Abstract
Objectives
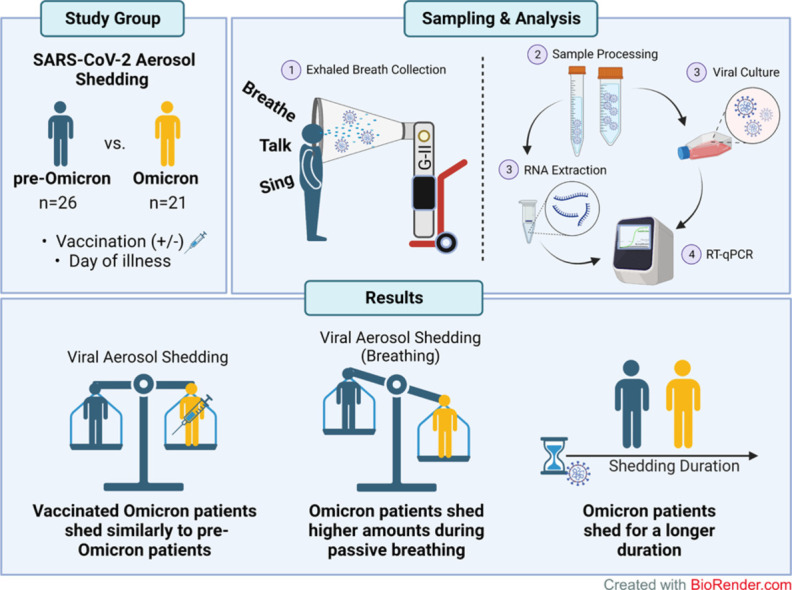
As the world transitions to COVID-19 endemicity, studies focusing on aerosol shedding of highly transmissible SARS-CoV-2 variants of concern (VOCs) are vital for the calibration of infection control measures against VOCs that are likely to circulate seasonally. This follow-up Gesundheit-II aerosol sampling study aims to compare the aerosol shedding patterns of Omicron VOC samples with pre-Omicron variants analyzed in our previous study.
Design
Coarse and fine aerosol samples from 47 patients infected with SARS-CoV-2 were collected during various respiratory activities (passive breathing, talking, and singing) and analyzed using reverse transcription-quantitative polymerase chain reaction and virus culture.
Results
Compared with patients infected with pre-Omicron variants, comparable SARS-CoV-2 RNA copy numbers were detectable in aerosol samples of patients infected with Omicron despite being fully vaccinated. Patients infected with Omicron also showed a slight increase in viral aerosol shedding during breathing activities and were more likely to have persistent aerosol shedding beyond 7 days after disease onset.
Conclusion
This follow-up study reaffirms the aerosol shedding properties of Omicron and should guide continued layering of public health interventions even in highly vaccinated populations.
Keywords: Aerosol transmission, Airborne transmission, COVID-19, Omicron variant of concern, SARS-CoV-2
Graphical abstract
Introduction
The emergence of SARS-CoV-2 causing the prolonged COVID-19 pandemic has had profound public health and socioeconomic consequences worldwide [1]. Throughout the course of the pandemic, the rapid mutation of SARS-CoV-2 has spawned numerous novel strains, including variants of concern (VOCs), further complicating infection control measures with altered transmissibility, virulence, and immune evasion [2,3]. Currently, the world is gradually transitioning to endemicity with the rapid spread and dominance of the Omicron VOC and its subvariants. As SARS-CoV-2 continues to adapt to the human airway [4], there are concerns as to whether Omicron has enhanced aerosol transmission potential that could further accelerate its spread.
We previously demonstrated that SARS-CoV-2 viral particles are shed through aerosols emitted during breathing, talking, and singing [5]. Other studies also showed that infectious virus can be cultured from similar samples, albeit from a small number of subjects [6,7]. However, most were conducted before the global dominance of Omicron and before widespread vaccination. Lai et al. has most recently showed that highly transmissible variants, such as Omicron, shed higher amounts of virus in exhaled breath aerosols [7]. However, questions remained on the factors contributing to the increased aerosol shedding in Omicron VOCs compared with pre-Omicron variants. Whether these differences would necessitate further updates in public health interventions should be investigated in detail.
Therefore, we conducted a follow-up study using the Gesundheit-II (G-II) exhaled breath sampler to collect aerosol samples from patients infected with Omicron. We analyzed the aerosol carriage of Omicron VOCs in breathing, talking, and singing activities from vaccinated patients and compared its aerosol shedding properties with that of preceding SARS-CoV-2 variants.
Material and methods
Patient recruitment and data collection
Participants were recruited from February 2021 through May 2022 at the National Centre for Infectious Diseases, Singapore. The inclusion criteria were age 21 years or older, positive for COVID-19 through reverse transcription-quantitative polymerase chain reaction (RT-qPCR), and index nasopharyngeal swab qPCR cycle threshold (Ct) value of <25.0. Patients early into the infection phase were preferentially selected; whereas patients with co-infections, with severe disease, or requiring supplemental oxygen were excluded because they were expected to be unable to the complete study procedures. The data from our previous study were included to facilitate comparison with non-Omicron variants (n = 23) [5]. The recruitment and sampling pertaining to this study followed the same procedures to generate comparable data. This study was approved by the National Healthcare Group Domain Specific Review Board (reference number 2020/01113). All study participants provided written informed consent. In total, accounting for two withdrawals, 47 participants had samples collected and analyzed for the study. The summary of recruitment numbers is illustrated in Figure S1.
Clinical sample collection
Coarse (>5 µm) and fine (≤5 µm) fractions of aerosol samples were collected using the G-II exhaled breath sampler, as previously described [5]. Participants performed one to three separate expiratory activities in the same sitting, i.e., 30 minutes of tidal breathing, 15 minutes of talking, 15 minutes of singing, with 30 minutes of rest between activities. For the talking activity, the participants repeated passages read to them from Dr Seuss’ “Green Eggs and Ham.” For the singing activity, participants sang along to “Happy Birthday,” “ABC Song,” “Twinkle Twinkle Little Star,” and “We Wish You A Merry Christmas.” Nasal swab samples were collected for comparison (for participants enrolled from November 2021 onward).
Clinical sample processing
The samples were transported to and processed in the National University of Singapore Biosafety Level 3 Laboratory on the same day as the collection. The coarse fraction and nasal swab samples were vortexed and transferred into screw-capped tubes. Fine fraction samples were concentrated with Amicon Ultra-15 100-kDa cut-off centrifugal filter units (Millipore) and filtered through a 0.22-μm centrifuge tube filter (Corning). The concentrate was topped up with Dulbecco Modified Eagle Medium, supplemented with 2% fetal bovine serum and 1 × antibiotic-antimycotic, and transferred into screw-capped tubes. Aliquots of nasal swab and fine fraction samples were also subjected to virus culture on the same day.
Virus culture
VeroE6-transmembrane protease serine 2 (VeroE6-TMPRSS2) (BPS Bioscience; #78081) or A549-angiotensin-converting enzyme 2 (A549-ACE2) cells were inoculated with samples from nasal swabs and fine fractions. The presence of cytopathic effect was monitored every 3-5 days, and a second passage was performed on either 3 or 7 days after infection, depending on the presence of cytopathic effect. Supernatants were harvested for RT-qPCR at the end of each passage (about 14 days).
Viral RNA quantification by RT-qPCR
RNA extraction and RT-qPCR were performed as previously described [5]. RNA was extracted using the QIAamp MinElute Virus Spin Kit (Qiagen), according to the manufacturer's instructions. The US Centers for Disease Control N1 assay (Integrated DNA Technologies) was used for the quantification of SARS-CoV-2. Viral RNA copies in each original sample were calculated from a standard curve constructed with the nucleocapsid gene positive control plasmid (Integrated DNA Technologies).
Classical RT-qPCR and Sanger sequencing
Due to the high prevalence and number of Omicron infections during the study period, not all samples collected were sequenced using next-generation sequencing. Hence, Sanger sequencing was performed for samples collected from patients with presumed Omicron infections. A549-ACE2 cells were inoculated with nasal swab samples before RNA extraction using the Direct-zol RNA Miniprep kit (Zymo Research), according to the manufacturer's instructions. Partial length SARS-CoV-2 spike (S) gene fragment (733-bp target) was then amplified from extracted viral RNA using SuperScript™ III One-Step RT-PCR System with Platinum™ Taq High Fidelity DNA Polymerase (Invitrogen) and S gene primers [8]. The thermal cycling conditions were as follows: 60°C for 1 minute, 50°C for 45 minutes, 94°C for 2 minutes; 40 cycles each, consisting of 95°C for 15 seconds, 55°C for 30 seconds, 68°C for 1 minute; followed by a final extension step at 68°C for 7 minutes. RT-PCR products were visualized by 1% agarose gel electrophoresis, and target amplicon bands purified using the QIAquick Gel Extraction Kit (Qiagen) according to the manufacturer's instructions. The purified amplicons were subjected to Sanger sequencing, and the resultant sequence data were deposited in GenBank.
Sequence construction and bioinformatics analysis
Sequencing chromatograms were visualized using FinchTV software (Version 1.4.0). Partial S gene sequences were constructed, and pairwise alignment was carried out to screen for relevant mutations with reference to the full-length S gene sequence of the Wuhan-Hu-1 isolate (NC_045512.2).
Statistical analysis
Statistical analyses and generation of graphs were conducted using GraphPad Prism version 8.0.2. For the multigroup comparison across the variants, Kruskal-Wallis test with Dunn correction for multiple comparison was used. For the two-sample comparison, the Mann-Whitney U test was performed. Individual statistical analyses are denoted in the figure legends. Statistical significance was defined as a P-value of <0.05.
Results
Fine and coarse aerosol fractions emitted by patients were collected across three activities, i.e., breathing (n = 31), talking (n = 47), and singing (n = 29). Not all participants completed every activity; hence, there were different sample sizes for each activity (Figure S1). The data collected from these samples (n = 26) were then combined with our previous study (n = 23) using the same methodology [5] to analyze and compare the aerosol carriage of SARS-CoV-2 across variants (Table S1). Among the 21 Omicron samples collected, four were sequenced by next-generation sequencing and the rest by Sanger sequencing. From the sequencing, three samples were confirmed to be Omicron BA.1 subvariant, nine were Omicron BA.2 subvariant, and three were Omicron with unconfirmed subvariant. Six of the sequencing assays were unsuccessful, but these samples were considered as Omicron because they were collected during a period (March 17 to May 21, 2022) when 100% of SARS-CoV-2 infections in Singapore were Omicron infections [9]. The GenBank accession numbers of Omicron samples that successfully completed an extra round of Sanger sequencing for confirmation are listed in Table S2.
SARS-CoV-2 RNA can be detected in aerosols of all three activities (breathing, talking, singing)
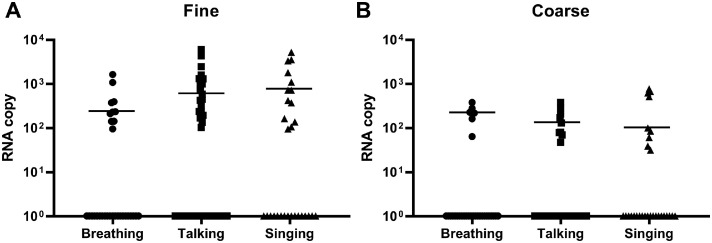
The viral RNA loads reported as viral copy numbers in fine and coarse fractions across the different activities were collated (Figure 1 ). All fine and coarse fraction samples passed the initial quality control, except for one coarse fraction sample from a talking activity, which yielded unusually low Ct value (lower than nasal swab Ct value), likely due to an artifact in detection. The latter sample was thus removed from the subsequent analyses. Similar to our previous study [5], higher viral RNA positivity was detected in the fine fraction (32.3% breathing, 40.4% talking, 44.8% singing) (Figure 1a) than the coarse fraction (22.6% breathing, 19.6% talking, 31.0% singing) (Figure 1b). Among the positive samples, talking and singing generated slightly higher viral copy numbers than breathing in the fine fraction, whereas the reverse was observed for the coarse fraction. The SARS-CoV-2 copy numbers ranged from 93.9 to 5964.9 for the fine fraction and 31.0 to 764.0 for the coarse fraction. All aerosol samples were negative for infectious viruses after the A549-ACE2 cell culture. However, infectious viruses could be cultured from the nasal swab samples collected from the same patients on the same day, prior to performing the G-II activities. The nasal swab samples from patients infected with Omicron yielded higher viral copy numbers (median copy number = 8.36 × 106) than those infected with the Delta variant (median copy number = 2.62 × 104). However, it should be noted that this comparison was made between 20 Omicron samples and three Delta samples, when nasal swabs were available during the current round of collection (n = 26). Therefore, only the median copy number is reported here as an observation, and no further statistical analyses were performed.
Figure 1.
Detection of SARS-CoV-2 RNA copy numbers in all SARS-CoV-2 infected patients’ aerosol samples from different respiratory activities. (a) SARS-CoV-2 RNA copy numbers in fine aerosol fractions from breathing, talking, and singing. (b) SARS-CoV-2 RNA copy numbers in coarse aerosol fractions from breathing, talking, and singing. All plots are shown as median values of SARS-CoV-2 RNA copy numbers from detected samples only. Statistical significance was calculated using Kruskal-Wallis test with Dunn correction for multiple comparison. A P-value of P <0.05 is considered statistically significant.
Viral RNA in aerosols from patients infected with Omicron are comparable to earlier VOCs despite widespread vaccination
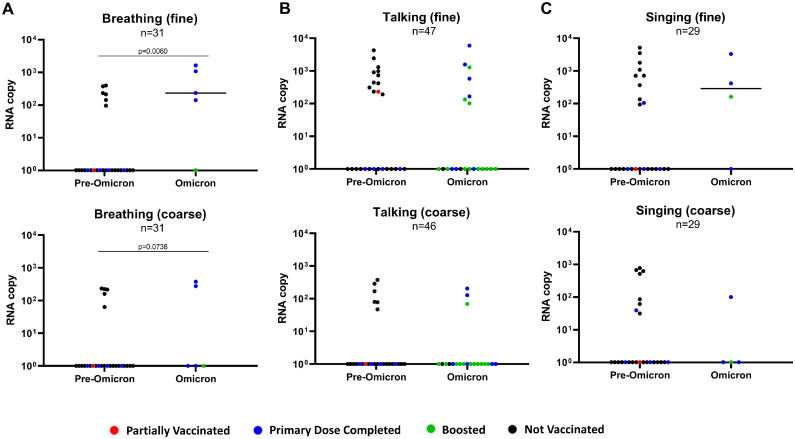
The collated data were then compared between all patients infected with pre-Omicron variants (pooled) against patients infected with Omicron (Figure 2 ). It was observed that the SARS-CoV-2 RNA copy numbers were comparable between the Omicron and pre-Omicron variants among the positive aerosol fractions (Figure 2a-c). Similar viral RNA levels were detected despite the widespread vaccine coverage of the Singapore population during the second sampling period [10]. In our study, only 15.4% (4 of 26) of patients not infected with Omicron had received the primary doses of SARS-CoV-2 vaccine, whereas 90.5% (19 of 21) of patients infected with Omicron had received the primary doses, with 57.1% (12 of 21) boosted. In addition, comparing pre-Omicron and Omicron copy numbers in aerosol emissions, the breathing activity yielded a significantly higher SARS-CoV-2 RNA copy number in the Omicron fine fraction (Figure 2a; P = 0.006). The samples were then further categorized according to their VOC to assess whether the differences in aerosol shedding of SARS-CoV-2 are due to specific variants (Figure S2). Each of the variants could be detected in either aerosol fraction from at least one activity evaluated, with the lowest detection rate coming from the ancestral SARS-CoV-2, suggesting potential adaptation favoring aerosol shedding of the virus (Figure S2A-C). It is also interesting to note that among the small number of vaccinated patients (i.e., at least the primary dose was completed) infected with the pre-Omicron variants, all of them did not have detectable viral RNA in their aerosol emissions, except for one singing emission sample from a patient infected with the Kappa variant.
Figure 2.
Comparison of SARS-CoV-2 RNA copy numbers from patients infected with pre-Omicron vs Omicron variant of concerns. Comparison of viral RNA copy numbers in fine and coarse fractions of aerosol samples from (a) breathing, (b) talking, and (c) singing activities. All non-Omicron variants were consolidated as the pre-Omicron group for comparison with the Omicron group. The color of each datapoint indicates the patients’ SARS-CoV-2 vaccination status. All plots are shown as median values of SARS-CoV-2 RNA copy numbers. Statistical significance was calculated using the Mann-Whitney U test. A P-value of P <0.05 is considered statistically significant.
Aerosol detection of SARS-CoV-2 RNA extends beyond 7 days from disease onset, particularly for Omicron VOC
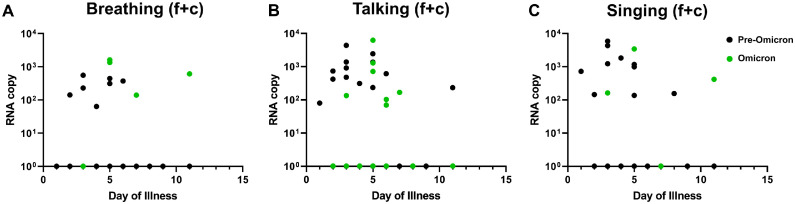
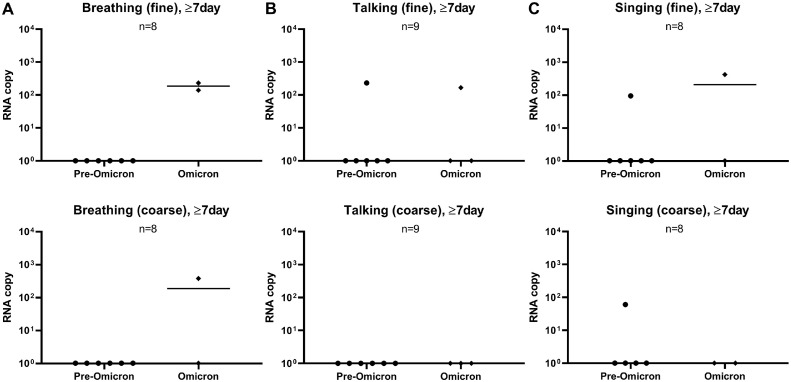
Given that the days of illness of patient samples differed greatly, we further generated a plot comparing between pre-Omicron and Omicron samples on their aerosol shedding over day of illness (Figure 3 ). Although we observed decreasing trends of SARS-CoV-2 copy numbers with time, some subjects continued to shed virus in their fine or coarse aerosol fractions for all three activities up to 11 days after the illness (Figure 3a-c). Interestingly, patients infected with Omicron generally exhibited detection positivity (for all three activities tested) that continued later into day of illness based on the pooled RNA copy number of both fine and coarse (f + c) fractions. As such, the data points from samples collected on or after day 7 of illness were extracted to compare late viral shedding (Figure 4 ). Among the positive samples at 7 days or later (2 of 8 for breathing, 2 of 9 for talking, 2 of 8 for singing), it was found that the positive detection rate of Omicron VOC was higher than that of pre-Omicron VOCs (Figure 4a-c), especially in the fine fraction. The samples collected on or after day 7 of illness were further stratified according to their SARS-CoV-2 variants to analyze the rate of viral detection for late illness shedding. It was observed that, apart from Omicron, other VOCs with viral RNA-positive samples belonged to the later-emerging Beta and Delta variants (Figure S3). Interestingly, positive samples were detected from patients with partial vaccination for Beta and Delta VOCs and patients who were not boosted for the Omicron VOC (Table S1). This suggests that the duration of aerosol shedding of SARS-CoV-2 may be longer, especially for the Omicron variant. Therefore, further investigations are warranted to determine if such aerosol shedding late into the disease process is attributable to viable virus because this will influence public health measures to mitigate aerosol transmission of SARS-CoV-2.
Figure 3.
SARS-CoV-2 RNA copy numbers over day of illnesses between pre-Omicron and Omicron variant of concerns. Total RNA copy numbers from both fine and coarse (f + c) fractions are consolidated and plotted against day of illness for (a) breathing, (b) talking and (c) singing activities. The black data points represent the pre-Omicron samples, whereas the green data points represent the Omicron samples.
Figure 4.
Comparison of SARS-CoV-2 RNA copy numbers of pre-Omicron versus Omicron variants in fine and coarse fractions at late phase of illness (≥7 days). Comparison of viral RNA copy numbers in both fine and coarse fractions of aerosol samples from (a) breathing, (b) talking, and (c) singing activities. The day of illness denotes the number of days that elapsed since positive diagnosis of SARS-CoV-2 infection. All non-Omicron variants were consolidated as the pre-Omicron group for comparison with the Omicron group. All plots are shown as median values of SARS-CoV-2 RNA copy numbers. Statistical significance was calculated using Kruskal-Wallis test with Dunn correction for multiple comparison. A P-value of P <0.05 is considered statistically significant.
Discussion
The emergence and current predominance of the Omicron VOC has ushered in a new normal, where SARS-CoV-2 is being recognized as endemic in most human populations [11,12]. Because SARS-CoV-2 would most likely join the ranks of seasonal respiratory viruses, it is critical to understand the role of aerosol shedding of SARS-CoV-2 (especially the Omicron VOC) to devise appropriate control measures against virus transmission [11]. In our study, we used the G-II apparatus to collect aerosols in fine and coarse fractions to detect SARS-CoV-2 using RT-qPCR and compared them across the variants to assess if differential aerosol carriage occurs.
After analysis, it was observed that patients infected with SARS-CoV-2 generated detectable viral RNA in respiratory aerosols (expressed as viral RNA copy numbers). This observation is similar to our previous findings in patients infected with previous SARS-CoV-2 variants [5,6]. Our data also reaffirmed that the fine aerosol fraction carried more virus particles, corroborating recent studies [7,13], and that talking and singing generate more viral RNA in aerosols than breathing. However, culturing of infectious viruses from aerosol samples continues to be a challenge [5,14], with numerous steps in the collection process that subject viruses to environmental degradation and mechanical stress during sampling that may impact in vitro viability. Therefore, these reasons may explain why all the collected samples were negative for virus culture, despite the positive virus cultures obtained from direct nasal swabs from the same patients. Notwithstanding, there are a few reports where infectious viruses can indeed be isolated from aerosol samples even through masks [6,7], indicating that aerosol shedding remains an important transmission pathway that necessitates proper controls in appropriate settings.
As the world shifts toward Omicron predominance, we extended our previous study and evaluated the aerosol shedding of the Omicron VOC. In addition, with widespread vaccination that precedes Omicron emergence, this follow-up study was able to compare the responses in a vaccinated versus a largely unvaccinated cohort. Omicron VOC-specific vaccines were not yet available in Singapore at the time of this study, and to date, most variant-specific vaccines tend to lag behind the currently circulating strains. Despite the high vaccination rates, the Omicron VOC exhibited comparable aerosol shedding as the pre-Omicron variants, similar to another study conducted by Lai et al. [7]. This suggests that although vaccination protects against severe disease, the current vaccines that induce systemic immunity do not prevent aerosol shedding, especially of the highly mutated Omicron VOC, which may be related to their relatively poorer immunogenicity for the development of mucosal antibodies [15]. However, it was observed, in limited capacity, that vaccination may reduce the length of aerosol shedding because patients with longer aerosol shedding detection were those who were partially vaccinated (Beta and Delta VOCs) or not boosted (Omicron VOC). Nevertheless, this finding suggested that the prevention of aerosol shedding may require mucosal protection against viral infections. As such, mucosal vaccines, similar to that of FLUMIST® live attenuated influenza vaccine used in influenza infections [16,17], should be developed and investigated for their efficacy in preventing the aerosol-mediated transmission of SARS-CoV-2 [18,19].
Furthermore, compared with the pre-Omicron VOCs, Omicron exhibited a significant increase in aerosol shedding during breathing. This difference may be attributed to greater tropism of Omicron for the upper respiratory tract and/or bronchial tissue [4,20,21]. With this change in tropism, the greater concentration of virus in the upper respiratory tract is more likely to be detected during tidal breathing, as opposed to the virus concentrating in the lower respiratory tract that may show up in greater numbers when deep breaths are taken for talking or singing. In addition, it is also noted that a fraction of the samples tested positive for viral RNA despite being collected at 7 or more days from illness onset. This finding is congruent with the clinical and epidemiological studies that indicate a more prolonged virus shedding in patients infected with Omicron, even among those who have received vaccination [22,23]. This may also help to explain the current global dominance of the Omicron variant.
Therefore, the findings of our study continue to highlight the importance of other public health interventions that complement the broad community vaccination. This strategy can be achieved, along with updating of vaccine formulations, to mitigate the impact of future COVID-19 waves on the health care systems and community [24]. Our study also observed significant heterogeneity of viral RNA copy numbers between patients, with some subjects shedding higher viral RNA loads for longer periods—a potential feature that may contribute to super-spreading aerosol-associated transmission [25]. Future studies should focus on identifying the risk factors that contribute to this heterogeneity leading to super-spreading events to better ring-fence the infections and transmission, particularly during seasonal outbreaks of SARS-CoV-2 infection.
Our study does have certain limitations. First, to make our analysis more meaningful, the analyses were pooled as Omicron compared with pre-Omicron infections due to the relatively small sample sizes across the VOCs, which may dilute the specific differences found in some VOCs. However, the rationale of the data consolidation was that the only strain circulating in most parts of the world has been Omicron and its subvariants for most of 2022. Second, although the sampling methodology was identical for both cohorts, there may have been differences in terms of community exposures, time from vaccination, and other seasonal factors, which may have affected the comparison. Nevertheless, the data from this study revealed that patients infected with Omicron emit similar aerosol viral loads as other variants, despite widespread vaccination coverage during the Omicron wave. Therefore, our data indicate that the global dominance of Omicron may not only be due to the multiple mutations in Omicron culminating in a more easily aerosolized virus per se. The more persistent and longer duration of shedding of Omicron also suggests enhanced adaptation to human hosts, thus sustaining the effective generation of viral aerosols. This, coupled with greater immune evasion and vaccine escape of the Omicron VOC [26], may better explain its enhanced transmissibility [27,28] and global dominance.
Conclusion
Our study underscores the transmission potential of the Omicron variants through aerosols during normal respiratory activities. The persistent and prolonged periods of aerosol shedding of high viral loads by individuals infected with Omicron (despite being fully vaccinated and largely asymptomatic) may contribute to its greater transmissibility. With most countries recognizing that the virus is endemic, it is vital to remain vigilant with continued public health interventions, such as adequate ventilation, disinfection technologies, and high efficiency particulate air (HEPA) filters, moving away from original strategies, such as quarantine and contact tracing. This applies not only for SARS-CoV-2 but also other respiratory viruses. Future detailed studies are warranted to explore the improved building designs, as well as physical, molecular, and host risk factors, that contribute to the transmissibility of SARS-CoV-2 and other respiratory viruses. Such studies will be instrumental in formulating targeted public health interventions to ameliorate the socioeconomic impact of these viral diseases.
Declaration of competing interest
PAT reports receiving grants from Roche, Arcturus, Johnson and Johnson, and Sanofi Pasteur and personal fees from AJ Biologicals outside the submitted work. The remaining authors have no competing interests to declare. All authors have completed the ICMJE Form for Disclosure of Potential Conflicts of Interest.
Funding
This work was supported by the Singapore National Medical Research Council (MOH-000443 to KWT, and NMRC/CG/M009/2017 NUH/NUHS to JJHC) and the National University of Singapore (NUS Reimagine Research Grant to JJHC).
Author contributions
Conceptualization: KWT, VTKC, PAT, KST, SWXO; investigation and methodology: KST, SWXO, MHK, DJWT, DZHA, YWN, MRBA; formal analysis: KST, SWXO, MHK, DJWT, DZHA, YWN, KWT, KKC, DKM; resources: KST, SWXO, MRBA, JJHC, KWT, VTKC; writing: KST, SWXO, MHK, VTKC, PAT, KWT, KKC, DKM; funding acquisition and supervision: JJHC, PAT, VTKC, KWT. All authors have read and agreed to the final version of the manuscript.
Acknowledgments
The authors thank the NUS Medicine BSL-3 Core Facility team for their support in BSL-3 procedures. The authors gratefully acknowledge the University of Maryland for the availability of the G-II equipment. The authors also thank Lin Cui and Raymond Lin from the National Public Health Laboratory for sharing whole genome sequence data for VOC identification. The authors thank Linfa Wang from Duke-NUS Medical School for providing the A549-ACE2 cell line. The authors are grateful to Mark Chen, Margaret Soon, LY Phoon, KY Loh, JX Pang, and all the nursing, infection control, and operational staff at the National Centre for Infectious Diseases for their support.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2023.03.029.
Appendix. Supplementary materials
Table S1. Details of subject information and their individual SARS-CoV-2 RNA copy numbers.
Table S2. GenBank accession numbers of partial SARS-CoV-2 spike sequences of samples confirmed by additional Sanger sequencing.
Figure S1. Flowchart of patient sampling workflow. Flowchart detailing the recruitment for each respiratory activity, including the number of samples for each activity.
Figure S2. Comparison of SARS-CoV-2 RNA copy numbers across viral variants. Comparison of viral RNA copy numbers in fine and coarse fractions of aerosol samples from (A) breathing, (B) talking, and (C) singing activities. The number of samples for each viral variant is shown. The color of each data point indicates the patient's SARS-CoV-2 vaccination status. All plots are shown as median values of SARS-CoV-2 RNA copy numbers. Statistical significance was calculated using Kruskal-Wallis test with Dunn correction for multiple comparison. A P-value of P <0.05 is considered statistically significant.
Figure S3. Comparison of SARS-CoV-2 RNA copy number by SARS-CoV-2 variant at late phase of illness (≥7 days). Comparison of viral RNA copy numbers in fine and coarse aerosol fractions from (A) breathing, (B) talking, and (C) singing activities. All plots are shown as median values of SARS-CoV-2 RNA copy number. Statistical analyses were not performed due to the small number of available samples for each activity.
References
- 1.Guo YR, Cao QD, Hong ZS, Tan Y, Chen S, Jin H, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boehm E, Kronig I, Neher RA, Eckerle I, Vetter P, Kaiser L, et al. Novel SARS-CoV-2 variants: the pandemics within the pandemic. Clin Microbiol Infect. 2021;27:1109–1117. doi: 10.1016/j.cmi.2021.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plante JA, Mitchell BM, Plante KS, Debbink K, Weaver SC, Menachery VD. The variant gambit: COVID-19′s next move. Cell Host Microbe. 2021;29:508–515. doi: 10.1016/j.chom.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hui KPY, Ng KC, Ho JCW, Yeung H, Ching RHH, Gu H, et al. Replication of SARS-CoV-2 omicron BA.2 variant in ex vivo cultures of the human upper and lower respiratory tract. EBiomedicine. 2022;83 doi: 10.1016/j.ebiom.2022.104232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coleman KK, Tay DJW, Tan KS, Ong SWX, Than TS, Koh MH, et al. Viral load of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in respiratory aerosols emitted by patients with coronavirus disease 2019 (COVID-19) while breathing, talking, and singing. Clin Infect Dis. 2022;74:1722–1728. doi: 10.1093/cid/ciab691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adenaiye OO, Lai J, Bueno de Mesquita PJ, Hong F, Youssefi S, German J, et al. Infectious SARS-CoV-2 in exhaled aerosols and efficacy of masks during early mild infection. Clin Infect Dis. 2021;75:e241–e248. doi: 10.1093/cid/ciab797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai J, Coleman KK, Tai SHS, German J, Hong F, Albert B, et al. Exhaled breath aerosol shedding by highly transmissible versus prior SARS-CoV-2 variants. Clin Infect Dis. 2023;76:786–794. doi: 10.1093/cid/ciac846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloemen M, Rector A, Swinnen J, Ranst MV, Maes P, Vanmechelen B, et al. Fast detection of SARS-CoV-2 variants including Omicron using one-step RT-PCR and Sanger sequencing. J Virol Methods. 2022;304 doi: 10.1016/j.jviromet.2022.114512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng OT, Marimuthu K, Lim N, Lim ZQ, Thevasagayam NM, Koh V, et al. Analysis of COVID-19 incidence and severity among adults vaccinated with 2-dose mRNA COVID-19 or inactivated SARS-CoV-2 vaccines with and without boosters in Singapore. JAMA Netw Open. 2022;5 doi: 10.1001/jamanetworkopen.2022.28900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biancolella M, Colona VL, Mehrian-Shai R, Watt JL, Luzzatto L, Novelli G, et al. COVID-19 2022 update: transition of the pandemic to the endemic phase. Hum Genomics. 2022;16:19. doi: 10.1186/s40246-022-00392-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.COVID is here to stay: countries must decide how to adapt. Nature. 2022;601:165. doi: 10.1038/d41586-022-00057-y. [DOI] [PubMed] [Google Scholar]
- 13.Alsved M, Nygren D, Thuresson S, Fraenkel CJ, Medstrand P, Löndahl J. Size distribution of exhaled aerosol particles containing SARS-CoV-2 RNA. Infect Dis (Lond) 2023;55:158–163. doi: 10.1080/23744235.2022.2140822. [DOI] [PubMed] [Google Scholar]
- 14.Ong SWX, Tan YK, Coleman KK, Tan BH, Leo Y, Wang DL, et al. Lack of viable severe acute respiratory coronavirus virus 2 (SARS-CoV-2) among PCR-positive air samples from hospital rooms and community isolation facilities. Infect Control Hosp Epidemiol. 2021;42:1327–1332. doi: 10.1017/ice.2021.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kar S, Devnath P, Emran TB, Tallei TE, Mitra S, Dhama K. Oral and intranasal vaccines against SARS-CoV-2: current progress, prospects, advantages, and challenges. Immun Inflamm Dis. 2022;10:e604. doi: 10.1002/iid3.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.FluMist: an intranasal live influenza vaccine. Med Lett Drugs Ther. 2003;45:65–66. [PubMed] [Google Scholar]
- 17.Sun K, Ye J, Perez DR, Metzger DW. Seasonal FluMist vaccination induces cross-reactive T cell immunity against H1N1 (2009) influenza and secondary bacterial infections. J Immunol. 2011;186:987–993. doi: 10.4049/jimmunol.1002664. [DOI] [PubMed] [Google Scholar]
- 18.Hassan AO, Shrihari S, Gorman MJ, Ying B, Yuan D, Raju S, et al. An intranasal vaccine durably protects against SARS-CoV-2 variants in mice. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Afkhami S, D'Agostino MR, Zhang A, Stacey HD, Marzok A, Kang A, et al. Respiratory mucosal delivery of next-generation COVID-19 vaccine provides robust protection against both ancestral and variant strains of SARS-CoV-2. Cell. 2022;185:896–915. doi: 10.1016/j.cell.2022.02.005. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hui KPY, Ho JCW, Cheung MC, Ng K, Ching RHH, Lai K, et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature. 2022;603:715–720. doi: 10.1038/s41586-022-04479-6. [DOI] [PubMed] [Google Scholar]
- 21.Eales O, de Oliveira Martins L, Page AJ, Wang H, Bodinier B, Tang D, et al. Dynamics of competing SARS-CoV-2 variants during the Omicron epidemic in England. Nat Commun. 2022;13:4375. doi: 10.1038/s41467-022-32096-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi K, Ishikane M, Ujiie M, Iwamoto N, Okumura N, Sato T, et al. Duration of infectious virus shedding by SARS-CoV-2 omicron variant-infected vaccinees. Emerg Infect Dis. 2022;28:998–1001. doi: 10.3201/eid2805.220197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boucau J, Marino C, Regan J, Uddin R, Choudhary MC, Flynn JP, et al. Duration of shedding of culturable virus in SARS-CoV-2 omicron (BA.1) Infection. N Engl J Med. 2022;387:275–277. doi: 10.1056/NEJMc2202092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prunas O, Warren JL, Crawford FW, Gazit S, Patalon T, Weinberger DM, et al. Vaccination with BNT162b2 reduces transmission of SARS-CoV-2 to household contacts in Israel. Science. 2022;375:1151–1154. doi: 10.1126/science.abl4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen PZ, Bobrovitz N, Premji Z, Koopmans M, Fisman DN, Gu FX. Heterogeneity in transmissibility and shedding SARS-CoV-2 via droplets and aerosols. eLife. 2021;10:e65774. doi: 10.7554/eLife.65774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan CW, Chia WN, Zhu F, Young B, Chantasrisawad N, Hwa SH, et al. SARS-CoV-2 Omicron variant emerged under immune selection. Nat Microbiol. 2022;7:1756–1761. doi: 10.1038/s41564-022-01246-1. [DOI] [PubMed] [Google Scholar]
- 27.Cao Y, Wang J, Jian F, Xiao T, Song W, Yisimayi A, et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meng B, Abdullahi A, Ferreira IATM, Goonawardane N, Saito A, Kimura I, et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature. 2022;603:706–714. doi: 10.1038/s41586-022-04474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Details of subject information and their individual SARS-CoV-2 RNA copy numbers.
Table S2. GenBank accession numbers of partial SARS-CoV-2 spike sequences of samples confirmed by additional Sanger sequencing.
Figure S1. Flowchart of patient sampling workflow. Flowchart detailing the recruitment for each respiratory activity, including the number of samples for each activity.
Figure S2. Comparison of SARS-CoV-2 RNA copy numbers across viral variants. Comparison of viral RNA copy numbers in fine and coarse fractions of aerosol samples from (A) breathing, (B) talking, and (C) singing activities. The number of samples for each viral variant is shown. The color of each data point indicates the patient's SARS-CoV-2 vaccination status. All plots are shown as median values of SARS-CoV-2 RNA copy numbers. Statistical significance was calculated using Kruskal-Wallis test with Dunn correction for multiple comparison. A P-value of P <0.05 is considered statistically significant.
Figure S3. Comparison of SARS-CoV-2 RNA copy number by SARS-CoV-2 variant at late phase of illness (≥7 days). Comparison of viral RNA copy numbers in fine and coarse aerosol fractions from (A) breathing, (B) talking, and (C) singing activities. All plots are shown as median values of SARS-CoV-2 RNA copy number. Statistical analyses were not performed due to the small number of available samples for each activity.