Abstract
Thousands of microbial species inhabiting the animal gut, collectively known as the gut microbiota, play many specific roles related to host nutrient metabolism and absorption, immune regulation, and protection from pathogenic bacteria. Gut microbiota composition is affected by several internal and external factors, such as the host genotype, dietary intake, breeding environment, and antibiotic exposure. As deer species are important members for maintaining ecosystem balance, understanding the effects of multiple factors on the gut microbiota of deer species, particularly endangered ones, is crucial. In this review, we summarize and discuss the factors that significantly affect the gut microbiota of deer and present the impacts of these factors on microbial composition. In particular, we focused on the changes in gut microbiota due to dietary differences under different conditions, including seasonal changes, different geographical locations, and captivity, as well as weaning and pathogen disturbance. Understanding the correlations between gut microbiota composition and its driving factors is important for evaluating and improving the captive breeding environment for better conservation of endangered deer species, and reintroducing wild deer populations in the future.
Keywords: deer species, gut microbiota, environmental factors, conservation, microbial composition
1. Introduction
Several species affiliated with Cervidae and the primitive deer Moschus spp. (collectively referred to as deer species hereafter) are known to play a vital role in enriching the dense forest biodiversity and maintaining ecosystem balance (Li et al., 2016; Spake et al., 2020). However, recent climate changes and human activities have led to the endangerment or even extinction of many wild animals, including deer, from forest systems (Cai et al., 2020). At present, numerous deer species are listed as endangered, vulnerable, or highly endangered by the International Union for Conservation of Nature Red List of Threatened Species and as critically endangered by the Red List of China’s Vertebrates (Jiang et al., 2016); these include musk deer (Moschus [M.] spp.) (Harris, 2016), Père David’s deer (Elaphurus [E.] davidianus) (Zhang et al., 2017), sika deer (Cervus [C.] nippon) (Guan et al., 2017), and white-lipped deer (C. albirostris) (Harris, 2016). Both captive breeding and ex situ conservation have been effectively applied for maintaining and restoring these endangered deer species, with considerable success being achieved (Wang et al., 2016; Sun et al., 2020). However, increasing numbers of studies have reported subhealth conditions of many captive animals, including deer, and even mass die-offs in protected areas, mainly resulting from gastrointestinal infections (Li et al., 2022).
The gut microbiota, which consists of trillions of microorganisms (including archaea, bacteria, fungi, and viruses), plays crucial roles in the health, physiology, and development of the host and is thus recognized as an integral part of the animal holobiont (Hugon et al., 2017). The gut microbiota is not constant and differs among individuals; it is susceptible to various internal and external factors, such as the host genotype, dietary intake, lifestyle, breeding environment, and antibiotic exposure (Li Y. et al., 2017; Wang et al., 2022). Notably, certain members of the gut microbiota play more important roles than the remaining commensal ones, e.g., conferring resistance to pathogens and facilitating food digestion; moreover, several isolates with desired beneficial functions have been obtained through omics-guided microbiota analysis and targeted microbial isolation approaches (e.g., ref. Zipperer et al., 2016; Yuan et al., 2022). Beneficial microbe administration-based microbiota manipulation approaches mentioned below have consequently been developed and have shown promising benefits in animal breeding and raising practices (Anee et al., 2021). With such rapid developments in omics technologies and their contributions to microbiota decryption and application, several omics-based studies have been conducted for analyzing the gut microbiota of deer species, particularly six species belonging to the family Cervidae [sika deer (C. nippon) (Guan et al., 2017; Wang et al., 2022), Père David’s deer (E. davidianus) (Zhang et al., 2018; Sun et al., 2019), red deer (C. elaphus) (Menke et al., 2019; Wang et al., 2019), white-lipped deer (C. albirostris) (Li J. G. et al., 2017; Li et al., 2022; You et al., 2022), Siberian roe deer (Capreolus pygargus) (Liu J. et al., 2019), and white-tailed deer (Odocoileus virginianus)] (Delgado et al., 2017; Minich et al., 2021) and three species belonging to the genus Moschus [alpine musk deer (M. chrysogaster) (Sun et al., 2020), Siberian musk deer (M. moschiferus) (Su et al., 2022), and forest musk deer (M. berezovskii)] (Li Y. et al., 2017). The predominant bacterial phyla in these deer species were found to be Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria; these findings are consistent with previous findings regarding the gut microbiota of ruminants (Tanca et al., 2017). Furthermore, these studies also demonstrated that the gut microbiota composition of deer was dramatically changed under the effects of environmental factors. However, comprehensive understanding of the effects of environmental factors on the gut microbiota of deer species is still lacking. In this minireview, we summarize and discuss recent findings regarding the gut microbiota of deer species using omics approaches, mainly focusing on how the gut microbiota structure is affected by captivity-, season-, and geographical location-related dietary changes; weaning; and the presence of pathogens. Our findings can benefit the development of microbiota optimization-based approaches to improve the captive breeding and raising processes of endangered deer populations.
2. Effects of diets on the gut microbiota of deer species
Among the factors that are known to dramatically affect the gut microbiota, particular attention has been paid to diet and its role in shaping the composition and function of the gut microbiota (Bibbò et al., 2016). Because of changes in dietary nutrients (e.g., fiber, starch, proteins, and fats), the taxa that prefer the given nutrients usually exhibit higher growth and proliferation rates, resulting in rapid alteration of the gut microbiota composition. However, a large fraction of microbes can still be remarkably stable in healthy individuals for years (Fassarella et al., 2021). In recent years, several studies have been conducted to understand the effects of diet changes on the gut microbiota of deer species, mainly by comparing the gut microbiota compositions of individuals in different seasons and geographical locations and those of captive and wild individuals, and are discussed below in detail.
2.1. Diet changes due to seasonal changes
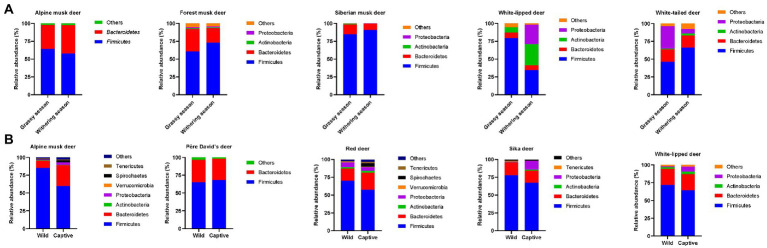
Food resources can change over temporal scales. For the deer population, sufficient and diverse fresh plant-derived food is available in the grassy season; however, food resources and choices are relatively limited in the withering season due to severe weather conditions (Hu et al., 2018; Li et al., 2022; Su et al., 2022; You et al., 2022). Dramatic seasonal variations in gut microbiota compositions are observed in deer species (Figure 1A). The relative abundance of Bacteroidetes is significantly higher in the grassy season than in the withering season. Members affiliated with Bacteroidetes are known to play a key role in degrading high-molecular-weight organic materials, including carbohydrates and proteins (Jami et al., 2013), thereby improving the nutritional composition of the host. This is consistent with the fact that the protein, starch, and lactate contents are higher in fresh leaves available in the grassy season than in limited foods available in the withering season (Hu et al., 2018). In contrast, the relative abundance of Firmicutes and the Firmicutes/Bacteroidetes (F/B) ratio are higher in the withering season than in the grassy season. Firmicutes can digest and absorb nutrients by degrading diverse substances, and the gut microbiota with a high F/B ratio can exhibit a higher fermentation efficiency and thus obtain more energy from food (Chevalier et al., 2015; Su et al., 2022). Moreover, a high F/B ratio can promote fat deposition in the host (Su et al., 2022), which is important for adapting to the cold withering season. However, these recent studies were conducted using 16S rDNA amplicon-based analyses, which hampered the identification of the key microbes and associated functions that are responsible for plant-derived substance degradation and energy conversion, and whole-genome-based metagenomics, metagenome-assembled-genome recovery and analysis, and culturomics can be performed to bridge these knowledge gaps (Stewart et al., 2019; Thomas et al., 2021; Whon et al., 2021).
Figure 1.
Microbial composition of the gut bacterial community of deer species at the phylum level in distinct seasons (A), and wild and captive environments (B), respectively.
2.2. Diet changes due to geographical locations
The climatic conditions, including temperature, precipitation, and vegetation, usually vary dramatically among different geographical locations. The gut microbiota compositions of forest musk deer species from Sichuan (subtropical monsoon climate) and Qinghai (highland continental climate, higher latitude, and lower temperature than Sichuan) differ significantly (Liu X. et al., 2019). Moreover, the Père David’s deer populations living in Shishou (subtropical monsoon climate) and Beijing (semihumid monsoon climate, higher latitude, and lower temperature than Shishou) harbor very different gut microbiota, with the gut microbiota of deer in Beijing exhibiting a higher F/B ratio than that of deer in Shishou (Zhang et al., 2018). These observed gut microbiota differences are probably associated with the available vegetation and temperature variations among geographic locations, the high abundance of Firmicutes and higher F/B ratio in the gut microbiota of deer species living in the geographic location with lower temperature may benefit the host given that the gut microbiota with a high F/B ratio usually exhibit a higher fermentation efficiency and thus the host can obtain more energy from food to maintain the body temperature (Chevalier et al., 2015; Su et al., 2022). Again, species- and strain-level resolution-based microbiota analysis can be performed to identify the key microbial members and their key functional traits involved in geographic location-associated diet and temperature adaptation of deer species.
2.3. Diet changes due to captivity
Captive breeding and raising have been implemented for several endangered deer species. The formulated forage provided to captive deer species is usually dramatically different from the food available in the wild (Li Y. et al., 2017). Consequently, the gut microbiota structure, particularly the F/B ratio, is very different between captive and wild deer populations (Guan et al., 2017; Sun et al., 2019, 2020, 2021; Jiang et al., 2021; Li et al., 2022). The relative abundance of Firmicutes is significantly higher in wild deer species than in captive ones, while the relative abundance of Bacteroidetes exhibits an opposite trend (Figure 1B). Therefore, the F/B ratio is higher in wild deer species than in captive ones. This difference in F/B ratio in the gut microbiota between wild and captive deer populations probably reflects the fact that a diverse diet spectrum, mostly consisting of various high-fiber leaves, is accessible to wild deer species, while a diet predominately consisting of fresh leaves supplemented with formulated foods containing high carbohydrate and protein concentrations is available to captive deer species. Notably, a higher relative abundance of Proteobacteria is found in captive sika deer and white-lipped deer than in wild ones (Li et al., 2022; Wang et al., 2022). Gut microbes belonging to Proteobacteria are known to degrade lignin and other various ingredients (Fang et al., 2012), further suggesting the effects of an artificially formulated diet on the gut microbiota structure of captive deer species. However, an increase in Proteobacteria in captive deer species could also indicate an increased risk of intestinal disorders because many Proteobacteria-affiliated gut bacteria are known pathogens or potential pathogens (Joat et al., 2021). Although the gut microbiota has certain plasticity that can help the host adapt to changes from natural to captive dietary supplies, some potential health risks, such as a decrease in nutrient absorption efficiency and an increase in potentially pathogenic bacteria, among captive populations cannot be ignored (Gogarten et al., 2012). Therefore, monitoring the digestive system of captive deer species and understanding whether the deer species have adapted to artificial diets and new environments are important for wildlife conservation.
3. Effects of weaning on the gut microbiota of deer species
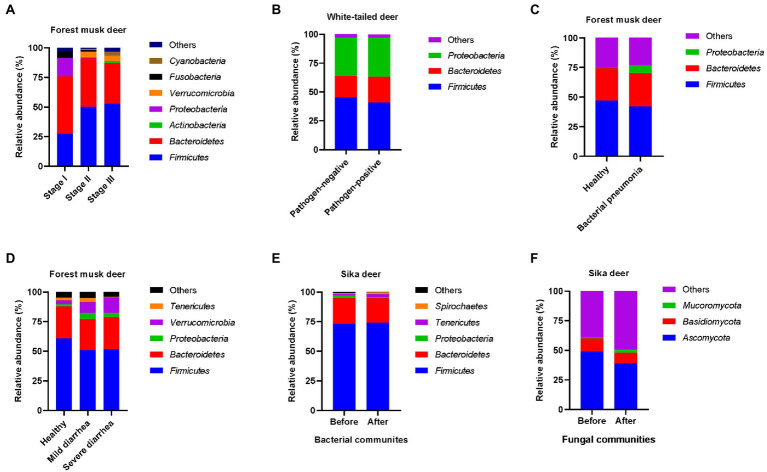
Weaning, a process of replacing milk feeding with an increasing range of ingested nutrients, is an important event in the early life of mammals (Li et al., 2020). It is usually accompanied by dramatic changes in the composition of the gut microbiota. Special interest has been paid to the gut microbiota dynamics of humans and several animals during the weaning period. Recent studies on forest musk deer revealed that Proteobacteria maintained a relatively high abundance when infants were solely fed milk (stage I), while its relative abundance decreased with an increase in the relative abundance of Firmicutes in the gut microbiota from stage I to stage II (when milk feeding reduced and plant leaves and feed concentrate were added) and stage III (when only leaves and feed concentrate were fed; Figure 2A; Li et al., 2020, 2021). Proteobacteria is ubiquitous and abundant in the intestines of breastfed infants, whereas Firmicutes are capable of digesting and absorbing nutrients from diverse substrates, including fiber-rich plant leaves. Notably, the gut microbiota composition does not differ significantly between stage II and stage III in weaning deer species, suggesting that the establishment of the gut microbiota is preliminarily completed before total weaning. Similar gut microbiota succession patterns have been observed in human infants (Bäckhed et al., 2015), pigs (Kim et al., 2012), and horses (Mach et al., 2017). Hence, careful attention should be paid to the formula of plant leaves and feed concentrate fed to deer species during the weaning process in order to enable a more healthy and mature gut microbial composition. However, the microbial members and functions important for gut microbiota reassembly and maturity during the weaning process need to be explored further.
Figure 2.
Microbial composition of the gut bacterial community of deer species at the phylum level in weaning (A), pathogen-positive or -negative (B-D), and antibiotic treatments (E,F), respectively.
4. Effects of pathogens and antibiotic treatments on the gut microbiota of deer species
The presence of pathogens in the gut microbiota usually alters the microbiota composition, even if the host is asymptomatic. White-tailed deer containing diarrheagenic Escherichia coli in their gut microbiota harbored an altered microbiota structure compared with those not containing E. coli (Delgado et al., 2017; Figure 2B). Moreover, Li et al. (2018) compared the gut microbiota of healthy forest musk deer and those with mild and severe diarrhea, and observed that their gut microbiota compositions differed, with a lower relative abundance of Firmicutes and higher relative abundance of Proteobacteria being noted in the gut microbiota of diseased individuals than in that of healthy ones (Li et al., 2018; Figure 2D). Their study also identified Escherichia/Shigella and Fusobacterium as the potential causal agents of diarrhea, suggesting that metagenomics-based microbiota profiling can be a powerful tool to identify the causal agents of infective diseases. Interestingly, forest musk deer with and without bacterial pneumonia were found to harbor different gut microbiota compositions (Zhao et al., 2021; Figure 2C), although the pathogen that causes pneumonia does not directly interact with the gut microbiota. The roles of gut microbes that exhibit positive and negative correlations with pneumonia can be further explored in order to develop gut microbiota manipulation approaches for preventing and/or treating pneumonia. Antibiotic administration is a widely used treatment for controlling bacterial infections. The relative abundance of gastrointestinal pathogenic bacteria together with that of Proteobacteria phyla in the gut microbiota of sike deer dramatically decreased after antibiotic treatment (Hu et al., 2020; Figure 2E). However, the fungal content (Hu et al., 2020; Figure 2F) in the gut microbiota significantly changed from that before antibiotic treatment. The effects of such changes in fungal community compositions in the gut microbiota on the hosts need to be studied further.
5. Conclusion and future outlook
The health of an animal is inevitably associated with the stability of its gut microbiota. Throughout the lifespan of deer, their gut microbiota composition is affected by various factors, such as diet, living environment, antibiotic use, and diseases. A thorough understanding of how the gut microbiota is affected by the given factors through high-throughput sequencing can enable a more reliable assessment of the effects of various factors on gut microbiota composition and on host development and health. The relative abundance of Firmicutes and Bacteroides and the F/B ratio in the gut microbiota are important for deer species to adapt to their habitats and are mainly determined by amplicon sequencing. Studies identifying the microbial members and metabolic functions that play key roles in host adaptation, including digestion of the given feed, low temperature adaptation, and health maintenance, are urgently needed. A few whole-genome metagenomic studies have assessed the gut microbiota of deer and identified several functional genes and genome sequences of beneficial microbes (Su et al., 2022; Wang et al., 2022). With the rapid development of high-throughput sequencing, bioinformatics, culturomics, and in situ microbiota editing technologies, the gut microbiota can be manipulated to help the host adapt to the environment (e.g., efficiently digest food and exhibit antagonistic effects against pathogens; Stewart et al., 2019; Thomas et al., 2021; Whon et al., 2021). The development of gut microbiota research will contribute to the conservation of deer species, particularly endangered ones, and benefit future wild population restoration programs.
Author contributions
YZ and YW conceived and revised the manuscript. BX, HC, and FY contributed reference download and its organization. JH, XJ, and YZ contributed funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Yangzhou University Interdisciplinary Research Foundation for Veterinary Medicine Discipline of Targeted Support (grant number yzuxk202003), the Postgraduate Research and Practice Innovation Program of Jiangsu Province (grant number KYCX21_3208), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) and 111 Project (D18007).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Anee I. J., Alam S., Begum R. A., Shahjahan R. M., Khandaker A. M. (2021). The role of probiotics on animal health and nutrition. JoBAZ 82:52. doi: 10.1186/s41936-021-00250-x [DOI] [Google Scholar]
- Bäckhed F., Roswall J., Peng Y., Feng Q., Jia H., Kovatcheva-Datchary P., et al. (2015). Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 690–703. doi: 10.1016/j.chom.2015.04.004, PMID: [DOI] [PubMed] [Google Scholar]
- Bibbò S., Ianiro G., Giorgio V., Scaldaferri F., Masucci L., Gasbarrini A., et al. (2016). The role of diet on gut microbiota composition. Eur. Rev. Med. Pharmacol. Sci. 20, 4742–4749. PMID: [PubMed] [Google Scholar]
- Cai Y., Yang J., Wang J., Yang Y., Fu W., Zheng C., et al. (2020). Changes in the population genetic structure of captive forest musk deer (Moschus berezovskii) with the increasing number of generation under closed breeding conditions. Animals 10:255. doi: 10.3390/ani10020255, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier C., Stojanović O., Colin D. J., Suarez-Zamorano N., Tarallo V., Veyrat-Durebex C., et al. (2015). Gut microbiota orchestrates energy homeostasis during cold. Cells 163, 1360–1374. doi: 10.1016/j.cell.2015.11.004, PMID: [DOI] [PubMed] [Google Scholar]
- Delgado M. L., Singh P., Funk J. A., Moore J. A., Cannell E. M., Kanesfsky J., et al. (2017). Intestinal microbial community dynamics of white-tailed deer (Odocoileus virginianus) in an agroecosystem. Microb. Ecol. 74, 496–506. doi: 10.1007/s00248-017-0961-7, PMID: [DOI] [PubMed] [Google Scholar]
- Fang W., Fang Z., Zhou P., Chang F., Hong Y., Zhang X., et al. (2012). Evidence for lignin oxidation by the giant panda fecal microbiome. PLoS One 7:e50312. doi: 10.1371/journal.pone.0050312, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassarella M., Blaak E. E., Penders J., Nauta A., Smidt H., Zoetendal E. G. (2021). Gut microbiome stability and resilience: elucidating the response to perturbations in order to modulate gut health. Gut 70, 595–605. doi: 10.1136/gutjnl-2020-321747, PMID: [DOI] [PubMed] [Google Scholar]
- Gogarten J. F., Brown L. M., Chapman C. A., Cords M., Doran-Sheehy D., Fedigan L. M., et al. (2012). Seasonal mortality patterns in non-human primates: implications for variation in selection pressures across environments. Evolution 66, 3252–3266. doi: 10.1111/j.1558-5646.2012.01668.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Yang H., Han S., Feng L., Wang T., Ge J. (2017). Comparison of the gut microbiota composition between wild and captive sika deer (Cervus nippon hortulorum) from feces by high-throughput sequencing. AMB Express 7, 212–213. doi: 10.1186/s13568-017-0517-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. (2016). Moschus chrysogaster. The IUCN Red List of Threatened Species 2016:e.T13895A61977139. doi: 10.2305/IUCN.UK.2016-1.RLTS.T13895A61977139.en [DOI] [Google Scholar]
- Hu X., Liu G., Li Y., Wei Y., Lin S., Liu S., et al. (2018). High-throughput analysis reveals seasonal variation of the gut microbiota composition within forest musk deer (Moschus berezovskii). Front. Microbiol. 9:1674. doi: 10.3389/fmicb.2018.01674, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Xu Y., Liu G., Hu D., Wang Y., Zhang W., et al. (2020). The impact of anthelmintic treatment on gut bacterial and fungal communities in diagnosed parasite-free sika deer Cervus nippon. Appl. Microbiol. Biotechnol. 104, 9239–9250. doi: 10.1007/s00253-020-10838-y, PMID: [DOI] [PubMed] [Google Scholar]
- Hugon P., Lagier J. C., Colson P., Bittar F., Raoult D. (2017). Repertoire of human gut microbes. Microb. Pathog. 106, 103–112. doi: 10.1016/j.micpath.2016.06.020 [DOI] [PubMed] [Google Scholar]
- Jami E., Israel A., Kotser A., Mizrahi I. (2013). Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 7, 1069–1079. doi: 10.1038/ismej.2013.2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Gao H., Qin W., Song P., Wang H., Zhang J., et al. (2021). Marked seasonal variation in structure and function of gut microbiota in forest and Alpine Musk Deer. Front. Microbiol. 12:699797. doi: 10.3389/fmicb.2021.699797, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Jiang J., Wang Y., Zhang E., Zhang Y., Li L., et al. (2016). Red list of China’s vertebrates. Biodivers. Sci. 24, 500–551. doi: 10.17520/biods.2016076 [DOI] [Google Scholar]
- Joat N. N., Khan S., Chousalkar K. (2021). Understanding the effects of intramuscular injection and feed withdrawal on Salmonella typhimurium shedding and gut microbiota in pullets. J. Anim. Sci. Biotechnol. 12, 78–15. doi: 10.1186/s40104-021-00597-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. B., Borewicz K., White B. A., Singer R. S., Sreevatsan S., Tu Z. J., et al. (2012). Microbial shifts in the swine distal gut in response to the treatment with antimicrobial growth promoter, tylosin. Proc. Natl. Acad. Sci. U. S. A. 109, 15485–15490. doi: 10.1073/pnas.1205147109, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Bleisch W. V., Jiang X. (2016). Effects of ethnic settlements and land management status on species distribution patterns: a case study of endangered musk deer (Moschus spp.) in Northwest Yunnan, China. PLoS One 11:e0155042. doi: 10.1371/journal.pone.0155042, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Gao H., Song P., Liang C., Jiang F., Xu B., et al. (2022). Captivity shifts gut microbiota communities in white-lipped deer (Cervus albirostris). Animals 12:431. doi: 10.3390/ani12040431, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Hu X., Yang S., Zhou J., Qi L., Sun X., et al. (2018). Comparison between the fecal bacterial microbiota of healthy and diarrheic captive musk deer. Front. Microbiol. 9:300. doi: 10.3389/fmicb.2018.00300, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Hu X., Yang S., Zhou J., Zhang T., Qi L., et al. (2017). Comparative analysis of the gut microbiota composition between captive and wild forest musk deer. Front. Microbiol. 8:1705. doi: 10.3389/fmicb.2017.01705, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Shi M., Zhang T., Hu X., Zhang B., Xu S., et al. (2020). Dynamic changes in intestinal microbiota in young forest musk deer during weaning. PeerJ 8:e8923. doi: 10.7717/peerj.8923, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. G., Wang C. D., Tang Z. H., Guo Y. Q., Zheng T. C., Li Y. Z., et al. (2017). The gut bacterial community composition of wild Cervus albirostris (white-lipped deer) detected by the 16S ribosomal RNA gene sequencing. Curr. Microbiol. 74, 1100–1107. doi: 10.1007/s00284-017-1288-9, PMID: [DOI] [PubMed] [Google Scholar]
- Li Y., Zhang T., Shi M., Zhang B., Hu X., Xu S., et al. (2021). Characterization of intestinal microbiota and fecal cortisol, T3, and IgA in forest musk deer (Moschus berezovskii) from birth to weaning. Integr. Zool. 16, 300–312. doi: 10.1111/1749-4877.12522, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Li D. Q., Zhao W., Yu D., Cheng J. G., Luo Y., et al. (2019) Sequencing and analysis of gut microbiota in forest musk deer from Qinghai and Sichuan. BIBE The Third International Conference on Biological Information and Biomedical Engineering, VDE, Hangzhou, China. 1–7. [Google Scholar]
- Liu J., Liang X., Liu Y. (2019). Comparison of the gut microbiota composition between captive and wild roe deer. bioRxiv. doi: 10.1101/831222 [DOI] [Google Scholar]
- Mach N., Foury A., Kittelmann S., Reigner F., Moroldo M., Ballester M., et al. (2017). The effects of weaning methods on gut microbiota composition and horse physiology. Front. Physiol. 8:535. doi: 10.3389/fphys.2017.00535, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke S., Heurich M., Henrich M., Wilhelm K., Sommer S. (2019). Impact of winter enclosures on the gut bacterial microbiota of red deer in the Bavarian Forest National Park. Wildlife. Biol. 2019, 1–10. doi: 10.2981/wlb.00503 [DOI] [Google Scholar]
- Minich D., Madden C., Evans M. V., Ballash G. A., Barr D. J., Poulsen K. P., et al. (2021). Alterations in gut microbiota linked to provenance, sex, and chronic wasting disease in white-tailed deer (Odocoileus virginianus). Sci. Rep. 11, 13212–13218. doi: 10.1038/s41598-021-89896-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spake R., Bellamy C., Gill R., Watts K., Wilson T., Ditchburn B., et al. (2020). Forest damage by deer depends on cross-scale interactions between climate, deer density and landscape structure. J. Appl. Ecol. 57, 1376–1390. doi: 10.1111/1365-2664.13622 [DOI] [Google Scholar]
- Stewart R. D., Auffret M. D., Warr A., Walker A. W., Roehe R., Watson M. (2019). Compendium of 4,941 rumen metagenome-assembled genomes for rumen microbiome biology and enzyme discovery. Nat. Biotechnol. 37, 953–961. doi: 10.1038/s41587-019-0202-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su R., Dalai M., Luvsantseren B., Chimedtseren C., Hasi S. (2022). Comparative study of the function and structure of the gut microbiota in Siberian musk deer and forest musk deer. Appl. Microbiol. Biotechnol. 106, 6799–6817. doi: 10.1007/s00253-022-12158-9, PMID: [DOI] [PubMed] [Google Scholar]
- Sun C. H., Liu H. Y., Liu B., Yuan B. D., Lu C. H. (2019). Analysis of the gut microbiome of wild and captive Père David’s deer. Front. Microbiol. 10:2331. doi: 10.3389/fmicb.2019.02331, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Sun Y., Shi Z., Liu Z., Zhao C., Lu T., et al. (2020). Gut microbiota of wild and captive alpine musk deer (Moschus chrysogaster). Front. Microbiol. 10:3156. doi: 10.3389/fmicb.2019.03156, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Yu Y., Guo J., Zhong L., Zhang M. (2021). Comparative analysis of gut microbiome between captive and wild red deer 1 (Cervus elaphus) in Inner Mongolia. Res. Square. doi: 10.21203/rs.3.rs-312376/v1 [DOI] [Google Scholar]
- Tanca A., Fraumene C., Manghina V., Palomba A., Abbondio M., Deligios M., et al. (2017). Diversity and functions of the sheep faecal microbiota: a multi-omic characterization. Microb. Biotechnol. 10, 541–554. doi: 10.1111/1751-7915.12462, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. A., Hitch A. A., Thomas R., Antonios K., Dirk H., Ilias L., et al. (2021). Recent advances in culture-based gut microbiome research. Int. J. Med. Microbiol. 311:151485. doi: 10.1016/j.ijmm.2021.151485, PMID: [DOI] [PubMed] [Google Scholar]
- Wang X., Chen Y., Shang Y., Wu X., Wei Q., Chen J., et al. (2019). Comparison of the gut microbiome in red deer (Cervus elaphus) and fallow deer (Dama dama) by high-throughput sequencing of the V3–V4 region of the 16S rRNA gene. Sci. Asia 45, 515–524. doi: 10.2306/scienceasia1513-1874.2019.45.515 [DOI] [Google Scholar]
- Wang Y. H., Liu S. Q., Yang S., Zhang T. X., Wei Y. T., Zhou J. T., et al. (2016). Determination of ovarian cyclicity and pregnancy using fecal progesterone in forest musk deer (Moschus berezovskii). Anim. Reprod. Sci. 170, 1–9. doi: 10.1016/j.anireprosci.2016.03.002, PMID: [DOI] [PubMed] [Google Scholar]
- Wang Y., Xu J., Chen H., Yu J. Y., Xu X. M., Sun L., et al. (2022). A balanced gut microbiota is essential to maintain health in captive sika deer. Appl. Microbiol. Biotechnol. 106, 5659–5674. doi: 10.1007/s00253-022-12111-w, PMID: [DOI] [PubMed] [Google Scholar]
- Whon T. W., Shin N. R., Kim J. Y., Roh S. W. (2021). Omics in gut microbiome analysis. J. Microbiol. 59, 292–297. doi: 10.1007/s12275-021-1004-0 [DOI] [PubMed] [Google Scholar]
- You Z., Deng J., Liu J., Fu J., Xiong H., Luo W., et al. (2022). Seasonal variations in the composition and diversity of gut microbiota in white-lipped deer (Cervus albirostris). PeerJ 10:e13753. doi: 10.7717/peerj.13753, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X. H., Xue H., Xu X. M., Jiao X. A., Pan Z. M., Zhang Y. Z. (2022). Closely related Salmonella Derby strains triggered distinct gut microbiota alteration. Gut Pathog. 14:6. doi: 10.1186/s13099-022-00480-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. M., Bai J. D., Li Y. P., Chen Q., Cheng Z. B., Meng Q. H., et al. (2017). Père David’s Deer’s ex-situ conservation status, conservation patterns and conservation suggestions in China. Forest. Res. Manag. 2:16. doi: 10.13466/j.cnki.lyzygl.2017.02.004, (English abstract and Chinese main-text) [DOI] [Google Scholar]
- Zhang M., Shi M., Fan M., Xu S., Li Y., Zhang T., et al. (2018). Comparative analysis of gut microbiota changes in Père David's deer populations in Beijing Milu Park and Shishou, Hubei Province in China. Front. Microbiol. 9:1258. doi: 10.3389/fmicb.2018.01258, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Ren Z., Luo Y., Cheng J., Wang J., Wang Y., et al. (2021). Metagenomics analysis of the gut microbiome in healthy and bacterial pneumonia forest musk deer. Genes Genom. 43, 43–53. doi: 10.1007/s13258-020-01029-0, PMID: [DOI] [PubMed] [Google Scholar]
- Zipperer A., Konnerth M. C., Laux C., Berscheid A., Janek D., Weiden-maier C., et al. (2016). Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535, 511–516. doi: 10.1038/nature18634, PMID: [DOI] [PubMed] [Google Scholar]