Abstract
The PHD fingers of the human MLL and Drosophila trx proteins have strong amino acid sequence conservation but their function is unknown. We have determined that these fingers mediate homodimerization and binding of MLL to Cyp33, a nuclear cyclophilin. These two proteins interact in vitro and in vivo in mammalian cells and colocalize at specific nuclear subdomains. Overexpression of the Cyp33 protein in leukemia cells results in altered expression of HOX genes that are targets for regulation by MLL. These alterations are suppressed by cyclosporine and are not observed in cell lines that express a mutant MLL protein without PHD fingers. These results suggest that binding of Cyp33 to MLL modulates its effects on the expression of target genes.
The myeloid lymphoid leukemia (MLL) gene (also known as HRX, HTRX, and ALL1) is involved in recurring chromosome translocations and duplications associated with human leukemia (33, 40). The MLL 430-kDa protein has two domains with extensive similarity to the trithorax (trx) Drosophila protein (4, 8, 13, 34). trx is involved in the process of maintenance of gene activity, an epigenetic mechanism that mediates the clonal inheritance of cell fates during development (17). The trx gene is the prototype of a group of genes (the trithorax group, or trx-G) which regulate the activity of Drosophila type I Hox genes and have related mutant homeotic phenotypes. Another group of genes, the polycomb group, or Pc-G, that mediates gene repression antagonizes the activating function of trx on its target genes. Pc-G and trx-G proteins are components of chromatin and modify gene activity by influencing chromatin structure at enhancers and promoters (17). A similar interaction between Pc-G and trx-G genes also regulates type I HOX gene expression in mammals (14). Disruption of the maintenance mechanism by mutation of MLL may contribute to the leukemogenic process.
One of the two main domains of similarity between MLL and trx is formed by four Zn fingers of a special type called PHD (for plant homeodomain) Zn fingers (4) and an atypical bromodomain (16) nested between the third and fourth fingers. PHD fingers occur in a large number of proteins, most of which seem to be components of chromatin and/or regulators of gene activity (1), but the PHD finger cluster of trx and MLL proteins has special features conserved throughout evolution that suggest it is a functional cassette.
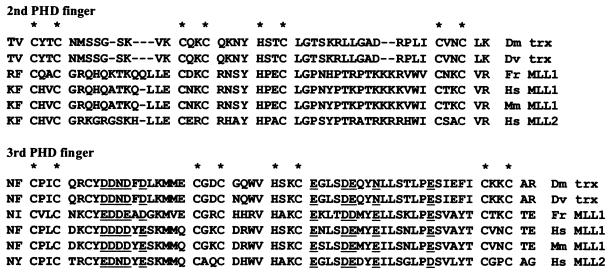
The PHD finger cassette of MLL is conserved not only with the Drosophila trx proteins but also with the mouse and puffer fish MLL proteins and with the putative product of the human MLL2 gene, a recently described close relative of MLL (11) (Fig. 1). Each one of the four PHD fingers within these proteins has distinctive characteristics. The third and fourth PHD fingers have an N-terminal extension (not shown in Fig. 1) including pairs of cysteines (CXXC) and/or histidine-cysteine pairs (HXXC) that may form an additional Zn finger or may form a more complex structure with the associated PHD finger. For that reason we call these two fingers extended PHD fingers (ePHD fingers).
FIG. 1.
Alignment of the amino acid sequences of the second and third PHD fingers of trx and MLL proteins. The N-terminal extension of the third PHD finger is not shown. Conserved acidic (or amphotropic) residues in the third PHD finger are underlined. Asterisks above the alignments mark the positions of conserved pairs of C and H residues that define the PHD fingers and, presumably, coordinate Zn++ ions.
Because of their strong evolutionary conservation, it is suspected that the PHD fingers play an important role in trx and MLL function. Missense mutations within one of the PHD fingers of trx have a mutant phenotype in Drosophila. Nevertheless, the function of the PHD fingers is unknown. The PHD fingers have similarity in structure to the RING finger, another motif frequently found in chromatin component proteins. Since the RING fingers are involved in protein-protein interactions, it has been proposed that the PHD fingers may have a similar general function (1). In order to search for such interactions we used the yeast two-hybrid system, finding that some of the MLL and trx PHD fingers homodimerize. In addition, the MLL third ePHD finger binds strongly to Cyp33, a nuclear cyclophilin with an amino terminal RNA binding domain of the RRM type (3). We have investigated the effects of Cyp33 on the gene regulatory functions of MLL and have found that overexpression of Cyp33 does modulate the transcriptional control of the human HOX genes by MLL.
MATERIALS AND METHODS
Yeast two-hybrid system.
The numbering of MLL amino acid residues is according to the HRX amino acid sequence published by Tkachuk et al. (34) (GenBank accession no. M31617). To generate constructs for the yeast two-hybrid system, the MLL sequences encoding PHD fingers 1, 2, and e3 (PHD1–3; amino acids [aa] 1394 to 1630) were cloned into the Gal4 DNA binding domain vector pGBT9 (Clontech). This construct is called pGBT9-PHD1–3. The same insert (MLL; aa 1394 to 1630) was cloned into the Gal4 activation domain vector pGAD424 (Clontech). The trx constructs pSN16 and pSN17 are described in Breen and Harte (4) and contain an insert encoding the trx PHD finger region (nucleotides 4508 to 6673; GenBank accession no. M31617). For the yeast two-hybrid library screen, the plasmid pGBT9-PHD1–3 and a cDNA library made from human HeLa cells in the Gal4 activation domain vector pGAD GH (Clontech) were cotransformed into the yeast strain Y190 (Trp− Leu− His− LacZ−). Approximately 10 million clones were screened on Trp/Leu/His dropout media containing 35 mM 3-aminotriazole. Plasmid DNA was isolated from 60 His+ LacZ+ yeast clones and was used to transform competent Escherichia coli strain KC8 (Trp− Leu− His−). Transformants were plated on leucine-deficient M9 media to isolate the library plasmid from positive clones. Plasmid DNA was extracted, was characterized, and then was retransformed back into yeast strain Y190 with pGBT9-PHD1–3 or the parental DNA binding domain vector pGBT9 alone. The 20 plasmids that passed through these tests were sequenced, and their sequences were compared to nucleotide and protein databases using the BLAST program. Specific two-hybrid assays for protein interactions were performed for yeast strain Y190. As controls for nonspecific yeast two-hybrid protein interactions, GBT-NonO and GAD-NonO plasmids were used (37). Tests for β-galactosidase activity were performed by filter lift assay by using X-Gal as a substrate or were quantitated from liquid culture using chlorophenol red β-d-galactopyranoside as a substrate.
Generation of deletion constructs.
Yeast two-hybrid constructs were generated encoding each one of the MLL PHD fingers 1 to 4 separately or encoding different domains of Cyp33. PCR primers were designed with ends that could be digested with BamHI to amplify each cDNA segment. The PCR products were digested with BamHI and cloned into pGAD424.
GST in vitro binding assays.
pGEX-KT-MLL clones contain either MLL cDNA aa 1392 to 1700 (GST-MLL PHD1–3) or aa 1392 to 2000 fused to a glutathione S-transferase (GST) open reading frame (ORF) (GST-MLL PHD1–4) (39). DNA encoding the Cyp33 ORF from pGAD GH Cyp33 was subcloned into the pGEX-4T-1 (Pharmacia) PspA1 and XhoI restriction sites. The GST fusion proteins were expressed in E. coli and purified according to Grieco et al. (12). Expression plasmids encoding the full-length MLL zinc finger region, aa 1392 to 2000, in pBluescript S/K+ (Stratagene) or the full-length CYP33 cDNA in pSP6 (Promega) were used as templates for coupled in vitro transcription/translation in a rabbit reticulocyte lysate system (Promega) containing [35S]methionine (Amersham). Radiolabeled protein was added directly to GST Sepharose beads loaded with equal amounts of fusion proteins or GST alone in a binding buffer (50 mM K phosphate, 150 mM KCl, 1 mM MgCl2, 5% glycerol, 0.01% Triton X-100). After 1 h of incubation at 4°C, the beads were washed five times in the binding buffer and finally were boiled for 5 min in loading buffer. The supernatant was loaded on a sodium dodecyl sulfate–15% polyacrylamide gel electrophoresis (SDS-15% PAGE) gel and was fractionated.
Mammalian two-hybrid system.
The cDNAs encoding MLL zinc fingers 1 to 3 or the entire Cyp33-encoding ORF were subcloned into the multiple cloning site of the mammalian expression vector pVP-HA1 or pVP-FLAG (35). The MLL zinc finger 1 to 3 cDNA was also subcloned into the mammalian expression vector pCMV-Gal4 (35). Human 293 cells, NIH 3T3 cells, or COS cells were cotransfected with the VP16 transactivation construct (6 μg), the Gal4 DNA binding domain construct (6 μg), a Gal4 responsive luciferase reporter gene (G5LUC) (4 μg), and a constitutively active Renilla reporter gene (ptkRL) (1 μg). Transfections were done using Superfect transfection reagent (Qiagen), and lysates were analyzed using the Dual-Luciferase Reporter assay system (Promega) and a luminometer (Turner Designs; 20/20).
Coimmunoprecipitation and Western analysis.
The GAL4-MLL expression plasmid was constructed as described above for the mammalian two-hybrid assay. FLAG-MLL and FLAG-Cyp33 were generated by excising the MLL PHD1–3 cDNA or the CYP33 cDNA, respectively, from the pVP-FLAG vector together with sequences that encode the FLAG epitope. The inserts were then cloned into the NotI and HindIII sites of pCMV-Not (35). Human 293 kidney cells were transiently transfected with 10 μg of each expression construct using SuperFect transfection reagent (Qiagen). After 48 h, cell lysates were prepared using a low-salt NP-40 buffer (10 mM HEPES buffer [pH 7.6], 250 mM NaCl, 0.1% Nonidet P-40, 5 mM EDTA). Polyclonal GAL4 antibody (Santa Cruz Biotech, Santa Cruz, Calif.) or polyclonal antiserum to Cyp33 (22) was added to the appropriate cell lysates with protein A agarose (Sigma) and was rocked at 4°C. Protein A agarose was then pelleted and washed four times with low-salt NP-40 buffer, was resuspended in Laemmli loading buffer, and was boiled for 5 min. Supernatants were fractionated on an SDS–15% PAGE gel and were transferred to an Immobilon-P membrane (Millipore) for Western blot analysis with the FLAG-specific M5 monoclonal antibody (Sigma).
Immunofluorescence and confocal microscopy.
The pFLAG-PHD-NTS-MLL was constructed in a multistep subcloning process. The MLL cDNA fragment containing aa 15 to 708, which include the AT hooks and all the elements involved in localization of MLL to nuclear speckles (38, 39), was subcloned into plasmid pSP6 containing FLAG epitope sequences (pSP6-FLAG-NTS). The MLL PHD1–3 sequences were PCR amplified from cDNA and were cloned into the EcoRI site of pSP6-FLAG-NTS. Then, the cDNA fragment containing PHD1–3, the nuclear targeting sequences (NTS), and the FLAG epitope was excised from pSP6 and was subcloned into the NotI and XbaI sites of pCMV-Not (Invitrogen). All junctions and MLL-amplified sequences were verified by dideoxy-nucleotide sequencing. The pHA-Cyp33 expression vector was generated by subcloning the CYP33 cDNA along with the influenza virus hemagglutinin (HA) epitope tag from pVP-HA-Cyp33 into the NotI and HindIII sites of pCMV-Not. Human HeLa cells were grown on coverslips overnight in six-well culture dishes. The pFLAG-PHD-NTS-MLL and pHA Cyp33 plasmids were transiently transfected together or each one separately into HeLa cells seeded on coverslips. After 36 h the cells were washed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde at room temperature for 20 min and then with 100% methanol for 10 min at −20°C. After rehydration, cells were incubated for 2 h at room temperature with mouse monoclonal M2 anti-FLAG antibody (Sigma) and rat monoclonal anti-HA antibody (Boehringer Mannheim). Following three washes, the cells were incubated with anti-mouse immunoglobulin G (IgG)–Texas red (Jackson Immunoresearch) and anti-rat IgG–fluorescein isothiocyanate (Boehringer Mannheim). Finally, the coverslips were washed and mounted in DABCO antifade medium (Sigma). Stained cell images were captured by using a Zeiss LSM510 confocal microscope with a ×60 objective and an optical slice set at 0.9 μm.
Overexpression of Cyp33 from a transfected expression construct.
The construction of the plasmid pHA-Cyp33 was described in the previous section. The deletion construct, pHA-RRM, which lacks the conserved cyclophilin domain, was generated using the restriction enzyme Bsu36I. Bsu36I cleaves 5′ and 3′ of the cyclophilin domain of Cyp33. The deletion construct pHA-Cyp was generated by amplifying the nucleotide sequence encoding the cyclophilin domain of Cyp33 by PCR with specific primers (forward, 5′-CCATGGGCCCGCTCAATCCTCAGTG-3′; reverse, 5′-CTGCAGCACGTACTCCCCACAGTCGG-3′). The amplified DNA fragment was cloned into the vector pGMT (Promega), was cut with the NcoI and PstI restriction enzymes, and was inserted into the pVP-HA vector.
The suspension cell lines K562, ML-1, and THP1 (5 × 106 cells) were transiently transfected with 8 μg of DNA from the above constructs or from the pCMV-Not vector using Superfect (Qiagen) transfection reagent. Cells were harvested after 48 h, and RNA was isolated using Trizol (Gibco/BRL). Reverse transcriptase PCR (RT-PCR) was performed using Superscript II/RNase H reverse transcriptase (Gibco/BRL) using the following reverse primers; the cDNA was amplified by using the following specific primers for the different genes, using HotStarTaq DNA polymerase (Qiagen): HOXC8 forward (5′-CCGCCAACACTAACAGTAGC-3′); HOXC8 reverse (5′-CAGTCCCAGGGCATGAGAG-3′); HOXC9 forward (5′-CCGGCAGCAAGCACAAAGA-3′); HOXC9 reverse (5′-CGCTCGGTGAGATTGAGAACC-3′); HOXC6 forward (5′-TAGTTCTGAGCAGGGCAGGACTGCG-3′); HOXC6 reverse (5′-CCGCTCCGTAGCCGACCCCACTGT-3′); GAPDH forward (5′-ACATCAAGAAGGTGAAGCAGG-3′); GAPDH reverse (5′-TCTTCCTCTTGTGCTCTTGCTGG-3′); HA epitope forward (5′-TACCCATATGACGTCCCAGAC-3′); RRM domain reverse (5′-TGGTTTGGCCAAATTGACACG-3′); and cyclophilin domain reverse (5′-CTGCAGCACGTACTCCCCACAGTCGG-3′).
The effect of cyclosporine was tested in a similar experiment with the following differences. At the time of transfection and 24 h later, cyclosporine dissolved in PBS-ethanol (50:50, vol/vol) was added to the cells to a final concentration of 1 μg/ml; an equivalent amount of PBS-ethanol (50:50) was added to the control cells. The cells were harvested 48 h posttransfection, and RNA was isolated using Trizol (Gibco/BRL). RT-PCR was performed using a Marathon cDNA kit (Clontech).
RESULTS
Protein interactions of the MLL PHD finger domain.
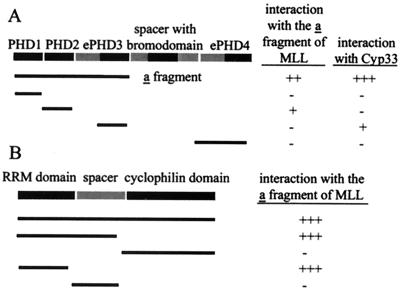
To determine whether the zinc finger region of MLL can mediate dimerization, we tested a series of hybrid GAL4-MLL fusion polypeptides for interaction in the yeast two-hybrid system. Pairs of the fusion constructs were cotransformed into the yeast strain Y190, and positive protein interactions were determined by growth on media lacking histidine and by positive β-galactosidase activity. A summary of the yeast two-hybrid results is presented in Table 1 and Fig. 2. Positive protein interactions occurred between pairs of hybrid constructs containing the first three complete PHD zinc fingers (PHD1–3) in both bait and prey vectors (Table 1; Fig. 2A). Positive interactions occurred as well between the first three complete zinc fingers in the bait vector and only the second PHD finger in the prey vector (PHD2) but not with the first (PHD1) or third (PHD3) PHD zinc fingers (Fig. 2A; Table 1). These results indicate that the PHD1–3 polypeptides can homodimerize and that the second PHD finger is necessary to mediate such interaction. Nevertheless, the interaction with PHD1–3 seems to be stronger than the interaction with PHD2 alone (Table 1).
TABLE 1.
Protein interactions in the yeast two-hybrid system
| GAL4-DNA binding domain fusions | GAL4-activation domain fusions | β-Galactosidase activity (Miller units ± SD)a |
|---|---|---|
| GAL4 DBD aloneb | GAL4 AD alonec | 0.0 |
| MLL PHD1–3 | GAL4 AD alone | 0.0 |
| MLL PHD1–3 | MLL PHD1–3 | 29.5 ± 3.0 |
| MLL PHD1–3 | MLL PHD2 | 8.6 ± 0.4 |
| MLL PHD1–3 | MLL PHD3 | 0.0 |
| Non-Od | Non-O | 206.8 ± 14.6 |
| MLL PHD1–3 | Non-O | 0.0 |
| MLL PHD1–3 | Complete Cyp33 | 133.5 ± 0.8 |
| MLL PHD1–3 | Cyp33 RRM + spacer | 185 ± 7.8 |
| MLL PHD1–3 | Cyp33 RRM | 164.6 ± 4.4 |
| Cyp33 | GAL4 AD alone | 0.0 |
| Cyp33 | MLL PHD2 | 0.0 |
| Cyp33 | MLL PHD3 | 10.5 ± 0.5 |
Average from three experiments ± the standard deviation.
DBD, DNA binding domain.
AD, activation domain.
Non-O is a protein with two RRM domains, used as a control.
FIG. 2.
Summary of the deletion analysis of protein-protein interactions involving the MLL PHD fingers. (A) Using the yeast two-hybrid system, the interactions of different MLL-PHD finger constructs containing the first three PHD fingers (fragment a) or interactions with the full-length Cyp33 protein were tested. +, ++, or +++ indicate a weaker or stronger positive interaction, respectively, detected by the induction of β-galactosidase activity. (B) The interactions of different fragments from the Cyp33 protein with the first three PHD fingers in MLL construct 1 were analyzed in the same way.
We have also tested the PHD zinc finger region of the Drosophila trx protein containing PHD fingers 1 to 3 for homodimerization by using the yeast two-hybrid system. The results were also positive. However, we could not detect heterodimerization between the PHD fingers of MLL and those of trx (Table 1).
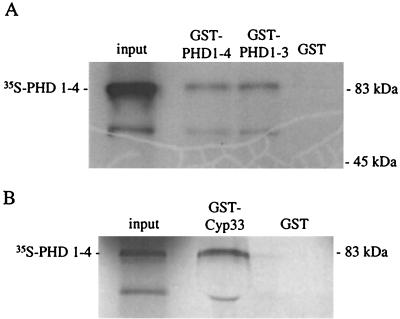
To test for homodimerization of the MLL PHD fingers in vitro, the first three PHD fingers (aa 1394 to 1700) as well as the full-length PHD finger cassette (aa 1394 to 2000) were fused to a GST ORF and were used in affinity precipitation experiments (Fig. 3A). The GST-MLL-PHD finger fusion proteins were expressed in bacteria and were purified by binding to glutathione-Sepharose beads. The full-length PHD zinc finger motif (aa 1394 to 2000) was synthesized and was 35S labeled in a coupled transcription-translation system and then was tested for binding to the GST-MLL-PHD finger fusion proteins or to GST alone. Radiolabeled MLL-PHD1–4 could be affinity purified by precipitation with GST-MLL PHD1–3 but not with GST alone. Similar results were obtained using the GST fusion protein containing PHD1–4. These results indicate that the MLL PHD zinc fingers can mediate homodimerization in vitro.
FIG. 3.
Affinity precipitation of 35S-labeled proteins containing the complete PHD finger cluster of MLL with different GST fusion proteins: (A) GST fusion proteins containing either the complete PHD finger cluster of MLL or PHD fingers 1 to 3; (B) GST-Cyp33 fusion protein. The input lane shows the migration of proteins from the supernatant from the in vitro transcription-translation reaction. The GST lane shows the lack of affinity precipitation with the GST protein alone.
A nuclear cyclophilin (Cyp33) interacts with the PHD finger domain of MLL.
To identify proteins that may interact with the MLL PHD fingers, we performed a yeast two-hybrid screen. We used the first three PHD zinc finger motifs of MLL in the plasmid pGBT9 as the bait. A HeLa cell cDNA library in the prey vector was used to search for interacting proteins. The GAL4 DBD-MLL plasmid and the human HeLa cDNA library in pGAD GH were expressed in the yeast strain Y190. Of approximately 107 transformants, 60 were isolated as positives from the initial screening. From these, 40 were considered false positives because they didn't induce β-galactosidase upon retransformation into yeast or because they interacted nonspecifically with the GAL4 DBD alone. The remaining 20 positives were isolated and sequenced, and their sequences were compared with the sequence databases using the BLAST program.
Eight of the 20 positive clones contained overlapping cDNAs from the same gene. The gene encodes a protein of about 30 kDa, with an RNA binding domain of the RRM type at its amino terminus. Separated by a spacer of 60 amino acids from the RRM amino terminal domain, there is a cyclophilin-like domain. A cDNA identical to the one from this gene was previously reported (18, 22) and was called CyP33 (or PPIE) (we will call the gene CYP33 and the encoded protein Cyp33). This protein was isolated from the Jurkat T-cell leukemia cell line and was found to be present in the nuclear fraction of Jurkat cells. The protein binds RNA, with a preference for A- or T-rich sequences, and according to in vitro enzymatic assays it possesses the prolyl-cis-trans-isomerase activity typical of cyclophilins, which can be inhibited by cyclosporine (22).
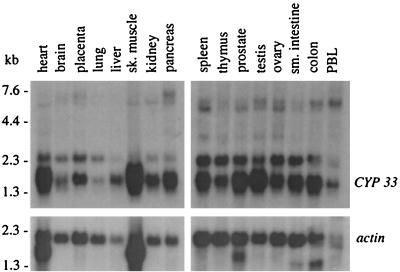
Hybridization of a cDNA probe encoding sequences from the Cyp33 spacer region to a Northern blot containing mRNAs from different human tissues revealed two main transcripts expressed in a variety of different tissues (Fig. 4). Comparison of the CYP33 cDNA sequence with nucleotide sequence databases revealed a strong match with a cyclophilin gene of the parasitic flatworm Schistosoma mansoni (7) and later on with cyclophilin genes from other animal species (M. Anderson, K. Fair, S. Amero, S. Nelson, P. J. Harte, and M. O. Diaz, submitted for publication). The S. mansoni sequence had a 5′ extension of its reported ORF that encodes an RRM domain and spacer, similar to Cyp33. The protein sequence encoded in the extended S. mansoni ORF has 60% identity and a similarity index of 61.6, as calculated by Lipman-Pearson Ktuple parameters, to our human Cyp33 sequence. Such high conservation across evolutionary boundaries suggests an important function for all domains of this protein.
FIG. 4.
Northern blot with mRNA from different human tissues hybridized to a Cyp33 spacer probe. This probe detects two main transcripts of 2.5 and 1.8 kb. An actin cDNA probe was hybridized to the same blot as a loading control shown in the lower panels.
The third PHD finger of MLL mediates binding of Cyp33.
To determine the strength and specificity of the interaction between the MLL zinc fingers and Cyp33 in the yeast two-hybrid system, bait constructs encoding each of the four MLL PHD fingers were made. We found that the PHD3 (aa 1555 to 1636) alone was necessary and sufficient to mediate interaction between Cyp33 and MLL (Fig. 2A). This motif is separate and distinct from the PHD2 motif that mediates dimerization of MLL polypeptides.
Cyp33 and the MLL PHD fingers interact in vitro.
We confirmed the yeast two-hybrid results by standard in vitro binding studies using purified, bacterially expressed GST-Cyp33 fusion proteins. An HA-MLL fusion protein containing PHD1–4 was transcribed and translated in vitro in the presence of [35S]methionine. GST-Cyp33 bound to glutathione Sepharose beads was able to affinity precipitate 35S-labeled HA-MLL, whereas GST alone did not bring down HA-MLL in these experiments (Fig. 3B). In addition, GST-Cyp33 did not interact with radiolabeled HA alone. These experiments show a direct and specific interaction between Cyp33 and the MLL PHD fingers in vitro.
Cyp33 and MLL interact in a mammalian two-hybrid system.
To determine whether the MLL PHD fingers interact with Cyp33 or mediate self binding in mammalian cells, we used a modified two-hybrid assay to examine protein interactions in the nucleus of mammalian cells (35). The PHD1–3 sequences were cloned into a mammalian expression vector containing the DNA-binding domain of GAL4. Other expression vectors were constructed containing the entire Cyp33 protein or the PHD1–3 sequences fused to the herpesvirus VP16 transactivation domain.
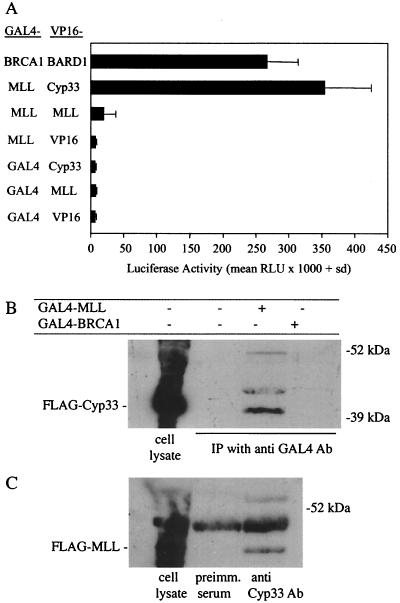
The mammalian two-hybrid assay was performed by transfecting NIH 3T3 or h293 cells with paired combinations of the appropriate expression constructs along with a GAL4-responsive luciferase reporter gene and a constitutively active Renilla luciferase reporter gene to normalize for transfection efficiency. Positive interactions were determined by evaluating transcriptional activation of the luciferase reporter gene. The strength of the interaction was quantitated by measuring the luciferase activity of lysates prepared from transfected cells (Fig. 5A). Control transfections show that coexpression of GAL4-MLL with the VP16 domain alone did not significantly induce luciferase activity. Likewise, VP16-MLL or VP16-Cyp33 in combination with the GAL4 DNA binding domain alone did not induce luciferase activity in mammalian cells. However, cotransfection of cells with GAL4-MLL and VP16-Cyp33 resulted in a large increase in luciferase activity that was similar to, or greater than, the luciferase activity induced by coexpression of GAL4-BRCA1 and VP16-B202, proteins that are known to interact in mammalian cells (35). Coexpression of GAL4-MLL and VP16-MLL resulted in a small increase in induced luciferase activity. These results suggest that the MLL polypeptide and the Cyp33 protein can form stable interactions in the nucleus of mammalian cells. In addition, the MLL PHD1–3 polypeptides also homodimerize, even if with low affinity, in mammalian cells.
FIG. 5.
(A) Results from the mammalian two-hybrid analysis of MLL-PHD finger protein interactions. Mouse NIH 3T3 cells were transiently transfected with the G5LUC reporter construct, the Renilla luciferase control expression plasmid under the control of the thymidine kinase gene promoter, and the two indicated expression vectors. GAL4 or VP16 indicates the DNA binding or activation domain expression vectors. The other constructs are denoted by the name of the domain fused to either the GAL4 DNA binding domain (first column) or the VP16 activation domain (second column). The BRCA1-BARD1 interaction was used as a positive control (22). The thick bars represent the average normalized luciferase activities from three experiments, and the error bars represent one standard deviation. (B) Immunoprecipitation and Western blotting of cell lysates from 293 human kidney cells transfected with an expression plasmid for a FLAG-Cyp33 fusion gene alone or plus expression plasmids for either a GAL4-MLL fusion protein containing PHD fingers 1 to 4 or a GAL4-BRCA1 fusion protein; the lysate was immunoprecipitated with an anti-GAL4 antibody. (C) Immunoprecipitation and Western blotting of cell lysates from 293 human kidney cells transfected with only an expression plasmid containing a FLAG-MLL fusion protein containing the PHD fingers 1 to 4; the immunoprecipitation was performed with an anti-Cyp33 polyclonal antibody or with preimmune rabbit serum. In both experiments the Western blotting was performed with an anti-FLAG antibody. The first lane in each experiment contains the whole cell lysate without immunoprecipitation.
MLL zinc fingers interact with endogenous Cyp33 in vivo.
We could also confirm by coimmunoprecipitation that the MLL zinc fingers and Cyp33 protein interact in mammalian cells. Human 293 kidney cells were transiently transfected with expression plasmids encoding both the MLL PHD1–3 polypeptide fused to an N-terminal GAL4 DNA binding domain and the full-length Cyp33 polypeptide with an N-terminal FLAG epitope. As controls, 293 cells were cotransfected with the GAL4-BRCA1 protein and FLAG-Cyp33 or with FLAG-Cyp33 alone. When cells were lysed 2 days after transfection and were immunoprecipitated with antibody to the GAL4 domain, we found that FLAG Cyp33 coprecipitated with GAL4-MLL polypeptide (Fig. 5B). This interaction was specific because Cyp33 could not be coprecipitated with GAL4-BRCA1.
To determine whether the MLL zinc fingers can interact with endogenous Cyp33 protein, 293 cells were transiently transfected with the MLL PHD1–3 polypeptide with an N-terminal FLAG epitope. Cell lysates were immunoprecipitated with a rabbit antiserum raised against the Cyp33 protein (22) or with preimmune rabbit serum. The precipitates were fractionated by SDS-PAGE, and the presence of FLAG-MLL in the precipitate was determined by Western blot analysis with a monoclonal antibody that recognizes the FLAG epitope. FLAG-MLL coimmunoprecipitated with antiserum specific for Cyp33 but not with preimmune rabbit serum (Fig. 5C). Therefore, endogenous Cyp33 can interact in vivo with the MLL PHD fingers. These experiments demonstrate that Cyp33 and the MLL PHD fingers interact in mammalian cells.
The RRM motif of Cyp33 is necessary and sufficient to mediate protein interaction.
To determine which domains from Cyp33 mediate the interaction with the MLL PHD fingers, deletion constructs of the different functional domains of Cyp33 were constructed and were tested for interaction with the MLL PHD fingers in the yeast two-hybrid assay. Deletions of Cyp33 were fused to the GAL4 activation domain and were tested for interaction with the GAL4-DNA binding domain-MLL PHD1–3 fusion (Fig. 2B). The strength of the interactions as quantitated by a liquid β-galactosidase assay for all protein interactions tested in the yeast two-hybrid system is summarized in Table 1. These results show that Cyp33 binds the MLL PHD fingers through its RRM domain.
Since the RRM domain is the only region of Cyp33 necessary for interaction with the MLL PHD fingers and this domain has been shown to bind RNA (22), it is possible that RNA mediates the protein interaction. To determine if RNA is necessary for the interaction between Cyp33 and MLL, we added RNase to the 35S-labeled Cyp33 product from our in vivo transcription translation reaction (which contains RNA) and tested for its ability to be precipitated with GST-PHD1–3. There was no difference in the amount of radiolabeled Cyp33 that could be precipitated with GST-MLL bound to glutathione beads in the presence or absence of RNase (data not shown). These results suggest that RNA is not necessary for and did not detectably enhance the protein interaction between Cyp33 and the MLL PHD fingers.
MLL and Cyp33 exhibit overlapping patterns of nuclear localization.
Previously, MLL has been localized to the nucleus in a speckled punctate pattern, a localization that is mediated by distinct N-terminal amino acid sequence elements (6, 38). Cyp33 has also been found in the nuclear fraction of cultured cells (22). To determine if the nuclear distribution of Cyp33 and MLL are overlapping, the two proteins were transiently expressed with specific epitope tags in Cos7 or HeLa cells and were localized by immunostaining. The entire Cyp33 protein tagged with HA or a truncated version without the cyclophilin domain was coexpressed in cells with the FLAG-MLL protein. The FLAG-MLL protein included the AT hooks, two N-terminal elements of the MLL protein necessary and sufficient for its nuclear localization, PHD1–3, and a FLAG epitope tag.
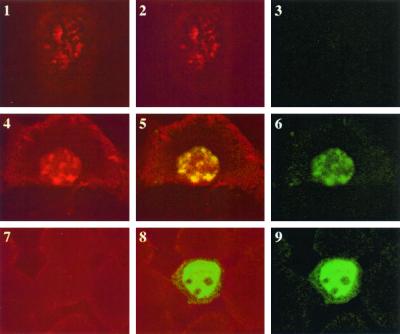
To characterize the nuclear localization pattern of MLL and Cyp33, fluorescent confocal microscopy was employed. When FLAG MLL was expressed alone in HeLa cells and was visualized by immunostaining, the protein distribution showed a distinct pattern of nuclear speckles as previously described (6, 38) (Fig. 6). However, the HA-Cyp33 protein alone, with or without the cyclophilin domain, was distributed uniformly throughout the nucleus but excluded from the nucleoli (Fig. 6). In the Cyp33 and MLL cotransfected cells, Cyp33 (or its RRM domain) and MLL always showed an overlapping pattern of nuclear distribution (Fig. 6). The two proteins colocalized to distinct nuclear speckles in about 30% of the double-positive cells. The remaining 70% of double-stained cells showed a nuclear speckled staining against a more diffuse pattern uniformly distributed throughout the nucleus. The distribution of the proteins to the nuclear speckles always depended on the presence of expressed MLL, since 100% of the cells that expressed Cyp33 alone showed a diffuse pattern of protein distribution throughout the nucleus.
FIG. 6.
HeLa cells transfected with an expression construct encoding a FLAG-MLL fusion protein containing the AT hooks, nuclear localization domains, and the first three PHD fingers of MLL and an expression construct encoding a HA-Cyp33 fusion protein. The fixed cells were stained with an anti-FLAG antibody coupled to Texas red (red fluorescence) and with an anti-HA antibody coupled to fluorescein isothiocyanate (FITC; green fluorescence). The cells were analyzed with a fluorescence confocal microscope, and the images show the fluorescent image from the anti-HA–FITC antibody (panels 3, 6, and 9), from the anti-FLAG–Texas red antibody (panels 1, 4, and 7), or the overlap of both fluorescent images. Three cells are shown, one expressing only the FLAG-MLL protein (panels 1 to 3), one expressing both proteins (panels 4 to 6), and one expressing only the HA-Cyp33 protein (panels 7 to 9). It can be seen that the HA-Cyp33 protein is diffusely distributed in the nucleus when expressed alone but localizes to the MLL-containing speckles when expressed with FLAG-MLL.
Effects of Cyp33 overexpression on the regulation of MLL target genes.
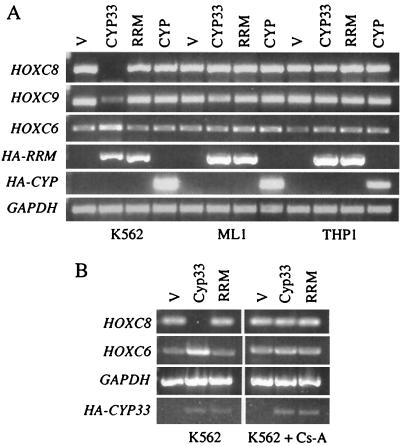
We wanted to study the possible effects of Cyp33 overexpression on the transcriptional regulation of target genes of MLL. For this purpose we set up the following experiment. We transfected a Cyp33-expressing plasmid into three leukemia cell lines: K562, ML-1, and THP1. K562 is derived from a patient with chronic myelogenous leukemia in blast crisis (21), which has two normal MLL alleles (13). ML-1 is derived from a patient with acute myeloblastic leukemia; these cells have two chimeric MLL-AF6 fusion genes without PHD fingers and with no normal MLL allele (31). THP1 is derived from a patient with monocytic acute myeloblastic leukemia, and it has an MLL-AF9 fusion gene and a normal MLL allele (23). The three cell lines express HOXC6, HOXC8, and HOXC9 mRNAs at levels detectable by RT-PCR. These three HOX genes are regulated by MLL in cells from MLL mutant mice (14). The overexpression of Cyp33 induced a marked down-regulation of HOXC8 and HOXC9 mRNA and induced an up-regulation of HOXC6 in the K562 cell line but produced no change in the expression of these genes in the ML-1 and THP1 cell lines (Fig. 7A). Overexpression either of the empty vector or of vectors expressing truncated forms of Cyp33 lacking either the cyclophilin domain or the RRM domain plus the spacer produced no significant effect in either of the three cell lines (Fig. 7A).
FIG. 7.
(A) RT-PCR products from mRNA of K562, ML-1, and THP1 human leukemia cells transfected with different plasmid constructs. V, the plasmid vector without an insert; Cyp33, an expression plasmid encoding Cyp33; RRM, an expression plasmid encoding a truncated version of Cyp33 that lacks the cyclophilin domain; Cyp, a truncated construct that lacks the RRM domain and the spacer but retains the cyclophilin domain. Different pairs of primers, specific for the genes HOXC8, HOXC9, HOXC6, and glyceraldehyde phosphate dehydrogenase (GAPDH), were used. In addition, a forward primer within the HA coding sequence and reverse primers within sequences encoding either the RRM domain or the cyclophilin domain of Cyp33 were used to detect the expression of mRNA encoding the fusion proteins by PCR. (B) RT-PCR products from the mRNA of K562 human leukemia cells transfected with the same plasmid constructs as in panel A and cultured with or without cyclosporine. The same PCR primers as in panel A were used for RT-PCR.
In order to determine if the effect of Cyp33 overexpression was dependent on the PPIase activity of Cyp33, we repeated the experiment in the presence of cyclosporine, a cyclophilin-specific PPIase inhibitor. Cyclosporine completely suppressed both the down-regulation of HOXC8 and the up-regulation of HOXC6 expression in the K562 cell line transfected with the Cyp33 expression construct. Nevertheless, cyclosporine had no effect on the expression of HOXC8 and HOXC6 in the controls transfected with empty vector or with the truncated version of Cyp33 without the cyclophilin domain (Fig. 7B) or on the ML-1 cells transfected with the same constructs (data not shown).
DISCUSSION
The PHD fingers mediate protein-protein interactions.
The presence of PHD fingers in many chromatin-associated proteins and their evolutionary conservation within different protein families suggested that they perform fundamental functions in these proteins. Nevertheless, it was not known if these domains interacted with nucleic acids, like many Krox-type Zn fingers, or with proteins. The results presented in this paper show that the PHD fingers could mediate protein-protein interactions.
The MLL and trx PHD fingers homodimerize.
The first three PHD fingers of both MLL and trx homodimerize in vitro in the yeast two-hybrid assay and more weakly in the mammalian cell two-hybrid assay. Even if the isolated PHD finger dimerization function is relatively weak, it may mediate dimerization of the intact protein in vivo, acting cooperatively with other dimerization domains like the SET domain (27). Dimerization of MLL and trx may have a role in the assembly of maintenance complexes after the components of such complexes are dispersed to the two daughter DNA strands during replication. Therefore, trx and MLL dimerization may be an important component of the mechanism of propagation of maintenance for active loci.
In all MLL fusion proteins associated with leukemia this dimerization domain is deleted from the functional product, as is the other known dimerization domain, the SET domain. Nevertheless, in some fusion proteins the partner protein contributes a dimerization domain (26, 32). It is possible that dimerization of the central region of MLL and trx, mediated by PHD2, allows molecules docked on the other PHD fingers, such as Cyp33, to target the partner MLL molecule in a way similar to that in which reciprocal phosphorylation of certain transmembrane receptors occurs after dimerization.
MLL binds Cyp33 through its third PHD finger.
The interaction of MLL and Cyp33 is conserved in Drosophila, where the third PHD finger of trx interacts with the Drosophila homologue of Cyp33 (Anderson et al., submitted). Our dissection of the interaction domains shows that the third PHD finger of MLL is necessary and sufficient for Cyp33 binding and that the first and second PHD fingers do not interact with Cyp33. The MLL PHD2 has, as a distinctive feature, a cluster of acidic amino acid residues in the first spacer and several acidic residues in the third spacer. These features are conserved with the third PHD fingers of the other sequenced proteins of the trx/MLL family (Fig. 1).
The interacting domain in Cyp33 was identified as the RRM domain. This interaction is specific to Cyp33, since the RRM domains of another RNA binding protein, Non-O (37), do not bind to the MLL PHD fingers. Even if RRM domains have been found to interact specifically with RNA, there are precedents for RRM domain proteins that interact without the intermediation of RNA, as in the case of the interaction between the Sxl and Sin Drosophila proteins (9). Sxl has two RRM domains while Sin, like MLL PHD2 (see Fig. 1), has two clusters of acidic residues in its sequence. It is possible that basic residues on the RRM domain interact electrostatically with the acidic residues in Sin and in MLL PHD3 and, alternatively, with the phosphate groups of RNA.
On the other hand, it has been reported that dimerization of Sxl through its RRM domains is mediated by RNA (28). Cyp33 has been reported to bind poly(A) or poly(U) polyribonucleotides (22) but our results suggest that the interaction between MLL and Cyp33 does not require RNA. Alternatively, it is possible that RNA competes with MLL for binding to the RRM domain of Cyp33. As proposed below, competition with nascent RNA transcripts could provide a mechanism for the recognition of transcriptionally active loci by the MLL complex.
Colocalization of MLL and Cyp33.
MLL has been shown to localize in nuclear subdomains described as irregular-size speckles identified by some authors as part of the nuclear matrix (6, 38). When Cyp33 is coexpressed in the same cells, it colocalizes with the MLL fragment. Nevertheless, for Cyp33 the discrete localization is dependent on simultaneous overexpression of MLL. In the absence of overexpressed MLL, Cyp33 localizes diffusely within the nucleus. This suggests that the localization of Cyp33 to specific nuclear subdomains is mediated by its binding to MLL. The concentration of endogenous MLL is too low to target detectable amounts of Cyp33 to the same domains. The nuclear sublocalization of Cyp33 does not seem to depend on its PPIase function, since a truncated form of the protein without the cyclophilin domain shows the same localization as the whole protein.
Overexpression of Cyp33 modulates the gene regulatory function of MLL.
The HOC8 and HOXC9 mRNA levels are down-regulated in Mll null fibroblasts. The HOXC6 mRNA is also down-regulated but to a lesser extent (14). Overexpression of Cyp33 from an expression vector results in down-regulation of the HOXC8 and HOXC9 mRNAs in the leukemia cell line K562, which has two normal MLL alleles. This down-regulation is inhibited by cyclosporine and is not observed in the leukemia cell lines ML-1 and THP1, which have mutant MLL alleles without PHD fingers and have either no normal MLL allele or one such allele, respectively. In addition, overexpression of truncated proteins that lack either the RRM domain and the spacer or the cyclophilin domain has no effect on the expression of these HOX genes. This is consistent with the observed down-regulation being mediated by the binding of Cyp33 to the PHD fingers of MLL and the targeting of the PPIase activity of its cyclophilin domain either to other domains of MLL or to associated proteins. The opposite effect, up-regulation, was observed for the HOXC6 mRNA, and this up-regulation was also eliminated by cyclosporine. It is possible that one of these two effects is indirect and could be explained if some of the HOX proteins act as repressors of the other HOX gene, i.e., if HOXC8 and/or HOXC9 repress the HOXC6 gene.
Since Cyp33 is present in the cell at a higher concentration than MLL, one can ask why a further increase in the concentration of Cyp33 would affect MLL function. The answer could be that Cyp33 can bind AU-rich RNA transcripts within the cell (22). Only when overexpressed will it titer all the RNA transcripts and then be able to bind to MLL.
The HOXC9 gene is an ortholog of the Drosophila gene AbdB. Down-regulation of AbdB expression after overexpression of the DmCyp33 protein in Drosophila SL1 cells has been reported (Anderson et al., submitted). abdB is a Hox gene regulated by Drosophila trithorax and polycomb group proteins. This observation suggests that modulation of Hox gene expression through the interaction of Cyp33 and trx and MLL proteins is a fundamental feature of epigenetic regulation across evolution.
The observation that the expression of HOXC8 is not altered by Cyp33 overexpression in ML-1 and THP1 cells, which have chimeric MLL genes, is consistent with the hypothesis previously put forward by Slany et al. (30) that the chimeric MLL proteins have constitutive transactivation activity. Nevertheless, our results suggest that this constitutive activity results not only from the possible contribution of an activation domain by the fusion partner (AF6 and AF9 in these cases) but also from the lack of negative regulation by factors normally bound to the PHD fingers, such as Cyp33.
There are precedents for the participation of cyclophilins in the modulation of gene regulation by transcription factors. Cyclophilin A has been reported to interact with the transcription factor YY1 and modulate its repressing or transactivating activity in a cyclosporine sensitive way (36). It is interesting that YY1 shares sequence similarity and similar DNA binding sites with Pho, a Drosophila Pc-G protein (5). Another cyclophilin, Cyp40, binds to the transcription factor c-myb and inhibits its DNA binding activity and transactivation activity in a cyclosporine dependent way (19). Cyp40 does not inhibit DNA binding of v-Myb, which has mutations in the Cyp40 binding domain. Cyp40 is also part of a chaperone complex associated with nuclear steroid receptors and is needed for its normal ligand interactions and consequent transactivation function (10, 24). A recent report (2) suggests that cyclophilin A and PIN, two prolyl isomerases, have a function in the regulation of repression by sin3-histone deacetylase complexes.
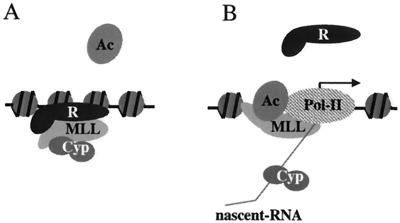
We do not know which domain of MLL, if any, is targeted by the cyclophilin domain of Cyp33, but the effect of Cyp33 on MLL or on its associated proteins may play a role in the initiation of maintenance by promoting repression by MLL complexes assembled on inactive loci. In one possible model, during the initiation of maintenance at inactive loci, Cyp33 would be bound to MLL, promoting repression and allowing the assembly of Pc-G complexes on such loci. At transcriptionally active loci, Cyp33 would be displaced from its association with MLL by AU-rich nascent RNA transcripts, allowing MLL to function as an activator that prevents the assembly of Pc-G repression complexes (Fig. 8). Consistent with this model, it has been reported that in Drosophila, noncoding RNA transcripts arise from most BX-C enhancer sequences, overlapping PREs and TREs, the sites of binding of trx-G and Pc-G protein complexes (20, 29); these transcripts have many A and U nucleotide stretches. Noncoding transcripts have been described also from at least one of the mammalian HOX gene clusters (15, 25). A prediction of this model is that the loss of the PHD fingers after splitting of the MLL gene by an 11q23 translocation would eliminate Cyp33 binding, turning MLL into a constitutive activator. This could result in alterations of MLL target gene expression, thus contributing to leukemia.
FIG. 8.
Model of the modulation of MLL effects on target genes by Cyp33. After binding the PHD fingers, Cyp33 may target activating or repressing domains of MLL to prevent binding of activators or to promote binding of repressors, thus enhancing the repressing functions of MLL. At transcriptionally active loci during early embryonic development, Cyp33 is sequestered onto the nascent transcripts and MLL is released from its regulation, thus becoming an activator.
ACKNOWLEDGMENTS
We are grateful to Richard Baer for providing the vector constructs for the mammalian two-hybrid system and to Yih-Sheng Yang for yeast two-hybrid control plasmids GBT-NonO and GAD-NonO. We thank Peter Harte and Stephanie L. Nelson for trithorax yeast two-hybrid constructs. We also thank Nancy Zeleznik-Le for GST-MLL fusion protein constructs and discussion of results and K. FitzGerald and U. Osmers for discussion and help in editing the manuscript.
This work was supported by U.S. Public Health Service Grant ROI CA38725 (to M.O.D.) and by Deutsche Forschungsgemeinschaft (SFB 388/B3) and the Fonds der Chemischen Industrie (to M.T.).
REFERENCES
- 1.Aasland R, Gibson T J, Stewart A F. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 2.Arevalo-Rodriguez M, Cardenas M E, Wu X, Hanes S D, Heitman J. Cyclophilin A and Essl interact with and regulate silencing by the Sin3-Rpd3 histone deacetylase. EMBO J. 2000;19:3739–3749. doi: 10.1093/emboj/19.14.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birney E, Kumar S, Krainer A R. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breen T R, Harte P J. Molecular characterization of the trithorax gene, a positive regulator of homeotic gene expression in Drosophila. Mech Dev. 1991;35:113–127. doi: 10.1016/0925-4773(91)90062-b. [DOI] [PubMed] [Google Scholar]
- 5.Brown J L, Mucci D, Whiteley M, Dirksen M L, Kassis J A. The Drosophila polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- 6.Butler L H, Slany R, Cui X, Cleary M L, Mason D Y. The HRX Proto-oncogene product is widely expressed in human tissues and localizes to nuclear structures. Blood. 1997;89:3361–3370. [PubMed] [Google Scholar]
- 7.Davis R E, Hardwick C, Tavernier P, Hodgson S, Singh H. RNA transplicing in flatworms. J Biol Chem. 1995;270:21813–21819. doi: 10.1074/jbc.270.37.21813. [DOI] [PubMed] [Google Scholar]
- 8.Djabali M, Selleri L, Parry P, Bower M, Young B D, Evans G A. A trithorax-like gene is interrupted by chromosome 11q23 translocations in acute leukemias. Nat Genet. 1992;2:113–118. doi: 10.1038/ng1092-113. [DOI] [PubMed] [Google Scholar]
- 9.Dong Z, Bell L R. SIN, a novel Drosophila protein that associates with the RNA binding protein sex-lethal. Gene. 1999;237:421–428. doi: 10.1016/s0378-1119(99)00303-0. [DOI] [PubMed] [Google Scholar]
- 10.Duina A A, Chang H-C J, Marsh J A, Lindquist S, Gaber R F. A cyclophilin function in Hsp90-dependent signal transduction. Science. 1996;274:1713–1715. doi: 10.1126/science.274.5293.1713. [DOI] [PubMed] [Google Scholar]
- 11.FitzGerald K T, Diaz M O. MLL2: a new mammalian member of the trx/MLL family of genes. Genomics. 1999;59:187–192. doi: 10.1006/geno.1999.5860. [DOI] [PubMed] [Google Scholar]
- 12.Grieco F, Hay J M, Hull R. An improved procedure for the purification of protein fused with glutathione S-transferase. BioTechniques. 1992;13:856–857. [PubMed] [Google Scholar]
- 13.Gu Y, Nakamura T, Alder H, Prasad R, Canaani O, Cimino G, Croce C, Canaani E. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to drosophila trithorax, to the AF-4 gene. Cell. 1992;71:701–708. doi: 10.1016/0092-8674(92)90603-a. [DOI] [PubMed] [Google Scholar]
- 14.Hanson R D, Hess J L, Yu B D, Ernst P, van Lohuizen M, Berns A, van der Lugt N M, Shashikant C S, Ruddle F H, Seto M, Korsmeyer S J. Mammalian Trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. Proc Natl Acad Sci USA. 1999;96:14372–14377. doi: 10.1073/pnas.96.25.14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh-Li H M, Witte D P, Weinstein M, Branford W, Li H, Small K, Potter S S. Hoxa11 structure, extensive antisense transcription, and function in male and female fertility. Development. 1995;121:1373–1385. doi: 10.1242/dev.121.5.1373. [DOI] [PubMed] [Google Scholar]
- 16.Jeanmougin F, Wurtz J-M, Le Douarin B, Chambon P, Losson R. The bromodomain revisited. Trends Biochem Sci. 1997;22:151–153. doi: 10.1016/s0968-0004(97)01042-6. [DOI] [PubMed] [Google Scholar]
- 17.Kennison J A. The polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu Rev Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- 18.Kim J O, Nau M M, Allikian K A, Makela T P, Alitalo K, Johnson B E, Kelley M J. Co-amplification of a novel cyclophilin-like gene (PPIE) with L-myc in small cell lung cancer cell lines. Oncogene. 1998;17:1019–1026. doi: 10.1038/sj.onc.1202006. [DOI] [PubMed] [Google Scholar]
- 19.Leverson J D, Ness S A. Point mutations in v-Myb disrupt a cyclophilin-catalyzed negative regulatory mechanism. Mol Cell. 1998;1:203–211. doi: 10.1016/s1097-2765(00)80021-0. [DOI] [PubMed] [Google Scholar]
- 20.Lipshitz H D, Peattie D, Hogness D S. Novel transcripts from the Ultrabithorax domain of the bithorax complex. Genes Dev. 1987;1:307–322. doi: 10.1101/gad.1.3.307. [DOI] [PubMed] [Google Scholar]
- 21.Lozzio C B, Lozzio B B. Human chronic myelogenous leukemia cell line with positive Philadelphia chromosome. Blood. 1975;45:321–334. [PubMed] [Google Scholar]
- 22.Mi H, Zimmerman E, Jäschke A, Tropschug M. A nuclear RNA binding cyclophilin in human T cells. FEBS Lett. 1996;398:201–205. doi: 10.1016/s0014-5793(96)01248-3. [DOI] [PubMed] [Google Scholar]
- 23.Odero M D, Zeleznik-Le N J, Chinwalla V, Rowley J D. Cytogenetic and molecular analysis of the acute monocytic leukemia cell line THP-1 with an MLL-AF9 translocation. Genes Chromosomes Cancer. 2000;29:333–338. [PubMed] [Google Scholar]
- 24.Owens-Grillo J K, Hoffmann K, Hutchison K A, Yem A W, Deibel M R, Handschumacher R E, Pratt W B. The cyclosporin A-binding immunophilin CyP-40 and the FK506-binding immunophilin hsp56 bind to a common site on hsp90 and exist in independent cytosolic heterocomplexes with the untransformed glucocorticoid receptor. J Biol Chem. 1995;270:20479–20484. doi: 10.1074/jbc.270.35.20479. [DOI] [PubMed] [Google Scholar]
- 25.Potter S S, Branford W W. Evolutionary conservation and tissue-specific processing of Hoxa11 antisense transcripts. Mammalian Genome. 1998;9:799–806. doi: 10.1007/s003359900870. [DOI] [PubMed] [Google Scholar]
- 26.Prasad R, Leshkowitz D, Gu Y, Alder H, Nakamura T, Saito H K, Berger R, Croce C M, Canaani E. Leucine-zipper dimerization motif encoded by the AF17 gene fused to ALL-1 (MLL) in acute leukemia. Proc Natl Acad Sci USA. 1994;91:8107–8111. doi: 10.1073/pnas.91.17.8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rozovskaia T, Rozenblatt-Rosen O, Sedkov Y, Burakov D, Yano T, Nakamura T, Petruk S, Ben-Simchon L, Croce C M, Mazo A, Canaani E. Self-association of the SET domains of human ALL-1 and of Drosophila TRITHORAX and ASH1 proteins. Oncogene. 2000;19:351–357. doi: 10.1038/sj.onc.1203307. [DOI] [PubMed] [Google Scholar]
- 28.Sakashita E, Sakamoto H. Protein-RNA and protein-protein interactions of the Drosophila sex-lethal mediated by its RNA-binding domains. J Biochem. 1996;120:1028–1033. doi: 10.1093/oxfordjournals.jbchem.a021495. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez-Herrero E, Akam M. Spatially ordered transcription of regulatory DNA in the bithorax complex of Drosophila. Development. 1989;107:321–329. doi: 10.1242/dev.107.2.321. [DOI] [PubMed] [Google Scholar]
- 30.Slany R K, Lavau C, Cleary M L. The oncogenic capacity of HRX-ENL requires the transcriptional transactivation activity of ENL and the DNA binding motifs of HRX. Mol Cell Biol. 1998;18:122–129. doi: 10.1128/mcb.18.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strout M O, Mrozek K, Heinonen K, Sait S N J, Shows T B, Aplan P D. ML-1 cell line lacks a germline MLL locus. Genes Chromosomes Cancer. 1996;16:204–210. doi: 10.1002/(SICI)1098-2264(199607)16:3<204::AID-GCC8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 32.Taki T, Hayashi Y, Taniwaki M, Seto M, Ueda R, Hanada R, Suzukawa K, Yokota J, Morishita K. Fusion of the MLL gene with two different genes, AF-6 and AF-5 alpha, by a complex translocation involving chromosomes 5, 6, 8 and 11 in infant leukemia. Oncogene. 1996;13:2121–2130. [PubMed] [Google Scholar]
- 33.Thirman M J, Gill H J, Burnett R C, Mbangkollo D, McCabe N R, Kobayashi H, Ziemin-van der Poel S, Kaneko Y, Morgan R, Sandberg A A, Chaganti R S K, Larson R A, Bean M L, Diaz M O, Rowley J D. Rearrangement of the MLL gene in acute lymphoblastic and acute myeloid leukemias with 11q23 chromosomal translocations. N Engl J Med. 1993;329:909–917. doi: 10.1056/NEJM199309233291302. [DOI] [PubMed] [Google Scholar]
- 34.Tkachuk D C, Kohler S, Cleary M L. Involvement of a homolog of Drosophila trithorax by 11q23 chomosomal translocations in acute leukemias. Cell. 1992;71:691–697. doi: 10.1016/0092-8674(92)90602-9. [DOI] [PubMed] [Google Scholar]
- 35.Wu L C, Wang Z W, Tsan J T, Spillman M A, Phung A, Xu X L, Yang M C, Hwang L Y, Bowcock A M, Baer R. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 36.Yang W-M, Inouye C J, Seto E. Cyclophilin A and FKBP12 interact with YY1 and alter its transcriptional activity. J Biol Chem. 1995;270:15187–15193. doi: 10.1074/jbc.270.25.15187. [DOI] [PubMed] [Google Scholar]
- 37.Yang Y S, Hanke J H, Carayannopoulos L, Craft C M, Capra J D, Tucker P W. NonO, a non-POU-domain-containing, octamer-binding protein, is the mammalian homolog of Drosophila nonAdiss. Mol. Cell Biol. 1993;13:5593–5603. doi: 10.1128/mcb.13.9.5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yano T, Nakamura T, Blechman J, Sorio C, Dang C V, Geiger B, Canaani E. Nuclear punctate distribution of ALL-1 is conferred by distinct elements at the N-terminus of the protein. Proc Natl Acad Sci USA. 1997;94:7286–7291. doi: 10.1073/pnas.94.14.7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeleznik-Le N J, Harden A M, Rowley J D. 11q23 translocations split the “AT-hook” cruciform DNA-binding region and the transcriptional repression domain from the activation domain of the mixed-lineage leukemia (MLL) gene. Proc Natl Acad Sci USA. 1994;91:10610–10614. doi: 10.1073/pnas.91.22.10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ziemin-van der Poel S, McCabe N, Gill H J, Espinosa III R, Patel Y, Harden A, Rubinelli P, Smith S D, Le Beau M M, Rowley J D, Diaz M O. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc Natl Acad Sci USA. 1991;88:10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]