Abstract
Nuclear export of ribosomes requires a subset of nucleoporins and the Ran system, but specific transport factors have not been identified. Using a large subunit reporter (Rpl25p-eGFP), we have isolated several temperature-sensitive ribosomal export (rix) mutants. One of these corresponds to the ribosomal protein Rpl10p, which interacts directly with Nmd3p, a conserved and essential protein associated with 60S subunits. We find that thermosensitive nmd3 mutants are impaired in large subunit export. Strikingly, Nmd3p shuttles between the nucleus and cytoplasm and is exported by the nuclear export receptor Xpo1p. Moreover, we show that export of 60S subunits is Xpo1p dependent. We conclude that nuclear export of 60S subunits requires the nuclear export sequence-containing nonribosomal protein Nmd3p, which directly binds to the large subunit protein Rpl10p.
Most steps in ribosome synthesis take place in the nucleolus, a specialized subnuclear region. This process starts with the synthesis of two pre-rRNA transcripts, 35S and pre-5S rRNA, which are processed and base modified to yield the mature 25S/28S, 18S, 5.8S, and 5S rRNAs, respectively (18). During these processes about 80 ribosomal proteins assemble onto the rRNAs to yield preribosomal particles, which are exported into the cytoplasm (41). In contrast to pre-rRNA processing and modification, very little is known about the assembly pathway for eukaryotic ribosomal subunits or the features that make them competent for nuclear exit (for recent reviews, see references 18 and 40).
The transport of macromolecules through the nuclear pores is thought to involve facilitated diffusion of soluble transport factors over the repeat sequences of the nuclear pore proteins (nucleoporins) that form and line the nuclear pore complex. Directionality of transport is provided by the small GTPase Ran, due to the presence of a step RanGTP/RanGDP gradient across the nuclear membrane (for a review, see reference 23). RanGTP binds with high affinity to nuclear import and export receptors (importins and exportins, respectively) of the karyopherin β superfamily (10). For nuclear exit, export cargoes, which harbor nuclear export sequences (NESs) (e.g., leucine-rich NESs), form an intranuclear complex with the NES receptor Xpo1p/Crm1 in the presence of RanGTP (8, 33). This trimeric complex is then exported from the nucleus into the cytoplasm.
Saccharomyces cerevisiae has been a useful system for the analysis of the nuclear pore complex as well as transport factors (6). We have reported an in vivo assay for ribosomal export in yeast that uses a fusion between green fluorescent protein (GFP) and ribosomal protein Rpl25p (15). Rpl25p is imported into the nucleus and assembles with ribosomes by direct binding to the rRNA inside the nucleolus (39). Passage of both the free Rpl25p-GFP and the preribosomal particles through the nucleoplasm appears to be rapid in wild-type cells, and GFP-labeled ribosomes were detected by fluorescence microscopy in the cytoplasm. Mutations causing defects in subunit export lead to a readily detectable nucleoplasmic GFP signal. This assay showed that a subset of the nucleoporins and the Ran system were required for 60S subunit export. An assay for the nuclear export of 40S subunits led to similar conclusions (26).
We have exploited the 60S export assay to screen a bank of randomly generated temperature-sensitive (ts) mutants for a ribosomal export defect (rix) phenotype. Here we report the characterization of the rix5-1 mutant, which is complemented by the RPL10 gene (also called QSR1 or GRC5). This gene encodes the ribosomal protein Rpl10p (21), which was reported to assemble late with 60S subunits. Since Rpl10p was reported to interact with Nmd3p, a nonribosomal protein that associates with 60S subunits (3, 12, 13, 16), we analyzed whether Nmd3p plays a role in nuclear export of ribosomes. We find that nmd3 ts mutants accumulate the large subunit reporter Rpl25-eGFP inside the nucleus at the restrictive temperature. Interestingly, Nmd3p has a C-terminal NES domain, which uses Xpo1p for nuclear exit. Since the large subunit reporter Rpl25-eGFP accumulates inside the nucleus when Xpo1p is inhibited, Nmd3p and Xpo1p are likely to be factors involved in nuclear export of 60S subunits. Recently, Ho et al. reported similar findings that a dominant negative allele of nmd3 impairs 60S subunit export and Nmd3p acts as an adapter for Xpo1p-mediated nuclear export of large subunits (14).
MATERIALS AND METHODS
Yeast strains, DNA recombinant work, and microbiological techniques.
Yeast strains used in this study are described in Table 1. Microbiological techniques, plasmid transformation and recovery, mating, sporulation of diploids, and tetrad analysis were done essentially as described elsewhere (30). DNA recombinant work was performed as described in reference 22.
TABLE 1.
Yeast strains used
| Strain | Genotype | Reference or origin |
|---|---|---|
| RS453 | MATa/α ade2/ade2 leu2/leu2 ura3/ura3 his3/his3 trp1/trp1 | 15 |
| RS453a | MATaade2 leu2 ura3 his3 trp1 | 15 |
| FY23 | MATaura3 trp1 leu2 | Derived from S288C |
| FY86 | MATα ura3 his3 leu2 | Derived from S288C |
| rix5-1 | MATα ura3 his3 leu2 rix5-1 | Isolated from ts collection (2) |
| RPL25-shuffle | MATα ade2 leu2 ura3 his3 trp1 rpl25Δ::HIS3 + pRS316-RPL25 (ARS/CEN RPL25 URA3) | Derived from RS453 |
| Rpl25p-GFP | MATα ade2 leu2 ura3 his3 trp1 rpl25Δ::HIS3 + YEplac195ADE-Rpl25p-GFP (2μm URA3 ADE2 RPL25-GFP) | 15 |
| Rpl25p-eGFP | MATα ade2 leu2 ura3 his3 trp1 rpl25Δ::HIS3 + pASZ11-Rpl25p-eGFP (ARS/CEN ADE2 RPL25-eGFP) | This study |
| NMD3-shuffle | MATahis3 leu2 lys2 ura3 nmd3::kanMX4 (ARS/CEN URA3 NMD3) | This study |
| nmd3-2 | MATahis3 leu2 lys2 ura3 nmd3::kanMX4 (ARS/CEN LEU2 nmd3-2) | This study |
| BY4743 | MATa/α his3Δ1/his3Δ1 leu2Δ0 leu2Δ0 lys2Δ0/LYS2 MET15Δ0 ura3Δ0 ura3Δ0 nmd3::kanMX4/NMD3 | EUROSCARF |
| kap123Δ | MATα ura3 leu2 his3 kap123Δ::HIS3 | 29 |
| JWY1457 | MATα can1 ade2 his3 lys2 trp1 leu2 ura3 rpl16aΔ::TRP1 rpl16bΔ::LEU2 +YCp50-RPL16b (ARS/CEN URA3 RPL16b) | 25 |
| JWY1460 | MATα can1 ade2 his3 lys2 trp1 leu2 ura3 rpl16aΔ::TRP1 rpl16bΔ::LEU2 +pHIS3CEN3-Rplb16b-2 (ARS/CEN HIS3 rpl16b-2) | 25 |
| MNY8 (xpo1 LMB sensitive) | MATaleu2 hs3 trp1 ura3 Δcrm1::KAN (ARS/CEN LEU2 pDC-CRM1T539C) | 27 |
| rrp44-1 | MATaura3 lys2 ade2 trp1 his3 leu2/rrp44-1 | Mitchell et al., submitted for publication |
| rsa1Δ | MATahis3 leu2 met15 ura3 rsa1Δ::kanMX4 | EUROSCARF |
| MNY7 (xpo1 LMB resistant) | MATaleu2 hs3 trp1 ura3 Δcrm1::KANr (ARS/CEN LEU2 pDC-CRM1) |
Plasmid constructions.
Plasmid YEplac195-ADE2-L25NLS-GFP was derived from YEplac195-ADE2-URA3-RPL25-GFP by removing most of the RPL25 coding sequence, leaving the 5′ end that encodes the first 52 residues of Rpl25p. YEplac195-ADE2-Rpl25p-GFP was previously described (15). This Rpl25p-GFP reporter was improved, and the derived Rpl25p-eGFP (enhanced GFP) construct complements the rpl25Δ::HIS3 disruption at all temperatures tested, including 37°C. The new Rpl25p-eGFP construct was tested in nucleoporin and Ran cycle mutants and shown to behave similar to the original Rpl25p-GFP construct except that nuclear accumulation can be directly seen at the restrictive temperature, without requiring a short regrowth phase at the permissive temperature. This pASZ11-RPl25p-eGFP construct was generated in three steps. The 3′ untranslated region of RPS20 was amplified by PCR on yeast genomic DNA using the primers CAA CGG ATC CTA AGC TGG TTC TAA CTG GAA ATA AT and TTT TGT CGA CAC ATT GCC TTA TTC AAA GCC GCC. The 386-bp-long fragment generated was cut with BamHI-SalI and cloned into pASZ11 at the same site, generating pASZ11-3′-RPS20. The eGFP was amplified by PCR on pEGFP-1 vector (Clontech, Palo Alto, Calif.) using the primers AAA GGA TCC ATG GTG AGC AAG GGC G and TTT TTT GTC GAC TTA AGA TCT CTT GTA CAG CTC GTC CAT GCC. The 745-bp-long generated fragment was cut with BamHI-SalI and cloned into pBluescript to generate a pBS-eGFP vector, and the sequence of the eGFP coding region was confirmed by DNA sequencing. A BamHI-BglII fragment from pBS-eGFP containing the eGFP was cloned into the correct orientation into the BamHI site of pASZ11-3′-RPS20. RPL25 including its authentic RPL25 5′ untranslated region was amplified by PCR on yeast genomic DNA using the primers TAG GAT CCA ATG TAA CCG ATT CTG TTA GCA ATG TCC and TTC CCG GGC AAT GAA GAG ATG AGG AGG CAT GG. The 1,293-bp-long generated fragment was cut with XmaI-BamHI and ligated into pASZ11-eGFP-3′-RPS20, generating the pASZ11-RPL25-eGFP. The open reading frame sequence of RPL25 was verified by DNA sequencing. Plasmids pRS314-RPL25-eGFP, pRS315-RPL25-eGFP, and pRS316-RPL25-eGFP were obtained by subcloning the XmaI-SalI 2,378-bp-long fragment derived from pASZ11-RPL25-eGFP into the XmaI-SalI sites of, respectively, pRS314, pRS315, and pRS316.
DsRed (red fluorescent protein) was amplified by PCR on plasmid pDsRed1-N1 (Clontech) using the primers AAA AAA GCA TGC GGA TCC ATG GTG CGC TCC TCC AAG AAC GTC A and TTT TTT GTC GAC CTA GCA TGC CAG GAA CAG GTG GTG GCG GCC CTC G. The derived 717-bp-long generated fragment was cut with SphI and cloned into the SphI site of pUN100-ProtA-NOP1 (4), replacing protein A (ProtA) with DsRed. pRS314-DsRed-NOP1 was generated by cloning the 2,323-bp-long SacI-SnaBI fragment from pUN100-DsRed-NOP1 into the SacI-SmaI site of pRS314.
Isolation of rix mutants.
A collection of 900 thermosensitive yeast mutants (2, 11) derived from haploid strains FY23 and FY86 used in the screen was previously generated in the laboratory of C. Cole (Dartmouth Medical School, Hanover, N.H.). The collection was kept in 96-well plates at −80°C and obtained from Ann Mutvei (University of Umeå, Umeå, Sweden). Mutants were plated on YPD (yeast-peptone-dextrose) medium, transformed with YEplac 195-ADE2-URA3-RPL25-GFP, plated on SDC-Ura for 5 days, then replica plated onto two YPD plates, and incubated at 37°C. After 2 h or overnight incubation at 37°C, each plate was transferred at 23°C for 1 h. Cells were mounted on a slide and inspected by fluorescence microscopy. When the wild-type strain showed Rpl25p-GFP cytoplasmic staining and control mutants (rna1-1, prp20-1 and nup49-313) showed nuclear accumulation of Rpl25p-GFP, the phenotype of the ts mutants was investigated. Mutants that accumulated Rpl25p-GFP in distinct regions were then rechecked by the same procedure but with subsequent DNA staining without fixation. Cells were resuspended in water containing Hoechst H33342 (50 μg/ml) for 20 min at room temperature under continuous shaking, then washed with 1 ml of water, mounted on a slide, and inspected by fluorescence microscopy. In 35 mutants, Rpl25p-GFP accumulation colocalizes with DNA staining. In these 35 mutants, the old reporter construct was lost and pRS315-RPL25-eGFP was introduced by transformation and selected on SDC-Leu medium. Individual transformants were picked, grown in liquid SDC-Leu medium at 23°C to an optical density at 600 nm (OD600) of about 1, and then shifted for 5 h to 37°C in liquid YPD medium. After centrifugation, cells were resuspended in water, mounted on a slide, and viewed in a fluorescence microscope. Fifteen mutants showing less than 50% of accumulation phenotype or accumulation only after reinduction of growth at the permissive temperature were not kept. In 20 mutants, more than 50% of the cells show nuclear accumulation of Rpl25p-eGFP under this condition. Accordingly, these mutants were defined as rix mutants.
Cloning of RIX5/RPL10.
A yeast genomic library in a LEU2-containing ARS/CEN plasmid was transformed into rix5-1 strain. From colonies growing at the restrictive temperature (37°C), plasmids with genomic inserts were isolated. Complementing plasmids always contained the RPL10 gene. pRS315-RPL10 harboring only the RPL10 gene was generated and shown to complement the thermosensitive growth defect of the rix5-1 mutant.
Generation of NMD3-shuffle and nmd3 mutant strains.
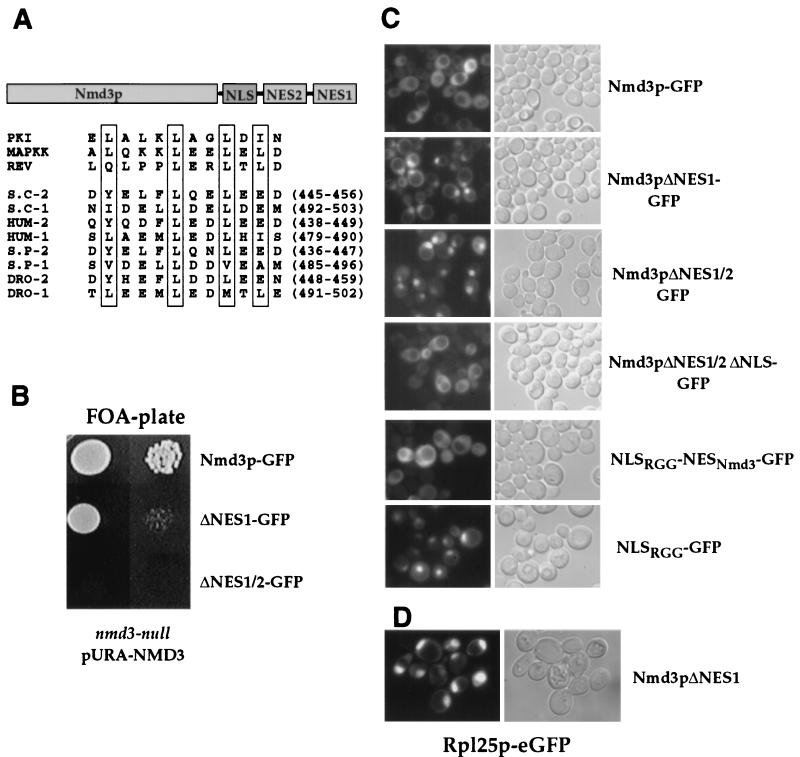
NMD3 was isolated by PCR from yeast genomic DNA. A haploid NMD3-shuffle strain was constructed from a heterozygous nmd3::kanMX4/NMD3 diploid. The nmd3 ts mutants were generated by random PCR-mediated mutagenesis as described elsewhere (35). nmd3-2 contains the mutations M141V, D255V, F318L, I328M, K413T, E440G, F449L, N464S, E479D, D498G, and D501G. Tagging of Nmd3p with GFP was performed by exchanging RPL25 from pRS315-RPL25-eGFP with the NMD3 gene including its authentic promoter. C-terminal deletions of NMD3 were generated by using NMD3 under its own promoter, but lacking 44 (ΔNES1), 79 (ΔNES1/2), and 116 (ΔNES1/2 ΔNLS) C-terminal residues. The protein kinase inhibitor (PKI)-NES attached to Nmd3pΔNES has the amino acid sequence NELALKLAGLDINKT. The nuclear localization signal (NLS) derived from Npl3p fused to GFP is described in reference 34; the NES domain from Nmd3p corresponding to residues 440 to 518 was attached to this reporter construct.
Expression of Nmd3p and Rpl10p in Escherichia coli.
NMD3 was cloned into the pET24d vector, harboring the kanamycin resistance marker and allowing C-terminal tagging of NMD3 with six histidines. NMD3 was amplified from genomic DNA with the primers 5′ aaa aaa gct agc ATG GAA TTC ACA CCT ATA GA and 3′ ttt ttt ttg cgg ccg cCT GCT GAG ATT CAA CGG GTG (where lowercase letters indicate the restriction site) before inserting it into pET-24d (Novagen), which was cut with NheI and NotI. pGEX-4T-3 (Amersham Pharmacia Biotech) harboring the ampicillin resistance marker was used for glutathione S-transferase (GST) expression. pHFF17, which also harbors the ampicillin resistance marker, contains a GST-RPL10 fusion construct (38). For coexpression, the E. coli BL21-codon plus-RIL strain (Stratagene) was transformed with (i) pHFF17, (ii) pHFF17 plus pET24d-NMD3, or (iii) pGEX-4T-3 plus pET24d-NMD3. It was inoculated in 0.5 liter of minimal medium plus ampicillin-kanamycin with 1/100 of overnight culture and grown to an OD600 of 0.6 at 16°C; recombinant proteins were induced for 6 h at 16°C with 0.1 mM isopropyl-β-d-thiogalactopyranoside. Purification of GST-tagged proteins was done as described elsewhere (20). The E. coli cell pellet was resuspended in MU buffer (0.5% Tween 20, 100 mM potassium acetate, 20 mM HEPES [pH 7.0], 2 mM magnesium acetate, 10% glycerol, 2.5 mM dithiothreitol), and cells were lysed by sonication. The supernatant was incubated with 100 μl of glutathione-Sepharose 4B (Amersham Pharmacia Biotech), the beads were washed with MU buffer, and the proteins bound to the beads were eluted with sample buffer. To detect Rpl10p and Nmd3p by Western blotting, QSR1-specific antibody (α-QSR1) and monoclonal antibody (α-His) (BAbCo, Richmond, Calif.), respectively, were used.
Miscellaneous.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis were performed as described elsewhere (32); isolation of ribosomes under low- and high-salt conditions by sucrose gradient centrifugation was done as described in reference 37. Whole-cell lysates and fractions from the sucrose gradient were separated by SDS-PAGE and analyzed by Western blotting using the indicated antibodies. Pulse-chase labeling of rRNA and analysis of rRNA processing by Northern hybridization were performed as described previously (36, 37). GFP-tagged fusion proteins were detected in vivo in a Zeiss Axioskop fluorescence microscope, and pictures were obtained with a Xillix Microimager charge-coupled device camera. For leptomycin B (LMB) treatment of the LMB-sensitive (CRM1-T539C) and LMB-resistant (CRM1) S. cerevisiae strains, 100 ng of LMB/ml was added to the culture medium (27).
RESULTS
Isolation of rix mutants.
To identify factors required for ribosomal export, we GFP tagged a 60S ribosomal subunit protein (Rpl25p-GFP) and used it in a fluorescence-based assay to monitor large subunit export. Here we screened a bank of 900 randomly generated ts mutants for a rix phenotype. We analyzed these mutants for nuclear accumulation of Rpl25p-GFP at the restrictive temperature followed by a short regrowth phase at the permissive temperature (15); 35 rix mutants were identified in this assay.
Since the Rpl25p-GFP construct did not fully complement the rpl25Δ::HIS3 disruption mutant, and the fluorescence signal of Rpl25p-GFP decreased when cells were incubated at higher temperatures (e.g., 37°C; see also reference 15), we constructed a modified Rpl25p reporter (see Materials and Methods). This new construct, Rpl25p-eGFP, complemented the rpl25Δ::HIS3 disruption at all temperatures tested, including 37°C, with no apparent growth difference compared to a wild-type strain (data not shown). Following subcellular fractionation under high salt, Rpl25p-eGFP could be detected only in the 60S ribosomal subunit fraction (data not shown). Important for the analysis of the ts mutants, the Rpl25p-eGFP fluorescence signal was readily detected at higher temperatures (e.g., 37°C), and nuclear accumulation of Rpl25p-eGFP was seen at the restrictive temperature in nucleoporin and Ran cycle mutants (data not shown).
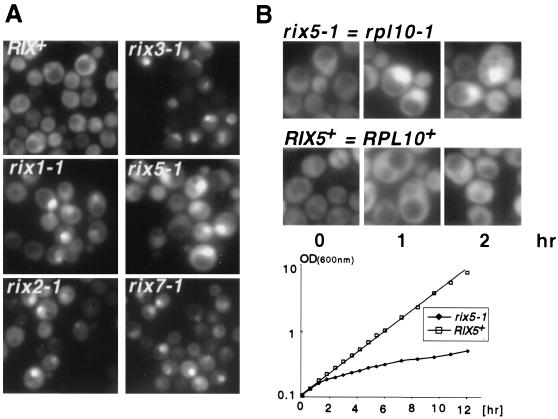
The 35 rix mutants were rescreened using the new Rpl25p-eGFP construct. Twenty strains exhibited nuclear accumulation of Rpl25p-eGFP at the restrictive temperature (Fig. 1A). The defects were manifested between 30 min to 5 h after shift to the restrictive temperature, depending on the rix mutant. Notably, the patterns of nuclear accumulation differed in the various rix mutants, independent of the Rpl25p-GFP reporter used, rix1-1 and rix5-1 mutants showed a generalized nuclear accumulation, as observed in nucleoporin mutants, whereas rix2-1, rix3-1, and rix7-1 mutants exhibited a stronger accumulation in the nucleolus than in the nucleoplasm (Fig. 1A).
FIG. 1.
A genetic screen for rix mutants identifies the ribosomal protein Rpl10p. (A) Five rix mutants (rix1-1, rix2-1, rix3-1, rix5-1, and rix7-1) and the isogenic wild-type RIX+ strain expressing Rpl25p-eGFP were grown at 23°C before a shift for 5 h to 37°C and inspection by fluorescence microscopy. (B) rix5-1 is complemented by RPL10. Top, time course of nuclear accumulation of Rpl25p-eGFP in rix5-1 (=rpl10-1) and RIX5+ (=RPL10+) cells shifted for 1 or 2 h to 37°C; bottom, growth curve of rix5-1 cells and rix-5 strain complemented by the cloned RIX5 (=RPL10) at 37°C. (C) Pre-rRNA processing in an rpl10 ts mutant shifted to 37°C for 6 h. rRNAs were pulse-labeled for 2 min with [methyl-3H] methionine followed by a 5-min chase. rRNA was analyzed by autoradiography. (D) Rpl10p contains an NLS in the N domain. Full-length Rpl10p (residues 1 to 221), Rpl10p-NLS (residues 1 to 64), and Rpl25p-NLS (residues 1 to 52), all tagged with GFP, were analyzed in KAP123+ and kap123Δ strains by fluorescence microscopy. (E) Nuclear accumulation of Rpl10p-eGFP and Rpl25p-eGFP in rrp44-1 and wild-type (wt) cells grown at 23°C before transfer to 37°C for 5 h.
The rix5-1 mutation alters the ribosomal protein Rpl10p.
In one mutant designated rix5-1, Rpl25p-eGFP was detected throughout the nucleoplasm after a 30 to 60-min shift to the restrictive temperature (Fig. 1B), indicating that the export of 60S subunit is blocked predominantly at a nuclear stage. The structure of the nucleolus was checked in a rix5-1 strain to ensure that the apparent nucleoplasmic accumulation of Rpl25p-eGFP is not due to enlargement or fragmentation of the nucleolus. At the restrictive temperature, Rpl25p-eGFP accumulated throughout the entire nucleus whereas DsRed-Nop1p (a nucleolar marker) exhibited the typical crescent staining of the nucleolus (data not shown), suggesting that the nucleolus remains intact in rix5-1 cells. To further characterize this putative ribosomal subunit export factor, the wild-type RIX5 gene was cloned by complementation of the ts phenotype of rix5-1 cells. Unexpectedly, we found that RIX5 corresponds to RPL10, a gene encoding the essential large ribosomal subunit protein Rpl10p. Accordingly, the rix5-1 mutant is also called rpl10-1 (Fig. 1B). Consistent with our findings, a number of qsr1 mutant alleles of RPL10, which have previously been identified (7), are all impaired in nuclear export of Rpl25p-eGFP (e.g., qsr1-2 [data not shown]). We also tested a ts mutation in another essential 60S subunit protein, Rpl16p (25) but could not detect nuclear accumulation of Rpl25p-eGFP at the restrictive temperature (data not shown). As anticipated, the rix5-1 mutant, when shifted to the restrictive temperature, exhibits a half-mer polysome profile like that seen in the previously characterized qsr1 mutants (data not shown).
In earlier studies, Rpl10p was reported to associate with 60S ribosomal subunits in the cytoplasm, where it functions to mediate joining of 60S and 40S subunits (7, 19). However, Rpl10p may have a nuclear function as well, as indicated by the observed defect in ribosome export in rpl10 ts mutants. As a further indication of a nuclear function for Rpl10p, we tested whether nucleolar pre-rRNA processing is defective in rpl10 ts mutants. Northern hybridization showed a rapid inhibition of pre-rRNA processing following transfer of rix5-1 cells to 37°C (data not shown). Pre-rRNA processing was also assessed by pulse-chase labeling with [methyl-3H]methionine (Fig. 1C). Consistent with the Northern analysis of rix5-1 cells, rpl10 ts cells showed processing defects of the 35S pre-rRNA, thereby retarding synthesis of the 27S pre-rRNAs. Production of the 20S pre-rRNA was also delayed and reduced, with appearance of the aberrant 23S rRNA.
Cytoplasmic Rpl10p contains an NLS and accumulates in the nucleus in an exosome mutant.
The observation that Rpl10p is required for efficient intranuclear pre-rRNA processing suggested that Rpl10p enters the nucleus to participate in preribosomal assembly. We therefore looked for an NLS in Rpl10p. At steady state, Rpl10p is located in the cytoplasm (Fig. 1D). However, we find that the first 64 amino acids of Rpl10p can target a GFP reporter to the nucleus (Fig. 1D). Kap123p is an import receptor for ribosomal proteins (29) and nuclear import of Rpl10p(1–64)-GFP, as the uptake of Rpl25p(1–52)-GFP is strongly inhibited in the kap123Δ mutant (Fig. 1D) but not in other karyopherin mutants that we have tested (data not shown). Next, we analyzed whether Rpl10p-eGFP accumulates in the nucleus in nucleocytoplasmic transport mutants such as rna1-1 or nup49-313 but found no accumulation (data not shown). Only in the exosome mutant rrp44/dis3-1 does Rpl10p-eGFP exhibit a distinct nuclear accumulation at the restrictive temperature similar to Rpl25p-eGFP (Fig. 1E). Rrp44p/Dis3p is component of the exosome, a large protein complex involved in the processing and degradation of many RNAs (24) including the 5.8S rRNA and other rRNAs (1). Thus, although Rpl25p-eGFP accumulates inside the nucleus in several classes of transport mutants, nuclear accumulation of Rpl10p was seen only in an exosome mutant (see Discussion).
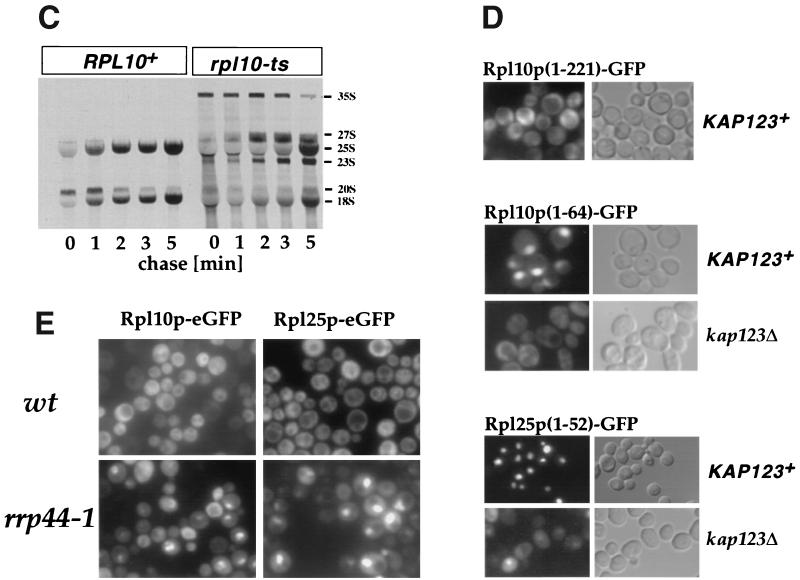
Two nonribosomal proteins, Rsa1p and Nmd3p, linked to Rpl10p are required for 60S subunit export.
Previous work showed that the two nonribosomal proteins Nmd3p and Rsa1p are genetically linked to Rpl10p (16, 17). Rsa1p is a nuclear protein implicated in facilitating the association of Rpl10p with the 60S ribosomal subunits (17). Nmd3p is an essential and conserved protein that is located in the cytoplasm and associated with 60S subunits (3, 13, 16). Rsa1p is nonessential, but rsa1 Δ strains exhibited retarded growth, particularly at 37°C (Fig. 2A; see also reference 17). Expression of Rpl25p-eGFP in the rsa1 Δ strain showed a clear nucleoplasmic accumulation at 37°C (Fig. 2A). To test whether Nmd3p functions in nuclear export of ribosomes, we generated nmd3 ts mutants (Fig. 2B). Strikingly, the large subunit reporter Rpl25p-eGFP significantly accumulates inside the nucleus in nmd3-2 ts cells after a shift to 37°C (Fig. 2C). Nmd3p and Rsa1p, as observed with Rpl10p, are required for nuclear export of 60S ribosomal subunits. This phenotype of intranuclear 60S subunit accumulation could also explain the half-mer polysome phenotype (polysomes containing a unjoined 40S subunits at the initiation site) previously observed in rsa1 (17) and nmd3 mutants (16).
FIG. 2.
The rsa1 disruption mutant and an nmd3 ts mutant accumulate Rpl25p-eGFP in the nucleus. (A) Left, growth of the rsa1Δ disruption and its isogenic wild-type strain on a YPD plate and in liquid medium. Equivalent amounts of cells (dilution in 10−1 steps) were grown at the indicated temperatures for 3 days. Right, nuclear accumulation of Rpl25p-eGFP in the rsa1Δ mutant. rsa1Δ and RSA1+ strains were transformed with RPL25-eGFP and grown at 23°C before incubation at 37°C for 8 h. Rpl25p-eGFP localization was analyzed by fluorescence microscopy. (B) Growth of the nmd3-2 ts mutant at 23 and 37°C. (C) Rpl25p-eGFP accumulates in the nucleus of the nmd3-2 ts mutant. Cells were shifted for 0, 30, 90, and 180 min to 37°C, and then the Rpl25p-eGFP signal was analyzed by fluorescence microscopy.
Recently, the nmd3-4 allele was tested for defects in 60S subunit export, but no nuclear accumulation of Rpl25p-GFP was observed in this nmd3 ts mutant at the restrictive temperature (14). However, other ts alleles of nmd3 generated in our laboratory all give rise to nuclear accumulation of Rpl25p-eGFP at the restrictive temperature (D. Strauß and O. Gadal, unpublished data). The reason for this difference is not clear, but Ho et al. used Rpl25p-GFP under the control of a GAL10 promoter (13, 14), whereas in our assay RPL125-GFP is under the control of its own promoter.
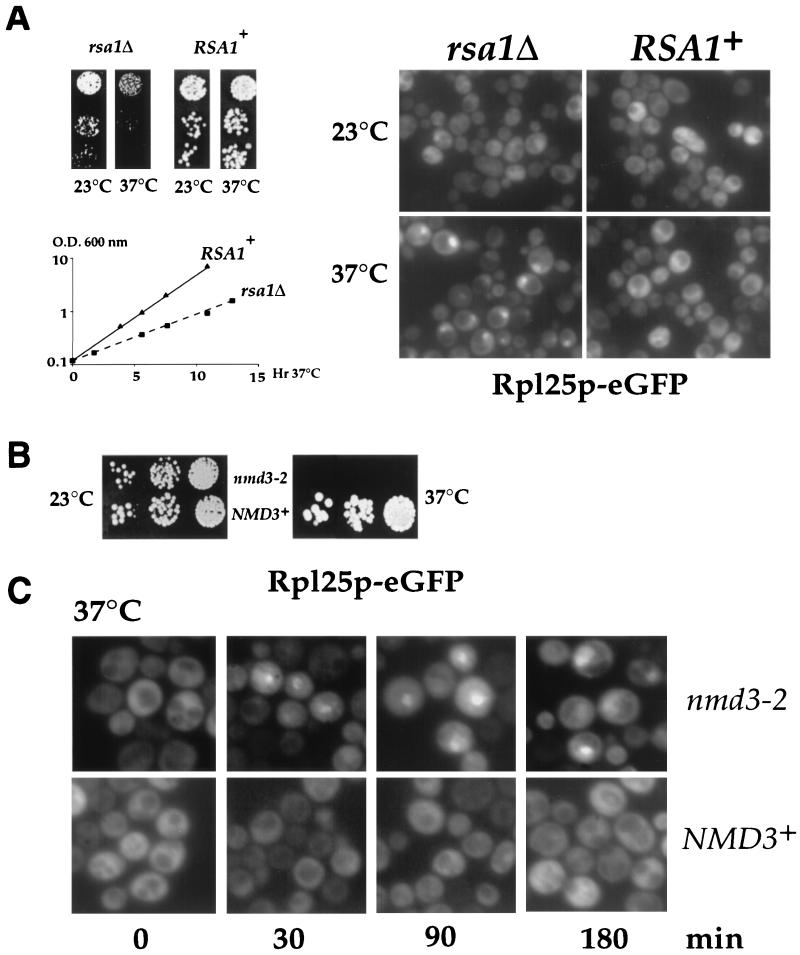
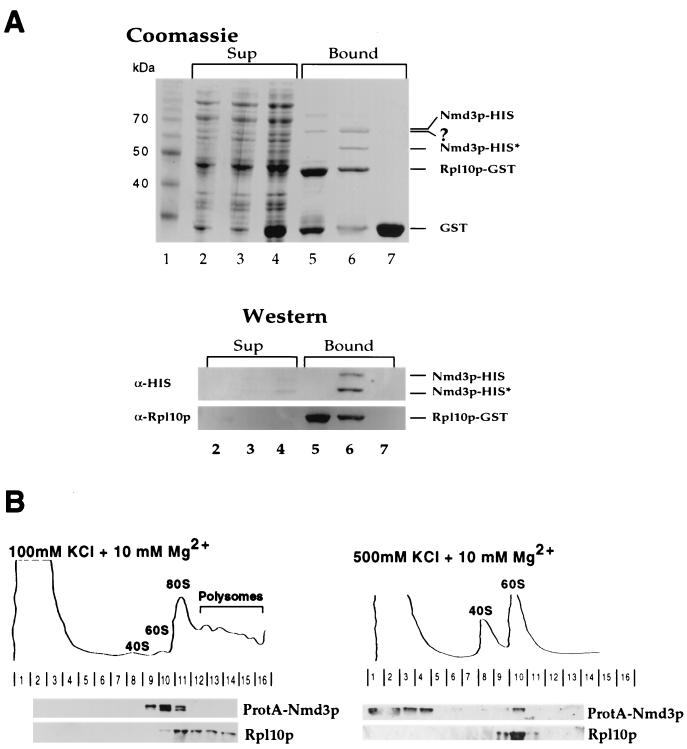
Nmd3p binds to Rpl10p and associates with 60S subunits in a salt-dependent manner.
Genetic evidence suggested that RPL10 and NMD3 are functionally linked (16). To test whether Nmd3p directly associates with Rpl10p, we coexpressed GST-tagged Rpl10p and His-tagged Nmd3p in E. coli and purified Rpl10p-GST on glutathione-Sepharose beads. Both full-length (65 kDa; Nmd3p-His6) and a prominent N-terminally cleaved (55 kDa; Nmd3p-His6*) Nmd3p copurify with GST-Rpl10p (Fig. 3A, lane 6). In contrast, GST alone when coexpressed with His-tagged Nmd3p does not reveal copurification (Fig. 3A, lane 7). Furthermore, GST-Rpl10p purification in the absence of His-tagged Nmd3p does not show Nmd3p copurification, but the E. coli contaminant of approximately 64 kDa is still evident (Fig. 3A, lane 5). Thus, Nmd3p and Rpl10p directly interact when coexpressed in E. coli (Fig. 3A). In yeast cells, Nmd3p is associated exclusively with free 60S subunits, not with 80S ribosomes or polysomes (Fig. 3B, left). In contrast, the ribosomal protein Rpl10p is in the 60S, 80S, and polysome fractions under low-salt conditions (Fig. 3B, left) and in the 60S peak under high-salt conditions (Fig. 3B, right), Nmd3p, however, is released from 60S subunits by high-salt treatment (Fig. 3B, right). We conclude that Nmd3p is a nonribosomal protein that loosely associates with 60S subunits and can directly bind to Rpl10p.
FIG. 3.
Nmd3p binds directly to Rpl10p and associates with 60S subunits in a salt-dependent manner. (A) Nmd3p binds to GST-tagged Rpl10p when expressed in E. coli. Expression and purification of recombinant Rpl10p-GST in the absence or presence of Nmd3p-His6 is described in Materials and Methods. The supernatants of E. coli cell lysates (Sup) were incubated with glutathione-Sepharose 4B beads. After washing, bound proteins were eluted with sample buffer (Bound), and the fractions were analyzed by SDS-PAGE and Coomassie blue staining (top) or Western blotting using α-Rpl10p and α-His (bottom). Lanes: 1, protein standard (10-kDa ladder); 2 to 4, supernatants; 5 to 7, bound fractions; 2 and 5, Rpl10p-GST in the absence of Nmd3p-His6; 3 and 6, Rpl10p-GST coexpressed with Nmd3p-His6; 4 and 7, GST coexpressed with Nmd3p-His6. Note that full-length Nmd3p-His6 is sensitive to proteolysis in E. coli, yielding a more stable breakdown product Nmd3p-His6*, which apparently lacks a short N-terminal part. Indicated is also an unknown E. coli contaminant (?) which coisolates with Rpl10p-GST. (B) Nmd3p, which was tagged with ProtA and is fully functional, is extracted from 60S ribosomes by high-salt (500 mM KCl) treatment. Ribosomes were analyzed by sucrose gradient centrifugation in low- and high-salt buffer, and ribosomal profiles were determined by OD260 measurement of the gradient fractions (top). Western blot analysis of these gradient fractions reveals ProtA-tagged Nmd3p and Rpl10p (bottom).
Nmd3p contains two NESs in its carboxy-terminal domain.
The results shown above suggest that Nmd3p may also enter the nucleus. To determine whether Nmd3p shuttles between the nucleus and cytoplasm, the protein was tagged with eGFP. Nmd3p-eGFP, which is fully functional (Fig. 4B), is located in the cytoplasm (Fig. 4C). Interestingly, the deletion of this C domain was previously shown to cause a dominant-negative phenotype in yeast when overproduced from the strong GAL promoter (3). This C-terminal domain of Nmd3p contains two conserved putative leucine-rich NESs (Fig. 4A). We deleted each of these NES-like sequences and expressed the truncated NMD3 construct under its authentic promoter. In this case, no dominant-lethal phenotype is observed, but a strong nuclear accumulation of these truncated Nmd3p proteins can be seen (Fig. 4C). Deleting both NESs results in no apparent growth of the nmd3-shuffle strain on 5-fluoroorotic acid-containing plates after 5 days of incubation. This shows that this region of Nmd3p is required for growth. However, the truncated form lacking only one NES (ΔNES1) is still able to complement nmd3 null strain, but cells grow very slowly at all temperatures (Fig. 4B). In addition, when the NES domain of Nmd3p is fused to the well-characterized NLS of Npl3p, the fusion protein is exported to the cytoplasm (Fig. 4C). Further deletion from the C terminus abolishes nuclear accumulation, indicating that this region contains the NLS (Fig. 4C). Together these data indicate that the C-terminal domain of Nmd3p exhibits both NES and NLS activities, which could account for shuttling of Nmd3p between the nucleus and cytoplasm.
FIG. 4.
Two NESs in the Nmd3p carboxy-terminal domain. (A) Schematic drawing of the Nmd3p domains. Sequence comparison of classical NESs (PKI, mitogen-activated protein kinase kinase [MAPKK], human immunodeficiency virus Rev) and NES-like sequences present in the Nmd3p C domains from S. cerevisiae (S.C-1 and S.C-2), humans (HUM-1, HUM-2), Schizosaccharomyces pombe (S.P-1 and S.P-2) and Drosophila (DRO-1 and DRO-2). (B) The nmd3 null strain is complemented by full-length Nmd3p-eGFP, by an Nmd3p truncation construct lacking one of the two C-terminal NESs (ΔNES1) but not by one lacking both NESs (ΔNES1/2) after 5 days on 5-fluoroorotic acid-containing plates. (C) Intracellular location of the indicated Nmd3p constructs tagged with GFP in yeast cells. The NLS of Npl3p (NLSRGG) tagged with GFP was used to test for NES activity of the Nmd3p C domain. (D) Nuclear accumulation of Rpl25p-eGFP expressed in the slow-growing nmd3ΔNES1 mutant grown at 30°C.
Previous work has shown that proteins harboring a leucine-rich NES are transported to the cytoplasm by the export receptor Xpo1p. To determine whether Xpo1p might be involved in the export of Nmd3p, we first examined whether the Nmd3p-NES could be replaced by the well-characterized Xpo1p-dependent NES from PKI. Significantly, the PKI-NES fused to the C-terminal end of Nmd3pΔNES1/2 not only restores nuclear export of Nmd3p but also complements the growth defect of the nmd3 ΔNES mutant (data not shown). Thus, the Nmd3p NES domain is important for cell growth as well as nuclear export of the protein.
Consistent with our findings, Ho et al. identified an NLS and essential NES in Nmd3p carboxy-terminal domain, and the nonfunctional nmd3Δ100 construct could be restored by attaching the heterologous PKI-NES (14). However, nmd3Δ50 described by Ho et al. is not functional (14), whereas our nmd3ΔNES construct lacking the last 44 residues of Nmd3p gives viable cells. Since Nmd3p contains two leucine-rich NESs in the C domain and deletion of one of them impairs nuclear export of Nmd3p, resulting in a slow-growth phenotype (Fig. 4B and C, Nmd3p-ΔNES1), we sought to test nuclear export of 60S subunits in this mutant. Therefore, the slow-growing Nmd3p-ΔNES1 strain was transformed with Rpl25p-eGFP. A striking nuclear accumulation of the large subunit reporter is seen in this mutant when grown at 30°C (Fig. 4D). Thus, mutations in the NES domain of Nmd3p and in the export receptor Xpo1p cause inhibition of 60S subunit export.
Both Nmd3p and 60S subunit export is inhibited when the Xpo1p function is blocked.
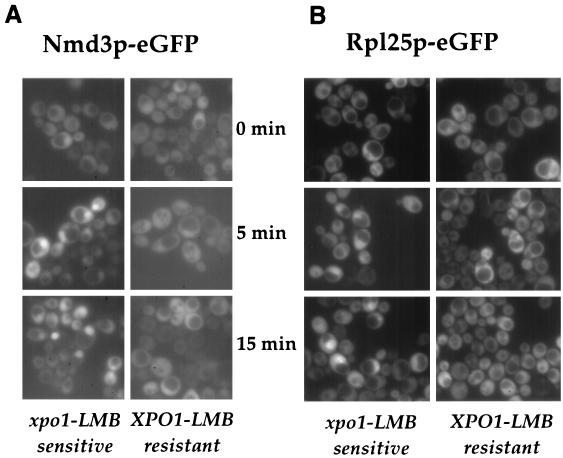
Next we sought to test directly whether Xpo1 is the export receptor for Nmd3p. Recently, an LMB-sensitive xpo1 mutant of S. cerevisiae was engineered by site-specific mutagenesis of a critical residue within Xpo1p (27). This mutant exhibits a strong and rapid inhibition of nuclear export of NES-containing cargoes upon addition of LMB. As shown in Fig. 5A, nuclear export of Nmd3p-eGFP is strongly inhibited in the LMB-sensitive xpo1 strain 5 min after addition of LMB. Strikingly, the large subunit reporter Rpl25p-eGFP also accumulates inside the nucleus within the same time range (Fig. 5B). These data show that a rapid and strong export defect of both Nmd3p-eGFP and Rpl25p-eGFP occurs in the LMB-sensitive xpo1 mutant, suggesting that Nmd3p and 60S ribosomal subunit export are coupled. We conclude that 60S ribosomal export requires Xpo1p. Our findings are different from those of previous studies which had shown that Rpl25p-GFP accumulates inside the nucleus in the LMB-sensitive xpo1 mutant only at later time points (i.e., 2 h) (14). Notably, Ho et al. used RPL25-GFP under the control of the inducible GAL10 promoter (14). Accordingly, the slow onset of Rpl25p-GFP nuclear accumulation in the xpo1 mutant may be due to the fact the GAL10 promoter must be switched on to allow for Rpl25p-eGFP expression. In our study the expression of RPL25-GFP is under its native promoter, leading to a nuclear accumulation 5 to 15 min after blocking export by LMB treatment.
FIG. 5.
Nmd3p and 60S large subunits are exported through the Xpo1p-dependent transport pathway. (A) Nmd3p-eGFP expressed in the LMB-sensitive xpo1 mutant and isogenic wild-type strain. (B) Rpl25p-eGFP expressed in the LMB-sensitive xpo1 mutant and isogenic wild-type strain. At time point 0 min, LMB (100 ng/ml) was added to the culture medium, and the fluorescence signals from GFP fusion proteins were observed by fluorescence microscopy.
In contrast, nuclear accumulation of Nmd3p-eGFP is less evident in the xpo1-1 ts mutant when shifted to the restrictive temperature and requires longer incubation at the restrictive temperature (data not shown). No nuclear accumulation of Nmd3p-eGFP is seen in the cse1-2 ts mutant (31), which is defective in nuclear export of Srp1p/importin α (data not shown). As reported earlier for Rpl25p-GFP (15) and also found here for Rpl25p-eGFP, both reporters do not significantly accumulate in the xpo1-1 mutant when shifted for 1 h to the restrictive temperature; nuclear accumulation of Rpl25p-eGFP is seen in some of the xpo1-1 cells only after prolonged incubation at 37°C (data not shown). It is possible that the observed inhibition of nuclear poly(A)+ RNA export in xpo1-1 cells when shifted to 37°C (33) impairs synthesis of ribosomal proteins, because mRNAs encoding ribosomal proteins are no longer exported to the cytoplasm.
DISCUSSION
The mechanism of ribosomal export from the nucleus to the cytoplasm is not understood. Compared to the nuclear export of other cargoes, e.g., small RNAs such as tRNA, nuclear export of ribosomes is likely to be more complicated, in view of an extremely complex biogenesis pathway of ribosomes. In particular, modification, processing, and conformational rearrangement steps, which take place during ribosomal assembly, could be a trigger for transport from the nucleolus to the nuclear pores and further to the cytoplasm. Here we report the identification of a novel class of mutants, defined as rix mutants, using a visual assay for the nuclear retention of a Rpl25p-eGFP reporter. Around 2% (20 of 900) of the ts mutant strains screened gave rise to a rix phenotype, comparable to levels of mutations found for other major pathways, e.g., cell cycle progression or pre-mRNA splicing. While we do not yet know how many complementation groups these mutants represent, the first five genes tested fell into different complementation groups (data not shown). We conclude that a substantial number of genes (probably at least 50) can give rise to a rix phenotype. Analysis of these should allow the genetic dissection of ribosomal assembly and transport and possibly their coupling.
The first mutant to be analyzed, rix5-1, was found to be due to an A68V mutation in the ribosomal protein Rpl10p. This suggests that late assembly steps during ribosome biogenesis, which occur inside the nucleus, are crucial for subsequent export to the cytoplasm. Rpl10p is conserved, having homologues in humans (28), archea, and E. coli (9). Since Rpl10p was absent from the preparation of the 66S nuclear preribosomal particle, it was proposed that Rpl10p assembles into 60S subunits in the cytoplasm (19). However, Rpl10p is relatively weakly associated with the 60S subunit (5) and thus may be lost from the preribosomal particles during purification.
Our findings that Rpl10p is required for nuclear pre-rRNA processing is another hint that it associates with preribosomes in the nucleus. Consistent with this interpretation, we find an NLS in Rpl10p that uses the import receptor Kap123p, which is one of two importins (Kap121p and Kap123p) involved in import of ribosomal proteins (29). On the other hand, we do not see nuclear accumulation of Rpl10p-eGFP in nucleoporin mutants. This is not understood, but Rpl10p-eGFP, in contrast to Rpl25p-eGFP, may be more sensitive to proteolysis inside the nucleus when attached to preribosomal particles that are either not fully assembled or blocked in export. It is well known that both rRNA processing intermediates and ribosomal proteins are quickly degraded when not assembled or assembled incorrectly into preribosomes. Interesting in this context is that newly synthesized 25S rRNA is unstable (half-life of 4 min) in nmd3 ts mutants at the restrictive temperature (13). Apparently, 25S rRNA-containing ribosomal particles that accumulate inside the nuclei of nmd3 mutants have a short lifetime. Thus, our observation that Rpl10p-eGFP does not accumulate in the nuclei of transport mutants could reflect a high turnover rate within transport-arrested precursor particles. On the other hand, the Rpl25p-eGFP reporter appears to have a longer half-life in nuclear ribosomal precursor particles and therefore may be observed by fluorescence microscopy. Thus, it remains to be shown why a nuclear form of Rpl10p-eGFP can be detected in the rrp44/dis3-1 strain. One possibility is that inactivation of the exosome system, which functions in degradation of (aberrant) pre-rRNAs (1), stabilizes pre-rRNA and as well ribosomal proteins bound to it.
The finding that Rpl10p is involved in a late ribosomal assembly step, which is coupled to nuclear export, together with the well-described genetic link between RPL10 and NMD3 (16) suggested that Nmd3p could play a role in large subunit export. Nmd3p has been implicated in translational control of gene expression (16) and in formation, function, or maintenance of stable 60S subunits (3, 13). Strikingly, we find nuclear accumulation of the large subunit reporter Rpl25p-eGFP in several nmd3 mutants, including a slow-growing mutant lacking one of the two NESs from the C domain. When both leucine-rich NESs are deleted, the corresponding Nmd3p construct is no longer functional. Furthermore, the NES domain of Nmd3p, when attached to another NLS, exhibits a distinct NES activity. All of these findings suggest that the essential role of the Nmd3p C domain is to function in nuclear export. This is consistent with the observation that archaebacterial homologues of Nmd3p lack the NES-containing C domain characteristic of eukaryotes (3).
Our data also strongly suggest that Xpo1p is the NES receptor for Nmd3p. Interestingly, an LMB-sensitive xpo1 mutant, which exhibits inhibition of nuclear export of NES-containing cargoes upon addition of LMB (27), allows us to observe a rapid nuclear accumulation of both Nmd3p-eGFP and Rpl25p-eGFP upon inhibition of Xpo1p. In contrast, the classical xpo1-1 ts mutant does not reveal this strong nuclear export defect of both Nmd3p and Rpl25p-eGFP. It is conceivable that the fast onset and strong inhibition of nuclear poly(A)+ RNA export in xpo1-1 cells (33) impairs ribosomal biogenesis at an early stage, because mRNAs encoding ribosomal proteins are not exported to the cytoplasm. In such a scenario, ribosomal proteins including the Rpl25p-eGFP reporter would no longer be synthesized at the restrictive temperature. What could be the reason why Nmd3p enters the nucleus and is subsequently exported to the cytoplasm by Xpo1p? A likely possibility is that Nmd3p participates in ribosomal subunit export, which correlates well with the observed concomitant nuclear accumulation of Rpl25p-eGFP and Nmd3p-eGFP in the LMB-sensitive XPO1 strain. Biochemical data further support a model in which Nmd3p binds to Rpl10p, thereby targeting Nmd3p to the 60S subunits. In the course of this work, Ho et al. characterized Nmd3p as a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit (14), which is in full agreement with the data presented here.
Remarkably, the nuclear protein Rsa1p was implicated in facilitating the association of Rpl10p with 60S ribosomal subunits (17). We found that like Rpl10p, Rsa1p is required for efficient nuclear export of 60S ribosomal subunits at 37°C. Interestingly, at the restrictive temperature free 60S ribosomal subunits of an rsa1 null mutant are depleted of Rpl10p (17). The accumulation of half-mer polysomes has been reported for many mutants that underaccumulate 60S subunits, due to the presence of polysomes carrying an initiation complex that awaits a 60S subunit. This phenotype is also seen in Rpl10p mutants and strains lacking Rsa1p, but these strains show no clear change in overall 40S/60S ratios. It appears possible that this phenotype is a consequence of the retention of 60S subunits within the nucleus. Thus, Rsa1p could be a factor that mediates assembly of Rpl10p onto 60S ribosomal subunits inside the nucleus (17).
In summary, we have identified several factors that function in nuclear export of 60S ribosomal subunits. We found that a protein of the 60S ribosomal subunit, Rpl10p, is required for large subunit export. This protein most likely participates in late intranuclear maturation steps leading to export-competent 60S subunits. Rpl10p interacts directly with Nmd3p, a NES-containing protein that is specifically associated with 60S subunits. Strikingly, Nmd3p shuttles between the nucleus and cytoplasm and uses the Xpo1p receptor for its export. Accordingly, the highly conserved Nmd3p could act as an adapter which requires the general NES receptor Xpo1p for export. As a consequence, 60S subunits could be exported by Xpo1p. Thus, an essential role of Xpo1p in yeast appears to be its involvement in ribosomal export. A similar conclusion that Nmd3p is an Xpo1p-dependent adapter protein for nuclear export of the large subunit has been recently published by another group (14). Finally, it is possible that other ribosomal proteins may bind to Nmd3p or that other adapter proteins similar to Nmd3p exist. By exploiting the powerful yeast genetics, it should be possible to find additional components of the ribosomal export machinery.
ACKNOWLEDGMENTS
We are grateful to C. Cole and A. Mutvei for the ts bank, J. Woolford for rpl16b-2, K. Weis for xpo1-1, and M. Rosbash for the LMB-sensitive xpo1 strain. We especially thank Robin Reed for critical reading of the manuscript.
E.H. was recipient of grants from the Deutsche Forschungsgemeinschaft (Schwerpunktprogramm “Funktionelle Architektur des Zellkerns”), and O.G. holds an HFSP fellowship. D.T. was funded by the Wellcome Trust, and research from the Trumpower laboratory was supported by The Hitchcock Foundation and by NIH grant GM 20379.
REFERENCES
- 1.Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28:1648–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amberg D C, Goldstein A L, Cole C N. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- 3.Belk J P, Jacobson A. Overexpression of truncated Nmd3p inhibits protein synthesis in yeast. RNA. 1999;5:1055–1070. doi: 10.1017/s1355838299990027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergès T, Petfalski E, Tollervey D, Hurt E C. Synthetic lethality with fibrillarin identifies NOP77p, a nucleolar protein required for pre-rRNA processing and modification. EMBO J. 1994;13:3136–3148. doi: 10.1002/j.1460-2075.1994.tb06612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dick F A, Karamanou S, Trumpower B L. QSR1, an essential yeast gene with a genetic relationship to a subunit of the mitochondrial cytochrome bc1 complex, codes for a 60 S ribosomal subunit protein. J Biol Chem. 1997;272:13372–13379. doi: 10.1074/jbc.272.20.13372. [DOI] [PubMed] [Google Scholar]
- 6.Doye V, Hurt E C. From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- 7.Eisinger D P, Dick F A, Trumpower B L. Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40S and 60S subunits. Mol Cell Biol. 1997;17:5136–5145. doi: 10.1128/mcb.17.9.5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 9.Franceschi F J, Nierhaus K H. Ribosomal proteins L15 and L16 are mere late assembly proteins of the large ribosomal subunit. Analysis of an Escherichia coli mutant lacking L15. J Biol Chem. 1990;265:16676–16682. [PubMed] [Google Scholar]
- 10.Görlich D. Transport into and out of the cell nucleus. EMBO J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorsch L C, Dockendorff T C, Cole C N. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J Cell Biol. 1995;129:939–955. doi: 10.1083/jcb.129.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- 13.Ho J H, Johnson A W. NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2389–2399. doi: 10.1128/mcb.19.3.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho J H, Kallstrom G, Johnson A W. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol. 2000;151:1057–1066. doi: 10.1083/jcb.151.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurt E, Hannus S, Schmelzl B, Lau D, Tollervey D, Simos G. A novel in vivo assay reveals inhibition of ribosomal nuclear export in Ran-cycle and nucleoporin mutants. J Cell Biol. 1999;144:389–401. doi: 10.1083/jcb.144.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karl T, Onder K, Kodzius R, Pichova A, Wimmer H, Hundsberger H, Loffle M, Klade T, Beyer A, Breitenbach M, Kolle L. GRC5 and NMD3 function in translational control of gene expression and interact genetically. Curr Genet. 1999;34:419–429. doi: 10.1007/s002940050416. [DOI] [PubMed] [Google Scholar]
- 17.Kressler D, Doere M, Rojo M, Linder P. Synthetic lethality with conditional dbp6 alleles identifies Rsa1p, a nucleoplasmic protein involved in the assembly of 60S ribosomal subunits. Mol Cell Biol. 1999;19:8633–8645. doi: 10.1128/mcb.19.12.8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kressler D, Linder P, De La Cruz J. Protein trans-acting factors involved in ribsome biogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7897–7912. doi: 10.1128/mcb.19.12.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruiswijk T, Planta R J, Knop J M. The course of the assembly of ribosomal subunits. Biochim Biophys Acta. 1978;517:378–389. doi: 10.1016/0005-2787(78)90204-6. [DOI] [PubMed] [Google Scholar]
- 20.Künzler M, Hurt E C. Cse1p functions as the nuclear export receptor for importin α in yeast. FEBS Lett. 1998;433:185–190. doi: 10.1016/s0014-5793(98)00892-8. [DOI] [PubMed] [Google Scholar]
- 21.Mager W H, Planta R J, Ballesta J P G, Lee J C, Mizuta K, Suzuki K, Warner J R, Woolford J. A new nomenclature for the cytoplasmic ribosomal proteins Saccharomyces cerevisiae. Nucleic Acids Res. 1997;25:4872–4875. doi: 10.1093/nar/25.24.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maniatis T, Fritsch E T, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 23.Mattaj I W, Englmeir L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 25.Moritz M, Pulaski B A, Woolford J L., Jr Assembly of 60S ribosomal subunits is perturbed in temperature-sensitive yeast mutants defective in ribosomal protein L16. Mol Cell Biol. 1991;11:5681–5692. doi: 10.1128/mcb.11.11.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moy T I, Silver P A. Nuclear export of the small ribosomal subunit requires the Ran-GTPase cycle and certain nucleoporins. Genes Dev. 1999;13:2118–2133. doi: 10.1101/gad.13.16.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neville M, Rosbash M. The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae. EMBO J. 1999;18:3746–3756. doi: 10.1093/emboj/18.13.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen Y H, Mills A A, Stanbridge E J. Assembly of the QM protein onto the 60S ribosomal subunit occurs in the cytoplasm. J Cell Biochem. 1998;68:281–285. [PubMed] [Google Scholar]
- 29.Rout M P, Blobel G, Aitchison J D. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- 30.Santos-Rosa H, Moreno H, Simos G, Segref A, Fahrenkrog B, Panté N, Hurt E. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol Cell Biol. 1998;18:6826–6838. doi: 10.1128/mcb.18.11.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schroeder A J, Chen X-H, Xiao Z, Fitzgerald-Hayes M. Genetic evidence for interactions between yeast importin a (Srp1p) and its nuclear export receptor, Cse1p. Mol Gen Genet. 1999;261:788–795. doi: 10.1007/s004380050022. [DOI] [PubMed] [Google Scholar]
- 32.Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt E C. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- 33.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 34.Sträßer K, Baßler J, Hurt E C. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J Cell Biol. 2000;150:695–706. doi: 10.1083/jcb.150.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sträßer K, Hurt E C. Yraip, a conserved nuclear RNA binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987;6:4169–4175. doi: 10.1002/j.1460-2075.1987.tb02763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tollervey D, Lehtonen H, Jansen R P, Kern H, Hurt E C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- 38.Tron T, Yang M, Dick F A, Schmitt M E, Trumpower B L. QSR1, an essential yeast gene with a genetic relationship to a subunit of the mitochondrial cytochrome bc1 complex, is homologous to a gene implicated in eukaryotic cell differentiation. J Biol Chem. 1995;270:9961–9970. doi: 10.1074/jbc.270.17.9961. [DOI] [PubMed] [Google Scholar]
- 39.Van Beekvelt C A, Kooi E A, de Graaff-Vincent M, Riet J, Venema J, Raue H A. Domain III of Saccharomyces cerevisiae 25 S ribosomal RNA: its role in binding of ribosomal protein L25 and 60 S subunit formation. J Mol Biol. 2000;296:7–17. doi: 10.1006/jmbi.1999.3432. [DOI] [PubMed] [Google Scholar]
- 40.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 41.Woolford J L., Jr The structure and biogenesis of yeast ribosomes. Adv Genet. 1991;29:63–118. doi: 10.1016/s0065-2660(08)60107-8. [DOI] [PubMed] [Google Scholar]