Abstract
The brain's functional architecture and organization undergo continual development and modification throughout adolescence. While it is well known that multiple factors govern brain maturation, the constantly evolving patterns of time‐resolved functional connectivity are still unclear and understudied. We systematically evaluated over 47,000 youth and adult brains to bridge this gap, highlighting replicable time‐resolved developmental and aging functional brain patterns. The largest difference between the two life stages was captured in a brain state that indicated coherent strengthening and modularization of functional coupling within the auditory, visual, and motor subdomains, supplemented by anticorrelation with other subdomains in adults. This distinctive pattern, which we replicated in independent data, was consistently less modular or absent in children and presented a negative association with age in adults, thus indicating an overall inverted U‐shaped trajectory. This indicates greater synchrony, strengthening, modularization, and integration of the brain's functional connections beyond adolescence, and gradual decline of this pattern during the healthy aging process. We also found evidence that the developmental changes may also bring along a departure from the canonical static functional connectivity pattern in favor of more efficient and modularized utilization of the vast brain interconnections. State‐based statistical summary measures presented robust and significant group differences that also showed significant age‐related associations. The findings reported in this article support the idea of gradual developmental and aging brain state adaptation processes in different phases of life and warrant future research via lifespan studies to further authenticate the projected time‐resolved brain state trajectories.
Keywords: brain aging, brain development, connectivity, functional connectivity, resting state fMRI, time‐resolved functional connectivity
We highlight the replicable time‐resolved developmental and aging functional brain patterns in a systematic evaluation of over 47,000 youth and adult brains. Our study captured the most contrastive difference between the two life stages in a brain state that indicated coherent strengthening and modularization of functional coupling within the auditory, visual, and motor sub‐domains, supplemented by anticorrelation with other subdomains in adults, a pattern that was consistently less modular or absent in adolescents and presented a negative association with age in adults, thus indicating an overall inverted U‐shaped trajectory. This indicates greater synchrony, strengthening, modularization, and integration of the brain's functional connections beyond adolescence, and the gradual decline of this pattern during the healthy aging process.

1. INTRODUCTION
The human brain is an organ that experiences continual adaptation through different life stages—prenatal, neonatal, infancy, toddlerhood, early childhood, middle childhood, adolescence, young adulthood, middle adulthood, and late adulthood (Menon, 2013). Although each life stage encompasses different developmental and aging adaptations, studying neurodevelopment in adolescence is particularly relevant as this phase is characterized by fundamental physical, cognitive, neurobiological, psychological, social, and emotional changes. Principally, adolescence is the transient developmental period between middle (school‐age) childhood and young adulthood and is divided into the early, middle, and late stages spanning from 10 to 24 years of age (Pringle et al., 2016; Sawyer et al., 2018). During this phase of brain development, distributed functional brain networks develop critical interactions in the maturation process to construct sophisticated cognitive systems. Understanding functional interactions in this developmental phase can inform scientists and clinicians about how the brain achieves complex and extraordinary cognitive abilities, understand aberrations in neuropsychiatric disorders, and thus design interventions to diminish the incidence of critical health problems. Accumulating evidence suggests that most psychopathologies find their onset and diagnosis in childhood or adolescence; therefore, it is vital to understand the brain developmental phases. While functional interactions in the brain undergo significant modifications with development, the patterns in which they evolve from adolescence to young adulthood are not yet clear.
Presently, researchers study neurodevelopmental changes extensively with brain imaging modalities, including the use of structural magnetic resonance imaging (Giedd, 2008) to highlight anatomical reorganization and functional MRI (fMRI) (Ernst et al., 2015) to study cognitive changes associated with a task or in the resting state of the brain. The resting‐state fMRI (rs‐fMRI) modality assumes that individuals engage in unrestrained cerebral activity that reveals the brain's functional organization. One approach to studying the large‐scale functional organization of the resting‐state brain is to evaluate it as an integrative network of various spatially distributed yet functionally interacting resting‐state networks (RSNs) that frequently process and distribute information. Modern fMRI research typically identifies the distributed brain regions and associated temporal signals by spatially reducing the high‐dimensional fMRI data via region of interest, standardized template/atlas‐based approaches, or adaptive parcellation techniques like group independent component analysis (gICA) (Calhoun & Adalı, 2012; Calhoun et al., 2001). The gICA approach quantifies the synchronous co‐activation or coupling patterns of brain regions (also termed functional connectivity, FC) by statistical covariation measures (e.g., correlation, coherence, mutual information, etc.), as they collectively support specific brain functions.
Functional connectivity (FC) has emerged as a promising tool for understanding the brain's functional architecture (Friston, 2011; van den Heuvel & Hulshoff Pol, 2010) and has been applied to understand several stages of development and adaptation in a human lifespan. FC characterization among networks at a whole‐brain level, termed functional network connectivity (FNC; Jafri et al., 2008), has been studied at a static (i.e., time‐averaged—by measuring FNC for the entire temporal signal) or a dynamic scale (i.e., time‐resolved—by evaluating FNC recurrently using short consecutive temporal frames of the entire temporal signal). Given the unconstrained nature of mental activity at rest, it is intuitive to presume that dynamic changes will be even more prominent in rs‐fMRI (Calhoun et al., 2014; Hutchison et al., 2013; Miller et al., 2018; Preti et al., 2017). As a result, the last decade has witnessed a significant paradigm shift in measuring FNC from a time‐averaged approach to a time‐resolved manner, as the latter provides additional access to the rich temporal structure of the FNC patterns (Lurie et al., 2020). Besides, one can probe the structure and frequency of the underlying temporal fluctuations in the FNC patterns by assessing brain network states and state summary measures (Allen et al., 2014). A significant body of the literature confirms the biological relevance of the time‐resolved FNC (trFNC) states, with studies validating their association with cognition, behavior, demographics, phenotypic traits, and neurological and psychiatric brain conditions (Lurie et al., 2020). Moreover, the canonical utility and robustness of the time‐resolved brain state‐based approach in short to moderate‐length resting‐state sessions (Abrol et al., 2016; Abrol et al., 2017) make it a rational choice to investigate the brain maturation process.
Age‐related differences in FC are evident in the early developmental phase of the brain, with significant rewiring and topological restructuring over time (Fair et al., 2007; Power et al., 2010; Supekar et al., 2009). Existing time‐resolved brain connectivity research in infants, children, and adolescents has shown that the dynamic interregional interactions accurately reflect neonatal maturity (Ma et al., 2020), predict brain maturity (Qin et al., 2015), present marked changes (Faghiri et al., 2018), and variability with maturity (Hutchison & Morton, 2015; López‐Vicente et al., 2021), differ in atypical development (Liu et al., 2019; Rashid et al., 2018), characterize risk for neurodevelopment disorders (Marusak et al., 2017) and provide unique insights regarding pathophysiology (Sato et al., 2015). In contrast, brain connectivity again evolves in middle and late adulthood and is accompanied by gradual cognitive decline. There is evidence that advancing age induces increased functional topological reorganization to establish compensatory mechanisms to counteract the aging process (Fjell et al., 2017; Grady et al., 2016; Li et al., 2015; Meunier et al., 2014; Sala‐Llonch et al., 2015; Sugiura, 2016). The underlying network mechanisms of brain plasticity are regional (Yin et al., 2016), and processes vary in the stages of healthy aging, with some evidence that aging‐associated neural changes can originate toward the end of early adulthood (Siman‐Tov et al., 2016). Relevant work has also demonstrated an age‐related decline in modularization and variation of the trFNC state patterns in late adulthood (Chen et al., 2017; Tian et al., 2018) and a lack of a metastable brain state responsible for efficient global communications in late adulthood, as compared to middle‐adulthood (Escrichs et al., 2021).
Most of the existing time‐resolved brain connectivity research has been conducted over short time spans in relatively small samples of children/adolescents and adults and/or unilaterally (i.e., separate analysis for both groups). However, the launch of mega‐scale long‐term studies such as the Adolescent Brain Cognitive Development (ABCD) study and the UK Biobank (UKB) study provides an unprecedented opportunity to address the limitations of sampling variability and permitting adequate statistical power and reproducible estimations of the studied effect sizes (Marek et al., 2022). The ABCD study is a large long‐term sample of pediatric data sets (Casey et al., 2018), including a diverse set of behavioral measures. In contrast, the UKB study (Miller et al., 2016) is a large long‐term biobank of middle and old‐age adults, including in‐depth genetic and health information.
In line with the hypothesis that the brain connectivity state patterns may vary in adolescence compared to adulthood, we probed these two large brain imaging adult and adolescent resting‐state fMRI repositories. We focused on using a fully automated and adaptive spatially constrained group independent component analysis (SC‐gICA) approach (Du et al., 2020) to estimate individual‐specific spatial maps and activity time‐courses corresponding to shared aggregate RSNs. Subsequently, we sought to access the intrinsic temporal dynamics of the trFNC patterns with a sliding window correlation approach (Allen et al., 2014; Sakoğlu et al., 2010), estimating the recurring, transient and replicable brain state profiles with unilateral and pooled k‐means clustering analyses on youth and adult data sets. We systematically probe the associated state summary measures to gain complementary insights into the significant trFNC differences in adults and adolescents, as detected by our time‐resolved analyses. Notably, the differences we seek are typically thought to reflect developmental and aging‐related brain adaptations and thus constitute a significant research problem. The following sections cover the materials and methods, a demonstration of noteworthy results, and a discussion of key outcomes of our article.
2. MATERIALS AND METHODS
2.1. Functional MRI data
We used rs‐fMRI scans (eyes open: passive crosshair viewing) from the UKB (n = 36,461, F:M = 19,474:16,987, Age = 44–82 years) (Miller et al., 2016) and the ABCD (n = 9617, F:M = 4722:4895, Age = 9–13 years; Casey et al., 2018) studies for the discovery (original) analyses in this article. Furthermore, we used the Human Connectome Project (HCP: n = 833, F:M = 439:394, Age ~ 22‐35y) (Van Essen et al., 2013) and the Developmental Chronnecto‐Genomics (Dev‐CoG: n = 191; F:M = 95:96, Age = 8–16 years; Stephen et al., 2021) studies for replication (confirmatory) analyses (see Table 1). Written informed consent was obtained from all human subjects participating in the several data repositories (UKB, ABCD, HCP, and Dev‐CoG). The study procedures were approved by the institutional review boards of all participating centers. The enrollment procedure of human subjects was inclusive of all participants without limitations by sex or gender, race, ethnicity, and age other than as scientifically justified and as specified in the enrollment inclusion and exclusion criteria of the respective data sets.
TABLE 1.
Data demographics (total and gender‐wise sample size and age range), repetition time (TR), scan lengths, and sliding window length
| Data set | Sample size (F/M) (post‐QC) | Age | TR (ms) | Total TRs/scan length (s) | Window width (#TRs/length[s]) |
|---|---|---|---|---|---|
| UKB | 36,461 (19,474/16987) | 44–82 years | 735 | 450/330.75 s | 40/29.4 s |
| ABCD | 9617 (4722/4895) | 9–13 years | 800 | 320/256 s | 40/32.0 s |
| HCP | 833 (439/394) | 22–35 years | 720 | 1200/864 s | 40/28.8 s |
| Dev‐CoG | 191 (95/96) | 8–16 years | 460 | 650/299 s | 60/27.6 s |
Abbreviations: ABCD, Adolescent Brain Cognitive Development; HCP, Human Connectome Project; UKB, UK Biobank.
The fMRI data sets available from the data repositories underwent rigid body motion correction, distortion correction, normalization to the Montreal Neurological Institute template using the old SPM12 normalization module, and smoothing using a Gaussian kernel with a full width at half maximum = 6 mm. We also implemented thorough quality control (QC) on the preprocessed fMRI images to discard the images that exhibited (1) poor correlation with individual and group data masks, (2) markedly briefer scan lengths, and (3) high head motion parameters (>3° rotations and >3 mm translations). We used only one image per subject in this analysis, and the reported numbers are post‐QC sample sizes.
2.2. Time‐resolved feature extraction
In the following data processing stage, we extracted time‐resolved rs‐fMRI features used in this article. We first estimated spatial maps and activity time courses for RSNs from the fMRI data. To that end, we decomposed the preprocessed and quality‐controlled fMRI time courses using the SC‐gICA approach, as explained next (Du & Fan, 2013). We applied our multidata set generated Neuromark_fMRI_1.0 template (included in the GIFT software at http://trendscenter.org/software/gift and also at http://trendscenter.org/data; Du et al., 2020) as a spatial reference for this decomposition (n = 53 brain regions), grouped in seven brain subdomains (see Figure 1).
FIGURE 1.
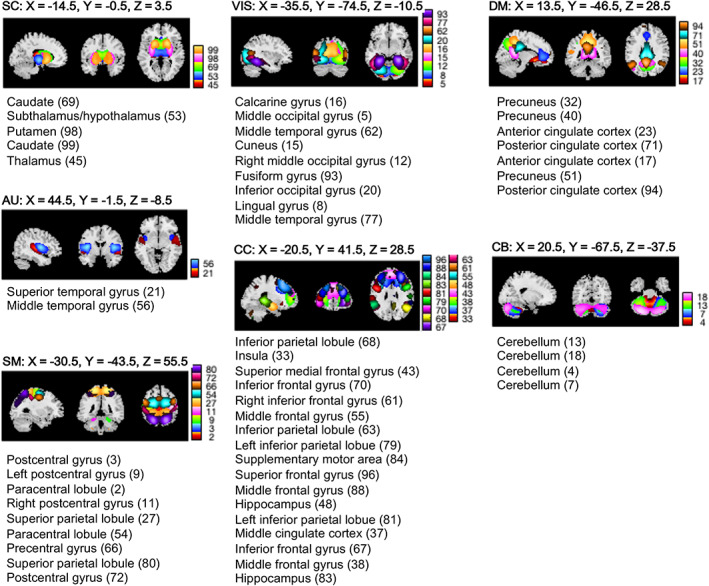
Resting state networks (RSNs). Spatial maps of the RSNs (n = 53) derived from a spatially constrained group independent component analysis are plotted at the exhibited sagittal, coronal, and axial slices. The RSNs are partitioned into brain sub‐domains based on anatomical and functional properties of the (n = 53) brain components: AUD, Auditory; CC, Cognitive control; CEREB, Cerebellum; DMN, Default mode network; SC, Subcortical; SM, Somatomotor; VIS, Visual
The Neuromark_fMRI_1.0 template provides spatial priors based on two independent resting‐state fMRI data sets—the Brain Genomics Superstruct Project (Holmes et al., 2015) and the HCP (Van Essen et al., 2013). We first decomposed these two data sets separately using standard gICA analysis (Calhoun et al., 2001) to identify independent sets of aggregate (i.e., group‐level) RSNs. Next, we matched the two sets of RSNs using greedy spatial correlation analysis to determine the set of replicable RSNs retained. This overall strategy successfully discovered key shared and distinct biomarkers in various clinical populations (Du et al., 2021), is generally quite robust to processing pipeline (DeRamus et al., 2021) and data length variation (Duda et al., 2022), and has been carefully evaluated in both adult and adolescent data (Fu et al., 2022). We could use another robust template of spatial priors, but we wanted to leverage the extensive amount of work already known about the current template to focus on the fundamental idea here, which is to evaluate RSNs specific to individual subjects that are also comparable across subjects and data sets.
Subsequently, for each subject, we calculated the trFNC features between all pairs of brain regions (i.e., for n = 53C2 = 1378 brain connections) using a tapered sliding window featuring convolution of a rectangular window (width = “n” TRs) with a Gaussian (σ = 3 TRs), where n is the number of TRs for each data set as listed in Table 1. We shifted this window in gradual steps of 1 TR, resulting in “w” windows, where w is the width of the window for each data set, as listed in Table 1. This procedure generated a temporal series of windowed correlations, for which we identified recurring, transient trFNC brain state patterns as multidimensional cluster centroids using a k‐means clustering algorithm (Allen et al., 2014). These brain state patterns are transient patterns of whole‐brain FC states that the brain navigates through during the entire scan length. A significant body of literature supports the biological relevance of brain state patterns, with studies validating their association with cognition, behavior, demographics, phenotypic traits, and neurological and psychiatric brain conditions (Lurie et al., 2020).
A first‐level clustering estimated an initial point input to stabilize the grouping in the second‐level clustering, while all windowed FNC data was clustered in the second‐level clustering (Abrol et al., 2017; Allen et al., 2014; Pascual‐Marqui et al., 1995). We found the initial point input to the second level clustering by estimating and clustering the subject exemplars corresponding to windows of subject‐level FNC data featuring the highest variance in FNC. The centroid connectivity patterns resulting from this first‐level clustering were then used to initialize the second‐level clustering of all FNC data. The initial and final clusterings were repeated 150 times to increase the likelihood of escaping local minima to derive stable aggregate connectivity patterns.
We conducted unilateral (i.e., independent), pooled (i.e., conjoint), and cross‐data unilateral (i.e., using ABCD and UKB pooled clustering states as a common basis to interrogate clustering summary measures of both groups in a unilateral manner) clustering. The unilateral clustering analysis provided independent estimates of the brain state patterns for each data set, thus yielding prominent insights into the expected group differences. While, pooled clustering of adult and adolescent data sets allowed us to contrast the brain state patterns more directly by providing a more equivalent basis for clustering the data sets. Likewise, the cross‐data unilateral analyses allowed us to evaluate both groups using an equivalent basis for clustering the data sets. The pooled and cross‐data unilateral analyses thus augment and further validate our findings from the unilateral clustering analyses.
The sliding window‐related parametrical choices were made to maximize correspondence in the window length across data sets used in the unilateral analyses, while the data were calibrated to the fastest TR in pooled and cross‐data unilateral clustering analyses. Furthermore, the unilateral clustering experiments in this article used the original data TR, whereas it was calibrated to the fastest TR for the pooled clustering cases to permit a consistent time‐resolved assessment. Finally, as detailed in the succeeding subsection, we computed the statistical state summary measures to gain further insights into the brain state development and adaption process in adolescence and adulthood.
2.3. Statistical measures
To compare brain state profiles between adolescents and adults, we sorted them using Pearson's correlation using a greedy matching approach. The greedy sorting approach used in this article generates a bijective mapping between the sets of states based on the correlation metric by iteratively matching states with the highest correlation value without replacement. This was done separately for all discovery and replication data sets for the unilateral clustering experiments. We sorted the state patterns using unilateral patterns as references for the pooled experiments.
Next, we computed several state‐based summary measures, comprising hard clustering metrics—mean dwell times and fractional occupancy times (frequency), and a fuzzy clustering metric—regression betas. We computed the mean dwell time as the mean period of a given state's temporally consecutive runs and fractional occupancy time as the percentage of windows apportioned to that state in the corresponding imaging session. We also recorded weighted state memberships in a fuzzy framework—regression betas, computed as the beta‐coefficient estimates of the multiple linear regression of the windowed correlations on the cluster centroids, averaged across all windows.
3. RESULTS
3.1. Unilaterally clustered brain state profiles in adulthood and adolescence
We estimated the brain state profiles for all four data sets independently—discovery: UKB (adults) and ABCD (adolescents); replication: HCP (adults) and DevCoG (adolescents) and sorted them across the data sets using Pearson's correlation metric. The lower triangular matrices in Figure 2 demonstrate the sorted brain state profiles for the optimal clustering model order (k = 4) modularized by the assessed seven brain subdomains. In contrast, the upper triangular matrices in this figure display this data averaged by subdomains to highlight the most dominant intra‐ and inter‐domain brain connectivity patterns. Notably, the most striking difference constitutes the coherent strengthening of brain connections within the auditory, visual, and motor subdomains, supplemented by anticorrelation with other subdomains in the adults' S1 profile.
FIGURE 2.
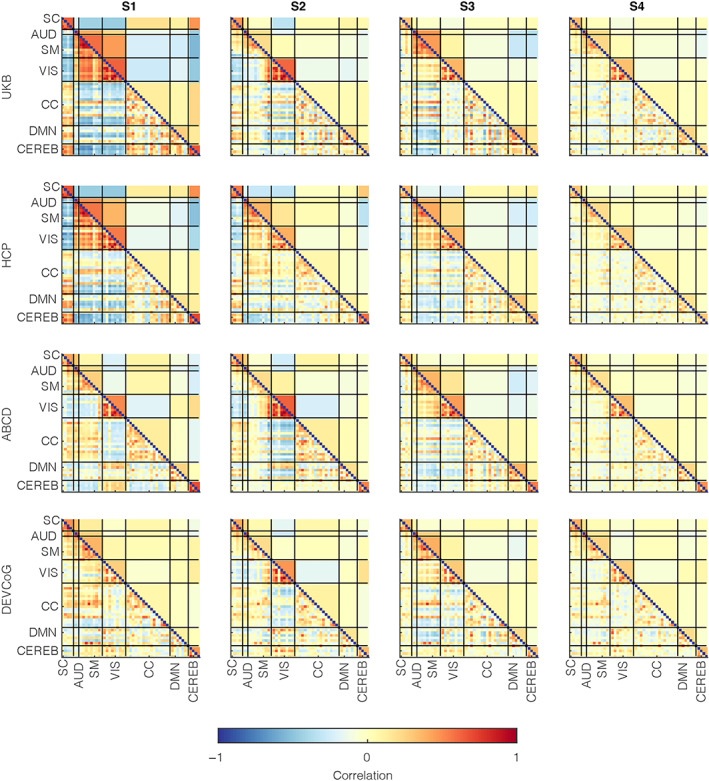
Time‐resolved functional network connectivity (trFNC) state patterns with unilateral clustering. Windowed functional connectivity features (i.e., windowed pair‐wise correlations estimated from the activity time‐courses of the brain components) were clustered unilaterally for discovery (UK Biobank [UKB] [n = 36,461] and Adolescent Brain Cognitive Development [n = 9617]) and replication (Human Connectome Project [HCP] [n = 833] and DEVCoG [n = 191]) data sets for a range of model orders (k = 2–5). The lower triangular parts of the matrices represent the cluster centroid patterns marking the quasistable trFNC state patterns of the human brain that reoccur over time. The trFNC state patterns were matched across the clustering model orders using greedy sorting with correlation as the similarity metric. The upper triangular parts of the matrices represent the matched trFNC state profiles averaged at the domain level to highlight dominant inter‐domain and intra‐domain brain state patterns. Vertical and horizontal black lines partition the brain state patterns by (n = 7) domains based on anatomical and functional properties of the (n = 53) brain components: AUD, Auditory; CC, Cognitive control; CEREB, Cerebellum; DMN, Default mode network; SC, Subcortical; SM, Somatomotor; VIS, Visual
This distinctive brain profile (S1) also featured stronger correlation patterns within the subcortical subdomain and between the subcortical and cerebellar subdomains and stronger anticorrelation patterns of the subcortical and cerebellar subdomains with the auditory, visual, and motor subdomains in adults. Additionally, this unique adult brain state profile (S1) presented a higher correlation strength among the subcortical regions and certain regions in the cognitive control domains (specifically, the medial cingulate cortex, inferior and middle frontal gyrus, and hippocampus), and similarly, some default‐mode network regions involving the anterior and posterior cingulate cortex. Generally, the corresponding patterns were less modular for adolescents, with a visibly lower correlation strength within the visual and motor subdomains and disjointed subcortical and cerebellum subdomains.
Remarkably, the distinctive pattern presented in S1 in adults was consistently absent in adolescents. Furthermore, we replicated these results for a range of clustering model orders (k = 2–5) in both discovery and replication data sets, wherein we observed similar trends across all model orders. Additionally, all results held for a matched sample size of UKB with respect to ABCD for repeated subsampling of the UKB data, that is, for 9617 UKB subjects, compared to using all UKB subjects. These results are included in supplementary data figures S1 (k = 2), S2 (k = 3), S3 (k = 4), and S4 (k = 5).
We also conducted a post hoc comparison to highlight group differences in the trFNC state profiles. For this analysis, we estimated the subject‐wise state representations as medians of subject‐wise windowed observations for a given state per the state membership functions. Finally, we evaluated the group‐level aggregate representations by computing the state‐wise medians of the subject‐level state patterns. Figure 3 illustrates the group‐level state‐wise medians for adults (UKB subjects: top panel) and adolescents (ABCD subjects: middle panel) while also plotting the difference between the two groups (UKB‐ABCD: bottom panel). This result underscores S1 as the most contrastive state between the two groups, thus further validating this observation from our previous results. Other brain states showed several modular differences that were not as contrastive as the S1 pattern.
FIGURE 3.
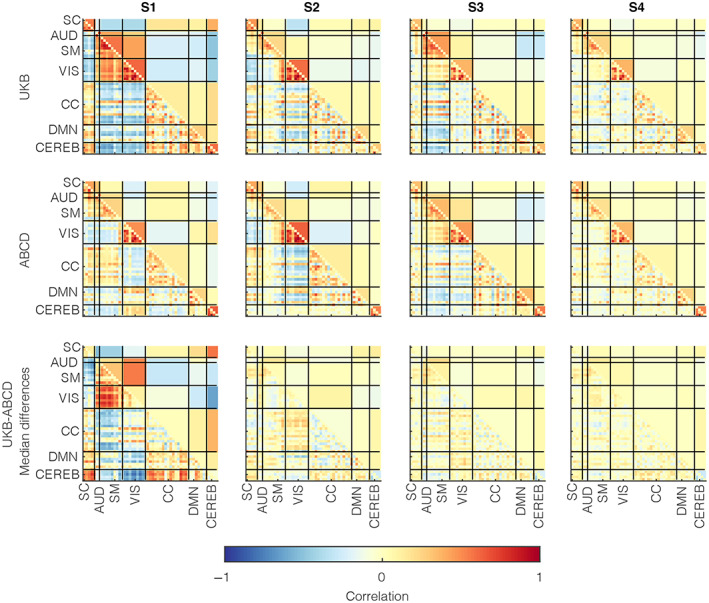
Group differences in time‐resolved functional network connectivity (trFNC) state patterns with unilateral clustering. We assessed the group‐level median patterns from the subject‐level median patterns in the trFNC state patterns. This figure demonstrates the group‐level data for adults (UK Biobank [UKB] subjects) in the top row and adolescents (Adolescent Brain Cognitive Development [ABCD] subjects) in the middle panel. In contrast, we plot the patterns in their group differences (UKB‐ABCD) in the bottom row. Consistent with previous results, the most contrastive pattern between the two groups is presented in the S1 brain state profile, though other states also differ marginally. Vertical and horizontal black lines partition the brain state patterns by (n = 7) domains based on anatomical and functional properties of the (n = 53) brain components: AUD, Auditory; CC, Cognitive control; CEREB, Cerebellum; DMN, Default mode network; SC, Subcortical; SM, Somatomotor; VIS, Visual
3.2. Conjointly clustered brain state profiles in adulthood and adolescence
To provide a more equivalent basis for the clustered data, we contrasted the brain state profiles estimated via conjoint (i.e., pooled) clustering of adult and adolescent data sets. In this analysis, we probed the structure of the state patterns and critical state summary measures assessed in previous literature. The lower triangular matrices in Figure 4 provide the trFNC patterns for the pooled clustering case with sample‐size matched UKB (n = 9617) and ABCD (n = 9617) data sets for a range of model orders (k = 2–5), while the upper triangular matrices provide the supplementary domain‐averaged result. We sorted the patterns presented in this result as per the brain state patterns from the unilateral clustering (i.e., as per patterns visualized in Figure 2). The central objective of testing different model orders here is to confirm the algorithm stability and verify if the contrastive pattern of significant interest appeared consistently across all model orders. Analyzing the brain state patterns across a range of clustering model orders involves multiple unsupervised learning analyses (e.g., one independent decomposition for each clustering model order). Thus, there may be uncertainty in the validated cluster centroids. Given that the underlying data distribution (in the windowed correlations) is identical, assuming a high similarity in the emergent states across different model orders is rational. As expected, the brain state patterns replicate across the range of model orders, but more interesting is the fact that the unique adult state pattern (S1) was prevalent for all model orders, and the other states emerged consistently with an increase in the clustering model order. Furthermore, repeated sub‐sampling of the UKB data successfully reproduced this result. We plot the equivalent brain connectograms for the optimal model order (k = 4), thresholded at a Pearson's correlation value of 0.5, in Figure 5.
FIGURE 4.
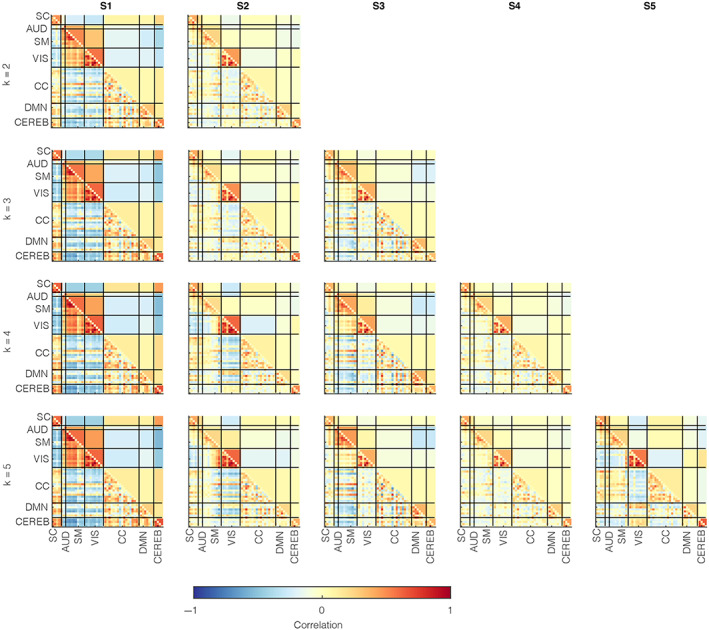
Time‐resolved functional network connectivity (trFNC) state patterns with pooled clustering. Windowed functional connectivity features (i.e., windowed pair‐wise correlations estimated from the activity time‐courses of the brain components) were clustered conjointly for sample‐size matched UK Biobank (UKB; n = 9617) and Adolescent Brain Cognitive Development (n = 9617) data sets for a range of model orders (k = 2–5). The lower triangular parts of the matrices represent the cluster centroid patterns, whereas the upper triangular elements represent the matched trFNC state profiles averaged at the domain level to highlight dominant inter‐domain and intradomain brain state patterns. Vertical and horizontal black lines partition the brain state patterns by (n = 7) domains based on anatomical and functional properties of the (n = 53) brain components: AUD, Auditory; CC, Cognitive control; CEREB, Cerebellum; DMN, Default mode network; SC, Subcortical; SM, Somatomotor; VIS, Visual
FIGURE 5.
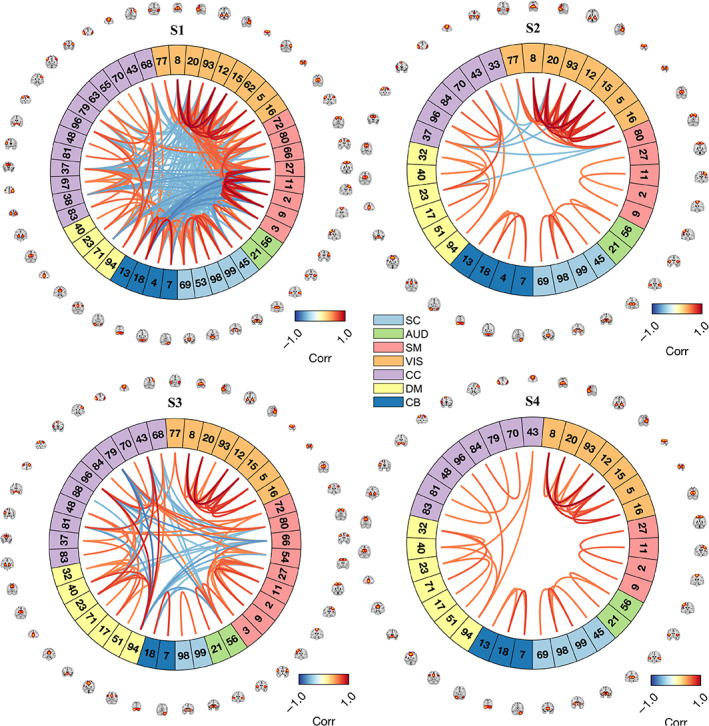
Connectogram plots for conjoint brain state patterns via pooled clustering. Here we highlight the connectivity strengths of the most significant brain state connections (thresholded at a correlation value of ±0.5) for the patterns illustrated in Figure 3 (for the optimal model order: k = 4), also plotting the peak activations in the spatial maps of the corresponding resting state brain networks on the connectogram ring periphery. The colors on the connectogram ring partition the brain state patterns by (n = 7) domains based on anatomical and functional properties of the (n = 53) brain components: AUD, Auditory; CC, Cognitive control; CEREB, Cerebellum; DMN, Default mode network; SC, Subcortical; SM, Somatomotor; VIS, Visual. Component names for the listed component indices are listed in Figure 1
3.3. Cross‐data unilateral state clustering analysis
We further examined the trends by fixing the unilateral ABCD and UKB states as a common basis to interrogate clustering summary measures of both groups in a unilateral manner. In this analysis, we regressed the data onto the predefined set of state patterns, assigned the state membership based on the highest regression beta coefficient, and measured the hard clustering metrics using the derived state memberships. For this analysis, we measured effect sizes of the difference in means of the derived state measures (fractional rates and mean dwell times) assessed via Cohen's d for independent samples for four cases, as shown in Figure 6: (1) comparing unilateral UKB states on UKB data and ABCD data (UsUd – UsAd), (2) comparing unilateral ABCD states on ABCD data and UKB data (AsAd – AsUd), (3) comparing unilateral UKB states and unilateral ABCD states on UKB data (UsUd – AsUd), and (4) comparing unilateral ABCD states and unilateral UKB states on ABCD data (AsAd – UsAd).
FIGURE 6.
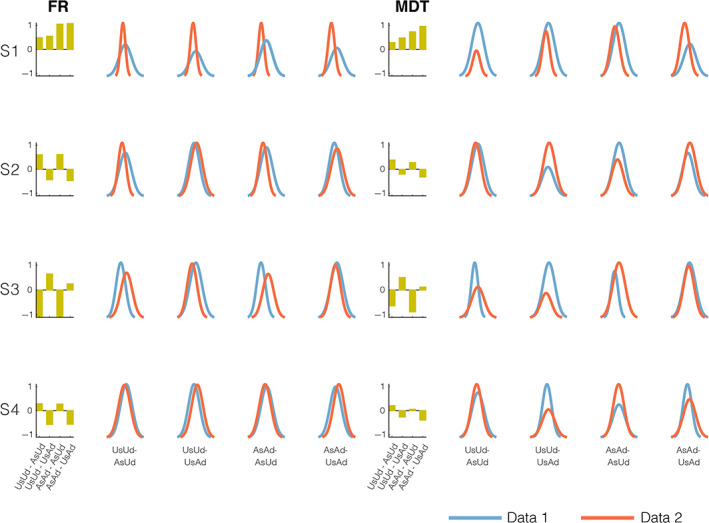
Cross‐data unilateral state clustering analysis: We used the time‐resolved functional network connectivity (trFNC) brain state patterns (S1: State 1, S2: State 2, S3: State 3 and S4: State 4) from unilateral UK Biobank (UKB) and Adolescent Brain Cognitive Development (ABCD) clustering to estimate state summary measures (frequency: FR and mean dwell times: MDT) in ABCD and UKB data, respectively. The bar plots (columns 1 and 6) show the effect size of the difference in means of the derived state measures assessed with Cohen's d. Column panels (2–5 and 7–10) plot the normalized histograms of the two state summary measures (FR and MDT on the x‐axis) for the four cross‐data unilateral state clustering cases. The S1 pattern always showed a positive effect size, implying the dominance of this pattern in adult data for all cases. In contrast, the other three patterns showed a mixed direction for both measures. In all tested cases, the S2 and S4 state patterns (most comparable to the static functional connectivity pattern) shared the same direction of the effect as anticipated, whereas the S3 pattern presented the exact opposite direction of the effect.
Note, for ease of interpretation of the tested cross‐data unilateral clustering cases in Figure 6, we intuitively set the order of the arguments (i.e., states and data sets), expecting a positive effect for the S1 brain pattern in all tested cases. If validated, this would imply a greater mean value of the assessed metrics for adults as compared to adolescents for state 1 (S1) while also providing a reference order to assess trends in other state patterns. Results for the S1 state pattern verify strong positive effects for all these tests, further validating the known robust contrast presented by this state as per earlier unilateral and pooled clustering results. In contrast, we observed a mixed pattern for the other three state patterns. While the S2 and S4 state patterns, which are most comparable to the static FC pattern, as expected, share the same direction of the effect, the S3 pattern presented the exact opposite direction of the effect in all tested cases. Taken together, the observations in the cross‐data unilateral clustering analyses strengthen our prior unilateral and pooled clustering outcomes on the contrastive S1 pattern, while also providing additional insights into the trends for other state patterns.
3.4. Replicability of brain state summary measures across a range of model orders
As expected, the trFNC brain state patterns estimated via the various clustering trials bear a strong resemblance. However, probing the state summary measures provides the appropriate tool to contrast the groups, as evaluated in this analysis. Figure 7a includes group differences in the pooled clustering assessed via the considered hard clustering summary measures—mean dwell times and fractional occupancy rates (state frequencies). Similarly, Figure 7b shows the group differences in the pooled clustering case assessed via the fuzzy clustering metric—regression betas, which are the beta‐coefficient estimates of multiple linear regression of the windowed FC features on the cluster centroids, averaged across all windows.
FIGURE 7.
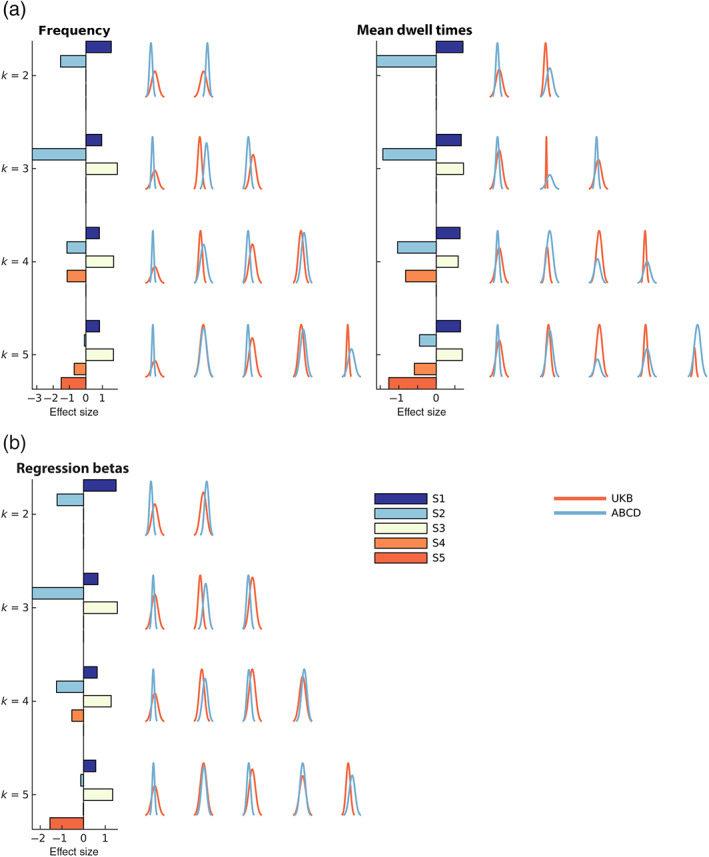
Group differences in pooled clustering. (a) Hard clustering metrics. We plot the distributions for the fractional occupancy rates (frequency) and the mean dwell times of the brain states for the UK Biobank (UKB) and Adolescent Brain Cognitive Development (ABCD) groups for a range of clustering model orders (k = 2–5) on the right of each top panel. (b) Fuzzy clustering metric. We plot the distribution for the fuzzy regression beta coefficients for the UKB and ABCD groups for a range of clustering model orders (k = 2–5) on the right side of the bottom left panel. The effect size of the difference in group means of these metrics, assessed via Cohen's d for independent samples, is shown in the horizontal bar plot on the left of each panel. All brain state patterns showed replicable effects across the model orders and metrics, with the S1 and S3 patterns consistently showing a robust positive effect size. In contrast, the S2 and S4 patterns revealed negative effect sizes.
The S1 pattern showed significantly lower frequency, mean dwell times, and regression beta values in adolescents than adults. This evidence further substantiates the strengthening of the S1 pattern in adults. We replicated this result across the model orders and replication data sets alike. Here we additionally display the effect size of the difference in means of these metrics (UKB‐ABCD), assessed via Cohen's d for independent samples in the horizontal bar plots on the left of each panel. In this figure, the histogram bar plots are colored by the state number, while the color of the metric distributions (histograms) signifies the corresponding data set. We report a consistently strong pattern in effect size that replicates across model orders, states, and metrics.
3.5. Projection analysis validates distinguished structure in the brain state patterns
We conducted a rigorous QC check to visualize, quantify and further validate the structure in the latent projection encodings of the trFNC features (Figure 8). This analysis uses the t‐distributed stochastic neighbor embedding (tSNE) algorithm to project the high‐dimensional (n = 1378) windowed correlations (i.e., trFNC features) to a two‐dimensional space for pooled clustering data from “All” groups (i.e., the UKB and ABCD groups), the UKB group and the ABCD group, for a range of clustering model order (k = 2–5).
FIGURE 8.
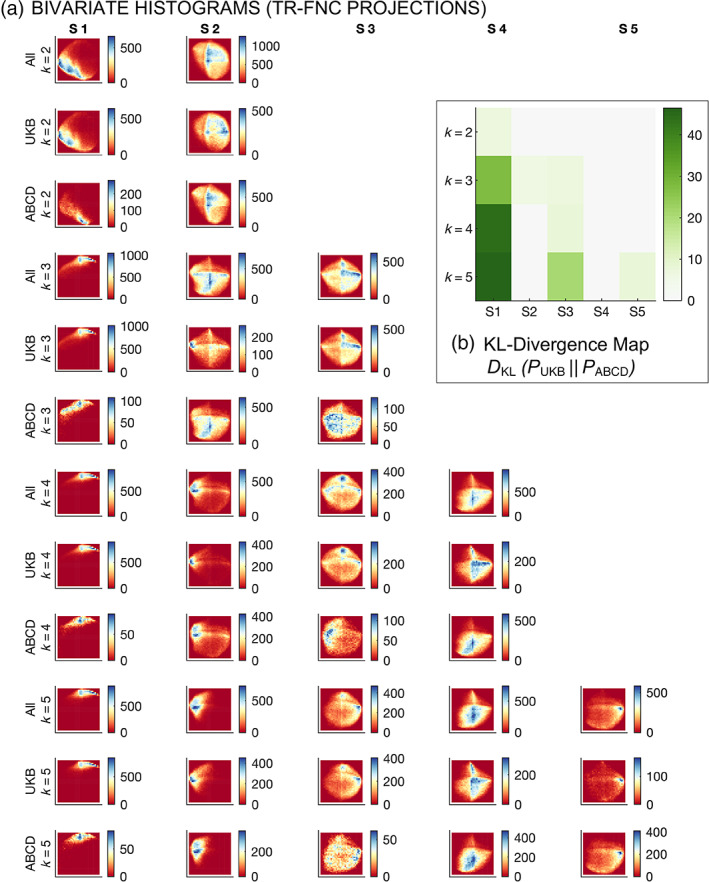
Bivariate projections of the windowed correlations for a range of clustering model orders. This analysis uses the t‐distributed stochastic neighbor embedding (tSNE) algorithm to project the high‐dimensional (n = 1378) windowed correlations to a two‐dimensional space for pooled clustering data from “All” groups (i.e., the UK Biobank [UKB] and Adolescent Brain Cognitive Development [ABCD] groups), the UKB group and the ABCD group, for a range of clustering model order (k = 2–5). (a) Bivariate histograms. Density peaks for the two‐dimensional projections of the windowed correlations are consistently localized across the groups and model orders, thus verifying the rationality of the detected clustering patterns. (b) KL‐divergence maps. The KL‐divergence metric highlights the differences in metric distributions/densities in the joint histograms. KL‐divergence is the highest for state one across all clustering model orders, underscoring S1 as the most contrastive pattern.
Figure 8a shows the bivariate histograms for the trFNC projections that reveal consistent localization of the density peaks across model orders and groups, verifying the clustering patterns revealed by our analysis. We further evaluate the trFNC data projections using the Kullback–Leibler metric to capture group differences in the densities of these joint histograms. As expected, the KL‐divergence was the highest for the S1 brain state pattern across all clustering model orders, as highlighted in Figure 8b, thus confirming S1 as the most contrastive pattern.
3.6. Dominant adult state pattern (S1) indicates an inverted U‐shaped association with age
Studying the association of age with the dynamic FC metrics can reflect gradual strengthening (e.g., positive correlation) or decline (e.g., negative correlation) in the brain state patterns with aging. To study that pattern in adults, we used the hard clustering (fractional occupancy rates and the mean dwell times) and the fuzzy clustering (regression betas) summary measures of the trFNC brain state patterns for the UKB subjects and evaluated the association of age with these three metrics using Pearson's correlation. We repeated this procedure for a range of clustering model orders (k = 2–5) for all metrics and plotted Pearson's correlation of the clustering metrics with age in Figure 9a.
FIGURE 9.
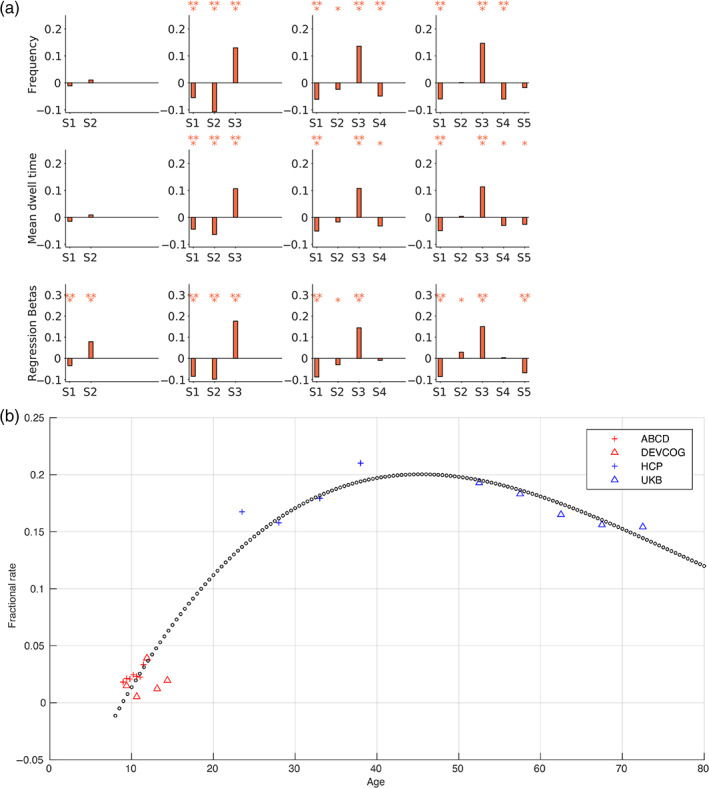
Association of dynamic metrics with age in pooled clustering. (a) We computed the fractional occupancy rates (frequency), the mean dwell times, and regression beta metrics for UK Biobank subjects for a range of clustering model orders (k = 2–5). This figure plots the Pearson's correlation of age with the considered three clustering metrics for all trFNC brains state patterns (S1: State 1, S2: State 2, S3: State 3, S4: State 4, and S5: State 5). The statistical significance of these associations is tested at two significance levels ‐ marked as * for α = .05 and ** for α = .05/m (Bonferroni correction for multiple comparisons, where m = 42 tests, comparing three metrics in states). The dominant adult state pattern (S1) presented a statistically significant negative association with age in adults across model orders k = 3–5. (b) Polynomial curve fitting for the optimal model order (k = 4) indicates an overall positively skewed inverted U‐shaped association with age for the most contrastive brain pattern.
We tested the statistical significance of these associations at two significance levels—(1) uncorrected (marked as *) and (2) Bonferroni‐corrected for multiple comparisons (marked as ** for , where m = 42 tests, comparing three metrics in = 14 states). We found a significant negative correlation between the hard clustering metrics and regression beta coefficients for the adult group in the dominant adult state pattern (S1) for the optimal model order (k = 4). The reported effect was consistent for higher clustering model orders k = 3–5. This observation suggests that although this unique (S1) pattern strengthened from adolescence into adulthood, the frequency dropped beyond a certain period in adulthood. Additionally, the dynamic metrics for the S3 pattern present the highest correlation with age across these clustering model orders (k = 3–5).
Polynomial curve fitting (in a least‐squares sense) for the state frequency versus age association for the most contrastive brain pattern revealed a positively skewed inverted U‐shaped relationship (Figure 9b). However, this result needs to be interpreted with caution, given the uncertainty of the polynomial relationship assumption and the missing samples in the functional estimation. This limitation can be addressed by incorporating additional data sets to fill the age gap (especially younger adults) or using adequately sampled lifespan studies to determine and corroborate the theorized time‐resolved brain state trajectories.
4. DISCUSSION
Studying the resting state FNC changes across the human lifespan can inform us how the underlying functional architecture and interrelationships develop or diminish over time. Motivated by that, we leveraged the two largest accessible adolescent (ABCD) and adult (UKB) brain data sets to understand the functional interrelationships in the resting state and evaluate group differences in the trFNC patterns and the associated state summary measures, reproducing them using external replication data sets (adolescent: Dev‐CoG and adult: HCP). We detected a robust contrastive pattern (S1) in one of the brain‐state patterns that presented a highly modular distinction between the two groups, characterized themes of elevated FC within the auditory, visual, and motor subdomains, and exhibited a consistent anticorrelation with all other domains in adult brains.
The detected S1 pattern was significantly weaker in the adolescent brains, thus underscoring the idea of strengthened functional integration in the motor‐visual interdomain networks as people mature. Of note, there is additional evidence from previous work that the static FC for the motor‐visual networks is reduced in youth (Jolles et al., 2011) and that time‐resolved FC is not elevated for these networks in any of the adolescent brain states (López‐Vicente et al., 2021). These previous results, together with our results, provide a cohesive view into the possible modularization, integration, and strengthening of the sensorimotor (Zuo et al., 2010) and visual resting‐state brain connections beyond childhood, signaling a change in the broader organization of the brain's functional architecture. Remarkably, the clustering metrics for this pattern are also negatively correlated with age in adults, suggesting an inverted‐U‐shaped lifespan pattern. We speculate that this negative correlation of the S1 pattern in adults may be interlinked to the redistribution of the restricted cognitive resources due to the systematic decline in the global brain processing capacity due to brain aging.
Group analyses of the brain state patterns most resembling the static FC pattern (S2 and S4) indicate that the occupancy of these patterns was significantly higher for adolescents compared to adults. This is in parallel with the observation of the negative association between age and dwell times reported by Faghiri et al. (2018) for the Pediatric Imaging, Neurocognition, and Genetics (PING) data set and Rashid et al. (2018) for the Generation R data set (White et al., 2013). As such, these strong positive effects imply that the developmental changes may also bring along a departure from the canonical static FC pattern to accommodate more efficient and modularized utilization of the vast brain interconnections. Our results also suggest that the estimated trFNC state profiles and summary measures are robust, as these are highly comparable within each group across different clustering model orders and replicate in the discovery and replication data sets.
We also performed an exploratory comparative analysis evaluating the relationship of age to the studied dynamic metrics. Interestingly, the occupancy rates (suggested via the frequency, mean dwell times, and regression betas metrics) in adults increased most consistently with age for the S3 pattern. The S3 brain state pattern varied from the unique S1 adult pattern in reduced correlation strengths from the visual domain to the auditory and subcortical brain domains. Additionally, the strong anticorrelated patterns observed in the S1 pattern faded for the S3 pattern for brain connections from the subcortical and cognitive control domains to the auditory, somatomotor, and visual domains. Likewise, we found a more neutral pattern in S3 compared to strong anticorrelations observed in S1 for the visual to default‐mode and cerebellum inter‐domain connectivity. The S3 pattern always exhibited positive correlations with age for adolescents, but we note to interpret this observation with caution given little variability in age for the adolescent group.
Our study inherits generic limitations of the fMRI data, including an indirect link to neural activity, measurement noise (e.g., head movement, cardiac, respiratory, etc.), and limited spatiotemporal resolution. However, we used a rigorous fMRI data preprocessing pipeline and QC checks based on years of experience to diminish the influence of measurement noise. Head motion can confound the interpretation of resting‐state fMRI (Power et al., 2012; Satterthwaite et al., 2012), and adolescents can certainly show greater movement levels. To minimize the effect of head motion, we used only the low‐motion subjects that passed our thorough QC check, additionally regressing out the head motion parameters from the windowed correlations. Yet, this does not safeguard our analyses from other known measurement confounds such as physiological noise, etc., which is an even more complex topic, warranting further research in this direction. While the group ICA technique is relatively effective at eliminating physiological noise, faster data sampling and physiological noise regression can help assess their contribution to the dynamic brain connectivity measurements. Nonetheless, we do not presume any systematic effects leading to the highlighted group differences, especially given the reproducible effects with out‐of‐sample replication data sets.
Here, we postulate the breakthroughs suggested by this article convergently indicate that the brain's FC patterns evolve systematically during adolescence, and these changes reflect the brain's intrinsic maturation process. While the included data sets yield rich insight into the time‐resolved FC differences in adults and adolescents, the extraction of more generalizable and normative age trajectories is partially hampered by the noninclusion of specific age groups such as infants, young children, and early middle‐aged adults. Future work may therefore incorporate other data sets that fill these age gaps and also expand current lifespan studies to further corroborate the theorized time‐resolved brain state trajectories.
Other topics to study include examining linkages of the time‐resolved states and summary measures with anatomical measures and clinical assessments in both groups and studying gender effects in such analyses. It is also of great significance to detect aging effects in time‐resolved FC in brain disorders and diseases and compare those patterns to the normative population, as various psychiatric illnesses typically emerge during the developmental phase of life. Lastly, there may be protective modifiable (e.g., physical activity, social interactions, calorific restrictions, etc.), nonprotective modifiable (e.g., smoking, alcohol, drug abuse, etc.), and nonmodifiable (e.g., sex, APOE status, etc.) factors that control aging‐related alterations in the functional connectome in adulthood (Jockwitz & Caspers, 2021) that must be examined with longitudinal research designs of lifespan studies.
5. CONCLUSION
Our findings suggest substantial and consistent evidence for accentuated temporal dynamics in a unique adult‐brain‐specific dynamic connectivity pattern showing elevated FC between the auditory, visual, and motor subdomains and a consistent anticorrelation with all other domains. The reproducible nature of our investigation provides supporting evidence for our interpretation of this pattern as a reflection of the human brain's evolving FNC and maturation process. We also note consistent group differences in other brain state patterns and associated summary measures. Overall, this article suggests that trFNC analysis holds great promise and can be a critical resource in identifying developmental changes in the human brain. Further investigation associating the examined dynamic measures with the anatomical and clinical assessments will augment the breadth of our findings. We conclude that future work in this direction has great potential to conceptualize and discover biomarkers crucial to understanding developmental aberrations and aging effects in spontaneous FC fluctuations.
Supporting information
Figure S1: Time‐resolved functional network connectivity (trFNC) state patterns with unilateral clustering (k = 2). Unilateral clustering results for discovery [UKB (n = 36,461), ABCD‐sample sized matched UKB (UKB‐sm: n = 9617), ABCD (n = 9617)] and replication [HCP (n = 833) and DEVCoG (n = 191)] data sets for model order 2 (k = 2). The lower triangular parts of the matrices represent the cluster centroid patterns marking the quasi‐stable trFNC state patterns of the human brain that reoccur over time. The trFNC state patterns (S1: State 1 and S2: State 2), were matched across the clustering model orders using greedy sorting with correlation as the similarity metric. The upper triangular parts of the matrices represent the matched trFNC state profiles averaged at the domain level to highlight dominant inter‐domain and intra‐domain brain state patterns. Vertical and horizontal black lines partition the brain state patterns by (n = 7) domains based on anatomical and functional properties of the (n = 53) brain components – SC, subcortical, AUD: Auditory, SM: Somatomotor, VIS: Visual, CC: Cognitive Control, DMN: Default Mode Network and CEREB: Cerebellum.
Figure S2: Time‐resolved functional network connectivity (trFNC) state patterns with unilateral clustering (k = 3). Unilateral clustering results for discovery [UKB (n = 36,461), ABCD‐sample sized matched UKB (UKB‐sm: n = 9617), ABCD (n = 9617)] and replication [HCP (n = 833) and DEVCoG (n = 191)] data sets for model order 3 (k = 3). The lower triangular parts of the matrices represent the cluster centroid patterns marking the quasi‐stable trFNC state patterns of the human brain that reoccur over time. The trFNC state patterns (S1: State 1, S2: State 2 and S3: State 3), were matched across the clustering model orders using greedy sorting with correlation as the similarity metric. The upper triangular parts of the matrices represent the matched trFNC state profiles averaged at the domain level to highlight dominant inter‐domain and intra‐domain brain state patterns. Vertical and horizontal black lines partition the brain state patterns by (n = 7) domains based on anatomical and functional properties of the (n = 53) brain components – SC: subcortical, AUD: Auditory, SM: Somatomotor, VIS: Visual, CC: Cognitive Control, DMN: Default Mode Network and CEREB: Cerebellum.
Figure S3: Time‐resolved functional network connectivity (trFNC) state patterns with unilateral clustering (k = 4). Unilateral clustering results for discovery [UKB (n = 36,461), ABCD‐sample sized matched UKB (UKB‐sm: n = 9617), ABCD (n = 9617)] and replication [HCP (n = 833) and DEVCoG (n = 191)] data sets for model order 4 (k = 4). The lower triangular parts of the matrices represent the cluster centroid patterns marking the quasi‐stable trFNC state patterns of the human brain that reoccur over time. The trFNC state patterns (S1: State 1, S2: State 2, S3: State 3, and S4: State 4), were matched across the clustering model orders using greedy sorting with correlation as the similarity metric. The upper triangular parts of the matrices represent the matched trFNC state profiles averaged at the domain level to highlight dominant inter‐domain and intra‐domain brain state patterns. Vertical and horizontal black lines partition the brain state patterns by (n = 7) domains based on anatomical and functional properties of the (n = 53) brain components – SC: subcortical, AUD: Auditory, SM: Somatomotor, VIS: Visual, CC: Cognitive Control, DMN: Default Mode Network and CEREB: Cerebellum.
Figure S4: Time‐resolved functional network connectivity (trFNC) state patterns with unilateral clustering (k = 5). Unilateral clustering results for discovery [UKB (n = 36,461), ABCD‐sample sized matched UKB (UKB‐sm: n = 9617), ABCD (n = 9617)] and replication [HCP (n = 833) and DEVCoG (n = 191)] data sets for model order 5 (k = 5). The lower triangular parts of the matrices represent the cluster centroid patterns marking the quasi‐stable trFNC state patterns of the human brain that reoccur over time. The trFNC state patterns (S1: State 1, S2: State 2, S3: State 3, S4: State 4, and S5: State 5), were matched across the clustering model orders using greedy sorting with correlation as the similarity metric. The upper triangular parts of the matrices represent the matched trFNC state profiles averaged at the domain level to highlight dominant inter‐domain and intra‐domain brain state patterns. Vertical and horizontal black lines partition the brain state patterns by (n = 7) domains based on anatomical and functional properties of the (n = 53) brain components – SC: subcortical, AUD: Auditory, SM: Somatomotor, VIS: Visual, CC: Cognitive Control, DMN: Default Mode Network and CEREB: Cerebellum.
ACKNOWLEDGMENTS
This research was supported by NIH grants (R01MH123610 and R01MH118695) and NSF grant 2112455 to Dr. Vince Calhoun, NIH grants (R01MH121101, R01MH116782, and P20GM144641) to Dr. Tony Wilson, and Natural Science Foundation of China grants (62076157 and 61703253) to Dr. Yuhui Du. The authors acknowledge the contribution of Helen Petropoulos and Jill Fries for preprocessing the Dev‐CoG fMRI data.
Abrol, A. , Fu, Z. , Du, Y. , Wilson, T. W. , Wang, Y.‐P. , Stephen, J. M. , & Calhoun, V. D. (2023). Developmental and aging resting functional magnetic resonance imaging brain state adaptations in adolescents and adults: A large N (>47K) study. Human Brain Mapping, 44(6), 2158–2175. 10.1002/hbm.26200
Funding information National Institutes of Health, Grant/Award Numbers: P20GM144641, R01MH116782, R01MH118695, R01MH121101, R01MH123610; National Science Foundation, Grant/Award Number: 2112455; Natural Science Foundation of China, Grant/Award Numbers: 62076157, 61703253
DATA AVAILABILITY STATEMENT
The fMRI data that support the findings of this article are available to researchers via the following data access procedures—(1) UKB (https://www.ukbiobank.ac.uk/enable-your-research), (2) ABCD (https://nda.nih.gov/abcd/request-access.html), (3) HCP (https://db.humanconnectome.org), and (4) DeVCOG (https://www.mrn.org/common/developmental-chronnecto-genomics). The fMRI data were preprocessed and quality controlled via customized SPM12‐based pipelines. The preprocessed and quality‐controlled fMRI data were decomposed using SC‐gICA using our multidata set generated Neuromark_fMRI_1.0 template. The SC‐gICA pipeline used spatial reference maps, and time‐resolved FC analyses are included in the GIFT software download available at http://trendscenter.org/software/gift and also at http://trendscenter.org/data. The custom scripts used for the time‐resolved analysis can also be made available upon request to the corresponding author.
REFERENCES
- Abrol, A. , Chaze, C. , Damaraju, E. , & Calhoun, V. D. (2016, August 16‐20). The chronnectome: Evaluating replicability of dynamic connectivity patterns in 7500 resting fMRI datasets [Conference presentation]. 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). Orlando, FL, USA. [DOI] [PubMed]
- Abrol, A. , Damaraju, E. , Miller, R. L. , Stephen, J. M. , Claus, E. D. , Mayer, A. R. , & Calhoun, V. D. (2017). Replicability of time‐varying connectivity patterns in large resting state fMRI samples. NeuroImage, 163, 160–176. 10.1016/j.neuroimage.2017.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, E. A. , Damaraju, E. , Plis, S. M. , Erhardt, E. B. , Eichele, T. , & Calhoun, V. D. (2014). Tracking whole‐brain connectivity dynamics in the resting state. Cerebral Cortex, 24(3), 663–676. 10.1093/cercor/bhs352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun, V. D. , & Adalı, T. (2012). Multisubject independent component analysis of fMRI: A decade of intrinsic networks, default mode, and neurodiagnostic discovery. IEEE Reviews in Biomedical Engineering, 5, 60–73. 10.1109/rbme.2012.2211076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun, V. D. , Adali, T. , Pearlson, G. D. , & Pekar, J. J. (2001). A method for making group inferences from functional MRI data using independent component analysis. Human Brain Mapping, 14(3), 140–151. 10.1002/hbm.1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun, V. D. , Miller, R. , Pearlson, G. , & Adalı, T. (2014). The chronnectome: Time‐varying connectivity networks as the next frontier in fMRI data discovery. Neuron, 84(2), 262–274. 10.1016/j.neuron.2014.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey, B. J. , Cannonier, T. , Conley, M. I. , Cohen, A. O. , Barch, D. M. , Heitzeg, M. M. , Soules, M. E. , Teslovich, T. , Dellarco, D. V. , Garavan, H. , Orr, C. A. , Wager, T. D. , Banich, M. T. , Speer, N. K. , Sutherland, M. T. , Riedel, M. C. , Dick, A. S. , Bjork, J. M. , Thomas, K. M. , … Dale, A. M. (2018). The adolescent brain cognitive development (ABCD) study: Imaging acquisition across 21 sites. Developmental Cognitive Neuroscience, 32, 43–54. 10.1016/j.dcn.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Wang, W. , Zhao, X. , Sha, M. , Liu, Y. , Zhang, X. , Ma, J. , Ni, H. , & Ming, D. (2017). Age‐related decline in the variation of dynamic functional connectivity: A resting state analysis. Frontiers in Aging Neuroscience, 9, 203. 10.3389/fnagi.2017.00203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRamus, T. , Iraji, A. , Fu, Z. , Silva, R. , Stephen, J. , Wilson, T. W. , Wang, Y. P. , Du, Y. , Liu, J. , & Calhoun, V. (2021, October 25‐27). Stability of functional network connectivity (FNC) values across multiple spatial normalization pipelines in spatially constrained independent component analysis [Conference presentation]. 2021 IEEE 21st International Conference on Bioinformatics and Bioengineering (BIBE). Kragujevac, Serbia.
- Du, Y. , & Fan, Y. (2013). Group information guided ICA for fMRI data analysis. NeuroImage, 69, 157–197. 10.1016/j.neuroimage.2012.11.008 [DOI] [PubMed] [Google Scholar]
- Du, Y. , Fu, Z. , Sui, J. , Gao, S. , Xing, Y. , Lin, D. , Salman, M. , Abrol, A. , Rahaman, M. A. , Chen, J. , Hong, L. E. , Kochunov, P. , Osuch, E. A. , & Calhoun, V. D. (2020). NeuroMark: An automated and adaptive ICA based pipeline to identify reproducible fMRI markers of brain disorders. NeuroImage: Clinical, 28, 102375. 10.1016/j.nicl.2020.102375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Y. , Fu, Z. , Xing, Y. , Lin, D. , Pearlson, G. , Kochunov, P. , Hong, L. E. , Qi, S. , Salman, M. , Abrol, A. , & Calhoun, V. D. (2021). Evidence of shared and distinct functional and structural brain signatures in schizophrenia and autism spectrum disorder. Communications Biology, 4(1), 1073. 10.1038/s42003-021-02592-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda, M. , Iraji, A. , & Calhoun, V. D. (2022). Spatially constrained ICA enables robust detection of schizophrenia from very short resting‐state fMRI. Annual International Conference of IEEE Engineering in Medicine and Biological Society, 2022, 1867–1870. 10.1109/embc48229.2022.9871305 [DOI] [PubMed] [Google Scholar]
- Ernst, M. , Torrisi, S. , Balderston, N. , Grillon, C. , & Hale, E. A. (2015). fMRI functional connectivity applied to adolescent neurodevelopment. Annual Review of Clinical Psychology, 11, 361–377. 10.1146/annurev-clinpsy-032814-112753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escrichs, A. , Biarnes, C. , Garre‐Olmo, J. , Fernández‐Real, J. M. , Ramos, R. , Pamplona, R. , Brugada, R. , Serena, J. , Ramió‐Torrentà, L. , Coll‐De‐Tuero, G. , Gallart, L. , Barretina, J. , Vilanova, J. C. , Mayneris‐Perxachs, J. , Essig, M. , Figley, C. R. , Pedraza, S. , Puig, J. , & Deco, G. (2021). Whole‐brain dynamics in aging: Disruptions in functional connectivity and the role of the rich club. Cerebral Cortex, 31(5), 2466–2481. 10.1093/cercor/bhaa367 [DOI] [PubMed] [Google Scholar]
- Faghiri, A. , Stephen, J. M. , Wang, Y. P. , Wilson, T. W. , & Calhoun, V. D. (2018, Mar). Changing brain connectivity dynamics: From early childhood to adulthood. Human Brain Mapping, 39(3), 1108–1117. 10.1002/hbm.23896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair, D. A. , Dosenbach, N. U. F. , Church, J. A. , Cohen, A. L. , Brahmbhatt, S. , Miezin, F. M. , Barch, D. M. , Raichle, M. E. , Petersen, S. E. , & Schlaggar, B. L. (2007). Development of distinct control networks through segregation and integration. Proceedings of the National Academy of Sciences, 104(33), 13507–13512. 10.1073/pnas.0705843104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjell, A. M. , Sneve, M. H. , Grydeland, H. , Storsve, A. B. , & Walhovd, K. B. (2017). The disconnected brain and executive function decline in aging. Cerebral Cortex, 27(3), 2303–2317. 10.1093/cercor/bhw082 [DOI] [PubMed] [Google Scholar]
- Friston, K. J. (2011). Functional and effective connectivity: A review. Brain Connectivity, 1(1), 13–36. 10.1089/brain.2011.0008 [DOI] [PubMed] [Google Scholar]
- Fu, Z. , Liu, J. , Salman, M. , Sui, J. , & Calhoun, V. (2022). Functional connectivity uniqueness and variability? A signature of cognitive and psychiatric problems in children . Research Square, rs‐1514598. 10.21203/rs.3.rs-1514598/v1 [DOI] [Google Scholar]
- Giedd, J. N. (2008). The teen brain: Insights from neuroimaging. The Journal of Adolescent Health, 42(4), 335–343. 10.1016/j.jadohealth.2008.01.007 [DOI] [PubMed] [Google Scholar]
- Grady, C. , Sarraf, S. , Saverino, C. , & Campbell, K. (2016). Age differences in the functional interactions among the default, frontoparietal control, and dorsal attention networks. Neurobiology of Aging, 41, 159–172. 10.1016/j.neurobiolaging.2016.02.020 [DOI] [PubMed] [Google Scholar]
- Holmes, A. J. , Hollinshead, M. O. , O'Keefe, T. M. , Petrov, V. I. , Fariello, G. R. , Wald, L. L. , Fischl, B. , Rosen, B. R. , Mair, R. W. , Roffman, J. L. , Smoller, J. W. , & Buckner, R. L. (2015). Brain genomics superstruct project initial data release with structural, functional, and behavioral measures. Scientific Data, 2(1), 150031. 10.1038/sdata.2015.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison, R. M. , & Morton, J. B. (2015, Apr 29). Tracking the brain's functional coupling dynamics over development. The Journal of Neuroscience, 35(17), 6849–6859. 10.1523/jneurosci.4638-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison, R. M. , Womelsdorf, T. , Allen, E. A. , Bandettini, P. A. , Calhoun, V. D. , Corbetta, M. , Della Penna, S. , Duyn, J. H. , Glover, G. H. , Gonzalez‐Castillo, J. , Handwerker, D. A. , Keilholz, S. , Kiviniemi, V. , Leopold, D. A. , de Pasquale, F. , Sporns, O. , Walter, M. , & Chang, C. (2013). Dynamic functional connectivity: Promise, issues, and interpretations. NeuroImage, 80, 360–378. 10.1016/j.neuroimage.2013.05.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafri, M. J. , Pearlson, G. D. , Stevens, M. , & Calhoun, V. D. (2008). A method for functional network connectivity among spatially independent resting‐state components in schizophrenia. NeuroImage, 39(4), 1666–1681. 10.1016/j.neuroimage.2007.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockwitz, C. , & Caspers, S. (2021, May). Resting‐state networks in the course of aging‐differential insights from studies across the lifespan vs. amongst the old. Pflügers Archiv, 473(5), 793–803. 10.1007/s00424-021-02520-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolles, D. D. , van Buchem, M. A. , Crone, E. A. , & Rombouts, S. A. (2011). A comprehensive study of whole‐brain functional connectivity in children and young adults. Cerebral Cortex, 21(2), 385–391. 10.1093/cercor/bhq104 [DOI] [PubMed] [Google Scholar]
- Li, H. J. , Hou, X. H. , Liu, H. H. , Yue, C. L. , Lu, G. M. , & Zuo, X. N. (2015). Putting age‐related task activation into large‐scale brain networks: A meta‐analysis of 114 fMRI studies on healthy aging. Neuroscience and Biobehavioral Reviews, 57, 156–174. 10.1016/j.neubiorev.2015.08.013 [DOI] [PubMed] [Google Scholar]
- Liu, C. , Xue, J. , Cheng, X. , Zhan, W. , Xiong, X. , & Wang, B. (2019). Tracking the brain state transition process of dynamic function connectivity based on resting state fMRI. Computational Intelligence and Neuroscience, 2019, 9027803. 10.1155/2019/9027803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Vicente, M. , Agcaoglu, O. , Pérez‐Crespo, L. , Estévez‐López, F. , Heredia‐Genestar, J. M. , Mulder, R. H. , Flournoy, J. C. , van Duijvenvoorde, A. C. K. , Güroğlu, B. , White, T. , Calhoun, V. , Tiemeier, H. , & Muetzel, R. L. (2021). Developmental changes in dynamic functional connectivity from childhood into adolescence [original research]. Frontiers in Systems Neuroscience, 15, 724805. 10.3389/fnsys.2021.724805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurie, D. J. , Kessler, D. , Bassett, D. S. , Betzel, R. F. , Breakspear, M. , Kheilholz, S. , Kucyi, A. , Liégeois, R. , Lindquist, M. A. , McIntosh, A. R. , Poldrack, R. A. , Shine, J. M. , Thompson, W. H. , Bielczyk, N. Z. , Douw, L. , Kraft, D. , Miller, R. L. , Muthuraman, M. , Pasquini, L. , … Calhoun, V. D. (2020). Questions and controversies in the study of time‐varying functional connectivity in resting fMRI. Network Neuroscience (Cambridge, Mass.), 4(1), 30–69. 10.1162/netn_a_00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X. , Wu, X. , & Shi, Y. (2020). Changes of dynamic functional connectivity associated with maturity in late preterm infants. Frontiers in Pediatrics, 8, 412. 10.3389/fped.2020.00412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek, S. , Tervo‐Clemmens, B. , Calabro, F. J. , Montez, D. F. , Kay, B. P. , Hatoum, A. S. , Donohue, M. R. , Foran, W. , Miller, R. L. , Hendrickson, T. J. , Malone, S. M. , Kandala, S. , Feczko, E. , Miranda‐Dominguez, O. , Graham, A. M. , Earl, E. A. , Perrone, A. J. , Cordova, M. , Doyle, O. , … Dosenbach, N. U. F. (2022). Reproducible brain‐wide association studies require thousands of individuals. Nature, 603(7902), 654–660. 10.1038/s41586-022-04492-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusak, H. A. , Calhoun, V. D. , Brown, S. , Crespo, L. M. , Sala‐Hamrick, K. , Gotlib, I. H. , & Thomason, M. E. (2017). Dynamic functional connectivity of neurocognitive networks in children. Human Brain Mapping, 38(1), 97–108. 10.1002/hbm.23346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon, V. (2013). Developmental pathways to functional brain networks: Emerging principles. Trends in Cognitive Sciences, 17(12), 627–640. 10.1016/j.tics.2013.09.015 [DOI] [PubMed] [Google Scholar]
- Meunier, D. , Stamatakis, E. A. , & Tyler, L. K. (2014). Age‐related functional reorganization, structural changes, and preserved cognition. Neurobiology of Aging, 35(1), 42–54. 10.1016/j.neurobiolaging.2013.07.003 [DOI] [PubMed] [Google Scholar]
- Miller, K. L. , Alfaro‐Almagro, F. , Bangerter, N. K. , Thomas, D. L. , Yacoub, E. , Xu, J. , Bartsch, A. J. , Jbabdi, S. , Sotiropoulos, S. N. , Andersson, J. L. R. , Griffanti, L. , Douaud, G. , Okell, T. W. , Weale, P. , Dragonu, I. , Garratt, S. , Hudson, S. , Collins, R. , Jenkinson, M. , … Smith, S. M. (2016). Multimodal population brain imaging in the UK biobank prospective epidemiological study. Nature Neuroscience, 19(11), 1523–1536. 10.1038/nn.4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, R. L. , Abrol, A. , Adali, T. , Levin‐Schwarz, Y. , & Calhoun, V. D. (2018). Resting‐state fMRI dynamics and null models: Perspectives, sampling variability, and simulations [perspective]. Frontiers in Neuroscience, 12, 551. 10.3389/fnins.2018.00551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual‐Marqui, R. D. , Michel, C. M. , & Lehmann, D. (1995). Segmentation of brain electrical activity into microstates: Model estimation and validation. IEEE Transactions on Biomedical Engineering, 42(7), 658–665. 10.1109/10.391164 [DOI] [PubMed] [Google Scholar]
- Power, J. D. , Barnes, K. A. , Snyder, A. Z. , Schlaggar, B. L. , & Petersen, S. E. (2012). Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage, 59(3), 2142–2154. 10.1016/j.neuroimage.2011.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power, J. D. , Fair, D. A. , Schlaggar, B. L. , & Petersen, S. E. (2010). The development of human functional brain networks. Neuron, 67(5), 735–748. 10.1016/j.neuron.2010.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preti, M. G. , Bolton, T. A. , & Van De Ville, D. (2017). The dynamic functional connectome: State‐of‐the‐art and perspectives. NeuroImage, 160, 41–54. 10.1016/j.neuroimage.2016.12.061 [DOI] [PubMed] [Google Scholar]
- Pringle, J. , Mills, K. , McAteer, J. , Jepson, R. , Hogg, E. , Anand, N. , & Blakemore, S.‐J. (2016). A systematic review of adolescent physiological development and its relationship with health‐related behaviour: A protocol. Systematic Reviews, 5(1), 3. 10.1186/s13643-015-0173-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, J. , Chen, S.‐G. , Hu, D. , Zeng, L.‐L. , Fan, Y.‐M. , Chen, X.‐P. , & Shen, H. (2015). Predicting individual brain maturity using dynamic functional connectivity [original research]. Frontiers in Human Neuroscience, 9, 418. 10.3389/fnhum.2015.00418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid, B. , Blanken, L. M. E. , Muetzel, R. L. , Miller, R. , Damaraju, E. , Arbabshirani, M. R. , Erhardt, E. B. , Verhulst, F. C. , van der Lugt, A. , Jaddoe, V. W. V. , Tiemeier, H. , White, T. , & Calhoun, V. (2018). Connectivity dynamics in typical development and its relationship to autistic traits and autism spectrum disorder. Human Brain Mapping, 39(8), 3127–3142. 10.1002/hbm.24064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoğlu, U. , Pearlson, G. D. , Kiehl, K. A. , Wang, Y. M. , Michael, A. M. , & Calhoun, V. D. (2010). A method for evaluating dynamic functional network connectivity and task‐modulation: Application to schizophrenia. Magma, 23(5–6), 351–366. 10.1007/s10334-010-0197-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala‐Llonch, R. , Bartrés‐Faz, D. , & Junqué, C. (2015). Reorganization of brain networks in aging: A review of functional connectivity studies. Frontiers in Psychology, 6, 663. 10.3389/fpsyg.2015.00663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, J. R. , Biazoli, C. E., Jr. , Salum, G. A. , Gadelha, A. , Crossley, N. , Satterthwaite, T. D. , Vieira, G. , Zugman, A. , Picon, F. A. , Pan, P. M. , Hoexter, M. Q. , Anés, M. , Moura, L. M. , Del'aquilla, M. A. , Amaro, E., Jr. , McGuire, P. , Lacerda, A. L. , Rohde, L. A. , Miguel, E. C. , … Bressan, R. A. (2015). Temporal stability of network centrality in control and default mode networks: Specific associations with externalizing psychopathology in children and adolescents. Human Brain Mapping, 36(12), 4926–4937. 10.1002/hbm.22985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite, T. D. , Wolf, D. H. , Loughead, J. , Ruparel, K. , Elliott, M. A. , Hakonarson, H. , Gur, R. C. , & Gur, R. E. (2012). Impact of in‐scanner head motion on multiple measures of functional connectivity: Relevance for studies of neurodevelopment in youth. NeuroImage, 60(1), 623–632. 10.1016/j.neuroimage.2011.12.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer, S. M. , Azzopardi, P. S. , Wickremarathne, D. , & Patton, G. C. (2018). The age of adolescence. The Lancet Child & Adolescent Health, 2(3), 223–228. 10.1016/S2352-4642(18)30022-1 [DOI] [PubMed] [Google Scholar]
- Siman‐Tov, T. , Bosak, N. , Sprecher, E. , Paz, R. , Eran, A. , Aharon‐Peretz, J. , & Kahn, I. (2016). Early age‐related functional connectivity decline in high‐order cognitive networks. Frontiers in Aging Neuroscience, 8, 330. 10.3389/fnagi.2016.00330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen, J. M. , Solis, I. , Janowich, J. , Stern, M. , Frenzel, M. R. , Eastman, J. A. , Mills, M. S. , Embury, C. M. , Coolidge, N. M. , Heinrichs‐Graham, E. , Mayer, A. , Liu, J. , Wang, Y. P. , Wilson, T. W. , & Calhoun, V. D. (2021). The developmental chronnecto‐genomics (Dev‐CoG) study: A multimodal study on the developing brain. NeuroImage, 225, 117438. 10.1016/j.neuroimage.2020.117438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura, M. (2016). Functional neuroimaging of normal aging: Declining brain, adapting brain. Ageing Research Reviews, 30, 61–72. 10.1016/j.arr.2016.02.006 [DOI] [PubMed] [Google Scholar]
- Supekar, K. , Musen, M. , & Menon, V. (2009). Development of large‐scale functional brain networks in children. PLoS Biology, 7(7), e1000157. 10.1371/journal.pbio.1000157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, L. , Li, Q. , Wang, C. , & Yu, J. (2018). Changes in dynamic functional connections with aging. NeuroImage, 172, 31–39. 10.1016/j.neuroimage.2018.01.040 [DOI] [PubMed] [Google Scholar]
- van den Heuvel, M. P. , & Hulshoff Pol, H. E. (2010). Exploring the brain network: A review on resting‐state fMRI functional connectivity. European Neuropsychopharmacology, 20(8), 519–534. 10.1016/j.euroneuro.2010.03.008 [DOI] [PubMed] [Google Scholar]
- Van Essen, D. C. , Smith, S. M. , Barch, D. M. , Behrens, T. E. J. , Yacoub, E. , & Ugurbil, K. (2013). The WU‐Minn human connectome project: An overview. NeuroImage, 80, 62–79. 10.1016/j.neuroimage.2013.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, T. , El Marroun, H. , Nijs, I. , Schmidt, M. , van der Lugt, A. , Wielopolki, P. A. , Jaddoe, V. W. , Hofman, A. , Krestin, G. P. , Tiemeier, H. , & Verhulst, F. C. (2013). Pediatric population‐based neuroimaging and the generation R study: The intersection of developmental neuroscience and epidemiology. European Journal of Epidemiology, 28(1), 99–111. 10.1007/s10654-013-9768-0 [DOI] [PubMed] [Google Scholar]
- Yin, D. , Liu, W. , Zeljic, K. , Wang, Z. , Lv, Q. , Fan, M. , Cheng, W. , & Wang, Z. (2016). Dissociable changes of frontal and parietal cortices in inherent functional flexibility across the human life span. The Journal of Neuroscience, 36(39), 10060–10074. 10.1523/jneurosci.1476-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, X. N. , Kelly, C. , Di Martino, A. , Mennes, M. , Margulies, D. S. , Bangaru, S. , Grzadzinski, R. , Evans, A. C. , Zang, Y. F. , Castellanos, F. X. , & Milham, M. P. (2010). Growing together and growing apart: Regional and sex differences in the lifespan developmental trajectories of functional homotopy. The Journal of Neuroscience, 30(45), 15034–15043. 10.1523/jneurosci.2612-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Time‐resolved functional network connectivity (trFNC) state patterns with unilateral clustering (k = 2). Unilateral clustering results for discovery [UKB (n = 36,461), ABCD‐sample sized matched UKB (UKB‐sm: n = 9617), ABCD (n = 9617)] and replication [HCP (n = 833) and DEVCoG (n = 191)] data sets for model order 2 (k = 2). The lower triangular parts of the matrices represent the cluster centroid patterns marking the quasi‐stable trFNC state patterns of the human brain that reoccur over time. The trFNC state patterns (S1: State 1 and S2: State 2), were matched across the clustering model orders using greedy sorting with correlation as the similarity metric. The upper triangular parts of the matrices represent the matched trFNC state profiles averaged at the domain level to highlight dominant inter‐domain and intra‐domain brain state patterns. Vertical and horizontal black lines partition the brain state patterns by (n = 7) domains based on anatomical and functional properties of the (n = 53) brain components – SC, subcortical, AUD: Auditory, SM: Somatomotor, VIS: Visual, CC: Cognitive Control, DMN: Default Mode Network and CEREB: Cerebellum.
Figure S2: Time‐resolved functional network connectivity (trFNC) state patterns with unilateral clustering (k = 3). Unilateral clustering results for discovery [UKB (n = 36,461), ABCD‐sample sized matched UKB (UKB‐sm: n = 9617), ABCD (n = 9617)] and replication [HCP (n = 833) and DEVCoG (n = 191)] data sets for model order 3 (k = 3). The lower triangular parts of the matrices represent the cluster centroid patterns marking the quasi‐stable trFNC state patterns of the human brain that reoccur over time. The trFNC state patterns (S1: State 1, S2: State 2 and S3: State 3), were matched across the clustering model orders using greedy sorting with correlation as the similarity metric. The upper triangular parts of the matrices represent the matched trFNC state profiles averaged at the domain level to highlight dominant inter‐domain and intra‐domain brain state patterns. Vertical and horizontal black lines partition the brain state patterns by (n = 7) domains based on anatomical and functional properties of the (n = 53) brain components – SC: subcortical, AUD: Auditory, SM: Somatomotor, VIS: Visual, CC: Cognitive Control, DMN: Default Mode Network and CEREB: Cerebellum.
Figure S3: Time‐resolved functional network connectivity (trFNC) state patterns with unilateral clustering (k = 4). Unilateral clustering results for discovery [UKB (n = 36,461), ABCD‐sample sized matched UKB (UKB‐sm: n = 9617), ABCD (n = 9617)] and replication [HCP (n = 833) and DEVCoG (n = 191)] data sets for model order 4 (k = 4). The lower triangular parts of the matrices represent the cluster centroid patterns marking the quasi‐stable trFNC state patterns of the human brain that reoccur over time. The trFNC state patterns (S1: State 1, S2: State 2, S3: State 3, and S4: State 4), were matched across the clustering model orders using greedy sorting with correlation as the similarity metric. The upper triangular parts of the matrices represent the matched trFNC state profiles averaged at the domain level to highlight dominant inter‐domain and intra‐domain brain state patterns. Vertical and horizontal black lines partition the brain state patterns by (n = 7) domains based on anatomical and functional properties of the (n = 53) brain components – SC: subcortical, AUD: Auditory, SM: Somatomotor, VIS: Visual, CC: Cognitive Control, DMN: Default Mode Network and CEREB: Cerebellum.
Figure S4: Time‐resolved functional network connectivity (trFNC) state patterns with unilateral clustering (k = 5). Unilateral clustering results for discovery [UKB (n = 36,461), ABCD‐sample sized matched UKB (UKB‐sm: n = 9617), ABCD (n = 9617)] and replication [HCP (n = 833) and DEVCoG (n = 191)] data sets for model order 5 (k = 5). The lower triangular parts of the matrices represent the cluster centroid patterns marking the quasi‐stable trFNC state patterns of the human brain that reoccur over time. The trFNC state patterns (S1: State 1, S2: State 2, S3: State 3, S4: State 4, and S5: State 5), were matched across the clustering model orders using greedy sorting with correlation as the similarity metric. The upper triangular parts of the matrices represent the matched trFNC state profiles averaged at the domain level to highlight dominant inter‐domain and intra‐domain brain state patterns. Vertical and horizontal black lines partition the brain state patterns by (n = 7) domains based on anatomical and functional properties of the (n = 53) brain components – SC: subcortical, AUD: Auditory, SM: Somatomotor, VIS: Visual, CC: Cognitive Control, DMN: Default Mode Network and CEREB: Cerebellum.
Data Availability Statement
The fMRI data that support the findings of this article are available to researchers via the following data access procedures—(1) UKB (https://www.ukbiobank.ac.uk/enable-your-research), (2) ABCD (https://nda.nih.gov/abcd/request-access.html), (3) HCP (https://db.humanconnectome.org), and (4) DeVCOG (https://www.mrn.org/common/developmental-chronnecto-genomics). The fMRI data were preprocessed and quality controlled via customized SPM12‐based pipelines. The preprocessed and quality‐controlled fMRI data were decomposed using SC‐gICA using our multidata set generated Neuromark_fMRI_1.0 template. The SC‐gICA pipeline used spatial reference maps, and time‐resolved FC analyses are included in the GIFT software download available at http://trendscenter.org/software/gift and also at http://trendscenter.org/data. The custom scripts used for the time‐resolved analysis can also be made available upon request to the corresponding author.


