Abstract
The Epstein-Barr virus (EBV) replicates once per cell cycle and segregates with high efficiency yet does not encode the enzymes needed for DNA replication or the proteins required to contact mitotic spindles. The virus-encoded EBNA-1 (EBV nuclear antigen 1) and latent replication origin (oriP) are required for both replication and segregation. We developed a sensitive and specific fluorescent labeling strategy to analyze the interactions of both EBNA-1 with viral episomes and viral episomes with host chromosomes. This enabled investigation of the hypothesis that replication and chromosome tethering are linked through the EBNA-1 protein. We show that deleting EBNA-1 or oriP disrupts mitotic chromosome tethering but removing the dyad symmetry element of oriP does not. Microscopic and biochemical approaches demonstrated that an EBNA-1 mutant lacking residues 16 to 372 bound to oriP plasmids but did not support their mitotic chromosome association and that the mutant lost the ability of wild-type EBNA-1 to associate with interphase chromatin. Importantly, the transient-replication abilities of various mutant forms of EBV plasmids, including the mutant form with the EBNA-1 internal deletion, correlated directly with their chromosome-tethering abilities. These data lead us to propose that EBNA-1 recruits oriP-containing plasmids into chromatin subdomains in interphase nuclei to both engage the host replication machinery and enable the plasmids to adhere to host chromosomes to increase their segregation efficiency.
Proper transmission of the cellular genome requires both accurate and complete replication and subsequent faithful segregation of replicated molecules to daughter cells during mitosis. It is becoming increasingly clear that replication and segregation are closely linked. For example, studies with yeast show that sister chromatid cohesion, which contributes to faithful chromosome segregation during mitosis, is established during DNA replication (44, 46). This indicates that molecular mechanisms governing replication and segregation overlap, at least in part.
Coordinated replication and segregation are also required for DNA tumor viruses to be maintained as stable episomes during latent infections in dividing host cells. Epstein-Barr virus (EBV) provides an excellent model for studying such coordination between replication and segregation. EBV exists as a 165-kb double-stranded circular episome in latently infected human B cells (for reviews, see references 26 and 27). Only two viral elements, the cis-acting latent origin of replication (oriP) and the trans-acting viral protein EBNA-1 (52), are required to propagate extrachromosomal EBV replicons. As EBNA-1 has no reported catalytic functions required for DNA replication (11) yet latent EBV episomes replicate only once per host cell cycle (51), it is likely that the host replication machinery controls viral replication. These features contrast with the replication of other DNA tumor viruses, such as simian virus 40, which occurs multiple times per host cell cycle and requires the helicase activity of the virus-encoded large T antigen (45).
oriP is composed of two clusters of EBNA-1 binding sites, referred to as the dyad symmetry (DS) element and the family-of-repeats (FR) element (38). The DS element consists of four low-affinity EBNA-1 binding sites, while the FR element contains 20 high-affinity EBNA-1 binding sites (11, 36, 38). Genetic analyses revealed that the DS and FR elements perform distinct, essential functions for plasmid replication and maintenance. The DS region comprises a minimal cis-acting sequence required for replication of EBV plasmids in EBNA-1-positive cells (40, 50), although the precise role of EBNA-1 in the initiation of replication in the DS region remains elusive (reviewed in reference 26). On the other hand, the FR element itself lacks the ability to initiate replication, but FR-containing plasmids have a nuclear retention ability in cells expressing EBNA-1 (23, 31). This nuclear retention function proved to be useful for cloning of human sequences that substitute for the DS element (23). These data have been interpreted as evidence that the DS element contributes to plasmid replication while the FR element contributes to nuclear retention.
Whether the FR element contributes to DS-dependent replication is a matter of debate. When plasmid replication was assayed soon (48 h) after transfection of 293 cells, the addition of the FR element showed a minimal effect on DS-dependent replication (50). In contrast, the FR element contributed significantly to DS-dependent replication when plasmid replication was assayed long (96 h) after transfection of HeLa cells (40). One interpretation of the data is that the combination of FR and EBNA-1 stabilizes the replicated plasmid molecules through the nuclear retention function of EBNA-1, and this effect does not become apparent until 96 h after transfection. An equally likely alternative is that FR–EBNA-1 can directly enhance plasmid replication, although the effect is not apparent at 48 h posttransfection. It is of note that deletion of DS from the EBV genome did not impair stable replication after infection (34), indicating that the DS region is not absolutely required for viral latent replication.
Accumulating evidence suggests that the nuclear retention of EBV plasmids requires their association with mitotic chromosomes. Chromosome-sorting and fluorescence in situ hybridization (FISH) experiments previously indicated that EBV plasmids were attached to mitotic chromosomes (9, 13, 42, 48). EBNA-1 protein is also known to associate with mitotic chromosomes (12, 35). Recent analysis of EBNA-1 deletion mutant proteins fused to green fluorescent protein (GFP) identified three chromosome binding domains in the N-terminal region of the EBNA-1 protein (29) which are distinct from the oriP binding domain in the C-terminal region (2). Taken together, these data raise the possibility that EBNA-1 provides a bridge between mitotic chromosomes and oriP-containing plasmids. Such chromosome tethering could increase the segregation efficiency of EBV plasmids into daughter nuclei when the nuclear membrane reforms at the end of mitosis.
As the EBNA-1–oriP combination appears to be involved in both viral replication and segregation, it is tempting to hypothesize that viral replication and chromosome tethering are functionally coupled. In order to investigate links between replication and chromosome tethering, it is critical to directly examine the effects of mutations in oriP and EBNA-1 on chromosome tethering. For example, if there is a direct link between tethering and replication, then oriP plasmids lacking the EBNA-1 gene should neither replicate autonomously nor associate with mitotic chromosomes. Another prediction is that a mutant form of the EBNA-1 protein lacking the chromosome binding domains should neither support replication nor recruit oriP plasmids onto mitotic chromosomes. It has been difficult to test these predictions rigorously because a direct and sensitive system for simultaneously analyzing chromosome tethering and replication of EBV plasmids has not been available.
We utilized a lac operator (lacO)/lac repressor (lacR)-GFP system (39) to directly examine the subcellular localization of various mutant forms of EBV plasmids. We provide microscopic evidence that chromosome tethering of EBV plasmids is dependent on both EBNA-1 protein and the FR region of oriP, while the DS region is dispensable. Deletion analyses of EBNA-1 revealed that the previously defined chromosome binding domains (29) are important for both mitotic chromosome binding and chromatin association in interphase nuclei. Importantly, the efficient transient replication of various mutant forms of the EBV-lacO plasmid correlates with their chromosome tethering competence. Taken together, these data lead us to propose that chromatin-associated EBNA-1 protein recruits oriP plasmids onto cellular chromatin, which then engages the host replication apparatus. This model suggests direct links among chromosome tethering, replication competence, and efficient mitotic segregation.
MATERIALS AND METHODS
Plasmid construction.
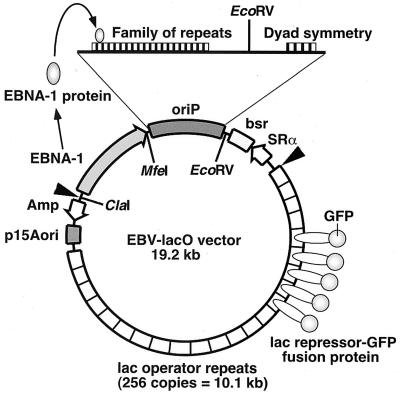
The EBV-lacO plasmid, which has been described previously (20), contains EBV oriP and EBNA-1 coding sequences derived from pCEP4 (Invitrogen, Carlsbad, Calif.). A fragment encoding a hygromycin resistance gene, a cytomegalovirus promoter, and a simian virus 40 polyadenylation signal was first deleted from pCEP4 by SalI (blunted by Klenow enzyme)-NruI digestion and then self-ligated to make pEP4, which recovered the SalI site after self-ligation. A blasticidin resistance gene (18), driven by the SRα promoter, was clipped out from pYN3215-bsr (kindly provided by Fumio Hanaoka, Osaka University) by NheI digestion and subcloned into the XbaI site of pEP4 to make pEPB. Subsequently, an EcoRI-SalI fragment of pEPB, containing a blasticidin resistance gene, oriP, and the EBNA-1-encoding gene, was subcloned into EcoRI-SalI-digested pMBL19 to make pMBL19EBVbsr. pMBL19, which has a bacterial p15A ori, was chosen for its ability to subclone unstable inserts (33). lac operator repeats (256 direct repeats) were obtained by SalI-XhoI digestion of pSV2-dhfr 8.32 (39). The repeats were subcloned into the SalI site of pMBL19EBVbsr to make the EBV-lacO vector. This step was carried out using STBL2 competent cells (Life Technologies, Grand Island, N.Y.), which were grown at 30°C (5).
The oriP sequence and the EBNA-1 gene in the EBV-lacO vector were deleted using the restriction enzyme recognition sites flanking these sequences (see Fig. 1). The EBV-lacO vector was digested with either MfeI-ClaI (blunted with Klenow enzyme), EcoRV-MfeI (blunted), EcoRV only, or EcoRV-ClaI (blunted) and then self-ligated to make ΔEBNA1, ΔoriP, ΔDS, and ΔoriP/EBNA1, respectively.
FIG. 1.
Experimental system for tracking of transfected EBV plasmids. The EBV-lacO vector encodes EBNA-1 and contains the latent replication origin oriP. oriP contains 24 EBNA-1 binding sites clustered in two functional elements, FR and DS. The plasmid has 256 tandem repeats of lacO, to which the lacR-GFP fusion protein binds with high affinity. The plasmid also encodes a blasticidin resistance gene (bsr) driven by the SRα promoter as a drug selection marker. The restriction enzyme recognition sites used to construct various deletion mutant plasmids are indicated. The arrowheads indicate the boundary of the EcoRI fragment, which was detected by Southern analyses of the transient-replication assay (see Fig. 7).
A BglII restriction sequence was introduced between codons 372 and 373 of the EBNA-1 coding sequence (AGAGGT and AGATCT) of the pEP4 vector using PCR-mediated mutagenesis to make pEP4-BglII. The AvrII site (codon 14) and the introduced BglII site (codon 372) of pEP4-BglII were used for further constructions. pEP4-BglII was digested with AvrII-BglII (blunted) and then self-ligated to make EBNA-1(Δ16–372). A DNA fragment encoding EBNA-1 (codons 14 to 89 and 328 to 372), with AvrII and BglII sites on its ends, was synthesized by two-step PCR using pEP4-BglII as a template, and the PCR product was subcloned into a pEP4-BglII vector digested with AvrII-BglII to make EBNA-1(Δ90–327). The MfeI-ClaI fragment of the EBV-lacO vector was swapped with the MfeI-ClaI fragments of EBNA-1(Δ16–372) and EBNA-1(Δ90–327) to make EBNA-1(Δ16–372)-lacO and EBNA-1(Δ90–327)-lacO vectors. The sequences of the primers used for the PCRs are available upon request.
Retroviral vectors expressing mutant EBNA-1 proteins were constructed using PCR products which had an engineered AflIII site overlapping the ATG start codon and a BamHI site just after the stop codon of EBNA-1(Δ90–327) and EBNA-1(Δ16–372). The AflIII-BamHI fragments containing the coding sequences were subcloned into an NcoI-BamHI-digested pCLMFGMCS vector (kindly provided by Nikunj Somia, Salk Institute). pCLMFGMCS is identical to the pMFG vector (10), except that it has a CMV promoter instead of the U3 region of the 5′ long terminal repeat and a multiple cloning site (NcoI-EcoRI-SalI-XhoI-NotI-BamHI).
A retroviral vector expressing the lac repressor-GFP (lacR-GFP) fusion has been described (20). Briefly, pCLMFGlacR-GFP was constructed by ligating the following three fragments; the NcoI-BsrGI fragment of pEGFPN1 (Clontech), the BsrGI-DraI fragment of p3′SSdimerClonEFP containing a gene encoding the lac repressor-nuclear localization signal (39), and XhoI (blunted by Klenow)-NcoI digested pCLMFGMCS.
Visualization of the EBV-lacO plasmid in HeLa cells.
Production of vesicular stomatitis virus G glycoprotein-pseudotyped retroviruses expressing LacR-GFP was performed by cotransfection of pCLMFGlacR-GFP and pMD.G (the plasmid encoding vesicular stomatitis virus envelope protein G glycoprotein) into 293gp/bsr cells as previously described (21, 32). A stable HeLa cell line expressing lacR-GFP (HeLa/lacR-GFP cell line) was established by infection of the lacR-GFP retrovirus, followed by single-colony isolation. The HeLa/lacR-GFP cells (4 × 105 cells) were plated in 6-cm dishes and transfected with various EBV-lacO plasmids (5 μg of each) using a modified calcium phosphate precipitation protocol (7). The transfected cells were split 1:6 at 3 days posttransfection. Mitotic cells were collected 5 days after transfection by gently washing the dishes with a pipette, stained with Hoechst 33342 (5 μg/ml), overlaid onto poly-l-lysine-coated slide glasses for 10 min, fixed with 3.7% formaldehyde for 10 min, washed once with phosphate-buffered saline (PBS), covered with Vectashield (Vector, Burlingame, Calif.), and then observed using fluorescence microscopy.
Immunostaining.
Mitotic cells overlaid onto slide glasses were fixed with 3.7% formaldehyde for 10 min, washed with PBS three times, and treated with blocking buffer (2.5% bovine serum albumin, 0.2 M glycine, 0.1% Triton X-100) for 30 min. Primary and secondary antibodies were diluted in the blocking buffer. The primary antibody used to detect the EBNA-1 protein was rabbit K67-3 serum (1:1,000 dilution; kindly provided by Jaap Middeldorp, Free University Hospital, Amsterdam, The Netherlands), which recognizes the DNA binding domain of the EBNA-1 protein. Following incubation for 60 min at room temperature, slides were washed three times with PBS. The secondary antibody was Texas red-conjugated donkey anti-rabbit immunoglobulin G (1:500). Following incubation for 60 min at room temperature, slides were washed three times with PBS and chromosomes were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) (1 μg/ml). Fluorescence of lacR-GFP was preserved well by this protocol.
Microscopy.
All of the images that appear in this article were collected using a DeltaVision microscope system (Applied Precision Inc., Issaquah, Wash.) with a 100×/NA 1.35 oil immersion objective (Olympus). Three-dimensional data sets were collected to visualize the fluorescent dots of EBV distributed in multiple focal planes. Optical sections were collected at 0.2-μm focal intervals; the pixel size was 0.0669 μm. Out-of-focus contamination was removed from each optical section via deconvolution processing, and two-dimensional images were created by projecting the three-dimensional data stacks using the software supplied with the DeltaVision system.
Cell fractionation and Western blotting.
Whole-cell extracts were prepared by direct lysis of cells (∼2 × 107 cells in 10-cm dishes) with 500 μl of lysis buffer (125 mM Tris [pH 6.7], 10% glycerol, 3% sodium dodecyl sulfate, 6% urea, 100 μg of phenylmethylsulfonyl fluoride per ml, 2 μg of aprotinin per ml, 1 mM sodium vanadate), followed by sonication. Chromatin was isolated from interphase nuclei essentially as previously described (30). Briefly, HeLa cells expressing EBNA-1(Δ90–327) or EBNA-1(Δ16–372) (2 × 107 of each) were trypsinized, harvested, and resuspended in 400 μl of buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM dithiothreitol, proteinase inhibitors). Triton X-100 (0.1%) was added, and the cells were incubated for 5 min on ice. Nuclei were separated from the supernatant (S2, soluble fraction), washed once with buffer A, resuspended in 500 μl of buffer A plus 1 mM CaCl2, and divided into two aliquots (250 μl each). One of them was incubated with 1 μl of micrococcal nuclease (200 U/ml; Sigma, St. Louis, Mo.) for 5 min at 37°C, and the other was incubated under the same conditions without nuclease. The nuclease reaction was stopped by the addition of 2 μl of 0.5 M EDTA. Nuclei were then collected by centrifugation and lysed in 400 μl of buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM dithiothreitol, proteinase inhibitors) for 30 min on ice. Insoluble chromatin fractions were separated from the supernatant (S3; solubilized nuclear proteins) by centrifugation and washed once in buffer B. The final chromatin (and nuclear matrix) pellet (P3) was resuspended in 400 μl of lysis buffer and sonicated. The aliquots of each fraction were analyzed by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis and Western blotting using anti-EBNA-1 serum K67-3 (1:1,000) as the primary antibody.
Transient-replication assay.
The ability of each plasmid to replicate transiently was determined as previously described (50). Briefly, HeLa cells (in 6-cm dishes) were transfected with 5 μg of test plasmids and replated into 10-cm dishes at 3 days posttransfection. Plasmids were extracted 96 h after transfection using alkaline lysis as previously described (50). The nucleic acid pellets were dissolved in 50 μl of Tris-EDTA containing RNase and then tested for resistance to DpnI in the presence of EcoRI digestion. The EBV-lacO vector has multiple EcoRI sites within the lacO repeats and an additional EcoRI site just upstream of the ClaI site. Therefore, a 6.4-kb fragment containing EBNA-1–oriP–bsr (the boundary of the fragment is shown by arrowheads in Fig. 1), a 2.7-kb fragment containing the ampicillin resistance gene and p15Aori, and multiple very small fragments derived from the sheared lacO repeats were to be generated by EcoRI digestion. The fragment containing the bsr gene, which has multiple DpnI sites, should become resistant to DpnI when test plasmids replicate in transfected cells. A 10-μl sample was mixed with an equal volume of an enzyme mixture containing either EcoRI (20 U/sample) alone or a mixture of EcoRI and DpnI (6 U/sample) and then incubated at 37°C for 6 h. The digested samples were electrophoresed on a 0.8% agarose gel and analyzed by Southern blotting using the 480-bp HindIII fragment of the pYN3215-bsr plasmid as a probe.
Colony formation assay.
Colony formation was assayed after blasticidin selection (5 μg/ml for the first 5 days and then 2 μg/ml for the following 9 days) of serially diluted (1:5, 1:50, and 1:500), transfected cells. Drug-resistant cells were fixed with 3.7% formaldehyde and stained with crystal violet, and the colonies were counted to calculate the transformation efficiency.
RESULTS
Visualization of transfected EBV plasmids with the lacO/lacR-GFP system.
The objective of this study was to investigate the relationships among EBNA-1, oriP, and tethering of EBV-based plasmids to host chromosomes. FISH was not optimal, as the background signals limited the ability to detect small circular DNA molecules. In order to provide a direct, rapid, and sensitive means of identifying small transfected DNAs, we added 256 direct repeats of the lac operator to the EBV plasmid (EBV-lacO) (Fig. 1). We reasoned that introducing the EBV-lacO plasmid into cells expressing a lacR-GFP fusion protein should enable detection of the plasmids as punctate green fluorescent signals (20, 39). A HeLa cell line that stably expresses a lacR-GFP fusion protein with a C-terminal nuclear localization signal was generated (HeLa/lacR-GFP cells). As expected, the expressed lacR-GFP fusion protein localized to nuclei (data not shown). In mitotic cells, the breakdown of the nuclear membranes resulted in cytoplasmic staining by the lacR-GFP fusion (Fig. 2a). In neither case was a punctate signal observed. By contrast, when HeLa/lacR-GFP cells were transiently transfected with the EBV-lacO plasmid, small but distinct fluorescent dots were detected in interphase nuclei at 5 days posttransfection (Fig. 2b). Since the fluorescent dots were only detected in HeLa/lacR-GFP cells after EBV-lacO transfection, we concluded that such dots derive from the interaction of lacR-GFP with EBV-lacO and that they provide a sensitive indication of the location of the EBV-lacO plasmids. In the remainder of this paper, lacR-GFP-labeled plasmids and lacR-GFP fluorescent dots are considered to be synonymous.
FIG. 2.
The EBV-lacO plasmids associate with mitotic chromosomes in HeLa cells. HeLa cells stably expressing lacR-GFP protein (HeLa/lacR-GFP cells) were transfected with EBV-lacO plasmids. Representative images of an untransfected cell (a) and transfected cells in various mitotic phases at 5 days posttransfection (b through f) are shown. Chromosomes were counterstained with Hoechst 33342 (blue). Note that no fluorescent dot is observed in an untransfected cell (a) and that fluorescent dots (green) associate with mitotic chromosomes in transfected cells (b through f). Scale bar, 10 μm.
We next examined the distribution of lacR-GFP-labeled plasmids, particularly focusing on whether they associated with mitotic chromosomes. We observed many mitotic cells in which lacR-GFP-labeled plasmids associated with chromosomes in various phases of mitosis (Fig. 2c through f). The lacR-GFP-labeled plasmids were associated with chromosomes in more than 50% of mitotic cells with at least one clear fluorescent dot. The fluorescent dots appeared to be distributed randomly on mitotic chromosomes, suggesting that the EBV-lacO plasmids did not bind to specific chromosomal regions, such as centromeres or telomeres. The remaining 50% of the dot-positive mitotic cells contained only a few fluorescent dots that were free in the cytoplasm, possibly representing nonfunctional plasmid DNAs undergoing degradation after transient transfection (data not shown). These data validate the previous FISH analyses indicating that the EBV genome and EBV-based plasmids associate with mitotic chromosomes (9, 42, 48).
EBNA-1 protein colocalizes with oriP-containing plasmids in interphase and mitotic cells.
The lacO/lacR-GFP strategy provides a sensitive and specific microscopic method to investigate directly whether EBNA-1 protein colocalizes with EBV-lacO plasmids. EBNA-1 protein was detected by indirect immunofluorescence under conditions that preserved lacR-GFP fluorescence. Cells with labeled EBV plasmids were always positive for EBNA-1 staining, which validated this labeling strategy. The amount of EBNA-1 protein, estimated by immunofluorescence, varied from cell to cell, probably due to the heterogeneous copy number of the EBV-lacO plasmids encoding EBNA-1. Cells expressing high levels of EBNA-1 protein typically exhibited diffuse EBNA-1 localization inside interphase nuclei (data not shown). However, in cells expressing less EBNA-1, the EBV-lacO plasmids and EBNA-1 protein were clearly observed to colocalize (Fig. 3a, b, and c). In mitotic cells in which lacR-GFP-labeled plasmids associated with mitotic chromosomes, EBNA-1 protein was most frequently observed to label mitotic chromosomes diffusely, confirming previous observations (12, 29, 35) (Fig. 3d, e, and f). Importantly, when less EBNA-1 was expressed, EBNA-1 protein colocalized with EBV-lacO plasmids on mitotic chromosomes (Fig. 3g, h, and i). These results are consistent with EBNA-1 protein constitutively binding to the oriP region of EBV plasmids throughout the cell cycle. Mitotic cells containing a few labeled EBV plasmids scattered in the cytoplasm were always negative for EBNA-1 staining (data not shown), suggesting that such plasmids were nonfunctional and might be undergoing cytoplasmic degradation. Interestingly, even in cells expressing such high levels of EBNA-1 that it was distributed ubiquitously inside the cells, lacR-GFP-labeled plasmids clearly localized to mitotic chromosomes (data not shown). This result indicates that EBV plasmids do not merely colocalize with EBNA-1 protein according to its distribution. Rather, it appears that mitotic chromosomes provide a preferred location for EBV plasmids in the presence of EBNA-1 protein.
FIG. 3.
EBNA-1 protein colocalizes with oriP-containing plasmids in interphase nuclei and on mitotic chromosomes. HeLa/lacR-GFP cells transfected with the EBV-lacO plasmid (at 5 days posttransfection) were processed for immunofluorescence analyses to detect EBNA-1 protein while keeping the fluorescence of lacR-GFP. The EBV-lacO plasmids are shown as green dots, and EBNA-1 protein is shown as red (middle panels). Merged images are shown in the rightmost panels. Chromosomes were counterstained with DAPI (blue). Note the different expression levels of EBNA-1 protein in panels e and h. Scale bar, 10 μm.
Chromosome tethering of EBV plasmids requires both EBNA-1 and oriP.
A previous FISH analysis demonstrated that an oriP-containing yeast artificial chromosome (YAC) associated with mitotic chromosomes in stably transfected EBNA-1-positive human cells, but a YAC lacking oriP did not exhibit chromosomal association in stably transfected EBNA-1-negative mouse cells (42). These data leave open the question of whether the failure to associate with chromosomes reflected a difference in the species of the host cell or the absence of EBNA-1. We used a direct microscopic assay to investigate the effects of deleting the EBNA-1-encoding gene and/or oriP from the EBV-lacO plasmid on chromosome tethering (Fig. 4A). Western blots showed that the EBV-lacO plasmid produced a high level of EBNA-1, while ΔΕBNA1 and ΔoriP/EBNA1, which both lack the EBNA-1 gene, expressed no detectable EBNA-1 protein (Fig. 4B). The ΔoriP plasmid expressed a low level of EBNA-1, even though it contains an intact EBNA-1 gene. This is likely due to the lack of transcriptional enhancement achieved by EBNA-1 binding to oriP, which increases EBNA-1 protein expression by a positive feedback mechanism (37).
FIG. 4.
Chromosome tethering of EBV plasmids depends on EBNA-1 gene and oriP sequence. (A) Schematic representations of various mutant forms of the EBV-lacO plasmid lacking the EBNA-1 gene and/or the oriP sequence. The FR, the DS, the blasticidin resistance gene (bsr), and the lacO repeats are shown. (B) Western blot showing the expression of EBNA-1 protein (arrow) from each mutated plasmid at 48 h posttransfection. (C, parts a, b, and c) Representative images showing the mitotic distribution of various plasmids at 5 days posttransfection. Chromosomes were counterstained with Hoechst 33342 (blue). (d, e, and f) HeLa/lacR-GFP cells transfected with the ΔDS plasmid were processed for immunofluorescence analyses to detect EBNA-1 localization (red in part e). Merged images are shown in part f. Chromosomes were counterstained with DAPI (blue). The strong green signals in parts d and f are derived from aggregated plasmid DNAs introduced into the cells. Scale bar, 10 μm.
The subcellular localization of each deletion mutant plasmid in mitotic cells was examined after transient transfection into HeLa/lacR-GFP cells. When ΔEBNA1 was transfected, few, if any, cells were detected in which the labeled lacO plasmid associated with mitotic chromosomes, although in some cells the plasmids were found near mitotic chromosomes. However, in the majority of cases, the ΔΕBNA1 plasmids were detected free in the cytoplasm (Fig. 4C, part a). Similarly, the ΔoriP plasmids were distributed randomly inside cells and did not manifest a specific chromosomal association (Fig. 4C, part b). Since the lack of a chromosomal association of ΔoriP plasmids could have been due to the low level of EBNA-1 expression (Fig. 4B), we transfected the ΔoriP/EBNA1 plasmid into EBNA-1-expressing HeLa/lacR-GFP cells. Even in these HeLa cells, expressing substantial EBNA-1, the ΔoriP/EBNA1 plasmids did not associate with mitotic chromosomes (data not shown).
Taken together, the results provide a direct demonstration that the chromosomal association of EBV plasmids requires both EBNA-1 and oriP and that deletion of either of these elements disrupts tethering of EBV plasmids to chromosomes.
The DS element is dispensable for chromosome tethering.
It is well known that the FR element is sufficient for nuclear retention of oriP plasmids in EBNA-1-positive cells (23, 31). Since chromosome tethering may contribute to nuclear retention, we next determined whether plasmids lacking the DS region but containing the intact EBNA-1-encoding gene and FR element (ΔDS, Fig. 4A) exhibit normal chromosome tethering. Expression of the EBNA-1 protein was not affected by deletion of the DS element (Fig. 4B). Like the wild-type plasmid, the ΔDS plasmid associated with mitotic chromosomes in a substantial fraction of cells (Fig. 4C, part c). Simultaneous immunofluorescence analyses revealed that ΔDS plasmids and EBNA-1 protein colocalize on mitotic chromosomes (Fig. 4C, parts d, e, and f). These results show that the FR element, together with EBNA-1, is sufficient for the chromosomal association of EBV plasmids.
EBNA1(Δ16–372) binds oriP plasmids but does not enable mitotic chromosome tethering.
Previous analyses indicated that EBNA-1 has a modular structure in which three distinct N-terminal domains mediate chromosomal association (29) while a C-terminal domain contributes to oriP binding (2). We used a microscopic approach to dissect the modular domain structure by deleting portions of the EBNA-1 gene from the EBV-lacO plasmid while leaving the other regions intact (Fig. 5A). One of the mutated plasmids, EBNA-1(Δ90–327)-lacO, encodes an EBNA-1 protein with a deletion of the entire Gly-Gly-Ala repeat region. According to previous studies, the EBNA-1(Δ90–327) protein should behave like the wild-type EBNA-1 protein (52). Another mutated plasmid, EBNA-1(Δ16–372)-lacO, encodes an EBNA-1 protein with an N-terminal deletion that removes the three identified chromosome binding domains (29). Both the EBNA-1(Δ16–372) and EBNA-1(Δ90–327) plasmids have intact DNA binding-dimerization domains and intact nuclear localization signals, and both can be recognized using an antiserum specific for the C-terminal region. Western blotting revealed that proteins with the expected sizes were expressed in cells transfected with these plasmids (Fig. 5B).
FIG. 5.
Deletion analyses of EBNA-1 protein. (A) Schematic representations of mutant EBNA-1 proteins encoded by the mutated EBV-lacO plasmids. The previously identified chromosome binding domains of the EBNA-1 protein are indicated at the bottom (29). The Gly-Gly-Ala repeats, Gly-Arg-rich regions, nuclear localization signal (NLS), and dimerization-DNA binding domain are indicated. Regions rich in basic residues (++) are also indicated. (B) Western blot showing the expression of mutated EBNA-1 protein at 48 h posttransfection. (C) EBNA-1(Δ16–372) can bind oriP plasmids but cannot recruit them onto mitotic chromosomes. HeLa/lacR-GFP cells transfected with either the EBNA-1(Δ90–327)-lacO plasmid (a, b, and c) or the EBNA-1(Δ16–372)-lacO plasmid (d, h, and i) were processed for immunofluorescence analyses to detect EBNA-1 localization (b and e) while keeping the fluorescence of lacR-GFP. Merged images are shown in parts c and f. Scale bar, 10 μm. a.a., amino acids.
The subcellular localization of each of the mutated plasmids was analyzed in transfected HeLa/lacR-GFP cells, and simultaneous immunostaining was used to examine the localization of each of the mutated EBNA-1 proteins. The EBNA-1(Δ90–327)-lacO plasmid exhibited a pattern of chromosome association similar to that of the wild-type EBV-lacO plasmid (Fig. 5C, part a). The EBNA-1(Δ90–327) protein clearly associated with mitotic chromosomes and colocalized with the lacR-GFP-labeled plasmids (Fig. 5C, parts b and c). In contrast, most EBNA-1(Δ16–372)-lacO plasmids were present in the cytoplasm (Fig. 5C, part d). Importantly, the EBNA-1(Δ16–372) protein colocalized with the scattered labeled plasmids in the cytoplasm (Fig. 5C, parts e and f), demonstrating that the EBNA-1(Δ16–372) protein binds to the oriP sequence. The colocalization of EBNA-1(Δ16–372) with the lacR-GFP-labeled plasmids in interphase nuclei (data not shown) supports the idea that the EBNA-1(Δ16–372) protein constitutively binds to oriP plasmids yet cannot support mitotic chromosome tethering. These results clearly demonstrate that the chromosome-tethering and oriP binding domains of EBNA-1 protein are separable.
EBNA-1(Δ90–327), but not EBNA-1(Δ16–372), is chromatin associated.
The results described above revealed a striking difference between EBNA-1(Δ90–327) and EBNA-1(Δ16–372) in the ability to recruit oriP plasmids onto mitotic chromosomes. In order to further investigate the difference between these EBNA-1 variants, we expressed them in HeLa cells in the absence of oriP plasmids. Retroviral vectors were successfully used to obtain bulk-infected cells, ∼70% of which were positive for EBNA-1. Immunostaining revealed that both mutant proteins, which contain nuclear localization signals, localized to interphase nuclei (Fig. 6A, parts a and c). However, they exhibited distinctly different patterns in mitotic cells. While EBNA-1(Δ90–327) associated with mitotic chromosomes, EBNA-1(Δ16–372) appeared to be excluded from chromosomal domains (Fig. 6A, parts b and d).
FIG. 6.
EBNA-1 is associated with cellular chromatin. (A) Subcellular localization of EBNA-1 deletion mutant proteins. HeLa cells expressing either EBNA-1(Δ90–327) (a and b) or EBNA-1(Δ16–372) (c and d) were processed for immunofluorescence analyses using anti-EBNA-1 serum. Representative images of interphase cells (a and c) and mitotic cells (b and d) are shown. Scale bar, 10 μm. (B) EBNA-1(Δ90–327), but not EBNA-1(Δ16–372), is chromatin associated. HeLa cells expressing either EBNA-1(Δ90–327) or EBNA-1(Δ16–372) were subjected to a chromatin binding assay (30). Whole-cell extracts (WCE), the soluble fraction obtained by detergent extraction (S2), the solubilized nuclear protein fraction (S3), and the chromatin-nuclear matrix-bound protein fraction (P3) are indicated. Half aliquots of the isolated nuclei were treated with micrococcal nuclease (MN′ase) to remove chromatin-bound proteins (lanes 5 and 6). The fractionated samples were analyzed by Western blotting using anti- EBNA-1 antibody.
The above observation prompted us to test the idea that the subnuclear localizations of EBNA-1(Δ90–327) and EBNA-1(Δ16–372) in interphase nuclei are different although they appeared to be similar by microscopic analyses. Specifically, we examined whether these mutant proteins associate with interphase chromatin. Interphase nuclei were isolated from asynchronously growing HeLa cells expressing each of the EBNA-1 mutant proteins and subjected to a chromatin binding assay (30). We found that EBNA-1(Δ16–372) was recovered exclusively in the soluble fraction (S2; Fig. 6B, lane 2). By contrast, EBNA-1(Δ90–327) was preferentially recovered in the chromatin-nuclear matrix fraction (P3; lane 4). Importantly, the EBNA-1(Δ90–327) protein present in the chromatin-nuclear matrix fraction (lane 4) was solubilized almost completely by treatment of nuclei with micrococcal nuclease (lanes 5 and 6), indicating that EBNA-1(Δ90–327) associates with chromatin and not with a nuclear matrix structure. We concluded that EBNA-1(Δ90–327), but not EBNA-1(Δ16–372), is chromatin associated and that the chromosome binding domains of EBNA-1 (29) are required for its association with interphase chromatin.
Chromosome tethering correlates with replication competence and efficient transformation.
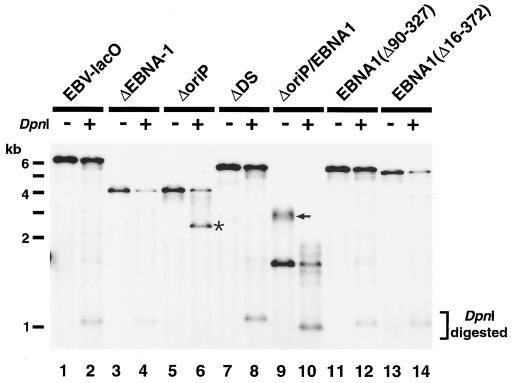
The data show that only EBNA-1(Δ90–327), which is chromatin associated, can mediate the mitotic chromosome tethering of oriP-containing plasmids, suggesting that the ability of EBNA-1 to associate with interphase chromatin could provide the basis for the mitotic chromosome tethering of oriP plasmids. Since the regions of EBNA-1 protein required for chromatin-chromosome association also appear to be needed for the replication of oriP-containing plasmids (28), it is possible that replication competence and chromosome tethering are directly linked. We evaluated this possibility by examining the replication competence of various EBV-lacO plasmids. Although the transient-replication abilities of various mutant forms of EBV plasmids have been extensively analyzed (1, 6, 14, 22, 28, 38, 40, 49, 50), it was essential to examine the replication competence of EBV-lacO variants due to the potential effects of the size increase created by the lacO repeats (10.1 kb). One representative result is shown in Fig. 7. As expected, the EBV-lacO plasmid replicated efficiently, as indicated by the majority of transiently transfected plasmids being DpnI resistant. In contrast, when ΔEBNA-1, ΔoriP, or ΔoriP/EBNA1 was transfected, little DpnI-resistant material was detected. Thus, these plasmids either do not replicate or do so inefficiently. Importantly, the ΔDS plasmid, which associated with mitotic chromosomes, showed significant replication activity, indicating that the DS region is dispensable for the replication of EBV-lacO plasmids. We also examined the effect of deleting the chromosome binding domains of EBNA-1 on transient DNA replication. EBNA-1(Δ90–327), which enables chromosome tethering of the oriP-containing plasmids, supported efficient replication. By contrast, EBNA-1(Δ16–372), which binds to oriP but not to mitotic chromosomes or interphase chromatin, did not support the replication of the oriP-containing plasmids. These data show that the replication competence of the EBV-lacO plasmids correlates directly with their ability to be tethered to chromosomes (Table 1).
FIG. 7.
Transient-replication abilities of various mutant forms of the EBV-lacO plasmid. Various mutant plasmids (indicated at the top) were transiently transfected into HeLa cells. Plasmids were harvested at 4 days posttransfection, and aliquots were digested by either EcoRI or EcoRI/DpnI (shown as minus or plus DpnI) and analyzed by Southern blotting. Replication activities can be estimated by comparing the band intensities of the EcoRI fragment containing the bsr gene (see the legend to Fig. 1) in the presence or absence of DpnI digestion. Note the significant amount of DpnI-resistant plasmids in the lanes containing EBV-lacO (lane 2), ΔDS (lane 8), and EBNA-1(Δ90–327) (lane 12). The largest band generated after DpnI digestion of the ΔoriP plasmid is indicated by an asterisk. Incompletely digested ΔoriP/EBNA1 plasmid remaining after EcoRI digestion is indicated by an arrow.
TABLE 1.
Frequencies of establishment of blasticidin-resistant colonies of HeLa cells after transfection of various mutant forms of the EBV-lacO plasmid
| Plasmid | Transformation efficiencya
|
Chromosome tetheringb | Transient replicationc | ||
|---|---|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | |||
| EBV-lacO | 4.5 × 10−2 | 2.1 × 10−2 | 8.0 × 10−3 | + | + |
| ΔEBNA-1 | 1.1 × 10−4 | 5.0 × 10−4 | 1.9 × 10−4 | − | − |
| ΔoriP | 5.1 × 10−4 | 6.2 × 10−4 | 1.3 × 10−4 | − | − |
| ΔDS | 1.2 × 10−2 | 3.2 × 10−3 | 1.6 × 10−3 | + | + |
| ΔoriP/EBNA1 | 2.0 × 10−5 | 6.2 × 10−5 | 6.2 × 10−5 | − | − |
| EBNA1(Δ90–327) | 1.1 × 10−2 | 3.1 × 10−3 | 3.6 × 10−3 | + | + |
| EBNA1(Δ16–372) | 5.6 × 10−4 | 6.2 × 10−4 | 2.5 × 10−5 | − | − |
Since all of the replicating plasmids examined associated with mitotic chromosomes, it is reasonable to predict that they should be maintained more stably than plasmids that neither replicate nor associate with mitotic chromosomes. Consistent with this idea, the frequency of cells with lacR-GFP-labeled plasmids was higher in the case of replicating (chromosome-associated) plasmids compared to nonreplicating plasmids that were not attached to mitotic chromosomes (data not shown). In order to estimate the efficiency of plasmid maintenance, we determined the transformation efficiency of each mutant plasmid by counting the number of blasticidin-resistant colonies generated after transfection and drug selection. The results reveal a strong correlation among replication competence, chromosome tethering, and transformation efficiency (Table 1). For example, the ΔEBNA-1 and ΔoriP plasmids showed severely impaired transformation efficiency compared to the wild-type EBV-lacO plasmid. However, the ΔDS plasmid, which replicates autonomously and associates with mitotic chromosomes, showed relatively high transformation efficiency. Similarly, the EBNA-1(Δ90–327)-lacO plasmid, which replicated and associated with mitotic chromosomes, showed a high level of transformation efficiency, while the EBNA-1(Δ16–372)-lacO plasmid had a low level of transformation efficiency.
Taken together, these results are consistent with the idea that replication capability is tightly linked with chromosome-tethering ability and results in efficient mitotic segregation and high transformation efficiency.
DISCUSSION
Stable episomal maintenance of EBV plasmids requires both efficient replication during S phase and faithful partitioning of the replicated progeny during mitosis. The mechanisms leading to each process have been studied independently in the past (for reviews, see references 26 and 27). However, it has not been possible to derive an integrated picture of the contributions of the cis elements and trans-acting factors involved in episome replication and segregation due to the lack of a single experimental paradigm capable of a direct and integrated analysis of each variable. Toward this end, we used the lacO/lacR-GFP system (20, 39) to develop a novel microscopic strategy to localize transiently transfected EBV plasmids. We integrated this strategy with immunofluorescence of the viral EBNA-1 protein, which was previously proposed to contribute to viral replication and segregation, to elucidate the protein-DNA interactions involved in replication and segregation. Theoretically, a similar strategy could be applied to track any DNA containing lacO repeat sequences.
The results show that EBNA-1 protein colocalizes with oriP plasmids in interphase nuclei, as well as on mitotic chromosomes, suggesting that the association is maintained throughout the cell cycle. This observation corresponds well to biochemical data showing that EBNA-1 binds to the oriP sequence throughout the cell cycle (16). Many studies show that EBNA-1 and oriP are both indispensable elements for the replication of EBV plasmids (28, 38, 40, 49). One implication of our and other studies is that EBNA-1 binding to the oriP sequence may have different roles in different cell cycle phases. While EBNA-1–oriP interaction in S phase seems to be required for replication, it may also tether the episomes to sister chromatids to increase segregation efficiency in mitosis.
We have begun to explore whether chromosome tethering mediated by EBNA-1 is required for episome replication. Host proteins alone cannot support chromosomal association of EBV plasmids, since a plasmid lacking the EBNA-1-encoding gene (ΔEBNA-1) neither replicated nor associated with chromosomes. Mutant plasmids were generated with alterations in different regions of the EBNA-1-encoding gene and in oriP to evaluate their effects on chromosome tethering, transient replication, and long-term maintenance, which was assayed by the ability of the test plasmids to engender stable drug resistance following transfection. The results revealed that chromosome tethering was clearly dependent on subdomains of the EBNA-1 protein and on specific regions of the oriP sequence. In good agreement with a recent study that identified the chromosome binding domains of EBNA-1 (29), we found that the EBNA-1(Δ16–372)-lacO plasmid, which encodes an EBNA-1 protein lacking all three chromosome binding domains, did not associate with mitotic chromosomes. In contrast, EBNA-1(Δ90–327), which only lacks the Gly-Ala repeat and is functionally wild type, did associate with chromosomes. Importantly, the chromosome-tethering abilities of these mutant plasmids correlated well to their transient-replication abilities. A plasmid lacking the oriP sequence (ΔoriP) neither replicated nor associated with mitotic chromosomes, even in the presence of EBNA-1. These data are consistent with the idea that there is a close link between replication and chromosome tethering, both of which are dependent on interactions between EBNA-1 and oriP.
Biochemical fractionation revealed that there is a relationship between EBNA-1-mediated chromosome tethering and binding to interphase chromatin. For example, an EBNA-1 variant lacking the Gly-Ala repeat [EBNA1(Δ90–327)] binds to interphase chromatin and tethers, while an EBNA-1 variant lacking three previously defined chromosome binding domains [EBNA1(Δ16–372)] does not. These data are consistent with the conclusion that EBNA-1 protein is in the chromatin fraction and not in the nuclear matrix fraction (35). Our data indicate that wild-type EBNA-1 associates with cellular chromatin throughout the cell cycle and is not selectively recruited onto chromosomes during mitosis.
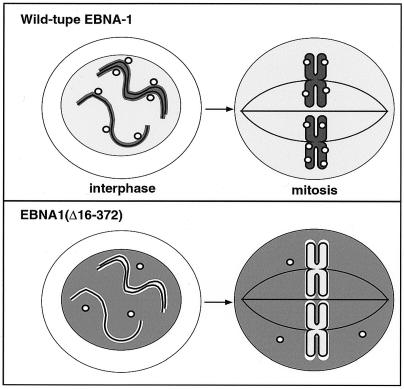
The association of the EBNA-1 protein with interphase chromatin may be related to the observed close link between EBV replication and chromosome tethering. Our abilities to study episome tethering directly using microscopy, to use biochemical fractionation to study EBNA-1–chromatin interactions, and to employ transient-replication and transformation analyses to evaluate replication and segregation have provided direct experimental evidence highlighting strong links between replication and chromosome tethering of oriP plasmids, both of which require the EBNA-1 protein. Our data suggest a model in which EBNA-1 associates with interphase chromatin and brings viral replicons to chromatin to allow episomal replication by the host machinery once per cell cycle (Fig. 8, top). This view is compatible with a previous proposal that EBNA-1 recruits oriP plasmids into specific subnuclear domains (28) and that EBNA-1 mediates the tethering of oriP plasmids to chromatin throughout the cell cycle (27). An important implication of this model envisioning “chromatin-associated replication” of oriP plasmids is that the newly replicated oriP plasmids can be evenly loaded onto newly replicated sister chromatids at the same time that sister chromatid cohesion is established during replication (44, 46). This is likely to increase the probability of efficient segregation of oriP plasmids during mitosis. Conversely, the model proposes that EBV episomes that cannot engage chromatin because they either lack EBNA-1 or encode EBNA-1 mutant proteins lacking chromatin binding domains or because they do not contain the relevant regions of oriP should neither replicate nor bind to mitotic chromosomes nor be able to segregate efficiently. These predictions are consistent with our observation that EBNA-1(Δ16–372), which cannot recruit oriP plasmids onto interphase chromatin, cannot support their replication and fails to mediate mitotic chromosomal association (Fig. 8, bottom).
FIG. 8.
Model for the link between replication and chromosome tethering of EBV plasmids. (Top) Wild-type EBNA-1 protein (in gray) mediates the association of oriP plasmids (circles) with interphase chromatin (solid lines). The chromatin-associated oriP plasmids efficiently replicate together with cellular chromatin using host replication machinery, and the replicated molecules are loaded onto sister chromatids (shown as duplicated solid lines). Such molecules give the appearance that they associate with mitotic chromosomes in subsequent mitosis. (Bottom) The truncated mutant form EBNA-1(Δ16–372) (in gray) does not associate with cellular chromatin, resulting in the inability to support replication, as well as mitotic chromosome tethering of oriP plasmids (circles).
Previous studies indicated that the combination of EBNA-1 and FR was not sufficient for the replication of EBV plasmids in the absence of DS (31, 38). By contrast, we found that the ΔDS plasmid used in this study, which contains only the FR and not DS, could both replicate and tether to mitotic chromosomes. This result demonstrates the ability of EBNA-1–FR to support replication and chromosome tethering and the independence of these processes from the DS element in the EBV-lacO system we employed. There are several possible explanations for how EBNA-1–FR enables replication in the absence of DS. First, it is conceivable that EBNA-1–FR, which has nuclear retention ability, indirectly stimulates replication by minimizing plasmid loss from the nucleus before and following replication. A second and more likely explanation is that EBNA-1–FR-mediated chromatin association, combined with the additional sequences in the ΔDS plasmid, complemented the DS deficiency. As we showed that EBNA-1–FR enables EBV plasmid association with cellular chromatin throughout the cell cycle, it is possible that the associations in S phase stimulate replication of the viral replicon by recruiting the host replication machinery to the sequences that substitute for DS. Indeed, previous studies showed that both human- and bacterium-derived sequences can complement DS deficiency and enable replication of FR-containing plasmids and that the probability of complementation increased with the length of the sequences cloned into the plasmids (15, 23). It is reasonable to propose, therefore, that the lacO repeat sequence (10.1 kb) could complement DS, as there appears to be little sequence specificity required for such complementation (15). Further experiments are required to test the idea that EBNA-1–FR-mediated chromatin association itself can convert nonreplicating plasmids into replication-competent molecules.
The link between replication and chromosome tethering raises the question of whether the DS element itself, which can mediate replication in a mutant lacking the FR element (40, 50), can tether. It is well established that a plasmid having only the DS element and lacking the FR element can replicate efficiently in EBNA-1-expressing cells but cannot be stably retained afterward (14, 40, 50). Based on our results, we provide the following explanation for this observation. We suggest that the lower binding affinity of EBNA-1 for DS may be sufficient to enable transient replication, but it may not be strong enough to maintain the association between replicated plasmids and the cellular chromatin through mitosis. Further improvement of our microscopic assay may enable the long-term analyses of plasmid segregation needed to evaluate the validity of such a proposal.
The molecular basis of EBNA-1 binding to cellular chromatin remains a mystery. We constructed expression vectors of EBNA-1 mutant forms tagged with GFP similar to those recently described (29) and found that the regions containing the three chromosome binding domains work cooperatively to achieve maximum mitotic chromosome association (unpublished results). A previous analysis indicated the insensitivity of this EBNA-1–chromosome interaction to RNase, suggesting that protein-protein-DNA or protein-DNA interactions are most likely (13). Since the chromatin binding domains of EBNA-1 are positively charged, electrostatic interactions may contribute to EBNA-1 binding to chromatin. Alternatively, EBNA-1 may bind specifically to one or more cellular proteins, which are presumably chromatin associated (41). Further deletion analyses or substitution mutation analyses of EBNA-1 should clarify the molecular mechanisms by which episomes tether to cellular chromatin.
The tethering of viral genomes to host chromosomes appears to be a common aspect of the life cycle of DNA viruses with a latent infection phase. These include other gammaherpesviruses, such as Kaposi's sarcoma-associated herpesvirus (3, 8) and herpesvirus saimiri (24), and bovine papillomavirus (4, 17, 25, 43). Like EBV, Kaposi's sarcoma-associated herpesvirus and bovine papillomavirus also contain cis-acting sequences for episomal maintenance, and they encode trans-acting viral proteins, latency-associated nuclear antigen, and E2, respectively, which mediate chromosome tethering. Therefore, chromosome tethering may be a common mechanism for enhancing the transmission of extrachromosomally replicating viruses into daughter nuclei. Interestingly, the 2μm plasmid of Saccharomyces cerevisiae (47) and bacterial plasmids (19) also contain cis-acting sequences and encode trans-acting factors, which colocalize with the plasmid DNAs. Therefore, this mode of plasmid segregation may be conserved during evolution. Our recent studies also demonstrate that cellular acentric, autonomously replicating chromatin bodies, called double minute chromosomes, tether to mitotic chromosomes and that they are preferential targets for integration of EBV plasmids (20). Examination of the cis- and trans-acting requirements for the replication and segregation of extrachromosomal replicons in bacterial, yeast, and mammalian cells should shed light on the mechanism by which they achieve efficient mitotic segregation in spite of their lack of functional centromeres. Equally importantly, such studies should provide clues for designing drugs to disrupt tethering and decrease the efficiency of transmission of such acentric replicons to daughter cells, which could generate novel antiviral and anticancer agents.
ACKNOWLEDGMENTS
We thank A. Belmont for generously providing the lac operator/lac repressor-GFP system. We also thank F. Hanaoka for pYN3215-bsr, J. L. Kolman for a pEPB construct, T. Koga for pMBL19, N. Somia for pCLMFG-MCS and 293gp/bsr cells, and J. Middeldorp and G. Miller for anti EBNA-1 serum. We thank H. Miyoshi for helpful suggestions about retroviral vectors and Wahl laboratory members for critically reading the manuscript.
This work was supported by a grant from the California Cancer Research Program (99-00573V-10074) and the Pioneer Fund Fellowship (T.K.).
REFERENCES
- 1.Aiyar A, Tyree C, Sugden B. The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J. 1998;17:6394–6403. doi: 10.1093/emboj/17.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambinder R F, Mullen M, Chang Y N, Hayward G S, Hayward S D. Functional domains of Epstein-Barr virus nuclear antigen EBNA-1. J Virol. 1991;65:1466–1478. doi: 10.1128/jvi.65.3.1466-1478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballestas M E, Chatis P A, Kaye K M. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 4.Bastien N, McBride A A. Interaction of the papillomavirus E2 protein with mitotic chromosomes. Virology. 2000;270:124–134. doi: 10.1006/viro.2000.0265. [DOI] [PubMed] [Google Scholar]
- 5.Belmont A S, Li G, Sudlow G, Robinett C. Visualization of large-scale chromatin structure and dynamics using the lac operator/lac repressor reporter system. Methods Cell Biol. 1999;58:203–222. doi: 10.1016/s0091-679x(08)61957-3. [DOI] [PubMed] [Google Scholar]
- 6.Ceccarelli D F, Frappier L. Functional analyses of the EBNA-1 origin DNA binding protein of Epstein-Barr virus. J Virol. 2000;74:4939–4948. doi: 10.1128/jvi.74.11.4939-4948.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotter M A, 2nd, Robertson E S. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology. 1999;264:254–264. doi: 10.1006/viro.1999.9999. [DOI] [PubMed] [Google Scholar]
- 9.Delecluse H-J, Bartnizke S, Hammerschmidt W, Bullerdiek J, Bornkamm G W. Episomal and integrated copies of Epstein-Barr virus coexist in Burkitt lymphoma cell lines. J Virol. 1993;67:1292–1299. doi: 10.1128/jvi.67.3.1292-1299.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dranoff G, Jaffee E, Lazenby A, Golumbek P, Levitsky H, Brose K, Jackson V, Hamada H, Pardoll D, Mulligan R C. Vaccination with irradiated tumor cells engineered to secrete murine granulocyte-macrophage colony-stimulating factor stimulates potent, specific, and long-lasting antitumor immunity. Proc Natl Acad Sci USA. 1993;90:3539–3543. doi: 10.1073/pnas.90.8.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frappier L, O'Donnell M. Overproduction, purification, and characterization of EBNA-1, the origin binding protein of Epstein-Barr virus. J Biol Chem. 1991;266:7819–7826. [PubMed] [Google Scholar]
- 12.Grogan E A, Summers W P, Dowling S, Shedd D, Gradoville L, Miller G. Two Epstein-Barr viral nuclear neoantigens distinguished by gene transfer, serology, and chromosome binding. Proc Natl Acad Sci USA. 1983;80:7650–7653. doi: 10.1073/pnas.80.24.7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris A, Young B D, Griffin B E. Random association of Epstein-Barr virus genomes with host cell metaphase chromosomes in Burkitt's lymphoma-derived cell lines. J Virol. 1985;56:328–332. doi: 10.1128/jvi.56.1.328-332.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison S, Fisenne K, Hearing J. Sequence requirements of the Epstein-Barr virus latent origin of DNA replication. J Virol. 1994;68:1913–1925. doi: 10.1128/jvi.68.3.1913-1925.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinzel S S, Krysan P J, Tran C T, Calos M P. Autonomous DNA replication in human cells is affected by the size and the source of the DNA. Mol Cell Biol. 1991;11:2263–2272. doi: 10.1128/mcb.11.4.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsieh D J, Camiolo S M, Yates J L. Constitutive binding of EBNA-1 protein to the Epstein-Barr virus replication origin, oriP, with distortion of DNA structure during latent infection. EMBO J. 1993;12:4933–4944. doi: 10.1002/j.1460-2075.1993.tb06187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ilves I, Kivi S, Ustav M. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J Virol. 1999;73:4404–4412. doi: 10.1128/jvi.73.5.4404-4412.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Izumi M, Miyazawa H, Kamakura T, Yamaguchi I, Endo T, Hanaoka F. Blasticidin S-resistance gene (bsr): a novel selectable marker for mammalian cells. Exp Cell Res. 1991;197:229–233. doi: 10.1016/0014-4827(91)90427-v. [DOI] [PubMed] [Google Scholar]
- 19.Jensen R B, Gerdes K. Mechanism of DNA segregation in prokaryotes: ParM partitioning protein of plasmid R1 colocalizes with its replicon during the cell cycle. EMBO J. 1999;18:4076–4084. doi: 10.1093/emboj/18.14.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanda T, Otter M, Wahl G M. Mitotic segregation of viral and cellular acentric extrachromosomal molecules by chromosome tethering. J Cell Sci. 2001;114:49–58. doi: 10.1242/jcs.114.1.49. [DOI] [PubMed] [Google Scholar]
- 21.Kanda T, Sullivan K F, Wahl G M. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 22.Kirchmaier A L, Sugden B. Dominant-negative inhibitors of EBNA-1 of Epstein-Barr virus. J Virol. 1997;71:1766–1775. doi: 10.1128/jvi.71.3.1766-1775.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krysan P J, Haase S B, Calos M P. Isolation of human sequences that replicate autonomously in human cells. Mol Cell Biol. 1989;9:1026–1033. doi: 10.1128/mcb.9.3.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kung S H, Medveczky P G. Identification of a herpesvirus saimiri cis-acting DNA fragment that permits stable replication of episomes in transformed T cells. J Virol. 1996;70:1738–1744. doi: 10.1128/jvi.70.3.1738-1744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehman C W, Botchan M R. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc Natl Acad Sci USA. 1998;95:4338–4343. doi: 10.1073/pnas.95.8.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leight E R, Sugden B. EBNA-1: a protein pivotal to latent infection by Epstein-Barr virus. Rev Med Virol. 2000;10:83–100. doi: 10.1002/(sici)1099-1654(200003/04)10:2<83::aid-rmv262>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 27.Mackey D, Sugden B. Applications of oriP plasmids and their mode of replication. Methods Enzymol. 1999;306:308–328. doi: 10.1016/s0076-6879(99)06020-6. [DOI] [PubMed] [Google Scholar]
- 28.Mackey D, Sugden B. The linking regions of EBNA-1 are essential for its support of replication and transcription. Mol Cell Biol. 1999;19:3349–3359. doi: 10.1128/mcb.19.5.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marechal V, Dehee A, Chikhi-Brachet R, Piolot T, Coppey-Moisan M, Nicolas J-C. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J Virol. 1999;73:4385–4392. doi: 10.1128/jvi.73.5.4385-4392.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Middleton T, Sugden B. Retention of plasmid DNA in mammalian cells is enhanced by binding of the Epstein-Barr virus replication protein EBNA-1. J Virol. 1994;68:4067–4071. doi: 10.1128/jvi.68.6.4067-4071.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyoshi H, Takahashi M, Gage F H, Verma I M. Stable and efficient gene transfer into the retina using an HIV-based lentiviral vector. Proc Natl Acad Sci USA. 1997;94:10319–10323. doi: 10.1073/pnas.94.19.10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakano Y, Yoshida Y, Yamashita Y, Koga T. Construction of a series of pACYC-derived plasmid vectors. Gene. 1995;162:157–158. doi: 10.1016/0378-1119(95)00320-6. [DOI] [PubMed] [Google Scholar]
- 34.Norio P, Schildkraut C L, Yates J L. Initiation of DNA replication within oriP is dispensable for stable replication of the latent Epstein-Barr virus chromosome after infection of established cell lines. J Virol. 2000;74:8563–8574. doi: 10.1128/jvi.74.18.8563-8574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petti L, Sample C, Kieff E. Subnuclear localization and phosphorylation of Epstein-Barr virus latent infection nuclear proteins. Virology. 1990;176:563–574. doi: 10.1016/0042-6822(90)90027-o. [DOI] [PubMed] [Google Scholar]
- 36.Rawlins D R, Milman G, Hayward S D, Hayward G S. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- 37.Reisman D, Sugden B. trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol Cell Biol. 1986;6:3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reisman D, Yates J, Sugden B. A. putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5:1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinett C C, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont A S. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shirakata M, Hirai K. Identification of minimal oriP of Epstein-Barr virus required for DNA replication. J Biochem (Tokyo) 1998;123:175–181. doi: 10.1093/oxfordjournals.jbchem.a021907. [DOI] [PubMed] [Google Scholar]
- 41.Shire K, Ceccarelli D F, Avolio-Hunter T M, Frappier L. EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J Virol. 1999;73:2587–2595. doi: 10.1128/jvi.73.4.2587-2595.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simpson K, McGuigan A, Huxley C. Stable episomal maintenance of yeast artificial chromosomes in human cells. Mol Cell Biol. 1996;16:5117–5126. doi: 10.1128/mcb.16.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skiadopoulos M H, McBride A A. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J Virol. 1998;72:2079–2088. doi: 10.1128/jvi.72.3.2079-2088.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skibbens R V, Corson L B, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stahl H, Droge P, Knippers R. DNA helicase activity of SV40 large tumor antigen. EMBO J. 1986;5:1939–1944. doi: 10.1002/j.1460-2075.1986.tb04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uhlmann F, Nasmyth K. Cohesion between sister chromatids must be established during DNA replication. Curr Biol. 1998;8:1095–1101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
- 47.Velmurugan S, Yang X M, Chan C S, Dobson M, Jayaram M. Partitioning of the 2-micron circle plasmid of Saccharomyces cerevisiae. Functional coordination with chromosome segregation and plasmid-encoded rep protein distribution. J Cell Biol. 2000;149:553–566. doi: 10.1083/jcb.149.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Westphal E M, Sierakowska H, Livanos E, Kole R, Vos J M. A system for shuttling 200-kb BAC/PAC clones into human cells: stable extrachromosomal persistence and long-term ectopic gene activation. Hum Gene Ther. 1998;9:1863–1873. doi: 10.1089/hum.1998.9.13-1863. [DOI] [PubMed] [Google Scholar]
- 49.Yates J L, Camiolo S M. Dissection of DNA replication and enhancer functions of Epstein-Barr virus nuclear antigen 1. In: Kelley T, Stillman B, editors. Eukaryotic DNA replication. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. pp. 197–205. [Google Scholar]
- 50.Yates J L, Camiolo S M, Bashaw J M. The minimal replicator of Epstein-Barr virus oriP. J Virol. 2000;74:4512–4522. doi: 10.1128/jvi.74.10.4512-4522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yates J L, Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol. 1991;65:483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yates J L, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]