Abstract
The sequencing of modern and ancient genomes from around the world has revolutionized our understanding of human history and evolution. However, the problem of how best to characterize ancestral relationships from the totality of human genomic variation remains unsolved. Here, we address this challenge with non-parametric methods that enable us to infer a unified genealogy of modern and ancient humans. This compact representation of multiple datasets explores the challenges of missing and erroneous data and uses ancient samples to constrain and date relationships. We demonstrate the power of the method to recover relationships between individuals and populations, as well as to identify descendants of ancient samples. Finally, we introduce a simple non-parametric estimator of ancestor geographical location that recapitulates key events in human history.
One-Sentence Summary:
The largest genealogy of modern and ancient genomes yet constructed delivers insights into human history and evolution.
Our ability to determine relationships among individuals, populations and species is being transformed by population-scale biobanks of medical samples (1, 2), collections of thousands of ancient genomes (3), and efforts to sequence millions of eukaryotic species for comparative genomic analyses (4). Such relationships, and the resulting distributions of genetic and phenotypic variation, reflect the complex set of selective, demographic and molecular processes and events that have shaped species such as our own (5–8).
However, learning about evolutionary events and processes from the totality of genomic variation, in humans or other species, is challenging. Combining information from multiple data sets, even within a species, is technically demanding: discrepancies between cohorts due to error (9), differing sequencing techniques (10, 11) and variant processing (12) can lead to noise that can easily obscure genuine signal. Furthermore, few tools can cope with the vast data sets that arise from the combination of multiple sources (13). Also, statistical analysis typically relies on data reduction techniques (14, 15) or the fitting of parametric models (16–19), which may provide an incomplete picture of the complexities of evolutionary history. Finally, data access and governance restrictions often limit the ability to combine data sources (20).
The succinct tree sequence data structure provides a potential solution to many of these problems (13, 21). Tree sequences extend the fundamental concept of a phylogenetic tree to multiple correlated trees along the genome, necessary when considering genealogies within recombining organisms (22). Importantly, the tree sequence, and the mapping of mutation events to it, reflects the totality of what is knowable about genealogical relationships and the evolutionary history of individual variants. A tree sequence is defined as a graph with a set of nodes representing sampled chromosomes and ancestral haplotypes, edges connecting nodes representing lines of descent, and variable sites containing one or more mutations mapped onto the edges (Fig. 1A). Recombination events in the ancestral history of the sample create different edges and thus distinct, but highly correlated trees along the genome. Tree sequences can not only be used to compress genetic data (13), but also lead to highly efficient algorithms for calculating population genetic statistics (23).
Fig. 1. Schematic overview and validation of the inference methodology.
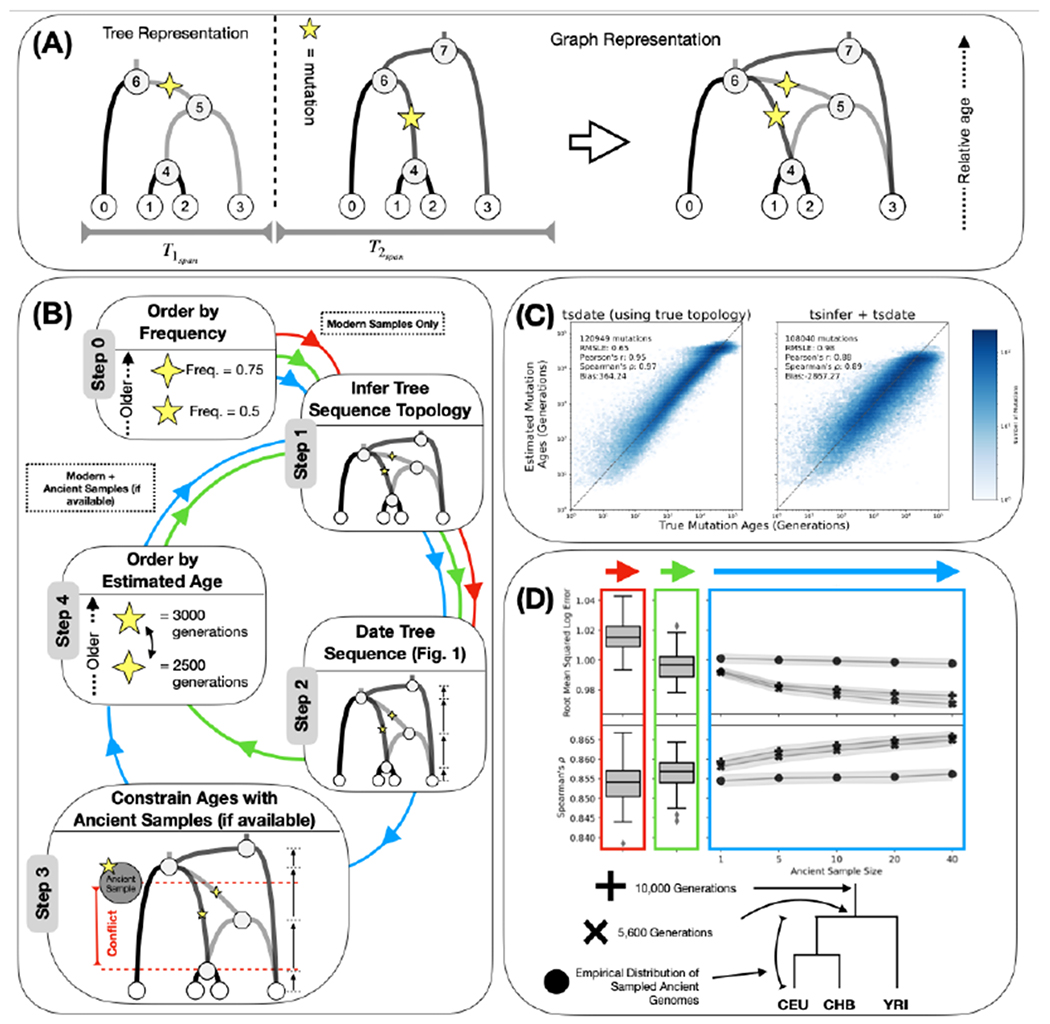
(A) An example tree sequence topology with four samples (nodes 0-3), two marginal trees, four ancestral haplotypes (nodes 4-7), and two mutations. Tspan measures the genomic span of each marginal tree topology, with the dotted line indicating the location of a recombination event. The graph representation is equivalent to the tree representation. (B) Schematic representation of the inference methodology. Step 0: alleles are ordered by frequency; the mutation represented by the four-point star is considered to be older. Step 1: the tree sequence topology is inferred with tsinfer using modern samples. Step 2: the tree sequence is dated with tsdate. Step 3: node date estimates are constrained with the known age of ancient samples. Step 4: ancestral haplotypes are reordered by the estimated age of their focal mutation; the five pointed star mutation is now inferred to be older. The algorithm returns to Step 1 to re-infer the tree sequence topology with ancient samples. Arrows refer to modes of operation: Steps 0, 1 and 2 only (red); Steps 0, 1, 2, 4, 1, and 2 (green) and Steps 0, 1, 2, 3, 4, 1, 2 (blue) (24). (C) Scatter plots and accuracy metrics comparing simulated (x-axis) and inferred (y-axis) mutation ages from msprime neutral coalescent simulations, using tsdate with the simulated topology (left) and inferred topology from tsinfer (right). (D) Accuracy metrics, root-mean squared log error (top) and Spearman rank correlation coefficient (bottom), with modern samples only (first panel), after one round of iteration (second panel) and with increasing numbers of ancient samples (colored arrows as in panel B). Ancient samples from three eras of human history are considered as in the schematic (24).
A unified genealogy of modern and ancient human genomes
Here, we introduce, validate and apply non-parametric methods for inferring time-resolved tree sequences from multiple heterogeneous sources to efficiently infer a single, unified tree sequence of ancient and contemporary human genomes. We note that while humans are the focus of this study, the methods and approaches we introduce are valid for most recombining organisms.
To generate a unified genealogy of modern and ancient human genomes, we integrated data from three modern datasets: the 1000 Genomes Project (TGP) which contains 2,548 sequenced individuals from 26 populations (6), the Human Genome Diversity Project (HGDP), which consists of 929 sequenced individuals from 54 populations (8), and the Simons Genome Diversity Project (SGDP) with 278 sequenced individuals from 142 populations (7). 154 individuals appear in more than one of these datasets (24). In addition, we included data from three high-coverage sequenced Neanderthal genomes (25–27), a single Denisovan genome (28), and high coverage whole genome data from a nuclear family of four (a mother, a father, and their two sons with average coverage of 10.8x, 25.8x, 21.2x, and 25.3x) from the Afanasievo Culture, who lived ~4.6 thousand years ago (kya) in the Altai Mountains of Russia (Table S1). Finally, we used 3,589 published ancient samples from over 100 publications compiled by the Reich Laboratory (24) and three sequenced ancient samples: Loschbour, LBK-Stuttgart, and Ust’-Ishim (5, 29) to constrain allele age estimates. These ancient genomes were not included in the final tree sequence due to the lack of reliable phasing for the majority of samples.
We built a unified genealogy from these datasets using an iterative approach (Fig. 1B). We first merged the modern datasets and inferred a tree sequence for each autosome using tsinfer version 0.2 (24, 30). We then estimated the age of ancestral haplotypes with tsdate, a Bayesian approach that infers the age of ancestral haplotypes with good accuracy and scaling properties (Fig. 1C and figs. S1–S5) (24, 31). Note that tsdate can be used to date any valid tree sequence, not only those inferred by tsinfer. tsdate can also use ancient samples to improve date estimates (Fig. 1D). We identified 6,412,717 variants present in both ancient and modern samples. A lower bound on variant age is provided by the estimated archaeological date of the oldest ancient sample in which the derived allele is found. Where this was inconsistent with the initial inferred value (559,431, or 8.7% of variants) we used the archaeological date as the variant age.
Finally, we integrated the Afanasievo family and four archaic sequences with the modern samples and re-inferred the tree sequence. The Afanasievo family have high coverage and comparably reliable haplotype phasing and were included to demonstrate the ability of our approach to incorporate high quality ancient samples.
The integrated tree sequences of each autosome together contain 26,958,720 inferred ancestral haplotype fragments, 231,073,278 edges, 91,172,114 variable sites, and 245,631,834 mutations. We infer that 38.7% of variant sites require more than one change in allelic state in the tree sequence to explain the data. This may indicate either recurrent mutations or errors, all of which are represented by additional mutations in the tree sequence. If we discount mutations that are likely indicative of sequencing errors (24), we find that 13,513,873 sites contain at least two mutations affecting more than one sample, implying that up to 17.5% of variable sites could result from more than one ancestral mutation. A high proportion of sites with over ~100 mutations on chromosome 20 have sequencing or alignment quality issues as defined by the TGP accessibility mask (6) or are in minimal linkage disequilibrium to their surrounding sites (fig. S6), suggesting they are largely erroneous. Moreover, analysis of data simulated with an empirically-calibrated error profile and evaluation of enrichment of multiple mutations at sites with known elevated mutation rates, suggests that the majority of the multiple mutations we identify are likely explained by error, but a minority (c. 20%) are the result of genuine recurrence or back mutation (24). We chose to retain such sites so that our inferred tree sequences are lossless representations of the original data sources; however, future iterative approaches to the removal of such probable errors are likely to improve use cases such as imputation.
To characterize fine-scale patterns of relatedness between the 215 populations of the constituent datasets, we estimated the time to the most recent common ancestor (TMRCA) between pairs of haplotypes from these populations at the 122,637 distinct trees in the tree sequence of chromosome 20 (~300 billion pairwise TMRCAs). In this and other analyses we present data from this chromosome as it is representative of genome-wide patterns. After performing hierarchical clustering on the average pairwise TMRCA values, we find that samples do not cluster by data source (which would indicate artifacts), but reflect patterns of global relatedness (Fig. 2 and Interactive fig.S1). We conclude that our method of integrating datasets is therefore robust to biases introduced by different datasets.
Fig. 2. Clustered heatmap showing the average time to the most recent common ancestor (TMRCA) on chromosome 20 for haplotypes within pairs of the 215 populations in the HGDP, TGP, SGDP, and ancient samples.
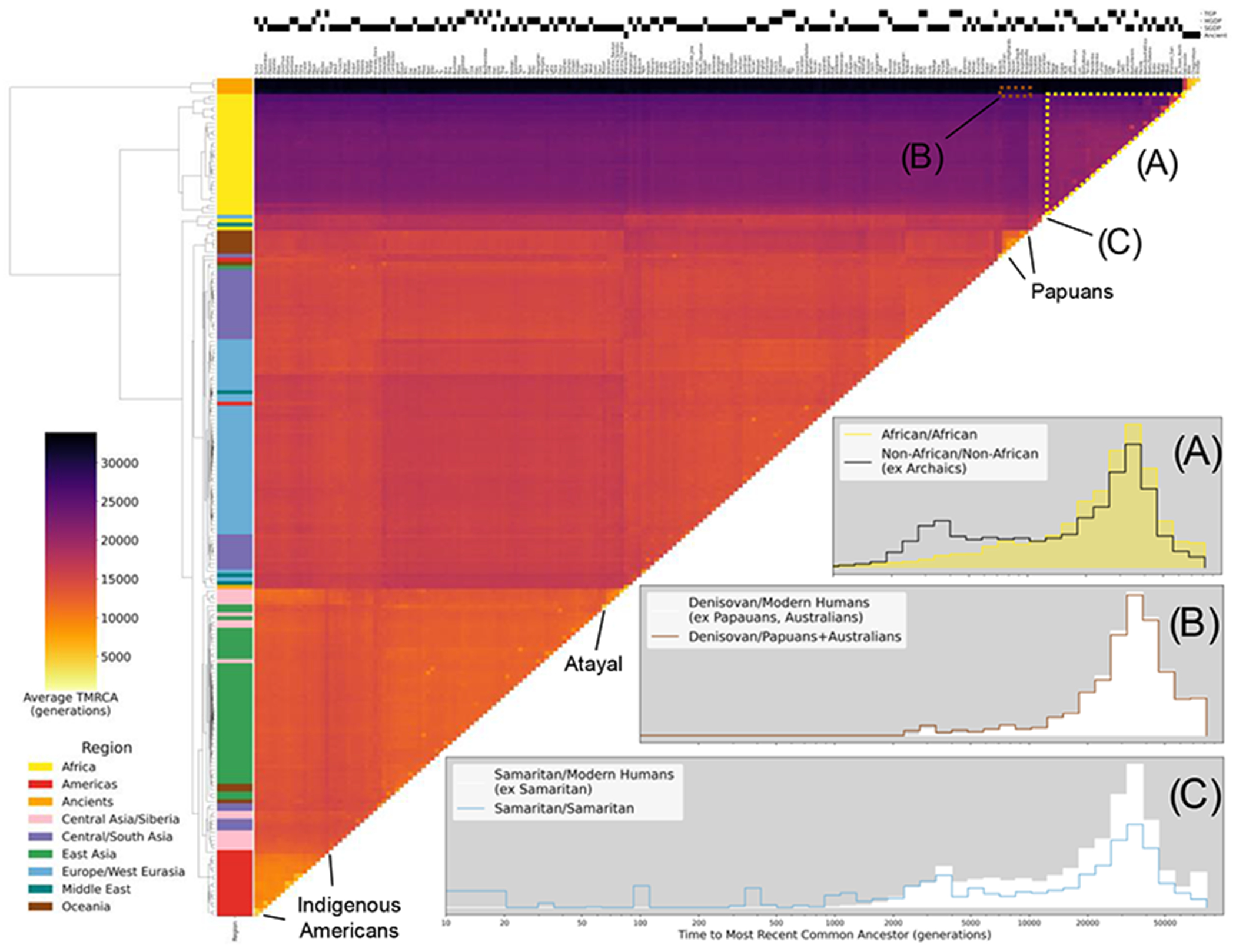
Each cell in the heatmap is colored by the logarithmic mean TMRCA of samples from the two populations. Hierarchical clustering of rows and columns has been performed using the UPGMA algorithm on the value of the pairwise average TMRCAs. Row colors are given by the region of origin for each population, as shown in the legend. The source of genomic samples for each population is indicated in the shaded boxes above the column labels. Three population relationships are highlighted using span-weighted histograms of the TMRCA distributions: (A) average distribution of TMRCAs between all non-African populations (black line) compared to African/African TMRCAs (solid yellow). (B) Denisovan and Papuan/Australian TMRCAs (solid line), compared to the Denisovan against all non- Archaic populations (solid white). This subtle but unique signal of elevated recent ancestry between the Denisovan and Papuans/Australians is particularly evident in Interactive fig. S1 at https://awohns.github.io/unified_genealogy/interactive_figure.html. (C) TMRCAs between the two Samaritan chromosomes (solid line), compared to the Samaritans/all other modern humans (solid white). Selected populations with particularly recent within-group TMRCAs are indicated. Duplicate samples appearing in more than one modern dataset are included in this analysis. Interactive Figure S1 is an interactive version of this figure and is available at: https://awohns.github.io/unified_genealogy/interactive_figure.html.
In this genealogy, numerous features of human history are immediately apparent, such as the deep divergence of archaic and modern humans, the effects of the Out of Africa event (Fig. 2A), and a subtle increase in Oceanian/Denisovan MRCA density from 2,000-5,000 generations ago (Fig. 2B). Multiple populations show recent within-group TMRCAs, suggestive of recent bottlenecks or consanguinity. The most extreme cases occur when a population consists of a single individual in our dataset, such as the Samaritan individual from the SGDP where we see a logarithmic average within-group TMRCA of ~ 1,000 generations, which is caused by multiple MRCAs at very recent times (Fig. 2C) and is consistent with a severe bottleneck and consanguinity in recent centuries (32). Indigenous populations in the Americas, an Atayal individual from Taiwan, and Papuans also exhibit particularly recent within-group TMRCAs (Fig. 2).
Tree sequence based analysis of descent from ancient sequences
To validate the dating methodology, we used simulations to show that the integration of ancient samples improves derived allele age estimates under a range of demographic histories (Fig. 1D). To provide empirical validation of the method, we tested how best to infer allele ages that are consistent with observations from ancient samples. Thus, we inferred and dated a tree sequence of TGP chromosome 20 (without using ancient samples) and compared the resulting point estimates and upper and lower bounds on allele age with results from GEVA (33) and Relate (34). This resulted in a set of 659,804 variant sites where all three methods provide an allele age estimate. Of these, 76,889 derived alleles are observed within the combined set of 3,734 ancient genome samples, thus putting a lower bound on allele age. The estimated allele ages from tsdate and Relate showed the greatest compatibility with ancient lower bounds, despite the fact that the mean age estimate from tsdate is more recent than that of Relate (Fig. 3A) (24).
Fig.3. Validation of inference methods using ancient samples.
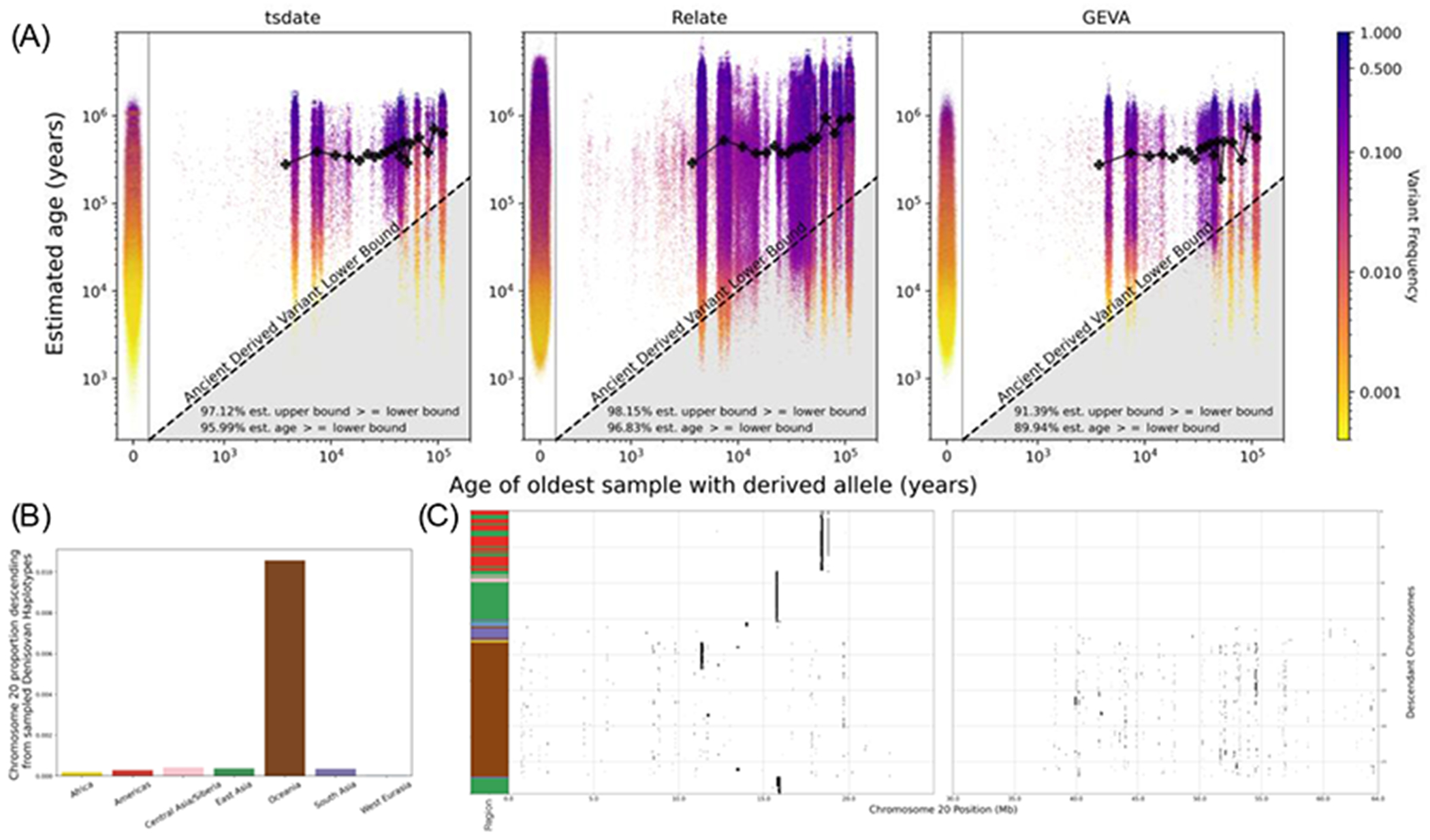
(A) Comparison of mutation age estimates from tsdate, Relate and GEVA with 3,734 ancient samples at 76,889 variants on chromosome 20 (note that Relate estimates ages separately for each population in which a variant is found). The radiocarbon- dated age of the oldest ancient sample carrying a derived allele at each variant site in the 1000 Genomes Project is used as the lower bound on the age of the mutation (diagonal lines). Mutations below this line have an estimated age that is inconsistent with the age of the ancient sample. Black lines on each plot show the moving average of allele age estimates from each method as a function of oldest ancient sample age. Plots to the left show the distribution of allele age estimates for modern-only variants from each respective method. Additional metrics are reported in each plot. (B) Percentage of chromosome 20 for modern samples in each region that is inferred to descend from Denisovan haplotypes, calculated with the genomic descent statistic (57). (C) Tracts of descent along chromosome 20 descending from Denisovan haplotypes in modern samples with at least 100 kilobases (kb) of total descent (colors as in Fig. 2).
Next, to assess the ability of the unified tree sequence to recover known relationships between ancient and modern populations, we considered the patterns of descent to modern samples from Archaic sequences on chromosome 20. Simulations indicate that this approach detects introgressed genetic material from Denisovans at a precision of ~86% with a recall of ~61% (24). We find descendants among non-archaic individuals, including both modern individuals and the Afanasievo, for 13% of the span of the Denisovan haplotypes on chromosome 20. The highest degree of descent among modern humans is in Oceanian populations as previously reported (28, 35–37) (Fig. 3B). However, the tree sequence also reveals how both the extent and nature of descent from Denisovan haplotypes varies greatly among modern humans. In particular, we find that Papuans and Australians carry multiple fragments of Denisovan haplotypes that are largely unique to the individual (Fig. 3C). In contrast, other modern descendants of Denisovan haplotypes have fewer blocks which are more widely shared, often between geographically distant individuals.
Examining the other ancient samples in the unified genealogy, we find the greatest amount of descent from the haplotypes of the Afanasievo family among individuals in Western Eurasia and South Asia (fig. S7A), consistent with findings from the genetically similar Yamnaya peoples and supporting a contemporaneous diffusion of Afanasievo-like genetic material via multiple routes (38). For the Neanderthals, where there are three samples of different ages, our simulations indicate that interpretation of the descent statistics is complicated by varying levels of precision and recall among lineages. Nevertheless, recall is highest at regions where introgressing and sampled archaic lineages share more recent common ancestry and precision is higher for the Vindija sample, which is more closely related to introgressing lineages. Examining patterns of descent from Vindija haplotypes across autosomes indicates that modern non-African groups carry similar levels of Vindija-like material (fig. S8), supporting suggestions that the proportions are similar between East Asians and West Eurasians (39) and inconsistent with other reports (26, 40).
Non-parametric inference of spatio-temporal dynamics in human history
Tree sequence based analysis of ancient samples demonstrates an ability to characterize patterns of recent descent. We developed a simple estimator of ancestor spatial location that uses the coordinates of descendants of a node, combined with the structure of the tree sequence, to provide an estimate of ancestors’ geographic position (24). Briefly, this is accomplished by determining the coordinates of a parent node in the tree sequence as the midpoint of its immediate children (24), an approach that performs well in simple simulations (fig. S9). The approach can use information on the location of ancient samples, though it does not attempt to capture the geographical plausibility of different locations and routes. The inferred locations are thus a model-free estimate of ancestors’ location, informed by the tree sequence topology and geographic distribution of samples.
We applied our method to the unified tree sequence of chromosome 20, excluding TGP individuals (which lack precise location information). We find that the inferred ancestor location recovers multiple key events in human history (Fig. 4 and Movie S1). Despite the fact that the geographic center of sampled individuals is in Central Asia, by 72 kya the average location of ancestral haplotypes is in Northeast Africa and remains there until the oldest common ancestors are reached. Indeed, the inferred geographic center of gravity of the 100 oldest ancestral haplotypes (which have an average age of ~2 million years) is located in Sudan at 19.4 N, 33.7 E. These findings reflect the depth of African lineages in the inferred tree sequence and are compatible with well-dated early modern human fossils from eastern and northern Africa (41, 42). We caution if we analyzed data from a grid sampling of populations in Africa the geographic center of gravity of independent lineages at different time depths would shift. In addition, migrations occurring within the last few thousand years (43, 44) mean that present day distributions of groups in Africa and elsewhere may not represent those of their ancestors, and thus we may have a distorted picture of ancient geographic distributions (45). Nevertheless, the deep tree structure is geographically centered in Africa in autosomal data, just as it is for mitochondrial DNA and Y chromosomes (46, 47).
Fig. 4. Visualization of the non-parametric estimator of ancestor geographic location for HGDP, SGDP, Neanderthal, Denisovan, and Afanasievo samples on chromosome 20.
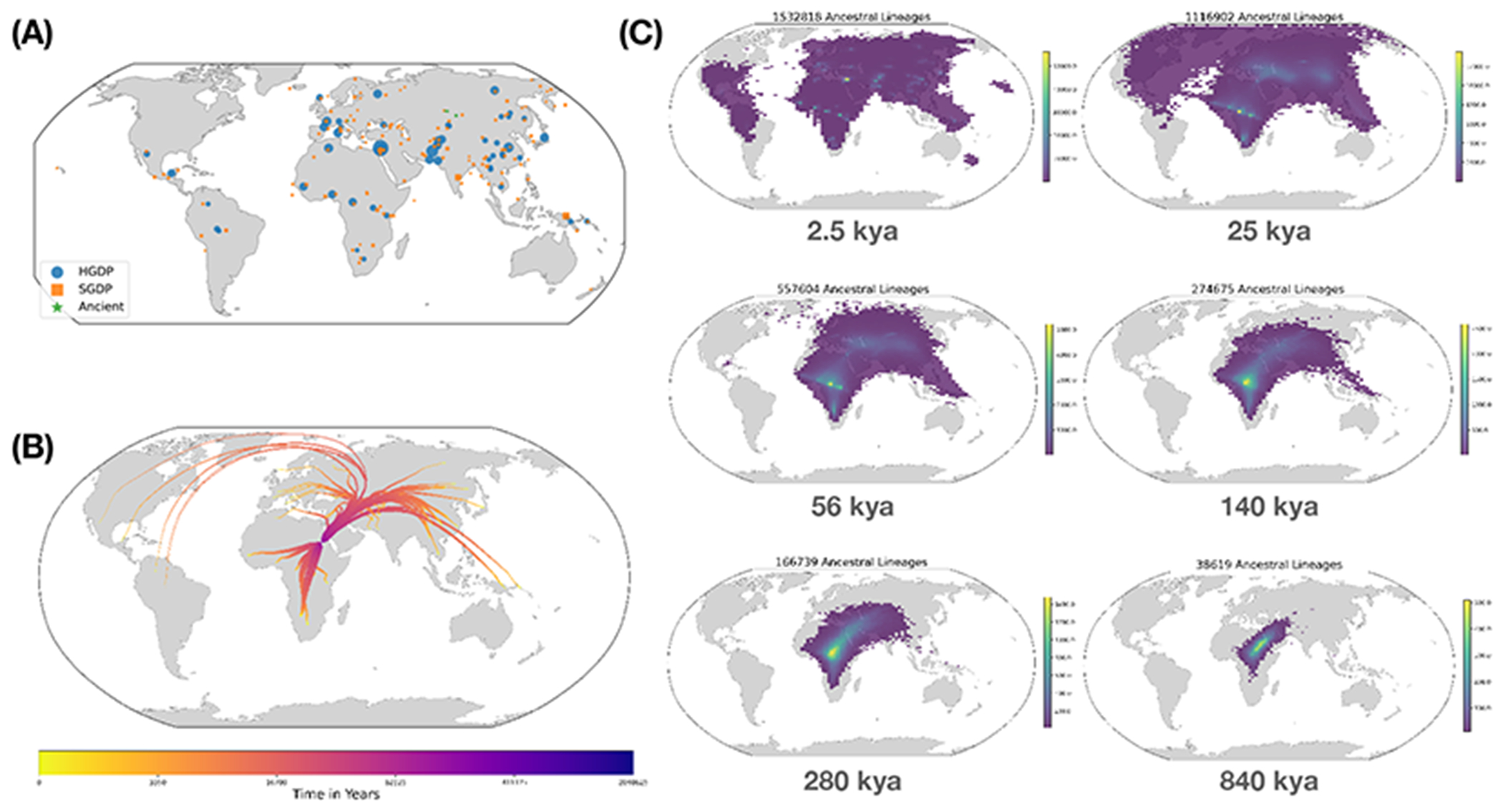
(A) Geographic location of samples used to infer ancestral geography. The size of each symbol is proportional to the number of samples in that population. (B) The average location of the ancestors of each HGDP population from time t=0 to ~2 million years ago. The width of lines is proportional to the number of ancestors of each population over time. The ancestor of a population is defined as an inferred ancestral haplotype with at least one descendant in that population. (C) 2d-histograms showing the inferred geographical location of HGDP ancestral lineages at six time-points. Histogram bins with fewer than 10 ancestors are not shown. Note that the geographic concentration of ancestors at more recent times is an artifact of uneven sampling and our geographic inference method.
By 280 kya, the estimated geographic center of human ancestors is still located in Africa, but many are also observed in the Middle East and Central Asia and a few are located in Papua New Guinea. At 140 kya, more ancestors are found in Papua New Guinea. This is almost 100 kya before the earliest documented human habitation of the region (48). However, our findings are potentially consistent with the proposed timescales of deeply diverged Denisovan lineages unique to Papuans (37) and possibly admixture with unsampled, “ghost” lineages. At 56 kya, some ancestral lineages are observed in the Americas, earlier than the estimated migration times to the Americas (49). This effect is possibly attributable to the presence of ancestors that predate the migration and did not live in the Americas, but whose descendants now exist solely in this region (50); the same effect may also explain observations from Papua New Guinea. Additional ancient samples and more sophisticated inference approaches are required to distinguish between these hypotheses, since there remains considerable uncertainty about the true age of any single ancestor (24). Nevertheless, these results demonstrate the ability of inference methods applied to tree sequences to capture key features of human history in a manner that does not require complex parametric modeling.
Discussion
A central theme in evolutionary biology is how best to represent and analyze genomic diversity to learn about the processes, forces and events that have shaped organismal history. Historically, many modeling approaches focus on the temporal behavior of individual mutation frequencies in idealized populations (51, 52). More recently, modelling techniques have shifted to focus on the genealogical history of sampled genomes and the correlation structures arising through recombination (22, 53). Critically, a single (albeit extremely complex) set of ancestral relationships exist that, coupled with how mutation events have altered genetic material through descent, describes what we observe today.
However, developing efficient methods for inferring the underlying genealogy has proved challenging (54, 55). The methods described here produce high quality dated genealogies that include thousands of modern and ancient samples. These genealogies cannot be entirely accurate, nevertheless, they enable a wealth of analyses that reveal features of human evolution (23, 56–60). That our highly simplistic geographic estimator captures key events suggests that more sophisticated approaches, coupled with the ongoing program of sequencing ancient samples, will continue to generate new insights into our history. Specifically, the methods developed here provide a framework for testing different models of human migration and demographic history, such as Neanderthal absorption models (61), using a parametric and explicitly spatial simulation framework. However, the accuracy of any ancestral geographic inference method will be limited when the distribution of sampled individuals does not reflect the location of the samples’ ancestors.
Our study also highlights the importance of accommodating genotype error and recurrent mutation in the analysis of genomic variation. While a large number of sites are inferred to carry multiple mutations, we find that the majority of these likely reflect genotype error and potentially errors arising from paralogy (particularly at sites requiring high numbers of mutations), though there remains a significant signal of recurrent mutation, as previously reported (62, 63). Similarly, we find some evidence for certain classes of error in ancient sequence leading to false “correction” of variant ages. We choose to retain all additional mutations in the analyses described in this paper, including those which are highly likely to reflect sequencing error, as this reflects the input data used to build the tree sequence and any effort to remove mutations corresponding to errors will itself introduce bias. We caution that the absolute ages we report have some degree of error, in part due to these errors in the sequencing datasets. Estimates from simulations show that genotype error may cause an upwards bias of up to 16% in age estimates derived from modern samples (fig. S3), but we also find that removing sites that are highly likely to be erroneous has a marginal effect on age estimates (fig. S10). Improving methods to detect and correct, or mitigate against the impact of genotype errors is an important direction for future research.
Because the tree sequence approach aims to capture the structure of human relationships and genomic diversity, it provides a principled basis for combining data from multiple different sources, not just correcting errors, but also enabling tasks such as imputing missing data. Although additional work is required to integrate other types of mutation, a reference tree sequence for human variation - along with the tools to use it appropriately (13, 23) - potentially represents a basis for harmonizing much larger and wider sets of genomic data sources and enabling cross data-source analyses. We note that reference tree sequences could also enable data sharing and preserve privacy in genomic analysis (20) through compression of cohorts against such a reference structure.
There exists room for improvement as well as new opportunities for genomic analyses that use the dated tree sequence structure. Our approach requires phased genomes, a particular challenge for ancient samples. However, it should be possible to use a diploid version of the matching algorithm in tsinfer to jointly solve phasing and imputation. This also has the potential to alleviate biases introduced by using modern and genetically distant reference panels for ancient samples (64). In addition, our approach to age inference within tsdate only provides an approximate solution to the cycles that are inherent in genealogical histories (65) and could be extended to model heterogeneity in mutation rates. There are also many possible approaches for improving the sophistication of spatio-temporal ancestor inference.
The unified genealogy presented in this work represents a foundation for building a comprehensive understanding of human genomic diversity, including both modern and ancient samples, which enables applications ranging from improving genome interpretation to deciphering our earliest roots. Although much work is still required to build the genealogy of everyone, the methods presented here provide a solution to this fundamental task.
Supplementary Material
Acknowledgments:
We thank the Oxford Big Data research computing team, specifically A. Huffman and R. Esnouf, and D. Lieberman, K. Lohse, and E. Castedo Ellerman for comments.
Funding:
This work was supported by Wellcome Trust grant 100956/Z/13/Z (to G.M.); the Li Ka Shing Foundation (to G.M.); the Robertson Foundation (to J.K.); the Rhodes Trust (to A.W.W.); NIH (NIGMS) grant GM100233 (to D.R.); the Paul Allen Foundation (to D.R.); the John Templeton Foundation grant 61220 (to D.R.), and the Howard Hughes Medical Institute (to D.R.). The computational aspects of this research were supported by the Wellcome Trust (Core Award 203141/Z/16/Z) and the NIHR Oxford BRC. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health.
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Competing interests: G.M. is a director of and shareholder in Genomics plc and a partner in Peptide Groove LLP.
Data and materials availability:
Newly reported sequencing data from the Afanasievo family is available from the European Nucleotide Archive, accession number PRJEB43093; phased variant data for the family is available from the European Variation Archive, accession number PRJEB46983. All publicly available datasets used in this paper are available from their original publications, tsinfer is deposited to Zenodo at doi:10.5281/zenodo.5168051 and available at https://tsinfer.readthedocs.io/ under the GNU General Public License v3.0 (30), tsdate is deposited to Zenodo at doi:10.5281/zenodo.5168040 and available at https://tsdate.readthedocs.io/ under the MIT License (31), and tskit is deposited to Zenodo at doi:10.5281/zenodo.5465773 and available at https://tskit.readthedocs.io/ under the MIT License (66). All code used to perform analyses in this paper is deposited to Zenodo at doi:10.5281/zenodo.5172104 and can be found at https://github.com/awohns/unified_genealogy_paper (67). Unified tree sequences of the HGDP, SGDP, and TGP autosomes are available from Zenodo at https://doi.org/10.5281/zenodo.5495535 (68). Unified tree sequences of the HGDP, SGDP, TGP, and high coverage ancient autosomes are available at https://doi.org/10.5281/zenodo.5512994 (69). Tree sequences were compressed using the tszip utility; see the documentation at https://tszip.readthedocs.io/ for further details.
References and Notes
- 1.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, Cortes A, Welsh S, Young A, Effingham M, McVean G, Leslie S, Allen N, Donnelly P, Marchini J, The UK Biobank resource with deep phenotyping and genomic data. Nature. 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, Taliun SAG, Corvelo A, Gogarten SM, Kang HM, Pitsillides AN, LeFaive J, Lee S, Tian X, Browning BL, Das S, Emde A-K, Clarke WE, Loesch DP, Shetty AC, Blackwell TW, Smith AV, Wong Q, Liu X, Conomos MP, Bobo DM, Aguet F, Albert C, Alonso A, Ardlie KG, Arking DE, Aslibekyan S, Auer PL, Barnard J, Barr RG, Barwick L, Becker LC, Beer RL, Benjamin EJ, Bielak LF, Blangero J, Boehnke M, Bowden DW, Brody JA, Burchard EG, Cade BE, Casella JF, Chalazan B, Chasman DI, Chen Y-DI, Cho MH, Choi SH, Chung MK, Clish CB, Correa A, Curran JE, Custer B, Darbar D, Daya M, de Andrade M, DeMeo DL, Dutcher SK, Ellinor PT, Emery LS, Eng C, Fatkin D, Fingerlin T, Forer L, Fornage M, Franceschini N, Fuchsberger C, Fullerton SM, Germer S, Gladwin MT, Gottlieb DJ, Guo X, Hall ME, He J, Heard-Costa NL, Heckbert SR, Irvin MR, Johnsen JM, Johnson AD, Kaplan R, Kardia SLR, Kelly T, Kelly S, Kenny EE, Kiel DP, Klemmer R, Konkle BA, Kooperberg C, Köttgen A, Lange LA, Lasky-Su J, Levy D, Lin X, Lin K-H, Liu C, Loos RJF, Garman L, Gerszten R, Lubitz SA, Lunetta KL, Mak ACY, Manichaikul A, Manning AK, Mathias RA, McManus DD, McGarvey ST, Meigs JB, Meyers DA, Mikulla JL, Minear MA, Mitchell BD, Mohanty S, Montasser ME, Montgomery C, Morrison AC, Murabito JM, Natale A, Natarajan P, Nelson SC, North KE, O’Connell JR, Palmer ND, Pankratz N, Peloso GM, Peyser PA, Pleiness J, Post WS, Psaty BM, Rao DC, Redline S, Reiner AP, Roden D, Rotter JI, Ruczinski I, Sarnowski C, Schoenherr S, Schwartz DA, Seo J-S, Seshadri S, Sheehan VA, Sheu WH, Shoemaker MB, Smith NL, Smith JA, Sotoodehnia N, Stilp AM, Tang W, Taylor KD, Telen M, Thornton TA, Tracy RP, Van Den Berg DJ, Vasan RS, Viaud-Martinez KA, Vrieze S, Weeks DE, Weir BS, Weiss ST, Weng L-C, Willer CJ, Zhang Y, Zhao X, Arnett DK, Ashley-Koch AE, Barnes KC, Boerwinkle E, Gabriel S, Gibbs R, Rice KM, Rich SS, Silverman EK, Qasba P, Gan W, Abe N, Almasy L, Ament S, Anderson P, Anugu P, Applebaum-Bowden D, Assimes T, Avramopoulos D, Barron-Casella E, Beaty T, Beck G, Becker D, Beitelshees A, Benos T, Bezerra M, Bis J, Bowler R, Broeckel U, Broome J, Bunting K, Bustamante C, Buth E, Cardwell J, Carey V, Carty C, Casaburi R, Castaldi P, Chaffin M, Chang C, Chang Y-C, Chavan S, Chen B-J, Chen W-M, Chuang L-M, Chung R-H, Comhair S, Cornell E, Crandall C, Crapo J, Curtis J, Damcott C, David S, Davis C, de las Fuentes L, DeBaun M, Deka R, Devine S, Duan Q, Duggirala R, Durda JP, Eaton C, Ekunwe L, El Boueiz A, Erzurum S, Farber C, Flickinger M, Frazar C, Fu M, Fulton L, Gao S, Gao Y, Gass M, Gelb B, Geng XP, Geraci M, Ghosh A, Gignoux C, Glahn D, Gong D-W, Goring H, Graw S, Grine D, Gu CC, Guan Y, Gupta N, Haessler J, Hawley NL, Heavner B, Herrington D, Hersh C, Hidalgo B, Hixson J, Hobbs B, Hokanson J, Hong E, Hoth K, Hsiung CA, Hung Y-J, Huston H, Hwu CM, Jackson R, Jain D, Jhun MA, Johnson C, Johnston R, Jones K, Kathiresan S, Khan A, Kim W, Kinney G, Kramer H, Lange C, Lange E, Lange L, Laurie C, LeBoff M, Lee J, Lee SS, Lee W-J, Levine D, Lewis J, Li X, Li Y, Lin H, Lin H, Lin KH, Liu S, Liu Y, Liu Y, Luo J, Mahaney M, N. T.-O. for P. M. (TOPMed) Consortium, Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature. 590, 290–299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reich D, Who We Are and How We Got Here: Ancient DNA and the New Science of the Human Past (Oxford University Press, Oxford, UK, 2018). [Google Scholar]
- 4.Lewin HA, Robinson GE, Kress WJ, Baker WJ, Coddington J, Crandall KA, Durbin R, Edwards SV, Forest F, Gilbert MTP, Goldstein MM, Grigoriev IV, Hackett KJ, Haussler D, Jarvis ED, Johnson WE, Patrinos A, Richards S, Castilla-Rubio JC, van Sluys M-A, Soltis PS, Xu X, Yang H, Zhang G, Earth BioGenome Project: Sequencing life for the future of life. Proc. Natl. Acad. Sci 115, 4325–4333 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lazaridis, Patterson N, Mittnik A, Renaud G, Mallick S, Kirsanow K, Sudmant PH, Schraiber JG, Castellano S, Lipson M, Berger B, Economou C, Bollongino R, Fu Q, Bos KI, Nordenfelt S, Li H, de Filippo C, Prüfer K, Sawyer S, Posth C, Haak W, Hallgren F, Fornander E, Rohland N, Delsate D, Francken M, Guinet J-M, Wahl J, Ayodo G, Babiker HA, Bailliet G, Balanovska E, Balanovsky O, Barrantes R, Bedoya G, Ben-Ami H, Bene J, Berrada F, Bravi CM, Brisighelli F, Busby GBJ, Cali F, Churnosov M, Cole DEC, Corach D, Damba L, van Driem G, Dryomov S, Dugoujon J-M, Fedorova SA, Gallego Romero I, Gubina M, Hammer M, Henn BM, Hervig T, Hodoglugil U, Jha AR, Karachanak-Yankova S, Khusainova R, Khusnutdinova E, Kittles R, Kivisild T, Klitz W, Kučinskas V, Kushniarevich A, Laredj L, Litvinov S, Loukidis T, Mahley RW, Melegh B, Metspalu E, Molina J, Mountain J, Näkkäläjärvi K, Nesheva D, Nyambo T, Osipova L, Parik J, Platonov F, Posukh O, Romano V, Rothhammer F, Rudan I, Ruizbakiev R, Sahakyan H, Sajantila A, Salas A, Starikovskaya EB, Tarekegn A, Toncheva D, Turdikulova S, Uktveryte I, Utevska O, Vasquez R, Villena M, Voevoda M, Winkler CA, Yepiskoposyan L, Zalloua P, Zemunik T, Cooper A, Capelli C, Thomas MG, Ruiz-Linares A, Tishkoff SA, Singh L, Thangaraj K, Villems R, Comas D, Sukernik R, Metspalu M, Meyer M, Eichler EE, Burger J, Slatkin M, Pääbo S, Kelso J, Reich D, Krause J, Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 513, 409–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.1000 Genomes Project Consortium, A global reference for human genetic variation. Nature. 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mallick S, Li H, Lipson M, Mathieson I, Gymrek M, Racimo F, Zhao M, Chennagiri N, Nordenfelt S, Tandon A, Skoglund P, Lazaridis I, Sankararaman S, Fu Q, Rohland N, Renaud G, Erlich Y, Willems T, Gallo C, Spence JP, Song YS, Poletti G, Balloux F, van Driem G, de Knijff P, Romero IG, Jha AR, Behar DM, Bravi CM, Capelli C, Hervig T, Moreno-Estrada A, Posukh OL, Balanovska E, Balanovsky O, Karachanak-Yankova S, Sahakyan H, Toncheva D, Yepiskoposyan L, Tyler-Smith C, Xue Y, Abdullah MS, Ruiz-Linares A, Beall CM, Di Rienzo A, Jeong C, Starikovskaya EB, Metspalu E, Parik J, Villems R, Henn BM, Hodoglugil U, Mahley R, Sajantila A, Stamatoyannopoulos G, Wee JTS, Khusainova R, Khusnutdinova E, Litvinov S, Ayodo G, Comas D, Hammer MF, Kivisild T, Klitz W, Winkler CA, Labuda D, Bamshad M, Jorde LB, Tishkoff SA, Watkins WS, Metspalu M, Dryomov S, Sukernik R, Singh L, Thangaraj K, Pääbo S, Kelso J, Patterson N, Reich D, The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 538, 201–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergström A, McCarthy SA, Hui R, Almarri MA, Ayub Q, Danecek P, Chen Y, Felkel S, Hallast P, Kamm J, Blanché H, Deleuze J-F, Cann H, Mallick S, Reich D, Sandhu MS, Skoglund P, Scally A, Xue Y, Durbin R, Tyler-Smith C, Insights into human genetic variation and population history from 929 diverse genomes. Science. 367, eaay5012 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belsare S, Levy-Sakin M, Mostovoy Y, Durinck S, Chaudhuri S, Xiao M, Peterson AS, Kwok P-Y, Seshagiri S, Wall JD, Evaluating the quality of the 1000 genomes project data. BMC Genomics. 20, 620 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi L, Guo Y, Dong C, Huddleston J, Yang H, Han X, Fu A, Li Q, Li N, Gong S, Lintner KE, Ding Q, Wang Z, Hu J, Wang D, Wang F, Wang L, Lyon GJ, Guan Y, Shen Y, Evgrafov OV, Knowles JA, Thibaud-Nissen F, Schneider V, Yu C-Y, Zhou L, Eichler EE, So K-F, Wang K, Long-read sequencing and de novo assembly of a Chinese genome. Nat. Commun. 7, 12065 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wenger M, Peluso P, Rowell WJ, Chang P-C, Hall RJ, Concepcion GT, Ebler J, Fungtammasan A, Kolesnikov A, Olson ND, Töpfer A, Alonge M, Mahmoud M, Qian Y, Chin C-S, Phillippy AM, Schatz MC, Myers G, DePristo MA, Ruan J, Marschall T, Sedlazeck FJ, Zook JM, Li H, Koren S, Carroll A, Rank DR, Hunkapiller MW, Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nature Biotechnology. 37, 1155–1162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang S, Kim E, Lee I, Marcotte EM, Systematic comparison of variant calling pipelines using gold standard personal exome variants. Scientific Reports. 5, 17875 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelleher J, Wong Y, Wohns AW, Fadil C, Albers PK, McVean G, Inferring whole-genome histories in large population datasets. Nature genetics. 51, 1330–1338 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cavalli-Sforza LL, Feldman MW, The application of molecular genetic approaches to the study of human evolution. Nature genetics. 33, 266–275 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Patterson N, Price AL, Reich D, Population Structure and Eigenanalysis. PLOS Genetics. 2, 1–20 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pritchard JK, Stephens M, Donnelly P, Inference of Population Structure Using Multilocus Genotype Data. Genetics. 155, 945–959 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawson DJ, Hellenthal G, Myers S, Falush D, Inference of Population Structure using Dense Haplotype Data. PLOS Genetics. 8, 1–16 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D, Ancient admixture in human history. Genetics. 192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pickrell JK, Pritchard JK, Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data. PLOS Genetics. 8, 1–17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonomi L, Huang Y, Ohno-Machado L, Privacy challenges and research opportunities for genomic data sharing. Nature Genetics. 52, 646–654 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelleher J, Etheridge AM, McVean G, Efficient Coalescent Simulation and Genealogical Analysis for Large Sample Sizes. PLOS Computational Biology. 12, 1–22 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hudson RR, Properties of a neutral allele model with intragenic recombination. Theoretical Population Biology 23, 183–201 (1983). [DOI] [PubMed] [Google Scholar]
- 23.Ralph P, Thornton K, Kelleher J, Efficiently Summarizing Relationships in Large Samples: A General Duality Between Statistics of Genealogies and Genomes. Genetics. 215, 779–797 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Materials and methods are available as supplementary materials at the Science website.
- 25.Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, Li H, Mallick S, Dannemann M, Fu Q, Kircher M, Kuhlwilm M, Lachmann M, Meyer M, Ongyerth M, Siebauer M, Theunert C, Tandon A, Moorjani P, Pickrell J, Mullikin JC, Vohr SH, Green RE, Hellmann I, Johnson PLF, Blanche H, Cann H, Kitzman JO, Shendure J, Eichler EE, Lein ES, Bakken TE, Golovanova LV, Doronichev VB, Shunkov MV, Derevianko AP, Viola B, Slatkin M, Reich D, Kelso J, Pääbo S, The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 505, 43–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prüfer K, de Filippo C, Grote S, Mafessoni F, Korlević P, Hajdinjak M, Vernot B, Skov L, Hsieh P, Peyrégne S, Reher D, Hopfe C, Nagel S, Maricic T, Fu Q, Theunert C, Rogers R, Skoglund P, Chintalapati M, Dannemann M, Nelson BJ, Key FM, Rudan P, Kućan Ž, Gušić I, Golovanova LV, Doronichev VB, Patterson N, Reich D, Eichler EE, Slatkin M, Schierup MH, Andrés AM, Kelso J, Meyer M, Pääbo S, A high-coverage Neandertal genome from Vindija Cave in Croatia. Science. 358, 655–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mafessoni F, Grote S, de Filippo C, Slon V, Kolobova KA, Viola B, Markin SV, Chintalapati M, Peyrégne S, Skov L, Skoglund P, Krivoshapkin AI, Derevianko AP, Meyer M, Kelso J, Peter B, Prüfer K, Pääbo S, A high-coverage Neandertal genome from Chagyrskaya Cave. Proc. Natl. Acad. Sci. 117, 15132–15136 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer M, Kircher M, Gansauge M-T, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, de Filippo C, Sudmant PH, Alkan C, Fu Q, Do R, Rohland N, Tandon A, Siebauer M, Green RE, Bryc K, Briggs AW, Stenzel U, Dabney J, Shendure J, Kitzman J, Hammer MF, Shunkov MV, Derevianko AP, Patterson N, Andrés AM, Eichler EE, Slatkin M, Reich D, Kelso J, Pääbo S, A High-Coverage Genome Sequence from an Archaic Denisovan Individual. Science. 338, 222–226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu Q, Li H, Moorjani P, Jay F, Slepchenko SM, Bondarev AA, Johnson PLF, Aximu-Petri A, Prüfer K, de Filippo C, Meyer M, Zwyns N, Salazar-García DC, Kuzmin YV, Keates SG, Kosintsev PA, Razhev DI, Richards MP, Peristov NV, Lachmann M, Douka K, Higham TFG, Slatkin M, Hublin J-J, Reich D, Kelso J, Viola TB, Pääbo S, Genome sequence of a 45,000-year-old modern human from western Siberia. Nature. 514, 445–449 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelleher J, Yan W, Jeffery B, Wohns AW, tsinfer (2021; https://github.com/tskit-dev/tsinfer). doi: 10.5281/zenodo.5168051. [DOI]
- 31.Wohns W, Wong Y, Ben J, tsdate (2021; https://github.com/tskit-dev/tsdate). doi: 10.5281/zenodo.5168040. [DOI]
- 32.Bonné, The Samaritans: a demographic study. Human biology. 35, 61–89 (1963). [PubMed] [Google Scholar]
- 33.Albers PK, McVean G, Dating genomic variants and shared ancestry in population-scale sequencing data. PLOS Biology. 18, 1–26 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Speidel L, Forest M, Shi S, Myers SR, A method for genome-wide genealogy estimation for thousands of samples. Nature Genetics. 51, 1321–1329 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PLF, Maricic T, Good JM, Marques-Bonet T, Alkan C, Fu Q, Mallick S, Li H, Meyer M, Eichler EE, Stoneking M, Richards M, Talamo S, Shunkov MV, Derevianko AP, Hublin J-J, Kelso J, Slatkin M, Pääbo S, Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 468, 1053–1060 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reich D, Patterson N, Kircher M, Delfin F, Nandineni MR, Pugach I, Ko AM-S, Ko Y-C, Jinam TA, Phipps ME, Saitou N, Wollstein A, Kayser M, Pääbo S, Stoneking M, Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am J Hum Genet. 89, 516–528 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacobs GS, Hudjashov G, Saag L, Kusuma P, Darusallam CC, Lawson DJ, Mondal M, Pagani L, Ricaut F-X, Stoneking M, Metspalu M, Sudoyo H, Lansing JS, Cox MP, Multiple Deeply Divergent Denisovan Ancestries in Papuans. Cell. 177, 1010–1021 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Narasimhan VM, Patterson N, Moorjani P, Rohland N, Bernardos R, Mallick S, Lazaridis I, Nakatsuka N, Olalde I, Lipson M, Kim AM, Olivieri LM, Coppa A, Vidale M, Mallory J, Moiseyev V, Kitov E, Monge J, Adamski N, Alex N, Broomandkhoshbacht N, Candilio F, Callan K, Cheronet O, Culleton BJ, Ferry M, Fernandes D, Freilich S, Gamarra B, Gaudio D, Hajdinjak M, Harney É, Harper TK, Keating D, Lawson AM, Mah M, Mandl K, Michel M, Novak M, Oppenheimer J, Rai N, Sirak K, Slon V, Stewardson K, Zalzala F, Zhang Z, Akhatov G, Bagashev AN, Bagnera A, Baitanayev B, Bendezu-Sarmiento J, Bissembaev AA, Bonora GL, Chargynov TT, Chikisheva T, Dashkovskiy PK, Derevianko A, Dobeš M, Douka K, Dubova N, Duisengali MN, Enshin D, Epimakhov A, Fribus AV, Fuller D, Goryachev A, Gromov A, Grushin SP, Hanks B, Judd M, Kazizov E, Khokhlov A, Krygin AP, Kupriyanova E, Kuznetsov P, Luiselli D, Maksudov F, Mamedov AM, Mamirov TB, Meiklejohn C, Merrett DC, Micheli R, Mochalov O, Mustafokulov S, Nayak A, Pettener D, Potts R, Razhev D, Rykun M, Sarno S, Savenkova TM, Sikhymbaeva K, Slepchenko SM, Soltobaev OA, Stepanova N, Svyatko S, Tabaldiev K, Teschler-Nicola M, Tishkin AA, Tkachev VV, Vasilyev S, Velemínský P, Voyakin D, Yermolayeva A, Zahir M, Zubkov VS, Zubova A, Shinde VS, Lalueza-Fox C, Meyer M, Anthony D, Boivin N, Thangaraj K, Kennett DJ, Frachetti M, Pinhasi R, Reich D, The formation of human populations in South and Central Asia. Science. 365 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen L, Wolf AB, Fu W, Li L, Akey JM, Identifying and Interpreting Apparent Neanderthal Ancestry in African Individuals. Cell. 180, 677–687.e16 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Wall JD, Yang MA, Jay F, Kim SK, Durand EY, Stevison LS, Gignoux C, Woerner A, Hammer MF, Slatkin M, Higher levels of Neanderthal ancestry in East Asians than in Europeans. Genetics. 194, 199–209 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McDougall, Brown FH, Fleagle JG, Stratigraphic placement and age of modern humans from Kibish, Ethiopia. Nature. 433, 733–736 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Hublin J-J, Ben-Ncer A, Bailey SE, Freidline SE, Neubauer S, Skinner MM, Bergmann I, Le Cabec A, Benazzi S, Harvati K, Gunz P, New fossils from Jebel Irhoud, Morocco and the pan-African origin of Homo sapiens. Nature. 546, 289–292 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Wang K, Goldstein S, Bleasdale M, Clist B, Bostoen K, Bakwa-Lufu P, Buck LT, Crowther A, Dème A, McIntosh RJ, Mercader J, Ogola C, Power RC, Sawchuk E, Robertshaw P, Wilmsen EN, Petraglia M, Ndiema E, Manthi FK, Krause J, Roberts P, Boivin N, Schiffels S, Ancient genomes reveal complex patterns of population movement, interaction, and replacement in sub-Saharan Africa. Science Advances. 6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prendergast ME, Lipson M, Sawchuk EA, Olalde I, Ogola CA, Rohland N, Sirak KA, Adamski N, Bernardos R, Broomandkhoshbacht N, Callan K, Culleton BJ, Eccles L, Harper TK, Lawson AM, Mah M, Oppenheimer J, Stewardson K, Zalzala F, Ambrose SH, Ayodo G, Gates HLJ, Gidna AO, Katongo M, Kwekason A, Mabulla AZP, Mudenda GS, Ndiema EK, Nelson C, Robertshaw P, Kennett DJ, Manthi FK, Reich D, Ancient DNA reveals a multistep spread of the first herders into sub-Saharan Africa. Science. 365 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalkauskas, Perron U, Sun Y, Goldman N, Baele G, Guindon S, De Maio N, Sampling bias and model choice in continuous phylogeography: Getting lost on a random walk. PLOS Computational Biology. 17, 1–27 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson A, African populations and the evolution of human mitochondrial DNA. Science. 253, 1503–1507 (1991). [DOI] [PubMed] [Google Scholar]
- 47.Underhill PA, Passarino G, Lin AA, Shen P, Mirazón Lahr M, Foley RA, Oefner PJ, Cavalli-Sforza LL, The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations. Annals of Human Genetics. 65, 43–62 (2001). [DOI] [PubMed] [Google Scholar]
- 48.O’Connell JF, Allen J, The process, biotic impact, and global implications of the human colonization of Sahul about 47,000 years ago. Journal of Archaeological Science. 56,73–84 (2015). [Google Scholar]
- 49.Llamas B, Fehren-Schmitz L, Valverde G, Soubrier J, Mallick S, Rohland N, Nordenfelt S, Valdiosera C, Richards SM, Rohrlach A, Romero MIB, Espinoza IF, Cagigao ET, Jiménez LW, Makowski K, Reyna ISL, Lory JM, Torrez JAB, Rivera MA, Burger RL, Ceruti MC, Reinhard J, Wells RS, Politis G, Santoro CM, Standen VG, Smith C, Reich D, Ho SYW, Cooper A, Haak W, Ancient mitochondrial DNA provides high-resolution time scale of the peopling of the Americas. Science Advances. 2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moreno-Mayar JV, Vinner L, de Barros Damgaard P, de la Fuente C, Chan J, Spence JP, Allentoft ME, Vimala T, Racimo F, Pinotti T, Rasmussen S, Margaryan A, Iraeta Orbegozo M, Mylopotamitaki D, Wooller M, Bataille C, Becerra-Valdivia L, Chivall D, Comeskey D, Devièse T, Grayson DK, George L, Harry H, Alexandersen V, Primeau C, Erlandson J, Rodrigues-Carvalho C, Reis S, Bastos MQR, Cybulski J, Vullo C, Morello F, Vilar M, Wells S, Gregersen K, Hansen KL, Lynnerup N, Mirazón Lahr M, Kjær K, Strauss A, Alfonso-Durruty M, Salas A, Schroeder H, Higham T, Malhi RS, Rasic JT, Souza L, Santos FR, Malaspinas A-S, Sikora M, Nielsen R, Song YS, Meltzer DJ, Willerslev E, Early human dispersals within the Americas. Science. 362 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Fisher RA, The Genetical Theory of Natural Selection (Clarendon, 1930). [Google Scholar]
- 52.Wright S, Evolution in Mendelian populations. Genetics. 16, 97–159 (1931). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kingman JFC, The coalescent. Stochastic processes and their applications. 13, 235–248 (1982). [Google Scholar]
- 54.McVean GAT, Cardin NJ, Approximating the coalescent with recombination. Philosophical Transactions of the Royal Society B: Biological Sciences. 360, 1387–1393 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rasmussen MD, Hubisz MJ, Gronau I, Siepel A, Genome-Wide Inference of Ancestral Recombination Graphs. PLOS Genetics. 10, 1–27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harris K, From a database of genomes to a forest of evolutionary trees. Nature Genetics. 51, 1306–1307 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scheib CL, Hui R, D’Atanasio E, Wohns AW, Inskip SA, Rose A, Cessford C, O’Connell TC, Robb JE, Evans C, Patten R, Kivisild T, East Anglian early Neolithic monument burial linked to contemporary Megaliths. Annals of Human Biology. 46, 145–149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stern J, Speidel L, Zaitlen NA, Nielsen R, Disentangling selection on genetically correlated polygenic traits via whole-genome genealogies. The American Journal of Human Genetics. 108, 219–239 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Speidel L, Cassidy L, Davies RW, Hellenthal G, Skoglund P, Myers SR, Inferring population histories for ancient genomes using genome-wide genealogies. bioRxiv (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schaefer NK, Shapiro B, Green RE, An ancestral recombination graph of human, Neanderthal, and Denisovan genomes. Science Advances. 7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nielsen R, Akey JM, Jakobsson M, Pritchard JK, Tishkoff S, Willerslev E, Tracing the peopling of the world through genomics. Nature. 541, 302–310 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Michaelson JJ, Shi Y, Gujral M, Zheng H, Malhotra D, Jin X, Jian M, Liu G, Greer D, Bhandari A, Wu W, Corominas R, Peoples A, Koren A, Gore A, Kang S, Lin GN, Estabillo J, Gadomski T, Singh B, Zhang K, Akshoomoff N, Corsello C, McCarroll S, Iakoucheva LM, Li Y, Wang J, Sebat J, Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell. 151, 1431–1442 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nesta V, Tafur D, Beck CR, Hotspots of Human Mutation. Trends in Genetics (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hui R, D’Atanasio E, Cassidy LM, Scheib CL, Kivisild T, Evaluating genotype imputation pipeline for ultra-low coverage ancient genomes. Scientific Reports. 10, 18542 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Murphy KP, Weiss Y, Jordan MI, Loopy belief propagation for approximate inference: An empirical study, in Proceedings of the Fifteenth conference on Uncertainty in artificial intelligence, Stockholm, Sweden, 30 July - 1 August 1999. [Google Scholar]
- 66.tskit developers, tskit (2021; https://github.com/tskit-dev/tskit). doi: 10.5281/zenodo.5465773. [DOI]
- 67.Code to reproduce the analyses presented in this paper can be found at https://github.com/awohns/unified_genealogy_paper. doi: 10.5281/zenodo.5172dl04. [DOI]
- 68.Unified tree sequences of the HGDP, SGDP, and TGP autosomes are available from Zenodo at https://zenodo.org/record/5495535. doi: 10.5281/zenodo.5495535. [DOI]
- 69.Unified tree sequences of the HGDP, SGDP, TGP and high-coverage sequenced ancient autosomes are available from Zenodo at https://zenodo.org/record/5512994. doi: 10.5281/zenodo.5512994. [DOI]
- 70.Lowy-Gallego E, Fairley S, Zheng-Bradley X, Ruffier M, Clarke L, Flicek P, The 1000 Genomes Project Consortium, Variant calling on the GRCh38 assembly with the data from phase three of the 1000 Genomes Project. Wellcome Open Res. 4, 50 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Allentoft ME, Sikora M, Sjögren K-G, Rasmussen S, Rasmussen M, Stenderup J, Damgaard PB, Schroeder H, Ahlström T, Vinner L, Malaspinas A-S, Margaryan A, Higham T, Chivall D, Lynnerup N, Harvig L, Baron J, Casa PD, Dąbrowski P, Duffy PR, Ebel AV, Epimakhov A, Frei K, Furmanek M, Gralak T, Gromov A, Gronkiewicz S, Grupe G, Hajdu T, Jarysz R, Khartanovich V, Khokhlov A, Kiss V, Kolář J, Kriiska A, Lasak I, Longhi C, McGlynn G, Merkevicius A, Merkyte I, Metspalu M, Mkrtchyan R, Moiseyev V, Paja L, Pálfi G, Pokutta D, Pospieszny \Lukasz, Price TD, Saag L, Sablin M, Shishlina N, Smrčka V, Soenov VI, Szeverényi V, Tóth G, Trifanova SV, Varul L, Vicze M, Yepiskoposyan L, Zhitenev V, Orlando L, Sicheritz-Pontén T, Brunak S, Nielsen R, Kristiansen K, Willerslev E, Population genomics of Bronze Age Eurasia. Nature. 522, 167–172 (2015). [DOI] [PubMed] [Google Scholar]
- 72.Amorim CEG, Vai S, Posth C, Modi A, Koncz I, Hakenbeck S, La Rocca MC, Mende B, Bobo D, Pohl W, Baricco LP, Bedini E, Francalacci P, Giostra C, Vida T, Winger D, von Freeden U, Ghirotto S, Lari M, Barbujani G, Krause J, Caramelli D, Geary PJ, Veeramah KR, Understanding 6th-century barbarian social organization and migration through paleogenomics. Nat. Commun. 9, 3547 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Antonio ML, Gao Z, Moots HM, Lucci M, Candilio F, Sawyer S, Oberreiter V, Calderon D, Devitofranceschi K, Aikens RC, Aneli S, Bartoli F, Bedini A, Cheronet O, Cotter DJ, Fernandes DM, Gasperetti G, Grifoni R, Guidi A, La Pastina F, Loreti E, Manacorda D, Matullo G, Morretta S, Nava A, Fiocchi Nicolai V, Nomi F, Pavolini C, Pentiricci M, Pergola P, Piranomonte M, Schmidt R, Spinola G, Sperduti A, Rubini M, Bondioli L, Coppa A, Pinhasi R, Pritchard JK, Ancient Rome: A genetic crossroads of Europe and the Mediterranean. Science. 366, 708–714 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brace S, Diekmann Y, Booth TJ, van Dorp L, Faltyskova Z, Rohland N, Mallick S, Olalde I, Ferry M, Michel M, Oppenheimer J, Broomandkhoshbacht N, Stewardson K, Martiniano R, Walsh S, Kayser M, Charlton S, Hellenthal G, Armit I, Schulting R, Craig OE, Sheridan A, Parker Pearson M, Stringer C, Reich D, Thomas MG, Barnes I, Ancient genomes indicate population replacement in Early Neolithic Britain. Nature Ecology & Evolution. 3, 765–771 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Broushaki F, Thomas MG, Link V, López S, van Dorp L, Kirsanow K, Hofmanová Z, Diekmann Y, Cassidy LM, Díez-del-Molino D, Kousathanas A, Sell C, Robson HK, Martiniano R, Blöcher J, Scheu A, Kreutzer S, Bollongino R, Bobo D, Davoudi H, Munoz O, Currat M, Abdi K, Biglari F, Craig OE, Bradley DG, Shennan S, Veeramah KR, Mashkour M, Wegmann D, Hellenthal G, Burger J, Early Neolithic genomes from the eastern Fertile Crescent. Science. 353, 499–503 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cassidy LM, Martiniano R, Murphy EM, Teasdale MD, Mallory J, Hartwell B, Bradley DG, Neolithic and Bronze Age migration to Ireland and establishment of the insular Atlantic genome. Proc Natl Acad Sci U S A. 113, 368–373 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.de P, Damgaard B, Marchi N, Rasmussen S, Peyrot M, Renaud G, Korneliussen T, Moreno-Mayar JV, Pedersen MW, Goldberg A, Usmanova E, Baimukhanov N, Loman V, Hedeager L, Pedersen AG, Nielsen K, Afanasiev G, Akmatov K, Aldashev A, Alpaslan A, Baimbetov G, Bazaliiskii VI, Beisenov A, Boldbaatar B, Boldgiv B, Dorzhu C, Ellingvag S, Erdenebaatar D, Dajani R, Dmitriev E, Evdokimov V, Frei KM, Gromov A, Goryachev A, Hakonarson H, Hegay T, Khachatryan Z, Khaskhanov R, Kitov E, Kolbina A, Kubatbek T, Kukushkin A, Kukushkin I, Lau N, Margaryan A, Merkyte I, Mertz IV, Mertz VK, Mijiddorj E, Moiyesev V, Mukhtarova G, Nurmukhanbetov B, Orozbekova Z, Panyushkina I, Pieta K, Smrčka V, Shevnina I, Logvin A, Sjögren K-G, Štolcová T, Taravella AM, Tashbaeva K, Tkachev A, Tulegenov T, Voyakin D, Yepiskoposyan L, Undrakhbold S, Varfolomeev V, Weber A, Wilson Sayres MA, Kradin N, Allentoft ME, Orlando L, Nielsen R, Sikora M, Heyer E, Kristiansen K, Willerslev E, 137 ancient human genomes from across the Eurasian steppes. Nature. 557, 369–374 (2018). [DOI] [PubMed] [Google Scholar]
- 78.de P, Damgaard B, Martiniano R, Kamm J, Moreno-Mayar JV, Kroonen G, Peyrot M, Barjamovic G, Rasmussen S, Zacho C, Baimukhanov N, Zaibert V, Merz V, Biddanda A, Merz I, Loman V, Evdokimov V, Usmanova E, Hemphill B, Seguin-Orlando A, Yediay FE, Ullah I, Sjögren K-G, Iversen KH, Choin J, de la Fuente C, Ilardo M, Schroeder H, Moiseyev V, Gromov A, Polyakov A, Omura S, Senyurt SY, Ahmad H, McKenzie C, Margaryan A, Hameed A, Samad A, Gul N, Khokhar MH, Goriunova OI, Bazaliiskii VI, Novembre J, Weber AW, Orlando L, Allentoft ME, Nielsen R, Kristiansen K, Sikora M, Outram AK, Durbin R, Willerslev E, The first horse herders and the impact of early Bronze Age steppe expansions into Asia. Science. 360, eaar7711 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ebenesersdóttir SS, Sandoval-Velasco M, Gunnarsdóttir ED, Jagadeesan A, Guðmundsdóttir VB, Thordardóttir EL, Einarsdóttir MS, Moore KHS, Sigurðsson Á, Magúsdóttir DN, Jónsson H, Snorradóttir S, Hovig E, Møller P, Kockum I, Olsson T, Alfredsson L, Hansen TF, Werge T, Cavalleri GL, Gilbert E, Lalueza-Fox C, Walser JW, Kristjánsdóttir S, Gopalakrishnan S, Árnadóttir L, Magnússon ÓÞ, Gilbert MTP, Stefánsson K, Helgason A, Ancient genomes from Iceland reveal the making of a human population. Science. 360, 1028–1032 (2018). [DOI] [PubMed] [Google Scholar]
- 80.Feldman M, Fernández-Domínguez E, Reynolds L, Baird D, Pearson J, Hershkovitz I, May H, Goring-Morris N, Benz M, Gresky J, Bianco RA, Fairbairn A, Mustafaoğlu G, Stockhammer PW, Posth C, Haak W, Jeong C, Krause J, Late Pleistocene human genome suggests a local origin for the first farmers of central Anatolia. Nat. Commun. 10, 1218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Feldman M, Master DM, Bianco RA, Burri M, Stockhammer PW, Mittnik A, Aja AJ, Jeong C, Krause J, Ancient DNA sheds light on the genetic origins of early Iron Age Philistines. Science Advances. 5, eaax0061 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de la Fuente C, Ávila-Arcos MC, Galimany J, Carpenter ML, Homburger JR, Blanco A, Contreras P, Cruz Dávalos D, Reyes O, San Roman M, Moreno-Estrada A, Campos PF, Eng C, Huntsman S, Burchard EG, Malaspinas A-S, Bustamante CD, Willerslev E, Llop E, Verdugo RA, Moraga M, Genomic insights into the origin and diversification of late maritime hunter-gatherers from the Chilean Patagonia. Proc. Natl. Acad. Sci. 115, E4006–E4012 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fernandes DM, Strapagiel D, Borówka P, Marciniak B, Żądzińska E, Sirak K, Siska V, Grygiel R, Carlsson J, Manica A, Lorkiewicz W, Pinhasi R, A genomic Neolithic time transect of hunter-farmer admixture in central Poland. Scientific Reports. 8, 14879 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Flegontov P, Altınışık NE, Changmai P, Rohland N, Mallick S, Adamski N, Bolnick DA, Broomandkhoshbacht N, Candilio F, Culleton BJ, Flegontova O, Friesen TM, Jeong C, Harper TK, Keating D, Kennett DJ, Kim AM, Lamnidis TC, Lawson AM, Olalde I, Oppenheimer J, Potter BA, Raff J, Sattler RA, Skoglund P, Stewardson K, Vajda EJ, Vasilyev S, Veselovskaya E, Hayes MG, O’Rourke DH, Krause J, Pinhasi R, Reich D, Schiffels S, Palaeo-Eskimo genetic ancestry and the peopling of Chukotka and North America. Nature. 570, 236–240 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fregel R, Méndez FL, Bokbot Y, Martín-Socas D, Camalich-Massieu MD, Santana J, Morales J, Ávila-Arcos MC, Underhill PA, Shapiro B, Wojcik G, Rasmussen M, Soares AER, Kapp J, Sockell A, Rodríguez-Santos FJ, Mikdad A, Trujillo-Mederos A, Bustamante CD, Ancient genomes from North Africa evidence prehistoric migrations to the Maghreb from both the Levant and Europe. Proc. Natl. Acad. Sci. 115, 6774–6779 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fu Q, Mittnik A, Johnson PLF, Bos K, Lari M, Bollongino R, Sun C, Giemsch L, Schmitz R, Burger J, Ronchitelli AM, Martini F, Cremonesi RG, Svoboda J, Bauer P, Caramelli D, Castellano S, Reich D, Pääbo S, Krause J, A Revised Timescale for Human Evolution Based on Ancient Mitochondrial Genomes. Curr. Biol. 23, 553–559 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fu Q, Hajdinjak M, Moldovan OT, Constantin S, Mallick S, Skoglund P, Patterson N, Rohland N, Lazaridis I, Nickel B, Viola B, Prüfer K, Meyer M, Kelso J, Reich D, Pääbo S, An early modern human from Romania with a recent Neanderthal ancestor. Nature. 524, 216–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fu Q, Posth C, Hajdinjak M, Petr M, Mallick S, Fernandes D, Furtwängler A, Haak W, Meyer M, Mittnik A, Nickel B, Peltzer A, Rohland N, Slon V, Talamo S, Lazaridis I, Lipson M, Mathieson I, Schiffels S, Skoglund P, Derevianko AP, Drozdov N, Slavinsky V, Tsybankov A, Cremonesi RG, Mallegni F, Gély B, Vacca E, Morales MRG, Straus LG, Neugebauer-Maresch C, Teschler-Nicola M, Constantin S, Moldovan OT, Benazzi S, Peresani M, Coppola D, Lari M, Ricci S, Ronchitelli A, Valentin F, Thevenet C, Wehrberger K, Grigorescu D, Rougier H, Crevecoeur I, Flas D, Semal P, Mannino MA, Cupillard C, Bocherens H, Conard NJ, Harvati K, Moiseyev V, Drucker DG, Svoboda J, Richards MP, Caramelli D, Pinhasi R, Kelso J, Patterson N, Krause J, Pääbo S, Reich D, The genetic history of Ice Age Europe. Nature. 534, 200–205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gamba C, Jones ER, Teasdale MD, McLaughlin RL, Gonzalez-Fortes G, Mattiangeli V, Domboróczki L, Kővári I, Pap I, Anders A, Whittle A, Dani J, Raczky P, Higham TFG, Hofreiter M, Bradley DG, Pinhasi R, Genome flux and stasis in a five millennium transect of European prehistory. Nat. Commun. 5, 5257 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.González-Fortes G, Jones ER, Lightfoot E, Bonsall C, Lazar C, Grandal-d’Anglade A, Garralda MD, Drak L, Siska V, Simalcsik A, Boroneant A, Romani JRV, Rodríguez MV, Arias P, Pinhasi R, Manica A, Hofreiter M, Paleogenomic Evidence for Multi-generational Mixing between Neolithic Farmers and Mesolithic Hunter-Gatherers in the Lower Danube Basin. Curr. Biol. 27, 1801–1810.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.González-Fortes G, Tassi F, Trucchi E, Henneberger K, Paijmans JLA, Díez-Del-Molino D, Schroeder H, Susca RR, Barroso-Ruíz C, Bermudez FJ, Barroso-Medina C, Bettencourt AMS, Sampaio HA, Grandal-d’Anglade A, Salas A, de Lombera-Hermida A, Fabregas Valcarce R, Vaquero M, Alonso S, Lozano M, Rodríguez-Alvarez XP, Fernández-Rodríguez C, Manica A, Hofreiter M, Barbujani G, A western route of prehistoric human migration from Africa into the Iberian Peninsula. Proc Biol Sci. 286, 20182288 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Günther T, Malmström H, Svensson EM, Omrak A, Sánchez-Quinto F, Kılınç GM, Krzewińska M, Eriksson G, Fraser M, Edlund H, Munters AR, Coutinho A, Simões LG, Vicente M, Sjölander A, Jansen Sellevold B, Jørgensen R, Claes P, Shriver MD, Valdiosera C, Netea MG, Apel J, Lidén K, Skar B, Storå J, Götherström A, Jakobsson M, Population genomics of Mesolithic Scandinavia: Investigating early postglacial migration routes and high-latitude adaptation. PLOS Biology. 16, 1–22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Günther T, Valdiosera C, Malmström H, Ureña I, Rodriguez-Varela R, Sverrisdóttir ÓO, Daskalaki EA, Skoglund P, Naidoo T, Svensson EM, Bermúdez de Castro JM, Carbonell E, Dunn M, Storå J, Iriarte E, Arsuaga JL, Carretero J-M, Götherström A, Jakobsson M, Ancient genomes link early farmers from Atapuerca in Spain to modern-day Basques. Proc. Natl. Acad. Sci. 112, 11917–11922 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Haber M, Doumet-Serhal C, Scheib C, Xue Y, Danecek P, Mezzavilla M, Youhanna S, Martiniano R, Prado-Martinez J, Szpak M, Matisoo-Smith E, Schutkowski H, Mikulski R, Zalloua P, Kivisild T, Tyler-Smith C, Continuity and Admixture in the Last Five Millennia of Levantine History from Ancient Canaanite and Present-Day Lebanese Genome Sequences. The American Journal of Human Genetics. 101, 274–282 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Haber M, Doumet-Serhal C, Scheib CL, Xue Y, Mikulski R, Martiniano R, Fischer-Genz B, Schutkowski H, Kivisild T, Tyler-Smith C, A Transient Pulse of Genetic Admixture from the Crusaders in the Near East Identified from Ancient Genome Sequences. The American Journal of Human Genetics. 104, 977–984 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hajdinjak M, Fu Q, Hübner A, Petr M, Mafessoni F, Grote S, Skoglund P, Narasimham V, Rougier H, Crevecoeur I, Semal P, Soressi M, Talamo S, Hublin J-J, Gušić I, Kućan Ž, Rudan P, Golovanova LV, Doronichev VB, Posth C, Krause J, Korlević P, Nagel S, Nickel B, Slatkin M, Patterson N, Reich D, Prüfer K, Meyer M, Pääbo S, Kelso J, Reconstructing the genetic history of late Neanderthals. Nature. 555, 652–656 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harney É, May H, Shalem D, Rohland N, Mallick S, Lazaridis I, Sarig R, Stewardson K, Nordenfelt S, Patterson N, Hershkovitz I, Reich D, Ancient DNA from Chalcolithic Israel reveals the role of population mixture in cultural transformation. Nat. Commun 9, 3336 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Harney É, Nayak A, Patterson N, Joglekar P, Mushrif-Tripathy V, Mallick S, Rohland N, Sedig J, Adamski N, Bernardos R, Broomandkhoshbacht N, Culleton BJ, Ferry M, Harper TK, Michel M, Oppenheimer J, Stewardson K, Zhang Z, Harashawaradhana, Bartwal MS, Kumar S, Diyundi SC, Roberts P, Boivin N, Kennett DJ, Thangaraj K, Reich D, Rai N, Ancient DNA from the skeletons of Roopkund Lake reveals Mediterranean migrants in India. Nat. Commun 10, 3670 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hofmanová Z, Kreutzer S, Hellenthal G, Sell C, Diekmann Y, Díez-del-Molino D, van Dorp L, López S, Kousathanas A, Link V, Kirsanow K, Cassidy LM, Martiniano R, Strobel M, Scheu A, Kotsakis K, Halstead P, Triantaphyllou S, Kyparissi-Apostolika N, Urem-Kotsou D, Ziota C, Adaktylou F, Gopalan S, Bobo DM, Winkelbach L, Blöcher J, Unterländer M, Leuenberger C, Çilingiroğlu Ç, Horejs B, Gerritsen F, Shennan SJ, Bradley DG, Currat M, Veeramah KR, Wegmann D, Thomas MG, Papageorgopoulou C, Burger J, Early farmers from across Europe directly descended from Neolithic Aegeans. Proc. Natl. Acad. Sci 113, 6886–6891 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Järve M, Saag L, Scheib CL, Pathak AK, Montinaro F, Pagani L, Flores R, Guellil M, Saag L, Tambets K, Kushniarevich A, Solnik A, Varul L, Zadnikov S, Petrauskas O, Avramenko M, Magomedov B, Didenko S, Toshev G, Bruyako I, Grechko D, Okatenko V, Gorbenko K, Smyrnov O, Heiko A, Reida R, Sapiehin S, Sirotin S, Tairov A, Beisenov A, Starodubtsev M, Vasilev V, Nechvaloda A, Atabiev B, Litvinov S, Ekomasova N, Dzhaubermezov M, Voroniatov S, Utevska O, Shramko I, Khusnutdinova E, Metspalu M, Savelev N, Kriiska A, Kivisild T, Villems R, Shifts in the Genetic Landscape of the Western Eurasian Steppe Associated with the Beginning and End of the Scythian Dominance. Curr. Biol 29, 2430–2441.e10 (2019). [DOI] [PubMed] [Google Scholar]
- 101.Jeong C, Balanovsky O, Lukianova E, Kahbatkyzy N, Flegontov P, Zaporozhchenko V, Immel A, Wang C-C, Ixan O, Khussainova E, Bekmanov B, Zaibert V, Lavryashina M, Pocheshkhova E, Yusupov Y, Agdzhoyan A, Koshel S, Bukin A, Nymadawa P, Turdikulova S, Dalimova D, Churnosov M, Skhalyakho R, Daragan D, Bogunov Y, Bogunova A, Shtrunov A, Dubova N, Zhabagin M, Yepiskoposyan L, Churakov V, Pislegin N, Damba L, Saroyants L, Dibirova K, Atramentova L, Utevska O, Idrisov E, Kamenshchikova E, Evseeva I, Metspalu M, Outram AK, Robbeets M, Djansugurova L, Balanovska E, Schiffels S, Haak W, Reich D, Krause J, The genetic history of admixture across inner Eurasia. Nature Ecology & Evolution. 3, 966–976 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jeong C, Ozga AT, Witonsky DB, Malmström H, Edlund H, Hofman CA, Hagan RW, Jakobsson M, Lewis CM, Aldenderfer MS, Di Rienzo A, Warinner C, Long-term genetic stability and a high-altitude East Asian origin for the peoples of the high valleys of the Himalayan arc. Proc. Natl. Acad. Sci 113, 7485–7490 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jeong C, Wilkin S, Amgalantugs T, Bouwman AS, Taylor WTT, Hagan RW, Bromage S, Tsolmon S, Trachsel C, Grossmann J, Littleton J, Makarewicz CA, Krigbaum J, Burri M, Scott A, Davaasambuu G, Wright J, Irmer F, Myagmar E, Boivin N, Robbeets M, Rühli FJ, Krause J, Frohlich B, Hendy J, Warinner C, Bronze Age population dynamics and the rise of dairy pastoralism on the eastern Eurasian steppe. Proc. Natl. Acad. Sci 115, E11248–E11255 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jones ER, Gonzalez-Fortes G, Connell S, Siska V, Eriksson A, Martiniano R, McLaughlin RL, Gallego Llorente M, Cassidy LM, Gamba C, Meshveliani T, Bar-Yosef O, Müller W, Belfer-Cohen A, Matskevich Z, Jakeli N, Higham TFG, Currat M, Lordkipanidze D, Hofreiter M, Manica A, Pinhasi R, Bradley DG, Upper Palaeolithic genomes reveal deep roots of modern Eurasians. Nat. Commun 6, 8912 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jones ER, Zarina G, Moiseyev V, Lightfoot E, Nigst PR, Manica A, Pinhasi R, Bradley DG, The Neolithic Transition in the Baltic Was Not Driven by Admixture with Early European Farmers. Carr. Biol 27, 576–582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kanzawa-Kiriyama H, Kryukov K, Jinam TA, Hosomichi K, Saso A, Suwa G, Ueda S, Yoneda M, Tajima A, Shinoda K, Inoue I, Saitou N, A partial nuclear genome of the Jomons who lived 3000 years ago in Fukushima, Japan. Journal of Human Genetics. 62, 213–221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Keller, Graefen A, Ball M, Matzas M, Boisguerin V, Maixner F, Leidinger P, Backes C, Khairat R, Forster M, Stade B, Franke A, Mayer J, Spangler J, McLaughlin S, Shah M, Lee C, Harkins TT, Sartori A, Moreno-Estrada A, Henn B, Sikora M, Semino O, Chiaroni J, Rootsi S, Myres NM, Cabrera VM, Underhill PA, Bustamante CD, Vigl EE, Samadelli M, Cipollini G, Haas J, Katus H, O’Connor BD, Carlson MRJ, Meder B, Blin N, Meese E, Pusch CM, Zink A, New insights into the Tyrolean Iceman’s origin and phenotype as inferred by whole-genome sequencing. Nat. Commun 3, 698 (2012). [DOI] [PubMed] [Google Scholar]
- 108.Kennett DJ, Plog S, George RJ, Culleton BJ, Watson AS, Skoglund P, Rohland N, Mallick S, Stewardson K, Kistler L, LeBlanc SA, Whiteley PM, Reich D, Perry GH, Archaeogenomic evidence reveals prehistoric matrilineal dynasty. Nat. Commun 8, 14115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kılınç GM, Omrak A, Özer F, Günther T, Büyükkarakaya AM, Bıçakçı E, Baird D, Doüertaş HM, Ghalichi A, Yaka R, Koptekin D, Açan SC, Parvizi P, Krzewińska M, Daskalaki EA, Yüncü E, Dağtaş ND, Fairbairn A, Pearson J, Mustafaoğlu G, Erdal YS, Çakan YG, Togan İ, Somel M, Storå J, Jakobsson M, Gütherstrüm A, The Demographic Development of the First Farmers in Anatolia. Curr. Biol 26, 2659–2666 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Korneliussen TS, Albrechtsen A, Nielsen R, ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics. 15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Krzewińska M, Kjellstrüm A, Günther T, Hedenstiema-Jonson C, Zachrisson T, Omrak A, Yaka R, Kılınç GM, Somel M, Sobrado V, Evans J, Knipper C, Jakobsson M, Storå J, Gütherstrüm A, Genomic and Strontium Isotope Variation Reveal Immigration Patterns in a Viking Age Town. Curr. Biol 28, 2730–2738.e10 (2018). [DOI] [PubMed] [Google Scholar]
- 112.Krzewińska M, Kılınç GM, Juras A, Koptekin D, Chyleński M, Nikitin AG, Shcherbakov N, Shuteleva I, Leonova T, Kraeva L, Sungatov FA, Sultanova AN, Potekhina I, Łukasik S, Krenz-Niedbała M, Dalén L, Sinika V, Jakobsson M, Storå J, Gütherstrüm A, Ancient genomes suggest the eastern Pontic-Caspian steppe as the source of western Iron Age nomads. Science Advances. 4, eaat4457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lamnidis TC, Majander K, Jeong C, Salmela E, Wessman A, Moiseyev V, Khartanovich V, Balanovsky O, Ongyerth M, Weihmann A, Sajantila A, Kelso J, Pääbo S, Onkamo P, Haak W, Krause J, Schiffels S, Ancient Fennoscandian genomes reveal origin and spread of Siberian ancestry in Europe. Nat. Commun 9, 5018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lazaridis, Nadel D, Rollefson G, Merrett DC, Rohland N, Mallick S, Fernandes D, Novak M, Gamarra B, Sirak K, Connell S, Stewardson K, Harney E, Fu Q, Gonzalez-Fortes G, Jones ER, Roodenberg SA, Lengyel G, Bocquentin F, Gasparian B, Monge JM, Gregg M, Eshed V, Mizrahi A-S, Meiklejohn C, Gerritsen F, Bejenaru L, Blüher M, Campbell A, Cavalleri G, Comas D, Froguel P, Gilbert E, Kerr SM, Kovacs P, Krause J, McGettigan D, Merrigan M, Merriwether DA, O’Reilly S, Richards MB, Semino O, Shamoon-Pour M, Stefanescu G, Stumvoll M, Tünjes A, Torroni A, Wilson JF, Yengo L, Hovhannisyan NA, Patterson N, Pinhasi R, Reich D, Genomic insights into the origin of farming in the ancient Near East. Nature. 536, 419–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lazaridis, Mittnik A, Patterson N, Mallick S, Rohland N, Pfrengle S, Furtwängler A, Peltzer A, Posth C, Vasilakis A, McGeorge PJP, Konsolaki-Yannopoulou E, Korres G, Martlew H, Michalodimitrakis M, Özsait M, Özsait N, Papathanasiou A, Richards M, Roodenberg SA, Tzedakis Y, Arnott R, Fernandes DM, Hughey JR, Lotakis DM, Navas PA, Maniatis Y, Stamatoyannopoulos JA, Stewardson K, Stockhammer P, Pinhasi R, Reich D, Krause J, Stamatoyannopoulos G, Genetic origins of the Minoans and Mycenaeans. Nature. 548, 214–218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lindo J, Achilli A, Perego UA, Archer D, Valdiosera C, Petzelt B, Mitchell J, Worl R, Dixon EJ, Fifield TE, Rasmussen M, Willerslev E, Cybulski JS, Kemp BM, DeGiorgio M, Malhi RS, Ancient individuals from the North American Northwest Coast reveal 10,000 years of regional genetic continuity. Proc. Natl. Acad. Sci 114, 4093–4098 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lindo J, Haas R, Hofman C, Apata M, Moraga M, Verdugo RA, Watson JT, Llave CV, Witonsky D, Beall C, Warinner C, Novembre J, Aldenderfer M, Rienzo AD, The genetic prehistory of the Andean highlands 7000 years BP though European contact. Science Advances. 4, eaau4921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lipson M, Skoglund P, Spriggs M, Valentin F, Bedford S, Shing R, Buckley H, Phillip I, Ward GK, Mallick S, Rohland N, Broomandkhoshbacht N, Cheronet O, Ferry M, Harper TK, Michel M, Oppenheimer J, Sirak K, Stewardson K, Auckland K, Hill AVS, Maitland K, Oppenheimer SJ, Parks T, Robson K, Williams TN, Kennett DJ, Mentzer AJ, Pinhasi R, Reich D, Population Turnover in Remote Oceania Shortly after Initial Settlement. Curr. Biol 28, 1157–1165.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lipson M, Szécsényi-Nagy A, Mallick S, Pósa A, Stégmár B, Keerl V, Rohland N, Stewardson K, Ferry M, Michel M, Oppenheimer J, Broomandkhoshbacht N, Harney E, Nordenfelt S, Llamas B, Gusztáv Mende B, Kühler K, Oross K, Bondár M, Marton T, Osztás A, Jakucs J, Paluch T, Horváth F, Csengeri P, Koós J, Sebők K, Anders A, Raczky P, Regenye J, Barna JP, Fábián S, Serlegi G, Toldi Z, Gyüngyvér Nagy E, Dani J, Molnár E, Pálfi G, Márk L, Melegh B, Bánfai Z, Domboróczki L, Fernández-Eraso J, Antonio Mujika-Alustiza J, Alonso Fernández C, Jiménez Echevarría J, Bollongino R, Orschiedt J, Schierhold K, Meller H, Cooper A, Burger J, Bánffy E, Alt KW, Lalueza-Fox C, Haak W, Reich D, Parallel palaeogenomic transects reveal complex genetic history of early European farmers. Nature. 551, 368–372 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lipson M, Ribot I, Mallick S, Rohland N, Olalde I, Adamski N, Broomandkhoshbacht N, Lawson AM, López S, Oppenheimer J, Stewardson K, Asombang RN, Bocherens H, Bradman N, Culleton BJ, Cornelissen E, Crevecoeur I, de Maret P, Fomine FLM, Lavachery P, Mindzie CM, Orban R, Sawchuk E, Semal P, Thomas MG, Van Neer W, Veeramah KR, Kennett DJ, Patterson N, Hellenthal G, Lalueza-Fox C, MacEachern S, Prendergast ME, Reich D, Ancient West African foragers in the context of African population history. Nature. 577, 665–670 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lipson M, Cheronet O, Mallick S, Rohland N, Oxenham M, Pietrusewsky M, Pryce TO, Willis A, Matsumura H, Buckley H, Domett K, Nguyen GH, Trinh HH, Kyaw AA, Win TT, Pradier B, Broomandkhoshbacht N, Candilio F, Changmai P, Fernandes D, Ferry M, Gamarra B, Harney E, Kampuansai J, Kutanan W, Michel M, Novak M, Oppenheimer J, Sirak K, Stewardson K, Zhang Z, Flegontov P, Pinhasi R, Reich D, Ancient genomes document multiple waves of migration in Southeast Asian prehistory. Science. 361, 92–95 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gallego Llorente M, Jones ER, Eriksson A, Siska V, Arthur KW, Arthur JW, Curtis MC, Stock JT, Coltorti M, Pieruccini P, Stretton S, Brock F, Higham T, Park Y, Hofreiter M, Bradley DG, Bhak J, Pinhasi R, Manica A, Ancient Ethiopian genome reveals extensive Eurasian admixture throughout the African continent. Science. 350, 820–822 (2015). [DOI] [PubMed] [Google Scholar]
- 123.Malaspinas A-S, Lao O, Schroeder H, Rasmussen M, Raghavan M, Moltke I, Campos PF, Sagredo FS, Rasmussen S, Gonçalves VF, Albrechtsen A, Allentoft ME, Johnson PLF, Li M, Reis S, Bernardo DV, DeGiorgio M, Duggan AT, Bastos M, Wang Y, Stenderup J, Moreno-Mayar JV, Brunak S, Sicheritz-Ponten T, Hodges E, Hannon GJ, Orlando L, Price TD, Jensen JD, Nielsen R, Heinemeier J, Olsen J, Rodrigues-Carvalho C, Lahr MM, Neves WA, Kayser M, Higham T, Stoneking M, Pena SDJ, Willerslev E, Two ancient human genomes reveal Polynesian ancestry among the indigenous Botocudos of Brazil. Curr. Biol 24, R1035–R1037 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Malmström H, Günther T, Svensson EM, Juras A, Fraser M, Munters AR, Pospieszny Ł, Tõrv M. Lindström J, Götherström A, Storå J, Jakobsson M, The genomic ancestry of the Scandinavian Battle Axe Culture people and their relation to the broader Corded Ware horizon. Proceedings of the Roval Society B: Biological Sciences. 286, 20191528 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Martiniano R, Caffell A, Holst M, Hunter-Mann K, Montgomery J, Müldner G, McLaughlin RL, Teasdale MD, van Rheenen W, Veldink JH, van den Berg LH, Hardiman O, Carroll M, Roskams S, Oxley J, Morgan C, Thomas MG, Barnes I, McDonnell C, Collins MJ, Bradley DG, Genomic signals of migration and continuity in Britain before the Anglo-Saxons. Nat. Commun 7, 10326 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Martiniano R, Cassidy LM, Ó’Maoldúin R, McLaughlin R, Silva NM, Manco L, Fidalgo D, Pereira T, Coelho MJ, Serra M, Burger J, Parreira R, Moran E, Valera AC, Porfirio E, Boaventura R, Silva AM, Bradley DG, The population genomics of archaeological transition in west Iberia: Investigation of ancient substructure using imputation and haplotype-based methods. PLOS Genetics. 13, 1–24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mathieson, Lazaridis I, Rohland N, Mallick S, Patterson N, Roodenberg SA, Harney E, Stewardson K, Fernandes D, Novak M, Sirak K, Gamba C, Jones ER, Llamas B, Dryomov S, Pickrell J, Arsuaga JL, de Castro JB, Carbonell E, Gerritsen F, Khokhlov A, Kuznetsov P, Lozano M, Meller H, Mochalov O, Moiseyev V, Guerra MAR, Roodenberg J, Vergès JM, Krause J, Cooper A, Alt KW, Brown D, Anthony D, Lalueza-Fox C, Haak W, Pinhasi R, Reich D, Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 528, 499–503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mathieson, Alpaslan-Roodenberg S, Posth C, Szécsényi-Nagy A, Rohland N, Mallick S, Olalde I, Broomandkhoshbacht N, Candilio F, Cheronet O, Fernandes D, Ferry M, Gamarra B, Fortes GG, Haak W, Harney E, Jones E, Keating D, Krause-Kyora B, Kucukkalipci I, Michel M, Mittnik A, Nägele K, Novak M, Oppenheimer J, Patterson N, Pfrengle S, Sirak K, Stewardson K, Vai S, Alexandrov S, Alt KW, Andreescu R, Antonović D, Ash A, Atanassova N, Bacvarov K, Gusztáv MB, Bocherens H, Bolus M, Boroncanţ A, Boyadzhiev Y, Budnik A, Burmaz J, Chohadzhiev S, Conard NJ, Cottiaux R, Čuka M, Cupillard C, Drucker DG, Elenski N, Francken M, Galabova B, Ganetsovski G, Gély B, Hajdu T, Handzhyiska V, Harvati K, Higham T, Iliev S, Janković I, Karavanić I, Kennett DJ, Komšo D, Kozak A, Labuda D, Lari M, Lazar C, Leppek M, Leshtakov K, Vetro DL, Los D, Lozanov I, Malina M, Martini F, McSweeney K, Meller H, Menđušić M, Mirea P, Moiseyev V, Petrova V, Price TD, Simalcsik A, Sineo L, šlaus M, Slavchev V, Stanev P, Starović A, Szeniczey T, Talamo S, Teschler-Nicola M, Thevenet C, Valchev I, Valentin F, Vasilyev S, Veljanovska F, Venelinova S, Veselovskaya E, Viola B, Virag C, Zaninović J, Zäuner S, Stockhammer PW, Catalano G, Krauß R, Caramelli D, Zariņa G, Gaydarska B, Lillie M, Nikitin AG, Potekhina I, Papathanasiou A, Borić D, Bonsall C, Krause J, Pinhasi R, Reich D, The genomic history of southeastern Europe. Nature. 555, 197–203 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McColl H, Racimo F, Vinner L, Demeter F, Gakuhari T, Víctor Moreno-Mayar J, Van Driem G, Wilken UG, Seguin-Orlando A, De la Fuente Castro C, Wasef S, Shoocongdej R, Souksavatdy V, Sayavongkhamdy T, Saidin MM, Allentoft ME, Sato T, Malaspinas AS, Aghakhanian FA, Komeliussen T, Prohaska A, Margaryan A, De Barros Damgaard P, Kaewsutthi S, Lertrit P, Nguyen TMH, chun Hung H, Tran TM, Truong HN, Nguyen GH, Shahidan S, Wiradnyana K, Matsumae H, Shigehara N, Yoneda M, Ishida H, Masuyama T, Yamada Y, Tajima A, Shibata H, Toyoda A, Hanihara T, Nakagome S, Deviese T, Bacon AM, Duringer P, Ponche JL, Shackelford L, Patole-Edoumba E, Nguyen AT, Bellina-Pryce B, Galipaud JC, Kinaston R, Buckley H, Pottier C, Rasmussen S, Higham T, Foley RA, Lahr MM, Orlando L, Sikora M, Phipps ME, Oota H, Higham C, Lambert DM, Willerslev E, The prehistoric peopling of Southeast Asia. Science. 361, 88–92 (2018). [DOI] [PubMed] [Google Scholar]
- 130.Mittnik, Wang C-C, Pfrengle S, Daubaras M, Zariņa G, Hallgren F, Allmäe R, Khartanovich V, Moiseyev V, Tõrv M, Furtwängler A, Andrades Valtueña A, Feldman M, Economou C, Oinonen M, Vasks A, Balanovska E, Reich D, Jankauskas R, Haak W, Schiffels S, Krause J, The genetic prehistory of the Baltic Sea region. Nat Commun. 9, 442 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mittnik, Massy K, Knipper C, Wittenborn F, Friedrich R, Pfrengle S, Burri M, Carlichi-Witjes N, Deeg H, Furtwängler A, Harbeck M, von Heyking K, Kociumaka C, Kucukkalipci I, Lindauer S, Metz S, Staskiewicz A, Thiel A, Wahl J, Haak W, Pernicka E, Schiffels S, Stockhammer PW, Krause J, Kinship-based social inequality in Bronze Age Europe. Science. 366, 731–734 (2019). [DOI] [PubMed] [Google Scholar]
- 132.Mondal M, Casals F, Xu T, Dall’Olio GM, Pybus M, Netea MG, Comas D, Laayouni H, Li Q, Majumder PP, Bertranpetit J, Genomic analysis of Andamanese provides insights into ancient human migration into Asia and adaptation. Nature Genetics. 48, 1066–1070 (2016). [DOI] [PubMed] [Google Scholar]
- 133.Moreno-Mayar JV, Potter BA, Vinner L, Steinrücken M, Rasmussen S, Terhorst J, Kamm JA, Albrechtsen A, Malaspinas A-S, Sikora M, Reuther JD, Irish JD, Malhi RS, Orlando L, Song YS, Nielsen R, Meltzer DJ, Willerslev E, Terminal Pleistocene Alaskan genome reveals first founding population of Native Americans. Nature. 553, 203–207 (2018). [DOI] [PubMed] [Google Scholar]
- 134.Moreno-Mayar JV, Vinner L, de Barros Damgaard P, de la Fuente C, Chan J, Spence JP, Allentoft ME, Vimala T, Racimo F, Pinotti T, Rasmussen S, Margaryan A, Iraeta Orbegozo M, Mylopotamitaki D, Wooller M, Bataille C, Becerra-Valdivia L, Chivall D, Comeskey D, Devièse T, Grayson DK, George L, Harry H, Alexandersen V, Primeau C, Erlandson J, Rodrigues-Carvalho C, Reis S, Bastos MQR, Cybulski J, Vullo C, Morello F, Vilar M, Wells S, Gregersen K, Hansen KL, Lynnerup N, Mirazón Lahr M, Kjær K, Strauss A, Alfonso-Durruty M, Salas A, Schroeder H, Higham T, Malhi RS, Rasic JT, Souza L, Santos FR, Malaspinas A-S, Sikora M, Nielsen R, Song YS, Meltzer DJ, Willerslev E, Early human dispersals within the Americas. Science. 362 (2018), doi: 10.1126/science.aav2621. [DOI] [PubMed] [Google Scholar]
- 135.Nikitin G, Stadler P, Kotova N, Teschler-Nicola M, Price TD, Hoover J, Kennett DJ, Lazaridis I, Rohland N, Lipson M, Reich D, Interactions between earliest Linearbandkeramik farmers and central European hunter gatherers at the dawn of European Neolithization. Sci Rep. 9, 19544 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ning, Wang C-C, Gao S, Yang Y, Zhang X, Wu X, Zhang F, Nie Z, Tang Y, Robbeets M, Ma J, Krause J, Cui Y, Ancient Genomes Reveal Yamnaya-Related Ancestry and a Potential Source of Indo-European Speakers in Iron Age Tianshan. Curr. Biol 29, 2526–2532 (2019). [DOI] [PubMed] [Google Scholar]
- 137.Olalde, Schroeder H, Sandoval-Velasco M, Vinner L, Lobón I, Ramirez O, Civit S, García Borja P, Salazar-García DC, Talamo S, María Fullola J, Xavier Oms F, Pedro M, Martínez P, Sanz M, Daura J, Zilhão J, Marquès-Bonet T, Gilbert MTP, Lalueza-Fox C, A Common Genetic Origin for Early Farmers from Mediterranean Cardial and Central European FBK Cultures. Mol Biol Evol. 32, 3132–3142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Olalde, Allentoft ME, Sánchez-Quinto F, Santpere G, Chiang CWK, DeGiorgio M, Prado-Martinez J, Rodríguez JA, Rasmussen S, Quilez J, Ramírez O, Marigorta UM, Femández-Callejo M, Prada ME, Encinas JMV, Nielsen R, Netea MG, Novembre J, Sturm RA, Sabeti P, Marquès-Bonet T, Navarro A, Willerslev E, Lalueza-Fox C, Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European. Nature. 507, 225–228 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Olalde, Brace S, Allentoft ME, Armit I, Kristiansen K, Booth T, Rohland N, Mallick S, Szécsényi-Nagy A, Mittnik A, Altena E, Lipson M, Lazaridis I, Harper TK, Patterson N, Broomandkhoshbacht N, Diekmann Y, Faltyskova Z, Fernandes D, Ferry M, Harney E, de Knijff P, Michel M, Oppenheimer J, Stewardson K, Barclay A, Alt KW, Liesau C, Ríos P, Blasco C, Miguel JV, García RM, Fernández AA, Bánffy E, Bemabò-Brea M, Billoin D, Bonsall C, Bonsall L, Allen T, Büster L, Carver S, Navarro LC, Craig OE, Cook GT, Cunliffe B, Denaire A, Dinwiddy KE, Dodwell N, Ernée M, Evans C, Kuchařík M, Farré JF, Fowler C, Gazenbeek M, Pena RG, Haber-Uriarte M, Haduch E, Hey G, Jowett N, Knowles T, Massy K, Pfrengle S, Lefranc P, Lemercier O, Lefebvre A, Martínez CH, Olmo VG, Ramírez AB, Maurandi JL, Majó T, McKinley JI, McSweeney K, Mende BG, Modi A, Kulcsár G, Kiss V, Czene A, Patay R, Endrődi A, Köhler K, Hajdu T, Szeniczey T, Dani J, Bernert Z, Hoole M, Cheronet O, Keating D, Velemínský P, Dobeš M, Candilio F, Brown F, Fernández RF, Herrero-Corral A-M, Tusa S, Carnieri E, Lentini L, Valenti A, Zanini A, Waddington C, Delibes G, Guerra-Doce E, Neil B, Brittain M, Luke M, Mortimer R, Desideri J, Besse M, Brücken G, Furmanek M, Ha\luszko A, Mackiewicz M, Rapiński A, Leach S, Soriano I, Lillios KT, Cardoso JL, Pearson MP, W\lodarczak P, Price TD, Prieto P, Rey P-J, Risch R, Rojo Guerra MA, Schmitt A, Serralongue J, Silva AM, Smrčka V, Vergnaud L, Zilhão J, Caramelli D, Higham T, Thomas MG, Kennett DJ, Fokkens H, Heyd V, Sheridan A, Sjögren K-G, Stockhammer PW, Krause J, Pinhasi R, Haak W, Barnes I, Lalueza-Fox C, Reich D, The Beaker phenomenon and the genomic transformation of northwest Europe. Nature. 555, 190–196 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Olalde, Mallick S, Patterson N, Rohland N, Villalba-Mouco V, Silva M, Dulias K, Edwards CJ, Gandini F, Pala M, Soares P, Ferrando-Bernal M, Adamski N, Broomandkhoshbacht N, Cheronet O, Culleton BJ, Fernandes D, Lawson AM, Mah M, Oppenheimer J, Stewardson K, Zhang Z, Jiménez Arenas JM, Toro Moyano IJ, Salazar-García DC, Castanyer P, Santos M, Tremoleda J, Lozano M, García Borja P, Fernández-Eraso J, Mujika-Alustiza J, Barroso C, Bermúdez FJ, Viguera Mínguez E, Burch J, Coromina N, Vivó D, Cebrià A, Fullola JM, García-Puchol O, Morales JI, Oms FX, Majó T, Vergès JM, Díaz-Carvajal A, Ollich-Castanyer I, López-Cachero FJ, Silva AM, Alonso-Fernández C, Delibes de Castro G, Jiménez Echevarría J, Moreno-Márquez A, Pascual Berlanga G, Ramos-García P, Ramos-Muñoz J, Vijande Vila E, Aguilella Arzo G, Esparza Arroyo Á, Lillios KT, Mack J, Velasco-Vázquez J, Waterman A, Benítez de Lugo Enrich L, Benito Sánchez M, Agustí B, Codina F, de Prado G, Estalrrich A, Fernández Flores Á, Finlayson C, Finlayson G, Finlayson S, Giles-Guzmán F, Rosas A, Barciela González V, García Atiénzar G, Hernández Pérez MS, Llanos A, Carrión Marco Y, Collado Beneyto I, López-Serrano D, Sanz Tormo M, Valera AC, Blasco C, Liesau C, Ríos P, Daura J, de Pedro Miché MJ, Diez-Castillo AA, Flores Fernández R, Francès Farré J, Garrido-Pena R, Gonçalvcs VS, Guerra-Doce E, Herrero-Corral AM, Juan-Cabanilles J, López-Reyes D, McClure SB, Merino Pérez M, Oliver Foix A, Sanz Borràs M, Sousa AC, Vidal Encinas JM, Kennett DJ, Richards MB, Werner Alt K, Haak W, Pinhasi R, Lalueza-Fox C, Reich D, The genomic history of the Iberian Peninsula over the past 8000 years. Science. 363, 1230–1234 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Omrak, Günther T, Valdiosera C, Svensson EM, Malmström H, Kiesewetter H, Aylward W, StorÅ J, Jakobsson M, Götherström A, Genomic Evidence Establishes Anatolia as the Source of the European Neolithic Gene Pool. Curr. Biol 26, 270–275 (2016). [DOI] [PubMed] [Google Scholar]
- 142.Pickrell JK, Patterson N, Barbieri C, Berthold F, Gerlach L, Güldemann T, Kure B, Mpoloka SW, Nakagawa H, Naumann C, Lipson M, Loh P-R, Lachance J, Mountain J, Bustamante CD, Berger B, Tishkoff SA, Henn BM, Stoneking M, Reich D, Pakendorf B, The genetic prehistory of southern Africa. Nat Commun. 3, 1143 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Posth, Nakatsuka N, Lazaridis I, Skoglund P, Mallick S, Lamnidis TC, Rohland N, Nägele K, Adamski N, Bertolini E, Broomandkhoshbacht N, Cooper A, Culleton BJ, Ferraz T, Ferry M, Furtwängler A, Haak W, Harkins K, Harper TK, Hünemeier T, Lawson AM, Llamas B, Michel M, Nelson E, Oppenheimer J, Patterson N, Schiffels S, Sedig J, Stewardson K, Talamo S, Wang C-C, Hublin J-J, Hubbe M, Harvati K, Nuevo Delaunay A, Beier J, Francken M, Kaulicke P, Reyes-Centeno H, Rademaker K, Trask WR, Robinson M, Gutierrez SM, Prufer KM, Salazar-García DC, Chim EN, Müller Plumm Gomes L, Alves ML, Liryo A, Inglez M, Oliveira RE, Bernardo DV, Barioni A, Wesolowski V, Scheifler NA, Rivera MA, Plens CR, Messineo PG, Figuti L, Corach D, Scabuzzo C, Eggers S, DeBlasis P, Reindel M, Méndez C, Politis G, Tomasto-Cagigao E, Kennett DJ, Strauss A, Fehren-Schmitz L, Krause J, Reich D, Reconstructing the Deep Population History of Central and South America. Cell. 175, 1185–1197 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Posth, Nägele K, Colleran H, Valentin F, Bedford S, Kami KW, Shing R, Buckley H, Kinaston R, Walworth M, Clark GR, Reepmeyer C, Flexner J, Maric T, Moser J, Gresky J, Kiko L, Robson KJ, Auckland K, Oppenheimer SJ, Hill AVS, Mentzer AJ, Zech J, Petchey F, Roberts P, Jeong C, Gray RD, Krause J, Powell A, Language continuity despite population replacement in Remote Oceania. Nat Ecol Evol. 2, 731–740 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Prendergast ME, Lipson M, Sawchuk EA, Olalde I, Ogola CA, Rohland N, Sirak KA, Adamski N, Bernardos R, Broomandkhoshbacht N, Callan K, Culleton BJ, Eccles L, Harper TK, Lawson AM, Mah M, Oppenheimer J, Stewardson K, Zalzala F, Ambrose SH, Ayodo G, Gates HLJ, Gidna AO, Katongo M, Kwekason A, Mabulla AZP, Mudenda GS, Ndiema EK, Nelson C, Robertshaw P, Kennett DJ, Manthi FK, Reich D, Ancient DNA reveals a multistep spread of the first herders into sub-Saharan Africa. Science. 365 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Raghavan M, Skoglund P, Graf KE, Metspalu M, Albrechtsen A, Moltke I, Rasmussen S, Stafford TWJ, Orlando L, Metspalu E, Karmin M, Tambets K, Rootsi S, Mägi R, Campos PF, Balanovska E, Balanovsky O, Khusnutdinova E, Litvinov S, Osipova LP, Fedorova SA, Voevoda MI, DeGiorgio M, Sicheritz-Ponten T, Brunak S, Demeshchenko S, Kivisild T, Villems R, Nielsen R, Jakobsson M, Willerslev E, Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature. 505, 87–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Raghavan M, DeGiorgio M, Albrechtsen A, Moltke I, Skoglund P, Korneliussen TS, Grønnow B, Appelt M, Gulløv HC, Friesen TM, Fitzhugh W, Malmström H, Rasmussen S, Olsen J, Melchior L, Fuller BT, Fahrni SM, Stafford TJ, Grimes V, Renouf MAP, Cybulski J, Lynnerup N, Lahr MM, Britton K, Knecht R, Arneborg J, Metspalu M, Cornejo OE, Malaspinas A-S, Wang Y, Rasmussen M, Raghavan V, Hansen TVO, Khusnutdinova E, Pierre T, Dneprovsky K, Andreasen C, Lange H, Hayes MG, Coltrain J, Spitsyn VA, Götherström A, Orlando L, Kivisild T, Villems R, Crawford MH, Nielsen FC, Dissing J, Heinemeier J, Meldgaard M, Bustamante C, O’Rourke DH, Jakobsson M, Gilbert MTP, Nielsen R, Willerslev E, The genetic prehistory of the New World Arctic. Science. 345, 1255832 (2014). [DOI] [PubMed] [Google Scholar]
- 148.Raghavan M, Steinrücken M, Harris K, Schiffels S, Rasmussen S, DeGiorgio M, Albrechtsen A, Valdiosera C, Ávila-Arcos MC, Malaspinas A-S, Eriksson A, Moltke I, Metspalu M, Homburger JR, Wall J, Cornejo OE, Moreno-Mayar JV, Korneliussen TS, Pierre T, Rasmussen M, Campos PF, de Barros Damgaard P, Allentoft ME, Lindo J, Metspalu E, Rodríguez-Varela R, Mansilla J, Henrickson C, Seguin-Orlando A, Malmstrüm H, Stafford TJ, Shringarpure SS, Moreno-Estrada A, Karmin M, Tambets K, Bergstrüm A, Xue Y, Warmuth V, Friend AD, Singarayer J, Valdes P, Balloux F, Leboreiro I, Vera JL, Rangel-Villalobos H, Pettener D, Luiselli D, Davis LG, Heyer E, Zollikofer CPE, Ponce de Léon MS, Smith CI, Grimes V, Pike K-A, Deal M, Fuller BT, Arriaza B, Standen V, Luz MF, Ricaut F, Guidon N, Osipova L, Voevoda MI, Posukh OL, Balanovsky O, Lavryashina M, Bogunov Y, Khusnutdinova E, Gubina M, Balanovska E, Fedorova S, Litvinov S, Malyarchuk B, Derenko M, Mosher MJ, Archer D, Cybulski J, Petzelt B, Mitchell J, Worl R, Norman PJ, Parham P, Kemp BM, Kivisild T, Tyler-Smith C, Sandhu MS, Crawford M, Villems R, Smith DG, Waters MR, Goebel T, Johnson JR, Malhi RS, Jakobsson M, Meltzer DJ, Manica A, Durbin R, Bustamante CD, Song YS, Nielsen R, Willerslev E, POPULATION GENETICS. Genomic evidence for the Pleistocene and recent population history of Native Americans. Science. 349, aab3884 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rasmussen M, Li Y, Lindgreen S, Pedersen JS, Albrechtsen A, Moltke I, Metspalu M, Metspalu E, Kivisild T, Gupta R, Bertalan M, Nielsen K, Gilbert MTP, Wang Y, Raghavan M, Campos PF, Kamp HM, Wilson AS, Gledhill A, Tridico S, Bunce M, Lorenzen ED, Binladen J, Guo X, Zhao J, Zhang X, Zhang H, Li Z, Chen M, Orlando L, Kristiansen K, Bak M, Tommerup N, Bendixen C, Pierre TL, Grønnow B, Meldgaard M, Andreasen C, Fedorova SA, Osipova LP, Higham TFG, Ramsey CB, Hansen TVO, Nielsen FC, Crawford MH, Brunak S, Sicheritz-Pontén T, Villems R, Nielsen R, Krogh A, Wang J, Willerslev E, Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 463, 757–762 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rasmussen M, Anzick SL, Waters MR, Skoglund P, DeGiorgio M, Stafford TWJ, Rasmussen S, Moltke I, Albrechtsen A, Doyle SM, Poznik GD, Gudmundsdottir V, Yadav R, Malaspinas A-S, White SS 5th, Allentoft ME, Cornejo OE, Tambets K, Eriksson A, Heintzman PD, Karmin M, Korneliussen TS, Meltzer DJ, Pierre TL, Stenderup J, Saag L, Warmuth VM, Lopes MC, Malhi RS, Brunak S, Sicheritz-Ponten T, Bames I, Collins M, Orlando L, Balloux F, Manica A, Gupta R, Metspalu M, Bustamante CD, Jakobsson M, Nielsen R, Willerslev E, The genome of a Late Pleistocene human from a Clovis burial site in western Montana. Nature. 506, 225–229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rasmussen M, Sikora M, Albrechtsen A, Korneliussen TS, Moreno-Mayar JV, Poznik GD, Zollikofer CPE, de León MP, Allentoft ME, Moltke I, Jónsson H, Valdiosera C, Malhi RS, Orlando L, Bustamante CD, Stafford TWJ, Meltzer DJ, Nielsen R, Willerslev E, The ancestry and affiliations of Kennewick Man. Nature. 523, 455–458 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rodríguez-Varela R, Günther T, Krzewińska M, Storå J, Gillingwater TH, MacCallum M, Arsuaga JL, Dobney K, Valdiosera C, Jakobsson M, Gütherstrüm A, Girdland-Flink L, Genomic Analyses of Pre-European Conquest Human Remains from the Canary Islands Reveal Close Affinity to Modem North Africans. Curr. Biol 27, 3396–3402 (2017). [DOI] [PubMed] [Google Scholar]
- 153.Saag L, Varul L, Scheib CL, Stendemp J, Allentoft ME, Saag L, Pagani L, Reidla M, Tambets K, Metspalu E, Kriiska A, Willerslev E, Kivisild T, Metspalu M, Extensive Farming in Estonia Started through a Sex-Biased Migration from the Steppe. Curr. Biol 27, 2185–2193 (2017). [DOI] [PubMed] [Google Scholar]
- 154.Saag L, Laneman M, Varul L, Malve M, Valk H, Razzak MA, Shirobokov IG, Khartanovich VI, Mikhaylova ER, Kushniarevich A, Scheib CL, Solnik A, Reisberg T, Parik J, Saag L, Metspalu E, Rootsi S, Montinaro F, Remm M, Mägi R, D’Atanasio E, Crema ER, Díez-del-Molino D, Thomas MG, Kriiska A, Kivisild T, Villems R, Lang V, Metspalu M, Tambets K, The Arrival of Siberian Ancestry Connecting the Eastern Baltic to Uralic Speakers further East. Curr. Biol 29, 1701–1711.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Scheib L, Li H, Desai T, Link V, Kendall C, Dewar G, Griffith PW, Mürseburg A, Johnson JR, Potter A, Kerr SL, Endicott P, Lindo J, Haber M, Xue Y, Tyler-Smith C, Sandhu MS, Lorenz JG, Randall TD, Faltyskova Z, Pagani L, Danecek P, O’Connell TC, Martz P, Boraas AS, Byrd BF, Leventhal A, Cambra R, Williamson R, Lesage L, Holguin B, Soto EY-D, Rosas J, Metspalu M, Stock JT, Manica A, Scally A, Wegmann D, Malhi RS, Kivisild T, Ancient human parallel lineages within North America contributed to a coastal expansion. Science. 360, 1024–1027 (2018). [DOI] [PubMed] [Google Scholar]
- 156.Sánchez-Quinto, Malmström H, Fraser M, Girdland-Flink L, Svensson EM, Simões LG, George R, Hollfelder N, Burenhult G, Noble G, Britton K, Talamo S, Curtis N, Brzobohata H, Sumberova R, Gütherstrüm A, Storå J, Jakobsson M, Megalithic tombs in western and northern Neolithic Europe were linked to a kindred society. Proc. Natl. Acad. Sci 116, 9469–9474 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Schuenemann VJ, Peltzer A, Welte B, van Pelt WP, Molak M, Wang C-C, Furtwängler A, Urban C, Reiter E, Nieselt K, Tcßmann B, Francken M, Harvati K, Haak W, Schiffels S, Krause J, Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods. Nat Commun. 8, 15694 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Schiffels S, Haak W, Paajanen P, Llamas B, Popescu E, Loe L, Clarke R, Lyons A, Mortimer R, Sayer D, Tyler-Smith C, Cooper A, Durbin R, Iron Age and Anglo-Saxon genomes from East England reveal British migration history. Nat. Commun 7, 10408 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Schlebusch M, Malmström H, Günther T, Sjödin P, Coutinho A, Edlund H, Munters AR, Vicente M, Steyn M, Soodyall H, Lombard M, Jakobsson M, Southern African ancient genomes estimate modern human divergence to 350,000 to 260,000 years ago. Science. 358, 652–655 (2017). [DOI] [PubMed] [Google Scholar]
- 160.Schroeder H, Sikora M, Gopalakrishnan S, Cassidy LM, Maisano Delser P, Sandoval Velasco M, Schraiber JG, Rasmussen S, Homburger JR, Ávila-Arcos MC, Allentoft ME, Moreno-Mayar JV, Renaud G, Gómez-Carballa A, Laffoon JE, Hopkins RJA, Higham TFG, Carr RS, Schaffer WC, Day JS, Hoogland M, Salas A, Bustamante CD, Nielsen R, Bradley DG, Hofman CL, Willerslev E, Origins and genetic legacies of the Caribbean Taino. Proc Natl Acad Sci U S A. 115, 2341–2346 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Schroeder H, Margaryan A, Szmyt M, Theulot B, W\lodarczak P, Rasmussen S, Gopalakrishnan S, Szczepanek A, Konopka T, Jensen TZT, Witkowska B, Wilk S, Przyby\la MM, \Lukasz Pospieszny, Sjögren K-G, Belka Z, Olsen J, Kristiansen K, Willerslev E, Frei KM, Sikora M, Johannsen NN, Allentoft ME, Unraveling ancestry, kinship, and violence in a Late Neolithic mass grave. Proc. Natl. Acad. Sci 116, 10705–10710 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Seguin-Orlando, Korneliussen TS, Sikora M, Malaspinas A-S, Manica A, Moltke I, Albrechtsen A, Ko A, Margaryan A, Moiseyev V, Goebel T, Westaway M, Lambert D, Khartanovich V, Wall JD, Nigst PR, Foley RA, Lahr MM, Nielsen R, Orlando L, Willerslev E, Genomic structure in Europeans dating back at least 36,200 years. Science. 346, 1113–1118 (2014). [DOI] [PubMed] [Google Scholar]
- 163.Shinde V, Narasimhan VM, Rohland N, Mallick S, Mah M, Lipson M, Nakatsuka N, Adamski N, Broomandkhoshbacht N, Ferry M, Lawson AM, Michel M, Oppenheimer J, Stewardson K, Jadhav N, Kim YJ, Chatterjee M, Munshi A, Panyam A, Waghmare P, Yadav Y, Patel H, Kaushik A, Thangaraj K, Meyer M, Patterson N, Rai N, Reich D, An Ancient Harappan Genome Lacks Ancestry from Steppe Pastoralists or Iranian Farmers. Cell. 179, 729–735.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sikora M, Pitulko VV, Sousa VC, Allentoft ME, Vinner L, Rasmussen S, Margaryan A, de Barros Damgaard P, de la Fuente C, Renaud G, Yang MA, Fu Q, Dupanloup I, Giampoudakis K, Nogués-Bravo D, Rahbek C, Kroonen G, Peyrot M, McColl H, Vasilyev SV, Veselovskaya E, Gerasimova M, Pavlova EY, Chasnyk VG, Nikolskiy PA, Gromov AV, Khartanovich VI, Moiseyev V, Grebenyuk PS, Fedorchenko AY, Lebedintsev AI, Slobodin SB, Malyarchuk BA, Martiniano R, Meldgaard M, Arppe L, Palo JU, Sundell T, Mannermaa K, Putkonen M, Alexandersen V, Primeau C, Baimukhanov N, Malhi RS, Sjögren K-G, Kristiansen K, Wessman A, Sajantila A, Lahr MM, Durbin R, Nielsen R, Meltzer DJ, Excoffier L, Willerslev E, The population history of northeastern Siberia since the Pleistocene. Nature. 570, 182–188 (2019). [DOI] [PubMed] [Google Scholar]
- 165.Sikora M, Seguin-Orlando A, Sousa VC, Albrechtsen A, Korneliussen T, Ko A, Rasmussen S, Dupanloup I, Nigst PR, Bosch MD, Renaud G, Allentoft ME, Margaryan A, Vasilyev SV, Veselovskaya EV, Borutskaya SB, Deviese T, Comeskey D, Higham T, Manica A, Foley R, Meltzer DJ, Nielsen R, Excoffier L, Mirazon Lahr M, Orlando L, Willerslev E, Ancient genomes show social and reproductive behavior of early Upper Paleolithic foragers. Science. 358, 659–662 (2017). [DOI] [PubMed] [Google Scholar]
- 166.Skoglund P, Thompson JC, Prendergast ME, Mittnik A, Sirak K, Hajdinjak M, Salie T, Rohland N, Mallick S, Peltzer A, Heinze A, Olalde I, Ferry M, Harney E, Michel M, Stewardson K, Cerezo-Román JI, Chiumia C, Crowther A, Gomani-Chindebvu E, Gidna AO, Grillo KM, Helenius IT, Hellenthal G, Helm R, Horton M, López S, Mabulla AZP, Parkington J, Shipton C, Thomas MG, Tibesasa R, Welling M, Hayes VM, Kennett DJ, Ramesar R, Meyer M, Pääbo S, Patterson N, Morris AG, Boivin N, Pinhasi R, Krause J, Reich D, Reconstructing Prehistoric African Population Structure. Cell. 171, 59–71 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Skoglund P, Mallick S, Bortolini MC, Chennagiri N, Hünemeier T, Petzl-Erler ML, Salzano FM, Patterson N, Reich D, Genetic evidence for two founding populations of the Americas. Nature. 525, 104–108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Skoglund P, Posth C, Sirak K, Spriggs M, Valentin F, Bedford S, Clark GR, Reepmeyer C, Petchey F, Fernandes D, Fu Q, Harney E, Lipson M, Mallick S, Novak M, Rohland N, Stewardson K, Abdullah S, Cox MP, Friedlaender FR, Friedlaender JS, Kivisild T, Koki G, Kusuma P, Merriwether DA, Ricaut F-X, Wee JTS, Patterson N, Krause J, Pinhasi R, Reich D, Genomic insights into the peopling of the Southwest Pacific. Nature. 538, 510–513 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Skoglund P, Malmström H, Omrak A, Raghavan M, Valdiosera C, Günther T, Hall P, Tambets K, Parik J, Sjögren K-G, Apel J, Willerslev E, Storå J, Götherström A, Jakobsson M, Genomic diversity and admixture differs for Stone-Age Scandinavian foragers and farmers. Science. 344, 747–750 (2014). [DOI] [PubMed] [Google Scholar]
- 170.Slon V, Mafessoni F, Vernot B, de Filippo C, Grote S, Viola B, Hajdinjak M, Peyrégne S, Nagel S, Brown S, Douka K, Higham T, Kozlikin MB, Shunkov MV, Derevianko AP, Kelso J, Meyer M, Prüfer K, Pääbo S, The genome of the offspring of a Neanderthal mother and a Denisovan father. Nature. 561, 113–116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Unterländer M, Palstra F, Lazaridis I, Pilipenko A, Hofmanová Z, Groß M, Sell C, Blöcher J, Kirsanow K, Rohland N, Rieger B, Kaiser E, Schier W, Pozdniakov D, Khokhlov A, Georges M, Wilde S, Powell A, Heyer E, Currat M, Reich D, Samashev Z, Parzinger H, Molodin VI, Burger J, Ancestry and demography and descendants of Iron Age nomads of the Eurasian Steppe. Nat. Commun. 8, 14615 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Valdiosera, Günther T, Vera-Rodríguez JC, Ureña I, Iriarte E, Rodríguez-Varela R, Simões LG, Martínez-Sánchez RM, Svensson EM, Malmström H, Rodríguez L, Bermúdez de Castro J-M, Carbonell E, Alday A, Hernández Vera J, Götherström A, Carretero J-M, Arsuaga JL, Smith CI, Jakobsson M, Four millennia of Iberian biomolecular prehistory illustrate the impact of prehistoric migrations at the far end of Eurasia. Proc Natl Acad Sci U S A. 115, 3428–3433 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.van de Loosdrecht M, Bouzouggar A, Humphrey L, Posth C, Barton N, Aximu-Petri A, Nickel B, Nagel S, Talbi EH, El Hajraoui MA, Amzazi S, Hublin J-J, Pääbo S, Schiffels S, Meyer M, Haak W, Jeong C, Krause J, Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations. Science. 360, 548–552 (2018). [DOI] [PubMed] [Google Scholar]
- 174.van den Brink CM, Beeri R, Kirzner D, Bron E, Cohen-Weinberger A, Kamaisky E, Gonen T, Gershuny L, Nagar Y, Ben-Tor D, Sukenik N, Shamir O, Maher EF, Reich D, A Late Bronze Age II clay coffin from Tel Shaddud in the Central Jezreel Valley, Israel: context and historical implications. Levant. 49, 105–135 (2017). [Google Scholar]
- 175.Veeramah KR, Rott A, Groß M, van Dorp L, López S, Kirsanow K, Sell C, Blöcher J, Wegmann D, Link V, Hofmanová Z, Peters J, Trautmann B, Gairhos A, Haberstroh J, Päffgen B, Hellenthal G, Haas-Gebhard B, Harbeck M, Burger J, Population genomic analysis of elongated skulls reveals extensive female-biased immigration in Early Medieval Bavaria. Proc Natl Acad Sci U S A. 115, 3494–3499 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Vyas N, Al-Meeri A, Mulligan CJ, Testing support for the northern and southern dispersal routes out of Africa: an analysis of Levantine and southern Arabian populations. Am J Phys Anthropol.. 164, 736–749 (2017). [DOI] [PubMed] [Google Scholar]
- 177.Wang C-C, Reinhold S, Kalmykov A, Wissgott A, Brandt G, Jeong C, Cheronet O, Ferry M, Harney E, Keating D, Mallick S, Rohland N, Stewardson K, Kantorovich AR, Maslov VE, Petrenko VG, Erlikh VR, Atabiev B. Ch., Magomedov RG, Kohl PL, Alt KW, Pichler SL, Gerling C, Meller H, Vardanyan B, Yeganyan L, Rezepkin AD, Mariaschk D, Berezina N, Gresky J, Fuchs K, Knipper C, Schiffels S, Balanovska E, Balanovsky O, Mathieson I, Higham T, Berezin YB, Buzhilova A, Trifonov V, Pinhasi R, Belinskij AB, Reich D, Hansen S, Krause J, Haak W, Ancient human genome-wide data from a 3000-year interval in the Caucasus corresponds with eco-geographic regions. Nat. Commun 10, 590 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yang MA, Gao X, Theunert C, Tong H, Aximu-Petri A, Nickel B, Slatkin M, Meyer M, Pääbo S, Kelso J, Fu Q, 40,000-Year-Old Individual from Asia Provides Insight into Early Population Structure in Eurasia. Curr. Biol 27, 3202–3208 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zalloua P, Collins CJ, Gosling A, Biagini SA, Costa B, Kardailsky O, Nigro L, Khalil W, Calafell F, Matisoo-Smith E, Ancient DNA of Phoenician remains indicates discontinuity in the settlement history of Ibiza. Sci Rep. 8, 17567 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Picard toolkit. Broad Institute, GitHnb repository (2019), (available at http://broadinstitute.github.io/picard/).
- 181.Kuhn RM, Haussler D, Kent WJ, The UCSC genome browser and associated tools. Briefings in Bioinformatics. 14, 144–161 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Browning SR, Browning BL, Rapid and Accurate Haplotype Phasing and Missing-Data Inference for Whole-Genome Association Studies By Use of Localized Haplotype Clustering. The American Journal of Human Genetics. 81, 1084–1097 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hunt SE, McLaren W, Gil L, Thormann A, Schuilenburg H, Sheppard D, Parton A, Armean IM, Trevanion SJ, Flicek P, Cunningham F, Ensembl variation resources. Database. 2018 (2018), doi: 10.1093/database/bayll9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Li N, Stephens M, Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics. 165, 2213–2233 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Shen Y, Sun W, Wang H, Wang Y, Wang Y, Xiong X, Xu L, Waye MMY, Tsui SKW, Xue H, Wong JT-F, Galver LM, Fan J-B, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier J-F, Phillips MS, Roumy S, Sallée C, Verner A, Hudson TJ, Kwok P-Y, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui L-C, Mak W, Qiang Song Y, Tam PKH, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, Deloukas P, Bird CP, Delgado M, Dermitzakis ET, Gwilliam R, Hunt S, Morrison J, Powell D, Stranger BE, Whittaker P, Bentley DR, Daly MJ, de Bakker PIW, Barrett J, Chretien YR, Maller J, McCarroll S, Patterson N, Pe’er I, Price A, Purcell S, Richter DJ, Sabeti P, Saxena R, Schaffner SF, Sham PC, Varilly P, Altshuler D, Stein LD, Krishnan L, Vernon Smith A, Tello-Ruiz MK, Thorisson GA, Chakravarti A, Chen PE, Cutler DJ, Kashuk CS, Lin S, Abecasis GR, Guan W, Li Y, Munro HM, Steve Qin Z, Thomas DJ, McVean G, Auton A, Bottolo L, Cardin N, Eyheramendy S, Freeman C, Marchini J, Myers S, Spencer C, Stephens M, Donnelly P, Cardon LR, Clarke G, Evans DM, Morris AP, Weir BS, Tsunoda T, Johnson T, Mullikin JC, Sherry ST, Feolo M, Skol A, Zhang H, Zeng C, Zhao H, Matsuda E, Fukushima Y, Macer DR, Suda E, Rotimi CN, Adebamowo CA, Ajayi F, Aniagwu T, Marshall PA, Nkwodimmah C, Royal CDM, Leppert MF, Dixon M, Peiffer A, Qiu R, Kent A, Kato K, Niikawa N, Adewole IF, Knoppers BM, Foster MW, Wright Clayton E, Watkin J, Gibbs RA, Belmont JW, Muzny D, Nazareth F, Sodergren E, Weinstock GM, Wheeler DA, Yakub F, Gabriel SB, Onofrio RC, Richter DJ, Ziaugra F, Birren BW, Daly MJ, Altshuler D, Wilson RK, Fulton LL, Rogers J, Burton J, Carter NP, Clee CM, Griffiths M, Jones MC, McLay K, Plumb RW, Ross MT, Sims SK, Willey DL, Chen Z, Han H, Kang L, Godbout M, Wallenburg JC, L’Archevêque P, Bellemare G, Saeki K, Wang H, An D, Fu H, Li Q, Wang Z, Wang R, Holden AL, Brooks LD, McEwen JE, Guyer MS, Ota Wang V, Peterson JL, Shi M, Spiegel J, Sung LM, Zacharia LF, Collins FS, Kennedy K, Jamieson R, A second generation human haplotype map of over 3.1 million SNPs. Nature. 449, 851–861 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Haldane J, The combination of linkage values and the calculation of distances between the loci of linked factors. J Genet. 8, 299–309 (1919). [Google Scholar]
- 187.Donnelly P, Leslie S, The coalescent and its descendants. arXiv (2010) (available at https://arxiv.org/abs/1006.1514). [Google Scholar]
- 188.Rosen YM, Paten BJ, An average-case sublinear forward algorithm for the haploid Li and Stephens model. Algorithms for Molecular Biology. 14, 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hudson RR, Testing the constant-rate neutral allele model with protein sequence data. Evolution, 203–217 (1983). [DOI] [PubMed] [Google Scholar]
- 190.Wiuf C, Donnelly P, Conditional genealogies and the age of a neutral mutant. Theoretical Population Biology!. 56, 183–201 (1999). [DOI] [PubMed] [Google Scholar]
- 191.Wright S, Isolation by distance. Genetics. 28, 114–138 (1943). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Malécot G, Mathématiques de l’hérédité (1948). [Google Scholar]
- 193.Felsenstein J, A pain in the torus: some difficulties with models of isolation by distance. The american naturalist. 109, 359–368 (1975). [Google Scholar]
- 194.Barton NH, Kelleher J, Etheridge AM, A new model for extinction and recolonization in two dimensions: quantifying phylogeography. Evolution: International journal of organic evolution. 64, 2701–2715 (2010). [DOI] [PubMed] [Google Scholar]
- 195.Battey CJ, Ralph PL, Kern AD, Space is the Place: Effects of Continuous Spatial Structure on Analysis of Population Genetic Data. Genetics. 215, 193–214 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lemey P, Rambaut A, Drummond AJ, Suchard MA, Bayesian Phylogeography Finds Its Roots. PLOS Computational Biology. 5, 1–16 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Osmond MM, Coop G, Estimating dispersal rates and locating genetic ancestors with genome-wide genealogies. bioRxiv (2021). [Google Scholar]
- 198.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD, Inferring the Joint Demographic History of Multiple Populations from Multidimensional SNP Frequency Data. PLOS Genetics. 5, 1–11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Adrion JR, Cole CB, Dukler N, Galloway JG, Gladstein AL, Gower G, Kyriazis CC, Ragsdale AP, Tsambos G, Baumdicker F, Carlson J, Cartwright RA, Durvasula A, Gronau I, Kim BY, McKenzie P, Messer PW, Noskova E, Ortega-Del Vecchyo D, Racimo F, Struck TJ, Gravel S, Gutenkunst RN, Lohmueller KE, Ralph PL, Schrider DR, Siepel A, Kelleher J, Kem AD, A community-maintained standard library of population genetic models. eLife. 9, e54967 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kendall M, Colijn C, Mapping phylogenetic trees to reveal distinct patterns of evolution. Molecular biology and evolution. 33, 2735–2743 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Robinson DF, Foulds LR, Comparison of phylogenetic trees. Mathematical biosciences. 53, 131–147 (1981). [Google Scholar]
- 202.Kuhner MK, Yamato J, Practical Performance of Tree Comparison Metrics. Systematic Biology. 64, 205–214 (2014). [DOI] [PubMed] [Google Scholar]
- 203.Douka K, Sion V, Jacobs Z, Ramsey CB, Shunkov MV, Derevianko AP, Mafessoni F, Kozlikin MB, Li B, Grün R, Comeskey D, Devièse T, Brown S, Viola B, Kinsley L, Buckley M, Meyer M, Roberts RG, Pääbo S, Kelso J, Higham T, Age estimates for hominin fossils and the onset of the Upper Palaeolithic at Denisova Cave. Nature. 565, 640–644 (2019). [DOI] [PubMed] [Google Scholar]
- 204.Sankararaman S, Mallick S, Dannemann M, Prüfer K, Kelso J, Pääbo S, Patterson N, Reich D, The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 507, 354–357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Vernot B, Akey JM, Resurrecting Surviving Neandertal Lineages from Modern Human Genomes. Science. 343, 1017–1021 (2014). [DOI] [PubMed] [Google Scholar]
- 206.Vernot B, Tucci S, Kelso J, Schraiber JG, Wolf AB, Gittelman RM, Dannemann M, Grote S, McCoy RC, Norton H, Scheinfeldt LB, Merriwether DA, Koki G, Friedlaender JS, Wakefield J, Pääbo S, Akey JM, Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science. 352, 235–239 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Skov L, Coll Macià M, Sveinbjörnsson G, Mafessoni F, Lucotte EA, Einarsdóttir MS, Jonsson H, Halldorsson B, Gudbjartsson DF, Helgason A, Schiemp MH, Stefansson K, The nature of Neanderthal introgression revealed by 27,566 Icelandic genomes. Nature. 582, 78–83 (2020). [DOI] [PubMed] [Google Scholar]
- 208.Hubisz MJ, Williams AL, Siepel A, Mapping gene flow between ancient hominins through demography-aware inference of the ancestral recombination graph. PLOS Genetics. 16, 1–24 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Harris CR, Millman KJ, van der Walt SJ, Gommers R, Virtanen P, Cournapeau D, Wieser E, Taylor J, Berg S, Smith NJ, Kern R, Picus M, Hoyer S, van Kerkwijk MH, Brett M, Haldane A, Fernández del Rio J, Wiebe M, Peterson P, Gérard-Marchant P, Sheppard K, Reddy T, Weckesser W, Abbasi H, Gohlke C, Oliphant TE, Array programming with NumPy. Nature. 585, 357–362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Virtanen P, Gommers R, Oliphant TE, Haberland M, Reddy T, Cournapeau D, Burovski E, Peterson P, Weckesser W, Bright J, van der Walt SJ, Brett M, Wilson J, Millman KJ, Mayorov N, Nelson ARJ, Jones E, Kern R, Larson E, Carey CJ, Polat I, Feng Y, Moore EW, VanderPlas J, Laxalde D, Perktold J, Cimrman R, Henriksen I, Quintero EA, Harris CR, Archibald AM, Ribeiro AH, Pedregosa F, van Mulbregt P, SciPy 1.0 Contributors, SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nature Methods. 17, 261–272 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Newly reported sequencing data from the Afanasievo family is available from the European Nucleotide Archive, accession number PRJEB43093; phased variant data for the family is available from the European Variation Archive, accession number PRJEB46983. All publicly available datasets used in this paper are available from their original publications, tsinfer is deposited to Zenodo at doi:10.5281/zenodo.5168051 and available at https://tsinfer.readthedocs.io/ under the GNU General Public License v3.0 (30), tsdate is deposited to Zenodo at doi:10.5281/zenodo.5168040 and available at https://tsdate.readthedocs.io/ under the MIT License (31), and tskit is deposited to Zenodo at doi:10.5281/zenodo.5465773 and available at https://tskit.readthedocs.io/ under the MIT License (66). All code used to perform analyses in this paper is deposited to Zenodo at doi:10.5281/zenodo.5172104 and can be found at https://github.com/awohns/unified_genealogy_paper (67). Unified tree sequences of the HGDP, SGDP, and TGP autosomes are available from Zenodo at https://doi.org/10.5281/zenodo.5495535 (68). Unified tree sequences of the HGDP, SGDP, TGP, and high coverage ancient autosomes are available at https://doi.org/10.5281/zenodo.5512994 (69). Tree sequences were compressed using the tszip utility; see the documentation at https://tszip.readthedocs.io/ for further details.


