Abstract
Glioblastoma (GBM) is characterized by exceptionally high intratumoral heterogeneity. However, the molecular mechanisms underlying the origin of different GBM cell populations remain unclear. Here, we found that the compositions of ribosomes of GBM cells in the tumour core and edge differ due to alternative RNA splicing. The acidic pH in the core switches before messenger RNA splicing of the ribosomal gene RPL22L1 towards the RPL22L1b isoform. This allows cells to survive acidosis, increases stemness and correlates with worse patient outcome. Mechanistically, RPL22L1b promotes RNA splicing by interacting with lncMALAT1 in the nucleus and inducing its degradation. Contrarily, in the tumour edge region, RPL22L1a interacts with ribosomes in the cytoplasm and upregulates the translation of multiple messenger RNAs including TP53. We found that the RPL22L1 isoform switch is regulated by SRSF4 and identified a compound that inhibits this process and decreases tumour growth. These findings demonstrate how distinct GBM cell populations arise during tumour growth. Targeting this mechanism may decrease GBM heterogeneity and facilitate therapy.
Glioblastoma (GBM) is a highly malignant tumour characterized by almost 100% recurrence resulting in virtually universal lethality1. This clinically unfavourable behaviour has been largely attributed to the exceptionally heterogeneous cancer cell population within the tumour. Morphological examination has focused on the microenvironment as a factor that can be responsible for the origin of diverse cell phenotypes2,3. In the GBM necrotic core, cells are exposed to hypoxia and acidic pH values as low as 5.9 (refs. 4,5). These conditions support stemness and ultimately promote an aggressive mesenchymal phenotype3,4. In contrast, cells located in microvascular proliferation zones maintain initial proneural properties3,6 and are affected by pro-oncogenic signals of bystander endothelial cells7. However, the molecular mechanisms underlying the origin of different GBM cell populations remain to be elucidated.
Numerous studies have revealed properties of GBM cells that correlate with patient outcome. Intratumoral and intertumoral profiling of the transcriptome2,8, proteome9,10 and metabolome11,12 allow for the separation of GBM into subgroups characterized by different levels of aggressiveness and survival. However, unexpectedly, little correlation was observed between proteome and transcriptome signatures13. Variable translation efficiency is one of the key mechanisms that explains the inconsistencies between the proteome and transcriptome14. Even minor changes in the ribosome may lead to a substantial alteration of the entire proteome during normal development15,16, as well as in tumorigenesis including epithelial-to-mesenchymal transition17,18. Substantial differences in ribosomal protein levels have also been detected in gliomas19,20. However, the role of ribosomal perturbations in GBM development and progression is still unknown.
Inclusion of additional (non-mandatory) proteins in the ribosome modulates overall protein synthesis as well as the affinity of the organelle to individual messenger RNAs (mRNAs)21. One of the proteins that can be co-purified with ribosomes is RPL22L1. It is highly conserved from humans to zebrafish22 and is critical for normal development23. RPL22L1 has also been shown to be important for prostate24, ovarian25 and colorectal26 cancers and neuroblastoma27. Outside the ribosome, RPL22L1 can directly interact with RNAs and has been shown to modulate the splicing of certain pre-mRNA molecules23,28. However, the molecular functions of this protein in GBM are still elusive.
In the present study, we investigate the ribosomal content in GBM cells isolated from different tumour regions and identify the molecular mechanisms by which RPL22L1 splicing isoforms regulate the phenotype of GBM. Finally, we elucidate the regulation of RPL22L1 splicing and identify a small molecule that impairs this process.
Results
The protein composition of ribosomes differs between the GBM core and edge
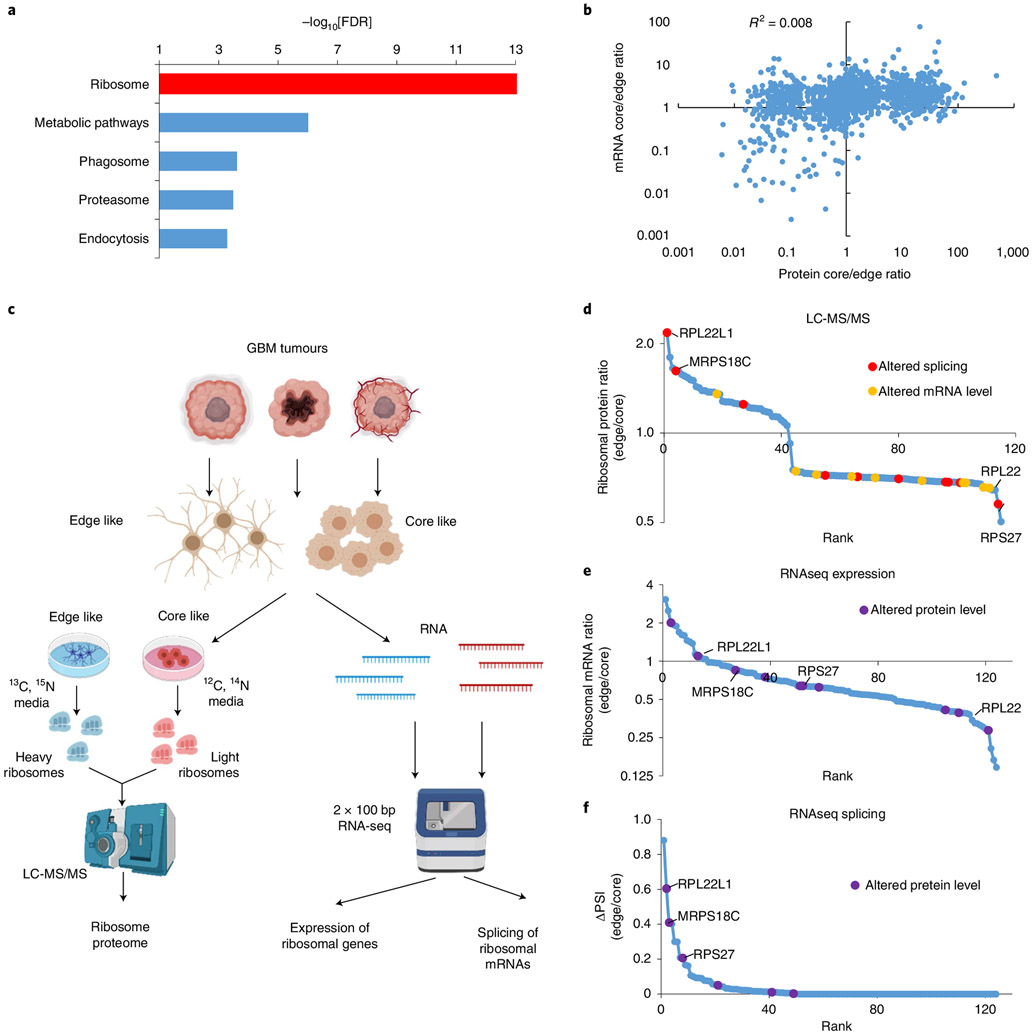
Previously, we reported that GBM cells isolated from the central (core) versus peripheral (edge) areas of the tumour demonstrate substantial differences in phenotype and transcriptome or metabolome profiles3,11,29. To elucidate the molecular mechanisms underlying these alterations, we compared the proteome and transcriptome in paired GBM tissue samples (core versus edge) as well as in primary neurosphere cultures established from each tumour zone. Analysis by liquid chromatography with tandem mass spectrometry (LC-MS/MS) revealed that the most significant differences between samples were attributed to the ribosomal protein cluster (Fig. 1a and Supplementary Table 1). Subsequent comparison of LC-MS/MS and RNA sequencing (RNA-seq) data showed that although both the proteome and transcriptome signatures could be used to differentiate between core and edge samples (Extended Data Fig. 1a), there was little or no correlation between differences detected on the proteomic and transcriptomic levels (Fig. 1b and Extended Data Fig. 1b). We hypothesized that these discrepancies could appear due to the altered properties of ribosomes between cells from the core and the edge of the tumour.
Fig. 1 ∣. GBM cells from the edge and core of the tumour have ribosomes with different protein compositions.
a, Kyoto Encyclopedia of Genes and Genomes enrichment analysis of proteins differentially present (fold change > 4; spectral count > 2) between the edge and core of the tumour. FDR, false discovery rate. b, Correlation between the proteome and transcriptome of edge and core GBM tissue. c, Experimental workflow used to study differences in the protein composition of ribosomes. bp, base pair. d, Relative levels of ribosomal proteins identified by SILAC LC-MS/MS in ribosomes purified from GBM sphere lines with edge versus core phenotypes. e, Relative mRNA levels of ribosomal genes in GBM sphere lines with edge versus core phenotypes, as identified by RNA-seq. f, Differences in the splicing of ribosomal genes between GBM sphere lines with edge and core phenotypes, as identified by RNA-seq. PSI, percentage spliced in index1. In d–f, the top ten differentially present proteins, differentially expressed mRNAs and differentially spliced pre-mRNAs are highlighted in violet, yellow and red, respectively. RNA-seq was performed for neurospheres obtained from n = 6 different patients.
To test this hypothesis, we analysed the protein composition of ribosomes using the stable isotope labelling by amino acids in cell culture (SILAC) LC-MS/MS approach (Fig. 1c). This experiment revealed substantial differences between edge- and core-like GBM neurospheres (Fig. 1d and Supplementary Table 2). However, no correlation was observed when proteomic differences were compared with levels of corresponding mRNAs (Fig. 1e and Extended Data Fig. 1c), indicating that transcription has little or no effect on the amount of ribosome-incorporated protein. Surprisingly, we observed differential pre-mRNA splicing for multiple proteins whose abundance altered most between ribosomes from the core and the edge cells (Fig. 1f, Extended Data Fig. 1d and Supplementary Table 3).
These data led us to speculate that alternative splicing may be responsible for the differential protein composition of ribosomes between GBM cell populations.
Alternative splicing generates two RPL22L1 protein isoforms
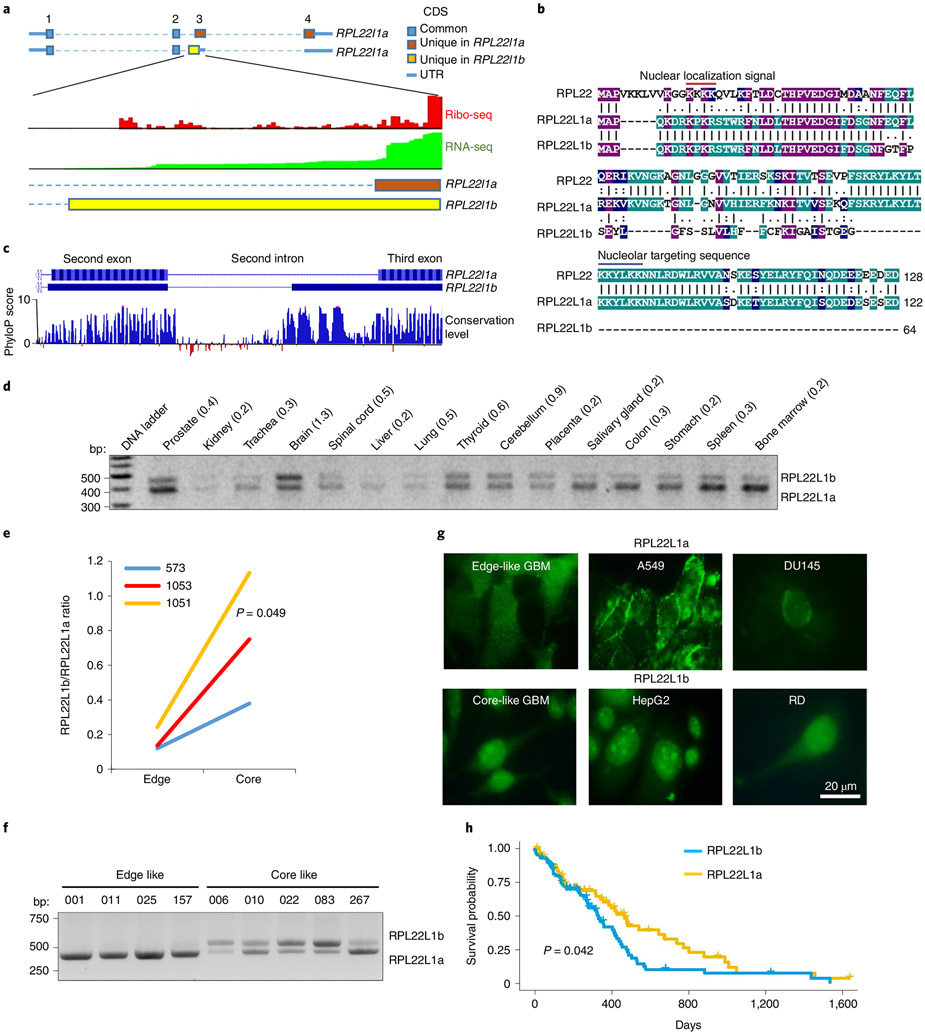
RPL22L1 showed the most pronounced differences in ribosomes isolated from GBM cells with core and edge phenotypes, as determined by both protein and splicing analyses. RNA-seq and reverse transcription PCR (RT-PCR) followed by Sanger sequencing revealed that alternative splicing gives rise to two RPL22L1 isoforms, one of which has not been described previously (Extended Data Fig. 1e,f). The isoform differed in the 5′ end of the third exon, which leads to a reading frame shift and gives rise to a new carboxy (C)-terminal amino acid sequence (Fig. 2a,b). We termed the known isoform RPL22L1a. The one discovered in this study was named RPL22L1b.
Fig. 2 ∣. Alternative splicing generates two different isoforms of RPL22L1.
a, Schematic of RPL22L1 isoforms together with ribo-seq and RNA-seq read densities aligned to the corresponding regions of the genome. The data were obtained from GWIPs-viz Browser37. The first, second, third and fourth exons are indicated by the numbers 1–4 at the top. CDS, coding sequence; UTR, untranslated region. b, Alignment of the amino acid sequences of RPL22, RPL22L1a and RPL22L1b (shadings indicate positions which have a single fully conserved residue in all three proteins (purple); in any two proteins (green) or residue with similar properties in two proteins (blue); number of amino acids in the protein sequence is indicated). c, Pattern of genome conservation among 100 vertebrates for the RPL22L1 region spanning from the second to the third exon. d, RT-PCR analysis of RPL22L1 splicing in different human tissues. Approximate RPL22L1b/RPL22L1a isoform ratios are indicated. e, Ratios of RPL22L1 isoforms in paired GBM tissue isolated from edge and core regions of tumours (tissue from n = 3 different patients; Wilcoxon signed rank-sum test). The data were obtained from RNA-seq. f, RT-PCR analysis of RPL22L1 splicing in GBM sphere lines with edge (001, 011, 025 and 157) and core (006, 010, 022, 083 and 267) phenotypes (spheres isolated from n = 9 different patients). g, Representative immunofluorescence stainings of cells predominantly expressing the RPL22L1a isoform (GBM spheres 157, A549 and DU145) or RPL22L1b isoform (GBM spheres 267, HepG2 and RD) with antibodies against the N-terminal part of RPL22L1. h, Kaplan–Meier curve showing the overall survival of patients with GBM, subdivided into two groups based on the splicing of RPL22L1 (n = 154 different patients; log-rank test). The data were obtained from the TCGA database.
PhyloP analysis30 revealed that the part of the intron that gives rise to RPL22L1b is highly conserved among vertebrates, in drastic contrast with the neighbouring 5′ sequence of the same intron, which is not incorporated into the RPL22L1b isoform (Fig. 2c). These data suggest that RPL22L1b could have appeared early in vertebrate evolution and may perform important functions in living cells. Next, we confirmed RPL22L1b expression in human tissue and standard cancer cell lines (Fig. 2d and Extended Data Fig. 1g). Interestingly, maximal expression of this isoform was detected in the brain. Analysis of RPL22L1b splicing in a broader cohort of samples from patients with GBM revealed at least fourfold higher levels of RPL22L1b in each of the tumour core specimens compared with the periphery (Fig. 2e). Consistent data were obtained using nine cancer stem cell-enriched GBM in vitro cultures with either the core or the edge phenotype (Fig. 2f and Extended Data Fig. 1h).
The use of an alternative 3′ splice site in RPL22L1b mRNA leads to the formation of a premature stop codon, which theoretically may induce nonsense-mediated decay (NMD)31. However, recent studies have shown that such predictions are not entirely accurate32-34. To test whether RPL22L1b undergoes NMD, we treated GBM neurospheres with the small-molecule NMD inhibitor NMDI-14 (ref. 35) and demonstrated that it has no effect on the abundance of RPL22L1b mRNA (Extended Data Fig. 1i). Consistent results were obtained when we analysed an RNA-seq dataset of cells with knockdown of the different components of NMD machinery36 (Extended Data Fig. 2a), indicating that RPL22L1b is not subject to NMD.
Next, we confirmed the presence of RPL22L1b on the protein level. First, we detected RPL22L1b in the global aggregate of human ribosome profiling (ribo-seq) data37 (Fig. 1a), as well as in a previously published ribo-seq dataset from GBM cells38 (Extended Data Fig. 2b). Second, we demonstrated that in our patient-derived GBM cells RPL22L1b mRNA is associated with polyribosome fraction and, therefore, is actively translated (Extended Data Fig. 2c). Third, we analysed multiple proteomics datasets39 and found the peptide SEYLGFSSLVLH, which corresponds uniquely to RPL22L1b (Extended Data Fig. 2d). Fourth, we performed a western blot analysis of GBM sphere lines with antibodies against full-length RPL22L1a and against the amino (N)-terminal fragment of RPL22L1, which is common for both isoforms. We demonstrated that GBM cells with an edge phenotype express the RPL22L1a isoform, while core-like cells have a smaller protein variant that is only detectable by the antibodies against the N-terminal part of RPL22L1, which probably corresponds to RPL22L1b (Extended Data Fig. 2e). Finally, using immunostaining with antibodies against the N-terminal part of RPL22L1, we showed that in cells that according to RT-PCR predominantly express RPL22L1a (A549, DU145, 001, 025 and 157), a fluorescence signal was detected mainly in the cytoplasm, while cells containing RPL22L1b mRNA (HepG2, RD, 006, 022 and 083) showed a nuclear staining pattern (Fig. 2g and Extended Data Fig. 3). Taken together, these data strongly argue for the presence of endogenous RPL22L1b protein.
Next, we addressed the association of RPL22L1 splicing with disease outcome. High abundance of RPL22L1b mRNA correlated with poor survival in patients with GBM (Fig. 2h). Interestingly, RPL22L1b was also associated with poor prognosis for patients with adrenocortical carcinoma, whereas in melanoma and kidney renal clear cell carcinoma it correlated with prolonged survival (Extended Data Fig. 4a). We detected no correlation of the RPL22L1 isoform ratio with any clinical prognostic factors for patients with GBM (age > 50 years; methylation of the MGMT promoter or mutations in the isocitrate dehydrogenase gene 40) (Extended Data Fig. 4b) or with the phase of the cell cycle (Extended Data Fig. 4c-e).
Altogether thus far, we have demonstrated that the RPL22L1b isoform is expressed both in normal tissue and in tumours and that the intracellular localization of this protein differs from that of RPL22L1a.
RPL22L1b is expressed in the acidic environment of the GBM core
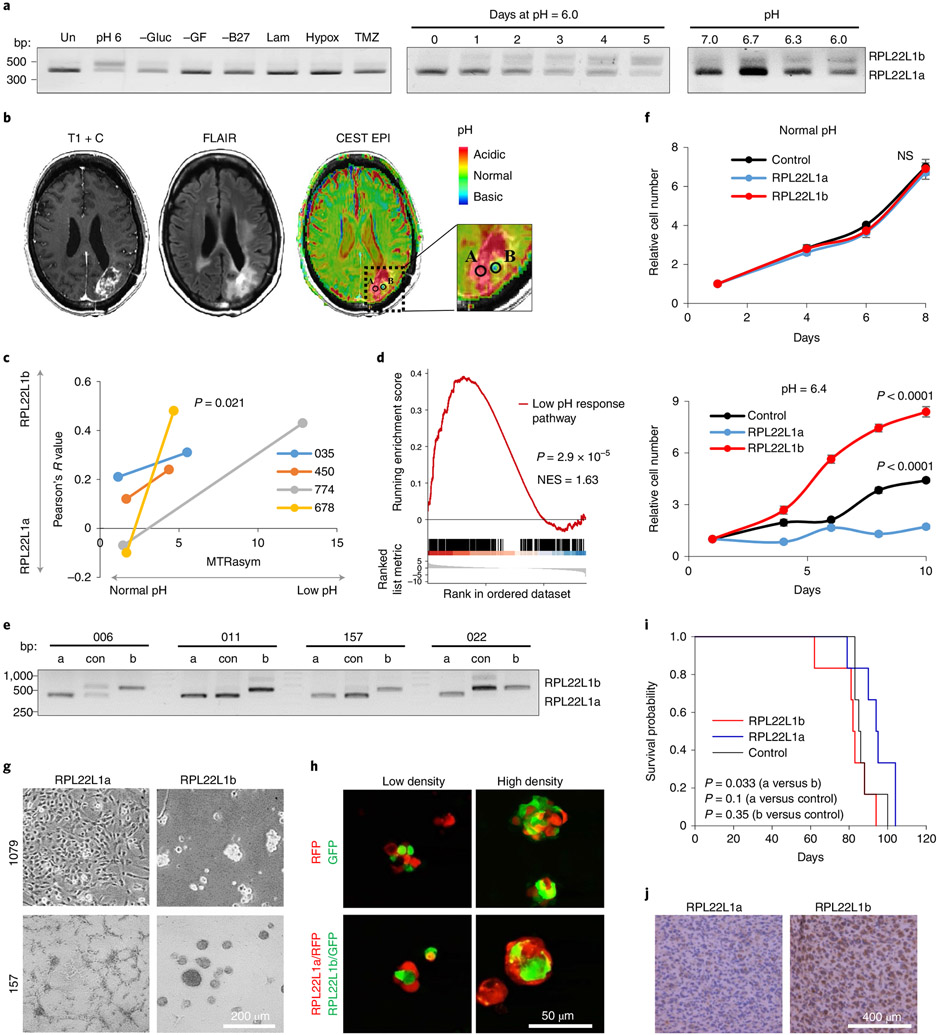
Currently, there are few if any data describing how multiple GBM cell populations may arise during tumour expansion. To investigate the role of RPL22L1 in this process, we first sought to determine how microenvironmental conditions may regulate RPL22L1 mRNA splicing in proneural GBM cells, which are believed to serve as a precursor for all other subtypes of GBM6. By cultivating neurospheres in vitro under different conditions, we demonstrated that only exposure to acidic pH upregulated RPL22L1b (Fig. 3a). These data were confirmed using cells isolated from four different patients (Extended Data Fig. 5a). Again, the NMD inhibitor had no effect on RPL22L1b mRNA abundance (Extended Data Fig. 1i).
Fig. 3 ∣. The RPL22L1 isoform ratio is regulated by extracellular pH.
a, RT-PCR analysis of RPL22L1 splicing in 157 GBM spheres cultivated under different conditions for the indicated periods of time. Un, untreated cells in control medium; pH 6, acidified medium; −Gluc, without glucose; −GF, without growth factors; −B27, without nutrition supplement; Lam, cultured as a monolayer on laminin; Hypox, cultured under hypoxia; TMZ, cultured in the presence of 50 μM temozolomide (similar data were obtained for cells isolated from n = 4 different patients; see Extended Data Fig. 3a). b, Representative contrast-enhanced T1-weighted (left), fluid attenuated inversion recovery (FLAIR; middle) and pH–weighted amine CEST–EPI (MTRasym at 3 ppm; right) MRI images of GBM tumours. Biopsy regions with low and normal pH are labelled A and B, respectively. c, Colocalization coefficient of RPL22L1 and DAPI staining plotted against MTRasym values. MTRasym represents the acidity of the tumour region. The colocalization coefficient reflects the RPL22L1b/RPL22L1a isoform ratio (samples were obtained from n = 4 different patients; Wilcoxon signed rank-sum test). d, GSEA of RNA-seq data from RPL22Lb/RPL22L1ahigh versus RPL22Lb/RPL22L1alow GBM tissue. The data were obtained from the TCGA database (n = 77 different patients). NES, normalized enrichment score. e, RT-PCR analysis of RPL22L1 splicing in GBM cells transduced with lentiviruses encoding RPL22L1a (a), RPL22L1b (b) or an empty vector (con) (n = 4 different patients). f, In vitro cell growth assay of 157 glioma spheres overexpressing RPL22L1a, RPL22L1b or an empty vector (control) and cultivated in normal (pH 7.4; top) or acidified medium (pH 6.4; bottom) (the experiment was performed in n = 6 biological replicates; unpaired two-tailed t-test). NS, not significant. g, Representative microscopic images of 1079 and 157 glioma spheres overexpressing RPL22L1a or RPL22L1b (similar data were obtained for cells isolated from n = 4 different patients). h, Representative fluorescence images of 1079 glioma spheres overexpressing GFP, RFP, RFP+ RPL22L1a or GFP+ RPL22L1b. The cells were mixed in different densities and imaged 24 h later (similar data were obtained for cells isolated from n = 2 different patients). i, Kaplan–Meier survival curves of mice injected with 3 × 105 luciferase-labelled 1051 glioma spheres overexpressing RPL22L1a, RPL22L1b or an empty vector (n = 6 mice per group; log-rank test). j, Representative immunohistochemical staining for CD109 of brain sections from mice injected as in i (n = 2 mice per group; see Extended Data Fig. 6d). All quantitative data are means ± s.d.
To study whether pH affects the RPL22L1 isoform ratio in vivo, we investigated the intracellular localization of this protein in patient-derived GBM tumour tissue. Staining of the large GBM section that contained multiple morphologically distinct zones demonstrated that in some areas of the tumour RPL22L1 was localized in the cytoplasm, while in other regions RPL22L1 had mainly nuclear localization (Extended Data Fig. 5b). These differences were quantified using a colocalization analysis of 4′,6-diamidino-2-phenylindole (DAPI) and RPL22L1 staining (Extended Data Fig. 5c). One may suggest that the tumour zones with nuclear localization of RPL22L1 correspond to the areas of low pH (the RPL22L1b isoform is present) while regions with cytoplasmic RPL22L1 correspond to areas of normal pH (the RPL22L1a isoform is expressed). To directly confirm this hypothesis, we used pH–weighted molecular MRI41 to obtain paired samples of GBM tissue from the tumour regions with different extracellular pH (Fig. 3b). Immunostaining demonstrated that areas of low pH contained higher levels of nuclear-localized RPL22L1 (Fig. 3c), indicating that the acidic microenvironment indeed shifts RPL22L1 splicing towards the RPL22l1b isoform in human gliomas in situ. Next, utilizing a previously published set of genes whose expression is altered by low-pH conditions42, we established the pathway for gene set enrichment analysis (GSEA). By applying it to the samples from The Cancer Genome Atlas (TCGA), we demonstrated that tumours with an increased RPL22L1b/RPL22L1a ratio show high activation of the acidic response pathway (P = 2.9 × 10−5; Fig. 3d).
Next, we established patient-derived GBM neurospheres with overexpression of RPL22L1 isoforms. Interestingly, ectopic expression of one RPL22L1 isoform resulted in diminished expression of the other, suggesting a mechanism of mutually negative influence (Fig. 3e). We confirmed these data by immunostaining (Extended Data Fig. 5d). Proliferation analysis revealed that established cell lines exhibited a similar growth rate in regular media; however, at pH 6.4 only cells overexpressing RPL22L1b maintained an unaltered proliferation speed, control cells grew twice slower, while cells overexpressing the RPL22L1a isoform were unable to proliferate in acidified medium (Fig. 3f). Consistently, expression of the RPL22L1a isoform significantly increased cell death upon exposure to low pH (Extended Data Fig. 5e). Interestingly, RPL22L1 isoforms dramatically affected the microscopic appearance of GBM cells. RPL22L1a-overexpressing cells tended to adhere to the plastic and formed compact clusters–a feature that is rather unusual for primary GBM cultures. In contrast, RPL22L1b-overexpressing counterparts formed dense neurospheres with an irregular shape resembling stem cell-enriched cultures (Fig. 3g). Moreover, cells derived from one patient after overexpression of RPL22L1b were substantially enlarged in size compared with control or RPL22L1a-overexpressing counterparts (Extended Data Fig. 6a).
Since RPL22L1a is characteristically expressed by peripheral GBM cells whereas RPL22L1b is a marker of core counterparts, we investigated the localization of cells that overexpress RPL22L1a and RPL22L1b in neurospheres. We showed that RPL22L1a/red fluorescent protein (RFP)-overexpressing cells were localized at the periphery of mixed neurospheres whereas RPL22L1b/green fluorescent protein (GFP)-positive cells were in the centre (Fig. 3h and Extended Data Fig. 6b). Importantly, this effect was not associated with the differential motility of the cells (Extended Data Fig. 6c). Finally, we studied the effect of RPL22L1 isoforms on tumour growth in vivo. Overexpression of RPL22L1b in intracranially injected patient-derived GBM cells led to accelerated death of the animals (Fig. 3i). Subsequent immunohistochemical examinations revealed a noticeable increase in CD109–a marker of GBM core region3,29–in RPL22L1b xenografts (Fig. 3j and Extended Data Fig. 6d).
Based on the data described above, we concluded that RPL22L1b appears under low-pH conditions in the tumour core, where it facilitates cell proliferation and survival upon acidosis, thereby contributing to GBM progression.
The interactome of RPL22L1 isoforms in GBM cells
To elucidate the functions of RPL22L1 isoforms, we identified proteins that interact with RPL22L1a and RPL22L1b using two complementary approaches. First, each isoform was expressed in Escherichia coli (Extended Data Fig. 6e) and immobilized on magnetic beads. The beads were incubated with GBM cell lysates and the proteins bound to the beads were identified using LC-MS/MS and subsequently subjected to enrichment analysis (Supplementary Table 4). We found that RPL22L1a interacted with proteins involved in L13a-mediated translational silencing, while RPL22L1b bound to the U1 small nuclear ribonucleoprotein particle (snRNP) complex, which is critical for pre-mRNA splicing (Fig. 4a and Extended Data Fig. 6f). Second, we generated GBM cells with stable expression of Fc-tagged RPL22L1 isoforms. Fc-RPL22L1a and Fc-RPL22L1b, together with their binding partners, were isolated from neurospheres using magnetic beads and analysed by LC-MS/MS (Supplementary Table 5). While this approach yielded smaller numbers of bound proteins, the results were consistent with the previous experiment. The data suggested that RPL22L1a interacts with the ribosome whereas RPL22L1b is involved in RNA decay (Fig. 4b and Extended Data Fig. 7a).
Fig. 4 ∣. Interactome of RPL22L1 isoforms.
a, Venn diagram representing proteins that were bound to recombinant His-tagged RPL22L1a (blue), RPL22L1b (red) or control beads (green) and subsequently identified by LC-MS/MS. Proteins that were detected only in RPL22L1a or only in RPL22L1b samples were subjected to enrichment analysis. The most significantly enriched terms and corresponding P values are indicated. b, Venn diagram representing proteins that were co-purified with Fc-tagged RPL22L1a (blue), RPL22L1b (red) or control protein (green) and subsequently identified by LC-MS/MS. Proteins that were present in the RPL22L1a sample or RPL22L1b sample and absent in the control sample were subjected to enrichment analysis. The most significantly enriched terms and corresponding P values are indicated. KEGG, Kyoto Encyclopedia of Genes and Genomes. c, Capillary electrophoresis of RNA that was co-purified with Fc-tagged RPL22L1a or RPL22L1b. d, Venn diagram representing RNAs that were co-purified with Fc-tagged RPL22L1a (blue), RPL22L1b (red) or control protein (green) and subsequently identified by RNA-seq. RNA was considered differentially present if the fold change was >6 and the number of transcripts per million (tpm) was >0.5. Differentially present RNAs were subjected to enrichment analysis. The most significantly enriched terms and corresponding P values are indicated. e, Hallmark database GSVA of RNA-seq data obtained from 157 cells overexpressing RPL22L1a, RPL22L1b or an empty vector (sequencing was performed in n = 2 biological replicates). IFN-α, interferon-α; NF-κB, nuclear factor-κB; TNF-α, tumour necrosis factor-α; TGF-β, transforming growth factor-β. f, Results of SILAC LC-MS/MS analysis of 157 cells overexpressing RPL22L1a (grown in heavy isotope medium) or RPL22L1b (grown in regular isotope medium). Proteins were considered differentially expressed if the difference between the intensity-based absolute quantification value for heavy and light peptides was >50%. Differentially present proteins were subjected to KEGG enrichment analysis. The graph indicates the ratio of proteins (in RPL22L1b versus RPL22L1a samples) related to the most significantly enriched clusters (the experiment was performed in n = 2 biological replicates).
RPL22L1 is capable of direct interaction with ribosomal RNA (rRNA) and mRNA23,28. Therefore, we next identified RNAs that were co-precipitated with Fc-RPL22L1a and Fc-RPL22L1b. Capillary electrophoresis of eluted RNA confirmed a strong interaction of RPL22L1a with rRNA (Fig. 4c). In contrast, RPL22L1b did not interact with rRNA, although it bound multiple other RNAs. Depletion of rRNA and subsequent sequencing showed that, in addition to rRNA, RPL22L1a interacted with mRNAs involved in stem cell differentiation and the AKT signalling pathway (Fig. 4d and Extended Data Fig. 7b), whereas RPL22L1b interacted with mRNAs that regulated splicing and apoptosis as well as with a noticeable number of different long non-coding RNAs (Supplementary Table 6).
Finally, we determined how RPL22L1a and RPL22L1b affect the transcriptome and proteome profiles of GBM cells. Total RNA-seq and gene set variation analysis (GSVA) pointed to the activation of p53 signalling by RPL22L1a. This finding is in line with the role of TP53 as a master regulator of the transcriptional network underlying the proneural phenotype43, which is typical for edge GBM cells3. Furthermore, RPL22L1a upregulated the angiogenesis pathway and Notch signalling, both of which are related to cells at the tumour edge7,44 (Fig. 4e). In contrast, RPL22L1b activates the spliceosome, oxidative phosphorylation and DNA repair pathways (Fig. 4e and Extended Data Fig. 7c). The two latter pathways have been shown to be upregulated in an aggressive mesenchymal GBM phenotype observed in the tumour core45,46.
Quantitative analysis of the proteome using SILAC LC-MS/MS (Supplementary Table 7) revealed that the most significant differences observed between cells overexpressing RPL22L1 isoforms were for the valine, leucine and isoleucine degradation pathway, lysosomes, the tricarboxylic acid (TCA) cycle and the cluster of spliceosomal proteins (Fig. 4f and Extended Data Fig. 7d). It is important to note that 5 of 16 proteins involved in the valine, leucine and isoleucine degradation pathway and differentially present between analysed samples belonged to the aldehyde dehydrogenase family, which is known to be important for stemness regulation47. In contrast, activation of the TCA cycle in RPL22L1b-overexpressing cells supports our RNA-seq data since the TCA is a major source of metabolites for oxidative phosphorylation. Finally, the lysosomal pathway, which is activated in RPL22L1a-expressing cells, has also been attributed to stem cell differentiation48.
Altogether, we can conclude that although RPL22L1 isoforms have identical N terminals, half of their interactomes are different. RPL22L1a interacts with ribosomes and is directly involved in the regulation of translation, whereas RPL22L1b performs extra-ribosomal functions and participates in RNA processing. Interestingly, both isoforms strongly affected GBM metabolism, but in a different manner: RPL22L1a activates the signalling pathways characteristic of peripheral GBM cells while RPL22L1b is related to the mesenchymal core GBM phenotype.
RPL22L1b promotes RNA splicing in GBM cells
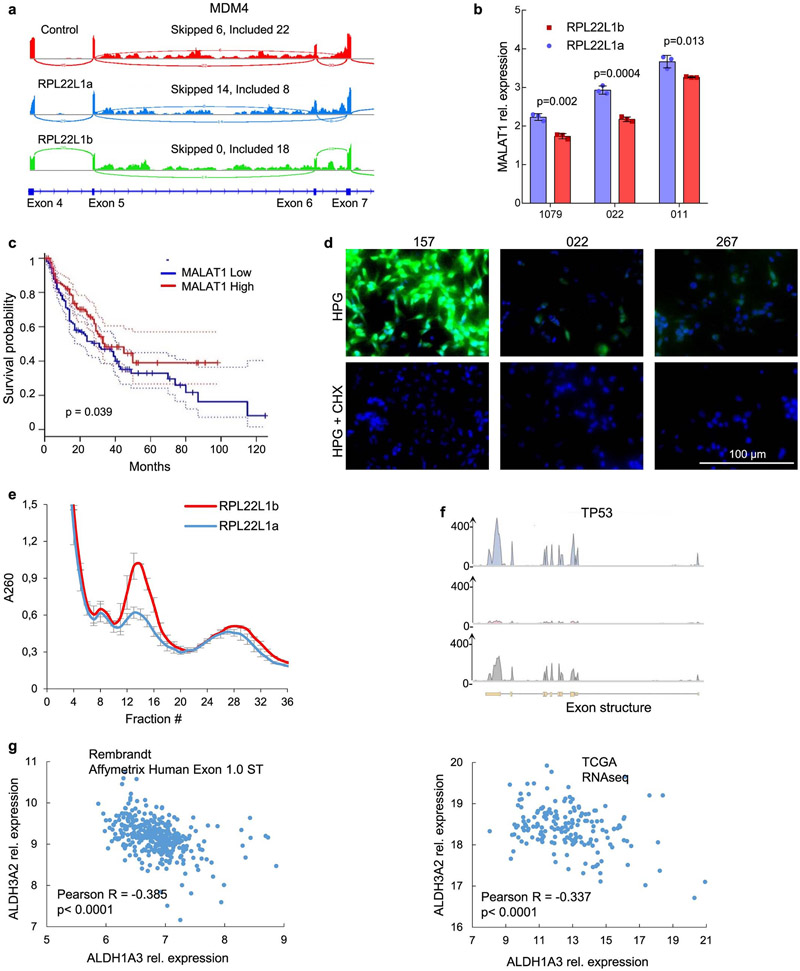
Based on our interactome and intracellular localization data, we hypothesized that RPL22L1b can regulate pre-mRNA splicing. Transcriptome analysis of RPL22L1-overexpressing cells revealed that RPL22L1b induces substantial changes in RNA splicing (Fig. 5a). This isoform promoted exon inclusion and attenuated intron retention, thereby increasing the efficacy of the spliceosomal complex49 (Fig. 5b). Enrichment analysis showed that RPL22L1b regulates the splicing of pre-mRNAs encoding DNA repair proteins as well as the proteins involved in stem cell proliferation (Extended Data Fig. 7e). These results corroborated our data on gene expression profiling by RPL22L1b (Fig. 4e). As a proof of concept, we demonstrated that RPL22L1b overexpression shifts the splicing of MDM4 from the non-functional MDM4 short isoform (exon VI is excluded) to the MDM4 full-length isoform (exon VI is included) (Fig. 5c and Extended Data Fig. 8a) that has previously been shown to protect cells from therapy and to promote proliferation50.
Fig. 5 ∣. Molecular functions of RPL22L1b.
a, Pie chart representing the number and type of alternative splicing events detected in 157 cells expressing RPL22L1b compared with control cells. b, Volcano plot showing significantly different splicing events related to exon skipping (left) and intron retention (right) detected in samples as in a (sequencing was performed in n = 2 biological replicates). c, qRT-PCR analysis of the MDM4 isoform expression ratios between 157 GBM spheres overexpressing RPL22L1a, RPL22L1b or an empty vector (the experiment was performed in n = 3 biological replicates; unpaired two-tailed t-test). d, RNA-IP enrichment profiles of RPL22L1a, RPL22L1b and a control protein for MALAT1. 157 cells overexpressing Fc-tagged proteins were used for the experiment. e, qRT-PCR analysis of samples as in d with primers for MALAT1 (the experiment was performed in n = 3 biological replicates; unpaired two-tailed t-test). f, qRT-PCR analysis of MALAT1 expression in 157 cells stably expressing different isoforms of RPL22L1 (the experiment was performed in n = 3 biological replicates). g, Analysis of MALAT1 RNA stability at different time points after actinomycin D treatment (10 μg ml−1) of cells as in f (similar data were obtained for cells isolated from n = 4 different patients; see Extended Data Fig. 7b; for each sphere line, the experiment was performed in n = 3 biological replicates; unpaired two-tailed t-test). h, Kaplan–Meier curve showing the overall survival of patients with glioma, subdivided based on the MALAT1 expression level (n = 338 different patients; log-rank test; the data were obtained from the TCGA database). i, FACS analysis for CD133 staining of GBM spheres expressing RPL22L1 isoforms (the cells were isolated from n = 2 different patients). j, qRT-PCR analysis of ALDH1A3, CD133, Nanog, Oct4, Sox2 and GFAP expression in 157 cells stably overexpressing RPL22L1a, RPL22L1b or an empty vector (the experiment was performed in n = 3 biological replicates; unpaired two-tailed t-test). k, Neurosphere formation assay of cells expressing RPL22L1a or RPL22L1b (the cells were isolated from n = 3 different patients). The line within each box represents the mean stem cell frequency. The top and bottom edges of each box are the upper and lower estimated confidence intervals for stem cell frequency. All quantitative data are means ± s.d.
Since RPL22L1 has not been detected in the spliceosome, we can propose that this protein regulates splicing indirectly. Our interactome analysis revealed binding of RPL22L1b to the long non-coding RNA MALAT1 (Fig. 5d,e), which interacts with the U1 snRNP spliceosomal complex51. This may explain why U1 snRNP-related proteins were co-precipitated with RPL22L1b (Fig. 4a). Interestingly, overexpression of RPL22L1b significantly decreased MALAT1 levels (Fig. 5f), suggesting that RPL22L1b might induce MALAT1 degradation. To test this hypothesis, we exposed GBM cells overexpressing RPL22L1b isoforms to the transcription inhibitor actinomycin D. Subsequent quantitative RT-PCR (qRT-PCR) analysis showed that RPL22L1b substantially decreased the lifetime of MALAT1 (Fig. 5g and Extended Data Fig. 8b). Based on these results, we can speculate that RPL22L1b affects RNA splicing by inducing MALAT1 degradation. This hypothesis is in good agreement with previous data showing that MALAT1 depletion increases exon inclusion due to the subsequent release of splicing-activating factors52.
Although in most cancers MALAT1 is known to be a factor of tumour progression, in GBM this RNA probably plays the role of tumour suppressor53. Consistent with these observations, our bioinformatics analysis of the TCGA database revealed that diminished expression of MALAT1 correlated with shorter overall survival (Fig. 5h) and disease-free survival (Extended Data Fig. 8c) in patients with GBM and low-grade glioma. Mechanistically, MALAT1 was shown to negatively regulate CD133 (ref. 54), a key marker of GBM stem cells. Using flow cytometry (Fig. 5i) and qRT-PCR (Fig. 5j), we found that RPL22L1b upregulates CD133 and elevates the expression of ALDH1A3, Nanog and Oct4, which serve as key markers of cancer stem cells55. These results are in line with our observation that RPL22L1b induces MALAT1 degradation. Interestingly, both isoforms of RPL22L1 decreased the expression of GFAP, which is known to be a marker of more differentiated GBM neurospheres. However, the effect of RPL22L1b was more pronounced. Finally, using the limiting dilution assay, we demonstrated that RPL22L1b markedly increased the frequency of GBM stem cells in in vitro cultures derived from three different patients (Fig. 5k).
Taken together, we provide evidence that RPL22L1b induces MALAT1 degradation, thereby promoting GBM stemness.
RPL22L1a activates translation in GBM cells
Since RPL22L1a is a component of the ribosome (Fig. 4), we investigated its role in the regulation of protein synthesis in GBM cells. Metabolic labelling of newly synthesized proteins with l-homopropargylglycine (HPG)–an analogue of methionine56–revealed that RPL22L1a overexpression substantially increased the rate of translation (Fig. 6a). We confirmed these data for endogenous RPL22L1a, demonstrating that GBM neurosphere lines that predominantly express RPL22L1a had elevated protein synthesis compared with cells expressing RPL22L1b (Fig. 6b and Extended Data Fig. 8d). To further verify these findings, we performed a ribosome fractionation experiment57, which showed that RPL22L1a overexpression decreased the number of monosomes and increased the polysomal content, pointing to the activation of protein synthesis (Fig. 6c and Extended Data Fig. 8e).
Fig. 6 ∣. Molecular functions of RPL22L1a.
a, FACS analysis of HPG incorporation into newly synthesized proteins in 022 cells overexpressing RPL22L1a, RPL22L1b or an empty vector. Cells pre-treated for 30 min with cycloheximide (100 μg ml−1) were used as a negative control. b, FACS analysis of HPG incorporation into newly synthesized proteins in 001, 025 and 157 GBM sphere lines (predominantly expressing RPL22L1a) and 006, 010 and 022 GBM sphere lines (predominantly expressing RPL22L1b) (the cells were isolated from n = 6 different patients). c, Polysome profiles of 1079 GBM spheres stably expressing RPL22L1a or RPL22L1b (similar data were obtained for cells isolated from n = 2 different patients; see Extended Data Fig. 7e). d, RNA-IP enrichment profiles of RPL22L1a, RPL22L1b and a control protein for the genes CDK5, ALDH3A2 and RPN2. 157 cells overexpressing Fc-tagged proteins were used for the experiment. e, qRT-PCR analysis of RNA-IP samples as in c with primers for TP53, CDK5, ALDH3A2 and RPN2 (the experiment was performed in n = 3 biological replicates; unpaired two-tailed t-test). f, qRT-PCR analysis of TP53, CDK5, ALDH3A2 and RPN2 expression in 157 cells expressing the different isoforms of RPL22L1 (the experiment was performed in n = 3 biological replicates). g, Western blotting analysis of GBM spheres overexpressing RPL22L1a, RPL22L1b or an empty vector (the cells were isolated from n = 3 different patients). The numbers below the panels indicate the approximate band intensity, which was first normalized to the actin β level and then to the intensity of the band in the corresponding control sample. All quantitative data are means ± s.d.
Next, we studied whether RPL22L1a can regulate the translation of its mRNA binding partners. Among the transcripts co-precipitated with RPL22L1a (Supplementary Table 8), we selected mRNAs involved in the p53 pathway, namely TP53 and CDK5 (ref. 58), as well as stemness-related ALDH3A2 and RPN2 (refs. 59,60) (Fig. 6d and Extended Data Fig. 8f). Using qRT-PCR, we confirmed the interaction of these transcripts with RPL22L1a but not with RPL22L1b (Fig. 6e). Importantly, we did not detect any changes in the abundance of the respective mRNAs in cells upon overexpression of RPL22L1a or RPL22L1b (Fig. 6f). However, western blot analysis of neurospheres derived from three different patients demonstrated that RPL22L1a overexpression was associated with elevated levels of TP53, CDK5, ALDH3A2 and RPN2 (Fig. 6g). At the same time, isoforms of RPL22L1 had little or no effect on RPL22 expression.
Interestingly, RPL22L1b activated ALDH1A3 expression (Fig. 5j), whereas RPL22L1a upregulated the closely related ALDH3A2 (Fig. 6g). Analysis of TCGA and REMBRANDT databases (Extended Data Fig. 8g) revealed that the genes ALDH3A2 and ALDH1A3 have a reciprocal expression pattern. The exact role in GBM of the aldehyde dehydrogenases that these genes transcribe is unclear; however, it was previously shown that in gastric adenocarcinoma ALDH3A2 is associated with prolongation of survival61 while ALDH1A3 is a predictor of poor prognosis in various malignancies62. Therefore, RPL22L1 isoform switching may induce a subsequent switch of the expression between ALDH family members.
Altogether, our data demonstrated that RPL22L1a can stimulate global protein synthesis as well as translation of specific mRNAs that interact with this protein.
SRSF4 regulates RPL22L1 mRNA splicing
Next, we aimed to dissect the mechanisms involved in RPL22L1 isoform switching. Analysis of the TCGA database identified serine/arginine-rich splicing factor 4 (SRSF4) as the most significant correlate of RPL22L1 splicing (Fig. 7a). Analysis of the previously published iCLIP (individual-nucleotide resolution cross-linking and immunoprecipitation) data for SRSF4 (ref. 63) confirmed that this protein binds to RPL22L1 pre-mRNA on the boundary of the second and third exons (Extended Data Fig. 9a)–the region that is different between RPL22L1a and RPL22L1b (Fig. 2a). To strengthen these data, we performed an RNA immunoprecipitation (RNA-IP) experiment and demonstrated relatively weak but statistically significant enrichment of RPL22L1 mRNA in samples eluted from the immobilized SRSF4 protein (Extended Data Fig. 9b).
Fig. 7 ∣. SRSF4 regulates the splicing of RPL22L1.
a, Correlation between the RPL22L1b/RPL22L1a isoform ratio and the expression levels of different splicing factors. The x axis shows the correlation coefficient for all cancer samples (n = 7,631 different patients). The y axis represents the correlation for GBM samples (n = 154 different patients). The data were obtained from the TCGA database. b, SRSF4 mRNA levels in paired edge and core GBM samples (tissue from n = 3 different patients; Wilcoxon signed rank-sum test). The data were obtained by RNA-seq. FPKM, fragments per kilobase of transcript per million mapped reads. c, Representative immunohistofluorescence staining with antibodies against SRSF4 (red) and DAPI (blue) of low- and normal-pH GBM tumour biopsy samples obtained using pH–weighted molecular MRI (from n = 2 different patients). d, qRT-PCR analysis of SRSF4 expression in 157 cells cultivated in acidified medium (pH 6.0) for 0–5 d (the experiment was performed in n = 3 biological replicates; unpaired two-tailed t-test). e, RPL22L1 isoform ratio in U87MG cells transfected with plasmids encoding GFP (control) or GFP-tagged SRSF1, SRSF2, SRSF3 or SRSF4 proteins and subsequently sorted for GFP using FACS (the experiment was performed in n = 3 biological replicates). f, RT-PCR analysis of RPL22L1 splicing in 157 glioma spheres overexpressing GFP or GFP-SRSF4. g, Representative microscopic images of 006 (left) and 030 (right) GBM cells transduced with lentiviruses encoding non-target small hairpin RNA (shNT) or small hairpin RNAs against SRSF4 (shSRSF4). The cells were attached to the laminin-coated surface. h, RT-PCR analysis of RPL22L1 splicing in 006, 030 and 157 GBM cells transduced with lentiviruses encoding shNT or two different small hairpin RNAs against SRSF4 (48shSRSF4 and 49shSRSF4). i, qRT-PCR analysis of CD133 and ALDH1A3 expression in 030 (top) and 157 (bottom) GBM spheres transduced with lentiviruses as in h (the experiment was performed in n = 3 biological replicates; unpaired two-tailed t-test). j, In vitro cell growth assay of cells as in i (the experiment was performed in n = 6 biological replicates; unpaired two-tailed t-test). k, Kaplan–Meier curve showing the overall survival of patients with glioma (n = 446) subdivided based on the SRSF4 expression level (log-rank test). The data were obtained from the REMBRANDT database. All quantitative data are means±s.d.
To study the intratumoral variation of SRSF4 expression, we analysed our RNA-seq data and demonstrated that SRSF4 is highly upregulated in the tumour core compared with the tumour edge in paired patient samples (Fig. 7b). Importantly, the SRSF4 expression pattern looked nearly identical to the RPL22L1b/RPL22L1a ratio in the corresponding samples (Fig. 2e). Consistent results were obtained by analysing the IVY Glioblastoma Atlas Project database (Extended Data Fig. 9c) and immunohistochemical staining of patient-derived GBM tissue samples collected with the help of pH–weighted molecular MRI (Fig. 7c). To validate these findings in vitro, we showed that propagation of GBM spheres in an acidic pH led to an increase in SRSF4 expression (Fig. 7d), supporting the role of acidosis in formation of the RPL22L1b isoform.
To directly determine whether SRSF4 regulates RPL22L1 splicing, we first overexpressed SRSF1, SRSF2, SRSF3 or SRSF4 proteins that belong to the same family of splicing factors and demonstrated that all of them indeed increase the RPL22L1b/RPL22L1a transcript ratio; however, SRSF4 evoked the strongest effect (Fig. 7e,f). Next, we performed knockdown of SRSF4 in GBM cells with core (006 and 030) and edge (157) phenotypes (Extended Data Fig. 9d). Excitingly, transduction with lentiviruses encoding shSRSF4 resulted in dramatic changes in the morphology of the core-like GBM cells (Fig. 7g). As expected, downregulation of SRSF4 shifted the splicing of RPL22L1 towards the RPL22L1a isoform (Fig. 7h and Extended Data Fig. 9e). In agreement with this observation, we also detected downregulation of the stem cell markers CD133 and ALDH1A3 in 030 cells after SRSF4 knockdown (Fig. 7i). Importantly, in 157 cells, which express RPL22L1b at a very low basal level, silencing of SRSF4 had no effect on the amount of CD133 and ALDH1A3 mRNAs (Fig. 7i). Finally, we tested the effect of SRSF4 knockdown on cell proliferation in vitro. Silencing of SRSF4 in GBM cells with the core phenotype nearly completely inhibited their proliferation (Fig. 7j), while in cells with the edge phenotype, knockdown of SRSF4 had a much less pronounced effect (Fig. 7j).
To determine the clinical relevance of SRSF4, we estimated its influence on patient survival. Our results revealed that SRSF4 expression was associated with poor prognosis in patients with GBM and low-grade glioma (Fig. 7k and Extended Data Fig. 9f).
Altogether, we have shown that the acidic conditions in the GBM core promote SRSF4 expression, which in turn switches RNA splicing towards RPL22L1b. This isoform is important in the maintenance of stemness of GBM cells with the core phenotype, while edge-like GBM cells may utilize an alternative signalling pathway.
A small-molecule Clk inhibitor impairs RPL22L1 splicing
Finally, we set out to determine whether pharmacological targeting of SRSF4 can impair RPL22L1 splicing and provide a therapeutic benefit for GBM. Currently, no direct SRSF inhibitors have been discovered. However, the functions of these proteins can be downregulated by attenuating the activity of upstream protein kinases, including the CDC-like kinase (Clk) family64 and NIMA-related kinase 2 (NEK2)65. Recently, the Nakano and Moreau groups have discovered a series of small molecular inhibitors of Clk and NEK2 kinases66-68. Comparison of these compounds with pladienolide B (a broad-spectrum splicing inhibitor69) and conventional antitumor agents revealed that FG1059 (a dual Clk1/4 inhibitor) induces the most substantial changes in RPL22L1 pre-mRNA splicing (Fig. 8a,b). This result was confirmed using GBM neurospheres from three different patients (Extended Data Fig. 9g). As shown in Fig. 8c, FG1059 caused complete skipping of the third exon (that is, the one that differs between RPL22L1a and RPL22L1b isoforms) (Fig. 2a).
Fig. 8 ∣. FG1059 impairs the splicing of RPL22L1.
a, RT-PCR analysis of RPL22L1 splicing in 157 glioma spheres that were left untreated (Un) or treated for 24 h with CMP3a, EY404 (EY), FG1059 (FG) or pladienolide B (Pl-B) (similar data were obtained for cells isolated from n = 3 different patients; see Extended Data Fig. 8f). b, Chemical structure of FG1059. c, Results of Sanger sequencing of the RPL22L1 isoform purified from FG1059-treated cells. d, Principal component analysis of the phosphoproteome of 157 GBM cells treated with 3 μM FG1059 for 0, 3, 6 or 12 h. e, Representative fluorescence images of 157 cells transfected with a plasmid encoding RFP-SRSF4 and subsequently treated with 5 μM FG1059 for 18 h. The DNA was stained with DAPI. DMSO, dimethyl sulfoxide. f, In vitro cell viability assay of GBM spheres obtained from n = 7 different patients and normal human astrocytes (NHAs) that were treated with various concentrations of FG1059 for 5 d (the experiment was performed in n = 6 biological replicates). Cells predominantly expressing the RPL22L1a (001, 025 and 157) and RPL22L1b (006, 022, 030 and 267) isoforms are indicated. g, Representative immunohistochemical staining of GBM intracranial xerographs for Ki-67 and ALDH1A3. Immunocompromised mice were injected with 5 × 105 patient-derived 1763 glioma cells and treated 1 month later with FG1059 (10 mg kg−1 tail vein injection five times every 3 d). Mice were sacrificed 3 d after the last injections to obtain brain slices (n = 5 mice in the control group and n = 3 mice in the FG1059-treated group). h, Quantification of immunohistochemical staining for Ki-67 in the samples as in g, using the German immunohistochemical scoring (GIS) system (unpaired two-tailed t-test). i, Same as in h, but for ALDH1A3 staining (unpaired two-tailed t-test). j, Top, experimental workflow used to study the effect of FG1059 on tumour growth in vivo. Bottom, Kaplan–Meier survival curves of mice intracranially injected with 5 × 105 1763 glioma cells and subsequently intraperitoneally injected with FG1059 or a solvent (n = 6 and n = 8 mice per treatment and control group, respectively; log-rank test). k, Proposed molecular mechanism of GBM spatial phenotype regulation by RPL22L1 isoforms. All quantitative data are means ± s.d.
To confirm that FG1059 affects RPL22L1 due to the inhibition of SRSF protein phosphorylation, we studied the phosphoproteome of patient-derived GBM neurospheres treated with FG1059 for 0, 3, 6 and 12 h (Supplementary Table 9). LC-MS/MS and subsequent principal component analysis revealed that the differences observed at 3 h were distinct and more substantial than those at the 6- and 12-h time points (Fig. 8d). This result is not surprising, since at the early time point we are probably observing a direct effect of FG1059, while longer incubation may induce activation of the compensatory signalling pathways and apoptotic cascades. Enrichment analysis revealed that at 3 h of FG109 treatment the most significant differences were observed for pre-mRNA processing and RNA splicing pathways and were detected in proteins containing an RNA recognition motif, including the SRSF4 protein (Extended Data Fig. 9h). Interestingly, at 12 h we observed an enrichment in Rho-GTPases and PH-like domain-containing proteins, which contribute to multiple processes such as proliferation, invasion and secretion70. Altogether, these results indicate that the primary process affected by FG1059 is RNA splicing and SRSF4 is one of its downstream targets.
Clk kinases control the function of SRSF proteins by regulating their intracellular localization71. Using a plasmid encoding the RFP-SRSF4 fusion protein, we found that treatment with FG1059 caused SRSF4 translocation from the nucleus to the cytoplasm, therefore preventing the splicing-related activity of SRSF4 (Fig. 8e).
Next, we tested the effect of FG1059 on the viability of patient-derived GBM neurospheres and normal human astrocytes in vitro. FG1059 was able to eliminate GBM cells with a half-maximal inhibitory concentration of <500 nM, whereas its cytotoxicity against normal astrocytes was noticeably weaker (Fig. 8f). Interestingly, RPL22L1b-expressing cells appeared to be more resistant to FG1059 than cells producing the RPL22L1a isoform. This observation is in good agreement with the fact that RPL22L1b increases RNA splicing efficiency and therefore may better protect GBM cells from FG1059, which impairs spliceosome functions. To confirm this hypothesis, we demonstrated that exogenous RPL22L1b enhances cell survival upon FG1059 treatment (Extended Data Fig. 10a).
Next, we studied possible synergy between FG1059 and temozolomide (Extended Data Fig. 10b). According to our data, FG1059 decreased the viability of GBM cells treated with temozolomide nearly twofold. Similar results were obtained when FG1059 was applied together with pladienolide B (Extended Data Fig. 10c). Finally, we evaluated the antitumor efficacy of FG1059 in vivo. This experiment revealed that intraperitoneal injection of FG1059 substantially decreased the percentage of proliferating GBM cells (Ki-67+) as well as putative core-like glioma stem cells (ALDH1A3+) within the patient-derived GBM xenografts (Fig. 8g,h,i) and significantly prolonged animal survival (Fig. 8j).
Altogether, our data show that FG1059 inhibits SRSF4 phosphorylation, impairs PRL22L1 splicing and ultimately attenuates GBM growth in vitro and in vivo. However, it is noteworthy that FG1059 probably affects a broad range of downstream targets and only a part of its effect is mediated by RPL22L1.
Discussion
In this study, we highlight a molecular mechanism for the emergence of intratumoral spatial heterogeneity within GBM. Acidification of the central zone of the tumour that normally occurs during GBM development serves as a driving force of this process. It upregulates expression of the splicing factor SRSF4, which switches splicing of the ribosomal protein RPL22L1 towards production of the RPL22L1b isoform. This isoform is incapable of binding to the ribosome due to the altered structure of the C terminus, although it retains the ability to interact directly or indirectly with other RNA molecules. Thus, RPL22L1b mediates degradation of the long non-coding RNA MALAT1. Diminished levels of MALAT1, together with a lowered intensity of translation and TP53 downregulation resulting from the lack of the normal RPL22L1a isoform, provokes a more aggressive GBM phenotype that is characteristic of the cells in the tumour core (Fig. 8k). As a result, phenotypically distinct GBM cell populations arise in different regions of the tumour, ultimately increasing GBM heterogeneity and promoting tumour malignancy.
Currently, the functions of RPL22L1 and its paralogue RPL22 remain poorly investigated. These proteins, being mutually exclusive components of the 60S ribosomal complex, participate in CAP-independent translation of certain mRNAs72,73. It was also demonstrated that RPL22 can inhibit RPL22L1 translation in normal murine cells via binding to the hairpin on RPL22L1 mRNA74. Interestingly, according to other authors75 and our analysis of the TCGA database (Extended Data Fig. 10d), RPL22 functions as a tumour suppressor and is therefore weakly expressed in cancer cells. Moreover, RPL22 deletion is the most frequent mutation among the ribosomal genes that was observed across multiple tumour types. Consistently, Rpl22 haploinsufficiency promotes the development and dissemination of murine T cell lymphoma76. One may suggest that the pro-oncogenic effect of RPL22 deletion can be associated with the compensatory increase of RPL22L1.
According to our results, the amount of RPL22L1 within ribosomes is regulated by alternative splicing of its pre-mRNA. This may lead to the formation of ribosomes with different protein composition and supposedly different specialization towards translated mRNA molecules. In recent years, ribosome specialization has become the topic of numerous studies, yet to the best of our knowledge only one group has demonstrated the role of alternative splicing in this process: Brumwell et al.77 reported that hypoxia alters RPS24 pre-mRNA splicing, giving rise to an additional isoform that may be incorporated into ribosomes. We have previously demonstrated an important role of splicing during phenotypic transformations of GBM45. However, in that work, we restricted our analysis to genes whose products were alternatively spliced. Our present results indicate that splicing might have a much broader effect on the proteome by altering the intensity of translation.
Our data also show an important extra-ribosomal function of ribosomal proteins. Herein, we have shown that long non-coding RNAs might be a downstream target of RPL22L1b, in particular MALAT1. The MALAT1 gene is frequently overexpressed in solid tumours and is associated with unfavourable outcomes78. However, in gliomas, the role of MALAT1 is controversial. On the one hand, MALAT1 abundance in GBM is lower compared with the normal brain and its overexpression correlates with prolonged patient survival79. On the other hand, knockdown of MALAT1 has also been shown to prevent glioma progression80. Our results are likely to reconcile this discrepancy. We speculate that the role of MALAT1 might be different depending on the stage of GBM development. During the early stages of tumour progression, MALAT1 can be pro-oncogenic, whereas in the course of formation of harsh microenvironmental conditions in the GBM core, MALAT1 may emerge as a tumour suppressor and therefore GBM cells may tend to decrease its expression.
We have shown the critical importance of low-pH conditions (acidosis) in the switching of RPL22L1 splicing. Acidosis represents an important hallmark of malignant tumours and probably occurs early during cancer development81. To adapt to low-pH microenvironments, cancer cells employ highly complicated mechanisms. Thus, several lysosomal proteins can be transported to the cell surface, thereby protecting the plasma membrane81. In addition, acidosis induces metabolic changes promoting low glucose consumption and reprogramming lipid metabolism82. In agreement with these observations are our findings of the differential influence of RPL22L1 isoforms on GBM cell metabolism. By shifting the balance between the isoforms, cells can optimize the energy supply. Furthermore, acidification is known to upregulate stemness-related proteins, as well as tumour-initiating and self-renewal potential4,83,84. We provide a mechanistic link between acidosis and the stemness of GBM cells. Thus, acidosis causes a decrease in ribosomal RPL22L1a and retardation of translation. Low ribosome activity has been shown to be a characteristic feature of tumour stem cells that allows them to promote the translation of a subpool of mRNAs essential for the maintenance of stemness while inhibiting differentiation76. Therefore, alternative splicing of RPL22L1 pre-mRNA under acidic conditions may promote stemness via two mechanisms: by lowering the rate of translation and by MALAT1 degradation. It is important to mention that the basal RPL22L1a/RPL22L1b ratio can differ greatly depending on the patient or tissue even if the cells were cultivated under the same pH conditions. Therefore, we can conclude that there are probably multiple intracellular factors that affect the splicing of RPL22L1. However, regardless of the RPL22L1a/RPL22L1b ratio, low pH promoted the production of the RPL22L1b isoform in all of our experiments, indicating that it might be one of the key microenvironmental factors controlling RPL22L1 splicing.
To confirm the therapeutic relevance of RPL22L1 splicing in GBM, we tested the small molecular agent FG1059, which inhibits Clk kinases and dysregulates RPL22L1 splicing. A number of small-molecule inhibitors of splicing have been described, including E7107, spliceostatin A, pladienolide B, H3B-8800, FD-895, GEX1, SM08502 and T-025 (refs. 64,85,86). Most of these compounds target the core spliceosomal protein SF3B1 (ref. 85) and are generally toxic for non-malignant cells due to the critical importance of splicing machinery for normal cell physiology87. Therefore, identifying inhibitors that target tumour-specific splicing-related proteins would be more promising for clinical application. In contrast, agents that increase the efficacy of translation and, as a result, induce differentiation of cancer stem cells have been proposed for the treatment of malignant tumours76. Our results further highlight that the compounds designed to target specific mechanisms of RNA splicing and/or oncogenic variants of ribosomes emerge as a promising therapeutic approach as they may prevent the appearance of certain cancer cell populations and thereby decrease tumour heterogeneity and facilitate treatment with standard anticancer drugs.
In conclusion, we have shown that the protein composition of ribosomes can be dictated by the tumour microenvironment and that alternative RNA splicing is involved in the regulation of ribosomal functions. One target of alternative splicing is RPL22L1. Both isoforms of this protein perform functions vital for GBM cells, although their effects are implemented in a spatially/temporarily different manner. We believe that our findings shed light on the mechanisms of GBM cell plasticity and might explain the basis for the emergence of an extremely heterogeneous pool of cancer cells from the initially homogenous glioma tumour. However, multiple questions remained to be addressed. Thus, more studies are needed to understand how SRSF4 is regulated by the acidic milieu. Furthermore, the mechanism whereby RPL22L1 isoforms affect their bound RNA partners is obscure. Finally, our bioinformatics analysis revealed a very strong correlation between RPL22L1 splicing and the survival of patients with kidney renal clear cell carcinoma and adrenocortical carcinoma. This observation presumes that RPL22L1 can be generally important for the biology of tumours beyond GBM.
Methods
Our research complies with all of the relevant ethical regulations and guidelines. The collection of glioma specimens was conducted under the approved Institutional Review Board protocols of the University of California Los Angeles (approved protocol 11-000432), N.N. Burdenko National Medical Research Center of Neurosurgery, Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry and First Affiliated Hospital of Xi’an Jiaotong University. Informed consent was obtained from the patients in all cases. No compensation was provided. Tissues were processed in the research laboratories after de-identification of the samples. All of the animal experiments were carried out under Institutional Animal Care and Use Committee protocols at the Shemyakin-Ovchinnikov Institute of Bioogranic Chemistry (approved protocol 266/220) and University of California Los Angeles (approved protocol ARC-1993-285) according to National Institutes of Health guidelines.
Amine CEST–SAGE–EPI and anatomic MRI acquisition
Simultaneous acquisition of pH–weighted amine chemical exchange saturation transfer (CEST) contrast and oxygen-sensitive R2′ mapping was performed using the CEST–spin-and-gradient echo (SAGE)–echoplanar imaging (EPI) pulse sequence, as previously described88. This sequence consists of a CEST saturation pulse train of three 100-ms Gaussian pulses with peak amplitude B1 = 6 μT and a SAGE–EPI readout consisting of two gradient echoes with echo times of 14.0 and 34.1 ms, one asymmetric spin echo with an echo time of 58.0 ms and one spin echo with an echo time of 92.4 ms. Additional acquisition parameters include a repetition time of >10,000 ms, a field of view of 217 mm × 240 mm, a matrix size of 116 × 128, a slice thickness of 4.0 mm (with no inter-slice gap), a partial Fourier encoding 6/8, GRAPPA = 3 and a bandwidth of 1,628 Hz per pixel. A total of 29 z-spectral points were acquired at offset frequencies from −3.5 to −2.5 parts per million (ppm), −0.3 to +0.3 ppm and +2.5 to +3.5 ppm, all with respect to the water proton resonance frequency. An additional reference (S0) scan was obtained with four averages using identical parameters and no saturation pulses. All MRI images were acquired on 3 T MR scanners (Prisma or Skyra; Siemens Healthcare). The total acquisition time for CEST–SAGE–EPI was 7 min and 30 s, benchmarked on a 3 T Siemens Prisma MR scanner (software versions VE11A–C). In addition to the CEST scan before contrast administration, all patients received anatomic images according to the standardized brain tumour imaging protocol89.
CEST–SAGE–EPI data post-processing
All CEST–SAGE–EPI and CEST–EPI images were motion corrected using affine transformation (mcflirt, FSL; FMRIB) and B0 correction via a z-spectra-based k-means clustering and Lorentzian-fitting algorithm90. Following motion and B0 correction, the integral of width of 0.4 ppm was quantified around both the −3.0 and +3.0 ppm spectral points (−3.2 to −2.8 ppm and +2.8 to +3.2 ppm, respectively). These data points were combined with the S0 image to calculate the magnetization transfer ratio with asymmetric analysis (MTRasym) at 3.0 ppm–a measure related to pH91–as defined using the equation: MTRasym(3.0 ppm) = S(−3.0 ppm)/S0 − S(+3.0 ppm)/S0, where S(ω) is the amount of bulk water signal available after the saturation pulse with offset frequency ω and S0 is the signal available without application of radiofrequency (RF) saturation. For the CEST–SAGE–EPI data, the average MTRasym at 3.0 ppm was calculated by averaging the first (echo time = 14.0 ms) and second (echo time = 34.1 ms) gradient echoes to increase the available signal-to-noise ratio.
Estimates of transverse relaxation rates R2, R2* and R2′ = R2* − R2, which is proportional to oxygen extraction, were obtained by solving a system of Bloch equations as detailed previously88. All post-processing was performed using MATLAB (release 2017b; MathWorks). All of the resulting maps were registered to high-resolution post-contrast T1-weighted images for subsequent analyses.
The correlation coefficient between the DAPI and anti-RPL22L1 staining patterns was calculated using ImageJ software (version 1.53).
Glioma sphere lines
Glioma specimens were mechanically dissociated into pieces with a diameter of 1–3 mm. The samples were then treated with trypsin for 30 min at 37 °C to obtain single cells. Cell suspensions were run through Lympholyte-H (Cedarlane Laboratories) separation to remove red blood cells and debris according to the manufacturer’s specifications. Established sphere lines were cultivated in serum-free neurosphere media for no longer than 30 passages. For all sphere lines, short tandem repeat (STR) analysis was performed to confirm cell identity. Detailed characteristics of the sphere lines and STR analysis data are presented in Supplementary Table 10.
Cell culture, transfection and lentiviral transduction
All cells were cultivated at 37°C under a humidified atmosphere with 5% CO2. GBM neurospheres were propagated in DMEM/F12 medium (Sigma–Aldrich) containing 2% MACS NeuroBrew-21 supplement (Miltenyi Biotec), 1% penicillin–streptomycin solution (Thermo Fisher Scientific), 20 ng ml−1 basic fibroblast growth factor (bFGF; Sigma–Aldrich) and 20 ng ml−1 epidermal growth factor (EGF; Sigma–Aldrich). bFGF and EGF were added twice a week and the culture medium was changed every 5–10 d. Spheres were dissociated using StemPro Accutase (Thermo Fisher Scientific).
Phoenix-GP (ATCC; CRL-3215), A549 (ATCC; CCL-185), DU145 (ATCC; HTB-81), HepG2 (ATCC; HB-8065) and RD (ATCC; CCL-136) cells were cultivated in DMEM/F12 medium (Sigma–Aldrich) containing 10% foetal bovine serum, 1% penicillin–streptomycin solution and 1 mM sodium pyruvate. Cells were passaged 2–3 times per week until 80–90% confluency was reached. Normal human astrocytes (Gibco; N7805200) were cultivated in DMEM medium (Thermo Fisher Scientific) containing 10% foetal bovine serum, 1% N-2 Supplement (Thermo Fisher Scientific) and 1% penicillin–streptomycin solution (Thermo Fisher Scientific). For all cell lines, Mycoplasma contamination was tested with a LookOut Mycoplasma PCR Detection Kit (Sigma–Aldrich). For microscopy purposes, cells were plated on a Lab-Tek II Chambered Coverglass (Nunc).
Plasmid DNA was purified from bacterial cells with a HiSpeed Plasmid Midi Kit (Qiagen). Transfection was performed using Lipofectamine LTX Reagent (Invitrogen) according to the manufacturer’s protocol. Cells were examined 48 h after transfection. Lentiviruses were produced as described previously92. Briefly, Phoenix-GP cells were co-transfected with pCDH-EF1-MCS-IRES-Puro (System Biosciences) or pLKO-1 (Sigma–Aldrich) lentiviral vectors and two packaging plasmids, psPAX2 and pMD2.G (Addgene). The media was changed the following day and lentivirus-containing supernatants were harvested 72 h later and filtered through a 0.45-μm syringe filter. On the day of transduction, GBM spheres were dissociated into single cells with StemPro Accutase (Thermo Fisher Scientific) and incubated with viral supernatants for 24 hr in the presence of 8 μg ml−1 polybrene (EMD Millipore). Two days after infection, transduced cells were selected with 1 mg ml−1 puromycin (Sigma–Aldrich) for 3 d.
Cell viability assay
GBM neurospheres were dissociated using StemPro Accutase (Thermo Fisher Scientific) and counted using a Countess II Automated Cell Counter (Thermo Fisher Scientific) with Trypan Blue reagent (Thermo Fisher Scientific). Cells were plated in a 96-well plate at 6,000 cells per well in 100 μl medium. The next day, the corresponding drug was added to each well in 50 μl medium. Five days later, the cell viability was assessed with alamarBlue reagent (Thermo Fisher Scientific). The fluorescence was measured using a Fusion α-FP HT Universal Microplate Analyzer (PerkinElmer) with an excitation filter for 535 nm and an emission filter for 620 nm.
Cell proliferation assay
Cells were plated in a 96-well plate at a density of 6,000 cells per well in 150 μl medium. The number of cells was assessed using alamarBlue reagent (Thermo Fisher Scientific). The fluorescence was measured using a Fusion α-FP HT Universal Microplate Analyzer (PerkinElmer) with an excitation filter for 535 nm and an emission filter for 620 nm. The measurements were taken on days 1, 4, 6 and 8.
Neurosphere formation assay
Cells were seeded into 96-well plates at 100, 50, 25, 12, 6, 3, 1.5 and 0.75 cells per well in 200 μl medium. After 14 d, the number of wells without spheres was counted. The data were analysed using Extreme Limiting Dilution Analysis software (https://bioinf.wehi.edu.au/software/elda/index.html)93.
Wound healing assay
A wound healing assay was performed as described previously68. Briefly, glioma spheres were dissociated into single cells using Accutase and plated on a laminin-coated 6-well plate at 2.5 × 106 cells per well. The next day, cell cultures were scratched using a 200-μl pipette tip. After 48 h, the cells were then observed under a microscope. The assay was performed three times.
Flow cytometry and cell sorting
For CD133 staining, glioma spheres were dissociated into single cells and stained with anti-CD133/2-APC antibody (Miltenyi Biotec) according to the manufacturer’s protocol. Cells without primary antibody were used as a negative control. For apoptosis, assay cells were stained with a CellEvent Caspase-3/7 Green Flow Cytometry Assay Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. After staining, the samples were analysed by NovoCyte Flow Cytometer (ACEA biosciences). Data were collected using NovoExpress Software (version 1.3.0) and processed with FlowJo 10 software (version X.0.7).
For cell sorting, cells with overexpression of GFP or RFP fusion proteins were dissociated with Accutase 2 d after the transfection, resuspended in 1 ml ice-cold phosphate-buffered saline (PBS) and sorted on a FACSAria III instrument (BD Biosciences). To obtain populations of cells at different stages of the cell cycle, neurospheres were dissociated and fixed with a mixture of ethanol/glacial acetic acid (95/5) as described previously94. Next, the cells were stained with propidium iodide95 and sorted on a FACSAria III instrument (BD Biosciences). Collected cell fractions were used for RNA purification. The samples were gated by forward scatter height and side scatter height to distinguish cells from debris (Extended Data Fig. 10e).
Cell line STR analysis
Services were provided by the company GORDIZ (https://gordiz.ru/). Genomic DNA was purified using a DNeasy Blood & Tissue Kit (Qiagen) according to the manufacturer’s protocol. PCR was performed and products were separated by capillary electrophoresis using an AB 3730 DNA Analyzer (Applied Biosystems). Samples were run on a 36-cm capillary array (Applied Biosystems). For data analysis, electropherograms were analysed from the .fsa files and allelic values were assigned using GeneMarker software version 1.85 (Soft Genetics). Alleles were matched to the STR profile recorded with German Collection of Microorganisms and Cell Cultures (DMSZ).
Immunofluorescence staining
Immunofluorescence staining was performed as described previously68. Briefly, cells were fixed with 4% paraformaldehyde (PFA), permeabilized with 0.2% Triton X, blocked with 1% bovine serum albumin and incubated with corresponding primary antibodies for 1 h at room temperature. Next, cells were incubated with Alexa Fluor-conjugated secondary antibody (Thermo Fisher Scientific) for 1 h at room temperature. Images were captured with the fluorescence microscope Nikon Diaphot 300 or the confocal microscope Leica DMIRE2.
Immunohistochemistry
Immunohistochemistry was performed as previously described68. Briefly, tumours embedded in paraffin blocks were deparaffinized and hydrated through an ethanol series. After microwave antigen retrieval in DakoCytomation target retrieval solution pH 6 (Dako), slides were incubated in 0.3% hydrogen peroxide solution in methanol for 15 min at room temperature to inhibit internal peroxidase activity. Next, the samples were blocked with serum-free protein block solution (Dako) and incubated with the corresponding primary antibodies overnight at 4°C. The next day, slides were stained with EnVision+ System HRP-Labelled Polymer (Dako) and visualized with a DAB peroxidase substrate kit (Vector Laboratories).
In vivo intracranial xenograft tumour model
Six-week-old female NOD SCID mice (Prkdcscid; The Jackson Laboratory) were used for intracranial implantation of patient-derived GBM cells. The mice were housed in groups of five animals per cage and had access to autoclaved water and pelleted feed. The cage environment was enriched with a mouse house. The mice were kept at a standard temperature of 22±2 °C and a relative humidity of 55% (45–70%) in a 12 h light/12 h dark cycle (lights on from 06:00 to 18:00). GBM neurospheres were dissociated using StemPro Accutase (Thermo Fisher Scientific) and counted using a Countess II Automated Cell Counter (Thermo Fisher Scientific) with Trypan Blue reagent (Thermo Fisher Scientific). The cell suspension (5 × 105 cells in 3 μl PBS) was injected into the brains of mice as previously described68. When neuropathological symptoms developed, mice were sacrificed and perfused with ice-cold PBS and 4% PFA. Mouse brains were dissected, fixed in 4% PFA for 24 h and then transferred to 10% formalin.
In vivo drug treatments
FG1059 was dissolved in a dimethyl sulfoxide:ethanol:water mixture (1:1:3). On day 15 after transplantation of GBM cells, mice were intraperitoneally injected with FG109 solution at a dose of 10 mg kg−1. Injections were repeated every 3 d, five times in total. Kaplan–Meier curves were generated from the mouse survival data.
HPG protein synthesis assay
GBM cells were labelled for nascent protein synthesis using a Click-iT HPG Alexa Fluor 488 Protein Synthesis Assay Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Briefly, spheres were dissociated into single cells and incubated in methionine-free medium (Thermo Fisher Scientific) for 30 min and subsequently incubated with HPG for 1 h. During pre-incubation, the control samples were treated with 100 μg ml−1 cycloheximide (Abcam) to inhibit de novo protein synthesis. The cells were fixed with 4% PFA in PBS for 15 min and permeabilized with 0.25% Triton X-100 in PBS for 20 min, followed by two washes with 3% bovine serum albumin. Cells were then stained using Alexa Fluor azide. After extensive washing, the cells were analysed by flow cytometry or imaged using a fluorescence microscope.
Ribosome isolation
Ribosome isolation was performed according to the previously published protocol96 with brief modifications. Cells were lysed with 1 ml lysis buffer (20 mM Tris pH 7.4, 150 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol (DTT), 0,5% Triton X-100, 0.1% sodium deoxycholate and 24 U ml−1 DNAse I (Thermo Fisher Scientific)) and incubated on ice for 20 min. The cell lysate was centrifuged for 10 min at 20,000g and 4 °C. After centrifugation, the supernatant was mixed with 9 ml lysis buffer and loaded slowly over 1 ml 1 M sucrose solution (20 mM Tris pH 7.5, 150 mM KCl, 10 mM MgCl2, 1 mM DTT, 0.5% Triton X-100, 0.1% sodium deoxycholate and 1 M sucrose) and subsequently centrifuged at 210,000g and 4 °C for 3 h. The pellet was washed carefully with 1 ml lysis buffer and then resuspended in 70 μl lysis buffer.
Polysome fractionation
GBM neurospheres were dissociated using StemPro Accutase (Thermo Fisher Scientific) and incubated in ice-cold PBS containing 100 μg ml−1 cycloheximide (Abcam) for 10 min. PBS was removed and 750 μl ice-cold polysome lysis buffer (20 mM Tris-HCl pH 7.5, 250 mM NaCl, 1.5 mM MgCl2, 1 mM DTT, 0.5% Triton X-100, 100 μg ml−1 cycloheximide (Abcam) and 20 U ml−1 DNAse I (Thermo Fisher Scientific)) was added. After 10 min incubation on ice, the cells were centrifuged for 10 min at 20,000g and 4 °C. The supernatant was loaded on a 10–60% (m/v) sucrose density gradient containing 20 mM Tris-HCl pH 7.5, 250 mM NaCl, 15 mM MgCl2 and 1 mM DTT and centrifuged at 210,000g and 4 °C for 3 h. After centrifugation, gradients were manually fractionated from the top to the bottom, followed by measurement of the optical density at 260 nm for each sample using a spectrophotometer.
Recombinant protein expression and purification
To produce His-tagged RPL22L1a and RPL22L1b proteins in bacterial cells, BL21-CodonPlus (DE3)-RIL E. coli cells (Agilent) were transformed with pET28-RPL22L1a or pET28-RPL22L1b plasmids. Bacteria were incubated at 37 °C in a shaker until the optical density at a wavelength of 600 nm reached 0.7. Next, isopropylthio-β-galactoside was added to a final concentration of 1 mM and the bacteria were incubated for an additional 4 h at 37 °C. Then, 200 ml media with bacteria was centrifuged for 15 min at 6,000g and 4 °C and the pellet was resuspended in 12 ml lysis buffer B (100 mM NaCl, 100 mM NaH2PO4, 10 mM Tris-HCl pH 8.0, 8 M urea and 0.5 mM DTT) and incubated for 1 h at room temperature with slow agitation. Next, the solution was centrifuged for 15 min at 20,000g and 4 °C and the supernatant was incubated with 2 ml Ni-NTA resin (Qiagen) for 1 h under constant agitation. The suspension was transferred to a column and washed with 20 ml buffer B and 10 ml buffer C (the same as buffer B but at pH 6.3). Bounded proteins were eluted with buffer D (buffer C with 250 mM imidazole) and dialysed overnight against PBS with 0.5 mM DTT. The purity of the obtained protein was assessed by electrophoresis and subsequent Coomassie blue staining.
Recombinant protein pull-down assay
GBM cells were incubated on ice for 30 min with lysis buffer (20 mM Tris pH 7.5, 150 mM KCl, 10 mM imidazole, 0.5 mM DTT, 0.5% NP-40, 0.1% sodium deoxycholate and protease inhibitor cocktail) and subsequently centrifuged for 15 min at 20,000g and 4 °C. Cleared lysate was used for the pull down with immobilized recombinant protein.
Recombinant His-tagged RPL22L1 isoforms were immobilized on 30 μl HisPur Ni-NTA Magnetic Beads (Thermo Fisher Scientific) according to the manufacturer’s protocol. The beads were washed three times with PBS and incubated for 1 h with cleared lysate under constant agitation. The beads were washed once with lysis buffer and three times with PBS. Bounded proteins were eluted with 300 mM imidazole in PBS and subjected to subsequent LC-MS/MS analysis.
Immunoprecipitation
GBM cells overexpressing Fc-RPL22L1a, Fc-RPL22L1b or Fc-tagged control protein were incubated on ice for 30 min with lysis buffer (20 mM Tris pH 7.5, 150 mM KCl, 10 mM MgCl2, 0.5 mM DTT, 0.5% NP-40, 0.1% sodium deoxycholate and protease inhibitor cocktail) and subsequently centrifuged for 15 min at 20,000g and 4 °C. Cleared lysate was used for immunoprecipitation.
To obtain a protein complex, 30 μl Protein A/G Magnetic Beads (Thermo Fisher Scientific) were incubated with GBM cell lysate for 1 h at room temperature under constant agitation. The beads were washed once with lysis buffer and three times with PBS. Bounded proteins were eluted with urea elution buffer (8 M urea, 2 M thiourea and 10 mM Tris-HCl pH 8) and subjected to subsequent LC-MS/MS analysis.
Western blotting
Cells were lysed for 30 min on ice in RIPA buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 1 mM ethylenediaminetetraacetic acid and 5% glycerol) containing 1% protease inhibitor cocktail (Sigma–Aldrich). Lysates were pre-cleaned by centrifugation at 20,000g and 4 °C for 15 min. The protein concentration was determined by bicinchoninic acid assay (Thermo Fisher Scientific). Equal amounts of protein lysates (10 μg per lane) were fractionated by Novex WedgeWell 4–20% Tris-Glycine Gel (Thermo Fisher Scientific) and transferred to a polyvinylidene difluoride membrane (Millipore). The membrane was subsequently blocked with 5% Blotting Grade Blocker Non Fat Dry Milk (Bio-Rad) for 1 h, incubated with the corresponding primary antibody overnight and next incubated with peroxidase-conjugated secondary antibodies (Thermo Fisher Scientific) for 1 h. Staining was visualized with a SuperSignal West Pico PLUS chemiluminescent substrate (Thermo Fisher Scientific) using ImageQuant LAS 500 (GE Healthcare). β-Actin served as a loading control. Unprocessed images of all western blots are provided as source data.
Antibody dilutions
The following antibodies were used in the study (supplier; catalogue number; experiment type; host; dilution): anti-ALDH3A2 (Abcam; ab250728; western blot; rabbit; 1:1,000), anti-CD109 (Santa Cruz Biotechnology; sc-271085; immunohistochemistry; mouse; 1:100), anti-CD133/2-APC (Miltenyi Biotec; 130-113-746; fluorescence-activated cell sorting (FACS); mouse; 1:20), anti-CDK5 (Abcam; ab40773; western blot; rabbit; 1:1,000), anti-RPL22 (Abcam; ab229458; western blot; rabbit; 1:1,000), anti-RPL22L1 (Abcam; ab188299; immunofluorescence; rabbit; 1:100), anti-RPL22L1 (Abcam; ab234792; western blot; rabbit; 1:500), anti-RPN2 (Novus Biologicals; 80836; western blot; rabbit; 1:1,000), anti-SRSF4 (Abnova; H00006429-A01; immunofluorescence; mouse; 1:200), anti-TP53 (Abcam; 131442; western blot; rabbit; 1:1,000), anti-β Actin (Cell Signaling Technology; 3700; western blot; mouse; 1:2,000), Goat anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (Thermo Fisher Scientific; A-11034; immunofluorescence; goat; 1:600), Goat anti-Rabbit IgG (H + L) Secondary Antibody, HRP (Thermo Fisher Scientific; 31460; western blot; goat; 1:5,000) and Goat anti-Mouse IgG (H + L) Cross-Adsorbed Secondary Antibody, HRP (Thermo Fisher Scientific; G-21040; western blot; goat; 1:5,000).
SILAC
SILAC was performed using a Pierce SILAC Protein Quantitation Kit (Trypsin) (Thermo Fisher Scientific). DMEM/F12 for the SILAC media (Thermo Fisher Scientific), supplemented with 2% MACS NeuroBrew-21 (Miltenyi Biotec), 20 ng ml−1 EGF (Sigma–Aldrich) and 20 ng ml−1 bFGF (Sigma–Aldrich), was used instead of DMEM. Cells were grown in SILAC medium lacking arginine and lysine and supplemented with light lysine and arginine or with heavy lysine and arginine (13C6 l-lysine-HCl and 13C6,15N4 l-arginine-HCl) according to the manufacturer’s protocol. Cells were kept in labelling medium for at least five cell passages to achieve >98% labelling.
Peptide preparation for LC-MS/MS analysis
Cells from the SILAC experiment were collected, washed three times with PBS and then lysed using 4% SDS and 50 mM Tris-HCl pH 8 with protease inhibitors, as described previously97. The cell lysates were subjected to sonication on ice (three cycles of 10-s on/off pulses with a 30% amplitude). Disulfide bonds were reduced with 5 mM DTT for 30 min at room temperature. Afterwards, iodoacetamide was added to a final concentration of 10 mM. The samples were incubated at room temperature for 20 min in the dark and the reaction was stopped by the addition of 5 mM DTT. After precipitation of proteins using methanol/chloroform, the semi-dry protein pellet was dissolved in 50 μl 8 M urea, 2 M thiourea and 10 mM Tris-HCl pH 8. The protein concentration was then measured using a Quick Start Bradford protein assay kit (Bio-Rad) and samples were diluted with ammonium bicarbonate solution to reduce the urea concentration to 2 M. Trypsin (Promega) was added at a ratio of 1:100 and the samples were incubated for 14 h at 37 °C.
Eluates from immunoprecipitations and fresh frozen tissue samples were first resuspended in solubilization buffer (8 M urea, 2 M thiourea and 10 mM Tris-HCl pH 8) and subsequently incubated with 5 mM DTT at room temperature for 40 min. Then proteins were alkylated with 10 mM iodoacetamide at room temperature for 20 min in the dark. Alkylated samples were diluted by the addition of 50 mM ammonium bicarbonate solution at a ratio of 1:4. Next, trypsin (0.01 μg per 1 μg protein) was added and the samples were incubated at 37 °C for 14 h.
After 14 h, the reaction was stopped by the addition of formic acid to a final concentration of 5%. The tryptic peptides were desalted using styrenedivinylbenzene–reverse phase sulfonate membrane (Sigma–Aldrich), vacuum dried and stored at −80 °C before LC-MS/MS analysis. Before LC-MS/MS analysis, the samples were redissolved in 5% acetonitrile with 0.1% trifluoroacetic acid solution and sonicated.
Phosphopeptide preparation for LC-MS/MS analysis
To enrich for phosphorylated peptides, we applied the High-Select TiO2 Phosphopeptide Enrichment Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. Briefly, 10 × 106 GBM cells were washed three times with PBS, lysed in buffer containing 4% SDS and 50 mM Tris-HCl pH 8.0 with phosphatase inhibitor cocktails 1 and 2 (Sigma–Aldrich) and subsequently incubated at 60 °C for 30 min. The cell lysates were subjected to sonication on ice (three cycles of 10-s on/off pulses with a 30% amplitude). Disulfide bonds were reduced with 5 mM DTT for 30 min at room temperature. Afterwards, iodoacetamide was added to a final concentration of 10 mM. The samples were incubated at room temperature for 20 min in the dark and the reaction was stopped by the addition of 5 mM DTT. After precipitation of the proteins using methanol/chloroform, the semi-dry protein pellets were dissolved in 50 μl 8 M urea, 2 M thiourea and 10 mM Tris-HCl pH 8. Then, the protein amounts were adjusted to 0.5 mg using the Quick Start Bradford Protein Assay (Bio-Rad) and the samples were diluted with ammonium bicarbonate solution to reduce the urea concentration to 2 M. Trypsin (Promega) was added at a ratio of 1:100 and the samples were incubated for 14 h at 37 °C. After 14 h, the reaction was stopped by the addition of formic acid to a final concentration of 5%. The tryptic peptides were desalted using styrenedivinylbenzene–reverse phase sulfonate membrane (Sigma–Aldrich) and vacuum dried. Then, a phosphoenrichment procedure was performed according to the manufacturer’s protocol. Before LC-MS/MS analysis, the samples were redissolved in 5% acetonitrile with 0.1% trifluoroacetic acid solution and sonicated.
LC-MS/MS analysis
Proteomic analysis was performed using a Q Exactive HF mass spectrometer. Samples were loaded onto 50-cm columns packed in house with a C18 3 μM Acclaim PepMap 100 (Thermo Fisher Scientific), with an UltiMate 3000 Nano LC System (Thermo Fisher Scientific) coupled to the mass spectrometer (Q Exactive HF; Thermo Fisher Scientific). Peptides were loaded onto a column thermostatically controlled at 40 °C in buffer A (0.2% formic acid) and eluted with a linear (120 min) gradient of 4–55% buffer B (0.1% formic acid and 80% acetonitrile) in buffer A at a flow rate of 350 nl min−1. Mass spectrometric data were stored during automatic switching between MS1 scans and up to 15 MS/MS scans (topN method). The target value for MS1 scanning was set to 3 × 106 in the range 300–1,200 m/z with a maximum ion injection time of 60 ms and a resolution of 60,000. The precursor ions were isolated at a window width of 1.4 m/z and a fixed first mass of 1,000 m/z. Precursor ions were fragmented by high-energy dissociation in a C-trap with a normalized collision energy of 28 eV. MS/MS scans were saved with a resolution of 15,000 at 400 m/z and at a value of 1 × 105 for target ions in the range of 200–2,000 m/z with a maximum ion injection time of 30 ms.
Protein identification and quantification
Raw LC-MS/MS data from the Q Exactive HF mass spectrometer were converted to .mgf peak lists with MSConvert (version 3). For this procedure, we used the following parameters: ‘−mgf–filter peakPicking true [1,2]’. For thorough protein identification in samples from immunoprecipitations, the generated peak lists were searched with the MASCOT (version 2.5.1) and X! Tandem (ALANINE, 2017.02.01) search engines against the UniProt human protein knowledgebase with the concatenated reverse decoy dataset. The precursor and fragment mass tolerance were set at 20 ppm and 0.04 Da, respectively. Database-searching parameters included the following: tryptic digestion with one possible missed cleavage, static modification for carbamidomethyl (C) and dynamic/flexible modifications for oxidation (M). For X! Tandem, we also selected parameters that allowed a quick check for protein N-terminal residue acetylation, peptide N-terminal glutamine ammonia loss or peptide N-terminal glutamic acid water loss. The resulting files were submitted to Scaffold 4 software (version 4.0.7) for validation and meta-analysis. We used the local false discovery rate scoring algorithm with standard experiment-wide protein grouping. For the evaluation of peptide and protein hits, a false discovery rate of 5% was selected for both. False positive identifications were based on reverse database analysis. We also set protein annotation preferences in Scaffold to highlight Swiss-Prot accessions among others in protein groups.
For protein quantification in samples of cancer cells, raw LC-MS/MS data were analysed with MaxQuant (version 1.6.10.43) against the UniProt human protein knowledgebase. For this procedure, we used the following parameters: Orbitrap instrument type, tryptic digestion with two possible missed cleavages, fixed modification for carbamidomethyl (C), variable modifications for oxidation (M) and acetyl (protein N-term) and LFQ label-free quantification. For the phosphoproteome analysis, we also added serine/threonine/tyrosine phosphorylation as a variable modification. For SILAC analyses, we also included the following parameters: Arg10 and Lys6 as heavy labels and a maximum of three labelled amino acids.
Plasmid construction
The DNA fragments encoding RPL22L1 isoforms were amplified from complementary DNA (cDNA) using the PCR technique and the primer pair BglII-RPL22L1/RPL22L1-SalI and cloned into the BamHI/SalI sites of the pET28a+ vector (Addgene). Sanger sequencing of the plasmids purified from 12 bacterial colonies was performed. All sequences were fully consistent with our RNA-seq data and revealed the presence of only two isoforms of RPL22L1. The resulting plasmids were named pET28-RPL22L1a and pET28-RPL22L1b, respectively. Next, the DNA fragments encoding RPL22L1a and RPL22L1b were amplified from pET28-RPL22L1a and pET28-RPL22L1b plasmids using the PCR technique and the primer pair EcoRI-RPL22L1/RPL22L1-BamHI and cloned into the EcoRI/BamHI sites of the pCDH-EF1-MCS-IRES-Puro vector (System Biosciences) to generate the pCDH-RPL22L1a and pCDH-RPL22L1b plasmids. The DNA fragment encoding Fc-fragment was amplified from the pFUSE-hIgG1-Fc1 vector (InvivoGen) using the PCR technique and the primer pair NheI-ATG-Fc/Fc-GS-EcoRI and cloned into the NheI/EcoRI sites of the pCDH-RPL22L1a or pCDH-RPL22L1b plasmids to generate the pCDH-Fc-RPL22L1a and pCDH-Fc-RPL22L1b plasmids, respectively. The DNA fragments encoding SRSF1, SRSF2, SRSF3 and SRSF4 were amplified from cDNA using the PCR technique and the primer pairs EcoRI-SRSF1/SRSF1-SalI, EcoRI-SRSF2/SRSF2-BamHI, EcoRI-SRSF3/SRSF2-SalI and EcoRI-SRSF4/SRSF4-SalI, respectively. The obtained DNA was cloned into the EcoRI/SalI or EcoRI/BamHI (for SRSF2) sites of the pTagRFP-C vector (Evrogen) to generate the pTagRFP-SRSF1, pTagRFP-SRSF2, pTagRFP-SRSF3 and pTagRFP-SRSF4 plasmids. The DNA fragment encoding SRSF4 was amplified from the pTagRFP-SRSF4 plasmid using the PCR technique and the primer pair EcoRI-SRSF4(2)/SRSF4-BamHI(2) and cloned into the EcoRI/BamHI sites of the pCDH-Fc-RPL22L1a plasmid to generate the pCDH-Fc-SRSF4 plasmid. The RPL22L1 PCR fragment amplified from the cDNA of cells treated with FG1059 was purified using the QIAquick Gel Extraction Kit (Qiagen) according to the manufacturer’s manual and ligated into the pKAN-T vector (Evrogen) to generate the pKAN-T-RPL22L1 plasmid that was subjected to Sanger sequencing. In all cases, the absence of unwanted mutations in the inserts and vector–insert boundaries was verified by sequencing.
RNA-IP
RPl22L1a, RPL22L1b and SRSF4 protein–RNA complexes were purified from glioma spheres stably expressing Fc-RPl22L1a, Fc-RPL22L1b or Fc-SRSF4 with the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (EMD Millipore) according to the manufacturer’s protocol. The eluted RNA was subjected to RNA-seq or qRT-PCR analysis.
RNA stability assay
To analyse the degradation rate of MALAT1, GBM spheres were incubated with 10 μg ml−1 actinomycin D (PanReac AppliChem) for 0, 3, 6 or 9 h. RNA was subsequently purified using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions and analysed using qRT-PCR.
RNA isolation and PCR
RNA was isolated using the RNeasy Mini Kit (Qiagen) or QIAzol Lysis Reagent (Qiagen) (for cells obtained after cell sorting). The RNA concentration was determined using a Nanodrop One C Spectrophotometer (Thermo Fisher Scientific). cDNA was synthesized using Maxima H Minus cDNA Synthesis Master Mix (Thermo Fisher Scientific) according to the manufacturer’s protocol. qPCR was performed on a LightCycler 96 (Roche) with qPCRmix-HS SYBR reagent (Evrogen). The cycling conditions were 95 °C for 150 s and then 45 cycles of 95 °C for 20 s, 60 °C for 20 s and 72 °C 20 s. The data were collected using LightCycler Software (version 4.1). The primer specificity was confirmed by visualizing DNA on an agarose gel following PCR. GAPDH or 18S RNA was used as an internal control. The primer sequences are shown in Supplementary Table 11. Unprocessed images of all DNA gels are shown in the source data.
RNA-seq
Libraries of cDNA for paired-end sequencing were prepared using the TruSeq Stranded mRNA-Seq Library Preparation Kit (Illumina) according to the manufacturer’s protocol. Samples were sequenced with an lllumina HiSeq 2500 system (Illumina) and 125 base pair paired-end reads were generated.
Analysis of gene expression and alternative splicing
RNA-seq reads were trimmed for quality and paired reads were cropped using Trimmomatic (version 0.35). Trimmed RNA-seq reads were quantified against Homo sapiens GRCh38 genome annotation at the transcript level using Salmon (version 1.4). The results were aggregated to the gene level using the R package tximport. Differentially expressed genes were identified using the R package DESeq2. To compare samples from different studies, we conducted surrogate variable analysis with ComBat using the R package sva to correct data from intra-study batch effects. Principal component analysis was used to compare differences between all samples using the FactoMineR package in R. Enrichment analysis by GSVA was performed using the GSVA package in R against the hallmark, ontology and canonical pathways gene sets from the Molecular Signatures Database (MSigDB). The gsva function in the GSVA package returns an enrichment score for each sample for the given signature. To compare enrichment scores between interesting groups for the given signature, we ran a t-test.
For alternative splicing analysis, trimmed RNA-seq reads were mapped to Homo Sapiens GRCh38 genome annotation using STAR. The STAR software (version 2.7.9) was used in a two-pass mode, where the first pass was used to identify non-annotated junctions in the input data. The genome index was generated using annotated junctions and the common set of junctions that were found during the first pass mode. During the second pass, we re-ran all of the samples with this genome. To compare splicing events between samples, we ran the rMATS splicing tool using the following parameters recommended by the developers: -c 0.0001 and -novel SS1 (multi-align reads were ignored as a default). Minor splicing differences were filtered out by thresholds of a false discovery rate of <0.05 and an IncLevelDifference value of >5%. Both annotated and unannotated splicing events were analysed.
RNA-IP sequencing and ribo-seq data analysis
To increase the read quality before mapping, RNA-IP sequencing data were trimmed using Trimmomatic (version 0.6.6). Trimmed RNA-seq reads were mapped against Homo Sapiens GRCh38 genome annotation using STAR (version 2.7.9a) according to recommendations from STAR manual 2.4.0.1 with the following parameters: the maximum number of mismatches for paired reads was 4% of the read length, the maximum number of multiple alignments allowed for a read was ten, non-canonical junctions were removed, the minimum number of allowed splice overhangs was eight for unannotated junctions and one for annotated junctions, the minimum intron length was 20, the maximum intron length was 1,000,000 and the number of spurious junctions was reduced. The transcript expression signal was quantified using RSEM.
Gene set enrichment analysis
We downloaded gene expression data and clinical information for the TCGA GBM cohort using the TCGAbiolinks R/Bioconductor package. Patients with GBM were divided into high and low RPL22L1b expression groups according to the percentage spliced in index value of the corresponding alternative 3′ splice site of the RPL22L1 gene. Next, the low-pH response pathway gene set for GSEA was created based on the previously published GSE116035 dataset42. Finally, GSEA was performed using clusterProfiler software.
Phylogenetic conservation analysis
The pattern of conservation of the RPL22L1 alternative splicing region spanning exon 2 and exon 3 was explored and visualized with UCSC Genome Browser98. The genomic sequence on chromosome 3 at base pairs 170,868,007–170,868,400 (GRCh38/hg38) was analysed with 100 vertebrates Basewise Conservation by PhyloP (phyloP100way track)99. Multiple sequence alignment of several vertebrates was performed using the Multiz aligner100.
Statistics and reproducibility
All of the data are presented as means ± s.d. The number of replicates for each experiment is stated in the figure caption and always refers to the independent biological replicates. Statistical differences between two groups were evaluated by unpaired two-tailed t-test unless mentioned otherwise in the figure caption. The statistical significance of Kaplan–Meier survival plots was determined by log-rank analysis. A statistical correlation was performed to calculate the regression coefficient of determination (R2) value and Pearson’s correlation coefficient. One-way analysis of variance was utilized in comparisons of more than two groups, following Dunnett’s/Tukey’s post-test. The differences between paired edge and core tissue samples were compared using the Wilcoxon signed rank-sum test. Statistical analysis was performed using Prism 6 (GraphPad Software). P < 0.05 was considered to be statistically significant. No statistical method was used to predetermine sample size. This (that is, the number of mice) was determined on the basis of our previous studies3,45. No blinding or randomization was used. No samples, mice or data points were excluded from the reported analyses. Unless otherwise noted in the figure caption, all of the DNA and protein gels, western blots, FACS data and microscopy images are representative of three biologically independent experiments.
Extended Data
Extended Data Fig. 1 ∣. Proteomic and transcriptomic intratumoral heterogeneity of glioblastoma.
a, The principal component analysis of LC-MS/MS (left panel) and RNAseq (right panels) data obtained from GBM sphere lines derived from core (n = 4 different clones) and edge (n = 3 different clones) of the 1051 tumor. b, Correlation analysis of protein-to-RNA abundance in GBM sphere lines as in ‘a’. c, Correlation of protein composition of ribosomes with mRNA levels of the corresponding ribosomal genes. d, Correlation of protein composition of ribosomes with the differences in pre-mRNA splicing of the corresponding ribosomal genes. Proteins that are differentially included into ribosomes of GBM sphere lines with edge and core phenotype were determined by SILAC LC-MS/MS. Differences in mRNA levels and in pre-mRNA splicing were determined by RNA sequencing of the corresponding cells. Correlation coefficient, trend line and the most differentially present proteins/mRNAs are indicated. e, RT-PCR analysis RPL22L1 splicing in GBM cells from n = 3 different patients. PCR products were separated using PAAG electrophoresis. f, Sashimi plots demonstrating differences in splicing of RPL22L1 between GBM sphere lines with edge (157, 011, 025) and core (083, 028, 006) phenotype (cells isolated from n = 6 different patients). g, RT-PCR analysis of RPL22L1 splicing in different human cell lines. h, The principal component analysis of RNAseq data obtained from GBM sphere lines used in this study. Red – previously characterized GBM spheres with core phenotype, blue - previously characterized GBM spheres with edge phenotype, gray – sphere lines established in this study (n = 14 different patients). i, RT-PCR analysis of RPL22L1 isoform abundance in 267 cells cultivated for 5 days in normal (pH 7.4) or acidified (pH 6.0) medium. NMD inhibitor (NMDI-14) was added to a final concentration of 5 μM 24 hours before cells were collected for RNA purification.
Extended Data Fig. 2 ∣. Expression of RPL22L1 isoforms in human cancers.
a, Relative expression of RPL22L1 isoforms and SETD4 (previously described NMD target36) in Hela cells depleted for different NMD factors (experiment was performed in n = 3 biological replicates; data are mean ± SD). Raw RNAseq data were obtained from GSE152437. b, Riboseq and RNAseq read densities for RPL22L1 in U251 GBM cells (data were obtained from GSE141013 dataset). c, Polysome profile of 083 GBM spheres (upper panel) and RT-PCR analysis of RPL22L1 splicing (lower panel). d, Mass spectrometry identification of the peptide related to RPL22L1b isoform (data were obtained from PXD004023 dataset). e, Western blotting analysis of GBM spheres with edge (157, 025) and core (022, 083) phenotypes with different antibodies against RPL22L1.
Extended Data Fig. 3 ∣. Representative immunofluorescent staining of GBM cells predominantly expressing RPL22L1a (001, 157, 025) or RPL22L1b (006, 022, 083) isoform with antibodies against N-terminal part of RPL22L1.
The red dotted line indicates areas of DAPI staining (borders of the nucleus).
Extended Data Fig. 4 ∣. Prognostic value of RPL22L1 isoforms.
a, Kaplan-Meier curve showing the overall survival of Kidney Renal Clear Cell Carcinoma (n = 532 different patients), Adrenocortical carcinoma (n = 79 different patients) and Uveal Melanoma (n = 80 different patients) patients subdivided based on the splicing of RPL22L1 (log-rank test). RNAseq data were obtained from TCGA database. b, Splicing of RPL22L1 in GBM patients with wild type and mutated IDH1 (left panel); in patients younger and older than 50 years (middle panel) and in patients with high and low level of MGMT promoter methylation (right panel). 14 different probes were used to assess methylation status, none of them showed statistically significant differences in RPL22L1 splicing. Date were obtained from TCGA database (n = 154 different patients; the line in the box is the median, the up and low of the box are the first and third quartiles, and the whiskers extend to 10th and 90th percentiles respectively). Higher psi values indicate higher percentage of RPL22L1b isoform. c, FACS analysis of cell cycle distribution of 267 cells stained with propidium iodide. Populations that were collected by cell sorting are indicated. d, qRT-PCR analysis of Ki67 expression in cell populations collected as in ‘c’. e, RT-PCR analysis of RPL22L1 splicing in cell populations collected as in ‘c’ (experiment was performed in n = 3 biological replicates). All quantitative data are mean ± SD, n.s. – non significant.
Extended Data Fig. 5 ∣. RPL22L1b facilitates GBM growth in low pH conditions.
a, RT-PCR analysis of RPL22L1 splicing in 001, 011 and 022 GBM spheres that were cultivated for 5 days in normal (pH 7.4) or acidified (pH 6.0) medium. b, Immunofluorescent staining of different areas of GBM tumor tissues from patient 1051 for RPL22L1 (green) and DNA (blue). Yellow and red arrows indicate cells with nuclear and cytoplasmic localization of RPL22L1 respectively. c, Colocalization analysis of RPL22L1 and DAPI staining for the same tumor areas as in ‘b’. Pearson’s R value is indicated. d, Representative immunofluorescent staining of 022 GBM cells overexpressing RPL22L1a, RPL22L1b or an empty vector with antibodies against N-terminal part of RPL22L1. e, FACS analysis of caspase 3/7 activity and SYTOX staining of 157 glioma spheres that were transduced with lentiviruses encoding RPL22L1a, RPL22L1b or an empty vector (control) and cultivated in normal (pH 7.4) or acidified medium (pH 6.4) for 8 days.
Extended Data Fig. 6 ∣. Differential splicing of RPL22L1 promotes GBM intratumoral heterogeneity.
a, Forward vs. side scatter plot of 020 GBM spheres overexpressing RPL22L1a or RPL22L1b. Cell size was determined using Countess II Automated Cell Counter (experiment was performed in n = 4 biological replicates). b, Representative confocal images of 157 glioma spheres that were first transduced with lentiviruses encoding RPL22L1a or RPL22L1b and subsequently transduced with lentiviruses encoding GFP or RFP. Next cells overexpressing RPL22L1a + RFP were mixed with cells overexpressing RPL22L1b + GFP and imaged 4 days later. c, Representative images of wound healing assay with 157 cells stably expressing RPL22L1a, RPL22L1b or an empty vector. d, Representative IHC staining for CD109 of mouse brain sections obtained 3 months after intracranial injection of 3·105 luciferase labeled 1051 glioma spheres overexpressing RPL22L1a or RPL22L1b (n = 2 mice per group). e, PAAG electrophoresis of recombinant RPL22L1a (a), or RPL22L1b (b), that were purified from E. coli. f, Enrichment analysis of proteins that were bound to recombinant His-tagged RPL22L1a (upper panel) or RPL22L1b (lower panel) and subsequently identified by LC-MS/MS.
Extended Data Fig. 7 ∣. Molecular functions RPL22L1 isoforms in GBM cells.
a, Enrichment analysis of proteins that were co-purified with Fc-tagged RPL22L1a (lower panel) or RPL22L1b (upper panel) and subsequently identified by LC-MS/MS. b, Enrichment analysis of mRNAs that were co-purified with Fc-tagged RPL22L1a (left panel) or RPL22L1b (right panel) and subsequently identified by RNA sequencing. c, KEGG database GSVA analysis of RNA sequencing data obtained from 157 cells overexpressing RPL22L1a, RPL22L1b or an empty vector (experiment was performed in two biological replicates). d, Enrichment analysis of proteins that were differentially present in 157 cells overexpressing RPL22L1a or RPL22L1b isoform as determined by SILAC LC-MS/MS. e, Enrichment analysis of alternative splicing events related to exon skipping (upper panel) or mutually exclusive exon inclusion (lover panel) detected in 157 cells stably expressing RPL22L1b compared to the control cells.
Extended Data Fig. 8 ∣. RPL22L1 isoforms regulate pre-mRNA splicing and mRNA translation in GBM cells.
a, Sashimi plots demonstrating differences in splicing of MDM4 between 157 GBM spheres overexpressing RPL22L1a, RPL22L1b or an empty vector. Number of reeds that confirms exon skipping or exon inclusion is indicated. b, qRT-PCR analysis of MALAT1 RNA stability in 1079, 022 and 011 GBM spheres overexpressing RPL22L1a or RPL22L1b isoforms. Cells were cultivated for 6 hours with Actinomycin D at final concentration of 10 μg/ml (experiment was performed in n = 3 biological replicates). c, Kaplan-Meier curve showing the disease-free survival of glioma patients subdivided based on the MALAT1 expression level (n = 338 different patients, log-rank test). Data were obtained from TCGA database. d, Representative fluorescence images demonstrating L-homopropargylglycine (HPG) incorporation into newly synthesized proteins in 157, 022 and 267 GBM sphere lines. HPG was detected by Alexa Fluor488 azide (green). Nucleus were visualized by DAPI (blue). Cells pretreated for 30 min with cycloheximide (100 μg/ml) were used as a control. e, Polysome profiles of 1079 GBM spheres stably expressing RPL22L1a or RPL22L1b (experiment was performed in n = 2 biological replicates). f, RNA-IP enrichment profiles of RPL22L1a, RPL22L1b and a control protein for TP53 gene. 157 cells overexpressing Fc-tagged proteins were used for the experiment. g, Correlation of ALDH3A2 and ALDH1A3 expression levels in glioma. Data were obtained from REMBRANDT (n = 354 different patients; left panel) or TCGA (n = 671 different patients; right panel) databases. All quantitative data are mean ± SD.
Extended Data Fig. 9 ∣. SRSF4 regulates RPL22L1 splicing.
a, SRSF4 CLIP-tag enrichment profile in Rpl22l1 RNA sequence. Data were obtained from E-MTAB-747 dataset. b, qRT-PCR analysis of RNAs that were co-purified with Fc-tagged SRSF4 (red), or a control protein (blue) with primers for RPL22L1 exon 3, RPL22L1 3’UTR or RPL22 (experiment was performed in n = 3 biological replicates). c, SRSF4 expression level in different regions of GBM tumor (n = 122 RNA samples obtained from n = 10 different patients, one-way ANOVA test, following Dunnett’s/Tukey’s posttest). Data were obtained from IVY GAP database. The line in the box is the median, the up and low of the box are the first and third quartiles, and the whiskers extend to 10th and 90th percentiles respectively. d, qRT-PCR analysis of SRSF4 expression in 006, 030 and 157 GBM cells transduced with lentiviruses encoding non-target shRNA (shNT) or two different shRNAs against SRSF4 (48shSRSF4 and 49shSRSF4). e, Quantification of RPL22L1 splicing differences in 030 (upper) and 157 (lower) GBM cells transduced with lentiviruses as in ‘d’ (experiment was performed in n = 3 biological replicates). Higher psi values indicate higher percentage of RPL22L1b isoform. f, Kaplan-Meier curve showing the overall survival of glioblastoma patients (n = 179) subdivided based on the SRSF4 expression level (log-rank test). Data were obtained from REMBRANDT database. g, RT-PCR analysis of RPL22L1 splicing in 011 and 1079 glioma spheres that were left untreated (Un) or treated for 24 hours with CMP3a (10 μM), EY404 (10 μM), Fg1059 (10 μM), Cisplatin (10 μM), TMZ (100 μM) or Pladienolide B (1 μM). h, Enrichment analysis of the proteins that were differentially phosphorylated (more than 10 fold differences) in 157 GBM cells after 3 and 12 hours of incubation with 3 μM of FG1059 as opposed to untreated cells (experiment was performed in n = 2 biological replicates). Reactom and InterPro databases were used to calculate enrichment. All quantitative data are mean ± SD.
Extended Data Fig. 10 ∣. FG1059 induce apoptosis of GBM cells.
a, In vitro cell viability assay of 022, 157 and 1079 GBM spheres stably expressing RPL22L1a, RPL22L1b or an empty vector. Cells were treated with different concentrations of FG1059 for 5 days (experiment was performed in n = 6 biological replicates; data are mean ± SD). b, FACS analysis of caspase 3/7 activity and SYTOX staining of 157 cells treated with DMSO; 0.2 μM of FG1059; 20 μM of TMZ or with both compounds simultaneously for 24 hours. c, FACS analysis of caspase 3/7 activity and SYTOX staining of 157 cells treated with DMSO; 50 nM of FG1059; 50 nM of Pladienolide B or with both compounds simultaneously for 24 hours. d, Kaplan-Meier curve showing the overall survival of glioma (n = 509 different patients; left panel) and glioblastoma (n = 152 different patients; right panel) patients subdivided based on the RPL22 expression level (log-rank test). Data were obtained from TCGA database. e, Flow cytometry gating used for apoptosis assay (left panel) and for CD133 or HPG staining (right panel).
Supplementary Material
Acknowledgements
We thank all of our respective laboratory colleagues for helpful discussion. We are grateful to M. A. Nakano and Y. D. Nakano for input on manuscript writing and editing. This work was supported by grants from the Ministry of Science and Higher Education of the Russian Federation (for cell culture, 075-15-2020-773 to M.S.P. and M.I.S. and for LC-MS/MS analyses, 075-15-2019-1669 to V.O.S., G.P.A., K.S.A. and P.V.S.), Russian Science Foundation (for bioinformatics analyses, 22-15-00462 to V.O.S., K.S.A., G.P.A., P.V.S. and A.N.K., for ribosome fractionation, 21-64-00006 to O.A.D. and for cell culture work, 22-14-00234 to M.I.S.), Russian Foundation for Basic Research (19-34-90193 to K.S.A., 20-04-00804 to M.S.P. and 19-34-90102 to T.D.L.), Russian Federation (MD-4501.2021.1.4 to M.S.P.), Scholarship of the Russian Federation (SP-3815.2021.4 to V.O.S.), National Natural Science Foundation of China (81802502 to J.W.), National Cancer Institute (P50CA211015 and R01CA241927 to H.I.K., R01CA270027 and R21CA223757 to B.M.E. and R01CA201402 to I.N.), National Institute of Neurological Disorders and Stroke (R01NS121617 to H.I.K., R01NS107071 to I.N. and R01NS113631 to I.N.), Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (to H.I.K.), Adelson Medical Research Foundation (to H.I.K.), American Cancer Society RSG-15-003-01-CCE (to B.M.E.) and Department of Defense (CA200290 to B.M.E.). We also thank the Center for Precision Genome Editing and Genetic Technologies for Biomedicine of Federal Research and Clinical Center of Physical-Chemical Medicine for RNA-seq. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Competing interests
The authors declare no competing interests.
Extended data is available for this paper at https://doi.org/10.1038/s41556-022-00994-w.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41556-022-00994-w.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41556-022-00994-w.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All of the proteomics data that were obtained during this study are presented in Supplementary Tables 1, 2, 4, 5, 7 and 9. The raw mass spectrometry data have been deposited in ProteomeXchange with the primary accession codes PXD035849, PXD035767 and PXD035855. All of the RNA-seq data that were obtained during this study are presented in Supplementary Tables 3, 6 and 8. The raw RNA-seq data of GBM neurospheres overexpressing different isoforms of RPL22L1 protein or an empty vector as a control have been deposited in the Gene Expression Omnibus (GEO) database under accession code GSE180465. The RNA-IP sequencing data for RNA that interacts with Fc-tagged RPL22L1a, RPL22L1b or a control protein have been deposited in the GEO database under accession code GSE180464. Previously published RNA-seq data for GBM neurospheres and GBM tissue isolated from the different regions of the tumours that were re-analysed here are available from the GEO database under accession codes PRJNA344648 and GSE153746. iCLIP data for the SRSF4 protein that were re-analysed here are available from the Array Express database under accession code E-MTAB-747. Gene expression data from the Ivy Glioblastoma Atlas Project database (https://glioblastoma.alleninstitute.org/), gene expression and survival data from the Repository for Molecular Brain Neoplasia Data (GSE108474 and GSE68848) and gene expression, DNA methylation, survival and phenotype data from The Cancer Genome Atlas database (https://tcga-data.nci.nih.gov/tcga/) were used in this study. Source data are provided with this paper.
References
- 1.Van Linde ME et al. Treatment outcome of patients with recurrent glioblastoma multiforme: a retrospective multicenter analysis. J. Neurooncol 135, 183–192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puchalski RB et al. An anatomic transcriptional atlas of human glioblastoma. Science 360, 660–663 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastola S et al. Glioma-initiating cells at tumor edge gain signals from tumor core cells to promote their malignancy. Nat. Commun 11, 4660 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hjelmeland AB et al. Acidic stress promotes a glioma stem cell phenotype. Cell Death Differ. 18, 829–840 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaupel P, Kallinowski F & Okunieff P Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a review. Cancer Res. 49, 6449–6465 (1989). [PubMed] [Google Scholar]
- 6.Ozawa T et al. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell 26, 288–300 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bastola S et al. Tumor edge architecture in glioblastoma is constructed by inter-cellular signals from vascular endothelial cells. Preprint at bioRxiv 10.1101/2020.10.12.335091 (2020). [DOI] [Google Scholar]
- 8.Phillips HS et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9, 157–173 (2006). [DOI] [PubMed] [Google Scholar]
- 9.De Aquino PF et al. A time-based and intratumoral proteomic assessment of a recurrent glioblastoma multiforme. Front. Oncol 6, 183 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yanovich-Arad G et al. Proteogenomics of glioblastoma associates molecular patterns with survival. Preprint at medRxiv 10.1101/2020.04.28.20083501 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Gularyan SK et al. Investigation of inter- and intratumoral heterogeneity of glioblastoma using TOF-SIMS. Mol. Cell. Proteom 19, 960–970 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heiland DH et al. The integrative metabolomic–transcriptomic landscape of glioblastome multiforme. Oncotarget 8, 49178–49190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemée JM et al. Integration of transcriptome and proteome profiles in glioblastoma: looking for the missing link. BMC Mol. Biol 19, 13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruggero D Translational control in cancer etiology. Cold Spring Harb. Perspect. Biol 5, a012336 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujii K, Shi Z, Zhulyn O, Denans N & Barna M Pervasive translational regulation of the cell signalling circuitry underlies mammalian development. Nat. Commun 8, 14443 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondrashov N et al. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145, 383–397 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guimaraes JC & Zavolan M Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biol. 17, 236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ebright RY et al. Deregulation of ribosomal protein expression and translation promotes breast cancer metastasis. Science 367, 1468–1473 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panda A et al. Tissue- and development-stage-specific mRNA and heterogeneous CNV signatures of human ribosomal proteins in normal and cancer samples. Nucleic Acids Res. 48, 7079–7098 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yong WH et al. Ribosomal proteins RPS11 and RPS20, two stress-response markers of glioblastoma stem cells, are novel predictors of poor prognosis in glioblastoma patients. PLoS ONE 10, e0141334 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi Z et al. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol. Cell 67, 71–83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y et al. Control of hematopoietic stem cell emergence by antagonistic functions of ribosomal protein paralogs. Dev. Cell 24, 411–425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y et al. Ribosomal proteins Rpl22 and Rpl22l1 control morphogenesis by regulating pre-mRNA splicing. Cell Rep. 18, 545–556 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang Z et al. Identification of candidate diagnostic and prognostic biomarkers for human prostate cancer: RPL22L1 and RPS21. Med. Oncol 36, 56 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Ma J, Jing X, Chen Z, Duan Z & Zhang Y MiR-361-5p decreases the tumorigenicity of epithelial ovarian cancer cells by targeting at RPL22L1 and c-Met signaling. Int. J. Clin. Exp. Pathol 11, 2588–2596 (2018). [PMC free article] [PubMed] [Google Scholar]
- 26.Rao S et al. RPL22L1 induction in colorectal cancer is associated with poor prognosis and 5-FU resistance. PLoS ONE 14, e0222392 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bell JL et al. Identification of RNA-binding proteins as targetable putative oncogenes in neuroblastoma. Int. J. Mol. Sci 21, 5098 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Leary MN et al. The ribosomal protein Rpl22 controls ribosome composition by directly repressing expression of its own paralog, Rpl22l1. PLoS Genet. 9, e1003708 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minata M et al. Phenotypic plasticity of invasive edge glioma stem-like cells in response to ionizing radiation. Cell Rep. 26, 1893–1905 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pollard KS, Hubisz MJ, Rosenbloom KR & Siepel A Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurosaki T & Maquat LE Nonsense-mediated mRNA decay in humans at a glance. J. Cell Sci 129, 461–467 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato H & Singer RH Cellular variability of nonsense-mediated mRNA decay. Nat. Commun 12, 7203 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoek TA et al. Single-molecule imaging uncovers rules governing nonsense-mediated mRNA decay. Mol. Cell 75, 324–339 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lindeboom RG, Supek F & Lehner B The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat. Genet 48, 1112–1118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin L et al. Identification and characterization of small molecules that inhibit nonsense-mediated RNA decay and suppress nonsense p53 mutations. Cancer Res. 74, 3104–3113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Longman D et al. Identification of a localized nonsense-mediated decay pathway at the endoplasmic reticulum. Genes Dev. 34, 1075–1088 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michel AM et al. GWIPS-viz: development of a ribo-seq genome browser. Nucleic Acids Res. 42, D859–D864 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choudhary S et al. Genomic analyses of early responses to radiation inglioblastoma reveal new alterations at transcription, splicing, and translation levels. Sci. Rep 10, 8979 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pearson H et al. MHC class I-associated peptides derive from selective regions of the human genome. J. Clin. Invest 126, 4690–4701 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brennan CW et al. The somatic genomic landscape of glioblastoma. Cell 155, 462–477 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ellingson BM et al. pH-weighted molecular MRI in human traumatic brain injury (TBI) using amine proton chemical exchange saturation transfer echoplanar imaging (CEST EPI). Neuroimage Clin. 22, 101736 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corbet C et al. TGFβ2-induced formation of lipid droplets supports acidosis-driven EMT and the metastatic spreading of cancer cells. Nat. Commun 11, 454 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sonabend AM et al. The transcriptional regulatory network of proneural glioma determines the genetic alterations selected during tumor progression. Cancer Res. 74, 1440–1451 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saito N et al. A high Notch pathway activation predicts response to γ secretase inhibitors in proneural subtype of glioma tumor-initiating cells. Stem Cells 32, 301–312 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pavlyukov MS et al. Apoptotic cell-derived extracellular vesicles promote malignancy of glioblastoma via intercellular transfer of splicing factors. Cancer Cell 34, 119–135 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, Zhang H, Xu S, Liu Z & Cheng Q The adaptive transition of glioblastoma stem cells and its implications on treatments. Signal Transduct. Target. Ther 6, 124 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moreb JS Aldehyde dehydrogenase as a marker for stem cells. Curr. Stem Cell Res. Ther 3, 237–246 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Julian LM & Stanford WL Organelle cooperation in stem cell fate: lysosomes as emerging regulators of cell identity. Front. Cell Dev. Biol 8, 591 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu G et al. Inhibition of SF3B1 by molecules targeting the spliceosome results in massive aberrant exon skipping. RNA 24, 1056–1066 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dewaele M et al. Antisense oligonucleotide-mediated MDM4 exon 6 skipping impairs tumor growth. J. Clin. Invest 126, 68–84 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCown PJ, Wang MC, Jaeger L & Brown JA Secondary structural model of human MALAT1 reveals multiple structure–function relationships. Int. J. Mol. Sci 20, 5610 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tripathi V et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baspinar Y, Elmaci I, Ozpinar A & Altinoz MA Long non-coding RNA MALAT1 as a key target in pathogenesis of glioblastoma. Janus faces or Achilles’ heal? Gene 739, 144518 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Latorre E et al. The ribonucleic complex HuR–MALAT1 represses CD133 expression and suppresses epithelial–mesenchymal transition in breast cancer. Cancer Res. 76, 2626–2636 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Mao P et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc. Natl Acad. Sci. USA 110, 8644–8649 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh K et al. c-MYC regulates mRNA translation efficiency and start-site selection in lymphoma. J. Exp. Med 216, 1509–1524 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pospísek M & Valásek L Polysome profile analysis–yeast. Methods Enzymol. 530, 173–181 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Lee JH, Kim HS, Lee SJ & Kim KT Stabilization and activation of p53 induced by Cdk5 contributes to neuronal cell death. J. Cell Sci 120, 2259–2271 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Chang PM et al. Transcriptome analysis and prognosis of ALDH isoforms in human cancer. Sci. Rep 8, 2713 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ono M et al. The expression and clinical significance of ribophorin II (RPN2) in human breast cancer. Pathol. Int 65, 301–308 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Yin Z et al. Identification of ALDH3A2 as a novel prognostic biomarker in gastric adenocarcinoma using integrated bioinformatics analysis. BMC Cancer 20, 1062 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duan JJ, Cai J, Guo YF, Bian XW & Yu SC ALDH1A3, a metabolic target for cancer diagnosis and therapy. Int J. Cancer 139, 965–975 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Änkö ML et al. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 13, R17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Iwai K et al. Anti-tumor efficacy of a novel CLK inhibitor via targeting RNA splicing and MYC-dependent vulnerability. EMBO Mol. Med 10, e8289 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Naro C et al. The centrosomal kinase NEK2 is a novel splicing factor kinase involved in cell survival. Nucleic Acids Res. 42, 3218–3227 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Esvan YJ et al. Discovery of pyrido[3,4-g]quinazoline derivatives as CMGC family protein kinase inhibitors: design, synthesis, inhibitory potency and X-ray co-crystal structure. Eur. J. Med. Chem 118, 170–177 (2016). [DOI] [PubMed] [Google Scholar]
- 67.Tazarki H et al. New pyrido[3,4-g]quinazoline derivatives as CLK1 and DYRK1A inhibitors: synthesis, biological evaluation and binding mode analysis. Eur. J. Med. Chem 166, 304–317 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Wang J et al. Spatiotemporal dynamics of intra-tumoral dependence on NEK2–EZH2 signaling in glioblastoma cancer progression. Preprint at bioRxiv 10.1101/2020.12.01.405696 (2020). [DOI] [Google Scholar]
- 69.Anufrieva KS et al. Therapy-induced stress response is associated with downregulation of pre-mRNA splicing in cancer cells. Genome Med. 10, 49 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Etienne-Manneville S & Hall A Rho GTPases in cell biology. Nature 420, 629–635 (2002). [DOI] [PubMed] [Google Scholar]
- 71.Zhou Z & Fu XD Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 122, 191–207 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lavergne JP, Conquet F, Reboud JP & Reboud AM Role of acidic phosphoproteins in the partial reconstitution of the active 60 S ribosomal subunit. FEBS Lett. 216, 83–88 (1987). [DOI] [PubMed] [Google Scholar]
- 73.Kim SJ & Strich R Rpl22 is required for IME1 mRNA translation and meiotic induction in S. cerevisiae. Cell Div. 11, 10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.O’Leary MN et al. The ribosomal protein Rpl22 controls ribosome composition by directly repressing expression of its own paralog, Rpl22l1. PLoS Genet. 9, e1003708 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goudarzi KM & Lindström MS Role of ribosomal protein mutations in tumor development (Review). Int. J. Oncol 48, 1313–1324 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bastide A & David A The ribosome, (slow) beating heart of cancer (stem) cell. Oncogenesis 7, 34 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brumwell A, Fell L, Obress L & Uniacke J Hypoxia influences polysome distribution of human ribosomal protein S12 and alternative splicing of ribosomal protein mRNAs. RNA 26, 361–371 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gutschner T, Hämmerle M & Diederichs S MALAT1—a paradigm for long noncoding RNA function in cancer. J. Mol. Med. (Berl.) 91, 791–801 (2013). [DOI] [PubMed] [Google Scholar]
- 79.Han Y et al. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by downregulation of MMP2 and inactivation of ERK/MAPK signaling. Cell Death Dis. 7, e2123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li Z et al. Long non-coding RNA MALAT1 promotes proliferation and suppresses apoptosis of glioma cells through derepressing Rap1B by sponging miR-101. J. Neurooncol 134, 19–28 (2017). [DOI] [PubMed] [Google Scholar]
- 81.Damaghi M et al. Chronic acidosis in the tumour microenvironment selects for overexpression of LAMP2 in the plasma membrane. Nat. Commun 6, 8752 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Corbet C et al. Acidosis drives the reprogramming of fatty acid metabolism in cancer cells through changes in mitochondrial and histone acetylation. Cell Metab. 24, 311–323 (2016). [DOI] [PubMed] [Google Scholar]
- 83.Andreucci E et al. The acidic tumor microenvironment drives a stem-like phenotype in melanoma cells. J. Mol. Med. (Berl.) 98, 1431–1446 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hu P et al. Acidosis enhances the self-renewal and mitochondrial respiration of stem cell-like glioma cells through CYP24A1-mediated reduction of vitamin D. Cell Death Dis. 10, 25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tan DQ et al. PRMT5 modulates splicing for genome integrity and preserves proteostasis of hematopoietic stem cells. Cell Rep. 26, 2316–2328 (2019). [DOI] [PubMed] [Google Scholar]
- 86.Tam BY et al. The CLK inhibitor SM08502 induces anti-tumor activity and reduces Wnt pathway gene expression in gastrointestinal cancer models. Cancer Lett. 473, 186–197 (2020). [DOI] [PubMed] [Google Scholar]
- 87.Eskens FA et al. Phase I pharmacokinetic and pharmacodynamic study of the first-in-class spliceosome inhibitor E7107 in patients with advanced solid tumors. Clin. Cancer Res 19, 6296–6304 (2013). [DOI] [PubMed] [Google Scholar]
- 88.Harris RJ et al. Simultaneous pH–sensitive and oxygen-sensitive MRI of human gliomas at 3 T using multi-echo amine proton chemical exchange saturation transfer spin-and-gradient echo echo-planar imaging (CEST-SAGE-EPI). Magn. Reson. Med 80, 1962–1978 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ellingson BM et al. Consensus recommendations for a standardized brain tumor imaging protocol in clinical trials. Neuro Oncol. 17, 1188–1198 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yao J et al. Improving B0 correction for pH–weighted amine proton chemical exchange saturation transfer (CEST) imaging by use of k-means clustering and Lorentzian estimation. Tomography 4, 123–137 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Harris RJ et al. Simulation, phantom validation, and clinical evaluation of fast pH–weighted molecular imaging using amine chemical exchange saturation transfer echo planar imaging (CEST-EPI) in glioma at 3 T. NMR Biomed. 29, 1563–1576 (2016). [DOI] [PubMed] [Google Scholar]
- 92.Pavlyukov MS et al. Survivin monomer plays an essential role in apoptosis regulation. J. Biol. Chem 286, 23296–23307 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hu Y & Smyth GK ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009). [DOI] [PubMed] [Google Scholar]
- 94.Esser C, Göttlinger C, Kremer J, Hundeiker C & Radbruch A Isolation of full-size mRNA from ethanol-fixed cells after cellular immunofluorescence staining and fluorescence-activated cell sorting (FACS). Cytometry 21, 382–386 (1995). [DOI] [PubMed] [Google Scholar]
- 95.Krishan A Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodide staining. Cell Biol. 66, 188–193 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Belin S et al. Purification of ribosomes from human cell lines. Curr. Protoc. Cell Biol 10.1002/0471143030.cb0340s49 (2010). [DOI] [PubMed] [Google Scholar]
- 97.Saei AA et al. ProTargetMiner as a proteome signature library of anticancer molecules for functional discovery. Nat. Commun 10, 5715 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kent WJ et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pollard KS, Hubisz MJ, Rosenbloom KR & Siepel A Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Blanchette M et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14, 708–715 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the proteomics data that were obtained during this study are presented in Supplementary Tables 1, 2, 4, 5, 7 and 9. The raw mass spectrometry data have been deposited in ProteomeXchange with the primary accession codes PXD035849, PXD035767 and PXD035855. All of the RNA-seq data that were obtained during this study are presented in Supplementary Tables 3, 6 and 8. The raw RNA-seq data of GBM neurospheres overexpressing different isoforms of RPL22L1 protein or an empty vector as a control have been deposited in the Gene Expression Omnibus (GEO) database under accession code GSE180465. The RNA-IP sequencing data for RNA that interacts with Fc-tagged RPL22L1a, RPL22L1b or a control protein have been deposited in the GEO database under accession code GSE180464. Previously published RNA-seq data for GBM neurospheres and GBM tissue isolated from the different regions of the tumours that were re-analysed here are available from the GEO database under accession codes PRJNA344648 and GSE153746. iCLIP data for the SRSF4 protein that were re-analysed here are available from the Array Express database under accession code E-MTAB-747. Gene expression data from the Ivy Glioblastoma Atlas Project database (https://glioblastoma.alleninstitute.org/), gene expression and survival data from the Repository for Molecular Brain Neoplasia Data (GSE108474 and GSE68848) and gene expression, DNA methylation, survival and phenotype data from The Cancer Genome Atlas database (https://tcga-data.nci.nih.gov/tcga/) were used in this study. Source data are provided with this paper.