Summary
Telomere maintenance 2 (TELO2), Tel2 interacting protein 2 (TTI2), and Tel2 interacting protein 1 (TTI1) are the three components of the conserved Triple T (TTT) complex that modulates activity of phosphatidylinositol 3-kinase-related protein kinases (PIKKs), including mTOR, ATM, and ATR, by regulating the assembly of mTOR complex 1 (mTORC1). The TTT complex is essential for the expression, maturation, and stability of ATM and ATR in response to DNA damage. TELO2- and TTI2-related bi-allelic autosomal-recessive (AR) encephalopathies have been described in individuals with moderate to severe intellectual disability (ID), short stature, postnatal microcephaly, and a movement disorder (in the case of variants within TELO2). We present clinical, genomic, and functional data from 11 individuals in 9 unrelated families with bi-allelic variants in TTI1. All present with ID, and most with microcephaly, short stature, and a movement disorder. Functional studies performed in HEK293T cell lines and fibroblasts and lymphoblastoid cells derived from 4 unrelated individuals showed impairment of the TTT complex and of mTOR pathway activity which is improved by treatment with Rapamycin. Our data delineate a TTI1-related neurodevelopmental disorder and expand the group of disorders related to the TTT complex.
Keywords: mendelian disorders, gene, pathogenic variants, autosomal recessive, TTI1 gene, consanguinity, neurodevelopment, microcephaly
Pathogenic mutations in TTI1 (encoding a protein of the TTT complex) cause a rare autosomal-recessive disorder with a neurodevelopmental phenotype. These findings are important for diagnosis and genetic counseling; in addition, they provide the opportunity for development of potential therapies.
Introduction
Telomere maintenance 2 (TELO2 [MIM: 611140]), TELO2 interacting protein 2 (TTI2 [MIM: 614426]), and TELO2 interacting protein 1 (TTI1 [MIM: 614425]) are the three components of the conserved Triple T (TTT) complex that regulates activities of phosphatidylinositol 3-kinase-related protein kinases (PIKKs).1,2,3,4,5 The three TTT proteins interact directly with the PIKK family, which includes MTOR (mammalian target of rapamycin [MIM: 601231]), ATM (ataxia telangiectasia mutated [MIM: 607585]), ATR (ATM- and Rad3-related [MIM: 601215]), SMG-1 (SMG1 Nonsense-Mediated mRNA Decay-Associated PI3K-related Kinase [MIM: 607032]), and TRRAP (transformation/transcription domain associated protein [MIM: 603015]). The TTT complex is involved in the regulation of mTOR complex 1 (mTORC1) assembly1 and plays an essential role in the protein accumulation, maturation, and stability of ATM and ATR in response to DNA damage.4,6,7,8
Pathogenic variants in TELO2 and TTI2 have been described in autosomal-recessive (AR) encephalopathies in individuals sharing a clinical picture of moderate to severe intellectual disability often associated with postnatal microcephaly. Ten individuals have been described so far with TELO2-related encephalopathy (MIM: 616954)9,10,11 and 10 others with TTI2-related disease (MIM: 615541).12,13,14,15,16 Short stature and movement disorder is described in both.
We present clinical and functional data in a series of 11 individuals from 9 unrelated families with neurodevelopmental disorder (NDD) phenotypes and TTI1 bi-allelic likely pathogenic variants. Functional studies were conducted to evaluate the consequences of TTI1 variants in HEK293T cell lines and functional impairment of the TTT complex and mTOR pathway in fibroblasts and lymphoblastoid cell lines derived from affected individuals. Taken together, these data establish a third TTT-related disorder resulting from pathogenic variants in TTI1.
Subjects and methods
Subjects
Clinical information was summarized from individuals with TTI1 bi-allelic pathogenic variants from France, England, Australia, and the United States ascertained via GeneMatcher.17 Variants were identified by exome sequencing (ES) for families 1, 2, 4–7, and 9; by genome sequencing (GS) for family 8, and by targeted Sanger sequencing in family 3. In all cases, the TTI1 variants identified in probands were searched for in their parents by targeted Sanger sequencing. The studies have been approved by the local Institutional Review Boards. Written informed consent was obtained for genetic testing, blood sampling, and skin biopsy for functional studies, publication, and use of photographs.
Exome sequencing and analysis
Genomic DNA was extracted from whole blood. TTI1 variants were detected by different genomic platforms in this cohort. In summary, the affected individuals underwent ES or GS in a clinical or research setting using either Illumina HiSeq or NovaSeq instrumentation and related chemistry. Bioinformatic pipelines annotated, filtered, and prioritized high-quality variants that differed from the reference sequence into a small number of clinically relevant variants as described previously.18
3D structure predictions of variants affecting TTI1
We used both the cryo-EM structure of TTI1, TTI2, and TELO2 complex19 stored under 7F4U code in the PDB and the AlphaFold220 derived model AF-O43156-F1. Figures were made with UCSF Chimera software.21 The protein stability loss was estimated using FoldX 5.0 software22 using AF2 derived model.
Expression of TTI1 mutants
The TTI1 variants were analyzed by over-expressing TTI1 in HEK293T cell line. TTI1 was expressed by transfecting HEK293T cells with the plasmid, p3xFLAG-CMV10-hTti1 (Addgene, Cat# 30213) that encodes human TTI1 with FLAG epitope tag at the N terminus (TTI1-Flag).1 The various TTI1 variants were generated using PCR site-directed mutagenesis and sequence confirmed at the Johns Hopkins DNA sequencing facility. Protein accumulation of TTI1-Flag was detected by immunoblotting with both anti-FLAG and anti-TTI1 antibodies. The immunoblot analysis further confirmed increased levels of the TTI1-Flag wild-type and mutants in HEK293T cells compared to control cells that were transfected with empty Flag plasmid vector. TTT and PIKK proteins immunoprecipitation (pulldown) of TTI1-Flag from HEK293T lysate was performed using anti-Flag antibody immobilized on beads.
SiMPull analyses
SiMPull (single-molecule pull-down) combines immunoprecipitation and immobilization of protein complexes from lysates that contain fluorescently tagged proteins with single-molecule fluorescent microscopy. The SiMPull imaging uses a total internal reflection fluorescent (TIRF) microscope and allows the study of organization, behavior, stoichiometry, and activity of protein complexes.23 The association of TTI1 missense variants with mTOR and mTORC1 was studied by SiMPull.24 Whole lysates from cells co-expressing TTI1-Flag, green fluorescent protein-tagged mTOR (GFP-mTOR), and red fluorescent protein-tagged Raptor (RFP-Raptor) were applied to anti-FLAG antibody-conjugated quartz slides prepared to pull down TTI1-Flag and analyzed by TIRF. Since TTI1 is critical for mTORC1 formation,1 we used SiMPull photobleaching to determine the stoichiometry of the mTOR oligomeric state and mTORC1 composition associated with the TTI1 variants.
mTORC1 activation assay
We analyzed the status of the mTOR signaling pathway in the HEK293 cells over-expressing TTI1-Flag. We determined the phosphorylation levels of mTORC1 proteins, mTOR, S6K1, 4EBP1, and PRAS40 under different nutrient (amino acids) status.
Evaluation of mTORC1 drug treatments
Treatment of starved cells by mTORC1 activator MHY1485 increases mTOR and S6K phosphorylation, whereas treatment by mTORC1 inhibitor rapamycin decreases the phosphorylation levels.25 Cells were treated with MHY1485 (10 μM) or rapamycin (0.1 μM) immediately after starvation prior to addition of amino acids. The drugs were left in media with amino acids for 30 min to evaluate their effects on mTORC1 activation in the presence of the TTI1 mutants.
Cell cycle analyses
We used a dual-reporter system with nuclear localized H2B-mCherry and a GFP-tagged Cdk2 substrate, DNA helicase B (DHB-GFP).26 Cells co-expressing Flag-TTI1 and CDK-DHB dual-reporter were serum deprived for 24 h to synchronize the cells in G0 phase followed by stimulation with serum for 12 h.26
Examination of TTI1-PIKK mediated UV-induced DNA damage response and repair
To evaluate DNA damage response, we exposed live cells expressing TTI1 mutants to UV radiation and then analyzed phosphorylation levels of ATM/ATR-mediated DNA damage response substrates at 60 min post-UV exposure.
To evaluate DNA repair, the live cells were exposed to UV radiation and then fixed at different times after exposure. The amount of 6-4PP produced immediately after exposure (UV 0 h) and amount remaining in the cells at 2 h after UV exposure (UV 2 h) were determined.
G2 chromosomal radio-sensitivity investigation, lymphoblastoid cell line generation, western blot
To assess chromosomal radiosensitivity, whole blood taken into lithium heparin was cultured in RPMI 1640 and FCS; these cultures were stimulated with PHA for 72 h and exposed to 1 Gy 137Cs γ-rays 4 h before harvesting (in G2 phase of the cell cycle). Colcemid was added 1 h prior to harvesting. Chromosome spreads were made using standard methods and chromosome damage analyzed by light microscopy.
Epstein-Barr virus (EBV)-transformed lymphoblastoid cell lines (LCL) were established from a range of individuals including normal control subjects, an individual with classic A-T, as well as the individual with TTI1 variants. Whole blood in lithium heparin from each individual was layered on to a blood separation medium (Cedarlane Lympholyte Separation medium, CL5020) and centrifuged for 30 min; the lymphocyte layer was removed and washed and supernatant from B95.8 cell cultures (producing EBV virus), first filtered through a Sartorius 0.45 μm filter, was added to the lymphocytes overnight. The lymphocytes were resuspended in RPMI medium containing Cyclosporin A to kill any T cells and left for 14 days after which they were transformed.
Immunoblotting for ATM levels and ATM activity assays were performed as previously described.27,28 In brief, after activation of ATM by exposure to 2 Gy ionizing radiation (137Cs γ-rays), LCL cells were harvested after 30 min and ATM present was tested for its ability to phosphorylate a range of target proteins. Antibodies used for immunoblotting were: ATM 11G12 (custom-made mouse monoclonal), anti-phospho ATM (S1981) (AF1655) (R&D Systems); anti-SMC1 (A300-055A), anti-phospho SMC1 (S966) (A300-050A), anti-KAP-1 (A300-275A), anti-phospho KAP-1 (S824) (A300-767A) (Bethyl Laboratories, Universal Biologicals); anti-NBN (ab23996), anti-phospho NBN (S343) (ab47272), anti-CREB (32,515) (Abcam); anti-phospho CREB (S121) (NB100-410; Novus Biologicals). An LCL derived from a normal individual and another derived from a known classical A-T individual were used as positive and negative controls for ATM activity/signaling. Cell lysates showed either a normal, reduced, or absent detectable ATM activity/signaling. For immunoblotting to detect presence or absence of TTT complex proteins and PIKK kinases, the following antibodies were used: anti-TTI1 (A303-451A-T) and anti-TTI2 (A303-476A-T) (Bethyl Laboratories); anti-Telo 2 (I5975-I-AP) (Proteintech); anti-ATM- custom made mouse monoclonal; anti-ATR (EIS3S) and anti-SMG1 (D42D5) (Cell Signaling Technology); anti-mTOR (A300-504A-T) and anti-DNA PKcs (A300-517A-T) (Bethyl Laboratories); anti-Tubulin (05-829) (Millipore).
Results
Clinical data
Clinical observations are summarized in Table 1, Figure 1, and Supplemental Note. Presenting features include developmental delay (11/11) with language impairment (11/11) ranging from delayed speech to non-verbal and variable motor impairment (7/11) from unsteady gait to non-ambulation. Moderate to severe microcephaly (OFC < −2 SD) was observed in 9/11 affected individuals with a mean OFC of −4.2 SD, ranging from −1.2 SD to −9.2 SD. Short stature (8/11) with a mean of −2.6 SD (range −1 to −4.7 SD), dysmorphic features (10/11), scoliosis (5/10), strabismus (5/8) hypogonadism (5/10), and brain malformations (6/10) were also present. The movement disorder described in six individuals (6/11) included ataxia, either in isolation or with chorea. Seizures are described in 4 cases (treatment-resistant seizures in one), and renal malformation is described in two individuals. Two individuals presented with mild intra-uterine growth restriction (IUGR) whereas all others were born with normal birth parameters.
Table 1.
Clinical features of TTI1 variants (GenBank: NM_014657.3) in the 11 affected individuals from 9 families
| Affected individuals | Family 1, Ind 1 (IV-2) | Family 1, Ind 2 (IV-3) | Family 2, Ind 3 (II-1) | Family 3, Ind 4 (II-2) | Family 4, Ind 5 (II-3) | Family 5, Ind 6 (III-5) | Family 5, Ind 7 (III-6) | Family 6, Ind 8 (II-1) | Family 7, Ind 9 (II-2) | Family 8, Ind 10 (II-2) | Family 9, Ind 11 (II-1) | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele 1 | c.2300T>C (p.Leu767Ser) | c.2300T>C (p.Leu767Ser) | c.2302+1G>T | c.835T>G (p.Trp279Gly) | c.2998+2T>C | c.2761G>A (p.Asp921Asn) | c.2761G>A (p.Asp921Asn) | c.1271A>G (p.His424Arg) | c.1204T>C (p.Ser402Pro) | c.3241G>A (p.Val1081Met) | c.1901T>C (p.Ile634Thr) | – |
| Allele 2 | c.2300T>C (p.Leu767Ser) | c.2300T>C (p.Leu767Ser) | c.835T>G (p.Trp279Gly) | c.2978T>G (p.Leu993Arg) | c.2513C>T (p.Ser838Leu) | c.2761G>A (p.Asp921Asn) | c.2761G>A (p.Asp921Asn) | c.629dup (p.Leu210Phefs∗17) | c.2998G>C (p.Gly1000Arg) | c.106C>T (p.Arg36Ter) | c.1468C>T (p.Gln490Ter) | – |
| Origin | Turkish | Turkish | Argentinian/Australian | British European | European | Turkish | Turkish | European | Mexican | European | North African | – |
| Consanguinity | yes (parents first cousins) | yes (parents first cousins) | No | no | no | yes (parents first cousins) | yes (parents first cousins) | no | no | no | no | – |
| Gender | male | male | female | female | male | female | male | male | female | male | female | – |
| Year of birth | 1997 | 2003 | 2012 | 2010 | 2005 | 2000 | 2012 | 2016 | 2011 | 2010 | 2016 | – |
| Age at last examination | 20 y 11 m | 14 y 4 m | 7 y 9 m | 10 y 9 m | 13 y 9 m | 19 y | 7 y 6 m | 2 y 9 m | 8 y 4 m | 10 y | 3 y 7 m | – |
| OFC at last examination (SD) | −2 SD | −1.5 SD | −2 SD | 47.2 cm (−5.3 SD) | 53 cm (−1.2 SD) | 44.5 cm (−9.2 SD) | 44.5 cm (−5.6 SD) | 45.5 cm (−2.39 SD) | 42 cm (−8.2 SD) | −3 SD | (−3 SD) | 9/11 (microcephaly) |
| Height at last examination (SD) | −3 SD | −2.5 SD | −1 SD | 131.4 cm (−1.7 SD) | 147 cm (−1.8 SD) | 116 cm (−4.7 SD) at 12 y | 106 cm (−2.9 SD) | 85.5 cm (−2.15 SD) | 112 cm (−3 SD) | −3.6 SD | (−1 SD) | 8/11 (short stature) |
| Weight at last examination | 25th centile | 10th centile | – | 28.3 kg (−1.2 SD) | 34.9 kg (−2.1 SD) | 22 kg (−5.3 SD) | 14 kg (−3.5 SD) | – | 18.5 kg, <1st%ile | −3.4 SD | +1 SD | – |
| Other | cervical hyperpigmentation, premature greying of hair, acrocyanosis | – | facial telangiectasia developed at age 4 | – | – | – | – | – | – | – | – | – |
| Developmental delay | moderate | moderate | moderate | severe | severe | severe | severe | severe | moderate | mild to moderate | moderate | 11/11 |
| Age of sitting | – | 6 m | 10 m | 8–9 m | 10 m (with support) | 1 y | 1 y | 11 m | 7–8 y | 9 m | – | – |
| Age of walking | 14 m, instability in childhood | 12 m, instability in childhood | 20 m | 23–24 m | 3 y 10 m with assistance, no independent walking | 5 y | 5 y | 2 y 2 m | non ambulatory | 24 m | 22 m | 9/11 |
| Age of first words/language abilities | delayed/few sentences | delayed/few sentences | delayed/simple conversational speech | delayed speech | non-verbal, 3 signs | 6 y, no sentences at 19 y | 1.5 y, no sentences at 7.5 y | 2.5 y (a few words) | non-verbal | 20 m/language delay, speak in short sentences | no language | 11/11 |
| Schooling | specialized structure | specialized structure | specialized | special education | special education | special education | special education | special education | special education | special education | N/A | 9/10 |
| Level of autonomy | works in a specialized structure | – | – | minimal | no autonomous feeding, no autonomous locomotion | – | – | minimal | none | – | – | – |
| Puberty | premature | – | – | no signs of puberty | normal | primary and secondary amenorrhea | prepubertal | prepubertal | precocious puberty at 6 y | no signs of puberty | prepubertal | 2/10 |
| Intellectual deficiency | moderate | mild | moderate | moderate to severe | severe | severe | severe | too young for dx | severe | mild to moderate | too young for dx | 9/9 |
| Dysmorphism | present | present | absent | present | present | present | present | present, nonspecific | present | mild dysmorphism | mild dysmorphism | 10/11 |
| Hypotonia | no | no | yes in infancy | no | yes in infancy | no | no | yes | poor head control | yes | no | 5/11 |
| Spasticity/hypertonia | no | no | no | no | lower limbs | spasticity and hypertonia | spasticity and hypertonia | – | lower extremities | no | no | 4/11 |
| Movement disorder | no | no | hyperkinetic choreiform movements, ataxic gait | choreo-athetoid movements in upper limbs, gait ataxia | choreo-athetoid movements in upper limbs | waddling gait | waddling gait | truncal ataxia, gait ataxia, hypotonia | choreo-athetoid movements in upper limbs | no | no | 6/11 |
| Behavior | mainly calm with rare aggressive outburst | no | – | poor concentration, excessive drooling, sleep problems | sporadic spontaneous laughter | self mutilation: biting, hitting her head | – | – | – | aggressivity hyperactivity, impulsivity, separation anxiety. | aggressivity, stereotypy, sleeping disorder | – |
| Seizures | no | no | no | no | yes | no | no | no | yes | yes | febrile seizures (at 6months) | 4/11 |
| Brain MRI | not done | not done | not done | abnormal | abnormal | none | abnormal | abnormal | abnormal | abnormal | normal | 6/8 |
| Abdominal ultrasound | vesico-ureteral reflux surgery at age 2 y | kidneys: ectopic right kidney | not done | normal | not done | normal | normal | not done | multicystic atrophic right kidney, vesicoureteral reflux | normal | not done | 2/7 |
| Skeletal findings | kyphoscoliosis, genu varum | coxa vara, scoliosis | not done | no | scoliosis, hip dysplasia, bilateral equinovalgus | scoliosis, delayed bone age 8–9 y | scoliosis | none (normal bone survey) | advanced bone age | external tibial torsion, femoral retrotorsion | no | 7/10 |
| Reproductive organs | premature puberty, regressive bilateral gynecomastia | right cryptorchidism mild regressive gynecomastia | normal | normal | normal | primary and secondary amenorrhea | hypoplastic genitalia | normal | small labia minora and clitoris | normal | normal | 5/11 |
| Ophthalmic features | strabismus surgery at age 12 | normal | bilateral esotropia | hypermetropia | normal | strabismus | strabismus | normal | cortical visual impairment, nystagmus, myopia both eyes | strabismus (right esotropia), post surgical repair | normal | 7/11 |
Abbreviations: OFC, occipital frontal circumference; SD, standard deviation.
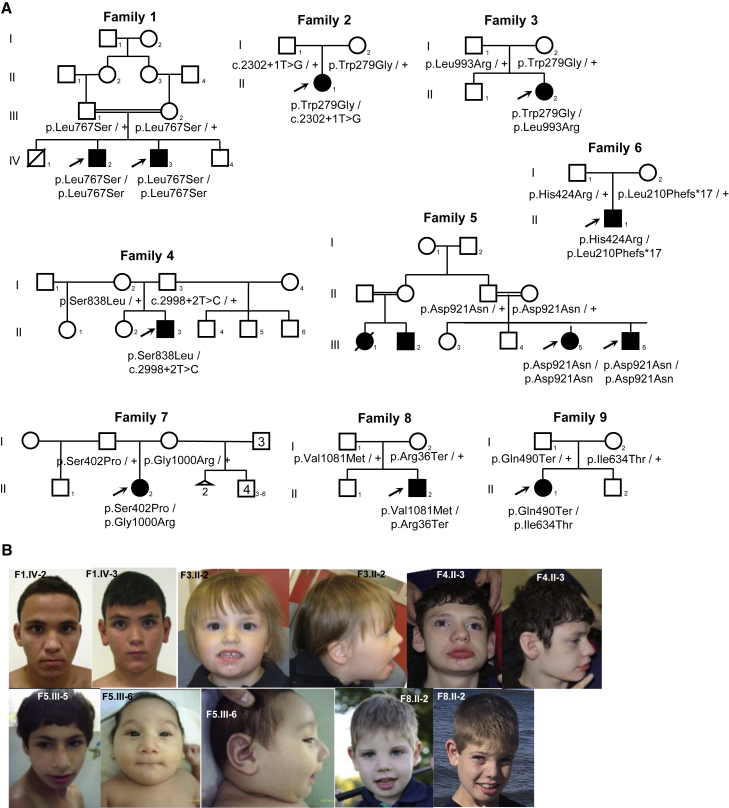
Figure 1.
Pedigrees and clinical photographs of the affected individuals
(A) Pedigrees of the 11 affected individuals with bi-allelic, likely pathogenic TTI1 variants.
(B) Clinical photographs of selected affected individuals. Individuals 1 (F1.IV-2) and 2 (F1.IV-3): smooth philtrum, thin upper lip. Individual 4 (F3.II-2): fine facial features with round face, low-set ears, and widely spaced teeth. Individual 5 (F4.II-3): lateral flare of eyebrows, upturned nose, wide mouth with fully everted lower lip, and large protruding ears with underfolded helices. Individual 6 (F5.III-5): narrow forehead, strabismus, upslanting palpebral fissures, blue sclerae, prominent nose, micrognathia, and short philtrum. Individual 7 (F5.III-6): narrow forehead, upslanting palpebral fissures, epicanthus, blue sclerae, prominent nose, micrognathia, and short philtrum. Individual 10 (F8.II-2): prominent metopic suture, upslanting palpebral fissures, infraorbital creases, and anteverted nares.
Molecular genetic findings
Molecular data, in silico analyses, and ACMG (American College of Medical Genetics) criteria for variant interpretation29 for each allele are summarized in Tables S1 and S2. Bi-allelic variants in TTI1 were present in compound heterozygous state in 7 individuals and in the homozygous state in 4 individuals. Each parent was confirmed to be a heterozygous carrier of one of the familial TTI1 variants. 15 variants were identified including 10 missense (p.Leu767Ser, p.Trp279Gly, p.Leu993Arg, p.Ser838Leu, p.Asp921Asn, p.His424Arg, p.Ser402Pro, p.Val1081Met, p.Ile634Thr and p.Gly1000Arg), 2 nonsense (p.Arg36Ter and p.Gln490Ter), 2 canonical splice variants (c.2302+1G>T and c.2998+2T>C), and 1 nucleotide insertion at codon Leu210 predicted to lead to a frameshift allele. The missense variant, p.Trp279Gly, was recurrent and identified in compound heterozygosity in two unrelated families (2 and 3).
Functional studies
Expression of TTI1 mutant constructs
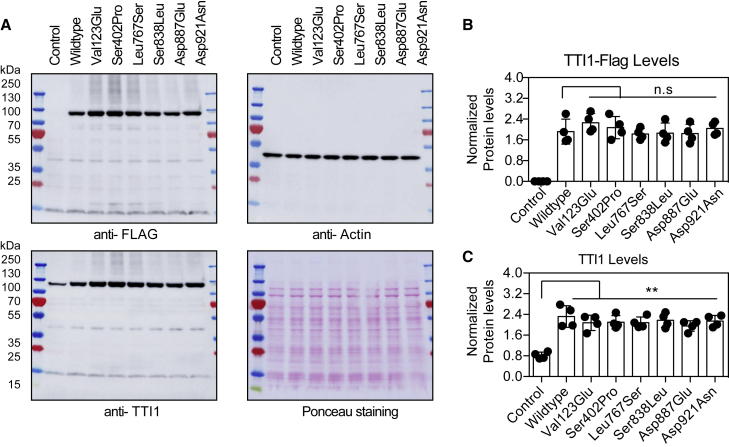
We evaluated he activity of six of the ten non-synonymous TTI1 substitution variant alleles. Four of these variants (p.Ser402Pro, p.Leu767Ser, p.Ser838Leu, p.Asp921Asn) were the initial ones identified by this collaborative effort. During the course of the study the other two variants (GenBank: NM_001303457.2; c.368T>A [p.Val123Glu] and GenBank: NM_001303457.2; c.2661T>A [p.Asp887Glu]) were identified as compound heterozygous variants in additional individuals and were included in the functional analyses. A TTI1 knockout (KO) cell line was generated to characterize all above-mentioned variants, but these cells were not viable. We therefore generated TTI1-FLAG plasmid DNA constructs encoding human TTI1 with a FLAG epitope tag at the N terminus (TTI1-Flag)1 to express the TTI1 variants in HEK293T cells (Figure S2). Protein levels of TTI1-Flag was detected by immunoblotting with both anti-FLAG and anti-TTI1 antibodies (Figure 2A). The immunoblot analyses showed substantial protein accumulation of the TTI1-Flag wild-type and mutants transfected in HEK293T cells compared to control cells that were transfected with empty Flag plasmid vector (Figures 2A–2C). No significant differences in the protein levels of the TTI1-flag mutants compared to the wild-type were observed (Figure 2C).
Figure 2.
Protein levels of TTI1 variants in HEK293T cells
(A) Immunoblot images of TTI1-Flag accumulation in HEK293T. Control cells were transfected with empty plasmid vector.
(B) Quantification of band intensity of anti-FLAG blot normalized to anti-Actin blots in (A) (n = 4).
(C) Quantification of band intensity of anti-TTI1 blot normalized to anti-Actin blots in (A) (n = 4).
TTI1 structural protein analysis
The structure of TTI1 was solved in the TTT complex with Cryo-EM technique as a curved α-solenoid domain consisting of 16 helical repeats.19 The TELO2 binding site extends from 5th to 10th repeat, in the middle of TTI1 structure.19 The missense variants described in this study are located on different helices and loops of the domain. Only the p.His424Arg variant is located directly on the TELO2 binding interface.
The variants p.Leu767Ser, p.Trp279Gly, p.Ser402Pro, p.Asp921Asn, p.Ile634Thr, p.Val1081Met, and p.Leu993Arg likely decrease the stability of the TTI1 complex and might each interfere to a different extent with its function. The p.His424Arg variant likely changes the interactions with TELO2. The p.Ser838Leu variant may influence the phosphorylation of TTI1 by the CK2 kinase, as it is located close to the phosphorylation site (Figure S1A), and the p.Gly1000Arg variant may disturb the interactions with other proteins as it points toward the solution. For details see the legend of Figure S1.
TTI1 variants interact with the TTT complex and PIKK proteins
TTI1 interacts with telomere maintenance 2 (Tel2) and Tel2 interacting protein 2 (TTI2) to form the TTT complex that regulates the activities of the phosphatidyl-inositol 3-kinase-related protein kinase (PIKK) family.1,3,4 We therefore evaluated the TTI1 mutant activity through components of the PIKK family, which include mammalian target of rapamycin (mTOR), ATM, and SMG-1. Overexpression of TTI1 wild-type or mutants did not alter the levels of the TTT and PIKK proteins (Figure S2A). We performed immunoprecipitation (pulldown) of TTI1-Flag from HEK293T lysate using anti-FLAG antibody immobilized on beads to evaluate the interaction of TTI1 variants with the TTT and PIKK proteins. Interaction data confirmed that wild-type Flag-TTI1 interacts with the TTT and PIKK proteins (Figure S2A). TTI2, Tel2, and mTOR coprecipitated with Flag-TTI1 mutants similar to wild-type (Figures S2B–S2D). Evaluation of variant-encoded TTI1 interactions with PIKK proteins suggests that the PIKK proteins coprecipitated with Flag-TTI1 mutants similar to wild-type (Figures S2E–S2G). The results are consistent with the mutations not significantly affecting TTI1 association with TTT or PIKK complexes.
TTI1 variants and SiMPull studies of mTOR complex 1 (mTORC1)
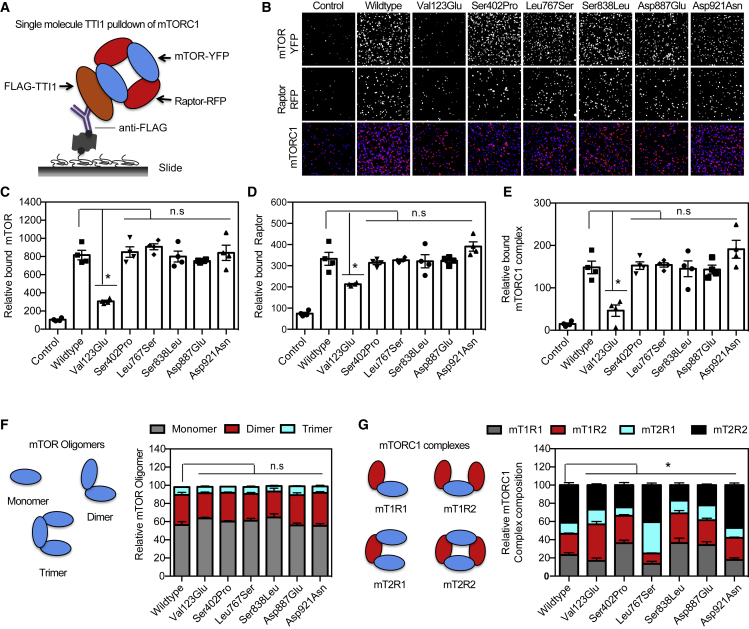
Although no significant alteration in the association of TTI1 encoded by variants with PIKK proteins was observed in the immunoprecipitation experiments, mTOR interactions at the single-molecule level were evaluated further. Studies have shown that the TTT complex including TTI1 regulates the assembly of mTOR complex 1 (mTORC1).1 The variants’ association with mTOR and mTORC1 (mTOR and Raptor complex) was further investigated employing an approach to interrogate protein-protein interaction termed single-molecule pull-down (SiMPull).23 SiMPull combines immunoprecipitation and immobilization of protein complexes from lysates that contain fluorescently tagged proteins with single-molecule fluorescent microscopy. The SiMPull imaging uses a total internal reflection fluorescent (TIRF) microscope and allows the study of organization, behavior, stoichiometry, and activity of protein complexes.23 Whole lysates from cells co-expressing TTI1-Flag, green fluorescent protein tagged mTOR (GFP-mTOR), and red fluorescent protein tagged Raptor (RFP-Raptor) were applied to anti-FLAG antibody conjugated quartz slides prepared to pull down TTI1-Flag and analyzed by TIRF (Figures 3A and 3B). The SiMPull analysis was consistent with wild-type TTI1-Flag associating strongly with the mTORC1 proteins, mTOR and Raptor (Figures 3C and 3E). The p.Val123Glu variant showed reduced association to the mTORC1 proteins, mTOR and Raptor, while there was no significant differences observed with bulk pull down compared to wild-type of the other 5 TTI1 variants tested.
Figure 3.
TTI1 variants alter mTOR complex 1 (mTORC1) composition
(A) Schematic depiction of mTORC1-TTI1-Flag SiMPull.
(B) Representative quartz slide images from SiMPull shown in (A) with the TTI1 variants. Control is similar experiments with cells without TTI1-Flag.
(C and D) Graphical representation of relative amount of mTOR (C) or of Raptor (D) bound to TTI1-Flag (n = 4 for each).
(E) Graphical representation of relative amount of mTORC1 (mTOR-Raptor) bound to TTI1-Flag (n = 4).
(F) Schematic depiction of different mTOR oligomeric states and graphical representation showing the distribution of the different mTOR complexes in the presence of the TTI1 variants (n = 4).
(G) Schematic depiction of different mTORC1 composition (mTOR, mT-Raptor,R) complexes and graphical representation showing the distribution of the different Raptor-mTOR complexes in the presence of the TTI1 variants (n = 4).
Data in (C)–(G) are mean ± SEM experiments, ∗p < 0.05; n.s., p > 0.05, ANOVA with Tukey-Kramer post-hoc test compared with wild-type.
The oligomeric state of free mTOR (not associated with Raptor) was not altered in the presence of TTI1 variants (Figure 3F). Wild-type TTI1 photobleaching distribution was observed to correspond to a mixture of monomers (mT1R1), heterodimers (mT1R2, mT2R1), and homodimers (mT2R2) of mTORC1, mTOR (mT), and Raptor (R). The wild-type TTI1 was predominantly associated with the homodimers of mTOR or Raptor, which are comprised of approximately 50% of the mTORC1 complexes consisting of dimers of mTOR or Raptor (Figure 3G). There is a significant increase in heterodimers (cumulative ∼35% of mT1R2 and mT2R1) in cells with p.Leu767Ser and increase in homodimers (∼60% mT2R2) associated with TTI1 p.Asp921Asn mutant, whereas a significant decrease in homodimers is associated with the other TTI1 variants, p.Ser402Pro and p.Ser838Leu. These variants are predominantly associated with mTORC1 monomers (mT1R1) and heterodimers (mT1R2). These results are consistent with activation of mTORC1 complex being altered in the presence of TTI1 variants due to changes in mTORC1 composition. Constructs containing the p.Leu767Ser and p.Asp921Asn variants form more stable active mTOR complexes, whereas the constructs containing the p.Ser402Pro and p.Ser838Leu variants exhibit less active mTOR complexes compared to wild-type. TTI1 therefore acts as a scaffold for the assembly of mTOR and Raptor, with the stoichiometric balance being distorted in the presence of the TTI1 variants.
TTI1 variants and mTORC1 activation by phosphorylation
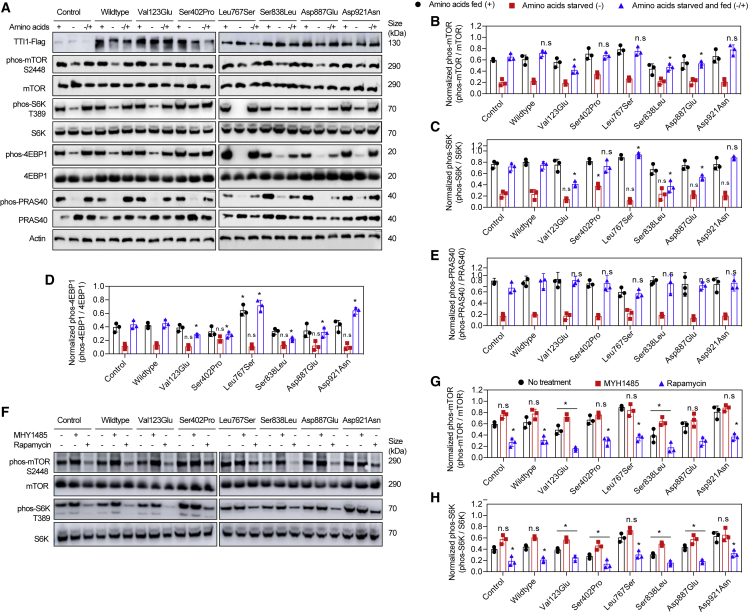
To further evaluate the effect of TTI1 variants on mTORC1, the levels of components of the mTOR signaling pathway were assessed in HEK293 cells over-expressing TTI1-Flag. mTORC1 promotes growth in response to the availability of nutrients, such as amino acids, which drive mTOR to phosphorylate several substrates, including S6K1, PRAS40, and 4EBP1.30,31,32,33,34 The phosphorylation of S6K1 stimulates the initiation of protein synthesis, whereas phosphorylation of translation inhibitor, 4EBP1, releases it from eukaryotic translation initiation factor 4E.30,31,33,34,35,36 The phosphorylation levels of mTORC1 proteins, mTOR, S6K1, 4EBP1, and PRAS40 were assessed under different nutrient (amino acids) status (Figure 4A). As previously reported in normally fed (amino acids +) cells, phosphorylation of the mTORC1 was moderate in control and wild-type Flag-TTI1 cells but decreased during prolonged (50 min) amino acid starvation.30,31,32,33,34
Figure 4.
TTI1 variants alter mTORC1 activation by amino acids
(A) Representative immunoblots of showing mTORC1 activation (phosphorylation) status during normal amino acids fed (+), starvation (−), and starvation followed by amino acid supplementation (−/+).
(B) Quantification of phosphorylated mTOR (phos-mTOR, S2448) normalized to total mTOR (mTOR) of images in (A) (n = 3).
(C) Quantification of phosphorylated S6K (phos-S6K, T389) normalized to total S6K (S6K) of images in (A) (n = 3).
(D) Quantification of phosphorylated 4EBP1 (phos-4EBP1) normalized to total 4EBP1 (4EBP1) of images in (A) (n = 3).
(E) Quantification of phosphorylated PRAS40 (phos-PRAS40) normalized to total PRAS40 (PRAS40) of images in (A) (n = 3).
QA (F) Representative immunoblots of showing mTORC1 activation (phosphorylation) status in the absence (− −) or presence (+ −) of MHY1485 or Rapamycin (− +).
(G) Quantification of phosphorylated S6K (phos-S6K, T389) normalized to total S6K (S6K) of images in (F) (n = 3).
Data in (B)–(E), (G), and (H) are mean ± SEM of experiments performed, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, n.s., p > 0.05, ANOVA with Tukey-Kramer post-hoc test compared with WT.
Upon re-stimulation of starved cells with amino acids, it was observed that mTORC1 was hyperactivated (as demonstrated by S6K1/4EBP1 phosphorylation) in cells expressing TTI1 variants (Figures 4B–4E). Among these, p.Leu767Ser and p.Asp921Asn resulted in hyperactivation of mTORC1, whereas p.Ser402Pro and p.Ser838Leu displayed mTORC1 hypoactivation compared to wild-type. These results are consistent with the TTI1 variants reported in this study increasing mTORC1 activation in physiological conditions that alter mTORC1 signaling such as nutrient deprivation, supplementation, or energy stress.
TTI1 variants and drug induced mTORC1 activation and inhibition
The effects of the mTORC1 activator MHY148525 and inhibitor rapamycin32,37 on mTOR activity were examined in cells expressing TTI1 mutations. As significant alteration in mTORC1 activity was detected upon stimulation of starved cells with amino acids, these cells were treated with MHY1485 or rapamycin immediately after starvation prior to addition of amino acids. The drugs were left in media with amino acids for 30 min to evaluate their effects on mTORC1 activation in the presence of the TTI1 mutants. As reported previously, MHY1485 (10 μM) treatment increased mTOR and S6K phosphorylation, whereas rapamycin (0.1 μM) decreased the phosphorylation levels25 in control and wild-type cells (Figures 4F–4H).
MHY1485 treatment increased mTOR/S6K phosphorylation in p.Ser838Leu variant HEK293T cells similar to control or wild-type levels. Rapamycin treatment decreased the high mTOR/S6K phosphorylation levels in p.Leu767Ser and p.Asp921Asn cells (Figures 4F–4H). Overall, treatments of TTI1 mutant cells with mTOR drugs, MHY1485, or rapamycin were able to partially normalize the altered mTOR/S6K phosphorylation levels observed in these cells.
TTI1 variants and cell cycle progression
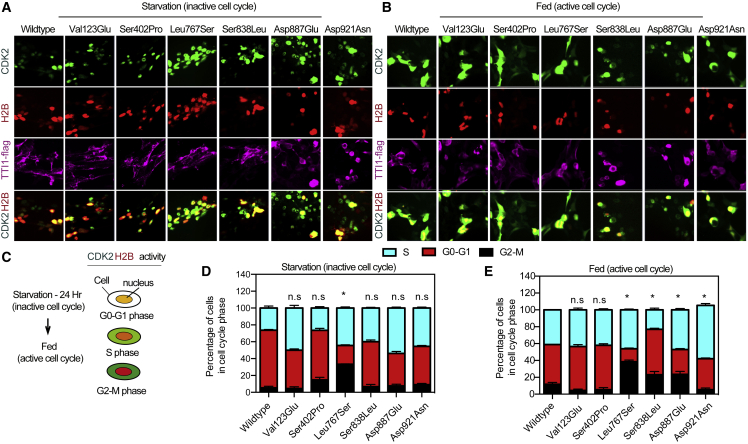
Several studies have reported that mTOR controls the G1-phase of the cell cycle progression and the G2/M phase through its cell growth effectors S6K1 and 4EBP1.38,39,40 mTORC1 is also hypothesized to regulate the activity of cyclin-dependent kinase (CDK) and glycogen synthase kinase 3 during cell cycle progression.40 A dual-reporter system with nuclear localized H2B-mCherry and a GFP-tagged CDK2 substrate, DNA helicase B (DHB-GFP),26 was used to address the effect of TTI1 variants on cell cycle progression. During the G1 to S transition, CDK2 phosphorylates DHB-GFP, which is subsequently translocated from the nucleus to the cytoplasm. The buildup of cytoplasmic DHB-GFP occurs until mitosis.41 Cells co-expressing Flag-TTI1 and CDK-DHB dual-reporter were serum deprived for 24 h to synchronize the cells in G0 phase (Figures 5A–5C) followed by stimulation with serum for 12 h.26
Figure 5.
TTI1 likely pathogenic variants affect cell cycle progression
(A) Representative images of starved cells co-expressing TTI1-flag (wild-type or variant, purple), CDK2 sensor-GFP (green), and H2B-RFP (red).
(B) Representative images of re-fed starved cells to stimulate cell cycle progression.
(C) Schematic of CDK2 activity and colocalization with H2B at different phases of the cell cycle.
(D and E) Graphical representative of the percentage of the different phases of the cell cycle in starved cells (D) and re-fed cells (E) of images in (A) and (B) (n = 4).
Data in (D) and (E) are mean ± SEM of experiments performed, ∗p < 0.05, n.s., p > 0.05, ANOVA with Tukey-Kramer post-hoc test compared with WT.
The analysis of different cell-cycle phases are consistent with cells with the p.Leu767Ser variant displaying a significantly reduced G0-G1 phase (25%) compared to wild-type (70%) (Figure 5D). After stimulation with serum following starvation, wild-type TTI1 cells displayed about 10% G2-M, 50% G0-G1 phase, and 40% S phase (Figure 5E). TTI1 p.Leu767Ser cells had significantly increased (40%) G2-M phase, consistent with an increase in mitotic phase compared to wild-type. Most cells with the p.Ser838Leu TTI1 variant were observed to be in the G0-G1 phase (∼60%), consistent with significant delays in G0-G1 phase progression. Most TTI1 p.Asp921Asn cells were observed in the S phase (70%), consistent with active progression from G1 to S phase but a possible delay in the G2 to M phase (∼7%). Cells expressing TTI1 variant p.Ser402Pro displayed similar cell cycle progression as in wild-type.
TTI1-PIKK mediated UV-induced DNA damage response and repair
The PIKK family proteins, including ATR and ATM kinases, are essential for cellular response to DNA damage.6,42 Phosphorylation of ATR and ATM is induced by the presence of short single-stranded DNA gaps caused by UV damage. The activated ATR and ATM phosphorylate numerous DNA damage response and repair proteins, including checkpoint kinase 1 (CHK1), histone H2AX (H2A.X), and p53.6,7,8,42,43 It has also been shown that the TTT complex is essential for the expression, maturation, and stability of ATM and ATR in response to DNA damage.4,6,7,8 To evaluate DNA damage response in cells harboring TTI1 mutants, live cells were exposed to UV radiation and then analyzed for phosphorylation levels of ATM/ATR-mediated DNA damage response substrates at 60 min post-UV exposure (Figure S3A).
Wild-type TTI1 cells exhibited increased phosphorylation levels of ATM/ATR and their substrates (Chk1, p53, and H2A.X) when exposed to UV (UV+) compared to control cells without UV treatment (UV−) (Figure S3A). There was a significant decrease in the levels of ATM/ATR phosphorylation in TTI1 p.Ser402Pro cells with reduced CHK1/H2A.X phosphorylation and an increase in p53 phosphorylation. Although the p.Leu767Ser cells showed reduced ATR phosphorylation levels, the overall response in these cells was similar to wild-type. Cells transfected with the variants p.Ser838Leu and p.Asp921Asn showed reduced ATM/ATR/CHK1/H2A.X phosphorylation levels and significantly increased p53 phosphorylation levels (Figures S3B–S3F).
The effects of whether the altered UV-induced DNA damage response observed in the cells reflected DNA repair was investigated to further understand the effect of the TTI1 variants on DNA. The mammalian nucleotide excision repair pathway removes 6-4PP during UV-induced DNA damage.44 Hence, 6-4PP is generated after the DNA damage response. As expected, 6-4PP levels increased in wild-type cells in comparison to variant cells after UV exposure, with the amount significantly decreasing by 60% after 2 h of DNA repair (Figures S3G–S3H). Cells with p.Ser402Pro, p.Leu767Ser, and p.Asp921Asn mutants exhibited significantly increased levels of 6-4PP when exposed to UV (UV 0 h) than wild-type, consistent with the variants increasing sensitivity to UV-induced DNA damage. This effect was not significant for p.Ser838Leu (55%).
Evaluation of PIKK levels and ATM activity in a lymphoblastoid cell line from individual 4 (F3.II-2)
A lymphoblastoid cell line was made from the blood of individual 4 and this showed both a low level of ATM and a reduced level of ATM activity/signaling compared to wild-type. The level of signaling was indicated by the ability of ATM to phosphorylate targets Smc1 Ser966, KAP-1 Ser824, Nbn Ser343, and CREB Ser121 and the reduced activity/signaling was particularly notable for autophosphorylation of ATM Ser1981 (Figure S4A). We made additional LCLs with further blood samples from individual 4 and obtained the same results (data not shown). Despite the low level of ATM, a radiosensitivity assay showed that the level of chromosome damage at G2 was normal in peripheral lymphocytes from individual 4. These findings were the functional consequence of individual 4 being compound heterozygous for TTI1 missense variants p.Trp279Gly and p.Leu993Arg. The individual 4 LCLs showed a reduced level of TTI1, TTI2, and Telo2 on western blotting (Figure S4B). ATM and ATR levels were reduced on this blot, DNA-PKcs showed no definitive alteration, and other PIKK protein levels (SMG1, mTOR) appeared normal (Figure S4C). An LCL made from the maternal blood (heterozygous for the p.Trp279Gly variant) showed normal levels of ATM signaling activity (data not shown).
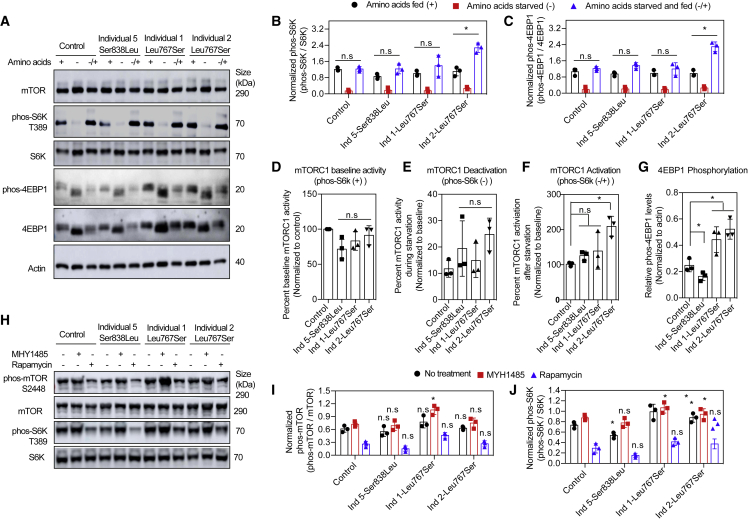
Evaluation of mTORC1 activity on cultured fibroblasts from individuals 1 (F1.IV-2), 2 (F1.IV-3), and 5 (F4.II-3)
mTORC1 activity was evaluated in fibroblasts of individuals 1 and 2 (siblings harboring the homozygous p.Leu767Ser missense variant) and those of individual 5 who was compound heterozygous for the missense variant p.Ser838Leu and the splice site variant c.2998+2T>C. mTORC1 activity was examined under different nutritional statuses (Figures 6A–6C) following the method described previously.30,31,33,34,35,36 Cells derived from individual 5 had a reduced mTORC1 basal activity (Figure 6D), whereas cells derived from individuals 1 and 2 had an activity similar to that of control cells under normal feeding (amino acids +) conditions. All three cell lines showed an equal deactivation of mTORC1 during amino acid starvation (amino acids −), similar to that observed in control cells (Figure 6E). Upon stimulation of starved cells with amino acids (amino acids −/+), cells from individuals 1 and 2 showed increased mTORC1 activation compared to the control subject (Figure 6F). Individual 5’s cells displayed similar mTORC1 activation as control cells. Overall, a decrease in phosphorylation of 4EBP1 (mTORC1 substrate) was observed in cells from individual 5, whereas cells from individuals 1 and 2 exhibited increased levels (Figure 6G). When treating these cells with the mTOR drug MHY1485, the low basal mTORC1 activity in individual’s 5 cells was reverted to normal levels, whereas there was an increase in basal mTORC1 activity in individuals 1 and 2. The treatment with rapamycin resulted in a decrease in mTORC1 activity levels in all three fibroblast lines (Figures 6H–6J).
Figure 6.
mTORC1 activation altered in cells from affected individuals
(A) Representative immunoblots showing mTORC1 activation (phosphorylation) status during normal amino acids fed (+), starvation (−), and starvation followed by amino acid supplementation (−/+).
(B and C) Quantification of phosphorylated S6K (phos-S6K, T389) and 4EBP1 (phos-4EBP1) normalized to total proteins of images in (A) (n = 3).
(D–G) Graphical representative of basal and amino acids stimulated mTORC1 activation.
(H) Representative immunoblots showing mTORC1 activation (phosphorylation) status in the absence (− −) or presence of MHY1485 (+ −) or Rapamycin (− +).
(I and J) Quantification of phosphorylated mTOR and S6K normalized to total proteins of images in (F) (n = 3).
Data in (B)–(G), (I), and (J) are mean ± SEM of experiments performed, ∗p < 0.05, n.s., p > 0.05, ANOVA with Tukey-Kramer post-hoc test compared with control.
Cell cycle progression in fibroblasts from the affected individuals was also examined using the CDK-H2B dual reporter. Time-lapse imaging of individual cells was performed with analysis of the different cell cycle phases over 24 h (Figure S5A). Quantification of the length between sequential mitoses showed an increase of the total cell-cycle length in cells of individual 5, shifting from 8–14 h to 16–24 h, but with no significant change in cells from individuals 1 and 2 compared to control cells (Figure S5B).
mTORC1 also regulates cell migration in addition to cell cycle and growth,25,39,45 and so evidence was sought as to whether alteration to mTORC1 levels had any impact on cell growth or migration. A scratch assay was performed to measure cell migration.46 The results were consistent with cells from individual 5 migrating more slowly than control cells (Figure S5C), whereas there was no significant change in the migration of cells from individuals 1 and 2 (Figure S5D). Treatment of individual 5 cells with MHY1485 increased the rate of cell migration (Figure S5E).
DNA damage response and repair were also evaluated in these fibroblasts. All 3 cell lines had reduced UV-induced DNA damage response compared to control cells (Figures S6A and S6B). Cells from individual 5 and individual 1 were more sensitive to UV damage as indicated by high levels of 6-4PP immediately after UV exposure (UV 0 h) (Figure S6C). Time course analysis of 6-4PP levels after UV exposure suggested that all cells have the ability to repair DNA damage. However, a significantly high residual level of 6-4PP was observed in individuals 1 and 5 cells after 2 h compared to control cells (Figures S6C and S6D); we concluded that DNA damage repair is delayed in cells from individuals 1 (Fam1.IV-2) and 5 (Fam4.II-3).
Mitochondrial activity
Studies have suggested that mTORC1 activation plays a role in mitochondria energy/ATP production for cell growth and the levels of ATP regulate the assembling of the ATP-dependent TTT-RUVBL complex interaction with mTORC1.31 We therefore examined mitochondrial activity, including mitochondrial oxygen consumption rate (OCR) and ATP production in the fibroblasts from affected individuals. A significant decrease in basal OCR and mitochondrial spare respiratory capacities was observed in cells from individuals 2 and 5 compared to controls (Figures S7A–S7C). ATP production was also significantly decreased in cells from individuals 2 and 5 cells but not in those from individual 1 (Figure S7D). Extracellular acidification rate (ECAR) was significantly increased in cells from individual 2 compared to control cells with no significant changes observed in cells from individuals 1 and 5 (Figure S7E). The decreased OCR and ATP production of the cells from individual 5 and individual 2 is consistent with these cells having diminished metabolic and respiratory capacity (summarized in Table S2).
Discussion
TELO2- and TTI2-related encephalopathies have been described in affected individuals presenting with autosomal-recessive moderate to severe ID and postnatal microcephaly, a movement disorder, or short stature. The third component of the TTT complex, TTI1, had been proposed as a potential candidate in two siblings with ID harboring the same homozygous TTI1 variant.47 This study brings together both clinical and functional details for 11 individuals inclusive of those from Karaca et al.47 (individuals 6 and 7) to provide functional evidence that pathogenic variants in TTI1 result in a rare AR syndromic ID, with ataxia and microcephaly. Postnatal microcephaly was present in 9 and short stature in 8 individuals with bi-allelic TTI1 missense variants. In 6 of 10 cases, a movement disorder including cerebellar ataxia and chorea involving the arms and legs is observed.
The clinical findings of the TTI1-related neurodevelopmental disorder were compared with those observed in the other two components of the TTT complex, TELO2 and TTI2.1,2,3,4,5 There were many features in common between the affected individuals described (Table 2). These include ID, microcephaly, and speech delay. Interestingly, the movement disorder (ataxia and chorea) observed in some of the affected individuals reported here is also prominent in TELO2-related neurodevelopmental disorder (You-Hoover-Fong syndrome; YHFS [MIM: 616954]). In the YHFS not only ataxia, but also paroxysmal movements of arms and feet have been described.
Table 2.
Comparison of clinical presentation of affected individuals with TELO2, TTI2, and TTI1 deficiencies and the AT phenotype
| YHFS | MRT39 | This study | AT | |
|---|---|---|---|---|
| Gene involved | TELO2 | TTI2 | TTI1 | ATM |
| MIM gene number | 611140 | 614426 | 614425 | 607585 |
| MIM phenotype number | 616954 | 615541 | – | 208900 |
| Mode of transmission | autosomal-recessive | autosomal-recessive | autosomal-recessive | autosomal-recessive |
| Number of individuals described | 10 | 10 | 11 | many |
| Gender | 3M, 7F | 5M, 5F | 6M, 5F | |
| ID (intellectual disability) | 100% (9/9) | 100% (9/9) | 100% (9/9) | – |
| Global delay | 100% (9/9) | 100% (8/8) | 100% (11/11) | – |
| language absence or delay | 100% (9/9) | 100% (8/8) | 100% (11/11) | – |
| locomotion absence or delay | 89% (8/9) | 80% (4/5) | 73% (8/11) | – |
| Microcephaly | 100% (10/10) | 100% (8/8) | 82% (9/11) | – |
| Short stature | 90% (9/10) | 87% (7/8) | 73% (8/11) | + |
| Dysmorphism | 90% (9/10) | 100% (8/8) | 91% (10/11) | – |
| Movement disorder | 89% (8/9) | 50% (4/8) | 64% (7/11) | + |
| Seizures | 30% (3/10) | 0% (0/8) | 30% (4/11) | + |
| Brain MRI anomalies | 14% (1/7) | 100% (4/4) | 60% (6/10) | + |
| Abnormal balance | 100% (8/8) | N/A | 22% (2/9) | + |
| Abnormal sleep pattern | 55% (5/9) | 42% (3/7) | 22% (2/9) | – |
| Behavioral abnomalities | 44% (4/9) | 87% (⅞) | 45% (6/11) | – |
| Congenital heart disease | 40% (4/10) | 0% (0/4) | 0% (0/11) | – |
| Scoliosis/kyphoscoliosis | 40% (4/10) | 85% (6/7) | 36% (5/11) | + |
| Brachydactyly and clinodactyly | 80% (8/10) | 100% (1/1) | 30% (3/9) | – |
| Toe syndactyly | 50% (5/10) | 100% (2/2) | 0% (0/11) | – |
| Hearing loss | 44% (4/9) | 0% (0/8) | 9% (1/11) | – |
| Cortical visual impairment | 50% (4/8) | 0% (0/2) | 9% (1/11) | + |
| Strabismus | 44% (4/9) | 50% (4/8) | 45% (5/11) | – |
| Reproductive organ anomalies | 42% (3/7) | 50% (1/2) | 50% (5/10) | + |
| Kidney malformation | 100% (1/1) | 0% (0/2) | 28% (2/7) | – |
| Cutaneous telangiectasia | N/A | N/A | 12.5% (⅛) | + |
| Café-au-lait spots | N/A | N/A | 18% (2/11) | + |
| Immune deficiencies | N/A | N/A | 0% (0/11) | + |
| Malignancies | not known | not known | not known | + |
TTI1 is part of the of the TTT complex that regulates the activity of the PIKK family of proteins. This complex is essential for mTORC1 assembly and plays an important role in the protein levels, maturation, and stability of ATM and ATR in response to DNA damage.6,7,8,42,43 mTOR alone is reported to exist in different oligomeric states, regulated by the TTT complex.23,31 The results of functional studies reported here are consistent with mTORC1 composition and activity being altered in the presence of TTI1 p.Val123Glu, p.Ser402Pro, p.Ser838Leu, and p.Asp887Glu.
mTORC1 promotes growth as a response to nutrient availability, specifically amino acids. mTORC1 phosphorylates several substrates including S6K1, PRAS40, and 4EBP1.30,31,34,35,36,45 In cells starved of amino acids, the normal phosphorylation of mTORC1 substrates decreases30,31,32,45 whereas the addition of amino acids to starved cells increases phosphorylation levels rapidly.30,31,32,45 The TTI1 variants identified in this cohort prevent the rapid recovery in cells grown in amino acid-depleted media after subsequent amino acid supplementation. These results are consistent with abnormal mTOR activation, either hyperactivation for p.Leu767Ser and p.Asp921Asn variants or hypoactivation for p.Ser402Pro and p.Ser838Leu variants. Treatment of TTI1 mutant cells with mTOR drugs, MHY1485 or rapamycin, corrected some of the mTORC1 alterations observed in these cells.
Several studies have suggested that mTOR controls cell cycles31,40,41 especially the G1-phase progression and G2/M phase through its cell growth effectors S6K1 and 4EBP1.38,39,40 Here we have shown that cell cycle progression is altered in half of the variants studied. UV-induced DNA damage response and repair is known to be mediated by TTT-PIKK activity. In the experiments reported here, all mutant cells demonstrated an oversensitivity to UV-induced DNA damage and exhibited a delayed DNA damage response. This raises the question of a possible cancer predisposition in affected individuals suffering from TTI1-related neurodevelopmental disorder. In this context, it is worth noting that no cancer has been diagnosed in our series.
The amount of the TTI1, TTI2, TELO2, ATR, and ATM assessed by plasma western blotting were decreased, similar to what it was observed in the other TTT-related disorders. Mutant TTI1 in HEK293T cells transfected with the missense mutations identified displayed significant impairment of TTI1 structure, mTOR interaction, mTORC1 or mTORC2 assembly, mTORC1 activation, cell cycle phase progression, UV DNA damage repair, cell migration, and/or mitochondria respiration (Table S2).
Functional studies on a lymphoblastoid cell line from individual 4, harboring p.Trp279Gly and p.Leu993Arg TTI1 variants, showed both a low level of ATM and a strongly reduced level of ATM activity compared to wild-type, alongside with reduced levels of TTI1, TTI2, and TELO2 accumulation.
Functional studies on fibroblasts from sibling individuals 1 and 2 with a homozygous p.Leu767Ser TTI1 variant showed increased mTORC1 activation upon stimulation of starved cells with amino acids, an increase in phosphorylation of mTORC1 substrate 4EBP1, reduced UV-induced DNA damage response, delayed DNA damage repair (only in individual 1), and diminished metabolic and respiratory capacity (only in individual 2).
Functional studies on fibroblasts from individual 5 with compound heterozygosity for p.Ser838Leu missense and c.2998+2T>C splicing variants showed significantly reduced mTORC1 basal activity, a decrease in phosphorylation of mTORC1 substrate 4EBP1, an increase of the total cell-cycle length, slower cell migration, reduced UV-induced DNA damage response, delayed DNA damage repair, and diminished metabolic and respiratory capacity. We cannot exclude that the level of signaling in individual 5’s cells may be proportional to the reduced level of ATM accumulation in those cells, i.e., a gene dosage effect.48 However, a lymphoblastoid cell line made from the maternal blood showed a normal level of ATM signaling and activity.
It is not clear how individuals with different TTI1 likely pathogenic variants that alter mTOR activation in opposite ways have overlapping phenotypes. Microcephaly linked to downregulation of mTOR is associated with changes in cell cycle length during late cortical development, increased cell death, altered differentiation of neural progenitors, and decreased Stat3 signaling, which is a target of mTORC1.49 Microcephaly caused by hyperactivation of mTORC1 is likely due to excessive apoptosis of cortical progenitors, accompanied by elevated expression of both hypoxia-inducible factor-1α and its downstream gene.50 Together, these studies and our results show that both inactivation and hyperactivation of mTOR signaling pathways alter brain development that is critically dependent on the TTT-mTORC1 functions. The detailed mechanism of how mTOR inactivation and hyperactivation contribute to microcephaly needs further investigations.
In conclusion, we report the observation of a TTI1-related neurodevelopmental disorder due to bi-allelic variants in TTI1. Functional analyses in transfected cells and in cell lines from some affected individuals support the pathogenicity of the described variant. We propose that individuals with bi-allelic pathogenic variants in TTI1, TTI2, and TELO2 genes have related autosomal-recessive disorders under the umbrella term of TTT-related disorders/syndromes or TTT-opathies.
Acknowledgments
The authors thank the affected individuals and their families for their participation in this study. This work was supported by UM1 HG006542 (J.R.L.) from the National Human Genome Research Institute (NHGRI)/National Heart, Lung, and Blood Institute (NHLBI) to the Baylor-Hopkins Center for Mendelian Genomics (BHCMG), by an Australian NHMRC Center for Research Excellence Grant APP 1117394 (Transforming the Genomic Diagnosis and Management of Severe Neurocognitive Disorders) to T.R., Y.Z., C.Y.N., C.-A.E., and M.F.B., and also by grants from the NIH/NINDS (NS099362 to G.K.E.U. and R35 NS105078 to J.R.L.). S.E.A. was partially supported by the ChildCare Foundation and an ERC grant. T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Disease. Data for family 4, individual 5 were obtained by the Duke Genome Sequencing clinic supported by the Duke University Health System (Duke Pro00032301 – Genomic Study of Disorders of Unknown Etiology). Sequencing and analysis for individual 10 was funded by the NHGRI, NHLBI, and National Eye Institute (NEI) grant UM1 HG008900, NHGRI grant R01 HG009141, and by the Chan Zuckerberg Initiative to the Rare Genomes Project. G.S.S. is funded by a CR-UK Programme grant (C17183/A23303). This study is dedicated to Professor Lionel van Maldergem, who died on July 10, 2021 during this ongoing international collaboration.
Declaration of interests
S.E.A. is a co-founder and CEO of Medigenome, Swiss Institute of Genomic Medicine, and serves in the Scientific Advisory Board of the “Imagine” Institute in Paris. The Department of Medical and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing conducted at Baylor Genetics Laboratories. J.R.L. has stock ownership in 23andMe; is a paid consultant for Regeneron Genetics Center; and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, genomic disorders, and bacterial genomic fingerprinting, and serves on the Scientific Advisory Board of Baylor Genomics.
Published: January 31, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2023.01.006.
Contributor Information
Margaux Serey-Gaut, Email: sereymargaux@gmail.com.
Stylianos E. Antonarakis, Email: stylianos.antonarakis@unige.ch.
Web resources
OMIM, https://www.omim.org/
Supplemental information
Data and code availability
The datasets (FASTQ files) supporting the current study have not been deposited in a public repository because of confidentiality of medical diagnostic tests, but are available from the corresponding author on request.
References
- 1.Kaizuka T., Hara T., Oshiro N., Kikkawa U., Yonezawa K., Takehana K., Iemura S.I., Natsume T., Mizushima N. Tti1 and Tel2 are critical factors in mammalian target of rapamycin complex assembly. J. Biol. Chem. 2010;285:20109–20116. doi: 10.1074/jbc.M110.121699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hurov K.E., Cotta-Ramusino C., Elledge S.J. A genetic screen identifies the Triple T complex required for DNA damage signaling and ATM and ATR stability. Genes Dev. 2010;24:1939–1950. doi: 10.1101/gad.1934210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rao F., Cha J., Xu J., Xu R., Vandiver M.S., Tyagi R., Tokhunts R., Koldobskiy M.A., Fu C., Barrow R., et al. Inositol pyrophosphates mediate the DNA-PK/ATM-p53 cell death pathway by regulating CK2 phosphorylation of Tti1/Tel2. Mol. Cell. 2014;54:119–132. doi: 10.1016/j.molcel.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goto G.H., Ogi H., Biswas H., Ghosh A., Tanaka S., Sugimoto K. Two separate pathways regulate protein stability of ATM/ATR-related protein kinases Mec1 and Tel1 in budding yeast. PLoS Genet. 2017;13:e1006873. doi: 10.1371/journal.pgen.1006873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugimoto K. Branching the Tel2 pathway for exact fit on phosphatidylinositol 3-kinase-related kinases. Curr. Genet. 2018;64:965–970. doi: 10.1007/s00294-018-0817-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abraham R.T. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 7.Bakkenist C.J., Kastan M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 8.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 9.You J., Sobreira N.L., Gable D.L., Jurgens J., Grange D.K., Belnap N., Siniard A., Szelinger S., Schrauwen I., Richholt R.F., et al. A syndromic intellectual disability disorder caused by variants in TELO2, a gene encoding a component of the TTT complex. Am. J. Hum. Genet. 2016;98:909–918. doi: 10.1016/j.ajhg.2016.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moosa S., Altmüller J., Lyngbye T., Christensen R., Li Y., Nürnberg P., Yigit G., Vogel I., Wollnik B. Novel compound heterozygous mutations in TELO2 in a patient with severe expression of You-Hoover-Fong syndrome. Mol. Genet. Genomic Med. 2017;5:580–584. doi: 10.1002/mgg3.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciaccio C., Duga V., Pantaleoni C., Esposito S., Moroni I., Pinelli M., Castello R., Nigro V., Chiapparini L., D'Arrigo S., TUDP Study Group Milder presentation of TELO2-related syndrome in two sisters homozygous for the p.Arg609His pathogenic variant. Eur. J. Med. Genet. 2021;64:104116. doi: 10.1016/j.ejmg.2020.104116. [DOI] [PubMed] [Google Scholar]
- 12.Najmabadi H., Hu H., Garshasbi M., Zemojtel T., Abedini S.S., Chen W., Hosseini M., Behjati F., Haas S., Jamali P., et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- 13.Langouët M., Saadi A., Rieunier G., Moutton S., Siquier-Pernet K., Fernet M., Nitschke P., Munnich A., Stern M.H., Chaouch M., Colleaux L. Mutation in TTI2 reveals a role for triple T complex in human brain development. Hum. Mutat. 2013;34:1472–1476. doi: 10.1002/humu.22399. [DOI] [PubMed] [Google Scholar]
- 14.Ziegler A., Bader P., McWalter K., Douglas G., Houdayer C., Bris C., Rouleau S., Coutant R., Colin E., Bonneau D. Confirmation that variants in TTI2 are responsible for autosomal recessive intellectual disability. Clin. Genet. 2019;96:354–358. doi: 10.1111/cge.13603. [DOI] [PubMed] [Google Scholar]
- 15.Picher-Martel V., Labrie Y., Rivest S., Lace B., Chrestian N. Whole-exome sequencing identifies homozygous mutation in TTI2 in a child with primary microcephaly: a case report. BMC Neurol. 2020;20:58. doi: 10.1186/s12883-020-01643-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang R., Han S., Liu H., Khan A., Xiaerbati H., Yu X., Huang J., Zhang X. Novel compound heterozygous mutations in TTI2 cause syndromic intellectual disability in a chinese family. Front. Genet. 2019;10:1060. doi: 10.3389/fgene.2019.01060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sundercombe S.L., Berbic M., Evans C.A., Cliffe C., Elakis G., Temple S.E.L., Selvanathan A., Ewans L., Quayum N., Nixon C.Y., et al. Clinically responsive genomic analysis pipelines: elements to improve detection rate and efficiency. J. Mol. Diagn. 2021;23:894–905. doi: 10.1016/j.jmoldx.2021.04.007. [DOI] [PubMed] [Google Scholar]
- 19.Kim Y., Park J., Joo S.Y., Kim B.G., Jo A., Lee H., Cho Y. Structure of the human TELO2-TTI1-TTI2 complex. J. Mol. Biol. 2022;434:167370. doi: 10.1016/j.jmb.2021.167370. [DOI] [PubMed] [Google Scholar]
- 20.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 22.Delgado J., Radusky L.G., Cianferoni D., Serrano L. FoldX 5.0: working with RNA, small molecules and a new graphical interface. Bioinformatics. 2019;35:4168–4169. doi: 10.1093/bioinformatics/btz184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain A., Arauz E., Aggarwal V., Ikon N., Chen J., Ha T. Stoichiometry and assembly of mTOR complexes revealed by single-molecule pulldown. Proc. Natl. Acad. Sci. USA. 2014;111:17833–17838. doi: 10.1073/pnas.1419425111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jain A., Liu R., Ramani B., Arauz E., Ishitsuka Y., Ragunathan K., Park J., Chen J., Xiang Y.K., Ha T. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473:484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rakhmanova V., Jin M., Shin J. Inhibition of mast cell function and proliferation by mTOR activator MHY1485. Immune Netw. 2018;18:e18. doi: 10.4110/in.2018.18.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spencer S.L., Cappell S.D., Tsai F.C., Overton K.W., Wang C.L., Meyer T. The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell. 2013;155:369–383. doi: 10.1016/j.cell.2013.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verhagen M.M.M., Last J.I., Hogervorst F.B.L., Smeets D.F.C.M., Roeleveld N., Verheijen F., Catsman-Berrevoets C.E., Wulffraat N.M., Cobben J.M., Hiel J., et al. Presence of ATM protein and residual kinase activity correlates with the phenotype in ataxia-telangiectasia: a genotype-phenotype study. Hum. Mutat. 2012;33:561–571. doi: 10.1002/humu.22016. [DOI] [PubMed] [Google Scholar]
- 28.Schon K., van Os N.J.H., Oscroft N., Baxendale H., Scoffings D., Ray J., Suri M., Whitehouse W.P., Mehta P.R., Everett N., et al. Genotype, extrapyramidal features, and severity of variant ataxia-telangiectasia. Ann. Neurol. 2019;85:170–180. doi: 10.1002/ana.25394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D.H., Sarbassov D.D., Ali S.M., King J.E., Latek R.R., Erdjument-Bromage H., Tempst P., Sabatini D.M. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 31.Kim S.G., Hoffman G.R., Poulogiannis G., Buel G.R., Jang Y.J., Lee K.W., Kim B.Y., Erikson R.L., Cantley L.C., Choo A.Y., Blenis J. Metabolic stress controls mTORC1 lysosomal localization and dimerization by regulating the TTT-RUVBL1/2 complex. Mol. Cell. 2013;49:172–185. doi: 10.1016/j.molcel.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarbassov D.D., Ali S.M., Sengupta S., Sheen J.H., Hsu P.P., Bagley A.F., Markhard A.L., Sabatini D.M. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 33.Saxton R.A., Sabatini D.M. mTOR Signaling in growth, metabolism, and disease. Cell. 2017;169:361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 34.Tang H., Hornstein E., Stolovich M., Levy G., Livingstone M., Templeton D., Avruch J., Meyuhas O. Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol. Cell Biol. 2001;21:8671–8683. doi: 10.1128/MCB.21.24.8671-8683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abu-Remaileh M., Wyant G.A., Kim C., Laqtom N.N., Abbasi M., Chan S.H., Freinkman E., Sabatini D.M. Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science. 2017;358:807–813. doi: 10.1126/science.aan6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sancak Y., Peterson T.R., Shaul Y.D., Lindquist R.A., Thoreen C.C., Bar-Peled L., Sabatini D.M. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schubert-Bast S., Rosenow F., Klein K.M., Reif P.S., Kieslich M., Strzelczyk A. The role of mTOR inhibitors in preventing epileptogenesis in patients with TSC: Current evidence and future perspectives. Epilepsy Behav. 2019;91:94–98. doi: 10.1016/j.yebeh.2018.05.039. [DOI] [PubMed] [Google Scholar]
- 38.Cuyàs E., Corominas-Faja B., Joven J., Menendez J.A. Cell cycle regulation by the nutrient-sensing mammalian target of rapamycin (mTOR) pathway. Methods Mol. Biol. 2014;1170:113–144. doi: 10.1007/978-1-4939-0888-2_7. [DOI] [PubMed] [Google Scholar]
- 39.Fingar D.C., Richardson C.J., Tee A.R., Cheatham L., Tsou C., Blenis J. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol. Cell Biol. 2004;24:200–216. doi: 10.1128/MCB.24.1.200-216.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramírez-Valle F., Badura M.L., Braunstein S., Narasimhan M., Schneider R.J. Mitotic raptor promotes mTORC1 activity, G(2)/M cell cycle progression, and internal ribosome entry site-mediated mRNA translation. Mol. Cell Biol. 2010;30:3151–3164. doi: 10.1128/MCB.00322-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoon K.J., Ringeling F.R., Vissers C., Jacob F., Pokrass M., Jimenez-Cyrus D., Su Y., Kim N.S., Zhu Y., Zheng L., et al. Temporal control of mammalian cortical neurogenesis by m(6)A methylation. Cell. 2017;171:877–889.e17. doi: 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maréchal A., Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013;5:a012716. doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ray A., Blevins C., Wani G., Wani A.A. ATR- and ATM-mediated DNA damage response is dependent on excision repair assembly during G1 but not in S phase of cell cycle. PLoS One. 2016;11:e0159344. doi: 10.1371/journal.pone.0159344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lo H.L., Nakajima S., Ma L., Walter B., Yasui A., Ethell D.W., Owen L.B. Differential biologic effects of CPD and 6-4PP UV-induced DNA damage on the induction of apoptosis and cell-cycle arrest. BMC Cancer. 2005;5:135. doi: 10.1186/1471-2407-5-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169:361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 46.Liang C.C., Park A.Y., Guan J.L. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 47.Karaca E., Harel T., Pehlivan D., Jhangiani S.N., Gambin T., Coban Akdemir Z., Gonzaga-Jauregui C., Erdin S., Bayram Y., Campbell I.M., et al. Genes that affect brain structure and function identified by rare variant analyses of mendelian neurologic disease. Neuron. 2015;88:499–513. doi: 10.1016/j.neuron.2015.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lupski J.R. Biology in balance: human diploid genome integrity, gene dosage, and genomic medicine. Trends Genet. 2022;38:554–571. doi: 10.1016/j.tig.2022.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cloëtta D., Thomanetz V., Baranek C., Lustenberger R.M., Lin S., Oliveri F., Atanasoski S., Rüegg M.A. Inactivation of mTORC1 in the developing brain causes microcephaly and affects gliogenesis. J. Neurosci. 2013;33:7799–7810. doi: 10.1523/JNEUROSCI.3294-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kassai H., Sugaya Y., Noda S., Nakao K., Maeda T., Kano M., Aiba A. Selective activation of mTORC1 signaling recapitulates microcephaly, tuberous sclerosis, and neurodegenerative diseases. Cell Rep. 2014;7:1626–1639. doi: 10.1016/j.celrep.2014.04.048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets (FASTQ files) supporting the current study have not been deposited in a public repository because of confidentiality of medical diagnostic tests, but are available from the corresponding author on request.