Abstract
The genomes of Archaea harbor homologs of the global bacterial regulator leucine-responsive regulatory protein (Lrp). Archaeal Lrp homologs are helix–turn–helix DNA-binding proteins that specifically repress the transcription of their own genes in vitro. Here, we analyze the interaction of Pyrococcus LrpA with components of the archaeal transcriptional machinery at the lrpA promoter. DNA–protein complexes can be isolated by electrophoretic mobility shift assays that contain both LrpA and the two archaeal transcription factors TBP and TFB. Phenanthroline–copper footprinting experiments showed that the DNA-binding sites of LrpA and TBP/TFB do not overlap. These results and the finding that association of RNA polymerase with the TBP–TFB promoter complex was inhibited in the presence of LrpA indicate that Pyrococcus LrpA interferes with RNA polymerase recruitment. A DNA motif required for the LrpA–DNA interaction was inferred from dimethylsulfate methylation interference experiments.
INTRODUCTION
The basal transcriptional machinery of Archaea consists of two cis- and three trans-elements that all share homology with components of the eucaryotic RNA polymerase (RNAP) II system. The archaeal TATA box (1,2) is recognized by the archaeal TATA-box-binding protein, TBP (3,4), an ortholog of eucaryotic TBP (5). This interaction is stabilized by TFB, the second archaeal transcription factor (6–8), which is a homolog of eucaryotic TFIIB. TFB binds to a purine-rich sequence immediately upstream of the archaeal TATA box, the B recognition element (BRE), and this TFB–BRE interaction defines the polarity of archaeal transcription (8). This binary TBP–TFB promoter complex recruits the RNAP that is similar in complexity and sequence to eucaryotic RNAP II (9).
In spite of this similarity of archaeal and eucaryotic transcriptional components, analyses of archaeal genome sequences revealed no evidence for the existence of eucaryotic-like transcriptional regulators (10). However, a number of putative homologs of bacterial-like regulators have been identified, posing the question of how these interact with the archaeal transcriptional machinery.
Two of these regulators have been initially characterized, MDR1 and Lrp. MDR1, an archaeal homolog of bacterial metal-dependent regulators represses expression of its own gene and of an ABC metal transporter encoded in the same polycistronic transcription unit in a metal-dependent manner (11). The DNA-binding site of this repressor was located downstream from that of TBP and TFB and the regulator was shown to inhibit RNAP binding to the promoter.
Archaeal homologs of the bacterial Lrp/AsnC family of regulators have been identified and initially characterized (12–18). Lrp is a global regulator of metabolism in Escherichia coli that can act as both repressor or activator. Lrp seems to stimulate expression of biosynthetic operons and to repress operons that function in catabolic pathways (reviewed in 19,20). The activity of Lrp as gene regulator is often modulated by l-leucine. Escherichia coli Lrp binds to one to six DNA sites depending on the promoter (21,22) and a consensus binding sequence of 15 nt has been identified. Archaeal Lrp homologs are also DNA-binding proteins, but thus far only binding of the protein to its own promoter has been studied. The Lrp homologs from Sulfolobus species, Lrs14 form S.solfataricus and Sa-Lrp from S.acidocaldarius, share only 10% amino acid sequence identity and are most likely not functionally equivalent (14–16). Sa-Lrp and Lrs14 protect a DNA segment of 150 and 60 nt from DNaseI digestion, respectively, indicating the existence of multiple binding sites (14–16). Lrs 14 was shown to inhibit cell-free transcription from the lrs14 promoter. The binding site of Lrs14 overlaps the TATA box and BRE of the lrs14 promoter and binding of Lrs14 and of TBP–TFB were mutually exclusive events, indicating that this autoregulation is brought about by inhibition of binding of TBP/TFB to its own promoter (16). By a modifed SELEX procedure, the consensus sequences for binding of Ptr1 and Ptr2, the putative transcriptional regulators of Methanococcus jannaschii related to the Lrp/AsnC family, were identified. These motifs consisted of 6 bp repeats separated by 1–3 bp (18). Footprinting experiments indicate symmetrical binding of protein dimers to palindromic sequences. Pyrococcus LrpA was also shown to bind to the promoter region and to inhibit cell-free transcription from its own promoter (17). The interaction of the Methanococcus and Pyrococcus Lrp homologs with the transcriptional machinery has not yet been studied.
In the experiments reported here, we demonstrate by mobility shift and in situ phenanthroline–copper (OP–Cu) footprinting that LrpA and TBP/TFB bind simultaneously to the lrpA promoter. Addition of LrpA to DNA-binding reactions prevented binding of RNAP to DNA, indicating that the mechanism of action of Sulfolobus Lrs14 and Pyrococcus LrpA differ strikingly. In addition, we identified by methylation interference studies the DNA sequences required for the binding of Pyrococcus LrpA and TBP/TFB to the promoter.
MATERIALS AND METHODS
Purification of RNAP
RNAP from Pyrococcus furiosus was purified as previously described by Hethke et al. (23).
Expression and purification of recombinant proteins
The transcription factors TBP and TFB from Pyrococcus woesei were overproduced in E.coli as previously described by Hausner et al. (6). The DNA sequences of P.woesei TBP and TFB show 100% identity to those of P.furiosus. LrpA from P.furiosus was cloned, overexpressed, and the heat-stable cell-free extract prepared as previously described by Brinkman et al. (17). A different purification protocol was used. The soluble fraction was applied to a 5 ml affinity chromatography column (Econopac® Heparin Cartridge; BioRad) that had been equilibrated with 125 mM Na-citrate pH 5.0. Proteins were eluted with a linear gradient (15 ml 0–1 M NaCl). Fractions containing LrpA as determined by SDS–gel electrophoresis and silver staining were pooled and dialyzed against 20 mM Tris–HCl pH 8.0 to remove citrate, which inhibits the in vitro transcription reactions.
Templates for in vitro transcription, electrophoretic mobility shift assay (EMSA) and footprinting reactions
The plasmid pLUW613 containing the lrpA promoter region was used for cell-free transcription (17). It was linearized with PstI (New England Biolabs), which cleaves the DNA in the region of the gene. Plasmid DNA was purified by repeated centrifugation in CsCl density gradients as previously described by Frey et al. (24). Templates for EMSA and footprinting reactions extending from base pair –109 to +80 (relative to the transcriptional start site as +1) of the P.furiosus lrpA promoter were amplified by PCR. To generate the fragment the plasmid pLUW613 was used as template for the primers lrp+1500 (5′-GCAATGCTT-GACCGTGGATGGG-3′) and lrp-CC (5′-AGTGAAGGGCGTTCTCGCATCTT-3′).
Cell-free transcription reactions
In vitro transcription experiments were performed essentially as previously described by Hethke et al. (23). A standard reaction mixture (50 µl) contained 1.0 µg linearized lrpA promoter DNA, 250 ng recombinant TBP, 280 ng recombinant TFB, 220 ng purified RNAP and 1.2 µg recombinant LrpA. The transcriptional components (TBP, TFB, RNAP) were preincubated with DNA for 0, 10 and 20 min at 70°C before LrpA and nucleotides were added to initiate transcription, followed by an incubation for 15 min at 70°C. The reaction was terminated and the RNA purified for electrophoresis on a denaturing polyacrylamide gel as previously described by Frey et al. (24). Quantification was done using a PhosphorImager and accompanying software (Molecular Dynamics).
Preparation of antisera against recombinant LrpA from P.furiosus
Purified protein (500 µg) was used for immunization of a rabbit (Eurogentec) following the standard immunization protocol. The anti-Pyrococcus LrpA and the preimmune IgG fractions were isolated from the serum using affinity chromatography on protein A–Sepharose (Amersham Pharmacia Biotech).
Electrophoretic mobility shift assay
The DNA probe containing the P.furiosus lrpA promoter was amplified by PCR and end-labeled. Protein–DNA complexes were assembled in 10 µl volume of 40 mM Na-HEPES, pH 7.3, 325 mM KCl, 2.5 mM MgCl2, 0.1 mM EDTA, 5% PEG, 1 µg of poly[d(IC)], varying concentrations of proteins and 0.5–1 ng radiolabeled DNA. Reactions were incubated for 30 min at 70°C and complexes and free DNA separated on a non-denaturing 5% polyacrylamide gel. For the TBP–TFB-binding reaction, we used 75 ng recombinant TBP, 300 ng recombinant TFB and recombinant LrpA (50 and 100 ng) as well as additional BSA (3 µg; Roche Molecular Biochemicals). Gel electrophoresis was performed in a 1× Tris borate EDTA buffer system (25). For binding reactions of TBP–TFB together with TBP–TFB–RNAP, 50 ng TBP, 40 ng TBP, 30 ng purified RNAP and LrpA (100, 200 and 400 ng) were used and the native gel was run in a 1× Tris glycine EDTA buffer system (25). For the antibody supershift assays the binding reactions were carried out for 30 min at 70°C and the reaction mixture was allowed to cool down for 2 min at 37°C. After addition of antibodies, samples were incubated for 15 min at 37°C. Polyclonal antisera were generated and purified as described previously (26). We used 1.6 and 3.2 µg of anti-TFB from P.woesei (26), 0.8 and 1.5 µg of antibodies against subunit A′ of the RNAP from Methanobacterium thermoautotrophicum (27) and 0.5 µg anti-LrpA from P.furiosus (described above) as well as equal amounts of the preimmune serum of each antibody as control. In an additional EMSA, shown in Figure 3B, transcriptional components were incubated with DNA before the addition of LrpA. The EMSA was performed as described above, except that LrpA (25, 50, 100, 200 and 400 ng) was added after preincubation of TBP (50 ng), TFB (40 ng) and RNAP (30 ng) with DNA for 30 min at 70°C. Then, all components were incubated for 30 min at 70°C. In a further approach shown in Figure 3C, LrpA or TBP/TFB were incubated with DNA prior to the addition of the other proteins. For this experiment, EMSA-binding reactions containing LrpA (100 ng) or TBP (75 ng) and TFB (300 ng) were preincubated with template DNA for 20 min at 70°C before the other components (TBP/TFB or LrpA, respectively) were added. Reactions were incubated again for 20 min and analyzed by EMSA.
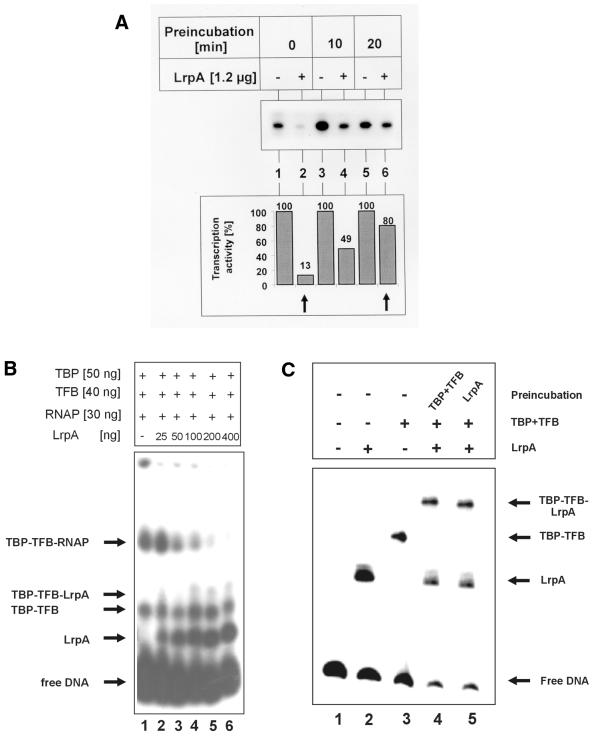
Figure 3.
Effects of preincubation of transcriptional components or LrpA with DNA. (A) Preformation of the PIC impedes the inhibitory effect of LrpA. Cell-free transcription was performed as described in Materials and Methods. TBP, TFB, RNAP and DNA were incubated at 70°C (as indicated) before the addition of LrpA (lanes 2, 4 and 6) and nucleotides. Cell-free transcription was performed for 15 min at 70°C. Transcriptional activities were quantified using a PhosphorImager. Transcription activity in the absence of LrpA equals 100% for each preincubation. (B) Preformed TBP–TFB–RNAP–DNA complexes are more stable against disruption of LrpA. In a standard EMSA, TBP, TFB and RNAP (specified above each lane) were allowed to interact with radiolabeled lrpA promoter DNA for 30 min at 70°C prior to the addition of increasing amounts of LrpA, as shown above. The binding reaction was carried out for 30 min at 70°C. Samples were separated by native gel electrophoresis and subjected to autoradiography. Arrows refer to free DNA and protein–DNA complexes. (C) Preincubation of LrpA or TBP–TFB results in the formation of identical protein–DNA complexes. A 75 ng aliquot of TBP and 300 ng TFB (lane 4) or 100 ng LrpA (lane 5) was assembled to lrpA promoter DNA for 20 min prior to the addition of the other protein(s). The samples were incubated again for 20 min at 70°C and analyzed by EMSA. The presence (+) or absence (–) of proteins or preincubation is indicated above. Free DNA and protein–DNA complexes are marked by arrows.
Phenanthroline–copper (Op–Cu) footprinting experiments
OP–Cu footprinting was performed with protein–DNA complexes purified by EMSA (28). TBP, TFB and LrpA were allowed to interact with the radiolabeled probe containing lrpA promoter DNA in a 5-fold scaled-up EMSA-binding reaction as described above. Complexed and free DNA were excised from the gel. The gel slices were immersed in 100 ml Tris–HCl pH 8 and subsequently subjected to the footprinting reaction using 10 µl of solution A [2 mM 1,10-phenanthroline (OP)/0.45 mM CuSO4] and 10 µl of solution B (58 mM mercaptopropionic acid). The samples were mixed gently, quenched after 1–5 min by the addition of 10 µl of 28 mM 2,9-dimethyl-OP and allowed to stand for 2 min. DNA was eluted from the gel overnight after the addition of 270 µl of elution buffer (0.5 mM NH4Ac pH 7.5, 1 mM EDTA, 0.1% SDS, 10 mM MgCl2). After phenol–chloroform extraction and precipitation with ethanol, the DNA fragments were separated on a 8% sequencing gel. Images of the footprints were analyzed by phosphoimaging using the Aida software package (Raytest).
Methylation interference footprinting assay
Methylation of the DNA probes at no more than one position per molecule was performed generally as described by Li and Wrage (29). In a standard reaction, 5′-end-labeled DNA containing the lrpA promoter was methylated for 2 min at 20°C with 50 mM dimethylsulfate (DMS) in 200 µl of DMS reaction buffer (120 mM NaCl, 20 mM Tris–HCl pH 8.0, 20 mM MgCl2, 2 mM EDTA). The reaction was terminated by the addition of 50 µl of 1 M NaAc pH 7.0/1 M 2-mercaptoethanol. After ethanol precipitation, transcriptional components and LrpA were assembled on the methylated probe (1 × 106 c.p.m.) and subjected to a 5-fold scaled-up EMSA as described above. Bands corresponding to free and bound DNA were excised from the gel. The eluted DNA was cleaved for 30 min at 90°C with piperidine, followed by phenol–chloroform extraction and precipitation with ethanol. Samples were applied to a 8% sequencing gel and analyzed by phosphoimaging (Raytest).
RESULTS
Lrp A interferes with RNA polymerase binding
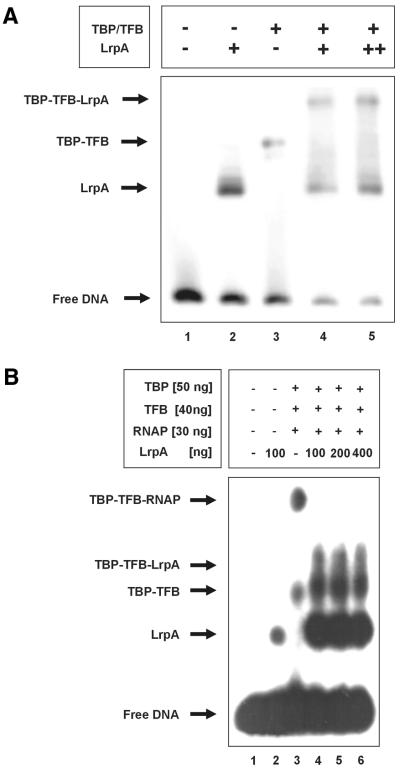
EMSAs were conducted to investigate the interaction of LrpA with Pyrococcus transcriptional machinery. An end-labeled DNA fragment comprising nucleotide –109 to +80 of the lrpA promoter was incubated with TBP/TFB and TBP/TFB and RNAP both in the presence and absence of LrpA (Fig. 1). In agreement with previous work (17), we found that LrpA binds to its own promoter in EMSA (Fig. 1A, lane 2). A single LrpA promoter complex was detected at a LrpA:DNA molar ratio of 650:1. The factors TBP and TFB formed with this probe a complex with lower electrophoretic mobility (Fig. 1A, lane 3). Addition of LrpA to binding reactions containing TBP and TFB resulted in the appearance of a third distinct complex with low mobility and the signal corresponding to the TBP/TFB shift disappeared (lane 5). An increase of the amount of LrpA in the binding reaction increased the intensity of the LrpA shifted signal and of the second signal with lower mobility. These findings suggested that the complex showing lower mobility contains both TFB/TFB and LrpA. When Pyrococcus RNAP was added to binding reactions containing TBP/TFB, a second complex with lower mobility was detected (Fig. 1B, lane 3). This complex was destroyed in the presence of 100 ng (LrpA:RNAP molar ratio of 81:1) or at higher concentrations of LrpA in binding reactions (Fig. 1B, lanes 4–6). This finding suggested that LrpA inhibits binding of RNAP to the TBP/TFB promoter complex.
Figure 1.
Effect of LrpA binding on binding of transcriptional components to DNA. The EMSA-binding reaction was performed on lrpA promoter DNA for 30 min at 70°C and complexes were analyzed by native gel electrophoresis (EMSA). (A) LrpA and TBP–TFB form a distinct complex. Fifty nanograms TBP, 300 ng TFB and/or 50 (+) or 100 ng (++) LrpA were assembled on 1 ng of radiolabeled DNA, as indicated. Free DNA and protein–DNA complexes are indicated to the left. (B) LrpA prevents the formation of the TBP–TFB–RNAP–DNA complex. TBP, TFB, RNAP and LrpA were added to DNA (specified above each lane). Arrows mark the position of free DNA, LrpA–, TBP–TFB–, TBP–TFB–LrpA– and TBP–TFB–RNAP–DNA complexes.
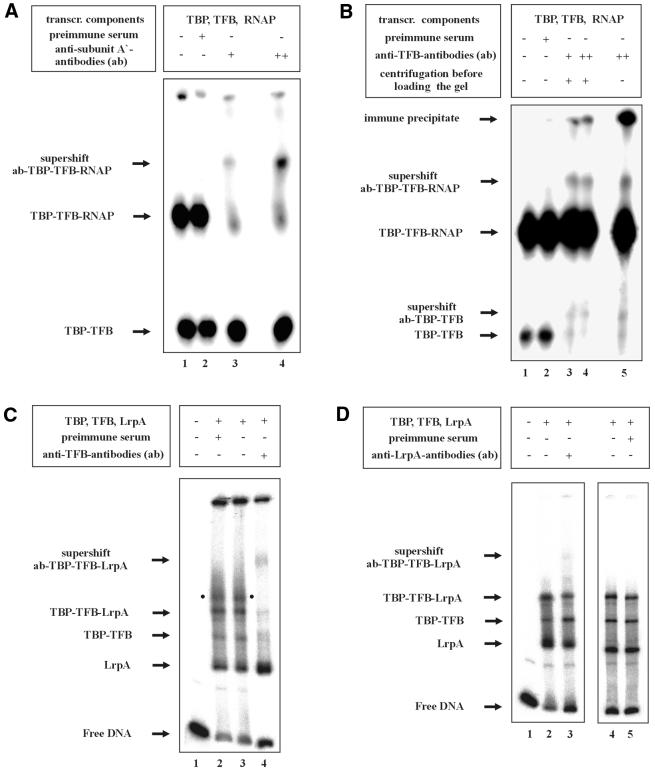
To provide additional evidence that LrpA can coexist with TBP/TFB in a DNA–protein complex but abrogates RNAP binding, the existence of individual components of the binding reactions was studied by the addition of antisera to binding reactions. If antibodies recognize epitopes of proteins present in a DNA–protein complex, the electrophoretic mobility of this complex is reduced due to the bound IgG molecules, and in such a way proteins in distinct DNA–protein complexes can be identified. After addition of antibodies raised against subunit A′ of RNAP, a supershift corresponding to an antibody-RNAP–TBP–TFB complex (Fig. 2A, lane 3) was observed. The intensity of this shifted band was increased at higher concentrations of antibodies (lane 4). The existence of this supershift indicated the presence of RNAP in the complex with lowest mobility in lanes 1 and 2 of Figure 2A. Similarly, the presence of TFB in the two complexes visible in Figure 2A and B, lanes 1 and 2, was shown by the addition of anti-TFB antibodies to binding reactions (Fig. 2B). Two distinct additional complexes of lower mobility were detected (Fig. 2B, lanes 3–5), indicating binding of antibodies to TFB in the TBP–TFB and TBP–TFB–RNAP complexes. No supershift was observed when preimmune serum was added to the binding reactions (Fig. 2A and B, lane 2). An analysis of the composition of DNA–protein complexes formed in the presence of TBP/TFB and LrpA is shown in Figure 2C and D. A complex with low electrophoretic mobility predicted to contain TBP/TFB and LrpA (Fig. 2C, lane 3) was shifted after addition of antibodies raised against TFB (Fig. 2C, lane 4) and when antisera against LrpA were added to binding reactions (Fig. 2D, lane 2). The preimmune serum did not cause a supershift (Fig. 2C, lane 2, and D, lane 5). These findings suggest that LrpA and the two archaeal transcription factors have different binding sites on the lrpA promoter and provide evidence that three distinct complexes were formed at the lrpA promoter in the presence of the three components of the transcriptional machinery and LrpA: DNA–LrpA, DNA–TBP–TFB and DNA–TBP–TFB–LrpA. These immunological studies also confirm the conclusion that the complex with the lowest electrophoretic mobility (Fig. 1B, lane 3), which cannot be detected in the presence of LrpA (Fig. 1B, lanes 4–6), contains the RNAP.
Figure 2.
Immunochemical analysis of protein–DNA complexes. Binding reactions with 50 ng of TBP, 300 ng of TFB and 30 ng of RNAP on 0.5 ng of lrpA promoter were incubated for 30 min at 70°C. The specific antibodies were added and allowed to bind for 15 min at 37°C. (A) Identification of RNAP in the protein–DNA complex. An aliquot of 0.8 (+) or 1.5 µg (++) of purified antiserum against subunit A′ of RNAP was added to the EMSA-binding reaction. A 1.5 µg aliquot of purified preimmune serum was used in a control reaction (lane 2). Arrows label shifted and supershifted protein–DNA complexes. (B) Identification of TFB in TBP–TFB and in TBP–TFB–RNAP–DNA complexes. The standard EMSA of TBP, TFB and RNAP was challenged with 1.6 (+) or 3.2 µg (++) of purified antibodies (ab) raised against TFB or 3.2 µg of purified preimmune serum in a control reaction (indicated above each lane). The samples shown in lanes 3 and 4 were centrifuged (lanes 1, 3 and 5) before loading the gel to remove large immune precipitates not entering the gel. The samples shown in lanes 1, 3 and 5 were not centrifuged. Positions of TBP–TFB–, TBP–TFB–RNAP–DNA complexes, of supershifted protein–DNA complexes and of the immune precipitate are indicated to the left. In the experiments shown in (A) and (B) it was necessary to run the free DNA out of the gel to obtain optimal resolution of complexes. (C) Identification of TFB in the TBP–TFB–LrpA complex. Antibodies raised against TFB (lane 4) or preimmune serum (lane 2) were added to binding reactions containing TBP, TFB and LrpA. EMSA and processing was as shown in (A). The dot inside the figure indicates an additional shift observed in this particular experiment that is most likely caused by binding of multiple copies of LrpA to the DNA fragment. Binding of multimers of LrpA to DNA has been described earlier (17). (D) Identification of LrpA in the TBP–TFB–LrpA complex. A 0.5 µg aliquot of purified antiserum against LrpA (lane 3) or preimmune serum (lane 5) was added to binding reactions containing TBP, TFB and LrpA. Only in lane 3 a clear supershifted DNA band was detected indicating the presence of LrpA in the complex of lowest electrophoretic mobility visible in lanes 2, 4 and 5.
Provided that binding of LrpA to DNA inhibts RNAP recruitment, it can be predicted that preinitiation complexes (PIC) assembled in the absence of LrpA are less sensitive to the inhibitory effects of LrpA. To investigate this the DNA template was preincubated with TBP, TFB and RNAP for varying periods prior to the additon of LrpA. Transcription reactions were started by the addition of ribonucleoside triphosphates and run-off transcripts from the lrpA promoter were analyzed. When Lrp was added at the same time as transcriptional components, transcriptional activity at a LrpA: DNA molar ratio of 160:1 was 13% compared with a reaction not containing LrpA (Fig. 3A, lanes 1 and 2). When LrpA was added 10 or 20 min after the formation of the PIC, the residual activity was 49 and 80%, respectively. In a further experiment, transcriptional components were preincubated with DNA for 10 min but increasing amounts of LrpA were added. Under these conditions more LrpA (molar ratio Lrp:DNA 347:1 instead of 160:1) was required to obtain an inhibition of transcription of 13% (data not shown). These data show that preformation of PIC impedes the inhibitory activity of LrpA.
To further investigate the interaction of LrpA with prefomed PIC, the transcriptional components were incubated for 30 min with DNA. Then, varying amounts of LrpA were added and the complexes analyzed by EMSA. At LrpA:DNA and LrpA:RNAP molar ratios of 650:1 and 81:1, respectively, the complex containing RNAP was still visible (Fig. 3B, lanes 2–4).
In contrast, when LrpA and transcriptional components were added simultaneously, the association of RNAP to the TBP–TFB complex was already inhibited at a LrpA:RNAP molar ratio of 81:1 (Fig. 1B, lane 4). At higher concentrations of LrpA, the preformed complexes containing RNAP were displaced by LrpA (Fig. 3B, lanes 5 and 6). As expected, TBP–TFB complexes were not destroyed at all concentrations of LrpA investigated. In addition, preincubation of LrpA with template DNA did not inhibit binding of TBP/TFB to the lrpA promoter (Fig. 3C). Taken together, these findings indicate that LrpA interferes with binding of RNAP to the TBP–TFB promoter complex.
The binding sites of LrpA and TBP/TFB do not overlap
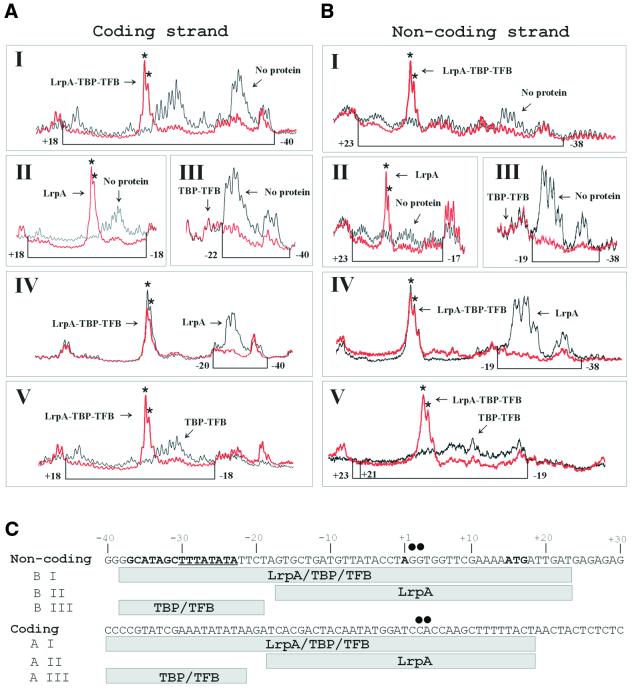
DNaseI and hydroxylradical footprinting has shown that LrpA protects the DNA region from –22 to +24 at the lrpA promoter (17). To investigate binding of LrpA to this promoter in the presence of TBP and TFB, distinct complexes isolated by EMSA were analyzed in the gel matrix by OP–Cu footprinting (28). On the coding DNA strand, the DNA region from –40 to –22 was protected by TBP/TFB (Fig. 4A, III). LrpA alone protected the region from –18 to +18 (Fig. 4A, II). Two strong hypersensitivity signals were generated by LrpA binding at positions +4/+3. In the complex with lower electrophoretic mobility (Fig. 1A, lanes 4 and 5), which was supposed to consist of TBP, TFB and LrpA, the OP–Cu footprint extended from position –40 to +18. The two hypersensitivity sites indicative for LrpA binding were also visible in this complex (Fig. 4A, I and II). The same protection patterns were observed when the lanes showing the LrpA–TBP–TFB footprint were aligned to that of the LrpA footprint (Fig. 4A, IV) or that of the TBP–TFB footprint (Fig. 4A, V), respectively. On the non-coding DNA strand, the protection patterns of transcriptional components were less pronounced. Two clear hypersensitivity signals were detected in the LrpA–DNA– and LrpA–TBP–TFB complexes at positions +3/+2. TBP/TFB protected the DNA region from –38 to –19 (Fig. 4B, III), LrpA from –17 to +23 (Fig. 4B, II). In the complex of higher molecular mass generated only in the presence of TBP/TFB and LrpA (Fig. 1A, lanes 4 and 5), the protection pattern differed somewhat from that of the individual components, but beside the hypersensitivity sites at positions +3/+2 a protection in the TBP–TFB-binding site extending from position –19 to –38 was detected (Fig. 4B, I). Also, at the non-coding strand which showed weaker signals, the protection patterns of the LrpA–TBP–TFB footprint compared with those from the individual footprints were shown to be the same (Fig. 4B, IV and V). These findings demonstrated clearly the coexistence of LrpA and TBP/TFB in a distinct isolated complex and show that the DNA-binding sites of the TBP–TFB complex and of LrpA do not overlap significantly at the lrpA promoter.
Figure 4.
DNA-bound LrpA and TBP–TFB can coexist at the lrpA promoter. LrpA promoter DNA was incubated with TBP–TFB and/or LrpA (as indicated below the panels) for 30 min at 70°C in a 5-fold scaled-up EMSA-binding reaction followed by the separation of free DNA from protein–DNA complexes through native gel electrophoresis. Total and bound DNA was excised from the gel, and the DNA was recovered and submitted to the OP–Cu cleavage reaction as described in Materials and Methods. DNA fragments were processed for electrophoresis and analyzed using a PhosphorImager and accompanying software. Densitometric plots of lanes of sequencing gels corresponding to standard cleavage reactions of gel-purified probe DNA and of DNA–protein complexes were compared in the individual panel. The red line in each panel refers to footprints, the black line to cleavage patterns of non-complexed DNA (I–III) or to footprints (IV and V) subjected to gel electrophoresis, as labeled by arrows. (I) LrpA–TBP–TFB footprint; (II) LrpA footprint; (III) TBP–TFB footprint; (IV) LrpA–TBP–TFB footprint plotted to the LrpA footprint; (V) LrpA–TBP–TFB footprint plotted to the TBP–TFB footprint. Hypersensitivity sites are indicated by asterisks. The boundaries of each binding site are marked by numbers relative to the start of transcription as +1. (A) Coding strand. (B) Non-coding strand. (C) Schematic depiction of the Cu–OP footprinting data. The lrpA sequence from –40 to +30 is given. The TATA box, the BRE and the start of transcription (+1) are indicated in bold letters on the top strand; and the TATA box is underlined in addition. The regions protected from Cu–OP cleavage of the non-coding (B) and the coding strand (A) are shown by gray rectangles below the sequence. The protected region refers to the number of the panel in (A) and (B), as indicated to the left. Hypersensitivity sites are marked by black dots.
Identification of a LrpA recognition motif by methylation interference
To identify nucleotides required for binding of LrpA and TBP/TFB at the lrpA promoter, we conducted in vitro DMS interference experiments. DMS methylates N-7 of guanine in the major groove of the B-form of DNA and to a lesser extent N-3 of adenine in the minor groove. To detect A and G residues in the lrpA promoter that are in contact with LrpA and the TBP–TFB complex, DMS-treated DNA was incubated with these proteins in binding reactions. Complexed DNA was purified by EMSA and the DNA in these complexes was isolated, subjected to chemical cleavage and analyzed by denaturing gel electrophoresis. DNA fragments cleaved at G and A residues, which were weaker in intensity than in the cleavage pattern of probe DNA, are required for DNA binding of the corresponding protein.
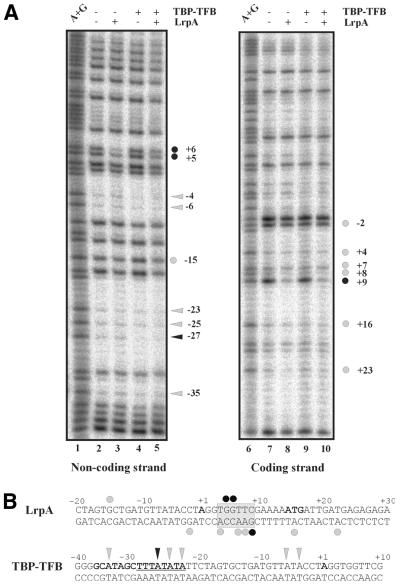
On the non-coding DNA strand, methylation of the G residues at positions +5, +6 and –15 interfered with LrpA binding (Fig. 5A, compare lanes 2 and 3), methylation of A residues at positions –4, –6, –23, –25, –27 and –35 with TBP/TFB binding (Fig. 5A, compare lanes 2 and 4, lanes 2 and 5). On the coding strand, G residues at positions –2 and +9, and A residues at positions +4, +7, +8, +16 and +23 inhibited DNA binding of LrpA (Fig. 5A, compare lanes 7 and 8). The G residues at positions +5, +6 and +9 (labeled by bold symbols in Fig. 5) were most important for LrpA binding. With the exception of the A residue at position –27, which interfered strongly with binding of TBP/TFB, the effects caused by modification of A residues were less pronounced but significant. At the coding DNA strand, G and A residues required for binding of TBP/TFB were not detected using this technique (Fig. 5A, lane 9). The data of the methylation interference experiments are summarized in Figure 5B.
Figure 5.
Identification of a LrpA recognition motif by methylation interference. DMS-treated probes of the lrpA promoter were subjected to a preparative EMSA with LrpA and TBP–TFB; unbound and bound DNA were separated through native gel electrophoresis. Bands corresponding to free and bound DNA were cut out. The recovered DNA was cleaved with piperidine and applied to a sequencing gel. (A) Denaturing gel electrophoresis of modified DNA in DNA–protein complexes. The presence (+) and absence (–) of proteins is indicated above. Positions of guanine or adenine residues that strongly interfere with protein binding when methylated are shown by black triangles (TBP–TFB) or circles (LrpA), respectively. Weaker interference is indicated by gray triangles and circles. Their position relative to the start site of transcription (+1) is shown by numbers. Each reaction was run in parallel with the Maxam Gilbert sequencing reaction A+G. (B) Summary of the methylation interference pattern. The upper panel presents the LrpA data. The lrpA sequences from –20 to +40 (LrpA) or from –40 to +10 (TBP–TFB) are given. Within the LrpA-binding site the start of transcription (+1) and translation is shown in bold letters on the top strand; within the TBP–TFB-binding site the start of transcription, the TATA box and BRE are indicated in bold and the TATA box is underlined in addition. Strong interferences are indicated by black circles (LrpA) or triangles (TBP–TFB), weak interferences by gray symbols. The DNA region where most interferences were located is highlighted by a gray box.
DISCUSSION
Pyrococcus LrpA, like Lrp from E.coli and Lrs14 from S.solfataricus, inhibits transcription of its own gene. The DNA-binding sites of Lrs14 and LrpA differ. Lrs14 protects the TATA box and BRE (16), whereas the LrpA footprint does not overlap with these promoter elements (17) (Fig. 4) but is centered around the transcription start site. The work presented here provides evidence that LrpA, in contrast to Lrs14, does not inhibit binding of TBP–TFB to the promoter but abrogates RNAP recruitment. First, TBP, TFB and LrpA formed a distinct DNA complex that can be isolated by EMSA (Fig. 1A). Secondly, formation of the TBP–TFB–RNAP ternary complex was inhibited in the presence of LrpA (Fig. 1B). Thirdly, preformed ternary complexes were more stable against disruption by LrpA (Fig. 3A and B), but preincubation of LrpA with the template did not inhibit the formation of TBP–TFB–DNA complexes (Fig. 3C). Furthermore, analysis of isolated DNA–protein complexes containing LrpA or TBP/TFB by OP–Cu footprinting revealed that the binding sites of the proteins did not overlap significantly (Fig. 4). However, this LrpA-protected DNA region extending to position +23 overlapped clearly with DNA regions protected by archaeal RNAPs bound to the TBP–TFB complex (16,30). Finally, isolated complexes of lower electrophoretic mobility that were supposed to contain TBP–TFB and LrpA showed at the non-coding strand identical (Fig. 4A), and on the coding strand similar (Fig. 4B), protection patterns to the individual components.
Thus, the mode of transcriptional repression by LrpA is highly different from that mediated by Lrs14 but akin to that of the archaeal metal-dependent repressor MDR1, which also binds downstream to the TATA box and prevents RNAP binding (11). An analysis of the DNA sequences encoding LrpA and Lrs14 revealed that the identity on the level of amino acids was only 16%; the similarity was 37% (data not shown). Our results show that two regulators of the Lrp/AsnC family, which are only distantly related, interact in a completely different manner with the archaeal transcriptional machinery. The sequence identity of Pyrococcus LrpA and Methanococcus Ptr1 and Ptr2 is much higher (43 and 55%) (18). The finding that the consensus sequences for binding of Ptr1 and Ptr2 do not show similarity to a TATA-box sequence might be considered as an indication that these proteins do not interfere with binding of TBP, but further work is required to clarify whether Ptr1 and Ptr2 use a similar or different mechanism to LrpA. It has been speculated that regulated promoters having prebound TBP/TFB can respond more rapidly to changing environmental conditions (16) than promoters that are repressed by inhibition of binding of TBP/TFB. The biological function of archaeal members of the Lrp/AsnC family is not clear. It has only been shown that Lrs14 and Sa-Lrp accumulate in late growth stages (14,15). Analysis of the action of archaeal Lrp homologs at different promoters both in vitro and in vivo under different physiological conditions is required to elucidate its function, but the findings reported here suggest that the physiological functions of Lrp homologs might be dissimilar in different genera. In the light of these results, it seems also possible that the 12 homologs of the Lrp/AsnC family detected in the genome of P.furiosus (http://www.genome.utah.edu/sequence.html) have different functions in the physiology of Pyrococcus and may also use dissimilar mechanisms to regulate transcription.
The DNA regions required for binding of Sulfolobus Lrs14 and Pyrococcus LrpA have been analyzed by mutational studies. The DNA region from –30 to –26 was most important for the stability of Lrs14 promoter interactions (16). This sequence is a part of the archaeal TATA box, the second DNA region from –25 to –22 involved in binding of Lrs14, is also AT-rich. In contrast, the most important DNA region for binding of LrpA (17), ranging from position –1 to +8, differs significantly in its DNA sequence from the major recognition signal for Lrs14 5′-TTTAT-3′ (16). The methylation interference experiments reported here demonstrate that each individual base pair of the sequence 5′-TGGTTC-3′, ranging from +4 to +9 (boxed in Fig. 5B), is important for binding of LrpA. As in the Sulfolobus Lrs14 DNA-binding site, a second DNA region is important for binding of the regulator, and these methylations upstream and downstream of the major recognition motif interfered only weakly with LrpA binding similar to the second –25 to –22 DNA region in the Lrs14-binding site identified by mutational studies, which is also less important for DNA binding (16). Also, the consensus recognition sequence motif inferred as a binding signal for Methanococcus Lrp homologs Ptr1 and Ptr2 consisted of two DNA elements separated by a short intervening sequence (1–3 bp in this case) (18). But the Methanococcus Ptr recognition sequences were two inverted repeats of hexanucleotides that showed no sequence similarity with the DNA regions important for LrpA binding. From the work reported here and previous studies (17), the sequence 5′-TGGTTC-3′ can be proposed as a major recognition signal and as a putative interaction site for the helix–turn–helix motif of LrpA (31) with DNA at the lrpA promoter. Two strong hypersensitivity sites induced by LrpA binding were located by OP–Cu footprints immediately upstream of the TGGTTC-hexanucleotide (Fig. 4A; see also summary in Fig. 4C). OP–Cu cleaves DNA in the minor groove and hypersensitivity sites are usually explained as a result of bending of DNA. Therefore, the existence of these hypersensitivity sites suggests that binding of LrpA changes the conformation of DNA. This bending of DNA most likely leads to an exposition of the recognition motif for LrpA and this might stabilize the interaction of DNA with the helix–turn–helix motif of the protein. Although a hypersensitivity site at position +7 of the non-coding DNA strand (see sequence of the lrpA promoter in Fig. 4C) has been detected in the DNaseI footprint of LrpA (17), this clustering of strong hypersensitivity sites on both strands in the DNA region upstream of the LrpA recognition signal was only observed in OP–Cu footprints. Further studies will show whether this sequence is also conserved among more Pyrococcus promoters regulated by LrpA.
Our methylation interference data show also that three A residues of the TATA box of the lrpA promoter and an A residue at position –35, located 5 bp upstream of the TATA box, are important for binding of TBP/TFB to DNA. Methylation of A residues of the TATA box and of a G residue 6 bp upstream of the TATA box of the T6 promoter of the Sulfolobus virus SSV had also a strong inhibitory effect on binding of Sulfolobus TBP and TFB (7). These findings suggest that the major structural determinants of promoters interacting with the two basal transcription factors are conserved between the Crenarchaeote Sulfolobus and the Euryarchaeote Pyrococcus. Modification of A residues at positions –6 and –4 also impaired binding of TBP/TFB at the lrpA promoter (Fig. 5). Interestingly, a protection of this DNA region was also observed in hydroxyl radical footprints of TBP/TFB at the Pyrococcus glutamate dehydrogenase promoter (S.Francois and M.Thomm, unpublished data). These findings suggest that an additional downstream TBP/TFB contact DNA site exists in Pyrococcus promoters that has not yet been detected in TBP–TFB–DNA crystals (32).
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Drs J. van der Oost and W. de Vos, University of Wageningen, for providing LrpA and antibodies against LrpA for initial experiments, Drs M. Ouhammouch, and P. Geiduscheck, University of California, for valuable advice with DMS interference experiments and B. Goede, University of Kiel, for help with bioinformatic work. This study was supported by a grant of the DFG, the priority program of the DFG ‘Genome regulation and genome organization in Archaea’ and by the Fonds der Chemischen Industrie.
REFERENCES
- 1.Thomm M. and Wich,G. (1988) An archaeal promoter element for stable RNA genes with homology to the TATA box of higher eukaryotes. Nucleic Acids Res., 16, 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reiter W.D., Hüdepohl,U. and Zillig,W. (1990) Mutational analysis of an archaeal promoter: essential role of a TATA box for transcription efficiency and start site selection in vitro. Proc. Natl Acad. Sci. USA, 87, 9509–9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rowlands T., Baumann,P. and Jackson,S.P. (1994) The TATA-binding protein: a general transcription factor in eukaryotes and archaeabacteria. Science, 264, 1326–1329. [DOI] [PubMed] [Google Scholar]
- 4.Gohl H.P., Gröndahl,B. and Thomm,M. (1995) Promoter recognition in archaea is mediated by transcription factors: identification of transcription factor aTFB from Methanococcus thermolithotrophicus as archaeal TATA-binding protein. Nucleic Acids Res., 23, 3837–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wettach J., Gohl,H.P., Tschochner,H. and Thomm,M. (1995) Functional interaction of yeast and human TATA-binding proteins with an archaeal RNA polymerase and promoter. Proc. Natl Acad. Sci. USA, 92, 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hausner W., Wettach,J., Hethke,C. and Thomm,M. (1996) Two transcription factors related with the eucaryal transcription factors TATA-binding protein and transcription factor IIB direct promoter recognition by an archaeal RNA polymerase. J. Biol. Chem., 271, 30144–30148. [DOI] [PubMed] [Google Scholar]
- 7.Qureshi S.A. and Jackson,S.P. (1998) Sequence-specific DNA binding by the S.shibatae TFB homolog, TFB, and its effect on promoter strength. Mol. Cell, 1, 389–400. [DOI] [PubMed] [Google Scholar]
- 8.Bell S.D., Kosa,P.L., Sigler,P.B. and Jackson,S.P. (1999) Orientation of the transcription preinitiation complex in archaea. Proc. Natl Acad. Sci. USA, 96, 13662–13667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langer D., Hain,J., Thuriaux,P. and Zillig,W. (1995) Transcription in archaea: similarity to that in eucarya. Proc. Natl Acad. Sci. USA, 92, 5768–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kyrpides N.C. and Ouzounis,C.A. (1999) Transcription in Archaea. Proc. Natl Acad. Sci. USA, 20, 8545–8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bell S.D., Cairns,S.S., Robson,R.L. and Jackson,S.P. (1999) Transcriptional regulation of an archaeal operon in vivo and in vitro. Mol. Cell, 4, 971–982. [DOI] [PubMed] [Google Scholar]
- 12.Kyrpides N.C. and Ouzounis,C.A. (1995) The eubacterial transcriptional activator Lrp is present in the archaeon Pyrococcus furiosus. Trends Biochem. Sci., 20, 140–141. [DOI] [PubMed] [Google Scholar]
- 13.Charlier D., Roovers,M., Thia-Thong,T.L., Durbecq,V. and Glansdorff,N. (1997) Cloning and identification of the Sulfolobus solfataricus lrp gene encoding an archaeal homologue of the eubacterial leucine-responsive global transcriptional regulator Lrp. Gene, 201, 63–68. [DOI] [PubMed] [Google Scholar]
- 14.Napoli A., van der Oost,J., Sensen,C.W., Charlebois,R.L., Rossi,M. and Ciaramella,M. (1999) An Lrp-like protein of the hyperthermophilic archaeon Sulfolobus solfataricus binds to its own promoter. J. Bacteriol., 181, 1474–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enuro-Etat J., Gigot,D. Thia-Thong,T.L., Glansdorff,N. and Charlier,D. (2000) Purification and characterization of Sa-Lrp, a DNA-binding protein from the extreme thermoacidophilic archaeon Sulfolobus acidocaldarius homologous to the bacterial global transcriptional regulator Lrp. J. Bacteriol., 182, 3661–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell S.D. and Jackson,S.P. (2000) Mechanism of autoregulation by an archaeal transcriptional repressor. J. Biol. Chem., 275, 31624–31629. [DOI] [PubMed] [Google Scholar]
- 17.Brinkman A.B., Dahlke,I., Tuininga,J.E., Lammers,T., Dumay,V., de Heus,E., Lebbink,J.H., Thomm,M., de Vos,W.M. and van der Oost,J. (2000) An Lrp-like transcriptional regulator from the archaeon Pyrococcus furiosus is negatively autoregulated. J. Biol. Chem., 275, 38160–38169. [DOI] [PubMed] [Google Scholar]
- 18.Ouhammouch M. and Geiduschek,P.E. (2001) A thermostable platform for transcriptional regulation: the DNA-binding properties of two Lrp homologs from the hyperthermophilic archaeon Methanococcus jannaschii. EMBO J., 20, 146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calvo J.M. and Matthews,R.G. (1994) The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev., 58, 466–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman E.B. and Lin,R. (1995) Leucine-responsive regulatory protein: a global regulator of gene expression in E.coli. Annu. Rev. Microbiol., 49, 747–775. [DOI] [PubMed] [Google Scholar]
- 21.Cui Y., Midkiff,M.A., Wang,Q. and Calvo,J.M. (1996) The leucine-responsive regulatory protein (Lrp) from Escherichia coli. Stoichiometry and minimal requirements for binding to DNA. J. Biol. Chem., 271, 6611–6617. [DOI] [PubMed] [Google Scholar]
- 22.Paul L., Blumenthal,R.M. and Matthews,R.G. (2001) Activation from a distance: role of Lrp and integration host factor in transcriptional activation of gltBDF. J. Bacteriol., 183, 3910–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hethke C., Geerling,A.C.M., Hausner,W., de Vos,W.M. and Thomm,M. (1996) A cell-free transcription system for the hyperthermophilic archaeon Pyrococcus furiosus. Nucleic Acids Res., 43, 2369–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frey G., Thomm,M., Brüdigam,B. and Hausner,W. (1990) An archaeal cell-free transcription system. The expression of tRNA genes from Methanococcus vannielii is mediated by a transcription factor. Nucleic Acids Res., 18, 1361–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J., Maniatis,T. and Fritsch,E.F. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Hausner W. and Thomm,M. (1995) The translation product of the presumptive Thermococcus celer TATA-binding protein sequence is a transcription factor related in structure and function to Methanococcus transcription factor B. J. Biol. Chem., 270, 17649–17651. [DOI] [PubMed] [Google Scholar]
- 27.Hausner W., Lange,U. and Musfeldt,M. (2000) Transcription factor S, a cleavage induction factor of the archaeal RNA polymerase. J. Biol. Chem., 275, 12393–12399. [DOI] [PubMed] [Google Scholar]
- 28.Kuwabara M.D. and Sigman,D.S., (1987) Footprinting DNA–protein complexes in situ following gel retardation assays using 1,10-phenanthroline-copper ion: Escherichia coli RNA polymerase–lac promoter complexes. Biochemistry, 26, 7234–7238. [DOI] [PubMed] [Google Scholar]
- 29.Li Q. and Wrage,O. (1997) Assays for transcription factors access to nucleosomal DNA. Methods, 12, 96–104. [DOI] [PubMed] [Google Scholar]
- 30.Hausner W and Thomm,M. (2000) Events during initiation of archaeal transcription: open complex formation and DNA–protein interactions. J. Bacteriol., 183, 3025–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leonard P.M., Smith,S.H., Sedelnikova,S.E., Brinkman,A.B., de Vos,W.M., van der Oost,J., Rice,D.W. and Rafferty,J.B. (2001) Crystal structure of the Lrp-like transcriptional regulator from the archaeaon Pyrococcus furiosus. EMBO J., 20, 990–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Littlefield O., Korkhin,Y. and Sigler,P.B. (1999) The structural basis for the oriented assembly of a TBP/TFB/promoter complex. Proc. Natl Acad. Sci. USA, 96, 13668–13673. [DOI] [PMC free article] [PubMed] [Google Scholar]