Abstract
HIV viral protein R (Vpr) contributes to HIV-associated nephropathy, induces cell cycle arrest, and alters expression of mineralocorticoid-responsive genes. To investigate Vpr modulation of aldosterone-mediated regulation of the expression of Na-Cl cotransporter Slc12a3 and sodium reabsorption in distal nephron segments, we performed single-nucleus RNA sequencing of kidney cortices of wild-type (WT) and Vpr transgenic (Vpr Tg) mice on low sodium diet and a WT mouse on regular chow. In Vpr Tg mouse, Slc12a3 expression was downregulated in connecting tubules (CNT) and late distal convoluted tubules (DCT2) but not in early DCT (DCT1). Mineralocorticoid receptor (Nr3c2) expression was higher in CNT and DCT2 than in DCT1. DCT1 cell percentage in salt-depleted Vpr Tg was lower (1.8%) compared with salt-depleted or salt-replete WT (3.7% and 3.6%, respectively). Sub-clustering confirmed that Pvalb+ DCT1 subcluster had fewer cells in Vpr Tg (0.95%) compared with salt-depleted or salt-replete WT (2.9% and 3.1%, respectively). Autophagy and protein ubiquitination genes were upregulated in DCT1 subcluster of Vpr Tg mice. In situ hybridization and immunohistochemistry demonstrated fewer Slc12a3+ Pvalb+ DCT1 segments in Vpr Tg mice. Downregulation of Slc12a3 and higher levels of Nr3c2 in the CNT/DCT2 suggested that the aldosterone-mediated upregulation of Slc12a3 with salt depletion was most prominently inhibited by Vpr in the CNT/DCT2. These observations demonstrate that the salt-wasting effect of Vpr in Vpr Tg mice is mediated by loss of Slc12a3+ Pvalb+ DCT1 segments. The downregulation of Slc12a3 occurred in more distal segments rather than in DCT1.
Keywords: viral protein R, mineralocorticoid receptor, distal convoluted tubule, connecting tubule, in situ hybridization
Introduction
The distal convoluted tubule (DCT) is the shortest segment of the distal nephron. Its physiological roles include maintaining sodium homeostasis, under the influence of aldosterone. The DCT specifically expresses the thiazide-sensitive protein NCC (sodium-chloride cotransporter) encoded by SLC12A3, which is responsible for reabsorption of luminal Na+ into DCT cells. The abundance and activity of NCC are influenced by several hormones, including aldosterone, angiotensin II, insulin, and vasopressin.1–8
The DCT shows vulnerability as well as adaptability to conditions that interfere with Na reabsorption. In a study by Kaissling and colleagues, minipumps containing furosemide were implanted in rats for six days. The rats drank large volumes of salt solution ad libitum to compensate for urine sodium losses. This treatment resulted in enlargement of kidney cortices with histologic evidence of proliferation of distal convoluted tubules.9
In 1996, Loffing and colleagues showed that in thiazide-treated rats, the DCT epithelium lost the structural characteristics of electrolyte transporting epithelia with cells in various stages of apoptosis. Transcripts of the thiazide-sensitive sodium-chloride cotransporter (rTSC1) were greatly reduced in kidney cortex homogenates and almost entirely absent in damaged DCT cells. Focal inflammatory infiltrates were localized to the DCT. All other tubular segments were unaffected by this intervention. Thus, the inhibition of apical NaCl entry into DCT cells by thiazides is associated with apoptosis of DCT cells and focal peritubular inflammation.10
HIV-associated nephropathy (HIVAN) involves dysfunction of both glomeruli and tubules. In the past, attention has been largely focused on HIVAN glomerulopathy. Recently, tubular injury has drawn increased attention because of hyponatremia due to sodium wasting11, is common in hospitalized HIV/AIDS patients.12
A recent study by our group demonstrated a reduction of NCC protein in the cortex of HIV viral protein R (Vpr) transgenic mice. These mice showed increased urine sodium loss despite being on a low sodium diet for 4 days. The NCC protein reduction in Vpr Tg mice was mediated by the downregulation of Slc12a3 mRNA expression in DCT cells.13
Since HIV-1 Vpr causes renal cell dysfunction and contributes to HIVAN11, we wished to explore mechanisms where by Vpr compromises DCT function. Therefore, we performed single-nucleus RNA sequencing (snRNA-seq) on kidney cortices of mice that were kept on a sodium-deficient diet to characterize the effects of Vpr across the nephron, particularly, on distal tubular cells at single-cell resolution.
Methods
Generation and maintenance of Vpr transgenic mice
The generation, characterization, and genotyping of tetracycline repressible Vpr transgenic mice have been described previously.13, 14 Detailed information is provided in Supplementary materials.
Preparation of mouse kidney cortex samples for snRNA-seq
Out of seven WT and thirteen Vpr Tg mice, we selected one representative sample from each group for single-nucleus RNA sequencing. We also included a salt-replete WT (WT_SR) kidney cortex sample from an FVB mouse for snRAN-seq. Nuclei from frozen mouse kidney outer cortex tissue samples were prepared following the protocol of Kirita et al.15 Briefly, ~ 8 mm3 tissue fragments were cut by razor blade in the EZlysis buffer (#NUC101–1KT, Sigma, Darmstadt, Germany) and homogenized 30 times using a Dounce homogenizer equipped with a loose pestle and 5 times with a tight pestle. After 5 min of incubation, the homogenate was passed through a 40 μm filter (PluriSelect, El Cajon, CA) and centrifuged at 500×g at 4°C for 5 min. The pellet was washed with EZlysis buffer (Sigma Aldrich, St Louis, MO) and again centrifuged at 500×g at 4°C for 5 min. The pellet was resuspended in Dulbecco’s phosphate-buffered saline supplemented with 1% fetal bovine serum and passed through a 5 μm filter (PuriSelect, El Cajon, CA) for obtaining the final nuclear suspension.
The prepared nuclei were loaded onto a 10x Chromium Chip G (10X Genomics, San Francisco, CA) for formation of gel beads in emulsion (GEM). Single nuclear isolation, RNA capture, cDNA preparation, and library preparation were accomplished following the manufacturer’s protocol (Chromium Next GEM Single Cell 3’ Reagent Kit, v3.1 chemistry, 10x Genomics, Pleasanton, CA).
The prepared cDNA libraries were sequenced at the Frederick National Laboratory for Cancer Research Sequencing Facility (NCI, Frederick, MD). Salt-depleted Vpr Tg and WT mouse renal cortical nuclear samples had sequencing yields of more than 630 million reads per sample with more than 95% of bases in the barcode regions had Q30 or above, and at least 93% of bases in the RNA read had Q30 or above, and more than 95% of bases in the unique molecular identifier (UMI) had Q30 or above. The salt-replete WT (WT_SR) sample had sequencing yield of more than 249 million reads with 96% of bases in the barcode regions had Q30 or above, and 90.9% of bases in the RNA region had Q30 and above. More than 95% of bases in WT_SR sample had Q30 or above. Analysis was performed with the Cell Ranger v5.0.0 software using the default parameters including the pre-mRNA analysis (10x Genomics, Pleasanton, CA). The reference was built from the mm10 (Mus musculus) reference genome complemented with reported HIV-1 viral sequences.
snRNA-seq data analysis
The cellranger output files of individual 10x Genomic datasets were used as input for SoupX16 (version 1.6.1) (https://github.com/constantAmateur/SoupX) to estimate and correct the contamination fraction for ambient or cell-free RNA released during the process of lysing cells for nuclear preparation. The 10x datasets corrected for ambient RNA were converted into Seurat objects by excluding nuclei with (a) less than 200 or more than 4000 genes, (b) total RNA count of more than 15000, and (c) more than 20% of mitochondrial transcripts (Seurat version 4.1.1)17. The data were adjusted using the mitochondrial transcript percentage of each nucleus, based on the 2000 most variable nuclear-encoded genes.
Potential cell doublets were predicted and removed by DoubletFinder18 (version 2.0.3) (https://github.com/chris-mcginnis-ucsf/DoubletFinder) by assuming a doublet rate of 7.6% and using ten principal components, pN and pK values of 0.25 and 0.005, respectively (Supplementary Figure 1). The three datasets (WT, WT_SR and Vpr Tg) were integrated using an anchor-based integration. After batch correction using Harmony19 (version 0.1.0) (https://github.com/immunogenomics/harmony) (Supplementary Figure 2), clustering was done with 30 principal components and a resolution of 0.5. Subclustering of distal tubule data was performed by subsetting three distal tubule cell clusters – DCT, CNT and principal cell (PC) through identifying the most variable genes in these three clusters in each sample, followed by anchor-based integration, and by repeating batch correction.
Details of tissue microarray preparation, in situ hybridization, confocal microscopy & image analysis, immunohistochemistry (IHC) of parvalbumin, TUNEL assay to detect apoptotic cells and quantification of IHC and TUNEL images are mentioned in supplementary materials.
Results
Reduced DCT1 cells in Vpr Tg mouse
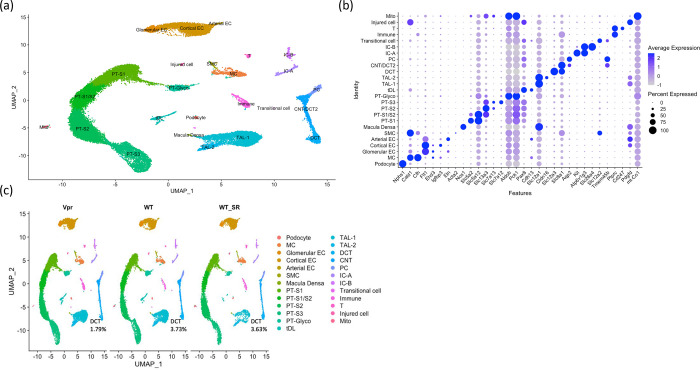
We performed snRNA-seq analysis of salt-depleted WT and Vpr Tg mouse kidney cortex samples and a salt-replete WT (WT_SR) mouse kidney cortex samples to aid clustering of cell types. Unsupervised clustering in Seurat gave 25 cell clusters which were annotated using known canonical cell type marker genes (Figure 1). We identified 25 cell types: six proximal tubule cell clusters (labeled as PT-S1, PT-S1/S2, PT-S2, PT-S3, PT-Glyco and Mito), three distal tubular cell clusters (DCT, CNT/DCT2 and PC), three endothelial cell clusters (glomerular EC, cortical EC and arterial EC), two intercalated cell clusters (IC-A and IC-B), two thick ascending limb clusters (TAL-1 and TAL-2), and one cluster each of podocytes, mesangial cells (MC), smooth muscle cells (SMC), injured cells (expressing injury marker genes), macula densa cells, thin descending limb cells, transitional cells, T cells and other immune cells.
Figure 1. snRNA-seq of mouse kidney cortex samples (salt-depleted WT, Vpr Tg and WT_SR).
(a) UMAP projection of 10x datasets from three samples after anchor-based integration.
(b) Dot plot showing the marker genes to identify the cell type for each cluster. MC = mesangial cell; Glomerular EC = glomerular endothelial cell; Cortical EC = cortical endothelial cell; Arterial EC = arterial endothelial cell; SMC = smooth muscle cell; PT-S1 = proximal tubule, S1 segment; PT-S1/S2 = proximal tubule, S1 and S2 segments; PT-S3 = proximal tubule, S3 segment; PT-Glyco = proximal tubule cells rich in glycolytic enzymes; tDL = thin descending limb; TAL-1 = thick ascending limb 1; TAL-2 = thick ascending limb 2; DCT = distal convoluted tubule; CNT/DCT2 = connecting tubule/type 2 distal convoluted tubule; PC = principal cell; IC-A = type A intercalated cell; IC-B = type B intercalated cell; Immune = immune cell; T = T cell; Mito = cells enriched in mitochondrial transcripts.
(c) UMAP projections of individual sample showing the percentages of DCT cells to overall cells in each sample.
For each sample, we calculated the percentages of each cell type within total cell number. The WT and Vpr Tg samples showed similar cell percentages across most of the identified renal epithelial cell clusters. A notable exception was found in the DCT cluster which showed a reduced fraction in the Vpr Tg sample (1.79 % as compared with 3.73 % and 3.63% in WT and WT_SR samples, respectively) (Figure 1c and Supplementary Table 1).
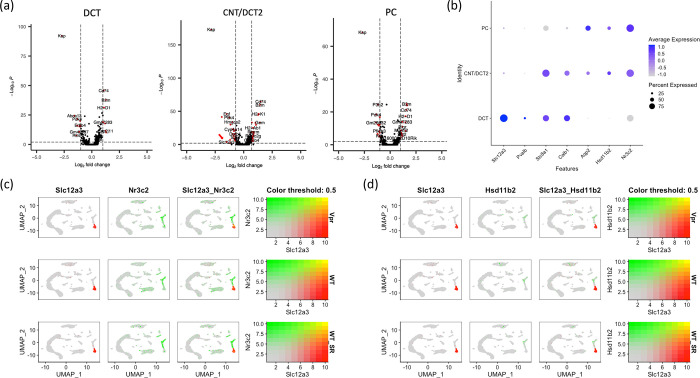
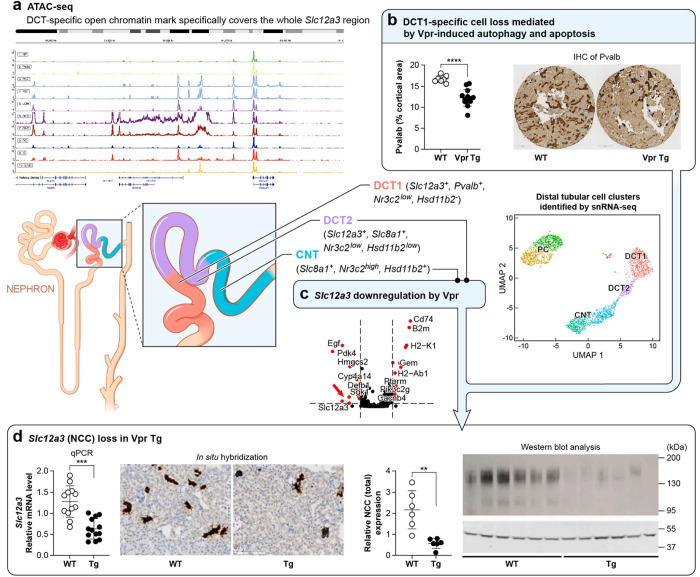
Slc12a3 was downregulated in CNT/DCT2 cells, but not in DCT1 cells of Vpr Tg mouse
Since Vpr binds the mineralocorticoid receptor (MR) and thus regulates aldosterone-responsive Slc12a3, an MR-responsive gene, we performed differential expression analysis between WT and Vpr Tg samples to evaluate changes in the expression of Slc12a3 in distal tubular cells. Slc12a3 was most significantly downregulated in the Slc8a1+ CNT/DCT2 cluster but not in the Pvalb+ DCT cluster, the latter of which is most likely composed of a majority of DCT1 cells. The CNT/DCT2 cluster showed strong expression of Slc8a1 and a weak expression of Slc12a3 in a relatively smaller fraction of cells compared with DCT cluster (15% of Vpr Tg cells and 27.5% of WT cells in CNT/DCT2 cluster compared with 95.9% of Vpr cells and 100% of WT cells in DCT cluster) (Table 1 and Figure 2a).
Table 1.
Statistics showing the significant downregulation of Slc12a3 in CNT/DCT2 cluster but not in DCT and PC clusters.
| Cluster | Percent of cells expressing Slc12a3 in Vpr Tg (%) | Percent of cells expressing Slc12a3 in WT (%) | average log2 fold change | adjusted P-value |
|---|---|---|---|---|
| DCT | 95.9 | 100 | 0.05 | 1 |
| CNT/DCT2 | 15 | 27.5 | −1.50 | 0.00018492 |
| PC | 15.6 | 23.2 | −0.78 | 1 |
Adjusted p-values were calculated by comparing WT and Vpr Tg data.
Figure 2. Expression pattern of Slc12a3, Nr3c2 and Hsd11b2 genes across distal tubular clusters.
(a) Volcano plots showing differentially expressed genes in DCT, CNT/DCT2 and PC clusters. Red arrow indicates Slc12a3 downregulation in CNT/DCT2 cluster.
(b) Dot plot showing the expression of genes distinguishing distal tubular cells.
(c) Feature plots of individual samples showing the expression overlap between Slc12a3 and Nr3c2.
(d) Feature plots showing the overlap of expression between Slc12a3 and Hsd11b2.
The CNT/DCT2 cluster showed the highest expression of Nr3c2 and Hsd11b2, genes essential for conferring aldosterone responsiveness
Vpr has been reported to bind to MR (Nr3c2) and to inhibit MR-mediated upregulation of Slc12a3 by acting as an MR corepressor. The Hsd11b2 gene encoding the enzyme 11-β-hydroxysteroid dehydrogenase 2, is required to metabolize glucocorticoids; these are present in higher concentrations within cells, compared with aldosterone, and this promotes specific binding of mineralocorticoids to the MR rather than the GR.
To investigate the cell types most effectively regulated by aldosterone, we examined the expression profiles of Nr3c2 and Hsd11b2 in distal tubular cell clusters. Expression levels of Nr3c2 and Hsd11b2 were highest in CNT/DCT2 and PC clusters while their expression levels were very low in the DCT cluster (Figure 2b). There was little overlap in the expression profiles of Slc12a3 and of these two genes (Figure 2c and 2d). This pattern is consistent with the finding that Slc12a3 downregulation occurred in the CNT/DCT2 cluster of salt-depleted Vpr Tg mice (Table 1).
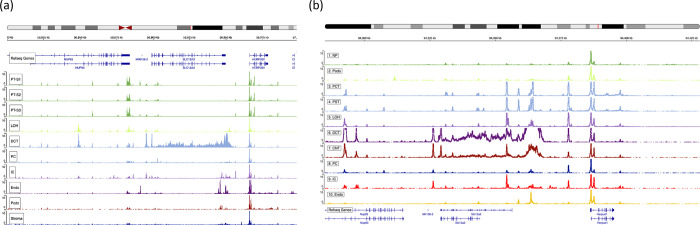
A DCT-specific open chromatin mark spans Slc12a3 region
Slc12a3 is a DCT-defining signature gene, and it is known from single-nucleus ATAC-seq data that there is an open chromatin area over this gene region in DCT cells. Using the human and developing mouse kidney ATAC-seq data from the Susztaklab Kidney Biobank (https://susztaklab.com/)20–28, we observed that the open chromatin feature spanning over a 50kb region is present in the Slc12a3 region in a DCT-specific manner in both human and mouse. This suggests that this region may qualify as a super-enhancer that ensures robust expression of lineage-defining Slc12a3 in DCT cells (Figure 3).
Figure 3. snATAC-seq data of kidney cells showing transposase-accessible open chromatin peaks around Slc12a3 regions retrieved from Susztaklab Kidney Biobank.
(a) Open chromatin regions around SLC12A3 region on chromosome 16 of the human kidney.
(b) Open chromatin regions around Slc12a3 region on chromosome 8 of the developing mouse kidney.
To look for the evidence of the regulatory association between the open chromatin region and Slc12a3 gene expression, we inspected the expression quantitative trait locus (eQTL) and the H3K27ac histone acetylation mark in human SLC12A3 region from Genotype Tissue Expression (GTEx) and candidate cis-regulatory elements (cCRE) data from the Encyclopedia of DNA Elements (ENCODE) databases. The GTEx data showed multiple eQTL SNPs associated with the SLC12A3 expression levels across the open chromatin data (Supplementary Table 2) and H3K27ac enhancer mark in frontal cortex, left ventricle, skeletal muscle and lung (Supplementary Figure 3). In ENCODE database, we looked for evidence of regulatory elements in this region with the focus on glomerular epithelial cells and kidney tissues from human male subjects (Supplementary Figure 4). There were multiple cCREs across the region from all registry samples. Furthermore, there were CTCF ChIP-seq signals in the kidney tissue data from three male adult subjects (Supplementary Figure 4b).
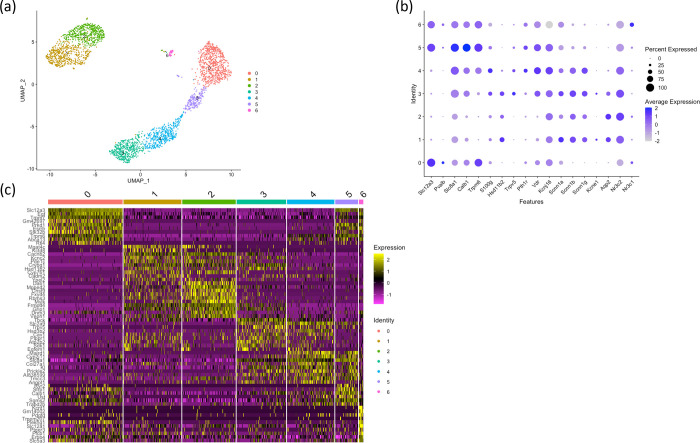
The Vpr Tg mice showed loss of Pvalb+ DCT1 cell fraction
We performed sub-clustering of the three distal tubule cell clusters (DCT, CNT/DCT2 and PC) to pinpoint the cell sub-populations that were lost in the Vpr Tg mouse and to identify the underlying pathways involved in the cell loss. This approach identified seven subclusters (three DCT clusters and two clusters each for CNT and PC) (Figure 4). Of the three DCT clusters in Vpr Tg sample, a decreased cell fraction was observed only in Pvalb+ DCT1 cluster (Cluster 0) but not in Slc8a1+ Calb1+ DCT2 cluster (Cluster 5) or in Nr3c1+ DCT cluster (Cluster 6) (Supplementary Table 1). The DCT subclusters we identified are consistent with those described in a recent report employing targeted single-cell RNA-seq on distal nephron cells.29
Figure 4. Subclustering of distal tubule clusters (DCT, DCT2/CNT and PC).
(a) UMAP plot showing 7 subclusters.
(b) Dot plot showing marker genes to identify sub-populations of distal tubular cells.
(c) Heatmap showing the ten most highly expressed genes in each subcluster compared to all remaining subclusters.
Pathway Analysis suggested autophagy in DCT1 cells
Ingenuity Pathway Analysis (IPA) software was used to identify differentially regulated pathways between WT and Vpr Tg mice. We found the protein ubiquitination and autophagy pathways among the top upregulated pathways in DCT1 cluster but not in DCT2 cluster or overall DCT, CNT/DCT2 and PC clusters (Table 2). Since autophagy has been reported to be associated with apoptosis,30 we hypothesized that apoptosis of DCT1 cells is the cause of reduced DCT cell percentage in our snRNA-seq results.
Table 2.
Ingenuity pathway analysis (IPA) of the differentially expressed genes between salt-depleted Vpr Tg and WT mouse samples showing upregulated genes in Vpr annotated as related to canonical pathways in DCT, CNT and PC clusters and DCT1 and DCT2 subclusters.
| Top canonical pathways | ||
|---|---|---|
| Pathway name | P-value | Overlap (%) |
| DCT | ||
| Insulin Receptor Signaling | 3.37E-11 | 39/140 (27.9%) |
| Estrogen Receptor Signaling | 1.40E-08 | 71/409 (17.4%) |
| Circadian Rhythm Signaling | 3.46E-08 | 52/269 (19.3%) |
| Molecular Mechanisms of Cancer | 5.33E-08 | 74/447 (16.6%) |
| mTOR Signaling | 1.27E-07 | 43/212 (20.3%) |
| DCT1 | ||
| Protein Ubiquitination Pathway | 9.17E-09 | 49/275 (17.8%) |
| Insulin Receptor Signaling | 3.85E-07 | 29/140 (20.7%) |
| Leucine Degradation I | 3.91E-07 | 7/9 (77.8%) |
| Autophagy | 5.06E-07 | 38/215 (17.7%) |
| AMPK Signaling | 7.30E-07 | 41/244 (16.8%) |
| DCT2 | ||
| AMPK Signaling | 8.86E-06 | 25/244 (10.2%) |
| Sirtuin Signaling Pathway | 9.29E-06 | 28/292 (9.6%) |
| RHOGDI Signaling | 9.91E-06 | 23/215 (10.7%) |
| Antigen Presentation Pathway | 1.32E-05 | 9/39 (23.1%) |
| TR/RXR Activation | 1.84E-05 | 13/84 (15.5%) |
| CNT | ||
| Protein Ubiquitination Pathway | 9.04E-13 | 64/275 (23.3%) |
| Senescence Pathway | 5.69E-09 | 59/298 (19.8%) |
| Protein Kinase A Signaling | 2.37E-08 | 72/409 (17.6%) |
| Valine Degradation I | 3.68E-08 | 12/21 (57.1%) |
| Molecular Mechanisms of Cancer | 4.45E-08 | 76/447 (17.0%) |
| PC | ||
| Protein Kinase A Signaling | 6.95E-10 | 56/409 (13.7%) |
| Antigen Presentation Pathway | 1.10E-07 | 13/39 (33.3%) |
| Huntington’s Disease Signaling | 2.23E-07 | 39/283 (13.8%) |
| Role of NFAT in Cardiac Hypertrophy | 3.26E-07 | 33/222 (14.9%) |
| Corona Virus Pathogenesis Pathway | 4.02E-07 | 31/203 (15.3%) |
Imaging confirmed the DCT1 cell loss and increased apoptosis in Vpr Tg mice
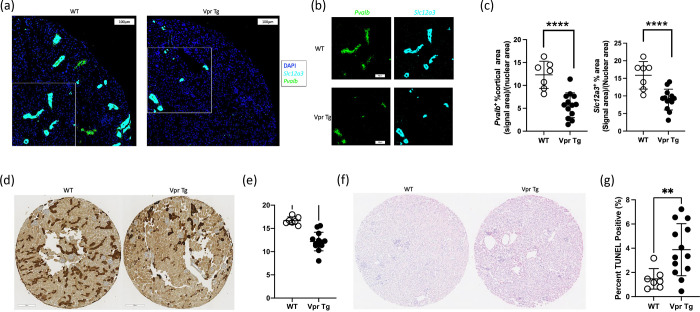
To further confirm the results obtained in the snRNA-seq analysis, in situ hybridization (ISH), immunohistochemistry (IHC) and terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) apoptosis assay were performed and the results were compared between WT and Vpr Tg mice. The ISH results showed marked reduction of Slc12a3 and Pvalb transcripts in Vpr Tg mouse cortex (Figures 5a–c and Supplementary Figure 5), suggesting the loss or dedifferentiation of DCT1 cells. The IHC staining and quantification of DCT1-specific parvalbumin showed a significant decrease in the percent of parvalbumin+ cortical tubular area in Vpr Tg samples (Figures 5d and 5e). The TUNEL assay showed that the Vpr Tg samples had more TUNEL-positive nuclei compared with WT samples (Figure 5f and 5g) although TUNEL-positivity is not specific to DCT1 cells. Thus, it is likely that reduced Slc12a3 and Pvalb expression observed in Vpr Tg mouse cortex is caused by activation of the apoptosis and subsequent loss of DCT1 cells.
Figure 5. Imaging results of Vpr and WT cortex samples.
(a) RNAScope images of a quarter of a tissue microarray section showing that Vpr mice had less Scl12a3 and Pvalb fluorescence signals than WT mice.
(b) Areas of images from Figure 5a marked by the white rectangles showing individual genes for Pvalb and Slc12a3.
(c) Dot plots showing unpaired t-tests comparing the cortical areas positive for fluorescence signals of Pvalb and Slc12a3 between WT and Vpr Tg mouse.
(d) Tissue microarray sections showing parvalbumin (Pvalb) staining of WT samples and Vpr Tg samples.
(e) Dot plot of the unpaired t-test comparing parvalbumin (+) cortical area between thirteen Vpr Tg and seven WT mouse samples.
(f) Tissue microarray sections showing TUNEL staining of WT and Vpr Tg samples.
(g) Dotplot showing the results of Mann-Whitney U test comparing the percent of TUNEL (+) nuclei between Vpr and WT samples. The Pvalb and TUNEL values of each sample on Y-axis were average values from triplicates.
Discussion
We recently reported that transgenic Vpr expression causes downregulation of NCC protein expression in renal distal tubules and proposed that binding of Vpr to the mineralocorticoid receptor (MR) blocks aldosterone/MR-mediated transcription of Slc12a3 in mice.13 Various interventions, such as thiazide administration,10 deletion of Slc12a3 or Pvalb,31, 33 over-expression of WNK4,33 or loss of SPAK34 have been shown to reduce the levels of NCC protein and shortening of the DCT length. In this study, we applied single-nucleus RNA sequencing to salt-depleted WT and Vpr Tg mouse kidney cortices to investigate the molecular mechanisms underlying the reduction of NCC expression, and found apoptosis to be a major factor contributing to the DCT1-specific cell loss, which contributed to the NCC loss and subsequent salt wasting observed in Vpr Tg mice.13
Sodium reabsorption is a highly regulated process in all nephron segments, including the distal nephron. Slc12a3 (NCC) is expressed in the DCT and is responsible for reabsorbing approximately 5–10% of Na+ from the glomerular filtrate.1 Aldosterone upregulates the expression of NCC through augmenting the transcriptional activity of the MR on Slc12a3. The aldosterone action is also dependent on the intracellular expression of the enzyme Hsd11b2 which metabolizes glucocorticoids (specifically corticosterone in mice) into inactive 11-dehydroxycortisone. Glucocorticoids which have higher intracellular concentrations than aldosterone and compete with aldosterone for binding to MR.35, 36
In the present study, the highest expression of MR (Nr2c3) and Hsd11b2 was found in the CNT/DCT2 cluster. These findings are consistent with those of Chen et al. who also observed higher expression of these genes in DCT2 and CNT than in other nephron segments.29 Using Susztaklab Kidney Biobank data, we noted that the Slc12a3 region spanning over 50kb is characterized as open chromatin in a DCT-specific fashion, suggesting a possibility that this genomic region harbors enhancer elements. The GTEx and ENCODE data showing the cis-regulatory elements, especially the CCCTC-binding factor (CTCF)-binding sites in adult kidney tissue further supports the likelihood that this region acts as a super-enhancer. The CTCF was reported to be bound near most and within some super-enhancers,37–40 and is critical for super-enhancer-mediated gene expression regulation.41–43
In DCT, Slc12a3 is one of the cell type-defining genes, therefore the open chromatin region around this gene may function as a super-enhancer. Super-enhancers are usually larger than other enhancer regions, facilitate the binding of multiple key transcription factors, and promote cellular development/differentiation into particular lineages.44–46 Taken together, these findings suggest that DCT cells have a strong baseline expression of Slc12a3 which is less dependent on aldosterone and more resistant to inhibitory factors such as Vpr. This scenario reflects the physiological necessity of the Slc12a3 super-enhancer in DCT cells to ensure the baseline salt reabsorption through NCC. Relative to DCT1 cells, DCT2 and CNT cells appear to be more dependent on aldosterone-mediated upregulation and likely more vulnerable to the inhibition by Vpr through its repression of MR transcriptional activity.
Compared with WT mice, Vpr Tg mice showed reduced fractions of DCT cells, specifically in Pvalb+ DCT1 cells. Pathway analysis and TUNEL assay suggested autophagy and apoptosis as a probable mechanism leading to DCT1 cell loss. Loffing and colleagues reported DCT cell apoptosis and the appearance of autophagosomes in rats treated with thiazide diuretics. The DCT-specific induction of apoptosis by thiazide in rats was attributed to inhibition of the apical NaCl reabsorption by thiazides.10 The exact mechanism underlying Vpr-associated DCT1 apoptosis remains unclear. It could be through the cell-cycle arresting action of Vpr and subsequent induction of apoptosis independent of its transcriptional activity on nuclear hormone receptors including MR.11 However, when we evaluated the aggregate scores of cell cycle phase-specific genes in WT and Vpr Tg samples, the scores of G2M and S phase-specific genes were similar between the WT and Vpr Tg mice (Supplementary Tables 3 and 4 and Supplementary Figure 6) suggesting that, at least in the present setting, the cell cycle arrest function of Vpr may not contribute to its transcriptional effects.
Autophagy is a cellular process that recycles and clears misfolded or toxic protein aggregates and damaged mitochondria and other organelles from the cells,47–49 with the aim of promoting cellular survival in response to stimuli such as starvation or viral infection.47, 48, 50 Pathway analysis showed activation of other autophagy-related pathways in DCT1 (Table 2). The AMPK pathway is known to activate autophagy,51 and the protein ubiquitination pathway could be related to protein degradation in autophagy.30, 52, 53 Insulin signaling also inhibits apoptosis,54–56 and as shown in Table 2, insulin receptor signaling pathway could be a part of cellular mechanisms to prevent apoptosis.
Particular renal epithelial cell types, such as podocytes and proximal tubular cells have high basal levels of autophagy, which is essential to keep these terminally differentiated and/or highly metabolically active cells in a healthy state. In mice, deficiency or inhibition of autophagy in proximal tubules leads to interstitial fibrosis and renal failure with aging,30, 57, 58 and inhibition in podocytes leads to podocyte loss, albuminuria, late onset glomerular sclerosis, and accumulation of glomerular aggregates with aging.52, 53, 59 Notably, however, in distal tubules, inhibition of autophagy did not show histologic abnormalities in mice without additional insults,60 suggesting that distal tubular cells do not rely on autophagy for survival under the conditions studied. However, surprisingly, we found that the aggregate scores of the 38 autophagy-related genes from the DCT1 autophagy pathway were higher in the distal tubular cell clusters compared with the podocyte and proximal tubular cell clusters (Supplementary Table 5 and Supplementary Figure 7). Further studies are needed to elucidate what causes DCT cell loss and why DCT1 cells are particularly vulnerable to autophagy and apoptosis.
In a previous study, we reported that the reduced expression of Slc12a3 in Vpr Tg mice was mediated by Vpr inhibition of the aldosterone/MR-mediated induction of this gene. These observations were supported by a reporter assay employing CV-1 African green monkey kidney cells, which lack an endogenous expression of Slc12a3. Although the results are valid in these cells, they do not represent the complex epigenomic regulatory mechanisms operating endogenously in the DCT cells in vivo. The findings from the current study suggest that Vpr-mediated downregulation of Slc12a3 may not be a principal mechanism for the NCC loss observed in the DCT1 segment, although this Vpr action appears to contribute to the loss of NCC observed in more distal segments, such as DCT2 and CNT.
The snRNA data presented here are from one representative WT (one out of seven) and one Vpr Tg (one out of thirteen) mouse kidney cortices. This may have limited the statistical power and the sensitivity to discover novel genes and pathways that are more mildly affected than those reported here. The results from the current snRNA-seq analysis, however, were independently validated and supported by applying imaging approaches in situ hybridization, immunohistochemistry and TUNEL assays performed in triplicate from seven WT and thirteen Vpr transgenic kidney cortex samples.
The lack of direct evidence of a DCT-specific super-enhancer in Slc12a3 gene could be viewed as a limitation of the current study. Although the ATAC-seq data clearly showed a DCT-specific open chromatin mark, the GTEx and ENCODE data are not specific to DCT. This is most likely due to the relatively small population of DCT cells in the kidneys compared with other cell types. We speculate that the enhancer and CTCF-binding signals would be much more prominent in a pure DCT cell population.
In summary, the DCT is one of the final segments in the nephron responsible for reabsorption of Na+ from the glomerular filtrate before it enters into the collecting duct. Furthermore, the DCT is known for possessing a high plasticity to adapt to changes in salt balance in the body.9 DCT function is also vulnerable to effects of HIV-1 Vpr. In the present study, we applied snRNA-seq to a Vpr Tg mouse as a model of salt wasting in HIV kidney disease, to gain insight into the DCT injury and NCC protein loss. Our findings show that the loss of DCT1 cells is a main mechanism for Vpr-induced reduction of NCC, through induction of DCT1 apoptosis. Slc12a3 downregulation via modulating the MR transcriptional activity on this gene by this viral protein as we previously reported13 is a minor contributing factor to NCC loss. Instead, the downregulation of Slc12a3 gene by Vpr may occur in more distal nephron segments, such as DCT2 and CNT as shown in the Figure 6. Further studies will be required to clarify detailed mechanisms underlying the induction of apoptosis in DCT by HIV-1 Vpr.
Figure 6. Diagram of the nephron showing mechanisms contributing to Slc12a3 (NCC) loss induced by Vpr in DCT1, DCT2 and CNT.
(a) ATAC-seq data of developing mouse kidney cortex shown in Figure 3b.
(b) loss of parvalbumin (+) DCT1 cells in Vpr Tg mouse samples as shown in Figure 5d and e.
(c) Slc12a3 downregulation in CNT and DCT2 cells as shown in Table 1.
(d) data showing NCC loss retrieved from Shrivastav et al, (PlosOne, 2022).
Supplementary Material
Supplementary Figure 1. Results of doublet detection analysis in each sample by DoubletFinder.
Supplementary Figure 2. SnRNA-seq analysis of the dataset after anchor-based integration and batch correction by harmony.
Supplementary Figure 3. H3K27ac enhancer mark on human chromosome 16 covering SLC12A3 region in frontal cortex, left ventricle, skeletal muscle and lung retrieved from GTEx database.
Supplementary Figure 4. Candidate cis-regulatory elements (cCREs) in human SLC12A3 region retrieved from ENCODE database showing multiple enhancer-like elements (yellow vertical bars) in all registry samples.
Supplementary Figure 5. Imaging results of Vpr and WT cortex samples.
Supplementary Figure 6. Violin plots showing the aggregate gene scores of cell cycle genes in distal tubular cell subclusters.
Supplementary Figure 7. Violin plots showing the aggregate scores of 38 autophagy-related genes upregulated in the DCT1 subcluster.
Supplementary Table 1. Metrics of the 25 cells clusters and 3 DCT sub-clusters that were identified in the unsupervised clustering of Vpr Tg transgenic, WT, and WT_SR mouse renal cortex samples.
Supplementary Table 2. Expression quantitative trait locus (eQTL) SNPs associated with SLC12A3 expression levels across different tissues retrieved from the Genotype Tissue Expression (GTEx) database.
Supplementary Table 3. The mouse G2M phase genes used in the aggregate gene scores shown in Supplementary Figure 5a.
Supplementary Table 4. The mouse S phase genes used in the aggregate gene scores shown in Supplementary Figure 5b.
Supplementary Table 5. Thirty-eight autophagy-related genes from the DCT1 autophagy pathway analysis results.
Acknowledgment
We thank the Sequencing Facility and Bioinformatics Group (Frederick National Laboratory for Cancer Research (FNLCR), NCI, NIH) for sequencing and informatics support. This work utilized the computational resources of the NIH HPC Biowulf cluster. (http://hpc.nih.gov) The authors also thank the NIDDK Advanced Light Microscopy & Image Analysis Core (ALMIAC) for the use of its resources. We acknowledge Huiyan Lu, Kris Ylaya for technical support, and Luis Fernando Menezes for critical manuscript review.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Funding
This Research was supported by the Intramural Research Program of the NIH, including the National Cancer Institute, Center for Cancer Research and the NIDDK. This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract 75N91019D00024.
Abbreviations used:
- DCT
(distal convoluted tubule)
- Vpr
(viral protein R)
- HIVAN
(HIV-associated nephropathy)
- snRNA-seq
(single nucleus RNA sequencing)
- CNT
(connecting tubule)
- PC
(principal cell)
Footnotes
Disclosure
The authors have no competing interest.
References
- 1.Subramanya AR, Ellison DH. Distal convoluted tubule. Clin J Am Soc Nephrol 2014; 9: 2147–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komers R, Rogers S, Oyama TT, et al. Enhanced phosphorylation of Na(+)-Cl− cotransporter in experimental metabolic syndrome: role of insulin. Clin Sci (Lond) 2012; 123: 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saritas T, Borschewski A, McCormick JA, et al. SPAK differentially mediates vasopressin effects on sodium cotransporters. J Am Soc Nephrol 2013; 24: 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez-Villalobos RA, Janjoulia T, Fletcher NK, et al. The absence of intrarenal ACE protects against hypertension. J Clin Invest 2013; 123: 2011–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiga M, Rai T, Yang SS, et al. Dietary salt regulates the phosphorylation of OSR1/SPAK kinases and the sodium chloride cotransporter through aldosterone. Kidney Int 2008; 74: 1403–1409. [DOI] [PubMed] [Google Scholar]
- 6.Pedersen NB, Hofmeister MV, Rosenbaek LL, et al. Vasopressin induces phosphorylation of the thiazide-sensitive sodium chloride cotransporter in the distal convoluted tubule. Kidney Int 2010; 78: 160–169. [DOI] [PubMed] [Google Scholar]
- 7.Sohara E, Rai T, Yang SS, et al. Acute insulin stimulation induces phosphorylation of the Na-Cl cotransporter in cultured distal mpkDCT cells and mouse kidney. PLoS One 2011; 6: e24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim GH, Masilamani S, Turner R, et al. The thiazide-sensitive Na-Cl cotransporter is an aldosterone-induced protein. Proc Natl Acad Sci U S A 1998; 95: 14552–14557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaissling B, Bachmann S, Kriz W. Structural adaptation of the distal convoluted tubule to prolonged furosemide treatment. Am J Physiol 1985; 248: F374–381. [DOI] [PubMed] [Google Scholar]
- 10.Loffing J, Loffing-Cueni D, Hegyi I, et al. Thiazide treatment of rats provokes apoptosis in distal tubule cells. Kidney Int 1996; 50: 1180–1190. [DOI] [PubMed] [Google Scholar]
- 11.Rednor SJ, Ross MJ. Molecular Mechanisms of Injury in HIV-Associated Nephropathy. Front Med (Lausanne) 2018; 5: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menon MC, Garcha AS, Khanna A. The management of hyponatremia in HIV disease. J Nephrol 2014; 27: 109. [DOI] [PubMed] [Google Scholar]
- 13.Shrivastav S, Lee H, Okamoto K, et al. HIV-1 Vpr suppresses expression of the thiazide-sensitive sodium chloride co-transporter in the distal convoluted tubule. PLoS One 2022; 17: e0273313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balasubramanyam A, Mersmann H, Jahoor F, et al. Effects of transgenic expression of HIV-1 Vpr on lipid and energy metabolism in mice. Am J Physiol Endocrinol Metab 2007; 292: E40–48. [DOI] [PubMed] [Google Scholar]
- 15.Kirita Y, Wu H, Uchimura K, et al. Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. Proc Natl Acad Sci U S A 2020; 117: 15874–15883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Young MD, Behjati S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. Gigascience 2020; 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell 2021; 184: 3573–3587 e3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst 2019; 8: 329–337 e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korsunsky I, Millard N, Fan J, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 2019; 16: 1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balzer MS, Doke T, Yang YW, et al. Single-cell analysis highlights differences in druggable pathways underlying adaptive or fibrotic kidney regeneration. Nat Commun 2022; 13: 4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dhillon P, Park J, Hurtado Del Pozo C, et al. The Nuclear Receptor ESRRA Protects from Kidney Disease by Coupling Metabolism and Differentiation. Cell Metab 2021; 33: 379–394 e378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doke T, Abedini A, Aldridge DL, et al. Single-cell analysis identifies the interaction of altered renal tubules with basophils orchestrating kidney fibrosis. Nat Immunol 2022; 23: 947–959. [DOI] [PubMed] [Google Scholar]
- 23.Liu H, Doke T, Guo D, et al. Epigenomic and transcriptomic analyses define core cell types, genes and targetable mechanisms for kidney disease. Nat Genet 2022; 54: 950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miao Z, Balzer MS, Ma Z, et al. Single cell regulatory landscape of the mouse kidney highlights cellular differentiation programs and disease targets. Nat Commun 2021; 12: 2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park J, Shrestha R, Qiu C, et al. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 2018; 360: 758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu C, Huang S, Park J, et al. Renal compartment-specific genetic variation analyses identify new pathways in chronic kidney disease. Nat Med 2018; 24: 1721–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheng X, Guan Y, Ma Z, et al. Mapping the genetic architecture of human traits to cell types in the kidney identifies mechanisms of disease and potential treatments. Nat Genet 2021; 53: 1322–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheng X, Qiu C, Liu H, et al. Systematic integrated analysis of genetic and epigenetic variation in diabetic kidney disease. Proc Natl Acad Sci U S A 2020; 117: 29013–29024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L, Chou CL, Knepper MA. Targeted Single-Cell RNA-seq Identifies Minority Cell Types of Kidney Distal Nephron. J Am Soc Nephrol 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Havasi A, Dong Z. Autophagy and Tubular Cell Death in the Kidney. Semin Nephrol 2016; 36: 174–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belge H, Gailly P, Schwaller B, et al. Renal expression of parvalbumin is critical for NaCl handling and response to diuretics. Proc Natl Acad Sci U S A 2007; 104: 14849–14854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loffing J, Vallon V, Loffing-Cueni D, et al. Altered renal distal tubule structure and renal Na(+) and Ca(2+) handling in a mouse model for Gitelman’s syndrome. J Am Soc Nephrol 2004; 15: 2276–2288. [DOI] [PubMed] [Google Scholar]
- 33.Lalioti MD, Zhang J, Volkman HM, et al. Wnk4 controls blood pressure and potassium homeostasis via regulation of mass and activity of the distal convoluted tubule. Nat Genet 2006; 38: 1124–1132. [DOI] [PubMed] [Google Scholar]
- 34.Grimm PR, Taneja TK, Liu J, et al. SPAK isoforms and OSR1 regulate sodium-chloride co-transporters in a nephron-specific manner. J Biol Chem 2012; 287: 37673–37690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bostanjoglo M, Reeves WB, Reilly RF, et al. 11Beta-hydroxysteroid dehydrogenase, mineralocorticoid receptor, and thiazide-sensitive Na-Cl cotransporter expression by distal tubules. J Am Soc Nephrol 1998; 9: 1347–1358. [DOI] [PubMed] [Google Scholar]
- 36.Campean V, Kricke J, Ellison D, et al. Localization of thiazide-sensitive Na(+)-Cl(−) cotransport and associated gene products in mouse DCT. Am J Physiol Renal Physiol 2001; 281: F1028–1035. [DOI] [PubMed] [Google Scholar]
- 37.Agrawal P, Rao S. Super-Enhancers and CTCF in Early Embryonic Cell Fate Decisions. Front Cell Dev Biol 2021; 9: 653669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dowen JM, Fan ZP, Hnisz D, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 2014; 159: 374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang J, Li K, Cai W, et al. Dissecting super-enhancer hierarchy based on chromatin interactions. Nat Commun 2018; 9: 943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ing-Simmons E, Seitan VC, Faure AJ, et al. Spatial enhancer clustering and regulation of enhancer-proximal genes by cohesin. Genome Res 2015; 25: 504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanssen LLP, Kassouf MT, Oudelaar AM, et al. Tissue-specific CTCF-cohesin-mediated chromatin architecture delimits enhancer interactions and function in vivo. Nat Cell Biol 2017; 19: 952–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hay D, Hughes JR, Babbs C, et al. Genetic dissection of the alpha-globin super-enhancer in vivo. Nat Genet 2016; 48: 895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin HY. The structural and functional roles of CTCF in the regulation of cell type-specific and human disease-associated super-enhancers. Genes Genomics 2019; 41: 257–265. [DOI] [PubMed] [Google Scholar]
- 44.Hnisz D, Abraham BJ, Lee TI, et al. Super-enhancers in the control of cell identity and disease. Cell 2013; 155: 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013; 153: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moorthy SD, Davidson S, Shchuka VM, et al. Enhancers and super-enhancers have an equivalent regulatory role in embryonic stem cells through regulation of single or multiple genes. Genome Res 2017; 27: 246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beatman E, Oyer R, Shives KD, et al. West Nile virus growth is independent of autophagy activation. Virology 2012; 433: 262–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olasunkanmi OI, Chen S, Mageto J, et al. Virus-Induced Cytoplasmic Aggregates and Inclusions are Critical Cellular Regulatory and Antiviral Factors. Viruses 2020; 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol 2007; 9: 1102–1109. [DOI] [PubMed] [Google Scholar]
- 50.Hafren A, Ustun S, Hochmuth A, et al. Turnip Mosaic Virus Counteracts Selective Autophagy of the Viral Silencing Suppressor HCpro. Plant Physiol 2018; 176: 649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang J, Shao SH, Xu ZX, et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol 2007; 9: 218–224. [DOI] [PubMed] [Google Scholar]
- 52.Hartleben B, Wanner N, Huber TB. Autophagy in glomerular health and disease. Semin Nephrol 2014; 34: 42–52. [DOI] [PubMed] [Google Scholar]
- 53.Huber TB, Edelstein CL, Hartleben B, et al. Emerging role of autophagy in kidney function, diseases and aging. Autophagy 2012; 8: 1009–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iida KT, Suzuki H, Sone H, et al. Insulin inhibits apoptosis of macrophage cell line, THP-1 cells, via phosphatidylinositol-3-kinase-dependent pathway. Arterioscler Thromb Vasc Biol 2002; 22: 380–386. [DOI] [PubMed] [Google Scholar]
- 55.Nakazawa T, Chiba T, Kaneko E, et al. Insulin signaling in arteries prevents smooth muscle apoptosis. Arterioscler Thromb Vasc Biol 2005; 25: 760–765. [DOI] [PubMed] [Google Scholar]
- 56.Tseng YH, Ueki K, Kriauciunas KM, et al. Differential roles of insulin receptor substrates in the anti-apoptotic function of insulin-like growth factor-1 and insulin. J Biol Chem 2002; 277: 31601–31611. [DOI] [PubMed] [Google Scholar]
- 57.Isaka Y, Kimura T, Takabatake Y. The protective role of autophagy against aging and acute ischemic injury in kidney proximal tubular cells. Autophagy 2011; 7: 1085–1087. [DOI] [PubMed] [Google Scholar]
- 58.Kimura T, Takabatake Y, Takahashi A, et al. Autophagy protects the proximal tubule from degeneration and acute ischemic injury. J Am Soc Nephrol 2011; 22: 902–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hartleben B, Godel M, Meyer-Schwesinger C, et al. Autophagy influences glomerular disease susceptibility and maintains podocyte homeostasis in aging mice. J Clin Invest 2010; 120: 1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu S, Hartleben B, Kretz O, et al. Autophagy plays a critical role in kidney tubule maintenance, aging and ischemia-reperfusion injury. Autophagy 2012; 8: 826–837. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Results of doublet detection analysis in each sample by DoubletFinder.
Supplementary Figure 2. SnRNA-seq analysis of the dataset after anchor-based integration and batch correction by harmony.
Supplementary Figure 3. H3K27ac enhancer mark on human chromosome 16 covering SLC12A3 region in frontal cortex, left ventricle, skeletal muscle and lung retrieved from GTEx database.
Supplementary Figure 4. Candidate cis-regulatory elements (cCREs) in human SLC12A3 region retrieved from ENCODE database showing multiple enhancer-like elements (yellow vertical bars) in all registry samples.
Supplementary Figure 5. Imaging results of Vpr and WT cortex samples.
Supplementary Figure 6. Violin plots showing the aggregate gene scores of cell cycle genes in distal tubular cell subclusters.
Supplementary Figure 7. Violin plots showing the aggregate scores of 38 autophagy-related genes upregulated in the DCT1 subcluster.
Supplementary Table 1. Metrics of the 25 cells clusters and 3 DCT sub-clusters that were identified in the unsupervised clustering of Vpr Tg transgenic, WT, and WT_SR mouse renal cortex samples.
Supplementary Table 2. Expression quantitative trait locus (eQTL) SNPs associated with SLC12A3 expression levels across different tissues retrieved from the Genotype Tissue Expression (GTEx) database.
Supplementary Table 3. The mouse G2M phase genes used in the aggregate gene scores shown in Supplementary Figure 5a.
Supplementary Table 4. The mouse S phase genes used in the aggregate gene scores shown in Supplementary Figure 5b.
Supplementary Table 5. Thirty-eight autophagy-related genes from the DCT1 autophagy pathway analysis results.