Abstract
In the yeast Saccharomyces cerevisiae, a and α mating-type information is stored in transcriptionally silenced cassettes called HML and HMR. Silencing of these loci, maintained by the formation of a specialized type of heterochromatin, requires trans-acting proteins and cis-acting elements. Proteins required for silencing include the Sir2 NAD+-dependent deacetylase, Sir3, and Sir4. Factors that bind to the cis elements at HMR and HML and that are important for silencing include the origin recognition complex (ORC). Mutations of any of these Sir proteins or combinations of cis elements result in loss of silencing. SUM1-1 was previously identified as a dominant mutation that restores silencing to HMR in the absence of either the Sir proteins or some of the cis elements. We have investigated the novel mechanism whereby Sum1-1 causes Sir-independent silencing at HMR and present the following findings: Sum1-1 requires the Sir2 homolog, Hst1, for silencing and most probably requires the NAD+-dependent deacetylase activity of this protein. Sum1-1 interacts strongly with ORC, and this strong interaction is dependent on HMR DNA. Furthermore, ORC is required for Sum1-1-mediated silencing at HMR. These observations lead to a model for Sum1-1 silencing of HMR in which Sum1-1 is recruited to HMR by binding to ORC. Sum1-1, in turn, recruits Hst1. Hst1 then deacetylates histones or other chromatin-associated proteins to cause chromatin condensation and transcriptional silencing.
The silent mating-type loci (HML and HMR) in the yeast Saccharomyces cerevisiae are maintained in a transcriptionally inactive state due to the formation of a specialized chromatin structure analogous to heterochromatin of higher eukaryotes (reviewed in references 24 and 25). Both cis-acting elements and trans-acting factors have been identified that are required for silencing of the mating-type loci. The transcription factors Rap1 and Abf1 and the multisubunit origin recognition complex (ORC), which is essential for replication, bind to sequences within the E and I silencers that flank the silent mating-type loci (24, 29). The role of these proteins is to recruit other silencing proteins, Sir1, Sir2, Sir3, and Sir4. Orc1, for example, binds to and recruits Sir1 to the silent mating loci (35), and Rap1 binds to and recruits Sir3 and Sir4 (26, 32). The Sir proteins then participate in the formation of heterochromatin.
Sir3 and Sir4 probably function as structural components of heterochromatin (9). Sir2 has recently been shown to be a novel NAD+-dependent protein deacetylase (12, 18, 30). It is the deacetylase activity of Sir2, most probably acting on histones, that is thought to be required for silencing (3, 8, 12, 18, 30). SIR2 is a member of a highly conserved, multigene family that in S. cerevisiae also includes HST1 through HST4 (2). Of the four Hst proteins, Hst1 is most similar to Sir2 (2, 6) and, when expressed from a high-copy-number plasmid, can partially suppress the silencing defects of a strain with SIR2 deleted (2, 6). However, Δhst1 mutants do not have silencing defects (2, 6).
Strains with a deletion of SIR2, SIR3, or SIR4, or of two of the three protein binding sites of the HMR-E silencer, cause a complete loss of silencing at this locus (24). The consequence of this is that a mating-type information is expressed and MATα strains become nonmaters. A number of years ago, a mutation called SUM1-1 was obtained that restored the mating ability to a Δsir2 MATα strain (16). The SUM1-1 mutation was subsequently found to be dominant in most strain backgrounds (19), to restore mating by causing repression of transcription at the silent mating-type loci (4, 19, 21), and to restore silencing at HMR more efficiently than at HML (4, 16, 19, 21). The silencing of HMR by SUM1-1 is novel in that it is independent of any of the Sir proteins (19). The function of the wild-type Sum1 protein in the cell remained unclear, since strains with SUM1 deleted have few or no silencing defects (4). Sum1-1 differs from Sum1 at a single amino acid (codon 988) near the carboxyl terminus, where a threonine residue is replaced by an isoleucine. This mutation seems to cause novel properties in Sum1-1, rather than increased levels, since SUM1 expressed from a high-copy-number plasmid does not suppress the mating defects of a Δsir2 strain (4).
Very recently, the wild-type Sum1 protein has been implicated in the transcriptional repression of certain sporulation-specific genes during vegetative growth (37). The promoters of genes that are repressed by Sum1 contain a cis-acting element called middle sporulation element (MSE) that is required not only for mitotic repression (37) but also for meiotic induction (5). During mitotic growth, strains with SUM1 deleted have elevated levels of expression of a number of genes containing this element, including SMK1 and SPR3. Sum1 binds to MSE DNA (37). Interestingly, strains with mutations in HST1, the SIR2 homolog, also have elevated levels of transcription of these MSE-controlled genes, although the effects are not as great as when SUM1 is deleted (37). Hst1 also coimmunoprecipitates with Sum1 (A. Vershon, personal communication). These data suggest that Sum1 and Hst1 interact to cause transcriptional repression of MSE-controlled genes.
Here we show that SUM1-1 requires HST1 for its role in repression of HMR transcription. We also provide evidence suggesting that Hst1, like Sir2, is an NAD+-dependent protein deacetylase and that the deacetylase activity of Hst1 is required for Sum1-1 silencing of HMR and for Sum1 repression of MSE-regulated genes. We further show that the novel silencing of HMR by Sum1-1 requires ORC and that components of ORC interact with Sum1-1 in vivo. The data presented suggest a model whereby ORC recruits Sum1-1 to the HMR locus and, in turn, Sum1-1 recruits Hst1. Hst1 then deacetylates either histones or other chromatin-associated proteins to form silenced heterochromatin.
MATERIALS AND METHODS
Strains, plasmids, and sequence analysis.
The yeast strains used in this study are listed in Table 1. The plasmids for the two-hybrid analysis were created as follows. A fragment of SUM1 (encoding amino acid 775-end) was amplified from the genome by PCR with a BamHI site at the 5′ end and a PstI site at the 3′ end. This fragment was cloned into the BamHI-PstI sites of pSTT91 (pBTM116 [1] with the ADE2 gene inserted) such that SUM1 is in frame with lexA to create pJL029. The analogous fragment of SUM1-1 was cloned into pSTT91 to create pJL030. This plasmid was used as the bait in the two-hybrid screen. A BamHI-PstI fragment of SUM1 from pJL029 was cloned into the BamHI-PstI sites of pGAD424 such that SUM1 is in frame with GAD to create pRH01. A BamHI-PstI fragment of SUM1-1 from pJL030 was cloned into the BamHI-PstI sites of pGAD424 such that SUM1-1 is in-frame with GAD to create pRH02. A fragment containing all of ORC5 was amplified from the genome with the addition of an EcoRI site at the 5′ end and a BamHI site at the 3′ end. This fragment was cloned into pBTM116 such that ORC5 is in frame with lexA to create pTT93. An EcoRI-HindIII fragment containing HMR from pLSD1 (HMR in pUC13 [obtained from D. Shore]) was cloned into pRS426 to create pRH34. pLP0349 is SIR2 cloned into YEp351 (obtained from L. Pillus). pGK16 contains SPR3:lacZ in YCp50 (10) and was obtained from A. Neiman. pJX43 contains the MSE from SMK1 cloned into a lacZ reporter plasmid (37) and was obtained from A. Vershon. A fragment containing all but the first 69 nucleotides of HST1 was amplified by PCR from the genome with a 5′ NcoI site and a 3′ XhoI site. The fragment was cloned into pET28c to create pAGN24, which encodes all of Hst1 except the first 23 amino acids and has a His6 tag at the carboxyl terminus.
TABLE 1.
S. cerevisiae strains used in this study
| Name | Genotype | Source or reference |
|---|---|---|
| W303-1b | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | R. Rothstein |
| YRH07 | MATα SUM1-1 sir2::HIS3 ura3 his3 leu2 trp1 ade2 lys hmr-I::kanMX6 | This study |
| YRH15 | W303-1b sir2::his5+ | This study |
| YRH20 | W303-1b SUM1-myc::TRP1 sir2::his5+ | This study |
| YRH21 | MATα SUM1-1-myc::TRP1 sir2::HIS3 ura3 his3 leu2 trp1 ade2 lys | This study |
| YRH34 | W303-1b SUM1-1 sir2::his5+ | This study |
| YRH35 | W303-1b SUM1-1 sir2::his5+ npt1::kanMX6 | This study |
| YRH36 | W303-1b SUM1-1-myc sir2::his5+ | This study |
| YRH38 | W303-1b SUM1-1 sir2::his5+ hst1::kanMX6 | This study |
| RS1056 | MATα SUM1-1 sir2::HIS3 ura3 his3 leu2 trp1 ade2 lys | |
| JLY04 | MATα SUM1-1 sir2::HIS3 ura3 his3 leu2 trp1 ade2 lys hst1::kanMX6 | This study |
| AY1283 | MATahis1 | K. Arndt |
| JXY3 | W303-1a sum1::kanMX4 | A. Verhon |
| JXY5 | W303-1a hst1::kanMX4 | A. Vershon |
| MC89 | W303-1b SUM1-1 | D. Shore |
| YB0057 | W303-1a orc5-1 | B. Stillman |
| L40 | MATahis3Δ200 trp1-901 leu2-3,112 ade2 lys2-801am URA3::(lexAop)8-lacZ LYS2: (lexAop)4-HIS3 | 11 |
A PatMatch search was done using the Sum1-1 sequence BZZBILLZBBBB, where ILL are the exact amino acids including I988 which is changed from T in Sum1, B is any hydrophobic residue, and Z is any hydrophilic residue.
Media and genetic methods.
All strains were grown in yeast extract-peptone-dextrose (YPD) medium supplemented with adenine at 25 μg/ml or in supplemented SD medium (drop-out medium) (15). Patch-mating assays were performed by growing patches of strains to be tested on YPD medium or the appropriate drop-out medium for 2 days at 30°C. Patches were then replica plated to SD plates onto which a mating tester strain had been spread. The plates were incubated for 2 days at 30°C before being photographed.
Mutagenesis of HST1.
A SacI fragment containing the HST1 gene and flanking sequences was isolated from plasmid pCAR158 [HST1 in pBS KS(−) from J. Boeke] and cloned into pRS314 to create pJC7a. pCAR158 was used as a template for in vitro mutagenesis to change the first pair of cysteines (C320 and C323) in the zinc finger of Hst1 to alanines. A PstI site was engineered into the mutated region to screen for the mutation. The oligonucleotides used were 5′CGGTTCATTCGCCACTGCATCTGCAGTGACTGCCCATTGGCAAAT ACCTGG3′ and 5′CCAGGTATTTGCCAATGGGCAGTCACTGCAGATGCAGTGGCGAATGAACCG3′. The mutagenesis was done with the QuikChange site-directed mutagenesis kit (Stratagene) as specified by the manufacturer. A StyI-BglII fragment, which included the mutated region, was used to replace the equivalent wild-type sequences in pJC7a to create pJC10a. The StyI-BglII DNA was checked by sequencing.
Gene deletions and epitope tagging.
The entire open reading frame of HST1 in strains YRH34 and RS1056 was replaced with the kanMX6 cassette using the method described previously (22) to create strains YRH38 and JLY04. The entire open reading frame except the sequence encoding the first 13 amino acids of NPT1 in YRH34 was replaced with the kanMX6 sequence to create strain YRH35. The SIR2 open reading frame in W303-1b and MC89 was replaced with the his5+ gene of Schizosaccharomyces pombe to create YRH15 and YRH34. The following oligonucleotides were used to replace HMR-I in strain RS1056 with the kanMX6 cassette: 5′CTAAAGGAAAAGAAGAGAGAAAATAGCT ATTTACCTCAACATTTAAAGGTGAATTCGAGCTCGTTTAAAC3′ and 5′GCGATAAAGTTATTATTTAGATTACATGTCACCAACATTTTCGTAT ATGGCGGATCCCCGGTTAATTAA3′. Thirteen myc tags were added to the carboxyl terminus of Sum1 in strain YRH15 using the pFA6a-13Myc-TRP1 module (22) to create YRH20 and to SUM1-1 in strains RS1056 and YRH34 to create YRH21 and YRH36.
Two-hybrid and β-galactosidase analyses.
To identify proteins that interact with Sum1-1, we used the version of the two-hybrid system previously described (11) and a yeast GAD library provided by P. James (14). The original clone obtained from the library encoded an in-frame fusion between GAD and amino acids 527 to 551 of Top3, followed by all but the first 6 amino acids of Orc5.
Filter assays for β-galactosidase for the two-hybrid analysis and for the MSE repression assays were as described previously (1) except that nitrocellulose filters were used. Quantitative β-galactosidase assays were done using three independent cultures for each strain and the permeabilized cell assay described previously (15).
Immunological analyses.
For the Western analysis in Fig. 3, extracts were prepared as described previously (33). Following electrophoresis, proteins were transferred to Immobilon-P membranes and detected using anti-Gal4-TA antibody (Santa Cruz) at 1:2,000 dilution followed by peroxidase-conjugated anti-mouse immunoglobulin G (IgG) secondary antibody at 1:5,000 dilution. The antibody complexes were detected using ECL-Plus reagents (Amersham Pharmacia) as specified by the manufacturer. The coimmunoprecipitation analyses were carried out essentially as described previously (31) with the following modifications. Extracts containing 3.7 mg of protein in 250 μl were diluted into 250 μl of 2× immunoprecipitation (IP) buffer without ammonium acetate. The extracts were incubated at 4°C overnight with 20 μl of cell culture supernatant from 9E10 anti-myc monoclonal antibodies (gift of F. McKenzie). Immunoprecipitates were collected following a 2-h incubation at 4°C with 30 μl of protein G-Sepharose beads and washed twice with IP buffer plus bovine serum albumin (BSA) and twice with IP buffer without BSA as described previously (31). Precipitates were resuspended in 30 μl of sodium dodecyl sulfate (SDS) sample buffer and run on an SDS–8% polyacrylamide gel. Following transfer to Immobilon-P, proteins were detected with either tissue culture supernatant from the 9E10 anti-myc antibody (1:100 dilution) or anti-Orc3 monoclonal antibody (1:50,000 dilution [a gift of B. Stillman]). The treatment of membranes with secondary antibody and detection with ECL reagents was as described above.
FIG. 3.
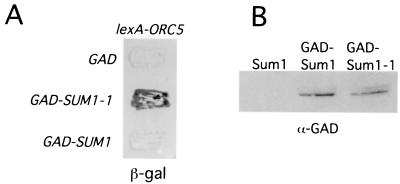
Sum1-1, but not Sum1, interacts with Orc5 in the two-hybrid assay. (A) Filter assay for lacZ expression of strain L40 transformed with a plasmid containing lexA-ORC5 (pTT93) and plasmids containing GAD (pGAD424), GAD-SUM1-1 (pRH02), or GAD-SUM1 (pRH01). (B) Western analysis of extracts from strains used in panel A probed with anti-GAD antibody. The significance of the faint upper band in panel B, which is seen only with GAD-Sum1 and GAD-Sum1-1 fusion proteins, is unknown. β-gal, β-galactosidase.
Protein expression and exchange assays.
Hst1 was expressed from pAGN24 in Escherichia coli strain BL21(DE3) with a 3-h induction with 0.35 mM isopropyl-β-d-thiogalactoside (IPTG) at room temperature. Extracts were prepared using BugBuster (Novagen) as specified by the manufacturer. Exchange reactions were performed as previously described (18) with the following modifications. Reactions were carried out for 45 min at 30°C, E. coli extracts instead of purified proteins were used in the assay, and the films were exposed for 5 to 7 days. The H4 peptide consisted of the first 20 amino acids of histone H4, with K16 acetylated.
RESULTS
Suppression of silencing defects at HMR by SUM1-1 requires HST1.
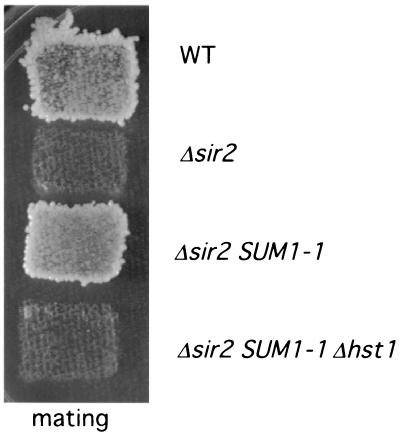
Because it had recently been shown that SUM1 requires HST1 for maximal repression of transcription from MSE-regulated promoters (37), we wondered whether SUM1-1 similarly requires HST1 for silencing of the HMR locus in sir mutants. To address this, we compared the mating ability of a Δsir2 SUM1-1 strain to that of a Δsir2 SUM1-1 Δhst1 strain. SUM1-1 restored mating to a MATα Δsir2 strain (Fig. 1). This effect of SUM1-1 has previously been shown to result from repression of HMRa1 transcription (4, 19, 21). However, SUM1-1 is unable to restore mating to a Δsir2 strain that also lacks HST1 (Fig. 1). Thus, repression of HMR by Sum1-1 absolutely requires Hst1.
FIG. 1.
SUM1-1 silencing requires HST1. Patches of the indicated strains were replica plated onto an SD plate containing a lawn of MATa his1 cells (AY1283). The plate was incubated for 2 days at 30°C. WT, strain W303-1b; Δsir2, strain YRH15; Δsir2 SUM1-1, strain YRH34; Δsir2 SUM1-1 Δhst1, strain YRH38.
Hst1 is an NAD+-dependent deactylase.
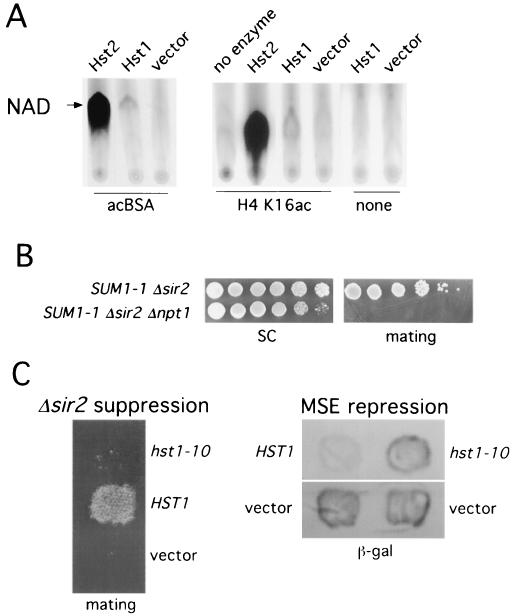
HST1 is a member of a highly conserved gene family that also consists of SIR2 and HST2-4 in S. cerevisiae. We and others have recently demonstrated that yeast Sir2 and Hst2, as well as Sir2 homologs from human, Salmonella enterica serovar Typhimurium, and Archaeoglobus fulgidus, are NAD+-dependent protein deacetylases in vitro, and this is most likely to be their function in vivo (12, 18, 30). To determine whether Hst1 is also an NAD+-dependent deacetylase, we expressed a His-tagged version of the protein in E. coli cells. In contrast to Sir2 and Hst2 (18), Hst1 made in E. coli is largely insoluble. Therefore, it has been very difficult to detect in vitro activity of this protein. Our most sensitive assay for NAD+-dependent deacetylases is a nicotinamide-NAD+ exchange reaction (18). This assay is based on the fact that the Sir2 family of deacetylases cleaves the glycosidic bond between nicotinamide and ADP-ribose in NAD+ (17, 34). This reaction leads to the establishment of the following equilibrium: NAD+ + enzyme ↔ enzyme-ADP ribose + nicotinamide. This reaction is absolutely dependent on the presence of an acetylated substrate (18). Incubating radioactive nicotinamide with NAD+ and enzyme leads to the NAD+ becoming radioactive. Fig. 2A shows the results of an exchange assay using E. coli extracts expressing Hst1. Weak exchange activity was found for Hst1. The activity was dependent on the presence of an acetylated substrate, either an H4 peptide acetylated on K16 or chemically acetylated BSA. For comparison, purified Hst2 showed much greater activity in this assay. The Hst1 activity was reproducibly greater than that seen for an E. coli extract not expressing Hst1 (vector, Fig. 2A). We think the latter activity is due to CobB, the E. coli Sir2 homolog, which we have previously shown to have exchange and deacetylation activity (18). Note that the putative CobB activity is also dependent on the presence of an acetylated substrate. Based on the tight coupling of NAD hydrolysis and deacetylation for this class of enzymes (17, 18, 34), these results demonstrate that Hst1 also is a protein deacetylase. We believe the low level of activity in the Hst1 extracts reflects the fact that almost all of the protein is insoluble. However, it may also result because we have not found the optimal acetylated substrate to drive the reaction.
FIG. 2.
Hst1 is an NAD+-dependent protein deacetylase. (A) NAD+-nicotinamide exchange reactions with purified Hst2 (∼200 ng) or 5 μl of total extracts prepared following induction of BL21(DE3) transformed with either pAGN24 (Hst1) or pET28c (vector). Reaction mixtures also contained acetylated BSA (0.2 mg/ml), H4 peptide acetylated at K16 (5 nM), or no acetylated substrate (none). The arrow points to the position of the NAD+ that has become radioactive during the exchange reaction. (B) npt1 mutants are defective in SUM1-1 silencing. Strains YRH34 and YRH35 were tested for mating by spotting 10-fold serial dilutions of exponentially growing cultures onto an SD plate containing a lawn of MATa his1 (AY1283) cells. The same strains were spotted onto a synthetic complete (SC) plate to show number of cells tested. The npt1 strain grows somewhat more slowly than the NPT1 strain does. (C) The left panel shows that the zinc finger mutant of Hst1 abolishes SUM1-1 silencing. Strain JLY04 (MATα SUM1-1 Δsir2 Δhst1) was transformed with either pJC10a (hst1-10), pJC7a (HST1), or pRS424 (vector). Transformed strains were tested for mating as described in the legend to Fig. 1. The right panel shows that the zinc finger mutant of Hst1 is defective in MSE-dependent repression. Patches of strain JXY5, transformed with an SPR3-lacZ reporter plasmid (pGK16) and the plasmids described in the left panel, were grown for 2 days on SC −Ura −Trp medium and then tested for β-galactosidase activity using a filter assay.
We have two lines of evidence to suggest that Hst1 is also an NAD+-dependent protein deacetylase in vivo and that this activity is required for its Sum1-1- and Sum1-dependent functions. Data showing that Hst1 deacetylase activity is required for Sum1-1 silencing come from our analysis of the effects of deletion of NPT1. NPT1 encodes a homolog of the serovar Typhimurium enzyme nicotinate phosphoribosyltransferase that participates in a salvage pathway for NAD+ synthesis (36). Strains with mutations in NPT1 have 2.6 times less NAD+ than do wild-type strains and are severely defective in Sir2-dependent silencing at rDNA and telomeres and slightly defective in silencing at HMR (30). These phenotypes presumably result because the NAD+-dependent deacetylase activity of Sir2 is reduced in the npt1 mutant. We deleted NPT1 in a MATα Δsir2 SUM1-1 strain to see whether the silencing at HMR by Sum1-1 and Hst1 is also sensitive to cellular NAD+ levels. The results of this analysis (Fig. 2B) show that the npt1 mutation abolishes the mating of the SUM1-1 Δsir2 strain. This suggests that Hst1, like Sir2, is dependent on NAD+ for activity and that npt1 mutants lack sufficient NAD+ for Hst1 to function with Sum1-1 in HMR silencing.
A second line of evidence that Hst1 functions as a protein deacetylase in vivo comes from our analysis of effects of mutations in Hst1 on Sum1-1 and Sum1 function. All of the members of the Sir2 family contain a core domain that includes four cysteines of a zinc finger. Mutation of either pair of these cysteines to alanines in Sir2 disrupts silencing (28), and we have found that mutations in the zinc finger cysteines also abolish the in vitro deacetylase activity of the Sir2 protein (S. Tafrov, R. Heller, and R. Sternglanz, unpublished results). We reasoned that if mutations in the zinc finger of Hst1 also abolish its functions, it is likely that Hst1 is also a deacetylase in vivo. Therefore, we used in vitro mutagenesis to change the first pair of cysteines (C320 and C323) in the Hst1 zinc finger to alanines. We tested the function of the HST1 C320A, C323A (hst1-10) mutant in both HMR silencing and MSE-mediated repression. In the first assay, a MATα Δhst1 Δsir2 SUM1-1 strain was transformed with either HST1 or hst1-10 on a low-copy-number plasmid and tested for mating. While the strain with wild-type HST1 mated well in this assay, the strain with hst1-10 mated no better than did a strain transformed with empty vector (Fig. 2C). Therefore, the cysteines of the zinc finger of Hst1 are essential for its function in SUM1-1 silencing. For the MSE-mediated repression assay, we tested the level of lacZ expression from a promoter-reporter construct containing the SPR3 MSE site upstream of lacZ. Strains with a chromosomal deletion of HST1, transformed with wild-type HST1 on a low-copy-number plasmid, showed strong repression of lacZ in this assay (Fig. 2C). The same strain transformed with a plasmid containing hst1-10 showed high levels of lacZ expression which were comparable to the levels produced in a strain transformed with vector alone (Fig. 2C). Therefore, mutation of two cysteines in the zinc finger of Hst1 also abolished its function in Sum1-mediated repression of an MSE-regulated gene. Because Hst1 is so similar to Sir2 and because mutation of the zinc finger of Sir2 abolished its NAD+-dependent deacetylase activity, it is most likely that the zinc finger mutant of Hst1 no longer functions due to loss of a similar deacetylase activity.
SUM1-1, but not SUM1, interacts with ORC5 in a two-hybrid assay.
To try to understand the mechanism by which SUM1-1, but not SUM1, suppresses the mating defect of sir mutants, we initiated a search for proteins that interact with Sum1-1. For the search, we used the two-hybrid system with a construct encoding a fusion between LexA and the carboxy-terminal 288 amino acids of Sum1-1 as the bait. This part of Sum1-1 contains the T988I mutation, which is responsible for the novel properties of the protein. We screened more than 1.6 × 106 library transformants with this bait and obtained two interacting clones. One clone included all but the first 6 amino acids of the Orc5 protein. To test whether this library clone also interacted with SUM1, we made a lexA-SUM1 fusion construct containing the region of SUM1 comparable to that used for lexA-SUM1-1. However, we found that this construct activated the transcription of the two-hybrid reporter genes in the absence of any other plasmid and thus could not be tested for interaction with Orc5. We therefore constructed new hybrid proteins in which the same carboxyl termini of Sum1 and Sum1-1 were fused to GAD. In this case, the GAD-Sum1 fusion did not activate transcription of the reporter genes. We then tested these constructs for two-hybrid interactions with lexA-ORC5 and found that GAD-SUM1-1, but not GAD-SUM1, interacted with lexA-ORC5 (Fig. 3A). Western analysis of the proteins produced from these constructs showed that GAD-Sum1 and GAD–Sum1-1 were produced in approximately equal amounts in the cell (Fig. 3B). Thus, the two-hybrid results show that Orc5 interacts specifically with Sum1-1 and not with Sum1.
Sum1-1 associates with ORC in vivo.
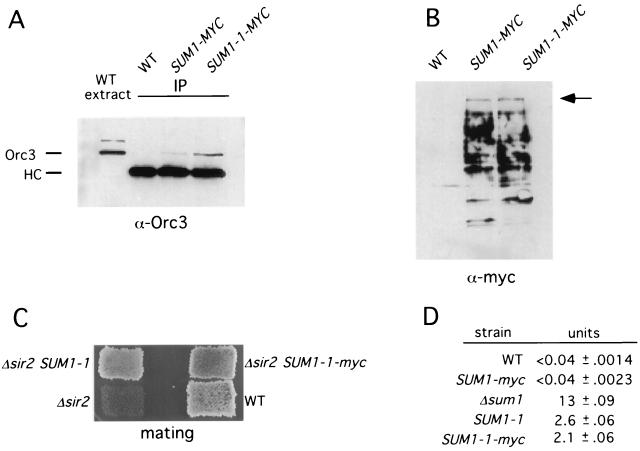
To establish that the interaction between Sum1-1 and Orc5 was physiologically significant, we carried out a co-IP analysis. To do this, we created strains in which chromosomally encoded Sum1 and Sum1-1 included 13 copies of the myc epitope at their carboxyl termini. We immunoprecipitated these proteins from cell extracts and tested whether we could detect Orc5 in either precipitate. Unfortunately, the Orc5 protein comigrates with the IgG heavy chain and was impossible to detect in our analyses. We reasoned that if Sum1-1 interacts with Orc5, it might also interact with other subunits of the ORC complex. Therefore, we tested the immunoprecipitates for the presence of Orc3. The results in Fig. 4A show that immunoprecipitates of Sum1-1–myc do contain easily detectable amounts of Orc3. This result is in contrast to that for Sum1-myc, which is approximately equally well precipitated by the myc antibody but brings down much less Orc3 (Fig. 4A and B). These results confirm and extend the results of the two-hybrid analysis and show that in vivo, ORC interacts much more strongly with Sum1-1 than with Sum1. The results shown in Fig. 4A are from strains that were transformed with HMR on a high-copy-number plasmid. We found that the co-IP between Orc3 and Sum1-1 was much more easily detected when extra copies of HMR were present. This result suggests that Sum1-1 interacts with ORC at the HMR locus and that HMR DNA somehow stabilizes the interaction.
FIG. 4.
Sum1-1 coimmunoprecipitates with Orc3. (A) IP analysis with Orc3. Strains YRH15 (WT), YRH20 (SUM1-myc), and YRH21 (SUM1-1-myc) were transformed with plasmid pRH34 (HMR in pRS426). Extracts were prepared and proteins were immunoprecipitated using anti-myc antibodies. Following SDS-polyacrylamide gel electrophoresis, the precipitates were analyzed by immunoblotting and probed with an anti-Orc3 antibody. A portion of the wild-type extract from YRH15 (with plasmid pRH34) was also run on the same gel to show the position of Orc3 as well as a slower-migrating protein that interacts with the Orc3 antibody (20). HC, heavy chain of IgG. (B) A portion (5%) of the precipitated material used in panel A was analyzed by immunoblotting with anti-myc antibodies. The Sum1-myc and Sum1-1–myc proteins always show multiple bands following immunoprecipitation, presumably due to proteolysis. The arrow points to the position of the full-length proteins. (C) Sum1-1–myc functions in vivo. Sum1-1–myc silencing in Δsir2 strains was measured by a patch-mating assay with WT (W303-1b), Δsir2 (YRH15), Δsir2 SUM1-1 (YRH34), or Δsir2 SUM1-1-myc (YRH36) strains. (D) Sum1-myc functions in vivo. Strains were transformed with plasmid pJX43 (SMK1 MSE-lacZ) and grown to early log phase in SC −Ura medium. β-Galactosidase activity was determined for three independent transformants for each strain. WT (YRH15), SUM1-myc (YRH20), Δsum1 (JXY3), SUM1-1 (YRH34), and SUM1-1-myc (YRH36) strains were used.
Because there was such a striking difference between the Sum1-1–Orc3 interaction and the Sum1-Orc3 interaction, it was important to test the Sum1-myc and Sum1-1–myc strains for in vivo function. To test the tagged Sum1-1 protein, we measured Sum1-1 silencing at HMR. This analysis revealed that a MATα Δsir2 SUM1-1–myc strain mated almost as well as a MATα Δsir2 SUM1-1 strain (Fig. 4C). Therefore, the myc tag does not significantly affect the function of Sum1-1 in silencing. To test the function of the tagged Sum1 protein, we measured Sum1 repression of MSE-regulated genes. For this analysis, we used a lacZ promoter-reporter plasmid with the MSE from SMK1. Using quantitative β-galactosidase assays, we found that the epitope-tagged SUM1-myc strain repressed the transcription of the MSE-lacZ reporter as well as the untagged SUM1 strain did (Fig. 4D). We also compared the repression of this reporter by Sum1-1 and Sum1-1–myc and found that epitope-tagged Sum1-1–myc repressed transcription of the reporter to the same extent as untagged Sum1-1 did (Fig. 4D). Therefore, the myc tag appears not to impair the function of either Sum1 or Sum1-1.
Interestingly, Sum1-1 did not function as well as Sum1 in repression of MSE-lacZ, although it showed more repression than a strain with SUM1 deleted (Fig. 4D). This result was somewhat surprising, since it has previously been reported that both proteins repress this reporter equally well (37). Our result is supported by results of chromatin IP experiments, where we found that Sum1-myc bound much better to the MSE of SMK1 than Sum1-1-myc did (data not shown). We do not understand the reason for the difference between our result and the one reported previously.
A silencing-defective orc mutation abolishes SUM1-1-dependent silencing at HMR.
ORC is essential for replication and important for Sir-dependent silencing at HMR and HML. Mutations have been isolated in ORC5 (orc5-1) that abolish Sir-dependent silencing from a weakened HMR-E silencer but not viability (replication) of cells grown at 24°C (7, 23). Because of the two-hybrid interaction between Orc5 and Sum1-1, we examined the effect of the orc5-1 mutation on HMR silencing by SUM1-1. We crossed a SUM1-1 Δsir2 strain with an orc5-1 strain. We then tested the Δsir2 ORC5 progeny for mating competence at 24°C. Of the Δsir2 ORC5 progeny, 27% (9 of 33) could mate and all were MATα. This result agrees well with the prediction that 25% of Δsir2 ORC5 progeny would be able to mate since 50% would have SUM1-1 and 50% of those would be MATα (SUM1-1 cannot suppress mating defects of Δsir2 MATa strains). In contrast, none (0 of 31) of the Δsir2 orc5-1 progeny could mate at 24°C. Since 25% of those progeny should also be MATα SUM1-1, this result shows that suppression of the mating defect of Δsir2 strains by SUM1-1 requires the silencing function of Orc5.
Strains lacking HMR-I are defective in SUM1-1 silencing.
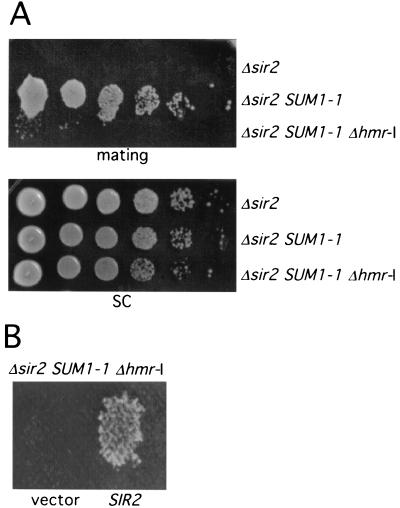
SUM1-1 has previously been found to suppress the silencing defects of a strain containing a synthetic silencer in which either the ORC binding site or the Rap1 and Abf1 binding sites at HMR-E are deleted (19). However, SUM1-1 is unable to restore repression to a strain in which the entire E site is deleted (19). In the constructs described above, the HMR-I site was present. Although HMR-I is dispensable for Sir-mediated silencing when HMR-E is intact, its role in SUM1-1 silencing has not been tested. To address this, we created a MATα Δsir2 SUM1-1 strain in which the HMR-I sequences were replaced with the kanMX6 casette. This strain mated very poorly (Fig. 5A), indicating that HMR-I is very important for Sum1-1-mediated silencing. To make sure that these manipulations did not affect regions important for silencing outside of HMR-I, we transformed this strain with SIR2 on a low-copy-number plasmid. SIR2 restored the mating ability of the Δhmr-I strain, confirming that the HMR-E silencer was still functional (Fig. 5B). Thus, HMR-I is much more important for silencing by Sum1-1 than for silencing by Sir2.
FIG. 5.
HMR-I is required for Sum1-1 silencing. (A) Strains YRH15 (Δsir2), RS1056 (Δsir2 SUM1-1), and YRH07 (Δsir2 SUM1-1 Δhmr-I) were tested for mating by plating 10-fold serial dilutions of saturated cultures onto an SD plate containing a lawn of MATa his1 (AY1283) cells. (B) Strain YRH07 was transformed with either vector (YEp351) or SIR2 (pLP0349) and tested for mating as in Fig. 1.
DISCUSSION
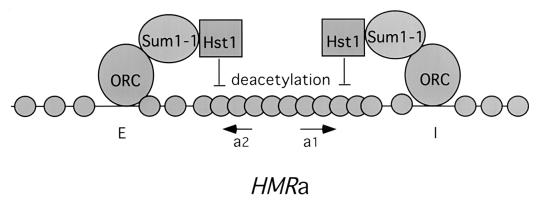
The results presented above demonstrate that Sum1-1-mediated repression of HMR requires Hst1. This is consistent with the importance of Hst1 in Sum1-mediated repression of MSE-regulated genes. We also show that the requirement for Hst1 by both Sum1 and Sum1-1 most probably is a requirement for an NAD+-dependent protein deacetylase activity. In addition, Sum1-1 interacts strongly with ORC, and this strong interaction is dependent on HMR DNA. Silencing of HMR by Sum1-1 is abolished by a mutation in a component of ORC. Finally, silencing of HMR by Sum1-1 requires the HMR-I sequences as well as HMR-E. The simplest model to explain the novel silencing function of the Sum1-1 protein is depicted in Fig. 6. ORC binds to autonomously replicating sequences found at both HMR-E and HMR-I. ORC then recruits Sum1-1 to these loci, and Sum1-1 in turn recruits Hst1. The Hst1 protein then deacetylates lysines, either on histones or on histone-associated proteins, to cause a condensed, silenced chromatin structure.
FIG. 6.
Model for Sum1-1 silencing at HMR.
Co-IP analysis revealed that although both Sum1 and Sum1-1 interact with ORC, the interaction of Sum1-1 is much stronger. We also found that only when the cell contained extra copies of HMR DNA could we easily detect co-IP between ORC and Sum1-1. This result suggests either that an interaction between Sum1-1 and HMR DNA is required for the Sum1-1-ORC interaction or that only when ORC is in the context of HMR is it recognized by Sum1-1. The Sum1-1 interaction with ORC at HMR is further supported by results of chromatin IP studies using HA epitope-tagged versions of Sum1-1 and Sum1. These studies revealed that Sum1-1–HA binds to HMR-E and HMR-I DNA and that it does so more efficiently than Sum1-HA. Unfortunately, the HA-tagged version of Sum1-1 does not suppress the mating defects of a Δsir2 strain, and we have been unable to obtain comparable chromatin IP results with the functional Sum1-1–myc strain. Therefore, we have not included these data here. The reason for the difference in behavior between the HA-and myc-tagged proteins is not clear; perhaps association of the functional Sum1-1–myc protein with HMR DNA is more transient and therefore more difficult to detect by chromatin IP, than is association of the nonfunctional Sum1-1-HA protein with HMR DNA.
It is remarkable that changing one amino acid, T988, to I can change the properties of the Sum1-1 protein so profoundly that it now interacts much more strongly than Sum1 with ORC. Interestingly, a PatMatch search (see Materials and Methods) of the yeast genome database using a stretch of amino acids of Sum1-1 surrounding the mutant I988 residue revealed similarity to a highly conserved region of two proteins, Mcm4 and Mcm6. These proteins are part of the MCM complex, which binds replication origins near ORC and may interact, directly or indirectly, with ORC (13). The similarity to Mcm4 and Mcm6 is not detected using the Sum1 T988 sequence. Perhaps the T988-to-I change in Sum1-1 mimics this Mcm motif and allows Sum1-1 to interact more strongly than Sum1 either with DNA sequences near ORC at HMR or with ORC.
The mutation in SUM1-1 appears to also alter its function in MSE-regulated repression. In our experiments, SUM1-1 strains are somewhat defective in repression of a lacZ reporter construct under control of the SMK1 MSE, and in chromatin IP experiments, Sum1-1–myc bound much more poorly to the MSE site than did Sum1-myc (data not shown). Either Sum1-1 no longer recognizes the MSE sequence as well as Sum1 does, or it has lost some protein-protein interaction at the locus critical for DNA binding and function.
We have also shown that the HMR-I site is much more important for Sum1-1 silencing than it is for Sir2 silencing. This may be because Sir2-mediated silencing is able to spread along the DNA from the initiation point, by virtue of the formation of polymers of Sir2, Sir3, and Sir4 along the chromatin (9). Sum1-1 silencing may be much more localized, and to achieve repression it may be necessary for Sum1-1 to bring Hst1 very close to the region of chromatin to be deacetylated. HMRa1 sequences, which must be repressed to allow mating of a MATα strain, are closer to HMR-I than to HMR-E (Fig. 6). Consistent with this hypothesis, the chromatin IP experiments described above using Sum1-1–HA also showed that Sum1-1 binding does not extend throughout the HMR locus, since no Sum1-1-specific precipitation of sequences between HMR-E and HMR-I was observed. Alternatively, the interaction of Sum1-1 with ORC at HMR-I may be stronger than that at HMR-E, making Sum1-1 silencing more dependent on the I site. Sum1-1 silencing has previously been shown to be independent of the ORC binding site (A site) in a synthetic HMR-E silencer (19). However, deletion of the entire natural HMR-E silencer abolishes Sum1-1 silencing (19). HMR-E DNA is flanked on both sides by additional ORC binding sites (24, 27); perhaps these sites are used for recruitment of Sum1-1 to HMR-E when the A site is deleted.
Our data suggest that Sum1-1 binds to ORC at HMR, which then presumably leads to deacetylation by Hst1 and repression of neighboring genes. Does Sum1-1 bind to ORC at all origins and cause the repression of many genes in the cell? Although SUM1-1 strains have a slight growth defect, if the Sum1-1 protein was causing widespread repression, one might expect more significant phenotypes. Perhaps, as postulated above, interaction of Sum1-1 with ORC also requires recognition by Sum1-1 of specific DNA sequences, and those sequences are not present near most origins.
The results that we present help to explain the novel Sir-independent mechanism of silencing by Sum1-1 at HMR. This mechanism provides another of a growing number of cases where transcriptional repression is achieved by recruitment of a protein deacetylase to chromatin. Silencing of HMR by Sum1-1 may be mechanistically more similar to the Sum1-mediated transcriptional repression of MSE-regulated genes than to Sir-mediated silencing. However, it is interesting that in both types of silencing at HMR, ORC plays a critical role in recruitment of the silencing machinery. In Sir-dependent silencing, ORC functions to recruit Sir1, which leads to recruitment of Sir4 and subsequently Sir3 and the Sir2 deacetylase. In our model of Sum1-1-dependent silencing, ORC recruits Sum1-1, which then brings the Hst1 deacetylase to the locus.
ACKNOWLEDGMENTS
We thank J. Boeke, L. Pillus, D. Shore, and A. Vershon for plasmids and strains and B. Stillman for the anti-Orc3 antibody. We also thank A. Vershon for communicating results prior to publication.
This work was supported by National Institutes of Health grants GM28220 and GM55641 to R.S. and a Beckman Foundation Scholarship to J.C.
REFERENCES
- 1.Bartel P L, Chien C, Sternglanz R, Fields S. Using the two-hybrid system to detect protein-protein interactions. In: Hartley D A, editor. Cellular interactions in development: a practical approach. Oxford, United Kingdom: IRL Press; 1993. pp. 153–179. [Google Scholar]
- 2.Brachmann C B, Sherman J M, Devine S E, Cameron E E, Pillus L, Boeke J D. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 3.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 4.Chi M H, Shore D. SUM1–1, a dominant suppressor of SIR mutations in Saccharomyces cerevisiae, increases transcriptional silencing at telomeres and HM mating-type loci and decreases chromosome stability. Mol Cell Biol. 1996;16:4281–4294. doi: 10.1128/mcb.16.8.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 6.Derbyshire M K, Weinstock K G, Strathern J N. HST1, a new member of the SIR2 family of genes. Yeast. 1996;12:631–640. doi: 10.1002/(SICI)1097-0061(19960615)12:7%3C631::AID-YEA960%3E3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 7.Foss M, McNally F J, Laurenson P, Rine J. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae. Science. 1993;262:1838–1844. doi: 10.1126/science.8266071. [DOI] [PubMed] [Google Scholar]
- 8.Gottschling D E. Gene silencing: two faces of SIR2. Curr Biol. 2000;10:R708–711. doi: 10.1016/s0960-9822(00)00714-4. [DOI] [PubMed] [Google Scholar]
- 9.Grunstein M. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- 10.Holaway B L, Kao G, Finn M C, Clancy M J. Transcriptional regulation of sporulation genes in yeast. Mol Gen Genet. 1987;210:449–459. doi: 10.1007/BF00327196. [DOI] [PubMed] [Google Scholar]
- 11.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imai S, Armstrong C M, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD- dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 13.Izumi M, Yanagi K, Mizuno T, Yokoi M, Kawasaki Y, Moon K Y, Hurwitz J, Yatagai F, Hanaoka F. The human homolog of Saccharomyces cerevisiae Mcm10 interacts with replication factors and dissociates from nuclease-resistant nuclear structures in G(2) phase. Nucleic Acids Res. 2000;28:4769–4777. doi: 10.1093/nar/28.23.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 16.Klar A J, Kakar S N, Ivy J M, Hicks J B, Livi G P, Miglio L M. SUM1, an apparent positive regulator of the cryptic mating-type loci in Saccharomyces cerevisiae. Genetics. 1985;111:745–758. doi: 10.1093/genetics/111.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landry J, Slama J T, Sternglanz R. Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem Biophys Res Commun. 2000;278:685–690. doi: 10.1006/bbrc.2000.3854. [DOI] [PubMed] [Google Scholar]
- 18.Landry J, Sutton A, Tafrov S T, Heller R C, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laurenson P, Rine J. SUM1–1: a suppressor of silencing defects in Saccharomyces cerevisiae. Genetics. 1991;129:685–696. doi: 10.1093/genetics/129.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livi G P, Hicks J B, Klar A J. The sum1–1 mutation affects silent mating-type gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:409–412. doi: 10.1128/mcb.10.1.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longtine M S, McKenzie III A, Demarini D J, Shah N G, Wach A, Brachat A, Philippsen P, Pringle J R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 23.Loo S, Fox C A, Rine J, Kobayashi R, Stillman B, Bell S. The origin recognition complex in silencing, cell cycle progression, and DNA replication. Mol Biol Cell. 1995;6:741–756. doi: 10.1091/mbc.6.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loo S, Rine J. Silencing and heritable domains of gene expression. Annu Rev Cell Dev Biol. 1995;11:519–548. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- 25.Lustig A J. Mechanisms of silencing in Saccharomyces cerevisiae. Curr Opin Genet Dev. 1998;8:233–239. doi: 10.1016/s0959-437x(98)80146-9. [DOI] [PubMed] [Google Scholar]
- 26.Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 27.Palacios DeBeer M A, Fox C A. A role for a replicator dominance mechanism in silencing. EMBO J. 1999;18:3808–3819. doi: 10.1093/emboj/18.13.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherman J M, Stone E M, Freeman-Cook L L, Brachmann C B, Boeke J D, Pillus L. The conserved core of a human SIR2 homologue functions in yeast silencing. Mol Biol Cell. 1999;10:3045–3059. doi: 10.1091/mbc.10.9.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shore D. RAP1: a protean regulator in yeast. Trends Genet. 1994;10:408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 30.Smith J S, Brachmann C B, Celic I, Kenna M A, Muhammad S, Starai V J, Avalos J L, Escalante-Semerena J C, Grubmeyer C, Wolberger C, Boeke J D. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spector M S, Raff A, DeSilva H, Lee K, Osley M A. Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol Cell Biol. 1997;17:545–552. doi: 10.1128/mcb.17.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 33.Sutton A, Immanuel D, Arndt K T. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol Cell Biol. 1991;11:2133–2148. doi: 10.1128/mcb.11.4.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanner K G, Landry J, Sternglanz R, Denu J M. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci USA. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Triolo T, Sternglanz R. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature. 1996;381:251–253. doi: 10.1038/381251a0. [DOI] [PubMed] [Google Scholar]
- 36.Vinitsky A, Grubmeyer C. A new paradigm for biochemical energy coupling. Salmonella typhimurium nicotinate phosphoribosyltransferase. J Biol Chem. 1993;268:26004–26010. [PubMed] [Google Scholar]
- 37.Xie J, Pierce M, Gailus-Durner V, Wagner M, Winter E, Vershon A K. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 1999;18:6448–6454. doi: 10.1093/emboj/18.22.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]