Abstract
SNF5/INI1 is a component of the ATP-dependent chromatin remodeling enzyme family SWI/SNF. Germ line mutations of INI1 have been identified in children with brain and renal rhabdoid tumors, indicating that INI1 is a tumor suppressor. Here we report that disruption of Ini1 expression in mice results in early embryonic lethality. Ini1-null embryos die between 3.5 and 5.5 days postcoitum, and Ini1-null blastocysts fail to hatch, form the trophectoderm, or expand the inner cell mass when cultured in vitro. Furthermore, we report that approximately 15% of Ini1-heterozygous mice present with tumors, mostly undifferentiated or poorly differentiated sarcomas. Tumor formation is associated with a loss of heterozygocity at the Ini1 locus, characterizing Ini1 as a tumor suppressor in mice. Thus, Ini1 is essential for embryo viability and for repression of oncogenesis in the adult organism.
The compact nature of chromatin structure presents a barrier to cellular processes that require access to DNA. A number of multiprotein complexes have been identified that share the ability to modify chromatin structure. These include the histone acetyltransferases and deacetylases, complexes which chemically modify the amino-terminal tails of histones by the addition or removal of acetyl groups, respectively, as well as a group of enzymes that utilize the energy derived from ATP hydrolysis to alter nucleosome structure (16, 20, 43, 44, 50). Included among these ATP-dependent chromatin remodeling enzymes is the SWI/SNF family of chromatin modifiers.
SWI/SNF enzymes are large multisubunit enzymes of ∼1 to 2 MDa. Yeast SWI/SNF genes were originally identified as being required for mating type switching or sucrose fermentation (4, 32, 42). Later work determined that SWI/SNF genes were required for the induction of a subset of yeast genes and that the SWI2/SNF2 protein possessed a DNA-stimulated ATPase activity (6, 22, 26, 33, 34, 54). Mutations in SWI/SNF genes could be suppressed by mutations altering histone gene expression, histone structure, or nonhistone chromatin proteins, leading to the suggestion that these gene products facilitated transcriptional activation by altering chromatin structure (15, 23, 24).
Human SWI/SNF (hSWI/SNF) complexes contain either the human BRM (hBRM) (hSNF2α) or BRG1 (hSNF2β) homologues of the yeast SWI2/SNF2 ATPase (7, 19, 30). Both yeast and human SWI/SNF complexes have been shown to possess nucleosome remodeling activity in vitro (8, 17, 25). Components of mammalian SWI/SNF complexes have been implicated in a variety of cellular processes, including gene activation and repression, development and differentiation, recombination and repair, and cell cycle control. There is evidence supporting a role for SWI/SNF in gene activation events mediated by nuclear hormone receptors, environmental stress, and viral infection (1, 7, 10, 13, 30). In contrast, SWI/SNF components also were shown to be involved in repression of c-fos and some E2F-regulated genes (31, 48). Both BRG1 and hBRM can interact with the retinoblastoma oncoprotein and induce cell cycle arrest, an effect that is abrogated by the association of BRG1 with cyclin E (11, 41, 45, 56). Evidence suggesting a role for hSWI/SNF in recombination and repair was provided by studies demonstrating an interaction of components of the hSWI/SNF complex with BRCA1, which is thought to be involved in DNA damage and repair pathways (3). Furthermore, members of the SWI/SNF complex are targets of viral regulatory proteins upon infection of cells by adenovirus, Epstein-Barr virus, human immunodeficiency virus, and human papillomavirus (18, 27, 28, 53).
The role of SWI/SNF enzymes in whole organisms is unclear. While homozygous disruption of Brg1 in mouse embryonic carcinoma cells resulted in lethality, disruption of Brm expression in mice produced only mild proliferative effects (35, 46). The upregulation of Brg1 in the Brm-deficient mice may provide a compensatory effect; however, one cannot rule out the possibility that these differences are due to distinct functions of Brm- or Brg1-containing complexes.
SNF5/INI1 is a member of both BRG1- and BRM-containing SWI/SNF complexes (29, 51). INI1 was shown to interact with ALL-1, translocations of which are associated with several types of human acute leukemias (37). Furthermore, INI1 has been found to be altered in malignant rhabdoid tumors, choroid plexus carcinomas, medullablastomas, and central primitive neuroectodermal tumors (2, 9, 39, 40, 49). Identification of constitutional mutations in a subset of these tumors indicates that INI1 is a tumor suppressor (2, 40). In an attempt to generate a mouse model that would allow further characterization of the mechanisms of Ini1 in tumorigenesis and to determine the role of the mammalian SWI/SNF complexes in development, we generated mice deficient for Ini1 expression. We show that Ini1-deficient mice die early in embryogenesis, likely due to an inability of the blastocysts to hatch, implant in the uterus, and continue development. In addition, we report that a subset of the Ini1-heterozygous mice present with a variety of tumors in the soft tissues of the head and neck and that loss of heterozygosity at the Ini1 locus is correlated with tumor formation.
MATERIALS AND METHODS
Ini1 targeting.
Embryonic stem (ES) cells (Omnibank no. OST32815) bearing a retroviral promoter trap that functionally inactivates one allele of Ini1 were generated as described previously (55). Analysis by rapid amplification of cDNA ends also is described. The site of insertion was determined using sequence analysis.
Creation of Ini1-null mice.
Ini1-targeted ES cells were injected into 3.5 days postcoitum (d.p.c.) C57BL/6 blastocysts. Male chimeric mice were mated with wild-type C57BL/6 or 129 females. Germ line transmission of the mutant allele was determined by PCR analysis of tail genomic DNA using the following primers: for Ini1, 5′-GCAAGCGCTCTGCCAATTGACC-3′ and 3′-CACACCCTATTGTCACTCTGGAA-5′; βgeo, 5′-CGGTATCGATAAGCTTGATGATC-3′ and 3′-GTCAACGCGTCGGACTTACCGC-5′. Ini1-heterozygous mice were intercrossed to generate Ini1-null mice. Embryos 6.5 d.p.c. and younger were prepared for genotyping by PCR as described previously (47). Nested PCR was done using the above primers for the first round of PCR (29 cycles) and the following intron 3-nested primers for the second round (29 cycles): 5′-GCGTGCGCCACCATGCCTGG-3′ and 3′-CTTCTGGAGACTTCACTTACGTCC-5′.
Blastocyst culture.
Blastocysts from heterozygous intercrosses were flushed from the uteri of Ini1in3/+ females 3.5 d.p.c. with M15 media (Dulbecco's minimal essential medium, 15% fetal calf serum, 100 μM β-mercaptoethanol, 2 mM glutamine, and 1× penicillin-streptomycin) and cultured in tissue culture plates for 96 h. Embryo cultures were genotyped as described above.
β-Galactosidase staining of cultured ES cells.
Wild-type AB2.2 ES cells and Ini1in3/+ ES cells were grown to near-confluency and fixed in 0.5% glutaraldehyde. Cells were then rinsed with phosphate-buffered saline and stained overnight in the dark at room temperature in a solution containing 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 2 mM MgCl2, and 1 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)/ml.
Whole mount staining of embryos for β-galactosidase activity.
Embryos were harvested at various time points postfertilization and fixed in 4% paraformaldehyde for 20 min at 4°C. Embryos were washed and then stained in an X-Gal histochemical reaction mixture (4 mM potassium ferrocyanide, 4 mM potassium ferricyanide, 2 mM MgCl2, 1 mg of X-Gal/ml) overnight at room temperature. Following staining, embryos were rinsed in phosphate-buffered saline and cleared in 30% sucrose.
Western analysis of tumor samples.
Control tissues and tumor samples were homogenized in lysis buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5% NP-40, 20% glycerol, 1 mM dithiothreitol, 1 μg of pepstatin A/ml, 4 μg of leupeptin/ml, and 1 mM phenylmethylsulfonyl fluoride. Extracts were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Western analysis for Ini1 protein was performed as described previously (10).
Histologic analysis of tumors in mice.
Tumor samples and selected tissues were fixed in 10% buffered formalin phosphate and processed for paraffin embedding as described previously (14). Sections were prepared, stained with hematoxylin and eosin, and examined under a microscope.
RESULTS AND DISCUSSION
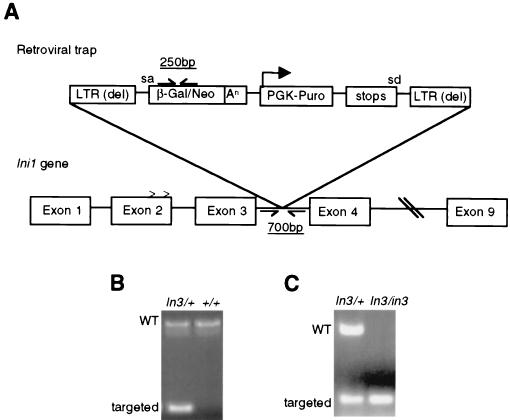
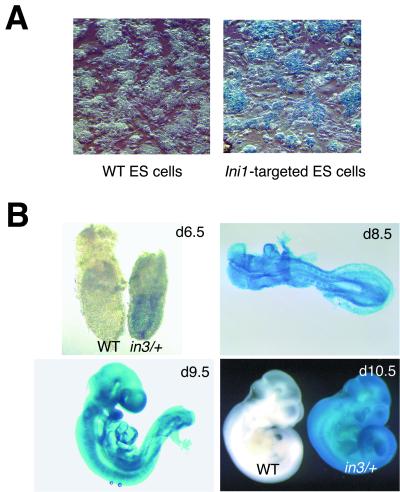
Mouse ES cells bearing a retroviral promoter trap that functionally inactivates one allele of Ini1 were constructed as described previously (55). Sequence analysis revealed that the promoter trap was inserted within intron 3 of Ini1 (Fig. 1A). The beta-galactosidase-neomycin (β-geo) gene fusion cassette within the retroviral insertion has a 5′ splice acceptor site; thus, β-geo expression is regulated by the native Ini1 promoter. We were able to utilize the β-geo gene cassette in a colorimetric assay to determine if Ini1 is normally expressed in ES cells. Ini1-targeted cells stained positive for β-galactosidase activity, indicating that Ini1 is expressed in ES cells (Fig. 2A). Northern analysis of ES cell total RNA confirmed Ini1 expression (data not shown). Furthermore, sequence data obtained from 5′ rapid amplification of cDNA ends analysis of the Ini1–β-galactosidase fusion mRNA revealed that transcripts utilizing either splice donor site in exon 2 spliced into the trap, indicating that both splice variants of Ini1 were inactivated (5).
FIG. 1.
Disruption of Ini1. (A) Targeting strategy for Ini1. A retroviral promoter trap vector was inserted in intron 3 of Ini1. sa and sd, the splice acceptor site in the β-geo gene cassette and the splice donator site in the puromycin selection marker, respectively. Carets represent the site of alternate splicing. Positions of the primers used for PCR analysis are shown. LTR (del), long terminal repeat deleted; An, polyadenylation signal; Puro, puromycin. (B) Genotyping of Ini1-targeted mice by PCR. Genomic DNA was harvested from tails and genotyped as described in Materials and Methods. The size and position of wild-type (WT) and targeted bands are indicated. (C) Genotyping of Ini1-targeted embryos. Embryos 6.5 d.p.c. and younger were genotyped by nested PCR. The size and position of wild-type and targeted bands are indicated.
FIG. 2.
Ini1 is expressed in ES cells and ubiquitously throughout development. (A) β-galactosidase staining of targeted ES cells showing expression of Ini1. Wild-type (WT) AB2.2 ES cells were used as a control. (B) Whole mount staining of Ini1in3/+ embryos showing ubiquitous expression of Ini1 at indicated time points. Wild-type embryos at 6.5 and 10.5 d.p.c. are shown as controls.
To determine the role of Ini1 in mammalian development and tumorigenesis, we used the targeted ES cells in blastocyst injection experiments to generate Ini1-heterozygous (Ini1in3/+) mice. In order to monitor expression of Ini1 during embryogenesis, we performed whole mount staining for β-galactosidase activity in embryos harvested from Ini1in3/+ matings at various times during development. We found that Ini1in3/+ embryos stained positive in all tissues at all time points examined, including 6.5, 8.5, 9.5, and 10.5 d.p.c., indicating that Ini1 is ubiquitously expressed during embryogenesis (Fig. 2B). Ini1 expression was also detected by Northern analysis in a wide range of adult tissues (35) (data not shown).
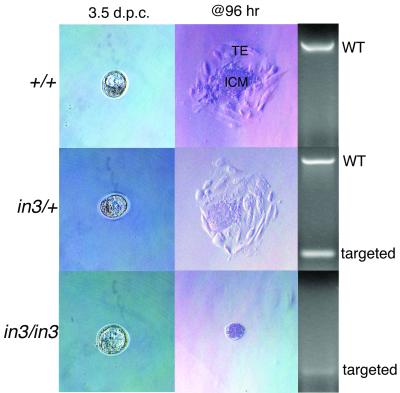
Chimeric mice generated from C57BL/6 strain blastocyst injections of the 129 strain-derived ES cells were bred to wild-type C57BL/6 or 129 mice in order to obtain Ini1in3/+ mice on either a mixed (C57BL/6 × 129) or pure (129) background. Intercrosses of Ini1in3/+ mice in both backgrounds yielded Ini1in3/+ offspring and wild-type offspring at a 2:1 ratio (63:26 in the mixed background, 34:17 in the pure background) and no Ini1-null offspring, indicating that disruption of Ini1 induces embryonic lethality (Fig. 1B). Timed matings of Ini1in3/+ mice were performed, and embryos were harvested at various time points in gestation for genotyping via PCR. Ini1-null embryos could be isolated at 3.5 d.p.c. and were normal in appearance (Fig. 1C and 3). However, no Ini1in3/in3 embryos were detected at 6.5 d.p.c. or later (Table 1). Dissection of maternal deciduae at 6.0 to 6.5 d.p.c. revealed no significant increase in the number of embryo reabsorptions, suggesting that Ini1in3/in3 lethality occurred between days 3.5 and 5.5 of gestation. These results indicate that Ini1-null embryos either failed to be implanted into the uterine wall or were implanted and were reabsorbed shortly thereafter. In order to examine further the developmental defect of Ini1in3/in3 embryos, we analyzed the ability of blastocysts from Ini1in3/+ intercrosses to expand in vitro. When 3.5-d.p.c. blastocysts were plated in culture, wild-type and Ini1in3/+ blastocysts hatched from the zona pellucida and were implanted onto the tissue culture plastic. Both wild-type and Ini1in3/+-implanted embryos formed the trophectoderm and expanded their inner cell mass (ICM). In contrast, no Ini1in3/in3 blastocysts hatched and were implanted in culture (Fig. 3). The results of these experiments suggest that the peri-implantation embryonic lethality of Ini1-null mice may be due to a defect in the hatching of the blastocyst from the zona pellucida, an obligate step for implantation of the embryo into the wall of the uterus during normal development. Manual disruption of the zona pellucida of 19 (C57BL/6 × 129) blastocysts harvested from Ini1in3/+ intercrosses did not result in expansion of the Ini1-null trophectoderm or ICM during in vitro culture, suggesting that growth of these tissues also is compromised (data not shown). Expression of Ini1 in ES cells, which are derived from the ICM of 3.5 d.p.c. blastocysts, is consistent with a gene crucial to the peri-implantation or preimplantation stage of embryogenesis.
FIG. 3.
Ini1-null mice are early embryonic lethal and fail to hatch in vitro. Blastocysts were harvested from C57BL/6 Ini1in3/+ females and plated in culture for 96 h, at which time outgrowths were processed for PCR. Blastocysts are shown before and after culturing. TE, trophectoderm; ICM, inner cell mass.
TABLE 1.
Genotyping of embryos from heterozygous intercrossesa
| Embryo category | No. of embryos
|
||
|---|---|---|---|
| WT | Het | Null | |
| 3.5 d.p.c. | 7 | 17 | 5 |
| In vitro culture | 11 | 14 | 1b |
| 6.5 d.p.c. | 9 | 23 | 0 |
| 8.5 d.p.c. | 4 | 14 | 0 |
| 13.5 d.p.c. | 5 | 6 | 0 |
WT, wild type; Het, heterozygous.
The embryo was dead.
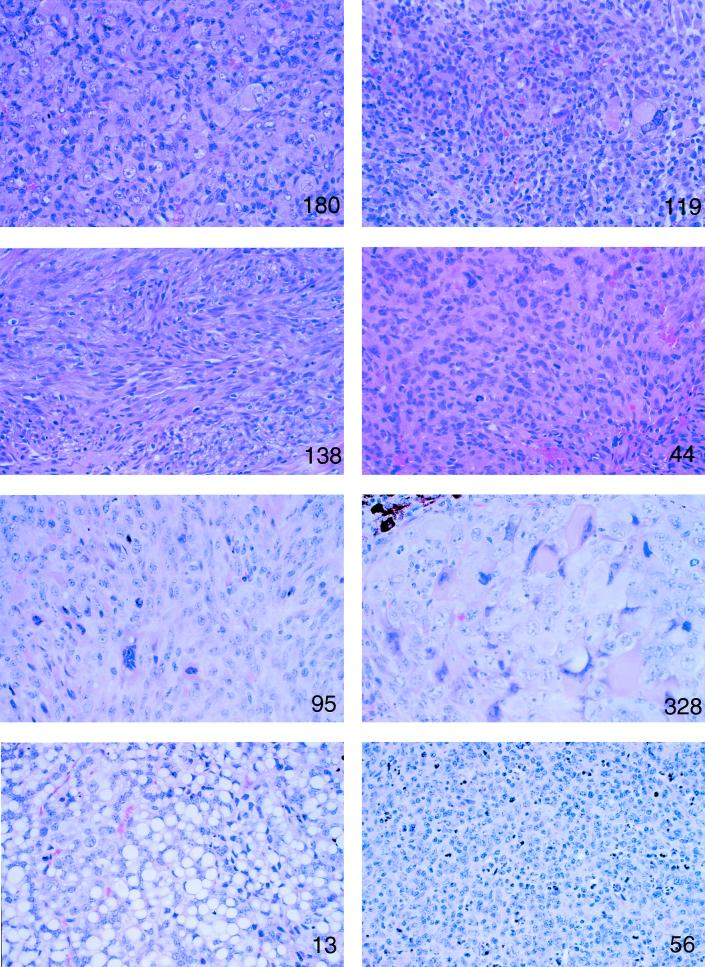
In humans, loss of INI1 is correlated with a variety of tumors, the vast majority of which are neuronal or renal in nature. To date, most human malignant rhabdoid tumors and choroid plexus carcinomas examined have deletions and/or mutations in INI1, as do a subset of central primitive neuroectodermal tumors and medullablastomas (39). In mice, we found that approximately 15% of Ini1 heterozygotes in both the mixed F1 (C57BL/6 × 129) or pure 129 backgrounds presented with tumors. All of these tumors arose in the head or neck regions of the mice, particularly in the soft tissue of the face (Table 2). While 2 of the 15 mouse tumors analyzed thus far had varying degrees of rhabdoid-like cells, none had the characteristic, monomorphous appearance of human rhabdoid tumors. Two Ini1in3/+ mice were found to have a lymphoproliferative disorder or lymphoma originating in an ill-defined region on the neck (Fig. 4). Two-thirds of the tumors originated on the faces of the mice. Interestingly, expression of Ini1 appears to be elevated during development in the branchial arch and in the frontonasal and maxillary processes (Fig. 2B), structures which contribute to formation of the face. While the majority of the facial tumors were poorly differentiated or undifferentiated sarcomas and not neuronal in origin, it is possible that the tumors arose in cells derived from neural crest progenitors, since neural crest cells, along with mesodermal cells, coordinate to form the facial primordia (12, 38).
TABLE 2.
Tumor occurrence
| Mouse no. | Age (wk) | Tumor site | Classification |
|---|---|---|---|
| Ini 119 | 36 | Face | Undifferentiated sarcoma |
| Ini 44 | 32 | Face | Undifferentiated sarcoma |
| Ini 138 | 23 | Face | Malignant fibrous histiocytoma |
| Ini 100 | 23 | Face | Malignant fibrous histiocytoma |
| Ini 180 | 31 | Face | Undifferentiated sarcoma |
| Ini 26a | 24 | Face | Malignant fibrous histiocytoma |
| Ini 262 | 16 | Face | Malignant fibrous histiocytoma |
| Ini 29a | 41 | Face | Malignant fibrous histiocytoma |
| Ini 13a | 59 | Face/eye | Liposarcoma |
| Ini 56 | 52 | Neck mass | Lymphoma |
| Ini 322 | 21 | Brain | Undifferentiated sarcoma |
| Ini 10 | 32 | Ventral to brain | Undifferentiated sarcoma |
| Ini 95a | 22 | Face mass | Malignant fibrous histiocytoma |
| Ini 127 | 12 | Neck mass | Lymphoproliferative disorder |
| Ini 328 | 25 | Eye | Undifferentiated sarcoma |
129 strain.
FIG. 4.
Ini1-heterozygous mice present with various tumors. Microscopic features of different tumors from Ini1in3/+ mice. Parafin-embedded tumors were sectioned, stained with hematoxylin and eosin, and examined under a microscope at magnification ×75. Mouse numbers corresponding to those presented in Table 2 are indicated in each panel.
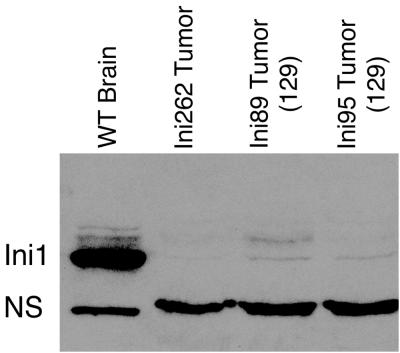
We have analyzed tumors in three representative mice. Northern analysis of total RNA harvested from tumor tissue indicated the presence of wild-type-length Ini1 message (data not shown). However, Western blot analysis of proteins harvested from these tumors revealed the absence of Ini1 protein in all three samples (Fig. 5). This indicates that loss of heterozygosity at the Ini1 locus is responsible for tumor formation in the Ini1in3/+ mice.
FIG. 5.
Loss of heterozygosity at the Ini1 locus results in tumor formation. Tumor samples and control tissue (wild-type [WT] male brain) were processed for Western analysis as detailed in Materials and Methods. The band corresponding to the Ini1 protein is indicated. NS represents a nonspecific band.
The mechanism of Ini1-mediated tumor suppression is unclear. Other subunits of the human and mouse SWI/SNF chromatin remodeling complexes have been reported to associate with known tumor suppressors, including Rb and Brca1 (3, 11, 48, 56), and several of the SWI/SNF subunits appear to be molecular targets of viral regulators of cell proliferation (18, 27, 28, 53). In addition, one of these subunits, BRG1, recently has been reported to be missing or mutated in a variety of human tumor cell lines, and reintroduction of BRG1 into these tumor cells reverses their transformed morphology (52). These findings suggest a role for chromatin remodeling in regulation of cell growth and/or in tumor suppression.
While this report was in preparation, Roberts et al. and Klochendler-Yeivin et al. published data consistent with our findings (21, 36). The fact that these results are reproducible in knockout lines generated by different targeting strategies confirms the importance of Ini1 in development and tumorigenesis. Klochendler-Yeivin et al. (21) further report in their study that Ini1-deficient embryos can induce the formation of maternal decidua, suggesting that Ini1-deficient embryos undergo hatching and implantation prior to their demise. In contrast, Ini1in3/in3 embryos fail to hatch from the zona pellucida, suggesting that subtle strain variations may influence the precise timing of embryonic lethality. In agreement with these other groups, a percentage of the Ini1-heterozygous mice in our colony presented with tumors that contained variable numbers of rhabdoid cells. However, we are hesitant to classify these undifferentiated sarcomas as true rhabdoid tumors, which are described as monomorphous tumors in the human population. Discrepancies between tumor types associated with disruption of Ini1 in humans and in mice may be due to differences in species-specific differentiation pathways. Regardless, the Ini1-heterozygous mice should provide a useful model for studying the general mechanisms involved in tumor suppression by Ini1.
ACKNOWLEDGMENTS
We thank J. Castillo and D. Hill for help in preparing the manuscript. We also thank A. Fraire and R. Hesselton for assistance with histopathology.
This work was supported in part by grants from the NIH to A.N.I. and S.N.J. A.N.I. is supported by a Scholar Award from the Leukemia and Lymphoma Society.
REFERENCES
- 1.Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 2.Biegel J A, Zhou J Y, Rorke L B, Stenstrom C, Wainwright L M, Fogelgren B. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59:74–79. [PubMed] [Google Scholar]
- 3.Bochar D A, Wang L, Beniya H, Kinev A, Xue Y, Lane W S, Wang W, Kashanchi F, Shiekhattar R. BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell. 2000;102:257–265. doi: 10.1016/s0092-8674(00)00030-1. [DOI] [PubMed] [Google Scholar]
- 4.Breeden L, Nasmyth K. Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell. 1987;48:389–397. doi: 10.1016/0092-8674(87)90190-5. [DOI] [PubMed] [Google Scholar]
- 5.Bruder C E, Dumanski J P, Kedra D. The mouse ortholog of the human SMARCB1 gene encodes two splice forms. Biochem Biophys Res Commun. 1999;257:886–890. doi: 10.1006/bbrc.1999.0563. [DOI] [PubMed] [Google Scholar]
- 6.Cairns B R, Kim Y-J, Sayre M H, Laurent B C, Kornberg R D. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci USA. 1994;91:1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiba H, Muramatsu M, Nomoto A, Kato H. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila Brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Côté J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 9.DeCristofaro M F, Betz B L, Wang W, Weissman B E. Alteration of hSNF5/INI1/BAF47 detected in rhabdoid cell lines and primary rhabdomyosarcomas but not Wilms' tumors. Oncogene. 1999;18:7559–7565. doi: 10.1038/sj.onc.1203168. [DOI] [PubMed] [Google Scholar]
- 10.de la Serna I L, Carlson K A, Hill D A, Guidi C J, Stephenson R O, Sif S, Kingston R E, Imbalzano A N. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol Cell Biol. 2000;20:2839–2851. doi: 10.1128/mcb.20.8.2839-2851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunaief J L, Strober B E, Guha S, Khavari P A, Ålin K, Luban J, Begemann M, Crabtree G R, Goff S P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 12.Francis-West P, Ladher R, Barlow A, Graveson A. Signalling interactions during facial development. Mech Dev. 1998;75:3–28. doi: 10.1016/s0925-4773(98)00082-3. [DOI] [PubMed] [Google Scholar]
- 13.Fryer C J, Archer T K. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 14.Harvey M, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A, Donehower L A. Spontaneous and carcinogen-induced tumorigenesis in p53-deficient mice. Nat Genet. 1993;5:225–229. doi: 10.1038/ng1193-225. [DOI] [PubMed] [Google Scholar]
- 15.Hirschhorn J N, Brown S A, Clark C D, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 16.Imbalzano A. ATP dependent chromatin remodelers: complex complexes and their components. Crit Rev Eukaryot Gene Expr. 1998;8:225–255. doi: 10.1615/critreveukargeneexpr.v8.i3-4.10. [DOI] [PubMed] [Google Scholar]
- 17.Imbalzano A N, Kwon H, Green M R, Kingston R E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 18.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 19.Khavari P A, Peterson C L, Tamkun J W, Crabtree G R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 20.Kingston R, Narlikar G. ATP dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 21.Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1:500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kodaki T, Hosaka K, Nikawa J, Yamashita S. The SNF2/SWI2/GAM1/TYE3/RIC1 gene is involved in the coordinate regulation of phospholipid synthesis in Saccharomyces cerevisiae. J Biochem (Tokyo) 1995;117:362–368. doi: 10.1093/jb/117.2.362. [DOI] [PubMed] [Google Scholar]
- 23.Kruger W, Herskowitz I. A negative regulator of HO transcription, SIN1 (SPT2), is a nonspecific DNA binding protein related to HMG1. Mol Cell Biol. 1991;11:4135–4146. doi: 10.1128/mcb.11.8.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kruger W, Peterson C L, Sil A, Coburn C, Arents G, Moudrianakis E N, Herskowitz I. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 1995;9:2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- 25.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 26.Laurent B C, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- 27.Lee D, Sohn H, Kalpana G V, Choe J. Interaction of E1 and hSNF5 proteins stimulates replication of human papillomavirus DNA. Nature. 1999;399:487–491. doi: 10.1038/20966. [DOI] [PubMed] [Google Scholar]
- 28.Miller M E, Cairns B R, Levinson R S, Yamamoto K R, Engel D A, Smith M M. Adenovirus E1A specifically blocks SWI/SNF-dependent transcriptional activation. Mol Cell Biol. 1996;16:5737–5743. doi: 10.1128/mcb.16.10.5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muchardt C, Sardet C, Bourachot B, Onufryk C, Yaniv M. A human protein with homology to Saccharomyces cerevisiae SNF5 interacts with the potential helicase hbrm. Nucleic Acids Res. 1995;23:1127–1132. doi: 10.1093/nar/23.7.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy D J, Hardy S, Engel D A. Human SWI-SNF component BRG1 represses transcription of the c-fos gene. Mol Cell Biol. 1999;19:2724–2733. doi: 10.1128/mcb.19.4.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in saccharomyces cerevisiae. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson C L, Dingwall A, Scott M P. Five SWI/SNF gene products are components of a large multiprotein complex required for transcriptional enhancement. Proc Natl Acad Sci USA. 1994;91:2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterson L, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 35.Reyes J C, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts C W M, Galusha S A, McMenamin M E, Fletcher C D M, Orkin S H. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc Natl Acad Sci USA. 2000;97:13796–13800. doi: 10.1073/pnas.250492697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rozenblatt-Rosen O, Rozovskaia T, Burakov D, Sedkov Y, Tillib S, Blechman J, Nakamura T, Croce C M, Mazo A, Canaani E. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc Natl Acad Sci USA. 1998;95:4152–4157. doi: 10.1073/pnas.95.8.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schilling T F. Genetic analysis of craniofacial development in the vertebrate embryo. Bioessays. 1997;19:459–468. doi: 10.1002/bies.950190605. [DOI] [PubMed] [Google Scholar]
- 39.Sévenet N, Lellouch-Tubiana A, Schofield D, Hoang-Xuan K, Gessler M, Birnbaum D, Jeanpierre C, Jouvet A, Delattre O. Spectrum of hSNF5/INI1 somatic mutations in human cancer and genotype-phenotype correlations. Hum Mol Genet. 1999;8:2359–2368. doi: 10.1093/hmg/8.13.2359. [DOI] [PubMed] [Google Scholar]
- 40.Sévenet N, Sheridan E, Amram D, Schneider P, Handgretinger R, Delattre O. Constitutional mutations of the hSNF5/INI1 gene predispose to a variety of cancers. Am J Hum Genet. 1999;65:1342–1348. doi: 10.1086/302639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shanahan F, Seghezzi W, Parry D, Mahony D, Lees E. Cyclin E associates with BAF155 and BRG1, components of the mammalian SWI-SNF complex, and alters the ability of BRG1 to induce growth arrest. Mol Cell Biol. 1999;19:1460–1469. doi: 10.1128/mcb.19.2.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stern M J, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 43.Sterner D E, Berger S L. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strahl B D, Allis C D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 45.Strobeck M W, Knudsen K E, Fribourg A F, DeCristoforo M F, Weissman B E, Imbalzano A N, Knudsen E S. BRG-1 is required for Rb-mediated cell cycle arrest. Proc Natl Acad Sci USA. 2000;97:7748–7753. doi: 10.1073/pnas.97.14.7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sumi-Ichinose C, Ichinose H, Metzger D, Chambon P. SNF2β-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol Cell Biol. 1997;17:5976–5986. doi: 10.1128/mcb.17.10.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takai H, Tominaga K, Motoyama N, Minamishima Y A, Nagahama H, Tsukiyama T, Ikeda K, Nakayama K, Nakanishi M, Nakayama K-I. Aberrant cell cycle checkpoint function and early embryonic death in Chk1−/− mice. Genes Dev. 2000;14:1439–1447. [PMC free article] [PubMed] [Google Scholar]
- 48.Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. Rb and hbrm cooperate to repress the activation functions of E2F1. Proc Natl Acad Sci USA. 1997;94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Versteege I, Sevenet N, Lange J, Rousseau-Merck M F, Ambros P, Handgretinger R, Aurias A, Delattre O. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 50.Vignali M, Hassan A H, Neely K E, Workman J L. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang W, Xue Y, Zhou S, Kuo A, Cairns B R, Crabtree G R. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 52.Wong A K, Shanahan F, Chen Y, Lian L, Ha P, Hendricks K, Ghaffari S, Iliev D, Penn B, Woodland A M, Smith R, Salada G, Carillo A, Laity K, Gupte J, Swedlund B, Tavtigian S V, Teng D H, Lees E. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 2000;60:6171–6177. [PubMed] [Google Scholar]
- 53.Wu D Y, Kalpana G V, Goff S P, Schubach W H. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J Virol. 1996;70:6020–6028. doi: 10.1128/jvi.70.9.6020-6028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshimoto H, Yamashita I. The GAM1/SNF2 gene of Saccharomyces cerevisiae encodes a highly charged nuclear protein required for transcription of the STA1 gene. Mol Gen Genet. 1991;228:270–280. doi: 10.1007/BF00282476. [DOI] [PubMed] [Google Scholar]
- 55.Zambrowicz B, Friedrich G, Buxton E, Lilleberg S, Person C, Sands A. Disruption and sequence identification of 2,000 genes in mouse embryonic stem cells. Nature. 1998;392:608–611. doi: 10.1038/33423. [DOI] [PubMed] [Google Scholar]
- 56.Zhang H S, Gavin M, Dahiya A, Postigo A A, Ma D, Luo R X, Harbour J W, Dean D C. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]