Abstract
Overexpression of the RAD52 epistasis group of gene products is a convenient way to investigate their in vivo roles in homologous recombination (HR) and DNA repair. Overexpression has the further attraction that any associated stimulation of HR may facilitate gene-targeting applications. Rad51p or Rad52p overexpression in mammalian cells have previously been shown to enhance some forms of HR and resistance to ionising radiation, but the effects of Rad52p overexpression on gene targeting have not been tested. Here we show that Rad52p overexpression inhibits gene targeting while stimulating extrachromosomal HR. We also find that Rad52p overexpression affects cell-cycle distribution, impairs cell survival and is lost during extensive passaging. Therefore, we suggest that excess Rad52p can inhibit the essential RAD51-dependent pathways of HR most likely to be responsible for gene targeting, while at the same time stimulating the RAD51-independent pathway thought to be responsible for extrachromosomal HR. The data also argue against Rad52p overexpression as a means of promoting gene targeting, and highlight the limitations of using a single HR assay to assess the overall status of HR.
INTRODUCTION
The RAD52 epistasis group of genes, including RAD51 and RAD52, was identified in yeast and is required for the accurate repair of DNA double-strand breaks (DSBs) by homologous recombination (HR) (1–3). This group of genes has been well conserved between yeast and man (4–7) and the basic mechanisms of HR are also thought to have been conserved. Broken DNA ends are processed to produce 3′-single-stranded tails that search for, invade and undergo strand exchange with an intact homologous DNA template. Subsequent migration and resolution of the junctions in such intermediates, combined with DNA synthesis and nick ligation, results in the accurate repair of the original break. The RAD51 gene product, Rad51p, is the central protein in the early steps of these mechanisms (8), and catalyses strand invasion in vitro, a reaction that is stimulated by Rad52p, the RAD52 gene product (9–11). Rad52p is one of many proteins known to interact with Rad51p (12,13) and can itself bind to DNA forming heptameric ring structures at DNA ends (14–17).
A second, less accurate, form of DSB repair by HR is the single-strand annealing (SSA) pathway which is best known for its action on DSBs situated between direct repeats (3). Repair is achieved by resection of the DNA ends to form single-stranded DNA, followed by annealing of the homologous repeats, trimming of extruded tails and nick ligation. In this non-conservative mechanism there is a net loss of one copy of the repeat sequence (and of the unique DNA between the repeats). Rad52p is known to promote the annealing of homologous single strands in vitro (17–20) and, in yeast at least, RAD52 is the only RAD52 epistasis group gene required for SSA (21,22).
DSB repair can also be achieved by non-homologous end joining (NHEJ), a pathway that is independent of the RAD52 epistasis group of genes, requiring the DNA end-binding Ku heterodimer and an associated protein kinase, DNA-PKcs (1,2,23). NHEJ does not necessarily restore the exact sequence existing before the DSB, but even inaccurate repair by NHEJ (or SSA) prevents genetic damage associated with broken chromosomes that undergo mitosis.
Assays for HR in mitotic cells are usually based on intrachromosomal HR, extrachromosomal HR or gene targeting (24). In intrachromosomal and extrachromosomal HR, the two homologous duplexes are situated in the same chromosome or on extrachromosomal DNA molecules, respectively. In gene targeting, one duplex is located at a chromosomal (target) locus and the other on an extrachromosomal DNA molecule (targeting construct). Typically, the latter is engineered and experimentally introduced into cells so as to produce a defined genetic alteration to the target locus (25). Despite the extensive use of gene targeting in ES cells to generate genetically modified mice (26), the low efficiency of gene targeting remains a significant barrier to its exploitation as a genetic tool and potential mode of gene therapy (27). Several lines of evidence, including its conservative nature (28), its dependence on RAD54 (29,30) (a RAD52 epistasis group gene whose product facilitates Rad51p activity) and its stimulation by Rad51p overexpression (31), suggest that gene targeting occurs by a strand-invasion type mechanism. In contrast, there is evidence that the predominant mechanism for extrachromosomal HR is SSA (32,33) whereas intrachromosomal HR occurs by both SSA and strand-invasion type mechanisms (34–36). Therefore, care must be taken not to overinterpret results based on only one type of HR assay.
Whereas biochemical analyses suggest an important degree of conservation in RAD protein function from yeast to human cells, genetic gain- or loss-of-function experiments indicate that there are some important differences in vivo (37). In yeast, loss of either RAD51 or RAD52 function seriously reduces resistance to ionising radiation (IR), but cells remain viable (3). However, in vertebrates, gene disruption experiments show that RAD51, and therefore presumably DSB repair by strand-invasion type mechanisms, is essential for viability (38–40), and it is thought that such repair is an essential part of DNA replication (41,42). On the other hand, disruption of RAD52 in vertebrate cells is not lethal, does not affect resistance to IR and results in only a small reduction in the frequency of gene targeting (43,44). In vertebrate cells, there is, therefore, at least a degree of redundancy for the role of Rad52p in DSB repair by HR. Nevertheless, overexpression of Saccharomyces cerevisiae Rad52p in human tumour cells caused a 2-fold rise in resistance to IR accompanied by a 12-fold rise in extrachromosomal HR (45). A similar increase in radiation resistance was reported for monkey cells overexpressing human Rad52p and this was accompanied by a 3-fold rise in the frequency of intrachromosomal HR (46). Together these studies provide clear evidence of an in vivo role, albeit non-essential, for Rad52p in DSB repair by HR.
Despite this progress, the HR pathway (SSA or strand invasion) most stimulated by Rad52p overexpression is unknown and the effects of Rad52p overexpression on gene targeting have yet to be tested. Given, as summarised above, that Rad52p overexpression stimulates other forms of HR in vivo, that Rad51p activity in vitro is stimulated by Rad52, and that gene targeting is stimulated by Rad51p overexpression, it seemed a reasonable possibility that Rad52p overexpression would stimulate gene targeting, especially if combined with Rad51p overexpression. Furthermore, it had been suggested that competition for DNA ends between Rad52p and Ku heterodimer may underlie the low frequency of gene targeting relative to random integration in higher eukaryotes, and that increasing the naturally low Rad52p:Ku ratio may promote gene targeting (14). Therefore, in the present study, we analysed the effects of Rad52p overexpression on our previously described system for gene targeting of the HPRT locus in the human fibrosarcoma cell line HT1080 (31). We found, unexpectedly, that gene targeting is inhibited by Rad52p overexpression while extrachromosomal HR is stimulated. On this basis we suggest that, in vivo, Rad52p is rate-limiting only for SSA pathways of HR and that Rad51p-dependent strand-invasion pathways of HR can be inhibited by excess Rad52p.
MATERIALS AND METHODS
Plasmids
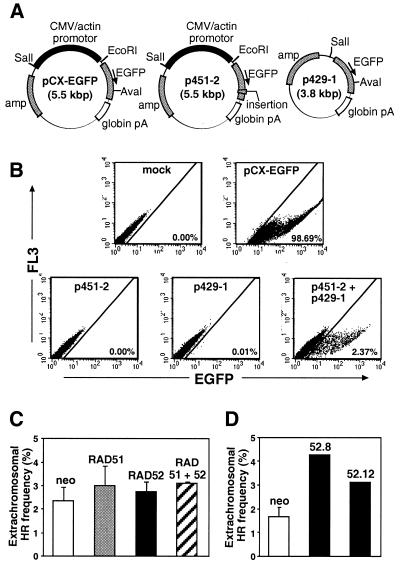
pBSneo, phRAD51neo and pHPRThyg have been previously described (31). phRAD52neo was produced by inserting a CMV-driven hRAD52 cDNA (a kind gift from F. Benson and S. West, ICRF, South Mimms; nucleotides 32–1291 in GenBank entry U27516) with an SV40 terminator in the XbaI site of pBSneo. phRAD51neo52 contains the same hRAD52 expression cassette cloned into a NotI site of phRAD51neo. The plasmids of the extrachromosomal recombination system [pCX-EGFP (47), p451-2 and p429-1] were kindly donated by M. Santibáñez-Koref (MRC Clinical Sciences Centre).
Cell culture and transfection
Human fibrosarcoma HT1080 cells (48) were grown and electroporated with 8 µg (unless stated otherwise) of plasmid DNA as described (31,49). Before electroporation, pBSneo, phRAD52neo and phRAD51neo52 were linearised by digestion with ScaI, phRAD51neo with Asp700I and pHPRThyg with SalI. Selective drugs were added 48 h after transfection and maintained throughout culture.
Polyclonal populations of control and RAD plasmid-transfected cells each contained 230–720 G418r colonies. G418 selection was started at 400 µg/ml and reduced to 200 µg/ml around 10 days later. Given the instability of hRad52p overexpression, all experiments with these polyclonal populations were done a maximum of 19 days after transfection of the RAD constructs, unless otherwise stated in the text. Polyclonal populations of hRAD51neo-transfected cells stably express hRad51p and have been previously described (31).
Stable transfection frequencies of pBSneo, phRAD51neo, phRAD52neo and phRAD51neo52 were measured by plating efficiency in triplicate samples (105 cells in 3.8 cm plates), counting the number of crystal violet-stained colonies after G418 (400 µg/ml) selection. Transfections of pHPRThyg for quantification of random integration and gene targeting were done in four independent polyclonal populations of control or RAD plasmid-transfected cells. For each population, the frequency of random integration of the targeting construct was measured in triplicate samples (each with 105 cells in a 3.8 cm plate), counting colonies after hygromycin (100 µg/ml) selection. Gene-targeting frequencies were measured in sextuplicate samples (each with 2.5 × 106 cells in a 15 cm plate) counting colonies after 3 days of hygromycin selection followed by 7 days in both 6-thioguanine (6-TG; 15 µg/ml) and hygromycin.
Extrachromosomal recombination experiments were done using a calcium phosphate transfection protocol (50). Briefly, HT1080 cells were seeded for either FACS analysis (3.5 × 105 per 9 cm plate) or fluorescence microscopy (5 × 104 on a coverslip in a 3.8 cm plate) and transfected 2 days later. We used equimolar amounts of all plasmids, equivalent to 10 µg of pCX-EGFP per 9 cm plate. Supercoiled pCX-EGFP and p451-2 were used for transfection, but p429-1 was linearised with SalI to reduce the background of enhanced green fluorescent protein (EGFP)-positive cells. The following day the monolayers were washed once with PBS-A and fresh medium was added. Forty-eight hours after transfection samples were processed for either flow cytometry or immunofluorescence. For flow cytometry, cells were trypsinised, resuspended in DMEM and analysed immediately. For immunofluorescence, cells were fixed with paraformaldehyde/methanol (see below) and a Texas red-conjugated secondary antibody (Amersham Biosciences N2034) was used for hRAD52 detection.
Western blots
Immunoblots were essentially as described previously (31); 3 µg of protein were loaded per lane and primary antisera FBE-2 (diluted 1:1000), FBE-3 (1:1000) and Sigma A2066 (1:250) were used to detect hRad51p, hRad52p and actin, respectively. FBE-2 and FBE-3 were kindly donated by F. Benson and S. West (ICRF, South Mimms). A secondary goat anti-rabbit immunoglobulin antiserum conjugated to horseradish peroxidase (DAKO P0448) was used at a dilution of 1:2000. Detection was by enhanced chemical luminescence (Amersham).
Flow cytometry
To study DNA content we stained nuclei with propidium iodide as previously described by Itzhaki et al. (51). Data were acquired in a FACSCalibur using CellQuest software (Becton–Dickinson). DNA content analyses were done with ModFit LT software (Verity Software House).
EGFP analyses were done with recently trypsinised cells resuspended in DMEM. The same results were obtained with paraformaldehyde-fixed cells (data not shown). Data were collected with the FACSCalibur and CellQuest. The proportion of EGFP-positive cells was scored in dot-plots of EGFP fluorescence versus FL3 autofluorescence, using the mock-transfected population to define the negative region. The percentage of extrachromosomal recombination was calculated as: [(% EGFP-positive cells after co-transfection of p451-2 and p429-1) – (% EGFP-positive cells after transfection of p429-1)] × 100 / (% EGFP-positive cells after transfection of positive control pCX-EGFP).
Immunofluorescence
Cells were grown on coverslips in 3.8 cm plates. Several fixation methods were used: paraformaldehyde/methanol (52), paraformaldehyde/Triton X-100 (as for paraformaldehyde/methanol but substituting 0.2% Triton X-100, 10 min at room temperature, for the methanol treatment) or methanol/acetone (53). After fixation, cells were washed four times with PBS-A, blocked with 0.5% BSA in PBS-A (30 min, room temperature), stained with anti-hRAD52 antiserum FBE-3 (1:1000 in BSA/PBS-A, 90 min, 37°C) and washed four times with PBS-A. Anti-rabbit immunoglobulin secondary antibodies were FITC- (Sigma F9887) or Texas red- (Amersham Biosciences N2034) conjugates, diluted 1:200 in BSA/PBS-A, and used for 30 min at room temperature. Samples were counterstained with 0.1 µg/ml DAPI, washed four times with PBS-A and mounted in Vectashield (Vector Laboratories). Samples were viewed on an Axiovert S100TV microscope (Zeiss) and monochrome pictures captured with a CCD camera (Princeton Instruments). False colour was added with Photoshop software (Adobe).
The percentage of cells overexpressing Rad52p was estimated by scoring more than 500 DAPI-stained cells for Rad52p staining. The percentage of EGFP-positive cells co-expressing Rad52p was estimated by scoring 100 EGFP-positive cells for Rad52p fluorescence. The subcellular distribution of Rad52p signal was scored in two to four samples of 100 hRad52p-positive cells each.
Statistical analyses
Analyses of variance and Student t-tests were carried out using Microsoft Excel software.
RESULTS
Overexpression of Rad52p (and to a lesser extent Rad51p) is harmful
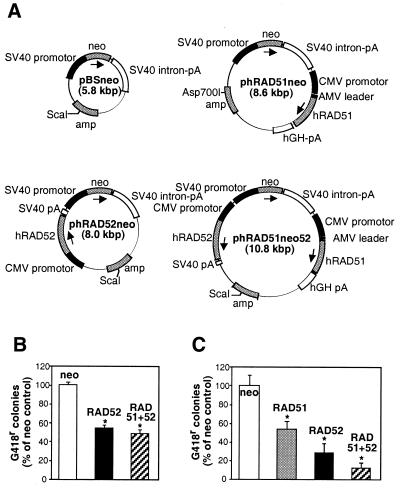
The plasmids phRAD51neo, for overexpression of Rad51p, and pBSneo, a vector control, have been previously described by Yáñez and Porter (31). We made two new plasmids, phRAD52neo and phRAD51neo52, for the overexpression of Rad52p alone or together with Rad51p, respectively. All four plasmids (Fig. 1A) carry the same neo cassette conferring resistance to the antibiotic G418. When equal weights (8 µg) of pBSneo, phRAD52neo or phRAD51neo52 were electroporated into HT1080 cells, G418r colonies were obtained from the latter two plasmids at ∼50% of the control (pBSneo) frequency (Fig. 1B). When equal molarities (equivalent to 8 µg of pBSneo) of these three plasmids or phRAD51neo were used, however, a 50% reduction in the control stable transfection frequency was observed for phRAD51neo, and the reductions observed for phRAD52neo and phRAD51neo52 became more marked (Fig. 1C). These data suggest that both Rad52p and Rad51p overexpression can compromise cell viability, that Rad52p has a more pronounced effect than Rad51p and that the two effects are synergistic.
Figure 1.
Stable transfection of Rad51p- and Rad52p-expressing plasmids in HT1080 cells. (A) Plasmid design. Shown are the control plasmid (pBSneo) and plasmids for expression of Rad51p (phRAD51neo), Rad52p (phRAD52neo) or both (phRAD51neo52). Promotors are shown as black boxes, open reading frames as stippled boxes and terminators as white boxes. The ScaI or Asp700I restriction sites used for linearisation before transfection are indicated. (B) Stable transfection frequency with equal amounts (8 µg) of pBSneo (open), phRAD52neo (black) and phRAD51neo52 (striped). The frequency for pBSneo is taken as 100%. (C) Stable transfection frequency with equimolar amounts (equivalent to 8 µg of pBSneo) of pBSneo (white), phRAD51neo (stippled), phRAD52neo (black) and phRAD51neo52 (striped). The frequency for pBSneo is taken as 100%. Asterisks indicate significant differences in Student t-tests compared with the control (P ≤ 0.002; n = 3–6).
Overexpression of Rad52p is unstable
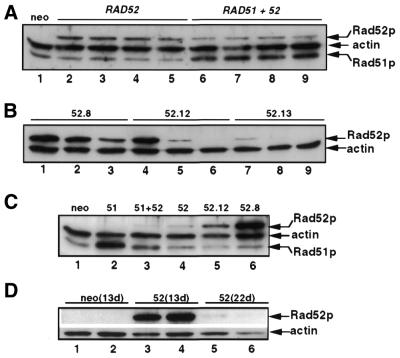
Individual clones or pools (>230 colonies) of G418r transfectants were expanded for further analyses. Western blots were used to study the overexpression patterns. Rad52p was undetectable in pBSneo-transfected cells but readily detected in phRAD52- and phRAD51neo52-transfected pools (Fig. 2A) and clones (Fig. 2B). Rad52p overexpression varied little between pools but greatly between clones, with some clones (e.g. clone 52.8) expressing much higher levels than the level detected in pools, which can be considered as the average level of overexpression in clones. Endogenous Rad51p is appreciable, but overexpression of Rad51p was detectable in pools of phRAD51neo52 transfectants (Fig. 2A), albeit at reduced levels compared with those in pools of phRAD51-transfectants (Fig. 2C) (31).
Figure 2.
Western blot analyses of Rad51p and Rad52p overexpression. (A) Comparison of independent polyclonal populations of HT1080 cells stably transfected with pBSneo (lane 1), phRAD52neo (lanes 2–5) and phRAD51neo52 (lanes 6–9). Cell lysates were made 14 days after transfection and were from cultures used for gene-targeting assays. (B) Rad52p levels in phRAD52neo-transfected clones 52.8 (lanes 1–3), 52.12 (lanes 4–6) and 52.13 (lanes 7–9) sampled 3.5 (lanes 1, 4 and 7), 8 (lanes 2, 5 and 8) and 12 (lanes 3, 6 and 9) weeks after transfection. (C) Analysis of polyclonal HT1080 populations transfected with pBSneo (lane 1), phRAD51neo (lane 2), phRAD51neo52 (lane 3) and phRAD52neo (lane 4), and of phRAD52neo-transfected clones 52.12 (lane 5) and 52.8 (lane 6). Samples were from cultures used for measuring extrachromosomal HR. (D) Loss of Rad52p overexpression in polyclonal populations. Two independent pools of pBSneo- (lanes 1–2) and phRAD52neo-transfectants (lanes 3–4) were sampled 13 days after transfection, and the latter two were re-sampled 9 days later (lanes 5–6).
Analysis of the Rad52p signal with increased passage number revealed that Rad52p overexpression is unstable in all samples tested, whether pools (Fig. 2D) or clones (Fig. 2B), becoming undetectable in some cases. This was so even though G418 selection was maintained at all times. Loss of expression in pools may indicate that clones overexpressing little or no Rad52p multiply faster than clones expressing larger amounts. The loss of Rad52p from clones suggests that a mechanism for silencing exogenous RAD52 genes is also at work. Silencing of the CMV promoter has been documented (54) and may be involved but cannot alone explain the loss of RAD52 expression because CMV promoter-driven RAD51 overexpression is stable in phRAD51neo-transfected HT1080 cells (31). Therefore, a combination of transgene silencing and impaired cellular proliferation in cells expressing high levels of Rad52p appears to explain the instability of Rad52p overexpression.
The cell cycle in Rad52p overexpressing cells is disturbed
To test the possibility that a negative effect of Rad52p overexpression on cellular proliferation might affect a particular phase of the cell cycle, we analysed DNA content by flow cytometry. We observed a significant increase (15%, P < 0.03; n = 4) in the proportion of diploid G1 cells in populations of phRAD52neo transfectants compared with control pBSneo transfectants, but not in phRAD51neo52 transfectants (data not shown). This increase was offset by smaller decreases in the other phases of the cell cycle. In principle, this redistribution could be caused by a delayed exit from G1, an accelerated transit through S/G2, or both. Given the apparent growth disadvantage of Rad52p overexpressing cells, delayed exit from G1 is likely to be the predominant cause.
Rad52p overexpression is detectable in a minority of cells and is mostly nuclear
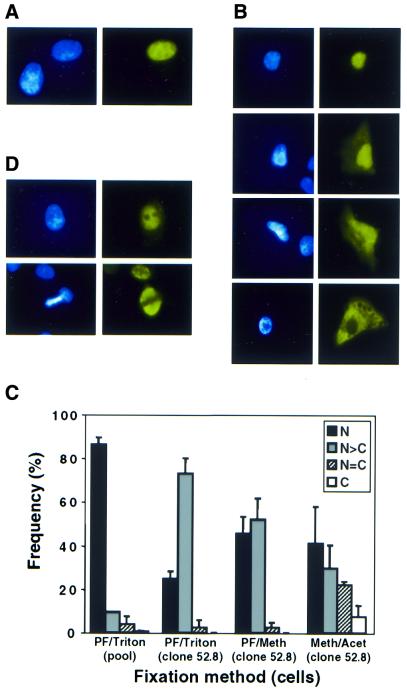
The distribution of Rad52p in cells transfected with phRAD52neo was studied by immunofluorescence (Fig. 3). Only a minority of cells showed staining above the endogenous background levels observed in untransfected cells. This minority ranged from ∼9% of cells, in pools of phRAD52neo transfectants, to 37% of cells in the clonal phRAD52neo transfectant (clone 52.8) expressing highest levels of Rad52p. Typical cells staining positively and negatively for Rad52p overexpression are shown in Figure 3A. Examples of the four classes of subcellular Rad52p distribution observed are shown in Figure 3B and their frequencies are summarised in Figure 3C. Staining was predominantly nuclear whether cells were fixed with paraformaldehyde/Triton X-100, paraformaldehyde/methanol or methanol/acetone. In pools of phRAD52neo transfectants, Rad52p overexpression was confined to the nucleus in the great majority (86%) of positively staining cells. When clone 52.8 was analysed in the same way, the staining pattern was still mostly nuclear, but with a greater proportion of cells in which cytoplasmic staining could also be detected. This might indicate that the Rad52p nuclear localisation mechanism becomes saturated when overexpression is high. When paraformaldehyde/saponin was use for fixation/permeabilisation the majority of Rad52p overexpression was excluded from the nucleus (not shown); the significance of this observation is unclear.
Figure 3.
Immunofluorescence analyses of cells overexpressing Rad52p. (A) Heterogeneity of Rad52p staining in a polyclonal population of hRAD52neo-transfected cells, 19 days after electroporation. Shown are a highly expressing cell and another cell in which Rad52p is undetectable (as it is in control cultures). (B) Classes of sub-cellular localisation of exogenous Rad52p in a polyclonal population, 19 days post-transfection. From top to bottom: nucleus-only signal, nucleus more intense than cytoplasm, nucleus and cytoplasm equally intense, cytoplasm-only signal. (C) Frequency of classes of Rad52p sub-cellular localisation using the indicated fixation methods. Black bars, nucleus-only signal; grey bars, nucleus > cytoplasm; striped bars, nucleus = cytoplasm; open bars, cytoplasm only. Two independent polyclonal pools were scored 19 days post-transfection; clone 52.8 was 8 weeks after transfection. (D) Exclusion of Rad52p from nucleoli (top panels) and mitotic chromosomes (bottom panels). Pictures are of clone 52.8, 8 weeks after transfection. In (A), (B) and (D), left- and right-hand panels show DAPI staining and Rad52p signal, respectively.
It has been previously reported that endogenous Rad52p in mouse cells has a dynamic localisation, being evenly distributed throughout the nucleus but excluded from nucleoli in the G1 phase of the cell cycle, restricted to the nucleoli in S-phase and spread throughout the cell in mitosis (52). In asynchronous HT1080 cells we observed patterns of overexpressed Rad52p consistent with nucleolar exclusion in G1 (Fig. 3D), but could find no evidence for a nucleolar-specific S-phase signal, even though we have analysed over 2000 cells of which ∼33% are expected to be in S-phase. We have also observed that Rad52p is excluded from mitotic chromosomes in metaphase (Fig. 3D) and anaphase (not shown).
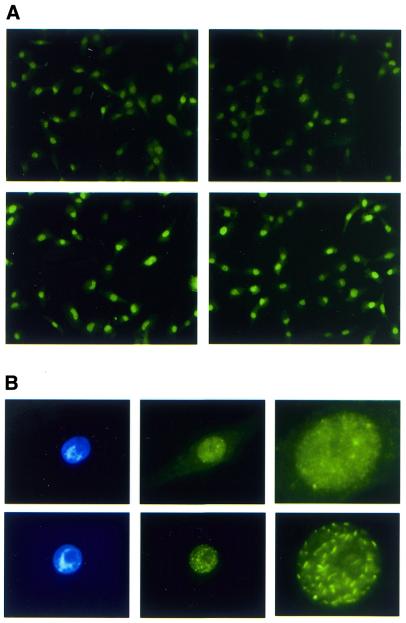
Rad51p is overexpressed in most cells and forms normal or elongated nuclear foci
It seemed likely that the small proportion of cells overexpressing Rad52p was a reflection of the harmful and unstable nature of Rad52p overexpression. To explore this further, we asked whether Rad51p overexpression, which appears to be less harmful and more stable than Rad52p overexpression, could be detected in a greater proportion of cells. We found that, indeed, the majority of the cells in a pool of phRAD51neo transfectants had immunofluorescence signals higher than the background signal due to endogenous Rad51p (Fig. 4A). Furthermore, of 23 phRAD51neo-transfected clones, 19 (83%) showed significant Rad51p overexpression in western blots (not shown). Therefore, the ∼4-fold increase in Rad51p expression detected by western blots in phRAD51neo-transfected pools (Fig. 2C) (31) represents Rad51p overexpression in the large majority of cells. The distribution of the Rad51p signal was predominantly nuclear although some accompanying cytoplasmic signal was sometimes detected (Fig. 4B). We observed nuclear foci similar to those described previously (55) for endogenous Rad51p in S-phase cells (Fig. 4B). However, in the cells expressing the highest amounts of Rad51p the foci were more numerous and pronounced and had an unusually elongated appearance (Fig. 4B). To our knowledge this pattern of nuclear staining has not been described before and its significance at this stage is unclear.
Figure 4.
Immunofluorescence for Rad51p detection. Cells were fixed with paraformaldehyde and permeabilised with Triton X-100. (A) Most cells transfected with phRAD51neo overexpress Rad51p. Shown are two fields from polyclonal populations transfected with pBSneo (top) or phRAD51neo (bottom). (B) Nuclear foci in Rad51p-overexpressing cells. Top, a cell expressing moderate levels of Rad51p has normal S-phase-type nuclear foci. Bottom, a cell with extremely high levels of Rad51p has elongated nuclear foci. Left, DAPI staining; middle, Rad51p signal; right, higher magnification of middle panels. Exposures for Rad51p detection in bottom panels were 10% of those for top panels.
Gene targeting is inhibited by Rad52p (but enhanced by Rad51p) overexpression
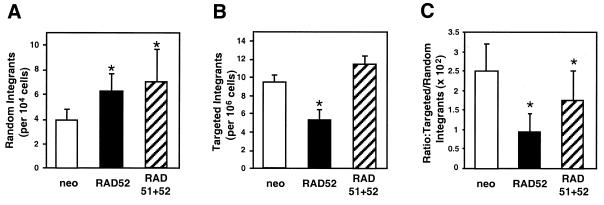
To test the effect of Rad52p overexpression on gene targeting we used our previously described targeting construct pHPRThyg (31). Random integration of this replacement-type targeting construct leads to hygromycin-resistance whereas targeted integration disrupts exon 2 of the X-chromosomal HPRT gene with the hygromycin-resistance cassette, generating colonies resistant to both hygromycin and 6-TG. Therefore, the frequencies of random and targeted integration can be estimated from the frequencies of colonies surviving single (hygromycin) and double (hygromycin and 6-TG) drug selections, respectively.
Four independent pools of pBSneo, phRAD52neo or phRAD51neo52 transfectants were electroporated with pHPRThyg and selected in the appropriate drug(s). The amount of Rad52p expressed in the populations at the time of electroporation was assessed by western blot (Fig. 2A) and showed very consistent levels of Rad52p overexpression. The entire experiment was repeated and the combined results of the experiments are summarised in Figure 5. The frequency of gene targeting was significantly less in phRAD52neo-transfected cells than in pBSneo transfectant controls. The decrease was measurable either as a 44% reduction in the absolute targeting frequency or as a 62% decrease in the targeting frequency relative to random integration. This inhibition of gene targeting following Rad52p overexpression is in marked contrast to the 2-fold stimulation that was observed in the same system following Rad51p overexpression (31). When Rad51p and Rad52p were simultaneously overexpressed (in phRAD51neo52 transfectants) the decrease in absolute gene-targeting frequency was not observed, and the decrease in gene targeting relative to random integration was less pronounced.
Figure 5.
Frequencies of gene targeting and random integration in HT1080 cells overexpressing Rad52p alone or together with Rad51p. Polyclonal transfectants of pBSneo (open bars), phRAD52neo (black bars) and phRAD51neo52 (striped bars) were transfected with the gene-targeting plasmid pHPRThyg and random integration and gene-targeting frequencies scored as described in Materials and Methods. Shown are plots of the frequencies of random integration (A), gene targeting (B) and the ratio of gene targeting to random integration (C). Asterisks indicate significant differences to control (pBSneo) values in an ANOVA analysis (P ≤ 0.04; n = 8).
We carried out equivalent experiments on pools of phRAD52neo-transfected cells that had been passaged to a point where Rad52p overexpression was barely detectable (22 days after transfection, including one freeze–thaw cycle; Fig. 2D). In this situation, no significant difference in gene targeting relative to pBSneo-transfected pools was detected (0.99 × 10–5 targeted cells per electroporated cell in pBSneo-transfected pools versus 1.1 × 10–5 in phRAD52neo-transfected cells; n = 4).
Extrachromosomal HR is enhanced by Rad52p (but not by Rad51p) overexpression
The decrease in gene targeting we observed following Rad52p overexpression was in marked contrast to the stimulation in extrachromosomal and intrachromosomal HR described in previous studies (45,46). We were interested to know whether this reflected a genuine difference in the two forms of HR or a difference in the experimental systems used. Therefore, we decided to measure extrachromosomal HR in our Rad52p overexpressing cells. For this we used two plasmids, p451-2 and p429-1, carrying defective EGFP expression cassettes (Fig. 6A). These plasmids are derived from pCX-EGFP, which encodes a functional EGFP. p451-2 contains an EGFP-inactivating 25 bp insertion close to the 3′ end of the gene, whereas p429-1 has a deletion of 1.7 kb at the 5′ end of the cassette and therefore lacks a promotor. Transient transfection of HT1080 cells with pCX-EGFP leads to EGFP expression in 55–100% of cells. After transfection of HT1080 cells with either p451-2 or p429-1, essentially no cells became EGFP-positive whereas 2–3% became positive after co-transfection of p451-2 and p429-1 (Fig. 6B). Thus extrachromosomal HR between p451-2 and p429-1 can generate a functional EGFP cassette and cells in which this has happened are readily detectable by flow cytometry (and fluorescence microscopy).
Figure 6.
Extrachromosomal HR assays. (A) Assay plasmids. pCX-EGFP expresses EGFP constitutively. p451-2 and p429-1 are mutant derivatives of pCX-EGFP that individually fail to express active EGFP but which, when co-transfected, can undergo intermolecular extrachromosomal HR to generate a functional EGFP gene. (B) Detection of extrachromosomal HR by FACS. HT1080 cells were mock-transfected or transfected with the indicated plasmids. Forty-eight hours later the cells were trypsinised and analysed by flow cytometry. The percentage of EGFP-positive cells (scored as described in Materials and Methods) is indicated. The data shown are from a polyclonal population of pBSneo-transfected cells. (C) Plots of extrachromosomal HR frequencies for polyclonal pools of pBSneo- (open), phRAD51neo- (stippled), phRAD52neo- (black) and phRAD51neo52- (striped) transfected cells (n = 4). (D) Plots of extrachromosomal HR frequencies for clones of pBSneo- (open, n = 2) and phRAD52neo- (black) transfected cells.
Plasmids p451-2 and p429-1 were transiently transfected into polyclonal pools of cells stably transfected with pBSneo, phRAD51neo, phRAD52neo or phRAD51neo52, and into clones 52.8 and 52.12. Expected levels of Rad51p and/or Rad52p overexpression were confirmed at the time of transfection by western blot (Fig. 2C). Flow cytometric analyses 48 h after transfection showed small increases (statistically not significant) in the frequency of EGFP-positive cells in the latter three pools relative to the former (Fig. 6C). Because Rad51p was overexpressed in most cells (see above), this result indicated that Rad51p overexpression had little or no effect on extrachromosomal HR. On the other hand, Rad52p overexpression was confined to only a fraction of the cells (see above), and it was possible that a significant increase in extrachromosomal HR in this subset of cells was masked by the much larger fraction of cells failing to overexpress Rad52p. In keeping with this, we observed considerably larger increases in extrachromosomal HR (up to 2.5-fold over control values) in clones 52.8 and 52.12 overexpressing the highest levels of Rad52p (Fig. 6D and see western blot in Fig. 2C).
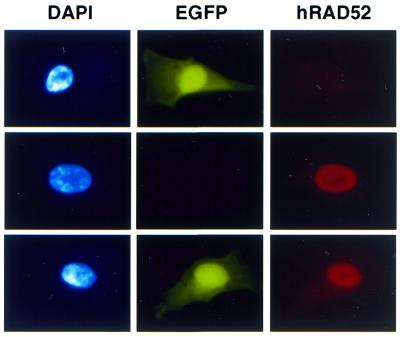
We used extrachromosomal HR experiments combined with immunofluorescence to test whether we could detect increased extrachromosomal HR in the minority of cells overexpressing Rad52p in phRAD52neo-transfected pools. We observed that the frequency of Rad52p-positive cells amongst EGFP-positive cells (31.8 ± 1.9%, n = 4) was significantly higher than the frequency of Rad52p-positive cells amongst the population of phRAD52neo-transfected cells as a whole (17 ± 1.6%, n = 4). This enrichment for Rad52p expression in the EGFP-positive population represents a significant (P < 0.0005) 1.9-fold stimulation of extrachromosomal HR in Rad52p-overexpressing cells. Examples of cells scored in this analysis are shown in Figure 7.
Figure 7.
Detection of extrachromosomal HR by immunofluorescence. A population of Rad52p overexpressing cells was co-transfected with the defective plasmids p451-2 and p429-1. Cells were fixed 48 h later and stained with DAPI and the antibody against hRAD52. Examples of cells positive for EGFP (top), Rad52p (middle) or both (bottom) are shown.
DISCUSSION
The aim of this study was to determine the consequences of Rad52p overexpression in human cells with particular reference to gene targeting. On the basis of previous studies showing that Rad52p overexpression can stimulate other forms of HR (45,46), that Rad51p overexpression can stimulate gene targeting (31) and that Rad52p stimulates Rad51p activity in vitro (10,11), it seemed reasonable to suppose that Rad52p overexpression might stimulate gene targeting, especially if combined with Rad51p overexpression. In the event, Rad52p overexpression caused an ∼2-fold inhibition of gene targeting. The inhibition of gene targeting by Rad52p overexpression was similar in magnitude to the stimulation caused by Rad51p overexpression (31). While these opposing effects were small and observed in separate studies, they were shown to be highly reproducible and statistically significant. Furthermore, the antagonistic effects on gene targeting of Rad51p and Rad52p overexpression were confirmed by our observation that the combined overexpression of both proteins had a negligible effect on gene targeting. Despite its negative effect on gene targeting, we were able to confirm that in our system, as in another (45), Rad52p overexpression stimulated extrachromosomal HR.
Although somewhat unexpected, these observations can be explained in terms of differences in the mechanisms of gene targeting and extrachromosomal HR. Gene targeting is thought to occur by a conservative strand-invasion type mechanism and therefore to be completely dependent on RAD51 (see Introduction). It is conceivable that unnaturally high levels of Rad52p, far from promoting gene targeting, actually inhibit it, perhaps by sequestering Rad51p away from the active complex of proteins required for strand-invasion type HR mechanisms. On the other hand, it has long been argued that extrachromosomal HR in mammalian cells occurs predominantly by a non-conservative mechanism, probably SSA (32,33). Furthermore, studies in yeast suggest that RAD52 is the only RAD52 epistasis group gene required for SSA (21,22). Therefore, it is possible that Rad52p is rate-limiting for SSA and that Rad52p overexpression can stimulate extrachromosomal HR despite its negative effect on gene targeting. If this model is correct, previously reported (45,46) increases in extrachromosomal HR, intrachromosomal HR and radiation resistance following Rad52p overexpression may also reflect a stimulation of SSA pathways rather than strand-invasion pathways of HR.
It remains to be explained how Rad52p overexpression in only a minority (∼10% as judged by immunofluorescence) of cells could cause a 2-fold reduction in the overall gene-targeting frequency, since in theory a maximum reduction of only 10% is predicted. It is possible that Rad52p expression is above background in the majority of cells but that only in a small fraction is there sufficient to be detected by immunofluorescence. In this case, one would have to suggest that Rad52p overexpression has to be high to have a stimulatory effect on extrachromosomal HR but that only a small amount is sufficient to have an inhibitory effect on gene targeting.
In contrast to the modest (2-fold) stimulation of extrachromosomal HR we detected after overexpression of human Rad52p in HT1080 cells, a previous study (45) described a 12-fold stimulation of extrachromosomal HR, also in HT1080 cells, after overexpression of S.cerevisiae Rad52p. It is possible that overexpression of the yeast protein is less harmful to human cells than overexpression of the human protein and that higher levels of overexpression were therefore achieved. Alternatively, yeast Rad52p may be more effective than the human protein in promoting SSA.
In addition to its negative effects on gene targeting we report that Rad52p overexpression can have adverse effects on cell viability or proliferation. An initial indication of this was the reduced efficiency of G418r colony formation after transfection with phRAD52neo compared with pBSneo (Fig. 1B and C). This negative effect on drug resistant colony formation is not maintained in G418r Rad52p-overexpressing cells which actually form hygror colonies more efficiently than controls after transfection with pHPRThyg (Fig. 5A). This apparent contradiction can be explained if the negative effect on colony formation requires particularly high levels of Rad52p overexpression that can only be achieved transiently. G418r Rad52p-overexpressing cells have been selected for their ability to form drug-resistant colonies, and the remaining exogenous Rad52p may be sufficient to promote transfection by binding to and stabilising the ends of transfected DNA. Though more tolerable, these levels of exogenous Rad52p are nevertheless unstable, presumably because their tendency to cause an accumulation in G1 places host cells at a proliferative disadvantage. That Rad51p overexpression exacerbates the inhibition of colony formation by Rad52p overexpression (Fig. 1C) but reverses the effect of Rad52p overexpression on cell-cycle distribution (not shown) suggests that these two effects occur via different mechanisms. If Rad52p, like Rad51p, interacts with a variety of cellular proteins, it would perhaps be unsurprising if the effects of exogenous Rad52p were to depend on the degree of overexpression.
Could it be that the effects of Rad52p overexpression on gene targeting and on cell-cycle distribution are related? Certainly both are reversed by co-overexpression of Rad51p. Furthermore, it is thought that DSB repair by HR, and therefore presumably gene targeting, is associated with S-phase (41,42), suggesting that the negative effect on gene targeting might be indirect, resulting from a primary effect on cell-cycle distribution. However, the 15% increase in the proportion of G1 cells appears to be too small to account for the 2-fold inhibition of gene targeting. More likely, perhaps, is the possibility that both effects, the inhibition of gene targeting and redistribution of the cell cycle, are a consequence of the same primary effect. As suggested above, the primary affect could be an inhibition of strand-invasion mechanisms of HR, although why this should cause an accumulation in G1, even in cells (including HT1080) with normal p53, is not clear.
Whatever the mechanism of the inhibition of gene targeting by Rad52p overexpression, the results presented here do not support the idea of Rad52p overexpression as a means of promoting gene targeting. Assuming the Ku heterodimer was unaffected by our procedures, our results also suggest that the ratio of Rad52p to Ku heterodimer is not a simple determinant of the ratio of gene targeting to random integration.
Acknowledgments
ACKNOWLEDGEMENTS
We are most grateful to Fiona Benson and Steve West for supplying antibodies and RAD52 cDNA and to Mauro Santibáñez-Koref for the extrachromosomal HR plasmids.
REFERENCES
- 1.Kanaar R., Hoeijmakers,J.H. and van Gent,D.C. (1998) Molecular mechanisms of DNA double strand break repair. Trends Cell Biol., 8, 483–489. [DOI] [PubMed] [Google Scholar]
- 2.Pastink A. and Lohman,P.H. (1999) Repair and consequences of double-strand breaks in DNA. Mutat. Res., 428, 141–156. [DOI] [PubMed] [Google Scholar]
- 3.Paques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shinohara A., Ogawa,H., Matsuda,Y., Ushio,N., Ikeo,K. and Ogawa,T. (1993) Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nat. Genet., 4, 239–243. [DOI] [PubMed] [Google Scholar]
- 5.Muris D.F., Bezzubova,O., Buerstedde,J.M., Vreeken,K., Balajee,A.S., Osgood,C.J., Troelstra,C., Hoeijmakers,J.H., Ostermann,K., Schmidt,H. et al. (1994) Cloning of human and mouse genes homologous to RAD52, a yeast gene involved in DNA repair and recombination. Mutat. Res., 315, 295–305. [DOI] [PubMed] [Google Scholar]
- 6.Kanaar R., Troelstra,C., Swagemakers,S.M., Essers,J., Smit,B., Franssen,J.H., Pastink,A., Bezzubova,O.Y., Buerstedde,J.M., Clever,B., Heyer,W.D. and Hoeijmakers,J.H. (1996) Human and mouse homologs of the Saccharomyces cerevisiae RAD54 DNA repair gene: evidence for functional conservation. Curr. Biol., 6, 828–838. [DOI] [PubMed] [Google Scholar]
- 7.Bezzubova O.Y., Schmidt,H., Ostermann,K., Heyer,W.D. and Buerstedde,J.M. (1993) Identification of a chicken RAD52 homologue suggests conservation of the RAD52 recombination pathway throughout the evolution of higher eukaryotes. Nucleic Acids Res., 21, 5945–5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumann P. and West,S.C. (1998) Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem. Sci., 23, 247–251. [DOI] [PubMed] [Google Scholar]
- 9.McIlwraith M.J., Van Dyck,E., Masson,J.Y., Stasiak,A.Z., Stasiak,A. and West,S.C. (2000) Reconstitution of the strand invasion step of double-strand break repair using human Rad51 Rad52 and RPA proteins. J. Mol. Biol., 304, 151–164. [DOI] [PubMed] [Google Scholar]
- 10.Benson F.E., Baumann,P. and West,S.C. (1998) Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature, 391, 401–404. [DOI] [PubMed] [Google Scholar]
- 11.Shinohara A. and Ogawa,T. (1998) Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature, 391, 404–407. [DOI] [PubMed] [Google Scholar]
- 12.Shen Z., Cloud,K.G., Chen,D.J. and Park,M.S. (1996) Specific interactions between the human RAD51 and RAD52 proteins. J. Biol. Chem., 271, 148–152. [DOI] [PubMed] [Google Scholar]
- 13.Shinohara A. and Ogawa,T. (1999) Rad51/RecA protein families and the associated proteins in eukaryotes. Mutat. Res., 435, 13–21. [DOI] [PubMed] [Google Scholar]
- 14.Van Dyck E., Stasiak,A.Z., Stasiak,A. and West,S.C. (1999) Binding of double-strand breaks in DNA by human Rad52 protein. Nature, 398, 728–731. [DOI] [PubMed] [Google Scholar]
- 15.Parsons C.A., Baumann,P., Van Dyck,E. and West,S.C. (2000) Precise binding of single-stranded DNA termini by human RAD52 protein. EMBO J., 19, 4175–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stasiak A.Z., Larquet,E., Stasiak,A., Muller,S., Engel,A., Van Dyck,E., West,S.C. and Egelman,E.H. (2000) The human Rad52 protein exists as a heptameric ring. Curr. Biol., 10, 337–340. [DOI] [PubMed] [Google Scholar]
- 17.Shinohara A., Shinohara,M., Ohta,T., Matsuda,S. and Ogawa,T. (1998) Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells, 3, 145–156. [DOI] [PubMed] [Google Scholar]
- 18.Mortensen U.H., Bendixen,C., Sunjevaric,I. and Rothstein,R. (1996) DNA strand annealing is promoted by the yeast Rad52 protein. Proc. Natl Acad. Sci. USA, 93, 10729–10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reddy G., Golub,E.I. and Radding,C.M. (1997) Human Rad52 protein promotes single-strand DNA annealing followed by branch migration. Mutat. Res., 377, 53–59. [DOI] [PubMed] [Google Scholar]
- 20.Sugiyama T., New,J.H. and Kowalczykowski,S.C. (1998) DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl Acad. Sci. USA, 95, 6049–6054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugawara N. and Haber,J.E. (1992) Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol. Cell. Biol., 12, 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanov E.L., Sugawara,N., Fishman-Lobell,J. and Haber,J.E. (1996) Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics, 142, 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Critchlow S.E. and Jackson,S.P. (1998) DNA end-joining: from yeast to man. Trends Biochem. Sci., 23, 394–398. [DOI] [PubMed] [Google Scholar]
- 24.Bollag R.J., Waldman,A.S. and Liskay,R.M. (1989) Homologous recombination in mammalian cells. Annu. Rev. Genet., 23, 199–225. [DOI] [PubMed] [Google Scholar]
- 25.Vasquez K.M., Marburger,K., Intody,Z. and Wilson,J.H. (2001) Manipulating the mammalian genome by homologous recombination. Proc. Natl Acad. Sci. USA, 98, 8403–8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller U. (1999) Ten years of gene targeting: targeted mouse mutants, from vector design to phenotype analysis. Mech. Dev., 82, 3–21. [DOI] [PubMed] [Google Scholar]
- 27.Yáñez R.J. and Porter,A.C.G. (1998) Therapeutic gene targeting. Gene Ther., 5, 149–159. [DOI] [PubMed] [Google Scholar]
- 28.Pennington S.L. and Wilson,J.H. (1991) Gene targeting in Chinese hamster ovary cells is conservative. Proc. Natl Acad. Sci. USA, 88, 9498–9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bezzubova O., Silbergleit,A., Yamaguchi-Iwai,Y., Takeda,S. and Buerstedde,J.-M. (1997) Reduced X-ray resistance and homologous recombination frequencies in a RAD54–/– mutant of the chicken DT40 cell line. Cell, 89, 185–193. [DOI] [PubMed] [Google Scholar]
- 30.Essers J., Hendriks,R.W., Swagemakers,S.M., Troelstra,C., de Wit,J., Bootsma,D., Hoeijmakers,J.H. and Kanaar,R. (1997) Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell, 89, 195–204. [DOI] [PubMed] [Google Scholar]
- 31.Yáñez R.J. and Porter,A.C.G. (1999) Gene targeting is enhanced in human cells overexpressing hRAD51. Gene Ther., 6, 1282–1290. [DOI] [PubMed] [Google Scholar]
- 32.Lin F.L., Sperle,K. and Sternberg,N. (1984) Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol. Cell. Biol., 4, 1020–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seidman M.M. (1987) Intermolecular homologous recombination between transfected sequences in mammalian cells is primarily nonconservative. Mol. Cell. Biol., 7, 3561–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fishman-Lobell J., Rudin,N. and Haber,J.E. (1992) Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol. Cell. Biol., 12, 1292–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang F., Han,M., Romanienko,P.J. and Jasin,M. (1998) Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc. Natl Acad. Sci. USA, 95, 5172–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lambert S. and Lopez,B.S. (2000) Characterization of mammalian RAD51 double strand break repair using non-lethal dominant-negative forms. EMBO J., 19, 3090–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonoda E., Takata,M., Yamashita,Y.M., Morrison,C. and Takeda,S. (2001) Homologous DNA recombination in vertebrate cells. Proc. Natl Acad. Sci. USA, 98, 8388–8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim D.S. and Hasty,P. (1996) A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol., 16, 7133–7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsuzuki T., Fujii,Y., Sakumi,K., Tominaga,Y., Nakao,K., Sekiguchi,M., Matsushiro,A., Yoshimura,Y. and Morita,T. (1996) Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl Acad. Sci. USA, 93, 6236–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sonoda E., Sasaki,M.S., Buerstedde,J.M., Bezzubova,O., Shinohara,A., Ogawa,H., Takata,M., Yamaguchi-Iwai,Y. and Takeda,S. (1998) Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J., 17, 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marians K.J. (2000) Replication and recombination intersect. Curr. Opin. Genet. Dev., 10, 151–156. [DOI] [PubMed] [Google Scholar]
- 42.Sonoda E., Morrison,C., Yamashita,Y.M., Takata,M. and Takeda,S. (2001) Reverse genetic studies of homologous DNA recombination using the chicken B-lymphocyte line, DT40. Philos. Trans. R. Soc. Lond. B. Biol. Sci., 356, 111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamaguchi-Iwai Y., Sonoda,E., Buerstedde,J.M., Bezzubova,O., Morrison,C., Takata,M., Shinohara,A. and Takeda,S. (1998) Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol. Cell. Biol., 18, 6430–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rijkers T., Van Den Ouweland,J., Morolli,B., Rolink,A.G., Baarends,W.M., Van Sloun,P.P., Lohman,P.H. and Pastink,A. (1998) Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol., 18, 6423–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johnson B.L., Thyagarajan,B., Krueger,L., Hirsch,B. and Campbell,C. (1996) Elevated levels of recombinational DNA repair in human somatic cells expressing the Saccharomyces cerevisiae RAD52 gene. Mutat. Res., 363, 179–189. [DOI] [PubMed] [Google Scholar]
- 46.Park M.S. (1995) Expression of human RAD52 confers resistance to ionizing radiation in mammalian cells. J. Biol. Chem., 270, 15467–15470. [DOI] [PubMed] [Google Scholar]
- 47.Ikawa M., Kominami,K., Yoshimura,Y., Tanaka,K., Nishimune,Y. and Okabe,M. (1995) A rapid and non-invasive selection of transgenic embryos before implantation using green fluorescent protein (GFP). FEBS Lett., 375, 125–128. [DOI] [PubMed] [Google Scholar]
- 48.Rasheed S., Nelson Rees,W.A., Toth,E.M., Arnstein,P. and Gardner,M.B. (1974) Characterization of a newly derived human sarcoma cell line (HT-1080). Cancer, 33, 1027–1033. [DOI] [PubMed] [Google Scholar]
- 49.Porter A.C.G. and Itzhaki,J.E. (1993) Gene targeting in human somatic cells. Complete inactivation of an interferon-inducible gene. Eur. J. Biochem., 218, 273–281. [DOI] [PubMed] [Google Scholar]
- 50.Dull T., Zufferey,R., Kelly,M., Mandel,R.J., Nguyen,M., Trono,D. and Naldini,L. (1998) A third-generation lentivirus vector with a conditional packaging system. J. Virol., 72, 8463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Itzhaki J.E., Gilbert,C.S. and Porter,A.C.G. (1997) Construction by gene targeting in human cells of a ‘conditional’ CDC2 mutant that rereplicates its DNA. Nat. Genet., 15, 258–265. [DOI] [PubMed] [Google Scholar]
- 52.Liu Y., Li,M., Lee,E.Y. and Maizels,N. (1999) Localization and dynamic relocalization of mammalian Rad52 during the cell cycle and in response to DNA damage. Curr. Biol., 9, 975–978. [DOI] [PubMed] [Google Scholar]
- 53.Haaf T., Golub,E.I., Reddy,G., Radding,C.M. and Ward,D.C. (1995) Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl Acad. Sci. USA, 92, 2298–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gerolami R., Uch,R., Jordier,F., Chapel,S., Bagnis,C., Brechot,C. and Mannoni,P. (2000) Gene transfer to hepatocellular carcinoma: transduction efficacy and transgene expression kinetics by using retroviral and lentiviral vectors. Cancer Gene Ther., 7, 1286–1292. [DOI] [PubMed] [Google Scholar]
- 55.Tashiro S., Kotomura,N., Shinohara,A., Tanaka,K., Ueda,K. and Kamada,N. (1996) S phase specific formation of the human Rad51 protein nuclear foci in lymphocytes. Oncogene, 12, 2165–2170. [PubMed] [Google Scholar]