Abstract
Mitochondrial DNA variants have previously associated with disease, but the underlying mechanisms have been largely elusive. Here, we report that mitochondrial SNP rs2853499 associated with Alzheimer’s disease (AD), neuroimaging, and transcriptomics. We mapped rs2853499 to a novel mitochondrial small open reading frame called SHMOOSE with microprotein encoding potential. Indeed, we detected two unique SHMOOSE-derived peptide fragments in mitochondria by using mass spectrometry – the first unique mass spectrometry-based detection of a mitochondrial-encoded microprotein to date. Furthermore, cerebrospinal fluid (CSF) SHMOOSE levels in humans correlated with age, CSF tau, and brain white matter volume. We followed up on these genetic and biochemical findings by carrying out a series of functional experiments. SHMOOSE acted on the brain following intracerebroventricular administration, differentiated mitochondrial gene expression in multiple models, localized to mitochondria, bound the inner mitochondrial membrane protein mitofilin, and boosted mitochondrial oxygen consumption. Altogether, SHMOOSE has vast implications for the fields of neurobiology, Alzheimer’s disease, and microproteins.
Introduction
Mitochondrial-encoded microproteins are a potential part of Alzheimer’s disease (AD) that have not been heavily studied. Microproteins are biologically active peptides encoded by small open reading frames (sORFs) of around 100 codons or fewer. Most microproteins have been missed by past genomic and proteomic pipelines, but now thousands have been identified due to refined techniques 1–4. Some of these techniques have been used to identify several mitochondrial-encoded microproteins. The most heavily studied mitochondrial-encoded microprotein is humanin, a 24 amino-acid peptide encoded by a sORF within 16S rRNA. Humanin was cloned from the occipital lobe of an AD patient and has attenuated AD-related pathology in vivo via trimeric receptor signaling and amyloid beta binding 5–10. Yen et al. reported that a single nucleotide polymorphism (SNP) within humanin predicted cognitive age and circulating humanin levels 11. A separate SNP in a different sORF that encodes the mitochondrial-encoded microprotein MOTS-c has associated with type 2 diabetes in men 12. These findings suggest mitochondrial DNA variation within sORFs that could reveal novel microproteins.
Mitochodnria-wide association studies (MiWAS) – in which mtDNA variation is associated with phenotypes, otherwise known as the mitochondrial version of a genome wide association study (GWAS) – have returned inconsistent results for a variety of reasons. For examples, certain analyses might contain cohort-allele specificity that is due to the sampled population and genome array. In addition, various analytic methods – such as classifying mtSNPs into haplogroups, principal components, or binary SNP variables – have been used to estimate the effects of mitochondrial SNPs with no gold-standard approach12–14. Furthermore, some analyses have studied community-based or convenience-based samples that may not be representative of general populations. Nevertheless, recent large-scale MiWAS approaches have been recently used to robustly identify SNPs that associate with a variety of phenotypes, and the capacity to generate whole genome sequencing data with efficiency has improved 15. These results, though, are not typically interpreted in the context of mitochondrial-encoded microproteins and the majority are rarely experimentally validated.
Here, we hypothesized a mitochondrial DNA variant that associates with neurobiological phenotypes would reveal a novel microprotein that can be detected experimentally.
Methods
Mitochondrial-wide association study (MiWAS) and mtSNP AD association analyses
Mitochondrial genotypes and diagnosis from ADNI1, ADNI GO, ADNI2, and ADNI3 were all merged for analysis. ADNI1 samples were genotyped using the Illumina 610-Quad BeadChip; ADNI GO/2 samples were genotyped using the Illumina HumanOmniExpress BeadChip; ADNI1/GO/2 samples were whole genome sequenced; and ADNI3 samples were genotyped using the Illumina Infinium Global Screening Array v2 (GSA2). Whole mitochondrial genotypes were processed with stringent quality controls and made available by Ridge et al. in variant call format 16. Mitochondrial whole genome sequencing data was converted to suitable format using PLINK (v1.9) and merged into bed/bam/bim format. We carried out principal component analysis on these merged data. Principal components were generated via singular value-decomposition of the combined mtSNP matrix, outputting eigenvectors that approximates the matrix with a minimal number of values (prcomp function in R), as portrayed elsewhere 13, 14, 17, 18. The degree of mitochondrial genetic admixture was low and ideal for a permutation approach, the same approach that Lakatos et al. previously conducted on ADNI1 alone19. To maximize number of SNPs in the MiWAS model, we included a minor allele frequency threshold of 5% and 80% complete individual genotypes among the merged data, leaving 29 mtSNPs qualified for permutation on 448 controls and 290 cases. A total of 957 permutations were conducted for the most significant mtSNP. Furthermore, we estimated the effect of the SHMOOSE mtSNP in the Rush Alzheimer’s Disease Center (RADC) comprised of Religious Orders Study (ROS) and Memory and Aging Project (MAP), Late-Onset Alzheimer’s Disease (NIA-LOAD), and NIA Alzheimer Disease Centers (ADC1 and ADC2) cohorts using logistic regression. ROSMAP samples were genotyped using whole genome sequencing, and mitochondrial genetic variants were made available to qualified users in VCF format. LOAD samples were genotyped using the Illumina 610-Quad BeadChip, and the ADC1 and ADC2 samples were genotyped using the Ilumina Human660W-Quad BeadChip. Mitochondrial genetic admixture in ROSMAP (n = 281 cases and n = 233 controls), LOAD (n = 993 cases and n = 883 controls) and ADC1/2 (n = 2261 cases and n = 654 controls) were also assessed by implementing mitochondrial principal component analysis (Supplemental Figure 4). We observed mitochondrial genetic heterogeneity in the form of individual clusters for SHMOOSE alternative allele carriers in LOAD and ADC1/2, for which we corrected by including including the first three mitochondrial principal components in the logistic regression model, with age and biological sex as additional covariates. For meta-analysis, age and biological sex were included in the ADNI model. After the effects of the SHMOOSE mtSNP were estimated in ROSMAP, LOAD, and ADC1/2, meta-analysis was conducted using a random effects model approach from the metafor R package 20. Work was conducted under approved NIAGADS project 162 - Mitochondrial Peptide DNA Variation in Alzheimer’s Disease with University of Southern California IRB approval ID UP-22-00231.
Neuroimaging phenome-wide association study (PheWAS)
This work was conducted using ADNI and the UK Biobank Resource (https://www.ukbiobank.ac.uk) under approved project 25641. We used brain MRI imaging (UK Biobank data-field: 110) from the 2018 August release of 22,392 participants (http://biobank.ctsu.ox.ac.uk/crystal/label.cgi?id=110). Details of the MRI acquisition is described in the UK Biobank Brain Imaging Documentation (http://biobank.ctsu.ox.ac.uk/crystal/refer.cgi?id=1977) and in a protocol form (http://biobank.ctsu.ox.ac.uk/crystal/refer.cgi?id=2367) 21, 22. This study discarded 1,002 participants whose MRI scans did not pass manual quality assessment, 45 participants due to data withdrawal or failed image processing, and 3,055 participants who did not have white British ancestry and/or did not pass the sample quality control for the genetic data (https://biobank.ctsu.ox.ac.uk/crystal/label.cgi?id=100313), resulting a sample of 18,330 individuals with age range from 45 to 81 years (mean age = 63.27 ± 7.45 years), 8,729 males (47.62%), and 4,680 SHMOOSE mtSNP carriers (25.53%). All MR images were processed using the FreeSurfer software package v6.0 (https://surfer.nmr.mgh.harvard.edu) to extract brain-wide morphological measures. The FreeSurfer workflow includes motion correction and averaging of volumetric T1-weighted images 23, removal of non-brain tissue,24 automated Talairach transformation, brain volume segmentation 25, 26, intensity normalization 27, tessellation of the boundary between gray matter (GM) and white matter (WM), automated topology correction 28, and surface deformation following intensity gradients to optimally place the GM/WM and GM/cerebrospinal fluid borders at the location where the greatest shift in intensity defines the transition to the other tissue class 29. Each hemispheric GM and WM surface is composed of 163,842 vertices arranged as 327,680 triangles. Once the surface models are complete, a number of deformable procedures were performed for further data processing and analysis, including surface inflation, registration to a spherical atlas using individual cortical folding patterns to match cortical geometry across subjects 30, and finally creation of a variety of surface-based brain morphological metrics. All the procedure for MRI processing were implemented on the LONI pipeline system (http://pipeline.loni.usc.edu) for high-performance parallel computing 31, 32. This study included 9 vertex-wise brain morphological measures: cortical thickness, volume, WM surface area, pial surface area, sulcal depth, WM surface Jacobian, GM/WM contrast, mean curvature and Gaussian curvature. Detailed information about these surface-based metrics is available at https://surfer.nmr.mgh.harvard.edu/fswiki/. Briefly, cortical thickness values were calculated as the shortest distance between the gray and white matter surfaces at each vertex. Vertex-wise volume is calculated by dividing each obliquely truncated trilateral pyramid between the GM and WM surfaces into three tetrahedra. Vertex-wise surface area measures on the pial and WM surfaces are estimated by assigning one third of the area of each triangle to each vertex. Sulcal depth conveys information on how far removed a particular vertex point on a surface is from a hypothetical mid-surface that exists between the gyri and sulci. It gives an indication of linear distance and displacements: how deep and high are brain folds. Surface Jacobian measures how much the surface is distorted to register to the spherical atlas. GM/WM contrast presents the vertex-by-vertex percent contrast between white and gray matter, where WM is sampled 1mm below the white surface, and GM is sampled 30% the thickness into the cortex. Mean curvature is the average of the two principal curvatures at a vertex. The Gaussian curvature is the product of the two principal curvatures at a vertex. Prior to statistical analysis, these surface-based data were smoothed on the tessellated surfaces using a Gaussian kernel with the full width half maximum of 20 mm to increase the signal-to-noise ratio and to reduce the impact of mis-registration. All UK Biobank participants were genotyped using the Affymetrix UK BiLEVE Axiom array (on an initial ~50,000 participants) and the Affymetrix UK Biobank Axiom array (on the remaining ~450,000 participants) were genotyped using the Affymetrix UK Biobank Axiom array. SHMOOSE genotype was extracted from the genotyping array using the PLINK2.0 software. To capture population structure, the UKB team computed the top 40 principal components (PCs) from the high-quality genotyping dataset 33. Furthermore, to capture the population structure hidden in the mitochondrial genome, we also computed mitochondrial PCs using a mitochondrial principal component analysis 34. To test effects of the SHMOOSE mtSNP on age-related brain structural differences, we assessed the interaction between the SHMOOSE mtSNP genotype and age by implementing linear mixed-effects regression at each cortical surface vertex i for a given morphological measure Yi with the model:
where G is the SHMOOSE mtSNP genotype, Age is individuals’ age in the scanner, e is the residual error, and the intercept and β terms are the fixed effects. Sex, intracranial volume (ICV), nuclear and mitochondrial PCs were added to the model as confounding variables. Statistical results at all vertices were corrected for the family-wise error rate (FWER) across the brain surface using the random field theory (RFT) method that adapts to spatial smoothness of the neuroimaging data 35. All surface-based analyses were conducted using our Neuroimaging PheWAS system, which is a cloud-computing platform for big-data, brainwide imaging association studies 36.
Cognitive decline analysis
In HRS, we assessed the effect of SHMOOSE genotype on longitudinal cognitive decline over the aging process by implementing a mixed effects regression approach. HRS used the HumanOmni2.5 array to directly genotype 256 mitochondrial SNPs. SHMOOSE genotype was extracted using PLINK2.0. The validated HRS cognitive score represents episodic memory learning, episodic memory retrieval, semantic fluency, and orientation 37. The mixed effects model included fixed effect terms for biological sex, linear and quadratic age, mitochondrial genetic ancestry, and SHMOOSE genotype for European-ancestral individuals. Subject-specific random effects contained between-individual variation at the age of 65 in addition to inter-individual variation in the rate of cognitive score change during aging (i.e., follow-up visits every two years). The lme4 package in R was used to carry out the analysis. A total of 8,072 individuals were individually assessed with 45,465 total data points. Data approved by University of Southern California IRB approval ID UP-19-00637.
SHMOOSE protein structure prediction
RoseTTAFold was used to predict the microprotein structure of SHMOOSE and was developed by Baker lab at the University of Washington. Full algorithm details have been comprehensively detailed elsewhere 38. In comparison to Alphafold2, RoseTTAFold achieved similar degree of accuracy for complex proteins. The wild-type version of SHMOOSE and SHMOOSE.D47N were modeled, and output files were downloaded into PDB format. Three additional protein domain tools were used: HeliQuest, idpr: Profiling and Analyzing Intrinsically Disordered Proteins in R, and IUPred3. HeliQuest was used to assess amphipathic potential for the predicted SHMOOSE 3–11 alpha-helix type 39. The R package idpr was used to calculate SHMOOSE charge across a 9-amino-acid window using the chargeCalculationLocal function 40. IUPred3 was used to assess long disordered protein regions 41.
SHMOOSE antibody production and ELISA development
Rabbit anti-SHMOOSE sera were produced by YenZym Antibodies (Brisbane, CA). SHMOOSE affinity antibody was purified from rabbit anti-SHMOOSE sera using CarboxyLink Immobilization kit with UltraLink Support (Thermo Scientific) according to manufacturer’s protocol. Briefly, anti-sera were applied onto the synthetic SHMOOSE peptide immobilized column and the eluted fractions were quantitated by UV absorbance at 280nM. Circulating levels of SHMOOSE were measured by in-house ELISA. Prior to assay, CSF was extracted with 90% acetonitrile and 10% 1N HCl. To measure endogenous SHMOOSE levels, synthetic SHMOOSE peptide was used as standard within range 100 pg/ml to 20,000 pg/ml. Briefly, 96-well microtiter plate was coated with anti-SHMOOSE polyclonal antibody for 3 hours followed by blocking the plate with SuperBlock buffer (Thermo Scientific). Next, standards, controls or extracted samples and pre-titered detection antibody were added to the appropriate wells and incubated overnight. Followed by 3 washes, wells were added streptavidin-HRP conjugate and incubated for 30 minutes. After four washes, ultra-sensitive TMB (Thermo Scientific) was added and incubated for 10–20 minutes. The reaction was stopped by the addition of 2N sulfuric acid and absorbance was measured on a plate spectrophotometer at 450 nm. The intra- and inter-assay coefficient variations (CV) of SHOOSE ELISA were less than 10%, respectively.
Correlation between CSF SHMOOSE levels and CSF tau, CSF p-tau 181, and brain DTI
We considered data for 79 subjects recruited through the University of Southern California Alzheimer Disease Research Center (ADRC) who had available diffusion MRI (dMRI) scans and CSF measures of SHMOOSE. Of those, one did not have a usable dMRI scan and 6 were excluded for preprocessing failures identified through quality assessment (see Diffusion MRI Preprocessing). Sample use was approved by University of Southern California IRB HS2000589. Our final sample included 72 non-demented older (mean 65.7; 47–82 years old) adults who had diffusion MRI scans that passed all quality checks and available CSF measures of SHMOOSE. Subjects had a clinical dementia rating (CDR) score of 0 (56 subjects) or 0.5 (16 subjects). Subject race/ethnicities were self-reported as: White (53), Asian (12), American Indian or Alaska Native (3), more than one race (4), race not reported (1); Hispanic/Latino (any race) (10), non-Hispanic/Latino (any race) 62. MR images were acquired on a 3 Tesla Siemens Prisma scanner at the University of Southern California Alzheimer’s Disease Research Center (ADRC). Anatomical sagittal T1-weighted magnetization prepared rapid acquisition gradient-echo (MPRAGE) scan parameters were acquired (TR 2300 ms; TE 2.95 ms; 1.2 × 1.0 × 1.0 mm3 voxel size). We also acquired a 64-direction (b=1000 s/mm2) diffusion MRI scan (TR 7100 ms; TE 71.0 ms; 2.5 × 2.5 × 2.5 mm3 voxel size). All scans were visually assessed for quality. For each subject, the diffusion images were denoised with MATLAB version R2014b software (MathWorks, Natick, MA) using a local primary components analysis (LPCA) tool with the Rician filter, with intensity bias correction 42. Distortion correction of DWI included correction for Gibbs ringing using MRtrix3 43 and eddy current correction using the eddy_correct tool in FSL utilities (FSL 5.0.9; (www.fmrib.ox.ac.uk/fsl; 44). We performed bias field correction using MRtrix3 43. Echo planar imaging (EPI) susceptibility artifacts were corrected using FSL and ANTS software 45, 46 to align the average b0 maps to subject-specific T1-weighted MPRAGE structural scan. Each step was visually quality checked. Fractional anisotropy (FA) maps – indicating diffusion restriction within a voxel – were created using FSL software. FA is a metric of microstructural integrity shown to augment the power to detect AD-specific deficits 47 with lower FA values typically representing poor white matter microstructural integrity in AD (as reviewed in 48. Voxelwise statistical analysis of DWI FA data was performed using the FSL-based tool49, tract based-spatial statistics (TBSS) 50. TBSS applies nonlinear registration to bring all FA maps into standard template space. A mean FA skeleton was created and then thresholded at 0.2, resulting in a 4D skeletonised FA image used in voxelwise statistical analyses detailed below. We used the general linear model (GLM) with FSL’s Threshold-Free Cluster Enhancement (TFCE) option to evaluate the relationship between SHMOOSE and voxelwise white-matter FA within the mean FA skeleton, covarying for CDR score, age, and reported sex 51. SHMOOSE values <100 were coded as 50 for this analysis (5 subjects). For CSF tau and p-tau 181, linear regression analysis was conducted with biological sex and age as a covariate, and SHMOOSE CSF levels as the dependent variable. Separately, we considered the effects for p-tau 181 were mediated through total tau levels. Hence, we used the mediation package in R and modeled the effect of p-tau 181, age, and biological sex on the mediator (i.e., tau); modeled the effect of tau, p-tau 181, biological sex, and age on SHMOOSE; and used these models to determine the indirect (ACME) and direct (ADE) effects using the mediate function.
Co-expression analysis
We utilized transcriptome data generated by Mayo (Synapse ID: syn5550404). SHMOOSE transcript count matrices were created from made-available bam files. This was done by constructing a sORF database in GTF format and implementing the summarizeOverlaps function of the GenomicAlignments package in R. Thereafter, normalized counts were used to conduct correlation between SHMOOSE counts and all nuclear-encoded gene counts, corrected for multiple hypotheses using a false discovery rate (FDR) of 0.05. Genes that statistically correlated with SHMOOSE expression were tested for enrichment using the enrichGo function from the clusterProfiler package, which returns enrichment of Gene Ontology (GO) categories after FDR control. Data output from the enrichGo function were used to generate plots using ggplot2 in R.
Cells
SH-SY5Y cells used in the study were purchased from ATCC (CRL-2266) without mycoplasma contamination. Cells were grown in DMEM/F12 with 10%FBS at 37°C with 5% CO2 and split every 4–7 days depending on confluency. In addition, for rho zero cells, SH-SY5Y cells were depleted of mitochondrial DNA by adding 5 ug/ml ethidium bromide, 50 ug/ml uridine, and 1mM pyruvate for approximately two months, as previously described 52. For all experiments, cells were differentiated by addition of 10uM retinoic acid in DMEM/F12 with 1%FBS, and the media was changed once every 48 hours for a total of two changes, as described previously. When indicated, cells were treated with chemically synthesized SHMOOSE, which was made by GenScript by solid-phase peptide synthesis methods. Triflouracetic acid (TFA) was used to cleave synthesized peptide from resin. After peptide synthesis, residual TFA was removed and the pH of reconstituted SHMOOSE was neutral.
Subcellular fractionation
Cytosolic, nuclear, and mitochondrial fractions were prepared from cultured SH-SY5Y cells. To extract nuclei, cells were washed in ice-cold DPBS and resuspended in fractionation buffer containing 10 mM HEPES pH 7.6, 3 mM MgCl2, 10 mM KCl, 5% (v/v) glycerol, 1% Triton-X100, and protease/phosphatase inhibitors for 15 minutes, followed by centrifugation for 5 minutes at 250 × g and 4°C. The resulting supernatant was further centrifuged once more at 18,000 × g for 10 minutes at 4°C to obtain a relatively pure cytoplasmic fraction. The original pellet prior to the 18,000 × g centrifugation was washed in 10mM HEPES pH 7.6, 1.5mM MgCl2, 10 mM KCl, and protease/phosphatase inhibitors and centrifuged at 250 × g and 4°C. The washed pellet was then resuspended in nuclear extraction buffer containing 20 mM HEPES pH 7.6, 1.5mM MgCl2, 420 mM NaCl, 25% (v/v) glycerol, 0.2 mM EDTA, and protease/phosphatase inhibitors, followed by three sonication periods of 5 seconds (separated by 10 seconds) with 30% amplitude on ice. The sonicated pellet was centrifuged at 18,000 × g for 10 minutes at 4°C to obtain a relatively pure nuclear lysate. To extract mitochondria, cells were washed in ice-cold DPBS and resuspended in 2 ml hypotonic buffer containing 10mM NaCl, 1.5mM MgCl2, and 10mM Tris-HCl pH 7.5 for 7.5 minutes. After the hypotonic incubation, cells were transferred to a glass homogenizer and homogenized by pressing straight down with the pestle 20 times, bursting the cells open while maintaining the integrity of mitochondria. Mitochondrial homogenization buffer (MHB) was then added to the 2ml homogenized sample to achieve a 1X concertation (210 mM mannitol, 70 mM sucrose, 20mM HEPES, and 2mM EGTA). The homogenate was then transferred to a clean 5 ml tube and centrifuged at 17,000 × g for 15 minutes at 4°C. The resulting pellet was washed in MHB buffer and centrifuged two more times, followed by a resuspension of the mitochondrial pellet in RIPA lysis buffer, and final centrifugation step of 14,000 × g for 10 minutes at 4°C to obtain a relatively pure mitochondrial lysate. For exogenous SHMOOSE administration, 1 uM of SHMOOSE was administered to cells for 30 minutes, washed twice in cold PBS, and fractionated. 5–15 mg of protein were reduced in NuPAGE sample buffer and run on NuPAGE 4–12% Bis-Tris gels. Proteins were transferred to PVDF membranes, blocked with 5% BSA in TBS 0.1% tween, and incubated with respective antibodies (GRSF1, Sigma HPA036985; Laminin B1, Cell Signaling D4Q4Z; GAPDH, Cell Signaling 5174S) at 1:1000 dilutions overnight at 4C. The next day, membranes were washed with TBST0.1% and incubated with 1:30,000 secondary antibody conjugated to HRP against the respective primary antibody species of origin, then excited using ECL reagent for 5 minutes.
Endogenous SHMOOSE detection using mass spectrometry
Eluents following SHMOOSE immunoprecipitation from mitochondrial fractions were precipitated by methanol/ chloroform and redissolved in 8 M urea/100 mM TEAB, pH 8.5. Proteins were reduced with 5 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP, Sigma-Aldrich) and alkylated with 10 mM chloroacetamide (Sigma-Aldrich). SHMOOSE was immunoprecipitated by coupling 10 ug of polyclonal SHMOOSE antibody to Dynabeads Protein A (Thermo). Approximately 5 × 107 differentiated SH-SY5Y cells were used for immunoprecipitation. Eluted immunoprecipitation sample was digested overnight at 37 C in 2 M urea/100 mM TEAB, pH 8.5, with trypsin (Promega). Digestion was quenched with formic acid, 5 % final concentration.
The digest was analyzed on a Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo), and the digest was injected directly onto a 25 cm, 100 um ID column packed with BEH 1.7um C18 resin (Waters). Samples were separated at a flow rate of 400 nl/min on an Easy nLC 1000 (Thermo). Buffer A and B were 0.1% formic acid in water and 0.1% formic acid in 90% acetonitrile, respectively. A gradient of 1–25% B over 100 min, an increase to 40% B over 20 min, an increase to 90% B over 10 min and held at 90%B for a final 10 min was used for 140 min total run time. Column was re-equilibrated with 15 ul of buffer A prior to the injection of sample. Peptides were eluted directly from the tip of the column and nanosprayed directly into the mass spectrometer by application of 2.5 kV voltage at the back of the column. The Lumos was operated in a data dependent mode. Full MS scans were collected in the Orbitrap at 120K resolution with a mass range of 400 to 1500 m/z and an AGC target of 4e5. The cycle time was set to 3 sec, and within this 3 sec the most abundant ions per scan were selected for CID MS/MS in the ion trap with an AGC target of 2e4 and minimum intensity of 5000. Maximum fill times were set to 50 ms and 35 ms for MS and MS/MS scans respectively. Quadrupole isolation at 1.6 m/z was used, monoisotopic precursor selection was enabled and dynamic exclusion was used with exclusion duration of 5 sec.
Protein and peptide identification were done with Integrated Proteomics Pipeline – IP2 (Integrated Proteomics Applications). Tandem mass spectra were extracted from raw files using RawConverter 53 and searched with ProLuCID 54 against Uniprot human database with the SHMOOSE sequence. The search space included all full-tryptic and half-tryptic peptide candidates. Carbamidomethylation on cysteine was considered as a static modification. Data was searched with 50 ppm precursor ion tolerance and 600 ppm fragment ion tolerance. Identified proteins were filtered to using DTASelect 55 and utilizing a target-decoy database search strategy to control the false discovery rate to 1% at the protein level 56.
ICV
Adult male Sprague-Dawley rats (Envigo, 250–275g on arrival) were anesthetized via an intramuscular injection of an anesthesia cocktail (ketamine 90mg/kg body weight [BW], xylazine, 2.8mg/kg BW and acepromazine and 0.72mg/kg BW) for implantation of a unilateral indwelling intracerebroventricular (ICV) cannula (26-gauge, Plastics One, Roanoke, VA) targeting the lateral ventricle (VL) at the following stereotaxic coordinates: −0.9mm AP defined at bregma, +1.8mm ML defined at bregma, −2.6mm DV defined at skull surface at site. Placement for the VL cannula was verified by elevation of blood glucose resulting from an injection of 210μg (2μL) of 5-thio-D-glucose (5TG) using an injector that extended 2mm beyond the end of the guide cannula (Ritter et al., 1981). All experiments were approved by the Animal Care and Use Committee at the University of Southern California and performed in accordance with the National Research Council Guide for the Care and Use of Laboratory Animals.
Rats received a 2uL ICV infusion of bolus of 5nmol SHMOOSE, DN47 or vehicle solution (10% dimethylsulfoxide in artificial cerebrospinal fluid) that was hand delivered via a 33-gauge micro syringe injector attached to a PE20 catheter and Hamilton syringe through the indwelling guide cannula. Rats weight did not differ among groups and conditions were randomized. The investigator was not blinded to the study. Dosing and sample size were derived from past mitochondrial-encoded microprotein ICV experiments 57. Samples were processed one-at-at-time by condition (i.e., Vehicle, SHMOOSE, D47N). The following day, brains were harvested from anesthetized rats and microdissections of the hypothalamus and dorsal hippocampus were flash frozen in isopentane surrounded by dry ice.
RNA was extracted from snap frozen tissue following a thaw cycle, with TRIzol (Thermo Scientific), and the Quick-RNA Miniprep Kit (Zymo Research). High quality RNA used for library preparation by the commercial company Novogene, which enriched poly-adenylated RNA, generated cDNA, and sequenced reads the Illumina NovaSeq platform. High quality fastq files were ensured using FastQC and mapped to the mRatBN7.2 reference transcriptome using kallisto. Normalized fold changes were then used to estimate differential gene expression between SHMOOSE and vehicle ICV using the DESeq2 package in R. Gene set enrichment (GSEA) was carried out on significantly different gene (FDR < 0.2) using the clusterProfiler package in R with the gseGO function 58.
SHMOOSE-mitofilin protein interaction assays
To identify the SHMOOSE interactome, multiple experiments were conducted. First, 1.5 nmol of SHMOOSE was spiked into 1mg SH-SY5Y cell lysate for 6 hours at 4 C. Lysates were prepared using Thermo Pierce CoIP lysis buffer with 1X Thermo protease inhibitor cocktail and 1mM PMSF. Briefly, cells were washed with ice-cold DPBS twice, lysed for 15 minutes on ice, and centrifuged for 10 minutes at 12,000 RCF at 4C. The rationale for conducting this experiment was to ensure identical protein amounts between conditions and avoid differential protein expression caused by SHMOOSE treatment to cells. After 6 hours, SHMOOSE was immunoprecipitated from samples using Dynabeads A conjugated to 5ug of custom c-terminus SHMOOSE antibody. As a negative control, SHMOOSE-spiked lysates were also immunoprecipitated using 5ug rabbit IgG. Proteins were eluted from beads using 50 mM Glycine pH 2.8, and eluents were pH neutralized using Tris HCl pH 7.5. Complete eluents were then processed for protein identification using LC-MS. Samples were mixed with same volume of digestion buffer (8M Urea, 0.1M Tris-HCl pH 8.5), then each sample was reduced and alkylated via sequential 20-minute incubations with 5 mM TCEP and 10 mM iodoacetamide at room temperature in the dark while being mixed at 1200 rpm in an Eppendorf thermomixer. 6 μl of carboxylate-modified magnetic beads (CMMB and widely known as SP3) was added to each sample. Ethanol was added to a concentration of 50% to induce protein binding to CMMB. CMMB were washed 3 times with 80% ethanol and then resuspended with 50μl 50mM TEAB. The protein was digested overnight with 0.1 μg LysC (Promega) and 0.8 μg trypsin (Pierce) at 37 °C. Following digestion, 1ml of 100% acetonitrile was added to each to sample to increase the final acetonitrile concentration to over 95% to induce peptide binding to CMMB. CMMB were then washed 3 times with 100% acetonitrile and the peptide was eluted with 50 μl of 2% DMSO. Eluted peptide samples were dried by vacuum centrifugation and reconstituted in 5% formic acid before analysis by LC-MS/MS. Peptide samples were separated on a 75μM ID, 25cm C18 column packed with 1.9 μM C18 particles (Dr. Maisch GmbH HPLC) using a 140-minute gradient of increasing acetonitrile concentration and injected into a Thermo Orbitrap-Fusion Lumos Tribrid mass spectrometer. MS/MS spectra were acquired using Data Dependent Acquisition (DDA) mode. MS/MS database searching was performed using MaxQuant (1.6.10.43) against the human reference proteome from EMBL (UP000005640_9606 HUMAN Homo sapiens, 20874 entries) 59. Statistical analysis of MaxQuant label-free quantitation data was performed with the artMS Bioconductor package, which performs the relative quantification of protein abundance using the MSstats Bioconductor package (default parameters) 60. The abundance of proteins missing from one condition but found in more than 2 biological replicates of the other condition for any given comparison were estimated by imputing intensity values from the lowest observed MS1-intensity across samples and p values were randomly assigned to those between 0.05 and 0.01 for illustration purposes 61. We chose to target mitofilin (IMMT) based on imputed fold change and p value thresholds.
Second, we validated the mitofilin interaction identified from MS using a series of reciprocal co-immunoprecipitation experiments with our SHMOOSE antibody and mitofilin antibody (Proteintech 10179). We treated differentiated SH-SY5Y cells for 30 minutes with 1 uM SHMOOSE and lysed cells using Thermo Pierce CoIP lysis buffer as mentioned above. Thereafter, we incubated samples with 5 ug of SHMOOSE antibody, mitofilin antibody, or negative rabbit IgG for 30 minutes at room temperature. Antibody-coupled Dynabeads A were washed 3 times with TBST0.1% and eluted using Glycine pH 2.8, NuPAGE LDS sample buffer, and NuPAGE sample reducing agent for 5 minutes at 95 C. Eluents were then loaded into NuPAGE 4–12% Bis-Tris gels for electrophoresis. Migrated proteins were transferred to PVDF membranes, blocked with 5% BSA in TBST0.1% tween, and incubated with respective antibodies at 1:1000 dilutions overnight at 4C. The next day, membranes were washed with TBST0.1% and incubated with 1:30,000 secondary antibody conjugated to HRP against the respective primary antibody species of origin, then excited using ECL reagent for 5 minutes.
Third, we conducted reciprocal dot blots for SHMOOSE, SHMOOSE.D47N, and mitofilin by immobilizing 140 ng of recombinant mitofilin (OriGene TP760022), SHMOOSE, or SHMOOSE.D47N on nitrocellulose membranes. After proteins were dried, membranes were blocked for 30 minutes with SuperBlock (PBS) blocking buffer (Thermo) at room temperature. Then, either SHMOOSE or mitofilin were flowed over blocked membranes at a concentration at 1 ug/ml for 30 minutes at room temperature in blocking buffer. Membranes were washed three times for five minutes each with TBST0.1% and then incubated with 0.5 ug/ml of respective antibodies for 30 minutes at room temperature in blocking buffer. Membranes were washed three times for five minutes each with TSBT 0.1% and incubated with 1:30,000 secondary antibodies against species of primary antibody origin. Membranes were washed three times for five minutes each with TBST 0.1%, followed by excitation using ECL reagent for 1 minute.
Mitofilin knockdown assay
Mitochondrial superoxide assays were used to measure the effect of mitofilin knockdown. SH-SY5Y cells were reverse transfected using RNAiMAX (Invitrogen) and 40 nM mitofilin siRNA (Horizon, SMARTpool) when plated into 96-well plates at a density of 10,000 cells. The following day, cells were differentiated for a total of 4 days and transfected with another 40 nM mitofilin siRNA. Two days later, differentiation medium was changed with an addition 40 nM mitofilin siRNA. 24 hours before the mitochondrial superoxide assay, cells were treated with 10 uM SHMOOSE or solvent control. The MitoSOX assay was performed according to manufacture instructions (Thermo Fisher Scientific; M36008).
MTT and Extracellular Flux (Seahorse) assays
SH-SY5Y cells were plated into 96-well plates at a density of 10,000 cells. The following day, cells were differentiated for a total of 4 days. Thereafter, SHMOOSE or SHMOOSE.D47N were incubated for 24 hours, followed by MTT metabolic activity assay and cell real-time oxygen consumption rate measurements using XF96 Extracellular Flux Analyzer (Seahorse Bioscience) at the USC Leonard Davis School of Gerontolgoy Seahorse Core. MTT (Sigma-Aldrich) reagent (5 mg/ml) was added to each well after treatments for four hours and lysed before absorbance values were read. ATP turnover and maximum respiratory capacity were calculated after challenging cells with oligomycin and FCCP (carbonyl cyanide 4-[trifluoromethoxy]phenylhydrazone). Additionally, glycolytic rate was determined using extracellular acidification rate (ECAR) and individually reported relative to basal level in percentage. All reading were normalized to total DNA content using Hoechst 33342.
SHMOOSE differential expression in iPSCs and AD brains
RNA-Seq data from neurons derived from iPSCs with FAD mutations were downloaded to test SHMOOSE expression as a function of FAD mutations (GEO: GSE128343). Fastq files were aligned to the human reference genome (GRCh38.p13) using STAR with default parameters 62. Aligned BAM files were loaded into R using the BioConductor package. A custom GTF file containing the SHMOOSE genomic coordinates and other mitochondrial genes were used for the differential expression analysis. Counts were normalized to mitochondrial read count. Counts were called using the “union” mode by the summarizeOverlaps function 63. Differential expression analysis was conducted using negative binomial regression by the DESeq2 package in R. We also used Mayo RNASeq data (Synapse ID: syn5550404) to assess SHMOOSE RNA differences by AD and by genotype, following the same processing workflow.
Amyloid beta toxicity assay
SH-SY5Y cells were differentiated for 4 days, incubated with 10 uM SHMOOSE or SHMOOSE.D47N for 24 hours followed by another incubation with SHMOOSE or SHMOOSE.D47N with or without oligomerized 1uM amyloid beta 42 (CPC Scientific), prepared as formerly described 64. A two-color fluorescence cell viability assay (LIVE/DEAD Viability/Cytotoxicity Kit; Invitrogen (cat. L3224) was used to distinguish live cells from dead cells after SHMOOSE and amyloid beta 42 treatment. The ratio of live to dead cells can be quantified since live cells retain the Calcein AM dye and dead cells with damaged membranes permit entry of the ethidium homodimer dye.
Human brain SHMOOSE mtSNP differential expression analysis
We utilized genotype and transcriptome data generated by Mayo (Synapse ID: syn5550404). “RNAseq TCX” data were analyzed by SHMOOSE genotype. Subject SHMOOSE genotype was extracted from Mayo LOAD GWAS data that was generated from the HumanHap300-Duo Genotyping BeadChips. A complete description of the processing and individual sub cohorts has been described previously 65. Briefly, gene expression fastq files from human brain temporal cortex were aligned using the Mayo MAP-RNAseq pipeline 66. Normalized read counts were then examined for differential expression by SHMOOSE genotype using multi-variable linear regression to adjust for age at death, biological sex, and RNA integrity. Source code in R provided by Mayo was modified to conduct the differential expression analysis by SHMOOSE genotype 67. Results contain all genes that have non-zero raw counts in at least 1 subject, and each gene contains a beta value representing the effect size by SHMOOSE mtSNP. Multiple hypothesis correction was performed using Benjamin Hochberg. Significant genes were included in gene enrichment analyses using the clusterProfiler package in R. By using the enrichGo and enrichWP functions, significantly enriched pathways were extracted according to a hypergeometric model 58. A total of 14 SHMOOSE mtSNP carriers were assessed against 55 reference allele SHMOOSE individuals. These samples were selected by extracting non-demented individuals at time of death that also contained SHMOOSE genotype data.
SHMOOSE-treated cell transcriptomics
Differentiated neural cells were incubated with 10uM SHMOOSE or SHMOOSE.D47N for 24 hours followed by rapid RNA extraction. Cells were washed once with ice-cold DPBS and immediately lysed with TRIzol (Thermo Scientific), and RNA was extracted using the Quick-RNA Miniprep Kit (Zymo Research). High quality RNA used for library preparation (mRNA-Seq Nu Quant), which captures poly-adenylated RNA. From there, prepped samples were sequenced on an Illumina NextSeq 550 platform for 75 single end cycles. Each sample achieved a read depth of nearly 25 million. High quality fastq files were ensured using FastQC and mapped to the human reference genome (GRCh38.p13) using kallisto. Normalized fold changes were then used to estimate differential gene expression between SHMOOSE and SHMOOSE.D47N treated cells using the DESeq2 package in R. Gene enrichment was carried out on significantly different gene (FDR < 0.2) using the clusterProfiler package in R with the enrichGO function. For the SHMOOSE vs. SHMOOSE.D47N comparison in vivo, the same gene enrichment techniques were used.
Results
Mitochondrial rs2853499 associates with AD and neuroimaging and occurs within a previously unannotated microprotein called SHMOOSE
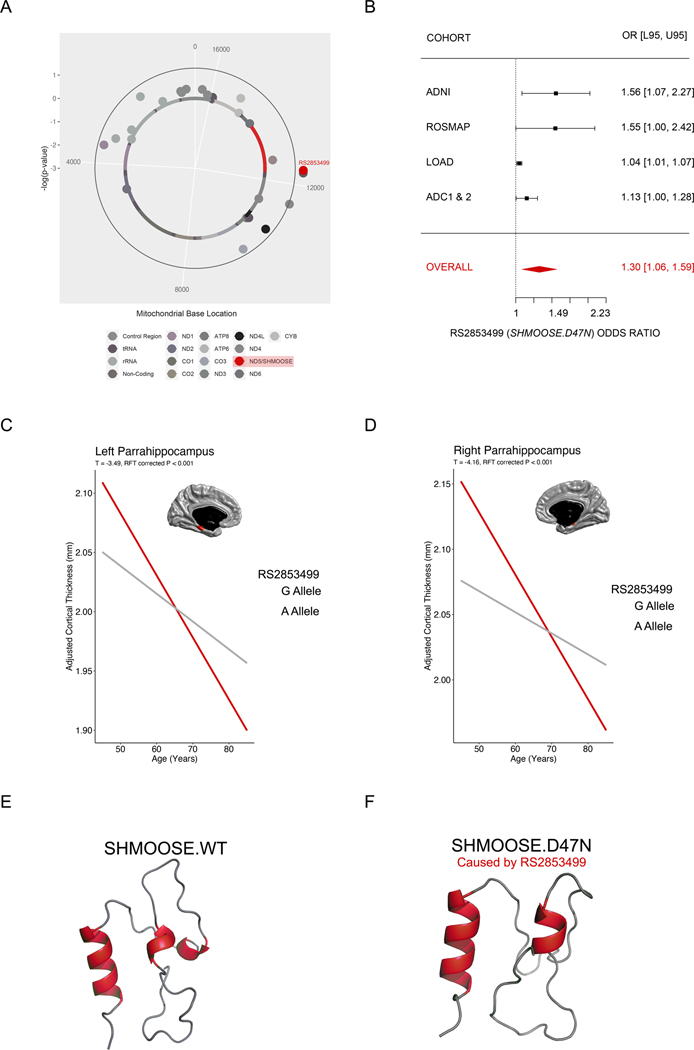
We tested the hypothesis that mitochondrial SNPs (mtSNPs) within sORFs would associate with AD. Previously, mitochondrial haplogroup UK associated with AD in the ADNI 1 cohort 19. ADNI has since expanded into multiple new phases, which we included in our MiWAS. We confirmed haplogroup UK SNPs associated with AD (Fig. 1A; full MiWAS results in Supplementary Tables 1–2). These three haplogroup SNPs at base pair positions 11467, 12308, and 12372 do not change the amino acid sequences of the mt-ND4 and mt-ND5 proteins. However, mtSNP 12372 (rs2853499) does change the amino acid sequence of a microprotein encoded by a sORF that we ultimately called SHMOOSE (Small Human Mitochondrial ORF Over SErine tRNA). In ADNI, SHMOOSE.D47N carriers presented an odds ratio of 1.56 (case frequency: 22.9%; control frequency: 15.7%; 95% CI: 1.06–2.30; permutation empirical p value < 0.03).
Fig. 1. Mitochondrial rs2853499 predicts AD risk and parahippocampus thickness.
A The GWAS Manhattan plot-equivalent of a MiWAS, called a Solar Plot. SNPs extending beyond the outer blue are statistically significant by a permutation empirical p value of 0.05. The most significant mtSNPs were rs2853498 and rs2853499 – both are haplogroup U determining, with the latter causing the missense change to SHMOOSE. B Meta-analysis forest plot of rs2853499 in ADNI, ROSMAP, LOAD, and ADC/1/2. Bars represent 95% confidence intervals. C Neuroimaging-based PheWAS in UK Biobank that illustrates the significant effects of SHMOOSE.D47N and age on the left parahippocampus. D Neuroimaging-based PheWAS in UK Biobank that illustrates the significant effects of SHMOOSE.D47N and age on the right parahippocampus E RosettaTTFold prediction of SHMOOSE. F RosettaTTFold prediction of SHMOOSE.D47N, caused by rs2853499.
Next, we examined the associative effect of SHMOOSE.D47N on AD in three additional cohorts: Religious Orders Study (ROS) and Memory and Aging Project (MAP), Late-Onset Alzheimer’s Disease (LOAD), and NIA Alzheimer Disease Centers (ADC1 and ADC2). In these three cohorts, the odds ratio of SHMOOSE.D47N was 1.55 (ROSMAP), 1.04 (LOAD) and 1.13 (ADC1 and ADC2). These three cohorts were analyzed with consideration to cohort-specific allele frequency differences (Supplemental Table 3). Mitochondrial genetic heterogeneity for SHMOOSE.D47N carriers was low in ADNI and ROSMAP and much greater in LOAD and ADC1/2, which was corrected in statistical models (Supplemental Fig. 3). Altogether, random effects meta-analysis estimated a SHMOOSE.D47N odds ratio of 1.30 (95% CI: 1.06–1.59; p value < 0.005; Fig. 1B; Supplemental Table 3). In addition, we estimated the effect of SHMOOSE.D47N associated with rapid cognitive decline in the Health and Retirement Study (HRS), a population-based study of US adults aged 50 years or older (n = ~15,000; Supplementary Fig. 4) 37.
Since GWAS and MiWAS are prone to spurious associations, we carried out a phenome wide association study (PheWAS) on 4,000 neuroimaging markers in UK Biobank (n = ~18,300 European-ancestral individuals). A significant advantage of PheWAS is the replication of SHMOOSE.D47N across related neurobiological phenotypes, which is both targeted and statistically robust due to abundant power. SHMOOSE.D47N (frequency: 25.5%) significantly associated with cortical thickness, volume, pial surface area, WM surface Jacobian, and GM/WM contrast in several paralimbic regions, including the parahippocampal gyri, the entorhinal cortex (EC), the anterior cingulate cortex (ACC), the posterior cingulate cortex (PCC), and the temporal pole (TPO) (clusterwise, RFT-corrected p value < 0.05; Supplementary Fig. 5). Over age-span, SHMOOSE.D47N carriers exhibited accelerated thinning of the parahippocampus (Fig. 1C), EC, and PCC. That is, across age trajectories of SHMOOSE.D47N showed inversed effects at younger and older ages. For instance, the SHMOOSE reference allele associated with smaller brain structural measures at middle (45–65 years) and/or young-old (65–75 years) ages, whereas the alternative allele (i.e., SHMOOSE.D47N) associated with structural loss at old ages (>75 years). In addition to the predominant results in the paralimbic areas, at a lenient threshold of uncorrected p value < 0.05, we observed distributed trends of SHMOOSE.D47N effects in the language centers (superior temporal and inferior frontal gyri), dorsolateral and medial prefrontal cortex, central motor, and occipital visual cortices (Supplemental Fig. 6). Separately, we observed similar trends in the ADNI PheWAS, which was conducted on the same sample used for our ADNI permutation MiWAS. As we observed in UK Biobank, SHMOOSE.D47N significantly associated with limbic regions such as the medial temporal cortex and posterior cingulate cortex at a lenient threshold of uncorrected p value < 0.05 (Supplemental Fig. 7).
rs2853499 (henceforth referred to as SHMOOSE.D47N) changes the 47th amino acid from glutamine to aspartic acid (Fig. 1D).
Endogenous SHMOOSE is detected in mitochondria
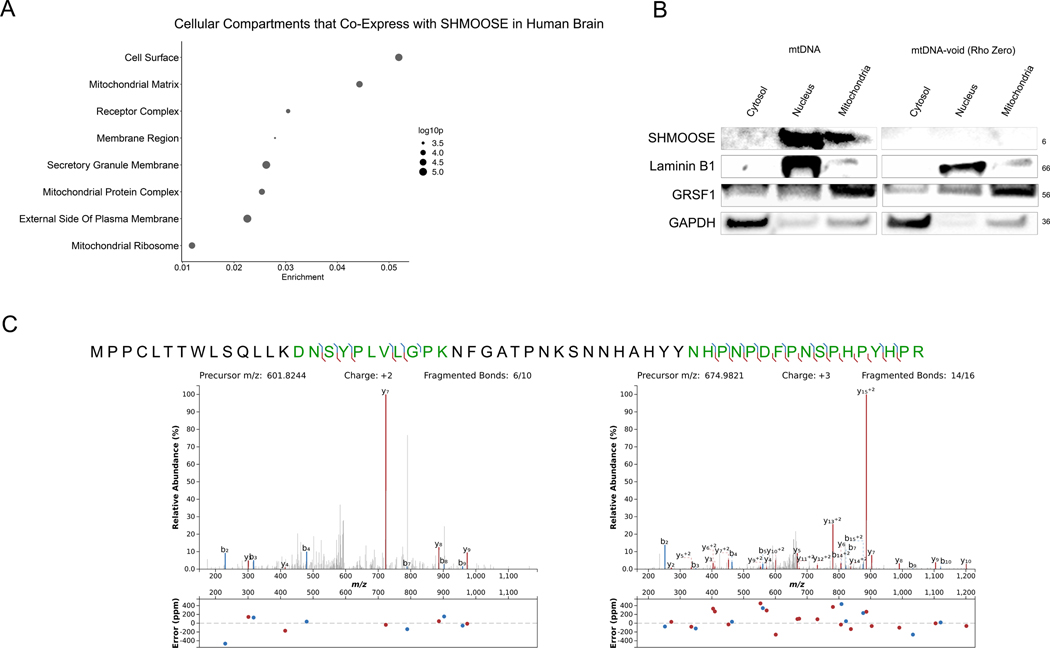
Endogenous SHMOOSE was detected using a variety of methods. To address cellular compartments to which SHMOOSE might localize, we analyzed RNA-Seq data from the temporal cortex of 69 human brains and observed mitochondria and ribosomal cellular compartment co-expressed with SHMOOSE (Fig. 2A). Indeed, with a custom antibody against the c-terminus of SHMOOSE, SHMOOSE was detected in neuronal mitochondria and nuclei fractions at the predicted ~6kDa molecular weight (Fig. 2B). Yet in rho zero cells (i.e., cells without mtDNA), SHMOOSE was not detected, further confirming SHMOOSE is derived from mitochondrial DNA. Notably, following precipitation of SHMOOSE from neuronal mitochondria, two unique SHMOOSE-derived peptides were confidently identified using mass spectrometry, one towards the N-terminus and the other at the C-terminus (Fig. 2C).
Fig. 2. Endogenous SHMOOSE is detected in mitochondria.
A GO cellular terms enriched by genes that co-express with SHMOOSE in the human temporal cortex (n = 69). B Western blot detection of ~6kDa SHMOOSE in cells containing mtDNA versus cells not containing mtDNA (i.e., rho zero cells). Laminin B1 is a nuclear marker; GRSF1 isoform is a mitochondrial marker; GAPDH is a cytosolic marker. C Unique mass spectrometry SHMOOSE-derived fragments detected in mitochondria fractions.
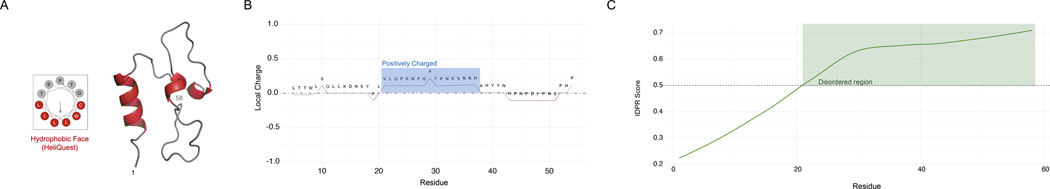
SHMOOSE has several potential biologically meaningful sequence features. First, the SHMOOSE n-terminus is predicted to contain a small alpha helix with an amphipathic feature, potentially adopting a confirmation with a hydrophobic face (Fig. 3A). Second, like other microproteins that localize to mitochondria, SHMOOSE contains a highly positively charged domain between amino acid residues 20–40, as predicted by the Henderson-Hasselbalch Equation (Fig. 3B) 68. Third, SHMOOSE is predicted to have a long-disordered protein region in the region of amino acid residues 20 to c-terminus 58 (Fig. 3C).
Fig. 3. Unique features of the SHMOOSE amino acid sequence.
A 3–11 amphipathic prediction output from HeliQuest that illustrates a potential hydrophobic face (red) derived from the RosettaFold structure model. B Positively charged domain of SHMOOSE that is highlighted in blue. C Disordered protein region that is highlighted in green as predicted by IUPred3.
Altogether, we targeted and detected the novel microprotein SHMOOSE after identifying an AD-related genetic variant within its sORF.
SHMOOSE levels in cerebrospinal fluid correlate with age, tau, and brain white matter microstructure
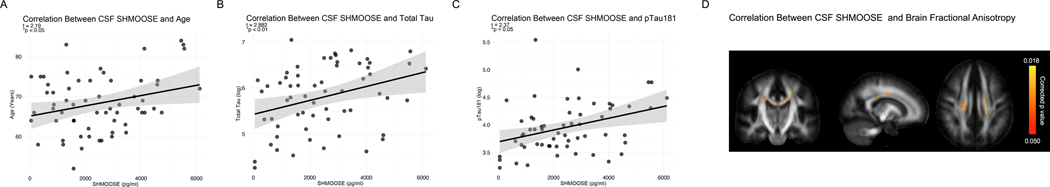
We developed a SHMOOSE enzyme-linked immunosorbent assay (ELISA) with sensitivity ranging from 100–250,000 pg/ml (Supplementary Fig. 8). In 79 cerebrospinal samples (CSF) without dementia, CSF SHMOOSE levels positively and significantly correlated with age, CSF total tau, and CSF phosphorylated tau 181 (p-tau) (Fig. 4A–C). To determine if the effect of p-tau 181 on SHMOOSE levels was mediated through total tau, mediation analyses were carried out in which total tau was considered the mediator. The indirect effect of this mediation was not considered statistically significant, and the direct effect of p-tau 181 on SHMOOSE levels was slightly attenuated when controlling for total tau (p value = 0.060). Together, the direct and indirect effect of p-tau 181 on SHMOOSE was significant (p value < 0.01). Therefore, p-tau 181 appears to be specifically driving the relationship with SHMOOSE and is not mediated by total tau (Supplementary Fig. 9). Separately, there was no significant correlations between SHMOOSE and CSF levels of amyloid beta 42. Finally, higher CSF SHMOOSE levels correlated with lower DTI fractional anisotropy FA in the body of the corpus callosum and bilateral superior corona radiata (Fig. 4D), even after controlling for age, biological sex, and cognitive status (CDR test score).
Fig. 4. SHMOOSE levels correlate with AD-related biomarkers and brain white matter.
A Human CSF SHMOOSE levels (pg/ml) correlation with age. Regression model includes biological sex as a covariate; p value < 0.001. B SHMOOSE correlation with CSF total tau (pg/ml). Regression model includes biological sex and age as covariates; p value < 0.05. C SHMOOSE correlation with CSF phosphorylated tau at residue 181 (p tau 181; pg/ml). Regression model includes biological sex and age as covariates; p value < 0.05. D Higher CSF SHMOOSE was significantly associated with lower DTI FA in the body of the corpus callosum and bilateral superior corona radiata in 72 non-demented older adults. Regression model included age, reported sex, and clinical dementia rating score. Colored voxels indicate FSLThreshold-Free Cluster Enhancement-derived p values < 0.05 after correction for voxelwise multiple comparisons. Presented in radiological orientation (L=R).
SHMOOSE acts directly on the brain when administered intracerebroventricularly
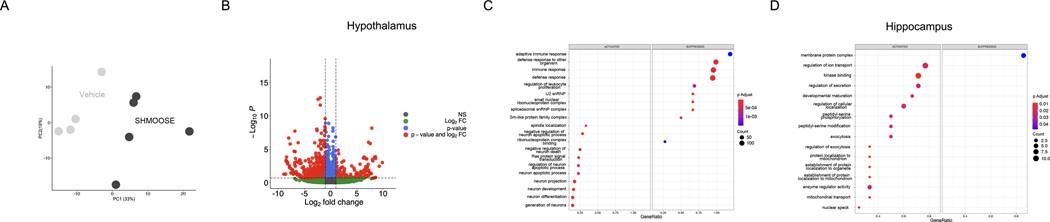
SHMOOSE was given directly to the rat brain via intracerebroventricular (ICV) administration for 24 hours followed by transcriptomics. Principal component analysis of the hypothalamus transcriptome revealed distinct clustering by SHMOOSE and vehicle (Fig. 5A), and 1698 transcripts were statistically differentially expressed under a p adjusted value of 0.2 between the two groups (Fig. 5B). These differential transcripts enriched multiple gene sets including, to name a few, the activation of neuron projection and suppression of immune responses (Fig. 5C). In the hippocampus of SHMOOSE-ICV rats, 93 transcripts were statistically differentially expressed under a p adjusted value of 0.2, enriching gene sets of activated mitochondrial transport and suppression of membrane protein complexes, among multiple other gene sets (Fig. 5D). These results highlight acute action of SHMOOSE on the brain when given ICV, most prominently in the hypothalamus.
Fig. 5. Direct actions of SHMOOSE on brain gene expression.
A. Principal component analysis for vehicle and SHMOOSE on hypothalamic gene expression following ICV administration. B Corresponding volcano plot that is color coded by significance (blue or red) and log fold change significance (red). C GSEA significant terms that were enriched by SHMOOSE-induced gene expression in the hypothalamus. D GSEA significant terms that were enriched by SHMOOSE-induced gene expression in the hippocampus.
SHMOOSE binds the mitochondrial inner membrane protein mitofilin and modifies mitochondrial biology
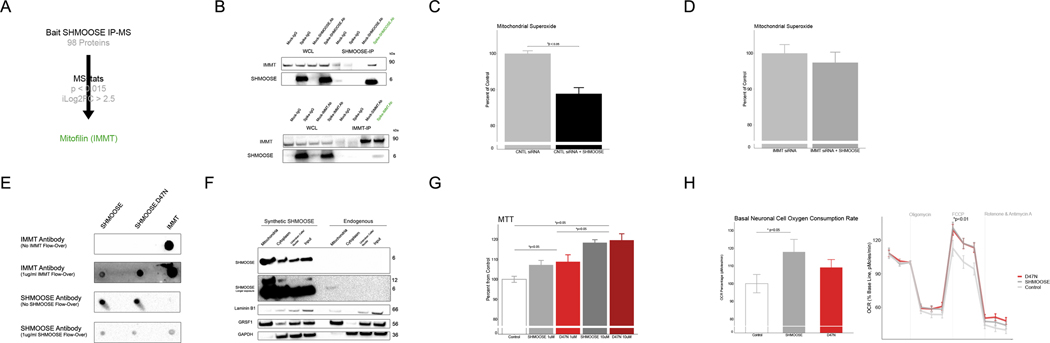
Co-immunoprecipitation strategies were used to detect SHMOOSE protein binding partners. Exogenous SHMOOSE was spiked in neuronal cell lysates followed by immunoprecipitation and mass spectrometry in triplicates. Mass spectrometry-based analysis suggested SHMOOSE bound 98 proteins; however, during protein quantification filtering and indexing by p value and fold change, we considered mitofilin a top SHMOOSE binding protein candidate (Fig. 6A). The SHMOOSE-mitofilin interaction was confirmed in vitro by treating cells with SHMOOSE followed by gentle lysis, reciprocal immunoprecipitation, and western blot at least three times (Fig. 6B). After knocking down mitofilin using siRNA, we observed no effect of SHMOOSE on mitochondrial superoxide activity (Fig. 6C–D). Likewise, reciprocal dot blots between recombinant SHMOOSE and mitofilin and SHMOOSE.D47N mitofilin showed binding (Fig. 6E).
Fig. 6. SHMOOSE interacts with mitofilin and modifies mitochondrial biology.
A Schematic proteomics analysis of SHMOOSE-spiked neural cell lysates that were immunoprecipitated using a custom SHMOOSE polyclonal antibody. B Reciprocal western blot validation of SHMOOSE/mitofilin interaction from SHMOOSE-treated cells. C SHMOOSE lowered mitochondrial superoxide levels, D yet SHMOOSE had no effect when mitofilin was knocked down with siRNA. Y axis is normalized to control baseline values. E SHMOOSE-treated differentiated SH-SY5Y cells (1 uM) localized to mitochondria after 15 minutes. Top portion of the blot represents a 5-second exposure. Second portion of the blot represents a 30-second exposure. SHMOOSE dimerized around 12 kDa in the SHMOOSE-treated conditions. Laminin, GRSF1, and GAPDH represent nuclear, mitochondrial, and cytosolic fractions, respectively. F Reciprocal dot blot illustrating recombinant mitofilin and SHMOOSE and SHMOOSE.D47N interact. G 1uM of both SHMOOSE (gray) and SHMOOSE.D47N (red) both increased metabolic activity (MTT) at 1uM and 10uM. Significant bars represent differences among dosage groups. H Normalized to the baseline third measurement, SHMOOSE increased oxygen consumption rate. SHMOOSE and SHMOOSE.D47N increased mitochondrial spare capacity. Significance defined as p value < 0.05 for independent t tests.
In vitro experiments were conducted to examine effects of SHMOOSE on mitochondrial biology. First, exogenous-administered SHMOOSE localize to mitochondria, as does endogenous SHMOOSE (Fig. 6F). Second, in a dose-dependent response from 1uM to 10uM, both SHMOOSE and SHMOOSE.D47N increased neural cell metabolic activity by 10% and 20%, respectively (Fig. 6G). Third, compared to SHMOOSE.D47N, wild type SHMOOSE significantly increased basal oxygen consumption rate by approximately 20% (Fig. 6H). Both SHMOOSE and SHMOOSE.D47N boosted mitochondrial spare capacity during mitochondrial stress in which maximum proton flow was permitted through the inner mitochondrial membrane following oligomycin and FCCP administration (Fig. 6H), suggesting general effects for both forms of SHMOOSE on mitochondrial biology.
SHMOOSE expression is elevated in Alzheimer’s disease patient brains and protects from amyloid beta pathology in vitro
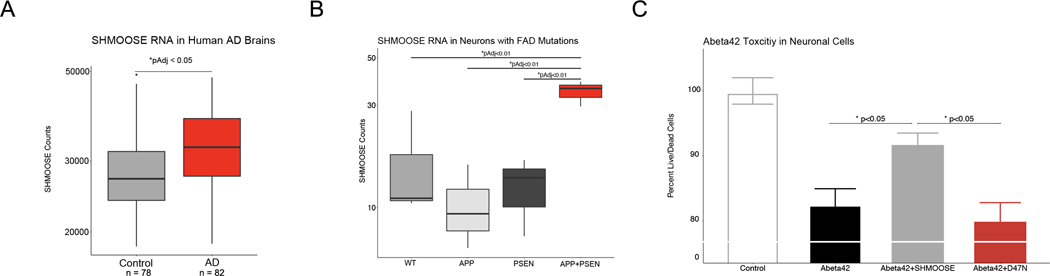
SHMOOSE expression was examined in the context of two AD-related data sets: human post-mortem brain samples and neurons derived from iPSCs with FAD mutations. In the temporal cortex of AD brains, SHMOOSE RNA was ~15% greater compared to controls (AD n = 82; control n = 78) (Fig. 7A). In neurons derived from iPSCs with both APP and PSEN1 mutations – two familial AD mutations – SHMOOSE RNA expression was three-fold higher than neurons derived with just one mutation, suggesting a role for SHMOOSE in amyloid beta biology (Fig. 7B). Experimentally, in neuronal cells stressed with oligomerized amyloid beta, SHMOOSE administration protected against cell death, but SHMOOSE.D47N did not similarly protect cells (Fig. 7C).
Fig. 7. SHMOOSE expression in human Alzheimer’s disease brains and activity in neuronal models of amyloid beta toxicity.
A SHMOOSE RNA expression in the temporal cortex of AD cases (red). Significance represented as pAdj < 0.05 following negative binomial regression on all normalized mitochondrial gene counts. B SHMOOSE expression in neurons derived from iPSCs with FAD APP mutation (light grey), FAD PSEN mutation (dark grey), and FAD APP plus PSEN mutations (red). SHMOOSE expression was highest in the latter cell type. Significance defined as pAdj < 0.05 following negative binomial regression on all normalized mitochondrial gene counts. C SHMOOSE-treated neural cells protects against amyloid beta 42-induced toxicity (grey). Significance defined as p value < 0.05 for independent t test.
Differential effects of wild type SHMOOSE and its D47N mutant on systematic gene expression
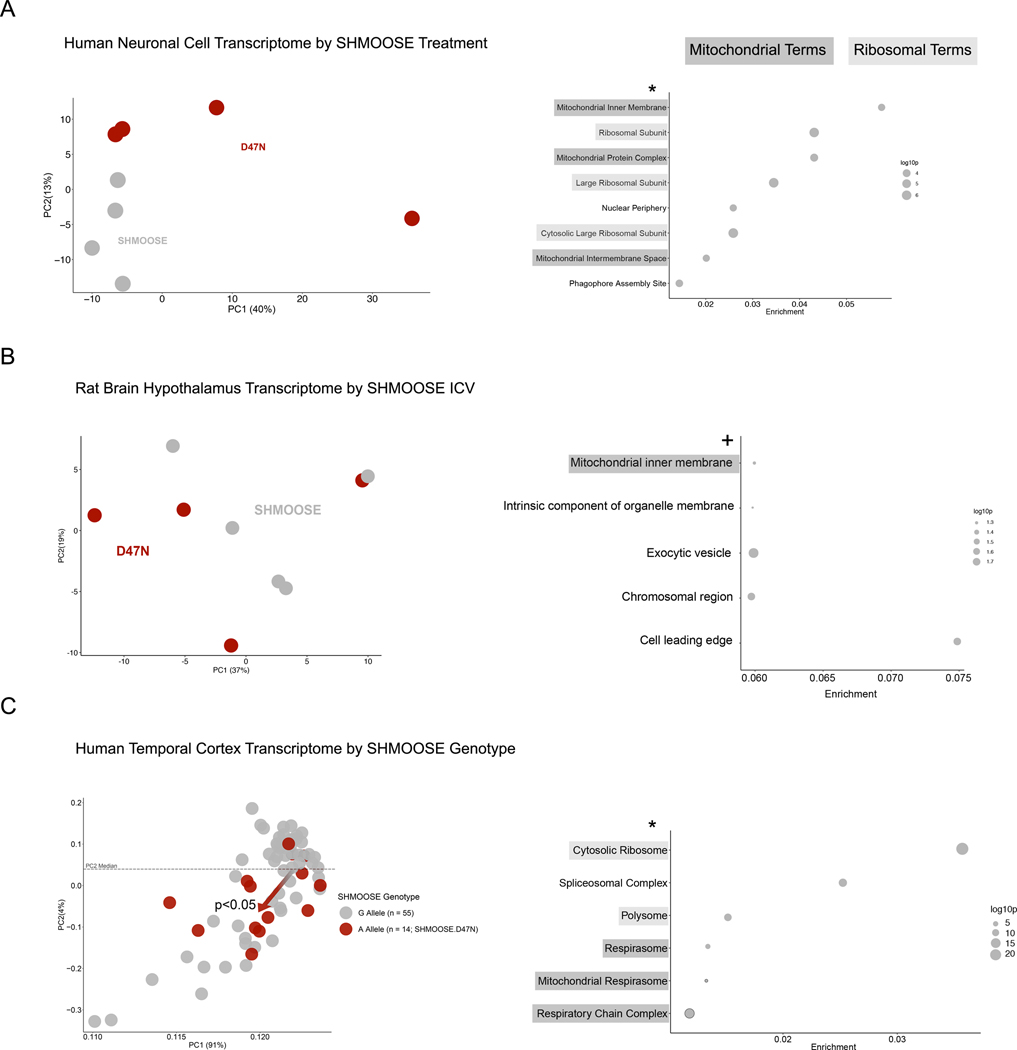
Bulk gene expression differences between SHMOOSE and SHMOOSE.D47N were examined using three models of transcriptomics: in vitro, in vivo, and human cohort based. First, SHMOOSE.D47N differentiated mitochondrial and ribosomal gene expression in neuronal cells compared to SHMOOSE. Following treatment of SHMOOSE or SHMOOSE.D47N to neuronal cells for 24 hours, SHMOOSE.D47N induced differential expression of 1,400 genes under a p adjusted value of 0.2. Subsequent gene enrichment analysis revealed a statistically significant enrichment of “Mitochondrial inner membrane” as the top enriched GO cellular compartment term (Fig. 8A). Second, SHMOOSE.D47N differentiated 82 transcripts in the hypothalamus of rats compared to SHMOOSE under a p adjusted value of 0.2. While no cellular compartment GO terms survived false discovery correction, nominally significant terms with a p value under 0.05 and a q value of 0.3 included “mitochondrial inner membrane” as one of the top terms with a few genes (Fig. 8B). Third, in human brains by SHMOOSE genotype, SHMOOSE.D47N associated with 2,122 differentially expressed genes under a p adjusted value of 0.05 (Alternative allele n = 14; reference allele n = 55). Notably, principal component analysis clustered SHMOOSE reference allele carriers, while SHMOOSE.D47N carriers drifted further from the reference cluster (Fig. 8C). This drifting was considered statistically significant when categorizing samples based on the median value of the second principal component (p value < 0.05). Considering that post-mortem brain gene expression is influenced by several factors (i.e., environment, etc.)69, we consider the significant effects of SHMOOSE.D47N noteworthy. As a contrast, we noticed a similar trend for APOE4 carriers, in which APOE4 carriers trended away from the population norm, although the trend was not statistically significant (Supplementary Fig. 10). These statistically differentially expressed genes by SHMOOSE.D47N carriers enriched GO cellular compartment terms for mitochondria and ribosomes.
Fig. 8. Bulk gene expression differences between SHMOOSE and mutant D47N indicate altered function.
Dark grey highlighting represents mitochondrial terms and light grey highlighting represents ribosomal terms. A Principal component analysis (PCA) of the in vitro gene expression signature for 10 uM SHMOOSE or SHMOOSE.D47N-treated neural cells after 24 hours. To the right of the PCA figure are GO cellular compartment terms enriched by SHMOOSE.D47N-treated neural cells, and the * indicates statistical significance following FDR. B PCA of the rat ICV gene expression signature for SHMOOSE or SHMOOSE.D47N after 24 hours. To the right of the PCA figure are GO cellular compartments that were nominally significant, and the + indicates nominal significance prior to FDR. C PCA for 14 SHMOOSE.D47N carriers (red) or 55 SHMOOSE reference allele carriers (grey). Dashed line represents the median value of PC2. Of the 14 SHMOOSE.D47N carriers, 11 fall below the median PC2 value (p value < 0.05; generalized linear model), which is represented by the red arrow at the center of the reference cluster. To the right of the PCA figure are GO cellular compartment terms significantly enriched by SHMOOSE.D47N carriers, and the * indicates statistical significance following FDR.
Discussion
In this report we identified a novel microprotein called SHMOOSE. We developed targeted detection strategies for SHMOOSE because of a series of large epidemiological genetic studies. These genetic studies revealed a mtSNP that changes the amino acid sequence of SHMOOSE and associated with AD and brain structure. In four cohorts, individuals with SHMOOSE.D47N had increased risk for AD (OR: 1.30; Fig. 1B). SHMOOSE.D47N carriers also showed greater atrophy over age in medial temporal areas such as the parahippocampus, entorhinal cortex, and anterior and parietal cingulate cortex. The medial temporal cortex and parietal cingulate cortex are known to be vulnerable in Alzheimer’s disease (AD), and the pathological atrophy likely appears years before clinical symptoms 70, 71. Similar effects in the limbic area were found in ADNI, but no results survived multiple correction, as we lacked the statistical power that UK Biobank provided. Therefore, we targeted and ultimately detected SHMOOSE in a variety of biological samples using immunological and mass spectrometry-based methods.
We detected SHMOOSE biochemically by developing a polyclonal antibody against amino acid residues 32–58. By immunoprecipitating SHMOOSE from neuronal mitochondria, two unique SHMOOSE-derived fragments were identified in mass spectrometry. In addition, via western blot, SHMOOSE was detected at the predicted ~6 kDa molecular weight in neuronal mitochondrial as well as nuclei. Furthermore, CSF SHMOOSE positively correlated with age, tau, and brain white matter. Since higher levels of CSF tau have previously predicted AD 72, correlations between SHMOOSE and tau suggest SHMOOSE could be involved in the progressive etiology of AD. SHMOOSE levels also associated with DTI FA in non-demented older adults. Various factors can contribute to lower DTI FA, but it may reflect lower levels of myelination and is often associated with a disease state. Myelin maintenance and repair is metabolically demanding and is particularly vulnerable to damage when energy deficits exist 73, providing a possible link between SHMOOSE and white matter microstructure. Although past MiWAS studies have examined the effects of mtSNPs on neurodegeneration, few have followed up experimentally 19, 74. One functional limitation of MiWAS is isolating the effects of individuals mtSNPs that define haplogroups 75, 76. As a result, we cannot rule out other SNPs within the SHMOOSE.D47N haplogroup (i.e., Haplogroup U) have independent effects. Nevertheless, we used MiWAS to identify a microprotein candidate for detection validation (i.e., SHMOOSE).
Notably, SHMOOSE contains several unique sequence features: (1) a positively charged domain, (2) potential amphipathic alpha-helix at the N-terminus, (3) disordered region towards the C-terminus, and (4) no mitochondrial targeting sequence. Many microproteins that localize to mitochondria also contain these features 68, 77, 78. A research area worth substantial consideration is whether SHMOOSE localizes to mitochondria following translation in cytoplasmic ribosomes or whether SHMOOSE is translated within the mitochondrial matrix by mitochondrial ribosomes. A separate mitochondrial-encoded microprotein called MOTS-c is obligatory translated in the cytosol due to a mitochondrial-specific stop codon within its second frame 79. SHMOOSE does not contain such mitochondrial-specific codon usage and is translated into identical sequences regardless of codon specificity. Techniques such as ribosome profiling might resolve whether mitochondrial-encoded microproteins are translated by cytoplasmic or mitochondrial ribosomes. However, improvements in ribosomal preparations, mitochondrial translation-initiation-specific inhibitors, and periodicity algorithms will likely need to be enhanced 80. For example, the mitochondrial transcriptome is complex and contains several small RNAs with unknown function that could affect the periodicity of existing ORF calling algorithms, and many mitochondrial sORFs overlap rRNA and tRNA regions 81.
After detecting SHMOOSE, we carried out a series of functional experiments. SHMOOSE was administered ICV to rats for 24 hours followed by transcriptomics of brain tissue. SHMOOSE significantly altered the hypothalamus transcriptome and to a lesser extent the hippocampus transcriptome. Gene sets that were enriched by SHMOOSE administration included neuronal-related processes, immune-related processes, and mitochondrial-related processes. While we observed significant effects at the gene transcription level at one dose and time point, future work might consider variable doses, multiple time points, and a variety of outcomes to better understand the biology of SHMOOSE. We sought to understand the precise biology of SHMOOSE by performing co-immunoprecipitation experiments. SHMOOSE bound the inner mitochondrial membrane mitofilin in multiple models including neuronal cells and dot blot. Mitofilin is a component of the MICOS complex that regulates mitochondrial crista junctions and inner membrane organization 82, 83. While we observed direct interaction between SHMOOSE and mitofilin, SHMOOSE might also be part of a larger mitochondrial protein complex that comprises the dynamic mitofilin protein.
Finally, we examined systematic differences between SHMOOSE and SHMOOSE.D47N in three separate models by implementing transcriptomics. In humans, remarkably, SHMOOSE.D47N alone differentiated the brain temporal cortex transcriptome. By using principal component analysis on bulk RNA expression data, reference allele carriers were visualized as one tight cluster, whereas alternative SHMOOSE.D47N carriers drifted from SHMOOSE reference allele cluster. This was surprising given the reported effects of environment, lifespan, etc. on human brain transcriptomics 69. In all our transcriptomics studies including in vitro, in vivo, and epidemiological models, we observed a common theme for mitochondrial enrichment.
Our results have several implications. First, we showed mitochondrial DNA variants can associate with several neurobiological phenotypes. Second, we revealed mitochondrial DNA variants can be mapped to sORFs that encode biologically functional microproteins. We identified SHMOOSE as the first biologically active mitochondrial-encoded microprotein detected by mass spectrometry, immunoblot, and ELISA. Moreover, as proteomics technology improves, additional mitochondrial-encoded microproteins might be detected 84. Third, the correlation among CSF levels of SHMOOSE, CSF AD-related biomarkers (e.g., tau), and brain white matter suggests SHMOOSE has potential as a biomarker. Finally, SHMOOSE is yet another microprotein of a growing number that modify mitochondrial biology 85.
Supplementary Material
Acknowledgements
The study was supported by NIH grants P30AG10161, P30AG072975, R01AG15819, R01AG17917, U01AG61356, R01AG069698, RF1AG061834, R01AG068405, P30AG068345, P01AG055369, DK118402, F31 AG059356, and T32 AG00037; as well as The Quebec Research Funds Postdoctoral Fellowship.
B.M. conceived and designed analyses, collected data, performed analyses, and wrote the paper. S.K. conceived experimental design. H.M. collected data. K.C. collected data. H.K. collected data. N.T. collected data. N.L conceived experimental design. H.J. contributed analysis tools. J.V. contributed experimental design and collected data. J.D. collected data. A.S. conceived experimental design. T.A contributed analysis tools. E.C. contributed analysis tools. N. E-T. contributed analysis tools. M. T carried out analyses. E. H carried out analyses. M.B conceived analysis design. L.D-S. carried out animal studies. S.K conceived experimental design. F.G collected data. D.B contributed analysis tools. L.Z. carried out analyses. A.T. contributed analysis tools. J.W designed experimental tools and carried out experiments. K.Y conceived analyses. P.C. conceived and designed analyses and contributed to manuscript preparation.
Footnotes
Competing Interests
Intellectual property related to SHMOOSE has been filed by the University of Southern California.
References
- 1.Saghatelian A, Couso JP. Discovery and characterization of smORF-encoded bioactive polypeptides. Nat Chem Biol 2015; 11(12): 909–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez TF, Chu Q, Donaldson C, Tan D, Shokhirev MN, Saghatelian A. Accurate annotation of human protein-coding small open reading frames. Nat Chem Biol 2020; 16(4): 458–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mudge JM, Ruiz-Orera J, Prensner JR, Brunet MA, Calvet F, Jungreis I et al. Standardized annotation of translated open reading frames. Nat Biotechnol 2022; 40(7): 994–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller B, Kim SJ, Kumagai H, Mehta HH, Xiang W, Liu J et al. Peptides derived from small mitochondrial open reading frames: Genomic, biological, and therapeutic implications. Exp Cell Res 2020: 112056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim SJ, Guerrero N, Wassef G, Xiao J, Mehta HH, Cohen P et al. The mitochondrial-derived peptide humanin activates the ERK1/2, AKT, and STAT3 signaling pathways and has age-dependent signaling differences in the hippocampus. Oncotarget 2016; 7(30): 46899–46912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo F, Jing W, Ma CG, Wu MN, Zhang JF, Li XY et al. [Gly(14)]-humanin rescues long-term potentiation from amyloid beta protein-induced impairment in the rat hippocampal CA1 region in vivo. Synapse 2010; 64(1): 83–91. [DOI] [PubMed] [Google Scholar]
- 7.Tajima H, Kawasumi M, Chiba T, Yamada M, Yamashita K, Nawa M et al. A humanin derivative, S14G-HN, prevents amyloid-beta-induced memory impairment in mice. J Neurosci Res 2005; 79(5): 714–723. [DOI] [PubMed] [Google Scholar]
- 8.Ikonen M, Liu B, Hashimoto Y, Ma L, Lee KW, Niikura T et al. Interaction between the Alzheimer’s survival peptide humanin and insulin-like growth factor-binding protein 3 regulates cell survival and apoptosis. Proc Natl Acad Sci U S A 2003; 100(22): 13042–13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsukamoto E, Hashimoto Y, Kanekura K, Niikura T, Aiso S, Nishimoto I. Characterization of the toxic mechanism triggered by Alzheimer’s amyloid-beta peptides via p75 neurotrophin receptor in neuronal hybrid cells. J Neurosci Res 2003; 73(5): 627–636. [DOI] [PubMed] [Google Scholar]
- 10.Hashimoto Y, Niikura T, Tajima H, Yasukawa T, Sudo H, Ito Y et al. A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer’s disease genes and Abeta. Proc Natl Acad Sci U S A 2001; 98(11): 6336–6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yen K, Wan J, Mehta HH, Miller B, Christensen A, Levine ME et al. Humanin prevents age-related cognitive decline in mice and is associated with improved cognitive age in humans. Scientific reports 2018; 8(1): 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zempo H, Kim SJ, Fuku N, Nishida Y, Higaki Y, Wan J et al. A pro-diabetogenic mtDNA polymorphism in the mitochondrial-derived peptide, MOTS-c. Aging (Albany NY) 2021; 13(2): 1692–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller B, Torres M, Jiang X, McKean-Cowdin R, Nousome D, Kim S-J et al. A Mitochondrial Genome-Wide Association Study of Cataract in a Latino Population. Translational Vision Science & Technology 2020; 9(6): 25–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller B, Arpawong TE, Jiao H, Kim S-J, Yen K, Mehta HH et al. Comparing the utility of mitochondrial and nuclear DNA to adjust for genetic ancestry in association studies. Cells 2019; 8(4): 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yonova-Doing E, Calabrese C, Gomez-Duran A, Schon K, Wei W, Karthikeyan S et al. An atlas of mitochondrial DNA genotype-phenotype associations in the UK Biobank. Nat Genet 2021; 53(7): 982–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ridge PG, Wadsworth ME, Miller JB, Saykin AJ, Green RC, Alzheimer’s Disease Neuroimaging I et al. Assembly of 809 whole mitochondrial genomes with clinical, imaging, and fluid biomarker phenotyping. Alzheimers Dement 2018; 14(4): 514–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller B, Haghani A, Ailshire J, Arpawong TE. Human Population Genetics in Aging Studies for Molecular Biologists. Aging. Springer; 2020, pp 67–76. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z, Castello A. Principal components analysis in clinical studies. Ann Transl Med 2017; 5(17): 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lakatos A, Derbeneva O, Younes D, Keator D, Bakken T, Lvova M et al. Association between mitochondrial DNA variations and Alzheimer’s disease in the ADNI cohort. Neurobiol Aging 2010; 31(8): 1355–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balduzzi S, Rucker G, Schwarzer G. How to perform a meta-analysis with R: a practical tutorial. Evid Based Ment Health 2019; 22(4): 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alfaro-Almagro F, Jenkinson M, Bangerter NK, Andersson JLR, Griffanti L, Douaud G et al. Image processing and Quality Control for the first 10,000 brain imaging datasets from UK Biobank. Neuroimage 2018; 166: 400–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller KL, Alfaro-Almagro F, Bangerter NK, Thomas DL, Yacoub E, Xu J et al. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat Neurosci 2016; 19(11): 1523–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reuter M, Rosas HD, Fischl B. Highly accurate inverse consistent registration: a robust approach. Neuroimage 2010; 53(4): 1181–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segonne F, Dale AM, Busa E, Glessner M, Salat D, Hahn HK et al. A hybrid approach to the skull stripping problem in MRI. Neuroimage 2004; 22(3): 1060–1075. [DOI] [PubMed] [Google Scholar]
- 25.Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 2002; 33(3): 341–355. [DOI] [PubMed] [Google Scholar]
- 26.Fischl B, Salat DH, van der Kouwe AJ, Makris N, Segonne F, Quinn BT et al. Sequence-independent segmentation of magnetic resonance images. Neuroimage 2004; 23 Suppl 1: S69–84. [DOI] [PubMed] [Google Scholar]
- 27.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 1998; 17(1): 87–97. [DOI] [PubMed] [Google Scholar]
- 28.Segonne F, Pacheco J, Fischl B. Geometrically accurate topology-correction of cortical surfaces using nonseparating loops. IEEE Trans Med Imaging 2007; 26(4): 518–529. [DOI] [PubMed] [Google Scholar]
- 29.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A 2000; 97(20): 11050–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischl B, Sereno MI, Tootell RB, Dale AM. High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum Brain Mapp 1999; 8(4): 272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dinov I, Lozev K, Petrosyan P, Liu Z, Eggert P, Pierce J et al. Neuroimaging study designs, computational analyses and data provenance using the LONI pipeline. PLoS One 2010; 5(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dinov ID, Van Horn JD, Lozev KM, Magsipoc R, Petrosyan P, Liu Z et al. Efficient, Distributed and Interactive Neuroimaging Data Analysis Using the LONI Pipeline. Front Neuroinform 2009; 3: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018; 562(7726): 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller B, Arpawong TE, Jiao H, Kim SJ, Yen K, Mehta HH et al. Comparing the Utility of Mitochondrial and Nuclear DNA to Adjust for Genetic Ancestry in Association Studies. Cells 2019; 8(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Worsley KJ, Evans AC, Marrett S, Neelin P. A three-dimensional statistical analysis for CBF activation studies in human brain. J Cereb Blood Flow Metab 1992; 12(6): 900–918. [DOI] [PubMed] [Google Scholar]
- 36.Zhao L, Batta I, Matloff W, O’Driscoll C, Hobel S, Toga AW. Neuroimaging PheWAS (Phenome-Wide Association Study): A Free Cloud-Computing Platform for Big-Data, Brain-Wide Imaging Association Studies. Neuroinformatics 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crimmins EM, Kim JK, Langa KM, Weir DR. Assessment of cognition using surveys and neuropsychological assessment: the Health and Retirement Study and the Aging, Demographics, and Memory Study. J Gerontol B Psychol Sci Soc Sci 2011; 66 Suppl 1: i162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021; 373(6557): 871–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gulsevin A, Meiler J. Prediction of amphipathic helix-membrane interactions with Rosetta. PLoS Comput Biol 2021; 17(3): e1008818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McFadden WM, Yanowitz JL. idpr: A package for profiling and analyzing Intrinsically Disordered Proteins in R. PLoS One 2022; 17(4): e0266929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Erdos G, Pajkos M, Dosztanyi Z. IUPred3: prediction of protein disorder enhanced with unambiguous experimental annotation and visualization of evolutionary conservation. Nucleic Acids Res 2021; 49(W1): W297–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manjón JV, Coupé P, Concha L, Buades A, Collins DL, Robles M. Diffusion weighted image denoising using overcomplete local PCA. PLoS One 2013; 8(9): e73021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tournier JD, Smith R, Raffelt D, Tabbara R, Dhollander T, Pietsch M et al. MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. Neuroimage 2019; 202: 116137. [DOI] [PubMed] [Google Scholar]
- 44.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. Neuroimage 2012; 62(2): 782–790. [DOI] [PubMed] [Google Scholar]
- 45.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal 2008; 12(1): 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 2011; 54(3): 2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nir TM, Jahanshad N, Villalon-Reina JE, Isaev D, Zavaliangos-Petropulu A, Zhan L et al. Fractional anisotropy derived from the diffusion tensor distribution function boosts power to detect Alzheimer’s disease deficits. Magn Reson Med 2017; 78(6): 2322–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lo Buono V, Palmeri R, Corallo F, Allone C, Pria D, Bramanti P et al. Diffusion tensor imaging of white matter degeneration in early stage of Alzheimer’s disease: a review. Int J Neurosci 2020; 130(3): 243–250. [DOI] [PubMed] [Google Scholar]
- 49.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 2004; 23 Suppl 1: S208–219. [DOI] [PubMed] [Google Scholar]
- 50.Smith SM, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols TE, Mackay CE et al. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage 2006; 31(4): 1487–1505. [DOI] [PubMed] [Google Scholar]
- 51.Smith SM, Nichols TE. Threshold-free cluster enhancement: addressing problems of smoothing, threshold dependence and localisation in cluster inference. Neuroimage 2009; 44(1): 83–98. [DOI] [PubMed] [Google Scholar]
- 52.Miller SW, Trimmer PA, Parker WD Jr., Davis RE. Creation and characterization of mitochondrial DNA-depleted cell lines with “neuronal-like” properties. J Neurochem 1996; 67(5): 1897–1907. [DOI] [PubMed] [Google Scholar]
- 53.He L, Diedrich J, Chu YY, Yates JR 3rd. Extracting Accurate Precursor Information for Tandem Mass Spectra by RawConverter. Anal Chem 2015; 87(22): 11361–11367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu T, Park SK, Venable JD, Wohlschlegel JA, Diedrich JK, Cociorva D et al. ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J Proteomics 2015; 129: 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tabb DL, McDonald WH, Yates JR, 3rd. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 2002; 1(1): 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res 2003; 2(1): 43–50. [DOI] [PubMed] [Google Scholar]
- 57.Gong Z, Su K, Cui L, Tas E, Zhang T, Dong HH et al. Central effects of humanin on hepatic triglyceride secretion. Am J Physiol Endocrinol Metab 2015; 309(3): E283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012; 16(5): 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008; 26(12): 1367–1372. [DOI] [PubMed] [Google Scholar]
- 60.Choi M, Chang CY, Clough T, Broudy D, Killeen T, MacLean B et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 2014; 30(17): 2524–2526. [DOI] [PubMed] [Google Scholar]
- 61.Webb-Robertson BJ, Wiberg HK, Matzke MM, Brown JN, Wang J, McDermott JE et al. Review, evaluation, and discussion of the challenges of missing value imputation for mass spectrometry-based label-free global proteomics. J Proteome Res 2015; 14(5): 1993–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29(1): 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chandramohan R, Wu PY, Phan JH, Wang MD. Benchmarking RNA-Seq quantification tools. Annu Int Conf IEEE Eng Med Biol Soc 2013; 2013: 647–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ryan TM, Caine J, Mertens HD, Kirby N, Nigro J, Breheney K et al. Ammonium hydroxide treatment of Abeta produces an aggregate free solution suitable for biophysical and cell culture characterization. PeerJ 2013; 1: e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Allen M, Carrasquillo MM, Funk C, Heavner BD, Zou F, Younkin CS et al. Human whole genome genotype and transcriptome data for Alzheimer’s and other neurodegenerative diseases. Sci Data 2016; 3: 160089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kalari KR, Nair AA, Bhavsar JD, O’Brien DR, Davila JI, Bockol MA et al. MAP-RSeq: Mayo Analysis Pipeline for RNA sequencing. BMC Bioinformatics 2014; 15: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Allen M, Wang X, Burgess JD, Watzlawik J, Serie DJ, Younkin CS et al. Conserved brain myelination networks are altered in Alzheimer’s and other neurodegenerative diseases. Alzheimers Dement 2018; 14(3): 352–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stein CS, Jadiya P, Zhang X, McLendon JM, Abouassaly GM, Witmer NH et al. Mitoregulin: A lncRNA-Encoded Microprotein that Supports Mitochondrial Supercomplexes and Respiratory Efficiency. Cell Rep 2018; 23(13): 3710–3720 e3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ng B, Casazza W, Patrick E, Tasaki S, Novakovsky G, Felsky D et al. Using Transcriptomic Hidden Variables to Infer Context-Specific Genotype Effects in the Brain. Am J Hum Genet 2019; 105(3): 562–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davatzikos C, Xu F, An Y, Fan Y, Resnick SM. Longitudinal progression of Alzheimer’s-like patterns of atrophy in normal older adults: the SPARE-AD index. Brain 2009; 132(Pt 8): 2026–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jack CR Jr., Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW et al. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol 2010; 9(1): 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grangeon L, Paquet C, Bombois S, Quillard-Muraine M, Martinaud O, Bourre B et al. Differential Diagnosis of Dementia with High Levels of Cerebrospinal Fluid Tau Protein. J Alzheimers Dis 2016; 51(3): 905–913. [DOI] [PubMed] [Google Scholar]
- 73.Bartzokis G Alzheimer’s disease as homeostatic responses to age-related myelin breakdown. Neurobiology of Aging 2011; 32(8): 1341–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hudson G, Nalls M, Evans JR, Breen DP, Winder-Rhodes S, Morrison KE et al. Two-stage association study and meta-analysis of mitochondrial DNA variants in Parkinson disease. Neurology 2013; 80(22): 2042–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McRae AF, Byrne EM, Zhao ZZ, Montgomery GW, Visscher PM. Power and SNP tagging in whole mitochondrial genome association studies. Genome Res 2008; 18(6): 911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Malhi RS, Eshleman JA, Greenberg JA, Weiss DA, Schultz Shook BA, Kaestle FA et al. The structure of diversity within New World mitochondrial DNA haplogroups: implications for the prehistory of North America. Am J Hum Genet 2002; 70(4): 905–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ge Q, Jia D, Cen D, Qi Y, Shi C, Li J et al. Micropeptide ASAP encoded by LINC00467 promotes colorectal cancer progression by directly modulating ATP synthase activity. J Clin Invest 2021; 131(22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang S, Reljic B, Liang C, Kerouanton B, Francisco JC, Peh JH et al. Mitochondrial peptide BRAWNIN is essential for vertebrate respiratory complex III assembly. Nat Commun 2020; 11(1): 1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee C, Zeng J, Drew BG, Sallam T, Martin-Montalvo A, Wan J et al. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab 2015; 21(3): 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller B, Kim SJ, Kumagai H, Yen K, Cohen P. Mitochondria-derived peptides in aging and healthspan. J Clin Invest 2022; 132(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM et al. The human mitochondrial transcriptome. Cell 2011; 146(4): 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Feng Y, Madungwe NB, Bopassa JC. Mitochondrial inner membrane protein, Mic60/mitofilin in mammalian organ protection. J Cell Physiol 2019; 234(4): 3383–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gieffers C, Korioth F, Heimann P, Ungermann C, Frey J. Mitofilin is a transmembrane protein of the inner mitochondrial membrane expressed as two isoforms. Exp Cell Res 1997; 232(2): 395–399. [DOI] [PubMed] [Google Scholar]
- 84.Miller B, Kim S-J, Kumagai H, Mehta HH, Xiang W, Liu J et al. Peptides derived from small mitochondrial open reading frames: Genomic, biological, and therapeutic implications. Experimental Cell Research 2020: 112056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wright BW, Yi Z, Weissman JS, Chen J. The dark proteome: translation from noncanonical open reading frames. Trends Cell Biol 2022; 32(3): 243–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.