Abstract
Background
The ongoing circulation of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) poses a diagnostic challenge because symptoms of coronavirus disease 2019 (COVID-19) are difficult to distinguish from other respiratory diseases. Our goal was to use statistical analyses and machine learning to identify biomarkers that distinguish patients with COVID-19 from patients with influenza.
Methods
Cytokine levels were analyzed in plasma and serum samples from patients with influenza and COVID-19, which were collected as part of the Centers for Disease Control and Prevention's Hospitalized Adult Influenza Vaccine Effectiveness Network (inpatient network) and the US Flu Vaccine Effectiveness (outpatient network).
Results
We determined that interleukin (IL)-10 family cytokines are significantly different between COVID-19 and influenza patients. The results suggest that the IL-10 family cytokines are a potential diagnostic biomarker to distinguish COVID-19 and influenza infection, especially for inpatients. We also demonstrate that cytokine combinations, consisting of up to 3 cytokines, can distinguish SARS-CoV-2 and influenza infection with high accuracy in both inpatient (area under the receiver operating characteristics curve [AUC] = 0.84) and outpatient (AUC = 0.81) groups, revealing another potential screening tool for SARS-CoV-2 infection.
Conclusions
This study not only reveals prospective screening tools for COVID-19 infections that are independent of polymerase chain reaction testing or clinical condition, but it also emphasizes potential pathways involved in disease pathogenesis that act as potential targets for future mechanistic studies.
Keywords: cytokine, human, machine learning, pneumonia, SARS-CoV-2
Influenza and COVID-19 can be separated by concentrations of serum inflammatory cytokines. IL-10 family cytokines differentiate inpatient and outpatient COVID-19 from influenza. Machine learning can define novel combinations of serum biomarkers that separate COVID-19 from influenza.
The continued global circulation of coronavirus disease 2019 (COVID-19) has created a diagnostic challenge for clinicians due to the broad spectrum of clinical presentations that arise in patients. Incidence of seasonal influenza has rebounded, complicating diagnosis because both COVID-19 and influenza have pulmonary involvement [1]. Coronavirus disease 2019 and influenza cause similar clinical manifestations with outcomes ranging from no symptoms (asymptomatic) to severe disease [1]. With both viruses circulating, it is becoming increasingly important to be able to distinguish between these 2 viruses and provide appropriate, early treatment for patients at high risk of developing complications from infection, which can be virus specific. Measuring levels of cytokines in COVID-19 and influenza patient plasma or serum will allow us to identify biomarkers for disease etiology and gain insights into how disease progression differs between these 2 viruses. These new findings will allow for improved stratification of patients based on the specific viral infection.
Early research during the COVID-19 pandemic proposed that infection leads to highly elevated levels of proinflammatory cytokines, termed cytokine storm syndrome (CSS), leading to harmful immunopathology [2]. Research has since contradicted this theory, highlighting that although levels of proinflammatory markers, such as interleukin (IL)-6, are typically higher in patients with severe COVID-19, they are only slightly higher (<2-fold) than patients with mild illness [3–5]. Meanwhile single-cell sequencing has revealed that patients with COVID-19 have inflammatory signatures distinct from those with influenza [6]. Patients with severe COVID-19 or influenza exhibit type I interferon (IFN)-driven inflammatory responses [5, 6]. Coronavirus disease 2019 is driven by upregulation of tumor necrosis factor (TNF)α/IL-1β inflammatory response, whereas influenza is characterized by upregulation of a variety of interferon-stimulated genes [6].
In addition, in many recent studies, researchers have investigated which cytokines contribute to CSS in patients with COVID-19. Interleukin-6 has been reported by several studies to be a hallmark of CSS and is associated with poor clinical outcomes [7–13]. In various studies, researchers have also found elevated levels of the proinflammatory markers IP-10 (CXCL10) [14], IL-1α/β [12, 15, 16], TNFα [8, 12, 13], and IL-8 [12, 15] in patients with severe COVID-19 infection. Several of these proinflammatory markers are also elevated in severe influenza infection. Interleukin-17 and type I/II/III IFNs are upregulated during influenza infection [17]. Interleukin-10 has been found to be elevated in patients with severe COVID-19 [9, 12, 13, 15] and influenza infection [18]. During viral infection, IL-10 primarily functions to modulate the antiviral response of immune cells and dampen immunopathology [19]. However, previous studies have not measured most other members of the IL-10 cytokine family (IL-10, IL-19, IL-20, IL-22, IL-26, IL-28A, IL-29). Due to their unknown role as biomarkers, in this study, we have included the IL-10 cytokine family members in our analysis to determine whether they are elevated in influenza or COVID-19 infection.
Despite the large number of published biomarker studies, few have characterized the breadth of immune modulators present in patient blood. To achieve this, we apply data-driven approaches to identify a specific set of biomarkers that strongly correlate with COVID-19 or influenza infection in inpatient and outpatient cohorts. Machine learning (ML) algorithms have been used in a wide variety of biomedical applications [20], such as (1) identifying biomarkers associated with aging [21] Alzheimer's disease [22] and depression [23] and (2) developing diagnostic, prognostic, and survival predictions for various types of cancer [24–28]. More recently, researchers have developed ML approaches to diagnosis COVID-19 in patients using clinical data (vital signs and symptoms), full blood counts, and radiographic patterns on computed tomography scans [29–31]. In this study, we assess the levels of 57 cytokines and other inflammatory markers from inpatient and outpatient blood samples of patients with influenza and patients with COVID-19 using statistical analyses and ML to predict SARS-CoV-2 and influenza infection.
METHODS
Cohort
Serum and plasma samples from COVID-19 and influenza patients were collected during the 2018 influenza season (December 2018–March 2019) and the 2019 influenza season (December 2019–March 2020) or the 2021 COVID-19 pandemic (November 2020–May 2021) as part of the University of Pittsburgh's site for the Centers for Disease Control and Prevention (CDC)'s Hospitalized Adult Influenza Vaccine Effectiveness Network (HAIVEN) and the CDC's US Flu Vaccine Effectiveness (FluVE) network. The HAIVEN focuses on inpatients, whereas FluVE is a network of outpatients. In FluVE, patients’ blood was sampled at the time of emergency department presentation and enrollment, and serum was isolated and banked. In HAIVEN, patients’ blood was retrieved from clinical residual samples after enrollment. All patients were enrolled within 3 days of admission and samples were collected as close to admission as possible. Upon collection, plasma was isolated and banked. Coronavirus disease 2019 and influenza infection were confirmed by molecular polymerase chain reaction (PCR) testing. For outpatients, PCR testing was performed at the time of sample collection. For inpatients, PCR testing may have preceded sample collection by up to 3 days. For both studies, inclusion criteria included ≥18 years of age with respiratory symptoms including cough or fever, and in the case of COVID-19, loss of sense of taste or smell. Before enrollment, preceding symptoms were limited to 7 days. Given the lack of circulating influenza virus during November 2020 through May 2021, it is unlikely that any patients with COVID-19 had coinfection with influenza.
Patient Consent Statement
All patients were enrolled with informed written consent approved by the University of Pittsburgh Institutional Review Board.
Cytokine Measurements
Inflammatory markers were measured with the MAGPIX (Bio-Rad) system. Twenty-seven cytokines and other inflammatory markers were measured with the Bio-Plex Pro Human Cytokine 27-plex Assay according to the manufacturer's instructions. Thirty-seven cytokines and other inflammatory markers were measured with the Bio-Plex Pro Human Inflammation Panel 1, 37-plex Assay according to the manufacturer's instructions. The protein targets measured between assays include APRIL (TNFSF13), BAFF (TNFSF13B), sCD30 (TNFRSF8), sCD163, Chitanase-3-like 1, gp130 (sIL-6Rβ), IFN-α2, IFN-β, IFN-γ, IL-1β, IL-1ra, IL-2, IL-4, IL-5, IL-6, sIL-6Rα, IL-7, IL-8, IL-9, IL-10, IL-11, IL-12 (p40), IL-12 (p70), IL-13, IL-15, IL-17, IL-19, IL-20, IL-22, IL-26, IL-27 (p28), IL-28A (IFN-λ2), IL-29 (IFN-λ1), IL-32, IL-34, IL-35, LIGHT (TNFSF14), MMP-1, MMP-2, MMP-3, osteocalcin, osteopontin, pentraxin-3, sTNF-R1, sTNF-R2, TSLP, TWEAK (TNFSF12), FGF basic, eotaxin, G-CSF, GM-CSF, IP-10, MCP-1, MIP-1α, MIP-1β, PDGF-BB, RANTES, TNF-α, and VEGF. Data were interpreted using xPONENT software, and concentrations are reported as picograms per milliliter. For analytes measured on both plates (IFNγ, IL-2, IL-8, IL-10, and IL-12p70), 37-plex assay data were utilized. A comparison of the correlation between 27-plex and 37-plex results is included in Supplementary Table 1.
Statistical Analysis
Measurements from 59 different cytokine and other inflammatory markers were obtained, 57of which were used for statistical analysis. RANTES and gp130 were excluded from analysis because most influenza patient samples were above the detectable threshold for the assay. For our statistical analyses, missing values from the datasets were excluded. To handle missing cytokine measurements for ML analyses, the missing value was replaced with the mean of the cytokine measurement among patients in the same clinical cohort (inpatient versus outpatient) with the same viral infection (SARS-CoV-2 versus influenza). Five values were replaced for 1 outpatient and 3 inpatients, where each patient only had 1 missing cytokine measurement except for 1 inpatient that had 2 missing cytokine values. The missing measurements were from the following cytokines: MCP-1, MIP-1b, IL-27 (p28), PDGF-BB, and IP-10. A Student t test was applied for the remaining 57 cytokines to determine whether there was a statistically significant difference in means between influenza and COVID-19 patient data within each clinical cohort using the stats.ttest_ind() function from the SciPy 1.7.1 package in Python v3. The Benjamini-Hochberg adjustment was used to control the false discovery rate (FDR) when considering multiple hypotheses. Fisher exact tests were used to determine whether there were significant differences in demographic data (age, sex, race, current smoker, smoker at home) from patients with COVID-19 and influenza within inpatient and outpatient cohorts using R (R version 4.1.2; R Foundation for Statistical Computing 2021). The FDR values less than 0.01 and P values less than .05 were considered to be statistically significant.
Data Cohort Analysis and Machine Learning
All data were standardized by taking the natural log of each measurement plus 1. Uniform Manifold Approximation and Projection (UMAP) was applied to the unlabeled cytokine and inflammatory marker data for dimensionality reduction so the high-dimensional data could be visualized in 2 dimensions. This was implemented using the default options in the umap-learn 0.5.1 package in Python 3. Support vector machine (SVM) models were applied to the data to predict patient virus infection using the default options from the sklearn 0.24.1 package in Python 3. The SVM models prediction performance was assessed using 5-fold cross-validation and the area under the receiver operating characteristics curve (AUC). Additional models were tested and compared with SVM including Random Forest, Logistic Regression, Decision Tree, and AdaBoost. The AUC for these models compared with SVM are shown in Supplementary Figures 1 and 2 and Supplementary Tables 2 and 3. The SVM outperformed the other algorithms and was chosen for the data analyses.
RESULTS
Cohort Characteristics
A total of 141 patients were enrolled through FluVE (n = 69) and HAIVEN (n = 72) studies and diagnosed with either influenza virus (n = 76) or SARS-CoV-2 virus (n = 65). Basic demographic information of inpatients can be seen in Table 1. Among inpatients, a significant difference (P ≤ .05) was seen between sexes: a greater ratio of females were diagnosed with influenza compared with COVID-19. In terms of age, race, and smoking status, no significant differences (P > .05) were seen between those diagnosed with influenza virus or SARS-CoV-2 virus. More importantly, there were no significant differences in the need for intensive care or duration of hospitalization between influenza and COVID-19 inpatients in the study. The basic demographic information of outpatients can be seen in Table 2. No significant demographic differences were observed in the outpatient cohort.
Table 1.
Inpatient Demographics
| Demographic Feature | Overall (n = 72) | COVID (n = 36) | Influenza (n = 36) | P Value |
|---|---|---|---|---|
| Gender (%) | .05054a | |||
| Female | 44 (61.1) | 18 (50) | 26 (72.2) | |
| Male | 27 (37.5) | 18 (50) | 9 (25) | |
| Unknown | 1 (1.4) | 0 (0) | 1 (2.8) | |
| Race | 1.0a | |||
| Black | 17 (23.6) | 9 (25) | 8 (22.2) | |
| White | 49 (68) | 24 (66.7) | 25 (69.4) | |
| Other/Unknown | 6 (8.3) | 3 (8.3) | 3 (8.3) | |
| Current Smoker | .7861 | |||
| Yes | 18 (25) | 8 (22.2) | 10 (27.8) | |
| No | 54 (75) | 28 (77.8) | 26 (72.2) | |
| Smoker at Home | .7531 | |||
| Yes | 12 (16.7) | 5 (13.9) | 7 (19.4) | |
| No | 60 (83.3) | 31 (86.1) | 29 (80.6) | |
| Age | .8113 | |||
| <65 years | 42 (58.3) | 22 (61.1) | 20 (55.6) | |
| 65 + years | 30 (41.7) | 14 (38.9) | 16 (44.4) | |
| ICU | .7861 | |||
| Yes | 3 (4.2) | 2 (5.6) | 1 (2.8) | |
| No | 69 (95.8) | 34 (94.4) | 35 (97.2) | |
| ICU Days | .5582 | |||
| Median (range) | 0 (0–12) | 0 (0–12) | 0 (0–2) | |
| Hospital Duration | .3869 | |||
| Median (range) | 4.0 (1–29) | 4.0 (1–29) | 3.5 (1–13) |
Abbreviations: COVID, coronavirus disease; ICU, intensive care unit.
Other/Unknown category excluded from comparison.
Table 2.
Outpatient Demographics
| Demographic Feature | Overall (n = 69) | COVID (n = 29) | Influenza (n = 40) | P Value |
|---|---|---|---|---|
| Gender (%) | .1275a | |||
| Female | 43 (62.32) | 15 (51.7) | 28 (70) | |
| Male | 25 (36.23) | 14 (48.3) | 11 (27.5) | |
| Unknown | 1 (1.45) | 0 | 1 (2.5) | |
| Race (%) | .7981a | |||
| Black | 27 (39.13) | 11 (37.9) | 16 (40) | |
| White | 35 (50.72) | 16 (55.2) | 19 (47.5) | |
| Other/Unknown | 7 (10.14) | 2 (6.9) | 5 (12.5) | |
| Current Smoker | 1.0a | |||
| Yes | 7 (10.1) | 1 (3.4) | 6 (15) | |
| No | 40 (58) | 7 (24.1) | 33 (82.5) | |
| Unknown | 22 (31.9) | 21 (72.4) | 1 (2.5) | |
| Smoker at Home | 1.0a | |||
| Yes | 9 (13.04) | 0 | 9 (22.5) | |
| No | 33 (47.83) | 3 (10.3) | 30 (75) | |
| Unknown | 27 (39.13) | 26 (89.7) | 1 (2.5) | |
| Age | .3017 | |||
| <65 years | 59 (85.5) | 23 (79.3) | 36 (90) | |
| +65 years | 10 (14.5) | 6 (20.7) | 4 (10) |
Abbreviations: COVID, coronavirus disease.
Other/Unknown category excluded from comparison.
Blood Cytokine Concentration Differs Between Influenza and Coronavirus Disease 2019
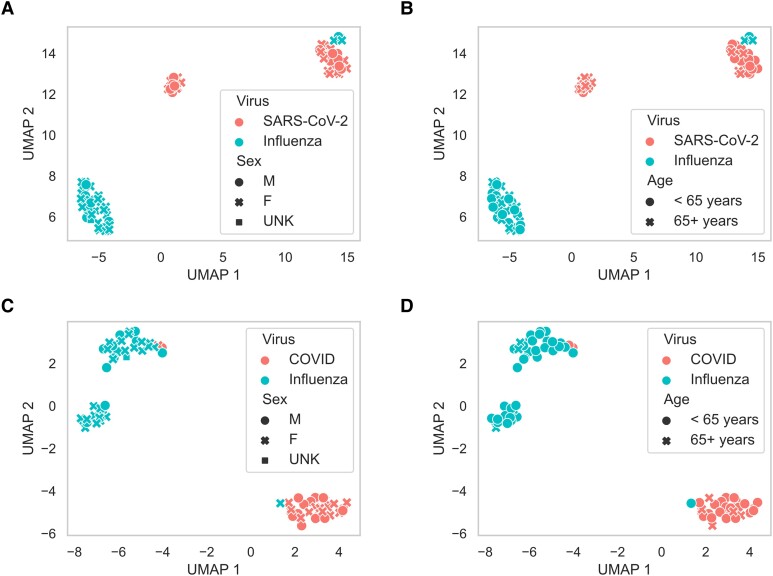
We compared the levels of 57 cytokines and other inflammatory markers between outpatients and inpatients with influenza or SARS-CoV-2 infection (Supplementary Figure 3). To determine whether patients with influenza and COVID-19 could be separable by blood cytokine concentrations, and to assess how demographic factors may impact observed differences between patients with influenza and patients with COVID-19, we performed a clustering analysis using UMAPs. Our analyses showed that both inpatients and outpatients were stratified due to virus type (influenza versus SARS-CoV-2) regardless of patient sex (Figure 1AandC) or age (Figure 1BandD). Although initially unclear why there were 2 clusters of inpatients with COVID-19, our analyses suggest that patients with COVID-19 can be further stratified according to age (Figure 1B) with one cluster being dominated by individuals over the age of 65 and the other COVID-19 cluster primarily consisting of patients under the age of 65. For outpatients with influenza, a second cluster was observed that was dominated by women under age 65.
Figure 1.
Clustering of inpatient and outpatient data using Uniform Manifold Approximation and Projections. (A and B) show how inpatient data clusters, with the color of each point (red, severe acute respiratory syndrome coronavirus 2 [SARS-CoV-2]; blue, influenza virus) indicating the type of viral infection and the shape indicating the sex (A) and age group (B) of each patient. (C and D) show the clustering results of the outpatient data, with the color of each point (red, SARS-CoV-2; blue, influenza virus) indicating the type of viral infection and the shape indicating the sex (C) and age group (D) of each patient.
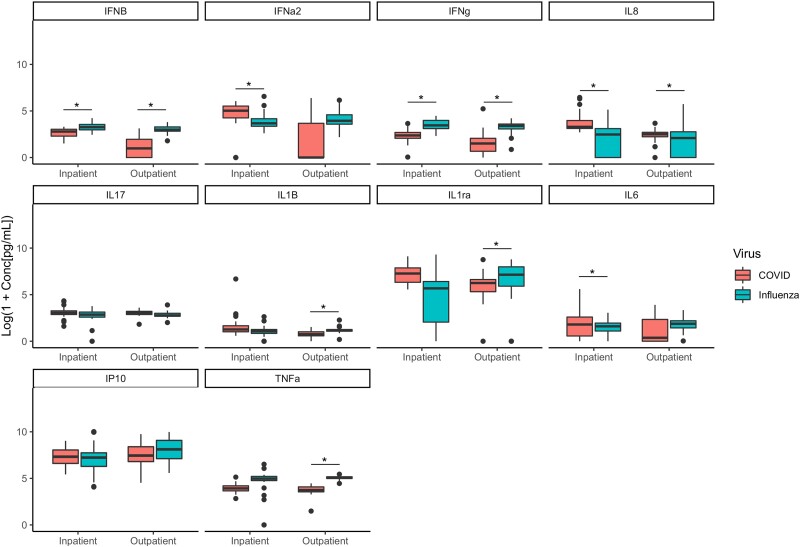
We examined 10 proinflammatory cytokines or associated molecules that differed between patients with COVID-19 and patients with influenza in the inpatient or outpatient context (Figure 2). Significant differences were seen in the expression of cytokines, including IFNβ (higher in influenza), IFNγ (higher in influenza), and IL-8 (higher in COVID-19), which were altered in both inpatients and outpatients. In addition, IFNα2, IL-1β, IL-1ra, IL-6, and TNFα were differentially induced by disease type depending on patient disease severity. Interleukin-6 and IFNα2 were elevated in inpatients with COVID-19 versus influenza, whereas IL-1β, IL-1ra, and TNFα were higher in outpatients with influenza versus COVID-19. Two previously described biomarkers, IL-17 and IP-10 (CXCL10), were not different between virus type in either inpatient or outpatient cohorts.
Figure 2.
Boxplots of biomarkers known to be associated with influenza and/or coronavirus disease 2019 (COVID-19) infection. * indicates a false discovery rate <0.01 and P < .01.
Interleukin-10 Family Cytokines Elevated in Patients With Severe Acute Respiratory Syndrome Coronavirus 2
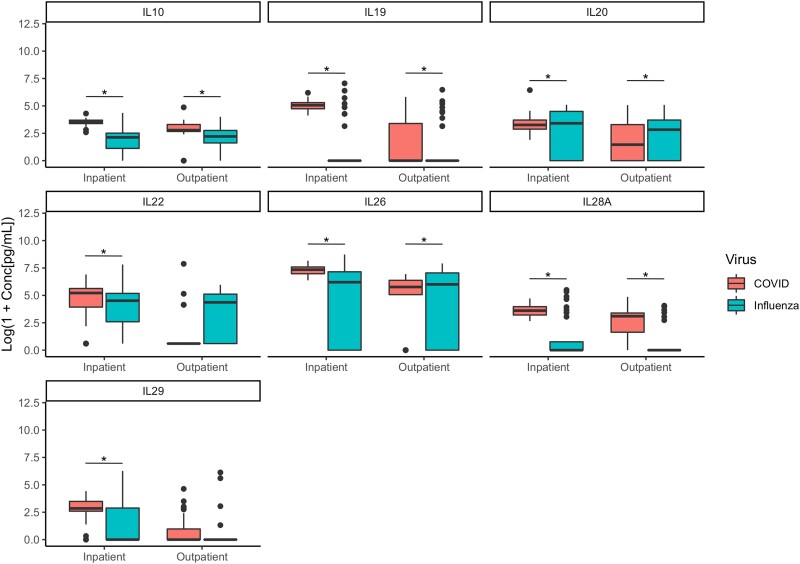
All 7 IL-10 cytokine family members (IL-10, IL-19, IL-20, IL-22, IL-26, IL-28A, and IL-29) were significantly different between patients diagnosed with influenza or COVID-19 (Figure 3). In the inpatient samples, a majority of the IL-10 family cytokines (IL-10, IL-19, IL-22, IL-28A, and IL-29) were significantly increased in patients diagnosed with COVID-19 compared with patients diagnosed with influenza virus. Interleukin-20 was elevated in inpatients and outpatients with influenza compared with inpatients and outpatients with COVID-19. It is interesting to note that IL-26 was elevated in outpatient influenza samples compared with COVID-19 samples, but it was lower in inpatient influenza samples than COVID-19 samples.
Figure 3.
Boxplots of the IL10 family. * indicates a false discovery rate <0.01 and P < .01.
Machine Learning Expands Diagnostic Options
Individual inflammatory biomarkers have been previously studied in the context of COVID-19 and influenza. In our study, the aforementioned cytokines sufficiently separate patients with influenza and COVID-19 in both inpatient and outpatient settings. To further expand our analyses, we used ML to identify combinations of cytokines, who alone are not linearly separable, that are capable of differentiating between influenza virus and SARS-CoV-2 virus infection in inpatients and outpatients. For this analysis, we only considered biomarkers that were nonsignificant (FDR > 0.01) between influenza and SARS-CoV-2 infection and individually produced an average AUC score between 0.4 and 0.6 using an SVM model in their respective inpatient and outpatient cohorts. The SVM model for the inpatient cohort produced an average AUC score up to 81% across the 5-fold cross-validation (Table 3), whereas the SVM model for the outpatient cohort generates an average AUC up to 84% (Table 4). These analyses reveal novel biomarker combinations that are predictive of influenza or COVID-19 disease.
Table 3.
Inpatient ML Results With Nonsignificant Biomarkers
| Biomarkers | Average AUC | AUC Standard Deviation |
|---|---|---|
| IL13, IL32, sIL-6Ra | 0.81122449 | 0.134939093 |
| Basic FGF, IL1B | 0.802040816 | 0.12586229 |
| IP10, PDGF-BB, pentraxin-3 | 0.781122449 | 0.116012824 |
| IL13, sIL-6Ra, sTNF-R2 | 0.758163265 | 0.141303505 |
| IP10, MMP3, PDGF-BB | 0.753571429 | 0.086668642 |
| IL13, sIL-6Ra | 0.753571429 | 0.143217476 |
| MMP3, PDGF-BB | 0.747959184 | 0.083296188 |
| MMP3, PDGF-BB, pentraxin-3 | 0.744897959 | 0.129032216 |
| Basic FGF, IL1B, PDGF-BB | 0.744897959 | 0.129676174 |
| PDGF-BB | 0.592346939 | 0.082274264 |
| Basic FGF | 0.555102041 | 0.181890438 |
| MMP3 | 0.53877551 | 0.104303984 |
| Pentraxin-3 | 0.534693878 | 0.097788968 |
| IP10 | 0.494387755 | 0.116080119 |
| sTNF-R2 | 0.460204082 | 0.12481348 |
| IL32 | 0.416326531 | 0.054076819 |
| sIL-6Ra | 0.402295918 | 0.264858826 |
| IL1B | 0.401785714 | 0.07827934 |
| IL13 | 0.400255102 | 0.117652663 |
Abbreviations: AUC, area under the receiver operating characteristics curve; FGF, fibroblast growth factor; IL, interleukin; IP10, interferon-inducible protein; ML, machine learning; MMP, matrix metalloproteinase; PDGF-BB, platelet-derived growth factor, two B subunits; sIL-6Ra, soluble interleukin 6 receptor alpha; sTNF, soluble tumor necrosis factor.
Table 4.
Outpatient ML Results With Nonsignificant Biomarkers
| Biomarkers | Average AUC | AUC Standard Deviation |
|---|---|---|
| IL8, MMP3, osteocalcin | 0.839583333 | 0.121048659 |
| IL35, MMP3, sIL-6Ra | 0.8225 | 0.116147655 |
| IL35, MMP3, osteocalcin | 0.815 | 0.079652021 |
| IL35, MMP3, sTNF-R1 | 0.815 | 0.108288452 |
| IL8, osteocalcin, sTNF-R1 | 0.811666667 | 0.10786437 |
| IL8, IL35, MMP3 | 0.808333333 | 0.153319746 |
| IL8, osteocalcin, sIL-6Ra | 0.8025 | 0.125626209 |
| IL8, osteocalcin | 0.795 | 0.118772659 |
| IL35, osteocalcin, sTNF-R1 | 0.789166667 | 0.067371276 |
| GM-CSF, IL8, osteocalcin | 0.7875 | 0.102401714 |
| IL35 | 0.560833333 | 0.210824275 |
| Osteocalcin | 0.56 | 0.233907033 |
| sIL-6Ra | 0.506666667 | 0.136437817 |
| MMP3 | 0.490833333 | 0.257099203 |
| IL8 | 0.486666667 | 0.14713467 |
| GM-CSF | 0.415 | 0.162373883 |
| sTNF-R1 | 0.405833333 | 0.123113543 |
Abbreviations: AUC, area under the receiver operating characteristics curve; FGF, fibroblast growth factor; GM-CSF, granulocyte-macrophage colony-stimulating factor; IL, interleukin; ML, machine learning; MMP, matrix metalloproteinase; sIL-6Ra, soluble interleukin 6 receptor alpha; sTNF, soluble tumor necrosis factor.
DISCUSSION
Pathogenesis of COVID-19 and influenza involves potent inflammatory responses, involving the secretion of cytokines and other inflammatory mediators. Using a cohort (n = 141) of outpatients and inpatients with confirmed influenza or COVID-19 virus, we sought to discover biomarkers correlated with the etiology of viral infection, allowing for insights into disease progression. Identifying a precise set of predictive biomarkers for influenza virus or SARS-CoV-2 infection in patients would significantly aid in diagnosis and clinical triage as well as provide insight into disparate pathogenic mechanisms. Large increases of systemic proinflammatory cytokines have been correlated with viral burden and clearance rates [32]. In our study, the expression of several cytokines and inflammatory biomarkers were found to be relatively conserved between outpatients and inpatients with influenza and COVID-19. We found that some cytokines and inflammatory markers, such as BAFF, chitinase-3-like-1, MMP2, eotaxin, GM-CSF, IL-9, IL-11, IL-15, IL-17, and IL-34 were detectable in influenza virus and SARS-CoV-2 virus infection similarly, regardless of etiology or clinical cohort (inpatient or outpatient), which indicates that a portion of inflammatory markers are induced regardless of viral pathogen (Supplementary Figure 1). However, several inflammatory markers differed by disease etiology. Clustering of responses by UMAP reveals clear separation of inpatients and outpatients with influenza and COVID-19. It is interesting to note that a small cluster of inpatients with COVID-19 was shown and characterized by age mostly older than 65. Outpatients with influenza were separated into 2 clusters as well, with a small group characterized mostly by females under the age of 65. These differences, although of interest, were not further analyzed due to a lack of sufficient power.
Consistent with previous studies, we confirmed that influenza infection is correlated with a significantly higher increase in type I/II interferons (IFNβ and IFNγ) compared with COVID-19 infection. Upregulation of type I/II interferons during influenza infection assists in viral control by restricting viral replication and promoting proinflammatory pathways. However, an overly robust type I/II interferon response can induce high levels of inflammation, apoptosis, and block proliferation of cells, leading to detrimental immunopathology [33]. In previous studies, researchers have shown that TNFα is strongly correlated with COVID-19 infection [8, 12, 13]; however, we found significantly higher increases of TNFα with influenza infection in the outpatients and similar expressions among the inpatients. Tumor necrosis factor α is a proinflammatory cytokine and has been shown to exacerbate airway inflammation and enhance morbidity in both influenza and COVID-19 infection [34, 35]. In previous studies, researchers have shown that COVID-19 infection is associated with an upregulation of IL-1, IL-6, and IL-8, which have all been implicated in the cause of COVID-19 cytokine storm [7–15]. In our analysis, IL-8 was found to be significantly higher in COVID-19 infection in both inpatient and outpatient cohorts. Interleukin-1β and IL-1ra were found to be significantly increased in the outpatients with influenza compared with COVID-19, but not in the inpatients. Interleukin-1β and IL-1ra have been shown to exacerbate disease during influenza infection by inducing lung tissue pathology [36]. Interleukin-6 was found to be significantly elevated during COVID-19 infection compared with influenza infection in inpatients but not outpatients.
Our analysis identified IL-10 and the other IL-10 family cytokines (IL-19, IL-20, IL-22, IL-26, IL-28A, and IL-29) as significantly different between SARS-CoV-2 and influenza virus infection; with all cytokines, except IL-20, statistically increased during SARS-CoV-2 infection compared with influenza infection in either the inpatient or outpatient cohorts or both. Therefore, our findings suggest that the IL-10 family cytokines are powerful diagnostic biomarkers for COVID-19 infection. Interleukin-10 is known to prevent immunopathology during viral infection. In several studies, we have found that IL-10 is elevated in patients with COVID-19 infection [9, 12, 13, 15]; however, data are limited regarding the correlation of the other IL-10 family cytokines with COVID-19 infection. Although many markers were analyzed in this study, we focused on the IL-10 family cytokines due to their known roles in antiviral response and prevention of tissue pathology. The type III IFNs, IL-28A and IL-29, are known to stimulate innate antiviral responses and prevent tissue damage from viral infections. Interleukin-19, IL-20, IL-22, and IL-26 are important in the induction of antimicrobial responses and enhancing tissue repair [37, 38]. The increased expression of members of the IL-10 cytokine family in patients with SARS-CoV-2 infection suggests that the immune response may be acting to dampen lung inflammation and enhance repair of damaged tissue to a greater extent than during influenza virus infection. However, increased expression of IL-10 family cytokines during COVID-19 infection could be acting in an immunosuppressive manner, impeding the development of an efficient antiviral response and subsequently prolonging viral replication. Further investigation is warranted to determine the roles of the IL-10 cytokine family during SARS-CoV-2 infection and whether their upregulation is beneficial or detrimental to COVID-19 severity. Interleukin-10 and the other members of the IL-10 cytokine family may be potential targets for reducing COVID-19 mortality or, conversely, are biomarkers of severe lung injury driven repair processes. Our analysis shows that members of the IL-10 cytokine family are powerful biomarkers for predicting SARS-CoV-2 infection as opposed to influenza virus infection.
In addition, we developed machine learning models that can predict SARS-CoV-2 and influenza virus infection among inpatients and outpatients. Although our previous analyses show that the aforementioned cytokines were linearly separable and able to easily and clearly identify influenza infection versus SARS-CoV-2 infection, our ML models reveal unique and novel cytokine combinations that are predictive of influenza and COVID-19 disease with an average AUC up to 0.81 for the inpatient cohort and 0.84 for the outpatient cohort. Our ML analyses focused solely on nonstatistically significant cytokines because including the statistically significant cytokines as inputs for the ML models does not demonstrate the power of ML algorithms to reveal hidden patterns in the data, because, on their own, these cytokines clearly show differences between patients with COVID-19 and patients with influenza infection. In our analyses that included statistically significant cytokines, we saw near perfect AUC scores, thus preventing the discovery of the novel diagnostic options revealed in Tables 3 and 4. The ML models illustrate that we can expand our diagnostic options that allow clinicians to classify influenza and SARS-CoV-2 infection among inpatient and outpatient cohorts. Thus, these models reveal the potential for an additional screening tool for SARS-CoV-2 and influenza infection using 2 or 3 cytokine measurements from blood samples taken during the initial stages of disease presentation.
A limitation of this study is sample size, decreasing the power of our analysis. Future analysis with a validation cohort needs to be conducted to confirm the accuracy of our results. In addition, there were differences in sample collection between our inpatient and outpatient cohorts, such as collection of plasma residual samples versus serum in the emergency department, respectively. Sampling methodology precludes comparisons of cytokine levels by disease severity and limits this study to cross-virus comparisons within each patient cohort. At the time of enrollment (December 2020–March 2021), there were no approved therapies and limited vaccines for COVID-19; however, our patients diagnosed with influenza virus (December–March of 2018 and 2019) may have had approved influenza vaccines. We are not aware of whether patients had previous exposures to influenza infection or vaccination or COVID-19 infection, so this study cannot infer how pre-existing immunity impacts cytokine expression during infection. In addition, we did not have access to our participant's full electronic medical records, including their drug histories for chronic comorbidities. However, these are not likely to differ significantly between patient groups because several comorbidities are common between influenza and COVID-19.
CONCLUSIONS
In summary, stratification and treatment of COVID-19 and influenza patients currently relies on clinical condition or PCR testing. Because of this, there is a great need to develop strategies to predict disease etiology during times of cocirculation of influenza virus and SARS-CoV-2. After analyzing numerous cytokines and inflammatory markers in the blood, our study has identified several powerful biomarkers that can categorize influenza or COVID-19 infection. These findings have highlighted potential pathways involved in disease pathogenesis, which serve as potential therapeutic targets for the reduction of disease burden.
Supplementary Material
Acknowledgments
We thank Dr. Louise Taylor and Dr. Fernanda Silveira for their contributions to patient study enrollment and sample acquisition.
Disclaimer. This work represents the views of the authors and not the Centers for Disease Control and Prevention or National Institutes of Health.
Financial support. The project described was supported by the National Institutes of Health through Grant Number UL1TR001857 and the DSF Charitable Foundation. This work was also supported in part by the Centers for Disease Control and Prevention (Grant Numbers U01IP001035 and U01IP000969).
Contributor Information
Lauren L Luciani, Department of Chemical and Petroleum Engineering, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Leigh M Miller, Department of Pediatrics, UPMC Children's Hospital of Pittsburgh, Pittsburgh, Pennsylvania, USA; Department of Immunology, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Bo Zhai, Department of Pediatrics, UPMC Children's Hospital of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Karen Clarke, Department of Family Medicine and Clinical Epidemiology, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Kailey Hughes Kramer, Department of Internal Medicine, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Lucas J Schratz, Department of Pediatrics, UPMC Children's Hospital of Pittsburgh, Pittsburgh, Pennsylvania, USA.
G K Balasubramani, Department of Epidemiology, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Klancie Dauer, Department of Epidemiology, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
M Patricia Nowalk, Department of Family Medicine and Clinical Epidemiology, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Richard K Zimmerman, Department of Family Medicine and Clinical Epidemiology, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Jason E Shoemaker, Department of Chemical and Petroleum Engineering, University of Pittsburgh, Pittsburgh, Pennsylvania, USA; Department of Computational and Systems Biology, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
John F Alcorn, Department of Pediatrics, UPMC Children's Hospital of Pittsburgh, Pittsburgh, Pennsylvania, USA; Department of Immunology, University of Pittsburgh, Pittsburgh, Pennsylvania, USA.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
References
- 1. Miller LM. Similarities and differences between flu and COVID-19. Available at: https://www.cdc.gov/flu/symptoms/flu-vs-covid19.htm. Accessed October 14, 2022.
- 2. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sinha P, Matthay MA, Calfee CS. Is a “cytokine storm” relevant to COVID-19? JAMA Intern Med 2020; 180:1152–4. [DOI] [PubMed] [Google Scholar]
- 4. Chen LYC, Quach TTT. COVID-19 cytokine storm syndrome: a threshold concept. Lancet Microbe 2021; 2:e49–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mudd PA, Crawford JC, Turner JS, et al. Distinct inflammatory profiles distinguish COVID-19 from influenza with limited contributions from cytokine storm. Sci Adv 2020; 6:eabe3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee JS, Park S, Jeong HW, et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci Immunol 2020; 5:eabd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Melo AKG, Milby KM, Caparroz ALMA, et al. Biomarkers of cytokine storm as red flags for severe and fatal COVID-19 cases: a living systematic review and meta-analysis. PLoS One 2021; 16:e0253894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Del Valle DM, Kim-Schulze S, Huang HH, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med 2020; 26:1636–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Han H, Ma Q, Li C, et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg Microbes Infect 2020; 9:1123–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cruz A S, Mendes-Frias A, Oliveira AI, et al. Interleukin-6 is a biomarker for the development of fatal severe acute respiratory syndrome coronavirus 2 pneumonia. Front Immunol 2021; 12:613422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Broman N, Rantasärkkä K, Feuth T, et al. IL-6 and other biomarkers as predictors of severity in COVID-19. Ann Med 2021; 53:410–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu BM, Martins TB, Peterson LK, Hill HR. Clinical significance of measuring serum cytokine levels as inflammatory biomarkers in adult and pediatric COVID-19 cases: a review. Cytokine 2021; 142:155478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Keddie S, Ziff O, Chou MKL, et al. Laboratory biomarkers associated with COVID-19 severity and management. Clin Immunol 2020; 221:108614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tamayo-Velasco Á, Peñarrubia-Ponce MJ, Álvarez FJ, et al. Evaluation of cytokines as robust diagnostic biomarkers for COVID-19 detection. J Pers Med 2021; 11:681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Herr C, Mang S, Mozafari B, et al. Distinct patterns of blood cytokines beyond a cytokine storm predict mortality in COVID-19. J Inflamm Res 2021; 14:4651–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tamayo-Velasco Á, Martínez-Paz P, Peñarrubia-Ponce MJ, et al. HGF, IL-1α, and IL-27 are robust biomarkers in early severity stratification of COVID-19 patients. J Clin Med 2021; 10:2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gu Y, Zuo X, Zhang S, et al. The mechanism behind influenza virus cytokine storm. Viruses 2021; 13:1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun J, Madan R, Karp CL, Braciale TJ. Effector T cells control lung inflammation during acute influenza virus infection by producing IL-10. Nat Med 2009; 15:277–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rojas JM, Avia M, Martín V, Sevilla N. IL-10: a multifunctional cytokine in viral infections. J Immunol Res 2017; 2017:6104054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mamoshina P, Vieira A, Putin E, Zhavoronkov A. Applications of deep learning in biomedicine. Mol Pharm 2016; 13:1445–54. [DOI] [PubMed] [Google Scholar]
- 21. Mamoshina P, Kochetov K, Putin E, et al. Population specific biomarkers of human aging: a big data study using South Korean, Canadian, and Eastern European patient populations. J Gerontol A Biol Sci Med Sci 2018; 73:1482–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chang CH, Lin CH, Lane HY. Machine learning and novel biomarkers for the diagnosis of Alzheimer's disease. Int J Mol Sci 2021; 22:2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dipnall JF, Pasco JA, Berk M, et al. Fusing data mining, machine learning and traditional statistics to detect biomarkers associated with depression. PLoS One 2016; 11:e0148195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dalal V, Carmicheal J, Dhaliwal A, Jain M, Kaur S, Batra SK. Radiomics in stratification of pancreatic cystic lesions: machine learning in action. Cancer Lett 2020; 469:228–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang B, He X, Ouyang F, et al. Radiomic machine-learning classifiers for prognostic biomarkers of advanced nasopharyngeal carcinoma. Cancer Lett 2017; 403:21–7. [DOI] [PubMed] [Google Scholar]
- 26. Xu W, Xu M, Wang L, et al. Integrative analysis of DNA methylation and gene expression identified cervical cancer-specific diagnostic biomarkers. Signal Transduct Target Ther 2019; 4:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rehman O, Zhuang H, Muhamed Ali A, Ibrahim A, Li Z. Validation of miRNAs as breast cancer biomarkers with a machine learning approach. Cancers (Basel) 2019; 11:431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xie Y, Meng WY, Li RZ, et al. Early lung cancer diagnostic biomarker discovery by machine learning methods. Transl Oncol 2021; 14:100907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou X, Wang Z, Li S, et al. Machine learning-based decision model to distinguish between COVID-19 and influenza: a retrospective, two-centered, diagnostic study. Risk Manag Healthc Policy 2021; 14:595–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Banerjee A, Ray S, Vorselaars B, et al. Use of machine learning and artificial intelligence to predict SARS-CoV-2 infection from full blood counts in a population. Int Immunopharmacol 2020; 86:106705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu X, Jiang X, Ma C, et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering (Beijing) 2020; 6:1122–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bhaskar S, Sinha A, Banach M, et al. Cytokine storm in COVID-19—immunopathological mechanisms, clinical considerations, and therapeutic approaches: the REPROGRAM consortium position paper. Front Immunol 2020; 11:1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Davidson S, Maini MK, Wack A. Disease-promoting effects of type I interferons in viral, bacterial, and coinfections. J Interferon Cytokine Res 2015; 35:252–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. DeBerge MP, Ely KH, Enelow RI. Soluble, but not transmembrane, TNF-α is required during influenza infection to limit the magnitude of immune responses and the extent of immunopathology. J Immunol 2014; 192:5839–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Montazersaheb S, Hosseiniyan Khatibi SM, Hejazi MS, et al. COVID-19 infection: an overview on cytokine storm and related interventions. Virol J 2014; 19:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schmitz N, Kurrer M, Bachmann MF, Kopf M. Interleukin-1 is responsible for acute lung immunopathology but increases survival of respiratory influenza virus infection. J Virol 2005; 79:6441–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol 2011; 29:71–109. [DOI] [PubMed] [Google Scholar]
- 38. Stephen-Victor E, Fickenscher H, Bayry J. IL-26: an emerging proinflammatory member of the IL-10 cytokine family with multifaceted actions in antiviral, antimicrobial, and autoimmune responses. PLoS Pathog 2016; 12:e1005624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.