EA; Townsend LK; Djordjevic D; Jorgensen SB; Steinberg GR GDF15: emerging biology and therapeutic applications for obesity and cardiometabolic disease. Nat. Rev. Endocrinol 2021, 17, 592–607. [DOI] [PubMed] [Google Scholar]
- (118).Goodman KM; Kjaer S; Beuron F; Knowles PP; Nawrotek A; Burns EM; Purkiss AG; George R; Santoro M; Morris EP et al. RET recognition of GDNF-GFRalpha1 ligand by a composite binding site promotes membrane-proximal self-association. Cell Rep. 2014, 8, 1894–1904. [DOI] [PubMed] [Google Scholar]
- (119).Amoresano A; Incoronato M; Monti G; Pucci P; de Franciscis V; Cerchia L Direct interactions among Ret, GDNF and GFRα1 molecules reveal new insights into the assembly of a functional three-protein complex. Cellular Signalling 2005, 17, 717–727. [DOI] [PubMed] [Google Scholar]
- (120).Kjaer S; Hanrahan S; Totty N; McDonald NQ Mammal-restricted elements predispose human RET to folding impairment by HSCR mutations. Nat. Struct. Mol. Biol 2010, 17, 726–731. [DOI] [PubMed] [Google Scholar]
- (121).Li J; Shang G; Chen YJ; Brautigam CA; Liou J; Zhang X; Bai XC Cryo-EM analyses reveal the common mechanism and diversification in the activation of RET by different ligands. Elife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (122).Bigalke JM; Aibara S; Roth R; Dahl G; Gordon E; Dorbeus S; Amunts A; Sandmark J Cryo-EM structure of the activated RET signaling complex reveals the importance of its cysteine-rich domain. Sci. Adv 2019, 5, eaau4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (123).Adams SE; Purkiss AG; Knowles PP; Nans A; Briggs DC; Borg A; Earl CP; Goodman KM; Nawrotek A; Borg AJ et al. A two-site flexible clamp mechanism for RET-GDNF-GFRalpha1 assembly reveals both conformational adaptation and strict geometric spacing. Structure 2021, 29, 694–708 e697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (124).Zhou M; Li Y; Hu Q; Bai XC; Huang W; Yan C; Scheres SH; Shi Y Atomic structure of the apoptosome: mechanism of cytochrome c- and dATP-mediated activation of Apaf-1. Genes Dev. 2015, 29, 2349–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (125).Scheres SH Processing of Structurally Heterogeneous Cryo-EM Data in RELION. Methods Enzymol. 2016, 579, 125–157. [DOI] [PubMed] [Google Scholar]
- (126).Richardson DS; Lai AZ; Mulligan LM RET ligand-induced internalization and its consequences for downstream signaling. Oncogene 2006, 25, 3206–3211. [DOI] [PubMed] [Google Scholar]
- (127).Organ SL; Tsao MS An overview of the c-MET signaling pathway. Ther. Adv. Med. Oncol 2011, 3, S7–S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (128).Park M; Dean M; Cooper CS; Schmidt M; O'Brien SJ; Blair DG; Vande Woude GF Mechanism of met oncogene activation. Cell 1986, 45, 895–904. [DOI] [PubMed] [Google Scholar]
- (129).Birchmeier C; Birchmeier W; Gherardi E; Vande Woude GF Met, metastasis, motility and more. Nat. Rev. Mol. Cell Biol 2003, 4, 915–925. [DOI] [PubMed] [Google Scholar]
- (130).Jung KH; Park BH; Hong SS Progress in cancer therapy targeting c-Met signaling pathway. Arch. Pharm. Res 2012, 35, 595–604. [DOI] [PubMed] [Google Scholar]
- (131).Eder JP; Vande Woude GF; Boerner SA; LoRusso PM Novel therapeutic inhibitors of the c-Met signaling pathway in cancer. Clin. Cancer. Res 2009, 15, 2207–2214. [DOI] [PubMed] [Google Scholar]
- (132).Liu X; Yao W; Newton RC; Scherle PA Targeting the c-MET signaling pathway for cancer therapy. Expert Opin. Investig. Drugs 2008, 17, 997–1011. [DOI] [PubMed] [Google Scholar]
- (133).Peruzzi B; Bottaro DP Targeting the c-Met signaling pathway in cancer. Clin. Cancer. Res 2006, 12, 3657–3660. [DOI] [PubMed] [Google Scholar]
- (134).Comoglio PM; Giordano S; Trusolino L Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nat. Rev. Drug Discov 2008, 7, 504–516. [DOI] [PubMed] [Google Scholar]
- (135).Cioce V; Csaky KG; Chan AM; Bottaro DP; Taylor WG; Jensen R; Aaronson SA; Rubin JS Hepatocyte growth factor (HGF)/NK1 is a naturally occurring HGF/scatter factor variant with partial agonist/antagonist activity. J. Biol. Chem 1996, 271, 13110–13115. [DOI] [PubMed] [Google Scholar]
- (136).Weidner KM; Arakaki N; Hartmann G; Vandekerckhove J; Weingart S; Rieder H; Fonatsch C; Tsubouchi H; Hishida T; Daikuhara Y et al. Evidence for the identity of human scatter factor and human hepatocyte growth factor. Proc. Natl. Acad. Sci. U S A 1991, 88, 7001–7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (137).Trusolino L; Comoglio PM Scatter-factor and semaphorin receptors: cell signalling for invasive growth. Nat. Rev. Cancer 2002, 2, 289–300. [DOI] [PubMed] [Google Scholar]
- (138).Nakamura T; Nishizawa T; Hagiya M; Seki T; Shimonishi M; Sugimura A; Tashiro K; Shimizu S Molecular cloning and expression of human hepatocyte growth factor. Nature 1989, 342, 440–443. [DOI] [PubMed] [Google Scholar]
- (139).Stamos J; Lazarus RA; Yao X; Kirchhofer D; Wiesmann C Crystal structure of the HGF beta-chain in complex with the Sema domain of the Met receptor. EMBO J. 2004, 23, 2325–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (140).Rubin JS; Day RM; Breckenridge D; Atabey N; Taylor WG; Stahl SJ; Wingfield PT; Kaufman JD; Schwall R; Bottaro DP Dissociation of heparan sulfate and receptor binding domains of hepatocyte growth factor reveals that heparan sulfate-c-met interaction facilitates signaling. J. Biol. Chem 2001, 276, 32977–32983. [DOI] [PubMed] [Google Scholar]
- (141).Sakata H; Stahl SJ; Taylor WG; Rosenberg JM; Sakaguchi K; Wingfield PT; Rubin JS Heparin binding and oligomerization of hepatocyte growth factor/scatter factor isoforms. Heparan sulfate glycosaminoglycan requirement for Met binding and signaling. J. Biol. Chem 1997, 272, 9457–9463. [DOI] [PubMed] [Google Scholar]
- (142).Zioncheck TF; Richardson L; Liu J; Chang L; King KL; Bennett GL; Fugedi P; Chamow SM; Schwall RH; Stack RJ Sulfated oligosaccharides promote hepatocyte growth factor association and govern its mitogenic activity. J. Biol. Chem 1995, 270, 16871–16878. [DOI] [PubMed] [Google Scholar]
- (143).Uchikawa E; Chen Z; Xiao GY; Zhang X; Bai XC Structural basis of the activation of c-MET receptor. Nat. Commun 2021, 12, 4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (144).Niemann HH; Jager V; Butler PJ; van den Heuvel J; Schmidt S; Ferraris D; Gherardi E; Heinz DW Structure of the human receptor tyrosine kinase met in complex with the Listeria invasion protein InlB. Cell 2007, 130, 235–246. [DOI] [PubMed] [Google Scholar]
- (145).Tolbert WD; Daugherty-Holtrop J; Gherardi E; Vande Woude G; Xu HE Structural basis for agonism and antagonism of hepatocyte growth factor. Proc. Natl. Acad. Sci. U S A 2010, 107, 13264–13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (146).Kovacs E; Zorn JA; Huang Y; Barros T; Kuriyan J A structural perspective on the regulation of the epidermal growth factor receptor. Annu. Rev. Biochem 2015, 84, 739–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (147).Wieduwilt MJ; Moasser MM The epidermal growth factor receptor family: biology driving targeted therapeutics. Cell. Mol. Life Sci 2008, 65, 1566–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (148).Sigismund S; Avanzato D; Lanzetti L Emerging functions of the EGFR in cancer. Mol. Oncol 2018, 12, 3–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (149).Ciardiello F; Tortora G EGFR antagonists in cancer treatment. N. Engl. J. Med 2008, 358, 1160–1174. [DOI] [PubMed] [Google Scholar]
- (150).Herbst RS Review of epidermal growth factor receptor biology. Int. J. Radiat. Oncol. Biol. Phys 2004, 59, 21–26. [DOI] [PubMed] [Google Scholar]
- (151).Singh B; Carpenter G; Coffey RJ EGF receptor ligands: recent advances. F1000Res 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (152).Freed DM; Bessman NJ; Kiyatkin A; Salazar-Cavazos E; Byrne PO; Moore JO; Valley CC; Ferguson KM; Leahy DJ; Lidke DS et al. EGFR ligands differentially stabilize receptor dimers to specify signaling kinetics. Cell 2017, 171, 683–695 e618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (153).Ogiso H; Ishitani R; Nureki O; Fukai S; Yamanaka M; Kim JH; Saito K; Sakamoto A; Inoue M; Shirouzu M et al. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell 2002, 110, 775–787. [DOI] [PubMed] [Google Scholar]
- (154).Dawson JP; Berger MB; Lin CC; Schlessinger J; Lemmon MA; Ferguson KM Epidermal growth factor receptor dimerization and activation require ligand-induced conformational changes in the dimer interface. Mol. Cell Biol 2005, 25, 7734–7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (155).Ferguson KM; Berger MB; Mendrola JM; Cho HS; Leahy DJ; Lemmon MA EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol. Cell 2003, 11, 507–517. [DOI] [PubMed] [Google Scholar]
- (156).Zhang X; Gureasko J; Shen K; Cole PA; Kuriyan J An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 2006, 125, 1137–1149. [DOI] [PubMed] [Google Scholar]
- (157).Bocharov EV; Lesovoy DM; Pavlov KV; Pustovalova YE; Bocharova OV; Arseniev AS Alternative packing of EGFR transmembrane domain suggests that protein-lipid interactions underlie signal conduction across membrane. Biochim. Biophys. Acta 2016, 1858, 1254–1261. [DOI] [PubMed] [Google Scholar]
- (158).Bocharov EV; Bragin PE; Pavlov KV; Bocharova OV; Mineev KS; Polyansky AA; Volynsky PE; Efremov RG; Arseniev AS The conformation of the epidermal growth factor receptor transmembrane domain dimer dynamically adapts to the local membrane environment. Biochemistry 2017, 56, 1697–1705. [DOI] [PubMed] [Google Scholar]
- (159).Huang Y; Ognjenovic J; Karandur D; Miller K; Merk A; Subramaniam S; Kuriyan J A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor. Elife 2021, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (160).Denisov IG; Sligar SG Nanodiscs in membrane biochemistry and biophysics. Chem. Rev. 2017, 117, 4669–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (161).Baselga J; Swain SM Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat. Rev. Cancer 2009, 9, 463–475. [DOI] [PubMed] [Google Scholar]
- (162).Hynes NE; MacDonald G ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol 2009, 21, 177–184. [DOI] [PubMed] [Google Scholar]
- (163).Yarden Y; Pines G The ERBB network: at last, cancer therapy meets systems biology. Nat. Rev. Cancer 2012, 12, 553–563. [DOI] [PubMed] [Google Scholar]
- (164).Jura N; Shan Y; Cao X; Shaw DE; Kuriyan J Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc. Natl. Acad. Sci. U S A 2009, 106, 21608–21613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (165).Sierke SL; Cheng K; Kim HH; Koland JG Biochemical characterization of the protein tyrosine kinase homology domain of the ErbB3 (HER3) receptor protein. Biochem. J 1997, 322 (Pt 3), 757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (166).Littlefield P; Liu L; Mysore V; Shan Y; Shaw DE; Jura N Structural analysis of the EGFR/HER3 heterodimer reveals the molecular basis for activating HER3 mutations. Sci. Signal 2014, 7, ra114. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- (167).van Lengerich B; Agnew C; Puchner EM; Huang B; Jura N EGF and NRG induce phosphorylation of HER3/ERBB3 by EGFR using distinct oligomeric mechanisms. Proc. Natl. Acad. Sci. U S A 2017, 114, E2836–E2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (168).Cho HS; Leahy DJ Structure of the extracellular region of HER3 reveals an interdomain tether. Science 2002, 297, 1330–1333. [DOI] [PubMed] [Google Scholar]
- (169).Cho HS; Mason K; Ramyar KX; Stanley AM; Gabelli SB; Denney DW Jr.; Leahy DJ Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature 2003, 421, 756–760. [DOI] [PubMed] [Google Scholar]
- (170).Hao Y; Yu X; Bai Y; McBride HJ; Huang X Cryo-EM Structure of HER2-trastuzumab-pertuzumab complex. PLoS. One 2019, 14, e0216095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (171).Diwanji D; Trenker R; Thaker TM; Wang F; Agard DA; Verba KA; Jura N Structures of the HER2–HER3–NRG1β complex reveal a dynamic dimer interface. Nature 2021, DOI: 10.1038/s41586-021-04084-z 10.1038/s41586-021-04084-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (172).Palovcak E; Wang F; Zheng SQ; Yu Z; Li S; Betegon M; Bulkley D; Agard DA; Cheng Y A simple and robust procedure for preparing graphene-oxide cryo-EM grids. J. Struct. Biol 2018, 204, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (173).Hota PK; Buck M Plexin structures are coming: opportunities for multilevel investigations of semaphorin guidance receptors, their cell signaling mechanisms, and functions. Cell Mol. Life Sci 2012, 69, 3765–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (174).Tran TS; Kolodkin AL; Bharadwaj R Semaphorin regulation of cellular morphology. Annu. Rev. Cell Dev. Biol 2007, 23, 263–292. [DOI] [PubMed] [Google Scholar]
- (175).Takahashi T; Fournier A; Nakamura F; Wang LH; Murakami Y; Kalb RG; Fujisawa H; Strittmatter SM Plexin-neuropilin-1 complexes form functional semaphorin-3A receptors. Cell 1999, 99, 59–69. [DOI] [PubMed] [Google Scholar]
- (176).He Z; Tessier-Lavigne M Neuropilin is a receptor for the axonal chemorepellent Semaphorin III. Cell 1997, 90, 739–751. [DOI] [PubMed] [Google Scholar]
- (177).Kolodkin AL; Levengood DV; Rowe EG; Tai YT; Giger RJ; Ginty DD Neuropilin is a semaphorin III receptor. Cell 1997, 90, 753–762. [DOI] [PubMed] [Google Scholar]
- (178).Yoo SK; Pascoe HG; Pereira T; Kondo S; Jacinto A; Zhang X; Hariharan IK Plexins function in epithelial repair in both Drosophila and zebrafish. Nat. Commun 2016, 7, 12282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (179).Antipenko A; Himanen JP; van Leyen K; Nardi-Dei V; Lesniak J; Barton WA; Rajashankar KR; Lu M; Hoemme C; Puschel AW et al. Structure of the semaphorin-3A receptor binding module. Neuron 2003, 39, 589–598. [DOI] [PubMed] [Google Scholar]
- (180).Love CA; Harlos K; Mavaddat N; Davis SJ; Stuart DI; Jones EY; Esnouf RM The ligand-binding face of the semaphorins revealed by the high-resolution crystal structure of SEMA4D. Nat. Struct. Biol 2003, 10, 843–848. [DOI] [PubMed] [Google Scholar]
- (181).Janssen BJ; Malinauskas T; Weir GA; Cader MZ; Siebold C; Jones EY Neuropilins lock secreted semaphorins onto plexins in a ternary signaling complex. Nat. Struct. Mol. Biol 2012, 19, 1293–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (182).Klostermann A; Lohrum M; Adams RH; Puschel AW The chemorepulsive activity of the axonal guidance signal semaphorin D requires dimerization. J. Biol. Chem 1998, 273, 7326–7331. [DOI] [PubMed] [Google Scholar]
- (183).Koppel AM; Raper JA Collapsin-1 covalently dimerizes, and dimerization is necessary for collapsing activity. J. Biol. Chem 1998, 273, 15708–15713. [DOI] [PubMed] [Google Scholar]
- (184).Adams RH; Lohrum M; Klostermann A; Betz H; Puschel AW The chemorepulsive activity of secreted semaphorins is regulated by furin-dependent proteolytic processing. EMBO J. 1997, 16, 6077–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (185).Parker MW; Hellman LM; Xu P; Fried MG; Vander Kooi CW Furin processing of semaphorin 3F determines its anti-angiogenic activity by regulating direct binding and competition for neuropilin. Biochemistry 2010, 49, 4068–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (186).Kong Y; Janssen BJ; Malinauskas T; Vangoor VR; Coles CH; Kaufmann R; Ni T; Gilbert RJ; Padilla-Parra S; Pasterkamp RJ et al. Structural Basis for Plexin Activation and Regulation. Neuron 2016, 91, 548–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (187).Lu D; Shang G; He X; Bai XC; Zhang X Architecture of the Sema3A/PlexinA4/Neuropilin tripartite complex. Nat. Commun 2021, 12, 3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (188).Nogi T; Yasui N; Mihara E; Matsunaga Y; Noda M; Yamashita N; Toyofuku T; Uchiyama S; Goshima Y; Kumanogoh A et al. Structural basis for semaphorin signalling through the plexin receptor. Nature 2010, 467, 1123–1127. [DOI] [PubMed] [Google Scholar]
- (189).Liu H; Juo ZS; Shim AH; Focia PJ; Chen X; Garcia KC; He X Structural basis of semaphorin-plexin recognition and viral mimicry from Sema7A and A39R complexes with PlexinC1. Cell 2010, 142, 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (190).Janssen BJ; Robinson RA; Perez-Branguli F; Bell CH; Mitchell KJ; Siebold C; Jones EY Structural basis of semaphorin-plexin signalling. Nature 2010, 467, 1118–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (191).Kuo YC; Chen H; Shang G; Uchikawa E; Tian H; Bai XC; Zhang X Cryo-EM structure of the PlexinC1/A39R complex reveals inter-domain interactions critical for ligand-induced activation. Nat. Commun 2020, 11, 1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (192).Wang Y; Pascoe HG; Brautigam CA; He H; Zhang X Structural basis for activation and non-canonical catalysis of the Rap GTPase activating protein domain of plexin. Elife 2013, 2, e01279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (193).He H; Yang T; Terman JR; Zhang X Crystal structure of the plexin A3 intracellular region reveals an autoinhibited conformation through active site sequestration. Proc. Natl. Acad. Sci. U S A 2009, 106, 15610–15615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (194).Tong Y; Hota PK; Penachioni JY; Hamaneh MB; Kim S; Alviani RS; Shen L; He H; Tempel W; Tamagnone L et al. Structure and function of the intracellular region of the plexin-b1 transmembrane receptor. J. Biol. Chem 2009, 284, 35962–35972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (195).Bell CH; Aricescu AR; Jones EY; Siebold C A dual binding mode for RhoGTPases in plexin signalling. PLoS biology 2011, 9, e1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (196).Tong Y; Chugha P; Hota PK; Alviani RS; Li M; Tempel W; Shen L; Park HW; Buck M Binding of Rac1, Rnd1, and RhoD to a novel Rho GTPase interaction motif destabilizes dimerization of the plexin-B1 effector domain. J. Biol. Chem 2007, 282, 37215–37224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (197).Liu Y; Ke P; Kuo YC; Wang Y; Zhang X; Song C; Shan Y A putative structural mechanism underlying the antithetic effect of homologous RND1 and RhoD GTPases in mammalian plexin regulation. Elife 2021, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (198).Pascoe HG; Gutowski S; Chen H; Brautigam CA; Chen Z; Sternweis PC; Zhang X Secondary PDZ domain-binding site on class B plexins enhances the affinity for PDZ-RhoGEF. Proc. Natl. Acad. Sci. U S A 2015, 112, 14852–14857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (199).Appleton BA; Wu P; Maloney J; Yin J; Liang WC; Stawicki S; Mortara K; Bowman KK; Elliott JM; Desmarais W et al. Structural studies of neuropilin/antibody complexes provide insights into semaphorin and VEGF binding. Embo J. 2007, 26, 4902–4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (200).Gaboriaud C; Gregory-Pauron L; Teillet F; Thielens NM; Bally I; Arlaud GJ Structure and properties of the Ca(2+)-binding CUB domain, a widespread ligand-recognition unit involved in major biological functions. Biochem J. 2011, 439, 185–193. [DOI] [PubMed] [Google Scholar]
- (201).Lee CC; Kreusch A; McMullan D; Ng K; Spraggon G Crystal structure of the human neuropilin-1 b1 domain. Structure 2003, 11, 99–108. [DOI] [PubMed] [Google Scholar]
- (202).Vander Kooi CW; Jusino MA; Perman B; Neau DB; Bellamy HD; Leahy DJ Structural basis for ligand and heparin binding to neuropilin B domains. Proc. Natl. Acad. Sci. U S A 2007, 104, 6152–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (203).Yelland T; Djordjevic S Crystal Structure of the Neuropilin-1 MAM Domain: Completing the Neuropilin-1 Ectodomain Picture. Structure 2016, 24, 2008–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (204).Parker MW; Linkugel AD; Vander Kooi CW Effect of C-terminal sequence on competitive semaphorin binding to neuropilin-1. J. Mol. Biol 2013, 425, 4405–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (205).Cai H; Reed RR Cloning and characterization of neuropilin-1-interacting protein: a PSD-95/Dlg/ZO-1 domain-containing protein that interacts with the cytoplasmic domain of neuropilin-1. J. Neurosci 1999, 19, 6519–6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (206).Salikhova A; Wang L; Lanahan AA; Liu M; Simons M; Leenders WP; Mukhopadhyay D; Horowitz A Vascular endothelial growth factor and semaphorin induce neuropilin-1 endocytosis via separate pathways. Circ. Res 2008, 103, e71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (207).Valdembri D; Caswell PT; Anderson KI; Schwarz JP; Konig I; Astanina E; Caccavari F; Norman JC; Humphries MJ; Bussolino F et al. Neuropilin-1/GIPC1 signaling regulates alpha5beta1 integrin traffic and function in endothelial cells. PLoS biology 2009, 7, e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (208).Shang G; Brautigam CA; Chen R; Lu D; Torres-Vazquez J; Zhang X Structure analyses reveal a regulated oligomerization mechanism of the PlexinD1/GIPC/myosin VI complex. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (209).Pascoe HG; Wang Y; Zhang X Structural mechanisms of plexin signaling. Prog. Biophys. Mol. Biol 2015, 118, 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (210).Seiradake E; Jones EY; Klein R Structural perspectives on axon guidance. Annu. Rev. Cell Dev. Biol 2016, 32, 577–608. [DOI] [PubMed] [Google Scholar]
- (211).Rozbesky D; Robinson RA; Jain V; Renner M; Malinauskas T; Harlos K; Siebold C; Jones EY Diversity of oligomerization in Drosophila semaphorins suggests a mechanism of functional fine-tuning. Nat. Commun 2019, 10, 3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (212).Rozbesky D; Verhagen MG; Karia D; Nagy GN; Alvarez L; Robinson RA; Harlos K; Padilla-Parra S; Pasterkamp RJ; Jones EY Structural basis of semaphorin-plexin cis interaction. EMBO J. 2020, 39, e102926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (213).Wang Y; He H; Srivastava N; Vikarunnessa S; Chen YB; Jiang J; Cowan CW; Zhang X Plexins are GTPase-activating proteins for Rap and are activated by induced dimerization. Sci. Signal 2012, 5, ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (214).Harburger DS; Calderwood DA Integrin signalling at a glance. J. Cell Sci 2009, 122, 159–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (215).Shattil SJ; Kim C; Ginsberg MH The final steps of integrin activation: the end game. Nat. Rev. Mol. Cell Biol 2010, 11, 288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (216).Abram CL; Lowell CA The ins and outs of leukocyte integrin signaling. Annu. Rev. Immunol 2009, 27, 339–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (217).Bokel C; Brown NH Integrins in development: moving on, responding to, and sticking to the extracellular matrix. Dev. Cell 2002, 3, 311–321. [DOI] [PubMed] [Google Scholar]
- (218).Barczyk M; Carracedo S; Gullberg D Integrins. Cell Tissue Res. 2010, 339, 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (219).Hynes RO Integrins: bidirectional, allosteric signaling machines. Cell 2002, 110, 673–687. [DOI] [PubMed] [Google Scholar]
- (220).Giancotti FG; Ruoslahti E Integrin signaling. Science 1999, 285, 1028–1032. [DOI] [PubMed] [Google Scholar]
- (221).Kechagia JZ; Ivaska J; Roca-Cusachs P Integrins as biomechanical sensors of the microenvironment. Nat. Rev. Mol. Cell Biol 2019, 20, 457–473. [DOI] [PubMed] [Google Scholar]
- (222).Humphries JD; Byron A; Humphries MJ Integrin ligands at a glance. J. Cell Sci 2006, 119, 3901–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (223).Luo BH; Carman CV; Springer TA Structural basis of integrin regulation and signaling. Annu. Rev. Immunol 2007, 25, 619–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (224).Takada Y; Ye X; Simon S The integrins. Genome Biol. 2007, 8, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (225).Campbell ID; Humphries MJ Integrin structure, activation, and interactions. Cold Spring Harb. Perspect. Biol 2011, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (226).Askari JA; Buckley PA; Mould AP; Humphries MJ Linking integrin conformation to function. J. Cell Sci 2009, 122, 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (227).Arnaout MA; Goodman SL; Xiong JP Structure and mechanics of integrin-based cell adhesion. Curr. Opin. Cell Biol 2007, 19, 495–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (228).Takagi J Structural basis for ligand recognition by integrins. Curr. Opin. Cell Biol 2007, 19, 557–564. [DOI] [PubMed] [Google Scholar]
- (229).Xiong JP; Mahalingham B; Alonso JL; Borrelli LA; Rui X; Anand S; Hyman BT; Rysiok T; Muller-Pompalla D; Goodman SL et al. Crystal structure of the complete integrin alphaVbeta3 ectodomain plus an alpha/beta transmembrane fragment. J. Cell Biol 2009, 186, 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (230).Zhu J; Luo BH; Xiao T; Zhang C; Nishida N; Springer TA Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol. Cell 2008, 32, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (231).Zhu J; Zhu J; Springer TA Complete integrin headpiece opening in eight steps. J. Cell Biol 2013, 201, 1053–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (232).Xiao T; Takagi J; Coller BS; Wang JH; Springer TA Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature 2004, 432, 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (233).Takagi J; Petre BM; Walz T; Springer TA Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell 2002, 110, 599–511. [DOI] [PubMed] [Google Scholar]
- (234).Xiong JP; Stehle T; Diefenbach B; Zhang R; Dunker R; Scott DL; Joachimiak A; Goodman SL; Arnaout MA Crystal structure of the extracellular segment of integrin alpha Vbeta3. Science 2001, 294, 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (235).Van Agthoven JF; Xiong JP; Alonso JL; Rui X; Adair BD; Goodman SL; Arnaout MA Structural basis for pure antagonism of integrin alphaVbeta3 by a high-affinity form of fibronectin. Nat. Struct. Mol. Biol 2014, 21, 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (236).Sen M; Yuki K; Springer TA An internal ligand-bound, metastable state of a leukocyte integrin, alphaXbeta2. J. Cell Biol 2013, 203, 629–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (237).Xie C; Zhu J; Chen X; Mi L; Nishida N; Springer TA Structure of an integrin with an alphaI domain, complement receptor type 4. EMBO J. 2010, 29, 666–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (238).Su Y; Xia W; Li J; Walz T; Humphries MJ; Vestweber D; Cabanas C; Lu C; Springer TA Relating conformation to function in integrin alpha5beta1. Proc. Natl. Acad. Sci. U S A 2016, 113, E3872–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (239).Schumacher S; Dedden D; Nunez RV; Matoba K; Takagi J; Biertumpfel C; Mizuno N Structural insights into integrin alpha5beta1 opening by fibronectin ligand. Sci. Adv 2021, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (240).Campbell MG; Cormier A; Ito S; Seed RI; Bondesson AJ; Lou J; Marks JD; Baron JL; Cheng Y; Nishimura SL Cryo-EM reveals Integrin-mediated TGF-beta activation without release from latent TGF-beta. Cell 2020, 180, 490–501 e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (241).Cormier A; Campbell MG; Ito S; Wu S; Lou J; Marks J; Baron JL; Nishimura SL; Cheng Y Cryo-EM structure of the alphavbeta8 integrin reveals a mechanism for stabilizing integrin extension. Nat. Struct. Mol. Biol 2018, 25, 698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (242).Ozawa A; Sato Y; Imabayashi T; Uemura T; Takagi J; Sekiguchi K Molecular basis of the ligand binding specificity of alphavbeta8 Integrin. J. Biol. Chem 2016, 291, 11551–11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (243).Shi M; Zhu J; Wang R; Chen X; Mi L; Walz T; Springer TA Latent TGF-beta structure and activation. Nature 2011, 474, 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (244).Arnold TD; Niaudet C; Pang MF; Siegenthaler J; Gaengel K; Jung B; Ferrero GM; Mukouyama YS; Fuxe J; Akhurst R et al. Excessive vascular sprouting underlies cerebral hemorrhage in mice lacking alphaVbeta8-TGFbeta signaling in the brain. Development 2014, 141, 4489–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (245).Qin Y; Garrison BS; Ma W; Wang R; Jiang A; Li J; Mistry M; Bronson RT; Santoro D; Franco C et al. A Milieu Molecule for TGF-beta Required for Microglia Function in the Nervous System. Cell 2018, 174, 156–171 e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (246).Dong X; Zhao B; Iacob RE; Zhu J; Koksal AC; Lu C; Engen JR; Springer TA Force interacts with macromolecular structure in activation of TGF-beta. Nature 2017, 542, 55–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (247).Wang J; Dong X; Zhao B; Li J; Lu C; Springer TA Atypical interactions of integrin alphaVbeta8 with pro-TGF-beta1. Proc Natl Acad Sci U S A 2017, 114, E4168–E4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (248).Erickson HP; Carrell N; McDonagh J Fibronectin molecule visualized in electron microscopy: a long, thin, flexible strand. J. Cell Biol 1981, 91, 673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (249).Engel J; Odermatt E; Engel A; Madri JA; Furthmayr H; Rohde H; Timpl R Shapes, domain organizations and flexibility of laminin and fibronectin, two multifunctional proteins of the extracellular matrix. J. Mol. Biol 1981, 150, 97–120. [DOI] [PubMed] [Google Scholar]
- (250).Sechler JL; Corbett SA; Schwarzbauer JE Modulatory roles for integrin activation and the synergy site of fibronectin during matrix assembly. Mol. Biol. Cell 1997, 8, 2563–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (251).Miyazaki N; Iwasaki K; Takagi J A systematic survey of conformational states in beta1 and beta4 integrins using negative-stain electron microscopy. J. Cell Sci 2018, 131. [DOI] [PubMed] [Google Scholar]
- (252).Takagi J; Strokovich K; Springer TA; Walz T Structure of integrin alpha5beta1 in complex with fibronectin. EMBO J. 2003, 22, 4607–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (253).Byron A; Humphries JD; Askari JA; Craig SE; Mould AP; Humphries MJ Anti-integrin monoclonal antibodies. J. Cell Sci 2009, 122, 4009–4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (254).Kawasaki T; Kawai T Toll-like receptor signaling pathways. Front Immunol. 2014, 5, 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (255).Medzhitov R Toll-like receptors and innate immunity. Nat. Rev. Immunol 2001, 1, 135–145. [DOI] [PubMed] [Google Scholar]
- (256).Nie L; Cai SY; Shao JZ; Chen J Toll-Like Receptors, Associated Biological Roles, and Signaling Networks in Non-Mammals. Front. Immunol 2018, 9, 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (257).Palm NW; Medzhitov R Pattern recognition receptors and control of adaptive immunity. Immunol. Rev 2009, 227, 221–233. [DOI] [PubMed] [Google Scholar]
- (258).Bell JK; Mullen GE; Leifer CA; Mazzoni A; Davies DR; Segal DM Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. 2003, 24, 528–533. [DOI] [PubMed] [Google Scholar]
- (259).Xu Y; Tao X; Shen B; Horng T; Medzhitov R; Manley JL; Tong L Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature 2000, 408, 111–115. [DOI] [PubMed] [Google Scholar]
- (260).Jin MS; Kim SE; Heo JY; Lee ME; Kim HM; Paik SG; Lee H; Lee JO Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 2007, 130, 1071–1082. [DOI] [PubMed] [Google Scholar]
- (261).Choe J; Kelker MS; Wilson IA Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science 2005, 309, 581–585. [DOI] [PubMed] [Google Scholar]
- (262).Kang JY; Lee JO Structural biology of the Toll-like receptor family. Annu. Rev. Biochem 2011, 80, 917–941. [DOI] [PubMed] [Google Scholar]
- (263).Jin MS; Lee JO Structures of the toll-like receptor family and its ligand complexes. Immunity 2008, 29, 182–191. [DOI] [PubMed] [Google Scholar]
- (264).Song DH; Lee JO Sensing of microbial molecular patterns by Toll-like receptors. Immunol. Rev 2012, 250, 216–229. [DOI] [PubMed] [Google Scholar]
- (265).Chang ZL Important aspects of Toll-like receptors, ligands and their signaling pathways. Inflamm. Res 2010, 59, 791–808. [DOI] [PubMed] [Google Scholar]
- (266).Trivedi S; Greidinger EL Endosomal Toll-like receptors in autoimmunity: mechanisms for clinical diversity. Therapy 2009, 6, 433–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (267).Ohto U Conservation and Divergence of Ligand Recognition and Signal Transduction Mechanisms in Toll-Like Receptors. Chem. Pharm. Bull. (Tokyo) 2017, 65, 697–705. [DOI] [PubMed] [Google Scholar]
- (268).Alexopoulou L; Holt AC; Medzhitov R; Flavell RA Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 2001, 413, 732–738. [DOI] [PubMed] [Google Scholar]
- (269).Heil F; Hemmi H; Hochrein H; Ampenberger F; Kirschning C; Akira S; Lipford G; Wagner H; Bauer S Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 2004, 303, 1526–1529. [DOI] [PubMed] [Google Scholar]
- (270).Diebold SS; Kaisho T; Hemmi H; Akira S; Reis e Sousa C Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 2004, 303, 1529–1531. [DOI] [PubMed] [Google Scholar]
- (271).Lund JM; Alexopoulou L; Sato A; Karow M; Adams NC; Gale NW; Iwasaki A; Flavell RA Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. U S A 2004, 101, 5598–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (272).Jurk M; Heil F; Vollmer J; Schetter C; Krieg AM; Wagner H; Lipford G; Bauer S Human TLR7 or TLR8 independently confer responsiveness to the antiviral compound R-848. Nat. Immunol 2002, 3, 499. [DOI] [PubMed] [Google Scholar]
- (273).Heil F; Ahmad-Nejad P; Hemmi H; Hochrein H; Ampenberger F; Gellert T; Dietrich H; Lipford G; Takeda K; Akira S et al. The Toll-like receptor 7 (TLR7)-specific stimulus loxoribine uncovers a strong relationship within the TLR7, 8 and 9 subfamily. Eur. J. Immunol 2003, 33, 2987–2997. [DOI] [PubMed] [Google Scholar]
- (274).Liu L; Botos I; Wang Y; Leonard JN; Shiloach J; Segal DM; Davies DR Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science 2008, 320, 379–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (275).Zhang Z; Ohto U; Shibata T; Krayukhina E; Taoka M; Yamauchi Y; Tanji H; Isobe T; Uchiyama S; Miyake K et al. Structural analysis reveals that Toll-like receptor 7 Is a dual receptor for guanosine and single-stranded RNA. Immunity 2016, 45, 737–748. [DOI] [PubMed] [Google Scholar]
- (276).Miyake K; Shibata T; Ohto U; Shimizu T; Saitoh SI; Fukui R; Murakami Y Mechanisms controlling nucleic acid-sensing Toll-like receptors. Int. Immunol 2018, 30, 43–51. [DOI] [PubMed] [Google Scholar]
- (277).Roers A; Hiller B; Hornung V Recognition of endogenous nucleic acids by the innate immune system. Immunity 2016, 44, 739–754. [DOI] [PubMed] [Google Scholar]
- (278).Ewald SE; Barton GM Nucleic acid sensing Toll-like receptors in autoimmunity. Curr. Opin. Immunol 2011, 23, 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (279).Majer O; Liu B; Barton GM Nucleic acid-sensing TLRs: trafficking and regulation. Curr. Opin. Immunol 2017, 44, 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (280).Lee BL; Barton GM Trafficking of endosomal Toll-like receptors. Trends Cell Biol. 2014, 24, 360–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (281).Yan N Structural Biology of the Major Facilitator Superfamily Transporters. Annu. Rev. Biophys 2015, 44, 257–283. [DOI] [PubMed] [Google Scholar]
- (282).Kim YM; Brinkmann MM; Paquet ME; Ploegh HL UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature 2008, 452, 234–238. [DOI] [PubMed] [Google Scholar]
- (283).Tabeta K; Hoebe K; Janssen EM; Du X; Georgel P; Crozat K; Mudd S; Mann N; Sovath S; Goode J et al. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat. Immunol 2006, 7, 156–164. [DOI] [PubMed] [Google Scholar]
- (284).Lee BL; Moon JE; Shu JH; Yuan L; Newman ZR; Schekman R; Barton GM UNC93B1 mediates differential trafficking of endosomal TLRs. Elife 2013, 2, e00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (285).Fukui R; Saitoh S; Kanno A; Onji M; Shibata T; Ito A; Onji M; Matsumoto M; Akira S; Yoshida N et al. Unc93B1 restricts systemic lethal inflammation by orchestrating Toll-like receptor 7 and 9 trafficking. Immunity 2011, 35, 69–81. [DOI] [PubMed] [Google Scholar]
- (286).Majer O; Liu B; Kreuk LSM; Krogan N; Barton GM UNC93B1 recruits syntenin-1 to dampen TLR7 signalling and prevent autoimmunity. Nature 2019, 575, 366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (287).Ishida H; Asami J; Zhang Z; Nishizawa T; Shigematsu H; Ohto U; Shimizu T Cryo-EM structures of Toll-like receptors in complex with UNC93B1. Nat. Struct. Mol. Biol 2021, 28, 173–180. [DOI] [PubMed] [Google Scholar]
- (288).Majer O; Liu B; Woo BJ; Kreuk LSM; Van Dis E; Barton GM Release from UNC93B1 reinforces the compartmentalized activation of select TLRs. Nature 2019, 575, 371–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (289).Deane JA; Pisitkun P; Barrett RS; Feigenbaum L; Town T; Ward JM; Flavell RA; Bolland S Control of toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity 2007, 27, 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (290).Marshak-Rothstein A Toll-like receptors in systemic autoimmune disease. Nat. Rev. Immunol 2006, 6, 823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (291).Tojo S; Zhang Z; Matsui H; Tahara M; Ikeguchi M; Kochi M; Kamada M; Shigematsu H; Tsutsumi A; Adachi N et al. Structural analysis reveals TLR7 dynamics underlying antagonism. Nat. Commun 2020, 11, 5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (292).Kurimoto A; Hashimoto K; Nakamura T; Norimura K; Ogita H; Takaku H; Bonnert R; McInally T; Wada H; Isobe Y Synthesis and biological evaluation of 8-oxoadenine derivatives as toll-like receptor 7 agonists introducing the antedrug concept. J. Med. Chem 2010, 53, 2964–2972. [DOI] [PubMed] [Google Scholar]
- (293).Ghosh P; Dahms NM; Kornfeld S Mannose 6-phosphate receptors: new twists in the tale. Nat. Rev. Mol. Cell Biol 2003, 4, 202–212. [DOI] [PubMed] [Google Scholar]
- (294).MacDonald RG; Pfeffer SR; Coussens L; Tepper MA; Brocklebank CM; Mole JE; Anderson JK; Chen E; Czech MP; Ullrich A A single receptor binds both insulin-like growth factor II and mannose-6-phosphate. Science 1988, 239, 1134–1137. [DOI] [PubMed] [Google Scholar]
- (295).Blyth AJ; Kirk NS; Forbes BE Understanding IGF-II Action through Insights into Receptor Binding and Activation. Cells 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (296).Furstenberger G; Senn HJ Insulin-like growth factors and cancer. Lancet Oncol. 2002, 3, 298–302. [DOI] [PubMed] [Google Scholar]
- (297).Bergman D; Halje M; Nordin M; Engstrom W Insulin-like growth factor 2 in development and disease: a mini-review. Gerontology 2013, 59, 240–249. [DOI] [PubMed] [Google Scholar]
- (298).Dynkevich Y; Rother KI; Whitford I; Qureshi S; Galiveeti S; Szulc AL; Danoff A; Breen TL; Kaviani N; Shanik MH et al. Tumors, IGF-2, and hypoglycemia: insights from the clinic, the laboratory, and the historical archive. Endocr. Rev 2013, 34, 798–826. [DOI] [PubMed] [Google Scholar]
- (299).Coutinho MF; Prata MJ; Alves S Mannose-6-phosphate pathway: a review on its role in lysosomal function and dysfunction. Mol. Genet. Metab 2012, 105, 542–550. [DOI] [PubMed] [Google Scholar]
- (300).Hawkes C; Kar S The insulin-like growth factor-II/mannose-6-phosphate receptor: structure, distribution and function in the central nervous system. Brain Res. Brain Res. Rev 2004, 44, 117–140. [DOI] [PubMed] [Google Scholar]
- (301).Wang R; Qi X; Schmiege P; Coutavas E; Li X Marked structural rearrangement of mannose 6-phosphate/IGF2 receptor at different pH environments. Sci. Adv 2020, 6, eaaz1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (302).Kaetzel CS The polymeric immunoglobulin receptor: bridging innate and adaptive immune responses at mucosal surfaces. Immunol. Rev 2005, 206, 83–99. [DOI] [PubMed] [Google Scholar]
- (303).Turula H; Wobus CE The role of the Polymeric Immunoglobulin Receptor and Secretory Immunoglobulins during mucosal infection and immunity. Viruses 2018, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (304).Asano M; Komiyama K Polymeric immunoglobulin receptor. J. Oral. Sci 2011, 53, 147–156. [DOI] [PubMed] [Google Scholar]
- (305).Kaetzel CS; Robinson JK; Chintalacharuvu KR; Vaerman JP; Lamm ME The polymeric immunoglobulin receptor (secretory component) mediates transport of immune complexes across epithelial cells: a local defense function for IgA. Proc. Natl. Acad. Sci. U S A 1991, 88, 8796–8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (306).Wei H; Wang JY Role of Polymeric Immunoglobulin Receptor in IgA and IgM Transcytosis. Int. J. Mol. Sci 2021, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (307).Mostov KE Transepithelial transport of immunoglobulins. Annu. Rev. Immunol 1994, 12, 63–84. [DOI] [PubMed] [Google Scholar]
- (308).Stadtmueller BM; Huey-Tubman KE; Lopez CJ; Yang Z; Hubbell WL; Bjorkman PJ The structure and dynamics of secretory component and its interactions with polymeric immunoglobulins. Elife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (309).Kumar N; Arthur CP; Ciferri C; Matsumoto ML Structure of the human secretory immunoglobulin M core. Structure 2021, 29, 564–571 e563. [DOI] [PubMed] [Google Scholar]
- (310).Li Y; Wang G; Li N; Wang Y; Zhu Q; Chu H; Wu W; Tan Y; Yu F; Su XD et al. Structural insights into immunoglobulin M. Science 2020, 367, 1014–1017. [DOI] [PubMed] [Google Scholar]
- (311).Tan YZ; Baldwin PR; Davis JH; Williamson JR; Potter CS; Carragher B; Lyumkis D Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 2017, 14, 793–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (312).Wang Y; Wang G; Li Y; Zhu Q; Shen H; Gao N; Xiao J Structural insights into secretory immunoglobulin A and its interaction with a pneumococcal adhesin. Cell Res. 2020, 30, 602–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (313).Kumar N; Arthur CP; Ciferri C; Matsumoto ML Structure of the secretory immunoglobulin A core. Science 2020, 367, 1008–1014. [DOI] [PubMed] [Google Scholar]
- (314).Weiser JN; Ferreira DM; Paton JC Streptococcus pneumoniae: transmission, colonization and invasion. Nat. Rev. Microbiol 2018, 16, 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (315).Hammerschmidt S; Talay SR; Brandtzaeg P; Chhatwal GS SpsA, a novel pneumococcal surface protein with specific binding to secretory immunoglobulin A and secretory component. Mol. Microbiol 1997, 25, 1113–1124. [DOI] [PubMed] [Google Scholar]
- (316).Hammerschmidt S; Tillig MP; Wolff S; Vaerman JP; Chhatwal GS Species-specific binding of human secretory component to SpsA protein of Streptococcus pneumoniae via a hexapeptide motif. Mol. Microbiol 2000, 36, 726–736. [DOI] [PubMed] [Google Scholar]
- (317).Dinarello CA Historical insights into cytokines. Eur J Immunol 2007, 37 Suppl 1, S34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (318).Zhang JM; An J Cytokines, inflammation, and pain. Int Anesthesiol Clin 2007, 45, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (319).Chung KF Cytokines. Asthma and COPD, 2nd Ed 2009, 327–341. [Google Scholar]
- (320).O'Shea JJ; Ma A; Lipsky P Cytokines and autoimmunity. Nat. Rev. Immunol 2002, 2, 37–45. [DOI] [PubMed] [Google Scholar]
- (321).Kelso A Cytokines: principles and prospects. Immunol. Cell Biol 1998, 76, 300–317. [DOI] [PubMed] [Google Scholar]
- (322).Feldmann M; Brennan FM; Maini R Cytokines in autoimmune disorders. Int. Rev. Immunol 1998, 17, 217–228. [DOI] [PubMed] [Google Scholar]
- (323).Ouyang W; Rutz S; Crellin NK; Valdez PA; Hymowitz SG Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu. Rev. Immunol 2011, 29, 71–109. [DOI] [PubMed] [Google Scholar]
- (324).Pestka S; Krause CD; Sarkar D; Walter MR; Shi Y; Fisher PB Interleukin-10 and related cytokines and receptors. Annu. Rev. Immunol 2004, 22, 929–979. [DOI] [PubMed] [Google Scholar]
- (325).Moore KW; de Waal Malefyt R; Coffman RL; O'Garra A Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol 2001, 19, 683–765. [DOI] [PubMed] [Google Scholar]
- (326).Wang X; Lupardus P; Laporte SL; Garcia KC Structural biology of shared cytokine receptors. Annu. Rev. Immunol 2009, 27, 29–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (327).Renauld JC Class II cytokine receptors and their ligands: key antiviral and inflammatory modulators. Nat. Rev. Immunol 2003, 3, 667–676. [DOI] [PubMed] [Google Scholar]
- (328).Yoon SI; Jones BC; Logsdon NJ; Harris BD; Deshpande A; Radaeva S; Halloran BA; Gao B; Walter MR Structure and mechanism of receptor sharing by the IL-10R2 common chain. Structure 2010, 18, 638–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (329).Josephson K; Logsdon NJ; Walter MR Crystal structure of the IL-10/IL-10R1 complex reveals a shared receptor binding site. Immunity 2001, 15, 35–46. [DOI] [PubMed] [Google Scholar]
- (330).Verma R; Balakrishnan L; Sharma K; Khan AA; Advani J; Gowda H; Tripathy SP; Suar M; Pandey A; Gandotra S et al. A network map of Interleukin-10 signaling pathway. J. Cell Commun. Signal 2016, 10, 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (331).Saraiva M; Vieira P; O'Garra A Biology and therapeutic potential of interleukin-10. J. Exp. Med 2020, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (332).Murray PJ Understanding and exploiting the endogenous interleukin-10/STAT3-mediated anti-inflammatory response. Curr. Opin. Pharmacol 2006, 6, 379–386. [DOI] [PubMed] [Google Scholar]
- (333).Shouval DS; Ouahed J; Biswas A; Goettel JA; Horwitz BH; Klein C; Muise AM; Snapper SB Interleukin 10 receptor signaling: master regulator of intestinal mucosal homeostasis in mice and humans. Adv. Immunol 2014, 122, 177–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (334).Engelhardt KR; Grimbacher B IL-10 in humans: lessons from the gut, IL-10/IL-10 receptor deficiencies, and IL-10 polymorphisms. Curr. Top. Microbiol. Immunol 2014, 380, 1–18. [DOI] [PubMed] [Google Scholar]
- (335).Glocker EO; Kotlarz D; Klein C; Shah N; Grimbacher B IL-10 and IL-10 receptor defects in humans. Ann. N. Y. Acad. Sci 2011, 1246, 102–107. [DOI] [PubMed] [Google Scholar]
- (336).Wang X; Wong K; Ouyang W; Rutz S Targeting IL-10 Family Cytokines for the Treatment of Human Diseases. Cold Spring Harb. Perspect. Biol 2019, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (337).O'Garra A; Barrat FJ; Castro AG; Vicari A; Hawrylowicz C Strategies for use of IL-10 or its antagonists in human disease. Immunol. Rev 2008, 223, 114–131. [DOI] [PubMed] [Google Scholar]
- (338).Saxton RA; Tsutsumi N; Su LL; Abhiraman GC; Mohan K; Henneberg LT; Aduri NG; Gati C; Garcia KC Structure-based decoupling of the pro- and anti-inflammatory functions of interleukin-10. Science 2021, 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (339).Saxton RA; Garcia KC Cryo-EM structure of the IL-10 receptor complex provides a blueprint for ligand engineering. FEBS J. 2021, DOI: 10.1111/febs.16207 10.1111/febs.16207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (340).Zimmerman B; Kelly B; McMillan BJ; Seegar TCM; Dror RO; Kruse AC; Blacklow SC Crystal Structure of a Full-Length Human Tetraspanin Reveals a Cholesterol-Binding Pocket. Cell 2016, 167, 1041–1051 e1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (341).Termini CM; Gillette JM Tetraspanins Function as Regulators of Cellular Signaling. Front Cell Dev. Biol 2017, 5, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (342).Hemler ME Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol 2005, 6, 801–811. [DOI] [PubMed] [Google Scholar]
- (343).Andreu Z; Yanez-Mo M Tetraspanins in extracellular vesicle formation and function. Front Immunol. 2014, 5, 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (344).Tedder TF; Inaoki M; Sato S The CD19-CD21 complex regulates signal transduction thresholds governing humoral immunity and autoimmunity. Immunity 1997, 6, 107–118. [DOI] [PubMed] [Google Scholar]
- (345).Scharenberg AM; Humphries LA; Rawlings DJ Calcium signalling and cell-fate choice in B cells. Nat. Rev. Immunol 2007, 7, 778–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (346).Carter RH; Barrington RA Signaling by the CD19/CD21 complex on B cells. Curr. Dir. Autoimmun 2004, 7, 4–32. [DOI] [PubMed] [Google Scholar]
- (347).Shoham T; Rajapaksa R; Boucheix C; Rubinstein E; Poe JC; Tedder TF; Levy S The tetraspanin CD81 regulates the expression of CD19 during B cell development in a postendoplasmic reticulum compartment. J. Immunol 2003, 171, 4062–4072. [DOI] [PubMed] [Google Scholar]
- (348).Mattila PK; Feest C; Depoil D; Treanor B; Montaner B; Otipoby KL; Carter R; Justement LB; Bruckbauer A; Batista FD The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity 2013, 38, 461–474. [DOI] [PubMed] [Google Scholar]
- (349).Susa KJ; Rawson S; Kruse AC; Blacklow SC Cryo-EM structure of the B cell co-receptor CD19 bound to the tetraspanin CD81. Science 2021, 371, 300–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (350).Oosterheert W; Xenaki KT; Neviani V; Pos W; Doulkeridou S; Manshande J; Pearce NM; Kroon-Batenburg LM; Lutz M; van Bergen En Henegouwen PM et al. Implications for tetraspanin-enriched microdomain assembly based on structures of CD9 with EWI-F. Life Sci. Alliance 2020, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (351).Umeda R; Satouh Y; Takemoto M; Nakada-Nakura Y; Liu K; Yokoyama T; Shirouzu M; Iwata S; Nomura N; Sato K et al. Structural insights into tetraspanin CD9 function. Nat. Commun 2020, 11, 1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (352).Hemler ME Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol 2003, 19, 397–422. [DOI] [PubMed] [Google Scholar]
- (353).Potel J; Rassam P; Montpellier C; Kaestner L; Werkmeister E; Tews BA; Couturier C; Popescu CI; Baumert TF; Rubinstein E et al. EWI-2wint promotes CD81 clustering that abrogates Hepatitis C Virus entry. Cell. Microbiol 2013, 15, 1234–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (354).Sala-Valdes M; Ursa A; Charrin S; Rubinstein E; Hemler ME; Sanchez-Madrid F; Yanez-Mo M EWI-2 and EWI-F link the tetraspanin web to the actin cytoskeleton through their direct association with ezrin-radixin-moesin proteins. J. Biol. Chem 2006, 281, 19665–19675. [DOI] [PubMed] [Google Scholar]
- (355).Stipp CS; Kolesnikova TV; Hemler ME EWI-2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J. Biol. Chem 2001, 276, 40545–40554. [DOI] [PubMed] [Google Scholar]
- (356).Charrin S; Le Naour F; Oualid M; Billard M; Faure G; Hanash SM; Boucheix C; Rubinstein E The major CD9 and CD81 molecular partner. Identification and characterization of the complexes. J. Biol. Chem 2001, 276, 14329–14337. [DOI] [PubMed] [Google Scholar]
- (357).Kolesnikova TV; Stipp CS; Rao RM; Lane WS; Luscinskas FW; Hemler ME EWI-2 modulates lymphocyte integrin alpha4beta1 functions. Blood 2004, 103, 3013–3019. [DOI] [PubMed] [Google Scholar]
- (358).Stipp CS; Kolesnikova TV; Hemler ME EWI-2 regulates alpha3beta1 integrin-dependent cell functions on laminin-5. J. Cell Biol 2003, 163, 1167–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (359).Aguila B; Morris AB; Spina R; Bar E; Schraner J; Vinkler R; Sohn JW; Welford SM The Ig superfamily protein PTGFRN coordinates survival signaling in glioblastoma multiforme. Cancer Lett. 2019, 462, 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (360).Yang XH; Kovalenko OV; Kolesnikova TV; Andzelm MM; Rubinstein E; Strominger JL; Hemler ME Contrasting effects of EWI proteins, integrins, and protein palmitoylation on cell surface CD9 organization. J. Biol. Chem 2006, 281, 12976–12985. [DOI] [PubMed] [Google Scholar]
- (361).Hong W; Mosca TJ; Luo L Teneurins instruct synaptic partner matching in an olfactory map. Nature 2012, 484, 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (362).Mosca TJ; Hong W; Dani VS; Favaloro V; Luo L Trans-synaptic Teneurin signalling in neuromuscular synapse organization and target choice. Nature 2012, 484, 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (363).Vysokov NV; Silva JP; Lelianova VG; Suckling J; Cassidy J; Blackburn JK; Yankova N; Djamgoz MB; Kozlov SV; Tonevitsky AG et al. Proteolytically released Lasso/teneurin-2 induces axonal attraction by interacting with latrophilin-1 on axonal growth cones. Elife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (364).Boucard AA; Maxeiner S; Sudhof TC Latrophilins function as heterophilic cell-adhesion molecules by binding to teneurins: regulation by alternative splicing. J. Biol. Chem 2014, 289, 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (365).Silva JP; Lelianova VG; Ermolyuk YS; Vysokov N; Hitchen PG; Berninghausen O; Rahman MA; Zangrandi A; Fidalgo S; Tonevitsky AG et al. Latrophilin 1 and its endogenous ligand Lasso/teneurin-2 form a high-affinity transsynaptic receptor pair with signaling capabilities. Proc. Natl. Acad. Sci. U S A 2011, 108, 12113–12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (366).Sando R; Jiang X; Sudhof TC Latrophilin GPCRs direct synapse specificity by coincident binding of FLRTs and teneurins. Science 2019, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (367).Bagutti C; Forro G; Ferralli J; Rubin B; Chiquet-Ehrismann R The intracellular domain of teneurin-2 has a nuclear function and represses zic-1-mediated transcription. J. Cell Sci 2003, 116, 2957–2966. [DOI] [PubMed] [Google Scholar]
- (368).Li J; Shalev-Benami M; Sando R; Jiang X; Kibrom A; Wang J; Leon K; Katanski C; Nazarko O; Lu YC et al. Structural basis for Teneurin function in circuit-wiring: A toxin motif at the synapse. Cell 2018, 173, 735–748 e715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (369).Oohashi T; Zhou XH; Feng K; Richter B; Morgelin M; Perez MT; Su WD; Chiquet-Ehrismann R; Rauch U; Fassler R Mouse ten-m/Odz is a new family of dimeric type II transmembrane proteins expressed in many tissues. J. Cell Biol 1999, 145, 563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (370).Feng K; Zhou XH; Oohashi T; Morgelin M; Lustig A; Hirakawa S; Ninomiya Y; Engel J; Rauch U; Fassler R All four members of the Ten-m/Odz family of transmembrane proteins form dimers. J. Biol. Chem 2002, 277, 26128–26135. [DOI] [PubMed] [Google Scholar]
- (371).Tucker RP; Beckmann J; Leachman NT; Scholer J; Chiquet-Ehrismann R Phylogenetic analysis of the teneurins: conserved features and premetazoan ancestry. Mol. Biol. Evol 2012, 29, 1019–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (372).Li J; Xie Y; Cornelius S; Jiang X; Sando R; Kordon SP; Pan M; Leon K; Sudhof TC; Zhao M et al. Alternative splicing controls teneurin-latrophilin interaction and synapse specificity by a shape-shifting mechanism. Nat. Commun 2020, 11, 2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (373).Del Toro D; Carrasquero-Ordaz MA; Chu A; Ruff T; Shahin M; Jackson VA; Chavent M; Berbeira-Santana M; Seyit-Bremer G; Brignani S et al. Structural basis of Teneurin-Latrophilin interaction in repulsive guidance of migrating neurons. Cell 2020, 180, 323–339 e319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (374).Jackson VA; Meijer DH; Carrasquero M; van Bezouwen LS; Lowe ED; Kleanthous C; Janssen BJC; Seiradake E Structures of Teneurin adhesion receptors reveal an ancient fold for cell-cell interaction. Nat. Commun 2018, 9, 1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (375).Wang L; Rotzinger S; Al Chawaf A; Elias CF; Barsyte-Lovejoy D; Qian X; Wang NC; De Cristofaro A; Belsham D; Bittencourt JC et al. Teneurin proteins possess a carboxy terminal sequence with neuromodulatory activity. Brain Res. Mol. Brain Res 2005, 133, 253–265. [DOI] [PubMed] [Google Scholar]
- (376).Qian X; Barsyte-Lovejoy D; Wang L; Chewpoy B; Gautam N; Al Chawaf A; Lovejoy DA Cloning and characterization of teneurin C-terminus associated peptide (TCAP)-3 from the hypothalamus of an adult rainbow trout (Oncorhynchus mykiss). Gen. Comp. Endocrinol 2004, 137, 205–216. [DOI] [PubMed] [Google Scholar]
- (377).Lovejoy DA; Al Chawaf A; Cadinouche MZ Teneurin C-terminal associated peptides: an enigmatic family of neuropeptides with structural similarity to the corticotropin-releasing factor and calcitonin families of peptides. Gen. Comp. Endocrinol 2006, 148, 299–305. [DOI] [PubMed] [Google Scholar]
- (378).Jackson VA; Mehmood S; Chavent M; Roversi P; Carrasquero M; Del Toro D; Seyit-Bremer G; Ranaivoson FM; Comoletti D; Sansom MS et al. Super-complexes of adhesion GPCRs and neural guidance receptors. Nat. Commun 2016, 7, 11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (379).Wucherpfennig KW; Gagnon E; Call MJ; Huseby ES; Call ME Structural biology of the T-cell receptor: insights into receptor assembly, ligand recognition, and initiation of signaling. Cold Spring Harb. Perspect. Biol 2010, 2, a005140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (380).Huang J; Brameshuber M; Zeng X; Xie J; Li QJ; Chien YH; Valitutti S; Davis MM A single peptide-major histocompatibility complex ligand triggers digital cytokine secretion in CD4(+) T cells. Immunity 2013, 39, 846–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (381).Brameshuber M; Kellner F; Rossboth BK; Ta H; Alge K; Sevcsik E; Gohring J; Axmann M; Baumgart F; Gascoigne NRJ et al. Monomeric TCRs drive T cell antigen recognition. Nat. Immunol 2018, 19, 487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (382).Samelson LE Signal transduction mediated by the T cell antigen receptor: the role of adapter proteins. Annu. Rev. Immunol 2002, 20, 371–394. [DOI] [PubMed] [Google Scholar]
- (383).Mariuzza RA; Agnihotri P; Orban J The structural basis of T-cell receptor (TCR) activation: An enduring enigma. J. Biol. Chem 2020, 295, 914–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (384).Dong; Zheng L; Lin J; Zhang B; Zhu Y; Li N; Xie S; Wang Y; Gao N; Huang Z Structural basis of assembly of the human T cell receptor-CD3 complex. Nature 2019, 573, 546–552. [DOI] [PubMed] [Google Scholar]
- (385).Chen Y; Zhu Y; Li X; Gao W; Zhen Z; Dong; Huang B; Ma Z; Zhang A; Song X et al. Cholesterol inhibits TCR signaling by directly restricting TCR-CD3 core tunnel motility. Mol. Cell 2022, 82, 1278–1287 e1275. [DOI] [PubMed] [Google Scholar]
- (386).Yin Y; Wang XX; Mariuzza RA Crystal structure of a complete ternary complex of T-cell receptor, peptide-MHC, and CD4. Proc Natl Acad Sci U S A 2012, 109, 5405–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (387).Garboczi DN; Ghosh P; Utz U; Fan QR; Biddison WE; Wiley DC Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature 1996, 384, 134–141. [DOI] [PubMed] [Google Scholar]
- (388).van Deventer SJ; Dunlock VE; van Spriel AB Molecular interactions shaping the tetraspanin web. Biochem. Soc. Trans 2017, 45, 741–750. [DOI] [PubMed] [Google Scholar]
- (389).Albrecht C; Appert-Collin A; Bagnard D; Blaise S; Romier-Crouzet B; Efremov RG; Sartelet H; Duca L; Maurice P; Bennasroune A Transmembrane peptides as inhibitors of protein-protein interactions: an efficient strategy to target cancer cells? Front Oncol. 2020, 10, 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (390).Frauenfeld J; Loving R; Armache JP; Sonnen AF; Guettou F; Moberg P; Zhu L; Jegerschold C; Flayhan A; Briggs JA et al. A saposin-lipoprotein nanoparticle system for membrane proteins. Nat Methods 2016, 13, 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (391).Morrison KA; Akram A; Mathews A; Khan ZA; Patel JH; Zhou C; Hardy DJ; Moore-Kelly C; Patel R; Odiba V et al. Membrane protein extraction and purification using styrene-maleic acid (SMA) copolymer: effect of variations in polymer structure. Biochem J. 2016, 473, 4349–4360. [DOI] [PubMed] [Google Scholar]
- (392).Himanen JP; Yermekbayeva L; Janes PW; Walker JR; Xu K; Atapattu L; Rajashankar KR; Mensinga A; Lackmann M; Nikolov DB et al. Architecture of Eph receptor clusters. Proc. Natl. Acad. Sci. U S A 2010, 107, 10860–10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (393).Poweleit N; Czudnochowski N; Nakagawa R; Trinidad DD; Murphy KC; Sassetti CM; Rosenberg OS The structure of the endogenous ESX-3 secretion system. Elife 2019, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (394).Lin S; Ke M; Zhang Y; Yan Z; Wu J Structure of a mammalian sperm cation channel complex. Nature 2021, 595, 746–750. [DOI] [PubMed] [Google Scholar]
- (395).Marx V Calling cell biologists to try cryo-ET. Nat. Methods 2018, 15, 575–578. [DOI] [PubMed] [Google Scholar]
- (396).Dunstone MA; de Marco A Cryo-electron tomography: an ideal method to study membrane-associated proteins. Philos. Trans. R. Soc. Lond. B Biol. Sci 2017, 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (397).Hampton CM; Strauss JD; Ke Z; Dillard RS; Hammonds JE; Alonas E; Desai TM; Marin M; Storms RE; Leon F et al. Correlated fluorescence microscopy and cryo-electron tomography of virus-infected or transfected mammalian cells. Nat. Protoc 2017, 12, 150–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (398).Villa E; Schaffer M; Plitzko JM; Baumeister W Opening windows into the cell: focused-ion-beam milling for cryo-electron tomography. Curr. Opin. Struct. Biol 2013, 23, 771–777. [DOI] [PubMed] [Google Scholar]
- (399).Wan W; Briggs JA Cryo-Electron tomography and subtomogram averaging. Methods Enzymol. 2016, 579, 329–367. [DOI] [PubMed] [Google Scholar]
- (400).Tran NH; Carter SD; De Maziere A; Ashkenazi A; Klumperman J; Walter P; Jensen GJ The stress-sensing domain of activated IRE1alpha forms helical filaments in narrow ER membrane tubes. Science 2021, 374, 52–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (401).Huppa JB; Davis MM T-cell-antigen recognition and the immunological synapse. Nat. Rev. Immunol 2003, 3, 973–983. [DOI] [PubMed] [Google Scholar]