Abstract
The FACT complex of vertebrate cells, comprising the Cdc68 (Spt16) and SSRP1 proteins, facilitates transcription elongation on a nucleosomal template and modulates the elongation-inhibitory effects of the DSIF complex in vitro. Genetic findings show that the related yeast (Saccharomyces cerevisiae) complex, termed CP, also mediates transcription. The CP components Cdc68 and Pob3 closely resemble the FACT components, except that the C-terminal high-mobility group (HMG) box domain of SSRP1 is not found in the yeast homolog Pob3. We show here that Nhp6a and Nhp6b, small HMG box proteins with overlapping functions in yeast, associate with the CP complex and mediate CP-related genetic effects on transcription. Absence of the Nhp6 proteins causes severe impairment in combination with mutations impairing the Swi-Snf chromatin-remodeling complex and the DSIF (Spt4 plus Spt5) elongation regulator, and sensitizes cells to 6-azauracil, characteristic of elongation effects. An artificial SSRP1-like protein, created by fusing the Pob3 and Nhp6a proteins, provides both Pob3 and Nhp6a functions for transcription, and competition experiments indicate that these functions are exerted in association with Cdc68. This particular Pob3-Nhp6a fusion protein was limited for certain Nhp6 activities, indicating that its Nhp6a function is compromised. These findings suggest that in yeast cells the Cdc68 partners may be both Pob3 and Nhp6, functioning as a bipartite analog of the vertebrate SSRP1 protein.
Transcription of eukaryotic protein-coding genes takes place in the context of chromatin. In chromatin, DNA is associated with histone proteins in a nucleosomal configuration and with other proteins that have structural and/or regulatory roles. This intricate assembly of structural and regulatory proteins with DNA is generally inhibitory for transcription through the interference with transcription activator proteins and general transcription factors (reviewed in reference 57) and obstruction of the RNA polymerase II (RNAP II) elongation complex by nucleosomes (8; reviewed in reference 57). The processivity of elongation complexes is also inhibited by several nonnucleosomal chromatin components (19, 50, 60). Among the proteins regulating both nucleosomal and nonnucleosomal transcriptional inhibition is one named FACT (51).
FACT is an abundant nuclear protein complex that has been purified and characterized based on its ability to allow transcription elongation along a nucleosomal template in vitro (27, 33, 34). In keeping with this activity, the FACT complex (also termed DUF [32]) has been shown to interact with nucleosomes and with histone H2A-H2B dimers and to have DNA-untwisting activity in vitro (32, 34). FACT has also been found to counteract the inhibitory effects on transcripton elongation imposed in vitro by the DSIF complex (51). In the budding yeast Saccharomyces cerevisiae, FACT has a structural, and possibly functional, homolog in the form of the CP complex, which has been implicated by genetic evidence in the interaction of chromatin with components of the transcription and replication machineries (5, 13, 29, 30, 39, 55, 56, 60) and in the process of transcription elongation in vivo (34). Thus, genetic and biochemical evidence suggests that FACT or CP facilitates these activities by regulating chromatin. The CP complex can be purified from yeast cells as a 1:1 complex of the Cdc68 protein and the Pob3 protein (5, 56). Similarly, FACT from vertebrate cells is composed of a 1:1 complex of the human version of the Cdc68 protein and SSRP1, the human structural analog of the yeast protein Pob3 (32, 34).
The yeast and human Cdc68 proteins are structurally related throughout their lengths (34). Likewise, throughout its length the yeast Pob3 protein is similar to SSRP1 proteins from various sources (55; our analysis). However, the Pob3 protein does not contain sequences related to the C-terminal portion of SSRP1. These C-terminal SSRP1 sequences absent from Pob3 include a high-mobility group (HMG) box domain, suggesting that HMG sequences not found in Pob3 may be provided to the CP complex by another protein.
Yeast cells do indeed contain an HMG protein that resembles the HMG box domain of SSRP1. This yeast protein, termed Nhp6, is present as two virtually identical isoforms, Nhp6a and Nhp6b, differing slightly in length and encoded by a gene pair (24). Nhp6 is small, consisting of only 93 (Nhp6a) and 99 (Nhp6b) residues, and highly abundant: the combined amounts of Nhp6a and Nhp6b are at least 50,000 copies per haploid nucleus (36), somewhat greater than the abundance of the CP complex (5, 56) and about equal to the number of nucleosomes.
In this report, we describe biochemical and genetic studies showing that the small HMG box protein Nhp6 associates and functions with the yeast complex of Pob3 and Cdc68 proteins and mediates CP-related transcriptional effects in vivo. Our finding that Pob3 and Nhp6 associate in the same complex suggests that these two yeast proteins, structurally analogous to different domains of the vertebrate SSRP1 protein, may function as a bipartite yeast analog of SSRP1.
MATERIALS AND METHODS
Growth conditions and extract preparation.
Yeast strains are listed in Table 1. Cells were routinely grown at 30°C or, if temperature sensitive, at 23°C, with orbital shaking in 50 to 100 ml of adenine-supplemented (20 mg/liter) rich medium to 1 × 107 to 5 × 107 cells/ml; plasmid-bearing cells were grown in enriched selective medium (5). Whole-cell extracts were prepared by pressurizing a suspension of cells (∼109 cells/ml) in extraction buffer (NaCl or potassium acetate at the indicated concentration, 50 mM HEPES [pH 7.4], 10% glycerol, 1 mM dithiothreitol, 5 mM EDTA, 0.1% Tween 20, protease-inhibitor cocktail [5]) to 16,000 lb/in2 in a French press, followed by slow release. The resultant suspension was centrifuged at 13,500 rpm for 10 min in an Eppendorf centrifuge, and the supernatant was saved as a whole-cell extract.
TABLE 1.
Yeast strains
| Strain | Genotype | Source or reference |
|---|---|---|
| W303-1a | MATaleu2-3,112 ura3-52 his3-11,15 ade2-1 trp1-1 can1-100 | |
| W303-1b | MATα leu2-3,112 ura3-52 his3-11,15 ade2-1 trp1-1 can1-100 | |
| NQ6811 | MATaleu2-3,112 ura3-52 trp1Δ1 his4-912δ lys2-128δ CDC68-200N POB3-200N, expressing HA-Cdc68 and HA-Pob3 proteins | 5 |
| NB6241 | W303-1b with NHP6A-V5-His6 replacing NHP6A | p303-NΔXN transformant |
| RJY6249 | MATaleu2-3,112 ura3-1 his3-11,15 ade2-1 trp1-1 can1-100 adl1 Δnhp6a::URA3 Δnhp6b::LEU2 | 36 |
| NB6040 | RJY6249 with his3::HIS3 | pRS303 transformant |
| BM404 | MATaura3-52 his4-912δ lys2-128δ suc2ΔUAS | F. Winston |
| DE4B-20a | MATα cdc68-Δ922 leu2-3,112 ura3-52 trp1-1 his4-912δ lys2-128δ | DE4B (13) segregant |
| DE4B-20b | MATaleu2-3,112 ura3-52 trp1-1 his4-912δ lys2-128δ | DE4B (13) segregant |
| NB4B-1c | MATα cdc68-Δ922 leu2-3,112 ura3 trp1-1 his4-912δ Δnhp6a::URA3 Δnhp6b::LEU2 | NB6040 × DE4B-20a segregant |
| NB4B-3b | MATα cdc68-Δ922 leu2-3,112 ura3 trp1-1 lys2-128δ Δnhp6a::URA3 Δnhp6b::LEU2 | NB6040 × DE4B-20a segregant |
| AW11-9a | MATα leu2-3,112 ura3-52 trp1-1 his4-912δ lys2-128δ suc2ΔUAS | BM404 × DE4B-20a segregant |
| NB39-11a | MATα leu2-3,112 ura3 trp1-1 ade2-1 his4-912δ lys2-128δ Δnhp6a::URA3 Δnhp6b::LEU2 suc2ΔUAS | NB6040 × AW11-9a segregant |
| NB39-18a | MATaleu2-3,112 ura3 trp1-1 ade2-1 his4-912δ lys2-128δ Δnhp6a::URA3 Δnhp6b::LEU2 | NB6040 × AW11-9a segregant |
| NB39-24b | MATaleu2-3,112 ura3 trp1-1 his4-912δ lys2-128δ Δnhp6a::URA3 Δnhp6b::LEU2 Ade+ | NB6040 × AW11-9a segregant |
| FY300 | MATaleu2Δ1 ura3-52 his4-912δ lys2-128δ spt5-194 Ade+ | 49 |
| FY1671 | MATα leu2Δ1 ura3-52 his4-912δ lys2-128δ Δppr2::hisG | 19 |
| NB2Δp-28b | MATα leu2 ura3-52 trp1-1 his4-912δ lys2-128δ Δppr2::hisG Δnhp6a::URA3 NHP6B Ade+ | (FY1671 × NB39-24a) × NB39-24a segregant |
| NB40 | NB39-18a × AW11-9a | |
| NB40-1aF | MATaleu2-3,112 ura3 trp1-1 his4-912δ lys2-128δ Δnhp6a::URA3 Δnhp6b::LEU2 with POB3-NHP6A-V5-His6 replacing POB3 | NB40 segregant transformed with pPNT |
| FY711 | MATα leu2-3,112 ura3-52 his4-912δ lys2-128δ snf5-5::URA3 | 4 |
| NB6244 | W303-1a with POB3-NHP6A-V5-His6 replacing POB3 | pPNT transformant |
| NB6249 | RJY6249 with POB3-NHP6A-V5-His6 replacing POB3 | pPNT transformant |
| NB6251 | NB6249 with trp1::HIS3 | pTH4 (11) transformant |
Immunoprecipitation and Western blots.
Immunoprecipitations were performed on 0.4-ml volumes (2 to 5 mg of soluble protein/ml) using 6 μl of anti-Cdc68 antiserum or preimmune serum, 5 μg of antihemagglutinin (anti-HA) monoclonal antibody (12CA5; Boehringer Mannheim) or 2 μg of anti-V5 monoclonal antibody (Invitrogen) and a 1-h incubation at 4°C. A second 1-h 4°C incubation was then carried out with 25 μl of protein A-agarose or G-Sepharose beads (Sigma) and constant mixing. The resultant bead-bound immunoprotein mix was washed twice with 10 volumes of extraction buffer and then denatured at 90°C for 5 to 10 min in 50 μl of electrophoresis sample buffer. Proteins were visualized by the above antibodies on Western blots (5) of samples resolved electrophoretically on 8, 15, or 6-10-15% (step-gradient) polyacrylamide-sodium dodecyl sulfate gels. Nhp6 proteins were detected using a 1/1,000 dilution of anti-Nhp6a antiserum (36) and overnight incubation at 4°C.
Gel filtration and metal affinity chromatography.
Fractionation of 200,000 × g supernatant material by gel filtration used S-300 Sephacryl as described previously (5). Batch purification of His6-tagged CP complex from whole-cell extract (50 mM potassium acetate extraction buffer without EDTA, pH 8.2) used Talon metal affinity resin (Clontech) according to the manufacturer's instructions. The elution buffer was extraction buffer containing 300 mM imidazole, pH 7.4. Protein concentrations were determined by the Bradford microassay (Bio-Rad).
Construction of a POB3-NHP6A fusion gene.
Plasmid pYESPN carries the POB3-NHP6A fusion gene under the control of the GAL1 promoter and was constructed by inserting a 300-bp PCR product containing the NHP6A open reading frame (ORF), minus the first three codons, into the unique BstEII site of pYESP, a 2μm-based URA3 plasmid carrying a C-terminally (V5+His6)-tagged POB3 ORF in the pYES2/GS vector (Invitrogen). The 300-bp NHP6A PCR product was generated from the plasmid pRJ1364 (61) template using primers 5′-AGCACAGTCCATATGGGTCACCCAAGAGAGAACCTAAGAAGAGAACC (containing NdeI and BstEII sites) and 5′-AAACGCGGGGCGGCCGCGGTGACCAGCCAAAGTGGCGTTATATAACTCCTT (containing BstEII and NotI sites). The POB3-NHP6A integration plasmid pYESPNΔC was derived from pYESPN by removing a 4-kbp ClaI fragment containing the 2μm sequences and the 5′ portion of the POB3 ORF. A TRP1 version of pYESPNΔC was constructed by ligating a 3.1-kbp ScaI-ClaI fragment of pYESPN with a 2.5-kbp ScaI-ClaI fragment of pRS304 (47), generating pPNT. Replacement of the chromosomal POB3 gene with the POB3-NHP6A fusion gene used pPNT linearized at a unique BclI site in the POB3 ORF and selection for Trp+ transformants.
NHP6A plasmid manipulations.
Plasmid p314NV5, a pRS314 (47) derivative carrying the NHP6A promoter and ORF modified to encode C-terminally (V5+His6)-tagged Nhp6a protein, was generated by replacing a 300-bp NdeI-BamHI fragment of pRJ1364 with an 800-bp PCR product from pYESPN, comprising the (V5+His6)-tagged NHP6A ORF and the CYC1 transcription terminator. PCR used primer 5′-TTCTGTGGATCCCCGTATTACCGCCTTTGAGTGAGC (encoding a BamHI site) and the above-described primer encoding NdeI and BstEII sites. The 1.3-kbp XhoI-BamHI fragment of p314NV5 containing the entire tagged NHP6A ORF, ∼500 bp of 5′ noncoding sequence, and ∼500 bp of 3′ plasmid sequences (including the CYC1 transcription terminator) was ligated into pRS424 and pRS423 (9) to generate p424NV5 and p423NV5, respectively. Plasmid pHAPNV5, which expresses HA-Pob3 and Nhp6a-V5-His6 proteins from independent genes, was generated by inserting a 1.3-kbp XhoI-BamHI p314NV5 fragment, blunt-ended with mung bean exonuclease, into the SmaI site of p424-HAPOB (see below). The NHP6A-V5-His6 integration plasmid p303-NΔXN was generated by blunt-end (exonuclease) religation after removing a 500-bp XhoI-NdeI sequence from p303-NV5, which was itself created by inserting a 1.3-kbp XhoI-BamHI NHP6A fragment from p314-NV5 into pRS303 (47). Replacement of the chromosomal NHP6A gene with the V5-tagged version used p303-NΔXN linearized with EcoRV and selection for His+ transformants.
POB3 plasmid manipulations.
Plasmid p424-HAPOB was generated by inserting a 3-kbp KpnI-EcoRI fragment of YEpHAPOB (5), comprising the HA-Pob3 ORF plus 5′ and 3′ noncoding sequences, into pRS424. The integration plasmid pPH6TI carrying the C-terminal half of the ORF encoding Pob3-V5-His6 was created by ligating a 2.8-kbp ClaI-ScaI fragment of pYESP with a 2.5-kbp ClaI-ScaI fragment of pRS304. Integration plasmid pYESPΔC was created by removing a 4-kbp ClaI fragment from pYESP. Replacement of the chromosomal POB3 gene with the tagged version used pYESPΔC (URA3) or pPH6TI (TRP1) linearized with BclI.
RESULTS
Nhp6a/b protein associates with the CP complex.
The close structural relationship between the yeast CP complex (Cdc68 plus Pob3) and the analogous vertebrate complexes comprising Cdc68 and the Pob3- and Nhp6-related protein SSRP1 (32, 34) suggests that the yeast Nhp6 and Cdc68 proteins may associate in the same complex. To determine if the Nhp6 proteins do indeed associate physically with Cdc68, Western blots of anti-Cdc68 immunoprecipitated protein (5) were probed for the presence of Nhp6 using polyclonal anti-Nhp6a antibody (36), which detects both Nhp6 isoforms. Nhp6a and/or Nhp6b (here termed Nhp6a/b) was not found in anti-Cdc68 immunoprecipitates using our previously reported conditions, and varying the pH (pH 7.4, 8.4, and 9.0) of the extraction buffer had little effect (data not shown). On the other hand, the use of 50 mM potassium acetate in place of 350 mM NaCl resulted in the presence of Nhp6a/b in the anti-Cdc68 immunoprecipitate (Fig. 1A). This coimmunoprecipitation of Nhp6a/b with Cdc68, and thus the CP complex, was unaffected over the pH range described above (data not shown) but was lost at potassium acetate concentrations ≥100 mM (Fig. 1A).
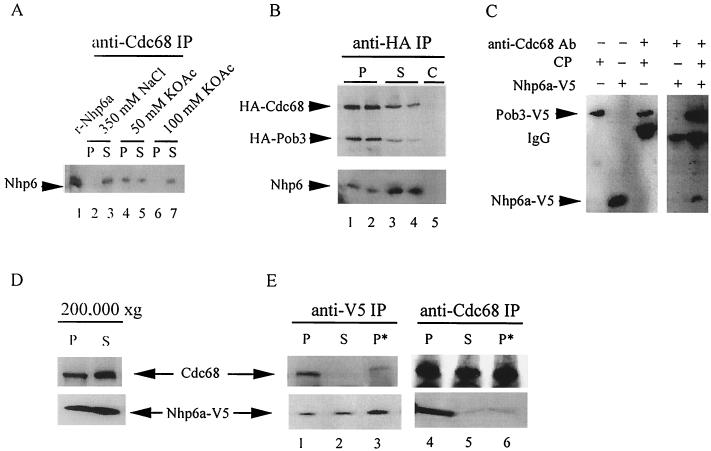
FIG. 1.
Nhp6 protein physically associates with the CP complex. (A) Material immunoprecipitated by anti-Cdc68 antibody (P) from a whole-cell extract (strain W303-1a) and that remaining in the supernatant (S) were resolved electrophoretically on a 15% polyacrylamide–sodium dodecyl sulfate gel and analyzed by Western blotting with anti-Nhp6a antibody. The buffer for extract preparation and immunoprecipitation contained 350 mM NaCl, 50 mM potassium acetate, or 100 mM potassium acetate as indicated. Recombinant Nhp6a protein (25 ng) served as the marker. (B) Material immunoprecipitated (P) or remaining in the supernatant (S) after treatment of whole-cell extracts of cells containing HA-tagged Cdc68 and Pob3 proteins (strain NQ6811) with anti-HA monoclonal antibody was resolved (8 and 15% gels) and analyzed by Western blotting with anti-HA and anti-Nhp6a antibodies. The negative control (designated C) comprised a whole-cell extract of untagged cells (strain W303-1a) treated as described above. Immunoprecipitations are shown for two independent experiments. (C) Nhp6a and the CP complex associate in vitro. Nhp6a-V5-His6 protein and CP containing Pob3-V5-His6 were separately purified (from strains RJY6249 harboring p314NV5 and NB6244) using metal affinity resin as described in the text, mixed, and immunoprecipitated with anti-Cdc68 antibody. Resulting Western blots of purified and immunoprecipitated material were probed with anti-V5 antibody. Data are from two independent experiments. (D) Nhp6-associated CP complex associates with high-molecular-weight material. A whole-cell extract from cells containing Nhp6a-V5-His6 protein (strain NB6241) made in 50 mM potassium acetate buffer was fractionated by centrifugation (1.5 h) at 200,000 × g to yield supernatant (S) and pellet (P) fractions. The pellet fraction was then resuspended in an original volume of 50 mM potassium acetate or 350 mM NaCl extraction buffer (P∗). Equal volumes were resolved (6-10-15% gel) and assayed by Western blotting with anti-Cdc68 or anti-V5 antibody. (E) Material immunoprecipitated from equal volumes of the indicated fractions with anti-Cdc68 or anti-V5 antibody was resolved (6-10-15% gels) and assayed by Western blotting using anti-Cdc68 and anti-V5 antibody.
Extracts were made, using 50 mM K acetate buffer, from cells containing either HA-tagged Pob3 (5) or HA-tagged Cdc68 (59). Using anti-HA monoclonal antibody, Nhp6a/b was found to coimmunoprecipitate with each of the two different HA-tagged versions of the CP complex (data not shown). From an extract of doubly tagged HA-Cdc68 HA-Pob3 cells (5), approximately 50% of both Cdc68 and Pob3 were immunoprecipitated by anti-HA antibody (Fig. 1B). In contrast, only 5% of total Nhp6a/b protein was present in the anti-HA immunoprecipitate (Fig. 1B). These values indicate that ∼10% of the total Nhp6a/b protein pool of 50,000 copies/cell (36) can be found in association with the CP complex, whose abundance is estimated at 10,000 to 50,000 copies/cell (5, 56). Based on these abundance estimates, from 10% to as much as 50% of the CP complexes are associated with Nhp6a/b protein.
The association of Nhp6a/b with CP was also assessed for CP complex affinity purified from cells in which Pob3 protein is linked to a V5 epitope and a run of six histidine residues. This tagged version of Pob3 provides essential Pob3 function in vivo, as indicated by robust cell growth (data not shown). An extract was prepared from these cells using 50 mM potassium acetate buffer and treated with metal affinity resin, which binds the His6 tag at the Pob3 C terminus. Western blot analysis of the bound versus unbound material again showed approximately 10% of the total amount of Nhp6a/b associated with CP (data not shown). We conclude that the CP complex physically associates with Nhp6a/b in vivo.
Association of Nhp6a with CP was reproduced in vitro. CP complex containing Pob3-V5-His6 was affinity purified using 350 mM NaCl buffer, eluted in 50 mM potassium acetate buffer, and mixed with (V5+His6)-tagged Nhp6a protein similarly purified. Under these buffer conditions, Nhp6a, identified by its V5 tag, was coimmunoprecipitated with anti-Cdc68 antibody (Fig. 1C).
Centrifugation at 200,000 × g was used to fractionate a whole-cell extract (50 mM potassium acetate buffer) of cells expressing the Nhp6a-V5-His6 protein from the chromosomal NHP6A locus into soluble and high-molecular-weight (pelleted) material; approximately 40% of the total amounts of Cdc68 and Nhp6a proteins were pelleted (Fig. 1D). A similar distribution of Cdc68 between soluble and pelleted, histone-containing material has been reported for a related fractionation procedure (56). In contrast, the CP complex in whole-cell extracts made using buffer containing 350 mM NaCl is at least 90% soluble, as defined by high-speed (100,000 × g) centrifugation (5). Therefore the 50 mM potassium acetate buffer, which allows the identification of CP-Nhp6 interactions, also alters the centrifugation characteristics of the CP complex. Association of Cdc68 with the high-molecular-weight material was also found for extracts of Δnhp6a/b mutant cells and is therefore independent of Nhp6 protein (data not shown). The soluble fraction and the high-molecular-weight material, resuspended in extraction buffer containing either 50 mM potassium acetate or 350 mM NaCl, were then subjected to immunoprecipitation. Similar amounts of Cdc68 protein immunoprecipitated with anti-Cdc68 antibody from all three fractions. Probing the same blot with anti-V5 antibody showed that Nhp6a coimmunoprecipitated with Cdc68 predominantly from the high-molecular-weight fraction resuspended in 50 mM potassium acetate buffer (Fig. 1E). The presence of ethidium bromide during resuspension and immunoprecipitation did not decrease this coimmunoprecipitation (data not shown), suggesting that protein-protein interactions rather than protein-DNA interactions mediate the association of Nhp6 with Cdc68 (25). As found when using whole-cell extracts, coimmunoprecipitation of Nhp6a protein with Cdc68 was markedly decreased when using high-molecular-weight material resuspended in 350 mM NaCl buffer, indicating that these conditions destabilize interactions responsible for Cdc68-Nhp6a association (Fig. 1E). Despite the similar amounts of Nhp6a in all three fractions, Cdc68 was effectively coimmunoprecipitated with Nhp6a (anti-V5 antibody) only from the high-molecular-weight fraction resuspended in 50 mM potassium acetate buffer, thus mirroring the anti-Cdc68 coimmunoprecipitation results (Fig. 1E). These findings verify the association of Nhp6a protein with the CP complex and suggest that this association may be stabilized by alterations to the CP complex and/or Nhp6a which facilitate CP complex and/or Nhp6a interactions with other proteins.
Nhp6 protein mediates promoter repression.
Genetic and biochemical assays have implicated both the CP complex and the Nhp6 proteins in transcriptional regulation (13, 29, 30, 36, 43). The Nhp6a and Nhp6b proteins are functionally redundant and important yet not essential for growth (10, 36), allowing the use of null alleles of the NHP6A and NHP6B genes (nhp6aΔ::URA3 and nhp6bΔ::LEU2) (36). Studies were carried out using nhp6aΔnhp6bΔ double-mutant null cells (designated here as Δnhp6a/b cells) and the plasmid-borne NHP6A gene, which encodes the more abundant Nhp6 isoform (36).
Two informative reporter genes for transcription have been his4-912δ and lys2-128δ (53, 54). Transcription from each of these genes is altered by the insertion of a Ty 1 retrotransposon long terminal repeat (δ element); as a result, cells harboring these mutant alleles are unable to make functional his4-912δ and lys2-128δ transcripts and are auxotrophic for histidine and lysine. Restoration of functional transcription at these alleles allows growth on medium lacking histidine and/or lysine. This effect (known as the Spt− phenotype) has been demonstrated for several mutations that affect chromatin function, including cdc68 and pob3 mutations (13, 30, 45, 58).
Growth of haploid Δnhp6a/b his4-912δ lys2-128δ segregants on medium lacking histidine was significant at 23°C, although less robust than the growth of Δnhp6a/b HIS4 segregants from the same cross. Similarly, Δnhp6a/b lys2-128δ segregants grew on medium lacking lysine but in a temperature-dependent fashion, showing better growth at 30 than at 23°C (data not shown). Suppression of the His− phenotype of his4-912δ was seen only in segregants containing both the nhp6aΔ and nhp6bΔ mutations (Fig. 2A). Transformation of Δnhp6a/b his4-912δ lys2-128δ cells with a low-copy-number NHP6A plasmid restored the histidine and lysine auxotrophies (data not shown), confirming that repression of functional transcription at his4-912δ and lys2-128δ is mediated by Nhp6 protein.
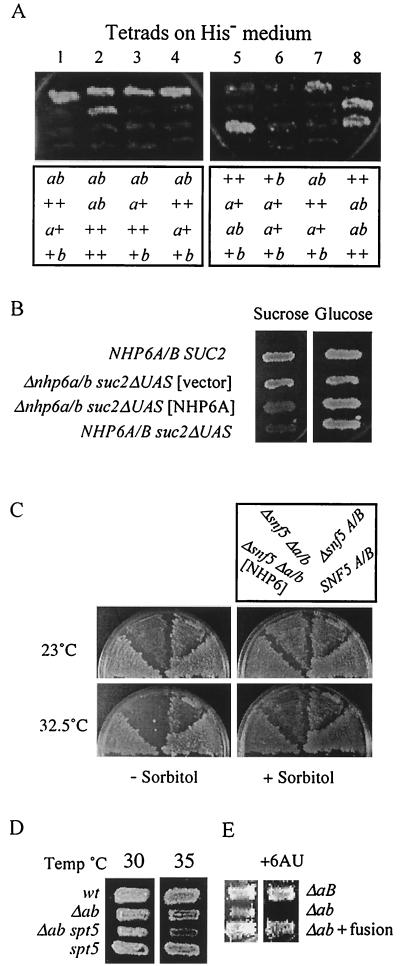
FIG. 2.
Nhp6 protein mediates transcription. (A) Haploid segregants from diploid strain NB40 (Δnhp6a::URA3/+ Δnhp6b::LEU2/+ his4-912δ/his4-912δ lys2-128δ/lys2-128δ) were grown on solid rich medium and replica plated to histidine-free medium to assess functional transcription from the his4-912δ reporter gene. The lower panels indicate NHP6A/B genotypes as indicated by Ura and Leu phenotypes. (B) Representative segregants from AW11-9a × NB6040 with genotypes suc2ΔUAS Δnhp6a/b harboring the NHP6A plasmid p314-NV5 or the pRS314 vector, SUC2 Δnhp6a/b, and suc2ΔUAS NHP6A NHP6B were grown on solid rich medium at 28°C, replica plated to rich medium containing glucose or sucrose (plus antimycin A at 1 μg/ml) as the carbon source, and incubated 3 days at 23°C. (C) Representative segregants from FY711 × NB39-18a with genotypes snf5-5::URA3 Δnhp6a/b with or without p314-NV5, SNF5 NHP6A NHP6B, and snf5-5 NHP6A NHP6B were grown on solid rich medium at 23°C, replica plated to rich medium, with or without 1 M sorbitol, containing glucose or sucrose (plus antimycin A) as the carbon source, and incubated for 6 days at 23 and 33°C. (D) Representative segregants from NB39-24b × (FY300 × NB39-11a segregant) were replica plated onto rich medium and incubated at the indicated temperatures. (E) Representative PPR2 segregants from NB2Δp-28b × NB40-1aF with genotypes Δnhp6a::URA3 Δnhp6b::LEU2, Δnhp6a::URA3 NHP6B, and Δnhp6a::URA3 Δnhp6b::LEU2 with POB3-NHP6A in place of POB3 were replica plated to uracil-free medium containing 6AU (100 μg/ml) and incubated at 28°C.
In cells that have Nhp6 protein, increased dosage of the CDC68 gene allows growth of his4-912δ lys2-128δ cells on medium lacking histidine and lysine (5, 30). In contrast, neither a high-copy NHP6A plasmid nor the increased dosage of both NHP6A and POB3 together had any effect on the His− or Lys− phenotype of his4-912δ lys2-128δ cells (data not shown). Furthermore, neither increased dosage of NHP6A nor increased dosage of both NHP6A and POB3 suppressed the Spt− (His+ Lys+) phenotype of his4-912δ lys2-128δ cells arising from increased CDC68 dosage (data not shown). The benign effect of excess Nhp6a protein is similar to the lack of effect of increased POB3 gene dosage on the his4-912δ and lys2-128δ reporter genes (5).
Chromatin is thought to maintain repression at the SUC2 gene promoter in the absence of SUC2 upstream activating sequences (UAS). Transcription from the core promoter of this suc2ΔUAS mutant allele, as indicated by growth on solid medium containing sucrose or raffinose as the sole carbon source, can be increased by mutating Cdc68 (13, 30, 39, 58) and proteins that alter chromatin (39). On sucrose medium at 30°C, Δnhp6a/b suc2ΔUAS segregants grew significantly better than did NHP6A NHP6B suc2ΔUAS segregants from the same genetic cross and showed only marginally less growth than NHP6A NHP6B SUC2 segregants. A low-copy NHP6A plasmid in Δnhp6a/b suc2ΔUAS cells prevented growth on sucrose medium, indicating that Nhp6a protein helps to maintain chromatin repression at the suc2ΔUAS core promoter (Fig. 2B). The degree of suppression of the suc2ΔUAS Suc− phenotype for cells lacking the Nhp6 proteins was similar to that for NHP6A NHP6B suc2ΔUAS cells harboring the cdc68-Δ922 mutation (13 and data not shown). Therefore Nhp6 protein, like Cdc68, helps to maintain repression at a core promoter.
Nhp6 protein facilitates transcription.
Like CP (13, 29, 43), Nhp6 protein facilitates activated transcription, in vivo and in vitro (36, 46). The interactions of Nhp6 with different transcription modulators were therefore investigated genetically.
Transcription of a subset of genes is brought about in part through the actions of the Swi-Snf complex (reviewed in reference 23), a chromatin-remodeling complex that interacts genetically with the CP complex (13, 30). Cells missing the Swi-Snf component Snf5 are unable to assemble a functional Swi-Snf complex (37); as a consequence, snf5 cells exhibit a mild growth impairment on rich medium. Using standard genetic techniques, the nhp6aΔ::URA3, nhp6bΔ::LEU2, and snf5-5::URA3 mutant alleles were reassorted in meiotic tetrads, and initial analysis focused on a tetrad in which the URA3 genes marking snf5 and nhp6aΔ cosegregated in two of the segregants, one of which also contained nhp6bΔ::LEU2. This Δnhp6a/b snf5 triple-mutant segregant grew poorly on solid rich medium and had the novel phenotype of temperature sensitivity at 32.5°C (data not shown). In contrast, the other segregants from the same tetrad and additional segregants that retained either Swi-Snf or Nhp6 (Nhp6a and/or Nhp6b) function grew much more effectively at this temperature, as did the NHP6A NHP6B snf5 and Δnhp6a/b SNF5 parental strains (Fig. 2C). Thus the combination of Δnhp6a/b and snf5 creates a synthetic lethality at 32.5°C. The NHP6A gene on a low-copy-number plasmid eliminated the 32.5°C growth defect of these Δnhp6a/b snf5 triple-mutant cells and allowed growth equivalent to that of the NHP6A NHP6B snf5 parental control cells (Fig. 2C). Furthermore, the temperature sensitivity of Δnhp6a/b snf5 cells was partially alleviated by the inclusion of sorbitol in the growth medium (Fig. 2C), a situation which also partially alleviates the high-temperature (38°C) growth defect of Δnhp6a/b mutant cells (10). These observations indicate that Nhp6 protein and the Swi-Snf complex similarly facilitate transcription important for growth at 32.5°C; consequently, the absence of Nhp6 protein imposes a need for the Swi-Snf complex at certain genes (and vice versa).
The Swi-Snf complex facilitates transcription of the SUC2 gene (1, 21), as evidenced by the poor growth of snf5 mutant cells using sucrose as a carbon source. We noted that Δnhp6a/b snf5 cells grow poorly on sucrose medium even at 23°C (data not shown), but the general growth impairment of these mutant cells, and its incomplete remediation by sorbitol, confounded our attempts to assess genetic interactions in SUC2 transcription. In cells with Swi-Snf function, the absence of Nhp6 protein did not impair growth on sucrose medium (data not shown), indicating that Nhp6 is dispensable for Swi-Snf-activated SUC2 transcription.
The synthetic lethality seen for Δnhp6a/b snf5 cells contrasts with the suppression of the effects of snf5 and another Swi-Snf mutation by alteration of the CP complex (13, 30). This difference suggests that the CP complex and Nhp6 interact functionally with the Swi-Snf complex in different ways. Gene dosage effects also indicate a functional difference. An increased dosage of the CDC68 and/or POB3 genes suppresses several effects of Swi-Snf dysfunction (5, 30), whereas an increased dosage of the NHP6A gene did not alleviate the Suc− and Ino− consequences of the snf5 mutation (data not shown).
Another transcription modulator is the complex containing the Spt4 and Spt5 proteins (19, 50). This complex, like CP, has both positive and negative effects on yeast cell growth (19), while the mammalian Spt4-Spt5 complex, termed DSIF, has been found to modulate the acquisition of elongation competence in vitro by the transcriptionally engaged RNAP II enzyme (50). The absence of Nhp6 protein produces a synthetic lethality with the spt5-194 mutation (19) (Fig. 2D), implicating Nhp6 in transcription elongation.
Mutations that affect transcription elongation, such as Δppr2, which eliminates sequences encoding transcription elongation factor TFIIS (15, 31), may confer sensitivity to the analog 6-azauracil (6AU). We found that Δnhp6a/b mutant cells were sensitive to 6AU (Fig. 2E), consistent with a role for Nhp6 protein in modulating transcription elongation. Over a range of 6AU concentrations, the sensitivity of Δnhp6a/b Δppr2 triple-mutant cells was no greater than that of Δnhp6a/b or Δppr2 cells (data not shown), consistent with Nhp6 modulation of 6AU resistance through TFIIS function. Thus, several indicators implicate Nhp6 in transcription, specifically in elongation.
cdc68-Δ922 has effects resembling those caused by the absence of Nhp6 protein.
Cells lacking both Nhp6a and Nhp6b proteins are temperature sensitive at 38°C, as are cells harboring the cdc68-Δ922 allele (10, 13, 36). Cells lacking Nhp6 are also sensitive to nitrogen starvation, as indicated by a loss of viability during incubation under nitrogen-deficient conditions (10); similarly, cdc68-Δ922 mutant cells were also found to be sensitive to nitrogen starvation. Cells were replica plated to synthetic medium without ammonium sulfate and incubated at 23°C for starvation, and then replica plated to rich medium for further incubation at 23°C to assess cell growth as a measure of cell survival. After 4 days of nitrogen starvation, the cdc68-Δ922 mutant cells exhibited significantly less growth on rich medium than did wild-type cells (Fig. 3A), indicating that many of the starved cdc68-Δ922 cells lost the capacity for colony formation. The cdc68-Δ922 colonies that did form were not the consequence of additional mutations, as cells from several cdc68-Δ922 colonies that formed after nitrogen starvation displayed the same overall sensitivity to nitrogen starvation as that of the original cdc68-Δ922 cells (data not shown). Survival decreased with starvation time, as indicated by the virtual absence of regrowth of cdc68-Δ922 cells, and of Δnhp6a/b cells, after 5 days of nitrogen starvation; in contrast, wild-type cells similarly starved showed significant regrowth (data not shown). Thus, cells with the cdc68-Δ922 mutant version of the CP complex display a growth phenotype resembling that caused by the absence of Nhp6 protein.
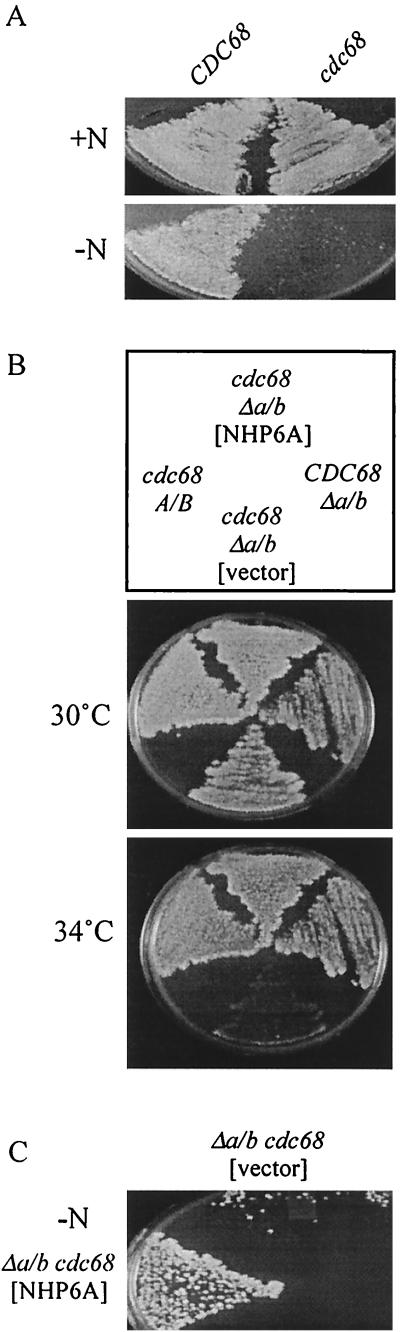
FIG. 3.
Nhp6 and Cdc68 proteins have related effects. (A) Cdc68 function helps to maintain viability upon nitrogen starvation. Cells of the cdc68-Δ922 strain DE4B-20a and the CDC68 strain DE4B-20b were grown on solid rich medium, replica plated to enriched synthetic medium lacking (NH4)2SO4, incubated 4 days, and then replica plated to rich medium and incubated 2 days to allow growth after starvation. All incubations were carried out at 23°C. (B) Nhp6 facilitates CP function. Cells of the cdc68-Δ922 Δnhp6a/b strain NB4B-3b harboring p314-NV5 or pRS314, the CDC68 Δnhp6a/b strain NB6040 harboring pRS314, and the cdc68-Δ922 NHP6A NHP6B strain DE4B-20a harboring pRS314 were grown on enriched selective medium at 23°C, replica plated to the same medium, and incubated at 30 and 34°C. (C) Cells of the cdc68-Δ922 Δnhp6a/b strain NB4B-1c harboring p314-NV5 or pRS314 were grown on solid enriched selective medium, replica plated to enriched selective medium lacking (NH4)2SO4, incubated 2 days, and then replica plated to rich medium and incubated 6 days to allow growth after starvation.
The temperature sensitivity and the starvation sensitivity of Δnhp6a/b mutant cells are partially alleviated by the presence of the osmotic stabilizer sorbitol (10; our observations). However, sorbitol (1 M) did not alleviate the temperature sensitivity or the starvation sensitivity of cdc68-Δ922 mutant cells (data not shown). This observation suggests that the temperature and starvation sensitivities of Δnhp6a/b and of cdc68-Δ922 are due to different effects.
Nhp6 protein facilitates CP activity in vivo.
A cross between Δnhp6a/b and cdc68-Δ922 parental strains showed that Δnhp6a/b cdc68-Δ922 triple-mutant segregants were compromised for growth and survival compared to cdc68-Δ922 segregants with either Nhp6a or Nhp6b function and to parental cells. The Δnhp6a/b cdc68-Δ922 triple-mutant cells were temperature sensitive for growth at 34°C, a temperature at which the Δnhp6a/b and cdc68-Δ922 parental strains and segregants with these genotypes show good growth (Fig. 3B), and lost viability more rapidly upon nitrogen starvation (Fig. 3C). The more severe temperature and starvation sensitivities of Δnhp6a/b cdc68-Δ922 cells were alleviated by a low-copy-number NHP6A plasmid (Fig. 3B and C) or a low-copy-number CDC68 plasmid (data not shown). Thus, the absence of Nhp6 protein is synthetically lethal with the cdc68-Δ922 mutation, a finding that strengthens the view that Nhp6 and the CP complex have related effects on transcription. In contrast to what is seen for the temperature sensitivity of Δnhp6a/b CDC68 cells, 1 M sorbitol did not alleviate the synthetically enhanced temperature sensitivity of Δnhp6a/b cdc68-Δ922 cells (data not shown). This synthetic enhancement is therefore a likely consequence of an impaired CP complex, rather than the absence of Nhp6 protein, so that CP-related temperature sensitivity is exacerbated in Δnhp6a/b cdc68-Δ922 triple-mutant cells. Thus Nhp6 facilitates high-temperature growth when the CP complex is compromised by mutation.
Pob3-Nhp6a fusion creates a yeast version of SSRP1 with Pob3 function.
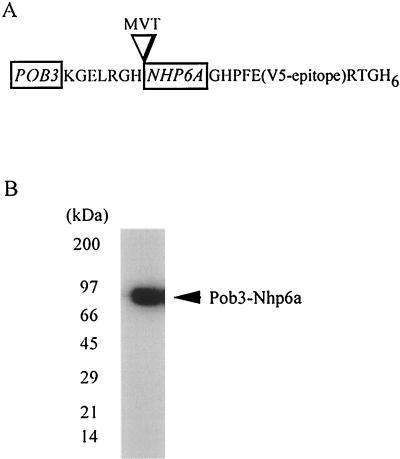
The biochemical and genetic findings discussed above suggest that the Nhp6 protein may function in association with the CP complex of Cdc68 and Pob3 and that the yeast Pob3 and Nhp6 proteins, structurally homologous to N-terminal and C-terminal regions, respectively, of the vertebrate SSRP1 protein, may function together. To test these ideas, the Nhp6a protein was covalently attached to the C terminus of Pob3, resulting in a Pob3-Nhp6a fusion protein that resembles SSRP1 (in primary sequence). The Pob3-Nhp6a fusion protein contains the entire sequence of Pob3, a 7-residue spacer, the entire Nhp6a sequence minus the first three amino acids (which are dispensable for Nhp6a function [61]), and the V5 epitope and His6 tag at its C terminus (Fig. 4A).
FIG. 4.
Pob3-Nhp6a fusion protein. (A) Schematic of the Pob3-Nhp6a protein tagged with V5 epitope and His6 tract; single-letter amino acid abbreviations are used. (B) The Pob3-Nhp6a-V5-His6 fusion protein is intact in vivo. A whole-cell extract (350 mM NaCl extraction buffer) from Δnhp6a/b POB3-NHP6A cells (strain NB6249) was resolved (6-10-15% gel) and analyzed by Western blotting with anti-V5 antibody.
To determine if the Pob3-Nhp6a fusion protein provides essential Pob3 function, the POB3-NHP6A gene was placed under the control of the regulated GAL1 promoter on a plasmid and transformed into Δpob3 mutant cells kept alive by the POB3 plasmid pGADPOB (5). Transformants were then grown in selective medium containing galactose and screened for leucine auxotrophy, indicating loss of the pGADPOB plasmid. The Δpob3 mutant cells that had lost pGADPOB but retained the POB3-NHP6A plasmid showed robust growth on galactose, indicating that the Pob3-Nhp6a fusion protein provides Pob3 function (data not shown). On glucose medium, which represses transcription of the plasmid-borne POB3-NHP6A gene, growth was greatly inhibited (data not shown), indicating that the Pob3-Nhp6a protein provides the essential function(s) of Pob3.
The chromosomal copy of the POB3 gene was replaced with the POB3-NHP6A gene under the control of POB3 transcription regulatory sequences. These transformants, containing Pob3-Nhp6a instead of Pob3 protein, grew as robustly as wild-type cells, indicating that the POB3-NHP6A gene, expressed at normal POB3 levels, provides wild-type Pob3 function (see Fig. 5). The Pob3-Nhp6a fusion protein in these cells migrated at an apparent size consistent with the addition of the Nhp6a-V5-His6 polypeptide to Pob3 protein (70 kDa) (5). Moreover, the Pob3-Nhp6a fusion protein is fully intact in vivo (Fig. 4B); therefore, biological activity of the POB3-NHP6A gene can be attributed to full-length Pob3-Nhp6a protein.
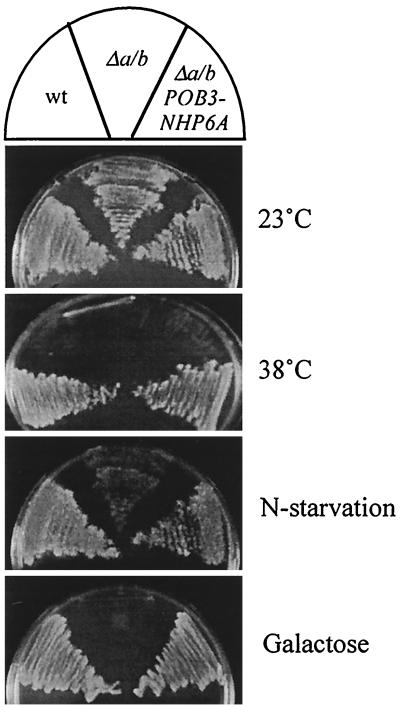
FIG. 5.
The Pob3-Nhp6a protein provides Pob3 and Nhp6a activity in vivo. Cells of the Δnhp6a/b POB3-NHP6A-V5-His6 strain NB6249 (designated Δa/b POB3-NHP6a), the Δnhp6a/b POB3 strain RJY6249 (designated Δa/b), and the NHP6A NHP6B POB3-V5-His6 strain NB6244 (designated wt) were grown on solid rich glucose medium at 23 and 38°C, on rich galactose medium at 23°C, and on rich glucose medium at 23°C following a 5-day incubation at 23°C on enriched medium lacking (NH4)2SO4 (designated N-starvation).
Pob3-Nhp6a protein provides Nhp6a activity.
The chromosomal replacement of POB3 with POB3-NHP6A was made in cells lacking Nhp6 protein (Table 1). The replacement of the POB3 gene with POB3-NHP6A in Δnhp6a/b mutant cells restores growth at 38°C (Fig. 5) and sensitivity to 6AU (Fig. 2E), while POB3-NHP6A expression in Δnhp6a/b cells alleviates synthetic interaction with spt5-194 (data not shown). The Pob3-Nhp6a protein also allows Δnhp6a/b cells to survive nitrogen starvation, although growth of Δnhp6a/b POB3-NHP6A cells after starvation was less substantial than that of wild-type cells (Fig. 5). Another test of Nhp6 function is growth on galactose. Cells lacking Nhp6a and Nhp6b are impaired for GAL1 induction (36) and unable to grow effectively using galactose as the carbon source (62). Similarly, we saw an extended (7-day) lag for Δnhp6a/b mutant cells on galactose solid medium at 23°C, and only low-level growth was ever achieved (Fig. 5). In contrast, robust galactose-dependent growth was evident after a 2-day incubation for Δnhp6a/b cells that contained the Pob3-Nhp6a protein in place of Pob3 (Fig. 5) and was indistinguishable from the growth of Δnhp6a/b cells harboring the wild-type NHP6A gene on a low-copy-number plasmid (data not shown). Thus, by several criteria the Pob3-Nhp6a fusion protein supplies both Pob3 and Nhp6a function in vivo.
Pob3-Nhp6a fusion protein associates with Cdc68 in the CP complex.
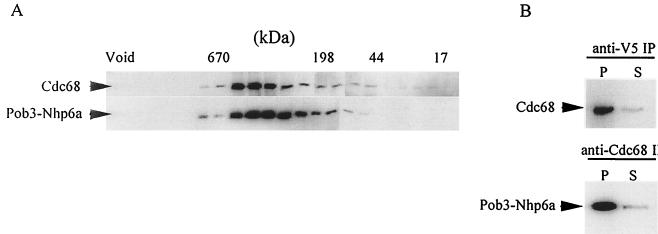
A high-speed supernatant fraction from Δnhp6a/b POB3-NHP6A cells was size-fractionated and assayed for the distributions of Cdc68 and Pob3-Nhp6a. As shown in Fig. 6A, both proteins cofractionated in the size range of the Cdc68 + Pob3 (CP) complex (5), suggesting that the Cdc68 and Pob3-Nhp6a proteins form an in vivo complex analogous to the CP complex. To verify this association, the proteins were separately immunoprecipitated from a clarified whole-cell extract using anti-V5 and anti-Cdc68 antisera. The anti-V5 antibody immunoprecipitated Pob3-Nhp6a, as expected, and coimmunoprecipitated Cdc68 (Fig. 6B). The reciprocal experiment using anti-Cdc68 antiserum to immunoprecipitate Cdc68 from the same clarified whole-cell extract showed that Pob3-Nhp6a coimmunoprecipitated with Cdc68 (Fig. 6B). Comparisons of the amounts of immunoprecipitated material versus residual soluble material (Fig. 6B) indicated that Cdc68 and Pob3-Nhp6a are present in the complex in equal stoichiometric amounts, as previously demonstrated for the CP complex of Cdc68 and wild-type Pob3 (5). These observations suggest that the Nhp6 activity attributed above to the Pob3-Nhp6a fusion protein is largely due to Nhp6a in covalent association with the CP complex.
FIG. 6.
The Pob3-Nhp6a protein forms a stable complex with Cdc68. (A) Material in the 200,000 × g supernatant fraction from cells of strain NB6249 was size fractionated by gel filtration (4) and analyzed by Western blotting using anti-Cdc68 and anti-V5 antibodies. Size markers (Bio-Rad) were cofractionated with the extract. (B) Equal volumes of the same 200,000 × g supernatant were treated with anti-Cdc68 or anti-V5 antibody and the resultant immunoprecipitates and corresponding supernatants were resolved (8% gel) and analyzed by Western blotting with anti-Cdc68 and anti-V5 antibody.
Nhp6 functions of Pob3-Nhp6a protein may depend on Cdc68 association.
Both Pob3 and the Pob3-Nhp6a fusion protein can associate with Cdc68; Pob3-Nhp6a protein, however, has the added ability to provide Nhp6a function. Pob3-Nhp6a protein may even be physically associated with Cdc68 for this Nhp6a function. To test this idea, the ability of Pob3 protein to displace Pob3-Nhp6a from Cdc68 was assessed under growth conditions that necessitate the Nhp6a activity of the Pob3-Nhp6a protein.
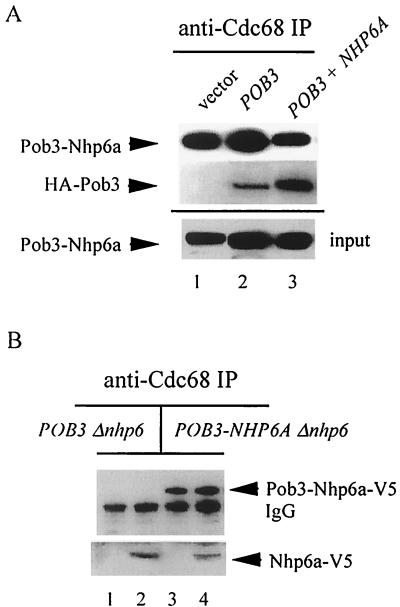
For this experiment, Δnhp6a/b POB3-NHP6A cells were transformed with a multicopy plasmid expressing HA-Pob3 from the POB3 promoter. In parallel, the same Δnhp6a/b POB3-NHP6A cells were transformed with a multicopy plasmid expressing both POB3 (HA-Pob3) and NHP6A (Nhp6a-V5-His6) as separate genes. Both transformant types, plus a vector-transformant control, were grown at 37°C, a temperature at which Nhp6 function is necessary for robust growth, and harvested in mid-log phase for the preparation of whole-cell extracts. Anti-Cdc68 antiserum was used to immunoprecipitate the CP complex, and levels of coimmunoprecipitating Pob3-Nhp6a protein were determined. Under these 37°C growth conditions, Pob3-Nhp6a was associated with Cdc68 in all cases but in amounts twofold greater in the absence of the individual Nhp6a and HA-Pob3 proteins than in their presence (Fig. 7A, compare lanes 1 and 3). The presence of HA-Pob3 alone did not decrease the amount of Pob3-Nhp6a associated with Cdc68 (Fig. 7A, lane 2). These observations indicate that growth at 37°C favors the association of Cdc68 with Pob3-Nhp6a rather than Pob3 (in the absence of native Nhp6 protein). To show that HA-Pob3 displaces Pob3-Nhp6a from Cdc68 when Nhp6a is present, the same blot was probed with anti-HA to assess the level of HA-Pob3 in the CP complex. Although at 37°C a small fraction of HA-Pob3 was incorporated into the CP complex in the absence of Nhp6a (Fig. 7A, lane 2), there was a marked increase in the level of HA-Pob3 in the CP complex when Nhp6a was present in the cell (Fig. 7A, lane 3). The increased amount of Cdc68-associated HA-Pob3 was consistent with the degree of displacement of Pob3-Nhp6a under those conditions. Similar experiments suggest that the association of Pob3-Nhp6a with Cdc68 also facilitates growth on galactose (data not shown). These findings therefore suggest that for some aspects of Nhp6a function, including 37°C growth and galactose catabolism, the Nhp6a protein may have to be associated with the CP complex, a situation that can be satisfied by either covalent (Pob3-Nhp6a) or noncovalent interactions.
FIG. 7.
Interactions of the complex containing Pob3-Nhp6a and Cdc68. (A) An Nhp6a function of the Pob3-Nhp6a protein depends on Cdc68 association. Cells of the Δnhp6a/b POB3-NHP6A-V5-His6 strain NB6251 harboring the pRS424 vector (lane 1) or the POB3 plasmid p424-HAPOB (lane 2) or the POB3 + NHP6A plasmid pHAPNV5 (lane 3) were grown at 37°C in enriched selective medium to ∼107 cells/ml. Whole-cell extracts were treated with anti-Cdc68 antibody, and the immunoprecipitates were resolved (8% gel) and analyzed for coimmunoprecipitation of Pob3-Nhp6-V5 and HA-Pob3 using anti-V5 and anti-HA antibodies. On the same blot (but in different lanes), whole-cell extracts (input) were probed with anti-V5 antibody to indicate the relative amounts of extract used for immunoprecipitation. (B) The assemblage of Cdc68 and Pob3-Nhp6a protein binds Nhp6a. Anti-Cdc68 immunoprecipitates from whole-cell extracts of Δnhp6a/b POB3 cells (strain RJY6249) and Δnhp6a/b POB3-NHP6A-V5-His6 cells (strain NB6251), each housing p314-NV5 (lanes 2 and 4) or the pRS314 vector (lanes 1 and 3), were resolved (6-10-15% gel) and analyzed by Western blotting with anti-V5 antibody.
Pob3-Nhp6a is compromised for certain Nhp6a functions.
Despite the above findings, Δnhp6a/b POB3-NHP6A cells do not grow as well as cells in which native Nhp6a protein is present, suggesting that the Pob3-Nhp6a protein lacks full Nhp6a activity. Inadequacy is also evident for Nhp6a activities related to chromatin repression: the Pob3-Nhp6a protein could not suppress the His+ Lys+ phenotype or the Suc+ phenotype of Δnhp6a/b his4-912δ lys2-128δ suc2ΔUAS mutant cells. On the other hand, the Pob3-Nhp6a protein in place of Pob3 does not diminish promoter repression at the his4-912δ, lys2-128δ, and suc2ΔUAS loci in cells containing the Nhp6a/b proteins (data not shown), and Nhp6a protein restores robust growth to Δnhp6a/b POB3-NHP6A cells (data not shown), indicating that the Pob3-Nhp6a protein does not interfere with the interactions of native Nhp6a protein. In Δnhp6a/b POB3 cells, expression of the POB3-NHP6A gene from the GAL1 promoter on a multicopy plasmid improves cell growth as expected but still does not suppress the His+ Lys+ phenotype (data not shown), making it unlikely that the chromosomal POB3-NHP6A gene simply produces insufficient protein for all Nhp6 activities. We considered the idea that the Nhp6a portion of the Pob3-Nhp6a fusion protein may not properly associate with the CP complex, in which case Nhp6a may be able to interact with the Pob3-Nhp6a version of the CP complex to restore full function. To test this idea, the CP complex was immunoprecipitated with anti-Cdc68 antibody from Δnhp6a/b POB3 cells and Δnhp6a/b POB3-NHP6A cells that also harbored a low-copy-number plasmid expressing (V5+His6)-tagged Nhp6a protein and the amount of tagged Nhp6a coimmunoprecipitating with Cdc68 was determined. These results showed that Nhp6a coimmunoprecipitates equally well with the (Cdc68 + Pob3) and the (Cdc68 + Pob3-Nhp6a) complexes (Fig. 7B). The amount of Nhp6a associated with each of these versions of the CP complex was not increased by an increased dosage of the NHP6A gene, suggesting that the Nhp6a protein interacts with a specific binding site associated with both versions of the complex (data not shown). The Nhp6a portion of Pob3-Nhp6a may not properly occupy an Nhp6-binding site associated with the CP complex, but can nonetheless provide certain Nhp6a functions by virtue of its physical attachment to Pob3.
DISCUSSION
The budding yeast S. cerevisiae contains a nuclear protein complex that is related structurally and functionally to the FACT complex of human cells and the DUF complex of Xenopus (32, 34). Both the yeast and vertebrate complexes contain the Cdc68 (Spt16) protein, and both also contain the structurally related proteins SSRP1 (vertebrate) and Pob3 (yeast). The yeast Pob3 protein does not contain the HMG box domain found at the C terminus of SSRP1; however, yeast cells have several small HMG box proteins. Here we provide evidence that members of a functionally redundant pair of small HMG box proteins, Nhp6a and Nhp6b, associate physically with the yeast complex of Pob3 and Cdc68 proteins and have effects on transcription that are related to those of the CP (Cdc68-Pob3) complex. These conclusions are derived in part from a functional analysis of a Pob3-Nhp6a fusion protein constructed to resemble an SSRP1 protein. Our findings suggest that Nhp6 protein (Nhp6a and/or Nhp6b) may associate with and function along with the Pob3 protein to make up a functional, but bipartite, yeast analog of the vertebrate SSRP1 protein.
Here we have demonstrated a physical association of Nhp6 with the CP complex by coimmunoprecipitation and affinity purification. Both procedures showed that a significant amount of Nhp6 protein could be isolated along with the CP complex, and that this CP-Nhp6 ensemble is for the most part associated with high-molecular-weight material that pellets at 200,000 × g; this material would include chromatin. In contrast, the CP that remained soluble during this fractionation associated poorly with Nhp6. Association of CP with Nhp6 was not improved by Nhp6a overexpression, suggesting that the affinity of Nhp6 for CP may be regulated and correlated with retention of CP and/or Nhp6 in high-molecular-weight material. Nhp6 has sequence-nonspecific DNA-binding ability (35, 61), but this DNA-binding activity is not solely responsible for the association of CP with high-molecular-weight material, for CP was also found in the 200,000 × g pellet made from extracts of Δnhp6a/b mutant cells. The molecular bases for the high-molecular-weight associations of CP and the differences in CP-Nhp6 affinity, and indeed whether the association between Nhp6 and CP is direct or indirect, remain to be determined.
The human FACT complex can facilitate transcription elongation on a nucleosomal template in vitro (27, 33), while genetic findings from yeast are consistent with an analogous role in vivo for the CP complex (34). The Cdc68 and Pob3 proteins constituting CP are both essential, as might be expected for global modulators of transcription (30, 55). In contrast, the Nhp6a-Nhp6b protein pair is dispensable for life and for the transcription of many genes (10, 36). Is this dispensability accounted for by the function of another homolog? Another small HMG box protein that resembles the C terminus of SSRP1 is Nhp10 (28). However, the Nhp10 protein is unlikely to have Nhp6-like function, because synthetic interactions between Δnhp6a/b and the complete absence of Nhp10 protein could not be detected (our unpublished observations). Likewise, no synthetic interactions were detected between Δnhp6a/b and the complete absence of Hmo1, another HMG box protein (28). These negative findings suggest that an HMG box domain is dispensable for some of the transcription functions of the CP complex.
Despite the dispensability of Nhp6 protein for some CP functions, our findings indicate a direct involvement of CP in Nhp6-mediated transcription. Nhp6 proteins do facilitate the transcription of a subset of genes (36, 46), and the galactose auxotrophy that we and others (62) find for Δnhp6a/b cells supports an important role for Nhp6 in GAL gene transcription. For effective growth on galactose, Nhp6 activity can be supplied by the Pob3-Nhp6a fusion protein, in association with Cdc68. Thus Nhp6 may function in a gene-specific manner through association with the CP complex.
The nature of one transcriptional role for Nhp6 may be indicated by the genetic relationship between Nhp6 and the Swi-Snf complex, a chromatin-remodeling machine that facilitates the transcription of several genes (reviewed in reference 23). We find that the absence of Nhp6 proteins increases the reliance on the Swi-Snf complex for overall growth, especially at elevated temperatures. These findings indicate that Nhp6 and the Swi-Snf complex affect the same process but in different ways.
Genetic interactions analogous to those reported here between Nhp6 and Swi-Snf are also found for Nhp6 proteins and the Gcn5 protein (62), which is the catalytic subunit of several histone acetyltransferase complexes (16, 38, 44), and for Gcn5 and Swi-Snf (3, 12, 18, 38, 41, 42, 48). Therefore, Nhp6 protein, the Swi-Snf complex, and Gcn5-mediated histone acetylation have mutually complementary effects that facilitate transcription. These common effects may assist transcriptional activators. Nhp6 can facilitate the binding of the TATA-binding protein component of TFIID to TATA sequences in vitro (36), and overexpression of Nhp6 protein increases transcription activation by mutated forms of the activator SBF (Swi4-Swi6) that retain sequence-specific DNA-binding ability (14, 46). Swi-Snf facilitates activator function through the remodeling of chromatin, whereas the chromatin-remodeling ability of Nhp6, in association with the CP complex or independently, is not known. Nhp6 proteins do, however, bend and partially unwind DNA by binding in a sequence-nonspecific manner in the minor groove (2, 35, 61). Negative supercoiling, as would result from Nhp6 binding, is correlated with disruption of nucleosome structure (26) and may therefore be involved in the facilitation of transcription by Nhp6 protein. Yeast Swi-Snf and mammalian Swi-Snf-like complexes can also bind the minor groove of DNA in a relatively sequence-nonspecific manner, with an affinity for bent and partially unwound DNA (40, 52), but this binding induces positive DNA supercoiling and may therefore have different consequences. Therefore, Nhp6 and Swi-Snf may facilitate the binding of general transcription factors by different mechanisms.
Our findings indicate that the Nhp6 protein has transcriptional effects similar to those of CP (13, 30) and can physically associate with CP. However, several Nhp6 functions are not provided by the Pob3-Nhp6a fusion protein. Our finding that native Nhp6a protein can associate with the Cdc68 + Pob3-Nhp6a complex is evidence that this particular fusion protein is not constructed to allow optimal Nhp6a interaction with CP-associated material, a situation which may prevent the full spectrum of Nhp6a activity. Alternatively, tethering Nhp6 to CP via Pob3-Nhp6a may interfere with other Nhp6 associations. Our biochemical findings are consistent with this possibility, in that not all native Nhp6 protein can be found to be associated with the CP complex. There are many examples of transcription-related proteins seen in multiple complexes. Those related to RNAP II activity include several components common to the SAGA, ADA and TFIID complexes (12, 17); the Anc1 (Swp29, Tfg3, or Taf30) protein that is a component of Swi-Snf (6), the NuA3 histone acetyltransferase (22), the mediator complex that binds RNAP II, and the general transcription factor TFIIG (20); the Arp7 and Arp9 proteins that are members of both the Swi-Snf and RSC chromatin-remodeling complexes (7); and undoubtedly others as well. The degree to which the CP complex and Nhp6 may be functionally congruent or distinct is open to further analysis.
ACKNOWLEDGMENTS
We thank Reid Johnson for strains, plasmids, and antibodies, Fred Winston for strains, Fred Cross for plasmids, and David Carruthers, Kendra Gillis, and Amy Wheeler Reich for technical assistance.
This work was supported by a grant to G.C.J. and R.A.S. from the Medical Research Council of Canada/Canadian Institutes of Health Research.
REFERENCES
- 1.Abrams E, Neigeborn L, Carlson M. Molecular analysis of SNF2 and SNF5, genes required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:3643–3651. doi: 10.1128/mcb.6.11.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allain F H-T, Yen Y-M, Masse J E, Schultze P, Dieckmann T, Johnson R C, Feigon J. Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding. EMBO J. 1999;18:2563–2579. doi: 10.1093/emboj/18.9.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biggar S R, Crabtree G R. Continuous and widespread roles for the Swi-Snf complex in transcription. EMBO J. 1999;18:2254–2264. doi: 10.1093/emboj/18.8.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bortvin A, Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- 5.Brewster N K, Johnston G C, Singer R A. Characterization of the CP complex, an abundant dimer of Cdc68 and Pob3 proteins that regulates yeast transcriptional activation and chromatin repression. J Biol Chem. 1998;273:21972–21979. doi: 10.1074/jbc.273.34.21972. [DOI] [PubMed] [Google Scholar]
- 6.Cairns B R, Henry N L, Kornberg R D. TFG3/TAF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9. Mol Cell Biol. 1996;16:3308–3316. doi: 10.1128/mcb.16.7.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cairns B R, Erdjument-Bromage H, Tempst P, Winston F, Kornberg R D. Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol Cell. 1998;2:639–651. doi: 10.1016/s1097-2765(00)80162-8. [DOI] [PubMed] [Google Scholar]
- 8.Chang C-H, Luse D S. The H3/H4 tetramer blocks transcript elongation by RNA polymerase II in vitro. J Biol Chem. 1997;272:23427–23434. doi: 10.1074/jbc.272.37.23427. [DOI] [PubMed] [Google Scholar]
- 9.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 10.Costigan C, Kolodrubetz D, Snyder M. NHP6A and NHP6B, which encode HMG1-like proteins, are candidates for downstream components of the yeast SLT2 mitogen-activated protein kinase pathway. Mol Cell Biol. 1994;14:2391–2403. doi: 10.1128/mcb.14.4.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cross F R. ‘Marker swap’ plasmids: convenient tools for budding yeast molecular genetics. Yeast. 1997;13:647–653. doi: 10.1002/(SICI)1097-0061(19970615)13:7<647::AID-YEA115>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 12.Eberharter A, Sterner D E, Schieltz D, Hassan A, Yates III J R, Berger S L, Workman J L. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:6621–6631. doi: 10.1128/mcb.19.10.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans D R H, Brewster N K, Xu Q, Rowley A, Altheim B A, Johnston G C, Singer R A. The yeast protein complex containing Cdc68 and Pob3 mediates core-promoter repression through the Cdc68 N-terminal domain. Genetics. 1998;150:1393–1405. doi: 10.1093/genetics/150.4.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewaskow S P, Sidorova J M, Hendle J, Emery J C, Lycan D E, Zhang K Y, Breeden L L. Mutation and modeling analysis of the Saccharomyces cerevisiae Swi6 ankyrin repeats. Biochemistry. 1998;37:4437–4450. doi: 10.1021/bi972652e. [DOI] [PubMed] [Google Scholar]
- 15.Exinger G, Lacroute F. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1992;22:9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- 16.Grant P A, Duggan L, Côté J, Roberts S M, Brownell J E, Candau R, Ohba R, Owens-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 17.Grant P A, Schieltz D, Pray-Grant M G, Steger D J, Reese J C, Yates III J R, Workman J L. A subset of TAFIIs are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- 18.Gregory P D, Schmid A, Zavari M, Münsterkötter M, Hörz W. Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J. 1999;18:6407–6414. doi: 10.1093/emboj/18.22.6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartzog G A, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henry N L, Campbell A M, Feaver W J, Poon D, Weil P A, Kornberg R D. TFIIF-TAF-RNA polymerase II connection. Genes Dev. 1994;8:2868–2878. doi: 10.1101/gad.8.23.2868. [DOI] [PubMed] [Google Scholar]
- 21.Hirschhorn J N, Brown S A, Clark C D, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 22.John S, Howe L, Tafrov S T, Grant P A, Sternglanz R, Workman J L. The Something About Silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAFII30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 2000;14:1196–1208. [PMC free article] [PubMed] [Google Scholar]
- 23.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 24.Kolodrubetz D, Burgum A. Duplicated NHP6 genes of Saccharomyces cerevisiae encode proteins homologous to bovine high mobility group protein 1. J Biol Chem. 1990;265:3234–3239. [PubMed] [Google Scholar]
- 25.Lai J-S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee M S, Garrard W T. Positive DNA supercoiling generates a chromatin conformation characteristic of highly active genes. Proc Natl Acad Sci USA. 1991;88:9675–9679. doi: 10.1073/pnas.88.21.9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LeRoy G, Orphanides G, Lane W S, Reinberg D. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282:1900–1904. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- 28.Lu J, Kobayashi R, Brill S J. Characterization of a high mobility group 1/2 homolog in yeast. J Biol Chem. 1996;271:33678–33685. doi: 10.1074/jbc.271.52.33678. [DOI] [PubMed] [Google Scholar]
- 29.Lycan D, Mikesell G, Bunger M, Breeden L. Differential effects of Cdc68 on cell cycle-regulated promoters in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:7455–7465. doi: 10.1128/mcb.14.11.7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malone E A, Clark C D, Chiang A, Winston F. Mutations in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5710–5717. doi: 10.1128/mcb.11.11.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakanishi T, Nakuno A, Nomura K, Sekimizu K, Natori S. Purification, gene cloning, and gene disruption of the transcription factor SII in Saccharomyces cerevisiae. J Biol Chem. 1992;267:13200–13204. [PubMed] [Google Scholar]
- 32.Okuhara K, Ohta K, Seo H, Shioda M, Yamada T, Tanaka Y, Dohmae N, Seyama Y, Shibata T, Murofushi H. A DNA unwinding factor involved in DNA replication in cell-free extracts of Xenopus eggs. Curr Biol. 1999;9:341–350. doi: 10.1016/s0960-9822(99)80160-2. [DOI] [PubMed] [Google Scholar]
- 33.Orphanides G, LeRoy G, Chang C-H, Luse D S, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 34.Orphanides G, Wu W-H, Lane W S, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400:284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- 35.Paull T T, Johnson R C. DNA looping by Saccharomyces cerevisiae high mobility group proteins NHP6A/B. J Biol Chem. 1995;270:8744–8754. doi: 10.1074/jbc.270.15.8744. [DOI] [PubMed] [Google Scholar]
- 36.Paull T T, Carey M, Johnson R C. Yeast HMG proteins NHP6A/B potentiate promoter-specific transcriptional activation in vivo and assembly of preinitiation complexes in vitro. Genes Dev. 1996;10:2769–2781. doi: 10.1101/gad.10.21.2769. [DOI] [PubMed] [Google Scholar]
- 37.Peterson C L, Dingwall A, Scott M P. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci USA. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pollard K J, Peterson C L. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol Cell Biol. 1997;17:6212–6222. doi: 10.1128/mcb.17.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prelich G, Winston F. Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics. 1993;135:665–676. doi: 10.1093/genetics/135.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quinn J, Fyrberg A M, Ganster R W, Schmidt M C, Peterson C L. DNA-binding properties of the yeast SWI/SNF complex. Nature. 1996;379:844–847. doi: 10.1038/379844a0. [DOI] [PubMed] [Google Scholar]
- 41.Recht J, Osley M A. Mutations in both the structured domain and N-terminus of histone H2B bypass the requirement for Swi-Snf in yeast. EMBO J. 1999;18:229–240. doi: 10.1093/emboj/18.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roberts S M, Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rowley A, Singer R A, Johnston G C. CDC68, a yeast gene that affects regulation of cell proliferation and transcription, encodes a protein with a highly acidic carboxyl terminus. Mol Cell Biol. 1991;11:5718–5726. doi: 10.1128/mcb.11.11.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saleh A, Schieltz D, Ting N, McMahon S B, Litchfield D W, Yates III J R, Lees-Miller S P, Cole M D, Brandl C J. Tra1p is a component of the yeast Ada-Spt transcriptional regulatory complexes. J Biol Chem. 1998;273:26559–26565. doi: 10.1074/jbc.273.41.26559. [DOI] [PubMed] [Google Scholar]
- 45.Schlesinger M B, Formosa T. POB3 is required for both transcription and replication in the yeast Saccharomyces cerevisiae. Genetics. 2000;155:1593–1606. doi: 10.1093/genetics/155.4.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sidorova J, Breeden L. The MSN1 and NHP6A genes suppress SW16 defects in Saccharomyces cerevisiae. Genetics. 1999;151:45–55. doi: 10.1093/genetics/151.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sudarsanam P, Cao Y, Wu L, Laurent B C, Winston F. The nucleosome remodeling complex, Snf/Swi, is required for the maintenance of transcription in vivo and is partially redundant with the histone acetyltransferase, Gcn5. EMBO J. 1999;18:3101–3106. doi: 10.1093/emboj/18.11.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swanson M S, Malone E A, Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol. 1991;11:3009–3019. doi: 10.1128/mcb.11.6.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog G A, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wada T, Orphanides G, Hasegawa J, Kim D-K, Shima D, Yamaguchi Y, Fukuda A, Hisatake K, Oh S, Reinberg D, Handa H. FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH. Mol Cell. 2000;5:1067–1072. doi: 10.1016/s1097-2765(00)80272-5. [DOI] [PubMed] [Google Scholar]
- 52.Wang W, Chi T, Xue Y, Zhou S, Kuo A, Crabtree G R. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc Natl Acad Sci USA. 1998;95:492–498. doi: 10.1073/pnas.95.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winston F. Analysis of SPT genes: a genetic approach toward analysis of TFIID, histones, and other transcription factors of yeast. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 1271–1293. [Google Scholar]
- 54.Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 55.Wittmeyer J, Formosa T. The Saccharomyces cerevisiae DNA polymerase α catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol Cell Biol. 1997;17:4178–4190. doi: 10.1128/mcb.17.7.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wittmeyer J, Joss L, Formosa T. Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase α. Biochemistry. 1999;38:8961–8971. doi: 10.1021/bi982851d. [DOI] [PubMed] [Google Scholar]
- 57.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 58.Xu Q, Johnston G C, Singer R A. The Saccharomyces cerevisiae Cdc68 transcription activator is antagonized by San1, a protein implicated in transcriptional silencing. Mol Cell Biol. 1993;13:7553–7565. doi: 10.1128/mcb.13.12.7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu Q, Singer R A, Johnston G C. Sug1 modulates yeast transcription activation by Cdc68. Mol Cell Biol. 1995;15:6025–6035. doi: 10.1128/mcb.15.11.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 61.Yen Y-M, Wong B, Johnson R C. Determinants of DNA binding and bending by the Saccharomyces cerevisiae high mobility group protein NHP6A that are important for its biological activities. J Biol Chem. 1998;273:4424–4435. doi: 10.1074/jbc.273.8.4424. [DOI] [PubMed] [Google Scholar]
- 62.Yu Y, Eriksson P, Stillman D J. Architectural transcription factors and the SAGA complex function in parallel pathways to activate transcription. Mol Cell Biol. 2000;20:2350–2357. doi: 10.1128/mcb.20.7.2350-2357.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]