Summary
Until menopause, women have a lower propensity to develop metabolic diseases than men, suggestive of a protective role for sex hormones. Although a functional synergy between central actions of estrogens and leptin has been demonstrated to protect against metabolic disturbances, the underlying cellular and molecular mechanisms mediating this crosstalk have remained elusive. By using a series of embryonic, adult-onset, and tissue/cell-specific loss-of-function mouse models, we document an unprecedented role of hypothalamic Cbp/P300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 (Cited1) in mediating estradiol (E2)-dependent leptin actions that control feeding specifically in pro-opiomelanocortin (Pomc) neurons. We reveal that within arcuate Pomc neurons, Cited1 drives leptin’s anorectic effects by acting as a co-factor converging E2 and leptin signaling via direct Cited1-ERα-Stat3 interactions. Together, these results provide new insights on how melanocortin neurons integrate endocrine inputs from gonadal and adipose axes via Cited1, thereby contributing to the sexual dimorphism in diet-induced obesity.
Keywords: leptin, estradiol, hypothalamus, ARC, Pomc, diet-induced obesity
Graphical abstract

Highlights
-
•
Global and hypothalamic Cited1 loss exacerbates diet-induced obesity in female mice
-
•
Cited1 is highly enriched in estradiol-sensitive melanocortin neurons
-
•
Hypothalamic Cited1 molecularly links the effects of E2 and leptin on food intake
-
•
Cited1 in PomcARC neurons acts as a downstream co-factor for E2-leptin signaling
Estradiol regulates energy homeostasis by altering feeding behavior, but how this sex hormone converges with the melanocortin system is not fully understood. Here, González-García et al. discover that Cited1 acts as a crucial hypothalamic co-factor mediating the anti-obesity effects of estradiol through potentiation of leptin’s anorectic actions. This neuroendocrine mechanism represents a mode by which melanocortin neurons integrate energy stores with reproductive signals for metabolic adaptation in diet-induced obesity.
Introduction
Multiple hormonal and nutritional cues converge in the hypothalamus to help determine and maintain an organism’s body weight. Via the exquisitely fine-tuned integration of endocrine signals, the melanocortin system, consisting of two antagonistic hypothalamic neuronal populations, the pro-opiomelanocortin (Pomc) and agouti-related protein (Agrp) neurons, senses and responds to circulating metabolites and hormones to regulate energy intake and expenditure.1,2,3,4 Unsurprisingly, genetic or acquired defects in the melanocortin system cause homeostatic impairments that can lead to the development of obesity.5,6,7 In particular, acquired leptin resistance in Pomc and Agrp neurons is considered a hallmark of diet-induced obesity,8 a finding that led to high interest in the identification of leptin sensitizers as potential anti-obesity drugs. Importantly, endogenous sex hormones, including estradiol (E2), have been reported to exert leptin-like anorectic effects9,10,11,12 by increasing the sensitivity of the organism to leptin’s actions within the melanocortin neurons to modulate energy homeostasis.13,14 Disruption of estrogen signaling via ovariectomy (OVX) or via estrogen receptor alpha (ERα) genetic ablation in many hypothalamic neuronal populations induces obesity by altering feeding behavior and energy expenditure,14,15,16 effects that strongly imply an interaction of E2 with leptin signaling.9,11,17 However, little is known about how E2 molecularly potentiates leptin’s ability to reduce food intake, an action that could explain why female mammals, including humans, are largely protected against metabolic diseases during reproductive age. A better understanding of how melanocortin neurons integrate and process sex-specific hormonal cues is crucial to comprehend how the hypothalamus is affected by diet-induced obesity, as well as to promote the development of sex-specific pharmacological or interventional therapies against obesity.
In these studies, we unravel a sex-specific weight-regulatory role for Cbp/P300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 (Cited1), a protein highly enriched in leptin-responsive melanocortin neurons specifically located in the arcuate nucleus of the hypothalamus (ARC). Cited1 was previously described as a transcriptional co-regulator of ERα in non-neuronal cells, displaying a gene-specific regulatory role.18,19 We further elucidate how Cited1 in ARC neurons plays a key role in mediating estrogen’s anti-obesity effect via a convergence with leptin signaling. Using global, constitutive or adult-onset hypothalamic, and Pomc-specific mouse models of Cited1 loss of function, we identified a sexually dimorphic estrogen-dependent phenotype in which females develop an exacerbated propensity to diet-induced obesity. Overall, we have document how Cited1, via ERα and Stat3 interactions, acts as a dual co-regulator of E2 and leptin signaling pathways in the hypothalamus, which allows E2 to regulate leptin sensitivity in Pomc neurons to protect against diet-induced obesity.
Results
Cited1 expression screening in a novel CRISPR-Cas9 knockin mouse model
To overcome the lack of specific Cited1 antibodies and to characterize the Cited1 protein expression profile, we generated a Cited1-HA knockin mouse model using CRISPR-Cas9 technology (Figure S1A). Using this mouse line, we found that the expression of Cited1 in the brain was restricted to the mediobasal hypothalamus (MBH), more specifically in the arcuate nucleus (ARC) (Figure 1A). In situ hybridization validated the model and expression pattern (Figure S1B). Cited1 expression was also observed in reproductive organs (uterus, ovary, epididymis, and testis), the kidney, the pituitary, the adrenal gland, and blood vessels from adipose tissue (Figure 1B), whereas it was not detected in the liver, skeletal muscle, adipocytes from subcutaneous white adipose tissue (WAT), gonadal WAT, or brown fat (Figure S1C).
Figure 1.
Central nervous system characterization and effect of Cited1 ablation on energy balance
(A) Left and lower right panels: representative images of a coronal brain section HA-Tag immunoreactivity (magenta) from Cited1-HA mice. Upper right panel: graphical representation of a mouse brain sagittal section. The dashed line indicates the anteroposterior location of the brain slice depicted in the immunofluorescence images from the same panel. Scale bars, 2 mm (left panel) and 200 μm (lower right panel).
(B) Representative confocal images depicting HA-Tag immunoreactivity (red) and DAPI (blue) in peripheral tissues of Cited1-HA mice. Scale bars, 500 μm (uterus, ovary, epididymis, testis, and kidney) and 200 μm (pituitary, adrenal gland, and blood vessels of the WAT).
(C and D) Body weight of WT or Cited1-KO male mice fed with chow or HFHS. n = 4–17 mice per group.
(E and F) Fat and lean mass, and relative scWAT, gWAT, and BAT depot weights of WT or Cited1-KO male mice fed with HFHS. n = 4–17 mice per group.
(G and H) Body weight of WT or Cited1-KO female mice fed with chow or HFHS. n = 3–16 mice per group.
(I and J) Fat and lean mass and relative scWAT, gWAT, and BAT depot weights of WT or Cited1-KO female mice fed with HFHS. n = 6–16 mice per group.
(K and L) Cumulative and total (light versus dark phase) food intake of WT or Cited1-KO female mice fed with HFHS. n = 5–7 mice per group.
(M and N) Time-dependent and total (light versus dark phase) locomotor activity of WT or Cited1-KO female mice fed with HFHS. n = 6–8 mice per group.
(O) ANCOVA of the total energy expenditure (72 h) versus body weight of WT or Cited1-KO female mice fed with HFHS. n = 6–8 mice per group.
(P) Relative body weight change (%) of WT or Cited1-KO male or female mice fed with HFHS. n = 6–16 mice per group.
(Q) Ratio of scWAT versus gWAT depot weights of WT or Cited1-KO male or female mice fed with HFHS. n = 5–17 mice per group.
(R) Relative body weight change (%) of WT, Cited1 (paternal heterozygous), Cited1 (maternal heterozygous), or Cited1-KO male mice fed with HFHS. n = 5–9 mice per group.
Data are expressed as mean ± SEM (C–N and P–R) and individual values (O). Statistical analyses include two-way ANOVA (C–N and P–R) and ANCOVA (O). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. 3V, third ventricle; ARC, arcuate nucleus of the hypothalamus.
Global loss of Cited1 elicits an obesity-prone phenotype in females, mimicking a “male-like” fat distribution
The Cited1 expression profile highlights its potential involvement in energy metabolism. To investigate this possibility, we obtained and validated a whole-body Cited1-knockout (Cited1-KO) mouse model (Figure S1D) and monitored several of its metabolic parameters over the course of 9 weeks under either a standard laboratory chow (chow) or a high-fat-high-sugar diet (HFHS) paradigm. Cited1-KO male and female mice had similar body weight to controls when fed the chow diet (Figures 1C and 1G). However, unlike what occurred in Cited1-KO males consuming HFHS (Figures 1D–1F and S1F–S1J), Cited1-KO female mice exhibited a significant susceptibility to develop obesity (Figure 1H), gaining more than twice as much body weight as the wild-type (WT) controls (Figure S1E). In vivo body composition analysis revealed that this increased body weight was exclusively due to higher fat mass (Figure 1I), and post-mortem analyses confirmed that both subcutaneous and gonadal fat pads were significantly heavier in Cited1-KO female mice (Figure 1J). This increased propensity for positive energy balance on HFHS was exclusive to Cited1-KO female mice, which also had increased food intake during the dark phase (Figures 1K and 1L), reduced ambulatory activity during the dark phase (Figures 1M and 1N), and diminished energy expenditure with relation to body weight (assessed by ANCOVA20,21) (Figure 1O). Interestingly, the relative weight curve of Cited1-KO female mice under HFHS completely overlapped those of male Cited1-KO and WT mice fed the same diet (Figure 1P). Importantly, the significantly altered weight ratio of the fat pads suggests that Cited1-KO female mice exhibit a “male-like” fat distribution pattern (Figure 1Q).
The obesity-prone phenotype of female Cited1-KO mice is not due to placental insufficiency
Cited1-KO mice have previously been suggested to suffer from placental insufficiency,22 a factor known to exacerbate the development of obesity in females.23 Thus, we next asked whether placental insufficiency is the underlying cause of the obesity-prone phenotype in Cited1-KO female mice. To address this, we took advantage of the specificity of X chromosome silencing (the chromosome on which Cited1 is located24), as the paternally inherited X chromosome is systematically the one silenced in the placenta of female mice. We thus generated two distinct cohorts of heterozygous female mice (Cited1-KO/+), with a transgene allele inherited either from the father (paternal heterozygous [pHet], WT placenta) or the mother (maternal heterozygous [mHet], Cited1-deficient placenta). Both groups of heterozygous female mice had a similar intermediate obese phenotype in comparison with homozygous KO or WT mice (Figure 1R), thus signifying that placental insufficiency does not underlie the obese phenotype of female Cited1-KO mice.
Cited1 regulation of energy metabolism is dependent on E2 signaling
The sexual dimorphism in response to HFHS exhibited by Cited1-KO mice with respect to weight gain and energy metabolism could be caused by a defect in ovarian hormone function, either via deficient hormone production or impaired hormonal response. To address this, we first collected plasma from HFHS-fed Cited1-KO mice and controls and measured several steroid hormones in both sexes. No changes in circulating steroid hormones were detected (Figures S2A–S2C), indicating proper hormone synthesis and release. To investigate whether the increased body weight of Cited1-KO female mice was linked with an altered response to ovarian hormones, we ovariectomized a cohort of Cited1-KO mice and controls and repeated the HFHS regimen. Ovariectomy completely abrogated the body weight differences between the two groups (Figures 2A and 2B), suggesting that the obesity-prone phenotype of ovary-intact Cited1-KO mice is estrogen-dependent. Conversely, E2 supplementation in males was less protective against weight gain in Cited1-KO compared with controls, suggesting specificity of impaired E2 signaling. Although differences in overall body weight did not reach statistical significance (Figures 2C and 2D), body composition analysis revealed a significantly higher fat mass in Cited1-KO males treated with E2 versus controls (Figure 2E). Using indirect calorimetry, the response of Cited1-KO male mice to E2 supplementation was diminished compared with controls, with Cited1-KO mice having lower physical activity and energy expenditure and no changes in food intake (Figures 2F–2J). Unchanged hypothalamic expression levels of estrogen receptors and aromatase (Figure S2D) in ovary-intact Cited1-KO females suggest that the impaired estrogen signaling lies downstream of hormonal generation and receptor expression. Collectively, these data strongly suggest that the role of Cited1 in the regulation of energy metabolism under HFHS is based on an E2-dependent mechanism and that it is conserved between sexes.
Figure 2.
Effect of estradiol manipulation on the Cited1 regulation in obesity
(A and B) Body weight and body weight gain of WT or Cited1-KO ovariectomized female mice fed with HFHS. n = 7–8 mice per group.
(C–E) Body weight, body weight gain, and fat and lean mass of WT or Cited1-KO male mice fed with HFHS and treated with E2 subcutaneously (s.c.) (2.8 μg/day). n = 6–9 mice per group.
(F and G) Cumulative and total (light versus dark phase) food intake of WT or Cited1-KO male mice fed with HFHS and treated with E2 s.c. (2.8 μg/day). n = 4–7 mice per group.
(H and I) Time-dependent and total (light versus dark phase) locomotor activity of WT or Cited1-KO male mice fed with HFHS and treated with E2 s.c. (2.8 μg/day). n = 4–6 mice per group.
(J) ANCOVA of the total energy expenditure (72 h) versus body weight of WT or Cited1-KO male mice fed with HFHS and treated with E2 s.c. (2.8 μg/day). n = 4–7 mice per group.
Data are expressed as mean ± SEM (A–I) and individual values (J). Statistical analyses include two-way ANOVA (A–I) and ANCOVA (J). ∗p < 0.05 and ∗∗p < 0.01.
Cited1 is highly and mainly expressed in E2-sensitive melanocortin neurons in the ARC
Given the remarkable abundance of Cited1 in the ARC (Figure 1A) and that E2 acts as an anti-obesity hormone mostly via its actions in the hypothalamus, we next investigated Cited1 expression levels under different conditions and successfully identified the specific Cited1-expressing cells in the ARC. First, we did not observe differences in Cited1 expression pattern, qualitative or quantitative, between biological sexes, suggesting that although the Cited1 gene is located on the X chromosome, it is not escaping the X-inactivation process in the hypothalamus (Figure S1D). Moreover, no differences in Cited1 expression in the hypothalamus were found to be following metabolic challenges such as a prolonged maintenance on HFHS (9 weeks) (Figure S4A), fasting, or refeeding at several time points (Figures S2E and S2F). Additionally, using C57Bl/6J WT mice, we observed that Cited1 was synchronized with the estrous cycle, showing a significant decrease in expression during proestrus (Figures S2G and S2H). To characterize Cited1-expressed cells, a publicly available single-cell sequencing database was reanalyzed to obtain further insight on Cited1 expression pattern in the ARC.25 With respect to cell type, Cited1 expression was observed in several neuronal populations and within a subset of tanycytes (Figure S3A). Using Cited1-HA mice, we confirmed that Cited1 was fundamentally restricted to neurons as no colocalization of Cited1 with either astrocyte or microglia markers was found, and minor co-expression being occasionally observed in tanycytes (Figure S3B). Stereological analysis revealed that Cited1-positive cells are predominantly located in the ARC (Figure S3C), and single-cell sequencing data indicated that Cited1 is highly expressed in select subpopulations of ARC neurons, with a marked expression in Agrp and Pomc neurons (Figure 3A). Importantly, we observed that Cited1 expression widely overlaps with ERα (Figures 3A–3C). We confirmed this using immunofluorescence and found that within the ARC, while not all Cited1-positive cells co-expressed ERα (54%), almost all (>99%) ERα-positive cells co-expressed Cited1 (Figures 3B and 3C).
Figure 3.
Effect of the loss of hypothalamic Cited1 on energy balance
(A) Violin plots depict the expression of Agrp, Pomc, Cited1, and Tubb3 genes across neuronal clusters identified by Campbell et al.25 Neurons were identified on the basis of expression of the canonical neuronal marker Tubb3 gene. The shape of the violin plot indicates the distribution of cell depending on their level of expression.
(B) Representative confocal micrographs depicting ERα (yellow) and HA-Tag (magenta) immunoreactivity in the ARC of Cited1-HA mice. Scale bars, 200 μm (upper panel) and 20 μm (lower panel).
(C) Upper panel: quantification of the relative number of hypothalamic ERα positive neurons which co-express Cited1 in Cited1-HA mice. Lower panel: quantification of the relative number of hypothalamic Cited1-positive neurons that co-express ERα in Cited1-HA mice. n = 4 mice per group; 6 sections/mouse.
(D and E) Body weight of control or HypΔCited1 male mice fed with chow or HFHS. n = 5–9 mice per group.
(F and G) Body weight of control or HypΔCited1 female mice fed with chow or HFHS. n = 6–13 mice per group.
(H and I) Relative scWAT, gWAT, and BAT weights, and fat and lean mass of control or HypΔCited1 female mice fed with HFHS. n = 8–13 mice per group.
(J and K) Cumulative and total (light versus dark phase) food intake of control or HypΔCited1 female mice fed with HFHS. n = 8–13 mice per group.
(L) Gene expression levels in the hypothalamus of control or HypΔCited1 female mice fed with HFHS. n = 6–11 mice per group.
(M) Upper panel: graphical representation of the experimental paradigm performed. Lower panel: food intake change percentage of ovariectomized control or HypΔCited1 female mice fed chow diet and treated either with vehicle or E2 s.c. (1 μg/mice). n = 5–6 mice per group.
(N) Schematic illustration and GFP immunofluorescence representative image depicting the stereotactical targeting of the mediobasal hypothalamus of Cited1loxP/loxP with AAV-GFP treatment. Scale bars, 200 μm.
(O–S) Body weight; relative scWAT, gWAT, and BAT depot weights; fat and lean mass; and cumulative and total (light versus dark phase) food intake of control or MBHΔCited1(AAV-Cre) female mice fed with HFHS. n = 4–10 mice per group.
Data are expressed as violin plot (A) mean ± SEM (D–M and O–S). Statistical analyses include two-way ANOVA (D–M and O–S). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, and ##p < 0.01. 3V, third ventricle; ARC, arcuate nucleus of the hypothalamus.
Deletion of Cited1 in hypothalamic neurons is sufficient to develop the obesity-prone phenotype in females
To investigate the role of hypothalamic Cited1 in energy metabolism, we obtained a mouse model of conditional Cited1 deletion from the EUCOMM (Cited1tm1c(EUCOMM)Hmgu, referred to as Cited1loxP/loxP) and crossed it with Nkx2.1-Cre mice to generate a neuronal and hypothalamus-specific Cited1 KO mouse (Nkx2.1-Cre;Cited1loxP/loxP, referred to as HypΔCited1, as validated in Figures S4A and S4B). The loss of Cited1 specifically within hypothalamic neurons replicated the phenotype observed in whole-body Cited1-KO mice, and it resulted in a sex- and diet-dependent obesity-prone phenotype. Male HypΔCited1 mice on chow or HFHS had similar body weight, food intake, fat pad weights, and body composition as their respective controls (Figures 3D, 3E, and S4C–S4L). Female HypΔCited1 mice on chow likewise had no metabolic phenotype apart from a slight increase in fat mass (Figures 3F, S4M, and S4N). In contrast, female HypΔCited1 mice on HFHS developed exacerbated obesity (Figure 3G), having increased subcutaneous and gonadal fat depot weight (Figure 3H), as well as higher total fat mass (Figure 3I). Their obese phenotype was linked to increased food intake, as observed in metabolic chambers over 72 h (Figures 3J and 3K) or throughout the whole diet protocol (Figure S4O). No changes in energy expenditure or locomotor activity were observed (Figures S4P–S4R). Interestingly, neuropeptides gene expression in the hypothalamus was characterized by a singular Pomc decrease under HFHS (Figure 3L), whereas Agrp, Npy, and Pomc were increased in mice fed chow (Figure S4S). Given that Nkx2.1-Cre-promoter-driven recombination can also be observed in the pituitary26 and that Cited1 expression was also observed in components of the hypothalamic-pituitary-adrenal (HPA) axis27 (Figure 1B), we measured HPA activation via circulating corticosterone levels and found no differences between HypΔCited1 mice and controls for both sexes and diets (Figure S4T). As Cited1 was found to be highly expressed in E2-sensitive ARC neurons (Figures 3A and 3B), and as E2 signaling in the hypothalamus has potent anorectic effects, we used OVX HypΔCited1 mice to determine whether Cited1 regulates the hypothalamic E2 effect on food intake. While OVX control mice displayed a clear reduction in food intake following E2 treatment, OVX HypΔCited1 mice had a blunted anorectic response to E2 (Figure 3M).
Next, we investigated whether the obesity-prone phenotype observed was consequent to the ablation of Cited1 specifically during the embryonic period,28 since Cited1 has been hypothesized to be implicated in the specification of ARC neuronal identities.29 We therefore generated an adult-onset hypothalamic KO mouse using a viral approach. An injection of adeno-associated viruses coding for a Cre recombinase successfully disrupted Cited1 expression in the MBH of Cited1loxP/loxP mice (referred to as MBHΔCited1(AAV-Cre); Figures 3N and S5A). When fed HFHS, the adult-onset deletion of Cited1 exclusively within MBH neurons led to exacerbated obesity in female mice as evidenced via body weight (Figure 3O), fat depot weight (Figure 3P), and body composition analysis (Figure 3Q). Using indirect calorimetry, we found increased food intake (Figures 3R and 3S) and reduced physical activity during the dark phase (Figures S5B and S5C). No significant differences were found in the relation between body weight and energy expenditure (Figure S5D). Interestingly, we also found exacerbated obesity in male MBHΔCited1(AAV-Cre) mice (Figures S5E–S5G). These results demonstrate that Cited1 disruption in mature and fully differentiated MBH neurons exacerbates metabolic disturbances associated with obesity.
Hypothalamic Cited1 is required for the food-intake-reducing effects of E2 and leptin, and potentiates the anorectic effect of estrogens upon HFHS-induced hyperleptinemia
Due to the importance of hypothalamic Cited1 for the anti-obesity effects of E2, we next assessed whether the deletion of Cited1 would impact leptin signaling as well, and thus, whether Cited1 in the hypothalamus exerts a protective role against obesity via estradiol-mediated leptin-sensitizing effects. In this regard, we first investigated the expression of the long leptin receptor isoform (LepR) in Cited1-expressing cells by generating Cited1-HA;LepR-Cre;Ai14-tdTomato mice. Using those mice, we confirmed the triple colocalization of LepR and ERα in arcuate Cited1-expressing neurons (Figure 4A). Intrigued by these results, we explored the possible role of Cited1 in mediating a crosstalk between E2 and leptin signaling by assessing the feeding response of chow-fed HypΔCited1 mice to exogenous leptin. While leptin induced an ∼20% decrease in food intake in both HypΔCited1 and control male mice (Figure 4B), it failed to suppress food intake in HypΔCited1 females (Figure 4C), indicative of hypothalamic defective leptin signaling.
Figure 4.
Cited1 mediates the crosstalk between estrogen and leptin signaling in the hypothalamus
(A) Representative confocal micrographs depicting HA-Tag (magenta), tdTomato (cyan), and ERα (yellow) immunoreactivity in the ARC of Cited1-HA;LepR-Cre;Ai14-tdTomato mice fed with chow diet. Scale bars, 200 μm (upper panel) and 20 μm (lower panel).
(B) Upper panel: graphical representation of the experimental paradigm performed. Lower panel: food intake change percentage of control or HypΔCited1 male mice fed chow diet and treated with vehicle and leptin intraperitoneally (i.p.) (3 mg/kg) (lower panel). n = 4 mice per group.
(C) Upper panel: graphical representation of the experimental paradigm performed. Lower panel: food intake change percentage of control or HypΔCited1 female mice fed chow diet and treated with vehicle and leptin i.p. (3 mg/kg). n = 5 mice per group.
(D) Upper panel: graphical representation of the experimental paradigm performed. Lower panel: food intake change percentage of ovariectomized control or HypΔCited1 female mice fed chow diet and treated either with vehicle s.c. and leptin i.p. (3 mg/kg) or E2 s.c. (1 μg/mice) and leptin i.p. (3 mg/kg) (lower panel). 10 = 12 mice per group.
(E) Upper panel: graphical representation of the experimental paradigm performed. Lower panel: food intake change percentage of control or HypΔCited1 male mice fed with chow diet and treated with E2 s.c. (1 μg/mice) and leptin i.p. (3 mg/kg). n = 4 mice per group.
(F) Serum leptin levels of control or HypΔCited1 female mice fed with chow or HFHS. n = 5–12 mice per group.
(G) Serum leptin levels versus fat mass correlation of control or HypΔCited1 female mice fed with chow or HFHS. n = 5–12 mice per group.
(H) Body weight of control or HypΔCited1 male mice fed with HFHS. The yellow bars indicate when the first and second E2-feeding behavior experiments were performed. n = 6–8 mice per group.
(I) Serum leptin levels of control or HypΔCited1 male mice fed with chow or HFHS for 1 week or 6-weeks HFHS. n = 6–8 mice per group.
(J) Upper panel: graphical representation of the experimental paradigm performed. Lower panel: food intake change percentage of control or HypΔCited1 male mice fed with HFHS and treated with E2 s.c. (1 μg/mice) after 1 week of HFHS exposure. n = 6–8 mice per group.
(K) Upper panel: graphical representation of the experimental paradigm performed. Lower panel: food intake change percentage of control or HypΔCited1 male mice fed with HFHS and treated with E2 s.c. (1 μg/mice) after 6 weeks of HFHS exposure. n = 6–8 mice per group.
Data are expressed as mean ± SEM (B–F and H–K) and individual values (G). Statistical analyses include two-way ANOVA (D, F, H, and I) and unpaired Student’s t tests (B, C, E, J, and K). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, and #p < 0.05. 3V, third ventricle.
To determine whether the sexually dimorphic effect of Cited1 on leptin action is mediated by E2, we next treated OVX HypΔCited1 female mice and OVX controls with both E2 and leptin. Unlike in the experiment performed in ovary-intact mice (Figure 4C), circulating E2 levels were this time controlled to maximize their effects by avoiding variations due to the estrous cycle. While exogenous E2 increased the anorectic effect of leptin in OVX controls, this potentiation effect was not observed in OVX HypΔCited1 female mice (Figure 4D). Similarly, male HypΔCited1 mice co-treated with E2 and leptin had attenuated decrease in food intake compared with controls (Figure 4E). Of note, no differences in circulating endogenous leptin levels were observed between HypΔCited1 mice and controls fed the chow diet (Figure 4F). Expectedly, as their fat mass is nearly doubled, the circulating leptin levels of HypΔCited1 mice on HFHS were significantly higher, leading us to use of chow-fed animals for these studies, in order to avoid this confounding factor (Figure 4F). In parallel, we observed a strong correlation (R2 = 0.82, p < 0.0001) between adiposity and leptin levels in HypΔCited1 and control female mice without any major group deviation, discarding a genotype-associated alteration in that regard (Figure 4G).
To further delineate the role of Cited1 with respect to E2 effects on feeding behavior in a context of normoleptinemia versus hyperleptinemia, we performed additional feeding assays in HypΔCited1 male mice treated with E2 after 1 week of HFHS (low leptin) and repeated the experimental paradigm after 6 weeks of HFHS (high leptin) (Figure 4H). First, we validated the expected diet-induced hyperleptinemia, which was proportional to the duration of HFHS feeding (Figure 4I). E2 treatment induced a weak but similar reduction in food intake of HypΔCited1 male mice and controls when fed a HFHS diet for 1 week (Figure 4J). Interestingly, the ability of E2 to decrease food intake was drastically increased in hyperleptinemic male mice fed HFHS for 6 weeks (−21% versus −3%) (Figures 4J and 4K). This synergy between E2 treatment and diet-induced hyperleptinemia was greatly reduced in HypΔCited1 male mice (−8% versus −3%) (Figures 4J and 4K). Overall, these findings support that Cited1 is involved in this hormonal crosstalk, which potentiates the capacity of E2 to decrease food intake in conditions of hyperleptinemia (Figure 4K).
Cited1 is a co-factor that converges hypothalamic E2 and leptin signaling
To verify that the hypothalamic response to leptin and estradiol is molecularly altered in the absence of Cited1, we quantified the number of neurons activated by E2-leptin co-treatment using a surrogate marker of neuronal activation, cFos, in our adult-onset hypothalamic Cited1 ablation model. Confirming the importance of Cited1 in the molecular integration of E2 and leptin signaling, there was a decreased number of cFos-positive cells in the ARC of MBHΔCited1(AAV-Cre) mice (Figure 5A).
Figure 5.
Molecular characterization of Cited1 as a co-factor of E2 and leptin signaling pathways in the hypothalamus
(A) Left: quantification of cFos+ neurons in the ARC of Cited1loxP/loxP female mice fed with chow diet and stereotactically injected with either AAV-Cre-GFP or AAV-GFP and treated with leptin i.p. n = 3–4 mice per group; 6 sections/mouse. Right: representative confocal micrographs depicting GFP (green) and cFOS immunoreactivity (magenta) in the ARC of control or MBHΔCited1(AAV-Cre) female mice fed with chow diet and treated with E2 s.c. (1 μg/mice) and leptin i.p. (3 mg/kg). Scale bars, 200 μm (right micrograph), 50 μm (left bottom micrograph), and 20 μm (left top micrograph).
(B and C) Western blot chemiluminescence images and protein level quantification of HA-Tag, Stat3, ERα, GAPDH, and histone H3 in hypothalamic nuclear and cytosolic fractions of Cited1-HA male mice fed with chow diet and treated either with vehicle s.c. or E2 s.c. (1 μg/mice) and vehicle i.p. or leptin i.p. (3 mg/kg). n = 3 mice per group.
(D and E) Western blot chemiluminescence representative images and protein level quantification of HA-Tag, Stat3, ERα, GAPDH, and histone H3 in hypothalamic nuclear and cytosolic fractions of Cited1-HA female mice fed with chow diet during metaestrus or proestrus and treated with either vehicle i.p. or leptin i.p. (3 mg/kg). n = 5 mice per group.
(F) Estradiol levels of Cited1-HA female mice during metaestrus or proestrus and Cited1-HA male mice treated with E2 s.c. (1 μg/mice) and fed with chow diet. n = 6–10 mice per group.
Data are expressed as mean ± SEM (A–F). Statistical analyses include two-way ANOVA (B–F) and unpaired Student’s t tests (A). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗∗p < 0.0001. 3V, third ventricle.
To further understand the molecular interactions between leptin and E2 signaling and the contribution of Cited1 to this process, we followed the intracellular dynamics of their main effectors, Stat3 and ERα. Using cellular fractionation, we tracked the translocation of Cited1, Stat3, and ERα from the cytosol to the nucleus in hypothalamic of Cited1-HA male mice treated with vehicle, E2, leptin, or E2 + leptin (Figures 5B and 5C). Strikingly, while modest translocation of all three proteins was observed with leptin or E2 treatment, a synergistic effect was seen in mice treated with E2 + leptin, where the observed translocation was considerably higher than the sum of the treatments performed separately. To better understand the implications in a more physiological context of fluctuating E2 plasma concentrations, we performed similar cellular fractionation studies in hypothalamic of female Cited1-HA mice treated with vehicle or leptin in different stages of their estrous cycle. Leptin treatment in the metaestrous stage, with low circulating E2 levels (Figures 5D and 5E), failed to induce the translocation of Cited1, Stat3, or ERα to the nucleus. In contrast, during the proestrous stage, with high circulating E2 levels (Figure 5F), leptin treatment induced the nuclear translocation of Cited1 and ERα (Figures 5D and 5E), as observed in E2-treated male mice (Figures 5B and 5C), which exhibited similar circulating E2 levels (Figure 5F).
Intrigued by these findings and considering the coherent dynamic of the leptin and estrogen signaling factors we observed (Figures 5B–5E), we further explored the molecular basis of this dynamic. Although Cited1 has been reported to directly bind to ERα in vitro18,19 and that Cited1 silencing is associated with reduced Stat3 activation,30 whether these proteins interact physically in the hypothalamus has never been assessed. To test whether Cited1 directly interacts with both Stat3 and ERα in vivo, we used hypothalami from Cited1-HA mice treated with vehicle, E2, leptin, or E2 + leptin to see whether Stat3 or ERα co-immunoprecipitate. Importantly, Cited1 physically interacts with both Stat3 and ERα (Figure S5H), suggesting a signaling integration via a molecular link between the signaling pathways of these two endocrine signals in the hypothalamus. Neither E2 nor leptin treatments altered the protein complex, suggesting that the physical interaction is stable under different hormonal conditions. We conducted similar Cited1-HA co-immunoprecipitation experiments in cycled Cited1-HA females (Figure S5I) with a similar outcome.
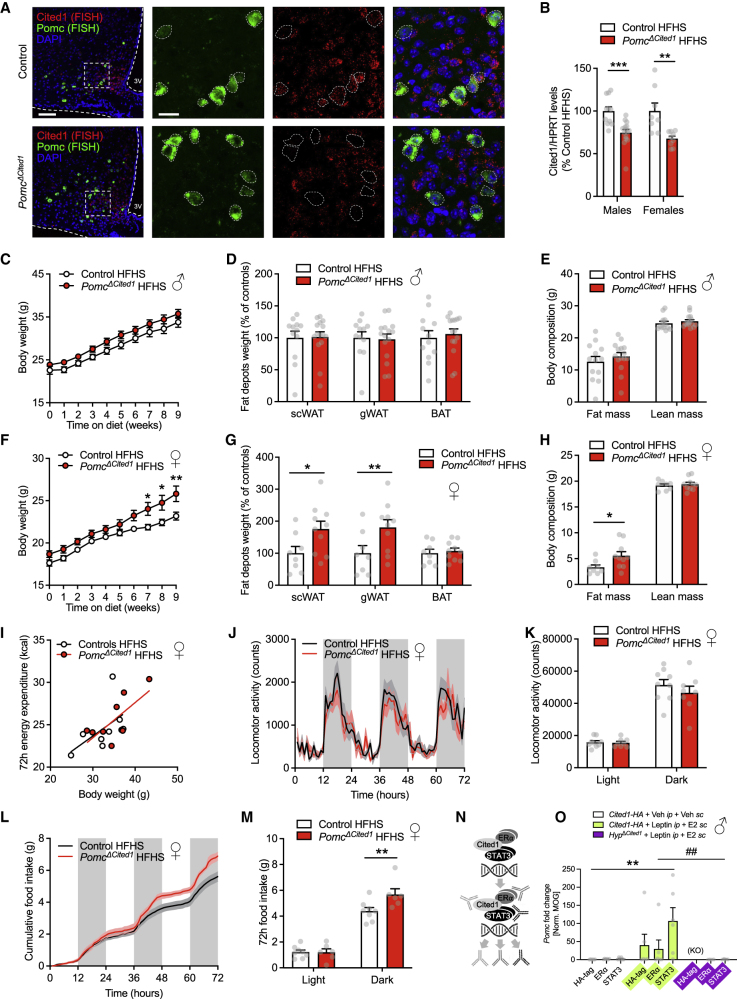
Cited1 ablation exclusively in Pomc neurons triggers an obesity-prone phenotype in females without affecting fertility
To delineate the neuronal population that drives the sexual dimorphism in energy homeostasis and convergence effects on E2 and leptin signaling, we next bred Cited1-HA mice with Pomc-GFP mice to assess the expression of Cited1 in Pomc neurons. Indeed, Pomc neurons are deeply regulated by leptin,8 are estrogen sensitive,31 and are tightly linked to energy balance.2 Thus, we hypothesized that they mediate the phenotype observed in our loss-of-function models. Histological analysis (Figure 6A) and iDisco 3D imaging reconstruction (Video S1) revealed that Cited1 was expressed in 87% and 78% of PomcARC neurons in males and females, respectively (Figure 6B), validating the expression pattern seen in single-cell sequencing reanalysis data (Figure 3A), and that Cited1-positive cells are anatomically positioned toward the lateral region of the ARC (Video S1). Moreover, we confirmed the colocalization of Cited1 and ERα in this neuronal population (Figure 6C). As only a subset of Pomc neurons express LepR, we next verified whether Cited1 was expressed in PomcLepR neurons by using a Cited1-HA;Pomc-GFP;LepR-Cre;Ai14-tdTomato mouse model and observed high colocalization of LepR with PomcCited1 neurons (Figure 6D). Roughly 10% of Cited1-expressing neurons in the ARC were both Pomc/LepR positive (Figure S5J). Looking specifically at the Pomc neuronal population, approximately 33% co-expressed LepR, consistent with the literature,32,33,34 and, interestingly, almost all of these Pomc/LepR-positive neurons co-expressed Cited1 (97%) (Figure S5K).
Figure 6.
Characterization of the Cited1 neuronal population and sex-dimorphic leptin action in the ARC hypothalamus
(A) Representative confocal micrographs depicting GFP (green) and HA-Tag (magenta) immunoreactivity in the ARC of Cited1-HA;Pomc-GFP mice. Scale bars, 200 μm (top) and 50 μm (bottom).
(B) Quantification of the relative number of hypothalamic Pomc neurons that co-express Cited1 in the Cited1-HA;Pomc-GFP male or female mice. n = 3 mice per group; 6 sections/mouse.
(C) Representative confocal micrographs depicting HA-Tag (magenta), GFP (cyan), and ERα (yellow) immunoreactivity in the ARC of Cited1-HA;Pomc-GFP;LepR-Cre;Ai14-tdTomato male mice fed with chow diet. Scale bars, 20 μm.
(D) Representative confocal micrographs depicting HA-Tag (blue), GFP (green), and tdTomato (red) immunoreactivity in the ARC of Cited1-HA;Pomc-GFP;LepR-Cre;Ai14-tdTomato male mice fed with chow diet. Scale bars, 200 μm (upper panel) and 20 μm (lower panel).
(E) Left: quantification of cFOS/GFP+ neurons in the ARC of Pomc-GFP male and female mice fed with chow diet and treated either with vehicle or leptin i.p. (3 mg/kg). n = 5–6 mice per group; 6 sections/mouse. Right: representative confocal micrographs depicting GFP (green) and cFOS immunoreactivity (magenta) in the ARC of Pomc-GFP male and female mice fed with chow diet and treated either with vehicle or leptin i.p. (3 mg/kg). Scale bars, 200 μm (left micrograph) and 20 μm (right micrographs).
(F) Quantification of GFP+ neurons in the ARC of Pomc-GFP male and female mice fed with chow diet. n = 9 mice per group; 6 sections/mouse.
Data are expressed as mean (B) and mean ± SEM (E and F). Statistical analyses include two-way ANOVA (E) and unpaired Student’s t tests (F). ∗∗p < 0.01, ∗∗∗∗p < 0.0001, and ##p < 0.01. 3V, third ventricle.
iDisco 3D-imaging reconstruction of the ARC showing Cited1-positive cells (magenta) and Pomc-GFP (green) neurons, related to Figure 6
In light of the sexually dimorphic response to leptin seen in our genetic models of Cited1 deletion, we then aimed to understand whether Pomc neurons exhibit an intrinsic differential response to leptin according to biological sex. In the Pomc-GFP reporter mouse model, female mice exhibit higher neuronal activation (cFos/Pomc-positive cells) upon leptin treatment than males, both in terms of absolute and relative numbers (Figures 6E and S5L). Furthermore, corroborating what others have reported,35 we also detected more Pomc-positive neurons in the hypothalamus of females in comparison with males (Figure 6F).
To test the functional importance of Cited1 in Pomc neurons, we next generated a constitutive Cited1 deletion mouse model exclusive to Pomc neurons (Pomc-Cre;Cited1loxP/loxP, referred to as PomcΔCited1). We validated the PomcΔCited1mouse model using fluorescence in situ hybridization and confirmed an absence of Cited1 mRNA in all ARC Pomc-positive neurons while the neighboring non-Pomc neurons still expressed it (Figure 7A). In addition, we found a 25.4% (males) and 32.3% (females) Cited1 reduction in the ARC by qPCR (Figure 7B), which closely matched the fraction of Cited1 neurons that are Pomc positive (26.7% [males] and 27.5% [females]; Figures S5M and S5N). Importantly, the PomcΔCited1 female mice recapitulated the female-specific diet-induced obese phenotype previously seen in the global, as well as in the hypothalamus-restricted, KO mice models, while no changes in metabolic parameters were observed in males (Figures 7C–7E and S6A–S6D). Female PomcΔCited1 developed a higher body weight, fat depot mass, global body fat mass content, and increased dark-phase food intake, with unchanged locomotor activity (Figures 7F–7M). To confirm the Pomc-specificity of the role of Cited1 in the melanocortin system, we generated two additional mouse models to first assess the expression of Cited1 in Agrp neurons (Cited1-HA;Npy-GFP mice, as Npy highly colocalizes with Agrp in the ARC36) and then to delete Cited1 exclusively in Agrp neurons (Agrp-Cre;Cited1loxP/loxP, referred to as AgrpΔCited1). Remarkably, although Cited1 is highly colocalized in Agrp neurons (Figures S6E–S6G), no metabolic phenotype was observed after its ablation in this neuronal population (Figures S6H and S6I) in males or females, suggesting that Cited1 expression specifically in Pomc neurons of the hypothalamus is key for E2-leptin signaling, and therefore in regulating energy homeostasis.
Figure 7.
Effect of Pomc neuron Cited1 KO in the development of diet-induced obesity
(A) Representative confocal micrographs depicting Cited1 mRNA (red), Pomc mRNA (green), and DAPI (blue) in the ARC of control or PomcΔCited1 mice using RNAscope. The dashed lines highlight the soma of Pomc-positive neurons. Scale bars, 200 μm (left micrograph) and 50 μm (right micrograph).
(B) Cited1 expression levels in the ARC of control or PomcΔCited1 male and female mice fed with HFHS. n = 8–15 mice per group.
(C–E) Body weight; relative scWAT, gWAT, and BAT depot weights; and fat and lean mass of control or PomcΔCited1 male mice fed with HFHS. n = 12–16 mice per group.
(F–M) Body weight; relative scWAT, gWAT, and BAT depot weights; fat and lean mass; ANCOVA of the total energy expenditure (72 h) versus body weight; time-dependent and total (light versus dark phase) locomotor activity; and cumulative and total (light versus dark phase) food intake of control or PomcΔCited1 female mice fed with HFHS. n = 6–10 mice per group.
(N and O) Schematic illustration of the HT-ChIPmentation assay, and DNA binding by HT-ChIPm-qPCR of HA-Tag, ERα, or Stat3 of hypothalamic samples from Cited1-HA and HypΔCited1 male mice fed with chow diet and treated either with vehicle s.c. or E2 s.c. (1 μg/mice) and vehicle i.p. or leptin i.p. (3 mg/kg). n = 4–6 mice per group.
Data are expressed as mean ± SEM (B–M and O). Statistical analyses include two-way ANOVA (B–M and O) and ANCOVA (I). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, and ##p < 0.01. 3V, third ventricle; FISH, fluorescence in situ hybridization; KO, knockout.
In order to exclude that pituitary expression of Cited1 (Figure 1B) may have an impact on the metabolic phenotype observed in PomcΔCited1 mice, we histologically assessed the pituitary from the Cited1-HA;Pomc-GFP mice. Although Cited1 and Pomc colocalize in the pars intermedia (Figure S6J), we did not find changes in circulating α-MSH (Figure S6K). Moreover, corticotrope cells of the pars distalis do not express Cited1 (Figure S6J), and no difference in plasma corticosterone was observed (Figure S6L).
As estrogen signaling in Pomc neurons is a crucial aspect of female reproduction,14 we assessed multiple parameters linked with fertility in all murine models used in this study (Figures S7A–S7H). We found that while litter size was reduced in whole-body Cited1-KO mice, which were reported to suffer from placental insufficiency,14 none of the constitutional hypothalamic/neuronal KO models showed changes in fertility markers, suggesting that hypothalamic Cited1 actions, although dependent on E2 signaling, are uncoupled from fertility.
Hypothalamic Cited1 is required for the Stat3 binding to the Pomc promoter upon E2 and leptin stimulation
Both Stat3 and ERα act as transcription factors that, upon their respective leptin and E2-induced activation and translocation, bind DNA at specific promotor regions to activate the transcription of target genes. As Cited1 effects on energy homeostasis and E2-leptin signaling appear to be mediated by Pomc neurons, and since one common target of Stat3 and ERα is the Pomc gene,37,38 we next proceeded to assess whether a Stat3-ERα-Cited1 complex could regulate the transcriptional effects of E2 and leptin. Thus, by interrogating whether the aforementioned transcriptional complex directly binds to a consensus sequence described for Stat3 at the Pomc promoter, we performed a modified high-throughput (HT) ChIPmentation-qPCR for Stat3, ERα, or Cited1-HA-Tag using hypothalami from Cited1-HA and HypΔCited1 mice treated with vehicle or with E2 + leptin (Figure 7N). We found no evidence of DNA binding for Stat3, ERα, or Cited1 at the Pomc promoter under basal unstimulated conditions (Figure 7O). However, upon E2 + leptin stimulation, binding to the consensus sequence was enriched in the ChIP-qPCR fractions. In the absence of Cited1, the same E2 + leptin treatment failed to increase Stat3 or ERα recruitment to the Pomc promoter, indicative of the formation of an active transcriptional complex with Cited1 as essential element (Figure 7O). Together, these results indicate that Cited1 acts as an essential co-factor in the crosstalk between E2 and leptin signaling within the hypothalamus, including its transcriptional effects.
Discussion
Although Cited1 has been previously reported to be highly enriched in melanocortin neurons of the ARC,25,39,40,41 its role in the hypothalamus in general and in the control of energy homeostasis specifically, has remained unknown until now. Here, we demonstrate that Cited1 in Pomc neurons is a key molecular driver of the sexual dimorphism in the control of feeding and response to hyperleptinemia in diet-induced obesity. Specifically, in this study, we highlight the role of Cited1 in the convergence of E2 and leptin signaling pathways in PomcARC neurons to induce the feeding responses required for the maintenance of energy homeostasis in a hypercaloric environment.
Multiple studies have pointed out that premenopausal females are largely protected against diet-induced obesity and have established a link between the vulnerability to develop metabolic disturbances with reproductive decline,42,43,44 suggesting that estrogens play a protective role in this context. Interestingly, chronic consumption of dense and palatable food rapidly and primarily affects hypothalamic feeding circuitry, including PomcARC neurons, by weakening their capacity to respond adequately to several metabolic hormones such as leptin.
Using multiple novel mouse models, we here identified Cited1 as a crucial factor in PomcARC neurons for integrating the anti-obesity effects of E2 and leptin signaling. We observed that global and hypothalamic (both in utero- and post-natal) ablation of Cited1 in mice consistently resulted in a diet- and sex-dependent obese phenotype, whereas only female mice developed the obesity-prone phenotype, and whereas this phenotype could only be observed in conditions of diet-induced hyperleptinemia. The mechanisms underlying Cited1 actions were subject to functional estrogen signaling and conserved between sexes, as evidenced by our studies in ovariectomized female mice combined with E2 supplementation and the diminished protective effect of exogenous E2 in Cited1-deficient male mice. Cited1 is highly expressed in both Pomc and Agrp neuronal populations, yet only the loss of Cited1 in Pomc neurons exhorts the obesity-prone phenotypes in females in response to diet-induced obesity observed in global and hypothalamic Cited1 loss-of-function models. These results, documenting that Cited1 acts as an E2-dependent downstream molecular link in the leptin signaling pathway, could explain why Cited1 deletion phenotypes are exclusively found in a hyperleptinemic state such as diet-induced obesity. Accordingly, we propose that Cited1 is the molecular downstream factor underlying the hormonal crosstalk between E2 and leptin, which potentiates the capacity of E2 to decrease food intake in conditions of hyperleptinemia. Consequently, both the presence of high E2 levels (occurring along the estrous cycle of reproductive females) together with high leptin levels (occurring with HFHS) are necessary neuroendocrine stimuli, which protect females against diet-induced obesity. Consequently, the lack of high circulating E2 levels in males justifies their higher susceptibility to diet-induced obesity and also explains the absence of an obese phenotype in HFHS-fed Cited1 KO males.
With respect to metabolic disturbances and E2 signaling, the importance of Cited1 specifically in Pomc neurons is consistent with previous studies, which have reported the presence of ERα only in Pomc neurons45 and whose ablation, specifically in this melanocortin population, promotes metabolic perturbances not found when deletion occurs in Agrp neurons.14 Importantly, deletion of Cited1 in Pomc neurons does not fully replicate the phenotype observed with ERα deletion,14 and particularly with respect to fertility abnormalities, and that only changes in bodyweight were observed upon a HFHS setting. Therefore, our findings support the concept that estradiol exerts its effects on energy balance and reproduction through non-necessary shared mechanisms, which could hold promising translational potential.
Recent insights have highlighted that Pomc neurons display a high level of heterogeneity where a diverse expression of receptors, neuropeptides, and neurotransmitters facilitate their key role as integrators for different neuroendocrine signals into a single coherent output.3,33,34 Consistent with reports of others who have found that Pomc-specific subtypes drive sexual dimorphism in energy homeostasis,35,46 our results progress forward, indicating that E2-sensitive Cited1-expressing Pomc neurons could represent a specialized population with another layer of complexity that integrates endocrine signals simultaneously from the gonadal and adipose axes to regulate food intake and body weight in response to calorie-dense diets.
The mechanisms described here offer the first molecular link between hypothalamic E2 signaling and leptin satiety effects. Consistent with previous evidence describing Cited1 as a selective ERα coactivator,18,47 we further explored how Cited1 exerts cell- and gene-specific regulation of estrogen-dependent transcription. Given that both Cited148,49,50 and Stat351,52 bind to CBP/p300 coactivator proteins to induce transcription, we used a novel mouse model with a HA-Tag peptide linked to Cited1 to demonstrate its interactions and cooperative effects with E2 on ERα and leptin on Stat3. The physical interaction of Cited-1 with both ERα and Stat3, together with the co-translocation to the nucleus and the DNA binding to the Stat3-consensus target sequence within the Pomc promoter proposed in this work, and in accordance with previous reports,12,18,19,47,48,50,51,52,53,54,55,56 suggest the existence of an integrating and activating a transcriptional complex formed by Stat3-ERα-Cited1. It remains unclear whether Cited1 contributes to the translocation of Stat3 and/or ERα and whether other factors such as CBP/p300 may be required for the initiation of transcription. Complexation of co-transcription factors is a frequent and highly conserved mechanism across signaling pathways which allows the fine-tuning of transcriptional events. Specific cooperation between co-factors and transcription factors dictates the expression of critical targets genes upon specific stimuli in a cell-dependent manner. Overall, our results indicate that Cited1 represents a transcriptional co-factor directing the molecular integrating scaffold between E2-ERα and leptin-Stat3 signaling pathways in Pomc neurons of the ARC, to mediate the converging effects of estradiol and leptin on food intake and body weight.
In summary, we here show that PomcARC neuronal Cited1 is a key molecular effector in the integration of endocrine signals from gonadal and adipose axes, which in turn is crucial for the appropriate control of feeding behavior in diet-induced obesity. This is accomplished via estradiol-dependent molecular interactions involving ERα and Stat3 that modulate the neuronal response to leptin and provides new insights into the pathophysiological role of both hormones in the development of obesity. Sexual dimorphism and female physiology are underrepresented research topics, and the findings reported here on the role of hypothalamic Cited1 significantly improve our understanding of sexual differences in the pathophysiology of obesity.
Limitations of study
In this study, we found two neuronal populations apart from Pomc that distinctly express both Cited1 and ERα in the ARC: Kisspeptin and Ghrh. Considering the functional interaction described, we cannot completely rule out some influence in the HypΔCited1 phenotype. The overall stronger phenotype seen in MBHΔCited1(AAV-Cre) mice, as well as the modest phenotype observed in HFHS-fed males, could be indicative of a role for Cited1 in energy metabolism in these neurons and of compensatory mechanisms. Interestingly, the presence of Cited1 in these neurons could suggest that this gene might participate in reproduction or growth. Despite tanycytes displaying a minor Cited1 expression, the biological meaning of this was not studied in the current project. Further studies should be conducted to address the potential role of Cited1 in these glial cells. On the other hand, the thorough evaluation of leptin and E2 crosstalk in vivo is challenging, in part due to endogenous hormones, and requires several mouse models. Our experiments demonstrate the presence of a leptin-sensitizing effect of E2 which was dependent on Cited1, but the contribution of this synergy to the observed phenotype remains unclear. The ideal paradigm to solve this question would require breeding a triple mutant mouse line deficient for leptin (i.e., ob/ob mice), together with the HypΔCited1 or PomcΔCited1, and perform ovariectomies. However, in addition to the breeding challenge, the massive obese phenotype that these mice would exhibit could hinder our ability to adequately measure their response to E2 and leptin. Future studies should be conducted to better understand the transcriptomic landscape influenced by Cited1 in Pomc neurons using cell-type-specific isolation approaches, RNA sequencing methods, and a careful combination of hormonal treatment paradigms. Finally, our findings were exclusively obtained via mouse models. Therefore, while Cited1 is also present in humans,50 it would be necessary to evaluate Cited1 genetic signatures and associated phenotypes to understand the full translational significance and clinical potential to humans.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-HA-Tag (C29F4) | Cell Signaling | #3724; RRID: AB_1549585 |
| Alexa Fluor 594 rabbit anti-HA-Tag (C29F4) | Cell Signaling | #45346; RRID: AB_2924897 |
| HA-Tag (C29F4) Rabbit mAb (HRP Conjugate) | Cell Signaling | #14031; RRID: AB_2798368 |
| Rabbit anti-cFos | Synaptic Systems | #226003; RRID: AB_2231974 |
| Chicken anti-GFP | Acris | #AP31791PU-N; RRID: AB_11145456 |
| Goat anti-GFAP | Sigma-Aldrich | #SAB2500462; RRID: AB_10603437 |
| Goat anti-Iba1 | Novus/Biotechne | #NB100-1028; RRID: AB_521594 |
| Chicken anti-Vimentin | Abcam | #ab24525; RRID: AB_778824 |
| Rabbit anti-ERα | Abcam | #ab32063; RRID: AB_732249 |
| Goat anti-Perilipin-1 | Abcam | #ab61682; RRID: AB_944751 |
| Mouse anti-Stat3 | Cell Signaling | #9139; RRID: AB_331757 |
| Rabbit anti-Stat3 (D3Z2G) | Cell Signaling | #12640; RRID: AB_2629499 |
| Mouse anti-β-actin | Santa Cruz Biotechnology | #sc-47778; RRID: AB_626632 |
| Mouse anti-GAPDH | Santa Cruz Biotechnology | #sc-166545; RRID: AB_2107299 |
| Rabbit anti-Histone3 | Abcam | #ab1791; RRID: AB_302613 |
| Alexa Fluor 647 donkey anti-rabbit | Invitrogen-Thermofisher | #A-31573; RRID: AB_2536183 |
| Alexa Fluor 488 donkey anti-goat | Invitrogen-Thermofisher | #A-11055; RRID: AB_2534102 |
| Alexa Fluor 488 goat anti-chicken | Invitrogen-Thermofisher | #A-11039; RRID: AB_2534096 |
| Alexa Fluor 488 donkey anti-chicken | Jackson ImmunoResearch | #703-545-155; RRID: AB_2340375 |
| HRP goat anti-mouse | Agilent-Dako | #P0447; RRID: AB_2617137 |
| HRP goat anti-rabbit | Agilent-Dako | #P0448; RRID: AB_2617138 |
| Bacterial and virus strains | ||
| AAV2/1-EF1a-EGFP-T2A-iCre | Vector Biolabs | VB2069 |
| AAV2/1-EF1a-EGFP | Vector Biolabs | VB2084 |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant mouse leptin | R&D systems | 498-OB |
| β-Estradiol 3-benzoate | Sigma-Aldrich | E8515 |
| 17β-Estradiol with biodegradable carrier binder | Innovative Research of America | NE-121 |
| Sesame oil | Sigma Aldrich | #S3547 |
| Paraformaldehyde | Carl Roth | 0335.2 |
| Bovine serum albumin | Sigma-Aldrich | A8806 |
| Gelatin from porcine skin (SUMI) | VWR International | SAFSG1890 |
| Triton X-100 | Roche Diagnostics | 11858620 |
| Phalloidin-iFluor 488 Reagent | Abcam | #ab176753 |
| Critical commercial assays | ||
| Mouse/Rat Leptin ELISA | Alpco | 22-LEPMS-E01 |
| RNeasy Mini Kit | QUIAGEN | 74104 |
| Parameter corticosterone assay | R&D systems | KGE009 |
| MSH, alpha (Human, Rat, Mouse) - EIA Kit | Phoenix Pharmaceuticals | EK-043-01 |
| Mouse/Rat Estradiol ELISA Kit | Calbiotech | ES180S-100 |
| Fast Panoptic kit | PanReac AppliChem | 254807 |
| RNA Scope Multiplex Fluorescent Reagent Kit v2 | Advanced Cell Diagnostic | #323100-USM |
| AbsoluteIDQTM Stero17 Kit | Biocrates | N/A |
| TaqMan Universal Master Mix II, no UNG | Thermo Fisher | 4440040 |
| RNA Scope Multiplex Fluorescent Reagent Kit v2 | Advanced Cell Diagnostic | #323100-USM |
| Deposited data | ||
| Data S1 | This paper | N/A |
| Experimental models: Organisms/strains | ||
| Cited1-KO mouse | EUCOMM (HMGU) | Cited1tm1b(EUCOMM)Hmgu |
| Cited1loxP/loxP mouse | EUCOMM (HMGU) | Cited1tm1c(EUCOMM)Hmgu |
| Nkx2.1-Cre mouse | Jackson Laboratory | #008661 |
| Pomc-Cre mouse | Jackson Laboratory | #5965 |
| Agrp-Cre mouse | Jackson Laboratory | #012899 |
| Pomc-GFP mouse | Jackson Laboratory | #009593 |
| Npy-GFP mouse | Jackson Laboratory | #006417 |
| LepR-Cre mouse | Jackson Laboratory | #008320 |
| Ai14-tdTomato mouse | Jackson Laboratory | #007914 |
| Wild-type mouse | Janvier | C57BL/6JRj |
| Cited1-HA mouse | This paper | N/A |
| Nkx2.1-Cre:Cited1loxP/loxP mouse | This paper | N/A |
| Pomc-Cre:Cited1loxP/loxP mouse | This paper | N/A |
| Agrp-Cre:Cited1loxP/loxP mouse | This paper | N/A |
| Cited1-HA;Pomc-GFP mouse | This paper | N/A |
| Cited1-HA;Npy-GFP mouse | This paper | N/A |
| Cited1-HA;LepR-Cre;Ai14-tdTomato mouse | This paper | N/A |
| Cited1-HA;Pomc-GFP;LepR-Cre;Ai14-tdTomato mouse | This paper | N/A |
| Oligonucleotides | ||
| Cited1-HA mice related oligonucleotides (See Table S1) | N/A | N/A |
| Agrp TaqMan Gene Expression Assay | Thermofisher | Mm00475829_g1 |
| Cited1 TaqMan Gene Expression Assay | Thermofisher | Mm01235642_g1 |
| Cyp19a1 TaqMan Gene Expression Assay | Thermofisher | Mm00484049_m1 |
| Esr1 TaqMan Gene Expression Assay | Thermofisher | Mm00433149_m1 |
| Esr2 TaqMan Gene Expression Assay | Thermofisher | Mm00599821_m1 |
| Hprt TaqMan Gene Expression Assay | Thermofisher | Mm01545399_m1 |
| Npy TaqMan Gene Expression Assay | Thermofisher | Mm03048253_m1 |
| Pomc TaqMan Gene Expression Assay | Thermofisher | Mm00435874_m1 |
| Socs3 TaqMan Gene Expression Assay | Thermofisher | Mm00545913_s1 |
| RNAscope Probe- Mm-Cited1 | Advanced Cell Diagnostic | #432471 |
| RNAscope Probe- Mm-Cited1-E2E3 | Advanced Cell Diagnostic | #400451 |
| Software and algorithms | ||
| Fiji 2.0.0 | NIH | https://imagej.net/Fiji/Downloads |
| GraphPad Prism 7.0 | GraphPad Software | N/A |
| IMARIS x64 9.1.2 | Oxford instruments | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Cristina García-Cáceres (garcia-caceres@helmholtz-muenchen.de).
Materials availability
Mouse lines generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Mice
Animal experiments were based on power analyses to assure adequate sample sizes, approved by the State of Bavaria, Germany and followed the guidelines of the Canadian Council on Animal Care. Male and female mice were group-housed on a 12:12-h light-dark cycle, under controlled temperature (23 °C) and humidity conditions with ad libitum access to standard chow diet (5.6% fat; LM-485, Harlan Teklad) and water unless indicated otherwise. The diet induced obesity paradigm was performed by switching mice aged 8 weeks old to a high-fat, high-sucrose (HFHS) diet (D12331; 58% of calories from lipids; Research Diets, New Brunswick, NJ). Weekly health status checks were performed for the mice.
Cited1 whole-body knockout mice (Cited1tm1d(EUCOMM)Hmgu), genetic background C57BL/6N, were obtained from InfraFrontier/EMMA. Conditional knockouts (Cited1loxP/loxP) were generated by crossing Cited1tm1a(EUCOMM)Hmgu mice, genetic background C57BL/6N, provided by InfraFrontier/EMMA with C57BL/6-Tg(CAG-Flpe)2Arte mice (Taconic model #7089, genetic background C57BL/6N). The Flp deleter transgene was subsequently bred out of the line. Wildtype C57BL/6J mice were obtained from Janvier, Le Genest-Saint-Isle, France.
Tissue specific Cited1 knockout mice were generated by crossing Nkx2.1-Cre (Jax mice stock #008661, genetic background C57BL/6J), Pomc-Cre (Jax mice stock #5965, genetic background C57BL/6J), and Agrp-Cre mice (Jax mice stock # 012899, genetic background C57BL/6J) with Cited1loxP/loxP mice to generate Nkx2.1-, Pomc-, and Agrp-specific Cited1-knockout mice (HypΔCited1, PomcΔCited1, or AgrpΔCited1). Littermate controls were used in all studies with the exception of whole-body hemi/homozygous Cited1 knockout mice vs. wildtype mice, due to breeding limitations linked with the location of Cited1 being on the X-chromosome.
For the neuronal visualization studies, Cited1-HA mice (generated ad hoc for this project) were crossed with mice that selectively express the green fluorescent protein (GFP) in Npy-expressing neurons (Npy-GFP, Jax mice stock #006417, genetic background C57BL/6J) or in Pomc-expressing neurons (Pomc-GFP Jax mice stock #009593, genetic background C57BL/6J). Colocalization studies of the long-form leptin receptor were conducted using the Cited1-HA;LepRb-Cre;Ai14-tdTomato and Cited1-HA;Pomc-GFP;LepRb-Cre;Ai14-tdTomato reporter mouse models, which were obtained from inter-crossing Cited1-HA mice (generated ad hoc for this project), Pomc-GFP mice (Jax mice stock #009593), B6.129(Cg)-LepRtm(cre)Rck/J mice (Jax mice stock #008320, genetic background C57BL/6J), and B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J mice (Jax mice stock #007914, genetic background C57BL/6J).
Generation of a Cited1-HA knock in mouse
To target a HA encoding sequence to the transcriptional start site of mouse Cited1, a gRNA was designed using CRISPOR tool (http://www.crispor.tefor.net/), in vitro transcribed (E3322, NEB), and purified using the MEGAclear Kit (ThermoFisher Scientific). The donor template for homologous recombination contained 53 and 59 nucleotides homology arms (5′ and 3′ respectively), between which the HA sequence was inserted. For generating the Cited1-HA knock-in mice, C57BL/6n females at 3 weeks of age were superovulated and bred with C57BL/6N males. Fertilized oocytes were prepared and electroporated (NEPA21) with 200 ng/μl of sgRNA, 200 ng/μl recombinant NLS-Cas9 protein (PNA Bio), and 30 ng/μl of ssODN templates (IDT) for homology directed repair. Electroporated zygotes were surgically implanted into recipient CD1 females. For genotyping, ear biopsies from the resulting mice were lysed using standard conditions and genomic DNA purified. The target gene was amplified using the primers listed (Table S1) and Sanger sequenced to confirm the nature of the HA-Tag insertion.
Method details
Physiological measures
Body composition (fat and lean mass) was measured using quantitative nuclear magnetic resonance technology (EchoMRI, Houston, TX). Food intake, energy expenditure, and respiratory exchange ratio were measured using metabolic cages from TSE Systems (TSE PhenoMaster, TSE Systems, Bad Homburg, Germany). The mice were habituated to the drinking bottles one week prior to the experiment. The animals spent 5 days in the metabolic cages, and data from the habituation period (first 48h) were discarded. O2 consumption, CO2 production, locomotor activity, and food intake were measured every 10 min. Energy expenditure (kcal over 72h) values were correlated to the body weight of the animals recorded at the end of the measurement using analysis of co-variance (ANCOVA).
Estrogen pellets supplementation
We implanted estradiol pellets in male mice at 6 weeks of age. Animals were anesthetized using isoflurane, a small subcutaneous incision was performed in the left mid-dorsal region, and an estradiol pellet (17β-estradiol pellets, 0.25 mg, 90 Days, Catalog #: NE-121, Innovative Research of America) was implanted subcutaneously. Sham-operated animals were similarly treated with the exclusion of pellet implantation.
Ovariectomy
Mice were bilaterally ovariectomized (OVX) or Sham-operated as previously described.57,58 Followingketamine–xylazine anesthesia (50 mg/kg, intraperitoneal, IP) both flanks were shaved, and mice were placed on their left side. Then, an incision through the skin and muscle layers was made in the right flank caudal to the last rib. The ovary was then carefully pulled out and removed after making an incision between the distal part of uterine horn and the ovary. After returning the horn into the abdominal cavity, the wound was closed by suturing the muscle layer and skin with surgical silk. The same procedure was applied to remove the left ovary. Sham surgeries were also performed, in which each ovary was exposed but not dissected. All treatments on OVX mice were carried out two weeks after surgery to ensure a total washout of ovarian hormones, as previously reported.
Determination of estrus cycle stages
Female mice were monitored for estrus cycle by daily vaginal cytology followed by staining with Fast Panoptic kit (254807, PanReac AppliChem) and posterior microscopic examination of the cells as previously reported.59
Leptin and estradiol feeding behavior studies
For the experiments of estradiol replacement, animals were individually housed and used for experimentation two days later. Male or OVX female mice were subjected to daily subcutaneous injections of vehicle (150 μl of sesame oil, Sigma Aldrich) over the baseline days followed by daily subcutaneous injections of estradiol (17-β-estradiol-3-benzoate; 1 μg dissolved in 150 μL of sesame oil; Sigma Aldrich) over the next five days.
Recombinant mouse leptin (498-OB, R&D systems, Minneapolis, US) was dissolved in ice-cold Tris-HCl buffer (20 mM, pH 8) to obtain a 1 mg/ml stock solution. This was further diluted with ice-cold PBS (pH 7.4) to get a 0.3 mg/mL working solution as previously described.60 Mice were subjected to vehicle intraperitoneal injections twice daily over three days to establish a feeding baseline followed by intraperitoneal injections of leptin (3 mg/kg BW, stock leptin solution diluted in ice-cold PBS, pH 7.4) or vehicle (equivalent volume of 20 mM Tris-HCl diluted in ice-cold PBS, pH 7.4) twice daily over three days.
For the leptin sensitivity test, animals were individually housed and used for experimentation two days later. Recombinant mouse leptin (498-OB, R&D systems, Minneapolis, US) was dissolved in ice-cold Tris-HCl buffer (20 mM, pH 8) to obtain a 1 mg/ml stock solution. This was further diluted with ice-cold PBS (pH 7.4) to get a 0.3 mg/mL working solution as previously described.60 Mice were subjected to a protocol of vehicle intraperitoneal injections twice daily over three days to establish a feeding baseline followed by intraperitoneal injections of leptin (3 mg/kg BW, stock leptin solution diluted in ice-cold PBS, pH 7.4) or vehicle (equivalent volume of 20 mM Tris-HCl diluted in ice-cold PBS, pH 7.4) twice daily over three days.
For the leptin sensitivity test coupled with estradiol administration, male or OVX female mice were individually housed and used for experimentation two days later. Mice were subjected to daily subcutaneous injections of vehicle (150 μl of sesame oil, Sigma Aldrich) over the baseline days followed by daily subcutaneous injections of estradiol (estradiol benzoate; 1 μg dissolved in 150 μL of sesame oil; Sigma Aldrich) over the three leptin-treatment days.
Leptin and estradiol treatment paradigms for molecular studies
For the molecular studies (i.e. cFOS immunohistochemistry, cell fractioning, Co-IP, and High-throughput ChIPmentation), food was removed and after one hour mice were subjected to subcutaneous injections of vehicle (150 μl of sesame oil, Sigma Aldrich) or estradiol (estradiol benzoate; 1 μg dissolved in 150 μL of sesame oil; Sigma Aldrich). Three hours later mice were subjected to intraperitoneal injections of vehicle (equivalent volume of 20 mM Tris-HCl diluted in ice-cold PBS, pH 7.4) or leptin (3 mg/kg BW, stock leptin solution diluted in ice-cold PBS, pH 7.4). One hour (cell fractioning, Co-IP and High-throughput ChIPmentation) or 90 minutes (cFOS immunohistochemistry) later mice were sacrificed and tissues were collected. When cycled female mice were used for the study, the same experimental paradigm was followed but estradiol injections were skipped.
Viral-mediated manipulation of Cited1
To ablate Cited1, adeno-associated viruses carrying the Cre recombinase (AAV2/1-EF1a-EGFP-T2A-iCre, Vector Biolabs, Cat. No. VB2069) or control viruses carrying GFP (AAV2/1-EF1a-EGFP, Vector Biolabs, Cat. No. VB2084) were injected bilaterally (0.5 μl/side; 1x1013 viral genomes/ml) into the MBH of Cited1loxP/loxP mice (8-wk-old), as specified, using a motorized stereotaxic system from Neurostar (Tubingen, Germany). Stereotaxic coordinates were −1.5 mm posterior and +/−0.25 mm lateral to bregma and −5.8 mm ventral from the dura. Surgeries were performed using a mixture of ketamine and xylazine (100 and 7 mg/kg, respectively) as anesthetic agents and Metamizol (200 mg/kg, subcutaneous), followed by Meloxicam (1 mg/kg, on 3 consecutive days, subcutaneous) for postoperative analgesia.
Immunohistochemistry
Adult mice were transcardially perfused with phosphate-buffered saline (PBS) followed by 4% neutral buffered paraformaldehyde (PFA). After dissection, brains and peripheral tissues were post-fixed for 24 h with 4% PFA, equilibrated in 30% sucrose for 24 h, and sectioned on a cryostat (Leica Biosystems) at 30 μm or 5 μm (adipose tissues). Brain sections were incubated with the primary antibodies overnight at 4°C in 0.1M TBS containing gelatin (0.25%) and Triton X100 (0.5%) or PBS containing FBS (5%) and BSA (1%) for adipose tissues. Sections were washed with 0.1M TBS and incubated with the secondary antibodies for 1 h at room temperature with 0.1 M TBS containing gelatin (0.25%) and Triton X100 (0.5%) or PBS containing BSA (1%) for adipose tissues. When Alexa-coupled antibodies were used to avoid host species cross-reactivity brain sections were incubated with the antibodies overnight after the previously described steps. Images were obtained using a Leica TCA SP-8-X Confocal Microscope (Leica Microsystems), and analysis was performed using Fiji 2.0.0 (ImageJ).
Fluorescence in situ hybridization
Fluorescence in situ hybridization was performed on brains from PomcΔCited1, HypΔCited1, and Cited1-HA mice using the RNA scope Multiplex Fluorescent Reagent Kit v2 (Advanced Cell Diagnostic, Cat. #323100-USM) following the manufacturer’s instructions. Sections were dried at 60°C for 30 minutes, dehydrated with an increasing ethanol gradient, pre-treated with hydrogen peroxide (Cat. #322381), and boiled in Target retrieval reagent (Cat. #322000). After dehydrating in pure ethanol, sections were surrounded by a hydrophobic barrier (ImmEdge hydrophobic barrier pen, Vector Lab, H-4000) and left to dry overnight. The following day slides were incubated in Protease III (Cat. #322340; 30 min at 40°C) followed by the target probe (Mm-Cited1-E2E3, Cat No. #400451 for PomcΔCited1, HypΔCited1 mice or Mm-Cited1, Cat. #432471 for Cited1-HA mice; 2 hours at 40°C) in a HybEZ oven. Signal amplification was achieved using amplifiers AMP1-3 and an Opal reagent (Opal570; Perkin-Elmer, Cat. #FP1488001KT). When combination with immunofluorescence was needed (Cited1-HA mice), staining of the HA-Tag was performed using an HRP-conjugated secondary antibody, followed by an Opal reagent (Opal690; Perkin-Elmer, Cat. #FP1497001KT). Sections were counterstained with DAPI (provided in kit) for 30 seconds, and mounted (ProLong Diamond Antifade Mountant; ThermoFisher, Cat. #P36961). Images were obtained using a Leica TCA SP-8 Confocal Microscope (Leica Microsystems) with 20x or 63x magnification.
iDISCO-assisted optical brain clearing
Cited1 HA;Pomc-GFP mice were transcardially perfused with 4% PFA and overnight post-fixated in 4% PFA, 4°C. Brains were washed three times in PBS, pH 7.4, before being cut on a 1 mm slice using a brain block. Brain slices were subjected to a pretreatment by washing in PTx.2, RT 1h x2, incubating in 1x PBS/0.2% TritonX-100/20% DMSO, 37°C overnight, incubating in 1x PBS/0.1% Tween-20/0.1% TritonX-100/0.1% Deoxycholate/0.1% NP40/20% DMSO, 37°C overnight and washing in PTx.2, RT 1h x 2. Brain slices were immunolabelled incubating samples in permeabilization solution, 37°C 2 days, blocking in blocking solution, 37°C, 2 days, incubating with primary antibody in PTwH/5% DMSO/3% donkey serum, 37°C, 4 days, washing in PTwH for 4-5 times until the next day, incubating with secondary antibody in PTwH/3% donkey serum, 37°C, 4 days and washing in PTwH for 4-5 times until the next day. Brain slices were subjected to the clearing protocol by dehydrating in methanol/H2O series: 20%, 40%, 60%, 80%, 100%, 100%; 1 hour each at room temperature, 3 hours incubation, with shaking, in 66% DCM / 33% methanol at room temperature, incubating in 100% DCM (Sigma 270997-12X100ML) 15 minutes twice with shaking to wash the MeOH, incubating in DiBenzyl Ether (DBE, Sigma 108014) and stored until image acquisition. Images were obtained using a Leica TCA SP-8 Confocal Microscope (Leica Microsystems) with 20x magnification and processed with Imaris 7.6.4 software.
Gene expression analysis by qRT-PCR
Dissected tissues were immediately frozen on dry ice, and RNA was extracted using RNeasy Mini Kits (Qiagen). cDNA was generated with reverse transcription QuantiTech reverse transcription kit (Qiagen). Quantitative Real-Time RT-PCR (qRT-PCR) was performed with a ViiA 7 Real-Time PCR System (Applied Biosystems) using the TaqMan probes listed in the key resources table. Target gene expression was normalized to reference genes Hprt. Calculations were performed by a comparative method (2−ΔΔCT).
Co-immunoprecipitation assay
Individual hypothalami were transferred to a dounce homogenizer containing 1mL of freshly prepared ice-cold immunoprecipitation (IP) buffer (50mM Tris pH 8.0, 1% IGEPAL-630, 15mM NaCl, 1mM PMSF, and 1x phosphatase and protease inhibitors). Homogenization was achieved by carefully douncing 20 strokes with the loose pestle, incubating on ice for 10 min and further douncing 20 more strokes with the tight pestle. The homogenate was filtered through a 20 μm cell strainer and incubated for 30 min in rotation at 4°C. Protein lysates were incubated with 2.0 μg of antibody (anti-HA-Tag or IgG-isotype control) in Lo binding tubes at 4°C while rotating for 1h. Subsequently, 50μl of μMACS protein-G μBeads were added and incubated for another 30 min at 4°C with rotation and IP was performed according to the manufacturer’s protocol. Proteins were eluted in 70 μl of preheated (95°C) sampling buffer, separated by SDS-PAGE and electrotransferred to a PVDF membrane for western blotting. Briefly, BSA-blocked membranes were incubated with anti-HA-Tag coupled to HRP, anti-ERα, anti-Stat3, and anti-β-Actin (1:1000 in blocking buffer) overnight at 4°C with agitation and further developed with appropriate HRP-secondary antibodies (1:5000).
Sub-cellular fractioning
For nuclei isolation, individual frozen hypothalami were transferred to a dounce homogenizer containing 700μl of freshly prepared ice-cold nuclei isolation buffer (0.25M sucrose, 25mM KCl, 5mM MgCl2, 20mM Tris pH 8.0, 0.4% IGEPAL 630, 1mM DTT, 0.15mM spermine, 0.5mM spermidine, and 1x phosphatase & protease inhibitors). Homogenization was achieved by carefully douncing 10 strokes with the loose pestle, incubating on ice for 5 min, and further douncing 15 more strokes with the tight pestle. The homogenate was filtered through a 20μm cell strainer and centrifuged at 1000 x g for 10min at 4°C. The supernatant was thoroughly collected on a new tube (cytosolic fraction) and the nuclei pellet resuspended in 300μl of nuclear lysis buffer (50mM Tris pH 8.0, 1% IGEPAL-630, 0.5mM sodium deoxycholate, 1mM PMSF, 1mM sodium butyrate, 1x phosphatase and protease inhibitors). Protein concentration measurement and subsequent steps were performed as previously described for SDS-PAGE and western blotting. HA-Tag, Stat3, and ERα were detected in both nuclear and cytosolic fractions using Histone 3 (H3) and GAPDH as loading controls, respectively.
High-throughput ChIPmentation
Individual hypothalami were homogenized as previously described for nuclei isolation in the previous section. After centrifugation, the nuclei pellet was thoroughly resuspended in 1mL of 1% ethanol-free formaldehyde and incubated at RT with agitation for 15 min to allow fixation. The fixation was quenched by adding 60μl of 2.5M glycine and further incubated under the same conditions for 5 min. Subsequently, 500μl of cold PBS were added and nuclei pelleted by centrifugation at 2000 x g for 10 min at 4°C. The pellet was resuspended in 200μl shearing buffer (1% vol/vol SDS, 10 mM EDTA, 50 mM Tris–HCl pH 8, and 1x phosphatase and protease inhibitors) and sonicated for 15 min x 30s ON/30s OFF on high power on a Bioruptor Plus. For HT-ChIPmentation we followed the protocol without modifications as previously described by Gustafsson et al.61 Per sample, the sheared chromatin was divided in 3 separate tubes with 2μg of anti-HA-Tag, -Stat3, or -Erα. An IgG-isotype negative control sample was included for each biological replicate. The specific Stat3 DNA-binding consensus sequence at the Pomc promoter was assessed using previously described primers.62
Serum analyses
Serum leptin levels were determined using a Leptin (Mouse/Rat) ELISA kit (22-LEPMS-E01, Alpco) following the manufacturer’s instructions. Serum corticosterone levels were determined using a Parameter corticosterone assay (KGE009, R&D systems) following the manufacturer’s instructions. Serum estradiol levels were determined using a Mouse/Rat ELISA Kit assay (ES180S-100, Calbiotech) following the manufacturer’s instructions. Serum α-MSH levels were determined using a Human, Rat, Mouse EIA Kit (#EK-043-01, Phoenix Pharmaceuticals) following the manufacturer’s instructions.
The analysis of steroids in plasma or serum have been performed in the Metabolomics Platform of the Genome Analysis Center, Helmholtz Zentrum München. The following 17 steroids were quantified using the AbsoluteIDQTM Stero17 Kit and LC-ESI-MS/MS: aldosterone, androstenedione (androst-4-en-3,17-dione), androsterone, corticosterone, cortisol, cortisone, 11-deoxycorticosterone, 11-deoxycortisol, dehydroepiandrosterone, dehydroepiandrosterone sulfate (DHEAS), dihydrotestosterone (DHT), estradiol, estrone, etiocholanolone, 17α-hydroxyprogesterone, progesterone, testosterone. Compound identification and quantification were based on scheduled multiple reaction monitoring measurements (sMRM). Sample preparation and LC-MS/MS measurements were performed as described by the manufacturer in manual UM-STERO17. The method of the AbsoluteIDQTM Stero17 Kit has been proven to be in conformance with the EMEA-Guideline "Guideline on bioanalytical method validation” (July 21st 2011)(CHMP, 2011),63 which implies proof of reproducibility within a given error range. Analytical specifications for LOD (limit of detection (CHMP, 2011)63), LLOQ and ULOQ (lower and upper limit of quantification), specificity, linearity, precision, accuracy, reproducibility, and stability were determined experimentally by Biocrates and are described manual AS-STERO17. A detailed method description has been published.64
Single-cell RNA sequencing analysis
Data for the scRNA-seq reanalysis was obtained from GEO Accession codes GSE93374, which is generated from the Arcuate-median eminence (Arc-ME) of the mouse hypothalamus by Campbell et al.25 We used Seurat software65 to perform the re-analysis. We first filter the dataset with Seurat. Only features expressed in at least three cells, only cells expressing at least 200 features and at most 8000 features and only cells' mitochondrial genes percentage below 15% were remained. The filtered dataset comprised of 20,700 cells and 22,746 genes. We further normalized the expression matrix with "logNormalize" method. Then we identified the 2,519 most-variable genes across the entire dataset controlling for the known relationship between mean expression and variance. During scaling of most-variable genes, we also removed the technical variances due to batch effects, mitochondrial gene percentages, and number of detected features. After that, we did the dimension reduction by performing the principal component analysis. We selected the top 25-significant PCs for cell neighbor determination and clustering. For neurons, based on metadata provided by GSE93374, we subset 12990 neuronal cells from above Seurat object with all cells. Similarly to above, we identified 2000 most-variable genes from the neuronal cell dataset, removed the technical variances, and scaled the most variable genes for PCA. We selected the top 30 PCs for neighbor cell determination and clustering. We further labeled neuronal clusters based metadata by GSE93374. Based on above analysis, we finally visualized candidate gene expression patterns with feature plot in tSNE or violin plot at all cell levels as well as neuronal cell levels.
Fertility assessments
We performed fertility assessments as previously described (Xu et al. Cell Metab 2011.). Briefly, using the historical records of our Wild-type, Cited1-KO, Cited1loxP/loxP, HypΔCited1, PomcΔCited1 and AgrpΔCited1 colonies we analyzed a) the average litter size considering all litters, b) the average size of the first litter, c) the average time period between mating day and delivery of pups, and d) the ratio of mating pairs that successfully delivered pups.
Quantification and statistical analysis
Statistical analyses were conducted using GraphPad Prism (version 7.0). Statistical significance was determined using unpaired 2-tailed Student’s t test (1 variable and 2 groups), 1-way ANOVA followed by Holm-Sidak’s post hoc test (1 variables and 2+ groups), 2-way ANOVA followed by Sidak’s post hoc test (2 variables), ChiSquare (expected vs. observed frequencies) or linear regression when appropriate. P ≤ 0.05 was considered statistically significant. Statistics information can be found in the figure legends and figures.
Acknowledgments
We thank Cassie Lynn Holleman, Clarita Layritz, Elisavet Lola, Nicole Klas, and Heidi Krause for technical assistance and assistance with animal studies. We thank Maria Kugler, Julia Scarpa, and Werner Römisch-Margl for metabolomics measurements performed at the Helmholtz Zentrum München, Genome Analysis Center, Metabolomics Core Facility. This research was funded by European Research Council ERC (StG AstroNeuroCrosstalk no. 757393 to C.G.-C. and AdG HypoFlam no. 695054 to M.H.T.), the German Research Foundation DFG under Germany’s Excellence Strategy within the framework of the Munich Cluster for Systems Neurology (EXC 2145 SyNergy – ID 390857198) and Helmholtz Excellence Network and the Deutsche Forschungsgemeinschaft (GS: SPP1757 – SA2114/2-2), and Helmholtz Association – Initiative and Networking Fund. I.G.-G. is a recipient of a fellowship from European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie actions (842080-H2020-MSCA-IF-2018), A.F. held a postdoctoral fellowship from the Canadian Institutes of Health Research (funding reference number: 152588), R.E.C. received funding from the European Union’s Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie grant agreement no. 675610, and P.T.P. is a recipient of the European Research Council ERC (CoG Yoyo-LepReSens no. 101002247). T.D.M. received funding from the German Research Foundation (DFG TRR296, TRR152, SFB1123, and GRK 2816/1), the German Center for Diabetes Research (DZD e.V.) and the European Research Council ERC-CoG trusted no. 101044445. The funders had no role in study design, data collection, analyses, decision to publish, or preparation of the manuscript.
Author contributions
A.F. and C.G.-C. initiated and supervised the project. I.G.-G., A.F., and C.G.-C. conceptualized all studies and designed all experiments. I.G.-G., A.F., and E.G.-C. (metabolic phenotyping); I.G.-G., E.G.-C., and O.L.T. (immunohistochemistry and confocal microscopy); I.G.-G. and T.G. (iDisco); I.G.-G., R.E.C., and S.C.S. (cellular fractioning); I.G.-G. (coIP); R.E.C. (ChIP); I.G.-G. and B.L. (sterotactic surgeries), A.C.-S. and W.W. (Cited1 HA-Tag mouse line generation); Y.X. (single-cell RNA sequencing analysis); and J.L. and J.A. (steroid hormones analysis) conducted the experiments and collected and analyzed the data. I.G.-G., A.F., and C.G.-C. wrote the manuscript in discussion with T.D.M., S.C.W., P.T.P., and M.H.T., who revised the article critically for important intellectual content. All authors have read and approved the final version of the manuscript.
Declaration of interests
M.H.T. is a member of the scientific advisory board of ERX Pharmaceuticals, Inc., Cambridge, MA. He was a member of the Research Cluster Advisory Panel (ReCAP) of the Novo Nordisk Foundation between 2017 and 2019. He attended a scientific advisory board meeting of the Novo Nordisk Foundation Center for Basic Metabolic Research, University of Copenhagen, in 2016. He received funding for his research projects by Novo Nordisk (2016–2020) and Sanofi-Aventis (2012–2019). He consulted for Böhringer Ingelheim Pharma GmbH & Co. KG (2020). He delivered a scientific lecture for Sanofi-Aventis Deutschland GmbH in 2020. He is an editorial board member of Cell Metabolism. He confirms that to the best of his knowledge none of the above were involved in the preparation of this paper.
Published: March 7, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cmet.2023.02.004.
Contributor Information
Alexandre Fisette, Email: alexandre.fisette@uqtr.ca.
Cristina García-Cáceres, Email: garcia-caceres@helmholtz-muenchen.de.
Supplemental information
Data and code availability
-
•
Data for creating all graphs in the paper are provided in Data S1.
-
•
No original code was generated in the study.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Sternson S.M., Atasoy D. Agouti-related protein neuron circuits that regulate appetite. Neuroendocrinology. 2014;100:95–102. doi: 10.1159/000369072. [DOI] [PubMed] [Google Scholar]
- 2.Andermann M.L., Lowell B.B. Toward a wiring diagram understanding of appetite control. Neuron. 2017;95:757–778. doi: 10.1016/j.neuron.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quarta C., Claret M., Zeltser L.M., Williams K.W., Yeo G.S.H., Tschöp M.H., Diano S., Brüning J.C., Cota D. POMC neuronal heterogeneity in energy balance and beyond: an integrated view. Nat. Metab. 2021;3:299–308. doi: 10.1038/s42255-021-00345-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim J.D., Leyva S., Diano S. Hormonal regulation of the hypothalamic melanocortin system. Front. Physiol. 2014;5:480. doi: 10.3389/fphys.2014.00480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams K.W., Elmquist J.K. From neuroanatomy to behavior: central integration of peripheral signals regulating feeding behavior. Nat. Neurosci. 2012;15:1350–1355. doi: 10.1038/nn.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Timper K., Brüning J.C. Hypothalamic circuits regulating appetite and energy homeostasis: pathways to obesity. Dis. Model. Mech. 2017;10:679–689. doi: 10.1242/dmm.026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maffei M., Giordano A. Leptin, the brain and energy homeostasis: from an apparently simple to a highly complex neuronal system. Rev. Endocr. Metab. Disord. 2022;23:87–101. doi: 10.1007/s11154-021-09636-2. [DOI] [PubMed] [Google Scholar]
- 8.Friedman J.M. Leptin and the endocrine control of energy balance. Nat. Metab. 2019;1:754–764. doi: 10.1038/s42255-019-0095-y. [DOI] [PubMed] [Google Scholar]
- 9.Ainslie D.A., Morris M.J., Wittert G., Turnbull H., Proietto J., Thorburn A.W. Estrogen deficiency causes central leptin insensitivity and increased hypothalamic neuropeptide Y. Int. J. Obes. Relat. Metab. Disord. 2001;25:1680–1688. doi: 10.1038/sj.ijo.0801806. [DOI] [PubMed] [Google Scholar]
- 10.Clegg D.J., Riedy C.A., Smith K.A., Benoit S.C., Woods S.C. Differential sensitivity to central leptin and insulin in male and female rats. Diabetes. 2003;52:682–687. doi: 10.2337/diabetes.52.3.682. [DOI] [PubMed] [Google Scholar]
- 11.Clegg D.J., Brown L.M., Woods S.C., Benoit S.C. Gonadal hormones determine sensitivity to central leptin and insulin. Diabetes. 2006;55:978–987. doi: 10.2337/diabetes.55.04.06.db05-1339. [DOI] [PubMed] [Google Scholar]
- 12.Gao Q., Mezei G., Nie Y., Rao Y., Choi C.S., Bechmann I., Leranth C., Toran-Allerand D., Priest C.A., Roberts J.L., et al. Anorectic estrogen mimics leptin's effect on the rewiring of melanocortin cells and Stat3 signaling in obese animals. Nat. Med. 2007;13:89–94. doi: 10.1038/nm1525. [DOI] [PubMed] [Google Scholar]
- 13.Gao Q., Horvath T.L. Cross-talk between estrogen and leptin signaling in the hypothalamus. Am. J. Physiol. Endocrinol. Metab. 2008;294:E817–E826. doi: 10.1152/ajpendo.00733.2007. [DOI] [PubMed] [Google Scholar]
- 14.Xu Y., Nedungadi T.P., Zhu L., Sobhani N., Irani B.G., Davis K.E., Zhang X., Zou F., Gent L.M., Hahner L.D., et al. Distinct hypothalamic neurons mediate estrogenic effects on energy homeostasis and reproduction. Cell Metab. 2011;14:453–465. doi: 10.1016/j.cmet.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martínez de Morentin P.B., González-García I., Martins L., Lage R., Fernández-Mallo D., Martínez-Sánchez N., Ruíz-Pino F., Liu J., Morgan D.A., Pinilla L., et al. Estradiol regulates brown adipose tissue thermogenesis via hypothalamic AMPK. Cell Metab. 2014;20:41–53. doi: 10.1016/j.cmet.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krause W.C., Rodriguez R., Gegenhuber B., Matharu N., Rodriguez A.N., Padilla-Roger A.M., Toma K., Herber C.B., Correa S.M., Duan X., et al. Oestrogen engages brain MC4R signalling to drive physical activity in female mice. Nature. 2021;599:131–135. doi: 10.1038/s41586-021-04010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matysková R., Zelezná B., Maixnerová J., Koutová D., Haluzík M., Maletínská L. Estradiol supplementation helps overcome central leptin resistance of ovariectomized mice on a high fat diet. Horm. Metab. Res. 2010;42:182–186. doi: 10.1055/s-0029-1243250. [DOI] [PubMed] [Google Scholar]
- 18.Yahata T., Shao W., Endoh H., Hur J., Coser K.R., Sun H., Ueda Y., Kato S., Isselbacher K.J., Brown M., et al. Selective coactivation of estrogen-dependent transcription by CITED1 CBP/p300-binding protein. Genes Dev. 2001;15:2598–2612. doi: 10.1101/gad.906301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McBryan J., Howlin J., Kenny P.A., Shioda T., Martin F. ERalpha-CITED1 co-regulated genes expressed during pubertal mammary gland development: implications for breast cancer prognosis. Oncogene. 2007;26:6406–6419. doi: 10.1038/sj.onc.1210468. [DOI] [PubMed] [Google Scholar]
- 20.Tschöp M.H., Speakman J.R., Arch J.R., Auwerx J., Brüning J.C., Chan L., Eckel R.H., Farese R.V., Jr., Galgani J.E., Hambly C., et al. A guide to analysis of mouse energy metabolism. Nat. Methods. 2011;9:57–63. doi: 10.1038/nmeth.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müller T.D., Klingenspor M., Tschöp M.H. Revisiting energy expenditure: how to correct mouse metabolic rate for body mass. Nat. Metab. 2021;3:1134–1136. doi: 10.1038/s42255-021-00451-2. [DOI] [PubMed] [Google Scholar]
- 22.Sparrow D.B., Boyle S.C., Sams R.S., Mazuruk B., Zhang L., Moeckel G.W., Dunwoodie S.L., de Caestecker M.P. Placental insufficiency associated with loss of Cited1 causes renal medullary dysplasia. J. Am. Soc. Nephrol. 2009;20:777–786. doi: 10.1681/ASN.2008050547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker M.S., Li G., Kohorst J.J., Waterland R.A. Fetal growth restriction promotes physical inactivity and obesity in female mice. Int. J. Obes. (Lond) 2015;39:98–104. doi: 10.1038/ijo.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fenner M.H., Parrish J.E., Boyd Y., Reed V., MacDonald M., Nelson D.L., Isselbacher K.J., Shioda T. MSG1 (melanocyte-specific gene 1): mapping to chromosome Xq13.1, genomic organization, and promoter analysis. Genomics. 1998;51:401–407. doi: 10.1006/geno.1998.5383. [DOI] [PubMed] [Google Scholar]
- 25.Campbell J.N., Macosko E.Z., Fenselau H., Pers T.H., Lyubetskaya A., Tenen D., Goldman M., Verstegen A.M., Resch J.M., McCarroll S.A., et al. A molecular census of arcuate hypothalamus and median eminence cell types. Nat. Neurosci. 2017;20:484–496. doi: 10.1038/nn.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Q., Tam M., Anderson S.A. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J. Comp. Neurol. 2008;506:16–29. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
- 27.Heck A.L., Handa R.J. Sex differences in the hypothalamic-pituitary-adrenal axis' response to stress: an important role for gonadal hormones. Neuropsychopharmacology. 2019;44:45–58. doi: 10.1038/s41386-018-0167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huisman C., Cho H., Brock O., Lim S.J., Youn S.M., Park Y., Kim S., Lee S.K., Delogu A., Lee J.W. Single cell transcriptome analysis of developing arcuate nucleus neurons uncovers their key developmental regulators. Nat. Commun. 2019;10:3696. doi: 10.1038/s41467-019-11667-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romanov R.A., Tretiakov E.O., Kastriti M.E., Zupancic M., Häring M., Korchynska S., Popadin K., Benevento M., Rebernik P., Lallemend F., et al. Molecular design of hypothalamus development. Nature. 2020;582:246–252. doi: 10.1038/s41586-020-2266-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cantelli G., Orgaz J.L., Rodriguez-Hernandez I., Karagiannis P., Maiques O., Matias-Guiu X., Nestle F.O., Marti R.M., Karagiannis S.N., Sanz-Moreno V. TGF-beta-induced transcription sustains amoeboid melanoma migration and dissemination. Curr. Biol. 2015;25:2899–2914. doi: 10.1016/j.cub.2015.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stincic T.L., Grachev P., Bosch M.A., Rønnekleiv O.K., Kelly M.J. Estradiol drives the anorexigenic activity of proopiomelanocortin neurons in female mice. eNeuro. 2018;5 doi: 10.1523/ENEURO.0103-18.2018. 0103–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams K.W., Margatho L.O., Lee C.E., Choi M., Lee S., Scott M.M., Elias C.F., Elmquist J.K. Segregation of acute leptin and insulin effects in distinct populations of arcuate proopiomelanocortin neurons. J. Neurosci. 2010;30:2472–2479. doi: 10.1523/JNEUROSCI.3118-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lam B.Y.H., Cimino I., Polex-Wolf J., Nicole Kohnke S., Rimmington D., Iyemere V., Heeley N., Cossetti C., Schulte R., Saraiva L.R., et al. Heterogeneity of hypothalamic pro-opiomelanocortin-expressing neurons revealed by single-cell RNA sequencing. Mol. Metab. 2017;6:383–392. doi: 10.1016/j.molmet.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biglari N., Gaziano I., Schumacher J., Radermacher J., Paeger L., Klemm P., Chen W., Corneliussen S., Wunderlich C.M., Sue M., et al. Functionally distinct POMC-expressing neuron subpopulations in hypothalamus revealed by intersectional targeting. Nat. Neurosci. 2021;24:913–929. doi: 10.1038/s41593-021-00854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang C., He Y., Xu P., Yang Y., Saito K., Xia Y., Yan X., Hinton A., Jr., Yan C., Ding H., et al. TAp63 contributes to sexual dimorphism in POMC neuron functions and energy homeostasis. Nat. Commun. 2018;9:1544. doi: 10.1038/s41467-018-03796-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen P., Li C., Haskell-Luevano C., Cone R.D., Smith M.S. Altered expression of agouti-related protein and its colocalization with neuropeptide Y in the arcuate nucleus of the hypothalamus during lactation. Endocrinology. 1999;140:2645–2650. doi: 10.1210/endo.140.6.6829. [DOI] [PubMed] [Google Scholar]
- 37.Münzberg H., Huo L., Nillni E.A., Hollenberg A.N., Bjørbaek C. Role of signal transducer and activator of transcription 3 in regulation of hypothalamic proopiomelanocortin gene expression by leptin. Endocrinology. 2003;144:2121–2131. doi: 10.1210/en.2002-221037. [DOI] [PubMed] [Google Scholar]
- 38.de Souza F.S., Nasif S., López-Leal R., Levi D.H., Low M.J., Rubinsten M. The estrogen receptor alpha colocalizes with proopiomelanocortin in hypothalamic neurons and binds to a conserved motif present in the neuron-specific enhancer nPE2. Eur. J. Pharmacol. 2011;660:181–187. doi: 10.1016/j.ejphar.2010.10.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knight Z.A., Tan K., Birsoy K., Schmidt S., Garrison J.L., Wysocki R.W., Emiliano A., Ekstrand M.I., Friedman J.M. Molecular profiling of activated neurons by phosphorylated ribosome capture. Cell. 2012;151:1126–1137. doi: 10.1016/j.cell.2012.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allison M.B., Pan W., MacKenzie A., Patterson C., Shah K., Barnes T., Cheng W., Rupp A., Olson D.P., Myers M.G., Jr. Defining the transcriptional targets of leptin reveals a role for Atf3 in leptin action. Diabetes. 2018;67:1093–1104. doi: 10.2337/db17-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allison M.B., Patterson C.M., Krashes M.J., Lowell B.B., Myers M.G., Jr., Olson D.P. TRAP-seq defines markers for novel populations of hypothalamic and brainstem LepRb neurons. Mol. Metab. 2015;4:299–309. doi: 10.1016/j.molmet.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carr M.C. The emergence of the metabolic syndrome with menopause. J. Clin. Endocrinol. Metab. 2003;88:2404–2411. doi: 10.1210/jc.2003-030242. [DOI] [PubMed] [Google Scholar]
- 43.Hong J., Stubbins R.E., Smith R.R., Harvey A.E., Núñez N.P. Differential susceptibility to obesity between male, female and ovariectomized female mice. Nutr. J. 2009;8:11. doi: 10.1186/1475-2891-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mauvais-Jarvis F., Clegg D.J., Hevener A.L. The role of estrogens in control of energy balance and glucose homeostasis. Endocr. Rev. 2013;34:309–338. doi: 10.1210/er.2012-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frank A., Brown L.M., Clegg D.J. The role of hypothalamic estrogen receptors in metabolic regulation. Front. Neuroendocrinol. 2014;35:550–557. doi: 10.1016/j.yfrne.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burke L.K., Doslikova B., D'Agostino G., Greenwald-Yarnell M., Georgescu T., Chianese R., Martinez de Morentin P.B., Ogunnowo-Bada E., Cansell C., Valencia-Torres L., et al. Sex difference in physical activity, energy expenditure and obesity driven by a subpopulation of hypothalamic POMC neurons. Mol. Metab. 2016;5:245–252. doi: 10.1016/j.molmet.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howlin J., McBryan J., Napoletano S., Lambe T., McArdle E., Shioda T., Martin F. CITED1 homozygous null mice display aberrant pubertal mammary ductal morphogenesis. Oncogene. 2006;25:1532–1542. doi: 10.1038/sj.onc.1209183. [DOI] [PubMed] [Google Scholar]
- 48.Bhattacharya S., Michels C.L., Leung M.K., Arany Z.P., Kung A.L., Livingston D.M. Functional role of p35srj, a novel p300/CBP binding protein, during transactivation by HIF-1. Genes Dev. 1999;13:64–75. doi: 10.1101/gad.13.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yahata T., de Caestecker M.P., Lechleider R.J., Andriole S., Roberts A.B., Isselbacher K.J., Shioda T. The MSG1 non-DNA-binding transactivator binds to the p300/CBP coactivators, enhancing their functional link to the Smad transcription factors. J. Biol. Chem. 2000;275:8825–8834. doi: 10.1074/jbc.275.12.8825. [DOI] [PubMed] [Google Scholar]
- 50.Shioda T., Lechleider R.J., Dunwoodie S.L., Li H., Yahata T., de Caestecker M.P., Fenner M.H., Roberts A.B., Isselbacher K.J. Transcriptional activating activity of Smad4: roles of SMAD hetero-oligomerization and enhancement by an associating transactivator. Proc. Natl. Acad. Sci. USA. 1998;95:9785–9790. doi: 10.1073/pnas.95.17.9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakashima K., Yanagisawa M., Arakawa H., Kimura N., Hisatsune T., Kawabata M., Miyazono K., Taga T. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- 52.Paulson M., Pisharody S., Pan L., Guadagno S., Mui A.L., Levy D.E. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J. Biol. Chem. 1999;274:25343–25349. doi: 10.1074/jbc.274.36.25343. [DOI] [PubMed] [Google Scholar]
- 53.Yamamoto T., Matsuda T., Junicho A., Kishi H., Saatcioglu F., Muraguchi A. Cross-talk between signal transducer and activator of transcription 3 and estrogen receptor signaling. FEBS Lett. 2000;486:143–148. doi: 10.1016/s0014-5793(00)02296-1. [DOI] [PubMed] [Google Scholar]
- 54.Ciana P., Ghisletti S., Mussi P., Eberini I., Vegeto E., Maggi A. Estrogen receptor alpha, a molecular switch converting transforming growth factor-alpha-mediated proliferation into differentiation in neuroblastoma cells. J. Biol. Chem. 2003;278:31737–31744. doi: 10.1074/jbc.M301525200. [DOI] [PubMed] [Google Scholar]
- 55.Gao Q., Wolfgang M.J., Neschen S., Morino K., Horvath T.L., Shulman G.I., Fu X.Y. Disruption of neural signal transducer and activator of transcription 3 causes obesity, diabetes, infertility, and thermal dysregulation. Proc. Natl. Acad. Sci. USA. 2004;101:4661–4666. doi: 10.1073/pnas.0303992101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Binai N.A., Damert A., Carra G., Steckelbroeck S., Löwer J., Löwer R., Wessler S. Expression of estrogen receptor alpha increases leptin-induced STAT3 activity in breast cancer cells. Int. J. Cancer. 2010;127:55–66. doi: 10.1002/ijc.25010. [DOI] [PubMed] [Google Scholar]
- 57.González-García I., Contreras C., Estévez-Salguero Á., Ruíz-Pino F., Colsh B., Pensado I., Liñares-Pose L., Rial-Pensado E., Martínez de Morentin P.B., Fernø J., et al. Estradiol regulates energy balance by ameliorating hypothalamic ceramide-induced ER stress. Cell Rep. 2018;25:413–423.e5. doi: 10.1016/j.celrep.2018.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.González-García I., Martínez de Morentin P.B., Estévez-Salguero Á., Contreras C., Romero-Picó A., Fernø J., Nogueiras R., Diéguez C., Tena-Sempere M., Tovar S., et al. mTOR signaling in the arcuate nucleus of the hypothalamus mediates the anorectic action of estradiol. J. Endocrinol. 2018;238:177–186. doi: 10.1530/JOE-18-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ajayi A.F., Akhigbe R.E. Staging of the estrous cycle and induction of estrus in experimental rodents: an update. Fertil. Res. Pract. 2020;6:5. doi: 10.1186/s40738-020-00074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gruber T., Pan C., Contreras R.E., Wiedemann T., Morgan D.A., Skowronski A.A., Lefort S., De Bernardis Murat C., Le Thuc O., Legutko B., et al. Obesity-associated hyperleptinemia alters the gliovascular interface of the hypothalamus to promote hypertension. Cell Metab. 2021;33:1155–1170.e10. doi: 10.1016/j.cmet.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gustafsson C., De Paepe A., Schmidl C., Månsson R. High-throughput ChIPmentation: freely scalable, single day ChIPseq data generation from very low cell-numbers. BMC Genomics. 2019;20:59. doi: 10.1186/s12864-018-5299-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kabra D.G., Pfuhlmann K., García-Cáceres C., Schriever S.C., Casquero García V., Kebede A.F., Fuente-Martin E., Trivedi C., Heppner K., Uhlenhaut N.H., et al. Hypothalamic leptin action is mediated by histone deacetylase 5. Nat. Commun. 2016;7:10782. doi: 10.1038/ncomms10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Committee for Medicinal Products for Human Use (CHMP) European Medicines Association; 2011. Guideline on bioanalytical method validation. [Google Scholar]
- 64.Breier M., Wahl S., Prehn C., Ferrari U., Sacco V., Weise M., Grallert H., Adamski J., Lechner A. Immediate reduction of serum citrulline but no change of steroid profile after initiation of metformin in individuals with type 2 diabetes. J. Steroid Biochem. Mol. Biol. 2017;174:114–119. doi: 10.1016/j.jsbmb.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 65.Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
iDisco 3D-imaging reconstruction of the ARC showing Cited1-positive cells (magenta) and Pomc-GFP (green) neurons, related to Figure 6
Data Availability Statement
-
•
Data for creating all graphs in the paper are provided in Data S1.
-
•
No original code was generated in the study.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.