Abstract
Purpose
Radioactive iodine (131I) therapy is a conventional post-surgery treatment widely used for papillary thyroid carcinoma (PTC). Since 131I is orally administered, we hypothesize that it may affect gut microbiome. This study aims to investigate alterations of intestinal microbiome caused by 131I therapy in PTC patients and explore its association with response to 131I therapy.
Methods
Fecal samples of 60 PTC patients pre- and post-131I therapy were collected to characterize the 131I therapy-induced gut microbiota alterations using 16S rRNA gene sequencing. According to the inclusion criteria, sequence data of 40 out of the 60 patients, divided into excellent response (ER) group and non-excellent response (NER) group, were recruited to investigate the possible connection between gut microbiota and response to 131I therapy. Multivariate binary logistic regression was employed to construct a predictive model for response to 131I therapy.
Results
Microbial richness, diversity, and composition were tremendously altered by 131I therapy. A significant decline of Firmicutes to Bacteroides (F/B) ratio was observed post-131I therapy. 131I therapy also led to changes of gut microbiome-related metabolic pathways. Discrepancies in β diversity were found between ER and NER groups both pre- and post-131I therapy. Furthermore, a predictive model for response to 131I therapy with a p value of 0.003 and an overall percentage correct of 80.0% was established, with three variables including lymph node metastasis, relative abundance of g_Bifidobacterium and g_Dorea. Among them, g_Dorea was identified to be an in independent predictor of response to 131I therapy (p = 0.04).
Conclusion
For the first time, the present study demonstrates the gut microbial dysbiosis caused by 131I therapy in post-surgery PTC patients and reveals a previously undefined role of gut microbiome as predictor for 131I ablation response. G_Dorea and g_Bifidobacterium may be potential targets for clinical intervention to improve response to 131I in post-operative PTC patients.
Trial registration
ChiCTR2100048000. Registered 28 June 2021.
Keywords: Gut microbiota, Radioactive iodine therapy, 131I therapy, 16s rRNA, Predictive model, 131I response
Introduction
Thyroid carcinoma (TC) is the commonest endocrine malignancy, the incidence of which is still increasing worldwide. Differentiated thyroid cancer (DTC) takes up over 90% of all TC, among which, papillary thyroid carcinoma (PTC) is the foremost histopathologic type, taking up more than 85% of TC [1].
Radioactive iodine (131I) therapy is the mainstay of treatment for PTC after surgery [2], which has been used for nearly 80 years and still plays a central role in the management of PTC today [3]. However, response to 131I therapy varies among patients. Distinct responses to 131I determine different subsequent clinical strategies and prognosis. According to the 2015 American Thyroid Association Management Guidelines for Adult Patients with Thyroid Nodules and Differentiated Thyroid Cancer [4], responses to 131I ablation can be categorized as excellent response (ER) or non-excellent response (NER). PTC patients with a negative imaging, negative thyroglobulin antibody (TgAb) and either suppressed thyroglobulin (Tg) < 0.2 ng/ml or thyroid stimulating hormone (TSH) stimulated Tg (sTg) < 1 ng/ml are evaluated as ER. Those who do not achieve these standards are defined as NER. ER to 131I indicates clinical cure of PTC, whereas patients with NER may require another course of 131I therapy. Hence, the need for a reliable tool predicting therapeutic response to 131I ahead of 131I therapy is vital and urgent, but has not been fulfilled yet.
131I is an orally administered radioactive nuclide used for internal-radiation therapy to treat PTC. After application, 131I stays and accumulates in the gastro-intestinal tract, thus is very likely to affect gut microbiome. Gut microbiota and its vital role in various diseases have drawn more and more attention. Accumulating evidences have indicated that gut microbiota plays a part in the pathophysiology of cancer [5]. Recently, a close connection between gut microbiome and thyroid carcinoma has been implicated [6]. Another study finds out that gut microbiome is tremendously altered in thyroid carcinoma patients compared to control subjects [7]. Thyroid function is influenced by gut microbiota [8]. Moreover, gut microbiota is also reported to be associated with radiation sensitivity and radiation-related toxicities [9–11]. Evidences from animal models have also shown that gut microbiome composition may predict radiation injury [9].
However, the alterations of gut microbiota after 131I therapy and the significance have not been elucidated yet. Whether difference in gut microbiome is related to distinct responses to 131I remains elusive. Can one or several gut genera be used as predictors of 131I response in PTC patients before the therapy is still unknown.
Given the paucity of gut microbiome studies in post operative PTC patients receiving 131I therapy, we reported the present prospective study.
Materials and methods
Study participants
This study was performed in agreement with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of the First Affiliated Hospital of Army Medical University (Third Military Medical University), China (Date Oct. 16th, 2020/No. KY2020214). The authors affirmed that all participants provided informed consents for clinical data and bio-sample use and publication of their basic characteristics and 16s rRNA sequence results of their stools.
All patients included in this study were from the southwest region of China (including Chongqing, Sichuan, and Guizhou), where the climate and residents’ eating habits were similar. 99mTcO4− thyroid imaging was negative before the 131I therapy on every single participant. Forty-eight hours after taking 131I orally at a dose of 150 mCi (5.55 GBq), a very small part of thyroid tissue was displayed in all of the patients by post radioactive iodine therapy whole-body scanning (RxWBS), with no significant difference in the amount of residual thyroid tissue among the patients. Serum TSH concentration of each participant before 131I therapy was over 30 mIU/L.
Self-controlled study cohort, recruitment of subjects, procedures of 131I therapy, and sampling
The inclusion criteria for TC patients were as follows: (1) patients were diagnosed as PTC; (2) patients had undergone complete thyroid resection procedure; (3) patients were scheduled to receive 131I therapy for the first time; (4) patients were willing to participate in this study and signed the informed consent forms; (5) patients promised to voluntarily accept and comply with this experimental protocol.
The exclusion criteria were as follows: (1) patients with known history of any other cancer besides PTC; (2) patients who had undergone prior 131I therapy or other radiotherapy; (3) patients with notable gastrointestinal disorder; (4) patients with known history of gastrointestinal surgery; (5) patients with long-term use of any probiotics or antibiotics, non-steroid anti-inflammatory drugs or proton pump inhibitors; (6) patients with an age < 18 years. Finally, a total of 60 subjects from the nuclear medicine department of the First Affiliated Hospital of the Army Medical University (Third Military Medical University), China, between October 2020 and March 2021, who fulfilled the inclusion criteria and provided the fecal and blood samples, were included in the self-controlled study to explore microbiota changes induced by 131I therapy.
Baseline characteristics and clinical parameters are listed in Table 1. Patients were given iodine-free diet for 4 weeks and underwent 3 weeks of Euthyrox withdrawal before each sampling. Two sequential peripheral blood and fecal samples were collected from patients at time points 1–2 days before and 5 months after 131I administration, respectively. Since the patients recruited in this study are either high-risk PTC, or intermediate-risk PTC with aggressive histology or with unexplained elevated sTg levels (sTg > 10 ng/ml) or with both, 131I was administered orally at a dose of 150 mCi (5.55 GBq) for adjuvant therapy. All samples were aliquoted and stored at − 80 °C for further use. TSH and sTg levels in the blood samples were assayed. DNA extraction and 16S rRNA gene sequencing were performed using the fecal samples.
Table 1.
Demographic and clinical characteristics of participants in self-controlled study
| Baseline characteristics | Participants (n = 60) |
|---|---|
| Age, years: mean ± SD | 40.02 ± 10.23 |
| Gender: N (%) |
Male: 19 (31.7%) Female: 41 (68.3%) |
| Risk level: N(%) | Low: 0 (0.0%) |
| Medium: 49 (81.7%) | |
| High: 11 (18.3%) | |
| Pathologic stage post-surgery: N (%) | I: 54 (90.0%) |
| II: 4 (6.7%) | |
| III: 2 (3.3%) | |
| IV: 0 (0.0%) | |
| Maximum tumor diameter, cm: mean ± SD | 1.38 ± 0.82 |
| Local invasion by pathology: N (%) | Without: 43 (71.7%) |
| With: 17 (28.3%) | |
| Lymph node metastasis by RxWBSa: N (%) | Without: 49 (81.7%) |
| With: 11 (18.3%) | |
| Distant metastasis by RxWBS: N (%) | Without: 59 (98.3%) |
| With: 1 (1.7%) | |
| sTgb or TgAbc level: | |
| sTg < 1 ng/ml: N (%) | 25 (41.7%) |
| sTg 1–10 ng/ml: N (%) | 22 (36.7%) |
| sTg > 10 ng/ml or rising TgAb level: N (%) | 13 (21.6%) |
aRxWBS, post radioactive iodine therapy whole-body scanning
bsTg, TSH-stimulated thyroglobulin
cTgAb, thyroglobulin antibody
Patient demographics and clinical parameters of 131I ER and NER patients
According to the definition of ER demonstrated previously, before 131I ablation, 20 out of the 60 patients already fulfilled all other criteria of ER (no metastases observed, sTg < 1 ng/ml and negative TgAb) except for very small amounts of thyroid remnant imaging as shown by RxWBS, indicating that after 131I ablation, it was very likely that these 20 patients would be classified as ER according to both our experiences and literatures [12]. As a matter of fact, after 131I ablation, they were actually evaluated as ER in this study. Thus, in order to reduce false-positive rate, these 20 patients were excluded from this part of study.
The rest 40 participants were divided into 131I ER group (24 cases) and NER group (16 cases) based on their assessment post 131I ablation according to the guidelines. Gut microbiota of the two groups both 1–2 days pre-treatment of 131I (Pre-) and 5 months post-treatment (Post-) were analyzed and compared.
DNA extraction and 16S rRNA sequencing analysis
Fecal DNA extraction was performed using CTAB method. The V3–V4 region of the bacterial 16S rRNA gene was amplified by Phusion® High-Fidelity PCR Master Mix (New England Biolabs, New England) using specific primers (515F-806R) with a barcode. PCR product purification was carried out with Qiagen Gel Extraction Kit (Qiagen, Germany). TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, USA) was used to generate sequencing libraries. The libraries were sequenced using the Illumina NovaSeq 6000 platform (Novogene Company, China). We took advantage of Microbial Ecology 2 (QIIME2, version 2020. 2) [13] platform to process the sequencing data in a conda environment. In brief, the V3–V4 primers of paired-end fastq format sequence files were trimmed using cutadapt 3.1 [14]. Next, trimmed fastq files were imported into QIIME2. After the DADA2 denoising, a feature table listing sequence number of all samples and features was analyzed. Samples with less than 8000 sampling depth were excluded. All samples were normalized to the same depth. After rarefaction, alpha diversity and beta diversity were calculated respectively. Then, the results of the principal coordinate analysis (PcoA analysis) was calculated. Taxonomic composition of the samples were also displayed. The discrepantly abundant bacterial taxa between two groups were analyzed and displayed by linear discriminant analysis effect size (LEfSe) [15].
Establishment of a response-prediction model for 131I therapy
Preliminary covariates were discriminatory gut taxa between ER and NER pre-131I therapy and some possibly associated clinical factors, including relative abundance of Tannerellacea, g_Parabacteroides, g_Dorea, g_Bifidobacterium, f_Bifidobacteriaceae, o_Bifidobacteriales, f_Erysipelotrichaceae, c_Erysipelotrichia, o_Erysipelotrichales, age, gender, risk level, pathologic stage, maximum tumor diameter, local invasion, lymph node metastasis, distant metastasis, sTg or TgAb level, and Firmicutes/Bacteroidetes ratio. Variable screening was carried out by determination method based on characteristic root. In short, 5 variables, including f_Tannerellaceae, f_Bifidobacteriaceae, o_Bifido-bacteriales, c_Erysipelot-richi, and o_Erysipelotrichales, with multicollinearity were eliminated from the model. It was validated that there was no multicollinearity among the left variables post screening by both the characteristic root determination method and calculation of the ranks of matrixes. The left variables, including age, gender, risk level, pathologic stage, maximum tumor diameter, local invasion, lymph node metastasis, distant metastasis, sTg or TgAb level, Firmicutes/Bacteroidetes ratio, relative abundance of g_Parabacteroides, g_Dorea, g_Bifidobacterium, and f_Erysipelotrichaceae, were analyzed by multivariate analysis using binary logistic regression with backward stepwise to establish the predictive model. After 12 rounds of conditional backward stepwise removal of covariates with non-significant predictive effect, ultimately 3 variables, consisting of lymph node metastasis, relative abundance of g_Bifidobacterium and g_Dorea, were screened out as the optimal set of characteristic variables to predict patients’ response to 131I therapy. The statistical software used was IBM SPSS Statistics 26.0.
Bioinformatics and statistical analysis
Alpha (α) diversity was evaluated by a set of indexes, including Shannon, Simpson, ACE, and CHAO1, while beta (β) diversity was assessed using Bray–Curtis distance-based non-metric multidimensional scaling (NMDS) analysis, supervised partial least squares-discriminant analysis (PLS-DA), PCoA based on unweighted unifrac distance matrix or based on Jaccard index. Taxa bar plot and LEfSe analysis were performed to distinguish discrepant abundant genera between the two groups. The predicted metabolic functional differences of gut microbiota between the two groups were compared using picrust2 to identify differentially involved Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
The normality of distribution was determined using Shapiro–Wilk test. Homogeneity of variance was tested by F test. Comparison of baseline characters was performed by χ2 or Student’s t-test. Paired t-test was used in the statistical analysis of self-controlled study. Significance between two groups was determined by Student’s t-test or Wilcoxon-Rank test. P values < 0.05 were considered significant.
Results
Study cohorts and clinical parameters of self-controlled study
Sixty post-operative PTC patients, who were planning for 131I therapy, were recruited in the self-controlled study to explore microbiota changes due to 131I therapy. Baseline characters are listed in Table 1.
Microbial richness, diversity, and composition alteration after 131I therapy
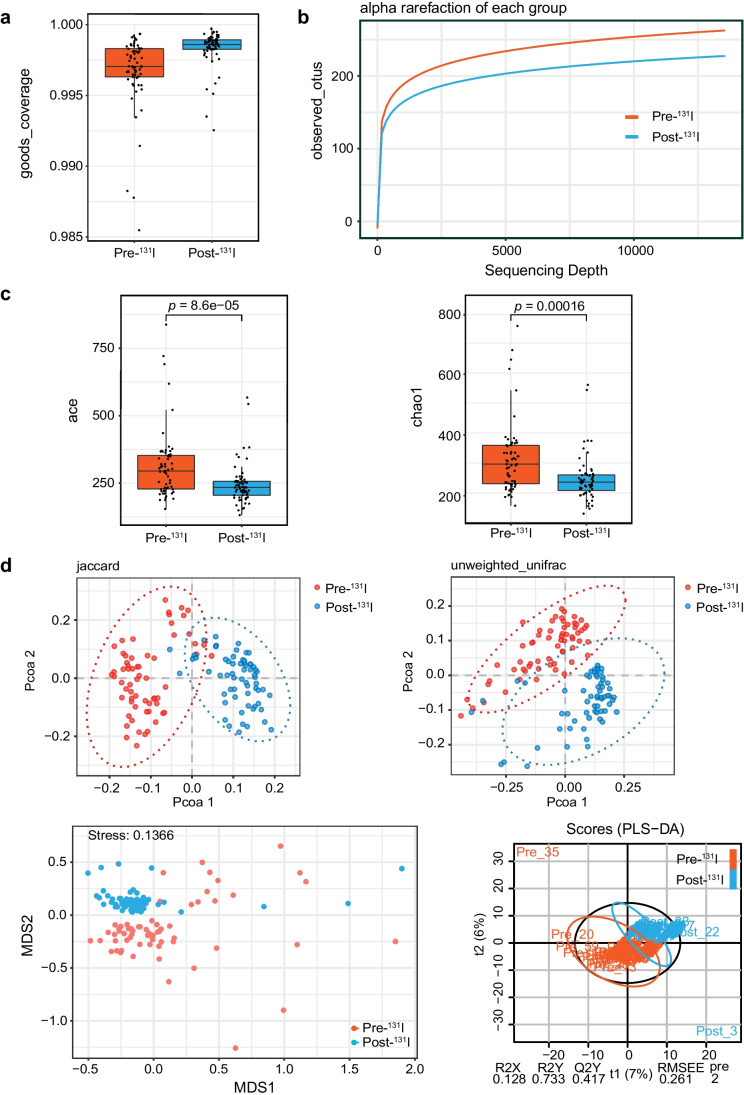
From the 60 self-controlled fecal samples, 7,540,436 high-quality sequences were used (average 59,845 per sample). The total number of ASV was 9583 at 99% similarity level. The Good’s coverage of each group was over 99% (Fig. 1a). In addition, alpha Rarefaction curve in each group is nearly smooth with a sufficient amount of sequencing data (Fig. 1b), indicating that the sequence depth represented the majority of gut bacteria in the samples.
Fig. 1.
Microbial richness and diversity alterations post-131I therapy. Pre-131I: 1–2 days before 131I administration; Post-131I: 5 months after.131I administration. a Goods_coverage indexes of two groups. b Alpha rarefaction of each group indicating sequencing depth. c Ace and Chao1 indexes of the taxonomic α diversities. Ace: paired t-test, p = 8.6e-05. Chao1: paired t-test, p = 0.00016. d PCoA plot, NMDS, and PLS-DA indexes of the β diversity (paired t-test, PCoA unweighted index: r = 0.2037, p = 0.001; PCoA Jaccard index: r = 0.2813, p = 0.001; NMDS: stress = 0.1366; PLS-DA: R2X = 0.128, R2Y = 0.733, Q2Y = 0.417)
The taxonomic α diversity indicated by ACE (Abundance-based Coverage Estimator) richness (p = 8.6e − 05) and Chao1 (p = 0.00016) indexes displayed significantly lower community richness and diversity in the fecal microbiota after 131I therapy (Fig. 1c).
Shifts in β diversity (composition and structure) from pre-131I therapy to post-131I therapy were also observed by PCoA based on unweighted unifrac distance matrix and Jaccard index, NMDS analysis, and PLS-DA (Fig. 1d). All results indicated that the samples within each group were clustered together, while the samples between groups were separated, illustrating prominent differences in bacterial structure pre- and post-131I therapy (PCoA unweighted index: r = 0.2037, p = 0.001; PCoA Jaccard index: r = 0.2813, p = 0.001; NMDS: stress = 0.1366; PLS-DA: R2X = 0.128, R2Y = 0.733, Q2Y = 0.417).
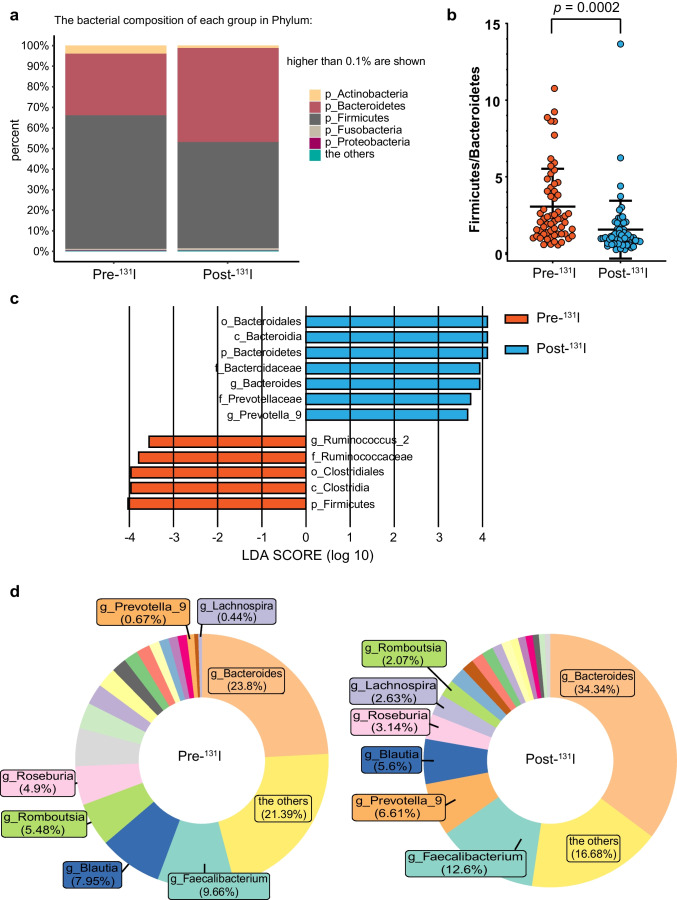
Taxa bar plot showed the bacterial composition of each group in Phylum (Fig. 2a). The most pronounced differences were decrease of Firmicutes and increase of Bacteroidetes, consequently leading to a significant decline in the Firmicutes to Bacteroides (F/B) ratio (p = 0.0002) after treatment (Fig. 2b), suggesting dysbiosis following 131I therapy. LEfSe analysis illustrated remarkably different microbes between the two groups with a LDA score over 3.5 (Fig. 2c), with notable increments in relative abundance of f_Bacteroidaceae and f_Prevotellaceae post-131I therapy, and reverse trend in f_Ruminococcaceae. Genus-level distribution alterations of fecal microbiota was demonstrated by pie plot (Fig. 2d), showing an overall pattern of an increase in “pathogenic microbiota,” for example, Bacteroides, Prevotella_9, and a decrease in “beneficial microbiota,” including Roseburia and Blautia.
Fig. 2.
Microbial composition alterations post-131I therapy. a Taxa bar plot illustrating the bacterial composition of each group in Phylum. b Firmicutes to Bacteroides (F/B) ratio of the two groups. Paired t-test. p = 0.0002. c Linear discriminant analysis effect size (LEfSe) analysis indicating differentially abundant bacterial taxa with a LDA score over 3.5 between the two groups. d Pie plot showing genus-level distribution of fecal microbiota
Predicted functional changes of microbiome induced by 131I therapy
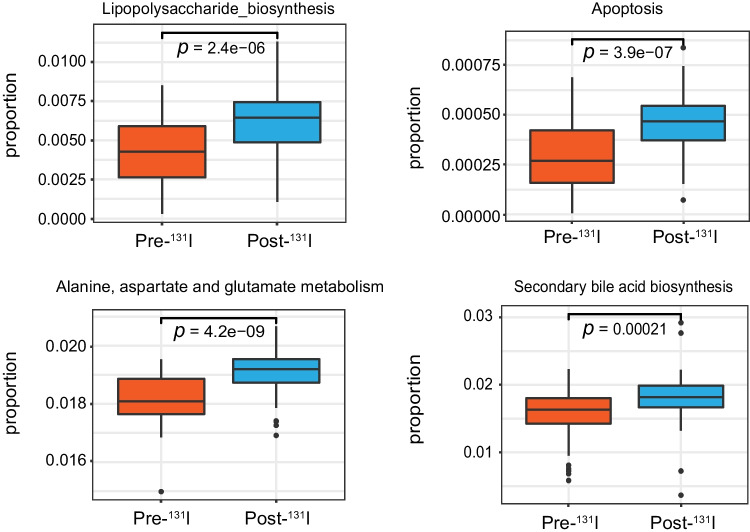
We further investigated the predicted functional alterations in gut microbiota owing to 131I therapy. Analysis of KEGG pathways showed that compared with microbiome before 131I therapy, after 131I therapy, pathways involved in lipopolysaccharide biosynthesis, apoptosis, alanine, aspartate, and glutamate metabolism, etc., were markedly altered (Fig. 3).
Fig. 3.
Significantly altered KEGG pathways of gut microbiome post-131I therapy
Study cohorts and clinical parameters of 131I ER and NER patients
After identifying the impact of 131I therapy on gut microbiota, we then explored whether microbiome dysbiosis would be correlated with therapeutic response to 131I therapy. After aforementioned exclusion of 20 patients, 40 participants were included and classified into 131I ER group or NER group. Patients’ demographic features and clinical parameters are shown in Table 2.
Table 2.
Demographic and clinical characteristics of participants in the curative effect-controlled study
| Baseline characteristics | Participants (n = 40) | Comparison between groups (p value) | |
|---|---|---|---|
| Excellent response (ER) |
Non-excellent response (NER) | ||
| Case: N (%) | 24 (60%) | 16 (40%) | |
| Age, years: mean ± SD | 38.67 ± 9.7 | 35.63 ± 9.0 | 0.324 |
| Gender: N (%) |
Male: 8 (33.3%) Female: 16 (66.7%) |
Male: 8 (50%) Female: 8 (50%) |
0.234 |
| Risk level: N (%) |
Medium: 20 (83.3%) High: 4 (16.7%) |
Medium: 14 (87.5%) High: 2 (12.5%) |
0.718 |
| Pathologic stage post-surgery: N (%) |
I: 22 (91.6) II: 1 (4.2%) III: 1 (4.2%) |
I: 16 (100%) II: 0 (0%) III: 0 (0%) |
1.000 |
| Maximum tumor diameter, cm: mean ± SD | 1.42 ± 0.95 | 1.48 ± 0.80 | 0.852 |
| Local invasion by pathology: N (%) |
Without: 18 (75%) With: 6 (25%) |
13 (81.3%) 3 (18.7%) |
0.717 |
| Lymph node metastasis by RxWBSa: N (%) |
Without:15 (62.5%) With: 9 (37.5%) |
14 (87.5%) 2 (12.5%) |
0.148 |
| Distant metastasis by RxWBS: N (%) |
Without: 23 (95.8%) With: 1 (4.2%) |
16 (100%) 0 (0%) |
1.000 |
| sTgb or TgAbc level: | |||
|
sTg < 1 ng/ml: N (%) sTg 1–10 ng/ml: N (%) sTg > 10 ng/ml or rising TgAb level: N (%) |
4 (16.7%) 15 (62.5%) 5 (20.8%) |
1 (6.3%) 7 (43.7%) 8 (50%) |
0.186 |
aRxWBS, post radioactive iodine therapy whole-body scanning
bsTg, TSH-stimulated thyroglobulin
cTgAb, thyroglobulin antibody
Gut microbiota richness, diversity, and composition differences between 131I ER and NER group pre- and post-131I therapy
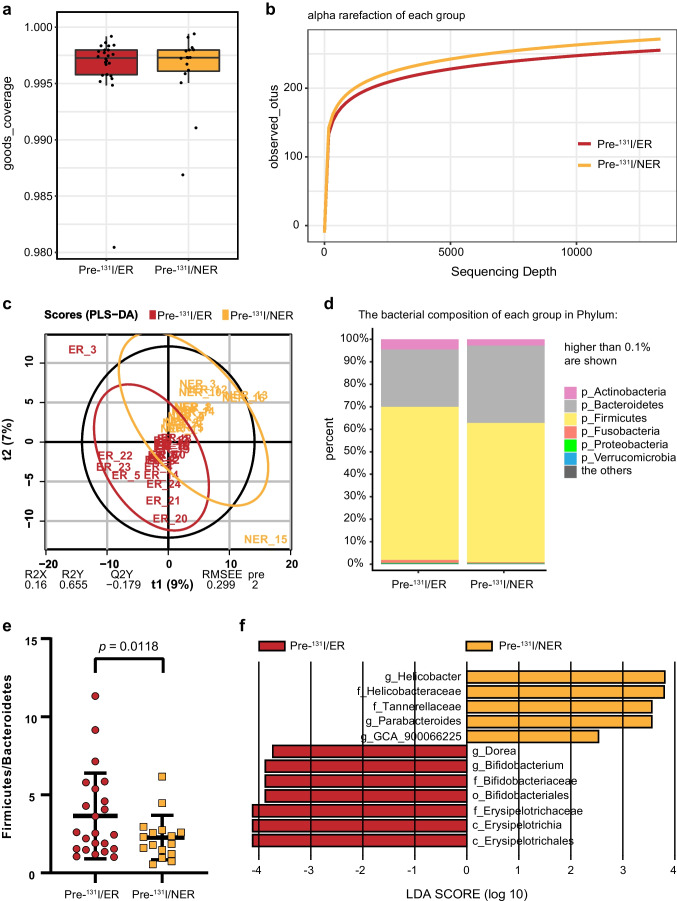
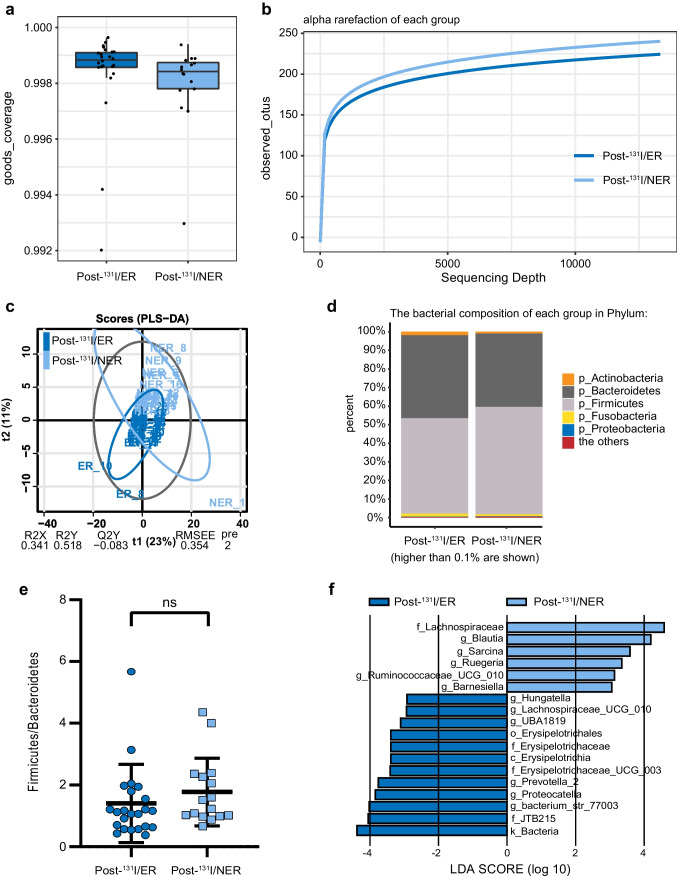
At 1–2 days prior to 131I radiotherapy, from the 40 participants’ samples, 2,268,985 high-quality sequences were used (average 56,725 per sample). The total number of ASV was 4958 at 99% similarity level. The Good’s coverage (over 99%) (Fig. 4a) and the observed species rarefaction curve (Fig. 4b) were used to characterize good sequencing depths. The microbiota of the two groups showed no significant difference in α diversity (data not shown). β diversity analysis represented by PLS-DA-plot illustrated that the microbiome samples were clustered by group (Fig. 4c; R2X = 0.16, R2Y = 0.655, Q2Y = − 0.179). Different bacterial composition of each group in Phylum was shown in taxa bar plot (Fig. 4d). A prominent decrease of F/B ratio was observed in the NER group as compared to ER group (Fig. 4e; p = 0.0118). LEfSe analysis illustrated substantially differently abundant bacterial genera, including Parabacteroides, Dorea, and Bifidobacterium and microbial family, such as Erysipelotrichaceae with a LDA score over 2.0 between the two groups (Fig. 4f).
Fig. 4.
Significant differences in microbial richness, diversity, and composition between excellent response (ER) and non-excellent response (NER) groups 1–2 days before 131I therapy. ER: excellent response to 131I therapy; NER: non-excellent response to 131I therapy. a Goods_coverage indexes of two groups. b Alpha rarefaction of each group indicating sequencing depth. c PLS-DA-plot of the two groups. d Taxa bar plot indicating the bacterial composition of each group in Phylum. e Firmicutes to Bacteroides (F/B) ratio of the two groups (t-test, p = 0.0002). f LEfSe analysis showing significantly different gut flora with a LDA score over 2 between the two groups
From the data of specimens collected at 5 months post-131I therapy, 2,253,269 high-quality sequences were used (average 60,832 per sample). The total number of ASV was 2,896 at 99% similarity level. The Good’s coverage of each group was over 99% (Fig. 5a). Rarefaction curve indicated good sequencing depths (Fig. 5b). No significant α diversity was noted between ER and NER groups. However, in line with the result of pre-131I therapy, a notable clustering effect of β diversity by response status post-131I was also revealed by PLS-DA-plot (Fig. 5c; R2X = 0.341, R2Y = 0.518, Q2Y = − 0.083). Taxa bar plot showed elevation of Firmicutes and reduction of Bacteroidetes composition in the NER group (Fig. 5d), but no significant statistic difference was found in F/B ratio (Fig. 5e). LEfSe analysis illustrated significantly different bacterial taxa with a LDA score over 2.0 between ER and NER groups (Fig. 5f), including f_Erysipelotrichaceae, f_Lachnospiraceae, etc.
Fig. 5.
Significant differences in microbial richness, diversity, and composition between excellent response (ER) and non-excellent response (NER) groups 5 months after 131I therapy. a Goods_coverage indexes of two groups. b Alpha rarefaction of each group indicating sequencing depth. c PLS-DA-plot of the two groups. d Taxa bar plot indicating the bacterial composition of each group in Phylum. e Firmicutes to Bacteroides (F/B) ratio of the two groups (t-test). f LEfSe analysis showing significantly different gut flora with a LDA score over 2 between the two groups
Establishment of a predictive model consisting of gut microbiome and clinical data for response to 131I therapy
Based on the results above, we speculated that the gut microbiome signatures of ER or NER pre-131I therapy, combined with some clinical data possibly affecting 131I response, might construct a predictive model for response to 131I therapy in post-surgery patients with PTC. After screening and elimination of variables with multicollinearity, a multivariate analysis using binary logistic regression analysis was performed to analyze the remaining 14 variables to establish a predictive model. Finally, lymph node metastasis and relative abundance of g_Bifidobacterium and g_Dorea were finally chosen as the optimal set to establish the predictive model, with a p value of 0.003 and an overall percentage correct of 80.0% (Table 3), suggesting that this model performed well on predicting response to 131I therapy. High abundance of g_Bifidobacterium and g_Dorea and no lymph node metastasis predicted the patient to be ER, and vice versa. Of note, with a p value of 0.04, g_Dorea was an independent predictor of 131I therapy response.
Table 3.
Summary of predictive model for response to 131I therapy by multivariate analysis using binary logistic regression
| Percentage correct | Chi-square | p | Variables | B | Wald | p | |
|---|---|---|---|---|---|---|---|
| Model | 80.0% | 13.795 | 0.003 | Lymph node metastasis | -1.658 | 2.832 | 0.092 |
| g_Bifidobacterium | 33.229 | 2.774 | 0.096 | ||||
| g_Dorea | 161.779 | 4.206 | 0.040 |
Discussion
Previous literatures have shown variable results in gut microbiome alterations induced by irradiation exposure [11, 16–18]. Nevertheless, the gut microbiota changes in the setting of 131I therapy have not been elucidated yet. Coinciding with some other findings in gut microbiota changes post radiation [19], our data illustrated that 131I therapy also resulted in deterioration of patients’ microbiome alpha diversity and alteration of microbial composition, with an enrichment of “harmful microbiota” and a reduction in “beneficial microbiota” on the whole. The impact of 131I therapy on gut microbiota was huge and durable, at least lasting until the end of our observation, 5 months post 131I administration. As shown by accumulating literatures, gut microbiota dysbiosis is involved in the development of a wide range of diseases, including diarrhea, allergy, cancer, aging, and diabetes [20]; thus, the dysbiosis after 131I therapy described in the present study has great influence on intestinal homeostasis and PTC patients’ health.
Among the changes of gut microbiome, alteration in F/B ratio is worth noting. In a healthy host, the majority of gut microbiome is typically dominated by four major phyla: Bacteroidetes, Firmicutes, Actinobacteria, and Proteobacteria [21]. Firmicutes and Bacteroidetes take up over 90% of the relative abundance of the gut microbiota, and their relationship plays a critical role in the maintenance of gut homeostasis. Aberrant ratio between the relative abundance of Firmicutes and Bacteroidetes (F/B ratio) has been found in a series of physiological and pathological conditions, including aging [22], tumor [23], obesity [24], type I diabetes [25], intestinal inflammation. In the present study, besides reduced F/B ratio, our data showed a prominent decline of Firmicutes post-131I therapy, which was in line with Firmicutes change after pelvic radiotherapy [17], indicating a comparable dysbiosis of gut microbiota after 131I therapy. Of note, F/B ratio was also found to be declined in NER group compared to ER group, indicating a possible connection between response to 131I therapy and microbial dysbiosis.
For the sake of understanding metabolic profile alterations of gut microbiota after 131I therapy, KEGG analysis of stool samples was performed. Some of the increases in metabolites after 131I therapy, such as bile acids and alanine, are in accordance with a previous reported characteristic elevation in radiation-induced acute intestinal symptoms in cervical cancer patients [26], indicating that metabolism of gut microbiome might share some common features post radiation, which warrants further study.
Numerous previous literatures have shown that gut microbiota exhibits value in predicting diseases [27, 28] and treatment response [29, 30]. In order to explore potential predictors of response to 131I therapy, gut microbiome differences were compared between 131I ER and NER groups both before and after the therapy. Different gut microbiota structures were identified as shown in LefSe analysis in Figs. 4f and 5f. Of note, in agreement with a previous study investigating gut microbiota signatures of distinct responses to neoadjuvant chemoradiotherapy [16], g_Dorea, a butyric acid-producing flora, regarded as an important component of functional microbiota in a healthy GI tract [31], was found to be significantly less abundant in 131I NER group pre-therapy in this study as well. Moreover, g_Dorea is later identified to be an independent predictor of response to 131I therapy, further suggesting its importance and predictive value. It is of interest that Erysipelotrichaceae, which belongs to the Firmicutes phylum, with multiple interactions with host immune response, gut inflammation, and lipid metabolism, etc. [32], was markedly decreased in NER patients in comparison with ER patients consistently before and after 131I therapy. Lachnospiraceae family, which is positively associated with protection against radiation-induced intestinal damages [33], was found to be remarkably increased in the NER group post-131I therapy, which awaits more study. The comparison of gut microbiota between groups with distinct responses to 131I may provide additional information on how the gut microbiome interacts with radiotherapy responses.
Moreover, a predictive model for response to 131I therapy of PTC patients was established in this study. Three variables, including lymph node metastasis, relative abundance of g_Bifidobacterium, and g_Dorea, for the first time, are identified to compose a model capable of predicting response to 131I therapy before the treatment initiation, which will be beneficial for customizing optimal therapeutic approaches for each PTC patient. The strategies to increase relative abundance of g_Dorea and g_Bifidobacterium in the gut microbiota may be promising in improving the response to 131I therapy in post-operative PTC patients. As stated before, g_Dorea, which produces butyric acid, is identified to be an independent predictor of response to 131I therapy in this study. Intriguingly, short-chain fatty acids (SCFAs), including butyric acid, are able to upregulate sodium/iodide (Na+/I−) symporter (NIS) expression through epigenetic modification, thus promote I− uptake and beneficially affect response to 131I therapy[34], which may be one of the underlying mechanisms and needs more investigation.
To sum up, the present study demonstrates gut microbiota changes after 131I therapy in post-surgery PTC patients. Differences in gut flora are also found between 131I ER and NER groups both pre- and post-131I therapy, suggesting microbiome may be associated with therapeutic responses. Hence, a predictive model is established to provide a noninvasive tool for predicting response to 131I therapy prior to treatment initiation.
These conclusions are based on analysis of data obtained from a small sample pool, and reported in the pilot study herein. The long-term systemic effects of alterations in gut microbiome after 131I therapy still need rigorous evaluation. With the aim to restore a healthy intestinal microbial ecosystem, which is beneficial for the response to 131I therapy, personalized modulation of gut microbiota of PTC patients during the therapy, including dietary changes, administration of antibiotics or probiotics, and fecal microorganism transfers, is promising. Since advanced PTC patients with distant metastasis often have unfavorable prognosis, future studies on gut microbiota with larger cohort of these patients and employing whole genome metagenomics approaches should be carried out. It will shed new light on prediction and promotion of ER to 131I therapy in high-risk PTC patients. Mice model of 131I therapy is needed to further explore the complicated association between gut microbiome and response to 131I therapy.
Author contribution
All authors contributed to the study conception and design. Material preparation and data collection were performed by Zheng, L; Zhang, LJ; Guo, W; He, S; and Huang, Y. Data were analyzed by Tang, B; Xiao, X; Tang, L; Zheng, L; Chen, J; Huang, DD; Pan, D; and Chen, Y. The draft of the manuscript was written and revised by Chen, J; Zheng, L; Tang, B; and Xiao, X. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by two fundings from Army Medical University, China, #2020–2017-018, #XZ-2019–505-035.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of the First Affiliated Hospital of Third Military Medical University, China (Date Oct. 16th, 2020/No. KY2020214).
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent for publication
The authors affirm that human research participants provided informed consent for publication of their data.
Competing interests
The authors declare no competing interests.
Footnotes
This article is part of the Topical Collection on Radiation biology.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lei Zheng and Linjing Zhang equally contributed to the article.
Contributor Information
Xu Xiao, Email: lanexiao@yeah.net.
Bo Tang, Email: botangxq@tmmu.edu.cn.
Jing Chen, Email: chenjingtmmu@tmmu.edu.cn.
References
- 1.Gu Y, Yu Y, Ai L, et al. Association of the ATM gene polymorphisms with papillary thyroid cancer. Endocrine. 2014;45(3):454–461. doi: 10.1007/s12020-013-0020-1. [DOI] [PubMed] [Google Scholar]
- 2.American Thyroid Association Guidelines Taskforce on Thyroid N, Differentiated Thyroid C, Cooper DS et al. Revised American Thyroid Association management guidelines for patients with thyroid nodules and differentiated thyroid cancer. Thyroid. 2009;19(11):1167–1214. doi: 10.1089/thy.2009.0110. [DOI] [PubMed] [Google Scholar]
- 3.Ciarallo A, Rivera J. Radioactive iodine therapy in differentiated thyroid cancer: 2020 update. AJR Am J Roentgenol. 2020;215(2):285–291. doi: 10.2214/AJR.19.22626. [DOI] [PubMed] [Google Scholar]
- 4.Haugen BR. 2015 American Thyroid Association Management Guidelines for Adult Patients with Thyroid Nodules and Differentiated Thyroid Cancer: what is new and what has changed? Cancer. 2017;123(3):372–381. doi: 10.1002/cncr.30360. [DOI] [PubMed] [Google Scholar]
- 5.Jaye K, Li CG, Bhuyan DJ. The complex interplay of gut microbiota with the five most common cancer types: from carcinogenesis to therapeutics to prognoses. Crit Rev Oncol Hemat. 2021;165:ARTN 10342910. doi: 10.1016/j.critrevonc.2021.103429. [DOI] [PubMed] [Google Scholar]
- 6.Yu X, Jiang W, Kosik RO, et al. Gut microbiota changes and its potential relations with thyroid carcinoma. J Adv Res. 2022;35:61–70. doi: 10.1016/j.jare.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X, Suo J, Huang X, et al. Whole grain Qingke attenuates high-fat diet-induced obesity in mice with alterations in gut microbiota and metabolite profile. Front Nutr. 2021;8:761727. doi: 10.3389/fnut.2021.761727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang JM, Zhang FH, Zhao CY, et al. Dysbiosis of the gut microbiome is associated with thyroid cancer and thyroid nodules and correlated with clinical index of thyroid function. Endocrine. 2019;64(3):564–574. doi: 10.1007/s12020-018-1831-x. [DOI] [PubMed] [Google Scholar]
- 9.Maier I, Schiestl RH. Evidence from animal models: is a restricted or conventional intestinal microbiota composition predisposing to risk for high-LET radiation injury? Radiat Res. 2015;183(6):589–593. doi: 10.1667/RR13837.1. [DOI] [PubMed] [Google Scholar]
- 10.Sims TT, El Alam MB, Karpinets TV, et al. Gut microbiome diversity is an independent predictor of survival in cervical cancer patients receiving chemoradiation. Commun Biol. 2021;4(1):ARTN 237. doi: 10.1038/s42003-021-01741-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira MR, Andreyev HJN, Mohammed K, et al. Microbiota- and Radiotherapy-Induced Gastrointestinal Side-Effects (MARS) study: a large pilot study of the microbiome in acute and late-radiation enteropathy. Clin Cancer Res. 2019;25(21):6487–6500. doi: 10.1158/1078-0432.Ccr-19-0960. [DOI] [PubMed] [Google Scholar]
- 12.Tian T, Kou Y, Huang R, et al. Prognosis of high-risk papillary thyroid cancer patients with pre-ablation stimulated Tg <1 Ng/Ml. Endocr Pract. 2019;25(3):220–225. doi: 10.4158/EP-2018-0436. [DOI] [PubMed] [Google Scholar]
- 13.Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37(8):852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcel M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17(1):10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 15.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yi Y, Shen L, Shi W, et al. Gut microbiome components predict response to neoadjuvant chemoradiotherapy in patients with locally advanced rectal cancer: a prospective, longitudinal study. Clin Cancer Res. 2021;27(5):1329–1340. doi: 10.1158/1078-0432.CCR-20-3445. [DOI] [PubMed] [Google Scholar]
- 17.Nam YDKH, Seo JG, Kang SW, et al. Impact of pelvic radiotherapy on gut microbiota of gynecological cancer patients revealed by massive pyrosequencing. PLoS ONE. 2013;8:e82659. doi: 10.1371/journal.pone.0082659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lam VMJ, Salzman NH, Dubinsky EA, et al. Intestinal microbiota as novel biomarkers of prior radiation exposure. Radiat Res. 2012;177:573–583. doi: 10.1667/RR2691.1. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Zhang Y, Wei K, et al. Review: Effect of gut microbiota and its metabolite SCFAs on radiation-induced intestinal injury. Front Cell Infect Microbiol. 2021;11:577236. doi: 10.3389/fcimb.2021.577236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jian Y, Zhang D, Liu M, et al. The impact of gut microbiota on radiation-induced enteritis. Front Cell Infect Microbiol. 2021;11:586392. doi: 10.3389/fcimb.2021.586392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spychala MS, Venna VR, Jandzinski M, et al. Age-related changes in the gut microbiota influence systemic inflammation and stroke outcome. Ann Neurol. 2018;84(1):23–36. doi: 10.1002/ana.25250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patrizz A, Dono A, Zorofchian S, et al. Glioma and temozolomide induced alterations in gut microbiome. Sci Rep-Uk. 2020;10(1):ARTN 2100210. doi: 10.1038/s41598-020-77919-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ley RE, Turnbaugh PJ, Klein S, et al. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 25.Bibbo S, Dore MP, Pes GM, et al. Is there a role for gut microbiota in type 1 diabetes pathogenesis? Ann Med. 2017;49(1):11–22. doi: 10.1080/07853890.2016.1222449. [DOI] [PubMed] [Google Scholar]
- 26.Chai Y, Wang J, Wang T, et al. Application of 1H NMR spectroscopy-based metabonomics to feces of cervical cancer patients with radiation-induced acute intestinal symptoms. Radiother Oncol. 2015;117(2):294–301. doi: 10.1016/j.radonc.2015.07.037. [DOI] [PubMed] [Google Scholar]
- 27.Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol. 2019;16(11):690–704. doi: 10.1038/s41575-019-0209-8. [DOI] [PubMed] [Google Scholar]
- 28.Iannone LF, Preda A, Blottiere HM, et al. Microbiota-gut brain axis involvement in neuropsychiatric disorders. Expert Rev Neurother. 2019;19(10):1037–1050. doi: 10.1080/14737175.2019.1638763. [DOI] [PubMed] [Google Scholar]
- 29.Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359(6371):97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou CB, Zhou YL, Fang JY. Gut microbiota in cancer immune response and immunotherapy. Trends Cancer. 2021;7(7):647–660. doi: 10.1016/j.trecan.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 31.Huang C, Li X, Wu L, et al. The effect of different dietary structure on gastrointestinal dysfunction in children with cerebral palsy and epilepsy based on gut microbiota. Brain Dev. 2021;43(2):192–199. doi: 10.1016/j.braindev.2020.09.013. [DOI] [PubMed] [Google Scholar]
- 32.Kaakoush NO. Insights into the role of Erysipelotrichaceae in the human host. Front Cell Infect Mi. 2015;5:ARTN 84. doi: 10.3309/fcimb.2015.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo H, Chou WC, Lai Y, et al. Multi-omics analyses of radiation survivors identify radioprotective microbes and metabolites. Sci. 2020;370(6516). 10.1126/science.aay9097 [DOI] [PMC free article] [PubMed]
- 34.Samimi H, Haghpanah V. Gut microbiome and radioiodine-refractory papillary thyroid carcinoma pathophysiology. Trends Endocrinol Metab. 2020;31(9):627–630. doi: 10.1016/j.tem.2020.03.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.