Abstract
Purpose:
Transcriptomic-based approaches are being developed to meet the needs for large-scale radiation dose and injury assessment and provide population triage following a radiological or nuclear event. This review provides background and definition of the need for new biodosimetry approaches, and summarizes the major advances in this field. It discusses some of the major model systems used in gene signature development, and highlights some of the remaining challenges, including individual variation in gene expression, potential confounding factors, and accounting for the complexity of realistic exposure scenarios.
Conclusions:
Transcriptomic approaches show great promise for both dose reconstruction and for prediction of individual radiological injury. However, further work will be needed to ensure that gene expression signatures will be robust and appropriate for their intended use in radiological or nuclear emergencies.
Keywords: ionizing radiation, gene expression, biodosimetry, radiological triage, dose reconstruction
Introduction – the need for biodosimetry
We live in an era of heightened concern over the possibility of large-scale radiological or nuclear events that may result from accidents, ecological disasters, or malicious intent. Damage to a power plant or detonation of a dirty bomb, in which conventional explosives are used to blow up and disperse a radioactive source, could result in potential widespread exposure to radionuclides in fallout. Detonation of an improvised nuclear device (IND) could result in radiological injuries from prompt radiation, contamination and ingestion of radionuclides from fallout, as well as burn and crush injuries and extensive damage to infrastructure. In preparing for the response to such large-scale events, it is clear there will be a critical need to rapidly assess the level of radiological injury to individuals in order to appropriately ration medical countermeasures from limited stockpiles. As the general populace will not have physical radiation dosimeters, we must rely on measurements of biological responses to radiation to provide markers of exposure, dose, or injury.
In a mass casualty event, it will be necessary to screen hundreds of thousands of people, possibly as many as a million, within a relatively short period of time (Waselenko et al., 2004). Thus, a stepwise approach to radiological triage is commonly envisioned, in which an initial rapid point-of-care screening will be used to separate the “worried well”, who are either unexposed or have only sub-clinical exposure, from those who may benefit from rapid treatment. The target for this initial determination is usually above or below 2 Gy, with false positive calls preferred over false negatives. Further testing, perhaps using a high-throughput platform, can then be performed on a greatly reduced number of individuals to provide a more accurate assessment of doses above 2 Gy (Sullivan et al., 2013; Wathen et al., 2020) in order to support the management of treatment. In most anticipated scenarios, the majority of those presenting for testing are expected to be unexposed, but rapid individualized testing can help to stem panic and prevent medical resources from being overwhelmed. As rapid field testing approaches are not yet available, a geo-statistical sampling approach has been proposed as a means to reduce the number of screenings required. In this scenario, dosimetry maps of likely exposure areas would first be modeled, and then sentinel individuals representing different projected exposure levels would be subjected to biodosimetry, as a means of reducing the numbers needing to be screened (Rogan et al., 2020).
In addition to high-throughput capacity and the speed of time to answer, a number of other factors are important to consider in the development of biodosimetry approaches for mass casualty events. Besides classifying exposures as above or below 2 Gy, a biodosimetry assay should ideally be able to accurately estimate dose up to 8 or 10 Gy, the range where bone marrow transplant may be considered, and above which casualties would currently be considered expectant (Wathen et al., 2020). The majority of radiation biodosimetry studies discussed here have thus focused on this dose range of interest, between 0.5 or 1 Gy on the low end and 8 or 10 Gy at the high end. A biodosimetry assay should also function during the window of opportunity for treatment, approximately 1-7 days after an event.
The inter-individual variability of both baseline and response is also important to understand, as well as the potential for confounding by sex, age, pre-existing conditions, medications, or non-radiation stresses. Furthermore, assays that can be run by minimally trained personnel and give a clear result as output are also preferred, to help avoid bottlenecks in sample acquisition, testing, and interpretation.
The gold standard of radiation biodosimetry has long been the dicentric chromosome assay (DCA). It remains the approach recommended for emergency use by the International Standards Organization (ISO) and the International Atomic Energy Agency (IAEA), as well as the United States Food and Drug Administration (FDA) reference method for development of new radiation biodosimetry approaches. It also appears to be specific for ionizing radiation exposure (Hoffmann and Schmitz-Feuerhake, 1999; Rozgaj et al., 2002). Despite the organization and testing of national and regional cytogenetic emergency networks (Wilkins et al., 2011; Maznyk et al., 2012) and steady advances in automation (Ramakumar et al., 2015; Royba et al., 2019; Shirley et al., 2020), the DCA cannot meet all potential biodosimetry needs. In particular, it still requires samples be cultured for several days and does not have good potential as a rapid point-of-care assay, as would be needed in a mass-casualty event. Therefore, a variety of alternative approaches to radiation biodosimetry are being developed, based on biomarkers of cytogenetic, protein, transcript, and metabolic origin. This review will focus on the use of radiation responsive gene transcripts for biodosimetry.
Transcriptomics for biodosimetry
The potential for gene expression signatures to detect and monitor exposure to radiation was recognized early in the days of transcriptomic microarrays (Amundson et al., 2000; Kang et al., 2003; Amundson et al., 2004). Focusing on measurements made in blood, these and subsequent studies have identified thousands of genes with transcript levels altered after radiation exposure. Sets of genes have been selected from among those responding to radiation to develop signatures with high accuracy of sample classification by dose level in the 0-10 Gy range (Dressman et al., 2007; Paul and Amundson, 2008; Filiano et al., 2011; Paul et al., 2011). Other studies have focused on dose reconstruction of unknown samples, with some reporting good accuracy within 1 Gy at higher doses (up to 10 Gy), or within 0.5 Gy at lower doses (up to 4.5 Gy) (Tucker et al., 2014; Park et al., 2017; Ghandhi et al., 2019a; Jacobs et al., 2020). Inter-laboratory comparisons have shown similar dose reconstruction accuracy with a shorter time to result of some gene expression based methods compared to DCA (Badie et al., 2013), although variations in performance between laboratories using different signatures and measurement methodologies have also been noted (Badie et al., 2013; Rothkamm et al., 2013; Abend et al., 2016; Ainsbury et al., 2017).
Model systems
The ultimate goal of radiation biodosimetry is the detection and characterization of radiation exposures in humans. Because the necessary experiments cannot be done directly in healthy people, surrogate model systems are needed for the development of biodosimetric approaches. Some studies have made use of blood samples from cancer patients undergoing radiotherapy (Amundson et al., 2004; Meadows et al., 2008; Paul et al., 2011; Lucas et al., 2014). Such work is limited by treatment protocols, however, with clinical exposures generally involving either a very limited area of the body, dose fractionation, or both. It is also unknown if the presence (and direct irradiation) of a tumor may alter the gene expression response measured in the peripheral blood. Interpretation of responses also may be complicated by other cancer treatments, although several studies have suggested that gene choice can circumvent this concern and allow discrimination between radiation and chemotherapy treatments (Meadows et al., 2008; Lucas et al., 2014; Zhao et al., 2018). More flexible model systems are generally required to establish the dose-response relationship of potential signature genes, or to enable the study of relevant factors such as dose rate or the effect of time since a radiological event. The two most commonly utilized models are in vivo irradiation of animals, or ex vivo irradiation of cultured blood obtained from healthy donors. While neither approach is perfect, the hope is that by understanding the similarities and differences between in vitro and in vivo responses, and between animal and human responses, appropriate translations may be made, enabling development of gene expression biodosimetry for practical human use (Figure 1).
Figure 1.
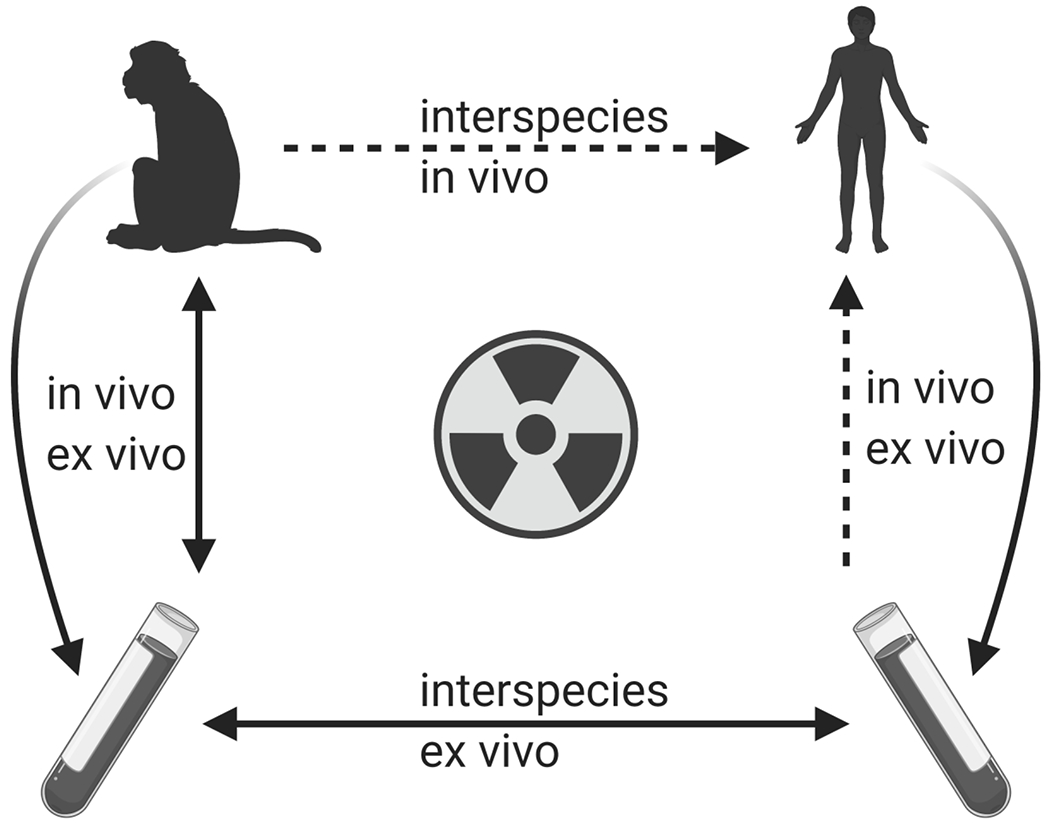
The use and intercomparison of multiple models is needed for the development and validation of gene expression signatures for radiation biodosimetry. As experiments cannot be performed directly in humans, much development must rely on the comparison of in vivo versus ex vivo models, and of human versus animal models. (Figure created with BioRender.com.)
Early ex vivo studies used isolated lymphocytes for biomarker discovery, and to investigate the potential effects of mitogenic stimulation or use of isolated CD3+, CD4+, CD8+, CD19+, or CD56+ cell subsets (Amundson et al., 2000; Mori et al., 2005; Mori et al., 2004; Pogosova-Agadjanyan et al., 2011; Kabacik et al., 2011; Riecke et al., 2012). Although some genes appeared to respond significantly to radiation only in a particular cellular subtype (Pogosova-Agadjanyan et al., 2011), the isolation of cellular subtypes would add to the cost and time requirements of a screening assay. Similarly, a large number of down-regulated genes were identified in response to radiation when blood cultures were stimulated to divide (Kabacik et al., 2011), but the majority of up-regulated genes were found to respond to radiation in both stimulated and quiescent blood cultures, and thus could be measured directly in the blood of potentially exposed individuals. More recent studies have used either whole blood or total peripheral blood mononuclear cells (PBMCs), and have identified large numbers of radiation responsive genes with potential for radiation biodosimetry, without the need for mitogenic stimulation or isolation of cell subsets (Paul and Amundson, 2008; Knops et al., 2012; Lucas et al., 2014; Cruz-Garcia et al., 2020a).
The gene expression response to radiation measured in blood cells irradiated ex vivo appears to represent a subset of the response that is seen in patients exposed in vivo, with informed selection of genes allowing signatures based on ex vivo exposures to reconstruct exposure levels in vivo (Amundson et al., 2004; Paul et al., 2011; Lucas et al., 2014; Port et al., 2018). In addition, many genes with potential for biodosimetry also respond to the stress of blood cells being cultured ex vivo, potentially complicating translation to in vivo exposures (Paul et al., 2011; Paul et al., 2013). One study has reported use of an ex vivo to in vivo translation factor for a single gene to allow prediction of exposure status in blood samples from prostate cancer patients (Abend et al., 2016). Another study reported variation in the response to different doses in the 0.1 Gy range given in vivo and ex vivo to blood from the same individual (O’Brien et al., 2018). Further work in both human and animal models is needed to establish the differences between ex vivo and in vivo gene expression responses in the dose range above 1 Gy, and to develop appropriate ex vivo to in vivo conversion approaches.
The mouse has been widely used as a model for in vivo radiation biodosimetry studies (Dressman et al., 2007; Filiano et al., 2011; Tucker et al., 2012; Lucas et al., 2014; Paul et al., 2019; Yamaguchi et al., 2020). Mice have also been used to test for possible confounding of the radiation response signature by administration of granulocyte colony-stimulating factor (G-CSF), the first FDA approved treatment for acute radiation syndrome, or by lipopolysaccharide treatment, a model for infection and inflammation (Meadows et al., 2008; Tucker et al., 2012). Importantly, the in vivo model has allowed the study of patterns of gene expression change over time since exposure (Meadows et al., 2008; Filiano et al., 2011; Paul et al., 2014; Ghandhi et al., 2015b; Paul et al., 2019) without confounding by the ex vivo effect. Although genes with invariant expression between one and seven days after irradiation would be ideal for use in biodosimetry, the extremely dynamic nature of gene expression responses as a function of time since exposure presents a challenge in this regard (Amundson et al., 2000; Macaeva et al., 2019; Paul et al., 2019; Ostheim et al., 2021). In situations where the time of an exposure event is known, inclusion of a time variable in the biodosimetry algorithm could provide more robust dose reconstructions. Gene expression dynamics can also complicate prediction of radiological injuries when exposures are protracted, or ongoing, such as from exposure to fallout (Ghandhi et al., 2020). Perhaps the greatest time-related challenge for gene expression would be an exposed source, where both time and duration of individual exposures may not be known.
Despite the usefulness of the mouse model for proof of concept studies, it is known that there are major differences between the radiation responses of mice and humans. Biodosimetric signatures developed in mice have not performed well for classifying levels of radiation exposure in human samples (Lucas et al., 2014). One factor contributing to this observation may be the central role of p53 as a prominent regulator of the gene expression response to radiation. Despite many conserved responses, a recent study has identified over 1000 genes with different p53-dependent expression in mice and humans (Fischer, 2019). Radiation experiments have also shown that not only are some genes significantly radiation responsive only in mouse or human, but some genes actually respond to radiation with significant up regulation in humans and significant down regulation in mice (Ghandhi et al., 2019b).
Non-human primates (NHP) are considered to be the best model for human exposures, as they are the most closely related to us. NHP studies, primarily using rhesus macaques or baboons, have been used to develop transcript signatures to detect radiation exposure, reconstruct dose, and predict the severity and outcome (survival or death) of hematopoietic syndrome (Menon et al., 2016; Port et al., 2016b; Fendler et al., 2017; Park et al., 2017; Port et al., 2017; Port et al., 2018; Port et al., 2018). A recent study comparing in vivo exposure of NHPs with patients undergoing total body irradiation found that gene sets selected based on the NHP radiation response could be used directly to reconstruct radiation dose to the human samples (Iversen et al., 2018). This conclusion is in contrast with an earlier ex vivo study that found it was necessary to select genes with correlated responses in NHP and humans, and then to apply a translation algorithm to NHP signatures in order to obtain accurate dose reconstruction in human samples (Park et al., 2017). Perhaps the starkest example of discordance between human and NHP radiation responses is seen in the widely used biodosimetry gene FDXR. This gene is among the most robust up regulated genes following irradiation of human samples with doses of 0.5 to 8 Gy (Paul and Amundson, 2008; O’Brien et al., 2018; Cruz-Garcia et al., 2020a), and shows no sign of a threshold for response down to 50 mGy or less (Manning et al., 2013; O’Brien et al., 2018). However, it shows no significant response to radiation in the range of 2-10 Gy in macaques (Park et al., 2017; Ghandhi et al., 2018), and is significantly down regulated in response to 2.5 or 5 Sv irradiation in baboons (Port et al., 2018). This suggests that some caution is still warranted when developing or validating radiation biodosimetry signatures in NHP for use in humans.
Signatures and consensus gene sets
With the increasing number of published radiation biodosimetry studies and the public availability of primary transcriptomic data in repositories such as the Gene Expression Omnibus (Barrett et al., 2009) and ArrayExpress (Brazma et al., 2003), several systematic literature reviews and meta-analyses have derived consensus human signatures for radiation biodosimetry. Despite using some of the same data sets for gene selection, their different approaches resulted in signatures with only moderate overlap (Figure 2). The three studies that used correlation with dose as one of their gene selection criteria (Li et al., 2017; Lacombe et al., 2018; Ghandhi et al., 2019a) had the most genes in common with each other, while the two meta-analyses (Zhao et al., 2018; Ghandhi et al., 2019a) that built classification or dose reconstruction models and tested them against independent data sets as part of the gene selection process included the most genes not selected by other approaches. Differences in the specific genes and models used by different groups are not unexpected in the field of gene signature selection, and should not undermine confidence in the robustness of individual approaches (Ein-Dor et al., 2006). Other factors besides those currently being used in gene selection may need to be considered, however. For instance, comparison of the genes from these four studies with the analysis by Park et al. (Park et al., 2017) shows that between 42 and 77% of the consensus signature genes did not show cross-species correlation between humans and NHPs. This may pose a potential obstacle for in vivo validation studies using a NHP model.
Figure 2.
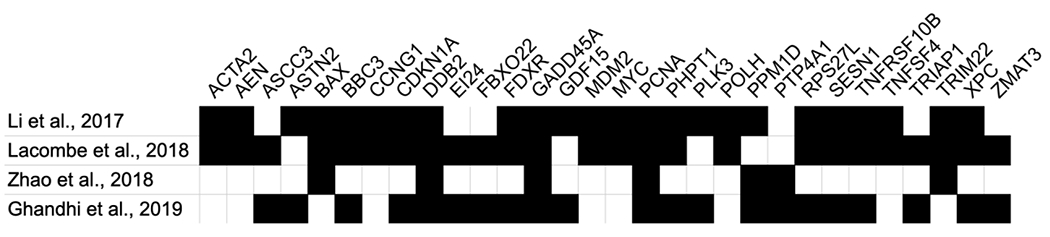
Genes appearing in two or more consensus signature studies. A black box indicates a gene selected by the corresponding study. An additional 11 genes were included only by Li et al. (2017), 6 by Lacombe et al. (2018), 19 by Zhao et al. (2018), and 16 by Ghandhi et al. (2019). The complete listing of all these genes is available in Supplementary File 1.
Individual variation and confounding factors
Gene expression is known to vary among individuals, both at baseline levels and in response to radiation (Cheung et al., 2003; Smirnov et al., 2009). Some of this variation may correlate with differences in individual radiosensitivity, with different gene expression shown to correlate with B-cell sensitivity to radiation killing in one study (Smirnov et al., 2012). In general, females tend to show heightened radiosensitivity compared with males, leading to concerns that sex may affect the performance of gene expression biodosimetry. Most studies have used both males and females in the development of signatures, and have not found significantly different performance in the two sexes (Meadows et al., 2008; Paul and Amundson, 2014; O’Brien et al., 2018). These studies have generally not been powered to detect sex effects on signature performance if they do exist, however. One study specifically testing for sex effects on a proposed radiation biodosimetry signature reported that 85% of the variance in gene expression was due to sex (Li et al., 2019). In a study of the baseline expression of a proposed radiation biodosimetry signature in 200 healthy donors, between 20 and 30% of the inter-individual variance was found to be due to the combined effects of age and sex (Agbenyegah et al., 2018). In neither case was the inter-individual variation found to degrade performance of the radiation signature, suggesting that although many genes show high variability between individuals or as a function of sex, selection of more stably expressed and radiation responsive genes allows for robust signature construction.
Similar to sex, age at exposure may contribute to radiosensitivity, with greater sensitivity to acute radiation syndrome and the lethal effects of radiation being reported for both children and the elderly (Crosfill et al., 1959; Garner et al., 1974; Adams et al., 2017; Stricklin et al., 2018). Changes in peripheral blood gene expression have also been found to occur with age (Peters et al., 2015; Irizar et al., 2015; Calabria et al., 2016), potentially altering the baseline for biodosimetry. The radiation response of some genes may also be affected by aging (Agbenyegah et al., 2018), and this remains an area in need of further study.
In addition to inter-individual variation, other sources of potential confounding are also of concern in signature development. As a large fraction of the in vivo radiation response is related to inflammation and immune system response (Amundson et al., 2004; Dressman et al., 2007; Paul et al., 2011), infection or chronic inflammatory conditions represent a potential source of confounding for radiation biodosimetry. Studies using lipopolysaccharide as a model of infection (Meadows et al., 2008; Budworth et al., 2012; Tucker et al., 2012), or infection status in patients (O’Brien et al., 2018) have shown only minimal effects on most biodosimetric signatures, although the expression levels of some individual radiation responsive genes were shown to be significantly affected. Studies have also shown clear discrimination between treatment of patients with radiation and chemotherapy agents (Lucas et al., 2014; Zhao et al., 2018), suggesting that radiation signatures can be robust against exposure to non-radiation DNA damaging agents. While rigorous testing of proposed signatures against potential confounders will still be needed, it appears that gene sets with radiation specificity and broad utility across a diverse population can be selected.
Addressing the complexity of realistic exposures
The majority of work on signature development for radiation biodosimetry has focused on the response to a single acute low linear energy transfer (LET) photon (gamma- or x-ray) exposure. In an actual large-scale radiological or nuclear event, additional factors are likely to produce more complex exposures that may vary with an individual’s location and circumstance. For instance, exposure to radionuclides dispersed by a dirty bomb or power plant accident, or from fallout from an IND, would result in a low dose rate exposure. Such dose protraction may complicate the interpretation of an acute exposure from the prompt radiation of an IND, or may continue past the time of screening if radionuclides are inhaled or ingested. Both dose rate sensitive and insensitive radiation response genes have been identified within a dose rate range of 3 orders of magnitude (Ghandhi et al., 2015a; Paul et al., 2015). Studies of common fallout radionuclides, water soluble 137Cs, the bone seeker 90Sr, or thyroid specific 131I have revealed complex patterns of gene expression changes dependent on radionuclide, dose, dose rate, and time, which are distinct from gene expression patterns occurring after acute exposure (Paul et al., 2014; Ghandhi et al., 2015b; Edmondson et al., 2016; Ghandhi et al., 2020). Taken together, this work suggests that signatures optimized for acute exposures may not be informative for protracted exposures.
One of the main planning scenarios for an IND involves a 10 kT “gun-type” device based on enriched uranium and detonated on the ground in an urban setting (Homeland Security Council, 2006). A ground burst would produce high levels of fallout, and urban infrastructure would partially shield the photons, but have much less effect on the neutron component of the radiation. This could result in up to 27% of the organ dose being due to neutrons at a survivable distance from the blast (Cullings et al., 2006; Kramer et al., 2016). Despite an acknowledged need to account for mixed neutron-photon exposures in radiation biodosimetry (Flood et al., 2016), relatively few studies have examined the effects of neutrons on biodosimetric signatures (Broustas et al., 2018; Ossetrova et al., 2018; Mukherjee et al., 2019). While most genes seem to show enhanced response to high-LET neutrons, some appear to respond without apparent modification by LET (Broustas et al., 2017). Further work will be needed to determine if these differences are large and robust enough to be exploited for characterization of radiation exposure details, or if they correlate to the relative level of radiation injury.
Shielding from urban infrastructure may result in partial bone marrow sparing, and is thus an important consideration for assessment and potential treatment of radiological injury. Studies have been conducted to develop signatures for distinguishing different patterns of partial or total body irradiation in mice (Meadows et al., 2010) and NHP (Ostheim et al., 2019; Ostheim et al., 2020a), with different gene sets possibly required at different times after exposure, or for the detection of specific partial body exposures. The measurement of miRNA in the blood is an especially promising approach for identifying radiological injury to specific organs. For instance, several miRNA species have been detected in the blood of irradiated rats, mice, and patients that appear to be specific to lung injury (Gao et al., 2017; Yadav et al., 2020).
Radiological injury versus physical dose
Although many radiation biodosimetry studies have focused on reconstruction of the radiation dose, physical dose is used as a surrogate for the expected radiation injury. In theory, biodosimetry should integrate the individual degree of injury into the readout, accounting both for the different characteristics of the radiation exposure, and for individual differences in sensitivity, as discussed above. For instance, the gene expression profile of a radiosensitive individual would be expected to report a higher dose than the actual physical dose received, while a protracted exposure to fallout would be expected to report a lower dose. It is not known, however, to what extent such differences in signature response actually correlate with the individual degree of radiological injury.
Some groups, however, are developing signatures to predict the severity or outcome of radiological injury rather than reconstructing the absorbed dose. Some studies of partial body irradiation in particular have focused on predicting the severity of ARS, or overall survival, regardless of the radiation dose or proportion of the body irradiated (Port et al., 2017; Port et al., 2016b; Port et al., 2016a). Others have compared responses to sublethal and lethal radiation doses to define signatures capable of predicting radiation fatalities early after exposure (Fendler et al., 2017; Acharya et al., 2015). Such studies, correlating gene expression markers with the degree of radiological injury, are also needed to incorporate the effects of low dose rate or mixed photon-neutron exposures into transcriptomic signatures.
New directions
Although considerable progress has been made in the development of gene expression approaches for radiation biodosimetry, additional work is clearly still needed for their application in a realistic mass radiation casualty scenario. In addition, several new directions also hold promise for improvements in the field.
Nearly all gene expression biodosimetry development has focused on the use of peripheral blood as a relatively easily biopsied biofluid, which contains some of the most radiation sensitive and responsive cells in the body. Several recent studies have also demonstrated altered expression of both mRNA (Lacombe et al., 2017) and miRNA (Greither et al., 2017) biomarkers in the saliva of patients following radiotherapy for head and neck cancer. While challenges remain, early results suggest the potential of saliva as the basis for a non-invasive biodosimetry approach (Ostheim et al., 2020b). Similarly, miRNA can also be detected in the urine (Cheng et al., 2014), and show promise as potential biomarkers of later radiation nephropathy, or as dose responsive biomarkers in the 2-8 Gy range (Bhayana et al., 2017), presenting another potential avenue for a non-invasive assay.
The use of exon arrays has revealed alternatively spliced transcripts of many radiation responsive genes as a consequence of radiation exposure (Sprung et al., 2011; Macaeva et al., 2016). One study has identified non-radiation responsive regions within two candidate biodosimetry genes, PPM1D and MDM2, and showed that normalization of the most dose-responsive exons of these genes to the levels of their non-responsive intragenic regions improved dose reconstruction compared with normalization to standard “housekeeping” genes (Forrester and Sprung, 2014). This approach has the potential to reduce the effects of inter-individual variation in situations where mass screening is needed and no individual pre-exposure samples are available. Many additional radiation responsive genes have shown differences in exon usage after radiation exposure, making this an approach that warrants additional investigation.
More recently, full-length nanopore sequencing has been applied to characterize the splice variants of one of the most widely used radiation responsive genes, FDXR. This study identified 14 variant transcripts, with differing magnitudes of radiation response, including 2 that were only detectable after radiation exposure (Cruz-Garcia et al., 2020b). Such off-or-on transcripts could circumvent the need for quantitative detection in point-of-care devices, allowing more straightforward adaption from methods being developed to detect RNA from a virus or parasite (Padhi et al., 2020; Li et al., 2020). Although the dose threshold for detection of these transcripts is not yet established, such an approach could potentially provide an important first step in radiological triage, by identifying individuals with no exposure, and allowing more detailed dosimetric efforts to focus on those with medically relevant exposures.
In conclusion, great progress has been made in defining transcriptomic signatures for dose reconstruction and prediction of radiological injury, but further work is still needed to ensure that signatures will be robust and appropriate for wide-scale population use in a realistic radiological or nuclear scenario.
Supplementary Material
Acknowledgements
The author would like to thank Dr. Shanaz Ghandhi for critical reading of the manuscript. The preparation of this manuscript was supported by the National Institute of Allergy and Infectious Diseases (NIAID grant no. U19-AI067773 to the Center for High-Throughput Minimally Invasive Radiation Biodosimetry).
Biographical Note:
Sally Amundson is an Associate Professor in the Center for Radiological Research at Columbia University Irving Medical Center in New York. She was graduate student #28 in Jack Little’s laboratory.
Footnotes
Declaration of interests:
The author reports no conflicts of interest. The author alone is responsible for the content and writing of this paper.
References
- Abend M, Badie C, Quintens R, Kriehuber R, Manning G, Macaeva E, Njima M, Oskamp D, Strunz S, Moertl S et al. 2016. Examining Radiation-Induced In Vivo and In Vitro Gene Expression Changes of the Peripheral Blood in Different Laboratories for Biodosimetry Purposes: First RENEB Gene Expression Study. Radiat Res. 185:109–123. [DOI] [PubMed] [Google Scholar]
- Acharya SS, Fendler W, Watson J, Hamilton A, Pan Y, Gaudiano E, Moskwa P, Bhanja P, Saha S, Guha C et al. 2015. Serum microRNAs are early indicators of survival after radiation-induced hematopoietic injury. Sci Transl Med. 7:287ra69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams TG, Sumner LE, Casagrande R. 2017. Estimating Risk of Hematopoietic Acute Radiation Syndrome in Children. Health Phys. 113:452–457. [DOI] [PubMed] [Google Scholar]
- Agbenyegah S, Abend M, Atkinson MJ, Combs SE, Trott KR, Port M, Majewski M. 2018. Impact of Inter-Individual Variance in the Expression of a Radiation-Responsive Gene Panel Used for Triage. Radiat Res. 190:226–235. [DOI] [PubMed] [Google Scholar]
- Ainsbury E, Badie C, Barnard S, Manning G, Moquet J, Abend M, Antunes AC, Barrios L, Bassinet C, Beinke C et al. 2017. Integration of new biological and physical retrospective dosimetry methods into EU emergency response plans - joint RENEB and EURADOS inter-laboratory comparisons. Int J Radiat Biol. 93:99–109. [DOI] [PubMed] [Google Scholar]
- Amundson SA, Do KT, Shahab S, Bittner M, Meltzer P, Trent J, Fornace AJJ. 2000. Identification of potential mRNA biomarkers in peripheral blood lymphocytes for human exposure to ionizing radiation. Radiat Res. 154:342–346. [DOI] [PubMed] [Google Scholar]
- Amundson SA, Grace MB, McLeland CB, Epperly MW, Yeager A, Zhan Q, Greenberger JS, Fornace AJ Jr. 2004. Human in vivo radiation-induced biomarkers: gene expression changes in radiotherapy patients. Cancer Res. 64:6368–6371. [DOI] [PubMed] [Google Scholar]
- Badie C, Kabacik S, Balagurunathan Y, Bernard N, Brengues M, Faggioni G, Greither R, Lista F, Peinnequin A, Poyot T et al. 2013. Laboratory intercomparison of gene expression assays. Radiat Res. 180:138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA et al. 2009. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 37:D885–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhayana S, Song F, Jacob J, Fadda P, Denko NC, Xu-Welliver M, Chakravarti A, Jacob NK. 2017. Urinary miRNAs as Biomarkers for Noninvasive Evaluation of Radiation-Induced Renal Tubular Injury. Radiat Res. 188:626–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, Parkinson H, Sarkans U, Shojatalab M, Vilo J, Abeygunawardena N, Holloway E, Kapushesky M, Kemmeren P, Lara GG et al. 2003. ArrayExpress--a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 31:68–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broustas CG, Harken AD, Gariy G, Amundson SA. 2018. Identification of differentially expressed genes and pathways in mice exposed to mixed field neutron/photon radiation. BMC Genomics. 19:504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broustas CG, Xu Y, Harken AD, Chowdhury M, Garty G, Amundson SA. 2017. Impact of Neutron Exposure on Global Gene Expression in a Human Peripheral Blood Model. Radiat Res. 187:433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budworth H, Snijders AM, Marchetti F, Mannion B, Bhatnagar S, Kwoh E, Tan Y, Wang SX, Blakely WF, Coleman M et al. 2012. DNA repair and cell cycle biomarkers of radiation exposure and inflammation stress in human blood. PLoS One. 7:e48619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabria E, Mazza EM, Dyar KA, Pogliaghi S, Bruseghini P, Morandi C, Salvagno GL, Gelati M, Guidi GC, Bicciato S et al. 2016. Aging: a portrait from gene expression profile in blood cells. Aging (Albany NY). 8:1802–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Sun X, Scicluna BJ, Coleman BM, Hill AF. 2014. Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine. Kidney Int. 86:433–444. [DOI] [PubMed] [Google Scholar]
- Cheung VG, Conlin LK, Weber TM, Arcaro M, Jen KY, Morley M, Spielman RS. 2003. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 33:422–425. [DOI] [PubMed] [Google Scholar]
- Crosfill ML, Lindop PJ, Rotblat J. 1959. Variation of sensitivity to ionizing radiation with age. Nature. 183:1729–1730. [DOI] [PubMed] [Google Scholar]
- Cruz-Garcia L, O’Brien G, Sipos B, Mayes S, Love MI, Turner DJ, Badie C. 2020a. Generation of a Transcriptional Radiation Exposure Signature in Human Blood Using Long-Read Nanopore Sequencing. Radiat Res. 193:143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Garcia L, O’Brien G, Sipos B, Mayes S, Tichý A, Sirák I, Davídková M, Marková M, Turner DJ, Badie C. 2020b. In Vivo Validation of Alternative FDXR Transcripts in Human Blood in Response to Ionizing Radiation. Int J Mol Sci. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullings HM, Fujita S, Funamoto S, Grant EJ, Kerr GD, Preston DL. 2006. Dose estimation for atomic bomb survivor studies: its evolution and present status. Radiat Res. 166:219–254. [DOI] [PubMed] [Google Scholar]
- Dressman HK, Muramoto GG, Chao NJ, Meadows S, Marshall D, Ginsburg GS, Nevins JR, Chute JP. 2007. Gene expression signatures that predict radiation exposure in mice and humans. PLoS Med. 4:e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DA, Karski EE, Kohlgruber A, Koneru H, Matthay KK, Allen S, Hartmann CL, Peterson LE, DuBois SG, Coleman MA. 2016. Transcript Analysis for Internal Biodosimetry Using Peripheral Blood from Neuroblastoma Patients Treated with (131)I-mIBG, a Targeted Radionuclide. Radiat Res. 186:235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ein-Dor L, Zuk O, Domany E. 2006. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc Natl Acad Sci U S A. 103:5923–5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendler W, Malachowska B, Meghani K, Konstantinopoulos PA, Guha C, Singh VK, Chowdhury D. 2017. Evolutionarily conserved serum microRNAs predict radiation-induced fatality in nonhuman primates. Sci Transl Med. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiano AN, Fathallah-Shaykh HM, Fiveash J, Gage J, Cantor A, Kharbanda S, Johnson MR. 2011. Gene expression analysis in radiotherapy patients and C57BL/6 mice as a measure of exposure to ionizing radiation. Radiat Res. 176:49–61. [DOI] [PubMed] [Google Scholar]
- Fischer M 2019. Conservation and divergence of the p53 gene regulatory network between mice and humans. Oncogene. 38:4095–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flood AB, Ali AN, Boyle HK, Du G, Satinsky VA, Swarts SG, Williams BB, Demidenko E, Schreiber W, Swartz HM. 2016. Evaluating the Special Needs of The Military for Radiation Biodosimetry for Tactical Warfare Against Deployed Troops: Comparing Military to Civilian Needs for Biodosimetry Methods. Health Phys. 111:169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester HB, Sprung CN. 2014. Intragenic controls utilizing radiation-induced alternative transcript regions improves gene expression biodosimetry. Radiat Res. 181:314–323. [DOI] [PubMed] [Google Scholar]
- Gao F, Liu P, Narayanan J, Yang M, Fish BL, Liu Y, Liang M, Jacobs ER, Medhora M. 2017. Changes in miRNA in the lung and whole blood after whole thorax irradiation in rats. Sci Rep. 7:44132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner RJ, Phemister RD, Angleton GM, Lee AC, Thomassen RW. 1974. Effect of age on the acute lethal response of the beagle to cobalt-60 gamma radiation. Radiat Res. 58:190–195. [PubMed] [Google Scholar]
- Ghandhi SA, Shuryak I, Morton SR, Amundson SA, Brenner DJ. 2019a. New Approaches for Quantitative Reconstruction of Radiation Dose in Human Blood Cells. Sci Rep. 9:18441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandhi SA, Sima C, Weber WM, Melo DR, Rudqvist N, Morton SR, Turner HC, Amundson SA. 2020. Dose and Dose-Rate Effects in a Mouse Model of Internal Exposure to 137 Cs. Part 1: Global Transcriptomic Responses in Blood. Radiat Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandhi SA, Smilenov L, Shuryak I, Pujol-Canadell M, Amundson SA. 2019b. Discordant gene responses to radiation in humans and mice and the role of hematopoietically humanized mice in the search for radiation biomarkers. Sci Rep. 9:19434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandhi SA, Smilenov LB, Elliston CD, Chowdhury M, Amundson SA. 2015a. Radiation dose-rate effects on gene expression for human biodosimetry. BMC Med Genomics. 8:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandhi SA, Turner HC, Shuryak I, Dugan GO, Bourland JD, Olson JD, Tooze JA, Morton SR, Batinic-Haberle I, Cline JM et al. 2018. Whole thorax irradiation of non-human primates induces persistent nuclear damage and gene expression changes in peripheral blood cells. PLoS One. 13:e0191402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandhi SA, Weber W, Melo D, Doyle-Eisele M, Chowdhury M, Guilmette R, Amundson SA. 2015b. Effect of 90 Sr internal emitter on gene expression in mouse blood. BMC Genomics. 16:586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greither T, Vorwerk F, Kappler M, Bache M, Taubert H, Kuhnt T, Hey J, Eckert AW. 2017. Salivary miR-93 and miR-200a as post-radiotherapy biomarkers in head and neck squamous cell carcinoma. Oncol Rep. 38:1268–1275. [DOI] [PubMed] [Google Scholar]
- Hoffmann W, Schmitz-Feuerhake I. 1999. How radiation-specific is the dicentric assay. J Expo Anal Environ Epidemiol. 9:113–133. [DOI] [PubMed] [Google Scholar]
- Homeland Security Council. National Planning Scenarios (Final version 21.3). Washington, DC: Homeland Security Council; 2006:1. [Google Scholar]
- Irizar H, Goñi J, Alzualde A, Castillo-Triviño T, Olascoaga J, Lopez de Munain A, Otaegui D. 2015. Age gene expression and coexpression progressive signatures in peripheral blood leukocytes. Exp Gerontol. 72:50–56. [DOI] [PubMed] [Google Scholar]
- Iversen ES, McCarthy JM, Bell Burdett K, Lipton G, Phillips G, Dressman H, Ross J, Chao N. 2018. Bridging the gaps: using an NHP model to predict single dose radiation absorption in humans. Int J Radiat Biol. 1–10. [DOI] [PubMed] [Google Scholar]
- Jacobs AR, Guyon T, Headley V, Nair M, Ricketts W, Gray G, Wong JYC, Chao N, Terbrueggen R. 2020. Role of a high throughput biodosimetry test in treatment prioritization after a nuclear incident. Int J Radiat Biol. 96:57–66. [DOI] [PubMed] [Google Scholar]
- Kabacik S, Mackay A, Tamber N, Manning G, Finnon P, Paillier F, Ashworth A, Bouffler S, Badie C. 2011. Gene expression following ionising radiation: identification of biomarkers for dose estimation and prediction of individual response. Int J Radiat Biol. 87:115–129. [DOI] [PubMed] [Google Scholar]
- Kang CM, Park KP, Song JE, Jeoung DI, Cho CK, Kim TH, Bae S, Lee SJ, Lee YS. 2003. Possible biomarkers for ionizing radiation exposure in human peripheral blood lymphocytes. Radiat Res. 159:312–319. [DOI] [PubMed] [Google Scholar]
- Knops K, Boldt S, Wolkenhauer O, Kriehuber R. 2012. Gene expression in low- and high-dose-irradiated human peripheral blood lymphocytes: possible applications for biodosimetry. Radiat Res. 178:304–312. [DOI] [PubMed] [Google Scholar]
- Kramer K, Li A, Madrigal J, Sanchez B, Millage K. Report DTRA-TR-13-045 (R1): Monte Carlo Modeling of the Initial Radiation Emitted by a Nuclear Device in the National Capitol Region. Fort Belvoir, VA: Defense Threat Reduction Agency; 2016:32. [Google Scholar]
- Lacombe J, Brooks C, Hu C, Menashi E, Korn R, Yang F, Zenhausern F. 2017. Analysis of Saliva Gene Expression during Head and Neck Cancer Radiotherapy: A Pilot Study. Radiat Res. 188:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe J, Sima C, Amundson SA, Zenhausern F. 2018. Candidate gene biodosimetry markers of exposure to external ionizing radiation in human blood: A systematic review. PLoS One. 13:e0198851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HY, Jia WN, Li XY, Zhang L, Liu C, Wu J. 2020. Advances in detection of infectious agents by aptamer-based technologies. Emerg Microbes Infect. 9:1671–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lu X, Feng JB, Tian M, Liu QJ. 2017. Identification and Validation of Candidate Radiation-responsive Genes for Human Biodosimetr. Biomed Environ Sci. 30:834–840. [DOI] [PubMed] [Google Scholar]
- Li S, Lu X, Feng JB, Tian M, Wang J, Chen H, Chen DQ, Liu QJ. 2019. Developing Gender-Specific Gene Expression Biodosimetry Using a Panel of Radiation-Responsive Genes for Determining Radiation Dose in Human Peripheral Blood. Radiat Res. 192:399–409. [DOI] [PubMed] [Google Scholar]
- Lucas J, Dressman HK, Suchindran S, Nakamura M, Chao NJ, Himburg H, Minor K, Phillips G, Ross J, Abedi M et al. 2014. A translatable predictor of human radiation exposure. PLoS One. 9:e107897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaeva E, Mysara M, De Vos WH, Baatout S, Quintens R. 2019. Gene expression-based biodosimetry for radiological incidents: assessment of dose and time after radiation exposure. Int J Radiat Biol. 95:64–75. [DOI] [PubMed] [Google Scholar]
- Macaeva E, Saeys Y, Tabury K, Janssen A, Michaux A, Benotmane MA, De Vos WH, Baatout S, Quintens R. 2016. Radiation-induced alternative transcription and splicing events and their applicability to practical biodosimetry. Sci Rep. 6:19251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Kabacik S, Finnon P, Bouffler S, Badie C. 2013. High and low dose responses of transcriptional biomarkers in ex vivo X-irradiated human blood. Int J Radiat Biol. 89:512–522. [DOI] [PubMed] [Google Scholar]
- Maznyk NA, Wilkins RC, Carr Z, Lloyd DC. 2012. The capacity, capabilities and needs of the WHO BioDoseNet member laboratories. Radiat Prot Dosimetry. 151:611–620. [DOI] [PubMed] [Google Scholar]
- Meadows SK, Dressman HK, Daher P, Himburg H, Russell JL, Doan P, Chao NJ, Lucas J, Nevins JR, Chute JP. 2010. Diagnosis of partial body radiation exposure in mice using peripheral blood gene expression profiles. PLoS One. 5:e11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows SK, Dressman HK, Muramoto GG, Himburg H, Salter A, Wei Z, Ginsburg GS, Chao NJ, Nevins JR, Chute JP. 2008. Gene expression signatures of radiation response are specific, durable and accurate in mice and humans. PLoS One. 3:e1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon N, Rogers CJ, Lukaszewicz AI, Axtelle J, Yadav M, Song F, Chakravarti A, Jacob NK. 2016. Detection of Acute Radiation Sickness: A Feasibility Study in Non-Human Primates Circulating miRNAs for Triage in Radiological Events. PLoS One. 11:e0167333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Benotmane MA, Tirone I, Hooghe-Peters EL, Desaintes C. 2005. Transcriptional response to ionizing radiation in lymphocyte subsets. Cell Mol Life Sci. 62:1489–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M, Benotmane MA, Vanhove D, van Hummelen P, Hooghe-Peters EL, Desaintes C. 2004. Effect of ionizing radiation on gene expression in CD4+ T lymphocytes and in Jurkat cells: unraveling novel pathways in radiation response. Cell Mol Life Sci. 61:1955–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Grilj V, Broustas CG, Ghandhi SA, Harken AD, Gariy G, Amundson SA. 2019. Human Transcriptomic Response to Mixed Neutron-Photon Exposures Relevant to an Improvised Nuclear Device. Radiat Res. 192:189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien G, Cruz-Garcia L, Majewski M, Grepl J, Abend M, Port M, Tichý A, Sirak I, Maikova A, Donovan E et al. 2018. FDXR is a biomarker of radiation exposure in vivo. Sci Rep. 8:684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossetrova NI, Stanton P, Krasnopolsky K, Ismail M, Doreswamy A, Hieber KP. 2018. Biomarkers for Radiation Biodosimetry and Injury Assessment after Mixed-field (Neutron and Gamma) Radiation in the Mouse Total-body Irradiation Model. Health Phys. 115:727–742. [DOI] [PubMed] [Google Scholar]
- Ostheim P, Coker O, Schüle S, Hermann C, Combs SE, Trott KR, Atkinson M, Port M, Abend M. 2021. Identifying a Diagnostic Window for the Use of Gene Expression Profiling to Predict Acute Radiation Syndrome. Radiat Res. 195:38–46. [DOI] [PubMed] [Google Scholar]
- Ostheim P, Haupt J, Herodin F, Valente M, Drouet M, Majewski M, Port M, Abend M. 2019. miRNA Expression Patterns Differ by Total- or Partial-Body Radiation Exposure in Baboons. Radiat Res. 192:579–588. [DOI] [PubMed] [Google Scholar]
- Ostheim P, Haupt J, Schüle S, Herodin F, Valente M, Drouet M, Majewski M, Port M, Abend M. 2020a. Differentiating Total- or Partial-Body Irradiation in Baboons Using mRNA Expression Patterns: A Proof of Concept. Radiat Res. 194:476–484. [DOI] [PubMed] [Google Scholar]
- Ostheim P, Tichý A, Sirak I, Davidkova M, Stastna MM, Kultova G, Paunesku T, Woloschak G, Majewski M, Port M et al. 2020b. Overcoming challenges in human saliva gene expression measurements. Sci Rep. 10:11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhi A, Gupta E, Singh G, Agarwal R, Sharma MK, Sarin SK. 2020. Evaluation of the Point of Care Molecular Diagnostic Genedrive HCV ID Kit for the detection of HCV RNA in clinical samples. Epidemiol Infect. 1–23. [DOI] [PubMed] [Google Scholar]
- Park JG, Paul S, Briones N, Zeng J, Gillis K, Wallstrom G, LaBaer J, Amundson SA. 2017. Developing Human Radiation Biodosimetry Models: Testing Cross-Species Conversion Approaches Using an Ex Vivo Model System. Radiat Res. 187:708–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Amundson SA. 2008. Development of gene expression signatures for practical radiation biodosimetry. Int J Radiat Oncol Biol Phys. 71:1236–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Amundson SA. 2014. Differential Effect of Active Smoking on Gene Expression in Male and Female Smokers. J Carcinog Mutagen. 5:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Barker CA, Turner HC, McLane A, Wolden SL, Amundson SA. 2011. Prediction of in vivo radiation dose status in radiotherapy patients using ex vivo and in vivo gene expression signatures. Radiat Res. 175:257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Ghandhi SA, Weber W, Doyle-Eisele M, Melo D, Guilmette R, Amundson SA. 2014. Gene expression response of mice after a single dose of 137 Cs as an internal emitter. Radiat Res. 182:380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Kleiman NJ, Amundson SA. 2019. Transcriptomic responses in mouse blood during the first week after in vivo gamma irradiation. Sci Rep. 9:18364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Smilenov LB, Amundson SA. 2013. Widespread Decreased Expression of Immune Function Genes in Human Peripheral Blood Following Radiation Exposure. Radiat Res. 180:575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Smilenov LB, Elliston CD, Amundson SA. 2015. Radiation Dose-Rate Effects on Gene Expression in a Mouse Biodosimetry Model. Radiat Res. 184:24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters MJ, Joehanes R, Pilling LC, Schurmann C, Conneely KN, Powell J, Reinmaa E, Sutphin GL, Zhernakova A, Schramm K et al. 2015. The transcriptional landscape of age in human peripheral blood. Nat Commun. 6:8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogosova-Agadjanyan EL, Fan W, Georges GE, Schwartz JL, Kepler CM, Lee H, Suchanek AL, Cronk MR, Brumbaugh A, Engel JH et al. 2011. Identification of radiation-induced expression changes in nonimmortalized human T cells. Radiat Res. 175:172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port M, Herodin F, Valente M, Drouet M, Lamkowski A, Majewski M, Abend M. 2016a. First Generation Gene Expression Signature for Early Prediction of Late Occurring Hematological Acute Radiation Syndrome in Baboons. Radiat Res. 186:39–54. [DOI] [PubMed] [Google Scholar]
- Port M, Hérodin F, Valente M, Drouet M, Lamkowski A, Majewski M, Abend M. 2017. Gene expression signature for early prediction of late occurring pancytopenia in irradiated baboons. Ann Hematol. 96:859–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port M, Hérodin F, Valente M, Drouet M, Ostheim P, Majewski M, Abend M. 2018. Persistent mRNA and miRNA expression changes in irradiated baboons. Sci Rep. 8:15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port M, Herodin F, Valente M, Drouet M, Ullmann R, Doucha-Senf S, Lamkowski A, Majewski M, Abend M. 2016b. MicroRNA Expression for Early Prediction of Late Occurring Hematologic Acute Radiation Syndrome in Baboons. PLoS One. 11:e0165307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port M, Majewski M, Herodin F, Valente M, Drouet M, Forcheron F, Tichy A, Sirak I, Zavrelova A, Malkova A et al. 2018. Validating Baboon Ex Vivo and In Vivo Radiation-Related Gene Expression with Corresponding Human Data. Radiat Res. 189:389–398. [DOI] [PubMed] [Google Scholar]
- Ramakumar A, Subramanian U, Prasanna PG. 2015. High-throughput sample processing and sample management; the functional evolution of classical cytogenetic assay towards automation. Mutat Res Genet Toxicol Environ Mutagen. 793:132–141. [DOI] [PubMed] [Google Scholar]
- Riecke A, Rufa CG, Cordes M, Hartmann J, Meineke V, Abend M. 2012. Gene expression comparisons performed for biodosimetry purposes on in vitro peripheral blood cellular subsets and irradiated individuals. Radiat Res. 178:234–243. [DOI] [PubMed] [Google Scholar]
- Rogan PK, Mucaki EJ, Lu R, Shirley BC, Waller E, Knoll JHM. 2020. Meeting radiation dosimetry capacity requirements of population-scale exposures by geostatistical sampling. PLoS One. 15:e0232008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothkamm K, Beinke C, Romm H, Badie C, Balagurunathan Y, Barnard S, Bernard N, Boulay-Greene H, Brengues M, De Amicis A et al. 2013. Comparison of established and emerging biodosimetry assays. Radiat Res. 180:111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royba E, Repin M, Pampou S, Karan C, Brenner DJ, Gariy G. 2019. RABiT-II-DCA: A Fully-automated Dicentric Chromosome Assay in Multiwell Plates. Radiat Res. 192:311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozgaj R, Kasuba V, Simić D. 2002. The frequency of dicentrics and acentrics and the incidence of rogue cells in radiation workers. Mutagenesis. 17:135–139. [DOI] [PubMed] [Google Scholar]
- Shirley BC, Knoll JHM, Moquet J, Ainsbury E, Pham ND, Norton F, Wilkins RC, Rogan PK. 2020. Estimating partial-body ionizing radiation exposure by automated cytogenetic biodosimetry. Int J Radiat Biol. 96:1492–1503. [DOI] [PubMed] [Google Scholar]
- Smirnov DA, Brady L, Halasa K, Morley M, Solomon S, Cheung VG. 2012. Genetic variation in radiation-induced cell death. Genome Res. 22:332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov DA, Morley M, Shin E, Spielman RS, Cheung VG. 2009. Genetic analysis of radiation-induced changes in human gene expression. Nature. 459:587–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprung CN, Li J, Hovan D, McKay MJ, Forrester HB. 2011. Alternative transcript initiation and splicing as a response to DNA damage. PLoS One. 6:e25758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricklin D, Prins R, Bellman J. 2018. Development of Age-Dependent Dose Modification Factors for Acute Radiation Lethality. Int J Radiat Biol. 1–44. [DOI] [PubMed] [Google Scholar]
- Sullivan JM, Prasanna PG, Grace MB, Wathen LK, Wallace RL, Koemer JF, Coleman CN. 2013. Assessment of biodosimetry methods for a mass-casualty radiological incident: medical response and management considerations. Health Phys. 105:540–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker JD, Grever WE, Joiner MC, Konski AA, Thomas RA, Smolinski JM, Divine GW, Auner GW. 2012. Gene expression-based detection of radiation exposure in mice after treatment with granulocyte colony-stimulating factor and lipopolysaccharide. Radiat Res. 177:209–219. [DOI] [PubMed] [Google Scholar]
- Tucker JD, Joiner MC, Thomas RA, Grever WE, Bakhmutsky MV, Chinkhota CN, Smolinski JM, Divine GW, Auner GW. 2014. Accurate gene expression-based biodosimetry using a minimal set of human gene transcripts. Int J Radiat Oncol Biol Phys. 88:933–939. [DOI] [PubMed] [Google Scholar]
- Waselenko JK, MacVittie TJ, Blakely WF, Pesik N, Wiley AL, Dickerson WE, Tsu H, Confer DL, Coleman CN, Seed T et al. 2004. Medical management of the acute radiation syndrome: recommendations of the Strategic National Stockpile Radiation Working Group. Ann Intern Med. 140:1037–1051. [DOI] [PubMed] [Google Scholar]
- Wathen LK, Eder PS, Horwith G, Wallace RL. 2020. Using biodosimetry to enhance the public health response to a nuclear incident. Int J Radiat Biol. 1–4. [DOI] [PubMed] [Google Scholar]
- Wilkins RC, Romm H, Oestreicher U, Marro L, Yoshida MA, Suto Y, Prasanna PG. 2011. Biological Dosimetry by the Triage Dicentric Chromosome Assay - Further validation of International Networking. Radiat Meas. 46:923–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav M, Bhayana S, Liu J, Lu L, Huang J, Ma Y, Qamri Z, Mo X, Jacob DS, Parasa ST et al. 2020. Two-miRNA-based finger-stick assay for estimation of absorbed ionizing radiation dose. Sci Transl Med. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Nishida T, Sato Y, Nakai Y, Kashiwakura I. 2020. Identification of Radiation-Dose-Dependent Expressive Genes in Individuals Exposed to External Ionizing Radiation. Radiat Res. 193:274–285. [DOI] [PubMed] [Google Scholar]
- Zhao JZL, Mucaki EJ, Rogan PK. 2018. Predicting ionizing radiation exposure using biochemically-inspired genomic machine learning. F1000Res. 7:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


