In a large cohort of patients, two thirds of individuals who had a noninvasive prenatal screening result concerning for maternal malignancy had a cancer diagnosis.
OBJECTIVE:
To evaluate the incidence and clinical outcomes of cell-free DNA results suspicious for maternal malignancy on prenatal cell-free DNA screening with single-nucleotide polymorphism (SNP)–based technology.
METHODS:
This retrospective cohort study included data from SNP-based, noninvasive prenatal screening samples from a commercial laboratory from January 2015 to October 2021. Maternal plasma was screened for trisomy 21, 18, and 13; monosomy X; and triploidy. Cases were considered suspicious for maternal malignancy if retrospective bioinformatics and visual inspection of the SNP plot were suggestive of multiple maternal copy number variants across at least two of the tested chromosomes. Clinical follow-up on patients was obtained by contacting individual referring clinician offices by telephone, facsimile, or email.
RESULTS:
A total of 2,004,428 noninvasive prenatal screening samples during the study period met criteria for inclusion in the analysis. Of these, 38 samples (0.002% or 1 in 52,748, 95% CI 1:74,539–1:38,430) had SNP-plot results that were suspicious for maternal malignancy. Maternal health outcomes were obtained in 30 of these patients (78.9%); eight were lost to follow-up. Maternal malignancy or suspected malignancy was identified in 66.7% (20/30) of the 30 patients with clinical follow-up provided by the clinic. The most common maternal malignancies were lymphoma (n=10), breast cancer (n=5), and colon cancer (n=3).
CONCLUSION:
Results suspicious for maternal malignancy are rare with SNP-based noninvasive prenatal screening (1:53,000), but two thirds of patients who had a noninvasive prenatal screening result concerning for malignancy in this study had a cancer diagnosis. Investigation for malignancy should be recommended for all pregnant patients with this type of result.
FUNDING SOURCE:
This study was funded by Natera, Inc.
Noninvasive prenatal screening, often called cell-free DNA screening, is the most sensitive and specific prenatal screening test for common fetal aneuploidies (trisomy 21, 18, and 13).1 In the United States, noninvasive prenatal screening is widely used as the first-line prenatal screening test for pregnant people of all ages.1 During pregnancy, circulating cell-free DNA is released from maternal tissues and the placenta into the plasma. It is recognized that malignancies can be another source of cell-free DNA in plasma.2–7 Rarely, cell-free DNA released by maternal cancer is detected on noninvasive prenatal screening and has led to reports of unexpected cancer diagnoses in otherwise asymptomatic pregnant people.8
Incidental discovery of maternal malignancy through cell-free DNA screening found to be discordant with the fetal karyotype has been reported after identification of aneuploidies and copy number variants involving multiple chromosomes.9–13 Identification of malignancy can have implications for medical care of the pregnant individual and for the management of their pregnancy.9 Existing population studies on this topic are limited and have been associated mainly with noninvasive prenatal screening with massively parallel shotgun sequencing or counting methods.10–21
Here we evaluated the incidence of cell-free DNA results suspicious for maternal malignancy on prenatal cell-free DNA screening using single-nucleotide polymorphism (SNP)–based technology in a large series of consecutive tests performed in a commercial laboratory. We provide follow-up on the clinical outcomes in this group and compare our findings with existing published studies.
METHODS
This retrospective cohort study included data from SNP-based noninvasive prenatal screening samples analyzed at a commercial laboratory in the United States from January 2015 to October 2021. Maternal plasma was screened for trisomy 21, 18, and 13; monosomy X; and triploidy with an SNP-based noninvasive prenatal screening method (Panorama, Natera, Inc) as described previously.22,23 Testing was subject to revisions in the protocols in April 2015 (version 2), January 2017 (version 3), and March 2021 (version 3.1).24–26
Samples were eligible for SNP-based noninvasive prenatal screening if the following criteria were met: the sample was of sufficient blood volume; the sample was drawn at gestational age greater than 9 weeks; the sample arrived in the laboratory within 8 days of blood collection; and the sample was collected in Streck tubes that were not damaged or hemolyzed on receipt. Only samples with sufficient fetal fraction for analysis (greater than or equal to 2.8% fetal fraction) on SNP-based noninvasive prenatal screening were included in the study cohort.24
Single-nucleotide polymorphism–based noninvasive prenatal testing involved the selective amplification and analysis of a panel of DNA sequences containing thousands of SNPs, many of which differ between mother and fetus. Chromosomal imbalances were detected by comparing observed SNP data with a set of hypothetical SNP distributions representing either euploid or aneuploid states across different fetal fractions. A maximum likelihood was then calculated that the fetus has the typical chromosome complement or has a chromosome abnormality for the conditions screened. This approach differs from massively parallel shotgun sequencing methods (DNA fragment counting) in that it can distinguish between maternal and fetal imbalances and detect chromosomal segregation errors that do not have proportional copy number differences between the chromosomes (eg, triploidy, complete mole, and uniparental disomy).27,28
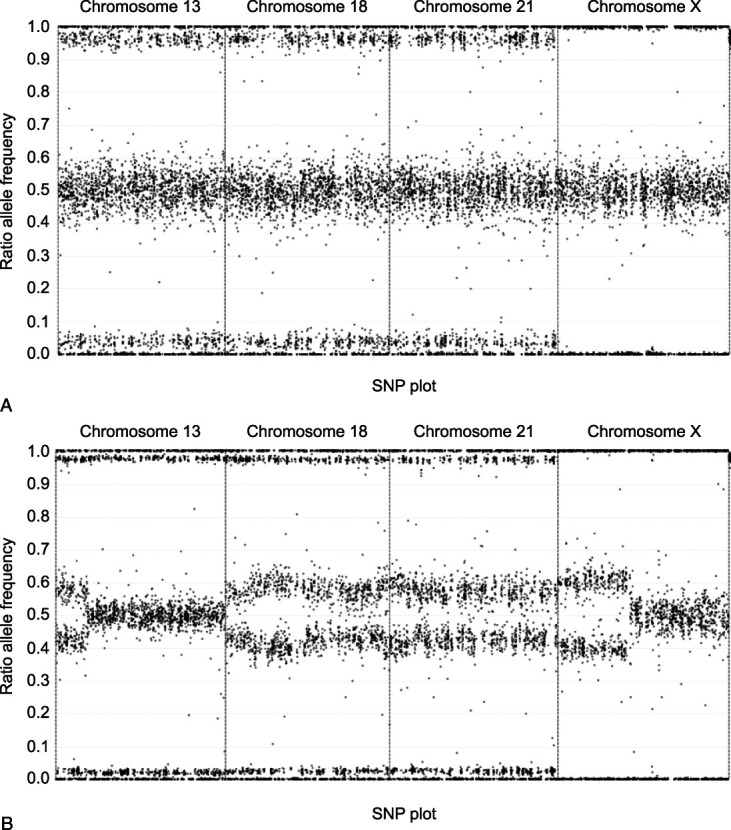
Cases were considered suspicious for maternal malignancy if retrospective bioinformatics and visual inspection of the SNP plot were suggestive of multiple maternal copy number variants across at least two of the tested chromosomes (Fig. 1). These cases were reported as a fetal uninterpretable result, and a redraw was not recommended or performed. Clinical follow-up on patients was obtained by contacting the referring clinician offices by telephone, facsimile, or email starting in April 2021. Information requested included the following: presence or absence of ultrasonographic anomalies; results of any prenatal diagnostic testing; any maternal blood karyotype or microarray results; maternal health conditions before pregnancy, during pregnancy, or reported after the pregnancy; presence or absence of leiomyomas; and presence or absence of maternal malignancy. If maternal malignancy was reported, the type of malignancy, timing of diagnosis, and staging were requested. This study was considered to qualify for exemption from IRB review under 45 CFR 46.104(d)(4) by the IRB at Salus Institutional Review Board (formerly Ethical and Independent Review Services). The study was conducted at Natera, Inc.
Fig. 1. Single-nucleotide polymorphism (SNP) plots representative of a normal pregnancy (A) and a pregnancy suspected of having atypical findings suggesting maternal copy number variants (B). The x axis of both plots shows the relative position of SNPs along the analyzed chromosomes. The y axis shows the relative amounts of the alleles (100% of allele A at the top of each plot and 100% of allele B at the bottom of each plot). In A, the relative vertical position of alleles shown is determined by the fetal fraction. For example, the upper band represents maternal BB genotype and fetus BB genotype, the band below it represents a maternal BB genotype and fetal AB genotype, and the spacing between the bands is determined by the relative amount of the A allele present (ie, the fetal fraction). In B, segments of maternal chromosomes have extra copies, indicative of the imbalances that would typically be seen when maternal cancer is present. The large spaces between the bands indicate that a substantial proportion of maternal DNA is abnormal. Note that this graphical representation of data does not in any way describe the functioning of Natera's technology.
Goldring. Maternal Malignancy and Cell-Free DNA Screening. Obstet Gynecol 2023.
Other noninvasive prenatal testing laboratories using a massively parallel shotgun sequencing methodology have previously reported maternal neoplasms in some patients with a high-risk cell-free DNA result for multiple aneuploidies and for single monosomy.17,29 However, internal data collected on patients using SNP-based methodology reported as high risk for two aneuploidies were not associated with an increased risk for maternal neoplasm (McKanna T, DiNonno W, Hook N, Maisenbacher MK, et al. High risk for double aneuploidy: outcome results from single nucleotide polymorphism-based noninvasive prenatal testing and incidence in products of conception testing. Poster presented at the National Society of Genetic Counselors 38th Annual Conference; November 5–8, 2019; Salt Lake City, Utah). Follow-up was collected for patients in whom only multiple maternal copy number variants were identified.
The outcome data obtained on study cohort patients were compelling enough to guide the laboratory to change verbiage on patient reports starting on May 30, 2021, to unequivocally report the concern of an incidental maternal finding that was strongly suggestive of maternal malignancy. In addition, any case that was suggestive of multiple copy number variants was reviewed to determine whether the patient had noninvasive prenatal screening in previous or subsequent pregnancies. For patients with suspicious results and SNP-based noninvasive prenatal screening in more than one pregnancy, results (when available) were compared with detect changes in maternal SNP data from pregnancy to pregnancy.
The prevalence of noninvasive prenatal screening results suspicious for malignancy was calculated. Patient characteristics, fetal anomalies, maternal malignancy, and other health outcomes were evaluated for noninvasive prenatal screening samples identified as suspicious for maternal malignancy. Clopper-Pearson two-sided 95% CIs were calculated for the percent of all patients with follow-up with cancer.
RESULTS
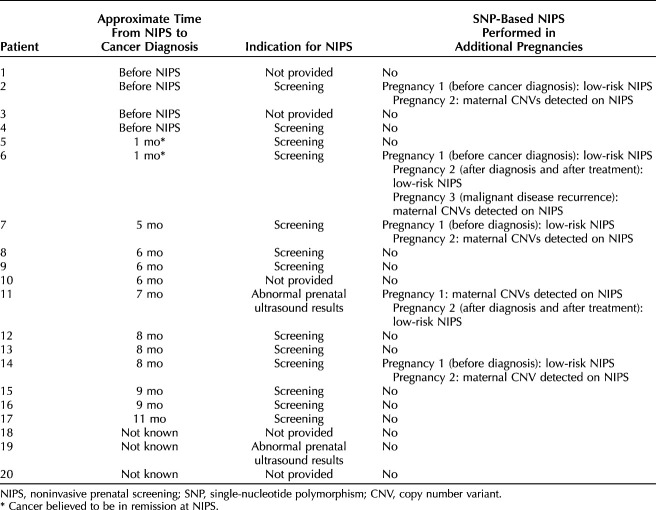
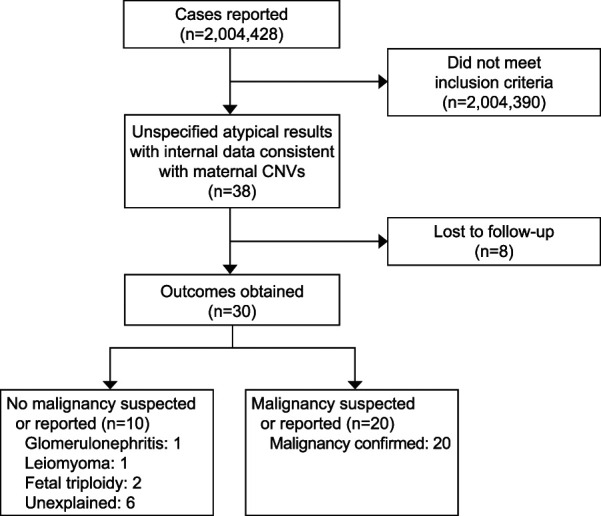
A total of 2,004,428 noninvasive prenatal screening samples during the study period met criteria for inclusion in the analysis. Of these, 38 samples (0.002% or 1 in 52,748, 95% CI 1:74,539-1:38,430) had SNP-plot results that were suspicious for maternal malignancy. Maternal and fetal health outcomes were obtained in 30 of these patients (78.9%); eight were lost to follow-up (Fig. 2). Maternal malignancy was identified in 20 patients: 52.6% (95% CI 35.8–69.0%) of all 38 noninvasive prenatal screenings with suspicious results and 66.7% (95% CI 47.2–82.7%) of the 30 patients with complete clinical follow-up. In patients with a noninvasive prenatal screening result suspicious for maternal copy number variants who were not diagnosed with a malignancy during the study period, the average follow-up period after noninvasive prenatal screening was 12 months, the shortest follow-up period was 1 month, and the longest follow-up period was 31 months after noninvasive prenatal screening. Details of the 20 patients who were found to have a malignancy are presented in Table 1. Median gestational age at noninvasive prenatal screening was 13 weeks. Median maternal age at the time of the patient's noninvasive prenatal screening was 33 years. Timing of cancer diagnosis with respect to the noninvasive prenatal screening sample was known in 17 of 20 patients: six patients had a preexisting cancer diagnosis at the time of undergoing noninvasive prenatal screening, and of these, two had a history of cancer and were thought to be in remission at the time of noninvasive prenatal screening (Table 1). The remaining 11 patients had occult tumors at the time of noninvasive prenatal screening. The time between the noninvasive prenatal screening sample result and reported diagnosis of malignancy ranged from 0 to 11 months. The most common maternal malignancies were lymphoma (n=10), breast cancer (n=5), and colon cancer (n=3) (Appendix 1, available online at http://links.lww.com/AOG/D58).
Fig. 2. Flow diagram of patients included in this cohort. CNV, copy number variant.
Goldring. Maternal Malignancy and Cell-Free DNA Screening. Obstet Gynecol 2023.
Table 1.
Details of Patients With Single-Nucleotide Polymorphism–Based Noninvasive Prenatal Screening and Confirmed or Suspicious Maternal Neoplasms
Information on fetal outcomes was available for 19 of the 20 pregnancies associated with maternal malignancy. No abnormalities were reported in 15. One pregnancy was complicated by a fetal abnormality, one had multiple soft markers on prenatal ultrasonogram, and two had fetal growth restriction. Four of the 19 cases associated with maternal malignancy for which information on fetal findings was available had prenatal invasive testing, the results of which were reported as normal in all cases.
Of the 10 patients with clinical information and no malignancy at the time of follow-up, two patients had fetal triploidy, one patient had maternal glomerulonephritis, and one patient had uterine leiomyomas. In all, 6 of the 30 patients (20%) had no reported fetal or maternal health issues at the time of follow-up (Fig. 2). Of the six patients with no reported maternal malignancies, we were able to obtain follow-up for them at 1, 4, 6, 14, 16, and 31 months after the noninvasive prenatal screening was collected, respectively. Limited follow-up was attributed to the patients no longer being seen at the ordering clinic. For one of the patients (6 months), the clinic reported that after her pregnancy the patient participated in a research study that included genome-wide analysis, which found multiple copy number variants across the genome with a concern for malignancy; however, to the best of our knowledge, follow-up testing for malignancy was not completed.
Seven patients in the cohort had SNP-based noninvasive prenatal screening in more than one pregnancy (Table 1). Four patients had low-risk results during a previous pregnancy and a sample collected during a subsequent pregnancy that was found to have multiple copy number variants. Of these four patients, three were diagnosed with a maternal malignancy after the second noninvasive prenatal screening result suspicious for maternal malignancy. The fourth patient had not been diagnosed with a maternal malignancy at the time the clinical follow-up was elicited. The fifth patient had a multiple copy number variant result and during a subsequent pregnancy had a noninvasive prenatal screening with an abnormal fetal result but was lost to follow-up. The sixth patient had a low-risk result during a previous pregnancy and afterward was diagnosed and treated for breast cancer. That patient had a subsequent low-risk noninvasive prenatal screening result after treatment. However, during a third pregnancy, multiple maternal copy number variants were identified, and after this result, metastatic cancer was diagnosed. The seventh patient was positive for multiple maternal copy number variants and afterward was diagnosed with cancer. After cancer treatment, this patient had a subsequent pregnancy with a low-risk SNP-based noninvasive prenatal screening result.
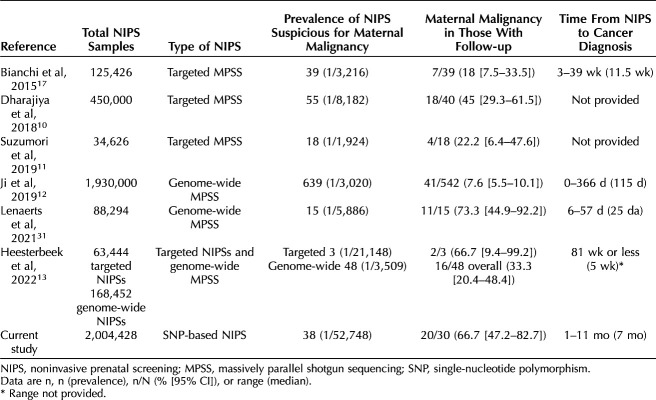
The cancers reported in our study are similar to the more than 100 maternal malignancies identified through noninvasive prenatal screening that have already been reported in the medical literature (Appendix 1, http://links.lww.com/AOG/D58). Table 2 provides a report of maternal malignancy after detection of multiple chromosomal aneuploidy or copy number variants across seven population-based studies with the prevalence of maternal malignancy in those with follow-up ranging from 7.6% (95% CI 5.5–10.1%) to 73.3% (95% CI 44.9–92.2%). Compared with other studies, the prevalence of noninvasive prenatal screening with findings suggestive of maternal malignancy is lower in our study (prevalence of approximately 1 in 53,000; Table 2). The percentage of patients found to have a malignancy after a suspicious noninvasive prenatal screening result in our study was one of the highest reported (66.7%; Table 2).
Table 2.
Population-Based Reports of Maternal Malignancy After Detection of Multiple Chromosomal Aneuploidy
DISCUSSION
This study evaluated the prevalence of maternal malignancy after atypical findings on SNP-based noninvasive prenatal screening. Our study demonstrates that although SNP-based noninvasive prenatal screening results suspicious for malignancy are rare (approximately 1 in 53,000), these results are associated with a high likelihood of maternal malignancy: 67% of patients with clinical follow-up and at least 53% of all patients in our cohort. The most common type of cancer identified by SNP-based noninvasive prenatal screening was lymphoma, followed by breast and colorectal cancer, findings that are consistent with existing noninvasive prenatal screening studies.30 Of note, two of the patients in our cohort had an active cancer diagnosis at the time of noninvasive prenatal screening.
Previous studies of targeted or genome-wide massively parallel shotgun sequencing noninvasive prenatal screening have reported variable prevalence of results suspicious for maternal cancer, ranging from approximately 1 in 2,000 to 1 in 21,000 (Table 2). Possible explanations for this variation include the noninvasive prenatal screening methodology (targeted vs genome-wide), the criteria used for reporting a result as suspicious (multiple copy number variants vs any apparent aneuploidy affecting multiple chromosomes), the population being screened, and change in laboratory experience over time. The likelihood of identifying a malignancy also varies among published studies, ranging from 8% to 73%. The high risk for maternal malignancy associated with suspicious SNP-based noninvasive prenatal screening results could reflect the ability of SNP-based technology to distinguish between maternal and fetal copy number variants. The lower incidence of results suspicious for maternal malignancy likely reflects the target-focused approach of SNP-based noninvasive prenatal screening (targeting only chromosomes 21, 18, 13, X, and Y as opposed to genome-wide noninvasive prenatal screening) and the ability to focus on maternal copy number variants rather than fetal aneuploidy. It is important to note that all of the published studies have relatively small numbers of patients with malignancy and that the 95% CIs around point estimates are wide. Therefore, it cannot be said with certainty whether these differences in reported cancer rates in the setting of suspicious prenatal cell-free DNA screening are truly different.
The clinical information obtained from patients with testing in more than one pregnancy in our study suggests that SNP-based noninvasive prenatal screening may be able to detect changes with malignancy diagnosis, remission, or recurrence from pregnancy to pregnancy. This has not been demonstrated in other published data sets and is an area for additional research. In the setting of SNP-based noninvasive prenatal screening, a finding of maternal copy number variants in patients with known risk factors for malignancy, such as a previous diagnosis or a known hereditary cancer condition, should be evaluated with particular care.
It is important to recognize that many pregnant people reported in the literature as having a noninvasive prenatal screening result suspicious for maternal malignancy have not had an identifiable cancer during the follow-up period. Some have been attributed to another reason (eg, uterine leiomyomas), and in some instances, no explanation has been identified. It is notable that the study with the highest maternal malignancy rate (73%) had a standardized, multidisciplinary approach to cancer investigation, including blood work and whole-body magnetic resonance imaging.31 Lower rates of malignancy in other studies may reflect a more ad hoc or less intensive approach to investigation and relatively short follow-up periods.
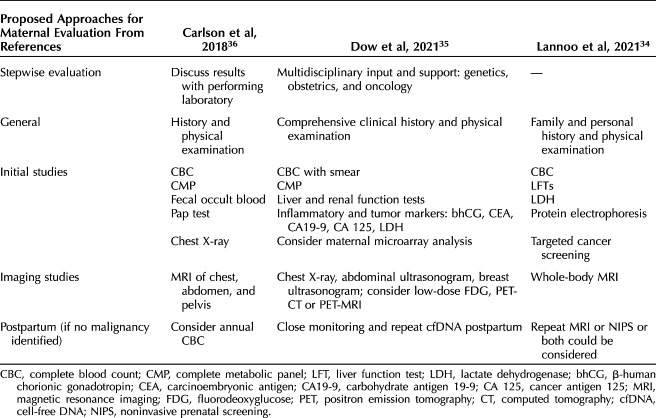
The strength of recommendations for malignancy investigation varies after atypical noninvasive prenatal screening results,9 but our study and other recent publications13,30–33 support investigation for malignancy in all patients. Currently, there are no national society guidelines in the United States for maternal malignancy investigation in patients with suspicious prenatal cell-free DNA screening results, and optimal management strategy remains unclear.9 The National Institutes of Health–sponsored IDENTIFY (Incidental Detection of Maternal Neoplasia Through Non-Invasive Cell Free DNA Analysis) study (ClinicalTrials.gov identifier NCT04049604) aims to answer this question, and patients can be referred to that study.33 We recommend evaluation for underlying maternal disease in all patients with multiple copy number variants of likely maternal origin on SNP-based noninvasive prenatal screening. Referral to a maternal–fetal medicine specialist or medical oncologist is ideal when access is available.13 At least three groups of authors have proposed specific algorithms for investigation (Table 3),34–36 and the recommendations of Carlson and colleagues36 are a reasonable approach. After a negative initial malignancy investigation, longitudinal screening of patients should be considered because some patients in our cohort were diagnosed up to 11 months after the noninvasive prenatal screening. It is also important to note that SNP-based identification of multiple maternal copy number variants was not associated with an increased risk in fetal aneuploidies in our study; however, in this setting, noninvasive prenatal screening cannot screen for these conditions, and patients should be offered the option of diagnostic testing in addition to detailed ultrasound evaluation. Finally, during the informed consent process, it is important that pregnant people are counseled about the potential for incidental maternal findings, including maternal chromosome abnormalities and health conditions, as well as the potential for failure to provide a fetal risk assessment.1
Table 3.
Summary From the Literature of Proposals for Maternal Evaluation After a Noninvasive Prenatal Screening Result Suspicious for Maternal Malignancy
This study reports outcomes after SNP-based noninvasive prenatal screening with results suspicious for maternal malignancy; however, it has several limitations. First, no standard protocol for malignancy evaluation was followed, and the follow-up time period varied. In addition, there was a change in reporting of cases with suspected maternal copy number variants in the last year of the study period. Therefore, there may have been differences in the follow-up offered to patients with the original report type compared with patients with the updated report type. These limitations may have led to under-reporting of malignancy diagnoses because patients may have been lost to follow-up or received an incomplete workup. Last, no follow-up data for patients with other SNP-based noninvasive prenatal screening result types were collected, and the cancer type, its stage, and the time of diagnosis were collected as a report from the clinic but not verified with medical records.
Our study and others highlight the importance of reporting suspicious cell-free DNA results given the high incidence of maternal malignancy when multiple copy number variants are present on noninvasive prenatal screening results. Reporting an incidental finding will allow the possibility of informed maternal clinical management and early cancer detection.
Footnotes
This study was funded by Natera, Inc.
Financial Disclosure Georgina Goldring, Cindy Trotter, Jeffrey T. Meltzer, Vivienne Souter, Wendy DiNonnno, Wenbo Xu, and Jeffrey N. Wetizel are employees of Natera, Inc with stocks or options to own stocks in the company. Neeta L. Vora receives supplies in kind from Illumina for an NIH funded grant on whole genome sequencing in fetal anomalies. Wendy DiNonno disclosed that Natera has applied for a patent directly related to this study. Natera also covers travel expenses to educational meetings. Neeta L. Vora disclosed that they receive supplies in kind from Illumina for an NIH funded grant that is unrelated to the work described in this article. Lynn Pais did not disclose any potential conflicts of interest.
Presented in part at the Society for Maternal-Fetal Medicine’s 42nd Annual Pregnancy Meeting, held virtually, January 31–February 5, 2022.
The authors thank Dusan Kijacic, MS, for his role in internal data review, Brittany Prigmore, MS, for her role in data analysis support, and Peter Benn, PhD, for his expertise and review of the manuscript. Editorial and medical writing support was provided by J. Bryce Ortiz, PhD, from Natera, Inc.
Each author has confirmed compliance with the journal's requirements for authorship.
Peer reviews and author correspondence are available at http://links.lww.com/AOG/D59.
Figure.
No available caption
REFERENCES
- 1.Screening for fetal chromosomal abnormalities. ACOG Practice Bulletin No. 226. American College of Obstetricians and Gynecologists. Obstet Gynecol 2020;136:e48–69. doi: 10.1097/AOG.0000000000004084 [DOI] [PubMed] [Google Scholar]
- 2.Kirkizlar E, Zimmermann B, Constantin T, Swenerton R, Hoang B, Wayham N, et al. Detection of clonal and subclonal copy-number variants in cell-free DNA from patients with breast cancer using a massively multiplexed PCR methodology. Transl Oncol 2015;8:407–16. doi: 10.1016/j.tranon.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keller L, Belloum Y, Wikman H, Pantel K. Clinical relevance of blood-based ctDNA analysis: mutation detection and beyond. Br J Cancer 2021;124:345–58. doi: 10.1038/s41416-020-01047-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early-and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campos-Carrillo A, Weitzel JN, Sahoo P, Rockne R, Mokhnatkin JV, Murtaza M, et al. Circulating tumor DNA as an early cancer detection tool. Pharmacol Ther 2020;207:107458. doi: 10.1016/j.pharmthera.2019.107458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azzi G, Krinshpun S, Tin A, Maninder M, Malashevich AK, Malhotra M, et al. Treatment response monitoring using a tumor-informed circulating tumor DNA test in an advanced triple-negative breast cancer patient: a case report. Case Rep Oncol 2022;15:473–9. doi: 10.1159/000524324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasi PM, Fehringer G, Taniguchi H, Starling N, Nakamura Y, Kotani D, et al. Impact of circulating tumor DNA–based detection of molecular residual disease on the conduct and design of clinical trials for solid tumors. JCO Precision Oncol 2022;6:e2100181. doi: 10.1200/PO.21.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bianchi DW. Unusual prenatal genomic results provide proof-of-principle of the liquid biopsy for cancer screening. Clin Chem 2018;64:254–6. doi: 10.1373/clinchem.2017.282459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rink BD, Stevens BK, Norton ME. Incidental detection of maternal malignancy by fetal cell-free DNA screening. Obstet Gynecol 2022;140:121–31. doi: 10.109710.1097/AOG.0000000000004833 [DOI] [PubMed] [Google Scholar]
- 10.Dharajiya NG, Grosu DS, Farkas DH, McCullough RM, Almasri E, Sun Y, et al. Incidental detection of maternal neoplasia in noninvasive prenatal testing. Clin Chem 2018;64:329–35. doi: 10.1373/clinchem.2017.277517 [DOI] [PubMed] [Google Scholar]
- 11.Suzumori N, Sekizawa A, Takeda E, Samura O, Sasaki A, Akaishi R, et al. Classification of factors involved in nonreportable results of noninvasive prenatal testing (NIPT) and prediction of success rate of second NIPT. Prenatal Diagn 2019;39:100–6. doi: 10.1002/pd.5408 [DOI] [PubMed] [Google Scholar]
- 12.Ji X, Li J, Huang Y, Sung P-L, Yuan Y, Liu Q, et al. Identifying occult maternal malignancies from 1.93 million pregnant women undergoing noninvasive prenatal screening tests. Genet Med 2019;21:2293–302. doi: 10.1038/s41436-019-0510-5 [DOI] [PubMed] [Google Scholar]
- 13.Heesterbeek CJ, Aukema SM, Galjaard R-JH, Boon EM, Srebniak MI, Bouman K, et al. Noninvasive prenatal test results indicative of maternal malignancies: a nationwide genetic and clinical follow-up study. J Clin Oncol 2022;40:2426–35. doi: 10.1200/JCO.21.02260 [DOI] [PubMed] [Google Scholar]
- 14.Osborne CM, Hardisty E, Devers P, Kaiser-Rogers K, Hayden MA, Goodnight W, et al. Discordant noninvasive prenatal testing results in a patient subsequently diagnosed with metastatic disease. Prenatal Diagn 2013;33:609–11. doi: 10.1002/pd.4100 [DOI] [PubMed] [Google Scholar]
- 15.Beamon CJ, Hardisty EE, Harris SC, Vora NL. A single center's experience with noninvasive prenatal testing. Genet Med 2014;16:681–7. doi: 10.1038/gim.2014.20 [DOI] [PubMed] [Google Scholar]
- 16.McCullough RM, Almasri EA, Guan X, Geis JA, Hicks SC, Mazloom AR, et al. Non-invasive prenatal chromosomal aneuploidy testing-clinical experience: 100, 000 clinical samples. PLoS One 2014;9:e109173. doi: 10.1371/journal.pone.0109173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bianchi DW, Chudova D, Sehnert AJ, Bhatt S, Murray K, Prosen TL, et al. Noninvasive prenatal testing and incidental detection of occult maternal malignancies. JAMA 2015;314:162–9. doi: 10.1001/jama.2015.7120 [DOI] [PubMed] [Google Scholar]
- 18.Romero R, Mahoney MJ. Noninvasive prenatal testing and detection of maternal cancer. JAMA 2015;314:131–3. doi: 10.1001/jama.2015.7523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amant F, Verheecke M, Wlodarska I, Dehaspe L, Brady P, Brison N, et al. Presymptomatic identification of cancers in pregnant women during noninvasive prenatal testing. JAMA Oncol 2015;1:814–9. doi: 10.1001/jamaoncol.2015.1883 [DOI] [PubMed] [Google Scholar]
- 20.Van Den Bogaert K, Lannoo L, Brison N, Gatinois V, Baetens M, Blaumeiser B, et al. Outcome of publicly funded nationwide first-tier noninvasive prenatal screening. Genet Med 2021;23:1137–42. doi: 10.1038/s41436-021-01101-4 [DOI] [PubMed] [Google Scholar]
- 21.Harasim T, Neuhann T, Behnecke A, Stampfer M, Holinski-Feder E, Abicht A. Initial clinical experience with NIPT for rare autosomal aneuploidies and large copy number variations. J Clin Med 2022;11:372. doi: 10.3390/jcm11020372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmermann B, Hill M, Gemelos G, Demko Z, Banjevic M, Baner J, et al. Noninvasive prenatal aneuploidy testing of chromosomes 13, 18, 21, X, and Y, using targeted sequencing of polymorphic loci. Prenatal Diagn 2012;32:1233–41. doi: 10.1002/pd.3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pergament E, Cuckle H, Zimmermann B, Banjevic M, Sigurjonsson S, Ryan A, et al. Single-nucleotide polymorphism–based noninvasive prenatal screening in a high-risk and low-risk cohort. Obstet Gynecol 2014;124:210–8. doi: 10.1097/AOG.0000000000000363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ravi H, McNeill G, Goel S, Meltzer SD, Hunkapiller N, Ryan A, et al. Validation of a SNP-based non-invasive prenatal test to detect the fetal 22q11.2 deletion in maternal plasma samples. Fetal Diagn Ther 2016;40:219–23. doi: 10.1159/000442931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravi H, McNeill G, Goel S, Meltzer SD, Hunkapiller N, Ryan A, et al. Validation of a SNP-based non-invasive prenatal test to detect the fetal 22q11. 2 deletion in maternal plasma samples. PLoS One 2018;13:e0193476. doi: 10.1371/journal.pone.0193476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dar P, Jacobsson B, MacPherson C, Egbert M, Malone F, Wapner RJ, et al. Cell-free DNA screening for trisomies 21, 18 and 13 in pregnancies at low and high risk for aneuploidy with genetic confirmation. Am J Obstet Gynecol 2022;227:259.e1–14. doi: 10.1016/j.ajog.2022.01.019 [DOI] [PubMed] [Google Scholar]
- 27.Martin KA, Samango-Sprouse CA, Kantor V, Dhamankar R, Valenti E, Trefogli MT, et al. Detection of maternal X chromosome abnormalities using single nucleotide polymorphism–based noninvasive prenatal testing. Am J Obstet Gynecol MFM 2020;2:100152. doi: 10.1016/j.ajogmf.2020.100152 [DOI] [PubMed] [Google Scholar]
- 28.Kantor V, Mo L, DiNonno W, Howard K, Palsuledesai CC, Parmar S, et al. Positive predictive value of a single nucleotide polymorphism (SNP)‐based NIPT for aneuploidy in twins: experience from clinical practice. Prenatal Diagn 2022;42:1587–93. doi: 10.1002/pd.6262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snyder HL, Curnow KJ, Bhatt S, Bianchi DW. Follow‐up of multiple aneuploidies and single monosomies detected by noninvasive prenatal testing: implications for management and counseling. Prenatal Diagn 2016;36:203–9. doi: 10.1002/pd.4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuo K, Klar M, Youssefzadeh AC, Mandelbaum RS, Roman LD, Ouzounian JG, et al. Assessment of severe maternal morbidity and mortality in pregnancies complicated by cancer in the US. JAMA Oncol 2022;8:1213–6. doi: 10.1001/jamaoncol.2022.1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lenaerts L, Brison N, Maggen C, Vancoillie L, Che H, Vandenberghe P, et al. Comprehensive genome-wide analysis of routine non-invasive test data allows cancer prediction: a single-center retrospective analysis of over 85, 000 pregnancies. EClinicalMedicine 2021;35:100856. doi: 10.1016/j.eclinm.2021.100856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Ju J, Zhao Q, Liu W, Yuan Y, Liu Q, et al. Effective identification of maternal malignancies in pregnancies undergoing noninvasive prenatal testing. Front Genet 2022;13:802865. doi: 10.3389/fgene.2022.802865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turriff AE, Annunziata CM, Bianchi DW. Prenatal DNA sequencing for fetal aneuploidy also detects maternal cancer: importance of timely workup and management in pregnant women. J Clin Oncol 2022;40:2398–401. doi: 10.1200/JCO.22.00733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lannoo L, Lenaerts L, Van Den Bogaert K, Che H, Brison N, Devriendt K, et al. Non‐invasive prenatal testing suggesting a maternal malignancy: what do we tell the prospective parents in Belgium? Prenatal Diagn 2021;41:1264–72. doi: 10.1002/pd.6031 [DOI] [PubMed] [Google Scholar]
- 35.Dow E, Freimund A, Smith K, Hicks RJ, Jurcevic P, Shackleton M, et al. Cancer diagnoses following abnormal noninvasive prenatal testing: a case series, literature review, and proposed management model. JCO Precision Oncol 2021;5:1001–12. doi: 10.1200/PO.20.00429 [DOI] [PubMed] [Google Scholar]
- 36.Carlson LM, Hardisty E, Coombs CC, Vora NL. Maternal malignancy evaluation after discordant cell-free DNA results. Obstet Gynecol 2018;131:464–8. doi: 10.1097/aog.0000000000002474 [DOI] [PMC free article] [PubMed] [Google Scholar]