Abstract
Here, we introduce a facile, scalable engineering approach to enable long-term development and maturation of organoids. We have redesigned the configuration of conventional organoid culture to develop a platform that converts single injections of stem cell suspensions to radial arrays of organoids that can be maintained for extended periods without the need for passaging. Using this system, we demonstrate accelerated production of intestinal organoids with significantly enhanced structural and functional maturity, and their continuous development for over 4 weeks. Furthermore, we present a patient-derived organoid model of inflammatory bowel disease (IBD) and its interrogation using single-cell RNA sequencing to demonstrate its ability to reproduce key pathological features of IBD. Finally, we describe the extension of our approach to engineer vascularized, perfusable human enteroids, which can be used to model innate immune responses in IBD. This work provides an immediately deployable platform technology toward engineering more realistic organ-like structures in a dish.
Stem cells in three-dimensional (3D) culture can self-organize into complex multicellular structures termed organoids that resemble the anatomical and functional units of the organ from which they are derived1,2. By providing a means to recapitulate the complexity of in vivo physiological systems at the convenience of traditional cell culture, organoids have emerged as an enabling research platform to study development, health and disease of various organs for biomedical applications3. In a conventional laboratory setting, the most common method to form organoids is to embed pluripotent or adult stem cells in sessile drops of extracellular matrix (ECM) hydrogels such as Matrigel4 (Fig. 1a). When supplied with culture media containing defined soluble factors, the 3D environment induces the differentiating cells to segregate into distinct domains and undergo fate specification, leading to their spontaneous organization into organ-like structures (Fig. 1b).
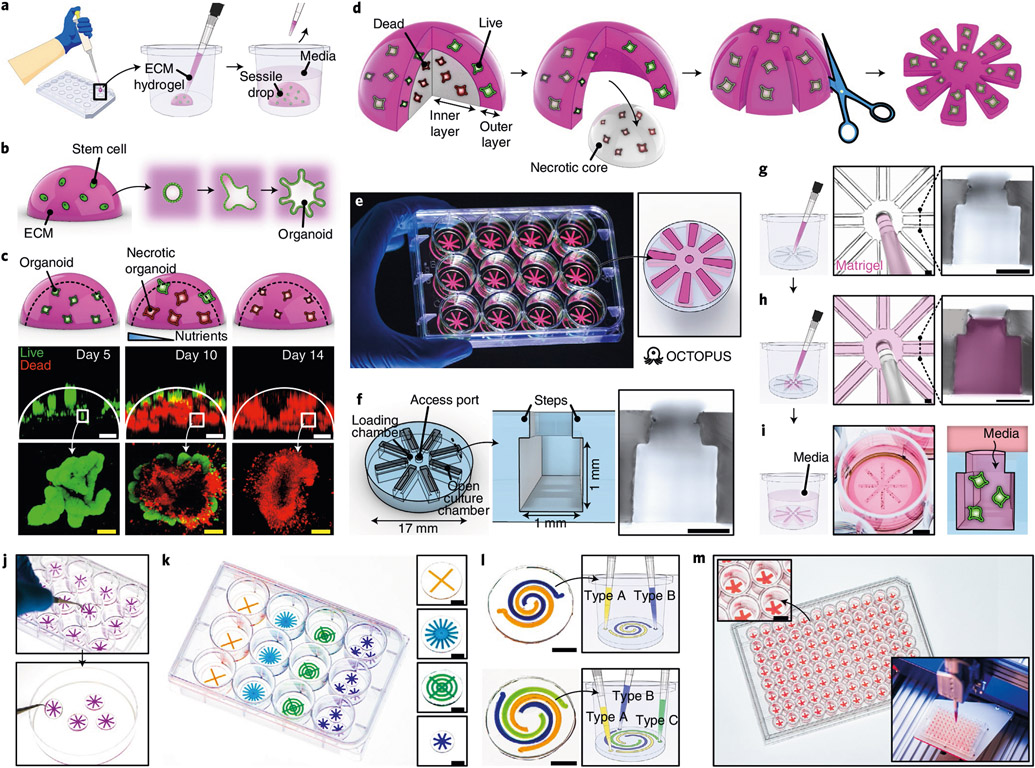
Fig. 1 ∣. Geometric engineering of conventional organoid culture using OCTOPUS.
a,b, Schematic showing conventional techniques that rely on 3D culture (a) and self-organization (b) of stem cells in sessile drops of ECM hydrogel to form organoids. c, Top row: schematic showing the formation of necrotic core in Matrigel drop culture. Middle and bottom rows: loss of viability and structural integrity in Matrigel drop culture of intestinal organoids over 10–14 d due to limited nutrient supply. Green and red show live and dead cells, respectively. Scale bars, 500 μm (top images) and 100 μm (bottom images). d, Conceptual diagram that shows the idea of reconfiguring the geometry of a conventional hydrogel drop scaffold to culture organoids without diffusion limitations. e, Photo of OCTOPUS inserts in a standard 12-well cell culture plate. f, Device design of OCTOPUS. The micrograph shows the cross-section of the culture chamber. Scale bar, 500 μm. g,h, Injection and distribution of stem-cell-containing Matrigel solution into the culture chambers of OCTOPUS. The micrographs show the top-down view of the access port with a 200-μl pipette tip (left) and the cross-section of the chamber (right) before (g) and after (h) gel injection. Pink shows Matrigel. Scale bars, 500 μm. i, OCTOPUS is kept submerged in media during culture. Developing organoids in the hydrogel are supplied with nutrients through the opening of the culture chambers. Scale bar, 5 mm. j, Facile handling and transferability of OCTOPUS. k, Examples of different chamber/device designs in OCTOPUS. Scale bars, 5 mm. l, Multi-chamber designs for co- and tri-culture in OCTOPUS. Scale bars, 5 mm. m, OCTOPUS can be deployed as a culture platform in a 96-well format coupled with a robotic fluid handling system to scale up the production of organoids. Scale bar, 3 mm.
The process of organoid formation in this conventional method relies on passive diffusion for nutrient supply and waste removal. As organoids continue to grow and become more metabolically demanding, however, limited diffusion of nutrients and oxygen into the inner regions of the 3D scaffold causes the formation of a necrotic core with greatly reduced cell viability, which eventually propagates through the entire hydrogel construct5. The rate at which this degenerative process occurs varies depending on the type of organoids and their growth rate. In the case of intestinal organoids, for instance, considerable cell death becomes evident within 10 d (Fig. 1c). In existing culture protocols, this problem is prevented by passaging the organoids every 5–7 d (refs. 4,6) but this short duration of each culture cycle is recognized as a critical issue because it poses fundamental challenges to establishing uninterrupted, continuous culture of organoids for prolonged periods, which is necessary for their sustained growth and maturation7.
To tackle this challenge, researchers have used bioreactors to improve diffusive transport and establish long-term culture of organoids to promote their continued development in vitro. Implementing this technique in routine laboratory settings, however, is burdened by the need for expensive capital equipment that is mechanically complex and requires specialized knowledge for operation and maintenance. While vascularization of organoids has been suggested as an alternative strategy8, the process of generating organoid models with controlled vascular perfusion is prohibitively complex and often requires advanced culture systems and specialized in vitro techniques that are not easily accessible to nonengineers.
Here we present a simple, immediately deployable device that makes it possible to reconfigure the geometry of the 3D culture environment of organoids to eliminate the problem of limited and nonuniform diffusion inherent in bulk hydrogel. This system is manufactured as simple culture inserts that are ready for use in standard cell culture plates without any modification of established protocols and workflow. We first describe the basic principle of our method and provide its proof-of-concept by demonstrating uninterrupted, long-term culture of human and mouse intestinal stem cells to generate enlarged intestinal organoids with significantly increased structural and functional maturity. We also show advanced capabilities of our technology by presenting (1) an organotypic model of human IBD using patient-derived enteroids and (2) microengineered organoids integrated with perfusable vasculature for in vitro modeling of immune-epithelial interactions in IBD.
Results
Design and production of 3D organoid culture platform
In essence, our approach can be conceptualized as (1) removing a necrotic core from a hydrogel drop while keeping the outer layer containing viable organoids and (2) radially segmenting the remaining layer and spreading it out to form a planar array of organoids (Fig. 1d). By decreasing the thickness of the culture scaffold, this array is designed to permit unrestricted, rapid diffusion and replenishment of nutrients, oxygen and other soluble factors.
To implement this idea, we created a disc-shaped device that enables the production of radially arranged organoid arrays in standard cell culture plates (Fig. 1e). This device, termed OCTOPUS (Organoid Culture-based Three-dimensional Organogenesis Platform with Unrestricted Supply of soluble signals), consists of eight identical culture chambers with cross-sectional dimensions of 1 mm (height) × 1 mm (width) that radiate from a central loading chamber with an open access port (Fig. 1f and Supplementary Fig. 1). Importantly, the culture chambers are open to the external environment and contain microscopic steps protruding from the edges of the opening (Fig. 1f).
Establishing organoid culture in this system only requires two simple steps. First, stem cells suspended in a hydrogel precursor are pipetted into the central chamber through the access port (Fig. 1g and Supplementary Video 1) and equally distributed to the culture chambers (Fig. 1h and Supplementary Video 2). During this process, surface tension acts to pin the liquid meniscus at the steps (Fig. 1h), allowing the injected solution to advance and fill the entire chamber without spillage through the open-top. After gelation, culture medium is added to the device-containing well to provide nutrient supply to the embedded cells through the exposed hydrogel surface (Fig. 1i).
The design of OCTOPUS as a removable culture insert makes this system easily transferable (Fig. 1j), facilitating handling and analysis of cultured organoids. It is also readily possible to change the number, size and shape of the culture chambers (Fig. 1k), which provides a means to control the volume and spatial organization of organoid-containing tissue constructs generated in this system. Similarly, the design of OCTOPUS can be modified to grow two or more types of organoids by creating individually accessible culture chambers on the same device (Fig. 1l and Extended Data Fig. 1). Furthermore, the overall size and shape of OCTOPUS can be tailored to different well sizes and formats of standard culture plates (Fig. 1m), providing opportunities for scalable organoid production and experimentation.
Long-term culture of intestinal organoids in OCTOPUS
First, we investigated whether OCTOPUS can extend the lifespan of organoids. This study was conducted using murine small intestinal organoids derived from commercially available stem cells as a model system. During the first 5d of culture, the cells in both OCTOPUS and Matrigel drops self-organized into highly viable intestinal organoids identified by their crypt–villus structures (Fig. 2a,d). When the culture period was extended beyond 5–7 d, however, a large fraction of organoids in Matrigel drops developed necrosis and disintegrated over time, whereas the organoids in OCTOPUS continued to grow and form buds without a measurable loss of viability or unwanted surface adhesion (Fig. 2b-e and Supplementary Figs. 2 and 3). This major difference was observed regardless of initial cell seeding density (Supplementary Fig. 4). Our data also showed that OCTOPUS culture yields higher long-term viability of organoids and significant increases in their growth and maturity when compared with other conventional techniques, such as 3D ‘on-top’ culture9 and monolayer culture of organoid-derived cells10 (Supplementary Figs. 5 and 6).
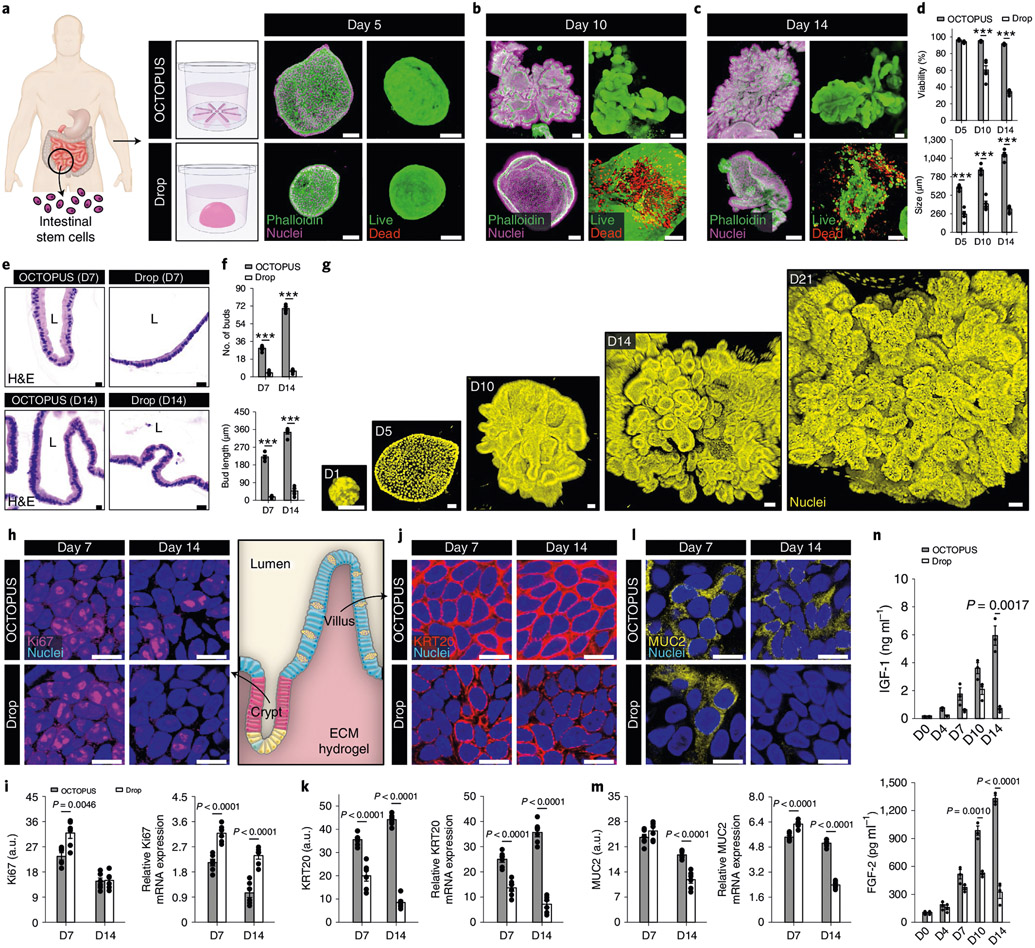
Fig. 2 ∣. Effects of long-term culture on the maturation of intestinal organoids in OCTOPUS.
a, Mouse intestinal adult stem cells in Matrigel self-assemble into intestinal organoids in both OCTOPUS and drop culture. Scale bars, 100 μm. b,c, When observed at 10 d (b) and 14 d (c), organoids in OCTOPUS continue their growth (top), whereas the ones in Matrigel drops rapidly lose viability and disintegrate (bottom). Scale bars, 100 μm. d,e, Quantification of organoid viability (n = 6; ***P < 0.0001) (d) and size (n = 5; *P = 0.0115, ***P < 0.0001) (e). f, Continuous enlargement of intestinal organoids in OCTOPUS over 21 d. Scale bars, 100 μm. g, OCTOPUS reduces variability in the size of organoids as evidenced by the substantially smaller coefficient of variation (CV). Scale bars, 100 μm. h, Bud formation is used as a metric for analyzing morphological development of intestinal organoids in 3D culture. i,j, Confocal micrographs of organoids in Matrigel drop (i) and OCTOPUS (j). Organoid budding is more pronounced in OCTOPUS. Scale bars, 100 μm. k, Quantification of bud number (n = 3) and length (n = 4). l, Confocal micrographs showing the spatial distribution of EdU+ cells (white) in OCTOPUS-generated organoids. The yellow lines in the close-up images outline organoid buds. Scale bars, 100 μm. m, Visualization and quantification of differentiation markers specific to enterocytes (villin, top), goblet cells (MUC2, middle) and enteroendocrine cells (somatostatin, bottom). Scale bars, 10 μm (n = 3). n, Imaging and quantification of intracellular calcium signaling in intestinal organoids treated with 50 mM glucose. Organoids in Matrigel drops at the maximum duration of culture (7 d) were compared with those maintained in OCTOPUS for 14 d to examine the contribution of extended culture. The time between stimulation and maximum fluorescence intensity is shaded in pink. Scale bars, 100 μm. All data are presented as mean ± s.e.m. and P values are from unpaired, two-sided t-test. a.u., arbitrary units; BF, bright field; D, day.
The more supportive environment of OCTOPUS permitted continuous growth of organoids for over 3 weeks without passaging (Fig. 2f), representing a more than threefold increase in their lifespan per culture cycle as compared with the typical duration of conventional intestinal organoid culture in Matrigel drops (5–7 d). Moreover, the enhanced longevity of culture in OCTOPUS was accompanied by reduced variability in the size of developing organoids (Fig. 2g). Our imaging analysis of diffusion provided further insight into these significant differences by showing rapid penetration and uniform spatial distribution of dye molecules and oxygen in OCTOPUS, which was in contrast to the development of concentration gradients along the scaffold thickness and limited molecular transport into the inner regions of the hydrogel in Matrigel drops (Extended Data Fig. 2, Supplementary Fig. 7 and Supplementary Video 3).
These results verify the design principle of OCTOPUS and suggest that long-term viability and sustained growth of organoids with reduced size variability in our system may be attributed to unrestricted and spatially uniform diffusive transport of nutrients, growth factors and oxygen, which is achieved by decreasing the distance between organoids and the Matrigel surface (Supplementary Fig. 8). While this demonstration focused on intestinal organoids, similar beneficial effects were observed in other types of organoids cultured in OCTOPUS (Extended Data Fig. 3).
Enhanced maturation of organoids in OCTOPUS
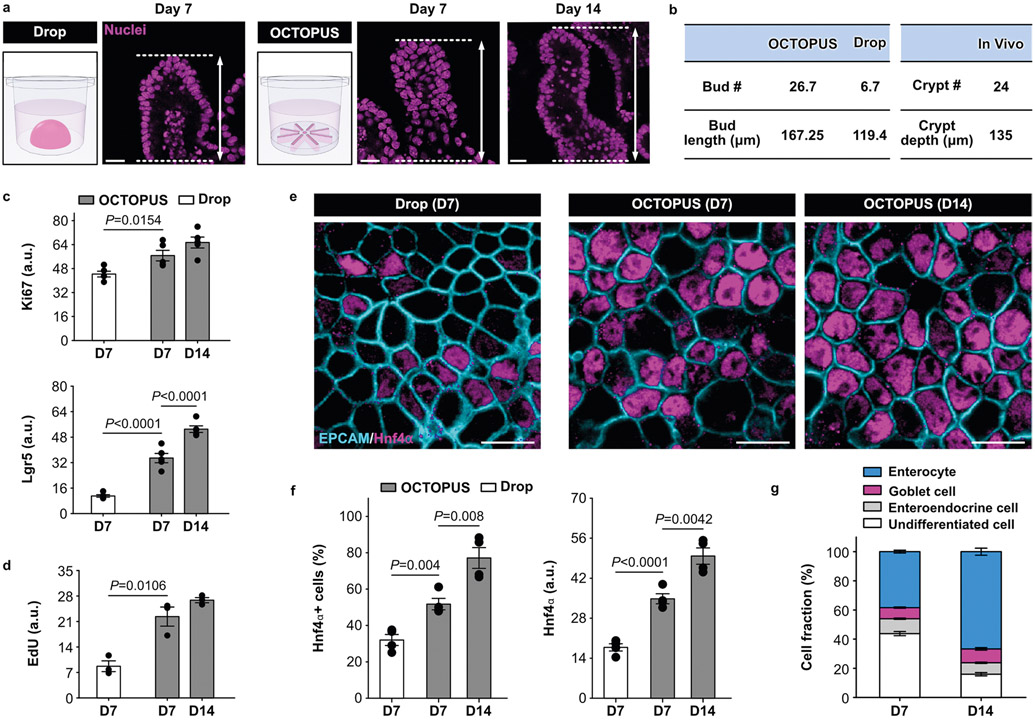
Next, we asked whether prolonged culture in OCTOPUS enhances structural and functional maturation of mouse intestinal organoids. Recognizing villus morphogenesis as the critical process of establishing the structural organization of the mature intestinal epithelium11, we first examined the formation of buds in the developing organoids that correspond to the crypt domain of the small intestine in vivo (Fig. 2h). During 7-d culture, budding was clearly visible in both Matrigel drops and OCTOPUS but the extent of this event as assessed by the bud number and length was significantly greater in OCTOPUS (Fig. 2i-k and Extended Data Fig. 4a). Our device allowed these organoids to continue their development beyond day 7 to form roughly three times as many buds by day 14 (Fig. 2j,k), at which point the number and length of buds approximated the average number and depth of crypts reported in the small intestine of healthy mice (Extended Data Fig. 4b)12,13.
For further characterization, we then conducted immunofluorescence analysis to examine the cellular phenotype of the developing organoids. Compared with drop culture, our results indicated increased activity of proliferative stem/progenitor cells in OCTOPUS as evidenced by higher expression of Lgr5, Ki67 and EdU (Extended Data Figs. 4c,d), which is known to be required for the development of the crypt–villus structures in intestinal organoids14,15. The EdU+ cells were initially distributed throughout the organoid, but at day 14, these cells were localized at the tip of the buds (Fig. 2l), recapitulating the restriction of cell proliferation to the crypt domains in vivo16. Importantly, OCTOPUS-produced organoids also displayed stronger immunofluorescence of hepatocyte nuclear factor 4α (Hnf4α), a transcription factor essential for intestinal maturation17 (Extended Data Fig. 4e,f). This result indicating enhanced and accelerated organoid maturation in our device was supported by significantly higher expression of specific markers of specialized intestinal epithelial cells, including villin (absorptive enterocytes), MUC2 (mucus-producing goblet cells) and somatostatin (enteroendocrine cells), all of which were further upregulated during prolonged culture in OCTOPUS (Fig. 2m). The number of enterocytes at the end of 14-d culture in the device was greater than that of the other cell populations (Extended Data Fig. 4g), reproducing the highest abundance of this cell type in the intestinal epithelium in vivo. Our data also suggested that the improved growth and maturation of intestinal organoids in our engineered system could be further enhanced by growing them with organ-specific stromal cells in coculture OCTOPUS devices (Extended Data Fig. 5).
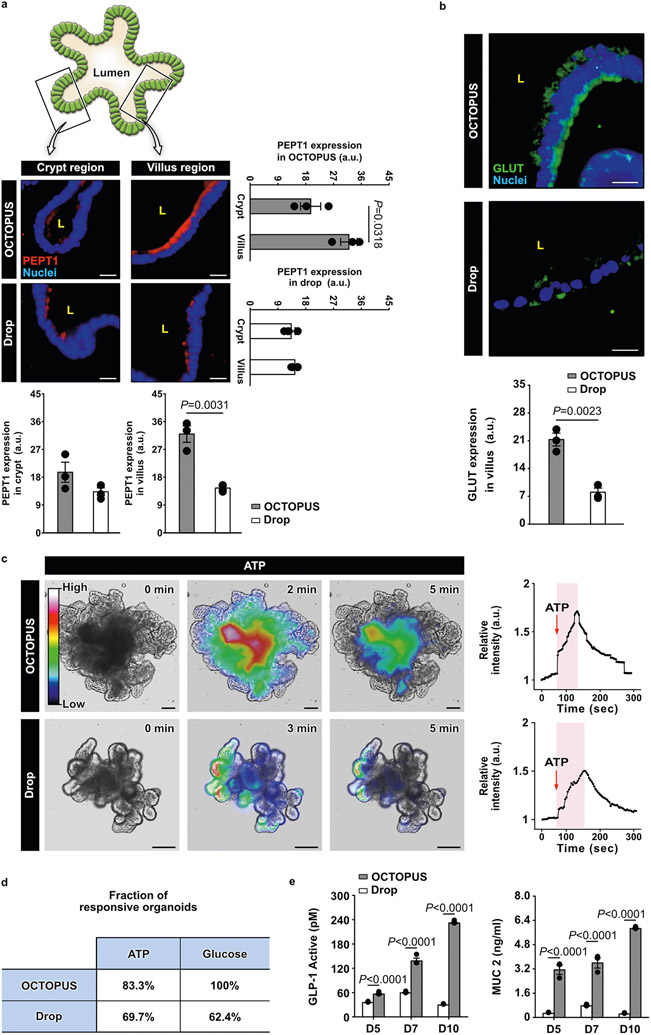
Importantly, immunofluorescence data from OCTOPUS also showed more robust, spatially defined expression of molecular transporters that regulate the physiological function of the intestinal epithelium to absorb nutrients, including peptide transporter 1 (PEPT1)18 and glucose transporter 2 (GLUT2)19,20 (Extended Data Fig. 6a,b). To characterize the functional capacity of the intestinal epithelium, we then harvested organoids from OCTOPUS and hydrogel drops, embedded them in a 1-mm-thick Matrigel layer and compared their intracellular calcium signaling, which is known to regulate the activity of nutrient transporters in the intestine20,21. When the organoids were stimulated with ATP or 50 mM d-glucose in this analysis, calcium responses measured in OCTOPUS were much more rapid and substantial than those in Matrigel drops (Extended Data Fig. 6c and Fig. 2n). Comparison of these two groups also revealed a substantially larger fraction of responsive organoids in OCTOPUS (Extended Data Fig. 6d). The increased functional maturity of organoids in OCTOPUS demonstrated by these results was further evidenced by significantly higher concentrations of glucose-induced release of glucagon-like peptide 1 (GLP-1) and MUC2 in the conditioned media which play a critical role in digestive and barrier function of the intestinal epithelium, respectively22,23 (Extended Data Fig. 6e).
Human intestinal organoids in OCTOPUS
Having demonstrated the proof-of-concept of our technology using mouse organoids, we then explored the production of human intestinal organoids in OCTOPUS using stem cells isolated from the terminal ileum of healthy donors (Fig. 3a). When seeded into our device, these cells formed spherical organoids with high cell viability within 5 d, but as culture progressed, the growing cystic organoids extended epithelial folds into the surrounding matrix (Fig. 3b-d). This developmental pattern was not observed in Matrigel drops, in which prolonged culture beyond 7 d resulted in arrested organoid growth and significant reduction in viability without the evidence of bud formation (Fig. 3b-d). Consistent with these results, histological analysis showed bud-like structures in the OCTOPUS enteroids, which continued to increase in both number and length during extended culture (Fig. 3e,f). In contrast, intestinal organoids in Matrigel drops remained mostly spherical with few buds throughout the culture period (Fig. 3e,f). Continuous culture in OCTOPUS over 1 month without passaging led to a more than 32-fold increase in size, yielding human enteroids with extensive epithelial folding that were as large as 2.6 mm in diameter (Fig. 3g).
Fig. 3 ∣. Prolonged culture of human intestinal organoids in OCTOPUS.
a, Human enteroids derived from human adult intestinal stem cells cultured in OCTOPUS and Matrigel drop for 5 d. Scale bars, 100 μm. b,c, Micrographs of enteroids at 10 d (b) and 14 d (c) show enteroids in OCTOPUS become larger and develop crypt/villus-like structures during 14-d culture (top), which is in contrast to arrested growth and decreased viability in Matrigel drop culture (bottom). Scale bars, 100 μm. d, Quantification of organoid size (n = 5; ***P < 0.0001) and viability (n = 5; ***P < 0.0001). e, Representative images of H&E-stained enteroid sections in OCTOPUS and Matrigel drop at days 7 (top) and 14 (bottom). Scale bars, 20 μm. f, Quantification of bud number (n = 5; ***P < 0.0001) and length (n = 5; ***P < 0.0001). g, Growth of human enteroids in OCTOPUS over 21 d. Scale bars, 50 μm. h–i, Immunofluorescence (h) and mRNA analysis (i) of Ki67+ proliferative cells in the crypt domain at days 7 and 14. Scale bars, 10 μm (n = 6). j, k, Immunofluorescence (j) and mRNA analysis (k) of differentiated KRT20+ absorptive enterocytes on the villus surface at days 7 and 14. Scale bars, 10 μm (n = 6). l, m, Immunofluorescence (l) and mRNA analysis (m) of differentiated MUC2+ goblet cells at days 7 and 14. Scale bars, 10 μm (n = 6). n, ELISA analysis of IGF-1 and FGF-2 in conditioned media collected from human enteroid culture in OCTOPUS and Matrigel drops (n = 3). All data are presented as mean ± s.e.m. and P values are from unpaired, two-sided t-test.
Closer examination of the cultured constructs revealed distinct cell populations and their spatial localization within the developing organoids. Found at the tip of the buds corresponding to the crypt region were Ki67+ proliferative cells (Fig. 3h,i), and their presence was further verified by PCR with reverse transcription (RT–PCR). Enhanced epithelial differentiation in OCTOPUS from day 7 to day 14 was evidenced by increasing induction of KRT20 and reduced expression of Ki67 and Cyclin D1 (Fig. 3i,k and Supplementary Fig. 9). The organoids in OCTOPUS also contained absorptive enterocytes identified by the expression of KRT20 (Fig. 3j). In Matrigel drop culture, KRT20 expression was significantly lower and also decreased over time (Fig. 3j,k). We observed similar trends in the induction of a goblet-cell-specific marker (MUC2) (Fig. 3l,m).
Interestingly, enzyme-linked immunosorbent assay (ELISA) analysis showed progressively increasing levels of growth factors that promote the differentiation of the human intestinal epithelium, such as insulin-like growth factor 1 (IGF-1) and fibroblast growth factor 2 (FGF-2)24 (Fig. 3n). These factors were also present in Matrigel drop culture, but their concentrations were substantially lower and decreased with time during 14-d culture (Fig. 3n). Considering that IGF-1 and FGF-2 were not included in the media formulation, this result suggests that prolonged culture in OCTOPUS can increase endogenous production of differentiation factors to enhance epithelial differentiation of intestinal organoids.
It should be noted that both human and mouse intestinal organoids grown in OCTOPUS could still be passaged using conventional subculture techniques, making it possible to expand mature organoids prepared by our device (Extended Data Fig. 7). Compared with drop culture, passaging of OCTOPUS-generated intestinal organoids resulted in higher yields (Extended Data Fig. 7b,c). This difference may be explained by the fact that (1) expansion of intestinal organoids is achieved by passaging the crypt regions of the epithelium and (2) intestinal organoids in OCTOPUS form larger numbers of crypts/buds, which suggests that OCTOPUS may be used to increase the yield of organoid expansion to permit more abundant and efficient tissue production.
Single-cell profiling of human enteroids in OCTOPUS
We next performed single-cell RNA sequencing (scRNA-seq) to examine cellular heterogeneity of human enteroids and their single-cell transcriptional profiles. Uniform manifold approximation and projection (UMAP) clustering yielded three groups of cells in OCTOPUS organoids at day 7-absorptive cells, secretory cells and stem cells–each of which contained multiple subpopulations identified by cell-type-specific genes25,26 (Fig. 4a,b and Supplementary Figs. 10-12). For example, the absorptive cell group was composed of five cell types, one of which represented absorptive enterocytes defined by high expression of KRT20, fatty acid binding protein 1 (FABP1) and other canonical markers (Fig. 4b and Supplementary Fig. 10). At both days 7 and 14, the absorptive cell lineage in OCTOPUS showed substantially higher abundance than that in Matrigel drops (Fig. 4a,c,d). Importantly, the sequencing of OCTOPUS enteroids also revealed unique cell populations not present in drop culture that play a critical role in homeostatic function of the intestine, including BEST4+ enterocytes (Fig. 4a) and lipid-absorbing enterocytes (Supplementary Fig. 13)27.
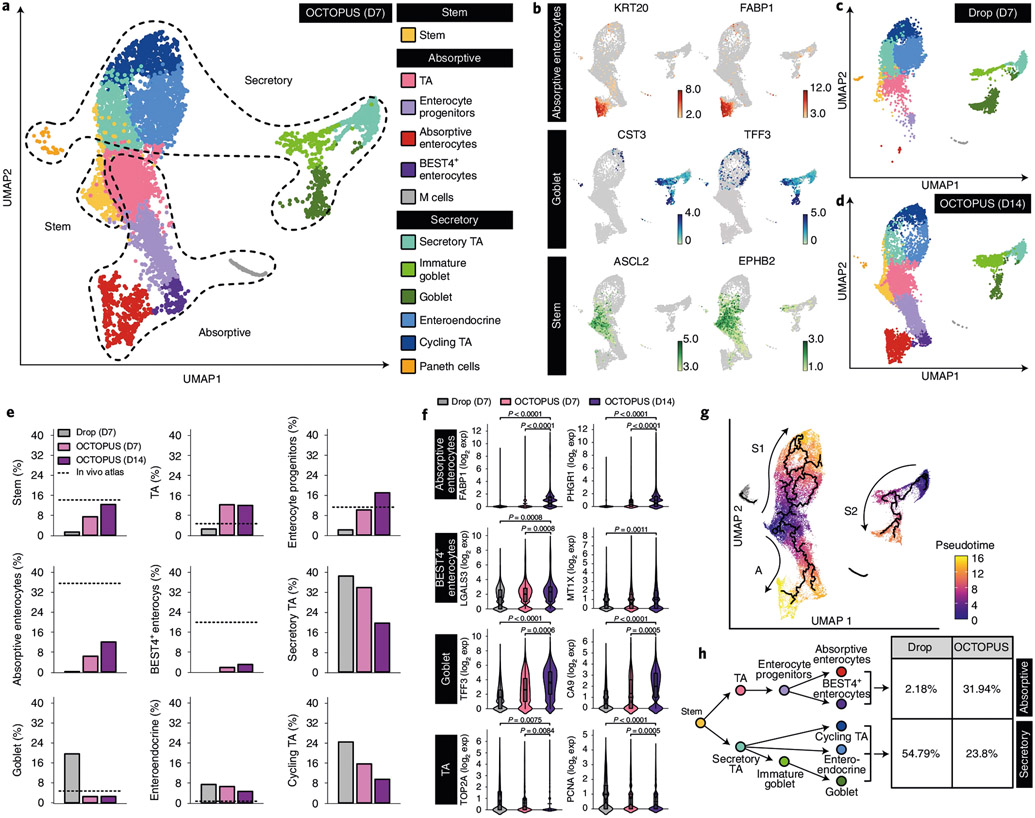
Fig. 4 ∣. scRNA-seq of human enteroids in OCTOPUS.
a, UMAP projection of 12 clusters representing distinct stem and intestinal epithelial cell populations in human enteroids produced by 7 d of culture in OCTOPUS. b, UMAP plots showing the expression of representative canonical genes specific to absorptive enterocytes, goblet cells and stem cells in log2 expression values. c,d, UMAP projection of cell clusters in human enteroids after 7-d culture in Matrigel drop (c) and 14 d of uninterrupted culture in OCTOPUS (d). e, Quantification of cellular compositions in human enteroids. Where available, the percentage of each cell type in the native human intestine was obtained from published in vivo atlases and shown with a dashed line. f, Violin plots comparing the expression of select cell-type-specific maturation markers between Matrigel drop culture and OCTOPUS. Violin plot elements showing unique molecular identifier (UMI) counts per cell represent the following values: center line, median; box limits, upper and lower quartiles; whiskers, 1.5-fold the interquartile range (n = 9,477 cells for Drop D7, 8,596 cells for OCTOPUS D7 and 1,1031 cells for OCTOPUS D14 examined over 3 independent experiments). g, Pseudotime trajectories (top) and branching plot (bottom) of intestinal stem cell differentiation into secretory and absorptive cell populations in human enteroids cultured in OCTOPUS for 14 d. h, Comparison of the fractions of differentiated epithelial cell types in OCTOPUS and Matrigel drop culture. All data are presented as mean ± s.e.m. and P values are from unpaired, two-sided t-test. exp, expression; M, microfold; TA, transit-amplifying.
Analysis of relative cellular abundance showed an increase in the proportion of stem cells from 7.4% (day 7) to 12.3% (day 14) in OCTOPUS, similar to the fraction of intestinal stem cells in vivo (14%) (Fig. 4e). Prolonged culture in this system also permitted expansion of enterocyte lineages, which was in contrast to the small or negligible fraction of these cells in Matrigel drops (Fig. 4e). The subpopulations of the secretory cell group in OCTOPUS displayed a general trend of decreasing abundance over time but their proportions at day 14 more closely approximated the cellular composition of the secretory epithelium in vivo25,26 than those evaluated in Matrigel drops (Fig. 4e). Our sequencing data also revealed increased expression of genes specific to mature absorptive enterocytes, BEST4+ enterocytes and the secretory cell populations (Fig. 4f and Supplementary Figs. 14-18) as a result of 14-d culture in OCTOPUS. Interestingly, in comparison with Matrigel drop culture, OCTOPUS enteroids showed significant downregulation of genes associated with the proliferative capacity of transit-amplifying cells (Fig. 4f and Supplementary Fig. 19), consistent with in vivo reports that cell proliferation in the transit-amplifying zone is suppressed with increasing tissue maturity during intestinal development28.
Single-cell trajectory analysis conducted to characterize the process of intestinal stem cell differentiation in OCTOPUS showed branching of the developmental trajectory into two distinct domains representing absorptive (A) and secretory cell (S1, S2) lineages (Fig. 4g). Quantification of cell types in each trajectory indicated that cell differentiation was directed more toward mature enterocytes to increase the abundance of enterocyte progenitors, absorptive enterocytes and BEST4+ enterocytes, which together constituted 31.94% of the epithelial population (Fig. 4h). In contrast, the development of stem cells was skewed heavily toward secretory cell lineages in Matrigel drops, accounting for 54.79% of the differentiated cells (Fig. 4h). Given the role of enterocytes as the most numerous cell type responsible for organ-specific function of the small intestine29, this comparison supports the advantage of OCTOPUS for engineering more physiological human enteroids.
OCTOPUS model of human IBD
Inspired by an emerging body of research highlighting the potential of organoids for the study of complex human diseases1-3, we set out to demonstrate the proof-of-principle of using OCTOPUS to construct organoid-based disease models with increased fidelity and physiological relevance. This study focused on modeling IBD using cells from two enteroid lines derived from pediatric patients with Crohn’s disease (Fig. 5a and Supplementary Table 1).
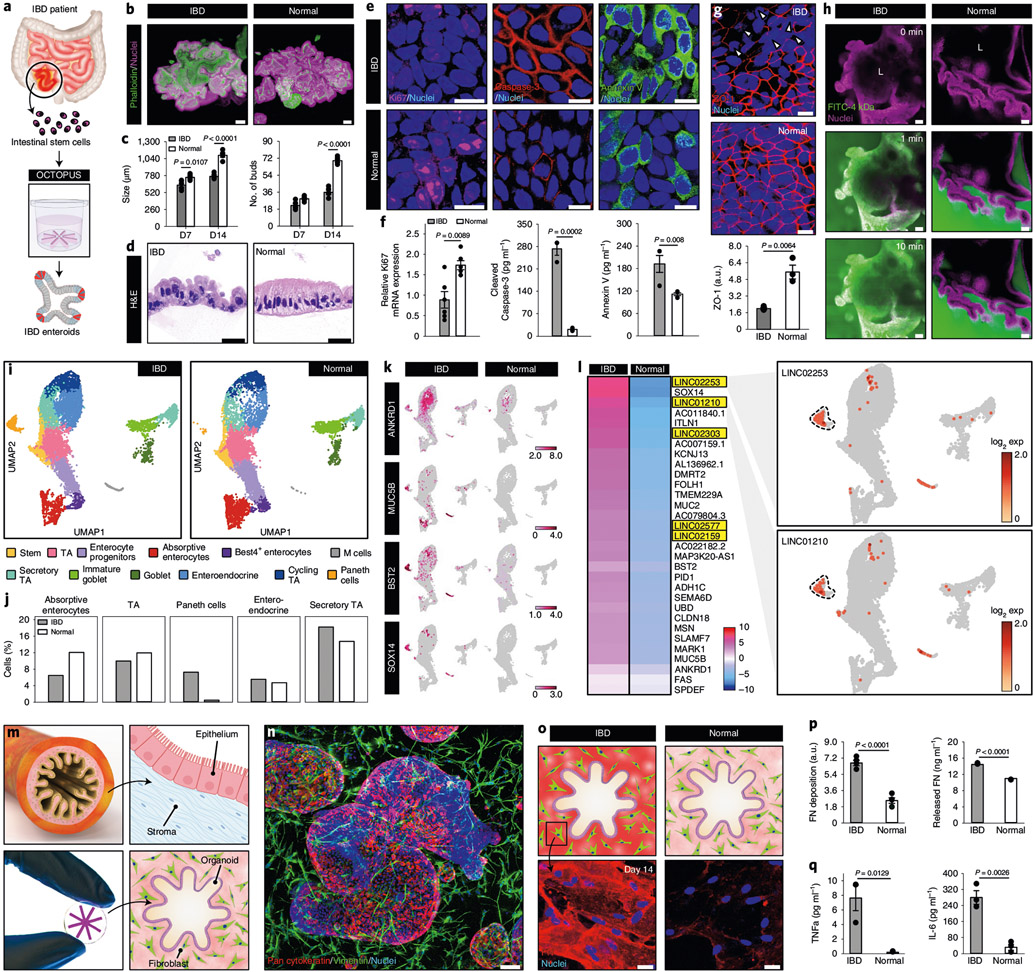
Fig. 5 ∣. Organoid-based model of human IBD in OCTOPUS.
a, Adult stem cells isolated from the intestine of patients with Crohn’s disease are used to form enteroids in OCTOPUS. b, Morphology of IBD and normal enteroids in OCTOPUS after 14-d culture visualized by immunofluorescence. Scale bars, 100 μm. c, Quantification of enteroid size and the number of buds at days 7 and 14 (n = 5). d, Histological sections of the intestinal epithelium after 14 d of culture. Scale bars, 5 μm. e,f, Immunofluorescence (e) and quantification (f) of cell proliferation (Ki67) (n = 6) and apoptosis (caspase-3 and annexin V) (n = 3) in IBD and normal enteroids. Scale bars, 10 μm. g, Confocal micrographs and quantification of ZO-1 expression in the villus domain of enteroids. Scale bars, 10 μm (n = 3). h, Visualization of 4-kDa dextran-FITC diffusion into the organoid lumen (L) to show epithelial permeability in the IBD enteroids. Scale bars, 50 μm. i,j, UMAP projection of distinct cell populations (i) and quantification of their proportions (j) in IBD and normal enteroids. k, Comparison of IBD-associated genes in log2 expression values. l, Heatmap showing the mean expression of transcription factors in IBD enteroids relative to that in normal enteroids. Upregulation of lncRNA genes in the IBD enteroids occurs mostly in Paneth cells shown with dashed lines in the UMAP plots. m, Coculture of human enteroids and primary human intestinal fibroblasts in OCTOPUS. n, Confocal micrograph of the coculture construct at day 14. Scale bar, 100 μm. o, Immunofluorescence micrographs of localized regions surrounding the enteroids after 14 d of culture. Scale bars, 25 μm. p, Quantification of FN production (n = 3). q, Quantification of cytokines (n = 3). All data are presented as mean ± s.e.m. and P values are from unpaired, two-sided t-test. ZO-1, Zonula occludens-1.
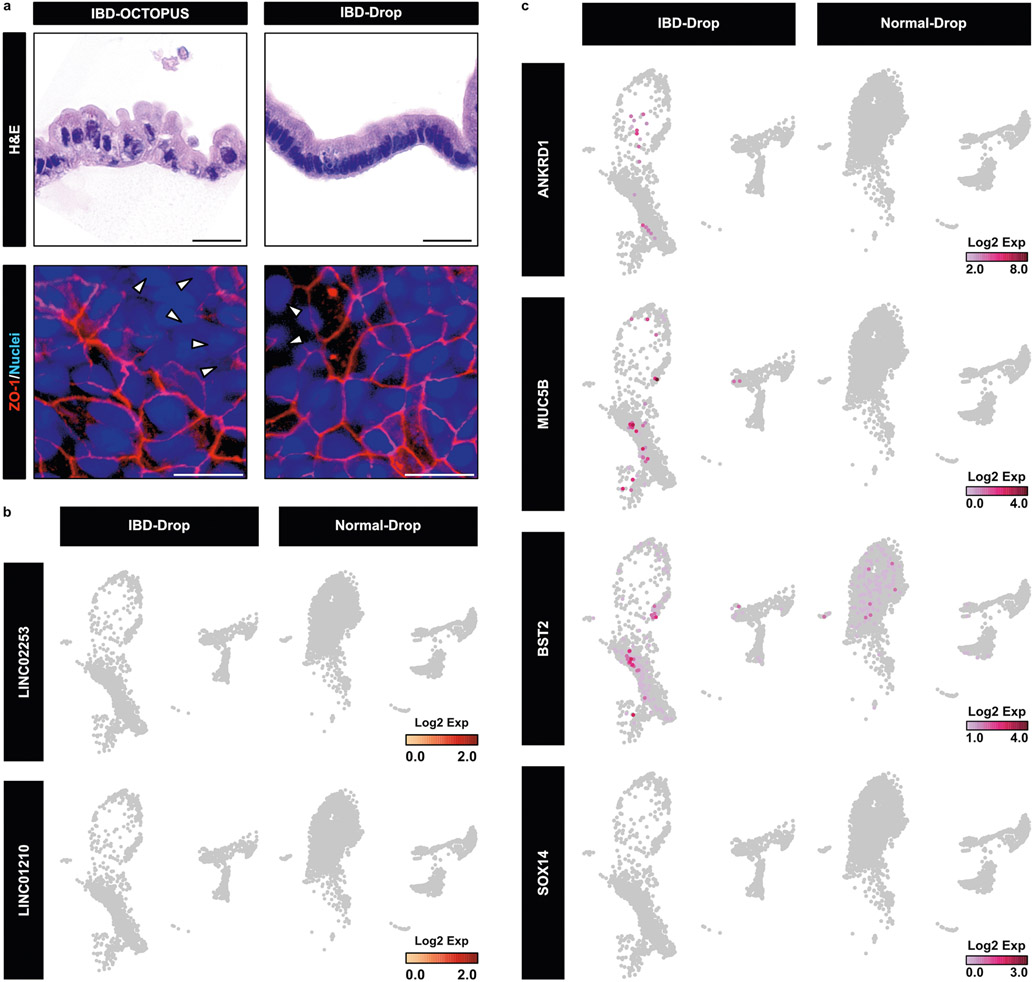
OCTOPUS allowed these cells to develop into organoids that appeared similar to those derived from healthy donors (Fig. 5b) but the IBD organoids grew more slowly and formed fewer buds (Fig. 5c), consistent with previous reports of intestinal growth deficiency and defective villus formation in Crohn’s disease30. Matching the histological abnormalities of the diseased intestinal epithelium in vivo31, the IBD organoids also contained enterocytes with enlarged, centrally located nuclei and a higher nuclear/cytoplasmic ratio (Fig. 5d). In addition to these morphological differences, the patient-derived enteroids showed reduced proliferative capacity, increased cell apoptosis and compromised integrity of the epithelial barrier in the villus domain (Fig. 5e-h and Supplementary Fig. 20). Interestingly, IBD organoids generated in Matrigel drops were unable to reproduce these disease phenotypes to the extent achieved in OCTOPUS (Extended Data Fig. 8a).
Further analysis of the IBD model using scRNA-seq showed reduced proportions of mature enterocytes and the other major subtypes of absorptive cell populations (Fig. 5i,j), which was in contrast to the expansion of secretory cell types. In particular, the Paneth cell population exhibited nearly 20-fold higher abundance in the IBD enteroids (Fig. 5i,j), similar to abnormal Paneth cell expansion known to occur in Crohn’s disease due to epithelial injury and inflammation32. At the transcriptomic level, our data revealed substantially increased expression of IBD-associated genes and the regulators of MAPK/ERK signaling pathways in the patient-derived enteroids (Fig. 5k and Supplementary Fig. 21). Importantly, we also discovered several long intergenic non-protein-coding (LINC) RNA genes upregulated in the Paneth cell and other secretory cell populations of these enteroids that have not been described in the context of IBD (Fig. 5l). This differential expression of the LINC and IBD-associated genes occurred to a significantly lesser extent in Matrigel drop culture of patient-derived enteroids (Extended Data Fig. 8b,c).
Having demonstrated the molecular and cellular features of OCTOPUS-generated IBD enteroids, we then explored their capacity to recapitulate intestinal abnormalities that develop at the tissue scale during the progression of IBD. Specifically, we aimed to model intestinal fibrosis, which is one of the complications of Crohn’s disease33. To this end, we cocultured patient-derived IBD enteroids with primary human intestinal fibroblasts to generate a multicellular construct reminiscent of the intestinal epithelium and the underlying stroma in vivo (Fig. 5m,n). When compared with the control group containing normal enteroids, the IBD constructs after 14 d of culture were seen with excessive fibronectin (FN) deposition in the hydrogel scaffold and significantly elevated levels of released FN in conditioned media (Fig. 5o,p), demonstrating spontaneous fibrotic tissue remodeling driven by the diseased epithelium. Similar differences were observed in interleukin-6 (IL-6) and tumor-necrosis factor-α (TNF-α), both of which have been shown to activate intestinal fibroblasts in Crohn’s disease-associated fibrosis34 (Fig. 5q).
Engineering vascularized, perfusable organoids in OCTOPUS
Integrating vasculature into organoid cultures is among the major goals of ongoing research efforts to advance organoid technology35. Vascularization of organoids is necessary for mimicking vascularity of native tissues and vascular contributions to parenchymal function but it also offers a promising strategy to improve nutrient and oxygen supply for enhanced organoid growth and maturation8.
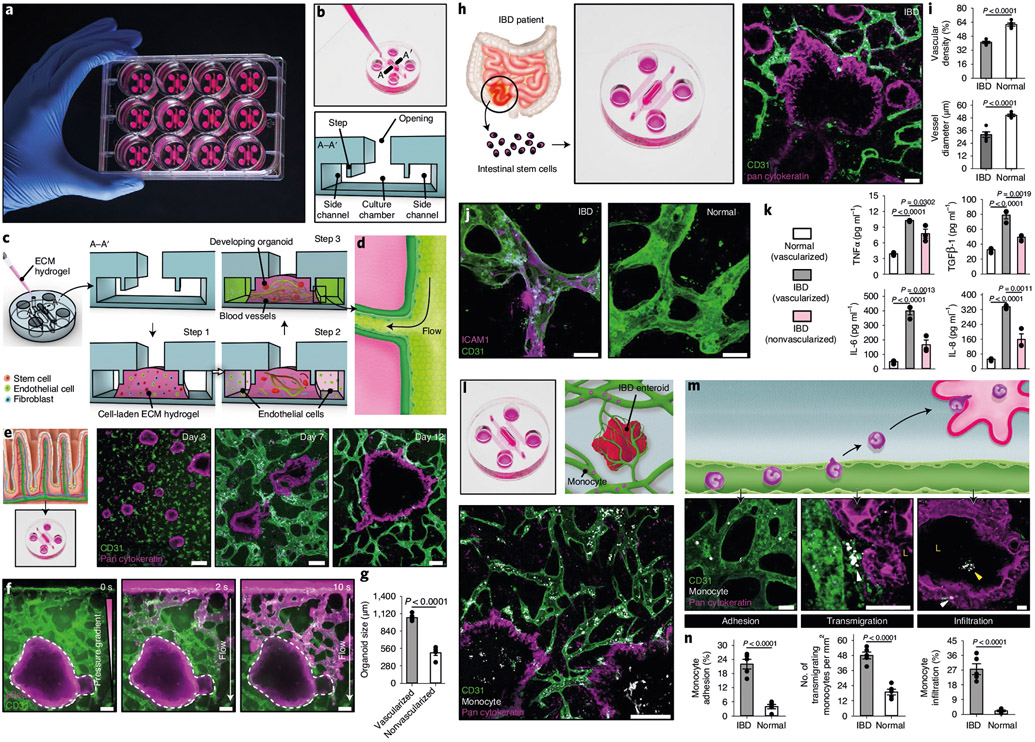
Motivated by this emerging trend, we created an advanced prototype of OCTOPUS, termed OCTOPUS-EVO (OCTOPUS for Engineering Vascularized Organoids), which provides capabilities to engineer vascularized, perfusable organoids while still offering the simplicity and convenience of the original platform (Fig. 6a). This device consists of an open cell culture chamber separated from two individually addressable side microchannels by a pair of microfabricated steps (Fig. 6b). To generate vascularized organoids, the culture chamber is injected with stem cells, endothelial cells and fibroblasts mixed in a hydrogel precursor solution to form a cell-laden ECM scaffold (Step 1, Fig. 6c). Subsequently, the side channels are seeded with endothelial cells to form a continuous endothelial lining (Step 2, Fig. 6c). During culture, stem cells in the hydrogel develop into organoids, while the endothelial cells embedded in the same gel self-assemble into a 3D network of interconnected blood vessels (Step 3, Fig. 6c). Over time, these vessels anastomose with the endothelium in the side channels, making the vascularized organoid construct directly accessible and perfusable from the side channels (Fig. 6d).
Fig. 6 ∣. Microengineering of vascularized human enteroids in OCTOPUS-EVO.
a, Photo of OCTOPUS-EVO devices in a standard 12-well cell culture plate. b, Device architecture of OCTOPUS-EVO. The device consists of an open cell culture chamber with cross-sectional dimensions of 3 mm (width) × 1 mm (thickness) flanked by two flow-through microchannels (1 × 1 mm2) on either side of the chamber. c,d, Sequential steps of microfluidic 3D culture (c) necessary for generating self-assembled and perfusable blood vessels (d) while supporting self-organization of stem cells into organoids in the same hydrogel scaffold. e, Micrographs demonstrating the concurrent development of human enteroids and microvasculature over the course of 12-d culture. Scale bars, 200 μm. f, Perfusability of the microengineered vascular network visualized by the flow of 1-μm fluorescent beads. Scale bars, 100 μm. g, Comparison of organoid size between vascularized and nonvascularized constructs (n = 5). h, Construction of vascularized, perfusable human IBD enteroids in OCTOPUS-EVO. Scale bars, 100 μm. i, Quantification of vascular density and vessel diameter (n = 5). j,k, Pro-inflammatory phenotype of the vascularized IBD model demonstrated by endothelial expression of ICAM-1 (j) and increased production of inflammatory mediators (k). Scale bars, 50 μm (n = 3). l, Micrograph of IBD enteroids perfused with peripheral blood monocytes. Scale bar, 200 μm. m,n, Confocal microscopy (m) and quantification (n) of sequential steps of monocyte recruitment to IBD enteroids. Scale bars, 50 μm (n = 5). All data are presented as mean ± s.e.m. and P values are from unpaired, two-sided t-test.
For proof-of-concept demonstration, we seeded OCTOPUS-EVO with a mixture of fibrin and Matrigel precursors containing human intestinal cells, endothelial cells and fibroblasts. This coculture system supported rapid self-organization of stem cells into enteroids and their continued growth, during which the self-assembled microvasculature emerged and remained stable and closely associated with the developing enteroids for prolonged periods (Fig. 6e). The vascularized organoid construct was perfusable by generating a pressure gradient across the hydrogel scaffold (Fig. 6f). Importantly, the vascularized, perfused human enteroids in OCTOPUS-EVO grew more than twice as large as nonvascularized ones in OCTOPUS cultured for the same duration (Fig. 6g).
Next, we explored whether our platform could be used to vascularize IBD organoids. A 12-d culture of patient-derived intestinal stem cells with endothelial cells and fibroblasts in OCTOPUS-EVO yielded IBD enteroids fully enveloped by the surrounding microvasculature (Fig. 6h). Interestingly, the blood vessels in this model showed reduced density and diameter compared with their counterparts formed around normal enteroids (Fig. 6i), matching vascular abnormalities of the chronically inflamed intestine in IBD36. Moreover, these vessels exhibited pro-inflammatory phenotype as illustrated by spontaneous endothelial expression of intercellular adhesion molecule-1 (ICAM-1) (Fig. 6j and Supplementary Fig. 22) and increased production of inflammatory cytokines (Fig. 6k). Of note, the levels of these cytokines in the vascularized enteroids were significantly higher than those in the nonvascularized IBD model (Fig. 6k).
When the vascularized construct was perfused with human peripheral blood monocytes (Fig. 6l), a large number of cells adhered to the enteroid-associated vessels and remained firmly attached during vascular perfusion (left panel, Fig. 6m). Some of these monocytes were observed to migrate across the endothelium into the perivascular space (middle panel, Fig. 6m). Also captured in this model were monocytes moving across the intestinal epithelium into the enteroid lumen (right panel, Fig. 6m). These complex events reproducing the sequential process of monocyte recruitment to the intestinal mucosa in IBD37 were observed in much fewer cells when monocytes were infused into vascularized normal enteroids (Fig. 6n).
Discussion
Here we presented a simple, ready-to-use microfabricated platform to reconfigure the three-dimensionality of conventional organoid models to engineer a more uniform, unrestricted soluble microenvironment beneficial for continuous long-term culture of organoids. OCTOPUS can increase organoid size and maturity beyond what is achievable using conventional techniques and may enable the production of more realistic multicellular constructs for in vitro modeling of organogenesis and disease development. Recent studies have demonstrated similar microengineering-based methods to control and manipulate the microenvironment of developing organoids and promote their maturation and biological complexity8,38, which has also led to commercial development of microfabricated organoid culture platforms39. We believe that OCTOPUS represents a promising approach that can add substantially to this emerging array of technologies. Although less sophisticated than many of the existing systems, our platform may have a lower entry barrier and reduce the perceived complexity and experimental burden of engineering organoid cultures, while still offering flexibility to accommodate more complex designs of organoid culture.
In addition to engineered culture systems, researchers have also shown that the composition of culture media can be modified to promote stem cell differentiation into more heterogenous and mature tissues, which has been successfully demonstrated in intestinal organoid cultures24,40. These organoids, however, still need to be passaged every 7 d or so because they become necrotic and terminally differentiated over time. This method can also be challenged by the experimental burden of optimizing complex, organoid-specific media formulations. OCTOPUS provides a different approach to resolving the issue of tissue immaturity by using a physical means to extend uninterrupted, continuous organoid culture. As shown by immunofluorescence and scRNA-seq analysis, OCTOPUS culture can accelerate the development of intestinal organoids and sustain their long-term growth. Our data also suggest that prolonged culture in this system leads to increased production of endogenous factors that can promote epithelial differentiation and maturation. It is not difficult to envision, however, that combing the two approaches may further increase the maturity and biological complexity of organoids beyond what has been demonstrated in this paper.
Our work also demonstrated a specialized organoid model to recapitulate the key features of the human intestinal epithelium in IBD. An important discovery from this study is increased expression of several long noncoding RNAs (lncRNAs) that have not been implicated in IBD. Supporting an emerging body of work demonstrating active involvement of lncRNAs in mediating key disease processes of IBD41-43, this finding warrants further investigation of the specific role of these genes in IBD, which may lead to identification of new therapeutic targets. For more complete investigation of IBD using our system, however, work remains to be done to capture other important aspects of the disease, such as the contribution of the immune system involving T cell activation and expansion44. Given the well-known variability of patient-derived organoids, the limited number of organoid lines used in this study is another limitation of the IBD model that needs to be addressed in future studies. As demonstrated in recent work by Clevers and colleagues45, efforts to overcome this limitation will greatly benefit from ongoing efforts to generate a large collection of patient-derived organoids and matching healthy organoids that can be biobanked.
The demonstration of intestinal organoid vascularization using OCTOPUS-EVO is another important outcome of this work. Researchers have demonstrated organoid vascularization through in vivo transplantation into vascular-rich organs such as the brain46, kidney8, lung47 and pancreas48, but generating such constructs with controlled vascular perfusion in vitro remains a major challenge. OCTOPUS-EVO provides an accessible means to tackle this challenge and increase the complexity of organoid models. Interestingly, vascularized enteroids in our device had significantly larger size compared with nonvascularized ones, supporting the notion that organoid vascularization is a promising strategy to facilitate organoid growth35. However, the mechanism underlying this difference remains to be elucidated. Presumably, vascularization increases nutrient and oxygen supply to permit more efficient and rapid organoid development. Based on a large body of evidence demonstrating endothelial interactions with parenchymal tissues49, it is also possible that biological crosstalk between the vasculature and organoids may be responsible for increased organoid growth.
Despite advantageous capabilities of OCTOPUS, we note that emulating the maturity of native organs in organoid models will require advanced approaches beyond the enhancement of nutrient supply and cell viability demonstrated here. Our study is based on the rationale that the short lifespan of organoids is the primary reason for their limited ability to reach later stages of development and acquire mature phenotype. From a developmental biology perspective, however, there is increasing recognition that the limited maturity of organoids in conventional models may also be due to the absence of the surrounding embryonic tissues of developing organs in vivo that provide instructive cues to guide the process of organ development and maturation2,50. Recapitulating this critical aspect of in vivo organogenesis may greatly enhance the ability of OCTOPUS to promote structural and functional maturation of organoids. The results from our coculture and vascularized organoid models indeed demonstrate the feasibility and potential benefit of this approach. Although preliminary, these findings call for further investigation of whether mimicking the collection of specialized tissues in native organs and their biological interactions can be leveraged as a complementary strategy for developing more mature and realistic organoid constructs in OCTOPUS.
To conclude, our work provides a good example of how rational design engineering of conventional organoid culture can advance the capacity of organoids to emulate the structural and functional complexity of their in vivo counterparts. We believe that OCTOPUS has the potential for substantial impact on organoid technology and may also provide a simple yet practical and powerful platform for various other applications that involve cell and tissue culture in 3D environments.
Methods
Device fabrication
We created OCTOPUS inserts (Supplementary Fig. 1) by casting poly(dimethylsiloxane) (PDMS) prepolymer against micropatterned 3D printed molds using standard soft lithography techniques. For device fabrication, PDMS (Sylgard 184, Dow Corning) monomer base was mixed with curing agent (10:1, w/w) and poured onto 3D printed molds (Protolabs). The casted molds were vacuum degassed in a desiccation chamber for 30 min and oven cured overnight at 65 °C to produce devices described in Fig. 1. The cured PDMS was removed from the molds and then sealed against a thin slab of cured PDMS (spin-coated onto a flat wafer at 1,500 r.p.m. for 5 min) and placed in a 24-well plate until use.
Mouse organoid culture
For organoid cultures, we used cryopreserved mouse intestinal organoids (70931, STEMCELL Technologies) and cryopreserved mouse hepatic progenitor organoids (70932, STEMCELL Technologies) derived from the small intestine and the liver of C57BL/6 mice, respectively. Intestinal and liver organoids were cultured in 24-well plates according to the manufacturer’s protocols using IntestiCult organoid growth medium (06005, STEMCELL Technologies) and Hepaticult organoid growth medium (06030, STEMCELL Technologies), respectively. To briefly explain, we injected 50 μl of the organoid/Matrigel mixture (1:1, v/v) into 24-well plates to form Matrigel drops, which were then incubated at 37°C and 5% CO2 for 10 min to allow gelation of Matrigel. Upon completion of this step, 750 μl of pre-warmed organoid growth medium was added to each well. Organoids were passaged every 5–7 d in fresh Matrigel until use as recommended by the manufacturer.
Generation of human enteroid lines
Enteroid lines generated from terminal ileum mucosal biopsies were provided by the Children’s Hospital of Philadelphia Gastrointestinal Epithelium Modeling Program under an Institutional Review Board-approved protocol (13042). All parents of patients provided written, informed consent. Patient demographics and enteroid passage details are listed in Supplementary Table 1. Briefly, two biopsy tissue fragments were rinsed three times in 1 ml of cold sterile PBS, then incubated in cold chelation buffer for 30 min on a turntable in a cold room, followed by mechanical dissociation (scraping) of epithelial layer. The fragments were strained through a 100-μm strainer to deplete the villi and resuspended in 80% Matrigel, then seeded at the density of 50–200 crypts per 30-μl drop. The droplets solidified at 37 °C for 30 min, and 500 μl of human IntestiCult (STEMCELL Technologies; complete when supplemented with Penicillin-Streptomycin (Gibco)) was added per well. Y-27632 (SelleckChem; final concentration 10 μM) was added to culture medium at seeding only.
Maintenance and passage of human enteroids
Enteroid medium was changed three times per week. To passage, the Matrigel droplet was dislodged by pipetting up and down through a P1000 tip and transferred into a 1.5-ml microfuge tube, followed by centrifugation and washing with ice-cold HBSS. Enteroids were mechanically dissociated into fragments by pipetting ten times through a P200 tip placed on top of a P1000 tip, followed by centrifugation. The pellet was reconstituted in 80% Matrigel and seeded as 30-μl drops at a split ratio of 1:4. Subsequent cultures are ready for passage and/or cryo-preservation on day 7.
Cell culture
For coculture demonstrations presented in Extended Data Fig. 5, we used primary mouse intestinal fibroblasts. For initial expansion from cryostorage, mouse intestinal fibroblasts were cultured in 75-cm2 flasks according to the manufacturer’s protocols using complete fibroblast medium (M2267, Cell Biologics) supplemented with growth factors. For modeling intestinal fibrosis (Fig. 5), we used primary human intestinal fibroblasts. In all experiments, we used cells between passages 3 and 6.
Formation of 3D organoid constructs in OCTOPUS
Before cell culture, the standard 24-well plates containing OCTOPUS inserts were sterilized by exposure to ultraviolet light (Electro-lite ELC-500) for 30 min. To form mouse intestinal organoids in our device, we first dissolved previously existing Matrigel by incubating Matrigel drops in Dispase. Cells were then transferred to a 15-ml falcon tube and centrifuged at 290g to obtain a stem cell pellet. We then added 100 μl of complete IntestiCult organoid growth medium to the pellet. After 100 μl of cold Matrigel was added, the suspension was gently pipetted up and down ten times for thorough mixing. For human intestinal enteroids, the cell pellet was resuspended in 80% Matrigel. Using a pre-wetted 200-μl tip, we injected 100 μl of the organoid/Matrigel mixture into OCTOPUS through the injection port. The OCTOPUS-containing well plates were then incubated at 37 °C and 5% CO2 for 10 min to allow gelation of Matrigel. Upon completion of this step, 750 μl of pre-warmed IntestiCult organoid growth medium was added to each well. The OCTOPUS plates were maintained in cell culture incubators at 37 °C and 5% CO2. During long-term culture, media exchange was conducted every other day.
Subculture of organoids in OCTOPUS
For both mouse and human intestinal organoid cultures in OCTOPUS, media were changed every other day. On day 7, organoids were washed with PBS and incubated with dispase for 30 min as shown in Extended Data Fig. 7 to dissolve Matrigel. The dissociated cells were then transferred to a 15-ml falcon tube and centrifuged at 290g. The pellet was reconstituted in Matrigel and seeded into new devices or wells at 200 crypts per 100 μl and 100 crypts per 60 μl for mouse and human intestinal organoids, respectively. For device seeding, we used a pre-wetted 200-μl tip to inject 100 μl of the crypt/Matrigel mixture into OCTOPUS through the loading port. The OCTOPUS-containing well plates were then incubated at 37 °C and 5% CO2 for 10 min to allow gelation of Matrigel. Upon completion of this step, 750 μl of pre-warmed IntestiCult organoid growth medium was added to each well. The OCTOPUS plates were maintained in cell culture incubators at 37 °C and 5% CO2. During long-term culture, media exchange was conducted every other day.
Assessment of organoid viability
To examine the viability of organoids in Figs. 1, 2 and 3, we used Live/Dead Viability/Cytotoxicity Kit, for mammalian cells (L3224, ThermoFisher Scientific). For this assay, a mixture of calcein AM (2 μM) and ethidium homodimer-1 (4 μM) in live cell imaging solution (LCIS) was introduced into the OCTOPUS-containing wells and incubated at room temperature for 30 min. Subsequently, the samples were washed with PBS three times, after which the labeled cells were examined using a laser scanning confocal microscope (LSM 800, Carl Zeiss). For quantitative analysis, we calculated the fractions of healthy and necrotic organoids from fluorescence generated by calcein AM and ethidium homodimer-1, respectively. In each device, 30 organoids were used for our analysis.
Evaluation of FITC-dextran diffusion
To examine the spatiotemporal patterns of diffusion in OCTOPUS and hydrogel drops (Extended Data Fig. 2a-c and Fig. 5), we used 4-kDa FITC–dextran (FD70S-100MG, Sigma) as a fluorescent tracer for visualization. For this assay, the organoid culture medium was replaced with a FITC-dextran solution (50 μg ml−1 in PBS). Dextran diffusion was monitored and visualized using a laser scanning confocal microscope (LSM 800, Carl Zeiss). Time-lapse images were acquired for 120 min and processed using ZEN software (Zeiss) to measure temporal changes in fluorescence intensity at defined locations within the hydrogel scaffolds.
Evaluation of oxygen diffusion
To examine oxygen diffusion in OCTOPUS and Matrigel drops (Extended Data Fig. 2d-f and Supplementary Fig. 7), we used Dichloritis(1,10-phenanthroline)ruthenium(II) hydrate (Ru(phen)3) (Sigma, Cat. no. 343714) as an oxygen indicator–dissolved oxygen molecules induce quenching of this fluorescent dye. Briefly, 15 μl of Ru(phen)3 (2 mM) and 270 μl of Matrigel were mixed with 15 μl of sodium sulfite (200 mM) (Sigma, Cat. no. S0505), which was used as an oxygen scavenging reagent to remove remaining aqueous oxygen in Matrigel (Supplementary Fig. 7). The mixture of Matrigel and sodium sulfite was then used to generate 3D tissue constructs in OCTOPUS and Matrigel drops. Theoxygen diffusion assay described in Extended Data Fig. 2d-f was performed based on the principle that when water is added, sulfite ions are released to water while dissolved oxygen diffuses into the hydrogel (Supplementary Fig. 7). Oxygen diffusion was monitored by confocal microscopy and images were acquired at defined time intervals and locations within the constructs. The captured images were analyzed using ImageJ to measure spatiotemporal changes in fluorescence intensity.
ELISA assays
To analyze albumin and urea production by liver organoids (Extended Data Fig. 3d,e), cell culture media in the wells were collected on days 5 and 10 of culture. We used Mouse Albumin ELISA Kit (abl08792, abcam) and Urea Assay Kit (ab83362, abcam) to measure the concentrations of albumin and urea in the collected media samples, respectively, and each assay was performed following the manufacturer’s protocol.
To analyze GLP-1 and mucin 2 secretion from the intestinal organoids (Extended Data Fig. 6e), the media in the wells were collected on days 5, 7 and 10 of culture. We used Multi-Species GLP-1 Total ELISA Kit (EZGLP1T-36K, Millipore Sigma), Glucagon Like Peptide-1 (active) ELISA Kit (EGLP-35K, Millipore Sigma) and MUC2 ELISA Kit (ABIN6730976, antibodies-online) to measure the concentrations of GLP-1 total, GLP-1 active and mucin 2, respectively, following the manufacturer’s protocol. For the analysis of FN production in the IBD model (Fig. 5), we used Human Fibronectin ELISA Kit (ab108849, abcam). The media in the wells were collected at specified time points in Fig. 5 and assayed using manufacturer-provided protocols.
To analyze the production of IGF-1 and FGF-2 by human enteroids (Fig. 3n), culture media in the wells were collected on days 0, 4, 7, 10 and 14. We used Human IGF1 SimpleStep ELISA Kit (ab211651, abcam) and Human FGF2 SimpleStep ELISA Kit (ab246531, abcam) to measure the concentrations of IGF-1 and FGF-2, respectively. Each assay was performed following the manufacturer’s protocol.
For measurement of caspase-3, annexin V, TNFα, TGFβ-1, IL-6 and IL-8 in the human enteroids shown in Figs. 5 and 6, conditioned media were collected on day 14 of culture and analyzed using Cleaved Caspase-3 (Asp175) ELISA Kit (ab220655, abcam), Human Annexin V ELISA Kit (ab223863, abcam), Human TNF Alpha ELISA Kit (ab181421, abcam), Human TGF Beta 1 ELISA Kit (ab100647, abcam), Human IL-6 ELISA Kit (ab178013, abcam) and Human IL-8 ELISA Kit (ab214030, abcam). Each assay was performed following the manufacturer’s protocol.
For all assays, we used a multimode plate reader (M200, Tecan, Switzerland) to measure theoptical density of samples. We generated a standard curve by plotting the mean optical density and concentration for each standard using a four-parameter logistic curve fitting method. Sample measurements were converted to target concentrations using the standard curves. All data were normalized by the total number of cells.
Proliferation assay
To detect proliferating cells within the intestinal organoids presented in Fig. 2, we used EdU Assay/EdU Staining Proliferation Kit iFluor 647 (ab222421, abcam). Briefly, the organoids were incubated with an EdU solution (20 μM in medium) for 3 h under normal culture conditions (5% CO2 at 37 °C). The organoids were then washed twice with PBS, fixed in 4% formaldehyde and permeabilized using a permeabilization buffer, according to the manufacturer’s protocol. Finally, the samples were stained with iFluor 647 azide dye and visualized using a confocal microscope (LSM 800, Carl Zeiss).
Calcium imaging
Before calcium imaging presented in Fig. 2, organoids in OCTOPUS and Matrigel drops were harvested using the protocol for passaging described above and embedded in a thin Matrigel layer with a thickness of 1 mm. After 24 h, media were removed from the culture wells, and the organoid constructs were washed once in LCIS. To examine cellular responses to glucose treatment, organoids were incubated in glucose-free DMEM medium for 1 h before assay. The organoids were then loaded with Fluo-4 calcium imaging solution (F10489, ThermoFisher Scientific) prepared according to the manufacturer’s protocol. The samples were incubated at 37 °C for 30 min, which was followed by another 30-min incubation at room temperature. Subsequently, Fluo-4 solution was removed, and the organoids were washed once with LCIS. All samples were kept in fresh LCIS until use. We used an inverted epi-fluorescence microscope (Axio Observer D1, Zeiss) to visualize calcium staining of organoids upon stimulation with 100 μM ATP (A1852, Sigma) and 50 mM glucose (G7021, Sigma) as described in Fig. 2.
For ratiometric analysis of changes in Ca2+ levels, fluorescence intensity (F) for each organoid was measured during an experiment and values were normalized by their resting intensities using the equation below.
H&E staining of human enteroid sections
For hematoxylin and eosin (H&E) staining of human enteroids, organoids were washed with cold PBS and fixed with 4% paraformaldehyde (Electron Microscopy Sciences). The organoids were then resuspended in embedding gel composed of 2% bacto-agar and 2.5% gelatin and transferred as a droplet onto the embedding rack. After the gel was solidified for 30 min, the organoid-embedded gel was placed in the pre-labeled tissue cassette and submerged in 70% ethanol. The slides containing paraffin sections were deparaffinized and rehydrated by immersing the slides sequentially into 3× xylene, 2× 100% ethanol, 95–95–80–70% ethanol and distilled water. Then, the slides were immersed in 10 mM citric acid buffer (pH 6.0) and incubated in a microwave oven for 15 min. After gently rinsing the slides, tissue sections were blocked with protein blocking agent. To perform H&E staining, the slides were immersed in hematoxylin followed by rinsing with deionized water. The slides were further immersed in eosin for 30 s and dehydrated in 95% ethanol–100% ethanol–xylene solutions. Tissue sections were covered with coverslip slides using Permount and stored until analysis.
Quantitative RT–PCR analysis
For RNA isolation, organoids were harvested by dissolving Matrigel including organoids with cold PBS. Following centrifugation at 300g for 5 min at 4 °C, the supernatant was removed and the pelleted organoids were resuspended in 350 μl of RLT buffer (QIAGEN). Total RNA was isolated using the RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions. Oligonucleotides to detect messenger RNA levels of human Cyclin D1 (fw-TCTACACCGACAACTCCATCCG; rv-TCTGGCATTTTGGAGAGGAAGTG), mouse Ki67 (fw-GAGGAGAAACGCCAACCAAGAG; rv-TTTGTCCTCGGTGGCGTTATCC), human Ki67 (fw-GAAAGAGTGGCAACCTGCCTTC; rv-GCACCAAGTTTTACTACATCTGCC), human KRT20 (fw-CTGAGGTTCAACTAACGGAGCTG; rv-AACAGCGACTGGAGGTTGGCTA) and human MUC2 (fw-GCCAGCTCATCAAGGACAG; rv-GCAGGCATCGTAGTAGTGCTG) were purchased from Integrated DNA Technologies. Complementary DNA was synthesized using iScript cDNA Synthesis Kit (Bio-Rad) following the manufacturer’s instructions. Quantitative RT–PCR was performed using TaqMan gene expression assays.
scRNA-seq and data processing
For single-cell sequencing analysis, collected organoids were incubated in trypsin for 10 min at 37°C and passed through a 20-μm cell strainer. Isolated single cells were resuspended at a density of 700 live cells per μl in DMEM with 5% FBS. The cells were then stained with trypan blue to check their viability and counted under the microscope twice to determine the average cell concentration. Sequencing data from 14-d Matrigel drop culture were excluded in our analysis to avoid confounding factors due to significant cell death observed in this group.
A single-cell suspension for each organoid sample was loaded onto a separate channel of a Chromium 10X Genomics Single Cell 3′ Reagent Kit v2 library chip (10X Genomics) according to the manufacturer’s protocol. RNA transcripts from single cells were uniquely barcoded and reverse-transcribed. cDNA sequencing libraries were prepared according to the manufacturer’s protocol (10X user guide for library prep) and sequenced on an Illumina NovaSeq 6000 using an S1100-cycle flow cell v1.5. Library quality control was done using Agilent TapeStation for sizing (bp) and KAPA quantitative PCR for concentration (nM).
Raw sequence reads data were processed using the CellRanger pipeline (10X Genomics, v.5.0.0) for demultiplexing and aligned to the human genome GRCh38 transcriptome. Sample data were aggregated using the CellRanger aggr pipeline and libraries were normalized for sequencing depth across the sample set. A total of five organoid sample count matrices were merged together for cell type identification and direct comparisons. For initial filtering, cells with a smaller number (<200) of uniquely detected genes were excluded. Then, cells with a proportion of unique molecular identifiers attributable to mitochondrial genes >25% were excluded from the analysis. Finally, highly variable genes were used for dimension reduction, and the results were visualized via UMAP. The numbers of cells captured per sample are listed in Supplementary Table 2.
Generation of human enteroids lines for modeling IBD
Intestinal stem cells isolated from the terminal ileum mucosal biopsies of patients with Crohn’s disease were provided by the Children’s Hospital of Philadelphia Gastrointestinal Epithelium Modeling Program under an Institutional Review Board-approved protocol (13042). All parents of patients provided written, informed consent–detailed information about patient demographics and enteroid passage numbers are provided in Supplementary Table 1. These cells were used to generate diseased enteroid lines used to model human IBD in OCTOPUS and Matrigel drops following the protocol described above.
Coculture of human enteroids with fibroblasts
To model intestinal fibrosis as a complication of IBD, human intestinal stem cells were mixed with 1 × 106 cells per ml of primary human intestinal fibroblasts in Matrigel (356255, Corning). This cell-containing hydrogel solution was injected into the device to form microtissue constructs in the organoid culture chambers. After gelation for 15 min in a regular cell culture incubator, 750 μl of IntestiCult organoid growth medium (06010, STEMCELL Technologies) was added into each well and maintained for 14 d to induce intestinal organoid development and fibroblast proliferation. During this period, the media were replenished every other day.
Formation of the vascularized human enteroids in OCTOPUS-EVO
Before cell culture, the fully assembled device was first sterilized by exposing it to ultraviolet light (Electro-lite ELC-500) for at least 30 min. To engineer vascularized organoids in OCTOPUS-EVO, we prepared 20 μl of cell suspension solution containing fibrinogen (5 mg ml−1; F8630, Sigma), thrombin (1 U ml−1; T7513, Sigma), aprotinin (0.15 U ml−1; A1153, Sigma), human intestinal stem cells, primary human umbilical vein endothelial cells (HUVECs) (5 × 106 cells per ml) and primary normal human lung fibroblasts (1 × 106 cells per ml), and injected it into the open cell culture chamber through its inlet access port. The device was then left in a cell culture incubator at 37 °C and 5% CO2 for 30 min. Upon gelation, IntestiCult media mixed with EGM-2 endothelial media were added to the medium reservoirs and the side microchannels.
Following the formation of a cell-laden hydrogel construct, the side microchannels were incubated with an FN solution (25 μg ml−1 in PBS; 356008, Corning) for 2 h at 37 °C to create an ECM coating on the channel surface. Then, the channels were washed once with IntestiCult/EGM-2, and 10 μl of HUVEC suspension (1 × 107 cells per ml) was introduced into both channels. The seeded cells were allowed to attach to the channel surface over a period of 1 h. Upon 1 h of incubation, pre-warmed medium was added to each medium reservoir.
Validation of vascular perfusability
To test the perfusability of the microengineered vascular network, we used fluorescently labeled 1-μm microbeads (FluoSpheres; F-8815, ThermoFisher) as flow tracers. To generate flow through the vasculature, medium in the reservoirs was aspirated and a bead solution was inserted into one of the side microchannels. This configuration created a gradient of hydrostatic pressures across the hydrogel scaffold and provided driving force for the flow of microbeads through the vessels. Vascular perfusion was monitored and visualized using a laser scanning confocal microscope (LSM 800, Carl Zeiss).
Monocyte infiltration assay
Human peripheral blood monocytes were obtained from the Human Immunology Core at the University of Pennsylvania. To test endothelial adhesion of monocytes in our model, we labeled the cells with a fluorescent dye (CellTracker Deep Red, ThermoFisher) and suspended them in IntestiCult/EGM-2 medium at the final concentration of 3 × 106 cells per ml. The cells were then injected into the vessels through one of the side microchannels and allowed to flow through the vasculature for 24 h in a cell culture incubator. At the completion of perfusion, the device was washed with DPBS three times and examined to analyze the numbers of adhered, transmigrated and infiltrated monocytes in our model.
Immunofluorescence analysis
For immunofluorescence staining, cells in OCTOPUS were washed with PBS twice, fixed with 4% paraformaldehyde (Electron Microscopy Sciences) for 15 min at room temperature and washed again twice with PBS. The cells were then permeabilized with 0.1% Triton X-100 (Sigma) in PBS for 3 min and exposed to blocking buffer composed of PBS and 3% BSA (Sigma) overnight at 4 °C. After washing with PBS twice, the cells were immunostained for actin filaments (Phalloidin-iFluor 488 Reagent, ab176753, 1:1,000, abcam; Phalloidin-iFluor 594 Reagent, ab176757, 1:1,000, abcam), mature epithelial cells (anti-EPCAM antibody, ab71916, 1:250, abcam; anti-HNF-4-alpha antibody [K9218]-ChIP Grade, ab41898, 1:500, abcam), stem cells (anti-Ki67 antibody, ab15580, 1:1,000, abcam; Lgr5 monoclonal antibody, MA5-25644, 1:1,000, ThermoFisher Scientific), enterocytes (anti-Villin antibody [3E5G11]-N-terminal, ab201989, 1:500, abcam), goblet cells (anti-MUC2 antibody, ab90007, 1:200, abcam), enteroendocrine cells (anti-Somatostatin antibody [M09204], ab30788, 1:100, abcam), peptide transporter 1 (anti-SLC15A1/PEPT1 antibody, ab203043, 1:100, abcam), glucose transporter 1 (anti-Glucose transporter GLUT1 antibody [SPM498], ab40084, 1:250, abcam), endothelial cells (anti-CD31 antibody [JC/70A] (Alexa Fluor 488), ab215911, 1:100, abcam), alpha smooth muscle actin (recombinant anti-alpha smooth muscle Actin antibody [E184], ab32575, 1:500, abcam), FN (anti-Fibronectin antibody [IST-9], ab6328, 1:200, abcam), cleaved caspase-3 (anti-cleaved caspase-3 antibody [E83-77], ab32042, 1:200, abcam), annexin V (anti-annexin V/ANXA5 antibody [EPR3980], ab108194, 1:500, abcam) or ICAM-1 (anti-ICAM1 antibody [EPR24639-3], ab282575, 1:500, abcam). After overnight incubation with primary antibody at 4 °C, the cells were washed twice with PBS and incubated with secondary antibody (Goat anti-Rabbit IgG H&L (Alexa Fluor 488), ab150077, 1:1,000, abcam; Goat anti-Mouse IgG H&L (Alexa Fluor 488), ab150113, 1:1,000, abcam; Goat anti-Mouse IgG H&L (Alexa Fluor 594), ab150116, 1:1,000, abcam; Goat anti-Rabbit IgG H&L (Alexa Fluor 594), ab150080, 1:1,000, abcam) overnight at 4 °C. For nuclear staining, we used DAPI (D1306, ThermoFisher Scientific) diluted at 1:1,000. Fluorescence images of the stained cells were acquired using a laser scanning confocal microscope (LSM 800, Carl Zeiss) and processed using ZEN software (Zeiss) and ImageJ software. Immunofluorescence data were presented on a per-cell basis by normalizing the mean fluorescence intensity by the total cell number, which was obtained by counting the number of DAPI-stained cell nuclei.
Statistical analysis and reproducibility
Sample size for each experiment was determined on the basis of a minimum of n = 3 independent devices for each experimental group. Data were analyzed with Student’s t-test using Origin PRO (OriginLab). All data are presented as mean ± s.e.m. and P values are from unpaired, two-sided t-test.
Extended Data
Extended Data Fig. 1 ∣. Co-culture of organoids in OCTOPUS.
Co-culture of mouse liver and intestinal organoids in OCTOPUS. The OCTOPUS insert shown in this example contains two juxtaposed spiral chambers with independent access ports. Scale bars, 300 μm (left image) and 100 μm (right images).
Extended Data Fig. 2 ∣. Diffusion of soluble factors in OCTOPUS.
a, b. Visualization of 70 kDa FITC-dextran diffusion into the inner and outer regions of the hydrogel scaffolds in Matrigel drop (a) and OCTOPUS (b). The organoids in the inner and outer regions were located 600 μm (OCTOPUS)/2400 μm (Matrigel drop) and 400 μm (both groups) from the hydrogel surface, respectively. Scale bars, 100 μm. c. Temporal profiles of mean fluorescence intensity (MFI) due to dextran diffusion. d, e. Visualization of temporal changes in oxygen concentration in the inner and outer regions of Matrigel scaffolds in conventional drop culture (d) and OCTOPUS (e). Quenching of blue fluorescence shown in the micrographs was caused by an increase in the level of oxygen. f. Temporal profiles of normalized fluorescence intensity due to oxygen diffusion into Matrigel (n=3 independent experiments).
Extended Data Fig. 3 ∣. Culture of mouse liver organoids in OCTOPUS.
a, b. Formation and extended culture of mouse liver organoids in OCTOPUS and conventional drop culture. Scale bars, 100 μm (n=5 biologically independent experiments). c. Confocal micro-graphs showing the expression of albumin (ALB) in mouse liver organoids. Scale bars, 10 μm. ELISA analysis of (d) albumin and (e) urea in conditioned media (n=3 biologically independent experiments). All data are presented as mean ± SEM and P values are from unpaired, two-sided t test.
Extended Data Fig. 4 ∣. Maturation of intestinal organoids in OCTOPUS.
a. Comparison of bud length. Confocal micrographs show the cross-section of buds extending from the main body of the organoids. In each image, the white dashed lines indicate the approximated positions of the base and tip of a bud. Organoids in OCTOPUS generate more elongated buds. Scale bars, 20 μm. b. Comparison of the number and size of organoid buds in vitro to those of mouse intestinal crypts in vivo. c, d. Quantification of Ki67, Lgr5, and EdU expression in mouse intestinal organoids (n = 5 and 3 biologically independent experiments for (c) and (d), respectively). e. Organoids developing in OCTOPUS exhibit increased expression of Hnf4α, a marker of mature intestinal epithelial cells, compared to the control group in Matrigel drop. Scale bars, 10 μm. f. Quantification of the fraction of Hnf4α+ cells and the level of Hnf4α expression. Immunofluorescence of Hnf4α was normalized with respect to the number of cells (n = 4). g. The graph shows the cellular composition of the intestinal epithelium in OCTOPUS. Quantification was performed by measuring the immunofluorescence of cell type-specific markers described in Fig. 2m. Undifferentiated cells were identified by positive DAPI staining without the expression of differentiation markers. The data indicate the abundance of enterocytes (79% of the differentiated cell population at day 14) (n = 3). All data are presented as meant SEM and P values are from unpaired, two-sided t test.
Extended Data Fig. 5 ∣. Effect of co-culture on organoid development in OCTOPUS.
a. Co-culture of intestinal organoids and fibroblasts to mimic the epithelial-stromal unit of the intestine. Scale bar, 300 μm. b, c. In comparison to monoculture, the co-culture organoids are larger and express higher levels of Hnf4α expression. Scale bars, 100 μm (b), and 10 μm (c). All data are presented as mean ± SEM and P values are from unpaired, two-sided t test (n = 5 biologically independent experiments).
Extended Data Fig. 6 ∣. Functional characterization of intestinal organoids in OCTOPUS.
a. Immunofluorescence analysis of PEPT1, a nutrient transporter responsible for intestinal uptake of peptides. In this analysis, organoids in Matrigel drops at the maximum duration of culture (7 days) were compared to those maintained in OCTOPUS for 14 days to examine the contribution of extended culture. Organoids in OCTOPUS exhibit robust immunostaining of PEPT1 predominantly in the villus domain of the organoids. PEPT1 expression in this region is restricted to the apical surface of the epithelium facing the organoid lumen (L), matching the localized distribution of PEPT1 on the brush border membrane of the native intestinal epithelium. This polarized expression of PEPT1 is also observed in the villus surface of the organoids in Matrigel drop culture but the level of expression is significantly lower than that in OCTOPUS. Scale bars, 20 μm. b. Comparison of GLUT immunofluorescence in the villus domain of organoids. GLUT is ex-pressed on both the apical and basal surfaces of the villus epithelium in OCTOPUS. This trans-porter is also present in drop culture but its immunofluorescence is much weaker. Scale bars, 20 μm (n = 3 biologically independent experiments). c. Imaging and quantification of intracellular calcium signaling in intestinal organoids treated with 100 μM ATP. in the plots of relative intensity, the time between organoid stimulation and maximum fluorescence intensity is shaded in pink. Data were normalized to fluorescence intensity at the initial time point (t = 0 min). Scale bars, 100 μm (n = 3). d. Fraction of responsive organoids. The fraction of organoids that respond to ATP and glucose stimulation is larger in OCTOPUS. e. ELISA analysis of an active form of GLP-1 and MUC2 secreted by intestinal organoids. Both analytes are produced in significantly higher concentrations in OCTOPUS, which also continue to increase over time during extended culture (n = 3 biologically independent experiments). All data are presented as mean ± SEM and P values are from unpaired, two-sided t test.
Extended Data Fig. 7 ∣. Passaging and expansion of intestinal organoids in OCTOPUS.
a. Experimental procedure for passaging organoids in OCTOPUS. Subculture and expansion of (b) mouse and (c) human intestinal organoids grown in OCTOPUS and Matrigel drop. For comparison, culture conditions (for example, seeding density, hydrogel volume, media composition) were kept the same between the two systems. After 7 days of culture, organoids at a given passage number (Passage N) were physically dissociated and then transferred to new OCTOPUS devices or Matrigel drops (Passage N+1) at densities of 200 crypts/100 μl and 100 crypts/60 μl for mouse and human intestinal organoids, respectively. White solid lines in the micrographs show the outline of culture chambers in the OCTOPUS device or a sessile drop of Matrigel. Scale bars, 1 mm.
Extended Data Fig. 8 ∣. IBD enteroids cultured in Matrigel drops.
a. When cultured in Matrigel drops, IBD enteroids show properly polarized epithelial cells (top right) that resemble those in the epithelium of normal enteroids. In comparison to IBD enteroids in OCTOPUS, they also retain the structural integrity of the epithelium as visualized by ZO-1 expression (bottom right). Scale bars, 5 μm. b. UMAP plots showing the expression of two representative LINC genes by IBD and normal enteroids cultured in Matrigel drops. c. UMAP plots showing the expression of 4 representative IBD-associated genes by IBD and normal enteroids cultured in Matrigel drops.
Supplementary Material
Acknowledgements
We thank R. Wells, W. Yang and G. Al for their input. This work was supported by the National Institutes of Health (NIH) (grant nos. 1DP2HL127720-01, 1UG3DK122644) (D.D.H.); NIH grant no. R01DK124369 (K.E.H.); Children’s Hospital of Philadelphia Institutional Development Funds (K.E.H.); the National Science Foundation (grant no. CMMI:15-48571) (D.D.H.); the Ministry of Trade, Industry & Energy of the Republic of Korea (D.D.H.); the Kwanjeong Educational Foundation (S.E.P.); the Children’s Hospital of Philadelphia Gastrointestinal Epithelium Modeling Program (K.E.H.); and the University of Pennsylvania (D.D.H.). D.D.H. is a recipient of the NIH Director’s New Innovator Award and the Cancer Research Institute Technology Impact Award.
Footnotes
Competing interests
D.D.H. is a cofounder of Vivodyne Inc. and holds equity in Vivodyne Inc. and Emulate Inc. The remaining authors declare no competing interests.
Extended data is available for this paper at https://doi.org/10.1038/s41592-022-01643-8.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41592-022-01643-8.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41592-022-01643-8.
Data availability
All the numeric data used in this study are included in the Source data files provided with this paper. The human enteroids scRNA-seq dataset analyzed during the current study is available at the NCBI Gene Expression Omnibus, under accession number GSE203380. The raw images are too large for public deposit and are available from the corresponding author on reasonable request.
References
- 1.Clevers H Modeling development and disease with organoids. Cell 165, 1586–1597 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Lancaster MA & Knoblich JA Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345, 1247125 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Dutta D, Heo I & Clevers H Disease modeling in stem cell-derived 3D organoid systems. Trends Mol. Med 23, 393–410 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Broutier L et al. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat. Protoc 11, 1724–1743 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Shin W et al. Spatiotemporal gradient and instability of Wnt induce heterogeneous growth and differentiation of human intestinal organoids. iScience 21, 101372 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu H et al. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell 175, 1591–1606 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Huch M & Koo B Modeling mouse and human development using organoid cultures. Development 142, 3113–3125 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Homan KA et al. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods 16, 255–262 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee GY, Kenny PA, Lee EH & Bissell MJ Three-dimensional culture models of normal and malignant breast epithelial cells. Nat. Methods 4, 359–365 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y et al. Long-term culture captures injury-repair cycles of colonic stem cells. Cell 179, 1144–1159 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y et al. Bioengineered systems and designer matrices that recapitulate the intestinal stem cell niche. Cell. Mol. Gastroenterol. Hepatol 5, 440–453 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walton KD et al. Hedgehog-responsive mesenchymal clusters direct patterning and emergence of intestinal villi. Proc. Natl Acad. Sci. USA 109, 15817–15822 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker EM, Thompson CA, Kohlnhofer BM, Faber ML & Battle MA Characterization of the developing small intestine in the absence of either GATA4 or GATA6. BMC Res. Notes 7, 902 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Serra D et al. Self-organization and symmetry breaking in intestinal organoid development. Nature 569, 66–72 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato T & Clevers H Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science 340, 1190–1194 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Sumigray KD, Terwilliger M & Lechler T Morphogenesis and compartmentalization of the intestinal crypt. Dev. Cell 45, 183–197 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cattin A et al. Hepatocyte nuclear factor 4α, a key factor for homeostasis, cell architecture, and barrier function of the adult intestinal epithelium. Mol. Cell. Biol 29, 6294–6308 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spanier B Transcriptional and functional regulation of the intestinal peptide transporter PEPT1. J. Physiol 592, 871–879 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferraris RP & Diamond J Regulation of intestinal sugar transport. Physiol. Rev 77, 257–302 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Tuo B & Dong H Regulation of intestinal glucose absorption by ion channels and transporters. Nutrients 8, 43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang X et al. Molecular mechanisms of calcium signaling in the modulation of small intestinal ion transporters and bicarbonate secretion. Oncotarget 9, 3727–3740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacDonald PE et al. The multiple actions of GLP-1 on the process of glucose-stimulated insulin secretion. Diabetes 51, S434–S442 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Kim YS & Ho SB Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr. Gastroenterol. Rep 12, 319–330 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuji M et al. Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell Stem Cell 23, 787–792 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Elmentaite R et al. Single-cell sequencing of developing human gut reveals transcriptional links to childhood Crohn’s disease. Dev. Cell 55, 771–783 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y et al. Single-cell transcriptome analysis reveals differential nutrient absorption functions in human intestine. J. Exp. Med 217, e20191130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito G et al. Lineage-specific expression of bestrophin-2 and bestrophin-4 in human intestinal epithelial cells. PLoS ONE 8, e79693 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sprangers J, Zaalberg IC & Maurice MM Organoid-based modeling of intestinal development, regeneration, and repair. Cell Death Differ. 28, 95–107 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peterson LW & Artis D Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat. Rev. Immunol 14, 141–153 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Modigliani R et al. Clinical, biological, and endoscopic picture of attacks of Crohn’s disease. Gastroenterology 98, 811–818 (1990). [DOI] [PubMed] [Google Scholar]
- 31.Guo C & Shen J Cytoskeletal organization and cell polarity in the pathogenesis in Crohn’s disease. Clin. Rev. Allergy Immunol 60, 164–174 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Schmitt M et al. Paneth cells respond to inflammation and contribute to tissue regeneration by acquiring stem-like features through SCF/c-Kit signaling. Cell Rep. 24, 2312–2328 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Spinelli A, Correale C, Szabo H & Montorsi M Intestinal fibrosis in Crohn’s disease: medical treatment or surgery? Curr. Drug Targets 11, 242–248 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Reimund JM et al. Increased production of tumour necrosis factor-α, interleukin-1β, and interleukin-6 by morphologically normal intestinal biopsies from patients with Crohn’s disease. Gut 39, 684–689 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grebenyuk S & Ranga A Engineering organoid vascularization. Front. Bioeng. Biotechnol 7, 1–12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deban L et al. Multiple pathogenic roles of microvasculature in inflammatory bowel disease: a Jack of all trades. Am. J. Pathol 172, 1457–1466 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones G et al. Dynamics of colon monocyte and macrophage activation during colitis. Front. Immunol 27, 2764 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Demers CJ et al. Development-on-a-chip: in vitro neural tube patterning with a microfluidic device. Development 143, 1884–1892 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasendra M et al. Development of a primary human small intestine-on-a-chip using biopsy-derived organoids. Sci. Rep 8, 2871 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barker N et al. Lgr5+ve stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 6, 25–36 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Wu F, Huang Y, Dong F & Kwon JH Ulcerative colitis-associated long noncoding RNA, BC012900, regulates intestinal epithelial cell apoptosis. Inflamm. Bowel Dis 22, 782–795 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Gu L et al. Identification of a 5-lncRNA signature-based risk scoring system for survival prediction in colorectal cancer. Mol. Med. Rep 18, 279–291 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Axelrad JE, Lichtiger S & Yajnik V Inflammatory bowel disease and cancer: the role of inflammation, immunosuppression, and cancer treatment. World J. Gastroenterol 22, 4794–4801 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jaeger N et al. Single-cell analyses of Crohn’s disease tissues reveal intestinal intraepithelial T cells heterogeneity and altered subset distributions. Nat. Commun 12, 1921 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van de Watering M et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mansour AA et al. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol 36, 432–441 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dye BR et al. A bioengineered niche promotes in vivo engraftment and maturation of pluripotent stem cell derived human lung organoids. eLife 5, e19732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahashi Y, Takebe T & Taniguchi H Methods for generating vascularized islet-like organoids via self-condensation. Curr. Protoc. Stem Cell Biol 45, e49 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Vargas-Valderrama A, Messina A, Mitjavila-Garcia MT & Guenou H The endothelium, a key factor in organ development and hPSC-derived organoid vascularization. J. Biomed. Sci 27, 67 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park SE, Georgescu A & Huh D Organoids-on-a-chip. Science 364, 960–965 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the numeric data used in this study are included in the Source data files provided with this paper. The human enteroids scRNA-seq dataset analyzed during the current study is available at the NCBI Gene Expression Omnibus, under accession number GSE203380. The raw images are too large for public deposit and are available from the corresponding author on reasonable request.