Abstract
Background
XPO1 mediates the nuclear export of several proteins, mainly tumor suppressors. KPT‐330 (Selinexor) is a selective inhibitor of XPO1 that has demonstrated good therapeutic effects in hematologic cancers.
Methods
We used TCGA and GTEx pan‐cancer database to evaluate XPO1 mRNA expression in various tumors. Cell proliferation assay and colony formation assay were used to analyze the in vitro antitumor effects of XPO1 inhibitor KPT‐330. Western blot was performed to explore the specific mechanisms.
Results
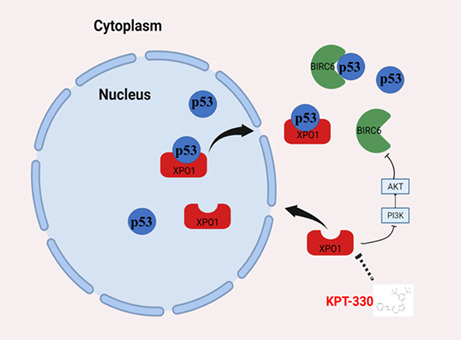
We found that XPO1 was highly expressed across a range of cancers and associated with poor prognosis in hepatobiliary and pancreatic tumors. We revealed that the XPO1 inhibitor KPT‐330 triggered the nuclear accumulation of the p53 protein and significantly disrupted the proliferation of cholangiocarcinoma cells. Mechanistically, the XPO1 inhibitor, KPT‐330, reduced BIRC6 expression by inhibiting the PI3K/AKT pathway to decrease p53 degradation and improve its stability.
Conclusion
Therefore, XPO1 may be a potential therapeutic target in cholangiocarcinoma, mediated by its effects on KPT‐330.
We found that XPO1 showed high expression in pan‐cancer and was associated with poor prognosis in hepatobiliary and pancreatic tumors. In CHOL, we revealed that XPO1 inhibitor KPT‐330 could trigger the nuclear accumulation of the p53 protein and in this way significantly disrupted the proliferation of cholangiocarcinoma cells. Mechanistically, XPO1 inhibitor KPT‐330 reduced BIRC6 expression by inhibiting PI3K/AKT pathway to decrease p53 degradation and improve its stability
Abbreviations
- XPO1
Exportin‐1
- FDA
United States Food and Drug Administration
- BLCA
Bladder Urothelial Carcinoma
- BRCA
Breast invasive carcinoma
- CESC
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- CHOL
Cholangiocarcinoma
- COAD
Colon adenocarcinoma
- DLBC
Lymphoid Neoplasm Diffuse Large B‐cell Lymphoma
- ESCA
Esophageal carcinoma
- GBM
Glioblastoma multiforme
- HNSC
Head and Neck squamous cell carcinoma
- KICH
Kidney Chromophobe
- KIRP
Kidney renal papillary cell carcinoma
- LAML
Acute Myeloid Leukemia
- LGG
Brain Lower Grade Glioma
- LIHC
Liver hepatocellular carcinoma
- LUAD
Lung adenocarcinoma
- LUSC
Lung squamous cell carcinoma
- OV
Ovarian serous cystadenocarcinoma
- PAAD
Pancreatic adenocarcinoma
- READ
Rectum adenocarcinoma
- STAD
Stomach adenocarcinoma
- TGCT
Testicular Germ Cell Tumors
- THCA
Thyroid carcinoma
- THYM
Thymoma
- TCGA
The Cancer Genome Atlas
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- CHX
cycloheximide
1. BACKGROUND
Cholangiocarcinoma is a highly aggressive malignancy, characterized by cholangiocyte differentiation. 1 The 5‐year survival rate for cholangiocarcinoma is 20%–40%, however, the only effective treatment is complete surgical resection. 2 Unfortunately, owing to the lack of typical clinical manifestations, many patients are diagnosed at an advanced stage and are not eligible for surgery. 3 Therefore, there is an urgent need to identify new diagnostic and therapeutic targets for the treatment of cholangiocarcinoma.
Exportin‐1 (XPO1) is a major nuclear export receptor protein that transports cargo proteins via a leucine‐rich nuclear export signal (NES) from the nucleus to the cytoplasm. 4 Cargo proteins of XPO1 are mainly tumor suppressors, including p21 and p53. 5 High expression of XPO1 has been reported in various tumors, including pancreatic adenocarcinoma, gastric cancer, neuroblastoma, and hematologic tumors, and is strongly associated with poor prognosis. 6 , 7 , 8 , 9 Therefore, the inhibition of XPO1 may be a therapeutic tool for tumors. Scientists have developed selective inhibitors for XPO1 over the years, and Selinexor (KPT‐330) has been clinically validated and approved by the United States Food and Drug Administration. 10 However, the role of XPO1 and its therapeutic effect on KPT‐330 on cholangiocarcinoma has yet to be elucidated.
In this study, we analyzed the information in the Cancer Genome Atlas (TCGA) database and found that XPO1 was highly expressed in 21 types of tumors and was clearly associated with poor patient prognosis. We used bioinformatics analysis to clarify that the genes positively associated with high expression of XPO1 were mainly associated with ubiquitin‐mediated proteolysis, RNA transport, and the spliceosome. We found that XPO1 inhibition by KPT‐330 significantly disrupted cholangiocarcinoma cell proliferations. Further experiments revealed that the XPO1 inhibitor KPT‐330 increased intranuclear accumulation of p53 and increased its protein stability. We discovered that BIRC6 could cause p53 degradation; however, KPT‐330 reduced BIRC6 expression by inhibiting PI3K/AKT pathway to alleviate p53 degradation. Therefore, XPO1 may be a potential therapeutic target and KPT‐330 may play a therapeutic role in cholangiocarcinoma.
2. MATERIALS AND METHODS
2.1. Cell culture and treatment
Cholangiocarcinoma cells (RBE and 9810) were purchased from the Cell Bank of the Shanghai Institute for Biological Sciences, Chinese Academy of Sciences. Cells were cultured in DMEM medium (Gibco) supplemented with 10% fetal bovine serum (Gibco) in a humid chamber at 37°C with 5% CO2. KPT‐330, cycloheximide (CHX), and 740 Y‐P were purchased from Selleck Chemicals.
2.2. RNA extraction and qRT‐PCR
Total RNAs were extracted from RBE and 9810 cells by using Trizol reagent (Invitrogen). cDNA was generated using the PrimeScript RT reagent kit with gDNA Eraser (TaKaRa) according to the manufacturer's instructions. The primers used for amplification were followed: GAPDH forward primer (5‐CAACAGCCTCAAGATCATCAGC‐3), GAPDH reverse primer (5‐TTCTAGACGGCAGGTCAGGTC‐3), p53 forward primer (5‐ CAGCACATGACGGAG GTTGT‐3), and p53 reverse primer (5‐TCATCCAAA TACTCCACACGC‐3).
2.3. Western blot
Total protein extraction and western blotting were conducted as previously described. 11 In brief, first, proteins were isolated with RIPA Lysis buffer (Beyotime). Next, proteins were separated by SDS‐PAGE and transferred onto PVDF membranes (Millipore). Then, 5% skim milk was used to block the blots for 1 h at room temperature. A series of primary antibodies (Abcam) was added to the appropriate position of the PVDF membranes and incubated overnight at 4°C. Finally, all blots reacted with the suitable Horseradish Peroxidase‐conjugated secondary antibody (Beyotime), and the immunoreactive bands were detected by chemiluminescence and visualized using a Gel Doc 2000 (Bio‐Rad). Antibodies against XPO1, p53, H3, BIRC6, PI3K, p‐PI3K(Tyr458), AKT, p‐AKT(Ser473), and GAPDH were purchased from Abcam.
2.4. Cell proliferation assays
Approximately 1500 cells of RBE and 9810 cells were seeded in 96‐well plates. Cell proliferation was assessed using a Cell Counting Kit‐8 assay (Yeasen). The cell proliferation curves were plotted using absorbance at 450 nm.
2.5. Colony formation assay
RBE and 9810 cells were seeded in 6‐well plates at a density of 1000 cells per well for 10 days. The cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet.
2.6. siRNA, plasmid construction, and transfection
The siRNA‐transfected cholangiocarcinoma cells used were as follows: p53‐siRNA: 5‐CGGCGCACAGAGGAAGAGAAUTT‐3. R‐fect (Baidai) was used to transfect siRNA. The plasmid pBIRC6, which encodes the full‐length cDNA of the BIRC6 (NM_001378125.1) and the XPO1 (NM_001410799.1) was constructed by Genomeditech. The empty vector (pCMV6) was used as a negative control. Viafect transfection reagent was used for plasmid transfection according to the protocol (Promega).
2.7. Immunofluorescence assay
RBE and 9810 cells were seeded on coverslips 1 day before and fixed using 4% paraformaldehyde for 15 min at room temperature. We implied Immunofluorescence Staining Kit (Beyotime) according to the protocol. A fluorescence microscope (Leica DM4B) was used to capture images.
2.8. Data collection and analysis
XPO1 expression profiles and TCGA and Genotype‐Tissue Expression (GTEx) clinical pan‐cancer data were downloaded from the University of California, Santa Cruz (UCSC) Xena database (https://xenabrowser.net/datapages/). For pan‐cancer data of TCGA and GTEx databases, differential XPO1 mRNA expression between various tumors and normal tissues was analyzed using t test, and their visualization was carried out by R software package “ggplot2.” For data transformation, RNAseq data in Fregments Per Kilobase per Million (FPKM) format were converted to transcripts per million reads (TPM) format and log2 transformed. In order to assess the expression of XPO1, tumor tissues were obtained from TCGA, and normal tissues were obtained from TCGA and the GTEx databases.
2.9. Correlation and enrichment analysis
Pearson correlation analysis of XPO1 mRNA and other mRNAs was conducted in cholangiocarcinoma using TCGA CHOL data. The 500 genes most positively associated with XPO1 were selected for enrichment analysis to determine the function of XPO1. Gene ontology (GO) analysis was performed using the EnrichGO function in the clusterProfiler R software package R with the following parameters: p value.adj <0.1 and Q value <0.2.
2.10. Statistical analysis
Statistical analyses were performed with Prism 8 (GraphPad Software) and SPSS 24 (SPSS Inc.). Student's t test was performed between two groups, and analysis between multiple groups was conducted by one‐way analysis of variance, data are recorded in the form of mean ± SD. p Values of <0.05 were considered statistically significant (*p < 0.05, **p < 0.01, ***p < 0.001).
3. RESULTS
3.1. Pan‐cancer expression of XPO1
Using TCGA and GTEx pan‐cancer database, we evaluated XPO1 mRNA expression in various tumors. The results of the analysis suggested that XPO1 showed high expression in 21 types of tumors, including bladder cancer (BLCA), breast cancer (BRCA), cervical cancer (CESC), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), diffuse large B‐cell lymphoma (DLBC), esophageal carcinoma (ESCA), glioblastoma (GBM), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal papillary cell carcinoma (KIRP), acute myeloid leukemia (LAML), brain lower grade glioma (LGG), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), pancreatic adenocarcinoma (PAAD), rectum adenocarcinoma (READ), stomach adenocarcinoma (STAD), testicular germ cell tumors (TGCT), and thymoma (THYM) (Figure 1A). Notably, in TCGA unpaired samples, XPO1 was highly expressed in hepatobiliary and pancreatic tumors (CHOL, LIHC, and PAAD) (Figure 1B). In the paired samples, we found that XPO1 was highly expressed in CHOL and LIHC compared with the corresponding normal tissues (pancreatic cancer pairing data lacked significance with only four pairs) (Figure 1C). These results suggest that XPO1 may play an important role in tumor development and warrant further investigation.
FIGURE 1.
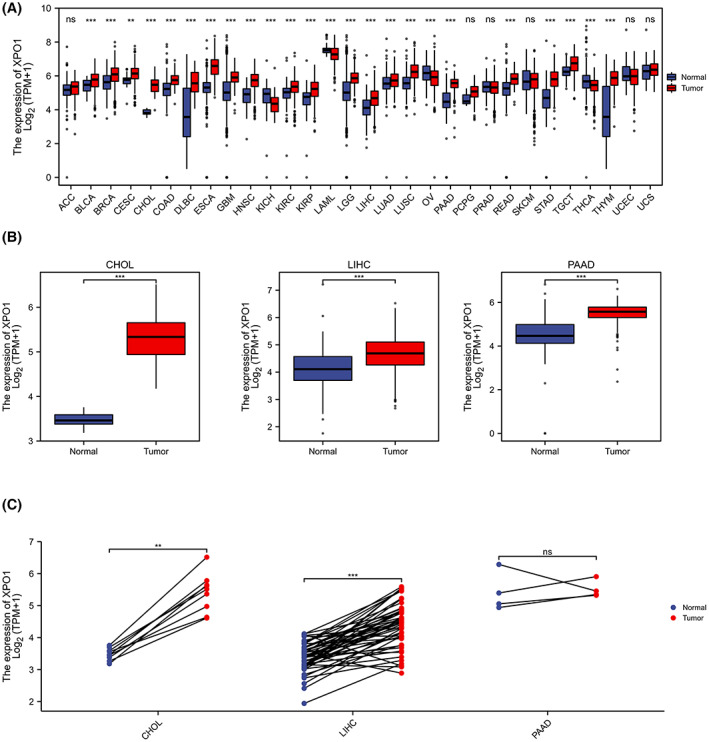
Pan‐cancer expression of XPO1. (A) XPO1 expression in tumor and normal tissues in pan‐cancer data of the Cancer Genome Atlas (TCGA) and GTEx. (B) XPO1 expression in tumor and normal tissues in CHOL, LIHC, and PAAD from TCGA. (C) XPO1 expression in paired tumor and normal tissues in CHOL, LIHC, and PAAD from TCGA. Data were shown as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. CHOL, cholangiocarcinoma; GTEx, genotype‐tissue expression; LIHC, liver hepatocellular carcinoma; PAAD, pancreatic adenocarcinoma; TCGA, the cancer genome atlas; XPO1, exportin‐1.
3.2. Association between high XPO1 expression and tumor prognosis in patients
Next, we focused on hepatobiliary and pancreatic tumors: CHOL, LIHC, and PAAD. To further investigate the relationship between high XPO1 expression and patient prognosis, we analyzed its association with “overall survival” and “progress‐free interval.” As shown in Figure 2A–F, higher XPO1 expression was significantly related to poor patient prognosis in LIHC and PAAD; however, we did not observe a significant difference in CHOL. We analyzed the prognosis of 30 patients with cholangiocarcinoma using GSE107943 from the GEO database and the results showed that higher expression of XPO1 was significantly associated with poor patient prognosis in CHOL (Figure 2G). In the data on “N stage” and “vascular invasion,” we found significant differences in CHOL (Figure 2H,I). These results suggest that high XPO1 expression in hepatobiliary and pancreatic tumors is strongly associated with poor prognosis.
FIGURE 2.
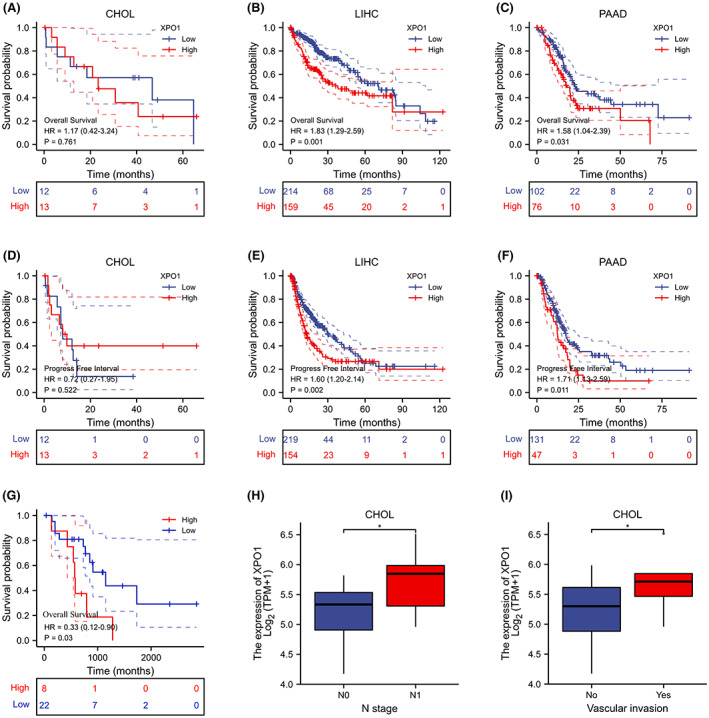
Association between high XPO1 expression and tumor prognosis in patients. (A–F) The correlation between XPO1 expression and the prognosis of CHOL, LIHC, and PAAD was analyzed using TCGA. (G) The correlation between XPO1 expression and the prognosis of CHOL was analyzed using the GEO database. (H) The correlation between XPO1 expression and “N stage” in CHOL. (I) The correlation between XPO1 expression and “Vascular invasion” in CHOL. Data were shown as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. CHOL, cholangiocarcinoma; LIHC, liver hepatocellular carcinoma; PAAD, pancreatic adenocarcinoma; TCGA, the cancer genome atlas; XPO1, exportin‐1.
3.3. Correlation and enrichment analyses in CHOL
To further elucidate the function of XPO1 in CHOL, we analyzed the genes that were positively correlated with XPO1 expression in the TCGA database. We selected the 500 genes that were most positively correlated with XPO1 for enrichment analysis and displayed the top 50 genes in a heat map (ranked according to correlation) (Figure 3A). Furthermore, we used the R clusterProfiler package to analyze the possible enrichment pathways and correlation of these 500 genes. GO functional enrichment analysis indicated that XPO1 was related to modified and binding functions, which corresponded to the transport function of XPO1 (Figure 3B–E). The Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed that spliceosome, RNA transport, ubiquitin‐mediated proteolysis, and cell cycle terms were significantly enriched (Figure 3A,F). These results indicate that high XPO1 expression is associated with multiple pathways related to cholangiocarcinoma progression which is closely linked to XPO1 transport function.
FIGURE 3.
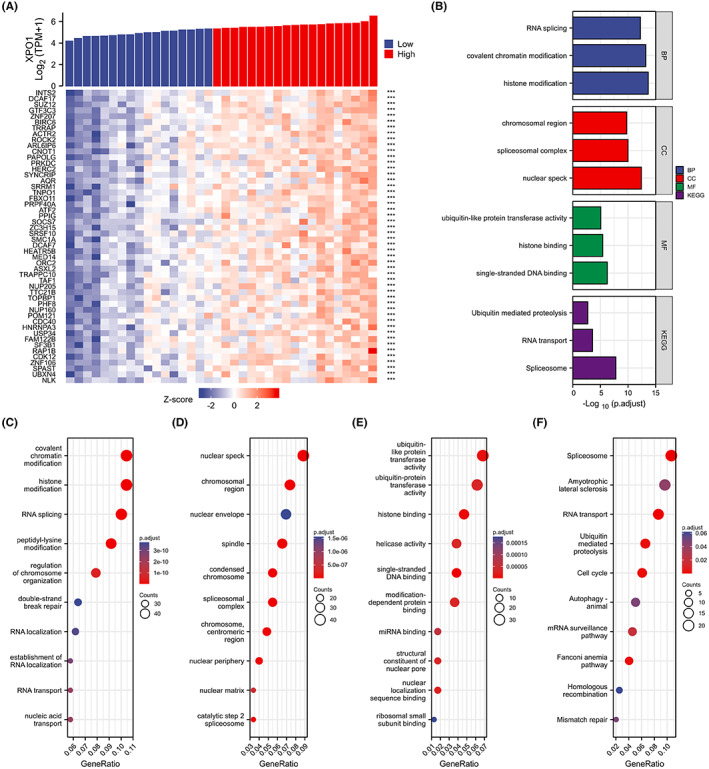
Correlation and enrichment analyses in CHOL. (A) The top 50 genes positively correlated with XPO1in a heat map (ranked according to correlation) from TCGA. (B–F) Significant Gene Ontology terms of the top 500 genes most positively associated with XPO1, including biological processes (C), cell component (D), molecular function (E), and KEGG (F). CHOL, cholangiocarcinoma; KEGG, Kyoto encyclopedia of genes and genomes; TCGA, the cancer genome atlas; XPO1, exportin‐1.
3.4. Effect of the XPO1 inhibitor KPT‐330 on cholangiocarcinoma cell proliferation
Based on bioinformatics analysis, we performed basic experiments to verify the effect of XPO1 on cholangiocarcinoma tumor cells. We found that the proliferation of RBE and 9810 cells was inhibited by the XPO1 inhibitor KPT‐330 in time‐ and concentration‐dependent manners (Figure 4A,B). We chose a KPT‐330 concentration of 2.5 μM and a treatment time of 48 hours for subsequent experiments. We conducted a western blot and found that XPO1 was effectively inhibited by KPT‐330 in RBE and 9810 cells (Figure 4C). The colony formation assay indicated that KPT‐330 significantly reduced the number and size of colonies formed by cholangiocarcinoma cells (Figure 4D), further confirming its antiproliferative effect. Further, EdU‐488 DNA synthesis assay experiments showed the inhibitory effect of KPT‐330 on the proliferation of RBE and 9810 (Figure 4E). These results indicate that XPO1 promotes cholangiocarcinoma cell proliferation.
FIGURE 4.
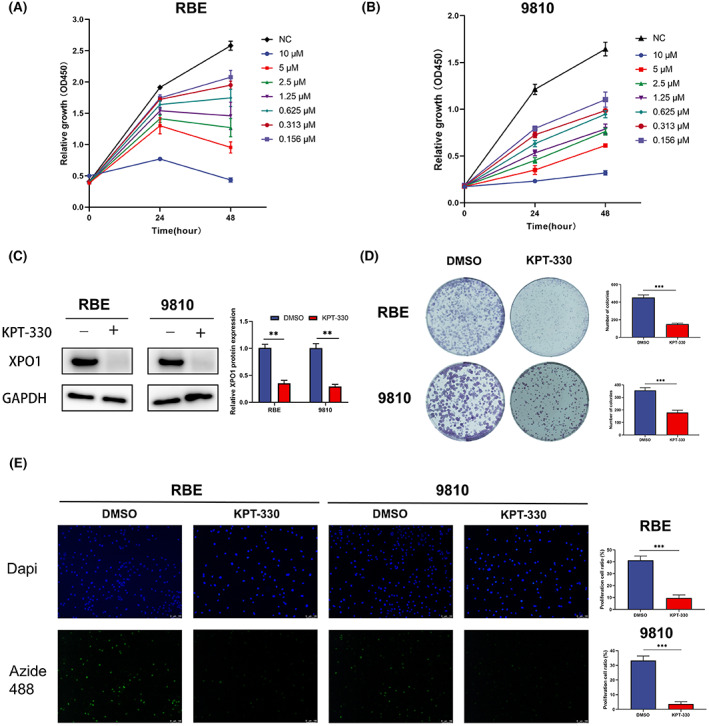
Effect of exportin‐1 (XPO1) inhibitor KPT‐330 on cholangiocarcinoma cell proliferation. (A, B) Cell proliferation of RBE and 9810 under KPT‐330 treatment. (C) Western blot of XPO1 under KPT‐330 treatment. (D) The colony formation of RBE and 9810 under KPT‐330 treatment. (E) EdU‐488 DNA synthesis assay of RBE and 9810 under KPT‐330 treatment. Data were shown as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
3.5. Overexpression of XPO1 promoted cholangiocarcinoma cell proliferation
We used XPO1 overexpression plasmid in RBE and 9810 cells to investigate the effect of XPO1 on cholangiocarcinoma cell proliferation (Figure 5A). CCK‐8 assays showed that overexpression of XPO1 significantly promoted the proliferation of cholangiocarcinoma cells (Figure 5B,C). In addition, the colony formation assay indicated that XPO1 overexpression significantly increased the number and size of colonies formed by cholangiocarcinoma cells (Figure 5D). In addition, the EdU‐488 DNA synthesis assay showed a significant pro‐proliferative effect of KPT‐330 on RBE and 9810 cells (Figure 5E). These results suggest that XPO1 promotes the proliferation of cholangiocarcinoma cells.
FIGURE 5.
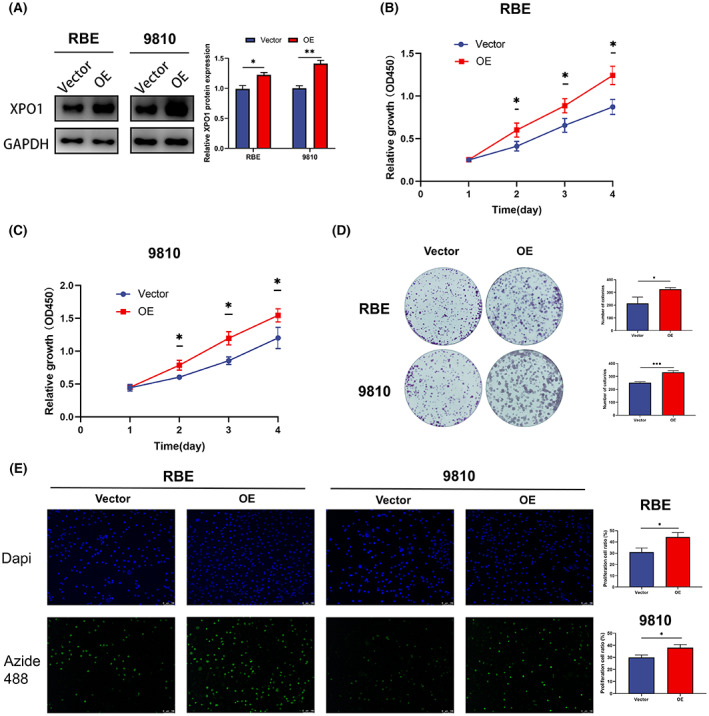
Overexpression of exportin‐1 (XPO1) promoted cholangiocarcinoma cell proliferation. (A) Western blot of XPO1 after XPO1 plasmid transfection in RBE and 9810 cells. (B) Cell proliferation of RBE after XPO1 plasmid transfection. (C) Cell proliferation of 9810 after XPO1 plasmid transfection. (D) The colony formation of RBE and 9810 after XPO1 plasmid transfection. (E) EdU‐488 DNA synthesis assay of RBE and 9810 after XPO1 plasmid transfection. Data were shown as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
3.6. Influence of XPO1 inhibitor KPT‐330 on p53
p53 is a carrier protein for XPO1. 12 , 13 Therefore, we conducted an immunofluorescence assay to clarify p53 status under KPT‐330 treatment. The results showed that XPO1 inhibition by KPT‐330 triggered the accumulation of p53 inside the nucleus (Figure 6A). Next, we analyzed the changes in the nuclear and cytoplasmic levels of p53 using a western blot, and the results suggested that p53 accumulated in the nucleus after KPT‐330 treatment (Figure 6B). Knockdown of p53 using siRNA rescued the inhibitory effect of KPT‐330 (Figure 6C). This indicates that XPO1 inhibition by KPT‐330 could activate the p53 function to exert anticancer effects. Furthermore, we found that the mRNA expression of p53 did not change significantly under the KPT‐330 treatment (Figure 6D). Next, we blocked the translation of p53 with CHX and detected protein expression of p53 at four time points: 0, 1, 2, and 4 h. Western blot results showed that KPT‐330 significantly reduced the degradation rate of p53 protein in RBE and 9810 cells (Figure 6E). In summary, XPO1 inhibition triggered the accumulation of p53 in the nucleus and improved its stability.
FIGURE 6.
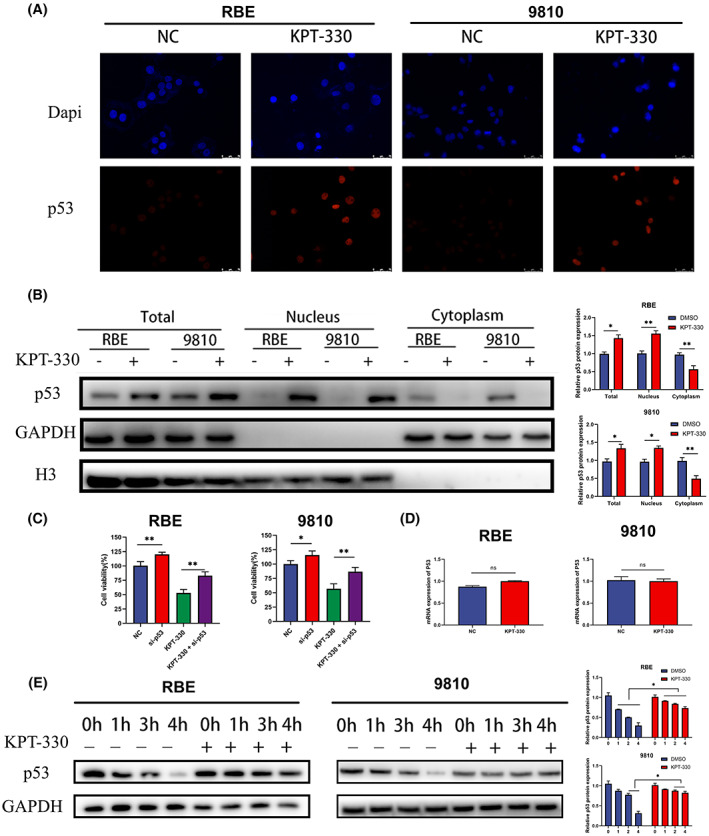
Influence of exportin‐1 (XPO1) inhibitor KPT‐330 on p53. (A) Immunofluorescence assay of p53. (B) Western blot results of the nucleus and cytoplasmic components after KPT‐330 treatment. (C) Cell proliferation of RBE and 9810 under KPT‐330 treatment and p53‐siRNA. (D) qRT‐PCR analysis of p53 mRNA under KPT‐330 treatment. (E) Western blot of the degradation rate of p53 protein after KPT‐330 treatment. Data were shown as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
3.7. Effect of BIRC6 on p53
Based on previous bioinformatics analysis, we screened the protein BIRC6, which may affect the progression of cholangiocarcinoma cells with XPO1. We first analyzed the correlation between XPO1 and BIRC6. The result suggested that the expression of BIRC6 was closely related to that of XPO1 (Figure 7A). Tang et al. reported that BIRC6 facilitates p53 degradation in hepatocellular carcinoma. 14 We detected the expression of BIRC6 and p53 after KPT‐330 treatment. Western blot results showed that BIRC6 expression was significantly decreased; however, p53 expression was significantly increased under KPT‐330 treatment (Figure 7B). Next, we overexpressed BIRC6 and assessed its effect on p53 expression. The results indicate that the overexpression of BIRC6 reduced the increase in p53 caused by XPO1 inhibition by KPT‐330 (Figure 7C). Furthermore, we used CCK‐8 to assess the effect of BIRC6 overexpression on the efficacy of KPT‐330. The results in Figure 7D show that overexpression of BIRC6 rescued the inhibitory effect on the proliferation of KPT‐330. XPO1 inhibition has been reported to inhibit the PI3K/AKT pathway, as demonstrated in our study (Figure 7E). Furthermore, we found that the addition of the PI3K agonist 740 Y‐P rescued the reduction in BIRC6 caused by XPO1 inhibition. Overall, XPO1 inhibition reduced the expression of BIRC6 by inhibiting the PI3K/AKT pathway and thus reducing p53 degradation.
FIGURE 7.
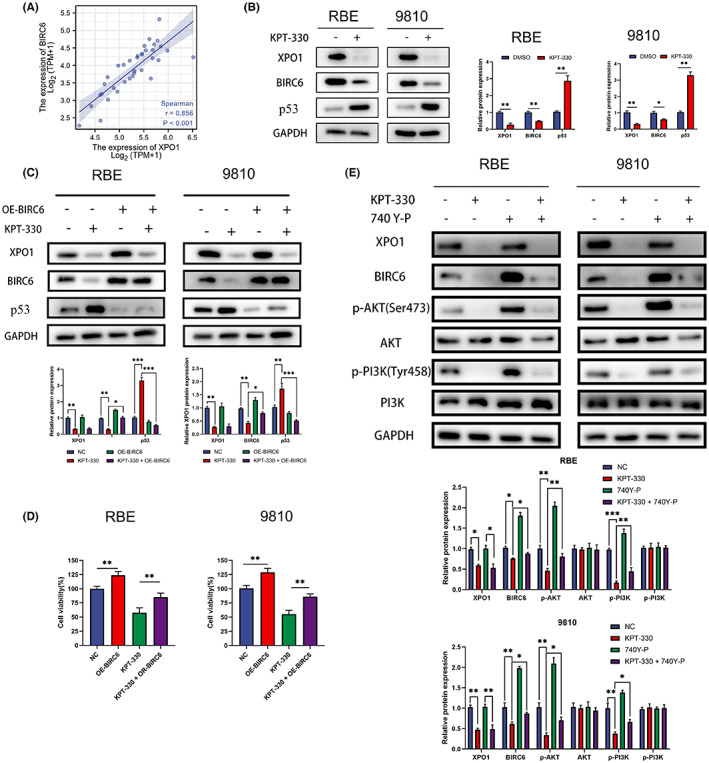
Effect of BIRC6 on p53. (A) Correlation between exportin‐1 (XPO1) and BIRC6 from TCGA. (B) Western blot of p53 and BIRC6 under KPT‐330 treatment. (C) Western blot of p53 and BIRC6 under KPT‐330 treatment and overexpression of BIRC6. (D) Cell proliferation of RBE and 9810 under KPT‐330 treatment and overexpression of BIRC6. (E) Western blot of PI3K/AKT pathway and BIRC6 under KPT‐330 treatment and 740 Y‐P. Data were shown as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
4. DISCUSSION
XPO1 carries NES‐containing cargo and transports it from the nucleus to the cytoplasm, thus playing an important role in maintaining cell survival. 15 , 16 XPO1 dysfunction leads to the mislocalization of cargo proteins, including many proteins associated with tumor progression, such as p53 and p27. 17 In normal cells, tumor suppressors function in the nucleus, but in tumor cells, XPO1 is overexpressed and transports tumor suppressors out of the nucleus, thereby promoting tumor progression. 18 , 19
We found that XPO1 was highly expressed in 21 tumor types, including BLCA, BRCA, CESC, CHOL, COAD, DLBC, ESCA, GBM, HNSC, KICH, KIRP, LAML, LGG, LIHC, LUAD, LUSC, PAAD, READ, STAD, TGCT, and THYM. In terms of prognosis, high XPO1 expression was significantly associated with overall survival and progress‐free interval in LIHC and PAAD. Although similar results were not observed in CHOL, high expression of XPO1 was significantly associated with the “N stage” and “vascular invasion” in CHOL. However, we found that the high expression of XPO1 was significantly associated with overall survival in patients with CHOL in the GEO database. These results suggest that high XPO1 expression is associated with the prognosis of patients with cholangiocarcinoma.
Using the R package clusterProfiler, GO enrichment analysis and KEGG pathway analysis were performed on the 500 genes positively associated with XPO1 expression in CHOL. GO functional enrichment analysis indicated that XPO1 was related to “modified and binding function,” and KEGG analysis revealed that genes involved in the spliceosome, RNA transport, ubiquitin‐mediated proteolysis, and cell cycle terms were significantly enriched. These results are similar to the mRNA‐seq results for neuroblastoma reported by Li‐jia Pan et al. 7 From the above analysis, we concluded that XPO1 is significantly associated with multiple tumor pathways in CHOL.
Furthermore, we found that inhibition of XPO1 significantly inhibited tumor cell proliferation. 20 , 21 , 22 Leptomycin B, the first specific XPO1 inhibitor, was discovered in the 1990s and has a hyperspecificity for XPO1. 23 However, its high toxicity limits its clinical application. 24 With time, several selective inhibitors of nuclear export (SINEs) have been developed. KPT‐330 (Selinexor), a novel oral SINE, has been approved by the United States Food and Drug Administration for the treatment of refractory multiple myeloma and relapsed/refractory diffuse large B‐cell lymphoma. Therefore, we selected KPT‐330 as an inhibitor of XPO1 for the subsequent experiments. The results showed that the XPO1 inhibitor KPT‐330 significantly disrupted CHOL cell proliferation.
p53 acts as a carrier protein for XPO1, and it naturally accumulates in the nucleus upon XPO1 inhibition. We demonstrated that KPT‐330 affects the proliferation of cholangiocarcinoma cells through the nuclear accumulation of p53. Based on the results of bioinformatics analysis, we noted that XPO1 might be associated with protein degradation; therefore, further experiments demonstrated that the XPO1 inhibitor KPT‐330 increased the stability of p53. We unearthed the key gene BIRC6, which can cause the degradation of p53. 14 BIRC6 carries an N‐terminal single baculovirus inhibition of apoptosis protein repeat (BIR) domain and a C‐terminal ubiquitin‐conjugating (UBC) enzyme domain, while through the UBC domain BIRC6 facilitates proteasomal degradation. 25 However, the XPO1 inhibitor KPT‐330 reduced the expression of BIRC6, which also explained how XPO1 inhibitor KPT‐330 increased p53 protein stability. The PI3K/AKT pathway plays an important role in the regulation of several cellular processes, including the maintenance of proliferative signaling. 26 We found that XPO1 inhibitor KPT‐330 inhibited the PI3K/AKT pathway and further inhibited BIRC6 expression.
5. CONCLUSION
In conclusion, we analyzed data from TCGA and found that XPO1 showed high expression across cancers and was associated with poor prognosis in hepatobiliary and pancreatic tumors. In CHOL, we revealed that XPO1 inhibitor KPT‐330 triggered the nuclear accumulation of the p53 protein and significantly disrupted the proliferation of cholangiocarcinoma cells. Mechanistically, XPO1 inhibitor KPT‐330 reduced BIRC6 expression by inhibiting the PI3K/AKT pathway to decrease p53 degradation and improve its stability (Figure 8). Therefore, XPO1 may be a potential therapeutic target, and KPT‐330 may play a therapeutic role in cholangiocarcinoma.
FIGURE 8.
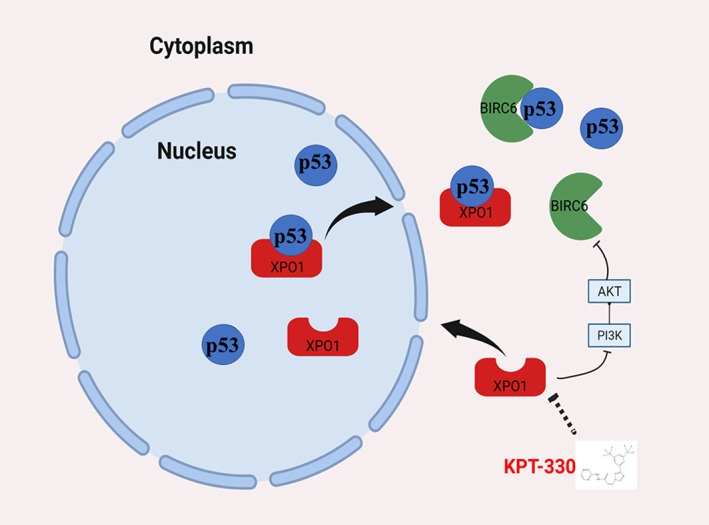
Schematic diagram of effects of XPO1 inhibition by KPT‐330 in CHOL cells. CHOL, cholangiocarcinoma; XPO1, exportin‐1.
AUTHOR CONTRIBUTIONS
Cheng Zhao: Conceptualization (lead); data curation (lead); formal analysis (lead); investigation (lead); project administration (lead); software (lead); validation (lead); visualization (lead); writing – original draft (lead); writing – review and editing (lead). Ben Ma: Conceptualization (equal); data curation (equal); investigation (equal); software (equal); supervision (equal); validation (equal); writing – original draft (supporting). Ziyi Yang: Conceptualization (equal); data curation (equal); formal analysis (equal); investigation (equal); methodology (equal); project administration (equal). Ou Li: Conceptualization (equal); methodology (equal); resources (equal); software (equal). Shilei Liu: Supervision (equal); visualization (equal). Lijia Pan: Data curation (equal); formal analysis (equal); funding acquisition (equal). Wei Gong: Conceptualization (equal); data curation (equal); formal analysis (equal); funding acquisition (equal); supervision (equal).
FUNDING INFORMATION
This study was supported by the National Natural Science Foundation of China (nos. 82172628, 81974371, 31701108, 82002503, and 82173081), The Shanghai Sailing Program (nos. 20YF1430200 and 22YF1427300), The Shanghai Anticancer Association Eyas Program (SACA‐CY21B05), and Shanghai scientific and technological innovation plan (22S31903600).
CONFLICT OF INTEREST
The authors have no conflict of interest to disclose.
Zhao C, Ma B, Yang Z‐y, et al. Inhibition of XPO1 impairs cholangiocarcinoma cell proliferation by triggering p53 intranuclear accumulation. Cancer Med. 2023;12:5751‐5763. doi: 10.1002/cam4.5322
Cheng Zhao, Ben Ma, Zi‐yi Yang are contributed equally.
Contributor Information
Ping Dong, Email: dongping@xinhuamed.com.cn.
Yi‐jun Shu, Email: shuyijun19881125@163.com.
DATA AVAILABILITY STATEMENT
The data sets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.
REFERENCES
- 1. Joechle K, Jumaa H, Thriene K, et al. Dual inhibition of mTORC1/2 reduces migration of cholangiocarcinoma cells by regulation of Matrixmetalloproteinases. Front Cell Dev Biol. 2021;9:785979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Han Y, Yang Y, Huang S, Yao L, Wu L. The miR‐34a/WNT7B modulates the sensitivity of cholangiocarcinoma cells to p53‐mediated photodynamic therapy toxicity. Biochem Biophys Res Commun. 2022;591:54‐61. [DOI] [PubMed] [Google Scholar]
- 3. Shen S, Wang J, Zheng B, et al. LINC01714 enhances gemcitabine sensitivity by modulating FOXO3 phosphorylation in cholangiocarcinoma. Mol Ther Nucleic Acids. 2020;19:446‐457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fisher JG, Walker CJ, Doyle AD, et al. Selinexor enhances NK cell activation against malignant B cells downregulation of HLA‐E. Front Oncol. 2021;11:785635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ishizawa J, Kojima K, Hail N Jr, Tabe Y, Andreeff M. Expression, function, and targeting of the nuclear exporter chromosome region maintenance 1 (CRM1) protein. Pharmacol Ther. 2015;153:25‐35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Birnbaum DJ, Finetti P, Birnbaum D, Mamessier E, Bertucci F. XPO1 expression is a poor‐prognosis marker in pancreatic adenocarcinoma. J Clin Med. 2019;8(5):596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pan L, Cheng C, Duan P, Chen K, Wu Y, Wu Z. XPO1/CRM1 is a promising prognostic indicator for neuroblastoma and represented a therapeutic target by selective inhibitor verdinexor. J Exp Clin Cancer Res. 2021;40:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sexton R, Mahdi Z, Chaudhury R, et al. Targeting nuclear exporter protein XPO1/CRM1 in gastric cancer. Int J Mol Sci. 2019;20(19):4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Govaerts I, Prieto C, Vandersmissen C, et al. PSEN1‐selective gamma‐secretase inhibition in combination with kinase or XPO‐1 inhibitors effectively targets T cell acute lymphoblastic leukemia. J Hematol Oncol. 2021;14:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Azmi AS, Uddin MH, Mohammad RM. The nuclear export protein XPO1 ‐ from biology to targeted therapy. Nat Rev Clin Oncol. 2021;18:152‐169. [DOI] [PubMed] [Google Scholar]
- 11. Li O, Zhao C, Zhang J, et al. UBAP2L promotes gastric cancer metastasis by activating NF‐κB through PI3K/AKT pathway. Cell Death Discov. 2022;8:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nguyen R, Wang H, Sun M, Lee DG, Peng J, Thiele CJ. Combining selinexor with alisertib to target the p53 pathway in neuroblastoma. Neoplasia. 2022;26:100776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shi Y, Xu S, Li S. Selinexor improves the anti‐cancer effect of tucidinostat on TP53 wild‐type breast cancer. Mol Cell Endocrinol. 2022;545:111558. [DOI] [PubMed] [Google Scholar]
- 14. Tang W, Xue R, Weng S, et al. BIRC6 promotes hepatocellular carcinogenesis: interaction of BIRC6 with p53 facilitating p53 degradation. Int J Cancer. 2015;136:E475‐E487. [DOI] [PubMed] [Google Scholar]
- 15. Lee Y, Pei J, Baumhardt JM, Chook YM, Grishin NV. Structural prerequisites for CRM1‐dependent nuclear export signaling peptides: accessibility, adapting conformation, and the stability at the binding site. Sci Rep. 2019;9:6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fung HYJ, Fu S‐C, Brautigam CA, Chook YM. Structural determinants of nuclear export signal orientation in binding to exportin CRM1. Elife. 2015;4:e10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu S, Qiao W, Sun Q, Luo Y. Chromosome region maintenance 1 (XPO1/CRM1) as an anticancer target and discovery of its inhibitor. J Med Chem. 2021;64:15534‐15548. [DOI] [PubMed] [Google Scholar]
- 18. Schmidt J, Braggio E, Kortuem KM, et al. Genome‐wide studies in multiple myeloma identify XPO1/CRM1 as a critical target validated using the selective nuclear export inhibitor KPT‐276. Leukemia. 2013;27:2357‐2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang AY, Liu H. The past, present, and future of CRM1/XPO1 inhibitors. Stem Cell Investig. 2019;6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ou L, Wang X, Cheng S, et al. Verdinexor, a selective inhibitor of nuclear exportin 1, inhibits the proliferation and migration of esophageal cancer via XPO1/c‐Myc/FOSL1 Axis. Int J Biol Sci. 2022;18:276‐291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang J, Sun T, Meng Z, et al. XPO1 inhibition synergizes with PARP1 inhibition in small cell lung cancer by targeting nuclear transport of FOXO3a. Cancer Lett. 2021;503:197‐212. [DOI] [PubMed] [Google Scholar]
- 22. Deng M, Zhang M, Xu‐Monette ZY, et al. XPO1 expression worsens the prognosis of unfavorable DLBCL that can be effectively targeted by selinexor in the absence of mutant p53. J Hematol Oncol. 2020;13:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;269:6320‐6324. [PubMed] [Google Scholar]
- 24. Newlands ES, Rustin GJ, Brampton MH. Phase I trial of elactocin. Br J Cancer. 1996;74:648‐649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen Z, Naito M, Hori S, Mashima T, Yamori T, Tsuruo T. A human IAP‐family gene, apollon, expressed in human brain cancer cells. Biochem Biophys Res Commun. 1999;264:847‐854. [DOI] [PubMed] [Google Scholar]
- 26. Lien EC, Dibble CC, Toker A. PI3K signaling in cancer: beyond AKT. Curr Opin Cell Biol. 2017;45:62‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.


