Abstract
OBJECTIVE—
Ketosis-prone diabetes (KPD) comprises four subgroups based on the presence or absence of β-cell autoantibodies (A+ or A−) and β-cell functional reserve (β+ or β−). Genetic factors could contribute to their distinctive phenotypes. Our aim was to specify the role of HLA class II alleles associated with susceptibility or resistance to autoimmune type 1 diabetes in determining KPD phenotypes.
RESEARCH DESIGN AND METHODS—
A total of 185 adults presenting with diabetic ketoacidosis were followed longitudinally for a mean of 5.5 years, with measurements of autoantibodies, β-cell functional reserve, insulin sensitivity, and insulin requirement. Frequencies of susceptibility and resistance alleles at HLA DQA1, DQB1, and DRB1 loci were correlated with clinical and phenotypic features of KPD subgroups and compared with those of ethnic-specific population control subjects.
RESULTS—
Susceptibility alleles were more frequent (P < 0.0001) in the two A+ than the two A− KPD subgroups; in the latter, the frequency was no greater than in population control subjects (except for DQB1*0302). Susceptibility alleles differentiated the two clinically similar β− subgroups (more frequent in A+β− than A−β− KPD; P < 0.01). Resistance alleles were more frequent in the two β+ than the two β− KPD subgroups (P < 0.01). The frequencies of certain susceptibility (e.g., DQB1*02) and resistance (DQB1*0602) alleles were higher in African-American A−β+ KPD patients than in African-American control subjects. DQB1*0302 was more frequent in all KPD subgroups compared with control subjects.
CONCLUSIONS—
HLA class II alleles associated with susceptibility or resistance to autoimmune type 1 diabetes help specify the four subgroups of KPD. Inheritance of these alleles may influence long-term β-cell functional reserve.
Ketosis-prone diabetes (KPD), an emerging form of diabetes defined by presentation with diabetic ketoacidosis (DKA) (1–7), is phenotypically heterogeneous (1–6). KPD patients may have variable β-cell function and clinical outcomes after the initial presentation with DKA. Of several methods to define its subgroups, the Aβ scheme (3,8), based on presence or absence of autoimmune markers (A+ or A−) and of β-cell functional reserve (β+ or β−), is the most accurate in predicting long-term β-cell function and clinical outcomes (8–10). A+β− and A+β+ KPD patients have autoantibodies against the 65-kDa isoform of GAD (GAD65) or tyrosine-phosphatase–like protein (insulinoma-associated protein-2), while A−β− and A−β+ KPD patients do not. A+β− and A−β− KPD patients lack β-cell functional reserve and remain totally insulin dependent, whereas A+β+ and A−β+ KPD patients retain β-cell functional reserve when assessed repeatedly, with variable long-term insulin dependency (3). We have previously reported the rationale underlying this classification and nomen-clature for KPD (3,8); briefly, they specify important differences in phenotype and natural history that are obscured in the current American Diabetes Association classification scheme, wherein both β− subgroups would be termed type 1 diabetes and both β+ groups would be termed type 2 diabetes. With the exception of the strong autoimmune basis for β-cell functional loss in A+β− KPD (11,12), the factors underlying irreversible or reversible severe β-cell dysfunction in the KPD subgroups remain unclear and may be complex, even for the other subgroup with an apparent autoimmune basis (A+β+ KPD) (13).
Genetic factors could contribute to the phenotypic differences between KPD subgroups. Here, we investigated whether HLA class II alleles associated with susceptibility or resistance to autoimmune type 1 diabetes (2,6,14–25) could play a role in specifying the phenotypes. We measured an array of HLA class II alleles at the DQA1, DQB1, and DRB1 loci (including those previously associated with susceptibility or resistance to autoimmune diabetes) in a prospective cohort of KPD patients, analyzed their relationship to the four phenotypes, and compared them with those reported for ethnic-specific normal control populations in Texas by the American Society of Histocompatibility and Immunogenetics (1998) (26–28).
RESEARCH DESIGN AND METHODS—
The protocol was approved by the institutional review boards for human studies of the Baylor College of Medicine and the Harris County Hospital District, Houston, Texas. Adult patients, sequentially admitted to Ben Taub General Hospital with DKA between June 1999 and April 2004, were followed prospectively till February 2006. Informed consent was obtained from all subjects.
DKA was defined as described previously by us (3). GAD65 and insulinoma-associated protein-2 autoantibodies were measured in sera as reported previously (29,30,31), using normal ranges established for each ethnic group (3). Patients were classified as β+ or β− by the presence or absence of β-cell functional reserve, as measured within 2 weeks after resolution of initial presentation with DKA and after the 12-month follow-up. The cutoffs and analysis to validate them have been published (3).
Genotyping for HLA class II alleles associated with susceptibility or resistance to autoimmune type 1 diabetes in previous reports (2,6,14–25) was performed by an oligoblot method using sequence-specific oligonucleotides on genomic DNA (18). The polymorphic second exons of DQA1, DQB1, DRB1, DRB3, DRB4, and DRB5 were PCR amplified and labeled by incorporation of digoxigenin-labeled 2ʹ-deoxyuridine 3ʹ-triphosphate. PCR products were hybridized to allele-specific probes selected from these exons. The blots were detected using anti-digoxigenin antibody conjugated to alkaline phosphatase followed by CDP-Star, (chemiluminescent substrate for alkaline phosphatase; Roche Applied Sciences, Indianapolis, IN) and visualized by auto-radiography. Long-term insulin dependence was assessed in all patients, and insulin was discontinued in patients initially categorized as β+, as previously described (3).
Homeostasis model assessment (HOMA) of insulin sensitivity and β-cell function were measured at baseline and periodically thereafter using the HOMA2 model (32). Descriptive statistics were used to characterize the four KPD subgroups. Contingency table analysis with likelihood ratio tests was used to assess frequencies of categorical variables (sex, ethnicity, family history of diabetes, DKA precipitant, proportion of obese patients, ability to discontinue insulin, autoantibody status, β-cell functional status, and HLA class II alleles). When this test indicated significant group differences, pairwise comparisons were made. ANOVA was used to compare continuous variables (age, age at diagnosis, years with diabetes, A1C on admission, BMI, A1C on follow-up, C-peptide levels, HOMA2 of insulin resistance, and HOMA2 of β-cell function). Pairwise testing for continuous measures using a post hoc multiple comparison procedure (Tukey-Kramer) and Fisher’s exact test or χ2 incorporating Yates’ correction for continuity for categorical variables were used when the four-group comparison indicated significant differences. Fisher’s exact test was used to compare allele frequencies between KPD groups and normal control subjects.
RESULTS—
In this analysis of 185 consecutive KPD patients, frequency distributions of the four KPD subgroups were 18% A+β−, 20% A−β−, 8% A+β+, and 54% A−β+. Their demographic and clinical characteristics are shown in Table 1. The mean age at diagnosis of diabetes was significantly higher and the duration of diabetes significantly shorter in the two β+ groups compared with the two β− groups (P < 0.0001). There were no significant differences in sex or ethnic distribution across the four groups, though African Americans were highly represented in the A+β− group (58%) and Hispanics in the A−β+ group (54%). BMI was significantly lower in the two β− groups compared with the β+ groups (P < 0.0001).
Table 1—
Clinical and biochemical characteristics of the four KPD subgroups
| A+β− KPD | A−β− KPD | A+β+ KPD | A−β+ KPD | P | |
|---|---|---|---|---|---|
| n | 33 (17.83) | 38 (20.54) | 14 (7.57) | 100 (54.1) | — |
| Age (years) | 35.9 ± 14.6 | 38.1 ± 13.7 | 40.65 ± 13.5 | 42.52 ± 11.8 | 0.05 |
| Age at diagnosis of diabetes (years) | 27.1 ± 13.4 | 28.4 ± 13.3 | 39.8 ± 11.7 | 40.34 ± 11.7 | <0.0001 |
| Years with diabetes | 8.8 ± 9.2 | 9.7 ± 9.0 | 0.9 ± 1.1 | 2.1 ± 4.1 | <0.0001 |
| Diabetes duration <6 months | 6 (18.2) | 5 (13.2) | 11 (78.6) | 65 (65) | <0.0001 |
| Men-to-women ratio | 1.36:1 | 1.53:1 | 0.75:1 | 1.7:1 | 0.5 |
| Ethnicity | |||||
| African American | 19 (57.6) | 15 (39.5) | 5 (35.7) | 37 (37) | 0.2 |
| Hispanic | 9 (27.3) | 16 (42.1) | 6 (42.9) | 54 (54) | |
| Caucasian | 4 (12.1) | 7 (18.4) | 3 (21.4) | 8 (8) | |
| Asian | 1 (3.0) | 0 | 0 | 1 (1) | |
| Family history of diabetes | 17 (51.5) | 31 (81.6) | 12 (85.7) | 85 (85) | 0.004 |
| BMI (kg/m2) | 24.5 ± 3.6 | 23.3 ± 2.6 | 31.3 ± 7.3 | 31.8 ± 9.1 | <0.0001 |
| A1C at baseline (%) | 13.0 ± 2.4 | 14.3 ± 2.5 | 13.0 ± 2.2 | 13.7 ± 2.5 | 0.1 |
| A1C at 12 months (%) | 10.8 ± 2.5 | 11.3 ± 2.1 | 7.5 ± 2.0 | 7.8 ± 2.4 | <0.0001 |
| HOMA2 of β-cell function at baseline | 5.4 ± 5.0 | 5.7 ± 4.8 | 30.8 ± 31.1 | 36.1 ± 41.2 | <0.0001 |
| HOMA2 of β-cell function at 12 months | 6.1 ± 5.8 | 6.5 ± 13.7 | 67.5 ± 57.9 | 97.9 ± 66.8 | <0.0001 |
| HOMA2 of insulin resistance at baseline | 0.46 ± 0.91 | 0.27 ± 0.29 | 1.98 ± 1.82 | 2.6 ± 5.6 | <0.0001 |
| HOMA2 of insulin resistance at 12 months | 0.17 ± 0.19 | 0.17 ± 0.19 | 1.94 ± 1.01 | 2.7 ± 2.2 | <0.0001 |
| Insulin discontinued | 0 | 0 | 7 (50) | 44 (44) | <0.0001 |
Data are means ± SD or n (%).
At baseline, A1C was not significantly different among the four groups. After 12 months of treatment in the dedicated research clinic, A1C was significantly lower in the two β+ groups than in the two β− groups (P < 0.0001). HOMA2 of β-cell function and HOMA2 of insulin resistance were significantly higher in the β+ groups than in the two β− groups, both at baseline and at 12 months (P < 0.0001) (Table 1). Insulin was successfully discontinued in 50% (7 of 14) of patients in the A+β+ subgroup and 44% of patients (44 of 100) in the A−β+ subgroup, whereas all patients in the A+β− and A−β− groups remained on insulin (P < 0.0001) after 12 months.
There were significant group differences in HLA class II susceptibility allele frequency (Fig. 1A) due to higher frequency in the two A+ compared with the two A− groups (P < 0.0001). The most significant difference occurred between the two most phenotypically divergent groups, A+β− KPD (highest) and A−β+ KPD (lowest). The two clinically similar β− groups, A+β− KPD and A−β− KPD, also differed, with a significantly lower frequency of susceptibility alleles in the latter (P = 0.004). Susceptibility allele frequencies were not different between the two A+ groups.
Figure 1—
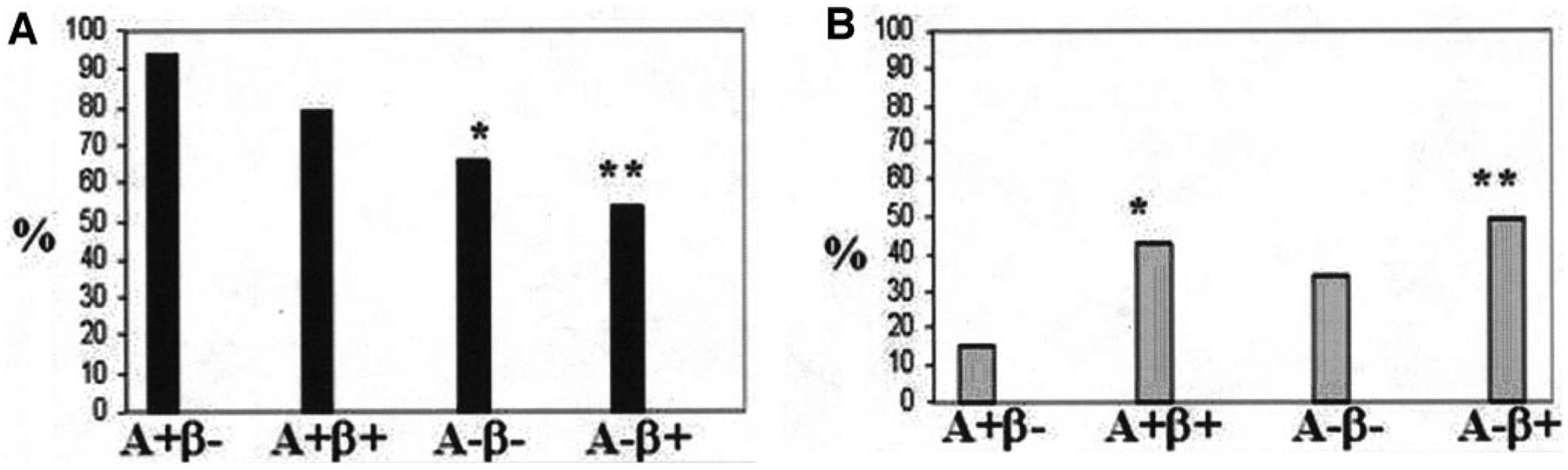
HLA class II allele frequencies in the KPD subgroups. A: Frequencies of autoimmune type 1 diabetes susceptibility alleles were 94, 66, 79, and 54% in the A+β−, A−β−, A+β+, and A−β+ KPD groups, respectively (P < 0.0001). *P = 0.004 for A+β− compared with A−β− KPD. **P < 0.0001 for A+β− compared with A−β+ KPD. B: Frequencies of autoimmune type 1 diabetes resistance alleles were 15, 34, 43, and 49% in the A+β−, A−β−, A+β+, and A−β+ KPD groups, respectively (P = 0.01). *P = 0.06 for A+β− compared with A+β+ KPD. **P < 0.01 for A+β− compared with A−β+ KPD.
There were also significant group differences among the resistance alleles (Fig. 1B), with significantly higher resistance allele frequency in the two A− groups collectively than the two A+ groups (45% compared with 23%, P = 0.01). The most significant difference occurred between the two most phenotypically divergent groups, A+β− KPD (lowest) and A−β+ KPD (highest).
Compared with the two β− groups, the two groups with preserved β-cell functional reserve had a higher frequency of a resistance allele (48% compared with 25%, P < 0.01) and a lower frequency of a susceptibility allele (57% compared with 79%, P < 0.01). β+ patients had a significantly higher frequency of the autoimmune type 1 diabetes resistance allele DQB1*0602 (18% among β+ patients compared with 3% among β− patients; P < 0.01). The frequency of DQB1*0602 was highest in A−β+ KPD, with 20% of patients possessing this allele compared with 4% in the other three groups combined (P < 0.001).
The susceptibility allele DRB1*03 was significantly higher in the two A+ groups, and DQB1*02 was significantly higher in the A+β− KPD group, than in a large control population sample of the same ethnicities in Texas (Table 2). However, the frequencies of these alleles were not different between the two A− groups and the control subjects. The frequencies of the resistance alleles DRB1*15, DQB1*0301, and DQB1*0303 were significantly lower in the two β− groups than among control subjects; the frequencies of these two DQB1*03 alleles were also lower in the A−β+ group. DQB1*0302, a susceptibility allele, had a significantly higher prevalence in the two β− groups and also in the A−β+ KPD group.
Table 2—
HLA class II allele frequencies in KPD subgroups compared with population control subjects
| HLA allele | Population control subjects | A+β− KPD | A−β− KPD | A+β+ KPD | A−β+ KPD |
|---|---|---|---|---|---|
| n | 561 | 33 | 38 | 14 | 100 |
| Susceptibility | |||||
| DRB1*03 | 97 (17) | 14 (42)* | 8 (21) | 6 (43)* | 19 (19) |
| DRB1*04 | 202 (36) | 17 (52) | 15 (39) | 6 (43) | 33 (33) |
| DRB1*07 | 104 (19) | 8 (24) | 7 (18) | 2 (14) | 15 (15) |
| DQB1*02 | 154 (27) | 22 (67)* | 14 (37) | 7 (50) | 28 (28) |
| DQB1*0302 | 96 (17) | 15 (45)* | 12 (32)* | 4 (29) | 29 (29)* |
| Resistance | |||||
| DRB1*15 | 137 (24) | 1 (3)* | 3 (8)* | 1 (7) | 16 (16) |
| DQB1*0301 and *0303 | 363 (65) | 13 (39)* | 16 (42)* | 8 (57) | 39 (39)* |
| DQB1*0602 | 77 (14) | 1 (3) | 2 (5) | 1 (7) | 20 (20) |
| Unknown | |||||
| DQB1*04 | 41 (7) | 0 | 4 (11) | 0 | 11 (11) |
| DQB1*05 | 138 (25) | 3 (9)* | 13 (34) | 3 (21) | 15 (15)* |
| DQB1*06 | 245 (44) | 4 (12)* | 6 (16)* | 2 (14)* | 18 (18)* |
Data are n (%). DRB1*0301 to DQB1*02 and DRB1*04 to DQB1*0302 have been reported previously to confer susceptibility to autoimmune type 1 diabetes (susceptibility alleles), while DRB1*15 to DQB1*0602 have been reported to confer protection against autoimmune type 1 diabetes (resistance alleles). HLA-DQB1*02 is linked to DRB1*07 and may be associated with susceptibility. The list of tested alleles includes DQB1 alleles that are linked to various DRB1 loci but have not been reported previously to be associated with autoimmune type 1 diabetes (DQB1*04 to DQB1*06).
P < 0.05 (KPD subgroup compared with population control subjects).
Since ethnicity affects HLA allele prevalence, we compared frequencies in the A+ (Table 3) and A− (Table 3) KPD groups to ethnic-matched population control subjects. Among the A+ KPD groups, frequencies of the susceptibility alleles were generally higher for all ethnicities compared with ethnic-specific control subjects (Table 3). DRB1*03 was significantly higher in Hispanic A+ KPD patients compared with Hispanic control subjects, and DRB1*04 was significantly higher in African-American A+β− KPD patients compared with African-American control subjects. DQB1*0302 was higher in Caucasian and African-American A+β− KPD patients, with a trend toward higher prevalence in Hispanic A+β− and in Caucasian A+β+ KPD patients, compared with their respective ethnic control subjects. DQB1*02 was also more frequent in the A+ KPD groups compared with ethnic-specific control subjects, although statistical significance was not achieved in some comparisons due to small patient sample sizes. DQB1*0301 and DQB1*0303 were significantly less frequent in Hispanic A+β− KPD patients than in Hispanic control subjects.
Table 3—
HLA class II allele frequencies in KPD subgroups compared with ethnicity-specific population control subjects
| HLA class II allele frequencies in autoantibody-positive (A+) KPD subgroups compared with ethnicity-specific population control subjects | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ethnic-specific control subjects | A+β− KPD | A+β+ KPD | |||||||
| Allele | Caucasian | African American | Hispanic | Caucasian | African American | Hispanic | Caucasian | African American | Hispanic |
| n | 414 | 69 | 78 | 4 | 19 | 9 | 3 | 5 | 6 |
| Susceptibility | |||||||||
| DRB1*03 | 80 (19) | 12 (17) | 5 (6) | 2 (50) | 6 (32) | 6 (67)* | 1 (33) | 2 (40) | 3 (50)* |
| DRB1*04 | 154 (37) | 7 (10) | 41 (53) | 3 (75) | 7 (37)* | 7 (78) | 2 (67) | 0 | 4 (67) |
| DQB1*02 | 130 (31) | 10 (14) | 14 (18) | 2 (50) | 14 (74)* | 5 (56)* | 2 (67) | 2 (40) | 3 (50) |
| DQB1*0302 | 59 (14) | 4 (6) | 33 (42) | 3 (75)* | 5 (26)* | 7 (78) | 2 (67) | 0 | 2 (33) |
| Resistance | |||||||||
| DQB1*0301 and *0303 | 258 (62) | 38 (55) | 67 (86) | 2 (50) | 8 (42) | 2 (22)* | 2 (67) | 2 (40) | 4 (67) |
| HLA class II allele frequencies in autoantibody-negative (A−) KPD subgroups compared with ethnicity-specific population control subjects | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ethnic-specific control subjects | A−β− KPD | A−β+ KPD | |||||||
| Allele | Caucasian | African American | Hispanic | Caucasian | African American | Hispanic | Caucasian | African American | Hispanic |
| n | 414 | 69 | 78 | 7 | 15 | 16 | 8 | 37 | 54 |
| Susceptibility | |||||||||
| DQB1*02 | 130 (31) | 10 (14) | 14 (18) | 3 (43) | 7 (47)* | 4 (25) | 1 (13) | 12 (32)* | 15 (28) |
| DQB1*0302 | 59 (14) | 4 (6) | 33 (42) | 2 (29) | 1 (7) | 9 (56) | 1 (13) | 1 (3) | 26 (48) |
| Resistance | |||||||||
| DQB1*0602 | 64 (15) | 7 (10) | 6 (8) | 0 | 1 (7) | 1 (6) | 1 (13) | 14 (38)* | 5 (9) |
| DQB1*0301 and *0303 | 258 (62) | 38 (55) | 67 (86) | 4 (57) | 6 (40) | 6 (38)* | 4 (50) | 8 (22)* | 26 (48)* |
| Unknown | |||||||||
| DQB1*05 | 116 (28) | 10 (14) | 12 (15) | 3 (43) | 7 (47)* | 3 (19) | 3 (38) | 7 (19) | 4 (7) |
| DQB1*06 | 203 (49) | 19 (28) | 23 (29) | 2 (29) | 2 (13) | 2 (13) | 4 (50) | 11 (30) | 3 (6)* |
Data are n (%).
P < 0.05 (KPD subgroup compared with ethnicity-specific control subjects).
Among A− KPD groups, frequency of the susceptibility allele DQB1*02 was significantly higher in African-American subjects than in control subjects of the same ethnic group. The frequency of the resistance allele DQB1*0602 was significantly higher in African-American A−β+ KPD patients than in African-American control subjects (Table 3). The resistance alleles DQB1*0301 and DQB1*0303 were significantly less frequent in Hispanics with A−β− and A−β+ KPD and African Americans with A−β+ KPD than among their respective ethnic control subjects. The control allele DQB1*05 was significantly higher in African Americans with A−β− KPD, and DQB1*06 was significantly lower in Hispanics with A−β+ KPD, compared with ethnic-specific control subjects.
CONCLUSIONS—
These results demonstrate that HLA class II alleles specify and further define the four subgroups of KPD and suggest that particular genotypes may contribute to distinct KPD phenotypes. There is a high prevalence of susceptibility alleles in the two A+ KPD subgroups. This differentiates the two sets of clinically and phenotypically similar groups: A+β− from A−β− KPD patients and A+β+ from A−β+ KPD patients (3). The ability of susceptibility allele frequency to distinguish A+β− from A−β− KPD is important for taxonomy and pathophysiology, as it diminishes the likelihood that A−β− KPD patients have been misclassified due to loss of circulating autoantibodies (33). The two β+ KPD groups (late onset and often insulin independent) also have a higher prevalence of resistance alleles and a lower prevalence of susceptibility alleles than the two β− groups (early onset and absolutely insulin dependent).
Previous investigators have described HLA allele associations with KPD. Banerji et al. (2) reported a higher frequency of HLA DR3/DR4 in GAD65 autoantibody-negative African-American patients with type 2 diabetes (65%) compared with nondiabetic control subjects (30%). Umpierrez et al. (6) reported that all those with GAD autoantibodies in a cohort of predominantly African-American KPD patients possessed the DQB1*0201 allele.
Wang et al. (34) found that the frequency of the autoimmune type 1 diabetes susceptibility genotype DR3-DQ2/DR4-DQ8 was 10-fold higher and that of the protective allele DQB1*0602 10-fold lower among newly diagnosed younger children with autoantibodies. Our data suggest that HLA class II alleles also modulate age at presentation and severity of β-cell functional loss in KPD patients.
DQB1*0602 is associated with dominant protection from diabetes among autoantibody-positive first-degree relatives of patients with autoimmune type 1 diabetes (20,25,35–38). DQB1*0602 is also significantly more frequent in β+ than in β− KPD patients. A total of 45–50% of patients in the β+ KPD groups became insulin independent over time, suggesting a possible effect of this allele in protecting β-cell function.
Other findings suggest potential roles for HLA class II alleles in the pathophysiology of β-cell dysfunction in autoantibody-negative forms of KPD. For example, African-American A−β+ KPD patients had a significantly higher prevalence of the protective allele DQB1*0602 and the susceptibility allele DQB1*02 than African-American control subjects. Higher frequencies of some susceptibility alleles in A−β+ KPD patients of ethnic minority groups may contribute to their predisposition to develop β-cell dysfunction severe enough to develop DKA, while concomitant inheritance of resistance alleles at a higher frequency than in β− KPD subgroups may contribute to their propensity to recover β-cell function sub-stantially following an episode of DKA.
Tuomi et al. (39) found that patients with latent autoimmune diabetes of adults, who are GAD antibody positive with a phenotype of type 2 diabetes, have a higher frequency of HLA-DQB1*0302–containing alleles than GAD antibody–negative type 2 diabetic patients and nondiabetic control subjects but a lower frequency than type 1 diabetic patients. Latent autoimmune diabetes of adults patients also had lower fasting C-peptide concentrations and diminished insulin responses to glucose than GAD antibody–negative type 2 diabetic patients. These latent autoimmune diabetes of adults patients would resemble A+β+ KPD, albeit without presentation with DKA. We found that the frequency of DQB1*0302 (and of another susceptibility allele DQB1*02) were higher in all KPD subgroups compared with population control subjects. These loci may contribute to the severe β-cell dysfunction resulting in ketoacidosis that defines all forms of KPD. Such a genetic background could contribute to a distinct pathophysiologic process in A−β+ KPD patients compared with nonketosis-prone patients with typical type 2 diabetes, such that β-cell dysfunction in the former is unmasked more abruptly by the development of insulin resistance.
In conclusion, HLA class II alleles associated with susceptibility or resistance to autoimmune type 1 diabetes further define the four KPD phenotypes. These alleles may also play a role in pathophysiology, as there is a significantly higher prevalence of susceptibility alleles in the autoantibody-positive groups as well as a higher prevalence of resistance alleles in the groups with preserved β-cell functional reserve and later onset of diabetes. Inheritance of specific alleles may serve as a marker of the evolution of β-cell function and clinical outcomes among KPD patients and identify appropriate candidates for immunomodulatory therapy. Although routine analysis of HLA alleles in KPD patients is not recommended at this time, these associations set the stage for identification of genetic mutations contributing to severe β-cell dysfunction in KPD.
Acknowledgments—
This article is supported by grants RO1-HL73696, RO1-HL79636 (both to A.B.), and P51-RR000166 (to L.K.G.) and an American Diabetes Association Career Development Award (to C.S.H.).
The authors thank Andrew Westmark (National Primate Research Center, University of Washington) for technical assistance in HLA analyses, Jesus Villanueva (Translational Metabolism Unit, Baylor College of Medicine) for meticulous maintenance of the KPD database, and the house officers of the Department of Medicine, Ben Taub General Hospital, for providing excellent inpatient care for the KPD patients.
Abbreviations:
- DKA
diabetic ketoacidosis
- GADA
GAD antibody
- HOMA
homeostasis model assessment
- KPD
ketosis-prone diabetes
References
- 1.Balasubramanyam A, Zern JW, Hyman DJ, Pavlik V: New profiles of diabetic ketoacidosis: type 1 vs type 2 diabetes and the effect of ethnicity. Arch Intern Med 159:2317–2322, 1999 [DOI] [PubMed] [Google Scholar]
- 2.Banerji MA, Chaiken RL, Huey H, Tuomi T, Norin AJ, Mackay IR, Rowley MJ, Zim-met PZ, Lebovitz HE: GAD antibody negative NIDDM in adult black subjects with diabetic ketoacidosis and increased frequency of human leukocyte antigen DR3 and DR4: Flatbush diabetes. Diabetes 43: 741–745, 1994 [DOI] [PubMed] [Google Scholar]
- 3.Maldonado M, Hampe CS, Gaur LK, D’Amico S, Iyer D, Hammerle LP, Bolgiano D, Rodriguez L, Rajan A, Lernmark A, Balasubramanyam A: Ketosis-prone diabetes: dissection of a heterogeneous syndrome using an immunogenetic and beta-cell functional classification, prospective analysis, and clinical outcomes. J Clin Endocrinol Metab 88:5090–5098, 2003 [DOI] [PubMed] [Google Scholar]
- 4.Umpierrez GE, Casals MM, Gebhart SP, Mixon PS, Clark WS, Phillips LS: Diabetic ketoacidosis in obese African-Americans. Diabetes 44:790–795, 1995 [DOI] [PubMed] [Google Scholar]
- 5.Umpierrez GE, Smiley D, Kitabchi AE: Narrative review: ketosis-prone type 2 diabetes mellitus. Ann Intern Med 144:350–357, 2006 [DOI] [PubMed] [Google Scholar]
- 6.Umpierrez GE, Woo W, Hagopian WA, Isaacs SD, Palmer JP, Gaur LK, Nepom GT, Clark WS, Mixon PS, Kitabchi AE: Immunogenetic analysis suggests different pathogenesis for obese and lean African-Americans with diabetic ketoacidosis. Diabetes Care 22:1517–1523, 1999 [DOI] [PubMed] [Google Scholar]
- 7.Westphal SA: The occurrence of diabetic ketoacidosis in non-insulin-dependent diabetes and newly diagnosed diabetic adults. Am J Med 101:19–24, 1996 [DOI] [PubMed] [Google Scholar]
- 8.Balasubramanyam AGG, Rodriguez L, Hampe C, Gaur L, Lernmark A, Maldonado M: Accuracy and predictive value of classification schemes for ketosis-prone diabetes mellitus (KPDM). Diabetes Care 29:2575–2579, 2006 [DOI] [PubMed] [Google Scholar]
- 9.Maldonado M, D’Amico S, Otiniano M, Balasubramanyam A, Rodriguez L, Cuevas E: Predictors of glycaemic control in indigent patients presenting with diabetic ketoacidosis. Diabetes Obes Metab 7:282–289, 2005 [DOI] [PubMed] [Google Scholar]
- 10.Maldonado MR, Otiniano ME, Cheema F, Rodriguez L, Balasubramanyam A: Factors associated with insulin discontinuation in subjects with ketosis-prone diabetes but preserved beta-cell function. Diabet Med 22:1744–1750, 2005 [DOI] [PubMed] [Google Scholar]
- 11.Bottazzo GF: Banting Lecture: On the honey disease: a dialogue with Socrates. Diabetes 42:778–800, 1993 [DOI] [PubMed] [Google Scholar]
- 12.Eisenbarth GS: Type I diabetes mellitus: a chronic autoimmune disease. N Engl J Med 314:1360–1368, 1986 [DOI] [PubMed] [Google Scholar]
- 13.Hampe CS, Nalini R, Maldonado MR, Hall TR, Garza G, Iyer D, Balasubramanyam A: Association of amino-terminal-specific anti-glutamate decarboxylase (GAD65) autoantibodies with beta cell functional reserve and a milder clinical phenotype in patients with GAD65 antibodies and ketosis prone diabetes mellitus. J Clin Endocrinol Metab 92:462–467, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Agardh D, Gaur LK, Agardh E, Landin-Olsson M, Agardh CD, Lernmark A: HLA-DQB1*0201/0302 is associated with severe retinopathy in patients with IDDM. Diabetologia 39:1313–1317, 1996 [DOI] [PubMed] [Google Scholar]
- 15.Duquesnoy RJ, MacDonald MJ, Mullins P, Hackbarth SA, Traisman HS, Levitsky LL: Increased frequency of HLA-Dw3 in North-American black patients with juvenile onset diabetes. Tissue Antigens 13: 369–372, 1979 [DOI] [PubMed] [Google Scholar]
- 16.Eisenbarth GS, Srikanta S, Fleischnick E, Ganda OP, Jackson RA, Brink SJ, Soeldner JS, Yunis EJ, Alper C: Progressive autoimmune beta cell insufficiency: occurrence in the absence of high-risk HLA alleles DR3, DR4. Diabetes Care 8:477–480, 1985 [DOI] [PubMed] [Google Scholar]
- 17.Greenbaum CJ, Gaur LK, Noble JA: ICA+ relatives with DQA1*0102/DQB1*0602 have expected 0602 sequence and DR types. J Autoimmun 18:67–70, 2002 [DOI] [PubMed] [Google Scholar]
- 18.Leech NJ, Kitabchi AE, Gaur LK, Hagopian WA, Hansen J, Burghen GA, Palmer JP, Nepom GT: Genetic and immunological markers of insulin dependent diabetes in Black Americans. Autoimmunity 22: 27–32, 1995 [DOI] [PubMed] [Google Scholar]
- 19.Monos DS, Spielman RS, Gogolin KJ, Radka SF, Baker L, Zmijewski CM, Kamoun M: HLA-DQw3.2 allele of the DR4 haplotype is associated with insulin-dependent diabetes: correlation between DQ beta restriction fragments and DQ beta chain variation. Immunogenetics 26: 299–303, 1987 [DOI] [PubMed] [Google Scholar]
- 20.Morel PA, Dorman JS, Todd JA, McDevitt HO, Trucco M: Aspartic acid at position 57 of the HLA-DQ beta chain protects against type I diabetes: a family study. Proc Natl Acad Sci U S A 85:8111–8115, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Platz P, Jakobsen BK, Morling N, Ryder LP, Svejgaard A, Thomsen M, Christy M, Kromann H, Benn J, Nerup J, Green A, Hauge M: HLA-D and -DR antigens in genetic analysis of insulin dependent diabetes mellitus. Diabetologia 21:108–115, 1981 [DOI] [PubMed] [Google Scholar]
- 22.Rodey GE, White N, Frazer TE, Duquesnoy RJ, Santiago JV: HLA-DR specificities among black Americans with juvenile-onset diabetes. N Engl J Med 301: 810–812, 1979 [DOI] [PubMed] [Google Scholar]
- 23.Sheehy MJ, Rowe JR, Fuller TC, Yunis EJ, Gabbay KH: A minor subset of HLA-DR3 haplotypes is preferentially increased in type 1 (insulin-dependent) diabetes. Diabetologia 28:891–894, 1985 [DOI] [PubMed] [Google Scholar]
- 24.Thorsby E, Ronningen KS: Particular HLA-DQ molecules play a dominant role in determining susceptibility or resistance to type 1 (insulin-dependent) diabetes mellitus. Diabetologia 36:371–377, 1993 [DOI] [PubMed] [Google Scholar]
- 25.Todd JA, Bell JI, McDevitt HO: HLA-DQ beta gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature 329:599–604, 1987 [DOI] [PubMed] [Google Scholar]
- 26.McCormack J, Osowski LD: Caucasian US normal. In HLA 1998. Gjertson DW and Terasaki PI, Eds. American Society of Histocompatibility and Immunogenetics, Lenexa, Kansas, 1998, p. 258–59 [Google Scholar]
- 27.McCormack J, Osowski LD: Black US normal. In HLA 1998. Gjertson DW and Terasaki PI, Eds. American Society of Histocompatibility and Immunogenetics, Lenexa, Kansas, 1998, p. 232 [Google Scholar]
- 28.McCormack J, Osowski LD: Hispanic US normal. In HLA 1998. Gjertson DW and Terasaki PI, Eds. American Society of Histocompatibility and Immunogenetics, Lenexa, Kansas, 1998, p. 261 [Google Scholar]
- 29.Grubin CE, Daniels T, Toivola B, Landin-Olsson M, Hagopian WA, Li L, Karlsen AE, Boel E, Michelsen B, Lernmark Å: A novel radioligand binding assay to determine diagnostic accuracy of isoform-specific glutamic acid decarboxylase antibodies in childhood IDDM. Diabetologia 37:344–350, 1994 [DOI] [PubMed] [Google Scholar]
- 30.Falorni A,Örtqvist E, Persson B, Lernmark Å: Radioimmunoassays for glutamic acid decarboxylase (GAD65) and GAD65 autoantibodies using 35S or 3H recombinant human ligands. J Immunol Methods 186:89–99, 1995 [DOI] [PubMed] [Google Scholar]
- 31.Hampe CS, Hammerle LP, Bekris L, Ortqvist E, Kockum I, Rolandsson O, Landin-Olsson M, Torn C, Persson B, Lernmark A: Recognition of glutamic acid decarboxylase (GAD) by autoantibodies from different GAD antibody-positive phenotypes. J Clin Endocrinol Metab 85: 4671–4679, 2000 [DOI] [PubMed] [Google Scholar]
- 32.Levy JC, Matthews DR, Hermans MP: Correct homeostasis model assessment(HOMA) evaluation uses the computer program. Diabetes Care 21:2191–2192, 1998 [DOI] [PubMed] [Google Scholar]
- 33.Borg H, Marcus C, Sjoblad S, Fernlund P, Sundkvist G: Islet cell antibody frequency differs from that of glutamic acid decarboxylase antibodies/IA2 antibodies after diagnosis of diabetes. Acta Paediatr 89: 46–51, 2000 [DOI] [PubMed] [Google Scholar]
- 34.Wang J, Miao D, Babu S, Yu J, Barker J, Klingensmith G, Rewers M, Eisenbarth GS, Yu L: Prevalence of autoantibody-negative diabetes is not rare at all ages and increases with older age and obesity. J Clin Endocrinol Metab 92:88–92, 2007 [DOI] [PubMed] [Google Scholar]
- 35.Erlich HA, Zeidler A, Chang J, Shaw S, Raffel LJ, Klitz W, Beshkov Y, Costin G, Pressman S, Bugawan T, et al. : HLA class II alleles and susceptibility and resistance to insulin dependent diabetes mellitus in Mexican-American families. Nat Genet 3:358–364, 1993 [DOI] [PubMed] [Google Scholar]
- 36.Pugliese A, Gianani R, Moromisato R, Awdeh ZL, Alper CA, Erlich HA, Jackson RA, Eisenbarth GS: HLA-DQB1*0602 is associated with dominant protection from diabetes even among islet cell antibody-positive first-degree relatives of patients with IDDM. Diabetes 44:608–613, 1995 [DOI] [PubMed] [Google Scholar]
- 37.Schreuder GM, Tilanus MG, Bontrop RE, Bruining GJ, Giphart MJ, van Rood JJ, de Vries RR: HLA-DO polymorphism associated with resistance to type I diabetes detected with monoclonal antibodies, iso-electric point differences, and restriction fragment length polymorphism. J Exp Med 164:938–943, 1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomsen M, Platz P, Christy M, Nerup J, Ryder LP, Svejgaard A: HLA-D-associated resistance and susceptibility to insulin-dependent diabetes mellitus. Transplant Proc 11:1307–1308, 1979 [PubMed] [Google Scholar]
- 39.Tuomi T, Carlsson A, Li H, Isomaa B, Miet-tinen A, Nilsson A, Nissen M, Ehrnstrom BO, Forsen B, Snickars B, Lahti K, Fors-blom C, Saloranta C, Taskinen MR, Groop LC: Clinical and genetic characteristics of type 2 diabetes with and without GAD antibodies. Diabetes 48:150–157, 1999 [DOI] [PubMed] [Google Scholar]


